Tumour-associated macrophages promote tumour malignancy and immunosuppression. Using human patient tissue samples and immunocompetent mouse models, Ross et al. show that platelet-derived growth factor beta promotes macrophage infiltration and an immunosuppressive phenotype in paediatric high-grade glioma.
Keywords: paediatric glioma, PDGFB, macrophage, diffuse intrinsic pontine glioma, TAM, inflammation
Abstract
Paediatric high-grade gliomas (HGGs) account for the most brain tumour-related deaths in children and have a median survival of 12–15 months. One promising avenue of research is the development of novel therapies targeting the properties of non-neoplastic cell-types within the tumour such as tumour associated macrophages (TAMs). TAMs are immunosuppressive and promote tumour malignancy in adult HGG; however, in paediatric medulloblastoma, TAMs exhibit anti-tumour properties. Much is known about TAMs in adult HGG, yet little is known about them in the paediatric setting. This raises the question of whether paediatric HGGs possess a distinct constituency of TAMs because of their unique genetic landscapes. Using human paediatric HGG tissue samples and murine models of paediatric HGG, we demonstrate diffuse midline gliomas possess a greater inflammatory gene expression profile compared to hemispheric paediatric HGGs. We also show despite possessing sparse T-cell infiltration, human paediatric HGGs possess high infiltration of IBA1+ TAMs. CD31, PDGFRβ, and PDGFB all strongly correlate with IBA1+ TAM infiltration. To investigate the TAM population, we used the RCAS/tv-a system to recapitulate paediatric HGG in newborn immunocompetent mice. Tumours are induced in Nestin-positive brain cells by PDGFA or PDGFB overexpression with Cdkn2a or Tp53 co-mutations. Tumours driven by PDGFB have a significantly lower median survival compared to PDGFA-driven tumours and have increased TAM infiltration. NanoString and quantitative PCR analysis indicates PDGFB-driven tumours have a highly inflammatory microenvironment characterized by high chemokine expression. In vitro bone marrow-derived monocyte and microglial cultures demonstrate bone marrow-derived monocytes are most responsible for the production of inflammatory signals in the tumour microenvironment in response to PDGFB stimulation. Lastly, using knockout mice deficient for individual chemokines, we demonstrate the feasibility of reducing TAM infiltration and prolonging survival in both PDGFA and PDGFB-driven tumours. We identify CCL3 as a potential key chemokine in these processes in both humans and mice. Together, these studies provide evidence for the potent inflammatory effects PDGFB has in paediatric HGGs.
Introduction
Despite our continued understanding of high-grade gliomas (HGGs) and the molecular mechanisms driving their malignancy, paediatric HGGs remain one of the most difficult malignancies to treat. Brain tumours account for the most cancer-related deaths in the paediatric population, and paediatric HGGs account for the most deaths among this category which can be attributed to multiple defining features (Ostrom et al., 2014, 2015). Paediatric HGGs have a high frequency (∼50%) of arising in infra-tentorial regions of the brain such as the brainstem, rendering these tumours surgically inaccessible and limited to therapeutic intervention (Mackay et al., 2017). Additionally, until recently, therapeutics for the treatment of paediatric HGG were adapted from their adult counterparts and failed to consider the innate biological and molecular differences between the two (Jones and Baker, 2014; Vinci et al., 2018). Lastly, the dynamic tumour microenvironment, which has been shown to be critical for tumour progression in adult HGGs, remains highly understudied and unaccounted for in paediatric HGGs.
Paediatric HGGs are characterized by PDGFRA amplification, TP53 deletion/mutation, CDKN2A deletion/mutation, and histone mutations such as H3F3A and HIST1H3B (Mackay et al., 2017). As our understanding of the molecular alterations driving paediatric HGGs increases, targeted therapies against molecular alterations and immune processes are being increasingly investigated (Lasky et al., 2013; Souweidane et al., 2018; Tejada et al., 2018; Chi et al., 2019; Hall et al., 2019). However, therapies designed to target non-neoplastic infiltrating cell types including tumour associated macrophages (TAMs) are lacking, and may be worth investigating as these cell types constitute a large majority of cells within the tumour mass. The TAM population is composed of brain-resident microglia and infiltrating bone-marrow derived monocytes (BMDMs) that differentiate into macrophages upon extravasation into the brain parenchyma (Gutmann and Kettenmann, 2019). We previously demonstrated in adult HGGs that TAMs constitute up to 40% of the total tumour cell mass, and up to 80% of the TAM population are peripheral infiltrating monocytes (Chen et al., 2017, 2020). These cell populations are attracted to tumour sites via chemokines and growth factors secreted by tumour and stromal cells in the microenvironment and promote tumour growth (Hambardzumyan et al., 2016; Gutmann and Kettenmann, 2019). While they promote tumour growth in adult HGG, TAMs exhibit anti-tumour properties in medulloblastoma, which is the most common malignant paediatric brain tumour (Maximov et al., 2019). These data suggest the role of TAMs is tumour-type dependent and further investigation is required to determine their contribution to paediatric HGG.
Little is known about the factors promoting inflammation in paediatric HGGs, and there remains an inadequate understanding of how the TAM population contributes to, and is affected by, these processes. To date, there are limited studies on the immune microenvironment of paediatric HGGs. It has been reported that diffuse intrinsic pontine gliomas have an increase in IBA1+ TAMs compared to non-tumour brain tissue (Lin et al., 2018). However, when comparing paediatric HGGs to paediatric low-grade gliomas, low-grade gliomas possess greater CD45+ infiltrating immune cells and CD8+ T cells (Lieberman et al., 2019). They also have greater chemokine expression compared to paediatric HGGs (Plant et al., 2018). These studies are limited in number and scope, and fail to identify driving mechanisms promoting immune cell infiltration. This study was designed (i) to test whether TAMs display pro- or anti-tumour effects in paediatric HGG; and (ii) to develop a foundational understanding of the factors promoting TAM infiltration. We also sought to develop an understanding of whether different genetic drivers affect TAM infiltration and patient survival. Using human patient samples, we establish a link between PDGFB/PDGFRβ and TAM infiltration in paediatric HGG. We induced tumorigenesis in immunocompetent mice using the RCAS/tv-a somatic cell gene transfer system driven by RCAS-PDGFA or RCAS-PDGFB overexpression. We observed differential PDGF signalling activity in paediatric HGG leads to an increase in TAM infiltration and a decrease in survival, providing us with a system to examine the mechanisms behind these processes. Further, we identify a previously unrecognized chemoattractant in paediatric HGG, CCL3, as a key mediator of TAM infiltration. When Ccl3 is deleted in a murine model of paediatric HGG, TAM infiltration is inhibited and survival is extended.
Materials and methods
NanoString analysis
Human formalin-fixed paraffin-embedded (FFPE) tissue scrolls were cut in 10 µm sections for RNA extraction. RNA integrity was confirmed using a bioanalyzer and samples possessing a DV300% >30 were used. We used 50 ng of RNA for NanoString analysis with the Pan-Cancer Pathways Immune Panel (NanoString, XT-CSO-HIP1-12). For mouse samples, animals were perfused with ice-cold Ringers solution, brains were extracted, and tumour tissue was snap-frozen in liquid nitrogen for storage. RNA extraction was performed using the RNeasy® Lipid Tissue Mini Kit (Qiagen #74804) according to the manufacturer’s instructions. RNA integrity was confirmed as described above. The mouse Pan-Cancer Pathways Immune Panel (NanoString, XT-CSO-MIP1-12) was used. All gene expression data were processed using nSolver Advanced Analysis Software version 4.0 and data were normalized using the geNorm algorithm, which uses housekeeping genes to compute a normalization factor applied to all genes (Vandesompele et al., 2002). For pathway analysis, a z-transformation was applied to normalized gene scores. For all other analyses, normalized genes were log2 transformed and plotted using GraphPad Prism 8 (GraphPad Software, San Diego, CA).
Human paediatric glioma tissue staining
Use of de-identified human paediatric tissue samples (ages 0–21 years old) was Institutional Review Board approved by Emory University and the Ann and Robert H. Lurie Children’s Hospital of Chicago. Sample diagnoses were confirmed by a board-certified neuropathologist (M.S.) and board-certified paediatric pathologist (N.W.). All diffuse midline glioma (DMG) samples included in this study are diffuse intrinsic pontine gliomas. See Supplementary Table 1 for the clinical characteristics of these samples. FFPE tissue samples were sectioned at 5 µm per slide and mounted. Heat mediated antigen retrieval in Tris-EDTA buffer pH 7.8 was performed. Samples were then stained on a Discovery XT Platform with; IBA1 (Wako 019-19741, 1:500), PDGFA (sc-128, 1:200), PDGFB (sc-127, 1:200), PDGFRα (cs3174, 1:300), PDGFRβ (cs3169, 1:300), CD31 (Dako m0823, 1:50), CD3 (Dako A0452, 1:100), and CD8 (Dako M7103, 1:100).
Mice
All injections were performed in mouse pups aged 0–2 days old among equally distributed sexes for each genotype. Ntva;Ptenfl/fl;Cdkn2a−/−mice are in a mixed genetic background while Ntva;Ptenfl/fl mice are in a pure C57BL/6 background (Herting et al., 2017). Mice were housed in a climate-controlled, pathogen-free facility with access to food and water ad libitum under a 12-h light/dark cycle. Ccl2 (#004434) and Ccl7 (#017638) knockout mice were obtained from the Jackson Laboratory and crossed to C57BL/6 Ntva;Ptenfl/fl mice until homozygous knockout mice were obtained as previously described for the generation of Ntva;Ptenfl/fl;Ccl2−/− mice (Tsou et al., 2007; Chen et al., 2017). Ccl3 knockout mice were gifted by Dr C. K. Qu, Ccl8/12−/− mice were gifted by Dr Sabina Islam, and both were crossed with C57BL/6 Ntva;Ptenfl/fl mice until homozygous knockout mice were obtained. Genotyping of all mice was performed using Transnetyx. All experimental procedures were approved by the Institutional Animal Care and Use Committee of Emory University (Protocol #2003253, #201700633) and Icahn School of Medicine at Mount Sinai (Protocol #201900619).
Cell culture and injections
DF1 cells (ATCC) were grown at 39°C, expanded to passage 4 and transfected with RCAS-PDGFB-HA, RCAS-PDGFA-Myc, RCAS-shp53-RFP, RCAS-H3.3WT, or RCAS-H3.3K27M using a FuGENE® 6 Transfection kit (Roche, 11814443001) (Ozawa et al., 2014). Cells were cultured with DMEM media (Gibco, 11995-065) supplemented with 1× l-glutamine, 1× penicillin/streptomycin, and 10% foetal bovine serum (FBS) (ATCC). Transfected DF1 cells were used for injections before reaching passage 25. DF1 cells were injected as equal mixtures of 4 × 104 cells/µl, 1 µl per mouse, using a 30-gauge needle attached to a Hamilton syringe.
Immunohistochemical staining of murine high-grade gliomas
Mice were sacrificed as described above, and brains were fixed in 10% neutral buffered formalin for 72 h. The brains were then embedded in paraffin and sectioned at 5 µm and mounted. Haematoxylin and eosin staining was performed for each sample. Murine gliomas were graded by a board-certified neuropathologist (P.J.C.) according to the 2016 WHO Classification of Central Nervous System Tumors (Louis et al., 2016). Antibodies used for staining include; IBA1 (Wako 019-19741, 1:500), CD31 (Dianova Dia-310, 1:50), α-SMA (Dako M0851, 1:200), PDGFRα (CST 3174S, 1:500), PDGFRβ (CST 3169S, 1:100), and Phospho-histone H3 (CST 9701S, 1:200). For a complete list of antibodies used, see Supplementary Table 2. Staining was performed on the Discovery XT Platform (Ventana Medical Systems) at the defined dilutions in 2% BSA in PBS.
Image analysis
Stained samples were scanned at ×20 and converted to digital images using a Nanozoomer 2.0HT whole slide scanner (Hamamatsu Photonic K.K.). For each staining and sample, multiple representative images were acquired to accurately represent intra-tumour heterogeneity as previously described (Kaffes et al., 2019). For CD3 and CD8 staining, the number of positively stained cells per ×20 field were counted. For IBA1, PDGFRα, and PDGFRβ, the percentage of positively stained tumour area was calculated using the threshold function in ImageJ Fiji (Schindelin et al., 2012) by recording the difference between positive staining intensity above background staining intensity. For PDGFA, samples were scored as positively or negatively stained. For PDGFB, three independent reviewers (J.R., C.H., Z.C.) assigned a relative score of 0 (low) to 5 (high) to each image based on staining intensity and the average scores were calculated. For phospho-histone 3, the number of positively stained nuclei per square millimetre was quantified. All image analysis was performed using ImageJ Fiji.
Flow cytometry
Mice were anaesthetized with an overdose of ketamine/xylazine. Blood (400 µl) was drawn from the left ventricle of the heart before the mice were perfused with Ringer’s solution. The blood was examined with BD TrucountTM tubes (BD Biosciences) to quantify the absolute number of myeloid cells according to manufacturer’s instructions. Tumours were dissociated either using the Miltenyi Neural Tissue Dissociation kit (Miltenyi, 130-092-628) for the analysis of myeloid cells following manufacturer’s protocol, or with papain for lymphocyte analysis. Cells were blocked with a solution containing 2% FBS, 5% normal rat serum, 5% normal mouse serum, 5% normal rabbit serum, 10 µg/ml anti-FcR (BioLegend, 101319) and 0.2% NaN3 in DPBS. Cells were then stained for CD45-APC, CD11b-PerCP-Cy5.5, Ly6C-PE-Cy7, and Ly6G-V450 for myeloid cell detection. For lymphocyte analysis, the cells were additionally stained for CD3-PE-Dazzle, CD4-APC-Cy7, CD8-BV510, B220-BV605, NK1.1-BV711, PD1-BV786, MHCII-A700, CD49d-PE-Cy7, live/dead Zombie UV dye (BioLegend) before they were fixed and permeabilized with an intracellular staining kit (eBiosciences). The cells were further stained for Foxp3-FITC and Granzyme B-PE. Fifty thousand events were recorded on a BD-LSR II or Fortessa flow cytometer. Data were compensated and quantified in FlowJo 10.4.1.
Quantitative PCR
One microgram of RNA was used to synthesize cDNA using the SuperScriptTM III First-Strand Synthesis System (Invitrogen). Complementary DNA was used for quantitative PCR using the SsoAdvanced SYBR® Green Supermix (Bio-Rad) on an Applied Biosystems 7500 Fast Real-Time PCR System (Life Technologies). PrimePCR SYBR® Green Assay primers were obtained from Bio-Rad. For a full list of genes used, see Supplementary Table 3 and Supplementary Fig 9. β-Actin was used as an internal reference gene and an inter-run calibrator was used to normalize comparative CT values for the combination of plates when necessary. Hierarchical clustering was performed on log2 transformed, z-score standardized quantitative (q)PCR data using Morpheus (Broad Institute).
Bone marrow-derived macrophage and microglia cell culture
BMDMs and microglia were isolated and cultured as previously described (Herting et al., 2019; Chen et al., 2020). Cells were incubated in FBS deficient media (0.5%) for 3 h prior to stimulation with recombinant human PDGFA (0–100 ng/ml; R&D Systems) or PDGFB (0–100 ng/ml; R&D Systems) for 6 h. For a full description see the Supplementary material.
RNA sequencing
Human RNA sequencing data were generated from the assembled database as previously described by (Mackay et al., 2017). For a full description of data acquisition, quality control, and normalization methods, see the Supplementary material.
Data analysis
For qPCR data normalization, ΔΔCT values were log2 transformed and z-score standardized. Statistical tests used for each analysis are further explained in figure legends. GraphPad Prism 8 was used for all statistical analyses. Significance was set at *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Data availability
All data used for this manuscript are available upon reasonable request.
Results
Immune profiling of human paediatric high-grade gliomas
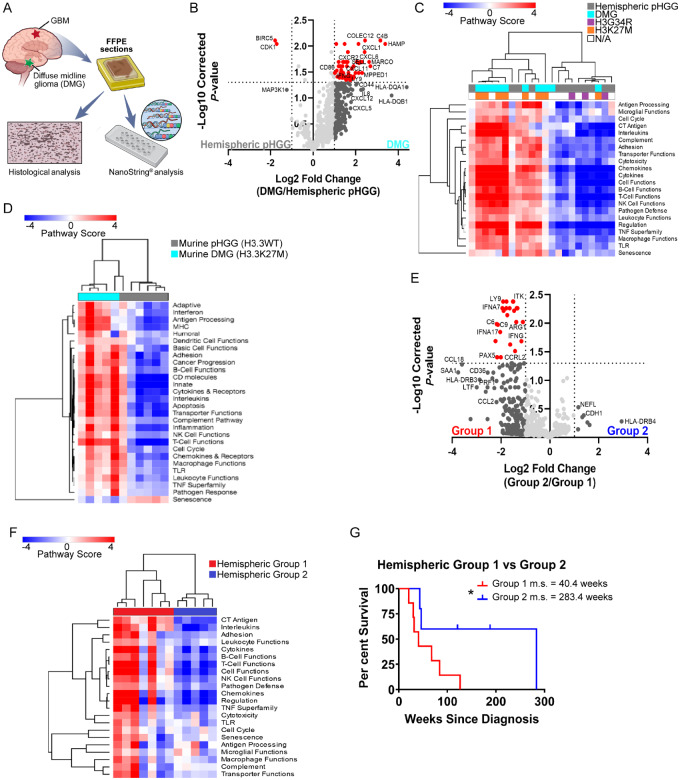
The presence of tumour infiltrating CD45+CD11b+ myeloid cells and the presence of infiltrating CD3+ and CD8+ lymphocytes has been reported in paediatric HGGs (Griesinger et al., 2014; Lin et al., 2018; Plant et al., 2018). However, little is known concerning the inflammatory microenvironment of these tumours. Further, there has been no comparison of hemispheric paediatric HGGs to diffuse midline gliomas (DMGs; including diffuse intrinsic pontine gliomas) in terms of their inflammatory microenvironments and infiltrating immune cells. To characterize these tumours, we obtained FFPE tissue blocks of 22 human paediatric HGGs and performed NanoString analysis (Fig. 1A). Twelve hemispheric paediatric HGGs (3/12 were H3G34R mutants), and 10 DMGs (8/10 were H3K27M mutants) were included. When comparing differentially expressed genes between the two subgroups, DMGs had higher expression of leucocyte-attracting chemokines CXCL2, CXCL5, CXCL6, and their receptor CXCR2. IL-8, a neutrophil chemotactic factor that strongly induces CXCR2 activity and expression was also highly expressed in these samples (Schinke et al., 2015). One of the most significantly differentially expressed genes between the two subgroups was CXCL1 (high in DMGs), which is a potent promoter of inflammation (Fig. 1B). CXCL1 has been shown to be produced by mast cells and macrophages, and plays a critical role in the recruitment and migration of immune cells to sites of inflammation (De Filippo et al., 2013). Further, in a model of pancreatic adenocarcinoma, high CXCL1 expression correlated with low T-cell infiltration, and deletion of CXCL1 rendered immunotherapy resistant tumours susceptible to CTLA-4 and PD1 blockade (Li et al., 2018). All these genes signal through the CXCR2 receptor, indicating a possible role for this signalling pathway in DMGs. When comparing the pathways represented by the expressed genes, unsupervised hierarchical clustering displayed separation between the two groups of paediatric HGG. Eight of ten DMG samples clustered together, seven of which were K27M mutant, while the hemispheric samples separately clustered together (Fig. 1C). DMGs had greater expression of genes involved in pathways including chemokine and cytokine signalling, leucocyte functions, and macrophage functions. We found similar clustering patterns between murine DMG (H3.3K27M) and hemispheric paediatric HGG (H3.3WT). This analysis again demonstrated DMGs were more inflammatory than their hemispheric counterparts (Fig. 1D). Additionally, in both human and murine DMG, there was greater representation of genes characterizing inflammatory cell types including T cells, macrophages, neutrophils, and CD45+ cells as opposed to hemispheric tumours (Supplementary Fig. 1A and B). Lastly, when clustering both human and murine tumours together by their expression of all genes, hemispheric paediatric HGGs were most similar to each other and DMGs were most similar to each other, regardless of species (Supplementary Fig. 1C). These results suggest DMGs have a greater inflammatory microenvironment when compared to hemispheric paediatric HGGs. This is intriguing since DMGs, specifically diffuse intrinsic pontine gliomas, have the shortest median survival yet have historically been considered ‘immune cold’ with respect to infiltrating lymphocytes.
Figure 1.
Immune profiling of human paediatric HGGs. (A) FFPE hemispheric paediatric HGG (n = 12) or DMG (n = 10) human tumour samples were used for NanoString analysis and histological characterization. (B) Volcano plot demonstrating differential gene expression between hemispheric samples and DMG samples. Vertical dashed line = fold change > 2. Horizontal dashed line = Benjamini-Hochberg corrected P < 0.05. (C) Unsupervised hierarchical clustering of human paediatric HGG samples based on pathway scores obtained from NanoString. n = 22 unique samples. (D) Unsupervised hierarchical clustering of murine hemispheric PDGFB-driven paediatric HGG (H3.3WT, n = 6) or DMG (H3.3K27M, n = 5) based on pathway scores obtained from NanoString. (E) Volcano plot demonstrating differential gene expression between human hemispheric samples. (F) Unsupervised hierarchical clustering of human hemispheric samples based on their pathway scores. (G) Survival analysis of human Group 1 hemispheric tumours (more inflammatory) versus Group 2 hemispheric tumours (less inflammatory). Group 1 m.s. = 40.4 weeks, Group 2 m.s. = 283.4 weeks. Log-rank Mantel-Cox test. P < 0.05.
When examining only human hemispheric samples, two distinct subgroups were observed; one with a high inflammatory signature (group 1) and one with a low inflammatory signature (group 2) (Fig. 1E and F). Group 1 samples had high expression of chemokines including CCL2, CCL5, CCL18, CCL21, and CXCL11. Markers of an activated immune response and complement system were also highly expressed including IFNG, IFNA17, PRF1 (perforin), HLA-DRB3 (MHC-II), C6, and C9 (Wherry et al., 2007; Hudson et al., 2019). Group 2 samples had unremarkable expression of genes involved in the immune response. Interestingly, patients from group 2 had a significantly longer median survival of 283.4 weeks from the time of diagnosis compared to patients from group 1 who had a median survival of only 40.4 weeks (Fig. 1G). Although group 2 contained two G34R tumours, there were no other defining clinical features of either group in terms of age, sex, or other defining mutations.
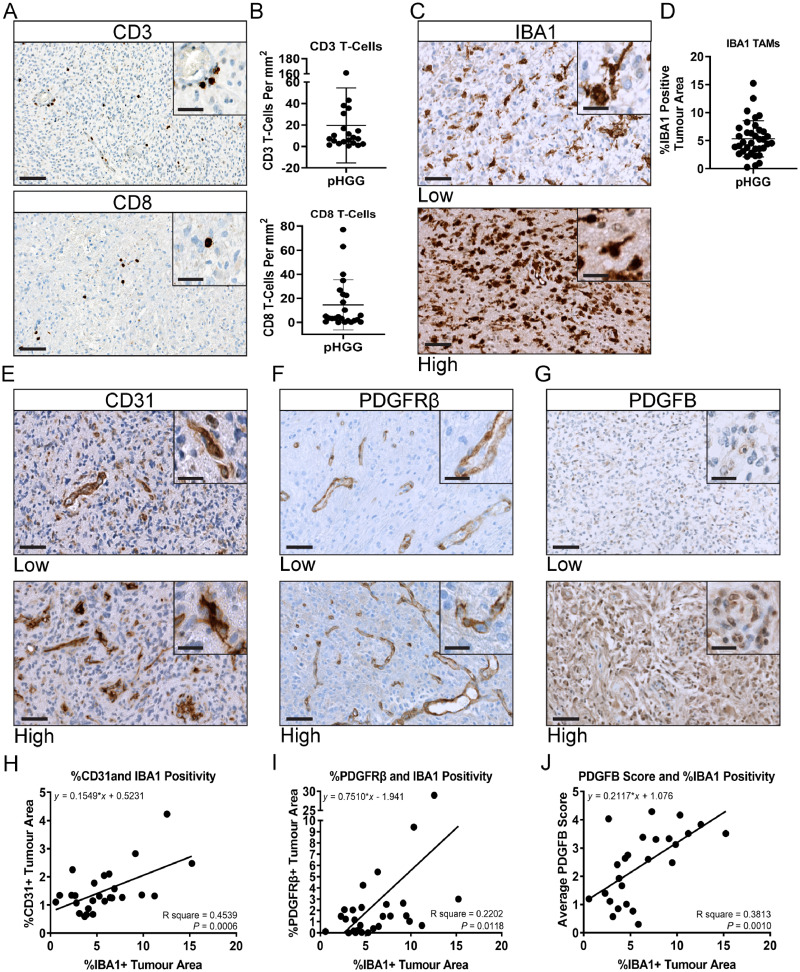
To determine whether the differential inflammatory signatures persist at the protein level we next evaluated a larger cohort of human paediatric HGG tumour samples by immunohistochemistry. CD3 and CD8 T-cell staining corroborated previous findings of sparse T-cell infiltrates in these tumours (Fig. 2A and B) (Lin et al., 2018). Most positively stained cells were found in perivascular spaces and around areas of necrosis. Because of the low number of lymphocytes in these tumours, we then stained for the pan-macrophage marker IBA1. Unlike T-cell staining, there was considerable sample to sample variability of IBA1+ infiltrating TAMs (Fig. 2C and D). There was no difference in IBA1+ TAM infiltration between DMG and glioblastoma samples (Supplementary Fig. 1D); however, samples with high IBA1 staining also had high macrophage and CD45 cell scores in NanoString (Supplementary Fig. 1E and F). Staining for CD31, an endothelial cell marker, and PDGFRβ, a stromal cell marker, demonstrates these tumours are highly vascularized with abnormal networks of blood vessels (Fig. 2E and F). Staining for the PDGFRβ ligand, PDGFB, also demonstrates variable staining from tumour to tumour (Fig. 2G). Interestingly, CD31, PDGFRβ, and PDGFB all strongly correlate with IBA1 positivity (Fig. 2H–J). Although there were few T cells in these tumours, there was still a positive correlation between CD3 and CD31, PDGFRβ, and IBA1 (Supplementary Fig. 2A–C). Additionally, samples with high PDGFB staining also had high CD31 and smooth muscle actin staining, implicating PDGFB in the reorganization of the tumour microenvironment (Supplementary Fig. 2D and E). Unlike staining for PDGFB (22/26 positively stained), the majority of tissue samples did not positively stain for PDGFA (7/33) (Supplementary Fig. 2F–H), and only four samples stained for both ligands, further highlighting the importance of the PDGFRβ/PDGFB pathway in paediatric HGG. Although limited by sample sizes, for those that NanoString were run on, we did observe a general trend of greater IBA1, PDGFRβ, and PDGFB staining in DMG samples compared to hemispheric tumour samples (Supplementary Fig. 2I). No correlations were found between these histological markers and molecular status, but this does not preclude the existence of correlation. Further studies with larger sample sizes are necessary to address this. Taken together, these results indicate a possible role for PDGFRβ/PDGFB pathway involvement in the infiltration of IBA1+ TAMs and other immune cell types in paediatric HGG. To directly test the role of PDGFRβ/PDGFB pathway involvement in the recruitment of TAMs in paediatric HGG we next generated hemispheric tumours using PDGFA and PDGFB ligands as oncogenic drivers in immunocompetent mice.
Figure 2.
Neuropathological characterization of human paediatric HGGs. FFPE tumour samples were stained for the following; (A) CD3 and CD8 T cells. (B) Quantification of CD3 (n = 21) and CD8 (n = 24) positivity. (C) IBA1. Representative images for low and high staining samples are displayed. (D) Quantification of IBA1 positivity (n = 36). (E) CD31 (n = 27). (F) PDGFRβ (n = 30). (G) PDGFB (n = 26). (H) Linear regression correlation of CD31+ tumour area with IBA1+ tumour area (n = 26). (I) Correlation of PDGFRβ+ tumour area with IBA1+ tumour area (n = 28). (J) Correlation of PDGFB score with IBA1+ tumour area (n = 25). All images ×40 magnification, ×80 insets. Scale bars = 100 μm and 20 μm, respectively.
PDGF-driven GEMM of hemispheric paediatric high-grade gliomas
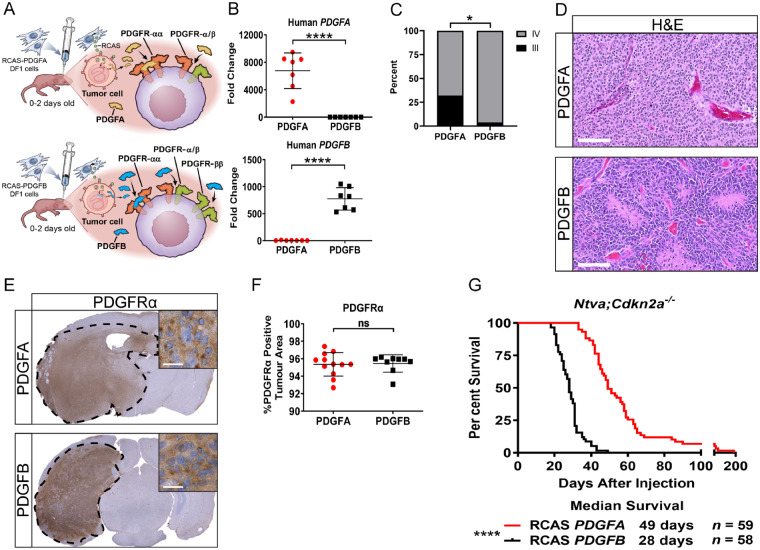
We used the RCAS/tv-a system to induce HGGs in nestin-positive brain cells in the cerebral hemispheres of immunocompetent newborn pups (postnatal Days 0–2). Ntva;Cdkn2a−/− mice received a single injection of DF1 cells producing either RCAS-PDGFA or RCAS-PDGFB. Ntva;Cdkn2awt/wt mice received a co-injection of DF1 cells consisting of RCAS-shP53 along with either RCAS-PDGFA or RCAS-PDGFB (Fig. 3A). PDGF signalling has been established as a potent inducer of cellular proliferation, survival, angiogenesis, and inflammation, thereby promoting tumorigenesis (Heldin and Westermark, 1999; He et al., 2015; Heldin et al., 2018; Papadopoulos and Lennartsson, 2018). Tumour cells co-express PDGF ligands and PDGFRα while stromal cells co-express PDGF ligands and both PDGFRα and PDGFRβ. The co-expression of ligands and receptors promotes autocrine and paracrine signalling, further promoting tumour growth, angiogenesis, and stromal recruitment (Calzolari and Malatesta, 2010). PDGFA has the ability to bind and activate PDGFRα, while PDGFB can bind and activate both PDGFRα and PDGFRβ (Westermark et al., 1995; Singh et al., 2002; Nordby et al., 2017). Freshly dissociated tumour tissue from these models was verified using qPCR for PDGFA and PDGFB (Fig. 3B). PDGFA-driven tumours produced a mix of grade III (32%) and grade IV (68%) tumours while PDGFB-driven tumours were all grade IV (96%), with the exception of one grade III tumour (4%) (Fig. 3C). These tumours displayed all major histological hallmarks of HGG including microvascular hyperplasia and pseudopalisading necrosis (Fig. 3D).
Figure 3.
Mouse modelling of PDGF-driven HGGs. (A) Schematic depicting the injection of newborn pups with RCAS-PDGFA or RCAS-PDGFB. Ligand-receptor binding interactions for PDGFA and PDGFB are highlighted. (B) Quantitative PCR validation for human PDGFA (n = 7) or PDGFB (n = 7) in their respective tumours, P < 0.0001. (C) Tumour grading on samples generated in Ntva;Cdkn2a−/− mice driven by PDGFA (n = 22, 7/22 grade III, 15/22 grade IV) or PDGFB (n = 26, 1/26 grade III, 25/26 grade IV). Fisher’s exact test, P = 0.0167. (D) Representative haematoxylin and eosin images of PDGFA and PDGFB-driven tumours. Scale bar = 100 μm. (E) Whole brain PDGFRα staining in PDGFA (n = 12) and PDGFB-driven (n = 9) tumours. Inset ×80, scale bar = 20 μm. (F) Quantification of PDGFRα staining. (G) Survival curves of Ntva;Cdkn2a−/− mice with tumours driven by RCAS-PDGFA or RCAS-PDGFB, P < 0.0001. Student’s t-test for quantifications, Log-rank Mantel-Cox test for survival curves.
Staining for PDGFRα in each tumour type displayed the presence of the receptor on all tumour cells and no differences in expression of this protein was observed (Fig. 3E and F). Conversely, PDGFB-driven tumours had greater tumour area covered by PDGFRβ and the pericyte marker smooth muscle actin (SMA) (Supplementary Fig. 3). Similar to our findings in human paediatric HGG samples, PDGFRβ positively correlated with IBA1 staining, suggesting the infiltration of TAMs is in part mediated through PDGFRβ/PDGFB signalling. Staining for CD31 demonstrated no significant difference in tumour area covered by blood vessels; however, PDGFA-driven tumours had smaller and more numerous blood vessels compared to PDGFB-driven tumours (Supplementary Fig. 3). Interestingly, PDGFB-driven tumours in Ntva;Cdkn2a−/− mice had a significantly decreased survival of 28 days compared to PDGFA-driven tumours whose median survival was 49 days (Fig. 3G). Taken together, PDGFB-driven tumours possessed irregular vasculature and decreased survival compared to PDGFA-driven tumours, thus implicating PDGFRβ/PDGFB signalling as an important regulator of the tumour microenvironment.
Immune profiling of PDGF-driven murine paediatric high-grade gliomas
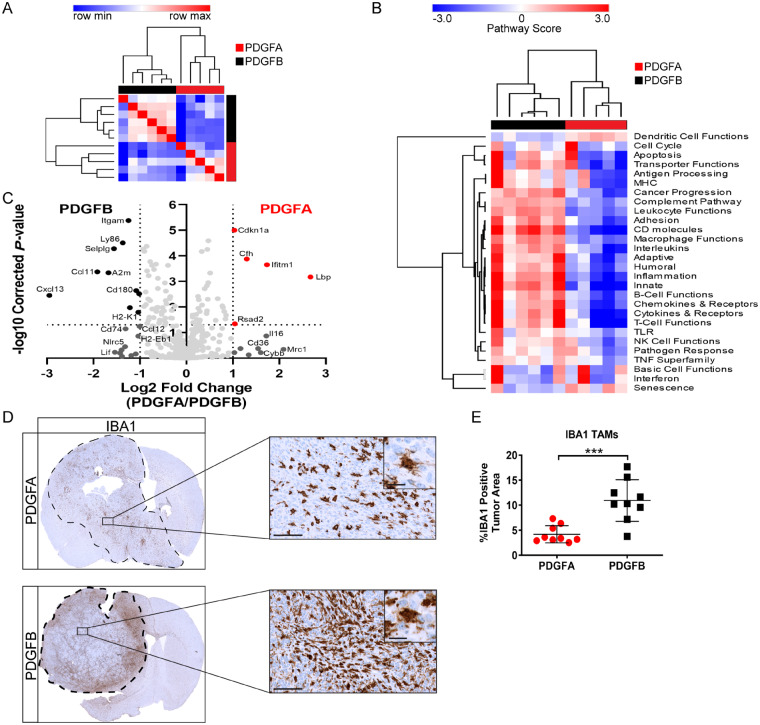
To complement our NanoString analysis of human paediatric HGGs, we also performed NanoString on PDGFA and PDGFB-driven tumours generated in Ntva;Cdkn2a−/− mice. Strong separation between the two tumour types was readily observed when unsupervised clustering on all 750 detected genes was performed (Fig. 4A). There were 15 statistically significant differentially expressed genes (Benjamini-Hochberg P < 0.05, fold change > 1) (Fig. 4B) and top differentially expressed genes were validated with qPCR (Supplementary Fig. 4A–D). PDGFB tumours possessed high expression of chemokines including Ccl11, Cxcl13, and Ccl12. PDGFB tumours also had significantly higher expression of inflammatory adhesion molecules Cd11b, Cd74, and Selplg (CD162), all of which play important roles in promoting and mediating leucocyte and macrophage trafficking. Genes involved in antigen processing and presentation were also highly expressed in PDGFB tumours including Ly86, Cd180, H2-K1, and H2-Eb1 (MHC-II). Conversely, PDGFA tumours had high expression of Cfh (complement factor H), which is an inhibitor of inflammation triggered by the complement system (Parente et al., 2017). Other highly expressed genes in PDGFA-driven tumours included Ifitm1, Rsad2, Cd36, and Il16, which are all important mediators of the innate immune response to foreign pathogens.
Figure 4.
Inflammatory microenvironment of PDGF-driven mouse tumours. (A) Similarity matrix performed on PDGFA (n = 5) and PDGFB (n = 6) tumour samples generated in Ntva;Cdkn2a−/− mice based on the expression of 750 genes detected using NanoString. (B) Unsupervised hierarchical clustering of PDGFA and PDGFB tumour samples based on pathway scores obtained from NanoString. (C) Volcano plot of differentially expressed genes detected using NanoString. (D) IBA1 staining on PDGFA and PDGFB tumours. Images magnified ×40 and ×80, scale bars = 100 μm and 20 μm, respectively. (E) Quantification of IBA1 staining. PDGFA n = 9, PDGFB n = 9. Student’s t-test, P = 0.0004.
Pathway analysis demonstrated exclusive clustering between PDGFA and PDGFB-driven tumours. PDGFB-driven tumours had demonstrably greater representation of genes involved in inflammatory processes including the complement pathway, leucocyte functions, CD molecules, macrophage functions, innate immune response, chemokines, and cytokines (Fig. 4C). These results mirror the two distinct subgroups observed in human paediatric HGG samples (Fig. 1F), with PDGFB-driven tumours closely matching the expression profiles of group 1 human paediatric HGGs (more inflammatory) and PDGFA-driven tumours closely matching group 2 human paediatric HGGs (less inflammatory). Cell cycle function and basic cell function pathways were not significantly differentially expressed between the two entities. Immunohistochemical staining for phospho-histone H3, qPCR for Ccnd2, and in vitro cell proliferation assays all confirmed these findings, demonstrating no major differences in proliferation between the two tumour types at end point (Supplementary Fig. 4E–H). These results suggest the differences in tumour malignancy may likely be derived from the tumour microenvironment and not necessarily tumour cell intrinsic factors. Although these PDGF ligands operate through the same pathways, it is clear PDGFB is a more potent inducer of the inflammatory response in the tumour microenvironment.
Next, to validate our NanoString findings, we performed IBA1 staining in FFPE tumour sections from PDGFA and PDGFB-driven tumours in Ntva;Cdkn2a−/− mice. There was a dramatic increase in the number of infiltrating TAMs observed in PDGFB tumours as shown by whole brain staining (Fig. 4D and E). TAMs were found throughout the entirety of both tumour types; however, PDGFB samples had twice as much tumour area covered by TAMs. Much like our findings in human paediatric HGGs, PDGFB, but not PDGFA, appears to be promoting a pro-inflammatory microenvironment and play a key role in TAM recruitment.
PDGFB-driven tumours have increased infiltration of peripheral monocytes
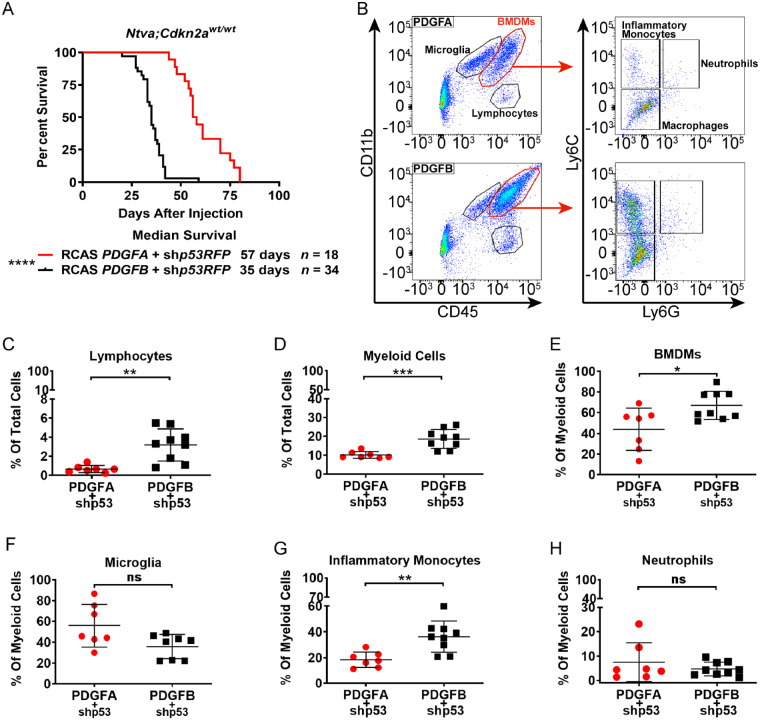
In adult glioblastoma and medulloblastoma, TAMs are composed of infiltrating BMDMs and brain-resident microglia; however, histological distinction between the two cannot be made because of a lack of distinct cell-type specific markers (Chen et al., 2019; Maximov et al., 2019). Since IBA1 stains both TAM populations, it is unclear what the identity is of the additional TAMs found in PDGFB-driven tumours. To address this, we induced tumours in Ntva;Cdkn2awt/wt mice with a co-injection of RCAS-shp53 along with either RCAS-PDGFA or RCAS-PDGFB. These mice are in a pure C57BL/6 background and are more suitable for myeloid cell lineage identification as compared to Ntva;Cdkn2a−/− mice which are in a mixed genetic background. As seen in Ntva;Cdkn2a−/− mice, PDGFB-driven tumours in the Ntva;Cdkn2awt/wt mice had a significantly decreased median survival (35 days) compared to PDGFA-driven tumours (57 days) (Fig. 5A).
Figure 5.
Flow cytometry of PDGF-driven mouse tumours. (A) Survival curves for Ntva;Cdkn2awt/wt mice with tumours induced by RCAS-PDGFA or RCAS-PDGFB with co-administration of RCAS-shP53. Log-rank Mantel-Cox test, P < 0.0001. (B) Representative flow cytometry dot plots of PDGFA (n = 7) and PDGFB-driven (n = 9) tumours generated in Ntva;Cdkn2awt/wt mice, gated for CD45, CD11b, Ly6C, and Ly6G. (C) %CD45hiCD11b− lymphocytes in the total tumour cell population. (D) %CD45+CD11b+ myeloid cells in the total tumour cell population, P = 0.0008. (E) %CD45hiCD11b+ BMDMs in the total myeloid cell population, P = 0.0171. (F) %CD45l°CD11b+ microglia in the total myeloid cell population. (G) %CD45hiCD11b+Ly6ChiLy6Glo inflammatory monocytes in the total BMDM population, P = 0.0030. (H) %CD45hiCD11b+Ly6ChiLy6Ghi neutrophils in the total BMDM population. Student’s t-test. ns = not significant.
Freshly dissociated tumour tissue stained for CD45, CD11b, Ly6C, and Ly6G demonstrated PDGFB tumours had a significant increase in the lymphocyte population (CD45+CD11b−) and in the total TAM population (CD45+CD11b+) (Fig. 5C and D). The increase in the total TAM population was not due to the number of microglia (CD45loCD11b+), but rather due to infiltrating BMDMs (CD45hiCD11b+). Further staining for Ly6C and Ly6G demonstrated the elevated BMDM population was from the number of inflammatory monocytes (Ly6ChiLy6G−) while there was no difference in neutrophils (Fig. 5E–H). These results indicate PDGFB/PDGFRβ signalling promotes the infiltration of circulating monocytes from the blood into the brain.
Inflammatory gene expression is upregulated in PDGFB-driven tumours
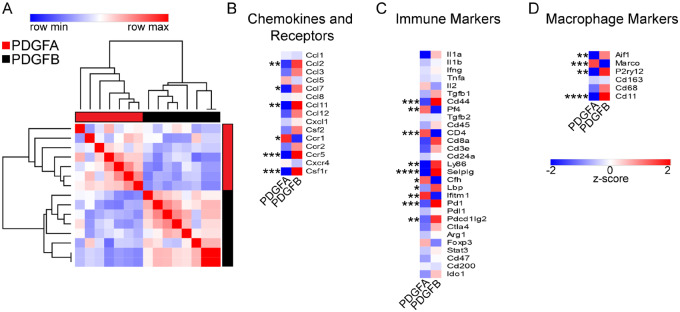
NanoString analysis demonstrated PDGFB-driven tumours are more inflammatory compared to PDGFA-driven tumours. To validate these findings and examine additional genes, we used a custom designed qPCR panel of 73 genes including chemokines and chemokine receptors, immune markers, macrophage markers, and cancer signalling markers. As found in our NanoString dataset, when clustering these tumours based on their expression profiles, PDGFA-driven tumours distinctly cluster apart from PDGFB-driven tumours (Fig. 6A). As expected, PDGFB-driven tumours had greater expression of Aif1 (IBA1) and Itgam1 (Cd11b). Further analysis revealed PDGFB tumours had greater expression of chemokines including Ccl2, Ccl3, Ccl7, and Ccl11. Ccr5 and Csf1r were also higher in PDGFB samples (Fig. 6B). A cytokine array and protein ELISA also confirmed results of increased chemokine expression in PDGFB tumours (Supplementary Fig. 5). Immunosuppressive genes including Pd1 (PD1), Pdcd1lg2 (PD-L2), and Ctla4 were highly expressed in PDGFB tumours, suggesting these tumours have a greater immunosuppressive phenotype compared to PDGFA tumours (Barber et al., 2006). There were only a few genes more highly expressed in PDGFA-driven tumours not previously examined in the NanoString analysis including Marco, a scavenger receptor, and chemokine receptor Ccr1 (Fig. 6C and D).
Figure 6.
Quantitative PCR molecular characterization of PDGF-driven mouse tumours. (A) Similarity matrix of PDGFA (n = 7) and PDGFB-driven (n = 7) tumours generated in Ntva;Cdkn2a−/− mice. Samples are clustered by their expression of 72 genes detected by qPCR. (B) Averaged z-scores for PDGFA and PDGFB-driven tumour samples based on their expression of chemokines and chemokine receptor genes. (C) Averaged z-scores for immune-related genes. (D) Averaged z-scores for macrophage related genes. Student’s t-test.
PDGFB stimulates chemokine production in bone marrow-derived monocytes
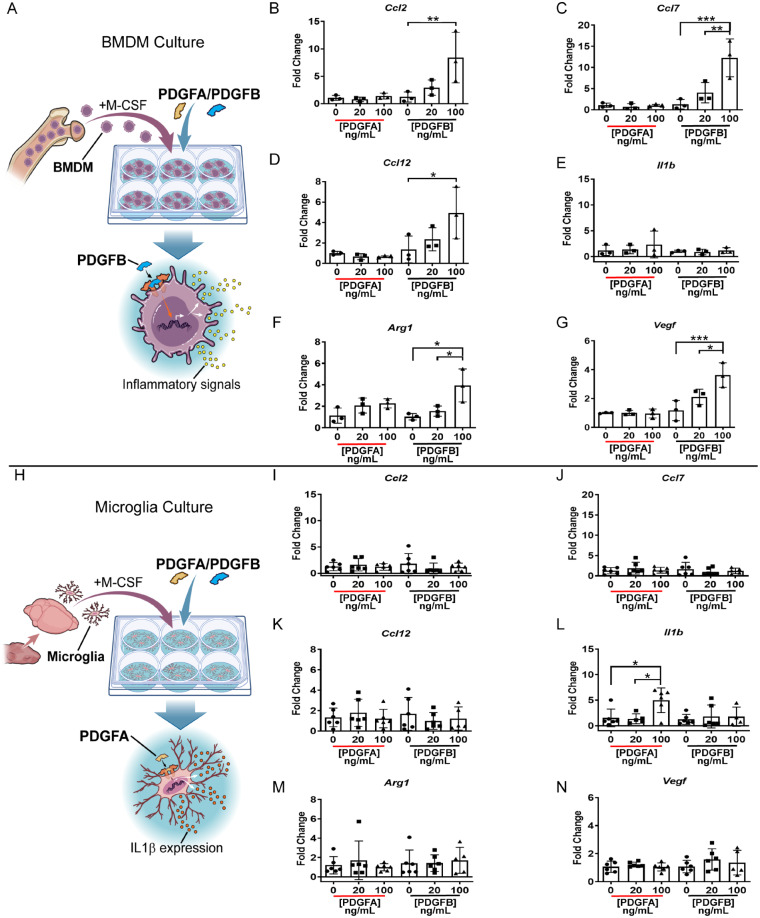
The function of the additional macrophages inside PDGFB-driven tumours remains unclear. To address this, whole bone marrow was isolated from naïve CL57/B6 adult mice and cultured in the presence of M-CSF to drive the cells towards a macrophage phenotype. After 7 days, cells were stimulated with increasing doses of either recombinant PDGFA or PDGFB protein (0–100 ng/ml) for 6 h (Fig. 7A). After stimulation, RNA was collected for qPCR. Quantitative PCR for chemokines Ccl2, Ccl7, Ccl12, and Vegf demonstrated in response to increasing doses of PDGFB but not PDGFA, BMDMs upregulate their expression of these genes (Fig. 7B–G). Interestingly, when the same experimental conditions and stimulations were performed using cultured microglia these responses were not observed (Fig. 7H–N). Additionally, in response to PDGFB stimulation, BMDMs also produced higher levels of Arg1, an enzyme which suppresses T-cell function in the tumour microenvironment (Munder et al., 2006). The only gene stimulated in microglia, Il1b, was in response to PDGFA stimulation and was not significant in stimulated BMDMs. These results support previous findings that PDGFB is a potent inducer of chemokines and growth factors in cells found in the tumour microenvironment, including CCL2 in astrocytes (Bethel-Brown et al., 2012), CCL7 in perivascular precursor cells (Au et al., 2009), and CCL12, which has not been previously implicated in glioma biology but shares the same receptor as CCL2 and CCL7 (CCR2) (Drevets et al., 2010).
Figure 7.
In vitro PDGF stimulation of BMDMs and microglia. (A) Schematic depicting the isolation and culture of BMDMs from naïve CL57BL/6 mouse bone marrow. BMDMs were stimulated with either PDGFA or PDGFB for 6 h prior to RNA isolation. (B–G) Quantitative PCR plots on BMDM samples stimulated with either PDGFA or PDGFB at 0 ng/ml, 20 ng/ml, or 100 ng/ml. n = 3 independent replicates per concentration. (H) Schematic depicting the isolation and culture of microglia from naïve newborn CL57BL/6 pup brains. Microglia were cultured with M-CSF and stimulated with either PDGFA or PDGFB for 6 h prior to RNA isolation. (I–N) Quantitative PCR plots on stimulated microglia. n = 5 independent replicates per concentration. One-way ANOVA multiple comparisons.
Chemokine knockout reduces TAM infiltration and extends survival
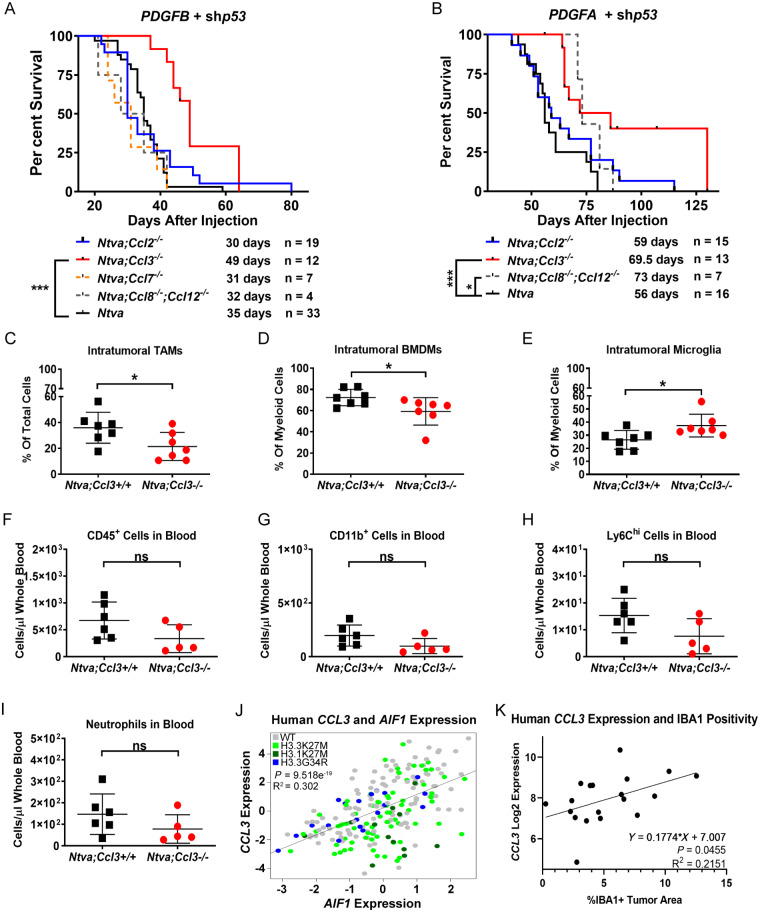
To address the question of whether reducing TAM infiltration can extend survival we used Ntva+ chemokine knockout mice and induced tumours with RCAS-shp53 and RCAS-PDGFA or RCAS-PDGFB. The chemokines we focused on were CCL2, CCL3, CCL7, and CCL8/12, as all of these were found to be highly expressed in murine PDGFB-driven tumours. Additionally, in vitro stimulation of BMDMs demonstrated PDGFB induces the expression of these chemokines. The role of these chemokines in leucocyte trafficking is well documented (Huang et al., 2001; Charo and Ransohoff, 2006; Tsou et al., 2007; Mitchell et al., 2015). Our lab and others have demonstrated the importance of CCL2 and CCL7 in attracting monocytes and other myeloid cells into the tumour microenvironment of adult glioblastoma (Okada et al., 2009; Chang et al., 2016; Chen et al., 2017; Takenaka et al., 2019). Unlike in adult glioblastoma, when PDGFB is the oncogenic driver in paediatric HGG, there is no survival benefit in Ccl2 knockout mice (Fig. 8A). Knockout of Ccl7 and Ccl8/12 also did not produce any survival benefit. The only chemokine that produced an increase in survival was Ccl3. These mice had a median survival of 49 days compared to 35 days in wild-type mice. Unlike in PDGFB-driven tumours, there was an extension in survival in Ccl8/12 knockout mice as well as Ccl3 knockout mice when tumours were driven by PDGFA (Fig. 8B). The mice with the greatest survival benefit was Ccl8/12 double knockout mice with a median survival of 73 days. Interestingly, homozygous deletion of Ccl2 did not result in any survival difference from wild-type tumours driven by PDGFA.
Figure 8.
Chemokine knockout extends survival. (A) Survival curves of RCAS-PDGFB + RCAS-shP53 driven tumours in Ntva;Ccl2−/−, Ntva;Ccl3−/− (P = 0.0004), Ntva;Ccl7−/−, Ntva;Ccl8/12−/−, and Ntva mice. (B) Survival curves of RCAS-PDGFA + RCAS-shP53 driven tumours in Ntva;Ccl2−/−, Ntva;Ccl3−/− (P = 0.0006), Ntva;Ccl8/12−/− (P = 0.0102), and Ntva mice. Log-rank Mantel-Cox test. (C) %CD45+CD11b+ TAMs in the total tumour cell population in Ntva;Ccl3+/+ mice (n = 7) or Ntva;Ccl3−/− mice (n = 7) driven by RCAS-PDGFB and RCAS-shP53. P = 0.0354. (D) %CD45hiCD11b+ BMDMs in the total myeloid cell population. P = 0.0409. (E) %CD45l°CD11b+ microglia in the total myeloid cell population. P = 0.0248. (F) Number of CD45+ cells in the blood of Ntva;Ccl3+/+ mice (n = 6) or Ntva;Ccl3−/− mice (n = 5). (G) CD11b+ cells in the blood. (H) Ly6chi cells in the blood. (I) Neutrophils in the blood. (J) CCL3 expression correlated with AIF1 in human paediatric HGG RNA sequencing data. n = 220. (K) CCL3 expression obtained from NanoString in human paediatric HGGs correlated with IBA1 positivity in matched samples. n = 19.
Since Ccl3 was the only chemokine to provide a survival benefit upon genetic deletion in PDGFB-driven tumours, IBA1 staining and flow cytometry was performed to determine if TAM infiltration was also inhibited. Staining for TAMs did not reveal any significant differences between wild-type and Ccl3 knockout mice (Supplementary Fig. 6A and B). However, through flow cytometry CD45 and CD11b staining indicated there was a significant reduction in the total TAM population (Fig. 8C). This reduction was due to a significant decrease in the CD45hiCD11b+ BMDM population (Fig. 8D). The decrease in BMDMs subsequently caused a significant increase in the microglial population within the total TAM population (Fig. 8E). The reduction in intra-tumoral BMDMs was not a result of decreased egress of myeloid cells from the bone marrow, as there were no significant differences observed in Ccl3 wild-type versus knockout mice in the blood (Fig. 8F–I). There were also no differences observed in PDGFA-driven tumours; however, Ccl3 knockout mice had significantly fewer numbers of phospho-histone H3-positive proliferating cells compared to wild-type tumours (Supplementary Fig. 6C–M). Lastly, in PDGFB-driven tumours, there were no significant differences observed in the number of intra-tumoral CD4 or CD8 T cells, PD1+, FOXP3+, or Granzyme B+ T cells, or natural killer (NK) cells in Ccl3 knockout mice (Supplementary Fig. 7A–D).
RNA sequencing data of histone wild-type human paediatric HGGs for chemokines revealed CCL3 and CCL4 cluster together, apart from all other chemokines (Supplementary Fig. 8A). Further, CCL3 mRNA expression strongly correlated with AIF1 (gene encoding IBA1) expression in both histone wild-type and histone mutant samples (Fig. 8J). CCL3 expression obtained from our NanoString dataset positively correlated with IBA1+ TAM infiltration in human paediatric HGGs, validating the RNA sequencing data (Fig. 8K). Additionally, CCL3 strongly correlated with PTPRC (CD45), ITGAM (CD11b), PDGFRB, PDL1, PD1, and CD3D in our dataset (Supplementary Fig. 8B–G). Interestingly, when murine BMDMs or microglia were stimulated with PDGFB, Ccl3 expression was not induced, indicating TAMs are not the source of Ccl3 and it is likely tumour or stromal cells producing the chemokine and attracting TAMs to the microenvironment (Supplementary Fig. 8H–I). Together, these results implicate PDGF-induced CCL3 as a key mediator of TAM infiltration in paediatric HGGs.
Discussion
In our study, we sought to define the inflammatory tumour microenvironment of paediatric HGGs and determine the factors promoting TAM infiltration. Corroborating a previous study on diffuse intrinsic pontine gliomas, we demonstrate paediatric HGGs, irrespective of their tumour location, have minimal T-cell infiltration (Lin et al., 2018). Despite low T-cell infiltration in our human dataset, we observed inflammatory differences between hemispheric paediatric HGGs and DMGs at the RNA level in both humans and mice. DMGs were found to express higher levels of pro-inflammatory transcripts compared to hemispheric paediatric HGGs. We also observed similar differences in the tumour microenvironment of murine tumours driven by PDGFB versus those driven by PDGFA. PDGFB-driven tumours were found to express transcripts relating to macrophage functions, inflammation, chemokines and receptors, and the innate immune response at higher levels than PDGFA-driven tumours.
PDGFB protein can be highly expressed despite low levels of transcript present (Brennan et al., 2009), therefore we performed a histological analysis of human paediatric HGG samples to identify relationships that would not be found using RNA sequencing databases. Although our human paediatric HGG samples were cold for T cells, akin to our findings in adult HGGs, we observed high infiltration of TAMs as demonstrated by IBA1 staining (Kaffes et al., 2019). We identified correlations between IBA1 and CD31, PDGFRβ, and PDGFB. Samples with high PDGFB staining had high levels of IBA1+ TAMs, independent of H3 status. Supporting these novel findings was the observation that PDGFB-driven murine HGGs had significantly higher levels of IBA1+ TAMs compared to PDGFA-driven tumours. Although our study does not directly reveal the identity of infiltrating TAMs in human samples, we demonstrate in our immunocompetent mouse models that peripheral BMDMs, not microglia, are the predominate TAM subpopulation in these tumours. Further molecular analysis of the murine tumours revealed PDGFB-driven tumours had greater expression of chemokines, chemokine receptors, immunosuppressive markers, and macrophage markers compared to PDGFA-driven tumours. Together with our human data, this raises the question of whether the molecular identity of the tumour or the tumour location itself is more important for determining the composition of the immune infiltrate and the factors governing these inflammatory processes.
To examine the properties of TAMs and their roles in the tumour microenvironment, we stimulated microglia and BMDMs with PDGFA or PDGFB and observed striking differences between the two cell types and between the two stimulants. When stimulated with PDGFB, BMDMs upregulated their expression of chemokines including Ccl2, Ccl7, and Ccl12. BMDMs also upregulated their expression of Vegf and Arg1, indicating these cells not only respond to tumour-derived signals by producing chemoattractant molecules, they also produce tumour microenvironment-modulating signals. These responses were not observed during PDGFA stimulation, nor were they observed in cultured microglia, suggesting PDGFB selectively induces an inflammatory response in infiltrating BMDMs. This is important because BMDMs are the predominant infiltrating immune cell-type within these tumours and possess clear differences in behaviour compared to microglia. These results indicate therapeutic strategies treating the TAM population as a homogenous entity need to be modified to account for cell type-specific differences, which we have previously shown in both humans and murine adult GBM (Chen et al., 2017, 2019, 2020; Herting et al., 2019).
Because of the significance of chemokine signalling in recruiting various immune cell types to tissues, we hypothesized genetic ablation of chemokines may result in reduced intra-tumoral TAM infiltration. As we observed PDGFB-driven murine paediatric HGGs have an increase in inflammatory monocytes, greater expression of chemokines, as well as a decreased median survival, we further postulated genetic deletion of chemokines would result in increased survival. Interestingly, the only chemokine to confer a survival benefit in PDGFB-driven tumours when knocked out was Ccl3. Flow cytometry analysis indicated this survival benefit was accompanied by a decrease in the BMDM population while having no effect on various T-cell populations. CCL3 binds to CCR1 and CCR4 to promote an inflammatory response characterized by neutrophil, monocyte, T cell, B cell, and dendritic cell chemoattraction (Castellino et al., 2006; Mitchell et al., 2015). CCL3 has been implicated in multiple sclerosis, a chronic CNS inflammatory disease characterized by demyelination of axons and accumulation of plaques containing microglia, macrophages, and other leucocytes (Simpson et al., 1998; Vogel et al., 2013). Treatment of mice with experimental allergic encephalomyelitis using antibodies against CCL3 prevented the infiltration of peripheral monocytes (Chui and Dorovini-Zis, 2010). Our results implicate CCL3 as an important driver of TAM recruitment and accumulation in paediatric HGGs and should be further investigated for its potential as a therapeutic target.
Unlike human tumours, which are variable in terms of treatment received, tumour location, genetic driver mutations, and tumour cell of origin, our murine studies are highly controlled for all these variables. Any differences observed between our mouse models is based purely on the oncogenic driver, PDGFA or PDGFB, allowing us to mechanistically dissect the inflammatory tumour microenvironment with greater resolution compared to solely relying on human sequencing datasets. Our results indicate PDGFB is a strong inducer of the inflammatory response in murine paediatric HGGs and establishes a link between TAM infiltration and PDGF signalling in human paediatric HGGs. Further, we establish a pro-tumour role for TAMs in paediatric HGG and provide supporting data for the notion paediatric HGGs are immunologically cold with respect to T cells. These studies should be used in future attempts to further characterize the paediatric HGG tumour microenvironment. More importantly, as the use of molecular and cellular targeted therapies become more popular, our findings will aid in the design of more effective therapeutic approaches which are desperately needed for treating paediatric HGGs.
Supplementary Material
Acknowledgements
Research reported in this publication was supported in part by the Emory Integrated Genomics Core and Cancer Tissue and Pathology Shared Resource of Winship Cancer Institute of Emory University and NIH/NCI under award number P30CA138292. We also thank the Emory Children’s Flow Cytometry Core and the Dean’s Flow Cytometry Core at Mount Sinai. We are grateful to Dr Alice Kamphorst for her excellent advice on flow cytometry; and to Dr Ruben Fernandez-Rodriguez for his technical support on immunohistochemistry. The authors thank Dave Schumick for professional illustrations.
Funding
This study was funded by National Institutes of Health 1F31CA232531-01 to J.R., R21 NS106554-01, R01NS10086 to D.H., and a CURE Childhood Cancer Award 642734 to Z.C.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- BMDM =
bone marrow-derived macrophage
- DMG =
diffuse midline glioma
- HGG =
high-grade glioma
- TAM =
tumour associated macrophage
References
- Au P, Tam J, Duda DG, Lin PC, Munn LL, Fukumura D, et al. Paradoxical effects of PDGF-BB overexpression in endothelial cells on engineered blood vessels in vivo. Am J Pathol 2009; 175: 294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 2006; 439: 682–7. [DOI] [PubMed] [Google Scholar]
- Bethel-Brown C, Yao H, Hu G, Buch S.. Platelet-derived growth factor (PDGF)-BB-mediated induction of monocyte chemoattractant protein 1 in human astrocytes: implications for HIV-associated neuroinflammation. J Neuroinflammation 2012; 9: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One 2009; 4: e7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzolari F, Malatesta P.. Recent insights into PDGF-induced gliomagenesis. Brain Pathol 2010; 20: 527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN.. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature 2006; 440: 890–5. [DOI] [PubMed] [Google Scholar]
- Chang AL, Miska J, Wainwright DA, Dey M, Rivetta CV, Yu D, et al. ccl2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res 2016; 76: 5671–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charo IF, Ransohoff RM.. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med 2006; 354: 610–21. [DOI] [PubMed] [Google Scholar]
- Chen Z, Feng X, Herting CJ, Garcia VA, Nie K, Pong WW, et al. Cellular and molecular identity of tumor-associated macrophages in glioblastoma. Cancer Res 2017; 77: 2266–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Herting CJ, Ross JL, Gabanic B, Puigdelloses Vallcorba M, Szulzewsky F, et al. Genetic driver mutations introduced in identical cell-of-origin in murine glioblastoma reveal distinct immune landscapes but similar response to checkpoint blockade. Glia 2020; 10: 2148–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Ross JL, Hambardzumyan D.. Intravital 2-photon imaging reveals distinct morphology and infiltrative properties of glioblastoma-associated macrophages. Proc Natl Acad Sci USA 2019; 116: 14254–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi AS, Tarapore RS, Hall MD, Shonka N, Gardner S, Umemura Y, et al. Pediatric and adult H3 K27M-mutant diffuse midline glioma treated with the selective DRD2 antagonist ONC201. J Neurooncol 2019; 145: 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui R, Dorovini-Zis K.. Regulation of CCL2 and CCL3 expression in human brain endothelial cells by cytokines and lipopolysaccharide. J Neuroinflammation 2010; 7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo K, Dudeck A, Hasenberg M, Nye E, van Rooijen N, Hartmann K, et al. Mast cell and macrophage chemokines CXCL1/CXCL2 control the early stage of neutrophil recruitment during tissue inflammation. Blood 2013; 121: 4930–7. [DOI] [PubMed] [Google Scholar]
- Drevets DA, Dillon MJ, Schawang JE, Stoner JA, Leenen PJ.. IFN-gamma triggers CCR2-independent monocyte entry into the brain during systemic infection by virulent Listeria monocytogenes. Brain Behav Immun 2010; 24: 919–29. [DOI] [PubMed] [Google Scholar]
- Griesinger AM, Donson AM, Foreman NK.. Immunotherapeutic implications of the immunophenotype of pediatric brain tumors. Oncoimmunology 2014; 3: e27256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann DH, Kettenmann H.. Microglia/brain macrophages as central drivers of brain tumor pathology. Neuron 2019; 104: 442–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MD, Odia Y, Allen JE, Tarapore R, Khatib Z, Niazi TN, et al. First clinical experience with DRD2/3 antagonist ONC201 in H3 K27M-mutant pediatric diffuse intrinsic pontine glioma: a case report. J Neurosurg Pediatr 2019; 23: 719–25. [DOI] [PubMed] [Google Scholar]
- Hambardzumyan D, Gutmann DH, Kettenmann H.. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci 2016; 19: 20–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Medley SC, Hu T, Hinsdale ME, Lupu F, Virmani R, et al. PDGFRbeta signalling regulates local inflammation and synergizes with hypercholesterolaemia to promote atherosclerosis. Nat Commun 2015; 6: 7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin CH, Lennartsson J, Westermark B.. Involvement of platelet-derived growth factor ligands and receptors in tumorigenesis. J Intern Med 2018; 283: 16–44. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Westermark B.. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev 1999; 79: 1283–316. [DOI] [PubMed] [Google Scholar]
- Herting CJ, Chen Z, Maximov V, Duffy A, Szulzewsky F, Shayakhmetov DM, et al. Tumour-associated macrophage-derived interleukin-1 mediates glioblastoma-associated cerebral oedema. Brain 2019; 12: 3834–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herting CJ, Chen Z, Pitter KL, Szulzewsky F, Kaffes I, Kaluzova M, et al. Genetic driver mutations define the expression signature and microenvironmental composition of high-grade gliomas. Glia 2017; 65: 1914–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DR, Wang J, Kivisakk P, Rollins BJ, Ransohoff RM.. Absence of monocyte chemoattractant protein 1 in mice leads to decreased local macrophage recruitment and antigen-specific T helper cell type 1 immune response in experimental autoimmune encephalomyelitis. J Exp Med 2001; 193: 713–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson WH, Gensheimer J, Hashimoto M, Wieland A, Valanparambil RM, Li P, et al. Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1(+) stem-like CD8(+) T cells during chronic infection. Immunity 2019; 51: 1043–58 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Baker SJ.. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer 2014; 14: 651–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffes I, Szulzewsky F, Chen Z, Herting CJ, Gabanic B, Velazquez Vega JE, et al. Human Mesenchymal glioblastomas are characterized by an increased immune cell presence compared to Proneural and Classical tumors. Oncoimmunology 2019; 8: e1655360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky JL 3rd, Panosyan EH, Plant A, Davidson T, Yong WH, Prins RM, et al. Autologous tumor lysate-pulsed dendritic cell immunotherapy for pediatric patients with newly diagnosed or recurrent high-grade gliomas. Anticancer Res 2013; 33: 2047–56. [PMC free article] [PubMed] [Google Scholar]
- Li J, Byrne KT, Yan F, Yamazoe T, Chen Z, Baslan T, et al. Tumor cell-intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity 2018; 49: 178–93 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman NAP, DeGolier K, Kovar HM, Davis A, Hoglund V, Stevens J, et al. Characterization of the immune microenvironment of diffuse intrinsic pontine glioma: implications for development of immunotherapy. Neuro Oncol 2019; 21: 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin GL, Nagaraja S, Filbin MG, Suva ML, Vogel H, Monje M.. Non-inflammatory tumor microenvironment of diffuse intrinsic pontine glioma. Acta Neuropathol Commun 2018; 6: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016; 131: 803–20. [DOI] [PubMed] [Google Scholar]
- Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, et al. Integrated Molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell 2017; 32: 520–37. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximov V, Chen Z, Wei Y, Robinson MH, Herting CJ, Shanmugam NS, et al. Tumour-associated macrophages exhibit anti-tumoural properties in Sonic Hedgehog medulloblastoma. Nat Commun 2019; 10: 2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Batich KA, Gunn MD, Huang MN, Sanchez-Perez L, Nair SK, et al. Tetanus toxoid and CCL3 improve dendritic cell vaccines in mice and glioblastoma patients. Nature 2015; 519: 366–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munder M, Schneider H, Luckner C, Giese T, Langhans CD, Fuentes JM, et al. Suppression of T-cell functions by human granulocyte arginase. Blood 2006; 108: 1627–34. [DOI] [PubMed] [Google Scholar]
- Nordby Y, Richardsen E, Rakaee M, Ness N, Donnem T, Patel HR, et al. High expression of PDGFR-beta in prostate cancer stroma is independently associated with clinical and biochemical prostate cancer recurrence. Sci Rep 2017; 7: 43378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Saio M, Kito Y, Ohe N, Yano H, Yoshimura S, et al. Tumor-associated macrophage/microglia infiltration in human gliomas is correlated with MCP-3, but not MCP-1. Int J Oncol 2009; 34: 1621–7. [DOI] [PubMed] [Google Scholar]
- Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro Oncol 2015; 17: iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007-2011. Neuro Oncol 2014; 16: iv1–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K, et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell 2014; 26: 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos N, Lennartsson J.. The PDGF/PDGFR pathway as a drug target. Mol Aspects Med 2018; 62: 75–88. [DOI] [PubMed] [Google Scholar]
- Parente R, Clark SJ, Inforzato A, Day AJ.. Complement factor H in host defense and immune evasion. Cell Mol Life Sci 2017; 74: 1605–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant AS, Koyama S, Sinai C, Solomon IH, Griffin GK, Ligon KL, et al. Immunophenotyping of pediatric brain tumors: correlating immune infiltrate with histology, mutational load, and survival and assessing clonal T cell response. J Neurooncol 2018; 137: 269–78. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9: 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinke C, Giricz O, Li W, Shastri A, Gordon S, Barreyro L, et al. IL8-CXCR2 pathway inhibition as a therapeutic strategy against MDS and AML stem cells. Blood 2015; 125: 3144–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JE, Newcombe J, Cuzner ML, Woodroofe MN.. Expression of monocyte chemoattractant protein-1 and other beta-chemokines by resident glia and inflammatory cells in multiple sclerosis lesions. J Neuroimmunol 1998; 84: 238–49. [DOI] [PubMed] [Google Scholar]
- Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 2002; 1: 203–9. [DOI] [PubMed] [Google Scholar]
- Souweidane MM, Kramer K, Pandit-Taskar N, Zhou Z, Haque S, Zanzonico P, et al. Convection-enhanced delivery for diffuse intrinsic pontine glioma: a single-centre, dose-escalation, phase 1 trial. Lancet Oncol 2018; 19: 1040–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka MC, Gabriely G, Rothhammer V, Mascanfroni ID, Wheeler MA, Chao CC, et al. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat Neurosci 2019; 22: 729–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejada S, Alonso M, Patino A, Fueyo J, Gomez-Manzano C, Diez-Valle R.. Phase I trial of DNX-2401 for diffuse intrinsic pontine glioma newly diagnosed in pediatric patients. Neurosurgery 2018; 83: 1050–6. [DOI] [PubMed] [Google Scholar]
- Tsou CL, Peters W, Si Y, Slaymaker S, Aslanian AM, Weisberg SP, et al. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J Clin Invest 2007; 117: 902–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinci M, Burford A, Molinari V, Kessler K, Popov S, Clarke M, et al. Functional diversity and cooperativity between subclonal populations of pediatric glioblastoma and diffuse intrinsic pontine glioma cells. Nat Med 2018; 24: 1204–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel DY, Vereyken EJ, Glim JE, Heijnen PD, Moeton M, van der Valk P, et al. Macrophages in inflammatory multiple sclerosis lesions have an intermediate activation status. J Neuroinflammation 2013; 10: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark B, Heldin CH, Nister M.. Platelet-derived growth factor in human glioma. Glia 1995; 15: 257–63. [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 2007; 27: 670–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used for this manuscript are available upon reasonable request.