Significance
Transcriptional activators represent a molecular recognition enigma. Their function in transcription initiation requires selective engagement of coactivators, yet the prevailing molecular recognition models propose this occurs via nonspecific intermolecular contacts. Here, mechanistic analysis of several related activator•coactivator complexes resolves this conundrum. In contrast to the expectations from nonspecific recognition models, even small sequence changes in the activators cause activator•coactivator complexes to undergo significant conformational redistribution, driven by specific intermolecular interactions and conformational changes in the coactivator itself. These unappreciated specific recognition mechanisms rationalize the high sequence variability of functional activators, opening new questions about the relationship between recognition and function.
Keywords: transcriptional activator, coactivator, Med25, ETV/PEA3, protein–protein interactions
Abstract
A key functional event in eukaryotic gene activation is the formation of dynamic protein–protein interaction networks between transcriptional activators and transcriptional coactivators. Seemingly incongruent with the tight regulation of transcription, many biochemical and biophysical studies suggest that activators use nonspecific hydrophobic and/or electrostatic interactions to bind to coactivators, with few if any specific contacts. Here a mechanistic dissection of a set of representative dynamic activator•coactivator complexes, comprised of the ETV/PEA3 family of activators and the coactivator Med25, reveals a different molecular recognition model. The data demonstrate that small sequence variations within an activator family significantly redistribute the conformational ensemble of the complex while not affecting overall affinity, and distal residues within the activator—not often considered as contributing to binding—play a key role in mediating conformational redistribution. The ETV/PEA3•Med25 ensembles are directed by specific contacts between the disordered activator and the Med25 interface, which is facilitated by structural shifts of the coactivator binding surface. Taken together, these data highlight the critical role coactivator plasticity plays in recognition of disordered activators and indicate that molecular recognition models of disordered proteins must consider the ability of the binding partners to mediate specificity.
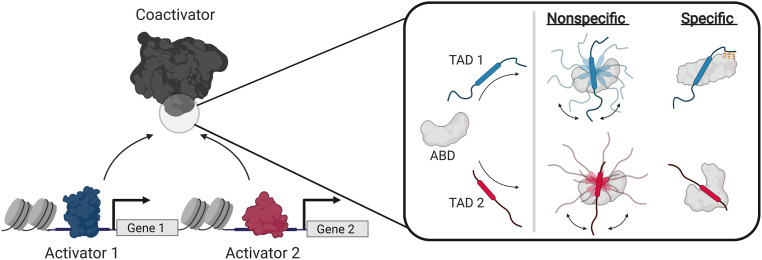
Protein–protein interactions (PPIs) formed between transcriptional activators and coactivators play a critical role in gene expression; these PPIs underpin colocalization of transcriptional machinery components and stimulate transcription initiation (1–8). A prevailing view of activator•coactivator PPIs is that they are largely nonspecific and, further, that the selectivity necessary for appropriate gene expression comes from other sources such as activator–DNA interactions and/or colocalization (3, 9–14). Indeed, there is considerable data suggesting activator•coactivator complexes form via almost entirely nonspecific intermolecular interactions, from early experiments demonstrating that a wide range of natural and nonnatural amphipathic molecules interact with coactivators to more recent structural studies indicating no fixed activator•coactivator binding mode (Fig. 1) (1, 9–11, 15–18).
Fig. 1.
Recognition models of activator function. (Left) Transcriptional activators regulate gene activity via protein–protein interactions with coactivators. (Right) Comparison of nonspecific (3, 11) and specific (this work) models of activator•coactivator recognition. Nonspecific models propose that the TADs of unique activators bind the ABDs of coactivators via nonspecific intermolecular interactions, forming complexes without fixed orientation or structure via “sequence-independent” recognition (11). Rounded boxes in the TADs represent the binding motif.
Nonspecific recognition models, while attractive in their simplicity, are inconsistent with the critical functional role that individual activator•coactivator PPIs play in gene expression. There are several examples of transcriptional activators that depend on interactions with specific activator binding domains (ABDs) of coactivators for function (19–23), e.g., the SREBP family of activators require the KIX ABD of the coactivator ARC105 to regulate fatty acid homeostasis (21) even though other coactivators such as CBP and p300 have structurally similar KIX motifs. Further, the biophysical studies that investigate how ABDs recognize diverse activators most often utilize qualitative equilibrium approaches (9–11) that are blind to critical mechanistic information (24–26) due to equilibrium averaging. It is therefore an open question whether there are other molecular recognition mechanisms at play; this would account for the diversity of functional activator sequences as well as the observed selectivity of activators in vivo. Because activator•coactivator complexes often represent promising therapeutic targets, developing a more detailed understanding of the molecular recognition mechanisms of these crucial PPIs is also essential for the development of small molecule modulators (4, 27–29).
Here, we take a critical look at activator•coactivator molecular recognition by mechanistically dissecting a representative set of dynamic complexes formed between the ABD of Mediator subunit Med25 and the amphipathic transcriptional activation domains (TADs) of the ETV/PEA3 family of Ets transcriptional activators (ETV1, ETV4, and ETV5) (23, 30–32). Previous biophysical studies indicated that the interaction of Med25 with family member ETV5 appears to be a prototypical nonspecific TAD•ABD complex: it occurs over a shallow surface, is driven by electrostatic and hydrophobic interactions, and forms a dynamic complex that is recalcitrant to structure determination (30, 32). We utilize a mechanistic and structural approach that combines quantitative data regarding activator•coactivator conformational states obtained via transient kinetic analysis with structural information obtained through mutagenesis and NMR spectroscopy. Our data reveal that the conformational ensembles of ETV/PEA3•Med25 PPIs are strikingly sensitive to slight changes in TAD sequence, despite being dynamic complexes with several well-populated conformational substates at equilibrium. Furthermore, the mechanism underlying this conformational sensitivity involves the ability of ordered and disordered regions of the TAD to participate in finite sets of specific interactions with the Med25 interface, as well as conformational changes in Med25 that remodel the TAD binding site. Together, these results reveal an unappreciated degree of specificity in the formation of activator•coactivator complexes that is in direct contrast to the prevailing nonspecific recognition models of these essential PPIs (Fig. 1) (3, 11).
Results
ETV/PEA3•Med25 PPIs as a Model System for Dynamic TAD•ABD Interactions.
ETV/PEA3•Med25 interactions represent an ideal system to study dynamic TAD•ABD interactions for several reasons. First, previous studies indicated that interaction of ETV/PEA3 family member ETV5 with the Med25 ABD is typical of a dynamic TAD•ABD complex (30, 32); the binding surface is shallow, both acidic and hydrophobic amino acids of the TAD determine affinity and activity, and multiple bound conformational states have been detected by both NMR and kinetic analyses. Second, our previous studies of the ETV5•Med25 complex showed that the bound conformational ensemble is directly accessible by transient kinetic analysis (32). The models of TAD•ABD molecular recognition can thus be dissected in this system without the loss of critical conformational information to equilibrium averaging. Third, the ETV/PEA3 family of transcription factors serves as an excellent natural system to test the relationship between TAD sequence and recognition; they contain almost identical arrangements of acidic and hydrophobic residues across the TAD sequence, especially within the helical binding region that undergoes coupled folding and binding with Med25 (30), but the identity of specific residues varies slightly (Fig. 2A). This system can therefore be used to test whether TAD•Med25 interactions are truly nonspecific and insensitive to variations in the TAD sequence (11), or if these interactions are affected by TAD sequence changes and thus have a degree of specificity to formation. Finally, the Med25 ABD is ligandable by small molecules (32), and therefore conclusions from mechanistic studies can be directly applied to guide and assess optimization of small molecule modulators of TAD•Med25 complex formation.
Fig. 2.
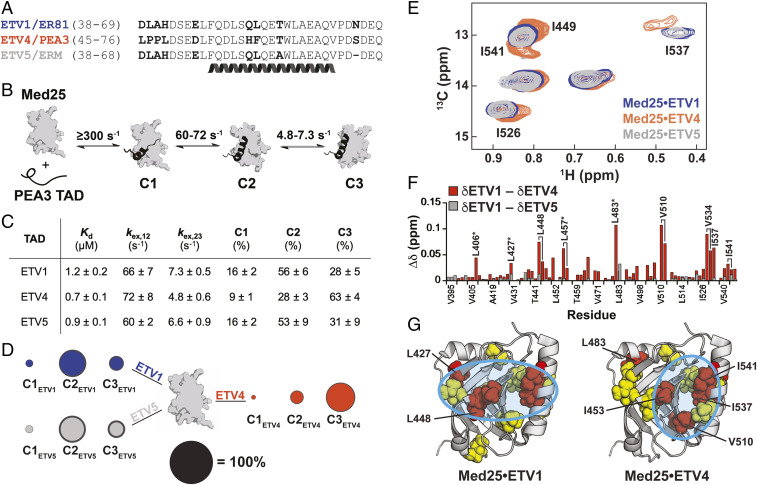
ETV/PEA3 activators differentially engage with Med25. (A) Alignment of ETV/PEA3 family activation domains. The helix denotes the residues that undergo coupled folding and binding with Med25, as determined by NMR chemical shift analysis (30). (B) Mechanism of binding of ETV/PEA3 activators to Med25, determined here for ETV1 and ETV4, and previously for ETV5 (32). The range of exchange rates between analogous steps for ETV/PEA3 TADs are shown. (C) Table of relevant binding parameters for each ETV/PEA3 TAD, including the equilibrium affinity, exchange rates between C1 and C2 (kex,12) and between C2 and C3 (kex,23), and equilibrium populations of each state. All values represent the average of three to four biological replicates, and the error is the SD. (D) Equilibrium populations of the three ETV/PEA3•Med25 conformations scaled relative to the diameter of the black circle. SDs of the values are shown as the dark gray outer circle. (E) Overlay of the Ile Cδ region of the 1H,13C-HSQC of Med25 in complex with 1.1 equivalents unlabeled ETV1 (blue), ETV4 (orange), or ETV5 (gray). Peaks that have chemical shift differences between complexes are labeled. Note: I449 and I541 form a single overlapped peak for the ETV1 and ETV5 complexes. Full spectra are in the supporting information. (F) Chemical shift differences of Med25 methyl resonances between ETV1 and ETV4 (orange) and ETV1 and ETV5 (gray). (G) Chemical shift perturbations induced by binding of ETV/PEA3 activators plotted on the structure of Med25 (PDB ID 2XNF) (34). Yellow = 0.040 to 0.080 ppm, red > 0.080 ppm. Cyan circles highlight general distinctions in perturbation patterns. Several residues with chemical shift differences >0.030 ppm between the ETV1•Med25 (or ETV5•Med25) and ETV4•Med25 complexes are labeled.
Small Sequence Differences between ETV/PEA3 Family Members Lead to Conformationally Distinct PPIs with Med25.
We first examined whether the slight sequence variations across the ETV/PEA3 family TADs affect the conformational ensembles of the individual ETV/PEA3•Med25 complexes. Stopped-flow fluorescence transient kinetic experiments were performed to directly evaluate conformational dynamics and equilibria, using TADs synthesized with the solvatochromic fluorophore 4-N,N-dimethylamino-1,8-naphthalimide (4-DMN) conjugated to the N terminus (33). We previously demonstrated with this approach that the ETV5•Med25 complex forms in a minimal three step linear mechanism (32): after an initial rapid association event that mostly occurs in the instrument dead time (∼2 to 4 ms), the complex undergoes two sequential conformational changes (Fig. 2B). At equilibrium, all three bound conformations of the ETV5•Med25 complex are well populated due to relatively small conformational equilibrium constants (32).
Application of the same experimental conditions to ETV1 and ETV4 indicated that the kinetic binding mechanism is conserved; for all ETV/PEA3 TADs a rapid binding step followed by two conformational change steps was observed. Each of these individual steps occurred with similar exchange rate constants (kex; the sum of the forward and reverse rate constants) for each ETV/PEA3•Med25 complex (Fig. 2 B and C); this suggests that the steps represent analogous conformational transitions in each complex. In addition, the equilibrium binding affinity between ETV/PEA3 TADs varied less than twofold (0.7 to 1.2 µM, Fig. 2C), consistent with the expectation from nonspecific models that minor substitutions in the TAD will not affect the overall stability of the activator•coactivator complex.
Despite a conserved binding mechanism and similar overall affinities, calculation of equilibrium conformational populations from the kinetic data revealed clear differences between the engagement modes of ETV/PEA3 family members (Fig. 2 C and D; for raw data and detailed kinetic analysis, see SI Appendix, Discussion of Kinetic Analysis). While the populations of analogous conformational states of the ETV1•Med25 and ETV5•Med25 complexes were essentially identical, the ETV4•Med25 complex populated the three analogous conformations in a unique manner (Fig. 2 C and D). Critically, this shift in conformational equilibria does not correlate with predicted structural propensity differences between the ETV/PEA3 TADs (SI Appendix, Fig. S1), which suggests that variable residues between ETV1/ETV5 and ETV4 alter the TAD•Med25 conformational ensemble via intermolecular interactions made in the bound state.
We next examined differences in the engagement modes of ETV/PEA3 TADs via NMR spectroscopy. Unique TADs with nonspecific engagement modes are expected to produce essentially identical binding signatures in NMR spectra of the partner ABD (11), and therefore comparison of chemical shift perturbation (CSP) patterns of labeled Med25 bound to individual ETV/PEA3 TADs provides an orthogonal source of insight into the specificity of the interaction. Sidechain methyl 1H,13C-HSQC (heteronuclear single quantum coherence) experiments were used as a primary method to enable direct detection of effects on both surface and buried residues of Med25.
Comparative analysis of Med25 1H,13C-HSQC spectra bound to different ETV/PEA3 family members was consistent with the expected engagement differences between the ETV/PEA3 TADs: spectra of ETV1- and ETV4-bound Med25 exhibited several large differences in CSP patterns, whereas the spectra of Med25 bound to ETV1 and ETV5 were essentially indistinguishable (Fig. 2 E and F; full spectral overlay shown in SI Appendix, Fig. S19). Inspection of the CSP data plotted on the structure of Med25 indicated that all ETV/PEA3 family members bind to a previously identified (30–32) core binding site formed between the central β-barrel and the C-terminal α3 helix (Fig. 2G), but ETV1/ETV5 and ETV4 produce unique perturbation patterns in the binding surface (Fig. 2G, cyan circles). In addition, several resonances representing buried and/or allosteric residues displayed significant CSP differences between the ETV1- and ETV4-bound complexes, suggesting that the conformation of the Med25 ABD may also be different between these complexes (Fig. 2F, starred). The NMR data therefore support the conclusion from kinetics experiments that ETV4 has a unique Med25 engagement mode as compared to ETV1 and ETV5; together, these biophysical and structural experiments suggest a model where one or more of the variable residues in the ETV/PEA3 TADs make distinct specific intermolecular interactions with the Med25 surface.
Ordered and Disordered Regions of the ETV/PEA3 TADs Dictate Conformational Differences between ETV/PEA3•Med25 PPIs.
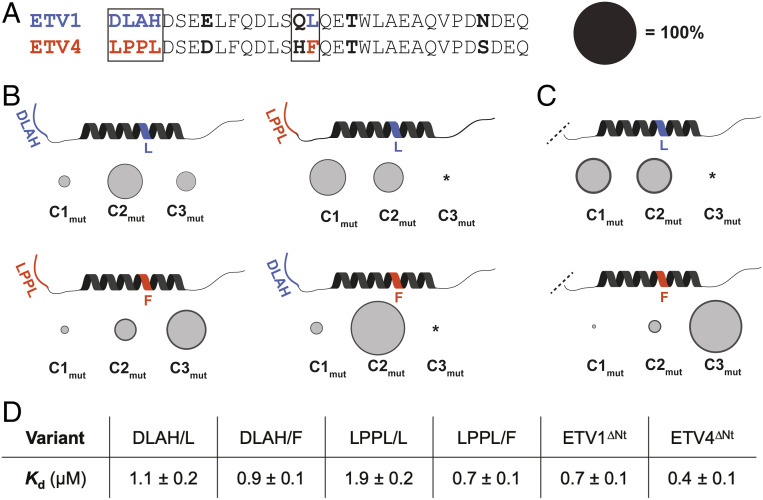
We next identified the TAD residues that bias ETV/PEA3•Med25 PPIs toward different conformational substates using a mutagenesis approach. Residues that are variable between ETV1 and ETV5 are not expected to contribute to these conformational differences, as these TADs have essentially identical conformational and structural properties (Fig. 2). Thus, this mutagenesis effort was constrained to residues that are conserved between ETV1 and ETV5, but not ETV4. Two regions of interest were evident (Fig. 3A, boxed): 1) a two amino acid “variable motif” in the helical binding region consisting of a polar residue followed by a hydrophobic residue (QL in ETV1/ETV5 and HF in ETV4), and 2) the four amino acid N terminus of the TAD (DLAH in ETV1/ETV5 and LPPL in ETV4), a region predicted to remain entirely disordered for all ETV/PEA3 TADs (SI Appendix, Fig. S1 and ref. 30). An additional variable Glu/Asp residue met the constraint we imposed on residue conservation (Fig. 3A) but was excluded from mutagenesis because acidic residues typically mediate long-range electrostatic interactions in TAD molecular recognition (35); thus this residue change was not anticipated to significantly affect the conformational ensemble.
Fig. 3.
Variable residues in the disordered N terminus and the helical binding region mediate differences in ETV/PEA3•Med25 conformational behavior. (A) Alignment of ETV1 and ETV4 activators with regions that were selected for mutational analysis boxed. Bolded residues are conserved between ETV1 and ETV5, but not ETV4. Regions/residues that affected the conformational ensemble are color coded to ETV1 (blue) or ETV4 (orange). Effects of the Gln/His residues in the variable motif were also tested but did not affect conformational populations and are thus omitted in B and C for clarity. Populations of conformational states in B and C are scaled relative to the diameter of the black circle. (B) Results from kinetics experiments of mutant TADs, for native (Left) and nonnative (Right) combinations of variable N termini and helical binding regions. Variants were made based on the ETV4 sequence. The data shown are the average across all of the variants tested from each group, with the error (dark gray outer circle) representing the SD. (C) Results from kinetics experiments with ETV1∆Nt (Top) and ETV4∆Nt (Bottom). (D) Average equilibrium Kd values of variants tested. *Conformer was undetectable in kinetics experiments (see SI Appendix, Discussion of Kinetic Data Analysis for further details).
A small library of mutant TADs varying residues in the two regions of interest of the ETV4 sequence—either the Leu or Phe residue in the variable motif and the DLAH or LPPL N terminus—was synthesized and assessed in stopped-flow kinetic assays (Fig. 3B; values for all variants in SI Appendix, Tables S2 and S3). The polar residue (Gln/His) in the variable motif was also tested but had no effect on conformational populations. Importantly, the equilibrium binding affinity varied only slightly across the mutants (Fig. 3D).
Consistent with the hypothesis that one or both of the variable regions dictate the conformational differences between the ETV/PEA3•Med25 complexes, ETV/PEA3 variants with “native” combinations of the N terminus and the hydrophobic residue of the variable motif exhibited similar conformational ensembles to the natural TADs (Fig. 3 B, Left). That is, combinations with DLAH at the N terminus and Leu in the variable motif (DLAH/L) populated three observable conformational states in a similar manner to ETV1 and ETV5 (Fig. 3 B, Top Left), and, analogously, combinations with LPPL at the N terminus and Phe in the variable motif (LPPL/F) populated three substates comparably to ETV4 (Fig. 3 B, Bottom Left).
Conversely, when nonnative combinations of the N terminus and variable motif were tested, unique conformational behavior was observed (Fig. 3 B, Right). Only two bound conformations of DLAH/F variants were detected in kinetics experiments and they displayed exchange kinetics similar to the C1–C2 transitions of the native complexes (Fig. 3 B, Top Right). Calculation of conformational populations indicated that the second conformation had a higher overall population (82 ± 3%) than the C2 conformations of the ETV1 (56 ± 6%), ETV5 (53 ± 9%), or ETV4 complexes (28 ± 3%). Similarly, two bound conformations with C1–C2-like exchange rates were detected with LPPL/L variants (Fig. 3 B, Bottom Right), but the initial bound substate was preferentially populated. We note that the C3 conformation of these complexes may be undetectable due to a low population (≤10%) or an increase in the exchange kinetics between C2 and C3; however, neither of these possibilities are inconsistent with the conclusion that these mutant complexes are conformationally distinct (SI Appendix, Discussion of Kinetic Data Analysis for further details). Together, these results indicate that the conformational differences between ETV/PEA3•Med25 complexes are dictated by the identity of both the hydrophobic residue in the variable motif and the disordered N terminus. The latter result is particularly striking, as disordered regions of TADs that do not undergo coupled folding and binding are often removed or ignored in biophysical and structural studies because they typically do not contribute to overall affinity (9–11, 25, 36–38, 35, 39, 40).
To obtain further evidence for the unexpected role of the disordered N terminus on the conformational behavior of ETV/PEA3•Med25 PPIs, we also tested the effects of removing the four variable N-terminal residues (∆Nt) of ETV1 and ETV4 in kinetics experiments. The resulting variants ETV1∆Nt and ETV4∆Nt displayed differential changes in conformational behavior from the parent TADs (Fig. 3C), in addition to a slight (∼1.7-fold) gain in affinity for both variants (Fig. 3D). Removal of the ETV1 N terminus resulted in significant conformational redistribution; kinetic analysis indicated that the ETV1∆Nt•Med25 complex exchanged between two equally populated conformational substates on a similar timescale to the C1–C2 transition of the parent ETV1•Med25 complex. On the other hand, the ETV4∆Nt•Med25 complex populated three conformational substates in an analogous manner to the parent ETV4•Med25 complex. These results therefore support a direct role of the N-terminal residues of the ETV1/ETV5 TADs, but not the ETV4 TAD, in biasing the conformational behavior of the native TAD•Med25 complexes.
Variable Regions of the ETV/PEA3 TADs Differentially Engage with the Med25 Surface.
We next directly examined the structural basis by which the two variable regions in the ETV/PEA3 TADs modulate the bound ETV/PEA3•Med25 conformations using NMR spectroscopy. A conservative mutagenesis strategy was pursued, where minimally perturbing mutations were individually introduced into unlabeled TADs, and then CSP analysis of 1H,13C-HSQC spectra of Med25 bound to the native or mutated TAD was performed to identify Med25 methyl groups affected by the mutation. Analysis of CSP differences in the mutant TAD•Med25 HSQC spectrum can therefore detect Med25 residues in direct proximity to the mutated site in the complex and also has the potential to reveal allosteric connections if the effects of the mutation are propagated from the interaction site (41). Furthermore, this strategy avoids the significant experimental challenge associated with NMR analysis of labeled ETV/PEA3 TADs bound to Med25, where a significant fraction of peaks are too broad to detect due to conformational exchange (30). Here, conservative mutations were introduced into the two key variable regions of the ETV1 and ETV4 TADs to detect differences in engagement modes that could explain the effects of these regions on ETV/PEA3•Med25 conformational states.
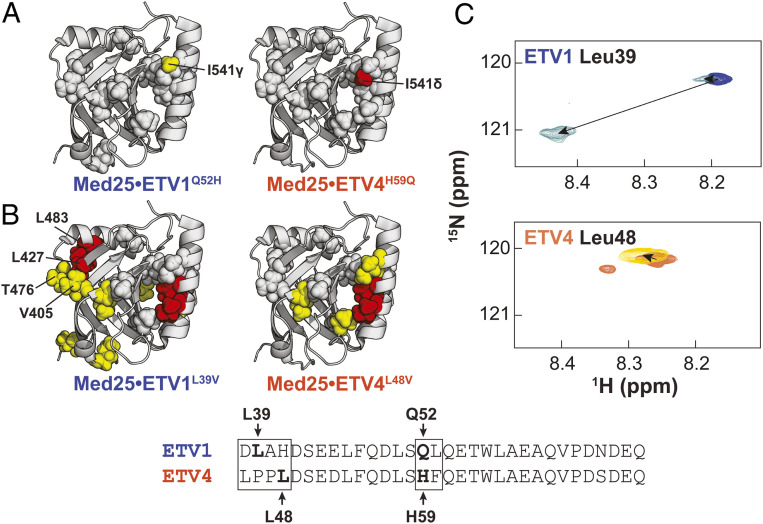
Mutations were first made within the variable motif of the helical binding region by swapping the variable polar residue between ETV1 and ETV4 to form ETV1Q52H and ETV4H59Q (note: residue numbers for ETV4 are shifted by +7 compared to ETV1 and ETV5), based on the observation that this change did not affect the populations of conformational states in kinetics experiments (SI Appendix, Table S3). Indeed, the 1H,13C-HSQC spectra of the mutant ETV/PEA3•Med25 complexes were almost identical to those of the native complexes except for single shifts in unique methyl peaks of Ile541 (Fig. 4A): the ETV4H59Q variant produced a large shift (0.032 ppm) in the Med25 Ile541δ peak, whereas the analogous ETV1Q52H variant produced a smaller perturbation (0.015 ppm) in the Ile541γ peak. These highly localized shifts are consistent with the mutations causing proximity-based perturbations near the mutation site in the complex. In addition, the fact that the individual mutations perturb unique methyl groups originating from the same residue suggest that the native ETV1 QL and ETV4 HF motifs are engaged in unique interactions in a similar interface.
Fig. 4.
ETV/PEA3 variable regions engage in unique interactions with the Med25 surface. Effects of conservative mutations in the (A) helical binding region and (B) N termini are plotted on the structure of Med25. Yellow = 0.015 to 0.030 ppm, red ≥ 0.030 ppm. Residues discussed in the text are labeled. Gray spheres denote residues that undergo identical perturbations in both parent and mutant complexes. Residues chosen for mutation are bolded and labeled in the alignment. (C) Chemical shift perturbations of 150 µM ETV1 (Top) and ETV4 (Bottom) TADs in the absence (blue and orange, respectively) and presence (light blue and maroon, respectively) of 280 µM unlabeled Med25. TADs were selectively 15N labeled at the positions noted. Small secondary peaks in free ETV4 spectra were observed and likely arose from isomerization of the two tandem Pro residues in the N-terminal region.
Next, we introduced mutations into the disordered N terminus. Specific sites for mutations in this region were not immediately obvious from previous data, so several point mutations were made; Leu to Val mutations ETV1L39V and ETV4L48V sufficed to produce measurable differences in bound 1H,13C-HSQC spectra compared to the native TADs, without significantly altering the overall spectra (Fig. 4B and SI Appendix, Figs. S22 and S23). In contrast to mutations in the helical binding region, ETV1L39V and ETV4L48V affected a larger overall site on the Med25 structure, indicating possible indirect effects stemming from these mutations. Indeed, both of these N-terminal mutants altered an overlapping subset of residues in the core binding site, which would be occupied by the TAD helical binding region. Nonetheless, evidence for a direct interaction of the ETV1 N terminus was also apparent from the ETV1L39V•Med25 spectrum; several shifts from the native ETV1•Med25 spectrum were observed in a cluster of residues in a distal site involving Val405, Leu427, Thr476, and Leu483. Conversely, this site was unaffected by the ETV4L48V variant, suggesting this interaction is made only by the ETV1 N terminus. Consistent with a functional role for differential engagement of this distal site, our kinetics data demonstrated that removal of the ETV1 N terminus significantly alters the ETV1•Med25 conformational ensemble, whereas removal of the ETV4 N terminus only slightly affected the ETV4•Med25 ensemble (Fig. 3C).
To further test this model, we synthesized ETV1 and ETV4 TADs that were selectively 15N labeled at single Leu residues and analyzed CSPs in 1H,15N-HSQC spectra upon binding of unlabeled Med25. Analysis of Leu residues in the helical binding region was attempted by this method; however, in almost all cases these peaks were too broad to detect when the TADs were bound to unlabeled Med25. In contrast, peaks for Leu residues in the N terminus remained relatively sharp upon binding to Med25, likely due to these regions retaining more structural disorder in the complex (30). Comparison of CSPs of Leu residues in the N termini of ETV1 and ETV4 were consistent with the proposed differential interaction with Med25: Leu39 in the ETV1 N terminus underwent a large (∼0.27 ppm) shift, whereas Leu48 in the ETV4 N terminus shifted only slightly (0.02 ppm) upon addition of unlabeled Med25 (Fig. 4C). Interestingly, we also observed a minor peak in spectra of bound ETV1 Leu39 that was slightly shifted from the unbound position, perhaps representing one of the lowly populated ETV1•Med25 conformations (Fig. 2D) where the N terminus is weakly bound to the Med25 surface.
ETV/PEA3•Med25 Conformational Changes Involve Shifts in Med25 Structure.
Altogether, these data support a model where the conformational differences between ETV/PEA3•Med25 complexes are caused by the ETV/PEA3 variable regions engaging with the Med25 surface in unique and sequence-dependent manners. While NMR and mutagenesis data revealed that the variable N terminus affects ETV/PEA3•Med25 conformation via differential engagement with the Med25 surface, the mechanism by which bound conformational behavior is affected by the variable motif in the helical binding region is unclear. Interestingly, NMR analysis demonstrated that the ETV1 and ETV4 variable motifs localize to a similar region (Fig. 4A), which indicates that conformational differences caused by this motif originate from distinct interactions in the same site. We thus reasoned that this would likely involve remodeling of the Med25 ABD. To test this hypothesis, we examined changes in the bound TAD•Med25 1H,13C-HSQC spectra produced by a point mutation in the variable motif that significantly redistributes the populations of conformational states. The ETV4F60L mutation was selected because this small change in residue identity caused a drastic conformational redistribution (Fig. 3B, compare Bottom Left and Top Right) to favor the initial bound substate (C1).
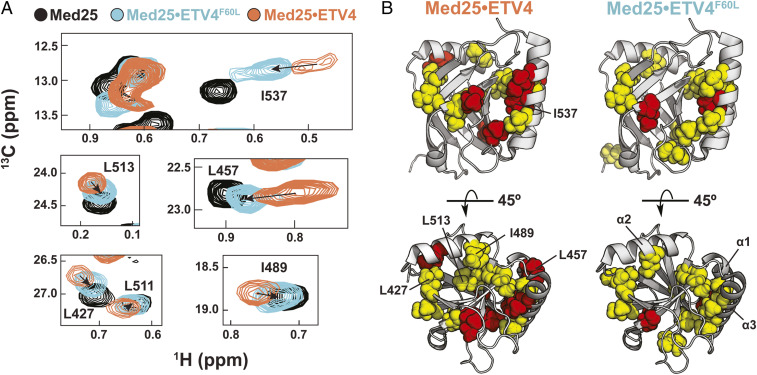
Comparison of the 1H,13C-HSQC spectra of ETV4•Med25 and ETV4F60L•Med25 revealed several Med25 peaks that are significantly perturbed when bound to native ETV4 shift less drastically when ETV4F60L is bound (Fig. 5A). This result is in striking contrast mutation of the polar residue in the variable motif (ETV4H59Q), which resulted in the movement of just a single Med25 peak (Fig. 4A). Significantly, the ETV4F60L variant elicited weaker CSPs around the core binding site than native ETV4 (Fig. 5B), suggesting that several of the large CSPs in this region are tied to the conformational changes. Furthermore, this behavior was observed for peaks representing residues that are buried or in allosteric regions of the protein, including the β-barrel core, the interface between the β-barrel and the allosteric α2 helix, and the interface between the C-terminal α3 helix and the allosteric α1 helix (Fig. 5B). These data are therefore consistent with a direct role for ABD conformational plasticity in molecular recognition, which likely enables specific interactions by revealing topology in the core binding site.
Fig. 5.
Structural shifts in the Med25 ABD accompany the conformational changes of ETV/PEA3•Med25 complexes. (A) Examples of Med25 resonances undergoing shifts toward the unbound position (black) upon ETV4F60L (cyan) mutation. (B) Comparison of chemical shift perturbations from binding of ETV4 (Left) and ETV4F60L (Right) demonstrate that conformational changes involve the binding site and allosteric regions of Med25. Residues shown in A are labeled on the structures. Yellow = 0.040 to 0.080 ppm, red > 0.080 ppm.
Discussion
The exceptional sequence variability of functional TADs—characterized only by a general preponderance of acidic and hydrophobic amino acids—has remained a molecular recognition enigma over the past several decades (1–5, 10, 12–14). There have been several recognition models advanced to account for the large variety of functional TADs, most of which propose that TAD•ABD recognition occurs via nonspecific intermolecular interactions (3, 9–12). The major driving force of association in these models is thus the stochastic burial of hydrophobic sidechains rather than the formation of defined intermolecular contacts typical for well-structured PPIs. However, there is limited direct biophysical evidence for nonspecific mechanisms, and the available biophysical data rely almost entirely on 1) the observation of similar CSP patterns of unique TAD sequences binding to the same ABD (11), and 2) NMR paramagnetic relaxation enhancement (PRE) patterns showing multiple bound orientations of the TAD on the ABD surface (9–11). Alternative explanations also exist for both of these observations: similar CSP trajectories are expected if the ABD undergoes a conserved conformational change upon binding (42–44), and significant PREs can be observed from lowly populated (<10%) substates that place the paramagnetic spin label in close proximity to the interacting partner (45, 46). The degree to which activator•coactivator recognition is truly nonspecific in such examples is therefore unclear. Unfortunately, this widely accepted view of activator•coactivator recognition also represents a primary reason why these PPIs have been traditionally considered “untargetable” (29).
Here, we scrutinized these recognition models by subjecting the dynamic PPIs formed between the ETV/PEA3 family of TADs and their binding partner Med25 to detailed mechanistic and structural dissection. We found that these interactions exhibited a striking degree of conformational sensitivity to small changes in TAD sequence, which is inconsistent with PPIs driven by nonspecific intermolecular interactions. Instead, both ordered and disordered regions of ETV/PEA3 TADs have the capacity to engage in a finite set of specific interactions with the Med25 surface. This recognition mechanism is further enabled by an underappreciated role of ABD plasticity in molecular recognition: in contrast to the shallow and largely featureless ETV/PEA3 binding surface presented by Med25 in the unbound state, the Med25 ABD undergoes significant remodeling upon complex formation and thus likely plays a direct role in enabling different interaction modes (47).
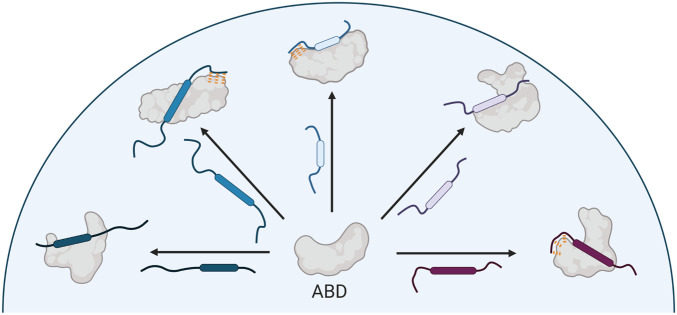
Taken together, these data reveal alternative specific mechanisms of TAD•ABD molecular recognition that rationalize the extreme sequence variability of functional TADs (Fig. 6), which has been proposed to be due to entirely nonspecific recognition mechanisms (3, 9–11). In contrast to nonspecific mechanisms, the model of TAD•ABD molecular recognition outlined here highlights a direct path to targeting strategies for small molecule therapeutics or probes of activator•coactivator complexes. Specifically, the role of ABD plasticity in recognition suggests that development of molecules that stabilize specific ABD conformational states may be a more effective targeting strategy than directly targeting the topologically challenging TAD•ABD interface. We previously obtained allosteric modulators of Med25 and other ABDs by covalent targeting of dynamic structural elements (32, 48), which supports the idea that ABD plasticity can be effectively exploited by small molecules.
Fig. 6.
Specific recognition model emerging from this work. Activator binding domains recognize a diversity of activators via conformational plasticity and unique specific intermolecular interactions. Rounded boxes in the TADs represent the “canonical” recognition motifs. Orange dashes indicate interactions made by disordered regions outside the recognition motifs.
A fundamental biological question that emerges from our study is how molecular recognition mechanisms affect function. In general, it is still exceptionally challenging to relate PPI function and affinity; in processes where dynamic PPIs serve as critical functional events, such as transcription and proteostasis, there is often little or no correlation between affinity and functional activity (49, 50). Several factors play into this observation, such as subcellular localization and concentration, but a potentially significant factor is the mechanism by which the complex forms. For example, in our current study we observed that the lifetimes of individual ETV/PEA3•Med25 conformations varied up to two orders of magnitude (SI Appendix, Table S2), and therefore shifting the conformational ensemble toward longer-lived substates could have significant functional outcomes without necessitating changes in affinity (51). Furthermore, the fact that the exceptionally dynamic interactions examined herein were nonetheless specific raises the question of whether molecular recognition in cognate activator•coactivator complexes are driven, like many traditional PPIs, by a small number of hotspot or anchoring residues (52–54). Future development of detailed structural models of these dynamic PPIs in combination with extensive mutagenesis and conformational analysis will facilitate a deeper understanding of the molecular drivers of specificity.
Altogether, our results implicate a critical need to reevaluate the nonspecific recognition models that are common for dynamic biomolecular interactions—such as those made by intrinsically disordered proteins and RNA (11, 55)—to develop a deeper understanding of the relationship between the function of biomolecular interactions and the mechanisms by which they are formed.
Methods
Protein Expression and Purification.
Med25 AcID was expressed and purified from a pET21b-Med25(394-543)-His6 plasmid from E. coli BL21 (DE3) cells as described previously (full details in SI Appendix) (32). Uniformly 13C,15N labeled Med25 for NMR experiments was expressed identically except using M9 minimal media supplemented with 1 g/L 15NH4Cl, 2 g/L 13C-D-glucose, and 0.5% 13C,15N-labeled Bioexpress media. Protein identity was confirmed by mass spectrometry (Agilent Q-TOF).
Peptide Synthesis.
The peptides used in this study were prepared using standard Fmoc solid-phase peptide synthesis on a Liberty Blue Microwave Peptide Synthesizer (CEM). 15N-Leu labeled peptides were synthesized using Fmoc-15N-Leu in place of unlabeled Fmoc-Leu at the specified positions. Details of synthesis and peptide characterization data are included in SI Appendix.
Stopped-Flow Kinetics.
Stopped-flow kinetic assays were performed using a Kintek SF-2001 stopped flow instrument equipped with a 100-W Xe arc lamp in two-syringe mode. All experiments were completed at 10 °C in stopped-flow buffer (10 mM sodium phosphate, 100 mM NaCl, 2% dimethylsulfoxide (DMSO), 1% glycerol, 0.001% Nonidet P-40, pH 6.8). All concentrations reported are after mixing. The 4-DMN fluorophore was excited at 440 nm, and fluorescence intensity was measured at wavelengths >510 nm using a long-pass filter (Corion). Further details, including an explanation of how reported values were determined, are in SI Appendix.
NMR Spectroscopy.
Constant time 1H,13C-HSQC experiments were performed with uniformly 13C,15N-labeled Med25 in NMR buffer (20 mM sodium phosphate pH 6.5, 150 mM NaCl, 3 mM dithiothreitol (DTT), 10% D2O, and 2% DMSO) on a Bruker 600 MHz instrument equipped with a cryoprobe. HSQC experiments were processed in NMRPipe (56) and visualized with NMRFAM-Sparky (57). Chemical shift perturbation analyses were performed on samples with 1.1 equivalents of unlabeled binding partner, which results in ≥96% bound Med25 based on measured Kd values. Assignments of sidechain methyl resonances of free Med25 were achieved through 3D H(CCCO)NH and (H)CC(CO)NH TOCSY (TOtal Correlation SpectroscopY) experiments (23 ms TOCSY mixing time) performed with a sample of 600 µM 13C,15N Med25 on a Bruker 800 MHz instrument equipped with a cryoprobe. Further details about assignment and data analysis can be found in SI Appendix.
Supplementary Material
Acknowledgments
Financial support for this work was received from NIH R01 GM65530, R35 GM136356 (to A.K.M.), CA207272 (to T.C.) and Welch Foundation Grant A-1987 (to C.A.F.). M.J.H. was supported by a fellowship from the Department of Education (Graduate Assistance in Areas of National Need), and B.S.M. by a fellowship from the Michigan Life Sciences May-Walt Fellowship Fund. We thank the A.K.M. laboratory for helpful comments and D. Sahu for assistance with performing the NMR assignment experiments. Figs. 1 and 6 were created using BioRender.com.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013244117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Ma J., Ptashne M., A new class of yeast transcriptional activators. Cell 51, 113–119 (1987). [DOI] [PubMed] [Google Scholar]
- 2.Hope I. A., Mahadevan S., Struhl K., Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature 333, 635–640 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Sigler P. B., Transcriptional activation. Acid blobs and negative noodles. Nature 333, 210–212 (1988). [DOI] [PubMed] [Google Scholar]
- 4.Mapp A. K., Ansari A. Z., A TAD further: Exogenous control of gene activation. ACS Chem. Biol. 2, 62–75 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Piskacek S., et al., Nine-amino-acid transactivation domain: Establishment and prediction utilities. Genomics 89, 756–768 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Krishnamurthy M., et al., Caught in the act: Covalent cross-linking captures activator-coactivator interactions in vivo. ACS Chem. Biol. 6, 1321–1326 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boija A., et al., Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabari B. R., et al., Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brzovic P. S., et al., The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol. Cell 44, 942–953 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warfield L., Tuttle L. M., Pacheco D., Klevit R. E., Hahn S., A sequence-specific transcription activator motif and powerful synthetic variants that bind Mediator using a fuzzy protein interface. Proc. Natl. Acad. Sci. U.S.A. 111, E3506–E3513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuttle L. M., et al., Gcn4-mediator specificity is mediated by a large and dynamic fuzzy protein-protein complex. Cell Rep. 22, 3251–3264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravarani C. N., et al., High-throughput discovery of functional disordered regions: Investigation of transactivation domains. Mol. Syst. Biol. 14, e8190 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnold C. D., et al., A high-throughput method to identify trans-activation domains within transcription factor sequences. EMBO J. 37, e98896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staller M. V., et al., A high-throughput mutational scan of an intrinsically disordered acidic transcriptional activation domain. Cell Syst. 6, 444–455.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minter A. R., Brennan B. B., Mapp A. K., A small molecule transcriptional activation domain. J. Am. Chem. Soc. 126, 10504–10505 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Rowe S. P., Mapp A. K., Assessing the permissiveness of transcriptional activator binding sites. Biopolymers 89, 578–581 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buhrlage S. J., et al., Amphipathic small molecules mimic the binding mode and function of endogenous transcription factors. ACS Chem. Biol. 4, 335–344 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majmudar C. Y., et al., Sekikaic acid and lobaric acid target a dynamic interface of the coactivator CBP/p300. Angew. Chem. Int. Ed. Engl. 51, 11258–11262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chrivia J. C., et al., Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365, 855–859 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Ruas J. L., Poellinger L., Pereira T., Functional analysis of hypoxia-inducible factor-1 α-mediated transactivation. Identification of amino acid residues critical for transcriptional activation and/or interaction with CREB-binding protein. J. Biol. Chem. 277, 38723–38730 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Yang F., et al., An ARC/mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442, 700–704 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Sela D., et al., Role for human mediator subunit MED25 in recruitment of mediator to promoters by endoplasmic reticulum stress-responsive transcription factor ATF6α. J. Biol. Chem. 288, 26179–26187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verger A., et al., The mediator complex subunit MED25 is targeted by the N-terminal transactivation domain of the PEA3 group members. Nucleic Acids Res. 41, 4847–4859 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boehr D. D., McElheny D., Dyson H. J., Wright P. E., The dynamic energy landscape of dihydrofolate reductase catalysis. Science 313, 1638–1642 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Sugase K., Dyson H. J., Wright P. E., Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447, 1021–1025 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Vallurupalli P., Hansen D. F., Kay L. E., Structures of invisible, excited protein states by relaxation dispersion NMR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 105, 11766–11771 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darnell J. E. Jr, Transcription factors as targets for cancer therapy. Nat. Rev. Cancer 2, 740–749 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Bushweller J. H., Targeting transcription factors in cancer - from undruggable to reality. Nat. Rev. Cancer 19, 611–624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mapp A. K., Pricer R., Sturlis S., Targeting transcription is no longer a quixotic quest. Nat. Chem. Biol. 11, 891–894 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landrieu I., et al., Characterization of ERM transactivation domain binding to the ACID/PTOV domain of the mediator subunit MED25. Nucleic Acids Res. 43, 7110–7121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Currie S. L., et al., ETV4 and AP1 transcription factors form multivalent interactions with three sites on the MED25 activator-interacting domain. J. Mol. Biol. 429, 2975–2995 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henderson A. R., et al., Conservation of coactivator engagement mechanism enables small-molecule allosteric modulators. Proc. Natl. Acad. Sci. U.S.A. 115, 8960–8965 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loving G., Imperiali B., A versatile amino acid analogue of the solvatochromic fluorophore 4-N,N-dimethylamino-1,8-naphthalimide: A powerful tool for the study of dynamic protein interactions. J. Am. Chem. Soc. 130, 13630–13638 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vojnic E., et al., Structure and VP16 binding of the mediator Med25 activator interaction domain. Nat. Struct. Mol. Biol. 18, 404–409 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Shammas S. L., Travis A. J., Clarke J., Allostery within a transcription coactivator is predominantly mediated through dissociation rate constants. Proc. Natl. Acad. Sci. U.S.A. 111, 12055–12060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radhakrishnan I., et al., Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell 91, 741–752 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Pomerantz W. C., et al., Profiling the dynamic interfaces of fluorinated transcription complexes for ligand discovery and characterization. ACS Chem. Biol. 7, 1345–1350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giri R., Morrone A., Toto A., Brunori M., Gianni S., Structure of the transition state for the binding of c-Myb and KIX highlights an unexpected order for a disordered system. Proc. Natl. Acad. Sci. U.S.A. 110, 14942–14947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang N., Lodge J. M., Fierke C. A., Mapp A. K., Dissecting allosteric effects of activator-coactivator complexes using a covalent small molecule ligand. Proc. Natl. Acad. Sci. U.S.A. 111, 12061–12066 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dahal L., Kwan T. O. C., Shammas S. L., Clarke J., pKID binds to KIX via an unstructured transition state with nonnative interactions. Biophys. J. 113, 2713–2722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhuravleva A., Gierasch L. M., Substrate-binding domain conformational dynamics mediate Hsp70 allostery. Proc. Natl. Acad. Sci. U.S.A. 112, E2865–E2873 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demarest S. J., et al., Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature 415, 549–553 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Lee C. W., Martinez-Yamout M. A., Dyson H. J., Wright P. E., Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein. Biochemistry 49, 9964–9971 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haberz P., Arai M., Martinez-Yamout M. A., Dyson H. J., Wright P. E., Mapping the interactions of adenoviral E1A proteins with the p160 nuclear receptor coactivator binding domain of CBP. Protein Sci. 25, 2256–2267 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clore G. M., Tang C., Iwahara J., Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement. Curr. Opin. Struct. Biol. 17, 603–616 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anthis N. J., Clore G. M., Visualizing transient dark states by NMR spectroscopy. Q. Rev. Biophys. 48, 35–116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schreiber G., Keating A. E., Protein binding specificity versus promiscuity. Curr. Opin. Struct. Biol. 21, 50–61 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang N., et al., Ordering a dynamic protein via a small-molecule stabilizer. J. Am. Chem. Soc. 135, 3363–3366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Z., et al., Targeting the transcriptional machinery with unique artificial transcriptional activators. J. Am. Chem. Soc. 125, 12390–12391 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Cesa L. C., et al., X-linked inhibitor of apoptosis protein (XIAP) is a client of heat shock protein 70 (Hsp70) and a biomarker of its inhibition. J. Biol. Chem. 293, 2370–2380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paakinaho V., et al., Single-molecule analysis of steroid receptor and cofactor action in living cells. Nat. Commun. 8, 15896 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clackson T., Wells J. A., A hot spot of binding energy in a hormone-receptor interface. Science 267, 383–386 (1995). [DOI] [PubMed] [Google Scholar]
- 53.Bogan A. A., Thorn K. S., Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280, 1–9 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Rajamani D., Thiel S., Vajda S., Camacho C. J., Anchor residues in protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 101, 11287–11292 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jankowsky E., Harris M. E., Specificity and nonspecificity in RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 16, 533–544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delaglio F., et al., NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 57.Lee W., Tonelli M., Markley J. L., NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.