Abstract
Mitochondrial ATP synthase is a reversible nanomotor synthesizing or hydrolyzing ATP depending on the potential across the membrane in which it is embedded. In the unicellular parasite Trypanosoma brucei, the direction of the complex depends on the life cycle stage of this digenetic parasite: in the midgut of the tsetse fly vector (procyclic form), the FoF1–ATP synthase generates ATP by oxidative phosphorylation, whereas in the mammalian bloodstream form, this complex hydrolyzes ATP and maintains mitochondrial membrane potential (ΔΨm). The trypanosome FoF1–ATP synthase contains numerous lineage-specific subunits whose roles remain unknown. Here, we seek to elucidate the function of the lineage-specific protein Tb1, the largest membrane-bound subunit. In procyclic form cells, Tb1 silencing resulted in a decrease of FoF1–ATP synthase monomers and dimers, rerouting of mitochondrial electron transfer to the alternative oxidase, reduced growth rate and cellular ATP levels, and elevated ΔΨm and total cellular reactive oxygen species levels. In bloodstream form parasites, RNAi silencing of Tb1 by ∼90% resulted in decreased FoF1–ATPase monomers and dimers, but it had no apparent effect on growth. The same findings were obtained by silencing of the oligomycin sensitivity-conferring protein, a conserved subunit in T. brucei FoF1–ATP synthase. However, as expected, nearly complete Tb1 or oligomycin sensitivity-conferring protein suppression was lethal because of the inability to sustain ΔΨm. The diminishment of FoF1–ATPase complexes was further accompanied by a decreased ADP/ATP ratio and reduced oxygen consumption via the alternative oxidase. Our data illuminate the often diametrically opposed bioenergetic consequences of FoF1–ATP synthase loss in insect versus mammalian forms of the parasite.
Keywords: Trypanosoma brucei, bioenergetics, mitochondria, electron transport, ATP synthase, oxidative phosphorylation, ATPase, alternative oxidase, respiration, mitochondrial membrane potential
Abbreviations: ΔΨm, mitochondrial membrane potential; AAC, ADP/ATP carrier; ACA, ε-aminocaproic acid; AOX, alternative oxidase; BNE, blue native electrophoresis; BSF, bloodstream form; cDKO, conditional double knock-out; DDM, dodecylmaltoside; H2DCFHDA, dichlorodihydrofluorescein; KCN, potassium cyanide; Mdm38, mitochondrial distribution and morphology protein 38 (aka YOL027C); mAb, monoclonal antibody; O2⋅−, superoxide; OSCP, oligomycin sensitivity-conferring protein; pAb, polyclonal antibody; PCF, procyclic form; ROS, reactive oxygen species; SHAM, salicylhydroxamic acid; Tb1, ATPaseTb1 (T. brucei FoF1–ATP synthase subunit 1); Tb2, ATPaseTb2 (T. brucei FoF1–ATP synthase subunit 2); TMRE, tetramethylrhodamine ethyl ester; WT, wild type
The FoF1–ATP synthase is a multisubunit protein complex capable of coupling ATP synthesis/hydrolysis with transmembrane proton translocation. In eukaryotes, this nanomachine is embedded in the inner mitochondrial membrane and consists of two parts, the matrix-facing F1 and the membrane-embedded Fo. The F1 domain, known as F1-ATPase, is responsible for the phosphorylation of ADP to ATP, and it consists of a heterohexamer of α and β subunits and a central stalk (subunits γ, δ, and ε) that connects the (αβ)3-headpiece to the Fo section. The core of the Fo section consists of a ring of c subunits that tightly interacts with subunit a, a highly hydrophobic subunit encoded by the mitochondrial genome in most eukaryotes, including trypanosomatids (1, 2). Aside from the central stalk, the interaction between the Fo and F1 domains is mediated by the peripheral stalk, an elongated structure that immobilizes the (αβ)3-headpiece during the rotation of the central rotor shaft (central stalk plus c-ring) by directly binding to subunits α and β (1). Despite the long period of evolutionary divergence of more than 2 billion years, the structure of prokaryotic and eukaryotic FoF1–ATP synthases is notably conserved, mainly at the level of tertiary and quaternary structures (2).
Nevertheless, in recent years, purifications and high-resolution structures of FoF1–ATP synthases from nonclassical model organisms revealed a wider variety in complex composition and structural organization than initially recognized (3, 4, 5, 6, 7). This includes the Trypanosoma brucei brucei FoF1–ATP synthase, an enzyme composed of 23 subunits, of which 14 are either lineage specific or highly divergent (8). For example, the lineage-specific subunits p18 and ATPaseTb2 (Tb2 in short) elaborate the otherwise conserved F1 domain (9, 10) and represent one of the largest peripheral stalk subunits found in FoF1–ATP synthases to date (11), respectively.
The peculiarities of T. brucei FoF1–ATP synthase are not restricted only to complex composition. A remarkable feature of this complex is that its activity depends on the parasite's life cycle. The procyclic form (PCF), also known as insect midgut stage, harbors a conventional mitochondrion where the FoF1–ATP synthase produces ATP (forward mode) using the electrochemical gradient across the inner mitochondrial membrane generated by the proton-pumping activity of respiratory complexes III and IV (8, 12, 13). In contrast, the infectious stage of the mammalian host, termed long slender bloodstream form (BSF), lacks a cytochrome-mediated electron transport chain and respires exclusively via the alternative oxidase (AOX) pathway (14). The mitochondrial membrane potential (ΔΨm) is generated by the proton-pumping activity (reverse mode) of the FoF1–ATP synthase (aka FoF1–ATPase) complex at the expense of ATP (15, 16). Hence, T. brucei represents a unique eukaryotic system that allows to study both modes of the FoF1–ATP synthase in physiological settings and the distinct bioenergetic consequences upon the loss of either of the activities.
The reverse mode of the FoF1–ATP synthase complex is used by some prokaryotes (17), but it is unusual in eukaryotes, where it occurs under rare nonphysiological and stress conditions, such as hypoxia or anoxia. In these cases, the respiratory arrest and subsequent collapse of the ΔΨm causes a reversal of the FoF1–ATP synthase to generate a modest ΔΨm (18, 19). The reversal of FoF1–ATP synthase also takes place in cells lacking mitochondrial DNA, which maintain ΔΨm by an electrogenic exchange of ATP4− for ADP3− by the ADP/ATP carrier (AAC) coupled to ATP hydrolysis by an incomplete FoF1–ATPase (20, 21, 22). The depletion of ATP due to the hydrolytic activity of the FoF1–ATP synthase during ischemic conditions is mitigated by a unidirectional inhibitor, the inhibitory factor 1 (23). Noteworthy, in T. brucei, the expression of inhibitory factor 1 is tightly regulated throughout the parasite's life cycle, and its experimental expression in BSF trypanosomes results in cell death (24, 25), highlighting the indispensability of the FoF1–ATPase complex for BSF T. brucei.
ATPaseTb1 (Tb1 in short; systematic TriTrypDB.org ID Tb927.10.520) (47 kDa) is one of the 14 lineage-specific subunits and the largest Fo subunit of the T. brucei FoF1–ATP synthase complex (8) (named Tb7760 in that study, after its previous systematic TriTrypDB ID TB10.70.7760). Downregulation of Tb1 in PCF trypanosomes inhibits cell growth, destabilizes FoF1–ATP synthase, and affects both the ATP synthetic and hydrolytic activities of the complex (8). Here, we studied in more detail the mitochondrial phenotypes associated with the downregulation of Tb1 in PCF cells and further explore the role of this subunit, as well as that of the peripheral subunit oligomycin sensitivity-conferring protein (OSCP), in the BSF stage.
Results
Tb1 is a membrane-bound subunit of the Fo moiety
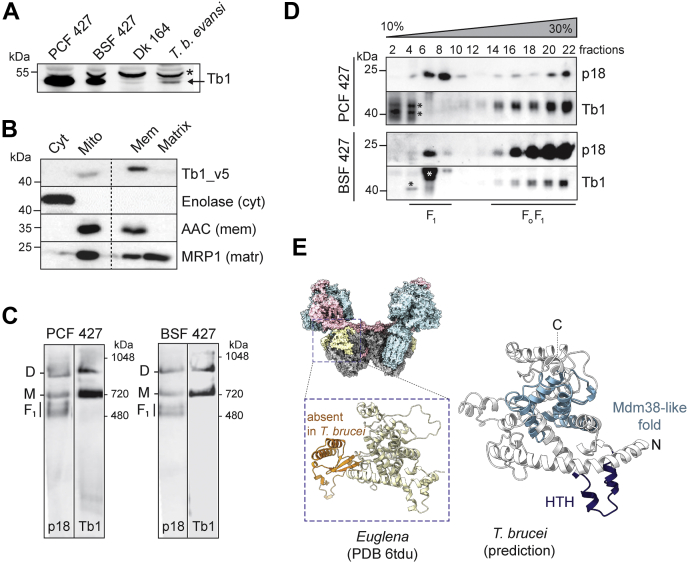
Tb1, the largest membrane-associated subunit of the T. brucei FoF1–ATP synthase, has homologs in representatives of the Euglenozoa group but appears to be absent from other eukaryotic lineages (8). In agreement with the reduced size and activity of the mitochondrion in the BSFs of T. brucei (26), Tb1 is less abundant in BSF cells than in PCF cells and barely detectable in the mitochondrial DNA-lacking (aka akinetoplastic) strains T. b. brucei Dk 164 and Trypanosoma brucei evansi AnTat 3/3 (Fig. 1A). To confirm the submitochondrial localization of Tb1 and determine how it is associated with the inner mitochondrial membrane, we performed carbonate extraction of mitochondria purified from T. brucei cells expressing C-terminally v5-tagged Tb1 (Fig. 1B). Tb1 is found exclusively in the membrane fraction, and the marker proteins, enolase, mitochondrial RNA-binding protein 1, and AAC, are detected in their expected compartments: cytosol, mitochondrial matrix, and mitochondrial membrane, respectively. This result suggests that Tb1 is an integral membrane protein. In PCF and BSF cells, Tb1 is present in fully assembled FoF1–ATP synthase monomers and dimers, as documented by high-resolution clear native electrophoresis (Fig. 1C) and sedimentation in glycerol gradient (Fig. 1D) followed by Western blot analyses with a specific anti-Tb1 antibody. In glycerol gradients, the Tb1 antibody detected, in addition to Tb1 migrating with the FoF1–ATP synthase, nonspecific bands of ∼40 kDa and 42 kDa in PCF and of 55 kDa in BSF, which are identified by the asterisk (Fig. 1D).
Figure 1.
Tb1 is a membrane-bound FoF1–ATP synthase subunit.A, Western blot analysis of whole-cell lysates prepared from 1 × 107T. brucei PCF, BSF, Dk 164, and Trypanosoma brucei evansi AnTat 3/3 cells probed with anti-Tb1 antibody. The laboratory-induced Dk 164 and the naturally occurring laboratory-adapted T. b. evansi AnTat 3/3 are BSF strains devoid of mitochondrial DNA. An asterisk points to a nonspecific band detected by the anti-Tb1 antibody. B, Western blot analysis of subcellular fractions obtained by carbonate extraction of mitochondria purified from PCF cells expressing v5-tagged Tb1 protein. Blots were probed with anti-v5 antibody, anti-enolase, anti-AAC, and anti-MRP1 antibodies to visualize Tb1, cytosolic enolase, inner mitochondrial membrane-bound AAC, and mitochondrial matrix–localized MRP1, respectively. C, High-resolution clear native electrophoresis of crude mitochondrial vesicles from PCF and BSF parasites. The F1-ATPase (F1) and the monomeric (M) and dimeric (D) forms of the complex were visualized using specific antibodies against subunits p18 and Tb1. D, glycerol gradient sedimentation of PCF and BSF lysed mitochondrial samples to determine the sedimentation profile of F1– and FoF1–ATP synthase complexes. Glycerol gradient fractions were analyzed by SDS-PAGE followed by Western blotting using antibodies against p18 and Tb1. The p18 antibody depicts the sedimentation profile of both F1–ATPase and the monomeric/dimeric states of the complex, whereas the Tb1 antibody detects only the monomeric/dimeric assemblies of the FoF1–ATP synthase. Asterisks represent nonspecific bands detected by the anti-Tb1 antibody. E, the structure of Tb1 from Euglena determined by cryo-EM (PDB ID 6TDU (6), light yellow) and the predicted structure of T. brucei Tb1. In the E. gracilis Tb1, the region absent in T. brucei is shown in orange. In the T. brucei Tb1, the Mdm38-like fold and the helix-turn-helix motif (HTH) intruding into the membrane are shown in light blue and dark blue, respectively. In the space-filling model of the E. gracilis FoF1–ATP synthase, the F1–ATPase and the c-ring are blue, the peripheral stalk subunits are pink, and all other membrane subunits are gray. AAC, ADP/ATP carrier; BSF, bloodstream form; cryo-EM, cryogenic electron microscopy; Cyt, cytosol; Matr, mitochondrial matrix; Mdm38, mitochondrial distribution and morphology protein 38; Mem, mitochondrial membranes; Mito, mitochondria; MRP1, mitochondrial RNA binding protein 1; PCF, procyclic form; Tb1, ATPaseTb1.
Our observations that Tb1 is present in fully assembled FoF1–ATP synthase are in agreement with a recent cryogenic electron microscopy structure of the FoF1–ATP synthase dimer from Euglena gracilis, a closely related free-living species within the Euglenozoa group, in which Tb1 is found on the matrix side of the membrane part of the complex at the peripheral stalk base (6). We predicted the structure of T. brucei Tb1 using E. gracilis Tb1 as a template (Fig. 1E). The modeling showed that both Tb1 orthologs share a fold similar to the mitochondrial distribution and morphology protein 38 (Mdm38) from Saccharomyces cerevisiae, a protein suggested to be involved in the assembly of the mitochondrial-encoded subunit a into the Fo moiety (27). Although several algorithms predicted one transmembrane helix in Tb1, the protein does not span the inner mitochondrial membrane. Instead, the presumptive transmembrane region occurs as a HTH motif intruding into the membrane (Fig. 1E), and therefore, Tb1 is a monotopic membrane protein.
Silencing of Tb1 in PCF cells leads to a transient increase of ΔΨm followed by redirection of respiration toward AOX
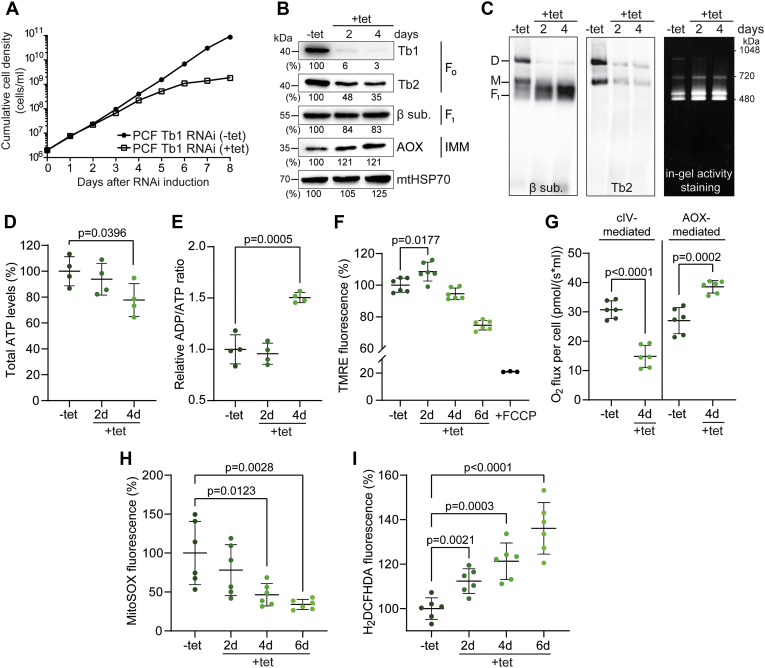
Previously, we showed that Tb1 silencing in PCF cells leads to decreased steady-state levels of fully assembled FoF1–ATP synthase complexes, and, therefore, to less ATP produced by substrate-stimulated oxidative phosphorylation in digitonin-permeabilized cells (8). Here, we explored in more detail the effect of Tb1 silencing on mitochondrial physiology and bioenergetics of PCF cells. As expected, Tb1 silencing caused a progressive growth phenotype detected first at day 4 of RNAi induction (Fig. 2A). Western blot analysis using a specific anti-Tb1 antibody showed that Tb1 protein expression was reduced to less than 10% at day 2 of RNAi induction (Fig. 2B). The downregulation of Tb1 was accompanied by a decrease in the steady-state level of the peripheral stalk subunit Tb2, whereas the abundance of the F1 moiety subunit β was not as strongly affected (Fig. 2B). The structural integrity and the activity of the FoF1–ATP synthase complex were assessed by blue native electrophoresis (BNE) followed by Western blotting and in-gel activity staining of the complex, respectively (Fig. 2C). Western blot analyses of native complexes showed a significant reduction of FoF1–ATP synthase monomers and dimers at days 2 and 4 of RNAi induction. In agreement with the steady-state levels of subunit β, the free F1-ATPase moiety was assembled and active, and it accumulated in Tb1-silenced cells. The decreased levels of fully assembled FoF1–ATP synthase complexes affected ATP production by oxidative phosphorylation (8), which was reflected by a ∼25% reduction in the total cellular ATP levels (Fig. 2D) and by a 50% increase in the ADP/ATP ratio (Fig. 2E) by day 4 of RNAi induction. Suppression of Tb1 expression also caused a mild, but statistically significant, increase in ΔΨm at day 2 of RNAi induction (Fig. 2F), but notably, this increase was only transient. In PCF cells, the ΔΨm is maintained by the activity of respiratory complexes III and IV, passing electrons from ubiquinol to molecular oxygen. However, electrons can be rerouted to another electron acceptor, a plant-like AOX, which reduces molecular oxygen to water without proton translocation (28). By doing so, PCF cells can uncouple cellular respiration from ΔΨm generation. To test if the cells responded to the hyperpolarization detected at day 2 by rewiring the electrons toward AOX, we measured the oxygen consumption rate in the presence of potassium cyanide (KCN) and salicylhydroxamic acid (SHAM), inhibitors of complex IV and AOX, respectively. Indeed, we detected that the respiration of Tb1-silenced cells is more sensitive to SHAM compared with KCN, confirming the higher proportion of AOX-mediated respiration (Fig. 2G). This rerouting of electrons was accompanied by an increase in AOX expression as determined by Western blot (Fig. 2B). Because complex III is one of the major producers of harmful superoxide (O2⋅−) molecules, an important subclass of reactive oxygen species (ROS) (29), we measured the mitochondrial concentration of O2⋅− before and after Tb1 RNAi in PCF cells (Fig. 2H). We found that O2⋅− decreased over time after Tb1 ablation, supporting the proposed rerouting of electrons from complexes III and IV toward AOX, perhaps as a protective mechanism against oxidative stress (30, 31, 32). Despite the lower levels of mitochondrial O2⋅−, the disruption of fully assembled FoF1–ATP synthase induced changes in cellular physiology that ultimately led to higher levels of various cytosolic ROS molecules (e.g., peroxyl, hydroxyl) as measured by dichlorodihydrofluorescein (H2DCFHDA), a fluorescent ROS-sensitive dye (Fig. 2I). This gradual increment of oxidative stress might have contributed to the growth phenotype observed in parasites with suppressed Tb1 expression.
Figure 2.
Loss of Tb1 in PCF cells affects the structural integrity of the FoF1–ATP synthase complex and induces changes in mitochondrial physiology.A, growth of noninduced (−tet) and Tb1 RNAi–induced (+tet) PCF cells measured for 8 days. Cumulative cell density was calculated from the cell counts adjusted by the dilution factor needed to seed the cultures at 2 × 106 cells/ml each day. B, Western blot analysis of PCF Tb1 RNAi trypanosomes grown in the absence (−tet) or presence (+tet) of tetracycline for 2 and 4 days. Whole-cell lysates were subjected to SDS-PAGE followed by immunostaining with antibodies against Tb1 and Tb2 (Fo moiety), subunit β (F1 moiety), and AOX. The numbers beneath each blot represent the abundance of immunodetected protein expressed as a percentage of the noninduced sample after normalizing to the signal intensity of the mitochondrial HSP70 probing (loading control). C, BNE of 4 μg of DDM-lysed mitochondria from PCF Tb1 RNAi–noninduced (−tet) and PCF Tb1 RNAi–induced (+tet, 2 and 4 days) cells followed by Western blot analysis using antibodies against subunit β and Tb2 to detect free F1 and monomeric (M) and dimeric (D) ATP synthase complexes (first two panels). BNE of 60 μg of DDM-lysed mitochondria from PCF Tb1 RNAi–noninduced (−tet) and PCF Tb1 RNAi–induced (+tet, 2 and 4 days) cells followed by in-gel staining of ATPase activity (rightmost panel). D, total cellular ATP levels of PCF Tb1 RNAi–noninduced cells (−tet) and cells induced for 2 and 4 days (+tet, 2 days and 4 days) (means ± SD, n = 4, Student’s unpaired t-test). E, relative ADP/ATP ratio of PCF Tb1 RNAi–noninduced cells (−tet) and cells induced for 2 and 4 days (+tet, 2 days and 4 days) (means ± SD, n = 4, Student’s unpaired t-test). F, flow cytometry analysis of TMRE-stained PCF Tb1 RNAi–noninduced cells (−tet) and cells induced for 2, 4, and 6 days (+tet, 2 days, 4 days, and 6 days) to measure ΔΨm. The addition of FCCP served as a control for mitochondrial membrane depolarization (+FCCP) (means ± SD, n = 6, Student’s unpaired t-test). G, the oxygen consumption rate of PCF Tb1 RNAi live cells in the presence of glycerol-3-phosphate. After the addition of the substrate, cells were consuming oxygen at the steady rate. Injection of SHAM inhibited AOX-mediated respiration. The difference between the original values and the values after addition of SHAM is graphed as AOX-mediated respiration. Additional injection of KCN inhibited complex IV–mediated respiration and ceased the oxygen consumption of the cells. The difference between the values after SHAM addition and after KCN addition is graphed as complex IV–mediated respiration (means ± SD, n = 6, Student’s unpaired t-test). H and I, flow cytometry analysis of MitoSOX-treated (H) and H2DCFHDA-treated (I) PCF Tb1 RNAi–noninduced cells (−tet) and cells induced for 2, 4, and 6 days (+tet, 2 days, 4 days, and 6 days) to measure mitochondrial O2⋅− and total cellular ROS levels, respectively (means ± SD, n = 6, Student’s unpaired t-test). ΔΨm, mitochondrial membrane potential; AOX, alternative oxidase; BNE, blue native electrophoresis; DDM, dodecylmaltoside; FCCP, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone; H2DCFHDA, dichlorodihydrofluorescein; KCN, potassium cyanide; O2⋅−, superoxide; PCF, procyclic form; ROS, reactive oxygen species; SHAM, salicylhydroxamic acid; Tb1, ATPaseTb1; Tb2, ATPaseTb2; TMRE, tetramethylrhodamine ethyl ester.
Stringent Tb1 silencing leads to a virtually complete loss of FoF1–ATPase complex in BSF parasites and mitochondrial membrane depolarization, followed by cell death
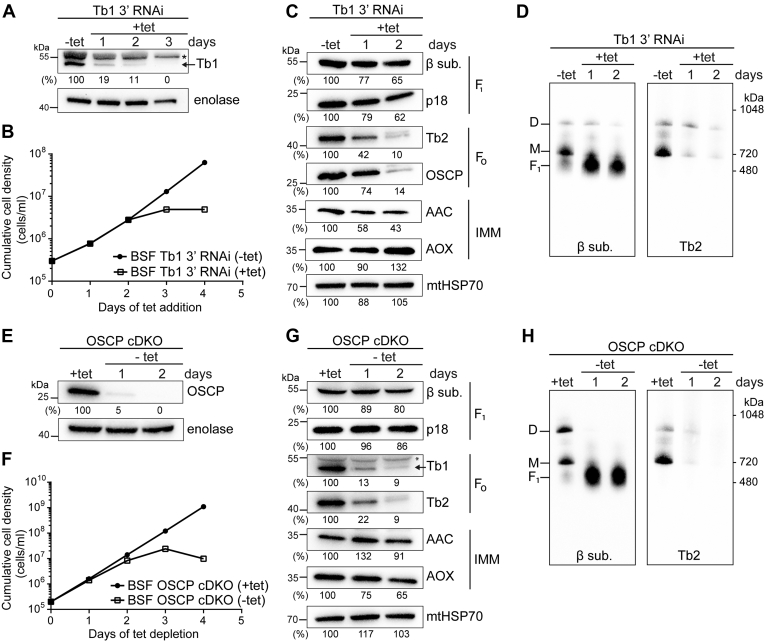
In contrast to the canonical role of the FoF1–ATP synthase in PCF cells, BSF cells use the reverse activity of this complex, which generates ΔΨm at the expense of ATP (15, 16). We silenced Tb1 in BSF cells by using an RNAi construct targeting the 3’ region of the Tb1 ORF. This caused a decrease of Tb1 to 11% at day 2 of RNAi induction, reaching undetectable levels of the protein at day 3 (Fig. 3A). As expected, the BSF Tb1 3’ RNAi cell line exhibited a strong growth phenotype and ceased growing entirely by day 4 of induction (Fig. 3B). The steady-state levels of F1–ATPase subunits β and p18 were decreased by ∼40% at day 2, whereas the steady-state levels of the Fo-associated subunits Tb2 and OSCP dropped to ∼10% (Fig. 3C). Moreover, the levels of AAC were decreased by 60% (Fig. 3C), suggesting that disruption of the FoF1–ATPase might affect the stability of this transporter. As in PCF cells, Tb1 downregulation caused a strong decrease of FoF1–ATPase monomers and dimers while there was a simultaneous increase in free F1 subcomplex (Fig. 3D, Fig. S1A).
Figure 3.
Virtually complete loss of FoF1–ATPase is lethal to BSF cells.A, Western blot of whole-cell lysates from BSF Tb1 3’ RNAi–noninduced (−tet) and BSF Tb1 3’ RNAi–induced (+tet, 1–3 days) cells using an antibody against Tb1. The numbers beneath the blot represent the abundance of immunodetected Tb1 expressed as a percentage of the noninduced samples after normalizing to the signal intensity of the enolase probing (loading control). An asterisk points to a nonspecific band detected by anti-Tb1 antibody. B, growth of BSF Tb1 3’ RNAi–noninduced (−tet) and BSF Tb1 3’ RNAi–induced (+tet) cells measured for 4 days. Cumulative cell density was calculated from the cell counts adjusted by the dilution factor needed to seed the cultures at 2 × 105 cells/ml each day. C, Western blot analysis of whole-cell lysates from BSF Tb1 3’ RNAi–noninduced (−tet) and cells induced for 1 and 2 days (+tet) using antibodies against the F1 moiety (anti-β and anti-p18), the Fo moiety (anti-Tb2 and anti-OSCP), and inner mitochondrial membrane proteins (anti-AAC and anti-AOX). The immunoblot probed with anti-mitochondrial HSP70 antibody served as loading control. The densitometric analysis is depicted by the percentages beneath each blot and was carried out as in Figure 2B. D, BNE of 20 μg of DDM-lysed mitochondria from BSF Tb1 3’ RNAi–noninduced cells (−tet) and cells induced for 1 and 2 days (+tet) followed by Western blot analysis using antibodies to detect free F1 (anti-subunit β) and monomeric (M) and dimeric (D) FoF1–ATPase complexes (anti-Tb2). E, Western blot of whole-cell lysates from BSF OSCP cDKO cells grown in the presence (+tet) or absence (−tet) of tetracycline for 1 and 2 days using an antibody against OSCP. The numbers beneath the blot represent the abundance of immunodetected OSCP expressed as a percentage of the +tet sample after normalizing to the signal intensity of the enolase probing (loading control). F, the growth curve of BSF OSCP cDKO cells cultured in the presence (+tet) or absence (−tet) of tetracycline for 4 days. Cumulative cell density was calculated as in Figure 3B. G, Western blot analysis of whole-cell lysates from BSF OSCP cDKO cells cultured in the presence (+tet) or absence (−tet) of tetracycline for 1 day and 2 days using antibodies against the F1 moiety (anti-β and anti-p18), the Fo moiety (anti-Tb1 and anti-Tb2) and inner mitochondrial membrane proteins (anti-AAC and anti-AOX). The immunoblot probed with antimitochondrial HSP70 antibody served as the loading control. The densitometric analysis is depicted by the percentages beneath each blot and was carried out as in Figure 2B. The asterisk points to a nonspecific band detected by anti-Tb1 antibody. H, BNE of 20 μg of DDM-lysed mitochondria from BSF OSCP cDKO cells grown in the presence (+tet) or absence (−tet) of tetracycline for 1 day and 2 days followed by Western blot analysis using antibodies to detect free F1 (anti-subunit β) and monomeric (M) and dimeric (D) FoF1–ATPase complexes (anti-Tb2). AAC, ADP/ATP carrier; AOX, alternative oxidase; BNE, blue native electrophoresis; BSF, bloodstream form; cDKO, conditional double knock-out; DDM, dodecylmaltoside; OSCP, oligomycin sensitivity-conferring protein; Tb1, ATPaseTb1; Tb2, ATPaseTb2.
Similarly, a conditional double knock-out (cDKO) of OSCP resulted in undetectable levels of OSCP at day 2 (Fig. 3E), which was accompanied by a strong growth phenotype (Fig. 3F). In this cell line, termed BSF OSCP cDKO, both OSCP alleles were replaced by drug resistance cassettes, and an ectopic copy of the OSCP gene, whose expression depends on the presence of tetracycline in the culture medium, was introduced (Fig. S2). The removal of tetracycline led to ablation of OSCP expression, reduced levels of F1 subunits to ∼80%, and Fo subunits to ∼10% (Fig. 3G), as well as virtually complete loss of the monomeric and dimeric forms of the complex (Fig. 3H, Fig. S1B), by day 2 of tetracycline removal.
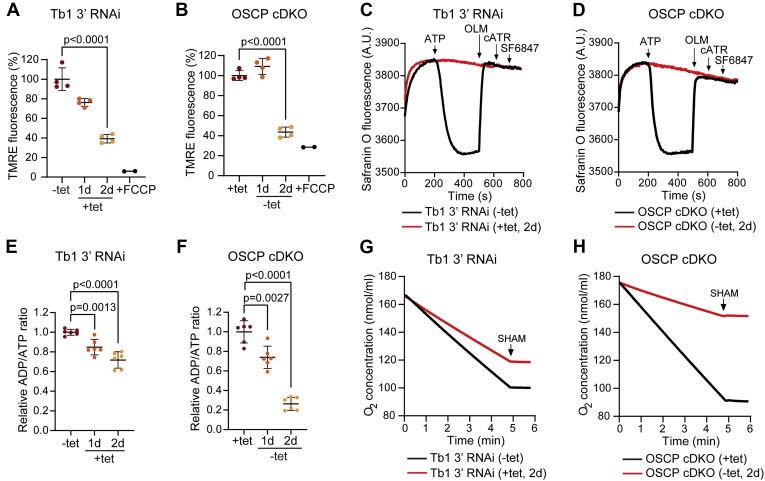
To investigate the effect of the FoF1–ATPase loss on ΔΨm, we used flow cytometry analysis of intact, live cells stained with tetramethylrhodamine ethyl ester (TMRE). In both BSF Tb1 3’ RNAi and OSCP cDKO cell lines, the ΔΨm was strongly compromised at day 2 of RNAi induction and tetracycline removal, respectively (Fig. 4, A and B), preceding the observed growth defect. To corroborate these results, we determined the ability of the mitochondrion to generate ΔΨm in digitonin-permeabilized cells using safranin O dye. Control BSF Tb1 3’ RNAi and OSCP cDKO cells were able to build up and retain a ΔΨm, as addition of ATP caused a decrease in safranin O fluorescence, indicating stacking of the dye within the energized organelle. This decrease was completely reversed by adding either oligomycin (Fig. 4, C and D, black lines) or carboxyatractyloside (Fig. S3, A and D, black lines), inhibitors of the FoF1–ATP synthase and the AAC, respectively. Subsequent addition of the uncoupler SF 6847 had no further effect on depolarization. In contrast, cells silenced for Tb1 or ablated for OSCP were unable to generate ΔΨm in situ (Fig. 4, C and D, red lines). No changes in fluorescence were detected when the addition of oligomycin or carboxyatractyloside preceded that of ATP (Fig. S3, B, C, E and F), confirming that the decrease in safranin O fluorescence observed in control cells indeed depends on the ATP-hydrolyzing activity of the FoF1–ATPase and on the ADP/ATP exchanging activity of the AAC. In summary, these measurements indicate that, in the absence of either Tb1 or OSCP, BSF cells cannot generate ΔΨm, presumably because the proton-pumping FoF1–ATPase is completely disrupted in these cells.
Figure 4.
Virtually complete loss of FoF1–ATPase leads to a sudden collapse of ΔΨm, lower ADP/ATP ratio, and reduced respiration rate in BSF parasites.A, flow cytometry analysis of TMRE-stained BSF Tb1 3’ RNAi–noninduced cells (−tet) and cells induced for 1 day and 2 days (+tet, 1 day and 2 days) to detect changes in the ΔΨm. The addition of FCCP served as a control for mitochondrial membrane depolarization (+FCCP) (means ± SD, n = 4, Student’s unpaired t-test). B, flow cytometry analysis of TMRE-stained BSF OSCP cDKO cells grown in the presence (+tet) or absence of tetracycline for 1 day and 2 days (−tet, 1 day and 2 days) to detect changes in the ΔΨm. The addition of FCCP served as a control for mitochondrial membrane depolarization (+FCCP) (means ± SD, n = 4, Student’s unpaired t-test). C, mitochondrial membrane polarization detected using safranin O dye in digitonin-permeabilized BSF Tb1 3’ RNAi–noninduced cells (−tet, black line) and cells induced for 2 days (+tet, 2 days, red line) in the presence of ATP. ATP, oligomycin (OLM), carboxyatractyloside (cATR), and SF 6847, an uncoupler, were added where indicated. cATR was added after OLM to test for any further depolarization of the mitochondrial membrane due to inhibition of the AAC, whose electrogenic activity can potentially contribute in the generation of ATP-stimulated ΔΨm. D, mitochondrial membrane polarization detected using safranin O dye in digitonin-permeabilized BSF OSCP cDKO cells grown in the presence (+tet, black line) or absence of tetracycline for 2 days (−tet, 2 days, red line) after the addition of ATP. ATP, oligomycin (OLM), carboxyatractyloside (cATR), and SF 6847, an uncoupler, were added where indicated. cATR was added after OLM to test for any further depolarization of the mitochondrial membrane due to inhibition of the AAC, whose electrogenic activity can potentially contribute in the generation of ATP-stimulated ΔΨm. E, relative ADP/ATP ratio in BSF Tb1 3’ RNAi–noninduced cells (−tet) and cells induced for 1 day and 2 days (+tet, 1 day and 2 days) (means ± SD, n = 6, Student’s unpaired t-test). F, relative ADP/ATP ratio in BSF OSCP cDKO cells cultured in the presence (+tet) or absence of tetracycline for 1 day and 2 days (−tet, 1 day and 2 days) (means ± SD, n = 6, Student’s unpaired t-test). G, oxygen consumption rates of digitonin-permeabilized BSF Tb1 3’ RNAi–noninduced cells (−tet, black line) and cells induced for 2 days (+tet, 2 days, red line) in the presence of glycerol-3-phosphate. Respiration was arrested by the addition of SHAM where indicated. H, oxygen consumption rates of digitonin-permeabilized BSF OSCP cDKO cells grown in the presence (+tet, black line) or absence of tetracycline for 2 days (−tet, 2 days, red line) after addition of glycerol-3-phosphate. Respiration was arrested by addition of SHAM where indicated. ΔΨm, mitochondrial membrane potential; AAC, ADP/ATP carrier; BSF, bloodstream form; cDKO, conditional double knock-out; FCCP, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone; OSCP, oligomycin sensitivity-conferring protein; SHAM, salicylhydroxamic acid; TMRE, tetramethylrhodamine ethyl ester.
Because the activity of the F1–ATPase is tightly connected to the cellular ADP/ATP pool (33), we measured the cellular ADP/ATP ratio in control BSF Tb1 3’ RNAi and OSCP cDKO cells and in cells with diminished Tb1 and OSCP expression. The ADP/ATP ratio was already significantly decreased at day 1 (Fig. 4, E and F), the opposite of the effect observed in PCF cells (compare Fig. 2E), likely because of the dysfunctional FoF1–ATPase. Higher levels of ATP than ADP may also affect the glycolytic flux, which is directly linked to respiration via the mitochondrial glycerol-3-phosphate dehydrogenase (34) and/or activity of AOX (35). Indeed, decreased levels of FoF1–ATPase caused lower glycerol-3-phosphate–stimulated oxygen consumption in digitonin-permeabilized BSF cells depleted of Tb1 or OSCP (Fig. 4, G and H). As expected, in both samples, the measured respiration was fully inhibited by addition of SHAM.
Suppression of Fo subunits expression to ∼10% is compatible with BSF cell viability in vitro
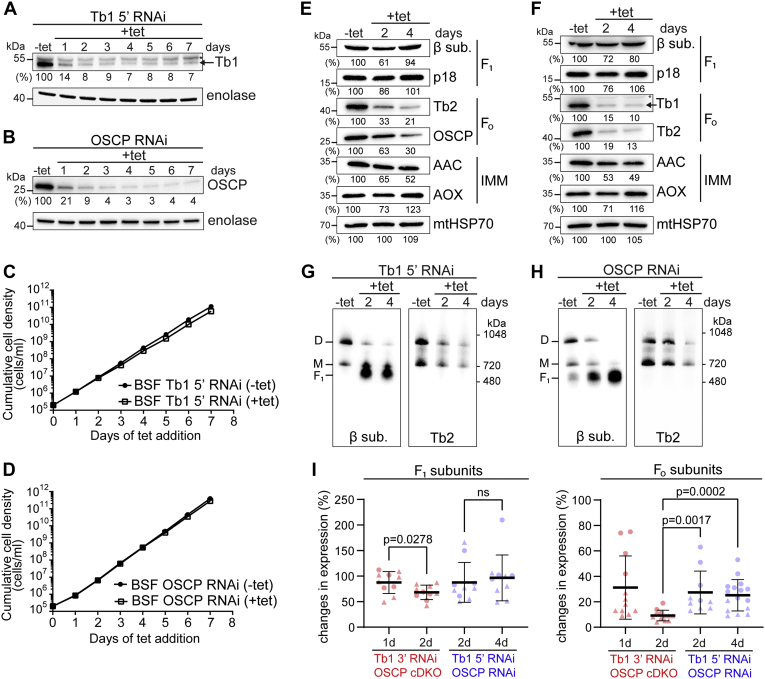
The virtually complete ablation of either Tb1 or OSCP (Fig. 3, A and E) is lethal for BSF trypanosomes, and this is in agreement with previous studies examining FoF1–ATPase subunits (9, 11, 16). Surprisingly, these parasites can withstand a 90 to 95% loss of the same proteins as revealed by analysis of two different cell lines: a BSF Tb1 5’ RNAi cell line, in which the dsRNA targets the 5’ region of the Tb1 ORF, and a BSF OSCP RNAi cell line. In these cell lines, Tb1 and OSCP expressions were stably downregulated to 7 to 9% and 3 to 4%, respectively, over the period of 7 days of RNAi induction (Fig. 5, A and B), with no obvious effect on their growth rate (Fig. 5, C and D). The different outcomes in the viability of the BSF Tb1 RNAi cell lines might be attributed to different efficiencies of the 3’ and 5’ targeted RNAi probes or to their distinctive genetic backgrounds (Tb1 3’ RNAi: EATRO 1125 AnTat 1.1 versus Tb1 5’ RNAi: BSF Lister 427). However, as the same difference in viability was observed for BSF OSCP cDKO and OSCP RNAi cells, which share the same genetic background, the distinct phenotypes were most likely due to the differences in stringency of suppression. Thus, we decided to further investigate the basis for the observed differences.
Figure 5.
BSF cells are able to tolerate suppression of Tb1 and OSCP expression by 90 to 95%.A and B, Western blot analysis of whole-cell lysates from BSF Tb1 5’ RNAi (A) and BSF OSCP RNAi (B) noninduced cells (−tet) and cells induced for 1 to 7 days (+tet) using antibodies against Tb1 and OSCP, respectively. The numbers beneath the blot represent the abundance of immunodetected Tb1 (A) or OSCP (B) expressed as a percentage of the noninduced sample after normalizing to the signal intensity of the enolase probing (loading control). In Figure 5A, an asterisk points to a nonspecific band detected by anti-Tb1 antibody. C and D, growth of BSF Tb1 5’ RNAi (C) and BSF OSCP RNAi (D) noninduced (−tet) and induced (+tet) cells measured for 7 days. Cumulative cell density was calculated as in Figure 3B. E and F, Western blot analysis of whole-cell lysates from BSF Tb1 5’ RNAi (E) and BSF OSCP RNAi (F) noninduced cells (−tet) and cells induced for 2 and 4 days (+tet) using antibodies against the F1 moiety (anti-β and anti-p18), the Fo moiety (anti-Tb1, anti-Tb2, and anti-OSCP), and inner mitochondrial membrane proteins (anti-AAC and anti-AOX). The immunoblots probed with anti-mitochondrial HSP70 antibody served as the loading control. The densitometric analysis is depicted by the percentages beneath each blot and was carried out as in Figure 2B. The asterisk points to a nonspecific band detected by anti-Tb1 antibody. G and H, BNE of 20 μg of DDM-lysed mitochondria from BSF Tb1 5’ RNAi (G) and BSF OSCP RNAi (H) noninduced cells (−tet) and cells induced for 2 and 4 days (+tet) followed by Western blot analysis using antibodies to detect free F1 (anti-subunit β) and monomeric (M) and dimeric (D) FoF1–ATPase complexes (anti-Tb2). I, comparison of changes in F1 and Fo subunit levels between BSF Tb1 3’ RNAi (red circles)/BSF OSCP cDKO cells (red triangles) and BSF Tb1 5’ RNAi (blue circles)/BSF OSCP RNAi (blue triangles) cells along the days of RNAi induction/tetracycline removal. The antibody signals of F1 (β and p18) and Fo (Tb1, Tb2, and OSCP) subunits at days 1 and 2 of RNAi induction/tetracycline removal (BSF Tb1 3’ RNAi and OSCP cDKO cells, respectively) and at days 2 and 4 of RNAi induction (BSF Tb1 5’ and OSCP RNAi cells) were quantified and normalized as in Figure 2B. In both Tb1 RNAi cell lines, the plotted values of Fo subunits correspond to the quantified signals of anti-Tb2 and anti-OSCP antibodies. In the OSCP cDKO and OSCP RNAi cell lines, the plotted values of Fo subunits correspond to the quantified signals of anti-Tb1 and anti-Tb2 antibodies. The values were analyzed statistically using GraphPad Prism 8.0 software (means ± SD, n ≥ 4, Student’s unpaired t-test). ΔΨm, mitochondrial membrane potential; AAC, ADP/ATP carrier; AOX, alternative oxidase; BSF, bloodstream form; cDKO, conditional double knock-out; ns, not significant; OSCP, oligomycin sensitivity-conferring protein; Tb1, ATPaseTb1.
Western blot analyses revealed a correlative decrease in Fo subunits in both BSF Tb1 5’ and OSCP RNAi cell lines, whereas F1 subunits stayed largely unaffected. Similar to BSF Tb1 3’ RNAi cells, the expression of AOX was slightly upregulated, whereas the levels of AAC were halved (Fig. 5, E and F). Next, we analyzed the assembly of FoF1–ATPase complexes in BSF Tb1 5’ and OSCP RNAi cells by BNE. Silencing of Tb1 and OSCP induced monomer and dimer instability as F1–ATPase subcomplexes accumulated in RNAi-induced cells, and less monomeric and dimeric forms were detected using antibodies against F1 and Fo moieties (Fig. 5, G and H, Fig. S1, C and D), but compared with BSF Tb1 3’ RNAi–induced cells and OSCP cDKO cells (Fig. 3, D and H), the decrease was not as profound.
We hypothesized that the observed differences in the viability of the examined cell lines may lay in the varying amounts of remaining active FoF1–ATPase complexes. Therefore, we quantified and compared the changes in the steady-state levels of F1 (β and p18) and Fo (Tb1, Tb2 and OSCP) subunits for the four BSF cell lines. At day 4 of RNAi induction, when the steady-state levels of the target proteins in BSF Tb1 5’ and OSCP RNAi cells were the lowest in their respective cell lines (Fig. 5, A and B), the steady-state levels of the tested Fo subunits remained significantly higher (25.1% ± 12.3%, means ± SD) than those in BSF Tb1 3’ RNAi and OSCP cDKO cells by day 2 of RNAi induction or tetracycline removal, respectively (9.2% ± 4.2%, means ± SD) (Fig. 5I). We propose that in the BSF Tb1 3’ RNAi and the OSCP cDKO cell lines, the levels of Fo subunits drop under a threshold that does not allow the parasite to assemble a sufficient amount of FoF1–ATPase complexes to maintain its viability. Assuming a direct relationship between the steady-state levels of individual Fo subunits and the total levels of FoF1 holocomplex, it would suggest that at least a 10% of assembled FoF1–ATPase complexes is necessary to maintain the viability of BSF T. brucei cells in vitro.
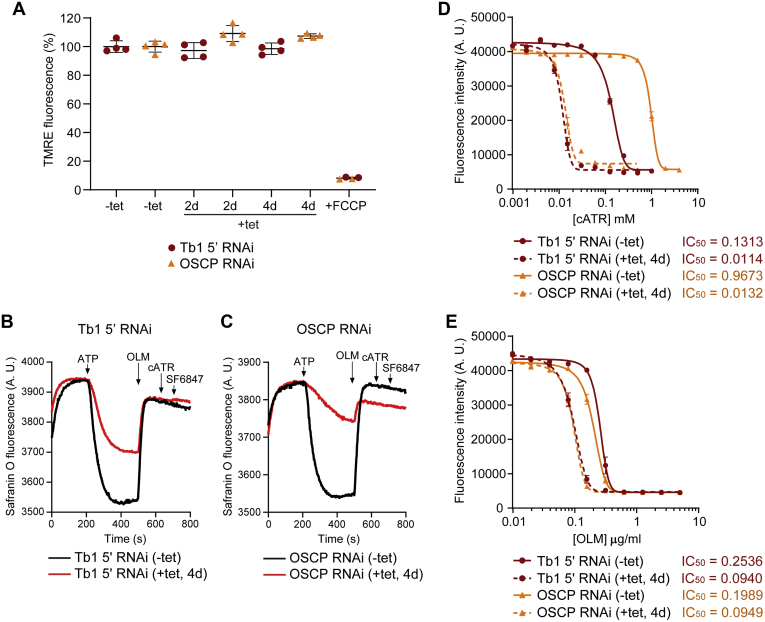
To test if the detected decrease in the levels of FoF1–ATPase monomers and dimers in BSF Tb1 5’ and OSCP RNAi cells affects ΔΨm, we measured TMRE fluorescence in live cells from both cell lines by flow cytometry. We did not detect any differences in ΔΨm between the measured time points (Fig. 6A). Furthermore, we assessed the ability of the FoF1–ATPase to polarize the mitochondrial membrane in digitonin-permeabilized BSF Tb1 5’ and OSCP RNAi cells using safranin O dye in the presence of ATP. Albeit reduced, compared with control cells (Fig. 6, B and C, Fig. S4, A and D, black lines), RNAi-induced cells from both cell lines were still able to generate a ΔΨm (Fig. 6, B and C, Fig. S4, A and D, red lines). In all four cases, the inner mitochondrial membrane was fully depolarized by the addition of oligomycin (Fig. 6, B and C) or carboxyatractyloside (Fig. S4, A and D). The results were further validated by measuring safranin O fluorescence when either oligomycin or carboxyatractyloside was present before ATP addition (Fig. S4, B, C, E and F). These observations show that the FoF1–ATPase activity is affected in BSF Tb1 5’ and OSCP RNAi–induced cells but is not fully abolished as observed for BSF Tb1 3’ RNAi and OSCP cDKO cells (Fig. 4, C and D, Fig. S3, A and D, red lines).
Figure 6.
BSF cells with 90 to 95% reduced expression of Tb1 and OSCP have unchanged ΔΨm but are more sensitive to AAC and FoF1–ATPase inhibitors.A, flow cytometry analysis of TMRE-stained BSF Tb1 5’ RNAi (brick-red circles) and BSF OSCP RNAi (orange triangles) noninduced cells (−tet) and cells induced for 2 and 4 days (+tet, 2 days and 4 days) to detect changes in ΔΨm. The addition of FCCP served as a control for mitochondrial membrane depolarization (means ± SD, n = 4, Student’s unpaired t-test). B and C, mitochondrial membrane polarization detected using safranin O dye in digitonin-permeabilized BSF Tb1 5’ RNAi (B) and BSF OSCP RNAi (C) noninduced cells (−tet, black lines) and cells induced for 4 days (+tet, 4 days, red lines) in the presence of ATP. ATP, oligomycin (OLM), carboxyatractyloside (cATR), and SF 6847, an uncoupler, were added where indicated. cATR was added after OLM to test for any further depolarization of the mitochondrial membrane due to inhibition of the AAC, whose electrogenic activity can potentially contribute in the generation of ATP-stimulated ΔΨm. D and E, sensitivity of BSF Tb1 5’ RNAi– and BSF OSCP RNAi–noninduced cells (−tet, brick-red and orange full lines, respectively) and cells induced for 4 days (+tet, 4 days, brick-red and orange dashed lines, respectively) to carboxyatractyloside (cATR) (D) and to oligomycin (OLM) (E) estimated by resazurin cell-viability assay. The dose–response curves were calculated using GraphPad Prism 8.0 software. The calculated IC50 values are shown beside the corresponding sample and are expressed in mM and in μg/ml for cATR and OLM, respectively. ΔΨm, mitochondrial membrane potential; AAC, ADP/ATP carrier; BSF, bloodstream form; FCCP, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone; OSCP, oligomycin sensitivity-conferring protein; TMRE, tetramethylrhodamine ethyl ester.
The difference between the ΔΨm measured in vivo (by flow cytometry) and in situ (by safranin O) may reflect the different aspects of ΔΨm generation interrogated by these assays: the former measures the actual magnitude of ΔΨm in living cells, and the latter, the capacity of cells to generate ΔΨm. The differences observed imply that BSF cells do not use FoF1–ATPase complexes at full capacity to generate ΔΨm, and therefore, a significant decrease in these assemblies has no effect on their total ΔΨm and the viability of cells grown in culture. The need for the ∼10% of remaining FoF1–ATPase complexes (Fig. 5I) in the mitochondria is corroborated by increased sensitivity of BSF Tb1 5’ and OSCP RNAi–induced cells to carboxyatractyloside (∼12 and ∼73 times, respectively) and to oligomycin (∼2.5 and ∼2.1 times, respectively) as measured by resazurin cell viability assay (Fig. 6, D and E).
In summary, our data provide insight into the distinct bioenergetic consequences of the loss of FoF1–ATP synthase/ATPase in PCF and BSF parasites. We showed that a decrease in the FoF1–ATP synthase levels in PCF cells led to lowered ATP levels and transiently increased ΔΨm, which manifested in the form of oxidative stress and slower cell growth. In BSF trypanosomes, as reported before, a full disruption of the FoF1–ATPase caused the loss of ΔΨm followed by cell death, but we observed that these parasites can withstand a substantial loss of the complex without an obvious effect on viability in culture.
Discussion
Tb1 is the largest membrane-associated subunit of the FoF1–ATP synthase in T. brucei. Inferring from the FoF1–ATP synthase structure of the related organism E. gracilis, Tb1 is located on the matrix side of the Fo periphery (6) and contains an Mdm38-like fold. Mdm38 is a component of the yeast mitochondrial cotranslational membrane insertion machinery, and it has been reported to assist in the assembly of the mitochondrial-encoded proton channel subunit a into the FoF1–ATP synthase (27). Despite the structural similarity between Mdm38 and Tb1, the former was never copurified with either electron transport chain complexes or FoF1–ATP synthase (27, 36, 37), whereas the latter is a bona fide component of the euglenozoan FoF1–ATP synthase (6, 8, 38). In addition, the E. gracilis Tb1 ortholog contacts nearly exclusively a species-specific extension of subunit a (6), suggesting that it is not involved in the incorporation of subunit a into the complex in the same way as in yeast. Based on the predicted structural conservancy of Tb1 and compositional similarity of the FoF1–ATP synthase in T. brucei and Euglena, it is reasonable to assume that Tb1 in T. brucei is located on the periphery of the dimer. Presumably, it establishes very few, or none at all, contacts with subunits involved directly in the proton translocation or dimerization of the complex. Yet, Tb1 is absolutely crucial for the FoF1–ATP synthase integrity (8), potentially by aiding in the stability or assembly of auxiliary Fo subunits.
Here, we silenced Tb1 subunit by RNAi with high efficiency in two cultivable forms of T. brucei to study phenotypes associated with FoF1–ATP synthase loss. In PCF cells, the severe loss of the monomeric and dimeric forms of the FoF1–ATP synthase after depletion of Tb1 causes a transient hyperpolarization of the inner mitochondrial membrane. This effect is then reversed by redirecting electrons from the conventional respiratory chain complexes III and IV toward AOX, as documented by the increased SHAM-sensitive respiration. The increased respiration through the AOX pathway is accompanied by a gradual but mild reduction in the ΔΨm (∼25% by day 6 of RNAi induction), consistent with the fact that AOX is incapable of pumping protons across the inner mitochondrial membrane, and therefore, does not contribute to the generation of ΔΨm (28). Moreover, the increase in AOX-mediated respiration led to lowered levels of mitochondrial O2⋅−, presumably produced to a large extent by complex III. The rerouting of electrons toward AOX is a phenomenon that was already reported in PCF T. brucei cells when complexes III and IV were downregulated (39, 40) and may reflect a protective mechanism also used by plants to cope with the increase in ROS generation associated with inhibition of the cytochrome pathway (39). Similarly, artificial expression of AOX in fruit fly and mouse cells was shown to limit mitochondrial ROS formation when respiration was inhibited by antimycin (inhibitor of complex III) or cyanide (inhibitor of complex IV) (31, 32).
Furthermore, we observed that the total cellular ATP levels decreased only by ∼25% at day 4 of RNAi induction, reflecting the ability of PCF cells to rely also on ATP production through both cytosolic and mitochondrial substrate-level phosphorylation via the pyruvate kinase and the succinyl-CoA synthetase, respectively, when grown in glucose-rich conditions (41, 42). Although the ATP levels were reduced just ∼25%, there was a 50% increase in the ADP/ATP ratio. This could be explained by the avid ATP consumption by the accumulated F1 subcomplexes, as reflected by the in-gel staining of ATPase activity. An increased ADP/ATP ratio could also reflect a higher intracellular ATP demand, namely by processes intended to restore homeostasis. Although the increased respiration through AOX contributed to the reduction of intramitochondrial O2⋅− levels, prolonged inhibition of the oxidative phosphorylation pathway due to Tb1 downregulation led to an increase of various cytosolic ROS, causing oxidative stress. Because the synthesis of certain ROS-detoxifying molecules requires ATP (43), we hypothesize that the inability of PCF Tb1 RNAi–induced cells to cope with the increasing concentrations of cytosolic ROS might be due to the lowered levels of intracellular ATP caused by inhibition of oxidative phosphorylation.
In BSF T. brucei, the FoF1–ATP synthase complex operates in reverse mode to generate the vital ΔΨm (15, 16). Therefore, the cell death that followed the collapse of the ΔΨm in parasites fully depleted of Tb1 or OSCP was expected and is consistent with the earlier studies. Nevertheless, while Tb1 and OSCP suppression down to 3 to 10% caused a corresponding loss of FoF1–ATPase complexes, these BSF trypanosomes were able to thrive in culture without any obvious effect on their viability. Similarly, perturbation of mitochondrial translation in BSF cells, manifested by decreased levels of FoF1–ATPase due to reduced production of its mitochondrial encoded subunit a, had no effect on growth in culture (44). Moreover, the ΔΨm of BSF Tb1 5’ and OSCP RNAi–induced cells was not affected when measured by flow cytometry of live cells, but there was a significant reduction in the ability of the FoF1–ATPase to generate a proton gradient when estimated in permeabilized cells by safranin O assay. This phenomenon can be explained by a combination of two factors: the higher sensitivity of safranin O to changes in ΔΨm compared with TMRE (45) and a possible overcapacity of the FoF1–ATPase complex in BSF trypanosomes. The overcapacity of certain enzymes has already been documented in T. brucei. For instance, the reduction of the maximum rate (Vmax) of AOX reaction by 50% was predicted to have no effect on oxygen consumption (46). Taken together, our observations would suggest that although the FoF1–ATPase is a promising drug target, the potential drug would have to inhibit the majority of the assembled complexes to exert cytotoxic effects, although the proportion of FoF1–ATPase complexes that is required to sustain the viability of the parasite in the host environment might be different compared with the situation in vitro.
Dyskinetoplastic trypanosomes are BSF cells that partially or entirely lack their mitochondrial DNA, termed kinetoplast DNA, and consequently, the mitochondrial-encoded subunit a (47, 48, 49). Because the vestigial F-ATPase in dyskinetoplastic cells is incapable of proton pumping, it is proposed that ΔΨm is maintained by the synergistic activities of the F1-ATPase and the electrogenic exchange of ADP for ATP mediated by the AAC (22). Unlike yeast “petite mutants” or mammalian ρ0 cells, dyskinetoplastic trypanosomes are successful in nature, where they are transmitted mechanically by bloodsucking insects among a vast range of mammals or by coitus in equids (50). It is striking that in African trypanosomes of the trypanozoon group, the kinetoplast DNA loss has occurred repeatedly and does not appear to impair cell viability of the replicative stage in the mammalian bloodstream (48). The loss of kinetoplast DNA in BSF trypanosomes is most likely facilitated by the facts that (i) these cells rely only on a single mitochondrial-encoded protein, the subunit a of the FoF1–ATP synthase, (ii) they do not use the FoF1–ATP synthase for energy production, as glycolysis fulfills their ATP demands, thanks to the abundant glucose in the mammalian host’s bloodstream, and (iii) these cells acquire a compensatory mutation that affects F1–ATPase function (22). We speculate that the long-term tolerance of reduced levels of FoF1–ATPase, exemplified by our BSF Tb1 5’ and OSCP RNAi cell lines, can provide BSF cells with a time frame to gain nuclear mutation(s) that allow ΔΨm generation in the absence of an intact FoF1–ATPase and consequently facilitate life without kinetoplast DNA (22). This has major implications, as a number of trypanocidal compounds, including those that constitute the currently available treatments for animal African trypanosomiasis, target the kinetoplast DNA network (51, 52, 53) and, indeed, the loss of dependence on kinetoplast DNA due to the compensatory mutations has been related to multidrug resistance in trypanosomes (54, 55, 56). Our results further highlight how the parasite’s unique biology helps this species to cope with the loss of mitochondrial DNA, an event that is deleterious, or even lethal, for the majority of eukaryotes (57).
A striking phenotype exhibited by BSF Tb1 3’ RNAi and OSCP cDKO trypanosomes is the reduced respiration rate upon Tb1 and OSCP depletion, respectively, as there is no direct link between the activities of AOX and the FoF1–ATPase. Rather, the oxygen consumption rate by AOX is a measure of glycolysis, which provides BSF trypanosomes with the majority, if not all, of the cellular ATP (34, 58). We hypothesize that disruption of the FoF1–ATPase complex leads to a lower consumption rate of intracellular ATP. In support of this argument, we observed a gradual reduction in the cellular ADP/ATP ratio after suppression of Tb1 or OSCP expression. A recent study showed that T. brucei AOX can be inhibited by a transient accumulation of intramitochondrial ATP caused by inhibition of the FoF1–ATPase hydrolytic activity with oligomycin (35), representing the first report proposing that the FoF1–ATPase activity can affect the glycolytic flux in BSF trypanosomes. Similarly, oligomycin treatment inhibits the rates of oxygen consumption and pyruvate production with a concomitant dissipation of ΔΨm when glucose is the substrate (15, 59). This effect that was originally contributed to the inhibition of glycolysis before triose phosphate oxidation (60) can now be linked to the inhibition of the FoF1–ATPase. According to the metabolic control theory (61), the flux control of a pathway can be exerted not solely by enzymes from the particular pathway but also by processes outside of it, such as the ATP-consuming processes. In T. brucei, the glycolytic flux is controlled mainly by enzymes outside of the pathway, specifically, by hexose transporters responsible for glucose uptake into the cell (62). The absence of regulation by components from the pathway is not restricted to this organism, as it was reported that overexpression of glycolytic enzymes does not exert significant control on the pathway in both yeast (63) and bacteria (64). Interestingly, a study in Escherichia coli demonstrated that incrementing the cytosolic ATP consumption by expression of free F1-ATPase resulted in a 70% increase in the glycolytic rate, concluding that enzymes consuming ATP can affect the glycolytic flux (65). Taken all together, our results further corroborate the idea that the levels of FoF1–ATPase activity can affect the respiration rate, playing an unexplored role in controlling the glycolytic rate in the BSF stage of the parasite.
Experimental procedures
Plasmid construction and generation of cell lines
The generation of the PCF Tb1 RNAi cell line was described in an earlier study (8). To generate the BSF Tb1 5’ and Tb1 3’ RNAi cell lines, Tb1 (Tb927.10.520) ORF fragments of 460 bp (between nucleotides 30 and 489) and 636 bp (between nucleotides 424 and 1060), respectively, were amplified from the genome of the wild type (WT) BSF T. brucei strain Lister 427 using primers 1 and 2 for the 5’ RNAi fragment, and primers 3 and 4 for the 3’ RNAi fragment. The resulting 5’ and 3’ RNAi amplicons were cloned into p2T7-177 vector (66) via BamHI and XhoI restrictions sites and transfected into the puromycin-resistant T. brucei SmOxB427 cell line (67) and the neomycin- and hygromycin-resistant T. b. brucei EATRO 1125 AnTat 1.1 90:13 cell line (68), respectively. For the inducible expression of Tb1 fused with a C-terminal 3x v5 tag, the Tb1 coding sequence was PCR-amplified from T. brucei strain Lister 427 genome using primers 5 and 6. Using the HindIII and BamHI restriction sites inherent in the primers, the fragment was cloned into the pT7_v5 vector (69). The construct was linearized with NotI and transfected into the neomycin-resistant transgenic BSF427 single-marker cell line as described previously (11).
To generate the BSF OSCP RNAi cell line, an OSCP (Tb927.10.8030) ORF fragment of 446 bp (between nucleotides 13 and 458) was amplified from the genome of the WT BSF T. brucei strain Lister 427 using primers 7 and 8. The resulting amplicon was cloned into p2T7-177 vector (66) via BamHI and XhoI restrictions sites, and the construct was linearized with NotI before transfection into the neomycin-resistant transgenic BSF427 single-marker cell line as described previously (11). For the generation of the BSF OSCP cDKO cell line, 5’ and 3’ intergenic region fragments from OSCP gene were amplified using primers 9 and 10 for the 5’ intergenic region, and primers 11 and 12 for the 3’ intergenic region. The resulting 5’ intergenic region amplicon of 452 bp was cloned into NotI and MluI restriction sites within pLEW13 vector to generate pLEW13::5’ IR452bp vector. Subsequently, the 3’ intergenic region amplicon of 414 bp was inserted into pLEW13::5’ IR452bp vector via XbaI and StuI restriction sites to obtain the OSCP single KO construct. This construct was linearized using NotI and transfected into the WT BSF T. brucei strain Lister 427 to knock out the first OSCP allele. To generate the construct for the KO of the second OSCP allele, the T7 RNA polymerase and neomycin cassette from the OSCP single KO construct was replaced by a cassette containing a 10% activity T7 promoter, the tetracycline repressor, and the hygromycin resistance gene from pLEW90 vector. For the generation of the tetracycline-inducible OSCP ectopic copy construct, the OSCP ORF was amplified from the genome of the WT BSF T. brucei strain Lister 427 using primers 13 and 14. The OSCP ORF was cloned into BamHI and XhoI restriction sites within pLEW79 vector, replacing the luciferase ORF. It is important to mention that the pLEW79 vector was mutagenized for the purpose of this study to include an XhoI restriction site downstream its HindIII site due to the existence of a HindIII site within the sequence of the OSCP ORF. BSF T. brucei OSCP single KO cells were transfected with the OSCP ectopic copy construct before knocking out the second OSCP allele. The strategy used for the conditional KO of OSCP in BSF cells was adapted from the reference (70). The correct integration of the OSCP single KO construct was verified by PCR using the following primer pairs: primers 15 (annealing upstream the 5’ intergenic region of the OSCP gene used as homologous recombination site) and 17 (binding the T7 RNA polymerase sequence); primers 18 (binding the neomycin resistance gene sequence) and 16 (annealing downstream the 3’ intergenic region of the OSCP gene used as homologous recombination site). The correct integration of the OSCP double KO construct was verified by PCR using the following primer pairs: primers 15 and 19 (which binds the tetracycline repressor sequence); primers 20 (binding the hygromycin resistance gene sequence) and 16. The presence of the OSCP inducible copy was verified by PCR using primers 21 (which anneals to the procyclic acidic repetitive protein promoter) and 22 (annealing downstream the aldolase 3’ UTR that follows the OSCP ORF). A schematic representation of the PCR verification is depicted on Fig. S2. The sequences of all the primers used in this study are found in Table S1.
T. brucei culture conditions
The PCF Tb1 RNAi cell line was grown at 27 °C in the glucose-rich medium SDM-79 (27) (Invitrogen 07490916N) supplemented with 10% fetal bovine serum (BioSera FB-1090/500) and 7.5 mg/ml hemin (Sigma H9039) and containing 15 μg/ml G418 (Sigma G8168), 25 μg/ml hygromycin B Gold (InvivoGen ant-hg-1), and 2.5 μg/ml phleomycin (InvivoGen ant-ph-2p).
The BSF T. b. brucei Lister 427 strain, EATRO 1125 AnTat 1.1 (68), stable acriflavine-induced dyskinetoplastic T. b. brucei EATRO 164 (71), and dyskinetoplastic T. b. evansi AnTat 3/3 that lost mitochondrial DNA upon culturing (16) were all grown at 37 °C and 5% CO2 in HMI-11 medium (Invitrogen 07490915N) supplemented with 10% fetal bovine serum. The BSF 5’ Tb1 RNAi cell line was grown in the presence of puromycin and phleomycin. The BSF 3’ Tb1 RNAi cell line was grown in the presence G418, hygromycin, puromycin, and phleomycin. The BSF OSCP RNAi cell line was grown in the presence of G418 and phleomycin. The BSF OSCP cDKO cell line was grown in G418, hygromycin, and phleomycin. The final concentrations of antibiotics in the culture medium were the following: 0.1 μg/ml puromycin, 2.5 μg/ml G418, 5 μg/ml hygromycin, and 2.5 μg/ml phleomycin.
The induction of RNAi and ectopically expressed tagged Tb1 was triggered by the addition of 1 μg/ml of tetracycline into the medium. BSF OSCP cDKO cells were constantly grown in the presence of 1 μg/ml of tetracycline to sustain OSCP expression. The expression of the OSCP ectopic copy was suppressed by cultivating BSF OSCP cDKO cells in the absence of tetracycline, preceded by a two-step washing of the cells with tetracycline-free medium. For all the experiments in this study, cells were maintained in a mid-late exponential growth phase, meaning 0.6 × 107 to 1.2 × 107 cells/ml in the case of PCF cells, and 0.6 × 106 to 1.2 × 106 cells/ml in the case of BSF cells.
SDS-PAGE and Western blotting
Appropriate volumes of PCF and BSF culture were spun down at 1400g for 10 min at 4 °C, and cell pellets were washed once with 1× PBS (10-mM phosphate buffer, 130-mM NaCl, pH 7.3). To prepare whole-cell lysates at a concentration of 1 × 107 cells in 30 μl, cell pellets were resuspended in 1× PBS before the addition of 3× Laemmli buffer (150-mM Tris-HCl, pH 6.8, 300-mM 1,4-dithiothreitol, 6% (w/v) SDS, 30% (w/v) glycerol, 0.02% (w/v) bromophenol blue). The mixture was boiled at 97 °C for 10 min and stored at −20 °C. For Western blot analysis, a volume of sample corresponding to 3 × 106 cells per well was separated by SDS-PAGE (Bio-Rad 4568094), blotted onto a polyvinylidene difluoride membrane (Pierce 88518), and probed with the appropriate monoclonal (mAb) or polyclonal (pAb) antibody. This was followed by incubation with a secondary HRP-conjugated anti-rabbit (Bio-Rad 1721019) or anti-mouse (Bio-Rad 1721011) antibody (1:2000), which immunoreacts, respectively, with the polyclonal or monoclonal primary antibodies. Proteins were visualized using the Clarity Western ECL substrate (Bio-Rad 1705060EM) on a ChemiDoc instrument (Bio-Rad). The PageRuler prestained protein ladder (Thermo Fisher Scientific 26617) was used to determine the size of the detected bands. The primary antibodies used in this study were the following: mAb anti-v5 epitope tag (1:2000, Invitrogen), mAb anti-mitochondrial HSP70 (1:5000; 72 kDa) (72), mAb anti-AOX (PCF 1:300, BSF 1:1000, kindly provided by Minu Chaudhuri; 33 kDa), pAb anti-mitochondrial RNA-binding protein 1 (1:1000; 23 kDa), pAb anti-enolase (1:1000; 47 kDa) (73), pAb anti-AAC (1:1000; 34 kDa), and antibodies against FoF1–ATP synthase subunits β (1:2000; 54 kDa), p18 (1:1000; 18 kDa), Tb1 (1:1000; 47 kDa), Tb2 (1:1000; 43 kDa), and OSCP (1:1000; 27 kDa). The latter antibodies were produced in the Zíková lab and are available upon request. The densitometric analysis of the bands was carried out using the ImageLab software by relating the signal intensity from the lanes corresponding to RNAi-induced/tetracycline-depleted cells to that of the lane pertinent to control cells. The percentage of downregulation relative to the control sample was then normalized to the corresponding signal intensity of the bands from the blots probed with anti-enolase or anti-mitochondrial HSP70 (loading controls).
Isolation of crude mitochondrial vesicles
Crude mitochondrial vesicles were obtained by hypotonic lysis as described earlier (11, 72). In summary, cell pellets from 3 × 108 BSF cells were washed once with a buffer I (150-mM NaCl, 20-mM glucose, 20-mM phosphate buffer, pH 7.9), resuspended in a buffer II (1-mM Tris-HCl, pH 8.0, 1-mM EDTA), and homogenized in a Dounce homogenizer. Alternatively, cell pellets from 1 × 109 PCF cells were washed in a buffer III (150-mM NaCl, 100-mM EDTA, 10-mM Tris-HCl, pH 8.0), resuspended in the buffer II, and homogenized by passing through a 25G needle. To restore the physiological isotonic conditions, 60% sucrose was promptly added to the cell lysate to attain a final concentration of 250 mM. Samples were spun down at 16,000g for 10 min at 4 °C to clear the soluble cytoplasmic material from the lysates. The organelle-enriched pellets were resuspended in STM (250-mM sucrose, 2-mM MgCl2, 20-mM Tris-HCl, pH 8.0) and supplemented with a final concentration of 3-mM MgCl2 and 0.3-mM CaCl2 before incubating with 5 μg/ml DNase I for 1 h on ice. Then, an equal volume of STE (250-mM sucrose, 2-mM EDTA, 20-mM Tris-HCl, pH 8.0) was added, and the material was centrifuged as before. Pellets enriched with the mitochondrial membrane vesicles were flash-frozen in liquid nitrogen and stored at −80 °C until their use.
Native electrophoresis and in-gel staining of FoF1–ATPase activity
The protocol for high-resolution clear native electrophoresis was adapted from published studies (74, 75). Briefly, crude mitochondrial vesicles from 5 × 108 cells were resuspended in a mitochondrial lysis buffer (2-mM ε-aminocaproic acid (ACA), 50-mM imidazole-HCl, 1-mM EDTA, 50-mM NaCl, pH 7.0) and lysed for 1 h on ice with 4 mg digitonin/1 mg protein. Samples were centrifuged at 16,000g for 30 min at 4 °C, and the protein concentrations of the cleared lysates were determined by bicinchoninic acid assay. Samples were mixed with 5× loading dye (0.1% (w/v) Ponceau-S, 50% (w/v) glycerol) and loaded onto a 3 to 12% native gradient gel. After electrophoresis (3 h, 100 V, 4 °C), the resolved mitochondrial proteins were transferred onto a nitrocellulose membrane (overnight, 20 V, 4 °C) and probed with selected antibodies (p18 1:1000 and Tb1 1:1000).
BNE was performed as described in an earlier study (9) with some modifications. Crude mitochondrial vesicles from 2 × 108 cells were resuspended in 1 M ACA and solubilized with either 2% (PCF) or 4% (BSF) dodecylmaltoside (DDM) for 1 h on ice. Samples were centrifuged at 16,000g for 30 min at 4 °C, and the protein concentrations of the cleared lysates were estimated using the bicinchoninic acid assay (Pierce 23225). Samples were mixed with 1.5 μl of the loading dye (500-mM ACA, 5% (w/v) Coomassie Brilliant Blue G-250) and loaded onto a 3 to 12% native gradient gel. After the electrophoresis (3 h, 140 V, 4 °C), proteins were blotted onto a polyvinylidene difluoride membrane (2 h, 100 V, 4 °C, stirring) and immunodetected using antibodies against different FoF1–ATP synthase subunits (subunit β 1:2000, p18 1:1000, Tb2 1:500, and OSCP 1:100). Alternatively, the gel was transferred into the ATPase reaction buffer (35-mM Tris-HCl, pH 8.0, 270-mM glycine, 19-mM MgSO4, 0.3% (w/v) Pb(NO3)2, 11-mM ATP) for overnight incubation under slow agitation (in-gel staining of FoF1–ATPase activity). Subsequently, the gel was soaked in 30% methanol to stop the reaction. The ATPase activity appears as a white precipitate.
Sodium carbonate submitochondrial fractionation
Sodium carbonate extraction of mitochondrial membranes was adapted from an earlier study (76). Mitochondrial vesicles from 3 × 108 cells were isolated by hypotonic lysis as described previously. The resulting supernatant from a 25G needle homogenization step was kept as a cytosolic fraction. The mitochondrial pellet was further treated with digitonin (80 μg/ml) for 15 min on ice to disrupt the mitochondrial outer membrane. The material was then cleared by centrifugation at 12,000g for 20 min at 4 °C and the pelleted mitoplasts were resuspended in 0.1 M Na2CO3 buffer (pH 11.5) before incubation on ice for 30 min. A final ultracentrifugation step at 100,000g for 1 h at 4 °C carried out in an SW50Ti rotor of a Beckman Instrument yielded a supernatant comprised of proteins from the mitochondrial matrix, including stripped peripheral membrane proteins, and a pellet containing integral proteins isolated from the mitochondrial membrane fraction.
Glycerol gradient sedimentation
Hypotonically purified mitochondrial vesicles from ∼2.5 × 109 cells were resuspended in glycerol gradient lysis buffer (10-mM Tris-HCl, pH 7.2, 10-mM MgCl2, 200-mM KCl, 1-mM 1,4-dithiothreitol) and solubilized with 1% Triton X-100 for 30 min on ice. The lysates were cleared by a centrifugation step (2× 16,000g, 30 min, 4 °C), and the protein concentration was determined by the Bradford assay. Cleared mitochondrial lysates were resolved by ultracentrifugation (Beckman Instrument, SW40 rotor) at 38,000g for 5 h on an 11- ml 10 to 30% glycerol gradient, which was poured using the Gradient Station (Biocomp) according to the manufacturer’s protocol. The glycerol gradients were then fractionated with the Gradient Station, and 500 μl fractions were stored at −80 °C.
ΔΨm measurement
The ΔΨm of live cells was estimated using the cell-permeant red-fluorescent dye TMRE (Thermo Fisher Scientific T669), whose fluorescence intensity is proportionally dependent on the ΔΨm values. Equal number of cells (3 × 106) were harvested for each time point and resuspended in the culture medium containing 60-nM TMRE. The staining of the cells was carried out for 30 min at the appropriate temperature for each life stage. Subsequently, cells were spun down at 1400g for 10 min at room temperature, resuspended in 1 ml of 1× PBS (see composition above) and immediately analyzed by flow cytometry using BD FACSCanto II instrument and its blue laser (488 nm) with the band pass PE filter (585/15). For each sample, 10,000 events were collected. Treatment with 20-μM carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (Sigma C2920) was used as a control for mitochondrial membrane depolarization. Data were evaluated using BD FACS Diva (BD Company) software. The TMRE signal corresponding to RNAi-induced/tetracycline-depleted cells was normalized to that of control cells and expressed in percentage. The values were plotted and analyzed statistically using GraphPad Prism 8.0 software.
In situ ΔΨm of permeabilized cells was determined fluorometrically using safranin O dye (Sigma S2255) (77). For each sample, 2 × 107 cells were harvested and washed once with ANT buffer (8-mM KCl, 110-mM K-gluconate, 10-mM NaCl, 10-mM free-acid Hepes, 10-mM K2HPO4, 0.015-mM EGTA potassium salt, 10-mM mannitol, 0.5 mg/ml fatty acid–free bovine serum albumin, 1.5-mM MgCl2, pH 7.25) (78). The cell pellet was resuspended in 2 ml of ANT buffer containing 5-μM safranin O and 4-μM digitonin. Fluorescence was recorded in a Hitachi F-7100 spectrofluorometer (Hitachi High-Technologies) at a 5-Hz acquisition rate, using 495 nm and 585 nm excitation and emission wavelengths, respectively. Substrates (1-mM ATP, PanReac AppliChem A13480025) and inhibitors (10 μg/ml oligomycin or 1-μM carboxyatractyloside, Sigma O4876 and Biorbyt orb259156-10, respectively) were added where indicated. Final addition of the uncoupler SF 6847 (250 nM; Enzo Life Sciences BML-EI215-0050) served as a control for maximal depolarization. All the experiments were performed at room temperature and constant stirring.
Mitochondrial O2⋅− and cellular ROS measurements
For the measurement of mitochondrial and cellular ROS molecules, the red mitochondrial O2⋅− indicator MitoSOX (Thermo Fisher Scientific M36008) and H2DCFHDA dye (Thermo Fisher Scientific) were used, respectively. The staining procedure followed was essentially the same as for the determination of ΔΨm in vivo except that TMRE was replaced by 5-μM MitoSOX or 10-μM H2DCFHDA in the corresponding assay. MitoSOX and H2DCFHDA fluorescence signals were recorded using BD FACSCanto II instrument and its blue laser (488 nm) with the band pass phycoerythrin (585/15 nm) and fluorescein isothiocyanate (530/30 nm) filters, respectively. The fluorescence signal of RNAi-induced/tetracycline-depleted cells was normalized to that of control cells and expressed in percentage. The values were plotted and analyzed statistically using GraphPad Prism 8.0 software.
High-resolution respirometry
The oxygen consumption rate was determined using the Oroboros Oxygraph-2K (Oroboros Instruments Corp). For each sample, 2 × 107 cells were harvested and washed once with Mir05 mitochondrial respiration medium (0.5-mM EGTA, 3-mM MgCl2, 60-mM lactobionic acid, 20-mM taurine, 10-mM KH2PO4, 20-mM Hepes, 110-mM sucrose, 1 mg/ml fatty acid–free bovine serum albumin, pH 7.1). The cell pellet was resuspended in 2.1 ml of Mir05 and transferred into the respiration chamber at the appropriate growth temperature for each life stage and under constant stirring. In the experiments carried out with intact PCF cells, 10-mM glycerol-3-phosphate (Sigma, 17766) was added, and complex IV- and AOX-mediated respirations were inhibited by injection of 1-mM KCN and 250-μM SHAM (Sigma S607), respectively. For the experiments performed with permeabilized BSF cells, the addition of 4-μM digitonin (Sigma D141) preceded the injection of 20-mM glycerol-3-phopshate into the chamber, and respiratory inhibition was achieved by addition of 250-μM SHAM. The most stable portion of either the oxygen consumption rate slope (PCF experiments) or the oxygen concentration in the chamber slope (BSF experiments) was determined for each biological replicate after the addition of substrates and inhibitors. The values were plotted and analyzed statistically using GraphPad Prism 8.0 software.
ADP/ATP ratio and total cellular ATP levels
Both the ADP/ATP ratio and the total cellular ATP were estimated using the bioluminescence-based ADP/ATP assay kit (Sigma MAK135) following the manufacturer's protocol. In brief, 1 × 106 cells per sample were harvested and washed once with PBS-G (1× PBS plus 6-mM glucose). Cells were resuspended in 10 μl of 1× PBS-G and transferred into a white flat-bottom 96-well microtiter plate. Luminescence was recorded in an Orion II microplate luminometer (Titertek-Berthold). The ATP levels and calculated ADP/ATP ratios of RNAi-induced/tetracycline-depleted cells were normalized to those of control cells and expressed in percentage. The values were plotted and analyzed statistically using GraphPad Prism 8.0 software.
Resazurin cell-viability assay
BSF cells were inoculated into a transparent flat-bottom 96-well microtiter plate at a density of 500 trypanosomes in a final volume of 200 μl of the culture medium per well. The cells were incubated in the presence of various drug concentrations (0.98- to 4000-μM carboxyatractyloside and 9.78–5000 ng/ml oligomycin) for 72 h at 37 °C. Wells without the drug served as the control for cell viability. Subsequently, 20 μl of a 125 μg/ml resazurin (Sigma R7017) stock solution was added and fluorescence was measured 24 h later (total drug incubation time of 96 h) in a Tecan Spark plate reader using 544-nm and 590-nm excitation and emission wavelengths, respectively. Data were analyzed with GraphPad Prism 8.0 software using a nonlinear regression and a sigmoidal dose–response analysis. All the experiments were performed in triplicate.
Modeling of Tb1 structure
The structure of Tb1 was predicted with I-TASSER (34) using the structure of E. gracilis Tb1 as a template (PDB ID: 6TDU (6)).
Data availability
All data discussed are presented in the article.
Conflict of interest
The authors declare that they have no conflicts of interest with the content of this article.
Acknowledgments
The authors thank Martina Slapničková for excellent technical support of the Zíková lab.
Author contributions
A. Z., B. P., C. H. -Y., K. S., O. G., A. S., and C. C., conceived and designed research, C. H.-Y. performed majority of experiments and data analysis, K. S. generated strains and conducted the initial analysis, C. D. generated strains and conducted growth assays, and A. Z., C. H.-Y., O. G., A. S., and C. C. wrote the manuscript.
Funding and additional information
This work was supported by the Czech Science Foundation (18-17529S) and European Regional Development Fund and Ministry of Education, Youth and Sport (CZ.02.1.01/0.0/0.0/16_019/0000759) to A. Z., a Senior Fellowship from the UK Medical Research Council, United Kingdom (MR/L019701/1) to A. S., and by grants from NKFIH [KH129567] and [K135027] to C. C.
Edited by John Denu
Footnotes
This article contains supporting information.
Present address for Caroline Dewar: Department of Chemistry and Biochemistry, University of Bern, Bern, Switzerland.
Supporting information
References
- 1.Walker J.E. The ATP synthase: The understood, the uncertain and the unknown. Biochem. Soc. Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 2.Kuhlbrandt W. Structure and mechanisms of F-type ATP synthases. Annu. Rev. Biochem. 2019;88:515–549. doi: 10.1146/annurev-biochem-013118-110903. [DOI] [PubMed] [Google Scholar]
- 3.Miranda-Astudillo H., Cano-Estrada A., Vazquez-Acevedo M., Colina-Tenorio L., Downie-Velasco A., Cardol P., Remacle C., Dominguez-Ramirez L., Gonzalez-Halphen D. Interactions of subunits Asa2, Asa4 and Asa7 in the peripheral stalk of the mitochondrial ATP synthase of the chlorophycean alga Polytomella sp. Biochim. Biophys. Acta. 2014;1837:1–13. doi: 10.1016/j.bbabio.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 4.van Lis R., Mendoza-Hernandez G., Groth G., Atteia A. New insights into the unique structure of the F0F1-ATP synthase from the chlamydomonad algae Polytomella sp. and Chlamydomonas reinhardtii. Plant Physiol. 2007;144:1190–1199. doi: 10.1104/pp.106.094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salunke R., Mourier T., Banerjee M., Pain A., Shanmugam D. Highly diverged novel subunit composition of apicomplexan F-type ATP synthase identified from Toxoplasma gondii. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2006128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muhleip A., McComas S.E., Amunts A. Structure of a mitochondrial ATP synthase with bound native cardiolipin. Elife. 2019;8 doi: 10.7554/eLife.51179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balabaskaran Nina P., Dudkina N.V., Kane L.A., van Eyk J.E., Boekema E.J., Mather M.W., Vaidya A.B. Highly divergent mitochondrial ATP synthase complexes in Tetrahymena thermophila. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zikova A., Schnaufer A., Dalley R.A., Panigrahi A.K., Stuart K.D. The F(0)F(1)-ATP synthase complex contains novel subunits and is essential for procyclic Trypanosoma brucei. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gahura O., Subrtova K., Vachova H., Panicucci B., Fearnley I.M., Harbour M.E., Walker J.E., Zikova A. The F1 -ATPase from Trypanosoma brucei is elaborated by three copies of an additional p18-subunit. FEBS J. 2018;285:614–628. doi: 10.1111/febs.14364. [DOI] [PubMed] [Google Scholar]
- 10.Montgomery M.G., Gahura O., Leslie A.G.W., Zikova A., Walker J.E. ATP synthase from Trypanosoma brucei has an elaborated canonical F1-domain and conventional catalytic sites. Proc. Natl. Acad. Sci. U. S. A. 2018;115:2102–2107. doi: 10.1073/pnas.1720940115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subrtova K., Panicucci B., Zikova A. ATPaseTb2, a unique membrane-bound FoF1-ATPase component, is essential in bloodstream and dyskinetoplastic Trypanosomes. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bringaud F., Riviere L., Coustou V. Energy metabolism of trypanosomatids: Adaptation to available carbon sources. Mol. Biochem. Parasitol. 2006;149:1–9. doi: 10.1016/j.molbiopara.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Hellemond J.J., Bakker B.M., Tielens A.G. Energy metabolism and its compartmentation in Trypanosoma brucei. Adv. Microb. Physiol. 2005;50:199–226. doi: 10.1016/S0065-2911(05)50005-5. [DOI] [PubMed] [Google Scholar]
- 14.Smith T.K., Bringaud F., Nolan D.P., Figueiredo L.M. Metabolic reprogramming during the Trypanosoma brucei life cycle. F1000Res. 2017;6 doi: 10.12688/f1000research.10342.2. F1000 Faculty Rev-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nolan D.P., Voorheis H.P. The mitochondrion in bloodstream forms of Trypanosoma brucei is energized by the electrogenic pumping of protons catalysed by the F1F0-ATPase. Eur. J. Biochem. 1992;209:207–216. doi: 10.1111/j.1432-1033.1992.tb17278.x. [DOI] [PubMed] [Google Scholar]
- 16.Schnaufer A., Clark-Walker G.D., Steinberg A.G., Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24:4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Futai M., Kanazawa H. Structure and function of proton-translocating adenosine triphosphatase (F0F1): Biochemical and molecular biological approaches. Microbiol. Rev. 1983;47:285–312. doi: 10.1128/mr.47.3.285-312.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chinopoulos C., Adam-Vizi V. Mitochondria as ATP consumers in cellular pathology. Biochim. Biophys. Acta. 2010;1802:221–227. doi: 10.1016/j.bbadis.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Chinopoulos C. Mitochondrial consumption of cytosolic ATP: Not so fast. FEBS Lett. 2011;585:1255–1259. doi: 10.1016/j.febslet.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Chen X.J., Clark-Walker G.D. The petite mutation in yeasts: 50 years on. Int. Rev. Cytol. 2000;194:197–238. doi: 10.1016/s0074-7696(08)62397-9. [DOI] [PubMed] [Google Scholar]
- 21.Appleby R.D., Porteous W.K., Hughes G., James A.M., Shannon D., Wei Y.H., Murphy M.P. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur. J. Biochem. 1999;262:108–116. doi: 10.1046/j.1432-1327.1999.00350.x. [DOI] [PubMed] [Google Scholar]
- 22.Dean S., Gould M.K., Dewar C.E., Schnaufer A.C. Single point mutations in ATP synthase compensate for mitochondrial genome loss in trypanosomes. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14741–14746. doi: 10.1073/pnas.1305404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campanella M., Casswell E., Chong S., Farah Z., Wieckowski M.R., Abramov A.Y., Tinker A., Duchen M.R. Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1. Cell Metab. 2008;8:13–25. doi: 10.1016/j.cmet.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Panicucci B., Gahura O., Zikova A. Trypanosoma brucei TbIF1 inhibits the essential F1-ATPase in the infectious form of the parasite. PLoS Negl. Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gahura O., Panicucci B., Vachova H., Walker J.E., Zikova A. Inhibition of F1 -ATPase from Trypanosoma brucei by its regulatory protein inhibitor TbIF1. FEBS J. 2018;285:4413–4423. doi: 10.1111/febs.14672. [DOI] [PubMed] [Google Scholar]
- 26.Vertommen D., Van Roy J., Szikora J.P., Rider M.H., Michels P.A., Opperdoes F.R. Differential expression of glycosomal and mitochondrial proteins in the two major life-cycle stages of Trypanosoma brucei. Mol. Biochem. Parasitol. 2008;158:189–201. doi: 10.1016/j.molbiopara.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Frazier A.E., Taylor R.D., Mick D.U., Warscheid B., Stoepel N., Meyer H.E., Ryan M.T., Guiard B., Rehling P. Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J. Cell Biol. 2006;172:553–564. doi: 10.1083/jcb.200505060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaudhuri M., Ott R.D., Hill G.C. Trypanosome alternative oxidase: From molecule to function. Trends Parasitol. 2006;22:484–491. doi: 10.1016/j.pt.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Brand M.D. The sites and topology of mitochondrial superoxide production. Exp. Gerontol. 2010;45:466–472. doi: 10.1016/j.exger.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maxwell D.P., Wang Y., McIntosh L. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. U. S. A. 1999;96:8271–8276. doi: 10.1073/pnas.96.14.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Ayala D.J., Sanz A., Vartiainen S., Kemppainen K.K., Babusiak M., Mustalahti E., Costa R., Tuomela T., Zeviani M., Chung J., O'Dell K.M., Rustin P., Jacobs H.T. Expression of the Ciona intestinalis alternative oxidase (AOX) in Drosophila complements defects in mitochondrial oxidative phosphorylation. Cell Metab. 2009;9:449–460. doi: 10.1016/j.cmet.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 32.El-Khoury R., Dufour E., Rak M., Ramanantsoa N., Grandchamp N., Csaba Z., Duvillie B., Benit P., Gallego J., Gressens P., Sarkis C., Jacobs H.T., Rustin P. Alternative oxidase expression in the mouse enables bypassing cytochrome c oxidase blockade and limits mitochondrial ROS overproduction. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chinopoulos C. The “B space” of mitochondrial phosphorylation. J. Neurosci. Res. 2011;89:1897–1904. doi: 10.1002/jnr.22659. [DOI] [PubMed] [Google Scholar]
- 34.Bakker B.M., Michels P.A., Opperdoes F.R., Westerhoff H.V. What controls glycolysis in bloodstream form Trypanosoma brucei? J. Biol. Chem. 1999;274:14551–14559. doi: 10.1074/jbc.274.21.14551. [DOI] [PubMed] [Google Scholar]
- 35.Alberto Luevano-Martinez L., Girard R., Alencar M.B., Silber A.M. ATP regulates the activity of an alternative oxidase in Trypanosoma brucei. FEBS Lett. 2020 doi: 10.1002/1873-3468.13790. [DOI] [PubMed] [Google Scholar]
- 36.Nowikovsky K., Reipert S., Devenish R.J., Schweyen R.J. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 2007;14:1647–1656. doi: 10.1038/sj.cdd.4402167. [DOI] [PubMed] [Google Scholar]
- 37.Bauerschmitt H., Mick D.U., Deckers M., Vollmer C., Funes S., Kehrein K., Ott M., Rehling P., Herrmann J.M. Ribosome-binding proteins Mdm38 and Mba1 display overlapping functions for regulation of mitochondrial translation. Mol. Biol. Cell. 2010;21:1937–1944. doi: 10.1091/mbc.E10-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez E., Lapaille M., Degand H., Cilibrasi L., Villavicencio-Queijeiro A., Morsomme P., Gonzalez-Halphen D., Field M.C., Remacle C., Baurain D., Cardol P. The mitochondrial respiratory chain of the secondary green alga Euglena gracilis shares many additional subunits with parasitic Trypanosomatidae. Mitochondrion. 2014;19 Pt B:338–349. doi: 10.1016/j.mito.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Horvath A., Horakova E., Dunajcikova P., Verner Z., Pravdova E., Slapetova I., Cuninkova L., Lukes J. Downregulation of the nuclear-encoded subunits of the complexes III and IV disrupts their respective complexes but not complex I in procyclic Trypanosoma brucei. Mol. Microbiol. 2005;58:116–130. doi: 10.1111/j.1365-2958.2005.04813.x. [DOI] [PubMed] [Google Scholar]
- 40.Gnipova A., Panicucci B., Paris Z., Verner Z., Horvath A., Lukes J., Zikova A. Disparate phenotypic effects from the knockdown of various Trypanosoma brucei cytochrome c oxidase subunits. Mol. Biochem. Parasitol. 2012;184:90–98. doi: 10.1016/j.molbiopara.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Coustou V., Besteiro S., Biran M., Diolez P., Bouchaud V., Voisin P., Michels P.A., Canioni P., Baltz T., Bringaud F. ATP generation in the Trypanosoma brucei procyclic form: Cytosolic substrate level is essential, but not oxidative phosphorylation. J. Biol. Chem. 2003;278:49625–49635. doi: 10.1074/jbc.M307872200. [DOI] [PubMed] [Google Scholar]
- 42.Lamour N., Riviere L., Coustou V., Coombs G.H., Barrett M.P., Bringaud F. Proline metabolism in procyclic Trypanosoma brucei is down-regulated in the presence of glucose. J. Biol. Chem. 2005;280:11902–11910. doi: 10.1074/jbc.M414274200. [DOI] [PubMed] [Google Scholar]
- 43.Krauth-Siegel R.L., Comini M.A. Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochim. Biophys. Acta. 2008;1780:1236–1248. doi: 10.1016/j.bbagen.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Prochazkova M., Panicucci B., Zikova A. Cultured bloodstream Trypanosoma brucei adapt to life without mitochondrial translation release factor 1. Sci. Rep. 2018;8:5135. doi: 10.1038/s41598-018-23472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chowdhury S.R., Djordjevic J., Albensi B.C., Fernyhough P. Simultaneous evaluation of substrate-dependent oxygen consumption rates and mitochondrial membrane potential by TMRM and safranin in cortical mitochondria. Biosci. Rep. 2015;36 doi: 10.1042/BSR20150244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helfert S., Estevez A.M., Bakker B., Michels P., Clayton C. Roles of triosephosphate isomerase and aerobic metabolism in Trypanosoma brucei. Biochem. J. 2001;357:117–125. doi: 10.1042/0264-6021:3570117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai D.H., Hashimi H., Lun Z.R., Ayala F.J., Lukes J. Adaptations of Trypanosoma brucei to gradual loss of kinetoplast DNA: Trypanosoma equiperdum and Trypanosoma evansi are petite mutants of T. brucei. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1999–2004. doi: 10.1073/pnas.0711799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schnaufer A. Evolution of dyskinetoplastic trypanosomes: How, and how often? Trends Parasitol. 2010;26:557–558. doi: 10.1016/j.pt.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carnes J., Anupama A., Balmer O., Jackson A., Lewis M., Brown R., Cestari I., Desquesnes M., Gendrin C., Hertz-Fowler C., Imamura H., Ivens A., Koreny L., Lai D.H., MacLeod A. Genome and phylogenetic analyses of Trypanosoma evansi reveal extensive similarity to T. brucei and multiple independent origins for dyskinetoplasty. PLoS Negl. Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brun R., Hecker H., Lun Z.R. Trypanosoma evansi and T. equiperdum: Distribution, biology, treatment and phylogenetic relationship (a review) Vet. Parasitol. 1998;79:95–107. doi: 10.1016/s0304-4017(98)00146-0. [DOI] [PubMed] [Google Scholar]
- 51.Motta M.C. Kinetoplast as a potential chemotherapeutic target of trypanosomatids. Curr. Pharm. Des. 2008;14:847–854. doi: 10.2174/138161208784041051. [DOI] [PubMed] [Google Scholar]
- 52.Shapiro T.A., Englund P.T. Selective cleavage of kinetoplast DNA minicircles promoted by antitrypanosomal drugs. Proc. Natl. Acad. Sci. U. S. A. 1990;87:950–954. doi: 10.1073/pnas.87.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giordani F., Morrison L.J., Rowan T.G., De Koning H.P., Barrett M.P. The animal trypanosomiases and their chemotherapy: A review. Parasitology. 2016;143:1862–1889. doi: 10.1017/S0031182016001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eze A.A., Gould M.K., Munday J.C., Tagoe D.N., Stelmanis V., Schnaufer A., De Koning H.P. Reduced mitochondrial membrane potential is a late adaptation of trypanosoma brucei brucei to Isometamidium preceded by mutations in the gamma subunit of the F1Fo-ATPase. PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gould M.K., Schnaufer A. Independence from kinetoplast DNA maintenance and expression is associated with multidrug resistance in Trypanosoma brucei in vitro. Antimicrob. Agents Chemother. 2014;58:2925–2928. doi: 10.1128/AAC.00122-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agbe A., Yielding K.L. Kinetoplasts play an important role in the drug responses of Trypanosoma brucei. J. Parasitol. 1995;81:968–973. [PubMed] [Google Scholar]
- 57.Young M.J., Copeland W.C. Human mitochondrial DNA replication machinery and disease. Curr. Opin. Genet. Dev. 2016;38:52–62. doi: 10.1016/j.gde.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghozlane A., Bringaud F., Soueidan H., Dutour I., Jourdan F., Thebault P. Flux analysis of the Trypanosoma brucei glycolysis based on a multiobjective-criteria bioinformatic approach. Adv. Bioinformatics. 2012;2012:159423. doi: 10.1155/2012/159423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiaira J.K., Njogu M.R. Oligomycin-sensitivity of hexose-sugar catabolism in the bloodstream form of Trypanosoma brucei brucei. Biotechnol. Appl. Biochem. 1994;20:347–356. doi: 10.1111/j.1470-8744.1994.tb00322.x. [DOI] [PubMed] [Google Scholar]
- 60.Miller P.G., Klein R.A. Effects of oligomycin on glucose utilization and calcium transport in African trypanosomes. J. Gen. Microbiol. 1980;116:391–396. doi: 10.1099/00221287-116-2-391. [DOI] [PubMed] [Google Scholar]
- 61.Heinrich R., Rapoport T.A. A linear steady-state treatment of enzymatic chains. General properties, control and effector strength. Eur. J. Biochem. 1974;42:89–95. doi: 10.1111/j.1432-1033.1974.tb03318.x. [DOI] [PubMed] [Google Scholar]
- 62.Bakker B.M., Walsh M.C., ter Kuile B.H., Mensonides F.I., Michels P.A., Opperdoes F.R., Westerhoff H.V. Contribution of glucose transport to the control of the glycolytic flux in Trypanosoma brucei. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10098–10103. doi: 10.1073/pnas.96.18.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schaaff I., Heinisch J., Zimmermann F.K. Overproduction of glycolytic enzymes in yeast. Yeast. 1989;5:285–290. doi: 10.1002/yea.320050408. [DOI] [PubMed] [Google Scholar]
- 64.Ruyter G.J., Postma P.W., van Dam K. Control of glucose metabolism by enzyme IIGlc of the phosphoenolpyruvate-dependent phosphotransferase system in Escherichia coli. J. Bacteriol. 1991;173:6184–6191. doi: 10.1128/jb.173.19.6184-6191.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koebmann B.J., Westerhoff H.V., Snoep J.L., Nilsson D., Jensen P.R. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J. Bacteriol. 2002;184:3909–3916. doi: 10.1128/JB.184.14.3909-3916.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wickstead B., Ersfeld K., Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 2002;125:211–216. doi: 10.1016/s0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- 67.Poon S.K., Peacock L., Gibson W., Gull K., Kelly S. A modular and optimized single marker system for generating Trypanosoma brucei cell lines expressing T7 RNA polymerase and the tetracycline repressor. Open Biol. 2012;2:110037. doi: 10.1098/rsob.110037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Engstler M., Boshart M. Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes Dev. 2004;18:2798–2811. doi: 10.1101/gad.323404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Flaspohler J.A., Jensen B.C., Saveria T., Kifer C.T., Parsons M. A novel protein kinase localized to lipid droplets is required for droplet biogenesis in trypanosomes. Eukaryot. Cell. 2010;9:1702–1710. doi: 10.1128/EC.00106-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wirtz E., Leal S., Ochatt C., Cross G.A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 71.Stuart K.D. Evidence for the retention of kinetoplast DNA in an acriflavine-induced dyskinetoplastic strain of Trypanosoma brucei which replicates the altered central element of the kinetoplast. J. Cell Biol. 1971;49:189–195. doi: 10.1083/jcb.49.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Panigrahi A.K., Zikova A., Dalley R.A., Acestor N., Ogata Y., Anupama A., Myler P.J., Stuart K.D. Mitochondrial complexes in Trypanosoma brucei: A novel complex and a unique oxidoreductase complex. Mol. Cell. Proteomics. 2008;7:534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- 73.Hannaert V., Albert M.A., Rigden D.J., da Silva Giotto M.T., Thiemann O., Garratt R.C., Van Roy J., Opperdoes F.R., Michels P.A. Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase. Eur. J. Biochem. 2003;270:3205–3213. doi: 10.1046/j.1432-1033.2003.03692.x. [DOI] [PubMed] [Google Scholar]
- 74.Wittig I., Karas M., Schagger H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell. Proteomics. 2007;6:1215–1225. doi: 10.1074/mcp.M700076-MCP200. [DOI] [PubMed] [Google Scholar]
- 75.Acestor N., Zikova A., Dalley R.A., Anupama A., Panigrahi A.K., Stuart K.D. Trypanosoma brucei mitochondrial respiratome: Composition and organization in procyclic form. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.006908. M110 006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Acestor N., Panigrahi A.K., Ogata Y., Anupama A., Stuart K.D. Protein composition of Trypanosoma brucei mitochondrial membranes. Proteomics. 2009;9:5497–5508. doi: 10.1002/pmic.200900354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Akerman K.E., Wikstrom M.K. Safranine as a probe of the mitochondrial membrane potential. FEBS Lett. 1976;68:191–197. doi: 10.1016/0014-5793(76)80434-6. [DOI] [PubMed] [Google Scholar]
- 78.Chinopoulos C., Vajda S., Csanady L., Mandi M., Mathe K., Adam-Vizi V. A novel kinetic assay of mitochondrial ATP-ADP exchange rate mediated by the ANT. Biophys. J. 2009;96:2490–2504. doi: 10.1016/j.bpj.2008.12.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed are presented in the article.