Abstract
An aberrant fusion of the DNAJB1 and PRKACA genes generates a chimeric protein kinase (PKA-CDNAJB1) in which the J-domain of the heat shock protein 40 is fused to the catalytic α subunit of cAMP-dependent protein kinase A (PKA-C). Deceivingly, this chimeric construct appears to be fully functional, as it phosphorylates canonical substrates, forms holoenzymes, responds to cAMP activation, and recognizes the endogenous inhibitor PKI. Nonetheless, PKA-CDNAJB1 has been recognized as the primary driver of fibrolamellar hepatocellular carcinoma and is implicated in other neoplasms for which the molecular mechanisms remain elusive. Here we determined the chimera’s allosteric response to nucleotide and pseudo-substrate binding. We found that the fusion of the dynamic J-domain to PKA-C disrupts the internal allosteric network, causing dramatic attenuation of the nucleotide/PKI binding cooperativity. Our findings suggest that the reduced allosteric cooperativity exhibited by PKA-CDNAJB1 alters specific recognitions and interactions between substrates and regulatory partners contributing to dysregulation.
Subject terms: Solution-state NMR, Solution-state NMR
Olivieri, Walker, Karamafrooz et al. show that the fusion of the dynamic J-domain to PKA-C (PKA-CDNAJB1) disrupts the internal allosteric network, attenuating the nucleotide/PKI binding cooperativity. This study suggests that the reduced allosteric cooperativity may contribute to the pathology that PKA-CDNAJB1 drives.
Introduction
Fibrolamellar hepatocellular carcinoma (FL-HCC) is a rare and aggressive form of liver cancer that predominantly affects young patients without underlying cirrhosis or disease1. Treatment is limited, as FL-HCC does not respond to chemotherapy2,3, and surgical excision remains the only therapy4,5. Sequencing the tumor genomes of FL-HCC patients identified a recurrent chimeric gene6–9 that originates from a ~400 kb deletion in chromosome 19. This deletion causes an in-frame fusion of exon 1 from the DNAJB1 gene, which encodes a member of the heat-shock protein 40 family, with exons 2–10 of the PRKACA gene, encoding for the catalytic subunit of cAMP-dependent protein kinase A (PKA-C)7. The resulting chimeric enzyme (PKA-CDNAJB1) is fully functional and comprises 405 residues with 69 amino acids of DNAJB1 (J-domain), replacing the first 14 residues of the N-terminal αA-helix of PKA-C7. The N-terminal deletion of part of the αA-helix prevents essential post-translational modifications such as N-myristoylation, deamidation, as well as phosphorylation at Ser10, which are responsible for the spatiotemporal localization of the kinase10,11.
The fusion of the J-domain does not alter the structure of the catalytic core of PKA-CDNAJB1, which remains virtually identical to the wild-type kinase (PKA-CWT). Instead, the αA-helix of PKA-CDNAJB1 is extended with the J-domain tucked under the large lobe of the enzyme (Fig. 1a)12. Moreover, PKA-CDNAJB1 retains the ability to form stable holoenzymes with all regulatory (R) subunit isoforms, although with non-canonical arrangements, and more importantly, responds to cAMP activation13,14. In vivo enzymatic assays, as well as conventional in vitro coupled assays, have shown that the kinetic parameters of PKA-CWT and PKA-CDNAJB1 are similar7,13,15. Furthermore, PKA-CDNAJB1 binds the endogenous inhibitor PKI (both full-length and derived peptides) with an affinity comparable to wild-type15.
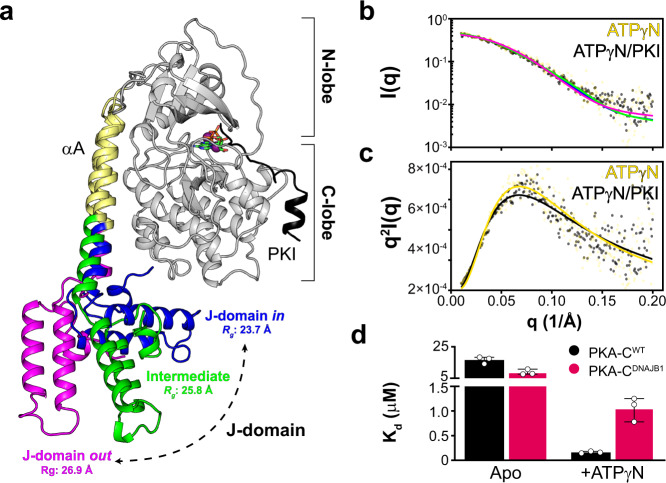
Fig. 1. Multiple conformations adopted by the dynamic J-domain alters allosteric binding cooperativity of PKA-C.
a Overlay of selected snapshots of PKA-CDNAJB1 from the MD trajectories highlighting the ensemble of conformers sampled by the J-domain. b SAXS profiles of PKA-CDNAJB1. Continuous lines show the structural fitting of the experimental SAXS data with the closed (J-domain in, blue), intermediate (green), and extended (J-domain out, magenta) conformations for binary (ATPγN-bound) and ternary (ATPγN/PKI-bound) forms. The conformations are extracted from the MD trajectories. c Corresponding Kratky plot of PKA-CDNAJB1 bound to ATPγN and ATPγN/PKI. d Bar graph showing the dissociation constants (Kd) calculated from the ITC titration curves of PKI5–24 binding to the apo and ATPγN-bound forms of PKA-CWT and PKA-CDNAJB1 (see also Supplementary Table 2).
Despite little differences conferred to the structure and activity of the kinase by the fusion of the J-domain, in vivo studies have shed light on the signaling changes induced by the chimeric fusion. Recently, Scott and co-workers suggested that the J-domain is able to recruit Hsp70, stabilizing the chimeric fusion protein16. Together with the higher expression levels due to the DNAJB1 gene7, the enhanced stability of the enzyme in complex with Hsp70 may explain the dominant expression in FL-HCC cells. Furthermore, these researchers showed that the phosphoproteomics of PKA-CDNAJB1 is different from that of the wild-type, implying that the J-domain skews the phosphorylation profile of cells toward alternative pathways16. According to this study, the J-domain is directly implicated in the oncogenicity of PKA-CDNAJB1. However, other evidence shows that the kinase domain is also required for tumorigenesis17. To date, it remains unclear how this aberrant kinase contributes to the progression of the disease.
A prominent feature of PKA-C is the alternation of positive and negative binding cooperativity that drives the enzymatic cycle18. Positive binding cooperativity induced by ATP-binding enhances the affinity for the substrate, whereas negative binding cooperativity of ADP facilitates the release of the phosphorylated product. Since binding cooperativity is essential to signal transduction amplification19, we surmised that the nucleotide/substrate-binding cooperativity of the aberrant kinase is disrupted, with implications for phosphorylation profiles and regulation by R-subunits. With this in mind, we investigated local and global responses of the kinase to nucleotide and pseudo-substrate binding using a combination of NMR spectroscopy, isothermal titration calorimetry (ITC), small-angle X-ray scattering (SAXS), and molecular dynamics (MD) simulations. We found that the J-domain fused to the N-terminus of PKA-C dramatically attenuates the canonical positive nucleotide/pseudo-substrate-binding cooperativity through alterations in the intramolecular network of communication. Analogous to other pathologic mutants of PKA-C20, the concomitant loss in binding cooperativity and intramolecular communication may render highly cooperative events, such as regulation, dysfunctional, contributing to disease progression.
Results
The dynamic J-domain of PKA-CDNAJB1 interconverts between in and out conformations
Previous X-ray crystallographic studies12 have shown that the core of PKA-CDNAJB1 adopts a conformation similar to the wild type. The catalytic domain in complex with an ATP analog and a pseudo-substrate peptide is virtually superimposable to the structure of the wild-type enzyme12,21, whereas the J-domain of the chimera is tucked tightly under the large lobe of the enzyme (Fig. 1a). In contrast, our computational and solution NMR studies suggest that PKA-CDNAJB1 spans a large ensemble of conformations, with the J-domain undergoing fast re-orientational dynamics relative to the core of the enzyme22. To provide orthogonal validation, we compared the SAXS profiles of PKA-CWT and PKA-CDNAJB1. For both binary (ATPγN-bound) and ternary (ATPγN/PKI-bound) complexes of PKA-CDNAJB1, we fit the SAXS profiles with the compact (J-domain in), intermediate, and extended (J-domain out) conformations extracted from the trajectories of MD simulations (Fig. 1b, c). We found all structures fit well with the SAXS profile with χ < 1.0 in the low q region (q < 0.2 Å−1). However, the radius of gyration (Rg) increases from 23.7 to 26.9 Å, indicating that the J-domain can dislodge from the kinase core and adopt an extended conformation. In contrast, the flexible N-terminus of PKA-CWT is much smaller and does not influence the Rg of the enzyme (21.7 Å) for both binary and ternary complexes (Supplementary Fig. 1). The addition of PKI to nucleotide-saturated PKA-CWT and PKA-CDNAJB1 does not change the SAXS profiles, suggesting that PKI binding does not alter the overall size and shape of the two kinases.
The J-domain of PKA-CDNAJB1 reduces the nucleotide/substrate degree of binding cooperativity (σ)
Previous studies from our group have shown that PKA-CWT behaves like a K-type enzyme, exhibiting positive binding cooperativity between nucleotide (ATPγN) and pseudo-substrate (PKI5–24). The latter property can be reduced by mutations, while maintaining a virtually constant maximal rate20,23,24. Thus, we sought to understand how this canonical positive binding cooperativity is altered in PKA-CDNAJB1, using isothermal titration calorimetry (ITC). For the binding of nucleotide alone, PKA-CDNAJB1 exhibits a fourfold higher binding affinity for ATPγN compared with PKA-CWT (Kd = 19 ± 4 μM versus 83 ± 8 μM; Supplementary Table 1 and Supplementary Fig. 2). To determine the degree of binding cooperativity (σ) for PKA-CDNAJB1, we monitored the binding affinity of PKI5–24 in the absence and presence of nucleotide. In the absence of nucleotide, PKA-CDNAJB1 binds PKI5–24 with a twofold higher affinity compared to wild-type (Kd = 9 ± 2 μM versus 17 ± 2 μM; Fig. 1d and Supplementary Fig. 2). In contrast, upon saturation with nucleotide, the PKA-CDNAJB1/ATPγN complex binds PKI5–24 with a sevenfold decrease in binding affinity (Kd = 1.1 ± 0.2 μM versus 0.16 ± 0.02 μM). Together, these data show that the addition of the J-domain causes a sizeable decrease (13-fold) in nucleotide/pseudo-substrate-binding cooperativity (Supplementary Table 2).
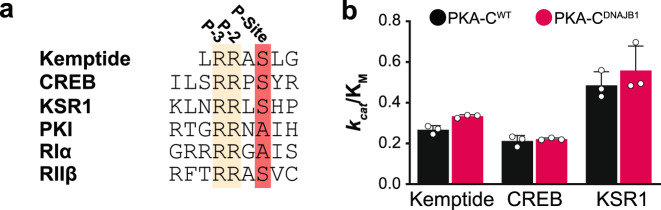
To evaluate the effect of the dramatic reduction in binding cooperativity on the ability of PKA-CDNAJB1 to perform phosphoryl transfer, we used steady-state coupled enzyme assays. Specifically, we evaluated the catalytic efficiencies of PKA-CWT and PKA-CDNAJB1 toward the standard peptide substrate, kemptide, physiological substrate, CREB, and a known hyper-phosphorylated substrate identified by Scott and co-workers, KSR116 (Fig. 2a). Though the substrates displayed different maximal velocities (Vmax = 0.21−0.35 μM/s) and Michaelis constants (KM = 29–56 μM), PKA-CDNAJB1 consistently displayed higher Vmax values resulting in slightly higher catalytic efficiencies (kcat/KM) compared with wild-type for all substrates tested (Fig. 2b, Supplementary Table 3, and Supplementary Fig. 3). Although the binding cooperativity is affected by the presence of the J-domain, these assays with peptides encompassing the recognition sequences of different substrates demonstrate that the kinetic parameters are only marginally altered as reported previously15.
Fig. 2. Attenuation in binding cooperativity does not influence phosphoryl transfer.
a Comparison of the substrate recognition sequences of substrates Kemptide, CREB, and KSR1, regulatory subunits, RIα and RIIβ, and endogenous inhibitor, PKI. b Bar graph displaying the catalytic efficiencies (kcat/KM) of PKA-CWT and PKA-CDNAJB1 towards Kemptide, CREB, and KSR1. Errors were calculated from triplicate measurements. All kinetics parameters are summarized in Supplementary Table 3 and shown in Supplementary Fig. 3.
The J-domain disrupts the internal allosteric network of PKA-CDNAJB1
To determine the origin of the decreased allosteric binding cooperativity, we analyzed the structural response of the kinases to ligand binding using solution NMR spectroscopy. Specifically, we mapped the amide fingerprints of both enzymes using [1H, 15N]-TROSY-HSQC experiments25 upon addition of nucleotide and PKI5–24 (Supplementary Fig. 4). The amide fingerprints of PKA-CWT and the PKA-CDNAJB1 kinase core are almost superimposable, reflecting their identical structures. This similarity enabled us to transfer the previously assigned PKA-CWT amide resonances23 to the PKA-CDNAJB1 spectrum (Supplementary Fig. 4a). To assign the remaining resonances of the J-domain, we expressed the 69-residue construct (DNAJB11–69) uniformly labeled with 15N and 13C in Escherichia coli bacteria and assigned the resonances in the [1H, 15N]-TROSY-HSQC spectrum using the classical out-and-back triple resonance experiments (Supplementary Fig. 5). A total of 340 amide resonances were assigned over the 388 expected. As previously reported26, the DNAJB11–69 domain folds autonomously, and its resonances overlay with those of the J-domain fused to PKA-CDNAJB1 (Supplementary Fig. 4b). To analyze the kinases response to ATPγN and PKI5–24 binding, we followed the 1H and 15N chemical shift perturbations (CSPs) of the amide fingerprints for the two kinases. We found that the fingerprints of both PKA-CWT and PKA-CDNAJB1 show similar CSPs upon binding ATPγN (Supplementary Fig. 6a). However, residues of PKA-CDNAJB1 encompassing the Gly-rich loop, activation loop, αF-helix, αG-helix, and the C-terminal tail show greater CSPs compared with PKA-CWT. Notably, these domains undergo the largest structural rearrangements from the open (apo) to closed (ternary) form. The J-domain is not directly involved in the binding events as it experiences CSPs well below one standard deviation. As for PKA-CWT, the binding of ATPγN causes broadening of amide resonances throughout the core of PKA-CDNAJB1, suggesting the presence of conformational dynamics in the μs–ms timescale27. Upon binding PKI5–24 to the ATPγN-saturated kinase, the CSPs of the kinase cores are similar, with effects propagated from the substrate-binding sites to both small and large lobes (Supplementary Fig. 6b). The J-domain remains structurally isolated from the core as we observe only a few chemical shift changes upon binding PKI5–24. As found for PKA-CWT, PKI5–24 binding sharpens the amide fingerprint resonances, indicating that the enzyme is trapped in a well-defined conformational minimum23. The linewidths of the J-domain, on the other hand, are unperturbed, suggesting that pseudo-substrate binding does not change its conformational dynamics.
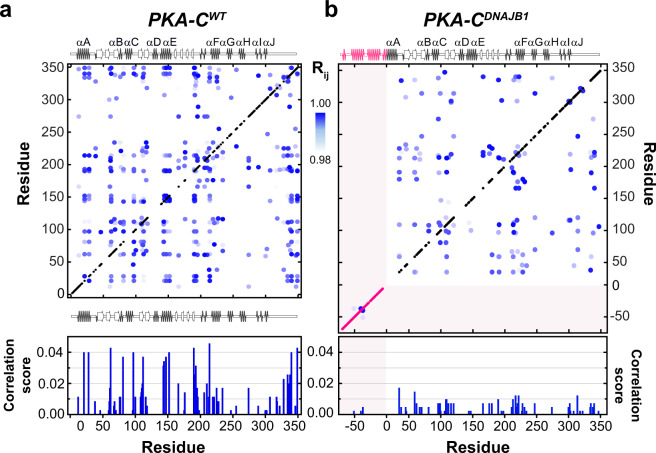
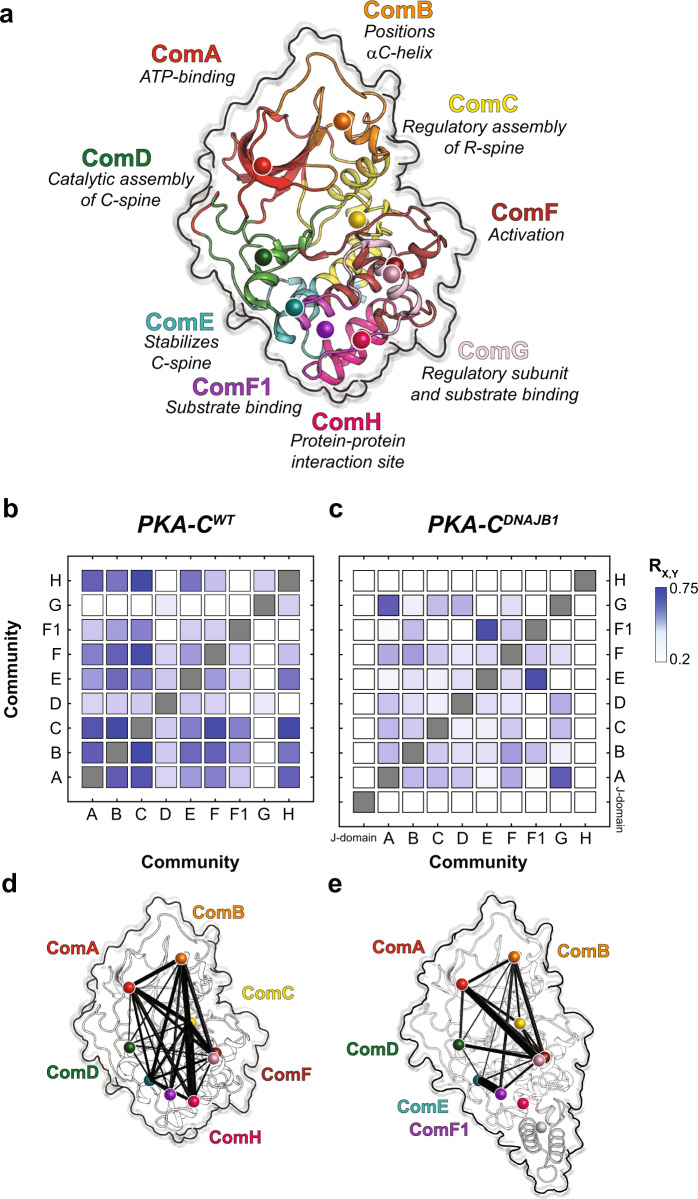
To analyze the internal network of communication of both PKA-CDNAJB1 and PKA-CWT, we used CHEmical Shift Covariance Analysis (CHESCA)28,29. This method identifies allosteric networks among the protein residues by tracking each residue’s coordinated changes of chemical shifts in response to ligand binding or mutations. For a binding process occurring in the fast NMR timescale, the resonances associated with allosterically coupled residues exhibit a correlated linear response. When plotted on a correlation matrix, changes in allosteric networks manifest as variations in the number and extent of inter-residue correlations29,30. We analyzed the chemical shift responses of four different forms of the kinase: apo, ATPγN-bound, ADP-bound, and ATPγN/PKI5–24bound. The CHESCA matrix of PKA-CWT shows a coordinated response for many residues spanning the entire enzyme, with a higher density of correlations in the N-lobe near the nucleotide-binding site and the substrate-binding groove at the interface between the N- and C-lobes (Fig. 3a). These correlations constitute central allosteric nodes necessary for binding cooperativity18,20,31. In addition, allosteric cross-talk exists between residues in the C-terminal tail and residues in the αA-helix (K21, K28), αE-helix (R144, A147, L152), activation loop (R190, V191, G193), and αF-helix (G214, G225). In contrast, the matrix of PKA-CDNAJB1 exhibits an overall attenuation in CHESCA correlations throughout the entire kinase, with a rewiring of the network between nucleotide and substrate-binding sites. Specific structural domains such as the αA-helix (K21, K27), αE-helix (A143, R144, A147, L152), αC-helix (Q96, A97), and C-terminal tail (I335, N340, E349) are more noticeably attenuated (Fig. 3b). The functional relevance of this extensive rewiring of intramolecular communication is apparent when we combined CHESCA with the definition of the structural and functional communities (community CHESCA)32. Upon dividing the kinase into nine different communities defined by McClendon et al.32 (Fig. 4a), each with different structural and functional roles, we observe a clear parallel between experimental data and theory31,32. The correlation matrix for community CHESCA for PKA-CWT (Fig. 4b) shows that structurally adjacent communities in the N-lobe and at the N/C lobe interface (ComA, ComB, ComC, and ComE) are strongly coupled with distal communities (ComH) as they exhibit high correlation coefficients (RX,Y). A structural representation of the extent of coupling is reported in Fig. 4d, where cross-talk between the nucleotide-binding (ComA) with the positioning of the αC-helix (ComB), the R-spine assembly (ComC) and the activation loop (ComF) is apparent. Although attenuated, there is a substantial coupling between these communities and the more peripheral communities that have structural roles, such as ComH. The cooperative response to nucleotide and PKI5–24 binding involves the entire enzyme, causing coordinated CSPs interspersed throughout its three-dimensional structure. In contrast, the community CHESCA for PKA-CDNAJB1 exhibits a dramatic decrease in inter-community coupling (Fig. 4c, e). The coupling network between the ATP-binding site (ComA) and the αC-helix (ComC) or the activation loop (ComF) is sparse. Notably, the correlations between the N-lobe communities with peripheral communities in the C lobe (ComD and ComE) are either notably reduced or lost. This phenomenon is especially evident for ComH located near the J-domain, which includes residues that are involved in docking of the R-subunits or kinase binding partners. Other correlations not found in the wild-type are observed between ComG and ComA, ComB, ComD, and ComF; and stronger correlations are observed between ComE and ComF1. Overall, the community CHESCA show that the reduced degree of cooperativity determined thermodynamically corresponds to a decrease in coordinated structural changes upon ligand binding in PKA-CDNAJB1.
Fig. 3. Defective internal allosteric network of PKA-CDNAJB1.
Chemical shift correlation matrices for a PKA-CWT and b PKA-CDNAJB1. The cross-peaks represent pairs of residues with the absolute value of the Pearson’s coefficient ≥0.98. The secondary structures for the two kinases are reported above and below the matrix. The bar graphs shown below each matrix report the average correlation scores per residue (see Methods section).
Fig. 4. Reduced cross-talk among structural communities of chimeric PKA-CDNAJB1 relative to PKA-CWT.
a Structure of PKA-C highlighting all communities and their roles in various functions and regulatory mechanisms. CHESCA community matrices for b PKA-CWT and c PKA-CDNAJB1 upon binding PKI5–24. d–e CHESCA community matrices plotted on their corresponding structures. The size of each node is independent of the number of residues it encompasses, meanwhile the weight of each line indicates the strength of coupling between the individual communities.
Global versus community-specific responses to ligand binding
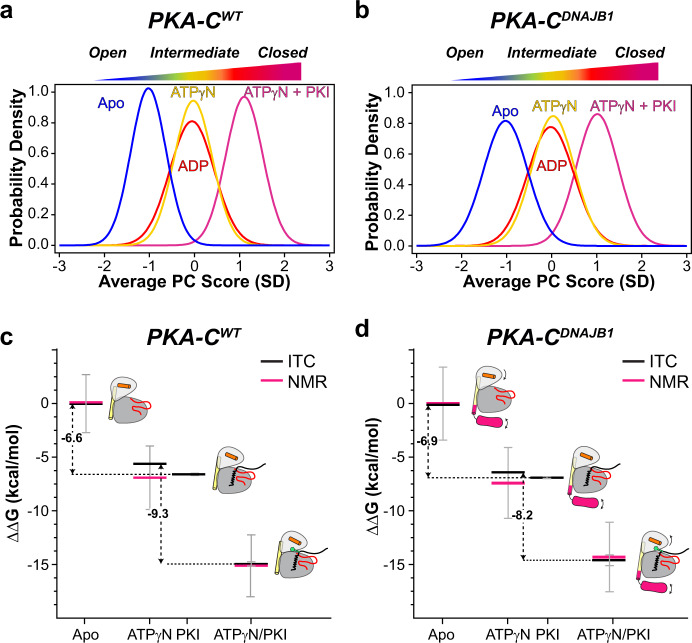
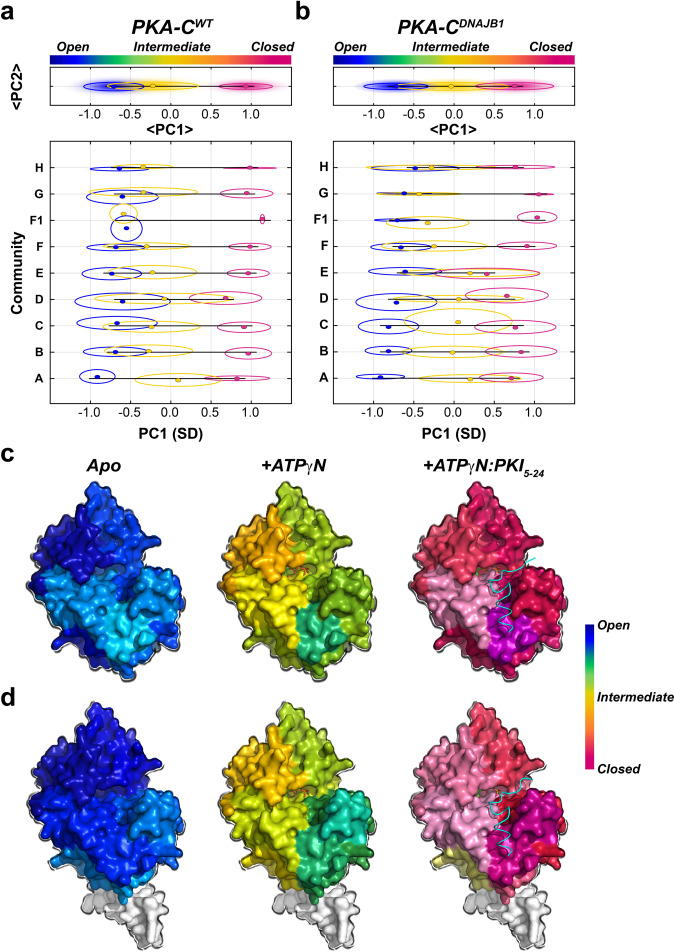
Although CHESCA provides an estimate of the correlated response for individual residues (or communities) to ligand binding, the analysis of the amide chemical shifts with CONCISE (COordiNated ChemIcal Shifts bEhavior)33 provides the probability density (population) of each state along the conformational equilibrium. According to CONCISE analysis, the probability density of the amide resonances for the apo form of the enzyme (open state) is clustered around an average principal component (PC) score (<PC>) of ~−1.2 (Fig. 5a)34. Upon binding ATPγN, ADP, and PKI5–24, the probability densities for the amide resonances of PKA-CWT progressively shift toward more positive <PC> values, with the density for the binary complex (intermediate state) showing a <PC> ~ 0, and the ternary complex an <PC> ~1.3 (closed state). A similar trend is observed for PKA-CDNAJB1, though the probability densities are broader, indicating that the amide resonances follow a less coordinated response33. Also, the maximum probability density for the intermediate state of PKA-CDNAJB1 is slightly shifted toward the closed conformation, whereas the probability density for the ternary complex is slightly more open than the corresponding wild-type (Fig. 5b). These probability densities or populations of each state can be converted into free energy, and the shift of the maxima from free to bound state along the <PC> axis reflects the free energy of binding33. Indeed, the ΔΔG values obtained from ITC data are very similar to those calculated from CONCISE (Fig. 5c, d and Supplementary Table 4). Consequently, the attenuation in the collective response of the amide resonances of the PKA-CDNAJB1 is correlated with the reduced extent of binding cooperativity. Interestingly, we found that the two enzymes respond to ligand binding in a community-specific manner (Fig. 6a, b). For PKA-CWT, the resonances associated with ComA have the most prominent response upon nucleotide binding. Also, large conformational transitions are observed for ComB, ComC, ComD, and ComE, whereas the remaining communities, located mostly in the C lobe, show only marginal changes, with an <PC1> of ~−0.4. Binding of PKI5–24 to the PKA-C/ATPγN complex shifts all the communities toward a fully closed state (<PC1> ~1) (Fig. 6c and Supplementary Fig. 7a). In contrast, the probability densities for ComA, ComB, ComC, ComD, and ComE for PKA-CDNAJB1 are shifted to a more closed intermediate state (<PC1> ~0). Moreover, PKI5–24 binding to PKA-CDNAJB1/ATPγN complex shifts the probability densities of the various communities toward the closed state to different extents (<PC1> ~0.75), with only ComF, ComF1, ComG, and ComH reaching a fully closed state. The remaining communities located in the N-lobe cluster around PC1 ~0.65, adopting a slightly more open conformation (Fig. 6d and Supplementary Fig. 7b). Overall, the J-domain fused to the catalytic core decreases the number of correlated chemical shift responses, i.e., the community of the kinase no longer transition in a correlated fashion, and prevents several communities (ComA, ComB, ComC, ComD, and ComE) from reaching a fully closed state, which explains the reduced extent of nucleotide/substrate-binding cooperativity24.
Fig. 5. Global coordinated response of PKA-CWT and PKA-CDNAJB1 to ligand binding.
Coordinated chemical shift changes analyzed with CONCISE for a PKA-CWT and b PKA-CDNAJB1. Changes in the free energy of binding of c PKA-CWT and d PKA-CDNAJB1 to ATPγN and PKI5–24. Black lines indicate the values obtained from ITC and pink lines correspond to values obtained from NMR measurements.
Fig. 6. Community-specific responses of PKA-CWT and PKA-CDNAJB1 upon ATPγN and PKI5–24 binding.
CONCISE analysis for each individual community for a PKA-CWT and b PKA-CDNAJB1. The top panel on the graphs shows the average PC1 (<PC1>) overall communities, whereas the bottom panel shows the PC1 scores of each individual community. CONCISE scores of each community mapped onto the surface of c PKA-CWT (PDB: 4WB5) and d PKA-CDNAJB1 (PDB: 4WB7) for each state (apo, +ATPγN, and +ATPγN/PKI5–24).
Discussion
There is mounting evidence that PKA-CDNAJB1 is the primary driver of FL-HCC6–8 and may be involved in other pathologies35,36. Despite its link to tumorigenesis, initial structural and functional studies have left more questions than answers regarding the molecular etiology of this rare liver carcinoma. Similar to other oncogenic kinases involved in Cushing’s syndrome, the X-ray structure of PKA-CDNAJB1 does not reveal any structural anomalies in the catalytic core (RMSD < 0.4 Å)12. In solution, however, NMR and SAXS have shown that the J-domain is flexible, assuming both in and out conformations22. The inherent flexibility of the J-domain enables the kinase to form hetero-tetrameric holoenzymes with RIα subunits, although with two alternative conformations14. Interestingly, the RIα subunit is overexpressed in carcinoma cells, presumably to “buffer” the PKA-CDNAJB1 overexpression6,13,37. In rare cases of FL-HCC, lack of PKA-C regulation by RIα may result in the progression of the disease38, like the dysregulation of PKA-C in Carney Complex39. These data alone do not adequately explain the aberrant function of PKA-CDNAJB1.
It is possible, however, that the functional repercussion of the J-domain fusion to PKA-C is multifaceted. First, the deletion of 14 residues in the αA-helix and N-terminus prevents post-translational modifications such as myristoylation, deamidation, and phosphorylation of Ser10. These modifications are essential for intracellular membrane localization40,41; therefore, the J-domain may interfere with the localization of the enzyme in the proximity of natural substrates. Second, the phosphorylation profile of PKA-CDNAJB1 is considerably different from the wild-type16. Third, the presence of the J-domain may interfere with the formation of localization complexes with A-kinase anchoring proteins (AKAPs)37. Fourth, PKA-CDNAJB1 may form stable complexes with Hsp70, with the J-domain acting as a scaffold for this chaperonin16.
Irrespective of these putative mechanisms, our studies reveal that the fusion of the J-domain to the kinase reduces canonical positive binding cooperativity between nucleotide and substrate24, and alters the kinases’ conformational equilibrium via disruptions of the intramolecular allostery. Indeed, cooperativity has an essential role in macromolecular assembly, regulation, and signal transduction19,42. Therefore, the attenuation in allosteric binding cooperativity is likely to be implicated in the altered interactome and phosphoproteome16. As the recognition sequences of R-subunits are highly homologous to PKI, the reduced nucleotide/PKI binding cooperativity of PKA-CDNAJB1 suggests an aberrant regulation of the holoenzyme as well. It is worth noting that a reduction in cooperativity is also a prominent feature of a single mutation in the P + 1 loop of the catalytic subunit (PKA-CL205R) in patients affected by Cushing’s syndrome20,43,44.
Consistent with our previous analyses of PKA-CL205R, the allosteric network of PKA-CDNAJB1 exhibits an overall attenuation in the density of correlations across the kinase with a marked reduction near the nucleotide-binding site and substrate-binding groove. The introduction of community-specific analyses assists in deciphering how the fusion of the J-domain perturbs the allosteric network and further underscores the linkage between the reduced binding cooperativity and the decrease in cross-talk among the structural communities32. The coupling between ComH, responsible for the docking of proteins such as PKI or R-subunits, to other communities is completely ablated, explaining the lower degree of binding cooperativity observed for PKA-CDNAJB1, and further suggesting that the R/C-interactions may be altered. Moreover, the dramatic decrease in the inter-community coupling of ComC, adjacent to the αA-helix, reflects the structural and/or dynamic changes caused by the J-domain fusion to the αA-helix. Overall, the similarities observed between PKA-CL205R and PKA-CDNAJB1 suggest a common mechanism, involving concomitant reductions in binding cooperativity and intramolecular allostery that may elicit their aberrant function.
Indeed, both computational and experimental methods have successfully elucidated allosteric pathways in a variety of different systems outside of protein kinase A45–52. Perhaps more importantly, many of these studies have shown how disruptions in these pathways act to change function. Taken with our previous studies, nucleotide/substrate-binding cooperativity emerges as a critical function in protein kinase A that arises from the structural changes of multiple domains (i.e., communities) that must be coordinated and synchronized to ensure a cooperative binding response18,20. Like other tumorigenic transcriptomes that result in a fully active PKA-C, it is likely that alterations in the allosteric network impart defective binding cooperativity and may play a role in their aberrant functions35,36,53. As for the chimeric PKA-CDNAJB1, attenuated or dysfunctional responses to allosteric effectors (such as ATP) may result in defective spatial and temporal control of phosphorylation-mediated signaling in other protein kinases, leading to disease.
Methods
Sample preparation of PKA-CWT and PKA-CDNAJB1
Recombinant PKA-CWT was expressed and purified as described in Walker et al.20. PKA-CDNAJB1 was cloned into a pET-28a (+) vector. A tobacco etch virus cleavage site was incorporated via mutagenesis into the vector between the cDNA coding for the kinase and a thrombin cleavage site. Transformed E. coli BL21 (DE3) pLyss (Agilent) cells were cultured in M9 minimal media supplemented with 15NH4Cl. Protein overexpression was induced with 0.4 mM isopropyl β-d-thiogalactopyranoside (IPTG) and carried out overnight at 20 °C. Following harvest, the cell pellet was resuspended in 50 mM Tris-HCl, 30 mM KH2PO4, 300 mM NaCl, 5 mM 2-mercaptoethanol, 0.15 mg/mL Lysozyme, 200 μM ATP, DNaseI, Roche ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor tablet (pH 8.0) and lysed using French press. The solution was cleared by centrifugation (16,000 rpm, 4 °C, 45 min) and the supernatant incubated with Ni2+ nitrilotriacetic acid resin (Thermo Scientific, 1 ml of resin per liter of culture) at 4 °C overnight. The resin was then washed with 50 mM Tris-HCl, 30 mM KH2PO4, 300 mM NaCl, 5 mM 2-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride (PMSF) (pH 8.0). The resin was further washed with the same buffer supplemented with 10 mM imidazole. PKA-CDNAJB1 was eluted using the same buffer with 250 mM imidazole. Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS–PAGE) electrophoresis samples were taken at each purification step listed and are shown in Supplementary Fig. 8a. Fractions containing PKA-CDNAJB1 were cleaved for 18 h at 4 °C in a dialysis buffer composed of 20 mM KH2PO4, 25 mM KCl, 5 mM 2-mercaptoethanol, 0.1 mM PMSF (pH 7.0). The phosphorylation states of PKA-CDNAJB1 were separated using a cation exchange column (HiTrap Q-SP, GE Healthcare Life Sciences using a linear gradient of KCl in 20 mM KH2PO4.
Sample preparation of DNAJB1
The 69 amino-acid sequences corresponding to the DNAJB1 heat-shock protein fragment, DNAJB11–69, was cloned into the pET-28a (+) vector. DNAJB11–69 was expressed in E. coli BL21 (DE3) (Agilent) in M9 minimal media containing 15NH4Cl and 13C-glucose as the sole source of nitrogen and carbon, respectively. Overexpression was induced with 0.4 mM IPTG and carried out for 5 h at 30 °C. The cell pellet was resuspended in 20 mM Tris-HCl, 300 mM NaCl, 5 mM 2-mercaptoethanol, 15 mg/mL Lysozyme, DNaseI, Roche EDTA-free protease inhibitor tablet (pH 8.0) and lysed using sonication. The lysate was cleared by centrifugation (18,000 rpm, 4 °C, 30 min,) and the supernatant was incubated with Ni2+ nitrilotriacetic acid resin at 4 °C overnight. Resin was washed with 20 mM Tris-HCl, 300 mM NaCl, 5 mM 2-mercaptoethanol, 0.5 mM PMSF (pH 8.0). DNAJB11–69 was eluted using the same buffer supplemented with 250 mM imidazole. SDS–PAGE electrophoresis samples were taken at each purification step listed as shown in Supplementary Fig. 8b. The elution was dialyzed for 3 h at 4 °C into 20 mM Tris-HCl, 150 mM NaCl and 5 mM 2-mercaptoethanol (pH 8.0). To cleave the His-tag, an appropriate amount of thrombin was added to the solution and allowed to occur at RT for 4 h. The cleavage reaction was monitored by SDS–PAGE, and upon completion, thrombin was inactivated by adding 1 mM PMSF. To purify the protein, the cleavage solution was passed through a Ni2+ nitrilotriacetic acid resin. The flowthrough was collected, concentrated using a 10 kDa, and subsequently a 3 kDa spin concentrator and stored at 4 °C in 20 mM Tris-HCl, 150 mM NaCl and 2 mM DTT supplemented with 0.5 mM of PMSF (pH 8.0).
Sample preparation of peptides
Kemptide (LRRASLG), CREB (123KRREILSRRPSYR135), and KSR1 (880LPKLNRRLSHPGHFWKS896), and PKI5–24 were synthesized on a CEM Liberty Blue microwave synthesizer. All peptides were purified using reverse-phase high-pressure liquid chromatography. The purified peptide was concentrated, lyophilized, and stored at −20 °C. All peptides molecular weight was verified by MALDI-TOF. Kemptide’s molecular weight quantity was verified by amino-acid analysis (Texas A&M University).
ITC measurements
ITC measurements were performed with a low-volume NanoITC (TA Instruments). PKA-CWT and PKA-CDNAJB1 were dialyzed into 20 mM MOPS, 90 mM KCl, 10 mM DTT, 10 mM MgCl2, and 1 mM NaN3 (pH 6.5). PKA-CWT and PKA-CDNAJB1 concentrations for ITC measurements were between 80 and 130 μM as confirmed by A280 = 53,860 M−1 cm−1 and A280 = 62,800 M−1 cm−1, respectively. Approximately 300 μL of the kinase was used for each experiment, with 50 μL of 2 mM ATPγN and/or 0.6–1 mM PKI5–24 in the titrant syringe. The heat of dilution of the ligand into the buffer was taken into account for all experiments and subtracted. For experiments with saturated nucleotide, 2 mM ATPγN was added. All measurements were performed at 300 K in triplicates. The binding was assumed to be 1:1, and curves were analyzed with the NanoAnalyze software (TA Instruments) using the Wiseman Isotherm43:
| 1 |
where d[MX] is the change in total complex relative to the change in total protein concentration, d[Xtot] is dependent on r, the ratio of Kd relative to the total protein concentration, and Rm, the ratio between total ligand and total protein concentration. The free energy of binding was determined using the following:
where R is the universal gas constant, and T is the temperature at measurement (300 K). The entropic contribution to binding was calculated using the following:
The degree of cooperativity (σ) was calculated as follows:
where Kd Apo is the dissociation constant of PKI5–24 binding to the apoenzyme, and Kd Nucleotide is the corresponding dissociation constant for PKI5–24 binding to the nucleotide-bound PKA-C.
Enzyme assays
Steady-state activity assays with Kemptide, CREB, and KSR1 were performed under saturating ATP concentrations and spectrophotometrically at 298 K as described by Cook et al.44. The values of Vmax and KM were obtained from a non-linear fit of the initial velocities to the Michaelis–Menten equation.
Small-angle X-ray scattering
Following the purification method outlined above, PKA-CDNAJB1 was buffer exchanged into 20 mM MOPS, 90 mM KCl, 60 mM MgCl2, 10 mM DTT, and 1 mM NaN3 (pH 6.5) and concentrated to 70 μM. For binary and ternary samples, 12 mM ATPγN and/or 70 μM PKI (1:1 ratio) was added to PKA-CDNAJB1. The fitting of structures from MD simulations against the experimental SAXS curves was performed using the FoXS web server54. One hundred random snapshots from MD simulations for PKA-CWT and PKA-CDNAJB1 were chosen as representative structures for the conformational ensembles, and the fitting of the SAXS data was carried out for a range of q values spanning from 0.01 to 0.2.
NMR spectroscopy
Uniformly 15N-labeled PKA-CWT and PKA-CDNAJB1 were overexpressed and purified as described above. NMR experiments were performed in 90 mM KCl, 20 mM KH2PO4, 10 mM dithiothreitol (DTT), 60 mM MgCl2, and 1 mM NaN3 at pH 6.5. Standard [1H-15N]-TROSY-HSQC experiments were carried out for PKA-CWT and PKA-CDNAJB1 on an 850-MHz Bruker Advance III spectrometer equipped with a TCI cryoprobe, respectively. Concentrations for samples were 0.1–0.3 mM as determined by A280 measurements, 12 mM ATPγN or ADP was added for the nucleotide-bound form, and 0.2–0.4 mM PKI5–24 for the ternary complex. Spectra were collected at 300 K, processed using NMRPipe55, and visualized using Sparky56.
All [1H-15N]-TROSY-HSQC experiments were acquired with 2048 (proton) and 256 (nitrogen) complex points. Combined CSPs were calculated using 1H and 15N chemical shifts according to the following:
| 2 |
Backbone resonance assignment of DNAJB11–69
Uniformly 13C,15N-labeled DNAJB11–69 was overexpressed and purified as described above. NMR experiments were performed in 90 mM KCl, 20 mM KH2PO4, 10 mM DTT, 60 mM MgCl2, and 1 mM NaN3 (pH 6.5). Standard 3D triple resonance experiments were carried out for DNAJB11–69 on a 600 MHz Bruker Avance NEO spectrometer equipped with a triple resonance cryogenic probe at 300 K. Concentrations for samples were 0.3 mM as determined by A280 measurements. All NMR data were processed using NMRPipe55 and visualized using Sparky56.
Chemical shift analyses
COordiNated ChemIcal Shift bEhavior (CONCISE)
CONCISE was used to measure the change in equilibrium position using the following PKA-C constructs: apo, ATPγN, ADP, and ATPγN/PKI5–24. This method identifies sets of residues whose chemical shifts respond linearly to the conformational transition using principal component analysis. Community-based and domain-based CONCISE analyses use similar methods using only select residues that comprise either an individual community or domain as defined by McClendon et al.32,57. ΔΔG values from NMR derived from CONCISE analysis and based on the extent of closure (% closed). Values used for the calculations can be found in Supplementary Table 4. PKA-CWT ITC values were used as a reference with apo PKA-CWT: ΔΔG = 0 and 0% closed; and +ATPγN/PKI-bound PKA-CWT: ΔΔG = −15 and 100% closed. The error was calculated by using the standard deviation of the gaussian fits from CONCISE analysis.
CHEmical Shift Covariance Analysis (CHESCA)
CHESCA was used to identify and functionally characterize allosteric networks of residues eliciting concerted responses to, in this case, nucleotide and pseudo-substrate. A total of four states were used to identify inter-residue correlations: apo, ATPγN-bound, ADP-bound, and ATPγN/PKI5–24. The identification of inter-residue correlations by CHESCA relies on agglomerative clustering and singular value decomposition. Pairwise correlations between chemical shift variations experienced by different residues were calculated to identify networks. When plotted on a correlation matrix, this allows for the identification of regions that are correlated to one another. For each residue, the max change in chemical shift was calculated in both the 1H (x) and 15N (y) dimensions (Δδx,y). Residues were included in CHESCA analysis only if they satisfied the following: Δδx,y > ½ ΔνxA,yA+½ Δν xB,yB, where A and B correspond to two different forms analyzed. (Note that there is no dependence on which two forms satisfied this statement) For every residue x, correlation scores were calculated by finding the total no. of residues correlated (with Rij > 0.98) to residue x and dividing by the total no. of residues in the kinase (350). Community CHESCA analysis is a chemical shift-based correlation map between various functional communities within the kinase. Each community is a group of residues (McClendon et al.35) associated with a function or regulatory mechanism. Mathematically, this community-based CHESCA analysis is a selective interpretation of CHESCA, where we evaluate a correlation score between residues in various communities, as shown below. To represent community-based CHESCA analysis, we lowered the correlation cutoff such that Rcutoff > 0.8. Suppose community X and community Y has nX and nY number of assigned residues respectively, the correlation score between X and Y is defined as,
Where Rij is the CHESCA correlation coefficient between residue i (belongs to community A) and residue j (belongs to community B). Rcutoff is the correlation value cutoff. RX,Y can assume values ranging from 0 (no correlation between residues in X and Y) to 1 (all residues in X are correlated with all residues in Y).
MD simulations
Parallel MD simulations were set up to compare PKA-CWT and PKADNAJB1. These simulations were repeated for the apo, the binary (ATP-bound), and the ternary complexes (ATP/PKI5–24). The systems with PKA-CWT and PKA-CDNAJB1 were built directly from the X-ray structures starting from the closed configurations (PDB ID: 1ATP and 4WB7, respectively)12,58. All simulations were performed using GROMACS 4.659 in the CHARMM36 force field60. The all-atom structures were solvated in a rhombic dodecahedron solvent box with a TIP3P61 water molecule layer extending ~10 Å away from the surface of the proteins. Counter ions (K+ and Cl−) were added to ensure electrostatic neutrality corresponding to an ionic concentration of ~150 mM. The LINCS62 algorithm was applied to constrain all covalent H-bonds to the equilibrium length, and particle-mesh Ewald63 was used to treat long-range electrostatic interactions with a real-space cutoff of 10 Å. All systems were minimized using steepest descent algorithm, and then were gradually heated to 300 K at a constant volume over 1 ns, using harmonic restraints with a force constant of 1000 kJ/(mol*Å2) on heavy atoms of both proteins and nucleotides. Over the following 12 ns of simulations at constant pressure (1 atm) and temperature (300 K), the restraints were gradually released. The systems were equilibrated for an additional 20 ns without positional constraints. A Parrinello–Rahman barostat64 was used to keep the pressure constant, while a V-rescale thermostat with a time step of 2 fs was used to maintain a constant temperature. Each system was simulated for 1.05 μs, with snapshots recorded every 20 ps. A total of 6.3 μs and 315,000 conformations were utilized for the analyses.
Statistics and reproducibility
Numerical ΔG, ΔH, −TΔS, and Kd values shown in Supplementary Tables 1 and 2 are averages of three independent ITC experiments with errors calculated as the standard deviation between these three measurements. Numerical values of Vmax and KM were determined with a non-linear regression function assuming Michaelis-Menten kinetics for substrate concentration versus velocity using GraphPad Prism 8.0.1. Error in kcat/KM values was propagated from error in kcat and KM.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of additional supplementary files
Acknowledgements
This work was supported by the National Institutes of Health, GM100310 (G.V.), S10 OD021536 (G.V.), HL139065 (D.D.T.), and AG26160 (D.D.T); and the American Heart Association, 20PRE35120253 (C.W.). NMR experiments were carried out at the Minnesota NMR Center and MD simulations using the Minnesota Supercomputing Institute. The SAXS instrument at the University of Utah was funded by the Department of Energy (Dr. J. Trewhella).
Author contributions
C.O. and C.W. collected and analyzed activity assays, NMR, and ITC data and contributed to the writing of the manuscript. A.K carried out preliminary experiments. Y.W. analyzed SAXS data and completed MD simulations. D.K.B collected the SAXS data. M.V.S. wrote all scripts for CHESCA and CONCISE analyses. F.P., D.D.T., D.A.B., S.M.S., and S.S.T. contributed to the critical analysis of the data and writing of the manuscript. G.V. conceived and directed the project, along with helping with data analysis and writing the manuscript.
Data availability
Raw data from steady-state activity assays and corresponding analysis is deposited on the Data Repository for the University of Minnesota (DRUM) and can be found at the following link: http://hdl.handle.net/11299/217205. Proton and amide chemical shift list files of wild-type PKA-C and the chimeric mutant (PKADNAJB1) are deposited on DRUM and can be found at the following link: http://hdl.handle.net/11299/217206. The source data for the graphs in the main figures are provided in Supplementary Data 1. All other data sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
MATLAB scripts used for the chemical shift analyses presented are deposited on DRUM and can be found at the following link: http://hdl.handle.net/11299/217206.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Cristina Olivieri, Caitlin Walker, Adak Karamafrooz.
Change history
4/6/2021
A Correction to this paper has been published: 10.1038/s42003-021-02006-3
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-01819-6.
References
- 1.Liu S, Chan KW, Wang B, Qiao L. Fibrolamellar hepatocellular carcinoma. Am. J. Gastroenterol. 2009;104:2617–2624. doi: 10.1038/ajg.2009.440. [DOI] [PubMed] [Google Scholar]
- 2.Weeda VB, et al. Fibrolamellar variant of hepatocellular carcinoma does not have a better survival than conventional hepatocellular carcinoma–results and treatment recommendations from the Childhood Liver Tumour Strategy Group (SIOPEL) experience. Eur. J. cancer. 2013;49:2698–2704. doi: 10.1016/j.ejca.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 3.Katzenstein HM, et al. Fibrolamellar hepatocellular carcinoma in children and adolescents. Cancer. 2003;97:2006–2012. doi: 10.1002/cncr.11292. [DOI] [PubMed] [Google Scholar]
- 4.Mavros MN, Mayo SC, Hyder O, Pawlik TM. A systematic review: treatment and prognosis of patients with fibrolamellar hepatocellular carcinoma. J. Am. Coll. Surg. 2012;215:820–830. doi: 10.1016/j.jamcollsurg.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 5.El-Serag HB, Davila JA. Is fibrolamellar carcinoma different from hepatocellular carcinoma? A US population-based study. Hepatol. (Baltim., Md.) 2004;39:798–803. doi: 10.1002/hep.20096. [DOI] [PubMed] [Google Scholar]
- 6.Simon EP, et al. Transcriptomic characterization of fibrolamellar hepatocellular carcinoma. Proc. Natl Acad. Sci. USA. 2015;112:E5916–5925. doi: 10.1073/pnas.1424894112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honeyman JN, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science. 2014;343:1010–1014. doi: 10.1126/science.1249484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham RP, et al. DNAJB1-PRKACA is specific for fibrolamellar carcinoma. Mod. Pathol. 2015;28:822–829. doi: 10.1038/modpathol.2015.4. [DOI] [PubMed] [Google Scholar]
- 9.Xu L, et al. Genomic analysis of fibrolamellar hepatocellular carcinoma. Hum. Mol. Genet. 2014;24:50–63. doi: 10.1093/hmg/ddu418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan M, Taylor SS. Dynamics of cAMP-dependent protein kinase. Chem. Rev. 2001;101:2243–2270. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- 11.Jedrzejewski PT, et al. A conserved deamidation site at asn 2 in the catalytic subunit of mammalian cAMP-dependent protein kinase detected by capillary LC-MS and tandem mass spectrometry. Protein Sci. 1998;7:457–469. doi: 10.1002/pro.5560070227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung J, et al. Structural insights into mis-regulation of protein kinase A in human tumors. Proc. Natl Acad. Sci. USA. 2015;112:1374–1379. doi: 10.1073/pnas.1424206112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riggle KM, et al. Enhanced cAMP-stimulated protein kinase A activity in human fibrolamellar hepatocellular carcinoma. Pediatr. Res. 2016;80:110–118. doi: 10.1038/pr.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao B, et al. Structures of the PKA RIα holoenzyme with the FLHCC Driver J-PKAcα or Wild-Type PKAcα. Structure. 2019;27:816–828. doi: 10.1016/j.str.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Averill AM, et al. Inhibition of the chimeric DnaJ-PKAc enzyme by endogenous inhibitor proteins. J. Cell Biochem. 2019;120:13783–13791. doi: 10.1002/jcb.28651. [DOI] [PubMed] [Google Scholar]
- 16.Turnham RE, et al. An acquired scaffolding function of the DNAJ-PKAc fusion contributes to oncogenic signaling in fibrolamellar carcinoma. eLife. 2019;8:e44187. doi: 10.7554/eLife.44187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kastenhuber ER, et al. DNAJB1–PRKACA fusion kinase interacts with β-catenin and the liver regenerative response to drive fibrolamellar hepatocellular carcinoma. Proc. Natl Acad. Sci. 2017;114:13076–13084. doi: 10.1073/pnas.1716483114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Globally correlated conformational entropy underlies positive and negative cooperativity in a kinase’s enzymatic cycle. Nat. Commun. 2019;10:799. doi: 10.1038/s41467-019-08655-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williamson JR. Cooperativity in macromolecular assembly. Nat. Chem. Biol. 2008;4:458–465. doi: 10.1038/nchembio.102. [DOI] [PubMed] [Google Scholar]
- 20.Walker C, et al. Cushing’s syndrome driver mutation disrupts protein kinase A allosteric network, altering both regulation and substrate specificity. Sci. Adv. 2019;5:eaaw9298. doi: 10.1126/sciadv.aaw9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knighton DR, et al. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 22.Tomasini MD, et al. Conformational landscape of the PRKACA-DNAJB1 chimeric kinase, the driver for fibrolamellar hepatocellular carcinoma. Sci. Rep. 2018;8:720. doi: 10.1038/s41598-017-18956-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masterson LR, Mascioni A, Traaseth NJ, Taylor SS, Veglia G. Allosteric cooperativity in protein kinase A. Proc. Natl Acad. Sci. USA. 2008;105:506–511. doi: 10.1073/pnas.0709214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Li G, Walters MA, Taylor SS, Veglia G. Uncoupling catalytic and binding functions in the cyclic AMP-dependent protein kinase A. Structure. 2016;24:353–363. doi: 10.1016/j.str.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by nitrogen-15 inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 26.Stark JL, et al. Structure and function of human DnaJ homologue subfamily a member 1 (DNAJA1) and its relationship to pancreatic cancer. Biochemistry. 2014;53:1360–1372. doi: 10.1021/bi401329a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava AtulK, et al. Synchronous opening and closing motions are essential for cAMP-dependent protein kinase A signaling. Structure. 2014;22:1735–1743. doi: 10.1016/j.str.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selvaratnam R, Chowdhury S, VanSchouwen B, Melacini G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proc. Natl Acad. Sci. USA. 2011;108:6133–6138. doi: 10.1073/pnas.1017311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boulton S, Akimoto M, Selvaratnam R, Bashiri A, Melacini G. A tool set to map allosteric networks through the NMR chemical shift covariance analysis. Sci. Rep. 2014;4:7306. doi: 10.1038/srep07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boulton S, Selvaratnam R, Ahmed R, Melacini G. Implementation of the NMR CHEmical Shift Covariance Analysis (CHESCA): a chemical biologist’s approach to allostery. Methods Mol. Biol. 2018;1688:391–405. doi: 10.1007/978-1-4939-7386-6_18. [DOI] [PubMed] [Google Scholar]
- 31.Ahuja LG, Kornev AP, McClendon CL, Veglia G, Taylor SS. Mutation of a kinase allosteric node uncouples dynamics linked to phosphotransfer. Proc. Natl Acad. Sci. 2017;114:E931–E940. doi: 10.1073/pnas.1620667114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClendon CL, Kornev AP, Gilson MK, Taylor SS. Dynamic architecture of a protein kinase. Proc. Natl Acad. Sci. USA. 2014;111:E4623–4631. doi: 10.1073/pnas.1418402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cembran A, Kim J, Gao J, Veglia G. NMR mapping of protein conformational landscapes using coordinated behavior of chemical shifts upon ligand binding. Phys. Chem. Chem. Phys. 2014;16:6508–6518. doi: 10.1039/c4cp00110a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, J. et al. Dysfunctional conformational dynamics of protein kinase A induced by a lethal mutant of phospholamban hinder phosphorylation. Proc. Natl Acad. Sci.112, 3716–3721 (2015). [DOI] [PMC free article] [PubMed]
- 35.Vyas, M. et al. DNAJB1-PRKACA fusions occur in oncocytic pancreatic and biliary neoplasms and are not specific for fibrolamellar hepatocellular carcinoma. Mod. Pathol.10.1038/s41379-019-0398-2 (2019). [DOI] [PMC free article] [PubMed]
- 36.Singhi AD, et al. Recurrent rearrangements in PRKACA and PRKACB in intraductal oncocytic papillary neoplasms of the pancreas and bile duct. Gastroenterology. 2020;158:573–582. doi: 10.1053/j.gastro.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riggle, K. M., Turnham, R., Scott, J. D., Yeung, R. S. & Riehle, K. J. Fibrolamellar hepatocellular carcinoma: mechanistic distinction from adult hepatocellular carcinoma. Pediatr. Blood Cancer10.1002/pbc.25970 (2016). [DOI] [PMC free article] [PubMed]
- 38.Graham RP, et al. Fibrolamellar carcinoma in the Carney complex: PRKAR1A loss instead of the classic DNAJB1-PRKACA fusion. Hepatology. 2018;68:1441–1447. doi: 10.1002/hep.29719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamilaris CDC, Faucz FR, Voutetakis A, Stratakis CA. Carney complex. Exp. Clin. Endocrinol. Diabetes. 2019;127:156–164. doi: 10.1055/a-0753-4943. [DOI] [PubMed] [Google Scholar]
- 40.Zhang P, et al. An isoform-specific myristylation switch targets type II PKA holoenzymes to membranes. Structure. 2015;23:1563–1572. doi: 10.1016/j.str.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tillo SE, et al. Liberated PKA catalytic subunits associate with the membrane via myristoylation to preferentially phosphorylate membrane substrates. Cell Rep. 2017;19:617–629. doi: 10.1016/j.celrep.2017.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitty A. Cooperativity and biological complexity. Nat. Chem. Biol. 2008;4:435–439. doi: 10.1038/nchembio0808-435. [DOI] [PubMed] [Google Scholar]
- 43.Wiseman T, Williston S, Brandts JF, Lin L-N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 44.Cook PF, Neville ME, Jr., Vrana KE, Hartl FT, Roskoski R., Jr. Adenosine cyclic 3’,5’-monophosphate dependent protein kinase: kinetic mechanism for the bovine skeletal muscle catalytic subunit. Biochemistry. 1982;21:5794–5799. doi: 10.1021/bi00266a011. [DOI] [PubMed] [Google Scholar]
- 45.Shi L, Kay LE. Tracing an allosteric pathway regulating the activity of the HslV protease. Proc. Natl Acad. Sci. 2014;111:2140–2145. doi: 10.1073/pnas.1318476111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, et al. Mapping allosteric communications within individual proteins. Nat. Commun. 2020;11:3862. doi: 10.1038/s41467-020-17618-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tong M, et al. Survey of solution dynamics in Src kinase reveals allosteric cross talk between the ligand binding and regulatory sites. Nat. Commun. 2017;8:2160. doi: 10.1038/s41467-017-02240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie T, Saleh T, Rossi P, Kalodimos CG. Conformational states dynamically populated by a kinase determine its function. Science. 2020;370:eabc2754. doi: 10.1126/science.abc2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu S, et al. The mechanism of ATP-dependent allosteric protection of Akt kinase phosphorylation. Structure. 2015;23:1725–1734. doi: 10.1016/j.str.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ni D, Song K, Zhang J, Lu S. Molecular dynamics simulations and dynamic network analysis reveal the allosteric unbinding of monobody to H-Ras triggered by R135K mutation. Int. J. Mol. Sci. 2017;18:2249. doi: 10.3390/ijms18112249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rivalta I, et al. Allosteric communication disrupted by a small molecule binding to the imidazole glycerol phosphate synthase protein–protein interface. Biochemistry. 2016;55:6484–6494. doi: 10.1021/acs.biochem.6b00859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marsiglia WM, et al. A conserved allosteric pathway in tyrosine kinase regulation. Structure. 2019;27:1308–1315. doi: 10.1016/j.str.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shibata T, Arai Y, Totoki Y. Molecular genomic landscapes of hepatobiliary cancer. Cancer Sci. 2018;109:1282–1291. doi: 10.1111/cas.13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 56.Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanks S, Quinn A, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 58.Zheng J, et al. 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr. Sect. D. 1993;49:362–365. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 59.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 60.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 62.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 63.Darden T, York D, Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089. [Google Scholar]
- 64.Parrinello M, Rahman A. Crystal structure and pair potentials: a molecular-dynamics study. Phys. Rev. Lett. 1980;45:1196–1199. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary files
Data Availability Statement
Raw data from steady-state activity assays and corresponding analysis is deposited on the Data Repository for the University of Minnesota (DRUM) and can be found at the following link: http://hdl.handle.net/11299/217205. Proton and amide chemical shift list files of wild-type PKA-C and the chimeric mutant (PKADNAJB1) are deposited on DRUM and can be found at the following link: http://hdl.handle.net/11299/217206. The source data for the graphs in the main figures are provided in Supplementary Data 1. All other data sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
MATLAB scripts used for the chemical shift analyses presented are deposited on DRUM and can be found at the following link: http://hdl.handle.net/11299/217206.