Abstract
Mammalian pregnancy evolved in the therian stem lineage, that is, before the common ancestor of marsupials and eutherian (placental) mammals. Ancestral therian pregnancy likely involved a brief phase of attachment between the fetal and maternal tissues followed by parturition—similar to the situation in most marsupials including the opossum. In all eutherians, however, embryo attachment is followed by implantation, allowing for a stable fetal–maternal interface and an extended gestation. Embryo attachment induces an attachment reaction in the uterus that is homologous to an inflammatory response. Here, we elucidate the evolutionary mechanism by which the ancestral inflammatory response was transformed into embryo implantation in the eutherian lineage. We performed a comparative uterine transcriptomic and immunohistochemical study of three eutherians, armadillo (Dasypus novemcinctus), hyrax (Procavia capensis), and rabbit (Oryctolagus cuniculus); and one marsupial, opossum (Monodelphis domestica). Our results suggest that in the eutherian lineage, the ancestral inflammatory response was domesticated by suppressing one of its modules detrimental to pregnancy, namely, neutrophil recruitment by cytokine IL17A. Further, we propose that this suppression was mediated by decidual stromal cells, a novel cell type in eutherian mammals. We tested a prediction of this model in vitro and showed that decidual stromal cells can suppress the production of IL17A from helper T cells. Together, these results provide a mechanistic understanding of early stages in the evolution of eutherian pregnancy.
Keywords: pregnancy, mucosal inflammation, interleukin-17, placental, marsupial, homology
Introduction
Embryo implantation is the process by which the blastocyst and the mother establish a sustained fetal–maternal interface for the maintenance of pregnancy. It begins with apposition of the blastocyst to the receptive endometrial luminal epithelium, followed by attachment via molecular interactions between the uterine epithelium and the trophoblast of the embryo, and in many eutherian (also known as placental mammals) species, embedding into the endometrium to establish a direct contact with the endometrial connective tissue and vasculature (Mossman 1937; Enders and Schlafke 1969; Schlafke and Enders 1975; Ashary et al. 2018).
Mammalian viviparity originated before the common ancestor of eutherian mammals and marsupials, that is, in the stem lineage of therian mammals. However, marsupial and eutherian pregnancies are different in many fundamental ways, including embryo implantation and the establishment of the fetal–maternal interface.
Marsupial pregnancy is very short—in most cases shorter than the luteal phase of the ovarian cycle (Renfree 1994; McAllan 2011). For most of the duration of marsupial pregnancy, the embryo is present inside of an eggshell (Selwood 2000; Griffith et al. 2017) that precludes a direct physical contact between the fetal and maternal tissues. Toward the end of the pregnancy, the eggshell breaks down and the fetal membranes attach to the uterine luminal epithelium. Despite some invasion of the luminal epithelium by the trophoblast (reviewed by Carter [2020]), the phase of attachment lasts a small fraction of the length of gestation and is soon followed by the birth of highly altricial neonates. For instance, pregnancy in the South American marsupial, the gray short-tailed opossum (Monodelphis domestica), lasts 14.5 days. Embryo attachment begins approximately on the 12th day postcopulation (dpc) and induces an inflammatory response in the uterus (Griffith et al. 2017). This inflammation presumably mediates parturition after embryo attachment (Chavan et al. 2017; Hansen et al. 2017; Stadtmauer and Wagner 2020).
Eutherian pregnancy is often much longer than the luteal phase of the ovarian cycle (Chavan et al. 2016) as a result of embryo attachment being followed by the establishment of a stable fetal–maternal interface that can sustain prolonged pregnancy. This is a key difference between marsupial and eutherian embryo attachment, the latter of which we consistently refer to as eutherian implantation in this article. Puzzlingly, in many eutherian mammals such as human, mouse, pig, and sheep, embryo implantation also shows signs of an inflammatory reaction in the uterus; some of these inflammatory processes are in fact necessary and beneficial for a successful implantation (Keys et al. 1986; Barash et al. 2003; Waclawik and Ziecik 2007; Plaks et al. 2008; Mor et al. 2011; Dekel et al. 2014; Robertson and Moldenhauer 2014; Chavan et al. 2017; Whyte et al. 2018). Resemblance of the physiological process of eutherian implantation to an inflammatory reaction appears paradoxical at first because inflammation in later stages of pregnancy leads to the termination of pregnancy. However, analysis of the evolutionary history of embryo implantation suggests that this resemblance is due to the evolutionary roots of implantation in an inflammatory response to embryo attachment (Finn 1986; Griffith et al. 2017). Griffith et al. (2017, 2018) argued, based on a comparison of embryo attachment in opossum to implantation in eutherian mammals, that eutherian implantation and the inflammatory marsupial attachment reaction are homologous processes. That is, these processes evolved from an inflammatory fetal–maternal attachment reaction that likely existed in the therian stem lineage.
The important difference between the fetal–maternal attachment in marsupials and eutherian implantation is thus in the outcome of the associated inflammatory process. In the opossum, the brief inflammatory attachment results in parturition, whereas in eutherians it results in the establishment of a sustained fetal–maternal interface.
Here, we elucidate the mechanism by which the ancestral attachment-induced inflammatory response was transformed into the process of embryo implantation in the eutherian lineage. We show that the origin of decidual stromal cells (DSCs)—a novel eutherian cell type—likely played a role in this transformation. First, we provide evidence to further support the homology between opossum attachment reaction and eutherian embryo implantation. Then, to explain the differences in the outcomes, we compare gene expression in the uterus of opossum during attachment to that in two eutherians during implantation: armadillo and rabbit. The key differences in gene expression suggest that embryo implantation evolved through suppression of a specific module of the ancestral mucosal inflammatory reaction—neutrophil recruitment mediated by the proinflammatory cytokine IL17A. We hypothesized that the origin of DSC, which occurred coincidentally with embryo implantation in the stem eutherian lineage, was responsible for the suppression of IL17A in members of this clade. We experimentally tested a prediction of this model using human cells, which showed that secretions from DSC can indeed inhibit the production of IL17A by TH17 cells (a subtype of helper T cells that is roughly defined as CD4+ and IL17A+), one of the primary producers of IL17A.
Results and Discussion
Inflammatory Implantation Is an Ancestral Eutherian Trait
The inference of homology between the opossum attachment reaction and eutherian embryo implantation is based on the comparison of opossum to species from Boreoeutheria (Griffith et al. 2017). Boreoeutheria is one of the three major clades that make up Eutheria and includes primates, rodents, ungulates, carnivores, bats, and their kin. However, Eutheria also contains two other major clades, Xenarthra and Afrotheria (dos Reis et al. 2012; Tarver et al. 2016), for which data on inflammatory gene expression at implantation were previously unavailable. Xenarthra includes armadillo, sloth, anteater, and others; and Afrotheria includes elephant, hyrax, tenrec, aardvark, dugong, and others. (See fig. 1D for the phylogenetic relationship among eutherian species.) To test the inference of homology, we investigated the hypothesis that inflammatory implantation is a shared eutherian character; that is, it is not limited to Boreoeutheria but is also observed in Xenarthra and Afrotheria. We did this by assessing the expression of marker genes of inflammation during embryo implantation in the nine-banded armadillo (Dasypus novemcinctus) and rock hyrax (Procavia capensis), as representatives of Xenarthra and Afrotheria, respectively.
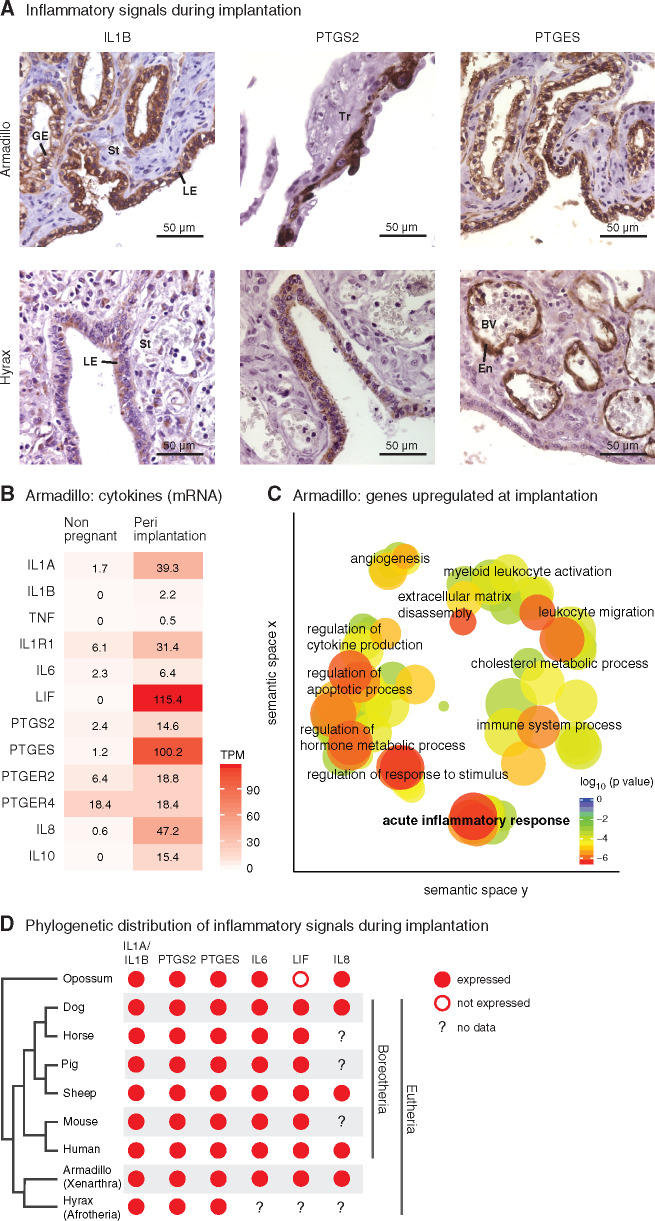
Fig. 1.
Inflammatory implantation is an ancestral eutherian character. (A) Immunohistochemistry for inflammation marker genes IL1B, PTGS2, and PTGES at the fetal–maternal interface in armadillo (peri-implantation stage) and hyrax (peri-implantation stage). Nuclei are blue due to hematoxylin counterstaining and the immunostaining signal is brown due to 3,3′-diaminobenzidine (DAB). GE, glandular epithelium; LE, luminal epithelium; St, stroma; Tr, trophoblast; BV, blood vessel; En, endothelium. (B) Abundance of mRNA transcripts (TPM = Transcripts per Million) of key inflammatory genes in armadillo uterus in nonpregnant and peri-implantation stage. Armadillo has two orthologs of IL6, ENSDNOG00000017107 and ENSDNOG00000023693, of which the former is shown here; the latter is not expressed. (C) Enriched GO categories among the genes that are upregulated at least 10-fold in armadillo uterus in transition from nonpregnant to peri-implantation stage. GO categories are clustered by semantic similarity. GO categories closer to each other are semantically similar; those represented by red circles are more significantly enriched than those in blue. (D) Comparison of expression of inflammatory genes during embryo attachment or implantation in therian mammals. Data for human: cytokines (reviewed by Van Sinderen et al. [2013]), PTGS2 (Marions and Danielsson 1999), and PTGES (Milne et al. 2001). Data for sheep, horse, pig, dog, and mouse (reviewed by Chavan et al. [2017]).
One of the earliest signals of inflammation, IL1B, is expressed during implantation in the luminal epithelium of the endometrium in armadillo and hyrax (fig. 1A). PTGS2 and PTGES, enzymes involved in the synthesis of prostaglandin E2 (PGE2), are also expressed during implantation in both species, although the tissues in which these genes are expressed differ between species. PTGS2 and PTGES are expressed on the two sides of the fetal–maternal interface in armadillo—PTGS2 in the trophoblast and PTGES in the endometrial luminal epithelium. In hyrax, they are both expressed on the maternal side, with PTGS2 in the luminal epithelium and PTGES in the endothelia within the endometrium. The expression of these two enzymes in different compartments of the fetal–maternal interface is also found during attachment in the opossum, where PTGS2 is expressed in the luminal epithelium and PTGES in the trophoblast (Griffith et al. 2017).
Next, we used transcriptomic data to test whether there is a broad signature of inflammation during implantation in armadillo uterus. A variety of inflammatory genes are upregulated during armadillo implantation: cytokines such as IL1A, IL1B, IL6, LIF, IL8 (CXCL8), and IL10; cytokine receptor IL1R1; prostaglandin synthesis enzymes PTGS2 and PTGES; and prostaglandin receptors PTGER2 and PTGER4 (fig. 1B). None of the cytokines shown in figure 1B is expressed in the nongravid uterus, that is, their mRNA abundance is below the operational threshold of 3 Transcripts per Million (TPM; Wagner et al. 2013), but most of them are expressed highly during implantation. Genes upregulated more than 10-fold in the transition from nonpregnant to implantation stage are significantly enriched in gene ontology (GO) categories related to inflammation (fig. 1C), such as acute inflammatory process, immune system process, leukocyte migration, regulation of cytokine production, and regulation of response to stimulus.
We have summarized the above-noted expression pattern of inflammation marker genes during implantation on a phylogeny of therian mammals, along with the expression patterns from other representative eutherian species, and from a marsupial, opossum, at the time of attachment between fetal and maternal tissues at 13.5 dpc (fig. 1D), but not during a sterile estrus cycle of the opossum, supporting the inference that the inflammatory reaction is induced by the embryo (Griffith et al. 2019). In all major clades of Eutheria, the uterine changes during embryo implantation closely resemble an acute inflammatory reaction. Parsimoniously, this suggests that embryo attachment-induced uterine inflammatory signaling is a plesiomorphic trait of eutherian mammals, that is, a trait that was inherited from an ancestral lineage and shared with the sister group, the marsupials, adding further support to the argument that attachment-induced inflammation of marsupials is homologous to eutherian embryo implantation (Griffith et al. 2017, 2018; Liu 2018). In other words, eutherian implantation likely evolved from, and through modification of, ancestral attachment-induced mucosal inflammation.
Differences in Uterine Gene Expression between Opossum and Eutherians
To understand how eutherian implantation evolved from the ancestral therian attachment-induced inflammation, we compared gene expression in the uterus (and the attached fetal membranes) during attachment-induced inflammation in opossum (M. domestica) to that during implantation in two eutherian mammals, armadillo (D. novemcinctus) and rabbit (Oryctolagus cuniculus) (7.25 dpc, i.e., implantation stage, rabbit uterine transcriptome data from Liu et al. [2016]). Gene expression patterns shared by armadillo and rabbit are likely to be shared by eutherians in general given that armadillo and rabbit are phylogenetically positioned so that their common ancestor is the common ancestor of all extant eutherian mammals.
In the uterine transcriptomes of opossum, armadillo, and rabbit, we classified each gene as either expressed or not expressed (see Materials and Methods). We then classified these genes as “opossum-specific” if they are expressed in opossum but not expressed in armadillo and rabbit, and “Eutheria-specific” if they are not expressed in opossum but are expressed in both armadillo and rabbit. There are 446 and 456 genes in these categories, respectively, among the total of 11,089 genes that have one-to-one orthologs in all three species.
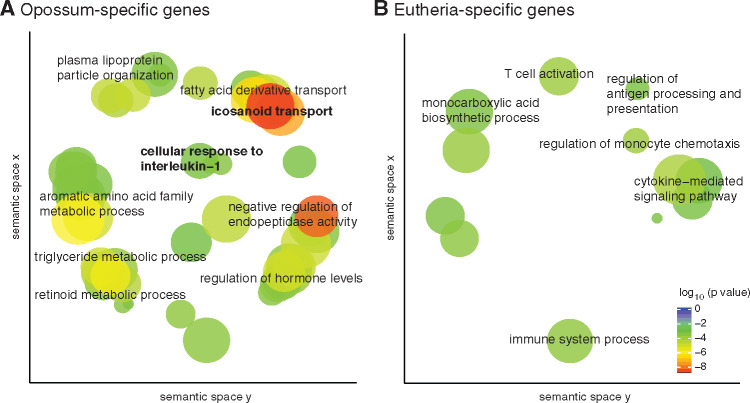
First, we identified enriched GO categories in the opossum-specific and Eutheria-specific lists of genes (fig. 2). To increase the specificity of GO enrichment analysis, we used only the subset of the opossum-specific expressed genes that have an at least 2-fold higher gene expression in the attachment phase compared with the nonpregnant stage (204 genes). Such refinement of the Eutheria-specific gene set was not possible since we did not have a transcriptome of nonpregnant rabbit uterus. The opossum-specific set is enriched for genes related to lipid metabolism, especially mobilization of fatty acids from cell membrane, and lipid transport. This is consistent with genes overrepresented in the opossum uterus during pregnancy compared with the equivalent time in the estrous cycle (Griffith et al. 2019). Lipid metabolism genes are also upregulated in the pregnant uterus of another marsupial, fat-tailed dunnart (Sminthopsis crassicaudata) (Whittington et al. 2018). These genes may have functions related to nutrient transfer or steroid metabolism, but lipid metabolism is also important in inflammation: for example, the first step in the synthesis of prostaglandins is to break down membrane triglycerides. The opossum-specific set is also enriched for GO category “cellular response to interleukin-1,” that is, genes downstream of IL1A and IL1B. This suggests that although inflammatory signaling is initiated by IL1A and/or IL1B upon embryo attachment in opossum as well as in eutherians, their downstream targets are preferentially activated in the opossum. This observation recapitulates at the molecular level a phenomenon at the organismal level—inflammatory signaling is observed in both but has different outcomes of parturition and implantation in opossum and eutherians, respectively.
Fig. 2.
Transcriptomic differences between marsupial and eutherian attachment reaction. GO categories enriched in each set are shown in the figures, where semantically similar categories are clustered together in space. Genes were classified as (A) opossum-specific (B) eutherian-specific.
IL17A Expression Was Suppressed in Eutheria
Differences in Cytokine Expression between Opossum and Eutherians
IL1A and IL1B set off a cascade of inflammatory signaling events mediated by cytokine molecules. Therefore, in order to identify the specific differences between opossum and eutherians in response to interleukin-1, we looked for differences in the expression pattern of cytokines (fig. 3A). Cytokines were identified as genes assigned to GO category “cytokine activity” (GO:0005125).
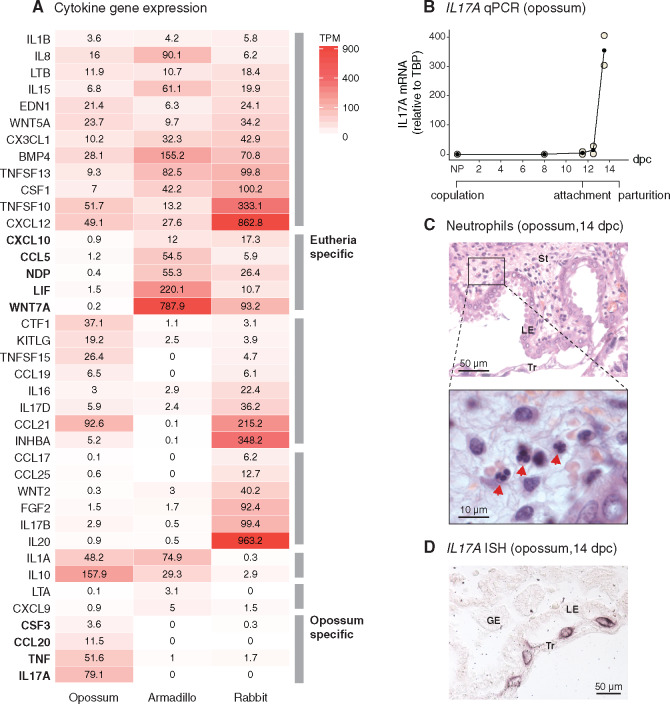
Fig. 3.
IL17A signaling is suppressed in eutherians. (A) Gene expression of cytokines in opossum (attachment stage, 13.5 dpc), armadillo (peri-implantation stage), and rabbit (implantation stage, 7.25 dpc) uterus. Intensity of red is proportional to the abundance of mRNA. Genes expressed below 3 TPM are considered unexpressed (Wagner et al. 2013). Rows are ordered by the gene expression patterns across species: expressed in all species; expressed only in Eutheria; expressed in opossum and rabbit, rabbit-specific; and expressed in opossum and armadillo, armadillo-specific, and opossum-specific. Cytokines not expressed in any of these species are not shown in the figure. (B) Expression of IL17A through the pregnancy of opossum, measured relative to TBP, by qPCR. Embryo attachment begins around 11.5 dpc, and pregnancy ends at 14.5 dpc (number of biological replicates: NP = 3, 8 dpc = 3, 11.5 dpc = 2, 12.5 dpc = 2, and 13.5 dpc = 2). (C) Neutrophil infiltration in H&E stained opossum uterus at 14 dpc, indicated by red arrows in the zoomed-in micrograph. (D) In situ hybridization for IL17A in opossum at 14 dpc. IL17A staining is detected in the trophoblast cells. LE, luminal epithelium; GE, glandular epithelium; St, stroma; Tr, trophoblast; TPM, Transcripts per Million.
Cytokines expressed in Eutheria-specific manner are CXCL10, CCL5, NDP, WNT7A, and LIF. The first two, CXCL10 and CCL5, attract leukocytes such as T cells, NK cells, and dendritic cells to the sites of inflammation (Schall 1991; Dufour et al. 2002). NDP and WNT7A are both members of the Wnt signaling pathway, which is important for communication between implanting blastocyst and endometrium (Wang and Dey 2006; Chen et al. 2009; Sonderegger et al. 2010); inhibition of this process prevents successful implantation in mouse (Mohamed et al. 2005). LIF is a critical signaling molecule in eutherian mammals for the differentiation of DSCs from endometrial stromal fibroblasts (ESFs), and its expression is, therefore, obligatory for successful implantation (Shuya et al. 2011; Filant and Spencer 2013; Kelleher et al. 2018). This set of genes represents cytokines and signaling molecules that were likely recruited within the eutherian lineage to enable decidual cell differentiation, embryo–uterine crosstalk, regulation of leukocyte traffic, and therefore implantation.
The inverse set, opossum-specific cytokines, includes CSF3, CCL20, TNF, and IL17A. TNF is a proinflammatory cytokine, whose elevated levels in the first trimester of human pregnancy are associated with pregnancy loss (Kaislasuo et al. 2020). CSF3, also known as Granulocyte Colony Stimulating Factor (G-CSF) attracts granulocytes (Lieschke et al. 1994; Panopoulos and Watowich 2008), such as neutrophils, and is positively regulated by IL17A (Ye et al. 2001; Onishi and Gaffen 2010). CCL20 attracts lymphocytes as well as neutrophils and also is a downstream gene of IL17A (Onishi and Gaffen 2010). This pattern is indicative of a mucosal inflammatory reaction, culminating in recruitment of effector cells such as neutrophils.
IL17A Induction and Signaling Coincide with Attachment in Opossum
Since, according to evidence in mouse and human, IL17A is known to regulate multiple inflammatory genes (Onishi and Gaffen 2010), found here to be specific to the opossum attachment, we then measured the expression of IL17A in opossum uterus and attached fetal membranes by quantitative polymerase chain reaction (qPCR) to test whether it is induced in response to the attaching embryo or expressed throughout pregnancy. Figure 3B shows that IL17A is not expressed in nonpregnant and 8 dpc uteri and only begins expression at 11.5 dpc, coincidental with the eggshell breakdown and beginning of fetal–maternal attachment. Its expression reaches a high level by 13.5 dpc; 79 TPM according to the transcriptomic data. The time-course expression data suggest that IL17A is induced specifically in response to embryo attachment in opossum, even though it is not detected in armadillo and rabbit during implantation (0 TPM in both species). Also, in human, IL17A expression is not seen at the fetal–maternal interface during implantation, neither in decidua nor in the villi (see supplementary fig. 1, Supplementary Material online, generated using single cell RNA-sequencing data from Suryawanshi et al. [2018]). One of the hallmarks of IL17A-mediated inflammation—through the action of cytokines like CXCL8, CSF3, and CCL20—is neutrophil infiltration into the inflamed tissue (Medzhitov 2007; Onishi and Gaffen 2010; Griffin et al. 2012; Pappu et al. 2012; Flannigan et al. 2017). Therefore, we tested whether IL17A expression in opossum is also accompanied by neutrophil infiltration. Neutrophils are not seen in early stages of opossum pregnancy, but at 14 dpc, neutrophil infiltration is clearly seen histologically (fig. 3C). Consistent with the pattern of IL17A expression, neutrophils are absent from the fetal–maternal interface at the time of implantation in eutherian mammals (evidence reviewed by Chavan et al. [2017]).
The molecular steps that lead to IL17A expression following the loss of the eggshell are unclear. The inflammatory cascade is likely initiated due to the tissue damage caused by the proteases helping dissolve the eggshell (Griffith et al. 2017), leading first to the induction of cytokines such as IL6 and TGFB1, which together are known to be essential for IL17A induction in mouse and human (Korn et al. 2009). IL6 and TGFB1 are indeed upregulated at late-gestation in opossum: IL6 expression increases from 0.46 TPM at 8 dpc to 178 TPM at 13.5 dpc, and TGFB1 from 13.9 TPM to 40.9 TPM (these cytokines are not shown in fig. 3A because they do not have 1:1 orthologs in rabbit and armadillo; see Materials and Methods).
Contrary to the data presented above for armadillo, rabbit, and healthy human pregnancy, some IL17A expression has been reported at the fetal–maternal interface in mouse and human, specifically in the γδ T cells in the mouse (Pinget et al. 2016). However, functional evidence suggests that even in mouse and human, an induction of IL17A expression at the fetal–maternal interface is detrimental to pregnancy. The strongest evidence perhaps comes from mouse, where injection of polyI:C to mimic viral infection during pregnancy induces the expression of IL17A in the decidua. Maternal IL17A, then, makes its way into the fetal circulation, interferes with brain development of the embryos, and leads to behavioral abnormalities resembling autism spectrum disorder in pups (Choi et al. 2016). Li et al. (2018) showed that in a mouse model of spontaneous abortion, TH17 cell numbers are higher in the decidua relative to normal pregnancy. In human, TH17 levels in peripheral blood are elevated in women with recurrent spontaneous abortions compared with women with healthy pregnancy (Wang et al. 2010). Nakashima et al. (2010) found that although IL17A-positive cells were only occasionally present in the deciduae of normal pregnancies, their numbers were significantly elevated in pregnancies with first trimester spontaneous abortions. These studies clearly indicate that IL17A signaling at the fetal–maternal interface is not conducive to successful pregnancy, and the cases in human and mouse where IL17A expression is observed, the downstream signaling is likely inhibited in some way.
Divergent Fates of IL17A Expression in Marsupial and Eutherian Lineages
The absence of IL17A expression in eutherian mammals at implantation is likely a result of its loss in the eutherian lineage rather than its recruitment in the marsupial lineage. The discovery of IL17 homologs as the early-responding cytokines in the sea urchin larva during gut infection (Buckley et al. 2017) suggests that it is an ancient mucosal inflammatory cytokine at least as old as deuterostomes. Since the endometrium is a mucosal tissue, IL17A—a key mucosal inflammatory cytokine—is likely to have been expressed in the ancestral therian endometrium during attachment-induced inflammation. Its expression was likely lost later during evolution in the eutherian lineage, whereas it was retained in the marsupial lineage.
In situ hybridization for IL17A mRNA on frozen sections of the fetal–maternal interface at 14 dpc shows that the cell type that produces IL17A in the opossum is the trophoblast (fig. 3D). For a generic maternal mucosal inflammatory response, IL17A expression is expected to be in the maternal tissue, as was likely the case in the therian ancestor. However, a shift in the cellular source of IL17A from maternal tissue to the fetal membranes during marsupial evolution is consistent with the “cooperative inflammation model” of marsupial pregnancy (Stadtmauer and Wagner 2020), which posits that the processes similar to inflammation in the opossum uterus likely benefit both the fetus and the mother. The exact biological role of this process has not been determined.
Although the opossum retains some features of the ancestral therian pregnancy, it is important to note that it has undergone ∼170 My of independent evolution during which the ancestral inflammatory response has likely been modified, just not in the same way as in the eutherian lineage. The cooperative inflammation model is based, in part, on the observation that during the brief attachment phase of the marsupial pregnancy, the growth rate of the fetus is notably accelerated (Rose 1989 and references therein), pointing to the beneficial effects of inflammatory process (e.g., increased blood flow and vascular permeability) on the efficiency of nutrient transfer. This suggests that the inflammatory response in the eutherian lineage was dampened in early pregnancy to support extended gestation, whereas in the marsupial lineage, the accidental benefits of the brief inflammatory period to fetal growth were integrated into the physiology of pregnancy. During the evolutionary transformation of an induced reaction into a physiological process, akin to genetic assimilation, the cell–cell network involved in the response is expected to be rewired. For IL17A signaling, this would imply the cellular source of IL17A switching from an unknown maternal cell type (ancestral therian) to the trophoblast (opossum) such that the fetal tissue becomes an active participant in the coordinated and cooperative physiological process. A similar rewiring is also observed in the case of prostaglandin E2 synthesis enzymes (Griffith et al. 2017). Such redistribution of cellular signals can occur by the process of “developmental systems drift” (True and Haag 2001) and may have contributed to the evolution of a physiological means of endometrial recognition of pregnancy that the maternal organism can distinguish from generic inflammation (Griffith et al. 2019).
The above-noted arguments support the model in which, ancestrally, IL17A was maternally expressed. The alternative model remains plausible, however, in which IL17A expression was trophoblastic in the therian stem lineage and was lost in the eutherian lineage from the trophoblast. A formal assessment of which of these models is more likely is difficult given that monotremes are oviparous. A phylogenetic test that does not rely on an outgroup can nevertheless be designed. On the one hand, if the switch from a maternal source to trophoblastic source indeed occurred by developmental systems drift in the marsupial lineage, we expect to see variation in the source of IL17A among different species across the marsupial clade. On the other hand, invariantly trophoblastic IL17A in the marsupial clade would support the alternative model. The lack of cell type-specific expression data in late-gestation, broadly sampled from across the marsupial lineage, however, leaves the ancestral therian cellular source of IL17A unresolved at this point.
Because IL17A is 1) the most highly expressed gene among opossum-specific cytokines; 2) an important regulator of mucosal inflammation; 3) known to regulate chemokines like CSF3; and 4) not expressed in armadillo, rabbit, and human at all, even in a leaky manner, we posited that the loss of IL17A expression—and consequently the loss of neutrophil infiltration—after embryo attachment was a key innovation that allowed the transformation of the ancestral inflammatory attachment reaction into embryo implantation.
DSCs Suppressed IL17A Expression at Implantation in Eutheria
DSCs are a novel cell type that originated in the stem lineage of eutherian mammals (Mess and Carter 2006; Wagner et al. 2014; Erkenbrack et al. 2018). They differentiate from ESFs in many eutherian mammals during pregnancy (Gellersen and Brosens 2014) and also during the menstrual cycle in primates, some bats, the elephant shrew (Emera et al. 2012), as well as—according to recent evidence—the spiny mouse (Bellofiore et al. 2017) in a process called decidualization. The evolution of DSC from ancestral therian ESF was associated with modulation of expression of genes involved in the innate immune response (Kin et al. 2016). DSCs perform many functions critical to the maintenance of pregnancy in human and mouse, for example, regulation of the traffic of leukocytes into the endometrium during pregnancy (Nancy et al. 2012; Erlebacher 2013), production of hormones like prolactin, regulation of invasiveness of the trophoblast (Gellersen and Brosens 2014), and regulation of the communication between cell types at the fetal–maternal interface (Pavlicev et al. 2017). Outside of euarchontoglirean mammals (primates, rodents, and their relatives), however, DSCs are not maintained throughout the pregnancy. In bats (Laurasiatheria), hyrax, tenrec (Afrotheria), and armadillo (Xenarthra), DSCs differentiate around the time of implantation but are often lost soon after implantation. This suggests that the ancestral function of DSCs when they originated was likely to have been limited to the time of implantation (Chavan et al. 2016).
Based on their inferred ancestral role at the time of implantation (Chavan et al. 2016), their ability to regulate the immune response during pregnancy (Erlebacher 2013), and their origin in the eutherian stem lineage (Mess and Carter 2006) coinciding with the evolution of suppression of IL17A expression (this study), we hypothesized that DSC played a role in the regulation of IL17A during implantation.
One prediction from the above-noted model is that DSC in extant eutherians should be able to regulate the expression of IL17A. We experimentally tested this using human cells in vitro. IL17A is typically produced by lymphocytes, such as TH17 cells, γδ T cells, and Innate Lymphoid Cells 3 (ILC3), and its expression in these lymphocyte types is driven by the transcription factor RORγt (Veldhoen 2017). We chose TH17 cells, which differentiate from naïve T cells upon exposure to IL6 and TGFB1 (Bettelli et al. 2006) (fig. 4A), as a model cell type for the experimental test. The choice of TH17 cells was pragmatic given that circulating levels of γδ T cells and ILC3 are much lower than naïve T cells, making the latter easier to isolate from blood samples and differentiate into TH17 cells in vitro. We isolated naïve T cells from human blood, differentiated them into TH17, and tested whether DSC can interfere with their expression of IL17A.
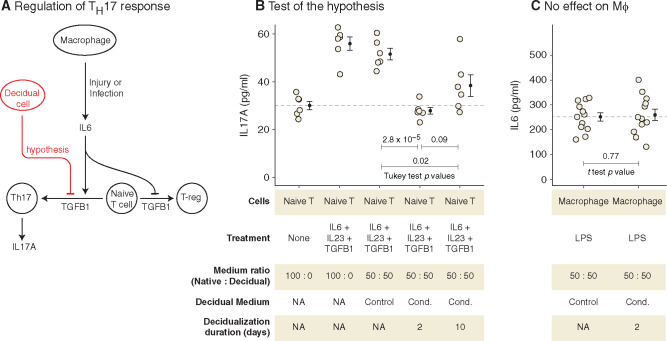
Fig. 4.
DSCs suppress IL17A production in TH17 cells. (A) Schematic showing how TH17 cells differentiate from naïve T cells, and the hypothesis for the role of DSC. (B) Test of the hypothesis using in vitro differentiation of human naïve T cells into TH17 cells. IL17A secretion (pg/ml) by TH17 cells is shown (mean and standard error of the mean). The first two samples are reference points, where TH17 cells were differentiated in their native medium, and the last three samples were differentiated in DSC control or conditioned medium. (See supplementary figure 2, Supplementary Material online, for mRNA levels of IL17A.) (C) DSC-conditioned medium has no effect on the activation of macrophages. IL6 secreted (pg/ml) by LPS-stimulated macrophages in the presence of control or DSC-conditioned medium is shown (mean and standard error of the mean). Cond, conditioned.
We differentiated primary human naïve T cells into TH17 in the presence of DSC-conditioned or control medium and measured IL17A secreted by T cells with enzyme-linked immunosorbent assay (ELISA). We collected DSC-conditioned medium from two time-points during decidualization, 2 and 10 days. Treatment of differentiating T cells with DSC-conditioned medium from both time-points decreased their IL17A production significantly (2 days: 0.54-fold, Tukey HSD test P = 2.8 × 10−5; 10 days: 0.74-fold, Tukey HSD test P = 0.02), whereas treatment with unconditioned DSC medium had no effect (0.92-fold, Tukey HSD test P = 0.80). The decreased level of IL17A due to DSC-conditioned medium from day 2 is not statistically distinguishable from that in unstimulated naïve T cells, suggesting that DSC-conditioned medium completely suppresses upregulation of IL17A production during TH17 differentiation (fig. 4B). A growing body of evidence suggests that decidualization is a biphasic process (Salker et al. 2012; Rytkönen et al. 2019). The early phase is proinflammatory, followed by a switch to the anti-inflammatory late phase that occurs after about 3–4 days of decidualization. The experiment, however, shows that even in the early proinflammatory phase of decidualization, at day 2, DSC-conditioned medium is able to suppress IL17A production, thus modifying a detrimental aspect of inflammatory reaction. This is consistent with the evolutionary history of DSC, as ancestrally in the eutherian lineage DSCs likely only existed around the time of implantation and were lost soon thereafter. That is, ancestrally they persisted only during the proinflammatory phase of pregnancy. If suppression of IL17A was indeed one of their ancestral functions, it must have evolved in the context of an otherwise proinflammatory milieu. Suppression of IL17A while simultaneously participating in a proinflammatory environment is thus likely what characterizes the early phase of decidualization, both in phylogeny and in ontogeny. The characteristic anti-inflammatory late stage of DSC differentiation is, we think, phylogenetically newer and perhaps limited to Euarchontoglires (Chavan et al. 2016).
The suppressive effect of DSC likely operates at the level of IL17A-producing cells, and not at the upstream level of macrophage activation. DSC-conditioned medium has no effect on the activation of human macrophages (differentiated from THP-1 monocytes in vitro), as measured by IL6 production, upon stimulation with lipopolysaccharides (LPSs) (fig. 4C).
Comparison of the transcriptomes of differentiating T cells in the presence or absence of DSC-conditioned medium (third and fourth samples from fig. 4B) suggests that the suppressive effect of DSC may be mediated by the activation of a pathway related to type 1 interferon signaling in the differentiating T cells, in turn leading to downregulation of protein synthesis (supplementary Results and fig. 3, Supplementary Material online). Further research into the identification of the soluble molecule(s) from DSC that mediate this effect, their evolutionary origin, and the molecular mechanisms by which they act will be important for better understanding the role of decidualization in the evolution of eutherian implantation as well as its homologous processes, menstruation and parturition (Pavlicev and Norwitz 2018; Critchley et al. 2020).
Wu et al. (2014) showed that CD45RO+ TH17 cells, that is, memory TH17 cells, are present in the human endometrium during the first trimester and that they are recruited there by DSC, which appears to contradict our results as well as those from aforementioned studies. Reconciling these observations will require further investigation into the possibility that different subtypes of TH17 cells have opposing effects on the maintenance of pregnancy.
A Model for the Evolution of Implantation
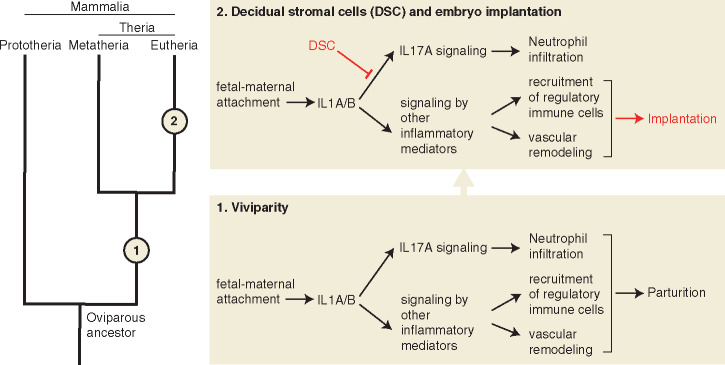
Placing the results from this study in the context of previous studies, the following model for the evolution of embryo implantation emerges (fig. 5).
Fig. 5.
Model for the evolution of eutherian implantation from attachment-induced inflammation. Evolutionary events in the therian and eutherian stem lineages are represented by numbers 1 and 2, respectively.
The ancestor of all mammals was an egg-laying amniote. Mammalian viviparity originated in the stem lineage of therian mammals by early “hatching” of the embryo while it was still within the uterus/shell gland, leading to a direct physical contact between fetal membranes and the uterine endometrial lining. This novel tissue interaction, and potentially an irritation of the endometrial lining from fetal proteases that help dissolve the shell (Griffith et al. 2017), induced an acute inflammatory response in the endometrium. An inflamed uterus, unable to maintain and nourish a live embryo within it, expelled the embryo, that is, parturition ensued as a consequence of attachment-induced inflammation.
Hence, we assume that the ancestral condition for therians was a typical mucosal inflammation in response to embryo attachment. However, in the stem lineage of eutherian mammals, this inflammatory response became evolutionarily modified such that the endometrium could allow embedding and nourishing of the embryo, even though it initiated an inflammation-like response. One of these modifications was the suppression of IL17A signaling, which resulted in the prevention of recruitment of neutrophils to the endometrium during implantation. This may have been a crucial modification to the inflammatory response since neutrophils are known to cause collateral tissue damage from the digestive enzymes they secrete, and the attaching embryo also would likely have not been spared. Turning off IL17A early in the evolution of the eutherian lineage may have allowed the endometrium to maintain the embryo for a prolonged period. In contrast, many other aspects of the ancestral inflammation may have been beneficial to the maintenance and nourishment of the embryo, for example, increased vascular permeability and angiogenesis may have helped nutrient transfer, therefore still maintained as a necessary part of implantation. The ancestral attachment-induced inflammation induced a stress reaction in the ESFs causing the death of most of these cells. In the eutherian lineage, however, ESF evaded the stress-induced cell death (Kajihara et al. 2006; Leitao et al. 2010; Muter and Brosens 2018) by evolving mechanisms to differentiate into a novel cell type, DSCs (Erkenbrack et al. 2018). It is this novel cell type that likely brought about the suppression of IL17A by inhibiting the expression of IL17A at the fetal–maternal interface, and thus enabled the evolution of embryo implantation and a sustainable fetal–maternal interface.
Materials and Methods
Animals and Tissue Samples
Nine-banded armadillos (D. novemcinctus) were collected in Centerville, TX. Two females were used in this study—one nonpregnant and one at the peri-implantation stage of pregnancy when the fetal membranes have begun invading the endometrium but no placental villi are yet formed. Rock hyrax (P. capensis) samples were collected at Bar-Ilan University, Israel. One of three females collected was in the implantation phase of pregnancy as determined by histological examination. The blastocyst was attached to the uterine lumen but had not started invading the endometrium. Opossum (M. domestica) tissues were collected from the colony housed at Yale University. Samples were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for histology and immunohistochemistry and saved in RNAlater (Ambion) for RNA extraction. For more information about armadillo samples, see Chavan and Wagner (2016) and about opossum samples, see Griffith et al. (2017). The list of animals used in this study is given in the Supplementary Material online.
Immunohistochemistry
Formaldehyde-fixed tissues were dehydrated in ethanol, cleared in toluene, and embedded in paraffin. Sections of 5-µm thickness were made on a microtome and placed on poly-l-lysine-coated glass slides. Immunohistochemistry was performed by following the protocol in Chavan and Wagner (2016). Briefly, slides were incubated at 60 °C for 30 min and allowed to cool at room temperature for 5 min. Paraffin was removed by dewaxing the slides in xylene. Slides were then rehydrated by successive washes in 100% ethanol, followed by running tap water. Sodium citrate buffer (pH 6.0) was used for heat-mediated antigen retrieval. After washing the slides in PBS, they were blocked in a 0.1% solution of bovine serum albumin (BSA) in PBS. Endogenous peroxidases were suppressed with Peroxidase Block (DAKO). Optimized dilutions of primary antibodies (see table 1) were added to the slides and were incubated overnight at 4 °C in a humidification chamber. Secondary antibody was added after washing the primary antibody off with PBS and 0.1% BSA–PBS, and incubated for 1 h at room temperature, and washed off with PBS and 0.1% BSA–PBS. Horseradish peroxidase (HRP)-tagged secondary antibodies were detected by 3,3′-diaminobenzidine and counterstained with hematoxylin.
Table 1.
Antibodies Used in This Study.
| Product ID | Company | Antigen | Antigen Species | Host Species | Label | Dilution Used |
|---|---|---|---|---|---|---|
| sc32294 | Santa Cruz | IL1B | Human | Mouse | None | 1:100 |
| M3210 | Spring | PTGS2 | Human | Rabbit | None | 1:1,000 |
| NB110-39058 | Novus | PTGES | Human | Rabbit | None | 1:100 |
| sc2005 | Santa Cruz | Mouse-IgG | Mouse | Goat | HRP | 1:200 |
| K4011 | DAKO | Rabbit-IgG | Rabbit | Goat | Polymer-HRP | Undiluted |
RNA-Seq Data
Among the animal tissue collected for this study, armadillo samples were used for RNA sequencing. RNA was extracted from whole uteri and the fetal membranes attached to them. Sequencing library for RNA from nonpregnant armadillo was prepared and sequenced at the Yale Center for Genome Analysis. Library preparation and sequencing of the RNA from implantation stage armadillo was performed at Cincinnati Children’s Hospital Medical Center. RNA from CD4+ naïve T cells differentiated into TH17 cells in the presence of DSC-conditioned medium was extracted using Qiagen RNeasy Micro Kit (74004, Qiagen). Libraries were prepared and sequenced at the Yale Center for Genome Analysis.
Data for rabbit uterus (O. cuniculus) (Liu et al. 2016) and gray short-tailed opossum (M. domestica) (Griffith et al. 2017) and human endometrial stromal cells (Kin et al. 2015) were downloaded from Gene Expression Omnibus (GEO) (Barrett et al. 2013). Accession numbers for the downloaded data sets as well as for those generated in this study are listed in Data Availability.
RNA-Seq Analysis
RNA-seq data were aligned to the following Ensembl genomes using TopHat2 (Kim et al. 2013): DasNov3 for armadillo, OryCun2 for rabbit, MonDom5 for opossum, and GRCh37 for human. The number of reads mapping to genes were counted with HTSeq (Anders et al. 2015). Read counts were normalized to TPM (Wagner et al. 2012), and 3 TPM was used as an operational threshold to call genes as expressed or unexpressed (Wagner et al. 2013). Median length of all transcripts of a gene was used as its “feature length” for TPM normalization. Differential gene expression analysis for human T cell and endometrial stromal cell data was performed on protein coding genes with the R package EdgeR (Robinson et al. 2010). For these samples, read counts are normalized by EdgeR to Counts per Million and are presented as such.
In analyses involving multiple species, only those genes were included that have one-to-one orthology among the species compared. Orthology data from Ensembl Compara database (Herrero et al. 2016) were used. In these analyses, read counts were renormalized to TPM using only the set of orthologous genes in order to make TPM values comparable between species (Musser and Wagner 2015).
Enriched GO categories in sets of genes were identified with the online tool GOrilla (Eden et al. 2009). Lists of enriched GO categories were visualized on REViGO (Supek et al. 2011), which clusters semantically similar GO terms in space, simplifying interpretation of long lists of redundant GO categories.
qPCR
Expression of IL17A through opossum pregnancy was measured using qPCR at a finer time scale than the transcriptomic data. RNA from nonpregnant, 8, 11.5, 12.5, and 13.5 dpc uteri was extracted and reverse transcribed to cDNA using High Capacity Reverse Transcriptase Kit (4368814, Thermo Fisher). Abundance of IL17A mRNA was measured relative to TBP (Tata Binding Protein) mRNA with a standard curve approach using Power SYBR Green PCR Master Mix (4367659, Applied Biosystems) on StepOne Plus Real Time PCR System (Applied Biosystems). Primer sequences are listed in table 2.
Table 2.
Primers Used for qPCR.
| Gene | Primer (5′–3′) | |
|---|---|---|
| IL17A (M. domestica) | Forward | TCTTCTCCAAGCAACTTGCCA |
| Reverse | AGAGCGGTTCTTGTAATCGGG | |
| TBP (M. domestica) | Forward | CTCTTCCATTCACAGACTCTTACC |
| Reverse | TCAAGTTTACAACCAAGATTCACG |
In Situ Hybridization
In situ hybridization on histological slides was performed using a protocol modified from (Abzhanov 2009). All aqueous solutions were prepared with diethyl pyrocarbonate-treated water when possible.
Uteri for in situ hybridization analysis were collected on separate occasions from two pregnant M. domestica animals, both at 14 dpc. Upon collection, the uteri were cut transversely and embryos were removed, although other fetal membranes and components of the yolk sac placenta remained attached to the uterine epithelium. Processed uteri were fixed in 4% paraformaldehyde in PBS overnight at −20 °C. The following day samples were transferred to 5% sucrose solution for 1 h and then 30% sucrose solution for 1 day at 4 °C on a rocking apparatus. The next day samples were embedded in block molds containing OCT mounting medium (TissueTek, Sakura Finetek, Japan) and frozen quickly by immersion in 2-methylbutane surrounded by dry ice. Frozen sections from 6- to 20-μm thickness were cut on a cryostat (Microm HM 500 OM) and stored at −80 °C.
Riboprobes were synthesized from DNA amplified by polymerase chain reaction (PCR) from late-gestation whole uterus cDNA of opossum. cDNA was prepared for this purpose by homogenization (TissueRuptor, Qiagen) of tissue preserved in RNAlater solution (Invitrogen), followed by RNA isolation (RNeasy Mini Kit, Qiagen) and reverse transcription (Applied Biosystems 4368814). A nested amplification approach was used whereby smaller probes were generated by secondary PCR from an initial larger amplicon from the target gene. Riboprobe synthesis was carried out by in vitro transcription starting at T7 RNA polymerase sequences incorporated into the probes by conjugation of corresponding T7 promoter sequences (5′-TAATACGACTCACTATAGG-3′) to either the reverse (antisense probe) or the forward (sense probe) PCR primer when generating the DNA template. The reaction was carried out in the presence of digoxigenin-UTP (Roche 11175025910) for labeling. Primer sequences are listed in tables 3 and 4.
Table 3.
Probes Used for IL17A In Situ Hybridization.
| Probe | Sense/Antisense | Forward Primer | Reverse Primer | Length |
|---|---|---|---|---|
| Full region | N/A | ARC101 | ARC102 | 875 |
| Upstream | Antisense | ARC101 | ARC105L | 404 |
| Upstream | Sense | ARC101L | ARC105 | 404 |
Table 4.
Primers Used for Probe Synthesis.
| Primer Name | Sequence (5′–3′) |
|---|---|
| ARC101 | TCGCCAGAAATGACAGGTAAG |
| ARC101L | TAATACGACTCACTATAGGTCGCCAGAAATGACAGGTAAG |
| ARC102 | CCCCAAGAAAACAAATTTACCC |
| ARC105 | GCTGTCTGTTTTCGTCCACA |
| ARC105L | TAATACGACTCACTATAGGGCTGTCTGTTTTCGTCCACA |
Thawed sections were incubated in 4% paraformaldehyde in PBS, washed in 0.1% Tween 20 in PBS, treated with 1 μg/ml proteinase K (Roche) for 10 min, washed again, and acetylated in 0.25% acetic anhydride in 0.1 M triethanolamine. Sections were then hybridized with riboprobes in hybridization solution at 37 °C overnight, then washed, blocked in heat-inactivated sheep serum (EMD Millipore S22-100mL) for 1 h, and incubated with anti-digoxigenin antibody Fab fragments (Roche 11093274910 lot 12486520). Immunoreactivity was visualized by incubation of slides covered with nitroblue tetrazolium and 5-bromo-4-chloro-3-indoyl phosphate alkaline phosphatase substrate (Sigma B1911) in humidification chamber for varying lengths of time, up to 3 days. Slides were then rinsed, coverslipped in a gelvatol (polyvinyl alcohol, glycerol) medium, and examined.
Cell Culture Experiments
DSC and DSC-Conditioned Medium
Immortalized human ESFs from ATCC (ATCC; cat. no. CRL-4003) were cultured in growth medium with the following contents per liter: 15.56 g Dulbecco's Modified Eagle Medium (DMEM) without phenol red (D2906, Sigma-Aldrich), 1.2 g sodium bicarbonate, 10 ml sodium pyruvate (11360, Thermo Fisher), 10 ml Antibiotic-Antimycotic (ABAM) (15240062, Gibco), 1 ml Insulin-Transferrin-Selenium (ITS) (354350, VWR), and 100 ml charcoal-stripped fetal bovine serum (FBS). ESFs were differentiated into DSCs in differentiation medium with the following contents per liter: 15.56 g DMEM (D8900, Sigma-Aldrich), 1.2 g sodium bicarbonate, 10 ml ABAM, 0.5 mM cyclic adenosine monophosphate (cAMP) analog 8-bromoadenosine 3′,5′-cyclic monophosphate sodium salt (B7880, Sigma-Aldrich), 1 µM progesterone analog medroxyprogesterone 17-acetate (M1629, Sigma-Aldrich), and 20 ml FBS.
Cells were differentiated for 10 days at 37 °C, conditioned medium was collected on day 2 and day 10 from at least six replicate flasks, filtered through sterile 0.45-μm filter to remove cell debris, aliquoted, and frozen at −80 °C until used. For control samples, decidualization medium was incubated at 37 °C without any cells in at least six replicate flasks, processed in the same way as conditioned medium, and frozen at −80 °C.
T Cells
Human peripheral blood mononuclear cells were isolated from whole blood (70501, StemCell Technologies) using Lymphoprep (07851, StemCell Technologies) and 50 ml SepMate tubes (85450, StemCell Technologies). These peripheral blood mononuclear cells were used to isolate CD4+ CD45RO− naïve T cells with EasySep Human Naïve CD4+ T Cell Isolation Kit (19555, StemCell Technologies). Naïve T cells were resuspended at 106 cells/ml in ImmunoCult-XF T Cell Expansion medium (10981, StemCell Technologies) in the presence of ImmunoCult CD3/CD28/CD2 T Cell Activator (10970, StemCell Technologies) for culture or TH17 differentiation. For TH17 differentiation, the following were added to the above cell suspension: 20 ng/ml IL6 (78050, StemCell Technologies), 5 ng/ml TGFB1 (78067, StemCell Technologies), and 50 ng/ml IL23 (14-8239-63, eBioScience). If conditioned medium was used, the above suspension was made in 1:1 solution of conditioned or control medium and the T-cell expansion medium. Cell suspension was then transferred to 24-well plates, with 106 cells per well. Samples with different treatments were placed in a Latin square design on 24-well plates to prevent systematic effects arising from the positions of the samples in the plate. Cells were incubated at 37 °C for 7 days for differentiation. Cells, which are in suspension, were spun down to collect the supernatant and cell-pellet. Secreted IL17A was measured in the supernatants with Quantikine ELISA kit for human IL17A (D1700, R&D Systems).
Macrophages
THP-1 monocytes were purchased from ATCC (ATCC TIB-202). They were grown in RPMI (90%; ATCC-30-2001), charcoal-stripped FBS (10%), and β-mercaptoethanol (0.05 mM). Macrophage differentiation was carried out in 24-well plates in growth medium supplemented with 100 nM phorbol 12-myristate 13-acetate for 72 h at 37 °C—during this time the cells become adherent. Differentiated cells were incubated in fresh growth medium for 12–24 h of “resting phase.” For induction of IL6 secretion, macrophages at the end of the resting phase were treated with 0.1 µg/ml LPS in growth medium. When testing the effect of DSC-conditioned medium, LPS treatment was carried out in a 1:1 mixture of macrophage growth medium and DSC-conditioned or control medium. Samples were arranged in a Latin square design to avoid positional effects. Cells were incubated for 8 h at 37 °C, supernatant was collected, and IL6 was measured in pg/ml with Quantikine ELISA kit for human IL6 (D6050, R&D Systems).
Animal Ethics Statement
Opossum (Monodelphis domestica) and armadillo (Dasypus novemcinctus) were handled according to protocols 2015-11313 and 2014-10906, respectively, approved by Yale University institutional animal care and use committee (IACUC). The hyrax (Procavia capensis) carcasses for this study were obtained as a secondary use of animals from governmental culling efforts, under permission from Israel Nature and National Parks Protection Authority (Permit Number: 41543). We exploited ongoing culling efforts and did not initiate any culling for this study.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This research was supported by the John Templeton Foundation (Grant Nos. 54860 and 61329 to G.P.W.); the National Cancer Institute of the National Institutes of Health (Grant No. U54-CA209992 to G.P.W.); in part, by the Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS); and, in part, with Federal funds from NICHD/NIH/DHHS under contract no. HHSN275201300006C. Dr Romero has contributed to this work as part of his official duties as an employee of the Unites States Federal Government. We would like to thank Haleigh Larson for assistance with histology and Gabriela Plucinska for assistance with data visualization.
Data Availability
Code used in this study is available at https://github.com/archavan/implantation-il17a. Previously published data were downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo) or SRA (https://www.ncbi.nlm.nih.gov/sra). Data generated in this study are available on GEO. Accession IDs for RNA-seq data sets used in this study are listed below.
| Species | Tissue/Cells | Source | Accession |
|---|---|---|---|
| Armadillo | Uterus | This study | GSE120510 |
| Human | TH17 cells | This study | GSE120380 |
| Rabbit | Uterus | Liu et al. (2016) | GSE76115 |
| Opossum | Uterus | Griffith et al. (2017) | SRP111668 |
| Human | ESF and DSC | Kin et al. (2015) | GSE63733 |
| Human | Decidua, villi | Suryawanshi et al. (2018) | PRJNA492902 |
References
- Abzhanov A. 2009. In situ hybridization analysis of embryonic beak tissue from Darwin’s finches. Cold Spring Harb Protoc. 2009(3):pdb.prot5175. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W.. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashary N, Tiwari A, Modi D.. 2018. Embryo implantation: war in times of love. Endocrinology 159(2):1188–1198. [DOI] [PubMed] [Google Scholar]
- Barash A, Dekel N, Fieldust S, Segal I, Schechtman E, Granot I.. 2003. Local injury to the endometrium doubles the incidence of successful pregnancies in patients undergoing in vitro fertilization. Fertil Steril. 79(6):1317–1322. [DOI] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. 2013. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 41(D1):D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellofiore N, Ellery SJ, Mamrot J, Walker DW, Temple-Smith P, Dickinson H.. 2017. First evidence of a menstruating rodent: the spiny mouse (Acomys cahirinus). Am J Obstet Gynecol. 216(1):40.e1–40.e11. [DOI] [PubMed] [Google Scholar]
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK.. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441(7090):235–238. [DOI] [PubMed] [Google Scholar]
- Buckley KM, Ho ECH, Hibino T, Schrankel CS, Schuh NW, Wang G, Rast JP.. 2017. IL17 factors are early regulators in the gut epithelium during inflammatory response to Vibrio in the sea urchin larva. eLife 6:e23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AM. 2020. The role of mammalian foetal membranes in early embryogenesis: lessons from marsupials. J Morphol. 10.1002/jmor.21140. [DOI] [PubMed] [Google Scholar]
- Chavan AR, Bhullar B-AS, Wagner GP.. 2016. What was the ancestral function of decidual stromal cells? A model for the evolution of eutherian pregnancy. Placenta 40:40–51. [DOI] [PubMed] [Google Scholar]
- Chavan AR, Griffith OW, Wagner GP.. 2017. The inflammation paradox in the evolution of mammalian pregnancy: turning a foe into a friend. Curr Opin Genet Dev. 47:24–32. [DOI] [PubMed] [Google Scholar]
- Chavan AR, Wagner GP.. 2016. The fetal–maternal interface of the nine-banded armadillo: endothelial cells of maternal sinus are partially replaced by trophoblast. Zool Lett. 2(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhang Y, Lu J, Wang Q, Wang S, Cao Y, Wang H, Duan E.. 2009. Embryo–uterine cross-talk during implantation: the role of Wnt signaling. MHR Basic Sci Reprod Med. 15(4):215–221. [DOI] [PubMed] [Google Scholar]
- Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, Hoeffer CA, Littman DR, Huh JR.. 2016. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 351(6276):933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley HOD, Babayev E, Bulun SE, Clark S, Garcia-Grau I, Gregersen PK, Kilcoyne A, Kim J-YJ, Lavender M, Marsh EE, et al. 2020. Menstruation: science and society. Am J Obstet Gynecol. 223(5):624–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel N, Gnainsky Y, Granot I, Racicot K, Mor G.. 2014. The role of inflammation for a successful implantation. Am J Reprod Immunol. 72(2):141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Inoue J, Hasegawa M, Asher RJ, Donoghue PC, Yang Z.. 2012. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc R Soc B. 279(1742):3491–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, Luster AD.. 2002. IFN-γ-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J Immunol. 168(7):3195–3204. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z.. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinf. 10(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emera D, Romero R, Wagner G.. 2012. The evolution of menstruation: a new model for genetic assimilation. BioEssays 34(1):26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders AC, Schlafke S.. 1969. Cytological aspects of trophoblast-uterine interaction in early implantation. Am J Anat. 125(1):1–29. [DOI] [PubMed] [Google Scholar]
- Erkenbrack EM, Maziarz JD, Griffith OW, Liang C, Chavan AR, Nnamani MC, Wagner GP.. 2018. The mammalian decidual cell evolved from a cellular stress response. PLoS Biol. 16(8):e2005594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlebacher A. 2013. Immunology of the maternal-fetal interface. Annu Rev Immunol. 31(1):387–411. [DOI] [PubMed] [Google Scholar]
- Filant J, Spencer TE.. 2013. Endometrial glands are essential for blastocyst implantation and decidualization in the mouse uterus. Biol Reprod. 88(4):93. [DOI] [PubMed] [Google Scholar]
- Finn CA. 1986. Implantation, menstruation and inflammation. Biol Rev. 61(4):313–328. [DOI] [PubMed] [Google Scholar]
- Flannigan KL, Ngo VL, Geem D, Harusato A, Hirota SA, Parkos CA, Lukacs NW, Nusrat A, Gaboriau-Routhiau V, Cerf-Bensussan N, et al. 2017. IL-17A-mediated neutrophil recruitment limits expansion of segmented filamentous bacteria. Mucosal Immunol. 10(3):673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellersen B, Brosens JJ.. 2014. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr Rev. 35(6):851–905. [DOI] [PubMed] [Google Scholar]
- Griffin GK, Newton G, Tarrio ML, Bu D-X, Maganto-Garcia E, Azcutia V, Alcaide P, Grabie N, Luscinskas FW, Croce KJ, et al. 2012. IL-17 and TNF-α sustain neutrophil recruitment during inflammation through synergistic effects on endothelial activation. J Immunol. 188(12):6287–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith OW, Chavan AR, Pavlicev M, Protopapas S, Callahan R, Maziarz J, Wagner GP.. 2019. Endometrial recognition of pregnancy occurs in the grey short-tailed opossum (Monodelphis domestica). Proc R Soc B Biol Sci. 286(1905):20190691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith OW, Chavan AR, Protopapas S, Maziarz J, Romero R, Wagner GP.. 2017. Embryo implantation evolved from an ancestral inflammatory attachment reaction. Proc Natl Acad Sci U S A. 114(32):E6566–E6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith OW, Chavan AR, Protopapas S, Maziarz J, Romero R, Wagner GP.. 2018. Reply to Liu: inflammation before implantation both in evolution and development. Proc Natl Acad Sci U S A. 115(1):E3–E4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen VL, Faber LS, Salehpoor AA, Miller RD.. 2017. A pronounced uterine pro-inflammatory response at parturition is an ancient feature in mammals. Proc R Soc B Biol Sci. 284(1865):20171694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero J, Muffato M, Beal K, Fitzgerald S, Gordon L, Pignatelli M, Vilella AJ, Searle SMJ, Amode R, Brent S, et al. 2016. Ensembl comparative genomics resources. Database 2016:baw053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaislasuo J, Simpson S, Petersen JF, Peng G, Aldo P, Lokkegaard E, Paidas M, Pal L, Guller S, Mor G.. 2020. IL-10 to TNFα ratios throughout early first trimester can discriminate healthy pregnancies from pregnancy losses. Am J Reprod Immunol. 83(1):e13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajihara T, Jones M, Fusi L, Takano M, Feroze-Zaidi F, Pirianov G, Mehmet H, Ishihara O, Higham JM, Lam EW, et al. 2006. Differential expression of FOXO1 and FOXO3a confers resistance to oxidative cell death upon endometrial decidualization. Mol Endocrinol. 20(10):2444–2455. [DOI] [PubMed] [Google Scholar]
- Kelleher AM, Milano-Foster J, Behura SK, Spencer TE.. 2018. Uterine glands coordinate on-time embryo implantation and impact endometrial decidualization for pregnancy success. Nat Commun. 9(1):2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys JL, King GJ, Kennedy TG.. 1986. Increased uterine vascular permeability at the time of embryonic attachment in the pig. Biol Reprod. 34(2):405–411. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kin K, Maziarz J, Chavan AR, Kamat M, Vasudevan S, Birt A, Emera D, Lynch VJ, Ott TL, Pavlicev M, et al. 2016. The transcriptomic evolution of mammalian pregnancy: gene expression innovations in endometrial stromal fibroblasts. Genome Biol Evol. 8(8):2459–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kin K, Nnamani Mauris C, Lynch Vincent J, Michaelides E, Wagner GP.. 2015. Cell-type phylogenetics and the origin of endometrial stromal cells. Cell Rep. 10(8):1398–1409. [DOI] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK.. 2009. IL-17 and Th17 cells. Annu Rev Immunol. 27(1):485–517. [DOI] [PubMed] [Google Scholar]
- Leitao B, Jones MC, Fusi L, Higham J, Lee Y, Takano M, Goto T, Christian M, Lam EWF, Brosens JJ.. 2010. Silencing of the JNK pathway maintains progesterone receptor activity in decidualizing human endometrial stromal cells exposed to oxidative stress signals. FASEB J. 24(5):1541–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Qu Q, Yan Q.. 2018. The role of Th17/Treg-mediated immunoregulation in abortion mice. Eur J Inflamm. 16:205873921876035. [Google Scholar]
- Lieschke GJ, Grail D, Hodgson G, Metcalf D, Stanley E, Cheers C, Fowler KJ, Basu S, Zhan YF, Dunn AR.. 1994. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood 84(6):1737–1746. [PubMed] [Google Scholar]
- Liu J-L. 2018. Implantation in eutherians: which came first, the inflammatory reaction or attachment? Proc Natl Acad Sci U S A. 115(1):E1–E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-L, Zhao M, Peng Y, Fu Y-S.. 2016. Identification of gene expression changes in rabbit uterus during embryo implantation. Genomics. 107(5):216–221. [DOI] [PubMed] [Google Scholar]
- Marions L, Danielsson KG.. 1999. Expression of cyclo-oxygenase in human endometrium during the implantation period. Mol Hum Reprod. 5(10):961–965. [DOI] [PubMed] [Google Scholar]
- McAllan BM. 2011. Chapter 10—reproductive endocrinology of prototherians and metatherians. In: Lopez DONH, editor. Hormones and reproduction of vertebrates. London: Academic Press. p. 195–214. [Google Scholar]
- Medzhitov R. 2007. Recognition of microorganisms and activation of the immune response. Nature 449(7164):819–826. [DOI] [PubMed] [Google Scholar]
- Mess A, Carter AM.. 2006. Evolutionary transformations of fetal membrane characters in Eutheria with special reference to Afrotheria. J Exp Zool. 306B(2):140–163. [DOI] [PubMed] [Google Scholar]
- Milne SA, Perchick GB, Boddy SC, Jabbour HN.. 2001. Expression, localization, and signaling of PGE(2) and EP2/EP4 receptors in human nonpregnant endometrium across the menstrual cycle. J Clin Endocrinol Metab. 86(9):4453–4459. [DOI] [PubMed] [Google Scholar]
- Mohamed OA, Jonnaert M, Labelle-Dumais C, Kuroda K, Clarke HJ, Dufort D.. 2005. Uterine Wnt/β-catenin signaling is required for implantation. Proc Natl Acad Sci U S A. 102(24):8579–8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor G, Cardenas I, Abrahams V, Guller S.. 2011. Inflammation and pregnancy: the role of the immune system at the implantation site. Ann N Y Acad Sci. 1221(1):80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman HW. 1937. Comparative morphogenesis of the fetal membranes and accessory uterine structures. Contrib Embryol. 26:133–137. [DOI] [PubMed] [Google Scholar]
- Musser JM, Wagner GP.. 2015. Character trees from transcriptome data: origin and individuation of morphological characters and the so-called “species signal”. J Exp Zool (Mol Dev Evol). 324(7):588–604. [DOI] [PubMed] [Google Scholar]
- Muter J, Brosens JJ.. 2018. Decidua. In: Skinner MK, editor. Encyclopedia of reproduction. 2nd ed.Oxford: Academic Press. p. 424–430. [Google Scholar]
- Nakashima A, Ito M, Shima T, Bac ND, Hidaka T, Saito S.. 2010. Accumulation of IL‐17‐positive cells in decidua of inevitable abortion cases. Am J Reprod Immunol. 64:4–11. [DOI] [PubMed] [Google Scholar]
- Nancy P, Tagliani E, Tay CS, Asp P, Levy DE, Erlebacher A.. 2012. Chemokine gene silencing in decidual stromal cells limits T cell access to the maternal–fetal interface. Science 336(6086):1317–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi RM, Gaffen SL.. 2010. Interleukin-17 and its target genes: mechanisms of interleukin-17 function in disease. Immunology 129(3):311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos AD, Watowich SS.. 2008. Granulocyte colony-stimulating factor: molecular mechanisms of action during steady state and ‘emergency’ hematopoiesis. Cytokine 42(3):277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu R, Rutz S, Ouyang W.. 2012. Regulation of epithelial immunity by IL-17 family cytokines. Trends Immunol. 33(7):343–349. [DOI] [PubMed] [Google Scholar]
- Pavlicev M, Norwitz ER.. 2018. Human parturition: nothing more than a delayed menstruation. Reprod Sci. 25(2):166–173. [DOI] [PubMed] [Google Scholar]
- Pavlicev M, Wagner GP, Chavan AR, Owens K, Maziarz J, Dunn-Fletcher C, Kallapur SG, Muglia L, Jones H.. 2017. Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Res. 27(3):349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinget GV, Corpuz TM, Stolp J, Lousberg EL, Diener KR, Robertson SA, Sprent J, Webster KE.. 2016. The majority of murine γδ T cells at the maternal–fetal interface in pregnancy produce IL-17. Immunol Cell Biol. 94(7):623–630. [DOI] [PubMed] [Google Scholar]
- Plaks V, Birnberg T, Berkutzki T, Sela S, BenYashar A, Kalchenko V, Mor G, Keshet E, Dekel N, Neeman M, et al. 2008. Uterine DCs are crucial for decidua formation during embryo implantation in mice. J Clin Invest. 118(12):3954–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree M. 1994. Endocrinology of pregnancy, parturition and lactation in marsupials. In: Lamming GE, editor. Marshall’s physiology of reproduction. Dordrecht (the Netherlands: ): Springer. p. 677–766. [Google Scholar]
- Robertson SA, Moldenhauer LM.. 2014. Immunological determinants of implantation success. Int J Dev Biol. 58(2–4):205–217. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose RW. 1989. Embryonic growth rates of marsupials with a note on monotremes. J Zool. 218(1):11–16. [Google Scholar]
- Rytkönen KT, Erkenbrack EM, Poutanen M, Elo LL, Pavlicev M, Wagner GP.. 2019. Decidualization of human endometrial stromal fibroblasts is a multiphasic process involving distinct transcriptional programs. Reprod Sci. 26(3):323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salker MS, Nautiyal J, Steel JH, Webster Z, Šućurović S, Nicou M, Singh Y, Lucas ES, Murakami K, Chan Y-W, et al. 2012. Disordered IL-33/ST2 activation in decidualizing stromal cells prolongs uterine receptivity in women with recurrent pregnancy loss. PLoS One 7(12):e52252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schall TJ. 1991. Biology of the RANTES/SIS cytokine family. Cytokine 3(3):165–183. [DOI] [PubMed] [Google Scholar]
- Schlafke S, Enders AC.. 1975. Cellular basis of interaction between trophoblast and uterus at implantation. Biol Reprod. 12(1):41–65. [DOI] [PubMed] [Google Scholar]
- Selwood L. 2000. Marsupial egg and embryo coats. Cells Tissues Organs. 166(2):208–219. [DOI] [PubMed] [Google Scholar]
- Shuya LL, Menkhorst EM, Yap J, Li P, Lane N, Dimitriadis E.. 2011. Leukemia inhibitory factor enhances endometrial stromal cell decidualization in humans and mice. PLoS One 6(9):e25288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonderegger S, Pollheimer J, Knöfler M.. 2010. Wnt signalling in implantation, decidualisation and placental differentiation—review. Placenta 31(10):839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtmauer DJ, Wagner GP.. 2020. Cooperative inflammation: the recruitment of inflammatory signaling in marsupial and eutherian pregnancy. J Reprod Immunol. 137:102626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T.. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6(7):e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryawanshi H, Morozov P, Straus A, Sahasrabudhe N, Max KEA, Garzia A, Kustagi M, Tuschl T, Williams Z.. 2018. A single-cell survey of the human first-trimester placenta and decidua. Sci Adv. 4(10):eaau4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarver JE, dos Reis M, Mirarab S, Moran RJ, Parker S, O’Reilly JE, King BL, O’Connell MJ, Asher RJ, Warnow T, et al. 2016. The interrelationships of placental mammals and the limits of phylogenetic inference. Genome Biol Evol. 8(2):330–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, Haag ES.. 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev. 3(2):109–119. [DOI] [PubMed] [Google Scholar]
- Van Sinderen M, Menkhorst E, Winship A, Cuman C, Dimitriadis E.. 2013. Preimplantation human blastocyst–endometrial interactions: the role of inflammatory mediators. Am J Reprod Immunol. 69(5):427–440. [DOI] [PubMed] [Google Scholar]
- Veldhoen M. 2017. Interleukin 17 is a chief orchestrator of immunity. Nat Immunol. 18(6):612–621. [DOI] [PubMed] [Google Scholar]
- Waclawik A, Ziecik AJ.. 2007. Differential expression of prostaglandin (PG) synthesis enzymes in conceptus during peri-implantation period and endometrial expression of carbonyl reductase/PG 9-ketoreductase in the pig. J Endocrinol. 194(3):499–510. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ.. 2012. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131(4):281–285. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ.. 2013. A model based criterion for gene expression calls using RNA-seq data. Theory Biosci. 132(3):159–164. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Muglia L, Pavlicev M.. 2014. Evolution of mammalian pregnancy and the origin of the decidual stromal cell. Int J Dev Biol. 58(2–4):117–126. [DOI] [PubMed] [Google Scholar]
- Wang H, Dey SK.. 2006. Roadmap to embryo implantation: clues from mouse models. Nat Rev Genet. 7(3):185–199. [DOI] [PubMed] [Google Scholar]
- Wang W-J, Hao C-F, Yi L, Yin G-J, Bao S-H, Qiu L-H, Lin Q-D.. 2010. Increased prevalence of T helper 17 (Th17) cells in peripheral blood and decidua in unexplained recurrent spontaneous abortion patients. J Reprod Immunol. 84:164–170. [DOI] [PubMed] [Google Scholar]
- Whittington CM, O’Meally D, Laird MK, Belov K, Thompson MB, McAllan BM.. 2018. Transcriptomic changes in the pre-implantation uterus highlight histotrophic nutrition of the developing marsupial embryo. Sci Rep. 8(1):2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte JJ, Meyer AE, Spate LD, Benne JA, Cecil R, Samuel MS, Murphy CN, Prather RS, Geisert RD.. 2018. Inactivation of porcine interleukin-1β results in failure of rapid conceptus elongation. Proc Natl Acad Sci U S A. 115(2):307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-X, Jin L-P, Xu B, Liang S-S, Li D-J.. 2014. Decidual stromal cells recruit Th17 cells into decidua to promote proliferation and invasion of human trophoblast cells by secreting IL-17. Cell Mol Immunol. 11(3):253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, Oliver P, Huang W, Zhang P, Zhang J, et al. 2001. Requirement of interleukin 17 receptor signaling for lung Cxc chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med. 194(4):519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code used in this study is available at https://github.com/archavan/implantation-il17a. Previously published data were downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo) or SRA (https://www.ncbi.nlm.nih.gov/sra). Data generated in this study are available on GEO. Accession IDs for RNA-seq data sets used in this study are listed below.
| Species | Tissue/Cells | Source | Accession |
|---|---|---|---|
| Armadillo | Uterus | This study | GSE120510 |
| Human | TH17 cells | This study | GSE120380 |
| Rabbit | Uterus | Liu et al. (2016) | GSE76115 |
| Opossum | Uterus | Griffith et al. (2017) | SRP111668 |
| Human | ESF and DSC | Kin et al. (2015) | GSE63733 |
| Human | Decidua, villi | Suryawanshi et al. (2018) | PRJNA492902 |