Abstract
Maize is one of the most important crops worldwide and is the number one arable crop in Portugal. A transition from the conventional farming system to organic agriculture requires optimization of cultivars and management, the interaction of plant–soil rhizosphere microbiota being pivotal. The objectives of this study were to unravel the effect of population genotype and farming system on microbial communities in the rhizosphere of maize. Rhizosphere soil samples of two open-pollinated maize populations (“SinPre” and “Pigarro”) cultivated under conventional and organic farming systems were taken during flowering and analyzed by next-generation sequencing (NGS). Phenological data were collected from the replicated field trial. A total of 266 fungi and 317 bacteria genera were identified in “SinPre” and “Pigarro” populations, of which 186 (69.9%) and 277 (87.4%) were shared among them. The microbiota of “Pigarro” showed a significant higher (P < 0.05) average abundance than the microbiota of “SinPre.” The farming system had a statistically significant impact (P < 0.05) on the soil rhizosphere microbiota, and several fungal and bacterial taxa were found to be farming system-specific. The rhizosphere microbiota diversity in the organic farming system was higher than that in the conventional system for both varieties. The presence of arbuscular mycorrhizae (Glomeromycota) was mainly detected in the microbiota of the “SinPre” population under the organic farming systems and very rare under conventional systems. A detailed metagenome function prediction was performed. At the fungal level, pathotroph–saprotroph and pathotroph–symbiotroph lifestyles were modified by the farming system. For bacterial microbiota, the main functions altered by the farming system were membrane transport, transcription, translation, cell motility, and signal transduction. This study allowed identifying groups of microorganisms known for their role as plant growth-promoting rhizobacteria (PGPR) and with the capacity to improve crop tolerance for stress conditions, allowing to minimize the use of synthetic fertilizers and pesticides. Arbuscular mycorrhizae (phyla Glomeromycota) were among the most important functional groups in the fungal microbiota and Achromobacter, Burkholderia, Erwinia, Lysinibacillus, Paenibacillus, Pseudomonas, and Stenotrophomonas in the bacterial microbiota. In this perspective, the potential role of these microorganisms will be explored in future research.
Keywords: maize, microbiota, rhizosphere, organic and conventional farming system, open-pollinated populations, next-generation sequencing
Introduction
Maize (Zea mays) is one of the most important cereal crops in human and animal diets worldwide. Together with rice and wheat, it provides at least 30% of the food calories to more than 4.5 billion people in 94 developing countries (Shiferaw et al., 2011). Aside from providing nutrients for humans and animals, maize serves as a basic raw material to produce starch, oil, protein, alcoholic beverages, food sweeteners, and fuel (Wu and Guclu, 2013).
According to FAOSTAT, from 2012 to 2017, the average world production per year of corn was around 1,036,263,896 t. In Portugal, maize is the most important arable crop, occupying an area of approximately 150,000 ha (ANPROMIS, 2019), producing an annual average of 826,417 Mg from 2012 to 2017 (FAOSTAT, 2019).
Due to new challenges related to the expansion of the human and animal populations allied with the global climatic changes, all opportunities for sustainably increasing the yield are relevant and should be developed. For example, improvements on both agronomic practices and breeding related with microorganism interactions (e.g., control of root diseases) and organic management are appellative proposals since the rhizosphere microbial communities are critically important for soil nitrogen cycling and plant productivity (Schmidt et al., 2016; Emmett et al., 2017; Wille et al., 2018).
In this context, alternatives that promote more resilient farming systems, increasing the economic activity of rural areas as well as preventing the significant loss of biodiversity in these areas, would contribute to more sustainable agriculture. In line with this thought, the traditional varieties are extremely important and should not be neglected (Altieri, 2004) since they present a genetic reservoir that can be used to select for advantages associated with the adaptability and resilience of cultivars to low-input and a permanently changing environment. One of the characteristics of interest in these varieties is the possibility of maintaining genetic diversity through open pollination. New genetic combinations may present new features or capabilities that will allow the plant population to respond with more resilience toward pests, diseases, or even the most adverse weather conditions (Garcia-Tejero et al., 2015). Therefore, traditional varieties can adapt themselves to the environment, having greater adaptability to external factors, increasing the crop population fitness (Altieri and Merrick, 1987; Lane and Jarvis, 2007).
Although traditional Portuguese varieties do not have high yields, they are still cultivated due to their high yield stability even under unfavorable conditions like drought (Garcia-Tejero et al., 2015; Leitão et al., 2019). It is important to note that these varieties also play an important role in the country’s rural economy, especially in the Central and Northern regions of Portugal, as their market value for bread making has increased thanks to their health benefits. This practice is also seen as a viable way to preserve the biodiversity of threatened farming systems (Vaz Patto et al., 2013).
Since 2002, knowledge about microbiota and the rhizosphere has increased exponentially; however, we are just beginning to understand the mechanisms of plant–microorganism interactions (Vandenkoornhuyse et al., 2015; Compant et al., 2019). According to Philippot et al. (2013), the rhizosphere is a hotspot of plant–microbe interactions with a profound influence on plant productivity, and all its functions are extremely important in terms of nutrition, health, and plant quality. Indeed, the rhizosphere is a critical interface that supports the exchange of resources between plants and their related soil environment. It is already known that several plants produce components that interact with the rhizosphere microbiota, thus forming a dynamic structure in which microbial diversity can be modified with soil composition, plant species, different genotypes within the same cultivar, and the development stage of the organism (Turner et al., 2013; Chaparro et al., 2014; Kandel et al., 2017). Moreover, the microbiota can help plants survive climate changes, modify tolerance to abiotic and biotic stresses, affect the plant–pathogen interactions, and change the nutrient contents inside the plant (Chadha et al., 2015). In addition to all these factors, management agricultural practices, the addition of fertilizers, the presence of pathogens, or extreme climatic conditions cause important effects on the microbial diversity composition (Andreote and Silva, 2017). All these factors are highly relevant to improve vigor, growth, and plant’s health (Muller et al., 2016).
In this context, this study aimed to unravel the effect of genotype and farming system on structural diversity and putative functions of the microbial communities in the rhizosphere of two open-pollinated maize populations (“SinPre” and “Pigarro”) cultivated under conventional and organic farming systems in Portugal. With this approach, groups of microorganisms with the potential to modulate the soil quality and fertility were identified and linked to specific conditions, thus potentially contributing to the increasing crop tolerance for stress conditions and to minimize the use of synthetic fertilizers and pesticides.
Materials and Methods
Germplasm Characteristics
A synthetic population of maize, “SinPre” (Sintético Precoce), was obtained through the crossing of 12 maize populations (10 Portuguese landraces and two American populations) using a polycross method based on “Nutica” experience (Mendes-Moreira et al., 2009). The 12 maize populations were open-pollinated in geographic isolation (from other maize) in 2009. The border was constituted by equal amounts of seeds of each population. Two equal sets of 12 rows were organized from the earliest to the latest flowering population and vice versa for the second set. With this spatial organization, the earliest populations were close to the latest populations. Per row, progenies were submitted to intensive selection among parents during continued cycles. Afterward, a bulk of the best ears was obtained and distributed to farms in different agroclimatic regions of Portugal. The purpose of this new synthetic population was to provide farmers with a new population, with high diversity that could be adapted to their needs, so that farmers can select the characteristics they appreciate the most (P. Mendes-Moreira, October 2019, personal communication). “Pigarro” is a Portuguese population used to produce maize bread that presents a strong emergency vigor, great tolerance at low temperatures, early flowering and fast drying of the grain, and with ripening group FAO 300. It has white and flint kernels and its ear is known for the high number of kernels per row (between 18 and 28) as well as a strong expression of fasciation (Mendes-Moreira et al., 2017).
Field Characterization and Agronomical Practices
Field trials were established at two locations in Coimbra, Portugal, that belong to Coimbra College of Agriculture: “Caldeirão” in organic (40°130′0.22″N, 8°260′47.69″W) and “Vagem grande” in conventional (40°13′16.2″N, 8°28′29.3″W), with a distance of approximately 2.4 km. Both locations have alluvial soils with a medium field texture; however, they can be differentiated between organic and conventional in the organic matter (1.8 vs. 0.8%), pH (6.4 vs. 6.7), available phosphorus (high vs. very high), and available potassium (very high vs. high) (Supplementary Table 1).
The maize seed used was produced in the organic location in 2018 and seeds were not treated. For both fields, the preceding crop was maize.
In conventional agriculture soil, tillage started on 15/05/2019 followed by the first fertilization on 21/05/2019. The fertilizer was distributed (12:20:12 NPK, 318 kg/ha) with a centrifugal fertilizer spreader and incorporated with a rotor tiller. On 24/05/2019, the fertilizer (12:20:12 NPK) was simultaneously incorporated at a rate of 592 kg/ha during sowing. Sowing was conducted with a single-seed sowing machine with nine plants per square meter, followed by a herbicide application (609.38 g/ha terbuthylazine + 121.88 g/ha mesotrione + 1,015.63 g/ha S-metolachlor). On 25/06/2019, a fertilizer application of 560 kg/ha with a composition of 40% nitrogen (N) and 14% sulfur (SO3) was done, followed by the application of herbicides (25.13 g/ha nicossulfuron + 375 g/ha terbuthylazine + 165 g/ha sulcotrione). Finally, a pilling up through mechanical weed control with a harrow helped to control weeds.
In organic and low-input farming, soil tillage was done on 13/05/2019. Sowing occurred on 14/05/2019. Sowing was conducted with a single-seed sowing machine with six plants per square meter. Weed control was carried out manually since sowing, until late June, when pilling up was finally done. No fertilizer was applied.
Germplasm Characterization
For 20 randomly chosen plants from each population, an adaptation of the HUNTERS descriptor was used based on the field data collected during the monitoring of the maize crop: height (H), height of first ear insertion (H1E), uniformity (U), root (R%), and stalk lodging percentage (S%). IBMSPSS® statistics program was used for phenotyping data analyses (IBM Corp, 2020).
Sampling of Maize Rhizosphere
The soil was collected at the flowering stage in the two maize populations: “SinPre” and “Pigarro”. For each population, samples were collected from the organic and conventional farming systems. Within each plot, three individual plants, separated by at least 5 m from each other, were selected. Entire plants were dug up with a soil monolith in the middle of them. Bulk soil was taken off the plant roots by vigorous shaking. Plant fine roots were collected from each plant, stored in cool temperature, and moved rapidly to the laboratory where rhizospheric soil samples (1–2 mm soil adhering to roots) were collected.
DNA Extraction and Sequencing
Total DNA was extracted using Nucleospin Soil Kit (Macherey Nagel, Düren, Germany) with Buffer SL1 in combination with Enhancer SX, according to manufacturer’s instructions. Internal transcribed spacer 2 (ITS2) region amplicon libraries and Illumina 16S ribosomal RNA (rRNA) genes were generated and sequenced at Genoinseq (Portugal). The DNA was amplified for the hypervariable regions with specific primers and further reamplified in a limited-cycle PCR reaction to add sequencing adapters and dual indexes. The first PCR reactions were performed using a pool of forward primers—ITS3NGS1_F-5′-CATCGATGAAGAACGCAG-3′, IT S3NGS2_F-5′-CAACGATGAAGAACGCAG-3′, ITS3NGS3_F-5′-CACCGATGAAGAACGCAG-3′, ITS3NGS4_F-5′-CATCG ATGAAGAACGTAG-3′, ITS3NGS5_F-5′-CATCGATGAAGA ACGTGG-3′, ITS3NGS10_F-5′-CATCGATGAAGAACGCTG-3′—and reverse primer ITS4NGS001_R-5′-TCCTSCGCTTA TTGATATGC-3′ for fungi (Tedersoo et al., 2014) and forward primer Bakt_341F-5′-CCTACGGGNGGCWGCAG-3′ and reverse primer Bakt_805R-5′-GACTACHVGGGTATCTAATCC-3′ for bacteria (Herlemann et al., 2011; Klindworth et al., 2013). The second PCR reaction added indexes and sequencing adapters to both ends of the amplified target region according to the manufacturer’s recommendations (Illumina, 2013). Negative PCR controls were included for all amplification procedures. PCR products were then one-step purified and normalized using a SequalPrep 96-well plate kit (Thermo Fisher Scientific, Waltham, United States) (Comeau et al., 2017), pooled, and pair-end sequenced in the Illumina MiSeq® sequencer with the V3 chemistry, according to the manufacturer’s instructions (Illumina, San Diego, CA, United States) at Genoinseq (Cantanhede, Portugal).
In silico Functional Analysis
Prediction of functional bacterial and fungal diversity within the 16S rRNA and ITS2 sequence libraries was performed using PICRUSt (Langille et al., 2013) and FUNGuild (Nguyen et al., 2015), respectively. PICRUSt predicts the potential metagenomic gene content of a 16S amplicon library based on genomic information of the bacteria represented within the Greengenes 16S database. To perform the process within the PICRUSt program, samples derived from the QIIME2 process, before taxonomic assignment, were selected and grouped into 97% operational taxonomic units (OTUs) against the Greengenes database v.13.8. The nearest sequence taxon index (NSTI) within the PICRUSt pipeline was also calculated as a quality control to validate the accuracy of the predicted functional annotations. FUNGuild assigns trophic modes to fungal taxa based on a comparison to a curated database of fungal lifestyles (sensu Tedersoo et al., 2014): pathotroph, symbiotroph, and saprotroph. Trophic mode refers to the mechanisms through which organisms obtain resources, providing putative information on the ecology of such organisms (Nguyen et al., 2015). Functional assignments through FUNGuild are based on taxonomy and are possible only if the taxa have been classified at the genus level or if the taxa belong to a fungal group with an exclusive lifestyle. Input data for FUNGuild was the OTU table.
Statistical and Bioinformatics Analysis
Raw reads were extracted from the IlluminaMiSeq® System in fastq format and quality-filtered with PRINSEQ version 0.20.4 to remove sequencing adapters, reads with less than 100 bases for the ITS2 region and 150 bases for the 16S rRNA gene, and trim bases with an average quality lower than Q25 in a window of 5 bases (Schmieder and Edwards, 2011). The forward and reverse reads were merged by overlapping paired-end reads with Adapter Removal version 2.1.5 using default parameters (Schubert et al., 2016). After sequencing, the bacterial and fungal communities were analyzed using the QIIME software package. Chimeric sequences were removed using the consensus method and clustered in OTUs at 99% using a reference. Taxonomy was assigned to bacterial and fungal OTU sequences using Greengenes v13.8 and UNITEv.7.2, respectively. The phylogenetic classification was performed to the genus level. The rarefaction curves obtained were saturated for each sample, demonstrating that the OTUs recovered were representative of the bacterial and fungi diversity, supporting a robust analysis.
The alpha diversity indexes Shannon index (H′), Simpson (D), and Chao1 were calculated with the Phyloseq package to include in MicrobiomeAnalyst (Dhariwal et al., 2017). The statistical significance of grouping based on experimental factor was estimated using t-test/analysis of variance (ANOVA, P < 0.05) to determine differences in the alpha diversity indexes among variables: “SinPre” and “Pigarro” populations and conventional and organic farming systems. A non-supervised principal component analysis (PCA) was performed to compare the bacterial and fungal community structures. Statistical analyses were performed with ANOVA at P < 0.05 using R software v.4.0. Venn diagrams were generated with Venny 2.1 (Oliveros, 2007–2015) to identify shared and unique taxa of each population according to the farming system. To identify fungal and bacterial taxa that differed in the relative abundance among population genotypes and farming systems in the rhizosphere of maize, a linear discriminant analysis (LDA) was performed combined with effect size (LEfSe) using a graphical interface in Galaxy version 1.0 (The Huttenhower Lab, 2018). A P-value of < 0.05 and a score ≥ 2.0 were considered significant in Kruskal–Wallis and pairwise Wilcoxon tests, respectively.
Results
Germplasm Agronomic Characterization
The averages of the data collected in the field according to the HUNTERS (Mendes-Moreira et al., 2017) descriptor for both populations (“SinPre” and “Pigarro”) in conventional and organic farming are detailed in Supplementary Table 2. The phenotypic characterization indicates that “SinPre” in conventional farming was significantly higher than in organic farming for plant and first ear height, but no differences were observed for “Pigarro.” Within farming systems (conventional and organic), there were no significant differences between the tested populations (“SinPre” × “Pigarro”) for the measured parameters. Both populations showed values of uniformity between 2 and 4, and for the parameters angle (N), tassel (T), and ear insertion position (E), the values variated from 5 to 6. Root lodging (R) ranged from 0% (“SinPre” conventional) to 4.1% (“Pigarro” conventional). Stalk lodging (S) ranged from 10.5% (“Pigarro” organic) to 17.1% (“SinPre” conventional).
Fungal and Bacterial Rhizosphere Microbiota Associated With Two Maize Populations
The structural compositions of the fungi and bacterial communities associated with the rhizosphere soils of two traditional populations of maize grown were grouped into 973 fungal and 4,051 bacterial OTUs.
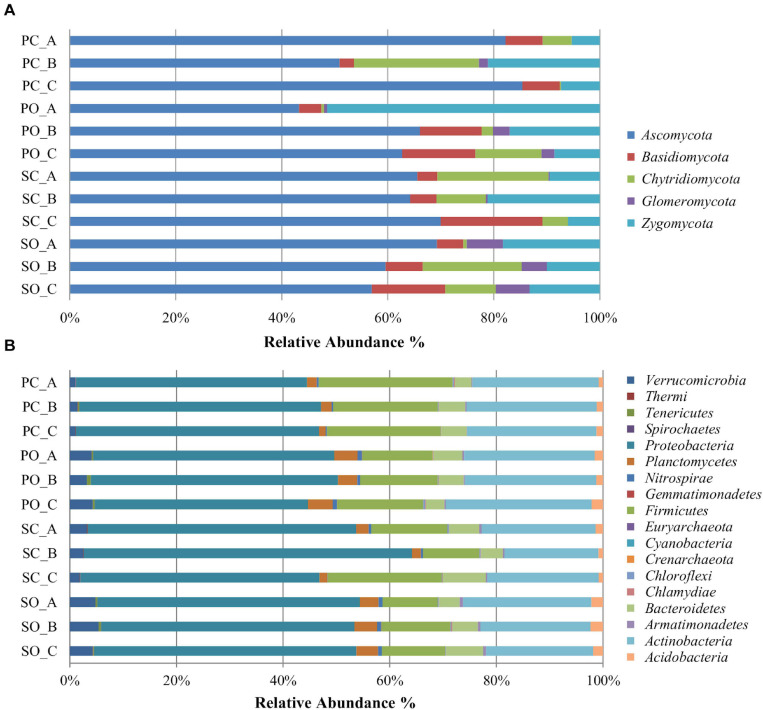
A total of five fungal phyla, 25 classes, 79, orders, 150 families, and 266 genera were identified (Supplementary Table 3). The most abundant and diverse phylum was Ascomycota (64%, comprising 144 genera), followed by Zygomycota (18%, comprising 12 genera), Basidiomycota (8%, comprising 87 genera), Chytridiomycota (8%, comprising 11 genera), and Glomeromycota (2% comprising 12 genera) (Figure 1A). The most abundant genera within Ascomycota were Odiodendron (13%), Fusarium (12%), and Aspergillus (11%); within Basidiomycota were Rhodotorula (18%), Puccinia (13%) Papiloterma (12%), and Conocybe (9%); within Chytridiomycota the genera Rhizoplictis (65%), Olpidium (17%), and Powellomyces (8%); within Glomeromycota were Dentiscutata (45%) Gigaspora (22%), and Paraglomus (15%); and within Zygomycota were Mortierella (39%), Rhizopus (39%), and Cunninghamella (13%) (Figure 2A). Overall, the most abundant genera were Oidiodendron and Fusarium (with 8% each), Rhizopus, Morteriella, and Aspergillus (with 7% each), Rhizophlyctis (5%), Alternaria and Penicillium (with 4% each), Podospora and Microdochium (with 3% each), and Cunninghamella, Coniochaeta, Exophiala, Talaromyces, Myrothecium, and Zopfiella (with 2% each), accounting for approximately 68% of the total diversity. Thirty-seven (14%) genera were shared by all samples; nevertheless, 22% of these bacterial taxa were rare since each represented less than 1% of the total diversity (Supplementary Table 3).
FIGURE 1.
Relative abundance of different fungal (A) and bacterial (B) phyla in the soil rhizosphere in both populations. PC_A, “Pigarro” conventional plant A; PC_B, “Pigarro” conventional plant B; PC_C, “Pigarro” conventional plant C; PO_A, “Pigarro” organic plant A; PO_B, “Pigarro” organic plant B; PO_C, “Pigarro” organic plant C; SC_A, “SinPre” conventional plant A; SC_B, “SinPre” conventional plant B; SC_C, “SinPre” conventional plant C; SO_A, “SinPre” organic plant A; SO_B, “SinPre” organic plant B; SO_C, “SinPre” organic plant C.
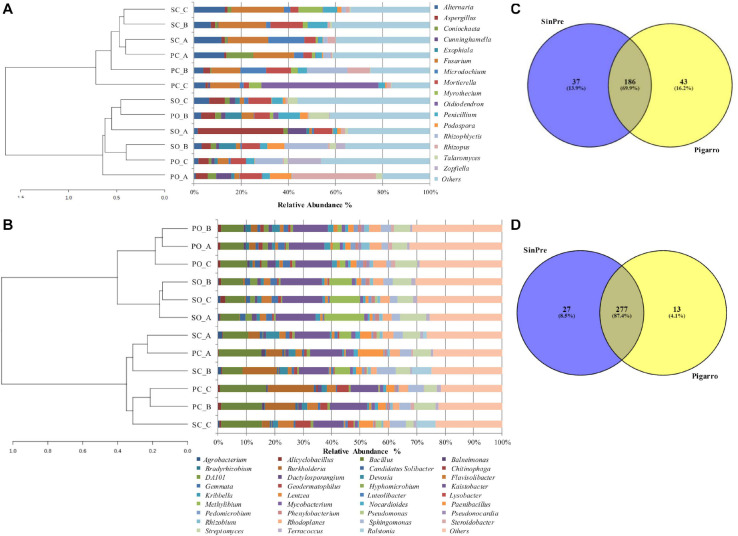
FIGURE 2.
Relative abundance of different fungal (A) and bacterial (B) genera in the soil rhizosphere in both populations representing the genera showing more than 1% relative abundance of all reads. The genera representing less than 1% of the total reads are grouped into “Others.” Venn diagrams showing the common and exclusive fungal (C) and bacterial (D) genera of the rhizosphere of the maize populations “SinPre” and “Pigarro.” PC_A, “Pigarro” conventional plant A; PC_B, “Pigarro” conventional plant B; PC_C, “Pigarro” conventional plant C; PO_A, “Pigarro” organic plant A; PO_B, “Pigarro” organic plant B; PO_C, “Pigarro” organic plant C; SC_A, “SinPre” conventional plant A; SC_B, “SinPre” conventional plant B; SC_C, “SinPre” conventional plant C; SO_A, “SinPre” organic plant A; SO_B, “SinPre” organic plant B; SO_C, “SinPre” organic plant C.
For the bacterial microbiota, a total of 18 phyla, 41 class, 72 orders, 158 families, and 317 genera were identified (Supplementary Table 4). The most abundant and diverse bacterial phylum was Proteobacteria (47%, comprising 119 genera), followed by Actinobacteria (23%, comprising 83 genera), Firmicutes (16%, comprising 59 genera), and Bacteroidetes (5%, comprising 36 genera) (Figure 1B). At the genus level, the most abundant by phylum were Kaistobacter (26%), Burkholderia (10%), Methylibium (9%), Sphingomonas (8%), and Rhodoplanes (7%) within Proteobacteria; Streptomyces (26%), Geodermatophilus (7%), and Mycobacterium (6%) within Actinobacteria; Bacillus (65%), Paenibacillus (18%), and Alicylobacillus (5%) within Firmicutes; and Flavisolibacter (51%) and Chitinophaga (14%) within Bacteroidetes (Figure 2B). Overall, the most abundant genera were Kaistobacter (12%), Bacillus (10%), Streptomyces (6%), Burkholderia (5%), Methylibium and Sphingomonas (with 4% each), Rhodoplanes, Paenibacillus, and Flavisolibacter (with 3% each), and Devosia, Geodermatophilus, and Gemmata (with 2% each), accounting for approximately 55% of the total diversity. One hundred and thirteen (36%) genera were shared by all samples, while only 20 of these bacterial taxa were restricted to one sample (Supplementary Table 4).
Core Rhizosphere Microbiota Associated With Different Maize Populations
The rhizosphere of the maize populations showed specific fungal and bacterial OTUs for each population and a cluster of shared OTUs. Comparing the fungal and bacterial microbiota of the “Pigarro” and “SinPre” populations, 69.9 and 87.4% of the fungal and bacterial genera, respectively, were shared between populations, demonstrating the existence of a “core” maize phylogeny (Figures 2C,D).
Eighty-seven fungal genera composed the core fungal rhizosphere microbiota, affiliated with Ascomycota (54 genera), Basidiomycota (20 genera), Chytridiomycota (three genera), Glomeromycota (three genera), and Zygomycota (seven genera) (Supplementary Table 5).
The core rhizosphere bacterial microbiota present in both maize populations was composed of 183 genera, affiliated with Proteobacteria (64 genera), followed by Actinobacteria (54 genera), Firmicutes (22 genera), Bacteroidetes (17 genera), Verrumicrobia (10 genera), Planctomycetes (four genera), Chlamydiae (three genera), Acidobacteria (three genera), Chloroflexi (two genera), and Armatimonadetes, Crenarchaeota, Nitrospirae, and Thermi, with one genus each (Supplementary Table 5).
Specific fungal genera were only associated with the “SinPre” population (37 genera, corresponding to 13.9%) and with the “Pigarro” population (43 genera, corresponding to 16.2%). Regarding the bacterial microbiota, 13 genera (4.1%) were exclusive to the “SinPre” population and 27 to the “Pigarro” population (8.5%) (Figures 2C,D). The population-specific genera are shown in Supplementary Table 6.
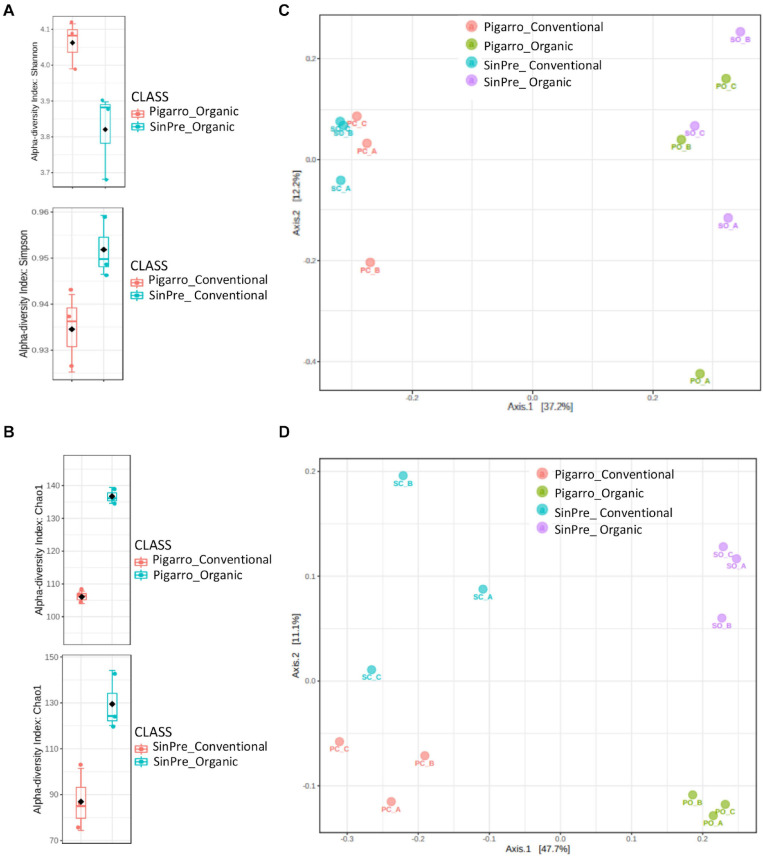
Fungal communities of the rhizospheric soil samples did not differ between populations. Bacterial communities were only significantly different in the rhizosphere associated with the “Pigarro” genotype in the organic farming system (F = 3.5, P < 0.05). The maize genotype was a significant factor structuring the bacterial community in both farming systems, supported by Shannon (t-test = 3.0627, P < 0.05) and Simpson (t-test = −2.7668, P < 0.05) α diversity indexes (Figures 3A,B and Table 1). Principal coordinate analysis (PCoA) further evidenced that most of the variations on the dataset could be attributed to the population genotype (Figures 3C,D).
FIGURE 3.
Box plot illustrating statistically significant α diversity measures of the effect of farming system in fungal (A) and bacterial (B) communities in the maize rhizosphere. Principal coordinate analysis (PCoA) was performed using the prcomp package to illustrate the effect of farming system on the fungal (C) and bacterial (D) communities in the maize rhizosphere. The calculation is based on singular value decomposition of the fungal and bacterial communities for the farming system. A greater distance between the two samples indicates low similarity. The percentage of variation is explained by component 1 (PC1) and by component 2 (PC2).
TABLE 1.
Experimental factors predicting α diversity of the rhizosphere-associated fungal and bacterial communities in maize.
| Chao1 |
Shannon |
Simpson |
|||||
| t-test | P | t-test | P | t-test | P | ||
| Fungi | |||||||
| Genotype | PC_PO | −17.014 | 0.0001 | −1.5369 | 0.1992 | −0.8259 | 0.4663 |
| SC_SO | −3.939 | 0.0171 | −2.1054 | 0.1503 | −0.4810 | 0.6759 | |
| Farming system | PC_SC | 2.4068 | 0.1330 | −0.5873 | 0.6123 | −0.6863 | 0.5625 |
| PO_SO | 0.4327 | 0.9591 | −0.1506 | 0.8880 | 0.0035 | 0.9974 | |
| Bacteria | |||||||
| Genotype | PC_SC | 0.3649 | 0.7338 | −2.1922 | 0.1001 | −2.7668 | 0.0540* |
| PO_SO | 0.9858 | 0.3956 | 3.0627 | 0.0530* | 2.8629 | 0.0623 | |
| Farming system | PC_PO | −0.48906 | 0.6513 | −7.3396 | 0.0036 | −5.1265 | 0.0171 |
| SC_SO | −0.20527 | 0.8150 | −0.50067 | 0.6437 | 0.5229 | 0.6288 | |
P, “Pigarro”; S, “SinPre”; C, conventional; O, organic. Statistics describe the linear random intercept models of Shannon and Simpson diversity and Chao1 richness in the rhizosphere. Alpha diversity analysis was performed using the phyloseq package. The statistical significance of grouping based on the experimental factor was estimated using t-test/ANOVA. Bold values indicate statistically significant results (P < 0.05). ∗Statistically significant.
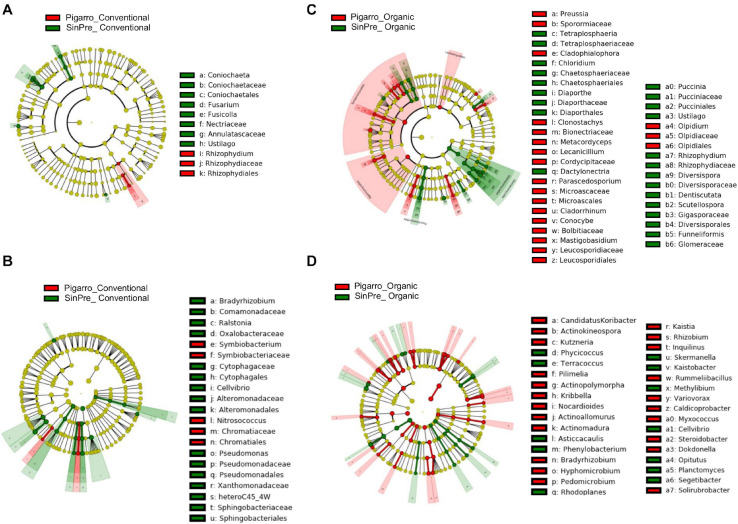
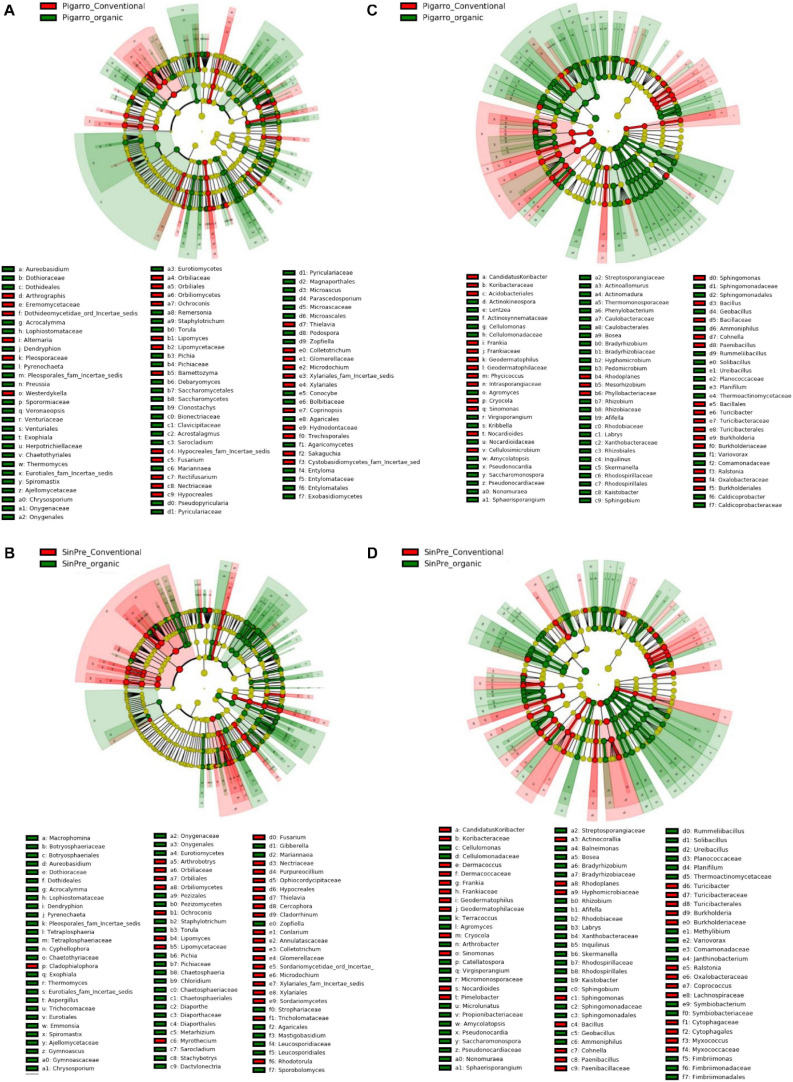
The LEfSe detected 64 fungal (12 PC_SC and 52 PO_SO) and 98 bacterial (24 PC_SC and 74 PO_SO) bacterial clades in the rhizosphere, which discriminated the microbial communities between populations (Figure 4). Annulatascaceae, Coniochaetaceae, Nectriaceae (class Sordariomycetes) were the major fungi families that contributed to differentiate the fungal communities associated with “Pigarro” and Rhizophydiaceae (class Chytridiomycetes) with “SinPre” under the conventional farming system. Under the organic farming system, the differences between genotypes were more pronounced due to the contributions of several other classes, namely, Agaricomycetes, Eurotiomycetes, Leotiomycetes, and Microbotryomycetes in “Pigarro” and Dothideomycetes, Glomeromycetes, Pucciniomycetes, and Ustilaginomycetes in “SinPre” (Figure 4A).
FIGURE 4.
Linear discriminant analysis (LDA) combined with effect size (LEfSe) was used to identify the most differentially abundant taxa among the population genotypes in the rhizosphere of maize. Cladogram generated by LEfSe indicating differences of fungi (A,B) and bacteria (C,D) at the phylum, class, family, and genus levels (relative abundance, ≤0.5%). Each successive circle represents a phylogenetic level. The red and green circles mean that “Pigarro” conventional and organic (red) and “SinPre” conventional and organic (green) showed differences in relative abundance; yellow circles mean non-significant differences. Differing taxa are listed on the right side of the cladogram. Bar graph showing the LDA scores for bacteria is represented in Supplementary Figure 1. Only taxa meeting an LDA significant threshold >2 are shown.
The main bacterial families that contributed to differentiate the communities associated with population genotypes in the conventional agricultural system were Alteromonadaceae, Bradyrhizobiaceae, Comamonadaceae, Cytophagaceae, Oxalobacteraceae, Pseudomonadaceae, Sphingobacteriaceae, and Xanthomonadaceae in “SinPre” and Chromatiaceae and Symbiobacteriaceae in “Pigarro.” The differences between populations were more pronounced in the organic farming system because of the presence of members of the classes Alcaligenaceae Caulobacteraceae, Cellvibrionaceae, Chitinophagaceae, Intrasporangiaceae, Hyphomicrobiaceae, Opitutaceae, Planctomycetaceae, Rhodospirillaceae, and Sphingomonadaceae in “SinPre” and Acidobacteriaceae, Bradyrhizobiaceae, Caldicoprobacteraceae, Comamonadaceae, Cystobacterineae, Hyphomicrobiaceae, Micromonosporaceae, Nocardioidaceae, Nocardiopsaceae, Planococcaceae, Pseudonocardiaceae, Rhizobiaceae, Rhodanobacteraceae, Rhodospirillaceae, Solirubrobacteraceae, and Steroidobacteraceae in “Pigarro” (Figure 4B). Detailed information can be found in Supplementary Figure 1. Several genera were significantly different between population genotypes, as detailed in Supplementary Table 7.
Specific fungal and bacterial OTUs associated with the “SinPre” population ranged from 9.4 and 13.8% in the conventional and between 45.3 and 17.2% in the organic farming system, respectively. The core microbiota of the “SinPre” population corresponded to 45.3% of the fungal and 69% of bacterial total diversity. A similar trend was observed in the dataset from the “Pigarro” population, with specific fungal and bacterial OTUs comprising 12.7 and 11.8% in conventional and 39.3 and 18.4% in the organic farming system, respectively. The core microbiota of the “Pigarro” population corresponded to 48% of the fungi and 69.7% of the bacteria total diversity. The farming system-specific genera are shown in Supplementary Table 8.
Core Rhizosphere Microbiota Associated With Farming Systems
The results obtained showed that rhizospheric soil harbored a distinct microbiota according to the farming system (P < 0.05; Table 1). This was visible in the cluster-based analysis of the fungal and bacterial community structure and composition, where maize populations were clustered by the farming system (Figure 2). Indeed, the rhizosphere showed specific fungal and bacterial OTUs for each farming type and a cluster of shared OTUs. Comparing the total dataset of the fungal and bacterial microbiota of the conventional and organic farming systems, 49.2 and 74.8% of fungal and bacterial OTUs, respectively, were shared between the two systems. Specific OTUs were associated with the conventional farming system, 25 fungal and 28 bacterial genera, representing 9.4 and 8.8% of the community, respectively. Regarding the organic farming system, the relative abundance of specific fungal (110, 41.4%) and bacterial (52, 16.4%) OTUs was considerably higher.
Core Rhizosphere Microbiota Across Both Maize Populations
The farming system was a significant factor structuring the fungal community predicted by Chao1 diversity in the “Pigarro” (t-test = −17 014, P < 0.05) and “SinPre” (t-test = −3,939, P < 0.05) genotypes (Figure 3A and Table 1). Besides, the mycobiota associated with “SinPre” was significantly different between the farming systems (F = 9.08, P < 0.05). However, the farming systems had a much weaker influence on the rhizosphere-associated bacterial communities. The PCoA further evidenced that most of the variations on the dataset could be related to the farming system (Figures 3C,D).
At the phylum level, the impact of the farming system on fungal diversity associated with the “Pigarro” rhizosphere translated into a higher dominance of Ascomycota in the conventional system, partially replacing the Zygomycota in the organic system. In the “SinPre” population, the relative abundance of Ascomycota was similar in both farming systems. Importantly, a notably higher presence of mycorrhizae (Glomeromycota) was observed in both populations cultivated in the organic farming system, in clear contrast to the conventional farming system (Figure 1A).
The LEfSe detected 255 fungal (124 PC_PO and 131 SC_SO) and 266 bacterial (138 PC_PO and 128 SO_SC) clades in the rhizosphere, which discriminated the microbial communities between the farming systems (Figure 5). Dothideomycetes, Orbiliomycetes, Sordariomycetes, and Tremellomycetes were the major fungi classes that contributed to differentiate the fungal communities associated with the conventional farming system despite the considered population, while under the organic farming system the differences were due to the contribution of several other classes, namely, Eurotiomycetes, Mortierellomycetes, Saccharomycetes, and Ustilaginomycetes. Worth noticing is that Glomeromycetes and Pucciniomycetes were highly abundant in the “SinPre” population cropped in organic farming (Figures 5A,B). The main bacterial classes that contributed to differentiate the communities associated with farming systems were Bacilli and Verrucomicrobiae in the conventional farming system and Alphaproteobacteria, Anaerolineae, Fimbriimonadia, Mollicutes, Nitrospira, Opitutae, Planctomycetia, Spartobacteria, and Solibacteres in the organic farming system (Figures 5C,D).
FIGURE 5.
Linear discriminant analysis (LDA) combined with effect size (LEfSe) was used to identify the most differentially abundant taxa among farming systems in the rhizosphere of maize. Cladogram generated by LEfSe indicating differences of fungi (A,B) and bacteria (C,D) at the phylum, class, family, and genus levels (relative abundance, ≤0.5%). Each successive circle represents a phylogenetic level. The red and green circles mean that conventional (red) and organic (green) showed differences in relative abundance; yellow circles mean non-significant differences. Differing taxa are listed on the right side of the cladogram. Bar graph showing the LDA scores for bacteria is represented in Supplementary Figure 1. Only taxa meeting an LDA significant threshold >2 are shown.
In silico Metagenome Analysis
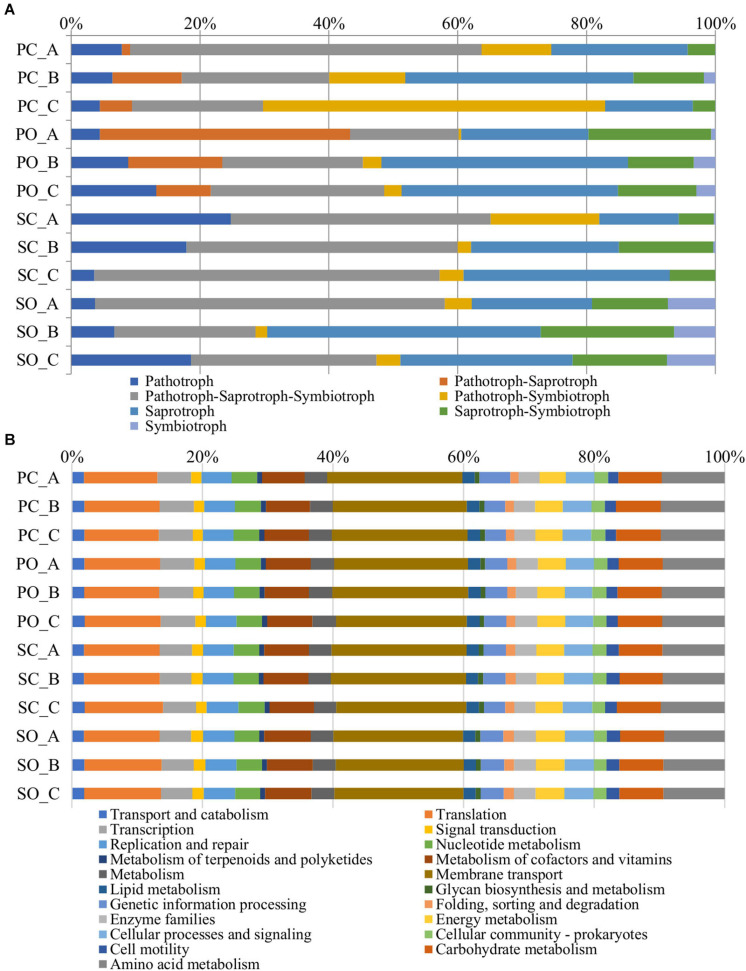
The functional predictions at the fungal level in both populations and both farming systems are shown in Figure 6A. An increase in the trophic modes defined as pathotroph–saprotroph, saprotroph-symbiotroph, and symbiotroph was observed in the “Pigarro” population under the organic farming system. The same population in the conventional farming system experienced an increase in the pathotroph–saprotroph–symbiotroph and pathotroph–symbiotroph trophic modes. In the “SinPre” population under the organic farming system, the trophic modes defined as pathotroph–saprotroph–symbiotroph, saprotroph, saprotroph–symbiotroph, and symbiotroph were more abundant, while the latter was practically non-existent in the conventional farming system.
FIGURE 6.
Metagenome predicted functions classified using FUNGuild for fungi (A) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) level 2 database in PICRUSt software for bacteria (B) showing the most abundant functions throughout the rhizospheric soil samples.
In detail, the trophic mode pathotroph comprised 32 genera of plant pathogens. In detail, 26 were detected in “Pigarro,” five specific to the conventional farming system (Chalastospora, Curvularia, Lectera, Rhizophydium, and Volutella) and 10 specific to the organic farming system (Cercospora, Kochiomyces, Dactylaria, Entyloma, Macrophomina, Sporisorium, Ramularia, Sclerotinia, Thanatephorus, and Gjaerumia), while the remaining 11 were shared between farming systems (Dendryphion, Farysia, Ilyonectria, Olpidium, Anthracocystis, Clonostachys, Drechslera, Gibberella, Monographella, Powellomyces, and Puccinia). Similarly, in the “SinPre” population, 32 genera of fungal plant pathogens were present, three specific to the conventional farming system (Olpidium, Lectera, and Thanatephorus), 13 related to the organic farming system (Protomyces, Dendryphion, Rhizophydium, Exserohilum, Macrophomina, Sphacelotheca, Entyloma, Microstroma, Glomosporium, Sporisorium, Taphrina, Ilyonectria, and Ramularia), and eight shared between farming systems (Monographella, Puccinia, Powellomyces, Clonostachys, Gibberella, Curvularia, Anthracocystis, and Drechslera). Both populations shared seven plant pathogens despite the farming system: Monographella, Puccinia, Powellomyces, Clonostachys, Gibberella, Anthracocystis, and Drechslera. Another important observation from the dataset was the increase in abundance and diversity of arbuscular mycorrhiza in the organic farming system.
A total of 6,886 functional orthologs were predicted to be present in the analyzed bacterial communities, corresponding to 21 level 2 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Figure 6B). The results showed nine pathways for metabolism, four for genetic information processing and unclassified, three pathways for cellular processes, and two for environmental information processing. The average relative abundance of the metabolism function was 33.95%, followed by genetic information processing (21.13%), environmental information processing (20.37%), cellular processes (5.34%), and unassigned with 19.22%. The most abundant KEGG pathway belonged to membrane transport, followed by translation, amino acid metabolism, carbohydrate metabolism, and metabolism of cofactors and vitamins.
Discussion
The rhizosphere is a specialized region where interactions between plant roots and the surrounding soil-associated microorganisms take place. Rhizospheric soil differs from bulk soil not only due to the direct effect of these microorganisms but also because root growth modifies the composition of the soil. Roots can release rhizodeposits, a wide range of substances containing carbon (e.g., root cells, mucilage, volatiles, and exudates), selecting and enhancing groups of microorganisms (Harkes et al., 2019), which, in addition to modifying some soil characteristics, can play a role in the plant health status.
This work intends to characterize the structural compositions of fungal and bacterial communities present in the rhizospheric soil associated with two maize populations cultivated in different farming systems. Although it is difficult to draw robust conclusions, significant interactions between the microbial diversity, farming systems, and genotype were obtained. Moreover, our results are in line with previous studies (Granzow et al., 2017; Ginnan et al., 2018; Berlanas et al., 2019) including those on maize crop (Galazka and Grzadziel, 2018; Brisson et al., 2019; Kong et al., 2020), shedding some light into the potential beneficial impacts of maize landraces and organic cultivation techniques into the rhizosphere microbial communities and network assembly.
Impact of Maize Populations and Farming System on the Associated Rhizosphere Mycobiota
The microbial community analysis revealed significant differences in the relative abundance of OTUs between maize populations, evidencing that plant genotype has a significant effect on the structure of the rhizosphere-associated microbiota. In our dataset, a higher portion of the differentially selected microbial community by genotypes was discovered compared with previous studies (Brisson et al., 2019). Indeed, 30.1% of the fungal community and 12.6% of the bacterial community were significantly associated with a specific population.
The novel landrace “SinPre” was derived from 12 populations, while the traditional landrace “Pigarro” encompassed less diversity. We expected that this intraspecific diversity would be translated into more heterogeneous rhizosphere mycobiota, but this was not the case. The rhizosphere mycobiota associated with the “Pigarro” and “SinPre” populations were similar, but significantly influenced by the farming systems. A higher heterogenicity was found in the rhizosphere mycobiota under the organic farming system, in line with previous works (Hartmann et al., 2015; Figure 3C). Also, high fertilization regimes are known to select and enrich fungi populations (Enebe and Babalola, 2020), aiding to explain the previously described diversity.
The fungal community was dominated by the Ascomycota phylum, which agrees with previous studies on the maize rhizosphere (Galazka and Grzadziel, 2018). The impact of the farming system in fungal diversity associated with the “Pigarro” rhizosphere translated into a higher dominance of Ascomycota in the conventional system, replaced in the organic system by Zygomycota. Our results follow the study by Galazka and Grzadziel (2018), where the abundance of the order Hypocreales (Ascomycota) was associated with a conventional farming system. Moreover, Hypocreales, Sordariales, and Helotiales were among the most abundant classes in our dataset, similar to the results described by Klaubauf et al. (2010) on the fungal diversity in arable soils. On the contrary, our results did not support the previously described relationship between the order Sordariales and the conventional farming system since, in our dataset, it was more abundant in the organic farming system.
This study also showed that the farming system was a significant factor structuring the maize-associated fungal community. This influence was less pronounced in the associated bacterial community. A similar trend was described by Galazka and Grzadziel (2018) in a study combining different cultivation practices, reinforcing the impact that these practices have on the microbial community and soil function. Our results showed that rhizospheric soil harbored a distinct mycobiota according to the farming system. Dothideomycetes, Orbiliomycetes, Sordariomycetes, and Tremellomycetes were the main classes within the Ascomycota phylum that contributed to differentiate the fungal communities associated with the conventional agricultural system. The Dothideomycetes class includes several endophytic and plant saprobic taxa, as well as crop-related pathogens. Both Alternaria sp. and Cladosporium sp. are examples of the latter that were associated with the conventional farming in our dataset. The class Orbiliomycetes includes most of the known nematode-trapping fungi constituting an important part of the subsoil ecosystem, among which Arthrobotrys sp. was associated in this study with conventional farming. Fusarium was the most abundant genus of the class Sordariomycetes and has been commonly associated with the maize rhizosphere. This genus includes saprotrophic and plant pathogenic species causing significant economic losses in cereals, especially in maize (McMullen et al., 2012; Salgado et al., 2015). Also, Fusarium can produce phytotoxins that inhibit the growth of infected plants and can act as virulence factors (Salgado et al., 2015). Tremellomycetes belong to Basidiomycota, the second most abundant phylum of our dataset, including Cryptococcus sp., known for the ability to aid plants to extract nutrients from the soil (Wolf et al., 2014).
Classes Eurotiomycetes, Mortierellomycetes, Saccharomycetes, Ustilaginomycetes, Glomeromycetes, and Pucciniomycetes shaped the rhizospheric mycobiota associated with maize cropped in the organic system. Interestingly, the latter two were more abundant in “SinPre” than in the “Pigarro” maize population. Eurotiomycetes and Saccharomycetes belong to Ascomycota, including two of the most important and problematic genera in the cultivation of maize, Penicillium and Aspergillus, present in our dataset. Penicillium species are ubiquitous in the soil wherever organic material is available (Duniere et al., 2017), while Aspergillus sp., in spite being involved in lignin degradation and (or) melanin synthesis (Levasseur et al., 2008), crucial to improving the amount of organic matter available in the soil, can also produce the mycotoxin deoxynivalenol (DON), which is harmful to humans and livestock. Ustilaginomycetes and Pucciniomycetes belong to Basidiomycota, the second most abundant phylum comprising some genera potentially pathogenic to maize plants, namely, Ustilago sp., where it includes the species Ustilago maydis, known for the ability to cause tumors in maize by redirecting vegetative and floral development (Walbot and Skibbe, 2009). In the class Pucciniomycetes, the genus Puccinia includes species responsible for the foliar disease of maize (Medina and López, 2007). Within the class Mortierellomycetes (phylum Zygomycota), the genus Morteriella was present in our dataset and was correlated with the organic farming system. In terms of its potential ecological role, this genus has been implicated in preventing soil degradation, improvement of soil health, and stimulating the production of plant growth hormones (Li et al., 2009; Zhang et al., 2011). Finally, the class Glomeromycete (phylum Glomeromycota) included an important functional group of organisms, the arbuscular mycorrhizae, which, according to the bibliography, can establish beneficial symbiosis with maize in various geographic distributions and different environments (Miransari et al., 2009).
Our dataset comprised 12 genera belonging to this class in the rhizosphere of both crop populations (“Pigarro” and “SinPre”) mostly associated with the organic farming system and lacking in most samples derived from the conventional farming system. Among the plant-associated mycobiota, these fungi are the most widespread (Desirò et al., 2014), and approximately 80% of the existing plant families have the potential to form this type of association (Trappe, 1987). Among the different alternatives toward sustainable agriculture, increasing arbuscular mycorrhizae is an important strategy with beneficial effects for the plant nutritional status by increasing plant nutrient uptake, in particular phosphorus (Smith and Jakobsen, 2013), improving the phytosanitary status (Ahmed et al., 2013; Hohmann and Messmer, 2017), expanding root range, improving tolerance to biotic and abiotic stresses, and inducing plant defense response (Oehl and Sieverding, 2004; Ismail et al., 2013). From our results, the organic farming system promotes these symbioses in the associated rhizosphere of these two Portuguese landraces, aiding the resilience of the crop without the insertion of synthetic fertilizers. Dentiscutata, Gigaspora, and Paraglomus were the most representative genera in the mycorrhizae community in our dataset and, along with Acaulospora, Cetraspora, Diversispora, Glomus, and Scutellospora, were restricted to the rhizosphere in the organic farming system; Septoglomus was genotype-specific in “SinPre.” Glomus belongs to the important group of arbuscular mycorrhizal (AM) fungi involved in the exchange with the plant of carbon, phosphorus, and other physiologically significant particles. As mentioned above, this interaction of mycorrhizae with other soil microorganisms promotes beneficial cooperation enhancing the competition against phytopathogenic microorganisms. Relating the rhizosphere fungal communities with their trophic mode, as expected, the abundance of taxa with saprotrophic and pathotroph modes was higher in the organic farming system than in the conventional farming system.
Impact of Maize Populations and Farming System on the Associated Rhizosphere Bacterial Communities
There was also a clear population–farming system interaction concerning bacterial communities. A higher heterogenicity was found in the rhizosphere bacteriota under the organic farming system, in line with previous works (Hartmann et al., 2015; Lupatini et al., 2017; Fernandez et al., 2020; Lee et al., 2020; Figure 3D). Differences between populations were more pronounced in the organic farming system due to the presence of members from the classes Alcaligenaceae, Caulobacteraceae, Cellvibrionaceae, Chitinophagaceae, Intrasporangiaceae, Hyphomicrobiaceae, Opitutaceae, Planctomycetaceae, Rhodospirillaceae, and Sphingomonadaceae in the “SinPre” population, while Acidobacteriaceae, Bradyrhizobiaceae, Caldicoprobacteraceae, Comamonadaceae, Cystobacterineae, Hyphomicrobiaceae, Micromonosporaceae, Nocardioidaceae, Nocardiopsaceae, Planococcaceae, Pseudonocardiaceae, Rhizobiaceae, Rhodanobacteraceae, Rhodospirillaceae, Solirubrobacteraceae, and Steroidobacteraceae were present in “Pigarro.” These results are in line with the recent studies of Beirinckx et al. (2020), confirming the existence of a core bacteriome in the maize rhizosphere composed of Hyphomicrobiacea, Streptomycetaceae, Comamonadaceae, Cytophagaceae, Oxalobacteraceae, Rhizobacteraceae, Xanthomonadaceae, and Caulobacteraceae. Also, several taxa from these families were recently associated with the maize rhizosphere soils of various farming systems and growth stages.
The family Oxalobacteraceae was specific to the conventional farming system and has been associated with nitrogen fixation activities. However, in this study, two of the most abundant genera of this family were Ralstonia, a well-known plant pathogen responsible for wilting in numerous plants, and Janthinobacterium, previously reported in maize residues and stalks and the rhizospheric soil of maize. The importance of the latter resides in the effective reduction of maize stalk colonization by Fusarium spp. (Cobo-Díaz et al., 2019).
Genera from the families Xanthomonadaceae (Luteimonas sp.) and Comamonadaceae (Methylibium sp.) were associated with the conventional farming system in our dataset, which have been previously used in the bioremediation of hydrocarbon-contaminated soil (Kane et al., 2007; Mu et al., 2016). The latter can also induce disease resistance through the production of antimicrobial compounds (Lagos et al., 2015). In line with this, it is important to highlight that we identified in our dataset families that include genera with reported biocontrol activity, such as Sphingobacteriaceae (Pedobacter sp.), Pseudomonadaceae (Pseudomonas sp.), Sphingomonadaceae (Sphingobium sp.), Xhantomonadaceae (Luteibacter sp.), Cytophagaceae (Dyadobacter sp.), and Rhizobiaceae (Rhizobium sp.) (Cobo-Díaz et al., 2019).
The main bacterial classes that contributed to differentiate the communities associated with farming systems were Bacilli and Verrucomicrobiae in the conventional farming system and Alphaproteobacteria, Anaerolineae, Fimbriimonadia, Mollicutes, Nitrospira, Opitutae, Planctomycetia, Spartobacteria, and Solibacteres in the organic farming system. Several bacterial genera of these classes were recently associated with maize rhizosphere-associated microbiota. At the genus level, Bacillus, Erwinia, Pseudomonas, Stenotrophomonas, Achromobacter, Lysinibacillus, and Paenibacillus were reported by Pereira et al. (2011) as the most common taxa in the maize rhizosphere. These genera, along with Burkholderia, were described as having plant growth-promoting rhizobacteria common in the maize-associated rhizosphere (Yang et al., 2017). Also, the antagonist potential of the genus Bacillus against maize pathogens has been addressed (Figueroa-López et al., 2016; Cheng et al., 2019; Hazarika et al., 2019; Liu et al., 2020). In terms of putative functions, Burkholderia has been linked with antifungal activity (Stopnisek et al., 2016), while some strains were described to produce ACC deaminase and siderophores important for maize growth promotion (Byrt et al., 2011). On the other hand, Erwinia is the causal agent of the bacterial wilt in a large range of hosts and the bacterial stalk rot disease of maize. Finally, the genus Kaistobacter is common in soils with atrazine (a compound present in some herbicides) (Lin et al., 2018), while Rhodoplanes can increase the fertility of soils (Sun et al., 2015).
Conclusion
In the current context of climate change, environmental degradation, and misuse of natural resources, the transition from conventional to organic farming requires the optimization of cultivars and management, with the rhizosphere microbiota playing an important role. Since maize is one of the main crops in the world, the adoption of more sustainable practices will contribute decisively to this strategy. In this study, the analysis of the microbial community revealed significant differences among maize populations, showing that the plant genotype has a significant effect on the structure of the microbiota associated with the rhizosphere. Additionally, the farming system had a statistically significant impact on rhizosphere-associated microbiota, and several taxa were found to be specific to the agricultural system. The presence of arbuscular mycorrhizae (Glomeromycota), known for the potential to establish a beneficial symbiosis with maize, was mainly detected in the microbiota of the “SinPre” population in the organic farming system, being very rare under the conventional system. The diversity of the rhizosphere-associated microbiota in the organic farming system was significantly higher than that in the conventional system in both varieties, shedding some light into the potential beneficial impacts of maize landraces and organic cultivation techniques into the rhizosphere microbial communities and network assembly. The role of some species as plant growth-promoting rhizobacteria and with the ability to improve the tolerance of crops to stress conditions (biotic and abiotic) has been previously described, including in some of the genera detected in this study, namely, Achromobacter, Burkholderia, Lysinibacillus, Paenibacillus, and Stenotrophomonas. The role of these organisms in the sustainability and production of maize will be evaluated in future studies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA675280.
Author Contributions
AA, CJ, DP, and DS helped in the investigation. AA, JC, CJ, and DP did the formal analysis. JC and DP helped in funding acquisition, project administration, resources, and supervision. AA and JC contributed to visualization and writing of the original draft. AA, JC, MM, and DP contributed to the conceptualization, reviewed, edited, and wrote the final manuscript. All authors contributed to the article and approved the submitted version.
Disclaimer
The opinions expressed and the arguments employed herein do not necessarily reflect the official views of the EC and the Swiss government. Neither the European Commission/SERI nor any person acting on behalf of the Commission/SERI is responsible for the use which might be made of the information provided on this website.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
AA acknowledges financial support by postdoctoral grants from FCT/MEC through national funds and the co-funding by the FEDER, within the PT2020 Partnership Agreement, and COMPETE 2020, within the project UID/BIA/04004/2013.
Footnotes
Funding. This work was made in the scope of CULTIVAR project (CENTRO-01-0145-FEDER-000020), co-financed by the Regional Operational Programme Centro 2020, Portugal 2020, and the European Union, through the European Fund for Regional Development (ERDF), Collaborative Project “LIVESEED: Improving the performance of organic agriculture by boosting organic seed and plant breeding efforts across Europe” is supported by the European Union’s HORIZON 2020 Research and IPN – Financiamento Base FITEC approved under the National Call with reference no. 01/FITEC/2018 to obtain multi-year base financing under the INTERFACE Program, Innovation Programme under grant agreement no. 727230, and by the Swiss State Secretariat for Education, Research and Innovation (SERI) under contract number 17.00090.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.636009/full#supplementary-material
LEfSe was used to identify the most differentially abundant taxa among population genotypes in the rhizosphere of maize. Bar graph showing LDA scores for fungi (A,B) and bacteria (C,D) are represented. The length of the bar column represents the LDA score. The figure shows the microbial taxa with significant differences between the “Pigarro” (red) and “SinPre” (green). Only taxa meeting an LDA significant threshold > 2 are shown.
Heat-map of the relative abundance with distance measure using Euclidean, at the level class for fungal (A) and bacterial (B) community. The relative abundance is scaled by a color gradient bar.
Soil characterization of the trial locations in august 2019.
Characterization of maize populations using HUNTERS descriptors in conventional and organic farming systems.
Abundance of fungal genera detected in each sample and the respective taxonomic assignment.
Abundance of bacterial genera detected in each sample and the respective taxonomic assignment.
Fungal and bacterial core rhizosphere microbiota associated with maize.
Fungal and bacterial genera specific to the rhizosphere microbiota in each population.
Fungal and bacterial genera specific to the rhizosphere microbiota of maize in distinct farming system.
Genera relative abundance significantly different between “Pigarro” and “SinPre” populations (P < 0.05).
References
- Ahmed M. S., Sallam N. M., Mohamed A. A., Hassan M. H. (2013). Effect of mycorrhiza and biofertilisers on reducing the incidence of Fusarium root and pod rot diseases of peanut. Arch. Phytopathol. 46 868–881. 10.1080/03235408.2012.753707 [DOI] [Google Scholar]
- Altieri M. (2004). Linking ecologists and traditional farmers in the search for sustainable agriculture. Front. Ecol. Environ. 2 35–42. 10.1890/1540-9295(2004)002[0035:leatfi]2.0.co;2 [DOI] [Google Scholar]
- Altieri M., Merrick L. (1987). In situ conservation of crop genetic resources through maintenance of traditional farming systems. Econ. Bot. 41 86–96. 10.1007/BF02859354 [DOI] [Google Scholar]
- Andreote F., Silva M. C. P. (2017). Microbial communities associated with plants: learning from nature to apply it in agriculture. Curr.Opin. Microbiol. 37 29–34. 10.1016/j.mib.2017.03.011 [DOI] [PubMed] [Google Scholar]
- ANPROMIS (2019). Associação Nacional de Produtores de Milho e Sorgo. Available online at: http://www.anpromis.pt/o-milho.html (access August 13, 2019). [Google Scholar]
- Beirinckx S., Viaene T., Haegeman A., Debode J., Amery F., Vandenabeele S., et al. (2020). Tapping into the maize root microbiome to identify bacteria that promote growth under chilling conditions. Microbiome 8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanas C., Berbegal M., Elena G., Laidani M., Cibriain J. F., Sagües A., et al. (2019). The fungal and bacterial rhizosphere microbiome associated with grapevine rootstock genotypes in mature and young vineyards. Front. Microbiol. 10:1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson V. L., Schmidt J. E., Northen T. R., Vogel J. P., Gaudin A. C. M. (2019). Impacts of maize domestication and breeding on rhizosphere microbial community recruitment from a nutrient depleted agricultural soil. Sci. Rep. 9:15611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrt C. S., Grof C. P., Furbank R. T. (2011). C4 Plants as biofuel feedstocks: optimizing biomass production and feedstock quality from a lignocellulosic perspective free access. J. Integr. Plant Biol. 53 120–135. 10.1111/j.1744-7909.2010.01023.x [DOI] [PubMed] [Google Scholar]
- Chadha N., Mishra M., Rajpal K., Bajaj R., Choudhary D. K., Varma A. (2015). An ecological role of fungal endophytes to ameliorate plants under biotic stress. Arch. Microbiol. 197 869–881. 10.1007/s00203-015-1130-3 [DOI] [PubMed] [Google Scholar]
- Chaparro J., Badri D., Vivanco J. (2014). Rhizosphere microbiome assemblage is affected by plant development. ISME J. 8 790–80396. 10.1038/ismej.2013.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Ji X., Ge Y., Li J., Qi W., Qiao K. (2019). Characterization of antagonistic Bacillus methylotrophicus isolated from rhizosphere and its biocontrol effects on maize stalk rot. Phytopathology. 109 571–581. 10.1094/phyto-07-18-0220-r [DOI] [PubMed] [Google Scholar]
- Cobo-Díaz J. F., Legrand F., Le Floch G., Picot A. (2019). Maize residues changes soil fungal composition and decrease soil microbial co-occurrence networks complexity. bioRxiv [preprint] 10.1101/703967 [DOI] [Google Scholar]
- Comeau A. M., Douglas G. M., Langille M. G. I. (2017). Microbiome Helper: a Custom and streamlined workflow for microbiome research. mSystems 2:e127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant S., Samad A., Faist H., Sessitsch A. (2019). A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19 29–37. 10.1016/j.jare.2019.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desirò A., Salvioli A., Ngonkeu E., Mondo S. J., Epis S., Faccio A., et al. (2014). Detection of a novel intracellular microbiome hosted in arbuscular mycorrhizal fungi. ISME J. 8 257–270. 10.1038/ismej.2013.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhariwal A., Chong J., Habib S., King I. L., Agellon L. B., Xia J. (2017). Microbiomeanalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 45 W180–W188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duniere L., Xu S., Long J., Elekwachi C., Wang Y., Turkington K., et al. (2017). Bacterial and fungal core microbiomes associated with small grain silages during ensiling and aerobic spoilage. BMC Microbiol. 17:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmett B. D., Youngblut N. D., Buckley D. H., Drinkwater L. E. (2017). Plant phylogeny and life history shape rhizosphere bacterial microbiome of summer annuals in an agricultural field. Front. Microbiol. 8:2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enebe M. C., Babalola O. O. (2020). Effects of inorganic and organic treatments on the microbial community of maize rhizosphere by a shotgun metagenomics approach. Ann. Microbiol. 70:49. [Google Scholar]
- FAOSTAT (2019). Agriculture Organization of the United Nations. Available online at: http://www.fao.org/faostat/en/#data/QC (access August 13, 2020). [Google Scholar]
- Fernandez A. L., Sheaffer C. C., Wyse D. L., Sadowsky M. J. (2020). Bacterial community composition in agricultural soils under long-term organic and conventional management. Agrosyst. Geosci. Environ. 3:e20063. [Google Scholar]
- Figueroa-López A. M., Cordero-Ramírez J. D., Martínez-Álvarez J. C., Lopez-Meyer M., Lizarraga-Sanchez G. J., Felix-Gastelum R., et al. (2016). Rhizospheric bacteria of maize with potential for biocontrol of Fusarium verticillioides. SpringerPlus 5:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galazka A., Grzadziel J. (2018). Fungal genetics and functional diversity of microbial communities in the soil under long-term monoculture of maize using different cultivation techniques. Front. Microbiol. 9:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Tejero I., Costa J., Lima R., Zuazo V., Chaves M., Vaz-Patto M. (2015). Thermal imaging to phenotype traditional maize land races for drought tolerance. Comunicata Scientiae 6 34–343. [Google Scholar]
- Ginnan N. A., Dang T., Bodaghi S., Ruegger P. M., Peacock B. B., McCollum G., et al. (2018). Bacterial and fungal next generation sequencing datasets and metadata from citrus infected with ‘Candidatus Liberibacter asiaticus’. Phytobiomes J. 2 64–70. 10.1094/pbiomes-08-17-0032-a [DOI] [Google Scholar]
- Granzow S., Kaiser K., Wemheuer B., Pfeiffer B., Daniel R., Vidal S., et al. (2017). The effects of cropping regimes on fungal and bacterial communities of wheat and faba bean in a greenhouse pot experiment differ between plant species and compartment. Front. Microbiol. 8:902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkes P., Suleiman A. K. A., van den Elsen S. J. J., de Haan J. J., Holterman M. (2019). Conventional and organic soil management as divergent drivers of resident and active fractions of major soil food web constituents. Sci. Rep. 9:13521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M., Frey B., Mayer J., Mader P., Widmer F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 9 1177–1194. 10.1038/ismej.2014.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazarika D. J., Goswami G., Gautom T., Parveen A., Das P., Barooah M., et al. (2019). Lipopeptide mediated biocontrol activity of endophytic Bacillus subtilis against fungal phytopathogens. BMC Microbiol. 19:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlemann D. P., Labrenz M., Jürgens K., Bertilsson S., Waniek J. J., Andersson A. F. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5 1571–1579. 10.1038/ismej.2011.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann P., Messmer M. M. (2017). Breeding for mycorrhizal symbiosis: focus on disease resistance. Euphytica 213:113. [Google Scholar]
- IBM Corp. (2020). IBM SPSS Statistics for Windows, Version 27.0. Armonk, NY: IBM Corp. [Google Scholar]
- Illumina (2013). 16s-Metagenomic-Library-Prep-Guide-15044223-b. (s.f.). Available online at: https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (access August 10, 2020). [Google Scholar]
- Ismail Y., McCormick S., Hijri M. (2013). The arbuscular mycorrhizal fungus, Glomus irregulare, controls the mycotoxin production of Fusarium sambucinum in the pathogenesis of potato. FEMS Microbiol. Lett. 348 46–51. 10.1111/1574-6968.12236 [DOI] [PubMed] [Google Scholar]
- Kandel S., Joubert P., Doty S. (2017). Bacterial endophyte colonization and distribution within plants. Microorganisms 5:77. 10.3390/microorganisms5040077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S. R., Chakicherla A. Y., Chain P. S. G., Schmidt R., Shin M. W., Legler T. C., et al. (2007). Whole-genome analysis of the methyl tert-butyl ether-degrading beta-proteobacterium Methylibiumpetroleiphilum PM1. J. Bacteriol. 189 1931–1941. 10.1128/jb.01259-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaubauf S., Inselsbacher E., Zechmeister-Boltenstern S., Wanek W., Gottsberger R., Strauss J., et al. (2010). Molecular diversity of fungal communities in agricultural soils from lower Austria. Fungal Divers. 44 65–75. 10.1007/s13225-010-0053-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X., Han Z., Tai X., Jin D., Ai S., Zheng X., et al. (2020). Maize (Zea mays L. Sp.) varieties significantly influence bacterial and fungal community in bulk soil, rhizosphere soil and phyllosphere. FEMS Microbiol. Ecol. 96:fiaa020. [DOI] [PubMed] [Google Scholar]
- Lagos M. L., Maruyama F., Nannipieri P., Mora M. L., Ogram A., Jorquera M. A. (2015). Current overview on the study of bacteria in the rhizosphere by modern molecular techniques: a mini-review. J. Soil Sci. Plant Nut. 15 504–523. [Google Scholar]
- Lane A., Jarvis A. (2007). Changes in climate will modify the geography of crop suitability: agricultural biodiversity can help with adaptation. SAT eJournal, 4 1–12. [Google Scholar]
- Langille M. G. I., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31 814–821. 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. A., Kim J. M., Kim Y., Joa J. H., Kang S. S. (2020). Different types of agricultural land use drive distinct soil bacterial communities. Sci. Rep. 15:17418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitão S., Ferreira E., Bicho M., Alves M., Pintado D., Santos D., et al. (2019). Maize open-pollinated populations physiological improvement: validating tools for drought response. Sustainability 11:6081. 10.3390/su11216081 [DOI] [Google Scholar]
- Levasseur A., Piumi F., Coutinho P., Rancurel C., Asther M., Delattre M., et al. (2008). FOLy: an integrated database for the classification and functional annotation of fungal oxidoreductases potentially involved in the degradation of lignin and related aromatic compounds. Fungal Genet. Biol. 45 638–645. 10.1016/j.fgb.2008.01.004 [DOI] [PubMed] [Google Scholar]
- Li G., Yang S., Li M., Qiao Y., Wang J. (2009). Functional analysis of an Aspergillus ficuum phytase gene in Saccharomyces cerevisiae and its root-specific, secretory expression in transgenic soybean plants. Biotechnol. Lett. 31 1297–1303. 10.1007/s10529-009-9992-6 [DOI] [PubMed] [Google Scholar]
- Lin Z., Zhen Z., Ren L., Yang J., Lou C., Zhong L., et al. (2018). Effects of two ecological earthworm species on atrazine degradation performance and bacterial community structure in red soil. Chemosphere 196 467–475. 10.1016/j.chemosphere.2017.12.177 [DOI] [PubMed] [Google Scholar]
- Liu Y., Teng K., Wang T., Dong E., Zhang M., Tao Y., et al. (2020). Antimicrobial Bacillus velezensis HC6: production of three kinds of lipopeptides and biocontrol potential in maize. J. Appl. Microbiol. 128 242–254. 10.1111/jam.14459 [DOI] [PubMed] [Google Scholar]
- Lupatini M., Korthals G. W., de Hollander M., Janssens T. K., Kuramae E. E. (2017). Soil microbiome is more heterogeneous in organic than in conventional farming system. Front. Microbiol. 7:2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen M., Bergstrom G., De Wolf E., Dill-Macky R., Hershman D., Shaner G., et al. (2012). A unified effort to fight an enemy of wheat and barley: fusarium head blight. Plant Dis. 96 1712–1728. 10.1094/pdis-03-12-0291-fe [DOI] [PubMed] [Google Scholar]
- Medina J., López N. (2007). Resistance to Puccinia polysora in maize accessions. Plant Dis. 91 1489–1495. 10.1094/pdis-91-11-1489 [DOI] [PubMed] [Google Scholar]
- Mendes-Moreira P., Satovic Z., Mendes-Moreira J., Santos J. P. P., Santos J. P. N. S., Pêgo S., et al. (2017). Maize participatory breeding in Portugal: Comparison of farmer’s and breeder’s on-farm selection. Plant Breed. 136 861–871. 10.1111/pbr.12551 [DOI] [Google Scholar]
- Mendes-Moreira P., VazPatto M. C., Mota M., Mendes-Moreira J., Santos J. P. P., Santos J. P. N. S., et al. (2009). ‘Fandango’: long term adaptation of exotic germplasm to a Portuguese on-farm-conservation and breeding project. Maydica 54 269–285. [Google Scholar]
- Miransari M., Bahrami H., Rejali F., Malakouti M. (2009). Effects of soil compaction and arbuscular mycorrhiza on corn (Zea mays L.) nutrient uptake. Soil Tillage Res. 103 282–290. 10.1016/j.still.2008.10.015 [DOI] [Google Scholar]
- Mu Y., Pan Y., Shi W., Liu L., Jiang Z., Luo X., et al. (2016). Luteimonasarsenica sp. nov., an arsenic-tolerant bacterium isolated from arsenic-contaminated soil. Int. J. Syst. Evol. Micr. 66 145–156. [DOI] [PubMed] [Google Scholar]
- Muller D., Vogel C., Bai Y., Vorholt J. (2016). The plant microbiota: systems-level insights and perspectives. Annu. Rev. Genet. 50 211–234. 10.1146/annurev-genet-120215-034952 [DOI] [PubMed] [Google Scholar]
- Nguyen N. H., Smith D., Peay K. G., Kennedy P. G. (2015). Parsing ecological signal from noise in next generation amplicon sequencing. New. Phytol. 205 1389–1393. 10.1111/nph.12923 [DOI] [PubMed] [Google Scholar]
- Oehl F., Sieverding E. (2004). Pacispora, a new vesicular arbuscular mycorrhizal fungal genus in the Glomeromycetes. J. Appl. Bot. 78 78–82. [Google Scholar]
- Oliveros J. C. (2007–2015). Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online at: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed September 2020). [Google Scholar]
- Pereira P., Ibáñez F., Rosenblueth M., Etcheverry M., Martínez-Romero E. (2011). Analysis of the bacterial diversity associated with the roots of Maize (Zea mays L.) through culture-dependent and culture-independent methods. ISRN Ecol. 2011 1–10. 10.5402/2011/938546 [DOI] [Google Scholar]
- Philippot L., Raaijmakers J., Lemanceau P., van der Putten W. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11 789–799. 10.1038/nrmicro3109 [DOI] [PubMed] [Google Scholar]
- Salgado J. D., Madden L. V., Paul P. A. (2015). Quantifying the effects of Fusarium head blight on grain yield and test weight in soft red winter wheat. Phytopathology 105 295–306. 10.1094/phyto-08-14-0215-r [DOI] [PubMed] [Google Scholar]
- Schmidt J., Bowles T., Gaudin A. (2016). Using ancient traits to convert soil health into crop yield: impact of selection on maize root and rhizosphere function. Front. Plant Sci. 7:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R., Edwards R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27 863–864. 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M., Lindgreen S., Orlando L. (2016). Adapter removal v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes 9:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiferaw B., Prasanna B., Hellin J., Bãnzinger M. (2011). Crops that feed the world. Past successes and future challenges to the role played by maize in global food security. Food Security 3:307. 10.1007/s12571-011-0140-5 [DOI] [Google Scholar]
- Smith S. E., Jakobsen I. (2013). Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol. 133 16–20. 10.1104/pp.103.024380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopnisek N., Zühlke D., Carlier A., Barberán A., Fierer N., Becher D., et al. (2016). Molecular mechanisms underlying the close association between soil Burkholderia and fungi. ISME J. 10 253–264. 10.1038/ismej.2015.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Xiao T., Ning Z., Xiao E., Sun W. (2015). Microbial community analysis in rice paddy soils irrigated by acid mine drainage contaminated water. Appl. Microbiol. Biotechnol. 99 2911–2922. 10.1007/s00253-014-6194-5 [DOI] [PubMed] [Google Scholar]
- Tedersoo L., Bahram M., Polme S., Koljalg U., Yorou N. S., Wijesundera R., et al. (2014). Global diversity and geography of soil fungi. Science 346 1256688–1256688. [DOI] [PubMed] [Google Scholar]
- The Huttenhower Lab (2018). Department of Biostatistics. Boston, MA: Harvard T.H. [Google Scholar]
- Trappe J. M. (1987). “Phylogenetic and ecologic aspects of mycotrophy in the angiospems from an evolutionary viewpoint,” in Ecophysiology of VA Mycorrhizal Plants, ed. Safir G. R. (Boca Raton, Fla: CRC; ), 5–25. [Google Scholar]
- Turner T., Ramakrishnan K., Walshaw J., Heavens M., Swarbreck D., Osbourn A., et al. (2013). Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 7 2248–2258. 10.1038/ismej.2013.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenkoornhuyse P., Quaiser A., Duhamel M., Le Van A., Dufresne A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206 1196–1206. 10.1111/nph.13312 [DOI] [PubMed] [Google Scholar]
- Vaz Patto M. C., Mendes-Moreira P. M., Alves M. L., Mecha E., Brites C., do Rosário, et al. (2013). “Participatory plant quality breeding: an ancient art revisited by knowledge sharing. the portuguese experience,” in Plant Breeding from Laboratories to Fields, ed. Andersen S. B. (Germany: BoD – Books on Demand; ). [Google Scholar]
- Walbot V., Skibbe D. (2009). Maize host requirements for Ustilago maydis tumor induction. Sex. Plant Reprod. 23 1–130. 10.1007/s00497-009-0109-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille L., Messmer M. B. S., Hohmann P. (2018). Insights to plant-microbe interactions provide opportunities to improve resistance breeding against root diseases in grain legumes. Plant Cell Environ. 42 20–40. 10.1111/pce.13214 [DOI] [PubMed] [Google Scholar]
- Wolf J., Espadas-Moreno J., Luque-Garcia J., Casadevall A. (2014). Interaction of Cryptococcus neoformans extracellular vesicles with the cell wall. Eukaryot. Cell 13 1484–1493. 10.1128/ec.00111-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Guclu H. (2013). Global maize trade and food security: implications from a social network model. Risk Anal. 33 2168–2178. 10.1111/risa.12064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Wang N., Guo X., Zhang Y., Te B. (2017). Comparative analysis of bacterial community structure in the rhizosphere of maize by high-throughput pyrosequencing. PLoS One 12:e0178425. 10.1371/journal.pone.0178425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wu X., Li G., Qin P. (2011). Interactions between arbuscular mycorrhizal fungi and phosphate-solubilizing fungus (Mortierella sp.) and their effects on Kostelelzkya virginica growth and enzyme activities of rhizosphere and bulk soils at different salinities. Biol. Fert. Soils 47:543. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LEfSe was used to identify the most differentially abundant taxa among population genotypes in the rhizosphere of maize. Bar graph showing LDA scores for fungi (A,B) and bacteria (C,D) are represented. The length of the bar column represents the LDA score. The figure shows the microbial taxa with significant differences between the “Pigarro” (red) and “SinPre” (green). Only taxa meeting an LDA significant threshold > 2 are shown.
Heat-map of the relative abundance with distance measure using Euclidean, at the level class for fungal (A) and bacterial (B) community. The relative abundance is scaled by a color gradient bar.
Soil characterization of the trial locations in august 2019.
Characterization of maize populations using HUNTERS descriptors in conventional and organic farming systems.
Abundance of fungal genera detected in each sample and the respective taxonomic assignment.
Abundance of bacterial genera detected in each sample and the respective taxonomic assignment.
Fungal and bacterial core rhizosphere microbiota associated with maize.
Fungal and bacterial genera specific to the rhizosphere microbiota in each population.
Fungal and bacterial genera specific to the rhizosphere microbiota of maize in distinct farming system.
Genera relative abundance significantly different between “Pigarro” and “SinPre” populations (P < 0.05).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA675280.