Abstract
Study Objectives:
Nocturnal blood pressure (BP) profile shows characteristic abnormalities in OSA, namely acute postapnea BP surges and nondipping BP. These abnormal BP profiles provide prognostic clues indicating increased cardiovascular disease risk. We developed a deep neural network model to perform computerized analysis of polysomnography data and predict nocturnal BP profile.
Methods:
We analyzed concurrently performed polysomnography and noninvasive beat-to-beat BP measurement with a deep neural network model to predict nocturnal BP profiles from polysomnography data in 13 patients with severe OSA.
Results:
A good correlation was noted between measured and predicted postapnea systolic and diastolic BP (Pearson r ≥ .75). Moreover, Bland-Altman analyses showed good agreement between the 2 values. Continuous systolic and diastolic BP prediction by the deep neural network model was also well correlated with measured BP values (Pearson r ≥ .83).
Conclusions:
We developed a deep neural network model to predict nocturnal BP profile from clinical polysomnography signals and provide a potential prognostic tool in OSA. Validation of the model in larger samples and examination of its utility in predicting CVD risk in future studies is warranted.
Citation:
Prasad B, Agarwal C, Schonfeld E, Schonfeld D, Mokhlesi B. Deep learning applied to polysomnography to predict blood pressure in obstructive sleep apnea and obesity hypoventilation: a proof-of-concept study. J Clin Sleep Med. 2020;16(10):1797–1803.
Keywords: deep neural network, blood pressure, OSA
INTRODUCTION
OSA is characterized by acute surges in systemic blood pressure (BP), following apneas and hypopneas, mediated by sympathetic hyperactivity and blunted baroreflex function. The acute surges in sympathetic activity and BP during sleep are hypothesized to persist during wakefulness, thereby causing hypertension in OSA.1 Nondipping, an absence of the normal 10%–20% reduction in BP during sleep, is another common BP abnormality in OSA.2 Therefore, nocturnal BP profiles in OSA can provide important phenotypic information regarding cardiovascular disease (CVD) risk.3–5
The AHI fails to fully explain the risk of CVD or predict response to OSA treatment.6 Novel prognostic biomarkers are needed for OSA treatment to be an effective strategy for CVD prevention. Recent software-aided analyses of polysomnography data have yielded clinically useful metrics for defining OSA endotypes.7 Polysomnography captures key physiological signals that play a role in the pathogenesis of hypertension in OSA: respiration and oxygenation (airway obstruction with intermittent hypoxia and intrathoracic pressure changes), electroencephalography (sleep depth and continuity), and electrocardiography (heart rate variability indicators of autonomic dysfunction). Noninvasive beat-to-beat BP monitoring by finger plethysmography during polysomnography can provide information on temporal changes in BP with respiratory events and during sleep. However, clinical polysomnography does not include noninvasive beat-to-beat BP but measures physiological signals from organ systems that drive hypertension in OSA. We hypothesized that physiological signals from polysomnography could be used to predict BP after respiratory events and during sleep using a machine learning framework. Such characterization of the nocturnal BP profile can provide early clues to the risk of hypertension and CVD in OSA.
Recent advancements in machine learning tools such as deep learning have led to its application in personalized medicine.8 Data-driven deep-learning algorithms implicitly extract features and patterns from the data, thus precluding the need to generate detailed models and solve complex mathematical optimization problems. Building on these benefits of the data-driven model, we developed a novel deep-learning framework (deep neural network [DNN]) to predict acute BP surges after respiratory events and continuously using standard clinical polysomnography signals. Here we present the methods and results of a proof-of-concept study in 13 patients with severe OSA and obesity hypoventilation syndrome.
METHODS
Sample
We conducted a secondary analysis of an institutional review board–approved published study,9 which was a 2-center randomized trial testing the efficacy of 3 different modalities of PAP therapy in patients with OSA and obesity hypoventilation syndrome overlap (ClinicalTrials.gov Identifier: NCT01368614). These PAP modalities included average volume assured pressure support, ontinuous PAP, and bilevel PAP. The inclusion criteria were ages 18–75 years, diagnosis of obesity hypoventilation syndrome, and AHI > 5 events/h. Exclusion criteria were chronic obstructive pulmonary disease (confirmed by spirometry), history of unstable cardiorespiratory comorbidity, history of PAP or surgical OSA treatment, and contraindication to PAP interface use. Each participant underwent a diagnostic, titration, and final polysomnography on therapeutic PAP after 6 weeks. We included only the diagnostic and titration studies because the polysomnography at 6 weeks showed minimal residual respiratory events (and postevent BP acute BP surges). The baseline characteristics of the participants included are presented in Table 1. Concurrent noninvasive beat-to-beat BP (Nexfin HD device, BMEYE B.V., Amsterdam, the Netherlands) and polysomnography (Nihon Kohden, Foothill Ranch, CA) included recordings of 6 electroencephalographic channels, bilateral electrooculograms, chin and tibialis electromyogram, electrocardiogram, airflow by nasal pressure transducer and oronasal thermocouples, chest and abdominal effort by respiratory inductance plethysmography belts, oxygen saturation by finger pulse oximeter, and transcutaneous carbon dioxide monitoring. Four of 17 participants in the study were excluded because of significant technical artifacts in the beat-to-beat BP signal. Polysomnography was scored by the American Academy of Sleep Medicine 2007 criteria (4% desaturation for hypopneas).10 Two polysomnography (diagnostic and PAP titration) results from each of the 13 participants were included in the analysis.
Table 1.
Baseline characteristics.
| Category | Value |
|---|---|
| Age, y | 51.8 ± 11.1 |
| Sex (men/women) | 6/7 |
| Hypertension | 12/13 |
| Number of antihypertensives | 1.6 ± 1.1 |
| SBP, mm Hg | 137.9 ± 23.2 |
| DBP, mm Hg | 85.6 ± 15.4 |
| Body mass index, kg/m2 | 46.7 ± 7.5 |
| PaCO2, mm Hg | 51.1 ± 7.9 |
| Epworth Sleepiness Scale | 12.5 ± 5.9 |
| Total recording time, min | 446 ± 66.8 |
| Total sleep time, min | 333.8 ± 85.6 |
| AHI, events/h | 85.6 ± 31.3 |
| T < 90 (SaO2), min | 219 ± 106 |
Values are proportions or mean ± standard deviation. PaCO2 = partial pressure of carbon dioxide in arterial blood gas obtained while sitting and breathing room air during wakefulness, SaO2 = oxygen saturation, T < 90 = time below 90% oxygen saturation during polysomnography.
Dataset generation for signal processing
We computed the respiratory events for each patient using the American Academy of Sleep Medicine definitions of apneas and hypopneas (> 30% flow reduction plus 4% desaturation for hypopneas) using scripts written in the Python program. The computed number of apneas and hypopneas (mean ± standard deviation = 66.4 ± 24.6) and manually scored events (85.6 ± 30.1) were not significantly different (t statistic = 1.78; P = .08) and correlated well (Pearson r = .69; P = .02). We generated 2 datasets using the diagnostic and titration polysomnography, which comprised 6,660 and 4,254 apnea and hypopnea events, respectively. As expected, a significant number of residual apnea and hypopnea events were noted at subtherapeutic pressures before optimal pressure was reached during the titration study, providing a sufficient number of respiratory events to train and validate the model. A third dataset was constructed by combining the events from both polysomnographies. For each respiratory event, we used 17 polysomnography signals during the respiratory event as the input to the model. We predicted the systolic and diastolic BP at 10 seconds following the respiratory event (average time for postevent BP peak). We assumed the respiratory event duration to be between 10 and 120 seconds. The data were split into training (90%) and testing (10%) sets, and a k-fold cross-validation technique was used to check the model’s robustness (k = 5). The statistical assumptions for parametric testing were met, and the Pearson product-moment correlation was used to examine the association between measured and predicted BP.
For the continuous BP prediction, we generated a supervised dataset using a rolling-window technique. A rolling-window analysis of a time-series model determines the model’s stability over time—that is, the model predicts BP instability over time. The rolling-window technique uses a time-window parameter (W)—the number of consecutive observations per rolling window and the number of increments between successive windows (WI) for sampling data from the respective signals. We used W = 30 seconds and WI = 1 second for sampling data in our experiments. We chose W = 30 seconds to be consistent with the conventional polysomnography epoch duration and WI to generate larger samples for precision. Similar to postrespiratory event BP prediction, we used the 17 polysomnography signals and systolic and diastolic BP signals for creating our training and testing datasets. For each window W, we took 30 seconds of the 17 polysomnography signals (input), and the average BP in the W + 10 seconds was predicted (output). Following this technique, we generated a total of 259,658 and 605,776 samples for the diagnostic and titration study datasets, respectively.
Proposed model
Broadly, there are 2 categories of learning algorithms: (1) unsupervised and (2) supervised. Although supervised machine-learning methods rely on labeled data to train models, unsupervised methods learn features without any guidance.
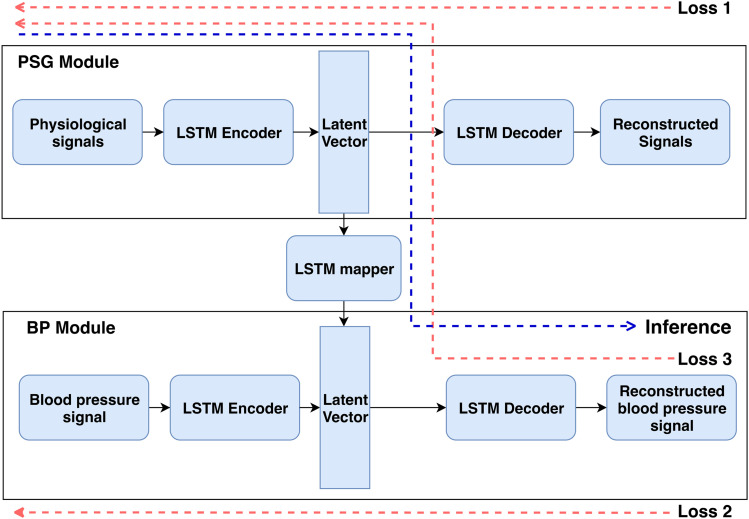
In this study, we used a simple neural network designed for time-series prediction11 that eliminates problems arising in recurrent neural networks while learning long-term dependencies in a timed event. We used an autoencoder-style neural network (Figure 1) for predicting BP from polysomnography physiological signals. An autoencoder model is a type of DNN that learns efficient data representation using a semisupervised technique, which leverages the advantages of both supervised and unsupervised learning. It learns feature representation for both polysomnography (Figure 1, PSG module) and BP signals (Figure 1, BP module) individually in an unsupervised manner and concurrently learns to map the features from polysomnography signals to BP signals in a supervised fashion. The framework comprises 3 main blocks. The first autoencoder neural network block reconstructs polysomnography signals by learning an intermediate data representation. The second autoencoder neural network block is used for learning the BP signals. Finally, a third mapper neural network block converts the polysomnography data to the BP data representation. The encoder and decoder neural network blocks have 1 layer of 128 hidden nodes each, and the neural network mapper block has 2 layers of 128 hidden nodes. The hidden nodes are densely connected neuron models. By this method, we trained multiple models using diagnostic and titration polysomnography for systolic and diastolic BP prediction (detailed description in the supplemental material).
Figure 1. Model architecture.
The proposed deep learning architecture for reconstructing BP signals using PSG signals. The model was trained using 3 separate reconstruction losses (red arrows). During inference (blue arrow), the input was a 1 × 17 set of physiological signals to obtain systolic and diastolic BP values as output. BP = blood pressure, LSTM = long short-term memory, PSG = polysomnography.
For the continuous BP prediction, we used a standard neural network with 1 layer of 32 hidden nodes. The input to our model was the 17 polysomnography signals from a given 30-second window, and the output layer was composed of 1 hidden node representing the average systolic and diastolic BP in the subsequent 10 seconds.
Training and inference
All models were trained end-to-end using the RMSProp optimizer for 2,000 iterations.12 An iteration was defined as a single pass of the entire dataset, both forward and backward through the neural network. At each iteration, we calculated 3 reconstruction losses (red arrows in Figure 1)—that is, reconstruction losses for the polysomnography signals, BP signal, and BP derived from polysomnography signals. In the inference stage (blue arrow in Figure 1), we used the polysomnography signals as input to the trained long short-term memory–based DNN model to obtain predicted systolic and diastolic BP values.
RESULTS
Postrespiratory event BP predictions
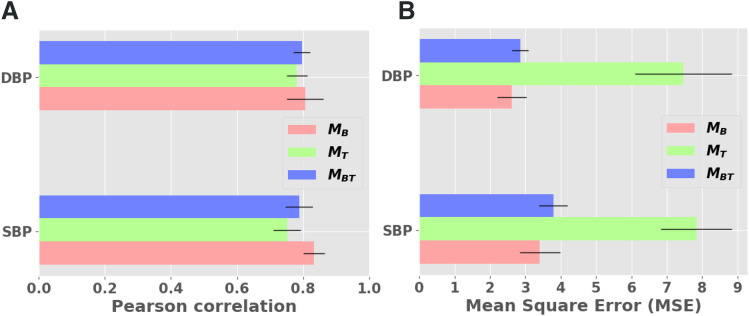
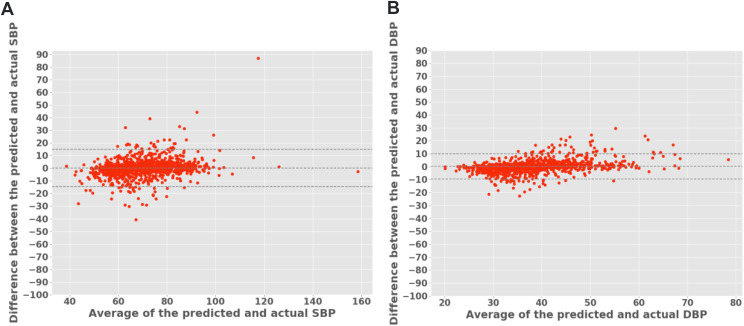
The models trained on the diagnostic, titration, and diagnostic + titration datasets were denoted as MB, MT, and MBT, respectively. The data distribution for measured and systolic BP were checked (Figure S2 in the supplemental material). We found a good correlation (Pearson r ≥ .75) between the measured and predicted systolic and diastolic BP, for 10 seconds after a respiratory event, with small standard deviations (Table 2) for all 3 models. The coefficient of determination (r2) was mostly ≥ .65, indicating that at least two-thirds of the variance in BP was shared by the predicted BP. As expected, because of a greater number of respiratory events (larger dataset), the diagnostic model MB showed the best performance between the predicted and measured BP (r = 0.83; Figure 2A). The correlation plots are provided in the supplemental material (Figure S1B). The maximum mean-squared error for the diagnostic MB and combined MBT model was ± 3.79 mm Hg and ± 2.85 mm Hg for systolic and diastolic BP, respectively (Figure 2B). The Bland-Altman plots using the combined dataset MBT showed good agreement between the predicted and measured systolic BP (Figure 3A) and diastolic BP (Figure 3B). A few average systolic and diastolic BP values were lower than the expected physiological range (40 mm Hg and 20 mm Hg, respectively), as depicted in Figure 3. This result was because of artifacts in the beat-to-beat signal generated when participants suddenly raised or lowered their hand (it takes a few heartbeats for the device to recalibrate to the level of the heart).
Table 2.
Correlations between measured and predicted BP values with postrespiratory event surge in BP.
| Dataset | SBP | DBP | ||
|---|---|---|---|---|
| r | r2 | r | r2 | |
| Baseline | .86 ± .03 | .75 ± .05 | .81 ± .05 | .67 ± .08 |
| Titration | .75 ± .04 | .56 ± .07 | .80 ± .03 | .63 ± .04 |
| Baseline plus titration | .84 ± .03 | .70 ± .04 | .83 ± .02 | .69 ± .04 |
Pearson correlations (r) with standard deviations (from 5 model runs) and coefficient of determination (r2). BP = blood pressure, DBP = diastolic blood pressure, SBP = systolic blood pressure.
Figure 2. Pearson correlation and MSE of postrespiratory BP predictions.
Error plots for correlation of predicted mean SBP and DBP 10 seconds after respiratory events (A) and MSE of SBP and DBP signal reconstruction (B). The correlation was approximately 0.8 across both SBP and DBP for the respiratory events in the baseline study. Although diagnostic and titration datasets had different distributions, with less frequent respiratory events in the latter, the model predicted that BP continued to be highly correlated (r = .79 ± .04) to measured BP in the combined dataset. A low MSE of ∼3.79 mm Hg and ∼2.85 mm Hg was observed for SBP and DBP, respectively, using our MBT model. MT had the highest MSE, likely because of a lower number of respiratory events (there was a smaller sample for training and testing). BP = blood pressure, DBP = diastolic blood pressure, MSE = mean-squared error, SBP = systolic blood pressure.
Figure 3. Bland-Altman plots for mean SBP and DBP values 10 seconds after obstructive respiratory events (ie, apneas and hypopneas) for the MBT model.
Most predictions of both SBP (A) and DBP (B) lay within the ± 1.96 SD of the distribution, indicating good agreement between predicted and measured blood pressure. DBP = diastolic blood pressure, SBP = systolic blood pressure, SD, standard deviation.
Continuous BP predictions
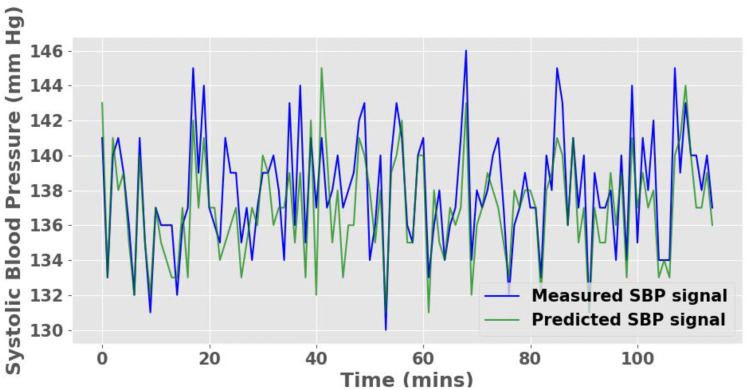
Pearson correlations were ≥ .83 between measured and predicted continuous systolic and diastolic BP (Table 3). Notably, a Pearson correlation of ∼.9 was obtained for the continuous BP prediction during the titration study. The correlation plots are included in the supplemental material (Figure S1A). Figure 4 shows the prediction of the model qualitatively compared to the measured noninvasive beat-to-beat BP for a randomly selected 120-minute period in a participant. The mean-squared error was ± 2.24 mm Hg between the measured and the predicted continuous BP. These results highlight both the potential efficiency and the robustness of the proposed model.
Table 3.
Correlations between measured and predicted BP values with continuous BP prediction.
| Dataset | SBP | DBP | ||
|---|---|---|---|---|
| r | r2 | r | r2 | |
| Baseline | .83 | .69 | .84 | .71 |
| Titration | .91 | .83 | .89 | .79 |
Pearson correlations (r) for single model run and coefficient of determination (r2). BP = blood pressure, DBP = diastolic blood pressure, SBP = systolic blood pressure.
Figure 4. Continuous SBP signal prediction for a randomly selected 120-minute period in a participant using diagnostic polysomnography signals.
We observed a small MSE of 2.24 mm Hg between the predicted and measured SBP. MSE = mean-squared error, SBP = systolic blood pressure.
We explored the relative importance of each polysomnography signal with an ablation approach (Table S1). Overall, respiratory flow, oximetry, electrooculograph, and posterior electroencephalograph had a significant role in BP prediction.
DISCUSSION
We developed a novel DNN model that can predict BP from clinical polysomnography signals, both at specific time points (postrespiratory event) and continuously throughout the recording, with acceptable accuracy. Contrary to other sequence-learning methods, the proposed DNN model can process long time sequences and is insensitive to gap length (the model remembers values over arbitrary time intervals).
Previous studies have used pulse transit time (from the finger plethysmography signal and electrocardiography) to estimate resting and exercising awake BP,13,14 but few studies have used polysomnography signals to predict BP during sleep.15 A recent study used pulse transit time from the finger plethysmography signal of polysomnography to estimate systolic BP after respiratory events.16 The study found several significant differences between patients with a ≥ 10 mm Hg rise in BP vs a < 10 mm Hg rise. Notable among these differences was a higher resting awake BP, lower AHI, and longer apnea duration in those with a ≥ 10 mm Hg rise in BP. The study illustrates the importance of examining the nocturnal BP profile in OSA because it is predictive of daytime BP and that AHI is insufficient to determine the risk of hypertension in OSA.
Given that the cardiovascular (heart rate and vascular tone), neurocardiac (sleep stage and autonomic system), and cardiorespiratory (intrathoracic pressure and vagal) coupling that influences nocturnal BP in OSA is captured by polysomnography signals, it is plausible to predict BP from polysomnography. We explored the contribution of each polysomnography signal to the prediction model and noted that respiratory flow, oximetry, electrooculograph, and occipital electroencephalograph were important elements of the model. These findings must be interpreted with caution in this small sample of patients with severe OSA complicated by obesity hypoventilation syndrome. A validated BP prediction tool that relies on polysomnography data has several apparent applications in practice and research. Polysomnography is the gold standard diagnostic test for OSA; therefore, sleep-related BP profiles could be generated for large clinical datasets efficiently, providing a scalable tool for population studies. Longitudinal studies could then be used to examine the utility of polysomnography-derived BP profiles vs conventional metrics, such as the AHI, in CVD risk assessment. If the DNN model of predicted BP proves to be a superior or complementary prognostic tool to AHI, then it could guide future OSA treatment trials, especially as additional therapeutic options such as the hypoglossal nerve stimulator become available.
A strength of this study is the application of advanced data-driven learning algorithms to develop a robust DNN model that uses all physiological data from both diagnostic and titration polysomnography. Such a model could be used not only to predict abnormal nocturnal profiles of BP but also to noninvasively determine acute effects of PAP on BP. The main limitation of our study is the small sample size, which restricts the accuracy and generalizability of the model. In addition to training the model with larger datasets, further refinement of the model could include identification of the minimum set of physiological polysomnography signals necessary for accurate BP prediction and sleep stage effects on model performance. A trained DNN model for BP prediction will need to be validated in independent samples of different OSA endotypes (eg, high upper airway collapsibility, high loop gain, or low arousal threshold) and phenotypes (eg, sleepy vs nonsleepy). This small sample of convenience was made up of complex patients with both severe OSA and obesity hypoventilation. Chronic hypercapnia may affect nocturnal BP profile, and the inclusion of transcutaneous carbon dioxide in the prediction input may affect model performance. These issues need to be resolved in larger studies, including patients with uncomplicated OSA of all severities.
In summary, we have developed a DNN model to predict the nocturnal BP profile from clinical polysomnography signals and provide a potential prognostic tool for hypertension in OSA. Validation of the model in larger samples and examination of its utility in predicting CVD risk in future studies is warranted.
DISCLOSURE STATEMENT
All authors have seen and approved the manuscript. Work for this study was performed at the University of Illinois at Chicago. This study was funded by Veterans Affairs Clinical Science Research and Development (IK2CX001026 PI Prasad). The authors report no conflicts of interest.
EDITOR'S NOTE
The Emerging Technologies section focuses on new tools and techniques of potential utility in the diagnosis and management of any and all sleep disorders. The technologies may not yet be marketed, and indeed may only exist in prototype form. Some preliminary evidence of efficacy must be available, which can consist of small pilot studies or even data from animal studies, but definitive evidence of efficacy will not be required, and the submissions will be reviewed according to this standard. The intent is to alert readers of Journal of Clinical Sleep Medicine of promising technology that is in early stages of development. With this information, the reader may wish to (1) contact the author(s) in order to offer assistance in more definitive studies of the technology; (2) use the ideas underlying the technology to develop novel approaches of their own (with due respect for any patent issues); and (3) focus on subsequent publications involving the technology in order to determine when and if it is suitable for application to their own clinical practice. The Journal of Clinical Sleep Medicine and the American Academy of Sleep Medicine expressly do not endorse or represent that any of the technology described in the Emerging Technologies section has proven efficacy or effectiveness in the treatment of human disease, nor that any required regulatory approval has been obtained.
SUPPLEMENTARY MATERIAL
ABBREVIATIONS
- BP
blood pressure
- CVD
cardiovascular disease
- DNN
deep neural network
REFERENCES
- 1.Somers VK, Dyken ME, Clary MP, Abboud FM. Sympathetic neural mechanisms in obstructive sleep apnea. J Clin Invest. 1995;96(4):1897–1904. 10.1172/JCI118235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mokhlesi B, Hagen EW, Finn LA, Hla KM, Carter JR, Peppard PE. Obstructive sleep apnoea during REM sleep and incident non-dipping of nocturnal blood pressure: a longitudinal analysis of the Wisconsin Sleep Cohort. Thorax. 2015;70(11):1062–1069. 10.1136/thoraxjnl-2015-207231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seif F, Patel SR, Walia HK, et al. Obstructive sleep apnea and diurnal nondipping hemodynamic indices in patients at increased cardiovascular risk. J Hypertens. 2014;32(2):267–275. 10.1097/HJH.0000000000000011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martínez-García MA, Capote F, Campos-Rodriguez F, et al. Effect of CPAP on blood pressure in patients with obstructive sleep apnea and resistant hypertension: the HIPARCO randomized clinical trial. JAMA. 2013;310(22):2407–2415. 10.1001/jama.2013.281250 [DOI] [PubMed] [Google Scholar]
- 5.Sapiña-Beltrán E, Torres G, Benítez I, et al. Differential blood pressure response to continuous positive airway pressure treatment according to the circadian pattern in hypertensive patients with obstructive sleep apnoea. Eur Respir J. 2019;54(1):1900098. 10.1183/13993003.00098-2019 [DOI] [PubMed] [Google Scholar]
- 6.McEvoy RD, Antic NA, Heeley E, et al. CPAP for prevention of cardiovascular events in obstructive sleep apnea. N Engl J Med. 2016;375(10):919–931. 10.1056/NEJMoa1606599 [DOI] [PubMed] [Google Scholar]
- 7.Mazzotti DR, Lim DC, Sutherland K, et al. Opportunities for utilizing polysomnography signals to characterize obstructive sleep apnea subtypes and severity. Physiol Meas. 2018;39(9):09TR01. 10.1088/1361-6579/aad5fe [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. doi: 10.2147/NSS.S220716. Khademi A, El-Manzalawy Y, Master L, Buxton OM, Honavar VG. Personalized sleep parameters estimation from actigraphy: a machine learning approach. Nat Sci Sleep . December 11 2019;11:387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter JR, Fonkoue IT, Grimaldi D, et al. Positive airway pressure improves nocturnal beat-to-beat blood pressure surges in obesity hypoventilation syndrome with obstructive sleep apnea. Am J Physiol Regul Integr Comp Physiol. 2016;310(7):R602–R611. 10.1152/ajpregu.00516.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iber C, Ancoli-Israel S, Chesson AL Jr, Quan SF; for the American Academy of Sleep Medicine. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications. 1st ed. Westchester, IL: American Academy of Sleep Medicine; 2007. [Google Scholar]
- 11.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. 10.1162/neco.1997.9.8.1735 [DOI] [PubMed] [Google Scholar]
- 12.Hinton G, Srivastava N, Swersky K. Neural networks for machine learning. Coursera. 2012;Video Lectures (264). https://www.cs.toronto.edu/~tijmen/csc321/slides/lecture_slides_lec6.pdf.
- 13.Payne RA, Symeonides CN, Webb DJ, Maxwell SR. Pulse transit time measured from the ECG: an unreliable marker of beat-to-beat blood pressure. J Appl Physiol. 2006;100(1):136–141. 10.1152/japplphysiol.00657.2005 [DOI] [PubMed] [Google Scholar]
- 14.Gesche H, Grosskurth D, Kuchler G, Patzak A. Continuous blood pressure measurement by using the pulse transit time: comparison to a cuff-based method. Eur J Appl Physiol. 2012;112(1):309–315. 10.1007/s00421-011-1983-3 [DOI] [PubMed] [Google Scholar]
- 15.Pépin JL, Tamisier R, Borel JC, Baguet JP, Levy P. A critical review of peripheral arterial tone and pulse transit time as indirect diagnostic methods for detecting sleep disordered breathing and characterizing sleep structure. Curr Opin Pulm Med. 2009;15(6):550–558. 10.1097/MCP.0b013e3283318585 [DOI] [PubMed] [Google Scholar]
- 16.Gehring J, Gesche H, Drewniok G, Kuchler G, Patzak A. Nocturnal blood pressure fluctuations measured by using pulse transit time in patients with severe obstructive sleep apnea syndrome. Sleep Breath. 2018;22(2):337–343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.