Abstract
The serine-rich repeat (SRR) glycoproteins of gram-positive bacteria are a family of adhesins that bind to a wide range of host ligands, and expression of SRR glycoproteins is linked with enhanced bacterial virulence. The biogenesis of these surface glycoproteins involves their intracellular glycosylation and export via the accessory Sec system. Although all accessory Sec components are required for SRR glycoprotein export, Asp2 of Streptococcus gordonii also functions as an O-acetyltransferase that modifies GlcNAc residues on the SRR adhesin gordonii surface protein B (GspB). Because these GlcNAc residues can also be modified by the glycosyltransferases Nss and Gly, it has been unclear whether the post-translational modification of GspB is coordinated. We now report that acetylation modulates the glycosylation of exported GspB. Loss of O-acetylation due to aps2 mutagenesis led to the export of GspB glycoforms with increased glucosylation of the GlcNAc moieties. Linkage analysis of the GspB glycan revealed that both O-acetylation and glucosylation occurred at the same C6 position on GlcNAc residues and that O-acetylation prevented Glc deposition. Whereas streptococci expressing nonacetylated GspB with increased glucosylation were significantly reduced in their ability to bind human platelets in vitro, deletion of the glycosyltransferases nss and gly in the asp2 mutant restored platelet binding to WT levels. These findings demonstrate that GlcNAc O-acetylation controls GspB glycosylation, such that binding via this adhesin is optimized. Moreover, because O-acetylation has comparable effects on the glycosylation of other SRR adhesins, acetylation may represent a conserved regulatory mechanism for the post-translational modification of the SRR glycoprotein family.
Keywords: accessory, glycoprotein:glucosylation, O-acetylation, Streptococcus gordonii
Abbreviations: aSec, accessory Sec; Asp2, aSec protein 2; BR, binding region; DPBS, Dulbecco's PBS; GspB, gordonii surface protein; GtfAB, glycosyltransferase AB complex; Gtfs, glycosyltransferases; PMAA, partially methylated alditol acetate; SRR, serine-rich repeat
The serine-rich repeat (SRR) glycoproteins are a family of adhesins found on the surface of many gram-positive bacteria. Originally identified in oral streptococci, they have subsequently been found in several other genera and numerous species. Their expression has been directly correlated with enhanced bacterial virulence in a number of disease settings, such as endocarditis, pneumonia, and meningitis (1, 2, 3, 4, 5). More recently, the SRR glycoproteins have been found in nonpathogenic commensal bacteria, promoting bacterial adherence to epithelial surfaces of the vaginal mucosa, gastrointestinal tract, lung epithelia, and oral surfaces, as well as mediating biofilm formation (6, 7). These findings underscore the importance of the SRR glycoproteins as bacterial-host attachment factors.
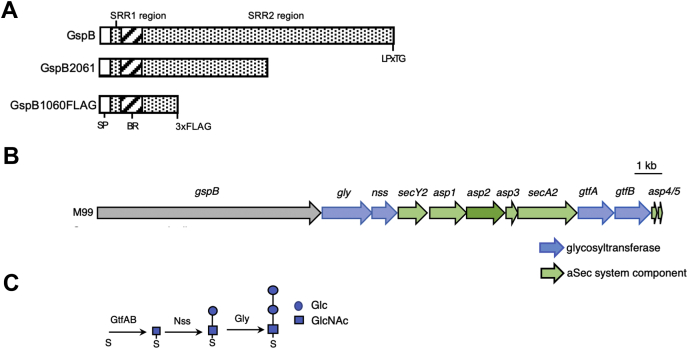
The domain organization of all the SRR glycoproteins follows a conserved architecture (Fig. 1A). The N-terminus contains an extended signal peptide that is necessary for preprotein targeting and export via the accessory Sec (aSec) system (8). This region is followed by two SRR domains (SRR1 and SRR2) that are extensively glycosylated and that flank a ligand binding region (BR). The structure of the BR domain can significantly vary between species, resulting in binding to a variety of host targets exposed on mucosal and epithelial surfaces, including sialoglycans, fibrinogen, and keratin-10 (9, 10, 11, 12, 13, 14). A LPxTG sequence at the C-terminus covalently attaches the SRR glycoprotein to the cell wall (15).
Figure 1.
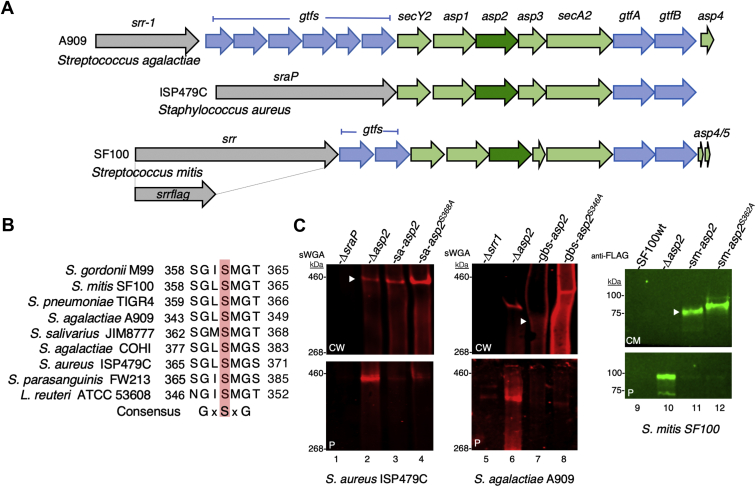
Gordonii surface protein (GspB) and accessory Sec operon.A, the schematic of WT GspB and the recombinant constructs, GspB2061 and GspB1060FLAG. B, the gspB-aSec operon. Components of the accessory aSec (aSec) system are shown in green, and glycosyltransferases are in blue. C, sequential steps in the modification of GspB by GtfAB, Nss, and Gly. GspB glycan structures were determined previously by MALDI-TOF mass spectrometry analysis (27). SP is the signal peptide; SRR1 and SRR2 are the serine-rich repeat regions that undergo post-translational modification; Asp, accessory Sec protein; BR, ligand-binding region; GtfAB, glycosyltransferase complex; LPxTG, cell wall anchoring domain.
The SRR adhesins are glycosylated by a number of glycosyltransferases (Gtfs) encoded within the gspB-aSec operon (Fig. 1B). A two-protein glycosyltransferase complex (GtfAB) O-glycosylates the SRR protein by transferring GlcNAc to serine and threonine residues within the SRR domains (16, 17), (Fig. 1C). The remaining Gtfs further extend the glycan by adding other sugar moieties to the GlcNAc core (18, 19, 20, 21). The number and types of these additional Gtfs vary considerably between species, as do the resulting glycan structures (6, 20, 22).
The SRR glycoprotein gordonii surface protein (GspB) of Streptococcus gordonii is among the most extensively studied SRR glycoproteins. GspB is a Siglec-like adhesin that binds human platelets in vitro through its interaction with sialyl T-antigen moieties on the platelet GPIbα receptor (23, 24). This interaction is thought to contribute to the pathogenesis of infective endocarditis, where the expression of GspB has been associated with increased virulence in animal models of this disease (1). Biochemical and genetic studies of GspB biogenesis have identified two additional Gtfs (Nss and Gly) that further modify the GlcNAc core of GspB (17, 25) (Fig. 1C). Nss first transfers Glc to GlcNAc, allowing Gly to transfer a second Glc residue, thereby generating an O-linked trisaccharide (Fig. 1C).
Glycosylation is in part required for GspB stability, in that nonglycosylated GspB variants (produced by ΔgtfA or ΔgtfB strains) are rapidly degraded in vivo as compared with glycosylated forms (26). In addition, deletion of the secondary gtf genes can result in the export of stable but altered SRR glycoforms with impaired bacterial binding to their host substrates. Deletion of nss or gly in S. gordonii M99 results in WT levels of GspB surface expression but is associated with slightly reduced bacterial binding to human platelets (25, 27). Comparable results have been seen in Streptococcus agalactiae, where deletion of six Gtfs (gtfC-gtfH) that modify the adhesin Srr1 significantly reduced bacterial binding to human lung and intestinal epithelial cells (4). Similarly, individual deletion of the gtfs (gtf3-gly) that modify Fap1 of Streptococcus parasanguinis led to the formation of biofilms with altered biomass (21). Thus, maintaining a specific SRR glycoform appears to be essential for optimal SRR glycoprotein function.
In addition to glycosylation, we recently found that GlcNAc residues on GspB can undergo O-acetylation (27). This modification is carried out by the aSec protein 2 (Asp2), a member of the aSec system, a specialized transporter that mediates the export of SRR adhesins (8, 28). Acetylation of the GspB glycan was shown to be coupled to aSec transport, such that rerouting the substrate to the general Sec system prevented GlcNAc modification (27). Thus, the post-translational modification and export of SRR adhesins appear to be coordinated steps in the biogenesis of these glycoproteins, such that the adhesin is modified to maximize its binding properties.
Asp2 homologues are found in all aSec systems, and O-acetylated GlcNAc residues have been described on the SRRs of S. agalactiae and Streptococcus salivarius (29, 30). Analysis of glycopeptide fragments derived from exported GspB and Srr1 showed that O-acetylation and glucosylation of the GlcNAc core only occurred on a fraction of GlcNAc residues throughout the SRR domains (17, 27, 29). The cause of this glycosylation heterogeneity has been unclear. Of note, however, S. gordonii expressing an Asp2 mutant lacking acetyltransferase activity exported a GspB glycoform of increased mass, as compared with the WT glycoprotein, suggesting a significant change in its glycan composition (31). Moreover, isogenic variants of S. gordonii expressing this altered GspB glycoform showed reduced binding to human platelets (27). However, the precise changes in SRR glycan composition of this glycoform were unknown. We now show that a loss of GlcNAc O-acetylation leads to increased glycosylation of GspB by Nss and Gly and that this excessive addition of glycan on the SRR regions impairs GspB-mediated binding to platelets. The finding that GlcNAc O-acetylation modulates GspB glycosylation indicates that O-acetylation is a novel control mechanism that regulates both the extent of glycosylation and the binding properties of the SRR adhesins.
Results
GlcNAc O-acetylation limits glucose deposition by Nss and Gly on exported GspB
Asp2 acetylates GlcNAc moieties on GspB, and the loss of O-acetylation leads to the export of altered GspB glycoforms of increased mass, as compared with the WT (27, 31). This change in mass is less evident when nss and gly are deleted, suggesting that O-acetylation may interfere with Nss and Gly-mediated glucosylation of GspB. To test this possibility, we evaluated the changes in glycosylation of two truncated variants of GspB (GspB1060FLAG and GspB2061) containing 417 aa and 1428 aa of the SRR2 domain, respectively (Fig. 1A). Each protein was expressed in isogenic variants of S. gordonii M99 with different Gtf and O-acetylation backgrounds (Table 1).
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | Reference |
|---|---|---|
| S. gordonii | ||
| M99 | Endocarditis causing parental strain | (51) |
| PS1225 | M99 expressing gspB736FLAG | (26) |
| PS3539 | PS1225 asp2S362A | (27) |
| PS921 | M99 expressing gspB1060FLAG | (26) |
| PS3874 | PS921 Δgly; SpecR | This study |
| PS3318 | PS921 Δgly-nss; SpecR | (27) |
| PS3541 | PS921 asp2S362A | (27) |
| PS3875 | PS3541 Δgly asp2S362A; SpecR | This study |
| PS3542 | PS3541 Δgly-nss asp2S362A; SpecR | (27) |
| PS1064 | PS921 ΔgtfA; SpecR | (27) |
| PS497 | M99 gspB ::pB2061Hi s6; CmR | (52) |
| PS612 | PS497 Δgly; SpecR | This study |
| PS3876 | PS497 Δgly-nss; SpecR | This study |
| PS3877 | PS497 asp2S362A | This study |
| PS3878 | PS497 Δgly asp2S362A | This study |
| PS3879 | PS497 Δgly-nss asp2S362A | This study |
| PS666 | M99 ΔgtfA; SpecR | (25) |
| PS3319 | M99 Δgly-nss; SpecR | (27) |
| PS3536 | M99 asp2S362A | (27) |
| PS3880 | M99 Δgly-nss asp2S362A; SpecR | This study |
| S. aureus | ||
| PS767 | ISP479C sraP::ermB | (48) |
| PS1280 | ISP479C asp2::spec | (48) |
| PS3855 | PS1280 (pKS80); ErmR | This study |
| PS3856 | PS1280 (pKS H6saAsp2); ErmR | This study |
| PS3857 | PS1280 (pKS H6saAsp2S368A); ErmR | This study |
| S. agalactiae | ||
| A909 | Serotype Ia clinical isolate | (43) |
| PS3204 | A909Dsrr1 | (10) |
| PS3860 | A909Dasp2 | This study |
| PS3862 | PS3860 (pDE H6gbsAsp2) | This study |
| PS3833 | PS3860 (pDE H6gbsAsp2S346A) | This study |
| S. mitis | ||
| SF100 | endocarditis causing parental strain | (53) |
| PS3397 | SF100 expressing SRRFLAG | This study |
| PS3870 | PS3397Dasp2 | This study |
| PS3871 | PS3397Dasp2 (pDE H6smAsp2) | This study |
| PS3872 | PS3397Dasp2 (pDE H6smAsp2S362A) | This study |
| E. coli | ||
| TOP10 | Host cell for cloning | |
| BL21 | Host cell for protein expression | |
| Plasmids | ||
| pET28Nss H6 | E. coli expression vector encoding Nssh6; KanR | This study |
| pC326 | E. coli vector with MCS::cat cassette; CmR | This study |
| pCasp2-KO[A909] | Group B streptococcus A909 asp2 KO vector; CmR | This study |
| pS326 | E. coli vector with MCS::spec cassette; SpecR | This study |
| pSasp2-KO[SF100] | S. mitis SF100 asp2 KO vector; SpecR | This study |
| pKS80 | S. aureus expression vector; ErmR | (47) |
| pKSH6saAsp2 | S. aureus expression vector encoding H6saAsp2; ErmR | This study |
| pKSH6saAsp2S368A | S. aureus expression vector encoding H6saAsp2S368A; ErmR | This study |
| pDE123 | Streptococcal expression vector; ErmR | (44) |
| pDEH6gbsAsp2 | Streptococcal expression vector encoding H6gbsAsp2; ErmR | This study |
| pDEH6gbsAsp2S346A | Streptococcal expression vector encoding H6gbsAsp2S346A; ErmR | This study |
| pDEH6smAsp2 | Streptococcal expression vector encoding H6smAsp2; ErmR | This study |
| pDEH6smAsp2S362A | Streptococcal expression vector encoding H6smAsp2S362A; ErmR | This study |
CmR, chloramphenicol resistant; ErmR, erythromycin resistant; SpecR, spectinomycin resistant; KanR, kanamycin resistant.
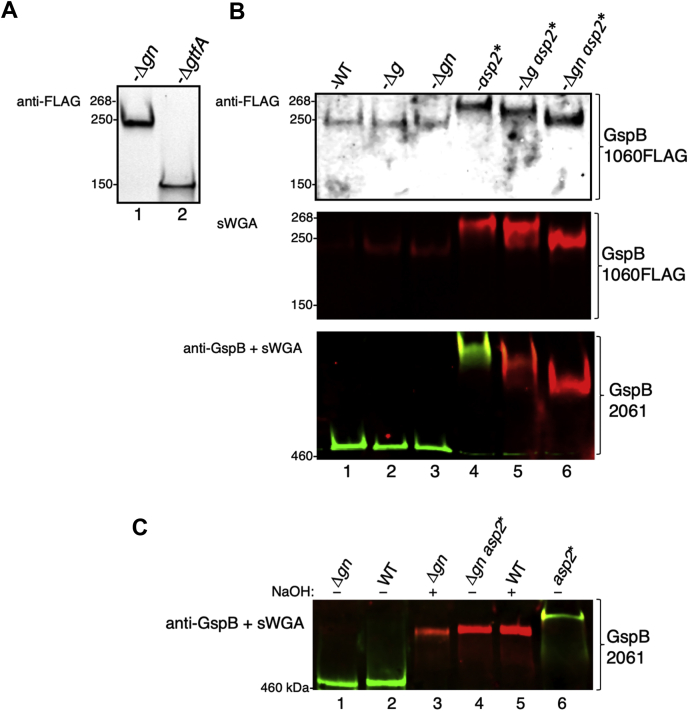
We first assessed the individual contribution of each Gtf to the composition of the glycan on exported GspB. Deletion of gtfA resulted in a large reduction in the mass of GspB1060FLAG (Fig. 2A, lane 1 versus 2), demonstrating that GlcNAc is a major component of the GspB glycan. In contrast, the deletion of gly (Δg) or gly and nss (Δgn) produced only minor changes in the electrophoretic mobilities of GspB1060FLAG and GspB2061, as compared with the WT glycoforms (Fig. 2B, lane 1 versus lanes 2 and 3), indicating a low abundance of Glc in the WT glycan.
Figure 2.
Loss of GlcNAc O-acetylation leads to an increase in glucosylation of exported GspB by Nss and Gly.A and B, western blot analysis of GspB1060FLAG and GspB2061 exported by isogenic variants of M99 containing a deletion in gtfA (ΔgtfA), gly (Δg), gly and nss (Δgn), and/or an asp2S362A mutation (asp2∗). Samples of culture media were separated by SDS-PAGE and probed with anti-FLAG antibodies and biotinylated sWGA, to detect levels of GspB and core GlcNAc, respectively. Exported GspB2061 was simultaneously probed with biotinylated sWGA (700-nm channel red) and anti-GspB polyclonal antisera raised against the WT GspB glycan (800-nm channel-green in overlay). C, saponification of GspB glycoforms. Exported GspB2061 glycoforms were treated with 100-mM NaOH at 37 °C for 1 h, to remove O-acetyl moieties. Changes to the GspB glycan were assessed by GspB2061 binding to both sWGA (700-nm channel red) and anti-GspB (800-nm channel green in overlay) glycan probes. GspB, gordonii surface protein.
To examine the effect of O-acetylation upon GspB glycan composition, we introduced an asp2S362A mutation into the genome of M99, producing a catalytic site mutation that abolishes GlcNAc O-acetyltransferases activity (27). This mutation was associated with an increase in apparent mass for both GspB1060FLAG and GspB2061 as compared with the WT strain (Fig. 2B, lane 1 versus lane 4). The increase in mass was most evident with the longer GspB2061 variant, likely reflecting the larger number of serine residues available for modification. In addition, binding of sWGA to both GspB variants was increased, consistent with our previous observation that this lectin has higher affinity for GlcNAc, as compared with O-acetylated GlcNAc (Fig. 2B, lane 1 versus lane 4) (27). Deletion of gly, or gly and nss, in an aps2S362A mutant resulted in a progressive reduction in the apparent mass of GspB (Fig. 2B, lane 4 versus lanes 5 and 6), further indicating that the increased mass of GspB associated with the loss of GlcNAc O-acetylation is at least in part due to increased Glc deposition by Gly and Nss on the GspB glycan.
O-acetylation alters the electrophoretic mobility of GspB
GspB2061 modified with O-acetylated GlcNAc (exported by M99Δgn) migrated with an apparent mass that was less than that of GspB2061 modified by nonacetylated GlcNAc (exported by M99Δgn asp2S362A) (Fig. 2B, lane 3 versus lane 6). Because an acetyl group will add 43 Da to the mass of GlcNAc, it was unclear why acetylation would lead to an apparent decrease in mass. To address this issue, we exposed different glycoforms of GspB2061 to mild-base ester hydrolysis (saponification), which removes O-acetyl groups (27). Saponification of WT GspB2061 increased the apparent mass of the glycoprotein to that of nonacetylated GspB2061 (exported from M99Δgn asp2S362A) (Fig. 2C, lanes 2 versus lane 5). Saponification of GspB2061 modified with GlcNAc alone produced a similar increase in mass (Fig. 2C, lane 3). These findings confirm that O-acetylation of GspB reduces its apparent mass on SDS-PAGE, which could be due to changes in hydrophobicity or viscosity (32). Importantly, we also found that saponified WT GspB2061 migrated at a lower mass than nonacetylated GspB2061 (exported by M99 asp2S362A) (Fig. 2C, lane 5 versus lane 6), suggesting that the increased mass of the latter is also due to increased glucose deposition by Nss and Gly.
O-acetyl moieties and glucose residues modify core GlcNAc at the same position
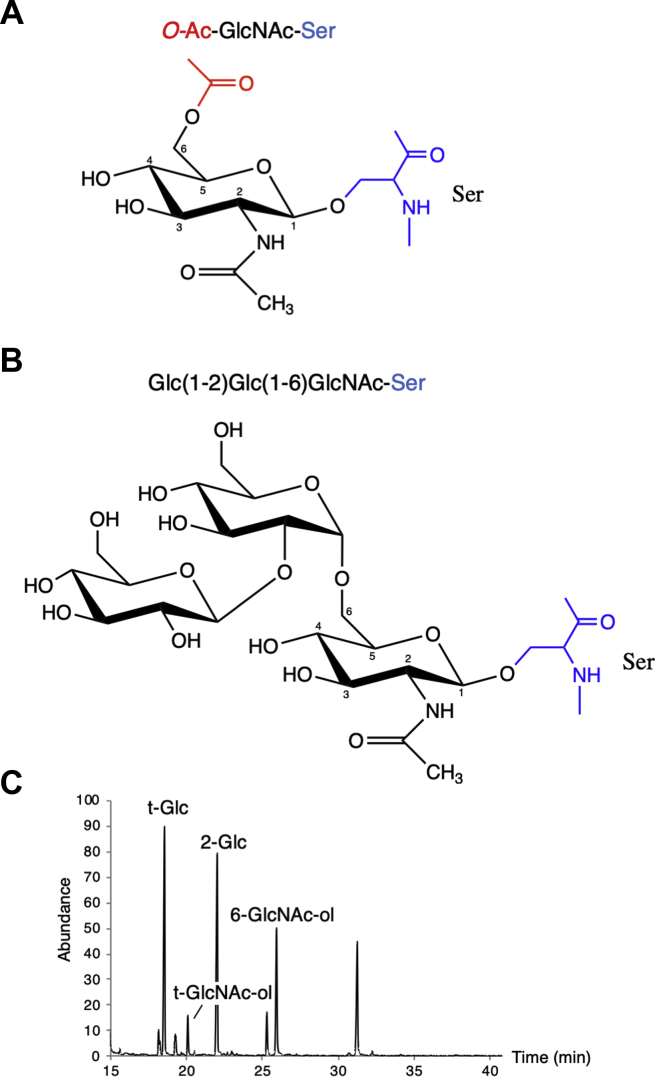
O-acetylation of the GspB glycan occurs at the C6-OH of core GlcNAc residues (27) (Fig. 3A). This is consistent with the location of the O-acetyl moiety on GlcNAc in the glycans of Srr1 of S. agalactiae, SrpA of S. salivarius, and O-acetylated forms of peptidoglycan (29, 30, 33, 34). In addition to O-acetylation, the GlcNAc core can be further modified through its sequential glycosylation by Gtfs such as Nss and Gly. To better assess how O-acetylation and glucosylation can simultaneously occur, we determined the structure of the glycans covalently attached to the SRR regions of GspB, by GC-MS analysis of partially methylated alditol acetates (PMAAs) or sugar linkage analysis of the GspB glycan.
Figure 3.
Structures of O-acetylated and Nss and Gly modified glycans of GspB.A, The structure of O-acetylated GlcNAc was obtained from published glycopeptide CID fragmentation analysis of GspB and the SRR adhesin Srr1 of S. agalactiae (27). B, Structure of GlcNAc modified by both Nss and Gly was obtained by GC-MS profiling of partially methylated alditol acetates (PMAA) derived from glycans released from a recombinant GST-SRR protein co-expressed with GtfAB, Nss, and Gly. GlcNAc carbon positions are numbered. C, PMAA derivatives of the O-glycans released from GST-SRR1. Terminally linked glucose (t-Glc), terminally linked GlcNAc (t-GlcNAc), 2-linked glucose, and 6-linked GlcNAc were identified based on retention times in reference to carbohydrate standards. GspB, gordonii surface protein; GtfAB, glycosyltransferase complex; SRR, serine-rich repeat.
To obtain sufficient quantities of the GspB glycan for linkage analysis, we used a recombinant GST–GspB fusion protein encompassing the SRR1 region, which undergoes glycosylation by all the Gtfs encoded within the gspB-aSec operon, generating a trisaccharide glycan (27). Analysis of the PMAA derivatives from the GspB glycan found the linkage of the trisaccharide to be Glc(1–2)Glc(1–6)GlcNAc (Fig. 3, B and C), where GlcNAc was identified as the reducing-end sugar. Resolution of the glycan structure revealed that Glc transferred by Nss is attached to GlcNAc at the C6 position, the same location that undergoes O-acetylation by Asp2. Collectively, these findings indicate that GlcNAc O-acetylation and glucosylation must be mutually exclusive events because they compete for the same site for GlcNAc modification.
O-acetylation of GlcNAc directly blocks subsequent glucosylation
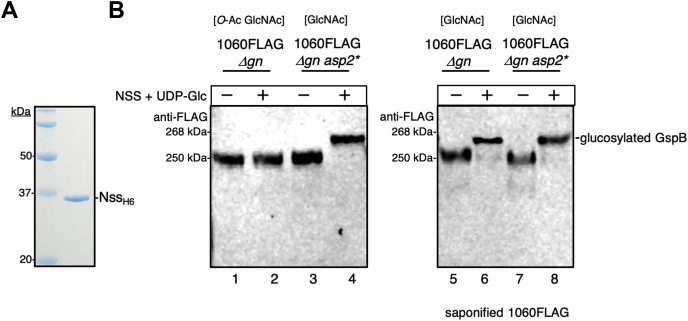
The finding that O-acetyl groups and Glc cannot modify the same GlcNAc residue, in conjunction with our observation that a loss of O-acetylation led to an increase in glucosylation of GspB, suggested that GlcNAc O-acetylation could serve to inhibit Glc deposition on GspB. To evaluate whether GlcNAc O-acetylation could control subsequent glucosylation of GspB, we tested whether O-acetylated and nonacetylated GspB glycoforms could be further modified by Nss in vitro. We purified the above glycoforms of GspB1060FLAG from M99Δgn and M99Δgn asp2S362A, respectively, mixed them individually with purified Nss and UDP-Glc, and assessed for the addition of Glc. O-acetylated GspB1060FLAG showed no obvious glucosylation, as indicated by no change in mass (Fig. 4B, lane 1 versus lane 2). In contrast, a 10-kDa increase in mass was seen with nonacetylated GspB1060FLAG after incubation with Nss and UDP-Glc, indicating that this glycoform was further modified by the addition of Glc to the GlcNAc core (Fig. 4B, lane 3 versus lane 4). Removing the O-acetyl groups on GspB1060FLAG by saponification before incubation with Nss and UDP-Glc produced a similar increase in mass (Fig. 4B, lane 5 versus 6, lane 7 versus 8), indicating further modification with Glc. These findings demonstrate that O-acetylation of GlcNAc residues prevents the subsequent Glc deposition on GspB. Moreover, the inability to detect in vitro glucosylation of exported WT GspB suggest GlcNAc residues are extensively O-acetylated in the WT glycan.
Figure 4.
Effect of O-acetylation on the subsequent glycosylation of GspB1060FLAG.A, SDS-PAGE analysis of purified NssH6 after expression in E. coli BL21 and purification on Ni-NTA resin. B, glucosylation of GspB1060FLAG before and after saponification. Acetylated (Δgn) or nonacetylated (Δgn asp2∗) GspB1060FLAG was combined with NssH6 and UDP-Glc as indicated. Glucosylation reactions were carried out at 37 °C for 1 h and samples analyzed by SDS-PAGE and Western blot analysis with anti-FLAG antibodies. Samples were saponified by incubation with 100-mM NaOH for 1 h, to release O-acetyl moieties.
Increased glucose deposition on GspB impairs platelet binding by bacteria
O-acetylation of the GspB glycan is essential for the binding of strain M99 to human platelets via this adhesin (27). Because O-acetylation inhibits glucosylation of GspB, we hypothesized that the loss of platelet binding associated with the asp2S362A mutation could be due to increased glycosylation of GspB with Glc. To address this possibility, we directly compared platelet binding by M99 and isogenic variants that differed in their post-translational modifications of GspB.
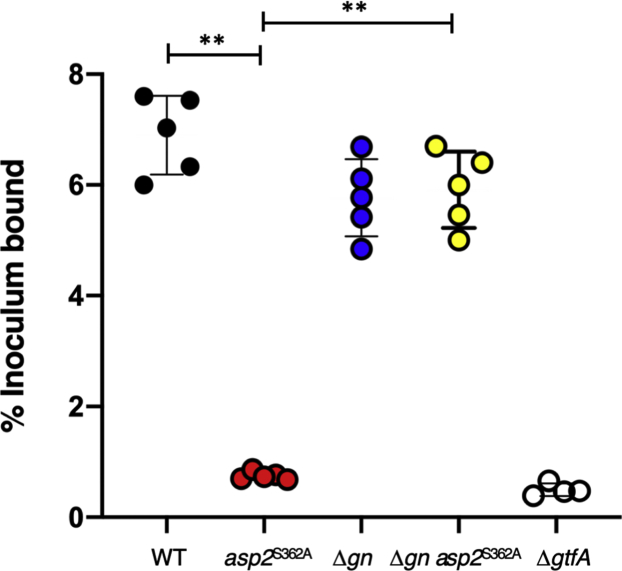
As seen in previous studies (25), WT M99 exhibited high levels of binding to human platelets, while a M99ΔgtfA strain, which does not express GspB because of protein instability, exhibited low levels of binding (Fig. 5). Deletion of gly and nss produced only a small decrease in platelet binding (strain M99 versus M99Δgn) that was not statistically significant. As expected, M99 expressing Asp2S362A showed significantly reduced binding to platelets (p < 0.001 versus WT M99), to levels comparable to the ΔgtfA strain. However, deletion of gly and nss in combination with an asp2S362A mutation (M99Δgn asp2S362A) restored platelet binding to levels similar to those seen with the WT strain (p = 0.437). These results suggest that increased Glc deposition on GspB, and not the loss of the O-acetyl moiety, impairs the ability of GspB to bind to human platelets.
Figure 5.
Effect of O-acetylation on the binding of Streptococcus gordonii to human platelets. WT strain M99 and mutants PS3536 (M99 asp2S362A), PS3319 (M99 Δgn), PS3880 (M99 Δgn asp2S362A), and PS666 (M99 ΔgtfA) expressing glycovariants of GspB were assessed for their binding to immobilized human platelets. Binding is expressed as the percent of input bacteria that remained bound to platelets after repeated washing of the wells (n = 5 + S.D. of triplicate results from a representative experiment). ∗∗p < 0.01, compared with WT M99 or PS3880 as indicated. Of note, no significant differences in platelet binding were seen between M99 and PS388.
Acetylation of SRR glycoproteins by Asp2 occurs in other species
Asp2 is an essential component of the aSec system (Fig. 6A) and is present in all bacteria expressing an SRR adhesin (8). Its acetyltransferase activity is mediated by a conserved Ser-Asp-His catalytic triad, where the serine residue is found within a GxSxG motif present in all Asp2 homologues (Fig. 6B). These findings suggest that other SRR glycoproteins undergo O-acetylation by Asp2. To test this possibility, we assessed the effect of mutating the catalytic site of Asp2 in Staphylococcus aureus ISP49C, S. agalactiae A909, and Streptococcus mitis SF100. Of note, each SRR adhesin is predicted to be glycosylated differently, based on the varying number of gtfs encoded within their respective srr-aSec operon (Fig. 6A). For each strain, we targeted the catalytic serine residue within the GxSxG motif of its Asp2 homologue (Fig. 6B) for alanine substitution. The resulting mutated asp2 orfs were used to complement a variant of the parent strain containing an asp2 deletion. Changes in the glycan of the SRR glycoproteins were assessed by sWGA binding and electrophoretic mobility.
Figure 6.
Asp2 mediates the O-acetylation of SRR adhesins from other gram-positive pathogens.A, organization of the srr-aSec loci of strain M99 and the homologous regions of S. agalactiae strain A909, Staphylococcus aureus strain ISP479C, and S. mitis strain SF100. The genes encoding the SRR proteins are shown in gray, the aSec system components are in green, and the glycosyltransferases in blue. B, amino acid alignment of the conserved GxSxG motif in Asp2 homologues. The catalytic Ser residues are highlighted in pink. C, western blot analysis of full-length SRR proteins exported by S. aureus and S. agalactiae and truncated SF100-SRRFLAG expressed by S. mitis SF100. Cell wall (CW), culture medium (CM), and protoplast (P) fractions were prepared from strains in the exponential phase of growth. Proteins were separated by SDS-PAGE (3–8%) and analyzed by Western blotting using either biotinylated sWGA or anti-FLAG antibodies. White arrows indicate the position of the WT SRR glycoform. Asp2, aSec protein 2; SRR, serine-rich repeat.
Lectin blotting with sWGA of cell wall fractions from S. aureus ISP479C and S. agalactiae A909 revealed a single band in both WT strains that was lost with deletion of sraP or srr1, respectively (Fig. 6C, lane 1 versus 3 and lane 5 versus 7). Deletion of asp2 in both WT strains reduced but did not abolish SRR glycoprotein export, with an intracellular accumulation of the preprotein in the Δasp2 deletion strains (Fig. 6C, lane 2 versus 3 and lane 6 versus 7). Complementation in trans of strains ISP49CΔasp2 and A909Δasp2 with their respective asp2 variants sa-asp2S368A and gbs-asp2S346A resulted in the export of altered SraP and Srr1 glycoforms, (Fig. 6C, lane 3 versus 4 and lane 7 versus 8). Both SRR proteins showed increased binding to sWGA, and Srr1 migrated with an increase in mass, indicating a change to the SRR glycan. Of note, no differences in mass were seen between acetylated and nonacetylated SraP glycoforms (Fig. 6C, lane 3 versus lane 4), likely reflecting the lack of other gtfs within the sraP-aSec operon (Fig. 6A).
To more precisely assess the role of Asp2 in the glycosylation and export of SRR adhesins, we examined the extracellular transport of a truncated SRR glycoprotein (SRRFLAG) by S. mitis SF100. Deletion of aps2 abolished SRRFLAG export, with a clear intracellular accumulation of the substrate (Fig. 6C, lane 10). Complementation with asp2 led to the efficient export of SRRFLAG, where a ∼75 kDa protein was detected in the culture medium, and only trace amounts in the protoplasts. Similarly, complementation with sm-asp2S362A restored glycoprotein export, but with SRRFLAG, now migrating with a higher molecular mass (∼85 kDa), indicating of loss of O-acetylation and increased glycosylation (Fig. 6C, lane 11 versus 12). Collectively, these findings demonstrate that the O-acetyltransferase activity of Asp2 is conserved in streptococci and staphylococci, and the loss of O-acetylation leads to the export of altered SRR glycoforms.
Discussion
The SRR glycoproteins are a family of surface adhesins that are increasingly recognized as important for host colonization and virulence. Their biogenesis involves the intracellular glycosylation of the preprotein, followed by their transport to the bacterial surface by the aSec system (6, 8, 21). All SRR glycoproteins undergo the transfer of GlcNAc moieties to serine and threonine residues within the SRR domains (17). Analogous to the synthesis of mucins (35), the GlcNAc residues can be further modified by other Gtfs to generate a range of O-linked glycan structures, which can vary between species (6, 18, 20, 22). Changes in these O-linked glycans on SRR adhesins are correlated with impaired binding to host targets and biofilm formation, emphasizing the importance of glycosylation for optimal SRR glycoprotein function (4, 25).
Early reports on the glycans of Fap1 and PsrP examined the in vitro properties of the Gtfs that modified theses adhesins, using recombinant SRR proteins as Gtf substrates, or by coexpressing the Gtfs and adhesins in Escherichia coli (20, 21). These studies were key in determining the specific enzymatic functions of the Gtfs and in identifying the glycan structures of these adhesins (20, 21). More recent direct analysis of the native glycan of the SRR glycoproteins Srr1 and SrpA revealed heterogeneity of the glycan, where not all of the GlcNAc core is further modified (29, 30), indicating that the glycan is not fully extended at all glycosylation sites under normal conditions.
An additional modification of the SRR adhesins is the O-acetylation of GlcNAc residues at the C6 hydroxyl position by Asp2 (27, 29, 30). Our results indicate that for GspB, this is the same position that can be further glucosylated to generate the Glc(1–2)Glc(1–6)GlcNAc trisaccharide. O-acetylation and glucosylation compete for the same site to modify GlcNAc residues, such that O-acetylation can block glucosylation of the GlcNAc core. Thus, GlcNAc O-acetylation is a regulatory modification preventing high levels of glucosylation of the GspB SRR domains, which can hinder ligand binding. Although we did not examine this directly, the blocking of glucosylation of the GlcNAc core by O-acetylation could also serve as a mechanism for generating glycan heterogeneity throughout the SRR regions of GspB.
Limiting the glycosylation of the GlcNAc core had a profound effect upon the binding properties of GspB. We have previously shown that the loss of O-acetylation markedly reduced GspB-mediated streptococcal binding to human platelets, to levels comparable with a gspB-deletion strain (27). Our finding that the deletion of gly and nss in an O-acetylation mutant restored platelet-binding indicates that the reduced binding is not directly due to the loss of acetylation but instead stems from the additional glucosylation of GlcNAc. How increased glucosylation of the GspB SRR domains can adversely affect platelet-binding function remains unclear. High levels of glucosylation of the SRR regions could either induce a conformation change within the BR or occlude its access to the GPIbα receptor on platelets. We recently found that, when tested under flow conditions, streptococcal binding to immobilized platelet GPIbα increased with shear force (36). This finding suggests that under static conditions, the flanking glycosylated SRR regions may occlude the BR, whereas under shear force, the SRR regions undergo a conformational change that allows the BR to interact with GPIbα (36). If this model proves to be correct, increased glucosylation may hinder the flexibility of GspB, preventing subsequent presentation of the BR domain to the host receptors.
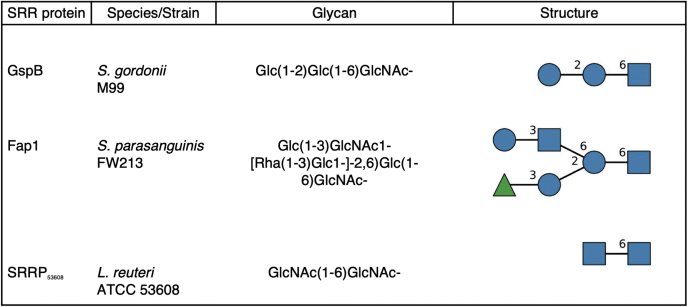
The ability to regulate further glycosylation of the GlcNAc core via O-acetylation may be a common mechanism in the biogenesis of SRR adhesins. As detailed above, catalytic mutants of Asp2 in S. agalactiae, S. mitis, and S. aureus all had altered glycosylation of their respective SRR adhesins, indicating that Asp2-mediated O-acetylation is a highly conserved regulatory mechanism for the post-translational modification of these adhesins. Moreover, analysis of the glycan of Fap1 of S. parasanguinis and SRRP53608 of Lactobacillus reuteri revealed that their glycans were also attached to core GlcNAc via the C-6 position (Fig. 7), indicating that O-acetylation of these SRR adhesins also serves to regulate modification of the GlcNAc core. Of note, the glycan of SraP from S. aureus is composed of GlcNAc monosaccharides (37), suggesting that O-acetylation may have additional roles in SRR glycoprotein function.
Figure 7.
Determined glycan linkages from known SRR protein glycan structures. Glycan structures of Fap1 and SRRP53608 as reviewed by Latousakis et al. (6). Monosaccharide symbols follow the Symbol Nomenclature for Glycans system (50). GlcNAc (blue square), glucose (Glc: blue circle), rhamnose (Rha; green triangle).
O-acetylation of bacterial glycans have been shown to control the deposition of additional sugars in other bacterial species. O-antigen of the lipopolysaccharide of Shigella flexneri consists of a tetrasaccharide [-GlcNAc(1–2)Rha(1–2)Rha(1–3)Rha-] repeat that can be further modified by bacteriophage-encoded glucosyltransferase and O-acetyltransferases, generating antigenic determinants that define serotype specificity (38, 39). Interestingly, the introduction of oacD (the acetyltransferase-mediating O-antigen GlcNAc O-acetylation) via integration of bacteriophage SfII abolished prior GlcNAc glucosylation at the C-6 position, converting the O-antigen serotype (40). These findings, in combination with our own on GspB glucosylation, suggest that GlcNAc O-acetylation may serve to prevent further GlcNAc modification at the C6 position for a broad range of bacterial glycans.
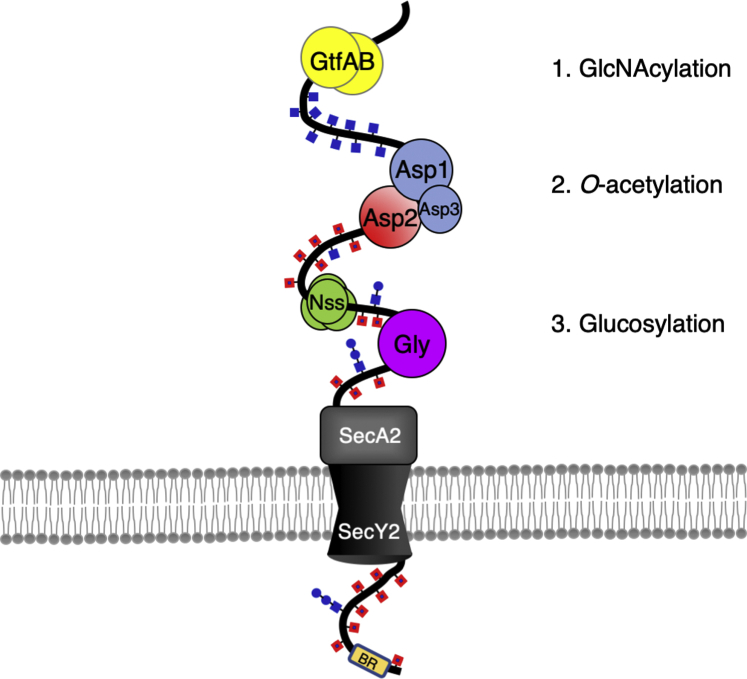
The finding that O-acetylation of GlcNAc by Asp2 controls glucose deposition suggests that Asp2 must engage with the SRR preprotein before Nss and Gly. How O-acetylation is prioritized over glucosylation is unknown. We have shown previously that O-acetylation by Asp2 is coupled to aSec transport (27), although it is unclear how these two processes could be coordinated. One possibility is that signal peptide–mediated targeting of SRR adhesins to the aSec translocon brings the preprotein into contact with all aSec components, thereby facilitating O-acetylation by Asp2. Such substrate targeting could serve to prioritize O-acetylation after GtfAB glycosylation during aSec export. In addition, Asp2 forms a complex with Asp1 and Asp3 that is necessary for GspB export (18, 41). Although a direct role of Asp1 and Asp3 in O-acetylation has not been demonstrated, it is conceivable that the Asps are also involved in coordinating aSec transport and O-acetylation of SRR glycoproteins (Fig. 8).
Figure 8.
Model for GspB glycoprotein biogenesis. GspB is sequentially glycosylated by the GtfAB complex that deposits GlcNAc along the SRR region. GlcNAc-modified GspB engages with the aSec system where upon GlcNAc residues are O-acetylated. Before full engagement with SecA2 at the cell membrane, GspB is further modified by Nss and Gly. GtfAB, glycosyltransferase AB complex; GspB, gordonii surface protein B; SRR, serine-rich repeat.
Our findings provide new insights into the importance of O-acetylation and Asp function in the biogenesis of SRR glycoproteins. Although it was previously proposed that Nss and Gly glucosylation occurred shortly after GtfAB glycosylation (42), our data indicate that GlcNAc O-acetylation modulates subsequent Glc addition. Therefore, we propose a revised model of SRR biogenesis, where the SRR preprotein is modified with GlcNAc by GtfAB, followed by Asp-mediated O-acetylation of the SRR glycan before further glycosylation by additional Gtfs and export through the aSec translocon (Fig. 8). Delineating the consequences of these glycan changes in relation to aSec component engagement will be key to better understanding the need for a dedicated SRR export system.
Experimental procedures
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Table 1. S. gordonii, S. agalactiae, and S. mitis strains were grown in the Todd-Hewitt broth (Becton, Dickinson and Company) or on 5% sheep blood agar (Hardy Diagnostics) at 37 °C in a 5% CO2 environment. S. aureus strains were grown at 37 °C under aeration in the Tryptic soy broth (Becton, Dickinson and Company). For some experiments, antibiotics were added to the media at the following concentrations: 50 μg/ml erythromycin and 100 μg/ml spectinomycin for S. gordonii and S. mitis strains; 15 μg/ml erthromycin for S. aureus strains; 5 μg/ml erythromycin for S. agalactiae strains; 5 μg/ml chloramphenicol for S. mitis and S. agalactiae strains. E. coli strain TOP10 was grown in LB broth or on LB agar containing 50 μg/ml kanamycin, 300 μg/ml erythromycin, 50 μg/ml spectinomycin and 15 μg/ml chloramphenicol when appropriate.
Construction of M99 strains expressing Asp2 catalytic mutants
An asp2S362A point mutation was introduced into S. gordonii strain M99 through allelic replacement as previously described (27). In brief, plasmid pCOLAH6asp123 (encoding asp123) containing a Ser-362-Ala codon replacement within asp2 was used as a template for heterologous recombination within the genome of M99 Δasp2::spec strains. Plasmids were used to transform M99 Δasp2::spec strains by natural transformation resulting in a replacement of a spectinomycin cassette with the mutated asp2 orf. Transformants were plated on sheep blood agar plates and scored for the loss of spectinomycin resistance. Chromosomal DNA was isolated from spectinomycin-sensitive clones according to Madoff et al. (43), and the asp2 gene was PCR-amplified and sequenced to confirm the correct mutation had occurred.
Deletion of gly and nss
Deletion of gly or nss genes in the gspB-aSec locus of M99 variants was performed by allelic replacement as previously described (25).
Construction of S. mitis strains expressing variants of SRRFLAG
A variant of S. mitis SF100 expressing a FLAG-tagged version of its SRR adhesin was constructed as described previously for S. gordonii M99 expressing GspB736FLAG (26). In brief, the suicide vector p736BflagC containing codons 604 to 736 of the GspB SRR2 region fused to a 3xFLAG sequence was used to transform S. mitis SF100 by natural transformation as described previously (44). The resulting strain (PS3397), encoding the SF100 SRR protein with a truncated SRR2 region and C-terminal 3xFLAG tag, was then used to express variant forms of SRRFLAG in S. mitis strain SF100.
Deletion of asp2 in S. mitis and S. agalactiae
To generate an asp2 deletion in either S. mitis SF100 or S. agalactiae A909, a gene replacement cassette was constructed by cloning the chromosomal regions flanking asp2 upstream and downstream of the chloramphenicol (cat) gene in pC326 (45). The resulting plasmids, pSF100asp2-KO and pA909asp2-KO, were introduced into the above strains by natural transformation and electroporation, respectively (44, 46). asp2-deletion mutants were selected by plating onto blood agar containing chloramphenicol.
Asp2 complementation
asp2 homologues were amplified by PCR from the genome of S. aureus ISP479C, S. agalactiae A909, or S. mitis SF100 and cloned as N-terminal 6xHis-tag fusions into the expression plasmids pKS80 (47) or pDE123 (44). Plasmids were introduced into staphylococcal and streptococcal strains by electroporation, as described previously (46, 48).
Analysis of secreted cell wall and protoplast proteins
Overnight cultures of S. gordonii, S. mitis, and S. agalactiae were diluted 1:6 in fresh Todd-Hewitt broth, grown at 37 °C, and cells were harvested at mid-logarithmic growth by centrifugation. For analysis of secreted proteins, samples of clarified culture media were mixed with protein sample buffer (Novagen) before SDS-PAGE separation and Western blot analysis or used directly for saponification analysis. For analysis of cell wall proteins, pelleted cells were resuspended in spheroplast buffer (60-mM Tris, pH 7, 150-mM NaCl, 25% raffinose, and 0.5 U/μl mutanolysin). The suspensions were incubated for 1 h at 37 °C and centrifuged at 10,000g for 10 min at room temperature (RT). The supernatants containing the cell wall extracts were mixed and boiled in protein sample buffer, followed by SDS-PAGE and Western blot analysis. The remaining pellets containing protoplasts were resuspended and lysed in protein sample buffer before gel electrophoresis. Preparation of cell wall and protoplast extracts from S. aureus were carried out as described previously (48). Anti-FLAG and anti-GspB antibodies were detected using fluorescent anti-mouse IRDye 800 and anti-goat IRDye 800 secondary antibodies, respectively. sWGA was detected with streptavidin IRDye 680 (red). Immunoreactive bands were visualized using the LI-COR Odyssey Infrared Imaging System.
Linkage analysis of glycosylated GST-SRR1
Glycan linkages were determined through the GlycoAnalytics Core center at the University of California, San Diego. Glycans were liberated from purified GST-SRR1 glycosylated by GtfAB, Nss, and Gly by reductive beta elimination using base-borohydride treatment (27). The O-glycans were then purified and partially methylated according to the method of Ciucannu and Kerek (49). O-glycans were dried and dissolved in anhydrous dimethyl sulfoxide, followed by adding NaOH slurry in dimethyl sulfoxide and methyl iodide. Partially methylated glycans were extracted by partitioning in water–chloroform mixture, and the chloroform layer was dried and used for linkage analysis. The partially methylated glycans were hydrolyzed using 4 N TFA at 100 °C for 6 h, followed by removal of the acid, reduced with sodium borodeuteride and acetylated to generate the PMAA derivatives, which were then analyzed by GC-MS using Restek-5ms capillary column for inter-residue linkages. Identifications are achieved by using a combination of retention times and electron ionization mass fragmentation pattern.
Cloning, overexpression, and purification of Nss
nss was cloned as a C-terminal 6xHis-tag fusion within the expression plasmid pET28b (Novagen). The pETnss construct was introduced to E. coli BL21 (Lucigene), and expression was induced by the addition of 1-mM IPTG. NssH6 was purified from clarified lysates under native conditions by Ni-NTA–affinity chromatography (Qiagen). Purified NssH6 was concentrated by ultrafiltration using an Amicon Ultra centrifugal filter (10 kDa cutoff) and dialyzed against 25-mM Tris, pH 7, and 150-mM NaCl overnight at 4 °C before use.
In vitro glucosylation assays
GspB1060FLAG secreted from M99Δgn or M99Δgn asp2S362A served as a substrate for Nss in vitro glucosylation of GspB. Cultures of each strain were grown to the mid-log phase, and 15 ml of the culture media were concentrated 50-fold by ultrafiltration as described above and reconstituted in 50-mM Tris, pH 7. Some samples of GspB1060FLAG in culture media were saponified as described below, before concentration and reconstitution. In vitro glucosylation assays were performed at 37 °C for 1 h. To initiate glucosylation of GspB1060FLAG, 10 μM of Nss and 20-mM UDP-Glc (Sigma) were mixed with 15 μl of GspB1060FLAG solution in 50-mM Tris, pH 7, with a final reaction volume of 20 μl.
GspB saponification
Removal of glycan O-acetyl groups was achieved by base-promoted ester hydrolysis. Culture supernatants containing secreted GspB variants were buffered in 10-mM Tris, pH 9, and incubated with 100-mM NaOH for 1 h at 37 °C.
Binding of S. gordonii to platelet monolayers
The binding of S. gordonii to immobilized platelets was performed as described previously (15). In brief, strains were grown for 18 h, washed twice in Dulbecco's PBS (DPBS), sonicated briefly to disrupt aggregated cells and diluted to approximately 2 × 107 cfu/ml. Bacterial suspensions were applied to wells of a microtiter plate coated with human platelets and blocked with a casein solution [blocking reagent (Roche) in DPBS] to prevent nonspecific bacterial adherence. After 2-h incubation at RT, the unbound bacteria were removed by aspiration. Wells were washed three times with DPBS, and the bound bacteria were released by trypsin treatment (1-h incubation with 50 μl of 1 mg/ml trypsin at RT). The number of input and bound bacteria was determined by plating serial dilutions of bacterial suspensions on sheep blood agar plates, and the binding was expressed as the percent of the input bound to human platelets. Differences in binding were compared by one-way ANOVA with post hoc Tukey's honestly significant difference (https://astatsa.com/OneWay_Anova_with_TukeyHSD/). p < 0.01 was considered statistically significant.
Data availability
Data sets associated with GC-MS linkage analysis of the GspB trisaccharide, together with MS spectra of the partially methylated alditol acetate (PMAA) derivatives, reported here have been deposited in the publicly accessible repository figshare (https://doi.org/10.6084/m9.figshare.13198340.v2). All other data are available in the manuscript.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
R. S. conceptulization; R. S., B. P. C., and P. M. S. data curation; R. S., A. C. A., and B. P. C. formal analysis; R. S. investigation; R. S., B. A. B., A. C. A., and B. P. C. methodology; R. S. writting original draft; R. S., B. A. B., and P. M. S. writting-review and editing; R. S., A. J. C., and P. M. S. supervision; A. J. A., and P. M. S. funding acquisition; P. M. S. project administration.
Funding and additional information
This work was supported by the Department of Veteran Affairs, NIH grants RO1-AI041513 and RO1-AI106987, and by the University of Guelph and the Natural Sciences and Engineering Research Council of Canada grant PGPIN3215-11. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Gerald Hart
References
- 1.Xiong Y.Q., Bensing B.A., Bayer A.S., Chambers H.F., Sullam P.M. Role of the serine-rich surface glycoprotein GspB of Streptococcus gordonii in the pathogenesis of infective endocarditis. Microb. Pathog. 2008;45:297–301. doi: 10.1016/j.micpath.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Sorge N.M., Quach D., Gurney M.A., Sullam P.M., Nizet V., Doran K.S. The group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J. Infect. Dis. 2009;199:1479–1487. doi: 10.1086/598217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rose L., Shivshankar P., Hinojosa E., Rodriguez A., Sanchez C.J., Orihuela C.J. Antibodies against PsrP, a novel Streptococcus pneumoniae adhesin, block adhesion and protect mice against pneumococcal challenge. J. Infect. Dis. 2008;198:375–383. doi: 10.1086/589775. [DOI] [PubMed] [Google Scholar]
- 4.Mistou M.-Y., Dramsi S., Brega S., Poyart C., Trieu-Cuot P. Molecular dissection of the secA2 locus of group B Streptococcus reveals that glycosylation of the Srr1 LPXTG protein is required for full virulence. J. Bacteriol. 2009;191:4195–4206. doi: 10.1128/JB.01673-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lizcano A., Sanchez C.J., Orihuela C.J. A role for glycosylated serine-rich repeat proteins in gram-positive bacterial pathogenesis. Mol. Oral Microbiol. 2012;27:257–269. doi: 10.1111/j.2041-1014.2012.00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Latousakis D., MacKenzie D.A., Telatin A., Juge N. Serine-rich repeat proteins from gut microbes. Gut Microbes. 2020;11:102–117. doi: 10.1080/19490976.2019.1602428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu F., Zhang H., Wu H. Glycosyltransferase-mediated sweet modification in oral streptococci. J. Dent. Res. 2015;94:659–665. doi: 10.1177/0022034515574865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bensing B.A., Seepersaud R., Yen Y.T., Sullam P.M. Selective transport by SecA2: An expanding family of customized motor proteins. Biochim. Biophys. Acta. 2014;1843:1674–1686. doi: 10.1016/j.bbamcr.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pyburn T.M., Bensing B.A., Xiong Y.Q., Melancon B.J., Tomasiak T.M., Ward N.J., Yankovskaya V., Oliver K.M., Cecchini G., Sulikowski G.A., Tyska M.J., Sullam P.M., Iverson T.M. A structural model for binding of the serine-rich repeat adhesin GspB to host carbohydrate receptors. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seo H.S., Minasov G., Seepersaud R., Doran K.S., Dubrovska I., Shuvalova L., Anderson W.F., Iverson T.M., Sullam P.M. Characterization of fibrinogen binding by glycoproteins Srr1 and Srr2 of Streptococcus agalactiae. J. Biol. Chem. 2013;288:35982–35996. doi: 10.1074/jbc.M113.513358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garnett J.A., Simpson P.J., Taylor J., Benjamin S.V., Tagliaferri C., Cota E., Chen Y.-Y.M., Wu H., Matthews S. Structural insight into the role of Streptococcus parasanguinis Fap1 within oral biofilm formation. Biochem. Biophys. Res. Commun. 2012;417:421–426. doi: 10.1016/j.bbrc.2011.11.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y.-H., Jiang Y.-L., Zhang J., Wang L., Bai X.-H., Zhang S.-J., Ren Y.-M., Li N., Zhang Y.-H., Zhang Z., Gong Q., Mei Y., Xue T., Zhang J.-R., Chen Y. Structural insights into SraP-mediated Staphylococcus aureus adhesion to host cells. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulte T., Lofling J., Mikaelsson C., Kikhney A., Hentrich K., Diamante A., Ebel C., Normark S., Svergun D., Henriques-Normark B., Achour A. The basic keratin 10-binding domain of the virulence-associated pneumococcal serine-rich protein PsrP adopts a novel MSCRAMM fold. Open Biol. 2014;4:130090. doi: 10.1098/rsob.130090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sequeira S., Kavanaugh D., MacKenzie D.A., Suligoj T., Walpole S., Leclaire C., Gunning A.P., Latousakis D., Willats W.G.T., Angulo J., Dong C., Juge N. Structural basis for the role of serine-rich repeat proteins from Lactobacillus reuteri in gut microbe-host interactions. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E2706–E2715. doi: 10.1073/pnas.1715016115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bensing B.A., Sullam P.M. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 2002;44:1081–1094. doi: 10.1046/j.1365-2958.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- 16.Bu S., Li Y., Zhou M., Azadin P., Zeng M., Fives-Taylor P., Wu H. Interaction between two putative glycosyltransferases is required for glycosylation of a serine-rich streptococcal adhesin. J. Bacteriol. 2008;190:1256–1266. doi: 10.1128/JB.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y., Seepersaud R., Bensing B.A., Sullam P.M., Rapoport T.A. Mechanism of a cytosolic O-glycosyltransferase essential for the synthesis of a bacterial adhesion protein. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E1190–E1199. doi: 10.1073/pnas.1600494113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y., Bensing B.A., Seepersaud R., Mi W., Liao M., Jeffrey P.D., Shajahan A., Sonon R.N., Azadi P., Sullam P.M., Rapoport T.A. Unraveling the sequence of cytosolic reactions in the export of GspB adhesin from Streptococcus gordonii. J. Biol. Chem. 2018;293:5360–5373. doi: 10.1074/jbc.RA117.000963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latousakis D., Nepravishta R., Rejzek M., Wegmann U., Le Gall G., Kavanaugh D., Colquhoun I.J., Frese S., MacKenzie D.A., Walter J., Angulo J., Field R.A., Juge N. Serine-rich repeat protein adhesins from Lactobacillus reuteri display strain specific glycosylation profiles. Glycobiology. 2019;29:45–58. doi: 10.1093/glycob/cwy100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Y.-L., Jin H., Yang H.-B., Zhao R.-L., Wang S., Chen Y., Zhou C.-Z. Defining the enzymatic pathway for polymorphic O-glycosylation of the pneumococcal serine-rich repeat protein PsrP. J. Biol. Chem. 2017;292:6213–6224. doi: 10.1074/jbc.M116.770446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu F., Zhang H., Yang T., Haslam S.M., Dell A., Wu H. Engineering and dissecting the glycosylation pathway of a streptococcal serine-rich repeat adhesin. J. Biol. Chem. 2016;291:27354–27363. doi: 10.1074/jbc.M116.752998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H., Zhu F., Yang T., Ding L., Zhou M., Li J., Haslam S.M., Dell A., Erlandsen H., Wu H. The highly conserved domain of unknown function 1792 has a distinct glycosyltransferase fold. Nat. Commun. 2014;5:4339. doi: 10.1038/ncomms5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takamatsu D., Bensing B.A., Cheng H., Jarvis G.A., Siboo I.R., Lopez J.A., Griffiss J.M., Sullam P.M. Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Ibalpha. Mol. Microbiol. 2005;58:380–392. doi: 10.1111/j.1365-2958.2005.04830.x. [DOI] [PubMed] [Google Scholar]
- 24.Bensing B.A., Khedri Z., Deng L., Yu H., Prakobphol A., Fisher S.J., Chen X., Iverson T.M., Varki A., Sullam P.M. Novel aspects of sialoglycan recognition by the Siglec-like domains of streptococcal SRR glycoproteins. Glycobiology. 2016;26:1222–1234. doi: 10.1093/glycob/cww042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takamatsu D., Bensing B.A., Sullam P.M. Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol. Microbiol. 2004;52:189–203. doi: 10.1111/j.1365-2958.2004.03978.x. [DOI] [PubMed] [Google Scholar]
- 26.Bensing B.A., Takamatsu D., Sullam P.M. Determinants of the streptococcal surface glycoprotein GspB that facilitate export by the accessory Sec system. Mol. Microbiol. 2005;58:1468–1481. doi: 10.1111/j.1365-2958.2005.04919.x. [DOI] [PubMed] [Google Scholar]
- 27.Seepersaud R., Sychantha D., Bensing B.A., Clarke A.J., Sullam P.M. O-acetylation of the serine-rich repeat glycoprotein GspB is coordinated with accessory Sec transport. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braunstein M., Bensing B.A., Sullam P.M. The two distinct types of SecA2-dependent export systems. Microbiol. Spectr. 2019 doi: 10.1128/microbiolspec.PSIB-0025-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaze T., Guillot A., Valot B., Langella O., Chamot-Rooke J., Di Guilmi A.-M., Trieu-Cuot P., Dramsi S., Mistou M.-Y. O-Glycosylation of the N-terminal region of the serine-rich adhesin Srr1 of Streptococcus agalactiae explored by mass spectrometry. Mol. Cell. Proteomics. 2014;13:2168–2182. doi: 10.1074/mcp.M114.038075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Couvigny B., Lapaque N., Rigottier-Gois L., Guillot A., Chat S., Meylheuc T., Kulakauskas S., Rohde M., Mistou M.-Y., Renault P., Dore J., Briandet R., Serror P., Guedon E. Three glycosylated serine-rich repeat proteins play a pivotal role in adhesion and colonization of the pioneer commensal bacterium, Streptococcus salivarius. Environ. Microbiol. 2017;19:3579–3594. doi: 10.1111/1462-2920.13853. [DOI] [PubMed] [Google Scholar]
- 31.Seepersaud R., Bensing B.A., Yen Y.T., Sullam P.M. The accessory Sec protein Asp2 modulates GlcNAc deposition onto the serine-rich repeat glycoprotein GspB. J. Bacteriol. 2012;194:5564–5575. doi: 10.1128/JB.01000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pauly M., Ramírez V. New insights into wall polysaccharide O-acetylation. Front. Plant Sci. 2018;9:1210. doi: 10.3389/fpls.2018.01210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moynihan P.J., Clarke A.J. O-acetylation of peptidoglycan in gram-negative bacteria: Identification and characterization of peptidoglycan O-acetyltransferase in Neisseria gonorrhoeae. J. Biol. Chem. 2010;285:13264–13273. doi: 10.1074/jbc.M110.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moynihan P.J., Sychantha D., Clarke A.J. Chemical biology of peptidoglycan acetylation and deacetylation. Bioorg. Chem. 2014;54:44–50. doi: 10.1016/j.bioorg.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 35.Hang H.C., Bertozzi C.R. The chemistry and biology of mucin-type O-linked glycosylation. Bioorg. Med. Chem. 2005;13:5021–5034. doi: 10.1016/j.bmc.2005.04.085. [DOI] [PubMed] [Google Scholar]
- 36.Yakovenko O., Nunez J., Bensing B., Yu H., Mount J., Zeng J., Hawkins J., Chen X., Sullam P.M., Thomas W. Serine-rich repeat adhesins mediate shear-enhanced streptococcal binding to platelets. Infect. Immun. 2018;86 doi: 10.1128/IAI.00160-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y., Huang X., Li J., Zeng J., Zhu F., Fan W., Hu L. Both GtfA and GtfB are required for SraP glycosylation in Staphylococcus aureus. Curr. Microbiol. 2014;69:121–126. doi: 10.1007/s00284-014-0563-2. [DOI] [PubMed] [Google Scholar]
- 38.Perepelov A.V., Shekht M.E., Liu B., Shevelev S.D., Ledov V.A., Senchenkova S.N., L’vov V.L., Shashkov A.S., Feng L., Aparin P.G., Wang L., Knirel Y.A. Shigella flexneri O-antigens revisited: Final elucidation of the O-acetylation profiles and a survey of the O-antigen structure diversity. FEMS Immunol. Med. Microbiol. 2012;66:201–210. doi: 10.1111/j.1574-695X.2012.01000.x. [DOI] [PubMed] [Google Scholar]
- 39.West N.P., Sansonetti P., Mounier J., Exley R.M., Parsot C., Guadagnini S., Prévost M.-C., Prochnicka-Chalufour A., Delepierre M., Tanguy M., Tang C.M. Optimization of virulence functions through glucosylation of Shigella LPS. Science. 2005;307:1313–1317. doi: 10.1126/science.1108472. [DOI] [PubMed] [Google Scholar]
- 40.Sun Q., Knirel Y.A., Wang J., Luo X., Senchenkova S.N., Lan R., Shashkov A.S., Xu J. Serotype-converting bacteriophage SfII encodes an acyltransferase protein that mediates 6-O-acetylation of GlcNAc in Shigella flexneri O-antigens, conferring on the host a novel O-antigen epitope. J. Bacteriol. 2014;196:3656–3666. doi: 10.1128/JB.02009-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seepersaud R., Bensing B.A., Yen Y.T., Sullam P.M. Asp3 mediates multiple protein-protein interactions within the accessory Sec system of Streptococcus gordonii. Mol. Microbiol. 2010;78:490–505. doi: 10.1111/j.1365-2958.2010.07346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yen Y.T., Cameron T.A., Bensing B.A., Seepersaud R., Zambryski P.C., Sullam P.M. Differential localization of the streptococcal accessory sec components and implications for substrate export. J. Bacteriol. 2013;195:682–695. doi: 10.1128/JB.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madoff L.C., Michel J.L., Gong E.W., Kling D.E., Kasper D.L. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4131–4136. doi: 10.1073/pnas.93.9.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seo H.S., Xiong Y.Q., Mitchell J., Seepersaud R., Bayer A.S., Sullam P.M. Bacteriophage lysin mediates the binding of streptococcus mitis to human platelets through interaction with fibrinogen. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell J., Siboo I.R., Takamatsu D., Chambers H.F., Sullam P.M. Mechanism of cell surface expression of the Streptococcus mitis platelet binding proteins PblA and PblB. Mol. Microbiol. 2007;64:844–857. doi: 10.1111/j.1365-2958.2007.05703.x. [DOI] [PubMed] [Google Scholar]
- 46.Framson P.E., Nittayajarn A., Merry J., Youngman P., Rubens C.E. New genetic techniques for group B streptococci: High-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 1997;63:3539–3547. doi: 10.1128/aem.63.9.3539-3547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartford O., O’Brien L., Schofield K., Wells J., Foster T.J. The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiol. Read. Engl. 2001;147:2545–2552. doi: 10.1099/00221287-147-9-2545. [DOI] [PubMed] [Google Scholar]
- 48.Siboo I.R., Chaffin D.O., Rubens C.E., Sullam P.M. Characterization of the accessory Sec system of Staphylococcus aureus. J. Bacteriol. 2008;190:6188–6196. doi: 10.1128/JB.00300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ciucanu I., Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 1984;131:209–217. [Google Scholar]
- 50.Varki A., Cummings R.D., Aebi M., Packer N.H., Seeberger P.H., Esko J.D., Stanley P., Hart G., Darvill A., Kinoshita T., Prestegard J.J., Schnaar R.L., Freeze H.H., Marth J.D., Bertozzi C.R. Symbol nomenclature for graphical representations of glycans. Glycobiology. 2015;25:1323–1324. doi: 10.1093/glycob/cwv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sullam P.M., Valone F.H., Mills J. Mechanisms of platelet aggregation by viridans group streptococci. Infect. Immun. 1987;55:1743–1750. doi: 10.1128/iai.55.8.1743-1750.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bensing B.A., Gibson B.W., Sullam P.M. The Streptococcus gordonii platelet binding protein GspB undergoes glycosylation independently of export. J. Bacteriol. 2004;186:638–645. doi: 10.1128/JB.186.3.638-645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bensing B.A., Rubens C.E., Sullam P.M. Genetic loci of Streptococcus mitis that mediate binding to human platelets. Infect. Immun. 2001;69:1373–1380. doi: 10.1128/IAI.69.3.1373-1380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sets associated with GC-MS linkage analysis of the GspB trisaccharide, together with MS spectra of the partially methylated alditol acetate (PMAA) derivatives, reported here have been deposited in the publicly accessible repository figshare (https://doi.org/10.6084/m9.figshare.13198340.v2). All other data are available in the manuscript.