Summary
Aminoacyl-tRNA synthetases (AARS) participate in decoding the genome by catalyzing conjugation of amino acids to their cognate tRNAs. During evolution, biochemical and environmental conditions markedly influenced the sequence and structure of the 20 AARSs, revealing adaptations dictating canonical and orthogonal activities. Here, we investigate the function of the appended Zn2+-binding domain (ZBD) in the bifunctional AARS, glutamyl-prolyl-tRNA synthetase (GluProRS). We developed GluProRS mutant mice by CRISPR-Cas9 with a deletion of 29 C-terminal amino acids, including two of four Zn2+-coordinating cysteines. Homozygous ZBD mutant mice die before embryonic day 12.5, but heterozygous mice are healthy. ZBD disruption profoundly reduces GluProRS canonical function by dual mechanisms: it induces rapid proteasomal degradation of the protein and inhibits ProRS aminoacylation activity, likely by sub-optimal positioning of ATP in the spatially adjacent catalytic domain. Collectively, our studies reveal the ZBD as a critical determinant of ProRS activity and GluProRS stability in vitro and in vivo.
Subject areas: Biological Sciences, Biochemistry, Molecular Biology
Graphical abstract
Highlights
-
•
Conserved zinc-binding domain (ZBD) of GluProRS is required for Pro-tRNA charging
-
•
ZBD stabilizes GluProRS and positions C-terminal carboxylate in the catalytic site
-
•
Embryonic lethality in mice with defective GluProRS ZBD reveals in vivo essentiality
-
•
Locked nucleic acid qPCR assay for CRISPR-mediated screening of chimeric mutant mice
Biological Sciences; Biochemistry; Molecular Biology
Introduction
Aminoacyl tRNA synthetases (AARS) are essential enzymes that decode genetic information by recognizing specific tRNAs harboring appropriate anti-codon triplets and catalyzing ATP-dependent ligation to the cognate amino acid. Twenty AARSs, one for each proteinaceous amino acid, are found in the three kingdoms of life with limited exceptions in certain bacteria and archaea. Each AARS contains an anticodon-binding domain for tRNA recognition and binding, and a core catalytic domain for amino acid activation and transfer. In addition to the principal aminoacylation activity, eukaryotic AARS have acquired appended domains during evolution, generally at the N or C terminus, that display additional non-canonical functions beyond translation of the genetic code (Guo et al., 2010). The non-canonical activities are diverse in scope and contribute to development as well as pathology, e.g., tumorigenesis, obesity, angiogenesis, and inflammation (Guo and Schimmel, 2013; Kwon et al., 2019; Lee et al., 2018; Yao and Fox, 2013).
Of the 20 AARSs, the genes encoding GluRS and ProRS, i.e., EARS and PARS, are fused in metazoans to express the bifunctional GluProRS. The synthetase domains are joined by a non-catalytic linker containing a variable number of WHEP repeated domains: three in humans and mice (Ray and Fox, 2014). The fusion might be an evolutionary strategy to overcome adverse metabolic and environmental conditions (Eswarappa et al., 2018). Human GluProRS resides in a large, cytoplasmic multi-tRNA synthetase complex (MSC) containing seven other AARSs and three non-catalytic proteins. GluProRS is present as an obligate dimer (Khan et al., 2020; Quevillon et al., 1999). Several mutations in human EPRS1 (the nuclear gene encoding GluProRS) cause severe, debilitating pathologies. For example, compound heterozygous (p.Pro1160Ser and p.Thr1223LeufsX3) and homozygous (p.Pro1115Arg and p.Met1126Thr) mutations in EPRS1 cause juvenile-onset hypomyelinating leukodystrophy (Mendes et al., 2018). A recent study links a missense EPRS1 mutation to a Parkinson disease (Yemni et al., 2019). In mice, a mutation in the GluProRS linker that abrogates phosphorylation leads to low body weight and increased lifespan (Arif et al., 2017). Eprs1-haploid mice exhibit enhanced viremia, inflammation, and delayed viral clearance (Lee et al., 2016). Cardiac-specific, Eprs1 conditional knockout mice show reduced cardiac fibrosis under stress, suggesting that mild reduction of GluProRS expression is cardioprotective (Wu et al., 2020). At the cellular level, GluProRS exhibits several non-canonical activities. In interferon-γ-activated myeloid cells, GluProRS forms a heterotetrameric, interferon-γ-activated inhibitor of translation (GAIT) complex that binds and silences the translation of multiple mRNAs encoding inflammation-related proteins (Mukhopadhyay et al., 2009; Sampath et al., 2004). In adipocytes, insulin induces binding of GluProRS to fatty acid transport protein 1 and consequent transport to the plasma membrane for enhanced long-chain fatty acid uptake (Arif et al., 2017). With respect to therapeutic approaches, halofuginone, a plant alkaloid isolated from Dichroa febrifuga inhibits ProRS catalytic activity, and exhibits potent anti-fibrotic activity (Keller et al., 2012).
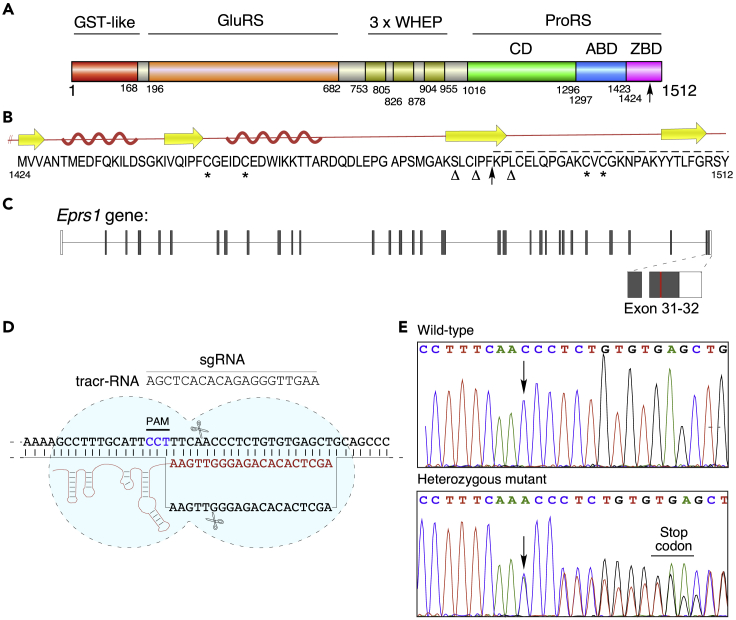
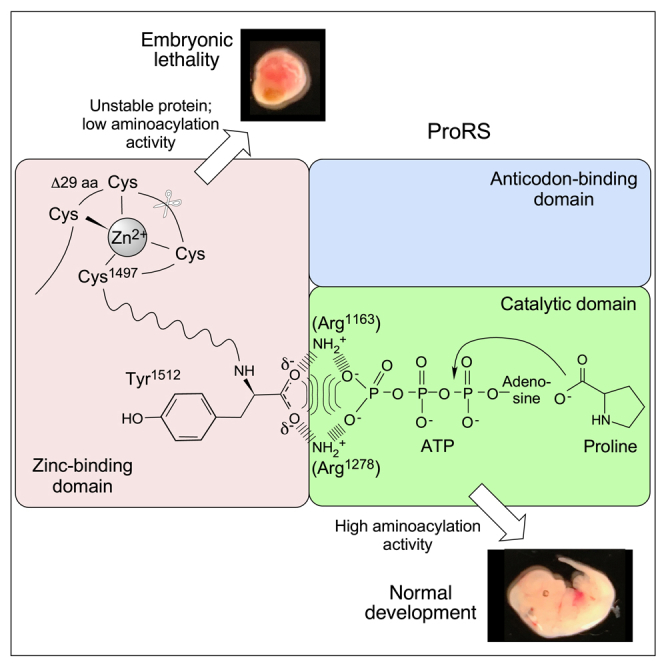
Mammalian bifunctional GluProRS has three appended domains lacking catalytic or substrate-binding activities: an N-terminal GST-like domain, three repeated WHEP domains in the linker joining the catalytic GluRS and ProRS domains, and the C-terminal ZBD (Figure 1A). A major function of the GST-like domain is to maintain structural integrity of the MSC by binary interactions with GST-like domains in other MSC constituents (Cho et al., 2015, 2019; Khan et al., 2020). In addition, following viral infection, the GST-like domain interacts with PCBP2, a negative regulator of MAVS, thereby enhancing antiviral activity (Lee et al., 2016). Several non-canonical functions of GluProRS are attributed to the WHEP domain-containing linker, including binding GAIT RNA elements and fatty acid transport protein 1 (Arif et al., 2017; Jia et al., 2008). In the C-terminal ZBD, Zn2+ is coordinated by four highly conserved Cys residues (Figures 1B and S1A). However, the functional significance of the mammalian ZBD remains unclear (Son et al., 2013; Yaremchuk et al., 2000; Zhou et al., 2013). Here, we have used CRISPR-Cas9 technology to generate a targeted mutation in the GluProRS ZBD causing a domain truncation that eliminates the terminal 29 amino acids and two of the four conserved Zn2+-binding cysteine residues (Figures 1A and 1B). Homozygous mice carrying the mutant allele are embryonic-lethal, revealing the essentiality of the ZBD. Structural and biochemical analysis reveal that the ZBD is a critical determinant of ProRS catalytic activity and GluProRS stability.
Figure 1.
Generation of Eprs1 ZBD-defective mutant mice
(A) Domain organization of human GluProRS; deletion start site (arrow).
(B) Sequence and secondary structure of human GluProRS ZBD. Horizontal arrows, β-sheets; “squiggly” lines, α-helices; ∗, Zn2+-coordinating Cys residues; Δ, identified SNPs; vertical arrow and dashed line indicate deletion.
(C) Design strategy for substitution in mouse Eprs1 exon 32; red line indicates mutation site.
(D) Detailed schematic of region targeted by sgRNA 483.
(E) Sanger sequencing of genomic DNA from wild-type and Eprs1+/ΔZ mice (arrow, insertion site).
Results
Generation of Eprs1 mutant mice with a partial Zn2+-binding domain deletion
The physiological significance of the GluProRS/ProRS ZBD has not been investigated in any model organism. The mouse and human ZBD amino acid sequences are homologous, with 88% identity and 95% similarity (Figure S1A). We generated an Eprs1 CRISPR mutant with a frameshift mutation leading to premature termination of translation of the Eprs1 gene. In mice, the ZBD occupies exons 31 and 32 at the terminus of the unspliced mRNA (Figure 1C). Targeting the 3′ end of the Eprs1 gene was accomplished by screening guide RNAs directed against the sequence. To prevent nonsense-mediated decay, exon 32 was selected for guide RNA screening. sgRNA 483 efficiently targeted the GluProRS C-terminal domain and was selected (Figures S2A and 1D). Interestingly, multiple non-synonymous SNPs were reported in human exon 32 that could potentially disrupt the ZBD (Figure 1B). Nuclear injection of CRISPR gRNA and Cas9 generated multiple allelic variants at the cut site with variable efficiency. Miseq analysis of the region surrounding the Eprs1 target site c.4449 was performed in 23 founder chimeric mice to comprehensively profile mosaicism (Figure S2B). Non-homologous end joining repair resulted in insertion of a single adenine nucleotide at c.4449 that introduces a frameshift and an in-frame stop codon 11 nucleotides downstream (Figure 1E). The mutation caused deletion of the terminal 29 amino acids and loss of two of the four conserved Cys residues essential for binding one Zn2+ ion per monomer (Figure 1B). This truncated protein (ΔZ) harbors the entire catalytic and anti-codon binding domains. Two founder mice with high mutagenesis efficiency, and without other secondary mutations, were selected for breeding and experiments.
To detect the desired allele in F1 mice, we designed a novel locked nucleic acid (LNA)-containing fluorescent probe set to distinguish between the wild-type (WT) and mutant alleles that differ by a single nucleotide. Two primers that spanned the mutation site in Eprs1 exon 32 resulted in amplification of a 103-bp segment (Figure S3A). One probe was conjugated with 5′ HEX fluorophore and 3′ IBFQ (Iowa Black fluorescent quencher) to detect the WT allele and another with 5′ 6-FAM and 3′ IBFQ to detect the mutant allele (Figure S3B). qPCR permitted definitive identification of all three mouse genotypes from tail DNA (Figure S3C). This qPCR design of LNA-based fluorescent probes can be generalized by others for distinguishing between alleles differing by a single nucleotide for routine screening of CRISPR-targeted mice.
Homozygous partial deletion of Eprs1 ZBD is embryonically lethal
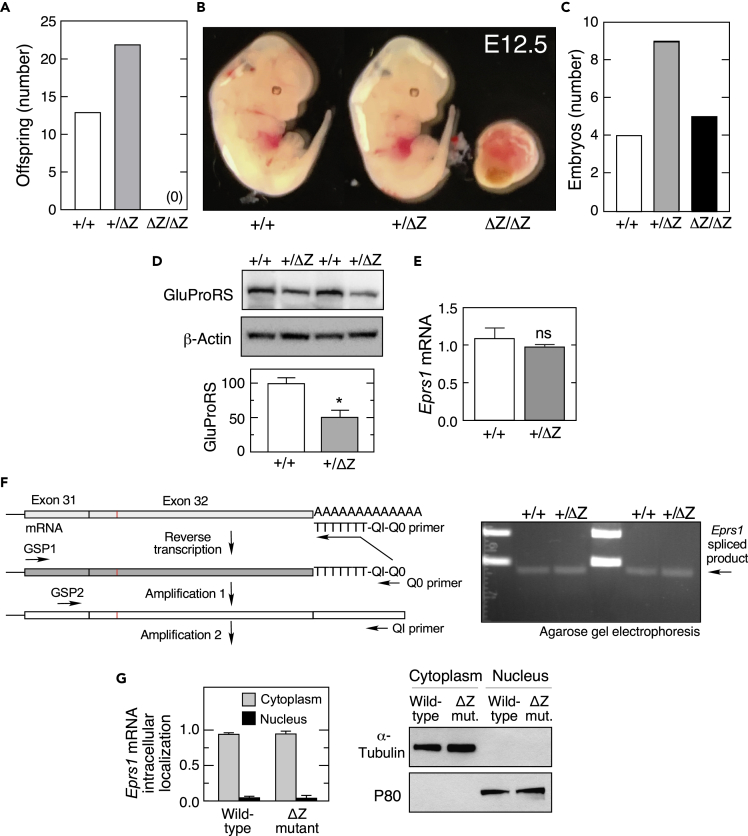
Heterozygous Eprs1+/ΔZ breeding pairs produced only Eprs1+/+ and Eprs1+/ΔZ offspring indicating that homozygous Eprs1 deletion is embryonic-lethal (Figure 2A). To investigate the stage of lethality, timed mating of heterozygous breeding pairs was initiated, and the pregnancy was terminated at embryonic day 12.5. Embryos genotyped as Eprs1+/+ and Eprs1+/ΔZ appeared virtually identical and healthy; however, severely degenerate embryos were observed in utero and genotyped as homozygous Eprs1ΔZ/ΔZ mutants (Figure 2B). Embryonic lethality was observed in two independently derived mouse lines, providing evidence that the result was not due to an off-target effect of CRISPR-Cas9 gene editing. The number of embryos of each genotype revealed a near-Mendelian distribution, indicating a normal fertilization rate in the null mutant (Figure 2C). Owing to the profoundly abnormal condition of homozygous embryos, heterozygous mutant mice were selected for molecular analysis.
Figure 2.
Reduced GluProRS expression and embryonic lethality of Eprs1ΔZ/ΔZ mice
(A) Live births from matings of Eprs1+/ΔZ mice.
(B and C) Embryo visualization (B) and genotype number (C) at E12.5 dpc following timed matings of Eprs1+/ΔZ mice.
(D) GluProRS protein in wild-type and Eprs1+/ΔZ mice from brain lysates was determined by immunoblot (above) and quantitated by densitometry (below; mean ± SEM, n = 8).
(E) qPCR analysis of Eprs1 mRNA levels in brain lysates from wild-type and Eprs1+/ΔZ mice (ns = not significant; mean ± SEM, n = 3).
(F) 3′ RACE analysis to determine splicing of Eprs1 mRNA in brain from wild-type and Eprs1+/ΔZ mice: schematic of reverse transcription and amplification (left); visualization of splice product by agarose gel electrophoresis (right).
(G) Intracellular distribution of EPRS1 mRNA (left) was determined by fractionation of lysates from HEK293 expressing wild-type or ΔZ mutant, followed by RT-qPCR (mean ± SEM, n = 3); lysates were subjected to immunoblot analysis for cytoplasmic and nuclear markers, α-tubulin, and P80, respectively (right).
Partial ZBD deletion reduces GluProRS expression
To investigate the mechanism underlying embryonic lethality accompanying ZBD disruption, GluProRS was determined in 3-week-old mouse brain lysates by immunoblot using an antibody targeting the linker region. GluProRS expression in Eprs1+/ΔZ mice was about 50% of that in WT littermates, indicating that the mutant allele does not generate protein, or generates a rapidly degraded truncated protein (Figure 2D). To determine the potential role of defective transcription, Eprs1 mRNA levels in brain tissue from WT and Eprs1+/ΔZ mice were compared by qPCR. Essentially identical mRNA levels were observed in both genotypes indicating comparable transcription, and that the mutant transcript is not subject to nonsense-mediated decay (Figure 2E). Likewise, 3′ RACE (rapid amplification of cDNA ends) analysis of Eprs1 mRNA from WT and Eprs1+/ΔZ mutant littermates, spanning exon 31 to the poly(A) tail, which includes the c.4449 insertion mutation in exon 32, showed identical splicing (Figure 2F). To assess a possible defect in nuclear export of mutant mRNA, we determined the intracellular distribution of the transcripts by fractionation of lysates followed by qPCR; fractionation efficacy was shown by immunoblot with antibodies against α-tubulin and P80, markers of cytoplasm and nucleus, respectively (Figure 2G, right). The cytoplasmic-to-nuclear Eprs1 mRNA ratios were similar, indicating that nuclear export of mutant mRNA is not defective (Figure 2G, left) and that altered expression must be post-transcriptional.
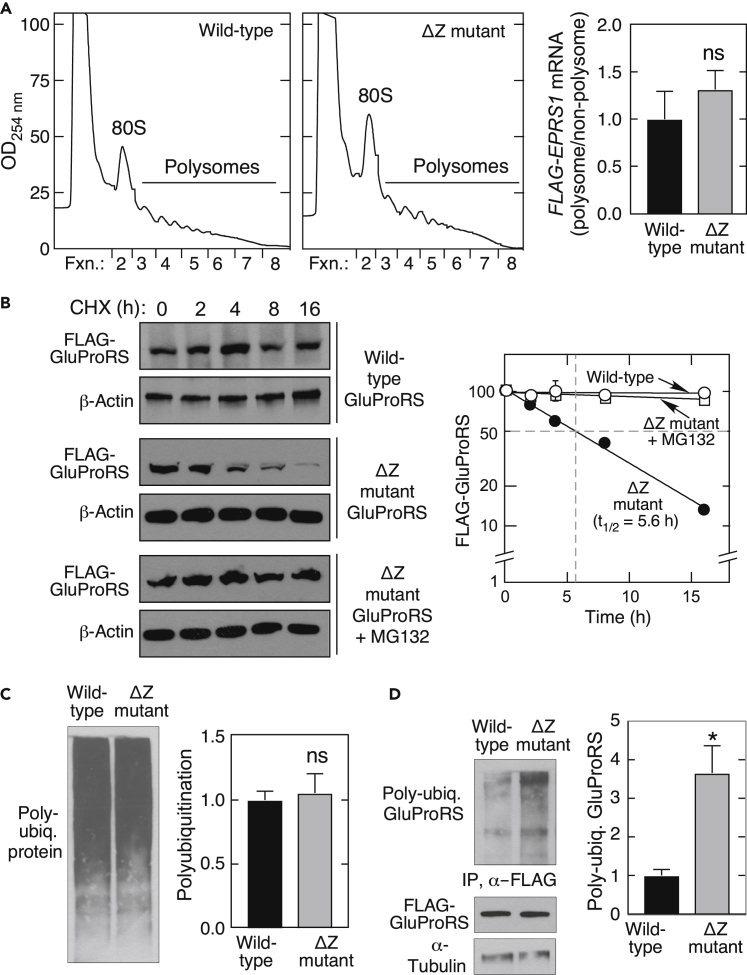
To assess the role of translation, polysome-profiling was done with HEK293F cells stably transfected with FLAG-tagged human EPRS1, or a mutant encoding a 29-amino acid C-terminal deletion. Expression of mutant EPRS1 mRNA did not affect global translation as shown by similar polysome loading (Figure 3A, left). FLAG-EPRS1 mRNA in polysomal and non-polysomal fractions was quantitated by RT-qPCR using primers specific for chimeric FLAG-EPRS1. Comparable ribosome loading was observed, indicating that translation of mutant EPRS1 mRNA was not defective (Figure 3A, right). GluProRS half-life was measured in the presence of cycloheximide to inhibit protein synthesis. In HEK293F cells stably expressing WT FLAG-tagged GluProRS, the protein was stable for at least 16 h; in contrast, mutant GluProRS exhibited rapid turnover, with a half-life of about 5.6 h (Figure 3B). The proteasome inhibitor MG132 completely blocked degradation of the mutant. To further explore the mechanism underlying degradation of the ΔZ mutant, ubiquitination was determined. Total cell accumulation of poly-ubiquitinated protein was similar in HEK293F cells transfected with either GluProRS forms (Figure 3C). However, in the presence of MG132 to block proteasomal degradation, the ZBD mutant exhibited ~3.5-fold more ubiquitination compared to WT (Figure 3D), suggestive of a newly exposed site or an altered conformation in the truncated protein. Interestingly, despite the deleted domain, and the very low amount of ZBD mutant in cells, a co-immunoprecipitation experiment revealed normal interaction of the remnant with multiple other MSC constituents, indicating similar MSC incorporation of WT and ZBD-defective GluProRS (Figure S4).
Figure 3.
Mutation in ZBD reduces GluProRS stability
(A) Polysome profiling of HEK293 cells expressing wild-type (left) or ΔZ mutant (center) EPRS1 mRNA. Distribution of EPRS1 mRNA in polysome and non-polysome fractions determined by RT-qPCR (right, ns = non-significant).
(B) To determine protein degradation rate, HEK293 cells expressing wild-type or ΔZ mutant FLAG-GluProRS were treated with cycloheximide (CHX) in the presence or absence of MG132 for up to 16 h and lysates subjected to immunoblot analysis (left) and densitometric quantitation (right) (mean ± SD, n = 3).
(C) Poly-ubiquitination of total protein in HEK293 cells expressing wild-type or ΔZ mutant EPRS1 was detected with anti-ubiquitin antibody (left) and quantitated by densitometry (right, mean ± SEM, n = 3).
(D) Poly-ubiquitination of wild-type FLAG-GluProRS and ΔZ mutant was determined in HEK293 cells treated with MG132 for 18 h. Lysates were subjected to immunoprecipitation (IP) with anti-FLAG antibody, followed by immunoblot with anti-ubiquitin, anti-FLAG, and anti-α-tubulin antibodies (left), and densitometric quantification (right; mean ± SEM, n = 3; ∗p < 0.05).
Proximity of the ProRS ZBD to the ATP-binding domain influences aminoacylation activity
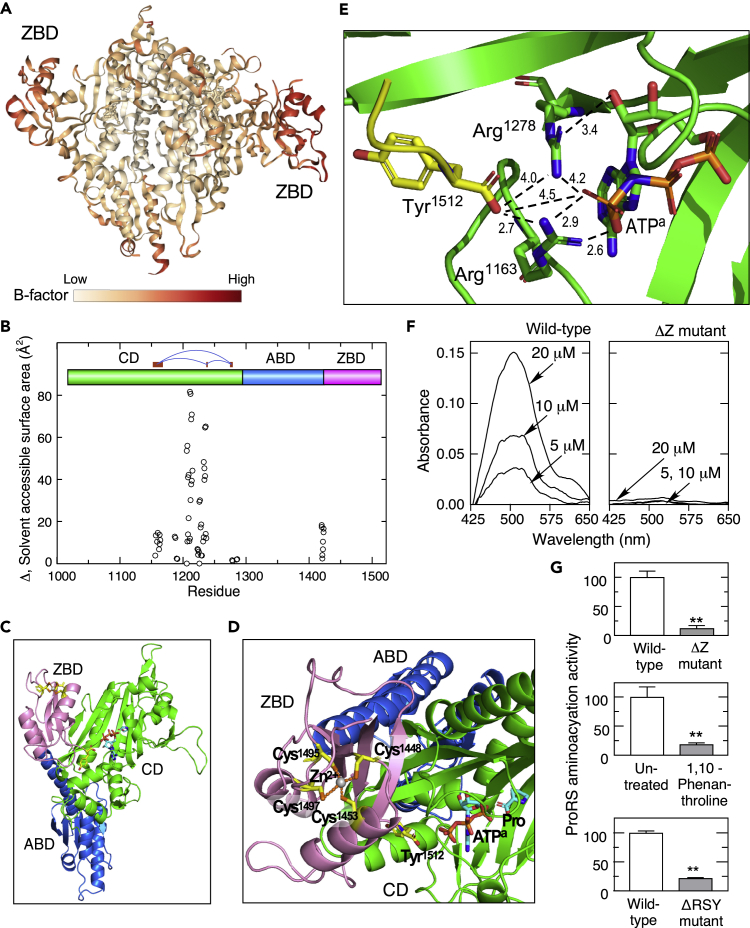
The B-factor of a protein structure describes the degree of electron density spread for each atom, and is a measure of conformational flexibility (Ringe and Petsko, 1985); however, static disorder due to relative positional differences of atoms within a crystal can also contribute to high local B-factor. According to the crystal structure of human ProRS (PDB: 4HVC) the ZBD exhibits the relatively highest B-factor, indicative of conformational flexibility or flexible linkage to the body of the protein (Figure 4A). To visualize the altered structure of the mutant protein, theoretical solvent-accessible surface area (SASA) was determined for ProRS WT and ΔZ structures in polar solvent. The SASA “rolling ball” algorithm rolls a theoretical sphere, with 1.4 Å radius, approximating a water molecule, to probe a macromolecule surface (Shrake and Rupley, 1973). ZBD deletion markedly increases solvent accessibility in the ATP-binding region of the catalytic domain, and to a lesser extent, a narrow region joining the anticodon-binding domain and ZBD (Figure 4B). These results are consistent with the position of the ZBD adjacent to the catalytic domain, despite its direct link in primary sequence to the anticodon-binding domain (Figure 4C). The human ZBD consists primarily of four β strands and two α helices connected by unstructured loops (Figures 1B and 4D). The carboxylate group of Tyr1512 is part of a network of charged interactions that includes Arg1163, Arg1278, and the γ-phosphate of the ATP analog (Figure 4E). Deletion of 29 C-terminal amino acids results in loss of the C-terminal-most β sheet and two of the four conserved Cys residues essential for tetrahedral coordination of Zn2+ (Figures 1B and S5A).
Figure 4.
Zn2+ and the ZBD are critical for ProRS aminoacylation activity
(A) B-factor analysis of human ProRS dimer (PDB: 4HVC).
(B) SASA measurements were made using human ProRS dimer coordinates with and without the C-terminal 29-amino acids and probed with sphere of radius 1.4 Å. Difference in accessible surface area (mutant minus wild-type) at each residue is shown. Regions forming the ATP-binding pocket are indicated (red bars).
(C) Crystal structure of human ProRS monomer (PDB: 4HVC) highlighting ZBD (magenta), anticodon binding domain (ABD, blue), and catalytic domain (CD, green).
(D) Crystal structure of the ZBD showing Zn2+ coordinated by four Cys residues and the C-terminal Tyr1512 aligned with ATP analog (ATPa) and Pro substrates.
(E) Expanded view of human ProRS region surrounding C-terminal Tyr1512 and ATPa. Charge interactions between Tyr1512 carboxylate, Arg1278, Arg1163, and ATPa terminal phosphate are highlighted (dashed lines, distances in Å).
(F) Zn2+ in wild-type (left) and ΔZ mutant (right) recombinant human ProRS was determined by Zn2+ release with methyl methanethiosulfonate (MMTS) and spectroscopic detection with 4-(2-pyridylazo)resorcinol (PAR).
(G) Aminoacylation activity of wild-type and ΔZ mutant ProRS was determined by incorporation of 14C-labeled Pro into yeast tRNA (top), following treatment of wild-type ProRS with the Zn2+ chelator, 1,10 phenanthroline (center), and in a mutant with deletion of the three terminal amino acids Arg-Ser-Tyr (bottom, ΔRSY) (mean ± SD, n = 3; ∗∗p < 0.001).
Zn2+ content of recombinant human wild-type and ΔZ mutant ProRS was determined by Zn2+ release by methyl methanethiosulfonate and spectroscopic detection with 4-(2-pyridylazo)resorcinol (Chongdar et al., 2015; Hunt et al., 1985). Unlike substantial Zn2+ content of wild-type enzyme (Figure 4F, left), Zn2+ in the ΔZ mutant was virtually undetectable, consistent with the absence of two coordinating cysteine ligands (Figure 4F, right). AARSs catalyze tRNA aminoacylation in two steps. The amino acid is initially activated by ATP, generating a high-energy aminoacyl-adenylate intermediate. Subsequently, the aminoacyl-adenylate attacks the 3′ OH tRNA terminus to form charged aminoacyl-tRNA. To determine the role of the ProRS ZBD in tRNA charging, we measured in vitro prolyl adenylation activity of recombinant proteins as transfer of L-[14C]proline to yeast tRNA. The ΔZ mutant exhibited about a 90% loss of aminoacylation activity compared with wild-type ProRS (Figure 4G, top). We considered the possibility that the zinc ion was essential for ZBD conformation and aminoacylation activity. Chelation of Zn2+ with 1,10-phenanthroline resulted in an ~80% loss of aminoacylation activity (Figure 4G, center). Based on proximity of the C terminus to the ATP-binding site (Figure 4D), we hypothesized that the ZBD was essential for positioning and orientation of the extreme C terminus in juxtaposition to ATP in the catalytic domain. Recombinant ProRS lacking the conserved Arg-Ser-Tyr from the C terminus (ΔRSY) exhibited an ~80% reduction in aminoacylation activity compared with wild-type ProRS, nearly the same activity loss due to zinc depletion or due to the larger deletion in the ΔZ mutant (Figure 4E, bottom). These results suggest that the primary role of Zn2+ and the ZBD in catalysis is to direct orientation of the C terminus adjacent to the ATP-binding site of the catalytic domain.
Discussion
Domains appended to AARSs during evolution generate functional complexity, particularly in eukaryotes where they exhibit multiple non-canonical activities unrelated to their primary function in the interpretation of the genetic code (Guo et al., 2010). However, these apparently non-catalytic appendages can also modulate the canonical activity, for example, by regulating transferase activity or by facilitating amino acid editing activity (Bullwinkle and Ibba, 2014; Chang et al., 2016; Guo et al., 2009; He et al., 2009; Raben et al., 1994). Our experiments show the GluProRS ZBD is an appended domain essential for optimal transferase activity. In addition, the domain protects the protein from rapid degradation. In primary sequence space, the ProRS ZBD is an extension of the anticodon-binding domain, flexibly linked via a long α helix (Figure 4C, left). Structurally, however, the primary interaction of the ZBD is with the ProRS catalytic domain.
Phylogenetic analysis showed segregation of C-terminal-bearing ZBD from ProRS (and GluProRS in species bearing the fused protein (Berthonneau and Mirande, 2000; Ray and Fox, 2014; Ray et al., 2011) into distinct classes. ZBD-bearing species form three major clusters bearing C-terminal-type ZBDs, comprising nearly all archaeal and eukaryal species and a subset of bacteria, likely sharing a common origin (Figure S1B). The vast majority of bacteria lack the ProRS C-terminal ZBD including gram-negative bacteria, e.g., Escherichia, and nonproteobacteria, e.g., Chlamydiae. In archaea, the ProRS ZBDs are clustered in the four major phyla, namely, DPANN, TACK, Euryarchaeota, and Asgard. In eukaryotes, the ZBD is present in both free-standing ProRS and in the fused GluProRS. Eukaryotic mitochondrial ProRS does not contain a ZBD, consistent with the endosymbiont theory of evolution.
Among the mammalian AARSs, GluProRS is likely to have the most complex evolutionary history. The earliest appended domain in ProRS appears to be the C-terminal ZBD, likely appearing before the divergence of the three kingdoms (Figure S1B). GluRS gained an N-terminus GST-like domain in the fungal eukaryote, S. cerevisiae (Havrylenko and Mirande, 2015). This domain directs interactions with other proteins bearing GST-like domain and facilitates the formation of a 3-protein multi-tRNA synthetase complex in S. cerevisiae, and the 11-protein MSC in animals. The most recent addition is the WHEP domain, a helix-turn-helix structure appended to the GluRS C terminus in Sphaeroforma arctica, an early unicellular eukaryote closely related to animals (Ray and Fox, 2014). Remarkably, a fusion event between GluRS and ProRS occurred in an as-yet unidentified unicellular eukaryote, forming the heterobifunctional GluProRS, presumably bearing a single WHEP domain joining the AARSs. Nearly all animal species contain a fused GluProRS bearing zero to six WHEP domains formed by discrete duplication and loss events; humans and mice have three WHEP repeats (Ray et al., 2011).
Our results show that the ZBD is both essential for ProRS catalytic activity and contributes to stability. Its presence in all four major archaeal sub-clusters and certain bacterial phyla is potentially instructive with respect to environmental or other forces driving its origin. Hyperthermic conditions and lateral gene transfer might have driven adaptations that enhanced stability (Groussin and Gouy, 2011; Reed et al., 2013). The tetrahedral interaction of Zn2+ with four cysteines renders the cation nearly inert chemically, and commonly, Zn2+-cysteine complexes perform structural functions within proteins (Pace and Weerapana, 2014). Both Mg2+ and Zn2+ are frequently observed in AARS crystal structures. Mg2+ generally functions to stabilize the transition state of ATP during hydrolysis, whereas the role of Zn2+ in AARS function is variable. In AARSs that ligate Ile, Leu, Val, Glu, Gln, Trp, and Met, Zn2+ contributes to efficient catalysis. TrpRS contains a consensus ZBD, and is activated by Zn2+; however, Zn2+ has not been observed in multiple crystal structures suggesting weak interaction with the ZBD (Doublie et al., 1995; Xu et al., 2018; Yang et al., 2007). Zn2+ can contribute to AARS discrimination between cognate and non-cognate amino acids. For example, when bound to E. coli ThrRS, the hydroxyl side chain of Thr switches Zn2+ geometry from tetrahedral to the active pentacoordinate intermediate (Sankaranarayanan et al., 2000). The methyl group side chain of non-cognate Val cannot co-ordinate with Zn2+ and is rejected. Similar amino acid selectivity is exhibited by E. coli CysRS where Cys forms a Zn2+-thiolate bond that directs a 20,000-fold discrimination against Ser (Zhang et al., 2003). SerRS of methanogenic bacteria exhibits a similar discrimination mechanism: Zn2+ coordination with Ser facilitates activation, but the longer side chain of Thr induces a steric clash that prevents activation (Bilokapic et al., 2006).
Our experiments with the recombinant ProRS region of human GluProRS show that a 29-amino acid deletion at the C terminus, that removes two of the four Zn2+-coordinating ligands, inhibits Zn2+-binding and ProRS aminoacylation activity. We also show that Zn2+ depletion with a chelator or deletion of three C-terminal amino acids, Arg-Ser-Tyr, inhibits aminoacylation activity. Together, these results reveal a key role of Zn2+ and the ZBD in catalysis. Importantly, the terminal three amino acids are highly conserved, and a consensus sequence of Arg/Lys-Ser/Thr/Ala-Tyr are present at the C terminus of ProRS enzymes containing a ZBD from bacteria to humans (Yaremchuk et al., 2000) (Figure S1A).
The ProRS ZBDs of mammals and Thermus thermophilus exhibit considerable sequence similarity, including a conserved terminal Tyr residue (Figure S1A). Likewise, overlay of the crystal structure of T. thermophilus (Crepin et al., 2006; Yaremchuk et al., 2000, 2001) and human ProRS reveals nearly identical structures in the active site region surrounding the terminal Tyr and ATP (Figure S5B). In T. thermophilus, based on the proximity of the terminal Tyr477 to Arg264, Arg152, and the γ-phosphate of ATP, a network of attractive electrostatic interactions was proposed to maintain a bent conformation of ATP for optimal reaction with the adjacent Pro substrate (Yaremchuk et al., 2001) (Figure S5B). The function of the terminal Tyr was clarified by finding that mutation to Phe or Ala in T. thermophilus only slightly reduced aminoacylation activity, suggesting the carboxylate group, rather than the amino acid side chain, is effective (Yaremchuk et al., 2000). A similar charge network in human ProRS consists of the C-terminal Tyr1512, Arg1163, Arg1278, and ATP (Figure 4E). The carboxylate group of Tyr1512 is 4.5 Å from the γ-phosphate group of ATP and forms a linear axis with the Pro substrate (Figures 4D and 4E). We propose that, in addition to the attractive interactions of Tyr carboxylate with the Arg amines, an electrostatic repulsion with the γ-phosphate drives ATP toward Pro for more efficient reaction to form the prolyl-adenylate intermediate. A second ZBD function is GluProRS stabilization, possibly by masking an ubiquitination site. These structure-activity relationships raise the possibility that the ZBD might contribute to physiological regulation of the catalytic activity of the ProRS domain, or pathological dysregulation of protein synthesis due to deficient aminoacylation activity or high enzyme turnover.
Limitations of the study
We have investigated the role of the ZBD in the canonical function of human ProRS, i.e., charging to the tRNA synthetase. We have not considered non-canonical functions potentially exhibited by the ZBD, a possibility given the structural similarity studies of the T. thermophilus ZBD with the C-terminal domain of translation initiation factor 3 from Bacillus stearothermophilus. Likewise, our study focuses on the effect of ZBD on prolyl-tRNA synthetase function, and does not consider potential long-range interactions that extend to the GluRS region of the bifunctional enzyme. We show that ZBD-deleted GluProRS is incorporated into the MSC; however, its influence on the structure of the MSC has not been investigated. CRISPR-mediated mutation induced a frameshift that appended four amino acids not present in mouse ProRS, i.e., Lys-Pro-Ser-Val, before a stop codon terminated translation. The present study proposes a reaction mechanism based on X-ray crystallography and limited mutagenesis studies by us and others. A more detailed study involving molecular dynamics simulation and guided mutagenesis would give more insights into the role of the C-terminal ZBD in the reaction mechanism.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Paul Fox (foxp@ccf.org).
Materials availability
All plasmids and mouse lines generated in this study will be made available on request, but we may require a completed Materials Transfer Agreement if there is potential for commercial application.
Data and code availability
This study did not generate datasets or analyze code.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank the Case Western Reserve University Transgenic and Targeting Facility and the Genomics Core, both of Case Western Reserve University, for their services in generating and validating the mutant mouse. This project was supported by NIH grants P01 HL076491, R01 DK123236, and R01 AG067146 (to P.L.F.), by a Clinical and Translational Science Collaborative (CTSC) Core Utilization Pilot Grant Award from Case Western Reserve University School of Medicine (to P.L.F.), and by American Heart Association (AHA) Postdoctoral Fellowships, Nationwide (19POST34380725 to I.R. and 19POST34380687 to D.K.).
Author contributions
K.V.: conceptualization, methodology, investigation, analysis, writing – original draft, review & editing; I.R.: investigation, methodology; D.K.: methodology, resources, investigation; A.C.: methodology, resources, investigation; F.T.: methodology, resources, investigation; A.C.: methodology, investigation; K.K.: methodology, resources, investigation; C.B.G.: visualization and formal analysis; V.G.: visualization, formal analysis; P.L.F.: conceptualization, supervision, project administration, funding Acquisition, writing – review & editing.
Declaration of interests
The authors declare no competing interests.
Published: March 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102215.
Supplemental information
References
- Arif A., Terenzi F., Potdar A.A., Jia J., Sacks J., China A., Halawani D., Vasu K., Li X., Brown J.M. EPRS is a critical mTORC1-S6K1 effector that influences adiposity in mice. Nature. 2017;542:357–361. doi: 10.1038/nature21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthonneau E., Mirande M. A gene fusion event in the evolution of aminoacyl-tRNA synthetases. FEBS Lett. 2000;470:300–304. doi: 10.1016/s0014-5793(00)01343-0. [DOI] [PubMed] [Google Scholar]
- Bilokapic S., Maier T., Ahel D., Gruic-Sovulj I., Soll D., Weygand-Durasevic I., Ban N. Structure of the unusual seryl-tRNA synthetase reveals a distinct zinc-dependent mode of substrate recognition. EMBO J. 2006;25:2498–2509. doi: 10.1038/sj.emboj.7601129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullwinkle T.J., Ibba M. Emergence and evolution. Top. Curr. Chem. 2014;344:43–87. doi: 10.1007/128_2013_423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.Y., Chien C.I., Chang C.P., Lin B.C., Wang C.C. A WHEP domain regulates the dynamic structure and activity of Caenorhabditis elegans glycyl-tRNA synthetase. J. Biol. Chem. 2016;291:16567–16575. doi: 10.1074/jbc.M116.730812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.Y., Lee H.J., Choi Y.S., Kim D.K., Jin K.S., Kim S., Kang B.S. Symmetric assembly of a decameric subcomplex in human multi-tRNA synthetase complex via interactions between glutathione transferase-homology domains and aspartyl-tRNA synthetase. J. Mol. Biol. 2019;431:4475–4496. doi: 10.1016/j.jmb.2019.08.013. [DOI] [PubMed] [Google Scholar]
- Cho H.Y., Maeng S.J., Cho H.J., Choi Y.S., Chung J.M., Lee S., Kim H.K., Kim J.H., Eom C.Y., Kim Y.G. Assembly of multi-tRNA synthetase complex via heterotetrameric glutathione transferase-homology domains. J. Biol. Chem. 2015;290:29313–29328. doi: 10.1074/jbc.M115.690867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chongdar N., Dasgupta S., Datta A.B., Basu G. Dispensability of zinc and the putative zinc-binding domain in bacterial glutamyl-tRNA synthetase. Biosci. Rep. 2015;35:e00184. doi: 10.1042/BSR20150005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepin T., Yaremchuk A., Tukalo M., Cusack S. Structures of two bacterial prolyl-tRNA synthetases with and without a cis-editing domain. Structure. 2006;14:1511–1525. doi: 10.1016/j.str.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Doublie S., Bricogne G., Gilmore C., Carter C.W., Jr. Tryptophanyl-tRNA synthetase crystal structure reveals an unexpected homology to tyrosyl-tRNA synthetase. Structure. 1995;3:17–31. doi: 10.1016/s0969-2126(01)00132-0. [DOI] [PubMed] [Google Scholar]
- Eswarappa S.M., Potdar A.A., Sahoo S., Sankar S., Fox P.L. Metabolic origin of the fused aminoacyl-tRNA synthetase, glutamyl-prolyl-tRNA synthetase. J. Biol. Chem. 2018;293:19148–19156. doi: 10.1074/jbc.RA118.004276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groussin M., Gouy M. Adaptation to environmental temperature is a major determinant of molecular evolutionary rates in archaea. Mol. Biol. Evol. 2011;28:2661–2674. doi: 10.1093/molbev/msr098. [DOI] [PubMed] [Google Scholar]
- Guo M., Chong Y.E., Beebe K., Shapiro R., Yang X.L., Schimmel P. The C-Ala domain brings together editing and aminoacylation functions on one tRNA. Science. 2009;325:744–747. doi: 10.1126/science.1174343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Schimmel P. Essential nontranslational functions of tRNA synthetases. Nat. Chem. Biol. 2013;9:145–153. doi: 10.1038/nchembio.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Yang X.L., Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Biol. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havrylenko S., Mirande M. Aminoacyl-tRNA synthetase complexes in evolution. Int. J. Mol. Sci. 2015;16:6571–6594. doi: 10.3390/ijms16036571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Zu L.D., Yao P., Chen X., Wang E.D. Two non-redundant fragments in the N-terminal peptide of human cytosolic methionyl-tRNA synthetase were indispensable for the multi-synthetase complex incorporation and enzyme activity. Biochim. Biophys. Acta. 2009;1794:347–354. doi: 10.1016/j.bbapap.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Hunt J.B., Neece S.H., Ginsburg A. The use of 4-(2-pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal. Biochem. 1985;146:150–157. doi: 10.1016/0003-2697(85)90409-9. [DOI] [PubMed] [Google Scholar]
- Jia J., Arif A., Ray P.S., Fox P.L. WHEP domains direct noncanonical function of glutamyl-prolyl tRNA synthetase in translational control of gene expression. Mol. Cell. 2008;29:679–690. doi: 10.1016/j.molcel.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T.L., Zocco D., Sundrud M.S., Hendrick M., Edenius M., Yum J., Kim Y.J., Lee H.K., Cortese J.F., Wirth D.F. Halofuginone and other febrifugine derivatives inhibit prolyl-tRNA synthetase. Nat. Chem. Biol. 2012;8:311–317. doi: 10.1038/nchembio.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan K., Beleanu Gogonea C., Willard B., Gogonea V., Fox P.L. 3-Dimensional architecture of the human multi-tRNA synthetase complex. Nucleic Acids Res. 2020;48:8740–8754. doi: 10.1093/nar/gkaa569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon N.H., Fox P.L., Kim S. Aminoacyl-tRNA synthetases as therapeutic targets. Nat. Rev. Drug Discov. 2019;18:629–650. doi: 10.1038/s41573-019-0026-3. [DOI] [PubMed] [Google Scholar]
- Lee E.Y., Kim S., Kim M.H. Aminoacyl-tRNA synthetases, therapeutic targets for infectious diseases. Biochem. Pharmacol. 2018;154:424–434. doi: 10.1016/j.bcp.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.Y., Lee H.C., Kim H.K., Jang S.Y., Park S.J., Kim Y.H., Kim J.H., Hwang J., Kim J.H., Kim T.H. Infection-specific phosphorylation of glutamyl-prolyl tRNA synthetase induces antiviral immunity. Nat. Immunol. 2016;17:1252–1262. doi: 10.1038/ni.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes M.I., Gutierrez Salazar M., Guerrero K., Thiffault I., Salomons G.S., Gauquelin L., Tran L.T., Forget D., Gauthier M.S., Waisfisz Q. Bi-allelic mutations in EPRS, encoding the glutamyl-prolyl-aminoacyl-tRNA synthetase, cause a hypomyelinating leukodystrophy. Am. J. Hum. Genet. 2018;102:676–684. doi: 10.1016/j.ajhg.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R., Jia J., Arif A., Ray P.S., Fox P.L. The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem. Sci. 2009;34:324–331. doi: 10.1016/j.tibs.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N.J., Weerapana E. Zinc-binding cysteines: diverse functions and structural motifs. Biomolecules. 2014;4:419–434. doi: 10.3390/biom4020419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon S., Robinson J.C., Berthonneau E., Siatecka M., Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein-protein interactions and characterization of a core protein. J. Mol. Biol. 1999;285:183–195. doi: 10.1006/jmbi.1998.2316. [DOI] [PubMed] [Google Scholar]
- Raben N., Nichols R., Dohlman J., McPhie P., Sridhar V., Hyde C., Leff R., Plotz P. A motif in human histidyl-tRNA synthetase which is shared among several aminoacyl-tRNA synthetases is a coiled-coil that is essential for enzymatic activity and contains the major autoantigenic epitope. J. Biol. Chem. 1994;269:24277–24283. [PubMed] [Google Scholar]
- Ray P.S., Fox P.L. Origin and evolution of glutamyl-prolyl tRNA synthetase WHEP domains reveal evolutionary relationships within Holozoa. PLoS One. 2014;9:e98493. doi: 10.1371/journal.pone.0098493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P.S., Sullivan J.C., Jia J., Francis J., Finnerty J.R., Fox P.L. Evolution of function of a fused metazoan tRNA synthetase. Mol. Biol. Evol. 2011;28:437–447. doi: 10.1093/molbev/msq246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed C.J., Lewis H., Trejo E., Winston V., Evilia C. Protein adaptations in archaeal extremophiles. Archaea. 2013;2013:373275. doi: 10.1155/2013/373275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringe D., Petsko G.A. Mapping protein dynamics by X-ray diffraction. Prog. Biophys. Mol. Biol. 1985;45:197–235. doi: 10.1016/0079-6107(85)90002-1. [DOI] [PubMed] [Google Scholar]
- Sampath P., Mazumder B., Seshadri V., Gerber C.A., Chavatte L., Kinter M., Ting S.M., Dignam J.D., Kim S., Driscoll D.M., Fox P.L. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell. 2004;119:195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan R., Dock-Bregeon A.C., Rees B., Bovee M., Caillet J., Romby P., Francklyn C.S., Moras D. Zinc ion-mediated amino acid discrimination by threonyl-tRNA synthetase. Nat. Struct. Biol. 2000;7:461–465. doi: 10.1038/75856. [DOI] [PubMed] [Google Scholar]
- Shrake A., Rupley J.A. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J. Mol. Biol. 1973;79:351–371. doi: 10.1016/0022-2836(73)90011-9. [DOI] [PubMed] [Google Scholar]
- Son J., Lee E.H., Park M., Kim J.H., Kim J., Kim S., Jeon Y.H., Hwang K.Y. Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone. Acta Crystallogr. D Biol. Crystallogr. 2013;69:2136–2145. doi: 10.1107/S0907444913020556. [DOI] [PubMed] [Google Scholar]
- Wu J., Subbaiah K.C.V., Xie L.H., Jiang F., Khor E.S., Mickelsen D., Myers J.R., Tang W.H.W., Yao P. Glutamyl-prolyl-tRNA synthetase regulates proline-rich pro-fibrotic protein synthesis during cardiac fibrosis. Circ. Res. 2020;127:827–846. doi: 10.1161/CIRCRESAHA.119.315999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhou H., Zhou Q., Hong F., Vo M.N., Niu W., Wang Z., Xiong X., Nakamura K., Wakasugi K. An alternative conformation of human TrpRS suggests a role of zinc in activating non-enzymatic function. RNA Biol. 2018;15:649–658. doi: 10.1080/15476286.2017.1377868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.L., Guo M., Kapoor M., Ewalt K.L., Otero F.J., Skene R.J., McRee D.E., Schimmel P. Functional and crystal structure analysis of active site adaptations of a potent anti-angiogenic human tRNA synthetase. Structure. 2007;15:793–805. doi: 10.1016/j.str.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao P., Fox P.L. Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol. Med. 2013;5:332–343. doi: 10.1002/emmm.201100626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaremchuk A., Cusack S., Tukalo M. Crystal structure of a eukaryote/archaeon-like protyl-tRNA synthetase and its complex with tRNAPro(CGG) EMBO J. 2000;19:4745–4758. doi: 10.1093/emboj/19.17.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaremchuk A., Tukalo M., Grotli M., Cusack S. A succession of substrate induced conformational changes ensures the amino acid specificity of Thermus thermophilus prolyl-tRNA synthetase: comparison with histidyl-tRNA synthetase. J. Mol. Biol. 2001;309:989–1002. doi: 10.1006/jmbi.2001.4712. [DOI] [PubMed] [Google Scholar]
- Yemni E.A., Monies D., Alkhairallah T., Bohlega S., Abouelhoda M., Magrashi A., Mustafa A., AlAbdulaziz B., Alhamed M., Baz B. Integrated analysis of whole exome sequencing and copy number evaluation in Parkinson's disease. Sci. Rep. 2019;9:3344. doi: 10.1038/s41598-019-40102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.M., Christian T., Newberry K.J., Perona J.J., Hou Y.M. Zinc-mediated amino acid discrimination in cysteinyl-tRNA synthetase. J. Mol. Biol. 2003;327:911–917. doi: 10.1016/s0022-2836(03)00241-9. [DOI] [PubMed] [Google Scholar]
- Zhou H., Sun L., Yang X.L., Schimmel P. ATP-directed capture of bioactive herbal-based medicine on human tRNA synthetase. Nature. 2013;494:121–124. doi: 10.1038/nature11774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets or analyze code.