Abstract
Background & Aims:
Advances in cross-sectional imaging have resulted in increased detection of intraductal papillary mucinous neoplasms (IPMNs) and their management remains controversial. At present, there is no reliable non-invasive method to distinguish between indolent and high risk IPMNs. We performed extracellular vesicle (EV) analysis to identify markers of malignancy in an attempt to better stratify these lesions.
Methods:
Using a novel ultrasensitive digital extracellular vesicle screening technique (DEST) we measured putative biomarkers of malignancy (MUC1, MUC2, MUC4, MUC5AC, MUC6, Das-1, STMN1, TSP1, TSP2, EGFR, EpCAM, GPC1, WNT-2, EphA2, S100A4, PSCA, MUC13, ZEB1, PLEC1, HOOK1, PTPN6, and FBN1) in EV from patient-derived cell lines and then on circulating EV obtained from peripheral blood drawn from patients with IPMNs. We enrolled a total of 133 patients in two separate cohorts: a clinical discovery cohort (n=86) and a validation cohort (n=47).
Results:
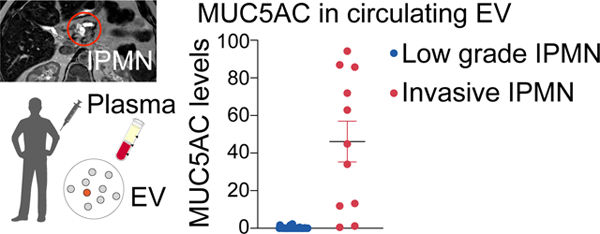
From 16 validated EV proteins in plasma samples collected from the discovery cohort, only MUC5AC showed significantly higher levels in high grade lesions. Of the 11 patients with invasive IPMN (inv/HG), 9 had high MUC5AC expression in plasma EV and of the 11 patients with high grade dysplasia alone, only 1 had high MUC5AC expression (specificity of 82%, sensitivity of 100%). These findings were corroborated in a separate validation cohort. The addition of MUC5AC as a biomarker to imaging and high risk stigmata allowed detection of all cases requiring surgery, whereas imaging and high risk stigmata alone would have missed 5/14 cases (36%).
Conclusions:
MUC5AC in circulating EV can predict the presence of invasive carcinoma within IPMN. This approach has the potential to improve the management and follow-up of patients with IPMN including avoiding unnecessary surgery.
Keywords: Pancreatic cancer, IPMN, pre-cancer, early detection, exosome
Lay summary:
Blood based analysis of circulating vesicles expressing MUC5AC enables the detection of invasive IPMNs.
Graphical Abstract
Introduction
The detection of intraductal papillary mucinous neoplasms (IPMNs), is rising due to the increasing use and improved quality of cross-sectional imaging1. These cystic neoplasms have been shown to evolve from low-grade dysplasia (LG) to high-grade dysplasia (HG) to invasive carcinoma (Inv/HG). This pathway of progression is believed to account for up to a quarter of all pancreatic cancers2. The timing and frequency of malignant progression is unknown, and therefore the management of patients with some forms of IPMN, in particular branch-duct IPMN, remains controversial3–7. This is in large part because current laboratory, endoscopic, cytologic, and imaging technologies are unable to reliably distinguish between low and high risk IPMN. While analysis of cyst fluid has shown some promise8, it requires repeat interventional endoscopy and the amounts of aspirated fluid in IPMN can be too small for analysis. Even though the majority of IPMNs will not progress, there is no reliable way to predict which ones will, and currently most of these patients undergo periodic surveillance with either MRI or CT. Some guidelines recommend stopping surveillance at 5 or 10 years, but other studies have shown that progression can occur beyond this time frame9, and therefore lifelong surveillance may be required5. This implies an enormous cost given the number of patients with incidentally-discovered pancreatic cysts and the fact that incidence increases with age10.
We hypothesize that analysis of circulating extracellular vesicles (EV) originating from (pre)malignant cells represents an opportunity for analyzing IPMNs and early pancreatic cancers. Several challenges exist, including the absence of known biomarkers that can identify high risk IPMN. Indeed a recent consensus conference concluded that “there are no available DNA, RNA or protein biomarkers in blood for clinical use to differentiate pancreatic cyst type or identify high grade dysplasia or cancer”6. We thus first set out to identify and measure EV concentrations of known protein biomarkers that have been associated with pancreatic cancer in the literature. To enable more rapid and sensitive measurements, we developed a digital EV screening technique (DEST) optimized for the analysis of scant surface and intravesicular EV proteins. The DEST method is a near single EV analytical technique with wide dynamic range and allows high throughput digital analysis of clinical samples (Fig. 1). Here we show proof-of-principle of this approach, including rapid isolation that does not require lengthy ultracentrifugation and is thus clinically practical. Following validation in cell lines and PDX models, we applied these technical innovations to a discovery cohort of 86 patients and a validation cohort of 47 patients.
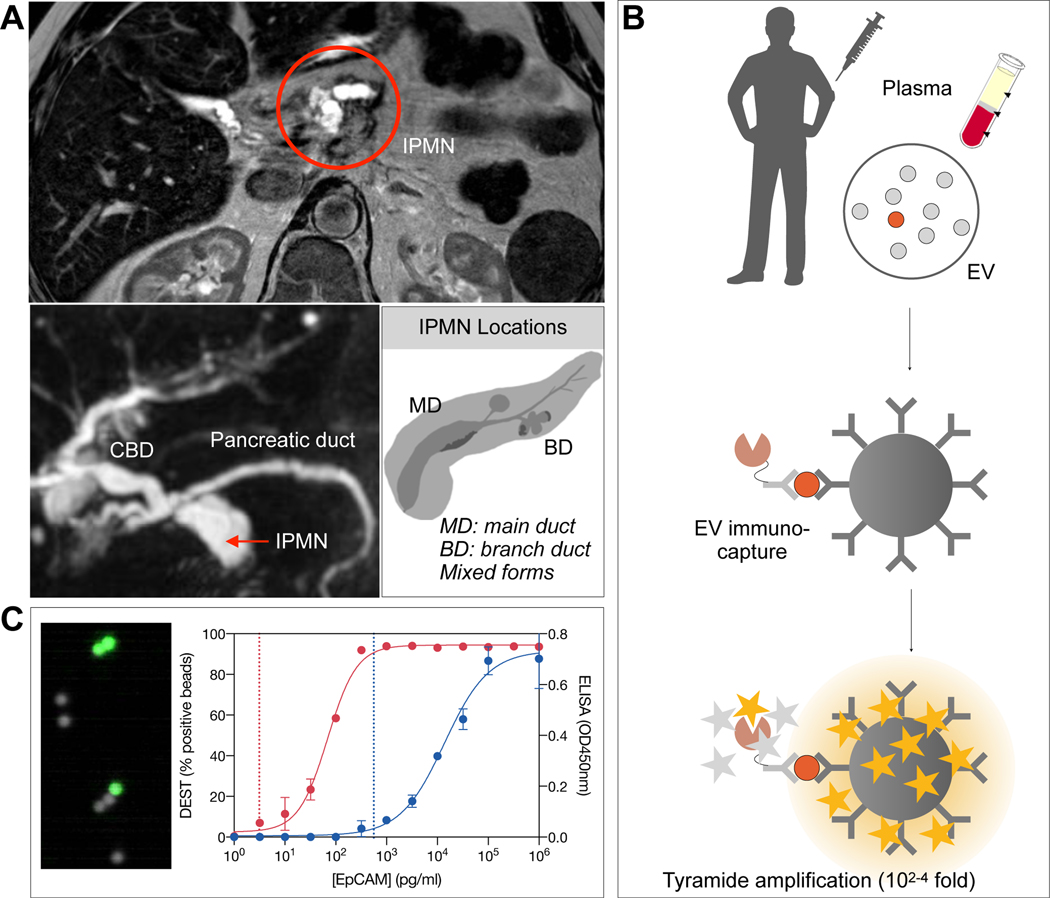
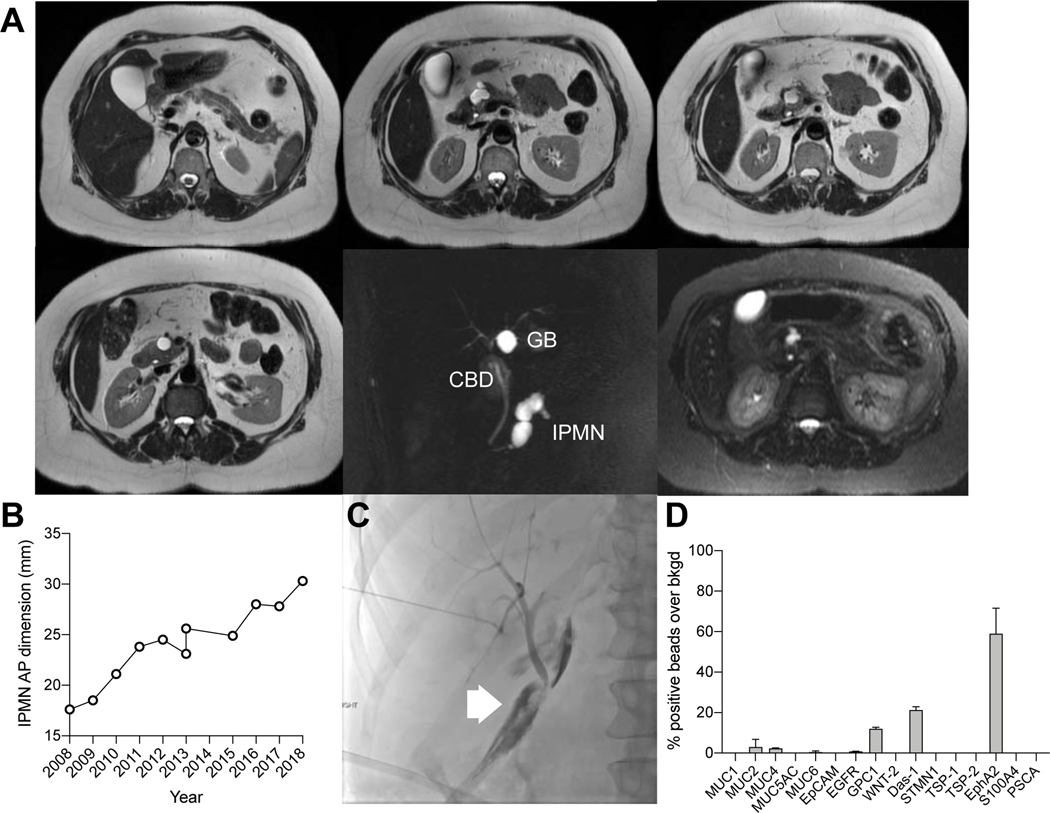
Figure 1. EV analysis for IPMN characterization.
(A) IPMNs are often incidentally detected by abdominal imaging. Repeat MRI is used to follow these T2-hyperintense lesions. Imaging alone has limited accuracy in separating benign from premalignant lesions, requiring resection. (B) Schematic diagram of the digital enzyme-linked immunosorbent assay (DEST) assay. EV are first captured on micron-sized beads via specific antibodies (see Table S3) and their presence is then detected via complementary antibodies followed by a tyramide signal amplification step. Presence of individual or multiple EV on a bead renders the entire bead as fluorescent. (C) Analysis is done in high throughput by counting the number of fluorescent beads. Overall the method is 3–4 orders of magnitude more sensitive than ELISA and uniquely suited to analyze rare EV subpopulations. Dashed lines represent the limit of detection for 1617 PDAC PDX EV in either the DEST (~100EV; left dashed line) or ELISA (~105 EV; right dashed line) assay.
Methods
Specimen acquisition
This study was approved by the institutional review board of Massachusetts General Hospital. Written informed consent was obtained from study participants. All specimens were collected from patients referred to Massachusetts General Hospital for surgical management. Participants were enrolled into one of two cohorts: a clinical discovery cohort and a validation cohort. The discovery cohort included healthy controls and patients with high or low grade IPMNs. The validation cohort was comprised of patients with IPMNs diagnosed on imaging. All of the patients in the validation cohort were followed for at least 1 year, during which time unresected pancreatic lesions were serially surveilled according to established clinical criteria including repeat imaging7,8. Cohort demographics and imaging features were recorded.
Clinical sample preparation
Blood collection was optimized for plasma EV analysis as described in Lobb et al11 and all samples were de-identified and analyzed in blinded fashion. Whole blood was collected in one 10-mL purple-top EDTA tube and was inverted 10 times to mix. Whole blood was stored upright at 4°C and processed within 1 hour of collection. To process blood for plasma isolation, the tube was centrifuged for 10 min at 400 × g (4°C). The plasma layer was collected in a 15mL tube using a pipette so as not to disturb the buffy coat. Plasma was then centrifuged for 10 min at 1100 × g (4°C). The plasma was then aliquoted into 1mL aliquots and stored at −80°C.
EV isolation from plasma
Optimized plasma EV isolation was done according to Lobb et al11. Briefly, plasma was thawed on ice and 500μL was centrifuged at 10,000 × g for 20 min at 4°C. A qEV original size exclusion column (IZON Science, SP1) was equilibrated to room temperature and then flushed with 10mL of 0.22μm filtered PBS. Plasma supernatant was loaded onto the column and 0.5mL fractions were immediately collected. Fractions 0–6 (dead volume) were discarded, while fractions 7–9 were collected (1.5mL). Combined fractions 7–9 were concentrated using an Amicon 10K (15mL, Millipore) and centrifuging at 3100 × g for 15 min. Total EV protein was measured using the Qubit protein assay (Thermo Scientific, Q33211). Final experiments tailored for a clinical workflow, used immunopurification inherent to the DEST assay to isolate EV proteins from unpurified plasma (see below). To reduce background in the DEST assay from plasma, we employed various blocking buffers (2% BSA, UltraBlock, HAMA blocker; see Table S4 and DEST assay methods below).
Plasma preparation
Unpurified plasma was lysed in RIPA lysis buffer for use in the DEST assay to lyse EVs for intra- and extravesicular analysis. Plasma was thawed at 4°C and then lysed in 1X RIPA buffer (Cell Signaling Technology, 9806S, 6X stock) for 15 min on ice. Lysed plasma was aliquoted and stored at −80°C until use.
EV isolation from cell culture
Cell-derived EV were initially isolated by ultracentrifugation as the gold standard for validating both the IZON and direct method (Fig. S1). Cells were grown for 48 – 72 hours in normal growth medium supplemented with 5% exosome-depleted FBS (Thermo, A2720801). Conditioned media was collected and centrifuged at 300 × g for 10 min to remove dead cells and debris, followed by filtration through a 0.22-μm cellulose acetate vacuum filter (430767, Corning). Media was then aliquoted to ultracentrifuge tubes (344058, Beckman) and centrifuged at 100,000 × g for 70 min to pellet EVs. Media was removed and EV pellets were combined in a single ultracentrifuge tube in PBS, which was then centrifuged again at 100,000 × g for 70 min. The final EV pellet was resuspended at a volume of ~100μl in PBS and stored at −80°C until use.
DEST assay
Experimental steps, including incubation times and concentrations, are outlined in Table S1. Reagents needed for the assay are listed in Table S2. Purified protein positive controls were used for TSP1 (R&D 3074-TH-050), TSP2 (R&D 1635-T2–050), GPC1 (R&D 4519-GP-050), WNT-2 (MyBioSource 2104322), S100A4 (MyBioSource 2089230), and PSCA (Fitzgerald 30R-3214). Bead readout was done using a CytoFlex flow cytometer (Beckman Coulter) with the following settings: FSC 201V, SSC 90V, PB450 (BV421 detection) 40V. A gate was drawn around single beads and 10,000 events were recorded for each sample and marker combination. Data was analyzed using the FlowJo software (BD, version 10.6.1). Briefly, pooled normal human plasma was used as a background control for the assay. A gate was drawn using the bisector tool to delineate positive from negative beads. All data are reported as percentage of positive beads out of 10,000 total beads measured and are the average of duplicate measurements. Error is represented as the standard error of the mean. False positives were identified using either HAMA blocker (Abcam) during the plasma incubation step for mouse antibody pairs (MUC1, MUC5AC) or isotype control beads (all other antibody pairs).
Statistics
Biomarkers and imaging comparison groups were categorized as (i) low vs non-invasive high grade (ii) low vs invasive high grade, and (iii) low vs high grade IPMNs. For the discovery cohort, a Mann-Whitney test was used to compare individual biomarker levels between IPMN groups. Subsequently, classifiers were built on individual biomarkers using the continuous values, and with the following combinations: 1) PDACEV signature (MUC1 + GPC1 + EGFR + EpCAM + WNT-2), and 2) DEST MUC5AC + imaging values. Diagnostic performance of these classifiers was assessed by a receiver operating characteristic curve, 95% Confidence Intervals (CIs), F1 scores and Chi-square tests for discovery cohorts. The optimum cutoff value(s) for each classifier was calculated based on Youden’s Index and the final value was manually selected when multiple optimal cutoffs were found. 95% CIs were estimated by bootstrapping (n = 5000 bootstrapped samples). For validation, only MUC5AC, imaging, and MUC5AC + imaging were included. Logistic regression equations derived in the discovery cohort was applied to assess diagnostic performance. Classifier performance metrics were calculated the same way as described for the discovery cohort. Specificity, sensitivity and positive predictive value (PPV) reported for the validation cohorts were based on the optimal thresholds chosen from the discovery cohort. Main analyses were performed using the pROC package on R 3.6.212.
Results
Clinical and pathologic features
133 participants were enrolled. The discovery cohort (n=86) was comprised of healthy controls (n =10), age matched healthy controls (benign) undergoing abdominal surgery but without evidence of any pancreatic lesion (n=14), patients with IPMNs harboring low grade dysplasia (n=40) and patients with high grade IPMN (11 with high-grade dysplasia and 11 with invasive carcinoma) (n=22). The validation cohort was divided into patients with low grade dysplasia, either confirmed at surgery or on the basis of imaging findings and temporal stability (n= 35) or high grade dysplasia, confirmed at resection (n= 12). In the validation cohort, 19 patients underwent surgical resection which yielded 9 lesions with high grade dysplasia and 3 lesions with invasive IPMN. The remaining patients were low grade IPMNs (n=7). Clinical and imaging features for each cohort are summarized in Table 1.
Table 1. Summary of patient cohorts and clinical findings.
Control: healthy control patients; Benign: age-matched control patients undergoing abdominal surgery but without evidence for any pancreatic lesions; LG: low grade IPMN; HG-IPMN: high grade IPMN. The HG-IPMN are divided into invasive and non-invasive forms. IQR: Interquartile range.
| Patients |
Discovery cohort |
Validation cohort |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HG-IPMN |
HG-IPMN |
|||||||||||
| Characteristic | All | All | Control | Benign | LG | Inv | Noninv | Comb | All | LG | Inv | Noninv |
| Total cases | 133 | 86 | 10 | 14 | 40 | 11 | 11 | 22 | 47 | 35 | 3 | 9 |
| Age | ||||||||||||
| Median (yrs) | 71 | 70 | 33 | 54 | 72 | 79 | 71 | 73 | 74 | 71 | 76 | 76 |
| IQR | 63–77 | 59–76 | 27–47 | 40–62 | 68–78 | 67–84 | 68–74 | 68–80 | 67–77 | 65–76 | 73–83 | 75–78 |
| Sex | ||||||||||||
| Male | 55 | 38 | 6 | 7 | 15 | 5 | 5 | 10 | 17 | 16 | 0 | 1 |
| Female | 78 | 48 | 4 | 7 | 25 | 6 | 6 | 12 | 30 | 19 | 3 | 8 |
| Clinical | ||||||||||||
| Symptoms | 43 | 38 | NA | 12 | 15 | 6 | 5 | 11 | 5 | 1 | 1 | 3 |
| Weight loss | 23 | 17 | NA | 2 | 9 | 3 | 3 | 6 | 6 | 1 | 3 | 2 |
| Pain | 31 | 28 | NA | 11 | 12 | 2 | 3 | 5 | 3 | 1 | 0 | 2 |
| Jaundice | 2 | 2 | NA | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| Lab values | ||||||||||||
| CA19-9 >37U/mL | 13 | 11 | NA | 3 | 4 | 3 | 1 | 4 | 2 | 0 | 2 | 0 |
| Imaging | ||||||||||||
| Duct > 10mm | 12 | 9 | NA | 0 | 2 | 3 | 4 | 7 | 3 | 0 | 2 | 1 |
| Duct 5–9 mm | 26 | 17 | NA | 1 | 7 | 3 | 6 | 9 | 9 | 2 | 1 | 6 |
| Cyst > 3cm | 43 | 32 | NA | 6 | 18 | 2 | 6 | 8 | 11 | 6 | 2 | 3 |
| Thickened wall | 7 | 4 | NA | 1 | 1 | 0 | 2 | 2 | 3 | 1 | 1 | 1 |
| Enhancing nodule | 13 | 8 | NA | 2 | 1 | 3 | 2 | 5 | 5 | 0 | 2 | 3 |
| Pancreatitis | 21 | 20 | NA | 9 | 7 | 1 | 3 | 4 | 1 | 0 | 0 | 1 |
| Adenopathy | 0 | 0 | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Surgery | ||||||||||||
| None | 44 | 16 | 10 | 0 | 15 | 1 | 0 | 1 | 28 | 28 | 0 | 0 |
| Whipple | 32 | 22 | NA | 4 | 12 | 6 | 10 | 16 | 10 | 2 | 2 | 6 |
| Distal pancreatectomy | 22 | 13 | NA | 4 | 8 | 1 | 1 | 2 | 9 | 5 | 1 | 3 |
| Middle pancreatectomy | 6 | 6 | NA | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total pancreatectomy | 2 | 2 | NA | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| Puestow | 2 | 2 | NA | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Other | 4 | 4 | NA | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
DEST analysis has near single EV sensitivity
Analysis of scant circulating EV produced by small tumors requires new ultrasensitive diagnostic assays as bulk detection methods are unlikely to detect human tumors < 1 cm3. While some single EV analytical methods have been described as research tools13–15, they often have limited clinical applications as the methods are too labor-intensive, costly or have limited multiplexing capability. To enable detection of HG-IPMN and early invasive pancreatic cancers we thus set out to first develop and optimize a bead based digital ELISA assay adopted for EV analysis. The DEST method utilizes magnetic beads coated with capture antibodies, biotinylated detection antibody, and intact or lysed EV to capture surface or intravesicular proteins respectively (Fig. 1B). To ensure the most sensitive readout, the DEST assay includes a tyramide signal amplification step that catalyzes the addition of biotin groups on/near HRP from tyramide-biotin radicals16. Brilliant violet 421 conjugated to streptavidin labels all free biotin molecules for ultra-bright readout of micron-sized beads by cytometry. As shown in Fig. 1C, bead readout is digital with a bead either being fluorescent or not. To compare the sensitivity of the DEST assay to traditional ELISA, we compared EpCAM analysis in EV from a low-passage patient-derived PDAC cell line (#1617). Traditional ELISA has a limit of detection of ~ 1 million EV, whereas DEST has a limit of detection of ~100 EV based on EpCAM analysis (Fig. 1C). This 10,000-fold increase in sensitivity may enable detection of ultra-rare EV and low abundance proteins. Additional benefits of the DEST method include analysis times of < 2 hours from start to finish, the ability to process hundreds of samples per day and the relatively low cost.
Choice of EV biomarkers and correlation to cellular signatures
To determine which EV biomarkers could be useful in differentiating LG and HG-IPMN we first surveyed the literature on biomarkers17–41. We identified 22 putative biomarkers, some of which had been used to analyze pancreatic cyst fluid obtained by interventional endoscopy18,19,22,24,25,29,35. Circulating TSP2 and MUC5AC levels have been tested in pancreatic cancer, but not for the ability to distinguish LG and HG-IPMN21,28. The chosen 22 biomarkers for which antibody pairs were commercially available include: MUC1, MUC2, MUC4, MUC5AC, MUC6, Das-1, STMN1, TSP1, TSP2, EGFR, EpCAM, GPC1, WNT-2, EphA2, S100A4, PSCA, MUC13, ZEB1, PLEC1, HOOK1, PTPN6, and FBN1. For each of these targets we had to identify reliable antibody pairs for protein capture and detection in order to establish the assay. We tested most commercially available antibodies to find suitable pairs but not all of them worked well. This may be due to steric hindrance, low affinity or polyclonality of some commercial products. Fig. S2 and Table S3 summarizes the 16 antibody pairs that ultimately proved reliable, while Table S5 lists pairs unsuitable for DEST. For each antibody pair we next determined positivity (against control matched IgG) and thus detection sensitivity. To enable clinical measurements we optimized the method so that only 1–10μL of plasma was necessary per measurement. To minimize the loss of potential very rare IPMN EV, we also compared different EV purification steps (Fig S1). We found that many traditional methods, such as ultracentrifugation and IZON column purification, resulted in considerable loss or shift of EV subpopulations and also required large amounts of plasma. We thus settled on simple direct EV processing from unpurified plasma, since the DEST assay includes an immunocapture “purification” as the first step (Fig. S1). Combined, this workflow is suited for the clinical setting where sample volume is limited and throughput is an important consideration. Thus established and validated DEST assays were next tested in whole cell lysates and EV of IPMN and PDAC PDX models. As shown in Fig. S2, DEST signal from EV generally correlated with whole cell signal for a given biomarker (or its absence). We were particularly interested in the signals from two invasive high grade IPMN PDX cell lines, #1966 and #1505, as these may inform on high grade IPMN patient samples. In the IPMN PDX EV, the most abundant markers were MUC5AC and MUC6.
Analysis of EV biomarkers in clinical IPMN discovery cohort
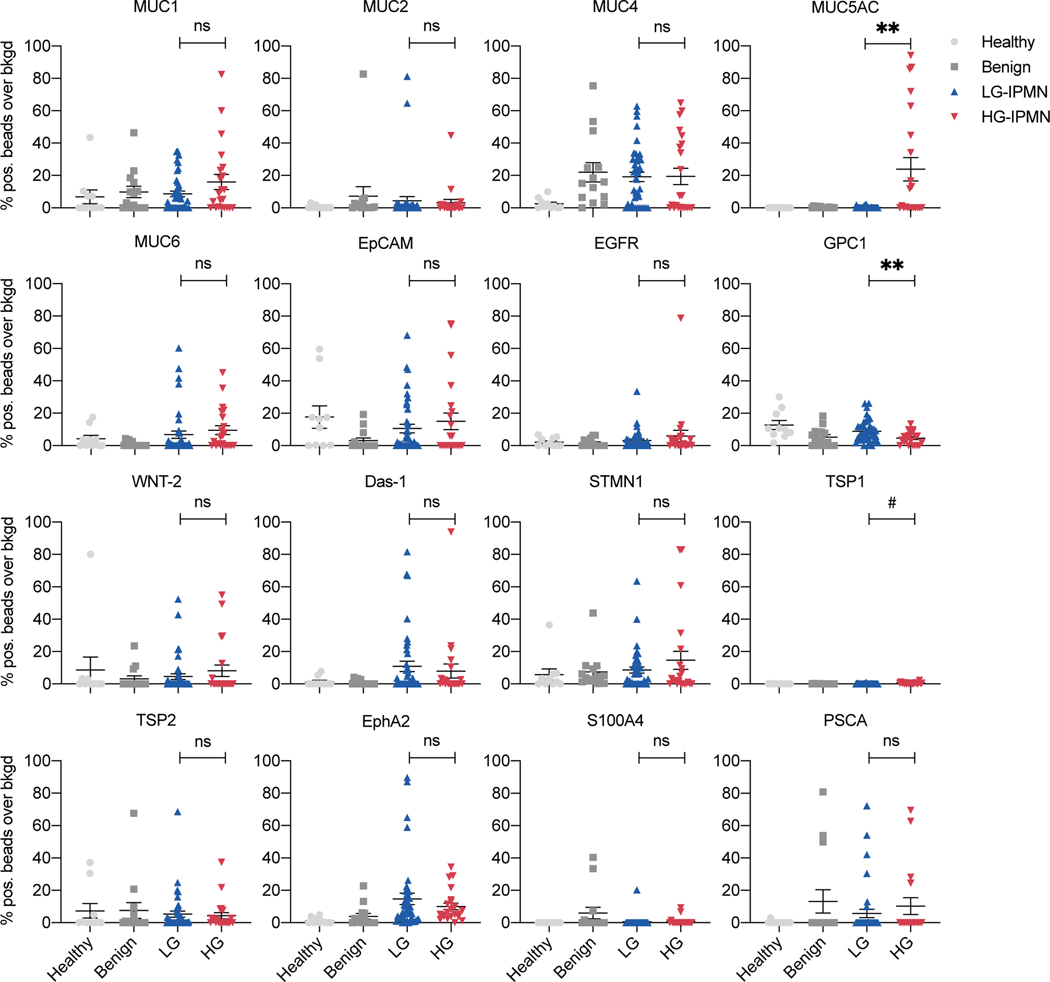
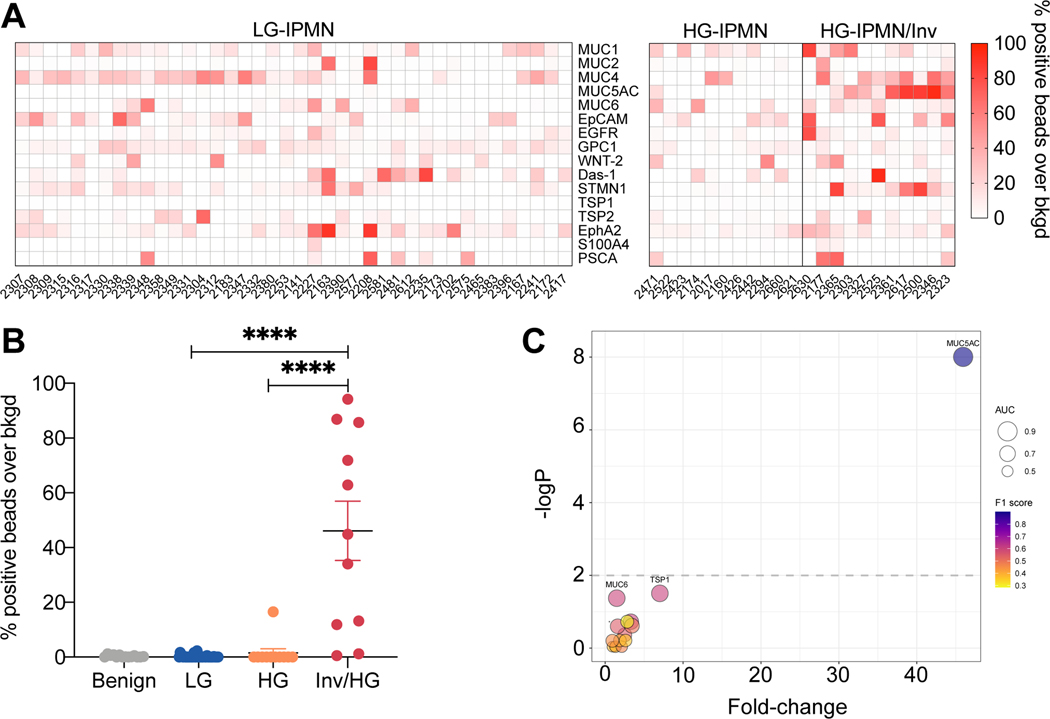
We next measured the 16 validated EV proteins in plasma samples collected from a clinical patient cohort (n = 86 patients, Table 1) that included healthy controls (n = 10), age-matched benign controls (n = 14), LG-IPMN (n = 40) and HG-IPMN (n = 22). These groups were chosen not based on imaging findings but rather to reflect the expected spectrum of EV profiles from clearly negative (healthy) to positive (HG-IPMN). Fig. 2 and Fig. 3A summarize the biomarker expression in the four patient categories for all 16 biomarkers. For the majority of the markers tested, we did not see a significant difference between LG-IPMN and HG-IPMN (Fig. 2, blue vs red, Mann-Whitney test, p<0.05; Table 2, Fig. S3 ROC analysis). This negative result was of interest because we had expected to be able to differentiate mucinous neoplasm subtypes by pan MUC-EV analysis that had first been proven to be useful in cyst fluid analysis25,36. We found that only MUC5AC showed significantly higher levels in high grade lesions (Table 2). MUC5AC is a high-molecular-weight secreted glycoprotein that has been associated with certain pulmonary diseases and malignant transformation in tissue sections of IPMN42. However, pancreatic MUC5AC had not been shown to circulate in EV. The HG-IPMN group had higher MUC5AC levels but interestingly there was a bimodal distribution (Table 1). Further analysis of pathology data showed that of the 11 patients with invasive carcinoma arising in IPMN with high grade, referred to hereafter as invasive HG-IPMN (inv/HG), 9 had high MUC5AC expression in plasma EV and of the 11 patients with high grade dysplasia alone, only 1 had high MUC5AC expression (Fig. 3B, Fig. S4). This resulted in a sensitivity of 100%, a specificity of 82%, with an overall diagnostic accuracy of 96% for differentiating invasive IPMN from LG-IPMN by MUC5AC measurements alone (Table 2). This suggests that MUC5AC in circulating EV may predict invasiveness of high grade IPMN and identify a patient cohort requiring surgical intervention. Future studies are required to determine the specificity in the presence of other co-morbidities.
Figure 2. Analysis of 16 prototypical biomarkers in EV from different patient groups.
Each datapoint represent an EV sample from a single patient. Data shown are from healthy controls (light grey dots, n= 10); benign: age-matched control patients undergoing abdominal surgery but without evidence for any pancreatic lesions (dark grey squares, n=14 patients); low grade IPMN (blue triangles, n=40 patients) and high grade IPMN (red triangles, n=22 patients). See Table 1 for cohort demographics. Differences between HG-IPMN and LG-IPMN are shown by asterisk (**** p <0.0001, * p <0.05, ns = not statistically significant, p > 0.05;). #TSP1 is statistically significant but the extremely low signal over background makes the results clinically unreliable.
Figure 3. Discovery cohort analysis.
(A) EV biomarker analysis across all patients with IPMN. Each column refers to a single patient. Patients are grouped into low grade, high grade IPMN and high grade IPMN with invasive features determined by pathology. Notable differences between the groups can be seen for MUC5AC. (B) Subgroup comparison of MUC5AC positive EV. Note the significantly higher levels in invasive HG-IPMN against noninvasive HG-IPMN or LG-IPMN (Mann-Whitney, p < 0.0001 two-tailed). (C) Bubble plot of each molecular EV biomarker tested. The graph summarizes the following descriptive statistics: fold-change over LG-IPMN (x-axis), -log p-value by Mann-Whitney test (y-axis), and classifier metrics: AUC (bubble size), and F1 score (bubble color). Highly predictive EV biomarkers reside in the upper right hand corner. Only MUC5AC EV analysis stood out in the discovery cohort.
Table 2. EV biomarker accuracy for the diagnosis of invasive and non-invasive HG-IPMN as compared to LG-IPMN in the discovery cohort.
All numbers for sensitivity (sens), specificity (spec) and accuracy (acc) are in fractions.
| Invasive HG-IPMN | Noninvasive HG-IPMN | Combined HG-IPMN | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Biomarker(s) | Sens | Spec | Acc | Sens | Spec | Acc | Sens | Spec | Acc |
| MUC1 | 0.45 (0.09, 0.91) | 0.90 (0.43, 1.00) | 0.80 (0.51, 0.90) | 0.45 (0.00, 1.00) | 0.80 (0.00, 1.00) | 0.68 (0.55, 0.77) | 0.45 (0.09, 0.82) | 0.80 (0.45, 1.00) | 0.68 (0.55, 0.77) |
| MUC2 | 0.73 (0.36, 1.00) | 0.55 (0.03, 0.83) | 0.59 (0.24, 0.76) | 0.73 (0.36, 1.00) | 0.53 (0.03, 0.83) | 0.58 (0.37, 0.71) | 0.32 (0.00, 1.00) | 0.83 (0.00, 1.00) | 0.65 (0.35, 0.73) |
| MUC4 | 0.55 (0.00, 1.00) | 0.65 (0.00, 1.00) | 0.61 (0.22, 0.78) | 0.82 (0.55, 1.00) | 0.75 (0.63, 0.88) | 0.66 (0.53, 0.77) | 0.36 (0.09, 0.59) | 0.9 (0.78, 1.00) | 0.69 (0.61, 0.79) |
| MUC5AC | 0.82 (0.55, 1.00) | 1.00 (1.00, 1.00) | 0.96 (0.90, 1.00) | 0.09 (0.00, 1.00) | 1.00 (0.00, 1.00) | 0.81 (0.73, 0.87) | 0.45 (0.27, 0.68) | 1.00 (1.00, 1.00) | 0.81 (0.74, 0.89) |
| MUC6 | 0.91 (0.64, 1.00) | 0.70 (0.53, 0.85) | 0.75 (0.61, 0.86) | 0.27 (0.00, 1.00) | 0.90 (0.00, 1.00) | 0.68 (0.56, 0.79) | 0.59 (0.27, 0.82) | 0.73 (0.55, 0.95) | 0.68 (0.56, 0.79) |
| EpCAM | 0.64 (0.00, 1.00) | 0.60 (0.00, 1.00) | 0.59 (0.22, 0.78) | 1.00 (0.55, 1.00) | 0.28 (0.10, 0.73) | 0.58 (0.35, 0.69) | 0.59 (0.00, 1.00) | 0.6 (0.00, 1.00) | 0.58 (0.35, 0.69) |
| EGFR | 0.82 (0.27, 1.00) | 0.60 (0.40, 1.00) | 0.65 (0.51, 0.86) | 0.55 (0.00, 1.00) | 0.55 (0.00, 1.00) | 0.61 (0.50, 0.73) | 0.64 (0.05, 0.86) | 0.6 (0.38, 1.00) | 0.61 (0.50, 0.71) |
| GPC1 | 0.73 (0.36, 1.00) | 0.70 (0.23, 0.95) | 0.71 (0.39, 0.86) | 0.91 (0.45, 1.00) | 0.58 (0.30, 0.93) | 0.68 (0.53, 0.81) | 0.82 (0.41, 1.00) | 0.60 (0.30, 0.95) | 0.68 (0.53, 0.81) |
| WNT-2 | 0.36 (0.00, 1.00) | 0.90 (0.00, 1.00) | 0.78 (0.22, 0.84) | 0.18 (0.00, 1.00) | 0.98 (0.00, 1.00) | 0.68, 0.61, 0.76) | 0.23 (0.05, 0.5) | 0.95 (0.75, 1.00) | 0.68 (0.61, 0.76) |
| Das-1 | 0.91 (0.64, 1.00) | 0.43 (0.18, 0.60) | 0.51 (0.33, 0.67) | 1.00 (0.73, 1.00) | 0.30 (0.08, 0.58) | 0.55 (0.42, 0.660 | 0.82 (0.64, 1.00) | 0.40 (0.10, 0.60) | 0.55 (0.40, 0.66) |
| STMN1 | 0.55 (0.00, 1.00) | 0.55 (0.00, 1.00) | 0.55 (0.22, 0.78) | 1.00 (0.91, 1.00) | 0.58 (0.40, 0.75) | 0.60 (0.39, 0.71) | 0.64 (0.23, 1.00) | 0.58 (0.00, 0.90) | 0.60 (0.35, 0.71) |
| TSP1 | 0.64 (0.27, 0.91) | 0.88 (0.63, 1.00) | 0.82 (0.65, 0.92) | 0.82 (0.55, 1.00) | 0.95 (0.85, 1.00) | 0.84 (0.73, 0.92) | 0.68 (0.45, 0.91) | 0.93 (0.73, 1.00) | 0.82 (0.73, 0.90) |
| TSP2 | 0.36 (0.00, 0.91) | 0.88 (0.50, 1.00) | 0.76 (0.53, 0.84) | 0.55 (0.00, 1.00) | 0.63 (0.00, 1.00) | 0.63 (0.35, 071) | 0.45 (0.00, 1.00) | 0.70 (0.00, 1.00) | 0.63 (0.35, 071) |
| EphA2 | 0.73 (0.18, 1.00) | 0.65 (0.22, 0.95) | 0.65 (0.37, 0.84) | 1.00 (0.73, 1.00) | 0.30 (0.08, 0.55) | 0.53 (0.42, 0.69) | 0.86 (0.18, 1.00) | 0.35 (0.13, 0.95) | 0.53 (0.42, 0.69) |
| S100A4 | 0.18 (0.00, 1.00) | 0.98 (0.00, 1.00) | 0.80 (0.22, 0.86) | 0.09 (0.00, 1.00) | 0.98 (0.00, 1.00) | 0.68 (0.35, 0.74) | 0.14 (0.00, 1.00) | 0.98 (0.00, 1.00) | 0.68 (0.35, 0.74) |
| PSCA | 0.27 (0.00, 1.00) | 0.95 (0.00, 1.00) | 0.80 (0.22, 0.86) | 0.18 (0.00, 1.00) | 0.93 (0.00, 1.00) | 0.66 (0.35, 0.73) | 0.18 (0.00, 1.00) | 0.95 (0.00, 1.00) | 0.66 (0.35, 0.73) |
| PDACEV | 0.70 (0.55, 0.88) | 1.00 (0.82, 1.00) | 0.76 (0.65, 0.88) | 0.72 (0.32, 0.98) | 0.82 (0.45, 1.00) | 0.75 (0.47, 0.90) | 0.68 (0.40, 0.85) | 0.86 (0.68, 1.00) | 0.74 (0.60, 0.84) |
| Imaging alone | 0.27 (0.1, 0.57) | 0.88 (0.74, 0.94) | 0.65 (0.53, 0.75) | 0.55 (0.28, 0.79) | 0.88 (0.74, 0.95) | 0.77 (0.65, 0.87) | 0.41 (0.23, 0.64) | 0.88 (0.78, 0.98) | 0.71 (0.61, 0.81) |
| DEST+Imaging | 0.95 (0.64, 1.00) | 1.00 (0.71, 1.00) | 0.93 (0.70, 1.00) | 1.00 (1.00, 1.00) | 1.00 (1.00, 1.00) | 1.00 (1.00, 1.00) | 0.73 (0.45, 0.91) | 0.88 (0.78, 1.00) | 0.82 (0.74, 0.92) |
Two other markers showed small differences between cohorts. GPC1 was slightly lower in HG-IPMN compared to LG-IPMN (Fig. 2, Fig. S4). This was an interesting inverse finding, as GPC1 has been associated with malignancy in some studies43 but not in others44. However, previous pathological studies of IPMN tissue also suggest that GPC1 is slightly higher in LG-IPMN vs HG-IPMN (56% positive rate vs 46%, respectively)45, consistent with our findings here. Another marker that was slightly higher in HG-IPMN was TSP1 although the overall TSP1 levels in HG-IPMN were very low and near background with considerable overlap to be clinically useful (Fig. 2, Fig. S4). TSP1 by itself only had moderate sensitivity (68%) and specificity (93%) for diagnosing invasive HG-IPMN (Table 2, Fig. S3). TSP1 is a large adhesive glycoprotein involved in cell-to-cell and cell-to-matrix interactions. Previous studies had suggested that circulating TSP1 expression decreases in PDAC compared to normal and benign pancreatic controls 46,47, while another study suggested stromal TSP1 tissue expression was an indicator of IPMN invasiveness26. Surprisingly, none of the other markers tested here showed a significant difference in EV between LG-IPMN and HG-IPMN (Fig. 2 and 3, Table 2). This included Das-1, a protein shown in pancreatic cyst fluid and histologic sections to differentiate between HG and LG-IPMN25. The bubble plot in Fig. 3C compares four different statistical parameters (fold-change in biomarker signal, -log p-value, AUC and F1 score), demonstrating that MUC5AC is superior to the other biomarkers tested in identifying invasive/HG-IPMN from indolent LG-IPMN.
Analysis of EV biomarkers in clinical IPMN validation cohort
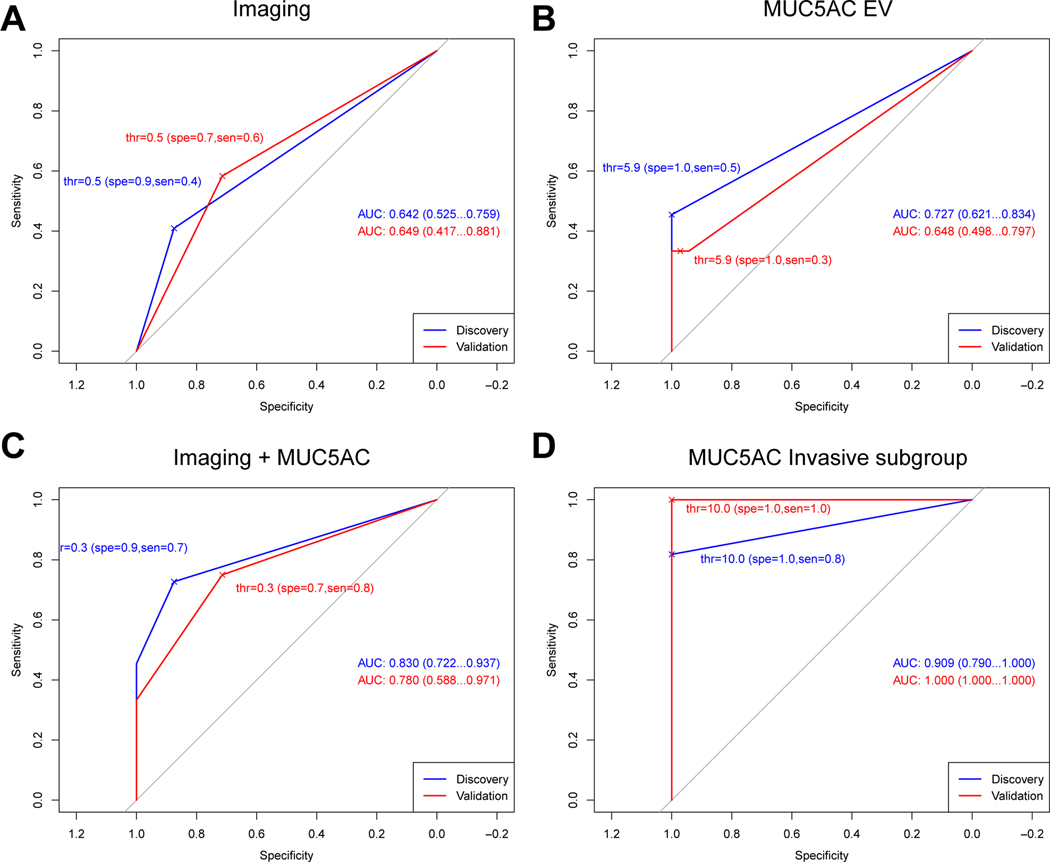
We next re-measured the 16 validated EV proteins in plasma samples collected in the validation cohort (Fig. S5). Our results show that of the 3 patients with invasive high grade IPMN, all had high MUC5AC expression in plasma EV (Table S6). None of the patients in the low grade dysplasia cohort had high MUC5AC expression. MUC5AC EV levels were highest in invasive and in high grade lesions and low in low grade lesions. In the combined cohorts of high grade and invasive lesions, the specificity of this biomarker for identifying invasive disease is 97–100%, the sensitivity 33–50%, and the AUC 0.648–0.727 (Table 2, Fig. 4B, Fig. S3). The values for differentiating low and high-grade (non-invasive) lesions was specificity 91–100%, sensitivity 11–32%, and AUC 0.545–0.648. Interestingly, neither GPC1 or TSP1 expression showed a significant difference between LG-IPMN and HG-IPMN in the validation cohort.
Figure 4. ROC analyses of imaging and MUC5AC-EV in discovery and validation cohorts.
(A) ROC analysis for differentiating high grade from low grade IPMN by imaging alone. The Area under the curves (AUC) are shown (95% confidence interval). Thr: threshold; spe: specificity; sen: sensitivity. (B) AUC for MUC5AC EV. (C) Combined imaging and MUC5AC analysis shows an improved AUC in both cohorts. (D) MUC5AC EV analysis for inv/HG-IPMN (without imaging combination). Note the high AUC in this important sub-group. Optimal cut-off thresholds were chosen from discovery and applied to the validation cohort analyses to display specificity and sensitivity at the chosen thresholds.
Integrating EV testing with imaging improves IPMN analysis
Magnetic resonance cholangiopancreatography (MRCP) is an established and non-invasive method for the initial evaluation and surveillance of IPMNs48 but lacks specificity for reliably distinction of low from high-grade IPMN. IPMNs are morphologically divided into main-duct (MD), branch duct (BD) or mixed-type (MT). Resection is indicated upon diagnosis of MD and MT-IPMNs, defined as main pancreatic duct diameter exceeding 10 mm, due to their high malignant potential. However, the management of BD-IPMNs is more nuanced as their incidence of malignancy is significantly lower. Surgical resection is indicated in the presence of high risk features including jaundice, an enhancing nodule > 5mm or dilated pancreatic duct above 10 mm. When worrisome features are identified (a cyst diameter >/=3 cm or >5 mm growth over 2 years, thickened cyst walls, enhancing septations or nodules < 5 mm, main duct diameter 5–9mm with pancreatic atrophy or lymphadenopathy), further evaluation with endoscopic ultrasound is then required5. Patients who do not meet requirements for surgical resection undergo imaging surveillance at varying time intervals. The performance profile of these diagnostic imaging features reported elsewhere was similar in our cohort with a low specificity at ROC curve analysis shown in Fig. 4A and Tables 2 and S5. Given the modest specificity of imaging, we asked whether combined analyses with EV testing would improve diagnostic performance.
Fig. 4 summarizes the ROC of imaging alone and combined imaging and DEST analysis (MUC5AC EV), while Table 2 and Table S6 provide additional metrics on accuracy for the different parameters. Our results indicate the following: i) HG-IPMN can be distinguished from LG-IPMN with high sensitivity and specificity when MUC5AC and imaging are combined (Fig. 4C vs Fig. 4A and B) and ii) MUC5AC alone identifies inv/HG-IPMN with high sensitivity and specificity (Fig. 4D). If currently established tools for stratifying IPMN risk had been used in isolation to guide surgical intervention, including imaging features or high risk stigmata, then 5/14 cases (36%) would not have proceeded to surgery. When MUC5AC was added, all patients would have been correctly identified (Fig. S6). Overall, our results suggest that the likelihood of invasive HG-IPMN in the absence of elevated MUC5AC levels is low. Fig. 5 summarizes this in one clinical example. A patient with an incidentally detected IPMN had been followed with yearly MRI for over a decade showing progressive increase in IPMN size. A clinical decision was made to resect the growing IPMN but this led to peri-operative complications requiring an extended hospital stay. Pathologic analysis confirmed a LG-IPMN without evidence for any invasive features. In this case, MUC5AC was not detectable in EV and therefore, this lesion would have been correctly classified as LG-IPMN through EV analysis.
Figure 5. Enlarging IPMN concerning for high grade dysplasia in a 57 year old patient.
(A) A predominantly branch duct IPMN was monitored annually with MRI over a decade. (B) During this time, the size of the IPMN increased. (C) The patient underwent a pancreaticoduodenectomy. The postoperative course was prolonged by a bile leak which was treated by percutaneous biliary drainage. The pathology results showed a low grade IPMN. (D) Note that the MUC5AC EV levels at the time of surgery were low indicative of a low grade lesion, suggesting that surgery and complicated postoperative course could have been avoided by EV analysis.
Discussion
The prevalence of pancreatic cysts in the general population has been found to be unexpectedly high and increases in number and size with age10,49. As discussed, the increased detection of IPMNs at cross-sectional imaging warrants a burdensome program of surveillance which potentially places enormous constraints on modern healthcare systems. Furthermore, the possible requirement for ancillary invasive testing, including endoscopic ultrasound, or surgical resection are often undertaken with an appreciation of the poor diagnostic performance of imaging features alone to distinguish low grade, indolent subtypes from high grade dysplastic lesions. While pancreatic cyst fluid can be analyzed it requires sampling by endoscopic ultrasound, but this is invasive and costly, and therefore not practical as a multiple repeated test25. Blood based tests are thus of high medical interest to manage these potentially large patient populations in a cost-effective manner, and more accurate markers of high-grade dysplasia or early invasive carcinoma could support potentially curative resection on the one hand and obviate the need for unnecessary resection on the other. Ideally, one would like to have blood biomarkers that could be serially analyzed.
To date, most biomarkers have focused on mutational aberrations8,36,50, protein33,51,52, and extracellular vesicle (EV) analysis. Aberrant protein/EV markers indicative of malignancy often include panels of proteins such as CA 125, EGFR, MUC1, GPC1, WNT2, EpCAM, mutant KRAS37,53,54, as well as carbohydrate antigen CA 19–9 (and related glycans/isoforms)55,56. CA 19–9 is the only FDA approved biomarker for PDAC, however, its utility in identifying high grade IPMN is poor as shown in a recent study of >500 IPMN patients by Ciprani et al57. Similar results are described here for the discovery cohort (Fig. S7). CA 19–9 is moderately useful in identifying invasive cancer with a sensitivity of 84.5%, but the specificity is only 40.8%57. Conversely, a much larger panel of proteins have been associated with PDAC in smaller studies and their utility remains to be validated in larger trials (e.g. CD73, TIMP1, LRG1, MSLN, EphA2, GNAS, RNF43)52,58. Many of the published studies are retrospective, comprised of small cohorts, relied on surgically resected specimen and required cyst fluid rather that peripheral blood and analyzed mutations. In general, results have not been universally reproducible. Finally, most series do not differentiate between PDAC arising from IPMN (~25%) vs independently (~75%).
During several recent consensus meetings the guidelines for predicting invasive carcinoma and high-grade dysplasia (HG), surveillance, and postoperative follow-up of IPMN have been revised5,6,59. In spite of this, identifying which patients are at high risk of harboring or developing HG or invasive PDAC is challenging. Ideally, these patients should undergo resection and, as yet, the continued management and surveillance of those who do not undergo resection is controversial. More recently, nomograms have been developed to aid in the clinical decision making8,60. Despite these recommendations and tools, differences remain in practice patterns around the world, and there is uncertainty on best approaches. What is very clear, however, is that i) missed HG and early invasive PDAC will become unresectable if not operated upon in a timely fashion, ii) long term follow-up of LG-IPMN is expensive, burdensome and often ambiguous and iii) better diagnostics are urgently needed to improve clinical decision making, prevent unnecessary surgeries and, thus, advance the field.
Tumor-derived EV and potentially host cell derived EV represent promising biomarkers in the analysis of pancreatic precursor lesions. Technological advances have improved our ability to measure rare EV populations in plasma and/or cyst fluid61. These advances are in part due to miniaturization of detection technology61, integrated sensor platforms capable of point-of-care testing in a clinical environment, digital sensing approaches62, and single vesicle analysis platforms13,15 among others. It is increasingly clear that EV are secreted from cancer cells at higher rates than normal cells and can be identified in the blood of patients with pancreatic cancer53,63,64. The challenge however, is to develop analytical methods with near single EV capabilities to detect rare tumor EV from small lesions against a high background of normal host cell EV. With such technologies in hand, we estimate that human cancers < 1 mm3 could be detectable65. Furthermore, any such assay has to be practical and high throughput so that it can be deployed clinically.
In the current study we developed a digital ELISA approach for EV analysis (DEST), that allows high throughput measurements of clinical samples. We show that high MUC5AC measurement in EV has a high predictive power to detect invasive HG-IPMN but was not elevated in LG-IPMN (AUC 0.9, adjusted p-value = 4.4×10−10 for discovery; AUC 1.0, adjusted p-value = 5.63×10−8 for validation cohort). This measurement was markedly better than imaging alone (AUC 0.6, adjusted p-value = 0.23 for discovery; AUC 0.5, adjusted p-value = 0.28 for validation cohort), and has a high-predictive power.
In summary, we show that a simple blood based test can effectively identify IPMNs with invasive carcinoma. EV profiling has the potential to improve triage of patients with worrisome lesions identified by imaging or endoscopic ultrasound, and therefore avoid unnecessary surgeries, but also could eventually simplify the care of all patients with pancreatic cysts that are currently managed with recurrent imaging. If larger prospective studies show that serial EV profiling can identify when an IPMN has become malignant, this test could have a major impact in the detection and care of pancreatic cancer, particularly if the addition of new EV biomarkers emerging from proteomic studies permit for the identification of high-grade dysplasia66–68.
Supplementary Material
WHAT YOU NEED TO KNOW.
BACKGROUND AND CONTEXT:
increasing detection of intraductal papillary mucinous neoplasms (IPMNs) from cross-sectional imaging is a problem for clinicians since these patients will require prolonged surveillance. A non-invasive method for the distinction of indolent from invasive subtypes is an unmet clinical need.
NEW FINDINGS:
Blood based analysis of extracellular vesicles (EV) permits the distinction of invasive IPMNs from low grade and non-invasive subtypes.
LIMITATIONS:
133 patients were examined in this study, of which 83 ultimately underwent surgical resection with histopathologic correlation. Confirmation of our study findings will require studies in larger cohorts inclusive of both surgical candidates and non-operative candidates undergoing surveillance.
IMPACT:
Our results show that MUC5AC EV profiling reliably identifies patients with invasive IPMN. When combined with imaging and clinical findings, the DEST method has the potential to transform IPMN/early PDAC cancer detection and surgical evaluation.
Acknowledgments
We thank the following individuals for helpful discussions: Scott Ferguson, Hyungsoon Im, Xandra Breakefield, Hakho Lee and Jonathan Carlson. We are grateful to the following individuals who contributed to preliminary feasibility data: Nidhi Shanakar and Alexandra Dibrindisi.
GRANT SUPPORT: This work was supported in part by the following NIH grants: R01CA237332 (NCI), R01CA204019 (NCI), R21CA236561 (NCI), P01CA069246 (NCI); the Andrew L. Warshaw, MD Institute for Pancreatic Cancer Research; the William F. Milton Fund.
ABBREVIATIONS USED IN THIS PAPER:
- DEST
digital extracellular vesicle screening technique
- EV
extracellular vesicle
- HG
high grade
- IPMN
intraductal papillary mucinous neoplasm
- inv/HG
invasive high grade
- LG
low grade
Footnotes
Disclosures: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gaujoux S, Brennan MF, Gonen M et al. Cystic lesions of the pancreas: changes in the presentation and management of 1,424 patients at a single institution over a 15-year time period. J Am Coll Surg. 2011;212:590–600; discussion 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maitra A, Fukushima N, Takaori K et al. Precursors to invasive pancreatic cancer. Adv Anat Pathol. 2005;12:81–91. [DOI] [PubMed] [Google Scholar]
- 3.Allen PJ. The management of intraductal papillary mucinous neoplasms of the pancreas. Surg Oncol Clin N Am. 2010;19:297–310. [DOI] [PubMed] [Google Scholar]
- 4.Allen PJ. The diagnosis and management of cystic lesions of the pancreas. Chin Clin Oncol. 2017;6:60. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka M, Fernández-Del Castillo C, Kamisawa T et al. Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology. 2017;17:738–753. [DOI] [PubMed] [Google Scholar]
- 6.European, SGOCTOTP. European evidence-based guidelines on pancreatic cystic neoplasms. Gut. 2018;67:789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buscail E, Cauvin T, Fernandez B et al. Intraductal papillary mucinous neoplasms of the pancreas and European guidelines: importance of the surgery type in the decision-making process. BMC Surg. 2019;19:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Springer S, Masica DL, Dal Molin M et al. A multimodality test to guide the management of patients with a pancreatic cyst. Sci Transl Med. 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pergolini I, Sahora K, Ferrone CR et al. Long-term Risk of Pancreatic Malignancy in Patients With Branch Duct Intraductal Papillary Mucinous Neoplasm in a Referral Center. Gastroenterology. 2017;153:1284–1294.e1. [DOI] [PubMed] [Google Scholar]
- 10.Kromrey ML, Bülow R, Hübner J et al. Prospective study on the incidence, prevalence and 5-year pancreatic-related mortality of pancreatic cysts in a population-based study. Gut. 2018;67:138–145. [DOI] [PubMed] [Google Scholar]
- 11.Lobb RJ, Becker M, Wen SW et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. 2015;4:27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robin X, Turck N, Hainard A et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee K, Fraser K, Ghaddar B et al. Multiplexed Profiling of Single Extracellular Vesicles. ACS Nano. 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Görgens A, Bremer M, Ferrer-Tur R et al. Optimisation of imaging flow cytometry for the analysis of single extracellular vesicles by using fluorescence-tagged vesicles as biological reference material. J Extracell Vesicles. 2019;8:1587567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko J, Wang Y, Gungabeesoon J et al. Droplet-based single EV sequencing for rare immune subtype discovery. μ-TAS 2019. 2019;T104c:85. [Google Scholar]
- 16.Akama K, Shirai K, Suzuki S. Droplet-Free Digital Enzyme-Linked Immunosorbent Assay Based on a Tyramide Signal Amplification System. Anal Chem. 2016;88:7123–7129. [DOI] [PubMed] [Google Scholar]
- 17.Furukawa T, Klöppel G, Volkan Adsay N et al. Classification of types of intraductal papillary-mucinous neoplasm of the pancreas: a consensus study. Virchows Arch. 2005;447:794–799. [DOI] [PubMed] [Google Scholar]
- 18.Maker AV, Katabi N, Gonen M et al. Pancreatic cyst fluid and serum mucin levels predict dysplasia in intraductal papillary mucinous neoplasms of the pancreas. Ann Surg Oncol. 2011;18:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gbormittah FO, Haab BB, Partyka K et al. Characterization of glycoproteins in pancreatic cyst fluid using a high-performance multiple lectin affinity chromatography platform. J Proteome Res. 2014;13:289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H, Shi J, Anandan V et al. Reevaluation and identification of the best immunohistochemical panel (pVHL, Maspin, S100P, IMP-3) for ductal adenocarcinoma of the pancreas. Arch Pathol Lab Med. 2012;136:601–609. [DOI] [PubMed] [Google Scholar]
- 21.Kaur S, Smith LM, Patel A et al. A Combination of MUC5AC and CA19–9 Improves the Diagnosis of Pancreatic Cancer: A Multicenter Study. Am J Gastroenterol. 2017;112:172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinha J, Cao Z, Dai J et al. A Gastric Glycoform of MUC5AC Is a Biomarker of Mucinous Cysts of the Pancreas. PLoS One. 2016;11:e0167070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basturk O, Khayyata S, Klimstra DS et al. Preferential expression of MUC6 in oncocytic and pancreatobiliary types of intraductal papillary neoplasms highlights a pyloropancreatic pathway, distinct from the intestinal pathway, in pancreatic carcinogenesis. Am J Surg Pathol. 2010;34:364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das KK, Xiao H, Geng X et al. mAb Das-1 is specific for high-risk and malignant intraductal papillary mucinous neoplasm (IPMN). Gut. 2014;63:1626–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das KK, Geng X, Brown JW et al. Cross Validation of the Monoclonal Antibody Das-1 in Identification of High-Risk Mucinous Pancreatic Cystic Lesions. Gastroenterology. 2019;157:720–730.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okada K, Hirabayashi K, Imaizumi T et al. Stromal thrombospondin-1 expression is a prognostic indicator and a new marker of invasiveness in intraductal papillary-mucinous neoplasm of the pancreas. Biomed Res. 2010;31:13–19. [DOI] [PubMed] [Google Scholar]
- 27.Tobita K, Kijima H, Dowaki S et al. Thrombospondin-1 expression as a prognostic predictor of pancreatic ductal carcinoma. Int J Oncol. 2002;21:1189–1195. [PubMed] [Google Scholar]
- 28.Kim J, Bamlet WR, Oberg AL et al. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19–9 blood markers. Sci Transl Med. 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jabbar KS, Arike L, Verbeke CS et al. Highly Accurate Identification of Cystic Precursor Lesions of Pancreatic Cancer Through Targeted Mass Spectrometry: A Phase IIc Diagnostic Study. J Clin Oncol. 2018;36:367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bausch D, Mino-Kenudson M, Fernández-Del Castillo C et al. Plectin-1 is a biomarker of malignant pancreatic intraductal papillary mucinous neoplasms. J Gastrointest Surg. 2009;13:1948–54; discussion 1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen R, Yi EC, Donohoe S et al. Pancreatic cancer proteome: the proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology. 2005;129:1187–1197. [DOI] [PubMed] [Google Scholar]
- 32.Jang JY, Park YC, Song YS et al. Increased K-ras mutation and expression of S100A4 and MUC2 protein in the malignant intraductal papillary mucinous tumor of the pancreas. J Hepatobiliary Pancreat Surg. 2009;16:668–674. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki K, Watanabe A, Araki K et al. High STMN1 Expression Is Associated with Tumor Differentiation and Metastasis in Clinical Patients with Pancreatic Cancer. Anticancer Res. 2018;38:939–944. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe A, Araki K, Yokobori T et al. Stathmin 1 promotes the proliferation and malignant transformation of pancreatic intraductal papillary mucinous neoplasms. Oncol Lett. 2017;13:1783–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Do M, Han D, Wang JI et al. Quantitative proteomic analysis of pancreatic cyst fluid proteins associated with malignancy in intraductal papillary mucinous neoplasms. Clin Proteomics. 2018;15:17:doi: 10.1186/s12014-018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Springer S, Wang Y, Dal Molin M, Masica DL et al. A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology. 2015;149:1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer GARNEAA, Cancer GARN. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32:185–203.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mito K, Saito M, Morita K et al. Clinicopathological and prognostic significance of MUC13 and AGR2 expression in intraductal papillary mucinous neoplasms of the pancreas. Pancreatology. 2018;18:407–412. [DOI] [PubMed] [Google Scholar]
- 39.Stiles ZE, Khan S, Patton KT et al. Transmembrane mucin MUC13 distinguishes intraductal papillary mucinous neoplasms from non-mucinous cysts and is associated with high-risk lesions. HPB (Oxford). 2019;21:87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lahat G, Lubezky N, Loewenstein S et al. Epithelial-to-mesenchymal transition (EMT) in intraductal papillary mucinous neoplasm (IPMN) is associated with high tumor grade and adverse outcomes. Ann Surg Oncol. 2014;21 Suppl 4:S750–7. [DOI] [PubMed] [Google Scholar]
- 41.Chang YR, Park T, Park SH et al. Prognostic significance of E-cadherin and ZEB1 expression in intraductal papillary mucinous neoplasm. Oncotarget. 2018;9:306–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanno A, Satoh K, Kimura K et al. The expression of MUC4 and MUC5AC is related to the biologic malignancy of intraductal papillary mucinous neoplasms of the pancreas. Pancreas. 2006;33:391–396. [DOI] [PubMed] [Google Scholar]
- 43.Melo SA, Luecke LB, Kahlert C et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frampton AE, Prado MM, López-Jiménez E et al. Glypican-1 is enriched in circulating-exosomes in pancreatic cancer and correlates with tumor burden. Oncotarget. 2018;9:19006–19013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka M, Ishikawa S, Ushiku T et al. EVI1 modulates oncogenic role of GPC1 in pancreatic carcinogenesis. Oncotarget. 2017;8:99552–99566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nie S, Lo A, Wu J et al. Glycoprotein biomarker panel for pancreatic cancer discovered by quantitative proteomics analysis. J Proteome Res. 2014;13:1873–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jenkinson C, Elliott VL, Evans A et al. Decreased Serum Thrombospondin-1 Levels in Pancreatic Cancer Patients Up to 24 Months Prior to Clinical Diagnosis: Association with Diabetes Mellitus. Clin Cancer Res. 2016;22:1734–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang HJ, Lee JM, Joo I et al. Assessment of Malignant Potential in Intraductal Papillary Mucinous Neoplasms of the Pancreas: Comparison between Multidetector CT and MR Imaging with MR Cholangiopancreatography. Radiology. 2016;279:128–139. [DOI] [PubMed] [Google Scholar]
- 49.Matsuda Y, Kimura W, Matsukawa M et al. Association Between Pancreatic Cystic Lesions and High-grade Intraepithelial Neoplasia and Aging: An Autopsy Study. Pancreas. 2019;48:1079–1085. [DOI] [PubMed] [Google Scholar]
- 50.Cohen JD, Li L, Wang Y et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y, Yan S, Tian H et al. Macrophage inhibitory cytokine-1 versus carbohydrate antigen 19–9 as a biomarker for diagnosis of pancreatic cancer: A PRISMA-compliant meta-analysis of diagnostic accuracy studies. Medicine (Baltimore). 2018;97:e9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan S, Chen R, Crispin DA et al. Protein alterations associated with pancreatic cancer and chronic pancreatitis found in human plasma using global quantitative proteomics profiling. J Proteome Res. 2011;10:2359–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang KS, Im H, Hong S et al. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci Transl Med. 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Yang J, Li H et al. Tumor markers CA19–9, CA242 and CEA in the diagnosis of pancreatic cancer: a meta-analysis. Int J Clin Exp Med. 2015;8:11683–11691. [PMC free article] [PubMed] [Google Scholar]
- 55.Tang H, Partyka K, Hsueh P et al. Glycans related to the CA19–9 antigen are elevated in distinct subsets of pancreatic cancers and improve diagnostic accuracy over CA19–9. Cell Mol Gastroenterol Hepatol. 2016;2:201–221.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh S, Pal K, Yadav J et al. Upregulation of glycans containing 3’ fucose in a subset of pancreatic cancers uncovered using fusion-tagged lectins. J Proteome Res. 2015;14:2594–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ciprani D, Morales-Oyarvide V, Qadan M et al. An elevated CA 19–9 is associated with invasive cancer and worse survival in IPMN. Pancreatology. 2020;20:729–735. [DOI] [PubMed] [Google Scholar]
- 58.Young MR, Wagner PD, Ghosh S et al. Validation of Biomarkers for Early Detection of Pancreatic Cancer: Summary of The Alliance of Pancreatic Cancer Consortia for Biomarkers for Early Detection Workshop. Pancreas. 2018;47:135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scholten L, van Huijgevoort NCM, Bruno MJ et al. Surgical management of intraductal papillary mucinous neoplasm with main duct involvement: an international expert survey and case-vignette study. Surgery. 2018 [DOI] [PubMed] [Google Scholar]
- 60.Attiyeh MA, Fernández-Del Castillo C, Al Efishat M et al. Development and Validation of a Multi-institutional Preoperative Nomogram for Predicting Grade of Dysplasia in Intraductal Papillary Mucinous Neoplasms (IPMNs) of the Pancreas: A Report from The Pancreatic Surgery Consortium. Ann Surg. 2018;267:157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shao H, Im H, Castro CM et al. New Technologies for Analysis of Extracellular Vesicles. Chem Rev. 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cohen L, Walt DR. Single-Molecule Arrays for Protein and Nucleic Acid Analysis. Annu Rev Anal Chem (Palo Alto Calif). 2017;10:345–363. [DOI] [PubMed] [Google Scholar]
- 63.Madhavan B, Yue S, Galli U, Rana S, Gross W, Müller M, Giese NA, Kalthoff H, Becker T, Büchler MW, Zöller M. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. 2015;136:2616–2627. [DOI] [PubMed] [Google Scholar]
- 64.Liang K, Liu F, Fan J et al. Nanoplasmonic Quantification of Tumor-derived Extracellular Vesicles in Plasma Microsamples for Diagnosis and Treatment Monitoring. Nat Biomed Eng. 2017;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferguson S, Weissleder R. Are current EV diagnostics sensitive enough for early cancer detection? Adv Biosystems. 2020 (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aronsson L, Andersson R, Bauden M et al. High-density and targeted glycoproteomic profiling of serum proteins in pancreatic cancer and intraductal papillary mucinous neoplasm. Scand J Gastroenterol. 2018;53:1597–1603. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y, Sun Y, Feng J et al. Glycopatterns and Glycoproteins Changes in MCN and SCN: A Prospective Cohort Study. Biomed Res Int. 2019;2019:2871289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park J, Han D, Do M et al. Proteome characterization of human pancreatic cyst fluid from intraductal papillary mucinous neoplasm by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2017;31:1761–1772. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.