Abstract
Caenorhabditis elegans has emerged as a major model in biomedical and environmental toxicology. Numerous papers on toxicology and pharmacology in C. elegans have been published, and this model has now been adopted by investigators in academic toxicology, pharmacology, and drug discovery labs. C. elegans has also attracted the interest of governmental regulatory agencies charged with evaluating the safety of chemicals. However, a major, fundamental aspect of toxicological science remains underdeveloped in C. elegans: xenobiotic metabolism and transport processes that are critical to understanding toxicokinetics and toxicodynamics, and extrapolating to other species. The aim of this review was to initially briefly describe the history and trajectory of the use of C. elegans in toxicological and pharmacological studies. Subsequently, physical barriers to chemical uptake and role of the worm microbiome in xenobiotic transformation were described. Then a review on what is and is not known regarding the classic Phase I, Phase II, and Phase III processes was examined. In addition, the following were discussed (1) regulation of xenobiotic metabolism; (2) review of published toxicokinetics for specific chemicals; and (3) genetic diversity of these processes in C. elegans. Finally, worm xenobiotic transport and metabolism was placed in an evolutionary context; key areas for future research highlighted; and implications for extrapolating C. elegans toxicity results to other species discussed.
Keywords: Caenorhabditis elegans, toxicokinetics, pharmacokinetics, xenobiotic metabolism, xenobiotic transport, microbiome, nuclear hormone receptor, genetic diversity, evolutionary toxicology
Introduction
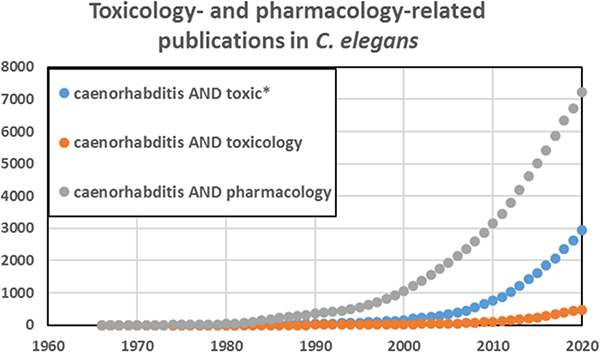
Caenorhabditis elegans has emerged as an important model in biomedical and environmental toxicology. C. elegans was first described over 100 years ago by Maupas (1900) and was intermittently studied thereafter until coming to prominence as an exceptionally powerful model organism for developmental biology, neurobiology, and genetics, as a result of pioneering efforts by Sydney Brenner and colleagues in the 1970s (Nigon and Felix 2017). Although there was some early research in toxicologically relevant areas such as DNA damage and repair (Hartman and Herman 1982) and antioxidant defenses (Blum and Fridovich 1983), the first explicit efforts to develop C. elegans as a model for toxicological research were carried out by Phil Williams, David Dusenberry, and colleagues beginning in the 1980s (Williams and Dusenbery 1987; 1988; 1990a; 1990b). In the early 1990s, Jonathan Freedman’s lab worked on heavy metal response (Slice et al 1990, Freedman et al. 1993), toxicogenomic analysis (Cui et al. 2007), and medium-throughput toxicity testing (Boyd et al 2007), and went on to establish a worm toxicology lab at the United States National Toxicology Program (Xia et al. 2018; Behl et al. 2015; Boyd et al. 2009; 2010; 2015). Starting in the mid-1990s, Christian Sternberg’s group carried out ecotoxicological studies with aquatic and sediment exposures (Hoss et al. 1997; 1999; Traunspurger et al. 1997), ultimately leading to a number of academic (Hoss et al. 2009; Hagerbaumer et al. 2015) and two standardized toxicology testing protocols (International Standard Organization (ISO) 2020, (ASTM) 2001). Richard Nass established the use of C. elegans for chemical-induced neurodegeneration (Nass et al 2001b; 2002). Further, C. elegans was used to study transgenerational and environmental epigenetics (Weinhouse et al. 2018; Kelly 2014). C. elegans is now a well-established model for human and ecological toxicology employed by many labs (a non-comprehensive sampling identifies approximately two dozen: (Leung et al. 2008; Boyd et al. 2010; Steinberg et al 2008, Helmcke et al 2010; Meyer and Williams 2014; Tejeda-Benitez and Olivero-Verbel 2016; Hunt 2017, Choi et al. 2014; Honnen 2017, Ferreira and Allard 2015, Allard et al. 2013; Lenz et al 2017, Nass et al 2001a; Harrington et al. 2010; Cooper and Van Raamsdonk 2018; Liao and Yu 2005; Fitsanakis et al 2014; Menzel et al. 2005; Harlow et al. 2018; Zhao et al. 2013; Brady et al. 2019; Anbalagan et al. 2013; Clavijo et al. 2016; Hoss et al. 2009; Horsman and Miller 2016; Shomer et al. 2019; Zhang et al. 2020; Haegerbaeumer et al. 2018b; Lee et al. 2020; Shen et al. 2019; Harlow et al. 2016; Dietrich et al. 2016; Xiong et al 2017)). The number of publications on toxicology and related fields in C. elegans has grown rapidly in recent years, with an even more rapid growth in pharmacology-related publications (Figure. 1).
Figure 1. There has been a rapid increase in the use of C. elegans for pharmacology and toxicology research.
Figure generated based on Pubmed searches using the terms in the legend, January 2021.
This review is complemented by extensive investigations of other stress-response pathways by non-toxicologists, including heat shock unfolded protein, DNA damage, antioxidant defense, osmotic stress, hormesis, insulin-responsive pathways, hypoxia, caloric stress, regulation of apoptosis and necrosis, and autophagy (Rodriguez et al. 2013; Prahlad and Morimoto 2009; Rieckher et al. 2018; Baugh 2013; Ristow and Schmeisser 2011; Murphy 2006; Baumeister et al 2006; Blackwell et al. 2015; Nikoletopoulou and Tavernarakis 2014; Hengartner and Horvitz 1994; Melendez and Levine 2009). Importantly, as reviewed by Hunt (2017) of the U.S. Food and Drug Administration (Center for Food Safety and Applied Nutrition), there is a significant correspondence between C. elegans and higher eukaryotes in rank-order acute toxicity of chemicals as noted in multiple studies in several labs.
Explicit discussion of the strengths and limitations of C. elegans as a model for toxicological research may be found in several review papers and books (Maurer et al 2018; Queiros et al. 2019; Weinhouse et al. 2018; Hunt 2017; Wang 2019a; 2019b; 2020), and thus will not be repeated here. The focus of this review will be on one very important and oft-cited limitation: the absence of detailed understanding of xenobiotic transport and metabolism processes that regulate concentrations of chemicals and metabolites that reach molecular targets. The study of these processes is fundamental to toxicology, pharmacology, and drug discovery. A truism in toxicology is that “the dose makes the poison,” and it is not possible to truly understand the toxicity of a chemical without understanding how much of it (or its metabolites) has reached specific molecular sites of action. Similarly, it is not possible to extrapolate effects observed in one species (e.g., worms) to another (e.g., humans) without being able to compare internal toxicant concentrations. In toxicology, a distinction is made between toxico(pharmaco)kinetics (understanding the absorption, distribution, metabolism, and excretion (ADME) processes that regulate xenobiotic transport and transformation processes) and toxico(pharmaco)dynamics (understanding the interactions of xenobiotics with molecular targets including receptors, DNA, and proteins).
Toxicodynamics (TD) appear to be relatively similar between C. elegans and higher eukaryotes basedup on conservation of genes that encode molecular targets such as proteins and signaling pathways, as reviewed in some detail previously (Leung et al. 2008; Wang 2019a; Hunt 2017). Further, the great majority of worm toxicology papers to date studied TD, and these investigations generally support similar mechanisms of action for many chemicals in worms compared to higher eukaryotes. However, genetic, biochemical, and other differences that qualitatively alter potential chemical toxicity exist (reviewed in Maurer et al 2018; Queiros et al. 2019; Weinhouse et al. 2018)) and need to be considered. Worms (1) do not synthesize heme or cholesterol; (2) lack an adaptive immune system; and (3) lack homologs of some important receptors such as estrogen receptors. Therefore, chemicals that act directly on proteins in these pathways in higher eukaryotes might exert differing or no effects in worms.
Toxicokinetics (TK), in contrast, the focus of this review, is relatively poorly studied in C. elegans. Further, based upon the large physiological differences between worms and humans TK are expected to be different in C. elegans. It is known the nematode cuticle and eggshell are effective barriers to some chemicals. In addition the ADME are likely different in an organism that has a simple gut, but lacks a circulatory system and many discrete organs such as the liver. Significant uncertainty also exists in our understanding of the regulation and catalytic activity of xenobiotic metabolic enzymes and transporters. This review is intended to highlight what is or is not known regarding these processes in C. elegans, with the goal of stimulating research to fill in these knowledge gaps and improve the utility of this powerful model organism for toxicology and pharmacology.
Uptake of chemicals by the nematode occurs predominantly either (1) across the cuticle, which has few openings and thus presents an effective barrier against many chemicals and particles, or (2) via ingestion. Description of these barriers is provided in the next section. Emerging evidence suggests that the worm’s microbiome exhibits the potential to alter chemicals prior to intracellular uptake, and the following section summarizes our nascent understanding of the role of the worm microbiome in xenobiotic and drug metabolism. The state of knowledge of the biochemical toxicological processes classically referred to as “Phase I, II, and III” (Casarett et al 2008) are present in worms. These processes, which are critically important for organic xenobiotics, but also in some cases important for metals and metalloids, are conceptualized as:
Phase I: enzymatically exposing or adding reactive moieties in parent xenobiotic;
Phase II: conjugating the Phase I-modified (or in some cases, parent) xenobiotic to large, water-soluble molecules, which facilitates excretion;
Phase III: transport of these metabolized compounds out of the cell (some transporters may act to import a parent compound, sometimes described as “Phase 0”).
The known transcriptional regulations of these processes, including the role of the large number of nuclear hormone receptors present in C. elegans are described below. An important caveat to keep in mind in the context of our discussion of specific genes is that most of what is presented is for the historically heavily-studied N2 Bristol strain. Subsequently a comprehensive catalog from the literature in which actual chemical measurements have been made in worms that can inform us about chemical uptake, transformation, and excretion is provided.. The impacts of genetic diversity present across C. elegans populations (C. elegans is found globally), its use to identify sources of natural variation in toxicological processes, and its role in an evolutionary toxicology context are discussed. Finally, knowledge gaps are summarized and future directions discussed.
Physical barriers and tissue-specific xenobiotic transport and metabolism
A simple cell membrane consists of a lipid bilayer composed primarily of phospholipids, glycolipids, and cholesterol. The phospholipids arrange in such a way that their hydrophobic tails are pointed inwards and their hydrophilic heads are oriented towards the outer and inner membrane surfaces. The membrane barrier is differentially permeable to various xenobiotics based upon their physicochemical properties (movement by transporters is addressed below). Some xenobiotics might passively diffuse across cell membranes; hydrophilic molecules might enter through aqueous pores in the membrane, and hydrophobic molecules might diffuse directly through the lipid domain of the membrane (Benz et al 1980). The smaller the molecule, the more rapidly it moves across a membrane, either by aqueous pores or simple diffusion. For large organic molecules, the octanol/water partition coefficient P dictates the rate at which the molecule moves across the membrane, with higher lipophilicity (positive log P) corresponding to higher membrane permeability, except at extreme levels of lipophilicity. In the case of weak organic acids and bases, ionic compounds move slowly and inefficiently through aqueous pores whereas non-ionized forms diffuse across the lipid membrane. Thus, the rate at which non-ionized organic acids and bases permeate the membrane depends on pKa/pKb of the compound and the pH of the surrounding environment (Avdeef 2001; Manallack et al. 2013; Lehman-McKeeman 2013). However, C. elegans is not just a simple cell model system with a single membrane for protection; it is a complex organism with many physical barriers that prevent the passive transport of molecules into its body. In this section, the physical barriers that may impact uptake and release of xenobiotics by C. elegans are presented (Figure 2).
Figure 2.
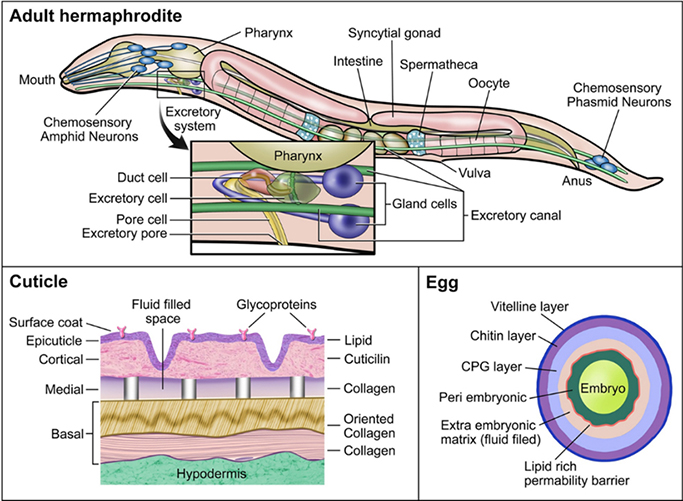
Schematic of physical barriers to xenobiotic diffusion in C. elegans.
The worm’s principle physical barrier is its cuticle (Figure 2). The cuticle of the nematode provides structure to maintain worm body shape, along with protection from its environment (Johnstone 1994; Page 2007; Lints 2009). The cuticle consists of a complex matrix of collagen and serves as a physical barrier against chemicals and pathogens. The cuticle is well known for protecting the worm from uptake of xenobiotics, and often viewed as a drawback to using C. elegans in high-throughput toxicity assays (Xiong et al 2017). Mutants with defective cuticle collagen proteins, of which there are more than 150, were employed to increase uptake in pharmacological screens (Xiong et al 2017; Burns et al. 2010; Johnstone 2000). In adult C. elegans, the cuticle is roughly 0.5 μm thick and contains 5 distinct zones: surface coat, epicuticle layer, cortical zone, medial zone, and basal zone (Cox et al 1981a; 1981b). The width of the cuticle increases with the age of the worm as the basal zone expands, and in older worms it often wrinkles, potentially due to weakening of the muscles and hypodermis (Herndon et al. 2002). The cuticle might also be damaged from physical interactions with the environment and the process of mating (Woodruff et al. 2014). Each cuticle layer has a distinct composition, suggesting a high degree of specialization between layers (Johnstone 2000). The structure of the cuticle varies at each larval stage along with the composition of each of the layers (Lints 2009; Page 2007). C. elegans undergoes molting periodically until adulthood. Molting occurs in two stages: lethargus, a period of relative behavioral dormancy and extracellular matrix remodeling in preparation for cuticle shedding, and ecdysis, when the cuticle is undergoing a molt cycle (Singh and Sulston 1978; Lažetić and Fay 2017). At each larval stage, C. elegans undergoes ecdysis, in which it sheds the current cuticle and forms a new one. The process of molting enables rapid growth and potentially depuration of any xenobiotic compounds on or within the current cuticle. The existence of varying cuticle structures throughout C. elegans development also suggests that critical windows of sensitivity to cuticular xenobiotic uptake exist. Dauer larvae, which are highly resistant to many stressors (Erkut et al. 2011; Fielenbach and Antebi 2008), possess thickened basal and epicuticle layers that might offer enhanced protection from their environment (Wolkow 2011; Cassada and Russell 1975). Early developmental exposures may initate more uptake of xenobiotics because of the difference in protein composition within the cuticle and thinness. Uptake might also be greater in older worms as the cuticle begins to wrinkle and weaken, although little research has been done to address alterations in uptake kinetics throughout the lifespan of C. elegans. It is also important to note that many of these physical processes and barriers have been extensively studied in C. elegans hermaphrodites but are rarely studied in males. Given the differences in size and morphology of male C. elegans, particularly around the tail region, it is possible that the different sexes might have altered uptake kinetics of xenobiotics, but this hypothesis has not apparently been addressed (Sulston et al 1980; Emmons 2005).
Another important barrier is the eggshell, which in C. elegans begins to form as soon as the oocyte enters the spermatheca (Figure 2) and is fertilized (Stein and Golden 2015). The eggshell of C. elegans is the dominant physical barrier when the nematode is in utero and externally (after the egg is laid at approximately 30-cell stage), prior to hatching. The trilaminar outer eggshell is made of an outer vitelline layer, a chitin layer, and chondroitin proteoglycan (CPG) layer (Olson et al. 2012; Johnston and Dennis 2012). Beneath these layers resides the fluid-filled extra-embryonic matrix (EEM), a lipid-rich layer that acts as a permeability barrier to the developing embryo, and an amorphous space referred to as the peri-embryonic layer (Olson et al. 2012). Maternal environment and food supply impact the structure and size of the eggshells produced (Harvey and Orbidans 2011). In producing the eggshell, it is postulated that it is possible that the mother might use this process as a way to excrete xenobiotics, offering a direct source of exposure to the offspring, although no apparent research has been done to address this. Similarly, transfer of xenobiotics from mother to egg via other maternally-loaded components, such as vitellogenin is likely to occur. Vitellogenin contains both hydrophobic and hydrophilic domains and is an important vector for the transfer of xenobiotics to eggs in other species (Monteverdi and Di Giulio 2000). After egg laying occurs, the eggshell is exposed to the environment where it serves to protect the developing worm. Molecules that are able penetrate the various layers of the eggshell may be taken up by the developing worm.
C. elegans might also be exposed to xenobiotic compounds through ingestion. C. elegans feeds on microbes in the lab, on bacterial lawns and suspension in liquid cultures through pharyngeal pumping. The pharynx is the neuromuscular pump that connects the mouth to the intestine and contracts and relaxes in order to take in bacteria and expel liquids (Avery 2012). The pharynx is lined with a specialized cuticle that helps to form structural elements of the pharynx such as the flaps, sieve, and the grinder, all of which are necessary for initial processing as well as transport of food to the intestine, and protection against diffusion of compounds in the pharynx (Page 2007; Altun 2009a). The pharyngeal cuticle and the pumping mechanism that expels liquids might serve as a protective barrier from xenobiotic uptake. However, xenobiotics that are bound to or taken up by bacterial food source would still be ingested by the worm. For chemicals that make their way into the intestine, uptake through epithelial cells lining the intestine might occur, as well as transport and trafficking to other locations in the organism. The intestine exhibits high expression of P-glycoproteins, involved in trafficking hydrophobic molecules, as well as cytochrome P450s, metallothioneins, and other phase I-III enzymes (McGhee 2007). It is important to note the availability and uptake of xenobiotics in the intestine varies by physicochemical properties and the chemical properties of the gut. As previously noted, membrane permeability to these compounds in the gut is determined by chemical structure, pKa, and pH of the environment (Casarett et al 2008). Alterations in the C. elegans diet and microbiome also lead to changes in intestine pH or metabolites, which might alter the availability of certain compounds (Höss et al 2011). Other cell types may also play specialized roles; for example, the 6 coelomocytes are macrophage-like cells that occupy the body cavity, are highly endocytotic, and may, similar to intestinal cells, play a “liver-like” role. Investigators demonstrated their importance in metal detoxification via endocytosis and metal transport and binding proteins (Tang et al. 2020; Schwartz et al. 2010; Maurer et al. 2016).
C. elegans possesses an excretory system comprising of 4 main cell types. The excretory system includes one pore cell, one duct cell, one canal cell (excretory cell), and a fused pair of gland cells (Altun 2009b) (Figure 2). The excretory system plays a role in osmoregulation and waste elimination, somewhat similar to the renal system in vertebrates (Sundaram and Buechner 2016). In the adult worm, the excretory canal cell extends from the head to the tail of the animal with the cell body located in the head region. The excretory canal cell is connected to the excretory gland cells, duct cell, and pore cell. The canal cell collects and guides fluids from within the worm towards the cell body for excretion through the duct and pore cells (Nelson et al 1983; Nelson and Riddle 1984; Stone et al 2009). The duct cell and pore cell form a channel that opens to the environment, through which fluid might be excreted. Both the duct and pore cells possess a specialized cuticle that provides structure and protects it from the surrounding environment (Lints 2009; Sundaram and Buechner 2016). Nonetheless, the excretory system might also permit entry as well as exit for xenobiotics.
C. elegans also have a sophisticated chemosensory system that mediates detection and avoidance of noxious conditions. Chemosensation is vital to survival of C. elegans, enabling them to find food, mate, avoid harmful environments, and enter or exit the dauer stage (Bargmann 2006). Chemosensory neurons are found in the head and tail regions of the worm. There are 16 pairs of bilaterally symmetric neurons (approximately10% of the total number of neurons, with neurons comprising approximately 1/3 of all somatic cells) that are involved with chemosensation (Bargmann 2006; Sengupta 2007). These neurons are bipolar, containing a single axon, a single dendrite, and cilia, which are exposed to the surrounding environment through small openings in the cuticle (Sengupta 2007; Ware et al. 1975; Ward et al. 1975). The ability for C. elegans to sense and respond to chemical stimuli enables them to avoid potentially toxic environments in the wild and protect themselves from exposures to xenobiotic compounds via chemotaxis. At the same time, the exposure of cilia to the external environment may also be a route of exposure for xenobiotics to infiltrate the neurons (Perkins et al. 1986).
Overall, physical and behavioral barriers play key roles in protecting C. elegans from xenobiotic uptake and accumulation, but knowledge of the manner by which most specific xenobiotics and even classes of xenobiotics cross these barriers and alter behavior is lacking.
C. elegans microbiome
The gut microbiome is a complex mixture of microorganisms (Koontz et al. 2019). Changes and alterations to the composition of the gut microbiota are associated with several diseases, including irritable bowel disease, obesity, type 2 diabetes, and neurodegenerative disorders in humans (Scotti et al. 2017). Recently, more research attempted to elucidate the complex interactions between host and the associated gut microbiome. This theme has also increased in toxicology research with many review papers emphasizing the importance and significance of understanding the role of the gut microbiome in toxicology (Koontz et al. 2019;National Academies of Sciences and Medicine 2018; Pryor et al. 2019; Mesnage et al. 2018; Koppel et al 2017; Claus et al 2016).
Humans possess several distinct microbiomes, including the skin, respiratory, reproductive system, and gut (National Academies of Sciences and Medicine 2018). The gut microbiome is the most abundant (National Academies of Sciences and Medicine 2018; Sender et al 2016). When chemicals enter the gut, bidirectional interactions occur: the chemicals may be transformed by microbiota present to be more or less toxic or effective, or chemicals might alter the composition of the microbiome (Koontz et al. 2019; National Academies of Sciences and Medicine 2018). Microbes within the gut primarily use hydrolytic and reductive reactions but are capable of utilizing a wide range of metabolic enzymes including azoreductases, nitroreductases, beta-glucuronidases, sulfatases, and beta-lyases (Koontz et al. 2019; Koppel et al 2017) to metabolize or transform environmental pollutants such as some heavy metals, pesticides, polycyclic aromatic hydrocarbons, and polychlorinated biphenyls. Conversely, toxicant exposure might lead to dysbiosis by altering abundance and diversity of the microbiota present (Claus et al 2016; Tsiaoussis et al. 2019).
Investigating the human microbiota may be challenging and complex; model organisms provide the opportunity to simplify these studies. C. elegans is particularly useful for studying the microbiome because of the ease of creating germ-free progeny through bleaching ability to (1) grow and develop on a monoculture of bacteria, (2) grow worms on bacteria-free media, and (3) use mutant strains (Gerbaba et al 2017; Zhang et al 2017). Although, as described above, C. elegans has been increasingly employed to study toxicological impacts of xenobiotics, most of these studies were performed without considering what role their gut microbiome might play (Dirksen et al. 2016). Further, most experiments to date were conducted by feeding only with standard strains of bacteria such as the common lab food source, Escherichia coli strain OP50. None of the commonly used lab food strains are found in nature; thus, these food sources presumably generate an artificial microbiome, although as discussed below, there is some limited evidence for non-food source bacteria colonizing the worm intestine in the lab.
A few studies examined gut microbiomes in C. elegans found in the wild. Zhang et al. (2017) presented the first three papers that document non-lab microbiomes in C. elegans. The dominant taxa across varying substrates were Proteobacteria (approximately80%) while the remaining approximately 20% were Bacteriodetes, Firmicutes, and Actinobacteria. These investigators also showed that the composition of the worm gut microbiome was distinct from and to a large extent independent of their environment (Zhang et al. 2017; Berg et al. 2016). The level worm microbiomes vary by geography has yet to be determined. Lee et al (2020) examined gut microbiome of N2 Bristol worms grown in lab conditions with OP50 and found that despite the lab culture conditions, these worms also contained a complex microbiome that consisted mainly of Proteobacteria, Firmicutes, and Actinobacteria (Stenotrophomonas, Bacillus, Microbacterium). Non-OP50 bacteria might be present because typical worm culture lab techniques are not sterile, such that other bacteria may be found and able to colonize the gut of the worm. However, this study contrasts with other reports that noted that when grown under lab conditions, the N2 strain with OP50 bacteria did not contain microbes other than OP50 within their gut (Dirksen et al. 2016). Clarifying this discrepancy, and the degree to which it depends upon lab culture conditions, is important, because certain bacterial strains influence worm development and reproduction (Dirksen et al. 2016; 2020).
The relationship of C. elegans with standard OP50 changes throughout aging. During development, bacteria consumed are destroyed in the worm’s grinder, leaving them inactivated in their intestine. However, during middle age, a small number of bacteria consumed pass through the grinder and colonize the intestine. As the worm ages, live bacteria found in the intestine begin to proliferate, eventually contributing to their death (Cabreiro and Gems 2013). Therefore, the microbiome may exert a greater effect on xenobiotic metabolism in older versus younger worms. Presumably, other bacteria might colonize the worm gut at earlier and later ages.
Tools for studying the worm microbiome
With advancing sequencing technology, analyzing the worm’s gut microbiome has become more rapid and cost-efficient. However, understanding how differences in diversity and abundance directly affect the worm’s health is more challenging. Recently, tools were developed that might help elucidate how microbes impact the health of C. elegans.
Dirksen et al. (2020) created CeMbio, a kit of 12 bacterial strains that colonize the gut of worms in one native European environment. Worms may be grown on individual strains or on a strain community. When worms were grown on different individual strains, growth rates changed. Compared to OP50, 9 of the strains initiated worms to develop faster, two strains significantly delayed growth, and one strain resulted in similar growth rates. Quantification of bacteria within the gut of worms might be accomplished by PCR, using strain-specific primers. Thus, this resource facilitates testing how exposures influence single or combined microbial populations, and the manner by which these microbiome changes affect host health.
Another tool, developed by Rutter et al (2019) permits an in vivo analysis of worm-bacterial-chemical interactions by using a bacterial food source engineered to serve as a sensor This sensor is a strain of E. coli carrying a plasmid encoding constitutively-expressed mCherry, plus GFP expressed upon exposure to isopropyl β-D-1-thiogalactopyranoside (IPTG). This enables simultaneous visualization of bacteria in the intestine via mCherry, and GFP fluorescence to detect response to IPTG. The ratio of GFP to mCherry therefore provides information both on the bacterial population present, and the response of the bacteria to chemical exposure.
The effect of specific bacterial genes on microbiome-chemical interactions might also be tested via the use of libraries of bacterial mutants. For example for E. coli K-12 there are 3985 mutant strains available (Baba et al. 2006); for E. coli OP50 (Govindan et al. 2015), approximately 2000; for Pseudomonas aeruginosa, approximately 4901 (Jacobs et al. 2003); and for Comamonas aquatica DA1877, approximately 5760 (Watson et al. 2014).
Finally, several methods have been described to reduce or eliminate the influence of bacteria on chemical toxicity. These include heat-killing bacteria (Powell and Ausubel 2008) inactivating bacteria with ultraviolet radiation, sometimes using a DNA-damage-deficient strain of bacteria (Meyer et al. 2010b); feeding standardized, lyophilized bacteria lysate (Garcia-Gonzalez et al. 2017); and culturing in axenic media, for which explicit toxicological protocols were provided (Sprando et al. 2009; Nass and Hamza 2007). It is important to note that each of these methods has its own caveats including in many cases reducing caloric intake and/or food quality (axenic media, heat-killed bacteria) or incomplete abrogation of biochemical metabolism (ultraviolet light inactivation).
Gut microbiome and toxicological studies using C. elegans
To date, relatively few studies explored how xenobiotics alter the C. elegans gut microbiome, or if transformation of xenobiotics occurs within the worm gut. Garcia-Gonzalez et al. 2017) investigated how microbiota transformed drugs, and how this affected worm health, in order to better understand drug efficacy variability often seen in clinical studies. Other studies examined the manner in which the worm microbiome was aletered after exposure to environmental pollutants.
Cabreiro et al (2013) using standard lab strain OP50 found that metformin decreased microbial folate production and altered methionine synthesis, which led to an elevation in worm lifespan. However, if worms were grown on metformin-resistant bacteria, or no bacteria, metformin exerted no significant effect on folate production, and reduced lifespan, indicating toxicity. Moreover, worms grown with metformin and OP50 displayed a significant reduction in S-adenosyl methionine levels, which led to diminished in methionine biosynthesis. Since C. elegans main source of methionine is the diet, a decrease in microbial folate and methionine biosynthesis led to a dietary deficiency in methionine. Cabreiro et al (2013) postulated that deficiency in methionine is the underlying mechanism attributed to metformin-mediated extended lifespan. Scott et al (2017) found that different strains of bacteria significantly influenced the minimum inhibitory range of 5 different fluoropyrimidines and that live bacteria were necessary to activate fluoropyrimidines. Supplementation with a vitamin B6 precursor, pyridoxal enhanced the efficacy of 5-FU in worms (Scott et al 2017). The addition of pyridoxal to E. coli enhances ribonucleotide metabolism of 5-FU thereby increasing the efficacy to the host. Further, data demonstrated that exposure to metformin altered bacterial one-carbon metabolism which is believed to modulate fluoropyrimidines activation. If worms were given both drugs concurrently, activation of 5-FU was inhibited, resulting in a decreased efficacy. These findings show the importance of understanding how the host microbiome, diet, and drug exposure alter drug efficacy and impact host health.
Lee et al (2020) examined how environmental chemicals alter gut microbiome. OP50-fed worms were exposed to cadmium (Cd) and marked effects on gut microbial composition were noted.Without Cd exposure, Proteobacteria dominated the gut, followed by Firmicutes and Actinobacteria. After Cd exposure, Firmicutes dominated the microbiota. Firmicutes are postulated to be resistant to Cd, which enables them to proliferate within the gut once the Proteobacteria and Actinobacteria were reduced (Lee et al 2020). However, when fed a more diverse population that was isolated from an organic farm, Cd exposure resulted in non-significant alterations in taxa by increasing the amount of Actinobacteria present. Lee et al (2020) demonstrated that worms that possessed a more diverse gut microbiome were more resistant to Cd exposure as evidenced by having a longer lifespan, more progeny, and fewer changes to gut microbial communities compared to worms fed OP50.
Arsenic (As) is biotransformed by microorganisms in the environment where reduction and methylation reactions are particularly important in altering transport and toxicity. In order to investigate how the microbiome might alter As speciation (redox status and methylation) and toxicity, Zhou et al (2020) used three different types of E. coli that either lacked the ability to reduce or methylate arsenic, or might reduce but not methylate As, or might both reduce and methylate As. Zhou et al (2020) found that bacteria biotransformed As, and that the worm responses (gene expression and reproduction) varied depending upon the strain of bacteria, supporting the importance of the microbiome in mediating As-induced toxicity.
Thus data demonstrated how C. elegans might be used as a powerful tool to elucidate the manner in which microbiota alter chemicals to which worms are exposed, as well as how environmental chemicals impact worm microbiota, potentially impacting worm physiology and stress response. It is noteworthy there are caveats associated with differences in C. elegans biology that need to be taken into account. For example, the above-mentioned lack of an adaptive immune system in C. elegans limits the ability to model mammalian mucosal-microbiota interactions, which when perturbed subsequently might alter xenobiotic metabolism, transport, and gut permeability.
Xenobiotic detoxification in C. elegans - Phase I
Phase I metabolic enzymes perform biotransformation of a parent organic compound to introduce or expose functional groups. This typically increases polarity and thus water solubility and reactivity, ultimately permitting conjugation of the compound (Ioannides 2001). Phase I reactions are broadly grouped into three categories: oxidation, reduction, and hydrolysis, with oxidation reactions being the most common and well-studied (Ioannides 2001). In C. elegans, all three types of phase I reactions are represented (Lindblom and Dodd 2006), and phase I enzymes are broadly expressed in somatic cells with some cell type specificity for particular isoforms likely driven by exogenous roles such as intestinal expression and endogenous roles such as hormone and/or neurotransmitter synthesis in neurons and other tissues.
Oxidative reactions are mainly carried out by the cytochrome P450 superfamily of monooxygenase enzymes, but also by flavin-containing monooxygenases, alcohol and aldehyde dehydrogenases, monoamine oxidase, and by peroxidases that perform co-oxidation (Ioannides 2001). These oxidation reactions are presumed to have evolved to increase water solubility and thus reduce toxicity from accumulation of chemicals in hydrophobic biological environments and structures. However, many oxidative reactions of benign hydrophobic substrates paradoxically result in formation of a toxic reactive metabolite (also termed ‘bioactivation’)(Guengerich 2006). A well-known example of this phenomenon is the pollutant benzo[a]pyrene (BaP), which is bioactivated by CYP1A/1B enzymes to a reactive epoxide metabolite that ultimately forms carcinogenic DNA adducts (Shiizaki et al 2017).
Reduction reactions are carried out by cellular reductases and have not been well-characterized for their xenobiotic metabolizing activities, even in mammalian systems. In C. elegans, several genes have been predicted to exhibit reductase activity but have not been closely studied. Finally, hydrolytic metabolism also occurs by enzymes termed hydrolases. These enzymes cleave their substrates using hydrolysis at functional groups, commonly at esters, amides, and epoxides.
Cytochromes P450 in C. elegans
Cytochrome P450 enzymes carry out the majority of oxidative Phase I reactions and, as illustrated by the example of BaP, are responsible for the majority of bioactivation reactions (McDonnell and Dang 2013; Guengerich 2008). Cytochromes P450 are monooxygenases that consume one molecule of molecular oxygen and one molecule of NAD(P)H to add one oxygen atom to a substrate to release water and NADP+ as byproducts. Cytochromes P450 are localized in the endoplasmic reticulum, where they associate with the co-enzyme cytochrome P450 reductase (emb-8 in C. elegans) to carry out their reactions. In mammals, P450s also localize to the mitochondria where they associate with adrenodoxin/adrenodoxin reductase (Ahn and Yun 2010; McDonnell and Dang 2013) (possibly Y73F8A.27 and Y62E10A.6 in C. elegans) although the presence of mitochondrial P450s in C. elegans is relatively understudied.
The C. elegans genome contains genes for at least 86 cytochromes P450 (compared to 60 in humans), and 8 of those are predicted to be pseudogenes (Menzel et al 2001). Three genome-wide RNAi screens collectively identified roles for several of these P450 enzymes (Table 1). Perhaps the most well-studied P450 is daf-9, an orthologue of human CYP2S1 that is involved in the modification of hormones involved in dauer signaling (Jia et al 2002).
Table 1.
C. elegans P450 genes with reported human orthologues, phenotypes, substrates, inhibitors, and/or expression patterns
| C. elegans gene | Human orthologue | Knockdown phenotype | Substrates | Inducers (mRNA expression) | Tissue Expression | Reference |
|---|---|---|---|---|---|---|
| daf-9 | CYP2S1 | Abnormal dauer formation | 3-keto-sterols | nervous system, intestine, spermatheca, vulval muscle | (Gill et al. 2004; Mak and Ruvkun 2004; Motola et al. 2006) | |
| cyp-13A1 | CYP3A4/TBXAS1 | None reported | coelomocyte, gonad | |||
| cyp-13A4 | CYP3A4/5/43 | Growth delay, Uncoordinated movement | broad somatic expression | (Simmer et al. 2003a; Kamath et al. 2003) | ||
| cyp-13A7 | CYP3A4/TBXAS1 | None reported | tetrabromobisphenol A, rifampcin | intestine | (Liu et al. 2020; Chakrapani et al 2008) | |
| cyp-13A8 | CYP3A7/51P | Clear body cavity, Sluggish, some Embryonic lethality | (Kamath et al. 2003) | |||
| cyp-13A10 | CYP3A4/7/43 | None reported | tetrabromobisphenol A | neuron, germline, embryos | (Liu et al. 2020) | |
| cyp-13A12 | CYP3A4/5/7 | None reported | tetrabromobisphenol A, acrylamide | marginal cells | (Liu et al. 2020; Hasegawa et al. 2007) | |
| cyp-14A5 | CYP2U1 | Short defecation cycle | PCB52 | GABAergic neuron, somatic gonad precursor, intestine | (Schafer et al. 2009) | |
| cyp-29A3 | CYP4V2 | Short defecation cycle | eicosapentaenoic acid | 1-methylnicotinamide, chlorpyrifos, 4-bromodiphenyl ether | neurons, intestine, body wall muscle, hypodermis, coelomocytes | (Kulas et al. 2008; Schmeisser et al. 2013; Viñuela et al. 2010) |
| cyp-31A1 (pseudogene) | N/A | Embryonic lethality | (Simmer et al. 2003b) | |||
| cyp-31A2 | CYP4V2 | Embryonic lethality | hypodermis, germline, neurons | (Simmer et al. 2003a) | ||
| cyp-31A5 | CYP4V2 | Embryonic lethality | germline, AFD neurons | (Simmer et al. 2003a) | ||
| cyp-33E2 | CYP2J2 | Reduced pharyngeal pumping, Extended lifespan, Reduced brood size | Long chain fatty acids, eicosapentaenoic acid | pharynx, intestine, neurons | (Deline et al. 2015; Kulas et al. 2008) | |
| cyp-34A9 | tolbutamide | (Harlow et al. 2018) | ||||
| cyp-35A1 | CYP2C18 CYP2D7 CYP2E1 |
Drug resistant1 | 3-bromopyruvate, β-naphthoflavone, PCB52, lansoprazole, primaquine | intestine | (Gu et al. 2020; Menzel et al 2001) | |
| cyp-35A2 | CYP2C18 CYP2D7 CYP2E1 |
Drug resistant1 | 3-bromopyruvate, β-naphthoflavone, PCB52, lansoprazole, atrazine | intestine, neurons (PVD/OLL) | (Gu et al. 2020; Menzel et al 2001; Chakrapani et al 2008) | |
| cyp-35A3 | CYP2C18 CYP2D7 CYP2E1 |
Reduced fat storage | chlorpyrifos, β-naphthoflavone, PCB52, lansoprazole, primaquine | intestine, AFD neurons | (Ashrafi et al. 2003; Viñuela et al. 2010; Menzel et al 2001) | |
| cyp-35A4 | CYP2C18 CYP2D7 CYP2E1 |
Short defecation cycle, Chemical response variant | 3-bromopyruvate, β-naphthoflavone, PCB52, lansoprazole, primaquine | muscle, intestine, neurons | (Gu et al. 2020; Menzel et al 2001) | |
| cyp-35A5 | CYP2C18 CYP2D7 |
Reduced fat storage | 3-bromopyruvate (weak), β-naphthoflavone, PCB52, lansoprazole, primaquine, atrazine | intestine | (Ashrafi et al. 2003; Gu et al. 2020; Kulas et al. 2008; Menzel et al 2001) | |
| cyp-35B1 | CYP2C18 CYP2D7 CYP2E1 |
Reduced fat storage | β-naphthoflavone, ethanol | intestine, hypodermis, dopaminergic neurons | (Ashrafi et al. 2003; Menzel et al 2001) | |
| cyp-35B2 | β-naphthoflavone, lansoprazole, primaquine, ethanol | (Menzel et al 2001) | ||||
| cyp-35C1 | CYP2C18 CYP2D7 CYP2E1 |
Drug resistant1, Oxidative stress resistant | 3-bromopyruvate, tetrabromobisphenol A, fluoranthene, β-naphthoflavone, PCB52, lansoprazole, primaquine | intestine, neurons (AFD/OLL/PVD) | (Gu et al. 2020; Liu et al. 2020; Menzel et al 2001; Taubert et al. 2008) | |
| cyp-42A1 | CYP4V2 | Uncoordinated movement | hypodermis, intestine | (Kamath et al. 2003) | ||
Term “Drug resistant” indicates reducing defects produced by a particular drug treatment (e.g. reproduction decline after exposure to xenobiotics)
Of the 86 cytochrome P450 genes in the C. elegans genome, many are induced upon exposure to xenobiotics (Table 1, ‘Inducers’), suggesting that at least some of the P450 isoforms encoded in the genome are xenobiotic metabolizing enzymes. For the vast majority of these enzymes, the evidence for induction was based upon mRNA levels quantified by microarray, RNA-seq, and/or qPCR or GFP signals in a strain expressing GFP under the control of the CYP promoter. Due to the lack of availability of antibodies for most C. elegans proteins, there have not been any apparent studies to the authors’ knowledge where P450 induction was quantified on the protein level. This is potentially problematic, because some P450 isoforms are highly regulated post-transcriptionally and mRNA levels often do not correlate well with protein and activity (Chang et al. 2003; Song et al. 1986; Sumida et al. 1999). Further recently Abbass et al (2021) provided indirect evidence that C. elegans might metabolize the commonly studied polycyclic aromatic hydrocarbon benzo[a]pyrene to more-reactive forms, despite confirming previous phenotypic and sequence homology-based evidence (Harris et al. 2020; Leung et al. 2010) that this species lacks the family 1 P450s that canonically produce metabolites that generate bulky DNA adducts. This result highlights the need to study the catalytic activity of C. elegans xenobiotic transformation enzymes.
There are a few examples, however, of more direct measurement of P450 metabolism of xenobiotics in the literature (Table 1, ‘Substrates’). In those cases, the expected metabolite from P450 monooxygenase activity was detected and attributed to a particular isoform. In addition to those examples, P450 metabolites of phenacetin (metabolized by human CYP1A2, abbreviated hCYP1A2), diclofenac (hCYP2C9 substrate), amitriptyline (hCYP2C19/2D6 substrate), clomipramine (hCYP2C19 substrate), dextromethorphan (hCYP2D6/3A4 substrate), and nifedipine (hCYP3A4 substrate) were detected in C. elegans but not attributed to a particular isoform (Harlow et al. 2018). Unfortunately, none of those studies compared metabolic activity with and without an inducer, such that the inducibility of C. elegans P450 enzyme activity is unclear. In addition, it is interesting that in the case of tolbutamide metabolism, the isoforms identified by RNAi to be metabolizers of tolbutamide were not the closest isoforms identified by sequence homology to the human tolbutamide hydroxylases (CYP2C8/9/19)(Harlow et al. 2018). This observation is an illustrative example of how sequence homology is a poor predictor of substrate specificity for P450 enzymes, and is supported by our observations that although several P450 isoforms have sequence homology to CYP2E1 (Table 1), one is not able to detect any CYP2E1-like activity for multiple substrates in C. elegans (unpublished observations, JHH and JNM).
Cytochrome P450 enzymes require a heme cofactor (bound to iron) and a coenzyme (cytochrome P450 reductase) to carry out their reactions (McDonnell and Dang 2013). Unlike humans, C. elegans cannot synthesize heme, and need to scavenge this component from their diet (bacteria) to incorporate it into newly synthesized cytochrome P450 polypeptides (Sinclair and Hamza 2015). C. elegans have a suite of specialized enzymes that deliver environmental heme into the intestine and into other tissues (Chen et al. 2012). Heme availability during xenobiotic challenge might limit the xenobiotic response, but this hypothesis has not been explored experimentally. Cytochrome P450 enzymes also require co-expression of cytochrome P450 reductase also termed P450 oxidoreductase (POR), which is encoded by emb-8 gene in worms. The emb-8 protein is expressed in highest levels in intestine, neurons, and pharynx and is also expressed in coelomocytes, hypodermis, and germline (essentially the same tissues where P450 enzymes are expressed). Loss of emb-8 produces temperature-sensitive embryonic lethality (Miwa et al. 1980), likely due to its role in embryonic polarization (Rappleye et al. 2003).
Finally, longevity associated with reduced mitochondrial function in C. elegans requires induction of cytochrome P450 enzymes through Kruppel-like factor 1, suggesting that protection from endobiotic and/or xenobiotic insults is part of the longevity mechanism (Herholz et al. 2019). This hypothesis fits with data from other species illustrating upregulation of xenobiotic detoxification programs in long-lived mutants. An open question is which endo- and xenobiotic compounds underlie this longevity phenotype.
Flavin Monooxygenases
Similar to humans, C. elegans possess 5 identified flavin monooxgenase (FMO) genes (fmo-1 to fmo-5). FMO enzymes consume oxygen and NADPH to incorporate a single oxygen atom into substrates, usually soft nucleophiles at nitrogen or sulfur atoms. C. elegans FMO enzymes are expressed mainly in two patterns: fmo-1, −2, and −5 in intestinal cells and excretory gland cells, and fmo-3 and −4 in the hypodermis (Petalcorin et al. 2005). In addition, expression was reported in duct cells (fmo-4), pore cells (fmo-4), body-wall muscle (fmo-3,), germline (fmo-4), hypodermal cells (fmo-3, −4), pharyngeal muscle (fmo-1), ventral nerve cord (fmo-4, −5) and neurons (fmo-2, −3, −4, −5).
Similar to cytochromes P450, FMOs contain both endogenous and exogenous substrates and exhibit functions. Nematodes lacking fmo-1, −4, and −5 induce neurodevelopmental defects (Gujar et al 2017). Further, animals deficient in fmo-4 are extremely sensitive to hypoosmotic stress (Petalcorin et al. 2005; Hirani et al. 2016). C. elegans fmo-4 is orthologous to human FMO4, and both contain similar protein structures. However, human FMO4 were not able to rescue hypoosmotic stress sensitivity phenotype in the fmo-4 deletion strain (Hirani et al. 2016). In addition, C. elegans fmo-2 is upregulated by starvation (Goh et al. 2018) and dietary restriction (Leiser et al. 2015), is involved in dietary restriction-induced lifespan extension, and might extend lifespan when overexpressed (Leiser et al. 2015). In addition, both oxidative stress (Goh et al. 2018) and stress from infection with Pseudomonas aeruginosa (PA14)(Dasgupta et al. 2020) or Staphylococcus aureus (Wani et al. 2020) induce fmo-2, which is required for pathogen resistance (Wani et al. 2020). Further, both fmo-2 and fmo-4 are upregulated by hypoxia (Shen et al. 2005).
In terms of catalytic activity, C. elegans FMOs (fmo-1 and −4) are functional for S-oxidation of the prototypical FMO substrate methimazole when expressed recombinantly in insect cells. The overall specific activities (per mg microsomal protein) were approximately10-fold lower than human enzymes expressed in the same background. However, lack of antibodies for C. elegans FMOs precluded estimation of protein expression efficiency in each case (Hirani et al. 2016). To date, this study is the only one that has functionally characterized C. elegans FMO activities, and further studies are needed to determine endogenous and xenobiotic substrates of each isoform.
Alcohol and Aldehyde Dehydrogenases
C. elegans possesses two alcohol dehydrogenases, sodh-1 and sodh-2. Both isoforms are widely expressed in the worm including in neurons, muscle, hypodermis, germline, and intestine. In biochemical assays, alcohol dehydrogenase activity appeared as a single band on a polyacrylamide gel. However, the two isoforms are close in sequence and size (349 vs. 351 amino acids for sodh-1 and sodh-2, respectively) and thus may be expected to co-migrate on a gel (Williamson et al 1991). C. elegans ADH activity is higher for longer-chain alcohols (propanol and butanol) compared to ethanol, and similar to the human enzyme, the worm has a preference for primary over secondary alcohols (Williamson et al 1991).
C. elegans contains several aldehyde dehydrogenase genes (alh-1 through alh-13), most of which have not been functionally characterized and do not have an observable knockdown phenotype (Alaimo et al. 2012). Using gas chromatographic methods to measure internal ethanol accumulation, Alaimo et al (2012) indicated that alh-6 and alh-13 metabolize the ethanol metabolite acetaldehyde to acetic acid.
Monoamine Oxidases
Monoamine neurotransmitters such as dopamine and serotonin are degraded in humans by monoamine oxidases MAO-A and MAO-B. These enzymes also act on xenobiotics that are structurally similar to their natural substrates. C. elegans has a single ortholog of these genes, amx-2 (Schmid et al. 2015), which was demonstrated to metabolize serotonin in the worm (Wang et al. 2017) and presumably would have overlapping specificity with MAO-metabolized xenobiotics identified in mammalian systems.
Hydrolases
Hydrolysis reactions use water to break a chemical bond. Enzymes that perform hydrolysis reactions are referred to as hydrolases. Xenobiotic-metabolizing hydrolases include esterases, amidases, and epoxide hydrolases. Of those, only epoxide hydrolases were studied in detail in C. elegans. Although humans contain 4 epoxide hydrolase isoforms including both membrane-bound (microsomal) and soluble (cytosolic), C. elegans possesses only two isoforms, ceeh-1 and ceeh-2 (Harris et al. 2008). These are most orthologous with the EH3 and EH4 human isoforms, which are the most recently discovered and least well characterized among human isoforms but are postulated to predominantly metabolize lipids. The C. elegans enzymes were confirmed to exhibit endobiotic and xenobiotic metabolizing activities, with ceeh-1 displaying higher activity toward substrates compared to ceeh-2 (Harris et al. 2008). Further studies are needed to establish the substrate specificity of both isoforms, particularly for xenobiotic substrates.
Phase II in C. elegans
Phase II reactions involve conjugation of a substrate with a large, water-soluble group, facilitating excretion. There are 4 major families of phase II enzymes: UDP-glucuronosyltransferases (UGTs), sulfotransferases (SULTs), glutathione S-transferases (GSTs), and N-acetyltransferases (NATs). Except in rare cases, phase II reactions produce safe and easily excreted metabolites, and therefore this phase of metabolism mainly impacts toxicity by facilitating clearance of the compound. Mutations in phase II enzymes in humans result in various phenotypes, including hyperbilirubinemia from mutations in the UGT1 family and elevated risk of cancers from mutations in various UGT and GST isozymes. In C. elegans, Phase II enzymes, similar to phase I, are expressed broadly in somatic cells and enriched in intestine, neurons, and other specialized cells depending upon isoform.
UDP-glucuronosyltransferases (UGTs) are endoplasmic reticulum-localized enzymes that catalyze the conjugation of pollutants, drugs, and endogenous compounds to a sugar group, glucuronic acid. The site of metabolism for UGTs is usually at hydroxyl, carboxyl, or amine functional groups. Individual UGT isoforms are classified into families based upon sequence identity, and substrate specificity for individual UGTs tends to be broad compared to Phase I enzymes (Burchell et al. 1997). Mammals express 22 UGTs from 4 families: UGT1, UGT2, UGT3, and UGT8 (Meech et al. 2019). In contrast, C. elegans possess 66 UGT genes (named ugt-1 through ugt-66) which have all been classified into families UGT1, UGT2, UGT3, and UGT8 based upon sequence homology, and most of the C. elegans UGT genes are each homologous to multiple human isoforms. Knockouts of individual UGT isoforms did not produce phenotypes other than drug hypersensitivity in some cases (Fontaine and Choe 2018; Cui et al. 2007).
A few studies reported induction of UGT after chemical exposures. Hasegawa et al (2010) found that allyl isothiocyanate exposure induces expression of ugt-13 (ortholog of human UGT1A1/2B28/2B7). Di et al (2018) noted that the mycotoxin deoxynivalenol induced ugt-26 (ortholog of human UGT1A4/1A6/2B10) and ugt-28 (ortholog of human UGT1A10/1A6/1A8). The insecticide albendazole also stimulated ugt-16 (ortholog of human UGT3A1/3A2)(Laing et al. 2010), ugt-22 (ortholog of human UGT1A5/1A6/2B17) and ugt-63 (ortholog of human UGT1A10/1A5/1A8)(Fontaine and Choe 2018). The latter study found that ugt-22 upregulation was downstream of the transcription factor skn-1 and noted that mutation of ugt-22 markedly protected against albendazole-mediated toxicity. The chemical acrylamide was also reported to induce several UGT isoforms in C. elegans (Hasegawa et al. 2007). The chemical inducers fluoranthene and β-naphthoflavone also stimulated several UGT isoforms including ugt-13 and ugt-63, as well as other phase I and II xenobiotic metabolizing enzymes (Taubert et al. 2008). Notably, induction was only detected at the transcriptional level, likely due in part to lack of validated antibodies against C. elegans UGT proteins. Further, biochemical activities of UGT enzymes have not been directly measured in C. elegans to date.
C. elegans exposed to the insecticide albendazole also produced conjugated albendazole-glucose metabolites, which Laing et al (2010) suggested may have been formed by UGT enzymes utilizing UDP-glucose as a substrate. The same glucose-conjugated albendazole metabolites were also detected in the helminth H. contortus (Cvilink et al. 2008). Further, glycosylated metabolites of the bacterial toxins 1-hydroxyphenazine and indole were also reported in C. elegans (Stupp et al. 2013). Taken together, these studies provide support to the postulation that UDP-glucose may be utilized by invertebrate UGT enzymes, although this substrate is uncommon in mammals. A similar concept was previously suggested based upon sequence analysis, that is, some C. elegans isoforms that were identified as UGTs may be more correctly termed glycosyltransferases that catalyze the transfer of galactose, glucose, or glucuronic acid (Kapitonov and Yu 1999).
Sulfotransferase enzymes (SULTs) catalyze the conjugation of a sulfonate group from a donor molecule, typically 3-phosphoadenosine-5’-phosphosulfate or PAPS, to a substrate (at a hydroxyl or amino functional group). SULTs might either be membrane-bound in the Golgi apparatus, where they metabolize endogenous molecules, or soluble in the cytosol, where they metabolize xenobiotic and endogenous substrates. The human genome encodes up to 14 SULTs, while the C. elegans genome encodes a single cytosolic sulfotransferase enzyme, ssu-1 (also termed Y113G7A.11 and ceST1)(Hattori et al. 2006). There is no apparent knockdown phenotype, although it is a suppressor of unc-1 and unc-24 phenotypes (thus the naming ssu, or suppressor of stomatin mutant uncoordination)(Carroll et al. 2006). Its enzymatic properties were investigated biochemically by recombinant protein expression, and C. elegans SULT enzyme sulfonated prototypical hydroxylated substrates including 4-nitrophenol and 2-naphthol as well as bisphenol A but did not metabolize monoamines or hydroxysteroids. Hattori et al (2006). created antibodies against the recombinant protein and were able to detect expression in cytosolic fractions of C. elegans lysate and induction by the substrate isophenylpropanol. The mRNA is also highly induced in dauer larvae, indicating a potential role for sulfonation in dauer signaling (Hattori et al. 2006).
Glutathione S-transferases (GSTs) conjugate glutathione to a wide variety of endogenous and xenobiotic substrates and are divided into two types: membrane-bound and soluble family members. Membrane-bound GSTs are found in the endoplasmic reticulum and mitochondria and form homo- and hetero-trimers with a single active site, and typically metabolize endogenous leukotrienes and prostaglandins. Human cytosolic GSTs (11 total) are expressed at high levels, are highly polymorphic, and classified into 6 groups: α, μ, ω, π, ς, θ, and ζ. The C. elegans genome contains 56 GST genes with most categorized within the ς family, which are the most abundant and conserved within insects. However, there are also GSTs in the C. elegans genome classified in the α, π, ζ, and θ subfamilies.
Induction of GSTs by xenobiotics was reported. Wu et al (2015) noted that BaP exposure resulted in induction of SKN-1 as well as gst-24 (homolog of human GST ς enzyme hematopoietic prostaglandin D synthase or HPGDS). The toxin indole up-regulated gst-5, gst-6, and gst-33 (all orthologous to human HPGDS)(Lee et al. 2017), and the natural compound ursolic acid upregulated gst-7 (also orthologous to human HPGDS)(Negi et al. 2017). Interestingly, exogenous heme induced expression of gst-19, gst-7, and gst-5, and these isoforms were proposed to play an endogenous role in heme trafficking (Perally et al. 2008).
Humans have two N-acetyltransferase enzymes that are xenobiotic-metabolizing, NAT1 and NAT2. To date, no N-acetyltransferase genes in C. elegans that are homologous to the human xenobiotic-metabolizing NATs have been identified. In addition to the 4 major families of phase II enzymes described here, it is also possible to conjugate xenobiotics to other cellular substrates including amino acids, but these reactions are less studied and less common.
Phase III in C. elegans
Phase III of xenobiotic metabolism is the export of xenobiotics and metabolites out of the cell, typically via transporters, processes that are especially critical for those xenobiotics whose physicochemical properties make them unable to pass the cell membrane itself (Xu et al 2005; Kell 2020a). Transporters vary vastly in mechanism, function, substrate, and sequence, and are classified largely along those lines into the families of ATP-binding cassette (ABC) transporters, solute carriers (SLCs), pumps, ion channels, and water channels or aquaporins (Hediger et al. 2013). Mutations in each of these transporter families were associated with altered individual sensitivity to xenobiotic or drug exposures and various diseases (DeGorter et al. 2012).
ATP-binding cassette (ABC) transporters comprise a broad family of transmembrane proteins defined by their two nucleotide binding domains and two transmembrane domains. Serving as both importers and exporters in prokaryotes and predominantly exporters in eukaryotes, ABC transporters bind and hydrolyze ATP to move substrates across the membrane (Rees et al 2009). ABC transporters have been implicated in drug resistance, including to chemotherapeutic drugs, efflux of many xenobiotics including phase I and especially phase II metabolites, as well as endogenous processes such as lipid and cholesterol transport, pathogen response, and mitochondrial iron homeostasis (Rees et al 2009; Jin et al. 2012; DeGorter et al. 2012; Glavinas et al. 2004; Klaassen and Casarett 2007).
With 60 sequences encoding ABC transporters, compared to only 49 in humans, these are the most prevalent family of transporters in C. elegans representing 0.3% of the N2 reference gene content (Sheps et al. 2004). Of the 60 identified genes, 15 encode full P-glycoproteins (PGPs), 8 encode PGP half-molecules, 8 encode other multidrug resistance proteins (MRPs), 7 encode members of the abt subfamily, and 1 encodes a cystic fibrosis transporter homolog. The remaining 21 were classified according to HUGO nomenclature into groups D-H but lack a corresponding subfamily name in C. elegans and are not discussed in depth. Eight are apparently not expressed, and one may be a pseudogene (Sheps et al. 2004). Though 16 ABC transporter encoding genes are found in tandem duplications, promoter linked fluorescent protein detection revealed differential expression patterns for 15 of the 16 duplicates, indicating they have different roles (Zhao et al. 2004). Notably, these patterns can overlap. Although PGPs are expressed in various cell types, many are expressed more or solely in intestinal cells, highlighting the intestine as a critical detoxification organ (Lincke et al. 1993).
In examining orthology to human ABC transporters, only 8 were orthologous (Sheps et al. 2004). With such low conservation, it seems logical that those transporters that are conserved play critical roles in biological functions. Thus, unsurprisingly, two are mitochondrial half transporters that support iron homeostasis, and subfamilies E and F, with the highest conservation between humans, Saccharomyces cerevisiae, and Drosophila melanogaster, play roles in formation of ribosome associated proteins (subfamily F) and RNase L inhibition (subfamily E) (Sheps et al. 2004).
Knockouts of several of these ABC transporters were examined, and led to increased sensitivity to heavy metals, xenobiotics, and bacterial toxins (Broeks et al. 1995; 1996; Mahajan-Miklos et al. 1999; Peng et al. 2018). Deletion of pgp-3 enhanced sensitivity to colchicine and chloroquine, and simultaneous knockout of pgp-1 and pgp-3 elevated sensitivity to bacterial toxins, Cd, and arsenite (Mahajan-Miklos et al. 1999, Broeks et al. 1995; 1996). pgp-13 inhibition by RNAi showed that it played a significant role in resistance to imidazolium-based ionic liquids with a suspected similar role for pgp-14 (Peng et al. 2018). pgp-12 knockouts are more susceptible to phenazine 14 exposure (Stupp et al. 2013). pgp-1 and mrp-1 were both upregulated in ivermectin resistant C. elegans, with greater PGP upregulation at higher resistance levels (James and Davey 2009). pgp-12 and pgp-13 are upregulated in ivermectin resistance, with pgp-12 silencing conferring sensitivity (Figueiredo et al. 2018). mrp-5 is implicated in embryonic vitamin B12 transport from mother to offspring (Na et al. 2018). pgp-5 plays roles in both heavy metal and bacterial infection resistance (Kurz et al. 2007). ced-7 promotes the engulfment of apoptotic cells and redistribution of phosphatidylserine (Hamon et al. 2000). pgp-2 contributes to lysosome formation and lipid storage within the intestine, as well as communication between intestine and AWA neurons.
SLCs are a family of more than 300 membrane-bound, ATP-independent transporter proteins, including vesicular and mitochondrial transporters, passive transporters, coupled transporters, and exchangers (Hediger et al. 2013). Their role in xenobiotic transport is increasingly recognized in humans, with a particular focus in their impact on drug pharmacokinetics (Kell 2020b; Colas et al 2016). SLCs are responsible for the membrane transport of metformin, antineoplastics such as tyrosine-kinase inhibitors, and angiotensin converting enzyme inhibitors such as captopril and quinaprilat, and might also play a major role in xenobiotic transport (DeGorter et al. 2012).
Interestingly, of 17 investigated species across 4 basal eukaryotic branches, C. elegans contained the largest number of conserved SLC families compared to H. sapiens, with 43 of 46 total present (Höglund et al. 2011). In the same analysis, 31 SLCs were identified in the C. elegans genome that could not be classified into known human families, highlighting the lack of understanding of the divergent evolution of these key proteins (Höglund et al. 2011). In total, the C. elegans genome encodes 348 SLCs, in comparison to 400 in the human genome (Höglund et al. 2011).
Functionally, SLCs are understudied in humans and even more so in nematodes (César-Razquin et al. 2015; Kell 2020a). It is established that SLC subfamilies 8 and 24 (sodium/calcium exchangers), 14 and 4 (chloride transporters), 17 (vesicular transporters), 1, 5, and 6 (neurotransmitter reuptake) play key roles in neuronal function, but these are not well-characterized functionally in worms. The SLC17 subfamily in particular has 51 members in C. elegans, a large expansion compared to the meager 9 in humans, but few have been characterized (Hobert 2005). SLC17 member eat-4 is involved in glutamatergic neurotransmission, and more recently that vglu-2 may play a role in collagen trafficking in the cuticle (Serrano-Saiz et al.2013; 2020). MISC-1, a mitochondrial solute carrier that plays a role in apoptosis (Gallo et al. 2011). Slc17.9, a.k.a., droe-5, is upregulated upon dietary restriction (Ludewig et al 2014). hut-1 maintains endoplasmic-reticulum homeostasis and is essential for larval development (Dejima et al. 2009). snf-5 assists in sensing in neurons and transport in intestinal cells of L-aspartate and L-glutamate (Metzler et al. 2013).
The flux of xenobiotics via SLCs and ABCs, in addition to transporters that are less understood or could not be addressed, often serves as the final step of xenobiotic metabolism prior to excretion from the organism. Though this step is increasingly examined, many transporters remain orphan and uncharacterized, providing ample opportunity for future exploration. Similarly, as for phase I and II processes, cell-specific expression of genes and isoforms is also in need of better description.
Transcriptional regulation of stress responses in C. elegans
Given the paramount role of phase I-III detoxification genes, it is not surprising that their transcriptional regulation is subject to myriad of inputs, not all of which are extensively discussed herein. Specifically, the regulation of such genes by metals and metalloids including Cd, iron, zinc, copper or selenium, metal-containing nanoparticles, exposure to reactive oxygen species(ROS) initiating oxidative stress, mitochondrial dysfunction, genetically or pharmacologically induced longevity, advanced glycation end products such as α-dicarbonyls, and/or various diets or dietary components were described elsewhere and not covered in detail herein (Shore and Ruvkun 2013; Hoffmann and Partridge 2015; Gonzalez-Moragas et al. 2017; Chaudhuri et al. 2018; Tejeda-Benitez and Olivero-Verbel 2016; Blackwell et al. 2015; Ferguson and Bridge 2019; Olsen and Gill 2017). The focus of this review is primarily regulation of phase I, II, and III detoxification genes by organic xenobiotic compounds including antihelminthic drugs and compounds used for crop control.
Evolutionary conservation and diversification of transcriptional regulators mediating xenobiotic detoxification
Transcription factors (TFs) and regulatory mechanisms that orchestrate the coordinated induction of a tailor-made detoxification response when an animal encounters a xenobiotic compound are fairly well conserved throughout evolution. Key regulators that control the induction of detoxification genes across species include nuclear hormone receptors (NHRs), the aryl hydrocarbon receptor (AHR; ahr-1 in C. elegans), and the basic leucine zipper (bZIP) protein nuclear factor erythroid 2-related factor 2 (NRF2; skn-1 in C. elegans) (Lindblom and Dodd 2006; Gracida and Eckmann 2013; Mackowiak and Wang 2016; Blackwell et al. 2015; Hoffmann and Partridge 2015; Tonelli et al 2018; Brinkmann et al. 2019)). In addition, the GATA-type TF ELT-2 is required to express most C. elegans intestinal genes, which includes many phase I, II, and III detoxification genes (McGhee 2013); as such, ELT-2 may play a general, permissive role in the expression of many detoxification gene programs. Recently, Herholz et al (2019) suggested new roles for additional TFs, such as KLF-1, although these are less well understood, especially in their relevance to defence against acute xenobiotic stress. Below, the current understanding of the roles of these regulators, highlighting emerging mechanistic insights as well as conserved and non-conserved functions is described
C. elegans NHRs are involved in xenobiotic detoxification
Based upon their overall architecture, NHRs are grouped into the NR1-NR6 classes (Nuclear Receptors Nomenclature 1999; Weikum et al 2018). The NHRs most prominently involved in detoxification gene regulation belong to the NR1J groups in C. elegans and Drosophila melanogaster and to the NR1I and H classes in mammals. The latter group includes several NHRs with important roles in detoxification such as: the pregnane X receptor (PXR; also known as the steroid and xenobiotic sensing nuclear receptor, SXR, NR1I2); constitutive androstane receptor (CAR, NR1I3); liver X receptor (LXR, NR1H3), farnesoid X receptor (FXR, NR1H4); and vitamin D receptor (VDR, NR1I1) (Mackowiak and Wang 2016; Hoffmann and Partridge 2015; Oladimeji and Chen 2018)). However, other NHRs also regulate detoxification genes in various stuations, including the peroxisome proliferator-activated receptors (PPARs) and the Hepatocyte Nuclear Factor 4 (HNF4) type NHRs (Wallace and Redinbo 2013)). The latter are especially notable as the C. elegans N2 reference genome features a large group of approximately 265 NHRs that appear to have descended and diversified from an HNF4-like ancestor (Taubert et al 2011). Most of these remain uncharacterized, but recent studies implicated several as putative xenobiotic response regulating NHRs.
Side note on endocrine disruption in C. elegans: lack of evidence for conserved receptors at present time
Although vertebrate endocrine-related receptors are not generally thought of as major regulators of xenobiotic transport and metabolism, a digression on the potential for C. elegans to be used as a model organism for endocrine disruption is warranted. Endocrine disruptors are molecules that interfere with an organism’s intrinsic endocrine systems, which often act by targeting NHRs that regulate endogenous endocrine signals such as estrogen and mammalian NHR estrogen receptor. As such, endocrine disruptors have the potential to disturb the normal physiology and development of an organism. Endocrine disruption is a major concern in environmental toxicology with great relevance for human and wildlife health (National Academies of Sciences and Medicine 2017; Hotchkiss et al. 2008). There are some investigations reporting results of using C. elegans to study endocrine disruption, and clearly chemicals that are agonists of vertebrate endocrine receptors exert effects in C. elegans (Chen et al. 2019; Mimoto et al. 2007; Jeong et al 2019; Fischer et al. 2012; Custodia et al. 2001; Cao et al. 2020). However, it needs to be emphasized that conclusions regarding the mechanism by which these effects are mediated be interpreted with great caution. It is far from clear that responses result from presumed receptor agonist or antagonist binding to a worm homolog of the vertebrate receptor. As noted above, sequence comparisons suggest that the NR1I group of classical detoxification NHRs is apparently absent in C. elegans. In addition, the families encoding the classical mammalian steroid/thyroid receptors: NR1A thyroid hormone receptors (TRs), NR3A estrogen receptors (ERs), and NR3C 3-ketosteroid receptors including glucocorticoid receptor (GR), mineralocorticoid receptor (MR), progesterone receptor (PR), and androgen receptor (AR) are also absent(Nuclear Receptors Nomenclature 1999, Weikum et al 2018, Taubert et al 2011). Accordingly, to our knowledge, no broad evidence base supports the concept that any C. elegans NHR is (in)activated by (ant)agonists of any of these vertebrate NHR, which would render it and the nematode susceptible to endocrine disruption by the presumed pathway, such as by a xenoestrogens acting on a worm “estrogen receptor.” Therefore, although it is not impossible that functional homologs of vertebrate endocrine receptors exist, one can argue that the burden of proof is on demonstrating such functional homology, which needs to be tested rigorously. On the other hand, it is entirely possible that C. elegans may serve as a useful model for the influence of NHR-activating xenobiotics in invertebrates (Hoss and Weltje 2007). Although evidence for such events is currently also lacking, one cannot rule out that endocrine disruption of DAF-12 driven developmental pathways may occur.
C. elegans NR1J group NHRs
DAF-12
In mammals, the NR1I group NHRs (PXR, CAR, VDR) play central roles in detoxification. The apparent absence of NR1I-type NHRs in the C. elegans genome thus suggests that other, perhaps closely related NHRs, might have adopted these roles. The closest C. elegans homologs to NR1I class NHRs are the NR1J group NHRs: DAF-12, NHR-8, and NHR-48. Although little is known regarding NHR-48, the other two have been widely studied for their functions in development and physiology.
DAF-12 is perhaps the best understood NHR in C. elegans, with important roles in development, aging, and metabolism. To date evidence for a direct link between DAF-12 and the detoxification of organic xenobiotics is lacking. This is curious because (i) some detoxification genes, including cyp, gst, and ugt genes, are down-regulated in long-lived daf-12 mutants; and (ii) the CYP450, DAF-9, is a key enzyme in the synthesis of the dafachronic acids (DA), a group of molecules that serve as DAF-12 ligands and modulate its activity; and (iii) reduced DA signaling increases the resistance of C. elegans to various types of stress (Motola et al. 2006; Gerisch et al. 2001; Hoffmann and Partridge 2015; Fisher and Lithgow 2006). Possibly, the pleiotropic roles of DAF-12 in aging and development accompanied by lack of experiments directly assessing consequences of its loss on xenobiotic sensitivity and induced gene regulation have obscured a role in systemic detoxification for this NHR.
NHR-8
In contrast to DAF-12, NHR-8 has emerged as one of the most important regulators of xenobiotic detoxification in C. elegans, as it controls gene activation in response to xenobiotic exposure and is required for organismal resistance to some compounds. Lindblom et al (2001) first showed that nhr-8 mutation or RNAi rendered worms sensitive to the toxins colchicine and chloroquine. Menez et al (2019) demonstrated that nhr-8 mutants displayed hypersensitivity to the anthelmintic ivermectin, with nhr-8 silencing in ivermectin-resistant worms enhancing drug efficacy. At the gene regulation level, nhr-8 mutant worms exhibited reduced expression of several phase I, II, and III detoxification genes, including pgp and cyp genes known to impact ivermectin tolerance in C. elegans, with correspondingly diminished ABC transporter-mediated drug efflux activity (Menez et al. 2019). Importantly, re-expression of the ABC transporter pgp-6 in nhr-8 mutant worms elevated tolerance to ivermectin, implicating NHR-8 control of ABC drug efflux transporters as a likely mechanism for drug resistance. Support for this model is provided by Guerrero et al. (https://www.biorxiv.org/content/10.1101/823302v1.full), who noted that induction of three pgp phase III detoxification genes by glycosylation inhibitor tunicamycin required nhr-8; that nhr-8 loss-mediated acute tunicamycin sensitivity while NHR-8 overexpression produced tunicamycin resistance; and that chemical inhibition of pgp glycoproteins suppressed tunicamycin resistance. Collectively, evidence suggests that NHR-8 is essential to induce phase I, II, and III detoxification genes in worms exposed to various toxins, perhaps especially phase III drug efflux transporters. Notably, the role of NHR-8 is xenobiotic-specific, as Lindblom et al. (2001) found that loss of nhr-8 rendered worms sensitive to only some, but not all tested xenobiotics. Indeed, NHR-8 likely possesses redundant functions with other transcriptional regulators in regulating xenobiotic detoxification genes, as other studies also showed that NHR-8 depletion alone did not broadly abrogate the expression of select phase I and II genes (Chamoli et al. 2014; Jones et al. 2013). Rather, Chamoli et al (2014) reported that PHA-4/FoxA, NHR-8, and AHR-1 cooperatively induce various cyp and ugt genes, albeit in a long-lived mutant background, rather than in acute exposure to xenobiotic compounds.
Although clearly important for the response to various xenobiotics, the mechanisms of NHR-8 activation by such compounds is not clear. Chloroquine, colchicine, and several sterol-derived small molecules that bind mammalian LXR failed to activate NHR-8 in a ligand-sensor screen (Magner et al. 2013); thus, evidence that NHR-8 may act analogously to PXR and CAR as a xenobiotic sensor remains lacking. An alternative mechanism of NHR-8 activation was suggested at by Verma et al. (2018), who showed that, in phosphatase vhp-1 or flr-4 kinase dead mutants, nhr-8 loss abrogates the expression of cytoprotective genes. As vhp-1 is an important negative regulator of mitogen-activated protein kinase (MAPK) activity, NHR-8 may thus require MAPK signaling to activate phase I, II, and III detoxification genes, although it is not clear whether and how this classical stress sensing kinase signalling pathway is activated by xenobiotic exposure.
C. elegans HNF4-like NHRs
NHR-86
Besides the NR1J group NHRs, several HNF4-related NHRs have recently emerged as regulators of detoxification gene programs in C. elegans. NHR-86 is required to express phase I detoxification in worms exposed to the immunomodulatory toxin RPW24, including 4 cyp genes, two gst genes, and 14 ugt genes. As in the case of NHR-8, however, the requirement for nhr-86 is not universal, as the highly RPW24-inducible cyp-35A2, cyp-35A3, cyp-35B1, and cyp-35B2 genes remained toxin chemical-responsive in nhr-86 mutants; whether these genes are instead regulated by other NHRs such as NHR-8 which was not tested (Peterson et al. 2019). It is noteworthy that Peterson et al (2019) used chromatin immunoprecipitation followed by sequencing to identify direct NHR-86 targets. The 32 genes whose promoters were bound by this TF and that required its activity for RPW24 induced expression included cyp-35A1 as the most strongly NHR-86 bound gene, as well as three ugt genes and gst-5. This provides first evidence that detoxification regulatory NHRs might directly bind critical response genes in live worms. Further, NHR-86 binding to several promoters increased upon exposure to RPW24. This suggests that NHR-86, and perhaps other NHRs, might display toxin-induced binding to relevant detoxification promoters as a mechanism to enhance their expression on demand.
NHR-114
Another HNF4-like NHR that is required to express detoxification genes is NHR-114 (Gracida and Eckmann 2013). In this case, the exposure of nematodes to a bacterial diet containing high levels of the amino acid tryptophan induced a broad detoxification program including cyp, ugt, and gst genes. Notably, some of these genes were nhr-114 dependent, suggesting that adaptation to a specific bacterial diet involves NHR-114-driven transcriptome rewiring to protect the organism from toxic effects. This scenario highlights that NHR (and TF) dependent rewiring of the detoxification pathways is not exclusive to exposure to “foreign”, potentially dangerous, substances, but represents an integrative component of the organism’s response to specific diets and the corresponding (endo-) metabolic modulation that their consumption invokes. Although it is not currently known whether NHR-114 binds any endo- or xenobiotic compound, it is tempting to speculate that it, and perhaps other diet-sensing NHRs, might undertake this to adapt xenobiotic and other metabolic pathways.
Other HNF4-like NHRs
Several other HNF4-like NHRs have also emerged as potential regulators of xenobiotic detoxification. Using high-throughput yeast-one-hybrid assays to study TF-promoter interactions, Fuxman-Bass et al (2016) found a significant overrepresentation of phase I, II, and III detoxification genes, including cyp, gst, ugt, and sdr genes, amongst promoters bound by NHRs. RNAi against 26 NHRs that bound the promoter of at least one detoxification gene in combination with exposure to 16 molecules that are toxic for C. elegans, including some xenobiotics, revealed new roles in xenobiotic resistance for 9 NHRs (NHR-42, −66, −72, −102, −109, −142, −178, −216, and −273). This unbiased study strongly supports the notion that xenobiotic resistance is an important role of HNF4-like NHRs in C. elegans.
Targeted investigations of individual NHRs also support such roles for NHRs. Jones et al (2013) showed that, despite its role in the response to several toxins, nhr-8 is not universally essential for xenobiotic detoxification (nor were skn-1, ahr-1, hif-1, and mdt-15). Studying chloroquine, dazomet, imidacloprid, and thiabendazole, and performing candidate RNAi screens of 387 TFs, Jones et al (2013) found 12 genes required for activation of some of 12 phase I, II, and III detoxification genes in response to toxin exposure. All 12 genes were NHRs. Most prominently, nhr-176 depletion enhanced susceptibility to the anthelminthic thiabendazole. A follow-up study demonstrated that NHR-176 directly binds thiabendazole and is required for thiabendazole induced cyp-35 gene transcription (Jones et al 2015). In contrast, nhr-176 is dispensable for resistance to 5-hydroxythiabendazole. Thus, Jones et al (2015) proposed that the xenobiotic response of C. elegans is rather specialized and might involve combinatorial actions of multiple regulators.
Several other NHRs also regulate cyp genes. However, these P450 enzymes may not be phase I detoxification genes, but rather participate in lipid synthesis and modification, or act dually in both functions. These functions include the aforementioned regulation of DAF-9 by DAF-12, a feedback loop wherein DAF-9 contributes to the synthesis of the DAF-12 ligands; in this context, DAF-9 is not known to play roles in detoxification. Similarly, the HIF-1 regulated P450, cyp-36A1, is required for the downstream activity of NHR-46 and might contribute NHR-46 ligand synthesis (Pender and Horvitz 2018). CYP-36A1 is related to the CYP2 family, which function in both detoxification of xenobiotics and metabolism of endogenous molecules (Nebert et al 2013), and as such might also perform detoxification reactions. Similarly, although initially characterized as a regulator of lipid metabolism, NHR-49 also regulates cyp and other detoxification genes in long-lived mutants and when worms experience oxidative stress (Chamoli et al. 2014; Hu et al. 2018; Goh et al. 2018). Whether or not NHR-49 is directly involved in xenobiotic compound sensing and adaptation is not clear.
In sum, phase I, II, and II detoxification genes appear to be common targets for C. elegans NHRs, especially NHR-8 and a growing number of HNF4-related NHRs. These TFs likely act in a combinatorial fashion to induce detoxification gene programs and consequently organismal resistance and tolerance. Underlying mechanisms may involve combinatorial promoter binding, heterodimer formation, or sequential action to evoke a customized response to a toxin(s) an individual worm may encounter. The extraordinarily large number of NHRs in the C. elegans genome, combined with similar, albeit smaller expansion of the cyp and ugt families, suggests that individual NHRs, or NHR dimer pairs, might help C. elegans fine tune its detoxification system in the face of ever changing environmental xenobiotic diversity.
7.2.3. Mechanistic considerations of xenobiotic sensing by C. elegans NHRs
The mammalian NHRs that orchestrate the response to xenobiotics are activated via direct and indirect mechanisms. That is, their activity is increased both by their ability to directly bind xenobiotic compounds that then act as ligand agonists, and also through additional independent mechanisms not involving direct physical contact between the molecule in question and an NHR. Such indirect signaling involving kinases and phosphatases alters NHR activity by inducing nuclear translocation, protein stabilization, and/or interaction with transcriptional coregulators (reviewed in (Mackowiak and Wang 2016)).
Unlike mammalian PXR and CAR, C. elegans NHRs have not been studied in as much detail for putative molecular mechanisms underlying their regulation of detoxification gene programs. However, collectively, the studies described above hint that direct and indirect mechanisms might contribute to xenobiotic sensing and signaling by C. elegans NHRs. Specifically, evidence exists suggesting that (i) NHRs can bind some xenobiotic compounds; (ii) their chromatin occupancy in the regulatory regions near detoxification genes increases when xenobiotics are sensed; and (iii) they activate the expression of said genes, thus promoting xenobiotic resistance and tolerance. Together, these studies agree with a model of direct, ligand-like activation of C. elegans NHRs by xenobiotics. Indirect signaling, possibly by the MAPK pathway, could also be required for NHR activation.
MDT-15 – a co-regulator shared between NHRs that control detoxification gene programs
To exert effects on gene regulation, NHRs (and other TFs) need to interact with co-regulators, which help activate and repress gene expression as necessary. An important co-regulator in eukaryotes is the Mediator complex, which is composed of 25–30 subunits, some of which serve as direct docking sites for TFs (Grants et al 2015). Notably, one such subunit, MDT-15, interacts physically with several of the above noted TFs, including NHR-8, NHR-49, NHR-86, and SKN-1 (see below) (Arda et al. 2010; Taubert et al. 2006; Reece-Hoyes et al. 2013). Accordingly, mdt-15 is required to induce numerous phase I, II, and III detoxification genes in worms exposed to xenobiotic molecules, and its mutation or deletion results in sensitivity to xenobiotics such as fluoranthene and RPW24 (Taubert et al. 2008; Pukkila-Worley et al. 2014). A model whereby MDT-15 binds NHRs that have been activated in direct or indirect fashion by the presence of a xenobiotic molecule and co-activates the expression of pertinent detoxification genes is thus plausible. As co-regulators might be rate-limiting for expression of inducible gene programs, it would be interesting to further investigate the role of MDT-15 in NHR driven detoxification responses.
AHR-1 is unlikely to perform major xenobiotic detoxification functions in C. elegans
In addition to NHRs, mammalian cyp and ugt genes are regulated by the aryl hydrocarbon receptor (AHR) in response to xenobiotics, especially aromatic (aryl) hydrocarbons including polycyclic aromatic hydrocarbons, dioxins, and polychlorinated biphenyls for which AHR is an important sensor (Nebert et al. 2004; Bock 2014). The C. elegans genome also encodes an AHR gene, ahr-1, as well as a gene encoding its obligate partner, the AHR nuclear translocator (ARNT) aha-1 (Powell-Coffman et al 1998). However, whereas mammalian AHR directly binds several aromatic xenobiotic compounds, C. elegans ahr-1 does not bind the classical AHR ligand, dioxin (Powell-Coffman et al 1998). Further, most functional investigations on ahr-1 to date suggested that it is predominantly involved in development, especially of the nervous system (Huang et al 2004; Zhang et al. 2013). Similarly, transcriptome analyses in two ahr-1 mutants revealed a gene program largely pertinent to neuronal function and development (Aarnio et al. 2014), although some genes identified as ahr-1 dependent did feature regulatory elements annotated as xenobiotic responsive elements (XREs). Recently Brinkmann et al (2019) suggested that ahr-1 may modulate life span in C. elegans, perhaps by modulating the effects of metabolites from commensal bacteria. In any case, the central role of mammalian AHR in the defence against certain aromatic hydrocarbons does not appear to be recapitulated by C. elegans ahr-1, and thus might arise later in evolution.
SKN-1
SKN-1 is a TF that belongs to the basic leucine zipper (bZIP) family of TFs that regulates stress response pathways across species (Blackwell et al. 2015). In C. elegans, SKN-1 is especially important for oxidative stress and starvation adaptation and is induced by complex regulatory pathways involving MAPK and insulin signaling as well as negative regulation by the WDR-23 protein, which appears to promote SKN-1 for degradation, an inhibitory response that is alleviated by oxidative stress (Blackwell et al. 2015). In addition to its role as an antioxidant regulator, SKN-1 also plays important roles in xenobiotic detoxification. Specifically, skn-1 loss sensitizes worms to the common benzimidazole albendazole, and skn-1 gain-of-function mutations increase tolerance to this drug. skn-1 regulated genes include albendazole induced cyp, gst, and ugt genes, of which ugt-22 loss also enhances albendazole efficacy (Fontaine and Choe 2018). Similarly, induction of phase I and II detoxification genes by acrylamide also requires skn-1, at least in part, and unbiased genetic screens for genes involved in stimulation of gst genes by acrylamide identified known components of the SKN-1 pathway, including the negative regulator wdr-23, and the metabolic enzyme alh-6 (Hasegawa and Miwa 2010; Fukushige et al. 2017). The Skp1 homologs skr-1/2, components of Skp-Cullin-F box ubiquitin ligase (SCF) complexes that regulate SKN-1, are also essential for the SKN-1 detoxification response to acrylamide (Wu et al. 2016). The regulation of SKN-1 during xenobiotic stress is thus apparently similar as that seen in oxidative stress, involving derepression from WDR-23 and SKR-1/2 mediated degradation. How xenobiotic molecules act to relieve WDR-23 action on SKN-1 is not known at this time, but the effects of skr-1/2 are notably independent of classical stress activated MAPK signaling, implicating alternative pathways. Indeed, SKN-1 activity is highly regulated, and studies on its activity in oxidative stress conditions revealed numerous novel regulators (Crook-McMahon et al. 2014). It would be interesting to test whether any of these factors are necessary for elevated SKN-1 activity in response to xenobiotic exposure.
Conclusions: transcriptional regulation of xenobiotic responsive genes
In sum, the transcriptional response of C. elegans to xenobiotic molecules is both similar and distinct from that found in other animals. Although NHRs and SKN-1 clearly play central roles in toxin resistance and the underlying gene regulation, these NHRs are distinct, belonging to NR1J and HNF4-like groups rather than the mammalian NR1I group, which appears to be absent from the C. elegans genome. Similarly, the role of AHR appears to be marginally conserved with regard to xenobiotic metabolism. Future investigations need to focus on identifying mechanisms pertaining to how xenobiotics activate C. elegans xenobiotic responsive TFs and what specificity or partnerships exist between different TFs to enable tailor-made detoxification and resistance in this worm. Attention also needs to be paid to cell-specific expression to inform our understanding of which cells contribute to xenobiotic metabolism and transport; the existence of publically-available datasets on cell-specific gene expression should facilitate this effort.
Empirical toxicokinetics (TK)
Toxicokinetics(TK) encompass uptake, distribution, metabolism, and excretion of xenobiotics. Understanding the rates at which these processes occur is a critical part of toxicological assessments, because these dictate the actual doses at internal sites of molecular interaction that initiate toxicity. Comparison of internal doses across lifestages, tissues, and species is critical to intraspecies extrapolations (e.g., why does toxicity occur in one cell type and not another?) as well as interspecies extrapolations (e.g., is the toxic effect observed relevant in another species?). Empirical studies of the kinetics of uptake, metabolism, and excretion are an important first step. For example, the absorption rate might be estimated by incubating the animals for different time periods and subsequently measuring the internal doses. Ultimately, TK models are developed, as has been done in many other species, including humans, rats, mice, and zebrafish. To date, a full TK model has not been developed for C. elegans, but some aspects of TK were determined and reported. A variety of methods were used to empirically measure xenobiotic uptake (and in some cases clearance) in C. elegans. The concept of internal doses as defined by the U.S. EPA Exposure Factor Handbook (2011; see https://www.epa.gov/expobox/about-exposure-factors-handbook) is described here.
Burns et al (2010) systematically investigated the uptake of drugs by C. elegans by surveying accumulation of 387 compounds. Unfortunately, Burns et al (2010) needed to set a high detection limit (19 μM, or roughly half of the external dose of 40 μM), which limited their ability to determine average uptake across the chemical space. However, under these conditions, only 6.7% of the compounds were detected above the detection limit in worm lysates after 6 hr incubation. Burns et al (2010) went on to construct a structure-activity relationship to predict which physicochemical properties would enable significant accumulation of a compound within C. elegans. Extension of this analysis to additional chemicals and classes of chemicals such as (1) both inorganic and organic, as well as molecules with a wide range of Kow and pKa values would be beneficial. Knowledge of additional lifestages and exposure times would also be of great value to the field.
Unfortunately, despite the thousands of toxicology- and pharmacology-related publications in C. elegans, the number that report internal doses appears to be quite limited. In Supplemental Table 1, results from publications are summarized that were identified that report analysis of xenobiotic uptake in C. elegans (Luz et al. 2016; 2017; Au et al. 2009; Yang et al. 2014; Maurer et al. 2015; Crone et al. 2015; Wyatt et al. 2016b; Helmcke et al. 2009; Allard and Colaiacovo 2010; Chen et al. 2016; Zheng et al. 2013; Roh et al 2016). While it may be challenging to draw broad conclusions from these studies because of variability in experimental design such as liquid versus agar plate culture, lifestage, or exposure time course, there are a few general patterns and several important limitations to the current state of the literature can be identified.
Some combination of poor absorption through the cuticle, and perhaps behavioral changes that reduce oral intake, appear to comprise a fairly effective barrier to uptake of many chemicals. For example, Luz et al (2017) found that uptake of arsenite in 8-day adults was approximately 1% in 24 hr (1 μM internal dose, 100 μM exposure concentration). However, this is not always the case: cisplatin uptake in 2 hr in L4s was higher, approximately 10–15% (Crone et al. 2015), and a 48hr developmental exposure to methylmercury, which biomagnifies in aquatic food webs, was present internally at >100-fold higher concentration compared to the exposure medium (Wyatt et al. 2016b). In general, it is probably not surprising that uptake varies significantly by chemical, which underscores the importance of such measurements in future research. It would also be helpful to distinguish, as much as possible, absorption through the gut from cuticular uptake.
Perhaps the more critical question for human health risk assessment is not how the internal dose in C. elegans is compared to the exposure concentration in the well, but how the measured internal dose in C. elegans is compared to exposure concentrations in other lab models (e.g., cell culture), or internal doses in the human population. Data to date show that for many chemicals, internal doses are reasonable compared to other systems. For example, the internal dose of cisplatin with doses between 150–350 μM result in internal dose less than 1 μM (Crone et al. 2015), and patients undergoing cisplatin infusion have peak plasma levels of around 3–5 μM (Hanada et al. 2001; Rajkumar et al. 2016). Internal doses in C. elegans approximating human blood levels were also been reported for ethanol (Alaimo et al. 2012). However, it needs to be emphasized that the literature to date is quite limited. Important areas for future research beyond analysis of additional chemicals include detailed time courses to establish not just internal doses but actual kinetics of uptake and excretion.
A second major limitation even when uptake has been measured is uncertainty regarding target tissue concentration within the worm, and cell-and tissue-specific TK. For example, analytical chemistry techniques are typically quantitative but do not usually provide fine-scale information regarding distribution (i.e., inside the worm versus on the cuticle, and which cells and subcellular organelles have the highest concentrations). Conversely, microscopic or X-ray spectroscopy methods provide more spatial distribution information, but are rarely quantitative. Interestingly, these methods revealed striking spatial variation in concentration, often with higher concentrations in the gut than elsewhere (Gonzalez-Moragas et al. 2017; Brinkhaus et al. 2014). On the other hand, Jackson et al (2005), using synchrotron X-ray spectroscopy, found large differences in the intra-organismal distribution of copper (which was fairly evenly distributed) and lead (which located mostly to the head region). Jackson et al (2005) were also able to demonstrate that the uptake of lead was almost certainly entirely via ingestion, not across the cuticle. Further, the physicochemical properties of the compound determine how quickly it is taken up into the worm, particularly to peripheral tissues beyond the gut, and how quickly it might be lost after removing the compound. For example, chlorpyrifos reaches peak internal doses and remains stable after approximately one hr exposure, and when the pesticide is removed it is quickly cleared with more than half disappearing within one hr and the remaining chlorpyrifos clearing by 20 hr (Roh et al 2016). In contrast, ethanol was reported to diffuse readily through the cuticle and might be lost even during wash steps, making accurate estimates of internal doses and TK difficult (Mitchell et al. 2007).
Finally, for chemicals that are taken up poorly, even if internal doses comparable to other systems can be achieved with high exposure concentrations, there are practical issues with the low uptake of some chemicals by C. elegans. Higher exposure concentrations require using more chemical, which may be costly, and often is limited by the solubility of the drug or toxicant, even with the use of carriers such as DMSO. A solution that was proposed is to use cuticle-defective mutants such as bus-5, which were demonstrated to display increased permeability to drugs. However, this presents additional challenges including needing this mutation in the background of all other genetic/transgenic strains used, reduced fitness resulting from the compromised cuticle, and possible compensatory changes in gene expression in response to the mutation that may have unintended consequences in the study (Xiong et al 2017). As a further aid to experimental design, Tables 2 and 3 provide factors that affect uptake of chemicals and nanoparticles in C. elegans, including both aspects of the worm’s biology (lifestage and genetics) and the worm’s environment (type of medium, and presence of food or other organic matter).
Table 2.
Factors impacting toxicokinetics in C. elegans
| Factor | Process(es) Likely Impacted | References |
|---|---|---|
| Worm Age | Uptake (due to cuticle thickness), Metabolism (age-related changes in gene expression), Sensitivity to toxicity | (Helmcke et al. 2009) |
| Worm strain | Uptake (cuticle integrity, transporter expression), Distribution (transporter expression), Metabolism (gene expression), Excretion (transporter expression), Sensitivity to toxicity | (Crone et al. 2015; Davies et al. 2003) |
| Delivery Method or Medium | Apparent External Dose, Uptake (pharyngeal pumping rate, cuticle diffusion, co-solvent delivery) | (Zheng et al. 2013; Alaimo et al. 2012; Burns et al. 2010) |
| Presence or Absence of Bacteria | Apparent External Dose, Uptake (delivery of compound by adsorption/internalization by bacteria, pharyngeal pumping rate) | (Yang et al 2017; Spann et al 2015; Höss et al 2011; Offermann et al 2009) |
| Bacterial Density | Apparent External Dose, Uptake (delivery of compound by adsorption/internalization by bacteria, pharyngeal pumping rate) | (Offermann et al 2009; Höss et al 2011) |
| Presence of Organic Matter | Apparent External Dose, Uptake (co-solvent delivery) | (Hoss et al. 2009; Haitzer et al. 1999) |
| Washing Worms Before Extraction | Apparent Internal Concentration | (Mitchell et al. 2007) |
| Co-Exposures | Uptake (competition for transport, co-solvent delivery), Metabolism (competition for metabolic enzymes), Sensitivity to toxicity | (Wyatt et al. 2016a) |
Table 3.
Factors impacting nanoparticle toxicokinetics in C. elegans
| Factor | Process(es) Likely Impacted | References |
|---|---|---|
| Presence of Food | Apparent External Dose, Uptake (delivery of compound by adsorption/internalization by bacteria, pharyngeal pumping rate) | (Yang et al 2014; 2017; Collin et al. 2014) |
| Environmental Transformation | Uptake, Distribution | (Starnes et al. 2015) |
| Presence of Organic Matter | Apparent External Dose, Uptake (co-solvent delivery) | (Yang et al. 2014; Collin et al. 2014) |
| Particle Charge and Coating | Uptake, Sensitivity to toxicity | (Ahn et al. 2014; Gonzalez-Moragas et al. 2017, Meyer et al. 2010a) |
| Particle Size | Uptake, Sensitivity to toxicity | (Ahn et al. 2014; Gonzalez-Moragas et al. 2017; Meyer et al. 2010a; Roh et al 2016) |
9. Genetic diversity: C. elegans as a model for natural population variation in xenobiotic response
Different individuals respond to xenobiotics in diverse ways. The same exposure might lead to extreme toxicity or to mild effects depending upon the genetics of the individual. For example, toxicants that pose high risks to biological systems are ubiquitous in the environment, yet individuals vary greatly in the degree to which these exposures initiate diseases. Knowledge of specific xenobiotic responses is limited to cases with a high known exposure source, which is not typical of a daily exposure scenario. A major missing link between individual responses and exposures comes from a limited understanding of the role of genetic variation in responses to xenobiotics. Toxicity assessment techniques are lacking that properly account for genetic variation in toxicological studies because sample sizes are too limited to accurately reflect true human genetic diversity. Similarly, there is also an absence of methods to correlate genotypes to disease phenotypes in human populations that thoroughly account for heterogeneous environmental exposures because this knowledge is essentially impossible to capture in an informative manner. Using a model system, one can address both weaknesses in xenobiotic assessments ethically and robustly. However, a gap exists in the translation of xenobiotic toxicity mechanisms in most model organisms because most assessments of xenobiotic responses use only one genetic background. This situation is akin to making xenobiotic-induced disease risk assessments from a single subject. Therefore, in order to define population-level risk factors, one requires a discovery platform that determines whether xenobiotic response pathways vary across genetically diverse individuals, and C. elegans is ideally suited for this purpose.
The C. elegans species has genetic diversity similar to humans and genetically distinct individuals are found worldwide (Andersen et al. 2012). Recent samples collected from nature have levels of genetic diversity from before the domestication of the lab strain N2 (Sterken et al. 2015; Cook et al. 2017; Crombie et al. 2019). Using these strains, one can begin to characterize the spectrum of xenobiotic responses present in the C. elegans species to make broader conclusions regarding how xenobiotics affect diverse individuals. Quantitative genetics studies correlations between phenotypic differences (e.g. xenobiotic responses) and genotypic differences (e.g. genetic backgrounds) to discover the genes that might underlie population-wide differences. C. elegans offers a unique opportunity to leverage natural genetic variation to identify novel genetic sources of variability in xenobiotic metabolism and transport. Diverse C. elegans populations encounter many xenobiotics in their natural rotting substrates, soil, and groundwater ecosystems. For this reason, subpopulations of C. elegans have likely adapted to either periodic or constant toxicant exposure and retained advantageous genetic differences that produced increased survival rates in these conditions. Alleles that have arisen naturally might be easily translated to human populations because several xenobiotic response pathways are conserved between C. elegans and humans (Shaye and Greenwald 2011; Ardelli 2013; Jones et al. 2013; Harlow et al. 2016). Powerful insights into the genetic basis of xenobiotic responses n may therefore be derived from C. elegans as a sentinel of toxic exposure.
Only the beginning of understanding mechanisms of xenobiotic resistance across natural C. elegans populations is known. Although drugs are inarguably essential, chemotherapeutic medications may be classified as xenobiotic compounds with known molecular targets and conserved TK properties (Zdraljevic and Andersen 2017). A relatively rare allele of the scb-1 gene induces bleomycin susceptibility in a large recombinant C. elegans population (Brady et al. 2019). This allele also confers resistance to several other chemotherapeutic drugs, particularly drugs that produce DNA double-stranded breaks (Evans et al. 2018; Evans and Andersen 2020). The results are less straightforward with heavy metals and pesticides, two common types of agricultural and industrial xenobiotics. Differential responses to these compounds might be explained by genetic differences among individuals. In particular, three regions of the genome were linked to variation in susceptibility to the pesticides paraquat, chlorpyrifos, and chlorothalonil, as well as the heavy metals silver and copper. However, when one of these regions was isolated through a series of genetic crosses, it alone could only explain variation in one specific trait in response to silver (Evans et al. 2018; Evans and Andersen 2020). Therefore, resistance to heavy metals and pesticides resistance is a complex trait governed by a set of genes with varying functions. As is the case with the majority of complex traits, individual resistance loci typically exert relatively small effects in isolation and are problematic to characterize. Further, these loci might participate in either antagonistic or synergistic interactions, making individual genetic effects even more difficult to detect and interpret. Nevertheless, some individual genes have outsized influences on xenobiotic resistance, even though the entire set of genes is difficult to infer.
One unique example of the power of C. elegans for translational TK insights is the discovery of a novel pathway to As resistance among natural populations. Genome-wide association (GWA) mapping involves measuring a phenotype of interest among a collection of genetically diverse individuals. The unique advantage of GWA compared to lab crosses is that it leverages past relatedness to detect resistance alleles with potentially low representation in traditional lab strains. In C. elegans, the molecular genetic toolkit is broad and tractable so that causal relationships between genetic variants and phenotypic variation may be established. GWA mapping, followed by a series of metabolomic experiments in edited C. elegans strains and human cell lines, confirmed that one cysteine-to-serine missense variant within the dbt-1 gene enhanced resistance to As by modulating branched chain fatty acid biosynthesis (Zdraljevic et al. 2019). Arsenic sensitivity mediated by differential metabolism of branched-chain amino acids is a novel observation that is consistent with other features of As-mediated toxicity in human populations. Most critically, these results demonstrate the unique opportunity for the toxicology community to leverage natural genetic variation in C. elegans to identify novel mechanisms of differential toxicodynamics in the human population and learn how individuals differ in xenobiotic metabolism.
Natural C. elegans populations likely encounter many xenobiotics for which there are poor assessments of human health risk. Nematodes need to find a way to survive in environments contaminated by industrial byproducts such as Cd and Pb. For this reason, C. elegans is sometimes considered a useful environmental indicator (Custodia et al. 2001: Hoss et al. 2009) and, in turn, lab assessments have important metal resistance genes (Swain et al. 2004: Broeks et al. 1996), including phytochelatins (Vatamaniuk et al. 2001), which play a key role in C. elegans but are not expressed in higher eukaryotes. Genetically diverse wild isolates can be sampled from many environments and screened against the battery of toxicants for which detailed human assessments are scant. C. elegans isolates can be collected from polluted sites and their xenobiotic responses compared to more traditional lab strains, offering a unique way to take advantage of natural populations. High-throughput phenotyping platforms (Andersen et al. 2015) enabled precise and rapid phenotypic characterization of thousands of individuals. Highly scalable xenobiotic screening efforts enable us to continue to explore C. elegans natural variation and expand the scope of large-scale quantitative genetics analyses (Cook et al. 2017). This endeavor might undoubtedly reveal a rich set of genomic regions linked to variation in xenobiotic metabolic and transport capacity (Evans et al. 2018), and provide new opportunities to discover drug-initiated cellular targets for xenobiotic-induced disease treatment.
Evolutionary toxicology: applications of inter-species variation to understanding stressor responses
Intraspecific differences in toxicant responses across a single species might serve as a powerful tool to discover new mechanisms of toxicity and toxicity mitigation. This technique can identify natural variation in genes involved in toxin responses, but the genes might not be conserved with other species because C. elegans likely has experienced evolutionary selection by chemical exposure differently. By broadening the number of species where natural history differences in chemical responses are measured, it is possible to increase the likelihood the discovered genes and toxin responses mechanisms might be connected to other species, including humans. The underlying assumption of this hypothesis is that different species have experienced similar evolutionary pressures by chemical exposure, such that mechanisms that were used to adapt are similar. In other words, evolutionary trajectories in different species all lead to the same mechanisms of toxicity mitigation. This hypothesis is likely not universally true, but when it is, the statistical likelihood that genome-wide association studies identify conserved toxin response loci is higher. Thus, some researchers used comparison of toxicological responses of C. elegans to those of other nematode and invertebrate species to infer expected ecotoxicological responses in other invertebrates (Haegerbaeumer et al. 2018a; 2019; Peredney and Williams 2000; Boyd and Williams 2003). Similarly, others investigators previously advocated for using evolutionary genetics to inform human health risk assessment (Leung et al. 2017; Brady et al. 2017; Hahn 2019).
Nematodes are incredibly ecologically diverse, either living freely in soils, freshwater, or seawater or as a parasites in plants or animals (De Ley 2006). The chemical exposure of nematodes is as diverse as their natural habitats. It includes organic compounds and minerals in soils (Ekschmitt and Korthals 2006), pesticides used in agriculture (Rich et al 2004), as well as defensive compounds secreted by bacteria, fungi, plants, and animals (Williamson and Kumar 2006; de Veer et al 2007). Some nematodes have evolved to survive desiccation and high salinity in extreme arid environments such as deserts (Treonis and Wall 2005). The panoply of abiotic and biotic stresses has likely contributed to the high abundance and diversity of many of the xenobiotic metabolism and transport-related gene classes discussed above. Even within a single species like C. elegans, the diversity of xenobiotic response genes is far larger than the suite of genes found in the lab-adapted strain N2 (https://www.biorxiv.org/content/10.1101/2020.07.23.218420v1.abstract). The power of C. elegans GWA mapping approaches might be expanded to related self-fertilizing species like Caenorhabditis briggsae and Caenorhabditis tropicalis, when reference genomes are completed and species-wide natural variation has been characterized. Comparative genetics and genomics across these three species will discover conserved Caenorhabditis toxin responses along with species-specific toxin responses. The conserved responses are more likely to be conserved with humans. To further supplement the comparative genetics and genomics approach, natural variation in toxin responses across nematode species beyond Caenorhabditis or even other invertebrate species (e.g., Drosophila melanogaster) need to be studied. The “worm community” has begun in-depth investigations of the biology of multiple related Caenorhabditis species (Martin et al. 2015; Elsworth et al 2011; Fitch 2005). However, the total number of nematode species is estimated at approximately one million (Kiontke and Fitch 2013), and only approximately 30,000 species have been described. A limited number of nematode species important in agriculture, biology, and medicine were also examined and provide a basis for comparison, including Ancylostoma duodenale (hookworm), Brugia malayi (Lymphatic filarial nematode), Meloidogyne incognita (root-knot nematode), and Steinernema carpocapsae (entomopathogenic nematode). As xenobiotic response loci are discovered in additional species, comparative approaches might increase in power and enable connections of discovered genes, pathways, and mechanisms to humans.
Finally, although this review focuses on C. elegans as a toxicology model for human health, C. elegans and other nematodes may also serve broader ecological health goals. Nematodes, or communities of nematode species, may serve as useful bioindicator species in ecotoxicology. The species diversity of nematode communities is one measure of ecological integrity, and is sensitive to environmental changes (Franco et al. 2019). Morphological identification of nematode species may be time-consuming and requires expertise. However, recent studies demonstrated that metabarcoding samples of nematode communities is similarly efficient to morphological species identification, and may be implemented rapidly (Schenk et al. 2020; De Ley et al. 2005). Further, given relevance for both human health and ecotoxicology, and the ability to be studied both in the field and in the lab, experiments with C. elegans has strong potential to help bridge ecotoxicological and human health research. Research with nematodes might be used in frameworks that were developed for integrating data from different species from molecular (Mattingly et al. 2006), biochemical (Harborne 1988), and pathological (Snyder et al. 2010) to ecological (Monosson 2012) levels, approaches that more recently were incorporated into the EcoHealth and OneHealth concepts (Perez and Pierce Wise 2018; Brooks et al. 2020; Haschek et al. 2019).
Knowledge gaps, opportunities, and future directions
It is hoped that this review will serve to orient C. elegans researchers who are not toxicologists to these important pharmacological and toxicological parameters, and to orient toxicologists and pharmacologists who are new to C. elegans to what is known and not known regarding relevant aspects of C. elegans biology. This review is somewhat narrowly focused on biology relevant to TK, largely excluding some pathways that are nonetheless relevant, including non-enzymatic xenobiotic defenses (metallothioneins, phytochelatins, glutathione, antioxidant molecules), and antioxidant enzyme systems.
It is also hoped that this review will stimulate additional research into some of the less well-understood aspects of TK in this species. Because of the long history of genomic and genetic research in this species, a remarkable understanding of genes involved in TK-related processes exists. Unfortunately, our understanding of the function (e.g., substrate specificity, enzymology, etc.) of the gene products is less complete, and it is recommended more research be done in this area. There is also a limited understanding of which specific cell types are responsible for carrying out many xenobiotic transport and metabolism functions (i.e., which cells act as hepatocytes or renal cells). Similarly, although a handful of careful analyses of internal doses of chemicals in worm exposure studies were carried out, many more are needed. Work to date permits a tentative conclusion that internal doses of C. elegans are often comparable to exposure concentrations in (1) cell culture studies, (2) vertebrate model organisms, and (3) human tissues in exposed individuals. However, the database of chemicals for which internal doses and exposure concentrations have been quantified in C. elegans exposure studies is still far too small to make extrapolations with confidence, or to model likely uptake kinetics based upon chemical characteristics and first principles. Exposure studies need to routinely test internal doses. Finally, There is a remarkable opportunity to tie research in these traditional toxicological areas into more general aspects of the stress response (thermal stress, osmotic stress, hyperoxic and hypoxic stress, or caloric stress). Stress responses, broadly defined, are poorly integrated in any species; i.e., how do multiple different stress-response pathways integrate in the context of combined stressors, chemical and otherwise? Such exposures are the reality for individuals and species that one attempts to protect with toxicological studies, and are therefore critical to study. The opportunity to do combination studies and examine the role of multiple stress response pathways is great in C. elegans, in which the non-xenobiotic stress response pathways are exceptionally well-studied and tractable.
Better toxicological knowledge will be critical in permitting rigorous use of C. elegans for toxicological research that might be used to protect human and environmental health. This is true for in-depth, mechanistic toxicological research, such as on the role of specific enzymes in mediating toxicity of specific chemicals (Harris et al. 2020) and the ability to extrapolate worm studies to other species. It will also permit adoption of this powerful genetic model to “functional toxicology” studies (Gaytan and Vulpe 2014). Finally, it will be crucial to the adoption of the C. elegans model for New Approach Methodologies that seek to reduce time, cost, and vertebrate animal use in chemical safety regulation and risk assessment. The Frank R. Lautenberg Chemical Safety for the 21st Century Act (2016) mandates the U.S. Environmental Protection Agency (U.S. EPA) to explore the use of non-vertebrate models for toxicity testing (U.S. Congress 2016). In 2019, the U.S. EPA Administrator Andrew Wheeler signed a directive to reduce funding for animal research 30% by 2025 and eliminate it by 2035(U.S. Environmental Protection Agency (US EPA) 2019). C. elegans has attracted interest of a number of regulatory agencies and governmental labs (Boyd et al. 2016; Hunt et al. 2018; Gong et al. 2018). Yet, at the time of this publication, only two standardized testing documents were developed (International Standard Organization (ISO) 2020, (ASTM) 2001), and none by the U.S. EPA exist. Closing these knowledge gaps and developing standardized protocols will support the use of C. elegans model to address key knowledge gaps in risk assessment, such as developmental neurotoxicity (Hunt et al. 2018), neurodegeneration (Sammi et al 2018), and mixture effects (Wittkowski et al. 2019).
Supplementary Material
Acknowledgements
We thank Steven Nadler, Christopher Pagan, and Leona Scanlan for their help. This work was also supported by the National Institutes of Health (T32ES021432 supported KSM; R01ES028218 and P42ES010356 to JNM; K99ES029552 to JHH; NSF GRFP DGE-1644868 to DEK; R01ES029930 to ECA), the Canadian Institutes of Health Research (CIHR; PJT-153199 to ST; http://www.cihr-irsc.gc.ca), and the Natural Sciences and Engineering Research Council of Canada (NSERC; RGPIN-2018-05133 to ST; http://www.nserc-crsng.gc.ca). No potential competing interest was reported by the authors.
References
- 1996. Interconnections Between Human and Ecosystem Health. Edited by Di Giulio RT and Monosson E: Springer. [Google Scholar]
- (ASTM), American Society of Testing and Materials. 2001. Conducting Laboratory Soil Toxicity Tests with the Nematode Caenorhabditis elegans.
- Aarnio V, Heikkinen L, Peltonen J, Goldsteins G, Lakso M, and Wong G. 2014. Transcriptional profiling reveals differential expression of a neuropeptide-like protein and pseudogenes in aryl hydrocarbon receptor-1 mutant Caenorhabditis elegans. Comp Biochem Physiol Part D Genomics Proteomics 9:40–48. doi: 10.1016/j.cbd.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Abbass M, Chen Y, Arlt VM, and Sturzenbaum SR. 2021. Benzo[a]pyrene and Caenorhabditis elegans: Defining the genotoxic potential in an organism lacking the classical CYP1A1 pathway. Arch Toxicol. doi: 10.1007/s00204-020-02968-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JM, Eom HJ, Yang X, Meyer JN, and Choi J. 2014. Comparative toxicity of silver nanoparticles on oxidative stress and DNA damage in the nematode, Caenorhabditis elegans. Chemosphere 108:343–352. doi: 10.1016/j.chemosphere.2014.01.078. [DOI] [PubMed] [Google Scholar]
- Ahn T, and Yun CH. 2010. Molecular mechanisms regulating the mitochondrial targeting of microsomal cytochrome P450 enzymes. Curr Drug Metab 11:830–838. doi: 10.2174/138920010794479655. [DOI] [PubMed] [Google Scholar]
- Alaimo JT, Davis SJ, Song SS, Burnette CR, Grotewiel M, Shelton KL, Pierce-Shimomura JT, Davies AG, and Bettinger JC. 2012. Ethanol metabolism and osmolarity modify behavioral responses to ethanol in C. elegans. Alcohol Clin Exp Res 36: 1840–1850. doi: 10.1111/j.1530-0277.2012.01799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard P, and Colaiacovo MP. 2010. Bisphenol A impairs the double-strand break repair machinery in the germline and causes chromosome abnormalities. Proc Natl Acad Sci U S A 107: 20405–20410. doi: 10.1073/pnas.1010386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard P, Kleinstreuer NC, Knudsen TB, and Colaiacovo MP. 2013. A C. elegans screening platform for the rapid assessment of chemical disruption of germline function Environ Health Perspect 121: 717–724. doi: 10.1289/ehp.1206301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun ZF and Hall DH 2009a. Alimentary System, Pharynx. In WormAtlas. [Google Scholar]
- Altun ZF and Hall DH 2009b. Excretory system. In WormAtlas. [Google Scholar]
- Anbalagan C, Lafayette I, Antoniou-Kourounioti M, Gutierrez C, Martin JR, Chowdhuri DK, and De Pomerai DI. 2013. Use of transgenic GFP reporter strains of the nematode Caenorhabditis elegans to investigate the patterns of stress responses induced by pesticides and by organic extracts from agricultural soils. Ecotoxicology 22:72–85. doi: 10.1007/s10646-012-1004-2. [DOI] [PubMed] [Google Scholar]
- Andersen EC, Gerke JP, Shapiro JA, Crissman JR, Ghosh R, Bloom JS, Felix MA, and Kruglyak L. 2012. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet 44:285–90. doi: 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen EC, Shimko TC, Crissman JR, Ghosh R, Bloom JS, Seidel HS, Gerke JP, and Kruglyak L. 2015. A powerful new quantitative genetics platform, combining Caenorhabditis elegans high-throughput fitness assays with a large collection of recombinant strains. G3 (Bethesda) 5 :911–920. doi: 10.1534/g3.115.017178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Taubert S, MacNeil LT, Conine CC, Tsuda B, Van Gilst M, Sequerra R, Doucette-Stamm L, Yamamoto KR, and Walhout AJ. 2010. Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network. Mol Syst Biol 6:367. doi: 10.1038/msb.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardelli BF 2013. Transport proteins of the ABC systems superfamily and their role in drug action and resistance in nematodes. Parasitol Int 62:639–646. doi: 10.1016/j.parint.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, and Ruvkun G. 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421:268–272. [DOI] [PubMed] [Google Scholar]
- Au C, Benedetto A, Anderson J, Labrousse A, Erikson K, Ewbank JJ, and Aschner M. 2009. SMF-1, SMF-2 and SMF-3 DMT1 orthologues regulate and are regulated differentially by manganese levels in C. elegans. PLoS One 4:e7792. doi: 10.1371/journal.pone.0007792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avdeef A 2001. Physicochemical profiling (solubility, permeability and charge state). Curr Top Med Chem 1:277–351. doi: 10.2174/1568026013395100. [DOI] [PubMed] [Google Scholar]
- Avery L and You YJ. 2012. C. elegans feeding (May 21, 2012), WormBook, ed. In WormBook. The C. elegans Research Community; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, and Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI 2006. Chemosensation in C. elegans. WormBook:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR 2013. To grow or not to grow: Nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics 194: 539–555. doi: 10.1534/genetics.113.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister R, Schaffitzel E, and Hertweck M. 2006. Endocrine signaling in Caenorhabditis elegans controls stress response and longevity. J Endocrinol 190:191–202. doi: 10.1677/joe.1.06856. [DOI] [PubMed] [Google Scholar]
- Behl M, Hsieh JH, Shafer TJ, Mundy WR, Rice JR, Boyd WA, Freedman JH, Hunter ES 3rd, Jarema KA, Padilla S, and Tice RR. 2015. Use of alternative assays to identify and prioritize organophosphorus flame retardants for potential developmental and neurotoxicity. Neurotoxicol Teratol 52 :181–193. doi: 10.1016/j.ntt.2015.09.003. [DOI] [PubMed] [Google Scholar]
- Benz R, Janko K, and Läuger P. 1980. Pore formation by the matrix protein (porin) of Escherichia coli in planar bilayer membranes. Ann N Y Acad Sci 358:13–24. doi: 10.1111/j.1749-6632.1980.tb15382.x. [DOI] [PubMed] [Google Scholar]
- Berg M, Stenuit B, Ho J, Wang A, Parke C, Knight M, Alvarez-Cohen L, and Shapira M. 2016. Assembly of the Caenorhabditis elegans gut microbiota from diverse soil microbial environments. ISME J 10:1998–2009. doi: 10.1038/ismej.2015.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Steinbaugh MJ, Hourihan JM, Ewald CY, and Isik M. 2015. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic Biol Med 88 : 290–301. doi: 10.1016/j.freeradbiomed.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum J, and Fridovich I. 1983. Superoxide, hydrogen peroxide, and oxygen toxicity in two free-living nematode species. Arch Biochem Biophys 222 :35–43. [DOI] [PubMed] [Google Scholar]
- Bock KW 2014. Homeostatic control of xeno- and endobiotics in the drug-metabolizing enzyme system. Biochem Pharmacol 90:1–6. doi: 10.1016/j.bcp.2014.04.009. [DOI] [PubMed] [Google Scholar]
- Boyd WA, McBride SJ, and Freedman JH. 2007. Effects of genetic mutations and chemical exposures on Caenorhabditis elegans feeding: Evaluation of a novel, high-throughput screening assay. PLoS ONE 2 :e1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd WA, McBride SJ, Rice JR, Snyder DW, and Freedman JH. 2010. A high-throughput method for assessing chemical toxicity using a Caenorhabditis elegans reproduction assay. Toxicol Appl Pharmacol 245: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd WA, Smith MV, Co CA, Pirone JR, Rice JR, Shockley KR, and Freedman JH. 2015. Developmental effects of the ToxCast Phase I and II chemicals in and corresponding responses in zebrafish, rats, and rabbits. Environ Health Perspect. doi: 10.1289/ehp.1409645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd WA, Smith MV, Co CA, Pirone JR, Rice JR, Shockley KR, and Freedman JH. 2016. Developmental effects of the ToxCast™ Phase I and Phase II chemicals in Caenorhabditis elegans and corresponding responses in zebrafish, rats, and rabbits. Environ Health Perspect 124:586–593. doi: 10.1289/ehp.1409645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd WA, Smith MV, Kissling GE, and Freedman JH. 2009. Medium- and high-throughput screening of neurotoxicants using C. elegans. Neurotoxicol Teratol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd WA, and Williams PL. 2003. Comparison of the sensitivity of three nematode species to copper and their utility in aquatic and soil toxicity tests. Environ Toxicol Chem 22: 2768–2774. [DOI] [PubMed] [Google Scholar]
- Brady SC, Zdraljevic S, Bisaga KW, Tanny RE, Cook DE, Lee D, Wang Y, and Andersen EC. 2019. A novel gene underlies bleomycin-response variation in Caenorhabditis elegans. Genetics 212:1453–1468. doi: 10.1534/genetics.119.302286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SP, Monosson E, Matson CW, and Bickham JW. 2017. Evolutionary toxicology: Toward a unified understanding of life’s response to toxic chemicals. Evol Appl 10 :745–751. doi: 10.1111/eva.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkhaus SG, Bornhorst J, Chakraborty S, Wehe CA, Niehaus R, Reifschneider O, Aschner M, and Karst U. 2014. Elemental bioimaging of manganese uptake in C. elegans. Metallomics 6:617–621. doi: 10.1039/c3mt00334e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V, Ale-Agha N, Haendeler J, and Ventura N. 2019. The aryl hydrocarbon receptor (AhR) in the aging process: Another puzzling role for this highly conserved transcription factor. Front Physiol 10:1561. doi: 10.3389/fphys.2019.01561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeks A, Gerrard B, Allikmets R, Dean M, and Plasterk RH. 1996. Homologues of the human multidrug resistance genes MRP and MDR contribute to heavy metal resistance in the soil nematode Caenorhabditis elegans. EMBO J 15:6132–6143. [PMC free article] [PubMed] [Google Scholar]
- Broeks A, Janssen HW, Calafat J, and Plasterk RH. 1995. A P-glycoprotein protects Caenorhabditis elegans against natural toxins. EMBO J 14:1858–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BW, Sabo-Attwood T, Choi K, Kim S, Kostal J, LaLone CA, Langan LM, Margiotta-Casaluci L, You J, and Zhang X. 2020. Toxicology advances for 21st century chemical pollution. OneEarth 2:312–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchell B, Brierley CH, Monaghan G, and Clarke DJ. 1997. “The structure and function of the UDP-glucuronosyltransferase gene family.” In Advances in Pharmacology, edited by Goldstein DS, Eisenhofer G and McCarty R, Academic Press. pp 335–338 [DOI] [PubMed] [Google Scholar]
- Burns AR, Wallace IM, Wildenhain J, Tyers M, Giaever G, Bader GD, Nislow C, Cutler SR, and Roy PJ. 2010. A predictive model for drug bioaccumulation and bioactivity in Caenorhabditis elegans. Nat Chem Biol 6:5 49–557. doi: 10.1038/nchembio.380. [DOI] [PubMed] [Google Scholar]
- Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cocheme HM, Noori T, Weinkove D, Schuster E, Greene ND, and Gems D. 2013. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabreiro F, and Gems D. 2013. Worms need microbes too: Microbiota, health and aging in Caenorhabditis elegans. EMBO Mol Med 5:1300–1310. doi: 10.1002/emmm.201100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Yan C, Wu X, Zhou L, and Xiu G. 2020. Nonylphenol induced individual and population fluctuation of Caenorhabditis elegans: Disturbances on developmental and reproductive system. Environ Res 186:109486. doi: 10.1016/j.envres.2020.109486. [DOI] [PubMed] [Google Scholar]
- Carroll BT, Dubyak GR, Sedensky MM, and Morgan PG. 2006. Sulfated signal from ASJ sensory neurons modulates stomatin-dependent coordination in Caenorhabditis elegans. J Biol Chem 281:35989–35996. doi: 10.1074/jbc.M606086200. [DOI] [PubMed] [Google Scholar]
- Casarett LJ, Doull J, and Klaassen CD. 2008. Casarett and Doull’s Toxicology : the Basic Science of Poisons. 7th ed. New York: McGraw-Hill. [Google Scholar]
- Cassada RC, and Russell RL. 1975. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev Biol 46 :326–342. doi: 10.1016/0012-1606(75)90109-8. [DOI] [PubMed] [Google Scholar]
- César-Razquin A, Snijder B, Frappier-Brinton T, Isserlin R, Gyimesi G, Bai X, Reithmeier RA, Hepworth D, Hediger MA, Edwards AM, and Superti-Furga G. 2015. A call for systematic research on solute carriers. Cell 162: 478–487. doi: 10.1016/j.cell.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Chakrapani BP, Kumar S, and Subramaniam JR. 2008. Development and evaluation of an in vivo assay in Caenorhabditis elegans for screening of compounds for their effect on cytochrome P450 expression. J Biosci 33: 269–277. [DOI] [PubMed] [Google Scholar]
- Chamoli M, Singh A, Malik Y, and Mukhopadhyay A. 2014. A novel kinase regulates dietary restriction-mediated longevity in Caenorhabditis elegans. Aging Cell 13:641–655. doi: 10.1111/acel.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TK, Chen J, Pillay V, Ho JY, and Bandiera SM. 2003. Real-time polymerase chain reaction analysis of CYP1B1 gene expression in human liver. Toxicol Sci 71: 11–19. doi: 10.1093/toxsci/71.1.11. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Bains Y, Guha S, Kahn A, Hall D, Bose N, Gugliucci A, and Kapahi P. 2018. The role of advanced glycation end products in aging and metabolic diseases: Bridging association and causality. Cell Metab 28:337–352. doi: 10.1016/j.cmet.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Samuel TK, Krause M, Dailey HA, and Hamza I. 2012. Heme utilization in the Caenorhabditis elegans hypodermal cells is facilitated by heme-responsive gene-2. J Biol Chem 287:9601–9612. doi: 10.1074/jbc.M111.307694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wang C, Li H, Ma R, Yu Z, Li L, Xiang M, Chen X, Hua X, and Yu Y. 2019. A review of toxicity induced by persistent organic pollutants (POPs) and endocrine-disrupting chemicals (EDCs) in the nematode Caenorhabditis elegans. J Environ Manage 237:519–525. doi: 10.1016/j.jenvman.2019.02.102. [DOI] [PubMed] [Google Scholar]
- Chen Y, Shu L, Qiu Z, Lee DY, Settle SJ, Que Hee S, Telesca D, Yang X, and Allard P. 2016. Exposure to the BPA-substitute Bisphenol S causes unique alterations of germline function. PLoS Genet 12:e1006223. doi: 10.1371/journal.pgen.1006223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Tsyusko OV, Unrine JM, Chatterjee N, Ahn JM, Yang XY, Thornton BL, Ryde IT, Starnes D, and Meyer JN. 2014. A micro-sized model for the in vivo study of nanoparticle toxicity: what has Caenorhabditis elegans taught us? Environ Chem 11: 227–246. doi: Doi 10.1071/En13187. [DOI] [Google Scholar]
- Claus SP, Guillou H, and Ellero-Simatos S. 2016. The gut microbiota: A major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes 2:16003. doi: 10.1038/npjbiofilms.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo A, Kronberg MF, Rossen A, Moya A, Calvo D, Salatino SE, Pagano EA, Morabito JA, and Munarriz ER. 2016. The nematode Caenorhabditis elegans as an integrated toxicological tool to assess water quality and pollution. Sci Total Environ 569–570: 252–261. doi: 10.1016/j.scitotenv.2016.06.057. [DOI] [PubMed] [Google Scholar]
- Colas C, Man-Un Ung P, and Schlessinger A. 2016. SLC transporters: Structure, function, and drug discovery. Med Chem Comm 7:1069–1081. doi: 10.1039/C6MD00005C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin B, Oostveen E, Tsyusko OV, and Unrine JM. 2014. Influence of natural organic matter and surface charge on the toxicity and bioaccumulation of functionalized ceria nanoparticles in Caenorhabditis elegans. Environ Sci Technol 48 : 1280–1289. doi: 10.1021/es404503c. [DOI] [PubMed] [Google Scholar]
- Cook DE, Zdraljevic S, Roberts JP, and Andersen EC. 2017. CeNDR, the Caenorhabditis elegans natural diversity resource. Nucl Acids Res 45: D650–D657. doi: 10.1093/nar/gkw893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JF, and Van Raamsdonk JM. 2018. Modeling Parkinson’s disease in C. elegans. J Parkinsons Dis 8:17–32. doi: 10.3233/JPD-171258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GN, Kusch M, and Edgar RS. 1981a. Cuticle of Caenorhabditis elegans: Its isolation and partial characterization. J Cell Biol 90:7–17. doi: 10.1083/jcb.90.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GN, Staprans S, and Edgar RS. 1981b. The cuticle of Caenorhabditis elegans: II. Stage-specific changes in ultrastructure and protein composition during postembryonic development. Dev Biol 86 :456–470. doi: 10.1016/0012-1606(81)90204-9. [DOI] [PubMed] [Google Scholar]
- Crombie TA, Zdraljevic S, Cook DE, Tanny RE, Brady SC, Wang Y, Evans KS, Hahnel S, Lee D, Rodriguez BC, Zhang G, van der Zwagg J, Kiontke K, and Andersen EC. 2019. Deep sampling of Hawaiian Caenorhabditis elegans reveals high genetic diversity and admixture with global populations. Elife 8. doi: 10.7554/eLife.50465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crone B, Aschner M, Schwerdtle T, Karst U, and Bornhorst J. 2015. Elemental bioimaging of cisplatin in Caenorhabditis elegans by LA-ICP-MS. Metallomics 7:1189–1195. doi: 10.1039/c5mt00096c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook-McMahon HM, Olahova M, Button EL, Winter JJ, and Veal EA. 2014. Genome-wide screening identifies new genes required for stress-induced phase 2 detoxification gene expression in animals. BMC Biol 12:64. doi: 10.1186/s12915-014-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YX, McBride SJ, Boyd WA, Alper S, and Freedman JH. 2007. Toxicogenomic analysis of Caenorhabditis elegans reveals novel genes and pathways involved in the resistance to cadmium toxicity. Genome Biol 8. doi: 10.1186/gb-2007-8-6-r122|ISSN1465-6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodia N, Won SJ, Novillo A, Wieland M, Li C, and Callard IP. 2001. Caenorhabditis elegans as an environmental monitor using DNA microarray analysis. Ann N Y Acad Sci 948: 32–42. doi: 10.1111/j.1749-6632.2001.tb03984.x. [DOI] [PubMed] [Google Scholar]
- Cvilink V, Skálová L, Szotáková B, Lamka J, Kostiainen R, and Ketola RA. 2008. LC-MS-MS identification of albendazole and flubendazole metabolites formed ex vivo by Haemonchus contortus. Anal Bioanal Chem 391:337–343. doi: 10.1007/s00216-008-1863-9. [DOI] [PubMed] [Google Scholar]
- Dasgupta M, Shashikanth M, Gupta A, Sandhu A, De A, Javed S, and Singh V. 2020. “NHR-49 transcription factor regulates immunometabolic response and survival of Caenorhabditis elegans during Enterococcus faecalis infection. Infect Immun 88. doi: 10.1128/iai.00130-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AG, Pierce-Shimomura JT, Kim H, VanHoven MK, Thiele TR, Bonci A, Bargmann CI, and McIntire SL. 2003. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell 115: 655–666. doi: 10.1016/s0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- De Ley P 2006. A quick tour of nematode diversity and the backbone of nematode phylogeny WormBook:1–8. doi: 10.1895/wormbook.1.41.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ley P, De Ley IT, Morris K, Abebe E, Mundo-Ocampo M, Yoder M, Heras J, Waumann D, Rocha-Olivares A, Jay Burr AH, Baldwin JG, and Thomas WK. 2005. An integrated approach to fast and informative morphological vouchering of nematodes for applications in molecular barcoding. Phil Trans R Soc Lond B Biol Sci 360: 1945–1958. doi: 10.1098/rstb.2005.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Veer MJ, Kemp JM, and Meeusen EN. 2007. The innate host defence against nematode parasites. Parasite Immunol 29:1–9. doi: 10.1111/j.1365-3024.2006.00910.x. [DOI] [PubMed] [Google Scholar]
- DeGorter MK, Xia CQ, Yang JJ, and Kim RB. 2012. Drug transporters in drug efficacy and toxicity. Annu Rev Pharmacol Toxicol 52 :249–273. doi: 10.1146/annurev-pharmtox-010611-134529. [DOI] [PubMed] [Google Scholar]
- Dejima K, Murata D, Mizuguchi S, Nomura KH, Gengyo-Ando K, Mitani S, Kamiyama S, Nishihara S, and Nomura K. 2009. The ortholog of human solute carrier family 35 member B1 (UDP-galactose transporter-related protein 1) is involved in maintenance of ER homeostasis and essential for larval development in Caenorhabditis elegans. FASEB J 23: 2215–2225. doi: 10.1096/fj.08-123737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deline M, Keller J, Rothe M, Schunck WH, Menzel R, and Watts JL. 2015. Epoxides derived from dietary dihomo-gamma-linolenic acid induce germ cell death in C. elegans. Sci Rep 5:15417. doi: 10.1038/srep15417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di R, Zhang H, and Lawton MA. 2018. Transcriptome analysis of C. elegans reveals novel targets for DON cytotoxicity. Toxins 10. doi: 10.3390/toxins10070262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich N, Tan CH, Cubillas C, Earley BJ, and Kornfeld K. 2016. Insights into zinc and cadmium biology in the nematode Caenorhabditis elegans. Arch Biochem Biophys 611:120–133. doi: 10.1016/j.abb.2016.05.021. [DOI] [PubMed] [Google Scholar]
- Dirksen P, Assie A, Zimmermann J, Zhang F, Tietje AM, Marsh SA, Felix MA, Shapira M, Kaleta C, Schulenburg H, and Samuel B. 2020. CeMbio - The Caenorhabditis elegans microbiome resource. G3 (Bethesda). doi: 10.1534/g3.120.401309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen P, Marsh SA, Braker I, Heitland N, Wagner S, Nakad R, Mader S, Petersen C, Kowallik V, Rosenstiel P, Felix MA, and Schulenburg H. 2016. The native microbiome of the nematode Caenorhabditis elegans: Gateway to a new host-microbiome model. BMC Biol 14:38. doi: 10.1186/s12915-016-0258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekschmitt K, and Korthals GW. 2006. Nematodes as sentinels of heavy metals and organic toxicants in the soil. J Nematol 38 :13–19. [PMC free article] [PubMed] [Google Scholar]
- Elsworth B, Wasmuth J, and Blaxter M. 2011. NEMBASE4: the nematode transcriptome resource. Int J Parasitol 41:881–894. doi: 10.1016/j.ijpara.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Emmons SW 2005. Male development (November 10, 2005), WormBook, ed. In WormBook. The C. elegans Research Community. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkut C, Penkov S, Khesbak H, Vorkel D, Verbavatz J-M, Fahmy K, and Kurzchalia TV. 2011. Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Curr Biol 21 :1331–1336. doi: 10.1016/j.cub.2011.06.064. [DOI] [PubMed] [Google Scholar]
- Evans KS, and Andersen EC. 2020. The gene scb-1 underlies variation in Caenorhabditis elegans chemotherapeutic responses. G3 (Bethesda) 10: 2353–2364. doi: 10.1534/g3.120.401310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans KS, Brady SC, Bloom JS, Tanny RE, Cook DE, Giuliani SE, Hippleheuser SW, Zamanian M, and Andersen EC. 2018. Shared genomic regions underlie natural variation in diverse toxin responses. Genetics 210:1509–1525. doi: 10.1534/genetics.118.301311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson GD, and Bridge WJ. 2019. The glutathione system and the related thiol network in Caenorhabditis elegans. Redox Biol 24:101171. doi: 10.1016/j.redox.2019.101171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira DW, and Allard P. 2015. Models of germ cell development and their application for toxicity studies. Environ Mol Mutagen 56:637–649. doi: 10.1002/em.21946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N, and Antebi A. 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev 22 :2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo LA, Rebouças TF, Ferreira SR, Rodrigues-Luiz GF, Miranda RC, Araujo RN, and Fujiwara RT. 2018. Dominance of P-glycoprotein 12 in phenotypic resistance conversion against ivermectin in Caenorhabditis elegans. PloS one 13: e0192995–e0192995. doi: 10.1371/journal.pone.0192995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Regitz C, Kahl M, Werthebach M, Boll M, and Wenzel U. 2012. Phytoestrogens genistein and daidzein affect immunity in the nematode Caenorhabditis elegans via alterations of vitellogenin expression. Mol Nutr Food Res 56:957–965. doi: 10.1002/mnfr.201200006. [DOI] [PubMed] [Google Scholar]
- Fisher AL, and Lithgow GJ. 2006. The nuclear hormone receptor DAF-12 has opposing effects on Caenorhabditis elegans lifespan and regulates genes repressed in multiple long-lived worms. Aging Cell 5:127–138. doi: 10.1111/j.1474-9726.2006.00203.x. [DOI] [PubMed] [Google Scholar]
- Fitch DH 2005. Introduction to nematode evolution and ecology. WormBook:1–8. doi: 10.1895/wormbook.1.19.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitsanakis VA, Negga R, and Hatfield HE. 2014. Caenorhabditis elegans as a model for biomarkers of diseases and toxicities. Biomarker Toxicol: 113–128. doi: 10.1016/B978-0-12-404630-6.00006-3. [DOI] [Google Scholar]
- Fontaine P, and Choe K. 2018. The transcription factor SKN-1 and detoxification gene ugt-22 alter albendazole efficacy in Caenorhabditis elegans. Int J Parasitol Drugs Drug Resist 8: 312–319. doi: 10.1016/j.ijpddr.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco ALC, Gherardi LA, de Tomasel CM, Andriuzzi WS, Ankrom KE, Shaw EA, Bach EM, Sala OE, and Wall DH. 2019. Drought suppresses soil predators and promotes root herbivores in mesic, but not in xeric grasslands. Proc Natl Acad Sci U S A 116 :12883–12888. doi: 10.1073/pnas.1900572116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman JH, Slice LW, Dixon D, Fire A, and Rubin CS. 1993. The novel metallothionein genes of Caenorhabditis elegans. Structural organization and inducible, cell-specific expression. J Biol Chem 268:2554–2564. [PubMed] [Google Scholar]
- Fukushige T, Smith HE, Miwa J, Krause MW, and Hanover JA. 2017. A genetic analysis of the Caenorhabditis elegans detoxification response. Genetics 206: 939–952. doi: 10.1534/genetics.117.202515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxman Bass JI, Pons C, Kozlowski L, Reece-Hoyes JS, Shrestha S, Holdorf AD, Mori A, Myers CL, and Walhout AJ. 2016. A gene-centered C. elegans protein-DNA interaction network provides a framework for functional predictions. Mol Syst Biol 12: 884. doi: 10.15252/msb.20167131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo M, Park D, Luciani DS, Kida K, Palmieri F, Blacque OE, Johnson JD, and Riddle DL. 2011. MISC-1/OGC links mitochondrial metabolism, Apoptosis and insulin secretion. PLoS ONE 6:e17827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalez AP, Ritter AD, Shrestha S, Andersen EC, Yilmaz LS, and Walhout AJM. 2017. Bacterial metabolism affects the C. elegans response to cancer chemotherapeutics. Cell 169:431–441 e8. doi: 10.1016/j.cell.2017.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaytan BD, and Vulpe CD. 2014. Functional toxicology: Tools to advance the future of toxicity testing. Front Genet 5:110. doi: 10.3389/fgene.2014.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbaba TK, Green-Harrison L, and Buret AG. 2017. Modeling host-microbiome interactions in Caenorhabditis elegans. J Nematol 49 :348–356. [PMC free article] [PubMed] [Google Scholar]
- Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, and Antebi A. 2001. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell 1 :841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- Gill MS, Held JM, Fisher AL, Gibson BW, and Lithgow GJ. 2004. Lipophilic regulator of a developmental switch in Caenorhabditis elegans. Aging Cell 3:413–421. doi: 10.1111/j.1474-9728.2004.00126.x. [DOI] [PubMed] [Google Scholar]
- Glavinas H, Krajcsi P, Cserepes J, and Sarkadi B. 2004. The role of ABC transporters in drug resistance, metabolism and toxicity. Curr Drug Deliv 1: 27–42. doi: 10.2174/1567201043480036. [DOI] [PubMed] [Google Scholar]
- Goh GYS, Winter JJ, Bhanshali F, Doering KRS, Lai R, Lee K, Veal EA, and Taubert S. 2018. NHR-49/HNF4 integrates regulation of fatty acid metabolism with a protective transcriptional response to oxidative stress and fasting. Aging Cell 17 :e12743. doi: 10.1111/acel.12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P, Donohue KB, Mayo AM, Wang Y, Hong H, Wilbanks MS, Barker ND, Guan X, and Gust KA. 2018. Comparative toxicogenomics of three insensitive munitions constituents 2,4-dinitroanisole, nitroguanidine and nitrotriazolone in the soil nematode Caenorhabditis elegans. BMC Syst Biol 12 (Suppl 7): 92–92. doi: 10.1186/s12918-018-0636-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Moragas L, Maurer LL, Harms VM, Meyer JN, Laromaine A, and Roig A. 2017. Materials and toxicological approaches to study metal and metal-oxide nanoparticles in the model organism Caenorhabditis elegans.” Mater Horiz 4:719–746. doi: 10.1039/C7MH00166E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Moragas L, Berto P, Vilches C, Quidant R, Kolovou A, Santarella-Mellwig R, Schwab Y, Stürzenbaum S, Roig A, and Laromaine A. 2017. In vivo testing of gold nanoparticles using the Caenorhabditis elegans model organism. Acta Biomater 53:598–609. doi: 10.1016/j.actbio.2017.01.080. [DOI] [PubMed] [Google Scholar]
- Govindan JA, Jayamani E, Zhang X, Mylonakis E, and Ruvkun G. 2015. Dialogue between E. coli free radical pathways and the mitochondria of C. elegans. Proc Natl Acad Sci U S A 112:12456–12461. doi: 10.1073/pnas.1517448112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracida X, and Eckmann CR. 2013. Fertility and germline stem cell maintenance under different diets requires NHR-114/HNF4 in C. elegans. Curr Biol 23: 607–613. doi: 10.1016/j.cub.2013.02.034. [DOI] [PubMed] [Google Scholar]
- Grants JM, Goh GY, and Taubert S. 2015. The mediator complex of Caenorhabditis elegans: Insights into the developmental and physiological roles of a conserved transcriptional coregulator.” Nucl Acids Res 43:2442–2453. doi: 10.1093/nar/gkv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu QL, Zhang Y, Fu XM, Lu ZL, Yu Y, Chen G, Ma R, Kou W, and Lan YM. 2020. Toxicity and metabolism of 3-bromopyruvate in Caenorhabditis elegans. J Zhejiang Univ Sci B 21:77–86. doi: 10.1631/jzus.B1900370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP 2008. Cytochrome p450 and chemical toxicology. Chem Res Toxicol 21: 70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- Guengerich F, Peter. 2006. Cytochrome P450s and other enzymes in drug metabolism and toxicity. AAPS J 8 :E101–E111. doi: 10.1208/aapsj080112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujar MR, Stricker AM, and Lundquist EA. 2017. Flavin monooxygenases regulate Caenorhabditis elegans axon guidance and growth cone protrusion with UNC-6/Netrin signaling and Rac GTPases. PLoS Genet 13 :e1006998. doi: 10.1371/journal.pgen.1006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegerbaeumer A, Hoss S, Heininger P, and Traunspurger W. 2018a. Is Caenorhabditis elegans representative of freshwater nematode species in toxicity testing? Environ Sci Pollut Res Int 25:2879–2888. doi: 10.1007/s11356-017-0714-7. [DOI] [PubMed] [Google Scholar]
- Haegerbaeumer A, Hoss S, Heininger P, and Traunspurger W. 2018b. Response of nematode communities to metals and PAHs in freshwater microcosms. Ecotoxicol Environ Saf 148:244–253. doi: 10.1016/j.ecoenv.2017.10.030. [DOI] [PubMed] [Google Scholar]
- Haegerbaeumer A, Raschke R, Reiff N, Traunspurger W, and Hoss S. 2019. Comparing the effects of fludioxonil on non-target soil invertebrates using ecotoxicological methods from single-species bioassays to model ecosystems. Ecotoxicol Environ Saf 183:109596. doi: 10.1016/j.ecoenv.2019.109596. [DOI] [PubMed] [Google Scholar]
- Hagerbaumer A, Hoss S, Heininger P, and Traunspurger W. 2015. Experimental studies with nematodes in ecotoxicology: An overview. J Nematol 47:11–27. [PMC free article] [PubMed] [Google Scholar]
- Hahn ME 2019. Evolutionary concepts can benefit both fundamental research and applied research in toxicology (A comment on Brady et al. 2017). Evol Appl 12 :350–352. doi: 10.1111/eva.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitzer M, K Burnison B, Höss S, Traunspurger W, and Steinberg CEW. 1999. Effects of quantity, quality, and contact time of dissolved organic matter on bioconcentration of benzo[a]pyrene in the nematode Caenorhabditis elegans. Environ Toxicol Chem 18: 459–465. doi: 10.1002/etc.5620180314. [DOI] [Google Scholar]
- Hamon Y, Broccardo C, Chambenoit O, Luciani M-F, Toti F, Chaslin S, Freyssinet J-M, Devaux PF, McNeish J, Marguet D, and Chimini G. 2000. ABC1 promotes engulfment of apoptotic cells and transbilayer redistribution of phosphatidylserine. Nat Cell Biol 2: 399–406. doi: 10.1038/35017029. [DOI] [PubMed] [Google Scholar]
- Hanada K, Nishijima K, Ogata H, Atagi S, and Kawahara M. 2001. Population pharmacokinetic analysis of cisplatin and its metabolites in cancer patients: Possible misinterpretation of covariates for pharmacokinetic parameters calculated from the concentrations of unchanged cisplatin, ultrafiltered platinum and total platinum. Jap J Clin Oncol 31: 179–184. doi: 10.1093/jjco/hye040. [DOI] [PubMed] [Google Scholar]
- Harborne JB 1988. Introduction to Ecological Biochemistry. 3rd ed. London; San Diego: Academic Press. [Google Scholar]
- Harlow PH, Perry SJ, Stevens AJ, and Flemming AJ. 2018. Comparative metabolism of xenobiotic chemicals by cytochrome P450s in the nematode Caenorhabditis elegans. Rep 8:13333. doi: 10.1038/s41598-018-31215-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow PH, Perry SJ, Widdison S, Daniels S, Bondo E, Lamberth C, Currie RA, and Flemming AJ. 2016. The nematode Caenorhabditis elegans as a tool to predict chemical activity on mammalian development and identify mechanisms influencing toxicological outcome. Sci Rep 6:22965. doi: 10.1038/srep22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington AJ, Hamamichi S, Caldwell GA, and Caldwell KA. 2010. C. elegans as a model organism to investigate molecular pathways involved with Parkinson’s disease. Dev Dynamics 239:1282–1295. doi: Doi 10.1002/Dvdy.22231. [DOI] [PubMed] [Google Scholar]
- Harris JB, Hartman JH, Luz AL, Wilson JY, Dinyari A, and Meyer JN. 2020. Zebrafish CYP1A expression in transgenic Caenorhabditis elegans protects from exposures to benzo[a]pyrene and a complex polycyclic aromatic hydrocarbon mixture Toxicology 440:152473. doi: 10.1016/j.tox.2020.152473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TR, Aronov PA, Jones PD, Tanaka H, Arand M, and Hammock BD. 2008. Identification of two epoxide hydrolases in Caenorhabditis elegans that metabolize mammalian lipid signaling molecules. Arch Biochem Biophys 472 :139–149. doi: 10.1016/j.abb.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman PS, and Herman RK. 1982. Radiation-sensitive mutants of Caenorhabditis elegans. Genetics 102 :159–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SC, and Orbidans HE. 2011. All eggs are not equal: the maternal environment affects progeny reproduction and developmental fate in Caenorhabditis elegans. PLoS One 6 :e25840. doi: 10.1371/journal.pone.0025840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschek WM, Berenbaum M, Hinton DE, Cora M, Chernoff N, Travlos G, Liu CW, Lu K, and Law M. 2019. Pathology in ecological research with implications for one health: Session summary. Toxicol Pathol 47:1072–1075. doi: 10.1177/0192623319880530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, and Miwa J. 2010. Genetic and cellular characterization of Caenorhabditis elegans mutants abnormal in the regulation of many phase II enzymes. PLoS One 5: e11194. doi: 10.1371/journal.pone.0011194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Miwa S, Isomura K, Tsutsumiuchi K, Taniguchi H, and Miwa J. 2007. Acrylamide-responsive genes in the nematode Caenorhabditis elegans. Toxicol Sci 101: 215–225. doi: 10.1093/toxsci/kfm276. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Miwa S, Tsutsumiuchi K, and Miwa J. 2010. Allyl isothiocyanate that induces GST and UGT expression confers oxidative stress resistance on C. elegans, as demonstrated by nematode biosensor. PLOS ONE 5 :e9267. doi: 10.1371/journal.pone.0009267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori K, Inoue M, Inoue T, Arai H, and Tamura H. 2006. A novel sulfotransferase abundantly expressed in the dauer larvae of Caenorhabditis elegans. J Biochem 139: 355–362. doi: 10.1093/jb/mvj041. [DOI] [PubMed] [Google Scholar]
- Hediger MA, Clémençon B, Burrier RE, and Bruford EA. 2013. The ABCs of membrane transporters in health and disease (SLC series): Introduction. Mol Aspects Med 34: 95–107. doi: 10.1016/j.mam.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmcke KJ, Avila DS, and Aschner M. 2010. Utility of Caenorhabditis elegans in high throughput neurotoxicological research. Neurotoxicol Teratol 32: 62–67. doi: DOI 10.1016/j.ntt.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Helmcke KJ, Syversen T, Miller DM 3rd, and Aschner M. 2009. Characterization of the effects of methylmercury on Caenorhabditis elegans. Toxicol Appl Pharmacol 240: 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner MO, and Horvitz HR. 1994. Programmed cell death in Caenorhabditis elegans. Curr Opin Genet Dev 4: 581–586. [DOI] [PubMed] [Google Scholar]
- Herholz M, Cepeda E, Baumann L, Kukat A, Hermeling J, Maciej S, Szczepanowska K, Pavlenko V, Frommolt P, and Trifunovic A. 2019. KLF-1 orchestrates a xenobiotic detoxification program essential for longevity of mitochondrial mutants. Nat Commun 10: 3323. doi: 10.1038/s41467-019-11275-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, and Driscoll M. 2002. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature 419: 808–814. [DOI] [PubMed] [Google Scholar]
- Hirani N, Westenberg M, Seed PT, Petalcorin MIR, and Dolphin CT. 2016. C. elegans flavin-containing monooxygenase-4 is essential for osmoregulation in hypotonic stress. Biol Open 5: 537–549. doi: 10.1242/bio.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert Oliver. 2005. Specification of the nervous system. In WormBook, edited by The C. elegans Research Community. Internet: Wormbook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JM, and Partridge L. 2015. Nuclear hormone receptors: Roles of xenobiotic detoxification and sterol homeostasis in healthy aging. Crit Rev Biochem Mol Biol 50: 380–392. doi: 10.3109/10409238.2015.1067186. [DOI] [PubMed] [Google Scholar]
- Höglund PJ, Nordström KJV, Schiöth HB, and Fredriksson R. 2011. The solute carrier families have a remarkably long evolutionary history with the majority of the human families present before divergence of Bilaterian species. Mol Biol Evol 28:1531–1541. doi: 10.1093/molbev/msq350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnen S 2017. Caenorhabditis elegans as a powerful alternative model organism to promote research in genetic toxicology and biomedicine. Arch Toxicol 91:2029–2044. doi: 10.1007/s00204-017-1944-7. [DOI] [PubMed] [Google Scholar]
- Horsman JW, and Miller DL. 2016. Mitochondrial sulfide quinone oxidoreductase prevents activation of the unfolded protein response in hydrogen sulfide. J Biol Chem 291: 5320–5325. doi: 10.1074/jbc.M115.697102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoss S, HAitzer M, Traunspurger W, Gratzer H, Ahlf W, and Steinberg C. 1997. Influence of particle size distribution and content of organic matter on the toxicity of copper in sediment bioassays using Caenorhabditis elegans (nematoda). Water Air Soil Pollut 99: 689–695. [Google Scholar]
- Hoss S, Haitzer M, Traunspurger W, and Steinberg CEW. 1999. Growth and fertility of Caenorhabditis elegans (Nematoda) in unpolluted freshwater sediments: Response to particle size distribution and organic content. Environ Toxicol Chem 18 :2921–2925. [Google Scholar]
- Hoss S, Jansch S, Moser T, Junker T, and Rombke J. 2009. Assessing the toxicity of contaminated soils using the nematode Caenorhabditis elegans as test organism. Ecotoxicol Environ Saf 72:1811–1818. doi: 10.1016/j.ecoenv.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Höss S, Schlottmann K, and Traunspurger W. 2011. Toxicity of ingested cadmium to the nematode Caenorhabditis elegans. Environ Sci Technol 45 :10219–10225. doi: 10.1021/es2027136. [DOI] [PubMed] [Google Scholar]
- Hoss S, and Weltje L. 2007. Endocrine disruption in nematodes: Effects and mechanisms. Ecotoxicology 16:15–28. doi: 10.1007/s10646-006-0108-y. [DOI] [PubMed] [Google Scholar]
- Hotchkiss AK, Rider CV, Blystone CR, Wilson VS, Hartig PC, Ankley GT, Foster PM, Gray CL, and Gray LE. 2008. Fifteen years after “Wingspread”--environmental endocrine disrupters and human and wildlife health: Where we are today and where we need to go. Toxicol Sci 105:235–259. doi: 10.1093/toxsci/kfn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, D’Amora DR, MacNeil LT, Walhout AJM, and Kubiseski TJ. 2018. The Caenorhabditis elegans oxidative stress response requires the NHR-49 transcription factor. G3 (Bethesda) 8:3857–3863. doi: 10.1534/g3.118.200727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Powell-Coffman JA, and Jin Y. 2004. The AHR-1 aryl hydrocarbon receptor and its co-factor the AHA-1 aryl hydrocarbon receptor nuclear translocator specify GABAergic neuron cell fate in C. elegans. Development 131:819–828. doi: 10.1242/dev.00959. [DOI] [PubMed] [Google Scholar]
- Hunt PR 2017. The C. elegans model in toxicity testing. J Appl Toxicol 37:50–59. doi: 10.1002/jat.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PR, Olejnik N, Bailey KD, Vaught CA, and Sprando RL. 2018. C. elegans development and activity test detects mammalian developmental neurotoxins. Food Chem Toxicol 121:583–592. doi: 10.1016/j.fct.2018.09.061. [DOI] [PubMed] [Google Scholar]
- International Standard Organization (ISO). 2020. ISO 10872 - Water and soil quality - Determination of the toxic effect of sediment and soil samples on growth, fertility and reproduction of Caenorhabditis elegans (Nematoda). Switzerland: ISO. [Google Scholar]
- Ioannides C 2001. Xenobiotic metabolism: An overview. In Enzyme Systems that Metabolise Drugs and Other Xenobiotics, pp. 1–32. [Google Scholar]
- Jackson BP, Williams PL, Lanzirotti A, and Bertsch PM. 2005. Evidence for biogenic pyromorphite formation by the nematode Caenorhabditis elegans. Environ Sci Technol 39:5620–5625. doi: 10.1021/es050154k. [DOI] [PubMed] [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, and Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James CE, and Davey MW. 2009. Increased expression of ABC transport proteins is associated with ivermectin resistance in the model nematode Caenorhabditis elegans. Int J Parasitol 39:213–220. doi: 10.1016/j.ijpara.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Jeong J, Kim H, and Choi J. 2019. In silico molecular docking and in vivo validation with Caenorhabditis elegans to discover molecular initiating events in adverse outcome pathway framework: Case study on endocrine-disrupting chemicals with estrogen and androgen receptors. Int J Mol Sci 20. doi: 10.3390/ijms20051209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Albert PS, and Riddle DL. 2002. DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development 129: 221–231. [DOI] [PubMed] [Google Scholar]
- Jin MS, Oldham ML, Zhang Q, and Chen J. 2012. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490: 566–569. doi: 10.1038/nature11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston WL, and Dennis JW. 2012. The eggshell in the C. elegans oocyte-to-embryo transition. Genesis 50:333–349. doi: 10.1002/dvg.20823. [DOI] [PubMed] [Google Scholar]
- Johnstone IL 1994. The cuticle of the nematode Caenorhabditis elegans: A complex collagen structure. Bioessays 16:171–178. doi: 10.1002/bies.950160307. [DOI] [PubMed] [Google Scholar]
- Johnstone IL 2000. Cuticle collagen genes: Expression in Caenorhabditis elegans. Trends Genet 16:21–27. doi: 10.1016/S0168-9525(99)01857-0. [DOI] [PubMed] [Google Scholar]
- Jones LM, Flemming AJ, and Urwin PE. 2015. NHR-176 regulates CYP-35d1 to control hydroxylation-dependent metabolism of thiabendazole in Caenorhabditis elegans. Biochem J 466: 37–44. doi: 10.1042/BJ20141296. [DOI] [PubMed] [Google Scholar]
- Jones LM, Rayson SJ, Flemming AJ, and Urwin PE. 2013. Adaptive and specialised transcriptional responses to xenobiotic stress in Caenorhabditis elegans are regulated by nuclear hormone receptors. PLoS One 8: e69956. doi: 10.1371/journal.pone.0069956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, and Ahringer J. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kapitonov D and Yu RK. 1999. Conserved domains of glycosyltransferases. Glycobiology 9:961–978. doi: 10.1093/glycob/9.10.961. [DOI] [PubMed] [Google Scholar]
- Kell D 2020a. How drugs really get into cells: Why passive bilayer diffusion is a myth.
- Kell DB 2020b. Hitchhiking into the cell. Nat Chem Biol 16:367–368. doi: 10.1038/s41589-020-0489-x. [DOI] [PubMed] [Google Scholar]
- Kelly WG 2014. Transgenerational epigenetics in the germline cycle of Caenorhabditis elegans. Epigenet Chromatin 7: 6. doi: 10.1186/1756-8935-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K, and Fitch DH. 2013. Nematodes. Curr Biol 23:R862–R864. doi: 10.1016/j.cub.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, and Casarett LJ. 2007. Casarett and Doull’s Toxicology. Blacklick, USA, United States: McGraw-Hill Professional Publishing. [Google Scholar]
- Koontz JM, Dancy BCR, Horton CL, Stallings JD, DiVito VT, and Lewis JA. 2019. The role of the human microbiome in chemical toxicity. Int J Toxicol 38: 251–264. doi: 10.1177/1091581819849833. [DOI] [PubMed] [Google Scholar]
- Koppel N, Maini Rekdal V, and Balskus EP. 2017. Chemical transformation of xenobiotics by the human gut microbiota. Science 356. doi: 10.1126/science.aag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulas J, Schmidt C, Rothe M, Schunck WH, and Menzel R. 2008. Cytochrome P450-dependent metabolism of eicosapentaenoic acid in the nematode Caenorhabditis elegans. Arch Biochem Biophys 472 :65–75. [DOI] [PubMed] [Google Scholar]
- Kurz CL, Shapira M, Chen K, Baillie DL, and Tan M-W. 2007. Caenorhabditis elegans pgp-5 is involved in resistance to bacterial infection and heavy metal and its regulation requires TIR-1 and a p38 map kinase cascade. Biochem Biophys Res Commun 363: 438–443. doi: 10.1016/j.bbrc.2007.08.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing ST, Ivens A, Laing R, Ravikumar S, Butler V, Woods DJ, and Gilleard JS. 2010. Characterization of the xenobiotic response of Caenorhabditis elegans to the anthelmintic drug albendazole and the identification of novel drug glucoside metabolites. Biochem J 432: 505–516. doi: 10.1042/BJ20101346. [DOI] [PubMed] [Google Scholar]
- Lažetić V, and Fay DS. 2017. Molting in C. elegans. Worm 6 :e1330246–e1330246. doi: 10.1080/21624054.2017.1330246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim YG, Kim M, Kim E, Choi H, Kim Y, and Lee J. 2017. Indole-associated predator-prey interactions between the nematode Caenorhabditis elegans and bacteria. Environ Microbiol 19:1776–1790. doi: 10.1111/1462-2920.13649. [DOI] [PubMed] [Google Scholar]
- Lee MH, Luo HR, Bae SH, and San-Miguel A. 2020. Genetic and chemical effects on somatic and germline aging. Oxid Med Cell Longev 2020:4684890. doi: 10.1155/2020/4684890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kim Y, and Choi J. 2020. Effect of soil microbial feeding on gut microbiome and cadmium toxicity in Caenorhabditis elegans. Ecotoxicol Environ Saf 187:109777. doi: 10.1016/j.ecoenv.2019.109777. [DOI] [PubMed] [Google Scholar]
- Lehman-McKeeman LD 2013. Chapter 5: Absorption, distribution, and excretion of toxicants.” In Casarett And Doull’s Toxicology : the Basic Science of Poisons, edited by Klaassen Curtis D.. New York: McGraw-Hill Education. [Google Scholar]
- Leiser SF, Miller H, Rossner R, Fletcher M, Leonard A, Primitivo M, Rintala N, Ramos FJ, Miller DL, and Kaeberlein M. 2015. Cell nonautonomous activation of flavin-containing monooxygenase promotes longevity and health span. Science 350:1375–1378. doi: 10.1126/science.aac9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz KA, Pattison C, and Ma H. 2017. Triclosan (TCS) and triclocarban (TCC) induce systemic toxic effects in a model organism the nematode Caenorhabditis elegans. Environ Pollut 231: 462–470. doi: 10.1016/j.envpol.2017.08.036. [DOI] [PubMed] [Google Scholar]
- Leung MC, Goldstone JV, Boyd WA, Freedman JH, and Meyer JN. 2010. Caenorhabditis elegans generates biologically relevant levels of genotoxic metabolites from aflatoxin B1 but not benzo[a]pyrene in vivo. Toxicol Sci 118:444–453. doi: 10.1093/toxsci/kfq295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung MCK, Procter AC, Goldstone JV, Foox J, DeSalle R, Mattingly CJ, Siddall ME, and Timme-Laragy AR. 2017. Applying evolutionary genetics to developmental toxicology and risk assessment. Reprod Toxicol 69:174–186. doi: 10.1016/j.reprotox.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung MC, Williams PL, Benedetto A, Au C, Helmcke KJ, Aschner M, and Meyer JN. 2008. Caenorhabditis elegans: An emerging model in biomedical and environmental toxicology. Toxicol Sci 106 :5–28. doi: 10.1093/toxsci/kfn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao VH, and Yu CW. 2005. Caenorhabditis elegans gcs-1 confers resistance to arsenic-induced oxidative stress. Biometals 18 :519–528. [DOI] [PubMed] [Google Scholar]
- Lincke CR, Broeks A, The I, Plasterk RH, and Borst P. 1993. The expression of two P-glycoprotein (pgp) genes in transgenic Caenorhabditis elegans is confined to intestinal cells. EMBO J 12:1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblom TH, and Dodd AK. 2006. Xenobiotic detoxification in the nematode Caenorhabditis elegans. J Exp Zool A Comp Exp Biol 305:720–730. doi: 10.1002/jez.a.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblom TH, Pierce GJ, and Sluder AE. 2001. A C. elegans orphan nuclear receptor contributes to xenobiotic resistance. Curr Biol 11: 864–868. doi: 10.1016/s0960-9822(01)00236-6. [DOI] [PubMed] [Google Scholar]
- Lints R and Hall DH 2009. The cuticle “ In WormAtlas [Google Scholar]
- Liu F, Zhang Y, Zhang M, Luo Q, Cao X, Cui C, Lin K, and Huang K. 2020. Toxicological assessment and underlying mechanisms of tetrabromobisphenol A exposure on the soil nematode Caenorhabditis elegans. Chemosphere 242: 125078. doi: 10.1016/j.chemosphere.2019.125078. [DOI] [PubMed] [Google Scholar]
- Ludewig AH, Klapper M, and Döring F. 2014. Identifying evolutionarily conserved genes in the dietary restriction response using bioinformatics and subsequent testing in Caenorhabditis elegans. Genes Nutr 9:363–363. doi: 10.1007/s12263-013-0363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz AL, Godebo TR, Bhatt DP, Ilkayeva OR, Maurer LL, Hirschey MD, and Meyer JN. 2016. From the over: Arsenite uncouples mitochondrial respiration and induces a Warburg-like effect in Caenorhabditis elegans. Toxicol Sci 152:349–362. doi: 10.1093/toxsci/kfw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz AL, Godebo TR, Smith LL, Leuthner TC, Maurer LL, and Meyer JN. 2017. Deficiencies in mitochondrial dynamics sensitize Caenorhabditis elegans to arsenite and other mitochondrial toxicants by reducing mitochondrial adaptability. Toxicology. doi: 10.1016/j.tox.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackowiak B, and Wang H. 2016. Mechanisms of xenobiotic receptor activation: Direct vs. indirect. Biochim Biophys Acta 1859:1130–1140. doi: 10.1016/j.bbagrm.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magner DB, Wollam J, Shen Y, Hoppe C, Li D, Latza C, Rottiers V, Hutter H, and Antebi A. 2013. The NHR-8 nuclear receptor regulates cholesterol and bile acid homeostasis in C. elegans. Cell Metab 18:212–224. doi: 10.1016/j.cmet.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Tan MW, Rahme LG, and Ausubel FM. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96 :47–56. [DOI] [PubMed] [Google Scholar]
- Mak HY, and Ruvkun G. 2004. Intercellular signaling of reproductive development by the C. elegans DAF-9 cytochrome P450. Development 131:1777–1786. doi: 10.1242/dev.01069. [DOI] [PubMed] [Google Scholar]
- Manallack DT, Prankerd RJ, Yuriev E, Oprea TI, and Chalmers DK. 2013. The significance of acid/base properties in drug discovery. Chem Soc Rev 42:485–496. doi: 10.1039/c2cs35348b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J, Rosa BA, Ozersky P, Hallsworth-Pepin K, Zhang X, Bhonagiri-Palsikar V, Tyagi R, Wang Q, Choi YJ, Gao X, McNulty SN, Brindley PJ, and Mitreva M. 2015. Helminth.net: Expansions to nematode.net and an introduction to trematode.net. Nucl Acids Res 43 (Database issue): D698–D06. doi: 10.1093/nar/gku1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly CJ, Rosenstein MC, Davis AP, Colby GT, Forrest JN Jr., and Boyer JL. 2006. The comparative toxicogenomics database: A cross-species resource for building chemical-gene interaction networks. Toxicol Sci 92:587–595. doi: 10.1093/toxsci/kfl008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupas E 1900. Modes et formes de reproduction des nématodes. Arch. Zool. Exp. 8:463–624. [Google Scholar]
- Maurer LL, Luz AL, and Meyer JN. 2018. Detection of mitochondrial toxicity of environmental pollutants using Caenorhabditis elegans. In Drug-Induced Mitochondrial Dysfunction, edited by Dykens JA and Will Y. Wiley. [Google Scholar]
- Maurer LL, Yang X, Schindler AJ, Taggart RK, Jiang C, Hsu-Kim H, Sherwood DR, and Meyer JN. 2015. Intracellular trafficking pathways in silver nanoparticle uptake and toxicity in Caenorhabditis elegans. Nanotoxicology:1–5. doi: 10.3109/17435390.2015.1110759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer LL, Yang X, Schindler AJ, Taggart RK, Jiang C, Hsu-Kim H, Sherwood DR, and Meyer JN. 2016. Intracellular trafficking pathways in silver nanoparticle uptake and toxicity in Caenorhabditis elegans. Nanotoxicology 10:831–835. doi: 10.3109/17435390.2015.1110759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell AM, and Dang CH. 2013. Basic review of the cytochrome p450 system. J Adv Practitioner Oncol 4 :263–268. doi: 10.6004/jadpro.2013.4.4.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD 2013. The Caenorhabditis elegans intestine. Wiley Interdiscip Rev Dev Biol 2: 347–367. doi: 10.1002/wdev.93. [DOI] [PubMed] [Google Scholar]
- McGhee JD 2007. The C. elegans intestine (March 27, 2007), WormBook, ed. In WormBook. The C. elegans Research Community. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meech R, Hu DG, McKinnon RA, Mubarokah SN, Haines AZ, Nair PC, Rowland A, and Mackenzie PI. 2019. The UDP-glycosyltransferase (UGT) superfamily: New members, new functions, and novel paradigms. Physiol Rev 99:1153–1222. doi: 10.1152/physrev.00058.2017. [DOI] [PubMed] [Google Scholar]
- Melendez A, and Levine B. 2009. Autophagy in C. elegans. WormBook:1–26. [DOI] [PubMed] [Google Scholar]
- Menez C, Alberich M, Courtot E, Guegnard F, Blanchard A, Aguilaniu H, and Lespine A. 2019. The transcription factor NHR-8: A new target to increase ivermectin efficacy in nematodes. PLoS Pathog 15 :e1007598. doi: 10.1371/journal.ppat.1007598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R, Bogaert T, and Achazi R. 2001. A systematic gene expression screen of Caenorhabditis elegans cytochrome P450 genes reveals CYP35 as strongly xenobiotic inducible. Arch Biochem Biophys 395:158–168. [DOI] [PubMed] [Google Scholar]
- Menzel R, Sturzenbaum S, Barenwaldt A, Kulas J, and Steinberg CE. 2005. Humic material induces behavioral and global transcriptional responses in the nematode Caenorhabditis elegans. Environ Sci Technol 39 :8324–8332. [DOI] [PubMed] [Google Scholar]
- Mesnage R, Antoniou MN, Tsoukalas D, Goulielmos GN, and Tsatsakis A. 2018. Gut microbiome metagenomics to understand how xenobiotics impact human health Curr Opin Toxicol 11–12:51–58. [Google Scholar]
- Metzler R, Meleshkevitch EA, Fox J, Kim H, and Boudko DY. 2013. An SLC6 transporter of the novel B0,− system aids in absorption and detection of nutrient amino acids in Caenorhabditis elegans. J Exp Biol 216:2843–2857. doi: 10.1242/jeb.081497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, and Williams PL. 2014. Toxicity testing of neurotoxic pesticides in Caenorhabditis elegans. J Toxicol Environ Health B 17: 284–306. doi: 10.1080/10937404.2014.933722. [DOI] [PubMed] [Google Scholar]
- Meyer JN, Lord CA, Yang XY, Turner EA, Badireddy AR, Marinakos SM, Chilkoti A, Wiesner MR, and Auffan M. 2010. Intracellular uptake and associated toxicity of silver nanoparticles in Caenorhabditis elegans. Aquat Toxicol 100:140–150. doi: 10.1016/j.aquatox.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Mimoto A, Fujii M, Usami M, Shimamura M, Hirabayashi N, Kaneko T, Sasagawa N, and Ishiura S. 2007. Identification of an estrogenic hormone receptor in Caenorhabditis elegans. Biochem Biophys Res Commun 364:883–888. doi: 10.1016/j.bbrc.2007.10.089. [DOI] [PubMed] [Google Scholar]
- Mitchell PH, Bull K, Glautier S, Hopper NA, Holden-Dye L, and O’Connor V. 2007. The concentration-dependent effects of ethanol on Caenorhabditis elegans behaviour. Pharmacogenomics J 7 :411–417. doi: 10.1038/sj.tpj.6500440. [DOI] [PubMed] [Google Scholar]
- Miwa J, Schierenberg E, Miwa S, and von Ehrenstein G. 1980. Genetics and mode of expression of temperature-sensitive mutations arresting embryonic development in Caenorhabditis elegans. Dev Biol 76:160–174. doi: 10.1016/0012-1606(80)90369-3. [DOI] [PubMed] [Google Scholar]
- Monosson E 2012. Evolution in a Toxic World: How Life Responds to Chemical Threats Island Press. [Google Scholar]
- Monteverdi GH, and Di Giulio RT. 2000. In vitro and in vivo association of 2,3,7,8-tetrachlorodibenzo-p-dioxin and benzo[a]pyrene with the yolk-precursor protein vitellogenin. Environ Toxicol Chem 19: 2502–2511. doi: 10.1002/etc.5620191016. [DOI] [Google Scholar]
- Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, Suino-Powell K, Xu HE, Auchus RJ, Antebi A, and Mangelsdorf DJ. 2006. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell 12: 1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- Murphy CT 2006. The search for DAF-16/FOXO transcriptional targets: Approaches and discoveries. Exp Gerontol 41:910–921. doi: 10.1016/j.exger.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Na H, Ponomarova O, Giese GE, and Walhout AJM. 2018. C. elegans MRP-5 exports vitamin B12 from mother to offspring to support embryonic development. Cell Rep 22: 3126–3133. doi: 10.1016/j.celrep.2018.02.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nass R, Hall DH, Miller DM, and Blakely RD. 2002. Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc Nat Acad Sci USA 99:3264–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nass R, and Hamza I. 2007. The nematode C. elegans as an animal model to explore toxicology in vivo: Solid and axenic growth culture conditions and compound exposure parameters. Curr Protoc Toxicol Chapter 1:Unit1 9. doi: 10.1002/0471140856.tx0109s31. [DOI] [PubMed] [Google Scholar]
- Nass R, Miller DM, and Blakely RD. 2001. C-elegans: A novel pharmacogenetic model to study Parkinson’s disease. Parkinsonism Related Disorders 7 :185–191. [DOI] [PubMed] [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine. 2017. Application of Systematic Review Methods in an Overall Strategy for Evaluating Low-Dose Toxicity from Endocrine Active Chemicals. Washington, DC: The National Academies Press. [PubMed] [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine. 2018. Environmental Chemicals, the Human Microbiome, and Health Risk: A Research Strategy. Washington, DC: The National Academies Press. [PubMed] [Google Scholar]
- Nebert DW, Dalton TP, Okey AB, and Gonzalez FJ. 2004. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem 279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Wikvall K, and Miller WL. 2013. Human cytochromes P450 in health and disease. Phil Trans R Soc Lond B Biol Sci 368: 20120431. doi: 10.1098/rstb.2012.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi H, Saikia SK, Kanaujia R, Jaiswal S, and Pandey R. 2017. 3β-Hydroxy-urs-12-en-28-oic acid confers protection against ZnONPs induced adversity in Caenorhabditis elegans. Environ Toxicol Pharmacol 53:105–110. doi: 10.1016/j.etap.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Nelson FK, Albert PS, and Riddle DL. 1983. Fine structure of the Caenorhabditis elegans secretory-excretory system. J Ultrastruct Res 82:156–1571. doi: 10.1016/s0022-5320(83)90050-3. [DOI] [PubMed] [Google Scholar]
- Nelson FK, and Riddle DL. 1984. Functional study of the Caenorhabditis elegans secretory-excretory system using laser microsurgery. J Exp Zool 231 :45–56. doi: 10.1002/jez.1402310107. [DOI] [PubMed] [Google Scholar]
- Nigon VM, and Felix MA. 2017. History of research on C. elegans and other free-living nematodes as model organisms. WormBook:1–91. doi: 10.1895/wormbook.1.181.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoletopoulou V, and Tavernarakis N. 2014. Necrotic cell death in Caenorhabditis elegans. Meth Enzymol 545:127–55. doi: 10.1016/B978-0-12-801430-1.00006-8. [DOI] [PubMed] [Google Scholar]
- Nuclear Receptors Nomenclature, Committee. 1999. “A unified nomenclature system for the nuclear receptor superfamily.” Cell 97 (2):161–3. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- Offermann K, Matthäi A, and Ahlf W. 2009. Assessing the importance of dietborne cadmium and particle characteristics on bioavailability and bioaccumulation in the nematode Caenorhabditis elegans. Environ Toxicol Chem 28:1149–1158. doi: 10.1897/08-272.1. [DOI] [PubMed] [Google Scholar]
- Oladimeji PO, and Chen T. 2018. PXR: More than just a master xenobiotic receptor. Mol Pharmacol 93:119–127. doi: 10.1124/mol.117.110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A, and Gill M. 2017. Ageing: Lessons from C. elegans. Cham, Switzerland: Springer. [Google Scholar]
- Olson SK, Greenan G, Desai A, Müller-Reichert T, and Oegema K. 2012. Hierarchical assembly of the eggshell and permeability barrier in C. elegans. J Cell Biology 198: 731–748. doi: 10.1083/jcb.201206008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AP and Johnstone IL 2007. The cuticle (March 19, 2007), WormBook, ed. In WormBook. The C. elegans Research Community. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pender CL, and Horvitz HR. 2018. Hypoxia-inducible factor cell non-autonomously regulates C. elegans stress responses and behavior via a nuclear receptor. Elife 7. doi: 10.7554/eLife.36828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Tong Z-H, Chong H-J, and Shao X-Y. 2018. Toxic effects of prolonged exposure to [C14mim]Br on Caenorhabditis elegans. Chemosphere 208:226–232. doi: 10.1016/j.chemosphere.2018.05.176. [DOI] [PubMed] [Google Scholar]
- Perally S, LaCourse EJ, Campbell AM, and Brophy PM. 2008. Heme transport and detoxification in nematodes: Subproteomics evidence of differential role of glutathione transferases. J Proteome Res 7:4557–4565. doi: 10.1021/pr800395x. [DOI] [PubMed] [Google Scholar]
- Peredney CL, and Williams PL. 2000. Utility of Caenorhabditis elegans for assessing heavy metal contamination in artificial soil. Arch Environ Contam Toxicol 39:113–118. doi: 10.1007/s002440010086. [DOI] [PubMed] [Google Scholar]
- Perez A, and Pierce Wise J Sr. 2018. One environmental health: An emerging perspective in toxicology. F1000Res 7. doi: 10.12688/f1000research.14233.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, and Culotti JG. 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol 117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Petalcorin MI, Joshua GW, Agapow PM, and Dolphin CT. 2005. The FMO genes of Caenorhabditis elegans and C. briggsae: Characterisation, gene expression and comparative genomic analysis. Gene 346:83–96. doi: 10.1016/j.gene.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Peterson ND, Cheesman HK, Liu P, Anderson SM, Foster KJ, Chhaya R, Perrat P, Thekkiniath J, Yang Q, Haynes CM, and Pukkila-Worley R. 2019. The nuclear hormone receptor NHR-86 controls anti-pathogen responses in C. elegans. PLoS Genet 15:e1007935. doi: 10.1371/journal.pgen.1007935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell-Coffman JA, Bradfield CA, and Wood WB. 1998. Caenorhabditis elegans orthologs of the aryl hydrocarbon receptor and its heterodimerization partner the aryl hydrocarbon receptor nuclear translocator. Proc Natl Acad Sci U S A 95:2844–2849. doi: 10.1073/pnas.95.6.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, and Ausubel FM. 2008. Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. MethMol Biol 415:403–427. doi: 10.1007/978-1-59745-570-1_24. [DOI] [PubMed] [Google Scholar]
- Prahlad V, and Morimoto RI. 2009. Integrating the stress response: Lessons for neurodegenerative diseases from C. elegans. Trends Cell Biol 19:52–61. doi: 10.1016/j.tcb.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor R, Norvaisas P, Marinos G, Best L, Thingholm LB, Quintaneiro LM, De Haes W, Esser D, Waschina S, Lujan C, Smith RL, Scott TA, Martinez-Martinez D, Woodward O, Bryson K, Laudes M, Lieb W, Houtkooper RH, Franke A, Temmerman L, Bjedov I, Cocheme HM, Kaleta C, and Cabreiro F. 2019. Host-microbe-drug-nutrient screen identifies bacterial effectors of metformin therapy. Cell 178: 1299–1312 e29. doi: 10.1016/j.cell.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R, Feinbaum RL, McEwan DL, Conery AL, and Ausubel FM. 2014. The evolutionarily conserved mediator subunit MDT-15/MED15 links protective innate immune responses and xenobiotic detoxification. PLoS Pathog 10 :e1004143. doi: 10.1371/journal.ppat.1004143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiros L, Pereira JL, Goncalves FJM, Pacheco M, Aschner M, and Pereira P. 2019. Caenorhabditis elegans as a tool for environmental risk assessment: Emerging and promising applications for a “nobelized worm”. Crit Rev Toxicol 49:411–429. doi: 10.1080/10408444.2019.1626801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar P, Mathew BS, Das S, Isaiah R, John S, Prabha R, and Fleming DH. 2016. Cisplatin concentrations in long and short duration infusion: Implications for the optimal time of radiation delivery. J Clin Diagnostic Res 10: XC01–XC04. doi: 10.7860/JCDR/2016/18181.8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye CA, Tagawa A, Le Bot N, Ahringer J, and Aroian RV. 2003. Involvement of fatty acid pathways and cortical interaction of the pronuclear complex in Caenorhabditis elegans embryonic polarity. BMC Dev Biol 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Pons C, Diallo A, Mori A, Shrestha S, Kadreppa S, Nelson J, Diprima S, Dricot A, Lajoie BR, Ribeiro PS, Weirauch MT, Hill DE, Hughes TR, Myers CL, and Walhout AJ. 2013. Extensive rewiring and complex evolutionary dynamics in a C. elegans multiparameter transcription factor network. Mol Cell 5:116–127. doi: 10.1016/j.molcel.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees DC, Johnson E, and Lewinson O. 2009. ABC transporters: The power to change. Nat Rev Mol Cell Biol 10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich JR, Dunn RA, and Noling JW. 2004. Nematicides: Past and present uses. In Nematology : Advances and Perspectives, edited by Chen ZX, Chen SY and Dickson DW. Cambridge, MA: CABI Publishing. pp. 1179–1200. [Google Scholar]
- Rieckher M, Bujarrabal A, Doll MA, Soltanmohammadi N, and Schumacher B. 2018. A simple answer to complex questions: Caenorhabditis elegans as an experimental model for examining the DNA damage response and disease genes. J Cell Physiol 233: 2781–2790. doi: 10.1002/jcp.25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, and Schmeisser S. 2011. Extending life span by increasing oxidative stress. Free Radic Biol Med 51:327–336. doi: 10.1016/j.freeradbiomed.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Snoek LB, De Bono M, and Kammenga JE. 2013. Worms under stress: C. elegans stress response and its relevance to complex human disease and aging. Trends Genet 29:367–374. doi: 10.1016/j.tig.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Roh JY, Lee HJ, and Kwon JH. 2016. Internal concentration and time are important modifiers of toxicity: The case of chlorpyrifos on Caenorhabditis elegans. Environ Sci Technol 50:9689–9696. doi: 10.1021/acs.est.6b02751. [DOI] [PubMed] [Google Scholar]
- Rutter JW, Ozdemir T, Galimov ER, Quintaneiro LM, Rosa L, Thomas GM, Cabreiro F, and Barnes CP. 2019. Detecting changes in the Caenorhabditis elegans intestinal environment using an engineered bacterial biosensor. A CS Synth Biol 8: 2620–2628. doi: 10.1021/acssynbio.9b00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammi SR, Agim ZS, and Cannon JR. 2018. From the cover: Harmane-induced selective dopaminergic neurotoxicity in Caenorhabditis elegans. Toxicol Sci 161:335–348. doi: 10.1093/toxsci/kfx223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer P, Muller M, Kruger A, Steinberg CE, and Menzel R. 2009. Cytochrome P450-dependent metabolism of PCB52 in the nematode Caenorhabditis elegans. Arch Biochem Biophys 488:60–68. [DOI] [PubMed] [Google Scholar]
- Schenk J, Hoss S, Brinke M, Kleinbolting N, Bruchner-Huttemann H, and Traunspurger W. 2020. Nematodes as bioindicators of polluted sediments using metabarcoding and microscopic taxonomy. Environ Int 143:105922. doi: 10.1016/j.envint.2020.105922. [DOI] [PubMed] [Google Scholar]
- Schmeisser K, Mansfeld J, Kuhlow D, Weimer S, Priebe S, Heiland I, Birringer M, Groth M, Segref A, Kanfi Y, Price NL, Schmeisser S, Schuster S, Pfeiffer AF, Guthke R, Platzer M, Hoppe T, Cohen HY, Zarse K, Sinclair DA, and Ristow M. 2013. Role of sirtuins in lifespan regulation is linked to methylation of nicotinamide Nat Chem Biol 9: 693–700. doi: 10.1038/nchembio.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid T, Snoek LB, Fröhli E, Leontien van der Bent M, Kammenga J, and Hajnal A. 2015. Systemic regulation of RAS/MAPK signaling by the serotonin metabolite 5-HIAA. PLOS Genet 11:e1005236. doi: 10.1371/journal.pgen.1005236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MS, Benci JL, Selote DS, Sharma AK, Chen AG, Dang H, Fares H, and Vatamaniuk OK. 2010. Detoxification of multiple heavy metals by a half-molecule ABC transporter, HMT-1, and coelomocytes of Caenorhabditis elegans. PLoS One 5: e9564. doi: 10.1371/journal.pone.0009564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott TA, Quintaneiro LM, Norvaisas P, Lui PP, Wilson MP, Leung KY, Herrera-Dominguez L, Sudiwala S, Pessia A, Clayton PT, Bryson K, Velagapudi V, Mills PB, Typas A, Greene NDE, and Cabreiro F. 2017. Host-microbe Co-metabolism dictates cancer drug efficacy in C. elegans. Cell 169 :442–456.e18. doi: 10.1016/j.cell.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti E, Boué S, Lo Sasso G, Zanetti F, Belcastro V, Poussin C, Sierro N, Battey J, Gimalac A, Ivanov NV, and Hoeng J. 2017. Exploring the microbiome in health and disease:Implications for toxicology. Toxicol Res Appl 1:2397847317741884. doi: 10.1177/2397847317741884. [DOI] [Google Scholar]
- Sender R, Fuchs S, and Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta Piali. 2007. Generation and modulation of chemosensory behaviors in C. elegans. Pflügers Archiv - Eur J Physiol 454:721. doi: 10.1007/s00424-006-0196-9. [DOI] [PubMed] [Google Scholar]
- Serrano-Saiz E, Vogt MC, Levy S, Wang Y, Kaczmarczyk KK, Mei X, Bai G, Singson A, Grant BD, and Hobert O. 2020. SLC17A6/7/8 vesicular glutamate transporter homologs in nematodes. Genetics 214:1 63–178. doi: 10.1534/genetics.119.302855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E, Poole RJ, Felton T, Zhang F, De La Cruz ED, and Hobert O. 2013. Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell 155:659–673. doi: 10.1016/j.cell.2013.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye DD, and Greenwald I. 2011. OrthoList: A compendium of C. elegans genes with human orthologs. PLoS One 6: e20085. doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Nettleton D, Jiang M, Kim SK, and Powell-Coffman JA. 2005. Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J Biol Chem 280:20580–20588. doi: 10.1074/jbc.M501894200. [DOI] [PubMed] [Google Scholar]
- Shen P, Zhang R, McClements DJ, and Park Y. 2019. Nanoemulsion-based delivery systems for testing nutraceutical efficacy using Caenorhabditis elegans: Demonstration of curcumin bioaccumulation and body-fat reduction. Food Res Int 120:157–166. doi: 10.1016/j.foodres.2019.02.036. [DOI] [PubMed] [Google Scholar]
- Sheps JA, Ralph S, Zhao Z, Baillie DL, and Ling V. 2004. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol 5:R15. doi: 10.1186/gb-2004-5-3-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiizaki K, Kawanishi M, and Yagi T. 2017. Modulation of benzo[a]pyrene-DNA adduct formation by CYP1 inducer and inhibitor. Genes Environ 39:14. doi: 10.1186/s41021-017-0076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomer N, Kadhim AZ, Grants JM, Cheng X, Alhusari D, Bhanshali F, Poon AF, Lee MYY, Muhuri A, Park JI, Shih J, Lee D, Lee SV, Lynn FC, and Taubert S. 2019. Mediator subunit MDT-15/MED15 and nuclear receptor HIZR-1/HNF4 cooperate to regulate toxic metal stress responses in Caenorhabditis elegans. PLoS Genet 15: e1008508. doi: 10.1371/journal.pgen.1008508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore DE, and Ruvkun G. 2013. A cytoprotective perspective on longevity regulation. Trends Cell Biol 23:409–420. doi: 10.1016/j.tcb.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, and Plasterk RH. 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol 1:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair J, and Hamza I. 2015. Lessons from bloodless worms: Heme homeostasis in C. elegans. Biometals 28:481–489. doi: 10.1007/s10534-015-9841-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RN, and Sulston JE. 1978. Some observations on moulting in Caenorhabditis elegans. Nematologica 24:63. doi: 10.1163/187529278X00074. [DOI] [Google Scholar]
- Slice LW, Freedman JH, and Rubin CS. 1990. Purification, characterization, and cDNA cloning of a novel metallothionein-like, cadmium-binding protein from Caenorhabditis elegans. J Biol Chem 265: 256–263. [PubMed] [Google Scholar]
- Snyder PW, Mann P, Bolon B, Elmore S, Haschek-Hock W, McDorman K, Morton DM, Ochoa R, and Ryan AM. 2010. The Society of Toxicologic Pathology and the “One Health” initiative Toxicol Pathol 38:521. [Google Scholar]
- Song BJ, Gelboin HV, Park SS, Yang CS, and Gonzalez FJ. 1986. Complementary DNA and protein sequences of ethanol-inducible rat and human cytochrome P-450s. Transcriptional and post-transcriptional regulation of the rat enzyme. J Biol Chem 261: 16689–16697. [PubMed] [Google Scholar]
- Spann N, Goedkoop W, and Traunspurger W. 2015. Phenanthrene bioaccumulation in the nematode Caenorhabditis elegans. Environ Sci Technol 49:1842–1850. doi: 10.1021/es504553t. [DOI] [PubMed] [Google Scholar]
- Sprando RL, Olejnik N, Cinar HN, and Ferguson M. 2009. A method to rank order water soluble compounds according to their toxicity using Caenorhabditis elegans, a complex object parametric analyzer and sorter, and axenic liquid media. Food Chem Toxicol 47: 722–728. doi: 10.1016/j.fct.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Starnes DL, Unrine JM, Starnes CP, Collin BE, Oostveen EK, Ma R, Lowry GV, Bertsch PM, and Tsyusko OV. 2015. Impact of sulfidation on the bioavailability and toxicity of silver nanoparticles to Caenorhabditis elegans. Environ Pollut 196:239–246. doi: 10.1016/j.envpol.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Stein KK, Golden A 2015. The C. elegans eggshell. (December 30, 2015), WormBook, ed In WormBook. The C. elegans Research Community. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg CE, Sturzenbaum SR, and Menzel R. 2008. Genes and environment - striking the fine balance between sophisticated biomonitoring and true functional environmental genomics. Sci Total Environ 400:142–161. doi: 10.1016/j.scitotenv.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Sterken MG, Snoek LB, Kammenga JE, and Andersen EC. 2015. The laboratory domestication of Caenorhabditis elegans. Trends Genet 31:224–231. doi: 10.1016/j.tig.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone CE, Hall DH, and Sundaram MV. 2009. Lipocalin signaling controls unicellular tube development in the Caenorhabditis elegans excretory system. Dev Biol 329:201–211. doi: 10.1016/j.ydbio.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp GS, von Reuss SH, Izrayelit Y, Ajredini R, Schroeder FC, and Edison AS. 2013. Chemical detoxification of small molecules by Caenorhabditis elegans ACS Chem Biol 8:309–313. doi: 10.1021/cb300520u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Albertson DG, and Thomson JN. 1980. The Caenorhabditis elegans male: Postembryonic development of nongonadal structures. Dev Biol 78:542–576. doi: 10.1016/0012-1606(80)90352-8. [DOI] [PubMed] [Google Scholar]
- Sumida A, Kinoshita K, Fukuda T, Matsuda H, Yamamoto I, Inaba T, and Azuma J. 1999. Relationship between mRNA levels quantified by reverse transcription-competitive PCR and metabolic activity of CYP3A4 and CYP2E1 in human liver. Biochem Biophys Res Commun 262 :499–503. doi: 10.1006/bbrc.1999.1233. [DOI] [PubMed] [Google Scholar]
- Sundaram MV, and Buechner M. 2016. The Caenorhabditis elegans excretory system: A model for tubulogenesis, cell fate specification, and plasticity. Genetics 203:35. doi: 10.1534/genetics.116.189357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SC, Keusekotten K, Baumeister R, and Sturzenbaum SR. 2004. C. elegans metallothioneins: New insights into the phenotypic effects of cadmium toxicosis. J Mol Biol 341:951–959. doi: 10.1016/j.jmb.2004.06.050. [DOI] [PubMed] [Google Scholar]
- Tang B, Williams PL, Xue KS, Wang JS, and Tang L. 2020. Detoxification mechanisms of nickel sulfate in nematode Caenorhabditis elegans. Chemosphere 260:127627. doi: 10.1016/j.chemosphere.2020.127627. [DOI] [PubMed] [Google Scholar]
- Taubert S, Hansen M, Van Gilst MR, Cooper SB, and Yamamoto KR. 2008. The mediator subunit MDT-15 confers metabolic adaptation to ingested material. PLoS Genet 4: e1000021. doi: 10.1371/journal.pgen.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S, Van Gilst MR, Hansen M, and Yamamoto KR. 2006. A mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev 20:1137–1149. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S, Ward JD, and Yamamoto KR. 2011. Nuclear hormone receptors in nematodes: Evolution and function. Mol Cell Endocrinol 334:49–55. doi: 10.1016/j.mce.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejeda-Benitez L, and Olivero-Verbel J. 2016. Caenorhabditis elegans, a biological model for research in toxicology. Rev Environ Contam Toxicol 237:1–35. doi: 10.1007/978-3-319-23573-8_1. [DOI] [PubMed] [Google Scholar]
- Tonelli C, Chio IIC, and Tuveson DA. 2018. Transcriptional regulation by Nrf2. Antioxid Redox Signal 29:1727–1745. doi: 10.1089/ars.2017.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traunspurger W, Haitzer M, Hoss S, Beier S, Ahlf W, and Steinberg C. 1997. Ecotoxicological assessment of aquatic sediments with Caenorhabditis elegans (nematoda) - A method for testing liquid medium and whole-sediment samples. Environ Toxicol Chem 16:245–250. [Google Scholar]
- Treonis AM, and Wall DH. 2005. Soil nematodes and desiccation survival in the extreme arid environment of the antarctic dry valleys. Integr Comp Biol 45:741–750. doi: 10.1093/icb/45.5.741. [DOI] [PubMed] [Google Scholar]
- Tsiaoussis J, Antoniou MN, Koliarakis I, Mesnage R, Vardavas CI, Izotov BN, Psaroulaki A, and Tsatsakis A. 2019. Effects of single and combined toxic exposures on the gut microbiome: Current knowledge and future directions. Toxicol Lett 312:72–97. doi: 10.1016/j.toxlet.2019.04.014. [DOI] [PubMed] [Google Scholar]
- U.S. Congress. 2016. Frank R. Lautenberg Chemical Safety for the 21st Century Act. In 130 Stat. 448. Washington, DC: U.S. Government Publishing Office. [Google Scholar]
- U.S. Environmental Protection Agency (US EPA). 2019. Directive to Prioritize Efforts to Reduce Animal Testing. Washington, DC: U.S. EPA [Google Scholar]
- Vatamaniuk OK, Bucher EA, Ward JT, and Rea PA. 2001. A new pathway for heavy metal detoxification in animals. Phytochelatin synthase is required for cadmium tolerance in Caenorhabditis elegans. J Biol Chem 276 : 20817–20820. [DOI] [PubMed] [Google Scholar]
- Verma S, Jagtap U, Goyala A, and Mukhopadhyay A. 2018. A novel gene-diet pair modulates C. elegans aging. PLoS Genet 14: e1007608. doi: 10.1371/journal.pgen.1007608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viñuela A, B Snoek L, Riksen JAG, and Kammenga JE. 2010. Genome-wide gene expression analysis in response to organophosphorus pesticide chlorpyrifos and diazinon in C. elegans. PloS one 5:e12145–e12145. doi: 10.1371/journal.pone.0012145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BD, and Redinbo MR. 2013. Xenobiotic-sensing nuclear receptors involved in drug metabolism: a structural perspective. Drug Metab Rev 45: 79–100. doi: 10.3109/03602532.2012.740049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D 2019a. Molecular Toxicology in Caenorhabditis elegans: Springer. [Google Scholar]
- Wang D 2019b. Target organ toxicology in Caenorhabditis elegans: Springer. [Google Scholar]
- Wang D 2020. Exposure Toxicology in Caenorhabditis elegans: Springer. [Google Scholar]
- Wang J, Luo J, Aryal DK, Wetsel WC, Nass R, and Benovic JL. 2017. G protein-coupled receptor kinase-2 (GRK-2) regulates serotonin metabolism through the monoamine oxidase AMX-2 in Caenorhabditis elegans. J Biol Chem 292:5943–5956. doi: 10.1074/jbc.M116.760850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani KA, Goswamy D, Taubert S, Ratnappan R, Ghazi A, and Irazoqui JE. 2020. NHR-49/PPAR-α and HLH-30/TFEB promote C. elegans host defense via a flavin-containing monooxygenase. BioRxiv: 2020.09.03.282087. doi: 10.1101/2020.09.03.282087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, and Brenner S. 1975. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans.?2UU. J Comp Neurol 160: 313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- Ware RW, Clark D, Crossland K, and Russell RL. 1975. The nerve ring of the nematode Caenorhabditis elegans: Sensory input and motor output. J Comp Neurol 162 :71–110. doi: 10.1002/cne.901620106. [DOI] [Google Scholar]
- Watson E, MacNeil LT, Ritter AD, Yilmaz LS, Rosebrock AP, Caudy AA, and Walhout AJM. 2014. Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell 156 :1336–1337. doi: 10.1016/j.cell.2014.02.036. [DOI] [PubMed] [Google Scholar]
- Weikum ER, Liu X, and Ortlund EA. 2018. The nuclear receptor superfamily: A structural perspective. Protein Sci 27 :1876–1892. doi: 10.1002/pro.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhouse C, Truong L, Meyer JN, and Allard P. 2018. Caenorhabditis elegans as an emerging model system in environmental epigenetics. Environ Mol Mutagen 59:560–575. doi: 10.1002/em.22203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PL, and Dusenbery DB. 1987. Screening test for neurotoxins using Caenorhabditis elegans. Prog Clin Biol Res 253:163–170. [PubMed] [Google Scholar]
- Williams PL, and Dusenbery DB. 1990a. A promising indicator of neurobehavioral toxicity using the nematode Caenorhabditis elegans and computer tracking. Toxicol Ind Health 6: 425–440. [DOI] [PubMed] [Google Scholar]
- Williams PL, and Dusenbery DB. 1988. Using the nematode Caenorhabditis elegans to predict mammalian acute lethality to metallic salts. Toxicology and Industrial Health 4: 469–478. [DOI] [PubMed] [Google Scholar]
- Williams PL, and Dusenbery DB. 1990b. Aquatic toxicity testing using the nematode, Caenorhabditis elegans. Environ Toxicol Chem 9:1285–1290. [PubMed] [Google Scholar]
- Williamson VM, and Kumar A. 2006. Nematode resistance in plants: The battle underground. Trends Genet 22: 396–403. doi: 10.1016/j.tig.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Williamson Valerie M., Long Manyuan, and Theodoris George. 1991. Isolation of Caenorhabditis elegans mutants lacking alcohol dehydrogenase activity. Biochem Genet 29: 313–323. doi: 10.1007/BF00554139. [DOI] [PubMed] [Google Scholar]
- Wittkowski P, Marx-Stoelting P, Violet N, Fetz V, Schwarz F, Oelgeschläger M, Schönfelder G, and Vogl S. 2019. Caenorhabditis elegans as a promising alternative model for environmental chemical mixture effect assessment-A comparative study. Environ Sci Technol 53: 12725–12733. doi: 10.1021/acs.est.9b03266. [DOI] [PubMed] [Google Scholar]
- Wolkow CA and Hall DH 2011. The dauer cuticle.” In WormAtlas. [Google Scholar]
- Woodruff GC, Knauss CM, Maugel TK, and Haag ES. 2014. Mating damages the cuticle of C. elegans hermaphrodites. PloS one 9: e104456–e104456. doi: 10.1371/journal.pone.0104456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CW, Deonarine A, Przybysz A, Strange K, and Choe KP. 2016. The Skp1 homologs SKR-1/2 are required for the Caenorhabditis elegans SKN-1 antioxidant/detoxification response independently of p38 MAPK. PLoS Genet 12 : e1006361. doi: 10.1371/journal.pgen.1006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Huang C, Taki FA, Zhang Y, Dobbins DL, Li L, Yan H, and Pan X. 2015. Benzo-α-pyrene induced oxidative stress in Caenorhabditis elegans and the potential involvements of microRNA. Chemosphere 139:496–503. doi: 10.1016/j.chemosphere.2015.08.031. [DOI] [PubMed] [Google Scholar]
- Wyatt LH, Diringer SE, Rogers LA, Hsu-Kim H, Pan WK, and Meyer JN. 2016. Antagonistic growth effects of mercury and selenium in Caenorhabditis elegans are chemical-species-dependent and do not depend on internal Hg/Se ratios. Environ Sci Technol. doi: 10.1021/acs.est.5b06044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Huang R, Shi Q, Boyd WA, Zhao J, Sun N, Rice JR, Dunlap PE, Hackstadt AJ, Bridge MF, Smith MV, Dai S, Zheng W, Chu PH, Gerhold D, Witt KL, DeVito M, Freedman JH, Austin CP, Houck KA, Thomas RS, Paules RS, Tice RR, and Simeonov A. 2018. Comprehensive analyses and prioritization of Tox21 10K chemicals affecting mitochondrial function by in-depth mechanistic studies. Environ Health Perspect 126:077010. doi: 10.1289/EHP2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Pears C, and Woollard A. 2017. An enhanced C. elegans based platform for toxicity assessment. Sci Rep 7: 9839. doi: 10.1038/s41598-017-10454-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Li CY, and Kong AN. 2005. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res 28:249–268. doi: 10.1007/bf02977789. [DOI] [PubMed] [Google Scholar]
- Yang XY, Jiang CJ, Hsu-Kim H, Badireddy AR, Dykstra M, Wiesner M, Hinton DE, and Meyer JN. 2014. Silver nanoparticle behavior, uptake, and toxicity in Caenorhabditis elegans: Effects of natural organic matter. Environ Sci Technol 48: 3486–3495. doi: Doi 10.1021/Es404444n. [DOI] [PubMed] [Google Scholar]
- Yang YF, Lin YJ, and Liao CM. 2017. Toxicity-based toxicokinetic/toxicodynamic assessment of bioaccumulation and nanotoxicity of zerovalent iron nanoparticles in Caenorhabditis elegans. Int J Nanomed 12: 4607–4621. doi: 10.2147/ijn.S138790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdraljevic S, and Andersen EC. 2017. Natural diversity facilitates the discovery of conserved chemotherapeutic response mechanisms. Curr Opin Genet Dev 47:41–47. doi: 10.1016/j.gde.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdraljevic S, Fox BW, Strand C, Panda O, Tenjo FJ, Brady SC, Crombie TA, Doench JG, Schroeder FC, and Andersen EC. 2019. Natural variation in C. elegans arsenic toxicity is explained by differences in branched chain amino acid metabolism. Elife 8. doi: 10.7554/eLife.40260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Berg M, Dierking K, Felix MA, Shapira M, Samuel BS, and Schulenburg H. 2017. Caenorhabditis elegans as a model for microbiome research. Front Microbiol 8:485. doi: 10.3389/fmicb.2017.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Holdorf AD, and Walhout AJ. 2017. C. elegans and its bacterial diet as a model for systems-level understanding of host-microbiota interactions. Curr Opin Biotechnol 46:74–80. doi: 10.1016/j.copbio.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li X, Jevince AR, Guan L, Wang J, Hall DH, Huang X, and Ding M. 2013. Neuronal target identification requires AHA-1-mediated fine-tuning of Wnt signaling in C. elegans. PLoS Genet 9: e1003618. doi: 10.1371/journal.pgen.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhong HQ, Chu ZW, Zuo X, Wang L, Ren XL, Ma H, Du RY, Ju JJ, Ye XL, Huang CP, Zhu JH, and Wu HM. 2020. Arsenic induces transgenerational behavior disorders in Caenorhabditis elegans and its underlying mechanisms. Chemosphere 252:126510. doi: 10.1016/j.chemosphere.2020.126510. [DOI] [PubMed] [Google Scholar]
- Zhao YL, Wu QL, Li YP, and Wang DY. 2013. Translocation, transfer, and in vivo safety evaluation of engineered nanomaterials in the non-mammalian alternative toxicity assay model of nematode Caenorhabditis elegans. RSC Adv 3 :5741–5757. doi: 10.1039/c2ra22798c. [DOI] [Google Scholar]
- Zhao Z, Sheps JA, Ling V, Fang LL, and Baillie DL. 2004. Expression Analysis of ABC transporters reveals differential functions of tandemly duplicated genes in Caenorhabditis elegans. J Mol Biol 344: 409–417. doi: 10.1016/j.jmb.2004.09.052. [DOI] [PubMed] [Google Scholar]
- Zheng S-Q, Ding A-J, Li G-P, Wu G-S, and Luo H-R. 2013. Drug absorption efficiency in Caenorhbditis elegans delivered by different methods. PloS one 8:e56877–e56877. doi: 10.1371/journal.pone.0056877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou GW, Yang XR, Zheng F, Zhang ZX, Zheng BX, Zhu YG, and Xue XM. 2020. Arsenic transformation mediated by gut microbiota affects the fecundity of Caenorhabditis elegans. Environ Pollut 260: 113991. doi: 10.1016/j.envpol.2020.113991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.