Abstract
The Neotropical Albitarsis Group is a complex assemblage of essentially isomorphic species which currently comprises eight recognized species—five formally described (Anopheles albitarsis Lynch-Arribalzaga, An. deaneorum Rosa-Freitas, An. janconnae Wilkerson and Sallum, An. marajoara Galvao and Damasceno, An. oryzalimnetes Wilkerson and Motoki) and three molecularly assigned (An. albitarsis F, G & I)—and one mitochondrial lineage (An. albitarsis H). To further explore species recognition within this important group, 658 base pairs of the mitochondrial DNA cytochrome oxidase subunit I (COI) were analyzed from 988 specimens from South America. We conducted statistical parsimony network analysis, generated estimates of haplotype, nucleotide, genetic differentiation, divergence time, and tested the effect of isolation by distance (IBD). Ten clusters were identified, which confirmed the validity of the eight previously determined species, and confirmed the specific status of the previous mitochondrial lineage An. albitarsis H. High levels of diversity were highlighted in two samples from Pará (= An. albitarsis J), which needs further exploration through additional sampling, but which may indicate another cryptic species. The highest intra-specific nucleotide diversity was observed in An. deaneorum, and the lowest in An. marajoara. Significant correlation between genetic and geographical distance was observed only in An. oryzalimnetes and An. albitarsis F. Divergence time within the Albitarsis Group was estimated at 0.58–2.25 Mya, during the Pleistocene. The COI barcode region was considered an effective marker for species recognition within the Albitarsis Group and a network approach was an analytical method to discriminate among species of this group.
Keywords: Albitarsis Group, barcoding, network, cryptic diversity, new lineage
Delineation of taxa within essentially isomorphic cryptic species groups is a significant challenge (de Queiroz 2007), leading to the increased reliance of molecular tools to identify and delimit species (Leache and Fujita 2010, Fujita et al. 2012). To explore relationships between cryptic taxa, tree-based phylogenetic approximations such as maximum parsimony, maximum likelihood, and distance methods are often used. However, gene evolution, which may be reticulate, is not always best represented in this way. To resolve such relationships, network approaches have been developed to estimate genealogies. These include statistical geometry (Eigen et al. 1988), likelihood networks (Strimmer and Moulton 2000), median networks (Bandelt et al. 2000), and median-joining network approaches (Bandelt et al. 1999). Hart and Sunday (2007) observed a strong association between breaks in network connectivity and species-level separation. The method of Templeton, Crandall, and Sing (1992) (TCS) has been used extensively to infer genealogies when divergences are low (Gerber and Templeton 1996, Vila et al. 1999, Gomes-Zurita et al. 2000). In addition, using a phylogenetic network, Chen et al. (2010) were able to predict species limits within nemertean species complexes (Cephalothrix rufifrons, C. major, and C. spiralis) that lacked sufficient morphological characters to separate individual taxa.
Many investigations have been undertaken to assess the taxonomic status of the component members of the Albitarsis Group, some of which play a significant role in malaria transmission in regions of South America (Klein et al. 1991a,b; Rubio-Palis 1994; Chadee et al. 2006; Povoa et al. 2006; Gutierrez et al. 2010). Based on phylogenetic analyses of mitochondrial protein-coding genes, Foster et al. (2017) suggested the elevation of Nyssorhynchus, Kerteszia, Lophopodomyia, and Stethomyia to a generic level. As the ranking of taxa as genera or subgenera is subjective, the traditional recognition of Anopheles as a genus (Harbach 2018) is retaining herein. According to the latest review (Ruiz-Lopez et al. 2012), the Albitarsis Group was understood to comprise eight formally recognized species: Anopheles albitarsis Lynch-Arribálzaga, An. deaneorum Rosa-Freitas, An. janconnae Wilkerson and Sallum (= An. albitarsis E) (Lehr et al. 2005, Motoki et al. 2009), An. marajoara Galvão and Damasceno, An. oryzalimnetes Wilkerson and Motoki (= An. albitarsis B) (Motoki et al. 2009), three molecularly determined taxa, An. albitarsis F (Brochero et al. 2007), An. albitarsis G (McKeon et al. 2010, Krzywinski et al. 2011, Ruiz-Lopez et al. 2012), and An. albitarsis I (Gutierrez et al. 2010, Ruiz-Lopez et al. 2012), and the mitochondrial lineage, An. albitarsis H (Ruiz-Lopez et al. 2012). Only An. deaneorum can reliably be separated from species in the Albitarsis Group and then only in the larval stage, based on seta 3-C of the larvae (Rosa-Freitas 1989). Seta 3-C is branched in An. deaneorum, whereas An. albitarsis, An. marajoara, and An. oryzalimnetes exhibit single, simple, or aciculate seta.
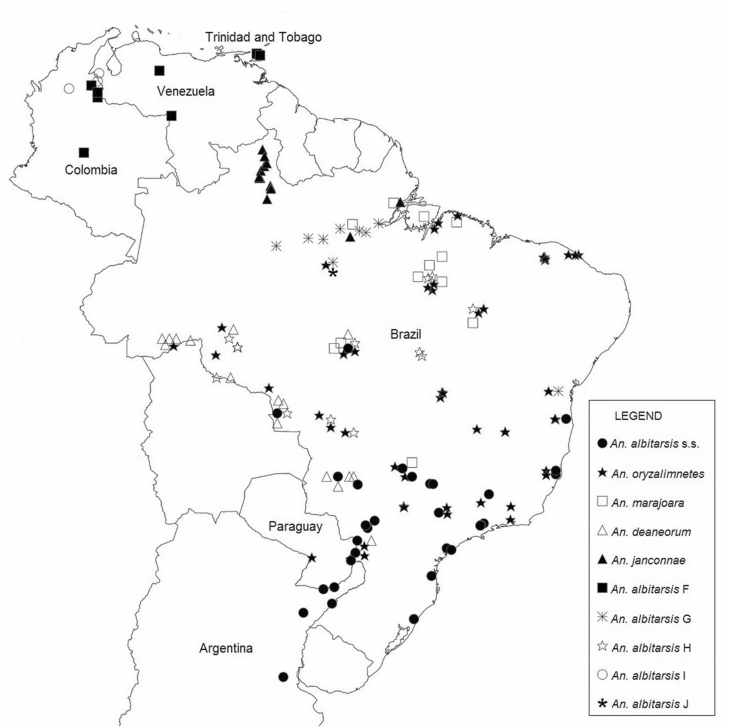
The Albitarsis Group has a large distribution in South America. Anopheles albitarsis is found in northern Argentina, southern Paraguay, and Brazil (Wilkerson et al. 1995a, Lehr et al. 2005, Li and Wilkerson 2005, Ruiz-Lopez et al. 2012, Foley et al. 2014); An. albitarsis F and An. albitarsis I are found in northern South America (Colombia, Trinidad and Tobago, and Venezuela) (Brochero et al. 2007, Gutierrez et al. 2010, Ruiz-Lopez et al. 2012); while An. oryzalimnetes is widely distributed in the central region of Brazil (Wilkerson et al. 1995a,b; Lehr et al. 2005; Ruiz-Lopez et al. 2012; Foley et al. 2014; herein), and An. marajoara in the northern and central regions of Brazil (Ruiz-Lopez et al. 2012, Foley et al. 2014). Anopheles deaneorum is present in the western regions of Brazil (Wilkerson et al. 1995b, Li and Wilkerson 2005, Ruiz-Lopez et al. 2012, Foley et al. 2014); An. janconnae is restricted to the northern region (Lehr et al. 2005, Povoa et al. 2006, Ruiz-Lopez et al. 2012); An. albitarsis G in the Amazon and east region (Ruiz-Lopez et al. 2012, Foley et al. 2014; herein); and An. albitarsis H in the central and western regions (Ruiz-Lopez et al. 2012, Foley et al. 2014).
Using COI sequences generated from specimens from Brazil, Argentina, Colombia, Paraguay, Trinidad and Tobago, and Venezuela, Ruiz-Lopez et al. (2012) proposed eight species in the Albitarsis Group (An. albitarsis, An. deaneorum, An. janconnae, An. marajoara, An. oryzalimnetes, and An. albitarsis F, G & I), and recognized the novel mitochondrial lineage An. albitarsis H. In addition, Foley et al. (2014) examined environmental and ecological divergence events for these nine taxa from additional localities. The present study examines the COI sequences from 988 individuals, using the dataset of Ruiz-Lopez et al. (2012) and Foley et al. (2014), including 20 new COI sequences, to test whether network analysis further supports Ruiz-Lopez et al.’s eight purported taxa, and whether additional support could be garnered for the taxonomic status of An. albitarsis H.
Materials and Methods
Sampling and Data Access
Full specimen records (collection locality, coordinates, specimen identifiers, location of voucher specimens etc.) and all genetic data (edited chromatograms, consensus COI sequence files, and corresponding GenBank numbers) are publicly accessible under the project container code MBIAA (Anopheles albitarsis complex) on the BOLD website (http://www.boldsystems.org), as part of efforts of the Mosquito Barcoding Initiative (MBI). Detailed collection data are lodged in VectorMap (vectormap.si.edu) where distribution maps can be readily visualized. Where available, voucher specimens and / or DNA extracts of specimens used in this study are stored in the archive collections of the Walter Reed Biosystematics Unit, Smithsonian Institution–National Museum of Natural History, Museum Support Center (MSC), Suitland, Maryland, United States, or in the Frozen Tissue Collection at the Natural History Museum, London, United Kingdom.
Specimens utilized in the molecular study were all morphologically verified as belonging to the Albitarsis Group using the available keys (Linthicum 1988, Gonzalez and Carrejo 2009) and original species descriptions. Specimens sequenced include topotypic material of An. albitarsis s.s., An. deaneorum, An. marajoara, and An. oryzalimnetes, and type series material of An. janconnae. In this dataset, a total of 988 COI sequences generated from specimens collected from all over Latin America over a period of 20 yr by RCW, FRL, MTM, and JEC and collaborators were analyzed. Data used in this analysis 564 COI sequences first reported in the phylogenetic treatment of Ruiz-Lopez et al. (2012)—An. albitarsis s.s. [JQ615201–JQ615309], deaneorum [JQ615310–JQ615345], An. janconnae [JQ615346–JQ615441], An. marajoara [JQ615442–JQ615511], An. oryzalimnetes [JQ615512–JQ615562], albitarsis F [JQ614998–JQ615041], An. albitarsis G [JQ615042–JQ615146], An. albitarsis H [JQ615147–JQ615188], and An. albitarsis I [JQ615189–JQ615200]—and 404 specimens included in the spatial evolutionary and ecological vicariance analysis (SEEVA) of Foley et al. (2014)—An. albitarsis s.s. [KJ492701–KJ492726, KJ492728–KJ492738, KJ012833], An. oryzalimnetes [KJ012000–KJ012004, KJ012015–KJ012065, KJ012834–KJ012861, KJ492780–KJ492894], An. marajoara [KJ011926–KJ011999, KJ492772–KJ492775, KJ492777–KJ492779], An. deaneorum [KJ492739–KJ492771], albitarsis G [KJ012832, KJ492676, KJ492776], An. albitarsis H [KJ011904–KJ011925, KJ492677–KJ492696, KJ492698–KJ492700]. To these previously published datasets, we added a further 20 new samples, including 2 An. oryzalimnetes [MT231277, MT231279], 16 An. albitarsis G [MT231262-MT231273, MT231275, MT231276, MT231278, MT231281], and two specimens of the newly defined An. albitarsis J [MT231274, MT231280]), and revisited the taxonomic status of An. albitarsis H.
DNA Extraction and Sequencing
A fragment of the mtDNA COI gene corresponding to the barcode region (658-bp) was amplified using the LCO1490 and HCO2198 primers of Folmer et al. (1994). The PCR amplification protocol utilized is detailed explicitly in Ruiz-Lopez et al. (2010). All sequencing reactions were carried out in both directions using ABI Big Dye Terminator Kit v.3.1 (PE Applied Biosystems, Warrington, United Kingdom), analyzed on an ABI Prism 3730—Avant Genetic Analyzer (Applied Biosystems, Foster City, CA), edited using Sequencher version 5.0.1 (Gene Codes Corporation, Ann Arbor, MI) and automatically aligned in CLUSTAL X (Thompson et al. 1997).
Data Analysis
To infer haplotype relationships within the data sets, a statistical parsimony network was performed using TCS 1.21 (Clement et al. 2000). Homoplasies were resolved using the algorithm estimation rules in Crandall and Templeton (1993).
Haplotype and nucleotide diversities were generated using DnaSP version 5.0 (Librado and Rozas 2009). Genetic differentiation was evaluated using the FST approach with significance determined by permutation test (10,000 replicates). FST values were generated using Arlequin 3.0 (Excoffier et al. 2005), based on the clusters determined by the phylogenetic network.
The Isolation By Distance Web Service version 3.23 (Jensen et al. 2005) was used to analyze the correlation between genetic distance estimated and geographic distances between collection sites of each species. Significance of the correlation coefficient was assessed applying the Mantel test (10,000 randomizations) (Mantel 1967).
Divergence times within the COI genealogy were estimated using the BEAST software package v1.8.0 (Drummond et al. 2012). The sequence data were summarized by making a set of haplotypes of each taxon. The standard arthropod mtDNA mutation rate of 2.3% per million years (Brower 1994) was set, giving a value of 0.0115 substitutions/site/lineage. The HKY model (Hasegawa et al. 1985) was applied with gamma distribution assuming constant size and relaxed molecular clock. The analysis was repeated five times, with 30 million generations of each run, sampling every 1,000 generations. The analysis results were combined in LogCombiner v1.8.0 with the initial 10% of the trees discarded as burn-in, and the results were averaged using Tracer v1.6 (Rambaut et al. 2013). The consensus tree was compiled with TreeAnnotator v1.8.0 and displayed in FigTree v1.3.1 (Rambaut 2009).
Results
Sample information is described in Fig. 1, Table 1, and Supp Table S1 (online only). Of the 415 haplotypes identified, 107 (26%) were shared within species and 308 (74%) were unique (Supp Table S1 [online only]; Table 2).
Fig. 1.
Map showing the distribution of member species in the Albitarsis Group based on the 988 specimens used in this study.
Table 1.
Origin by country and states of the 988 Albitarsis Group specimens used in this study
| Country | N | States | Species |
|---|---|---|---|
| Argentina | 38 | Buenos Aires (21), Corrientes (15), Misiones (2) | An. albitarsis s.s. |
| Brazil | 57 | Paraná (17), Santa Catarina (4), São Paulo (36) | An. albitarsis s.s. |
| 38 | Paraná (7), São Paulo (10), Rio Grande do Sul (4), Espírito Santo (4), Mato Grosso (3), Mato Grosso do Sul (1), Goiás (2), Minas Gerais (6), Bahia (1) |
An. albitarsis s.s. An. albitarsis s.s. |
|
| 42 | Bahia (4), Ceará (18), Mato Grosso (12), Pará (6), Rio de Janeiro (1), Rondônia (1), São Paulo (4) | An. oryzalimnetes | |
| 201 | Bahia (2), Ceará (33), Mato Grosso (3), Pará (67), Rondônia (20), São Paulo (12), Acre (2), Espírito Santo (11), Paraná (9), Maranhão (4), Piauí (7), Minas Gerais (18), Goiás (13) |
An. oryzalimnetes
An. oryzalimnetes |
|
| 69 | Amapá (27), Mato Grosso (3), Pará (39) | An. marajoara | |
| 86 | Amapá (4), Pará (80), Minas Gerais (1), Maranhão (1) | An. marajoara | |
| 36 | Mato Grosso (2), Rondônia (32), Paraná (2) | An. deaneorum | |
| 33 | Mato Grosso (10), Acre (13), Mato Grosso do Sul (10) | An. deaneorum | |
| 96 | Roraima (93), Amapá (2), Pará (1) | An. janconnae | |
| 105 | Amazonas (57), Pará (48) | An. albitarsis G | |
| 19 | Pará (18), Bahia (1) | An. albitarsis G | |
| 42 | Mato Grosso (39), Rondônia (3) | An. albitarsis H | |
| 45 | Mato Grosso (6), Rondônia (5), Pará (25), Tocantins (8), Maranhão (1) | An. albitarsis H | |
| 2 | Pará (2) | An. albitarsis J | |
| Colombia | 10 | Meta (2), Norte de Santander (6), Vichada (2) | An. albitarsis F |
| 11 | Antioquia (9), Norte Santander (2) | An. albitarsis I | |
| Paraguay | 14 | Alto Paraná (14) | An. albitarsis s.s. |
| 9 | Alto Paraná (9) | An. oryzalimnetes | |
| Trinidad | 19 | St. George East, St. Andrew/St. David (19) | An. albitarsis F |
| Venezuela | 15 | Cojedes (6), Zulia (9) | An. albitarsis F |
| 1 | Zulia (1) | An. albitarsis I |
N = total sample size. Numbers in parenthesis indicate samples size from each state.
Table 2.
Summary of haplotypes and diversity measures of the 988 COI gene sequences for members of the Albitarsis Group collected in six countries across South America (Argentina, Brazil, Colombia, Paraguay, Trinidad and Tobago, and Venezuela)
| Species | N | Haplotypes | h (SD) | π | ||
|---|---|---|---|---|---|---|
| Unique (%) | Shared (%) | Total | ||||
| An. albitarsis I | 12 | 5 (71) | 2 (29) | 7 | 0.864 (0.0780) | 0.00855 |
| An. albitarsis F | 44 | 26 (81) | 6 (19) | 32 | 0.970 (0.0160) | 0.01244 |
| An. deaneorum | 69 | 49 (86) | 8 (14) | 57 | 0.993 (0.0040) | 0.01076 |
| An. albitarsis G | 124 | 20 (71) | 8 (29) | 28 | 0.802 (0.0260) | 0.01040 |
| An. janconnae | 96 | 25 (64) | 14 (36) | 39 | 0.920 (0.0180) | 0.00890 |
| An. albitarsis H | 87 | 34 (72) | 13 (28) | 47 | 0.966 (0.0090) | 0.00806 |
| An. marajoara | 155 | 19 (68) | 9 (32) | 28 | 0.520 (0.0050) | 0.00171 |
| An. oryzalimnetes | 252 | 52 (63) | 30 (37) | 82 | 0.970 (0.0030) | 0.00687 |
| An. albitarsis s.s. | 147 | 76 (82) | 17 (18) | 93 | 0.982 (0.0050) | 0.01097 |
| An. albitarsis J | 2 | 2 (100) | – | 2 | 1.000 (0.0500) | 0.00354 |
| Total | 988 | 308 | 107 | 415 | 0.982 (0.0020) | 0.03129 |
N = number of individuals analyzed; Unique = unique haplotypes, the number in parentheses indicates the percentage of unique haplotypes from the total haplotypes; Shared = shared haplotypes, the number in parentheses indicates the percentage of shared haplotypes from the total numbers of haplotypes; Total = the total numbers of haplotypes detected; h = haplotype diversity; π = nucleotide diversity.
Genetic Variation
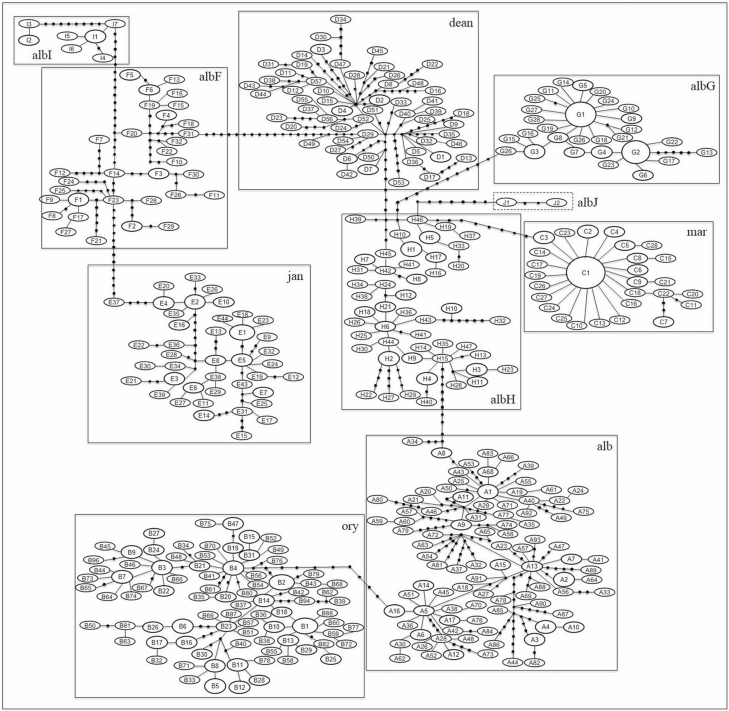
The statistical parsimony network illustrates the relationship among haplotypes of Albitarsis Group. The ten taxa differed by 8–18 mutational steps and could be connected parsimoniously (Fig. 2). This suggests substantial haplotype partitioning among species of the Albitarsis Group. Among the 10 taxa identified, a novel lineage was detected and designated as An. albitarsis J (Fig. 2).
Fig. 2.
Phylogenetic network of 415 COI haplotypes (658-bp) representing the 988 specimens of the Albitarsis Group collected from six countries across South America (Argentina, Brazil, Colombia, Paraguay, Trinidad and Tobago, and Venezuela). The area of each circle is proportional to the frequency of the haplotype. Black circle represents missing or unsampled haplotypes and each segment connecting haplotypes represents one mutational step. Each box depicts one taxa of the Albitarsis Group and the dashed box indicates a new lineage (albJ). albI = An. albitarsis I; albF = An. albitarsis F; dea = An. deaneorum; albG = An. albitarsis G; jan = An. janconnae; albH = An. albitarsis H; mar = An. marajoara; ory = An. oryzalimnetes; alb = An. albitarsis s.s.; albJ = An. albitarsis J.
Of the 155 specimens of An. marajoara confirmed from Pará, Amapá, Mato Grosso, Maranhão, and Minas Gerais states in Brazil, the most commonly observed haplotype was C1 (n = 107), found only in Pará and Amapá (Fig. 2; Supp Table 1 [online only]). The highest percentages (37, 36, and 32%, respectively) of shared haplotypes were found in An. oryzalimnetes, An. janconnae, and An. marajoara (Table 2). Conversely, a relatively high proportion of unique haplotypes (86, 82, and 81%) were noted in An. deaneorum, An. albitarsis s.s. and An. albitarsis F, respectively (Table 2). The most closely connected clusters were An. albitarsis H and An. albitarsis J, with the fewest mutational steps (eight steps) between them (Supp Table 1 [online only]).
The most diverse species—An. deaneorum—showed 57 haplotypes in 69 individuals (75%), and consequently the highest haplotype diversity (h = 0.993); An. marajoara presented the lowest haplotype diversity (h = 0.520) with only 28 haplotypes in 155 individuals (Table 2).
Genetic Differentiation
The genetic differentiation, FST, was significant among all the taxa (P < 0.001), except between An. albitarsis J and the remaining species (Table 3). The lowest but still significant FST value was between An. deaneorum and An. albitarsis s.s. (FST = 0.0127) and the highest significant value was between An. albitarsis I and An. marajoara (FST = 0.3723) (Table 3). The FST between An. albitarsis J and An. marajoara was not significant (FST = 0.4076) (Table 3). A significant positive correlation was found with the Mantel test for An. oryzalimnetes (P = 0.005, R2 = 0.0483) and An. albitarsis G (P = 0.023, R2 = 0.306) (Table 4).
Table 3.
Estimate of genetic differentiation (FST) based on pairwise estimates of COI haplotype frequencies of ten taxa (988 specimens) in the Neotropical Albitarsis Group
| alb | ory | mar | dea | jan | albF | albG | albH | albI | |
|---|---|---|---|---|---|---|---|---|---|
| An. albitarsis s.s. (alb) | |||||||||
| An. oryzalimnetes (ory) | 0.0242* | ||||||||
| An. marajoara (mar) | 0.2510* | 0.2417* | |||||||
| An. deaneorum (dea) | 0.0127* | 0.0186* | 0.2698* | ||||||
| An. janconnae (jan) | 0.0489* | 0.0543* | 0.2949* | 0.0440* | |||||
| An. albitarsis F (albF) | 0.0238* | 0.0297* | 0.2951* | 0.0181* | 0.0558* | ||||
| An. albitarsis G (albG) | 0.1075* | 0.1106* | 0.3450* | 0.1061* | 0.1405* | 0.1200* | |||
| An. albitarsis H (albH) | 0.0261* | 0.0319* | 0.2754* | 0.0205* | 0.0572* | 0.0318* | 0.1181* | ||
| An. albitarsis I (albI) | 0.0696* | 0.0755* | 0.3723* | 0.0644* | 0.1042* | 0.0775* | 0.1738* | 0.0786* | |
| An. albitarsis J (albJ) | 0.0138 | 0.0226 | 0.4076 | 0.0051 | 0.0612 | 0.0222 | 0.1560 | 0.0260 | 0.1016 |
The significance was tested by permutation tests (10,000 replicates).
*Significant P-value (<0.001).
Table 4.
Correlation between geographic and genetic distances among species of the Albitarsis Group by the Mantel test
| Species | P-value | R 2 |
|---|---|---|
| An. albitarsis s.s. | 0.4240 | 5.994 |
| An. oryzalimnetes | 0.0050 | 0.0483 |
| An. marajoara | 0.7330 | 0.0267 |
| An. deaneorum | 0.7950 | 0.180 |
| An. janconnae | 0.1980 | 0.0294 |
| An. albitarsis F | 0.0550 | 0.0885 |
| An. albitarsis G | 0.0230 | 0.306 |
| An. albitarsis H | 0.1340 | 0.107 |
| An. albitarsis I | 0.2550 | 0.105 |
The numbers in bold are significant (P < 0.05).
Estimation of Divergence Times
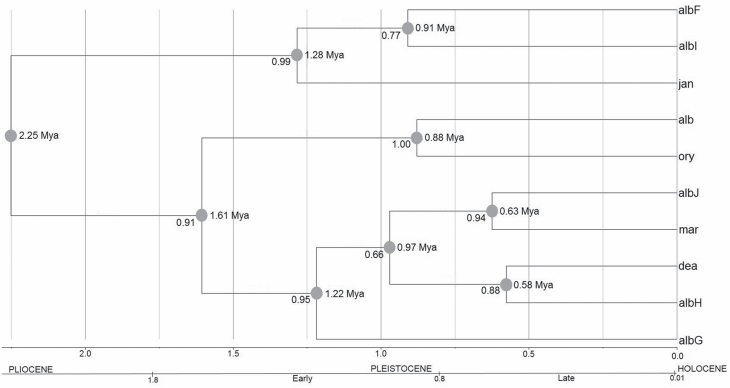
A phylogenetic topology is shown in Fig. 3, with the divergence times using the rate of 2.3% per million years under the relaxed molecular clock model. Using BEAST, the estimate of divergence time among species of the Albitarsis Group was 0.58–2.25 million of years ago, in the Pleistocene, with speciation events occurring between 0.58–1.61 Mya (Fig. 3).
Fig. 3.
Phylogenetic consensus tree obtained from COI gene sequences (658-bp) from 988 specimens collected in six countries across South America. Numbers to the left side of each node indicate posterior probabilities (PP). Numbers to right are estimates of mean divergence times (Mya) from the Beast calibrating with a rate of 0.0115 substitutions/site/Mya (albF = An. albitarsis F; albI = An. albitarsis I; jan = janconnae; alb = An. albitarsis s.s.; ory = An. oryzalimnetes; albJ = An. albitarsis J; mar = marajoara; dea = An. deaneorum; albH = An. albitarsis H; albG = An. albitarsis G).
Discussion
South American forests were substantially altered by climate and ecological oscillations that occurred in the Pleistocene (Colinvaux and Oliveira 2001). In this epoch, high levels of species divergence have been detected for many insect species (Lee and Li 2012, Li et al. 2012, Schultheis et al. 2012), including malaria vectors (Pedro and Sallum 2009, Loaiza et al. 2010, McKeon et al. 2010, Scarpassa and Conn 2011), suggesting climatic change as an important driver of speciation in the Neotropics. In the present study, the divergence time estimated among all taxa of the Albitarsis Group supported diversification during the Pleistocene.
The haplotype network identified 10 major clusters within the Albitarsis Group COI sequences (Fig. 2). Eight are formerly designated species: An. albitarsis s.s., An. oryzalimnetes, An. marajoara, An. deaneorum, An. janconnae, An. albitarsis F, An. albitarsis G, An. albitarsis I, one herein we confirm as a new species, An. albitarsis H, and here we add two samples of a hitherto undetermined lineage—An. albitarsis J. The clusters correspond partially to those supported by phylogenetic analyses in Wilkerson et al. (2005) and in Lehr et al. (2005); and totally supported in Ruiz-Lopez et al. (2012) with the additional lineage, An. albitarsis J (Fig. 2).
Ruiz-Lopez et al. (2012) first identified An. albitarsis H from Mato Grosso and Rondônia states in Brazil. Here we added several additional locations in Mato Grosso and Rondônia, and increased its known distribution to include Pará, Tocantins, and Maranhão states. Based on our haplotype network analysis, An. albitarsis H formed a distinct cluster from the remaining undisputed eight species within the Albitarsis Group, and with respect to our novel lineage (An. albitarsis J). The COI sequences representing An. albitarsis H separated from An. albitarsis J, An. marajoara, An. deaneorum, An. albitarsis s.s. and An. albitarsis G by 8, 9, 9, 11, and 13 mutational steps (Fig. 2), respectively, suggesting substantial haplotype partitioning.
Ruiz-Lopez et al. (2012) indicated highest genetic similarity of An. albitarsis H with An. marajoara and An. deaneorum using K2P distance method (0.020 and 0.024, respectively), but herein, with additional samples, we found that the estimate of genetic differentiation for An. albitarsis H from the remaining species of the Albitarsis Group was significant (FST = 0.0205–0.2754; P < 0.001; except for An. albitarsis J) (Table 3). The highest value was found between An. albitarsis H and An. marajoara, indicating that adding a further 86 specimens of An. marajoara to those examined in Ruiz-Lopez et al. (2012) supported the high value for genetic differentiation observed. The lowest differentiation but still significant value was between An. albitarsis H and An. deaneorum (FST = 0.0205; P < 0.001). In this case, the additional 33 specimens of An. deaneorum did not make any difference in the FST estimates and support the original result of Ruiz-Lopez et al. (2012). The relatively recent divergence between An. albitaris H and An. deaneorum explain the low differentiation between them. In addition, both species are sympatric in Mato Grosso and Rondônia states in Brazil, and their close relationship (sister species) corresponds with the geographic and ecological proximity detected previously (Foley et al. 2014).
In summary, Ruiz-Lopez et al. (2012) identified An. albitarsis H as a new lineage and, based on genetic approaches (i.e., network analysis, significant genetic differentiation), we confirm An. albitarsis H as a new species within the Albitarsis Group. The novel lineage detected, named here as An. albitarsis J, was separated from An. albitarsis H by 8 mutational steps (Fig. 2). Morphologically, An. albitarsis J is similar to the primary malaria vector An. marajoara. It was collected in Itaituba, Pará state, Brazil, in sympatry with An. albitarsis G and An. oryzalimnetes.
The FST was not significant for any comparison of An. albitarsis J with the other nine Albitarsis Group taxa; however, the network analysis supported An. albitarsis J as a distinct cluster (Fig. 2). The small sample size was a limitation; further detailed morphological, genetic, ecological studies and a large sample size are necessary before concluding if this lineage represents a new species within the Albitarsis Group.
A significant correlation between genetic and geographical distances was observed in An. albitarsis G and An. oryzalimnetes. A possible explanation for this, is the large distribution of both species; An. albitarsis G is present in Brazilian Amazon, while An. oryzalimnetes is widely distributed in Brazil, mainly in the central region (Fig. 1). Analysis of the ecological niche for An. oryzalimnetes revealed that its climatic and environmental profiles are very distinct from the other species of the Albitarsis Group (Foley et al. 2014). In addition, ecological and environmental determinants (e.g., mountains, rivers, climatic changes, ecoregions, demographic history) may also influence the genetic divergence of the Albitarsis Group, e.g., Amazon River appears to be important in the speciation of this group (Foley et al. 2014).
Lehr et al. (2005) using the COI gene sequence data and Wilkerson et al. (2005) using the COI, ND4, ITS2, and D2 data of An. albitarsis, An. albitarsis B (= An. oryzalimnetes), An. deaneorum and An. marajoara found similar results; however, Lehr et al. (2005) also detected paraphily of An. deaneorum and An. marajoara, suggestive of potential introgression or perhaps a recent speciation event. Lehr et al. (2005) hypothesized that An. marajoara may comprise two or more phylogenetic species. Four “An. marajoara” outliers in their Bayesian topology were later confirmed as An. albitarsis F in Ruiz-Lopez et al. (2012).
The undescribed species—An. albitarsis F, An. albitarsis G, An. albitarsis H, An. albitarsis I, and An. albitarsis J—are morphologically very similar to An. marajoara. Based on the white gene, ITS2, and COI sequences, An. albitarsis F (Brochero et al. 2007, Ruiz-Lopez et al. 2012) and An. albitarsis I (Gutierrez et al. 2010, Ruiz-Lopez et al. 2012) from Colombia were identified as new taxa. McKeon et al. (2010) analyzed COI gene sequences of An. marajoara from 14 populations in the Brazilian Amazon and detected a new species, which Ruiz-Lopez et al. (2012) later also found, and called An. albitarsis G. Ruiz-Lopez et al. (2012) discussed the distributions and taxonomic positions of the above species in relation to the pertinent literature for An. marajoara and concluded that An. albitarsis domesticus—the current synonym of An. marajoara—(Rios et al. 1984) did not refer to any of the newly recognized species (An. albitarsis F, An. albitarsis G, An. albitarsis I) /lineage (An. albitarsis H).
The only species of the Albitarsis Group that can be recognized morphologically is An. deaneorum (only in the larval stage based on seta 3-C) (Rosa-Freitas 1989), and its distinction from some species (An. albitarsis s.s., An. oryzalimnetes, An. marajoara) in the Albitarsis Group was supported by early allozyme loci, mtDNA and hybridization studies (Rosa-Freitas et al. 1990, Narang et al. 1993, Lima et al. 2004). The phylogenetic analysis of the mtDNA COI barcode gene further supported the separation of all species of the Albitarsis Group (Ruiz-Lopez et al. 2012). The current network analysis clearly showed distinct clusters of An. marajoara and An. deaneorum, corroborating the findings of Ruiz-Lopez et al. (2012).
The network analysis of the COI barcode region sequences further supports the finding of Ruiz-Lopez et al. (2012), confirming the status of the eight previously proposed species, establishes An. albitarsis H as a new species in the Albitarsis Group and identifies a novel lineage named An. albitarsis J. Based on our results, the Albitarsis Group includes: An. albitarsis s.s., An. oryzalimnetes, An. marajoara, An. deaneorum, An. janconnae, An. albitarsis F, An. albitarsis G, An. albitarsis H, An. albitarsis I and likely a novel lineage, An. albitarsis J. The COI barcode region is an important marker for species delimitation (Hebert et al. 2003, Cywinska et al. 2006, Kumar et al. 2007, Dai et al. 2012) in the Albitarsis Group and the parsimony network approach was shown to be a useful tool for species delineation in this highly diverse group, comprising essentially isomorphic taxa.
Supplementary Material
Acknowledgments
We are grateful to all collaborators and co-authors of Ruiz-Lopez et al. (2012) and Foley et al. (2014), upon which this manuscript builds. We also appreciate the two reviewers for providing helpful comments and suggestions. This investigation received financial support from the National Institutes of Health, United States (grant R01 AI50139-2 to J.E.C.). This research was performed under a Memorandum of Understanding between the Walter Reed Army Institute of Research and the Smithsonian Institution, with institutional support provided by both organizations. The published material reflects the views of the authors and should not be construed to represent those of the Department of the Army or the Department of Defense.
References Cited
- Bandelt, H J, Forster P, and Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16: 37–48. [DOI] [PubMed] [Google Scholar]
- Bandelt, H J, Macaulay V, and Richards M. 2000. Median networks: speedy construction and greedy reduction, one simulation, and two case studies from human mtDNA. Mol. Phylogenet. Evol. 16: 8–28. [DOI] [PubMed] [Google Scholar]
- Brochero, H H, Li C, and Wilkerson R C. 2007. A newly recognized species in the Anopheles (Nyssorhynchus) albitarsis complex (Diptera: Culicidae) from Puerto Carreno, Colombia. Am. J. Trop. Med. Hyg. 76: 1113–1117. [PubMed] [Google Scholar]
- Brower, A V Z. 1994. Rapid morphological radiation and convergence among races of the butterfly Heliconius-erato inferred from patterns of mitochondrial-DNA evolution. PNAS. 91: 6491–6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadee, D D, and Wilkerson R C. 2006. Ecology of the malaria vector, Anopheles (Nyssorhynchus) marajoara Galvao and Damasceno in Trinidad, West Indies. J. Am. Mosq. Control. Assoc. 22: 22–28. [DOI] [PubMed] [Google Scholar]
- Chen, H, Strand M, Norenburg J L, Sun S, Kajihara H, Chernyshev A V, Maslakova S A, and Sundberg P. 2010. Statistical parsimony networks and species assemblages in Cephalotrichid Nemerteans (Nemertea). PLoS One 5: e12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement, M, Posada D, and Crandall K A. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9: 1657–1659. [DOI] [PubMed] [Google Scholar]
- Colinvaux, P, and de Oliveira P. 2001. Amazon plant diversity and climate through the Cenozoic. Palaeogeogr. Palaeoclimatol. Palaeoecol. 166: 51–63. [Google Scholar]
- Crandall, K A, and Templeton A R. 1993. Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics. 134: 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cywinska, A, Hunter F F, and Hebert P D. 2006. Identifying Canadian mosquito species through DNA barcodes. Med. Vet. Entomol. 20: 413–424. [DOI] [PubMed] [Google Scholar]
- Dai, Q Y, Gao Q, Wu C S, Chesters D, Zhu C D, and Zhang A B. 2012. Phylogenetic reconstruction and DNA barcoding for closely related pine moth species (Dendrolimus) in China with multiple gene markers. PLoS One 7: e32544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Queiroz, K. 2007. Species concepts and species delimitation. Syst. Biol. 56: 879–886. [DOI] [PubMed] [Google Scholar]
- Drummond, A J, Suchard M A, Xie D, and Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen, M, Winkler-Oswatisch R, and Dress A. 1988. Statistical geometry in sequence space: a method of quantitative sequence analysis. Proc. Natl. Acad. Sci. USA 85: 5913–5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier, L, Laval G, and Schneider S. 2005. Arlequin v3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. Online. 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- Foley, D H, Linton Y M, Ruiz-Lopez F, Conn J E, Sallum M A M, Povoa M M, Bergo E S, Oliveira T M P, and Wilkerson R C. 2014. Geographic distribution, evolution and disease importance of species within the Neotropical Anopheles albitarsis group (Diptera: Culicidae). J. Vector. Ecol. 39: 168–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer, O, Black M, Hoeh W, Lutz R, and Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3: 294–299. [PubMed] [Google Scholar]
- Foster, P G, de Oliveira T M P, Bergo E S, Conn J E, Sant’Ana D C, Nagaki S S, Nihei S, Lamas C E, González C, Moreira C C, . et al. 2017. Phylogeny of Anophelinae using mitochondrial protein coding genes. R. Soc. Open Sci. 4: 170758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, M K, Leaché A D, Burbrink F T, McGuire J A, and Moritz C. 2012. Coalescent-based species delimitation in an integrative taxonomy. Trends Ecol. Evol. 27: 480–488. [DOI] [PubMed] [Google Scholar]
- Gerber, A S, and Templeton A R. 1996. Population sizes and within-deme movement of Trimerotropis saxatilis (Acrididae), a grasshopper with a fragmented distribution. Oecologia. 105: 343–350. [DOI] [PubMed] [Google Scholar]
- Gómez-Zurita, J, Petitpierre E, and Juan C. 2000. Nested cladistic analysis, phylogeography and speciation in the Timarcha goettingensis complex (Coleoptera, Chrysomelidae). Mol. Ecol. 9: 557–570. [DOI] [PubMed] [Google Scholar]
- Gonzalez, R, and Carrejo N. 2009. Introducción al estudio taxónomico de Anopheles de Colombia: Claves y notas de distribución Colombia. Programa Editorial Universidad del Valle, Cali, Colombia. [Google Scholar]
- Gutiérrez, L A, Orrego L M, Gómez G F, López A, Luckhart S, Conn J E, and Correa M M. 2010. A new mtDNA COI gene lineage closely related to Anopheles janconnae of the Albitarsis complex in the Caribbean region of Colombia. Mem. Inst. Oswaldo Cruz. 105: 1019–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbach, R E. 2018. Anopheles Meigen, 1818. Mosquito Taxonomic Inventory.http://mosquito-taxonomic-inventory.info/simpletaxonomy/term/6047
- Hart, M W, and Sunday J. 2007. Things fall apart: biological species form unconnected parsimony networks. Biol. Lett. 3: 509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, M, Kishino H, and Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22: 160–174. [DOI] [PubMed] [Google Scholar]
- Hebert, P D, Cywinska A, Ball S L, and deWaard J R. 2003. Biological identifications through DNA barcodes. Proc. Biol. Sci. 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, J L, Bohonak A J, and Kelley S T. 2005. Isolation by distance, web service. BMC Genet. 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, T A, Lima J B, and Tada M S. 1991a. Comparative susceptibility of anopheline mosquitoes to Plasmodium falciparum in Rondonia, Brazil. Am. J. Trop. Med. Hyg. 44: 598–603. [DOI] [PubMed] [Google Scholar]
- Klein, T A, Lima J B, Tada M S, and Miller R. 1991b. Comparative susceptibility of anopheline mosquitoes in Rondonia, Brazil to infection by Plasmodium vivax. Am. J. Trop. Med. Hyg. 45: 463–470. [DOI] [PubMed] [Google Scholar]
- Krzywinski, J, Li C, Morris M, Conn J E, Lima J B, Povoa M M, and Wilkerson R C. 2011. Analysis of the evolutionary forces shaping mitochondrial genomes of a Neotropical malaria vector complex. Mol. Phylogenet. Evol. 58: 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, N P, Rajavel A R, Natarajan R, and Jambulingam P. 2007. DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae). J. Med. Entomol. 44: 1–7. [DOI] [PubMed] [Google Scholar]
- Leache, A D, and Fujita M K. 2010. Bayesian species delimitation in West African forest geckos Hemidactylus fasciatus. Proc. R. Soc. Biol. 277: 3071–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y H, and Lin C P. 2012. Pleistocene speciation with and without gene flow in Euphaea damselflies of subtropical and tropical East Asian islands. Mol. Ecol. 21: 3739–3756. [DOI] [PubMed] [Google Scholar]
- Lehr, M A, Kilpatrick C W, Wilkerson R C, and Conn J E. 2005. Cryptic species in the Anopheles (Nyssorhynchus) albitarsis (Diptera: Culicidae) complex: incongruence between random amplified polymorphic DNA-polymerase chain reaction identification and analysis of mitochondrial DNA COI gene sequences. Ann. Entomol. Soc. Am. 98: 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C, and Wilkerson R C. 2005. Identification of Anopheles (Nyssorhynchus) albitarsis complex species (Diptera: Culicidae) using rDNA internal transcribed spacer 2-based polymerase chain reaction primes. Mem. Inst. Oswaldo Cruz. 100: 495–500. [DOI] [PubMed] [Google Scholar]
- Li, M, Liu Q, Ke Y, Tian Y, Zhu G, Xie Q, and Bu W. 2012. Biogeographical origin and speciation of the Anthocoris nemorum group. J. Insect Sci. 12: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado, P, and Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lima, J B, Valle D, and Peixoto A A. 2004. Analysis of reproductive isolation between sibling species Anopheles albitarsis sensu stricto and An. deaneorum, two malaria vectors belonging to the Albitarsis complex (Diptera: Culicidae). J. Med. Entomol. 41: 888–893. [DOI] [PubMed] [Google Scholar]
- Linthicum, K J. 1988. A revision of the Argyritarsis Section of the subgenus Nyssorhynchus of Anopheles (Diptera: Culicidae). Mosq. Syst. 20: 98–270. [Google Scholar]
- Loaiza, J R, Scott M E, Bermingham E, Rovira J, and Conn J E. 2010. Evidence for Pleistocene population divergence and expansion of Anopheles albimanus in Southern Central America. Am. J. Trop. Med. Hyg. 82: 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel, N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27: 209–220. [PubMed] [Google Scholar]
- McKeon, S N, Lehr M A, Wilkerson R C, Ruiz J F, Sallum M A, Lima J B, Povoa M M, and Conn J E. 2010. Lineage divergence detected in the malaria vector Anopheles marajoara (Diptera: Culicidae) in Amazonian Brazil. Malar. J. 9: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoki, M T, Wilkerson R C, and Sallum M A. 2009. The Anopheles albitarsis complex with the recognition of Anopheles oryzalimnetes Wilkerson and Motoki, n. sp. and Anopheles janconnae Wilkerson and Sallum, n. sp. (Diptera: Culicidae). Mem. Inst. Oswaldo Cruz. 104: 823–850. [DOI] [PubMed] [Google Scholar]
- Narang, S K, Klein T A, Perera O P, Lima J B, and Tang A T. 1993. Genetic evidence for the existence of cryptic species in the Anopheles albitarsis complex in Brazil: allozymes and mitochondrial DNA restriction fragment length polymorphisms. Biochem. Genet. 31: 97–112. [DOI] [PubMed] [Google Scholar]
- Pedro, P M, and Sallum M A M. 2009. Spatial expansion and population structure of the Neotropical malaria vector, Anopheles darlingi (Diptera: Culicidae). Biol. J. Linn. Soc. 97: 854–866. [Google Scholar]
- Póvoa, M M, de Souza R T, Lacerda R N, Rosa E S, Galiza D, de Souza J R, Wirtz R A, Schlichting C D, and Conn J E. 2006. The importance of Anopheles albitarsis E and An. darlingi in human malaria transmission in Boa Vista, state of Roraima, Brazil. Mem. Inst. Oswaldo Cruz. 101: 163–168. [DOI] [PubMed] [Google Scholar]
- Rambaut, A. 2009. FigTree v1.3.1 2006–2009 [Readme file]. Available at http://tree.bio.ed.ac.uk/software/figtree
- Rambaut, A, Suchard M, and Drummond A. 2013. Tracer v.1.6. Available at http://beast.bio.ed.Ac.uk/Tracer.
- Rios, R I, Nascimento L Z, and de Oliveira A C. 1984. Complexo Anopheles (Nyssorhynchus) albitarsis: impossibilidade de separa-los em duas subspecies, A. albitarsis albitarsis e A. albitarsis domesticus (Diptera: Culicidae). Rev. Brasil. Bio. 44: 461–465. [Google Scholar]
- Rosa-Freitas, M G. 1989. Anopheles (Nyssorhynchus) deaneorum: a new species in the Albitarsis complex (Diptera: Culicidae). Mem. Inst. Oswaldo Cruz. 84: 535–543. [Google Scholar]
- Rosa-Freitas, M G, Deane L M, and Momen H. 1990. A morphological, behavioural and isoenzymatic study in Anopheles albitarsis from 10 populations. Mem. Inst. Oswaldo Cruz. 85: 275–289. [Google Scholar]
- Rubio-Palis, Y. 1994. Variation of the vectorial capacity of some anophelines in western Venezuela. Am. J. Trop. Med. Hyg. 50: 420–424. [DOI] [PubMed] [Google Scholar]
- Ruiz-Lopez, F, Linton Y M, Ponsonby D J, Conn J E, Herrera M, Quiñones M L, Vélez I D, and Wilkerson R C. 2010. Molecular comparison of topotypic specimens confirms Anopheles (Nyssorhynchus) dunhami Causey (Diptera: Culicidae) in the Colombian Amazon. Mem. Inst. Oswaldo Cruz. 105: 899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Lopez, F, Wilkerson R C, Conn J E, McKeon S N, Levin D M, Quiñones M L, Póvoa M M, and Linton Y M. 2012. DNA barcoding reveals both known and novel taxa in the Albitarsis Group (Anopheles: Nyssorhynchus) of Neotropical malaria vectors. Parasit. Vectors. 5: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpassa, V M, and Conn J E. 2011. Mitochondrial DNA detects a complex evolutionary history with Pleistocene Epoch divergence for the neotropical malaria vector Anopheles nuneztovari sensu lato. Am. J. Trop. Med. Hyg. 85: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheis, A S, Booth J Y, Perlmutter L R, Bond J E, and Sheldon A L. 2012. Phylogeography and species biogeography of montane Great Basin stoneflies. Mol. Ecol. 21: 3325–3340. [DOI] [PubMed] [Google Scholar]
- Strimmer, K, and Moulton V. 2000. Likelihood analysis of phylogenetic networks using directed graphical models. Mol. Biol. Evol. 17: 875–881. [DOI] [PubMed] [Google Scholar]
- Templeton, A R, Crandall K A, and Sing C F. 1992. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 132: 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J, Gibson T, Plewniak F, Jeanmougin F, and Higgins D G. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila, C, Amorim I R, Leonard J A, Posada D, Castroviejo J, Petrucci-Fonseca F, Crandall K A, Ellegren H, and Wayne R K. 1999. Mitochondrial DNA phylogeography and population history of the grey wolf Canis lupus. Mol. Ecol. 8: 2089–2103. [DOI] [PubMed] [Google Scholar]
- Wilkerson, R C, Parsons T J, Klein T A, Gaffigan T V, Bergo E, and Consolim J. 1995a. Diagnosis by random amplified polymorphic DNA polymerase chain reaction of four cryptic species related to Anopheles (Nyssorhynchus) albitarsis (Diptera: Culicidae) from Paraguay, Argentina, and Brazil. J. Med. Entomol. 32: 697–704. [DOI] [PubMed] [Google Scholar]
- Wilkerson, R C, Gaffigan T V, and Bento Lima J. 1995b. Identification of species related to Anopheles (Nyssorhynchus) albitarsis by random amplified polymorphic DNA-polymerase chain reaction (Diptera: Culicidae). Mem. Inst. Oswaldo Cruz. 90: 721–732. [DOI] [PubMed] [Google Scholar]
- Wilkerson, R C, Foster P G, Li C, and Sallum M A. 2005. Molecular phylogeny of neotropical Anopheles (Nyssorhynchus) Albitarsis species complex (Diptera: Culicidae). Ann. Entomol. Soc. Am. 98: 918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.