Abstract
Background/Aims
South Korean soldiers are exposed to similar environmental factors. In this study, we sought to evaluate the gut microbiome of healthy young male soldiers (HYMS) and to identify the primary factors influencing the microbiome composition.
Methods
We prospectively collected stool from 100 HYMS and performed next-generation sequencing of the 16S rRNA genes of fecal bacteria. Clinical data, including data relating to the diet, smoking, drinking, and exercise, were collected.
Results
The relative abundances of the bacterial phyla Firmicutes, Actinobacteria, Bacteroidetes, and Proteobacteria were 72.3%, 14.5%, 8.9%, and 4.0%, respectively. Fifteen species, most of which belonged to Firmicutes (87%), were detected in all examined subjects. Using cluster analysis, we found that the subjects could be divided into the two enterotypes based on the gut microbiome bacterial composition. Compared with enterotype 2 subjects, subjects classified as enterotype 1 tended to be characterized by higher frequencies of potentially harmful lifestyle habits (current smoker 55.6% vs 36.6%, p=0.222; heavy drinker 16.7% vs 3.7%, p=0.120; insufficient physical activity 27.8% vs 14.6%, p=0.318). We identified a significant difference in the microbiome compositions of current and noncurrent smokers (p=0.008); the former differed from the latter mainly in a relatively lower abundance of Bifidobacterium species and a higher abundance of Negativicutes.
Conclusions
A high abundance of Actinobacteria and low abundance of Bacteroidetes were the main features distinguishing the gut microbiomes of HYMS, and current smokers could be differentiated from noncurrent smokers by their lower abundance of Bifidobacterium and higher abundance of Negativicutes.
Keywords: Microbiota, Health, Smokers, Military personnel
INTRODUCTION
The development of next-generation sequencing technology has led to an exponential growth in the number of studies focusing on the gut microbiome, in many of which the gut microbiomes of patients with disease are compared with those of healthy control subjects. Numerous such studies have examined the composition and characteristics of gut microbiomes in healthy populations worldwide, including the Human Microbiome Project in the United States1 and the MetaHIT project in Europe.2 In contrast, there have been relatively few comparable investigations, particularly large-scale studies, in the Korean population. Therefore, in 2016, with financial support from the Ministry of Science and ICT, we inaugurated the Korean gut microbiome bank. The initial step in this huge project entailed collecting stool samples from healthy Korean adults and analyzing the gut microbiomes. During this phase, we also collected the stools of healthy young male Korean soldiers serving military duty. There is currently little data regarding the gut microbiomes of healthy young Koreans, and given that Korean soldiers represent a unique group in which individuals are exposed to a similar range of major environmental factors, we believe that this project might provide valuable insights regarding the factors that influence the gut microbiomes of healthy young adults.
Against this background, we sought in the present study to evaluate the composition of the gut microbiomes of healthy young male Korean soldiers and to determine the main factors contributing to differences in the composition of these gut microbiomes.
MATERIALS AND METHODS
1. Subjects
We recruited subjects from among male soldiers who were working in the Korean Armed Forces Capital Hospital in Seongnam, South Korea. All subjects were serving in the hospital as part of the compulsory military duty imposed on young Korean males. The inclusion criteria stipulated that the males should be healthy and without any of the specified exclusion criteria (see below). We performed history taking, physical examinations, and laboratory tests to screen volunteers. If laboratory test results obtained within a 1-year period prior to enrollment were available, these data were used to evaluate whether the subject met the inclusion criteria. The exclusion criteria were as follows: (1) the presence of acute illness; (2) a history of chronic illness, including hypertension, diabetes, angina, acute myocardial infarction, stroke, dyslipidemia, more than a moderate degree of fatty liver, cardiopulmonary disease, chronic liver disease, chronic renal disease, thyroid disease, asthma, or allergy; (3) a history of cancer within 5 years; (4) a history of abdominal surgery within 5 years; (5) a history of antibiotic or probiotic use within 3 months; (6) an abnormal stool form (a Bristol stool form score of 1, 2, 6, or 7); (7) body mass index <18.5 kg/m2 or ≥25 kg/m2; (8) systolic and diastolic blood pressure ≥140/90 mm Hg; (9) fasting glucose ≥126 mg/dL or random glucose ≥200 mg/dL; (10) creatinine ≥2-fold of the normal upper limit; (11) aspartate transaminase or alanine transaminase ≥3-fold of the normal upper limit; and (12) infection with hepatitis B virus, hepatitis C virus, or human immune deficiency virus.
2. Collection of stool and clinical data
Stool samples were collected from those subjects who met the inclusion criteria. In addition, we used questionnaires to collect clinical data relating to diet, smoking, drinking, physical activity, and socioeconomic status. To assess the intake of energy, carbohydrate, fat, protein, minerals, and vitamins, we used a semi-quantitative food frequency questionnaire that was developed and validated for Korean adults.3 For alcohol intake, subjects who drank more than eight standard drinks per week was classified as heavy drinkers.4 To access physical activity, we used a validated Korean version of the International Physical Activity Questionnaire Short Form.5 To analyze levels of physical activity, we followed guidelines for data processing and analysis of the International Physical Activity Questionnaire Short Form (www.ipaq.ki.se). Among the three designated levels of physical activity, we considered “inactive” individuals as being “insufficiently active,” whereas subjects who were “minimally active” or undertook “health-enhancing physical activity” were classified as being “sufficiently active.” All subjects provided informed consent and the study was approved by the Institutional Review Board of the Seoul National University Bundang Hospital (IRB number: B-1701-380-304).
3. PCR amplification and Illumina sequencing
Total DNA was extracted from stool samples using a PowerSoil DNA Isolation Kit (MoBio, Solana Beach, CA, USA) according to the manufacturer’s instructions. Polymerase chain reaction (PCR) amplification was performed using primers targeting the V3 to V4 regions of the 16S rRNA gene. For bacterial 16S rRNA amplification, we used the primer pair 341F (5ʹ-TCGTCGGCAGCGTC-AGATGTGTATAAGAGACAG-CCTACGGGNGGCWGCAG-3ʹ; underlining sequence indicates the target region primer) and 805R (5ʹ-GTCTCGTGGGCTCGG-AGATGTGTATAAGAGACAzG-GACTACHVGGGTATCTAATCC-3ʹ). Amplifications were carried out under the following conditions: an initial denaturation at 95°C for 3 minutes, followed by 25 cycles of denaturation at 95°C for 30 seconds, primer annealing at 55°C for 30 seconds, and extension at 72°C for 30 seconds, with a final elongation at 72°C for 5 minutes. Secondary amplification for attaching the Illumina Nextera barcode was performed using the i5 forward primer (5ʹ-AATGATACGGCGACCACCGAGATCTACAC-XXXXXXXX-TCGTCGGCAGCGTC-3ʹ; where X indicates the barcode region) and i7 reverse primer (5ʹ-CAA GCAGAAGACGGCATACGAGAT-XXXXXXXX-AGT CTCGTGGGCTCGG-3ʹ). The conditions used for secondary amplification were the same as those used for primary amplification, with the exception that the number of amplification cycle was set to eight.
The PCR products were assessed using 2% agarose gel electrophoresis and visualized using a Gel Doc system (Bio-Rad, Hercules, CA, USA). The amplified products were purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA, USA). Equal concentrations of purified products were pooled together and short fragments (non-target products) were removed using an Ampure beads kit (Agencourt Bioscience, Beverly, MA, USA). The quality and product size were assessed with an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA) using a DNA 7500 chip. Mixed amplicons were pooled and sequencing was carried out by ChunLab Inc. (Seoul, Korea), using an Illumina MiSeq Sequencing system (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions.
4. Microbiome data analysis
Basic analyses were conducted according to previously described procedures.6-8 The reads obtained from different samples were sorted based on the unique barcodes of each PCR product. Barcode, linker, and primers sequences were subsequently removed from the original sequencing reads, and any reads with two or more ambiguous nucleotides, a low-quality score (average score <25), or sequences shorter than 300 bp were discarded. Potential chimeric sequences were detected using the Bellerophon method, which compares the BLASTN search results obtained for forward half and reverse half sequences.9 After removing chimeric sequences, the taxonomic classification of each read was assigned against the EzBioCloud database (http://ezbiocloud.net),10 which contains the 16S rRNA gene sequences of type strains that have valid published names and representative species-level phylotypes of either cultured or uncultured entries in the GenBank database, with complete hierarchical taxonomic classification from the phylum to the species level.
For analysis of alpha-diversity, the richness and diversity of samples were determined by abundance-based coverage estimators, Chao1. and Jackknife estimation. In addition, the Simpson and Shannon diversity indices at a 3% distance were calculated using the CL community program (ChunLab Inc.). Good’s method was used to calculate sequencing coverage.11 For comparisons of the composition of a selected taxon, we used the Wilcoxon rank-sum test.
For analysis of beta-diversity, the overall phylogenetic distance between communities was estimated and visualized using Jensen–Shannon-based principal coordinates analysis. We performed Permanova analysis to evaluate the set-difference between groups and linear discriminant analysis effect size (LEfSe) analysis to determine the features most likely to explain differences between classes by coupling standard tests for statistical significance with additional tests encoding biological consistency and effect relevance.12 Cluster analysis based on bacterial species composition was performed to classify the subjects into enterotypes.13
5. Statistical analysis
R version 3.3.2 (R Foundation for Statistical Computing, Vienna, Austria) was used for the statistical analysis of clinical data. Continuous variables were analyzed using Student t-test. The chi-square test or Fisher exact test was used to analyze categorical variables. All results were considered statistically significant when p-values were less than 0.05.
RESULTS
1. Basic characteristics of the gut microbiomes of healthy male soldiers
Between April and November 2017, we prospectively enrolled 100 healthy male soldiers as study subjects. Table 1 shows the baseline characteristics of the subjects. The median age of the subjects was 21 years and all were unmarried. The daily energy intake (total calories and the proportions of carbohydrate, fat, and protein) was found to be similar to that previously determined for 19- to 29-year-old males.14 The proportions of current smokers and heavy drinkers among the study subjects were 40.0% and 6.0%, respectively, whereas 17.0% of the subjects were adjudged to undertake insufficient levels of physical activity.
Table 1.
Baseline Characteristics of 100 Healthy Male Soldiers
| Variable | Value |
|---|---|
| Age, yr | 21.0 (20–22) |
| Body mass index, kg/m2 | 22.7 (21.2–23.9) |
| Systolic blood pressure, mm Hg | 120.0 (113.0–131.5) |
| Diastolic blood pressure, mm Hg | 69.9±8.4 |
| Laboratory results | |
| White blood cells, ×103/μL | 6.1 (5.4–7.4) |
| Hemoglobin, g/dL | 15.1±0.9 |
| Platelets, ×103/μL | 254.3±54.3 |
| Glucose, mg/dL | 92.9±6.9 |
| Creatinine, mg/dL | 0.9±0.1 |
| Total cholesterol, mg/dL | 159.0 (144.5–181.0) |
| Aspartate transaminase, IU/L | 20.0 (17.0–23.0) |
| Alanine transaminase, IU/L | 17.0 (14.0–24.0) |
| Bristol stool form scale | |
| 3 | 48 (48.0) |
| 4 | 52 (52.0) |
| Diet | |
| Energy intake, kcal/day | 2,242.9 (1,844.9–2,800.7) |
| Carbohydrate, % | 58.7 (53.1–63.1) |
| Fat, % | 23.9 (21.4–27.1) |
| Protein, % | 14.2 (13.3–15.3) |
| Smoking status | |
| Never | 52 (52.0) |
| Past | 8 (8.0) |
| Current | 40 (40.0) |
| Drinking status, standard drinks/wk | |
| Never | 10 (10.0) |
| 1–4 | 61 (61.0) |
| 5–8 | 23 (23.0) |
| >8 | 6 (6.0) |
| Exercise, MET-min/wk | 3,773.0 (2,541.0–5,933.0) |
| Levels of physical activity | |
| Inactive | 17 (17.0) |
| Minimally active | 21 (21.0) |
| Health-enhancing physical activity | 62 (62.0) |
| Education level | |
| High school | 82 (82.0) |
| College & university or more | 18 (18.0) |
| Income, dollars/mo | |
| <270 | 20 (20.0) |
| 270–540 | 49 (49.0) |
| >540 | 31 (31.0) |
Data are presented as median (interquartile range), mean±SD, or number (%). International Physical Activity Questionnaire Short Form.
MET, metabolic equivalent of task.
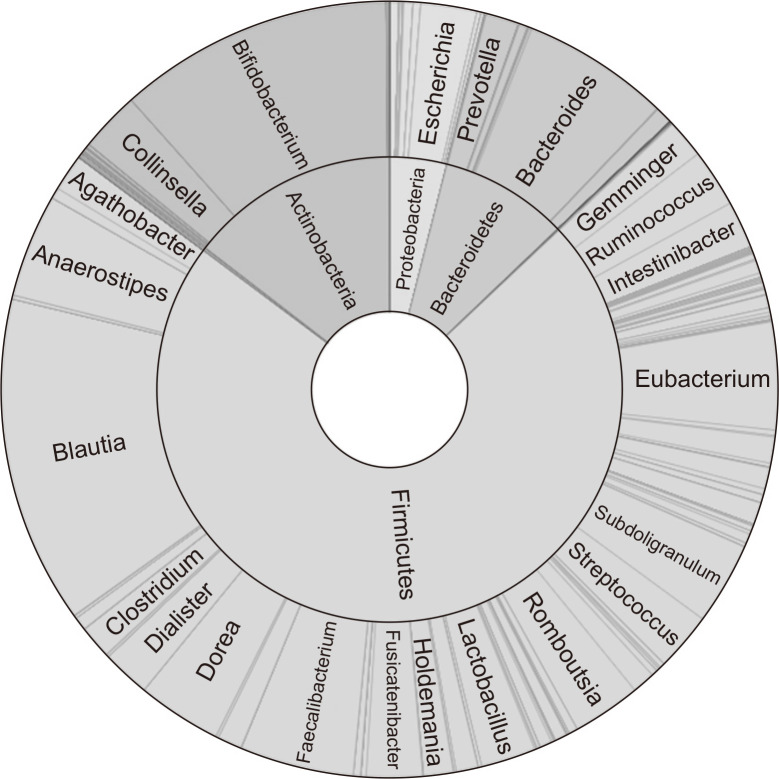
The mean number of valid 16S rRNA reads and operational taxonomic units for each sample was 82,937 and 312, respectively. The median value for Good’s library coverage was 99.9. Fig. 1 shows the taxonomic composition of the gut microbiome. At the phylum level, the relative abundances of Firmicutes, Actinobacteria, Bacteroidetes, and Proteobacteria were 72.3%, 14.5%, 8.9%, and 4.0%, respectively. The three genera showing the highest relative abundance were Blautia (13.6%), Bifidobacterium (11.4%), and Bacteroides (6.2%). Fifteen species, most of which belong to the Firmicutes (87%), were detected in all examined subjects (Table 2). Common species found in more than 90% of the subjects represented 1.9% of the total operational taxonomic units (45/2,313). The specific species are listed in Supplementary Table 1.
Fig. 1.
Taxonomic composition of the gut microbiome of 100 healthy male soldiers; double pie chart (inner circle: phylum level, outer circle: genus level).
Table 2.
Taxonomy of the 15 Species Found in 100% of Healthy Male Soldiers
| Phylum | Family | Species | Relative abundance, % |
|---|---|---|---|
| Actinobacteria | Bifidobacteriaceae | Bifidobacterium adolescentis | 5.76 |
| Firmicutes | Lachnospiraceae | Agathobacter rectalis | 1.53 |
| Firmicutes | Lachnospiraceae | Anaerostipes hadrus | 4.29 |
| Firmicutes | Lachnospiraceae | Blautia wexlerae | 7.49 |
| Firmicutes | Lachnospiraceae | LN913006_s | 2.67 |
| Firmicutes | Lachnospiraceae | Eubacterium hallii | 2.86 |
| Firmicutes | Lachnospiraceae | Fusicatenibacter saccharivorans | 2.33 |
| Firmicutes | Ruminococcaceae | Ruminococcus faecis | 2.12 |
| Firmicutes | Ruminococcaceae | Faecalibacterium prausnitzii | 4.54 |
| Firmicutes | Ruminococcaceae | Gemmiger formicilis | 1.86 |
| Firmicutes | Clostridiaceae | Clostridium celatum | 0.90 |
| Firmicutes | Peptostreptococcaceae | Romboutsia timonensis | 2.81 |
| Firmicutes | Streptococcaceae | Streptococcus pneumoniae | 0.07 |
| Firmicutes | Streptococcaceae | Streptococcus salivarius | 1.96 |
| Proteobacteria | Enterobacteriaceae | Escherichia coli | 2.33 |
2. Analysis of factors influencing the composition of the gut microbiome
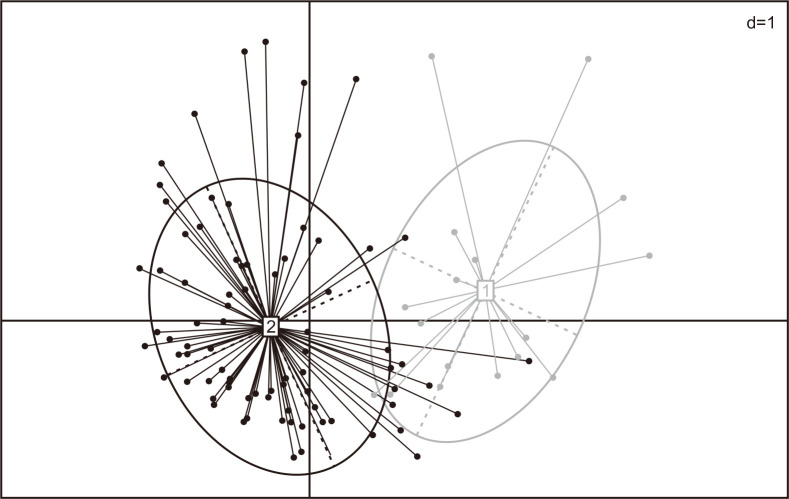
Using cluster analysis, we found that we could divide the subjects into two enterotypes based on bacterial species compositions (Fig. 2). Table 3 shows the clinical characteristics of healthy male soldiers according to enterotype. Compared with enterotype 2 subjects, those classified as having enterotype 1 tended to be characterized by a greater frequency of potentially harmful lifestyle habits (current smoker: 55.6% vs 36.6%, heavy drinker: 16.7% vs 3.7%, insufficient physical activity: 27.8% vs 14.6%); however, the differences did not attain statistical significance.
Fig. 2.
Cluster analysis based on the bacterial species compositions of 100 healthy male soldiers (1: Enterotype 1, 2: Enterotype 2).
Table 3.
Clinical Characteristics of Healthy Male Soldiers According to Enterotype
| Variable | Enterotype 1 (n=18) | Enterotype 2 (n=82) | p-value |
|---|---|---|---|
| Age, yr | 21.0 (21–22) | 21.0 (20–22) | 0.085 |
| Body mass index, kg/m2 | 21.8 (20.5–23.6) | 22.8 (21.3–23.9) | 0.216 |
| Systolic blood pressure, mm Hg | 115.0 (110.0–130.0) | 121.0 (114.0–132.0) | 0.106 |
| Diastolic blood pressure, mm Hg | 69.0±9.6 | 70.1±8.1 | 0.616 |
| Laboratory results | |||
| White blood cells, ×103/μL | 5.7 (5.0–7.0) | 6.2 (5.4–7.6) | 0.207 |
| Hemoglobin, g/dL | 15.2±0.8 | 15.1±1.0 | 0.748 |
| Platelets, ×103/μL | 251.3±52.7 | 254.9±55.0 | 0.797 |
| Glucose, mg/dL | 91.8±7.5 | 93.1±6.8 | 0.473 |
| Creatinine, mg/dL | 0.9±0.1 | 0.9±0.1 | 0.739 |
| Total cholesterol, mg/dL | 169.0 (146.0–179.0) | 158.0 (144.0–181.0) | 0.481 |
| Aspartate transaminase, IU/L | 20.0 (19.0–24.0) | 19.0 (17.0–23.0) | 0.324 |
| Alanine transaminase, IU/L | 20.0 (16.0–26.0) | 17.0 (14.0–23.0) | 0.191 |
| Bristol stool form scale | 0.942 | ||
| 3 | 8 (44.4) | 40 (48.8) | |
| 4 | 10 (55.6) | 42 (51.2) | |
| Diet | |||
| Energy intake, kcal/day | 2,126.6 (1,653.9–2,531.0) | 2,257.1 (1,930.4–2,821.7) | 0.335 |
| Carbohydrate, % | 62.2 (54.8–66.3) | 58.4 (53.0–62.3) | 0.159 |
| Fat, % | 22.4 (18.2–25.9) | 24.4 (21.6–27.4) | 0.201 |
| Protein, % | 13.8 (13.0–16.1) | 14.3 (13.4–15.3) | 0.427 |
| Smoking status | 0.222 | ||
| Never or past | 8 (44.4) | 52 (63.4) | |
| Current | 10 (55.6) | 30 (36.6) | |
| Drinking status | 0.120 | ||
| Non-heavy drinker | 15 (83.3) | 79 (96.3) | |
| Heavy drinker* | 3 (16.7) | 3 (3.7) | |
| Levels of physical activity | 0.318 | ||
| Insufficiently active | 5 (27.8) | 12 (14.6) | |
| Sufficiently active | 13 (72.2) | 70 (85.4) | |
| Education level | 0.616 | ||
| High school | 16 (88.9) | 66 (80.5) | |
| College & university or more | 2 (11.1) | 16 (19.5) | |
| Income, dollars/mo | 0.911 | ||
| <270 | 4 (22.2) | 16 (19.5) | |
| 270–540 | 8 (44.4) | 41 (50.0) | |
| >540 | 6 (33.3) | 25 (30.5) |
Data are presented as median (interquartile range), mean±SD, or number (%).
*Drinking status, more than 8 standard drinks/wk.
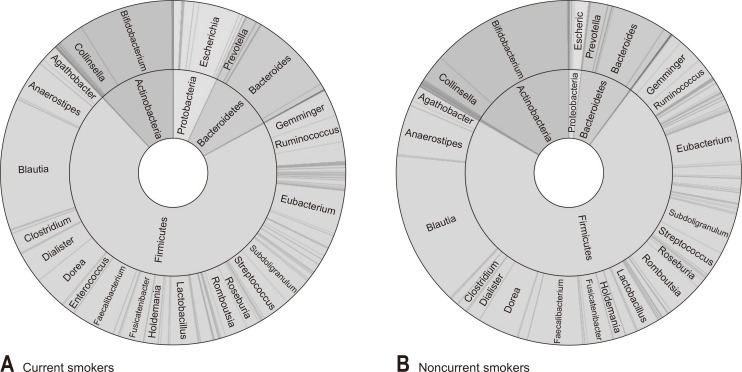
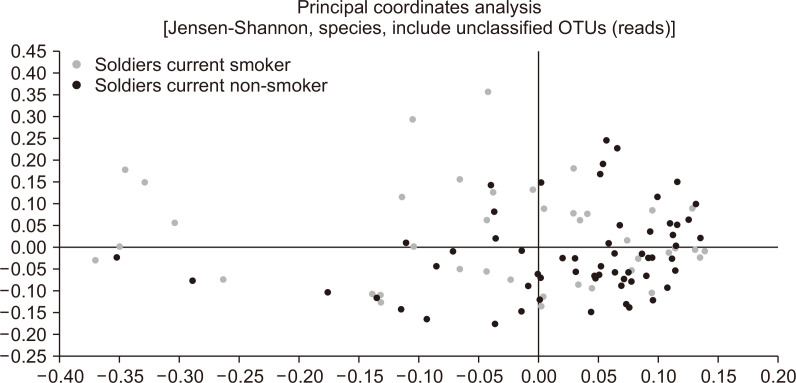
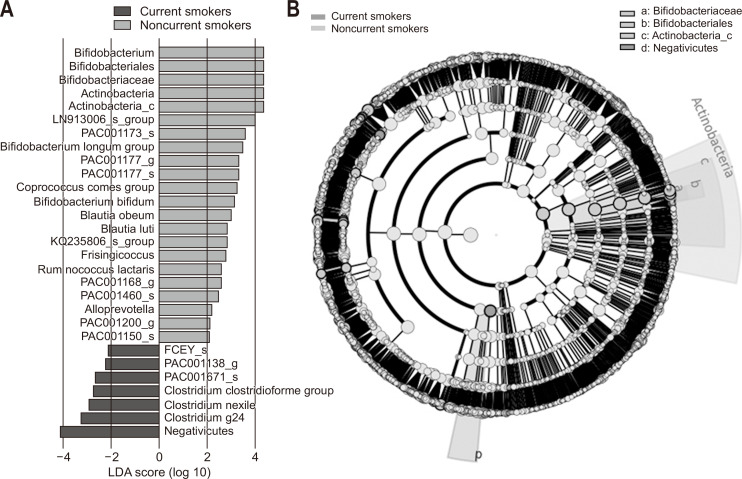
We subsequently analyzed the gut microbiome according to the smoking and drinking habits of the subjects and their levels of physical activity. Among current smokers, we found that the abundance of Actinobacteria was significantly lower than that in noncurrent smokers (11.9% vs 16.3%, p=0.026) (Fig. 3). Similarly, the abundance of Bifidobacterium, which is the main genus of Actinobacteria, was also lower in current smokers than in noncurrent smokers (8.8% vs 13.2%, p=0.008) (Fig. 3). Furthermore, the abundance of Proteobacteria was found to be higher in current smokers, although in this case the difference was not significant (7.0% vs 2.0%, p=0.072) (Fig. 3). In contrast, we detected no significant difference between smokers and non-smokers with respect to alpha-diversity evaluated in terms of species richness and diversity indices. Fig. 4 shows the results of Jensen–Shannon-based principal coordinates analysis of gut microbiome at the species level according to smoking status, which revealed a significant difference in microbial composition between current and noncurrent smokers (Permanova: p=0.008). On the basis of LEfSe analysis, we found that a relatively lower abundance of Bifidobacterium species and a higher abundance of bacteria in the class Negativicutes were main the determinants differentiating smokers from non-smokers (Fig. 5). In contrast to the effects of smoking, we detected no significant difference between heavy and non-heavy drinkers with respect to the beta-diversity of gut microbial composition (Permanova: p=0.238) or between subjects who undertook insufficient and sufficient physical activity (Permanova: p=0.135).
Fig. 3.
Taxonomic composition of the gut microbiome according to the smoking status (A: current smokers, B: noncurrent smokers); double pie chart (inner circle: phylum level, outer circle: genus level).
Fig. 4.
Jensen–Shannon-based principal coordinates analysis of the gut microbiome of 100 healthy male soldiers according to smoking status.
OTU, operational taxonomic unit.
Fig. 5.
(A) Linear discriminant analysis (LDA) effect size and (B) cladogram of the gut microbiome of 100 healthy male soldiers according to smoking status. A relatively lower abundance of Bifidobacterium species and a higher abundance of bacteria in the class Negativicutes were the main determinants differentiating current smokers from noncurrent smokers.
DISCUSSION
Previous studies have indicated that Firmicutes and Bacteroidetes typically comprise more than 90% of the adult human gut microbiome,15 and that the average abundances of Proteobacteria and Actinobacteria are approximately 4% and 8%, respectively.16,17 In the present study, we demonstrated that the four most abundant bacterial phyla in healthy young male Korean soldiers were Firmicutes (72.3%), Actinobacteria (14.5%), Bacteroidetes (8.9%), and Proteobacteria (4.0%). Taken together, these findings indicate that the most prominent character of the gut microbiome composition of soldiers recruited in the present study is a high abundance of Actinobacteria, which in a large part can be attributed to the high abundance of species in the genus Bifidobacterium (11.4%). In the Japanese population, it has been reported that the relative abundances of Actinobacteria and Bifidobacterium decrease substantially after weaning and continue to decrease with age.18 Therefore, in the present study, we may have obtained a snapshot of these bacterial taxa at an inflection point during early adulthood in the Korean population. In order to confirm this supposition, it will be necessary to evaluate changes in gut microbiomes within different age groups of Korean subjects. As mentioned in the Introduction section, this project is currently ongoing, and in the near future we indeed intend to analyze gut microbiomes across a range of age groups in the Korean population.
Of the 15 bacterial species detected in each of the study subjects, all, with the exception of Bifidobacterium adolescentis and Escherichia coli, belong to the Firmicutes. Among these, species in the genera Bifidobacterium, Agathobacter, Anaerostipes, Blautia, Eubacterium, Fusicatenibacter, and Faecalibacterium are all short-chain fatty acid producers,19-21 indicating the importance of short-chain fatty acid production in healthy adults. In addition, it has been reported that the abundance of Fusicatenibacter saccharivorans decreases in patients with active ulcerative colitis,19 whereas Gemmiger formicilis has a 100% prevalence in healthy Thais, regardless of their diet.20 Therefore, further research on the roles of these key players in healthy young adults are required. In the present study, we detected 45 operational taxonomic units in more than 90% of the subjects, which is similar to the findings of the aforementioned European human microbiome project.2 Other studies have also reported that the number of core taxa comprising the gut microbiome in healthy adults lies within the range of 40 to 87.21,22
Sex and age are invariably among the major determinants contributing to the differences observed in biological studies. Given that these variables were fixed in the subjects enrolled in the present study, who were also exposed to similar environments, we hypothesized that by classifying these subjects according to enterotype, we might obtain particularly meaningful information regarding the gut microbiome in healthy young adults. When we compared clinical factors between those classified as enterotype 1 and 2, it became apparent that a combination of potentially harmful lifestyle habits was a main major determining the differences in their gut microbiomes. However, because these differences did not attain a level of statistical significance, we performed further analyses of the relationship between gut microbiome composition and different lifestyle habits. When we compared stool microbiomes according to the smoking status of subjects, we obtained several interesting results. First, we found that the abundance of Actinobacteria in the stools of current smokers was significantly lower than that in the stools of noncurrent smokers, and that the abundance of Proteobacteria also tended to be higher in current smokers. These findings are consistent with those reported previously,23,24 and several mechanisms underlying this relationship have been proposed.25 Second, we found that there was a significant difference in the beta-diversity of stool microbial composition between current and noncurrent smokers. In this regard, a relatively low abundance of Bifidobacterium species and a high abundance of Negativicutes were the main determinants differentiating smokers from non-smokers. Whereas a decrease in Bifidobacterium has previously been reported in smokers,25 an increase in Negativicutes in current smokers is a new finding. To date, however, there have been relatively few studies that have examined the stool microbiomes in smokers, and the fact that the bacterial class Negativicutes includes several families, orders, and genera makes it difficult to interpret our results. Nevertheless, it has been observed that Negativicutes are abundant in the sub-gingiva of patients with chronic periodontitis, and species of Dialister, which was the most abundant genus among the Negativicutes detected in the present study, are known as periodontopathogens.26 Although a previous Chinese study found Dialister to be more commonly detected in current smokers than in those who have never smoked,27 these results were based on an analysis of the microbiome of the upper gastrointestinal tract, and we were unable to find any studies that have examined Dialister in the lower gastrointestinal tract of smokers. Given that periodontitis and smoking are both associated with poor oral hygiene, it remains uncertain as to whether an increase in the abundance of Negativicutes, including Dialister, in the gut microbiome of smokers is directly related to smoking. For example, it is conceivable that these bacteria gain a competitive advantage in a smoky environment. Further studies are accordingly required to clarify this point.
The present study has several strengths. Notably, the study subjects were recruited from a population in which numerous factors that could potentially affect the composition of the gut microbiome are well controlled. All subjects were male and aged between 20 and 22 years old. Moreover, these individuals were exposed to similar environmental factors. During the daytime, they worked in a military hospital as administrative clerks and ate the same meals three times a day in a cafeteria. At nights, they slept together in barracks. The duration between the date on which they started their military service and that on which they provided a stool sample was on average 9 months, and therefore we can assume that these young men had been exposed to the same environment for a sufficient length of time. Furthermore, when we did subgroup analysis according to the length of military service in the hospital (less vs more than 9 months), we could not detect significant difference in the beta-diversity of gut microbial composition (Permanova: p=0.181). In addition, we selected the subjects using very strict inclusion and exclusion criteria in order to ensure that they were all very healthy. This could be verified to a certain extent by our observation that Blautia was the most abundant bacterial genus detected in their stool samples, as an abundance of these bacteria is considered a strong indicator of a healthy gut.28,29 Outside business hours, however, they were free to partake in private activities such as exercise and smoking. When on holiday, they were even permitted to drink alcohol outside the military hospital. We thus speculate that it was these activities undertaken away from the work environment that contributed to observed differences in the gut microbiomes of these subjects. Accordingly, we can assume that the findings of the present study present a snapshot of the stool microbiome in healthy young Korean males.
There are, nevertheless, also certain limitations to the present study. First, the study was performed on a very specific group of individuals, and therefore it might be difficult to generalize these findings with respect to the Korean population as a whole. Second, although the daily energy intake of macronutrients was similar to that previously determined for males of the same age range, we could not include micronutrients data such as vitamins, minerals, and trace elements in the present analysis. Because the number and level of variables are too high, to perform it, more complex statistical methods are required. We are planning to perform it when we analyze gut microbiomes across a range of age groups in the Korean population. Third, we did not collect data on dental health of the subjects. Therefore, we cannot rule out the possibility that poor oral hygiene acted as a confounding factor in the relationship between smoking and increased abundance of Negativicutes.
In conclusion, a high abundance of Actinobacteria and a low abundance of Bacteroidetes are the main features characterizing the gut microbiomes of healthy young male soldiers in Korea. Further, a relatively lower abundance of Bifidobacterium species and a higher abundance of bacteria in the class Negativicutes were the main determinants differentiating the gut microbiomes of smokers from those of non-smokers.
Supplemental Materials
ACKNOWLEDGEMENTS
This study was supported by the Bio & Medical Technology Development Program of the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT (MIST) of the Republic of Korea (Project number: 2016M3A9F3947027).
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
AUTHOR CONTRIBUTIONS
Conceptualization: D.H.L., B.Y.K. Subjects enrollment: J.E.K. Microbiome analysis: J.H.L., S.J.Y., H.Y. Data and statistical analysis: H.Y., C.M.S. Writing- original draft: H.Y. Writing- review & editing: S.H.P., J.H.L., S.W.K., J.S.L. Funding acquisition: D.H.L., B.Y.L., J.S.L. Approval of final manuscript: all authors.
REFERENCES
- 1.Human Microbiome Project Consortium, author. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S, Lee JS, Hong KH, et al. Development and relative validity of semi-quantitative food frequency questionnaire for Korean adults. J Nutr Health. 2018;51:103–119. doi: 10.4163/jnh.2018.51.1.103. [DOI] [Google Scholar]
- 4.Kim EC, Kim JS, Jung JG, Kim SS, Yoon SJ, Ryu JS. Effect of alcohol consumption on risk of hyperhomocysteinemia based on alcohol-related facial flushing response. Korean J Fam Med. 2013;34:250–257. doi: 10.4082/kjfm.2013.34.4.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh JY, Yang YJ, Kim BS, Kang JH. Validity and reliability of Korean version of International Physical Activity Questionnaire (IPAQ) Short Form. J Korean Acad Fam Med. 2007;28:532–541. [Google Scholar]
- 6.Chun J, Kim KY, Lee JH, Choi Y. The analysis of oral microbial communities of wild-type and toll-like receptor 2-deficient mice using a 454 GS FLX Titanium pyrosequencer. BMC Microbiol. 2010;10:101. doi: 10.1186/1471-2180-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hur M, Kim Y, Song HR, Kim JM, Choi YI, Yi H. Effect of genetically modified poplars on soil microbial communities during the phytoremediation of waste mine tailings. Appl Environ Microbiol. 2011;77:7611–7619. doi: 10.1128/AEM.06102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim BS, Kim JN, Yoon SH, Chun J, Cerniglia CE. Impact of enrofloxacin on the human intestinal microbiota revealed by comparative molecular analysis. Anaerobe. 2012;18:310–320. doi: 10.1016/j.anaerobe.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 10.Yoon SH, Ha SM, Kwon S, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li XX, Wong GL, To KF, et al. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One. 2009;4:e7985. doi: 10.1371/journal.pone.0007985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kweon S, Kim Y, Jang MJ, et al. Data resource profile: the Korea National Health and Nutrition Examination Survey (KNHANES) Int J Epidemiol. 2014;43:69–77. doi: 10.1093/ije/dyt228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 17.Binda C, Lopetuso LR, Rizzatti G, Gibiino G, Cennamo V, Gasbarrini A. Actinobacteria: a relevant minority for the maintenance of gut homeostasis. Dig Liver Dis. 2018;50:421–428. doi: 10.1016/j.dld.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Odamaki T, Kato K, Sugahara H, et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16:90. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeshita K, Mizuno S, Mikami Y, et al. A single species of Clostridium subcluster XIVa decreased in ulcerative colitis patients. Inflamm Bowel Dis. 2016;22:2802–2810. doi: 10.1097/MIB.0000000000000972. [DOI] [PubMed] [Google Scholar]
- 20.Ruengsomwong S, La-Ongkham O, Jiang J, Wannissorn B, Nakayama J, Nitisinprasert S. Microbial community of healthy Thai vegetarians and non-vegetarians, their core gut microbiota, and pathogen risk. J Microbiol Biotechnol. 2016;26:1723–1735. doi: 10.4014/jmb.1603.03057. [DOI] [PubMed] [Google Scholar]
- 21.Martínez I, Muller CE, Walter J. Long-term temporal analysis of the human fecal microbiota revealed a stable core of dominant bacterial species. PLoS One. 2013;8:e69621. doi: 10.1371/journal.pone.0069621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 23.Biedermann L, Rogler G. Environmental factors and their impact on the intestinal microbiota: a role for human disease? Dig Dis. 2012;30 Suppl 3:20–27. doi: 10.1159/000342590. [DOI] [PubMed] [Google Scholar]
- 24.Biedermann L, Zeitz J, Mwinyi J, et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One. 2013;8:e59260. doi: 10.1371/journal.pone.0059260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savin Z, Kivity S, Yonath H, Yehuda S. Smoking and the intestinal microbiome. Arch Microbiol. 2018;200:677–684. doi: 10.1007/s00203-018-1506-2. [DOI] [PubMed] [Google Scholar]
- 26.Matthews CR, Joshi V, de Jager M, Aspiras M, Kumar PS. Host-bacterial interactions during induction and resolution of experimental gingivitis in current smokers. J Periodontol. 2013;84:32–40. doi: 10.1902/jop.2012.110662. [DOI] [PubMed] [Google Scholar]
- 27.Vogtmann E, Flores R, Yu G, et al. Association between tobacco use and the upper gastrointestinal microbiome among Chinese men. Cancer Causes Control. 2015;26:581–588. doi: 10.1007/s10552-015-0535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenq RR, Taur Y, Devlin SM, et al. Intestinal Blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant. 2015;21:1373–1383. doi: 10.1016/j.bbmt.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luu TH, Michel C, Bard JM, Dravet F, Nazih H, Bobin-Dubigeon C. Intestinal proportion of Blautia sp. is associated with clinical stage and histoprognostic grade in patients with early-stage breast cancer. Nutr Cancer. 2017;69:267–275. doi: 10.1080/01635581.2017.1263750. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.