SUMMARY
Mutations in the genes encoding the lysine demethylase 5 (KDM5) family of histone demethylases are observed in individuals with intellectual disability (ID). Despite clear evidence linking KDM5 function to neurodevelopmental pathways, how this family of proteins impacts transcriptional programs to mediate synaptic structure and activity remains unclear. Using the Drosophila larval neuromuscular junction (NMJ), we show that KDM5 is required presynaptically for neuroanatomical development and synaptic function. The Jumonji C (JmjC) domain-encoded histone demethylase activity of KDM5, which is expected to be diminished by many ID-associated alleles, is required for appropriate synaptic morphology and neurotransmission. The activity of the C5HC2 zinc finger is also required, as an ID-associated mutation in this motif reduces NMJ bouton number, increases bouton size, and alters microtubule dynamics. KDM5 therefore uses demethylase-dependent and independent mechanisms to regulate NMJ structure and activity, highlighting the complex nature by which this chromatin modifier carries out its neuronal gene-regulatory programs.
Graphical Abstract

In Brief
Mutations in the KDM5 family of histone demethylases are observed in individuals with intellectual disability (ID). Belalcazar et al. show that KDM5-regulated transcription is necessary in Drosophila for proper neuroanatomical development and neurotransmission at the glutamatergic larval neuromuscular junction.
INTRODUCTION
Dysregulation of gene expression in the central nervous system (CNS) has profound effects on cognitive and other neurological functions (Ronan et al., 2013). The recent expansion of genomic analyses in patients with neurodevelopmental disorders (NDDs) has dramatically increased our understanding of the transcriptional regulators that might contribute to cognitive impairment. These studies have revealed 122 distinct variants in the lysine demethylase 5 (KDM5) family genes KDM5A, KDM5B, and KDM5C that encode chromatin modifier proteins in individuals with NDDs, particularly intellectual disability (ID) (Kim et al., 2017; Vallianatos and Iwase, 2015).
KDM5 proteins are gene expression regulators that enzymatically remove di- and/or trimethylated lysine 4 of histone H3 (H3K4me2/3) through the activity of a conserved Jumonji C (JmjC) domain (Hyun et al., 2017). The H3K4me2/3 chromatin marks are predominantly found at the promoter region of transcriptionally active genes and changes to these histone modifications can impact transcriptional consistency (Barski et al., 2007; Benayoun et al., 2014). Prevailing models of KDM5-induced ID suggest that it is the loss of histone demethylase activity that drives the neuropathologies observed in patients. In support of this, in vitro studies using recombinant KDM5C harboring ID-associated variants showed reduced activity toward a histone peptide substrate (Iwase et al., 2007; Rujirabanjerd et al., 2010; Tahiliani et al., 2007). Consistent with this observation, a Drosophila mutant specifically lacking KDM5 demethylase activity shows learning and memory deficits (Zamurrad et al., 2018). Additionally, the cognitive deficits observed in Kdm5c knockout mice can be partially ameliorated by genetically lowering levels of the H3K4 methyltransferase KMT2A (Kmt2a heterozygotes) (Vallianatos et al., 2020). Contrasting these findings, some ID-associated mutations in KDM5C do not cause deficits to in vitro demethylase activity (Tahiliani et al., 2007; Vallianatos et al., 2018). Both non-enzymatic and enzymatic mechanisms of gene regulation by KDM5 family proteins may therefore be important for neuronal function.
There is still much to be learned regarding the cellular consequences of mutations in KDM5 family genes. Structural changes in neuronal morphology and circuitry are often observed in postmortem brain samples from individuals with neurodevelopmental and neuropsychiatric disorders (Forrest et al., 2018). These analyses have yet to be performed in patients with mutations in KDM5 family genes. However, model organisms have provided insights about the consequences of KDM5 loss of function on neuronal morphology. For example, knockdown of Kdm5c in cultured rat cerebellar granular neurons results in changes in dendritic arborization (Iwase et al., 2007) and Kdm5c knockout mice show decreased cortical neuron spine density (Iwase et al., 2016; Scandaglia et al., 2017). Although dendritic spine morphology can be altered by neuronal activity (Nägerl et al., 2004; Verpelli et al., 2010), the extent to which synaptic function is disrupted by mutations in KDM5 genes remains uncharacterized.
To further our understanding of the link between KDM5-regulated transcription and neuronal development, we examined its function at the Drosophila neuromuscular junction (NMJ). This glutamatergic synapse is a well-established model for addressing questions of neural development, morphology, and synaptic function, as it requires proteins homologous to those found in mammalian CNS excitatory synapses (Koh et al., 2000; Li et al., 2007; Rasse et al., 2005; Roos et al., 2000). Mutations in the orthologs of human ID-associated genes that encode synaptic proteins found at glutamatergic terminals such as ionotropic and metabotropic glutamate receptors, the scaffold protein PSD95/Dlg, the phosphoprotein synapsin, and the cell-adhesion proteins neurexin and neuroligin (Moretto et al., 2018; Volk et al., 2015) cause structural and functional alterations at the Drosophila NMJ (Budnik et al., 1996; DiAntonio, 2006; Vasin et al., 2014; Xing et al., 2018). Drosophila encodes a single KDM5 ortholog (also known as Lid) that shares homology with all four mammalian KDM5 proteins. Thus, in vivo analyses of KDM5 are expected to reveal activities relevant to KDM5A-, KDM5B-, and KDM5C-induced ID. In this study, we show that KDM5 is required presynaptically within motor neurons for proper NMJ morphology and function. The histone demethylase activity of KDM5 is a key contributor to its synaptic functions, as animals lacking this activity show a reduced number of synaptic boutons and a decrease in evoked glutamate release. Additionally, we find that KDM5’s neuroregulatory functions require the C5HC2 zinc-finger motif, which has no currently known function. This is based on our analyses of a variant within the C5HC2 motif of KDM5, equivalent to an ID-associated variant in KDM5C, that reduces the number of synaptic boutons and increases their size. Interestingly, this ID-associated allele does not appear to dramatically alter histone demethylase activity, suggesting that KDM5 uses enzymatic and non-enzymatic mechanisms of gene regulation to properly form and maintain NMJs. Consistent with these data, RNA-sequencing (RNA-seq) analyses of the ventral nerve cord (VNC), where the nuclei of motor neurons reside, revealed that mutations in the JmjC domain or C5HC2 motif cause distinct transcriptional changes. Combined, these data advance our understanding of the neuronal activities of KDM5 and highlight the importance of discrete domains that mediate its gene-regulatory programs in neurons.
RESULTS
KDM5 is required presynaptically for proper NMJ structure
During Drosophila embryonic development, motor neuron axons establish connections with target muscles forming branches of synaptic boutons, which are constantly added in order to maintain synaptic strength as the larva grows (Gramates and Budnik, 1999; Menon et al., 2013). Changes to bouton size and/or number are indicative of neurodevelopmental defects that are often observed in Drosophila models of NDDs (Bellosta and Soldano, 2019; Bodaleo and Gonzalez-Billault, 2016). To characterize the role of KDM5 at the synapse, we compared the NMJ structure in kdm5140-null mutant animals to controls (w1118). NMJs were visualized by immunolabeling third-instar larval NMJs with the neuronal membrane marker horseradish peroxidase (HRP) and the postsynaptic scaffold protein Discs large (Dlg), which is the Drosophila homolog of PSD95. We then quantified the number and size of type Ib synaptic boutons, which are predominantly responsible for muscle contraction (Newman et al., 2017), at muscle 4 of the third abdominal segment. Because kdm5140 mutants show a slow-growth phenotype that results in larvae taking longer to develop to the same size as wild-type (WT) animals, we matched animals developmentally rather than chronologically, as we have done previously (Drelon et al., 2018, 2019). In addition, we normalized the number of boutons to muscle size to account for any slight differences in larval size. These analyses revealed that loss of kdm5 alters NMJ morphology by decreasing the number and increasing the size of synaptic boutons (Figures 1A, 1B, 1F, and 1H).
Figure 1. kdm5 expression is required in motor neurons for normal NMJ development.

(A1–A3) NMJ morphology at muscle 4 of abdominal segment 3 (NMJ4-A3) of control (w1118) third-instar larvae labeled with the presynaptic marker HRP (magenta; A1) and postsynaptic marker Dlg (green; A2), and merged (A3).
(B1–B3) kdm5140 homozygous larva.
(C1–C3) kdm5 knockdown larva (Act5C>kdm5-shRNA).
(D1–D3) kdm5140; elav>UAS-kdm5.
(E1–E3) kdm5140; mef2>UAS-kdm5.
(A3′–E3′) Boxed areas in (A3)–(E3) showing higher magnification of type Ib boutons. Scale bars, 10 μm (E3) and 5 μm (E3′).
(F) Quantification of type Ib bouton number normalized to muscle surface area. ***p = 0.0001, **p = 0.0041. Control (w1118) n = 25, kdm5140 n = 20, Act5C>kdm5-shRNA n = 14, control (+; shRNA) n = 14. Error bars: mean ± SEM
(G) Quantification of type Ib bouton number normalized to muscle surface area. ****p < 0.0001, **p = 0.0029, *p = 0.0036 (OK6), *p = 0.0282 (da); ns, not significant. Control (w1118) n = 16, no driver (kdm5140; UAS-kdm5) n = 14, kdm5140; elav>kdm5 n = 12, kdm5140; OK6>kdm5 n = 10, kdm5140; mef2>kdm5 n = 10, kdm5140; repo>kdm5 n = 12, kdm5140; Act5C>kdm5 n = 10, kdm5140; da>kdm5 n = 11. Error bars: mean ± SEM
(H) Quantification of type Ib bouton size in kdm5140 larvae and ubiquitous knockdown (Act5C>kdm5-shRNA). Violin plots show the frequency distribution of bouton surface area indicating the median and quartiles. **p = 0.0020. Control (w1118) n = 327, kdm5140 n = 280, Act5C>kdm5-shRNA n = 198, control (+; shRNA) n = 281.
(I) Quantification of type Ib bouton size in rescue experiments using the genotypes shown. ****p < 0.0001, p = 0.064. Control (w1118) n = 340, no driver (kdm5140; UAS.kdm5) n = 149, kdm5140; elav>kdm5 n = 245, kdm5140; OK6>kdm5 n = 191, kdm5140; mef2>kdm5 n = 123, kdm5140; repo>kdm5 n = 254, kdm5140; Act5C>kdm5 n = 181, kdm5140; da>kdm5 n = 201.
(J1–K2) Futsch (white) immunolabeling in segments of NMJ4-A3 showing microtubules within synaptic boutons and HRP (magenta) from control (J1 and J2) and kdm5140 (K1 and K2). Green triangles indicate punctate signal and yellow triangles indicate looped boutons. Scale bar, 10 μm.
(L) Quantification of unbundled boutons, punctate and looped, as the percentage of total boutons in NMJ4-A3 of control and kdm5140. *p = 0.0137, **p = 0.0047. Control n = 20, kdm5140 n = 21. Error bars: mean ± SEM
To confirm the kdm5140-null mutant phenotype and rule out the possibility that the altered larval growth rate of kdm5140 animals indirectly caused the NMJ phenotypes observed, we used an inducible kdm5 short hairpin RNA (shRNA) transgene (Chen et al., 2019; Liu and Secombe, 2015). Ubiquitous knockdown of kdm5 using Act5C-Gal4 reduced KDM5 protein levels by ~75%, which is not sufficient to delay larval growth in the same manner as the null allele (Figure S1). kdm5 knockdown animals recapitulated the reduced NMJ bouton number phenotype that we observed in kdm5140 larvae (Figures 1C, 1F, and 1H). Reducing kdm5 also led to a modest increase in NMJ bouton size (p = 0.05) that was less pronounced but consistent with the kdm5140-null phenotype. Combined, these data suggest that KDM5 is required to promote NMJ bouton number and to restrict bouton size.
Changes in bouton size correlate with changes in microtubule (MT) stability, which can be visualized by examining the distribution of the MT-binding protein Futsch (Nechipurenko and Broihier, 2012; Viquez et al., 2006). We therefore stained the NMJs of control and kdm5140 animals with anti-Futsch and quantified the number of boutons containing unbundled MTs, which are characterized as looped or punctate staining (Packard et al., 2002). kdm5140 mutant larvae displayed a higher proportion of boutons with unbundled MTs (punctate 24.6%, looped 9%) compared to controls (punctate 17.6%, looped 3.2%) (Figures 1J–1L). Thus, the increased proportion of unbundled MTs caused by loss of KDM5 may contribute to the increase in bouton size observed in these animals.
To determine where KDM5 function is required to mediate its effects on NMJ morphology, we restored kdm5 expression in specific cell types within kdm5140 protein-null animals. To do this, we used a UAS-kdm5 transgene that drives low levels of expression (~2-fold increase over endogenous levels) (Drelon et al., 2019; Li et al., 2010; Secombe et al., 2007) and a range of well-characterized Gal4 drivers. Re-expression of kdm5 ubiquitously (Act5C-Gal4 and da-Gal4), pan-neuronally (elav-Gal4), or in motor neurons (OK6-Gal4) significantly rescued the bouton number deficit of kdm5140 larvae (Figures 1D and 1G). Restoring kdm5 expression in muscles (Mef2-Gal4) or glia (repo-Gal4), however, did not (Figures 1E and 1G). Similar findings were observed for bouton size phenotype in kdm5140 larvae. Neuronal re-expression of kdm5 using elav-Gal4 and OK6-Gal4 restored bouton size similar to WT, whereas muscle or glial expression did not (Mef2-Gal4 and repo-Gal4, respectively) (Figure 1I). These data suggest that KDM5 impacts NMJ growth by regulating presynaptic gene expression.
The JmjC domain and the C5HC2 motif of KDM5 are required for NMJ structure
Approximately half of the 122 reported NDD-associated variants in KDM5A, KDM5B, or KDM5C are predicted to compromise histone demethylase activity because they are expected to reduce or eliminate protein expression (Brookes et al., 2015; De Rubeis et al., 2014; Gonçalves et al., 2014; Najmabadi et al., 2011). We therefore examined the contribution of this chromatin-modifying activity to NMJ morphology using a fly strain with two point mutations in the JmjC domain that abolish enzymatic function (kdm5JmjC* allele) (Drelon et al., 2018; Navarro-Costa et al., 2016; Zamurrad et al., 2018) (Figure 2A). kdm5JmjC* homozygous larvae exhibited a decrease in bouton number but no change in bouton size compared to the isogenic control strain for this mutation (kdm5WT) (Figures 2B–2E). Thus, the catalytic activity of KDM5 is required for only one of the phenotypes observed in the kdm5-null mutant, indicating that non-enzymatic activities are also required for proper NMJ structure.
Figure 2. The demethylase activity of KDM5 is required to regulate NMJ bouton number.

(A) Schematic of Drosophila KDM5 showing the position of the mutations in the JmjC domain that abolish enzymatic activity.
(B1–C3) NMJ4-A3 of wild-type (kdm5WT; B1–B3) and demethylase-inactive (kdm5JmjC*; C1–C3) third-instar larvae stained with HRP (B1 and C1) and Dlg (B2 and C2), and merged (B3 and C3). Scale bar, 10 μm.
(D) Quantification of type Ib bouton number normalized to muscle surface area in kdm5WT and kdm5JmjC*. ****p < 0.0001. kdm5WT n = 15, kdm5JmjC* n = 31. Error bars: mean ± SEM
(E) Quantification of type Ib bouton size in kdm5WT and kdm5JmjC*. kdm5WT n = 234, kdm5JmjC* n = 388. ns, not significant.
To identify potential non-enzymatic means by which KDM5 could function in motor neurons, we surveyed known genetic alterations to human KDM5 genes that are associated with ID. Interestingly, 29% of ID-associated missense variants in KDM5C occur within the helical region harboring the uncharacterized C5HC2 zinc-finger motif (Human Gene Mutation Database; HGMD) (Stenson et al., 2017), suggesting a key functional role for this domain. We therefore generated three fly strains harboring ID-associated mutations in the C5HC2 domain: kdm5L854F, kdm5R873W, and kdm5Y874C, which are equivalent to KDM5C (p.L731F), (p.R750W), and (p.Y751C), respectively (Figure 3A) (Jensen et al., 2005; Tzschach et al., 2006). These alleles were chosen because they result in moderate to severe ID, suggesting they may more dramatically change gene expression in neuronal lineages.
Figure 3. Fly strains harboring ID-associated mutations are viable and developmentally similar to controls.
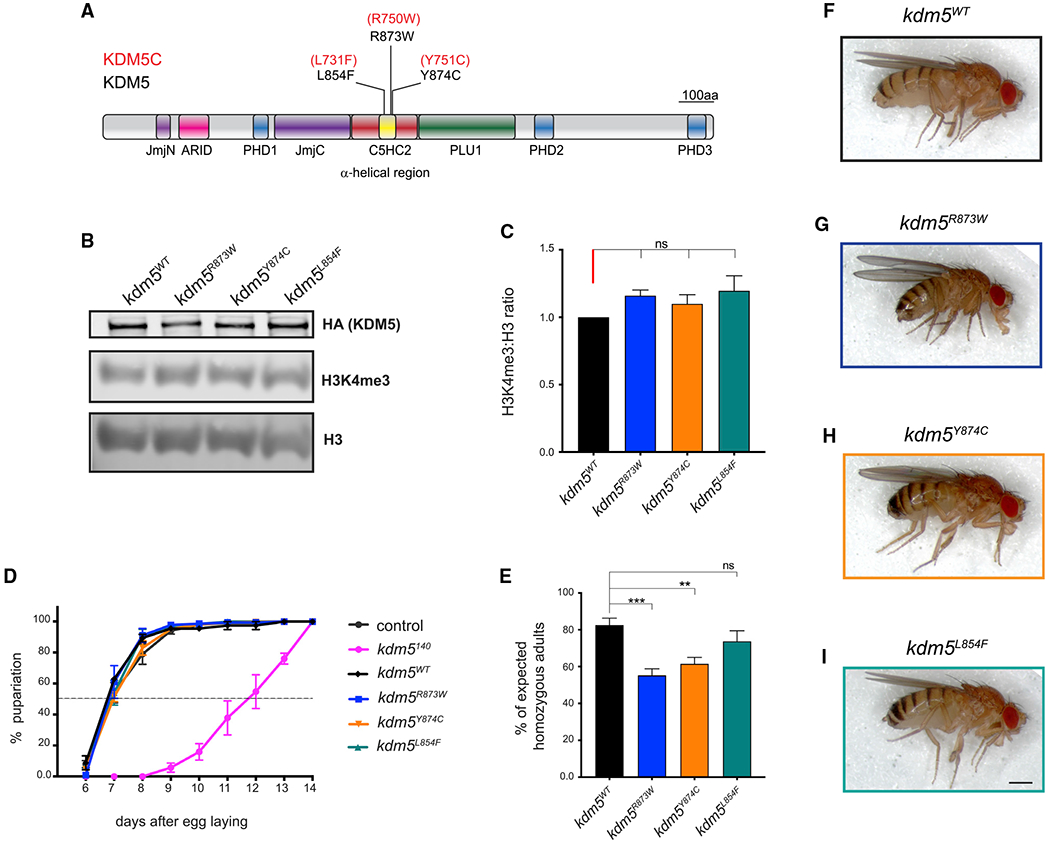
(A) Schematic of Drosophila KDM5 showing missense mutations that are equivalent ID-associated variants in human KDM5C.
(B) Western blot of larval CNS from kdm5WT, kdm5R873W, kdm5Y874C, and kdm5L854F showing expression of HA-tagged KDM5 (top), H3K4me3 (middle), and total histone H3 (bottom).
(C) Triplicate quantification of H3K4me3 levels relative to total histone H3 in kdm5R873W, kdm5Y874C, and kdm5L854F compared to the ratio observed in kdm5WT. ns, not significant. Error bars: mean ± SEM
(D) Developmental timing of control (w1118), kdm5WT, kdm5140, kdm5R873W, kdm5Y874C, and kdm5L854F. The dashed line indicates 50% pupariation. Control n = 88, kdm5140 n = 52, kdm5WT n = 398, kdm5R873W n = 216, kdm5Y874C n = 294, kdm5L854F n = 137. Error bars: SEM
(E) Adult survival of homozygous kdm5WT, kdm5R873W, kdm5Y874C, and kdm5L854 adult flies from intercrossed heterozygous parents as a percentage of the number expected based on Mendelian expectations. ***p = 0.0001, **p = 0.0057. Total scored flies: kdm5WT n = 359, kdm5R873W n = 1,164, kdm5Y874C n = 1,193, kdm5L854F n = 544. Error bars: mean ± SEM
(F–I) kdm5WT (F), kdm5R873W (G), kdm5Y874C (H), and kdm5L854F (I) female flies. Scale bar, 50 μm.
Similar to our approach for generating the kdm5JmjC* allele, we used a genomic rescue strategy to generate ID-associated alleles (Drelon et al., 2018; Li et al., 2010; Liu and Secombe, 2015; Navarro-Costa et al., 2016; Zamurrad et al., 2018). ID mutant fly strains lack the endogenous kdm5 locus and have the entire 11-kb kdm5 genomic region containing the point mutation and three tandem copies of the hemagglutinin (HA) epitope tag in an attP site at cytological location 86F. The WT control strain for these alleles is homozygous for the kdm5140 mutation and the HA-tagged WT locus at 86F (kdm5WT) (Drelon et al., 2018; Zamurrad et al., 2018).
To determine whether the ID-associated alleles affected KDM5 protein levels, we performed a western blot using dissected larval CNS from kdm5L854F, kdm5R873W, or kdm5Y874C animals and found no change compared to kdm5WT (Figure 3B). Because a key feature of kdm5140-null mutant larvae is slowed larval growth that delays pupariation, we quantified the developmental timing of kdm5L854F, kdm5R873W, and kdm5Y874C animals. Similar to our previous analyses of homozygous kdm5JmjC* animals (Drelon et al., 2018), ID mutant strains showed an identical developmental profile to kdm5WT (Figure 3D). Thus, we were able to examine the NMJ phenotypes of these animals without the complication of altered larval growth rate. All three ID-associated mutant fly strains were homozygous viable and visibly morphologically normal, although homozygous kdm5R873W and kdm5Y874C adult flies occurred slightly less frequently than expected (Figures 3E–3I). These alleles, therefore, do not dramatically alter the developmental functions of KDM5 that are required for adult viability.
Alleles of kdm5 that abolish enzymatic activity cause a global 2-fold increase in the ratio of H3K4me3 to total histone H3 (Drelon et al., 2018; Navarro-Costa et al., 2016; Secombe et al., 2007; Zamurrad et al., 2018). To examine the effect of the KDM5L854F, KDM5R873W, and KDM5Y874C mutations on histone demethylase activity in vivo, we similarly quantified levels of H3K4me3 using dissected larval CNS tissue. In contrast to kdm5JmjC*, kdm5L854F, kdm5R873W, and kdm5Y874C did not affect H3K4me3 levels compared to total histone H3 (Figures 3B and 3C). Although it remains possible that these mutations have modest or gene-specific effects that are undetectable by western blot, these alleles do not abolish demethylase activity in vivo.
To further characterize these ID-associated mutations, we examined their thermodynamic effect on KDM5 using the web server DynaMut, which calculates the change in folding Gibbs free energy (ΔΔG) by combining normal-mode analysis and graph-based signatures. We also used DynaMut to measure the vibrational entropy (ΔΔSVib), a measure of molecule flexibility (Rodrigues et al., 2018). Because there is no crystal structure of Drosophila KDM5, we analyzed the KDM5A structure PDB: 5CEH, which covers a broader N-terminal region and C-terminal helical zinc-binding domain compared to the structures available for KDM5C (Vinogradova et al., 2016) (Figure S2A). All residues investigated are conserved between Drosophila KDM5, human KDM5A, and human KDM5C. All three mutations were predicted to thermodynamically stabilize the protein with the maximum effect shown by Y720C (Y874C in Drosophila) (ΔΔG: 1.759 kcal/mol), followed by L700F (L854F in Drosophila) (ΔΔG: 1.624 kcal/mol) (Figures S2B–S2D). The latter is predicted to generate new hydrophobic contacts with residues localized in the adjacent helix and increase hydrogen bonds with a residue inside the first zinc-binding site (Figure S2B). DynaMut also predicted the greatest decrease in molecule flexibility for L700F/L854F using the Elastic Network Contact Model (ΔΔSVib ENCoM: −5.152 kcal·mol−1¸K−1). These results, together with the position of the L700/L854 residue in the middle of the first zinc-binding site, suggest that this amino acid change may have the most dramatic impact on KDM5 function compared to the other amino acid substitutions (Figure S2A).
To determine the effect of kdm5L854F, kdm5R873W, and kdm5Y874C on NMJ morphology, we quantified type Ib synaptic bouton number and size in homozygous mutant larvae. kdm5L854F larvae had significantly fewer muscle 4 synaptic boutons compared to kdm5WT. In contrast, kdm5Y874C animals exhibited a mild reduction (p = 0.077) whereas kdm5R873W bouton number was indistinguishable from kdm5WT animals (Figures 4A–4E). We additionally found an increase in kdm5L854F bouton size whereas kdm5R873W or kdm5Y874C mutants were unaffected compared to kdm5WT (Figure 4F). Based on our observation that kdm5140 mutant larvae show a change in MT dynamics (Figures 1J–4L), we carried out anti-Futsch staining of the two mutants that showed morphological NMJ phenotypes, kdm5L854F and kdm5JmjC*. kdm5L854F mutant larvae showed a higher proportion of boutons with looped MTs compared to kdm5WT (36.6% versus 15.7%, respectively). In contrast, no difference was observed in the proportion of boutons with unbundled MTs in kdm5JmjC* animals (Figures 4G–4J). Together, our analyses of Futsch staining in kdm5140, kdm5L854F, and kdm5JmjC* mutants suggest a link between increased bouton size and altered MT dynamics at the synapse. In addition, these data suggest a key role for the C5HC2 motif, but not the JmjC domain, in this process.
Figure 4. The L854F mutation in the C5HC2 domain of KDM5 affects NMJ bouton size and number.
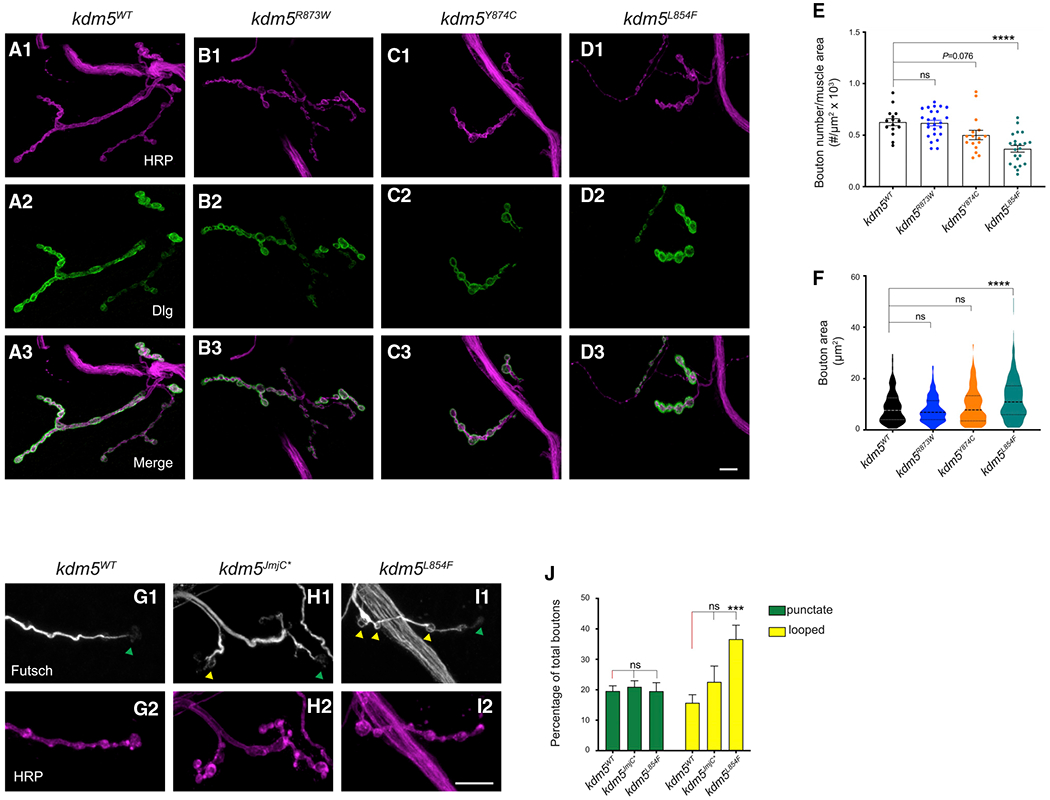
(A1–D3) NMJ morphology at muscle 4-A3 of third-instar larvae labeled with HRP (magenta; A1, B1, C1, and D1) and Dlg (green; A2, B2, C2, and D2), and merged (A3, B3, C3, and D3).
(A1–A3) kdm5WT.
(B1–B3) kdm5R873W.
(C1–CC3) kdm5Y874C.
(D1–D3) kdm5L854F. Scale bar, 10 μm.
(E) Quantification of type Ib bouton number normalized to muscle surface area from kdm5WT, kdm5R873W, kdm5Y874C, and kdm5L854 larvae. ****p < 0.0001. kdm5WT n = 15, kdm5R873W n = 25, kdm5Y874C n = 16, kdm5L854F n = 22. ns, not significant. Error bars: mean ± SEM
(F) Quantification of type Ib bouton size from kdm5WT, kdm5R873W, kdm5Y874C, and kdm5L854 larvae. ****p<0.0001. kdm5WT n = 309, kdm5R873W n = 214, kdm5Y874C n = 97, kdm5L854F n = 176.
(G–I) Futsch (white) immunolabeling and HRP (magenta) in NMJ4-A3 from kdm5WT (G1 and G2), kdm5JmjC* (H1 and H2), and kdm5L854 (I1 and I2) larvae. Green triangles indicate boutons with punctate signal and yellow triangles indicate looped boutons. Scale bar, 10 μm.
(J) Quantification of unbundled boutons, punctate (green) and looped (yellow), as the percentage of total boutons in NMJ4-A3 of kdm5WT, kdm5JmjC*, and kdm5L854 larvae. ***p = 0.0007. kdm5WT n = 19, kdm5JmjC* = 12, kdm5L854F n = 14. Error bars: mean ± SEM
Mutations in the JmjC domain and the C5HC2 motif of KDM5 show distinct effects on NMJ function
Because the kdm5JmjC* demethylase-dead strain and the kdm5L854F ID mutant showed NMJ phenotypes without altering larval growth rate, we chose to focus further analyses on these two strains. In addition, kdm5L854F does not increase global levels of H3K4me3 (Figures 3B and 3C) and its NMJ phenotypes were not identical to the demethylase-dead kdm5JmjC*. Comparing these two strains therefore allows us to examine KDM5’s demethylase activity-dependent and independent functions at the NMJ. To test whether the structural defects observed in kdm5JmjC* and kdm5L854F were associated with larval movement deficits, we quantified the crawling behavior of mutant third-instar larvae compared to control kdm5WT (Kashima et al., 2017; Li et al., 2007). There was a significant decreasein locomotor performance, as both genotypes traveled a shorter total distance (path length) compared to kdm5WT (Figures 5A and 5E). The shorter distance traveled was likely due to reduced locomotor speed as kdm5JmjC* and kdm5L854F animals showed lower average and maximal velocities as well as average speed normalized to larval size (Figures 5B–5D). Disrupting the function of the JmjC domain orthe C5HC2 motif therefore affects functional motor outputs.
Figure 5. KDM5 mutations in the JmjC and C5HC2 domains affect larval locomotion.
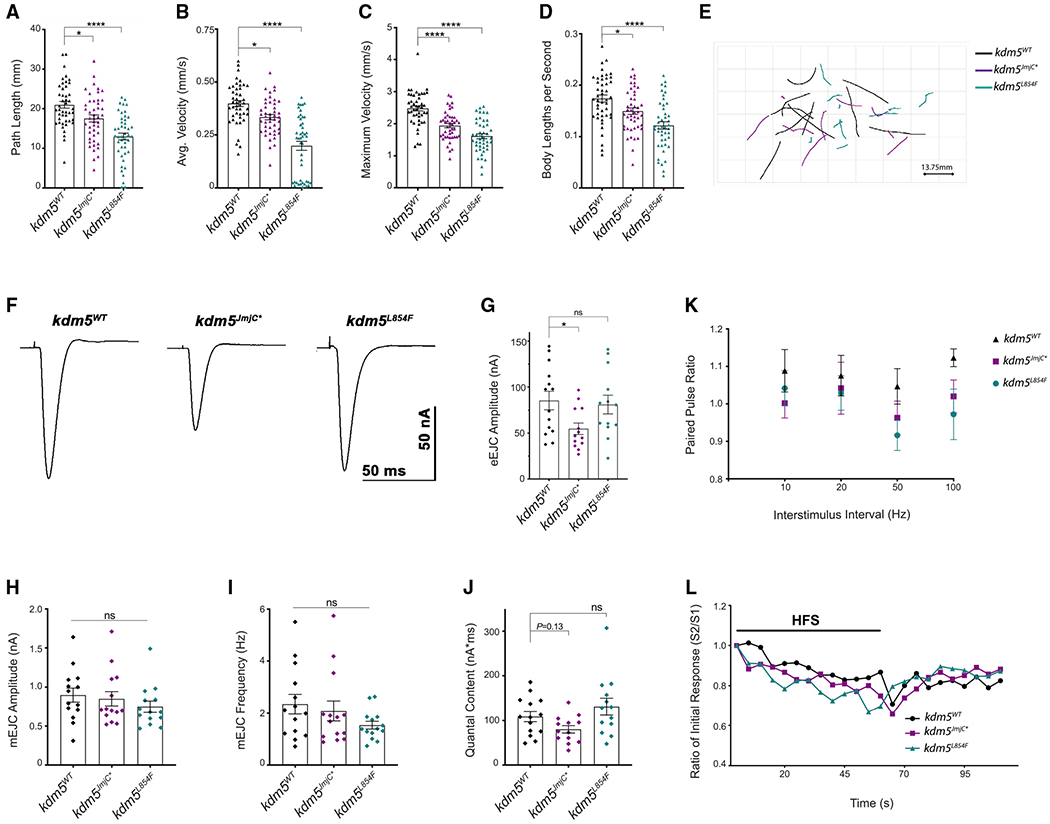
(A–E) Movement of third-instar kdm5WT, kdm5JmjC*, and kdm5L854F larvae. kdm5WT n = 45, kdm5JmjC* n = 45, kdm5L854F n = 45.
(A) Traveled distance. *p = 0.0114, ****p < 0.0001. Error bars: mean ± SEM
(B) Average speed. *p = 0.0105, ****p < 0.0001. Error bars: mean ± SEM
(C) Maximum speed. ****p < 0.0001. Error bars: mean ± SEM
(D) Average speed normalized to larval size. *p = 0.0248, ****p < 0.0001. Error bars: mean ± SEM
(E) Representative path trajectories of 10 larvae from each genotype.
(F) Representative evoked excitatory junctional currents (eEJCs) recorded from muscle 6 in kdm5WT, kdm5JmjC*, and kdm5L854F larvae.
(G) Quantification of eEJC amplitudes. *p = 0.0368; ns, not significant. Error bars: mean ± SEM
(H and I) Quantification of miniature excitatory junctional current (mEJC) amplitude (H) and frequency (I). Error bars: mean ± SEM
(J) Quantification of quantal content (eEJC area (nA*ms)/mEJC area (nA*ms)).Error bars: mean ± SEM
(K) Paired-pulse ratio (eEJC amplitude second response/eEJC amplitude first response) across multiple interstimulus intervals. Error bars: SEM
(L) eEJC amplitudes, normalized to the first response, during HFS (20 Hz × 60 s) followed by a recovery period of stimulation (0.2 Hz × 50 s). kdm5WT n = 12, kdm5JmjC n = 14, kdm5L854F n = 13. Error bars representing SEM are not displayed here to facilitate visualization.
To directly measure neuronal activity in kdm5JmjC* and kdm5L854F mutants, we examined evoked and spontaneous synaptic transmission at the glutamatergic NMJ of larval muscle 6, which displayed similar bouton phenotypes to those observed in muscle 4 (Figure S3). Compared to kdm5WT, evoked excitatory junctional currents (eEJCs) were reduced in kdm5JmjC* but not in kdm5L854F larvae (Figures 5F and 5G). The impaired neurotransmission observed in kdm5JmjC* animals could be caused by defects in synaptic excitation, neurotransmitter release, or postsynaptic response. Analyses of miniature excitatory junctional current (mEJC) amplitudes did not show any differences between kdm5JmjC* and kdm5WT animals (Figure 5H). These data suggest that the reduction in eEJCs was caused by neither a defect in the response of postsynaptic glutamatergic receptors to glutamate nor the amount of glutamate released by a single vesicle. mEJC frequency in kdm5JmjC* mutant larvae was also unaltered compared to kdm5WT, indicating that there were no changes in the number of functional active release sites (Figure 5I) (Long et al., 2010). Furthermore, immunohistochemical analysis of kdm5JmiC* NMJs with an antibody to Bruchpilot (Brp) revealed that active zones show no changes in density or defects in apposition to postsynaptic glutamatergic receptors (GluRIIC) relative to kdm5WT (Figures S3F and S3G). We also examined quantal content (ratio of eEJC area/mEJC area), which reflects the number of vesicles released per stimulus, and observed a slight decrease in kdm5JmjC* larvae compared to kdm5WT (p = 0.13) (Figure 5J). kdm5L854F mutant larvae did not show any differences in mEJC amplitude or frequency, quantal content, or active zone density or apposition (Figures 5 and S3). Taken together, these data indicate that loss of demethylase activity and the ID-associated mutation in the C5HC2 domain similarly impairs larval locomotion but only loss of demethylase activity reduces eEJC amplitudes.
To evaluate whether the deficiency in neurotransmission observed in kdm5JmjC* mutants was due to differences in synaptic vesicle release probability, we analyzed the responses to paired-pulse stimuli given at variable intervals (Figure 5K). The paired-pulse ratio (PPR), calculated by dividing the eEJC of the second response by the first, did not exhibit differences at any interstimulus interval for kdm5JmjC* or kdm5L854F mutants compared to kdm5WT control larvae. We also assessed the dynamics of synaptic vesicle recycling by recording eEJCs during a period of high-frequency stimulation (HFS) (20 Hz × 60 s) followed by a recovery period (0.2 Hz × 50 s) (Long et al., 2010). Similar to kdm5WT, both mutants showed a decrease in eEJCs during HFS, when the pool of readily releasable vesicles is depleted, and an increase in eEJCs during the recovery period when the pool is replenished (Figure 5L). Together, these data show that the reduction in eEJC amplitudes caused by loss of demethylase activity was not likely the result of perturbations in vesicle release probability nor endocytic recycling.
Mutations in KDM5 that affect NMJ structure alter transcriptional programs in the VNC
KDM5 is likely to exert its effect on synaptic structure and function by regulating gene expression. Because KDM5 functions presynaptically to regulate bouton number and size (Figure 1), we conducted transcriptomic analyses of dissected VNCs where the soma of motor neurons reside. Quadruplicate RNA-seq from kdm5JmjC* and kdm5L854F mutant larvae revealed that both mutations alter gene expression relative to kdm5WT. Using a false discovery rate (FDR) cutoff of 5%, kdm5JmjC* mutants showed 1,656 differentially expressed genes (DEGs), 858 of which were upregulated and 798 of which were downregulated (Figure 6A; Table S1). The kdm5L854F allele also altered gene expression, although fewer genes were differentially expressed than in kdm5JmjC*, with 284 downregulated and 334 upregulated genes relative to kdm5WT (5% FDR) (Figure 6B; Table S2). Comparing the genes that were dysregulated in the two genotypes uncovered a significant number of genes that were dysregulated by both alleles (273; p = 2.15e-37) in addition to many genes that were uniquely altered in either kdm5JmjC* or kdm5L854F (Figure 6C). There was a strong correlation between commonly dysregulated genes, 242 of which behaved similarly in the two genotypes (r = 0.77; Figure 6D), suggesting that these two mutations affect one or more KDM5 functions required at a subset of target genes. KDM5 therefore uses both its demethylase and its C5HC2 zinc-finger activity to regulate presynaptic expression programs.
Figure 6. kdm5JmjC* and kdm5L854F affect gene expression programs in the larval VNC.
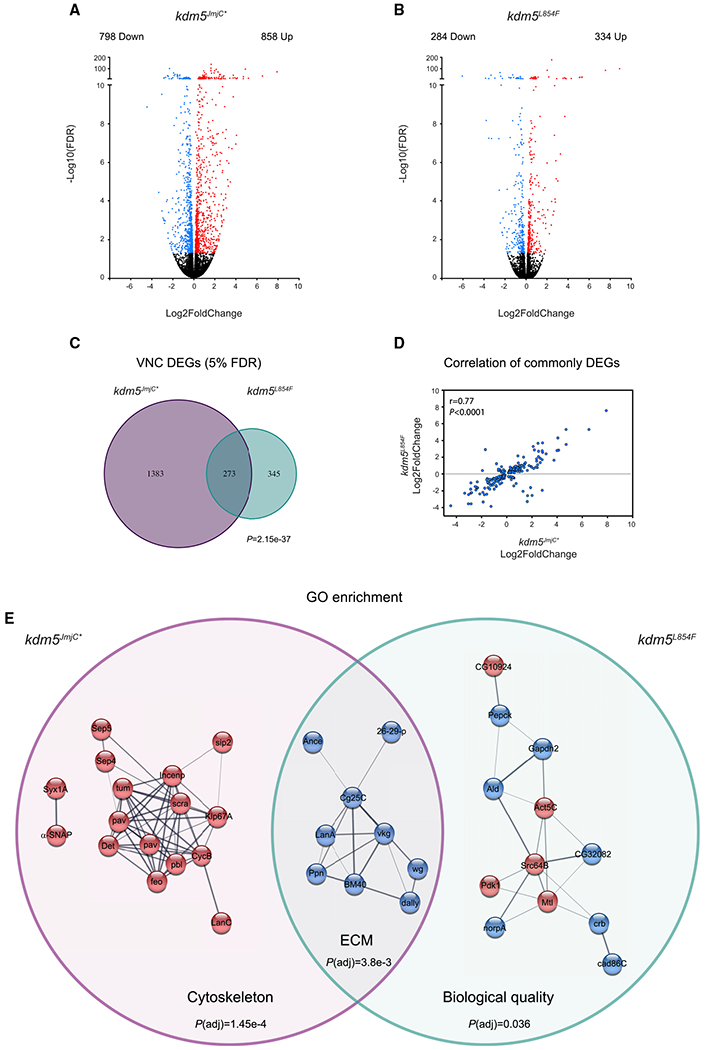
(A and B) Volcano plots showing dysregulated genes (5% FDR; red) in the VNC of kdm5JmjC* (A) and kdm5L854F (B).
(C) Venn diagram showing overlap between kdm5JmjC* and kdm5L854F RNA-seq data. p = 2.15e-37.
(D) Correlation between overlapping dysregulated genes from kdm5L854F and kdm5JmjC*.
(E) Networks from enrichedGOcategories affected in kdm5JmjC* (GO:0005856), kdm5JmjC* and kdm5L854F (GO:0005576), and kdm5L854F (GO:0065008). Red color indicates upregulation and blue indicates downregulation compared to kdm5WT.
Similar to previous studies of KDM5 in Drosophila and other species (Chen et al., 2019; Drelon et al., 2018; Iwase et al., 2017; Mariani et al., 2016; Scandaglia et al., 2017; Zamurrad et al., 2018), the changes in gene expression in the VNCs of kdm5JmjC* and kdm5L854F mutants were modest, averaging ~2-fold (Figures 6A and 6B). KDM5 is therefore likely to function in motor neurons by regulating sets of target genes within critical pathways, which can be highlighted by interrogating Gene Ontology (GO) and protein-interaction pathway databases. Examining genes that were dysregulated only in kdm5JmiC* revealed an enrichment for the GO cellular component category of cytoskeleton (P(adj) = 1.45e-4) (Figure 6E). All genes within this group were upregulated, consistent with the canonical function of the demethylase activity of KDM5 in transcriptional repression. Interestingly, in addition to well-characterized roles during mitosis, many of these genes play additional roles in post-mitotic neurons, particularly in the regulation of synaptic function. For example, genes encoding the septin proteins Septin4 and Septin5 and the synaptic SNARE complex component α-SNAP, upregulated in kdm5JmjC* mutant CNS, are all linked to the negative regulation of neurotransmitter release (Brunger et al., 2019; Marttinen et al., 2015). This observation is in keeping with the electrophysiological defect observed in kdm5JmjC* mutant larvae, which showed a slight decrease in the number of vesicles being released (quantal content; Figure 5J).
Similar GO analyses of the genes that were dysregulated in both kdm5JmjC* and kdm5L854F mutant VNCs identified a single enriched category of extracellular matrix (ECM) organization (P(adj) = 3.8e-3). All of the genes in this category, which encode proteins that function in the extracellular space, showed reduced expression compared to kdm5WT, implicating KDM5 in their activation. This included components of the basement membrane such as laminin A (lanA), collagen type IV alpha 1 (col4α1/Cg25c), and Viking (Vkg) (collagen IV), which form a complex (Guruharsha et al., 2011) (Figure 6E). In addition to playing structural roles, these proteins contribute to signaling, as emphasized by the presence of the Wnt/Wg pathway ligand Wingless, a well-established regulator of NMJ bouton growth, within this network (Figure 6E) (Packard et al., 2002). We also examined the 345 genes that were uniquely altered in kdm5L854F mutants and found a single enriched GO category of regulation of biological quality (P(adj) = 0.036) that comprised a combination of up- and downregulated genes that modulate qualitative or quantitative traits (Figure 6E). This group included proteins that influence synaptic bouton size such as phosphoinositide-dependent kinase 1 (Pdk1) (Cheng et al., 2011). In addition, genes encoding proteins that interact with, or regulate, the microtubule and actin cytoskeletons such as the Rac family protein Mtl (Hakeda-Suzuki et al., 2002; Trogden and Rogers, 2015) and the kinase Src64B (Feuillette et al., 2020) were dysregulated (Figure 6E; Table S2). Combined, these RNA-seq data provide molecular insights into potential mechanisms by which KDM5 carries out its neuromorphological and synaptic functions (Figure 7).
Figure 7. Model of KDM5 function at the NMJ.
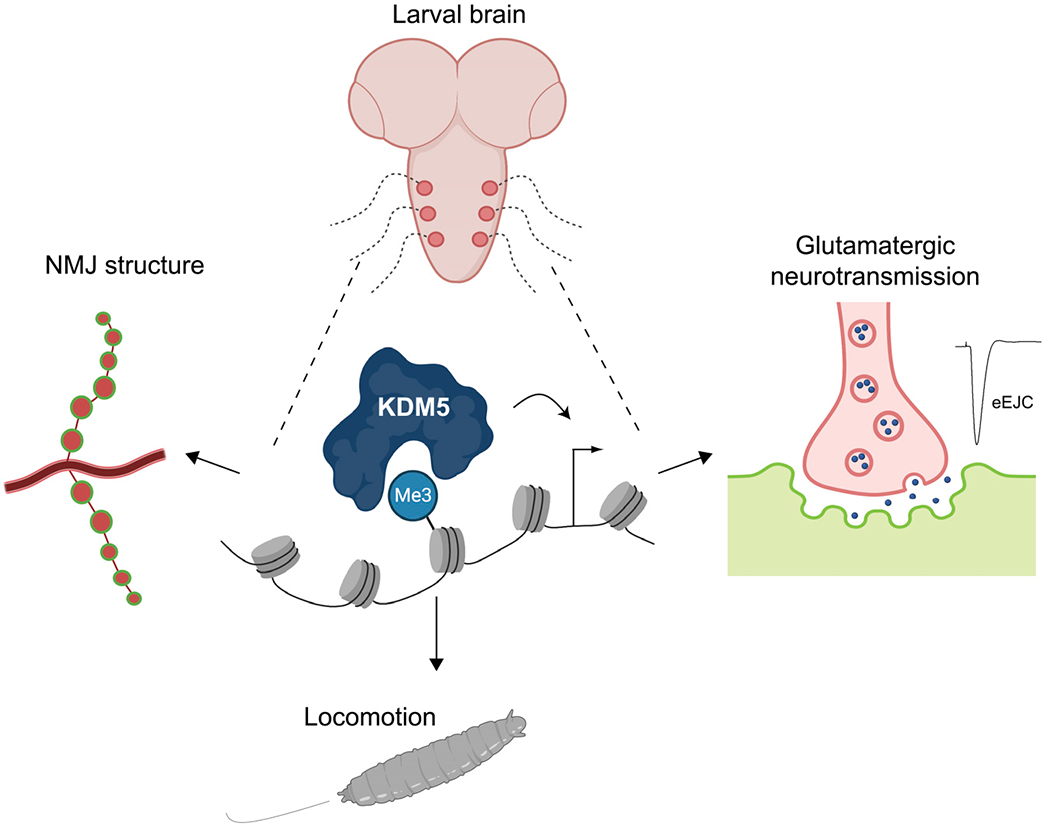
KDM5 regulates gene expression programs in motor neurons that are required for normal morphology and synaptic function of the NMJ.
DISCUSSION
Mutations in three of the four KDM5 paralogs in humans are associated with NDDs, suggesting that these proteins are key contributors to neuronal development and/or function. The roles that KDM5 proteins play in regulating synaptic function, however, remain largely uncharacterized. Here we show that KDM5 regulates the expression of genes within the motor neuron that are required for the growth and function of the Drosophila larval NMJ. This synapse serves as a model for mammalian CNS glutamatergic synapses (Coll-Tanée et al., 2019; Koh et al., 2000; Menon et al., 2013), which are altered in a number of inherited forms of cognitive impairment, including fragile X syndrome, Angelman syndrome, and Rett syndrome (Volket al., 2015). Our experiments demonstrate that the mutations in different domains of KDM5 can cause both overlapping and distinct morphological and functional phenotypes at the NMJ. This is likely to be, at least partially, due to the JmjC and C5HC2 domains facilitating both shared and unique aspects of gene regulation in the larval VNC. Combined, our data lead us to propose that KDM5 uses several distinct gene-regulatory functions in NMJ glutamatergic neurons to influence functional and structural neuronal characteristics.
Our analyses establish the importance of the histone demethylase activity of KDM5 at the larval NMJ, providing further support for the model that loss of enzymatic activity contributes to the cognitive phenotypes observed in patients with mutations in KDM5 genes. The kdm5JmjC* fly strain, but not the ID allele kdm5L854F, exhibited an evoked neurotransmission defect that correlated with a mild deficit in the number of vesicles being released per stimulus. The demethylase activity of KDM5 is therefore needed for appropriate glutamatergic neurotransmission. Although this allele is not a patient-associated mutation, at least half of the known ID-associated variants in KDM5 family genes are expected to affect histone demethylase activity (Brookes et al., 2015; Lebrun et al., 2018). Thus, phenotypic analyses of this allele are likely to have implications for a number of patients. Based on our analyses of genes that were dysregulated only in the VNC of kdm5JmjC* mutant larvae, we propose that the upregulation of cyto-skeletal genes that negatively regulate pre-synaptic neurotransmitter release plays a key role in the eEJC phenotype observed. For example, the α-SNAP gene has an evolutionarily conserved function in the inhibition of synaptic vesicle fusion through its interaction with the SNARE complex component syntaxin (Brunger et al., 2019). The expression of genes encoding the GTP-binding septins (Septin4 and Septin5), which can also impact vesicle release, was also upregulated in kdm5JmjC* One of the mammalian paralogs of Septin4 can inhibit synaptic vesicle release by forming a filament barrier (Yang et al., 2010)and, like α-SNAP, can interact with syntaxin to inhibit exocytosis (Amin et al., 2008). Notably, because mutations in KDM5 genes cause modest changes in gene expression (Iwase et al., 2016; Scandaglia et al., 2017; Zamurrad et al., 2018), it is likely that the dysregulation of several genes in kdm5JmiC* mutant neurons contributes to the reduced synaptic transmission detected. Defining the precise role that these proteins play in mediating this synaptic phenotype will require further analyses of existing and additional ID-associated missense alleles that do, or do not, alter demethylase activity.
Our studies also emphasize the importance of other domains of KDM5, particularly the C5HC2 zinc-finger motif that is immediately adjacent to the JmjC domain and is affected by the L854F mutation. This motif has been suggested to be structurally required for JmjC domain function based on the crystal structure of KDM5A (Horton et al., 2016). It is also possible that the converse is true and that the JmjC domain contributes to C5HC2 function. Consistent with the JmjC and C5HC2 domains acting in concert with one another in some contexts, the kdm5JmjC* and kdm5L854F alleles showed a similar reduction in the number of NMJ synaptic boutons that contribute to muscle contraction and slowed larval movement. It is therefore possible that the bouton number and larval movement changes in kdm5JmjC* and kdm5L854F mutant larvae are functionally related, as similar correlations have been observed previously (Kashima et al., 2017). Moreover, a subset of the transcriptomic defects seen in these two genotypes was shared, suggesting a possible molecular driver of the structural and functional phenotypes observed. Notably, this included diminished expression of wingless and ECM components that facilitate Wg signaling, which can result in reduced bouton number and slow larval locomotion (Kim and Cho, 2020). Alternatively, the bouton and movement phenotypes could have separate causes. The locomotor phenotype observed in kdm5JmjC* and kdm5L854F larvae may not be caused by NMJ perturbations but may instead be indirectly caused by aberrant neurotransmission in upstream motor networks or muscle deficits. Larval locomotion is a complex behavior that relies on linking appropriate sensory inputs with a wave of peristaltic muscle contractions that mediate larval movement (Kohsaka et al., 2017). As such, it is possible that the motor deficits displayed by kdm5JmjC* and kdm5L854F mutants are caused by synaptic or morphological changes in body-wall sensory neurons (Hughes and Thomas, 2007; Song et al., 2007) or defects in the premotor inhibitory interneurons (Kohsaka et al., 2014). Additional studies are needed to discern the functional links between KDM5-regulated transcription, changes in bouton number, and larval movement.
Our findings that kdm5L854F mutants possess larger boutons, which is not observed in kdm5JmjC*, and displayed a number of unique gene expression defects suggest that the C5HC2 motif can also function independent of the JmjC domain. One key group of KDM5-regulated genes whose expression was disrupted by the L854F mutation were those involved in the regulation of the MT dynamics, which play key structural roles and transport roles in neurons (Conde and Caceres, 2009). This observation may be important for the neuropathology of the equivalent mutation in human KDM5C (p.L731F) as changes to presynaptic MT physiology are observed in other ID disorders such fragile X syndrome (Bodaleo and Gonzalez-Billault, 2016; Lu et al., 2004). Moreover, restoring homeostasis to MT dynamics can attenuate morphological and functional phenotypes associated with mutations in the fragile X gene fmr1 (Zhang et al., 2001). Further exploration of this potential link could highlight a means to attenuate the cognitive deficits for a subset of mutations associated with ID. Because no independent molecular function has been attributed to C5HC2 zinc-finger motifs, defining the precise molecular defect caused by changes to this domain in KDM5 family proteins will require additional analyses. In addition to being required for JmjC domain function, this motif might act as a protein-protein interaction motif and/or bind to nucleic acids (Riechmann et al., 2000; Secombe et al., 2007). Consequently, mutations in this region could therefore alter a range of functions that are critical for gene regulation, such as recruitment of KDM5 to its target genes and/or its ability to mediate activation or repression of transcription. Interestingly, the three mutations in the C5HC2 zinc finger examined do not result in the same NMJ phenotypes, suggesting that they may differentially affect KDM5 transcriptional regulation on synaptic target genes. Whereas kdm5L854F affected bouton number and size, kdm5Y874C appeared to modestly decrease bouton number and kdm5R873W NMJs were indistinguishable from WT. The more severe phenotypes of animals expressing the KDM5L854F mutant protein are consistent with the amino acid substitution affecting one of the Zn2+-binding sites in the C5CH2 domain. Indeed, DynaMut predictions suggested that this amino acid variation leads to significant alterations in the dynamics and stability around the C5HC2 motif, which could cause more dramatic changes both to transcription and to NMJ morphology.
In conclusion, we show that KDM5 is a key transcription regulator of genes important for synaptic structure and function. Our observations that KDM5 plays critical roles at the NMJ are consistent with accumulating data linking disruption of glutamatergic synapsis to human cognitive disorders, including ID (Volk et al., 2015). Future studies will provide additional insights into how the individual domains of KDM5 contribute to its synaptic functions and how these activities are altered by ID-associated mutations.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Julie Secombe (Julie. Secombe@einsteinmed.org).
Materials availability
All fly strains described in this manuscript are available upon request.
Data and code availability
The accession number for the RNA-seq data reported in this paper is NCBI Gene Expression Omnibus GEO:GSE159298. A list of differentially expressed genes (and log2 fold change) observed compared to wild-type are provided in Tables S1 and S2 (5% FDR) for kdm5JmjC* and kdm5L854F, respectively.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Care of fly strains and crosses
Fly crosses were maintained at 25°C with 50% humidity and a 12-hour light/dark cycle. Food (per liter) contained 18 g yeast, 22 g molasses, 80 g malt extract, 9g agar, 65 cornmeal, 2.3g methyl para-benzoic acid, 6.35ml propionic acid. The number of male and female larvae were equal across the genotypes examined. For studies comparing wild-type and kdm5 mutant larvae, animals were matched for developmental stage based on the morphology of larval mouth hooks, not chronological age, similar to our previously reported analyses (Drelon et al., 2018,2019). Control wandering third instar larvae were therefore ~120 hours after egg laying (AEL) while kdm5140 larvae were 8-10 days old.
Fly strains
The kdm5140 null and kdm5JmjC* alleles have been previously described (Drelon et al., 2018; Navarro-Costa et al., 2016; Zamurrad et al., 2018). UASp-kdm5 transgene was validated in numerous rescue experiments (Drelon et al., 2019; Li et al., 2010; Secombe et al., 2007). The UAS-shRNA transgene to knockdown kdm5 was generated as part of the TRiP project and has been well-validated by us and others (BL #35706) (Liu et al., 2014; Navarro-Costa et al., 2016). To generate ID alleles, we took a similar approach as our previous analyses of kdm5JmjC* (Drelon et al., 2018, 2019; Navarro-Costa et al., 2016). Briefly, the 11kb genomic region encompassing kdm5 was amplified by PCR from w1118 and cloned into the pattB vector (Bischof et al., 2007) using the In-Fusion cloning system (Takara). A 3xHA tag was included in-frame at the 3′end of the kdm5 ORF. Point mutations (L854F, R873W, Y874C) were introduced by PCR-mediated site-directed mutagenesis. Sequenced constructs were sent to BestGene for injection into FlyC31 embryos (BL #24749). Transformed flies were crossed into the kdm5140 null background and homozygous stocks were established. Wild-type and demethylase dead transgenes and resulting fly strains are published (Navarro-Costa et al., 2016; Zamurrad et al., 2018). Gal4 driver fly strains used included OK6-Gal4 (BL #64199), elav-Gal4 (BL#25750), Act5C-Gal4 (BL #3954), da-Gal4 (BL #55851), Mef2-Gal4 (BL #27390), repo-Gal4 (BL #7415). Gal4 expression was validated by crossing each Gal4 strain with the UAS.GFPnls reporter line (Neufeld et al., 1998).
METHOD DETAILS
RNA-seq
Pooled male and female CNSsfrom wild-type (kdm5WT), kdm5JmjC*, and kdm5L854F third instar larvae were dissected Hemolymph-Like Solution (HL-3: NaCl 70mM, KCl 5mM, MgCl2 20mM, NaHCO3 10mM, trehalose 5mM, sucrose 115mM, HEPES 5mM, pH7.1) without CaCl2 and brain lobes were separated to enrich for VNC cells. Total RNA was isolated withTrizol (Invitrogen) and quality was assessed by Fragment Analyzer (Advanced Analytical, Ankeny, IA, USA) before sending to the Beijing Genomics Institute (BGI) for library preparation and sequencing. cDNA libraries were prepared using TruSeq Stranded mRNA Library. mRNAs were isolated from total RNA with oligo(dT) method. After mRNA fragmentation, first strand cDNA and second strand cDNA were synthesized, and cDNA fragments were purified and resolved with EB buffer for end reparation and single nucleotide A (adenine) addition. cDNA fragments were linked with adapters and those with suitable sizes were selected for PCR amplification. Libraries were sequenced on Illumina NovaSeq 6000 platform. Alignment of raw reads to the reference genome (dm6) was performed using Bowtie2 (v2.2.5) (Langmead and Salzberg, 2012), normalized and differential expression determined with DESeq2 package (Love et al., 2014; Soneson and Delorenzi, 2013).
Gene Ontology (GO) enrichment analysis of protein-coding genes found to be dysregulated in kdm5JmjC* and/or kdm5L854F RNA-seq data (5% FDR cutoff) was carried out using PANTHER overrepresentation test (http://geneontology.org/) (Mi et al., 2010) and String (http://string-db.org) v11.0b (Szklarczyk et al., 2019). Interaction networks were determined using String (Szklarczyk et al., 2019) and visualized using Cytoscape (Shannon et al., 2003).
Western blot
Third instar larval brains were dissected in ice-cold 1xPBS. Samples were stored in 2x NuPAGE LDS Buffer at −20°C until use. After a round of sonication and treatment with DTT, samples were subjected to SDS-PAGE and transferred to a PVDF membrane for immunoblotting. Membranes were incubated with primary antibodies at 4°C overnight, washed and incubated with secondary antibodies at room temperature for 30min. Mouse anti-histone H3,1:2000 (Abcam #24834); rabbit anti-H3K4me3 1:2000 (Active Motif #39159); mouse anti-HA, 1:500 (Cell Signaling #2367S); rabbit anti-KDM5, 1:250 (Secombe et al., 2007); anti-gamma-tubulin, 1:1000 (4D11; Invitrogen MA1-850); and mouse anti alpha-Tubulin, 1:10000 (12G10, Developmental Studies Hybridoma Bank, University of Iowa) were used as primary antibodies. IRDye® 800CW Donkey anti-Rabbit IgG (92632213) and IRDye® 680RD Donkey anti-Mouse IgG (925-68072) from LI-COR were used as secondary antibodies. Western blots were quantified using LI-COR Image Studio software.
Immunostaining
Third instar larvae were dissected in ice-cold 1xPBS and fillets were fixed in 4%PFA in PBS at room temperature for 30min. For Futsch staining, fillets were fixed in ice-cold methanol for 10min at −20°C. Samples were washed in 1xPBS for 10 min followed by two washes in 1xPBST (PBS + 0.1% Triton) for 10min each. Fillets were transferred to 0.5μl tubes for primary antibody incubation overnight at 4°C with agitation. After three 15 min washes in 1xPBST, samples were incubated in secondary antibodies for 1 h at room temperature. Samples were then washed three times for 10 min each and mounted with Fluoromount-G DAPI (SouthernBiotech) or SlowFade Glass Antifade Mountant (Invitrogen). Mouse anti-Dlg (4F3) 1:1000, anti-Futsch (22C10) 1:100, and anti Brp (nc82) 1:100 were obtained from Developmental Studies Hybridoma Bank (DSHB, University of Iowa). Rabbit anti- GluRIIC (1:5000) was a kind gift from Aaron DiAntonio. Alexa 647-conjugated goat anti-HRP was used at 1:200 (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Goat anti-mouse Alexa 488 (Invitrogen) was used at 1:400 while goat anti-mouse TRITC and anti-rabbit FITC (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) were used at 1:400.
Electrophysiology
Third instar larvae were fillet dissected in ice cold HL-3.1 (Feng et al., 2004) containing 0.25 mM Ca2+. Larvae were glued (Vetbond Tissue Adhesive, World Precision Instruments) to Sylgard-coated coverslips, the VNC was cut out of the animal, and the HL-3.1 was replaced with room temperature HL-3.1 containing 1.0 mM Ca2+ for recordings. Muscle 6 in segments A3 or A4 was voltage clamped at −60 mV using an Axoclamp 900A amplier (Molecular Devices). Clamp and recording electrodes with 10-20 MΩ of resistance were filled with 3 mM KCl. Supra threshold stimuli were delivered via stimulating electrodes filled with bath solution using a Grass S88 stimulator with a SIU5 isolation unit (Grass Technologies). Evoked EJCs were measured after stimulating segmental nerves at 0.2 Hz for 50 s to establish baseline responses. Endocytosis was assessed by stimulating by 20 Hz for 60 s. Recovery of the cycling pool of vesicles was assessed by stimulating at 0.2 Hz for 50 s. Paired pulse amplitudes were measured after delivering two 10 Hz, two 20 Hz, two 50 Hz, and two 100 Hz pulses, each of which were separated by a 20 s intertrial interval. Recordings were digitized with a Digidata 1443 digitizer (Molecular Devices). Quantal content was calculated by dividing the eEJC area (nA × ms) by the mEJC area (nA × ms) for each animal. 180 s of spontaneous activity was used to quantify mEJC frequency and amplitude. An equal number of recordings from controls and experimental animals were obtained each day. Data were analyzed in pClamp (v11.0, Molecular Devices).
Larval movement assays
Third instar larvae were placed on Petri dishes containing 1.6% agar in double distilled water. Larvae were allowed to crawl on the agar dish for one minute to remove excess food and then transferred to the crawling arena, which also consisted of 1.6% agar. Larvae were allowed to acclimate for one minute then larval locomotion was recorded using a Cannon EOS M50 camera at 29.97 frames per second for 30 s. Five larvae were recorded per video. Videos were opened in ImageJ (NIH) and 899 frames were analyzed with the publicly available wrMTrck plugin (Jesper S. Pedersen; https://www.phage.dk/plugins/wrmtrck.html).
Developmental timing quantification
Female and male flies were placed in a vial and allowed to lay eggs for 24 hours. Starting at day 5 after egg laying the number of animals that had pupariated were scored twice per day.
Image acquisition and processing
Images of NMJs from muscles 4 and 6 of abdominal segment 3 were acquired using Leica Sp8 Confocal Microscope, 63x/1.4NA and Zeiss Axioimager Z1 Apotome. Quantification of muscle area and type Ib bouton number and area were performed in the Dlg channel using ImageJ. The number of boutons was normalized to muscle surface area similar to previous studies (Banovic et al., 2010; Nechipurenko and Broihier, 2012). The Threshold and Measure commands were used to quantify bouton surface areas, similar to previous studies (Wagner et al., 2015). Adult fly images were obtained using a stereomicroscope Carl Zeiss Stereo Discovery V12 with 14X magnification and captured using AxioVision Release 4.8 software. Brp density was calculated by dividing the total Brp puncta by the NMJ area. Colocalization of Brp and GluRIIC was quantified using Fiji (NIH ImageJ) by outlining the NMJ in a max-projected image. Pearson’s R values were obtained for each image after splitting the channels using the Coloc2 plugin. All images were processed and assembled into figure format using Adobe Illustrator 2021. Images of adult head and ventral nerve cord and icons used in the model were created with BioRender.com.
In silico mutation analysis
Normal mode analysis was used to predict the thermodynamic changes upon mutations in the human KDM5A structure (PDB: 5CEH) via the DynaMut server (Rodrigues et al., 2018). Files generated by DynaMut were visualized using PyMol (The PyMOL Molecular Graphics System, Version 2.4 Schrödinger, LLC). UCSF’s Chimera (Pettersen et al., 2004) was used to modify the KDM5A structure.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses
All experiments were performed in biological triplicate (minimum) and numbers (n) are provided for each experiment either in the figure or the legend. Student’s t tests were used for pairwise comparisons while one-way ANOVAs were used for three or more groups, followed by Dunn’s or Dunnett’s multiple comparisons tests. Fisher’s exact test was used to analyze contingency tables (Survival adult flies and overlapping genes). To test the directionality of expression changes in overlapping genes, Spearman correlation was used. Two-way ANOVAs were used to analyze high frequency stimulation. GraphPad Prism (v8.4) or R version 3.6.2 was used for analyses. Error bars on graphs represent mean ±SEM.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-histone H3 | Abcam | Cat#24834; RRID: AB_470335 |
| Rabbit polyclonal anti-H3K4me3 | Active motif | Cat#39159; RRID: AB_2615077 |
| Mouse monoclonal anti-HA | Cell signaling | Cat#2367S; RRID: AB_10691311 |
| Rabbit polyclonal anti-KDM5 | Secombe et al., 2007 | N/A |
| Anti-gamma-tubulin (Clone 4D11) | Invitrogen | Cat#MA1-850; RRID: AB_2211249 |
| Mouse monoclonal anti alpha-Tubulin | Developmental Studies Hybridoma Bank, University of Iowa | Cat#12G10; RRID: AB_1157911 |
| Mouse monoclonal anti Dlg | Developmental Studies Hybridoma Bank, University of Iowa | Cat#4F3; RRID: AB_528203 |
| Mouse monoclonal anti-Futsch | Developmental Studies Hybridoma Bank, University of Iowa | Cat#22C10; RRID: AB_528403 |
| Mouse monoclonal anti Brp | Developmental Studies Hybridoma Bank, University of Iowa | Cat#nc82; RRID: AB_2314866 |
| Alexa 647-conjugated goat anti-HRP | Jackson ImmunoResearch Laboratories | Cat# 123-605-021; RRID: AB_2338967 |
| Rabbit polyclonal anti- GluRIIC | Aaron DiAntonio | N/A |
| IRDye® 800CW Donkey anti-Rabbit IgG | LI-COR | Cat#92632213; RRID: AB_621848 |
| IRDye® 680RD Donkey anti-Mouse IgG | LI-COR | Cat#925-68072; RRID: AB_10953628 |
| Goat anti-Mouse IgG (H+L) Alexa Fluor Plus 488 |
ThermoFisher | Catalog# A32723 |
| Goat anti-mouse TRITC | Jackson ImmunoResearch Laboratories | 111-025-003 RRID:AB_2337926 |
| Goat anti-rabbit FITC | Jackson ImmunoResearch Laboratories | 111-095-003 RRID:AB_2337972 |
| Chemicals, peptides, and recombinant proteins | ||
| SlowFade Glass Antifade Mountant | Invitrogen | Cat#S36918 |
| Fluoromount-G DAPI | SouthernBiotech | Cat#0100.01 |
| Trehalose | Sigma | Cat# PHR1344 |
| TRIZOL RNA Isolation Reagent | Invitrogen | Cat#15596026 |
| Critical commercial assays | ||
| In-Fusion® HD Cloning Plus | Takara | Cat# 638909 |
| TruSeq Stranded mRNA Library | Illumina | Cat#20020594 |
| Deposited data | ||
| Third instar VNC-kdm5WT RNA-seq | This paper | GEO: GSE159298 |
| Third instar VNC-kdm5JmjC* RNA-seq | This paper | GEO: GSE159298 |
| Third instar VNC-kdm5L854F RNA-seq | This paper | GEO: GSE159298 |
| Experimental models: organisms/strains | ||
| UASp-kdm5 | Secombe et al., 2007 | N/A |
| UAS-shRNA-kdm5 | Bloomington Drosophila Stock Center (BDSC) | BL35706 |
| kdm5140/Cyo-GFP | Drelon et. al, 2018 | N/A |
| kdm5140/Cyo-GFP; UASp-kdm5 | Drelon et. al, 2019 | N/A |
| kdm5140,OK6-Gal4/Cyo-GFP | This study | N/A |
| kdm5140/Cyo-GFP;repo-Gal4/TM6B | This study | N/A |
| kdm5140/Cyo-GFP;Mef2-Gal4 | This study | N/A |
| elav-Gal4; kdm5140/Cyo-GFP | This study | N/A |
| kdm5140/Cyo-GFP;actin-Gal4/TM6B | This study | N/A |
| kdm5140/Cyo-GFP;da-Gal4/TM6B | This study | N/A |
| Kdm5140; kdm5WT in attP86F | Navarro-Costa et al., 2016 | N/A |
| kdm5140;kdm5JmjC* in attP86F | Navarro-Costa et. al, 2016 | N/A |
| kdm5140;kdm5L854F in attP86F | This study | N/A |
| kdm5140;kdm5R873W in attP86F | This study | N/A |
| kdm5140;kdm5Y874C in attP86F | This study | N/A |
| w1118 | BDSC | BL5905 |
| FlyC31 | BDSC | BL24749 |
| OK6-Gal4 | BDSC | BL64199 |
| Elav-Gal4 | BDSC | BL25750 |
| Act5C-Gal4 | BDSC | BL3954 |
| da-Gal4 | BDSC | BL55851 |
| Mef2-Gal4 | BDSC | BL27390 |
| repo-Gal4 | BDSC | BL7415 |
| Recombinant DNA | ||
| pattB | Drosophila Genomics Resource Center | Cat 1420 |
| Software and algorithms | ||
| LI-COR Image Studio | https://www.licor.com | RRID:SCR_015795 |
| Adobe Illustrator CC 2021 | https://www.adobe.com/ | RRID: SCR_010279 |
| AxioVision Release 4.8 | Zeiss | RRID:SCR_002677 |
| UCSF Chimera | UCSF Resource for Biocomputing, Visualization, and Informatics | RRID:SCR_004097 |
| Pymol | http://www.pymol.org/ | RRID:SCR_000305 |
| GraphPad Prism 8 | GraphPad Software, Inc. | RRID: SCR_002798 |
| FIJI (ImageJ) | National Institutes of Health | RRID:SCR_003070 |
| pClamp | v11.0, Molecular Devices | RRID:SCR_011323 |
| Bowtie2 (v2.2.5) | Langmead and Salzberg, 2012 | RRID:SCR_005476 |
| R package DESeq2 | Love et al., 2014 | RRID:SCR_015687 |
| Other | ||
| DynaMut server | Rodrigues et al., 2018 | N/A |
| Biorender | http://biorender.com | RRID:SCR_018361 |
| String | http://string.embl.de/ | RRID:SCR_005223 |
| GeneOntology | (Ashburner et al., 2000) | N/A |
Highlights.
Drosophila KDM5 regulates transcriptional programs vital to synaptic development
KDM5 uses demethylase-dependent and independent means to regulate NMJ development
The demethylase activity of KDM5 promotes bouton number and neurotransmission
The C5HC2 motif of KDM5 is needed for neuroanatomical development
ACKNOWLEDGMENTS
We thank members of the Secombe, Liebl, and Baker labs for their insights and comments on the manuscript. We also thank Jacqueline Tobin for her help as a summer student. We are grateful for the GluRIIC antibody from Dr. Aaron DiAntonio, fly strains from the Bloomington Drosophila Stock Center (NIH P400D018537), and antibodies from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at the University of Iowa. We also thank the Analytical Imaging Facility and the Genomics Core at Einstein for their technical support. This work was supported by NIH funding to J.S. (R01 GM112783) and F.L.W.L. (1R15NS101608-01A1), a shared instrument grant (1S10OD023591-01), and an Einstein Cancer Center support grant (P30 CA013330).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2021.108753.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Amin ND, Zheng YL, Kesavapany S, Kanungo J, Guszczynski T, Sihag RK, Rudrabhatla P, Albers W, Grant P, and Pant HC (2008). Cyclindependent kinase 5 phosphorylation of human septin SEPT5 (hCDCrel-1) modulates exocytosis. J. Neurosci 28, 3631–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banovic D, Khorramshahi O, Owald D, Wichmann C, Riedt T, Fouquet W, Tian R, Sigrist SJ, and Aberle H (2010). Drosophila neuroligin 1 promotes growth and postsynaptic differentiation at glutamatergic neuromuscular junctions. Neuron 66, 724–738. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, and Zhao K (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. [DOI] [PubMed] [Google Scholar]
- Bellosta P, and Soldano A (2019). Dissecting the genetics of autism spectrum disorders: a Drosophila perspective. Front. Physiol. 10, 987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, et al. (2014). H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 158, 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, and Basler K (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodaleo FJ, and Gonzalez-Billault C (2016). The presynaptic microtubule cytoskeleton in physiological and pathological conditions: lessons from Drosophila fragile X syndrome and hereditary spastic paraplegias. Front. Mol. Neurosci. 9, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes E, Laurent B, Õunap K, Carroll R, Moeschler JB, Field M, Schwartz CE, Gecz J, and Shi Y (2015). Mutations in the intellectual disability gene KDM5C reduce protein stability and demethylase activity. Hum. Mol. Genet. 24, 2861–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Choi UB, Lai Y, Leitz J, White KI, and Zhou Q (2019). The pre-synaptic fusion machinery. Curr. Opin. Struct. Biol. 54, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik V, Koh YH, Guan B, Hartmann B, Hough C, Woods D, and Gorczyca M (1996). Regulation of synapse structure and function by the Drosophila tumor suppressor gene dlg. Neuron 17, 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Luan X, Liu Q, Wang J, Chang X, Snijders AM, Mao JH, Secombe J, Dan Z, Chen JH, et al. (2019). Drosophila histone demethylase KDM5 regulates social behavior through immune control and gut microbiota maintenance. Cell Host Microbe 25, 537–552.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Locke C, and Davis GW (2011). S6 kinase localizes to the pre-synaptic active zone and functions with PDK1 to control synapse development. J. Cell Biol. 194, 921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll-Tané M, Krebbers A, Castells-Nobau A, Zweier C, and Schenck A (2019). Intellectual disability and autism spectrum disorders ‘on the fly’: insights from Drosophila. Dis. Model. Mech. 12, dmm039180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde C, and Cáceres A (2009). Microtubule assembly, organization and dynamics in axons and dendrites. Nat. Rev. Neurosci. 10, 319–332. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. ; DDD Study; Homozygosity Mapping Collaborative for Autism; UK10K Consortium (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAntonio A (2006). Glutamate receptors at the Drosophila neuromuscular junction. Int. Rev. Neurobiol. 75, 165–179. [DOI] [PubMed] [Google Scholar]
- Drelon C, Belalcazar HM, and Secombe J (2018). The histone demethylase KDM5 is essential for larval growth in Drosophila. Genetics 209, 773–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drelon C, Rogers MF, Belalcazar HM, and Secombe J (2019). The histone demethylase KDM5 controls developmental timing in Drosophila by promoting prothoracic gland endocycles. Development 146, dev182568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Ueda A, and Wu CF (2004). A modified minimal hemolymph-like solution, HL3.1, for physiological recordings at the neuromuscular junctions of normal and mutant Drosophila larvae. J. Neurogenet. 18, 377–402. [DOI] [PubMed] [Google Scholar]
- Feuillette S, Charbonnier C, Frebourg T, Campion D, and Lecourtois M (2020). A connected network of interacting proteins is involved in human-Tau toxicity in Drosophila. Front. Neurosci. 14, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest MP, Parnell E, and Penzes P (2018). Dendritic structural plasticity and neuropsychiatric disease. Nat. Rev. Neurosci. 19, 215–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves TF, Gonçalves AP, Fintelman Rodrigues N, dos Santos JM, Pimentel MM, and Santos-Rebouças CB (2014). KDM5C mutational screening among males with intellectual disability suggestive of X-linked inheritance and review of the literature. Eur. J. Med. Genet. 57, 138–144. [DOI] [PubMed] [Google Scholar]
- Gramates LS, and Budnik V (1999). Assembly and maturation of the Drosophila larval neuromuscular junction. Int. Rev. Neurobiol. 43, 93–117. [DOI] [PubMed] [Google Scholar]
- Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, et al. (2011). A protein complex network of Drosophila melanogaster. Cell 147, 690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakeda-Suzuki S, Ng J, Tzu J, Dietzl G, Sun Y, Harms M, Nardine T, Luo L, and Dickson BJ (2002). Rac function and regulation during Drosophila development. Nature 416, 438–442. [DOI] [PubMed] [Google Scholar]
- Horton JR, Engstrom A, Zoeller EL, Liu X, Shanks JR, Zhang X, Johns MA, Vertino PM, Fu H, and Cheng X (2016). Characterization of a linked Jumonji domain of the KDM5/JARID1 family of histone H3 lysine 4 demethylases. J. Biol. Chem. 291, 2631–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CL, and Thomas JB (2007). A sensory feedback circuit coordinates muscle activity in Drosophila. Mol. Cell. Neurosci. 35, 383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun K, Jeon J, Park K, and Kim J (2017). Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 49, e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, and Shi Y (2007). The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128, 1077–1088. [DOI] [PubMed] [Google Scholar]
- Iwase S, Brookes E, Agarwal S, Badeaux AI, Ito H, Vallianatos CN, Tomassy GS, Kasza T, Lin G, Thompson A, et al. (2016). A mouse model of X-linked intellectual disability associated with impaired removal of histone methylation. Cell Rep. 14, 1000–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Bérubé NG, Zhou Z, Kasri NN, Battaglioli E, Scandaglia M, and Barco A (2017). Epigenetic etiology of intellectual disability. J. Neurosci. 37, 10773–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LR, Amende M, Gurok U, Moser B, Gimmel V, Tzschach A, Janecke AR, Tariverdian G, Chelly J, Fryns JP, et al. (2005). Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 76, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima R, Redmond PL, Ghatpande P, Roy S, Kornberg TB, Hanke T, Knapp S, Lagna G, and Hata A (2017). Hyperactive locomotion in a Drosophila model is a functional readout for the synaptic abnormalities underlying fragile X syndrome. Sci. Signal. 10, eaai8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, and Cho KO (2020). POU domain motif3 (Pdm3) induces wingless (wg) transcription and is essential for development of larval neuromuscular junctions in Drosophila. Sci. Rep. 10, 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee JH, Lee IS, Lee SB, and Cho KS (2017). Histone lysine methylation and neurodevelopmental disorders. Int. J. Mol. Sci. 18, 1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh YH, Gramates LS, and Budnik V (2000). Drosophila larval neuromuscular junction: molecular components and mechanisms underlying synaptic plasticity. Microsc. Res. Tech. 49, 14–25. [DOI] [PubMed] [Google Scholar]
- Kohsaka H, Takasu E, Morimoto T, and Nose A (2014). A group of segmental premotor interneurons regulates the speed of axial locomotion in Drosophila larvae. Curr. Biol. 24, 2632–2642. [DOI] [PubMed] [Google Scholar]
- Kohsaka H, Guertin PA, and Nose A (2017). Neural circuits underlying fly larval locomotion. Curr. Pharm. Des. 23, 1722–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun N, Mehler-Jacob C, Poirier K, Zordan C, Lacombe D, Carion N, Billuart P, and Bienvenu T (2018). Novel KDM5B splice variants identified in patients with developmental disorders: functional consequences. Gene 679, 305–313. [DOI] [PubMed] [Google Scholar]
- Li J, Ashley J, Budnik V, and Bhat MA (2007). Crucial role of Drosophila neurexin in proper active zone apposition to postsynaptic densities, synaptic growth, and synaptic transmission. Neuron 55, 741–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Greer C, Eisenman RN, and Secombe J (2010). Essential functions of the histone demethylase Lid. PLoS Genet. 6, e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, and Secombe J (2015). The histone demethylase KDM5 activates gene expression by recognizing chromatin context through its PHD reader motif. Cell Rep. 13, 2219–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Greer C, and Secombe J (2014). KDM5 interacts with Foxo to modulate cellular levels of oxidative stress. PLoS Genet. 10, e1004676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AA, Mahapatra CT, Woodruff EA III, Rohrbough J, Leung HT, Shino S, An L, Doerge RW, Metzstein MM, Pak WL, and Broadie K (2010). The nonsense-mediated decay pathway maintains synapse architecture and synaptic vesicle cycle efficacy. J. Cell Sci. 123, 3303–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Wang H, Liang Z, Ku L, O’Donnell WT, Li W, Warren ST, and Feng Y (2004). The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc. Natl. Acad. Sci. USA 101, 15201–15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani L, Lussi YC, Vandamme J, Riveiro A, and Salcini AE (2016). The H3K4me3/2 histone demethylase RBR-2 controls axon guidance by repressing the actin-remodeling gene wsp-1. Development 143, 851–863. [DOI] [PubMed] [Google Scholar]
- Marttinen M, Kurkinen KM, Soininen H, Haapasalo A, and Hiltunen M (2015). Synaptic dysfunction and septin protein family members in neurodegenerative diseases. Mol. Neurodegener. 10, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon KP, Carrillo RA, and Zinn K (2013). Development and plasticity of the Drosophila larval neuromuscular junction. Wiley Interdiscip. Rev. Dev. Biol. 2, 647–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, and Thomas PD (2010). PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 38, D204–D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretto E, Murru L, Martano G, Sassone J, and Passafaro M (2018). Glutamatergic synapses in neurodevelopmental disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry 84, 328–342. [DOI] [PubMed] [Google Scholar]
- Nägerl UV, Eberhorn N, Cambridge SB, and Bonhoeffer T (2004). Bidirectional activity-dependent morphological plasticity in hippocampal neurons. Neuron 44, 759–767. [DOI] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, Hosseini M, Behjati F, Haas S, Jamali P, et al. (2011). Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478, 57–63. [DOI] [PubMed] [Google Scholar]
- Navarro-Costa P, McCarthy A, Prudêncio P, Greer C, Guilgur LG, Becker JD, Secombe J, Rangan P, and Martinho RG (2016). Early programming of the oocyte epigenome temporally controls late prophase I transcription and chromatin remodelling. Nat. Commun. 7, 12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechipurenko IV, and Broihier HT (2012). FoxO limits microtubule stability and is itself negatively regulated by microtubule disruption. J. Cell Biol. 196, 345–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AF, Johnston LA, and Edgar BA (1998). Coordination of growth and cell division in the Drosophila wing. Cell 93, 1183–1193. [DOI] [PubMed] [Google Scholar]
- Newman ZL, Hoagland A, Aghi K, Worden K, Levy SL, Son JH, Lee LP, and Isacoff EY (2017). Input-specific plasticity and homeostasis at the Drosophila larval neuromuscular junction. Neuron 93, 1388–1404.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard M, Koo ES, Gorczyca M, Sharpe J, Cumberledge S, and Budnik V (2002). The Drosophila Wnt, wingless, provides an essential signal for pre- and postsynaptic differentiation. Cell 111, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Rasse TM, Fouquet W, Schmid A, Kittel RJ, Mertel S, Sigrist CB, Schmidt M, Guzman A, Merino C, Qin G, et al. (2005). Glutamate receptor dynamics organizing synapse formation in vivo. Nat. Neurosci. 8, 898–905. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. [DOI] [PubMed] [Google Scholar]
- Rodrigues CH, Pires DE, and Ascher DB (2018). DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 46, W350–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan JL, Wu W, and Crabtree GR (2013). From neural development to cognition: unexpected roles for chromatin. Nat. Rev. Genet. 14, 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos J, Hummel T, Ng N, Klämbt C, and Davis GW (2000). Drosophila Futsch regulates synaptic microtubule organization and is necessary for synaptic growth. Neuron 26, 371–382. [DOI] [PubMed] [Google Scholar]
- Rujirabanjerd S, Nelson J, Tarpey PS, Hackett A, Edkins S, Raymond FL, Schwartz CE, Turner G, Iwase S, Shi Y, et al. (2010). Identification and characterization of two novel JARID1C mutations: suggestion of an emerging genotype-phenotype correlation. Eur. J. Hum. Genet. 18, 330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandaglia M, Lopez-Atalaya JP, Medrano-Fernandez A, Lopez-Cascales MT, Del Blanco B, Lipinski M, Benito E, Olivares R, Iwase S, Shi Y, and Barco A (2017). Loss of Kdm5c causes spurious transcription and prevents the fine-tuning of activity-regulated enhancers in neurons. Cell Rep. 21, 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombe J, Li L, Carlos L, and Eisenman RN (2007). The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 21, 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS,Wang JT, Ramage D,Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, and Delorenzi M (2013). A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics 14, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Onishi M, Jan LY, and Jan YN (2007). Peripheral multidendritic sensory neurons are necessary for rhythmic locomotion behavior in Drosophila larvae. Proc. Natl. Acad. Sci. USA 104, 5199–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, and Cooper DN (2017). The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 136, 665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, and Shi Y (2007). The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 447, 601–605. [DOI] [PubMed] [Google Scholar]
- Trogden KP, and Rogers SL (2015).TOG proteins are spatially regulated by Rac-GSK3β to control interphase microtubule dynamics. PLoS One 10, e0138966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzschach A, Lenzner S, Moser B, Reinhardt R, Chelly J, Fryns JP, Kleefstra T, Raynaud M, Turner G, Ropers HH, et al. (2006). Novel JARID1C/SMCX mutations in patients with X-linked mental retardation. Hum. Mutat 27, 389. [DOI] [PubMed] [Google Scholar]
- Vallianatos CN, and Iwase S (2015). Disrupted intricacy of histone H3K4 methylation in neurodevelopmental disorders. Epigenomics 7, 503–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallianatos CN, Farrehi C, Friez MJ, Burmeister M, Keegan CE, and Iwase S (2018). Altered gene-regulatory function of KDM5C by a novel mutation associated with autism and intellectual disability. Front. Mol. Neurosci. 11, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallianatos CN, Raines B, Porter RS, Bonefas KM, Wu MC, Garay PM, Collette KM, Seo YA, Dou Y, Keegan CE, et al. (2020). Mutually suppressive roles of KMT2A and KDM5C in behaviour, neuronal structure, and histone H3K4 methylation. Commun. Biol 3, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasin A, Zueva L,Torrez C, Volfson D, Littleton JT, and Bykhovskaia M (2014). Synapsin regulates activity-dependent outgrowth of synaptic boutons at the Drosophila neuromuscular junction. J. Neurosci. 34, 10554–10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpelli C, Piccoli G, Zibetti C, Zanchi A, Gardoni F, Huang K, Brambilla D, Di Luca M, Battaglioli E, and Sala C (2010). Synaptic activity controls dendritic spine morphology by modulating eEF2-dependent BDNF synthesis. J. Neurosci. 30, 5830–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova M, Gehling VS, Gustafson A, Arora S, Tindell CA, Wilson C, Williamson KE, Guler GD, Gangurde P, Manieri W, et al. (2016). An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat. Chem. Biol. 12, 531–538. [DOI] [PubMed] [Google Scholar]
- Viquez NM, Li CR, Wairkar YP, and DiAntonio A (2006). The B′ protein phosphatase 2A regulatory subunit well-rounded regulates synaptic growth and cytoskeletal stability at the Drosophila neuromuscular junction. J. Neurosci. 26, 9293–9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk L, Chiu SL, Sharma K, and Huganir RL (2015). Glutamate synapses in human cognitive disorders. Annu. Rev. Neurosci 38, 127–149. [DOI] [PubMed] [Google Scholar]
- Wagner N, Laugks U, Heckmann M, Asan E, and Neuser K (2015). Aging Drosophila melanogaster display altered pre- and postsynaptic ultrastructure at adult neuromuscular junctions. J. Comp. Neurol. 523, 2457–2475. [DOI] [PubMed] [Google Scholar]
- Xing G, Li M, Sun Y, Rui M, Zhuang Y, Lv H, Han J, Jia Z, and Xie W (2018). Neurexin-neuroligin 1 regulates synaptic morphology and functions via the WAVE regulatory complex in Drosophila neuromuscular junction. eLife 7, e30457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YM, Fedchyshyn MJ, Grande G, Aitoubah J, Tsang CW, Xie H, Ackerley CA, Trimble WS, and Wang LY (2010). Septins regulate developmental switching from microdomain to nanodomain coupling of Ca(2+) influx to neurotransmitter release at a central synapse. Neuron 67, 100–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamurrad S, Hatch HAM, Drelon C, Belalcazar HM, and Secombe J (2018). A Drosophila model of intellectual disability caused by mutations in the histone demethylase KDM5. Cell Rep. 22, 2359–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, and Broadie K (2001). Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell 107, 591–603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq data reported in this paper is NCBI Gene Expression Omnibus GEO:GSE159298. A list of differentially expressed genes (and log2 fold change) observed compared to wild-type are provided in Tables S1 and S2 (5% FDR) for kdm5JmjC* and kdm5L854F, respectively.


