Abstract
Poor sleep quality can have harmful health consequences. Although many aspects of sleep are heritable, the understandings of genetic factors involved in its physiology remain limited. Here, we performed a genome-wide association study (GWAS) using the Pittsburgh Sleep Quality Index (PSQI) in a multi-ethnic discovery cohort (n = 2868) and found two novel genome-wide loci on chromosomes 2 and 7 associated with global sleep quality. A meta-analysis in 12 independent cohorts (100 000 individuals) replicated the association on chromosome 7 between NPY and MPP6. While NPY is an important sleep gene, we tested for an independent functional role of MPP6. Expression data showed an association of this locus with both NPY and MPP6 mRNA levels in brain tissues. Moreover, knockdown of an orthologue of MPP6 in Drosophila melanogaster sleep center neurons resulted in decreased sleep duration. With convergent evidence, we describe a new locus impacting human variability in sleep quality through known NPY and novel MPP6 sleep genes.
Keywords: sleep quality, genome-wide association study, MPP6, sleep centers
Statement of Significance.
Although many aspects of sleep are heritable, the genetic architecture of sleep quality remains poorly understood. Here, we conduct a genome-wide association study using the Pittsburgh Sleep Quality Index (PSQI) in a multi-ethnic discovery cohort. We discovered and replicated a locus on chromosome 7 between NPY and MPP6 with polymorphisms associated with poor sleep quality. Expression data suggests a higher expression of NPY and MPP6 in the brain. NPY, which codes for neuropeptide Y, has been found to promote sleep in humans and was identified as an important candidate gene in sleep regulation in Drosophila melanogaster. We tested the functional role of MPP6 in lateral ventral neurons and confirmed that MPP6 increases sleep duration in D melanogaster.
Introduction
Sleep is essential for brain homeostasis and optimal functioning [1]. Poor sleep has been shown to have a negative impact on multiple biological processes and can have harmful health consequences [2]. There is an intrinsic consequence to poor sleep in the risk of disease; for instance, circadian disruption in prolonged night shift workers increases risk of mortality from heart disease and cancers [3]. An overwhelming majority of chronic pain patients also suffer from poor sleep. The complaints of poor sleep and pain usually co-occur and lead to deteriorating quality of life [4]. Furthermore, poor sleep is associated with major depressive disorders and increased anxiety [5]. The term poor sleep encompasses a wide range of sleep disorders that can include, but are not limited to, insomnia, sleep-related breathing disorders, circadian rhythm disorders, and sleep quality disturbances. Sleep quality is a complex phenotype that is defined as a construct of sleep duration, sleep latency, number of arousals during sleep, and sleep restfulness [6, 7]. Laboratory sleep assessment is difficult and costly, but validated questionnaires like the Pittsburgh Sleep Quality Index (PSQI) help capture sleep quality in healthy and clinical populations [6]. Self-perceived poor sleep quality can be difficult to assess. The PSQI captures various components that can potentially affect sleep quality, such as sleep latency, sleep duration, and sleep efficiency. The content of PSQI has been validated against measures taken from polysomnography and covers multiple aspects relevant to the sleep quality construct [7]. Although many aspects of sleep are heritable (genetic factors explaining an estimated 17%–45% of phenotypic variance), the understanding of genetics involved in its physiology remains limited [8].
Genetic factors have previously been shown to influence multiple sleep traits like circadian rhythms, sleep duration, sleep latency, sleep apnea, and restless leg syndrome [8–14]. For instance, MEIS1 has been repeatedly associated with restless leg syndrome, whether idiopathic or familial [15, 16]. Another gene, PAX8 has been associated with sleep duration and was replicated in independent cohorts [17]. Recently, genome-wide association studies (GWAS) in large datasets on self-reported insomnia and excessive daytime sleeping also identified MEIS1 and PAX8 genes, but with further analyses it became clear that the observed signal was likely to be driven by another sleep disorder, namely, restless leg syndrome [9, 18]. Overall, we are at the beginning of our understanding of the genotypic architecture of sleep phenotypes, and there is an unmet need to perform standardized GWAS analysis of sleep using validated methodology. Based on the overall heritability of sleep in human populations, we would predict small effects from multiple genetic variants. Despite substantial evidence for the heritability of sleep quality using the PSQI (37%) [19, 20], to date, no GWAS has been reported using this tool.
In order to identify genetic factors implicated in sleep quality, here we present results of GWAS using the PSQI among US adults in the Orofacial Pain: Prospective Evaluation and Risk Assessment (OPPERA) cohort [21]. Genome-wide significant loci were then carried forward for replication across 12 independent cohorts, combining more than 100 000 individuals. Finally, we performed functional validation of a replicated locus through analysis of expression and by monitoring behavioral sleep patterns in transgenic Drosophila melanogaster. Together, our systematic genetic analysis of sleep has identified a novel conserved sleep locus, and these data help provide a better understanding of the underlying biological mechanisms contributing to sleep.
Methods
Cohort description
Orofacial Pain Prospective Evaluation and Risk Assessment
Study participants were selected from the OPPERA study, described in detail elsewhere [22]. In brief, the OPPERA cohort is a large population-based study designed to identify the psychological and physiological risk factors, clinical characteristics, and associated genetic mechanisms that influence the development of temporomandibular disorders (TMD) and related phenotypes. Individuals aged 18 to 44 years were recruited from four demographically diverse US locations (Buffalo, New York; Gainesville, Florida; Baltimore, MD; Chapel Hill, North Carolina). Over 200 pain phenotypes and pain-related comorbidities were collected within this study. For the current analysis, the phenotype of interest is the global score of the PSQI. The PSQI is a 19-item standardized validated instrument that assesses subject sleep quality over the last month [6]. Global score is derived from the sum of seven sub-components, namely subjective sleep quality, sleep latency, sleep duration, habitual sleep efficiency, sleep disturbances, use of sleep medication, and daytime sleepiness. PSQI global score ranges from 0 to 21, where lower scores denote good sleep quality and higher scores denote poor sleep quality. Other sleep phenotypes, such as insomnia and restless leg syndrome were not assessed in OPPERA.
Ethics statement
All OPPERA participants provided informed, signed consent for all study procedures. The OPPERA study was approved by institutional review boards at each of the four study sites (Buffalo, New York; Gainesville, Florida; Baltimore, MD; Chapel Hill, North Carolina) and at McGill University, Montreal, Canada.
Genotyping, quality control and imputation
DNA samples were extracted from whole blood following purification using a Qiagen Extraction Kit. A total of 3161 samples were genotyped for discovery using the Illumina HumanOmni2.5Exome-8v1A array (Illumina, Inc., San Diego, CA) at the Center for Inherited Disease Research (Johns Hopkins University, Baltimore, MD); from those, there were 2150 healthy controls and 1011 chronic TMD cases. Genetic data cleaning was done by the Genetics Coordinating Center at the University of Washington following their established pipeline [23]. Batch effects were assessed by comparing missing call rate and chi-square test for allelic frequency between genotyping batches. Sample identity and sample quality analyses included checks for missing call rate, chromosomal anomalies, cryptic relatedness, autosomal heterozygosity outliers, gender mismatch, and genetic ancestry. The median call rate was 99.9%. Cryptic relatedness was calculated using kinship coefficient with the R package SNPRelate. Samples were excluded if there was a discrepancy between annotated and genetic sex, the presence of chromosomal abnormalities, and higher than second-degree relatedness (19 parent-offspring pairs, 8 full siblings, and 11 second-degree relatives). With these criteria, 57 subjects were excluded for a total of 3104 samples that passed QC. Upon merging with available PSQI scores, a total of 2868 individuals (1092 males and 1776 females) were retained for the analysis.
Consistency of genotyping calls was assessed using 68 duplicates of study samples and 66 stock samples from HapMap reference subjects. SNP quality checks included assessments of missing call rate, duplicate discordance, and Mendelian errors. Because of the mixed population structure of the OPPERA cohort, Hardy-Weinberg equilibrium (HWE) was tested in homogenous European- and African-ancestry groups separately.
Cleaned genotypes were then imputed to the 1000 Genomes Project phase 3 reference panel, [24] using the software packages SHAPEIT2 [25] for pre-phasing and IMPUTE2 [26] for imputation. The IMPUTE2 algorithm was selected because it was recommended for use in a genetically diverse study sample using a worldwide reference panel. The IMPUTE2 algorithm uses a “k_hap” value to specify which number of reference haplotypes should be used to impute each study sample. The implementation of this parameter is one of the ways imputation with a worldwide reference panel is made computationally feasible: i.e. by choosing a subset of reference haplotypes to impute each study sample based on perceived genetic similarity [27]. The default k_hap value is 500; however, higher values are recommended when imputing into admixed populations. Thus for this project, we set k_hap to 2,000. Following SNP QC and imputation, 35 million high-quality SNPs were retained for the analysis.
Power calculation
We used the method described in [28] that allows estimating the expected proportion of false positives (expected False Discovery Rate, or “expected FDR”) among a specified number of the smallest p-values, U. The expected number of true positives among U top-scoring SNPs is given by (1-eFDR)*U. We set U = 10, N = 2868, the SNP frequency equal to 0.15 (the minimum frequency used in the present study for association testing was 0.05), and varied the assumed actual number of true positives among about 1.8 million tested SNPs as M = (10, 50, 100, 250, 500). To relate the effect size for the continuous standardized outcome to a commonly used measure for binary outcomes, the odds ratio, we took these effect sizes to correspond to three different values of the odds ratio, 1.2, 1.25, and 1.3. These values give the upper bound for FDR if at least M SNPs have the assumed odds ratio.
Statistical analysis
PLINK v.1.90 software was used for genome-wide association analysis under an additive model of inheritance [29]. Global PSQI scores were used as a dependent variable in a linear regression model. Covariates included in the equation were age, gender, dummy-coded recruitment sites, three principal components to account for population stratification, and TMD case status to account for recruitment bias. The three principal components were used as they account for the majority of the variance (Supplementary Figure 1). Furthermore, we also repeat the analysis using five principle components to further increase stringency of a control for population stratification. Genome-wide statistical significance threshold was set at p < 5 × 10−8.
In addition to the analysis in the full cohort, we also performed a stratified analysis by sex and by genetically defined race to account for multi-ancestry. For the race stratified analysis, one principle component generated using each race separately was used in the model, along with age, gender, dummy-coded recruitment sites and TMD case status. Manhattan and quantile-quantile (Q-Q) plots were generated using the R-package qqman [30]. Heritability of PSQI global score was computed using GCTA [31]. A conditional analysis was performed by adjusting for the other SNP that passed genome-wide significance by using its minor allelic count as a co-variable in the linear regression model.
Expression Quantitative trait loci (eQTL) analysis
Gene expression was explored using publicly available eQTL online resources. The BRAINEAC database [32], which contains gene expression data across 10 brain regions (cerebellar cortex, frontal cortex, hippocampus, medulla, occipital cortex, putamen, substantia nigra, temporal cortex, thalamus, and white matter) was used to identify eQTL with an averaged brain tissues expression. Averaged brain tissues expression was obtained by simply averaging expression values across all 10 brain regions. In addition to BRAINEAC, the GTEx portal version 6 [33] was used in 12 selected brain tissues (amygdala, anterior cingulate cortex, caudate, cerebellar hemisphere, cerebellum, cortex, frontal cortex, hippocampus, hypothalamus, nucleus accumbens, putamen, and substantia nigra).
GO enrichment pathway analysis in biological process
SNPs were assigned to genes based on distance. For cis-effects, we considered distances between the SNP and the gene locus up to 10 000 nucleotides, on both positive and negative genomic DNA strands. Analyses were performed using the Gene Ontology (GO) biological processes pathways definitions (file retrieved May 24, 2016 [34]). To generate a p-value for each pathway, we collected all SNP p-values in cis for all genes pertaining to the pathway. We compared the distribution of PSQI GWAS p-values among SNPs belonging to the pathway with the distribution of p-values for SNPs not belonging to the pathway. Comparisons were made with the Kolmogorov-Smirnov test using the ks.test function implemented in R version 3.6.0. The test was performed in a 1-sided fashion, because we searched for pathways enriched with SNPs of lower p-values. A total of 1133 pathways were inspected for enrichment. Adjustment for multiple testing was performed using Bonferroni correction.
Data availability
Study data have been deposited and made publicly available at the Database of Genotypes and Phenotypes (dbGaP) public repository (accession number phs000796.v1.p1).
Replication cohorts’ description
UK Biobank
The UK biobank is a prospective study that includes more than 500 000 people living in the United Kingdom [35]. In total, over 9.2 million invitations to participate in the study were sent, from which 503 325 individuals were recruited between 2006 and 2010. Participants were part of the National Health Service registry, aged between 40–69 years and living less than 25 miles from a study center. Recruited study participants gave informed consent and completed questionnaires; underwent a range of physical measures; and blood, urine, and saliva were collected for genetic data. Samples were genotyped on the UK BiLEVE array (~50 000 samples) and the UK Biobank Axiom array (~450 000 samples). Analysis was performed on the interim release of genotype data of 152 000 samples. Arrays contain around 800 000 markers. Following standard QC described elsewhere (http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=155580), SNPs were imputed for a total of 73 355 667 SNPs after phasing the autosomes using a modified version of the SHAPEIT3 program modified to allow for very large sample sizes. Imputation was then carried out using IMPUTE3 according to UK biobank standard described elsewhere (http://www.ukbiobank.ac.uk/wpcontent/uploads/2014/04/imputation_documentation_May2015.pdf). Sleep quality was assessed using a self-reported question: “Do you have trouble falling asleep at night or do you wake up in the middle of the night?” with four possible answers: “never/rarely,” “sometimes,” “usually,” or “prefer not to answer.” This phenotype was selected because it is a proxy to the global PSQI score with sensitivity of 0.94, specificity of 0.89, and ROC AUC of 0.947 [12]. The phenotype was dichotomized by using only “usually” as cases and “never/rarely” as controls. Association testing was performed using dosage data with SNPTEST v2.5.2 threshold method. Age, gender, genotyping array, and five principal components (PC) to account for population stratification were used as covariates. The current study was conducted under UK biobank application number 20802.
Hispanic Community Health Study /Study of Latinos (HCHS/SOL)
HCHS/SOL is a longitudinal multicenter cohort study of the Hispanic/Latino community in the United States with initial visits between 2008 and 2011. Participants were recruited in a two-stage sampling scheme of individuals from the Bronx, NY; Chicago, IL; Miami, FL; and San Diego, CA for a total of 16 415 individuals enrolled [36]. Of these, 12 803 individuals consented to participate in genetic studies. In the current analysis, n = 10 610 individuals participated after applying exclusion criteria. Genotyping was performed with an Illumina custom array (15041502 B3), which consists of the Illumina Omni 2.5 M array (HumanOmni2.5-8v1-1) plus approximately 150k custom SNPs. Genome-wide imputation was carried out using the 1000 Genomes Project phase 1 reference panel, SHAPEIT2, and IMPUTE2 software. The quantitative phenotypic outcome is the WHIIRS (Women’s Health Initiative Insomnia Rating Scale), which is derived from form code SLE – HCHS/SOL Sleep questionnaire:
where SLE4 is “Did you have trouble falling asleep?”; SLE5 is “Did you wake up several times at night?”’ SLE6 is “Did you wake up earlier than you planned to?”; SLE7 is “Did you have trouble getting back to sleep after you woke up too early?” with possible answers being “No, not in the past 4 weeks,” “Yes, less than once a week,” “Yes, 1 or 2 times a week,” “Yes, 3 or 4 times a week” and “Yes, 5 or more times a week.” SLEA11 asks “Overall, was your typical night’s sleep during the past 4 weeks” with possible answers “Very sound or restful,” “sound or restful,” “average quality,” “restless,” “very restless.”
High WHIIRS scores represent poor sleep quality. For this analysis, WHIIRS score was dichotomized with disturbed sleep defined as WHIIRS score of at least 9. Sex, age, recruitment center, five PC, and TMD status were used as covariates in the analysis. Subjects were excluded from the analysis if they presented any other sleep disorder, such as restless leg syndrome, narcolepsy, or sleep apnea, or if they were taking sleeping pills for the past 4 weeks.
EA-CRASH
This is a prospective cohort study of adults of European ancestry (EA) who presented to an emergency department (ED) following a motor vehicle collision (MVC) [37]. Patients were enrolled at nine study sites across Eastern United States. From a total of 10 629 patients screened, 1416 were eligible, 969 consented to study participation, and 948 were enrolled. Data and blood samples were collected at the initial emergency department visit, and follow-up data was collected at 6-week, 6-month, and 1-year assessments. Sleep was assessed using the question: “Prior to the accident, in the past month, please rate your insomnia or difficulty sleeping” with possible answers coded using a 0–10 scale were 0 denotes “no problems” and 10 denotes “major problems” with sleep. DNA was extracted from PAXgene blood DNA tubes and SNPs were genotyped using Sequenom technology. Due to the low frequency of discovery SNPs in EA, minor allele frequency of rs11976703, rs73284230 and rs60869707 was 1%, 0.1%, and 6%, respectively. Moreover, genotyping rate of rs73284230 was 92.2%, which is lower than the accepted threshold of 95%. The regression analysis included age and gender as covariates in the model.
AA-CRASH
The African American (AA) CRASH study (n=915) is a sister study of EA CRASH. Similar to EA CRASH, AA CRASH is a prospective multicenter observational cohort study of AA individuals ≥18 and ≤65 years of age who presented within 24 hours of MVC to one of 11 EDs in six states/districts (Michigan, Pennsylvania, Florida, Alabama, Massachusetts, and Washington, DC). The full details of this study have been described previously [38]. DNA was collected in the ED using PAXgene DNA tubes. Sleep quality in the month prior to MVC was assessed with the same question as in the EA CRASH study (above). Following DNA purification (PAXgene blood DNA kit, QIAGEN), genotyping using the Infinium Multi-Ethnic Global (MEG) Array (Illumina) was performed. DNA from an individual with known genotype (NA19819, 1000 genomes) and two repeat samples were included in each genotyping batch (96 samples) to ensure genotypic accuracy and reliability. SNPs rs11976703, rs73284230, and rs60869707 were not included in the MEGA array and were thus imputed using available genotyping data. Following, stringent QC and accounts of relatedness, the regression analysis included age, gender, and study site as covariates in the model.
The Finnish BrePainGen cohort
The BrePainGen cohort consists of 1000 Finnish women (aged 18–75 years) who had unilateral non-metastasized breast cancer and received surgery at the Breast Surgery Unit, Helsinki University Hospital, between August 2006 and December 2010. The day before surgery, following informed consent acquisition, medical and medication demographic history was taken and background data collected. Patients also underwent experimental cold and heat pain tests and answered psychological questionnaires that included questions about insomnia. This prospective study cohort has been described in detail earlier [39]. Genotyping was done at the Wellcome Trust Sanger Institute (Hinxton, United Kingdom) using the Human OmniExpress Illumina BeadChip (Illumina, Inc., San Diego, CA). After stringent sample quality control procedures, a total of 926 samples passed QC. An MDS plots (12 dimensions) was used to remove outliers (N = 4), when the rest of the subjects represented a homogeneous population. SNPs were filtered based on minor allele frequency (MAF > 0.005), Hardy-Weinberg equilibrium (HWE p > 1 × 10−6) and success rate (>0.97). Insomnia data was available for 823 participants. The sub-sample used in this study consists of the 757 individuals with both genotype and insomnia questionnaire data. Participants who stated that they never had insomnia were considered as unaffected (n = 399) and those who said that they have insomnia at least once a week (n = 237) or every night (n = 121) were considered affected, during which also MDS.
Post-mastectomy pain syndrome cohort
This cohort (N = 1200) was recruited from the Comprehensive Breast Cancer Program’s registry of breast cancer patients undergoing total or partial mastectomy at Magee Women’s Hospital of University of Pittsburgh Medical Center. University of Pittsburgh Institutional Review Board approval was obtained prior to all data collection, and all patients gave informed consent before participation in the study. The majority were postmenopausal. Ethnic group was primarily white/Caucasian. The percentage of women of ethnic groups other than white/Caucasian in the study was limited, making an assessment of racial/ethnic differences inappropriate. Patients completed study questionnaires a mean of 38.3 ± 35.4 months (range, 2 months–10 years) after surgery. Full cohort description was reported elsewhere [40]. Sleep disturbance was assessed using a short-form instrument from the National Institutes of Health roadmap initiative, Patient Reported Outcome Measurement Information System (PROMIS) [41]. Genotyping was done using the UK Biobank Axiom platform on samples derived from lymph node tissue, blood or saliva. Genotyping was done using the UK Biobank Axiom platform on samples derived from lymph node tissue, blood, or saliva at the Genome center at McGill University. Following QC and imputation, a total of 665 samples were used in the replication. Age and 3 principal components were used in the regression model.
Complex persistent pain conditions
The Complex Persistent Pain Conditions (CPPC): Unique and Shared Pathways of Vulnerability study included 745 participants enrolled in a case control study of overlapping pain conditions conducted at UNC Chapel Hill. Subjects were aged 18–64, and included both sexes (86% female) and major ethnic and racial groups (68% non-Hispanic white). All subjects had at least one of four index CPPCs (episodic migraine, irritable bowel syndrome, fibromyalgia, or vulvar vestibulitis), or were otherwise healthy controls with none of these conditions. Sleep quality was assessed using the PSQI. DNA from all subjects was genotyped using the Axiom Precision Medicine Research Array by Genome center at McGill University. PSQI was used as a dependant variable in a linear regression model with age, gender, three PCs, and CPPC dummy-coded.
OPPERA-S
The OPPERA–Subset (OPPERA–S) cohort was a subset of the original OPPERA cohort that consists of 973 healthy controls that were not genotyped as part of the initial project as they were saved for further replication. Sleep was assessed using the PSQI and the same analytical plan was used as in the discovery cohort. DNA from all subjects was genotyped using the Axiom Precision Medicine Research Array. Genotyping data was cleaned and imputed in the same manner as the discovery cohort. Covariates included in the analysis included age, gender, dummy coded recruitment sites, and three PCs.
OPPERA-R
The OPPERA-Replication (OPPERA–R) case-control study of chronic TMD (NIDCR protocol 12-052-E) was designed as a replication study from the initial OPPERA discovery GWAS. Recruitment was independent from the discovery cohort as it was done in 2016–2017. Potential subjects were recruited by telephone screening of 166 062 phone numbers listed in counties surrounding the four OPPERA initial recruitment sites, of which, 2430 were eligible and 1342 subjects (66% female, age 18–74) returned complete phenotype and genotype information and were included in the replication analysis. Phenotype was assessed using the question “please rate the quality of your sleep in the past 3 months on a 0–10 scale” where 0 represented the worst sleep quality and 10 represented the best sleep quality. Saliva samples for DNA genotyping were obtained using Oragene collection tubes (DNA Genotek Inc., Kanata, ON, Canada). DNA from all subjects was genotyped using the Axiom Precision Medicine Research Array. Sleep scale scores were used as a dependent variable in a linear regression. Covariates included in the equation were age, gender, dummy coded recruitment sites, and three PCs.
Jackson Heart Study
The Jackson heart study (JHS) is a populational-based longitudinal prospective cohort aiming to investigate cardiovascular disease among AAs. Recruitment is community-based in the Jackson Mississippi metropolitan area. This study recruited 5302 AA adults between 2012 and 2016 [42]. For this analysis, we used a subset of 2999 individuals that had both sleep assessment and genetic data. Sleep quality was determined at Exam 1 using the question “How do you rate your overall sleep quality?” with possible answers: Excellent, very good, good, fair, and poor. This cohort was genotyped using Affymetrix and imputed to a 1000 Genomes phase 1 version 3 template [43]. A univariate linear mixed model adjusted for genetic relatedness, age, and sex was used for each analysis using GEMMA version 0.94.1 [44].
Multi-Ethnic Study of Atherosclerosis
The multi-ethnic study of atherosclerosis (MESA) is a multicenter prospective cohort study aiming to study the development of cardiovascular disease. A total of 6,814 individuals between the ages of 45 and 65 were recruited for the first examination between 2000 and 2002. Participants were recruited in six US cities (Baltimore, MD; Chicago, IL; Forsyth County, NC; Los Angeles County, CA; Northern Manhattan, NY; and St. Paul, MN) [45]. In the present analysis, 518 participants from AA ancestry with complete data were used for replication (28%). Sleep quality was assessed using the question “In the past 4 weeks, rate the overall typical night of sleep,” extracted from the Center for Epidemiologic Studies Depression Scale (CES-D). Possible answers were: Very sound or restful, sound and restful, average quality, restless, very restless. This cohort was genotyped using the Affymetrix 6.0 array, phased using SHAPEIT and imputed using IMPUTE2 with the Haplotype Reference Consortium version 1.1 template [46]. A univariate linear mixed model adjusted for genetic relatedness, age, and sex was used for each analysis using GEMMA version 0.94.1 [44].
Cleveland Family Study
The Cleveland Family Study (CFS) aims to examine the genetic and familial basis of sleep apnea and consists of 2534 AAs and European Americans from 356 families. Index probands with sleep apnea were recruited from northern Ohio sleep centers [47]. Sleep was assessed using the question, “During the last month, have you had, or have you been told you do the following while asleep or trying to sleep? Toss, turn, or thrash frequently over the night.” Values ranged from 0 (Never) to 4 (always or almost always, or 5–7 times per week). A total of 719 AA individuals who had genotype data available and non-null values for the outcome variable were used for analysis. This cohort was genotyped using the Affymetrix 6.0 array, phased using SHAPEIT and imputed using IMPUTE2 with the Haplotype Reference Consortium version 1.1 template [46]. A univariate linear mixed model adjusted for genetic relatedness, age, and sex was used for each analysis using GEMMA version 0.94.1 [44].
Meta-analysis
An additive model of inheritance was used to generate summary statistics for each replication cohort. The direction of phenotype scale in each cohort was reverted if necessary to be consistent across all cohorts included in the meta-analysis, i.e. higher score represents poor sleep. Next, genotypic effect statistics were corrected to reflect the effect of the same allele in each cohort. Because sleep was assessed differently in each of the replication cohorts, genotypic effects were standardized according to the method described elsewhere [48]. Briefly, if the phenotype was binary, the effect size was converted to a continuous standardized scale using the following formula:
If the phenotype was continuous, the regression coefficient or the difference in mean generated was converted to a standardized effect size by dividing the effect size by the residual standard deviation [49].
Meta-analysis of replication studies, excluding OPPERA discovery was computed using the R-package metafor using a fixed-effect method. The p-value shown is a two-sided p-value. Heterogeneity of effects was verified using the heterogeneity coefficient Q for each SNP. A race-specific meta-analysis was also computed to account for ethnicity.
D melanogaster assay
Fly strain
PDF-Gal4 has been previously described [50]. Varicose RNAi hairpins 1&2 (GD #24157, KK104548) was from the Vienna Drosophila RNAi Center VDRC. Flies were reared on a standard cornmeal–yeast–agar medium at 25°C and 70% relative humidity in a 12 hour L:12 hour D cycle. Flies were placed in glass tubes containing standard fly food (2% agar and 5% sucrose). Flies were acclimated for at least 18 hour at 25°C in Light and Dark (LD) conditions, and then data were collected in LD for 7 days with the Drosophila Activity Monitoring (DAM) System (Trikinetics, Waltham, MA) in 5-minute bins. Sleep parameters were measured by averaging 5 days of LD [51]. The sleep parameters tested were: circadian patterns of sleep, percentage of sleep, number of sleep episodes, and the duration of sleep episodes in minutes. These parameters were measured separately for L and D intervals. Group comparison was done between parental controls (PDF-Gal4/+, UASVariIR1-2/+) and varicose knockdown flies (DF-Gal4>UAS-variIR1-2). All statistical analysis was performed in Prism 7.0. Significance levels were determined by one-way ANOVA with Dunnett’s multiple comparisons test.
Results
GWAS in OPPERA
We first performed a primary discovery GWAS of sleep quality measured by the PSQI in the OPPERA cohort. The OPPERA cohort was comprised of self-declared non-Hispanic whites (NHW, 58.4%), AAs (25.8%), and other ethnic/racial groups, including Asians, Pacific Islanders, Native Americans, and individuals with mixed races (15.8%). Racial differences in sleep quality were observed in our data, with AA reporting lower sleep quality compared to NHW (Supplementary Table 1), which is consistent with previous reports [52].
Genome-wide analysis using the PSQI global score identified two loci on chromosomes 2 and 7 at genome-wide significance (p ≤ 5 × 10−8), and one locus at suggestive significance (p ≤ 5 × 10−7), on chromosome 13 (Table 1, Figure 1, Supplementary Figure 2, Supplementary Table 2). PSQI global score was associated with rs11976703 (effect allele C, β = 0.78, standardized effect size = 0.23, p = 3.78 × 10−8) and rs73284230 (effect allele G, β = 0.95, standardized effect size = 0.28, p = 4.76 × 10−8) on chromosome 7 and rs60869707 (effect allele G, β = 1.09, standardized effect size = 0.32, p = 5.03 × 10−8) on chromosome 2. The effect allele (the major allele) in each of these three SNPs was associated with higher global PSQI scores, hence worse sleep quality. Genome-wide significant SNPs were also analyzed for association with four PSQI subscales: subjective sleep quality, sleep efficiency, sleep disturbance, and daytime dysfunction. All SNPs showed the strongest association with subjective sleep quality, followed by daytime dysfunction, in the same direction as the global score (Supplementary Table 3).
Table 1.
Genome-wide (p ≤ 5 × 10−8) and suggestive (p ≤ 5 × 10−7) loci associated with PSQI in the OPPERA discovery cohort
| SNP | Chr:position | *Nearest genes | EA/OA | EAF | INFO | β | SE | p |
|---|---|---|---|---|---|---|---|---|
| rs11976703 | 7:24549526 | NPY/MPP6 | C/A | 0.886 | 1.059 | 0.780 | 0.141 | 3.78 × 10 −8 |
| rs73284230 | 7:24548947 | NPY/MPP6 | G/A | 0.920 | 1.122 | 0.949 | 0.173 | 4.76 × 10 −8 |
| rs60869707 | 2:86023846 | ATOH8 | G/A | 0.930 | 1.171 | 1.086 | 0.199 | 5.03 × 10 −8 |
| rs151181914 | 7:24550927 | NPY/MPP6 | TATC/T | 0.921 | 1.126 | 0.917 | 0.173 | 1.27 × 10−7 |
| rs376585198 | 13:20529278 | ZMYM2 | ATTATT/- | 0.904 | 0.999 | 0.776 | 0.150 | 2.48 × 10−7 |
| rs78633772 | 13:20671080 | ZMYM2 | T/G | 0.909 | 1.000 | 0.789 | 0.154 | 3.31 × 10−7 |
| rs9579769 | 13:20598284 | ZMYM2 | A/G | 0.919 | 1.024 | 0.809 | 0.159 | 3.79 × 10−7 |
| rs9579744 | 13:20529559 | ZMYM2 | A/C | 0.911 | 1.024 | 0.781 | 0.154 | 4.08 × 10−7 |
| rs7318279 | 13:20602148 | ZMYM2 | C/T | 0.911 | 1.028 | 0.776 | 0.154 | 4.66 × 10−7 |
| rs115462079 | 13:20586370 | ZMYM2 | C/T | 0.911 | 1.028 | 0.776 | 0.154 | 4.66 × 10−7 |
| rs9578239 | 13:20539229 | ZMYM2 | A/T | 0.911 | 1.028 | 0.776 | 0.154 | 4.67 × 10−7 |
| rs9315234 | 13:20541404 | ZMYM2 | C/G | 0.911 | 1.023 | 0.777 | 0.154 | 4.99 × 10−7 |
| rs9579762 | 13:20584060 | ZMYM2 | T/C | 0.925 | 1.017 | 0.827 | 0.164 | 5.10 × 10−7 |
| rs71803599 | 13:20549173 | ZMYM2 | CATTT/C | 0.912 | 1.024 | 0.777 | 0.154 | 5.14 × 10−7 |
Chr: Chromosome; EA: Effect allele; OA: Other allele; EAF: Effect allele frequency; INFO: imputation quality from IMPUTE2. Position is based on NCBI Build 37 (hg19). Genome-wide significant results are shown in bold. *Intragenic for ZMYM2.
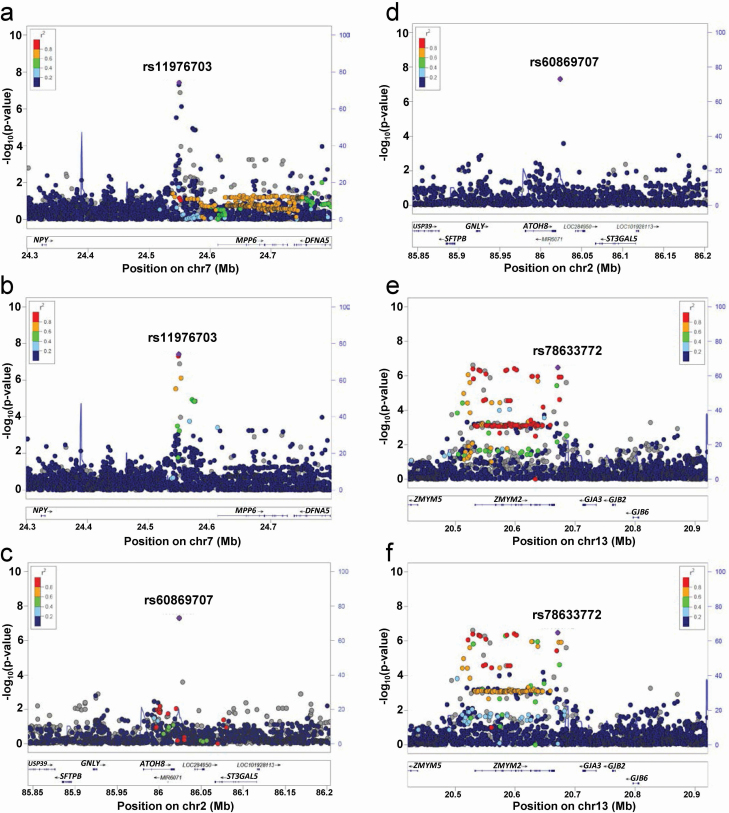
Figure 1.
Regional association plots of discovery GWAS for PSQI global score in OPPERA. (a–b) Regional association plots for genome-wide significant loci at chromosome 7 using EUR as a reference panel (a) and AFR panel (b). (c–d) Regional association plot at chromosome 2 using EUR as reference panel (c) and AFR panel (d). (e–f) Regional association plots for suggestive loci at chromosome 13 using EUR as reference panel (e) and AFR panel (f) Chromosomal position (Mb) is indicated on the x axis, and the –log10 p-value is indicated on the y axis. Each SNP is plotted as filled circle and the lead SNP is shown in purple. The genes within each region are shown in the lower panel. Recombination sites and rates are shown in blue. Additional SNPs in the locus are colored according to linkage disequilibrium (r2) with the lead SNP. rs78633772 instead of rs376585198 as the latter is not in the reference panel.
The top SNPs on chromosome 7 were in linkage disequilibrium (D′ = 0.99; R2 = 0.83) in all populations, highest in Africans (D′ = 0.99; R2 = 0.95) and lowest in Europeans (D′ = 1; R2 = 0.15). The minor allelic frequencies for genome-wide significant SNPs were much higher in AA than in NHW (26% vs. 6% for rs11976703, 25% vs. 1% for rs73284230, and 23% vs. 0.2% for rs60869707 in AA and NHW, respectively). In analyses stratified by self-declared race, these SNPs showed robust effects in AA (rs11976703: β = 0.95, p = 2.29 × 10−6; rs73284230: β = 0.99, p = 1.54 × 10−6; and rs60869707: β = 1.04, p = 6.17 × 10−7) but no effects in NHW (rs11976703: β = 0.52; p = 0.028; rs73284230: β = 0.59; p = 0.24; and rs60869707: β = 0.99, p = 0.99). A meta-analysis of ancestry specific results showed an association for rs11976703 (β = 0.75, p = 1.03 × 10−6; Q(2.56, p = 0.11, Ihet = 61), rs73284230 (β = 0.92, p = 2.6 × 10−6; Q(0.35, p = 0.55, Ihet = 0), and rs60869707 (β = 1.11, p = 1.36 × 10−7; Q(0.06, p = 0.81, Ihet = 0).
Genome-wide analyses stratified by race did not show any SNP above genome-wide significance in NHW, whereas two loci on chromosomes 8 and 16 near TSNARE1 and FAM234A, respectively, were significant in AA only (Supplementary Table 4 and Supplementary Figure 3). A sex-stratified analysis did not identify any significant SNPs in females, whereas in males rs28483449, on chromosome 15, located near ZNF710, was associated with PSQI (Supplementary Table 5).
Heritability and pathway analysis of sleep quality
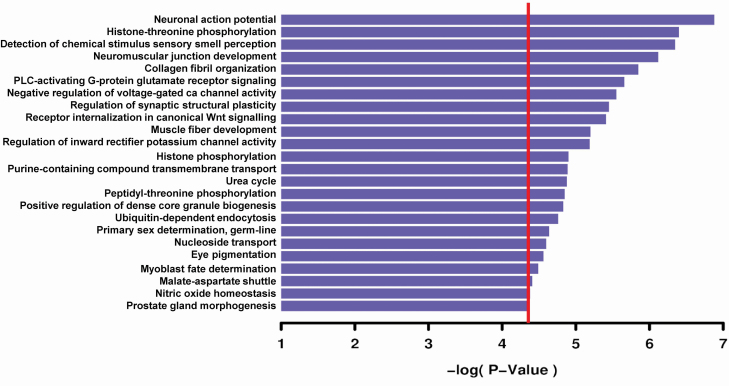
Next, GWAS results were used to measure heritability of sleep quality. It was estimated at 14.37%, using GCTA [31], which was consistent with other heritability estimates of sleep traits [9]. Furthermore, pathway analysis of the full GWAS results using gene ontologies (GO) for “biological process” class function identified many significant biological pathways, of which more than half reflected different aspects of neuronal action potential and activities at the synapse, whether receptor signaling, receptor internalization or the neuromuscular junction (Figure 2). These analyses are consistent with the current understanding of sleep processes, which largely depend on synaptic plasticity occurring during sleep, reinforcing the validity of results obtained from PSQI GWAS data [53].
Figure 2.
Pathway analysis of sleep GWAS using Gene ontology’s biological process. Horizontal bar plots represent –log10 p-value enrichment of pathways. The red line represents Bonferroni threshold for statistical significance.
Power analysis
In order to assess if the discovery OPPERA cohort was powered to detect genome-wide significant hits, we modeled the proportion of true hits given our sample size (2868), the number of SNPs tested (1.83M), and three values of the standardized effect size (0.33, 0.40, and 0.48). These values of effect sizes approximately correspond to the odds ratios 1.2, 1.25, and 1.3 in a case-control design [48]. The model showed that among 10 top-scoring SNPs, the expected false discovery rate (eFDR) [28] is high for the lowest standardized effect size (0.33) and reached only about 0.42 if the total number of tested SNPs carrying such effect size is 500. Thus, about six true positives were expected, i.e. (1-eFDR)*10 = 5.8. One would need about 200 true positive SNPs in the overall GWAS to bring eFDR below 0.05. However, larger standardized effect sizes (0.40 and 0.48) required smaller numbers of true positives. For example, about eight out of 10 top-scoring SNPs are expected to be true positives (eFDR = 0.153) for the standardized effect size equivalent to 0.48, assuming 100 true positive SNPs in total (Supplementary Figure 4). Hence, the discovery OPPERA cohort was well powered to detect true positives with the assumed effect sizes and densities in a GWAS using PSQI.
Replication and meta-analysis
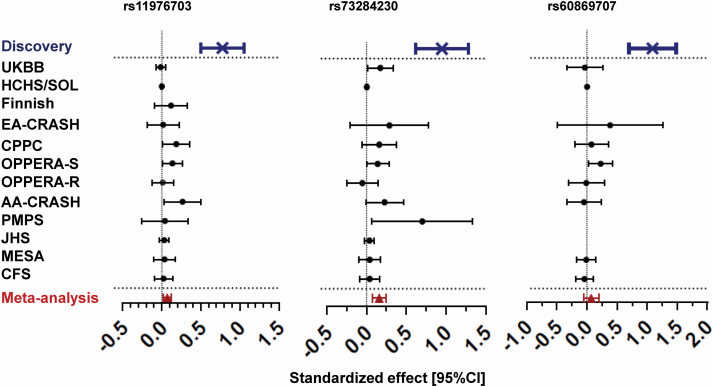
To replicate our genome-wide significant associations, we used 12 independent cohorts that assessed sleep quality (Supplementary Table 6). The closest phenotype that captures sleep quality in the absence of PSQI was tested for association in each replication cohort with rs11976703, rs73284230, and rs60869707. Each phenotype was normalized to account for different measures of sleep (see Methods). The replication studies’ association results were then combined using a fixed-effect weighted meta-analysis for a total sample size of 100 805. Both rs11976703 and rs73284230 had a p-value < 0.05 in more than one individual replication study, whereas rs60869707 was only statistically significant in one replication cohort. In the overall meta-analysis, all three SNPs showed an effect size that is in the same direction as the discovery cohort, but only rs11976703 and rs73284230, on chromosome 7 replicated in a meta-analysis combining all replication studies (rs11976703: standardized effect [95% CI] = 0.07 [0.02; 0.12]; p = 3.50 × 10−3 and, rs73284230 standardized effect [95% CI] = 0.16 [0.08; 0.25]; p = 2.0 × 10−4) (Table 2, Figure 3).
Table 2.
Association of genome-wide significant SNPs in independent replication cohorts and meta-analysis
| Sample size | Effect allele frequency | effect size (se) | p | ||
|---|---|---|---|---|---|
| rs11976703 | OPPERA-Discovery | 2,868 | 0.89 | 0.78 (0.14) | 3.78 × 10−8 |
| Effect allele (C) | UKBB | 79,947 | 0.95 | −0.01 (0.03) | 6.70 × 10−1 |
| HCHS/SOL | 7,247 | 0.87 | 0.002 (0.013) | 8.87 × 10−1 | |
| Finnish BrePainGen* | 800 | 0.95 | 0.12 (0.11) | 3.54 × 10−1 | |
| EA-CRASH | 894 | 0.95 | 0.018 (0.10) | 8.64 × 10−1 | |
| CPPC | 641 | 0.90 | 0.19 (0.09) | 3.60 × 10 −2 | |
| OPPERA-S | 929 | 0.87 | 0.14 (0.06) | 3.20 × 10 −2 | |
| OPPERA-R | 1,297 | 0.92 | 0.01 (0.07) | 8.45 × 10−1 | |
| AA-CRASH | 906 | 0.70 | 0.27 (0.12) | 2.10 × 10 −2 | |
| PMPS | 399 | 0.94 | 0.04 (0.15) | 7.87 × 10−1 | |
| JHS | 2,999 | 0.73 | 0.03 (0.03) | 2.76 × 10−1 | |
| MESA | 518 | 0.74 | 0.04 (0.07) | 5.99 × 10−1 | |
| CFS | 719 | 0.74 | 0.02 (0.06) | 7.04 × 10−1 | |
| Meta-analysis | 97,296 | 0.07 (0.03) | 3.6 × 10 −3 | ||
| rs73284230 | OPPERA-Discovery | 2,868 | 0.92 | 0.95 (0.17) | 4.76 × 10−8 |
| Effect allele (G) | UKBB | 79,947 | 0.99 | 0.18 (0.08) | 3.50 × 10 −2 |
| HCHS/SOL | 7,247 | 0.90 | 0.007 (0.014) | 6.17 × 10−1 | |
| EA-CRASH | 895 | 0.99 | 0.29 (0.25) | 2.52 × 10−1 | |
| CPPC | 657 | 0.93 | 0.16 (0.11) | 1.50 × 10−1 | |
| OPPERA-S | 944 | 0.90 | 0.14 (0.07) | 4.90 × 10 −2 | |
| OPPERA-R | 1,328 | 0.96 | −0.05 (0.10) | 6.04 × 10−1 | |
| AA-CRASH | 915 | 0.71 | 0.23 (0.12) | 7.80 × 10−2 | |
| PMPS | 399 | 0.99 | 0.70 (0.32) | 3.02 × 10 −2 | |
| JHS | 2,999 | 0.74 | 0.04 (0.03) | 2.16 × 10−1 | |
| MESA | 518 | 0.75 | 0.04 (0.07) | 5.65 × 10−1 | |
| CFS | 719 | 0.75 | 0.04 (0.06) | 5.25 × 10−1 | |
| Meta-analysis | 96,568 | 0.16 (0.04) | 2.0 × 10 −4 | ||
| rs60869707 | OPPERA-Discovery | 2,868 | 0.93 | 1.09 (0.20) | 5.03 × 10−8 |
| Effect allele (G) | UKBB | 79,947 | 0.99 | −0.03 (0.15) | 8.28 × 10−1 |
| HCHS/SOL | 7,247 | 0.96 | 0.011 (0.025) | 6.67 × 10−1 | |
| EA-CRASH | 896 | 0.99 | 0.39 (0.45) | 3.88 × 10−1 | |
| CPPC | 622 | 0.94 | 0.08 (0.14) | 5.98 × 10−1 | |
| OPPERA-S | 877 | 0.92 | 0.23 (0.10) | 2.10 × 10 −2 | |
| OPPERA-R | 1,282 | 0.97 | −0.01 (0.15) | 9.40 × 10−1 | |
| AA-CRASH | 915 | 0.76 | −0.05 (0.15) | 6.32 × 10−1 | |
| MESA | 518 | 0.79 | −0.01 (0.08) | 8.89 × 10−1 | |
| CFS | 719 | 0.78 | −0.04 (0.07) | 5.67 × 10−1 | |
| Meta-analysis | 93,023 | 0.07 (0.06) | 2.48 × 10−1 |
*All directions are presented with respect to the effect allele. SNPs rs73284230 and rs60869707 were not genotyped in the Finnish and rs60869707 was not genotyped in the PMPS and the JHS cohorts. The OPPERA Discovery cohort was excluded from the meta-analysis calculation. OPPERA: Orofacial Pain: Prospective Evaluation and Risk Assessment; UKBB: UK biobank; HCHS/SOL: Hispanic Community Health Study/Study of Latinos; EA: European American; AA: African American; PMPS: Post-mastectomy pain syndrome. CPPC: Complex Persistent Pain Conditions. OPPERA-S: Subset; OPPERA-R: Replication; JHS: Jackson Heart Study; MESA: Multi-Ethnic Study of Atherosclerosis CFS: Cleveland Family Study. Statistically significant results are shown in bold. OPPERA-Discovery cohort was not included in the meta-analysis.
Figure 3.
Forest plots in meta-analysis. Forest plots of standardized effect size with 95% confidence interval for each replication study as well as for the fixed-effect meta-analysis for genome-wide significant SNPs. Higher effect sizes represent worse sleep quality. The discovery cohort was excluded from the meta-analysis calculation. The test of heterozygosity for each SNP was Q(df 11) = 16.29 p = 0.13 for rs11976703; Q(df 10) = 20.52 p = 0.03 for rs73284230 and Q(df 8) = 6.74 p = 0.57 for rs60869707. The sample sizes for the meta-analysis is 100659, 99931, and 96386 for rs11575542, rs73284230, and rs60869707, respectively.
In order to account for the large difference in allelic frequency between Europeans and African ancestries, we undertook a separate race-stratified meta-analysis in racially homogeneous cohorts. In Non-Hispanic whites ancestry only replication cohorts, both rs11976703 and rs73284230 replicated in the same direction as the discovery cohort (effect = 0.07 and 0.16; p = 0.047 and 2.0 × 10−4, respectively). The same was also true in African ancestry replication cohorts (effect = 0.09 and 0.087; p = 0.017 and 0.027, respectively). Overall, we concluded that both SNPs rs11976703 and rs73284230 have a significant effect on sleep quality in both ancestries (Supplementary Table 7).
The following analyses will focus solely on the locus of chromosome 7 given that it is the only locus that replicated in an independent meta-analysis.
Genetic analysis of locus chromosome 7
Using a probabilistic identification of causal SNP (PICS) approach, we determined that the probability for causality was distributed as 51.33% for rs11976703 and 26.53% for rs73284230. A conditional analysis showed that both SNPs are not independently associated with global PSQI score (rs73284230condit11976703; β = 0.5147; p = 6.9 × 10−2). The combined annotation-dependent depletion (CADD) scores for rs11976703 and rs73284230 were 0.19 and 1.718, respectively, which does not indicate high deleteriousness [54].
The locus on chromosome 7 is situated between neuropeptide Y (NPY) and membrane palmitoylated protein 6 (MPP6) genes with a surrounding LD structure that differs between ancestries (Figure 1, a and b). Using EUR and AFR as reference panels, it can be observed that SNPs in high LD with rs11976703 are located in MPP6 (EUR), or upstream of it (AFR). All highly associated SNPs (p < 1 × 10−4) around rs11976703 were upstream of MPP6 and downstream of NPY. Nevertheless, the associated locus is substantially closer to the promoter of MPP6 and separated by the presence of a recombination hotspot from NPY.
SNP association with other phenotypes
According to the Genome-wide Repository of Associations between SNPs and Phenotypes (GRASP) [55], rs11976703 was previously reported to be associated with coronary artery disease, Parkinson’s disease and body mass index (BMI) with p-values of 6.33 × 10−3, 1.2 × 10−2, and 3.0 × 10−2, respectively [56–58]. By contrast, rs73284230 was not associated with any phenotype in GRASP.
We next evaluated whether the effects of SNPs associated with sleep quality were mediated by phenotypes known to influence sleep quality and available in the OPPERA cohort. Six clinically relevant phenotypes were tested for correlation with the PSQI score: two clinical pain conditions (painful temporomandibular disorders [TMD] and low back pain), two measures of psychological distress (trait anxiety and depression), and two measures of sensitivity to experimental pain (heat pain tolerance and threshold). All six phenotypes correlated with PSQI. SNPs rs11976703, rs73284230, and rs60869707 were also associated with depression, anxiety, and experimental pain (Supplementary Tables 8 and 9). After adjustment for each potential mediator individually, the allelic effect was slightly attenuated but remained statistically significant. After inclusion of all potential mediators simultaneously, the effect size for each genome-wide significant SNP was attenuated by around 45% but remained statistically significant (p < 0.05) (Supplementary Table 10), even after correcting for seven tests. Consequently, the effect of SNPs on sleep is unlikely to be fully mediated by pain states or psychological distress.
Furthermore, using Genome-wide Complex Trait Analysis (GCTA), we calculated the genetic correlation (Rg) between PSQI and the above-mentioned phenotypes. We did not find evidence for genetic correlation of PSQI with TMD, back pain, anxiety, and depression, whereas heat pain threshold and tolerance approached statistical significance of such correlation (heat pain threshold, Rg = 0.68, p = 0.09 and heat pain tolerance Rg = 0.53, p = 0.07) (Supplementary Table 11a). Furthermore, a LDHub [59] screen with 173 disease/traits from publicly available summary GWAS did not show any genetic correlations, with inflammatory bowel disease and Crohns disease showing nominal significance at p = 0.05 (Supplementary Table 11b). This might be due to the fact that we were underpowered to detect any association. Because the findings are not sufficiently robust, we hesitate to draw firm conclusions.
eQTL analysis
The two SNPs on chromosome 7 that replicated in the meta-analysis were tested for evidence of functional effects through the expression quantitative trait loci (eQTL) analysis using BRAINEAC and Genotype-Tissue Expression (GTEx) databases in 12 brain tissues. At p = 0.05, rs73284230 was not an eQTL in any brain tissues in any dataset. However, because of its low allelic frequency this analysis was underpowered. In BRAINEAC, the effect allele (C) of rs11976703 was associated with lower mRNA levels of MPP6 in brain tissues averaged, with a p-value of 3.6 × 10−4 [32]. Furthermore, rs11976703 was an MPP6 eQTL in the GTEx dataset in the same direction in the frontal cortex with p-values of 9.56 × 10−3 (Supplementary Figure 5). Although this association p-value did not cross the strict Bonferroni correction for 12 tissues (p = 4.2 × 10−3), substantial correlation of expression in GTEx brain tissues suggests employment of this threshold to be very conservative [33]. Additionally, some brain tissues are duplicates of each other in the GTEx dataset (e.g. cortex and frontal cortex) [33]. Finally, in the GTEx dataset this same allele (C) of rs11976703 is also associated with lower mRNA levels of NPY in the anterior cingulate cortex, the cerebral hemisphere, the cerebellum, and the frontal cortex with p-values ranging from 0.05 to 1.4 × 10−4 (Supplementary Figure 6).
Functional validation in vivo
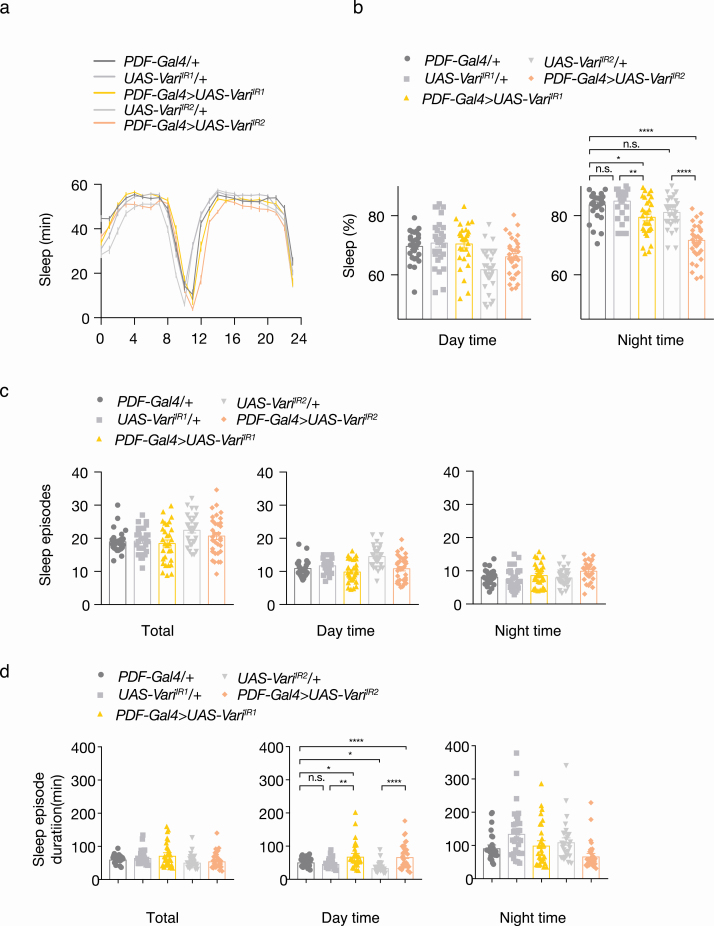
To investigate the functional role of genes on chromosome 7 in regulating sleep quality in vivo, we used the fruit fly D melanogaster [60]. In previous studies, NPF, the fly ortholog for NPY, has been implicated in suppression of sleep during starvation [61]; however, nothing is known about the role of Drosophila ortholog of MPP6 (varicose) in sleep. In the fruit fly, sleep quality is controlled by lateral ventral neurons (LNv) in the brain, which can be specifically manipulated using the driver PDF-Gal4 [50]. Using transgenic RNAi (UAS-Inverted repeat; UAS-IR), we generated varicose knockdown flies specifically within LNv neurons (PDF-Gal4> UAS-variIR1/2). Sleep patterns for these flies were then compared to parental controls (PDF-Gal4/+ or UAS-variIR1/2/+). Firstly, we did not observe any difference in circadian behavior between parental controls and transgenic flies (Figure 4a). Next, although no difference was observed in daytime sleep behavior, PDF-Gal4>UAS-variIR1/2 flies showed a marked reduction in overall nighttime sleep duration (Figure 4b) without a difference in sleep fragmentation assessed by the total number of sleep episodes (Figure 4c). Accordingly, we observed longer sleep episode duration in day time in PDF-Gal4>UAS-variIR1/2 animals (Figure 4d). Together, our results show reduced sleep time during the night and increased durations of sleep episodes during day in MPP6 RNAi flies, which represents poor sleep homeostasis [62] and this may be a proxy for poor sleep quality. The poor sleep during the night appears to be compensated by longer sleep duration bouts during the day that can serve as consolidation. This effect supports the GWAS finding since the effect allele associated with worse sleep was an eQTL with lower mRNA levels of both MPP6 and NPY. Overall, we concluded that varicose (MPP6) expression plays a major role in nighttime sleep maintenance in vivo.
Figure 4.
LNv-specific knockdowns of varicose in D melanogaster. PDF-Gal4 is a neuron-specific driver. UAS-IR represents transgenic RNAi inverted repeats. PDF-Gal4/+, UAS-VariIR1-2/+ are parental control flies. PDF-Gal4>UAS-variIR1-2 are varicose knockdown flies within LNv neurons. (a) Circadian pattern of sleep (n = 28–32). (b) Percentage of sleep in day time and night time (n = 28–32). (c) Number of sleep episode in day time and night time (n = 28–32). (d) Sleep episode duration in minutes in day time and night time (n = 28–32). Data presented as mean ± SEM. Statistics were determined one-way ANOVA with Tukey’s multiple comparisons test. n.s., not significant, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.001.
Conclusion/Discussion
In this study, a GWAS analysis revealed novel loci and genes associated with sleep quality, measured by the validated PSQI questionnaire. Overall, 15% of the variation in the global score of PSQI was explained by the combined additive effects of assessed SNPs. Previous twin studies showed that the PSQI global score is highly heritable (34–37%) [19, 63]. In this study, the heritability estimate for PSQI global score was lower, commonly observed when comparing SNP-based heritability estimates with twin studies. However, the reported SNP-based heritability estimate was within the range of what was previously reported from GWAS for other sleep traits; i.e. 11.5% for insomnia disorder [64] and 10.3% for self-reported sleep duration [9].
The two loci reported here, on chromosomes 2 and 7, were not previously reported to be linked to any sleep phenotype in human association studies. ATOH8 has been implicated in the development of the nervous system and muscles but has not been reported to be related to sleep [65]. ATOH8 is a transcription factor that recognized an E-box element and regulates transcription of genes [66]. More than nine circadian clock genes are E-box-regulated genes and occur in a rhythmic fashion [67]. Whether ATOH8 specifically binds a circadian clock gene is unknown, but our genetic association could open the door to new research avenues, though we recognize that this finding was not replicated in independent cohorts.
The locus situated downstream of NPY and upstream of MPP6 was replicated in independent cohorts. The NPY gene codes for Neuropeptide Y, the most abundant peptide of the central nervous system and a master regulator of stress response, circadian and feeding rhythms through afferent projections from the hypothalamus [68, 69]. Neuropeptide Y has been found to promote sleep in humans and zebrafish [70, 71]. NPY was also identified as an important candidate gene in sleep regulation in D melanogaster and Caenorhabditis elegans [72, 73]. Through a genetic screen in zebrafish, it was reported that the overexpression of NPY increases sleep by inhibiting noradrenergic signaling [71], while in the fly NPF integrates feeding and sleep behavior [61]. Importantly, no genetic association between NPY and sleep quality in humans has been reported previously.
MPP6 codes for membrane palmitoylated protein 6, a member of the peripheral membrane-associated guanylate kinase (MAGUK) family. This family includes synaptic scaffolding proteins DLG1 and PSD95 [74], which have never been previously associated with any sleep-related phenotype. In D melanogaster, this gene was previously shown to regulate the formation of the fly respiratory system [75] but has not yet been implicated in regulating sleep.
Interestingly, the collective evidence suggests that the locus on chromosome 7 identified in our study affect functions of two genes simultaneously, NPY and MPP6, where NPY is a known sleep gene and MPP6 is a novel sleep gene. This possibility would be in line with the relatively strong effect of the identified locus on sleep phenotypes. In tissue-expression analysis, the effect allele of the top SNP was found to be associated with lower MPP6 and NPY expressions in many brain tissues, including the cerebral hemispheres, the anterior cingulate cortex, the cerebellum and the frontal cortex. The direction of association was consistent between brain tissues and validated in two independent datasets. Taking into account our genetic findings, where the effect allele was associated with higher PSQI global scores, hence worse sleep quality, our genetic results suggest that lower levels of MPP6 and NPY expression are associated with worse sleep, which is in agreement with what is known for NPY [68, 71].
We then decided to take an additional step and test the functionality of MPP6 in vivo, demonstrating that genetic knockdown of the fly MPP6 ortholog varicose resulted in altered sleep homeostasis (worse sleep) compared to parental controls. Although the behavioral translation between species is difficult to interpret, the direction of the effect is consistent. The association of MPP6 RNA with chromosome 7 locus can be a consequence of genetic co-regulation of these two genes resulting from the physical proximity in the human genome, but in D melanogaster these two genes are not situated at the same gene locus. Thus, our fly experiments reemphasize the independent functional role of the MPP6 gene in sleep.
MPP6 acts centrally as the conditional knockdown was done in the sleep center neurons (LNv) of the fly brain. In humans, MPP6 is known to be expressed in the central nervous system; however, its function is still unknown. MPP6, previously named VAM-1, is expressed across the central nervous system, with highest expression in the cerebellum, the caudate and in the pituitary gland. Its sequence is predicted to contain a conserved PDZ domain that binds to veli-1. Veli-1 is a protein that helps couple synaptic vesicle exocytosis with neuronal cell adhesion, suggesting that it promotes the assembly of veli-1 containing protein complex in neurons [76]. Veli-1 also binds PDZ-motif containing proteins that are known to contribute to receptor clustering complexes at the post-synaptic levels [77].
Our pathway analysis of the full GWAS revealed pathways that are in line with MPP6’s function. Pathways such as regulation of synaptic structural plasticity and receptor internalization point towards an important genetic contribution of processes occurring at the synaptic level and towards a role for receptor anchoring at the synapse. Many previously identified molecules such as neuroligin, neuropeptides, ion channels, vesicle proteins, and scaffolding molecules have been shown to be regulated by sleep homeostasis and circadian rhythms [78]. Indirect evidence from our pathway analysis, the role of MPP6 in synaptic receptor clustering and ATOH8 as a transcription factor for clock genes, support this hypothesis. These findings open the door to future research that should focus on the role of MPP6 in neuronal communication in brain structures that are important in sleep processes as well as sleep homeostasis using complementary methods such as electroencephalography in mouse models.
In this study, in line with many other reports [4], we show that there is a significant relationship between the report of pain conditions, like TMD and back pain, with poor sleep quality. Moreover, we show that there is a positive correlation between psychological factors, like anxiety and depression, with poor sleep quality, as well as a positive correlation between experimental heat pain sensitivity and poor sleep. These epidemiological associations did not translate into genetic correlation findings in our study, probably due to modest size of a discovery cohort. On the other hand, based on publicly available data, our lead SNP was previously shown to be associated with cardiovascular disease and BMI. This finding is in line with poor sleep being a predisposing factor to poor cardiovascular health. The pathway by which NPY and MPP6 participate in the manifestation of cardiovascular disease remains unknown. Further work is needed to determine if variations in NPY and MPP6 are independent or show causal links between poor sleep quality and cardiovascular disease.
This study presents some limitations that should be addressed. First, even though we demonstrated that we can identify true positives in our discovery cohort, we have to acknowledge that the sample size of our discovery cohort is small. A larger cohort would have more power to identify additional loci and stronger associations. Second, the phenotype that we used to assess sleep quality, the PSQI, is based on self-report, although through a validated questionnaire and not on objective assessment of sleep with polysomnography. Our study’s phenotype is a compromise between the use of a self-reported unspecific question derived from a large dataset of hundreds of thousands of individuals and a smaller study with comprehensively assessed polysomnography but lacking power for a genome-wide analysis. In an ideal scenario, our results should be validated with more objective measures, including actigraphy and polysomnography. Third, in a majority of replication cohorts, the phenotypes chosen for replication were closer to insomnia than sleep quality. While the PSQI has a number of questions related to insomnia, it also has non-insomnia questions and has only modest correlations with insomnia questionnaires. The heterogeneity in the phenotypes might explain why replication was weaker in certain cohorts.
In summary, we performed a discovery GWAS of sleep quality and identified two novel sleep loci. In a meta-analysis of 12 independent cohorts, we replicated the association with one locus on chromosome 7, situated between two genes, NPY and MPP6. Our eQTL analysis established the association of this locus in an allelic-dependent expression of both NPY and MPP6. While NPY is thought to be important for sleep [68], MPP6 has not previously been implicated in regulation of sleep. Using sleep center-specific gene knockdown in the fly, we showed that decreasing levels of fly ortholog of MPP6 leads to altered sleep homeostasis (i.e. worse sleep) in vivo, establishing its functionality. Overall, these data are consistent with the observed allelic association between MPP6 and human sleep quality. Our results have broad biological significance by recognizing the role of MPP6 as a novel sleep gene, potentially involved in synaptic processing in sleep centers. This work provides new insights into sleep biology and our findings should spur future investigation of mechanisms involving MPP6 in sleep.
Supplementary material
Supplementary material is available at SLEEP online.
Supplementary Figure 1: Scree plot of the OPPERA discovery cohort. Twelve eigenvectors are represented on the x-axis and the percentage of variance accounted for by each eigenvector is shown on the y-axis.
Supplementary Figure 2: Manhattan plot and Q-Q plot of discovery GWAS for PSQI global score in OPPERA. (a) Manhattan plot: The x axis shows chromosomes, and the y axis shows –log10 p-value association. Red line marks the threshold for genome-wide significance (p=5x10-8), and the lower blue line marks the threshold for suggestive significance at p=5x10-7. Nearest gene names are annotated for genome-wide significant SNPs. Heritability estimates were calculated using BOLT-REML. (b) QQ-plot: Quantile-Quantile plot shows the observed versus expected p-values from PSQI association analysis.
Supplementary Figure 3: Manhattan plot and QQ-plots stratified by race for PSQI global score in OPPERA. (a) Manhattan plot in African Americans (AA) only. (b) Manhattan plot in Non-Hispanic whites (NHW) only. The x axis shows chromosomes, and the y axis shows –log10 p-value association. Red line marks the threshold for genome-wide significance at p=5x10-8, and the lower blue line marks the threshold for suggestive significance at p=5x10-7. Nearest gene names are annotated for genome-wide significant SNPs. (c) QQ-plot: Quantile-Quantile plot in AA only. (d) QQ-plot: Quantile-Quantile plot in NHW only. QQ-plot shows the observed versus expected p-values from PSQI association analysis.
Supplementary Figure 4: Graph plot of expected false discovery rate (eFDR) among top 10 SNPs. The x axis shows the number of true associations. The y axis shows the eFDR among the top 10 SNPs having the smallest p=value of association. The blue dots represent SNPs with an OR of 1.2; Green dots represent SNPs with an OR of 1.25 and red dots represent SNPs with an OR of 1.3.
Supplementary Figure 5: Violin plots for mRNA expression per rs11976703 genotype (MPP6). Expression analysis eQTL of MPP6 extracted from GTEx for rs11976703 in twelve brain tissues. Note that rs73284230 is not an eQTLs for MPP6 in any brain tissue. The scale represents normalized expression values.
Supplementary figure 6: Violin plots for mRNA expression per rs11976703 genotype (NPY). Expression analysis eQTL of NPY extracted from GTEx for rs11976703 in twelve brain tissues. Note that rs73284230 is not an eQTLs for NPY in any brain tissue. The scale represents normalized expression values.
Supplementary Table 1: Demographic details of the OPPERA discovery cohort.
Supplementary Table 2: Genome-wide (P ≤ 5x10-8) and suggestive (P ≤ 5x10-7) loci associated with PSQI in the OPPERA discovery cohort with five principle components.
Supplementary Table 3: Association results of genome-wide significant SNPs with PSQI subscales.
Supplementary Table 4: Race stratified genome-wide results for global PSQI score in OPPERA.
Supplementary Table 5: Sex stratified genome-wide results for global PSQI score in OPPERA.
Supplementary Table 6: Replication cohorts’ description.
Supplementary Table 7: Ancestry-stratified fixed-effect meta-analysis.
Supplementary Table 8: Correlation between PSQI global scores and related phenotypes.
Supplementary Table 9: Association results of genome-wide significant SNPs with other phenotypes.
Supplementary Table 10: Multivariate regression analysis adjusting for related phenotypes.
Supplementary Table 11: a) Genetic correlation of PSQI with related OPPERA phenotypes. b) Genetic correlation of PSQI with other phenotypes.
Funding
This work was supported by the Canadian Excellence Research Chairs (CERC) Program grant (http://www.cerc.gc.ca/home-accueileng.aspx, CERC09). OPPERA was supported by NIDCR (grant number U01DE017018). Funding for genotyping was provided by NIDCR through a contract to the Center for Inherited Disease Research at Johns Hopkins University (HHSN268201200008I). The Complex Persistent Pain Conditions: Unique and Shared Pathways of Vulnerability Program Project was supported by NIH/NINDS grant NS045685 to the University of North Carolina at Chapel Hill, and genotyping was funded by the Canadian Excellence Research Chairs (CERC) Program (grant CERC09). The OPPERA-R study was supported by the NIDCR under Award Number U01DE017018, and genotyping was funded by the Canadian Excellence Research Chairs (CERC) Program (grant CERC09). The NIH grant for AA CRASH was R01AR060852. The NIH grant for EA CRASH was R01AR056328. G.G.N. is supported through NHMRC project grants APP1026310, APP1046090, APP1107514 and an NHMRC career development fellowship II CDF1111940. Q.P.W. was supported by National Natural Science Foundation of China (31800993,31970937) and Natural Science Foundation of Guangdong, China (2018B030306002). S.B.S. was supported by K12DE022793. The authors thank the staff and participants of HCHS/SOL for their important contributions. Investigators website - http://www.cscc.unc.edu/hchs/. The Hispanic Community Health Study/Study of Latinos is a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (HHSN268201300001I / N01-HC-65233), University of Miami (HHSN268201300004I / N01-HC-65234), Albert Einstein College of Medicine (HHSN268201300002I / N01-HC-65235), University of Illinois at Chicago (HHSN268201300003I / N01-HC-65236 Northwestern Univ), and San Diego State University (HHSN268201300005I / N01-HC-65237). The following Institutes/Centers/Offices have contributed to the HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, NIH Institution-Office of Dietary Supplements. The Genetic Analysis Center at the University of Washington was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). MESA research was supported by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168 and N01-HC-95169 from the National Heart, Lung, and Blood Institute, and by grants UL1-TR-000040, UL1-TR-001079, and UL1-TR-001420 from NCATS. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, CA) and the Broad Institute of Harvard and MIT (Boston, MA) using the Affymetrix Genome-Wide Human SNP Array 6.0. The Finnish BrePainGen study was supported by the European Union Seventh Framework Programme (FP7/2007 - 2013) under grant agreement no 602919 and Governmental Funds to Helsinki University Hospital TYH2012212. The current study was conducted under UK Biobank application number 20802. The Jackson Heart Study (JHS) is supported and conducted in collaboration with Jackson State University (HHSN268201800013I), Tougaloo College (HHSN268201800014I), the Mississippi State Department of Health (HHSN268201800015I/HHSN26800001), and the University of Mississippi Medical Center (HHSN268201800010I, HHSN268201800011I and HHSN268201800012I) contracts from the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute for Minority Health and Health Disparities (NIMHD). The authors also wish to thank the staffs and participants of the JHS.
Author contributions
S.K., L.D.: scientific conception, drafted manuscript, analyzed data. Q.P.W., M.P., P.G., A.V.B., S.L., A.T., T.S., T.L., J.L., M.A.K., H.M.M., S.B.S., G.G.N.: analyzed data. S.A.M., E.A.K., S.R., I.B., A.N., G.D.S., R.B.F., R.O., J.D.G., W.M.: data acquisition and interpretation. All authors revised the manuscript.
Conflict of interest statement. None declared.
References
- 1. Hobson JA. Sleep is of the brain, by the brain and for the brain. Nature. 2005;437(7063):1254–1256. [DOI] [PubMed] [Google Scholar]
- 2. Siegel JM. Clues to the functions of mammalian sleep. Nature. 2005;437(7063):1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lin X, et al. Night-shift work increases morbidity of breast cancer and all-cause mortality: a meta-analysis of 16 prospective cohort studies. Sleep Med. 2015;16(11):1381–1387. [DOI] [PubMed] [Google Scholar]
- 4. Andersen ML, et al. Sleep disturbance and pain: a tale of two common problems. Chest. 2018;154(5):1249–1259. [DOI] [PubMed] [Google Scholar]
- 5. Hertenstein E, et al. Insomnia as a predictor of mental disorders: a systematic review and meta-analysis. Sleep Med Rev. 2019;43:96–105. [DOI] [PubMed] [Google Scholar]
- 6. Buysse DJ, et al. The Pittsburgh Sleep Quality Index: a new instrument for psychiatric practice and research. Psychiatry Res. 1989;28(2):193–213. [DOI] [PubMed] [Google Scholar]
- 7. Mollayeva T, et al. The Pittsburgh Sleep Quality Index as a screening tool for sleep dysfunction in clinical and non-clinical samples: a systematic review and meta-analysis. Sleep Med Rev. 2016;25:52–73. [DOI] [PubMed] [Google Scholar]
- 8. Marinelli M, et al. Heritability and genome-wide association analyses of sleep duration in children: the EAGLE consortium. Sleep. 2016;39(10):1859–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lane JM, et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet. 2017;49(2):274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lane JM, et al. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK Biobank. Nat Commun. 2016;7:10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jones SE, et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet. 2016;12(8):e1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hammerschlag AR, et al. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat Genet. 2017;49(11):1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen H, et al. Multi-ethnic meta-analysis identifies RAI1 as a possible obstructive sleep apnea related quantitative trait locus in men. Am J Respir Cell Mol Biol. 2018;58(3):391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Amin N, et al. Genetic variants in RBFOX3 are associated with sleep latency. Eur J Hum Genet. 2016;24(10):1488–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Winkelmann J, et al. Genetics of restless legs syndrome. Sleep Med. 2017;31:18–22. [DOI] [PubMed] [Google Scholar]
- 16. Winkelmann J, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39(8):1000–1006. [DOI] [PubMed] [Google Scholar]
- 17. Gottlieb DJ, et al. Novel loci associated with usual sleep duration: the CHARGE Consortium Genome-Wide Association Study. Mol Psychiatry. 2015;20(10):1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. El Gewely M, et al. Reassessing GWAS findings for the shared genetic basis of insomnia and restless legs syndrome. Sleep. 2018;41(11). doi: 10.1093/sleep/zsy164 [DOI] [PubMed] [Google Scholar]
- 19. Zhang J, et al. Insomnia, sleep quality, pain, and somatic symptoms: sex differences and shared genetic components. Pain. 2012;153(3):666–673. [DOI] [PubMed] [Google Scholar]
- 20. Gasperi M, et al. Genetic and environmental influences on sleep, pain, and depression symptoms in a community sample of twins. Psychosom Med. 2017;79(6):646–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Slade GD, et al. Study methods, recruitment, sociodemographic findings, and demographic representativeness in the OPPERA study. J Pain. 2011;12(11 Suppl):T12–T26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maixner W, et al. Orofacial Pain Prospective Evaluation and Risk Assessment Study - the OPPERA study. J Pain. 2011;12(11 Suppl. 3):T4–T11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laurie CC, et al. ; GENEVA Investigators. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010;34(6):591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. 1000 Genomes Project Consortium; Auton A, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Delaneau O, et al. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9(2):179–181. [DOI] [PubMed] [Google Scholar]
- 26. Howie BN, et al. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Howie B, et al. Genotype imputation with thousands of genomes. G3 (Bethesda). 2011;1(6):457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vsevolozhskaya OA, et al. The more you test, the more you find: the smallest P-values become increasingly enriched with real findings as more tests are conducted. Genet Epidemiol. 2017;41(8):726–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. J Open Source Softw. 2018;3(25):731. [Google Scholar]
- 31. Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramasamy A, et al. ; UK Brain Expression Consortium; North American Brain Expression Consortium. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17(10):1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Consortium G. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sorlie PD, et al. Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol. 2010;20(8):629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Platts-Mills TF, et al. Using emergency department-based inception cohorts to determine genetic characteristics associated with long-term patient outcomes after motor vehicle collision: methodology of the CRASH study. BMC Emerg Med. 2011;11:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Linnstaedt SD, et al. Methodology of AA CRASH: a prospective observational study evaluating the incidence and pathogenesis of adverse post-traumatic sequelae in African-Americans experiencing motor vehicle collision. BMJ Open. 2016;6(9):e012222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaunisto MA, et al. Pain in 1,000 women treated for breast cancer: a prospective study of pain sensitivity and postoperative pain. Anesthesiology. 2013;119(6):1410–1421. [DOI] [PubMed] [Google Scholar]
- 40. Belfer I, et al. Persistent postmastectomy pain in breast cancer survivors: analysis of clinical, demographic, and psychosocial factors. J Pain. 2013;14(10):1185–1195. [DOI] [PubMed] [Google Scholar]
- 41. Buysse DJ, et al. Development and validation of patient-reported outcome measures for sleep disturbance and sleep-related impairments. Sleep. 2010;33(6):781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taylor HA Jr, et al. Toward resolution of cardiovascular health disparities in African Americans: design and methods of the Jackson Heart Study. Ethn Dis. 2005;15(4 Suppl 6):S6–S4. [PubMed] [Google Scholar]
- 43. Auton A, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou X, et al. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44(7):821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bild DE, et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol. 2002;156(9):871–881. [DOI] [PubMed] [Google Scholar]
- 46. McCarthy S, et al. ; Haplotype Reference Consortium. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Redline S, et al. The familial aggregation of obstructive sleep apnea. Am J Respir Crit Care Med. 1995;151(3 Pt 1):682–687. [DOI] [PubMed] [Google Scholar]
- 48. Chinn S. A simple method for converting an odds ratio to effect size for use in meta-analysis. Stat Med. 2000;19(22):3127–3131. [DOI] [PubMed] [Google Scholar]
- 49. Hasselblad V, et al. A survey of current problems in meta-analysis. Discussion from the Agency for Health Care Policy and Research inter-PORT Work Group on Literature Review/Meta-Analysis. Med Care. 1995;33(2):202–220. [PubMed] [Google Scholar]
- 50. Park JH, et al. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A. 2000;97(7):3608–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pfeiffenberger C, et al. Processing sleep data created with the Drosophila Activity Monitoring (DAM) System. Cold Spring Harb Protoc. 2010;2010(11):pdb.prot5520. [DOI] [PubMed] [Google Scholar]
- 52. Chen X, et al. Racial/ethnic differences in sleep disturbances: the Multi-Ethnic Study of Atherosclerosis (MESA). Sleep. 2015;38(6):877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tononi G, et al. Sleep function and synaptic homeostasis. Sleep Med Rev. 2006;10(1):49–62. [DOI] [PubMed] [Google Scholar]
- 54. Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Eicher JD, et al. GRASP v2.0: an update on the genome-wide repository of associations between SNPs and phenotypes. Nucleic Acids Res. 2015;43(Database issue):D799–D804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deloukas P, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45(1):25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pankratz N, et al. ; PD GWAS Consortium. Meta-analysis of Parkinson’s disease: identification of a novel locus, RIT2. Ann Neurol. 2012;71(3):370–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Speliotes EK, et al. ; MAGIC; Procardis Consortium. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zheng J, et al. ; Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33(2):272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sehgal A, et al. Genetics of sleep and sleep disorders. Cell. 2011;146(2):194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chung BY, et al. Drosophila neuropeptide F signaling independently regulates feeding and sleep-wake behavior. Cell Rep. 2017;19(12):2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. van Alphen B, et al. A dynamic deep sleep stage in Drosophila. J Neurosci. 2013;33(16):6917–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Genderson MR, et al. Genetic and environmental influences on sleep quality in middle-aged men: a twin study. J Sleep Res. 2013;22(5):519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stein MB, et al. Genome-wide analysis of insomnia disorder. Mol Psychiatry. 2018;23(11):2238–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen J, et al. Diversification and molecular evolution of ATOH8, a gene encoding a bHLH transcription factor. PLoS One. 2011;6(8):e23005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ejarque M, et al. Characterization of the transcriptional activity of the basic helix-loop-helix (bHLH) transcription factor Atoh8. Biochim Biophys Acta. 2013;1829(11):1175–1183. [DOI] [PubMed] [Google Scholar]
- 67. Matsumura R, et al. Multiple circadian transcriptional elements cooperatively regulate cell-autonomous transcriptional oscillation of Period3, a mammalian clock gene. J Biol Chem. 2017;292(39):16081–16092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dyzma M, et al. Neuropeptide Y and sleep. Sleep Med Rev. 2010;14(3):161–165. [DOI] [PubMed] [Google Scholar]
- 69. Shimizu N, et al. Refeeding after a 24-hour fasting deepens NREM sleep in a time-dependent manner. Physiol Behav. 2011;104(3):480–487. [DOI] [PubMed] [Google Scholar]
- 70. Held K, et al. Neuropeptide Y (NPY) shortens sleep latency but does not suppress ACTH and cortisol in depressed patients and normal controls. Psychoneuroendocrinology. 2006;31(1):100–107. [DOI] [PubMed] [Google Scholar]
- 71. Singh C, et al. Neuropeptide Y regulates sleep by modulating noradrenergic signaling. Curr Biol. 2017;27(24):3796–3811.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. He C, et al. Regulation of sleep by neuropeptide Y-like system in Drosophila melanogaster. PLoS One. 2013;8(9):e74237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Huang H, et al. Combining human epigenetics and sleep studies in Caenorhabditis elegans: a cross-species approach for finding conserved genes regulating sleep. Sleep. 2017;40(6). doi: 10.1093/sleep/zsx063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhu J, et al. Mechanistic basis of MAGUK-organized complexes in synaptic development and signalling. Nat Rev Neurosci. 2016;17(4):209–223. [DOI] [PubMed] [Google Scholar]
- 75. Wu VM, et al. Drosophila Varicose, a member of a new subgroup of basolateral MAGUKs, is required for septate junctions and tracheal morphogenesis. Development. 2007;134(5):999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tseng TC, et al. VAM-1: a new member of the MAGUK family binds to human Veli-1 through a conserved domain. Biochim Biophys Acta. 2001;1518(3):249–259. [DOI] [PubMed] [Google Scholar]
- 77. Jo K, et al. Characterization of MALS/Velis-1, -2, and -3: a family of mammalian LIN-7 homologs enriched at brain synapses in association with the postsynaptic density-95/NMDA receptor postsynaptic complex. J Neurosci. 1999;19(11):4189–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. El Helou J, et al. Neuroligin-1 links neuronal activity to sleep-wake regulation. Proc Natl Acad Sci U S A. 2013;110(24):9974–9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Study data have been deposited and made publicly available at the Database of Genotypes and Phenotypes (dbGaP) public repository (accession number phs000796.v1.p1).