Abstract
Sterol biosynthesis, primarily associated with eukaryotic kingdoms of life, occurs as an abbreviated pathway in the bacterium Methylococcus capsulatus. Sterol 14α-demethylation is an essential step in this pathway and is catalyzed by cytochrome P450 51 (CYP51). In M. capsulatus, the enzyme consists of the P450 domain naturally fused to a ferredoxin domain at the C-terminus (CYP51fx). The structure of M. capsulatus CYP51fx was solved to 2.7 Å resolution and is the first structure of a bacterial sterol biosynthetic enzyme. The structure contained one P450 molecule per asymmetric unit with no electron density seen for ferredoxin. We connect this with the requirement of P450 substrate binding in order to activate productive ferredoxin binding. Further, the structure of the P450 domain with bound detergent (which replaced the substrate upon crystallization) was solved to 2.4 Å resolution. Comparison of these two structures to the CYP51s from human, fungi, and protozoa reveals strict conservation of the overall protein architecture. However, the structure of an “orphan” P450 from nonsterol-producing Mycobacterium tuberculosis that also has CYP51 activity reveals marked differences, suggesting that loss of function in vivo might have led to alterations in the structural constraints. Our results are consistent with the idea that eukaryotic and bacterial CYP51s evolved from a common cenancestor and that early eukaryotes may have recruited CYP51 from a bacterial source. The idea is supported by bioinformatic analysis, revealing the presence of CYP51 genes in >1,000 bacteria from nine different phyla, >50 of them being natural CYP51fx fusion proteins.
Keywords: sterol biosynthesis, evolution, cytochrome P450, CYP51 redox partner, crystallography
Introduction
Cytochrome P450 enzymes (CYP; P450) are heme-containing monooxygenases that play essential roles in sterol/steroid biosynthesis and degradation, vitamin biosynthesis, detoxification and activation of many drugs and environmental pollutants, enzymatic activation of carcinogens, and the biosynthesis of vast arrays of secondary metabolites (Guengerich 2001). One such P450 is sterol 14α-demethylase (CYP51), an enzyme found in all kingdoms of life, catalyzing the same three-step monooxygenation reaction that removes the 14α-methyl group from the nucleus of the sterol precursor molecule (fig. 1). Ultimately, this reaction is required for the biosynthesis of cholesterol in animals, ergosterol in fungi and protozoa, and stigmasterol in plants and algae. The sterols function as integral components of eukaryotic cell membranes and also serve as precursors for a multitude of different regulatory molecules, such as hormones and brassinosteroids (Lepesheva et al. 2018). Sterols are often considered as a defining signature (biomarkers) of eukaryotic organisms and are rarely found in bacteria, which usually employ hopanoids to facilitate cell membrane integrity (Jackson et al. 2002). Like sterols, hopanoids are cyclic isoprenoidal lipids, and are cyclized by closely related cyclase enzymes (Abe 2007), but their synthesis does not require atmospheric molecular oxygen. Thus, hopanoids have been used as molecular proxies for ancient microbial life predating the enrichment of oxygen in Earth’s atmosphere (Ourisson and Albrecht 1992; Sáenz et al. 2015).
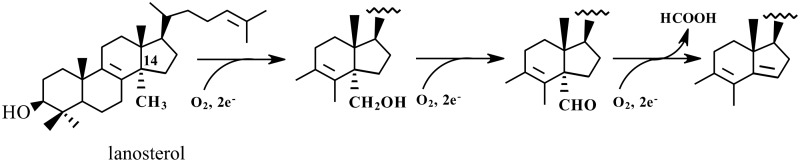
Fig. 1.
CYP51 reaction. The 14α-methyl group is first converted to the alcohol, then to the aldehyde intermediate, and at the third step is removed as formic acid with the introduction of the double bond into the sterol core.
In 1971, production of sterols in bacteria was first discovered biochemically in the aerobic methanotroph Methylococcus capsulatus Bath. A truncated postsqualene sterol pathway was predicted because only the modified lanosterol molecules were detected and identified experimentally (fig. 2) (Bird et al. 1971). These observations were verified following the complete sequencing of the M. capsulatus genome, in which only a small number of genes encoding sterol enzyme orthologs were seen and mapped to the sterol biosynthetic pathway (Ward et al. 2004). Subsequently, further examples of sterol-producing bacteria were identified, including Methylosphaera hansonii (Schouten et al. 2000), Stigmatella aurantiaca, Nannocystis excedens (Bode et al. 2003), Gemmata obscuriglobus (Pearson et al. 2003), Plesiocystis pacifica (Desmond and Gribaldo 2009), Methylomicrobium alcaliphilum (Banta et al. 2015). With the rapid advances in genomic and bioinformatic analysis, the numbers of bacteria known to possess genes for sterol synthesis are continually increasing (Wei et al. 2016).
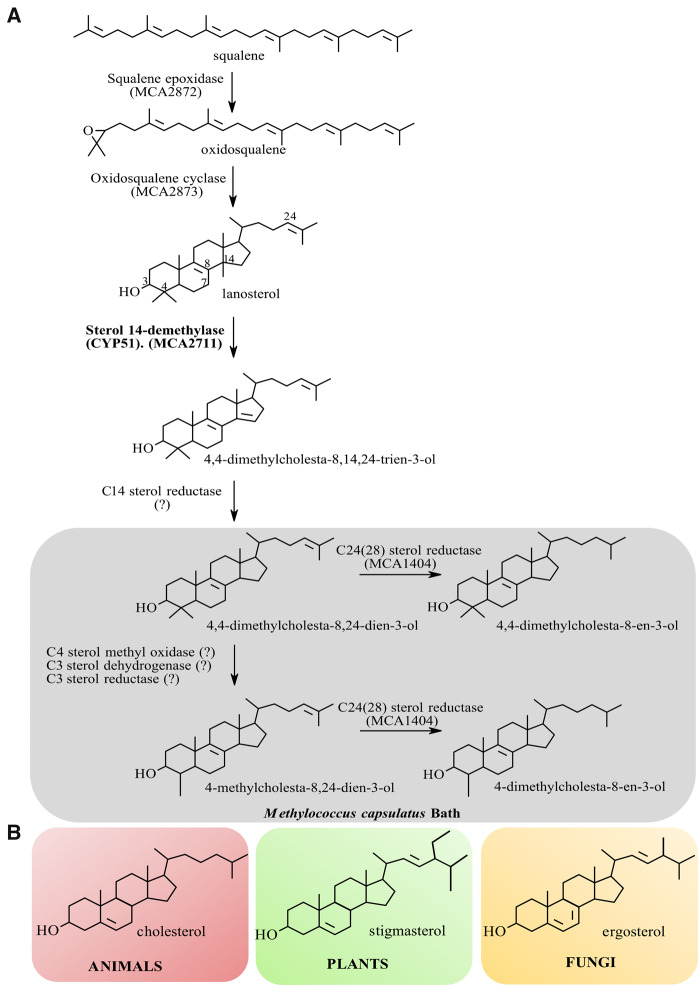
Fig. 2.
Sterol biosynthesis and final sterol products in Methylococcus capsulatus (A) versus eukaryotes (B). (A) Methylococcus capsulatus was shown to encode a truncated postsqualene sterol pathway that synthesizes the modified lanosterol molecules 4,4-dimethylcholesta-8,24-dien-3-ol, 4,4-dimethylcholesta-8-en-3-ol, 4-methylcholesta-8,24-dien-3-ol, and 4-methylcholesta-8-en-3-ol. Subsequent studies have demonstrated the production of similar sterols in other aerobic methanotrophs of the Methylococcales order within the γ-Proteobacteria. (B) Chemical structures of the major eukaryotic sterol molecules: cholesterol in animals; stigmasterol in plants and algae; and ergosterol in fungi and protozoa.
Questions of the evolutionary origin of bacterial sterol biosynthetic machinery, as well as structural relationship of bacterial water-soluble CYP51 proteins to eukaryotic CYP51s (which are membrane-bound microsomal proteins) remain unanswered. Mycobacterium tuberculosis is the only bacterial species with a P450 with CYP51 activity whose 3D crystal structure has been resolved (Podust et al. 2001). However, that structure differs profoundly from bone fide eukaryotic CYP51 crystal structures, both in the composition of the substrate-binding cavity/active site volume and in the location of the substrate access channel (Lepesheva et al. 2010). Moreover, the CYP51 gene was shown to be not essential in My. tuberculosis physiology (McLean et al. 2006) nor essential for cholesterol metabolism (van Wyk et al. 2019), whereas the CYP51 gene is essential in eukaryotes. Thus, two opposite and contradicting hypotheses on CYP51 evolution exist and remain unresolved to-date: 1) CYP51 and sterol biosynthesis has a bacterial origin with soluble (nonmembrane-bound) enzymes that are possible evolutionary ancestors for the membrane-bound eukaryotic CYP51 P450 family (Yoshida et al. 1997; Nelson 1999; Yoshida et al. 2000) or 2) bacterial CYP51s are the result of horizontal gene transfer from some early eukaryote (Debeljak et al. 2003; Rezen et al. 2004). Although the origin and evolution of the cytochrome P450 superfamily as a whole is unknown, current thinking suggests a role for CYP51 as the ancestral P450 progenitor with its ability to fix atmospheric oxygen resulting in increased P450 importance in generating metabolites for membrane integrity. Subsequently, this was followed by gene duplication and evolutionary diversification leading to broad P450 functionality in many roles (Sezutsu et al. 2013).
To address these fundamental questions regarding P450 evolution and sterol biosynthesis, the bacterium M. capsulatus is an excellent organism of choice because it has a CYP51 gene and it makes sterols. Furthermore, M. capsulatus is one of a handful of organisms discovered to-date carrying a gene encoding P450 heme domain fused to a ferredoxin (fx) redox partner domain (Jackson et al. 2002; Hannemann et al. 2007; McLean et al. 2007).
Herein, we report: 1) CYP51 genes are found in >1,000 bacteria from nine different phyla establishing that horizontal gene transfer to bacteria is extremely unlikely and supporting a prokaryotic origin (and thus ancestral role) of CYP51 in P450 evolution; 2) the existence of CYP51 ferredoxin fusion proteins (CYP51fx) in >50 bacterial organisms, suggesting that early cytochrome P450s may have initially been fusion proteins; and 3) the first crystal structure of a bacterial sterol biosynthetic enzyme. The P450 domain of M. capsulatus CYP51fx is similar in protein architecture to eukaryotic CYP51s, and not to My. tuberculosis CYP51, indicating an evolutionary linkage between bacterial and eukaryotic sterol 14α-demethylases.
Results and Discussion
In this study, we first performed biochemical characterization of M. capsulatus CYP51fx to confirm its enzymatic competency. Then we crystallized the fusion protein, determined the crystal structure of its P450 domain, in the ligand-free and a detergent-bound state, and compared these with the known eukaryotic and My. tuberculosis CYP51 structures. Finally, we used bioinformatic approaches to complement our structure-functional data and provide further support for a bacterial origin of CYP51.
Biochemical Characterization of M. capsulatus CYP51fx
Optical Properties
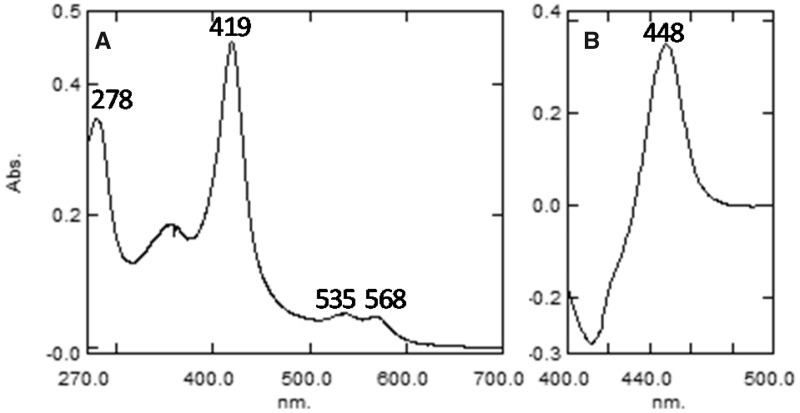
The absolute absorbance spectrum of the purified protein was typical of ferric, low-spin P450 with the maxima of the α-, β-, and γ (Soret) bands at 568, 535, and 419 nm, respectively, the spectrophotometric index A419/A278 = 1.33, and the ratio ΔA393–470/ΔA419–470 = 0.38 (fig. 3A). The heme iron was readily reduced by sodium dithionite and the difference spectrum of the ferrous CO-complex had an absorbance maximum at 448 nm (a hallmark of the cysteinate-coordinated heme iron) with no quantifiable denatured cytochrome P420 form (fig. 3B) (Omura and Sato 1964).
Fig. 3.
The UV-visible absolute (A) and reduced carbon monoxide complex difference (B) absorbance spectra of purified Methylococcus capsulatus CYP51fx. The P450 concentration was ∼3.8 µM, the optical path length was 1 cm. The specific heme content of the preparation was 15.6 nmol per mg protein (98% of the ideal specific content [15.9 nmol per mg protein] calculated from the predicted molecular weight of the M. capsulatus CYP51 sequence [63 kDa]).
Binding of Sterol Substrates
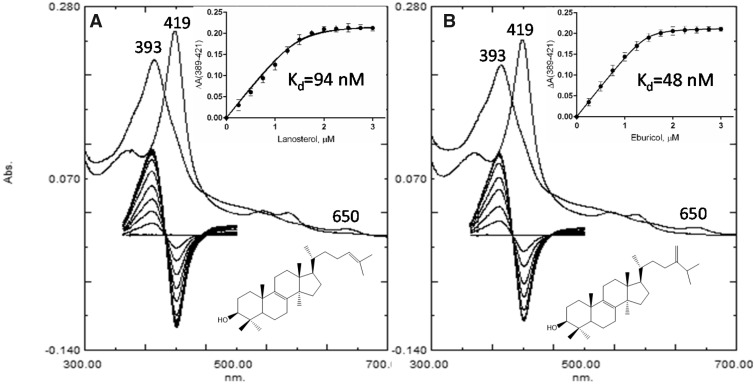
Substrate titration experiments were first conducted with lanosterol and eburicol to clarify which of the two sterols has a higher affinity for M. capsulatus CYP51fx (and therefore should be selected for cocrystallization). Though M. capsulatus is known to synthesize lanosterol in vivo (Bird et al. 1971), the CYP51 family amino acid sequence alignment suggested that the enzyme might prefer eburicol, due to the isoleucine residue at position 81. The I81 in M. capsulatus CYP51fx corresponds to I105 in CYP51 from the protozoan pathogen Trypanosoma cruzi, (the only eukaryotic sterol 14α-demethylase having Ile in this position), and we have previously found that T. cruzi CYP51 prefers eburicol over lanosterol (Lepesheva et al. 2006). Titration with both sterols produced typical Type I-binding spectra, reflecting changes in the spin state of the heme Fe atom, with a blue shift in the Soret band maximum from 419 to 393 nm, and a ΔA393–470/ΔA418–470 ratio (2.12–2.14), indicating >95% heme iron transition from low- to high-spin form, and the charge transfer band at 650 nm (fig. 4). The apparent spectral dissociation constants (Kd) derived from the titration plots were also similar, although binding of eburicol appeared to be about twice as tight (fig. 4, Inset, Kd=48 ± 4 vs. 94 ± 11 nM for lanosterol).
Fig. 4.
Spectral changes observed during titration of Methylococcus capsulatus CYP51fx with (A) lanosterol and (B) eburicol. Absolute (top) and difference (bottom) spectra. The P450 concentration was ∼2 µM. Inset: The titration curves with hyperbolic fitting (quadratic Morrison equation). The experiments were performed in duplicates, the results are presented as means ± SD.
Enzymatic Activity
Both lanosterol and eburicol were metabolized by M. capsulatus CYP51fx to the 14α-demethylated products, 3β-hydroxy-4,4-dimethyl-cholesta-8,14,24-triene, and 3β-hydroxy-4,4-dimethyl-24-methylene-cholesta-8,14-diene, with the same efficiency and comparable rates, 1.8 and 1.6 min−1, respectively. The turnover of obtusifoliol was 1.2 min−1 (supplementary table S1, Supplementary Material online). By comparison, the turnover of obtusifoliol by My. tuberculosis CYP51 (its preferred substrate) was 0.5 min−1, when its activity was reconstituted under the same conditions (Lepesheva et al. 2004), that is, using Escherichia coli flavodoxin/flavodoxin reductase as electron donor partners, as detailed in Materials and Methods. Thus, M. capsulatus CYP51fx catalyzes 14α-demethylation of both C4-dimethylated sterols and C4-monomethylated obtusifoliol at appreciable rates, perhaps contrary to suggestions based on the presence of Ile81 and the apparent binding affinities.
Because lanosterol is the M. capsulatus CYP51fx substrate in vivo (Bird et al. 1971), this substrate was next used to verify the ability of the fused ferredoxin domain to transfer electrons to the P450 heme iron. There was no activity observed at 2 µM M. capsulatus CYP51fx (5 µM spinach ferredoxin reductase) under these reaction conditions, but increasing the concentration to 10 µM (25 µM ferredoxin reductase) resulted in ∼25% of lanosterol conversion in a 2-h reaction (supplementary fig. S1, Supplementary Material online), suggesting a possibility of both inter- and intrareduction, as was proposed for flavocytochrome P450 BM3 (CYP102) from the soil bacterium Bacillus megaterium (a natural fusion of a P450 to a cytochrome P450 reductase) (Kitazume et al. 2007).
Structural Characterization of M. capsulatus CYP51fx
Crystal Structure of the P450 Domain with the Bound Water Molecule
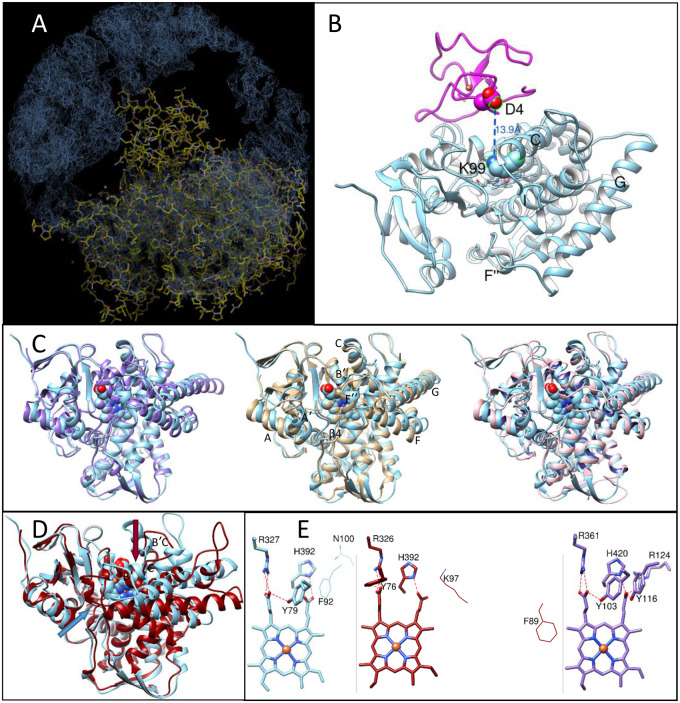
We first crystallized M. capsulatus CYP51fx in the resting (ferric) ligand-free form. The structure was solved to 2.7 Å resolution (supplementary table S2, Supplementary Material online) and contained one P450 molecule per asymmetric unit. However, no electron density for the ferredoxin domain was observed. Instead, the space above the P450 proximal surface (the region where ferredoxin is expected to bind) looked like a “black hole” (fig. 5A and B). We believe this is because, in the absence of the substrate, the P450/ferredoxin complex, even if formed, is weak, likely representing multiple “encounter states” (also called transient complexes) where ferredoxin can have many orientations that it adopts before forming a final, “productive” complex (Schilder and Ubbink 2013; Bowen et al. 2018) with the P450 domain. Physiologically, the electron transfer process between redox partner proteins and P450s takes place after a P450 enzyme binds its substrate (Guengerich and Yoshimoto 2018). The large-scale conformational switch that accompanies binding of the substrate in eukaryotic CYP51 structures (both protozoan, Hargrove et al. 2018 and human, Hargrove et al. 2020) alters the topology of the P450 proximal surface that is known to be involved in the interaction with the electron donor protein partners. We conclude that the presence of substrate in the M. capsulatus CYP51fx active site must be required for the formation of a “productive” P450/ferredoxin complex, one which might be tight enough to be detected crystallographically. Indeed, the P450 domain in the 6MI0 structure (fig. 5) has a typical substrate-free (i.e., water-/inhibitor-bound like) CYP51 conformation (Hargrove et al. 2018): the substrate channel is open, the proton delivery route is closed, and helix C is not crossing the heme plane.
Fig. 5.
Crystal structure of ligand-free Methylococcus capsulatus CYP51 (PDB ID 6mi0). (A) A Coot snapshot of the 2F0–Fc electron density map (contoured at 1.5 σ), map radius 50 Å. The density for the near P450 chains from the neighboring unit cells is seen at the top, left and right, the density for the fx domain is missing. (B) Ribbon representation of the P450 domain structure (blue, upper P450 face) and the fx domain model (magenta). Orientation is about the same as in (A). The residues that are expected to be involved in the (productive) protein–protein interaction are shown and labeled. (C) Superimposition of the M. capsulatus P450 domain with the Trypanosoma brucei (plum, PDB ID 3g1q, Cα RMSD, 1.6 Å), human (tan, PDB ID 4uhi, Cα RMSD, 1.7 Å), and Candida albicans (pink, PDB ID 5tz1, Cα RMSD, 1.9 Å) CYP51 orthologs. (D) Superimposition with CYP51 from Mycobacterium tuberculosis (red, PDB ID 1e9×, Cα RMSD, 5.4 Å). The substrate entrances are marked with the arrow of the corresponding color. (E) Heme support in M. capsulatus (blue), My. tuberculosis (red), and T. brucei (plum) structures. Y103, R361, and H420 (T. brucei numbering) are invariant across the whole CYP51 family, Y116 corresponds to F in bacterial and plant sequences, and R124 is protozoa-specific, its role being played by a Lys (located one turn downstream of helix C) in animal and fungal CYP51. Distal P450 face. The residues that correspond to T. brucei Y116 and R124 in the bacterial structures are shown in wire representation and labeled.
When overlaid with the other substrate-free CYP51 structures, the P450 domain of M. capsulatus CYP51fx revealed the highest similarity with protozoan T. brucei CYP51, Cα root-mean-square deviation (Cα RMSD) of 1.6 Å. The Cα RMSDs with human and fungal (Candida albicans) CYP51s are only slightly larger, 1.7 and 1.9 Å, respectively (fig. 5C). The greatest differences were observed with CYP51 from the nonsterol-producing bacterium My. tuberculosis (Cα RMSD of 5.4 Å), both in the conformation of the secondary structural elements that define the active site topology (fig. 5D), and in the “eukaryotic” location of the substrate channel entrance, on the distal P450 face, bordered by the A′ and F″ helices and the tip of the β4 hairpin, the same as in eukaryotic CYP51 (Lepesheva et al. 2010) and unlike My. tuberculosis CYP51, which has the channel entrance on the upper P450 face, bordered by helix B′, B′C loop, and helix C (Podust et al. 2001).
The protein support for the heme propionates, however, in both bacterial CYP51 structures is the same and includes the H-bonds from only three residues: Tyr (Y79 and Y76 [M. capsulatus and My. tuberculosis, respectively], B′ helix) and Arg (R327 and R326, β strand 1–4) to the ring A propionate and His (H392, the heme bulge) to the ring D propionate (fig. 5E). These three residues are invariant in all CYP51 sequences, whereas in protozoan, fungal, and animal CYP51s, two more amino acid side chains are involved in the interaction with the ring D propionate (Lepesheva et al. 2010; Hargrove et al. 2017). One of them is a Tyr on the B″ turn (Y116 in fig. 5E), the residue conserved across all these three kingdoms, but always represented by Phe in plants and in most bacteria (F92 in M. capsulatus and F89 in My. tuberculosis CYP51, fig. 5E), although some bacterial CYP51s, carry a Tyr in this position (table 1 andfig. 8B), like fungal, animal, and protozoan orthologs. The other residue is an Arg/Lys in helix C (R124 in fig. 5E). There is no conservation of the corresponding amino acid among plant and bacterial CYP51 sequences (N100 in M. capsulatus and K97 in My. tuberculosis; fig. 5E, see also supplementary fig. S3A, Supplementary Material online).
Table 1.
Taxonomy of Selecteda Bacteria Containing a CYP51 Gene, Sequence Identity, and “Phylum-Marker” Residues.
| Bacteria |
CYP51 |
||||||
|---|---|---|---|---|---|---|---|
| Group | Phylum (Class) | Order | Family | Examples | Sequence Identity, % P450/Fx | I81/F105b | F92/Y116 |
| Terrabacteria | Actinobacteria | Corynebacteriales | Dietziaceae | Dietzia timorensis | 43c | F | F |
| Gordoniaceae | Gordonia sp. HY186 | 43 | F | F | |||
| Skermania piniformis | 46 | F | F | ||||
| Nocardiaceae | Nocardia paucivorans | 47 | F | F | |||
| Williamsia limnetica | 45 | F | F | ||||
| Rhodococcus opacus | 45 | F | F | ||||
| Mycobacteriaceae | Mycobacteroides abscessus | 45 | F | F | |||
| Mycolicibacter algericus | 47 | F | F | ||||
| Mycobacterium tuberculosis | 47 | F | F | ||||
| Propionibacteriales | Nocardioidaceae | Marmoricola ginsengisoli | 42 | F | F | ||
| Pimelobacter simplex | 44 | F | F | ||||
| Pseudomonas sp. SLBN-26 | 42 | F | F | ||||
| Streptosporangiales | Streptosporangiaceae | Planobispora rosea | 46 | F | F | ||
| Nonomuraea sp. ATCC 55076 | 46 | F | F | ||||
| Thermomonosporaceae | Actinocorallia populi | 43 | F | F | |||
| Spirillospora albida | 46 | F | F | ||||
| Actinomadura kijaniata | 46 | F | F | ||||
| Pseudonocardiales | Pseudonocardiaceae | Amycolatopsis halophila | 46 | F | F | ||
| Amycolatopsis rhizosphaerae | 47 | F | F | ||||
| Amycolatopsis kentuckyensis | 45 | F | F | ||||
| Saccharopolyspora dendranthemae | 48 | F | F | ||||
| Acidimicrobiales | Acidimicrobiaceae | Acidimicrobiaceae bacterium | 48 | F | F | ||
| Microthrixaceae | Candidatus Microthrix d | ||||||
| parvicella Bio17-1 | 45/37 | F | F | ||||
| parvicella RN1 | 44/37 | F | F | ||||
| Streptomycetales | Streptomycetaceae | Streptomyces curacoi | 38 | F | Y | ||
| Cyanobacteria | Oscillatoriales | Oscillatoriaceae | Moorea sp. SIOASIH | 40 | F | Y | |
| Nostocales | Calotrichaceae | Calothrix sp. NIES-4071 | 38 | F | Y | ||
| Calothrix rhizosoleniae | 43 | F | v | ||||
| Oscillatoriales | Gomontiellaceae | Hormoscilla sp. GUM202 | 44 | F | Y | ||
| Chloroflexi (Dehalococcoidia) | Unclassified | Unclassified | Chloroflexi bacterium | 36 | F | Y | |
| Unclassified | Unclassified | Dehalococcoidia bacterium | 34 | F | Y | ||
| Firmicutes | Bacillales | Thermoactinomycetaceae | Kroppenstedtia sanguinis | 42 | F | F | |
| — | Proteobacteria (Gammaproteobacteria) | Methylococcales | Methylococcaceae | Methylococcus capsulatus d | 100/100 | I | F |
| Methylomagnum ishizawai d | 69/47 | L | F | ||||
| Methylocaldum marinum d | 65/55 | I | F | ||||
| Methylotetracoccus oryzae d | 64/42 | I | F | ||||
| Methylocaldum szegediense d | 63/48 | I | F | ||||
| Methylomicrobium alcaliphilum d | 58/42 | I | F | ||||
| Methylomicrobium kenyense d | 58/42 | I | F | ||||
| Methylobacter whittenburyi d | 57/40 | I | F | ||||
| Methylobacter luteus d | 56/40 | I | F | ||||
| Methylomicrobium buryatense d | 53/42 | I | F | ||||
| Methylomonas lenta d | 52/38 | I | F | ||||
| Methylobacter marinus d | 53/38 | I | |||||
| Methylosarcina lacus d | 52/40 | I | F | ||||
| Methylovulum sp.d | 51/40 | I | F | ||||
| Methylicorpusculum oleiharenae d | 55/33 | I | F | ||||
| Nevskiales | Sinobacteraceae | Stenotrophob. rhamnosiphilum d | 46/32 | L | F | ||
| Sinimarinibacterium sp. NLF-5-8d | 49/37 | L | F | ||||
| Algiphilaceae | Algiphilus aromaticivorans d | 45/35 | L | F | |||
| Oceanospirillales | Alcanivoracaceae | Ketobacter alkanivorans d | 48/30 | L | F | ||
| Alcanivoracaceae TS13_700 d | 55/35 | L | F | ||||
| Oceanospirillaceae | Oceanospirillaceae bacterium | 41 | A | F | |||
| Unclassified | Unclassified | Gammaproteobacteria-HGW d | 63/38 | I | F | ||
| Gammaproteobacteria WB5_2B d | 61/36 | I | F | ||||
| Cellvibrionales | Halieaceae | Halioglobus japonicus | 45 | A | F | ||
| Parahaliea mediterranea | 42 | F | Y | ||||
| Spongiibacteraceae | Spongiibacter tropicus d | 50/33 | A | F | |||
| Oceanicoccus sp. KOV_DT_Chl | 44 | A | F | ||||
| Unclassified | Cellvibrionales bacteriumd | 44/22 | L | F | |||
| Chromatiales | Unclassified | Chromatiales bacterium | 39 | F | Y | ||
| Pseudomonadales | Unclassified | Pseudomonadales bacteriumd | 47/36 | L | F | ||
| (Deltaproteobacteria) | Myxococcales | Nannocystaceae | Nannocystis exedens | 40 | L | Y | |
| Plesiocystis pacifica | 43 | L | Y | ||||
| Sandaracinaceae | Sandaracinus amylolyticus d | 35/30 | I | Y | |||
| Sandaracinus sp. NAT8d | 37/31 | L | F | ||||
| Polyangiaceae | Polyangium sp. SDU3-1 | 35 | F | Y | |||
| Unclassified | Enhygromyxa salina | 41 | L | Y | |||
| Minicystis rosea d | 42/37 | I | Y | ||||
| Myxococcales bacteriumd | 37/30 | I | Y | ||||
| (Alphaproteobacteria) | Rhodospirillales | Acetobacteraceae | Zavarzinia sp. HR-ASd | 50/33 | L | F | |
| Zavarzinia compransoris d | 49/31 | L | F | ||||
| Oleomonas sp. K1W22B-8d | 46/30 | L | F | ||||
| Sphingomonadales | Sphingomonadaceae | Novosphingobium tardaugens | 43 | L | Y | ||
| PVCe | Plantomyces | Unclassified | Planctomycetes bacterium | 36 | F | Y | |
| FCBf | Gemmatimonadetes | Gemmatimonadales; | Unclassified | Gemmatimonadales bacterium | 36 | F | Y |
| — | Spirochetes | Spirochaetales | Spirochaetaceae | Spirochaeta sp.d | 41/36 | F | F |
| — | Nitrospirae | Nitrospirales | Nitrospiraceae | Nitrospira sp. SB0677_bin_15 | 36 | F | Y |
| Unclassified | Candidatus Rokubacteria | 41 | Y | Y | |||
Representative species from each family and genus are shown, including bacteria that do not make sterols, for example, Actinobacteria.
Methylococcus capsulatus/Trypanosoma brucei numbering.
Identity to M. capsulatus CYP51 (H260).
In these organisms, CYP51 is naturally fused to ferredoxin (CYP51fx).
Named after the phyla Planctomycetes, Verrucomicrobia, and Chlamydiae.
Named after the main member phyla Fibrobacteres, Chlorobi, and Bacteroidetes (NCBI taxonomy).
Fig. 8.
CYP51 sequences from bacterial genomes. For simplicity, only one species per genus is shown. Trypanosoma brucei, T. cruzi, and human orthologs are used as references. (A) Phylogenetic tree (rendered in TreeDyn 198.3). Colored branches represent different bacterial phyla: Actinobacteria (blue), Cyanobacteria (cyan), Firmicutes (sky-blue), Chloroflexi (navy), Proteobacteria, α- (saddle brown), γ- (dark red), δ- (coral), Planctomycetes (seagreen), Gemmatomonadales (limegreen), Nitrospirae (spring green), and Spirochetes (olive). Species that make sterols are highlighted in bold. (B) Two CYP51 signature motives in the aligned bacterial sequences. Trypanosoma brucei numbering is presented on the top. Residues crucial for the CYP51 function and absolutely conserved in all biological domains are in yellow. Alignment and phylogenetic tree of 150 bacterial CYP51 proteins are available as supplementary figure S3A and B, Supplementary Material online, respectively.
Crystal Structure of CYP51fx with the Bound Detergent Anapoe-X-114
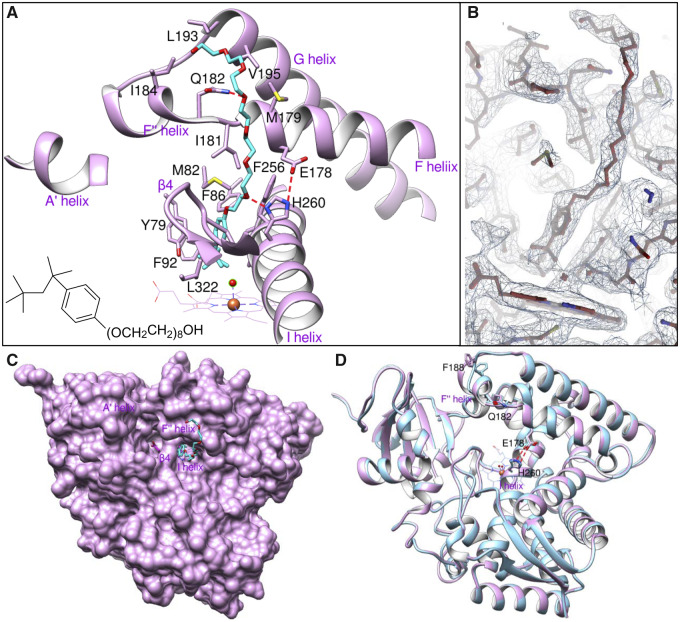
The crystal structure was solved to 2.4 Å resolution and also contained one P450 molecule per asymmetric unit with no electron density for the observed ferredoxin domain. Although addition of substrate (either lanosterol or eburicol) produced >95% high-spin P450 (fig. 3), and the sample remained in the high-spin state at least until the crystal plates were set, upon crystallization the substrate was expelled from the CYP51 active site by the detergent: the spectra of the dissolved crystals revealed low-spin P450, with the Soret band maximum returned to 417 nm. In the detergent-bound structure (fig. 6A and B), the heme iron is indeed hexa-coordinated: the detergent (23-(4-(2,4,4-trimethylpentan-2-yl)phenoxy)-3,6,9,12,15,18,21-heptaoxatricosan-1-ol) lies above the water molecule, which serves as the sixth (distal) ligand, like it does in structure 6MI0, though here it is shifted 0.7 Å toward the I-helix, and its distance to the iron is 2.4 Å (vs. 2.6 Å in 6mi0). The proximal methyl group of the detergent 2,4,4-trimethylpentan moiety is hanging 4.2 Å above the iron, the phenoxy ring plane is perpendicular to the heme plane, with the oxygen atom directed toward the I helix. The largest portion of the 3,6,9,12,15,18,21-heptaoxatricosan-1-ol arm is exposed to the bulk solvent, protruding above the protein surface. This explains its higher B-factor (supplementary table S2, Supplementary Material online), though it anchors to the outer wall of the F″ helix through a weak (3.7 Å) H-bond, linking oxygen atom 9 to the nitrogen of the Gln182 side chain. Thus, contrary to all other known CYP51-bound small molecules, the detergent must have entered the active site not through the substrate access channel, between helices A′, F″, and the tip of the β4 hairpin (although it probably expelled the substrate via this channel), but through the “water channel” that is sometimes seen between helices I, F, and the tip of the β4 hairpin in the CYP51 structures (Lepesheva et al. 2010; Lepesheva and Waterman 2011) (fig. 6A and C). It appears that the detergent reorients the imidazole ring of His260 (the CYP51 family signature histidine) involved in the proton delivery network (Hargrove et al. 2018; Hargrove et al. 2020), and its oxygen 18 forms a 3.0-Å H-bond with the imidazole N1 atom (fig. 6A), thus interfering with the “proper” closing of the conserved His260/Glu178 salt bridge (3.4 Å bond length), the event expected in substrate-free/inhibitor-bound CYP51 molecules (fig. 6D). Overall, except for the ∼2 Å rearrangements in helix F″, the ligand-free and detergent-bound structures are very similar (Cα RMSD of 0.37 Å), indicating that the closed (active) CYP51 conformation that is observed while the substrate is productively bound in the active site does not last when it is replaced by the detergent.
Fig. 6.
Crystal structure of the detergent-bound P450 domain of Methylococcus capsulatus CYP51fx (PDB ID 6mcw). (A) Interaction of Anapoe X 114 (cyan) with surrounding residues (within 4.5 Å, shown in plum sticks and labeled). Red dotted lines are H-bonds. Selected red sphere is a water molecule. (B) The 2F0–Fc electron density map (1.2 σ) for the detergent area. Orientation is similar to that in (A). (C) Surface representation. Heme ring A propionate (magenta) is seen through the substrate access channel. (D) Overlaid detergent-bound and ligand-free (blue, PDB ID 6mi0) structures, Cα RMSD of 0.37 Å. Distal P450 face in both cases.
Structural Insights into the Broad Substrate Tolerance of M. capsulatus CYP51fx
A structural explanation for the lack of obvious M. capsulatus CYP51fx substrate preferences toward eburicol over lanosterol or obtusifoliol (supplementary table S1, Supplementary Material online) could be that Ile81 in M. capsulatus CYP51, probably due to some structural disorder observed in this region, is positioned more like L134 in human CYP51 (fig. 7), which also has similar preference for lanosterol, eburicol, or obtusifoliol (Hargrove et al. 2016). This is unlike the I105 in T. cruzi CYP51, which prefers eburicol over lanosterol and even more over obtusifoliol (Lepesheva et al. 2006), or the F105 in T. brucei CYP51, which has a strict requirement for obtusifoliol (Lepesheva et al. 2004). To further test M. capsulatus CYP51fx substrate tolerance, we spectrally titrated the enzyme with three naturally occurring lanosterol isomers, cycloartenol (the initial sterol in plants and some bacteria), parkeol (one of the sterol end-products in an abbreviated pathway in bacterium G. obscuriglobus [Planctomyces]; Pearson et al. 2003), and cucurbitadienol (a phytosterol precursor found in some plant families; Shibuya et al. 2004). The classical type I spectral response was observed for cycloartenol and parkeol, showing 88% and 73% low- to high-spin transition of the heme iron with the calculated Kds of 0.46 and 0.26 µM, respectively (supplementary fig. S2A and B, Supplementary Material online). Additionally, a modified type I response to cucurbitadienol was seen. Based on the Soret band maximum at 417 nm, this was probably the result of an unproductive binding mode via a water molecule; Kd = 0.67 µM (supplementary fig. S2C, Supplementary Material online). As no spectral response to these sterols is produced by eukaryotic orthologs (human and T. cruzi CYP51; data not shown), these results support our structural findings. Consequently, we propose that ancestral bacterial CYP51 may have had broad substrate specificity, and this may be a reflection of various substrates made available to the enzyme from preceding steps. Bacteria encode for oxidosqualene cyclases (OSC) that act either as a lanosterol synthase or a cycloartenol synthase whereas in eukaryotes generally only a single class of OSC is found in any specific phylum (cycloartenol synthase in plants and algae vs. lanosterol synthase in animals, fungi, and protozoa). This may be of importance because the final products of many eukaryotic sterol biosynthetic pathways are utilized by other biochemical pathways to produce other important molecules, for example, hormones, brassinosteroids, signaling molecules, etc. Bacteria, on the other hand, may have relaxed CYP51 substrate specificity because the pathways are abbreviated and the sterol end-product(s) is not part of downstream enzymatic modifications.
Fig. 7.
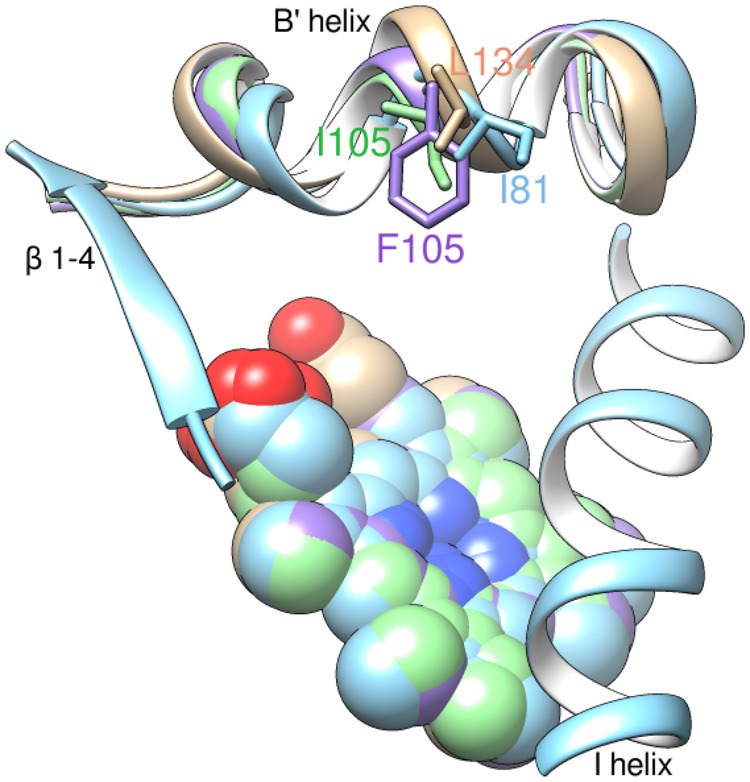
Ile81 in Methylococcus capsulatus CYP51 (blue, PDB ID 6mi0). A flexible loop-like turn can be seen in the middle of the B′ helix. Overlaid with Trypanosoma brucei (plum, PDB ID 3g1q), human (tan, PDB ID 4uhi), and T. cruzi (green, PDB ID 4ck8) CYP51 structures.
Unfortunately, determining the complete structure of M. capsulatus CYP51fx (which would be P450–ferredoxin complex) by X-ray crystallography remains problematic due to the lack of electron density for tracing the ferredoxin domain. The complex, at least in the absence of the substrate in the P450 active site, is either not strong enough or (more likely, in our opinion) the density for the small (8.4 kDa) ferredoxin is not seen because it can form multiple intermediate (transient) interactions (Schilder and Ubbink 2013) with the P450 molecule (52 kDa), reaching the/a productive binding mode only when the substrate is bound, and the electron transfer is expected. Indeed, to date only the X-ray structures of two artificial (genetically engineered) P450–ferredoxin complexes have been resolved: the cross-linked CYP101(P450cam)-putidoredoxin (Tripathi et al. 2013) and the partial structure of CYP11A1(P450scc)-adrenodoxin, a synthetic fusion (Strushkevich et al. 2011). In both of these cases, the P450 domain was substrate-bound. The most recently reported structure of a self-sufficient P450 monooxygenase CYP116B46 (a P450-reductase-ferredoxin in one polypeptide chain), although outlining the interaction between the FMN of the reductase domain and the Fe-S cluster of the ferredoxin domain, does not clearly reveal the productive interaction between the ferredoxin and the P450 domains (Zhang et al. 2020). Most likely this is because imidazole and not the substrate is bound in the P450 active site. The continued work on cocrystallization of M. capsulatus CYP51fx with bound lanosterol/eburicol is currently in progress, the spin state of the P450 heme iron in the crystals being monitored prior to data collection, as described in Materials and Methods.
Phylogenetic Analysis, Sequence Conservation, and the Evolutionary Origins of CYP51
Strikingly, bioinformatic analysis of >247,000 bacterial genomes (https://www.ncbi.nlm.nih.gov/genome/browse#!/prokaryotes/) reveals that >1,000 bacteria possess a CYP51 gene even though the vast majority of them do not biosynthesize sterols. Specifically, these CYP51 genes are found both in Gram-positive and in Gram-negative bacteria that include the Terrabacteria group [phyla Actinobacteria, Firmicutes, Chloroflexi, and Cyanobacteria], the PVC group [phylum Planctomycetes], the FCB group [phyla Gemmatimonadetes, Fibrobacteres, Chlorobi, and Bacteroidetes], and ungrouped phyla Nitrospirae, Spirochetes, and Proteobacteria (classes Gamma, Alpha, and Delta) (table 1 and fig. 8A). Alignment of 150 CYP51 representatives from nine bacterial phyla (including CYP51fx fusion proteins) and their phylogenetic tree can be seen as supplementary figure S3A and B, Supplementary Material online, respectively. These organisms are evolutionarily very distant, and, accordingly, their CYP51 sequence identities across bacterial phyla are low (30–45%). Even among order Methylococcales (where M. capsulatus belongs) the identity varies from 98% to 52%, the mean being ∼70%. The average CYP51 sequence identity in order Myxococcales is ∼45% (between 32% and 79%). Nevertheless, all these sequences possess the unique CYP51 signature motifs (fig. 8B). The B′ helix/B′C loop conservation is essential for the interaction with the sterol substrates, and the I-helical –HTt(s)– triad is required for the CYP51-specific proton delivery network (Lepesheva and Waterman 2011; Hargrove et al. 2020). Our finding of CYP51 sequences in four alpha-proteobacteria, Zavarzinia sp. HR‐AS (NCBI protein accession number WP_109905014.1), Zavarzinia compransoris (WP_109920848.1), Oleomonas sp. K1W22B‐8 (WP_119777160.1), Novosphingobium tardaugens (WP_021691104.1) (table 1), may indicate an alpha proteobacterial ancestor of mitochondria as a source of the first eukaryotic P450 given that Lokiarcheaum, the closest known living archaeon relative of eukaryotes, encodes no P450 genes (Spang et al. 2015).
Furthermore, >50 bacterial CYP51s (mostly Proteobacteria [including three alpha-proteobacteria, family Acetobacteraceae] but also Actinobacteria and Spirochaeta) (noted in table 1), like M. capsulatus CYP51, are native fusion proteins, where the P450 domain (N-terminus) is connected, via a 20–30 residues linker, to a ferredoxin domain (C-terminus). Methylococcus capsulatus-based alignment of ferredoxin domains from bacterial CYP51fx proteins is shown as supplementary figure S4, Supplementary Material online. These ferredoxins are 64–82 amino acid residues long, possessing three invariant cysteines that form the sulfur–iron cluster. The FeS cluster is responsible for the functioning of ferredoxins as electron transfer protein partners of most bacterial and all eukaryotic mitochondrial P450s (accepting electrons from NADPH/NADH and transferring to the P450 heme iron). The average sequence identity of the ferredoxin domains is 45%, ranging between 22% and 98%. Existence of such a large number of CYP51fx fusion proteins in different bacterial species is unprecedented to date, suggesting that it is unlikely to be a coincidence but rather provided an evolutionary advantage, for example, guaranteed electron transfer in conditions when molecular oxygen (dioxygen) was scarce in the burgeoning Earth atmosphere, the event preceding the formation of sterol-enforced membranes and leading to the appearance of eukaryotic forms of life (Nes and McKean 1977).
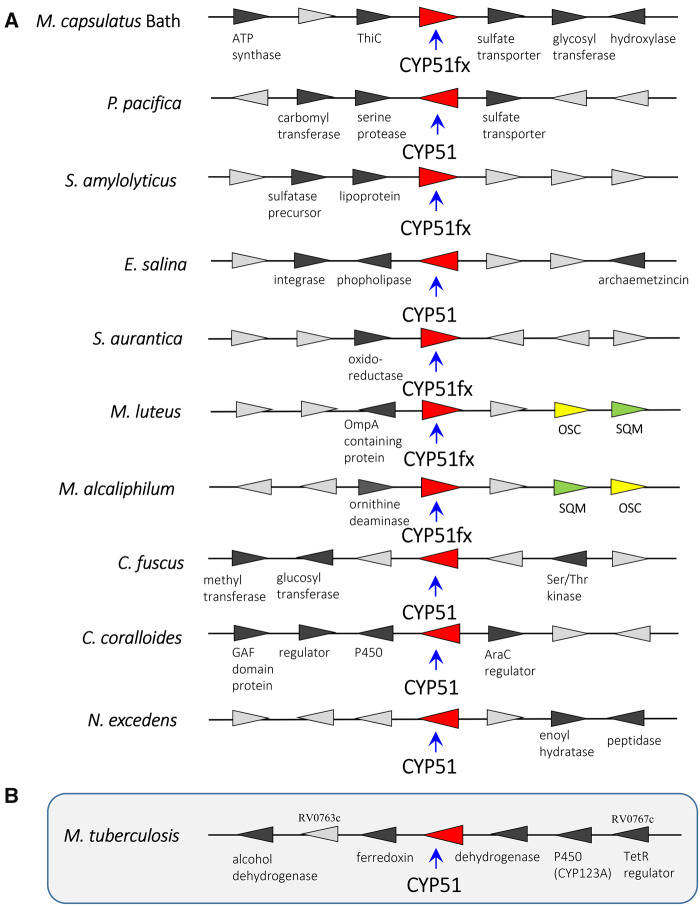
A separate root for bacterial sequences seen in the phylogenetic tree that includes CYP51 representatives from each biological kingdom (supplementary fig. S5, Supplementary Material online) is also consistent with common ancestry for eukaryotic CYP51s and CYP51s in modern bacteria. Different and variable gene synteny around CYP51fx and CYP51 genes present in the genomes of sequenced and annotated sterol and nonsterol-producing bacteria (fig. 9 and supplementary fig. S6, Supplementary Material online) provides additional evidence that bacterial CYP51s are not a result of horizontal gene transfer. On the contrary, bioinformatics analysis suggests a divergence hypothesis, that is, modern bacteria that do not make sterols but whose genomes still possess a CYP51 gene (or rather did not lose the CYP51 gene) are derived from ancestors that used to make sterols but evolved to stop producing them for some reasons, for example, as symbionts, parasites, etc.
Fig. 9.
Examples of bacterial CYP51 gene synteny. (A) CYP51fx and CYP51 genes found in sequenced and annotated genomes of sterol-producing bacteria (M. capsulatus Bath, Methylococcus capsulatus Bath; P. pacifica, Plesiocystis pacifica; S. amylolyticus, Sandaracinus amylolyticus; E. salina, Enhygromyxa salina; S. aurantica, Stigmatella aurantica; M. luteus, Methylobacter luteus; M. alcaliphilum, Methylomicrobium alcaliphilum; C. fuscus, Cystobacter fuscus; C. coralloides, Corallococcus coralloides; N. excedens, Nannocystis excedens) and (B) synteny of CYP51 in Mycobacterium tuberculosis, a representative bacterium that possess a CYP51 gene but does not make sterol. As predicted at the BioCyS Database (https://biocyc.org/gene? orgid=MTBH37RV&id=G185E-4912#tab=TU), genes RV0767c–RV0763c form a putative 6-gene operon and may act in a pathway with CYP123.
It is possible that within some species the resulting gene product evolved to new biological function(s). Such P450s could be recruited to other lipid (nonsterol) biochemical pathways involved in membrane biogenesis given that many of the nonsterol-producing bacteria synthesize complex lipids, for example, Mycobacteria sp. CYP51 is the third postsqualene step of sterol biosynthesis, preceded by squalene monooxygenase (SQM) and OSC reactions. In a similar fashion, an analogous scenario seems likely for the gene encoding SQM in bacteria. In sterol-producing bacteria, the SQM, which utilizes atmospheric oxygen, is fixed in sterol biosynthesis. However, it is observed that SQM genes are also distributed and retained in nonsterol-producing bacteria, also suggesting that these enzymes have evolved to new function(s) as sterol biosynthesis was lost over time. In contrast, the OSC enzyme is seemingly only found in sterol-producing organisms (supplementary fig. S6B and C, Supplementary Material online). This is probably due to the highly selective nature of the OSC catalytic reaction, being one of the most complex biochemical reactions known (the formation of four rings along the long chain of the substrate [oxidosqualene], producing lanosterol). It is likely that genes encoding OSC were not retained following loss of sterol biosynthesis because the complexity of its chemistry and lack of ease of application to new substrates and functions. Conversely, SQM and CYP51, which fix atmospheric oxygen, could use their chemistry efficiently with other molecules in evolving to new function(s), an example being reported for a CYP51 evolved to new function in the biosynthesis of avenacin in the roots of oat plants (Qi et al. 2006). We await crystal structural analysis of bacterial SQMs (from sterol and nonsterol-producing species) and subsequent comparison to the recent eukaryotic SQM structures (Padyana et al. 2019). Such comparisons may also reveal alterations in the enzyme SQM structural architecture through evolving to new functionality, as we have described herein for CYP51.
Conclusions
Our crystallographic data showed that the P450 domain of the sterol-producing M. capsulatus CYP51fx enzyme displays a greater overall structural similarity to eukaryotic CYP51s (20–26% amino acid sequence identity) than to nonsterol-producing My. tuberculosis CYP51, even though they share 47% amino acid sequence identity, and yet the H-bond network between the apoprotein and the heme propionates in M. capsulatus CYP51fx is the same as in My. tuberculosis CYP51 (and presumably in plant CYP51 orthologs [no structure available yet]). This finding strongly reinforces the notion that sterol 14α-demethylases, an essential enzyme of sterol biosynthesis, most probably derived from a bacterial origin and may indeed have been the ancestral progenitor for the whole P450 superfamily (Yoshida et al. 2000). The M. capsulatus CYP51fx enzyme may be evolutionarily closer to a predecessor from some ancient sterol-making bacteria that was cenancestor for: 1) all eukaryotic CYP51s (which further diverged to enable faster sterol flow), 2) modern bacterial CYP51s, and 3) possibly for other bacterial P450s (which evolved to acquire new, mostly simplified functions, such as one-step vs. three-step monooxygenation reactions, abilities to accommodate and metabolize compounds of various chemotypes or to accept electrons from different protein donors).
Finally, a wide distribution of CYP51 genes across bacterial phyla, a broad variability of bacterial CYP51 sequences, a separate cluster in the phylogenetic tree, and particularly the fact that bacterial CYP51 carry all of the amino acid residues that serve as phyla-specific markers for eukaryotic CYP51 ((table 1 and fig. 8), for example, in bacterial sequences there can be I (T. cruzi only), Y (plants/other protozoa), or L(animals/fungi) and more rarely, M, V, or A at the position that aligns with I81 in M. capsulatus CYP51, also, there can be either F or Y (invariant F in plants but always Y in animals/fungi/protozoa) that aligns with F92 (see also fig. 5E)), implies that both plant and animal/fungi sterol biosynthesis pathways could be derived from pathways initially developed in ancient aerobic bacteria.
Materials and Methods
Bioinformatic Analysis
The search for bacterial CYP51 sequences in the National Center for Biotechnology Information sequence databases was performed using NCBI protein–protein BLAST (TBlast suite) (Altschul et al. 1997) and M. capsulatus CYP51 as a protein query. Multiple sequence alignment was carried out in Clustal Omega and analyzed in GeneDoc. The phylogenetic tree was rendered in TreeDyn; branch support values (%) were generated in phylogeny analysis (one-click mode: MUSCLE-Gblocks-PyML) (http://www.phylogeny.fr/index.cgi) (Dereeper et al. 2010).
Protein Expression and Purification
Methylococcus capsulatus CYP51fx (P450Fx native fusion) in the pET17-b plasmid (Novagen) (Jackson et al. 2002) was expressed in E. coli. HMS-174 (DE3) (Novagen). The expression level was 300–400 nmol l−1. A single colony of bacteria was used to inoculate 5 ml of LB media containing 0.1 mg ml−1 ampicillin. An overnight culture was incubated at 37 °C and 250 rpm for 16 h and then diluted into 500 ml of TB media supplemented with 100 mM potassium phosphate buffer (pH 7.4) containing 0.1 mg ml−1 ampicillin and 125 µl of trace elements salt solution. After incubation at 37 °C and 250 rpm for 5 h, the flasks were cooled to 22 °C, the cultures were supplemented with 1 mM δ-aminolevulinic acid, 1 mM IPTG, and 1 mM FeCl3 (to ensure proper FeS cluster formation) and incubated for 20 h at 22 °C and 180 rpm. The cells were harvested by centrifugation at 1,850 × g for 15 min, the pellet was resuspended in 50 mM potassium phosphate buffer (pH 7.4) containing 0.1 mM EDTA, 100 mM NaCl, 10% glycerol (v/v), and 0.1% Triton X-100 (v/v) (buffer A). Lysozyme (25 mg ml−1) was added, and the suspension was placed on ice for 15 min and then, after addition of 0.5 mM PMSF, frozen at −80 °C. All purification steps were done at 4 °C, and all buffers contained 0.1 mM PMSF and 0.1 mM dithiothreitol, which were added fresh daily. The pellet was homogenized in buffer A, the suspension was sonicated on ice (Sonic Dismembrator model 500, Fisher Scientific), and stirred with 0.2% Triton X-100 (v/v) at 4 °C for 1 h. The solubilized protein was separated from the insoluble material by centrifugation at 82,000 × g for 30 min. The supernatant was frozen in liquid nitrogen and stored at −80 °C. Methylococcus capsulatus CYP51fx fusion was purified in two steps, including affinity chromatography on Ni2+-NTA agarose, and anion exchange chromatography on Q-Sepharose. The thawed supernatant was diluted 2-fold with buffer A and applied to Ni2+-NTA agarose equilibrated with the same buffer and the (NTA-) bound protein was washed with equilibration buffer and then with 50 mM potassium phosphate buffer (pH 7.4) containing 500 mM NaCl, 10% glycerol (v/v), and 1 mM imidazole until the Triton X-100 was eliminated (as judged by A280 measurements).Then the protein was washed with 20 mM potassium phosphate buffer (pH 7.4) containing 200 mM NaCl, 10% glycerol (v/v), 5, 10, and then 15 mM imidazole, and eluted with a linear gradient of imidazole (20–200 mM) in 20 mM potassium phosphate buffer (pH 7.4) containing 500 mM NaCl, 10% glycerol (v/v). The fractions with a spectrophotometric index (A425/A280) ≥1 were pooled and concentrated using an Amicon Ultra 50K (Millipore) to a volume of 2–4 ml. The protein was then diluted 25-fold with 20 mM potassium phosphate buffer (pH 8.0) containing 10% glycerol (v/v), 0.1 mM EDTA (Q buffer), and applied to a Q-Sepharose column equilibrated with Q buffer containing 20 mM NaCl. The column was washed with the equilibration buffer, and the protein was eluted with 20 mM potassium phosphate buffer (pH 7.6) containing 180 mM NaCl, 10% glycerol (v/v), and 0.1 mM EDTA, pooled and concentrated to ∼500 μM. The purity and molecular weight of the protein (63 kDa, 561 amino acid residues in length, including 451 residues of the P450 domain, 22 residues of the linker, 78 residues of the ferredoxin domain, and a 10 His tag at the C-terminus; Jackson et al. 2002) was confirmed by SDS–PAGE. The correctness of the genes was confirmed by DNA sequencing. Escherichia coli flavodoxin and flavodoxin reductase were expressed and purified as described (Jenkins and Waterman 1998). Spinach ferredoxin reductase was purchased from Sigma.
Spectroscopic Characterization
UV-visible spectra were recorded at ambient temperature in a dual-beam Shimadzu UV-240IPC spectrophotometer. P450 concentrations were determined from the Soret band absorbance in the absolute spectrum, using an absolute molar extinction coefficient (ε417) of 117 mM−1 cm−1 for the low-spin oxidized form of the protein or a difference molar extinction coefficient (Δε446–490) of 91 mM −1 cm−1 for the reduced carbon monoxide complex in the difference spectra. The spin states of P450 samples were estimated from the absolute spectra as the ratio ΔA393–470/ΔA418–470 (Lepesheva et al. 1999).
Substrate Binding Assay
Binding of lanosterol, eburicol, cycloartenol, parkeol, and cucurbitadienol to 2 μM M. capsulatus CYP51fx was monitored in a 50-mM phosphate buffer, pH 7.4, containing 100 mM NaCl and 0.1 mM EDTA. The sterols were dissolved in 45% (w/v) 2-hydroxypropyl-β-cyclodextrin (HPCD) to 0.5 mM concentration and added in 1 µl aliquots to 2 ml protein samples (concentration range 0.25–3 µM). Equal amounts of 45% HPCD solution were added to the reference cuvette to correct for the solvent-induced spectral perturbations. The Kd values were calculated by fitting the data for the substrate-induced absorbance changes in the difference spectra Δ(Amax−Amin) versus substrate concentration to the quadratic Morrison equation (ΔA=(ΔAmax/2E)((L + E+Kd)−((L + E+Kd)2−4LE)0.5)) using GraphPad Prism 6 (GraphPad, La Jolla, CA).
Reconstitution of Enzymatic Activity
The sterol 14α-demethylase activity of M. capsulatus CYP51fx was reconstituted with radiolabeled (3-3H) lanosterol, eburicol, and obtusifoliol, specific activity ∼4,000 dpm nmol−1, as described previously for CYP51 from My. tuberculosis (Lepesheva et al. 2001), except that the sterols were added from a 0.5-mM solution in 45% (w/v) HPCD. Escherichia coli flavodoxin and flavodoxin reductase (molar excess over P450 18 and 2, respectively), served as electron donor partners. The enzyme/substrate molar ratio was 1/25 (2/50 µM). The final reaction volume was 500 µl and contained 20 mM MOPS (pH 7.4), 50 mM KCl, 5 mM MgCl2, 10% (v/v) glycerol, 0.4 mg ml−1 isocitrate dehydrogenase, and 25 mM sodium isocitrate. The reaction was initiated by addition of 5 mM NADPH and stopped by extraction of the sterols with ethyl acetate. The extracted sterols were analyzed by a reversed-phase HPLC system (Waters) equipped with a β-RAM detector (INUS Systems). In order to verify the ability of the fusion M. capsulatus ferredoxin domain to transfer electrons to the P450 domain, we used spinach ferredoxin reductase (Jackson et al. 2002). In this case, the CYP51fx concentration was increased to 10 µM, the concentration of ferredoxin reductase was 25 µM. At these conditions, 25% of lanosterol conversion was observed in a 1-h reaction.
Crystallization and Determination of the X-ray Structures
Crystals were obtained at 24 °C by vapor diffusion in hanging drops containing equal volumes of the protein and well solution. The crystals of ligand-free M. capsulatus CYP51fx were grown from a 250-µM protein sample in 10 mM potassium phosphate buffer (pH 7.4) containing 100 mM NaCl, 5.6 mM tris(2-carboxyethyl)phosphine (TCEP), 5% glycerol (v/v) and 2% ANAPOE X-114 (α-[(1,1,3,3-tetramethylbutyl)phenyl]-w-hydroxy-poly(oxy-1,2-ethanediyl)) (Anatrace) mixed with 0.2 M lithium acetate (pH 7.5) and 18% PEG 3,350 (w/v). Methylococcus capsulatus CYP51fx was crystallized in the presence of the detergent since it could not be crystallized in its absence. The crystals of M. capsulatus CYP51fx in complex with the detergent were obtained in an attempt to cocrystallize substrate-bound protein, because we expected that binding of the substrate should induce the large-scale conformational rearrangement in the CYP51 molecule, which should strengthen P450–ferredoxin interaction (Hargrove et al. 2018; Hargrove et al. 2020). The M. capsulatus CYP51fx sample (5 μM in 20 mM potassium phosphate buffer [pH 7.4] containing 200 mM NaCl, 5.6 mM TCEP, and 10% glycerol [v/v]) was gradually saturated with eburicol (using a 0.5-mM stock solution in 45% [w/v] HPCD, because of the limited solubility of this highly hydrophobic sterol, Log P 8.6; Hargrove et al. 2018), incubated for 20 min at room temperature, concentrated using an Amicon Ultra 50K (Millipore) to 500 µM, diluted 2-fold with 5 mM phosphate buffer (pH 7.4), and mixed with ANAPOE X-114 (final concentration 2%). The high-spin (substrate-bound) state of the P450 heme in the mixture was spectrally confirmed. The well solution consisted of 0.2 M lithium acetate (pH 7.5) and 15% PEG 6,000 (w/v). After harvesting, crystals were cryoprotected with 25% (v/v) glycerol and frozen in liquid nitrogen. The X-ray diffraction data were collected at the Advanced Photon Source, Argonne National Laboratory, 21-ID-F (0.97872 nm), and 21-ID-D (1.07807 nm) beamlines, respectively. The diffraction images were processed with HKL-2000, and crystal structures solved by molecular replacement with Phaser MR (CCP4 Program Suite; Potterton et al. 2003) using T. brucei CYP51 (PDB ID 3g1q) as a search model. The structure was built with Coot (Emsley et al. 2010) and refined with Refmac5 (CCP4 Suite). Details of the data collection and refinement statistics are listed in supplementary table S2, Supplementary Material online. After determining the structure and observing ANAPOE X-114 instead of the substrate in the P450 active site, the crystals were reproduced at the same conditions, harvested, dissolved in 10 mM potassium phosphate buffer (pH 7.4) containing 100 mM NaCl, 5.6 mM TCEP, and 5% glycerol (v/v), and the absorbance spectra (2 µl samples) were taken using a Nanovue 4282 V1.7.3 spectrophotometer as described previously (Hargrove et al. 2018). The Soret maximum of the enzyme was 417 nm, exactly corresponding to the low-spin water coordinated P450 (see fig. 6A) and indicating that the detergent replaced the substrate upon crystallization. Structure superimposition and RMSD calculation was performed in lsqcab (CCP4 Suite). Molecular graphics were rendered using Chimera. The model of M. capsulatus ferredoxin was built in Modeler (CCP4 Suite) based on the structure of adrenodoxin (PDB ID 1AYF) with 32 truncated amino acid residues at the N-terminus.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The study was supported by National Institutes of Health (Grant No. R01 GM067871 to G.I.L.) and by a UK-USA Fulbright Scholarship and the Royal Society (to D.C.L.).
Data Availability
The atomic coordinates and structure factors (accession codes 6MI0 and 6MCW) have been deposited in the Protein Data Bank (http://wwpdb.org/), and all other data underlying this article and in its online Supplementary Matarial .
References
- Abe I. 2007. Enzymatic synthesis of cyclic triterpenes. Nat Prod Rep. 24(6):1311–1331. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta AB, Wei JH, Welander PV.. 2015. A distinct pathway for tetrahymanol synthesis in bacteria. Proc Natl Acad Sci U S A. 112(44):13478–13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird CW, Lynch JM, Pirt FJ, Reid WW, Brooks CJW, Middleditch BS.. 1971. Steroids and squalene in Methylococcus capsulatus grown on methane. Nature 230(5294):473–474. [DOI] [PubMed] [Google Scholar]
- Bode HB, Zeggel B, Silakowski B, Wenzel SC, Reichenbach H, Müller R.. 2003. Steroid biosynthesis in prokaryotes: identification of myxobacterial steroids and cloning of the first bacterial 2,3(S)-oxidosqualene cyclase from the myxobacterium Stigmatella aurantiaca. Mol Microbiol. 47(2):471–481. [DOI] [PubMed] [Google Scholar]
- Bowen AM, Johnson EOD, Mercuri F, Hoskins NJ, Qiao R, McCullagh JSO, Lovett JE, Bell SG, Zhou W, Timmel CR, et al. 2018. A structural model of a P450-ferredoxin complex from orientation-selective double electron-electron resonance spectroscopy. J Am Chem Soc. 140(7):2514–2527. [DOI] [PubMed] [Google Scholar]
- Debeljak N, Fink M, Rozman D.. 2003. Many facets of mammalian lanosterol 14α-demethylase from the evolutionarily conserved cytochrome P450 family CYP51. Arch Biochem Biophys. 409(1):159–171. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie J-M, Blanc G.. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 10(1):8–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond E, Gribaldo S.. 2009. Phylogenomics of sterol synthesis: insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biol Evol. 1:364–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K.. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 66(4):486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP. 2001. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 14(6):611–650. [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Yoshimoto FK.. 2018. Formation and cleavage of C-C bonds by enzymatic oxidation-reduction reactions. Chem Rev. 118(14):6573–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannemann F, Bichet A, Ewen KM, Bernhardt R.. 2007. Cytochrome P450 systems—biological variations of electron transport chains. Biochim Biophys Acta. 1770(3):330–344. [DOI] [PubMed] [Google Scholar]
- Hargrove TY, Friggeri L, Wawrzak Z, Qi A, Hoekstra WJ, Schotzinger RJ, York JD, Guengerich FP, Lepesheva GI.. 2017. Structural analyses of Candida albicans sterol 14α-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis. J Biol Chem. 292(16):6728–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove TY, Friggeri L, Wawrzak Z, Sivakumaran S, Yazlovitskaya EM, Hiebert SW, Guengerich FP, Waterman MR, Lepesheva GI.. 2016. Human sterol 14α-demethylase as a target for anticancer chemotherapy: towards structure-aided drug design. J Lipid Res. 57(8):1552–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove TY, Wawrzak Z, Fisher PM, Child SA, Nes WD, Guengerich FP, Waterman MR, Lepesheva GI.. 2018. Binding of a physiological substrate causes large-scale conformational reorganization in cytochrome P450 51. J Biol Chem. 293(50):19344–19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove TY, Wawrzak Z, Guengerich FP, Lepesheva GI.. 2020. A requirement for an active proton delivery network supports a Compound I mediated C-C bond cleavage in CYP51 catalysis. J Biol Chem. 295(29):9998–10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CJ, Lamb DC, Marczylo TH, Warrilow AG, Manning NJ, Lowe DJ, Kelly DE, Kelly SL.. 2002. A novel sterol 14α-demethylase/ferredoxin fusion protein (MCCYP51FX) from Methylococcus capsulatus represents a new class of the cytochrome P450 superfamily. J Biol Chem. 277(49):46959–46965. [DOI] [PubMed] [Google Scholar]
- Jenkins CM, Waterman MR.. 1998. NADPH-flavodoxin reductase and flavodoxin from Escherichia coli: characteristics as a soluble microsomal P450 reductase. Biochemistry 37(17):6106–6113. [DOI] [PubMed] [Google Scholar]
- Kitazume T, Haines DC, Estabrook RW, Chen B, Peterson JA.. 2007. Obligatory intermolecular electron-transfer from FAD to FMN in dimeric P450BM-3. Biochemistry 46(42):11892–11901. [DOI] [PubMed] [Google Scholar]
- Lepesheva GI, Friggeri L, Waterman MR.. 2018. CYP51 as drug targets for fungi and protozoan parasites: past, present and future. Parasitology 145(14):1820–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesheva GI, Nes WD, Zhou W, Hill GC, Waterman MR.. 2004. CYP51 from Trypanosoma brucei is obtusifoliol-specific. Biochemistry 43(33):10789–10799. [DOI] [PubMed] [Google Scholar]
- Lepesheva GI, Park HW, Hargrove TY, Vanhollebeke B, Wawrzak Z, Harp JM, Sundaramoorthy M, Nes WD, Pays E, Chaudhuri M, et al. 2010. Crystal structures of Trypanosoma brucei sterol 14α-demethylase and implications for selective treatment of human infections. J Biol Chem. 285(3):1773–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesheva GI, Podust LM, Bellamine A, Waterman MR.. 2001. Folding requirements are different between sterol 14α-demethylase (CYP51) from Mycobacterium tuberculosis and human or fungal orthologs. J Biol Chem. 276(30):28413–28420. [DOI] [PubMed] [Google Scholar]
- Lepesheva GI, Strushkevich NV, Usanov SA.. 1999. Conformational dynamics and molecular interaction reactions of recombinant cytochrome P450scc (CYP11A1) detected by fluorescence energy transfer. Biochim Biophys Acta. 1434(1):31–43. [DOI] [PubMed] [Google Scholar]
- Lepesheva GI, Waterman MR.. 2011. Structural basis for conservation in the CYP51 family. Biochim Biophys Acta. 1814(1):88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesheva GI, Zaitseva NG, Nes WD, Zhou W, Arase M, Liu J, Hill GC, Waterman MR.. 2006. CYP51 from Trypanosoma cruzi: a phyla-specific residue in the B′ helix defines substrate preferences of sterol 14α-demethylase. J Biol Chem. 281(6):3577–3585. [DOI] [PubMed] [Google Scholar]
- McLean KJ, Dunford AJ, Sabri M, Neeli R, Girvan HM, Balding PR, Leys D, Seward HE, Marshall KR, Munro AW.. 2006. CYP121, CYP51 and associated redox systems in Mycobacterium tuberculosis: towards deconvoluting enzymology of P450 systems in a human pathogen. Biochem Soc Trans. 34(6):1178–1182. [DOI] [PubMed] [Google Scholar]
- McLean KJ, Girvan HM, Munro AW.. 2007. Cytochrome P450/redox partner fusion enzymes: biotechnological and toxicological prospects. Expert Opin Drug Metab Toxicol. 3(6):847–863. [DOI] [PubMed] [Google Scholar]
- Nelson DR. 1999. Cytochrome P450 and the individuality of species. Arch Biochem Biophys. 369(1):1–10. [DOI] [PubMed] [Google Scholar]
- Nes WR, McKean MR.. 1977. Biochemistry of steroids and other isopentenoids. Baltimore: University Park Press. [Google Scholar]
- Omura T, Sato R.. 1964. The carbon monoxide-binding pigment of liver microsomes. J Biol Chem. 239:2379–2385. [PubMed] [Google Scholar]
- Ourisson G, Albrecht P.. 1992. Hopanoids. 1. Geohopanoids: the most abundant natural products on Earth? Acc Chem Res. 25(9):398–402. [Google Scholar]
- Padyana AK, Gross S, Jin L, Cianchetta G, Narayanaswamy R, Wang F, Wang R, Fang C, Lv X, Biller SA, et al. 2019. Structure and inhibition mechanism of the catalytic domain of human squalene epoxidase. Nat Commun. 10(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson A, Budin M, Brocks JJ.. 2003. Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci U S A. 100(26):15352–15357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust LM, Poulos TL, Waterman MR.. 2001. Crystal structure of cytochrome P450 14α-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc Natl Acad Sci U S A. 98(6):3068–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E.. 2003. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 59(7):1131–1137. [DOI] [PubMed] [Google Scholar]
- Rezen T, Debeljak N, Kordis D, Rozman D.. 2004. New aspects on lanosterol 14α-demethylase and cytochrome P450 evolution: lanosterol/cycloartenol diversification and lateral transfer. J Mol Evol. 59(1):51–58. [DOI] [PubMed] [Google Scholar]
- Qi X, Bakht S, Qin B, Leggett M, Hemmings A, Mellon F, Eagles J, Werck-Reichhart D, Schaller H, Lesot A, et al. 2006. A different function for a member of an ancient and highly conserved cytochrome P450 family: from essential sterols to plant defense. Version 2. Proc Natl Acad Sci U S A. 103(49):18848–18853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáenz JP, Grosser D, Bradley AS, Lagny TJ, Lavrynenko O, Broda M, Simons K.. 2015. Hopanoids as functional analogues of cholesterol in bacterial membranes. Proc Natl Acad Sci U S A. 112(38):11971–11976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilder J, Ubbink M.. 2013. Formation of transient protein complexes. Curr Opin Struct Biol. 23(6):911–918. [DOI] [PubMed] [Google Scholar]
- Schouten S, Bowman JP, Rijpstra WI, Sinninghe Damsté JS.. 2000. Sterols in a psychrophilic methanotroph, Methylosphaera hansonii. FEMS Microbiol Lett. 186(2):193–195. [DOI] [PubMed] [Google Scholar]
- Sezutsu H, Le Goff G, Feyereisen R.. 2013. Origins of P450 diversity. Philos Trans R Soc B. 368(1612):20120428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya M, Adachi S, Ebizuka Y.. 2004. Cucurbitadienol synthase, the first committed enzyme for cucurbitacin biosynthesis, is a distinct enzyme from cycloartenol synthase for phytosterol biosynthesis. Tetrahedron 60(33):6995–7003. [Google Scholar]
- Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema T.. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521(7551):173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strushkevich N, MacKenzie F, Cherkesova T, Grabovec I, Usanov S, Park H-W.. 2011. Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system. Proc Natl Acad Sci U S A. 108(25):10139–10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S, Li H, Poulos TL.. 2013. Structural basis for effector control and redox partner recognition in cytochrome P450. Science 340(6137):1227–1230. [DOI] [PubMed] [Google Scholar]
- van Wyk R, van Wyk M, Mashele SS, Nelson DR, Syed K.. 2019. Comprehensive comparative analysis of cholesterol catabolic genes/proteins in Mycobacterial species. Int J Mol Sci. 20(5):1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward N, Larsen Ø, Sakwa J, Bruseth L, Khouri H, Durkin AS, Dimitrov G, Jiang L, Scanlan D, Kang KH, et al. 2004. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2(10):e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei JH, Yin X, Welander PV.. 2016. Sterol synthesis in diverse bacteria. Front Microbiol. 7:990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Aoyama Y, Noshiro M, Gotoh O.. 2000. Sterol 14-demethylase P450 (CYP51) provides a breakthrough for the discussion on the evolution of cytochrome P450 gene superfamily. Biochem Biophys Res Commun. 273(3):799–804. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Noshiro M, Aoyama Y, Kawamoto T, Horiuchi T, Gotoh O.. 1997. Structural and evolutionary studies on sterol 14-demethylase P450 (CYP51), the most conserved P450 monooxygenase: II. Evolutionary analysis of protein and gene structures. J Biochem. 122(6):1122–1128. [DOI] [PubMed] [Google Scholar]
- Zhang L, Xie Z, Liu Z, Zhou S, Ma L, Liu W, Huang JW, Ko TP, Li X, Hu Y, et al. 2020. Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase. Nat Commun. 11(1):2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors (accession codes 6MI0 and 6MCW) have been deposited in the Protein Data Bank (http://wwpdb.org/), and all other data underlying this article and in its online Supplementary Matarial .