Abstract
Monomethylamine (MMA) is an important climate-active oceanic trace gas and ubiquitous in the oceans. γ-Glutamylmethylamide synthetase (GmaS) catalyzes the conversion of MMA to γ-glutamylmethylamide, the first step in MMA metabolism in many marine bacteria. The gmaS gene occurs in ∼23% of microbial genomes in the surface ocean and is a validated biomarker to detect MMA-utilizing bacteria. However, the catalytic mechanism of GmaS has not been studied because of the lack of structural information. Here, the GmaS from Rhodovulum sp. 12E13 (RhGmaS) was characterized, and the crystal structures of apo-RhGmaS and RhGmaS with different ligands in five states were solved. Based on structural and biochemical analyses, the catalytic mechanism of RhGmaS was explained. ATP is first bound in RhGmaS, leading to a conformational change of a flexible loop (Lys287-Ile305), which is essential for the subsequent binding of glutamate. During the catalysis of RhGmaS, the residue Arg312 participates in polarizing the γ-phosphate of ATP and in stabilizing the γ-glutamyl phosphate intermediate; Asp177 is responsible for the deprotonation of MMA, assisting the attack of MMA on γ-glutamyl phosphate to produce a tetrahedral intermediate; and Glu186 acts as a catalytic base to abstract a proton from the tetrahedral intermediate to finally generate glutamylmethylamide. Sequence analysis suggested that the catalytic mechanism of RhGmaS proposed in this study has universal significance in bacteria containing GmaS. Our results provide novel insights into MMA metabolism, contributing to a better understanding of MMA catabolism in global carbon and nitrogen cycles.
Keywords: γ-glutamylmethylamide synthetase (GmaS), crystal structure, enzyme mechanism, monomethylamine (MMA) metabolism, enzyme catalysis, protein complex, bacterial metabolism
Abbreviations: AMPPCP, phosphomethylphosphonic acid-adenylate ester; AMPPNP, phosphoaminophosphonic acid-adenylate ester; GMA, γ-glutamylmethylamide; GmaS, γ-glutamylmethylamide synthetase; GS, glutamine synthetase; MetSox, methionine sulfoximine; MetSox-P, methionine sulfoximine phosphate; MMA, monomethylamine; MRC, marine Roseobacter clade; NMG, N-methylglutamate; PDB, protein data bank; RhGmaS, GmaS from Rhodovulum sp. 12E13
Methylated amines, such as monomethylamine (MMA), dimethylamine, trimethylamine, and trimethylamine N-oxide, are widely distributed in marine environments and play important roles in marine carbon (C) and nitrogen (N) cycles (1, 2, 3). In the ocean, the major source of methylated amines is likely the breakdown of quaternary amine osmolytes, such as betaine, choline, and carnitine (4, 5). MMA, an ammonium analog, is an important component of marine one-carbon (C1) compounds (i.e., compounds with no carbon–carbon bond) (6, 7); MMA provides carbon and energy sources as well as a nitrogen source for many microorganisms (2, 8, 9). In addition, the volatile MMA can enter the atmosphere from the oceans and has impacts on the global climate through participation of the formation of marine aerosols (3, 10).
There are two pathways identified from bacteria for aerobic MMA metabolism, a direct MMA-oxidation pathway and an indirect MMA-oxidation pathway (6). The direct MMA-oxidation pathway is only found in methylotrophic bacteria, through which MMA is metabolized to formaldehyde and ammonium by a single enzyme (4, 6). This enzyme is an MMA oxidase in gram-positive bacteria such as Arthrobacter (11) or an MMA dehydrogenase in gram-negative bacteria such as Methylobacterium extorquens and Paracoccus denitrificans (12, 13). In the indirect MMA-oxidation pathway, MMA is converted by γ-glutamylmethylamide synthetase (GmaS) to γ-glutamylmethylamide (GMA), which is further converted to N-methylglutamate (NMG) by NMG synthase, and finally to 5,10-methylenetetrahydrofolate (CH2 = THF) by NMG dehydrogenase (4, 8). Because NMG is an essential intermediate of the indirect MMA-oxidation pathway, this pathway is also termed as the NMG pathway.
Unlike the direct MMA-oxidation pathway which is only found in methylotrophic bacteria to date, the NMG pathway is adopted by both methylotrophic and nonmethylotrophic bacteria, in particular, by many bacteria of the marine Roseobacter clade (MRC) (14). MRC bacteria are ubiquitous and numerically abundant in marine environments (15, 16) and are important participants in MMA metabolism (14, 17, 18). It is estimated that half of the genomes of MRC strains contain gmaS, and this gene has been chosen as a biomarker to detect MMA-utilizing bacteria in the environment (6, 18). The gmaS gene occurs in ∼23% of microbial genomes in the surface ocean (17), suggesting that GmaS plays an important role in marine N and C cycles. However, despite that the biochemical characteristics of GmaS from several strains have been reported (19, 20, 21, 22), little is known about the molecular mechanism of GmaS catalyzing the conversion of MMA to GMA.
Sequence analysis indicates that GmaS is closely related to, but distinct from, the glutamine synthetase (GS) family, one of the oldest and most ubiquitously existing families of enzymes in biota (4, 23, 24). GmaS is more specific to MMA, whereas the natural substrate of GS enzymes is ammonia (19, 20, 25). This is consistent with the results of sequence analysis, showing that GmaS lacks key ammonia-binding residues that are conserved in GS enzymes (4, 21, 24, 26). GS enzymes can be divided into three distinct types, GSI, GSII, and GSIII (27, 28), and the crystal structures of GS have been determined (29, 30, 31). However, no crystal structure of GmaS is available to date, and the structural basis of GmaS to convert MMA to GMA remains obscure.
In this study, we report the structure of a GmaS enzyme and its molecular mechanism for the conversion of MMA to GMA. The gmaS gene from strain Rhodovulum sp. 12E13 (RhgmaS) was expressed, and the recombinant RhGmaS was purified and characterized. Six crystal structures of RhGmaS in different states were solved. Based on structural and mutational assays, we proposed the molecular mechanism of RhGmaS for the conversion of MMA to GMA. The results provide novel insights into MMA metabolism, leading to a better understanding of MMA catabolism in global C and N cycles.
Results and discussion
Characterization of RhGmaS
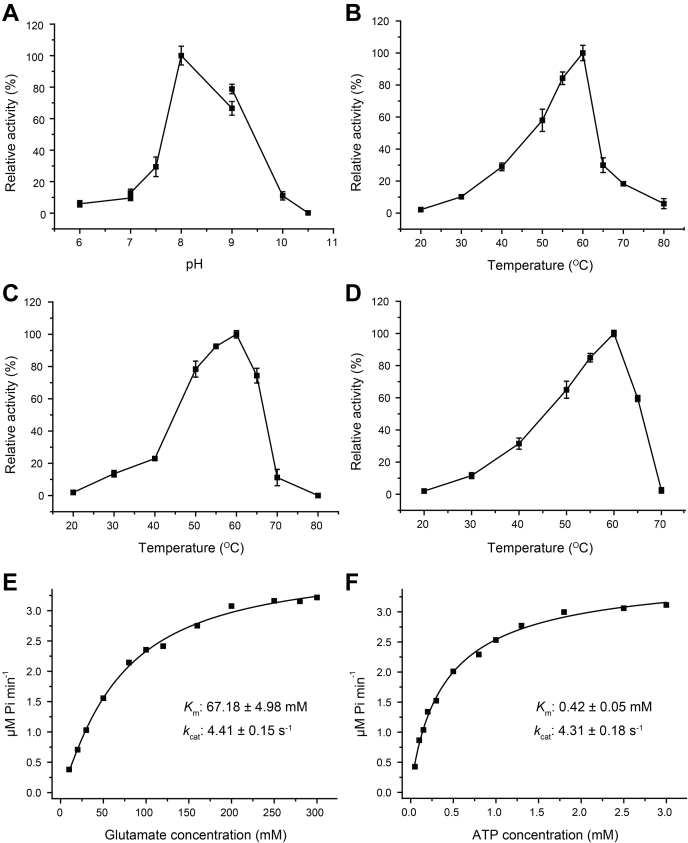
Full-length RhgmaS of R. sp. 12E13 contains 1293 nucleotides and encodes a 430-amino-acid polypeptide, which shows 61% identity to the functional GmaS from a type strain of MRC, Ruegeria pomeroyi DSS-3 (18, 32). We chemically synthesized RhgmaS, expressed it in Escherichia coli BL21 (DE3), and characterized the recombinant RhGmaS. The recombinant RhGmaS was active to catalyze the ligation of MMA and glutamate to produce GMA, with ATP and Mg2+ as cofactors. The optimal pH for RhGmaS enzymatic activity was ∼8.0 (Fig. 1A), similar to that of Methylovorus mays No.9 GmaS (7.5–8.0) (21). The optimal temperature of RhGmaS was 60 °C (Fig. 1B), which is higher than that of Methylophaga sp. AA-30 GmaS (40 °C) (19) and of M. mays No.9 GmaS (50 °C) (22). We noticed that the optimal temperatures of these GmaS enzymes are much higher than those of typical marine surface water column. Then, we purified two other GmaS homologs from MRC strains R. pomeroyi DSS-3 and Dinoroseobacter shibae DFL12. These two GmaS homologs also presented a high optimal temperature of 60 °C (Fig. 1, C–D), indicating that the relatively high optimal temperature is a common trait of GmaS proteins. Even so, RhGmaS still maintains a specific activity of ∼0.71 μmol min−1 mg−1 at 20 °C, suggesting that RhGmaS could work properly under the physiological temperature. The Km of RhGmaS for glutamate was 67.18 mM (Fig. 1E), and that for ATP was 0.42 mM (Fig. 1F). RhGmaS exhibited a Km value of 26.94 μM for MMA (Table 1), which is threefold lower than that of M. sp. AA-30 GmaS (89 μM) (19) and sixfold lower than that of M. mays No.9 GmaS (180 μM) (20). The micromolar level of Km values of GmaS enzymes for MMA indicates that GmaS enzymes possess high affinities to MMA.
Figure 1.
Characterization of recombinant GmaS. The error bar represents the SD of triplicate experiments.A, the effect of pH on the enzymatic activity of RhGmaS. B, the effect of temperature on the enzymatic activity of RhGmaS. C, the effect of temperature on the enzymatic activity of GmaS from R. pomeroyi DSS-3. D, the effect of temperature on the enzymatic activity of GmaS from Dinoroseobacter shibae DFL12. E, Kinetic parameters of RhGmaS for glutamate. F, kinetic parameters of RhGmaS for ATP. GmaS, γ-glutamylmethylamide synthetase; RhGmaS, GmaS from Rhodovulum sp. 12E13.
Table 1.
Kinetic parameters for recombinant RhGmaS with different substratesa
| Substrate | Km (μM) | Vmax (μM min−1) | kcat (s−1) |
|---|---|---|---|
| MMA | 26.94 ± 1.73 | 3.58 ± 0.11 | 4.18 ± 0.13 |
| Ethylamine | 58.10 ± 4.49 | 3.51 ± 0.11 | 4.09 ± 0.13 |
| Hydroxylamine | 211.03 ± 9.87 | 3.56 ± 0.05 | 4.15 ± 0.06 |
| Propylamine | 1.79 × 103 ± 0.12 × 103 | 3.63 ± 0.08 | 4.23 ± 0.09 |
| NH4Cl | 8.72 × 103 ± 0.97 × 103 | 3.65 ± 0.17 | 4.26 ± 0.20 |
| DMA | 16.07 × 103 ± 1.15 × 103 | 2.20 ± 0.09 | 2.57 ± 0.10 |
| TMA | 22.91 × 103 ± 1.50 × 103 | 2.13 ± 0.06 | 2.48 ± 0.07 |
DMA, dimethylamine; MMA, monomethylamine; TMA, trimethylamine.
The experiments were performed at pH 8.0, 30 °C. The data shown in the table are from triplicate experiments (means ± SDs).
Amine donors, such as hydroxylamine and ethylamine, can replace ammonia as substrate for some GS enzymes (20, 24, 30). To determine whether RhGmaS can catalyze different ammonia analogs, we analyzed the substrate specificity of RhGmaS (Table 1). In addition to MMA, RhGmaS can accept ethylamine, hydroxylamine, propylamine, ammonium chloride, dimethylamine, or trimethylamine as a substrate, indicating that this enzyme has a relatively broad substrate specificity. The Km of RhGmaS for MMA was the lowest among the tested ammonia analogs, while the Km for NH4Cl is much higher (Table 1), suggesting that MMA is likely the natural substrate of RhGmaS. This result is consistent with the previous reports that GmaS prefers MMA as its substrate rather than ammonia (4, 19, 20, 24). To investigate whether the N-terminal His-tag would affect the kinetic properties of the recombinant RhGmaS, we used thrombin to cut off the His-tag and measured the kinetic parameters of RhGmaS without His-tag (Fig. S1), which were similar to RhGmaS with His-tag. This result indicates that the presence of the His-tag has little effect on the kinetic properties of the enzyme. The RhGmaS proteins used in this study all contained the His-tag, unless otherwise noted.
Overall structure of RhGmaS
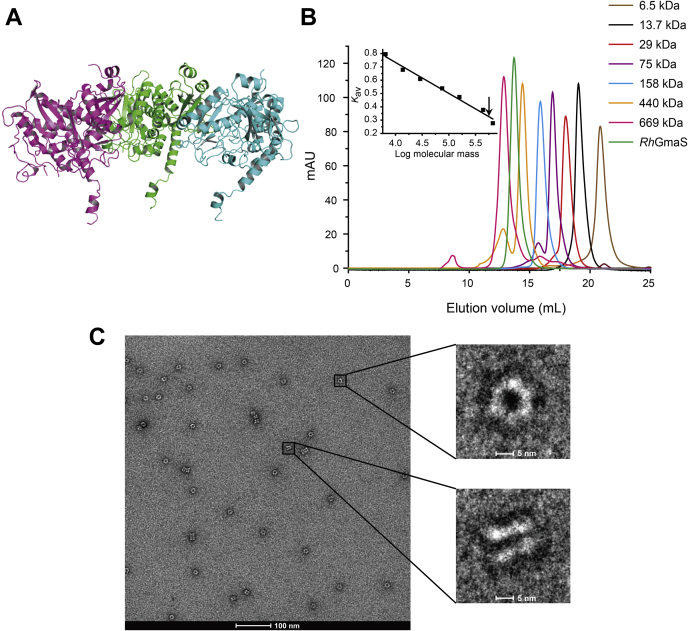
To gain insight into the catalytic mechanism of RhGmaS, the crystal structure of apo-RhGmaS was solved (Table 2). There are three monomers arranged as a trimer in an asymmetric unit (Fig. 2A), with each monomer composed of 15 α-helices and 13 β-strands. However, gel filtration analysis demonstrated that RhGmaS is a dodecamer in solution (Fig. 2B), which is consistent with the result of electron microscopic analysis (Fig. 2C). The negative staining electron micrograph clearly showed that RhGmaS consists of two hexameric rings, with each ring containing six monomers (Fig. 2C). Thus, RhGmaS should function as a dodecamer in the solution. During structural refinement, we could not find the electron densities of the N-terminal His-tag, suggesting that this tag is flexible. The electron microscopic analysis showed that RhGmaS without His-tag still maintains a dodecamer containing two hexameric rings in the solution (Fig. S2). These data suggest that the presence of the His-tag has little effect on the structural properties of RhGmaS. The overall structure of RhGmaS is similar to the structure of a GS enzyme from Bacillus subtilis (protein data bank [PDB] code: 4LNI), with an RMSD of 0.86 Å between these two structures. The B. subtilis GS also functions as a dodecamer (33).
Table 2.
Crystallographic data collection and refinement of RhGmaS
| Parameters | Apo-RhGmaS | RhGmaS–AMPPCP | RhGmaS–AMPPNP–MetSox | RhGmaS–ADP–MetSox-P | RhGmaS–ADP-C1 | RhGmaS–ADP-C2 |
|---|---|---|---|---|---|---|
| Diffraction data | ||||||
| Space group | I222 | I222 | I222 | I222 | I222 | I222 |
| Unit cell | ||||||
| a, b, c (Å) | 115.9, 179.0, 192.2 | 110.9, 176.7, 191.8 | 115.6, 174.5, 190.1 | 116.6, 174.7, 190.6 | 114.7, 176.0, 190.5 | 113.4, 177.6, 190.5 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution range (Å) | 50.0–2.8 (2.90–2.80) a | 50.0–1.96 (1.99–1.96) | 50.0–2.3 (2.34–2.30) | 50.0–2.1 (2.18–2.10) | 50.0–2.3 (2.34–2.30) | 50.0–2.3 (2.38–2.30) |
| Redundancy | 11.3 (10.7) | 4.6 (2.9) | 6.4 (6.0) | 4.0 (3.0) | 11.5 (10.5) | 13.4 (12.6) |
| Completeness (%) | 98.6 (99.1) | 98.1 (87.7) | 100.0 (100.0) | 97.5 (94.5) | 99.5 (100.0) | 100.0 (100.0) |
| Rmergeb | 0.1 (0.6) | 0.1 (0.4) | 0.1 (0.4) | 0.1 (0.5) | 0.2 (0.4) | 0.1 (0.5) |
| I/σI | 54.0 (14.7) | 14.2 (1.8) | 18.6 (2.9) | 21.8 (3.3) | 34.9 (6.4) | 22.6 (5.4) |
| Refinement statistics | ||||||
| Rwork/Rfree | 0.20/0.25 | 0.17/0.20 | 0.16/0.20 | 0.16/0.20 | 0.17/0.22 | 0.18/0.21 |
| RMSD from ideal geometry | ||||||
| Bond lengths (Å) | 0.009 | 0.007 | 0.007 | 0.007 | 0.007 | 0.007 |
| Bond angles (°) | 1.2 | 1.1 | 1.1 | 1.1 | 1.1 | 1.1 |
| Ramachandran plot (%) | ||||||
| Favored | 93.6 | 97.2 | 97.1 | 97.6 | 95.7 | 96.3 |
| Allowed | 6.2 | 2.8 | 2.9 | 2.3 | 4.1 | 3.7 |
| Outliers | 0.2 | 0 | 0 | 0.1 | 0.2 | 0 |
| Overall B-factors (Å2) | 57.1 | 18.9 | 37.2 | 24.3 | 35.7 | 27.1 |
Numbers in parentheses refer to data in the highest resolution shell.
Rmerge=∑hkl∑i|I(hkl)i -<I(hkl)>|/∑hkl∑iI(hkl)i, where I is the observed intensity, and I(hkl)i represents the observed intensity of each unique reflection.
Figure 2.
Overall structural analysis of RhGmaS.A, the overall structure of RhGmaS. There are three RhGmaS monomers arranged as a trimer in an asymmetric unit. The three monomers are colored in purple, green, and blue, respectively. B, gel filtration analysis of RhGmaS. Inset, semilog plot of the molecular mass of all standards used versus their Kav values. The black arrow indicates the position of RhGmaS Kav value interpolated in the regression line. The predicted molecular mass of RhGmaS monomer is 46.64 kDa. C, the negative-staining electron micrograph of RhGmaS. RhGmaS consists of two hexameric rings, with each ring containing six monomers. RhGmaS, GmaS from Rhodovulum sp. 12E13.
We further cocrystallized RhGmaS with ATP/ADP, glutamate, and MMA. However, after we solved the structures, we could not find glutamate or MMA bound in RhGmaS, and ATP was hydrolyzed to ADP during crystallization. We then tried to cocrystallize RhGmaS with substrate analogs. Phosphoaminophosphonic acid-adenylate ester (AMPPNP) and phosphomethylphosphonic acid-adenylate ester (AMPPCP) are nonhydrolyzable ATP analogs. Methionine sulfoximine (MetSox) is an analog of glutamate, with a methylsulfoximine group replacing the carboxyl group of glutamate. MetSox can be phosphorylated to form a transition analog, methionine sulfoximine phosphate (MetSox-P) (33). Finally, the crystal structures of the RhGmaS–AMPPCP complex, RhGmaS–AMPPNP–MetSox complex, RhGmaS–ADP–MetSox-P complex, and RhGmaS–ADP complexes in two conformations (RhGmaS–ADP-C1 and RhGmaS–ADP-C2) were solved (Table 2), which provide us abundant structural information for further mechanistic analysis. All the solved RhGmaS structures contain three molecules arranged as a trimer in an asymmetric unit, suggesting that the dodecameric RhGmaS has a relatively high symmetry.
Residues involved in the binding of ATP, glutamate and Mg2+
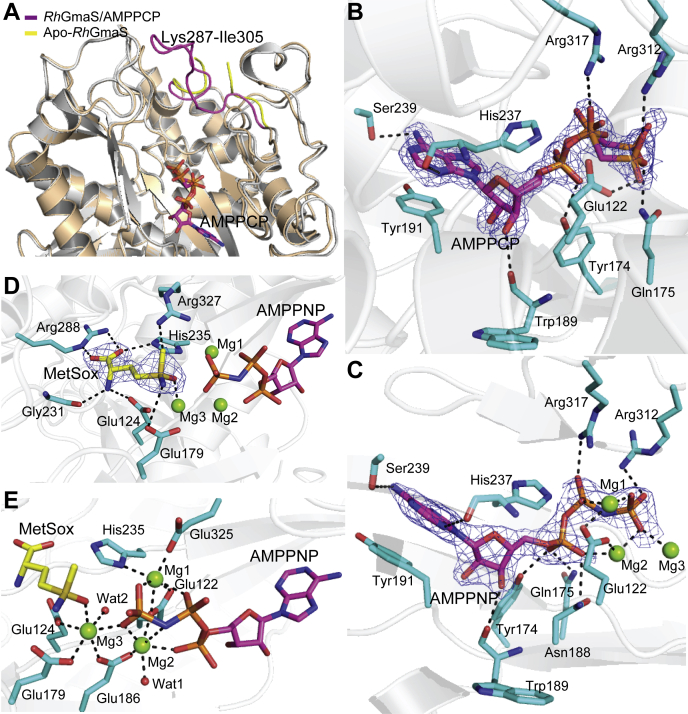
In the crystal structure of apo-RhGmaS, the electron densities of residues Pro292-Trp300 are rather poor, and we could not place these residues during structural refinement, indicating a relatively high flexibility of this region. However, in the crystal structure of the RhGmaS–AMPPCP complex, the electron densities of this region are clear, and the conformation of the loop Lys287-Ile305 is different from that in the apo-RhGmaS structure (Fig. 3A), suggesting that the binding of the ATP molecule will lead to the conformation change of the loop Lys287-Ile305.
Figure 3.
Structural analysis of RhGmaS–AMPPCP and RhGmaS–AMPPNP–MetSox complexes.A, structural alignment of RhGmaS–AMPPCP and apo-RhGmaS. The loop regions (Lys287-Ile305) of RhGmaS–AMPPCP (magenta) and that of apo-RhGmaS (yellow) are highlighted. B, residues involved in binding AMPPCP. AMPPCP is colored in magenta, and RhGmaS residues are colored in cyan. The 2Fo - Fc densities for AMPPCP are contoured in blue at 1.5σ. The possible hydrogen bonds are represented by dashed lines. C, residues and atoms involved in binding AMPPNP. AMPPNP is colored in magenta, and RhGmaS residues are colored in cyan. The 2Fo - Fc densities for AMPPNP are contoured in blue at 2.0σ. D, RhGmaS residues involved in binding MetSox. MetSox is colored in yellow. The 2Fo - Fc densities for MetSox are contoured in blue at 2.0σ. E, residues and ligands involved in binding Mg2+. The possible hydrogen bonds are represented by dashed lines. GmaS, γ-glutamylmethylamide synthetase; RhGmaS, GmaS from Rhodovulum sp. 12E13; MetSox, methionine sulfoximine.
Residues involved in binding ATP are identified based on the crystal structure of the RhGmaS–AMPPCP complex. The residue Tyr191 forms pi–pi stacking interaction with the adenine moiety of ATP, and residues His237 and Ser239 interact with the adenine moiety through hydrogen bonds (Fig. 3B). Trp189 forms a hydrogen bond with the ribose moiety, and residues Glu122, Tyr174, Gln175, Arg312, and Arg317 participate in binding the phosphate moieties of ATP (Fig. 3B). We noticed that the AMPPCP molecule exhibits two conformations in the RhGmaS–AMPPCP complex (Fig. 3B), suggesting that the binding of ATP without glutamate and Mg2+ may not be tight.
Despite many attempts, we did not obtain the crystal structure of the RhGmaS–glutamate complex, probably due to the low affinity of RhGmaS for glutamate (Fig. 1E). Instead, we solved the crystal structure of the RhGmaS–AMPPNP–MetSox complex (Table 2), which can reflect the binding mode of ATP and glutamate. The overall structure of the RhGmaS–AMPPNP–MetSox complex is similar to that of the RhGmaS–AMPPCP complex, with an RMSD of 0.19 Å between these two structures. Three Mg2+ were also identified in the crystal structure of the RhGmaS–AMPPNP–MetSox complex. The locations of AMPPNP, MetSox, and three Mg2+ are well defined based on the electron density map. All three Mg2+ participate in binding AMPPNP–ATP, which partially changes the binding mode of ATP. Compared with the RhGmaS–AMPPCP complex, the residue Glu122 does not interact with ATP directly but through Mg1 and Mg2, and Asn188 forms a new hydrogen bond with ATP (Fig. 3C).
The binding of MetSox in RhGmaS mainly depends on hydrogen bonds between its hydrophilic atoms and RhGmaS residues Glu124, Glu179, Gly231, His235, Arg288, and Arg327 (Fig. 3D). Interestingly, Arg288 is located in the flexible loop Lys287-Ile305, and the conformations of this loop in the RhGmaS–AMPPCP complex and the RhGmaS–AMPPNP–MetSox complex are identical. This result indicates that the conformational change of loop Lys287-Ile305 due to ATP binding will facilitate the subsequent binding of glutamate.
Three Mg2+ are coordinated by MetSox, AMPPNP, two water molecules, and RhGmaS residues Glu122, Glu124, Glu179, Glu186, His235, and Glu325 (Fig. 3E). In fact, when we cocrystallized RhGmaS and AMPPCP, we also added Mg2+ in the protein solution, but the fact that no Mg2+ presents in the RhGmaS–AMPPCP complex suggests that Mg2+ is bound after the binding of glutamate.
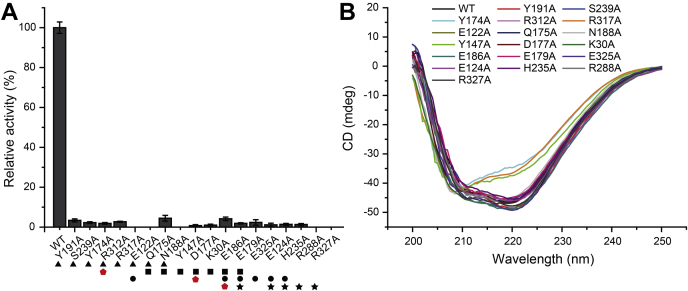
To confirm the importance of RhGmaS residues involved in binding ATP, glutamate, and Mg2+, we generated site-directed mutations to these residues and quantified the enzymatic activities of the mutants. All the mutations dramatically decreased the activity of RhGmaS (Fig. 4A), suggesting that these residues play important roles during the catalysis of RhGmaS. Circular-dichroism (CD) spectroscopy analysis showed that the secondary structures of the mutants exhibited little deviation from that of WT RhGmaS with the exceptions of Tyr174Ala and Arg317Ala (Fig. 4B), indicating that the decreases in the enzymatic activities of most mutants are caused by residue replacement rather than structural changes. Mutation of Tyr174 or Arg317 to Ala led to a change of the secondary structure of RhGmaS (Fig. 4B), which may be caused by the differences of side-chain properties between Tyr or Arg and Ala.
Figure 4.
Mutational analysis of important residues of RhGmaS that may participate in binding substrate and catalysis.A, the enzymatic activities of WT RhGmaS and its mutants. The activity of WT RhGmaS is taken as 100%. Residues involved in binding ATP, MMA, Mg2+, and glutamate are marked with black triangles, squares, dots, and stars, respectively. Red pentagons indicate the catalytic residues of RhGmaS. The error bar represents SD of triplicate experiments. B, CD spectra of WT RhGmaS and its mutants. MMA, monomethylamine; RhGmaS, GmaS from Rhodovulum sp. 12E13; CD, circular-dichroism.
The catalytic residues of RhGmaS
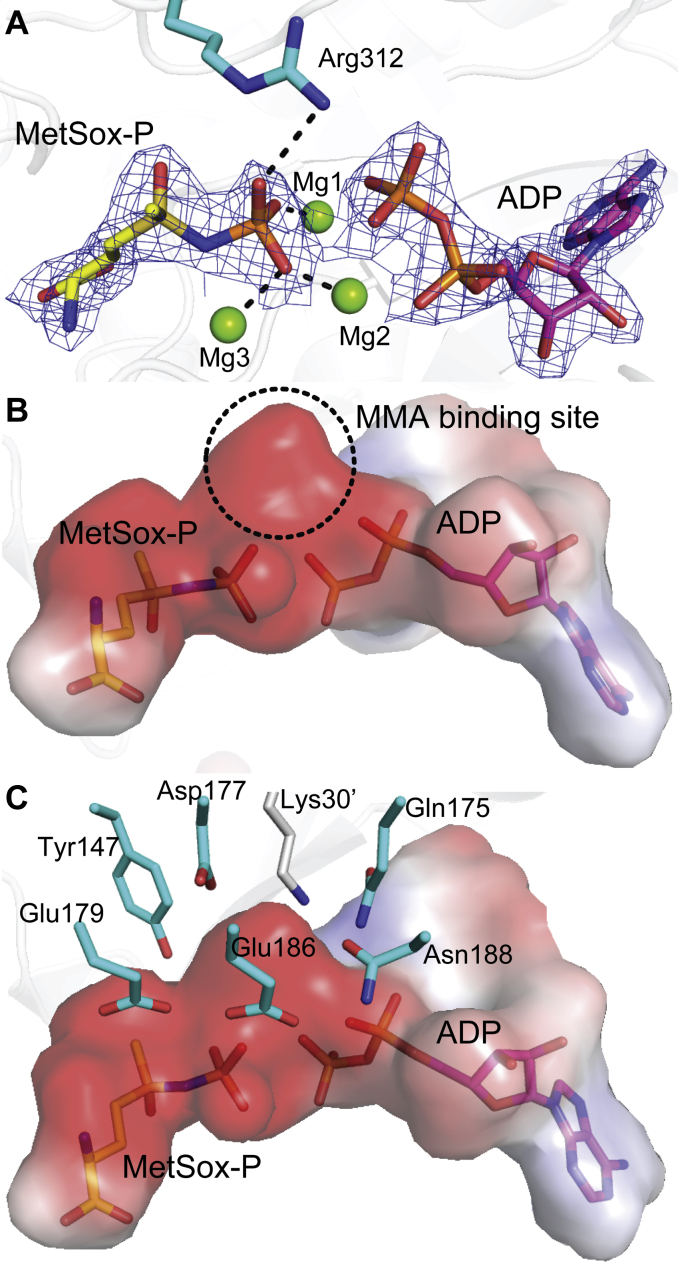
Based on the catalytic mechanism of GS enzymes (33), it is likely that RhGmaS catalyzes a two-step reaction: in the first step, glutamate is phosphorylated, leading to the formation of γ-glutamyl phosphate, which is subsequently attacked by MMA to yield GMA in the second step. To obtain a transition state of RhGmaS catalysis, we cocrystallized RhGmaS with ATP, MgCl2, and MetSox and obtained the crystal structure of the RhGmaS–ADP–MetSox-P complex (Table 2). In this structure, Arg312 and Mg2+ interact with the phosphate group of MetSox-P (Fig. 5A), indicating that Arg312 and Mg2+ are involved in stabilizing the transition state.
Figure 5.
Structural analysis of the RhGmaS–ADP–MetSox-P complex.A, the positions of MetSox-P and ADP. The 2Fo - Fc densities for MetSox-P and ADP are contoured in blue at 2.0σ. B, the MMA binding site of RhGmaS. C, RhGmaS residues composing the negative-charged MMA binding site. MMA, monomethylamine; RhGmaS, GmaS from Rhodovulum sp. 12E13; MetSox, methionine sulfoximine.
To obtain the crystal structure of RhGmaS in complex with MMA, we tried cocrystallization and soaking methods, but all attempts failed. However, structure analysis of the RhGmaS–ADP–MetSox-P (the γ-glutamyl phosphate mimic) complex provided us some clues of the MMA binding site. A negative-charged pocket close to the ADP and MetSox-P molecules is observed in the complex (Fig. 5B), which is mainly formed by hydrophilic residues, including Tyr147, Gln175, Asp177, Glu179, Glu186, Asn188, and Lys30' (where a prime indicates the neighboring subunit) (Fig. 5C). Because MMA is alkaline, it is likely that RhGmaS attracts MMA through electrostatic interactions. After MMA enters the active site, it will be deprotonated before its attack on the γ-glutamyl phosphate of ATP. Structure analysis suggests that Asp177 or Glu186 is the potential residue to deprotonate MMA (Fig. 5C). Previous studies on GS enzymes indicate that the attack of ammonia on γ-glutamyl phosphate will generate a tetrahedral intermediate, which is further deprotonated before the final formation of glutamine (33, 34). In the case of RhGmaS, the attack of MMA on γ-glutamyl phosphate may generate a similar tetrahedral intermediate. Because Glu186 is closer to the phosphoryl group of MetSox-P than Asp177 in the RhGmaS–ADP–MetSox-P complex, we deduced that Glu186 deprotonates the tetrahedral intermediate while Asp177 deprotonates MMA during RhGmaS catalysis.
Mutations of residues composing the MMA binding site and directly participating in the catalysis decreased the activity of RhGmaS significantly (Fig. 4A), suggesting a crucial role of these residues. CD spectroscopy analysis suggested that the decreases in the enzymatic activities of most mutants resulted from residue replacements, while the mutation Tyr147Ala changed the secondary structure of RhGmaS (Fig. 4B).
Conformational change of RhGmaS after catalysis
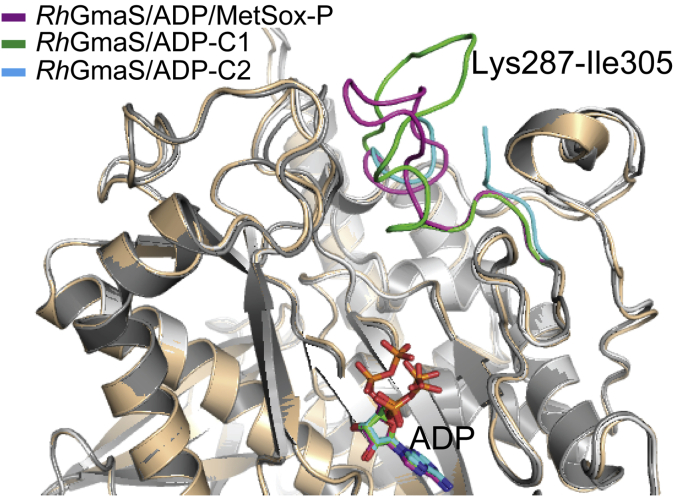
After GMA is generated, GMA, ATP, and Mg2+ will be released from the catalytic pocket of RhGmaS, enabling RhGmaS to be ready for the next catalytic cycle. We determined the crystal structures of two RhGmaS–ADP complexes in different conformations (RhGmaS–ADP-C1 and RhGmaS–ADP-C2), which may reflect the events after catalysis. When structures of RhGmaS–ADP–MetSox-P, RhGmaS–ADP-C1, and RhGmaS–ADP-C2 are aligned (Fig. 6), it can be seen that the conformation of ADP in RhGmaS–ADP–MetSox-P is different from those in RhGmaS–ADP-C1 and RhGmaS–ADP-C2, and the conformations of the loop region Lys287-Ile305 in these three structures are different. Because Arg288 in the loop region participates in binding glutamate (Fig. 3D), the conformational change of the loop region indicates that GMA and Mg2+ are released firstly after the catalysis. Without the binding of GMA and Mg2+, the binding of ADP becomes relatively loose and will be finally released from the active site of RhGmaS.
Figure 6.
Analysis of the conformational change of RhGmaS after catalysis. The loop regions (Lys287-Ile305) of RhGmaS–ADP–MetSox-P (magenta), of RhGmaS–ADP-C1 (green), and of RhGmaS–ADP-C2 (blue) are highlighted. ADP molecules are shown as sticks. RhGmaS, GmaS from Rhodovulum sp. 12E13; MetSox, methionine sulfoximine.
The catalytic cycle of RhGmaS
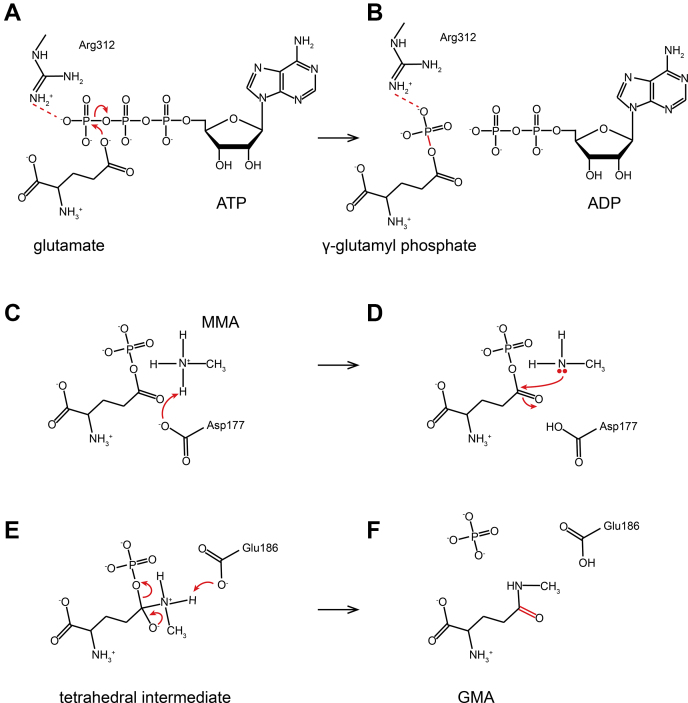
Based on structural and biochemical assays, the catalytic cycle of RhGmaS converting MMA to GMA is proposed (Fig. 7). In the first step of RhGmaS catalysis, ATP is first bound in RhGmaS, which leads to a conformational change of the loop Lys287-Ile305, assisting the subsequent binding of glutamate. The residue Arg312 and Mg2+ directly interact with ATP γ-phosphate (Fig. 3C), which helps polarize the γ-phosphate and facilitates the nucleophilic attack of glutamate (33) (Fig. 7A). After glutamate is phosphorylated, Arg312 forms a hydrogen bond with the phosphate group of γ-glutamyl phosphate to stabilize the transition state (Fig. 7B). In the second step, MMA is attracted by RhGmaS and enters into the binding pocket. The residue Asp177 deprotonates MMA (Fig. 7C), enabling MMA to attack γ-glutamyl phosphate (Fig. 7D). A tetrahedral intermediate is then produced with the attack of MMA (Fig. 7E). Finally, Glu186 abstracts a proton from the tetrahedral intermediate, and GMA is generated (Fig. 7F). Then RhGmaS releases GMA and ADP and is ready for the next catalytic cycle.
Figure 7.
The catalytic mechanism of RhGmaS converting MMA to GMA.A, the residue Arg312 directly interacts with ATP γ-phosphate, which helps polarize the γ-phosphate and facilitates the nucleophilic attack of glutamate. B, Arg312 interacts with the phosphate group of γ-glutamyl phosphate to stabilize the transition state. C, MMA is deprotonated by the residue Asp177. D, MMA attacks γ-glutamyl phosphate to produce a tetrahedral intermediate. E, Glu186 abstracts a proton from the tetrahedral intermediate. F, GMA is generated. GMA, γ-glutamylmethylamide; MMA, monomethylamine; RhGmaS, GmaS from Rhodovulum sp. 12E13.
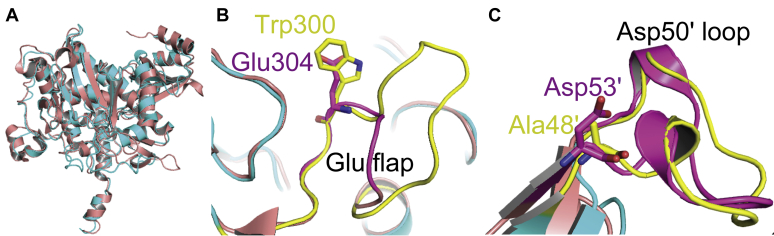
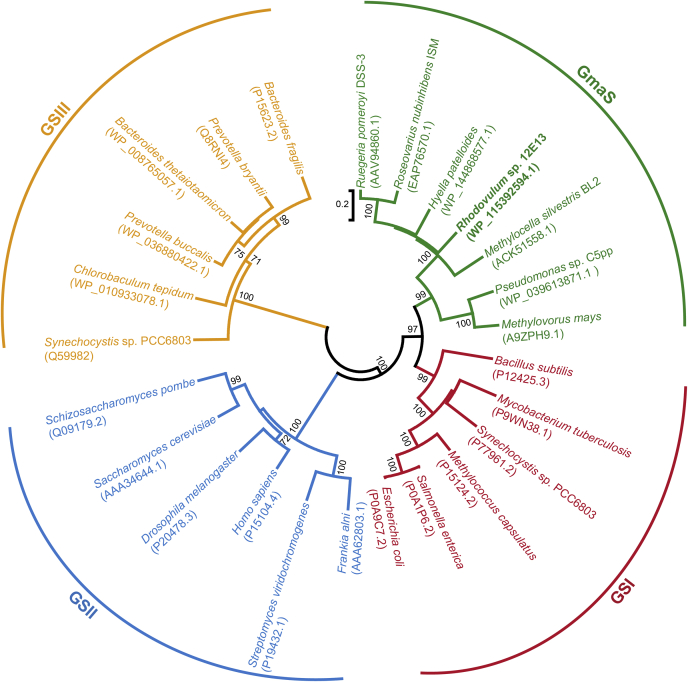
Despite that the overall structure of RhGmaS is similar to that of the GS enzyme from B. subtilis (PDB code: 4LNI) (33) (Fig. 8A), the catalytic processes of RhGmaS and GS are different. Two loops (the Glu flap and the Asp50' loop) play key roles in the catalytic process of GS (25, 33). The Glu flap of GS possesses several functions: (i) guarding substrate entry and product release; (ii) shielding the γ-glutamyl phosphate from aberrant hydrolysis; and more importantly, (iii) harboring the conserved glutamate residue (Glu304 in B. subtilis GS), which acts as the catalytic base for the final attack on the tetrahedral intermediate (25, 33). Structural analysis suggests that the loop Lys287-Ile305 of RhGmaS corresponds to the Glu flap in GS (Fig. 8B). However, there is no glutamate residue in this loop, and the corresponding residue of Glu304 in B. subtilis GS is Trp300 in RhGmaS (Fig. 8B). For RhGmaS, this loop region only acts as a gate for substrate entry and product release, and Glu186 is the catalytic residue to attack the tetrahedral intermediate. The aspartate residue in the Asp50' loop is strictly conserved in GS, which has two functions: (i) participating in binding ammonium and (ii) deprotonating the ammonium in the active site to create ammonia (25, 33). The corresponding region of the Asp50' loop in RhGmaS is Phe47'-Ala61', and the corresponding residue of Asp50' in RhGmaS is Ala48' (Fig. 8C). Structural analysis suggests that no residue of RhGmaS in the region Phe47'-Ala61' participates in binding MMA, and Lys30' is the only residue from the neighboring subunit involved in composing the MMA binding site (Fig. 5C). The result of structural analysis confirmed the previous sequence analysis that GmaS lacks the key ammonia-binding residue (4, 21, 24, 26), highlighting the differences between GmaS and GS. Moreover, phylogenetic analysis indicated that GmaS proteins form a separate clade from GSI, GSII, and GSIII enzymes (Fig. 9), which is consistent with previous studies (4, 32), suggesting the divergent evolution of GmaS from GS.
Figure 8.
Structural comparison between RhGmaS and the GS enzyme from Bacillus subtilis (PDB code: 4LNI).A, superimposition of the RhGmaS structure (cyan) onto the GS structure (red). B, the Glu flap of GS (purple) and the corresponding loop in RhGmaS (yellow). The conserved glutamate residue (Glu304 in B. subtilis GS) and the corresponding residue in RhGmaS (Trp300) are shown as sticks. C, the Asp50' loop of GS (purple) and the corresponding loop in RhGmaS (yellow). The conserved aspartate residue (Asp53' in B. subtilis GS) and the corresponding residue in RhGmaS (Ala48') are shown as sticks. GS, glutamine synthetase; PDB, protein data bank; RhGmaS, GmaS from Rhodovulum sp. 12E13.
Figure 9.
The neighbor-joining phylogenetic tree of GSI, GSII, GSIII, and GmaS. Phylogenetic analyses were carried out using the MEGA, version 6.0 (39). GmaS, γ-glutamylmethylamide synthetase.
Universality of the catalytic mechanism of RhGmaS
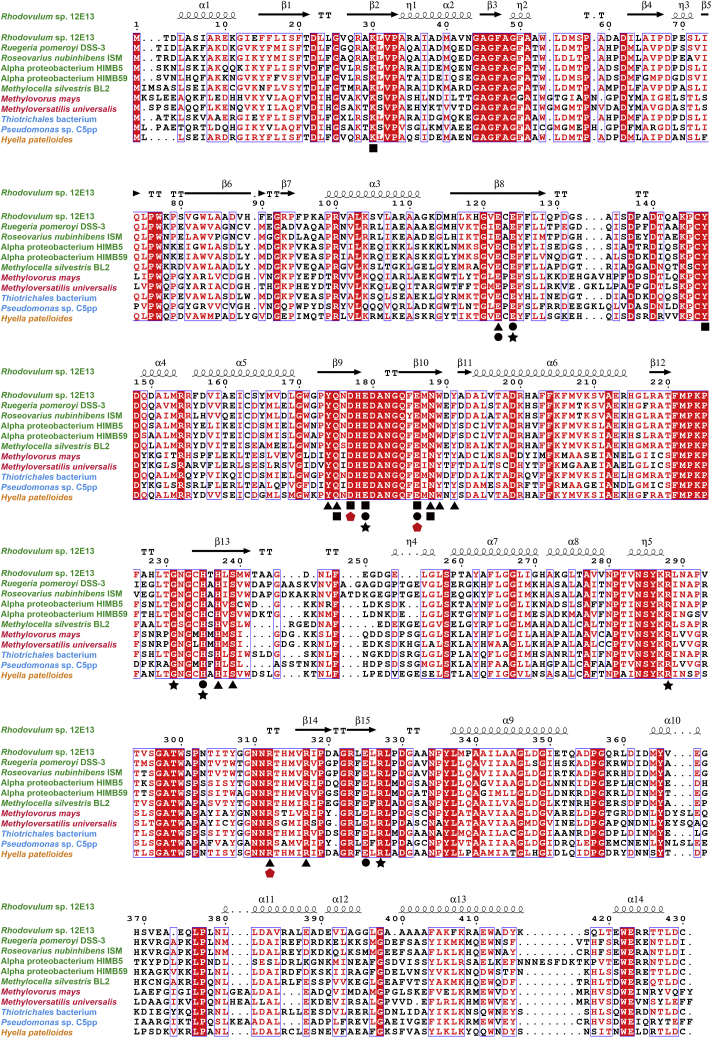
GmaS homologs widely occur in marine bacteria, including MRC and SAR11 clade (17), supporting a significant role of MMA metabolism in marine C and N cycles. Moreover, many soil bacteria are also reported to possess GmaS homologs, such as Methylocella silvestris BL2 (4) and Pseudomonas sp. C5pp (24). To study the ubiquity of the RhGmaS catalytic mechanism, we carried out sequence alignment of GmaS homologs from different strains (Fig. 10). The result showed that most of the residues involved in binding ATP, glutamate, MMA, and Mg2+ are highly conserved in GmaS homologs (Fig. 10). In particular, the catalytic residues Arg312, Asp177, and Glu186 are strictly conserved in all GmaS homologs (Fig. 10), suggesting that the molecular mechanism of RhGmaS to convert MMA to GMA is likely adopted by most of other bacterial GmaSs.
Figure 10.
Sequence alignment of GmaS proteins. Residues involved in binding ATP, MMA, Mg2+, and glutamate are marked with black triangles, squares, dots, and stars, respectively. Red pentagons indicate the catalytic residues. GmaS proteins from strains of different classes are colored as follows: green for Alphaproteobacteria, red for Betaproteobacteria, blue for Gammaproteobacteria, and yellow for Cyanophyceae. GmaS, γ-glutamylmethylamide synthetase; MMA, monomethylamine.
Conclusion
In this study, we solved the crystal structures of RhGmaS in different states for the first time. Based on structural and mutational analysis, we explained the catalytic mechanism of RhGmaS for the conversion of MMA to GMA in detail. Sequence alignment indicated that the proposed molecular mechanism of RhGmaS has universal significance. Our results provide novel insights into marine MMA metabolism and broaden our understanding of the biogeochemical cycles of C and N.
Experimental procedures
Bacterial strains and growth conditions
The E. coli strains DH5α and BL21 (DE3) were grown in the lysogeny broth medium at 37 °C.
Gene synthesis, point mutation, and protein expression and purification
The full-length RhgmaS gene from R. sp. 12E13 was synthesized by the Beijing Genomics Institute (China). The gene was then cloned into the pET-28a vector containing an N-terminal His-tag (Novagen). All point mutations of RhGmaS were created by the PCR-based method and confirmed by DNA sequencing. The RhGmaS protein and all the mutants were expressed in E. coli BL21 (DE3). The recombinant strains were grown at 37 °C in the lysogeny broth medium containing 30 μg/ml kanamycin and then were induced by 0.5-mM IPTG at 18 °C for 16 h. All proteins were purified first by Ni2+ nitrilotriacetic acid resin (Qiagen, Germany) and then by anion-exchange chromatography on a Source 15Q column (GE Healthcare) with 0- to 1-M NaCl in 50-mM Tris-HCl (pH 8.0). The eluted proteins were further fractionated by gel filtration on a Superose 6 column (GE Healthcare) with the buffer containing 10-mM Tris HCl (pH 8.0) and 100-mM NaCl. To prepare RhGmaS protein without His-tag, we digested the purified RhGmaS by thrombin (Solarbio, China) at 4 °C overnight, and then the mixtures were fractionated by gel filtration on a Superose 6 column.
Gel filtration analysis
The Superose 6 column was calibrated in the buffer containing 10-mM Tris HCl (pH 8.0) and 100-mM NaCl using the following standards from GE Healthcare: thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), carbonic anhydrase (29 kDa), ribonuclease A (13.7 kDa), and aprotinin (6.5 kDa). The void volume of Superose 6 column was determined with Blue Dextran 2000 (2000 kDa).
Negative-stain electron microscopy
RhGmaS proteins (∼6 μg/ml) were applied to glow-discharged electron microscopy grids for 1 min and then stained with 2% (w/v) uranyl acetate twice for 30 s and 1 min, respectively. Negatively stained grids were imaged on a Talos L120C transmission electron microscope. (Thermo Fisher Scientific) operated at 120 kV. Images were recorded at ×73,000 magnification with a Ceta 16M camera (Thermo Fisher Scientific).
Enzymatic activity assays
The enzymatic activity of GmaS was measured by determining the production of inorganic phosphate using a PiColorLock reagent kit (Expedeon, UK). The reaction system contained 200-mM glutamate, 25-mM MgCl2, 3-mM ATP, 1-mM MMA, and 100-mM Tris HCl (pH 8.0) and 2.5-nM RhGmaS, 41-nM R. pomeroyi DSS-3 GmaS, or 2.8-nM D. shibae DFL12 GmaS. The reaction mixture was incubated at 60 °C for 10 min, and then, 20% (v/v) hydrochloric acid was added into the mixture to stop the reaction. The amount of inorganic phosphate in the mixture was determined by monitoring the absorbance at 635 nm. The optimal temperature for GmaS enzymatic activity was measured in a range from 20 to 80 °C at pH 8.0. The optimal pH was examined at 60 °C using Bis-Tris buffer for pH 6–7, Tris-HCl buffer for pH 7–9, and glycine-NaOH buffer for pH 9–10.5. The kinetic parameters of RhGmaS (with and without the His-tag) for glutamate, ATP, and ammonia analogs were determined with the reaction system containing 14.3-nM RhGmaS and different concentrations of substrates. All the measurements were carried out in 100-mM Tris-HCl buffer (pH 8.0) at 30 °C.
Crystallization and data collection
Purified RhGmaS protein was concentrated to ∼11 mg/ml in the buffer containing 10 mM Tris HCl (pH 8.0) and 100-mM NaCl. All of the crystals were obtained at 18 °C after 1–2 weeks using the sitting-drop vapor diffusion method (the ratio of protein to reservoir solution was 1:1). Apo-RhGmaS crystals were obtained in the buffer containing 0.1-M MES (pH 6.5) and 1-M lithium sulfate. To obtain RhGmaS–AMPPCP crystals, AMPPCP and MgCl2 were added to RhGmaS with a final concentration of 2 mM, and the solution was mixed 1:1 with the crystallization buffer containing 0.1-M HEPES (pH 7.5), 0.2-M ammonium acetate, and 25% (v/v) isopropanol. To obtain the crystals of the RhGmaS–AMPPNP–MetSox complex, RhGmaS was cocrystallized with AMPPNP (2 mM), MetSox (2 mM), and MgCl2 (2 mM), and the crystals were obtained in the crystallization buffer containing 0.1 M bicine (pH 8.5), 10% (v/v) 2-propanol, and 30% (v/v) polyethylene glycol 1500. RhGmaS–MetSox-P–ADP crystals were obtained under the same condition to the RhGmaS–AMPPNP–MetSox crystals, with AMPPNP replaced by ATP. When RhGmaS was cocrystallized with ATP (2 mM), glutamate (2 mM), and MgCl2 (2 mM), two structures of RhGmaS in complex with ADP were obtained. The crystals of RhGmaS–ADP-C1 were obtained in the crystallization buffer containing 1.1-M ammonium tartrate dibasic (pH 7.0), and the crystals of RhGmaS–ADP-C2 were obtained in the crystallization buffer containing 0.1-M MES (pH 6.5) and 1-M lithium sulfate.
All X-ray diffraction data were collected on the BL18U1 beamline at the Shanghai Synchrotron Radiation Facility. The initial diffraction data sets were processed by the HKL3000 program with its default settings (35).
Structure determination and refinement
The crystal structures of apo-RhGmaS, RhGmaS–AMPPCP, RhGmaS–AMPPNP/MetSox, RhGmaS–ADP–MetSox-P, RhGmaS–ADP-C1, and RhGmaS–ADP-C2 all belong to the I222 space group. The crystal structure of apo-RhGmaS was determined by molecular replacement using the CCP4 program Phaser (36), with the structure of a GS enzyme from B. subtilis (PDB code: 4LNI) as the search model. All the other structures were determined by molecular replacement with the structure of apo-RhGmaS as the search model. The refinements of these structures were performed using Coot (37) and Phenix (38). All the structure figures were processed using the program PyMOL (http://www.pymol.org/).
Bioinformatics
Multiple sequence alignment was performed using CLC sequence viewer 6.5.3 (CLC Bio A/S). Phylogenetic tree was conducted using the MEGA, version 6.0 (39), with the neighbor-joining algorithms.
CD spectroscopic assays
CD spectroscopic assays for RhGmaS and all its mutants were carried out on a J-1500 Spectrometer (Jasco, Japan) at 25 °C. The concentration of the proteins was 8.5 μM in the buffer of 10-mM Tris HCl (pH 8.0) containing 100-mM NaCl. The spectra were collected from 250 to 200 nm at a scan speed of 500 nm min−1 with a band width of 1 nm.
Data availability
The structures of apo-RhGmaS, RhGmaS–AMPPCP, RhGmaS–AMPPNP–MetSox, RhGmaS–ADP–MetSox-P, RhGmaS–ADP-C1, and RhGmaS–ADP-C2 have been deposited in PDB under accession codes 7CQL, 7CQN, 7CQQ, 7CQU, 7CQW, and 7CQX, respectively. All data are contained within the manuscript and the supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank the staffs from BL17U1, BL18U1 and BL19U1 beamlines of the National Facility for Protein Science Shanghai (NFPS) and Shanghai Synchrotron Radiation Facility, for assistance during data collection.
Author contributions
C.-Y. L. and Y.-Z. Z. designed the study. N. W. performed the majority of the biochemical experiments. C.-Y. L. solved all the crystal structures and performed data interpretation. X.-L. C. directed the study. C. G., M. P., P. W., N. Z., and Q.-T. S. helped in experiments. N. W., C.-Y. L., and X.-L. C. wrote the manuscript. Y. C. and F. L. did critical revision of the manuscript for important intellectual content.
Funding and additional information
This work was supported by the National Science Foundation of China (grants 91851205, 31630012, U1706207, 42076229, 31800107, and 41706152), the National Key Research and Development Program of China (2018YFC1406700, 2016YFA0601303, 2018YFC1406504), the Major Scientific and Technological Innovation Project (MSTIP) of Shandong Province (2019JZZY010817), the Program of Shandong for Taishan Scholars (tspd20181203), AoShan Talents Cultivation Program supported by Qingdao National Laboratory for Marine Science and Technology (2017ASTCP-OS14), the grant of Laboratory for Marine Biology and Biotechnology (OF2019NO02), Pilot National Laboratory for Marine Science and Technology (Qingdao), and the Fundamental Research Funds for the Central Universities.
Biography

Ning Wang is currently studying for a PhD in Microbiology at Shandong University. Their research interests are in the biochemical processes and molecular mechanisms of methylated amines metabolism driven by marine bacteria. They hope to keep working in this field, answering the unsolved questions of how marine bacteria transport and metabolize these small organic molecules.
Edited by Chris Whitfield
Footnotes
This article contains supporting information.
Supporting information
References
- 1.Gibb S.W., Mantoura R.F.C., Liss P.S., Barlow R.G. Distributions and biogeochemistries of methylamines and ammonium in the Arabian Sea. Deep Sea Res. Part II. 1999;46:593–615. [Google Scholar]
- 2.Stein L.Y. Methylamine: a vital nitrogen (and carbon) source for marine microbes. Environ. Microbiol. 2017;19:2117–2118. doi: 10.1111/1462-2920.13716. [DOI] [PubMed] [Google Scholar]
- 3.Carpenter L.J., Archer S.D., Beale R. Ocean-atmosphere trace gas exchange. Chem. Soc. Rev. 2012;41:6473–6506. doi: 10.1039/c2cs35121h. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y., Scanlan J., Song L., Crombie A., Rahman M.T., Schäfer H., Murrell J.C. Gamma-glutamylmethylamide is an essential intermediate in the metabolism of methylamine by Methylocella silvestris. Appl. Environ. Microbiol. 2010;76:4530–4537. doi: 10.1128/AEM.00739-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poste A.E., Grung M., Wright R.F. Amines and amine-related compounds in surface waters: a review of sources, concentrations and aquatic toxicity. Sci. Total Environ. 2014;481:274–279. doi: 10.1016/j.scitotenv.2014.02.066. [DOI] [PubMed] [Google Scholar]
- 6.Wischer D., Kumaresan D., Johnston A., El Khawand M., Stephenson J., Hillebrand-Voiculescu A.M., Chen Y., Colin Murrell J. Bacterial metabolism of methylated amines and identification of novel methylotrophs in Movile Cave. ISME J. 2015;9:195–206. doi: 10.1038/ismej.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhuang G.C., Peña-Montenegro T.D., Montgomery A., Hunter K.S., Joye S.B. Microbial metabolism of methanol and methylamine in the Gulf of Mexico: insight into marine carbon and nitrogen cycling. Environ. Microbiol. 2018;20:4543–4554. doi: 10.1111/1462-2920.14406. [DOI] [PubMed] [Google Scholar]
- 8.Latypova E., Yang S., Wang Y.S., Wang T., Chavkin T.A., Hackett M., Schäfer H., Kalyuzhnaya M.G. Genetics of the glutamate-mediated methylamine utilization pathway in the facultative methylotrophic beta-proteobacterium Methyloversatilis universalis FAM5. Mol. Microbiol. 2010;75:426–439. doi: 10.1111/j.1365-2958.2009.06989.x. [DOI] [PubMed] [Google Scholar]
- 9.Taubert M., Grob C., Howat A.M., Burns O.J., Pratscher J., Jehmlich N., von Bergen M., Richnow H.H., Chen Y., Murrell J.C. Methylamine as a nitrogen source for microorganisms from a coastal marine environment. Environ. Microbiol. 2017;19:2246–2257. doi: 10.1111/1462-2920.13709. [DOI] [PubMed] [Google Scholar]
- 10.Lidbury I., Mausz M.A., Scanlan D.J., Chen Y. Identification of dimethylamine monooxygenase in marine bacteria reveals a metabolic bottleneck in the methylated amine degradation pathway. ISME J. 2017;11:1592–1601. doi: 10.1038/ismej.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X., Fuller J.H., McIntire W.S. Cloning, sequencing, expression, and regulation of the structural gene for the copper/topa quinone-containing methylamine oxidase from Arthrobacter strain P1, a gram-positive facultative methylotroph. J. Bacteriol. 1993;175:5617–5627. doi: 10.1128/jb.175.17.5617-5627.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chistoserdov A.Y., Chistoserdova L.V., McIntire W.S., Lidstrom M.E. Genetic organization of the mau gene cluster in Methylobacterium extorquens AM1: complete nucleotide sequence and generation and characteristics of mau mutants. J. Bacteriol. 1994;176:4052–4065. doi: 10.1128/jb.176.13.4052-4065.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Husain M., Davidson V.L. Purification and properties of methylamine dehydrogenase from Paracoccus denitrificans. J. Bacteriol. 1987;169:1712–1717. doi: 10.1128/jb.169.4.1712-1717.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lidbury I.D.E.A., Murrell J.C., Chen Y. Trimethylamine and trimethylamine N-oxide are supplementary energy sources for a marine heterotrophic bacterium: implications for marine carbon and nitrogen cycling. ISME J. 2015;9:760–769. doi: 10.1038/ismej.2014.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selje N., Simon M., Brinkhoff T. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature. 2004;427:445–448. doi: 10.1038/nature02272. [DOI] [PubMed] [Google Scholar]
- 16.González J.M., Moran M.A. Numerical dominance of a group of marine bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y., Patel N.A., Crombie A., Scrivens J.H., Murrell J.C. Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc. Natl. Acad. Sci. U. S. A. 2011;108:17791–17796. doi: 10.1073/pnas.1112928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y. Comparative genomics of methylated amine utilization by marine Roseobacter clade bacteria and development of functional gene markers (tmm, gmaS) Environ. Microbiol. 2012;14:2308–2322. doi: 10.1111/j.1462-2920.2012.02765.x. [DOI] [PubMed] [Google Scholar]
- 19.Kimura T., Sugahara I., Hanai K., Tonomura Y. Purification and characterization of gamma-glutamylmethylamide synthetase from Methylophaga sp. AA-30. Biosci. Biotechnol. Biochem. 1992;56:708–711. doi: 10.1271/bbb.56.708. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto S., Wakayama M., Tachiki T. Characterization of theanine-forming enzyme from Methylovorus mays no. 9 in respect to utilization of theanine production. Biosci. Biotechnol. Biochem. 2007;71:545–552. doi: 10.1271/bbb.60590. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto S., Wakayama M., Tachiki T. Cloning and expression of Methylovorus mays No. 9 gene encoding gamma-glutamylmethylamide synthetase: an enzyme usable in theanine formation by coupling with the alcoholic fermentation system of baker's yeast. Biosci. Biotechnol. Biochem. 2008;72:101–109. doi: 10.1271/bbb.70462. [DOI] [PubMed] [Google Scholar]
- 22.Yang S.Y., Han Y.H., Park Y.L., Park J.Y., No S.Y., Jeong D., Park S., Park H.Y., Kim W., Seo S.O., Yang Y.H. Production of L-Theanine Using Escherichia coli whole-cell overexpressing gamma-glutamylmethylamide synthetase with baker’s yeast. J. Microbiol. Biotechnol. 2020;30:785–792. doi: 10.4014/jmb.1910.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumada Y., Benson D.R., Hillemann D., Hosted T.J., Rochefort D.A., Thompson C.J., Wohlleben W., Tateno Y. Evolution of the glutamine synthetase gene, one of the oldest existing and functioning genes. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3009–3013. doi: 10.1073/pnas.90.7.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamini, Sharma R., Punekar N.S., Phale P.S. Carbaryl as a carbon and nitrogen source: an inducible methylamine metabolic pathway at the biochemical and molecular levels in Pseudomonas sp. strain C5pp. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.01866-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisenberg D., Gill H.S., Pfluegl G.M., Rotstein S.H. Structure-function relationships of glutamine synthetases. Biochim. Biophys. Acta. 2000;1477:122–145. doi: 10.1016/s0167-4838(99)00270-8. [DOI] [PubMed] [Google Scholar]
- 26.Liaw S.H., Kuo I., Eisenberg D. Discovery of the ammonium substrate site on glutamine synthetase, a third cation binding site. Protein Sci. 1995;4:2358–2365. doi: 10.1002/pro.5560041114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woods D.R., Reid S.J. Recent developments on the regulation and structure of glutamine synthetase enzymes from selected bacterial groups. FEMS Microbiol. Rev. 1993;11:273–283. doi: 10.1111/j.1574-6976.1993.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 28.Brown J.R., Masuchi Y., Robb F.T., Doolittle W.F. Evolutionary relationships of bacterial and archaeal glutamine synthetase genes. J. Mol. Evol. 1994;38:566–576. doi: 10.1007/BF00175876. [DOI] [PubMed] [Google Scholar]
- 29.Almassy R.J., Janson C.A., Hamlin R., Xuong N.H., Eisenberg D. Novel subunit—subunit interactions in the structure of glutamine synthetase. Nature. 1986;323:304–309. doi: 10.1038/323304a0. [DOI] [PubMed] [Google Scholar]
- 30.Unno H., Uchida T., Sugawara H., Kurisu G., Sugiyama T., Yamaya T., Sakakibara H., Hase T., Kusunoki M. Atomic structure of plant glutamine synthetase: a key enzyme for plant productivity. J. Biol. Chem. 2006;281:29287–29296. doi: 10.1074/jbc.M601497200. [DOI] [PubMed] [Google Scholar]
- 31.van Rooyen J.M., Abratt V.R., Belrhali H., Sewell T. Crystal structure of type III glutamine synthetase: surprising reversal of the inter-ring interface. Structure. 2011;19:471–483. doi: 10.1016/j.str.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y., McAleer K.L., Murrell J.C. Monomethylamine as a nitrogen source for a nonmethylotrophic bacterium, Agrobacterium tumefaciens. Appl. Environ. Microbiol. 2010;76:4102–4104. doi: 10.1128/AEM.00469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray D.S., Chinnam N., Tonthat N.K., Whitfill T., Wray L.V., Fisher S.H., Schumacher M.A. Structures of the Bacillus subtilis glutamine synthetase dodecamer reveal large intersubunit catalytic conformational changes linked to a unique feedback inhibition mechanism. J. Biol. Chem. 2013;288:35801–35811. doi: 10.1074/jbc.M113.519496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krajewski W.W., Jones T.A., Mowbray S.L. Structure of Mycobacterium tuberculosis glutamine synthetase in complex with a transition-state mimic provides functional insights. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10499–10504. doi: 10.1073/pnas.0502248102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 36.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structures of apo-RhGmaS, RhGmaS–AMPPCP, RhGmaS–AMPPNP–MetSox, RhGmaS–ADP–MetSox-P, RhGmaS–ADP-C1, and RhGmaS–ADP-C2 have been deposited in PDB under accession codes 7CQL, 7CQN, 7CQQ, 7CQU, 7CQW, and 7CQX, respectively. All data are contained within the manuscript and the supporting information.