Abstract
The ability to measure total and phosphorylated tau levels in clinical samples is transforming the detection of Alzheimer’s disease (AD) and other neurodegenerative diseases. In particular, recent reports indicate that accurate detection of low levels of phosphorylated tau (p-tau) in plasma provides a reliable biomarker of AD long before sensing memory loss. Therefore, the diagnosis and monitoring of neurodegenerative diseases progression using blood samples is becoming a reality. These major advances were achieved by using antibodies specific to p-tau as well as sophisticated high-sensitivity immunoassay platforms. This review focuses on these enabling advances in high-specificity antibody development, engineering, and novel signal detection methods. We will draw insights from structural studies on p-tau antibodies, engineering efforts to improve their binding properties, and efforts to validate their specificity. A comprehensive survey of high-sensitivity p-tau immunoassay platforms along with sensitivity limits will be provided. We conclude that although robust approaches for detecting certain p-tau species have been established, systematic efforts to validate antibodies for assay development is still needed for the recognition of biomarkers for AD and other neurodegenerative diseases.
Keywords: phosphorylated tau, antibody specificity, antibody validation, Alzheimer’s disease, neurodegeneration
Statement of Significance: Levels of total and phosphorylated tau protein are believed to correlate with the onset of Alzheimer’s disease. Detection of this protein is achieved through the use of antibodies, and it is important to understand how these antibodies distinguish phosphorylated tau from non-phosphorylated tau. Understanding antibody binding mechanisms and validating their specificity are crucial in the design of sensitive diagnostic assays. Such validation is essential to realize the full potential of recently developed high-sensitivity assay platforms.
INTRODUCTION
The observation of neurofibrillary tangles (NFTs) in neurons is a defining pathological feature of Alzheimer’s disease (AD). These tangles consist of paired helical filaments (PHFs) of microtubule-associated protein tau [1,2]. Such tau inclusions are also observed in other neurodegenerative diseases including Pick’s disease, progressive supranuclear palsy, chronic traumatic encephalopathy, and corticobasal degeneration. Dominantly inherited mutations in MAPT encoding tau have been discovered in genetically predisposed patients with frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) [3], indicating that abnormal forms of tau are sufficient to cause neurodegeneration. At the molecular level, a common signature of NFTs is the hyperphosphorylation of tau [4,5]. It is also hypothesized that tau posttranslational modifications such as hyperphosphorylation cause conformational variants that lead to tau inclusions [6].
Although a disease-modifying therapy for AD is not currently available, major advances in the diagnosis of AD have been made in the past few years. Many reports support that measuring the concentrations of amyloid β, tau and p-tau in the cerebrospinal fluid (CSF) can differentiate AD from normal aging and lead to the detection of AD many years prior to onset of cognitive impairment [7–9]. The most recent advances in this area are immunoassays capable of detecting p-tau at pg/mL (femtomolar) concentrations in plasma [10–12]. The levels of tau phosphorylated at threonine 181 (pThr181) in plasma correlated with CSF pThr181 tau and made it possible to differentiate AD from non-AD neurodegenerative diseases [10,13]. Plasma levels of pThr217 also emerged as a highly accurate biomarker for the diagnosis of AD [11], and for monitoring AD progression [14]. Considering the wide diversity of p-tau sites [15,16] and their high relevance to tau pathology [6,17,18], plasma p-tau biomarkers are expected to rapidly expand in the near future. Since obtaining plasma is far less invasive and cost-effective, these findings point to major advances in biomarker development for AD that will support early intervention strategies and drug efficacy assessment.
An essential component of high-sensitivity immunoassays is antibodies that selectively recognize the target in complex samples [19–24]. In identifying high-quality antibodies, much focus has been given to affinity due to the low concentrations of p-tau. However, antibody specificity—the ability to discriminate the target from other proteins—is as important as the tightness of binding. Achieving p-tau specificity is particularly challenging since the antibodies need to distinguish the presence of a single phosphorylated residue. This review will draw insights from structural studies on how specificity is achieved and engineering efforts to improve the affinity and specificity of p-tau antibodies. Findings from efforts to validate the specificity of p-tau antibodies, along with approaches used will be introduced. Finally, the new high-sensitivity detection methods that resulted in major improvements in p-tau detection sensitivity will be summarized.
INSIGHTS FROM STRUCTURAL ANALYSIS AND ENGINEERING OF ANTIBODIES TARGETING PHOSPHORYLATED TAU
Since the first report on the structure of an antibody fragment bound to a phosphorylated epitope [25], several following studies expanded our understanding on how antibodies recognize phosphorylation sites [26–31]. The majority of these structures are that of p-tau antibodies, reflecting interest in the target. The main feature of these antibodies is their tight association with the phosphate group of the modified residues (Fig. 1). In antibodies that bind promiscuously to the non-phosphorylated target site, the phosphate group faces away from the antibody–antigen interface (Fig. 1a) [29] or a free phosphate molecule was bound near the phosphorylation site (Fig. 1b) [32]. The fact that these antibodies were raised using phosphorylated peptides as antigens illustrates the importance of performing negative selections to remove nonselective binders during the antibody screening process [33,34]. The structural analyses revealed that the antibodies engage the phosphate group using diverse complementarity determining region (CDR) residues including those in CDR H1, H2, H3, or L1 (Fig. 1c–f). A majority of antibodies use a single CDR loop for phosphate recognition, but for the antibody dmCBTAB-22.1 (targeting phospho-serine (pSer) 422 tau), residues in CDR H1 and H3 both form hydrogen bonds with the phosphate (Fig. 1f) [35]. A unique example is the antibody AT8, which binds to three phosphate groups from pSer202, pThr 205, and pSer208 of tau (Fig. 1g) [27]. Residues from CDR L1, L2, H1, and H3 interact with the three phosphates. Another key aspect of phosphate binding sites is the frequent presence of positively charged residues (i.e., lysine and arginine) (see surface charge in Fig. 1), glycine (indicated as “G” in Fig. 1), tyrosine, threonine, and histidine. Most of the antibodies possess one or more lysine or arginine residues that form a salt bridge with the phosphate group (Fig. 1c–e and g), although the antibody dmCBTAB-22.1 relies only on hydrogen bonds (Fig. 1f). Glycine within the CDR often forms hydrogen bonds with the phosphate group (Fig. 1c, e, and f) and is commonly observed in other phosphate binding proteins [36–38].
Figure 1.
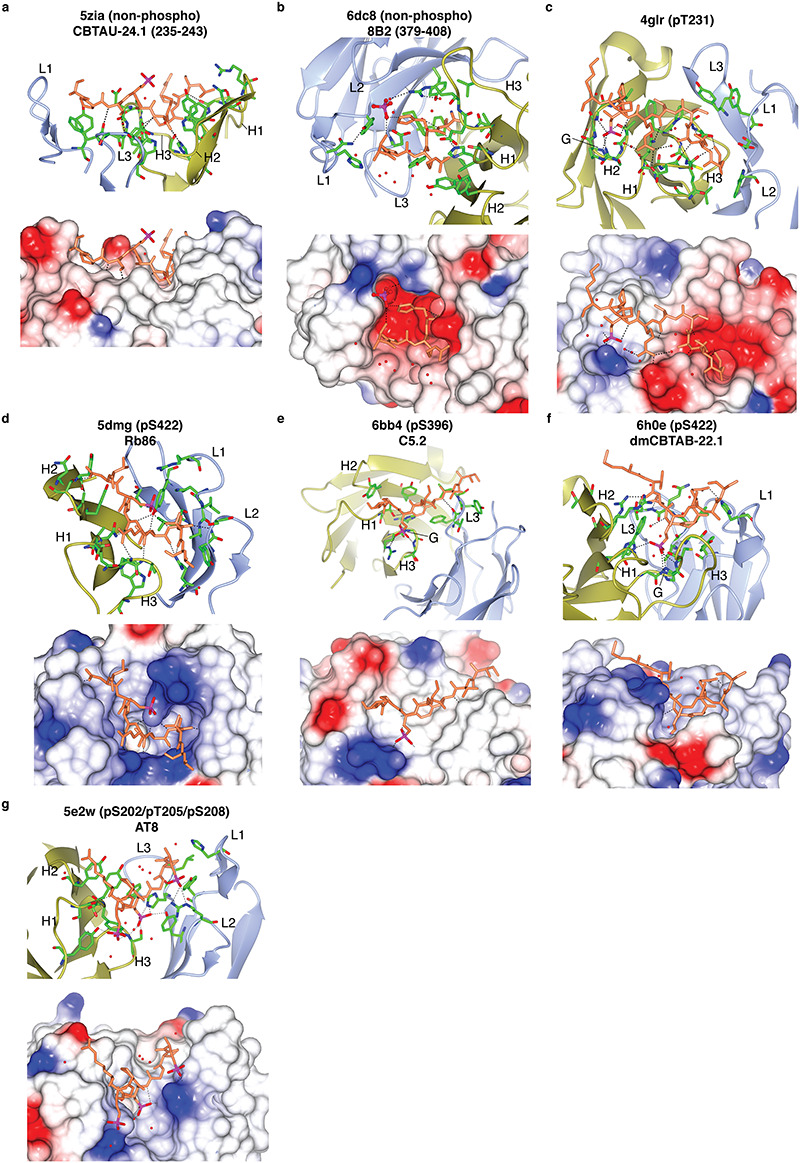
Structural analysis of p-tau antibodies. Each panel is labeled with the corresponding PDB ID, name of antibody clone, and phospho-site recognized by the antibody. The panels were generated using CCP4MG [91]. Antibody complementarity determining regions L1-3 and H1-3 that interact with the p-tau peptide are indicated. The lower half of each panel shows surface charge (blue—positive, red—negative). The phosphorus in the phosphate group is indicated in magenta. Antibodies in panels (a-b) are not specific to p-tau. Antibodies in panels (c–f) interact with a single phosphorylated residue. Antibodies in panel (g) interact with three phosphorylated residues.
Remarkably, Koerber et al. [26] demonstrated that phospho-specific antibodies can be designed by incorporating a phosphate-binding motif into an antibody CDR. First, they identified an antibody structure with CDR H2 that possesses a conformation similar to that of an anion-binding motif. After finding antibodies containing such CDRs that interact with an aspartate or glutamate in the antigen, they diversified the sequence of the CDR site to tune the antibody binding toward pSer, pSer/pThr, or phospho-tyrosine. They then expanded the pSer and pSer/pThr binding antibodies by diversifying non-phospho interacting residues in CDR H2, H3, and L3. These pools of antibodies were screened against 10 distinct phospho-sites leading to successful identification of phospho-specific binders for 7 of the 10. This study pioneered the idea of modular antibody paratopes—a region that binds the phosphate group (phospho-recognition) and another region that binds the surrounding amino acids (sequence recognition). Whether the modularity will prevail in targeting other posttranslational modifications remains to be tested.
These studies suggest that a balance in stability of phospho-recognition and sequence recognition is critical for specificity. A single CDR that captures the phosphate group imparts the ability to weakly interact with a wide range of phospho-peptides to antibodies [26]. In contrast, strong sequence recognition is likely to result in promiscuous binding to non-phosphorylated epitopes. This knowledge was highly valuable in engineering high-affinity p-tau antibodies [34]. Since the phosphate group is small relative to the surrounding amino acid residues in the phospho-epitope, a hypothesis emerged that antibody affinity maturation may over-stabilize the sequence recognition interactions. To test this, Li et al. [34] performed directed evolution of a high-specificity pThr231 tau antibody for improved affinity and assessed the binding specificity of the identified variants. Although the wild-type antibody did not bind to the non-phospho-peptide, over half of the high-affinity variants showed detectable binding to a non-phospho-peptide with the target sequence. None of the high-affinity variants showed binding to a phospho-peptide containing scrambled sequence, indicating that over-stabilization of phosphate recognition did not occur. Based on the fact that not all variants with improved affinity showed binding to the non-phospho-peptide, a second stage of screening was conducted for the absence of non-phospho-peptide binding. This led to the identification of a high-specificity pThr231 tau antibody with a picomolar dissociation constant. These results again highlight the importance of specificity validation in developing phospho-specific antibodies.
APPROACHES FOR ANTIBODY SPECIFICITY VALIDATION
Although the need for antibody specificity validation is widely recognized, published data on antibody validation have been scarce. Across different areas of research, antibody validation has been identified as a major bottleneck in improving reproducibility of research outcomes [39–41]. However, the very aspect that makes antibodies so useful—a wide range of applications—presents a formidable challenge in standardizing their validation. For example, Kalina et al. [42] and Pillai-Kastoori et al. [43] express the importance of validating antibodies using their intended experiment. Furthermore, comprehensive antibody validation has been demonstrated in the case of histone modification targeting antibodies [44–48]. Due to the importance of combinatorial recognition of multiple nearby modification sites [46,49], peptide microarrays were used to systematically validate site-specificity [44,46,50]. The collective dataset is a rare example of a quantitative assessment of antibody specificity [44]. Other studies validated histone antibody binding in the context of cellular background proteins using Western blotting and chromatin immunoprecipitation [47]. These studies also reported data on antibodies that lack specificity or failed to bind, providing a valuable guide for selecting antibodies. Acknowledging these aspects, we herein focus our attention to antibodies that target p-tau.
Though few validation studies have been published for p-tau antibodies, they provide insight into the types of nonspecific binding observed. Methods used for specificity validation include immunoblotting approaches and a whole cell immunocytochemistry assay [51–53]. Immunoblotting has been performed using synthetic peptide sequences, cell lysate, and extracted PHF tau [52,53]. This method allows characterization of nonspecific binding to non-tau proteins (using cell lysate), non-phosphorylated tau (using phosphatase treated PHF tau), and binding to other known phospho-sites (using synthetic peptides). Ercan et al. [52] observed that, in addition to some antibodies not binding to their specific sites, other antibodies may be specific but their binding could be inhibited by other modifications in the vicinity of the binding site, which could lead to false negative signal. Another important finding was the frequent nonspecific binding to unmodified peptides. As an example, all tested antibodies that claimed to target pSer262 also bound to unmodified peptide, leading to no antibody validated for the pSer262 site [52]. Cell lysates and tissue from tau knockout (TKO) mice [54] provide an effective means to assess nonspecific binding of p-tau antibodies to other nontarget proteins. Petry et al. [53] showed that p-tau antibodies show nonspecific binding to TKO mouse brain lysate proteins via Western blotting. Non-specific binding observed in certain p-tau antibodies could be greatly reduced by using heat stable fractions of the lysates [53]. Heating complex samples such as cell lysates can deplete other proteins that cause nonspecific binding since tau is highly heat stable [55–57]. These results highlight the importance of validating the specificity of p-tau antibodies and demonstrate how assay conditions impact apparent nonspecific binding.
Li et al. [51] developed a whole cell immunocytochemistry assay which utilizes human embryonic kidney (HEK) 293FT cells and flow cytometry to provide a quantitative measurement of specificity (Φ). The approach measures the fraction of specific and nonspecific cell staining intensity within a single sample using flow cytometry. This is achieved by quantifying both the binding signal to cells expressing the wild-type tau and cells expressing tau with an alanine point mutation at the target phospho-site. Since the point mutation itself may disrupt antibody binding, the authors also measured binding to cells expressing wild-type tau treated with a phosphatase. This method measures nonspecific binding to different p-tau sites across the entire tau protein, binding to non-phospho tau, and binding to other cellular proteins. Li et al. [51] found that some p-tau antibodies showed nonspecific binding to irrelevant cellular proteins in HEK293FT cells and mouse primary hippocampal neurons. By performing confocal microscopy and Western blot experiments, Li et al. confirmed antibodies AT270 (targeting pThr181) and 1H6L6 (targeting pThr231) bound to cells not expressing tau.
These studies provide us with a collection of antibodies tested for their specificity. Taken together, they reveal that several commonly used p-tau antibodies either were not able to detect their site-specific modification or they bound to non-phosphorylated tau. Notably, the majority of commercially available p-tau antibodies are generated by rabbit immunization, and many of them remain polyclonal. Since the identity of polyclonal antibodies varies between batches, repeated validation is required. The list of validated monoclonal p-tau antibodies remains limited (Table 1). Given the fact that tau has 85 total serine, threonine, and tyrosine sites available for phosphorylation, 45 of which have been detected in neurons [58,59], systematic development and validation of p-tau antibodies is necessary. It is notable that antibody validation efforts have not yielded high-specificity antibodies for critical tau phosphorylation sites such as pThr181 and pSer262 (Table 1) [51,52].
Table 1.
Validated monoclonal p-tau antibodies
| Tau phospho-site | Antibodies | Validation method | References |
|---|---|---|---|
| pSer198 | pSer198 (ab79540) | Peptide Array, Immunoblotting | Ercan et al. [52] |
| pSer199 | 2H23L4 | Peptide Array, Immunoblotting | Ercan et al. [52] |
| pSer202/pThr205 | AT8 | Phi | Li et al. [51] |
| pThr231 | AT180, PHF6, TG-3 | Phi | Li et al. [51] |
| pSer396/pSer404 | PHF1 | Phi | Li et al. [51] |
| pSer404 | pSer404 (ab92676) | Phi | Li et al. [51] |
| pSer422 | pSer422 (ab79415) | Peptide Array, Immunoblotting | Ercan et al. [52] |
APPROACHES FOR HIGH-SENSITIVITY DETECTION OF PHOSPHORYLATED TAU
Since the concentration of tau and p-tau in clinical samples is estimated to be in the femtomolar (pg/mL) range, many efforts have been made to improve assay sensitivity. In the past few years, several major improvements in immunoassays have enabled the detection of total and p-tau in human CSF as well as in plasma. Table 2 lists the performance of assays for detecting total tau and p-tau. Many different studies have used an enzyme-linked immunosorbent assay (ELISA) to measure tau and p-tau in human CSF targeting a variety of phospho-sites [60–66]. However, the reported sensitivity differs between studies and largely depends on the antibodies used (Table 2). Although ELISA was the first platform systematically developed for p-tau detection, the need for more sensitive detection methods has pushed researchers to develop alternative assays with ultra-sensitivity.
Table 2.
Performances of assays for detecting total tau (t-tau) and phosphorylated tau (p-tau)
| Assay | Capture Antibody | Binding Sites | Detection Antibody | Binding Sites | Reported Sensitivity | Sample | Reference |
|---|---|---|---|---|---|---|---|
| ELISA Tyramide Signal Amplification t-tau | HT7 | aa 159-163 | tau antiserum 92e | N/A | 14 pg/ml | CSF | Yamamori et al. [85] |
| R134d | N/A | ||||||
| ELISA t-tau (INNOGENETICS) | AT120 | N/A | HT7 | aa 159-163 | LoD1: 34 pg/ml LoQ2: 57 pg/ml (INNOTEST hTau User Manual) | CSF | Blennow et al. [62] |
| BT2 | aa 194-198 | ||||||
| ELISA p-tau (INNOGENETICS) | AT180 | pThr231 | HT7 | aa 159-163 | N/A | ||
| AT270 | pThr181 | AT120 | N/A | ||||
| ELISA p-tau (INNOGENETICS) User Manual | HT7 | aa 159-163 | AT270 | pThr181 | LoD1: 13 pg/ml LoQ2: 20 pg/ml | CSF | INNOTEST User Manual [63] |
| Sandwich EIA (Ishiguro) | anti-tau mAb | N/A | anti-PT231PS235 | pThr231/pSer235 | N/A | CSF | Ishiguro et al. [60] |
| anti-PS199 | pSer199 | ||||||
| Sandwich ELISA (Kohnken) | PC1C6/Tau-1 | aa 196-205 | CP9 | pThr231 | N/A | CSF | Kohnken et al. [61] |
| CP27 | aa 130-150 | ||||||
| Sandwich ELISA (Vandermeeran) t-tau | AT120 | N/A | rabbit anti-normal tau | N/A | < 5 pg/ml | CSF | Vandermeeren et al. [65] |
| Sandwich ELISA (Vandermeeran) p-tau | AT8 | pSer202/pThr205 | rabbit anti-normal tau | N/A | < 20 pg/ml | ||
| Bienzyme-Substrate-Recycle ELISA t-tau | tau antiserum 92e | N/A | PC1C6/Tau-1 | aa 196-205 | 0.75-200 pg (7.5 pg/ml) | CSF | Hu et al. [64] |
| Bienzyme-Substrate-Recycle ELISA p-tau | tau antiserum 92e | N/A | PHF-1 | pSer396/404 | 0.5-50 pg (5 pg/ml) | ||
| Overlapping ELISA p-tau | Tau12 | aa 9-18 | AT270 | pThr181 | LoQ2: 6 pg/ml | CSF | Meredith Jr. et al. [66] |
| HT7 | aa 159-163 | AT270 | pThr181 | LoQ2: 2 pg/ml | |||
| HT7 | aa 159-163 | PHF-6 | pThr231 | LoQ2: 7.8 pg/ml | |||
| Overlapping ELISA t-tau | Tau12 | aa 9-18 | HT7 | aa 159-163 | LoQ2: 3.9 pg/ml | CSF | |
| Tau12 | aa 9-19 | BT2 | aa 194-198 | LoQ2: 1.6 pg/ml | |||
| HT7 | aa 159-163 | BT2 | aa 194-198 | LoQ2: 7.8 pg/ml | |||
| HT7 | aa 159-164 | Tau5 | aa 218-225 | LoQ2: 7.8 pg/ml | |||
| HT7 | aa 159-165 | 77G7 | aa 316-335 | LoQ2: 16 pg/ml | |||
| ELISA p-tau Kawayabarashi | AntihTau441-E22A3 Rat IgG mAb | N/A | anti-hTau p181-Rk27A6 Rat monoclonal IgG Fab | pThr181 | 3.06 pg/ml | CSF | Kawarabayashi et al. [86] |
| a-EIMAF p-tau | RZ3 | pThr231 | DA9 | aa 102-140 | LoQ2: 0.002 fg/ml LoD1: 0.00001 fg/ml | Plasma, CSF, Serum | Rubenstein et al. [68] |
| SiMoA | Tau5 | aa 218-225 | HT7 | aa 159-164 | 0.02 pg/ml | Plasma | Zetterberg et al. [74] |
| BT2 | aa 194-198 | ||||||
| SiMoA p-tau | Tau5 | aa 218-225 | AT270 | pThr181 | LoD1: 0.0090 pg/ml | Plasma | Tatebe et al. [13] |
| SiMoA p-tau User Manual pThr181 | N/A | N/A | N/A | N/A | LoQ2: 1.204, 2.64 pg/ml LoD1: 0.756, 0.724 pg/ml |
CSF | SiMoA User Manuals [87] |
| SiMoA p-tau User Manual pThr231 | N/A | N/A | N/A | N/A | LoQ2: 1.23, 1.83 pg/ml LoD1: 0.284, 0.621 pg/ml |
CSF | SiMoA User Manuals [88] |
| ELECSYS t-tau | 5.28.464 | aa 150-230 | PC1C6/Tau-1 | aa 196-205 | LoD1: 18.6 pg/ml LoQ3: 62.6 pg/ml | CSF | Lifke et al. [75] |
| 4.35.411 | |||||||
| ELECSYS p-tau | 11H5V1 | aa 170-205 pThr181 | PC1C6/Tau-1 | aa 196-205 | LoD1: 1.96 pg/ml LoQ2: 3.9 pg/ml | ||
| MSD p-tau | AT270 | pThr181 | SULFO-TAG-LRL | N/A | N/A | Plasma | Mielke et al. [76] |
| xMAP Technology t-tau | HT7 | aa 159-163 | AT120 | N/A | 45-1500 pg/ml (Bjornstal et al.) | CSF | Olsson et al. [79] |
| xMAP Technology p-tau | HT7 | aa 159-163 | AT270 | pThr181 | 10-225 pg/ml (Bjornstal et al.) | ||
| SQUID IMR t-tau | HT7 | aa 159-164 | < 1 pg/ml | Plasma | Chiu et al. [89] | ||
| SQUID IMR p-tau | AT270 | pThr181 | 0.0196 - 10000 pg/ml | Plasma | Yang et al. [81] | ||
| AlphaLISA t-tau | (Donor): HT7 | aa 159-163 | (Acceptor): BT2 | aa 194-198 | N/A | Dujardin et al. [18] | |
| AlphaLISA p-tau | (Donor): AT8 | pSer202/pThr205 | (Acceptor): HT7 | aa 159-163 | N/A | ||
| AlphaLISA p-tau | (Donor): HT7 | aa 159-163 | (Acceptor): PHF-6 | pThr231 | N/A |
Several different methods use the principles of sandwich ELISA coupled with a highly sensitive detection method, including enhanced immunoassay using multi-arrayed fiberoptics (EIMAF), single-molecule array (SiMoA), the ELECSYS platform, and the Meso Scale Discovery (MSD) platform.
Previously known as surround optical fiber immunoassay (SOFIA), the enhanced immunoassay using multi-arrayed fiberoptics (EIMAF) uses the principle of a sandwich ELISA coupled with a highly sensitive detection method [67]. The EIMAF instrumentation collects the emission of a fluorescent protein using an assembly of optical fibers positioned to cover the entire optical radiation pattern of the sample. Scattered light is eliminated and the light from the sample is focused to a single optical fiber and detected using an amplifier [67]. Rubenstein et al. [68] used EIMAF coupled with rolling circle amplification (a-EIMAF). In rolling circle amplification (RCA), a circular DNA hybridizes to an oligonucleotide primer, which results in synthesis of a long repetitive linear DNA upon addition of DNA polymerases [69]. Subsequently, multiple fluorescent DNA probes can be hybridized in situ to the long synthesized DNA, resulting in signal amplification. In immunoassays, RCA can amplify the antibody-binding signal by conjugating an oligonucleotide primer to the detection antibody [70]. In a-EIMAF, the detection antibody is biotinylated and streptavidin is added, which allows a biotinylated DNA primer to initiate RCA. This enabled the detection of pThr181 tau levels in plasma, CSF, and serum with tremendously low reported limits of detection and quantification (Table 2) [68,71]. SiMoA uses paramagnetic beads conjugated with antibodies mixed with the protein of interest. Following a Poisson distribution, the number of the protein molecules is small enough, typically at least 10 times less than the number of paramagnetic beads, so that a singular bead is either bound to an individual molecule or not bound [72], allowing for “digital” detection of target molecules. The formed immunocomplexes are loaded into micro-fabricated wells specifically sized to hold individual beads and sealed with a substrate of reporter enzyme. The assay produces a “digitized” signal, with the wells being either “on” (fluorescent) or “off” (not fluorescent) [72,73]. Zetterberg et al. [74] were the first to use SiMoA to detect both normal and p-tau protein in CSF and plasma samples. Since then, the assay technology has been commercialized by the company Quanterix. Additionally, Tatebe et al. [13] employed the SiMoA technology using different antibodies to recognize tau phosphorylated at Thr181 (Table 2) with an impressive limit of detection of 0.0090 pg/mL.
Examples of immunoassays that are based on electrochemiluminescence to produce a sensitive signal include ELECSYS and MSD. The ELECSYS immunoassay platform uses an antibody labeled with a ruthenium complex that can be magnetically captured onto the surface of an electrode of a measuring cell. A voltage is then applied to induce chemiluminescent emission, which is measured and correlated with the concentration of target molecules [75]. Lifke et al. [75] used the ELECSYS platform to detect both total tau and pThr181 tau in CSF and found that its automated nature makes it a more reliable alternative to other ELISA assays (Table 2). MSD also uses electrochemiluminescence but employs plates that are precoated with certain biomarkers or other proteins. Since they can be spot coated with working electrodes, each well can have a different number of spots. Having more spots increases the assay sensitivity, and the different plate designs allow for more customizability of the assay. Mielke et al. [76] used a small spot streptavidin plate from MSD to detect pThr181 tau in plasma samples (Table 2). A biotinylated capture antibody was added to the streptavidin MSD plate and a SULFO-TAG conjugated detection antibody allowed for the production of electrochemiluminescent signal.
Other developed methods that differ from the ELISA include Luminex xMAP technology, the superconducting quantum interference device (SQUID) immunomagnetic reduction assay (IMR), and the AlphaLISA. The Luminex xMAP technology is a microsphere-based flow cytometric method that features beads covalently coupled with different antibodies to capture target proteins [77,78]. This allows the assay to measure several different target proteins in one test since each microsphere has spectrally specific fluorescence. Olsson et al. [79] used this technology to design a multiplex assay that measures pThr181 tau in CSF with a sensitivity of 10 pg/mL. The IMR utilizes the magnetic properties of magnetic nanoparticles with a sensor known as the SQUID to measure concentration-dependent signal [80]. This assay takes advantage of a magnetic property known as multiple-frequency alternating current (AC) magnetic susceptibility, XAC, which changes when antibody-coated magnetic nanoparticles interact with the target antigen. The change in XAC is measured by the SQUID and then correlated with concentration [80]. Yang et al. [81] have used IMR to detect pThr181 tau in plasma successfully with a limit of detection as low as 0.0196 pg/mL (Table 2). AlphaLISA, which depends on luminescent oxygen channeling chemistry and was initially described for its use in the luminescent oxygen channeling immunoassay (LOCI) [82,83], has also been adopted for the detection of total and p-tau. This method uses a “donor” bead and an “acceptor” bead. The donor bead donates a singlet oxygen to the acceptor bead after excitation at a wavelength of 680 nm [82]. The singlet oxygen then reacts with the acceptor bead to emit a signal with a wavelength of 615 nm. For this reaction to occur, the two beads need to be in close proximity to one another. To ensure this, both beads are coated with antibodies that bind specifically to the antigen of interest. For its use to detect total tau and p-tau, Dujardin et al. [18] used three separate antibody variations with AlphaLISA to detect total tau, pSer202/pThr205, and pThr231.
These advanced assay technologies enable the detection of total tau and p-tau in human CSF and plasma samples without tenuous enrichment steps. Since antibodies are essential in these assays, antibody validation should accompany their development. Accurate measurement of total and p-tau has great potential to track biomarkers and monitor disease progression in AD and other neurodegeneration. Moreover, they may enable early detection of AD long before symptom onset [84], which will open a new window for therapeutic intervention.
CONCLUSIONS
Plasma biomarkers are transforming our ability to detect AD and other forms of neurodegeneration early and to monitor the disease progression. This new capability will enable clinical trials during early stages of neurodegeneration and the assessment of drug efficacy in delaying or preventing its progression. P-tau biomarkers are especially valuable, given their diversity and relevance in pathology of neurodegeneration. Now that platforms with sensitivities able to detect p-tau in plasma have been developed (Table 2), efforts to validate assay specificity and expand the panel of p-tau biomarkers are in critical need. Antibodies are an essential part of highly sensitive immunoassays for the recognition of AD biomarkers, including p-tau. In addition to affinity, the specificity of the antibodies used is just as important in discriminating p-tau species. Through the analysis of p-tau antibody structures and molecular engineering efforts, it is becoming clear that optimizing affinity and specificity should go hand in hand. A major bottleneck is the lack of antibody specificity validation, and a number of studies have been carried out that show some widely used p-tau antibodies do not specifically bind to the intended phospho-site. While some important phospho-site specificity has been validated in monoclonal antibodies (pS198, pS199, pS202/pT205, pT231, pS396, pS404, pS422) (Table 1), many phospho-sites remain without validated antibodies. These results point to the importance of validating antibody specificity when choosing which antibodies to use in diagnostic immunoassays. Although the methods differ, antibody validation should accompany all assay developments to ensure robust detection of the target p-tau species.
ACKNOWLEDGEMENTs
We thank Erik Ammermann for helpful comments on the manuscript. This work was funded by the National Science Foundation grant 1706743.
Contributor Information
Monika Arbaciauskaite, Department of Chemical and Biomolecular Engineering, University of Connecticut, Storrs, CT 06269, USA.
Yu Lei, Department of Chemical and Biomolecular Engineering, University of Connecticut, Storrs, CT 06269, USA.
Yong Ku Cho, Department of Chemical and Biomolecular Engineering, University of Connecticut, Storrs, CT 06269, USA; Institute for Systems Genomics, Connecticut Institute for the Brain and Cognitive Sciences, University of Connecticut, Storrs, CT 06269, USA.
CONFLICT OF INTEREST STATEMENT
None declared.
References
- 1. Kidd, M. Paired helical filaments in electron microscopy of Alzheimer’s disease. Nature 1963; 197: 192–3. [DOI] [PubMed] [Google Scholar]
- 2. Fitzpatrick, AWP, Falcon, B, He, S et al. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 2017; 547: 185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ghetti, B, Oblak, AL, Boeve, BF et al. Invited review: Frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: a chameleon for neuropathology and neuroimaging. Neuropathol Appl Neurobiol 2015; 41: 24–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Šimić, G, Babić Leko, M, Wray, S et al. Tau protein hyperphosphorylation and aggregation in alzheimer’s disease and other tauopathies, and possible neuroprotective strategies. Biomolecules 2016; 6: 2–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mandelkow, EM, Mandelkow, E. Biochemistry and cell biology of tau protein in neurofibrillary degeneration. Cold Spring Harb Perspect Med 2012; 2: 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arakhamia, T, Lee, CE, Carlomagno, Y et al. Posttranslational modifications mediate the structural diversity of Tauopathy strains. Cell 2020; 180: 633–644.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bateman, RJ, Xiong, C, Benzinger, TLS et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med 2012; 367: 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roe, CM, Fagan, AM, Grant, EA et al. Amyloid imaging and CSF biomarkers in predicting cognitive impairment up to 7.5 years later. Neurology 2013; 80: 1784–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vos, SJB, Xiong, C, Visser, PJ et al. Preclinical Alzheimer’s disease and its outcome: a longitudinal cohort study. Lancet Neurol 2013; 12: 957–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thijssen, EH, La Joie, R, Wolf, A et al. Diagnostic value of plasma phosphorylated tau181 in Alzheimer’s disease and frontotemporal lobar degeneration. Nat Med 2020; 26: 387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palmqvist, S, Janelidze, S, Quiroz, YT et al. Discriminative accuracy of plasma Phospho-tau217 for Alzheimer disease vs other neurodegenerative disorders. JAMA 2020; 324: 772–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janelidze, S, Mattsson, N, Palmqvist, S et al. Plasma P-tau181 in Alzheimer’s disease: relationship to other biomarkers, differential diagnosis, neuropathology and longitudinal progression to Alzheimer’s dementia. Nat Med 2020; 26: 379–86. [DOI] [PubMed] [Google Scholar]
- 13. Tatebe, H, Kasai, T, Ohmichi, T et al. Quantification of plasma phosphorylated tau to use as a biomarker for brain Alzheimer pathology: pilot case-control studies including patients with Alzheimer’s disease and down syndrome. Mol Neurodegener 2017; 12: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mattsson-Carlgren, N, Janelidze, S, Palmqvist, S et al. Longitudinal plasma p-tau217 is increased in early stages of Alzheimer’s disease. Brain 2020; 143: 3234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hanger, DP, Betts, JC, Loviny, TLF et al. New phosphorylation sites identified in hyperphosphorylated tau (paired helical filament-tau) from Alzheimer’s disease brain using nanoelectrospray mass spectrometry. J Neurochem 1998; 71: 2465–76. [DOI] [PubMed] [Google Scholar]
- 16. Spires-Jones, TL, Stoothoff, WH, de Calignon, A et al. Tau pathophysiology in neurodegeneration: a tangled issue. Trends Neurosci 2009; 32: 150–9. [DOI] [PubMed] [Google Scholar]
- 17. Wesseling, H, Mair, W, Kumar, M et al. Tau PTM profiles identify patient heterogeneity and stages of Alzheimer’s disease. Cell 2020; 183: 1699–1713.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dujardin, S, Commins, C, Lathuiliere, A et al. Tau molecular diversity contributes to clinical heterogeneity in Alzheimer’s disease. Nat Med 2020; 26: 1256–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lewis, J, McGowan, E, Rockwood, J et al. Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L)tau protein. Nat Genet 2000; 25: 402–5. [DOI] [PubMed] [Google Scholar]
- 20. Barthélemy, NR, Bateman, RJ, Hirtz, C et al. Cerebrospinal fluid phospho-tau T217 outperforms T181 as a biomarker for the differential diagnosis of Alzheimer’s disease and PET amyloid-positive patient identification. Alzheimer’s Res Ther 2020; 12: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barthélemy, NR, Mallipeddi, N, Moiseyev, P et al. Tau phosphorylation rates measured by mass spectrometry differ in the intracellular brain vs. extracellular cerebrospinal fluid compartments and are differentially affected by Alzheimer’s disease. Front Aging Neurosci 2019; 11: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buerger, K, Teipel, SJ, Zinkowski, R et al. CSF tau protein phosphorylated at threonine 231 correlates with cognitive decline in MCI subjects. Neurology 2002; 59: 627–9. [DOI] [PubMed] [Google Scholar]
- 23. Neddens, J, Temmel, M, Flunkert, S et al. Phosphorylation of different tau sites during progression of Alzheimer’s disease. Acta Neuropathol Commun 2018; 6: 52. doi: 10.1186/s40478-018-0557-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Venkataraman, L, He, P, Schulz, P et al. Isolation and characterization of antibody fragment selective for human Alzheimer’s disease brain-derived tau variants. Neurobiol Aging 2020; 94: 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shih, HH, Tu, C, Cao, W et al. An ultra-specific avian antibody to phosphorylated tau protein reveals a unique mechanism for phosphoepitope recognition. J Biol Chem 2012; 287: 44425–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koerber, JT, Thomsen, ND, Hannigan, BT et al. Nature-inspired design of motif-specific antibody scaffolds. Nat Biotechnol 2013; 31: 916–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malia, TJ, Teplyakov, A, Ernst, R et al. Epitope mapping and structural basis for the recognition of phosphorylated tau by the anti-tau antibody AT8. Proteins Struct Funct Bioinforma 2016; 84: 427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chukwu, JE, Pedersen, JT, Pedersen, L et al. Tau antibody structure reveals a molecular switch defining a pathological conformation of the tau protein. Sci Rep 2018; 8: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang, H, Zhu, X, Pascual, G et al. Structural basis for recognition of a unique epitope by a human anti-tau antibody. Structure 2018; 26: 1–9. [DOI] [PubMed] [Google Scholar]
- 30. Bujotzek, A, Lipsmeier, F, Harris, SF et al. VH-VL orientation prediction for antibody humanization candidate selection: a case study. MAbs 2016; 8: 288–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pascual, G, Wadia, JS, Zhu, X et al. Immunological memory to hyperphosphorylated tau in asymptomatic individuals. Acta Neuropathol 2017; 133: 767–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chukwu, JE, Congdon, EE, Sigurdsson, EM et al. Structural characterization of monoclonal antibodies targeting C-terminal Ser 404 region of phosphorylated tau protein. MAbs 2019; 11: 477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Olson, CA, Liao, HI, Sun, R et al. mRNA display selection of a high-affinity, modification-specific phospho-iκbα-binding fibronectin. ACS Chem Biol 2008; 3: 480–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li, D, Wang, L, Maziuk, BF et al. Directed evolution of a picomolar-affinity, high-specificity antibody targeting phosphorylated tau. J Biol Chem 2018; 293: 12081–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Ameijde, J, Crespo, R, Janson, R et al. Enhancement of therapeutic potential of a naturally occurring human antibody targeting a phosphorylated Ser 422 containing epitope on pathological tau. Acta Neuropathol Commun 2018; 6: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Möller, W, Amons, R. Phosphate-binding sequences in nucleotide-binding proteins. FEBS Lett 1985; 186: 1–7. [DOI] [PubMed] [Google Scholar]
- 37. Bossemeyer, D. The glycine-rich sequence of protein kinases: a multifunctional element. Trends Biochem Sci 1994; 19: 201–5. [DOI] [PubMed] [Google Scholar]
- 38. Copley, RR, Barton, GJA. Structural analysis of phosphate and sulphate binding sites in proteins. Estimation of propensities for binding and conservation of phosphate binding sites. J Mol Biol 1994; 242: 321–9. [DOI] [PubMed] [Google Scholar]
- 39. Bordeaux, J, Welsh, AW, Agarwal, S et al. Antibody validation. Biotechniques 2010; 48: 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Andersson, S, Sundberg, M, Pristovsek, N et al. Insufficient antibody validation challenges oestrogen receptor beta research. Nat Commun 2017; 8: 15840. doi: 10.1038/ncomms15840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Uhlen, M, Bandrowski, A, Carr, S et al. A proposal for validation of antibodies. Nat Methods 2016; 13: 823–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kalina, T, Lundsten, K, Engel, P. Relevance of antibody validation for flow Cytometry. Cytom Part A 2020; 97: 126–36. [DOI] [PubMed] [Google Scholar]
- 43. Pillai-Kastoori, L, Heaton, S, Shiflett, SD et al. Antibody validation for western blot: by the user, for the user. J Biol Chem 2020; 295: 926–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rothbart, SB, Dickson, BM, Raab, JR et al. An interactive database for the assessment of histone antibody specificity. Mol Cell 2015; 59: 502–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nishikori, S, Hattori, T, Fuchs, SM et al. Broad ranges of affinity and specificity of anti-histone antibodies revealed by a quantitative peptide immunoprecipitation assay. J Mol Biol 2012; 424: 391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fuchs, SM, Krajewski, K, Baker, RW et al. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol 2011; 21: 53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Egelhofer, TA, Minoda, A, Klugman, S et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol 2011; 18: 91–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bock, I, Dhayalan, A, Kudithipudi, S et al. Detailed specificity analysis of antibodies binding to modified histone tails with peptide arrays. Epigenetics 2011; 6: 256–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Seet, BT, Dikic, I, Zhou, MM et al. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol 2006; 7: 473–83. [DOI] [PubMed] [Google Scholar]
- 50. Cornett, EM, Dickson, BM, Rothbart, SB. Analysis of histone antibody specificity with peptide microarrays. JoVE 2017; 126: e55912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li, D, Cho, YK. High specificity of widely used phospho-tau antibodies validated using a quantitative whole-cell based assay. J Neurochem 2020; 152: 122–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ercan, E, Eid, S, Weber, C et al. A validated antibody panel for the characterization of tau post-translational modifications. Mol Neurodegener 2017; 12: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Petry, FR, Pelletier, J, Bretteville, A et al. Specificity of anti-tau antibodies when analyzing mice models of Alzheimer’s disease: problems and solutions. PLoS One 2014; 9: e94251. doi: 10.1371/journal.pone.0094251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dawson, HN, Ferreira, A, Eyster, MV et al. Inhibition of neuronal maturation in primary hippocampal neurons from tau deficient mice. J Cell Sci 2001; 114: 1179–87. [DOI] [PubMed] [Google Scholar]
- 55. Cleveland, DW, Hwo, SY, Kirschner, MW. Physical and chemical properties of purified tau factor and the role of tau in microtubule assembly. J Mol Biol 1977; 116: 227–47. [DOI] [PubMed] [Google Scholar]
- 56. Cleveland, DW, Hwo, SY, Kirschner, MW. Purification of tau, a microtubule-associated protein that induces assembly of microtubules from purified tubulin. J Mol Biol 1977; 116: 207–25. [DOI] [PubMed] [Google Scholar]
- 57. Goedert, M, Crowther, RA, Garner, CC. Molecular characterization of microtubule-associated proteins tau and map2. Trends Neurosci 1991; 14: 193–9. [DOI] [PubMed] [Google Scholar]
- 58. Hanger, DP, Seereeram, A, Noble, W. Mediators of tau phosphorylation in the pathogenesis of Alzheimer’s disease. Expert Rev Neurother 2009; 9: 1647–66. [DOI] [PubMed] [Google Scholar]
- 59. Morris, M, Knudsen, GM, Maeda, S et al. Tau post-translational modifications in wild-type and human amyloid precursor protein transgenic mice. Nat Neurosci 2015; 18: 1183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ishiguro, K, Ohno, H, Arai, H et al. Phosphorylated tau in human cerebrospinal fluid is a diagnostic marker for Alzheimer’s disease. Neurosci Lett 1999; 270: 91–4. [DOI] [PubMed] [Google Scholar]
- 61. Kohnken, R, Buerger, K, Zinkowski, R et al. Detection of tau phosphorylated at threonine 231 in cerebrospinal fluid of Alzheimer’s disease patients. Neurosci Lett 2000; 287: 187–90. [DOI] [PubMed] [Google Scholar]
- 62. Blennow, K, Wallin, A, Agren, H et al. Tau protein in cerebrospinal fluid: a biochemical marker for axonal degeneration in Alzheimer disease? Mol Chem Neuropathol 1995; 26: 231–45. [DOI] [PubMed] [Google Scholar]
- 63. Fujirebio . Innotest Phospho-Tau(181P), 2016, 1–15
- 64. Hu, YY, He, SS, Grundke-iqbal, I et al. Levels of nonphosphorylated and phosphorylated tau in cerebrospinal fluid of Alzheimer’s disease patients: an ultrasensitive Bienzyme-substrate-recycle enzyme-linked Immunosorbent assay. Am J Pathol 2002; 160: 1269–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vandermeeren, M, Mercken, M, Vanmechelen, E et al. Detection of T proteins in normal and Alzheimer’s disease cerebrospinal fluid with a sensitive sandwich enzyme-linked Immunosorbent assay. J Neurochem 1993; 61: 1828–34. [DOI] [PubMed] [Google Scholar]
- 66. Meredith, JE Jr, Sankaranarayanan, S, Guss, V et al. Characterization of novel CSF tau and ptau biomarkers for Alzheimer’s disease. PLoS One 2013; 8: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chang, B, Gray, P, Piltch, M et al. Surround optical fiber immunoassay (SOFIA): an ultra-sensitive assay for prion protein detection. J Virol Methods 2009; 159: 15–22. [DOI] [PubMed] [Google Scholar]
- 68. Rubenstein, R, Chang, B, Davies, P et al. A novel, ultrasensitive assay for tau: potential for assessing traumatic brain injury in tissues and biofluids. J Neurotrauma 2015; 32: 342–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nilsson, M, Malmgren, H, Samiotaki, M et al. Padlock probes : circularizing oligonucleotides for localized DNA detection. Science (80- ) 1994; 265: 2085–8. [DOI] [PubMed] [Google Scholar]
- 70. Schweitzer, B, Wiltshire, S, Lambert, J et al. Immunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection. Proc Natl Acad Sci U S A 2000; 97: 10113–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rubenstein, R, Chang, B, Yue, JK et al. Comparing plasma phospho tau, total tau, and phospho tau–total tau ratio as acute and chronic traumatic brain injury biomarkers. JAMA Neurol 2017; 74: 1063–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rissin, DM, Kan, CW, Campbell, TG et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol 2010; 28: 595–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rissin, DM, Kan, CW, Song, L et al. Multiplexed single molecule immunoassays. Lab Chip 2013; 13: 2902–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zetterberg, H, Wilson, D, Andreasson, U et al. Plasma tau levels in Alzheimer’ s disease. Alzheimers Res Ther 2013; 5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lifke, V, Kollmorgen, G, Manuilova, E et al. Elecsys ® Total-tau and Phospho-tau (181P) CSF assays : analytical performance of the novel , fully automated immunoassays for quantification of tau proteins in human cerebrospinal fluid. Clin Biochem 2019; 72: 30–8. [DOI] [PubMed] [Google Scholar]
- 76. Mielke, MM, Hagen, CE, Xu, J et al. Plasma phospho-tau181 increases with Alzheimer’s disease clinical severity and is associated with tau- and amyloid-positron emission tomography. Alzheimers Dement 2018; 14: 989–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fulton, RJ, McDade, RL, Smith, PL et al. Advanced multiplexed analysis with the FlowMetrix TM system. Clin Chem 1997; 43: 1749–56. [PubMed] [Google Scholar]
- 78. Gordon, RF, McDade, RF. Multiplexed quantification of human IgG, IgA, and IgM with the FlowMetrixTM system. Clin Chem 1997; 43: 1799–801. [PubMed] [Google Scholar]
- 79. Olsson, A, Vanderstichele, H, Andreasen, N et al. Total tau, and phosphorylated tau (Thr 181) in cerebrospinal fluid by the xMAP technology. Clin Chem 2005; 51: 336–45. [DOI] [PubMed] [Google Scholar]
- 80. Chieh, JJ, Yang, SY, Jian, ZF et al. Hyper-high-sensitivity wash-free magnetoreduction assay on biomolecules using high-Tc superconducting quantum interference devices. J Appl Phys 2008; 103: 014703. [Google Scholar]
- 81. Yang, C-C, Chiu, M-J, Chen, T-F et al. Assay of plasma phosphorylated tau protein (threonine 181) and total tau protein in early-stage Alzheimer’s disease. J Alzheimers Dis 2018; 61: 1323–32. [DOI] [PubMed] [Google Scholar]
- 82. Ullman, EF, Kirakossian, H, Singh, S et al. Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence. Proc Natl Acad Sci U S A 1994; 91: 5426–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ullman, EF, Kirakossian, H, Switchenko, AC et al. Luminescent oxygen channeling assay (LOCI(TM)): sensitive, broadly applicable homogeneous immunoassay method. Clin Chem 1996; 42: 1518–26. [PubMed] [Google Scholar]
- 84. Long, JM, Holtzman, DM. Alzheimer’s disease: an update on pathobiology and treatment strategies. Cell 2019; 179: 312–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yamamori, H, Khatoon, S, Grundke-Iqbal, I et al. Tau in cerebrospinal fluid: a sensitive sandwich enzyme-linked immunosorbent assay using tyramide signal amplification. Neurosci Lett 2007; 418: 186–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kawarabayashi, T, Nakamura, T, Miyashita, K et al. Novel ELISAs to measure total and phosphorylated tau in cerebrospinal fluid. Neurosci Lett 2020; 722: 134826. [DOI] [PubMed] [Google Scholar]
- 87. Quanterix . Simoa ® pTau-181 Advantage Kit HD-1 / HD-X Data Sheet, 2019, 1–2
- 88. Quanterix . Simoa ™ pTau-231 Advantage Kit SR-X ™ Data Sheet, 2017, 1–2
- 89. Chiu, M, Yang, S, Horng, H et al. Combined plasma biomarkers for diagnosing mild cognition impairment and Alzheimer’s disease. ACS Chem Nerosci 2013; 4: 1530–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Armbruster, DA, Pry, T. Limit of blank, limit of detection, and limit of quantification. Clin Biochem Rev 2008; 29: S49–52. [PMC free article] [PubMed] [Google Scholar]
- 91. McNicholas, S, Potterton, E, Wilson, KS et al. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr Sect D Biol Crystallogr 2011; 67: 386–94. [DOI] [PMC free article] [PubMed] [Google Scholar]


