Abstract
Objectives
Prediction of post-transplant health outcomes and identification of key factors remain important issues for pediatric transplant teams and researchers. Outcomes research has generally relied on general linear modeling or similar techniques offering limited predictive validity. Thus far, data-driven modeling and machine learning (ML) approaches have had limited application and success in pediatric transplant outcomes research. The purpose of the current study was to examine ML models predicting post-transplant hospitalization in a sample of pediatric kidney, liver, and heart transplant recipients from a large solid organ transplant program.
Materials and Methods
Various logistic regression, naive Bayes, support vector machine, and deep learning (DL) methods were used to predict 1-, 3-, and 5-year post-transplant hospitalization using patient and administrative data from a large pediatric organ transplant center.
Results
DL models did not outperform traditional ML models across organ types and prediction windows with area under the receiver operating characteristic curve values ranging from 0.50 to 0.593. Shapley additive explanations (SHAP) were used to increase the interpretability of DL model results. Various medical, patient, and social variables were identified as salient predictors across organ types.
Discussion
Results showed that deep learning models did not yield superior performance in comparison to models using traditional machine learning methods. However, the potential utility of deep learning modeling for health outcome prediction with pediatric patients in the presence of large number of samples warrants further examination.
Conclusion
Results point to DL models as potentially useful tools in decision-support systems assisting physicians and transplant teams in identifying patients at a greater risk for poor post-transplant outcomes.
Keywords: pediatric organ transplantation, machine learning, united network for organ sharing, UNOS
BACKGROUND AND SIGNIFICANCE
Rates of survival continue to be high and are improving for pediatric patients after undergoing an organ transplantation procedure. Overall, 5-year patient survival rates for patients transplanted between 2009 and 2013 were 98.4% for pediatric kidney transplant recipients [1] and 83.2% for pediatric liver transplant recipients [2]. In pediatric heart transplant recipients who underwent transplant procedure from 2006 to2013, 1- and 5-year patient survival rates were 90.1% and 81.5% [3]. Despite these improvements, ongoing concerns remain regarding the rate of hospitalization for these children [4–8] and especially for adolescent patients who experience higher rates of complications and nonadherence to immunosuppressive medication [4, 6–12].Data-driven approaches to identify unique risk factors, at-risk patients, and development of strategies to support clinical care and enhance decision-making within pediatric organ transplant centers are highly desired.
Increased number and frequency of hospitalizations, increase stress, and burden on patients and families represent a loss of quality of life for these pediatric patients [13, 14]. Yet, the prediction of hospitalization has been limited in pediatric transplant recipients [15, 16]. Notably, prior research has various methodological limitations, limited sample sizes, sample bias, and lack of rigorous statistical approaches [11]. Nationally, predictive modeling has relied on general linear modeling or Cox proportional hazards regression approaches, which offer limited predictive validity [17–19]. Data-driven modeling and machine learning (ML) analytic approaches offer an opportunity to use ubiquitous and abundant electronic health records (EHRs) and longitudinal data in pediatric transplantation[20–22] to make a significant advancement over the extant research and increase in the clinical usefulness of patient data sources.
ML approaches over EHR data lend themselves to translation into clinical care with the potential to improve the information available to multidisciplinary transplant teams, patients, and their families. These advanced analytic approaches can accommodate many different relationships among patient variables, assess the relative predictive utility of these complex relationships, and reach a final prediction model with an estimated value of predictive validity [23]. ML has been used to examine health outcomes in adult kidney [23, 24], liver [25–28], and cardiothoracic (heart and heart–lung) transplantation [29–33]. In pediatric transplant populations, use of ML and results have been more mixed. ML has shown limited predictive validity and sensitivity when examining mortality as an outcome in heart transplant recipients [16] using classification and regression trees, random forest (RF), and artificial neural network (ANN) approaches. ANN has been used in modeling to predict recipients’ mortality at 6-month post-transplant for pediatric liver transplantation. Over previous standards, ANN offered superior predictive utility when including 21 predictors in the analyses[34]. Similarly, RF identified key factors in predicting ideal post-transplant outcomes 3years after liver transplantation[15].
Numerous assessments and evaluations of pediatric transplant candidates pretransplant and recipients post-transplant are available, but many lack evidence of reliability and validity [11, 12]. The use of ML approaches in examination of post-transplant outcomes in pediatric care is limited [15, 17], yet applying ML and deep learning (DL) to large data repositories and patient records has great potential to inform care and decision-making within multidisciplinary transplant teams. Therefore, the purpose of the current study was to test and examine ML and DL models predicting the experience of hospitalization in samples of pediatric kidney, liver, and heart transplant recipients from a large solid organ transplant program. Additionally, the current study included longitudinal data from the transplant center to examine 1-, 3-, and 5-year outcomes to predict patient hospitalization over the post-transplant period. The inclusion of multiple windows of post-transplant and medical outcomes data represents an important advancement in the study and ML modeling of pediatric post-transplant outcomes.
DL models are often referred to as black-box implying the fact that their underlying mechanisms are not easy to understand by the end user [35]. Some application areas of DL, such as medicine, involve high risk and serious consequences. The lack of transparency in these models is a barrier towards their utilization in real-world medical settings. Thus, considering the importance of interpretability enhancement for DL models in medicine, in this study, an additional step was taken to interpret the results of DL models for each organ type.
METHODS AND MATERIALS
Patients
Data for the study were United Network for Organ Sharing (UNOS, U.S. Department of Health and Human Services) data from a pediatric transplant population at a large transplant center in the southwestern United States. UNOS data include individual, family, and medical variables which were used to predict long-term health and transplant outcomes. The sample included pediatric kidney, heart, or liver transplant recipients (ages 0–18). Data used in the analyses included medical and UNOS data from the patients transplanted at this center between 1988 and May 31, 2017.
UNOS data
The UNOS data include medical data and long-term health outcome variables with the main outcome in the current analyses being post-transplant hospitalization. UNOSmaintains national and center level data on organ transplant patients beginning at listing as a candidate for transplantation. Each transplant center is required to maintain pretransplant and post-transplant medical and health data on all transplant patients. Variables related to pretransplant illness severity, transplant procedure, postoperative data, post-transplant complications, and health outcomes are included on these forms. UNOS forms for each patient are completed at three points in the transplant process and aftercare: (1) candidate registration for an organ transplant (TCR), (2) recipient registration at the time of transplant procedure (TRR), (3) recipient follow-up completed annually post-transplant (TRF) (https://www.transplantpro.org/technology/data-collection-forms/).
Data preprocessing
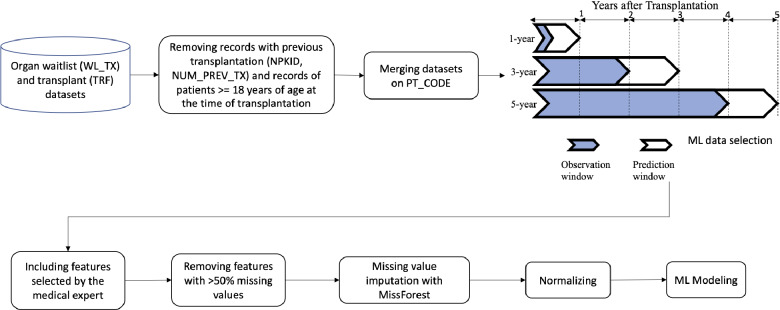
For each of the three organ types in this study, we considered three different outcome prediction windows of 1, 3, and 5years (Figure 1), similar to other studies predicting outcomes in pediatric organ transplantation [16, 36]. The binary prediction outcome was hospitalization within the prediction window after transplantation. In this study, the observation window of 1-year outcome prediction included the initial follow-up information of each patient (farthest from the prediction window) in addition to variables included in the TCR and TRR forms. The observation window of 3-year outcome prediction was the most recent TFR information of each patient within 2 years after transplantation. The observation window of 5-year outcome prediction was the most recent TRF follow-up information of each patient within 4 years after transplantation. In other words, the prediction task in each of 1-, 3-, and 5-year windows is hospitalization within a year after the observation window.
Figure 1.
The overall workflow of data processing.
In each of the datasets, we excluded patients over 18 years of age and those with a prior transplantation procedure. The samples with unknown values in their outcome variable (hospitalization) were also excluded. A medical expert (D.G.) identified clinically relevant variables from all available sets of variables for each organ type to be included in the final predictive models. Variables with >50% missing values were excluded. Finally, some decisions about inclusion and exclusion of certain variables for different organ types and prediction windows were taken by the authors to ensure the validity of prediction models. “Recipient Graft Status” across the TCR, TRR, and TRF forms, for instance, was excluded from analyses due to the known high correlation with hospitalization as a prediction outcome. “Treated for Rejection within 1 Year”, on the other hand, was excluded from the 1-year prediction in the analyses but was included for the 3- and 5-year prediction. Table 1 summarizes the number of variables included for each organ type in the final predictive models. Table 2 summarizes the demographic information in each prediction window for each organ type. A complete list of variables for each organ type is reported in Supplementary Material.
Table 1.
The summary of number of variables included in each dataset for each organ type
| Organ type | # total variables | # clinically relevant variables | # clinically relevant variables with <50% missing value |
|---|---|---|---|
| Kidney | 614 | 108 | 39 |
| Liver | 523 | 93 | 37 |
| Heart | 758 | 117 |
38 (1y) 43 (3y) 41 (5y) |
Table 2.
Demographic summary of samples in each dataset
| Organ type | Prediction window | Gender | Initial age | Education | # total samples |
|---|---|---|---|---|---|
| Kidney | 1y |
F: 125 M: 182 |
Up to 2: 12 >2–10: 115 >10–14: 82 >14–17: 93 18: 5 |
Attended college/technical school: 1 Grade school (0–8): 155 High school (9–12) or GED: 103 N/A (<5 y old): 41 None: 7 |
307 |
| 3y |
F: 130 M: 179 |
Up to 2: 13 >2–10: 126 >10–14: 78 >14–17: 87 18: 5 |
Attended college/technical school: 1 Grade school (0–8): 156 High school (9–12) or GED: 97 N/A (<5 y old): 48 None: 7 |
309 | |
| 5y |
F: 130 M: 179 |
Up to 2: 14 >2–10: 126 >10–14: 77 >14–17: 88 18: 4 |
Attended college/technical school: 1 Grade school (0–8): 157 High school (9–12) or GED: 96 N/A (<5 y old): 48 None: 7 |
309 | |
| Liver | 1y |
F: 175 M: 140 |
Up to 2: 178 >2–10: 80 >10–14: 35 >14–17: 19 18: 3 |
Grade school (0–8): 77 High school (9–12): 28 N/A (<5 y old): 207 None: 3 |
315 |
| 3y |
F: 178 M: 139 |
Up to 2: 178 >2–10: 82 >10–14: 36 >14–17: 19 18: 2 |
Grade school (0–8): 81 High school (9–12): 26 N/A (<5 y old): 207 None: 3 |
317 | |
| 5y |
F: 181 M: 131 |
Up to 2: 178 >2–10: 78 >10–14: 33 >14–17: 20 18: 3 |
Grade school (0–8): 77 High school (9–12): 25 N/A (<5 y old): 207 None: 3 |
312 | |
| Heart | 1y |
F: 13 M:19 |
Up to 2: 8 >2–10: 9 >10–14: 6 >14–17: 6 18: 3 |
Attended college/technical school: 2 Grade school (0–8): 12 High school (9–12): 6 N/A (<5 y old): 11 None: 1 |
32 |
| 3y |
F: 87 M:102 |
Up to 2: 82 >2–10: 45 >10–14: 27 >14–17: 28 18: 7 |
Attended college/technical school: 2 Grade school (0–8): 58 High school (9–12): 32 N/A (<5 y old): 96 None: 1 |
189 | |
| 5y |
F: 89 M:104 |
Up to 2: 84 >2–10: 46 >10–14: 29 >14–17: 27 18: 7 |
Attended college/technical school: 2 Grade school (0–8): 60 High school (9–12): 31 N/A (<5 y old): 99 None: 1 |
193 |
Since our datasets included a mixture of categorical and numerical variables, we used MissForest imputation method. This imputation method has been found effective in comparison to other methods with medical data [37]. MissForest benefits from the built-in routine in the RF algorithm to handle missing values [38, 39]. In this imputation method, for each variable, an initial guess for the missing values is considered. Then, the variables were sorted according to their amount of missing values starting with the lowest number of missing values. Then, an RF algorithm was trained on the observed data and then missing values were predicted using the trained RF model. This imputation procedure was repeated until stopping criteria were met. Further, the data sets were normalized between 0 and 1. Figure 1 illustrates the data preprocessing for this study. For data management and preprocessing, we used SAS, R, and Python.
Predictive modeling
We built several ML models using WEKA software [40] as well as DL models [41] using Python with Sklearn, Keras, and Tensorflow libraries. Four ML algorithms were tested using WEKA and their default setting. In a previous study [16], it was shown that RF outperformed other ML methods. In this work, in addition to RF, we built models based on other popular ML methods such as logistic regression, multilayer perceptron (MLP), and support vector machine with sequential minimal optimization algorithm (SMO) for comparison. DL is an algorithm biologically inspired by the way the human brain functions. Similar to the human brain, the information is processed through neurons activation from one layer to another[42]. Since the learning process happens based on a loss function, optimizers are needed to choose the parameters of the model in a way that reduces this loss. After hyperparameter tuning for the DL model, it included two hidden layers with 100 neurons each. Batch size was set to 32 with 50 epochs. Batch normalization was used for each hidden layer. We used rectified linear unit (ReLU) [43] as the activation function, adaptive gradient algorithm (Adagrad) [44] as the optimizer with an initial learning rate set to 0.01, and cross-entropy as the loss function. ReLU is a piecewise linear activation function that outputs zero if the input is either negative or zero and outputs the same input otherwise. Adagrad optimizes the model parameters based on their update frequency during training allowing for adaptivity of learning rate during training. Since the samples in this study were obtained from a single transplant center, the sample size was limited. On the other hand, DL models can easily overfit the data they are trained on. In the presence of high dimensionality and low sample size, DL models are more likely to suffer from overfitting [45]. To reduce the risk of overfitting and improve the generalization of the DL model, we employed early-stopping. Further, for both shallow and deep learners, we evaluated the performance using 10-fold cross-validation. We considered the area under the receiver operating characteristic curve (AUROC) as the performance metric. We also reported other performance metrics including precision (true positives/[true positives + false positives]), recall (true positives/[true positives + false positives]), F-measure (harmonic mean of precision and recall), and area under the precision-recall curve (AUPRC).
Interpretability enhancement of DL models using Shapley additive explanations
Prediction models based on DL algorithms are inherently complex and challenging to interpret, despite their superior predictive power [41]. The underlying reasoning behind their predictions is more difficult to extract in comparison to traditional ML models [46]. Thus, a large body of research is dedicated to explainable artificial intelligence (XAI) to explore possible methods for interpreting complex AI models to humans [47]. Shapley additive explanations (SHAP) area state-of-the-art unified framework for XAI [48]. SHAP reports variable or feature importance in a complex model based on Shapley values (i.e., function for numerically evaluating the “value” of a game in the cooperative game theory) of a conditional expectation function of that model [49]. Shapely values are one of the leading approaches to attribution problem (i.e., the distribution of the prediction score of a model over the input variables). Thus, values can represent the influence of a variable on the prediction outcome of the model. Since computing the exact SHAP values is challenging, we introduced Kernel SHAP [48] which benefits from the known additive variable attribution methods (i.e., LIME) [50] to approximate actual SHAP values. We refer the interested audience to the original paper for more details [48]. The implementation of SHAP is made publicly available by its authors on GitHub (https://github.com/slundberg/shap).
RESULTS
The performance of different traditional ML and DL models is reported in Tables 3–5. Each uses different datasets corresponding to the various outcome prediction windows by organ-types. Since the classification task is binary (0 or negative result: patient will not be hospitalized and 1 or positive result: patient will be hospitalized), the performance is measured using precision, recall, F-measure, AUROC, and AUPRC metrics.
Table 3.
The performance of different machine learning and DL models for hospitalization prediction using the kidney datasets
| Prediction window | Algorithm | Precision | Recall | F-Measure | AUROC | AUPRC |
|---|---|---|---|---|---|---|
| 1y | Logistic | 0.615 | 0.616 | 0.615 | 0.62 | 0.605 |
| MLP | 0.513 | 0.515 | 0.513 | 0.544 | 0.544 | |
| SMO.PolyKernel | 0.561 | 0.56 | 0.56 | 0.559 | 0.535 | |
| RandomForest | 0.562 | 0.564 | 0.563 | 0.612 | 0.623 | |
| DL | 0.546 | 0.553 | 0.530 | 0.557 | 0.542 | |
| 3y | Logistic | 0.517 | 0.528 | 0.518 | 0.506 | 0.525 |
| MLP | 0.449 | 0.45 | 0.449 | 0.45 | 0.483 | |
| SMO.PolyKernel | 0.493 | 0.511 | 0.492 | 0.487 | 0.501 | |
| RandomForest | 0.574 | 0.583 | 0.568 | 0.559 | 0.559 | |
| DL | 0.533 | 0.531 | 0.523 | 0.523 | 0.522 | |
| 5y | Logistic | 0.563 | 0.57 | 0.562 | 0.545 | 0.556 |
| MLP | 0.525 | 0.528 | 0.526 | 0.514 | 0.529 | |
| SMO.PolyKernel | 0.509 | 0.524 | 0.507 | 0.501 | 0.508 | |
| RandomForest | 0.621 | 0.625 | 0.611 | 0.609 | 0.587 | |
| DL | 0.568 | 0.563 | 0.553 | 0.555 | 0.538 |
Table 4.
The performance of different machine learning and DL models for hospitalization prediction using the liver datasets
| Prediction window | Algorithm | Precision | Recall | F-Measure | AUROC | AUPRC |
|---|---|---|---|---|---|---|
| 1y | Logistic | 0.523 | 0.524 | 0.523 | 0.531 | 0.517 |
| MLP | 0.574 | 0.575 | 0.574 | 0.6 | 0.586 | |
| SMO.PolyKernel | 0.584 | 0.581 | 0.571 | 0.577 | 0.545 | |
| RandomForest | 0.545 | 0.546 | 0.545 | 0.596 | 0.61 | |
| DL | 0.561 | 0.556 | 0.544 | 0.554 | 0.534 | |
| 3y | Logistic | 0.601 | 0.609 | 0.601 | 0.639 | 0.634 |
| MLP | 0.552 | 0.552 | 0.552 | 0.574 | 0.592 | |
| SMO.PolyKernel | 0.64 | 0.644 | 0.641 | 0.628 | 0.592 | |
| RandomForest | 0.623 | 0.631 | 0.612 | 0.619 | 0.602 | |
| DL | 0.551 | 0.555 | 0.536 | 0.530 | 0.525 | |
| 5y | Logistic | 0.662 | 0.676 | 0.664 | 0.645 | 0.667 |
| MLP | 0.574 | 0.577 | 0.575 | 0.574 | 0.606 | |
| SMO.PolyKernel | 0.674 | 0.686 | 0.644 | 0.593 | 0.598 | |
| RandomForest | 0.65 | 0.67 | 0.625 | 0.645 | 0.66 | |
| DL | 0.562 | 0.618 | 0.56 | 0.523 | 0.517 |
Table 5.
The performance of different machine learning and DL models for hospitalization prediction using the heart datasets
| Prediction window | Algorithm | Precision | Recall | F-Measure | AUROC | AUPRC |
|---|---|---|---|---|---|---|
| 1y | Logistic | 0.777 | 0.75 | 0.76 | 0.695 | 0.774 |
| MLP | 0.705 | 0.656 | 0.676 | 0.726 | 0.833 | |
| SMO.PolyKernel | 0.688 | 0.688 | 0.688 | 0.543 | 0.674 | |
| RandomForest | 0.599 | 0.719 | 0.653 | 0.56 | 0.71 | |
| DL | 0.611 | 0.781 | 0.685 | 0.50 | 0.50 | |
| 3y | Logistic | 0.588 | 0.587 | 0.588 | 0.597 | 0.589 |
| MLP | 0.62 | 0.619 | 0.619 | 0.66 | 0.647 | |
| SMO.PolyKernel | 0.64 | 0.64 | 0.636 | 0.633 | 0.587 | |
| RandomForest | 0.607 | 0.608 | 0.603 | 0.673 | 0.68 | |
| DL | 0.505 | 0.576 | 0.50 | 0.593 | 0.564 | |
| 5y | Logistic | 0.654 | 0.658 | 0.654 | 0.66 | 0.641 |
| MLP | 0.649 | 0.648 | 0.648 | 0.68 | 0.675 | |
| SMO.PolyKernel | 0.671 | 0.668 | 0.646 | 0.631 | 0.598 | |
| RandomForest | 0.715 | 0.71 | 0.696 | 0.741 | 0.745 | |
| DL | 0.537 | 0.554 | 0.499 | 0.565 | 0.547 |
Predictive models
Models based on RF algorithm and logistic regression were persistently the best performing models based on traditional ML algorithms across three organ types when considering the AUROC as the performance measure. The only exception was the 1-year prediction window in heart dataset with MLP demonstrating the best performing models. Focusing on the ML models based on kidney datasets suggested that long-term hospitalization prediction was more difficult than shortterm. The average AUROC of models predicting 1-year hospitalization after kidney transplantation was 0.58 ± 0.03. Average AUROC was 0.50 ± 0.037 and 0.54 ± 0.048 for 3- and 5-year outcomes, respectively. However, this was not the case for the models based on liver datasets. For predicting hospitalization after liver transplantation, in this study, the 1-year outcome prediction showed a performance of 0.57 ± 0.031 on average. Average AUROC was 0.61 ± 0.028 and 0.61 ± 0.036 for 3- and 5-year outcomes, respectively. A similar trend was observed for the models based on heart datasets: 0.631 ± 0.092, 0.64 ± 0.033, and 0.678 ± 0.046 for 1-, 3-, and 5-year outcomes, respectively. Thus, overall organ types and outcome prediction windows, the models based on long-term (5years) hospitalization prediction for heart transplantation, on average, resulted in a better performance.
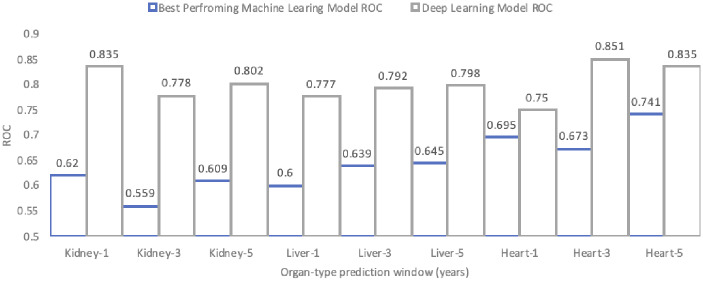
DL models did not outperform traditional ML models across organ types and prediction windows (Figure 2). The average AUROC of DL models across all three prediction windows for kidney transplantation was 0.5455 ± 0.018, for liver transplantation was 0.5358 ± 0.016, and for heart transplantation was 0.5528 ± 0.047. The same trend observed in the ML models’ performance was observed in the DL models. In case of the kidney datasets, DL models demonstrated better prediction of short-term hospitalization outcomes. In case of the liver and heart datasets, longer prediction windows from the initial transplantation enabled more accurate predictive modeling. The best performing DL model across all organ types and prediction windows was for 3-year hospitalization after heart transplantation with an AUROC of 0.593.
Figure 2.
The comparison between best performing machine learning models and DL models in terms of AUROC across all organ types and prediction windows.
Interpreting DL models using SHAP
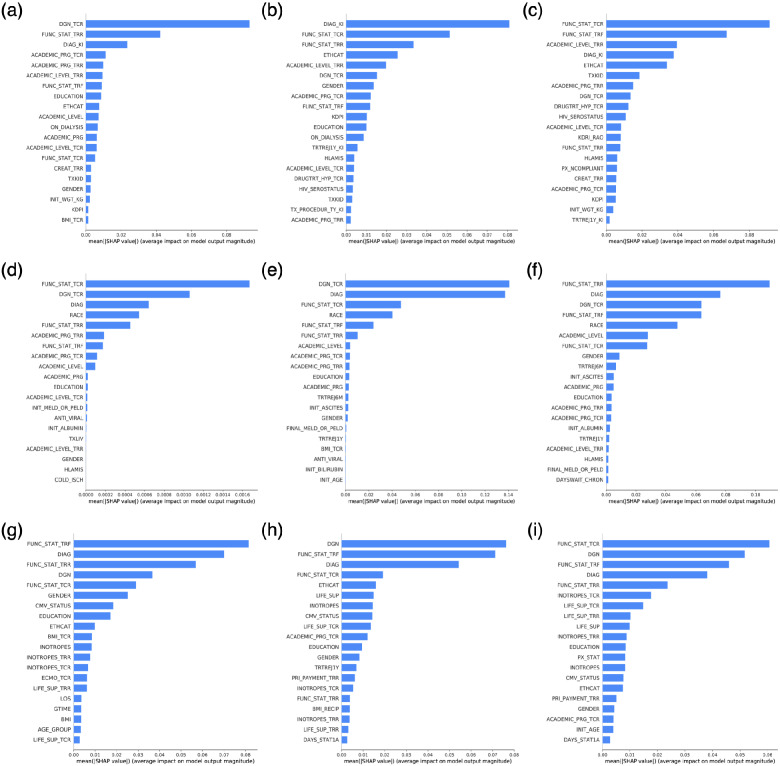
The result of applying SHAP on DL models in this study is demonstrated in terms of variable importance (Figure 3). The bar plots in each row correspond to an organ type: kidney, liver, and heart. The bar plots in each column refer to the prediction of 1-, 3-, and 5-year outcomes. Variables in these plots are ranked (descending) based on importance. The more important a variable is, the larger magnitude of impact on the model’s output it is associated with. Note that the produced SHAP explanations in this study are global (i.e., the explanation is provided regarding the whole samples in the dataset rather than a specific sample) by summing SHAP value magnitudes over all samples in the dataset. Also, to lower the computational cost, we summarized the samples to 50 weighted samples using the k-means algorithm (25 for heart 1-year dataset) [51]. The corresponding SHAP summary plots are available in Supplementary Material. Considering all nine models across organ types and prediction windows suggests that, in general, the functional status of the recipient (at the time of listing, transplantation, and follow-up) along with the primary diagnoses (at the time of listing and transplantation), ethnicity, and race are among the most important variables.
Figure 3.
SHAP results of deep learning models across all organ types and prediction windows: (A) kidney 1-y prediction window, (B) kidney 3-y prediction window, (C) kidney 5-y prediction window, (D) liver 1-y prediction window, (E) liver 3-y prediction window, (F) liver 5-y prediction window, (G) heart 1-y prediction window, (H) heart 3-y prediction window, and (I) heart 5-y prediction window.
DISCUSSION
Based on the current analyses, although deep learning did not outperform traditional methods, its potential predictive utility should not be underestimated. The data sets in this study were small with a mixed of numeric and categorical variables. Deep learning is known to be “data-hungry” and might not be the best approach for all predictive modeling problems. In a follow-up study on whole UNOS data, we are re-examining the performance of deep learning models in comparison to traditional ML methods in the presence of larger number of samples. Results also represent an important development in the use of ML models with pediatric transplant data as prior research found only poor to fair predictive utility and sensitivity [16]. Similar studies in adult populations have similarly reported lower predictive utility[29–31, 52–54]. Accuracy of DL models here outperformed a recent examination of pediatric liver transplantation using the Studies of Pediatric Liver Transplantation data and a RF decision tree approach to ML [15]. Importantly, DL models offer increased predictive utility through the examination and evaluation of complex relationships among variables as important pathways (e.g., neurons activation) for improving outcome prediction [42]. With gains in the complexity and predictive accuracy, DL models become increasingly complex and challenging to interpret the influence of individual variables, for example. In the current study, SHAP values [48] were examined to aid in the interpretability of DL models and the influence of individual variables within the models.
Medical variables
Primary pretransplant diagnosis and functional status of the patient at TCR and TRR were consistently the most predictive medical factors across prediction windows and organ types. Functional status across organ types and UNOS data collection forms is a measure of the health–related quality of life of the patient. This variable is recorded as a percentage ranging from 10% to 100% with responses related to inpatient status, ability to carry out daily living activities, and evidence of the impact of disease symptoms on daily activities. Primary pretransplant diagnosis includes hundreds of possible diagnoses across the three organ types.
Predictive medical variables for kidney recipients included dialysis status, creatinine levels, received treatment for hypertension at TCR, Kidney Donor Risk Index, and treatment for rejection episodes within 1-year post-transplant. Hospitalization for liver transplant recipients was predicted by the initial or final model for end-stage liver disease (patients age 12 years and older) or pediatric end-stage liver disease (younger than 12 years of age) scores, ascites at TCR, albumin levels at TCR, and treatment of rejection within 6 and 12 months post-transplant. Body mass index, days status 1A (most urgent status on transplant waiting list), use of inotropes with acute decompensated heart failure, and other forms of life support including ventricular assist devices during the pretransplant period predicted hospitalization for heart transplant patients. Most of these variables indicate severity of disease at the time of listing and/or transplantation however use of ML brings a unique set of variables out of the many which demonstrate good predictive ability and can help prognosticate the risk of complications in the post-transplant period.
Importantly, physician- or transplant team-reported noncompliance (UNOS variable of PX_NCOMPLIANT, “Recipient Noncompliant During this Follow-Up Period”) was a variable included on UNOS TRF forms from late 1999 to 2007. Within our data, this variable did not have >50% values and therefore had missing values imputed through MissForest, consistent with bestpractices in ML approaches [37–39]. The removal of this variable despite apparent predictive utility in this data and prior studies [11], and inclusion of adherence in other bigdata studies of pediatric post-transplant outcomes [17], points to the need for inclusion of an adherence measure within UNOS data. Nonadherence is associated with increased number and frequency of hospitalizations, the need for biopsies testing for organ rejection, and even mortality [4, 6–10, 55]. Although in need of continued validation across organ types, the Medication Level Variability Index (MLVI) has shown promise as a measure of nonadherence as higher MLVI scores have predicted poor post-transplant outcomes in samples of pediatric heart, liver, and kidney transplant recipients [4, 56–60]. Inclusion of measures of nonadherence in UNOS TRF forms, such as yearly patient MLVI and other measures of medication adherence, would provide a leap forward in post-transplant monitoring, care, and identification of high-risk patients.
Psychosocial variables
Numerous social, familial, and patient variables were found to be predictive of outcomes through DL models, across prediction windows, and within organ types. Across organ types and prediction windows, patient age, gender, race, and ethnicity each provided predictive utility to the DL models. Age and gender were both identified in numerous post-transplant models as predictive of hospitalizations. Older age at transplant and current patient age have been found to predict lower post-transplant health–related quality of life [61], more rejection episodes (both acuteand chronic) [62–64], and greater mortality riskwhen compared toyounger children across organ type [55, 65]. Female patients have reported poor post-transplant outcomes in studies of pediatric kidney [55, 66, 67] and heart [68] recipients.
Academic progress and level were important predictors in DL models. While these variables are likely highly correlated with functional status, their inclusion in the models suggests additional predictive utility beyond a proxymeasure of health–related quality of life. Social factors identified as predictive of hospitalizations in the current study included race, ethnicity, and socio-economic status (SES). Race/ethnicity and SES have been among the most studied factors predicting post-transplant outcomes and medication adherence [11], and numerous studies have identified significant associations between these social factors and medical outcomes in pediatric kidney, liver, and heart patients [13, 66–68]. Correlates of lower SES such as single-parent households, lower parental education, and receipt of public insurance have been reported as associated with poorer post-transplant outcomes [13, 22, 66, 69].
Limitations and future direction
One of the most important limitations of this study was the sample size. After preprocessing the data and only including the features with lower rates of missing values in the models in this study, the number of remained records in each prediction window of each organ type is pretty limited, especially in the case of heart data sets. DL models usually have more trainable parameters in comparison to the number of samples they are trained on. Thus, in theory, it is suggested to use some form of regularization to control the generalization error and reduce complexity [70]. However, building DL models on larger number of data points can potentially enhance the reliability and generalizability of the models. Also, the majority of features included in the predictive models in this study were categorical features. For the means of predictive modeling, we converted these categorical variables to numerical ones by assigning numbers to each category of each feature. Although the one-hot coding scheme might have been a better choice for the goal of this study, especially in terms of interpretations, the small sample size, the number of categorical variables, and the number of categories in each of these variables would not allow for inducing more sparsity. This would in turn lower the predictive accuracy of the models. Also, for some of the patients a complete record of follow-up data for a specific prediction window might have not been available. Thus, we assumed the outcome will not change in the subsequent years. While the current study serves as a proof of concept and informs the potential advantages of predictive modeling for future works, to address the aforementioned limitations, we are extending this research by modeling the complete UNOS data where we have more data points available. This will allow us to exclude those patients with incomplete follow-up data in each prediction window and have separate training, validation, and test sets for the learning process, hyperparameter tuning, and performance analysis.
CONCLUSIONS
Patients and their families experience varying challenges throughout the pre- and post-transplant period. The pediatric transplant service environment is rich in health, medical, patient, and family data, which can be collected from traditional EHR, administrative data (e.g., UNOS), unstructured clinical data (e.g., text from clinical notes), and even prospective data collection from patients and families. ML techniques and a DL approach to modeling have the ability to model evolving and heterogeneous patient-level data over time, identify predictors of poor outcomes, and inform care. Uses of ML in pediatric transplantation have numerous applications including decision-support systems and even development of data-driven assessments. A foundation for a physician and transplant team decision-support system is needed and will assist in identifying patients at greatest risk, the optimal time to intervene, and modifiable post-transplant factors [15]. These approaches can be powerful tools to aid in the mission of the transplant team to enhance and sustain health–related quality of life for these children and their families directly affecting patient and allograft survival.
FUNDING
This work was supported by University of Florida and Florida State University Clinical and Translational Science Institute with the National Center for Translational Science of the National Institutes of Health grant number 2UL1TR001427.
SUPPLEMENTARY MATERIAL
Supplementary material is available at JAMIA Open online.
AUTHOR CONTRIBUTIONS
All authors contributed significantly to the conceptualization, design, data collection and analysis, interpretation of results, and/or the development of the manuscript. All the authors of this publication have met the International Committee of Medical Journal Editors (ICMJE) criteria for authorship. M.O.K., Z.H., and D.D.contributed to study conceptualization and design. M.O.K., S.N.P., D.G., Z.H., and D.D. contributed to data acquisition, analysis, and/or interpretation. M.O.K., S.N.P., D.G., and Z.H. contributed to manuscript drafting.
CONFLICT OF INTEREST
The authors have no competing interests to declare.
DATA AVAILABILITY
The data underlying this article cannot be shared publicly due to privacy concerns and private health information of individuals receiving care from a single transplant center in the southwestern United States. However, this data is contained within the national United Network for Organ Sharing (UNOS, U.S. Department of Health and Human Services) database found at https://optn.transplant.hrsa.gov/data/.
Supplementary Material
REFERENCES
- 1.Hart A, Smith JM, Skeans MA, et al. OPTN/SRTR 2018 annual data report: kidney.Am J Transplant 2020; 20 (s1): 20–130. [DOI] [PubMed] [Google Scholar]
- 2.Kwong A, Kim WR, Lake JR, et al. OPTN/SRTR 2018 annual data report: liver.Am J Transplant 2020; 20 (s1): 193–299. [DOI] [PubMed] [Google Scholar]
- 3.Colvin M, Smith JM, Hadley N, et al. OPTN/SRTR 2018 annual data report: heart.Am J Transplant 2020; 20 (s1): 340–426. [DOI] [PubMed] [Google Scholar]
- 4.Shemesh E, Bucuvalas JC, Anand R, et al. The medication level variability index (MLVI) predicts poor liver transplant outcomes: a prospective multi-site study.Am J Transplant 2017; 17 (10): 2668–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molmenti E, Mazariegos G, Bueno J, et al. Noncompliance after pediatric liver transplantation. Transplant Proc 1999; 31 (1-2): 408. [DOI] [PubMed] [Google Scholar]
- 6.Shemesh E, Shneider BL, Emre S. Adherence to medical recommendations in pediatric transplant recipients: time for action. Pediatr Transplant 2008; 12 (3): 281–3. [DOI] [PubMed] [Google Scholar]
- 7.Oliva M, Singh TP, Gauvreau K, et al. Impact of medication non-adherence on survival after pediatric heart transplantation in the U.S.A. J Heart Lung Transplant 2013; 32 (9): 881–8. [DOI] [PubMed] [Google Scholar]
- 8.Kelly DA. Current issues in pediatric transplantation. Pediatr Transplant 2006; 10 (6): 712–20. [DOI] [PubMed] [Google Scholar]
- 9.Shemesh E, Annunziato RA, Shneider BL, et al. Improving adherence to medications in pediatric liver transplant recipients. Pediatr Transplant 2008; 12 (3): 316–23. [DOI] [PubMed] [Google Scholar]
- 10.Shemesh E, Shneider BL, Savitzky JK, et al. Medication adherence in pediatric and adolescent liver transplant recipients. Pediatrics 2004; 113 (4): 825–32. [DOI] [PubMed] [Google Scholar]
- 11.Killian MO. Psychosocial predictors of medication adherence in pediatric heart and lung organ transplantation.Pediatr Transplant 2017; 21 (4): e12899. [DOI] [PubMed] [Google Scholar]
- 12.Killian MO, Schuman DL, Mayersohn GS, et al. Psychosocial predictors of medication non‐adherence in pediatric organ transplantation: a systematic review.Pediatr Transplant 2018; 22 (4)p. [DOI] [PubMed] [Google Scholar]
- 13.Alonso EM, Martz K, Wang D, The Studies of Pediatric Liver Transplantation (SPLIT) Functional Outcomes Group (FOG), et al. Factors predicting health‐related quality of life in pediatric liver transplant recipients in the functional outcomes group.Pediatr Transplant 2013; 17 (7): n/a–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarwal MM, Bagga A. Quality of life after organ transplantation in children.Curr Opin Organ Transplant 2013; 18 (5): 563–8. [DOI] [PubMed] [Google Scholar]
- 15.Wadhwani SI, Hsu EK, Shaffer ML, et al. Predicting ideal outcome after pediatric liver transplantation: an exploratory study using machine learning analyses to leverage studies of pediatric liver transplantation data.Pediatr Transplant 2019; 23 (7): e13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller R, Tumin D, Cooper J, et al. Prediction of mortality following pediatric heart transplant using machine learning algorithms.Pediatr Transplant 2019; 23 (3): e13360. [DOI] [PubMed] [Google Scholar]
- 17.Srinivas TR, Taber DJ, Su Z, et al. Big data, predictive analytics, and quality improvement in kidney transplantation: a proof of concept.Am J Transplant 2017; 17 (3): 671–81. [DOI] [PubMed] [Google Scholar]
- 18.Weiss ES, Allen JG, Arnaoutakis GJ, et al. Creation of a quantitative recipient risk index for mortality prediction after cardiac transplantation (IMPACT). AnnThoracSurg 2011; 92 (3): 914–22. [DOI] [PubMed] [Google Scholar]
- 19.Dickinson DM, Shearon TH, O'Keefe J, et al. SRTR center‐specific reporting tools: posttransplant outcomes. Am J Transplant 2006; 6 (5p2): 1198. [DOI] [PubMed] [Google Scholar]
- 20.Dharnidharka VR, Lamb KE, Zheng J, et al. Across all solid organs, adolescent age recipients have worse transplant organ survival than younger age children: a US national registry analysis.Pediatr Transplant 2015; 19 (5): 471–6. [DOI] [PubMed] [Google Scholar]
- 21.Dharnidharka VR, Lamb KE, Zheng J, et al. Lack of significant improvements in long-term allograft survival in pediatric solid organ transplantation: a US national registry analysis. Pediatr Transplant 2015; 19 (5): 477–83. [DOI] [PubMed] [Google Scholar]
- 22.Tumin D, McConnell PI, Galantowicz M, et al. Reported nonadherence to immunosuppressive medication in young adults after heart transplantation: a retrospective analysis of a National Registry. Transplantation 2017; 101 (2): 421–9. [DOI] [PubMed] [Google Scholar]
- 23.Yoo KD, Noh J, Lee H, et al. A machine learning approach using survival statistics to predict graft survival in kidney transplant recipients: a multicenter cohort study.Sci Rep 2017; 7 (1): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravikumar A, Saritha R, Chandra V. Recent trends in computational prediction of renal transplantation outcomes. IJCA 2013; 63 (12): 33–7. [Google Scholar]
- 25.Lau L, Kankanige Y, Rubinstein B, et al. Machine-learning algorithms predict graft failure after liver transplantation. Transplantation 2017; 101 (4): e125–e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrero JI, Lucena JF, Quiroga J, et al. Liver transplant recipients older than 60 years have lower survival and higher incidence of malignancy. Am J Transplant 2003; 3 (11): 1407–12. [DOI] [PubMed] [Google Scholar]
- 27.Hong Z, Wu J, Smart G, et al. Survival analysis of liver transplant patients in Canada 1997–2002. TransplantProc 2006; 38 (9): 2951–6. [DOI] [PubMed] [Google Scholar]
- 28.Raji C, Chandra SV. Artificial neural networks in prediction of patient survival after liver transplantation. J Health Med Inform 2016; 7 (1): 1–7. [Google Scholar]
- 29.Dag A, Oztekin A, Yucel A, et al. Predicting heart transplantation outcomes through data analytics. Decis Support Syst 2017; 94: 42–52. [Google Scholar]
- 30.Dag A, Topuz K, Oztekin A, et al. A probabilistic data-driven framework for scoring the preoperative recipient-donor heart transplant survival. Decis Support Syst 2016; 86: 1–12. [Google Scholar]
- 31.Medved D, Nugues P, Nilsson J. Selection of an optimal feature set to predict heart transplantation outcomes. In: 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); 2016;IEEE. [DOI] [PubMed]
- 32.Oztekin A. An analytical approach to predict the performance of thoracic transplantations. J CENTRUM Cathedra 2012; 5 (2): 185–206. [Google Scholar]
- 33.Oztekin A, Delen D, Kong ZJ. Predicting the graft survival for heart–lung transplantation patients: an integrated data mining methodology. Int J Med Inform 2009; 78 (12): e84–e96. [DOI] [PubMed] [Google Scholar]
- 34.Rajanayagam J, Frank E, Shepherd RW, et al. Artificial neural network is highly predictive of outcome in paediatric acute liver failure. Pediatr Transplant 2013; 17 (6): 535–42. [DOI] [PubMed] [Google Scholar]
- 35.Castelvecchi D. Can we open the black box of AI? Nat News 2016; 538 (7623): 20–3. [DOI] [PubMed] [Google Scholar]
- 36.LaRosa C, Baluarte HJ, Meyers KEC. Outcomes in pediatric solid-organ transplantation. Pediatr Transplant 2011; 15 (2): 128–41. [DOI] [PubMed] [Google Scholar]
- 37.Stekhoven DJ, Bühlmann P. MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics 2012; 28 (1): 112–8. [DOI] [PubMed] [Google Scholar]
- 38.Breiman L. Random forests. Mach Learn 2001; 45 (1): 5–32. [Google Scholar]
- 39.Payrovnaziri SN, Xing A, Shaeke S, et al. The impact of missing value imputation on the interpretations of predictive models: a case study on one-year mortality prediction in ICU patients with acute myocardial infarction. medRxiv 2020.06.06.20124347, 2020. doi: 10.1101/2020.06.06.20124347.
- 40.Hall M, Frank E, Holmes G, et al. The WEKA data mining software: an update. Sigkdd Explor Newsl 2009; 11 (1): 10–8. [Google Scholar]
- 41.Miotto R, Wang F, Wang S, et al. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform 2018; 19 (6): 1236–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau MM, Lim KH. Review of adaptive activation function in deep neural network.In:2018 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES); 2018;IEEE.
- 43.Nair V, Hinton GE. Rectified linear units improve restricted Boltzmann machines. In: Proceedings of the 27th International Conference on Machine Learning; 2010; Haifa, Israel.
- 44.Duchi J, Hazan E, Singer Y. Adaptive subgradient methods for online learning and stochastic optimization. J Mach Learn Res 2011; 12 (7): 2121–59. [Google Scholar]
- 45.Liu B, Wei Y, Zhang Y, et al. Deep neural networks for high dimension, low sample size data. In: IJCAI; August 2017; Melbourne, Australia. [Google Scholar]
- 46.Samek W, Montavon G, Andrea A, et al. Explainable AI: Interpreting, Explaining and Visualizing Deep Learning. Vol. 11700. Switzerland: Springer Nature; 2019. [Google Scholar]
- 47.Payrovnaziri SN, Chen Z, Rengifo-Moreno P, et al. Explainable artificial intelligence models using real-world electronic health record data: a systematic scoping review. J Am Med Inform Assoc 2020; 27 (7): 1173–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. In: Advances in neural information processing systems. Cambridge, MA: MIT Press; 2017.
- 49.Shapley LS. A value for n-person games. Contribut Theory Games 1953; 2 (28): 307–17. [Google Scholar]
- 50.Ribeiro MT, Singh S, Guestrin C.Why should I trust you? Explaining the predictions of any classifier. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; August 2016; San Francisco, CA.
- 51.MacQueen J. Some methods for classification and analysis of multivariate observations. In: Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability;1967;Oakland, CA, USA.
- 52.Nilsson J, Ohlsson M, Höglund P, et al. The international heart transplant survival algorithm (IHTSA): a new model to improve organ sharing and survival. PLoS One 2015; 10 (3): e0118644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoon J, Zame WR, Banerjee A, Cadeiras M, Alaa AM, van der Schaar M. Personalized survival predictions for cardiac transplantation via trees of predictors. 2017; arXiv preprint arXiv:1704.03458. [DOI] [PMC free article] [PubMed]
- 54.Senanayake S, White N, Graves N, et al. Machine learning in predicting graft failure following kidney transplantation: a systematic review of published predictive models. Int J Med Inform 2019; 130: 103957. [DOI] [PubMed] [Google Scholar]
- 55.Bobanga ID, Vogt BA, Woodside KJ, et al. Outcome differences between young children and adolescents undergoing kidney transplantation. JPediatrSurg 2015; 50 (6): 996–9. [DOI] [PubMed] [Google Scholar]
- 56.Annunziato RA, Emre S, Shneider B, et al. Adherence and medical outcomes in pediatric liver transplant recipients who transition to adult services. Pediatr Transplant 2007; 11 (6): 608–14. [DOI] [PubMed] [Google Scholar]
- 57.de Oliveira JTP, Kieling CO, da Silva AB, et al. Variability index of tacrolimus serum levels in pediatric liver transplant recipients younger than 12 years: non-adherence or risk of non-adherence? Pediatr Transplant 2017; 21 (8): e13058. [DOI] [PubMed] [Google Scholar]
- 58.Fredericks EM, Lopez MJ, Magee JC, et al. Psychological functioning, nonadherence and health outcomes after pediatric liver transplantation. Am J Transplant 2007; 7 (8): 1974–83. [DOI] [PubMed] [Google Scholar]
- 59.Fredericks EM, Magee JC, Opipari-Arrigan L, et al. Adherence and health‐related quality of life in adolescent liver transplant recipients. Pediatr Transplant 2008; 12 (3): 289–99. [DOI] [PubMed] [Google Scholar]
- 60.Shemesh E, Fine RN. Is calculating the standard deviation of tacrolimus blood levels the new gold standard for evaluating non-adherence to medications in transplant recipients? Pediatr Transplant 2010; 14 (8): 940–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parmar A, Vandriel SM, Ng VL. Health‐related quality of life after pediatric liver transplantation: a systematic review. Liver Transplant 2017; 23 (3): 361. [DOI] [PubMed] [Google Scholar]
- 62.Berquist RK, Berquist WE, Esquivel CO, et al. Adolescent non-adherence: prevalence and consequences in liver transplant recipients. Pediatr Transplant 2006; 10 (3): 304–10. [DOI] [PubMed] [Google Scholar]
- 63.Berquist RK, Berquist WE, Esquivel CO, et al. Non-adherence to post-transplant care: prevalence, risk factors and outcomes in adolescent liver transplant recipients. Pediatr Transplant 2008; 12 (2): 194–200. [DOI] [PubMed] [Google Scholar]
- 64.Shaw RJ, Palmer L, Blasey C, et al. A typology of non-adherence in pediatric renal transplant recipients. Pediatr Transplant 2003; 7 (6): 489–93. [DOI] [PubMed] [Google Scholar]
- 65.Tosi L, Federman M, Markovic D, et al. The effect of gender and gender match on mortality in pediatric heart transplantation. Am J Transplant 2013; 13 (11): 2996–3002. [DOI] [PubMed] [Google Scholar]
- 66.Laskin BL, Mitsnefes MM, Dahhou M, et al. The mortality risk with graft function has decreased among children receiving a first kidney transplant in the United States. Kidney Int 2015; 87 (3): 575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Foster BJ, Dahhou M, Zhang X, et al. Change in mortality risk over time in young kidney transplant recipients. Am J Transplant 2011; 11 (11): 2432–42. [DOI] [PubMed] [Google Scholar]
- 68.Schumacher KR, Almond C, Singh TP, et al. Predicting graft loss by 1 year in pediatric heart transplantation candidates: an analysis of the Pediatric Heart Transplant Study database. Circulation 2015; 131 (10): 890–8. [DOI] [PubMed] [Google Scholar]
- 69.Schaeffner ES, Mehta J, Winkelmayer WC. Educational level as a determinant of access to and outcomes after kidney transplantation in the United States. Am J Kidney Dis 2008; 51 (5): 811–8. [DOI] [PubMed] [Google Scholar]
- 70.Zhang C, Bengio S, Hardt M, Recht B, Vinyals O. Understanding deep learning requires rethinking generalization. 2016; arXiv:1611.03530.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publicly due to privacy concerns and private health information of individuals receiving care from a single transplant center in the southwestern United States. However, this data is contained within the national United Network for Organ Sharing (UNOS, U.S. Department of Health and Human Services) database found at https://optn.transplant.hrsa.gov/data/.