Significance
A high frequency of missense mutations was recently discovered at histone H3.3 glycine 34 in 92% of giant cell tumors of the bone and 17% of high-grade astrocytomas. The molecular mechanism by which G34 mutations drive these tumors remains unclear. Here, we demonstrate that the G34-mutated “oncohistones” misregulate the negative cross-talk between two histone methyltransferase enzymes, Polycomb Repressive Complex 2 (PRC2) and SETD2. G34 mutations uniquely promote PRC2 activity by blocking SETD2-mediated H3K36 methylation at active enhancers and drive a gene expression program that enhances tumor growth. We propose that G34 oncohistones exploit the regulatory mechanisms that fine tune PRC2 activity in human malignancies.
Keywords: oncohistones, H3.3 G34 mutations, PRC2, NSD1/2, SETD2
Abstract
A high percentage of pediatric gliomas and bone tumors reportedly harbor missense mutations at glycine 34 in genes encoding histone variant H3.3. We find that these H3.3 G34 mutations directly alter the enhancer chromatin landscape of mesenchymal stem cells by impeding methylation at lysine 36 on histone H3 (H3K36) by SETD2, but not by the NSD1/2 enzymes. The reduction of H3K36 methylation by G34 mutations promotes an aberrant gain of PRC2-mediated H3K27me2/3 and loss of H3K27ac at active enhancers containing SETD2 activity. This altered histone modification profile promotes a unique gene expression profile that supports enhanced tumor development in vivo. Our findings are mirrored in G34W-containing giant cell tumors of bone where patient-derived stromal cells exhibit gene expression profiles associated with early osteoblastic differentiation. Overall, we demonstrate that H3.3 G34 oncohistones selectively promote PRC2 activity by interfering with SETD2-mediated H3K36 methylation. We propose that PRC2-mediated silencing of enhancers involved in cell differentiation represents a potential mechanism by which H3.3 G34 mutations drive these tumors.
Highly recurrent missense mutations in genes encoding histone H3 have been found in a variety of human tumors (1–4). Previous studies have led to biochemical and functional insights of the H3 K27M and H3 K36M “oncohistones” that are found in 84% of diffuse intrinsic pontine gliomas and 95% of chondroblastomas, respectively. The G34 class of oncohistones occur exclusively in the histone variant H3.3 and are found in 92% of giant cell tumors of the bone (GCTB; G34W/L), 17% of high-grade astrocytomas (HGA; G34R/V) and 5% of osteosarcomas (G34R/W) (1, 3, 5).
Histone H3 is encoded by 13 copies of histone H3.1/H3.2 and 2 copies of the H3.3 variant (H3F3A and H3F3B). Nucleosomes containing histone H3.3 are assembled at promoters, gene bodies, and enhancers of transcriptionally active genes by the HIRA histone chaperone complex and at telomeres and pericentric repeats by ATRX-DAXX (6). Despite constituting a small fraction of total H3 in the nucleus, recent reports found that incorporation of H3.3 at enhancers is required to maintain high levels of H3K27ac (7, 8). Interestingly, unlike the “K-to-M” mutations found in gliomas and chondroblastoma that affect both canonical and variant histone H3, the G34 mutations are only found in the histone variant H3.3 (1, 9, 10). Functional or mechanistic explanations for the remarkable specificity of G34 mutations for histone H3.3 remain unclear.
Polycomb group (PcG) proteins are essential for proper development and are frequently misregulated in human cancers (11). Previous studies have highlighted a central role for misregulation of polycomb silencing in the context of cancers harboring lysine-to-methionine oncohistones (10, 12–14). Polycomb Repressive Complexes (PRC1 and PRC2) function in a collaborative chromatin-based cross-talk with H3K27me3 to initiate and maintain transcriptional silencing of developmentally regulated genes (15). While only H3K27me3 interacts with PRC1 to mediate local chromatin compaction, PRC2-catalyzed H3K27me1/2/3 are proposed to serve as molecular “placeholders” that antagonize H3K27ac and prevent spurious enhancer activation (16). Several mechanisms have evolved to fine tune the activity of EZH1/2, the catalytic subunits of PRC2, including posttranslational modifications (PTMs) on the histone H3 tail. For example, PRC2 binds its own product H3K27me3 through the EED subunit, resulting in an ∼10-fold increase in its catalytic activity on the neighboring nucleosomes that is thought to promote spreading of Polycomb domains in cis (17). This spreading of PRC2 and H3K27me3 is impeded by modifications associated with active chromatin. In particular, nucleosomes containing H3K36me2/3, modifications catalyzed by SETD2, NSD1/2, and ASH1L enzymes, are poor substrates for PRC2 (18–20). Some human cancers exploit these regulatory mechanisms to disrupt normal PRC2 activity to drive tumor-specific transcriptomes (10, 21–24).
In this study, we describe a mechanism by which H3.3 G34 mutations alter gene expression through perturbation of local chromatin modifications. We find that G34W and other tumor-associated G34 mutations impede SETD2-dependent H3K36 methylation in cis, while allowing NSD2 activity. The reduction of SETD2-dependent H3K36 methylation promotes aberrant PRC2-mediated H3K27me2/3 on nucleosomes containing the G34W-mutated H3.3 at enhancers. Expression of H3.3 G34W or G34R transgenes in cell lines drives a unique gene expression profile that is dependent on histone variant H3.3 and PRC2-mediated methylation in cis. Finally, we find that H3.3 G34 oncohistones likely promote tumorigenesis in vivo by stimulating proinflammatory gene expression associated with early osteoblastic differentiation.
Results
G34 Mutations Promote H3K27me2/3 by Impeding SETD2 Activity.
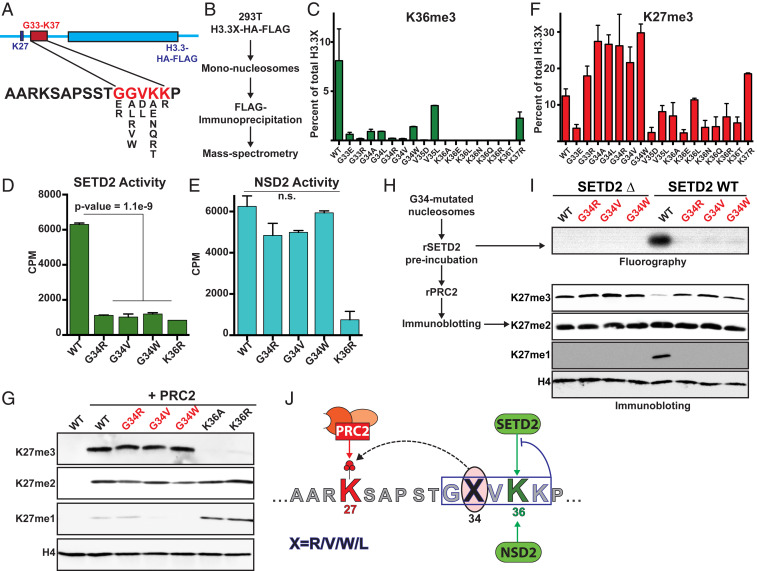
Glycine 34 on histone H3 lies in close proximity to the K27 and K36 residues that are methylated and acetylated in vivo. Previously, we found that histones containing G34R/V substitutions exhibited lower H3K36me3 in vivo (12). We hypothesized that G34 mutations function in tumorigenesis by promoting a unique PTM profile on nucleosomes containing the mutant histone and altering gene expression. Because G34 mutations decrease H3K36 methylation, we were curious to know if G34 substitutions affect other PTMs on nucleosomes containing the mutant histone. To this end, we generated a series of histone H3.3 transgenes that each contained an amino acid substitution that could be achieved by single nucleotide mutations between residues G33 and K37 (Fig. 1A). We expressed these transgenes in human HEK293T cells and purified mononucleosomes containing the mutant histones using FLAG affinity immunoprecipitation (Fig. 1B). Using bottom-up quantitative mass spectrometry, we observed that all mutant H3.3 histones exhibited lower H3K36me3 in cis relative to wild-type (WT) H3.3 (Fig. 1C and SI Appendix, Fig. S1A).
Fig. 1.
G34 mutated oncohistones exhibit increased H3K27me2/3. (A) Schematic displaying the histone H3.3 mutant transgenes used to assess PTM changes in cis using the purification strategy depicted in B. (C) Mass spectrometry data displaying the abundance of H3K36me3 on the mutated H3.3 peptides. (D and E) SETD2 and NSD2 in vitro methyltransferase assays using recombinant mononucleosome substrates containing point mutations on H3.3 as labeled. CPM: counts per minute. P value for reduced counts on the G34 mutated substrates was determined using t test. n.s.: not significant. (F) Mass spectrometry data displaying the abundance of H3K27me3 on the mutated peptides. (G) Immunoblots of in vitro PRC2 methyltransferase reactions using recombinant mononucleosome substrates containing point mutations in H3.3. (H and I) Schematic for sequential methyltransferase reactions using recombinant mononucleosomes containing WT or G34-mutated H3.3. (I) Immunoblots of reactions preincubated with WT or catalytically dead SETD2 and 50 µM S-adenosyl-methionine depicted in H. PRC2 and additional 50 µM SAM were added to the reaction and incubated for 3 h. (J) Model depicting the effect of G34 mutations. Missense mutations on H3.3 G33-K37 block SETD2 activity in cis. However, only mutations at G34 position permit PRC2 activity at K27 and hence, uniquely increase H3K27me2/3 in cis by decreasing H3K36me3. Error bars represent SD.
H3K36 methylation is catalyzed by SETD2, NSD1, NSD2, and ASH1L in mammals. While all these enzymes can catalyze H3K36me1/2, SETD2 is the sole enzyme responsible for H3K36me3 (21, 22). A recent co-crystal structure of the SETD2 SET domain with the H3 K36M peptide revealed that the side chains of amino acids G33 and G34 lie in a constrained substrate tunnel (21), and substitutions at these residues would likely obstruct SETD2 activity by sterically clashing with the substrate channel. To directly test this hypothesis, we determined SETD2 activity on recombinant nucleosomes containing G34 mutations using in vitro methyltransferase assays. Mutations at the G34 position reduced SETD2 activity to a similar extent as a K36R negative control in cis (Fig. 1D and SI Appendix, Fig. S1B). Surprisingly, we found that nucleosomes containing G34 mutations did not negatively affect NSD2-dependent K36 methylation in vitro (Fig. 1E). The SETD2 residues that lie in close proximity to G34 are substituted with smaller side chain amino acids in NSD1/2 SET domains (SI Appendix, Fig. S1C). We speculate that these smaller residues reduce steric clashing with the bulky R/W/V found on the mutant H3.3 and allow NSD2 to effectively catalyze H3K36 methylation on G34 mutant histones.
Although all H3.3 substitutions between G33 and K37 reduced H3K36me3 in vivo, our mass spectrometry analyses revealed that peptides containing G34 substitutions exhibited the highest levels of H3K27me3 (Fig. 1F and SI Appendix, Fig. S1 D and E). These data suggest that H3.3 G34 nucleosomes are excellent substrates for PRC2. In our in vitro methyltransferase assays using recombinant nucleosomes, we found that wild-type H3.3 and G34 mutant nucleosomes are equally good substrates for PRC2-mediated H3K27 methylation (Fig. 1G and SI Appendix, Fig. S1 B and F). We and others have found that nucleosomes containing H3K36me2/3 serve as poor substrates for PRC2 (18–20). Because G34 histones exhibit low H3K36me3 and high K27me2/3 levels, we hypothesized that G34 mutations uniquely promote H3K27me2/3 in cis through reduction of SETD2-mediated H3K36 methylation and toleration by PRC2. To directly test this proposition, we assessed PRC2 activity on nucleosomes preincubated with wild-type SETD2 or a catalytically dead mutant SETD2 (Fig. 1 H and I and SI Appendix, Fig. S1 G–I). We found high levels of PRC2-mediated H3K27me3 on H3.3 G34 mutant nucleosomes as compared to wild-type nucleosomes after pretreatment with SETD2. These data directly link the gain of H3K27me2/3 on G34 mutant nucleosomes in vivo with their ability to impede SETD2, but not PRC2, activity in vitro (Fig. 1J).
Histone H3.3 G34 mutations are common in GCTB and HGAs, whereas H3.3 K36 mutations are not found in these tumors. Interestingly, we found that nucleosomes containing K36A/R substitutions exhibited reduced conversion of H3K27me1 to H3K27me3 (Fig. 1 F and G), suggesting that PRC2 requires an unmodified lysine at the 36th position of the H3 substrate for its maximal activity (Fig. 1 F, G, and I). These data also provide a biochemical explanation for the coenrichment of H3K36me3 and H3K27me1 in cells (16). Taken together, these findings suggest that H3.3 G34 mutations uniquely promote increased H3K27me2/3 by preventing SETD2 activity, while allowing maximal PRC2 activity in cis (Fig. 1J).
Local Loss of H3K36me3 and Gain of H3K27me3 in Cells Expressing H3.3 G34W.
We generated immortalized mesenchymal stem cells (mMSCs) that expressed FLAG-HA-tagged H3.3 WT or G34W mutant transgenes to define how H3.3 G34 mutations affect histone PTMs in vivo. The H3.3 G34W histones in our cell lines constituted 8 to 9% of total histone H3, which is in the range of mutant histone H3.3 proteins found in human tumors (SI Appendix, Fig. S2A) (12). Expression of H3.3 G34W did not affect overall levels of H3K27me3 in cells (SI Appendix, Fig. S2B). However, we found that G34W mutant H3 histones exhibited both decreased H3K36me3 and increased H3K27me3 (SI Appendix, Fig. S2C). These findings are consistent with our in vitro experiments where we found that G34 mutations give rise to changes in H3K27me3 in cis by impeding SETD2 activity.
Because the G34 mutations work in cis, we employed sequential chromatin immunoprecipitation (reChIP) sequencing assays in order to assess changes in K27 and K36 methylation on nucleosomes containing the mutant histone. First, we performed FLAG ChIP to purify nucleosomes containing ectopically expressed H3.3 histones and subjected the eluate to reChIP using H3K27me3 or H3K36me3 antibodies (Fig. 2A and SI Appendix, Fig. S2D). As predicted, the G34W substitution did not affect the deposition pattern of histone H3.3 (SI Appendix, Fig. S2 E and F), which is instead dependent on key amino acids in the histone core (22). Next, we validated the efficiency of our reChIP protocol by comparing the distribution of H3K27me3 and H3K36me3 reChIP and ChIP intensities relative to H3.3. Importantly, we found that reChIP sequencing reads were substantially enriched within the genomic loci containing H3.3 and depleted from loci lacking H3.3 (SI Appendix, Fig. S2 G–I), confirming that the immunoprecipitated K36me3 and K27me3 were from nucleosomes containing the epitope-tagged H3.3.
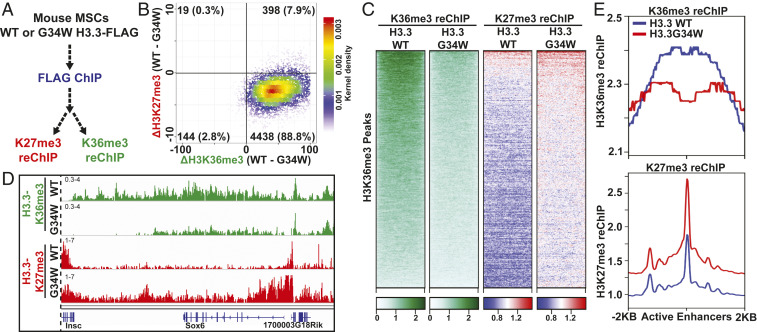
Fig. 2.
Local loss of H3K36me3 and gain of H3K27me3 in cells expressing H3.3G34W. (A) Strategy used to identify local changes in H3K27me3 and H3K36me3 in cells expressing H3.3 WT or G34W in vivo. (B) Scatterplot displaying the correlation between loss of H3.3X-K36me3 and gain of H3.3X-K27me3 at H3K36me3-enriched peaks. Each point represents a H3K36me3 peak, and the difference in the number of normalized ChIP-Seq reads was plotted. (C) Heatmap displaying the enrichment of H3.3-K36me3 and H3.3-K27me3 reChIP enrichment at H3K36me3 peaks in cells expressing H3.3 WT or G34W. (D) Genome browser view of the Sox6 gene displaying the loss of H3.3X-K36me3 and gain of H3.3X-K27me3. (E) Metagene analysis showing the average profile of H3.3X-K36me3 and H3.3X-K27me3 reChIP at active enhancers (promoter-distal H3K27ac peaks) in cells expressing H3.3 WT or G34W.
We mapped loci containing both H3.3 and H3K36me3 in order to identify genomic regions where the H3.3 G34W mutation would directly affect K36 methylation levels (SI Appendix, Fig. S3A). Consistent with previous studies, we found H3.3 enriched in genomic regions containing dynamic chromatin including active genes (transcript per kilobase million > 10) and enhancers (nonpromoter-associated H3K27ac peaks) (6, 23). While direct mapping of SETD2 occupancy has been elusive due to a lack of a ChIP-grade SETD2 antibody, we used quantitative ChIP sequencing (ChIP-Seq) from Nsd1/2 double knockout and Nsd1/2/Setd2 triple knockout cell lines to identify SETD2-mediated H3K36me2. By comparing the H3K36me2 ChIP-Seq signals from these knockout cell lines, we found that SETD2-mediated H3K36me2 and H3K36me3 were enriched at enhancers and gene bodies of active genes (SI Appendix, Fig. S3 A and D), in agreement with previous studies (24–26). Also consistent with the known overlap of H3.3 and H3K36me3, our H3.3-K36me3 reChIP sequencing reads primarily mapped to gene bodies and active enhancers (SI Appendix, Fig. S4A).
Notably, we observed a reduction of H3.3-K36me3 and a concomitant gain of H3.3-K27me3 in cells expressing H3.3 G34W at H3K36me3-enriched loci (Fig. 2 B–D and SI Appendix, Fig. S4B). This result is consistent with our in vitro finding that the G34W substitution blocks SETD2 but permits PRC2 activity. The gain of H3.3-K27me3 occurred primarily at active genes and enhancers (Fig. 2E and SI Appendix, Fig. S4 C and E). Using standard ChIP-Seq, we found a modest reduction of H3K36me3 and gain of H3K27me3 at active enhancers and H3K36me3 peaks (SI Appendix, Fig. S4D). Taken together, our in vitro and in vivo results demonstrate that deposition of H3.3 G34W leads to increased H3K27me3 by reducing H3K36 methylation.
Previously, we found that H3 K36M transgenes reduced global levels of H3K36me2/3 and increased H3K27me3 in intergenic regions (10). This genome-wide gain of H3K27me3 led to a dilution of PRC1 and derepression of polycomb-silenced genes. In contrast, we did not detect substantial changes in the EZH2 occupancy upon expression of H3.3 G34W (SI Appendix, Fig. S4F). Importantly, polycomb-silenced genes, which were derepressed in cells expressing H3 K36M, remained silent in cells expressing H3.3 G34W (SI Appendix, Fig. S4G). These results highlight important distinctions between the global effect of H3 K36M and local changes mediated by H3.3 G34W.
Reduction of H3K27ac at Enhancers in Cells Expressing H3.3 G34W.
Previous studies have found that H3K27 methylation (H3K27me1/2/3) can regulate enhancer activity by acting as a chemical group placeholder to antagonize acetylation at this residue (16, 27). We used ChIP-Seq in EED-depleted cells to test the role of PRC2 activity in regulating enhancer silencing. We found that loss of EED by transient expression of Cre-recombinase in EEDflox/flox mouse embryonic fibroblasts led to global loss of H3K27me2/3 and a concomitant gain of H3K27ac (Fig. 3A) (28). Importantly, upon genetic depletion of PRC2, we found a substantial gain of H3K27ac at promoter-distal sites (Fig. 3A), supporting the model that PRC2 activity influences H3K27ac levels at enhancers.
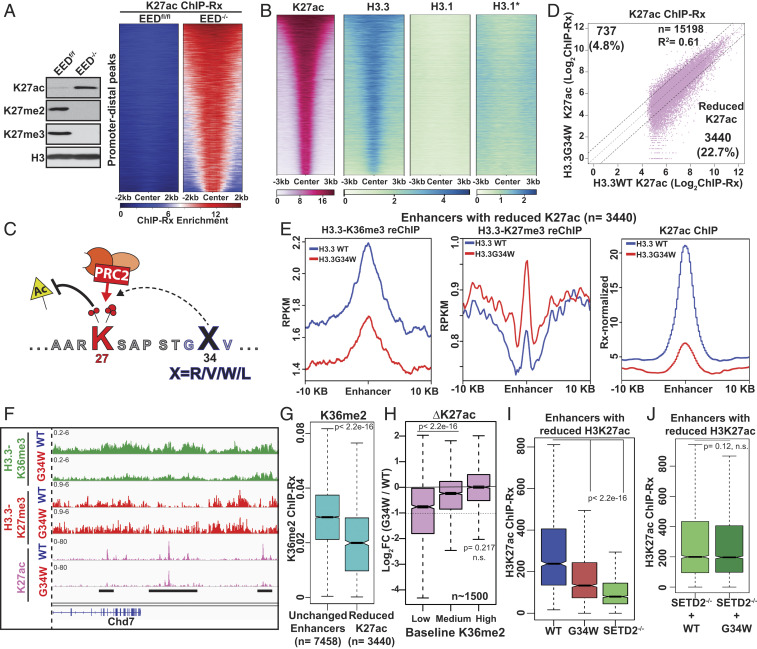
Fig. 3.
Aberrant reduction of H3K27ac at enhancers in cells expressing H3.3G34W. (A) Immunoblots of whole cell extracts from EEDf/f and EED−/− cells (Left). Heatmap displaying the H3K27ac ChIP enrichment at promoter-distal sites (n = 14,420) that gained H3K27ac in EED−/− cells (Right). (B) Heatmaps displaying the ChIP enrichment profiles of H3K27ac and HA-tagged H3.3 or H3.1 at all promoter-distal active enhancers in mMSCs (n = 15,198). The color scale of H3.1* is modified from 0 to 5. (C) Model depicting the effect of H3.3 G34W mutation at enhancers. Increased PRC2-mediated H3K27me1/2/3 on H3.3 G34W oncohistones antagonizes H3K27ac by providing a placeholder methyl group(s). (D) Scatterplot displaying the correlation between reference-normalized H3K27ac intensities at active enhancers in cells expressing H3.3 WT or G34W. (E) Metaplot displaying the normalized H3K27ac ChIP, H3.3-K36me3 reChIP, H3.3-K27me3 reChIP enrichments at a 10-kb region around enhancers with reduced H3K27ac as defined in B. (F) Genome browser view displaying enrichment of H3.3X-K36me3, H3.3X-K27me3 reChIPs, and H3K27ac ChIPs in cells expressing H3.3WT or G34W. (G) Boxplot displaying the reference-normalized H3K36me2 enrichment at unchanged enhancers (0.9 ≤ ∆K27ac ≤ 1.1, n = 7,458) and enhancers with decreased H3K27ac (∆K27ac ≤ 0.5, n = 3,440). (H) Boxplots displaying the fold change in reference-normalized H3K27ac intensities at enhancers containing low (1st to 10th quantile), medium (45th to 55th quantiles), and high levels of H3K36me2 (90th to 100th quantile). P values for loss of H3K27ac were determined using Wilcoxon’s rank sum test. Dashed line represent a twofold loss. (I) Boxplots displaying the H3K27ac ChIP enrichment at enhancers with reduced H3K27ac in cells expressing H3.3 WT or H3.3 G34W or SETD2−/− cells. P value was determined using Wilcoxon’s rank sum test. (J) Boxplots displaying the H3K27ac ChIP enrichment at enhancers with reduced H3K27ac in SETD2−/− cells overexpressing H3.3 WT or G34W. P values for loss of H3K27ac were determined using Wilcoxon’s rank sum test. n.s.: not significant.
Histone H3.3 is enriched at active enhancers where canonical histones H3.1/2 are depleted (Fig. 3B), and recent reports suggest that H3.3 is responsible for most H3K27 acetylation at enhancers (7, 8). Because G34 mutations in human cancers occur exclusively in H3.3 (1, 3), we hypothesized that a localized gain of H3K27 methylation on histone H3.3 G34W at enhancers would antagonize H3K27ac levels (Fig. 3C). We profiled active enhancers in cells expressing H3.3 WT or G34W using H3K27ac quantitative ChIP-Seq. Overall, 3,440 (∼23%) active enhancers exhibited decreased H3K27ac enrichment in cells expressing H3.3 G34W (Fig. 3D). Importantly, loss of H3K27ac at these enhancers correlated with a reduction in H3.3-K36me3 and a gain of H3.3-K27me3 as measured by reChIP-Seq (Fig. 3 E and F and SI Appendix, Fig. S5 A and B). Histone H3.3 is enriched at both active and poised enhancers (6, 29), and we observed only a small decrease in the occupancy of H3.3 G34W at these enhancers (SI Appendix, Fig. S5C). Moreover, intra- or intergenic enhancers were not enriched within the set with lowered H3K27ac, indicating that G34W-mediated changes at active enhancers are uncoupled from gene body transcription (SI Appendix, Fig. S5D).
Non-SETD2-Catalyzed H3K36me2 Prevents Enhancer Inactivation by H3.3 G34W.
Unlike SETD2, we found that NSD2-mediated catalysis of H3K36 methylation was unaffected by G34 mutations in vitro (Fig. 1E). Because we found that enhancers were not equally affected by the incorporation of H3.3 G34W histones, we hypothesized that NSD2-mediated H3K36me2 may protect a subset of enhancers from PRC2-mediated inactivation. In line with our hypothesis, we observed that enhancers with reduced H3K27ac contained lower baseline H3K36me2 levels (Fig. 3G). We measured changes in H3K27ac at active enhancers containing low, medium, and high levels of H3K36me2 in wild-type mMSCs. We found that enhancers with high levels of non-SETD2-catalyzed H3K36me2 were unaffected by H3.3 G34W (Fig. 3H and SI Appendix, Fig. S5E). In contrast, enhancers with low H3K36me2 levels exhibited a significant reduction in H3K27ac by H3.3 G34W (Fig. 3H and SI Appendix, Fig. S5 E and F).
Based on our in vitro findings and mass spectrometry data, we propose that the G34W substitution reduces SETD2 activity in cis on nucleosomes at enhancers. In support of this model, we found that SETD2−/− cells and H3.3 G34W-expressing cells exhibited a remarkably similar reduction of H3K27ac at enhancers (Fig. 3I). Furthermore, expression of H3.3 G34W in SETD2−/− cells did not affect H3K27ac at enhancers (Fig. 3J). These results support our model that reduced SETD2 activity on H3.3 G34W histones is a necessary intermediate step toward reduction of H3K27ac at enhancers.
Reduced H3K27ac by H3.3 G34W Correlates with Down-Regulation of Corresponding Genes.
We found that expression of H3.3 G34W led to increased H3K27me3 and loss of H3K27ac at a subset of enhancers and active genes. The recruitment of BRD4 and Mediator complex to enhancer elements is a major step toward activating associated genes, and previous studies have found that BRD4 and Mediator ChIP-Seq intensities correlate positively with enhancer activity (30, 31). We mapped BRD4 and Mediator complex subunit, Med1 occupancies in cells expressing H3.3 WT and G34W by ChIP-Seq. The majority of enhancers with reduced H3K27ac also exhibited decreased BRD4 and Med1 occupancy, while enhancers exhibiting no change in H3K27ac retained these proteins (Fig. 4 A and B). These data link the reduction of H3K27ac to enhancer inactivation by H3.3 G34W. Using RNA sequencing, we found that the majority of differentially expressed genes in H3.3 G34W cells became down-regulated (SI Appendix, Fig. S6A). Moreover, most genes (73%) proximal to enhancers with decreased H3K27ac were down-regulated in H3.3 G34W cells (SI Appendix, Fig. S6 B and C). Together, these findings suggest that H3.3 G34W decreases enhancer activity that leads to lower expression of associated genes. Some genes located near G34W-affected enhancers showed no change in expression (SI Appendix, Fig. S6C), and we speculate that enhancer redundancy might maintain expression of these genes (32).
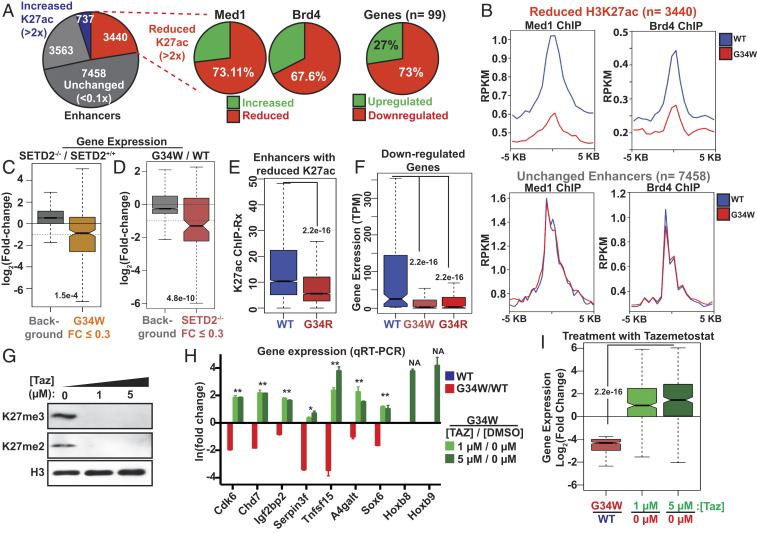
Fig. 4.
Reduced gene enhancer activity in cells expressing H3.3 G34W. (A) Pie chart displaying the proportion of enhancers with reduced H3K27ac (∆K27ac ≤ 2), unchanged (0.9 ≤ ∆K27ac ≥ 1.1) and increased H3K27ac (∆K27ac ≥ 2) in H3.3 G34W-expressing cells. (Right) Pie chart displaying the changes in Med1/Brd4 occupancy at enhancers with reduced H3K27ac and expression of genes associated with them. Gene-enhancer associations were identified using GREAT and genes associated with at least three enhancers with reduced H3K27ac (n = 99) were used for the analyses. (B) Average profiles of Med1 (Left) and Brd4 ChIP-Seq (Right) displaying their average profiles at enhancers with decreased H3K27ac (Top, n = 3,440) and unchanged enhancers (Bottom, n = 7,458). (C) Boxplot displaying the change in the expression of H3.3 G34W down-regulated genes (fold change ≤ 0.3, n = 279) in SETD2−/− cells. Dashed line displays a twofold change. (D) Boxplot displaying the change in the expression of genes down-regulated by SETD2 knockout (fold change ≤ 0.3, n = 132) in cells expressing H3.3G34W. Dashed line displays a twofold change. (E) Boxplot displaying the H3K27ac enrichment at enhancers with reduced H3K27ac (as defined in Fig. 3A) in cells expressing H3.3 WT or G34R. (F) Boxplot displaying the expression of H3.3 G34W down-regulated genes (fold change ≤ 0.3, n = 279). TPM, transcripts per million. (G) Immunoblots of whole cell extracts from cells expressing H3.3G34W treated with 0 µM, 1 µM, or 5 µM tazemetostat. (H) Fold change in the expression of genes down-regulated by the expression of H3.3G34W. Samples represent G34W/WT (red), cells expressing H3.3G34W treated with tazemetostat 1 µM/0 µM (light green) and 5 µM/0 µM (dark green) as measured by RT-qPCR. (I) Boxplots displaying the fold change in the expression of genes down-regulated by the expression of H3.3G34W (FC ≤ 0.3, n = 279) in the same samples as shown in H. Center line in the boxplots represents the median; Bottom and Top of the box represent 25th and 75th quartiles; whiskers extend to 1.5× interquartile range. Background includes all genes with posterior probability of differential expression (PPDE) ≥ 0.95. P value was determined using Wilcoxon’s rank sum test.
Based on our biochemical and ChIP-Seq studies, we hypothesized that H3.3 G34W at enhancers reduce H3K27ac by obstructing SETD2 activity on the mutant nucleosomes. Therefore, we compared gene expression changes in cells expressing H3.3 G34W to SETD2−/− cells. Overall, we found significant overlap between genes down-regulated by the expression of H3.3 G34W or the loss of SETD2 (Fig. 4 C and D and SI Appendix, Fig. S6D). These data support a model that H3.3 G34W alters gene expression primarily by interfering with SETD2 activity. SETD2 is thought to deposit H3K36me2/3 at transcribed genes through interaction with the phosphorylated C-terminal domain of RNA polymerase II (33). Surprisingly, we observed only a small increase of H3K27me3 at active gene bodies in cells expressing H3.3 G34W, likely reflecting a robust SETD2/NSD1/2 activity at these sites (SI Appendix, Fig. S4F). Importantly, we did not detect gene expression differences between exogenous reference normalized or internally median normalized expression values (SI Appendix, Fig. S6E), suggesting that the small increase of H3K27me3 in gene bodies has little effect on global gene expression.
While the G34W mutation is generally found in GCTBs, the H3.3 G34R/V mutations are most commonly found in HGA tumors (1, 3). Even though these specific G34 mutations are associated with different tumor types, we observed remarkable mechanistic similarities. For example, we observed similar gains of H3K27me3 on H3.3 G34R/W/V in our biochemical experiments (Fig. 1 F, G, and I). Moreover, we found that expression of H3.3 G34R in cells also reduced H3K27ac at enhancers that were sensitive to H3.3 G34W (Fig. 4E), and that genes proximal to enhancers with reduced H3K27ac were similarly affected by H3.3 G34R (SI Appendix, Fig. S6F). Importantly, expression of H3.3 G34R or H3.3 G34W led to remarkably similar gene expression profiles (Figs. 4F and 5A). These data are consistent with our biochemical findings that both G34R and G34W mutations act similarly by blocking SETD2 activity and consequently permitting H3K27 methylation.
Fig. 5.
H3.3G34W generates a unique gene expression profile in a PRC2-dependent manner. (A) Heatmap displaying the expression profiles of 180 differentially expressed genes (PPDE > 0.95; fold change > 3) in mMSCs expressing H3.3 G34W relative to H3.3 WT. Unguided hierarchical clustering was used to plot the divergence between samples. Data displays two independent biological replicates for each samples. (B) Schematic displaying the effect of K36R and K27R mutations in cis with the G34W mutation. G34W mutation allows increased PRC2 activity by blocking SETD2. PRC2 does not tolerate arginine substitution at the 36th position in vitro and in vivo; therefore, G34W-K36R is unable to increase G34W-dependent PRC2 activity in cis. K27R-G34W substitution disrupts K27ac/K27me3 on G34W oncohistone, without affecting G34W-dependent changes at K36 methylation. (C) Gene set enrichment analysis of ostoblastic differentiation signature in mMSCs expressing H3.3 G34W compared to H3.3 WT. (D) Fold change in the expression of genes associated with osteoblastic (green) and adipogenic differentiation (blue) in mMSCs expressing H3.3G34W. Genes implicated in promoting cell motility and chemotaxis are shown in red. (E) Heatmap displaying the expression profiles of genes associated with adipogenic and osteoblastic differentiation in human GCTB-derived cells. (F and G) Kaplan–Meier survival curve displaying the survival of mice injected with mesenchymal stem cells overexpressing H3.3WT (n = 9, 10), H3.3G34W (n = 10, 9), or H3.3G34W;K27R (n = 5, 10) double mutant. P values for significant difference for H3.3G34W samples were determined using the log rank test with respect to WT (in blue) or G34W;K27R double mutant (in green).
Our results suggest that incorporation of H3.3 G34W histones at enhancers favors PRC2-mediated H3K27me3 which leads to down-regulation of associated genes. To directly test this hypothesis, we treated cells expressing H3.3 G34W with tazemetostat, an EZH1/2 inhibitor, and probed the expression of down-regulated genes. Treatment of cells with tazemetostat led to reduced H3K27me2/3 in cells expressing H3.3 G34W and restored the expression of down-regulated genes near enhancers with reduced H3K27ac (Fig. 4 G–I). These results further link PRC2 activity with aberrant gene repression by H3.3 G34W.
The G34W Gene Expression Profile Requires PRC2 Activity on the Mutant H3.3.
In contrast to the K-to-M oncohistones, G34 mutations are exclusively found in genes encoding histone H3.3 (1, 3). Previous studies have found that histone H3.3 is enriched at transcriptionally active chromatin including enhancers (6). In line with these observations, our findings presented here suggest that H3.3 G34W functions locally at a subset of active enhancers to increase H3K27 methylation and reduce H3K27ac. To test the importance of H3.3 in mediating the G34 phenotype, we generated a cell line expressing H3.1 G34W transgene (SI Appendix, Fig. S6G). We found that expression of H3.1 G34W, which was depleted from active enhancers as measured by ChIP-Seq (Fig. 3B), failed to recapitulate the gene expression changes caused by the H3.3 G34W/R transgenes (Fig. 5A).
PRC2 activity is allosterically stimulated through binding of its own catalytic product, histone H3K27me3 or Jarid2 K116me3, to the EED subunit (17, 34). This feed-forward allosteric activation mechanism is proposed to help generate large H3K27me3 domains. To test the importance of H3K27 modifications in mediating the G34R/W gene expression, we used a G34W-K27R mutant H3.3 transgene that blocks K27me3 and hence, fails to stimulate PRC2 in trans (SI Appendix, Fig. S6 H and I). Interestingly, the K27R mutation on the H3.3 G34W oncohistone failed to recapitulate the gene expression profile mediated by G34W mutation alone. We suggest that these data show that allosteric activation of PRC2 by H3K27me3 on G34W oncohistones may amplify PRC2 activity locally. Collectively, our results indicate that reduced SETD2 activity on G34 oncohistones followed by PRC2-mediated methylation are intermediate steps in H3.3 G34W-driven gene expression changes.
In an alternative model, we supposed that the G34W mutation could reduce H3K36me3 and alter gene expression by negatively affecting recruitment of chromatin readers that bind to H3K36me3. To test this hypothesis, we used a H3.3 G34W-K36R transgene, which would continue to block recruitment of H3K36me3 readers while also negatively affecting PRC2 activity due to the K36R mutation (Figs. 1G and 5B). Notably, this G34W-K36R transgene also suppressed the gene expression profiles mediated by H3.3 G34W/R (Fig. 5A). These data indicate that loss of H3K36 methylation, alone, is insufficient to drive H3.3 G34W-mediated gene expression changes and provide an explanation for the absence of loss-of-function H3.3 K36 mutations (e.g., K36A and K36R) in GCTBs and HGAs. The inability of the H3.3 G34W-K36R double mutant to mimic the H3.3 G34W gene expression profile is consistent with our model that increased PRC2 activity on G34-mutated histones is important for generating the unique G34W/R gene expression profile.
H3.3 G34W Promotes Osteoblastic Gene Expression and Enhances Tumor Growth In Vivo.
We found that mMSCs expressing H3.3 G34W displayed down-regulation of several genes, including transcription factors and proteins involved in regulating cell fate commitment and up-regulation of genes associated with osteoblastic differentiation (Fig. 5 C and D and SI Appendix, Fig. S7A). This expression profile is similar to changes observed in H3.3 G34W-containing mesenchymal stromal cells in GCTBs. GCTB stromal cells retain their proliferative capacity and can be differentiated into osteoblastic, chondroblastic, and adipocytic lineages in vitro (35). Additionally, these malignant cells exhibit aberrant expression of osteoblastic factors, such as RANKL and CSF1, that are proposed to promote excessive recruitment of monocytes and subsequent tumor-associated osteoclastogenesis (36, 37).
The curious adipogenic and osteoblastic gene expression profile of mMSCs expressing H3.3 G34W led us to analyze RNA-Seq data from patient-derived giant cell tumor of the bone (38). Genes involved in promoting MSC differentiation, specifically toward osteoblastic and adipogenic lineages, were differentially expressed in human GCTBs containing H3.3 G34W mutations (Fig. 5E and SI Appendix, Fig. S7E). Importantly, master regulators of osteoblast and adipocytic lineages, RUNX2 and PPARG, respectively, were up-regulated in H3.3 G34W samples (Fig. 5E). As described previously, expression of the osteoblast-secreted proinflammatory factors, which aid matrix degradation and may contribute to local invasiveness in GCTBs, was significantly increased in G34W tumors (Fig. 5E) (39, 40). Taken together, these results indicate that H3.3 G34 mutations promote preosteoblastic gene expression profile to facilitate tumor growth.
Osteoblastic and adipogenic cells express genes implicated in tumor invasion and angiogenesis (39, 41–43). Based on this gene expression signature, we sought to determine if mMSCs expressing H3.3 G34W exhibited tumorigenic capacity. Subcutaneous and intratibial xenograft experiments revealed that expression of H3.3 G34W significantly promoted tumor progression of mMSCs in vivo (Fig. 5 F and G and SI Appendix, Fig. S7 B–D). Importantly, the H3.3 G34W-K27R double mutant alleviated H3.3 G34W-mediated tumor progression, highlighting the importance of lysine 27 in H3.3 G34W-mediated tumorigenesis.
Discussion
Our biochemical and cell-based studies have identified a mechanism by which H3.3 G34-mutated oncohistones disrupt the homeostatic balance of H3K36- and H3K27-modifying enzymes at gene enhancers and promotes aberrant gene expression and tumorigenesis (Fig. 6). Consistent with our biochemical data and those from a recent study, we found that expression of H3.3 G34W in cells resulted in increased H3K27me2/3 at and a concomitant reduction in H3K27ac levels (44). In specific cell contexts, these histone modification changes at enhancers promote a gene expression profile that reflects immature differentiation and simulates tumor progression in vivo.
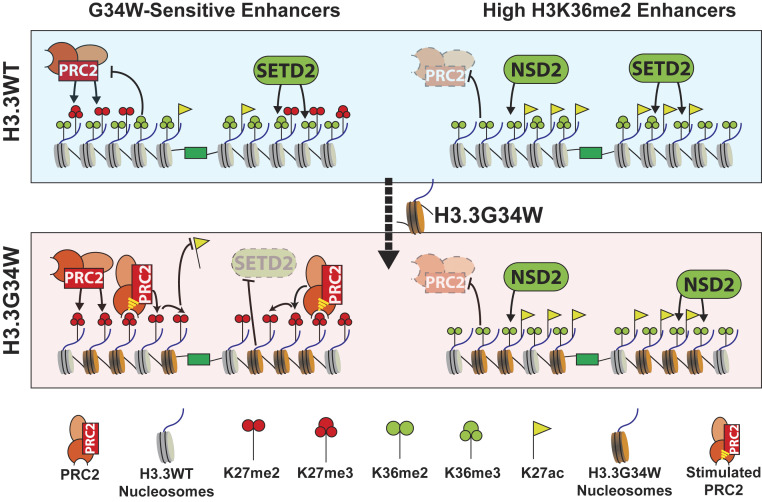
Fig. 6.
H3.3 G34W alters enhancer landscapes by boosting the “hit-and-run” mechanism of PRC2 recruitment. A balance between the activities of K36 methyltransferases, SETD2 and NSD2, and PRC2 exists at enhancers. “Robust” enhancers have high levels of NSD2-mediated H3K36me2 while other enhancers contain low NSD2 activity. Likewise, robust enhancers have little H3K27me2/3 while low-H3K36me2 enhancers exhibit higher PRC2 activity. The H3.3 G34W oncohistone disrupts the balance by decreasing SETD2 activity and promoting spreading of PRC2-mediated H3K27 methylation at low-H3K36me2 “G34W-sensitive” enhancers. However, the high NSD2 activity at robust enhancers prevents additional PRC2 activity. As a result of the increased H3K27 methylation, H3.3 G34W decreases H3K27ac specifically at enhancers containing low NSD2 activity. H3.3 G34W suppresses enhancers associated with genes involved in the maintenance of cell identity.
Unlike the K27 and K36 oncohistone mutations that have high preference for long aliphatic residues such as methionine or isoleucine, the substitutions at G34 residues—R, V, W, L—share few chemical similarities. The dissimilar biochemical properties of G34 mutations argue against a model that these substitutions generate a novel binding site for a chromatin modifier. Instead, we propose that these mutations work, in part, by generating steric hindrance that opposes SETD2 activity. In line with this hypothesis, bulky side chains of mutations at G34 position clashed with residues within the substrate channel of SETD2 SET domain and prevent its activity (21, 45). Interestingly, we find that substitutions at G33, V35, and K37 positions also reduce H3K36me3. However, mutations at these residues, or loss-of-function mutations at K36 position itself, are not prevalent in human cancers. The simple loss of H3K36me3 or disruption of H3K36me3-binding proteins, despite possibly promoting a selective advantage (45, 46), is not a compelling explanation for the high selectivity of tumor-associated mutations at glycine 34.
Our study uncovered the unique ability of G34 mutations to promote PRC2 activity in cis by impeding SETD2 activity. Strikingly, we found that no other residue in the H3.3 tail blocked SETD2 and was simultaneously tolerated by PRC2. Based on our data presented here, we propose that increased H3K27 methylation on G34 oncohistones is a critical step for their oncogenic function. Current high-resolution cocrystal structures of PRC2 and the H3 substrate peptide do not contain electron density maps for residues beyond P30 of the histone H3 tail. However, we predict that most H3 tail residues in this region are either sterically constrained or make critical contacts with PRC2 since substitutions at these sites are not tolerated by PRC2. Thus, our investigation has illuminated important sequence features of the H3 tail involved in the regulatory cross-talk between PRC2 and its substrates. Moreover, our study revealed how cancer-associated mutations modulate PRC2 activity to fine tune gene expression.
The G34 mutations stand apart from the K27M and K36M oncohistones for their preferential occurrence in the histone variant H3.3. Previous studies have characterized the K-to-M mutations as the specific inhibitors of their corresponding methyltransferases (12). Consistent with their dominant negative mode of action, the K-to-M oncohistones lead to a global reduction of corresponding lysine methylation irrespective of the histone variant affected (9, 10). Moreover, EZHIP, the recently characterized K27M mimic found in posterior fossa type-A ependymomas reduces H3K27me3 without their incorporation into the nucleosomes (28, 47). In contrast, our study argues that the localization of G34 oncohistones to H3.3-containing active genes and enhancers is critical for their oncogenic function and might explain their preference of histone variant H3.3.
While different G34 mutations (e.g., G34R and G34W) are associated with specific tumor types, our data suggest that the G34W and G34R mutations work through a common mechanism and promote similar gene expression changes. The recent discovery that G34W- and G34R-containing osteosarcomas coclustered within the same group by DNA methylation profile supports our conclusion that these mutations share common underlying biochemical mechanisms (5). Despite these similarities, the preference for specific substitutions in these distinct tumors is intriguing. Future studies in genetic model systems will explore possible phenotypic differences between the different G34 substitutions.
Tissue-specific and temporal regulation of PRC2 activity is critical for normal development (48). However, the cis regulatory elements that could account for the global pattern of PRC2 activity in mammals have not been identified. Instead, a recent model proposes that PRC2 adopts a hit-and-run strategy where factors that locally modulate PRC2 activity are responsible for the ultimate distribution of H3K27 methylation (49). Features associated with heterochromatin such as H3K27me3 and densely packed nucleosomes stimulate PRC2 activity, whereas active chromatin associated factors such as RNA, polyacetylated histones, H3K4me3 and H3K36me2/3 block PRC2 activity. The importance of these regulatory mechanisms for proper PRC2 recruitment and activity is most evident in human cancers driven by histone mutations. For example, the chondroblastoma-associated H3 K36M mutation is a competitive inhibitor of SETD2 and NSD1/2 enzymes and is associated with globally reduced levels of H3K36me2/3. This global loss of H3K36me2/3 provides novel substrates for PRC2, resulting in a genome-wide gain of H3K27me3, dilution of PRC1 recruitment, and subsequent derepression of silenced genes (10).
In contrast to the global changes in histone PTMs by H3 K36M mutations, we discovered that the H3.3 G34 mutations drive local changes of H3K27me2/3 at a subset of active enhancers. We find that G34 oncohistones exploit the ability of PRC2 to precisely “sense” local chromatin and drive localized gain of PRC2 activity. Ultimately, this increased PRC2 activity leads to down-regulation of specific genes and immature differentiation of mesenchymal stem cells. Overall, our studies have demonstrated that both H3 K36M and H3.3 G34 modulate finely tuned Polycomb activity to drive distinct cancers.
Materials and Methods
Transgenic Cell Line Generation.
Human 293T or immortalized mMSC C3H10T1/2 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and passaged at subconfluency. Cells were regularly tested for mycoplasma. Cells were transduced with recombinant lentiviruses produced using pCDH-EF1a-MCS-Puro expression vector containing histone transgene as describes previously (12). Transduced cells were selected with 1 µg/mL of puromycin for 4 d. SETD2, NSD1, and NSD2 knockout C3H10T1/2 cells used in this study to map contributions of these histone methyltransferases at various genomic loci were generated as previously described (50).
Purification of Mononucleosome from HEK293T Cells.
A total of ∼3 × 108 cells were lysed by resuspending in buffer-A (15 mM Hepes, pH 7.9, 10 mM KCl, 5 mM MgCl2, 0.5 mM ethylenediaminetetraacetic acid [EDTA], 1 mM β-mercaptoethanol) and nuclei were collected by centrifugation at 3,000 rpm for 5 min. Nuclei was resuspended in MNase digestion buffer (15 mM Hepes, pH 7.9, 30 mM KCl, 5 mM MgCl2, 3 mM CaCl2, 1× protease inhibitor mixture [Roche], 1 mM dithiothreitol [DTT], 0.4 mM phenylmethylsulfonyl fluoride [PMSF]). Mononucleosomes were prepared by incubating the nuclei with 1,500 units of MNase for 20 min at 37 °C. MNase reaction was quenched and mononucleosomes were solubilized by resuspending the nuclei in 3 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), 5 mM EDTA, 0.1% Triton, and 150 mM KCl. Cell debris were removed by centrifugation at 14,000 rpm for 10 min. Supernatant was incubated with FLAG M2 agarose beads to capture mononucleosomes containing FLAG-epitope- tagged mutated histone. Beads were washed five times with 1 mL of wash buffer (15 mM Hepes, pH 7.9, 300 mM KCl, 1 mM EDTA, 10% glycerol) and mononucleosomes were eluted using wash buffer containing 0.3 mg/mL of 3× FLAG peptide. Nucleosomes were dialyzed against 15 mM Hepes, pH 7.9, 150 mM KCl, 1 mM EDTA, and 10% glycerol for 2 h to remove FLAG peptides.
Histone Methyltransferase (HMT) Assays.
For a typical methyltransferase reaction, 300 nM recombinant nucleosome substrates were incubated with 40 nM SETD2 or NSD2 enzymes and 4 µM S-adenosyl methionine (1 µM 3H-SAM (Perkin-Elmer); 3 µM cold SAM) in 2× reaction buffer (50 mM Tris pH 8.0, 4 mM MgSO4, 10 mM DTT, and 8 mM PMSF) for 60 min. For scintillation counting, reactions were spotted on phosphocellulose filters (Whatman p81) and dried for 10 min. Filters were washed three times with 0.1 M NaHCO3, rinsed with acetone, and dried for 10 min. Scintillation counting was performed using a Perkin-Elmer Tri-Carb 2910 TR liquid scintillation analyzer. Appropriate negative controls were used for background correction. For PRC2 assays, similar reaction conditions were used except with 20 nM enzyme and 20 µM H3K27me3 peptide. Reaction mixtures were resolved by polyacrylamide gel electrophoresis gel, transferred on nitrocellulose membrane, and probed with antibodies for immunoblotting. For sequential HMT assays, 300 nM nucleosome substrates were incubated with 40 nM SETD2 and 50 µM cSAM at 30 °C for 3 h. An additional 40 nM SETD2 was added to the reaction for 12 h. Reaction products were incubated with 20 nM rPRC2, 20 nM rJarid2, 20 µM H3K27me3 peptide, and 50 µM cSAM for 3 h at 30 °C, followed by immunoblotting.
ChIP-Seq Analysis.
ChIP-Seq data were analyzed as previously described (10, 47). Briefly, reads that passed quality score were aligned to mouse mm9 and reference genomes (dm6 or hg19) using bowtie1 (parameters: -q -v 2 -m 2 -a -best -strata). Sample normalization factor was determined as 106/(reads mapped to reference genome) or reads per kilobase per million (RPKM) factor = 106/total aligned reads. H3K27me3 data were normalized by median normalization using RPKM normalized intensities at H3K27me3 peaks. Peaks were determined using Mosaics-HMM (broad peaks) for H3.3, H3K36me3, and H3K27me3; and Mosaics for K27ac ChIP to determine sharp peaks (51). Input samples were used to estimate background during peak calling. Active enhancers were defined as H3K27ac peaks excluded from the transcription start site (TSS) (5 kb upstream, 2 kb downstream). Bigwig files and average profiles were generated using Deeptools and visualized using the Integrative Genome Browser (IGV). Bigwig files, bed files containing peaks, etc., are deposited at Gene Expression Omnibus (GEO). Statistical analysis and graphs were generated using R. Enhancer-gene association was determined using GREAT (52, 53). ChIP-Seq results were also validated through correlations from previously published studies from several laboratories such as anticorrelation between H3K27me3 and H3K36me3, enrichment of H3.3 at enhancers, enrichment of H3K36me3 at gene bodies, etc. ChIP-Seq results were validated by qPCR at select enhancers.
Tumor Xenograft Study A: s.c. Injection.
Cells were prepared in single cell suspension in phosphate-buffered saline 50% Matrigel, and a volume of 200 µL containing 1.2 to 1.5 × 106 cells was injected s.c. into the left flank of the NOD scid gamma mice. Tumor formation was monitored weekly by palpation starting at 4 wk postinjection. Tumor diameters were measured with digital calipers, and the tumor volume in mm3 was calculated. Mice were killed when tumor volume reached ≥2,000 mm3 (maximum size) and the tumors were collected for the following experiments. Parental C3H10T1/2 cells used in this study or expression of H3.1 did not form tumors in vivo.
Intratibial (Orthotopic) Implantation.
The NRG mouse leg cleaned with 70% ethanol and a predrilled hole was created in the tibia plateau using a 25-gauge needle by rotating movements and 3 µL of PBS-20% Matrigel containing 1.8 to 2 × 105 cells was injected down into the diaphysis of the tibia with a Hamilton syringe (27-G needle). Tumor formation was monitored weekly and tumor volume was measured with digital calipers, and mice were killed when the tumor volume reached the maximum size.
Supplementary Material
Acknowledgments
This research was supported by funding from NIH P01CA196539 (to P.W.L., N. Jabado, C.D.A., and B.A.G.); the Greater Milwaukee Foundation (to P.W.L.), the RIDE Foundation for Cancer Research (to P.W.L.), the Sidney Kimmel Foundation (Kimmel Scholar Award to P.W.L.), and the Carbone Cancer Center (P30CA014520). P.W.L. is a Pew Scholar in the Biomedical Sciences. Innovator grant DP2OD007447, R01GM110174 and Robert Arceci Scholar Award from the Leukemia and Lymphoma Society (to B.A.G.). This work was performed within the context of the I-CHANGE consortium and supported by funding from Genome Canada, Genome Quebec, The Institute for Cancer Research of the Canadian Institutes for Health Research, McGill University, and the Montreal Children’s Hospital Foundation. D.M.M. is supported by T32GM008275 and TL1TR001880. D.N.W. is supported by T32GM007739 and F30CA224971. C.L. acknowledges support from NIH (R00CA212257), Damon Runyon Cancer Research Foundation (DFS-28-18), Pew-Stewart Scholar Award for Cancer Research, and an American Association for Cancer Research Gertrude B. Elion Cancer Research Award. N. Jabado. is a member of the Penny Cole laboratory and the recipient of a Chercheur Clinician Senior Award. We thank Joseph Cesare for his help with mass spectrometry data analyses; Dr. Jason Karamchandani, pathologist at Department of Pathology, McGill University, and McGill University Health Center, who characterized the tumors from the xenograft study. All histological staining was performed with the support of the Research Institute of the McGill University Health Centre Histopathology Platform.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006076117/-/DCSupplemental.
Data Availability.
The next-generation sequencing data generated in this study are deposited at GEO database with accession nos. GSE133722 (54) and GSE118785 (55). Published data used in this paper were downloaded from GEO (accession no. GSE103559) (56).
References
- 1.Schwartzentruber J.et al., Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Wu G.et al.; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project , Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44, 251–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behjati S.et al., Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 45, 1479–1482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackay A.et al., Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell 32, 520–537.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koelsche C.et al., Histone 3.3 hotspot mutations in conventional osteosarcomas: A comprehensive clinical and molecular characterization of six H3F3A mutated cases. Clin. Sarcoma Res. 7, 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldberg A. D.et al., Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martire S.et al., Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. Nat. Genet. 51, 941–946 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang T., Zhang Z., Dong Q., Xiong J., Zhu B., Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells. Genome Biol. 21, 45–47 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagaraja S.et al., Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell 31, 635–652.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu C.et al., Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352, 844–849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hock H., A complex polycomb issue: The two faces of EZH2 in cancer. Genes Dev. 26, 751–755 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis P. W.et al., Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohammad F.et al., EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat. Med. 23, 483–492 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Chan K.-M.et al., The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 27, 985–990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Margueron R., Reinberg D., The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrari K. J.et al., Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol. Cell 53, 49–62 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Margueron R.et al., Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan W.et al., H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 286, 7983–7989 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitges F. W.et al., Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42, 330–341 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Jani K. S.et al., Histone H3 tail binds a unique sensing pocket in EZH2 to activate the PRC2 methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 116, 8295–8300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang S.et al., Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase. Genes Dev. 30, 1611–1616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banaszynski L. A., Allis C. D., Lewis P. W., Histone variants in metazoan development. Dev. Cell 19, 662–674 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calo E., Wysocka J., Modification of enhancer chromatin: What, how, and why? Mol. Cell 49, 825–837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner E. J., Carpenter P. B., Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13, 115–126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henriques T.et al., Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 32, 26–41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang A. H.et al., The elongation factor Spt6 maintains ESC pluripotency by controlling super-enhancers and counteracting polycomb proteins. Mol. Cell 68, 398–413.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavarone E., Barbieri C. M., Pasini D., Dissecting the role of H3K27 acetylation and methylation in PRC2 mediated control of cellular identity. Nat. Commun. 10, 1679 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain S. U.et al., PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat. Commun. 10, 2146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banaszynski L. A.et al., Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whyte W. A.et al., Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovén J.et al., Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osterwalder M.et al., Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venkatesh S., Workman J. L., Set2 mediated H3 lysine 36 methylation: Regulation of transcription elongation and implications in organismal development. Wiley Interdiscip. Rev. Dev. Biol. 2, 685–700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanulli S.et al., Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol. Cell 57, 769–783 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wülling M., Delling G., Kaiser E., The origin of the neoplastic stromal cell in giant cell tumor of bone. Hum. Pathol. 34, 983–993 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Atkins G. J.et al., Expression of osteoclast differentiation signals by stromal elements of giant cell tumors. J. Bone Miner. Res. 15, 640–649 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Murata A., Fujita T., Kawahara N., Tsuchiya H., Tomita K., Osteoblast lineage properties in giant cell tumors of bone. J. Orthop. Sci. 10, 581–588 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Lim J.et al., Transcriptome and protein interaction profiling in cancer cells with mutations in histone H3.3. Sci. Data 5, 180283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghert M., Simunovic N., Cowan R. W., Colterjohn N., Singh G., Properties of the stromal cell in giant cell tumor of bone. Clin. Orthop. Relat. Res. 459, 8–13 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Tanaka Y., Nakayamada S., Okada Y., Osteoblasts and osteoclasts in bone remodeling and inflammation. Curr. Drug Targets Inflamm. Allergy 4, 325–328 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Divella R., De Luca R., Abbate I., Naglieri E., Daniele A., Obesity and cancer: The role of adipose tissue and adipo-cytokines-induced chronic inflammation. J. Cancer 7, 2346–2359 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kessenbrock K., Plaks V., Werb Z., Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 141, 52–67 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonnans C., Chou J., Werb Z., Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 15, 786–801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi L., Shi J., Shi X., Li W., Wen H., Histone H3.3 G34 mutations alter histone H3K36 and H3K27 methylation in cis. J. Mol. Biol. 430, 1562–1565 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang J.et al., Cancer-driving H3G34V/R/D mutations block H3K36 methylation and H3K36me3-MutSα interaction. Proc. Natl. Acad. Sci. U.S.A. 115, 9598–9603 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim J.et al., The histone variant H3.3 G34W substitution in giant cell tumor of the bone link chromatin and RNA processing. Sci. Rep. 7, 13459 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain S. U., et al., H3 K27M and EZHIP impede H3K27-methylation spreading by inhibiting allosterically stimulated PRC2. Mol. Cell., 10.1016/j.molcel.2020.09.028 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deevy O., Bracken A. P., PRC2 functions in development and congenital disorders. Development 146, dev181354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiles E. T., Selker E. U., H3K27 methylation: A promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 43, 31–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinberg D. N.et al., The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 573, 281–286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun G., Chung D., Liang K., Keleş S., Statistical analysis of ChIP-seq data with MOSAiCS. Methods Mol. Biol. 1038, 193–212 (2013). [DOI] [PubMed] [Google Scholar]
- 52.McLean C. Y.et al., GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rada-Iglesias A.et al., Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell 11, 633–648 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jain S. U., Lewis P. W., Histone H3.3 G34 mutations promote aberrant PRC2 activity and drive tumor progression. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133722. Deposited 2 July 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinberg D. N., et al. , H3K36me2 recruits DNMT3A and shapes intergenic DNA methylation landscapes. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118785. Deposited 20 August 2018. [Google Scholar]
- 56.Lim J., The histone variant H3.3 G34W substitution in giant cell tumor of the bone link chromatin and RNA processing [RNA-seq]. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE103559. Deposited 6 September 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The next-generation sequencing data generated in this study are deposited at GEO database with accession nos. GSE133722 (54) and GSE118785 (55). Published data used in this paper were downloaded from GEO (accession no. GSE103559) (56).