Abstract
There is a tremendous current interest in measuring multiple types of omics features (e.g., DNA sequences, RNA expressions, methylation profiles, metabolic profiles, protein expressions) on a large number of subjects. Although genotypes are typically available for all study subjects, other data types may be measured only on a subset of subjects due to cost or other constraints. In addition, quantitative omics measurements, such as metabolite levels and protein expressions, are subject to detection limits in that the measurements below (or above) certain thresholds are not detectable. In this article, we propose a rigorous and powerful approach to handle missing values and detection limits in integrative analysis of multi-omics data. We relate quantitative omics variables to genetic variants and other variables through linear regression models and relate phenotypes to quantitative omics variables and other variables through generalized linear models. We derive the joint likelihood for the two sets of models by allowing arbitrary patterns of missing values and detection limits for quantitative omics variables. We carry out maximum likelihood estimation through computationally fast and stable algorithms. The resulting estimators are approximately unbiased and statistically efficient. An application to a major study on chronic obstructive lung disease (COPD) yielded new biological insights.
Keywords: complex diseases, data integration, detection limits, genetic association, mediation analysis, missing data, quantitative trait loci, trans-omics studies
Introduction
Recent technological advances have led to a proliferation of genetics studies involving multiple types of omics data. For example, The Cancer Genome Atlas (TCGA) measured point mutation, copy number aberration, DNA methylation, and mRNA, micro RNA, and protein expressions on tumor tissues and matched normal tissues from >11,000 patients with 33 forms of cancer. As a second example, the Trans-Omics for Precision Medicine (TOPMed) program is generating deep whole-genome sequencing (WGS) and other omics data (e.g., metabolic profiles, protein and RNA expression patterns) on a large number of human subjects with rich phenotypic characterization and environmental exposure data. Such multi-omics studies provide unprecedented opportunities to explore the biological relationships among different types of omics variables, evaluate the direct and indirect effects of genetic variants on complex diseases and traits, and investigate the interplay between omics variables and clinical outcomes [Schadt et al., 2005; Hamid et al., 2009; Vaske et al., 2010; Xiong et al., 2012; Wang et al., 2012; Huang et al., 2014; Zhao et al., 2014; Huang et al., 2016].
A major challenge in integrative analysis of multi-omics data is the problem of missing values. In TCGA, for instance, RNA sequencing and protein expressions are not available for all patients due to cost and lack of tissue samples. In TOPMed, WGS data are currently available for >150,000 subjects, whereas other omics data are collected on much smaller scales due to budget constraints. Another major challenge is that quantitative omics measurements, such as metabolite levels and protein expressions, are subject to detection limits in that measurements below (or above) certain thresholds are not detectable.
There is a large body of statistical literature on missing data [Little and Rubin, 2014]. In the genetics context, single imputation for untyped variants works remarkably well [Marchini et al., 2007; Browning and Browning, 2007; Lin et al., 2008; Li et al., 2010], but the problem of missing data on quantitative omics measurements has received little attention. The prevailing approach to the problem of detection limits is to remove the unknown values or replace them by the detection limit or another value [Yu et al., 2014]. This approach is statistically inefficient and possibly biased [Helsel, 2006; Nie et al., 2010]. Although statistical methods have been developed to handle cases where the variable with a detection limit is either a dependent variable [Epstein et al., 2003; Diao and Lin, 2006] or an independent variable [Cole et al., 2009; Nie et al., 2010], no methods are available to deal with detection limits in joint modeling of multiple data types, where the variable with a detection limit is an independent variable in one model and a dependent variable in another model, or where both the dependent and independent variables are subject to detection limits. In addition, the situation in which a variable is subject to both missingness and detection limit has not been investigated before.
In this article, we propose a valid and efficient approach to handle missing values and detection limits in integrative analysis of multi-omics data. We relate the quantitative omics variable of interest to genetic variants and other variables through a linear regression model, and we relate the phenotype of interest to the quantitative omics variable and other variables through a generalized linear model. Indeed, we consider a very general setting with multi-dimensional quantitative omics variables and multivariate phenotypes by formulating the joint distribution of multi-dimensional quantitative omics variables through a multivariate linear regression model and the joint distribution of multiple phenotypes through a generalized linear mixed model. We derive the joint likelihood for the model parameters by allowing quantitative omics variables to be potentially missing and subject to lower or upper detection limits. We carry out maximum likelihood estimation through efficient expectation-maximization (EM) algorithms [Little and Rubin, 2014]. The resulting estimators are approximately unbiased and statistically efficient with a readily estimated covariance matrix. We demonstrate the advantages of the proposed methods over complete-case analysis and imputation methods through extensive simulation studies. Finally, we provide an application to the SubPopulations and InteRmediate Outcome Measures In COPD Study (SPIROMICS) [Couper et al., 2014], which measured ∼100 blood proteins and >600,000 SNPs on ∼3,000 patients.
Methods
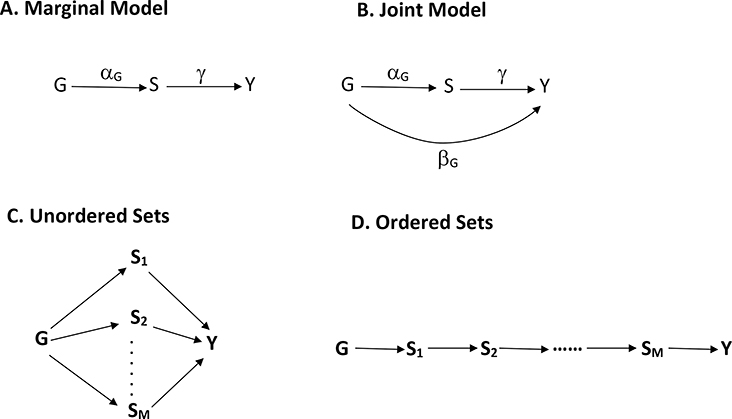
Let Y denote the phenotype of interest, G the genotype of a SNP, and S the quantitative omics variable of interest, which is potentially missing and may have a lower or an upper detection limit. Investigators are typically interested in studying the effects of G on S and S on Y, as shown in Figure 1 (A). They may also be interested in studying the direct and indirect effects of G on Y, as shown in Figure 1 (B). We will refer to these two scenarios as the Marginal Model and the Joint Model, respectively. In either scenario, we may include covariates (e.g., race, gender, age, and principal components for ancestry) in the model.
Figure 1.
Statistical models for integrative analysis of multi-omics data: (A) Marginal Model relating genotypes to quantitative omics variables and relating quantitative omics variables to phenotypes; (B) Joint Model relating genotypes to quantitative omics variables and relating genotypes and quantitative omics variables to phenotypes; (C) unordered relationships among M sets of quantitative omics variables; and (D) ordered relationships among M sets of quantitative omics variables.
We relate S (after an appropriate transformation) to a vector of independent variables X through the linear regression model
| (1) |
where X typically includes G and covariates, α is a vector of regression parameters, and ϵ is zero-mean normal with variance σ2. In addition, we relate Y to S and a vector of independent variables Z through the generalized linear model with density function f(Y|Z,S;η), where Z may overlap with X, and η is a set of unknown parameters. For a quantitative trait, we specify the linear regression model
| (2) |
where β and γ are regression parameters, and ϵ is zero-mean normal with variance τ2, such that η = (βT,γ,τ2)T; for a binary trait, we specify the logistic regression model
| (3) |
such that η = (βT,γ)T. As mentioned earlier, Z excludes G under the Marginal Model and includes G under the Joint Model. We set the first components in both X and Z to 1, such that the first components of α and β are the intercepts. We denote the components of α and β corresponding to G as αG and βG, respectively; see Figure 1 (A) and (B). We are primarily interested in parameter estimation and hypothesis testing on αG, βG, and γ.
To provide a general framework for integrative analysis of multi-omics data, we consider multi-SNP genotypes G and multi-dimensional quantitative omics variables S, together with possibly multivariate traits Y. We relate S to X through the (multivariate) density function f(S|X;ξ) indexed by a vector of unknown parameters ξ, where X is a function of G and covariates. It is convenient to specify f(S|X;ξ) through the multivariate linear regression model
where α is a matrix of regression parameters, and ϵ is a zero-mean normal random vector with covariance matrix Σ. That is,
where d is the dimension of S, and ξ consists of α and the upper triangle elements of Σ. In addition, we relate Y to S and other independent variables Z through the generalized linear (mixed) model with (multivariate) density function f(Y |Z,S;η), where Z may overlap with X, and η is a vector of unknown parameters. If we know a priori that certain components of S do not directly impact Y, then we set the corresponding regression parameters to zero.
There is considerable flexibility in specifying the relationships among different components of S. Suppose that S consists of S1,…,SM, which may pertain to M different types of omics variables or M omics variables of the same type. Figure 1 (C) and (D) show two types of relationships among S1,…,SM. In Figure 1 (C), the M sets of omics variables are unordered. In Figure 1 (D), there is a directed pathway from S1 to S2, from S2 to S3, …, and from SM−1 to SM. In the latter case, the density function f(S|X;ξ) can be specified through the sequential linear regression models
where (α1,…,αM) and (δ1,…,δM−1) are regression parameters, and ϵ1,… ϵM are independent zero-mean normal random vectors with covariance matrices Σ1,…, ΣM. That is,
where δ0 = 0 and S0 = 0.
Suppose that the study contains n unrelated subjects. For i = 1,…,n, let Y i, Si, Xi, and Zi denote the values of Y, S, X, and Z for the ith subject. In addition, let Rij indicate, by the values 1 versus 0, whether or not Sij, the jth quantitative omics variable for the ith subject, is observed. When Rij = 0, the measurement on Sij belongs to the interval , where if Sij is below the lower detection limit Lij, if Sij is above the upper detection limit Uij, and if Sij is not measured at all. The observed data consist of (j = 1,…,d;i = 1,…,n).
Under the missing-at-random assumption [Little and Rubin, 2014], the observed-data likelihood for the vector of parameters θ = (ηT,ξT)T is
where the integration is taken over the unknown Sij. To maximize this likelihood, we adopt the EM algorithm by treating the Si as potentially missing data. The log-likelihood function with the complete data (Y i,Xi,Zi,Si) (i = 1,…,n) is
In the M-step of the EM algorithm, we set the conditional expectation of the complete-data score function given the observed data to zero; in the E-step, we calculate the conditional expectation.
To be specific, let g1(Y i|Zi,Si;η) and g2(Y i|Zi,Si;η) denote, respectively, the first and second derivatives of log f(Y i|Zi,Si;η) with respect to η, and let h1(Si|Xi;ξ) and h2(Si|Xi;ξ) denote, respectively, the first and second derivatives of log f(Si|Xi;ξ) with respect to ξ. In the E-step, we evaluate the conditional expectations of gk(Y i|Zi,Si;η) and hk(Si|Xi;ξ) (k = 1,2) given the observed data and current parameter values, which are denoted by and (k = 1,2). For any function g(Si), the conditional expectation takes the form
Both the numerator and denominator on the right side can be evaluated through numerical integration. In the special case of , and (k = 1, 2).
In the M-step, we obtain the conditional expectation of the complete-data score equation given the observed data
and
We update η and ξ through the one-step Newton-Raphson algorithm
and
We iterate between the above E-step and M-step until convergence, i.e., the change of the parameter values at two successive iterations is less than 10−4. We denote the estimator of θ as , which is consistent and asymptotically multivariate normal.
We use the Louis-formula [Little and Rubin, 2014] to estimate the covariance matrix of . Specifically, we compute the complete-data information matrix
We also compute
In addition, we evaluate and . Finally, we calculate the observed-data information matrix
Then the covariance matrix of is estimated by Ω−1.
The above description is very general. Working out the details of the EM algorithm is not a trivial matter. We show how to implement the EM algorithm efficiently for the combination of models (1) and (2) and models (1) and (3) in Appendices A and B, respectively. We also show how to calculate Q, Ui(Si), , and efficiently in those two situations. The results in the next section are based on the algorithm given in Appendix A.
Results
Simulation Studies
We conducted extensive simulation studies to evaluate the performance of the proposed and existing methods. We generated a quantitative omics variable S from equation (1), in which X consists of five components: X1 = 1; X2 = 0, 1, and 2 with probabilities p2, 2p(1−p), and (1−p)2, respectively; X3 is standard normal; X4 is Bernoulli with 0.5 success probability; and X5 is Uniform(0,1). In this set-up, X2 represents the genotype of a SNP (i.e., G) with minor allele frequency (MAF) p under the Hardy-Weinberg equilibrium, X3 represents the first principal component for ancestry, X4 represents gender, and X5 represents (normalized) age. To create population stratification, we let . We generated a quantitative trait Y from equation (2), in which Z is X minus X2 under the Marginal Model and is the same as X under the Joint Model. We set the two intercepts to 1 and also the two error variances, σ2 and τ2, to 1. In addition, we set αG = 0.25, γ = 0.15, and βG = 0 or 0.2. Finally, we set all other regression parameters to 0.5.
We considered n = 1,000 and set the values of S to be missing (i.e., not measured at all) in a completely random manner for half of the subjects. For the remaining 500 subjects, we varied the lower detection limit from −1 to 1 (with 0.1 increment), such that the proportion of the omics measurements below the detection limit varied from 6.3% to 58.7%, and the number of subjects with detectable values of S (i.e., complete observations) decreased from 468 to 207. We set the nominal significance level at 10−3. We simulated 10,000 replicates under the Marginal Model and under the Joint Model with βG = 0.2, and we simulated 100 million replicates under the Joint Model with βG = 0.
In addition to the proposed method, we evaluated “complete-case analysis”, which removes subjects with missing or non-detectable values, as well as the imputation approach, which removes subjects with missing values and replaces the measurement below the detection limit by the detection limit L or the mid-point L − log2. We will refer to these two imputation methods as “imputation at limit” and “imputation at mid-point”, respectively. (We have implicitly assumed that S is the log-transformation of the original omics measurement. On the original measurement scale, the detection limit is eL, such that the mid-point between 0 and the detection limit is eL/2, which becomes L – log 2 after the log transformation.)
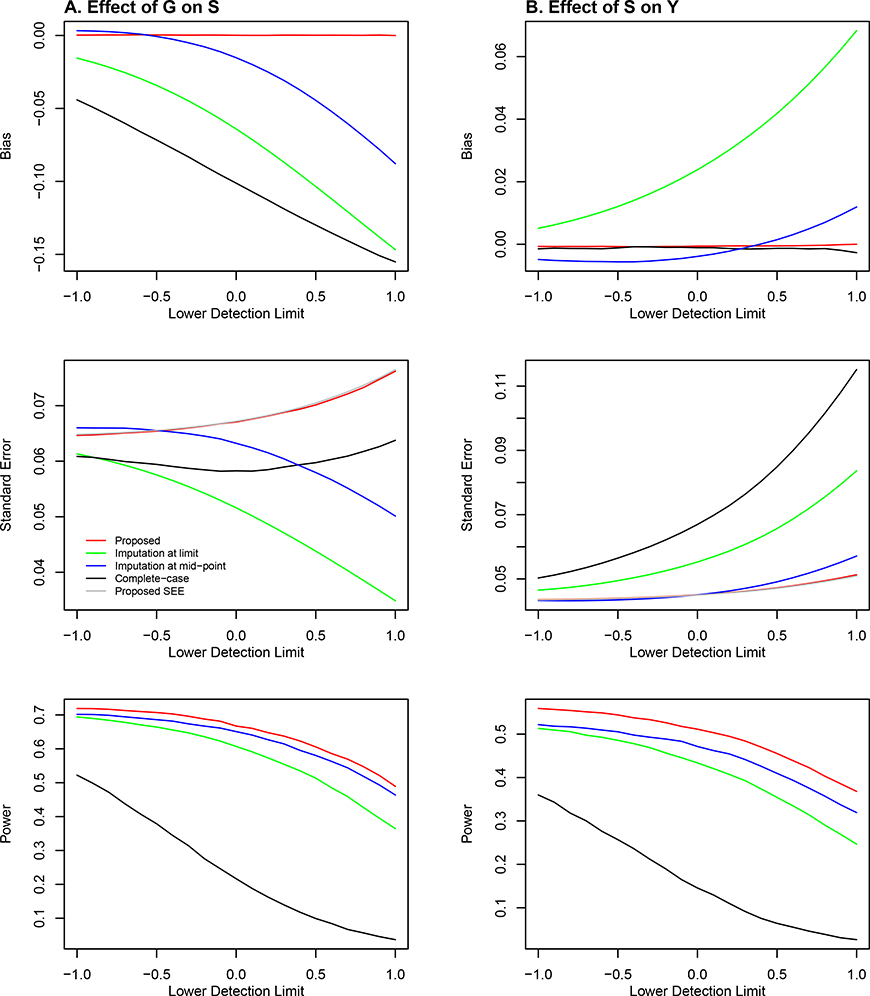
The results under the Marginal Model are shown in Figure 2. The proposed estimators for αG and γ are virtually unbiased, and the corresponding standard error estimators are accurate. Complete-case analysis yields a severely biased estimator of αG and much lower power for testing αG than the proposed method. It yields an approximately unbiased estimator of γ; however, it has a much higher standard error, and thus much lower power for testing γ, than the proposed method. Note that complete-case analysis should yield a biased estimator of αG and an approximately unbiased estimator of γ because S is the dependent variable under model (1) and an independent variable under model (2). (Complete-case analysis removes not only the subjects with missing values but also those with non-detectable values. Although data are missing completely at random, only the values below the detection limit are non-detectable, such that complete-case analysis is biased when S is the dependent variable in the model.) Imputation at limit yields a negatively biased estimator of αG and a positively biased estimator of γ, and it is substantially less powerful than the proposed method in testing αG and γ; the bias and power loss become more severe as the detection limit increases. (The power differences are approximately 0.13 when the detection limit is 1.) Imputation at mid-point also yields seriously biased estimators of αG and γ, with much lower power than the proposed method, although the bias and power loss are less severe than in the case of imputation at limit.
Figure 2.
Simulation results under the Marginal Model: (A) effect of the SNP genotype on the quantitative omics variable (i.e., αG); and (B) effect of the quantitative omics variable on the phenotype (i.e., γ). The bias and standard error of the parameter estimator and the power of the association test are plotted against the detection limit of the quantitative omics variable. The red, black, green, and blue curves pertain to the proposed method, complete-case analysis, imputation at limit, and imputation at mid-point, respectively. The silver curve pertains to the mean of the standard error estimator for the proposed method.
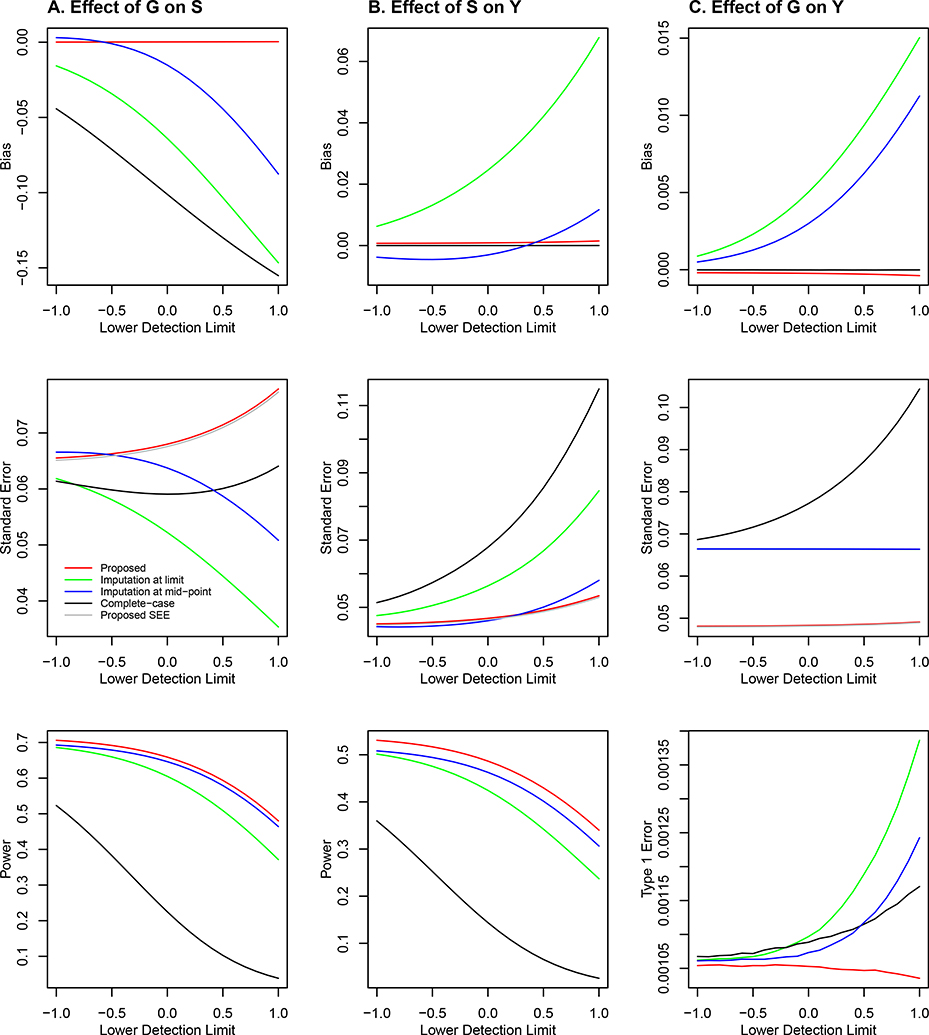
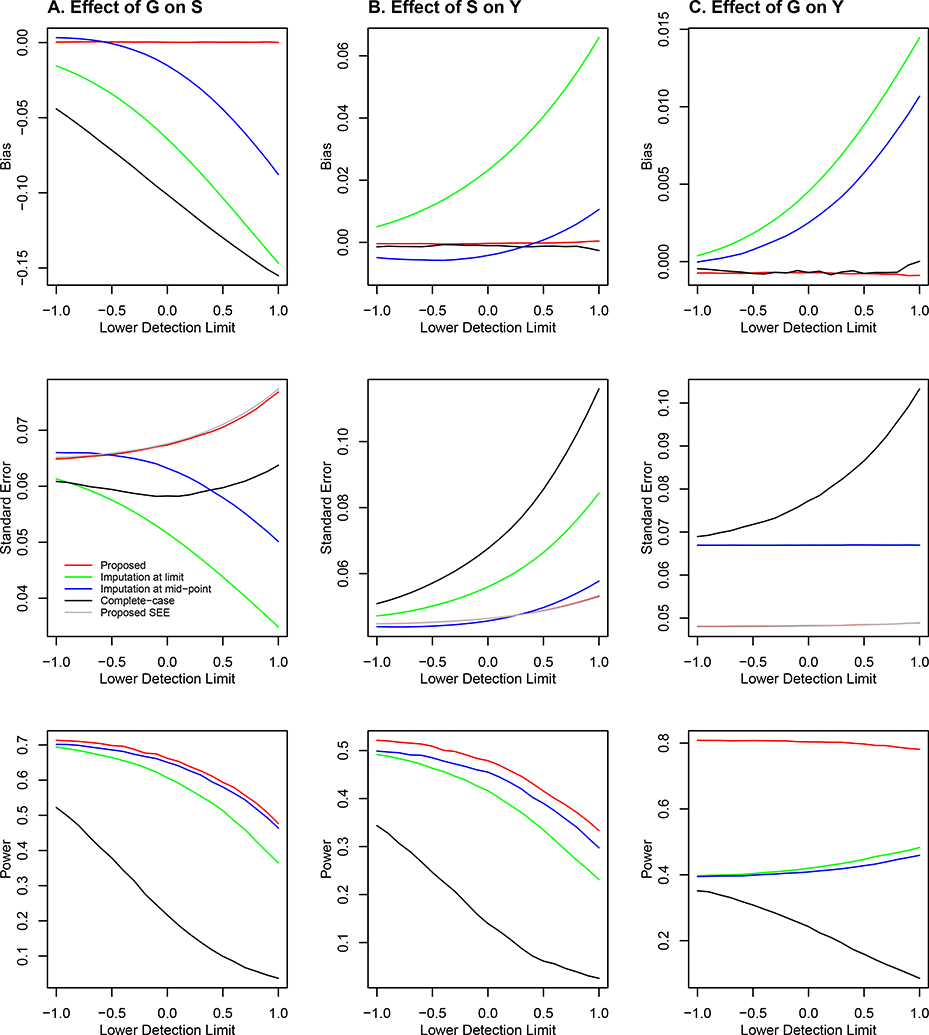
The results for the scenarios of βG = 0 and 0.2 under the Joint Model are displayed in Figures 3 and 4, respectively. The conclusions regarding the bias and power for αG and γ are the same as in the case of the Marginal Model. With regard to the inference on βG, the proposed method yields a virtually unbiased estimator of βG, an accurate standard error estimator, and correct type I error. Complete-case analysis also yields an approximately unbiased estimator of βG, but it has a much larger standard error and thus much lower power than the proposed method. The imputation approach, especially imputation at limit, yields severe bias of the parameter estimator for βG, serious inflation of the type I error (under the null hypothesis), and drastic loss of power (under the alternative hypothesis). The quantile-quantile plots in Figure S1 of the Supplementary Material show the patterns of the type I error inflation for the imputation methods on a broader scale. The poor performance of the imputation approach for making inference about βG is attributed to the bias in the estimation of γ and the correlation between S and G.
Figure 3.
Simulation results under the Joint Model with β = 0: (A) effect of the SNP genotype on the quantitative omics variable (i.e., αG); (B) effect of the quantitative omics variable on the phenotype (i.e., γ); and (C) effect of the genotype on the phenotype (i.e., βG). The bias and standard error of the parameter estimator and the power or type I error of the association test are plotted against the detection limit of the quantitative omics variable. The red, black, green, and blue curves pertain to the proposed method, complete-case analysis, imputation at limit, and imputation at mid-point, respectively. The silver curve pertains to the mean of the standard error estimator for the proposed method.
Figure 4.
Simulation results under the Joint Model with β = 0.2: (A) effect of the SNP genotype on the quantitative omics variable (i.e., αG); (B) effect of the quantitative omics variable on the phenotype (i.e., γ); and (C) effect of the genotype on the phenotype (i.e., β). The bias and standard error of the parameter estimator and the power of the association test are plotted against the lower detection limit of the quantitative omics variable. The red, black, green, and blue curves pertain to the proposed method, complete-case analysis, imputation at limit, and imputation at mid-point, respectively. The silver curve pertains to the mean of the standard error estimator for the proposed method.
To assess the robustness of the proposed method to the non-normal distributions of quantitative omics measurements, we conducted additional simulation studies under the Joint Model in which the error term of model (1) has a t-distribution with 5 degrees of freedom or is the logarithm of a standard exponential random variable. The rest of the simulation set-up was unchanged. As the lower detection limit varied from −1 to 1, the proportion of the omics measurements below the detection limit decreased from 8.9% to 58.1% for the t-distribution and from 17.8% to 70.8% for the logarithm of the exponential variable. In both situations, we performed the inverse-normal transformation on the observed values of S, such that the values above the detection limit follow approximately a truncated normal distribution. The results on the type I error and power are shown in Figures S2 and S3 of the Supplementary Material. The proposed method continues to have proper type I error and tends to be more powerful than complete-case analysis and imputation methods. We also performed the inverse-normal transformation in the original simulation set-up, where is standard normal. As shown in Figure S4, the use of the inverse-normal transformation does not affect the type I error or power of the proposed method when the omics measurements are normally distributed. Thus, we recommend the use of the inverse-normal transformation on the omics measurements.
In the above simulation studies, 50% of S were missing. We conducted another simulation study by setting only 30% of S to be missing. As shown in Figures S5 and S6, the relative performance of various methods remains the same. Finally, we conducted a simulation study by adding a SNP with MAF of 0.4. As shown in Figure S7, the basic conclusions are unchanged.
SPIROMICS
SPIROMICS is a multi-center observational study of COPD designed to guide future development of therapies by providing robust criteria for classification of COPD patients into groups that are most likely to benefit from a certain therapy and identification of biomarkers that can be used as intermediate outcomes to reliably predict clinical benefits [Couper et al., 2014]. The study enrolled 2,974 subjects in four strata (severe COPD, mild/moderate COPD, smokers without airflow obstruction, and non-smoking controls) between November 2011 and January 2015. Participants underwent a baseline visit that included morphometric measures, spirometry, six-minute walk, inspiratory and expiratory chest computed tomography, and standardized questionnaires. Biospecimens, including plasma, serum, DNA, urine, and induced sputum, were collected and stored.
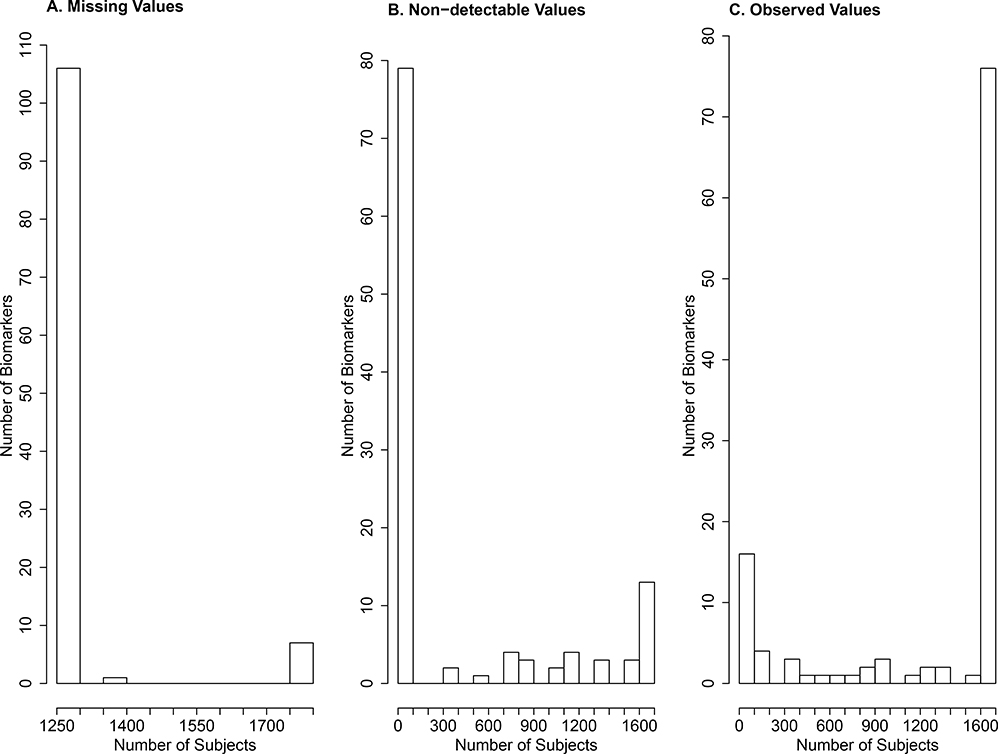
A custom biomarker panel assay was created for 114 blood proteins using 13 Myriad-RBM multiplex assays. The biomarkers were selected on the basis of known or putative links to COPD pathophysiology [O’Neal et al., 2014; Sun et al., 2016]. The assays produced varying levels of missing and non-detectable values among the 114 biomarkers; see Figure 5. For 104 of the biomarkers, 1,280 (43%) subjects have missing values; for the remaining 10 biomarkers, the number of subjects with missing values ranges from 1,281 to 1,763. Out of the 114 biomarkers, 24 have no measurements that are beyond detection limits, 85 have measurements that are below lower detection limits, and 8 have measurements that are above upper detection limits. (Three of the biomarkers have both lower and upper detection limits.) We removed 24 biomarkers with an excessive number of missing or non-detectable values. Each of the remaining 90 biomarkers has over 500 observed values. (Our simulation studies showed that the proposed method is highly reliable with such proportions of observed values.) We performed the inverse-normal transformation on all 90 biomarkers.
Figure 5.
Frequencies of missing values, non-detectable values, and observed values for 114 biomarkers among 2,794 patients in SPIROMICS.
Genotype data on 2,714 participants were derived from Illumina OmniExpress plus Exome GeneChip. There are a total of 673,688 SNPs. After removing those with missing rates > 10% or MAFs < 0.01, we were left with 615,535 autosomal SNPs. Principal component analysis was conducted using common SNPs to identify subjects of divergent ancestry.
We focused on the phenotype emphysema, which is quantified by the percentage of lung voxels ≤ 950 Hounsfield Units on the full inspiratory CT scans. A total of 2,672 subjects have both the phenotype and genotype information. We used the proposed method to handle the missing and non-detectable values in the biomarkers. For comparisons, we also adopted imputation at mid-point (i.e., removing subjects with missing biomarker values and replacing the value below the detection limit by half of the detection limit on the original measurement scale), which was shown in our simulations studies to perform the best among all existing methods. We included age, gender, body mass index, smoking pack years, and current smoking status, together with the top 5 principal components, as covariates in all the models.
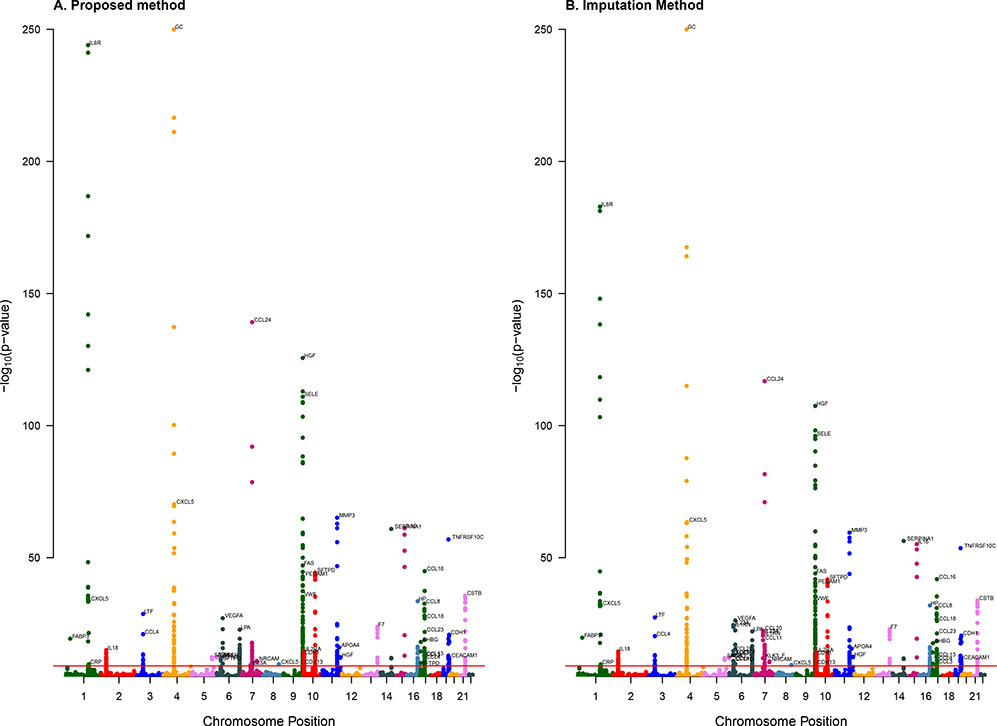
We first fit the Marginal Model; the main results are displayed in Figure 6(A). The quantile-quantile plots (not shown) are well behaved, indicating that the proposed method (with the inverse-normal transformation) is robust to non-normality of biomarker measurements. The combined Manhattan plots in Figure 6(A) show the locations of the SNPs that affect protein variation, which are referred to as protein quantitative trait loci (pQTLs). For the pQTL analysis, the genomewide significance threshold based on the Bonferroni correction for multiple testing of 615,535 SNPs and 90 biomarkers is approximately 9.03 × 10−10. A total of 493 pQTL SNPs in 42 (47%) of the biomarkers achieved the genomewide significance. The most significant pQTL SNPs are rs222047 and rs705120 in GC (vitamin D binding protein) on chromosome 4, with p-values < 10−250. The next most significant pQTL SNPs are rs8192284 and rs4129267 in IL6R (interleukin 6 receptor) on chromosome 1, with p-values of 1.02 × 10−244 and 6.83 × 10−242, respectively.
Figure 6.
Combined Manhattan plots for the associations between SNPs and blood biomarkers in SPIROMICS according to the proposed and imputation methods. The biomarkers with at least one SNP passing the genomewide significance threshold (red line) are marked at the chromosome locations of the top pQTLs. The −log10(p-values) >250 are truncated at 250.
The pQTL analysis results based on the imputation method are displayed in Figure 6(B). The imputation method also identified pQTL SNPs in GC and IL6R, but with less extreme p-values. Indeed, for most of the top pQTL SNPs, the proposed method yielded stronger evidence of association than the imputation method, reflecting the fact that the proposed method makes more efficient use of the data. On the other hand, the imputation method identified pQTL SNPs in CCL11 (chemokin c-c motif ligand 11), CCL20, and KLK3-F (kallikrein 3-F), whereas the proposed method did not. The fact that the same two SNPs are highly significant for three different biomarkers makes the findings of the imputation method dubious. Each of the three biomarkers has a relatively large number of measurements below detection limits (with 881, 1,103, and 895 non-detectable values for CCL11, CCL20, and KLK3-F, respectively).
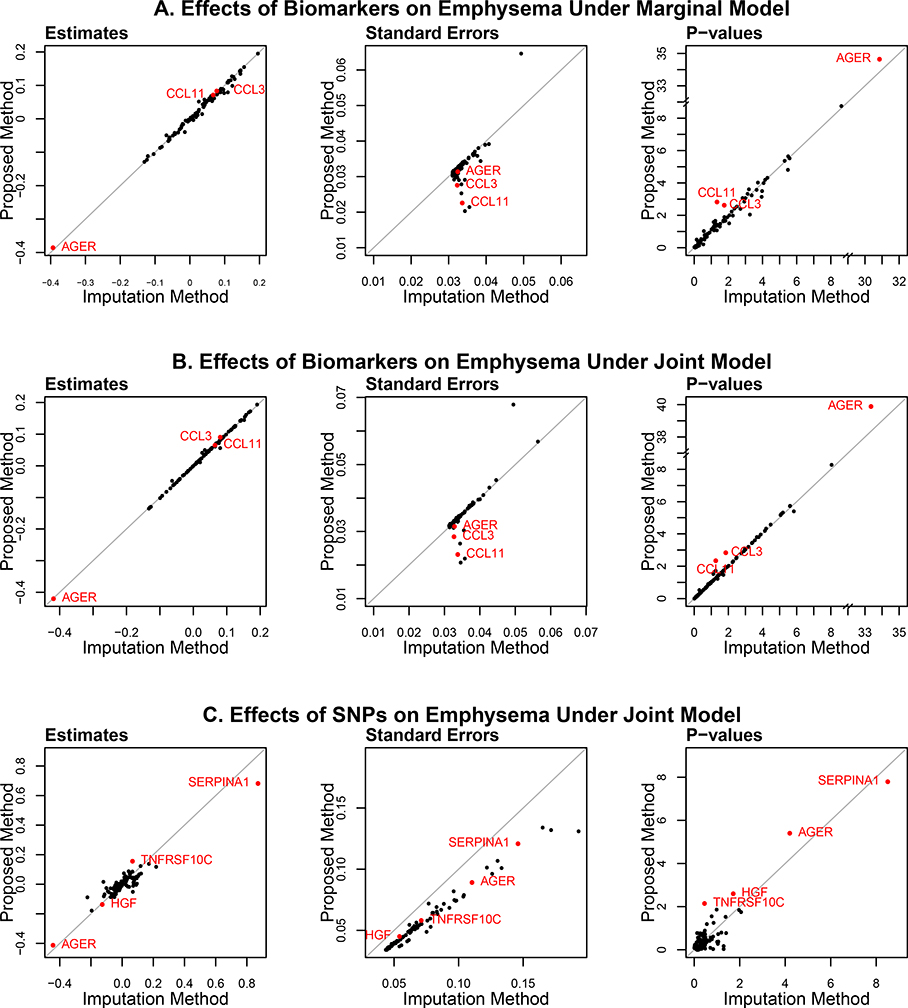
Figure 7 (A) compares the results between the proposed and imputation methods for estimation of the effects of the biomarkers on emphysema under the Marginal Model. For the proposed method, the EM algorithm was applied to the combination of models (1) and (2) for each biomarker, where X contains the genotype of the top pQTL SNP for that biomarker. For the imputation method, standard linear regression was performed on model (2) for each biomarker after mid-point imputation. There are noticeable differences between the effect-size estimates of the two methods. Compared to the imputation method, the proposed method yielded substantially smaller standard errors for the biomarkers with a small number of observed values and more extreme p-values for some of those biomarkers. Specifically, there are 1,351 missing values and 17 non-detectable values for AGER (advanced glycosylation end product-specific receptor); and there are 519 and 881 non-detectable values for CCL3 and CCL11, respectively, along with 1,280 missing values each. For these three biomarkers, the effect size estimates are similar between the two methods, but the proposed method yielded smaller standard errors and thus more extreme p-values.
Figure 7.
Results from the analysis of the SPIROMICS data by the proposed versus imputation methods: (A) effects of biomarkers on emphysema under the Marginal Model; (B) effects of biomarkers on emphysema under the Joint Model; and (C) effects of top pQTL SNPs on emphysema under the Joint Model. Biomarkers with major differences between the proposed and imputation methods are shown in red.
We also fit the Joint Model for each biomarker by adding the top pQTL SNP for that biomarker to equation (2). Figure 7 (B) and (C) compare the results between the proposed and imputation methods: 7 (B) pertains to the effects of the biomarkers on emphysema; 7 (C) pertains to the effects of the corresponding top pQTL SNPs on emphysema. The results for the effects of the biomarkers on emphysema are fairly similar to those of the Marginal Model. For AGER, the difference between the p-values of the proposed and imputation methods is more profound than before.
As shown in Figure 7 (C), the proposed and imputation methods produced appreciably different estimates for the effects of the top pQTL SNPs on emphysema. The proposed method yielded much smaller standard errors for all SNPs and smaller p-values for some SNPs than the imputation method. Specifically, the p-values for the top pQTL SNPs of AGER, HGF (hepatocyte growth factor), and TNFRSF10C (tumor necrosis factor receptor superfamily member 10C) are 3.92 × 10−6, 0.0025, and 0.0071, respectively, according to the proposed method, whereas the corresponding p-values from the imputation method are 6.3 × 10−5, 0.019, and 0.35. The pQTL SNPs associated with AGER, HGF, and TNFRSF10C are rs2070600, rs505922, and rs4760, respectively.
As mentioned previously, the top pQTL for AGER is rs2070600, which has an MAF of 3.9%, with G as the reference allele and A as the alternate allele. Under the Joint Model, the proposed method estimated the direct effect of this SNP on emphysema at −0.412, with standard error of 0.089 and 95% confidence interval of (−0.586, −0.238). The effect of this SNP on AGER was estimated at −0.659, with standard error of 0.084, and the effect of AGER on emphysema was estimated at −0.420, with standard error of 0.031, such that the indirect effect of rs2070600 SNP on emphysema was estimated at 0.277, with standard error of 0.041 and 95% confidence interval of (0.197,0.357).
Discussion
There is a worldwide proliferation of multi-omics studies, which provide unparalleled opportunities to understand the biological processes that underlie complex diseases and traits. It is economically impossible to measure all types of omics features on a large number of subjects and technically infeasible to detect quantitative values below (or above) certain thresholds. Thus, incompleteness of data is inevitable in any multi-omics study.
We have presented a very general approach for integrative analysis of multi-omics data with missing values and detection limits and implemented it through computationally efficient algorithms. The novelty of our work lies not only in the creation of the statistical framework but also in the construction of the EM algorithms. We note that genomewide analysis for one biomarker in the SPIROMICS data (containing 615,535 SNPs on 2,672 subjects) took ∼ 16 hours on an IBM Blade HS20 processor. We have posted our code at http://dlin.web.unc.edu/software/iMODA/.
The current approach to analysis of incomplete multi-omics data is to fit each model separately after removing subjects with missing values and imputing values below or above detection limits. This strategy is highly inefficient and potentially biased, and it may yield qualitatively different results than the proposed method, as shown with the SPIROMICS data. Specifically, removing subjects with missing values is inefficient, and it will also bias the analysis if the data are not missing completely at random. The imputation approach biases parameter estimation and reduces statistical power; it can also inflate the type I error. We note that unbiased parameter estimation is of great importance in integrative analysis of multi-omics data, especially when quantifying direct and indirect effects.
Recently, several authors [Cai et al., 2016; Lin et al., 2016; Voillet et al., 2016] proposed imputation methods for incomplete multi-omics data. In general, the imputation approach does not provide unbiased estimation of regression parameters. In addition, the recently proposed imputation methods [Cai et al., 2016; Lin et al., 2016; Voillet et al., 2016] deal with missing data only and do not allow non-detectable values. By contrast, our framework accommodates both missing and non-detectable values.
We have derived the covariance matrix for all model parameters, making it possible to make joint inference on multiple parameters. However, joint inference may require special care. Suppose, for example, that we are interested in estimating the indirect effect of a SNP on a phenotype, i.e., the product of αG and γ. Although the estimators for αG and γ are asymptotically normal, the normal approximation to the product of the two estimators is inaccurate unless the sample size is very large. We are currently exploring a resampling approach to construct confidence intervals for indirect effects. We should also note that standard statistical methods cannot be used to test the null hypothesis of no indirect effect, i.e., αGγ = 0, because the asymptotic null distribution of depends on whether both αG and γ are zero or only one of them is zero. We are currently developing proper tests for indirect effects.
Our current framework formulates the conditional distribution of Y given S. Some omics measurements may be influenced by diseases. In that case, the regression parameters in our phenotype models do not have causal interpretations. However, our likelihood approach can be applied to other formulations of the relationships between Y and S.
A previous meta-analysis of the SPIROMICS and COPDGene [Regan et al., 2011] data identified 527 pQTLs in 38 blood proteins [Sun et al., 2016]. Our analysis of the (new) SPIROMICS data confirmed pQTLs in 34 of those 38 blood proteins. In addition, we identified novel pQTLs in 9 blood proteins: A2M (alpha-2-macroglobulin), CCL13, CCL3, CDH13 (Cadherin-13), CEACAM1 (Carcinoembryonic antigen-related cell adhesion molecule 1), CRP (C-reactive protein), FABP3 (fatty acid binding protein 3), IL15, and IL1RN. Most of those biomarkers had relatively large detection limits, such that the proposed method was particularly effective.
In the SPIROMICS data, there was only one type of quantitative omics variables, namely blood proteins, and we considered one protein at a time, such that the analysis was straightforward. Because X and Z can be any vectors, the inclusion of multiple SNPs in the analysis is as easy as the use of a single SNP. Indeed, we considered multiple SNPs in our analysis of the SPIROMICS data, although the results were not shown. Our framework can be applied to multiple types of quantitative omics variables. However, modeling their relationships requires considerable biological expertise.
The TOPMed program has generated WGS data for >150,000 subjects. Other omics data (e.g., methylation profiles, metabolic profiles, RNA expression patterns) are being collected through TOPMed and ancillary studies, but for only a few thousand subjects. We are currently applying the proposed methodology to those data, and the results will be communicated in separate reports.
We have implicitly assumed that multi-omics data are collected from cohort studies and thus used the standard prospective likelihood. If the data are collected from case-control studies and there are missing and non-detectable values, then the retrospective likelihood conditional on the case-control status should be used instead. The corresponding EM algorithms can be constructed by combining the ideas of this article with those of Lin and Zeng [2006].
In microbiome studies, the abundance of the bacteria may be truly zero rather than below a detection limit. Then it is sensible to use a mixture distribution allowing a proportion of measurements to be zero. In addition, the relative microbiomial abundance pertains to compositional data, which require special treatments. It would be worthwhile to extend our framework to incorporate microbiome data.
We have assumed that investigators are interested in analyzing a small subset of multi-omics features at a time, such that the dimension of S is fixed and relatively small. We are currently exploring a penalized-likelhood approach to variable selection with multiple types of potentially missing features. The results will be communicated in due course.
Supplementary Material
Acknowledgments
This work was supported by NIH awards R01HG009974 and P01CA142538. The authors thank Dr. Christopher Sheldahl for programming assistance. They also thank the SPIROMICS participants and participating physicians, investigators, and staff for making this research possible. More information about the study and how to access SPIROMICS data is at www.spiromics.org. We would like to acknowledge the following current and former investigators of the SPIROMICS sites and reading centers: Neil E Alexis, MD; Wayne H Anderson, PhD; Mehrdad Arjomandi, MD; Igor Barjaktarevic, MD, PhD; R Graham Barr, MD, DrPH; Lori A Bateman, MSc; Surya P Bhatt, MD; Eugene R Bleecker, MD; Richard C Boucher, MD; Russell P Bowler, MD, PhD; Stephanie A Christenson, MD; Alejandro P Comellas, MD; Christopher B Cooper, MD, PhD; David J Couper, PhD; Gerard J Criner, MD; Ronald G Crystal, MD; Jeffrey L Curtis, MD; Claire M Doerschuk, MD; Mark T Dransfield, MD; Brad Drummond, MD; Christine M Freeman, PhD; Craig Galban, PhD; MeiLan K Han, MD, MS; Nadia N Hansel, MD, MPH; Annette T Hastie, PhD; Eric A Hoffman, PhD; Yvonne Huang, MD; Robert J Kaner, MD; Richard E Kanner, MD; Eric C Kleerup, MD; Jerry A Krishnan, MD, PhD; Lisa M LaVange, PhD; Stephen C Lazarus, MD; Fernando J Martinez, MD, MS; Deborah A Meyers, PhD; Wendy C Moore, MD; John D Newell Jr, MD; Robert Paine, III, MD; Laura Paulin, MD, MHS; Stephen P Peters, MD, PhD; Cheryl Pirozzi, MD; Nirupama Putcha, MD, MHS; Elizabeth C Olsner, MD, MPH; Wanda K ONeal, PhD; Victor E Ortega, MD, PhD; Sanjeev Raman, MBBS, MD; Stephen I. Rennard, MD; Donald P Tashkin, MD; J Michael Wells, MD; Robert A Wise, MD; and Prescott G Woodruff, MD, MPH. The project officers from the Lung Division of the National Heart, Lung, and Blood Institute were Lisa Postow, PhD, and Lisa Viviano, BSN; SPIROMICS was supported by contracts from the NIH/NHLBI (HHSN268200900013C, HHSN268200900014C, HHSN268200900015C, HHSN268200900016C, HHSN268200900017C, HHSN268200900018C, HHSN268200900019C, HHSN268200900020C), and a grant from the NIH/NHLBI (U01 HL137880, and supplemented by contributions made through the Foundation for the NIH and the COPD Foundation from AstraZeneca/MedImmune; Bayer; Bellerophon Therapeutics; Boehringer-Ingelheim Pharmaceuticals, Inc..; Chiesi Farmaceutici S.p.A.; Forest Research Institute, Inc.; GlaxoSmithKline; Grifols Therapeutics, Inc.; Ikaria, Inc.; Novartis Pharmaceuticals Corporation; Nycomed GmbH; ProterixBio; Regeneron Pharmaceuticals, Inc.; Sanofi; Sunovion; Takeda Pharmaceutical Company; and Theravance Biopharma and Mylan.
References
- Browning SR and Browning BL. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. The American Journal of Human Genetics 81: 1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T, Cai TT, and Zhang A. 2016. Structured matrix completion with applications to genomic data integration. Journal of the American Statistical Association 111: 621–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SR, Chu H, Nie L, and Schisterman EF. 2009. Estimating the odds ratio when exposure has a limit of detection. International Journal of Epidemiology 38: 1674–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couper D, LaVange LM, Han M, Barr RG, Bleecker E, Hoffman EA, Kanner R, Kleerup E, Martinez FJ, Woodruff PG, et al. 2014. Design of the subpopulations and intermediate outcomes in COPD study (SPIROMICS). Thorax 69: 492–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao G and Lin DY. 2006. Semiparametric variance-component models for linkage and association analyses of censored trait data. Genetic Epidemiology 30: 570–581. [DOI] [PubMed] [Google Scholar]
- Epstein MP, Lin X, and Boehnke M. 2003. A Tobit variance-component method for linkage analysis of censored trait data. The American Journal of Human Genetics 72: 611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid JS, Hu P, Roslin NM, Ling V, Greenwood CM, and Beyene J. 2009. Data integration in genetics and genomics: Methods and challenges. Human Genomics and Proteomics: HGP 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsel DR. 2006. Fabricating data: how substituting values for nondetects can ruin results, and what can be done about it. Chemosphere 65: 2434–2439. [DOI] [PubMed] [Google Scholar]
- Huang YT, Cai T, and Kim E. 2016. Integrative genomic testing of cancer survival using semiparametric linear transformation models. Statistics in Medicine 35: 2831–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YT, VanderWeele TJ, and Lin X. 2014. Joint analysis of SNP and gene expression data in genetic association studies of complex diseases. The Annals of Applied Statistics 8: 352–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, and Abecasis GR. 2010. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic Epidemiology 34: 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Hu Y, and Huang B. 2008. Simple and efficient analysis of disease association with missing genotype data. The American Journal of Human Genetics 82: 444–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D and Zeng D. 2006. Likelihood-based inference on haplotype effects in genetic association studies. Journal of the American Statistical Association 101: 89–104. [Google Scholar]
- Lin D, Zhang J, Li J, Xu C, Deng HW, and Wang YP. 2016. An integrative imputation method based on multi-omics datasets. BMC bioinformatics 17: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little RJ and Rubin DB. 2014. Statistical Analysis With Missing Data. John Wiley & Sons. [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, and Donnelly P. 2007. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genetics 39: 906–913. [DOI] [PubMed] [Google Scholar]
- Nie L, Chu H, Liu C, Cole SR, Vexler A, and Schisterman EF. 2010. Linear regression with an independent variable subject to a detection limit. Epidemiology 21: S17–S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neal WK, Anderson W, Basta PV, Carretta EE, Doerschuk CM, Barr RG, Bleecker ER, Christenson SA, Curtis JL, Han MK, et al. 2014. Comparison of serum, EDTA plasma and P100 plasma for luminex-based biomarker multiplex assays in patients with chronic obstructive pulmonary disease in the SPIROMICS study. Journal of Translational Medicine 12: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, and Crapo JD. 2011. Genetic epidemiology of COPD (COPDGene) study design. COPD: Journal of Chronic Obstructive Pulmonary Disease 7: 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, GuhaThakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. 2005. An integrative genomics approach to infer causal associations between gene expression and disease. Nature Genetics 37: 710–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Kechris K, Jacobson S, Drummond MB, Hawkins GA, Yang J, Chen TH, Quibrera PM, Anderson W, Barr RG, et al. 2016. Common genetic polymorphisms influence blood biomarker measurements in COPD. PLoS Genetics 12: e1006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, Haussler D, and Stuart JM. 2010. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics 26: 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voillet V, Besse P, Liaubet L, San Cristobal M, and González I. 2016. Handling missing rows in multi-omics data integration: multiple imputation in multiple factor analysis framework. BMC bioinformatics 17: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Baladandayuthapani V, Morris JS, Broom BM, Manyam G, and Do KA. 2012. iBAG: Integrative Bayesian analysis of high-dimensional multiplatform genomics data. Bioinformatics 29: 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q, Ancona N, Hauser ER, Mukherjee S, and Furey TS. 2012. Integrating genetic and gene expression evidence into genome-wide association analysis of gene sets. Genome Research 22: 386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Zheng Y, Alexander D, Morrison AC, Coresh J, and Boerwinkle E. 2014. Genetic determinants influencing human serum metabolome among African Americans. PLoS Genetics 10: e1004212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao SD, Cai TT, and Li H. 2014. More powerful genetic association testing via a new statistical framework for integrative genomics. Biometrics 70: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.