Graphical abstract
Keywords: Dysbiosis, Host-microbe interactions, Complex diseases, Oral microbiome, Oral biofilm
Abstract
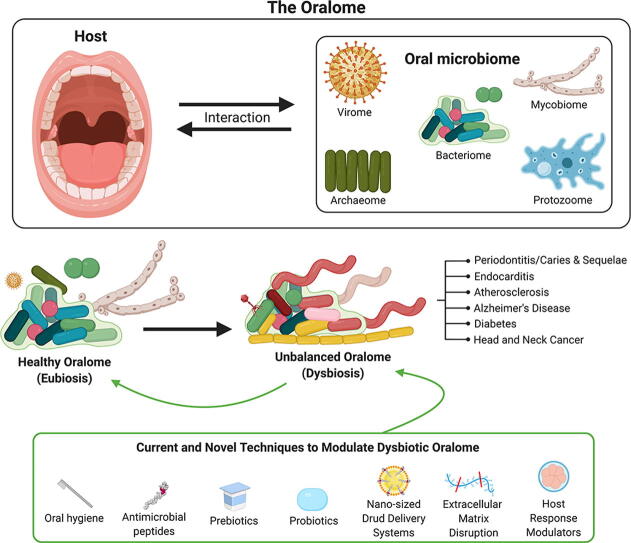
The oralome is the summary of the dynamic interactions orchestrated between the ecological community of oral microorganisms (comprised of up to approximately 1000 species of bacteria, fungi, viruses, archaea and protozoa - the oral microbiome) that live in the oral cavity and the host. These microorganisms form a complex ecosystem that thrive in the dynamic oral environment in a symbiotic relationship with the human host. However, the microbial composition is significantly affected by interspecies and host-microbial interactions, which in turn, can impact the health and disease status of the host. In this review, we discuss the composition of the oralome and inter-species and host-microbial interactions that take place in the oral cavity and examine how these interactions change from healthy (eubiotic) to disease (dysbiotic) states. We further discuss the dysbiotic signatures associated with periodontitis and caries and their sequalae, (e.g., tooth/bone loss and pulpitis), and the systemic diseases associated with these oral diseases, such as infective endocarditis, atherosclerosis, diabetes, Alzheimer’s disease and head and neck/oral cancer. We then discuss current computational techniques to assess dysbiotic oral microbiome changes. Lastly, we discuss current and novel techniques for modulation of the dysbiotic oral microbiome that may help in disease prevention and treatment, including standard hygiene methods, prebiotics, probiotics, use of nano-sized drug delivery systems (nano-DDS), extracellular polymeric matrix (EPM) disruption, and host response modulators.
1. Introduction
More than three centuries ago, Antony van Leeuwenhoek was the first person to observe microbes, possibly bacteria, from his own dental plaque with the use a microscope that he constructed [1]; thereby laying down the foundations of microbiology. Since then, new discoveries have highlighted that these microbes interact with their human host. A recent estimate shows that 3.8 × 1013 microorganisms colonize the human body, accounting for about half of the total cells on a human body [2]. In aggregate, these organisms are known as the human microbiome [3], [4].
2. Microbial habitats in the human body
The NIH Human Microbiome Project (HMP), which was launched in 2007, comprised 18 different studies to map and characterize the human microbiome and its role in human health and disease. It is well appreciated that the human microbiome is present in nearly every human body site [5], and the HMP has established, thus far, that there are 48 main microbial habitats in the human body [6]. According to the HMP [6], 34% of all primary microbial habitats were associated with human skin, 25% of all habitats were related to the gastrointestinal tract, and 20% of all habitats were associated with cavities of the head and neck region.
Among these main human habitats, the oral cavity is a challenging environment for microbial survival, since it undergoes high daily fluctuations in nutrient supply, temperature, pH, sheer and mechanical forces from mastication and hygiene practices, and chemical exposure from hygiene, pharmaceutical or toxic/smoking products [7]. Yet it retains a rich and complex ecosystem, harboring different micro-colonizers that thrive in this dynamic environment – the oral microbiome [8]
3. The oral microbiome and the oralome
The oral microbiome is defined as a community of microorganisms of up to 1000 total microbial species, comprising bacteria, fungi, viruses, archaea and protozoa that live in the oral cavity [1], [9], [10], [11], [12]. A bacterial predominance in the oral microbiome has hindered and limited the term “core microbiome” to that of a bacterial microbiome [9]. Thus, most reports usually focus only on the oral bacteria within the microbiome, while other oral biomes (i.e. fungi, archaea, protozoa and the viral biomes) receive much less attention. Although there is limited literature on these other biomes, they remain relevant [1], [9], [13], [14]. Thus, we will discuss each of these biomes in the context of the oral cavity.
3.1. Oral bacterial microbiome – the bacteriome
Due to changes in shear forces, nutrient/energy supply, temperature, pH, and oxygen content in different environments, bacteria have evolved by leveraging specific survival strategies, including co-aggregation of different cells into communities embedded in extracellular matrices –biofilms [10]. Despite the existence of planktonic bacteria in the oral cavity, most of the oral microbiome exists in a biofilm state, known as the oral biofilm.
3.1.1. The oral bacterial biofilm
Biofilms have been defined as a structured community of aggregated bacterial cells (either from the same species or multi-species) embedded and enclosed in a self-produced extracellular polymeric matrix (EPM) and adherent to an inert or living surface [15], [16], [17], [18]. Specifically, for oral bacterial biofilms (commonly referred to as “oral biofilms”), features of the oral cavity have shaped bacterial communities to adapt to high cell density, which, in turn create a micro-environment capable of modulating the pH, redox, and oxygen levels in its core [7], [16], [19].
Oral biofilms are formed by an initial adhesion of planktonic bacteria, known as “early colonizers” [20]. Typically, these early colonizers are saccharolytic aerobes and facultative anaerobes that primarily feed on oral glycoproteins and salivary mucins, with 80% of the early colonizers being represented by Streptococcus species [7], [20], [21]. Surface colonization occurs by bacterial attachment to an oral surface (e.g. dental surface) via specific surface adhesins [7]. After the initial colonization, the surface-attached bacteria change their metabolic and gene expression profiles to produce and secrete EPMs, which, for oral biofilms, is made up of polysaccharides, proteins, lipids and extracellular DNA (eDNA) [7], [22], [23], [24], [25], [26]. Interestingly, different microorganisms have been shown to depend on eDNA to form biofilms in monocultures, which may indicate that deoxyribonucleases (DNases) can be used to disrupt and control biofilm growth [26], [27], [28]. Production and secretion of EPMs provides several advantages to the oral microbiota, such as enhanced nutrient availability and protection from environmental stresses [29]. For instance, cells embedded in biofilm matrices are up to 1000-fold more tolerant to antibiotics compared to their planktonic counterparts, and therefore more difficult to disperse [30].
Overall, the oral bacterial biofilm experiences different levels of oxygenation within its own structure [31], [32], [33], [34]. For instance, analyzing oxygenation levels of ex vivo supragingival plaque biofilms (~300 µm thickness), von Ohle et al. [34] found anaerobic conditions (<0.5% oxygen [35]) in layers deeper than 220 µm. In this context, ex vivo biofilms exhibit beneficial conditions for the growth of different microbes that thrive under different levels of oxygenation. Thus, within these biofilm structures, aero-tolerant taxa are present on the exterior, whereas the interior anaerobic compartments of the biofilm provide the environmental conditions for attachment and growth of proteolytic obligate anaerobes, including methanogens and sulfate-reducers, also known as “late colonizers” [7], [20], [21]. This oxygenation gradient also suggests the existence of potential nutrient gradients [33].
Using oligotyping analysis, Eren et al. [36] demonstrated that oral bacterial biofilm genera have different distribution/abundance relative to the three distinct habitat zones - dental plaque, tongue dorsum, and keratinized gingiva. This finding led to the site-specialist hypothesis for oral bacterial biofilms, which predicts that a particular strain will actively find its preferential growth site (e.g., dental plaque, tongue dorsum, and keratinized gingiva) and thereby thrive [33]. Further discussing this hypothesis, Mark Welch et al. [33] point out that some genera can be either generalists, such as the Veillonella genus (e.g. Veillonella atypica seems to be a tongue dorsum specialist, Veillonella parvula/dispar 2 seems to be a dental plaque specialist, and Veillonella rogosae seems to be a keratinized gingiva specialist) or specialists that strongly specialize to one site, such as Capnocytophaga and Corynebacterium genera that are specialized to dental plaque sites.
In general, bacterial biofilms display a mushroom-shaped, whereas under turbulent flow, biofilms display a more elongated structure that is capable of rapid oscillations [15], [37], [38]. However, recent studies on dental plaque and togue dorsum biofilms demonstrated different shapes.
For supragingival biofilms, the “corncob” structure - long filamentous structures formed by Corynebacterium and/or actinomyces coated with cocci at the edges - has been reported for over 40 years [33], [39]. Recently, Mark Welch et al. [32] demonstrated the “hedgehog” structure, in which, filaments of Corynebacterium spp. radiate outward from the dental surface, forming a structured habitat inhabited by other taxa at specific positions. For example, Streptococcus spp. occupy an outer shell forming the “corncob” structure and they share the habitat with Porphyromonas spp. and Haemophilus spp. or Aggregatibacter spp. Microaerophilic taxa, such as Fusobacterium spp. and Leptotrichia spp., on the other hand, possibly occupy the anaerobic layers close to the base of the structure.
For subgingival biofilms, four different layers were found by Zijnge et al. [40]. The basal layer (close to the dental surface) was formed by Actinomyces spp. and other unidentified species (due to low fluorescence outcome), followed by a second layer comprised of spindle-like bacteria, such as F. nucleatum and Tannarella spp. The third layer was formed by filamentous, rod-shaped and coccoid-shaped cells belonging to the Cytophaga-Flavobacterium-Bacteroides cluster. Finally, on the top of the biofilm, the authors found a ‘palisade’-like lining layer, which was in close contact with eukaryotic cells. Unfortunately, subgingival biofilms are not easily analyzed without the loss of structural integrity [40]. Thus, more studies using novel strategies are still necessary to better understand the structure of subgingival plaque biofilms and to specifically unveil how shear forces and different flow parameters in the mouth shape both the supragingival and subgingival dental plaque bacterial biofilm structure.
For the tongue dorsum, Mark Welch et al. [33] found a third different structure, but with similar degree of organization as the other two structures. The human epithelial tissue occupied the central core of the biofilm followed by a layer of Actinomyces spp.. Streptococcus spp. were located in the exterior layer and in stripes in the interior of the biofilm. Other taxa, such as Rothia spp., Neisseria spp., Veillonella spp. were present in clusters and stripes in the interior of the biofilm as well, suggesting that the biofilm grew outwardly from the central core.
Unfortunately, we were unable to find any articles describing biofilm structures on keratinized gingiva; thus, more studies are required in the field. For more details on the biogeography of oral bacterial biofilms and the site-specialist hypothesis, please refer to Mark Welch et al. [32], [33].
Approximately 700 bacterial species have been identified in the oral cavity [7], [41], [42], making the oral cavity the second largest bacterial community in the human body, after the gut [1]. Aas et al. [41] detected 141 bacterial species in the oral cavity; among them, the most common species belonged to the Gemella, Granulicatella, Streptococcus, and Veillonella genera.
3.2. Oral viral biome – the virobiome/virome
Human oral viral biome, also known as the “oral virome” or “oral virobiome”, is a highly conserved [43] and highly personalized community; to such a degree that its composition can vary depending on the host’s sex [44]. The vast majority of viruses in the oral cavity are bacteriophages [44], [45], [46], which exhibit a very stable lytic/lysogenic cycle. This lytic process has the potential to exterminate bacterial species in the community or impart new functions on the oral bacteria, and thereby completely change these human bacterial communities [44]. Some of these bacteriophages are associated with Veillonella spp. and Streptococcus spp., two of the main commensal bacterial genera in the oral cavity [44].
In healthy patients, the main bacteriophage families found in the oral cavity were Siphoviridae, Myoviridae, and Podoviridae; all belonging to the Caudovirus order [45], [46]. Myoviridae and Podoviridae are predominantly lytic viruses that rapidly eliminate their bacterial hosts, while Siphoviridae are largely lysogenic viruses, establishing a dynamic equilibrium with their associated host species [47]. Interestingly, this suggests that the oral virome may play a big role in controlling the bacterial population in the oral cavity, since this bacteriophage dominance in the virome seems to be correlated with the bacterial dominance of the oral microbiome, and phages are thought to account for 20–80% of total bacterial death; thus, representing a profound bacterial growth limiting factor. Also, in 2018, de la Cruz Peña [48] found eight new viruses that are naturally abundant in saliva of healthy patients, such as the unculturable virus 92-C13, which seems to belong to the Caudovirales order as well.
So far, eukaryotic viruses, such as Herpesviridae, Papillomaviridae, and Anelloviridae are among the most common eukaryotic virus families present in healthy patients. Among them, Human Papillomavirus (HPV), Human cytomegalovirus (CMV), Herpes simplex virus type-1 (HSV-1), and e Epstein-Barr Virus (EBV), have been found in asymptomatic healthy individuals [47].
However, only few studies have evaluated the oral viral community [43], [44], [45], [48], [49] and, thus, more studies are still necessary to evaluate the role of the virome in the oral microbiome.
3.3. Oral fungal microbiome - the mycobiome
The human oral cavity is also colonized by fungi/yeast species [50]. Ghannoum el al. [13], in 2010, first described the “basal oral mycobiome”. The authors found a total of 101 oral fungal species, among 74 culturable- and 11 nonculturable-genera. Among those identified, three (i.e. Aspergillus, Fusarium and Cryptococcus) are known to be pathogenic in humans and are not known to be oral colonizers. The authors proposed that the pathogenicity of these fungal species might be controlled by other commensal oral fungi.
Following this study, Dupuy et al [14], in 2014, identified 5 additional fungal genera in the oral mycobiome. Among those, the Malassezia genera has been described as a skin commensal and an opportunistic pathogen associated with scalp disorders [51] and, now, is being considered a predominant member of the commensal “basal oral mycobiome” [52], [53]. Recent research by Peters et al [54], in 2017, increased the total number of commensal fungal species to 154.
The most common commensal fungi and members of the basal oral mycobiome are the Candida genera [55], which are found in 70% of healthy patients [13] as confirmed by Ghannoum el al. [13] and Peters et al [54]. The most abundant species in this genera is C. albicans, which is found in 40–80% of healthy individuals, followed by C. glabrata, C. parapsilosis, C. tropicalis, C. krusei, C. stellatoidea, C. kefyr, C. khmerensis and C. metapsilosis [55], [56].
C. albicans and C. glabrata are also important pathogens under certain conditions. The difference between commensalism and pathogenicity for C. albicans seems to be the result of a fine balance between fungal virulence and host defense mechanisms, and C. albicans is one of the most prevalent pathogens in mucosal and systemic fungal infections [57], [58]. In contrast, C. glabrata, which has been historically considered as a commensal saprophyte, has been reported as an opportunistic oral pathogen. Specifically, it has been identified as a co-infecting agent along with C. albicans in oral candidiasis in immunocompromised populations, and these co-infections tend to be more severe and more difficult to treat than single infections of C. albicans [58], [59], [60].
However, little is still known about fungal colonization succession and fungal biogeography in the oral cavity [50], [53] and their role in host health and disease. Also, further evaluation is still required to determine whether the species found so far are indeed functional residents or mere transient species [53]. Diaz et al. [53] recommend large longitudinal studies that include seasonal changes to help separate transient environmental fungi from permanent residents. For more details on the current and future perspectives on the oral mycobiome, please refer to Diaz et al. [53].
3.4. Oral protozoa biome – the protozoome
Historically, protozoa have been considered parasitic and assumed to have detrimental effects on the host [61]. However, a recent analysis revealed the presence of more than 15 known commensal protozoa genera in the human intestinal tract [62]. In the oral cavity, however, their roles are still unclear. Kofoid [63], in 1928, reported Trichomonas tenax and Entamoeba gingivalis as mouth parasites. However, in 1956, preliminary studies found these oral species frequently rated as “clean and healthy”. T. tenax and E. gingivalis have been found in increased numbers in subjects with poor oral hygiene [21], [64] and in patients with gingivitis and periodontal disease [65], [66], [67], [68]. Recently, T. tenax has also been associated with P. gingivalis, T. denticola and Eubacterium nodatum [68]. However, these results have recently been explained as related to protozoal nutrient factors, and with no impact on the host’s health. Specifically, it’s thought that poor oral hygiene and periodontitis increase protozoal nutrient sources - the amount of bacteria and food debris in the mouth [21], [69], and, thus, it is reasonable to find them associated with proteolytic bacteria. Currently, these species are regarded as harmless saprophytes and part of the oral microbiome [21], [65], [69].
3.5. Oral archaea biome – the archaeome
Archaea are organisms that are morphologically similar to bacteria but they share more molecular features with eukaryotes and they comprise the third domain of life [70], [71]. Some archaea species, classified as methanogenic, produce methane from carbon dioxide as an energy source for growth [72]. Five different methanogenic archaea genera have been identified in the oral cavity of healthy patients - Methanobrevibacter, Methanosphaera, Methanosarcina, Thermoplasmata and Methanobacterium [73]. Among these, Methanobrevibacter oralis, Methanobacterium congelense/curvum and Methanosarcina mazeii are the main archaea species found in healthy patients [73], [74], [75], [76], [77], with M. oralis dominating over other species at a prevalence of 40% [72], [76], [78]. Archaea are members of the oral microbiome, however, they are considered less abundant and diverse than oral bacteria [79]. More studies are needed to evaluate archaea’s role in the oral microbiome of healthy patients.
Archaea species have also been implicated in oral diseases, as they are known to form biofilms, and can interact with the human immune system [80]. Several studies identified increased numbers of M. oralis in periodontitis, peri-implantitis and root canal necrosis cases [73], [75], [76], [78], [81], possibly implicating the species in those diseases. Further, studies have demonstrated that archaea coexist with periodontal pathogens, such as Treponema denticola, Tannarella forsythia and Porphyromonas gingivalis [72], [75], [78]. These findings suggest a possible role for archaea as terminal degraders of host components, favoring the continuation of the catabolic cascade [72], [79]. However, their specific role in disease pathogenesis remains unclear and more studies are necessary to evaluate archaea’s pathogenic potential. For more details on the oral archaeome, please refer to Belmok et al. [79]
3.6. Interspecies interactions
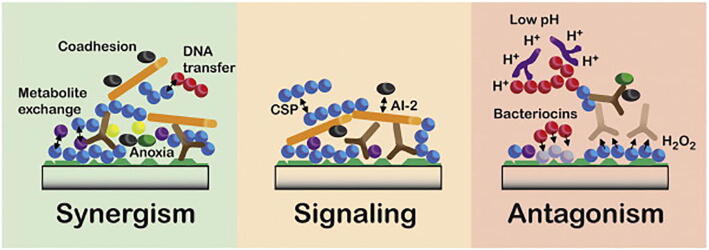
All of these micro-organisms live in close proximity, forming a complex relationship among them. These relationships establish a unique microbiome, known as the oral microbiome [82]. This close-proximity results in a wide range of interspecies interactions, which can be categorized into synergistic, signaling, or antagonistic interactions [19], [82], [83], as shown in Fig. 1.
Fig. 1.
Interactions between microorganisms that may drive the community assembly of oral biofilms. Different interactions are designated as having a primary function in synergy, signaling or antagonism. However, in many cases, components may have multiple roles with both positive and negative impacts depending on the situation. Reprinted from Jakubovics [83].
Oral microorganisms have a natural tendency to adhere to other microbes, facilitating the formation of multi-species biofilms [82], [83]. This process, called coadhesion, significantly changes the gene expression of both cells involved, usually enhancing biofilm formation [82], [84]. For instance, yeasts, such as C. albicans, can also coaggregate with oral streptococci, forming a synergistic partnership in which the bacteria enhance C. albicans’s invasive properties, while the yeast enhances streptococci biofilm formation [85], [86].
These coaggregated cells and the EPM around them obstruct the free movement of molecules, slowing down the diffusion of nutritional factors and oxygen, resulting in nutritional gradients throughout the biofilm [83]. Interestingly, these nutritional gradients also promote nutritional interactions when the metabolic byproducts of one organism become the food source for another organism. These interactions can even develop whole food webs in the biofilm, resulting in efficient and complete catabolism of complex molecules, such as glycoproteins, to their simplest metabolic end-products, such as CO2, CH4 and H2S [82]. Another functional consequence of the coaggregation is the protection of facultative and oxygen-tolerant anaerobes in anoxic pockets inside of the biofilm that result from neighboring oxygen-consuming species [31], [82].
Horizontal gene transfer (HGT) is a process that many bacteria use to gain access to novel genes, enabling them to acquire new traits, such as virulence factors and antibiotic resistance [87]. There are three canonical mechanisms that bacteria use to transfer genes to other individuals – conjugation, transformation, and transduction [88], [89], [90]. Briefly, conjugation is the direct transfer of DNA through cell surface pili or adhesins between two cells, in a process requiring cell-to-cell contact; transformation is the uptake and incorporation of free extracellular fragments of genetic material; and transduction is the gene transfer via bacteriophages [89], [90]. HGT is a common occurrence in the oral microbiome [90], [91], [92]. For instance, it has been revealed that S. mutans cells transform 10- to 600-fold more in biofilms than in their planktonic state [87]. Also, Tribble et al. [93] revealed the natural competence for transformation by several Porphyromonas gingivalis strains, which plays a major role in the overall survival and persistence of the species in the oral cavity, with conjugation playing a minor role. Besides, Chi et al. [94] demonstrated that regions of the pbp2x gene are shared between Streptococcus pneumoniae and other oral streptococci, such as Streptococcus oralis and Streptococcus mitis, thereby increasing penicillin resistance of these species. This finding indicates that there is both intra-species and interspecies HGT in the oral cavity.
Regarding signaling interactions, there are two major quorum sensing signaling systems in the oral microbiome – Autoinducer-2 (AI-2) and the competence-stimulating peptide (CSP).
Autoinducers are a group of molecules capable of eliciting phenotypic changes in bacteria. These molecules are produced by many bacterial species and they accumulate in the extracellular environment until a critical concentration threshold for detection is reached, after which downstream signaling and effector responses develop [95]. These molecules can induce bacterial group behaviors, known as bacterial quorum sensing [95]. Among these molecules, AI-2, a molecule derived from (S)-4,5-dihydroxypentane-2,3-dione and synthesized by the enzyme LuxS, appears widespread among prokaryotes. It is produced by over 50% of all sequenced bacterial species, including both Gram-positive and Gram-negative, so it has been described as the “common-language” among bacterial species [82], [95], [96]. In the oral cavity, AI-2 seems to regulate several phenotypes, including virulence and biofilm formation [96], [97]. Intriguingly, this molecule seems to induce biofilm formation in some species, whereas, in others, it inhibits formation. For instance, knocking out the AI-2 gene from S. oralis prevents Actinomyces naeslundii biofilm formation, whereas purified AI-2 from Veillonella tobetsuensis prevents Streptococcus gordonii biofilm formation [98], [99]. Further, Jang et al. [100] reported that AI-2 produced by a strain of Fusobacterium nucleatum had very different outcomes in two different species of the same genera – it enhanced biofilm formation in S. gordonii but inhibited biofilm formation in S. oralis. These data indicate that there are differences not only in the species that release AI-2 molecules, but also in how the molecules are perceived by each species. Remarkably, AI-2 may even participate in inter-kingdom communication, since the molecule is able to regulate fungal morphogenesis, germination, apoptosis, biofilm development, and pathogenicity [96]. For instance, Bachtiar et al. [101] demonstrated that AI-2 produced by Aggregatibacter actinomycetemcomitans inhibits both hyphae and biofilm formation of C. Albicans. Despite many studies on AI-2, it is still poorly understood and thus further studies are needed to better elucidated its biology [83], [96].
Another quorum sensing system is based on the competence-stimulating peptide (CSP) molecule. CSP is a peptide encoded by the comC gene. CSP is expressed then cleaved and exported out of the bacterial cell by the ABC transporter ComAB. Once CSP concentration reaches a certain threshold, the molecule binds to and activates the transmembrane histidine kinase receptor ComD. The activated ComD receptor can alter several bacterial transcription factors and protein synthesis related to biofilm formation, cell competence, bacteriocin synthesis, stress resistance, and autolysis [82], [102]. In the oral cavity, some streptococci can inhibit S. mutans biofilm formation by inactivating S. mutans CSP [103], [104]. Interestingly, CSP was found to be ubiquitous in streptococci, and important for controlling the acquisition of genetic material from the environment, biofilm formation, bacteriocin production, and, in pathogenic streptococci, virulence factor production [102]. Recently, Nagasawa et al. [105] demonstrated that CSP may be involved into releasing eDNA in S. mutans biofilms, contributing to firmer and more stable S. mutans biofilms. Counterintuitively, S. gordonii mutants, lacking the comC gene, produced more eDNA and more stable biofilms in the presence of C. albicans than the isogenic wild-type counterpart [106]. Thus, the regulatory pathways underpinning these interactions are far from clear [83],
In terms of antagonistic interactions, microbes use bacteriocins or hydrogen peroxide (H2O2) or they decrease a niche’s pH to gain a competitive advantage or even create a selective pressure on other species [82], [83].
Bacteriocins are part of a larger group of molecules named antimicrobial peptides (AMP). AMPs are peptides consisting of 12–100 amino acids that are synthesized by almost every form of life on earth (from bacteria to amphibians and humans) and they are released extracellularly to inhibit the growth of microbes [107]. Bacteriocins are AMPs produced by both gram-positive and gram-negative bacteria to kill or inhibit the growth of other prokaryotes, especially different bacterial strains in the same environment, thereby conferring to the bacteriocin-producing bacteria a competitive survival advantage over other strains in the same niche [82], [107].
Similarly, hydrogen peroxide (H2O2) can be produced to kill or inhibit the growth of other species. The deleterious effects of H2O2 on bacterial cells arises from the generation of hydroxyl radicals in the presence of Fe(II). This leads to a reaction of the hydroxyl radicals with other molecules, resulting in cellular damage by degradation of iron-sulfur clusters on enzymes, leading to their inactivation, or by oxidation of macromolecules, including DNA and proteins [108]. Certain oral Streptococci species can produce H2O2 at concentrations that inhibit the growth of a range of gram-positive bacteria in vitro and under aerobic conditions [82], [83], [108], [109]. Despite hydrogen peroxide being one of the most well studied agents in dental biofilms, its impact on the oral microbiota is complex and difficult to predict [82].
Sugar-fermenting species can create an antagonistic environment for other strains by decreasing the local pH, thus selecting strains that are either tolerant to low pH (aciduric) or even acid-producing (acidogenic) bacteria [83], [110]. Nevertheless, several commensal species of the oral cavity are well adapted to live in low pH microenvironments [83].
For more details on oral microbiome interspecies interactions, please refer to Bowen et al. [19], Jakubovics [83], and Marsh & Zaura [82]. The oral microbiome also forms a close symbiotic relationship with human host cells in the oral cavity and this will be discussed in the next section.
3.7. Oral host-microbial interactions – the oralome
The oral microbiome with all its interspecies interactions forms a complex ecosystem that thrives in a very dynamic environment that is the oral cavity. Thus, the oral microbiome not only mediates microbial interspecies interactions, but also interactions with the oral cavity and, thus, it creates a symbiotic relationship with the human host [4], [8] - the microbial composition is significantly affected by interspecies and host-microbial interactions, which in turn, impact the health and disease status of the host [7]. We propose the term Oralome to describe all the interactions that take place between the oral microbiome and the host. For instance, it is well stablished that the oral microbiota are important for the maturation and development of an appropriate oral immune response, as the host immune system has to defend the host against pathogenic microbes, but also harmonize and protect commensal oral microbes [111], [112], [113]. For example, EBV stimulates a strong host immune response by eliciting the production of antibodies, such as immunoglobulin (Ig) G, IgM and IgA. These immunoglobulins recognize some EBV targets, such as EBV glycoproteins, as well as lytic and latent antigens. However, both CD4 + helper and CD8 + cytotoxic T cells mediate the robust response against EBV [114]. At the same time, the immune system has to avoid producing antibodies against oral commensal microbes.
Nevertheless, the host immune response must balance between inflammation for pathogen eradication and prevention of an unwanted immune response against the host’s own tissue and commensal organisms, as mounting an aggressive immune response against microbes that pose no threat would be unnecessary, metabolically wasteful, and potentially damaging to host tissues [115]. In exchange, some oral commensals can act as a pathogen ‘Sensor’, ‘Mediator’ and ‘Killer’ and have coordinated roles in initiating the antagonistic action against a pathogen to prevent the colonization and integration of pathogens, a phenomenon referred to as colonization resistance [116]. For instance, Streptococcus salivarius antagonizes the main etiological agent of pharyngitis, Streptococcus pyogenes, preventing its colonization and growth in the pharyngeal mucosa, thus preventing pharyngitis [117], [118], [119]. This mutual protection is one indication that the host immune system evolved to tolerate and maintain some beneficial bacteria [120]. Also, some mucosally-adherent species can protect the host from carcinogenic metabolites [121], [122].
In addition to benefitting the oral cavity, the oral microbiome also exerts an important role on the systemic health of the host [123]. For instance, some oral bacteria also participate in an entero-salivary nitrate‐nitrite‐nitric oxide cycle in which dietary nitrate in the saliva is reduced to nitrite and nitric oxide [124], which may assist in cytoprotection, vasodilation, antithrombotic effects, immune modulatory effects, blood pressure modulation, and may improve outcomes of myocardial infarction, heart failure, pulmonary hypertension, and vascular hypertrophy in terms of infarct size, cardiac function and hypertrophy [125], [126]. Interestingly, high abundances of Rothia and Neisseria genera were shown to be beneficial for the maintenance of nitric oxide host homeostasis and cardiovascular health, while high abundances of Prevotella and Veillonella genera were detrimental to homeostasis.
Moreover, 16S rDNA studies have shed light on the existence of specific microbial patterns that are considered “healthy oral microbiota” [127], [128]. In this sense, a healthy oralome (i.e. symbiotic host-microbiome interactions between humans and these microorganisms) would be an an example of eubiosis [4], [8].
3.7.1. Eubiosis
Although the classification of ecological relationships (commensalism, parasitism and mutualism) is well defined, the boundaries between each category are fluid [129]. In this context, the oral microbiome, in a healthy host, under normal conditions, maintains balanced symbiotic/commensal relationships, which have been defined as a “microbial homeostasis” or “eubiosis” [4], [8], [130], [131]. This balance promotes beneficial mutualism and/or commensalism without causing harm to either the microorganisms or the host. Despite this homeostasis, this does not mean homogeneity of the oralome in all individuals, as recent studies report that up to five different clusters can be found in the salivary oralome of different healthy individuals [132], [133], [134]. However, the largest populational-based study to establish the salivary microbiome (with more than 2,300 individuals) reported only two salivary community types, one associated with oral health and the other associated to oral diseases [135]. These results suggest that the difference between health (eubiosis) and disease (dysbiosis) may be complex and more than just natural heterogeneity in healthy individuals.
3.7.1.1. Dysbiosis
Although the oral microbiome has resilience (i.e. capacity of an ecosystem to deal with perturbations without shifting its state of symbiosis [136]), insults or changes, such as tobacco use, can shift the eubiotic balance from mutualism/commensalism to a unbalanced parasitic/pathogenic state, thus, promoting disease in the host [129], [137]. This specific parasitic/pathogenic state wherein microbials promote disease in the host is known as “dysbiosis” [111]. This is also known as an “unbalanced microbiome” [138], [139]. According to Peterson et al [111], dysbiosis can be characterized by three different scenarios that are not mutually exclusive and may occur simultaneously. i) the overall loss of microbial diversity; ii) losing the beneficial microbes; and iii) expansion of the pathogenic microbes.
3.7.1.2. Loss of microbial diversity
The general ecological concept of loss of biodiversity within a community indicates a decline in the number, genetic variability, and/or variety of species of a biological community at a determined location [111]. This loss of biodiversity can lead to the breakdown of that ecosystem. In the context of caries, several reports indicate a loss of diversity as the severity of the disease increased, suggesting that increased acidification of the oral micro-environment is accompanied by loss of diversity and a reduction in the levels and metabolic activity of beneficial bacteria, leading to the rise of cariogenic bacteria [140], [141], [142], [143], [144], [145].
In terms of periodontal disease, changes in microbial diversity remain controversial, with some reports indicating loss of microbial diversity with periodontitis [146], [147], [148], [149], and others indicating the opposite (i.e., that periodontitis is associated with increased levels of microbial diversity compared to healthy control levels; as a consequence of the increased amount of nutrients derived from host’s tissue degradation [135], [139], [140], [149], [150], [151]; and even others reporting no significant difference between the groups (i.e., periodontitis vs. healthy patients) [152], [153]. For instance, Almeida et al. [148] reported significantly lower taxonomic diversity in subgingival microbial samples from both gingivitis and chronic periodontitis patients compared to healthy individuals (total of 110 participants). However, Griffen et al [150] reported a significantly higher taxonomic diversity for chronic periodontitis individuals compared to healthy controls (58 participants in total). Interestingly, this discrepancy may be due to sampling from different probing depths, as Ge et al.[154] indicates that deeper pockets (>5 mm) contain significantly higher richness and diversity levels compared to shallower (≤3 mm) pockets in patients with chronic periodontitis. Thus, further studies are needed to clarify these contradictory findings.
Interestingly, a meta-transcriptomic study of mature oral microbiomes demonstrated an over-expression of genes related to natural genetic transformation, indicating high functional redundancy in the oral microbiome [155]. However, under dysbiosis, this redundancy may be lost due to loss of beneficial microbes or growth of pathogenic bacteria, suggesting a general frailty in the oral microbiome related to the diversity and composition of its microbiota [156].
3.7.1.3. Loss of beneficial microbes
One of the main characteristics of dysbiosis is the loss of some of the benefits acquired from an established healthy oralome. As discussed before, the oral microbiota are important for the maturation and development of an appropriate oral immune response [111], [112]; protecting the host against oral pathogens [117] and from carcinogenic metabolites [121], [122]; and are part of the nitrate-nitrite-nitric oxide pathway [126], thereby offering several benefits to the host. Thus, losing those beneficial microbes may lower the host’s ability to fight off pathogenic bacteria and respond to an excessive immune response against the host’s own tissue, exposing the host to carcinogenic metabolites and detrimental vascular changes [111], [112]. This loss is particularly important in the context of periodontal disease, wherein excessive chronic inflammation leads to loss of supporting tissues around teeth, including alveolar bone loss, which leads to tooth loss in the severe forms of the disease [157].
3.7.1.4. Expansion of pathogenic microbes
In a eubiotic environment, the oral microbiome contains opportunistic pathogens at such low levels that they do not cause any issues to an immune-competent host [111], [122]. However, an outgrowth of these pathogens represents a risk for the host. Particularly, this may increase the risk for dental caries, periodontal disease and may even be correlated to several systemic diseases, such as atherosclerosis, Alzheimer’s disease, and cancer. Specifically, P. gingivalis and/or F. nucleatum have been associated with periodontal disease [56], [130], [139], head and neck cancer [130], [158], pancreatic cancer [159], colorectal cancer [160], [161], [162], [163], Alzheimer’s disease [164], [165], [166], [167], atherosclerosis [168] and pre-term births [169], [170].
In the next section, we will discuss the oral dysbiosis signatures of some these diseases.
4. Oral biofilm dysbiosis signatures of common diseases
4.1. Dental caries and periodontal disease
The search for the cause of tooth decay dates back to 5000 BCE, when Sumerian texts described a “tooth worm” as the causative agent of dental caries. In the late 1600s, Anthony van Leeuwenhoek was the first person to report microorganisms living in his own dental plaque; laying down the foundation for microbiology. In 1890, W. D. Miller proposed the “chemoparasitic” theory for the origin of caries, which explained that “in susceptible hosts who frequently consumed fermentable carbohydrates, oral microorganisms would convert these carbohydrates into acid, which would result in the demineralization of teeth”; thereby laying down the foundation for modern dental research [171], [172]. However, due to limitations in culturing bacteria in the 19th century, Miller was unable to identify any specific causative agents of caries. Based on the work of Miller and G. V. Black, it was believed, at the time, that the quantity and not any specific pathogens were responsible for periodontitis. In this sense, the disease would only develop if the bacteria were able to surpass the threshold capacity of the body to detoxify bacterial products. Thus, idea has been known as the “non-specific plaque hypothesis” [173].
Interestingly, the turn of the century brought new techniques to isolate and identify bacteria. In 1924, J. K. Clarke identified a caries-causative agent – Streptococcus mutans. Unfortunately, Clarke was unable to directly demonstrate that S. mutans caused caries; this was later demonstrated by R. J. Fitzgerald and P. H. Keyes in the 1960′s.
In the 1970′s, W. J. Loesche and colleagues reported that the antibiotic kanamycin was particularly effective against cariogenic bacteria [173], [174]. In 1976, the “Specific Plaque Hypothesis”, was postulated, which holds that dental caries is an infection of specific bacteria present in dental plaque, namely “mutans streptococci” (including S. mutans and Streptococcus sobrinus) and lactobacilli. At the same time, several studies started comparing health- and disease-associated plaque [173], [174]. Among those, M. A. Listgaten, in 1976, for instance, observed distinct qualitative differences between them using electron microscopy [175]. These studies identified several different species related to periodontal disease, leading some researchers to the conclusion that specific bacteria (i.e., periopathogens) could initiate and drive the disease. Thus, extending the “Specific Plaque Hypothesis” to periodontitis [173], [174], [176], [177].
In 1978, the term “biofilm” gained importance after J. W. Costerton’s publication described how bacteria stick [178], referring to the matrix-enclosed bacterial communities that are key to understanding how bacteria interact with the environment [179]. After this, other studies have shown that bacteria within biofilms usually display different phenotypes compared to their planktonic counterparts [30], [179], [180]. Within biofilms, there are extensive metabolites exchanged, signal trafficking, and different levels of interactions among different species [179]. The introduction of biofilm theory into oral microbiology provided an impetus for researchers to take a closer look at dental plaque [172], [179].
Recent studies have also shed light on possible interdomain interactions between bacterial and fungal species that can drive periodontitis. For instance, C. albicans can interact with and adhere to P. gingivalis; thereby potentially increasing P. gingivalis’s virulence [181]. Furthermore, the fungus can enhance P. gingivalis invasion of epithelial and gingival fibroblast cells via an unknown mannoprotein-b-glucan complex-dependent mechanism [182]. Another example is the interaction between bacterial and viral species. Several studies have reported a higher P. gingivalis abundance in EBV-positive patients [183], [184], [185], [186], [187]. In addition, P. gingivalis [187] and Porphyromonas endodontalis [188] can reactivate latent EBV and induce its lytic cycle via histone epigenetic modifications [114].
Recently, our group introduced a new polymicrobial mouse model of periodontal disease in a common mouse strain (BALB/cBy), which revealed a widening of the periodontal ligament space, alveolar bone loss, and an increased host immune response after a polymicrobial infection [49]. This model may be useful for assaying microbial biofilm interactions in vivo. Interestingly, using murine models, Payne et al. [189] demonstrated the natural transmission of the dysbiotic oral microbiome from a periodontally-diseased animal into a healthy one, leading to establishment of dysbiosis in the recipient, and suggesting that a dysbiosis may act as a conventional transmissible infectious disease agent by transmitting and stablishing dysbiosis in healthy individuals, with concomitant effects on its pathology.
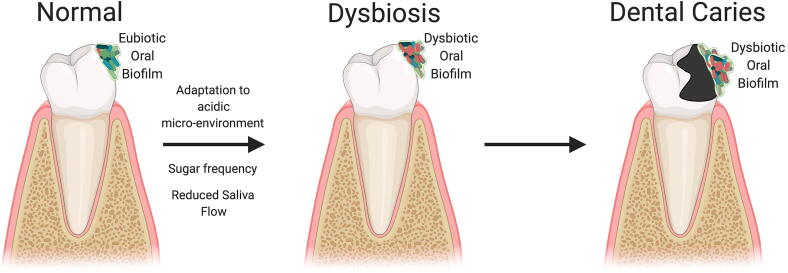
With regards to dental caries, one of the most accepted theories about its etiology is known as the “ecological plaque hypothesis” [190]. This hypothesis states that frequent sugar ingestion drives an adaptation of the commensal bacteria to a more acidic micro-environment, favoring the succession of aciduric bacterial species (especially streptococci and lactobacilli), while inhibiting beneficial organisms that preferentially grow at neutral pH [141], [191], [192]. This adaptation to a dysbiotic microbiome is enabled by an enrichment with certain species of Streptococcus, Lactobacillus, and Actinomycetes genera, which are believed to be responsible for the formation of dental caries in adults (Fig. 2). For instance, compared to caries-free children, children with severe early childhood caries exhibit enhanced levels of Streptococcus (specially S. mutans), Leptotrichia, Bifidobacterium, Porphyromonas, Leptotrichia, Stomatobaculum, Prevotella and Selenomas genera. Specifically, S. mutans has been heavily studied for its cariogenic properties and is, today, regarded as one of the main pathogens associated with caries [56], [130]. Yet, it should be noted that other species may also drive the disease, as S. mutans can persist in the oral cavity without evidence of detectable demineralization and caries, and caries can occur in the apparent absence of the species [190].
Fig. 2.
Oral biofilm dysbiotic signature leading to dental caries.
Recently, Hong et al. [52] found a high association between a Candida mycotype and aciduric bacterial species, whereas genera correlated with periodontitis, such Fusobacterium, Porphyromonas and Treponema, were highly associated to the Malassezia mycotype. Interestingly, the authors also found that the Candida mycotype was positively correlated with both caries and plaque, which suggests that both mycotypes may play a role in the link between periodontitis and caries. Thus, more studies are needed to better understand the role of both mycotypes in both diseases. Interestingly, these species have also been associated with caries sequelae, such as irreversible pulpitis. Recently, Zargar et al. [193] found 18 species in 41 root canals from patients with irreversible pulpitis. Among them, Dialister invisus, P. gingivalis, S. salivarius, T. denticola, C. albicans and HSV-1 were the ones with highest prevalence; Lysinibacillus fusiformis was detected for the first time in the root canals. This suggests a potential ecological succession of these species following caries progression.
The dysbiotic microbiome, led by S. mutans, would lead to the production of a wide range acids (predominantly lactic acid [194]) that partially demineralize the surface of the tooth. This mineral loss increases enamel porosity, widening the spaces between the enamel crystals and softening the surface, which allows the acids to diffuse even deeper into the tooth. This results in further demineralization of the layers below the surface, producing tooth decay. Once enamel is compromised, the pathogenic bacteria can eventually invade tooth dentin and pulp, causing pulpitis [195], as S. mutans has been frequently reported to be isolated in inflamed pulp associated with severe dental caries [196], [197].
Once inside of the pulp, the dysbiotic bacteria have access to the circulatory system, which could lead to a transient bacteremia. Interestingly, oral streptococci, especially S. mutans, Streptococcus sanguinis and Streptococcus mitis, are thought to be important causative agents in infective endocarditis [196], [197]. For instance, Nomura et al. [197] recently found significantly higher rates of S. mutans in the heart tissue of rats with ≥5 M containing dental caries extending to the pulp compared to rats with ≤4 M with similar dental caries. However, the authors reported that there is no direct evidence supporting a causative relationship between S. mutans from dental caries and the onset of infective endocarditis, as it remains unknown whether S. mutans can reach heart tissues through the bloodstream from advanced dental caries lesions [197].
Unlike caries, there is not a single hypothesis for the etiology of periodontitis [173] and this may explain why the initiation of periodontal disease and the microbes that drive periodontitis are still not completely established, despite being examined for decades [114], [173].
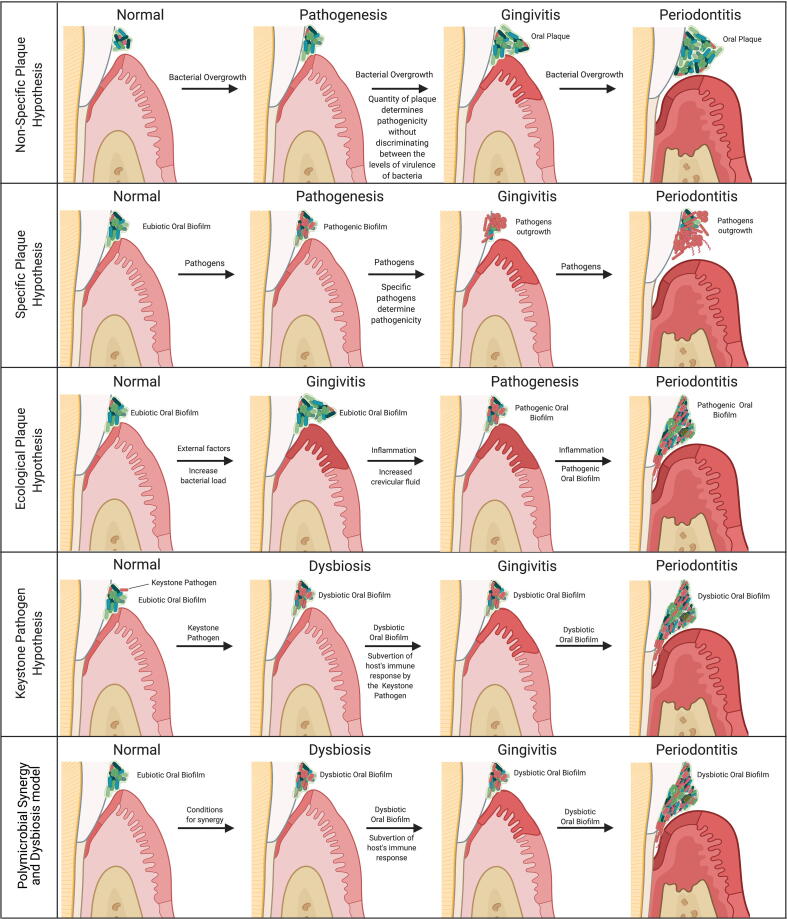
To date, there have been five main hypotheses proposed for the initiation and pathogenesis of periodontitis (Fig. 3). We briefly discussed the first two previously above (i.e. the non-specific and the specific plaque hypotheses). The third is the “ecological plaque hypothesis” developed by Marsh in 1994 [190], [198]. Marsh combined key concepts from the Specific and the Non-specific Plaque Hypothesis and concluded that the disease is the result of an imbalance in the microflora promoted by ecological stress, resulting in an enrichment of some periopathogens. Briefly, the hypothesis holds that the eubiotic microbiome can only harbor extremally low levels of periodontal pathogens, as the pathogens are unable to outcompete the predominant eubiotic saccharolytic bacteria. Due to external factors, the bacterial load in the subgingival microbiome can become incompatible with health and resulting in the activation of the host inflammatory response (gingivitis). This inflammatory response causes an increased flow of gingival crevicular fluid (a serum-like exudate), altering the subgingival nutrient status. This alteration drives an enrichment of proteolytic Gram-negative bacteria, such as Prevotella intermedia, F. nucleatum, P. gingivalis, T. forsythia, and T. denticola [56], [130], [139]. This enrichment drives host tissue degradation, promoting an even greater host inflammatory response and the cycle of destruction continues with the deregulation of the host inflammatory response, leading to periodontitis [139], [190], [198]. However, this hypothesis does not address the role of host genetic factors that significantly contribute to the composition of dental plaque and to susceptibility to disease [173].
Fig. 3.
Main hypotheses proposed so far for the initiation of periodontitis.
The fourth hypothesis is the “keystone pathogen hypothesis” proposed by Hajishengallis and colleagues in 2012 [199]. This hypothesis is based upon the ecological concept of “keystone species” coined by Robert Paine in his seminal work in the 1960’s [200], [201], which propose that some species are capable of holding their community together, despite their lower abundance and higher hierarchy in the food web; thus, having a disproportionately large effect relative to their abundance in their communities. Briefly, the “keystone pathogen hypothesis” holds that a certain low-abundance microbial pathogen can orchestrate a complete oralome remodulation into a dysbiotic state and attack the hosts defenses, including epithelial cells (as a nutritional source).
An important factor in this hypothesis is that the keystone pathogen be able to subvert the role of the immune system, as a wall of neutrophils stand between the plaque and the epithelial cell surface. In this context, Hajishengallis and colleagues [199], [202] argue that the “keystone pathogen” for periodontitis is P. gingivalis since the bacterium has developed sophisticated strategies to subvert and impair the host immune response via several mechanisms, including Toll-like receptor response manipulation and interleukin 8 subversion.
Others have argued that the keystone pathogen is, in fact, a viral infection that disturbs the oralome into a dysbiotic state [203]. For instance, it has been demonstrated that CMV and EBV can inhibit macrophages by significantly downregulating their TNF-α production and TLR-9 expression, and inhibiting macrophage phagocytotic activity [204]; indicating that these viruses may inhibit the initial macrophage response, allowing the pathogenic bacteria enough time and space to initiate host aggregation and, thus formation of periodontitis. Interestingly, Slots [205] found CMV, EBV and HSV-1 in 3%, 7% and 12% of healthy periodontal sites, whereas their prevalence increased to 40%, 32% and 45% in subgingival chronic periodontitis sites and 49%, 45% and 63% in subgingival aggressive periodontitis sites. However, the viral keystone pathogen concept remains contested in the literature as several studies failed to find an association between these viruses and severe periodontal diseases [46], [47], [206].
Yet, no specific pathogen has been significantly associated with gingivitis, but the amount of plaque present and its bacterial load has been correlated with disease severity [21], [177]; pointing out that the trigger may be more related to the abundance of organisms and their ecological succession rather than a specific pathogen driving the whole process from the beginning. In addition, certain periopathogens, such as P. gingivalis, can also be detected in periodontally healthy individuals, though less frequently, but without driving the disease [207], [208], [209]. Thus, Hajishengallis and Lamont updated the “Keystone Pathogen Hypothesis” to the “Polymicrobial Synergy and Dysbiosis” hypothesis in 2012 [140], [202], [210]. The concept of polymicrobial synergy in periodontitis has been established by animal models, which consistently demonstrated that significantly higher pathogenicity was found when several periopathogens were combined compared to the monospecies infection [140]. Interestingly, several important pathogenic functions require the expression of specific molecules, such as the appropriate adhesins, receptors, proteolytic enzymes and proinflammatory surface ligands, which cannot be found in one specific keystone pathogen, rather, the combination of these molecules act as community virulence factors to sustain a heterotypic, proinflammatory and dysbiotic microbial community that elicits a non-resolving and tissue-destructive host response [210], [211]. Consequently, it has become apparent that pathogenic periodontal microbes only exert their fully pathogenicity when conditions favor synergism [210], [211].
This model is consistent with the participation of both gram-negative and gram-positive bacteria in the pathogenesis of the disease, as long as they can provoke and/or tolerate inflammation [140], [202], [210]. Also, mixed microbial communities may provide opportunities for competitive and cooperative interspecies interactions, and such interactions can shape the nature and function of the entire microbiome [140], [202], [210]. For example, Passariello et al. [212] found a positive association between CMV, EBV and HSV-1 and several periodontal pathogens, including P. gingivalis, T. forsythia, Fusobacterium periodonticum and Staphylococcus aureus. Recently, our group, demonstrated an association between increased viral diversity and periodontal disease. Indeed, our data demonstrated that several viruses, such as Gammaretrovirus and Porcine type-C oncovirus, were also associated with alveolar bone loss and widening of the periodontal ligament in vivo, thereby implicating them in periodontitis [49]. Interestingly, Zhao et al. [213] demonstrated that hepatitis B virus (HBV) may also contribute to oral dysbiosis, as they found that a higher Neisseriaceae family abundance was positively associated with higher HBV titers on the tongue dorsum, whereas the virus was not present in healthy patients. Taken together, these data indicate a positive role for the virome in periodontal disease, but further studies are needed to determine all the critical triggers for periodontitis.
Currently, the subgingival dysbiotic microbiome signature most frequently identified includes a higher abundance of the red complex triad (Treponema denticola, Tannarella Forsythia, and Porphyromonas gingivalis) [140], as well as, the orange complex triad (Fusobacterium nucleatum, Prevotella intermedia, and Parvimonas micra), Actinobacillus actinomycetemcomitans, Campylobacter rectus, Eikenella corrodens, Bacteroides forsythus [114], Filifactor alocis, Peptoanaerobacter stomatis, Firmicutes phylum [140], Methanobrevibacter oralis [214], the archeon phylotype Thermoplasmata [81], C. albicans [54], [215], CMV and EBV [205]. Thus far, no protozoan has been associated with the periodontitis signature.
The dysbiotic bacteria associated with periodontitis have access to the bloodstream, thereby creating the possibility for bacteremias. Bacteremia is defined as a transient or continuous presence of bacteria in the bloodstream [216]. In a double-blind, placebo-controlled study with 290 subjects, Lockhart et al. [217] found a positive bacteremia in 56% of patients after tooth extraction and 32% after toothbrushing. A similar rate of bacteremias was found in a more recent systematic review that included 9 observational studies, which showed that the most frequently identified bacteria in bacteremia were Streptococcus viridans, A. actinomycetemcomitans, P. gingivalis, Micromonas micros, and Actinomyces species [216]. These bacteria in the circulatory system could lead to the colonization of other hosts tissues and, thus, the association of the oral microbiome with systemic diseases.
4.2. Atherosclerosis
The link between dental disease and cardiovascular diseases was first established in 1993, when De Stefano et al [218] reported an increased risk (25% higher) of atherosclerotic plaque formation in patients with periodontitis. A recent study [168] demonstrated the presence of 23 oral pathogenic bacteria within atherosclerotic plaques in patients undergoing carotid endarterectomy, catheter-based atherectomy, or similar procedures. Of these 23 bacteria, 5 (Campylobacter rectus, P. gingivalis, Porphyromonas endodontalis, P. intermedia, Prevotella nigrescens) were unique to coronary plaques, while the other 18 were additionally present in non-cardiac organs and associated with over 30 non-cardiac disorders.
At least five mechanisms have been proposed for the contribution of oral dysbiosis to atherosclerotic plaque formation - i. bacteremia from the pathogenic oral microbiome invades the arterial wall and promotes plaque formation. For example, it has been demonstrated that oral bacteria can invade endothelial cells and phagocytic cells in the atheroma, leading to pathogenic changes and progression of the atheroma lesion [219]; ii. oral sites with inflammation release inflammatory mediators into the blood stream, which promote plaque formation; iii. autoimmunity to host proteins caused by the host immune response to specific components of oral pathogens promote plaque formation; and iv. oral pathogenic bacteria release specific bacterial toxins with pro-atherogenic effects [220]; v. via dyslipidemia, since patients with chronic or aggressive periodontitis have elevated serum levels of low-density lipoprotein and triglycerides, and decreased levels of high-density lipoprotein [219]. For a detailed review on this subject, please refer to Aarabi et al [220] and Schenkein et al. [219].
4.3. Alzheimer’s disease
The first hint of a possible link between oral pathogens and Alzheimer’s disease was shown in 1993, when Miklossy reported the presence of spirochetes in the blood, cerebrospinal fluid, and brain of 14 histopathologically confirmed Alzheimer’s disease cases, but not in controls [164]. In 2002, Riviere et al confirmed this and reported the presence of 6 out of 7 Treponema species in the brain of Alzheimer’s disease postmortem tissues. Interestingly, not only was the prevalence of Treponema DNA in brain cortex much higher among Alzheimer’s disease samples than in controls, but Alzheimer’s disease subjects also had more Treponema species present. In 2012, a study that examined a longitudinal cohort found that poor oral hygiene was strongly linked to the development of dementia [221].
Recently, Borrelia genera and T. denticola, P. gingivalis and Escherichia coli [164], [165], [166] bacterial species and Fusarium, Alternaria, Botrytis, Candida, and Malassezia fungi genera [222] have been implicated in the etiology of Alzheimer’s disease.
In 2019, P. gingivalis DNA was found in 7 out of 10 patients diagnosed with Alzheimer’s disease, and a P. gingivalis gingipain protease was co-localized with Tau proteins in these patients’ tissues. The authors further showed that mice orally infected with P. gingivalis also demonstrated brain infection and induction of the stereotypical Alzheimer’s disease marker, amyloid beta 1–42 oligomers [167], suggesting that a bacteremia might have driven these pathogens to the brain. However, the role of these pathogens in this neurodegenerative disease remains unclear [223].
4.4. Diabetes
Type 1 and type 2 diabetes affects the periodontium of both children and adults, with an increase in periodontal inflammation and enhanced periodontal bone loss. The mechanism of action includes alterations in osteoclasts and osteoblasts of the periodontium by increasing the expression of inflammatory mediators, such as tumor necrosis factor (TNF), by increasing the receptor activator of nuclear factor kappa-B ligand (RANKL)/osteoprotegerin (OPG) ratios, and by enhancing advanced glycation end product (AGE) and oxygen reactive species (ROS) levels [224], [225].
In 2001, a “two-way” relationship between diabetes and periodontal disease was proposed. This hypothesis was supported by investigations of individuals in the Gila River Indian community, where severe periodontitis at baseline was associated with an increased risk of poor glycemic control (HbA1c > 9.0%) at follow-up (minimum 2 years), suggesting that severe periodontitis was a risk factor for compromised diabetes management [226].
In 2007, Hintao et al [227] found an increased frequency of T. denticola, Streptococcus sanguinis, Prevotella nigrescens, Staphylococcus intermedius, and S. oralis in the supragingival plaque of individuals with type 2 diabetes.
Furthermore, several studies in 2011 showed that the treatment of periodontal disease influenced glycemic control in individuals with both type 1 and type 2 diabetes [228]. In addition, shifts in the oralome of patients with diabetes compared to healthy controls have also been reported [229]. Oral microbial dysbiosis is highly associated with the development of periodontal disease, which can induce higher levels of inflammation locally and systemically, and in turn, aggravate hyperglycemia [229].
In 2013, a consensus report from the European Federation of Periodontology and the American Academy of Periodontology found significant evidence demonstrating that mechanical periodontal therapy was associated with a 0.4% reduction in HbA1C levels after 3 months. The authors argued that this reduction was clinically equivalent to adding a second drug to the pharmacologic regime of diabetes patients [230]. Since the report found inconsistent evidence that diabetes type 2 significantly impacts the oral microbiota [225], this diminished the concept of the “two-way” relationship between diabetes and periodontal disease.
A 2019 study [229] demonstrated that Leptotrichia, Staphylococcus, Catonella, and Bulleidia genera were significantly enriched in patients with diabetes with very high glucose levels, suggesting that dysbiosis of the oral microbiota may be a typical feature of hyperglycemia and a potential contributor to progression of hyperglycemia. However, the oral microbial signatures that mediate the progression of hyperglycemia are not clear. Adding complexity to the issue, growing evidence suggests that gut microbiome dysbiosis may also play a role in diabetes [231], [232]. For instance, recent metagenomic studies demonstrated a lower abundance of butyrate‐producing microbes and an enrichment of Bacteroides caccae, Clostridium hathewayi, Clostridium ramosum, Clostridium symbiosum, Eggerthella lenta and Escherichia coli; the latter are known gut pathogens that cause intra-abdominal infections and bacteremias in type 2 diabetes patients. Thus, butyrate‐producing bacteria may play a potential protective role.
Interestingly, Xiao et al. [233] demonstrated that diabetes can increase oral microbiome pathogenicity through IL-17 in vivo, bringing back the possibility of the “two-way” relationship between diabetes and periodontal disease. The pathogenic oral microbiome from diabetic mice was able to significantly increase periodontal inflammation and bone loss when transferred to germ-free mice, compared to an oral microbiome from healthy mice. Taken together, these data even suggest a possible “three-way” relationship where the oral microbiome, the gut microbiome dysbiosis, and diabetes exacerbate each other.
However, more studies are needed to identify the specific oral microbes that contribute to diabetes pathogenesis and to determine the underlying mechanisms by which both an oral and gastrointestinal microbial dysbiosis relate to diabetes. For more details on how diabetes can affect the oral microbiome, please refer to Graves et al. [225]
4.5. Pregnancy complications
The placenta is usually considered to be sterile, however, recent studies have shed light on a possible placental microbiome [169], [234], and this microbiome might be involved in pregnancy complications. For instance, preterm birth rates were significantly higher when bacterial invasion and an IL-6 immune response was present [169]. However, no association has been found between preterm birth and a “healthy” placental microbiome (i.e. without histologic evidence of infection) [169]. Combs et al [169] investigated the microbes found in preterm births and found that oral microbes, such as F. nucleatum, Bergeyella sp., Clostridium sp., Actinomyces sp., Peptostreptococcus spp., and Candida albicans were present in half of the woman diagnosed with preterm labor and infection. Aagaard et al [234] compared the placental microbiome to the oral, skin, nasal, vaginal, and gastrointestinal microbiome of nonpregnant women and reported that the bacterial taxonomic profile of the placental microbiome was more similar to the oral microbiome, suggesting a possible bacteremia from the oral cavity to the uterus. However, comparing the microbiome of pregnant subjects to that of nonpregnant subjects may not be an ideal comparison, since a previous study from the same group found a significant difference in the vaginal microbiome between pregnant and nonpregnant subjects [235].
A significant association has also been established between microbes in the amniotic fluid and previous pregnancy complications, such as miscarriage, intrauterine death, neonatal death, preterm delivery and premature rupture of membranes. In all of these cases where F. nucleatum was detected, previous pregnancies resulted in one or more miscarriages [170].
FadA adhesin is a small (111 amino acids) helical peptide from F. nucleatum that has been proposed as a virulence factor involved in F. nucleatum-mediated cell attachment and invasion of host cells [236], [237]. Ikegami et al. demonstrated that the deletion of the fadA gene resulted in >1000-fold lower bacterial titer in mice placenta compared to the control after 24 h of infection in vivo. Interestingly, restoration of the deleted gene on the strain also restored F. nucleatum’s ability to invade and colonize placental tissue, indicating that the peptide is highly involved F. nucleatum’s pathogenicity. However, fadA deletion did not completely eliminate placental invasion, thus suggesting the involvement of other proteins in the process.
The existence of a placental microbiome, however, has been recently scrutinized. Some studies demonstrated that the bacteria found in the uterus may have been due to contamination of laboratory reagents with bacterial DNA and lack of appropriate controls [238], [239] and, thus, the concept of a uterine/placental microbiome has become controversial.
4.6. Head and neck cancer
Head and neck cancer (HNC) has a complex etiology. Risk factors for HNC include alcohol and tobacco use and betel quid chewing with or without tobacco [240], [241]. Furthermore, alcohol consumption and smoking have a synergetic effect and these increase HNC risk, especially for oral and pharyngeal cancer [242]. In addition, alcohol [243] and tobacco [244] use negatively influence oral microbiome composition. For instance, Kumar et al [244] observed higher persistent rates of oral pathogens from Fusobacterium, Cardiobacterium, Synergistes, and Selenomonas genera in oral biofilms of smokers compared to non-smokers.
Molecular studies and meta-analysis have also pointed out that some viruses, such as Human Papillomavirus and Epstein-Barr virus, are associated with HNC progression [242]. Several molecular studies have found associations between HPV and oropharyngeal cancer (OPC) [245], [246]. Thus, HPV was recognized in 2005 as a risk factor for OPC by the International Agency for Research on Cancer (IARC) and the World Health Organization (WHO) [246]. It has been estimated that HPV accounts for 30–60% of OPC and 12% of pharyngeal cancer [245], [247]. Mechanistically, in HPV-induced OPC, p53 is present in both upstream and downstream pathways, but at low levels (10% or less) [245], [248]. This is mainly due to p53 degradation by the HPV E6 enzyme [248], [249], [250].
Epstein-Barr Virus (EBV), a member of the herpesvirus family, was the first virus to be directly associated with carcinogenesis. It is transmitted through saliva and replicates in the epithelial cells of the oropharynx [251]. A recent meta-analysis demonstrated a significant association between EBV and HNC [252], [253], although there has been no evidence of a direct role for EBV in HNC progression [253], [254]. A higher frequency of oral sex and a greater number of sex partners are thought to be the reason for this increased risk for viral-related HNC, especially related to the oropharynx [245]. Mechanistically, EBV upregulates programmed cell death protein 1 ligand (PD-L1) via CTAR family proteins; decreasing p53 stability and increasing secretion of matrix metalloproteases (MMPs), thus upregulating the expression of cancer stem cell markers and facilitating cancer cell invasion and tumorigenesis. However, little is known about the potential carcinogenic role of EBV in HNC [253].
The specific link between viral infections and HNC led to a paradigm shift in understanding HNC risk, especially when HPV infection was previously associated with alterations of the oral microbiome in both infants [255] and adults [256]. This is particularly noteworthy, as recent cohort studies demonstrated that poor oral health affects the survival of patients with HNC [257] and good oral hygiene, such as annual dental visits and daily tooth brushing, may reduce the risk of HNC [258].
Culture-based studies revealed an increase in salivary bacterial counts in oral cancer patients, such Capnocytophaga gingivalis, Prevotella melaninogenica and S. mitis. In addition, DNA sequencing studies have demonstrated that different bacterial species colonize oral tumors compared to healthy sites, and high fusobacterial and low streptococcal levels may be a potential signature for the transition from health to HNC [259], [260], [261], [262], [263]. These findings suggest that unique microbial signatures may be potential diagnostic indicators for HNC, although they do not directly address the relationship of the oralome to the development of HNC.
In 2016, Guerrero-Preston et al [260] demonstrated that increases in Lactobacillus and loss of Haemophilus, Neisseria, Gemellaceae and Aggregatibacter in saliva may be biomarkers for HNC compared to healthy controls. Also, the authors demonstrated a shift in the microbial community of HPV-positive HNC tumors, with an enrichment in certain Lactobacillus and Weeksellaceae strains, and an abundance of Eikenella, Neisseria, and Leptotrichia in the HPV-negative tumors.
In 2018, a nested case-control study with 129 HNC patients [264] demonstrated that commensal bacteria, Corynebacterium and Kingella, were associated with a reduced risk for HNC, and data trends tended to be stronger in cases with a history of tobacco use. The authors also reported that oral Corynebacterium and Kingella genera were potentially mediating xenobiotic biodegradation, including metabolism of toxicants found in cigarette smoke. This finding suggests that a potential HNC prevention strategy may include promotion of these bacterial strains, especially for those with a history of tobacco use.
P. gingivalis has been associated with a variety of cancers, such as esophageal squamous cell carcinoma, colorectal, pancreatic and oral cancer [130]. For oral cancer, P. gingivalis infection was positively associated with late TNM stages, poor differentiation and lymph node metastasis [265]. One study showed that P. gingivalis elevated the level of Cyclin A, diminished the level and activity of p53 and activated the PI3K pathway to promote the proliferation of gingival epithelial cells [266]. Another study demonstrated that incubation with P. gingivalis for 72 h led to a dramatic increase in α-defensin, thus boosting oral squamous cell carcinoma (OSCC) cell proliferation by 125% [158]. Furthermore, P. gingivalis can induce activation of β-catenin and disassociation of the β-catenin destruction complex by gingipain-dependent proteolytic processing, thereby contributing to cancer pathogenesis [267].
A study by Kamarajan et al [268] demonstrated that multiple periodontal pathogens (P. gingivalis, T. denticola, and F. nucleatum) promote oral cancer aggressivity in vivo. The mechanisms involved included TLR/MyD88-Integrin/FAK crosstalk, which promoted cancer cell migration and invasion. Furthermore, this periodontal pathogen-mediated carcinogenesis was therapeutically responsive to treatment with a probiotic bacteriocin. Therefore, this study established a potential paradigm shift in the treatment of cancers by focusing on antimicrobial-based therapeutics for cancer.
Recently, Hong et al. [52] showed a significant increase in the Candida mycotype with a significant reduction of the Malassezia mycotype in patients with cancer (most frequently OSCC) compared to control patients. Although the patients age and recent history of chemotherapy did not correlate with any mycotypes, the Candida mycotype did emerge as a potential player in OSCC tumorigenesis and even chemotherapy resistance and recurrence. However, the authors also showed that receiving steroid premedication (administered to subjects prior to cancer sampling), and the subsequent higher neutrophil counts were both also correlated to higher Candida counts. This could indicate that higher Candida counts were present before the steroid administration or that the steroid administration and the higher neutrophil counts could be driving the higher Candida counts. Interestingly, when the authors normalized the data for steroid administration, the difference in Candida counts became non-significant, indicating that the later hypothesis (steroid administration drives the higher Candida count) may be the correct explanation of the results. Thus, further studies are needed to test this possibility.
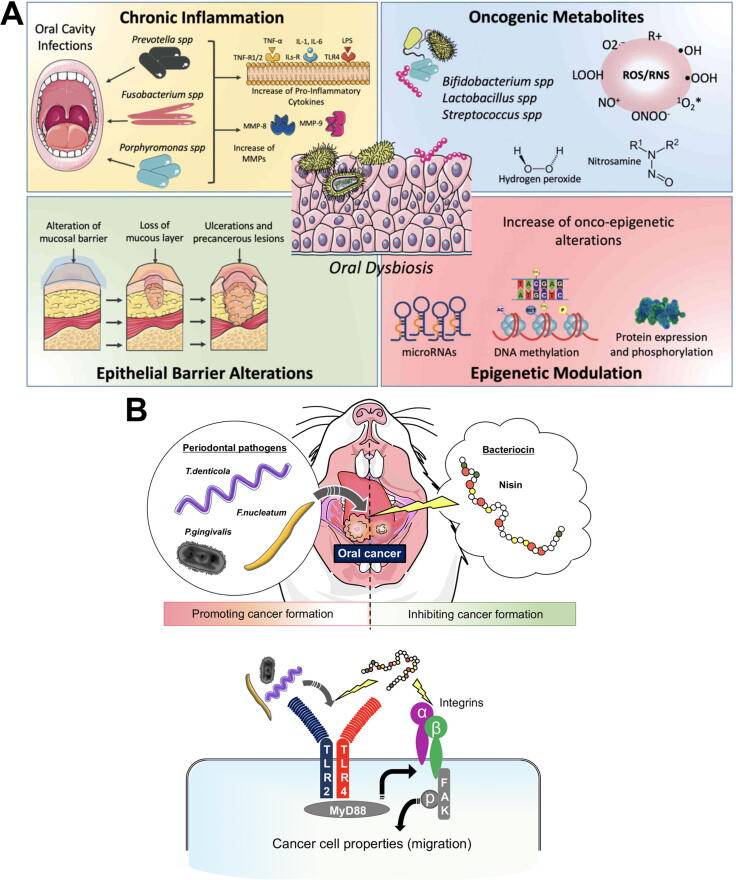
Several mechanisms have been proposed to explain how carcinogenesis might be mediated by oral microbial dysbiosis [121] (Fig. 4). i. pathogenic bacteria, such as Porphyromonas and Fusobacterium species can upregulate cytokines and inflammatory factors (e.g. IL-6, matrix-metalloproteinases and TNF-α), leading to chronic inflammation and alterations of different molecular pathways responsible for cell metabolism and proliferation [269], [270], [271], [272]; ii. Several substances produced by pathogenic oral bacteria, such as ROS, sulfides and nitrosamines induce pro-tumoral genetic damage [273], [274], [275], [276], [277], [278]; iii. oral biofilm dysbiosis can alter the homeostasis of epithelial barriers, leading to barrier dysfunction [279], [280], [281]; iv. recent studies have demonstrated the ability of the microbiota to modulate hosts gene expression via miRNAs; these epigenetic changes were associated with the development and progression of tumors [282], [283], [284]. For a more detailed review of these mechanisms, please refer to La Rosa et al [121] and Radaic et al. [12]. Although there is evidence that different oral pathogenic species promote carcinogenesis, direct evidence of the mechanisms involved is still emerging [127], [268].
Fig. 4.
Oral microbial dysbiosis is associated with oral cancer development through different mechanisms. A - Oral infections and dysbiosis are responsible for promoting a pro-inflammatory microenvironment, wherein inflammatory cytokines and matrix metalloproteinases favor the development and progression of tumors. Furthermore, the bacteria in the oral cavity produce oxygen and nitrogen reactive species, as well as oncogenic metabolites (e.g., nitrosamines) to induce genetic damage to cells within the oral mucosa. Another mechanism by which neoplastic transformation is mediated by oral dysbiosis is via the alteration of epithelial barriers, which predispose the oral mucosa to the development of chronic pre-cancerous lesions. Oral dysbiosis is responsible for several epigenetic alterations, which promote the development of tumors (e.g., alteration of onco-miR or DNA methylation phenomena). Figure reprinted from “Association of oral dysbiosis with oral cancer development” by La Rosa et al[121] licensed under CC BY 4.0; no changes were made to the figure. B - A recent report by Kamarajan et al[268] documents an in vivo model in which periodontal pathogens, i.e. Treponema denticola (ATCC 35405), Porphyromonas gingivalis (ATCC 33277) and Fusobacterium nucleatum (ATCC 25586) and, their lipopolysaccharides activate TLR/MyD88-Integrin/FAK cross talk to promote cancer cell migration and invasion. Interestingly, nisin Z (Handary, Belgium), a bacteriocin produced by Lactococcus lactis inhibits the cancer formation. Image courtesy of Drs. Pachiyappan Kamarajan and Ryutaro Kuraji.
4.7. Other cancers
Periodontal pathogens have also been associated with pancreatic and colorectal cancer. Next, we will briefly discuss these associations. For a more detailed review of the current epidemiological and microbiological evidence linking a dysbiotic oral microbiome to these and other types of cancer, please refer to Radaic et al. [12].
4.7.1. Pancreatic cancer
Fan et al. [285] reported that the presence of P. gingivalis and A. actinomycetemcomitans in the oral cavity was associated with a 60% increased risk of pancreatic cancer (hazard ratio = 1.60–95% confidence interval 1.15 to 2.22) in a nested case-control study with 732 participants. Interestingly, the authors reported that patients positive for P. gingivalis in the oral cavity have a 59% greater risk of developing pancreatic cancer than those who were negative for the pathogen.
Remarkably, Pushalkar et al. [286] demonstrated that pancreatic tumors harbor a specific microbiome, distinct from a normal pancreatic microbiome. Specifically, there is an increase in the genus Brevibacterium and order Chlamydiales in pancreatic cancers compared to normal controls. Despite a lack of direct evidence of oral microbial pathogens in pancreatic cancer, P. gingivalis is able to invade and survive inside of pancreatic cancer cells, enhance pancreatic tumor growth in vivo and cause higher rates of mutation in the tumor suppressor protein p53 and Kirsten rat sarcoma viral oncogene homolog (KRAS) gene in pancreatic cancer in vitro [159], [287], [288], [289], [290]. Thus, more studies are needed to evaluate whether P. gingivalis can directly colonize pancreatic tissue in vivo.
4.7.2. Colorectal cancer
Among all species in the gut microbiome found in the large intestine, F. nucleatum seems to be increasingly reported in gut infections, such as intestinal abscesses and acute appendicitis [291], with a high prevalence F. nucleatum infections in colorectal cancer [292]. In addition, F. nucleatum was found to enhance colorectal cancer growth and progression in vitro and in vivo via Toll like receptor 4 (TLR4) and myeloid differentiation primary response 88 (MyD88) protein activation and upregulation of microRNA 18a and microRNA 4802 [160], [161], [291] and is able to inhibit immune natural killer cells [293]. All these findings support that F. nucleatum may also play a major role in colorectal tumor growth and progression.
5. Current computational techniques to evaluate oral biofilm dysbiosis
Computational techniques, such as in silico models, can be used to understand biological systems as well as to select, complement, and inspire laboratory experiments [294], [295], [296]. For example, certain computational approaches can be used to estimate the immunogenic response of an antibody to a particular virulence factor [297] or evaluate the competition between two different bacterial species [298].
Dos Santos-Lima et al [297] used an in silico approach to analyze peptide epitopes from P. gingivalis virulence proteins for their potential immunogenic/IgG-mediated host response with relevance to periodontitis. Through this analysis, the authors were able to predict and select immunoreactive peptides from the pathogenic enzymes – lysine gingipain (KgP) and neuraminidase (also known as sialidase) before synthesizing them. Although, the authors confirmed the IgG immunoreactivity of the synthetized proteins, both peptides elicited very different immunoreactivity levels; KgP showed a high immunoreactivity, whereas neuraminidase showed very low immunoreactivity.
In another study, Valdebenito et al [298] also used an in silico approach to analyze the effects of competition between Streptococcus sanguinis and S. mutans in dental biofilms. The study focused on characterizing whole-genome differences between the species to explore potential metabolic advantages in a direct competition. The authors found that S. sanguinis had competitive advantages over S. mutans, primarily due to glutathione peroxidase activity and the capacity to undergo gluconeogenesis via genes present in the former but not in the later species.
Head et al [141] used in silico modelling to investigate how the frequency of and total sugar intake can promote the growth of more cariogenic species within oral biofilms. The authors concluded that total sugar intake was a determining factor in the promotion of cariogenic bacteria; with a low sugar intake, plaque was never cariogenic, whereas with a high sugar intake, oral plaque always promoted cariogenic bacteria, independent of frequency (Fig. 5). Furthermore, Marsh et al [299] also used in silico modelling to investigate how small effects, such as fluctuations in pH after sugar ingestion, can affect oral bacterial competitiveness. Their results demonstrate that variations in the buffering capacity of the plaque fluid modulate biofilm composition from a “healthy” state (i.e. low fraction of aciduric bacteria) to a dysbiotic state (i.e. dominated by acidogenic bacteria). This change resulted in a lower pH in the plaque fluid and an increased risk for enamel demineralization.
Fig. 5.
Snapshots of biofilm composition at t = 60d receiving 6 glucose pulses per day, and a total daily amount of (a) 10 g/L/d and (b) 20 g/L/d. (a) Green spheres represent particles of a non-aciduric cell type (NA), and (b) red spheres represent those of an aciduric type (A). The computer simulations were initiated with 5% particles of type A and the remaining of type NA. Reproduced from Head et al[141]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Computational techniques are not limited to in sillico models. Meta-analyses have revealed that metagenomics, metabolic networks, and interactomes have also been used to study the oralome.
Miossec et al. [300] evaluated 8 popular taxonomic profiling pipelines (i.e. MetaPhlAn2, metaMix, PathoScope 2.0, Sigma, Kraken, ConStrains, Centrifuge and Taxator-tk) that use different strategies for taxonomic profiling (e.g. marker-based, k-mer search, and read reassignment) to determine which factors or combinations affect time and robustness for perfect base calling. The different taxonomic profiling strategies were tested against 426 complete genomes stored in the Human Oral Microbiome Database to simulate various experimental conditions, including read length (75–1000 bp reads), sequence depth (100 K–10 M), metagenomic composition, number of species present (10, 100, 426), and species distribution. The authors found that taxonomic profilers, such as PathoScope 2.0 and Metamix, provided the most sensitive profiles but these took more computational time to execute the analysis. Other tools, such as Kraken and Centrifuge, required significantly less computational time, but found a lower microbial abundance. Thus, the more time-intensive pipelines may be the most beneficial tools.
Bernstein et al. [301] used a percolation theory to metabolic network analysis to estimate which metabolites, such as biomass components, are synthesized by the oral microbiome to better understand the metabolic interactions within the oral microbiome. To do so, the authors reconstructed the metabolic networks from the genome of 456 strains (371 different species) from the oral microbiome and generated an atlas of 88 different essential biomass metabolites from these organisms. This analysis can be further used in the field to estimate inter-microbial metabolic distances and correlate them with microbial co-occurrences.
Recently, Rosa et al. [302] developed a computational pipeline to specifically evaluate the oralome interactome – a specific network of protein–protein interactions (PPI) within a cell or even an organism. The authors reported that most PPI studies focus on the interactome of a particular organism, however, this analysis can be used to evaluate inter-organism interactions, such as PPI related to dysbiosis and symbiosis. Using the proposed pipeline and the SalivaTecDB, a database dedicated to annotated proteins, miRNAs and microorganisms present in the oral cavity and associated with oral or systemic mechanisms, the authors found 697 human-C. albicans protein interactions predicted with high confidence (score >0.9) (Fig. 6). The protein interactions found were primarily involved with cellular processes, such as metabolism, regulation of gene expression and cell cycle, and with pathogenesis, such as the phospholipase B enzyme, which is known for its role in virulence in mouse models of systemic infection [303]. Upon further analysis of the 317 human proteins that interact with C. albicans proteins, the authors also found an enrichment in leukocyte and neutrophil activation, inflammatory response, cytokine production and signaling, immune cell vesicle-mediated transport, secretion and exocytosis, as well as cell migration, which highlights a potential functional role for immune system interaction networks. Interestingly, this analysis suggests that both the yeast and host are aware of each other and that the host immune system is prepared to protect itself in case of a yeast infection.
Fig. 6.
Networks of protein–protein interactions (PPIs) between Candida albicans (orange triangles), human oral proteins (blue diamonds), and proteins from other oral microorganisms (gray rectangles) are identified by gene name. Reprinted from Rosa et al. [302]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
6. Current and novel techniques for modulating oral biofilm dysbiosis
Despite the harmful effects that can be mediated by a dysbiotic oralome within the host, a complete elimination of the oralome is not the answer, since a eubiotic oralome provides health benefits to the host. Rather, techniques to modulate the dysbiotic oralome to re-establish a eubiotic state would be preferred. Among the current and novel techniques being studied, we discuss the use of traditional oral hygiene techniques, antimicrobial peptides (AMPs), nano-sized drug delivery systems, prebiotics, probiotics, EPM disruption techniques, and modifiers of the inflammatory response.
6.1. Oral hygiene
Although there are no specific pathogens associated with the early stages periodontal inflammation (i.e. gingivitis), the bacterial load and the amount of plaque present and its maturity have been correlated with disease severity [21], [177]. Thus, frequent oral hygiene is an important technique for controlling the microbial load on the dentition and oral cavity and to prevent periodontitis and bacteremias [304], [305].
A variety of oral hygiene measures have been advocated for removing dental plaque, however, brushing with toothpaste is the most highly recommended and primary method for oral hygiene in industrialized countries [306]. Substantial data supports that individual plaque control techniques, such as conventional tooth brushing with a fluoridated dentifrice and the use of chemical anti-plaque mouthrinses, significantly improves gingival inflammation and lowers plaque scores, provided that cleaning is sufficiently thorough and performed at appropriate time intervals [306], [307]. When practiced regularly, oral hygiene keeps dental plaque in an immature state and in relatively small amounts that significantly decrease the chances of an bacteremia event compared to conditions where dental plaque is not effectively removed, as in patients with periodontal disease [21]; thereby, potentially avoiding the additional risks for systemic diseases associated with periodontal disease.
However, the frequency and duration of tooth brushing that is required to eliminate plaque and prevent periodontal disease is still not entirely clear, since longitudinal randomized controlled clinical trials are needed to fully evaluate this, yet these are challenging to carry out. Thus, we have to rely on information gained from either observational studies or from short-term studies conducted on gingivitis patients [306]. In general, the recommendation is to brush twice daily with a fluoride-containing toothpaste for 2 min, and the literature seems to endorse this [306]. For instance, a recently published 11-year-long prospective study with more than 1000 adults demonstrates a dose–response relationship between toothbrushing frequency and reduction in number of teeth with periodontal pocket depths (PPD) ≥ 4 mm - participants who reported brushing twice or more a day developed fewer teeth with PPD ≥ 4 mm than those who reported brushing less [308]. However, Chapple et al. [307] pointed out that expert opinion indicates that brushing for 2 min is likely insufficient for periodontitis patients, especially when considering the need for additional use of interdental cleaning devices.
Interestingly, a meta‐analysis of 56 trials found in the Cochrane Central Registry of Controlled Trials and MEDLINE databases demonstrated a significant reduction in plaque index scores in both short‐term (11% in 1‐3 months) and long-term studies (21% >3 months) when using mechanical toothbrushes compared to manual toothbrushes [309], suggesting that mechanical toothbrushing may be helpful in reducing plaque-associated sequelae. However, the authors point out that there is a possible risk of bias, due to a great heterogeneity among the studies regarding brushing duration and frequency. Also, the effects of mechanical toothbrushing on periodontal disease progression remains unclear [306].
In terms of caries, a recent meta-analysis of three randomized trials, including more than 700 early teens (age 10–13), concluded that the oral hygiene technique by itself (without fluoride) may not prevent or reduce carious lesions [310]. Although the conclusion may be not be directly extrapolated to adults, since adults may have a different dental restorative status, gingival recession, and differences in saliva flow or systemic diseases [311], this analysis indicates that oral hygiene techniques may have a lesser role in preventing caries compared to fluoride. It is widely known that fluoride favorably shifts the balance towards enamel remineralization [312], whereas oral hygiene may only temporarily disrupt the acidogenic biofilm; and cariogenic dysbiosis is majorly driven by lifestyle factors, such as frequent sugar ingestion [195], [311].
6.2. Antimicrobial peptides (AMPs)
Oral biofilm dysbiosis can be modulated through the use of antibiotics, however, antibiotics eliminate both pathogenic and commensal bacteria [313]. Also, due to their incomplete absorption by humans and animals, a large amount of ingested antibiotics are excreted into the environment via the feces or urine, contributing to environmental and multi-drug resistance concerns [314]. Therefore, new antimicrobial molecules are needed to effectively modulate microbial dysbiosis to address these concerns. In this regard, antimicrobial peptides (AMP) have shown promise in modulating oral biofilm dysbiosis. Due to their broad-spectrum antimicrobial activity, some AMP have the potential of becoming the next generation of antibiotics, and they may be useful in dealing with the crisis of multi antibiotic-resistant bacteria [107].
Among AMPs produced by bacteria, bacteriocins, like nisin, which is produced by Lactococcus lactis, are active against both gram-positive and gram-negative bacteria, including S. aureus, and Listeria monocytogenes. Due to its broad anti-microbial potential, nisin was approved as a food additive by the U.S. Food and Drug Administration (FDA) in 1988. To date, nisin is the most utilized bacteriocin and is the only bacteriocin used in food preservation in more than 50 countries. Our group has demonstrated that nisin disrupts oral biofilms without exhibiting cytotoxicity to oral cells [315]. Specifically, we demonstrated that nisin (from 1 to 50 µg/mL) inhibits the planktonic growth of oral bacteria, retards the development of multi-species biofilms, and disrupts biofilm biomass and thickness in a dose-dependent manner. These same concentrations did not affect primary periodontal ligament cells, gingival fibroblasts, primary oral keratinocytes, and osteoblast-like cells; showing no signs of apoptotic changes up to 200 µg/ml for 24 h.
Among AMPs found in amphibians, K4-S4 (1–15) showed bactericidal effects against S. mutans, Aggregatibacter actinomycetemcomitans, and F. nucleatum (the latter two related to periodontal disease), both in their planktonic and biofilm forms [316].
Among human AMPs, LL-37 is expressed by several immune and epithelial cells and is directly involved in the host cellular response to microbial attacks. LL-37 has anti-fungal, antimicrobial and anti-biofilm properties, and can act as a chemoattractant for human peripheral blood neutrophils, monocytes, and T-cells, and is even capable of inhibiting Kaposi's sarcoma-associated-herpesvirus. Recent research demonstrates that LL-37 opsonizes A. actinomycetemcomitans and inhibits its formation into biofilms [317], plus inhibits the growth of F. nucleatum [316].
However, AMPs face some challenges. For example AMP resistance mechanisms develop within bacteria and these mechanisms have been associated with the virulence potential of pathogens [318]. These mechanisms include proteolytic degradation or sequestration by secreted proteins, exopolymer and biofilm matrix molecule inactivation, alterations of the cell wall and/or membrane composition via cell envelope regulatory networks (e.g. VanRS and LiaR), and AMP removal by efflux pumps [107], [318]. In addition, AMPs can act as host sensitizers/allergens, which may lead to immunogenicity, especially after repeated exposure [107]. In this context, one alternative is to combine AMPs with nanoscale drug delivery systems (nano-DDS) [107], [318], [319], [320], [321].
6.3. Use of nanosized-drug delivery systems
Nanoparticles are colloidal particles made out of a variety of materials, such as lipids, metals, and polymers, that have sizes between 1 and 1000 nm [321], [322]. At this size range, these nanoparticles possess unique features compared with their bulkier counterparts – they can offer a high surface area, increased reactivity, reduction in thermal resistivity, intracellular delivery of therapeutics depending on the particle shape, size, surface area and charge, and release of high levels of ions at low incorporated amounts, distinguishing them from their bulkier size and micro-sized materials [16], [321], [322]. Indeed, an inverse relationship has been demonstrated between the size of the metallic nanoparticles and their antimicrobial activity; particles in the size range of 1–10 nm have the greatest biocidal activity against bacteria [323], [324], with silver nanoparticles, ranging from 5 − 40 nm being able to inactivate most microorganisms, including HIV-1 [325]. Thus, due to their antimicrobial ability, nanoparticles made of silver, copper, titanium oxide, and zinc oxide have been incorporated into polymer matrices as filler particles to control biofilm growth [16].
These unique features have also been used to improve therapeutic agents, which led to the development of nanomedicine [107], [322]. Nanomedicine seeks to improve the use of proteins, genetic materials, and low-weight molecular agents for diagnostics and treatment of diseases through the use of nano-sized drug delivery systems (Nano-DDS) [322]. Specifically, this improvement is related to more specific targeting, controlled release of the compound, lower toxicity and better bioavailability [321]. For instance, Horev et al. [326] developed a polymeric nano-DDS capable of releasing farnesol in a pH-dependent manner, releasing the drug only at acidic pHs (4.5–5.5). This is very relevant to caries biology, as this mechanism may specifically disrupt only the aciduric dysbiotic biofilm. The nano-DDS formulation did disrupt S. mutans biofilms 4-fold more effectively than the free drug in vitro, and it significantly reduced both the number and severity of carious lesions in vivo, compared to free farnesol, which presented no in vivo effects.
With regards to AMPs, Nano-DDS can potentially protect AMPs from degradative enzymes and improve AMP immunogenicity by helping to evade the host immune response when the AMP is incorporated inside of the nano-DDS matrix, thus making AMP more efficient. For instance, Bernegossi et al. [327] encapsulated the synthetic AMP KSL-W in a lipid liquid crystalline nano-DDS made of oleic acid, polyoxypropylene-polyoxyethylene-cetyl alcohol and poloxamer 407 and they found higher growth inhibition of human saliva-derived oral biofilms using the KSL-W-loaded nano-DDS compared to the free peptide and the nano-DDS alone; reaching up to 100% growth inhibition after 7 days. Thus, the use nano-DDS show promise, since they can overcome the main drawbacks of AMP, and thereby making them more relevant for clinical practice [107], [321]. However, very little has been tested in the area of oralome dysbiosis, as most of the studies focused on testing AMP-loaded nanoparticles against food-spoilage bacteria. For instance, of all the studies in the latest 4 reviews evaluating AMP-loaded nano-DDS [107], [319], [320], [321], only one study, Bernegossi et al.[327] tested the efficacy of AMP-loaded nano-DDS in a saliva-derived oral biofilm model in vitro and none used in vivo models.
However, there are still some challenges and limitations that need to be addressed. For instance, some nanoparticle synthesis methods can expose AMPs to procedures that could modify and/or damage their structure and, thus, reduce their potential activity [107]. Also, nanoparticles are inheritably variable and extremely dynamic and, thus, are difficult to synthesize uniformly, creating batch differences [107], [328], [329]. Despite these challenges, more studies are needed to test the efficacy and safety of AMP-loaded nano-DDS, namely, studies demonstrating the in vivo use of nano-DDS to deliver AMP, especially studies demonstrating that nano-DDS can overcome AMP pharmacokinetics, pharmacodynamics and immunogenicity issues [107].
For a more details on the recent advances on nano-DDS for AMPs delivery, please refer to Radaic et al. [107], Martin-Serrano et al. [319], Makowski et al. [321] and Teixeira et al. [320]. For more details on the recent use of nanoparticles in dentistry, please refer to Jiao et al. [16] and Allaker &Yuan [325].
6.4. Prebiotics
Prebiotics are natural or synthetic food ingredients or supplements that modulate the microbiome to benefit the host [120]. This concept has been applied to the gut microbiome over the last decade, and, only recently, has it been applied to the oral microbiome. In this context, arginine has been shown to prevent dental caries by buffering the acids produced by a dysbiotic oralome [330], [331]. The amino acid is metabolized by commensal arginolytic species, producing ATP and ammonia. ATP gives the arginolytic commensals a bioenergetic advantage over the acidogenic/aciduric bacteria [19], [331], and ammonia raises the pH of the biofilm, preventing acidic damage to enamel [120], [331]. Interestingly, Agnello et al. demonstrated by 16S sequencing that arginine pretreatment 20 h before sucrose addition resulted in a shift in the oral microbiome towards “health”, with increased Prevotella genus and Veillonella dispar and Streptococcus anginosus species, and an increased biofilm diversity compared to the non-pretreated biofilms.
Nitrate is an ecological factor that can induce rapid changes in the structure and function of polymicrobial communities, thus it may be a potential prebiotic. Rosier et al. [332] demonstrated that supplementation with nitrate can be useful against both caries and periodontitis. Despite not affecting oral biofilm growth, nitrate supplementation (6.5 mM) was able to significantly increase ammonium production and alter the pH of the oral biofilm, as well as significantly reduce periodontitis-associated species.
Recently, Slomka et al. screened for 742 nutritional compounds and identified β-methyl-D-galactoside and N-acetyl-D-mannosamine as promising oral prebiotics, as they selectively triggered the growth of commensal oral bacteria and shifted biofilm communities towards a eubiosis in vitro [333]. Next, the group compared N-acetyl-D-mannosamine with two other promising prebiotics (succinic acid and Met-Pro) and found that N-acetyl-D-mannosamine was the most promising prebiotic among them, by showing a clear dose-dependent effect on the oral biofilms, such that treatment with1.0 and 1.5 M resulted in biofilms with 97% beneficial species [334].
Despite these advances, prebiotics for the oralome are still very limited, with limited evidence showing prebiotic effects against oralome dysbiosis. Thus, additional studies are needed to clarify the potential benefits of prebiotics [335].
6.5. Probiotics
Probiotics are live microorganisms which, when administered in adequate amounts, confer a health benefit for the host. In the case of the oralome, oral administration of probiotics may also benefit oral health by preventing the growth of harmful microbiota and/or by modulating mucosal immunity in the oral cavity [336], [337].
In vitro studies have demonstrated that Lactobacilli and Streptococci strains isolated from healthy oral cavities have antibacterial activity against P. gingivalis, P. intermedia, A. actinomycetemcomitans and F. nucleatum [338], [339], [340], [341], which could lead to potential treatments for periodontal disease. However, Lactobacilli and Streptococci seem to use different antibacterial mechanisms. Streptococci may inhibit anaerobic species through the production of lactic acid and other organic acids [342], whereas Lactobacilli generate hydrogen peroxide via several different enzymes, including pyruvate oxidase [343], lactate oxidase, NADH oxidase [344] and NADH flavin-dependent reductase [345]. Interestingly, some studies have shown that different lactobacilli strains can inhibit the ability of multiple microbial species. For example, L. fermentum is active against microaerophiles, and L. gaseri is active against anaerobes [340].
Recently, our group demonstrated that a nisin-producing probiotic, Lactococcus lactis, prevents and disrupts oral biofilm formation in vitro. We also demonstrated that the probiotic modulated oral biofilm composition, by reducing the levels of Heamophillus influenzae, F. nucleatum, T. forsythia and Klebsiella pneumoniae (bacteria related to periodontal disease), while enriching for Neisseria flava (an oral commensal) within dysbiotic in vitro human saliva-derived oral biofilms; driving the composition of these biofilms back to control levels [31].
Pre-clinical and clinical studies have shown that probiotics are able to reduce gingivitis, plaque, alveolar bone loss, and modulate pro-inflammatory effects [346]. Specifically, chewing gum containing Lactobacillus reuteri led to reductions in gingivitis and plaque in patients with moderate to severe gingivitis [347], whereas Bacillus subtilis (CH201) reduced bone loss and attachment loss in rats with ligature-induced periodontitis [348]. Interestingly, Chatterjee et al. [349] suggested that the mechanisms of probiotic action are due to a combination of both local and systemic effects, including modulation of the local and systemic host immune response, antibacterial effects via different mechanisms, and stimulation of osteoblastic function.
Although probiotics have many health benefits, these benefits can only be realized if a high number of probiotic cells reach the intended target site, the periodontal pocket or caries surface. However, many probiotic cells die in food products after exposure to low pH and oxygen during refrigeration, distribution and storage, and the viability/survival rate of probiotic bacteria is strain-specific [350], [351], [352], [353]. Thus, encapsulation of probiotics in micro- and nano-sized drug delivery systems (nano-DDS) may be an alternative that has been shown to protect cells from environmental factors [107], [350]. For a more detailed review about nanoparticles and nano-DDS for targeting microbial biofilms, please refer to Koo et al. [354].
Also, one important consideration regarding probiotics is that changing the biofilm composition might alter the normal oral flora that is beneficial and/or critical for oral health [346]. However, studies detailing how probiotics interact with other microbes and how they interact with food in the oral cavity are still lacking [350]. Thus, further studies on long-term microbial changes in the oral cavity and probiotic interactions with food are still required. For a more detailed review of the current findings of probiotics as relates to periodontal health, please refer to Nguyen et al.[337], [346].
6.6. Extracellular polymeric matrix (EPM) disruption
As previously discussed, EPM is the matrix produced by the oral bacteriome that forms the oral biofilm. This matrix is made up of polysaccharides, proteins, lipids and eDNA [7], [22], [23], [24], [25], [26], and it protects the microbes from environmental stresses[29], such as from antibiotics. Nonetheless, controlling biofilm formation by disrupting the EPM can prevent the formation of dysbiotic biofilms and/or assist other therapies in biofilm disruption. Currently, there are two main strategies to disrupt EPM - disrupting EPS synthesis and secretion and targeting its composition and structure.
Disrupting EPS synthesis and secretion can be achieved by the use of cyclic-di-GMP or cyclic -di-AMP, as there is evidence that these molecules are responsible for governing the transition from planktonic state to a biofilm state [355], [356], [357]. For instance, Karaolis et al. showed that these compounds inhibited S. aureus cell-to-cell adhesion and reduced more than 50% of the species ability to form a biofilm. Similarly, Yan et al. [357] found that exogenous cyclic-di-GMP inhibit S. mutans adhesion and biofilm formation. Interestingly, deleting the gene that produces the molecule exacerbates matrix production. Taken together, these reports indicate the bacteria sense cyclic dinucleotide levels in the environment and use these as signaling molecules to decide whether to remain in a planktonic state or adhere to each other and promote biofilm formation [358]. Another approach is through the inhibition of glucosyltransferases, the enzymes responsible for initiation and elongation of matrix glycan chains [359]. From a virtual screening of about 150,000 molecules, Ren et al. [360] found that a particular quinoxaline derivative was a potential glucosyltransferase inhibitor. The authors then demonstrated that this compound could selectively antagonize glucosyltransferase in S. mutans by inhibiting its biofilm formation. Moreover, the authors evaluated the compound in an in vivo caries model and found that the molecule significantly reduced the incidence and severity of caries.
Disrupting EPS composition/structure can be achieved by the use of exopolysaccharide-degrading enzymes [361], [362], [363]. Some examples include glucanohydrolases, such as dextranase, mutanase, and dispersin B. Glucanohydrolases have been used successfully to degrade mixed-species S. aureus and Pseudomonas aeruginosa biofilms both in vitro and in vivo in models of chronic wounds by significantly dissoluting the biofilm matrix and reducing its biomass, thereby increasing the efficacy of subsequent antibiotic treatment [362]. Similar results has also been reported for dispersin B [361].
Despite the progress in the field, there is still limited knowledge about these techniques, with very few reports focused on the oralome. Thus, more studies are still needed [364]. For a more detailed review EPM disruption in microbial biofilms, please refer to Koo et al. [354]. For more details on cyclic di-nucletides in the oral microbiome, please refer to Gürsoy et al. [364]
6.7. Host response modulators (HRM)
Since the host response mediates periodontal destruction through inflammation, use of host response modulators (HRM) may be an important technique against the disease. Currently, there is one commercially available product, called Periostat®. The main active ingredient in Periostat® is doxycycline, which downregulates the activity of matrix metalloproteinases (MMPs). MMPs are key destructive enzymes in periodontal disease, thus Periostat works by inhibiting collagenase activity. This is the first FDA approved systemic drug for host response modulation and it is currently indicated as an adjunct treatment to scaling and root planning in periodontitis [365]. In the same category of HRM, another heavily studied compound is the chemically modified tetracycline-3. So far, tetracycline-3 has been shown to reduce the collagenolytic activity induced by P. gingivalis in vitro [366] and it inhibits alveolar bone loss by inhibiting MMPs in vivo [367].
Another category of HRM is anti-inflammatory drugs, such as steroidal (SAID) or nonsteroidal anti-inflammatory drugs (NSAID). While, NSAID have been extensively studied in the field, evidence of the use of SAID in periodontitis are still scarce [368]. In 2016, Cavagni et al. [368] reviewed most of the current evidence for both anti-inflammatory drugs, including in vitro and in vivo assays and human clinical trials. For NSAID, the authors concluded that, despite the drugs ability to improve inflammatory parameters for both gingivitis and periodontitis, evidence showing that these drugs can indeed treat or prevent periodontal diseases is still lacking. For SAID, the authors concluded that most reports found no significant difference between SAID and controls, and effects on the periodontium remained controversial, thus further evidence is needed.
As previously discussed, the oral microbiome has an immunomodulatory effect on the host, such as priming immune responses to ensure rapid and efficient defenses against pathogens [118]. In this context, the oral microbiome produces metabolites that can immunomodulate the host response and contribute to the balance of pro- and anti-inflammatory responses, such as short chain fatty-acids (SCFA), proteins and even nucleic acids [118]. For instance, Santos Rocha et al. [369] demonstrated that a Lactobacillus delbrueckii surface protein/peptide has an anti-inflammatory effect. The authors found that the peptide was able to do this by either preventing IκB phosphorylation or promoting IκB dephosphorylation and, thus, reducing NF-κB activation. Recently, Corrêa et al. [370] demonstrated that SCFA can significantly reduce the expression of TNF-α, Cxcl2, and IL-10 in vitro and in vivo in A. actinomycetemcomitans-stimulated neutrophils.
Despite their potential in modulating the inflammatory host response and suppressing tissue destruction [371], HRM are limited in that they do not directly affect the oral dysbiosis. Thus, recurrence of periodontitis is always possible. Also, these molecules may be detrimental to the host, if not used properly, as they inhibit neutrophil effector functions, thus inhibiting inflammation [370]. Consequently, it is important to apply these locally. Finally, only limited studies have investigated possible side effects of HRM agents in humans, thus further studies are needed [372]. For more information on HRM therapy in periodontal disease, please refer to Preshaw [371] and Bartold & Van Dyke [373].
7. Clinical trials
Although the oral microbiome has been implicated in all these diseases, there are few clinical trials on this topic. Searching the clinical trials database ClinicalTrials.gov for the term “dysbiosis” in the field “condition or disease”, we found 109 clinical trials in the database. Upon closer inspection, we found that the majority of these clinical trials were focused on the gut microbiome. When limiting the search to “oral dysbiosis”, we found only 4 clinical trials that are actively evaluating the dysbiotic oral microbiome - two observational studies and two interventional studies. One observational study (NCT03225950), resulted in the publication by Damgaard et al. [374], which demonstrated that P. gingivalis was significantly associated with aggressive and chronic periodontitis. Although the relative abundance of the pathogen was different between the conditions, this difference was not sufficient to discriminate both diseases. A second observational study (NCT04251650) was trying to define the oral microbiome profiles of periodontitis patients then correlate this with the host inflammatory profile using crevicular gingival fluid. However, no results have been reported. One interventional study (NCT03863249) is trying to develop a microelectronic biosensor system (xCELLigence) to monitor and select effective antibiotic treatment for severe/chronic periodontitis patients. The other interventional study (NCT04027179) is looking to evaluate the effects of mouthwashes on the arterial blood pressure of periodontitis patients by measuring nitrite levels in saliva, bacterial levels on the tongue, arterial blood pressure over 6 months. No results have been reported for either trial.
Nguyen et al. [337], recently reviewed clinical trials using probiotics as both monotherapy and adjunctive treatment for periodontitis and gingivitis. The authors reported that the majority of clinical trials tested probiotics as monotherapy for treatment and prevention of periodontitis and gingivitis, whereas the majority of adjunctive therapy studies focused on using probiotics with scaling and root planning in the treatment of chronic periodontitis.
For probiotics as monotherapy, the authors reported mixed results from the clinical trials. Some studies demonstrated that probiotics improve several periodontal parameters, such as bleeding on probing, plaque and gingival indices, and gingival crevicular fluid volume, whereas others found no significant improvement on these periodontal clinical parameters. Thus, further clinical trials are needed to address these different findings.
In terms of adjunctive use of probiotics for periodontitis, most studies found significant clinical improvements in periodontal pocket depths, especially in the deeper periodontal pocket depths, clinical attachment levels, bleeding on probing, sulcus bleeding index, gingival and plaque indices, and/or percentage of sites with plaque. Also, several studies demonstrated that adjunctive probiotic treatment lowered the levels of matrix metalloproteinase-8, tissue inhibitor of metalloproteinase-1, and pro-inflammatory cytokines, including tumor necrosis factor-alpha, interleukin-1beta, and interleukin-17. However, the authors advised caution in extrapolating or generalizing these results, as some of these clinical trials had several limitations, such as small sample size, lack of appropriate randomization, blinding, appropriate controls, and significant heterogeneity in terms of probiotic strains, doses, duration of use, mode of delivery, and patient population studied [337].
Greber and Dawgul [375], Costa et al. [376] and Koo & Seo [377] have recently reviewed clinical trials using AMPs. The studies found a plethora of clinical trials using AMP from spermicide to infections in skin, ear canal and eyes. Among them, only one clinical trial has evaluated AMPs for any of the diseases discussed in this work - The AMP C16G2 has been investigated for dental caries by C3 Jian Corporation in phase I and II clinical trials. C16G2 is a Specifically Targeted AMP (STAMP), synthesized specifically to kill S. mutans, by selectively disrupting the bacterial membrane, without having an effect on other streptococci [377], [378]. According to Baker et al. [379], C16G2 was first tested in a randomized, double-blind, placebo-controlled phase I clinical trial that focused on evaluating safety and pharmacokinetics. The study enrolled a total of 127 subjects and tested several formulations, including a mouth rinse, an oral gel, a toothpaste, a varnish, and a tooth strip (NCT02509845, NCT03004365, and NCT03052842). The study reported no severe adverse events and that a single varnish application outperformed the other applications, followed by the toothpaste. The varnish formulation is a product candidate for in-office treatment, similar to fluoride varnishes commonly applied by dentists or hygienists for the treatment of high-risk, caries-prone populations. The phase II trial (NCT02594254) enrolled 135 patients and focused on the effectiveness of the C16G2 varnish. It showed a significant reduction in S. mutans abundance and S. mutans remained low even after the cessation of the treatment. Also, no adverse effects were reported, thereby validating the previous results. An additional phase II clinical trial of C16G2 is ongoing and it’s goal is to determine the long-term effectiveness of the treatment.
Costa et al. [376] reported on a phase I/II clinical study evaluating whether hLF1-11, a synthetic antimicrobial peptide derived the human lactoferrin [380], can be used as a treatment for Staphylococcus epidermidis bacteremia from the oral cavity. However, the trial was closed prior to enrolment.
8. Summary and future outlook
Since the first observation of microbes in dental plaque by Antony van Leeuwenhoek in the early 1670’s, the role of oral microbes has evolved from being considered simply “passengers” on the human host to one of benefiting and promoting diseases in their human hosts. The balance between health and disease has been explained by the eubiosis-dysbiosis hypothesis, which suggests that the oralome can shift from a healthy to diseased state with destructive host-pathogen interactions. Although the reasons for this shift are still unclear, in silico studies have shed light on this, indicating that small changes, such as pH alterations in specific oral sites, might drive this imbalance.
Oral dysbiosis has been linked to several host diseases, such as diabetes, atherosclerosis, Alzheimer’s disease, and Head and Neck Cancer. These reports demonstrate the presence of oral microbes in several parts of the human body, linking these systemic diseases to oral dysbiosis and, ultimately, leading to new insights into those diseases.
Finally, the development and discovery of novel antimicrobial peptides and probiotics may help modulate the oralome from a diseased state towards a healthy state, and thereby, potentially improve the host’s health or prevent diseases, such as head and neck cancer or Alzheimer’s disease.
CRediT authorship contribution statement
Allan Radaic: Conceptualization, Investigation, Writing - original draft. Yvonne L. Kapila: Conceptualization, Funding acquisition, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by funding from an AAP Sunstar Innovation Grant, and NIH R01 DE025225 grant to YLK. All original figures in this article were created with BioRender.com. The authors acknowledge the courtesy of Ryutaro Kuraji, DDS, PhD (University of California, San Francisco and Nippon Dental University) and Pachiyappan Kamarajan, PhD (University of California, San Francisco) to make available Fig. 3B.
References
- 1.Deo P.N., Deshmukh R. Oral microbiome: unveiling the fundamentals. J Oral Maxillofac Pathol. 2019;23:122–128. doi: 10.4103/jomfp.JOMFP_304_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14 doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg R.D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 4.Dovrolis N., Kolios G., Spyrou G.M., Maroulakou I. Computational profiling of the gut-brain axis: microflora dysbiosis insights to neurological disorders. Brief Bioinf. 2019;20:825–841. doi: 10.1093/bib/bbx154. [DOI] [PubMed] [Google Scholar]
- 5.Proal A.D., Lindseth I.A., Marshall T.G. Microbe-microbe and host-microbe interactions drive microbiome dysbiosis and inflammatory processes. Discov Med. 2017;23:51–60. [PubMed] [Google Scholar]
- 6.NIH Human Microbiome Project Data Portal.
- 7.Kreth J., Merritt J., Qi F. Bacterial and host interactions of oral streptococci. DNA Cell Biol. 2009;28:397–403. doi: 10.1089/dna.2009.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avila M., Ojcius D.M., Yilmaz O. The oral microbiota: living with a permanent guest. DNA Cell Biol. 2009;28:405–411. doi: 10.1089/dna.2009.0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bandara H.M.H.N., Panduwawala C.P., Samaranayake L.P. Biodiversity of the human oral mycobiome in health and disease. Oral Dis. 2019;25:363–371. doi: 10.1111/odi.12899. [DOI] [PubMed] [Google Scholar]
- 10.Berger D, Rakhamimova A, Pollack A, Loewy Z (2018) Oral biofilms: development, control, and analysis. High-Throughput 7:. [DOI] [PMC free article] [PubMed]
- 11.Morse D.J., Wilson M.J., Wei X., Lewis M.A.O., Bradshaw D.J., Murdoch C. Denture-associated biofilm infection in three-dimensional oral mucosal tissue models. J Med Microbiol. 2018;67:364–375. doi: 10.1099/jmm.0.000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radaic A, Ganther S, Kamarajan P, Yom SS, Grandis JR, Kapila YL (2021) Paradigm Shift in the Pathogenesis and Treatment of Oral Cancer Focused on the Oralome and Antimicrobial-Based Therapeutics . Periodontol 2000. [DOI] [PMC free article] [PubMed]
- 13.Ghannoum M.A., Jurevic R.J., Mukherjee P.K., Cui F., Sikaroodi M., Naqvi A. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupuy A.K., David M.S., Li L., Heider T.N., Peterson J.D., Montano E.A. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One. 2014;9 doi: 10.1371/journal.pone.0090899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saini R., Saini S., Sharma S. Biofilm: a dental microbial infection. J Nat Sci Biol Med. 2011;2:71–75. doi: 10.4103/0976-9668.82317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiao Y., Tay F.R., Niu L.-N., Chen J.-H. Advancing antimicrobial strategies for managing oral biofilm infections. Int J Oral Sci. 2019;11:28. doi: 10.1038/s41368-019-0062-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donlan R.M. Biofilms: microbial life on surfaces. Emerging Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamal M., Ahmad W., Andleeb S., Jalil F., Imran M., Nawaz M.A. Bacterial biofilm and associated infections. J Chin Med Assoc. 2018;81:7–11. doi: 10.1016/j.jcma.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Bowen W.H., Burne R.A., Wu H., Koo H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 2018;26:229–242. doi: 10.1016/j.tim.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velsko I.M., Fellows Yates J.A., Aron F., Hagan R.W., Frantz L.A.F., Loe L. Microbial differences between dental plaque and historic dental calculus are related to oral biofilm maturation stage. Microbiome. 2019;7:102. doi: 10.1186/s40168-019-0717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wade W.G. The oral microbiome in health and disease. Pharmacol Res. 2013;69:137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Jenkinson H.F. Adherence and accumulation of oral streptococci. Trends Microbiol. 1994;2:209–212. doi: 10.1016/0966-842x(94)90114-k. [DOI] [PubMed] [Google Scholar]
- 23.Jenkinson H.F. Cell surface protein receptors in oral streptococci. FEMS Microbiol Lett. 1994;121:133–140. doi: 10.1111/j.1574-6968.1994.tb07089.x. [DOI] [PubMed] [Google Scholar]
- 24.Jenkinson H.F. Genetic analysis of adherence by oral streptococci. J Ind Microbiol. 1995;15:186–192. doi: 10.1007/BF01569824. [DOI] [PubMed] [Google Scholar]
- 25.Davey M.E. Costerton JW (2006) Molecular genetics analyses of biofilm formation in oral isolates. Periodontol. 2000;42:13–26. doi: 10.1111/j.1600-0757.2006.00052.x. [DOI] [PubMed] [Google Scholar]
- 26.Jakubovics N.S., Burgess J.G. Extracellular DNA in oral microbial biofilms. Microbes Infect. 2015;17:531–537. doi: 10.1016/j.micinf.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 27.Jakubovics N.S., Shields R.C., Rajarajan N., Burgess J.G. Life after death: the critical role of extracellular DNA in microbial biofilms. Lett Appl Microbiol. 2013;57:467–475. doi: 10.1111/lam.12134. [DOI] [PubMed] [Google Scholar]
- 28.Tang L., Schramm A., Neu T.R., Revsbech N.P., Meyer R.L. Extracellular DNA in adhesion and biofilm formation of four environmental isolates: a quantitative study. FEMS Microbiol Ecol. 2013;86:394–403. doi: 10.1111/1574-6941.12168. [DOI] [PubMed] [Google Scholar]
- 29.Nadell C.D., Drescher K., Wingreen N.S., Bassler B.L. Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME J. 2015;9:1700–1709. doi: 10.1038/ismej.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kouidhi B., Al Qurashi Y.M.A., Chaieb K. Drug resistance of bacterial dental biofilm and the potential use of natural compounds as alternative for prevention and treatment. Microb Pathog. 2015;80:39–49. doi: 10.1016/j.micpath.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 31.Radaic A., Ye C., Parks B., Gao L., Kuraji R., Malone E. Modulation of pathogenic oral biofilms towards health with nisin probiotic. J Oral Microbiol. 2020;12:1809302. doi: 10.1080/20002297.2020.1809302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mark Welch J.L., Rossetti B.J., Rieken C.W., Dewhirst F.E., Borisy G.G. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA. 2016;113:E791–800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mark Welch J.L., Dewhirst F.E., Borisy G.G. Biogeography of the oral microbiome: the site-specialist hypothesis. Annu Rev Microbiol. 2019;73:335–358. doi: 10.1146/annurev-micro-090817-062503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Ohle C., Gieseke A., Nistico L., Decker E.M., DeBeer D., Stoodley P. Real-time microsensor measurement of local metabolic activities in ex vivo dental biofilms exposed to sucrose and treated with chlorhexidine. Appl Environ Microbiol. 2010;76:2326–2334. doi: 10.1128/AEM.02090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loesche W.J. Oxygen sensitivity of various anaerobic bacteria. Appl Microbiol. 1969;18:723–727. doi: 10.1128/am.18.5.723-727.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eren A.M., Borisy G.G., Huse S.M., Mark Welch J.L. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA. 2014;111:E2875–84. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoodley P., Dodds I., Boyle J.D., Lappin-Scott H.M. Influence of hydrodynamics and nutrients on biofilm structure. J Appl Microbiol. 1998;85(Suppl 1):19S–28S. doi: 10.1111/j.1365-2672.1998.tb05279.x. [DOI] [PubMed] [Google Scholar]
- 38.Socransky S.S., Haffajee A.D. Dental biofilms: difficult therapeutic targets. Periodontol. 2002;28:12–55. doi: 10.1034/j.1600-0757.2002.280102.x. [DOI] [PubMed] [Google Scholar]
- 39.Zijnge V., van Leeuwen M.B.M., Degener J.E., Abbas F., Thurnheer T., Gmür R. Oral biofilm architecture on natural teeth. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zijnge V., Ammann T., Thurnheer T., Gmür R. Subgingival biofilm structure. Front Oral Biol. 2012;15:1–16. doi: 10.1159/000329667. [DOI] [PubMed] [Google Scholar]
- 41.Aas J.A., Paster B.J., Stokes L.N., Olsen I., Dewhirst F.E. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kulik E.M., Sandmeier H., Hinni K., Meyer J. Identification of archaeal rDNA from subgingival dental plaque by PCR amplification and sequence analysis. FEMS Microbiol Lett. 2001;196:129–133. doi: 10.1111/j.1574-6968.2001.tb10553.x. [DOI] [PubMed] [Google Scholar]
- 43.Robles-Sikisaka R., Ly M., Boehm T., Naidu M., Salzman J., Pride D.T. Association between living environment and human oral viral ecology. ISME J. 2013;7:1710–1724. doi: 10.1038/ismej.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pride D.T., Salzman J., Haynes M., Rohwer F., Davis-Long C., White R.A. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012;6:915–926. doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ly M., Abeles S.R., Boehm T.K., Robles-Sikisaka R., Naidu M., Santiago-Rodriguez T. Altered oral viral ecology in association with periodontal disease. MBio. 2014;5:e01133–14. doi: 10.1128/mBio.01133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pérez-Brocal V., Moya A. The analysis of the oral DNA virome reveals which viruses are widespread and rare among healthy young adults in Valencia (Spain) PLoS One. 2018;13 doi: 10.1371/journal.pone.0191867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker J.L., Bor B., Agnello M., Shi W., He X. Ecology of the oral microbiome: beyond bacteria. Trends Microbiol. 2017;25:362–374. doi: 10.1016/j.tim.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de la Cruz Peña M.J., Martinez-Hernandez F., Garcia-Heredia I., Lluesma Gomez M., Fornas Ò., Martinez-Garcia M. Deciphering the human virome with single-virus genomics and metagenomics. Viruses. 2018;10 doi: 10.3390/v10030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao L., Kang M., Zhang M.J., Reza Sailani M., Kuraji R., Martinez A. Polymicrobial periodontal disease triggers a wide radius of effect and unique virome. npj Biofilms Microbiomes. 2020;6:10. doi: 10.1038/s41522-020-0120-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward TL, Dominguez-Bello MG, Heisel T, Al-Ghalith G, Knights D, Gale CA (2018) Development of the Human Mycobiome over the First Month of Life and across Body Sites. mSystems 3:. [DOI] [PMC free article] [PubMed]
- 51.Park H.K., Ha M.-H., Park S.-G., Kim M.N., Kim B.J., Kim W. Characterization of the fungal microbiota (mycobiome) in healthy and dandruff-afflicted human scalps. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hong B.Y., Hoare A., Cardenas A., Dupuy A.K., Choquette L., Salner A.L. The salivary mycobiome contains 2 ecologically distinct mycotypes. J Dent Res. 2020;99:730–738. doi: 10.1177/0022034520915879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diaz P.I., Hong B.-Y., Dupuy A.K., Strausbaugh L.D. Mining the oral mycobiome: methods, components, and meaning. Virulence. 2017;8:313–323. doi: 10.1080/21505594.2016.1252015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peters B.A., Wu J., Hayes R.B., Ahn J. The oral fungal mycobiome: characteristics and relation to periodontitis in a pilot study. BMC Microbiol. 2017;17:157. doi: 10.1186/s12866-017-1064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naglik J.R., Tang S.X., Moyes D.L. Oral colonization of fungi. Curr Fungal Infect Rep. 2013;7:152–159. [Google Scholar]
- 56.He J., Li Y., Cao Y., Xue J., Zhou X. The oral microbiome diversity and its relation to human diseases. Folia Microbiol (Praha) 2015;60:69–80. doi: 10.1007/s12223-014-0342-2. [DOI] [PubMed] [Google Scholar]
- 57.Kirchner F.R., Littringer K., Altmeier S., Tran V.D.T., Schönherr F., Lemberg C. Persistence of Candida albicans in the oral mucosa induces a curbed inflammatory host response that is independent of immunosuppression. Front Immunol. 2019;10:330. doi: 10.3389/fimmu.2019.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li L., Redding S., Dongari-Bagtzoglou A. Candida glabrata: an emerging oral opportunistic pathogen. J Dent Res. 2007;86:204–215. doi: 10.1177/154405910708600304. [DOI] [PubMed] [Google Scholar]
- 59.Redding S.W., Dahiya M.C., Kirkpatrick W.R., Coco B.J., Patterson T.F., Fothergill A.W. Candida glabrata is an emerging cause of oropharyngeal candidiasis in patients receiving radiation for head and neck cancer. Oral Surgery, Oral Med, Oral Pathol, Oral Radiol, Endodontol. 2004;97:47–52. doi: 10.1016/j.tripleo.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 60.Masiá Canuto M., Gutiérrez Rodero F., de la Tabla O., Ducasse V., Hernández Aguado I., Martín González C. Determinants for the development of oropharyngeal colonization or infection by fluconazole-resistant Candida strains in HIV-infected patients. Eur J Clin Microbiol Infect Dis. 2000;19:593–601. doi: 10.1007/s100960000323. [DOI] [PubMed] [Google Scholar]
- 61.Chabé M., Lokmer A., Ségurel L. Gut protozoa: friends or foes of the human gut microbiota? Trends Parasitol. 2017;33:925–934. doi: 10.1016/j.pt.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 62.Hamad I., Raoult D., Bittar F. Repertory of eukaryotes (eukaryome) in the human gastrointestinal tract: taxonomy and detection methods. Parasite Immunol. 2016;38:12–36. doi: 10.1111/pim.12284. [DOI] [PubMed] [Google Scholar]
- 63.Kofoid CA (1929) The protozoa of the human mouth. presidential address, american society of parasitologists, new york, december 29, 1928. J Parasitol 15: 151.
- 64.Wantland W.W., Wantland E.M., Remo J.W., Winquist D.L. Studies on human mouth protozoa. J Dent Res. 1958;37:949–950. doi: 10.1177/00220345580370052601. [DOI] [PubMed] [Google Scholar]
- 65.Rashidi Maybodi F., Haerian Ardakani A., Fattahi Bafghi A., Haerian Ardakani A., Zafarbakhsh A. The effect of nonsurgical periodontal therapy on trichomonas tenax and entamoeba gingivalis in patients with chronic periodontitis. J Dent (Shiraz) 2016;17:171–176. [PMC free article] [PubMed] [Google Scholar]
- 66.Ibrahim S., Abbas R. Evaluation of Entamoeba gingivalis and Trichomonas tenax in patients with periodontitis and gingivitis and its correlation with some risk factors. J Bagh Coll Dent. 2012;24:158–162. [Google Scholar]
- 67.Albuquerque Junior R.L.C., Melo C.M., Santana W.A., Ribeiro J.L., Silva F.A. Incidence of Entamoeba gingivalis and Trichomonas tenax in samples of. Rev Gaucha Odontol. 2011;59:35–40. [Google Scholar]
- 68.Dubar M., Zaffino M.-L., Remen T., Thilly N., Cunat L., Machouart M.-C. Protozoans in subgingival biofilm: clinical and bacterial associated factors and impact of scaling and root planing treatment. J Oral Microbiol. 2020;12:1693222. doi: 10.1080/20002297.2019.1693222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nagarajan M., Prabhu V.R., Kamalakkannan R. Metagenomics. Elsevier; 2018. Metagenomics: implications in oral health and disease; pp. 179–195. [Google Scholar]
- 70.Eme L., Spang A., Lombard J., Stairs C.W., Ettema T.J.G. Archaea and the origin of eukaryotes. Nat Rev Microbiol. 2017;15:711–723. doi: 10.1038/nrmicro.2017.133. [DOI] [PubMed] [Google Scholar]
- 71.Bang C., Schmitz R.A. Archaea associated with human surfaces: not to be underestimated. FEMS Microbiol Rev. 2015;39:631–648. doi: 10.1093/femsre/fuv010. [DOI] [PubMed] [Google Scholar]
- 72.Matarazzo F., Ribeiro A.C., Faveri M., Taddei C., Martinez M.B., Mayer M.P.A. The domain Archaea in human mucosal surfaces. Clin Microbiol Infect. 2012;18:834–840. doi: 10.1111/j.1469-0691.2012.03958.x. [DOI] [PubMed] [Google Scholar]
- 73.Nguyen-Hieu T., Khelaifia S., Aboudharam G., Drancourt M. Methanogenic archaea in subgingival sites: a review. APMIS. 2013;121:467–477. doi: 10.1111/apm.12015. [DOI] [PubMed] [Google Scholar]
- 74.Ferrari A., Brusa T., Rutili A., Canzi E., Biavati B. Isolation and characterization of Methanobrevibacter oralis sp. nov. Curr Microbiol. 1994;29:7–12. [Google Scholar]
- 75.Faveri M., Gonçalves L.F.H., Feres M., Figueiredo L.C., Gouveia L.A., Shibli J.A. Prevalence and microbiological diversity of Archaea in peri-implantitis subjects by 16S ribosomal RNA clonal analysis. J Periodontal Res. 2011;46:338–344. doi: 10.1111/j.1600-0765.2011.01347.x. [DOI] [PubMed] [Google Scholar]
- 76.Sogodogo E., Doumbo O., Aboudharam G., Kouriba B., Diawara O., Koita H. First characterization of methanogens in oral cavity in Malian patients with oral cavity pathologies. BMC Oral Health. 2019;19:232. doi: 10.1186/s12903-019-0929-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mosaddad S.A., Tahmasebi E., Yazdanian A., Rezvani M.B., Seifalian A., Yazdanian M. Oral microbial biofilms: an update. Eur J Clin Microbiol Infect Dis. 2019;38:2005–2019. doi: 10.1007/s10096-019-03641-9. [DOI] [PubMed] [Google Scholar]
- 78.Matarazzo F., Ribeiro A.C., Feres M., Faveri M., Mayer M.P.A. Diversity and quantitative analysis of Archaea in aggressive periodontitis and periodontally healthy subjects. J Clin Periodontol. 2011;38:621–627. doi: 10.1111/j.1600-051X.2011.01734.x. [DOI] [PubMed] [Google Scholar]
- 79.Belmok A., de Cena J.A., Kyaw C.M., Damé-Teixeira N. The oral archaeome: a scoping review. J Dent Res. 2020;99:630–643. doi: 10.1177/0022034520910435. [DOI] [PubMed] [Google Scholar]
- 80.Bang C., Schmitz R.A. Archaea: forgotten players in the microbiome. Emerg Top Life Sci. 2018;2:459–468. doi: 10.1042/ETLS20180035. [DOI] [PubMed] [Google Scholar]
- 81.Li C.L., Jiang Y.T., Liu D.L., Qian J., Liang J.P., Shu R. Prevalence and quantification of the uncommon Archaea phylotype Thermoplasmata in chronic periodontitis. Arch Oral Biol. 2014;59:822–828. doi: 10.1016/j.archoralbio.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 82.Marsh P.D., Zaura E. Dental biofilm: ecological interactions in health and disease. J Clin Periodontol. 2017;44(Suppl 18):S12–S22. doi: 10.1111/jcpe.12679. [DOI] [PubMed] [Google Scholar]
- 83.Jakubovics N.S. Intermicrobial interactions as a driver for community composition and stratification of oral biofilms. J Mol Biol. 2015;427:3662–3675. doi: 10.1016/j.jmb.2015.09.022. [DOI] [PubMed] [Google Scholar]
- 84.Wright C.J., Burns L.H., Jack A.A., Back C.R., Dutton L.C., Nobbs A.H. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu H., Jenkinson H.F., Dongari-Bagtzoglou A. Innocent until proven guilty: mechanisms and roles ofStreptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol. 2014;29:99–116. doi: 10.1111/omi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Diaz P.I. Microbial diversity and interactions in subgingival biofilm communities. Front Oral Biol. 2012;15:17–40. doi: 10.1159/000329669. [DOI] [PubMed] [Google Scholar]
- 87.Li Y.H., Lau P.C., Lee J.H., Ellen R.P., Cvitkovitch D.G. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol. 2001;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sun D. Pull in and push out: mechanisms of horizontal gene transfer in bacteria. Front Microbiol. 2018;9:2154. doi: 10.3389/fmicb.2018.02154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.von Wintersdorff C.J.H., Penders J., van Niekerk J.M., Mills N.D., Majumder S., van Alphen L.B. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front Microbiol. 2016;7:173. doi: 10.3389/fmicb.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Olsen I., Tribble G.D., Fiehn N.-E., Wang B.-Y. Bacterial sex in dental plaque. J Oral Microbiol. 2013;5 doi: 10.3402/jom.v5i0.20736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu L., Chen X., Skogerbø G., Zhang P., Chen R., He S. The human microbiome: a hot spot of microbial horizontal gene transfer. Genomics. 2012;100:265–270. doi: 10.1016/j.ygeno.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 92.Smillie C.S., Smith M.B., Friedman J., Cordero O.X., David L.A., Alm E.J. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241–244. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 93.Tribble G.D., Rigney T.W., Dao D.-H.-V., Wong C.T., Kerr J.E., Taylor B.E. Natural competence is a major mechanism for horizontal DNA transfer in the oral pathogen Porphyromonas gingivalis. MBio. 2012;3 doi: 10.1128/mBio.00231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chi F., Nolte O., Bergmann C., Ip M., Hakenbeck R. Crossing the barrier: evolution and spread of a major class of mosaic pbp2x in Streptococcus pneumoniae, S. mitis and S. oralis. Int J Med Microbiol. 2007;297:503–512. doi: 10.1016/j.ijmm.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 95.Pereira C.S., Thompson J.A., Xavier K.B. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev. 2013;37:156–181. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- 96.Tourneroche A., Lami R., Hubas C., Blanchet E., Vallet M., Escoubeyrou K. Bacterial-fungal interactions in the kelp endomicrobiota drive autoinducer-2 quorum sensing. Front Microbiol. 2019;10:1693. doi: 10.3389/fmicb.2019.01693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ryu E.-J., Sim J., Sim J., Lee J., Choi B.-K. D-Galactose as an autoinducer 2 inhibitor to control the biofilm formation of periodontopathogens. J Microbiol. 2016;54:632–637. doi: 10.1007/s12275-016-6345-8. [DOI] [PubMed] [Google Scholar]
- 98.Mashima I., Nakazawa F. Role of an autoinducer-2-like molecule from Veillonella tobetsuensis in Streptococcus gordonii biofilm formation. J Oral Biosci. 2017;59:152–156. [Google Scholar]
- 99.Rickard A.H., Palmer R.J., Blehert D.S., Campagna S.R., Semmelhack M.F., Egland P.G. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol. 2006;60:1446–1456. doi: 10.1111/j.1365-2958.2006.05202.x. [DOI] [PubMed] [Google Scholar]
- 100.Jang Y.-J., Sim J., Jun H.-K., Choi B.-K. Differential effect of autoinducer 2 of Fusobacterium nucleatum on oral streptococci. Arch Oral Biol. 2013;58:1594–1602. doi: 10.1016/j.archoralbio.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 101.Bachtiar E.W., Bachtiar B.M., Jarosz L.M., Amir L.R., Sunarto H., Ganin H. AI-2 of Aggregatibacter actinomycetemcomitans inhibits Candida albicans biofilm formation. Front Cell Infect Microbiol. 2014;4:94. doi: 10.3389/fcimb.2014.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang Y., Lin J., Harrington A., Cornilescu G., Lau G.W., Tal-Gan Y. Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 2020;117:1689–1699. doi: 10.1073/pnas.1915812117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang B.Y., Deutch A., Hong J., Kuramitsu H.K. Proteases of an early colonizer can hinder Streptococcus mutans colonization in vitro. J Dent Res. 2011;90:501–505. doi: 10.1177/0022034510388808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marsh P.D., Head D.A., Devine D.A. Ecological approaches to oral biofilms: control without killing. Caries Res. 2015;49(Suppl 1):46–54. doi: 10.1159/000377732. [DOI] [PubMed] [Google Scholar]
- 105.Nagasawa R., Yamamoto T., Utada A.S., Nomura N., Obana N. Competence-stimulating-peptide-dependent localized cell death and extracellular DNA production in Streptococcus mutans biofilms. Appl Environ Microbiol. 2020;86 doi: 10.1128/AEM.02080-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jack A.A., Daniels D.E., Jepson M.A., Vickerman M.M., Lamont R.J., Jenkinson H.F. Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology (Reading, Engl) 2015;161:411–421. doi: 10.1099/mic.0.000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Radaic A., de Jesus M.B., Kapila Y.L. Bacterial anti-microbial peptides and nano-sized drug delivery systems: the state of the art toward improved bacteriocins. J Control Release. 2020;321:100–118. doi: 10.1016/j.jconrel.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 108.Jakubovics N.S., Gill S.R., Vickerman M.M., Kolenbrander P.E. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol Ecol. 2008;66:637–644. doi: 10.1111/j.1574-6941.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu L., Tong H., Dong X. Function of the pyruvate oxidase-lactate oxidase cascade in interspecies competition between Streptococcus oligofermentans and Streptococcus mutans. Appl Environ Microbiol. 2012;78:2120–2127. doi: 10.1128/AEM.07539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Simón-Soro A., Belda-Ferre P., Cabrera-Rubio R., Alcaraz L.D., Mira A. A tissue-dependent hypothesis of dental caries. Caries Res. 2013;47:591–600. doi: 10.1159/000351663. [DOI] [PubMed] [Google Scholar]
- 111.Petersen C., Round J.L. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014;16:1024–1033. doi: 10.1111/cmi.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gao L., Xu T., Huang G., Jiang S., Gu Y., Chen F. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. 2018;9:488–500. doi: 10.1007/s13238-018-0548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Idris A., Hasnain S.Z., Huat L.Z., Koh D. Human diseases, immunity and the oral microbiota—insights gained from metagenomic studies. Oral Sci Int. 2017;14:27–32. [Google Scholar]
- 114.Tonoyan L., Vincent-Bugnas S., Olivieri C.-V., Doglio A. New viral facets in oral diseases: the EBV paradox. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20235861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sultan A.S., Kong E.F., Rizk A.M., Jabra-Rizk M.A. The oral microbiome: a Lesson in coexistence. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.He X., McLean J.S., Guo L., Lux R., Shi W. The social structure of microbial community involved in colonization resistance. ISME J. 2014;8:564–574. doi: 10.1038/ismej.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guglielmetti S., Taverniti V., Minuzzo M., Arioli S., Stuknyte M., Karp M. Oral bacteria as potential probiotics for the pharyngeal mucosa. Appl Environ Microbiol. 2010;76:3948–3958. doi: 10.1128/AEM.00109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Devine D.A., Marsh P.D., Meade J. Modulation of host responses by oral commensal bacteria. J Oral Microbiol. 2015;7:26941. doi: 10.3402/jom.v7.26941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cosseau C., Devine D.A., Dullaghan E., Gardy J.L., Chikatamarla A., Gellatly S. The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host-microbe homeostasis. Infect Immun. 2008;76:4163–4175. doi: 10.1128/IAI.00188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Baker J.L., Edlund A. Exploiting the oral microbiome to prevent tooth decay: has evolution already provided the best tools? Front Microbiol. 2018;9:3323. doi: 10.3389/fmicb.2018.03323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.La Rosa G.R.M., Gattuso G., Pedullà E., Rapisarda E., Nicolosi D., Salmeri M. Association of oral dysbiosis with oral cancer development. Oncol Lett. 2020;19:3045–3058. doi: 10.3892/ol.2020.11441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.DeGruttola A.K., Low D., Mizoguchi A., Mizoguchi E. Current understanding of dysbiosis in disease in human and animal models. Inflamm Bowel Dis. 2016;22:1137–1150. doi: 10.1097/MIB.0000000000000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Duran-Pinedo A.E. (2021) Metatranscriptomic analyses of the oral microbiome. Periodontol 2000. 2021;85:28–45. doi: 10.1111/prd.12350. [DOI] [PubMed] [Google Scholar]
- 124.Marsh P.D. In sickness and in health-what does the oral microbiome mean to us? An ecological perspective. Adv Dent Res. 2018;29:60–65. doi: 10.1177/0022034517735295. [DOI] [PubMed] [Google Scholar]
- 125.Ahluwalia A., Gladwin M., Coleman G.D., Hord N., Howard G., Kim-Shapiro D.B. Dietary nitrate and the epidemiology of cardiovascular disease: report from a national heart, lung, and blood institute workshop. J Am Heart Assoc. 2016;5 doi: 10.1161/JAHA.116.003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vanhatalo A., Blackwell J.R., L’Heureux J.E., Williams D.W., Smith A., van der Giezen M. Nitrate-responsive oral microbiome modulates nitric oxide homeostasis and blood pressure in humans. Free Radic Biol Med. 2018;124:21–30. doi: 10.1016/j.freeradbiomed.2018.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mascitti M., Togni L., Troiano G., Caponio V.C.A., Gissi D.B., Montebugnoli L. Beyond head and neck cancer: the relationship between oral microbiota and tumour development in distant organs. Front Cell Infect Microbiol. 2019;9:232. doi: 10.3389/fcimb.2019.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.García-Castillo V., Sanhueza E., McNerney E., Onate S.A., García A. Microbiota dysbiosis: a new piece in the understanding of the carcinogenesis puzzle. J Med Microbiol. 2016;65:1347–1362. doi: 10.1099/jmm.0.000371. [DOI] [PubMed] [Google Scholar]
- 129.Méthot P.-O., Alizon S. What is a pathogen? Toward a process view of host-parasite interactions. Virulence. 2014;5:775–785. doi: 10.4161/21505594.2014.960726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang W.-L., Wang S.-S., Wang H.-F., Tang Y.-J., Tang Y.-L., Liang X.-H. Who is who in oral cancer? Exp Cell Res. 2019;384 doi: 10.1016/j.yexcr.2019.111634. [DOI] [PubMed] [Google Scholar]
- 131.Do T., Devine D., Marsh P.D. Oral biofilms: molecular analysis, challenges, and future prospects in dental diagnostics. Clin Cosmet Investig Dent. 2013;5:11–19. doi: 10.2147/CCIDE.S31005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ding T., Schloss P.D. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.De Filippis F., Vannini L., La Storia A., Laghi L., Piombino P., Stellato G. The same microbiota and a potentially discriminant metabolome in the saliva of omnivore, ovo-lacto-vegetarian and Vegan individuals. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zaura E., Brandt B.W., Prodan A., Teixeira de Mattos M.J., Imangaliyev S., Kool J. On the ecosystemic network of saliva in healthy young adults. ISME J. 2017;11:1218–1231. doi: 10.1038/ismej.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Takeshita T., Kageyama S., Furuta M., Tsuboi H., Takeuchi K., Shibata Y. Bacterial diversity in saliva and oral health-related conditions: the Hisayama Study. Sci Rep. 2016;6:22164. doi: 10.1038/srep22164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rosier B.T., Marsh P.D., Mira A. Resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J Dent Res. 2018;97:371–380. doi: 10.1177/0022034517742139. [DOI] [PubMed] [Google Scholar]
- 137.Kilian M., Chapple I.L.C., Hannig M., Marsh P.D., Meuric V., Pedersen A.M.L. The oral microbiome - an update for oral healthcare professionals. Br Dent J. 2016;221:657–666. doi: 10.1038/sj.bdj.2016.865. [DOI] [PubMed] [Google Scholar]
- 138.Nath S.G., Raveendran R. Microbial dysbiosis in periodontitis. J Indian Soc Periodontol. 2013;17:543–545. doi: 10.4103/0972-124X.118334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Van Dyke T.E., Bartold P.M., Reynolds E.C. The nexus between periodontal inflammation and dysbiosis. Front Immunol. 2020;11:511. doi: 10.3389/fimmu.2020.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lamont R.J., Koo H., Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 2018;16:745–759. doi: 10.1038/s41579-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Head D., Devine A.D., Marsh P.D. In silico modelling to differentiate the contribution of sugar frequency versus total amount in driving biofilm dysbiosis in dental caries. Sci Rep. 2017;7:17413. doi: 10.1038/s41598-017-17660-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gross E.L., Leys E.J., Gasparovich S.R., Firestone N.D., Schwartzbaum J.A., Janies D.A. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol. 2010;48:4121–4128. doi: 10.1128/JCM.01232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lu M., Xuan S., Wang Z. Oral microbiota: a new view of body health. Food Sci Hum Wellness. 2019;8:8–15. [Google Scholar]
- 144.Chen H., Jiang W. Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front Microbiol. 2014;5:508. doi: 10.3389/fmicb.2014.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gomar-Vercher S., Cabrera-Rubio R., Mira A., Montiel-Company J.M., Almerich-Silla J.M. Relationship of children’s salivary microbiota with their caries status: a pyrosequencing study. Clin Oral Investig. 2014;18:2087–2094. doi: 10.1007/s00784-014-1200-y. [DOI] [PubMed] [Google Scholar]
- 146.Ai D., Huang R., Wen J., Li C., Zhu J., Xia L.C. Integrated metagenomic data analysis demonstrates that a loss of diversity in oral microbiota is associated with periodontitis. BMC Genomics. 2017;18:1041. doi: 10.1186/s12864-016-3254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Deng Z.-L., Szafrański S.P., Jarek M., Bhuju S., Wagner-Döbler I. Dysbiosis in chronic periodontitis: key microbial players and interactions with the human host. Sci Rep. 2017;7:3703. doi: 10.1038/s41598-017-03804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Almeida V de S.M., Azevedo J., Leal H.F., de Queiroz A.T.L., da Silva Filho H.P., Reis J.N. Bacterial diversity and prevalence of antibiotic resistance genes in the oral microbiome. PLoS One. 2020;15 doi: 10.1371/journal.pone.0239664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liu B., Faller L.L., Klitgord N., Mazumdar V., Ghodsi M., Sommer D.D. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Griffen A.L., Beall C.J., Campbell J.H., Firestone N.D., Kumar P.S., Yang Z.K. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Abusleme L., Dupuy A.K., Dutzan N., Silva N., Burleson J.A., Strausbaugh L.D. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schulz S., Porsch M., Grosse I., Hoffmann K., Schaller H.-G., Reichert S. Comparison of the oral microbiome of patients with generalized aggressive periodontitis and periodontitis-free subjects. Arch Oral Biol. 2019;99:169–176. doi: 10.1016/j.archoralbio.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 153.Galimanas V., Hall M.W., Singh N., Lynch M.D.J., Goldberg M., Tenenbaum H. Bacterial community composition of chronic periodontitis and novel oral sampling sites for detecting disease indicators. Microbiome. 2014;2:32. doi: 10.1186/2049-2618-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ge X., Rodriguez R., Trinh M., Gunsolley J., Xu P. Oral microbiome of deep and shallow dental pockets in chronic periodontitis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Benítez-Páez A., Belda-Ferre P., Simón-Soro A., Mira A. Microbiota diversity and gene expression dynamics in human oral biofilms. BMC Genomics. 2014;15:311. doi: 10.1186/1471-2164-15-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ogawa T., Hirose Y., Honda-Ogawa M., Sugimoto M., Sasaki S., Kibi M. Composition of salivary microbiota in elderly subjects. Sci Rep. 2018;8:414. doi: 10.1038/s41598-017-18677-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hoare A., Soto C., Rojas-Celis V., Bravo D. Chronic inflammation as a link between periodontitis and carcinogenesis. Mediators Inflamm. 2019;2019:1029857. doi: 10.1155/2019/1029857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hoppe T., Kraus D., Novak N., Probstmeier R., Frentzen M., Wenghoefer M. Oral pathogens change proliferation properties of oral tumor cells by affecting gene expression of human defensins. Tumour Biol. 2016;37:13789–13798. doi: 10.1007/s13277-016-5281-x. [DOI] [PubMed] [Google Scholar]
- 159.Gnanasekaran J., Binder Gallimidi A., Saba E., Pandi K., Eli Berchoer L., Hermano E. Intracellular porphyromonas gingivalis promotes the tumorigenic behavior of pancreatic carcinoma cells. Cancers (Basel) 2020;12 doi: 10.3390/cancers12082331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rubinstein M.R., Wang X., Liu W., Hao Y., Cai G., Han Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kostic A.D., Chun E., Robertson L., Glickman J.N., Gallini C.A., Michaud M. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Olsen I., Yilmaz Ö. Possible role of Porphyromonas gingivalis in orodigestive cancers. J Oral Microbiol. 2019;11:1563410. doi: 10.1080/20002297.2018.1563410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu X.-B., Gao Z.-Y., Sun C.-T., Wen H., Gao B., Li S.-B. The potential role of P. gingivalis in gastrointestinal cancer: a mini review. Infect Agents Cancer. 2019;14:23. doi: 10.1186/s13027-019-0239-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Miklossy J. Alzheimer’s disease–a spirochetosis? NeuroReport. 1993;4:841–848. [PubMed] [Google Scholar]
- 165.Poole S., Singhrao S.K., Kesavalu L., Curtis M.A., Crean S. Determining the presence of periodontopathic virulence factors in short-term postmortem Alzheimer’s disease brain tissue. J Alzheimers Dis. 2013;36:665–677. doi: 10.3233/JAD-121918. [DOI] [PubMed] [Google Scholar]
- 166.Zhan X., Stamova B., Jin L.-W., DeCarli C., Phinney B., Sharp F.R. Gram-negative bacterial molecules associate with Alzheimer disease pathology. Neurology. 2016;87:2324–2332. doi: 10.1212/WNL.0000000000003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dominy S.S., Lynch C., Ermini F., Benedyk M., Marczyk A., Konradi A. Porphyromonas gingivalis in Alzheimer’s disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv. 2019;5:eaau3333. doi: 10.1126/sciadv.aau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chhibber-Goel J., Singhal V., Bhowmik D., Vivek R., Parakh N., Bhargava B. Linkages between oral commensal bacteria and atherosclerotic plaques in coronary artery disease patients. npj Biofilms Microbiomes. 2016;2:7. doi: 10.1038/s41522-016-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Combs C.A., Gravett M., Garite T.J., Hickok D.E., Lapidus J., Porreco R. Amniotic fluid infection, inflammation, and colonization in preterm labor with intact membranes. Am J Obstet Gynecol. 2014;210:125.e1–125.e15. doi: 10.1016/j.ajog.2013.11.032. [DOI] [PubMed] [Google Scholar]
- 170.Bearfield C., Davenport E.S., Sivapathasundaram V., Allaker R.P. Possible association between amniotic fluid micro-organism infection and microflora in the mouth. BJOG. 2002;109:527–533. doi: 10.1111/j.1471-0528.2002.01349.x. [DOI] [PubMed] [Google Scholar]
- 171.Ring M.E., Miller W.D. The pioneer who laid the foundation for modern dental research. N Y State Dent J. 2002;68:34–37. [PubMed] [Google Scholar]
- 172.He X.-S., Shi W.-Y. Oral microbiology: past, present and future. Int J Oral Sci. 2009;1:47–58. doi: 10.4248/ijos.09029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rosier B.T., De Jager M., Zaura E., Krom B.P. Historical and contemporary hypotheses on the development of oral diseases: are we there yet? Front Cell Infect Microbiol. 2014;4:92. doi: 10.3389/fcimb.2014.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Loesche W.J. The antimicrobial treatment of periodontal disease: changing the treatment paradigm. Crit Rev Oral Biol Med. 1999;10:245–275. doi: 10.1177/10454411990100030101. [DOI] [PubMed] [Google Scholar]
- 175.Listgarten M.A. Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. J Periodontol. 1976;47:1–18. doi: 10.1902/jop.1976.47.1.1. [DOI] [PubMed] [Google Scholar]
- 176.Socransky S.S., Haffajee A.D., Cugini M.A., Smith C., Kent R.L. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 177.Socransky S.S. Microbiology of periodontal disease – present status and future considerations. J Periodontol. 1977;48:497–504. doi: 10.1902/jop.1977.48.9.497. [DOI] [PubMed] [Google Scholar]
- 178.Costerton J.W., Geesey G.G., Cheng K.J. How bacteria stick. Sci Am. 1978;238:86–95. doi: 10.1038/scientificamerican0178-86. [DOI] [PubMed] [Google Scholar]
- 179.McLean R.J.C., Lam J.S., Graham L.L. Training the biofilm generation–a tribute to J. W. Costerton. J Bacteriol. 2012;194:6706–6711. doi: 10.1128/JB.01252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Davey M.E., O’toole G.A. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sztukowska MN, Dutton LC, Delaney C, Ramsdale M, Ramage G, Jenkinson HF, Nobbs AH, Lamont RJ (2018) Community Development between Porphyromonas gingivalis and Candida albicans Mediated by InlJ and Als3. MBio 9:. [DOI] [PMC free article] [PubMed]
- 182.Tamai R., Sugamata M., Kiyoura Y. Candida albicans enhances invasion of human gingival epithelial cells and gingival fibroblasts by Porphyromonas gingivalis. Microb Pathog. 2011;51:250–254. doi: 10.1016/j.micpath.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 183.Slots J., Saygun I., Sabeti M., Kubar A. Epstein-Barr virus in oral diseases. J Periodontal Res. 2006;41:235–244. doi: 10.1111/j.1600-0765.2006.00865.x. [DOI] [PubMed] [Google Scholar]
- 184.Chalabi M., Rezaie F., Moghim S., Mogharehabed A., Rezaei M., Mehraban B. Periodontopathic bacteria and herpesviruses in chronic periodontitis. Mol Oral Microbiol. 2010;25:236–240. doi: 10.1111/j.2041-1014.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- 185.Saygun I., Kubar A., Ozdemir A., Yapar M., Slots J. Herpesviral-bacterial interrelationships in aggressive periodontitis. J Periodontal Res. 2004;39:207–212. doi: 10.1111/j.1600-0765.2004.00728.x. [DOI] [PubMed] [Google Scholar]
- 186.Saygun I., Kubar A., Ozdemir A., Slots J. Periodontitis lesions are a source of salivary cytomegalovirus and Epstein-Barr virus. J Periodontal Res. 2005;40:187–191. doi: 10.1111/j.1600-0765.2005.00790.x. [DOI] [PubMed] [Google Scholar]
- 187.Imai K., Inoue H., Tamura M., Cueno M.E., Inoue H., Takeichi O. The periodontal pathogen Porphyromonas gingivalis induces the Epstein-Barr virus lytic switch transactivator ZEBRA by histone modification. Biochimie. 2012;94:839–846. doi: 10.1016/j.biochi.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 188.Makino K., Takeichi O., Imai K., Inoue H., Hatori K., Himi K. Porphyromonas endodontalis reactivates latent Epstein-Barr virus. Int Endod J. 2018;51:1410–1419. doi: 10.1111/iej.12959. [DOI] [PubMed] [Google Scholar]
- 189.Payne M.A., Hashim A., Alsam A., Joseph S., Aduse-Opoku J., Wade W.G. Horizontal and vertical transfer of oral microbial dysbiosis and periodontal disease. J Dent Res. 2019;98:1503–1510. doi: 10.1177/0022034519877150. [DOI] [PubMed] [Google Scholar]
- 190.Marsh P.D. Are dental diseases examples of ecological catastrophes? Microbiology (Reading, Engl) 2003;149:279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 191.Takahashi N., Nyvad B. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 2011;90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- 192.Takahashi N., Nyvad B. Caries ecology revisited: microbial dynamics and the caries process. Caries Res. 2008;42:409–418. doi: 10.1159/000159604. [DOI] [PubMed] [Google Scholar]
- 193.Zargar N., Ashraf H., Marashi S.M.A., Sabeti M., Aziz A. Identification of microorganisms in irreversible pulpitis and primary endodontic infections with respect to clinical and radiographic findings. Clin Oral Investig. 2020;24:2099–2108. doi: 10.1007/s00784-019-03075-9. [DOI] [PubMed] [Google Scholar]
- 194.Takahashi N. Microbial ecosystem in the oral cavity: metabolic diversity in an ecological niche and its relationship with oral diseases. Int Congr Ser. 2005;1284:103–112. [Google Scholar]
- 195.Pitts N.B., Zero D.T., Marsh P.D., Ekstrand K., Weintraub J.A., Ramos-Gomez F. Dental caries. Nat Rev Dis Primers. 2017;3:17030. doi: 10.1038/nrdp.2017.30. [DOI] [PubMed] [Google Scholar]
- 196.Nomura R., Ogaya Y., Nakano K. Contribution of the collagen-binding proteins of streptococcus mutans to bacterial colonization of inflamed dental pulp. PLoS One. 2016;11 doi: 10.1371/journal.pone.0159613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nomura R., Matayoshi S., Otsugu M., Kitamura T., Teramoto N., Nakano K. Contribution of severe dental caries induced by streptococcus mutans to the pathogenicity of infective endocarditis. Infect Immun. 2020;88 doi: 10.1128/IAI.00897-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Marsh P.D. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8:263–271. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- 199.Hajishengallis G., Darveau R.P., Curtis M.A. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Paine R. Food web complexity and species diversity. Am Nat. 1966;100:65–75. [Google Scholar]
- 201.Paine R. A note on trophic complexity and community stability. Am Nat. 1969;103:91–93. [Google Scholar]
- 202.Lamont R.J., Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015;21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Li N., Ma W.-T., Pang M., Fan Q.-L., Hua J.-L. The commensal microbiota and viral infection: a comprehensive review. Front Immunol. 2019;10:1551. doi: 10.3389/fimmu.2019.01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lin Y.L., Li M. Human cytomegalovirus and Epstein-Barr virus inhibit oral bacteria-induced macrophage activation and phagocytosis. Oral Microbiol Immunol. 2009;24:243–248. doi: 10.1111/j.1399-302X.2009.00504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Slots J. Periodontal herpesviruses: prevalence, pathogenicity, systemic risk. Periodontol 2000. 2015;69:28–45. doi: 10.1111/prd.12085. [DOI] [PubMed] [Google Scholar]
- 206.Stein J.M., Said Yekta S., Kleines M., Ok D., Kasaj A., Reichert S. Failure to detect an association between aggressive periodontitis and the prevalence of herpesviruses. J Clin Periodontol. 2013;40:1–7. doi: 10.1111/jcpe.12021. [DOI] [PubMed] [Google Scholar]
- 207.Haffajee A.D., Cugini M.A., Tanner A., Pollack R.P., Smith C., Kent R.L. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J Clin Periodontol. 1998;25:346–353. doi: 10.1111/j.1600-051x.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- 208.Mayanagi G., Sato T., Shimauchi H., Takahashi N. Detection frequency of periodontitis-associated bacteria by polymerase chain reaction in subgingival and supragingival plaque of periodontitis and healthy subjects. Oral Microbiol Immunol. 2004;19:379–385. doi: 10.1111/j.1399-302x.2004.00172.x. [DOI] [PubMed] [Google Scholar]
- 209.Diaz P.I., Chalmers N.I., Rickard A.H., Kong C., Milburn C.L., Palmer R.J. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol. 2006;72:2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hajishengallis G., Lamont R.J. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Shaikh H.F.M., Patil S.H., Pangam T.S., Rathod K.V. Polymicrobial synergy and dysbiosis: an overview. J Indian Soc Periodontol. 2018;22:101–106. doi: 10.4103/jisp.jisp_385_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Passariello C., Gigola P., Testarelli L., Puttini M., Schippa S., Petti S. Evaluation of microbiota associated with Herpesviruses in active sites of generalized aggressive periodontitis. Ann Stomatol (Roma) 2017;8:59–70. doi: 10.11138/ads/2017.8.2.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zhao Y., Mao Y.-F., Tang Y.-S., Ni M.-Z., Liu Q.-H., Wang Y. Altered oral microbiota in chronic hepatitis B patients with different tongue coatings. World J Gastroenterol. 2018;24:3448–3461. doi: 10.3748/wjg.v24.i30.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Huynh H.T.T., Pignoly M., Nkamga V.D., Drancourt M., Aboudharam G. The repertoire of archaea cultivated from severe periodontitis. PLoS One. 2015;10 doi: 10.1371/journal.pone.0121565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Nobile C.J., Johnson A.D. Candida albicans biofilms and human disease. Annu Rev Microbiol. 2015;69:71–92. doi: 10.1146/annurev-micro-091014-104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Horliana A.C.R.T., Chambrone L., Foz A.M., Artese H.P.C., de Rabelo M de S., Pannuti C.M., Romito G.A. Dissemination of periodontal pathogens in the bloodstream after periodontal procedures: a systematic review. PLoS One. 2014;9 doi: 10.1371/journal.pone.0098271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lockhart P.B., Brennan M.T., Sasser H.C., Fox P.C., Paster B.J., Bahrani-Mougeot F.K. Bacteremia associated with toothbrushing and dental extraction. Circulation. 2008;117:3118–3125. doi: 10.1161/CIRCULATIONAHA.107.758524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.DeStefano F., Anda R.F., Kahn H.S., Williamson D.F., Russell C.M. Dental disease and risk of coronary heart disease and mortality. BMJ. 1993;306:688–691. doi: 10.1136/bmj.306.6879.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Schenkein H.A., Papapanou P.N., Genco R. Sanz M (2020) Mechanisms underlying the association between periodontitis and atherosclerotic disease. Periodontol. 2000;83:90–106. doi: 10.1111/prd.12304. [DOI] [PubMed] [Google Scholar]
- 220.Aarabi G, Heydecke G, Seedorf U (2018) Roles of oral infections in the pathomechanism of atherosclerosis. Int J Mol Sci 19:. [DOI] [PMC free article] [PubMed]
- 221.Paganini-Hill A., White S.C., Atchison K.A. Dentition, dental health habits, and dementia: the Leisure World Cohort Study. J Am Geriatr Soc. 2012;60:1556–1563. doi: 10.1111/j.1532-5415.2012.04064.x. [DOI] [PubMed] [Google Scholar]
- 222.Alonso R., Pisa D., Fernández-Fernández A.M., Carrasco L. Infection of fungi and bacteria in brain tissue from elderly persons and patients with Alzheimer’s disease. Front Aging Neurosci. 2018;10:159. doi: 10.3389/fnagi.2018.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Pritchard A.B., Crean S., Olsen I., Singhrao S.K. Periodontitis, microbiomes and their role in Alzheimer’s disease. Front Aging Neurosci. 2017;9:336. doi: 10.3389/fnagi.2017.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wu Y.-Y., Xiao E., Graves D.T. Diabetes mellitus related bone metabolism and periodontal disease. Int J Oral Sci. 2015;7:63–72. doi: 10.1038/ijos.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Graves D.T., Corrêa J.D., Silva T.A. The oral microbiota is modified by systemic diseases. J Dent Res. 2019;98:148–156. doi: 10.1177/0022034518805739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Preshaw P.M., Alba A.L., Herrera D., Jepsen S., Konstantinidis A., Makrilakis K. Periodontitis and diabetes: a two-way relationship. Diabetologia. 2012;55:21–31. doi: 10.1007/s00125-011-2342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hintao J., Teanpaisan R., Chongsuvivatwong V., Ratarasan C., Dahlen G. The microbiological profiles of saliva, supragingival and subgingival plaque and dental caries in adults with and without type 2 diabetes mellitus. Oral Microbiol Immunol. 2007;22:175–181. doi: 10.1111/j.1399-302X.2007.00341.x. [DOI] [PubMed] [Google Scholar]
- 228.Al-Maskari A.Y., Al-Maskari M.Y., Al-Sudairy S. Oral manifestations and complications of diabetes mellitus: a review. Sultan Qaboos Univ Med J. 2011;11:179–186. [PMC free article] [PubMed] [Google Scholar]
- 229.Wang R.-R., Xu Y.-S., Ji M.-M., Zhang L., Li D., Lang Q. Association of the oral microbiome with the progression of impaired fasting glucose in a Chinese elderly population. J Oral Microbiol. 2019;11:1605789. doi: 10.1080/20002297.2019.1605789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Chapple I.L.C., Genco R., Working group 2 of joint EFP, AAP workshop Diabetes and periodontal diseases: consensus report of the Joint EFP/AAP Workshop on Periodontitis and Systemic Diseases. J Clin Periodontol. 2013;40(Suppl 14):S106–12. doi: 10.1111/jcpe.12077. [DOI] [PubMed] [Google Scholar]
- 231.Sohail M.U., Althani A., Anwar H., Rizzi R., Marei H.E. Role of the gastrointestinal tract microbiome in the pathophysiology of diabetes mellitus. J Diabetes Res. 2017;2017:9631435. doi: 10.1155/2017/9631435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Althani A.A., Marei H.E., Hamdi W.S., Nasrallah G.K., El Zowalaty M.E., Al Khodor S. Human microbiome and its association with health and diseases. J Cell Physiol. 2016;231:1688–1694. doi: 10.1002/jcp.25284. [DOI] [PubMed] [Google Scholar]
- 233.Xiao E., Mattos M., Vieira G.H.A., Chen S., Corrêa J.D., Wu Y. Diabetes enhances IL-17 expression and alters the oral microbiome to increase its pathogenicity. Cell Host Microbe. 2017;22:120–128.e4. doi: 10.1016/j.chom.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Aagaard K., Ma J., Antony K.M., Ganu R., Petrosino J., Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Aagaard K., Riehle K., Ma J., Segata N., Mistretta T.-A., Coarfa C. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Xu M., Yamada M., Li M., Liu H., Chen S.G., Han Y.W. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J Biol Chem. 2007;282:25000–25009. doi: 10.1074/jbc.M611567200. [DOI] [PubMed] [Google Scholar]
- 237.Han Y.W., Ikegami A., Rajanna C., Kawsar H.I., Zhou Y., Li M. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol. 2005;187:5330–5340. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Perez-Muñoz M.E., Arrieta M.-C., Ramer-Tait A.E., Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017;5:48. doi: 10.1186/s40168-017-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.de Goffau M.C., Lager S., Sovio U., Gaccioli F., Cook E., Peacock S.J. Human placenta has no microbiome but can contain potential pathogens. Nature. 2019;572:329–334. doi: 10.1038/s41586-019-1451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Jethwa A.R., Khariwala S.S. Tobacco-related carcinogenesis in head and neck cancer. Cancer Metastasis Rev. 2017;36:411–423. doi: 10.1007/s10555-017-9689-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Reichart P.A., Nguyen X.H. Betel quid chewing, oral cancer and other oral mucosal diseases in Vietnam: a review. J Oral Pathol Med. 2008;37:511–514. doi: 10.1111/j.1600-0714.2008.00669.x. [DOI] [PubMed] [Google Scholar]
- 242.Hashibe M., Brennan P., Chuang S.-C., Boccia S., Castellsague X., Chen C. Interaction between tobacco and alcohol use and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer Epidemiol Biomarkers Prev. 2009;18:541–550. doi: 10.1158/1055-9965.EPI-08-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Fan X., Peters B.A., Jacobs E.J., Gapstur S.M., Purdue M.P., Freedman N.D. Drinking alcohol is associated with variation in the human oral microbiome in a large study of American adults. Microbiome. 2018;6:59. doi: 10.1186/s40168-018-0448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kumar P.S., Matthews C.R., Joshi V., de Jager M., Aspiras M. Tobacco smoking affects bacterial acquisition and colonization in oral biofilms. Infect Immun. 2011;79:4730–4738. doi: 10.1128/IAI.05371-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Kobayashi K., Hisamatsu K., Suzui N., Hara A., Tomita H., Miyazaki T. A review of HPV-related head and neck cancer. J Clin Med. 2018;7 doi: 10.3390/jcm7090241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Gillison M.L., Chaturvedi A.K., Anderson W.F., Fakhry C. Epidemiology of human papillomavirus-positive head and neck squamous cell carcinoma. J Clin Oncol. 2015;33:3235–3242. doi: 10.1200/JCO.2015.61.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Parkin DM, Bray F (2006) Chapter 2: The burden of HPV-related cancers. Vaccine 24 Suppl 3: S3/11-25. [DOI] [PubMed]
- 248.Thomas M., Pim D., Banks L. The role of the E6–p53 interaction in the molecular pathogenesis of HPV. Oncogene. 1999;18:7690–7700. doi: 10.1038/sj.onc.1202953. [DOI] [PubMed] [Google Scholar]
- 249.Guerrero-Preston R., White J.R., Godoy-Vitorino F., Rodríguez-Hilario A., Navarro K., González H. High-resolution microbiome profiling uncovers Fusobacterium nucleatum, Lactobacillus gasseri/johnsonii, and Lactobacillus vaginalis associated to oral and oropharyngeal cancer in saliva from HPV positive and HPV negative patients treated with surgery and chemo-radiation. Oncotarget. 2017;8:110931–110948. doi: 10.18632/oncotarget.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Alsahafi E., Begg K., Amelio I., Raulf N., Lucarelli P., Sauter T. Clinical update on head and neck cancer: molecular biology and ongoing challenges. Cell Death Dis. 2019;10:540. doi: 10.1038/s41419-019-1769-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Thompson M.P., Kurzrock R. Epstein-Barr virus and cancer. Clin Cancer Res. 2004;10:803–821. doi: 10.1158/1078-0432.ccr-0670-3. [DOI] [PubMed] [Google Scholar]
- 252.She Y., Nong X., Zhang M., Wang M. Epstein-Barr virus infection and oral squamous cell carcinoma risk: a meta-analysis. PLoS One. 2017;12 doi: 10.1371/journal.pone.0186860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Broccolo F., Ciccarese G., Rossi A., Anselmi L., Drago F., Toniolo A. Human papillomavirus (HPV) and Epstein-Barr virus (EBV) in keratinizing versus non- keratinizing squamous cell carcinoma of the oropharynx. Infect Agents Cancer. 2018;13:32. doi: 10.1186/s13027-018-0205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Cruz I., Van Den Brule A.J., Brink A.A., Snijders P.J., Walboomers J.M., Van Der Waal I. No direct role for Epstein-Barr virus in oral carcinogenesis: a study at the DNA, RNA and protein levels. Int J Cancer. 2000;86:356–361. doi: 10.1002/(sici)1097-0215(20000501)86:3<356::aid-ijc9>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 255.Tuominen H., Rautava S., Collado M.C., Syrjänen S., Rautava J. HPV infection and bacterial microbiota in breast milk and infant oral mucosa. PLoS One. 2018;13 doi: 10.1371/journal.pone.0207016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Tuominen H., Rautava S., Syrjänen S., Collado M.C., Rautava J. HPV infection and bacterial microbiota in the placenta, uterine cervix and oral mucosa. Sci Rep. 2018;8:9787. doi: 10.1038/s41598-018-27980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Farquhar D.R., Divaris K., Mazul A.L., Weissler M.C., Zevallos J.P., Olshan A.F. Poor oral health affects survival in head and neck cancer. Oral Oncol. 2017;73:111–117. doi: 10.1016/j.oraloncology.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Hashim D., Sartori S., Brennan P., Curado M.P., Wünsch-Filho V., Divaris K. The role of oral hygiene in head and neck cancer: results from International Head and Neck Cancer Epidemiology (INHANCE) consortium. Ann Oncol. 2016;27:1619–1625. doi: 10.1093/annonc/mdw224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zhang L., Liu Y., Zheng H.J., Zhang C.P. The oral microbiota may have influence on oral cancer. Front Cell Infect Microbiol. 2019;9:476. doi: 10.3389/fcimb.2019.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Guerrero-Preston R., Godoy-Vitorino F., Jedlicka A., Rodríguez-Hilario A., González H., Bondy J. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment. Oncotarget. 2016;7:51320–51334. doi: 10.18632/oncotarget.9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Shin J.M., Luo T., Kamarajan P., Fenno J.C., Rickard A.H., Kapila Y.L. Microbial communities associated with primary and metastatic head and neck squamous cell carcinoma - a high fusobacterial and low streptococcal signature. Sci Rep. 2017;7:9934. doi: 10.1038/s41598-017-09786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Gaonkar P.P., Patankar S.R., Tripathi N., Sridharan G. Oral bacterial flora and oral cancer: the possible link? J Oral Maxillofac Pathol. 2018;22:234–238. doi: 10.4103/jomfp.JOMFP_89_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Mager D.L. Bacteria and cancer: cause, coincidence or cure? A review. J Transl Med. 2006;4:14. doi: 10.1186/1479-5876-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Hayes R.B., Ahn J., Fan X., Peters B.A., Ma Y., Yang L. Association of oral microbiome with risk for incident head and neck squamous cell cancer. JAMA Oncol. 2018;4:358–365. doi: 10.1001/jamaoncol.2017.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Chang C., Geng F., Shi X., Li Y., Zhang X., Zhao X. The prevalence rate of periodontal pathogens and its association with oral squamous cell carcinoma. Appl Microbiol Biotechnol. 2019;103:1393–1404. doi: 10.1007/s00253-018-9475-6. [DOI] [PubMed] [Google Scholar]
- 266.Kuboniwa M., Hasegawa Y., Mao S., Shizukuishi S., Amano A., Lamont R.J. P. gingivalis accelerates gingival epithelial cell progression through the cell cycle. Microbes Infect. 2008;10:122–128. doi: 10.1016/j.micinf.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Zhou Y., Sztukowska M., Wang Q., Inaba H., Potempa J., Scott D.A. Noncanonical activation of β-catenin by Porphyromonas gingivalis. Infect Immun. 2015;83:3195–3203. doi: 10.1128/IAI.00302-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Kamarajan P., Ateia I., Shin J.M., Fenno J.C., Le C., Zhan L. Periodontal pathogens promote cancer aggressivity via TLR/MyD88 triggered activation of Integrin/FAK signaling that is therapeutically reversible by a probiotic bacteriocin. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Leonardi R., Talic N.F., Loreto C. MMP-13 (collagenase 3) immunolocalisation during initial orthodontic tooth movement in rats. Acta Histochem. 2007;109:215–220. doi: 10.1016/j.acthis.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 270.Szkaradkiewicz A., Karpiński T. Microbiology of chronic periodontitis. J Biol Earth Sc. 2013;3:M14–M20. [Google Scholar]
- 271.Multhoff G., Molls M., Radons J. Chronic inflammation in cancer development. Front Immunol. 2011;2:98. doi: 10.3389/fimmu.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Katz J., Wallet S., Cha S. Periodontal disease and the oral-systemic connection: “is it all the RAGE?”. Quintessence Int. 2010;41:229–237. [PubMed] [Google Scholar]
- 273.Karpiński T.M. Role of oral microbiota in cancer development. Microorganisms. 2019;7 doi: 10.3390/microorganisms7010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Hussain S.P., Hofseth L.J., Harris C.C. Radical causes of cancer. Nat Rev Cancer. 2003;3:276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 275.Piao J.-Y., Lee H.G., Kim S.-J., Kim D.-H., Han H.-J., Ngo H.-K.-C. Helicobacter pylori Activates IL-6-STAT3 signaling in human gastric cancer cells: potential roles for reactive oxygen species. Helicobacter. 2016;21:405–416. doi: 10.1111/hel.12298. [DOI] [PubMed] [Google Scholar]
- 276.Shirazi M.S.R., Al-Alo K.Z.K., Al-Yasiri M.H., Lateef Z.M., Ghasemian A. Microbiome dysbiosis and predominant bacterial species as human cancer biomarkers. J Gastrointest Cancer. 2019 doi: 10.1007/s12029-019-00311-z. [DOI] [PubMed] [Google Scholar]
- 277.Carbonero F., Benefiel A.C., Alizadeh-Ghamsari A.H., Gaskins H.R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front Physiol. 2012;3:448. doi: 10.3389/fphys.2012.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Bhatt A.P., Redinbo M.R., Bultman S.J. The role of the microbiome in cancer development and therapy. CA Cancer J Clin. 2017;67:326–344. doi: 10.3322/caac.21398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Pang X., Tang Y.-J., Ren X.-H., Chen Q.-M., Tang Y.-L., Liang X.-H. Microbiota, epithelium, inflammation, and TGF-β signaling: an intricate interaction in oncogenesis. Front Microbiol. 2018;9:1353. doi: 10.3389/fmicb.2018.01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Schwabe R.F., Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Taur Y., Pamer E.G. Microbiome mediation of infections in the cancer setting. Genome Med. 2016;8:40. doi: 10.1186/s13073-016-0306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Yuan C., Subramanian S. microRNA-mediated tumor-microbiota metabolic interactions in colorectal cancer. DNA Cell Biol. 2019;38:281–285. doi: 10.1089/dna.2018.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Peck B.C.E., Mah A.T., Pitman W.A., Ding S., Lund P.K., Sethupathy P. Functional transcriptomics in diverse intestinal epithelial cell types reveals robust microrna sensitivity in intestinal stem cells to microbial status. J Biol Chem. 2017;292:2586–2600. doi: 10.1074/jbc.M116.770099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Dalmasso G., Nguyen H.T.T., Yan Y., Laroui H., Charania M.A., Ayyadurai S. Microbiota modulate host gene expression via microRNAs. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Fan X., Alekseyenko A.V., Wu J., Peters B.A., Jacobs E.J., Gapstur S.M. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67:120–127. doi: 10.1136/gutjnl-2016-312580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Pushalkar S., Hundeyin M., Daley D., Zambirinis C.P., Kurz E., Mishra A. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8:403–416. doi: 10.1158/2159-8290.CD-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Öğrendik M. Periodontal pathogens in the etiology of pancreatic cancer. Gastrointest Tumors. 2017;3:125–127. doi: 10.1159/000452708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Evans T., Faircloth M., Deery A., Thomas V., Turner A., Dalgleish A. Analysis of K-ras gene mutations in human pancreatic cancer cell lines and in bile samples from patients with pancreatic and biliary cancers. Oncol Rep. 1997;4:1373–1381. doi: 10.3892/or.4.6.1373. [DOI] [PubMed] [Google Scholar]
- 289.Barton C.M., Staddon S.L., Hughes C.M., Hall P.A., O’Sullivan C., Klöppel G. Abnormalities of the p53 tumour suppressor gene in human pancreatic cancer. Br J Cancer. 1991;64:1076–1082. doi: 10.1038/bjc.1991.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Prassolov V.S., Sakamoto H., Nishimura S., Terada M., Sugimura T. Activation of c-Ki-ras gene in human pancreatic cancer. Jpn J Cancer Res. 1985;76:792–795. [PubMed] [Google Scholar]
- 291.Sun C.-H., Li B.-B., Wang B., Zhao J., Zhang X.-Y., Li T.-T. The role of Fusobacterium nucleatum in colorectal cancer: from carcinogenesis to clinical management. Chronic Dis Transl Med. 2019;5:178–187. doi: 10.1016/j.cdtm.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Castellarin M., Warren R.L., Freeman J.D., Dreolini L., Krzywinski M., Strauss J. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Gur C., Ibrahim Y., Isaacson B., Yamin R., Abed J., Gamliel M. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Setty Y. In-silico models of stem cell and developmental systems. Theor Biol Med Model. 2014;11:1. doi: 10.1186/1742-4682-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Brodland G.W. How computational models can help unlock biological systems. Semin Cell Dev Biol. 2015;47–48:62–73. doi: 10.1016/j.semcdb.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 296.Kollmann M., Sourjik V. In silico biology: from simulation to understanding. Curr Biol. 2007;17:R132–4. doi: 10.1016/j.cub.2006.12.034. [DOI] [PubMed] [Google Scholar]
- 297.Dos Santos-Lima E.K.N., Cardoso K.A.P.A., de Miranda P.M., Pimentel A.C.M., de Carvalho-Filho P.C., de Oliveira Y.A. In silico analysis as a strategy to identify candidate epitopes with human IgG reactivity to study Porphyromonas gingivalis virulence factors. AMB Express. 2019;9:35. doi: 10.1186/s13568-019-0757-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Valdebenito B., Tullume-Vergara P.O., González W., Kreth J., Giacaman R.A. In silico analysis of the competition between Streptococcus sanguinis and Streptococcus mutans in the dental biofilm. Mol Oral Microbiol. 2018;33:168–180. doi: 10.1111/omi.12209. [DOI] [PubMed] [Google Scholar]
- 299.Marsh P.D., Head D.A., Devine D.A. Prospects of oral disease control in the future - an opinion. J Oral Microbiol. 2014;6:26176. doi: 10.3402/jom.v6.26176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Miossec M.J., Valenzuela S.L., Pérez-Losada M., Johnson W.E., Crandall K.A., Castro-Nallar E. Evaluation of computational methods for human microbiome analysis using simulated data. PeerJ. 2020;8 doi: 10.7717/peerj.9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Bernstein D.B., Dewhirst F.E., Segrè D. Metabolic network percolation quantifies biosynthetic capabilities across the human oral microbiome. Elife. 2019;8 doi: 10.7554/eLife.39733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Rosa N., Campos B., Esteves A.C., Duarte A.S., Correia M.J., Silva R.M. Tracking the functional meaning of the human oral-microbiome protein-protein interactions. Adv Protein Chem Struct Biol. 2020;121:199–235. doi: 10.1016/bs.apcsb.2019.11.014. [DOI] [PubMed] [Google Scholar]
- 303.Ying S., Chunyang L. Correlation between phospholipase of Candida albicans and resistance to fluconazole. Mycoses. 2012;55:50–55. doi: 10.1111/j.1439-0507.2011.02024.x. [DOI] [PubMed] [Google Scholar]
- 304.Han Y.W., Wang X. Mobile microbiome: oral bacteria in extra-oral infections and inflammation. J Dent Res. 2013;92:485–491. doi: 10.1177/0022034513487559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kilian M. The oral microbiome - friend or foe? Eur J Oral Sci. 2018;126(Suppl 1):5–12. doi: 10.1111/eos.12527. [DOI] [PubMed] [Google Scholar]
- 306.Sälzer S., Graetz C., Dörfer C.E., Slot D.E., Van der Weijden F.A. Contemporary practices for mechanical oral hygiene to prevent periodontal disease. Periodontol 2000. 2020;84:35–44. doi: 10.1111/prd.12332. [DOI] [PubMed] [Google Scholar]
- 307.Chapple I.L.C., Van der Weijden F., Doerfer C., Herrera D., Shapira L., Polak D. Primary prevention of periodontitis: managing gingivitis. J Clin Periodontol. 2015;42(Suppl 16):S71–6. doi: 10.1111/jcpe.12366. [DOI] [PubMed] [Google Scholar]
- 308.Joshi S., Suominen A.L., Knuuttila M., Bernabé E. Toothbrushing behaviour and periodontal pocketing: an 11-year longitudinal study. J Clin Periodontol. 2018;45:196–203. doi: 10.1111/jcpe.12844. [DOI] [PubMed] [Google Scholar]
- 309.Yaacob M., Worthington H.V., Deacon S.A., Deery C., Walmsley A.D., Robinson P.G. Powered versus manual toothbrushing for oral health. Cochrane Database Syst Rev. 2014 doi: 10.1002/14651858.CD002281.pub3. CD002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Hujoel P.P., Hujoel M.L.A., Kotsakis G.A. Personal oral hygiene and dental caries: a systematic review of randomised controlled trials. Gerodontology. 2018;35:282–289. doi: 10.1111/ger.12331. [DOI] [PubMed] [Google Scholar]
- 311.Innes N., Fee P.A. Is personal oral hygiene advice effective in preventing coronal dental caries? Evid Based Dent. 2019;20:52–53. doi: 10.1038/s41432-019-0028-3. [DOI] [PubMed] [Google Scholar]
- 312.ten Cate J.M., Featherstone J.D. Mechanistic aspects of the interactions between fluoride and dental enamel. Crit Rev Oral Biol Med. 1991;2:283–296. doi: 10.1177/10454411910020030101. [DOI] [PubMed] [Google Scholar]
- 313.Zhang M., Jiang Z., Li D., Jiang D., Wu Y., Ren H. Oral antibiotic treatment induces skin microbiota dysbiosis and influences wound healing. Microb Ecol. 2015;69:415–421. doi: 10.1007/s00248-014-0504-4. [DOI] [PubMed] [Google Scholar]
- 314.Jin Y., Wu Y., Zeng Z., Jin C., Wu S., Wang Y. From the cover: exposure to oral antibiotics induces gut microbiota dysbiosis associated with lipid metabolism dysfunction and low-grade inflammation in mice. Toxicol Sci. 2016;154:140–152. doi: 10.1093/toxsci/kfw150. [DOI] [PubMed] [Google Scholar]
- 315.Shin J.M., Ateia I., Paulus J.R., Liu H., Fenno J.C., Rickard A.H. Antimicrobial nisin acts against saliva derived multi-species biofilms without cytotoxicity to human oral cells. Front Microbiol. 2015;6:617. doi: 10.3389/fmicb.2015.00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Altman H., Steinberg D., Porat Y., Mor A., Fridman D., Friedman M. In vitro assessment of antimicrobial peptides as potential agents against several oral bacteria. J Antimicrob Chemother. 2006;58:198–201. doi: 10.1093/jac/dkl181. [DOI] [PubMed] [Google Scholar]
- 317.Sol A., Ginesin O., Chaushu S., Karra L., Coppenhagen-Glazer S., Ginsburg I. LL-37 opsonizes and inhibits biofilm formation of Aggregatibacter actinomycetemcomitans at subbactericidal concentrations. Infect Immun. 2013;81:3577–3585. doi: 10.1128/IAI.01288-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Joo H.-S., Fu C.-I., Otto M. Bacterial strategies of resistance to antimicrobial peptides. Philos Trans R Soc Lond B Biol Sci. 2016:371. doi: 10.1098/rstb.2015.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Martin-Serrano Á., Gómez R., Ortega P., de la Mata F.J. Nanosystems as vehicles for the delivery of antimicrobial peptides (amps) Pharmaceutics. 2019;11 doi: 10.3390/pharmaceutics11090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Teixeira MC, Carbone C, Sousa MC, Espina M, Garcia ML, Sanchez-Lopez E, Souto EB (2020) Nanomedicines for the delivery of antimicrobial peptides (amps). Nanomaterials (Basel) 10. [DOI] [PMC free article] [PubMed]
- 321.Makowski M., Silva Í.C., Pais do Amaral C., Gonçalves S., Santos N.C. Advances in lipid and metal nanoparticles for antimicrobial peptide delivery. Pharmaceutics. 2019;11 doi: 10.3390/pharmaceutics11110588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Radaic A., de Jesus M.B. Solid lipid nanoparticles release DNA upon endosomal acidification in human embryonic kidney cells. Nanotechnology. 2018;29 doi: 10.1088/1361-6528/aac447. [DOI] [PubMed] [Google Scholar]
- 323.Morones J.R., Elechiguerra J.L., Camacho A., Holt K., Kouri J.B., Ramírez J.T. The bactericidal effect of silver nanoparticles. Nanotechnology. 2005;16:2346–2353. doi: 10.1088/0957-4484/16/10/059. [DOI] [PubMed] [Google Scholar]
- 324.Verran J., Sandoval G., Allen N.S., Edge M., Stratton J. Variables affecting the antibacterial properties of nano and pigmentary Titania particles in suspension. Dyes Pigm. 2007;73:298–304. [Google Scholar]
- 325.Allaker R.P., Yuan Z. Nanobiomaterials in clinical dentistry. Elsevier; 2019. Nanoparticles and the control of oral biofilms; pp. 243–275. [Google Scholar]
- 326.Horev B., Klein M.I., Hwang G., Li Y., Kim D., Koo H. pH-activated nanoparticles for controlled topical delivery of farnesol to disrupt oral biofilm virulence. ACS Nano. 2015;9:2390–2404. doi: 10.1021/nn507170s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Bernegossi J., Calixto G.M.F., Sanches P.R. da S., Fontana C.R., Cilli E.M., Garrido S.S., Chorilli M. Peptide KSL-W-loaded mucoadhesive liquid crystalline vehicle as an alternative treatment for multispecies oral biofilm. Molecules. 2015;21:E37. doi: 10.3390/molecules21010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Baer D.R. The chameleon effect: characterization challenges due to the variability of nanoparticles and their surfaces. Front Chem. 2018;6:145. doi: 10.3389/fchem.2018.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Mülhopt S, Diabaté S, Dilger M, Adelhelm C, Anderlohr C, Bergfeldt T, Gómez de la Torre J, Jiang Y, Valsami-Jones E, Langevin D, et al. (2018) Characterization of Nanoparticle Batch-To-Batch Variability. Nanomaterials (Basel) 8. [DOI] [PMC free article] [PubMed]
- 330.Nascimento M.M. Potential uses of arginine in dentistry. Adv Dent Res. 2018;29:98–103. doi: 10.1177/0022034517735294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Agnello M., Cen L., Tran N.C., Shi W., McLean J.S., He X. Arginine improves ph homeostasis via metabolism and microbiome modulation. J Dent Res. 2017;96:924–930. doi: 10.1177/0022034517707512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Rosier B.T., Buetas E., Moya-Gonzalvez E.M., Artacho A., Mira A. Nitrate as a potential prebiotic for the oral microbiome. Sci Rep. 2020;10:12895. doi: 10.1038/s41598-020-69931-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Slomka V., Hernandez-Sanabria E., Herrero E.R., Zaidel L., Bernaerts K., Boon N. Nutritional stimulation of commensal oral bacteria suppresses pathogens: the prebiotic concept. J Clin Periodontol. 2017;44:344–352. doi: 10.1111/jcpe.12700. [DOI] [PubMed] [Google Scholar]
- 334.Slomka V., Herrero E.R., Boon N., Bernaerts K., Trivedi H.M., Daep C. Oral prebiotics and the influence of environmental conditions in vitro. J Periodontol. 2018;89:708–717. doi: 10.1002/JPER.17-0437. [DOI] [PubMed] [Google Scholar]
- 335.Ástvaldsdóttir Á., Naimi-Akbar A., Davidson T., Brolund A., Lintamo L., Attergren Granath A. Arginine and caries prevention: a systematic review. Caries Res. 2016;50:383–393. doi: 10.1159/000446249. [DOI] [PubMed] [Google Scholar]
- 336.Bosch M., Nart J., Audivert S., Bonachera M.A., Alemany A.S., Fuentes M.C. Isolation and characterization of probiotic strains for improving oral health. Arch Oral Biol. 2012;57:539–549. doi: 10.1016/j.archoralbio.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 337.Nguyen T., Brody H., Lin G.-H., Rangé H., Kuraji R., Ye C. Probiotics, including nisin-based probiotics, improve clinical and microbial outcomes relevant to oral and systemic diseases. Periodontol 2000. 2020;82:173–185. doi: 10.1111/prd.12324. [DOI] [PubMed] [Google Scholar]
- 338.Zhu Y., Xiao L., Shen D., Hao Y. Competition between yogurt probiotics and periodontal pathogens in vitro. Acta Odontol Scand. 2010;68:261–268. doi: 10.3109/00016357.2010.492235. [DOI] [PubMed] [Google Scholar]
- 339.Khalaf H., Nakka S.S., Sandén C., Svärd A., Hultenby K., Scherbak N. Antibacterial effects of Lactobacillus and bacteriocin PLNC8 αβ on the periodontal pathogen Porphyromonas gingivalis. BMC Microbiol. 2016;16:188. doi: 10.1186/s12866-016-0810-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Kõll-Klais P., Mändar R., Leibur E., Marcotte H., Hammarström L., Mikelsaar M. Oral lactobacilli in chronic periodontitis and periodontal health: species composition and antimicrobial activity. Oral Microbiol Immunol. 2005;20:354–361. doi: 10.1111/j.1399-302X.2005.00239.x. [DOI] [PubMed] [Google Scholar]
- 341.Terai T., Okumura T., Imai S., Nakao M., Yamaji K., Ito M. Screening of probiotic candidates in human oral bacteria for the prevention of dental disease. PLoS One. 2015;10 doi: 10.1371/journal.pone.0128657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Doran A., Kneist S., Verran J. Ecological control: In vitro inhibition of anaerobic bacteria by oral streptococci. Microb Ecol Health Dis. 2004;16:23–27. [Google Scholar]
- 343.Lorquet F., Goffin P., Muscariello L., Baudry J.-B., Ladero V., Sacco M. Characterization and functional analysis of the poxB gene, which encodes pyruvate oxidase in Lactobacillus plantarum. J Bacteriol. 2004;186:3749–3759. doi: 10.1128/JB.186.12.3749-3759.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Marty-Teysset C., de la Torre F., Garel J. Increased production of hydrogen peroxide by Lactobacillus delbrueckii subsp. bulgaricus upon aeration: involvement of an NADH oxidase in oxidative stress. Appl Environ Microbiol. 2000;66:262–267. doi: 10.1128/aem.66.1.262-267.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Hertzberger R., Arents J., Dekker H.L., Pridmore R.D., Gysler C., Kleerebezem M. H(2)O(2) production in species of the Lactobacillus acidophilus group: a central role for a novel NADH-dependent flavin reductase. Appl Environ Microbiol. 2014;80:2229–2239. doi: 10.1128/AEM.04272-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Nguyen T, Brody H, Radaic A, Kapila YL (2021) Probiotics for Periodontal Health -Current Molecular Findings. Periodontol 2000. [DOI] [PMC free article] [PubMed]
- 347.Twetman S., Derawi B., Keller M., Ekstrand K., Yucel-Lindberg T., Stecksen-Blicks C. Short-term effect of chewing gums containing probiotic Lactobacillus reuteri on the levels of inflammatory mediators in gingival crevicular fluid. Acta Odontol Scand. 2009;67:19–24. doi: 10.1080/00016350802516170. [DOI] [PubMed] [Google Scholar]
- 348.Messora M.R., Oliveira L.F.F., Foureaux R.C., Taba M., Zangerônimo M.G., Furlaneto F.A.C. Probiotic therapy reduces periodontal tissue destruction and improves the intestinal morphology in rats with ligature-induced periodontitis. J Periodontol. 2013;84:1818–1826. doi: 10.1902/jop.2013.120644. [DOI] [PubMed] [Google Scholar]
- 349.Chatterjee A., Bhattacharya H., Kandwal A. Probiotics in periodontal health and disease. J Indian Soc Periodontol. 2011;15:23–28. doi: 10.4103/0972-124X.82260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Evivie S.E., Huo G.-C., Igene J.O., Bian X. Some current applications, limitations and future perspectives of lactic acid bacteria as probiotics. Food Nutr Res. 2017;61:1318034. doi: 10.1080/16546628.2017.1318034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Kailasapathy K., Chin J. Survival and therapeutic potential of probiotic organisms with reference to Lactobacillus acidophilus and Bifidobacterium spp. Immunol Cell Biol. 2000;78:80–88. doi: 10.1046/j.1440-1711.2000.00886.x. [DOI] [PubMed] [Google Scholar]
- 352.Shah N.P. Functional cultures and health benefits. Int Dairy J. 2007;17:1262–1277. [Google Scholar]
- 353.Evivie S.E. Preliminary studies on pharmaceutical microencapsulation for synbiotic application. JANS. 2013;5:488–496. [Google Scholar]
- 354.Koo H., Allan R.N., Howlin R.P., Stoodley P., Hall-Stoodley L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol. 2017;15:740–755. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Ribbe J, Baker AE, Euler S, O’Toole GA, Maier B (2017) Role of Cyclic Di-GMP and Exopolysaccharide in Type IV Pilus Dynamics. J Bacteriol 199. [DOI] [PMC free article] [PubMed]
- 356.Cheng X., Zheng X., Zhou X., Zeng J., Ren Z., Xu X. Regulation of oxidative response and extracellular polysaccharide synthesis by a diadenylate cyclase in Streptococcus mutans. Environ Microbiol. 2016;18:904–922. doi: 10.1111/1462-2920.13123. [DOI] [PubMed] [Google Scholar]
- 357.Yan W., Qu T., Zhao H., Su L., Yu Q., Gao J. The effect of c-di-GMP (3’-5’-cyclic diguanylic acid) on the biofilm formation and adherence of Streptococcus mutans. Microbiol Res. 2010;165:87–96. doi: 10.1016/j.micres.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 358.Zheng Y., Tsuji G., Opoku-Temeng C., Sintim H.O. Inhibition of P. aeruginosa c-di-GMP phosphodiesterase RocR and swarming motility by a benzoisothiazolinone derivative. Chem Sci. 2016;7:6238–6244. doi: 10.1039/c6sc02103d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Zechel D.L., Withers S.G. Comprehensive natural products chemistry. Elsevier; 1999. Glycosyl Transferase Mechanisms; pp. 279–314. [Google Scholar]
- 360.Ren Z., Cui T., Zeng J., Chen L., Zhang W., Xu X. Molecule targeting glucosyltransferase inhibits streptococcus mutans biofilm formation and virulence. Antimicrob Agents Chemother. 2016;60:126–135. doi: 10.1128/AAC.00919-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Kaplan J.B. Biofilm matrix-degrading enzymes. Methods Mol Biol. 2014;1147:203–213. doi: 10.1007/978-1-4939-0467-9_14. [DOI] [PubMed] [Google Scholar]
- 362.Fleming D., Chahin L., Rumbaugh K. Glycoside hydrolases degrade polymicrobial bacterial biofilms in wounds. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.01998-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Pleszczyńska M., Wiater A., Janczarek M., Szczodrak J. (1→3)-α-D-Glucan hydrolases in dental biofilm prevention and control: a review. Int J Biol Macromol. 2015;79:761–778. doi: 10.1016/j.ijbiomac.2015.05.052. [DOI] [PubMed] [Google Scholar]
- 364.Gürsoy U.K., Gürsoy M., Könönen E., Sintim H.O. Cyclic dinucleotides in oral bacteria and in oral biofilms. Front Cell Infect Microbiol. 2017;7:273. doi: 10.3389/fcimb.2017.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Preshaw P.M., Hefti A.F., Jepsen S., Etienne D., Walker C., Bradshaw M.H. Subantimicrobial dose doxycycline as adjunctive treatment for periodontitis. A review. J Clin Periodontol. 2004;31:697–707. doi: 10.1111/j.1600-051X.2004.00558.x. [DOI] [PubMed] [Google Scholar]
- 366.Grenier D., Plamondon P., Sorsa T., Lee H.M., McNamara T., Ramamurthy N.S. Inhibition of proteolytic, serpinolytic, and progelatinase-b activation activities of periodontopathogens by doxycycline and the non-antimicrobial chemically modified tetracycline derivatives. J Periodontol. 2002;73:79–85. doi: 10.1902/jop.2002.73.1.79. [DOI] [PubMed] [Google Scholar]
- 367.Ramamurthy N.S., Rifkin B.R., Greenwald R.A., Xu J.-W., Liu Y., Turner G. Inhibition of matrix metalloproteinase-mediated periodontal bone loss in rats: a comparison of 6 chemically modified tetracyclines. J Periodontol. 2002;73:726–734. doi: 10.1902/jop.2002.73.7.726. [DOI] [PubMed] [Google Scholar]
- 368.Cavagni J., Muniz F.W.M.G., Rösing C.K. The effect of inflammatory response modulator agents on gingivitis and periodontitis. RGO, Rev Gaúch Odontol. 2016;64:312–319. [Google Scholar]
- 369.Santos Rocha C., Lakhdari O., Blottière H.M., Blugeon S., Sokol H., Bermúdez-Humarán L.G. Anti-inflammatory properties of dairy lactobacilli. Inflamm Bowel Dis. 2012;18:657–666. doi: 10.1002/ibd.21834. [DOI] [PubMed] [Google Scholar]
- 370.Corrêa RO, Vieira A, Sernaglia EM, Lancellotti M, Vieira AT, Avila-Campos MJ, Rodrigues HG, Vinolo MAR (2017) Bacterial short-chain fatty acid metabolites modulate the inflammatory response against infectious bacteria. Cell Microbiol 19. [DOI] [PubMed]
- 371.Preshaw P.M. Host modulation therapy with anti-inflammatory agents. Periodontol 2000. 2018;76:131–149. doi: 10.1111/prd.12148. [DOI] [PubMed] [Google Scholar]
- 372.Sulijaya B., Takahashi N., Yamazaki K. Host modulation therapy using anti-inflammatory and antioxidant agents in periodontitis: a review to a clinical translation. Arch Oral Biol. 2019;105:72–80. doi: 10.1016/j.archoralbio.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 373.Bartold P.M., Van Dyke T.E. Host modulation: controlling the inflammation to control the infection. Periodontol 2000. 2017;75:317–329. doi: 10.1111/prd.12169. [DOI] [PubMed] [Google Scholar]
- 374.Damgaard C., Danielsen A.K., Enevold C., Massarenti L., Nielsen C.H., Holmstrup P. Porphyromonas gingivalis in saliva associates with chronic and aggressive periodontitis. J Oral Microbiol. 2019;11:1653123. doi: 10.1080/20002297.2019.1653123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Greber K.E., Dawgul M. Antimicrobial peptides under clinical trials. Curr Top Med Chem. 2017;17:620–628. doi: 10.2174/1568026616666160713143331. [DOI] [PubMed] [Google Scholar]
- 376.Costa F., Teixeira C., Gomes P., Martins M.C.L. Clinical application of amps. Adv Exp Med Biol. 2019;1117:281–298. doi: 10.1007/978-981-13-3588-4_15. [DOI] [PubMed] [Google Scholar]
- 377.Koo H.B., Seo J. Antimicrobial peptides under clinical investigation. Peptide Sci. 2019:111. [Google Scholar]
- 378.Kaplan C.W., Sim J.H., Shah K.R., Kolesnikova-Kaplan A., Shi W., Eckert R. Selective membrane disruption: mode of action of C16G2, a specifically targeted antimicrobial peptide. Antimicrob Agents Chemother. 2011;55:3446–3452. doi: 10.1128/AAC.00342-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Baker J.L., He X., Shi W. Precision reengineering of the oral microbiome for caries management. Adv Dent Res. 2019;30:34–39. doi: 10.1177/0022034519877386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.van der Does A.M., Hensbergen P.J., Bogaards S.J., Cansoy M., Deelder A.M., van Leeuwen H.C. The human lactoferrin-derived peptide hLF1-11 exerts immunomodulatory effects by specific inhibition of myeloperoxidase activity. J Immunol. 2012;188:5012–5019. doi: 10.4049/jimmunol.1102777. [DOI] [PubMed] [Google Scholar]