Abstract
Aspergillus flavus (A. flavus), a pathogenic fungus, can produce carcinogenic and toxic aflatoxins that are a serious agricultural and medical threat worldwide. Attempts to decipher the aflatoxin biosynthetic pathway have been hampered by the lack of a high-quality genome annotation for A. flavus. To address this gap, we performed a comprehensive proteogenomic analysis using high-accuracy mass spectrometry data for this pathogen. The resulting high-quality data set confirmed the translation of 8724 previously predicted genes and identified 732 novel proteins, 269 splice variants, 447 single amino acid variants, 188 revised genes. A subset of novel proteins was experimentally validated by RT-PCR and synthetic peptides. Further functional annotation suggested that a number of the identified novel proteins may play roles in aflatoxin biosynthesis and stress responses in A. flavus. This comprehensive strategy also identified a wide range of posttranslational modifications (PTMs), including 3461 modification sites from 1765 proteins. Functional analysis suggested the involvement of these modified proteins in the regulation of cellular metabolic and aflatoxin biosynthetic pathways. Together, we provided a high-quality annotation of A. flavus genome and revealed novel insights into the mechanisms of aflatoxin production and pathogenicity in this pathogen.
Keywords: Aspergillus flavus, proteogenomics, genome annotation, aflatoxin production, pathogenicity
Abbreviations: AS, alternatively spliced; BCA, bicinchoninic acid; CA, czapek-dox agar; ESTs, expressed sequenced tags; GFF, general feature format; GSSPs, genome search-specific peptides; HCD, higher-energy collision dissociation; IDA, information dependent acquisition; IGV, Integrative Genomics Viewer; MM, minimal medium; PDA, potato dextrose agar; PMSF, phenylmethanesulfonyl fluoride; PTMs, posttranslational modifications; MS, mass spectrometry; NCE, normalized collision energy; ORF, open reading frame; SAAVs, single amino acid variants; YES, yeast extract-sucrose agar
Graphical Abstract

Highlights
-
•
Proteome data set confirms the translation of 8724 previously-predicted genes.
-
•
Proteogenomic strategy discovers novel, spliced, mutated and revised genes.
-
•
Proteome of A. flavus identifies a wide range of post-translational modifications.
In Brief
A. flavus is a pathogenic fungus capable of producing aflatoxin, which is harmful and carcinogenic to animals and human. In this study, we presented the first comprehensive draft map of the A. flavus proteome and a wide range of posttranslational modifications. This is the first study we are aware of to have employed such a proteomic approach to improve Aspergillus genomic annotation, which will serve as a valuable resource for future efforts to explore the synthesis and excretion of aflatoxins.
A. flavus (A. flavus) is one of the most important species in the Aspergillus genus that can cause both noninvasive and invasive systematic aspergillosis in immunocompromised individuals, animals, and economically important crops (1, 2). It can produce a range of potent carcinogens and toxins collectively referred to as aflatoxins, which represent a significant concern due to the potential for environmental contamination with these compounds (3). When foods contaminated with aflatoxin are ingested, or when A. flavus is able to grow without immunological constraint, this can lead to acute toxicity and a long-term increase in cancer risk in affected individuals (4). In recent years, the study of aflatoxin biosynthesis received a great deal of attention in attempts to reduce economic and human health impacts caused by A. flavus (5, 6). Despite advancements in the field, our understanding of the molecular mechanisms of aflatoxin production in A. flavus is still fragmentary.
The A. flavus NRRL3357 is one of the most widely utilized strains for studying the aflatoxin production and pathogenicity of A. flavus (7). Its genome was first sequenced in 2006 and was updated substantially in 2009, containing 19,618 expressed sequenced tags (ESTs) and 13, 485 putative protein-coding genes (8, 9, 10). A resequencing of the A. flavus genome in 2015 estimated the total genomic size to be ∼36.8 Mb, consisting of eight chromosomes and a number of additional unassembled sequences (10). Even in this most recent gene functional annotation effort (11, 12), however, roughly 5000 proteins remain to be characterized as hypothetical or uncharacterized in the NCBI (https://www.ncbi.nlm.nih.gov/genome/proteins/360?genome_assembly_id=28730&gi=-1) and Uniprot (https://www.uniprot.org/uniprot/?query=aspergillus+flavus+NRRL3357&sort=score) databases. It is important that the A. flavus genome be fully and accurately annotated in order to ensure a complete understanding of the molecular mechanisms governing aflatoxin production and associated pathogenicity. As such, there is a pressing need for a high-quality annotation of protein coding genes for this fungal pathogen.
Proteogenomics is referred as the use of mass spectrometry (MS)-derived proteomic data to annotate the protein coding genes and improve genome annotation quality (13, 14). In contrast to conventional MS data analysis, proteogenomic approach does not rely on an existing reference protein database, but uses the database translated from six-frame translation of the whole genome and three-frame translation of RNA-seq sequences. Thus, such complementary annotation analyses can not only provide valuable insights that offer direct experimental support for predicted protein-coding genes, thereby improving genomic annotation (15), but also have the potential to provide supporting evidence for the novel transcripts, alternative predictions, or single amino acid variants (SAAVs) at protein level (16, 17, 18, 19). Over the last few years, with the rapid advances in MS technology and computational tools, proteogenomic research has become a dramatically growing field (18, 19, 20). It has been successfully applied to improve the quality of genome annotation in many model organisms, including Homo sapiens (21, 22), Arabidopsis (23, 24, 25), Zea mays (26), Oryza sativa (27, 28), diatom (29), cyanobacteria (30) and bacteria (31, 32, 33, 34). These studies have provided invaluable information for genome annotation and the physiology of these model organisms. However, a high-quality annotation of the A. flavus genome is still not available, presenting a major obstacle for the understanding of the entire networks governing pathogenicity in this major fungal pathogen.
To address this gap, we performed high-quality comprehensive annotation of the A. flavus genome using an integrated proteogenomic approach. Through this approach, we were able to validate many predicted genes and identified novel protein-coding genes not currently represented in extant genomic annotations. We further provided a holistic view of posttranslational modifications (PTMs) events in A. flavus. Functional analysis suggested the involvement of these PTMs in the cellular metabolic and aflatoxin biosynthetic pathways. Our current results represent a significant advance in the accurate annotation of the A. flavus genome and provide new insights into the mechanisms of pathogenicity and aflatoxin biosynthesis in A. flavus.
Experimental Procedures
Cell Culture
A. flavus was cultured on liquid yeast extract-sucrose agar (YES) and potato dextrose agar (PDA) and the conidia were then grown under several conditions. For the low temperature and pH stresses, 106 conidia were resuspended in fresh YES media at 29 °C or YES media (pH 5, with hydrochloride) at 37 °C. For the oxidative and hyperosmotic stresses, 106 conidia were resuspended in fresh YES media containing additional H2O2 (0.8 M) and sodium chloride (1 M), respectively. For the cell wall stress and carbon substitution, A. flavus was inoculated into the fresh YES media supplied with additional congo red (200 μg/ml) and cultured in minimal medium (MM) (10 g/L glucose) and czapek-dox agar (CA) (6 g/L Ammonium tartrate) at 37 °C.
Protein Extraction, Proteolytic Digestion, and Offline High-pH Reversed-phase C18 Column Prefractionation
The cultures from different treatments were then collected by filtration and washed three times with PBS buffer, respectively. The mycelia were ground into powders and reconstituted in buffer [20 mM Tris-Cl (pH 7.5), 150 mM NaCl, 50 mM nicotinamide, pH 7.5, 5 mM β-glycero-phosphate, 10 mM Na4P2O7, 10 mM NaF, 1 mM Na3VO4, 1% Triton X-100, 1× phenylmethanesulfonyl fluoride (PMSF)]. The cultures were shaken at 30 °C for 1 h and the debris was removed by centrifugation at 5000g at 4 °C for 1 h. Finally, the protein concentration was measured by bicinchoninic acid (BCA) method (Tiangen).
The constant protein amounts from different stress conditions were combined and precipitated by using 80% ice-cold acetone. The whole lysates were then washed three times with ice-cold acetone to remove the pigment and other small molecules. The protein lysates were finally redissolved in 50 mM ammonium bicarbonate and in-solution/in-gel digested by trypsin (Promega) according to previously described (29, 30).
To reduce the complexity of tryptic digests, the in-solution proteolytic digested peptides were prefractionated on a self-packed SPE column as previously described (29, 30). Briefly, the peptides were loaded onto the column and then desalted with 20 mM ammonium formate. The columns were eluted with a series of elution buffers containing of 20 mM ammonium formate and different concentrations of acetonitrile (10, 13, 15, 18, 21, 25, 28, 35, 60, 80, and 100%, vol/vol). Collected fractions were completely dried with a vacuum centrifuge and stored at −80 °C for further use.
Nano-LC-MS/MS Analysis
The peptides were dissolved in 0.1% formic acid and separated on an online nano-flow EASY-nLC 1200 system with a 75 μm × 15 cm analytical column (C18, 3 μm, Thermo Fisher Scientific) and analyzed on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific). Peptides were eluted using a linear solvent gradient of 9–32% solvent B (0.1% formic acid/80% acetonitrile, v/v) over 100 min at 450 nl/min flow rate. The mass spectrometer was operated in data-dependent acquisition (DDA) mode with full scans (m/z range of 300–1800) at 120,000 mass resolution using an automatic gain control (AGC) target value of 3e6. The top 20 most intense precursor ions were selected for following MS/MS fragmentation by higher-energy collision dissociation (HCD) with normalized collision energy (NCE) of 27% and analyzed with 30,000 resolution in the Orbitrap. The dynamic exclusion was set to 20 s and the isolation width of precursor ion was set to 1.6 m/z. The maximum injection times were 20 ms and 50 ms for both MS and MS/MS, respectively. The intensity threshold was set to 10,000.
The digested peptides were also separated on an UltimateTM 3000 nano-LC System (Dionex) with a 75 μm × 15 cm analytical column (C18, 3 μm, Thermo Fisher Scientific) and analyzed on an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). Peptides were eluted at 300 nl/min flow rate with 70 min linear solvent gradient of 4–25% solvent B (0.1% formic acid/80% acetonitrile, v/v). A full MS scan from m/z 300 to 1800 was acquired at 60,000 resolution with a minimum signal intensity of 500. The top 15 most intense precursor ions were selected for following MS/MS fragmentation by HCD with NCE of 35% and analyzed with a resolution of 15,000 in the Orbitrap. The dynamic exclusion was set to 90 s and the isolation width of precursor ion was set to 2.0 m/z. The maximum injection times for both full MS and MS/MS were 120 ms and 120 ms, respectively.
To enhance coverage of the peptides and proteins, three in-gel digested peptides were separated on an Eksigent nanoLC 400 system with a 75 μm × 15 cm analytical column (ChromXP C18CL, 3 μm, 120 Å) and analyzed on a Triple TOF 6600 Mass Spectrometer (AB SCIEX). Peptides were eluted with 100 min linear solvent gradient of 5 to 45% solvent B (0.1% formic acid/80% acetonitrile, v/v) at 600 nl/min flow rate. The ion spray voltage was 2300 V, declustering potential 100 V, nebulizer gas 5 psi, curtain gas 35 psi, and interface heater temperature 150 °C. A full MS scan from 350 to 1500 m/z was acquired in an information dependent acquisition (IDA) mode. The top 40 most intense precursor ions were selected for following MS/MS fragmentation with a 2+ to 5+ charge state. The maximum injection times for both full MS and MS/MS were 250 ms and 50 ms, respectively. Dynamic exclusion was set for a period of 15 s and a tolerance of 50 ppm. Dynamic collision energy was used.
Retrieval of Data Sources
The A. flavus protein sequences, genome sequences, and general feature format (GFF) file were downloaded from NCBI database. The A. flavus protein sequence database contained 13,485 protein sequences (http://www.ncbi.nlm.nih.gov/; released 2013). The transcripts were retrieved from NCBI (SRX237459, SRX237295, SRX1330586, SRX396791) and the expressed sequence tags (ESTs) were downloaded from previously reported (35, 36). Furthermore, two large available proteomics data sets containing 46 raw files were also downloaded for further analysis (37, 38). These published MS sources (46 MS runs) can be downloaded the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD001296 and PXD000982.
Peptide Identification and Proteogenomic Analysis
All MS/MS data were processed with the GAPE tool (29). Briefly, the retrieved RNA-seq reads were firstly assembled into long transcripts by using Trinity (V2.8.4) with default parameters (39). Then, a customizable proteogenomic database was created with a six-frame translation of the complete genome and a three-frame translation of the assembled long transcripts. The sequence length of more than 20 amino acids was selected for further analysis. The existing reference protein sequences and the sequences of commonly observed laboratory contaminants were also added to the query databases (1,934,902 protein sequences).
After eliminating the potential redundant protein sequences, all the MS/MS spectra were searched against the minimally redundant protein databases using the search engines incorporated in GAPE, including Comet (40), MSGF+ (41) and X!Tandem (42). In order to achieve deep coverage, we also performed the pFind (version 3.1) (43, 44) and MASCOT (version 2.3) (45) searches as separate steps and added the results into the GAPE pipeline, with regard to the performance characteristics of the two proprietary and commercial search engines. The search parameters were as follows: trypsin digestion with maximum of two missed cleavages allowed; fixed modification was carbamidomethylation of cysteine, whereas acetylation (protein N-terminal), deamidation (Asn/Gln), and oxidation (Met) were set as dynamic modifications. The mass errors for precursor ions and fragment ions were set to 10 ppm and 0.05 Da, respectively. The filtering strategies in GAPE tool, including the target-decoy strategy for all identified peptides (46) and a more stringent filtering strategy (separated FDR) for the novel peptides (47, 48), were finally used to evaluate the identification error rates. Also, the identified spectra that were assigned different sequences in different searches were eliminated to filter the false discovery. All identified peptides were mapped to the existing reference protein database using BLASTP (49) and the remaining orphan peptides were designated as genome search-specific peptides (GSSPs).
All identified GSSPs were mapped to the A. flavus genome and the open reading frame (ORF) was predicted using GAPE tool (29). Novel protein-coding genes can be identified as the ORFs mapping to intergenic regions, while the ORFs partially overlapping within an annotated gene or exon will be reported as gene model revision. Splice variants can be found either by exon–exon spanning peptides or the GSSPs that partially overlapping with an existing exon. The predicted ORFs containing specific amino acids mutations were defined as SAAVs.
PTMs Identification
To gain a holistic view of PTMs events in A. flavus, the same experimental MS data were searched against the existing reference protein database (13,485 protein sequences) by using open-search algorithm in pFind (version 3.1) (43, 44) with a precursor ion mass tolerance of 20 ppm and fragment ion mass tolerance of 20 ppm. Two missed cleavages were allowed for trypsin. The target-decoy strategy based on score threshold was used to evaluate the error rates of identification. For the other parameters in pFind, we used the algorithm defaults.
We next performed restrictive database search strategy to confirm the localization accuracy of the modification sites by using MaxQuant program (version 1.6.2.4) (50). All MS/MS spectra were searched individually 23 times with different parameters as previously described (29, 30). In brief, the identical search parameters were as follows: the precursor and fragment ion mass tolerances were 10 ppm and 0.05 Da, respectively; enzyme specificity was set as full cleavage by trypsin; carbamidomethylation (Cys) was set as a fixed modification, whereas oxidation (Met) was set as dynamic modification. The other different dynamic modifications and enzyme missed cleavages as follows: (i) phosphorylation of Ser/Thr/Tyr and acetylation of protein N-terminus, deamidation of Asn/Gln, two missed; (ii) acetylation of Lys and protein N terminus, deamidation of Asn/Gln, six missed; (iii) succinylation of Lys and protein N terminus, deamidation of Asn/Gln, six missed; (iv) monomethylation of Cys and acetylation of protein N terminus, deamidation of Asn/Gln, two missed; (v) monomethylation of Glu/Gln, and acetylation of protein N terminus, two missed; (vi) monomethylation of Lys/Arg, deamidation of Asn/Gln and acetylation of protein N terminus, six missed; (vii) dimethylation of Lys/Arg, deamidation of Asn/Gln and acetylation of protein N terminus, six missed; (viii) trimethylation of Lys, deamidation of Asn/Gln and acetylation of protein N terminus, six missed; (ix) butyrylation, crotonylation, malonylation, propionylation of Lys, deamidation of Asn/Gln and protein N terminus, six missed; (x) biotinylation of Lys, deamidation of Asn/Gln and protein N terminus, six missed; (xi) GlyGly of Lys, deamidation of Asn/Gln and protein N terminus, six missed; (xii) persulfide of Cys/Asp, deamidation of Asn/Gln and protein N terminus, two missed; (xiii) oxidation to nitro of Trp/Tyr, deamidation of Asn/Gln and protein N terminus, two missed; (xiv) s-nitrosylation of Cys deamidation of Asn/Gln and protein N terminus, two missed; (xv) diphthamide of His, deamidation of Asn/Gln and protein N terminus, two missed; (xvi) farnesylation of Cys, deamidation of Asn/Gln and protein N terminus, two missed; (xvii) geranylation of Cys, deamidation of Asn/Gln and protein N terminus, two missed; (xviii) myristoylation of Cys/Lys, deamidation of Asn/Gln and protein N terminus, two missed; (xix) palmitoylation of Ser/Thr/Cys/Lys, deamidation of Asn/Gln and protein N terminus, two missed; (xx) ADP-ribose addition of Cys/Asp/Glu/Lys/Asn/Arg/Ser, deamidation of Asn/Gln and protein N terminus, two missed; (xxi) beta-methylthiolation of Asp, deamidation of Asn/Gln and protein N terminus, two missed; (xxii) hydroxymethylation of Asn and protein N terminus, two missed; (xxiii) hydroxytrimethylation of Lys, deamidation of Asn/Gln and protein N terminus, six missed. The identified spectra that were assigned different PTMs in different searches were eliminated to filter the false discovery. Finally, the estimated FDR thresholds for proteins, modification sites, and peptides were set at 1%, and the modified peptides with PTM score more than 40 were selected for further bioinformatics analysis.
Bioinformatics Analysis
The functional annotation of all the identified proteins was performed by using Blast2GO tool and the subcellular localization was analyzed by CELLO web tool (51). For the conservation analysis of all the predicted proteins in “non-coding genes analysis” section, we selected the protein databases of 811 fungi from NCBI (www.ncbi.nlm.nih.gov/), to provide evidence of the protein-coding ability as much as possible. For the conservation analysis of novel proteins identified in this work, the protein databases of 66 Aspergillus strains were retrieved from NCBI (www.ncbi.nlm.nih.gov/), JGI (www.jgi.doe.gov), AspGD (www.aspgd.org), and Ensemble (www.ensembl.org). Conservation analysis of the identified novel proteins was carried out by using reciprocal BLAST (49). The two-directional BLAST alignments were classified to be significant if the corresponding BLAST E values were lower than 1E-5. Finally, the best-scoring homologous protein was selected for further processing. The cluster analysis of identified proteins was performed by Cluster 3.0 and visualized by TreeView. The identified novel genes were visualized using the Integrative Genomics Viewer (IGV) browser (52). Potential noncoding genes analysis in A. flavus genome was performed as previous described (29). The homology analysis of novel protein was performed by ClustalW (53) and the novel protein was then mapped to KEGG pathway according to the known homologous proteins from other Aspergillus species. Signal peptide was predicted by SignalP (54) and PrediSi (55) tool, and the motif was visualized by using WebLogo tool (56). R scripts and Excel were used for the statistical analyses. Statistical significance was analyzed by Student's t-tests and expressed as a p value. p < 0.05 was considered to be statistically significant. For RNA-seq data analysis, the retrieved low-quality RNA reads from NCBI database were filtered and then mapped to the A. flavus genome using TopHat (version 2.1.1) (57). Transcripts per kilobase of exons per million (TPM) and fragments per kilobase of exons per million fragments mapped (FPKM) values were calculated by cufflinks (v2.2.1) (58). A protein–protein interaction map was constructed according to the known interactions among homologous proteins from other Aspergillus species in string database (https://string-db.org/). The novel proteins were substituted by the homologous proteins from other Aspergillus species if the bit score cutoff was not lower than 50, according to the string database.
Real-Time PCR
Total RNA was extracted from the mycelia grown in different conditions (37 °C, 29 °C, salt, and H2O2 stresses) using TRIzol reagent (Invitrogen) as the manufacturer's protocols. The concentration of total RNA was quantitated using Nanodrop 2000 spectrophotometer. RT-PCR validation of novel genes was performed using the SYBR Green PCR Master Mix (Applied Biosystems) and the LightCycler 480 Real-Time PCR System (Roche). The actin gene was used as the endogenous control and three independent biological replicates were carried out for duplicate samples. The gene-specific primers were compiled in supplemental Table S1.
Novel Peptides Validation Using Synthetic Peptides
The identified candidate novel peptides were randomly selected and synthesized by Bioyeargene Biotechnology (Wuhan, China). Approximately 1 pmol of synthetic peptide was analyzed on the LTQ-Orbitrap Elite mass spectrometers. The peptide fragments distributions of 49 synthetic peptides were then validated manually by aligning those from the identified peptides in proteogenomic analysis. To further ensure the reliability of the matching for all measured peptides, Cosine similarity analysis (Equation 1) of these spectra was performed and the corresponding score value >0.7 was considered reliable and selected as previously described (59, 60, 61, 62).
| (1) |
Experimental Design and Statistical Rationale
To achieve the in-deep protein coverage in this work, we performed the experiments for many times using different experimental procedures. The wild-type strain of A. flavus was grown under different treatment conditions, including different growth media (yeast extract-sucrose medium, MM, and czapek-dox agar medium) and stress conditions (H2O2, different pH, 28 °C, sodium chloride, congo red). The cultures from these different treatments were collected by filtration and the mycelia were ground into powders to extract the whole cell lysates, respectively. After measuring the protein concentration, the constant protein amounts from different cultures were mixed as previous reports (29, 30) and subsequently in-gel digested by trypsin and analyzed on the Q Exactive HF, Orbitrap Elite, and Triple TOF 6600 mass spectrometers, respectively. Furthermore, the lysates were also processed with in-solution tryptic digestion and prefractionated on a self-packed SPE column according to previously described (29, 30). All the eluted fractions were then analyzed on the Q Exactive HF and Orbitrap Elite mass spectrometers, respectively. The final proteomic data was combined based on these different experimental results. The GAPE tool was used for the identification of peptides, expressed proteins, and novel events. PTMs analysis was performed by using MODa and MaxQuant. Functional annotation of all proteins was performed by using Blast2GO tool and the subcellular localization was analyzed by CELLO web tool (51). Conservation analysis was carried out by using reciprocal BLAST (49) and ClustalW (53). The cluster analysis of identified proteins was performed by Cluster 3.0 and visualized by TreeView. A protein–protein interaction map was constructed according to the interaction data from string database (https://string-db.org/). Potential noncoding genes analysis in A. flavus genome was performed as previous described (29). Signal peptide was predicted by SignalP (54) and PrediSi (55) tool, and the motif was visualized by using WebLogo tool (56). RNA-seq data analysis was performed using TopHat (version 2.1.1) (57) and cufflinks (v2.2.1) (58). R scripts and Excel were used for the statistical analyses. Statistical significance was analyzed by Student's t tests and expressed as a p value. p < 0.05 was considered to be statistically significant.
Results
The A. flavus Proteomic Landscape
In order to ensure broad proteomic coverage, we began by preparing samples of A. flavus grown under eight different culture conditions, including different growth media (yeast extract-sucrose medium, MM, and czapek-dox agar medium) and stress conditions (H2O2, different pH, 28 °C, sodium chloride, congo red) (Fig. 1A). These samples were then further processed and subjected to high-resolution MS analysis (Fig. 1B). In addition to these data, additional MS sources from recently published large-scale MS experiments (37, 38) were also incorporated and analyzed using the GAPE tool, which facilitates automated genomic annotation and PTM discovery in eukaryotic organisms (29). By applying our proteomic strategy to 88 raw MS files, we identified total 840,095 significant peptide spectrum matches (PSMs), corresponding to 150,458 unique peptides using a stringent false discovery rate (FDR) filtering threshold (FDR < 1%) (29). All the peptides identified from the different search engines, along with charge, score, peptide mass, and mass error, were available at the iprox database (www.iprox.org) with the identifier IPX0001753001 (under the file name “Search_Results”). These peptides were then mapped to the A. flavus genome, with the resultant data allowing us to confirm the translation of 8724 previously predicted protein-coding genes of A. flavus. Among these proteins, our proteogenomic analysis has unambiguously identified 8411 proteins by at least two unique peptides or one uniquely identifying peptide using a manual validation method (supplemental Table S2). The spectra of all single-peptide identifications were also uploaded onto the iprox database (www.iprox.org) with the identifier IPX0001753001 (under the file name “Spectra of single-peptide identifications”). For all identified proteins, average sequence coverage was ∼25%, confirming the reliability of our proteomics datasets (supplemental Fig. S1A). Proteins that only matched a subset of peptides from another protein were designated as shared proteins as previous described (29). In addition to the 8411 clearly identified proteins, we additionally identified 313 shared proteins (supplemental Table S2), which only evidenced by the presence of a subset of peptides that mapped to two or more different proteins. Figure 1C provides a summary of the number of genes with all the identified features.
Fig. 1.
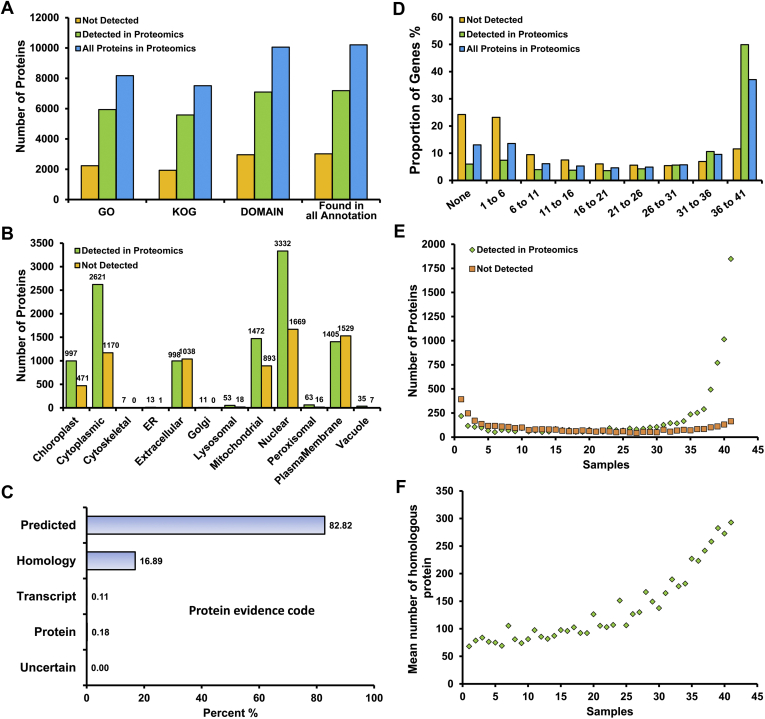
A. flavus proteogenomic workflow and results summary. A flowchart of the sample preparation (A) and MS analysis (B) involved in the present proteogenomic analyses. C, a high-level summary of findings from this study. D, features used to select potential noncoding gene sets, with feature sources indicated in brackets. The Not Detected Genes contains genes for which we found no peptide evidence in our MS data. The All Genes comprises all the predicted protein-coding genes in the NCBI database. B, Blastp; CA, czapek-dox agar; E, Ensembl; GO, Gene Ontology Consortium; I, InterPro; J, JGI; KOG, EuKaryotic Orthologous Groups; MM, minimal medium; N, NCBI; P, Pfam; U, Uniprot; YES, yeast extract-sucrose.
However, the remaining 4761 predicted genes were still not detected by any peptide evidence in this analysis, leading us to next determine whether they corresponded to noncoding genes in the A. flavus database according to previously described methods (63). Figure 1D exhibits the key details regarding the features of protein-coding genes identified in this analysis and summarized the identified result of each feature. We first noticed that the protein length of these unidentified proteins was shorter than those of detected proteins, suggesting that this may have influenced their proteomics-based detection owing to the relatively lower expression levels and number of peptides generated from these proteins (supplemental Fig. S1B). We next analyzed the functional features and localization of these predicted protein-coding genes based upon Gene Ontology (GO), KOG, and domain annotations, revealing that such functional annotations were present for many of these proteins (supplemental Table S3). However, roughly 3000 of these undetected proteins lacked any functional evidence (Fig. 2A), and roughly 50% of these undetected proteins were predicted to localize to the membrane (1529) or extracellular environment (1038) (Fig. 2B), potentially making them challenging to be detected via traditional MS approaches (63). With respect to the uniprot protein evidence pertaining to these genes, 82.82% of protein-coding genes were annotated based upon predictions and 16.89% were inferred from homology, with only 0.3% of proteins being annotated based upon protein and transcript-level experimental evidence (Fig. 2C and supplemental Table S4), suggesting the need for further proteomic analyses of A. flavus to ensure the robust and replicable characterization of this organism. We also found that there was a strong correlation between protein identification and the number of growth conditions in which a transcript was expressed (Fig. 2D and supplemental Table S5). Many more transcripts were detectable for identified proteins, with 7879 proteins having detected transcripts, whereas we could not detect any evidence of transcriptional expression for the ∼25% of undetected proteins. Of these identified proteins with transcript-level evidence, ∼89% (7022) had the evidence of transcriptional expression in ten or more samples, whereas only 334 (4%) were expressed in two or fewer samples. In contrast, many more of the undetected proteins (638) had transcripts that were detected in two or fewer samples (Fig. 2E). All the transcript abundances between protein-coding genes detected and notdetected in the proteomic experiments were provided in supplemental Table S5. Based upon correlation analyses between protein detection and conservation in fungi, we found that the more conditions under which a transcript is expressed, the more likely proteins tended to be conserved (Fig. 2F and supplemental Table S6). Also, the annotations of all predicted proteins from different databases were analyzed and we provided a complete annotation for the potential noncoding genes in supplemental Table S7, while we subsequently excluded the proteins supported with ESTs. Finally, after exclusion of the proteins supported with ESTs (supplemental Table S8), a list of all potential noncoding genes (3279) with at least one of above features is compiled in supplemental Table S9 and Figure 1D.
Fig. 2.
Proteomic data overview.A, functional analysis of A. flavus proteins. “All Proteins in Proteomics” corresponds to all predicted protein-coding genes in the NCBI database. B, A. flavus protein subcellular localization. C, UniProt protein evidence for the protein-coding genes predicted to be present in A. flavus. D, A. flavus gene transcript ubiquity, as determined based upon the number of stress conditions (x-axis) in which a minimum of five or more transcripts per million were detectable. E, the association between transcript expression and proteomic detection results, with the number of genes identified through proteomic analysis (y-axis) shown in comparison to the number of genes for which transcript expression was observed (x-axis). The number of samples where a minimum of five transcripts per million were counted. F, the relationship between sample expression and gene conservation. Gene conservation (y-axis) was compared with the number of samples wherein a given transcript was detected (at least five transcripts per million; x-axis).
Identification of Novel Events in the A. flavus Proteome
We next refined current gene predictions using GSSPs in order to account for any missing or incorrect genes or exons. A total of 72,879 unique GSSPs resulted in the identification of 732 novel protein-coding genes using the identification standard of two unique peptides per protein that were not initially identified during A. flavus genome assembly (supplemental Table S10). Additionally, besides identifying novel genes, the GSSPs were also used to revise the incorrectly predicted gene structures within the A. flavus proteome. We proposed the revision of an existing gene model and designated it as a “revised gene.” In total, 188 revised genes were identified in the current genomic annotation (supplemental Table S11). Moreover, the remaining novel peptides were then used to identify alternatively spliced (AS) proteins and proteins with SAAVs. Finally, we identified 269 AS proteins, of which 88 were novel and 181 were revised (supplemental Table S12). We additionally identified 447 SAAVs, of which 19 were novel, 7 were revised, and 421 were amino acid mutations in the known predicted protein-coding genes (supplemental Table S13). Each of the identified novel variants was supported by at least two unique GSSPs.
Figure 3A illustrated the identification of a novel gene in which the existing assembly did not have any predicted protein coding-gene. We were able to map eight unique intergenic peptides to the previously unannotated genomic region of the two existing exons of gi|238491824 and gi|238491826 exons, with corroborating transcriptomic data being shown. Many of the identified novel peptides could not be directly assigned to genomic sequences, possibly because they span exon–exon junctions corresponding to novel splice variants not currently reflected in genomic annotations. For example, we identified a novel AS variant represented by over 90 unique peptides, 16 of which spanned exon–exon junctions across four exons, with additional RNA-seq evidence supporting the identification of this protein (Fig. 3B). As eukaryotic gene models are often complex, bioinformatics approaches often incorrectly predict the boundaries of many protein-coding genes. We detected a predicted L-arabinofuranosidase precursor protein-coding gene represented by 32 peptides across two exons based on the current genome annotation (Fig. 3C). Notably, 19 GSSPs were clustered in the intronic regions of this gene and six spanned exon–intron boundaries, suggesting that the exons of this gene were extended relative to the current annotated gene model. The validity of this observed partially extended protein was also supported by transcriptomic evidence. Given the errors in and limited depth of whole-genome sequencing data, proteogenomic analyses such as those in the present study can offer direct evidence of translation for this type of gene. For example, besides the eight annotated peptides in the five exons, an additional three point-mutated GSSPs within the annotated gene of a short-chain dehydrogenases/reductase were observed, including the mutation of glycine to asparagic acid, valine to leucine, and isoleucine to valine (Fig. 3D). To further validate the four proteins, we finally used an alignment strategy to explore the conservation of these proteins more broadly, demonstrating orthologous proteins to be present across a range of other species (supplemental Fig. S2 and supplemental Table S14). The details of aligned information for the identified novel gene (Fig. 3A), revised gene (Fig. 3B), novel alternative splicing variant (Fig. 3C), and SAAV (Fig. 3D) in the other Aspergillus strains were uploaded onto the iprox database (www.iprox.org) with the identifier IPX0001753001 (under the file name “Results of sequence alignment”). In addition, to confirm the conservation of the putative mutation sites in the other Aspergillus strains, we performed sequence alignment analysis for all the identified SAAVs in this work and the conservation of the mutated amino acids that occurs in the other 66 different Aspergillus strains was analyzed (supplemental Table S15). Notably, the mutation of Val81 to Lue from the short-chain dehydrogenases/reductase (Fig. 3D) was conserved in 31 Aspergillus strains and the mutation of Ile169 to Val was detected in ten Aspergillus strains, while the third mutation site (Gly49 to Asp) was observed in four Aspergillus strains. For the other mutation sites, as depicted in supplemental Figure S3, 288 mutation sites identified in this study may also be detected in five or more Aspergillus strains, confirming the high conservation of these amino acids across a range of other Aspergillus species. While 114 sites were only observed in five or fewer Aspergillus strains and the remaining 45 mutation sites were not detected in the other Aspergillus strains. These findings suggested that most of the mutated residues identified in this work may have a potential function in Aspergillus. However, further experimental studies are needed to reveal the functional effects of these mutations in A. flavus.
Fig. 3.
Identification of novel, revised, and mutated genes.A, novel peptides mapped to intergenic regions, with eight novel peptides mapping to regions in the A. flavus genome lacking any current annotation. B, novel exon detection. Sixteen splice junction peptides and 74 intergenic peptides mapped to an intronic region in a novel locus. The transcripts also suggest the existence of a protein splice variant for this locus. C, revised gene model identification. A large proportion of novel peptides were partially mapped to the annotated exonic or intronic regions. D, single amino acid variant (SAAV) identification. Three SAAVs for existing genes were identified based upon five SAAV peptides in conjunction with RNA-seq evidence.
Functional Analysis of Novelties in A. flavus
In order to further gain insights into the identified novel proteins, we conducted a conservative analysis of these novel events more broadly across Aspergillus species via a two-directional protein BLAST approach (49). As shown in Figure 4A, while the majority of these proteins appeared to exhibit evolutionary conservation, several species-specific proteins were also detected lacking orthologs in other Aspergillus species indicating that they may play unique regulatory roles in A. flavus (supplemental Table S14). Additional functional annotation revealed that most of these evolutionary conserved novel proteins were associated with multiple metabolic processes, including biosynthetic and nitrogen metabolic processes (Fig. 4B and supplemental Table S16). Since the samples were corrected from different stress treatments, a large subset of these novel proteins were stress-associated and found on the membrane or lumen, suggesting they are important for Aspergillus stress responses and pathogenicity.
Fig. 4.
Overview of novel findings.A, a heatmap showing the conservation of identified novel genes among Aspergillus species, with colors determined based upon the log10 of E-value. B, GO classifications for the identified novel A. flavus proteome components based upon associated biological processes, cellular components, and molecular functions.
To investigate the potential function of these novel proteins, the novel proteins identified in this work were further mapped to the KEGG pathways by homology search strategy. Consistently, we were able to identify many novel proteins that involved in the important pathways (supplemental Table S17). Among these pathways, a large subset of these novel proteins were stress-associated, such as rho signaling pathway, G-protein signaling system, MAPK signaling pathway, and other stress-related signaling pathways, suggesting the potential role of these novel proteins in stress responses. Owing to the pivotal role of the signaling pathways in pathogenesis of Aspergillus (64), our results would facilitate the elucidation of signaling networks in A. flavus. Furthermore, we identified six novel peroxisomal proteins in early aflatoxin synthesis (4) and many novel proteins that involved in aflatoxin biosynthesis (5) and transport pathway (12), indicating the potential role of these novel proteins in secondary metabolism. Although other groups have focused on the role of fungal organelles (vacuole, Golgi apparatus, and endoplasmic reticulum) in growth, differentiation, secondary metabolism, and pathogenesis of Aspergillus (4, 65, 66), the detailed regulatory mechanism of these metabolism has not yet been reported, particularly the identification and functional analysis of the corresponding genes. As a result, these findings in this work could enable us to offer some potential insights into the important functional pathways and also provide a step tone for understanding the detailed regulatory mechanism of aflatoxin biosynthesis in A. flavus. However, further experimental procedures are needed to reveal how these novel events influence the synthesis and export of aflatoxin.
To further explore the biological roles of the novel proteins, we next constructed a protein–protein interaction network that can serve as an alternative strategy to analyze the physical and functional interactions, based upon known interactions among homologous proteins from other Aspergillus species (supplemental Table S18). The resultant network incorporated 115 novel proteins and 223 interactions (supplemental Fig. S4). While we do not have functional data to validate these interactions at present, this network highlights a potential framework for such interactions and offers some potential insights into the important physical and functional pathways in this pathogenic fungus. As expected, a total of 12 stress-associated novel proteins were identified in this network, suggesting they may play key roles in stress responses. Due to the fact that the stress conditions have been reported as an important factor for regulating growth and aflatoxins biosynthesis in Aspergillus (67, 68, 69, 70), we hypothesized that the identified novel proteins were likely to play an important role in regulating the pathogenicity of A. flavus.
Validation of Novelties in the A. flavus Proteome
In order to confirm that the identified novel proteins in this analysis were valid, we sought to independently validate our results via MS analysis of synthetic peptides and via RT-PCR. We first synthesized 49 random GSSPs across a range of peptide types and analyzed them via MS. When we compared the results of the mass spectra for these peptides, we found that the relative intensity and distribution of a series of b/y-ions of the synthetic peptide spectra were in high agreement with the selected GSSPs, strongly supporting the results of our proteogenomic analyses. Furthermore, we performed the Cosine similarity analysis to confirm the matching for all measured peptides. This method was interpreted as a cosine of the angle and chosen as a measure of similarity between different mass spectra in this work. It's calculated as normalized inner product, with score values ranging between 0 and 1. Based on this analysis, the corresponding scores for the 42 measured peptides were more than 0.9 and five were more than 0.8, while the scores of the remaining two peptides were 0.768867 and 0.758584, respectively. We then manually checked these spectra with scores between 0.7 and 0.9 and concluded that these spectra of synthetic peptides agreed with the proteomic results in both the peptide fragments distribution and the relative intensity. All the spectra were available at www.iprox.org with the identifier IPX0001753001 (under the file name “Dataset-MS validation of 49 random GSSPs by synthesized peptides”). Figure 5, A–C showed the representative MS/MS spectra of these synthesized peptides and GSSPs from novel protein, novel alternative splicing variant, revised exon, and SAAVs of our proteomic results.
Fig. 5.
Validation of novel genes.A–C, verification of the identified novel peptides by comparing the MS spectra of the peptides identified from proteogenomic analysis (Experiment) to those of synthetic peptides (Validated). The MS spectra of peptides from novel gene (A), novel alternative splicing variant (B), and SAAVs (C) are shown. Cosine similarity analysis of these spectra were performed and the corresponding score value >0.7 was considered reliable. D, RT-PCR validation of novel genes. RNAs was extracted from A. flavus grown under four different conditions, (37 °C, 29 °C, YES with additional NaCl or H2O2).
We additionally validated the expression of 23 identified novel genes via RT-PCR using a range of different stress conditions, including low temperature (29 °C), oxidative (H2O2), and hyperosmotic (NaCl) stresses. Among these differently expressed novel genes, five genes were upregulated under low temperature and two were upregulated under oxidative stress, while most genes were downregulated under hyperosmotic stress (Fig. 5D). The results of this analysis provided transcript-level results, suggesting that these genes may play an important role in stress responses.
PTM Analysis and Signal Peptide Prediction
We have previously conducted a specific PTM enrichment-based phosphoproteome and succinylome analysis of A. flavus (71, 72). In the current study, we next conducted a global systematic investigation of the most frequently detected PTMs in A. flavus based on open-search and restricted protein modification search approaches. By using these strategies, a total of 6147 proteins with different PTMs were identified among the 9047 proteins by the open-search strategy (supplemental Table S19). We further identified common intracellular PTMs in our data set as in previous reports (29), revealing 3461 modification sites from 1765 proteins in this study (supplemental Table S20). The original MaxQuant search results for different PTMs and the spectra of identified posttranslationally modified peptides were uploaded onto the iprox database (www.iprox.org) with the identifier IPX0001753001 (under the file name “Maxquant search results” and “Spectra of PTMs”). We next compared the score distribution of the different PTMs to the unmodified peptide identifications. We found that the MaxQuant Andromeda score of modified peptides was lower than that of unmodified peptides (supplemental Fig. S5 and supplemental Table S21). Consistent with our results, previous reports have shown that the stoichiometry of the modification is low, and the low stoichiometry of modified peptides in cell lysates may result in low identification score (73, 74). Additionally, we noticed that the four most common PTMs in this study were methylation, propionylation, malonylation, and crotonylation, with over 1000 modification sites (Fig. 6A). The in-depth identification of more than 20 PTMs also enabled us to analyze the function of these modified proteins based upon the NCBI eukaryotic orthologous groups (KOG), revealing these modified proteins to play diverse roles as regulators of many cellular processes (Fig. 6B and supplemental Table S22). We next investigated the different PTMs in A. flavus by mapping modified proteins to KEGG pathways. Consistent with the KOG annotations, most proteins were involved in metabolic pathways (225), the biosynthesis of secondary metabolites (104), and the biosynthesis of antibiotics (89) (supplemental Table S23).
Fig. 6.
Summary of modified proteins in A. flavus.A, distribution of the number of sites and proteins from 24 types of PTMs in A. flavus. B, functional annotations for modified proteins. (1) RNA processing and modification; (2) chromatin structure and dynamics; (3) energy production and conversion; (4) cell cycle control, cell division, chromosome partitioning; (5) amino acid transport and metabolism; (6) nucleotide transport and metabolism; (7) carbohydrate transport and metabolism; (8) coenzyme transport and metabolism; (9) lipid transport and metabolism; (10) translation, ribosomal structure and biogenesis; (11) transcription; (12) replication, recombination and repair; (13) cell wall/membrane/envelope biogenesis; (14) cell motility; (15) posttranslational modification, protein turnover, chaperones; (16) inorganic ion transport and metabolism; (17) secondary metabolites biosynthesis, transport and catabolism; (18) general function prediction only; (19) function unknown; (20) signal transduction mechanisms; (21) intracellular trafficking, secretion, and vesicular transport; (22) defense mechanisms; (23) extracellular structures; (24) nuclear structure; (25) cytoskeleton.
Based on our functional annotation of the identified predicted proteins (supplemental Table S3) and novel events (supplemental Tables S16 and S17), many proteins were likely to be involved in aflatoxin biosynthesis and export pathways and predicted to localize to the membrane and extracellular. It should be noted that the export of secreted proteins as well as proteins that are located in the inner or outer membrane or the periplasm usually requires an N-terminal signal sequence, while knowledge of signal peptides is important for understanding protein function. Additionally, signal peptide has been proved to play an important role in Aspergillus (75, 76). Thus, apart from the posttranslational chemical modifications, we next utilized two signal peptide prediction tools, PrediSi (55) and SignalP (54), to predict the presence of N-terminal signal peptides on identified proteins. In total, we identified 1116 and 757 signal peptides using PrediSi (supplemental Table S24) and SignalP (supplemental Table S25), respectively, corresponding with clear sequence motifs (supplemental Fig. S6A). The majority of these peptides were confirmed by both tools (supplemental Fig. S6B), providing a robust basis for their validity. Furthermore, the presence of N-terminal signal peptides on revised and novel proteins was also predicted by PrediSi and SignalP. As shown in supplemental Figure S6, there were similar motifs among the predicted, revised, and novel proteins. We detected the conserved alanine and valine at the -1 and -3 positions on predicted, revised, and novel proteins that predicted by SignalP and PrediSi, except the novel proteins from PrediSi (supplemental Tables S24 and S25). Because the prediction algorithms of PerdiSi and SignalP were trained with data sets of experimentally validated signal peptides and the moderate prediction accuracy could be a result of the lack of experimentally determined cleavage sites, so these features go some way toward explaining why we could not detect sequence motif for the novel proteins by PrediSi. Together, we provided a comprehensive map of PTMs in this pathogenic genome, including a large number of posttranslational chemical modifications and signal peptide cleavages. We anticipated that these identified signal peptides likely play a role in A. flavus secondary metabolite transport.
Discussion
In this study, we presented the first comprehensive draft map of the A. flavus proteome. This is the first study we are aware of to have employed such a proteomic approach to improve Aspergillus genomic annotation. Importantly, we were able to leverage these results to gain novel insights into the mechanisms of aflatoxin synthesis and A. flavus pathogenicity, thus potentially highlighting novel strategies for controlling aflatoxin contamination.
Through our proteogenomic analysis, we were able to definitively identify 732 novel protein-coding genes in A. flavus. Interestingly, some of these genes were relatively small in length, potentially explaining why they were overlooked in previous in silico analyses (8, 9, 10). Of these novel proteins, roughly 50% were conserved among 66 Aspergillus species (Fig. 4A and supplemental Table S14). The conserved novel genes (32) that could be detected having orthologs in 65 Aspergillus strains were selected for further functional annotation. We noted that these conserved proteins were involved in metabolic, reproductive, and developmental processes (supplemental Fig. S7). It seems that some of these proteins may play conserved roles in Aspergillus growth and survival, as such metabolic networks are critical in Aspergillus species (68, 69, 70, 77). In order to remain an effective pathogen, it is vital that Aspergillus be able to adapt to environmental stressors. In our proteogenomic analysis, the samples were prepared from eight different stress treatments and many novel genes were differently expressed in response to these conditions (Fig. 1). As such, we hypothesized that novel proteins identified in A. flavus cells under stress conditions may be related to the survival, growth, pathogenicity, and stress response behavior of A. flavus in the infected host cell. In addition, the novel proteins involved in aflatoxin synthesis and export were also identified in the present work (supplemental Table S17). For instance, the novel proteins involved in carbon metabolism would regulate the flux of acetyl CoA, which is the initial substrate of aflatoxins biosynthesis (78). The vacuole-related novel proteins may affect the biosynthesis of aflatoxins and their consequent export by regulating the vacuolar homeostasis (65, 66, 79). Although the detailed regulatory mechanism of these novel proteins remains unclear, the identified novel proteins in this work provide a rich source to facilitate our understanding of the stress responses and aflatoxins biosynthesis in A. flavus.
Despite widespread recognition of the importance of PTMs in various biological processes (80, 81, 82, 83), there is still lack of a holistic view of PTMs events on Aspergillus proteins and the functional roles of many types of PTMs have not yet been uncovered. In this study, we identified 8724 annotated protein-coding genes with high confidence and a significant number of these proteins were found to be modified with different types of PTMs. Although the phosphorylation, acetylation and succinylation have been reported from large-scale MS/MS studies (71, 72, 84), the current understanding of PTMs in Aspergillus is still limited and many important PTM events have not yet been discovered in this pathogenic fungus. Comprehensive analysis of these identified PTM events provided a deep coverage similar to that of individual PTM enrichment analysis (71) and a wide-ranging distribution of these PTMs in various cellular processes, including metabolic pathways and biosynthesis of secondary metabolites (supplemental Table S23). To the best of our knowledge, many important PTMs (such as acetylation, succinylation, propionylation, and malonylation) could provide a mechanism to respond to changes in the energy status of the cell and regulate metabolic pathways (85, 86, 87, 88, 89, 90, 91). In addition, we have previously provided the first evidence that phosphorylation, methylation, acetylation, succinylation, ubiquitination, and sumoylation may be a mechanism involved in aflatoxin biosynthesis by regulating the activities of enzymes in A. flavus (71, 72, 84, 92, 93, 94). Therefore, the identified PTM data sets from our work may provide a foundation for studying the biological functions of these PTM events in metabolic pathways and aflatoxin biosynthesis. Further experimental procedures are also needed to confirm the speculation and understand the potential regulatory mechanism of these PTMs in A. flavus. Moreover, secondary metabolites, produced by Aspergillus, exhibit harmful property for crops and humankind. Although only three secondary metabolite clusters have been characterized, those enzymes in the biosynthesis of secondary metabolites were also subjected to be modified with diverse PTMs, implying the importance of PTMs in the pathway. Together, our data sets provide an important resource for further functional analysis of these PTMs in A. flavus.
In summary, the proteogenomic strategy that we employed in this study allowed us to provide the high-quality comprehensive annotation of the A. flavus genome. The results of this study will serve as a valuable resource for future efforts to explore the entire networks governing the aflatoxin production in A. flavus.
Data availability
All of the raw MS data, the database search results, annotated spectra, the MS validation of 49 random GSSPs by synthesized peptides were uploaded to the public access iprox database (http://www.iprox.org) with the identifier IPX0001753001.
Conflict of interest
Authors declare no competing interests.
Acknowledgments
The authors would like to thank Hubei Proteingene Co, Ltd for help in proteomic experiments and the Wuhan Branch, Supercomputing Center, Chinese Academy of Sciences for help in data processing.
Author contributions
S. W., F. G., and M. Y. conceived and designed the study. M. Y. and Z. Z. performed the proteomics experiments. M. Y., Z. Z., Y. B., and Z. Z. analyzed and interpreted the data. The paper was written by M. Y., F. G., and S. W. and was edited by all the authors.
Funding and additional information
This work was supported by the National Key Research and Development Program of China (2016YFA0501304) and the National Natural Science Foundation of China (Grant No. 31570829), and CAS Key Technology Talent Program (to M. K. Y).
Footnotes
This article contains supplemental data.
Contributor Information
Shihua Wang, Email: wshyyl@sina.com.
Feng Ge, Email: gefeng@ihb.ac.cn.
Supplemental Data
References
- 1.Denning D.W., Follansbee S.E., Scolaro M., Norris S., Edelstein H., Stevens D.A. Pulmonary aspergillosis in the acquired immunodeficiency syndrome. N. Engl. J. Med. 1991;324:654–662. doi: 10.1056/NEJM199103073241003. [DOI] [PubMed] [Google Scholar]
- 2.Yu J., Cleveland T.E., Nierman W.C., Bennett J.W. Aspergillus flavus genomics: gateway to human and animal health, food safety, and crop resistance to diseases. Rev. Iberoam. Micol. 2005;22:194–202. doi: 10.1016/s1130-1406(05)70043-7. [DOI] [PubMed] [Google Scholar]
- 3.Banziger M. Workgroup report: public health strategies for reducing aflatoxin exposure in developing countries. Environ. Health Perspect. 2006;114:1898–1903. doi: 10.1289/ehp.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roze L.V., Hong S.Y., Linz J.E. Aflatoxin biosynthesis: current frontiers. Annu. Rev. Food Sci. Technol. 2013;4:293–311. doi: 10.1146/annurev-food-083012-123702. [DOI] [PubMed] [Google Scholar]
- 5.Rudramurthy S.M., Paul R.A., Chakrabarti A., Mouton J.W., Meis J.F. Invasive aspergillosis by Aspergillus flavus: epidemiology, diagnosis, antifungal resistance, and management. J. Fungi (Basel) 2019;5 doi: 10.3390/jof5030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shan X., Williams W.P. Toward elucidation of genetic and functional genetic mechanisms in corn host resistance to Aspergillus flavus infection and aflatoxin contamination. Front. Microbiol. 2014;5:364. doi: 10.3389/fmicb.2014.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amaike S., Keller N.P. Aspergillus flavus. Annu. Rev. Phytopathol. 2011;49:107–133. doi: 10.1146/annurev-phyto-072910-095221. [DOI] [PubMed] [Google Scholar]
- 8.Payne G.A., Nierman W.C., Wortman J.R., Pritchard B.L., Brown D., Dean R.A., Bhatnagar D., Cleveland T.E., Machida M., Yu J. Whole genome comparison of Aspergillus flavus and A. oryzae. Med. Mycol. 2006;44:S9–S11. doi: 10.1080/13693780600835716. [DOI] [PubMed] [Google Scholar]
- 9.Cleveland T.E., Yu J., Fedorova N., Bhatnagar D., Payne G.A., Nierman W.C., Bennett J.W. Potential of Aspergillus flavus genomics for applications in biotechnology. Trends Biotechnol. 2009;27:151–157. doi: 10.1016/j.tibtech.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Nierman W.C., Yu J., Fedorova-Abrams N.D., Losada L., Cleveland T.E., Bhatnagar D., Bennett J.W., Dean R., Payne G.A. Genome sequence of Aspergillus flavus NRRL 3357, a strain that causes aflatoxin contamination of food and feed. Genome Announc. 2015;3 doi: 10.1128/genomeA.00168-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang P.K., Scharfenstein L.L., Mack B., Hua S.S.T. Genome sequence of an Aspergillus flavus CA14 strain that is widely used in gene function studies. Microbiol. Resour. Announc. 2019;8 doi: 10.1128/MRA.00837-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang K.Y., Georgianna D.R., Heber S., Payne G.A., Muddiman D.C. Detection of alternative splice variants at the proteome level in Aspergillus flavus. J. Proteome Res. 2010;9:1209–1217. doi: 10.1021/pr900602d. [DOI] [PubMed] [Google Scholar]
- 13.Jaffe J.D., Berg H.C., Church G.M. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics. 2004;4:59–77. doi: 10.1002/pmic.200300511. [DOI] [PubMed] [Google Scholar]
- 14.Marx H., Hahne H., Ulbrich S.E., Schnieke A., Rottmann O., Frishman D., Kuster B. Annotation of the domestic pig genome by quantitative proteogenomics. J. Proteome Res. 2017;16:2887–2898. doi: 10.1021/acs.jproteome.7b00184. [DOI] [PubMed] [Google Scholar]
- 15.Menschaert G., Fenyo D. Proteogenomics from a bioinformatics angle: a growing field. Mass Spectrom. Rev. 2017;36:584–599. doi: 10.1002/mas.21483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renuse S., Chaerkady R., Pandey A. Proteogenomics. Proteomics. 2011;11:620–630. doi: 10.1002/pmic.201000615. [DOI] [PubMed] [Google Scholar]
- 17.Mertins P., Mani D.R., Ruggles K.V., Gillette M.A., Clauser K.R., Wang P., Wang X.L., Qiao J.W., Cao S., Petralia F., Kawaler E., Mundt F., Krug K., Tu Z.D., Lei J.T. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subbannayya Y., Pinto S.M., Gowda H., Prasad T.S.K. Proteogenomics for understanding oncology: recent advances and future prospects. Expert Rev. Proteomic. 2016;13:297–308. doi: 10.1586/14789450.2016.1136217. [DOI] [PubMed] [Google Scholar]
- 19.Nesvizhskii A.I. Proteogenomics: concepts, applications and computational strategies. Nat. Methods. 2014;11:1114–1125. doi: 10.1038/nmeth.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruggles K.V., Krug K., Wang X., Clauser K.R., Wang J., Payne S.H., Fenyo D., Zhang B., Mani D.R. Methods, tools and current perspectives in proteogenomics. Mol. Cell. Proteomics. 2017;16:959–981. doi: 10.1074/mcp.MR117.000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S., Thomas J.K., Muthusamy B., Leal-Rojas P., Kumar P., Sahasrabuddhe N.A. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilhelm M., Schlegl J., Hahne H., Gholami A.M., Lieberenz M., Savitski M.M., Ziegler E., Butzmann L., Gessulat S., Marx H., Mathieson T., Lemeer S., Schnatbaum K., Reimer U., Wenschuh H. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 23.Castellana N.E., Payne S.H., Shen Z.X., Stanke M., Bafna V., Briggs S.P. Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl. Acad. Sci. U. S. A. 2008;105:21034–21038. doi: 10.1073/pnas.0811066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baerenfaller K., Grossmann J., Grobei M.A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science. 2008;320:938–941. doi: 10.1126/science.1157956. [DOI] [PubMed] [Google Scholar]
- 25.Mergner J., Frejno M., List M., Papacek M., Chen X., Chaudhary A., Samaras P., Richter S., Shikata H., Messerer M., Lang D., Altmann S., Cyprys P., Zolg D.P., Mathieson T. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature. 2020;579:409. doi: 10.1038/s41586-020-2094-2. [DOI] [PubMed] [Google Scholar]
- 26.Castellana N.E., Shen Z.X., He Y.P., Walley J.W., Cassidy C.J., Briggs S.P., Bafna V. An automated proteogenomic method uses mass spectrometry to reveal novel genes in Zea mays. Mol. Cell Proteomics. 2014;13:157–167. doi: 10.1074/mcp.M113.031260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helmy M., Tomita M., Ishihama Y. OryzaPG-DB: rice proteome database based on shotgun proteogenomics. BMC Plant Biol. 2011;11 doi: 10.1186/1471-2229-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren Z., Qi D., Nina P., Li K., Wen B., Zhou R., Xu S., Liu S., Jones A.R. Improvements to the rice genome annotation through large-scale analysis of RNA-Seq and proteomics datasets. Mol. Cell Proteomics. 2019;18:86–98. doi: 10.1074/mcp.RA118.000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang M.K., Lin X.H., Liu X., Zhang J., Ge F. Genome annotation of a model diatom phaeodactylum tricornutum using an integrated proteogenomic pipeline. Mol. Plant. 2018;11:1292–1307. doi: 10.1016/j.molp.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Yang M.K., Yang Y.H., Chen Z., Zhang J., Lin Y., Wang Y., Xiong Q., Li T., Ge F., Bryant D.A., Zhao J.D. Proteogenomic analysis and global discovery of posttranslational modifications in prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E5633–E5642. doi: 10.1073/pnas.1412722111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matallana-Surget S., Werner J., Wattiez R., Lebaron K., Intertaglia L., Regan C., Morris J., Teeling H., Ferrer M., Golyshin P.N., Gerogiorgis D., Reilly S.I., Lebaron P. Proteogenomic analysis of epibacterium mobile BBCC367, a relevant marine bacterium isolated from the south pacific ocean. Front. Microbiol. 2018;9:3125. doi: 10.3389/fmicb.2018.03125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zai X., Yang Q., Liu K., Li R., Qian M., Zhao T., Li Y., Yin Y., Dong D., Fu L., Li S., Xu J., Chen W. A comprehensive proteogenomic study of the human Brucella vaccine strain 104 M. BMC Genomics. 2017;18:402. doi: 10.1186/s12864-017-3800-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christie-Oleza J.A., Miotello G., Armengaud J. High-throughput proteogenomics of Ruegeria pomeroyi: seeding a better genomic annotation for the whole marine Roseobacter clade. BMC Genomics. 2012;13:73. doi: 10.1186/1471-2164-13-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denef V.J., Kalnejais L.H., Mueller R.S., Wilmes P., Baker B.J., Thomas B.C., VerBerkmoes N.C., Hettich R.L., Banfield J.F. Proteogenomic basis for ecological divergence of closely related bacteria in natural acidophilic microbial communities. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2383–2390. doi: 10.1073/pnas.0907041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.OBrian G.R., Fakhoury A.M., Payne G.A. Identification of genes differentially expressed during aflatoxin biosynthesis in Aspergillus flavus and Aspergillus parasiticus. Fungal Genet. Biol. 2003;39:118–127. doi: 10.1016/s1087-1845(03)00014-8. [DOI] [PubMed] [Google Scholar]
- 36.Yu J.J., Whitelaw C.A., Nierman W.C., Bhatnagar D., Cleveland T.E. Aspergillus flavus expressed sequence tags for identification of genes with putative roles in aflatoxin contamination of crops. FEMS Microbiol. Lett. 2004;237:333–340. doi: 10.1016/j.femsle.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 37.Budak S.O., Zhou M.M., Brouwer C., Wiebenga A., Benoit I., Di Falco M., Tsang A., de Vries R.P. A genomic survey of proteases in Aspergilli. BMC Genomics. 2014;15 doi: 10.1186/1471-2164-15-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selvam R.M., Nithya R., Devi P.N., Shree R.S.B., Nila M.V., Demonte N.L., Thangavel C., Maheshwari J.J., Lalitha P., Prajna N.V., Dharmalingam K. Exoproteome of Aspergillus flavus corneal isolates and saprophytes: identification of proteoforms of an oversecreted alkaline protease. J. Proteomics. 2015;115:23–35. doi: 10.1016/j.jprot.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 39.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eng J.K., Jahan T.A., Hoopmann M.R. Comet: an open source tandem mass spectrometry sequence database search tool. Proteomics. 2012;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 41.Kim S., Pevzner P.A. MS-GF plus makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Craig R., Beavis R.C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 43.Wang L.H., Li D.Q., Fu Y., Wang H.P., Zhang J.F., Yuan Z.F., Sun R.X., Zeng R., He S.M., Gao W. PFind 2.0: a software package for peptide and protein identification via tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2007;21:2985–2991. doi: 10.1002/rcm.3173. [DOI] [PubMed] [Google Scholar]
- 44.Yagoub D., Tay A.P., Chen Z.L., Hamey J.J., Cai C., Chia S.Z., Hart-Smith G., Wilkins M.R. Proteogenomic discovery of a small, novel protein in yeast reveals a strategy for the detection of unannotated short open reading frames. J. Proteome Res. 2015;14:5038–5047. doi: 10.1021/acs.jproteome.5b00734. [DOI] [PubMed] [Google Scholar]
- 45.Perkins D.N., Pappin D.J.C., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 46.Elias J.E., Gygi S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 47.Karpova M.A., Karpov D.S., Ivanov M.V., Pyatnitskiy M.A., Chernobrovkin A.L., Lobas A.A., Lisitsa A.V., Archakov A.I., Gorshkov M.V., Moshkovskii S.A. Exome-driven characterization of the cancer cell lines at the proteome level: the NCI-60 case study. J. Proteome Res. 2014;13:5551–5560. doi: 10.1021/pr500531x. [DOI] [PubMed] [Google Scholar]
- 48.Wen B., Xu S., Zhou R., Zhang B., Wang X., Liu X., Xu X., Liu S. PGA: an R/Bioconductor package for identification of novel peptides using a customized database derived from RNA-Seq. BMC Bioinformatics. 2016;17:1. doi: 10.1186/s12859-016-1133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rastogi A., Maheswari U., Dorrell R.G., Vieira F.R.J., Maumus F., Kustka A., McCarthy J., Allen A.E., Kersey P., Bowler C., Tirichine L. Integrative analysis of large scale transcriptome data draws a comprehensive landscape of Phaeodactylum tricornutum genome and evolutionary origin of diatoms. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-23106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 51.Yu C.S., Lin C.J., Hwang J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004;13:1402–1406. doi: 10.1110/ps.03479604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 54.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 55.Hiller K., Grote A., Scheer M., Munch R., Jahn D. PrediSi: prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 2004;32:W375–W379. doi: 10.1093/nar/gkh378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14 doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chaze T., Guillot A., Langella O., Chamotrooke J., Guilmi A.M.D., Trieucuot P., Dramsi S., Mistou M. O-Glycosylation of the N-terminal region of the serine-rich adhesin Srr1 of Streptococcus agalactiae explored by mass spectrometry. Mol. Cell Proteomics. 2014;13:2168–2182. doi: 10.1074/mcp.M114.038075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin Y.M., Chen C.T., Chang J.M. MS2CNN: predicting MS/MS spectrum based on protein sequence using deep convolutional neural networks. BMC Genomics. 2019;20 doi: 10.1186/s12864-019-6297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu K., Li S., Wang L., Ye Y., Tang H. Full-spectrum prediction of peptides tandem mass spectra using deep neural network. Anal. Chem. 2020;92:4275–4283. doi: 10.1021/acs.analchem.9b04867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michal S., Tereza S., Petra J., Ondrej U. Whole-cell MALDI-TOF MS versus 16S rRNA gene analysis for identification and dereplication of recurrent bacterial isolates. Front. Microbiol. 2018;9:1294. doi: 10.3389/fmicb.2018.01294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ezkurdia I., Juan D., Rodriguez J.M., Frankish A., Diekhans M., Harrow J., Vazquez J., Valencia A., Tress M.L. Multiple evidence strands suggest that there may be as few as 19 000 human protein-coding genes. Hum. Mol. Genet. 2014;23:5866–5878. doi: 10.1093/hmg/ddu309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takagi H., Kitagaki H. Springer; Tokyo: 2015. Stress Biology of Yeasts and Fungi. [Google Scholar]
- 65.Veses V., Richards A., Gow N.A.R. Vacuoles and fungal biology. Curr. Opin. Microbiol. 2008;11:503–510. doi: 10.1016/j.mib.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 66.Chanda A., Roze L.V., Kang S., Artymovich K.A., Hicks G.R., Raikhel N.V., Calvo A.M., Linz J.E. A key role for vesicles in fungal secondary metabolism. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19533–19538. doi: 10.1073/pnas.0907416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.RL Buchanan H.S. Ability of various carbon sources to induce and support aflatoxin synthesis by Aspergillus parasiticus. J. Food Sci. 1984;6:271–279. [Google Scholar]
- 68.Davis N.D., Diener U.L. Growth and aflatoxin production by Aspergillus parasiticus from various carbon sources. Appl. Microbiol. 1968;16:158–159. doi: 10.1128/am.16.1.158-159.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mateles R.I., Adye J.C. Production of aflatoxins in submerged culture. Appl. Microbiol. 1965;13:208–211. doi: 10.1128/am.13.2.208-211.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiseman D.W., Buchanan R.L. Determination of glucose level needed to induce aflatoxin production in Aspergillus parasiticus. Can. J. Microbiol. 1987;33:828–830. doi: 10.1139/m87-144. [DOI] [PubMed] [Google Scholar]
- 71.Ren S.L., Yang M.K., Li Y., Zhang F., Chen Z., Zhang J., Yang G., Yue Y.W., Li S.T., Ge F., Wang S.H. Global phosphoproteomic analysis reveals the involvement of phosphorylation in aflatoxins biosynthesis in the pathogenic fungus Aspergillus flavus. Sci. Rep-uk. 2016;6 doi: 10.1038/srep34078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ren S.L., Yang M.K., Yue Y.W., Ge F., Li Y., Guo X.D., Zhang J., Zhang F., Nie X.Y., Wang S.H. Lysine succinylation contributes to aflatoxin production and pathogenicity in Aspergillus flavus. Mol. Cell Proteomics. 2018;17:457–471. doi: 10.1074/mcp.RA117.000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olsen J.V., Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell Proteomics. 2013;12:3444–3452. doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao Y., Jensen O.N. Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques. Proteomics. 2009;9:4632–4641. doi: 10.1002/pmic.200900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bat-Ochir C., Kwak J.Y., Koh S.K., Jeon M.H., Chung D., Lee Y.W., Chae S.K. The signal peptide peptidase SppA is involved in sterol regulatory element-binding protein cleavage and hypoxia adaptation in Aspergillus nidulans. Mol. Microbiol. 2016;100:635–655. doi: 10.1111/mmi.13341. [DOI] [PubMed] [Google Scholar]
- 76.Kmr K., Husaini A., Sing N.N., Tasnim T., Mohd S.F., Hussain H., Hossain M.A., Roslan H. Characterization and expression in Pichia pastoris of a raw starch degrading glucoamylase (GA2) derived from Aspergillus flavus NSH9. Protn. Expr. Purif. 2019:105462. doi: 10.1016/j.pep.2019.105462. [DOI] [PubMed] [Google Scholar]
- 77.Buchanan R.L., Stahl H.G. Ability of various carbon-sources to induce and support aflatoxin synthesis by aspergillus-parasiticus. J. Food Saf. 1984;6:271–279. [Google Scholar]
- 78.Yabe K., Nakajima H. Enzyme reactions and genes in aflatoxin biosynthesis. Appl. Microbiol. Biot. 2004;64:745–755. doi: 10.1007/s00253-004-1566-x. [DOI] [PubMed] [Google Scholar]
- 79.Lee L.W., Chiou C.H., Klomparens K.L., Cary J.W., Linz J.E. Subcellular localization of aflatoxin biosynthetic enzymes Nor-1, Ver-1, and OmtA in time-dependent fractionated colonies of Aspergillus parasiticus. Arch. Microbiol. 2004;181:204–214. doi: 10.1007/s00203-003-0643-3. [DOI] [PubMed] [Google Scholar]
- 80.Schwammle V., Verano-Braga T., Roepstorff P. Computational and statistical methods for high-throughput analysis of post-translational modifications of proteins. J. Proteomics. 2015;129:3–15. doi: 10.1016/j.jprot.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 81.Battchikova N., Angeleri M., Aro E.M. Proteomic approaches in research of cyanobacterial photosynthesis. Photosynth. Res. 2015;126:47–70. doi: 10.1007/s11120-014-0050-4. [DOI] [PubMed] [Google Scholar]
- 82.Ngounou Wetie A.G., Woods A.G., Darie C.C. Mass spectrometric analysis of post-translational modifications (PTMs) and protein-protein interactions (PPIs) Adv. Exp. Med. Biol. 2014;806:205–235. doi: 10.1007/978-3-319-06068-2_9. [DOI] [PubMed] [Google Scholar]
- 83.Chicooree N., Unwin R.D., Griffiths J.R. The application of targeted mass spectrometry-based strategies to the detection and localization of post-translational modifications. Mass Spectrom. Rev. 2015;34:595–626. doi: 10.1002/mas.21421. [DOI] [PubMed] [Google Scholar]
- 84.Yang G., Yue Y.W., Ren S.L., Yang M.K., Zhang Y., Cao X.H., Wang Y.C., Zhang J., Ge F., Wang S.H. Lysine acetylation contributes to development, aflatoxin biosynthesis and pathogenicity in Aspergillus flavus. Environ. Microbiol. 2019;21:4792–4807. doi: 10.1111/1462-2920.14825. [DOI] [PubMed] [Google Scholar]
- 85.Lin H.N., Su X.Y., He B. Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem. Biol. 2012;7:947–960. doi: 10.1021/cb3001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Olsen C.A. Expansion of the lysine acylation landscape. Angew. Chem. Int. Ed. 2012;51:3755–3756. doi: 10.1002/anie.201200316. [DOI] [PubMed] [Google Scholar]
- 87.He W.J., Newman J.C., Wang M.Z., Ho L., Verdin E. Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends Endocrinol. Metab. 2012;23:467–476. doi: 10.1016/j.tem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 88.Newman J.C., He W.J., Verdin E. Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. J. Biol. Chem. 2012;287:42436–42443. doi: 10.1074/jbc.R112.404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Colak G., Xie Z.Y., Zhu A.Y., Dai L.Z., Lu Z.K., Zhang Y., Wan X.L., Chen Y., Cha Y.H., Lin H.N., Zhao Y.M., Tan M.J. Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol. Cell Proteomics. 2013;12:3509–3520. doi: 10.1074/mcp.M113.031567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park J., Chen Y., Tishkoff D.X., Peng C., Tan M.J., Dai L.Z., Xie Z.Y., Zhang Y., Zwaans B.M.M., Skinner M.E., Lombard D.B., Zhao Y.M. SIRT5-Mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rardin M.J., He W.J., Nishida Y., Newman J.C., Carrico C., Danielson S.R., Guo A., Gut P., Sahu A.K., Li B., Uppala R., Fitch M., Riiff T., Zhu L., Zhou J. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18:920–933. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nie X.Y., Yu S., Qiu M., Wang X., Wang S. Aspergillus flavus SUMO contributes to the fungal virulence and toxin attributes. J. Agric. Food Chem. 2016;64:6772. doi: 10.1021/acs.jafc.6b02199. [DOI] [PubMed] [Google Scholar]
- 93.Nie X., Li B., Wang S. Epigenetic and posttranslational modifications in regulating the biology of Aspergillus species. Adv. Appl. Microbiol. 2018;105 doi: 10.1016/bs.aambs.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 94.Lan H., Wu L., Sun R., Keller N.P., Yang K., Ye L., He S., Zhang F., Wang S. The HosA histone deacetylase regulates aflatoxin biosynthesis through direct regulation of aflatoxin cluster genes. Mol. Plant Microbe Interact. 2019;32:1210–1228. doi: 10.1094/MPMI-01-19-0033-R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the raw MS data, the database search results, annotated spectra, the MS validation of 49 random GSSPs by synthesized peptides were uploaded to the public access iprox database (http://www.iprox.org) with the identifier IPX0001753001.