Key Points
The GMB differs between indolent lymphoma and DLBCL; pretreatment diversity and composition predicted treatment response.
Our results, if validated, could improve treatment outcomes by improving medication stewardship and informing novel GMB-targeted therapies.
Abstract
B-cell non-Hodgkin lymphoma cell survival depends on poorly understood immune evasion mechanisms. In melanoma, the composition of the gut microbiota (GMB) is associated with immune system regulation and response to immunotherapy. We investigated the association of GMB composition and diversity with lymphoma biology and treatment outcome. Patients with diffuse large B-cell lymphoma (DLBCL), marginal zone (MZL), and follicular lymphoma (FL) were recruited at Mayo Clinic, Minnesota, and Perlmutter Cancer Center, NYU Langone Health. The pretreatment GMB was analyzed using 16S ribosomal RNA gene sequencing. We examined GMB compositions in 3 contexts: lymphoma patients (51) compared with healthy controls (58), aggressive (DLBCL) (8) compared with indolent (FL, MZL) (18), and the association of GMB with immunochemotherapy treatment outcomes (8 responders, 6 nonresponders). Respectively, we found that the pretreatment GMB in lymphoma patients had a distinct composition compared with healthy controls (P < .001); GMB compositions in DLBCL patients were significantly different than indolent patients (P = .01) with a trend toward reduced microbial diversity in DLBCL patients (P = .08); and pretreatment GMB diversity and composition were significant predictors of treatment responses (P = .01). The impact of these pilot results is limited by our small sample size, and should be considered a proof of principle. If validated, our results could lead toward improved treatment outcomes by improving medication stewardship and informing which GMB-targeted therapies should be tested to improve patient outcomes.
Introduction
B-cell non-Hodgkin lymphoma (NHL) is highly dependent upon its ability to escape natural host defenses, yet the immune evasion strategies used are poorly understood. Growing evidence suggests that gut microbiota (GMB) play important roles in regulating innate and adaptive immunity.1-3 Murine experiments demonstrate the influence of the GMB on the development4 and homeostasis of the host immune system,5-9 and mechanistic preclinical studies show that introduction of specific bacterial populations directly affects endogenous immunity.10-12 This immune regulation is critical for the detection of nascent tumor cells.13,14 In melanoma patients, the GMB composition influences the clinical response to immunotherapy15-17; in allogeneic stem cell transplant, low GMB diversity and specific species are associated with increased risk of graft vs host disease and poor survival.18,19 However, the connections between the human GMB and lymphoma remain poorly understood. Compounding this, at least one-quarter of human-targeted medications collaterally affect the bacterial microbiota communities.20 It is not known to what degree lymphoma treatment affects the microbiota of patients and how this might hinder successful outcomes of therapy in the long term. Here, we investigated the relationship of GMB composition and specific taxon abundances with lymphoma biology and clinical outcome.
Methods
The Institutional Review Board of the Mayo Clinic approved the Mayo study, and the institutional review board of the Perlmutter Cancer Center (PCC) of NYU Langone Health approved the PCC study, which at both institutions was done according to the Declaration of Helsinki and International Harmonization Guidelines for Good Practice. Patients with diffuse large B-cell lymphoma (DLBCL), marginal zone (MZL), and follicular lymphoma (FL) were recruited at Mayo Clinic (Rochester, MN) or PCC (New York, NY). All enrolled patients provided written informed consent. All patients were recruited before initiating treatment or during expectant monitoring if they had indolent lymphoma. Patients who had used antibiotics within 30 days of screening were excluded. One patient failed screening for antibiotic use and was not included in the study. All registered patients completed a structured questionnaire, including travel, and medication history, and their clinical and medical information was extracted from electronic health records. The control subjects were selected from the Mayo Clinic Biobank and representative fecal samples were chosen according to sex, age, race, body mass index (BMI), alcohol use, and tobacco use. The collection and processing methods were the same as controls. The bacterial composition of the GMB, using pretreatment stools, was assessed using 16S ribosomal RNA gene sequencing of pretreatment fecal samples. Briefly, amplicon libraries covering the V4 region of the 16S ribosomal RNA gene were sequenced on Illumina MiSeq with a 2 × 150 bp paired-end kit, a highly reproducible assay,21 and sequence reads were grouped into operational taxonomic units (OTUs) and assigned microbial taxonomy using the Quantitative Insights Into Microbial Ecology (QIIME) 2 Deblur workflow. One replicate was studied using standard Earth Microbiome Project protocols for stool DNA extraction, amplification, and sequencing at the Argonne National Laboratory.22 We compared GMB overall structure and taxon abundance in relation to lymphoma biology (aggressive DLBCL) vs indolent (FL and MZL) and subsequent immunochemotherapy outcomes.
Results and discussion
Lymphoma patients have an altered GMB
To ascertain microbiota dysbiosis in lymphoma patients, we first sought to compare their GMB with nonlymphoma healthy controls. We analyzed an aliquot of the first 51 patient pretreatment fecal samples including DLBCL18 indolent B-cell NHL (FL 13, MZL 5, mantle cell lymphoma 3), and other or unclassified including HL,4 and T-cell3 patients from the University of Iowa/Mayo Clinic Lymphoma SPORE pilot study, and 58 age-matched Mayo Midwest Microbiome Clinic noncancer controls stool samples. The GMB of the lymphoma patients was significantly different from the controls (Figure 1A; P < .001, PERMANOVA of unweighted UniFrac). Specifically, the GMB of lymphoma patients showed an increase in Bacteroidetes with a concomitant decrease in Firmicutes at the phylum level (Figure 1B). We did not see significant differences in the distribution of demographic variables (age, sex, ethnicity, BMI) for the Mayo cohort, following the literature, which suggests the sex effect on the GMB is very moderate.23 These results suggest that the GMB is perturbed in lymphoma patients prior to initiation of their antilymphoma treatment.
Figure 1.
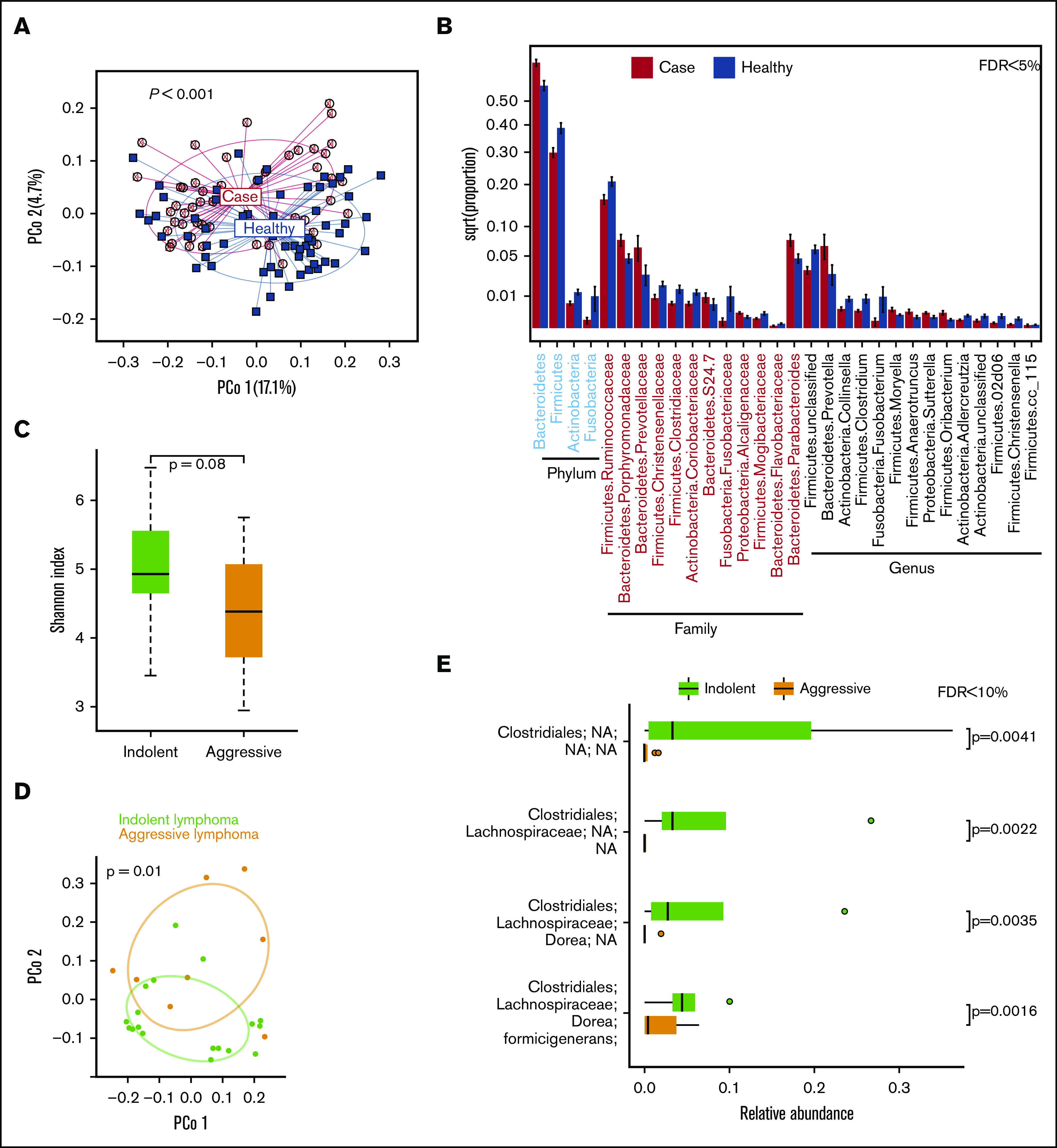
Gut microbiome in lymphoma patients and healthy controls. (A) Principal coordinate analysis of unweighted UniFrac distances. (B) Differentially abundant taxa. Gut microbiome and lymphoma aggressiveness: Shannon diversity index between indolent NHL (MZL, FL) and aggressive lymphoma (DLBCL) (C); principal coordinate analysis of Jensen Shannon divergence (D); and differentially abundant OTUs (E).
GMB and lymphoma aggressiveness
At the PCC, we compared the GMB of patients with indolent NHL (MZL, 8; FL, 10) (N = 18) and aggressive lymphoma (DLBCL) (N = 8) to test if the GMB is also associated with aggressive disease biology before treatment. PCC patients with DLBCL exhibited a trend toward further reduced GMB diversity than patients with indolent NHL (P = .08, Wilcoxon rank-sum test of Shannon index; Figure 1C). Additionally, patients with DLBCL had a distinct GMB composition compared with indolent patients (R2 = 7.1% and P = .01, PERMANOVA of Jensen Shannon divergence) (Figure 1D). Several OTUs of obligate anaerobe taxa were depleted in DLBCL patients (all false discovery rate–adjusted P < .10, Wilcoxon rank-sum test) (Figure 1E). The observed effect size in Shannon index is 0.7 standard deviations between indolent and aggressive patients (Figure 1D; supplemental Table 1).
GMB in response to subsequent immunochemotherapy outcomes
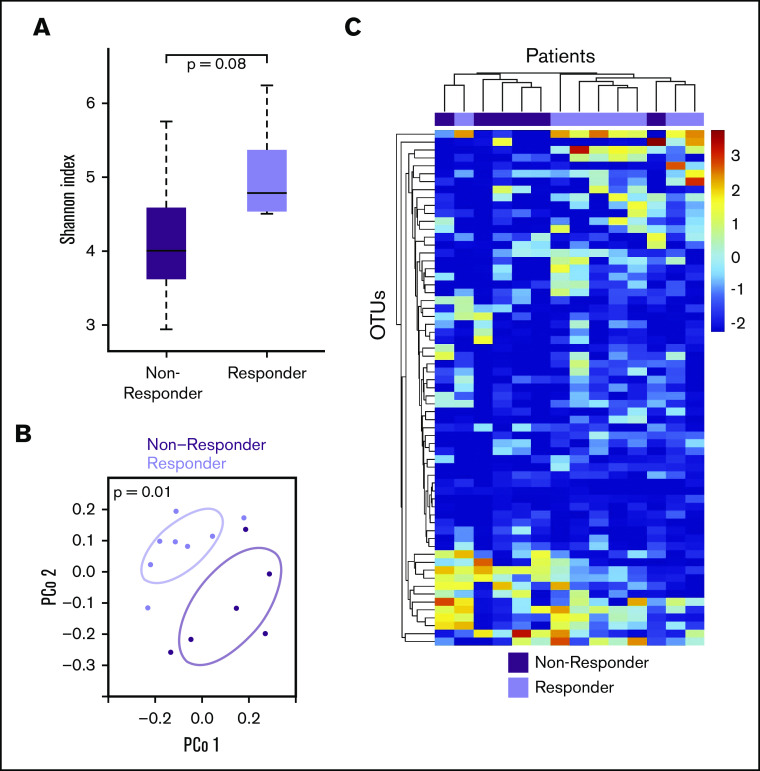
Recent evidence suggests that the GMB modulates response to checkpoint blockade immunotherapy in melanoma patients.16,24 We sought to explore whether the same may be true in lymphoma patients undergoing immunochemotherapy. Of the 26 NHL patients described previously, 14 received subsequent treatment (8 with DLBCL; 6 indolent lymphoma patients who transformed to aggressive lymphoma). There were 8 responders with complete or partial response and 6 nonresponders (NR). Responders tended to have higher pretreatment GMB diversity than NR (P = .08, Wilcoxon rank-sum test of Shannon index) (Figure 2A). Furthermore, pretreatment responders vs NR had distinct overall GMB composition (R2 = 14.6% and P = .01, PERMANOVA of Jensen Shannon divergence) (Figure 2B). Findings were similar after adjustment for age, sex, race, BMI, and history of treatment, stage, or tumor histology (P = .05). Unsupervised hierarchical clustering of all OTUs present in at least 50% of the treated patients clustered patients almost perfectly into NR and responders (Figure 2C), indicating differential abundance of specific OTU by therapeutic response status. Among others, the relative abundance of OTUs within Dorea formicigenerans (associated with indolent lymphoma; Figure 2C) and Faecalibacterium prausnitzii (associated with favorable melanoma immunotherapy response24) was higher in responders compared with NR patients (P < .05, Wilcoxon rank-sum test). The observed effect size in Shannon index is 1 standard deviation between NR and responders (Figure 2B; supplemental Table 1).
Figure 2.
Gut microbiome and lymphoma response to therapy. (A) Shannon diversity index. (B) Principal coordinate analysis of Jensen Shannon divergence. (C) OTU log-transformed relative abundance clustered heat map.
We found that lymphoma patients at diagnosis have altered GMB compositions; DLBCL patients have a less diverse overall microbiota diversity compared with patients with indolent NHL. Furthermore, we also found that the pretreatment microbiome was a significant predictor of lymphoma treatment response. The impact of these pilot results is limited by our small sample size and may be overestimated because of the Winner’s Curse effect25 and should be considered a proof of principle, which requires validation in large histology-specific data sets. However, if validated, these findings could have major implications for improving treatment outcomes by identifying predictive signatures that could facilitate personalized treatment decision-making by using easily obtainable stool samples and by improved medication stewardship seeking to reduce diversity loss. Importantly, our findings may inform the development of personalized, microbe-targeted therapies, a new minimally toxic treatment paradigm for aggressive and treatment refractory lymphomas, an unmet medical need.
Authorship
Contribution: C.S.D. conceived of and developed the study at Perlmutter Cancer Center (PCC); T.E.W. and N.N.B. conceived of and developed the study at Mayo Clinic; C.S.D., B.R., T.M., and K.H. recruited patients and collected clinical data at the PCC; T.E.W. and N.N.B. recruited patients and collected clinical data at Mayo Clinic; J.A. and B.A.P. performed the analysis at PCC; C.S.D., J.A., H.L., and B.A.P. analyzed the data at PCC; T.E.W. and J.C. performed the analysis at Mayo Clinic; T.E.W. and N.N.B. analyzed the data at Mayo Clinic; C.S.D., J.S., T.E.W., N.N.B., H.L., and J.A. wrote the manuscript; and all authors edited the manuscript and all authors approved a final version of the manuscript.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
C.S.D. is supported by American Cancer Society grant MRSG-14-052-01-LIB, and by the Doris Duke Charitable Foundation Fund to Retain Clinician Scientists (FRCS) through the NYU School of Medicine.
Footnotes
Data will be shared by the corresponding author, Catherine Diefenbach (catherine.diefenbach@nyulangone.org) in response to e-mail requests within the guidelines of institutional policy for data sharing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Catherine Diefenbach, Perlmutter Cancer Center, NYU Langone Health, 240 East 38th St, 19th Floor, New York, NY 10016; catherine.diefenbach@nyulangone.org.
References
- 1.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;33(4):570-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535(7610):75-84. [DOI] [PubMed] [Google Scholar]
- 3.Jones L, Ho WQ, Ying S, et al. A subpopulation of high IL-21-producing CD4(+) T cells in Peyer’s Patches is induced by the microbiota and regulates germinal centers [published correction appears in Sci Rep. 2016;6:34899]. Sci Rep. 2016;6(1):30784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107-118. [DOI] [PubMed] [Google Scholar]
- 5.Geva-Zatorsky N, Sefik E, Kua L, et al. Mining the human gut microbiota for immunomodulatory organisms. Cell. 2017;168(5):928-943.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balmer ML, Schürch CM, Saito Y, et al. Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J Immunol. 2014;193(10):5273-5283. [DOI] [PubMed] [Google Scholar]
- 7.Tan TG, Sefik E, Geva-Zatorsky N, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci USA. 2016;113(50):E8141-E8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atarashi K, Nishimura J, Shima T, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455(7214):808-812. [DOI] [PubMed] [Google Scholar]
- 9.Cahenzli J, Köller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14(5):559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov II, McKenzie BS, Zhou L, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126(6):1121-1133. [DOI] [PubMed] [Google Scholar]
- 11.Sivan A, Corrales L, Hubert N, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawahara T, Takahashi T, Oishi K, et al. Consecutive oral administration of Bifidobacterium longum MM-2 improves the defense system against influenza virus infection by enhancing natural killer cell activity in a murine model. Microbiol Immunol. 2015;59(1):1-12. [DOI] [PubMed] [Google Scholar]
- 13.Zitvogel L, Galluzzi L, Viaud S, et al. Cancer and the gut microbiota: an unexpected link. Sci Transl Med. 2015;7(271):271ps1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao Q, Jiang F, Yin R, et al. Interplay between the lung microbiome and lung cancer. Cancer Lett. 2018;415:40-48. [DOI] [PubMed] [Google Scholar]
- 15.Viaud S, Saccheri F, Mignot G, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342(6161):971-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinato DJ, Howlett S, Ottaviani D, et al. Association of prior antibiotic treatment with survival and response to immune checkpoint inhibitor therapy in patients with cancer. JAMA Oncol. 2019;5(12):1774-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holler E, Butzhammer P, Schmid K, et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant. 2014;20(5):640-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenq RR, Ubeda C, Taur Y, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med. 2012;209(5):903-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maier L, Pruteanu M, Kuhn M, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555(7698):623-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters BA, Dominianni C, Shapiro JA, et al. The gut microbiota in conventional and serrated precursors of colorectal cancer [published correction appears in Microbiome. 2017;5(1):29]. Microbiome. 2016;4(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao R, Boehnke M. Quantifying and correcting for the winner’s curse in genetic association studies. Genet Epidemiol. 2009;33(5):453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.