Abstract
Genome-scale mutagenesis screens for genes essential for apicomplexan parasite survival have been completed in three species: Plasmodium falciparum, the major human malaria parasite; P. berghei, a model rodent malaria parasite, and the more distantly related Toxoplasma gondii, the causative agent of toxoplasmosis. These three species share 2606 single-copy orthologs, 1500 of which have essentiality data in all three screens. In this review, we explore the overlap between these datasets to define the core essential genes of the phylum Apicomplexa. We further discuss the implications of these groundbreaking studies for understanding apicomplexan parasite biology, and we identify promising areas of focus for developing new pan-apicomplexan parasite interventions.
Keywords: malaria, Plasmodium falciparum, Plasmodium berghei, Toxoplasma gondii, Apicomplexa, essential genes
Genome-wide essentiality screens: A game-changer for studying apicomplexan parasite biology
Apicomplexan parasites are the causative agents of some of the world’s most devastating infectious diseases, including malaria (caused by Plasmodium), toxoplasmosis (Toxoplasma gondii) and numerous other deadly diseases of humans and animals. Though their importance to human health have made these parasites of sustained interest to the research community, many aspects of their biology remain largely mysterious compared to that of more experimentally tractable model organisms. Available tools for genetic manipulation that have worked well in model organisms to functionally characterize genes are typically much more limited in their application to apicomplexan parasites for a number of reasons (summarized in Table 1), leaving ~35% of malaria-parasite genes and ~50% of Toxoplasma genes with no functional annotation, even though sequenced genomes in some state of assembly have been available to explore for well over a decade (https://www.veupathdb.org; [1–3]). Overcoming these significant hurdles to explore parasite gene-function is of utmost importance, as a better understanding of parasite biology, and which processes and pathways are more essential for parasite survival, is needed to prioritize targets of more efficacious interventions, particularly in the face of rising parasite resistance to front-line antimalarial drugs.
Table 1.
General comparisons across apicomplexans.
| Characteristic | P. falciparum(3D7) | P. berghei (ANKA) | Toxoplasma gondii (GT1) |
|---|---|---|---|
| Primary host | mosquito | mosquito | cat |
| Secondary host | humans | mice | most mammals |
| infected cell type | red blood cell | red blood cell | epithelial cell |
| Genome size/protein-coding gene number | 23.33 MB/5460 genes | 18.78 MB/5067 genes | 63.95 MB/8460 genes |
| Advantages as experimental system | the deadliest human malaria parasite | relative genetic tractability; in vivo model for human malaria infection | most genetic tractability; the “model” apicomplexan, and important pathogen in its own right |
| Disadvantages as experimental system | difficult to genetically manipulate; very low transfection efficiency; high-throughput targeted mutagenesis unlikely due to repetitive genome composition | not the human malaria parasite; no in vitro system | far-removed from malaria parasites; limits to how much can be extrapolated to Plasmodium |
| Mutagenesis method | random transposon-mediated insertional mutagenesis [4] | targeted mutagenesis [5] | targeted mutagenesis [6] |
| % of genes assigned essentiality score | 87% | 50% | ~95% |
| Essentiality scoring metric | Mutagenesis Index Score (MIS) | Relative Growth Rate (RGR) | Phenotype score |
| studied stage | in vitro blood stage | in vivo blood stage | in vitro lytic cycle |
| Total shared genes* | |||
| Pf | -- | 4631 | 2928 |
| Pb | 4581 | -- | 2861 |
| Tg | 3044 | 3028 | -- |
Numbers of shared genes between species vary by direction of comparison (reflecting different numbers of within-species paralogs in ortholog groups).
Three studies in recent years have vastly accelerated gene-functional characterization by defining the essential genes of three related parasites: Plasmodium falciparum, the most deadly human malaria parasite [4]; Plasmodium berghei, a robust rodent malaria parasite that serves as an important model for human malaria [5], and Toxoplasma gondii, a distantly-related parasite responsible for toxoplasmosis whose relative experimental tractability has long led it to be known as the model apicomplexan [6]. Similar to other whole-genome essentiality screens in human cells [7–9], essential genes of these parasitic protists are defined by the inability to disrupt protein-coding sequences either by targeted or random mutagenesis methods (see Glossary). The small compact apicomplexan genomes are notable for their extensive signatures of horizontal gene transfer, especially as a result of the secondary endosymbiotic event giving rise to the plant-like apicoplast organelle, and allowing acquisition of new metabolic pathways [10–12]. The complete lack of functional characterization for significant proportions of apicomplexan genes has posed a formidable challenge to unraveling parasite biology; even the remaining genes with annotation deduced by orthology to model organisms largely have sparse functional annotation. Prior to these genome-wide studies, the numbers of genes confirmed to be essential to parasite survival, cumulatively over decades of research and laborious experimentation, numbered only in the 100s [13]. With the release of these genome-scale functional studies, researchers now have a clearer picture than ever before about what makes these parasites tick—critical knowledge of parasite biology to possibly weaponize against these diseases via improved interventions (Box 1).
Box 1. What biological properties of parasite genes make them good targets for intervention?
Drug targets
Good parasite drug targets should be essential to parasite survival, as perturbing their function should kill the parasite. Targets should be selective in that they have no or limited similarity to human genes to minimize the chance of harm to the host. Promising drug targets will also ideally operate in druggable pathways outside of those already targeted by frontline antimalarials to engineer multi-faceted therapies against which parasites will have difficulty evolving resistance [78].
Vaccine targets
Similar considerations for vaccine development in the post-essential-genome era have recently been discussed elsewhere [79] and thus will not be a focus of this review. We will mention only that the purpose of vaccines is to expose the immune system to a threat and prompt it to make inhibitory antibodies or effector immune cells that can be employed in the event of infection. In contrast to the comparatively recent selective pressures drugs have imposed—malaria parasites have co-evolved over millennia to evade the hominid immune system [27], and many potential epitope targets mislead or restrict broadly neutralizing immune inhibition, complicating identification of what is truly a conserved vaccine.
In this review, we compare the findings of these three groundbreaking studies to identify shared, essential genes across this diverse phylum. We further note the many shared essential genes that have no identified orthologs in human, making them parasite-specific potential therapeutic targets. Many of these essential genes of unknown function localize to the apicoplast and mitochondria—both of which have been repeatedly demonstrated to be “druggable”—underscoring the roles of these two organelles as rich hubs of essential parasite-specific biology.
Diverse parasites, one phylum
Malaria remains a leading global cause of death and severe disease even though rates have fallen dramatically in the past decade [14]. Plasmodium falciparum in particular demonstrates an unforeseen intransigence to being eliminated, as evident by its recent rebound; the tide of progress against the disease has plateaued at just over 200 million cases per year, with no significant reduction in the global malaria burden since 2015 [14]. Emerging drug-resistance in South-East Asia [15, 16] once again demonstrates this parasite’s ability to evolve resistance to any front-line antimalarial drug, even when given in combination therapies [17]. This recurring trend of resistance emphasizes the need for a better understanding of parasite biology to inform use of existing control measures to hold ground against a further malaria resurgence whilst new interventions are developed.
Targeted, gene-by-gene functional studies, many prior to availability of full parasite genome sequences and despite extreme limitations in P. falciparum in particular, formed the basis for mechanistic understanding of many critical parasite processes (reviewed in [18]). The advent of whole-genome sequencing technologies and publication of the first apicomplexan-parasite genomes enabled larger-scale characterization than ever before, particularly through comparative genomics, further driving the understanding that these parasites are in many ways in a league of their own biologically, far-removed from model-organisms, governed by different rules shaped by their parasitic lifestyles (for example, [19–23]). However, even with the tremendous advances in understanding the biology of this parasite through targeted gene characterization of important phenotypes (e.g., drug resistance), functional analysis of poorly characterized genes remains a significant challenge, especially for the many hypothetical genes and genes of unknown function. Our newfound ability to characterize targets based on their essentiality amongst diverse apicomplexan species will help prioritize targets and pathways that are most sensitive to functional disruption. In addition, mutant-parasite libraries can help define drug mechanisms of action by chemogenomic profiling of mutants [24, 25] and identifying underlying transcriptional changes associated with altered drug sensitivity [26].
Plasmodium berghei, the most tractable of the rodent models for malaria for experimental genetics is ~15 – 35 million years diverged from P. falciparum [27–29]. The genetic tool set developed by PlasmoGEM for studying gene function is more developed than that for P. falciparum [5, 30, 31]. The proportion of essential genes is surprisingly similar in both species, even though essentiality by in vivo infection for P. berghei would be presumably more stringent than the in vitro ideal culture conditions for P. falciparum. The overlap of orthologs also reflects the general level of homology between these species and is consistent with the accepted value of the rodent malaria model, as demonstrated by discoveries in malaria-parasite core biological functions originating from studies in P. berghei, then subsequently replicated with much more effort in P. falciparum (e.g., ApiAP2G [32, 33]).
Toxoplasmosis is a widespread zoonotic disease of world-wide social and economic importance, an opportunistic pathogen which can infect and destroy any nucleated cell, with dire consequences primarily for the immuno-compromised and unborn [34]. Widely viewed as the “model” apicomplexan parasite because of the sophistication of available genetic tools and ease of genetic manipulation, Toxoplasma gondii often serves as an experimental system for malaria parasites [35], even though T. gondii diverged ~500 – 900 million years from Plasmodium spp. [36]. While this considerable divergence in time is at a similar scale as ancestral chordates to modern humans [37], T. gondii shares core biological properties with Plasmodium spp.; both are obligate-intracellular parasites that undergo asexual replication within human cells and have common host-cell invasion, replication and egress processes (Table 1). Divergence does place limits on what can be extrapolated from this model to Plasmodium, especially in dissecting more recent evolutionary adaptations of malaria parasites. Nonetheless, the conserved aspects of biology and underlying genetics among apicomplexans in these evolutionarily distant organisms allow powerful insight into what it means to be an essential apicomplexan gene. In our three-way species comparison of essentiality data, we identify core, essential apicomplexan genes with the acknowledgement that many of these essential genes have likely taken on new, possibly distinct functions in the different species.
Comparing essentiality data across organisms/screens
The principles behind determining gene essentiality and dispensability from all three screens are similar. Genes that tolerate disruption of their protein-coding sequence (CDS) are dispensable whereas parasites die when the CDS of essential genes are disrupted, meaning parasites with mutations in these genes are not recovered. All the apicomplexan parasite screens involve large-scale mutagenesis to generate single-gene mutations per parasite, each with unique sequence signatures making them identifiable in a pool. Pooled mutants were then subjected to phenotypic screening via competitive growth assays. Analysis of all three assays utilized some form of high-throughput, Next-Gen sequencing to assess depletion in reads for mutants in the pool as a proxy for growth, with more reads indicating a more successful mutant (and therefore dispensable gene), and fewer reads/the absence of reads indicating sicker mutants and essential genes.
By definition, all genes in any orthogroup are descended from a single ancestral gene having the same sequence and function. As gene duplications and losses happen frequently over the course of evolution with complex outcomes for function that cannot be readily inferred, we limited our essentiality comparisons to orthologous genes with a single representative in each of the three parasites (single-copy orthologs) which had essentiality data in all three screens. Of the 2606 single-copy orthologs shared between these three parasites, 1500 had essentiality data in all three screens and were used for our comparative analyses (Supplementary Table S1).
Essentiality across genome-scale assays: what does it mean to be an essential apicomplexan gene?
Despite these commonalities, methodological differences in how the essentiality data were collected and interpreted for each apicomplexan screen necessitate defining a common language for assigning essentiality vs. dispensability for the purposes of comparison. All three assays assigned some sliding measure of confidence to assign essentiality category, with some genes falling clearly into the “essential” or “dispensable” categories for the tested conditions, and many more in the grey-area of uncertainty as we consider methodology/assay-specific caveats for the interpretation of each dataset. We sought to define the most high-confidence essential and dispensable genes from each assay for a conservative comparison of essentiality and dispensability across these parasites.
In the case of the P. falciparum screen, inherent properties of individual genes independent of essentiality can affect the likelihood of recovering a piggyBac transposon insertion in that gene, such as gene length and TTAA density. The piggyBac transposon preferentially inserts at TTAA sites, and while these sites are for the most part quite abundant and evenly distributed across the P. falciparum genome (averaging one site every ~80bp; [4]), we took caution in designating genes with low TTAA density (<7 TTAA sites/kb) as essential based on the absence of recovered insertions alone. Similarly, there may be decreased chances of recovering insertions from very short genes (<500bp) independent of essentiality. We therefore designated the <13% of genes falling in to either of these categories as “tentative” and did not consider them for comparison.
Methods of the P. berghei and T. gondii screens were similar, utilizing targeted, non-homologous recombination techniques to generate a library of mutants. The P. berghei screen is the most limited in terms of number of genes for which essentiality was characterized (at 2578 genes, >50% of the genome), through the use of targeted, non-homologous recombination techniques to excise the full coding-region of a gene. Mutants exhibited a range of growth phenotypes, and mutants characterized as having a less-conclusive “slow” growth phenotype as compared to wild-type parasites were not considered in our comparisons. The authors were additionally careful to note any “essential” designations resulting from mutants generated via vectors having particularly short homology-arms, which is known to decrease recombination efficiency, as less-conclusive [5, 38]. We filtered these genes from our comparisons. In T. gondii a CRISPR-based screen was feasible due to the parasite’s high rates of non-homologous end-joining, allowing for higher-throughput targeted mutagenesis methods. Individual in-depth follow-up phenotype studies of individual mutant parasites supported the overall conclusions of the whole-genome profiling.
Direct comparisons using simple essentiality-score cutoffs to assign gene essentiality/dispensability across assays suggested misleadingly low correlation in essentiality-classifications across these parasites than would be expected by overall orthology. We therefore used a ranking-method to binarily classify genes as essential or dispensable, ranking orthologous genes by sliding-scale essentiality score in each screen (Mutagenesis Index Score, or MIS, for P. falciparum; Relative Growth Rate, or RGR, for P. berghei, and Phenotype Score for T. gondii), then further reducing the dataset to genes ranking in the top- and bottom-quantiles for all three screens, ensuring a conservative comparison of only the most-confidently classified essential and dispensable genes. The final gene set for the three-species comparison comprised 816 genes with a high degree of correlation in essentiality between screens (ranging from 74.5% to 81.6%), more consistent with overall orthology (Figure 1A–C; Supplementary Table S2).
Figure 1. Genes included in essentiality comparisons for this review with added evolutionary context.
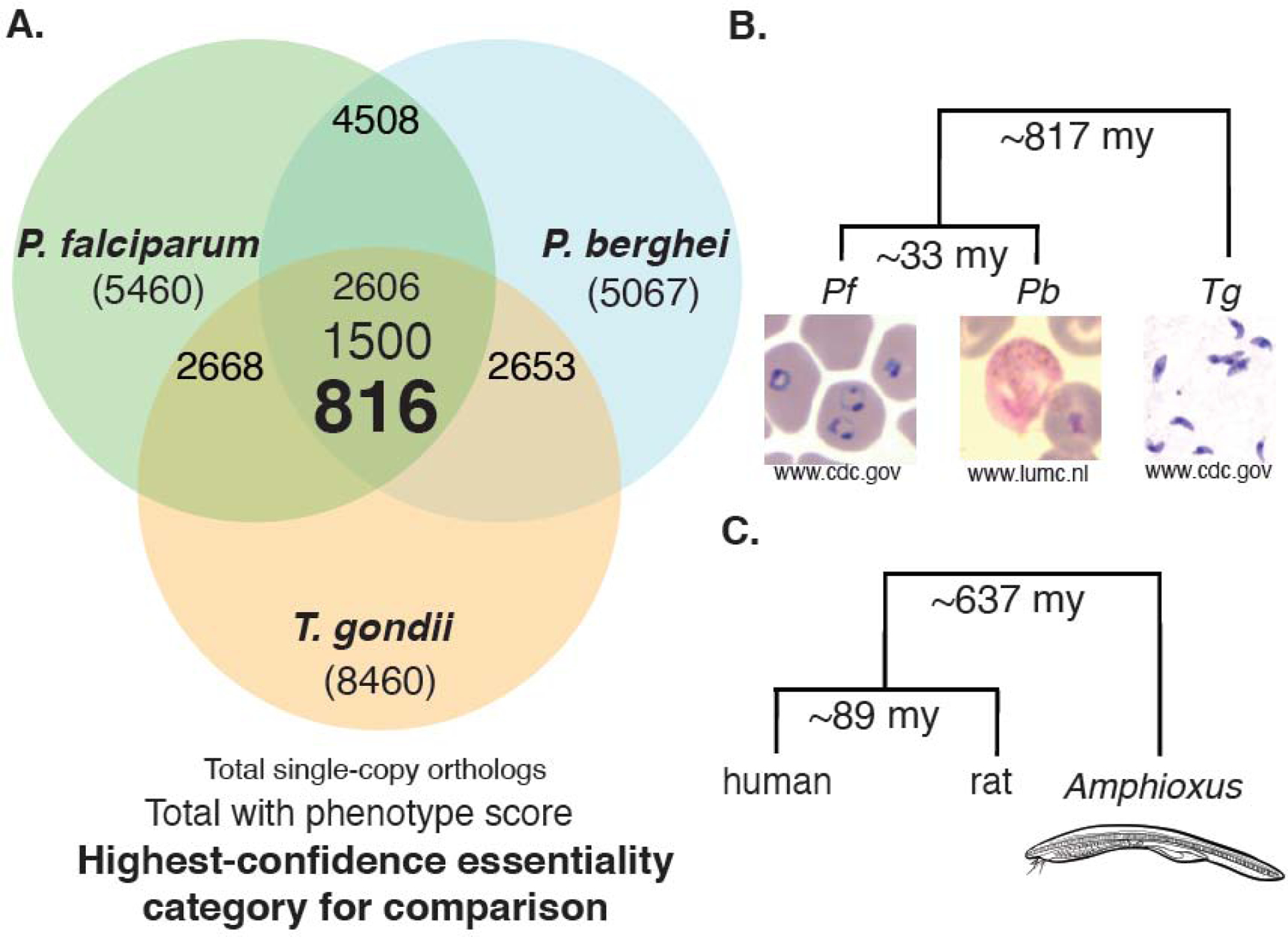
A. Species ortholog-overlaps and genes retained after each filtering step. Total gene-numbers for each organism are indicated in parentheses. Of 2606 single-copy orthologs shared between all three parasites, 1500 had phenotype data from all three essentiality screens. Our comparison-set comprised 816 genes confidently scored in all three screens. Single-copy orthologroups between each pair of species are also reported (Pf/Pb, 4508; Pf/Tg, 2668; Pb/Tg, 2653). B-C. Phylogenetic relatedness of P. falciparum, P. berghei, and T. gondii and their hosts, respectively. Amphioxus, a basal-branching marine chordate, has a similar scale of evolutionary distance from humans/rats as T. gondii has from Plasmodium (included for evolutionary reference). Evolutionary timescales are median estimates of divergence time based on several publications as calculated by timetree.org [28].
Specialized apicomplexan organelles are rich with essential, uncharacterized parasite biology
Broadly conserved categories of genes within each apicomplexan species tend to be essential in all three screens (Figure 2, Key Figure; Supplementary Table S3). Not surprisingly, many of the conserved apicomplexan essential processes have also been found to be essential in very distantly-related eukaryotes such as yeast, mice, and humans, particularly translation-associated functions and cellular components such as the ribosome [7–9, 39–42]. Similarly, core metabolic pathways for RNA metabolism appear highly conserved in all eukaryotic organisms and are unsurprisingly essential across apicomplexans, while motor-activity and signaling pathways are primarily dispensable. Essential vs dispensable gene-sets within each organism displayed the generally expected characteristics for these gene categories in agreement with other essentiality screens; essential genes compared to non-essential genes tend to be more deeply conserved, have lower occurrence of polymorphisms, and have lower redundancy (no paralogs that might compensate for loss-of-function) [4–6, 43]. The broad consensus across the growing number of essentiality-screens serves as confirmation of methodology for defining the apicomplexan essential-gene repertoire. Beyond these expected essential gene sets of broadly conserved genes are 132 apicomplexan-essential genes that have little or no functional annotation because they do not have enough similarity to studied genes in model organisms. Several of these essential unknowns have evidence for being targeted to the apicoplast (Supplementary Table S3). At least 10% of apicomplexan-essential genes are comprised of mitochondria and apicoplast-targeted genes.
Figure 2, Key Figure. Comparison and contrast between select essential pathways across parasites as determined by enrichment analyses of each essentiality category.
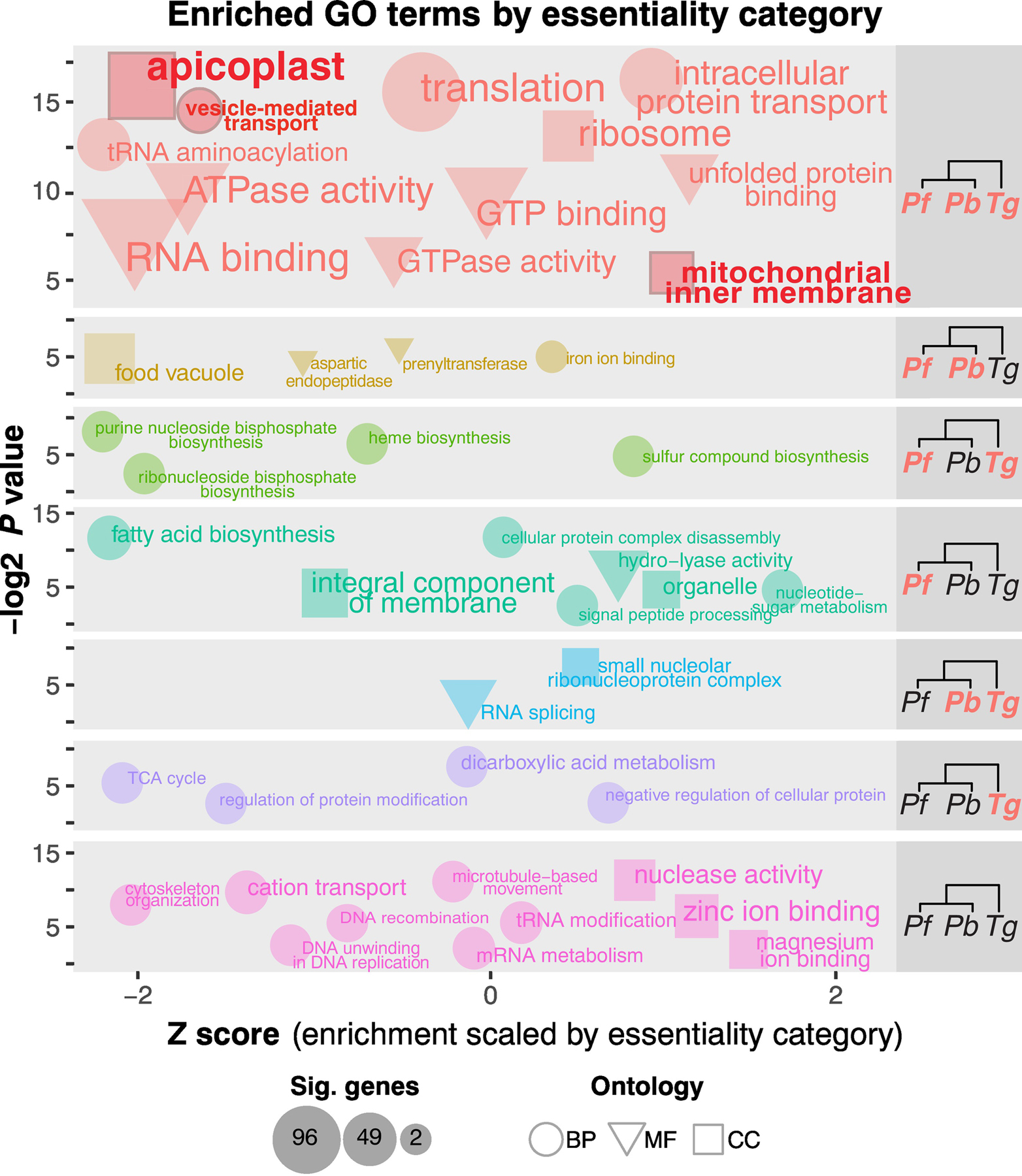
Cladograms on the right indicate each essentiality category, where orange indicates essentiality and black indicates dispensability for the given species. The y-axis (-log2 p-value) describes the absolute significance of a term’s enrichment, and the x-axis (Z-score) describes the significance of each term compared to every other significant term in that essentiality category. The upper left quadrant of each category indicates highest significance (lowest Z-score and highest –log2 p-value (weighted Fisher/elim hybrid test)). Only significant terms (p-value <=0.05) are plotted. Coordinates are approximate (points are staggered randomly for ease of viewing). Font-size and point-size indicate the number of significant genes in the essentiality category mapping to that GO-term. Expected highly conserved categories such as translation-associated terms are highly essential across all three parasites, as are terms related to endosymbiont-derived organelles (highlighted in red). BP: biological process. CC: cellular component. MF: molecular function.
The complex evolutionary origins of the apicomplexan mitochondrion and apicoplast (which arose from endosymbiosis of an alpha-proteobacterium and red-algal endosymbiont, respectively [21, 44] and thus have little overlap with human proteins) have long made these organelles promising drug targets. Both of these endosymbionts, over the course of hundreds of millions of years of evolution, became intimately integrated into parasite biology, housing some of the most highly essential pathways to parasite survival (Figure 2; Supplementary Table S3). Given the unusual origins of these pathways and adaptations the ancestral parasite had to make to interface with and regulate these new organelles—associated genes are rich with parasite-specific, essential yet unexplored functions, and processes mapping to these organelles are indeed overrepresented among shared-essential genes.
Essential apicomplexan-specialized mitochondrial processes
Many basic mitochondrial functions are conserved and essential across eukaryotes, including the generation of ATP, iron-sulfur cluster synthesis, or redox regulation [45]; indeed, we find key genes underlying these conserved processes among shared-essential genes in our comparison (Figure 2; Supplementary Table S2–S3). Though it is generally accepted that all eukaryotic mitochondria arose from the same endosymbiotic event, there are striking functional differences in mitochondrial functions and pathways across eukaryotes, with apicomplexan lineages having among the most-reduced mitochondrial genomes studied (comprising only three protein-coding genes, and highly fragmented bits of ribosomal RNA genes) [45–47]. The majority of mitochondrial proteins are thus nuclear-encoded, necessitating these parasites develop complex systems of signaling, protein-import and export to regulate this organelle [48].
There is strong evidence to suggest translation does happen within the parasite mitochondria, and that somehow the fragmented mitochondrially-encoded rRNAs get assembled into a functional ribosome [49, 50]. Factors regulating these complex processes were largely unknown. Plastid and mitochondrial proteins are known to be heavily post-transcriptionally regulated in plants, with a largely-expanded family of organelle-targeted, nuclear-encoded RNA-binding proteins, the pentatricopeptide-repeat (PPR) proteins, responsible for most of this regulation [51]. Plasmodium has two annotated PPR proteins, one of which is targeted to the apicoplast (PfPPR1; PF3D7_1406400/PBANKA_1035800) and the other does not have a clear targeting sequence for either the apicoplast or the mitochondria (PfPPR2; PF3D7_1233300/PBANKA_1448000/TGGT1_243530). Both are predicted to be highly essential in Plasmodium, and though PfPPR1 does not have a direct ortholog in T. gondii, T. gondii also appears to have a single apicoplast-targeted PPR protein (TGGT1_244050) that is essential for parasite growth [52]. Using models based on plant PPR-proteins and the related algal octatricopeptide-repeat (OPR) proteins (which do not have annotated representatives in the Apicomplexa), Hillebrand and colleagues recently discovered a novel family of related proteins they termed heptatricopeptide-repeat (HPR) proteins that they then confirmed to localize to mitochondria and have specific RNA-binding capability [53].
Subsequent searches for proteins containing HPR domains across the tree of life indicated representatives across green algae, alveolates (including most members of the Apicomplexa, their photosynthetic relatives the chromerids, and the more distantly-related dinoflagellates), as well as a small, more divergent handful in human, suggesting the family was an ancient acquisition of the eukaryotic ancestor; however the family has only expanded in green algae and chromalveolates. Phyletic distribution suggested lineage-specific expansion in lineages with more highly fragmented mitochondrial rRNA genes, such as Plasmodium and T. gondii, while representatives from the sister-alveolate ciliate clade, known to have a much more straightforward mitochondrial rRNA-organization, were much fewer. No HPR-domains were detected in basal-branching apicomplexan Cryptosporidium, which has only the remnants of a mitochondrion.
These findings taken together strongly suggest the apicomplexan HPR proteins are important regulatory components of the organellar (and particularly, mitochondrial) translation-machinery, and they provide another example of a plant-like innovation in these parasites that could possibly be exploited to human advantage. The approximately 20 HPR proteins in Plasmodium and the 25 in T. gondii for the most part did not meet the essentiality criteria to be included in our comparisons. However, several representatives were phenotypically characterized in each screen and strongly tend towards being essential. Further, four HPR proteins are shared-essential across all three screens with no human orthologs, while only one is shared-dispensable (Supplementary Table S4). One of four HPR proteins predicted to localize to the apicoplast at their initial discovery has since been verified (PF3D7_0930100), and it too was scored as highly essential in P. falciparum. The P. berghei ortholog (PBANKA_0830800) was not included in the essentiality screen, and there is no T. gondii ortholog. It is also interesting to note that the majority of these HPR proteins were annotated as “hypothetical” at the publication of the essentiality screens, further speaking to apicomplexan endosymbiotic-organelle biology as a frontier of essential functions awaiting characterization.
Essential apicoplast processes
The apicoplast has long been accepted as essential for parasite survival, though the reasons why were somewhat mysterious [12]. Why would a plastid organelle be essential for the survival of a non-photosynthetic, obligate-intracellular organism? Research over the last decade has illuminated that apicoplast-minus mutants can be completely chemically rescued by supplementation with a single intermediate molecule acting within the isoprenoid biosynthesis pathway, isopentenyl pyrophosphate (IPP) [54]. This discovery has since enabled development of small-scale phenotypic screening assays of chemically-generated mutant libraries for apicoplast-minus mutants, which were then rescued with IPP for sequencing to determine genes essential for apicoplast biogenesis [55]. Apicoplast-minus screening has also been employed to examine drug mechanism of action [56, 57].
Indispensable Conserved Apicomplexan Proteins (ICAPs): Functional studies.
While the essentiality classification provides one of the most useful basic metrics about these many apicomplexan-specific uncharacterized genes, further functional annotation will require assays to identify their essential functions.
Further such investigations to demonstrate the existence of apicomplexan-specific essential processes and to identify promising, parasite-specific drug targets were initiated in the T. gondii CRISPR-mutagenesis screen [6]. A set of ~200 genes, termed “indispensable conserved apicomplexan proteins (ICAPs)”, were identified to be apicomplexan-specific and essential, though the entire list of these genes was not explicitly delineated. Of the 17 ICAPs Sidik et al. specified, 14 have single-copy orthologs across P. falciparum and P. berghei, and half of them have essentiality predictions in all three species. Sidik et al. further molecularly characterized these 17 ICAPs, noting their localization and confirming essentiality by independently generated mutants (Supplementary Table S5). Interestingly, the majority of these proteins were localized to discrete compartments.
Temporal and spatial characterization of unknown proteins can be quite informative in apicomplexans, especially for the invasive motile stages, since their discrete organelles and other internal compartments often are associated with specialized functions (e.g., micronemes sequester invasion ligands [58]). For example, based on the initial T. gondii phenotype, secondary phenotyping with conditional knock-outs in P. falciparum using regulatable gene modifications provided verification of cross-species essentiality of a claudin-like apicomplexan microneme protein (CLAMP)[6]. Additional consensus-based P. falciparum essentiality data and experimental studies of ICAPS should broaden support for essentiality of potentially druggable targets ripe for investigation. Similarly, half of those ICAPs that could be phenotypically categorized in the P. berghei screen were essential or slow-growers, including the CLAMP ortholog [5].
Endosymbiont-derived organelles house a large proportion of apicomplexan-specific functions. Large-scale essentiality screens validate that these functions are highly enriched in shared, essential genes, many of which do not have human orthologs. Many of these shared, essential genes were annotated as “hypothetical” at publication of the large-scale essentiality screens, and even still at the time of this review. Apicomplexan-specific essential processes have been revealed several times to have emerged from co-opting distantly related protein-domains picked up along the way from their bacterial and algal endosymbionts (such as the HPR-family of organelle-targeted RNA-binding proteins, or the ApiAP2 family of master transcriptional regulatory proteins; [53, 59, 60] ). In light of these observations, it stands to reason that further experimental assessment of parasite-specific, shared-essential hypothetical genes will reveal even more organelle-specific functions and parasite biology.
Towards a nuanced interpretation of target-essentiality and specificity in rational, target-based anti-parasitic drug-development
A promising drug target does not necessarily have to be free of human orthologs; indeed there are several examples of promising drugs that were engineered to specifically target parasite orthologs of widely conserved essential proteins with no reported ill-effects on the orthologous human protein (for example, [61]). As there are no definitive guidelines regarding the acceptable level of conservation between host and parasite proteins before off-target effects become a concern in drug-development, genes with no identified orthologs in the human host are a conservative place to start in the search for viable targets. Endosymbiont-derived organelles are hotbeds of essential, apicomplexan-specific functions, many performed by genes without human orthologs (Supplementary Table S4, S6).
Several effective antimalarial drugs of the past have been found to target mitochondrial or apicoplast pathways; enthusiasm for these targets has waxed, waned, and waxed again as we assemble a more and more detailed puzzle of parasite biology necessitating an evolving, nuanced interpretation of just what makes a good target. Drugs known to target the housekeeping functions of mitochondrial or apicoplast pathways generally result in a “delayed death” phenotype, meaning parasites are able to undergo one round asexual replication before death, which made these drugs of lower priority as current drug-development guidelines prioritize fast-acting drugs that decrease parasite chances of developing resistance. Recent studies have identified promising apicoplast or mitochondrial targets that swiftly kill the parasite within a single cycle, however, which has revived enthusiasm for these organelles as potential sources of fast-killing targets [62].
Resistance against the mitochondrial electron transport chain (ETC) enzyme cytochrome b-targeted drug Atovaquone evolved alarmingly rapidly, particularly disappointing given its initial efficacy and highly safe toxicology profile. Atovaquone’s failure dulled excitement for other mitochondria-related drug-targets [63, 64]. That resistance, however, was not transmissible due to biological peculiarities of the parasite—functions of the mitochondria are more essential during mosquito-stage development, and the very characteristics that made resistance rapidly arise in blood-stage therapies made it an effective transmission-blocking possibility [63, 65]. Moreover, recent studies have found atovaquone to be effective in combinatorial therapy with another drug targeting the same component of the mitochondrial ETC in another active site [66], against which resistance did not evolve experimentally. Atovaquone-synergistic effects were not seen with another compound targeting cytochrome b [66]. These findings suggest not only that mitochondrial pathways retain value for antimalarial intervention—but that combinatorial therapies employing drugs acting synergistically against the same target may also prove to be worth-while. Given that vast compound-libraries screened for antimalarial activity and then interrogated for their mechanism of action have largely converged on the same handful of target-proteins, despite great chemodiversity in these libraries [67], further evaluation of these drugs in concert is warranted.
Mitochondrial-targeted proteins that are encoded in the single nuclear genome will have different properties to consider as potential drug-targets than mitochondrial genome-encoded proteins, and have other strengths—the single nuclear genome (as opposed to ~22 mitochondrial genomes per Plasmodium parasite) mean there is less likelihood that low-frequency population heterogeneity can allow a beneficial mutation to rise to fixation, as was the case with atovaquone [64]. The three essentiality screens targeted only nuclear-encoded genes, identifying many shared-essential, mitochondrial-targeted candidate genes (Figure 2; Supplementary Table S2, S3 & S4).
Notably, the most successful past and present front-line antimalarials (chloroquine and artemisinin, respectively) are themselves excellent examples of exploitation of parasite-specific biology/endosymbiont-derived organelles, with both drugs relying in part on vesicular trafficking of host-cell hemoglobin to the digestive vacuole for parasite-killing activity—a process ultimately dependent upon apicoplast-enabled post-translational modifications to Rab-family vesicular trafficking proteins [68–73].
Though concepts are emerging of what it means to be an “essential” gene with the availability of more and more screens across the tree of life (and even synthetic life; [74] )—it is clear that essentiality is context-dependent and cannot truly be binarily assigned [75], particularly in parasitic organisms where evolution by genome-reduction is the rule, and every gene is likely essential in an environment-dependent context (such as host, developmental stage, environmental stressors such as host fever or antimalarials). In addition to the conditional nature of essentiality, there are other properties of genes and genomes that necessitate a nuanced interpretation to these screens. Essentiality is not a fixed property of genes themselves; genes function as components in the larger service of metabolic pathways and biological processes, and evidence suggests these components can be and often are substituted[75]. Indeed there are only a few handful of genes themselves that are shared across all eukaryotes and can be traced back to the Last Eukaryotic Common Ancestor (LECA) [76], mostly related to translation; however essentiality of core processes necessary to sustain life remains conserved. Thus essentiality is a plastic characteristic of genes, and all essential genes are not created equal—defects in some essential genes can be compensated for more easily than defects in others [77].
Concluding remarks
We focused this review on shared characteristics across the three organisms, but contrasts in gene-essentiality between them can also be quite informative and reflect the vastly different niches these parasites have evolved to fill; for instance, the TCA cycle is essential in T. gondii, but dispensable during the Plasmodium blood stages, which instead relies on glycolytic metabolism to generate its energy (Supplementary Table S7–S12). Plasmodium-specific contrasts suggest the relative importance of apicoplast and mitochondrial pathways in these two parasites, with apicoplast fatty acid biosynthesis and a selection of mitochondrial genes essential in P. falciparum, yet dispensable in P. berghei (Supplementary Table S3 & S9). These apparent differences in essentiality of certain organellar pathways within Plasmodium spp. may possibly reflect differences in requirements for in vitro vs in vivo infection. Biological relevance of these and other indicated differences in essential processes require further investigation.
The essential/dispensable screens provide critical initial phenotype-data for most parasite genes under ideal growth conditions. However, the power of the resources generated in these screens—particularly the expansive libraries of uniquely identifiable, single-gene mutant parasites—extends far beyond answering the relatively broad question of parasite gene-essentiality during select developmental stages (see Outstanding Questions). These initial essentiality phenotypes will serve as the backbone for developing and interpreting screens for any number of phenotypes of interest, potentially allowing high-throughput functional characterization for thousands of genes at a time. These screens may enable the research-community to iteratively ascribe function to the vast majority of apicomplexan parasite genes over time, unlimited by a priori functional knowledge.
Outstanding Questions.
How important is predicted target-essentiality in the context of anti-parasitic drug development, and what is the value of genes in the “grey areas” of essentiality for possible drug targets?
What are the functions of the shared, unannotated apicomplexan-essential genes? Many of these proteins have been found to localize to the mitochondria, apicoplast or invasion machinery. Do these genes represent unexplored components of known pathways, or do these organelles harbor more as-yet unappreciated essential functions?
Mitochondrial and apicoplast pathways have proven to be druggable. Are the same mechanisms of resistance parasites have evolved to evade existing drugs that target components of these pathways effective against drugs designed to target “unknown” essential components of these pathways?
We are now in the post essential-genome era of apicomplexan parasite research. Forward-genetic screening utilizing these expansive mutant-parasite resources makes an accelerated pace for functional biological breakthroughs possible, or perhaps likely. Such insights have the potential to drive discovery of new druggable targets, or to enable thoughtful application of existing interventions as synergistic co-therapies.
Supplementary Material
Highlights.
Major breakthroughs in genome-scale screens for essential genes have recently been completed in the major human malaria parasite Plasmodium falciparum and two related parasites Plasmodium berghei and Toxoplasma gondii.
Many shared, parasite-specific genes are essential in all three parasites.
Shared-essential genes, particularly parasite-specific genes, provide a short-list of the most high-value targets for prioritization of new effective interventions against a range of related, important parasitic diseases.
Acknowledgements
We thank our colleagues Dr. Chengqi Wang, Dr. Rays Jiang, Swamy Rakesh Adapa, members of the Adams lab, and the USF Genomics Program Omics Hub for critical discussion that improved the quality of this work. We thank the Kissinger lab at the University of Georgia for extended methodological discussions and tips. Computational analyses were performed using resources provided or maintained by the USF Department of Research Computing. This work was supported by the National Institutes of Health grants F32-AI112271 (awarded to J.O.), R01 AI094973 and R01 AI117017 (awarded to J.H.A.)
Glossary
- Endosymbiont-derived organelles
the organelles arising from the endosymbiosis of an algal or bacterial cell in the apicomplexan parasite ancestor—the apicoplast and the mitochondria, respectively.
- Mutagenesis Index Score (MIS)
The sliding-scale score used as the primary metric for assessing comparative essentiality of P. falciparum genes.
- Orthologs
Genes in different species that originated by vertical descent from a single gene of the last common ancestor. All copies of genes descended from the same ortholog make up an ortholog group.
- Paralogs
Genes within the same genome that belong to the same ortholog group. Paralogs result from a gene duplication-event in evolutionary history somewhere downstream from the last eukaryotic common ancestor.
- Phenotype Score
The sliding-scale score used as the primary metric for assessing comparative essentiality of T. gondii genes.
- Random mutagenesis
Mutagenesis method usually utilizing a transposon that inserts preferentially into a single, random occurrence of its given recognition sequence per genome (in the case of the piggyBac transposon used for the P. falciparum essentiality screen, TTAA). Random mutagenesis is particularly useful for exploring highly repetitive genomes where large-scale utilization of targeted mutagenesis, which requires long stretches of homologous sequence for target-specificity and recombination efficiency, is impractical (such as the AT-rich P. falciparum genome).
- Relative Growth Rate (RGR)
The sliding-scale score used as the primary metric for assessing comparative essentiality of P. berghei genes.
- Shared-essential genes
Single-copy orthologs confidently scored as essential in all three essentiality screens.
- Single-copy orthologs
Also known as one-to-one orthologs. Single-copy ortholog groups have exactly one gene copy in each species being compared. As all genes in any orthogroup are descended from a single ancestral gene, with the same sequence and function, while duplications and loss happen comparatively frequently—single-copy orthologs are a less-complex subset of the wider dataset that still contains all the information of the complete dataset. Single-copy orthologs are the most useful for evolutionary comparisons, as we do not have to make any inferences about the ancestral state (or the order in which gene duplication or loss occurred based on presence or absence of paralogs).
- Targeted mutagenesis
Mutagenesis method utilizing plasmid vectors designed to edit a specific gene, usually by homologous recombination (e.g., CRISPR). Requires comparatively long homology regions with the target-gene for efficiency (~1kb on each side of the target-region), and as such has high target-specificity. Both the T. gondii and P. berghei essentiality screens utilized a form of targeted mutagenesis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gardner MJ, et al. (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kissinger JC, et al. (2003) ToxoDB: accessing the Toxoplasma gondii genome. Nucleic acids research 31, 234–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böhme U, et al. (2019) Progression of the canonical reference malaria parasite genome from 2002–2019 [version 2; peer review: 3 approved]. Wellcome Open Research 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang M, et al. (2018) Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushell E, et al. (2017) Functional Profiling of a Plasmodium Genome Reveals an Abundance of Essential Genes. Cell 170, 260–272.e268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidik SM, et al. (2016) A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell 166, 1423–1435.e1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang T, et al. (2015) Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blomen VA, et al. (2015) Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096 [DOI] [PubMed] [Google Scholar]
- 9.Hart T, et al. (2015) High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 163, 1515–1526 [DOI] [PubMed] [Google Scholar]
- 10.Huang J, et al. (2004) A first glimpse into the pattern and scale of gene transfer in Apicomplexa. Int J Parasitol 34, 265–274 [DOI] [PubMed] [Google Scholar]
- 11.Lim L and McFadden GI (2010) The evolution, metabolism and functions of the apicoplast. Philosophical transactions of the Royal Society of London. Series B, Biological sciences 365, 749–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Striepen B (2011) The apicoplast: a red alga in human parasites. Essays in biochemistry 51, 111–125 [DOI] [PubMed] [Google Scholar]
- 13.White J and Rathod PK (2018) Indispensable malaria genes. Science 360, 490–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO (2018) World Malaria Report. World Health Organization [Google Scholar]
- 15.Ariey F, et al. (2014) A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505, 50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miotto O, et al. (2013) Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nature genetics 45, 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bopp S, et al. (2018) Plasmepsin II-III copy number accounts for bimodal piperaquine resistance among Cambodian Plasmodium falciparum. Nat Commun 9, 1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sexton AE, et al. (2019) Post-Genomic Approaches to Understanding Malaria Parasite Biology: Linking Genes to Biological Functions. ACS Infectious Diseases 5, 1269–1278 [DOI] [PubMed] [Google Scholar]
- 19.DeBarry JD and Kissinger JC (2011) Jumbled genomes: missing Apicomplexan synteny. Mol Biol Evol 28, 2855–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeBarry JD and Kissinger JC (2014) A survey of innovation through duplication in the reduced genomes of twelve parasites. PLoS One 9, e99213–e99213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janouškovec J, et al. (2010) A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proceedings of the National Academy of Sciences 107, 10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo YH, et al. (2015) Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. Elife 4, e06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balaji S, et al. (2005) Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res 33, 3994–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pradhan A, et al. (2015) Chemogenomic profiling of Plasmodium falciparum as a tool to aid antimalarial drug discovery. Sci Rep 5, 15930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Voorhis WC, et al. (2016) Open Source Drug Discovery with the Malaria Box Compound Collection for Neglected Diseases and Beyond. PLoS Pathog 12, e1005763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibbons J, et al. (2018) Altered expression of K13 disrupts DNA replication and repair in Plasmodium falciparum. BMC Genomics 19, 849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacheco MA, et al. (2013) Malarial parasite diversity in chimpanzees: the value of comparative approaches to ascertain the evolution of Plasmodium falciparum antigens. Malaria Journal 12, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, et al. (2017) TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol Biol Evol 34, 1812–1819 [DOI] [PubMed] [Google Scholar]
- 29.Böhme U, et al. (2018) Complete avian malaria parasite genomes reveal features associated with lineage-specific evolution in birds and mammals. Genome Research [DOI] [PMC free article] [PubMed]
- 30.Sanderson T and Rayner JC (2017) PhenoPlasm: a database of disruption phenotypes for malaria parasite genes. Wellcome Open Res 2, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwach F, et al. (2015) PlasmoGEM, a database supporting a community resource for large-scale experimental genetics in malaria parasites. Nucleic Acids Res 43, D1176–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinha A, et al. (2014) A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature 507, 253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kafsack BF, et al. (2014) A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature 507, 248–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carruthers VB (2002) Host cell invasion by the opportunistic pathogen Toxoplasma gondii. Acta Tropica 81, 111–122 [DOI] [PubMed] [Google Scholar]
- 35.Kim K and Weiss LM (2004) Toxoplasma gondii: the model apicomplexan. International Journal for Parasitology 34, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Douzery EJ, et al. (2004) The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci U S A 101, 15386–15391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delsuc F, et al. (2018) A phylogenomic framework and timescale for comparative studies of tunicates. BMC Biol 16, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomes AR, et al. (2015) A genome-scale vector resource enables high-throughput reverse genetic screening in a malaria parasite. Cell Host Microbe 17, 404–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H, et al. (2019) New insights on human essential genes based on integrated analysis and the construction of the HEGIAP web-based platform. Briefings in Bioinformatics 21, 1397–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabir M, et al. (2017) Properties of genes essential for mouse development. PLoS One 12, e0178273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giaever G, et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 [DOI] [PubMed] [Google Scholar]
- 42.White Jacqueline K., et al. (2013) Genome-wide Generation and Systematic Phenotyping of Knockout Mice Reveals New Roles for Many Genes. Cell 154, 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen W-H, et al. (2012) Younger Genes Are Less Likely to Be Essential than Older Genes, and Duplicates Are Less Likely to Be Essential than Singletons of the Same Age. Molecular Biology and Evolution 29, 1703–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roger AJ, et al. (2017) The Origin and Diversification of Mitochondria. Current Biology 27, R1177–R1192 [DOI] [PubMed] [Google Scholar]
- 45.Vaidya AB and Mather MW (2009) Mitochondrial Evolution and Functions in Malaria Parasites. Annual Review of Microbiology 63, 249–267 [DOI] [PubMed] [Google Scholar]
- 46.Sheiner L, et al. (2013) The metabolic roles of the endosymbiotic organelles of Toxoplasma and Plasmodium spp. Current Opinion in Microbiology 16, 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santos HJ, et al. (2018) Reinventing an Organelle: The Reduced Mitochondrion in Parasitic Protists. Trends in Parasitology 34, 1038–1055 [DOI] [PubMed] [Google Scholar]
- 48.Mallo N, et al. (2018) Protein Import into the Endosymbiotic Organelles of Apicomplexan Parasites. Genes (Basel) 9, 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pino P, et al. (2010) Mitochondrial translation in absence of local tRNA aminoacylation and methionyl tRNA Met formylation in Apicomplexa. Mol Microbiol 76, 706–718 [DOI] [PubMed] [Google Scholar]
- 50.Sharma A and Sharma A (2015) Plasmodium falciparum mitochondria import tRNAs along with an active phenylalanyl-tRNA synthetase. Biochem J 465, 459–469 [DOI] [PubMed] [Google Scholar]
- 51.Colcombet J, et al. (2013) Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biol 10, 1557–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hicks JL, et al. (2019) An essential pentatricopeptide repeat protein in the apicomplexan remnant chloroplast. Cellular Microbiology 21, e13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hillebrand A, et al. (2018) Identification of clustered organellar short (cos) RNAs and of a conserved family of organellar RNA-binding proteins, the heptatricopeptide repeat proteins, in the malaria parasite. Nucleic Acids Res [DOI] [PMC free article] [PubMed]
- 54.Yeh E and DeRisi JL (2011) Chemical Rescue of Malaria Parasites Lacking an Apicoplast Defines Organelle Function in Blood-Stage Plasmodium falciparum. PLoS Biology 9, e1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang Y, et al. (2019) A mutagenesis screen for essential plastid biogenesis genes in human malaria parasites. PLOS Biology 17, e3000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu W, et al. (2015) A chemical rescue screen identifies a Plasmodium falciparum apicoplast inhibitor targeting MEP isoprenoid precursor biosynthesis. Antimicrobial agents and chemotherapy 59, 356–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amberg-Johnson K, et al. (2017) Small molecule inhibition of apicomplexan FtsH1 disrupts plastid biogenesis in human pathogens. eLife 6, e29865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gubbels M-J and Duraisingh MT (2012) Evolution of apicomplexan secretory organelles. International journal for parasitology 42, 1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oberstaller J, et al. (2014) The Cryptosporidium parvum ApiAP2 gene family: insights into the evolution of apicomplexan AP2 regulatory systems. Nucleic Acids Res 42, 8271–8284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Modrzynska K, et al. (2017) A Knockout Screen of ApiAP2 Genes Reveals Networks of Interacting Transcriptional Regulators Controlling the Plasmodium Life Cycle. Cell host & microbe 21, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gisselberg JE, et al. (2018) Specific Inhibition of the Bifunctional Farnesyl/Geranylgeranyl Diphosphate Synthase in Malaria Parasites via a New Small-Molecule Binding Site. Cell chemical biology 25, 185–193.e185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boucher MJ and Yeh E (2019) Disruption of Apicoplast Biogenesis by Chemical Stabilization of an Imported Protein Evades the Delayed-Death Phenotype in Malaria Parasites. mSphere 4, e00710–00718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goodman CD, et al. (2017) Is the Mitochondrion a Good Malaria Drug Target? Trends Parasitol 33, 185–193 [DOI] [PubMed] [Google Scholar]
- 64.Siegel S, et al. (2017) Mitochondrial heteroplasmy is responsible for Atovaquone drug resistance in Plasmodium falciparum. bioRxiv, 232033 [Google Scholar]
- 65.Goodman CD, et al. (2016) Parasites resistant to the antimalarial atovaquone fail to transmit by mosquitoes. Science 352, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stickles AM, et al. (2016) Atovaquone and ELQ-300 Combination Therapy as a Novel Dual-Site Cytochrome Inhibition Strategy for Malaria. Antimicrobial agents and chemotherapy 60, 4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White J and Rathod PK (2018) Indispensable malaria genes. Science 360, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Birnbaum J, et al. (2020) A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science 367, 51. [DOI] [PubMed] [Google Scholar]
- 69.Gnädig NF, et al. (2020) Insights into the intracellular localization, protein associations and artemisinin resistance properties of Plasmodium falciparum K13. PLoS Pathog 16, e1008482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suazo KF, et al. (2016) Global proteomic analysis of prenylated proteins in Plasmodium falciparum using an alkyne-modified isoprenoid analogue. Scientific Reports 6, 38615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Imlay L and Odom AR (2014) Isoprenoid Metabolism in Apicomplexan Parasites. Current Clinical Microbiology Reports 1, 37–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sullivan DJ Jr., et al. (1996) On the molecular mechanism of chloroquine’s antimalarial action. Proceedings of the National Academy of Sciences of the United States of America 93, 11865–11870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kennedy K, et al. (2019) Delayed death in the malaria parasite Plasmodium falciparum is caused by disruption of prenylation-dependent intracellular trafficking. PLOS Biology 17, e3000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Juhas M, et al. (2012) Essential genes as antimicrobial targets and cornerstones of synthetic biology. Trends in Biotechnology 30, 601–607 [DOI] [PubMed] [Google Scholar]
- 75.Rancati G, et al. (2017) Emerging and evolving concepts in gene essentiality. Nature Reviews Genetics 19, 34. [DOI] [PubMed] [Google Scholar]
- 76.Koonin EV (2003) Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol 1, 127–136 [DOI] [PubMed] [Google Scholar]
- 77.Liu G, et al. (2015) Gene Essentiality Is a Quantitative Property Linked to Cellular Evolvability. Cell 163, 1388–1399 [DOI] [PubMed] [Google Scholar]
- 78.Chaparro MJ, et al. (2018) Efforts Aimed To Reduce Attrition in Antimalarial Drug Discovery: A Systematic Evaluation of the Current Antimalarial Targets Portfolio. ACS Infectious Diseases 4, 568–576 [DOI] [PubMed] [Google Scholar]
- 79.Proietti C and Doolan DL (2014) The case for a rational genome-based vaccine against malaria. Front Microbiol 5, 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


