Abstract
Feed efficiency (FE) is an economically important trait in pig production. Gut microbiota plays an important role in energy harvest, nutrient metabolism, and fermentation of dietary indigestible components. Whether and which gut microbes affect FE in pigs are largely unknown. Here, a total of 208 healthy Duroc pigs were used as experimental materials. Feces and serum samples were collected at the age of 140 d. We first performed 16S rRNA gene and metagenomic sequencing analysis to investigate the relationship between the gut microbiome and porcine residual feed intake (RFI). 16S rRNA gene sequencing analysis detected 21 operational taxonomic units showing the tendency to correlation with the RFI (P < 0.01). Metagenomic sequencing further identified that the members of Clostridiales, e.g., Ruminococcus flavefaoiens, Lachnospiraceae bacterium 28-4, and Lachnospiraceae phytofermentans, were enriched in pigs with low RFI (high-FE), while 11 bacterial species including 5 Prevotella spp., especially, the Prevotella copri, had higher abundance in pigs with high RFI. Functional capacity analysis suggested that the gut microbiome of low RFI pigs had a high abundance of the pathways related to amino acid metabolism and biosynthesis, but a low abundance of the pathways associated with monosaccharide metabolism and lipopolysaccharide biosynthesis. Serum metabolome and fecal short-chain fatty acids were determined by UPLC-QTOF/MS and gas chromatography, respectively. Propionic acid in feces and the serum metabolites related to amino acid metabolism were negatively correlated with the RFI. The results from this study may provide potential gut microbial biomarkers that could be used for improving FE in pig production industry.
Keywords: feed efficiency, gut microbiota, 16S rRNA gene, metagenomic sequencing, metabolites, pigs
Introduction
Feed accounts for most costs of the pig industry (Jing et al., 2015). Therefore, improving feed efficiency (FE) will increase the economic benefits of the pig industry. FE can generally be expressed as residual feed intake (RFI) or feed conversion rate (FCR). RFI is also named net FE which is defined as the feed intake adjusted for the requirements of maintenance and body weight gain (DiGiacomo et al., 2018). Therefore, the pigs with lower RFI will have higher FE.
A variety of factors including genetics, diets, host health, and gut microbiota can affect FE. Ding et al. (2018) identified 2 quantitative trait loci on SSC1 and SSC7 that affect RFI and FCR. Gondret et al. (2017) showed that genes enriched in immune response, oxidative stress response, and protein metabolism were the main biological markers to distinguish high- and low-FE. The disease has also shown an adverse impact on reducing pig growth speed, which results in decreasing FE (Straw et al., 1990).
Gut microbiota can ferment and digest dietary polysaccharides and other substances (Zhang et al., 2018), promote nutrient absorption and energy acquisition (Hooper et al., 2002), and regulate physiological processes such as adaptive and innate immunity (Willing and Van Kessel, 2010). Some studies have revealed possible links between gut microbiota and FE in pigs. McCormack et al. (2019) indicated that gut microbiota might not be a dominant factor impacting the FE. Yang et al. (2017) found that 2 enterotype-like groups dominated by Treponema and Prevotella, respectively, had a significant association with FE. Tan et al. (2017)identified that the species Prevotella sp. CAG:604 had a higher abundance in the gut microbiome of pigs with low-FE, while pyruvate-related metabolic pathways were enriched in the gut microbiome of pigs with high-FE. Quan et al. (2019) showed that pigs with high FE had higher abundances of operational taxonomic units (OTUs) belonging to Lachnospiraceae and Prevotellaceae that had a strong ability to degrade dietary fiber, polysaccharide, and protein. However, these studies only focused on the association between the composition of gut microbiota and FE. In recent years, the relationship between the metabolites of gut microbiota and host phenotype has attracted comprehensive interest (Salek et al., 2007; Gong et al., 2020). The metabolites produced by the host or gut microbiota may be an effective means to regulate the phenotype (Clemmons et al., 2017; Wang and Kadarmideen, 2020).
In this study, we first used 16S rRNA gene and metagenomic sequencing methods to investigate the association of bacterial composition and functional capacity of the gut microbiome with porcine FE (RFI). Then, serum metabolome and fecal short-chain fatty acids (SCFAs) were determined to evaluate the correlation between RFI, fecal SCFAs, and serum metabolome. By combining these datasets, we investigate the possible mechanism of gut microbiota affecting pig FE.
Materials and Methods
All animal procedures involved in the experiment were conducted following the guidelines for the care and use of experimental animals established by the Ministry of Agriculture and Rural Affairs of China. This project was also approved by the Animal Care and Use Committee of Jiangxi Agricultural University.
Animal management
A total of 208 Duroc pigs from a commercial pig farm were used as experimental materials, including 70 gilts and 138 boars. All experimental pigs were weaned at 28 d of age and raised under similar management and feeding manner. When the body weight of piglets achieved 30 kg, the pigs were transferred to the fattening house. In the fattening stage, the same commercial formula feed was provided to all experimental pigs. The feed was mainly composed of corn, soybean meal, soybean oil, lysine, and calcium hydrogen phosphate. It contained about 15% crude protein, 1.5% crude fat, 5% crude fiber, 6% crude ash, 0.8% lysine, 0.9% calcium, 0.5% phosphorus, and 0.3% salt. Feed and water were available ad libitum. Osborne automatic feeders (Osborne Industries) were used to record feeding behaviors of pigs, including daily feed intake, daily body weight gain, and daily eating visiting times during the period from 30 to 100 kg (age about 90 to 170 d) in the fattening house. B-ultrasonoscope was used to measure the porcine backfat thickness. Those pigs treated with antibiotics a month before collecting fecal samples were removed from this study.
Phenotype measurement
The following model was used to calculate the RFI of the experimental pigs: where DFI refers to average daily feed intake, α is intercept, MBW represents metabolic body weight, ADG is average daily gain, β 1 and β 2 indicate the corresponding effects estimated for MBW and ADG, e represents the uncontrolled error. For calculating the MBW, we used the formula: where w0 is the body weight at the start of measurement and w1 represented the body weight at the end of the measurement (Haer et al., 1993). The RFI values obtained from the age of day 100 to day 160 (intermediate stage of phenotypic measurement) were used for the following analysis.
Sample collecting, amplicon sequencing of 16S rRNA gene, and data processing
Fecal samples were collected on March 20 and April 20 of 2017, respectively. At the age of about 140 d, fecal samples were collected from the anus of healthy experimental pigs at 9 to 11 a.m. after feeding. One gram of feces was put into a 1.5-mL Eppendorf tube, quickly dipped in liquid nitrogen, and stored at –80 °C until use. Fecal DNA was extracted using QIAamp Fast DNA Stool Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions (McOrist et al., 2002). The quality and quantity of DNA samples were checked using 0.8% agar gel electrophoresis and a Nanodrop-1000 (ThermoFisher). The DNA samples were then diluted at the proportion of 1:10, and 1 μL of the diluted DNA sample was used for PCR amplification. We used conservative primers 338F (5′-ACTCCTACGGGAGGCAGCA) and 806R (5′-GGACTACHVGGGTWTCTAAT) to amplify the V3–V4 hypervariable region of the 16S rRNA gene. PCR products were purified and used to construct libraries for sequencing on a MiSeq platform (Illumina). The primer and barcode sequences were removed from raw data using Trimmomatic (V.0.39; Bolger et al., 2014). FLASH (v.1.2.11) was used to merge paired-end clean sequence reads into tags (Magoč and Salzberg, 2011). Chimeric reads were removed using USEARCH (v.7.0.1090) (Edgar, 2010). To avoid the effect of sequencing depth on the microbial composition, sequence data were rarefied to 16,000 tags per sample. VSEARCH software (v.2.8.1) was used to cluster tags into OTUs based on 97% sequence identity (Rognes et al., 2016). Taxonomic assignments for the 16S rRNA gene sequences were performed using the RDP classifier program (v2.2; Wang et al., 2007). Alpha-diversity was calculated using MOTHUR (v1.31.2; Venkataraman et al., 2015). The corr.test in the R package (Version 4.0.0) was used to calculate the Spearman correlation between the α-diversity of gut microbiota and the RFI (Revelle, 2019). The 16S rRNA gene sequencing data were submitted to the China National GeneBank database (CNGBdb) with the accession number of CNP0000828.
Bacterial co-abundance groups (CAGs)
A total of 745 OTUs, all of which had an average of relative abundance greater than 1 ×10–4 and were presented in at least 20% of individuals were used for constructing CAGs. The correlations among OTUs were calculated by the SparCC method (Friedman and Alm, 2012). The correlation values were converted to the correlation distance matrix. The hclust function (ward. D2 algorithm) in R package was used to cluster the matrix into 8 CAGs (Murtagh and Legendre, 2014). These CAGs were further determined by permutational multivariate analysis of variance (PerMANOVA) with 999 permutations (P < 0.05; Zhang et al., 2016). We used Cytoscape (version 3.7.1) to visualize the network of OTUs under the weight value >0.5 (Shannon et al., 2003). The average abundance of all OTUs in each CAG was treated as the abundance of that CAG. Then, we computed the correlation between CAGs and the RFI values using the corr.test function in the R package (Fang et al., 2019).
The association between RFI and OTUs by a 2-part model
A 2-part model was applied to test the association between OTUs and the RFI (Fu et al., 2015). The advantage of this model is that it takes into account both binary and quantitative features of gut microbes and overcomes the problem of the non-normal distribution of microbial abundances. The binary analysis was to test for the effect of the presence/absence of a microbe on the RFI with the model where y is the RFI value of the tested samples after adjusting for sex, β 1 represents the estimated binary effect of a microbe, b refers to a binary feature, and e is the residual. The quantitative analysis was used to test the association between the abundance of the detected microbes and the RFI using the model where y refers to the RFI value per sample after adjusting for sex, β 2 represents the estimated effect of microbial abundance, q is the relative abundance of each OTU, and e is the residual. An unweighted Z-score method was used to obtain a meta P-value by combining the effect of both binary and quantitative analyses. The final P-value was derived from the minimum of P-values from binary analysis, quantitative analysis, and meta-analysis. The Z-score was calculated based on the Z distribution. Moreover, we performed 1,000× permutation tests to control the false discovery rate (FDR). The significance threshold was set at FDR < 0.05.
Metagenomic sequencing analysis
The RFI values of all 208 experimental pigs were adjusted to the effects of sex and batch. According to the residuals, the samples ranked the top 4 and the lowest 4 were selected for metagenomic sequencing (Supplementary Figure 1). DNA libraries were constructed according to the manufacturer’s instructions and sequenced on a Hiseq2500 platform (Illumina). We used the same procedures as described by Qin et al. (2012) to perform the bioinformatics analysis of the metagenomic sequencing data. Reads containing more than 10% of uncertain bases (N bases), adaptor sequences (15 bases or longer regions aligned to adaptor sequences), and sequencing reads with Q < 20 were removed. Moreover, the reads having more than 90 % sequence identity with the pig genome were excluded from further analyses by blasting with pig reference genome assembly Sscrofa 11.1. The high-quality sequence reads were assembled using SOAPdenovo2 (Luo et al., 2012). The assembly was grouped into longer contigs using Rabbit (You et al., 2013). Contigs <500 bp were filtered out from the subsequent analysis. We used MetaGeneMark (v2.10) to predict open reading frames (Zhu et al., 2010). The predicted genes were combined and clustered using CD-Hit software (v4.6.1) to remove redundant genes at the threshold of sequence identity >95% and >90% of sequence overlap (Li and Godzik, 2006). The non-redundant gene set was annotated to the CAZy database (carbohydrate-active enzymes; Lombard et al., 2014) and the KEGG database (Kyoto Encyclopedia of Genes and Genomes; Kanehisa et al., 2008) using BLAST software (Altschul et al., 1990). MEGAN (v4.6; Huson et al., 2007) was used for taxonomic assignment of metagenomic sequencing data by blasting the gene set against the NR database (non-redundant protein sequence database) with the LCA algorithm. LEfSe analysis was performed to compare the abundances of bacterial species and functional capacities of the gut microbiome between high and low RFI pigs. We calculated the Spearman correlation between differential bacterial species and differential functional genes with the corr.test function in the R package. Metagenomic sequencing data were submitted to the CNGB database with the accession number of CNP0000824.
Determination of serum metabolomic profiles
We collected 50 serum samples (20 from females and 30 from males) from the present experimental pig cohort when feces samples were collected at 9 to 11 a.m. after feeding. All samples were performed the untargeted metabolome analysis with ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UPLC-QTOF/MS). All serum samples were thawed on ice and precipitated with pre-cooled methanol. Three hundred microliter methanol was added into 100 μL serum, vortexed for 1 min and conserved at –20 °C for 20 min. The mixture was centrifuged at 14,000 rcf for 15 min at 4 °C. The supernatant was pipetted into a clean tube and dried in a savant vacuum evaporator. The dried supernatant was redissolved in 150-μL water: methanol (85%: 15%, v/v) and transferred to a sampling bottle for metabolite detection by UPLC-QTOF/MS (Waters Corp.; Dunn et al., 2011). An equal volume (15 μL) solution was taken from each sample and mixed to prepare a pool as quality control (QC) samples. In both positive and negative electrospray ion modes, serum samples were eluted using a linear gradient as described in our previous study (He et al., 2019). The collocation of mass spectrometric data was performed using a Waters Q-TOF Premier (Waters Corp.) coupled with an electrospray source operating in either ES+ or ES−. MassLynx (Waters Corp.) and Progenesis QI (v2.0; Nonlinear Dynamics) were used to process the preliminary data. MetaScope package in Progenesis QI was used to annotate serum metabolites according to the HMDB database based on neutral mass, isotope distribution, retention time, the collisional cross-sectional area, and MS/MS fragmentation data (Rusilowicz et al., 2016). When a feature was annotated to multiple metabolites in the HMDB database, we retained an optimal outcome based on mass error, match score, fragmentation score, and isotope similarity. The annotation results were further filtered by removing peaks with missing values (ion intensity = 0) in more than 80% of individuals and 50% of QC samples. Support vector regression algorithm of “MetNormalizer” in R package was applied to normalize the retained peaks to the QC samples (Shen et al., 2016). The relative standard deviation in the QC samples was set at a threshold of 30% to assess the repeatability of metabolomic data sets.
The Spearman correlation was calculated between serum metabolites and the RFI values by corr.test. The qvalue package in R (v4.0.0) was used to correct the multiple tests (Storey et al., 2020). Metabolic pathway analysis was performed for those serum metabolites showing the tendency to association with the RFI (P < 0.05).
Measurement of fecal SCFAs
A total of 162 feces samples (55 from females and 107 from males) were selected from 208 samples, which were performed 16S rRNA gene sequencing, and measured the concentration of SCFAs using a gas chromatograph (SHIMADZU, Japan). About 300-mg stool sample was mixed with DNase/RNase-Free water at the ratio of 1:5 in a 1.5-mL EP tube, homogenized by vortex for 30 s, and centrifuged at 5,000 rpm for 4 min. The supernatant was transferred to a new EP tube, mixed with the 240-μL mixture of metaphosphoric acid and crotonic acid, and centrifuged at 15,000 rpm for 15 min. The supernatant was collected and filtered through a 0.45-μm filter membrane. Finally, 1 µL standard sample and 1 µL of each supernatant sample were separately injected onto columns of the gas chromatograph for measuring SCFAs. The detecting temperature was set at 280 °C. The retention times of the standard sample were used for qualitative analysis. The concentrations of acetic acid, propionic acid, isobutyric acid, butyric, isovaleric acid, and valeric acid were calculated using the external reference method of peak area. To eliminate the effects of temperature during sample handling and storing time of fecal samples on SCFAs, relative ratios of SCFAs were calculated and used for subsequent statistical analysis (Cunningham et al., 2020). Spearman correlation test was used to investigate the association between the concentrations of SCFAs and the abundances of bacterial phyla and families. The multiple tests were corrected by the Benjamini–Hochberg FDR procedure.
Results
Gut microbial taxa associated with porcine RFI based on 16S rRNA gene sequencing data
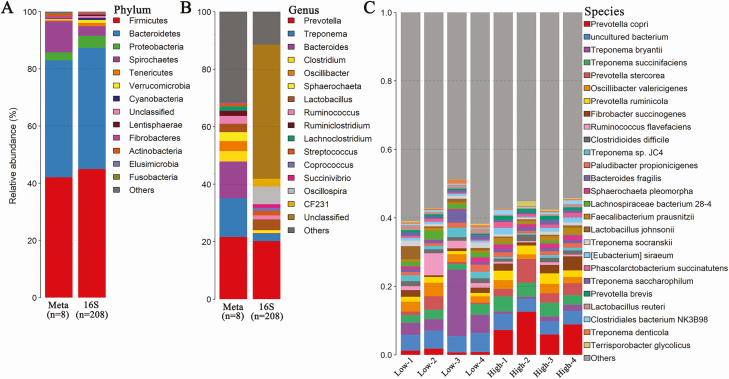
A total of 208 microbial DNA samples were performed 16S rRNA gene sequencing. After QC, an average of 34,382 high-quality tags per sample was obtained. Based on 97% of sequence identity, 872 OTUs on average were identified for each sample. We found that Firmicutes, Bacteroidetes, Spirochaetes, and Proteobacteria were the most abundant phyla in the gut microbiota of experimental pigs (Figure 1A). At the genus level, a total of 44 bacterial genera were detected in the tested samples (Figure 1B and Supplementary Table 1).
Figure 1.
Microbial composition of gut microbiota in experimental pigs at different taxonomic levels by 16S rRNA gene and metagenomic sequencing. (A and B) The distribution of bacterial taxa in the gut microbiota of experimental pigs at the phylum (A) and genus level (B) by shotgun metagenomic and 16S rRNA gene sequencing. (C) Microbial composition of the gut microbiome at the species level by shotgun metagenomic sequencing.
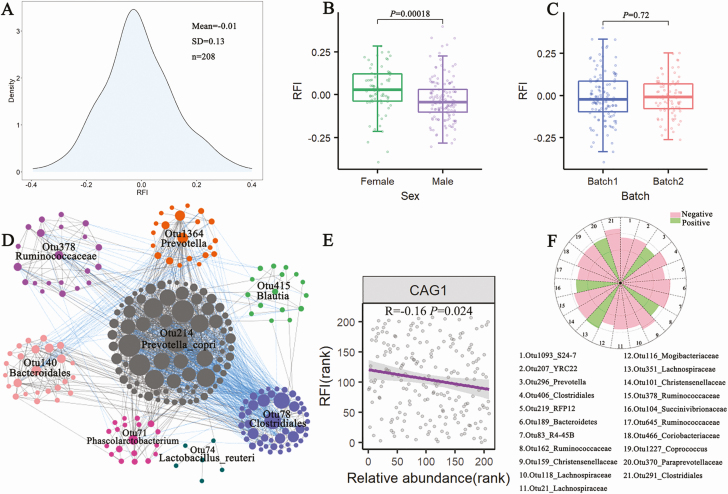
The phenotypical values of RFI in 208 experimental pigs followed the normal distribution (Figure 2A). The average RFI value in the experimental pig cohort was –0.01 ± 0.13 (mean ± SD). Wilcoxon–Rank test showed that sex had a significant effect on the RFI values, but the sampling batch (n = 2) showed no significant effect on the RFI (Figure 2B and C).
Figure 2.
The distribution and influencing factor of phenotypic values of the RFI, and the correlation between gut microbiota and the RFI based on 16S rRNA gene sequencing data. (A) The distribution of phenotypic values of the RFI in 208 experimental pigs. (B) and (C) The effects of sex (B) and batch (C) on phenotypic values of the RFI by Wilcox test. (D) CAG network of OTUs. The Spearman rank correlation was used. The OTUs with correlation coefficients > 0.5 are shown. Grey lines between nodes represent positive correlations, and blue lines refer to the negative correlations. The network was plotted by Cytoscape software (E) A negative correlation between CAG1 and the RFI values. (F) The 21 OTUs showing the tendency of correlation with the RFI by the 2-part model. The broken circles from the inside out indicate Z score = 1.0 and 4.0, respectively.
A correlation analysis was first performed between the RFI values and the α-diversity of the gut microbiota, but no significant correlation was identified (Supplementary Figure 2). We then performed a CAG analysis with 745 OTUs passed QC by the SparCC method. These OTUs were clustered into eight CAGs (Figure 2D). The CAG1 which was comprised of 112 OTUs showed a negative association with the phenotypic values of RFI (P = 0.024; Figure 2E). Otu378 annotated to Ruminococcaceae was the hub in the CAG1. Furthermore, we used a 2-part model to analyze the relationship between OTUs and the RFI. However, we only identified 21 OTUs showing the tendency to correlation with the RFI (P < 0.01; Figure 2F). Among these 21 OTUs, 12 OTUs were annotated to Clostridiales, such as Otu291 and Otu406 to Clostridiales, and Otu159 to Christensenellaceae (Figure 2F). Most members of these OTUs annotated to Clostridiales had a negative correlation with the RFI. In addition, 5 OTUs (e.g., Otu296_Prevotella and Otu370_Paraprevoaceae) showed the tendency to a positive correlation with the RFI.
Identification of potential bacterial species associated with porcine RFI with metagenomic sequencing data
Considering the limited resolution of bacterial taxa based on 16S rRNA gene sequencing data, we further performed metagenomic sequencing analysis in 8 feces samples from the pigs with extreme RFI values (4 samples for each of high and low RFI groups) to identify potential bacterial species associated with the RFI. We obtained a total of 3.2 million contigs with an average size of 1,233 bp, and an average N50 length of 1,406 bp (Supplementary Table 2). Similar to the results from 16S rRNA gene sequencing data, Firmicutes, Bacteroidetes, Spirochaetes, and Proteobacteria were the most abundant phyla in metagenomic sequencing data (Figure 1A). At the genus level, all contigs were assigned to 1,255 genera (Supplementary Table 1). A total of 4,442 bacterial species were detected in 8 samples, indicating that the phylotype resolution was largely improved using metagenomic sequencing at both genus and species levels. Prevotella copri, Treponema bryantii, and Treponema succinifaciens were the known species with the highest abundances in the tested samples (Figure 1C).
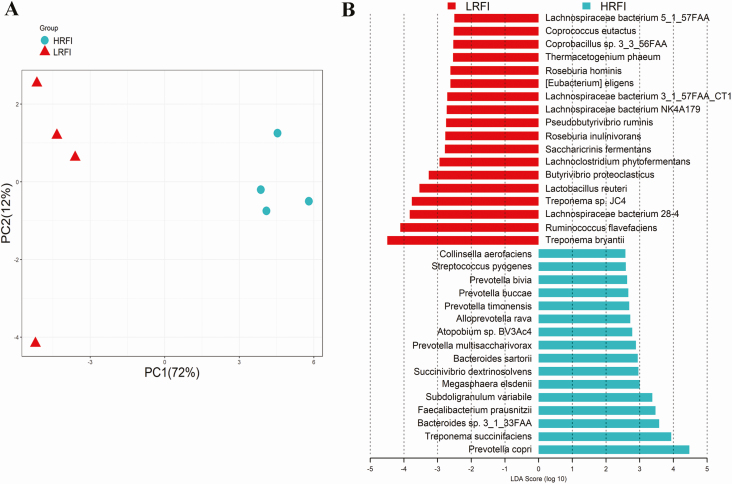
Metagenomic sequencing data were further used to identify the association between the RFI and bacterial species. PCA analysis revealed a clear structural difference of the gut microbiota between high and low RFI pigs (Figure 3A). LEfSe analysis identified 18 and 16 species that were enriched in low and high RFI pigs, respectively (Figure 3B). Among the 18 species enriched in low RFI pigs, 9 species belong to Clostridiales, including Lachnospiraceae bacterium 28-4, Butyrivibrio proteoclasticus, Roseburia inulinivorans, Lachnospiraceae bacterium NK4A179, Lachnospiraceae bacterium 3_1_57FAA_CT1, Roseburia hominis, Coprococcus eutactus, Lachnospiraceae bacterium 5_1_57FAA, and Ruminococcus flavefaoiens. Meanwhile, 6 out of the 16 species having a higher abundance in the high RFI pigs were annotated to Prevotellaceae, such as Prevotella copri, Prevotella multisaccharivorax, and Prevotella timonensis.
Figure 3.
Identification of gut bacterial species associated with porcine RFI with metagenomic sequencing data. (A) Principal component analysis of the gut microbiota between high and low RFI pigs. The result shows the distinct composition of gut microbiome between high and low RFI pigs. (B) The bacterial species associated with the RFI by the LEfSe analysis. LRFI: low RFI, HRFI: high RFI. The X-axis shows LAD scores.
Comparison of functional capacity of the gut microbiome between high and low RFI pigs
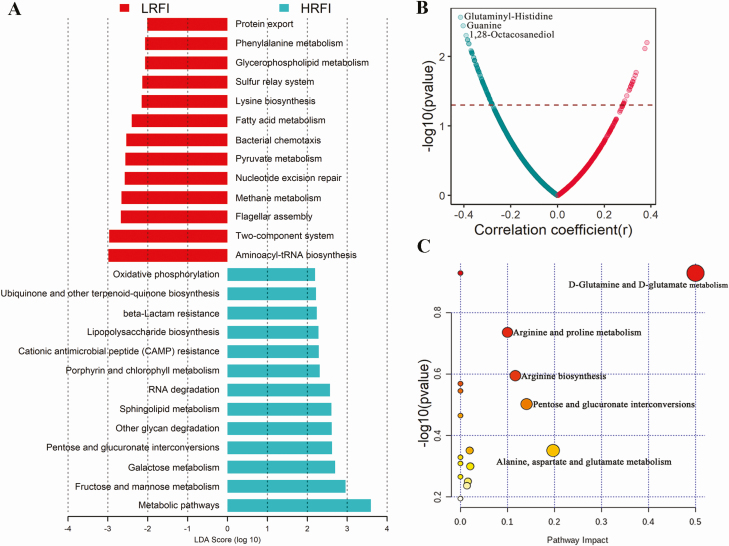
Based on the metagenomic sequencing data, we investigated the association of functional capacity of the gut microbiome with porcine RFI. LEfSe analysis identified 13 KEGG pathways having higher abundance in high RFI pigs, while 13 pathways were enriched in low RFI pigs (Figure 4A). The pathways related to carbohydrate metabolism and biosynthesis (e.g., fructose and mannose metabolism, galactose metabolism, pentose, and glucuronate interconversions, lipopolysaccharide biosynthesis, and other glycan degradation), the antimicrobial pathways (CAMP resistance and β-lactam resistance), and oxidative phosphorylation were significantly enriched in the gut microbiome of high RFI pigs. However, those pathways related to amino acid metabolism and biosynthesis (such as aminoacyl-tRNA biosynthesis, lysine biosynthesis, phenylalanine metabolism, and protein export), fatty acid metabolism, and 2-component system had higher abundances in the pigs with low RFI (Figure 4A).
Figure 4.
The KEGG pathways of the gut microbiome and the serum metabolites showing different abundances between high and low RFI pigs. (A) The KEGG pathways of the gut microbiome showing different abundances between high and low RFI pigs by the LEfSe analysis. (B) The Spearman rank correlations between serum metabolites and the RFI. (C) Pathways enriched by the 91 metabolites negatively correlated with the RFI. LRFI: low RFI, HRFI: high RFI.
Correlations between serum metabolites and the RFI values
A total of 2,347 serum metabolite features were detected by the UPLC-QTOF/MS. However, we did not detect the significant associations between serum metabolites and the RFI values at the significance threshold of q < 0.05. However, we found 110 metabolites showing the tendency to correlation with the RFI (P < 0.05 and q > 0.05; Figure 4B). Among these 110 metabolites, 91 metabolites had a tendency of negative correlations with the RFI, while 19 metabolites showed a tendency to positive correlations with the RFI. Pathway analysis further showed that the 91 metabolites negatively related to the RFI were mainly enriched in the pathways related to amino acid metabolism, including d-glutamine and d-glutamate metabolism, arginine and proline metabolism, alanine, aspartate and glutamate metabolism, and arginine biosynthesis (Figure 4C). However, the 19 metabolites positively related to the RFI were not enriched in any pathways.
Association of SCFAs of feces samples with the RFI
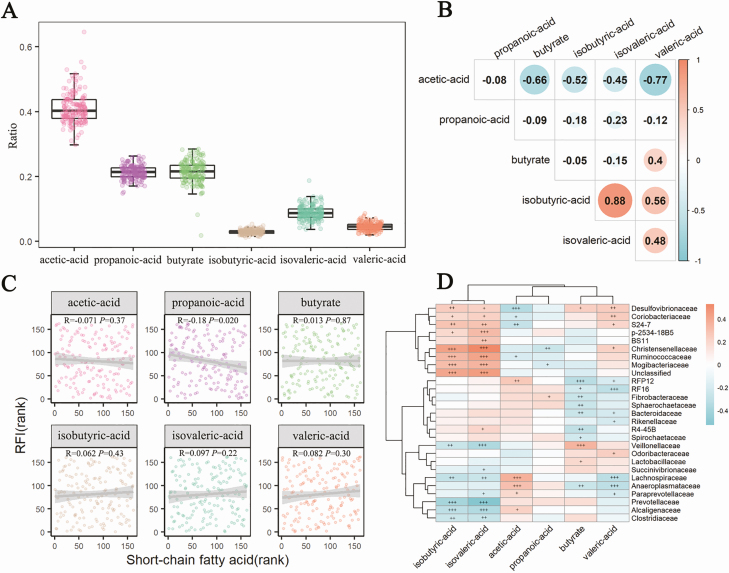
A total of 162 feces samples were determined the concentrations of SCFAs. Acetic acid, butyric acid, and propionic acid were the dominant SCFAs in feces, which accounted for 41%, 21.5%, and 21.3% of relative abundances, followed by isovaleric acid (8.8%), valeric acid (4.5%), and isobutyric acid (2.9%; Figure 5A). Among SCFAs, acetic acid was negatively correlated with nearly all other SCFAs, whereas, isobutyric acid and isovaleric acid had the strongest positive association (r = 0.88, P < 0.01; Figure 5B). Only propionic acid showed a significantly negative association with the RFI (P < 0.05, Figure 5C). However, the other SCFAs had no significant correlations with the RFI (P > 0.05). As for the correlation between gut bacterial composition and fecal SCFAs, Cyanobacteria showed the most significant correlation with most of SCFAs, while only Firmicutes had a significant association with propionic acid (Supplementary Figure 3). At the family level, 27 bacterial families had significant associations with 6 SCFAs. Especially, Desulfovibrionaceae, Coribobacteriaceae, S24-7, Christensenellaceae, and Lachnospiraceae exhibited significant correlations with more than 4 SCFAs (Figure 5D).
Figure 5.
The composition of SCFAs in feces samples and its association with the RFI. (A) The boxplot shows the distribution of 6 SCFAs. (B) Spearman rank correlation analysis between the ratio of 6 SCFAs and the RFI. (C) The Spearman rank correlations among 6 SCFAs. (D) The association of SCFAs with gut microbiota at the family level with the 16S rRNA gene sequencing data (+, P < 0.05, ++, P < 0.01, +++, P < 0.001). The associations were analyzed by Spearman rank correlation.
Discussion
In our previous study, we unraveled the fecal microbiota and metagenomic function capacity associated with FE in 280 Duroc pigs from Shahu farm (Yang et al., 2017). To further explore bacterial species and functional capacity of the gut microbiome associated with porcine FE in more experimental pig cohorts, in this study, we performed 16S rRNA gene and shotgun metagenomic sequencing analysis to detect bacterial taxa and functional capacity of the gut microbiome associated with porcine RFI in another cohort comprised of 208 Duroc pigs from Jiangyin farm. Especially, to our knowledge, for the first time, we measured the metabolomic profiles of serum samples and the concentrations of fecal SCFAs to isolate the metabolites and SCFAs associated with porcine FE. We also evaluated the correlation between gut microbiota, and the shifts of serum metabolites and fecal SCFAs.
Although the sample size used for metagenomic sequencing was quite small, the gut microbial composition was similar to that obtained from 16S rRNA gene sequencing data (Figure 1), indicating the repeatability of sequencing data. At the threshold of FDR < 0.05, we did not detect any associations between OTUs and the RFI by the 2-part model. However, we identified 21 OTUs showing the tendency to correlations with the RFI (P < 0.01). More than half of these OTUs belong to Clostridiales that could degrade cellulose and help the host to digest dietary polysaccharides. At the higher phylotype resolution, metagenomic sequencing analysis identified that the pigs with high FE had higher enrichment for Clostridiales, including Ruminococcus flavefaciens, Lachnospiraceae spp., Butyrivibrio proteoclasticus, Roseburia spp., Coprococcus eutactus, and Eubacterium eligens (Figure 3B). Eubacterium eligens can be promoted by pectin and competes with Bacteroides species under certain pH values (Chung et al., 2016). Ruminococcus flavefaciens is one of the predominant cellulolytic bacterial species (Julliand et al., 1999). The species from Clostridiales species were closely related to the production of SCFAs and had certain anti-inflammatory effects (Canani et al., 2011; Scott et al., 2011; Martin-Gallausiaux et al., 2020). Interestingly, we observed that SCFAs (propionic acid) had a positive correlation with FE (Figure 5C). This gave us a clue that the gut microbiota in pigs with high-FE had higher abundances of Clostridiales and might potentially be “healthier” (McCormack et al., 2017).
Consistent with the findings from our previous study (Yang et al., 2017) and McCormack (McCormack et al., 2017), those pigs with high-FE had higher abundances of the pathways related to amino acid metabolism and biosynthesis, such as phenylalanine metabolism and lysine biosynthesis in the gut microbiome (Figure 4A). The metabolic pathways of the metabolism of aromatic amino acids (e.g., phenylalanine metabolism), which were related to the synthesis of indole propionic acid, have also been associated with the species from Lachnospiraceae (Vacca et al., 2020). In animal models, indole propionic acid (IPA) could help to improve host metabolism by enhancing intestinal barrier function, showing anti-inflammatory properties, and strengthening immune function (Cani et al., 2019). Interestingly, the serum metabolites negatively related to the RFI were mainly enriched in the pathways of amino acid metabolism (Figure 4B and C). Particularly, the metabolite of glutaminyl-histidine that contains a sequence of glutamine and histidine joined by a peptide bond was most negatively correlated with the RFI. Glutamine is the most eager expendable for the cells of the intestine (Brosnan, 2003) and plays an important role in maintaining the normal integrity of the intestinal mucosa (Yamamoto et al., 2017). In this study, the main ingredient of the diet provided to experimental pigs was a soybean meal with high concentrations of proteins. These results indicated that the gut microbiota of pigs with high-FE might have a stronger ability in protein degradation, fermentation, and transportation than that of pigs with low-FE.
16S rRNA gene sequencing analysis detected 5 OTUs showing the tendency to a positive correlation with the RFI. Two out of these 5 OTUs belonged to the Prevotellaceae. Metagenomic sequencing analysis also identified that Prevotella spp. were the most abundant microbes in the pigs with low-FE, especially the Prevotella copri. Yang et al. (2018) found that Prevotella might be a keystone microbe increasing host feed intake. Correlation analysis indeed identified that the RFI had a significantly positive association with the feed intake and fat deposition (backfat thickness; Supplementary Figure 4). This result suggested that Prevotella could promote the host’s appetite and decrease FE. Prevotella has been reported to be associated with gut mucosal inflammation (Iljazovic et al., 2021). Comparison of KEGG pathways between pigs with high- and low-FE, we found that the pathways related to inflammation, e.g., lipopolysaccharide biosynthesis were enriched in the low-feed efficiency pigs. Prevotella-rich dysbiosis leads to the systemic release of lipopolysaccharides that promote obesity and systemic inflammation (Larsen, 2017). Moreover, the pathway related to monosaccharide metabolism had a higher abundance in the low-feed efficiency pigs (Figure 4A). Stanhope indicated that the consumption of excessive monosaccharide accelerated the development of cardiovascular disease and type 2 diabetes indirectly by increasing fat deposition (Stanhope, 2016). Besides, Zhou et al. (2017) found that mice injected with d-galactose showed significant fat deposition and oxidative damage.
In summary, we found that several members of Clostridiales were enriched in pigs with high-FE, while those bacteria related to inflammation such as Prevotella copri, had a higher abundance in pigs with low-FE. Propionic acid in feces and the metabolites related to amino acid metabolism in the serum had positive correlations with the FE. Nonetheless, the sample size used for metagenomic sequencing was quite small. The result was easily affected by an outlier. Further studies would be needed to confirm the causality of gut microbes with porcine FE and to elucidate the possible mechanism of gut microbiome affecting porcine FE. The results from this study also provide potential biomarkers of gut microbiota that may be used for improving pig FE.
Supplementary Material
Acknowledgments
We thank prof. Lusheng Huang for his initiation of the experiment and valuable supervision on the design of experiments, analysis of data, and revision of manuscript. We thank prof. Jun Gao for her supervision of H.J. This study was funded by National Natural Science Foundation of China (grant nos. 31702103 and 31472071). H.Y. was supported by National Postdoctoral Program for Innovative Talents (grant no. BX201700102).
Glossary
Abbreviations
- ADG
average daily gain
- CAG
co-abundance group
- CAZy
carbohydrate-active enzymes
- DFI
average daily feed intake
- FCR
feed conversion rate
- FE
feed efficiency
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LCA
lowest common ancestor
- LEfSe
linear discriminant analysis effect size
- MBW
metabolic body weight
- OTU
operational taxonomic units
- QC
quality control
- RFI
residual feed intake
- SCFA
short-chain fatty acid
Author contributions
H.J. and H.Y.: performed the experiments, analyzed the data, and wrote the manuscript; S.F.: collected the samples and performed parts of experiments; C.C.: discussed the results and revised the manuscript.
Conflict of interest statement
The authors declare no real or perceived conflicts of interest.
Literature Cited
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. . 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bolger, A. M., M. Lohse, and B. Usadel. . 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan, J. T. 2003. Interorgan amino acid transport and its regulation. J. Nutr. 133(6 Suppl. 1):2068S–2072S. doi: 10.1093/jn/133.6.2068S [DOI] [PubMed] [Google Scholar]
- Canani, R. B., M. D. Costanzo, L. Leone, M. Pedata, R. Meli, and A. Calignano. . 2011. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J. Gastroenterol. 17:1519–1528. doi: 10.3748/wjg.v17.i12.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani, P. D., M. Van Hul, C. Lefort, C. Depommier, M. Rastelli, and A. Everard. . 2019. Microbial regulation of organismal energy homeostasis. Nat. Metab. 1:34–46. doi: 10.1038/s42255-018-0017-4 [DOI] [PubMed] [Google Scholar]
- Chung, W. S., A. W. Walker, P. Louis, J. Parkhill, J. Vermeiren, D. Bosscher, S. H. Duncan, and H. J. Flint. . 2016. Modulation of the human gut microbiota by dietary fibres occurs at the species level. BMC Biol. 14:3. doi: 10.1186/s12915-015-0224-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmons, B. A., R. I. Mihelic, R. C. Beckford, J. B. Powers, E. A. Melchior, Z. D. McFarlane, E. R. Cope, M. M. Embree, J. T. Mulliniks, S. R. Campagna, . et al. 2017. Serum metabolites associated with feed efficiency in black angus steers. Metabolomics. 13(12):147. doi: 10.1007/s11306-017-1282-z [DOI] [Google Scholar]
- Cunningham, J. L., L. Bramstång, A. Singh, S. Jayarathna, A. J. Rasmusson, A. Moazzami, and B. Müller. . 2020. Impact of time and temperature on gut microbiota and SCFA composition in stool samples. PLoS One 15:e0236944. doi: 10.1371/journal.pone.0236944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiacomo, K., E. Norris, F. R. Dunshea, B. J. Hayes, L. C. Marett, W. J. Wales, and B. J. Leury. . 2018. Responses of dairy cows with divergent residual feed intake as calves to metabolic challenges during midlactation and the nonlactating period. J. Dairy Sci. 101:6474–6485. doi: 10.3168/jds.2017-12569 [DOI] [PubMed] [Google Scholar]
- Ding, R., M. Yang, X. Wang, J. Quan, Z. Zhuang, S. Zhou, S. Li, Z. Xu, E. Zheng, G. Cai, . et al. 2018. Genetic architecture of feeding behavior and feed efficiency in a duroc pig population. Front. Genet. 9:220. doi: 10.3389/fgene.2018.00220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, W. B., D. Broadhurst, P. Begley, E. Zelena, S. Francis-McIntyre, N. Anderson, M. Brown, J. D. Knowles, A. Halsall, J. N. Haselden, . et al. 2011. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 6:1060–1083. doi: 10.1038/nprot.2011.335 [DOI] [PubMed] [Google Scholar]
- Edgar, R. C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Fang, S., X. Chen, L. Zhou, C. Wang, Q. Chen, R. Lin, T. Xiao, and Q. Gan. . 2019. Faecal microbiota and functional capacity associated with weaning weight in meat rabbits. Microb. Biotechnol. 12:1441–1452. doi: 10.1111/1751-7915.13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J., and E. J. Alm. . 2012. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8:e1002687. doi: 10.1371/journal.pcbi.1002687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, J., M. J. Bonder, M. C. Cenit, E. F. Tigchelaar, A. Maatman, J. A. Dekens, E. Brandsma, J. Marczynska, F. Imhann, R. K. Weersma, . et al. 2015. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ. Res. 117:817–824. doi: 10.1161/CIRCRESAHA.115.306807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondret, F., A. Vincent, M. Houée-Bigot, A. Siegel, S. Lagarrigue, D. Causeur, H. Gilbert, and I. Louveau. . 2017. A transcriptome multi-tissue analysis identifies biological pathways and genes associated with variations in feed efficiency of growing pigs. BMC Genomics 18:244. doi: 10.1186/s12864-017-3639-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, H., S. Zhang, Q. Li, C. Zuo, X. Gao, B. Zheng, and M. Lin. . 2020. Gut microbiota compositional profile and serum metabolic phenotype in patients with primary open-angle glaucoma. Exp. Eye Res. 191:107921. doi: 10.1016/j.exer.2020.107921 [DOI] [PubMed] [Google Scholar]
- Haer, L., P. Luiting, and H. L. M. Aarts. . 1993. Relations among individual (residual) feed intake, growth performance and feed intake pattern of growing pigs in group housing. Livest. Prod. Sci. 36:233–253. doi: 10.1016/0301-6226(93)90056-N [DOI] [Google Scholar]
- He, M., J. Gao, J. Wu, Y. Zhou, H. Fu, S. Ke, H. Yang, C. Chen, and L. Huang. . 2019. Host gender and androgen levels regulate gut bacterial taxa in pigs leading to sex-biased serum metabolite profiles. Front Microbiol. 10:1359. doi: 10.3389/fmicb.2019.01359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper, L. V., T. Midtvedt, and J. I. Gordon. . 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259 [DOI] [PubMed] [Google Scholar]
- Huson, D. H., A. F. Auch, J. Qi, and S. C. Schuster. . 2007. MEGAN analysis of metagenomic data. Genome Res. 17:377–386. doi: 10.1101/gr.5969107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iljazovic, A., U. Roy, E. J. C. Gálvez, T. R. Lesker, B. Zhao, A. Gronow, L. Amend, S. E. Will, J. D. Hofmann, M. C. Pils, . et al. 2021. Perturbation of the gut microbiome by Prevotella spp. enhances host susceptibility to mucosal inflammation. Mucosal Immunol. 14:113–124. doi: 10.1038/s41385-020-0296-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, L., Y. Hou, H. Wu, Y. Miao, X. Li, J. Cao, J. M. Brameld, T. Parr, and S. Zhao. . 2015. Transcriptome analysis of mRNA and miRNA in skeletal muscle indicates an important network for differential residual feed intake in pigs. Sci. Rep. 5:11953. doi: 10.1038/srep11953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julliand, V., A. de Vaux, L. Millet, and G. Fonty. . 1999. Identification of Ruminococcus flavefaciens as the predominant cellulolytic bacterial species of the equine cecum. Appl. Environ. Microbiol. 65:3738–3741. doi: 10.1128/AEM.65.8.3738-3741.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M., M. Araki, S. Goto, M. Hattori, M. Hirakawa, M. Itoh, T. Katayama, S. Kawashima, S. Okuda, T. Tokimatsu, . et al. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36(Database issue):D480–D484. doi: 10.1093/nar/gkm882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, J. M. 2017. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 151:363–374. doi: 10.1111/imm.12760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., and A. Godzik. . 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 22(13):1658–1659. doi: 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- Lombard, V., H. Golaconda Ramulu, E. Drula, P. M. Coutinho, and B. Henrissat. . 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42(Database issue):D490–D495. doi: 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, R., B. Liu, Y. Xie, Z. Li, W. Huang, J. Yuan, G. He, Y. Chen, Q. Pan, Y. Liu, . et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč, T., and S. L. Salzberg. . 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 27(21):2957–2963. doi: 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Gallausiaux, C., L. Marinelli, H. M. Blottière, P. Larraufie, and N. Lapaque. . 2020. SCFA: mechanisms and functional importance in the gut. Proc Nutr Soc. 1–13. doi: 10.1017/s0029665120006916 [DOI] [PubMed] [Google Scholar]
- McCormack, U. M., T. Curião, S. G. Buzoianu, M. L. Prieto, T. Ryan, P. Varley, F. Crispie, E. Magowan, B. U. Metzler-Zebeli, D. Berry, . et al. 2017. Exploring a possible link between the intestinal microbiota and feed efficiency in pigs. Appl. Environ. Microbiol. 83(15):AEM.00380-00317. doi: 10.1128/aem.00380-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack, U. M., T. Curião, B. U. Metzler-Zebeli, T. Wilkinson, H. Reyer, F. Crispie, P. D. Cotter, C. J. Creevey, G. E. Gardiner, and P. G. Lawlor. . 2019. Improvement of feed efficiency in pigs through microbial modulation via fecal microbiota transplantation in sows and dietary supplementation of inulin in offspring. Appl. Environ. Microbiol. 85(22):e01255–01219. doi: 10.1128/aem.01255-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McOrist, A. L., M. Jackson, and A. R. Bird. . 2002. A comparison of five methods for extraction of bacterial DNA from human faecal samples. J. Microbiol. Methods 50:131–139. doi: 10.1016/s0167-7012(02)00018-0 [DOI] [PubMed] [Google Scholar]
- Murtagh, F., and P. Legendre. . 2014. Ward’s hierarchical agglomerative clustering method: which algorithms implement ward’s criterion? J. Classif. 31:274–295. doi: 10.1007/s00357-014-9161-z [DOI] [Google Scholar]
- Qin, J., Y. Li, Z. Cai, S. Li, J. Zhu, F. Zhang, S. Liang, W. Zhang, Y. Guan, D. Shen, . et al. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 490(7418):55–60. doi: 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- Quan, J., G. Cai, M. Yang, Z. Zeng, R. Ding, X. Wang, Z. Zhuang, S. Zhou, S. Li, H. Yang, . et al. 2019. Exploring the fecal microbial composition and metagenomic functional capacities associated with feed efficiency in commercial DLY pigs. Front. Microbiol. 10:52. doi: 10.3389/fmicb.2019.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revelle, W. 2019. psych: procedures for psychological, psychometric, and personality research[R package version 1.9.12]. Available from https://CRAN.R-project.org/package=psych
- Rognes, T., T. Flouri, B. Nichols, C. Quince, and F. Mahé. . 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusilowicz, M., M. Dickinson, A. Charlton, S. O’Keefe, and J. Wilson. . 2016. A batch correction method for liquid chromatography-mass spectrometry data that does not depend on quality control samples. Metabolomics 12:56. doi: 10.1007/s11306-016-0972-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salek, R. M., M. L. Maguire, E. Bentley, D. V. Rubtsov, T. Hough, M. Cheeseman, D. Nunez, B. C. Sweatman, J. N. Haselden, R. D. Cox, . et al. 2007. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiol. Genomics 29:99–108. doi: 10.1152/physiolgenomics.00194.2006 [DOI] [PubMed] [Google Scholar]
- Scott, K. P., J. C. Martin, C. Chassard, M. Clerget, J. Potrykus, G. Campbell, C. D. Mayer, P. Young, G. Rucklidge, A. G. Ramsay, . et al. 2011. Substrate-driven gene expression in Roseburia inulinivorans: importance of inducible enzymes in the utilization of inulin and starch. Proc. Natl. Acad. Sci. USA. 108(Suppl. 1):4672–4679. doi: 10.1073/pnas.1000091107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P., A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski, and T. Ideker. . 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X., X. Gong, Y. Cai, Y. Guo, J. Tu, H. Li, T. Zhang, J. Wang, F. Xue, and Z.-J. Zhu. . 2016. Normalization and integration of large-scale metabolomics data using support vector regression. Metabolomics. 12(5):89. doi: 10.1007/s11306-016-1026-5 [DOI] [Google Scholar]
- Stanhope, K. L. 2016. Sugar consumption, metabolic disease and obesity: the state of the controversy. Crit. Rev. Clin. Lab. Sci. 53:52–67. doi: 10.3109/10408363.2015.1084990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, J. D., A. J. Bass, A. Dabney, and D. Robinson. . 2020. qvalue: Q-value estimation for false discovery rate control[R package version 2.20.0]. Available from http://github.com/jdstorey/qvalue
- Straw, B., S. Shin, and A. Yeager. . 1990. Effect of pneumonia on growth rate and feed efficiency of minimal disease pigs exposed to Actinobacillus pleuropneumoniae and Mycoplasma hyopneumoniae. Prev. Vet. Med. 9:287–294. doi: 10.1016/0167-5877(90)90074-R [DOI] [Google Scholar]
- Tan, Z., T. Yang, Y. Wang, K. Xing, F. Zhang, X. Zhao, H. Ao, S. Chen, J. Liu, and C. Wang. . 2017. Metagenomic analysis of Cecal microbiome identified microbiota and functional capacities associated with feed efficiency in landrace finishing pigs. Front. Microbiol. 8:1546. doi: 10.3389/fmicb.2017.01546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca, M., G. Celano, F. M. Calabrese, P. Portincasa, M. Gobbetti, and M. De Angelis. . 2020. The controversial role of human gut lachnospiraceae. Microorganisms. 8(4):573. doi: 10.3390/microorganisms8040573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman, A., C. M. Bassis, J. M. Beck, V. B. Young, J. L. Curtis, G. B. Huffnagle, and T. M. Schmidt. . 2015. Application of a neutral community model to assess structuring of the human lung microbiome. mBio. 6(1):e02284-02214. doi: 10.1128/mBio.02284-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. . 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. doi: 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., and H. N. Kadarmideen. . 2020. Metabolite genome-wide association study (mGWAS) and gene-metabolite interaction network analysis reveal potential biomarkers for feed efficiency in pigs. Metabolites. 10(5):201. doi: 10.3390/metabo10050201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing, B., and A. Van Kessel. . 2010. Host pathways for recognition: establishing gastrointestinal microbiota as relevant in animal health and nutrition. Livest Sci. 133(1):82–91. doi: 10.1016/j.livsci.2010.06.031 [DOI] [Google Scholar]
- Yamamoto, T., T. Shimoyama, and M. Kuriyama. . 2017. Dietary and enteral interventions for Crohn’s disease. Curr. Opin. Biotechnol. 44:69–73. doi: 10.1016/j.copbio.2016.11.011 [DOI] [PubMed] [Google Scholar]
- Yang, H., X. Huang, S. Fang, M. He, Y. Zhao, Z. Wu, M. Yang, Z. Zhang, C. Chen, and L. Huang. . 2017. Unraveling the fecal microbiota and metagenomic functional capacity associated with feed efficiency in pigs. Front. Microbiol. 8:1555. doi: 10.3389/fmicb.2017.01555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., M. Yang, S. Fang, X. Huang, M. He, S. Ke, J. Gao, J. Wu, Y. Zhou, H. Fu, . et al. 2018. Evaluating the profound effect of gut microbiome on host appetite in pigs. BMC Microbiol. 18:215. doi: 10.1186/s12866-018-1364-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, M., Z. Yue, W. He, X. Yang, G. Yang, M. Xie, D. Zhan, S. W. Baxter, L. Vasseur, G. M. Gurr, . et al. 2013. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 45:220–225. doi: 10.1038/ng.2524 [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Y. Wu, J. Wang, G. Wu, W. Long, Z. Xue, L. Wang, X. Zhang, X. Pang, Y. Zhao, . et al. 2016. Accelerated dysbiosis of gut microbiota during aggravation of DSS-induced colitis by a butyrate-producing bacterium. Sci. Rep. 6:27572. doi: 10.1038/srep27572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T., Y. Yang, Y. Liang, X. Jiao, and C. Zhao. . 2018. Beneficial effect of intestinal fermentation of natural polysaccharides. Nutrients. 10(8):1055. doi: 10.3390/nu10081055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Q. Xu, Y. Dong, S. Zhu, S. Song, and S. Sun. . 2017. Supplementation of mussel peptides reduces aging phenotype, lipid deposition and oxidative stress in D-galactose-induce aging mice. J. Nutr. Health Aging 21:1314–1320. doi: 10.1007/s12603-016-0862-3 [DOI] [PubMed] [Google Scholar]
- Zhu, W., A. Lomsadze, and M. Borodovsky. . 2010. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 38:e132. doi: 10.1093/nar/gkq275 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.