Abstract
Purpose
SARS-CoV-2 engages human ACE2 through its spike (S) protein receptor binding domain (RBD) to enter the host cell. Recent computational studies have reported that withanone and withaferin A, phytochemicals found in Withania somnifera, target viral main protease (MPro) and host transmembrane TMPRSS2, and glucose related protein 78 (GRP78), respectively, implicating their potential as viral entry inhibitors. Absence of specific treatment against SARS-CoV-2 infection has encouraged exploration of phytochemicals as potential antivirals.
Aim
This study aimed at in silico exploration, along with in vitro and in vivo validation of antiviral efficacy of the phytochemical withanone.
Methods
Through molecular docking, molecular dynamic (MD) simulation and electrostatic energy calculation the plausible biochemical interactions between withanone and the ACE2-RBD complex were investigated. These in silico observations were biochemically validated by ELISA-based assays. Withanone-enriched extract from W. somnifera was tested for its ability to ameliorate clinically relevant pathological features, modelled in humanized zebrafish through SARS-CoV-2 recombinant spike (S) protein induction.
Results
Withanone bound efficiently at the interacting interface of the ACE2-RBD complex and destabilized it energetically. The electrostatic component of binding free energies of the complex was significantly decreased. The two intrachain salt bridge interactions (K31-E35) and the interchain long-range ion-pair (K31-E484), at the ACE2-RBD interface were completely abolished by withanone, in the 50 ns simulation. In vitro binding assay experimentally validated that withanone efficiently inhibited (IC50=0.33 ng/mL) the interaction between ACE2 and RBD, in a dose-dependent manner. A withanone-enriched extract, without any co-extracted withaferin A, was prepared from W. somnifera leaves. This enriched extract was found to be efficient in ameliorating human-like pathological responses induced in humanized zebrafish by SARS-CoV-2 recombinant spike (S) protein.
Conclusion
In conclusion, this study provided experimental validation for computational insight into the potential of withanone as a potent inhibitor of SARS-CoV-2 coronavirus entry into the host cells.
Keywords: ACE2-RBD complex, Withania somnifera, withanone, docking and MD simulation, ELISA, SARS-CoV-2 S-protein, humanized zebrafish model
Video abstract
Point your SmartPhone at the code above. If you have a QR code reader the video abstract will appear. Or use:
Introduction
The COVID-19 nightmare began with the report of a pneumonia case of unknown cause in Wuhan, China on December 31, 20191 and soon after, in March 2020, WHO declared the disease a pandemic. According to the weekly epidemiological update, until December 27, 2020, the total number of COVID-19 cases crossed 79.2 million with number of deaths being more than 1.7 million since the beginning of the pandemic (https://www.who.int/publications/m/item/weekly-epidemiological-update---29-december-2020).
SARS-CoV-2, the causative coronavirus responsible for COVID-19 outbreak, is a sister clade of SARS-CoV, that caused severe acute respiratory syndrome (SARS) in 2002.2 Resembling SARS-CoV, this virus also exploits ACE-2 receptors to invade host cell, thus, making the high level ACE2 expressing alveolar type II epithelial cells the primary target.3 Without encouraging outcomes from clinical trials on repurposed drugs,4 there is an unmet requirement for newer therapeutics which can specifically act against SARS-CoV-2.
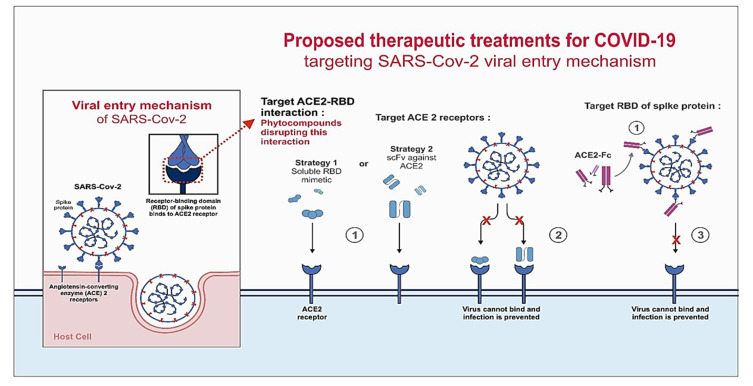
During viral infection, entry of the virus into the host cell is a critical step that can be exploited for antiviral therapy.5 Therefore, receptor binding domain (RBD) has been an attractive target for the researchers to abrogate coronavirus infection. Reports suggested that certain human antibodies recognized RBD on the S1 domain of SARS-CoV and inhibited the viral infection by blocking its attachment to ACE2.6,7 Three possible mechanisms, namely, targeting the ACE2 receptor, RBD of S protein, and the interaction between ACE2 and RBD have been schematically depicted in Figure 1 through which SARS-CoV-2 entry/infusion can be abrogated. Prime focus is on targeting RBD or ACE2 and less thought is given toward disrupting the RBD-ACE2 interaction. Nevertheless, a contingency approach to prevent viral entry, in case the first window of opportunity (before RBD gets to interact with ACE2) is missed, will be quite valuable. In fact, the observations reported in this study identify a potential candidate for this contingent approach. We are referring to Withania somnifera (L.) Dunal (Solanaceae), commonly known as Ashwagandha. It is one of the most valued medicinal plants of the traditional Indian systems of medicines, and has been used in more than 100 Ayurvedic formulations. It is therapeutically equivalent to ginseng, concordant with the observation that medicinal plants with similar bioactivity exhibit phylogenetic clustering.8,9 W. somnifera is used to treat genital infection by herpes simplex virus among African tribes.10 It also exhibits anti-influenza properties.11 In our preliminary study in March 2020, we observed that withanone binds at the center of the ACE2-RBD interface.12 A recent computational study has predicted that withanone can also interact with the main protease (MPro) of SARS-CoV-2, which is responsible for cleaving host transmembrane protease serine 2 (TMPRSS2) required for viral entry into host cell.13 In a follow-up study, the same group has shown that withanone and withaferin A can interact with TMPRSS2 and block viral entry into the host cell. Withanone was also shown to downregulate TMPRSS2 transcription.14 Likewise, withaferin A, the second major phytochemical with reported cytotoxicity against cancer cells that co-extracts with withanone from W. somnifera leaves, can bind to GRP78 as observed in another docking study.15 GRP78 was reported to be involved in the MERS infection. Blocking ACE2 with soluble antibodies or RBD of viral S protein with soluble ACE2 are identified as considerable options.16–18 Synthetic nanobodies/sybodies to block RBD of SARS-CoV-2 spike (S) protein are reported recently in a preprint.19 However, all these strategies target either the host cell or viral factors independently. Inhibition of specific host–pathogen interaction will be more preferable, but no report exists to show that this option has been explored yet. So, given the reported antiviral efficacy of W. somnifera, we evaluated whether withanone can disrupt host–viral interaction.
Figure 1.
Proposed mechanisms to block the entry of SARS-CoV-2 into host cells.
Notes: The mechanism behind SARS-CoV-2 entry into the host cell is schematically represented. Three proposed models are depicted where COVID-19 infection can be abrogated by blocking the interaction of RBD of spike (S) protein and ACE2. In one of the approaches, ACE2-RBD interaction can be destabilized by small molecules. In the second approach, ACE2 can be blocked with RBD mimetics or single-chain antibody fragment (scFv) against ACE2. In the third approach, RBD of SARS-CoV-2 S-protein can be blocked using the ACE-2 extracellular domain. An Fc domain fused to ACE-2 would facilitate prolonged circulation of the biologic (ACE2-Fc). The observations made in this study support the strategy to block or weaken the interaction between RBD and ACE2 by using phytocompounds of natural origin.
The objective of the present study was to evaluate the potential of withanone, the major phytocompound present in W. somnifera as an inhibitor of host–pathogen specific interaction between host ACE2 and viral RBD of SARS-CoV-2 S-protein and its efficacy in ameliorating S-protein-induced inflammatory pathologies in a humanized zebrafish model of the disease. We employed computational methods to evaluate the effects of withanone on interaction between viral S-protein RBD and host ACE2 receptor. Through an ELISA-based interaction study, we observed that in the presence of commercially procured withanone, interaction between RBD and ACE2 was significantly reduced. In this regard, it is pertinent to mention that co-extraction of withaferin A with withanone from leaves of W. somnifera is unavoidable.20 Withaferin A is cytotoxic and this property has been explored for treating different types of cancers.21 Human equivalent dose (HED) and maximum recommended starting dose (MRSD) for withaferin A have been worked out.22 But one would like to avoid its presence altogether, if possible, particularly when a severe viral infection that leaves the immune system vulnerable is being dealt with. Keeping this, and our subsequent plan for experimental validation of antiviral efficacy of withanone, in perspective we devised a method to prepare W. somnifera leaf extracts enriched only in withanone (WiNeWsE). Furthermore, the antiviral activity of WiNeWsE was checked in a humanized zebrafish (HZF) model of SARS-CoV-2, which was established according to earlier reports.23–25 WiNeWsE treatment was found to be effective in ameliorating the characteristic behavioral and phenotypic manifestations associated with SARS-CoV-2 disease induction in zebrafish.
Materials and Methods
Computational Study
Structures
We screened 200 different phytocompounds by in silico methods. The strategy was to find phytocompounds that bind at the RBD-ACE2 complex interface, and perturb their interactions. In this regard, the phytocompound from W. somnifera showed promising results. Nine structurally similar withanolides, namely, 27-hydroxy withanone, 17-hydroxy withaferin A, 17-hydroxy-27-deoxy withaferin A, withaferin A, withanolide D, 27-hydroxy withanolide B, withanolide A, withanone, and 27-deoxywithaferin A were identified through reverse-phase HPLC in W. somnifera root extracts.26 Unpublished data from our laboratory also showed significant concentration of withanolide A, withanolide B, withaferin A, and withanone in 15–20 days old W. somnifera sapling. We studied these compounds in detail after preliminary screening. The 3D structures of all the phytocompounds were retrieved from PubChem database (https://pubchem.ncbi.nlm.nih.gov).
Molecular Docking
The latest structure of SARS-CoV-2 RBD complexed with ACE2 receptor (PDB ID: 6M17, 2.9 Å resolution) was retrieved from protein data bank (http://rcsb.org). Clean protease domain of chain B (ACE2) and chain E (RBD) were selected after editing on PyMol.37 Energy minimization was performed by 100 sttif of steepest descent, followed by 500 sttif of conjugate gradient using UCSF Chimera-1.13.127 after adding hydrogens. The stereochemical quality of the energy minimized structure was checked using VERIFY3D, ERRAT, PROCHECK, and RAMPAGE (for Ramachandran plot).28–31
The SDF files of all the ligands downloaded from the PubChem database were converted into PDB files using Open Babel 2.4.1.32 Hydrogens were added using UCSF Chimera-1.13.1 (Pettersen et al), and docked against ACE2-RBD complex.27 There is still a challenge in identifying a putative binding site of a ligand on a protein. There are many reports on detecting protein ligand binding sites, but their accuracies only go up to 50–80%.33 Focused docking is supposed to be better than blind docking in this regard.34 The more accurate method is the molecular-docking binding-site finding (MolSite) method proposed by Fukunishi and Nakamura,33 which showed 80–99% accuracy in predicting ligand binding sites. In the current study, the ligand binding site in the ACE2-RBD complex was determined by the MolSite method.33 Briefly, the ACE2-RBD complex was mapped by five-grid center (including the protein interface) of grid size 30×30×30, and was subjected to AutoDock Vina calculations using the PyRx Virtual Screening Tool35 with default parameters. The grid was centered on Cα of five different amino acids (x=166.539, y=105.678, z=256.879; x=178.643, y=126.269, z=220.744; x=165.097, y=119.940, z=247.957; x=150.015, y=122.092, z=213.204; x=150.015, y=122.092, z=213.204; x=130.190, y=136.848, z=221.901); thus, the method helped to map the whole complex surface. The best binding pose was supposed to be the pose with the lowest energy, obtained after comparison of the five independent docking runs, and was aligned with the receptor for the ligand-receptor interaction analysis using Maestro-12.4 (Schrödinger Release 2020–2: Maestro, Schrödinger, LLC, New York, NY, USA). Discovery Studio 2017 R2 Client36 and PyMol37 were used to generate the graphics.
Molecular Dynamics (MD) Simulation
The ligand-ACE2-RBD complex obtained after molecular docking was subjected to MD simulation. The simulation systems for ACE2-RBD complex without or with withanone was prepared using the VMD software.38 Ligand parameterization was done with CHARMM-GUI web interface (http://www.charmm-gui.org)39 using CHARMM general force field.39 MD simulation was performed with CHARMM36 force field using the NAMD package.40 The protein complex without or with withanone was solvated with TIP3P water molecules 105 Å from the protein. The systems were ionized and neutralized with 145 mM of NaCl. The systems contained 69,051 and 69,003 water molecules in the protein complex without and with withanone, respectively. NPT ensemble was used with periodic boundary conditions. Pressure was fixed at 1.01325 bar and the temperature at 310 K. The particle-mesh Ewald method was used to evaluate the Coulomb interactions with a grid size of 1 Å. A 2 fs of time step was used in all MD simulations. Initially, water was equilibrated for 0.5 ns at 310 K after fixing the protein and ligand, and energy minimization of 1000 sttif. One thousand sttif of energy minimization of the whole system were performed, and further equilibration for 5 ns at 310 K, after releasing the protein and ligand, was done. The production run was of 50 ns. The trajectory data were saved at every 50 ps to analyze the change in the dynamics of the ACE2-RBD binding interface. The results for flexibility were analyzed by plotting the RMSD values of backbone atoms (C, Cα, N) against the 1000 conformations, and per residue RMSF was calculated for Cα atoms. For electrostatics calculation, trajectory clustering was performed by UCSF Chimera-1.13.127 using a step size of one with default parameters.
Salt Bridge Analysis
We considered ion pair as a salt bridge when side-chain charged group centroids and at least one pair of side-chain nitrogen and oxygen atoms is within 3.2 Å distance, or a longer-range ion pair (up to 7 Å), and was analyzed using VMD.38 Electrostatic free energy upon salt bridge formation was calculated in initial and final trajectories complexed with the withanone. All the computations were performed according to the protocol of Hendsch and Tidor,41 with slight modifications. Briefly, it was calculated relative to a mutation of its salt-bridging side-chains to their hydrophobic isosteres. They are identical with the charged residue side-chains, with the exception that their partial atomic charges were set to zero. The protonation states of all the charged residues were assigned at pH 7.4 using the ProteinPrepare module in PlayMolecule (https://www.playmolecule.org). Continuum electrostatic calculations were performed with the DelPhi v8.4.3.42 The PARSE partial atomic charges and atomic radii were used.43 The solvent probe radius used was 1.4 Å. The dielectric constants of the protein and the solvent were 4.0 and 80.0 respectively, and the ionic strength was 0.145 M. The Poisson–Boltzmann equation was solved using the iterative finite difference method, initially mapping the molecule on a 3D grid with a grid size of 165 and scale of one, and then focusing with a grid size of 65 and scale of four. DelPhi gives the energy values in units of kT, where k is the Boltzmann constant and T is absolute temperature, and the values were multiplied by 0.5922 to obtain the results in kcal/mol at room temperature (25°C).
Computation of Electrostatic Component of Binding Free Energy of the ACE2-RBD Complex
Quantification of electrostatic interactions is important to study protein–protein interactions in biomolecular systems. We solved the Poisson–Boltzmann equation (PBE) in the implicit solvent model to study the interaction between the receptor ACE2 and the viral RBD.44 The electrostatic component of ΔΔG, ΔΔGel can be quantified by solving the Poisson–Boltzmann equation (PBE) with the assumption that there are no conformational changes upon binding.
We used DelPhi v8.4.342 to calculate the electrostatic component of the binding free energies, and in the trajectories obtained after clustering. The term, ΔΔGel can be obtained by subtracting the grid energy of the binding partners from that of the complex (DelPhi manual). Grid size of 165, scale of one and ionic strength of 0.145 M were used for the calculation. Protein and solvent dielectric constants were 4 and 80, respectively.
In vitro Validation of the Inhibitory Effect of Withanone on ACE2-RBD Interaction
The effect of withanone on ACE2-RBD interaction was evaluated using the SARS-CoV-2 inhibitor screening kit (ACRObiosystems, Newark, DE, USA) according to the manufacturer’s protocol. Briefly, wells of the ELISA plate were coated with recombinant truncated SARS-CoV-2 S-protein expressing only RBD. Subsequently, biotinylated human ACE2 protein was allowed to bind to the immobilized RBD in the ELISA wells in the presence of 47 pg/mL (10 pM)–4.7 µg/mL (10 µM) and absence of commercially procured withanone (Sigma, USA). Efficiency of ACE2 binding to RBD was detected through HRP conjugated streptavidin by measuring the absorbance at 450 nm in EnVision microplate reader (Perkin Elmer Inc, Waltham, MA, USA). The absorbance was used to calculate percent inhibition of ACE2-RBD interaction, relative to the reaction mix without withanone. Reaction mix treated with a known inhibitor (provided with the kit) served as a positive control. The observation was represented as a dose-response curve and IC50 calculated using an option built into the GraphPad Prism version 7.0.0 for Windows (GraphPad Software, San Diego, California, USA).
Preparation of Withanone Rich Extract from Leaves of W. somnifera (WiNeWsE) and its Compositional Analysis
Fresh leaves of W. somnifera were collected from the medicinal herbal garden of the Patanjali Research Institute, Haridwar, India, washed with water and pulverized. Then, 100 g of this pulverized material was extracted in 1:10 volume of solvent (methanol:water in 80:20 ratio) and agitated three times at 70°C in reflux conditions. Each extraction lasted for two hours. Extracted layers were filtered, pooled, and concentrated to yield 2.7 g of powdered hydromethanolic extract. For compositional fingerprinting of the leaf extract through HPTLC, 500 mg of the sample was mixed in 5 mL of methanol:water (80:20), sonicated for 20 min and centrifuged. Chromatographic separation was done on silica gel 60 F254 plates in a mobile phase containing toluene, ethylacetate and formic acid in a 5:5:1 ratio. The plates were scanned at 230 nm and derivatized in an anisaldehyde sulfuric acid reagent. Withanone content in the leaf extract was quantified from the regression equation 234.190*X+3.043 using a linearity range from 400–1200 ng having a coefficient of correlation as 0.9997. Limits of detection per spot were 0.30 and 0.90 ng, respectively. Rf value used for detection was 0.40.
The compositional quantification of this extract was done through high performance liquid chromatography (HPLC) (Waters Corporation, USA) equipped with binary pump (1525), PDAD (2998) and autosampler (2707). Withanone standard was purchased from Natural Remedies Pvt. Ltd (Bangalore, India) and dissolved in methanol to get the appropriate concentration. Next, 0.25 g of W. somnifera leaf extract was diluted with 10 mL methanol:water (75:25), sonicated for 30 min and filtered using a 0.45 µm nylon filter before injecting 10 µL of this into the column. Separation was achieved using a Shodex C18-4E (5 µm, 4.6*250 mm) column and elution was carried out at a flow rate of 1.5 mL/min using a defined gradient program of mobile phases A (1 mM KH2PO4, 0.05% H3PO4) and B (acetonitrile). Column temperature was maintained at 27°C. Wavelength was set at 227 nm for analysis.
In vivo Preclinical Assessment of the Antiviral Activity of WiNeWsE in Humanized Zebrafish Model of SARS-CoV-2
Cell Line and Chemicals
Human alveolar epithelial cell line (A549) was procured from ATCC certified repository at the National Centre of Cell Science (NCCS), Pune, India. SARS-CoV-2 recombinant S-protein was obtained from Bioss Antibodies (Woburn, MA, USA). H&E stain and PBS were purchased from Sigma Aldrich (St Louis, MO, USA).
Zebrafish Husbandry, Animal Selection, and Group Setting
Zebrafish experiments were in line with the standards of Institutional Animal Ethics Committee (IAEC) in compliance with the guidelines of the Committee for the Purpose of Control and Supervision of Experiments (CPCSEA), India. All zebrafish experiments were conducted according to IAEC approved protocols (approval number 223/Go062062/IAEC). The fish were housed under 14 h light and 10 h dark cycle in 16 L tanks with water temperature maintained at 27±1°C and were fed with 5 mg of commercial feed (TetraBit, Blacksburg, VA, USA) per gram body weight once per day. One-year-old, 25–30 mm long zebrafish of both the genders, weighing 0.5 g were selected for the study. Selection criteria and housing arrangements are summarized in Table 1. The study subjects were divided into seven groups, each containing 24 fish, maintained in two clutches with 12 each. Different groups along with the corresponding treatments administered are provided in Table 2.
Table 1.
Zebrafish Selection Criteria and Housing Conditions
| Gender | Male (♂) and Female (♀) |
|---|---|
| Age | 1 year |
| Body weight | 0.5 g |
| Body length | 25–30 mm |
| Number of fish/group/endpoint | 24 |
| Number of fish per clutch | 12 |
| Tank capacity | 16 L |
| Ambient water temperature | 27±1°C |
| Light–dark cycle | 14 h light, 10 h dark |
| Feed frequency | 5 mg/g body weight/once/day |
Notes: The above inclusion conditions were considered while selecting the fish for the study. The housing conditions of the selected subjects are also mentioned above.
Table 2.
Experimental Groups, Group Size, and Administered Treatments
| Name of the Group | Group Sizea | Administered Treatment |
|---|---|---|
| NC | 72 | Zebrafish with no xenotransplantation and no SARS-CoV-2 induction |
| XMC | 72 | Zebrafish transplanted with human alveolar epithelial A549 cells on the posterior lobe of swim bladder |
| DC | 72 | XMC injected with SARS-CoV-2 recombinant S-protein |
| 1X-Dexa-HED | 72 | DC administered with 0.08 µg/kg body weight of dexamethasone through feed |
| 0.2X-HED | 72 | DC administered with 6 µg/kg body weight of WiNeWsE through feed |
| 1X-HED | 72 | DC administered with 28 µg/kg body weight of WiNeWsE through feed |
| 5X-HED | 72 | DC administered with 142 µg/kg body weight of WiNeWsE through feed |
Notes: aThere were 24 subjects/group/endpoint. There were three endpoints in this study.
Abbreviations: NC, normal control; XMC, xenotransplant model control; DC, disease control; 1X-Dexa-HED, human equivalent dose of dexamethasone; WiNeWsE, withanone enriched Withania somnifera extract; 0.2X-HED, 0.2X human equivalent dose of WiNeWsE; 1X-HED, human equivalent dose of WiNeWsE; 5X-HED, 5X human equivalent dose of WiNeWsE.
Xenotransplant Model Establishment
A549 cells, cultured in DMEM with 10% FBS and 1% penicillin-streptomycin at 37°C, were passaged continuously three times before using the third passage for transplantation. The 1X102 cells in PBS were injected intramuscularly at the junction of the trunk and the caudal region, along the midline to seed them at the posterior lobe of the swim bladder. Postinjection, the fish were transferred to their respective clutches and were observed for a period of seven days. Cytology of the swim bladder was studied on day 7 to confirm adherence of A549 cells and establishment of humanized zebrafish (HZF) model. A group of 72 HZF without injection of S-protein was maintained as the xenotransplant model control (XMC) and studied alongside the normal control (NC). The NC group contained 72 ordinary zebrafish selected as per the criteria mentioned in Table 1.
Induction of Pathological Milieu by SARS-CoV-2 S-protein
Seven days post xenotransplantation, the fish were given intramuscular injections at the site of xenotransplant, with 2.8 µL of 1 ng/µL SARS-CoV-2 recombinant S-protein in PBS using a Hamilton syringe. The fish were anesthetized using gradual cooling of the water to 12°C. Anesthetization was confirmed through reduced operculum movement and lack of response to touch by the caudal fin. Post recombinant S-protein injection, the zebrafish undergoes a series of immune responses. During this period the fish is housed at 27±1°C to aid the physiological changes in immunity.
Dosing and Screening
Translational human equivalent doses (HED) were optimized to be 1000X less than the human dose by body weight. Human dose equivalences are provided in Table 3. Oral dosing was adopted over a dilution range of 0.2, 1, 5 relative to HED for a period of 10 days. To prepare the infused oral feeds, a known quantity of the compound was mixed with fish food weighing 2.5 mg per pellet and were extruded in to uniform pellets. The study groups were fed on a 24-h cycle with an estimated number of pellets per fish. During the dosing session the fish were isolated from the respective study groups and were fed individually in the feeding tank. Control fish were fed with unmodified fish feed and were fed under conditions similar to that of study groups. Drug screening was done at two screening time points, after three and six days of treatment to understand the acute and subacute pathophysiological changes. Dexamethasone was administered as a positive control.
Table 3.
Human Dose Equivalence and Translational Dosing for Zebrafish in Present Study
| Human Dose | Translational Dose for Zebrafisha (µg/kg Body Weight) | ||||
|---|---|---|---|---|---|
| Dexamethasone | WiNeWsE | Dexamethasone | WiNeWsE | ||
| 1X-Dexa-HED | 0.2X-HED | 1X-HED | 5X-HED | ||
| 6 mg/day | 2 g/day | 0.08 | 6 | 28 | 142 |
Note: aZebrafish weighing 0.5 g were included in the study.
Endpoint Studies
Behavioral Fever
Zebrafish being an ectotherm exhibits behavioral fever as an adaptive immunity, that is, it migrates to the water at a temperature matching its body temperature. So, if a zebrafish is having a higher body temperature due to immunological reactions, it will migrate and spent more time in water at a higher temperature matching that of its body. To study the behavioral fever, an experimental tank with three chambers at different temperatures (23°C, 29°C, and 37°C) was used. The chambers were positioned left-to-right in order of increasing temperature. Continuous cooling and heating were maintained in the flanking chambers during the study period. The fish from the respective study clutches were introduced individually into the temperature gradient chamber at 29°C and given opportunity to choose the temperature that supports the physiological change. The time spent by the fish in the temperature gradient chamber was calculated for consecutive three minutes and the readings were recorded.
Screening for Presence of Skin Hemorrhage
Whole animal imaging was performed using a digital single lens reflex (DSLR) camera (D3100 Nikon Corporation, Tokyo, Japan). Fish with skin hemorrhages, demarcated as red spots, were enumerated.
Dissection of Swim Bladder for Anatomical and Histopathological Observations
The fish were euthanized by putting them in water at 2–4°C and dissected as per ethical guidlines by ventral incision in the skin from the lower jaw to the vent. The intestinal tract and gonads were removed to expose the swim bladder. The swim bladder, located ventral to the kidney, was isolated without any damage to the esophagus connected to it through pneumatic duct. Likewise, heart, intestinal cavity, and gonads were removed to expose the kidney attached to the dorsal body wall. The fish was pinned to the dissection board with ventral side up to isolate the kidney intact. The isolated swim bladder and kidney were washed in PBS and observed under the Labomed CM4 stereomicroscope at 1X magnification, using 14 MP Labomed camera. Cytology slides were prepared from whole swim bladder and kidney necropsies and were stained with H&E.
Kaplan–Meier Survival Curve
Mortality was counted on a daily basis and plotted as a Kaplan–Meier survival curve to understand the survival rate of the study groups over therapeutic intervention.
Data Analysis
All graphical data are displayed as mean ± standard error of the mean (SEM). Statistical significance of the variance observed among different means was determined through GraphPad Prism 7.0 software. Two-way ANOVA with Dunnett’s multiple comparison test was used to determine the statistical significance.
Result
Computational Studies
Docking and Molecular Dynamic (MD) Simulation
The stereochemical quality of the structure was checked and confirmed. Phytocompounds including withanolides present in roots and leaves of W. somnifera were docked with the ACE2-RBD complex. The phytocompounds were bound at the ACE2-RBD complex tightly (see Table 4 for Vina scores).
Table 4.
Names, Structure, Molecular Weight, and Binding Energies (AutoDock Vina Score) of Phytocompounds from W. somnifera Docked into ACE2-RBD Complex
| Phytocompounds in W. somnifera | Structure | Molecular Weight (g/mol) | Binding Energy in ACE-RBD Complex (kcal/mol) |
|---|---|---|---|
| Withanone | 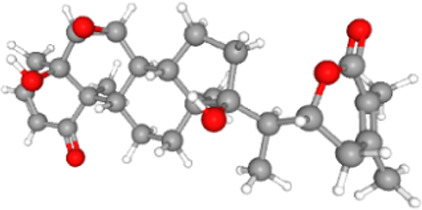 |
470.6 | −9.4 |
| Withanolide A | 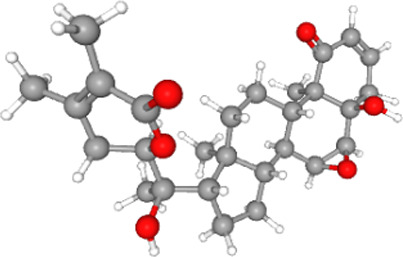 |
470.6 | −9.6 |
| Withanolide B | 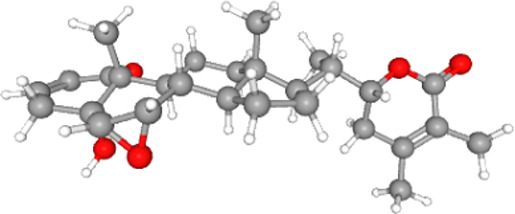 |
454.6 | −9.4 |
| Withaferin A | 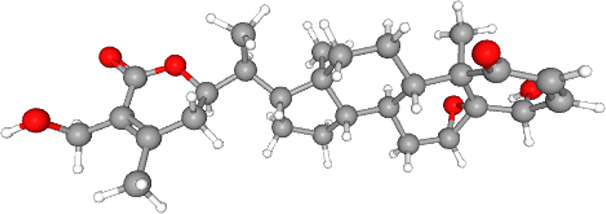 |
470.6 | −9.1 |
Notes: The structures, molecular weights and binding energies (with the ACE2-RBD complex) of major phytocompounds present in W. somnifera are provided in the table above. The structures and molecular weights of these phytocompounds were mined from PubChem data sources (https://pubchem.ncbi.nlm.nih.gov/). The binding energies were obtained from molecular docking studies reported here.
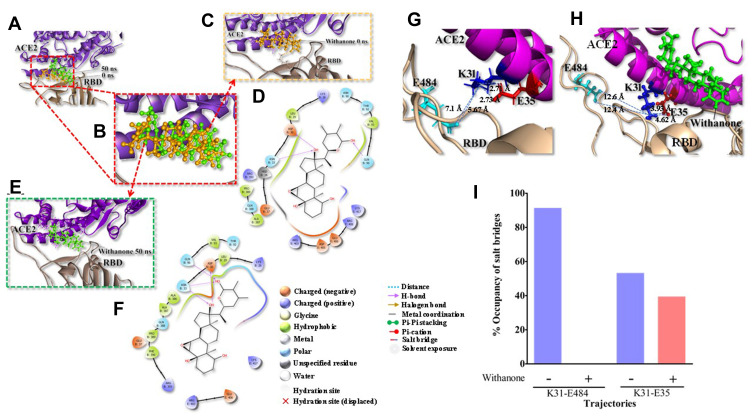
The phytocompound which bound to the interface was subjected to further investigation. Only withanone was found to be docked into the ACE2-RBD complex (Figure 2A). It bound at the interface of the ACE2 receptor and RBD, interacted with the residues from both ACE2 and RBD (Figure 2B) and was thus analyzed further to study its role in weakening or blocking the interactions between the ACE2 receptor and RBD. Docking showed efficient binding of withanone (ligand) through two H-bonds (D30 of ACE2 and R403 of RBD to withanone), alkyl, and van der Waals interactions at the ACE2-RBD interface (Figure 2C and D) (Table 5).
Figure 2.
Withanone docks at the interface of the ACE2-RBD complex and shifts slightly towards the center of the interface, modulating several molecular interactions in the process.
Notes: (A) Initial and final position of withanone in the ACE2-RBD complex (PDB ID: 6M17) and is predicted to move slightly toward the ACE2 side of the complex, as revealed by 50 ns molecular dynamic (MD) simulation. (B) The initial position of withanone (shown in golden yellow at 0 ns) and its final positioning as predicted from MD simulation after 50 ns (shown in green) is depicted as a magnified view. (C and D) Withanone at 0 ns (C), binds in the pocket forming three H-bonds, D30, N33, and Q96 of ACE2, in addition to alkyl and van der Waals interactions (D). (E and F) Withanone at 50 ns, (after MD simulation) with final trajectory zoomed in (E) and interactions of withanone within the ACE2-RBD complex as seen in the final trajectory (F). All atoms RMSD of withanone between initial and final positions are 2.166 Å. (G and H) Salt bridge interaction at the binding interface of ACE2-RBD in the final trajectory without withanone (G) and with withanone (H). (I) Percent occupancy of the salt bridge and long-range ion pair modulated by withanone incorporation as seen by analysis of the simulation trajectories.
Table 5.
Residues of ACE2-RBD Complex Involved Within 4 Å Range of Its Interaction with Withanone Before and After Simulation Runs
| Pre MD | Post MD | ||||
|---|---|---|---|---|---|
| H-Bond | Alkyl Interaction | Van der Waals | H-Bond | Alkyl Interaction | Van der Waals |
|
ACE2: D30 RBD: R403 |
ACE2: K26, L29, V93, P389 |
ACE2: T92, Q96 RBD: E405, E406, R408, Q409, G416, K417 |
ACE2: D30, N33, Q96 |
ACE2: K26, L29, V93, A387, P389 |
ACE2: E37, T92, A386, Q388, P390, R393 RBD: R403, E406, K417 |
Note: The participating residues of the ACE2-RBD complex within 4 Å radial range of its interaction with withanone identified through 50 ns molecular dynamic (MD) simulation of aforementioned complex docked with withanone are provided along with the type of interactions involved.
The RMSD of the protein complexes without or with withanone was 2.201 Å, and that of ACE2 alone was 1.476 Å, when final trajectories were compared. The RMSD of the simulated molecule (withanone) was 2.166 Å compared to its starting position. At the end of simulation, it moved slightly toward the ACE2 side at the ACE2-RBD interface (Figure 2C–F), revealed by losing contacts (within 4 Å) with the RBD (Figure 2E and F). On analyzing the ligand interaction, it was found that withanone still forms three H-bonds in the protein interface, but with ACE2 only. ACE2 D30, N33 and Q96 form H-bonds with withanone. In addition, withanone was stabilized by alkyl and van der Waals interactions (Figure 2E and F) (Table 5).
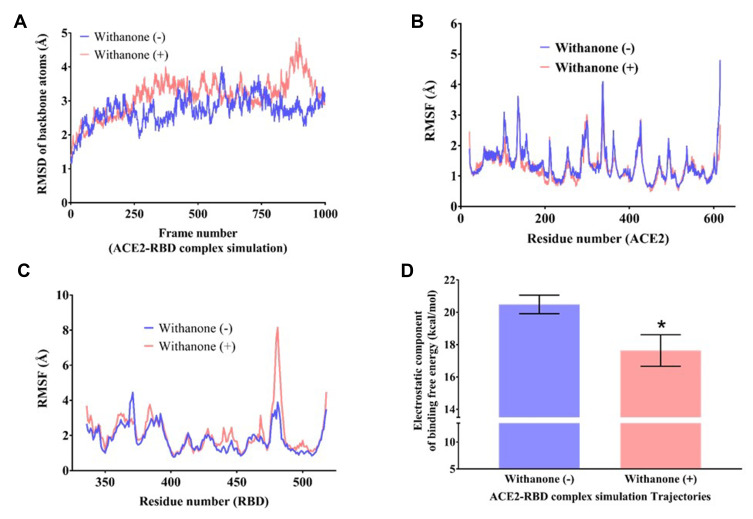
Figure 3.
Flexibility analysis of the ACE2-RBD complex docked with withanone.
Notes: RMSD changes of backbone atoms (C, CA, N) of (A) the complex, (B and C) per residue RMSF (Cα atom only) of ACE2 (C) and RBD (D) during 50 ns simulation time, as observed in the presence and absence of withanone. (D) Comparison of electrostatic component of binding free energies in the ACE2-RBD complexes with and without withanone. Statistical significance was analyzed through Welch’s t-test and represented as * p<0.05.
We observed that two intrachain (at the protein interface on ACE2 side) salt bridge interactions, K31 NZ-E35 OE1/OE2 (2.73 Å/2.71 Å), and one interchain long-range ion pair, K31 NZ-E484 OE2 (5.67 Å), were abolished upon incorporation of withanone (Figure 2G–I). The length of the two N-O bridges between K31 and E35 of ACE2 increased by 1.22 and 1.89 Å (Figure 2G and H). We also observed that the occupancy of these N-O bridges reduced by 13.8% upon incorporation of withanone (Figure 2I). The intrachain salt bridge (K31-E35) was destabilizing, as calculated in the trajectory without withanone (5 kcal/mol). This salt bridge was abolished in the presence of withanone. We detected that the length of the interchain salt bridge (ACE2 D30-RBD K417) was 2.6 Å, which increased by 0.4 Å in the final trajectory. Electrostatic contribution of this salt bridge (D30-K417) in the initial trajectory was −2.52 kcal/mol and in the final trajectory it was −1.32 kcal/mol. This salt bridge was not abolished in our study, however, it was found to be destabilizing at the end of 50 ns MD simulation trajectory.
Flexibility Analysis and Calculation of the Electrostatic Component of Binding Free Energy of ACE2-RBD Complex
For global flexibility analysis, total RMSDs of backbone atoms (C, CA, N) of the residues in the protein was determined in the trajectories without and with withanone (Figure 3A). The data showed a slight increase in RMSD simulated with the withanone. Figure 3B and C show a Cα atom fluctuations of the residues in ACE2 and RBD. Per residue RMSFs of ACE2 trajectories simulated with or without the withanone were almost similar except for some fluctuations in the non-interfacial region (Figure 3B). Per residue RMSF of RBD shows a sharp increase in the beta hairpin region (a part of the interface region aa468 to aa498), in the trajectories simulated with the withanone (Figure 3C). Other signatures were almost similar in the trajectories without or with the withanone.
In MD simulation, total 96 and 94 clusters were obtained upon clustering analysis of the trajectories with and without Withanone, respectively. The electrostatic component of the binding free energies of ACE2-RBD complex were estimated on the post 25 ns representative trajectories of the cluster (51 trajectories, with the ligand; 43 trajectories, without the ligand) using DelPhi v8.4.342 to assess the hypothesis that the proposed phytocompound weakens the interactions between ACE2 and RBD. The ΔΔGel of the complex simulated with the ligand (17.6±0.97 kcal/mol) was decreased by 2.85 kcal/mol compared to the ones without the ligand (20.49±0.57 kcal/mol) (Figure 3D), and was statistically significant at 5% confidence level (Welch’s t-test p-value=0.014). This indicates that the binding of withanone at the interface of ACE2 and RBD weakens their interactions.
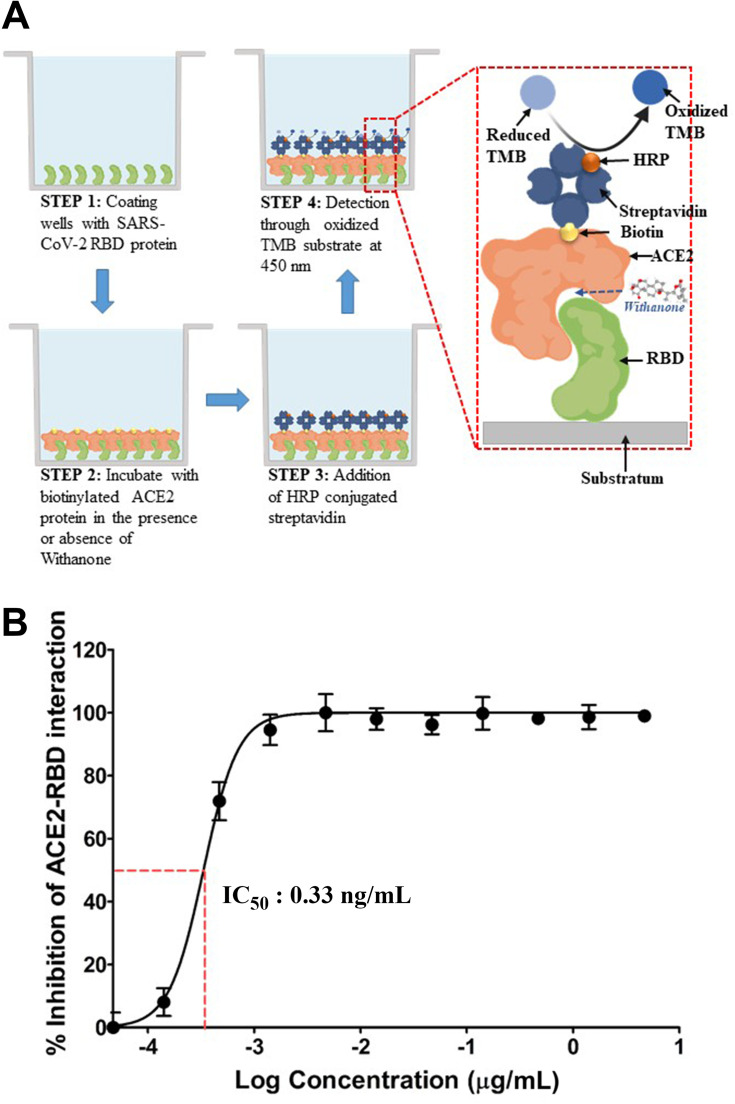
Withanone Inhibits Interaction Between ACE2 and RBD in vitro
Earlier studies have shown that Withanone can target SARS-CoV-2 main protease (Mpro) and TMPRSS2.14 However, experimental validation of that computational prediction has been lacking. In our study, we have checked the effect of commercially available withanone on the interaction between human ACE2 protein and RBD of SARS-CoV-2 viral spike (S) protein, through an ELISA-based biochemical method (Figure 4A). We noted that withanone, dose-dependently, inhibited ACE2-RBD interaction. The dose-response curve exhibited a steep rise over a withanone concentration range of 0.1–1 ng/mL. Concomitantly, the calculated IC50 fell in this range and was found to be 0.33 ng/mL. ~100% inhibition of ACE2-RBD interaction was observed with 4.7 ng/mL of withanone (Figure 4B). From these results, we can conclude that withanone efficiently disrupts the biochemical interaction between the host ACE2 and viral RBD.
Figure 4.
Experimental validation of the computational insights.
Notes: (A) Schematic representation of the experimental procedure employed in evaluating the inhibitory effect of withanone on the biochemical interaction between host ACE2 and viral RBD. Biotinylated ACE2 bound to RBD, immobilized to the substratum, is detectable through HRP-conjugated streptavidin due to oxidation of 3,3ʹ,5,5ʹ-tetramethylbenzidine (TMB). (B) Dose-response curve exhibiting the inhibitory effect of withanone on interaction between human ACE2 and RBD of SARS-CoV-2 S-protein. IC50 is found to be 0.33 ng/mL.
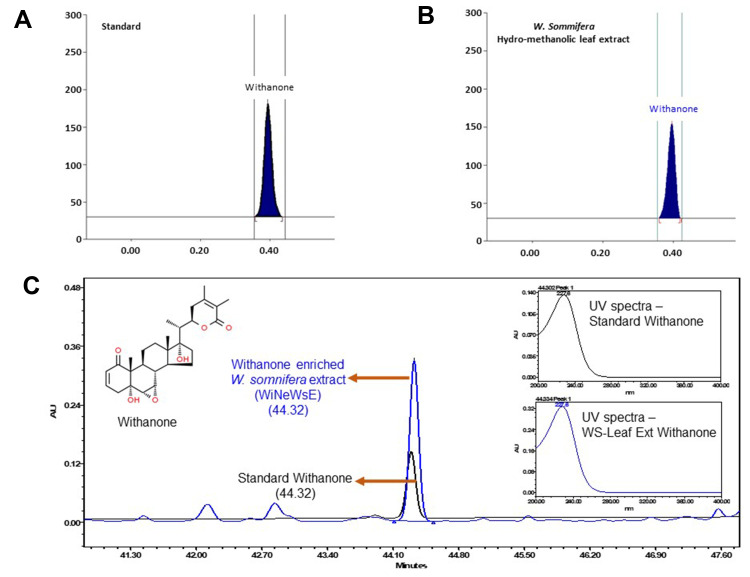
Leaf Extract from W. somnifera is Enriched in Withanone
As mentioned earlier, 200 phytocompounds were screened before settling down with withanone. We intend to validate the in silico findings through detailed in vitro and in vivo experiments for which we have optimized a protocol to prepare hydro-methanolic extract enriched in withanone from fresh leaves of W. somnifera (WiNeWsE). HPTLC analysis showed that WiNeWsE was indeed enriched with withanone (Figure 5A and B). Quantification of HPTLC data showed that 1.29% w/w of WiNeWsE contained withanone. From the compositional analysis of the extract through HPLC (Figure 5C) we found that 8.93 μg/mg of withanone was present in WiNeWsE. These observations confirmed the feasibility of preparing W. somnifera leaf extract enriched with only withanone. We further validated the antiviral efficacy of WiNeWsE in a humanized zebrafish model of SARS-CoV-2.
Figure 5.
Compositional analysis of extract prepared from leaves of W. somnifera.
Notes: (A and B) HPTLC spectra of withanone and W. somnifera hydro-methanolic leaf extract both showing peaks at 230 nm. (C) Overlap chromatograms of standard mix (in black) and hydro-methanolic extract of W. somnifera leaves (in blue) showing the peak for withanone eluted at 227 nm. Insets show the individual peaks of standard withanone and that present in W. somnifera leaf extracts.
In vivo Assessment of Antiviral Efficacy of WiNeWsE in Humanized Zebrafish Model Induced to Reproduce Human Pathological Reactions to SARS-CoV-2
Establishment of Humanized Zebrafish Model and Induction of SARS-CoV-2 Pathological Milieu
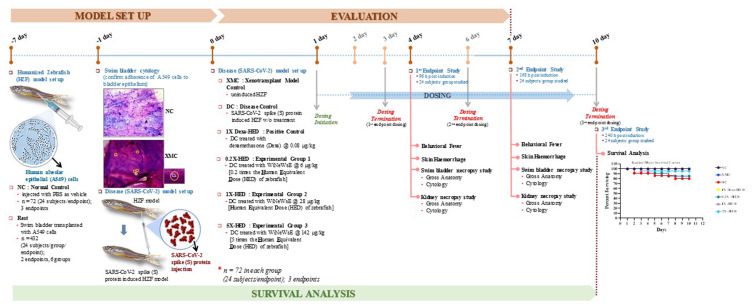
The in vivo study comprised of screening the antiviral efficacy of WiNeWsE and assessing its effect on survivability. Accordingly, three endpoints were included in the study plan. The first two were for screening the antiviral efficacy after 3 and 6 days of treatment, while, the last endpoint was to assess the survivability after 10 days of treatment. Posterior lobes of swim bladders of one-year-old adult zebrafish were transplanted with human alveolar epithelial A549 cells to develop the xenotransplant model (XMC). The A549 cells took seven days to adhere to the bladder epithelium and was confirmed through histological staining. Thereafter, in order to reproduce the human pathological reactions to SARS-CoV-2 in these XMCs, they were injected with recombinant S-protein. Dosing with positive control, dexamethasone, and WiNeWsE at human equivalent doses was administered for 3, 6, and 10 days. On days 4 and 7, designated groups of zebrafish were sacrificed to conduct the endpoint studies that included assessment of behavioral fever, skin hemorrhages, swim bladder, and kidney necropsy through gross anatomical and histopathological studies. Survival was analyzed through Kaplan–Meier survival curve. The study consisted of seven groups, namely, NC, XMC, DC, 1X-Dexa-HED, 0.2X-HED, 1X-HED, and 5X-HED. The details about these groups are provided in Table 3. Briefly, NC consisted of ordinary adult zebrafish, XMC had xenotransplanted zebrafish, also referred as humanized zebrafish (HZF), DC is the disease control group, in which the HZF subjects received injections of SARS-CoV-2 recombinant S-protein, 1X-Dexa-HED group subjects were essentially DCs receiving dexamethasone treatment, and serves as a positive control, 0.2X-HED, 1X-HED and 5X-HED are the three experimental groups that received WiNeWsE 0.2 times less than human equivalent dose, human equivalent dose and five times more than human equivalent dose, respectively. The entire experimental plan is depicted in Figure 6 together with the observation of Kaplan–Meier survival analysis. NC and XMC showed 100% survival, whereas, the survivability was reduced to 80% in DC group. Treatment with dexamethasone restored the survivability to 100%. 1X-HED and 0.2X-HED groups, respectively, receiving human equivalent dose of WiNeWsE (28 µg/kg body weight) and lesser than HED (6 µg/kg body weight), experienced only slightly better survival (84.8%) than the DC group. However, the survival was found to be 95.2% in the 5X-HED group which was administered with 142 µg/kg body weight of WiNeWsE.
Figure 6.
Experimental plan of the in vivo study conducted in xenotransplanted humanized zebrafish model.
Notes: Overall timeline of in vivo experiment has been schematically shown along with the major study sttif with corresponding time points. The study included two screenings (at days 4 and 7) and one survival assay (until day 10) conducted in parallel. The establishment and subsequent, cytological confirmation of the xenotransplanted humanized zebrafish model (HZF) took 7 days from the day of transplantation. Human alveolar epithelial (A549) cells were intramuscularly injected into the posterior lobe of the swim bladder of zebrafish, incubated for 7 days, and adherence of the injected human cells to the fish swim bladder epithelium was confirmed through gross cytology of the swim bladder. A group of HZF subjects were maintained throughout the experiment as xenotransplant model control (XMC). Likewise, a group of zebrafish without any xenotransplantation was taken as normal control (NC). Successive to the model confirmation, recombinant SARS-CoV-2 protein was injected at the site of xenotransplantation to establish the disease model. A group of HZF subjects injected with viral S-protein without any treatment was included as disease control (DC). Three experimental groups of DC subjects, namely, 0.2X-HED, 1X-HED and 5X-HED, respectively, received 6, 28 and 142 µg/kg of withanone enriched W. somnifera extract (WiNeWsE) for 3 (for 1st endpoint study), 6 (for 2nd endpoint study) and 10 (for 3rd endpoint study) days. The group of DC subjects (1X-Dexa-HED) receiving 0.08 µg/kg of dexamethasone for same period as experimental groups was included as a positive control. Doses used in all treatment are shown in Table 3. The screening endpoint studies included monitoring of behavioral fever, skin hemorrhage, and swim bladder and kidney necropsy studies. Survival assay endpoint study included Kaplan–Meier analysis, represented as survival curve.
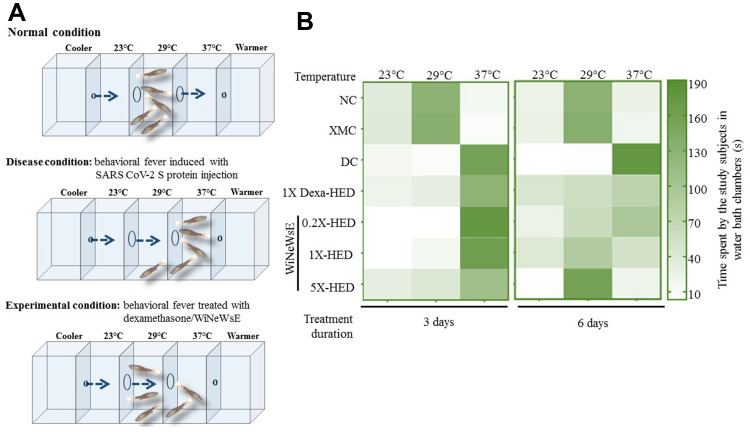
WiNeWsE Treatment Rescued Zebrafish from Behavioral Fever
The zebrafish elicits many immunological reactions that resemble the ones found in humans and therefore, is considered a more preferable model for immunity research, particularly that pertaining to viral infections.23–25,45–48 Zebrafish, being an ectotherm, exhibits behavioral fever as an adaptive immunity, that is, increase in their body temperature coaxes them to migrate to an environment at an elevated temperature. Therapeutics that can bring back the body temperature to normal will induce their migration back to the environment at physiological temperature (Figure 7A). An individual study subject from each group was placed in the 29°C water chamber and given sufficient time to migrate to chambers at 23°C and 37°C. Subsequently, durations of their stay in a particular chamber were monitored for three minutes. The recorded durations were plotted as a heat map; longer durations being represented as darker shades of the chosen green color palate (Figure 7B). Normal zebrafish from the NC group and HZFs from the XMC group dwelt in the chamber at 29°C in the case of both 3 and 6-day treatment durations. Zebrafish belonging to the DC group which were injected with SARS-CoV-2 recombinant S-protein migrated to the chamber at 37°C and spent the entire period monitored there, clearly, indicating that these study subjects were suffering from fever. This was observed for both 3 and 6-day treatments. With dexamethasone treatment for 6 days, the fish started spending almost equal duration in 29°C and 37°C chambers suggesting that dexamethasone could rescue behavioral fever with prolonged administration. But, its effect was not very obvious with 3-day treatment. This observation was more pronounced after 6 days of treatment. The fish treated with 6 µg/kg and 28 µg/kg body weight of WiNeWsE for 3 days did not show any amelioration in behavioral fever as they spent most of the monitored duration in the chamber at 37°C. However, after receiving 142 µg/kg of WiNeWsE for three days, the fish in the 5X-HED started spending more time at 29°C compared to those from groups receiving lower doses of WiNeWsE. The effect of WiNeWsE treatment became more prominent after 6 days of treatment and was clear across all the doses administered, although, the highest dose (142 µg/kg body weight) exhibited the best outcome, as was evident from the subjects in the 5X-HED group spending the entire duration of monitoring at 29°C. Body weight doses of 6 µg/kg and 28 µg/kg of WiNeWsE for 6 days could also ameliorate behavioral fever, but the effect was to a lesser extent compared to the highest dose. Taken together, these observations revealed that WiNeWsE was capable of eliminating one of the manifestations of adaptive immune response mounted against the recombinant S-protein of SARS-CoV-2 injected into these fish.
Figure 7.
WiNeWsE rescues zebrafish from SARS-CoV-2 S-protein induced behavioral fever.
Notes: (A) Pictorial depiction of the chambered water bath maintained at different temperatures with free access between these chambers for zebrafish subjects to migrate to conducive temperature zones. The anterior-most and posterior-most chambers are connected to cooling and heating facilities respectively to maintain the temperature gradient along the experimental chambers. In addition, these flanking chambers are designed to prevent access to fish from experimental chambers. As predictive outcome of the treatments, the picture depicts three scenarios. The first scenario pertains to normal condition where the subjects, could be humanized or not, but have not been induced with viral S-protein and therefore, populate the chamber at 29°C, matching the body temperature of the zebrafish. In the second scenario, where the HZF subjects, injected with recombinant viral S-protein, experience a rise in their body temperature and thus, migrate to the chamber at 37°C. This phenomenon in which the zebrafish with higher body temperature than their physiological one migrate to a warmer surrounding matching their body temperature is called behavioral fever. In the third scenario, the fevered subjects are treated either with dexamethasone or WiNeWsE, and therefore, are seen migrating back to the chamber at 29°C. The time spent by each treated or untreated subject in a specified chamber is recorded and represented as a color-coded matrix, called a heat map. Increase in time spent is depicted as corresponding darkening of the chosen green color palate. (B) Heat map representing the effect of treatments on behavioral fever.
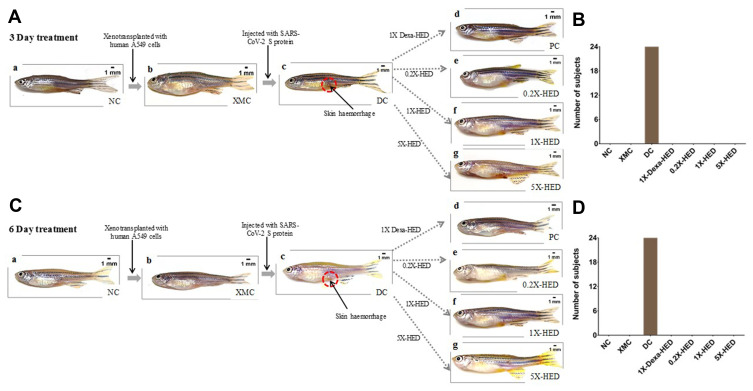
WiNeWsE Prevented SARS-CoV-2 S-protein Induced Skin Hemorrhaging in Zebrafish
Coronavirus infections, including that caused by the current SARS-CoV-2, are associated with blood coagulation.49,50 This phenomenon is closely reproduced in zebrafish injected with viral S-protein as visible skin hemorrhage owing to blood clotting under it. Like behavioral fever, this characteristic manifestation of adaptive immune response of humanized zebrafish to SARS-CoV-2 induction was monitored subsequent to treatments with dexamethasone and WiNeWsE for 3 and 6 days (Figure 8). To begin with, it was ensured that xenotransplantation with human cells did not evoke such outcomes in the zebrafish (Figure 8A and C, compare panels a and b for absence of skin hemorrhage). Distinct red spots pertaining to skin hemorrhage were visible in humanized zebrafish injected with SARS-CoV-2 recombinant S-protein, suggesting successful recapitulation of blood coagulation observed in humans with severe SARS-CoV-2 infections (Figure 8A and C, panel c). In fact, every individual subject of the DC group was found to develop such hemorrhagic spots (Figure 8B and D). SARS-CoV-2 S-protein-induced fish treated with dexamethasone and different doses of WiNeWsE did not develop these spots. Both 3 and 6-day treatments demonstrated relief from skin hemorrhaging in these groups (Figure 8A and B, panels d-g). Thus, WiNeWsE was able to erase yet another manifestation of adaptive immune reaction in zebrafish against SARS-CoV-2.
Figure 8.
WiNeWsE ameliorates SARS-CoV-2 S-protein induced skin hemorrhages in zebrafish.
Notes: (A and C) Pictorial flowchart depicting the establishment of HZF model (XMC) in normal zebrafish population (NC) and subsequent induction of skin hemorrhage (encircled with open red circles) with intramuscular injections of recombinant viral S-protein to develop the disease control (DC), and monitoring the effects of various treatments, namely, 0.08 µg/kg bodyweight of dexamethasone (1X-Dexa-HED) or different concentrations of WiNeWsE (6 µg/kg for 0.2X-HED, 28 µg/kg for 1X-HED and 142 µg/kg for 5X-HED) after 3 (A) and 6 (B) days. (B and D) Number of zebrafish subjects developing skin hemorrhages were counted and plotted separately for 3 (C) and 6-day (D) cohorts, for a quantitative understanding of the effect of treatments on this disease outcome.
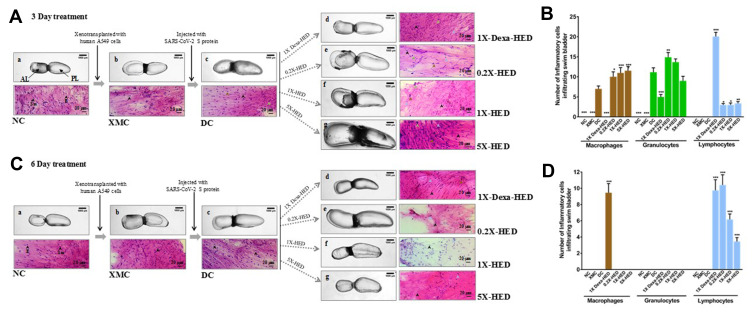
WiNeWsE Modulates Immune Response to SARS-CoV-2 Induction in Zebrafish Swim Bladder
Zebrafishes were humanized by seeding A549 cells on the posterior lobes of their swim bladders. The viral induction was also done at this site. Therefore, signs for anticipated morphological alterations and immune responses were studied in the swim bladders as gross anatomical visualization and tissue histopathology, respectively, for both 3 and 6-day treatment regimes. Zebrafish swim bladder is a gas filled sac comprising of an anterior lobe (AL) and a posterior lobe (PL) connected by a wide ductus communicans (Figure 9, panel a). The tunica externa envelops the inner gas gland made out of mesothelium comprising of epithelial cells and musculature formed by smooth muscle cells. The pneumatic duct connects the posterior lobe to the esophagus. Both lobes function under tight coordination with each other. Zebrafish in the NC group which were neither xenotransplanted nor induced with viral protein, had morphologically healthy swim bladders in terms of size, shape and color with continuous tunica externa and mesothelial lining of smooth muscles. Morphology of swim bladders in humanized zebrafish from XMC groups, from both 3 and 6-day treatment cohorts, was similar to the normal ones, thereby, confirming that xenotransplantation with human cells did not have any undesirable effects (Figure 9A and C, panel b). But, upon induction with SARS-CoV-2 recombinant S-protein, anterior lobes were inflated indicating edema, more prominent in the 3-day treatment cohort (Figure 9A and C, panel c). Dexamethasone treated 1X-Dexa-HED group subjects from 3-day treatment cohort had slightly inflated swim bladders (Figure 9A, panel d). The observed morphological anomaly in the 1X-Dexa-HED group from 6-day treatment cohort was more pronounced as enlarged anterior lobe and narrowed posterior lobe indicating edematous gas gland (Figure 9C, panel d). Subjects in the 0.2X-HED group from 3-day treatment cohort receiving 6 µg/kg body weight of WiNeWsE had more inflated lobes, both anterior and posterior when compared to their corresponding NC counterparts. This effect was more obvious in the 6-day treatment cohort (Figure 9A and C, panel e). A similar alteration occurred in the fish in the 1X-HED group from 3-day treatment cohort receiving 28 µg/kg body weight of WiNeWsE. The same group from 6-day cohort showed shrunken tunica externa and overall reduction of sizes of the bladder lobes (Figure 9A and C, panel f). Zebrafish in the 5X-HED group that received 142 µg/kg body weight of WiNeWsE for 3 days showed visibly significant enlargement of bladder lobes with anomalous tunica externa (Figure 9A, panel g). The identical group from the 6-day treatment cohort had overall shrunken swim bladders (Figure 9C, panel g). These observed morphological alterations, despite the fact that WiNeWsE treatment supported more than 95% survival (Kaplan–Meier curve in Figure 6) among the subjects, raise curiosity regarding the underlying possible reasons. Zebrafish, being a teleost, harbor the features of both innate and adaptive immune responses.25,45–48 Evoked immune reactions quite often manifest themselves as transient anomalies in the anatomy of affected organs. Our histological study revealed that indeed, these responses were triggered after inducing the humanized zebrafish with SARS-CoV-2 recombinant S-protein. Histological sections of the swim bladders of fish from the DC group showed infiltration of macrophages and granulocytes (Figure 9, panel DC; Figure 9B and D). The xenotransplanted zebrafish did not exhibit any such infiltration of inflammatory cells, thereby providing a clean control background for noise-free comparison of the observations from the DC and treatment groups (Figure 9A and C, compare panels NC and XMC; Figure 9B and D). In the 3-day WiNeWsE treatment cohort, number of macrophages found to infiltrate the swim bladders were comparable and, in some cases, more than that found in the DC group. Dexamethasone treatment for three days, however, brought down the number of infiltrating macrophages significantly compared to the DC group. A similar observation was noted for infiltrating granulocytes in the case of dexamethasone treatment. Although, infiltrating granulocytes were found to be trending towards reduction, but the observation was not statistically significant when compared to the DC group. Infiltrating lymphocytes were not observed in the case of the DC group for both 3 and 6-day treatments. However, in fish treated with dexamethasone over both these periods, significantly large numbers of lymphocytes were found to infiltrate when compared to the DC group (p<0.001). With WiNeWsE treatment for 3 days, the number of lymphocytes infiltrating the swim bladder were greatly reduced. However, a 6-day treatment with WiNeWsE showed an increase in infiltrating lymphocytes at lower doses (Figure 9A and C, panels 1X-Dexa-HED, 0.2X-HED, 1X-HED, 5X-HED; Figure 9B and D). Knowing that macrophages and granulocytes are part of innate immune response and lymphocytes that of an adaptive response, the overall trend surfacing from the quantification of these cells showed that in the initial phase (3-day treatment), WiNeWsE tends to reduce the innate immune response with complete success through 6-day treatment. This is implicative of the capability of WiNeWsE in preventing the innate immune response from going into overdrive. Likewise, WiNeWsE sustains the adaptive immune response in the form of lymphocytes suggesting that it might be helpful in refraining the adaptive immune response from becoming dysfunctional. Interestingly, SARS-CoV-2 infection affects both adaptive and innate immune responses to sustain itself. The inflammatory innate immune response is sent into overdrive and the adaptive immune response is made dysfunctional.51 So, WiNeWsE might be a solution to the complex immunomodulation associated with SARS-CoV-2 infection.
Figure 9.
WiNeWsE attenuates viral S-protein induced inflammation in zebrafish swim bladder.
Notes: (A and C) Effects of xenotransplantation, subsequent induction of HZF subjects with recombinant viral S-protein and successive different ameliorative therapeutic ministrations on the morphology of swim bladders and infiltration of inflammatory cells therein, as monitored through histopathology, are depicted through pictorial flowcharts for 3- (A) and 6-day (C) treatments. (B and D) Infiltration of different inflammatory cells [macrophages (mud brown arrowheads), granulocytes (green arrowheads) and lymphocytes (blue arrowheads)] in response to xenotransplantation, disease model development and subsequent, treatments were quantified and graphically, represented separately for 3- (B) and 6-day (D) treatments. Data plotted are mean ±SEM of counts obtained from 24 individual subjects in each group. Statistical significance of the means of different groups was analyzed through one-way ANOVA followed by Dunnett’s post hoc test and marked as *p<0.05, **p<0.01 and ***p-<0.001, when compared to disease control (DC).
Abbreviations: Sm, smooth muscle nuclei; E, epithelial nuclei.
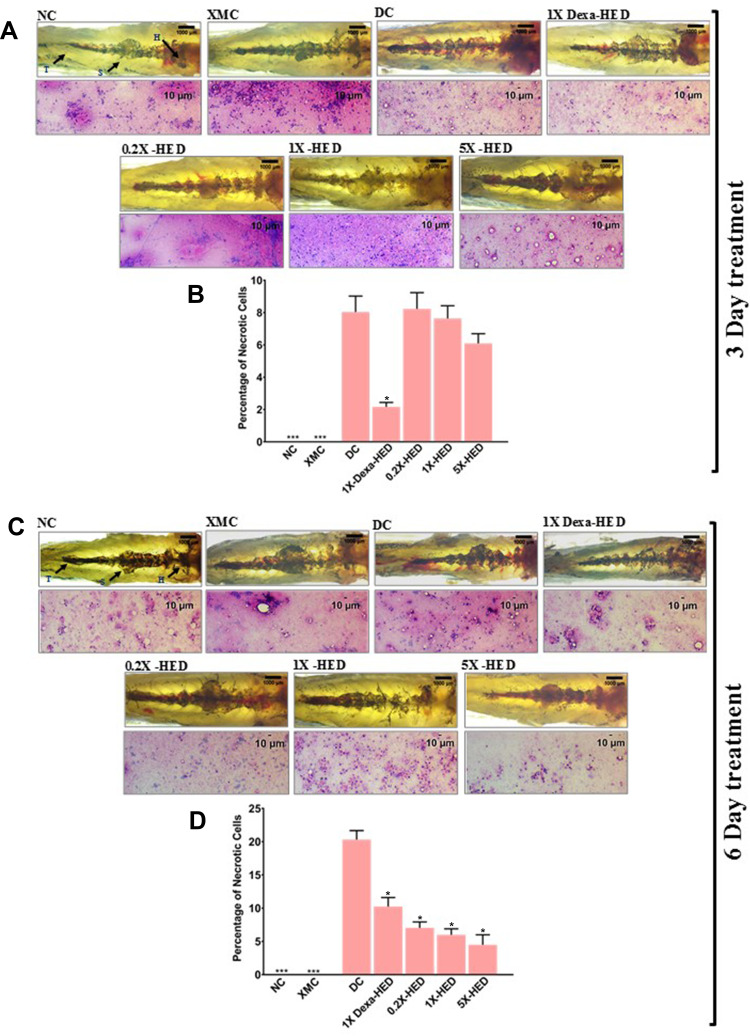
WiNeWsE Attenuated SARS-CoV-2 S-Protein-induced Kidney Necrosis
SARS-CoV-2 infection affects kidneys in humans.52 Therefore, the effect of WiNeWsE treatment on fish kidneys was assessed through gross anatomical and cytological studies. The zebrafish kidney is attached to the parenchymal layer of the dorsal wall of the viscera. It is divided into head (H), trunk or saddle (S), and the tail (T) and extends along the anterioposterior length of the body (Figure 10A and B, panel NC). After carefully dissecting out the kidney from the euthanized subject, the whole organ was observed microscopically for anatomical anomalies. In addition, alterations at the tissue level owing to any of the experimental procedure, particularly, administered treatments, was evaluated through cytology. Effects on the kidney were monitored after 3 and 6 days of treatments. Zebrafish from the NC groups of both the treatment cohorts, had defined internal arrangements of packed mesonephric nephrons in the head zone of the kidney with glomerular tufts followed by proximal and distal tubule segments. Highly pigmented mesonephrons were distributed from head to tail of the kidney (Figure 10A and B, panel NC). Similar arrangements were observed in the XMC groups with humanized zebrafish from both the treatment cohorts, thus, confirming that xenotransplantation did not affect the kidneys adversely (Figure 10A and B, panel XMC). However, humanized zebrafish induced with SARS-CoV-2 S-protein showed loss of tubular segments and vascular degeneration observed as dark staining bodies indicating renal necrosis (Figure 10A and B, panel DC). Percentage of necrotic cells were also high in this group compared to NC and XMC groups, with normal and humanized zebrafish, respectively (Figure 10B and D). The kidney anatomy after 3 days of dexamethasone treatment showed an arborized network with packed glomerular tufts and tubular segments indicating normal renal architecture (Figure 10A, panel 1X-Dexa-HED). However, prolonged dexamethasone treatment for six days, showed a disorganized kidney network with irregular glomerular tufts and melanocytic pigmentation indicating structural anomaly (Figure 10B, panel 1X-Dexa-HED). However, anatomical anomaly was not reflected as renal necrosis. In both 3 and 6-day treatment cohorts, dexamethasone significantly reduced the necrosis (p<0.001) (Figure 10B and D). For all the concentrations of WiNeWsE, in cases of both the treatment cohorts, renal anatomy was found to be anomalous (Figure 10A and C, panels 0.2X-HED, 1X-HED and 5X-HED). However, a 3-day WiNeWsE treatment started reducing the renal necrosis which was statistically significant in comparison to the DC group after 6-day treatments (Figure 10B and D). Structural anomaly of the kidney could be a manifestation of the SARS-CoV-2 infection at organ level, when the fish are left untreated. The fact that dexamethasone and WiNeWsE treatments were found to attenuate renal necrosis within the span of 6 days, is indicative of the anatomical rescuing of the kidneys as well, provided longer monitoring is done. Taken together, these observations showed that WiNeWsE is capable of preventing deleterious effects of SARS-CoV-2 infection that percolate to secondary organs, which were not infected directly. In a clinical context, this might be an indication of its ability to prevent multiorgan failure.
Figure 10.
WiNeWsE assuages detrimental effects of SARS-CoV-2 S-protein on zebrafish kidney.
Notes: (A and C) Effect of experimental model set-up and subsequent therapeutic treatments on zebrafish kidneys was monitored through gross anatomical study and corresponding histopathology for necrosis and degeneration after 3 (A) and 6 (C) days. Zebrafish kidney is anatomically divided into head (H), saddle (S) and tail (T). (B and D) Necrotic cells were quantified and separately, plotted as percentage for 3 (B) and 6 (D) days of treatments. Data plotted are mean ± SEM of counts obtained from 24 individual subjects in each group. Statistical significance of the means of different groups was analyzed through one-way ANOVA followed by Dunnett’s post hoc test and marked as *p<0.001, when the comparison was done with the disease control (DC).
Discussion
The S-protein of SARS-CoV interacts with its cellular receptor, ACE2 via receptor binding motif (RBM; aa 424–494) present in its RBD. Eighteen residues of the receptor make contact with 14 residues of the viral S-protein, mainly through hydrophilic interactions.53,54
Molecular dynamic (MD) simulation of the ACE2-RBD complex was performed using NAMD, in the presence and absence of the ligand molecule (withanone). Withanone moved slightly toward the ACE2 side of the protein interface, as revealed by losing H-bond and contact residues with the RBD upon MD simulation. Initially, docking showed 14 residues within 4 Å of withanone. Out of these, seven residues were from ACE2, and seven were from RBD (Table 5). Upon 50 ns MD simulation, R403, E406, and K417 from RBD were within 4 Å of withanone, and rest of the residues were from ACE2 (Figure 2C and D). Furthermore, we explored the ionic interaction at the binding interface of the ACE2 receptor and RBD complex of SARS-CoV-2. The intrachain salt bridge K31-E35 (at the interface on the ACE2 side) was completely abolished (Figure 2G and H). Similarly, the interchain long-range ion pair K31-E484 (between ACE2 and RBD) was also completely abolished, with a large swing of the residue K31. Percent occupancy of the intrachain salt bridge K31-E35 was also decreased in the simulation trajectories with withanone, compared to the trajectories without withanone (Figure 2I). Similarly, the percent occupancy of the interchain long-range ion pair K31-E484 was decreased dramatically. Electrostatic component of binding free energy of the ACE2-RBD complex was decreased in the simulation trajectories with withanone (Figure 3D). In simple words, this means withanone prevented stabilization of the interaction between ACE2 and RBD.
Protein surfaces have many hydrophilic residues, and salt bridges present in the surface play an important role in protein–protein association or binding.55 Hence the protein interface (binding interface) is generally more hydrophilic than the protein interiors. Electrostatic interactions are thought to be more important for protein–protein or protein-ligand interactions than protein folding.56 No wonder the interfacial salt bridges, being the major contributors to the electrostatic interactions between proteins, play a central role in binding events. Generally, the structures of the proteins do not change significantly upon complex formation, but some conformational rearrangements are observed, and most of these are due to side chain movements.57 Geometrical complementarity and stability in energetics are the two key driving factors for protein binding while hydrophobic effect, hydrogen bonds, and salt bridges are the key players in energetic. A salt bridge can provide favorable free energy to the binding,56 alternatively, an isolated charge without forming a salt bridge, when buried in the protein interface, could substantially destabilize binding through a sizeable desolvation cost. We calculated the energetics of the salt bridges at the interface of ACE2 and RBD. The salt bridge K31-E35 and ion-pair K31-E484 were abolished. These results clearly indicated that the incorporation of withanone is likely to decrease protein complex stability by disrupting the factors that build up the favorable binding energetics.
Global RMSD changes obtained from MD simulation of the ACE2-RBD complex with and without withanone provided a measure of flexibility (Figure 3A). Per residue RMSF was calculated to assess the flexibility at residue level (Figure 3B and C). Except for some minor fluctuations, a noticeable change was observed in the beta turn in the interfacial region in RBD (aa468-aa498). Generally, residues in the molecular recognition region have higher fluctuations. Reduced electrostatic component of the binding free energies of the ACE2-RBD complex suggests that withanone binding at the interface of ACE2 and RBD weakens the interaction between the latter two. Taken together, these observations reveal that withanone has the potential to disrupt the interaction between human ACE2 receptor and RBD of SARS-CoV-2 S-protein.
Computational observations on withanone targeting SARS-CoV-2 main protease (Mpro) and host transmembrane TMPRSS2 have been reported.13,14 Likewise, withaferin A is also reported to bind TMPRSS2 and GRP78.14,15 While targeting virus-specific factors is encouraging, the same cannot be said for host factors, since that would have associated side effects. Moreover, withaferin A is cytotoxic and this prospect has been widely explored for treating cancer. SARS-CoV-2 sends the immune response into overdrive. It is possible that such an altered physiological condition might not be able to handle cytotoxic assault and lead to other complications. According to the study by Kumar et al,13 binding score of withanone to Mpro is −4.42 kcal/mol, whereas we obtained a binding score of −9.4 kcal/mol when withanone was docked into the interacting interface of ACE2 and RBD. Kumar et al13 have justified the theoretical prospects of functional efficacy of withanone at inhibiting Mpro through a comparison with the binding score (−5.6 kcal/mol) of an already reported inhibitor N3 with Mpro.58,59 Similarly, binding of withanone to TMPRSS2 (−4.30 k/mol) was weaker when compared to withanone binding to the ACE2-RBD complex (−9.4 kcal/mol).14 Employing an identical approach, low binding score in this case was substantiated through comparison with known TMPRSS2 inhibitor camostat mesylate (−5.90 kcal/mol). However, disparity in the binding score calculations between these reported studies and our study, due to the use of different tools, is ruled out as the software programs used are reported to give comparable outputs,60 clearly reflecting that withanone shows a tighter binding with the ACE2-RBD complex than with either Mpro or TMPRSS2. Using an in silico peptide binding approach, Hamza et al attempted identifying the SARS-CoV-2 peptides which could be potential targets for the phytochemicals, hydroxychloroquine, kaempferol and anthraquinone from Moringa oleifera.61 The authors translated the SARS-CoV-2 genome in silico (using EMBOSS TRANSEQ server) into peptides which were then used for binding studies with the above mentioned phytochemicals. The maximum binding energies obtained from AutoDock Vina for hydroxychloroquine is −5.1 kcal/mol, kaempferol is −6.2 kcal/mol, and anthraquinone is −6 kcal/mol,61 which are again less than those observed in the case of withanone and the ACE2-RBD complex (−9.4 kcal/mol) in this study. Thus, it is possible that withanone might act specifically during viral–host interaction and not target the host factors, sparing the body from side effects. Moreover, the computational findings reported in these studies were not experimentally validated. Although, apparently, our study is on similar lines, however we have demonstrated that withanone targets the ACE2-RBD complex, and have validated our observations from computational studies by wet-lab experiments.
For experimental validation of our computational insights, overexpressed truncated S-protein of SARS-CoV-2 expressing only RBD was used while the human ACE2 (hACE2) used was full length. ELISA wells were coated with viral RBD into which biotinylated hACE2 was added along with different concentrations of withanone. An inhibitor of the ACE2-RBD interaction provided with the kit was used as the positive control, while the reaction without any inhibitor was taken as the negative control. Inhibition of ACE2-RBD interaction was calculated with respect to the negative control assuming the interaction to be 100%. Interaction was detected using HRP-conjugated streptavidin against biotinylated ACE2. Corroborating with the in silico observation, we found that in the presence of withanone, ACE2 binding with RBD was less efficient and that the effect was positively correlated to the concentration of withanone used. Withanone efficiently inhibited interaction of ACE2 with RBD (IC50=0.33 ng/mL) and this inhibition was found to be dose-dependent.
We have also developed a protocol to prepare leaf extracts from W. somnifera enriched in withanone (WiNeWsE). The reason behind choosing extract over isolated phytocompound is to ensure minimum modulation in the chemical milieu of withanone when it is being extracted from the plant. Co-extracting other phytocompound/s along with the one required ensures minimum deviation in the functional set-up of the compound from that present within the plant. Such an arrangement is likely to modulate the activity of the phytocompound in vitro or in vivo in a way similar to what actually happens in nature. To prove this point, we validated the antiviral activity of WiNeWsE in a humanized zebrafish xenotransplanted with human alveolar epithelial A549 cells on the posterior lobe of swim bladder and induced with SARS-CoV-2 recombinant S-protein to elicit pathological reactions similar to those observed in humans.
The zebrafish is a useful and attractive model for infectious disease and immunity research and is considered a refinement over the use of mammalian infection models.23 The zebrafish model allows the investigation of specific immune system components at various stages of immunologic development, and extensive molecular, genetic, and imaging tools are available for this species. Zebrafish exhibit both the innate and adaptive arms of the immune system, including leukocyte populations, inflammatory mediators, and signaling molecules that are similar to those of the mammalian immune system.24 Many components of the complement system have been identified in zebrafish. Similarly, homologs of many mammalian cytokines have been identified in zebrafish, including interleukin 1β,25 type I45 and type II46 interferons, tumor necrosis factor,25,47 and several interleukins.48 Therefore, zebrafish have also been used as models of mammalian viral infection. Zebrafish models for mammalian viral infections have the advantages of live imaging, whole-organism histopathology, and immunohistochemistry. The first zebrafish infection studies using a mammalian virus demonstrated dose-dependent infection of the adult zebrafish nervous system with herpes simplex virus type 1 (Herpesviridae) and its attenuation in response to antiviral acyclovir.23,62 This indicated that the zebrafish model of human viral diseases can be used to test the efficacies of antivirals. Observations from the studies conducted in humanized zebrafish model of SARS-CoV-2 pathology were compelling regarding the efficacy of WiNeWsE in relieving fever, managing the immune reactions to therapeutic advantage, and preventing spreading of pathological responses to secondary organs.
Thousands of chemical molecules, already existing as approved drugs in the market for other diseases, are being virtually screened for this approach.63 With this strategy of drug repurposing, the hunt for a COVID-19 cure can be significantly expedited by foregoing phases I and II of the clinical trials (the phases dedicated towards establishing safety and dose of a new drug).64 The time constraint pertaining to the regulatory procedure required for launching a new drug in the market might limit the scope of exhaustive exploration for the most suitable cure. However, cures based on phytochemicals follow naturopathy, thus eliminating the demerit of ensuing toxicity in the body. Nevertheless, preliminary studies are crucial to establish the potency of such a treatment. Hence, the significance of this study which serves as a prefatory for a phytochemical-based intervention against SARS-CoV-2 entry in the host cell.
Observations made in our present study, sheds light on our next course of action, a situation arguably better than repurposing approved drugs. To appreciate the essence of this study, we have to understand the potential demerits of repurposing of drugs approved for other diseases. While it is absolutely logical to adapt such an approach in the face of the current compounding crisis, yet, we are unaware of the side effects such repurposed drugs might bring along, particularly when time is too limited to explore all these details, which otherwise would have been the most obvious course of action. Without any intention of downplaying the contemporary attempts made at finding a cure for COVID-19, we would like to reiterate that if a cure from nature herself is showing promise, it is worthwhile by all means to give it recognition. This study highlights the importance of natural origin phytochemicals in controlling SARS-CoV-2 entry into host cells, and provides an attractive and alternative means for the management of COVID-19 infection.
Conclusion
Through this study we have demonstrated that withanone has the potential to inhibit the interaction between hACE2 receptor and RBD of SARS-CoV-2 S -protein. Since this interaction is crucial for viral entry into the host cell, thus, withanone being able to prohibit it implicates a virus-specific therapeutic option. More importantly, we have provided experimental evidence demonstrating the inhibitory effect of withanone against the biochemical interaction between hACE2 and viral RBD proteins. In addition, we have also shown that extracts enriched with withanone can be prepared from the leaves of W. somnifera. This report is the first of its kind amidst several others in which withaferin A unavoidably co-extracts with withanone. Last, but not the least, the potential of this extract as an antiviral was validated in a preclinical set-up using humanized zebrafish as a model that reliably reproduced the human pathological responses to SARS-CoV-2. In conclusion, this study lays the foundation for developing withanone in to a treatment for COVID-19 as an entry inhibitor against SARS-CoV-2 coronavirus.
Acknowledgments
We appreciate the zebrafish test facilities and experimentations at our CRO partner, Pentagrit Labs, Chennai, India. We thank Mr Sudeep Verma and Ms Meenu Tomer for their help with HPLC; Dr Pratima Singh, Dr Preeti Raj, Mr Ajeet Chauhan, Mr Shoor Singh and Mr Arun Raturi for their help with visual graphics. We extend our gratitude to Ms Priyanka Kandpal, Mr Tarun Rajput, Mr Gagan Kumar and Mr Lalit Mohan for their swift administrative support.
Funding Statement
This research received no external funding. This presented work has been conducted using internal research funds from the Patanjali Research Foundation Trust, Hardwar, India.
Abbreviations
ACE2, angiotensin-converting enzyme-2; COVID-19, coronavirus disease 2019; MERS, Middle East respiratory syndrome; MD, molecular dynamics; NAMD, nanoscale molecular dynamics; PBE, Poisson–Boltzmann equation; RBD, receptor binding domain; RBM, receptor binding motif; SARS-CoV, severe acute respiratory syndrome-coronavirus; SARS-CoV-2, severe acute respiratory syndrome-coronavirus 2; S-protein, spike protein; VMD, visual molecular dynamics; RMSD, root mean square deviation; WiNeWsE, withanone enriched Withania somnifera extract.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethics Approval
Animal ethics guidelines from the Committee for the Purpose of Control and Supervision of Experiments (CPCSEA), Government of India, were followed to conduct the zebrafish (Danio rerio) experiments. Established zebrafish protocols were duly approved by the Institutional Animal Ethics Committee (IAEC study number 223/Go062020/IAEC).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J Med Virol. 2020;92(4):401–402. doi: 10.1002/jmv.25678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorbalenya AE, Baker SC, Baric RS, et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ni W, Yang X, Yang D, et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit Care. 2020;24(1):1–10. doi: 10.1186/s13054-020-03120-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. Solidarity trial consortium. Repurposed antiviral drugs for COVID-19 – interim WHO SOLIDARITY trial results. medRxiv. 2020;(October 15). doi: 10.1101/2020.10.15.20209817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bupp K, Roth MJ. Alteration and analyses of viral entry with library-derived peptides. Adv Virus Res. 2005;65(5):147–172. doi: 10.1016/S0065-3527(05)65005-1 [DOI] [PubMed] [Google Scholar]
- 6.Dau B, Holodniy M. Novel targets for antiretroviral therapy: clinical progress to date. Drugs. 2009;69(1):31–50. doi: 10.2165/00003495-200969010-00003 [DOI] [PubMed] [Google Scholar]
- 7.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. doi: 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- 8.Sangwan RS, Chaurasiya ND, Misra LN, et al. Phytochemical variability in commercial herbal products and preparations of Withania somnifera (Ashwagandha). Curr Sci. 2004;86(3):461–465. [Google Scholar]
- 9.Zaman W, Saqib S, Ullah F, Ayaz A, Ye J. COVID-19: phylogenetic approaches may help in finding resources for natural cure. Phytother Res. 2020;34(11):2783–2785. doi: 10.1002/ptr.6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kambizi L, Goosen BM, Taylor MB, Afolayan AJ. Anti-viral effects of aqueous extracts of Aloe ferox and Withania somnifera on herpes simplex virus type 1 in cell culture. S Afr J Sci. 2007;103:(October):359–362. [Google Scholar]
- 11.Cai Z, Zhang G, Tang B, Liu Y, Fu X, Zhang X. Promising anti-influenza properties of active constituent of Withania somnifera ayurvedic herb in targeting neuraminidase of H1N1 influenza: computational study. Cell Biochem Biophys. 2015;72(3):727–739. doi: 10.1007/s12013-015-0524-9 [DOI] [PubMed] [Google Scholar]
- 12.Balkrishna A, Pokhrel S, Singh J, Varshney A. Withanone from Withania somnifera may inhibit novel coronavirus (COVID-19) entry by disrupting interactions between viral s-protein receptor binding domain and host ACE2 receptor. Res Sq. 2020. doi: 10.21203/rs.3.rs-17806/v1 [DOI] [Google Scholar]
- 13.Kumar V, Dhanjal JK, Kaul SC, Wadhwa R, Sundar D. Withanone and caffeic acid phenethyl ester are predicted to interact with main protease (Mpro) of SARS-CoV-2 and inhibit its activity. J Biomol Struct Dyn. 2020. doi: 10.1080/07391102.2020.1772108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar V, Dhanjal JK, Bhargava P, et al. Withanone and withaferin-A are predicted to interact with transmembrane protease serine 2 (TMPRSS2) and block entry of SARS-CoV-2 into cells. J Biomol Struct Dyn. 2020. doi: 10.1080/07391102.2020.1775704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sudeep H. Molecular docking analysis of Withaferin A from Withania somnifera with the glucose regulated protein 78 (GRP78) in comparison with the COVID-19 main protease. Bioinformation. 2020;16(5):411–417. doi: 10.6026/97320630016411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Case JB, Rothlauf PW, Chen RE, et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. SSRN Electron J. 2020. doi: 10.2139/ssrn.3606354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batlle D, Wysocki J, Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin Sci. 2020;134(5):543–545. doi: 10.1042/CS20200163 [DOI] [PubMed] [Google Scholar]
- 18.Inal JM. Decoy ACE2-expressing extracellular vesicles that competitively bind SARS-CoV-2 as a possible COVID-19 therapy. Clin Sci. 2020;134(12):1301–1304. doi: 10.1042/CS20200623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter JD, Hutter CAJ, Zimmermann I, et al. Sybodies targeting the SARS-CoV-2 receptor-binding domain. bioRxiv. 2020. doi: 10.1101/2020.04.16.045419 [DOI] [Google Scholar]
- 20.Kaul SC, Ishida Y, Tamura K, et al. Novel methods to generate active ingredients-enriched Ashwagandha leaves and extracts. PLoS One. 2016;11(12):1–15. doi: 10.1371/journal.pone.0166945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dutta R, Khalil R, Green R, Mohapatra SS, Mohapatra S. Withania somnifera (Ashwagandha) and withaferin a: potential in integrative oncology. Int J Mol Sci. 2019;20(21):21. doi: 10.3390/ijms20215310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pires N, Gota V, Gulia A, Hingorani L, Agarwal M, Puri A. Safety and pharmacokinetics of Withaferin-A in advanced stage high grade osteosarcoma: a Phase I trial. J Ayurveda Integr Med. 2020;11(1):68–72. doi: 10.1016/j.jaim.2018.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgos JS, Ripoll-gomez J, Alfaro JM, Sastre I, Valdivieso F. Zebrafish as a new model for herpes simplex virus type 1 infection. Zebrafish. 2008;5(4):323–333. doi: 10.1089/zeb.2008.0552 [DOI] [PubMed] [Google Scholar]
- 24.Crim MJ, Riley LK. Viral diseases in zebrafish: what is known and unknown. ILAR J. 2012;53(2):135–143. doi: 10.1093/ilar.53.2.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pressley ME, Phelan PE, Witten PE, Mellon MT, Kim CH. Pathogenesis and inflammatory response to Edwardsiella tarda infection in the zebrafish. Dev Comp Immunol. 2005;29(6):501–513. doi: 10.1016/j.dci.2004.10.007 [DOI] [PubMed] [Google Scholar]
- 26.Das CN, Uniyal GC, Lal P, et al. Analysis of withanolides in root and leaf of Withania somnifera by HPLC with photodiode array and evaporative light scattering detection. Phytochem Anal. 2008;19(2):148–154. doi: 10.1002/pca.1029 [DOI] [PubMed] [Google Scholar]
- 27.Pettersen EF, Goddard TD, Huang CC, et al. UCSF chimera - a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 28.Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356(6364):83–85. doi: 10.1038/356083a0 [DOI] [PubMed] [Google Scholar]
- 29.Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–1519. doi: 10.1002/pro.5560020916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26(2):283–291. doi: 10.1107/s0021889892009944 [DOI] [Google Scholar]
- 31.Lovell SC, Davis IW, Adrendall WB, et al. Structure validation by C alpha geom basic local alignment search tool. J Mol Biol. 2003;50(August2002):437–450. doi: 10.1002/prot.10286 [DOI] [Google Scholar]
- 32.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open babel: an open chemical toolbox - 1758-2946-3-33.pdf. J Cheminform. 2011;3(33):1–14. doi: 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukunishi Y, Nakamura H. Prediction of ligand-binding sites of proteins by molecular docking calculation for a random ligand library. Protein Sci. 2011;20(1):95–106. doi: 10.1002/pro.540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghersi D, Sanchez R. Improving accuracy and efficiency of blind protein-ligand docking by focusing on predicted binding sites. Proteins. 2009;74(2):417–424. doi: 10.1002/prot.22154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. In: Hempel JE, Al E, editors. Chemical Biology. Methods in Molecular Biology. Vol. 1263. New York, NY: Humana Press; 2015: 243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 36.Dassault. Dassault Systèmes BIOVIA, Discovery Studio 2017 R2 Client, Release 2017. San Diego: Dassault Systèmes; 2017. [Google Scholar]
- 37.DeLano WL. Pymol: an open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr. 2002;40:82–92. [Google Scholar]
- 38.Humphrey W, Dalke A, Theoretical KS. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/j.carbon.2017.07.012 [DOI] [PubMed] [Google Scholar]
- 39.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29(11):1859–1865. doi: 10.1002/jcc [DOI] [PubMed] [Google Scholar]
- 40.Phillips JC, Braun R, Wang W, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hendsch ZS, Tidor B. Do salt bridges stabilize proteins? A continuum electrostatic analysis. Protein Sci. 1994;3(2):211–226. doi: 10.1002/pro.5560030206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L, Li C, Sarkar S, et al. DelPhi: a comprehensive suite for DelPhi software and associated resources. BMC Biophys. 2012;5(1):9. doi: 10.1186/2046-1682-5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sitkoff D, Sharp KA, Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem. 1994;98(7):1978–1988. doi: 10.1021/j100058a043 [DOI] [Google Scholar]
- 44.Fogolari F, Brigo A, Molinari H. The Poisson-Boltzmann equation for biomolecular electrostatics: a tool for structural biology. J Mol Recognit. 2002;15(6):377–392. doi: 10.1002/jmr.577 [DOI] [PubMed] [Google Scholar]
- 45.Altmann SM, Mellon MT, Distel DL, Kim CH. Molecular and functional analysis of an interferon gene from the zebrafish, danio rerio. J Virol. 2003;77(3):1992–2002. doi: 10.1128/JVI.77.3.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Igawa D, Sakai M, Savan R. An unexpected discovery of two interferon gamma-like genes along with interleukin (IL) −22 and −26 from teleost: IL-22 and −26 genes have been described for the first time outside mammals. Mol Immunol. 2006;43(7):999–1009. doi: 10.1016/j.molimm.2005.05.009 [DOI] [PubMed] [Google Scholar]
- 47.Praveen K, Evans DL, Jaso-friedmann L. Constitutive expression of tumor necrosis factor-alpha in cytotoxic cells of teleosts and its role in regulation of cell-mediated cytotoxicity. Mol Endocrinol. 2006;43:279–291. doi: 10.1016/j.molimm.2005.01.012 [DOI] [PubMed] [Google Scholar]
- 48.Sullivan C, Kim CH. Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol. 2008;25(4):341–350. doi: 10.1016/j.fsi.2008.05.005 [DOI] [PubMed] [Google Scholar]
- 49.Vinayagam S, Sattu K. SARS-CoV-2 and coagulation disorders in different organs. Life Sci. 2020;260:118431. doi: 10.1016/j.lfs.2020.118431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giannis D, Ziogas IA, Gianni P. Coagulation disorders in coronavirus infected patients: COVID-19, SARS- CoV-1, MERS-CoV and lessons from the past. J Clin Virol. 2020;127:104362. doi: 10.1016/j.jcv.2020.104362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Catanzaro M, Fagiani F, Racchi M, Corsini E, Govoni S, Lanni C. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct Target Ther. 2020;5(84):1–10. doi: 10.1038/s41392-020-0191-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soleimani M. Acute kidney injury in SARS-CoV-2 infection: direct effect of virus on kidney proximal tubule cells. Int J Mol Sci. 2020;21(3275):2–6. doi: 10.3390/ijms21093275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prabakaran P, Gan J, Feng Y, et al. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J Biol Chem. 2006;281(23):15829–15836. doi: 10.1074/jbc.M600697200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li F, Li W, Farzan M, Harrison SC. Structural biology: structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480 [DOI] [PubMed] [Google Scholar]
- 55.Barlow DJ, Thornton JM. The distribution of charged groups in proteins. Biopolymers. 1986;25(9):1717–1733. doi: 10.1002/bip.360250913 [DOI] [PubMed] [Google Scholar]
- 56.Xu D, Lin SL, Nussinov R. Protein binding versus protein folding: the role of hydrophilic bridges in protein associations. J Mol Biol. 1997;265(1):68–84. doi: 10.1006/jmbi.1996.0712 [DOI] [PubMed] [Google Scholar]
- 57.Norel R, Wolfson HJ, Nussinov R. Small molecule recognition: solid angles surface representation and molecular shape complementarity. Comb Chem High Throughput Screen. 1999;2(4):223–236. [PubMed] [Google Scholar]
- 58.Jin Z, Du X, Xu Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- 59.Yang H, Xie W, Xue X, et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:10. doi: 10.1371/journal.pbio.0030324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castro-Alvarez A, Costa AM, Vilarrasa J. The performance of several docking programs at reproducing protein-macrolide-like crystal structures. Molecules. 2017;22(1):1. doi: 10.3390/molecules22010136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamza M, Ali A, Khan S, et al. nCOV-19 peptides mass fingerprinting identification, binding, and blocking of inhibitors flavonoids and anthraquinone of Moringa oleifera and hydroxychloroquine. J Biomol Struct Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1778534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hubbard S, Darmani NA, Thrush GR, Dey D, Burnham L. Zebrafish-encoded 3-O-sulfotransferase-3 isoform mediates herpes simplex virus type 1 entry and spread. Zebrafish. 2010;7(2):181–187. doi: 10.1089/zeb.2009.0621 [DOI] [PubMed] [Google Scholar]
- 63.Wu C, Liu Y, Yang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gautret P, Lagier J-C, Parola P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56(1):105949. doi: 10.1016/j.ijantimicag.2020.105949 [DOI] [PMC free article] [PubMed] [Google Scholar]