Abstract
The outbreak of Coronavirus Disease 2019 (COVID-19) has been declared as a Public Health Emergency of International Concern (PHEIC) by the World Health Organization (WHO), which is being rapidly spread by the extremely spreadable and pathogenic 2019 novel coronavirus (2019-nCoV), also known as SARS-CoV-2. Pandemic incidence of COVID-19 has created a severe threat to global public health, necessitating the development of effective drugs or inhibitors or therapeutics agents against SARS-CoV-2. Spike protein (S) of the SARS-CoV-2 plays a crucial role in entering viruses into the host cell by binding to angiotensin-converting enzyme 2 (ACE-2), and this specific interaction represents a promising drug target for the identification of potential drugs. This study aimed at the receptor-binding domain of S protein (RBD of nCoV-SP) and the ACE-2 receptor as a promising target for developing drugs against SARS-CoV-2. Over 100 different flavonoids with antioxidant, anti-inflammatory, and antiviral properties from different literatures were taken as a ligand or inhibitor for molecular docking against target protein RBD of nCoV-SP and ACE-2 using PyRX and iGEMDOCK. Top flavonoids based on docking scores were selected for the pharmacokinetic study. Selected flavonoids (hesperidin, naringin, ECGC, and quercetin) showed excellent pharmacokinetics with proper absorption, solubility, permeability, distribution, metabolism, minimal toxicity, and excellent bioavailability. Molecular dynamics simulation studies up to 100 ns exhibited strong binding affinity of selected flavonoids to RBD of nCoV-SP and ACE-2, and the protein-ligand complexes were structurally stable. These identified lead flavonoids may act as potential compounds for developing effective drugs against SARS-CoV-2 by potentially inhibiting virus entry into the host cell.
Keywords: SARS-CoV-2, COVID-19, S-glycoprotein RBD, Angiotensin-converting enzyme- 2, Flavonoids, Molecular docking, ADMET
SARS-CoV-2, COVID-19, S-glycoprotein RBD, angiotensin-converting enzyme- 2, Flavonoids, Molecular docking, ADMET.
1. Introduction
On 30th January 2020, WHO declared the outbreak of COVID-19 as a Public Health Emergency of International Concern (Rodríguez-Morales et al., 2020). The outbreak was first reported from Wuhan City, Hubei, China (Lee and Hsueh, 2020) as an unexplained case of pneumonia on Dec 31, 2019, and was published in The New York Times. It was soon identified as a new SARS strain and named it 2019-nCoVorSARS-CoV-2 (Zhou et al., 2020). As of December 09, 2020, WHO has reported 6,88,45,410 SARS-CoV-2 cases, including 15,68,876 deaths globally. The exponential growing cases worldwide indicate this outbreak as one of the worst pandemics. Unfortunately, there are no specific vaccines or therapies against the new virus.
SARS-CoV-2 is a positive-sense single-stranded RNA virus belonging to the subgenus Sarbecovirus (beta-CoV lineage B). It has been reported that this strain is quite divergent from the other betacoronavirus, including the SARS I virus (Walls et al., 2020). ACE-2 is the unique receptor responsible for the virus's attachment to the host cell membrane (Andersen et al., 2020; Huang and Herrmann, 2020). Recently published reports also suggest that the 2019-nCoV binds to the human angiotensin-converting enzyme 2 (ACE-2) receptor through densely glycosylated spike (S) protein to penetrate human cells (Hoffmann et al., 2020). The S protein binds ACE-2 through its receptor binding domains (RBD), and the RBD "up" conformation of the S protein is a prerequisite for the formation of the RBD-ACE-2 complex (Wrapp et al., 2020). ACE-2 is the only necessary and essential receptor for binding the receptor-binding domain of S protein (RBD of nCoV-SP) of 2019-CoV and was targeted in the present study. Due to the unavailability of a high-resolution crystal structure, the task of rapid virtual screening against highly essential target proteins is relatively lackluster.
Flavonoids are an extraordinary group of natural substances with variable phenolic structures. Over 4,000 varieties of flavonoids have been identified (Groot and Rauen, 1998) and possess various favorable biochemical and antioxidant effects associated with various diseases (Burak and Imen, 1999). Albert Szent-Gyorgyii reported the first evidence of flavonoids' biological activity in 1938 (Narayana et al., 2001). After that, a wide range of biological functions such as antioxidant, antiviral, anti-inflammatory, and anticancer has been described for flavonoids (Xia et al., 2010; Khan et al., 2013; Gorlach et al., 2015; Iranshahi et al., 2015; Nabavi et al., 2015). The concern of the antiviral effects of flavonoids have been the subject material of various reports (Gescher et al., 2011; Roh and Jo, 2011; Chen et al., 2012) with reported antiviral activities through blockage of cellular receptors, blocking viral antigenic determinants, loss of enzymatic activities and inhibition of particle biosynthesis (Bae et al., 2000; Calzada et al., 2001; Chang et al., 2003; Savi et al., 2010). The natural product has been used during two earlier coronavirus outbreaks of SARS-CoV and MERS-CoV as a medicinal treatment, which suggests the probable ability of flavonoids to be useful for the ongoing epidemic of 2019-CoV (Li et al., 2005; Lin et al., 2005; Lau et al., 2008).
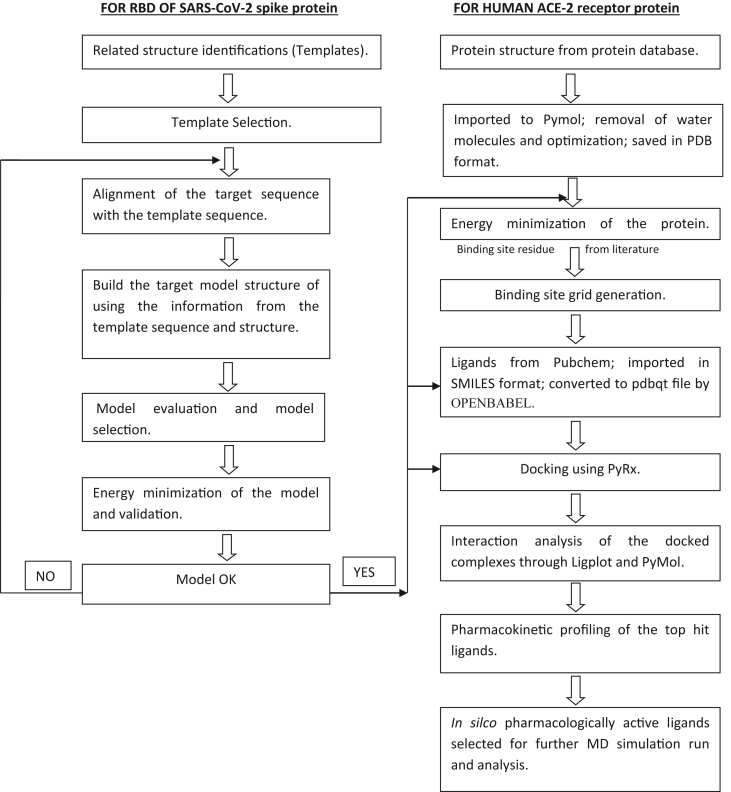
A flowchart of the methodology is depicted in Figure 1. In this study, different flavonoids and their subclasses with antioxidant, hepatoprotective, antibacterial, anti-inflammatory, anticancer, and antiviral properties from different literatures (Kumar and Pandey, 2013; Panche et al., 2016) were screened against our modeled structure of the receptor-binding domain of S protein (RBD of nCoV-SP) of 2019-CoV and human angiotensin-converting enzyme 2 (ACE-2). While examining the RBD of nCoV-SP and ACE-2 docking interactions and pharmacophore analysis, we have observed that flavonoids may inhibit SARS-CoV-2 cell entry into host cells.
Figure 1.
Flowchart of the methodology followed while docking target protein to the ligands.
2. Materials and methods
2.1. Sequence retrieval and homology modeling
Genbank ® of the National center for biotechnology information (NCBI) facilitates retrieving the biological information of the explored and assembled data (Sayers et al., 2019). The sequence for the receptor-binding domain (RBD) of the 2019-CoV spike protein (nCoV-SP) was collected from the NCBI database in FASTA format (GenBank accession number: MN908947.3). Low resolution and conformation of 3.5 Å of Cryo-EM structure (Wrapp et al., 2020) make it unreliable for virtual screening, and hence a homology model of the receptor-binding domain (RBD) of nCoV-SP was built with the help of MODWEB, an indispensable component of MODBASE (Eswar et al., 2003). This is followed by validation using PROCHECK at the EBI server (Laskowski et al., 1993) and saved in PDB format. The crystal structure of the human ACE-2 receptor protein (ACE-2) (PDB ID: 1R42) was also downloaded (Towler et al., 2004) and used for studying the interaction of our choice of inhibitors with the viral receptor.
2.2. Energy minimization and model validation
YASARA Energy Minimization Server (Krieger et al., 2009) was used for energy minimization of protein model for RBD of nCoV-SP and human ACE-2 receptor to obtain a high stable model with minimum energy. Structural quality and authenticity of the RBD of nCoV-SP were evaluated through ERRAT, Verify3D, and ProSa (Colovos and Yeates, 1993; Eisenberg et al., 1997; Wiederstein and Sippl, 2007).
2.3. Binding site identification
Previous studies on protein-protein docking, molecular dynamics, and virtual screening studies on SARS-CoV-2 spike glycoprotein (nCoV-SP) as well human ACE-2 receptor already evaluated the binding pockets and essential residues in the RBD and their interaction with the hotspots in ACE-2 receptor (Wu et al., 2011; Shang et al., 2020; Wan et al., 2020; Xu et al., 2020; Senathilake et al., 2020). Based on the above findings, the binding site residues for both RBD of nCoV-SP and the human ACE-2 receptor were selected in this study, and docking was carried out.
2.4. Ligand selection and ligand file preparation
Different flavonoids with antioxidant, hepatoprotective, antibacterial, anti-inflammatory, anticancer, and antiviral properties from different literatures were considered (Kumar and Pandey, 2013; Panche et al., 2016). Flavonoids possessing antiviral activity against some viruses, including coronavirus, were also taken into consideration (Kaul et al., 1985; Bae et al., 2000; Li et al., 2005; Kiat et al., 2006; Savi et al., 2010; Zandi et al., 2011; Frabasile et al., 2017; Yang et al., 2017; Zakaryan et al., 2017; Jucá et al., 2018; Sarwar et al., 2018; Song, 2018; Dai et al., 2019; Jo et al., 2020). However, no study on the molecular level of flavonoids has been reported against nCoV-SP, so over 100 different flavonoids were considered in this computational study against RBD of nCoV-SP affinity towards spike protein and probable interfere with the viral-receptor binding and decrease the viral load and infection. SMILES of the flavonoids were taken from the PubChem database, and the smi file of the compounds was generated and stored in a single file. The smi file was converted to pdbqt (AutoDock PDBQT format) and mol (MDL MOL format) file by OPENBABEL, a chemical file format converter according to our molecular docking protocols.
2.5. Molecular docking of flavonoids against RBD of nCoV-SP and human ACE-2 receptor (ACE-2)
For better reliability and precise results, two docking software, PyRx (Dallakyan and Olson, 2015) and iGEMDOCK (Hsu et al., 2011), utilizing two different algorithms, were used. PyRx uses AutoDock 4 and AutoDock Vina as docking software implying the Lamarckian Genetic Algorithm and Empirical Free Energy Scoring Function. On the other hand, iGEMDOCK implies the Generic Evolutionary Method for molecular docking, limiting the risk of false-positive results from a ligand library file efficiently (Raj et al., 2014). iGEMDOCK tool is utilized for docking purposes providing the interface for every single ligand and binding pocket of the targeted protein for interaction. Using the PyRx platform, macromolecule of the receptor-binding domain (RBD) of nCoV-SP as well as ACE-2 receptor protein and ligands were prepared. Docking was carried out taking the binding site residues for both RBD of nCoV-SP protein and ACE-2 inside a grid box with co-ordinates along X, Y, and Z-axis and dimensions conformed to 35.81 Å × 16.23 × Å ×31.14 Å and 23.13 Å × 12.03 Å × 8.22Å, respectively. On the other hand, 'Stable docking' with population size (N = 800), 80 generations, and ten solutions were carried out with our choice of ligands in iGEMDOCK against both the protein models (Raj et al., 2014; Kashif et al., 2017). Interactions between the RBD of nCoV-SP and ACE-2 with the ligands were studied using Ligplot (Wallace et al., 1995) and visualized in PyMol Molecular Visualization Software (Lill and Danielson, 2011).
2.6. Pharmacological analysis
The pharmacological analysis of the selected flavonoids included Lipinski's rule of five (Lipinski, 2004), drug-likeness, and ADMET studies. The analysis was carried with various software like Lipinski's rule of 5, Molsoft L.L.C.: Drug-Likeness and molecular property prediction for drug-likeness, and admetSAR prediction tool for ADMET studies.
2.7. Molecular dynamics (MD) simulation
The protein-ligand complexes were considered for Molecular dynamics simulation. GROMACS was used to do the MD simulation of RBD-Hesperidin, RBD-Naringin, RBD-EGCG, and ACE2-Quercetin (Van Der Spoel et al., 2005). Gromacs topology files were created individually. The topology and force field's parameter files were constructed by Automated Topology Builder (ATB v1.0) webserver (Malde et al., 2011), along with a 1.0 nm side's cubic box within which the RBD-ACE-flavonoid complexes were centered. Before the energy minimization of the protein-ligand complexes, the steepest descent and conjugate gradient methods were followed (Knyazev and Lashuk, 2008). AMBER force fields were used to detect all the bonded and non-bonded interactions of our query complex. The charge of the complete system was neutralized by the addition of ions before minimization. The particle mesh Ewald (PME) method was used to calculate the system's energy (Darden et al., 1993; Wang et al., 2010). Ewald summation was utilized at a reckoning cost computationally, while the Linear constraint solver (LINCS) (Hess et al., 1997) was applied for covalent bond constraints. Finally, the MD simulation was carried at 300K temperatures and one bar pressure, where Berendsen's weak coupling method regulated the system's pressure and temperature (Syed et al., 2017). MD simulation was done for 100 ns, and trajectories files were saved for further analysis using Xmgrace.
3. Results
3.1. Homology modeling and model validation
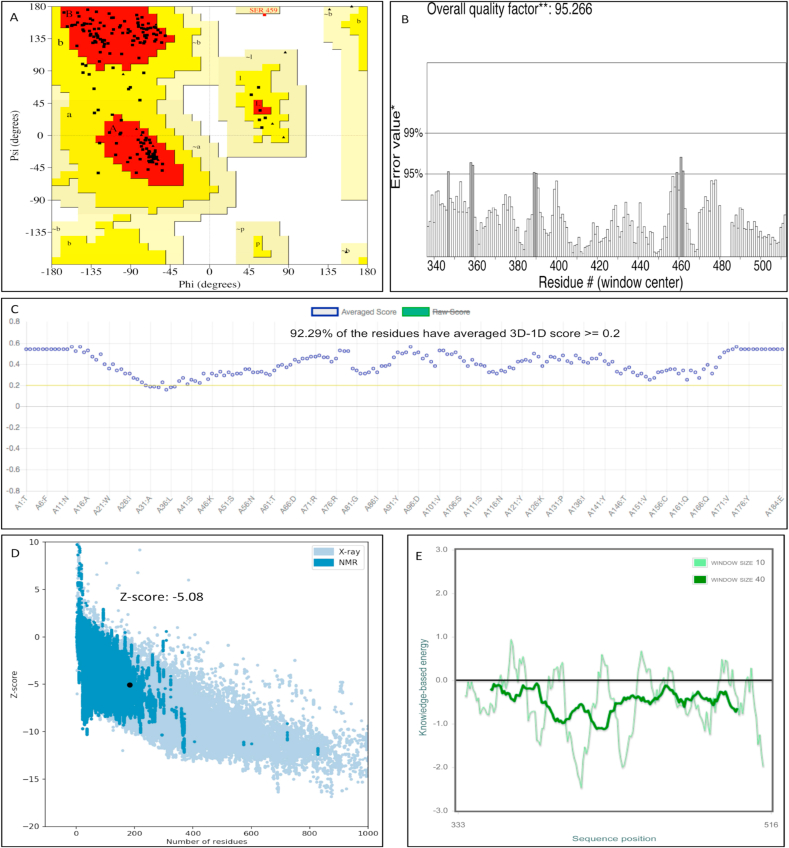
Sequence similarity between the template and the query sequence determines the modeled protein (Azam et al., 2014). Protein BLAST evaluated SARS-CoV spike glycoprotein (PDB ID: 2GHV) with a sequence similarity of 73% with our query sequence of RBD of nCoV-SP and, hence, was selected as the template to build the protein model of RBD of nCoV-SP. MODWEB (Eswar et al., 2003) was used to build the model and validated by the Ramachandran plot using PROCHECK (Laskowski et al., 1993). Model number 2 was selected for further studies as it had 99.4% residues covering favored, additionally allowed, and generously allowed regions while only 0.6 % in the disallowed region, as shown in Figure 2A.
Figure 2.
Modeled structure (RBD of nCoV-SP) validation by Procheck and ProSa. (A) Ramachandran plot of the optimized model. (B) ERRAT plot of RBD of the nCoV-SP model. Black bars represent a distantly located misfolded region from the active site; gray bars display the error region between 95% and 99%, and white bars represent regions with a lower rate of protein folding. (C) Verify3D plot of the RBD of nCoV-SP structure. (D) ProSA-web z-scores of all protein chains in PDB is determined by X-ray crystallography (light blue) or NMR spectroscopy (dark blue) regarding their length. The z-score of RBD of nCoV-SP is -5.08 and is highlighted with a black dot and (E) ProSA-web energy plot of RBD of nCov-SP. A thick line represents average energy over each 40-residue fragment. The thin line represents average energy with a window size of 10 residues in the plot's background.
YASARA was used for energy minimization for both model 2 and human ACE-2 receptors (PDB ID: 1R42). Verify3D, Errat, and ProSa further evaluated the optimized structure of and validated it (Colovos &Yeates, 1993; Eisenberg et al., 1997; Wiederstein and Sippl, 2007; Krieger et al., 2009). The overall quality of ERRAT and Verfy3D for the energy-minimized model was found to be 95.26% & 92.29%, respectively (Figure 2B and C) with a z-score of -5.08 (Figure 2D) and energy plots by ProSa (Figure 2E).
3.2. Binding site identification
Previous studies on RBD of nCoV-SP revealed TYR 449, GLN 493, GLN 498, THR 500, ASN 501, and GLY 502 to interact with human ACE-2 receptor with TYR 505 well exposed for initial contact with ACE-2 receptor (Xu et al., 2020; Senathilake et al., 2020). Other studies suggested LEU 455, PHE 486, GLN 493, and SER 494 residues of RBD of SARS-CoV/SARS-CoV-2 S protein to interact with hotspot 31 (LYS 31), whereas residues ASN 487 and SER 494 are described to interact with hotspot 353 (LYS 353) of human ACE-2 receptor (Wu et al., 2011; Shang et al., 2020; Wan et al., 2020). Previously published data suggest that hotspot 31 is made up of a salt bridge between LYS 31 and GLU 35, and hotspot 353 comprises of a salt bridge between LYS 353 and ASP 38, both buried in a hydrophobic environment (Wu et al., 2011). It is also reported that SER 494 of RBD of nCoV-SP reinforces the structural stability of hotspot 353 (LYS 353) of the ACE-2 receptor (Wu et al., 2011) and hence justified the importance of these residues for selection criteria as our binding site residues in this study (Table 1) and carry out our docking studies for interaction analysis.
Table 1.
Summary of binding sites of the model (RBD of nCoV-SP) as well as human ACE-2 receptor from literature.
| Binding site for RBD of CoV-SP and human ACE-2 | Residues at the binding site | References |
|---|---|---|
| RBD binding site 1 | TYR 449, GLN 493, GLN 498, THR 500, ASN 501, GLY 502, TYR 505 | (Xu et al., 2020) |
| RBD binding site 2 | LEU 455, PHE 486, GLN 493, ASN 487, SER 494 | (Wan et al., 2020) |
| RBD binding site 3 | TYR 449, GLN 493, SER 494, GLN 498, THR 500, ASN 501, GLY 502, TYR 505 | (Senathilake et al., 2020) |
| ACE-2 binding site | LYS 31, GLU 35, ASP 38, LYS 353 | (Wu et al., 2011) |
3.3. Docking result of the flavonoids with receptor binding domain (RBD) of nCoV-SP
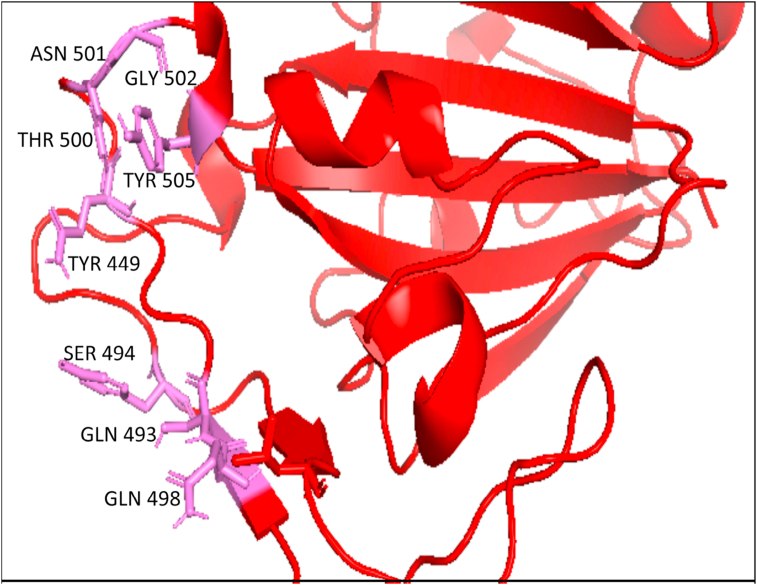
Computational chemistry holds a firm position in predicting and analyzing the correct mode of ligands' binding pattern into a protein-binding site (Azam et al., 2014). This study compiled the previous knowledge on the interaction of the RBD of nCoV-SP with the human ACE-2 receptor and indicated the residues TYR 449, GLN 493, SER 494, GLN 498, THR 500, ASN 501, GLY 502, and TYR 505 to be much more appealing toward the human receptor (Senathilake et al., 2020). The Pymol visualization of the selected binding site residues of RBD of nCoV-SP is depicted in Figure 3.
Figure 3.
Pymol visualization of the important residues (purple) in RBD of nCoV-SP (red) participates in interaction with the human ACE-2 receptor.
Docking with PyRx showed excellent and satisfactory results with the flavonoids. Hesperidin showed best-docked energy, followed by naringin and epigallocatechin gallate (EGCG). Hesperidin showed the lowest binding energy of -8.6 kcal/mol, followed by naringin and ECGC with energy -8.3 kcal/mol and -8.1 kcal/mol, respectively (Table 2), whereas docking with iGEMDOCK revealed EGCG to be the best flavonoid with binding energy -145.74 kcal/mol followed by hesperidin and naringin with energy -139.72 kcal/mol and -136.03 kcal/mol (Table 3).
Table 2.
Binding energy values (docking score) and interactions between the top 3 flavonoids and key residues of RBD of nCoV-SP in PyRx along with the respective H-bonds and bond lengths.
| Flavonoids | Binding energy (kcal/mol) | Key residues interraction | H-bonds | Bond length (Å) |
|---|---|---|---|---|
| Hesperidin | -8.6 | SER 494 | O6-O | 2.71 |
| TYR 505 | O11-OH | 3.05 | ||
| Naringin | -8.3 | GLN 493 | O14-OE1 | 3.09 |
| SER 494 | O11-O | 2.92 | ||
| Epigallocatechin gallate (EGCG) | -8.1 | GLN 498 | O6-NE2 | 2.81 |
| ASN 501 | O8-ND2 | 3.23 |
Table 3.
Docking results of top 3 flavonoids against RBD of nCoV-SP by iGEMDOCK tool.
| Flavonoids | Energy (kcal/mol) | VDW (kcal/mol) | H-Bond (kcal/mol) |
|---|---|---|---|
| Hesperidin | -139.72 | -102.32 | -37.40 |
| Naringin | -136.03 | -99.25 | -36.75 |
| Epigallocatechin gallate (EGCG) | -145.74 | -109.51 | -47.22 |
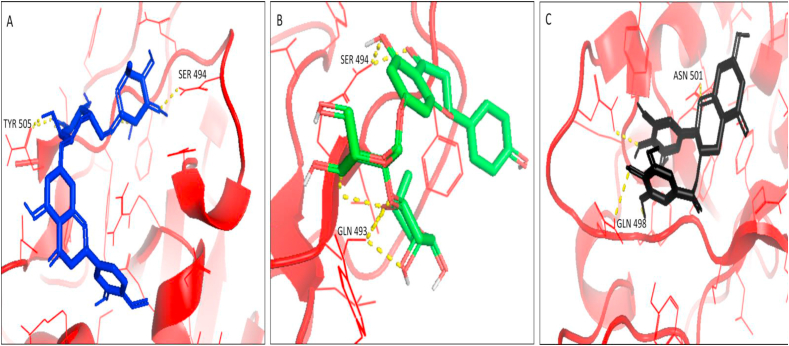
Thus, docking with PyRx and iGEMDOCK showed reliable outputs and indicated a strong affinity of these considered flavonoids with the RBD of nCoV-SP and indicator of a potential inhibitor of SARS-CoV-2. The best docking poses of Hesperidin, Naringin and ECGC were shown in Figures 4A, 4B, and 4C, respectively.
Figure 4.
Pymol visualization of interaction (Docking results of top 3 ligands). (A) flavonoid hesperidin (blue) with residues SER 494 and TYR 505. (B) flavonoid naringin (green) with residues SER 494 and GLN 493. (C) the flavonoid EGCG (black) with residues GLN 4948 and ASN 501 of RBD of nCoV-SP (red). H-bonds are depicted in yellow.
3.4. Interaction analysis of the flavonoids with receptor binding domain (RBD) of nCoV-SP
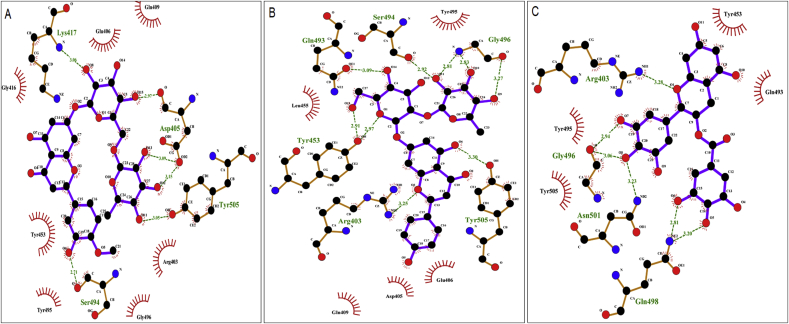
Interaction studies between flavonoids and RBD of nCoV-SP were carried out with Ligplot (Wallace et al., 1995) and visualized by PyMol (Lill and Danielson, 2011). Ligplot studies showed that hesperidin formed H-bonds with residues ASP405, LYS417, SER494, and TYR505 of RBD of nCoV-SP (Figure 5A). Naringin formed H-bonds with GLN 493 and SER 494, whereas EGCG formed H-bonds with the residues GLN 498 and ASN 501 apart from other residues in the RBD of nCoV-SP (Figures 5B and 5C, respectively). These data demonstrate the affinity of these flavonoids to RBD of nCoV-SP.
Figure 5.
Diagrammatic sketch illustrating the interaction between binding site residues of RBD of nCoV-SP and top 3 ligands of (A) flavonoid hesperidin (B) flavonoid naringin (C) the flavonoid EGCG by LigPlot. Ligand is shown in purple and: green dashed lines indicate hydrogen bonds with distance in angstrom (Å), spoked red arcs indicate hydrophobic contacts, atoms are shown in black for carbon, Ligand is shown in purple, and: green dashed lines indicate hydrogen blue for nitrogen and red represents oxygen.
Previous computational studies on RBD of nCoV-SP revealed the importance of amino acid residues and their interaction with ACE-2 receptor with residues TYR449, THR 500, GLN493, GLN498, ASN501, and GLY502 to interact with human ACE-2 with TYR505 well exposed for initial contact with ACE-2 receptor (Senathilake et al., 2020). Another study revealed the two hotspots' recognition in the human ACE-2 receptor by RBD of nCoV-SP by the residue GLN493 and LEU 455, which recognize the hotspot LYS 31 whereas residues LEU 455, PHE 486, and SER 494 recognize hotspot LYS 353 (Wu et al., 2011). It is also documented that SER494 of RBD of nCoV-SP strengthens the structural stability of hotspot LYS 353 of the human ACE-2 receptor (Wan et al., 2020). Thus, interaction analysis of our three best flavonoids selected for further pharmacological analysis indicated a precise and efficient mode of binding and utilizing critical residues, i.e., GLN 493, SER 494, GLN 498, ASN 501, and TYR 505 of RBD of nCoV-SP, and might interfere with the binding of the spike protein with human ACE-2 receptor and further loading of the virus on to its respective human receptors. might contribute to the inhibition of binding of the S protein with human ACE-2 receptor.
3.5. Docking result of the flavonoids with human ACE-2 receptor
Docking result of the flavonoids with ACE-2 receptor also showed an excellent result with quercetin to be the best docked among other flavonoids followed by epigallocatechin gallate (EGCG) and gallocatechin gallate (GCG) with the docked value of -6.0 kcal/mol, -4.5 kcal/mol and -4.3 kcal/mol respectively in PyRx (Table 4). Docking with the iGEMDOCK tool also revealed quercetin as the best flavonoid with binding energy -144.09 kcal/mol, followed by EGCG and GCG with energy -131.15 kcal/mol and -130.56 kcal/mol, respectively (Table 5). The best docking pattern of quercetin was shown in Figure 6. Interactions were further examined for bond lengths and Hydrogen bonds, as illustrated in Ligplot, as shown in Figure 7.
Table 4.
Binding energy values (docking score) and interactions between the top 3 flavonoids and key residue of human ACE-2 receptor in PyRx along with the respective H-bond and bond length.
| Flavonoids | Binding energy (kcal/mol) | Key residue interraction | H-bond | Bond length (Å) |
|---|---|---|---|---|
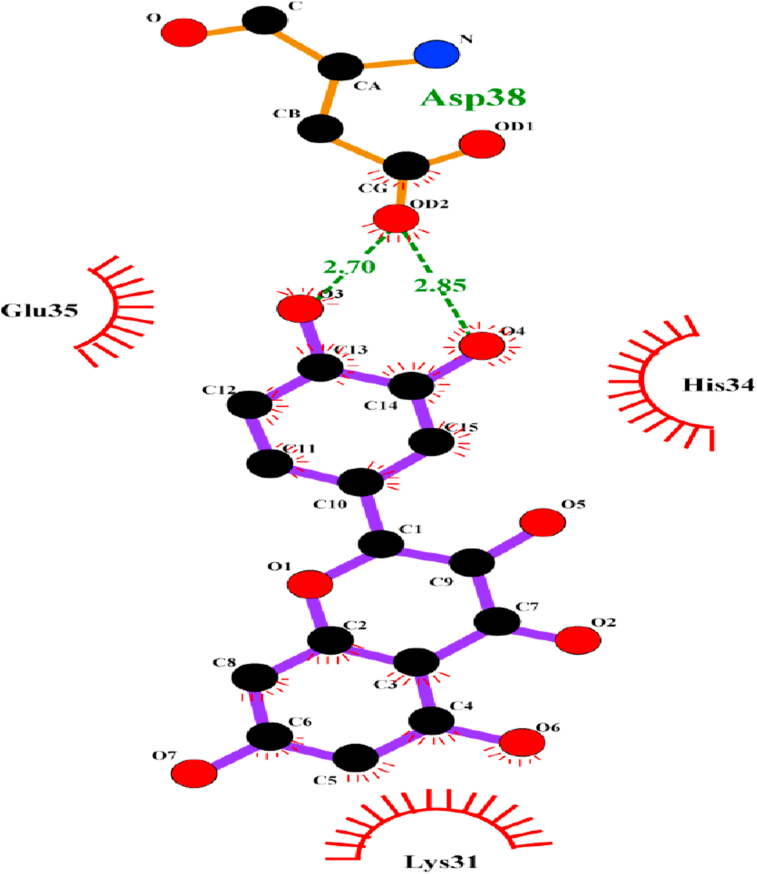
| Quercetin | -6.0 | ASP 38 | O3-OD2 | 2.70 |
| Epigallocatechin gallate (EGCG) | -4.5 | ASP 38 | O11-OD2 | 2.82 |
| Gallocatechin gallate (GCG) | -4.3 | ASP 38 | O11-OD2 | 2.73 |
Table 5.
Docking results of top 3 flavonoids against human ACE-2 receptor by iGEMDOCK tool.
| Flavonoids | Energy (kcal/mol) | VDW (kcal/mol) | H-Bond (kcal/mol) |
|---|---|---|---|
| Quercetin | -144.09 | -92.46 | -51.64 |
| Epigallocatechin gallate (EGCG) | -131.15 | -91.36 | -23.4 |
| Gallocatechin gallate (GCG) | -129.56 | 84.18 | -18.32 |
Figure 6.
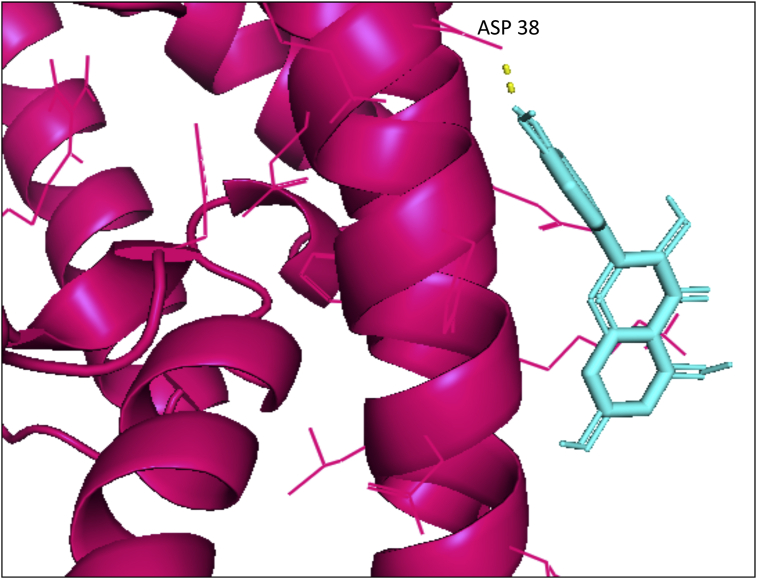
Pymol visualization of the interaction of the flavonoid quercetin (cyan) with the important residue ASP 38 of human ACE-2 receptor (pink). H-bonds are depicted in yellow.
Figure 7.
Diagrammatic sketch illustrating the interaction between binding site residues of ACE-2 receptor and flavonoid quercetin by LigPlot. Ligand is shown in purple and: green dashed lines indicate hydrogen, blue for nitrogen, and red represents oxygen.
Hesperidin and naringin did not show any efficiency and other flavonoids with no affinity or weak energy towards the ACE-2 receptor in both the docking software. Thus, the docking results indicated quercetin to be much more efficient towards the human ACE-2 receptor than RBD of nCoV-SP. The docking score of quercetin with RBD of nCoV-SP was -5.3 kcal/mol and -97.15 kcal/mol in PyRx and iGEMDOCK, respectively. This indicates that quercetin might act by blocking the human ACE-2 receptor, thus preventing interaction upon viral entry with their respective target.
3.6. Interaction analysis of flavonoids with human ACE-2 receptor
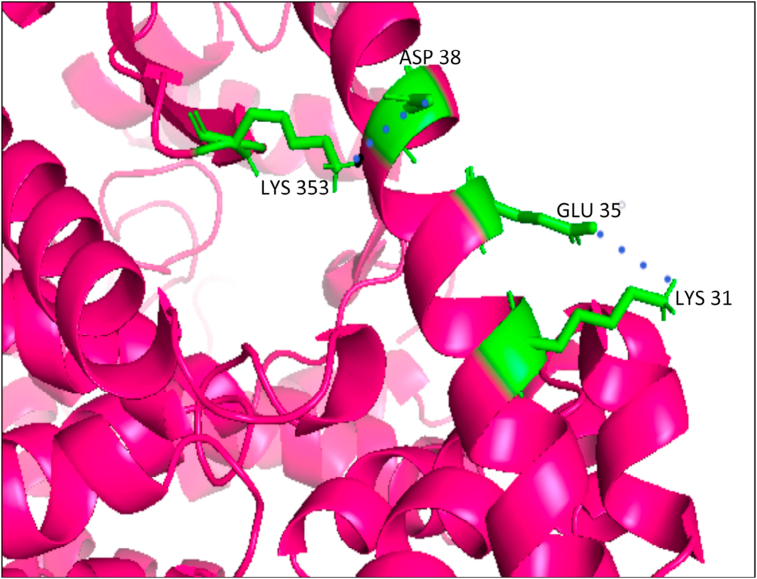
Interaction studies revealed quercetin to interact with the residue ASP38 of human ACE-2 that formed the salt bridge with LYS 353 and forms the hotspot LYS353, which is utilized by the residues LEU 455, PHE 486, and SER 494 of RBD of nCoV-SP, SER 494 additionally enhancing the stability of this hotspot (Figure 8) (Wu et al., 2011). In human ACE-2, without any viral control or hold, LYS353 does not form any salt bridge with ASP 38, clearly indicating that the virus binding hotspot does not pre-exist on human ACE-2, and the salt bridge is only induced upon viral entry and load (Wu et al., 2011). ECGC and GCG were also found to form H- bond with ASP 38 of the human ACE-2 receptor with a relatively low binding efficiency than that of their binding efficiency and affinity towards the RBD receptor, as noticed in PyRx and iGEMDOCK docking software. Though with their low binding efficiency, we do not nullify that they also bind with the human ACE-2 receptor, thus interfering with the viral load and interaction with the human receptor.
Figure 8.
Pymol visualization of the important residues (green) in human ACE-2 receptor (pink) with the residues participating in a salt bridge (shown in blue dots) formation.
3.7. Pharmacological analysis
Pharmacological analyses of the selected flavonoids based on Lipinski's rule of five were performed and are listed in Table 6. While EGCG and quercetin satisfied the Lipinski rule very well, violations were observed with hesperidin and naringin in properties such as molecular mass, hydrogen bond donor, hydrogen bond acceptor, and molar refractivity. Lipinski's rule of five indicated hesperidin and naringin to be a poor drugs for an oral route. Nevertheless, previous experiments suggested that both hesperidin and naringin possess inhibitory activity in vitro against infection with rotavirus, where hesperidin showed the maximum inhibition among the flavonoids followed by naringin with an IC50 value of 10μM and 25μM, respectively (Bae et al., 2000). Thus, the two flavonoids' strong antiviral efficacy could not be underestimated and, therefore, was considered for further pharmacological analysis. The water: octanol partition coefficient (LOGP) of the considered flavonoids suggested a good permeability across the membrane with a value of less than 5 (Table 6) (Lipinsk, 2004).
Table 6.
The ligand parameters to satisfy Lipinski's rule of 5 with a drug-likeness score by Molsoft L.L.C.: Drug-Likeness and molecular property prediction of the selected flavonoids after docking.
| Flavonoids | Mass (<500) | Hydrogen bond donor (<5) | Hydrogen bond acceptor(<10) | LOGP (<5) | Molar Refractivity (40–130) | Drug likeness (>0) |
|---|---|---|---|---|---|---|
| Hesperidin | 610.00 | 8 | 15 | -1.15 | 140.69 | 0.94 |
| Naringin | 580.00 | 8 | 14 | -1.16 | 134.14 | 1.05 |
| Epigallocatechin gallate (EGCG) | 458 | 8 | 11 | 2.23 | 108.92 | 0.23 |
| Quercetin | 302.00 | 5 | 7 | 2.01 | 74.05 | 0.52 |
Compounds with negative drug scores are generally not considered potential drug candidates (Chandrasekaran & Thilak Kumar, 2016). All the selected flavonoids showed outstanding drug-likeness drug scores in Molsoft L.L.C.: Drug-Likeness and molecular property prediction (http://www.molsoft.com/mprop/) and are listed in Table 6. Naringin possessed the best drug-likeness score of 1.05, followed by hesperidin and quercetin, respectively, of 0.94 and 0.52.
The ADMET properties of the flavonoid compounds revealed through admetSAR are presented in Table 7. The solubility of the flavonoids was checked and found that all the selected flavonoids possessed optimal water solubility (>-4) (Mokhnache et al., 2019). The selected flavonoids have excellent human intestinal absorption (HIA) and good permeability values in Caco2 permeability analysis (Caco-2 permeability is a technique that calculates the degree of a flux of a compound throughout polarized Caco-2 cell monolayers and it can be used to calculate in vivo absorption of drugs). They are expected to be highly absorbed in the intestine as they possessed an excellent score and indicated high absorption upon oral administration (Table 7) (Sood et al., 2018). Hesperidin showed the lowest HIA value and indicated a little poor absorption due to its high molecular weight. A lower molecular weight drug boosts the absorption rate, and thus, most of them are kept at their lowest possible molecular weight. Moreover, the flavonoids showed no characteristics of inhibiting P-glycoprotein (P-gp). The blood-brain barrier (BBB) value of the flavonoids was excellent, indicating better penetration and distribution across the barrier (Table 7).
Table 7.
ADMET profile of the selected flavonoids along with the controls performed in admetSAR.
| Inhibitors | Log S (>-4) | Blood Brain Barrier (BBB) | Human Intestinal Absorption (HIA) | P-glycoprotein substrate/inhibitor | CYP substrate/inhibitor | AMES toxicity | Carcinogenecity | LD50 (Rat Acute Toxicity) (mol/kg) |
|---|---|---|---|---|---|---|---|---|
| Hesperidin | -2.64 | 0.94 | 0.63 | Substrate/Non-inhibitor | Substrate/Non-inhibitor | Non-toxic | Non-carcinogen | 2.62 |
| Naringin | -2.53 | 0.84 | 0.86 | Substrate/Non-inhibitor | Non-substrate/Non-inhibitor | Non-toxic | Non-carcinogen | 2.26 |
| Quercetin | -2.99 | 0.57 | 0.96 | Substrate/Non-inhibitor | Non-substrate/Inhibitor | Non-toxic | Non-carcinogen | 3.02 |
| Epigallocatechin gallate (EGCG) | -3.31 | 0.60 | 0.88 | Substrate/Non-inhibitor | Non-substrate/Non-inhibitor | Non-toxic | Non-carcinogen | 2.66 |
| Gallocatechin gallate (GCG) | -3.31 | 0.60 | 0.88 | Substrate/Non-inhibitor | Non-substrate/Non-inhibitor | Non-toxic | Non-carcinogen | 2.66 |
| Remdevisir | -3.47 | 0.74 | 0.88 | Substrate/Inhibitor | Substrate/Non-inhibitor | Non-toxic | Non-carcinogen | 2.71 |
| Chloroquine | -4.34 | 0.74 | 0.99 | Substrate/Non-inhibitor | Substrate/Non-inhibitor | Non-toxic | Non-carcinogen | 2.95 |
| Hydroxychlhoroquine | -3.56 | 0.56 | 0.98 | Substrate/Inhibitor | Non-substrate/Non-inhibitor | Non-toxic | Non-carcinogen | 2.63 |
In terms of metabolism, we found that only hesperidin was a substrate (but non-inhibitor) of cytochrome P450 (CYP 450), indicating its proper metabolism by CYP450, while only quercetin was a non-substrate but inhibitor of CYP 450 (Table 7). A non-inhibitor of CYP450 means that the molecule will not hamper the biological transformation of drugs metabolized by the CYP 450 enzyme (Nisha et al., 2016). All the flavonoids were non-AMES toxic and non-carcinogens (Table 7).
Comparing the predicted LD50 doses, a compound with a lower dose is more lethal than the compound having a higher LD50 (Nisha et al., 2016). From our observation in admetSAR, we found that all the flavonoid compounds have optimal LD50 doses indicating they are nonlethal (Table 7).
All the ADMET parameters of the selected flavonoids were compared with the controls taken into consideration in this study, i.e., remdesivir, chloroquine, and hexochloroquine that are currently used to treat COVID-19 (Norkin, 1995; Wang et al., 2020). Our selected flavonoids showed excellent values in the ADMET studies, with some even fulfilling the parameters much better than the ongoing clinical drugs and are listed in Table 7.
3.8. Molecular dynamic (MD) simulation studies
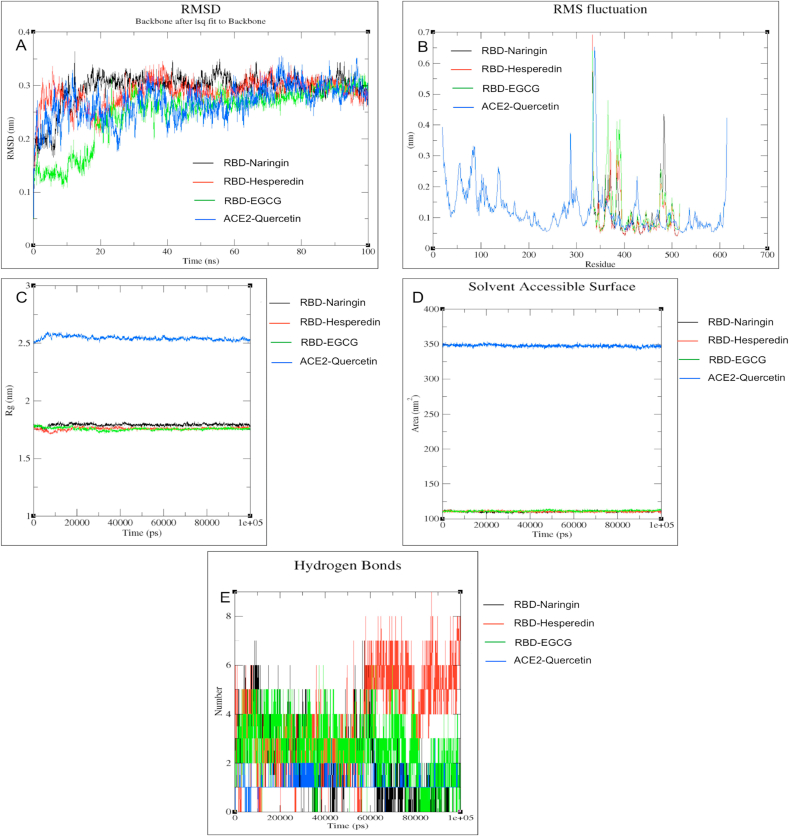
Based on the molecular docking and pharmacokinetics study, the top and the best flavonoids against RBD and ACE-2 proteins were considered for MD simulation investigations. MD simulation for the protein-ligand complex was carried out using GROMACS (Van Der Spoel et al., 2005). Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuations (RMSF) were examined for seeing the deviation of Cα atoms of the protein from its backbone and also the fluctuations correlated with the protein residues throughout the MD simulation (Malathi et al., 2019; Li et al., 2019). MD simulation results show that the RMSD for the protein-ligand complexes, fluctuations for RBD-EGCG were observed only during the initial 20ns MD simulation apart from the other protein-ligand complexes, which showed less fluctuation and more stability up to 100ns simulation (Figure 9A-B). RMS fluctuations for the protein-ligand complexes, including RBD-EGCG, were less than 0.4 nm, which ultimately showed the complexes' remarkable structural stability (Jeyaram et al., 2019; Zhang et al., 2020).
Figure 9.
The 100 ns MD results of four protein-ligand complexes (RBD-Hesperidin, RBD-Naringin, RBD-EGCG, and ACE2-Quercetin complex). (A) RMSD value of carbon alpha atoms of the complex. (B) RMSF of carbon alpha of complex structure. (C) Rg of backbone atoms. (D) SASA of the ligands. (E) Total number of H-bonds between ligands and Protein.
The RMSF analysis of the RBD and ACE-2 protein was studied to understand the protein-ligand complex's fluctuations and flexibility. The RMSF results showed the fluctuations of the protein's individual residues in the presence of the ligand in bound conformation. RMSF fluctuations were declined and less during the initial 50 residues for the protein-ligand complex for quercetin, which indicated ACE-2-Quercetin to be stable as the residue ASP 38 was the binding site at ACE-2 taken into account in this study (Figure 9A-B). The residues from 490-510, centered as the binding site residues for RBD protein that interact with the ACE-2 receptor protein, showed the lowest fluctuations for the complex RBD-Hesperidin followed by RBD-EGCG with fluctuations value less than 0.3 nm. Results showed that these residues and active site residues for ACE-2 are relatively rigid and stable and hence displayed outstanding stability in the dynamic environment (Figure 9B) (Bhowmik et al., 2020; Suvannang et al., 2011).
Further, all four complex conformation stability was determined by the radius of gyration (Rg), which defines the protein's structural compactness. The average Rg values for RBD-EGCG, RBD-naringin, and RBD-hesperidin complex were calculated as 1.75 nm, whereas Rg for ACE2-Quercetin complex was calculated as 2.6 nm (Figure 9C). The Rg plot shows no fluctuation throughout the simulation for all four complexes (Figure 9C).
The changes in complexes' structural features have also been analyzed by computing the solvent-accessible surface area (SASA). Thus, it helps to calculate protein-inhibitor complexes' structural stability under the solvent environment. Figure 9D shows that the RBD-EGCG, RBD-naringin, and RBD-hesperidin complexes have average SASA values around 110 nm2. Simultaneously, the SASA value for the ACE2-quercetin complex has average SASA values of 350 nm2 (Figure 9D).
The hydrogen bond investigation through the gmx H-bond revealed stable and effective hydrogen bond interaction of EGCG with RBD followed by the RBD-Hesperidin and RBD-Naringin complex. EGCG. Naringin and hesperidin could form a hydrogen bond with the binding site residues without leaving the pocket throughout the 100ns MD simulation run (Figure 9E). On the other hand, quercetin formed at least one H-bond with ACE-2 during the 100ns simulation run (Figure 9E). Thus, MD simulation of our RBD-ACE-2-flavonoids complexes leads to establishing our complexes to be basically and structurally stable and reliable (Bhowmik et al., 2020).
4. Discussions
The present study emphasizes modeling the receptor-binding domain (RBD) of nCoV-SP and generating a stable protein model. Flavonoids with their broad biological activity, including antiviral activity against other viruses as well as SARS (Kaul et al., 1985; Bae et al., 2000; Kiat et al., 2006; Zandi et al., 2011; Frabasile et al., 2017; Yang et al., 2017; Zakaryan et al., 2017; Jucá et al., 2018; Sarwar et al., 2018; Song., 2018; Dai et al., 2019; Jo et al., 2020) were taken under consideration in this study, as molecular works of flavonoids against SARS-CoV and SARS-CoV-2 have not been done yet.
The present study focused on the importance of densely glycosylated spike protein of the RBD of nCoV-SP that binds with the human ACE-2 receptor, ultimately forming the RBD-ACE-2 complex (Hoffmann et al., 2020; Wrapp et al., 2020). Viruses or viral proteins infect by attaching to their specific receptor on a susceptible host cell's surface and thus prepare their way for cell entry (Norkin, 1995). Thus, it led to the perspective of molecular docking and interaction analysis of the flavonoids against both RBD of nCoV-SP and human ACE-2 receptors. Homology model of RBD of nCoV-SP was prepared by MODWEB (Eswar et al., 2003) with PDB ID: 2GHV as a template for its high similarity sequence (73%) with our query protein while PDB ID: 1R42 for human ACE-2 receptor was considered, and the energy minimized YASARA generated model for both the protein.
A previous study revealed the essential residues in the RBD of nCoV-SP that interact with the viral hotspots in human ACE-2 receptors (Shang et al., 2020; Wan et al., 2020). It is also experimentally determined that the viral hotspots are formed only upon viral entry and are not preexistent (Wu et al., 2011), strongly favoring our consideration of these two proteins for docking analysis.
Docking studies with PyRx (Dallakyan and Olson, 2015) and iGEMDOCK (Hsu et al., 2011) provided a reliable and extremely satisfying result. Hesperidin, naringin, and EGCG showed the best docking result with good binding energy in both the docking software than other flavonoids under consideration, indicating their strong affinity against the RBD of nCoV-SP. Interaction analysis revealed hesperidin, naringin, and EGCG to form H-bond with some critical residues in the RBD of nCoV-SP that interact with the ACE-2 receptor indicated that these selected flavonoids might interfere with the viral binding onto its respective receptors.
On the other hand, quercetin was evaluated as the best-docked flavonoid against the human ACE-2 receptor (PDB ID: 1R42) in both the docking software. Thus, docking results indicated quercetin to have a high affinity to interact with ACE-2 receptor than RBD of nCoV-SP; thus, it indicated another probable route of viral inhibition by blocking the respective receptors for viral interaction and inhibit from cell entry.
Hesperidin and naringin were involved in Lipinski's violation of pharmacological analysis but were already experimented with antiviral activity against rotavirus infection (Bae et al., 2000). So their antiviral property and efficacy cannot be neglected, and further pharmacological analysis was concluded. EGCG and quercetin gently passed the Lipinski's violation with good drug scores listed in Table 6.
All the ADMET parameters of the selected flavonoids were compared with the controls taken into consideration in this study, i.e., remdesivir, chloroquine, and hexochloroquine that are currently used to treat COVID-19 (Gautret et al., 2020; Wang et al., 2020). Our selected flavonoids showed good values in the ADMET studies, with some even fulfilling the parameters much better than the ongoing clinical drugs and are listed in Table 7.
The selected flavonoids were even compared with some controls to better evaluate our selected flavonoids as potent drug candidates against nCoV-SP and quercetin to potentially block the ACE-2 receptor, which might interfere with the viral receptor interaction and inhibit viral from cell entry. Remdesivir, chloroquine, and hexochloroquine were taken under this study as the control for their current use in clinical trial treating COVID-19 (Norkin, 1995; Wang et al., 2020), and to our concern, our selected flavonoids showed good values in the ADMET studies, with some even fulfilling the parameters much better than the ongoing clinical drugs and were listed in Table 7.
Lastly, MD simulation results support that the strong interaction of EGCG followed by hesperidin and naringin with the RBD protein while quercetin with ACE-2 receptor through H-bonding. The protein-ligand complex was stable during the 100 ns simulation run as interpreted by the RMSD and RMSF plots (Figure 9A-B). The present in-silico study proposed that EGCG, hesperidin, and quercetin may be potent inhibitors against RBD of nCoV-SP and human ACE-2 receptors eventually prevent the 2019-nCoV entries into the human cell.
5. Conclusion
The present study demonstrated that the flavonoid hesperidin, naringin, and ECGC were potent inhibitors of RBD of nCoV-SP, while quercetin demonstrated an affinity towards the human ACE-2 receptor. While hesperidin, naringin, and ECGC found to be more efficient towards the viral spike glycoprotein, quercetin was demonstrated to efficiently block the human ACE-2 receptor by binding to a vital residue ASP38 and hinders the formation of the salt bridge with LYS353 and ultimately the hotspot LYS353 which is well recognized by the virus. Binding to the residues ASP38 also holds a strong point that the viral hotspot is not recognized and forms only upon viral load and entry (Wu et al., 2011). On the other hand, hesperidin, naringin, and ECGC were found to interact with some of the essential residues of RBD of nCoV-SP that interact with the human ACE-2 receptor. Our in silico results revealed that selected flavonoids have excellent ADMET properties and may have inhibitory effects against 2019-nCoV. The MD simulation results suggested EGCG and hesperidin have strong interaction with RBD while quercetin with host receptor ACE-2 without leaving the pocket during simulation, as seen from RMSD and RMSF graph. This indicated our stronghold on inhibitors' choice and their potential interaction with the RBD of nCoV-SP and human ACE-2 receptors that might inhibit viral interaction with its respective receptors and further viral load. Nevertheless, further in vivo work would be needed to establish these inhibitors' effectiveness and surety against SARS-CoV-2.
Declarations
Author contribution statement
Deep Bhowmik, Rajat Nandi, Amresh Prakash, Diwakar Kumar: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by SERB-ECR, GOI grant (FILE NO. ECR/2015//000155) and DBT-Twinning, GOI grant (GOI) (order no: BT/PR16224/NER/95/176/2015 & BT/PR24504/NER/95/746/2017) to Dr. Diwakar Kumar. Deep Bhowmik received funding from the SERB-ECR grant (FILE NO. ECR/2015//000155), Whereas Rajat Nandi received funding from the DBT-Twinning grant (order no: BT/PR24504/NER/95/746/2017).
Data availability statement
Data included in article/supp. material/referenced in article.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- Andersen K.G., Rambaut A., Lipkin W.I. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam S.S., Sarfaraz S., Abro A. Comparative modeling and virtual screening for the identification of novel inhibitors for myo-inositol-1-phosphate synthase. Mol. Biol. Rep. 2014;41(8):5039–5052. doi: 10.1007/s11033-014-3370-8. [DOI] [PubMed] [Google Scholar]
- Bae E.A., Han M.J., Lee M., Kim D.H. In vitro inhibitory effect of some flavonoids on rotavirus infectivity. Biol. Pharm. Bull. 2000;23(9):1122–1124. doi: 10.1248/bpb.23.1122. [DOI] [PubMed] [Google Scholar]
- Bhowmik D., Jagadeesan R., Rai P., Nandi R., Gugan K., Kumar D. Evaluation of potential drugs against leishmaniasis targeting catalytic subunit of Leishmania donovani nuclear DNA primase using ligand based virtual screening, docking and molecular dynamics approaches. J. Biomol. Struct. Dynam. 2020:1–15. doi: 10.1080/07391102.2020.1739557. Advance online publication. [DOI] [PubMed] [Google Scholar]
- Burak M., Imen Y. Flavonoids and their antioxidant properties. TurkiyeKlin Tip BilDerg. 1999;19:296–304. [Google Scholar]
- Calzada F., Cedillo-Rivera R., Bye R., Mata R. Geranins C and D, additional new antiprotozoal A-type proanthocyanidins from Geranium niveum. Planta Med. 2001;67(7):677–680. doi: 10.1055/s-2001-17358. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran K., Thilak Kumar R. Molecular properties prediction, docking studies and antimicrobial screening of ornidazole and its derivatives. J. Chem. Pharmaceut. Res. 2016;8(3):849–861. [Google Scholar]
- Chang L.K., Wei T.T., Chiu Y.F., Tung C.P., Chuang J.Y., Hung S.K., Li C., Liu S.T. Inhibition of Epstein-Barr virus lytic cycle by (-)-epigallocatechin gallate. Biochem. Biophys. Res. Commun. 2003;301(4):1062–1068. doi: 10.1016/s0006-291x(03)00067-6. [DOI] [PubMed] [Google Scholar]
- Chen C., Qiu H., Gong J., Liu Q., Xiao H., Chen X.W., Sun B.L., Yang R.G. (-)-Epigallocatechin-3-gallate inhibits the replication cycle of hepatitis C virus. Arch. Virol. 2012;157(7):1301–1312. doi: 10.1007/s00705-012-1304-0. [DOI] [PubMed] [Google Scholar]
- Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., Bi J., Li F., Wang S., Huang X., Meng X., Sun B., Wang D., Kong W., Jiang C., Su W. Antiviral efficacy of flavonoids against enterovirus 71 infection in vitro and in newborn mice. Viruses. 2019;11(7):625. doi: 10.3390/v11070625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015;263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98(12):10089–10092. [Google Scholar]
- Eisenberg D., Lüthy R., Bowie J.U. VERIFY3D: assessment of protein models with three-dimensional profiles. Macromolecular Crystallography Part B. 1997;20:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- Eswar N., John B., Mirkovic N. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 2003;31(13):3375–3380. doi: 10.1093/nar/gkg543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frabasile S., Koishi A.C., Kuczera D., Silveira G.F., Verri W.A., Jr., Duarte Dos Santos C.N., Bordignon J. The citrus flavanone naringenin impairs dengue virus replication in human cells. Sci. Rep. 2017;7:41864. doi: 10.1038/srep41864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P., Lagier J.C., Parola P. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020:105949. doi: 10.1016/j.ijantimicag.2020.105949. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gescher K., Hensel A., Hafezi W., Derksen A., Kühn J. Oligomeric proanthocyanidins from Rumex acetosa L. inhibit the attachment of herpes simplex virus type-1. Antivir. Res. 2011;89(1):9–18. doi: 10.1016/j.antiviral.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Groot H., Rauen U. Tissue injury by reactive oxygen species and the protective effects of flavonoids. Fund. Clin. Pharmacol. 1998;12(3):249–255. doi: 10.1111/j.1472-8206.1998.tb00951.x. [DOI] [PubMed] [Google Scholar]
- Gorlach S., Fichna J., Lewandowska U. Polyphenols as mitochondria-targeted anticancer drugs. Canc. Lett. 2015;366(2):141–149. doi: 10.1016/j.canlet.2015.07.004. [DOI] [PubMed] [Google Scholar]
- Huang Q., Herrmann A. Fast assessment of human receptor-binding capability of 2019 novel coronavirus (2019-nCoV) bioRxiv. 2020 2020.02.01.930537. [Google Scholar]
- Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18(12):1463–1472. [Google Scholar]
- Hsu K.C., Chen Y.F., Lin S.R., Yang J.M. iGEMDOCK: a graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinf. 2011;12(Suppl 1):S33. doi: 10.1186/1471-2105-12-S1-S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor [published online ahead of print. Cell. 2020;S0092–8674(20):30229–30234. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranshahi M., Rezaee R., Parhiz H., Roohbakhsh A., Soltani F. Protective effects of flavonoids against microbes and toxins: the cases of hesperidin and hesperetin. Life Sci. 2015;137:125–132. doi: 10.1016/j.lfs.2015.07.014. [DOI] [PubMed] [Google Scholar]
- Jeyaram R.A., Priyadarzini T., Anu Radha C., Siva Shanmugam N.R., Ramakrishnan C., Gromiha M.M., Veluraja K. Molecular dynamics simulation studies on influenza A virus H5N1 complexed with sialic acid and fluorinated sialic acid. J. Biomol. Struct. Dynam. 2019;37(18):4813–4824. doi: 10.1080/07391102.2019.1568304. [DOI] [PubMed] [Google Scholar]
- Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzym. Inhib. Med. Chem. 2020;35(1):145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucá M.M., Cysne Filho F., de Almeida J.C. Flavonoids: biological activities and therapeutic potential. Nat. Prod. Res. 2018;34(5):692–705. doi: 10.1080/14786419.2018.1493588. [DOI] [PubMed] [Google Scholar]
- Kashif M., Manna P.P., Akhter Y., Alaidarous M., Rub A. Screening of novel inhibitors against Leishmania donovani Calcium ion channel to fight Leishmaniasis. Infect. Disord. - Drug Targets. 2017;17(2):120–129. doi: 10.2174/1871526516666161230124513. [DOI] [PubMed] [Google Scholar]
- Knyazev A.V., Lashuk I. Steepest descent and conjugate gradient methods with variable preconditioning. SIAM J. Matrix Anal. Appl. 2008;29(4):1267–1280. [Google Scholar]
- Kaul T.N., Middleton E., Jr., Ogra P.L. Antiviral effect of flavonoids on human viruses. J. Med. Virol. 1985;15(1):71–79. doi: 10.1002/jmv.1890150110. [DOI] [PubMed] [Google Scholar]
- Khan N., Syed D.N., Ahmad N., Mukhtar H. Fisetin: a dietary antioxidant for health promotion. Antioxidant redox signal. 2013;19(2):151–162. doi: 10.1089/ars.2012.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiat T.S., Pippen R., Yusof R. Inhibitory activity of cyclohexenyl chalcone derivatives and flavonoids of fingerroot, Boesenbergia rotunda (L.), towards dengue-2 virus NS3 protease. Bioorg. Med. Chem. Lett. 2006;16(12):3337–3340. doi: 10.1016/j.bmcl.2005.12.075. [DOI] [PubMed] [Google Scholar]
- Krieger E., Joo K., Lee J., Raman S., Thompson J., Karplus K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins. Structure, Function, and Bioinformatics. 2009;77(Suppl 9):114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Pandey A.K. Chemistry and biological activities of flavonoids: an overview. Sci. World J. 2013;162750:1–16. doi: 10.1155/2013/162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereo chemical quality of protein structures. J. Appl. Crystallogr. 1993;26(2):283–291. [Google Scholar]
- Lau K.M., Lee K.M., Koon C.M., Cheung C.S., Lau C.P., Ho H.M., Lee M.Y., Au S.W., Cheng C.H., Lau C.B., Tsui S.K., Wan D.C., Waye M.M., Wong K.B., Wong C.K., Lam C.W., Leung P.C., Fung K.P. Immunomodulatory and anti-SARS activities of Houttuynia cordata. J. Ethnopharmacol. 2008;118(1):79–85. doi: 10.1016/j.jep.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.I., Hsueh P.R. Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. J. Microbiol. 2020:30011–30016. doi: 10.1016/j.jmii.2020.02.001. immunology, and infection = Wei mian yu gan ran za zhi. S1684-1182 (20) Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.Y., Chen C., Zhang H.Q., Guo H.Y., Wang H., Wang L., Zhang X., Hua S.N., Yu J., Xiao P.G., Li R.S., Tan X. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir. Res. 2005;67(1):18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.-Y., Duan Y.-Q., Lu X.-H., Ma Y., Wang R.-L. Exploring the cause of the inhibitor 4AX attaching to binding site disrupting protein tyrosine phosphatase 4A1 trimerization by molecular dynamic simulation. J. Biomol. Struct. Dyn. 2019:1–18. doi: 10.1080/07391102.2019.1567392. [DOI] [PubMed] [Google Scholar]
- Lill M.A., Danielson M.L. Computer-aided drug design platform using PyMOL. J. Comput. Aided Mol. Des. 2011;25(1):13–19. doi: 10.1007/s10822-010-9395-8. [DOI] [PubMed] [Google Scholar]
- Lin C.W., Tsai F.J., Tsai C.H., Lai C.C., Wan L., Ho T.Y., Hsieh C.C., Chao P.D. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antivir. Res. 2005;68(1):36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C.A. Lead and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Malde A.K., Zuo L., Breeze M., Stroet M., Poger D., Nair P.C., Oostenbrink C., Mark A.E. An automated force field topology builder (ATB) and repository: version 1.0. J. Chem. Theor. Comput. 2011;7(12):4026–4037. doi: 10.1021/ct200196m. [DOI] [PubMed] [Google Scholar]
- Malathi K., Anbarasu A., Ramaiah S. Identification of potential inhibitors for Klebsiella pneumoniae carbapenemase-3: a molecular docking and dynamics study. J. Biomol. Struct. Dyn. 2019:1–13. doi: 10.1080/07391102.2018.1556737. [DOI] [PubMed] [Google Scholar]
- Mokhnache K., Madoui S., Khither H., Charef N. Drug-likeness and pharmacokinetics of a bis-phenolic ligand: evaluations by computational methods. Scholars J. Appl. Med. Sci. 2019;1:167–173. [Google Scholar]
- Nabavi S.F., Braidy N., Habtemariam S. Neuroprotective effects of chrysin: from chemistry to medicine. Neurochem. Int. 2015;90:224–231. doi: 10.1016/j.neuint.2015.09.006. [DOI] [PubMed] [Google Scholar]
- Narayana K.R., Reddy M.S., Chaluvadi M.R., Krishna D.R. Bioflavonoids classification, pharmacological, Biochemical effects and therapeutic potential. Indian J. Pharm. 2001;33:2–16. [Google Scholar]
- Nisha C.M., Kumar A., Nair P., Gupta N., Silakari C., Tripathi T., Kumar A. Molecular docking and in silico ADMET study reveals acylguanidine 7a as a potential inhibitor of β-secretase. Advances in bioinformatics. 2016:9258578. doi: 10.1155/2016/9258578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norkin L.C. Virus receptors: implications for pathogenesis and the design of antiviral agents. Clin. Microbiol. Rev. 1995;8(2):293–315. doi: 10.1128/cmr.8.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panche A.N., Diwan A.D., Chandra S.R. Flavonoids: an overview. Journal of nutritional science. 2016;5:1–15. doi: 10.1017/jns.2016.41. e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj L.S.,M., J J., I K., Krishna P S., K A S. Molecular docking study for inhibitors of aggregatibacter actinomycetamcomitans toxins in treatment of aggressive perioodontitis. J. Clin. Diagn. Res. : J. Clin. Diagn. Res. 2014;8(11):ZC48–ZC51. doi: 10.7860/JCDR/2014/10067.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Morales A., MacGregor K., Kanagarajah S., Patel D., Schlagenhauf P. Going global – travel and the 2019 novel coronavirus. Trav. Med. Infect. Dis. 2020;33:101578. doi: 10.1016/j.tmaid.2020.101578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh C., Jo S.K. (-)-Epigallocatechin gallate inhibits hepatitis C virus (HCV) viral protein NS5B. Talanta. 2011;85(5):2639–2642. doi: 10.1016/j.talanta.2011.08.035. [DOI] [PubMed] [Google Scholar]
- Sarwar M.W., Riaz A., Dilshad S.M.R., Al-Qahtani A., Nawaz-Ul-Rehman M.S., Mubin M. Structure activity relationship (SAR) and quantitative structure activity relationship (QSAR) studies showed plant flavonoids as potential inhibitors of dengue NS2B-NS3 protease. BMC Struct. Biol. 2018;18(1):6. doi: 10.1186/s12900-018-0084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savi L.A., Caon T., de Oliveira A.P., Sobottka A.M., Werner W., Reginatto F.H., Schenkel E.P., Barardi C.R., Simões C.M. Evaluation of antirotavirus activity of flavonoids. Fitoterapia. 2010;81(8):1142–1146. doi: 10.1016/j.fitote.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E.W., Agarwala R., Bolton E.E. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2019;47(D1):D23–D28. doi: 10.1093/nar/gky1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senathilake K., Samarakoon S., Tennekoon K. 2020. Virtual Screening of Inhibitors against Spike Glycoprotein of 2019 Novel Corona Virus: A Drug Repurposing Approach. Preprints, 2020030042. [Google Scholar]
- Shang J., Ye G., Shi K. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.M. Anti-infective potential of catechins and their derivatives against viral hepatitis. Clinical and experimental vaccine research. 2018;7(1):37–42. doi: 10.7774/cevr.2018.7.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood D., Kumar N., Singh A., Sakharkar M.K., Tomar V., Chandra R. Antibacterial and pharmacological evaluation of fluoroquinolones: a chemoinformatics approach. Genomics & informatics. 2018;16(3):44–51. doi: 10.5808/GI.2018.16.3.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvannang N., Nantasenamat C., Isarankura-Na-Ayudhya C., Prachayasittikul V. Molecular docking of aromatase inhibitors. Molecules. 2011;16(5):3597–3617. [Google Scholar]
- Syed S.B., Arya H., Fu I.-H., Yeh T.-K., Periyasamy L., Hsieh H.-P., Coumar M.S. Targeting P-glycoprotein: investigation of piperine analogs for overcoming drug resistance in cancer. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-08062-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler P., Staker B., Prasad S.G. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279(17):17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. 1995. [DOI] [PubMed] [Google Scholar]
- Walls A.C. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;S0092–8674(20):30262–30272. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 2020;94(7):e00127–e00220. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Dommert F., Holm C. Optimizing working parameters of the smooth particle mesh Ewald algorithm in terms of accuracy and efficiency. J. Chem. Phys. 2010;133(3) doi: 10.1063/1.3446812. [DOI] [PubMed] [Google Scholar]
- Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three dimensional structures of proteins. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkm290. (Web Server issue), W407–W410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Chen L., Peng G., Zhou W., Pennell C.A., Mansky L.M., Li F. A virus-binding hot spot on human angiotensin-converting enzyme 2 is critical for binding of two different coronaviruses. J. Virol. 2011;85(11):5331–5337. doi: 10.1128/JVI.02274-10. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia E.Q., Deng G.F., Guo Y.J., Li H.B. Biological activities of polyphenols from grapes. Int. J. Mol. Sci. 2010;11(2):622–646. doi: 10.3390/ijms11020622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Lin J., Zhou B., Liu Y., Zhu B. Activity of compounds from Taxillus sutchuenensis as inhibitors of HCV NS3 serine protease. Nat. Prod. Res. 2017;31(4):487–491. doi: 10.1080/14786419.2016.1190719. [DOI] [PubMed] [Google Scholar]
- Zakaryan H., Arabyan E., Oo A., Zandi K. Flavonoids: promising natural compounds against viral infections. Arch. Virol. 2017;162(9):2539–2551. doi: 10.1007/s00705-017-3417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi K., Teoh B.T., Sam S.S., Wong P.F., Mustafa M.R., Abubakar S. Antiviral activity of four types of bioflavonoid against dengue virus type-2. Virol. J. 2011;8:560. doi: 10.1186/1743-422X-8-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yan J., Wang H., Wang Y., Wang J., Zhao D. Molecular docking, 3DQSAR, and molecular dynamics simulations of thieno[3,2-b]pyrrole derivatives against anticancer targets of KDM1A/LSD1. J. Biomol. Struct. Dyn. 2020:1–24. doi: 10.1080/07391102.2020.1726819. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supp. material/referenced in article.