SUMMARY
Optogenetics has revolutionized neuroscience in small laboratory animals, but its impact on animal models more closely related to humans, such as non-human primates (NHP), has been mixed. To make evidence-based decisions in primate optogenetics, the scientific community would benefit from a centralized database listing all attempts, successful and unsuccessful, at using optogenetics in the primate brain. We contacted members of the community to ask for their contribution to an open science initiative. As of this writing, 45 laboratories around the world contributed more than 1000 injection experiments, including precise details about their methods and outcomes. Of those entries, more than half had not been published. The resource is free for everyone to consult, and to contribute to, on the Open Science Framework website. Here we review some of the insights from this initial release of the database and discuss methodological considerations to improve the success of optogenetic experiments in NHP.
INTRODUCTION
Optogenetics is a revolutionary technique in neuroscience. By making neurons sensitive to light, the technique allows unprecedented control over neuronal activity in living biological systems (Boyden et al., 2005; Deisseroth, 2010). The use of optogenetics in small animal models such as mice and flies has boomed over the last decade, with hundreds of publications citing the technique every year (Figure 1). Major advances in our understanding of memory (Xu and Südhof, 2013), sleep (Tsunematsu et al., 2011), fear (Liu et al., 2012), decision making (Friedman et al., 2015), social behavior (Gunaydin et al., 2014), and other processes have resulted from the application of optogenetics to small animal models. The neuroscience community hopes to learn much more over the coming decades as the technique continues to fuel discovery.
Figure 1. Number of published papers using optogenetics.
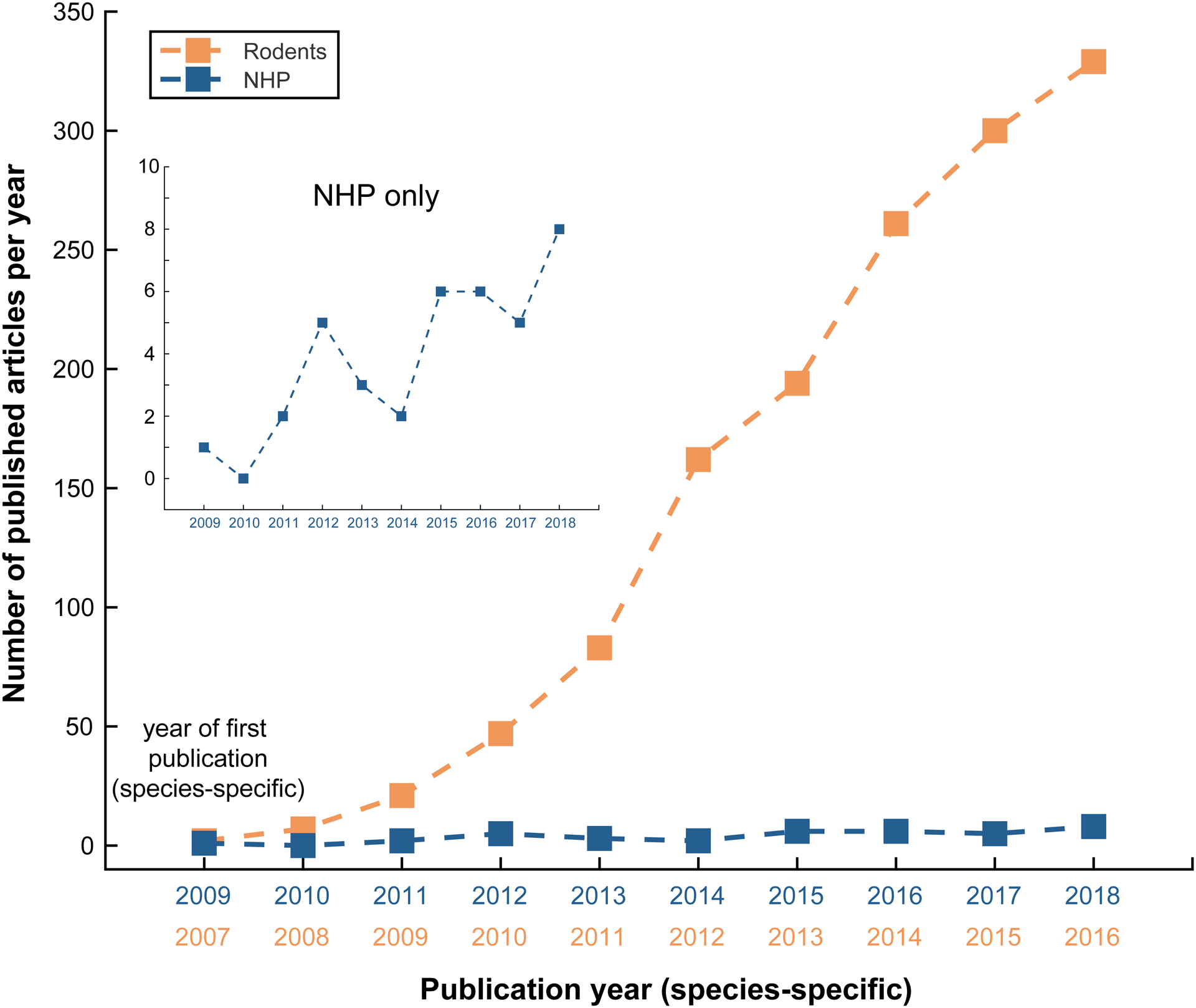
Number of new published articles per year using optogenetics in rodents (red) and in NHP (blue) since year of first species-specific publication. Review articles are excluded. Inset shows the same data but includes only NHP studies.
Optogenetics has the potential to become an important clinical tool for human patients affected by neurological and psychiatric disorders (Chow and Boyden, 2013; Deisseroth, 2012). Before the technique can be applied to humans, however, it is prudent to first demonstrate its safety and efficacy in closely related non-human primates (NHP). In addition to facilitating the development of new therapies for humans, NHP optogenetics could trigger a revolution in our fundamental understanding of the primate brain at the cellular and systems levels, similar to the impact on our understanding of neurobiological processes in rodents and flies. This is in part why, in 2009, the first report applying optogenetics to NHP was published (Han et al., 2009). Since then, however, optogenetics has not been adopted as rapidly in the NHP community as it has in smaller laboratory animals (Figure 1; blue line). Although the reasons behind this trend are complex (El-Shamayleh and Horwitz, 2019; Galvan et al., 2017), one can identify technical difficulties associated with the expression of opsin genes and delivery of light in behaviorally-relevant volumes of the primate brain as a significant challenge. Because the optogenetic technique was pioneered in mice, the genetic methods, injection protocols, and light stimulation apparatus were optimized for an animal model with a brain several orders of magnitude smaller than the most widely used NHP brain (i.e. the macaque) and with an immune system that differs in several important respects (Bjornson-Hooper et al., 2019). In addition, the absence of widely available genetically modified primates is another significant reason for the slow adoption of optogenetic techniques in NHP.
For NHP researchers who want to launch optogenetics experiments, there are few guidelines on how to proceed. The choice of viral vectors, genetic promoters, and opsin genes is often based on the limited information available in the methods section of published manuscripts or on casual conversations between scientists who share personal experiences from their laboratories. Although one can refer to the few published studies in NHP (Acker et al., 2016; Afraz et al., 2015; Amita et al., 2020; Andrei et al., 2019; Bohlen et al., 2019; Cavanaugh et al., 2012; Chernov et al., 2018; Dai et al., 2014; De et al., 2020; Diester et al., 2011; Ebina et al., 2019; El-Shamayleh et al., 2017; Fabbrini et al., 2019; Fetsch et al., 2018; Fortuna et al., 2020; Fredericks et al., 2020; Galvan et al., 2016; 2012; Gerits et al., 2012; 2015; Han et al., 2009; 2011; Inoue et al., 2015; Jazayeri et al., 2012; Ju et al., 2018; Khateeb et al., 2019; Klein et al., 2016; Komatsu et al., 2017; Ledochowitsch et al., 2015; Lu et al., 2015; MacDougall et al., 2016; May et al., 2014; McGregor et al., 2020; Mendoza et al., 2017; Nakamichi et al., 2019; Nandy et al., 2019; Nassi et al., 2015; Nurminen et al., 2018; Oguchi et al., 2015; Ohayon et al., 2013; Ozden et al., 2013; Ruiz et al., 2013a; Senova et al., 2018; Shea et al., 2018; Stauffer et al., 2016; Tamura et al., 2012; 2017; Watanabe et al., 2020; Williams et al., 2019; Yazdan-Shahmorad et al., 2016; 2018a; 2018b), there is a sense in the community that many results, especially negative ones, do not get published. Moreover, researchers who want to innovate with new combinations of opsins and vectors are left in the dark as to whether these tests have already been conducted by other members of the community.
To make evidence-based decisions in NHP optogenetics, the NHP user community would benefit from an open and exhaustive list of every attempt made using this technique in NHP, including the results of experiments deemed unsuccessful. This resource would need to include not only details about the viral construct used in experiments, but also details about the injection technique, surgical practice, the post-injection survival period, and the outcome measures used to assess the success of experiments. With sufficient data, this resource could reveal critical factors that influence the success or failure of experiments. With this objective in mind, we reached out to members of the NHP neuroscience community who had published optogenetics results in peer-reviewed journals, presented preliminary findings at conferences, or who were referred by colleagues as using optogenetics in NHP, to ask for their contributions to a new open science initiative. The response from the community was overwhelmingly positive. Here, we present the initiative and review some of the preliminary insights emerging from the database.
RESULTS
Descriptive statistics
We created a list of all known investigators who had attempted optogenetics in NHP around the world and searched for their contact information on personal and institutional websites. Through the published literature and referrals, 66 labs were identified in the USA, Canada, UK, Germany, France, Belgium, Switzerland, China and Japan. Each lab was sent an invitation to share published and unpublished data with the initiative, to which 52 labs responded positively (Figure 2). Of those that responded positively, 45 contributed data and 32 provided unpublished data. As of November 2019, 1042 viral vector injections in NHP were included in the database. Of those, 552 entries were unpublished results, effectively doubling the number of injection cases in the literature. Each entry in the database consists of a single injection of a viral vector. Multiple entries could be generated from the same animal. Injections at different depths along the same needle penetration tract were grouped into a single entry. The database includes experiments conducted in 6 different NHP species (Figure 3A). Although most injection experiments were done in rhesus macaques (71%), many were conducted in cynomolgus macaques (11%), Japanese macaques (8%), marmosets (6%), and squirrel monkeys (3%). The database accepts entries from all NHP species and welcomes new contributions from investigators.
Figure 2. Flow chart of laboratories’ participation in the NHP Optogenetics Open Database.
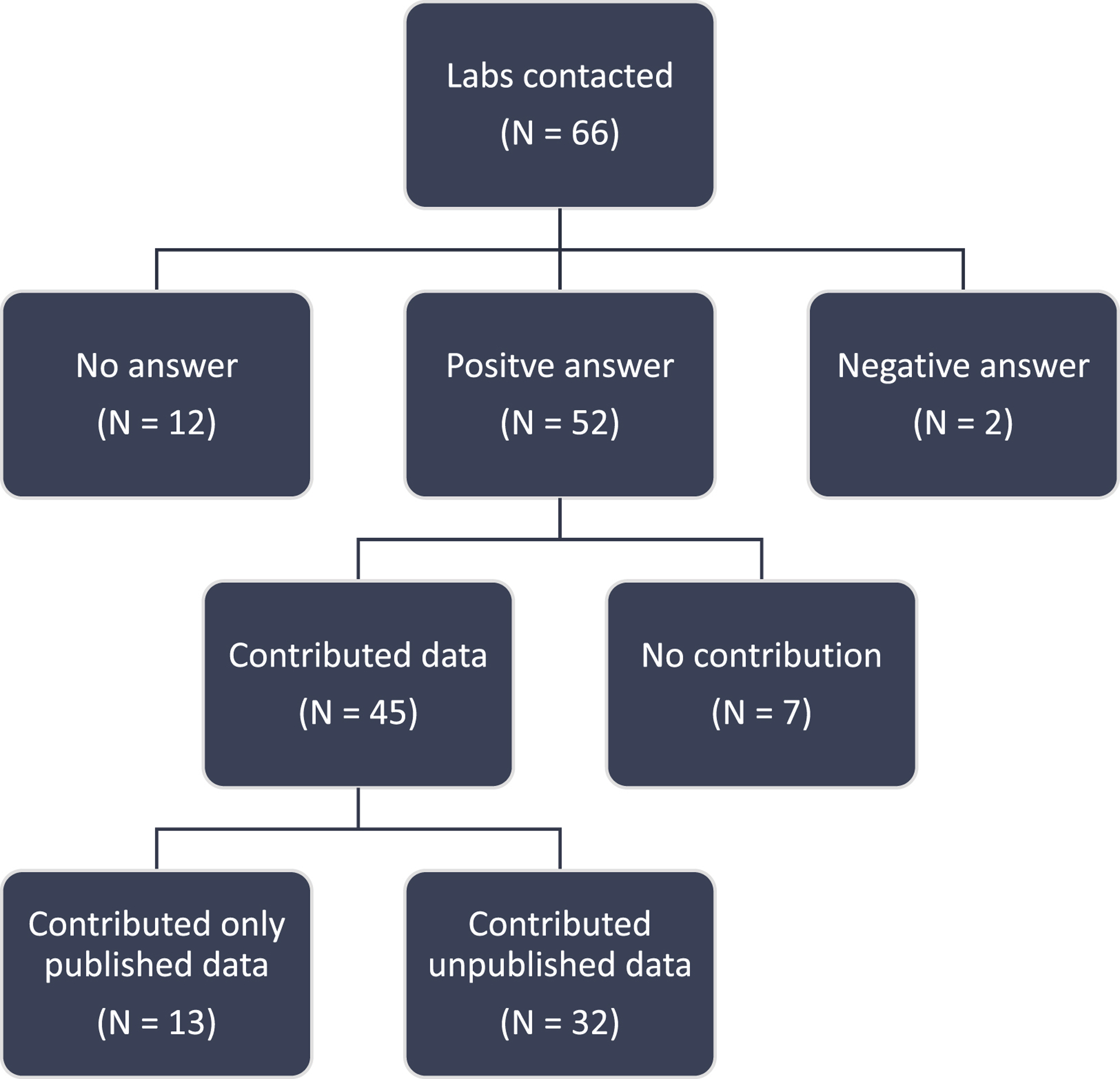
A “No answer” was considered after three repeated failed attempts at contacting the investigator. Overall, the response of the community was overwhelmingly positive.
Figure 3. Popularity of various resources for NHP optogenetics.
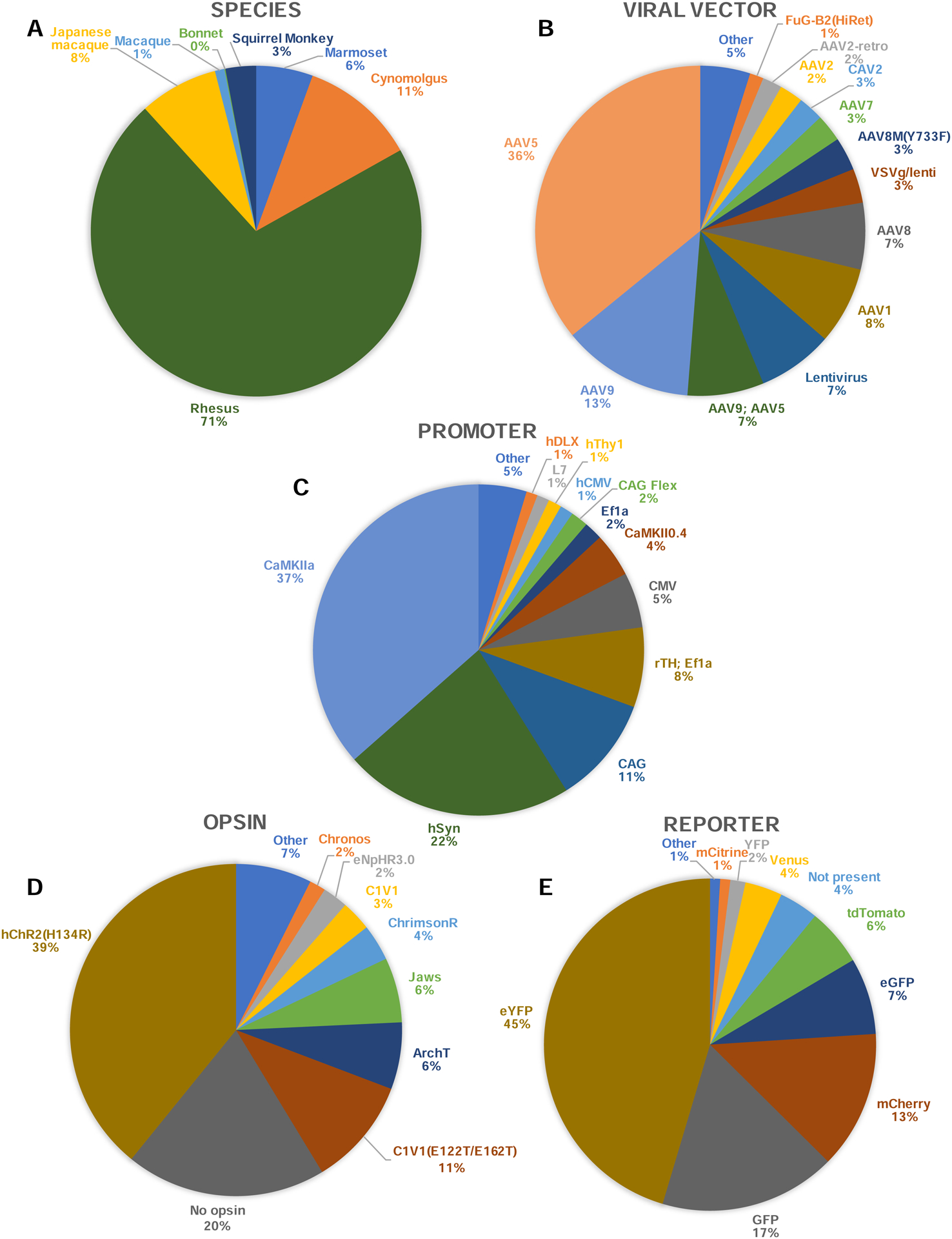
Frequency of each NHP species (A), viral vector (B), genetic promoter (C), opsin gene (D), and reporter protein (E) across the entire database. One entry corresponds to one injection. Variants with less than 10 entries were grouped in the category “Other”. “Macaque” refers to entries where the species of macaque was not specified. “AAV9; AAV5” refers to a mix of both capsids in the same viral solution.
Most injection protocols used an Adeno-Associated Virus (AAV; https://www.addgene.org/guides/aav/) as the viral vector for gene delivery (Figure 3B). The AAV5 serotype (see Note on AAV nomenclature) accounted for 36% of all viral injections, followed by AAV9 (13%). Lentiviruses were used in 7% of cases and the canine adenovirus type 2 (CAV-2) in 3% of cases. The most widely used promoter was the Ca2+/calmodulin-dependent protein kinase II (CaMKIIa, 37%), a promoter that preferentially targets excitatory neurons (Figure 3C). The second and third most used promoters are the human synapsin neuron-specific promoter (hSyn, 22%) and the fused cytomegalovirus (CMV) enhancer, chicken beta-Actin promoter and rabbit beta Globin splice acceptor site (CAG, 11%), which is a ubiquitous promoter expressed in all mammalian cell types. The most widely used opsin was the humanized ChR2 with H134R mutation for optogenetic activation through cation conductance (hChR2(H134R), 39%). Some experiments used inhibitory opsins (25% of experiments), including ArchT, an opsin acting as an outward proton pump, used in 6% of experiments, and Jaws, a halorhodopsin which acts through a chloride pump, also used in 6% of injections. A large number of tests logged in the database (20%) did not include an opsin gene (Figure 3D). These experiments included only a reporter gene (e.g. green fluorescent protein, GFP)to test the efficacy and spread of viral transduction in preparation for a later optogenetic experiment. To visualize the spread and efficiency of transduction in the targeted brain areas, most experiments included a fluorescent reporter gene in their cassette (Figure 3E). By far the most widely used reporter gene in NHP is the enhanced yellow fluorescent protein (eYFP), which accounted for 45% of tests. The GFP (17%) and mCherry (13%) reporters follow in second and third places.
Box 1 - AAV nomenclature clarifications.
The nomenclature used for adeno-associated viruses can be confusing. For example, when an investigator reports using an AAV5, is s/he referring to a serotype AVV5/5, with both rep and cap genes from AAV5, or is s/he referring to an AAV2/5, with the rep genes of AAV2 and the cap genes of AAV5? Following consultations with the Penn Vector Core, the UNC Vector Core, and the Stanford Vector Core, it was confirmed that all Cores use the AAV2 rep genes for recombinant AAV production and that only the capsid genes are swapped to create a specific serotype. In other words, an AAV1 is really an AAV2/1, an AAV2 is really an AAV2/2, an AAV9 is really and AAV2/9, and so on. Therefore, in the database, AAV5 and AAV2/5 were grouped together into a single category of AAV (i.e. AAV5). Same for AAV2/1 and AAV1 (i.e. AAV1), etc. In this article, when entries list “AAV2; AAV5”, it refers to two independent viral vectors (AAV2 & AAV5) mixed in the same viral solution before injection.
Outcome measures
Investigators included outcome measures to describe the success or failure of an optogenetic experiment in NHP. These outcomes were grouped into three broad categories: 1) Anatomy; referring to results involving the examination of tissue using histology (in vivo observation of epifluorescence is also included in this category); 2) Physiology; referring to assessments of the functionality of the opsin and its effect on neural activity, whether using electrophysiology, imaging, or other means; 3) Behavior; where behavioral variables, ranging from simple muscle contractions to complex decision making, were assessed in response to optogenetic stimulation. Contributing investigators had to include one answer for each of these outcome categories for each test they included in the database.
Because results could vary widely across experiments, investigators were free to answer in an unstructured way about the outcome of each test. For example, written answers like “No expression”, “Strong neural response”, or “Weak behavioral effects” were common. Investigators were also encouraged to provide additional details about the interpretation of each result in the “Notes” section beside the outcomes of each test. For the purpose of quantitative analyses, each written outcome was translated into one of four categories of effect strength: “Strong effect”, “Weak or mixed effect”, “No effect”, or “Not tested”. For example, written answers like “strong neural response” were considered “strong effect” for the “Physiological” outcome category, and “weak behavioral effects” translated to a “weak or mixed effect” in the “Behavior” outcome category. This translation depended on the investigators including details about the strength of the effect observed, which was not mandatory. In the absence of moderators like “low”, “weak”, “strong”, “robust”, the outcome was assumed to be a strong one (e.g. “GFP expression in pyramidal cells” would be considered a “strong effect” for “Anatomy”). For this reason, the readers should be careful not to over-interpret the difference between “strong” and “weak or mixed” effects, which can be influenced by the level of details offered by the investigators about their results.
Based on this categorization, we looked at the overall success of optogenetics experiments across all NHP species and methods. Across all 1042 vector injections, 55% tested the physiological effects of the opsin on neural activity. Of those that tested physiology, 69% reported a strong effect on neural activity, 16% reported a weak or mixed effect, and 15% no effect at all (Figure 4A). Notably, 45% of all experiments did not conduct a physiological examination of the opsin function, favoring other outcome measures such as histology/anatomy. The investigators conducted anatomical analyses of opsin expression in 68% of all experiments. Of those, they reported that 78% of injections led to a strong expression of the opsin/reporter gene in targeted neurons (Figure 4B). Thirty-two percent (32%) of all tests did not conduct histological analyses, often because the animals were still alive and involved in experiments at the time of contribution. In terms of behavioral outcomes, the majority of injections were not tested (80%). Of those that were tested, 55% found a strong effect of the optogenetic stimulation, while 15% and 30% found weak or no effect, respectively (Figure 4C).
Figure 4. Outcome of optogenetics experiments in NHP.
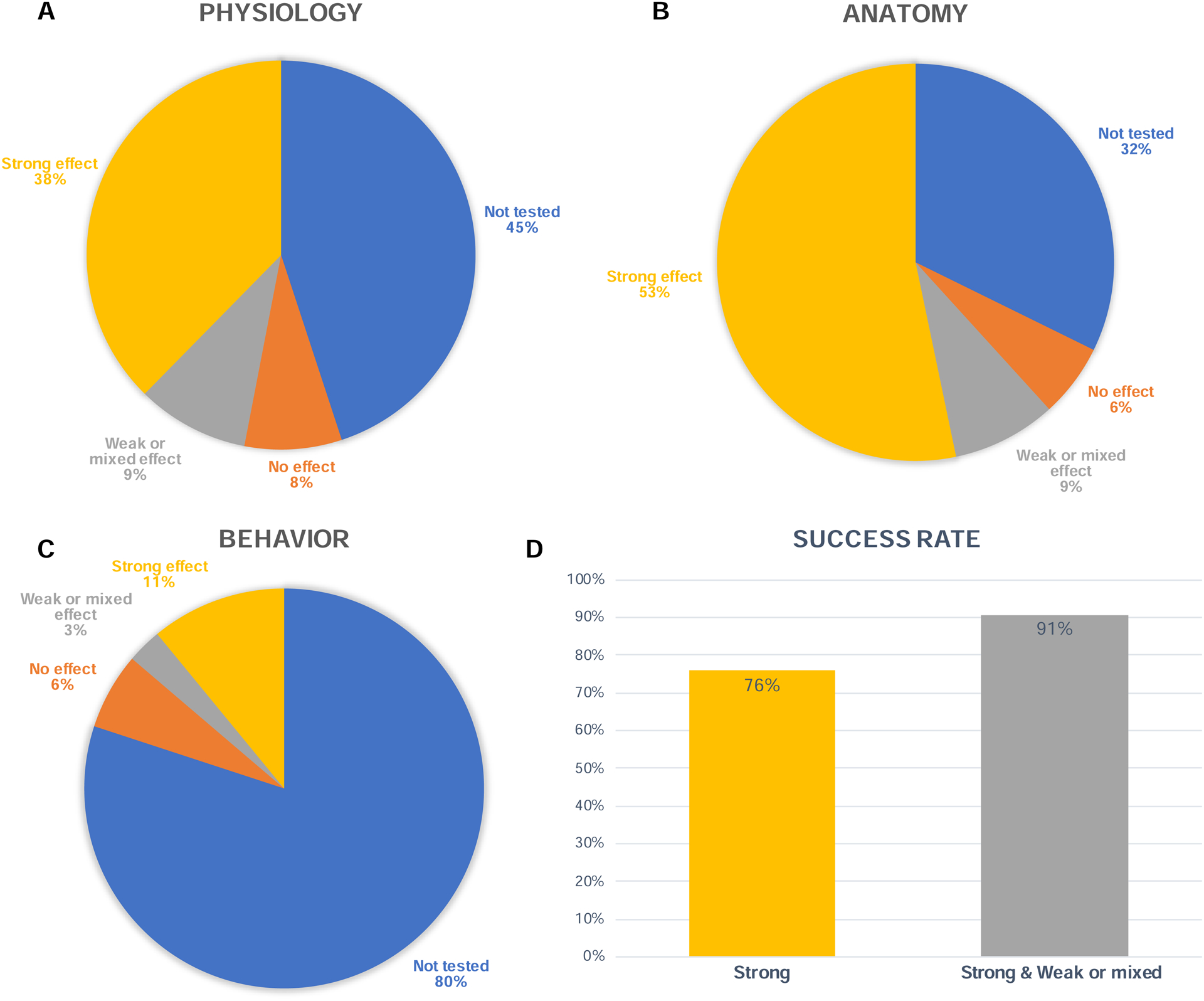
A) Proportion of injections that led to a strong physiological effect, a weak or mixed effect, or no effect. A weak or mixed effect was defined as per the investigators’ comments reporting either weak, unreliable, or mixed physiological response. White numbers represent absolute number of injection tests. B & C) Same as in A, but for anatomical and behavioral analyses, respectively. D) Left: Proportion of injections that led to at least one strong outcome across all three possible outcome modalities (physiology, anatomy, or behavior). Right: This bar also includes injections that led to at least one weak or mixed outcome (or better), across all three outcome modalities.
We sought to estimate the success rate of all injection experiments reported in the database. Because different experiments may seek different outcome measures (e.g. one experiment may measure physiology but not anatomy, and another one the reverse), we needed to choose a criterion for “success” that would be flexible enough to encompass all database entries. For the sake of analysis, the success of an optogenetic experiment in NHP was defined by the presence of at least one outcome measure demonstrating a strong effect, whether anatomical, physiological or behavioral. For example, an experiment that only tested physiology and obtained the expected positive results could be considered a successful injection experiment. We quantified the number of experiments that led to at least one strong effect, or at least one weak or mixed effect across the three possible outcomes (Figure 4D). Seventy-six percent (76%) of all experiments reported at least one strong effect, and 91% reported at least one weak or mixed effect, or a strong effect, across all outcome categories. As noted above, most of those positive outcomes came from anatomical or physiological observations since most experiments did not test behavioral effects.
Outcomes per viral construct
To identify important factors that may influence the success of optogenetic experiments in NHP, we examined the success rate of experiments as a function of specific methods and tools. For this analysis, “success” is defined (as above) as having at least one strong effect among the three possible outcome measures (anatomy, physiology, and behavior), which was the case in 76% of the tests reported (Figure 4D). We looked at each type of viral vector, promoter, opsin gene, and reporter protein and calculated the percentage of tests that were successful when using each variant. Since some variants were infrequently used in the database, we flagged those with fewer than 20 data entries or those tested in fewer than five different animals (dotted bars in Figure 5). We warn readers to be careful when interpreting the success rate of variants that have a low sample size (dotted bars). We hope that future additions to the database will improve the reliability of these interpretations. Importantly, all reported success rates are univariate analyses that need to be interpreted carefully in consideration of all other parameters that might influence the success of an injection experiment. Please refer directly to the database for a full list of the >30 parameters listed for each injection experiment (e.g. species, brain region, injection volume, immunosuppressive drug, etc.).
Figure 5. Success rate of experiments as a function of optogenetics resources used.
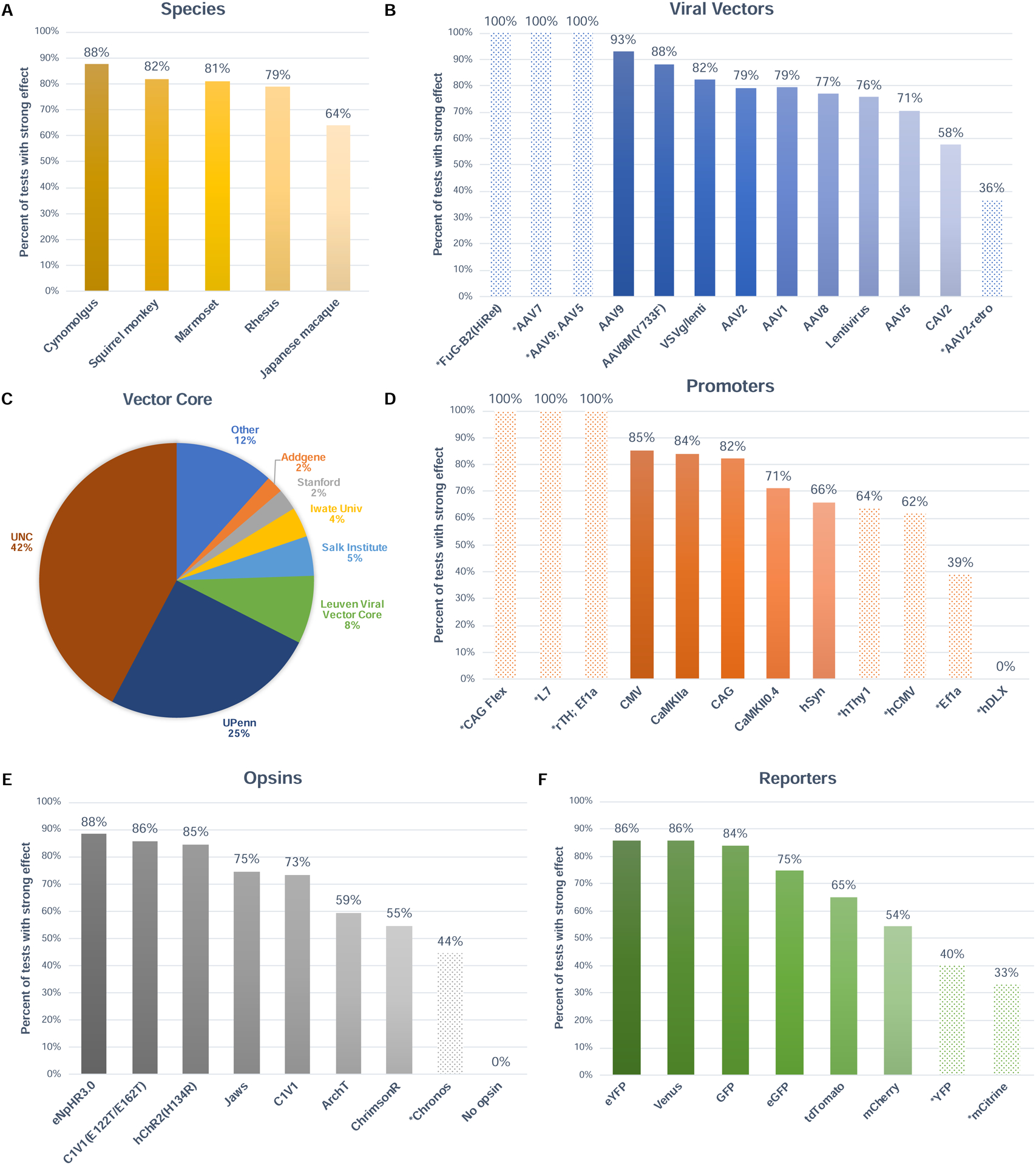
A) Percentage of injections that led to at least one strong effect across the three outcome modalities, as a function of NHP species. B) Same as in A, as a function of viral vector. C) Proportion of experiments that acquired their viral vector from each Vector Core. D) Same as in A, as a function of genetic promoter. E) Same as in A, as a function of opsin gene. F) Same as in A, as a function of reporter protein. Dotted bars represent variables that had low sample size in the database (i.e. tested in fewer than 20 injections or in fewer than 5 animals).
When examining the success rates of all experiments as a function of NHP species, we found that the highest success rates were achieved in cynomolgus monkeys (88%; 110 injection tests) (Figure 5A). Japanese macaques show the lowest success rates with 64% (75 injection tests). The database listed a total of 24 viral vectors that have been used across all injection experiments in the database (see Table 1). In Figure 5B, only those with more than 10 entries were included (N = 13). Of those vectors with adequate sample sizes, we found that AAV9 and AAV8M(Y733F) yielded the highest success rates, with 93% and 88%, respectively. At the other end of the spectrum, we find CAV2 (58%) and AAV5 (71%) have the lowest success rates across species. Overall, investigators tended to purchase their viral vectors either from the UNC Vector Core (42%) or the UPenn Vector Core (25%), with a total of 26 vector cores serving the international community (Figure 5C). The public plasmid repository Addgene is now offering ready-to-use viral vectors from a standard collection (https://www.addgene.org). Custom viral construct productions still need to be ordered from other vector cores.
Table 1 -.
List of all viral construct variants included in the database with the corresponding number of entries (i.e. injection experiments).
| N | |||||||
|---|---|---|---|---|---|---|---|
| AAV5 | 374 | CaMKIIα | 365 | hChR2(H134R)a | 397 | eYFP | 462 |
| AAV9 | 134 | hSyn | 224 | No opsin | 198 | GFP | 175 |
| AAV1 | 79 | CAG | 105 | C1V1(E122T/E162T)a | 107 | mCherry | 137 |
| AAV9; AAV5* | 78 | rTH; Ef1a* | 78 | ArchTb | 66 | eGFP | 76 |
| Lentivirus | 77 | CMV | 54 | Jawsb | 64 | tdTomato | 57 |
| AAV8 | 68 | CaMKII0.4 | 43 | ChrimsonRa | 36 | Not present | 39 |
| VSVg/lenti | 35 | Ef1a | 18 | C1V1a | 30 | Venus | 37 |
| AAV8M(Y733F) | 34 | CAG Flex | 17 | eNphR3.0b | 25 | YFP | 15 |
| AAV7 | 28 | hThy1 | 13 | Chronosa | 16 | mCitrine | 10 |
| CAV2 | 26 | hCMV | 13 | SwiChR++b | 12 | DsRedII | 7 |
| AAV2 | 24 | L7 | 12 | GtACR2b | 8 | BFP | 1 |
| AAV2-retro | 20 | hDLX | 11 | C1V1(E162T)a | 7 | RFP | 1 |
| FuG-B2(HiRet) | 14 | SAD | 7 | eArch3.0b | 7 | mNEon | 1 |
| RV | 7 | MmCaMKIIα0.4 | 7 | eNpHRb | 6 | ||
| AAV-retro | 5 | TH | 6 | ChETAa | 6 | ||
| EIAV-Rabies-VSVg | 5 | CBA | 4 | Archb | 4 | ||
| AAV6 | 5 | a-CaMKII | 4 | ArchT3.0b | 4 | ||
| HSV | 5 | PGK | 4 | oChlEFa | 4 | ||
| AAV2; AAV5* | 4 | GAD67 | 3 | ChR2(C128S/D156A)a | 3 | ||
| EIAV-Rabies | 3 | MmCaMKIIα1.3 | 3 | NpHRb | 3 | ||
| AAVDJ | 2 | phSyn1(S) | 2 | ChR2(E123A)a | 2 | ||
| LVV | 2 | dbh | 2 | SFOa | 1 | ||
| LT-HSV | 1 | hChAT | 2 | SFFOa | 1 | ||
| Rabies | 1 | EF1a/IRES | 1 | ||||
| pCAG | 1 | ||||||
| hCMV(en)hSyn | 1 |
indicates when two viral constructs were mixed in the same solution.
Excitatory opsin,
Inhibitory opsin.
Legend. LT-HSV: Long-term herpes simplex virus; AAV: Adeno-associated virus; LVV: Lentiviral vector; EIAV: Equine infectious anemia virus; VSV: Vesicular stomatitis virus; RV: Retroviral vector; FuG: Fusion envelope glycoprotein; CAV: canine adenovirus; EF1a: Elongation factor-1 alpha; IRES: Internal ribosome entry site; CAG: cytomegalovirus enhancer, chicken beta-actin promoter and rabbit beta globin splice acceptor site; CMV: Cytomegalovirus; hCMV(en)hSyn: human synapsin-1 promoter preceded by the CMV enhancer; dbh: dopamine beta-hydroxylase; hChAT: Human choline acetyltransferase; GAD67: Glutamic acid decarboxylase 67; CamKIIa: Ca2+/calmodulin-dependent protein kinase II; MmCAMKIIα: Mus musculus CamKIIa; CBA: Chicken β-actin; PGK: Phosphoglycerate kinase; TH: Tyrosine hydroxylase; SAD: stearoyl-acyl carrier protein desaturase; hDLX: human distal-less homeobox; L7: Purkinje cell protein 2; hThy1: human thymocyte-1; Flex: Flip-excision; SFO: Step-function opsin; SFFO: Stabilized step-function opsin; ChR2: Channelrhodopsin-2; NpHR: Natronomonas pharaonis halorhodopsin; Arch: Archaerhodopsin; GtACR2: Guillardia theta anion-conducting channelrhodopsin; SwiChR: Step-waveform inhibitory channelrhodopsin; BFP: Blue fluorescent protein; RFP: Red fluorescent protein; YFP: Yellow fluorescent protein; GFP: Green fluorescent protein.
Investigators have used 26 different genetic promoters in their experiments (Table 1). Figure 5D includes those that have more than 10 entries. Of those included, CMV and CaMKIIa yielded the most successful experiments, with 85% and 84% success rate, respectively. Human synapsin (hSyn), a promoter used in 224 experiments, yielded the lowest success rate of the five promoters that satisfy the sample size threshold, with 66%.
In terms of opsin genes, investigators used 24 different types (Table 1), including closely related variants of the same opsin. The eNpHR3.0 opsin, an inhibitory chloride pump, had the highest success rate with 88%. The E122T/E162T variant of C1V1 (also known as T/T), itself a red-shifted chimera of channelrhodopsin, yielded success rates around 86% (Figure 5E). The hChR2(H134R) opsin, the most widely used in the community with 397 injections, produced an 85% success rate. The K176R mutation to Chrimson (ChrimsonR) had the lowest yield, with 55%. Finally, investigators used several different reporter genes as a proxy to visualize the expression of the opsin (Table 1). The most widely used, eYFP, is also one with a high success rate at 86% (Figure 5F). The mCherry reporter yielded the lowest rates of success, with 54%. Again, all the reported success rates herein are to be interpreted carefully in consideration of other parameters that might also influence the success of an injection experiment. Please refer directly to the database for a full list of the >30 parameters listed for each injection.
Viral delivery methods
Injecting viral vectors into the brain parenchyma can be performed using several different surgical techniques (Figure 6A). For example, investigators have used an existing recording chamber to lower the injection needle through a grid under control of a micromanipulator (63% of database entries, 78% success rate). This technique has the advantage of using existing hardware to access the brain and may be performed without general anesthesia. It has the caveat of offering no visual access to the cannula’s penetration of the brain tissue, which is helpful to confirm that the cannula penetrated cortex and is not merely creating a dent in the pliant cortical surface. However, this limitation is surmountable with an “injectrode” that allows simultaneous injection and recording of neural activity to ascertain cannula placement (84 injections used an injectrode). Alternatively, investigators have performed injections in awake monkeys through a silicon artificial dura implant, as originally described by Ruiz et al. (4% of entries, 82% success rate; Ruiz et al., 2013a). Given the translucid appearance of the artificial dura, the cannula can be inserted into the cortex under visual guidance and expression can be confirmed with in vivo epifluorescence imaging. Penetration through the artificial dura is not always necessary. Within the same preparation, the artificial dura can be temporarily removed and replaced with a chamber insert that dampens cortical movement. This procedure allows for glass pipette injection and brain stabilization without the need to penetrate the artificial dura (Seidemann et al., 2016). For deep brain structures where this is not feasible, an in vivo fluorescence detector has been suggested (Diester et al., 2011). Investigators have also performed viral injections under general anesthesia in the operating room. When doing so, surgeons have either performed large craniotomies allowing dural flap and dura closure (30% of entries, 79% success rate), or smaller “burr hole” craniotomies offering just enough space to insert the injection cannula into the brain (3% of entries). In the latter case, the dura can be left in place and penetrated by the injection cannula, or a sharp needle can be used to nick the dura before inserting the injection cannula into the brain. Overall, 43.5% of optogenetics experiments performed some form of dura removal before inserting the injection needle into the brain (83% success rate with dura removal vs. 71% without).
Figure 6. Surgical and injection methods used across experiments.
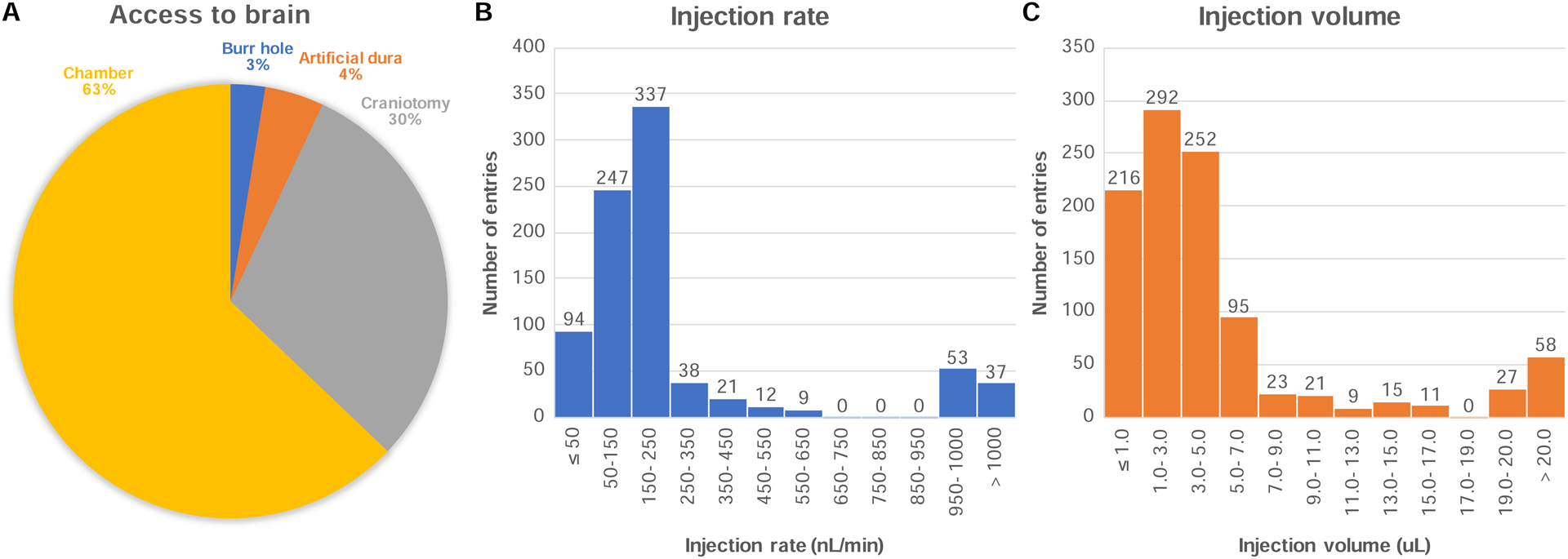
A) Proportion of each surgical method used to access the brain parenchyma for injection purposes. B) Histogram of the rate of injection of the viral solution across all experiments. C) Histogram of the volume of viral solution injected across all experiments.
In preparation for the injection surgery, some investigators chose to dilute their viral solution to a lower titer than the one provided by their vector core (e.g. using sterile phosphate-buffered saline). For this, the viral solution, which is usually shipped on dry ice by the vector core, needs to be thawed. The solution can be refrozen for storage and thawed again later for injection purposes. However, it is good practice to limit the number of freeze/thaw cycles to minimize cryoinjury to the viral particles that can alter the solution titer (addgene.org/recipient-instructions/myvirus/). Most experiments performed by contributing investigators complied with this recommendation (98% of entries with 1 or 2 freeze/thaw cycles). Titers for viral solutions can be calculated in several ways depending on the providing vector core. Please see https://www.addgene.org/viral-service/viral-production/#aav-titer for a detailed description of viral titer assays.
The injection of the viral solution can be made at various infusion speeds. When selecting a speed, the goal is to minimize trauma to the tissue and maximize the efficiency of delivery, while also minimizing the time required to perform the injection. Injection speed may also affect the size of the transduced area in a virus-specific manner (Lerchner et al., 2014). It is generally assumed that slower injection rates minimize trauma to the brain tissue. However, the database reports successful experiments with injection rates ranging from 6 nL/min to 5000 nL/min, with a median injection rate of 200 nL/min (Figure 6B). The higher end of this spectrum falls into the category of “convection-enhanced delivery” (CED) techniques, pioneered by Krystof Bankiewicz (UCSF) for gene therapy applications, and refined for use in NHP optogenetics by Dr. Yazdan-Shahmorad in the Sabes lab (Khateeb et al., 2019; Yazdan-Shahmorad et al., 2016; 2018b). The principle of CED is that higher flow rates drive bulk fluid flow through the tissue, allowing for much greater distribution of the vector from a single infusion. Earlier work from the Bankiewicz lab showed that infusion rates of up to 5000 nL/min do not cause detectable tissue damage (Kells et al., 2009; Varenika et al., 2009), and both labs have shown that the distribution volume of the vector is proportional to the infusion volume, with a constant of proportionality in the range of 1 to 4 (Krauze et al., 2005; 2008; Yazdan-Shahmorad et al., 2016). This allows for a 10 to 100-fold larger volume of brain tissue transduced compared to standard microinfusion techniques (Kells et al., 2009; Varenika et al., 2009). Note that reflux-resistant cannulas need to be used when delivering solutions at these high rates to prevent the solution from tracking up along the cannula during delivery (Krauze et al., 2005).
The volume of viral solution injected varied by several orders of magnitude across experiments. Some investigators injected as little as 0.2 μL, summing across all injection depths of the same needle penetration (Figure 6C). On the other end of the spectrum, CED techniques have injected up to 246 μL per needle penetration at a single depth (Yazdan-Shahmorad et al., 2018b). The median volume injected across experiments was 3.2 μL per needle penetration. The volume of viral solution will have an impact on the number of cells that will encounter viral particles, and therefore the density and volume of transduced tissue. However, transduction efficiency is also modulated by the tropism of the viral vector capsid, with some serotypes being more potent than others at transducing neurons (Dodiya et al., 2010; Markakis et al., 2010). Toxicity can occur as a result of high viral titers in combination with highly potent capsids; thus, both factors need to be considered together (among other parameters). Finally, most investigators waited several minutes before moving the injection cannula to a different position along the injection tract. The assumption is that if the cannula is removed too early, the vector solution will not have time to diffuse into the nearby tissue and might reflux along the injection tract with the cannula during removal. Investigators have waited between 1 and 30 minutes before moving the cannula to a different depth, with 10 minutes being a typical value.
Incubation and immune response
Following vector delivery, the rDNA will either integrate the genome (using lentiviruses) or remain episomal (using AAV) and persist in non-dividing cells for years without damaging the host cell (Deyle and Russell, 2009). The AAV family of vectors has become the preferred tool for viral transduction (>85% of tests) in part because of its episomal stability, enabling long-term transgene expression and its independence from local chromatin structure which can affect gene transcription (see addgene.org/viral-vectors/aav/aav-guide/ for a complete resource). Once synthetized by the cell’s innate machinery, the opsin protein needs to be trafficked to the cell membrane to play its role as an ion channel or pump. To accommodate these steps, most investigators wait several weeks before beginning their optogenetics experiments in NHP (median = 7.5 weeks). Some have waited for as little as one week and found successful labeling. In theory, opsin expression should peak after two weeks and will maintain a level of expression that depends on the immune response to the foreign opsin and reporter protein (Gurda et al., 2016).
The immune response to the viral transduction might alter the outcome of an experiment (Mendoza et al., 2017; Wang et al., 2011) (Maimon et al., 2018). First, an immune response could be mounted against the viral capsid itself. Many strains of AAVs are naturally-occurring and monkeys could have been in contact with those benign viruses and produced antibodies against them before the beginning of an experiment (Calcedo et al., 2015). Before choosing a specific serotype of AAV, some investigators elect to test their animal for the presence of neutralizing antibodies (NAbs) against AAV that could theoretically inhibit transduction. NAbs can be tested both in the serum and in the CSF, with important distinctions (see Dr. Calcedo’s advice below in the STAR Methods). Only 4.7% of all entries in the database reported having tested for NAbs to guide this decision. Unfortunately, the sample size was too low to determine whether this approach yielded better success rates. Finally, several labs used immunosuppressing drugs during the incubation period to dampen the innate and adaptive immune response to the transgene. Around 20% of experiments used the steroidal drug dexamethasone either after injection, or both before and after the injection of the viral vector. However, in the sample of entries where immunosuppressing drug use was reported (N=594), there was no difference in success rates between those who used (70.5%) versus those who did not use such drugs (73.6%, Chi-Square test P = .42). The choice of immunosuppressing drug and drug regimen can affect those results and was not accounted for in this preliminary analysis. Moreover, systemic immunosuppressing drugs may have other effects in the host such as making it more vulnerable to opportunistic infections, local or systemic (Youssef et al., 2016). Whether these drugs may be used locally at the site of injection has not been tested so far. As the database continues to grow, we hope that it will be possible to parse out those effects and provide better guidance on best immunosuppression strategies.
Publication bias
The current initiative collected a large amount of unpublished data (N=552 injections), effectively doubling the available literature on NHP optogenetics. This allowed us to quantify the success rates of experiments that have been published in comparison with those that have not been published in the literature. We tested the extent to which negative results go missing in the literature (aka the “file-drawer problem” (Editorial, 2019)). Again, we qualified a study as “successful” if it had at least one strong outcome across the measures provided by anatomy, physiology or behavior. Unsuccessful experiments were those where no effects were reported across all three modalities. Following this definition, we found that around 1% of the published literature presented negative results, while 18% of the unpublished results were negative (Figure 7). Notably, there was a large proportion of positive, strong results that were unpublished (65%), emphasizing the importance of the current initiative.
Figure 7. Publication bias in NHP optogenetics studies.
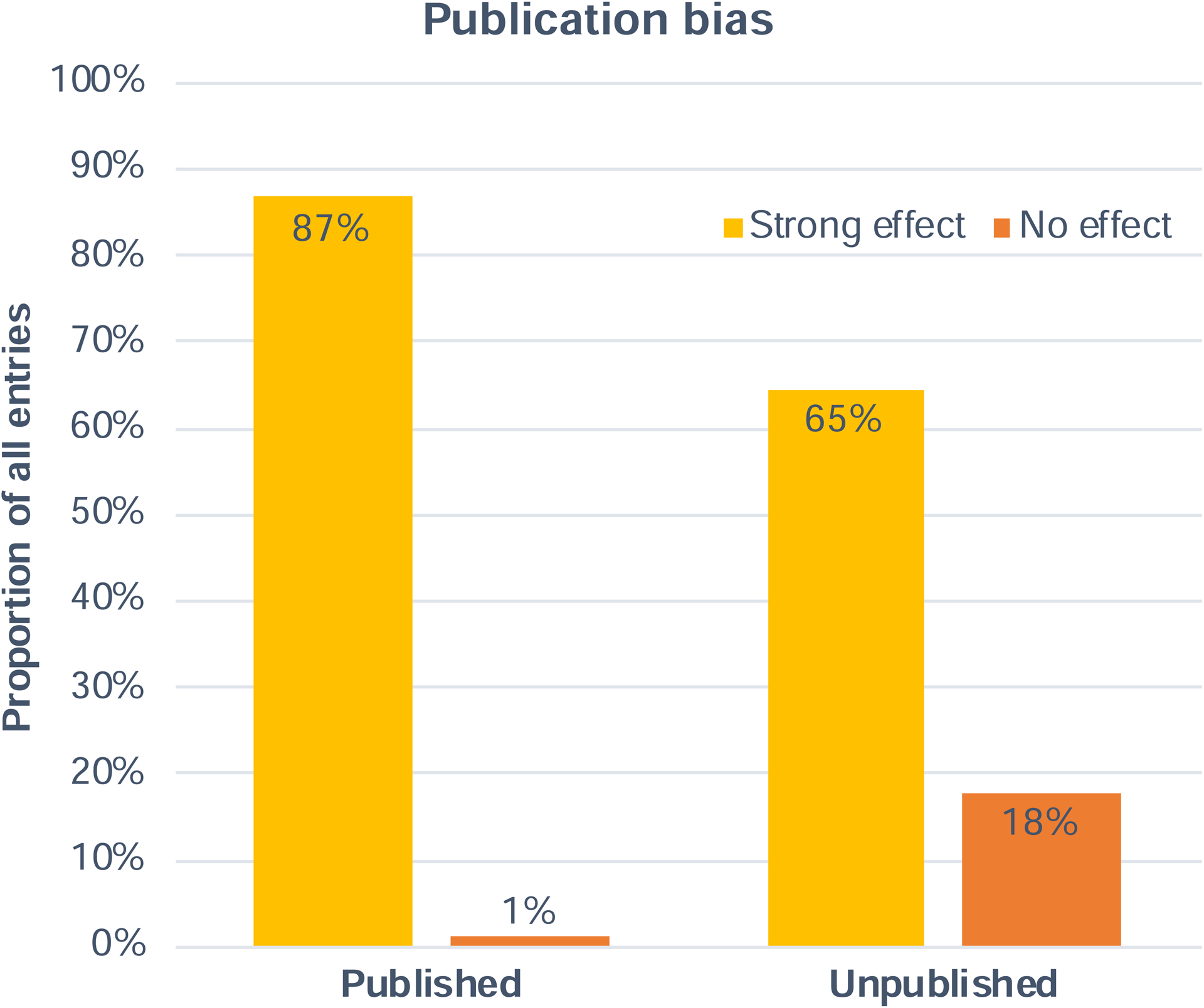
Comparison of the rate of successful (strong effect) and unsuccessful (no effect) experiments across the published and unpublished entries in the database.
DISCUSSION
The NHP Optogenetics Open Database is accessible for everyone to contribute to and to consult on the Open Science Framework website (https://osf.io/mknfu/). It continues to accept new contributions from investigators who would like to share both positive and negative results with the community (instructions are included on the website), and it encourages sharing information on a broad range of variables, which could be expanded as the community agrees in the future to include more parameters. As the database continues to accept new entries from contributing laboratories, the data will be curated and quality-checked by Dr. Tremblay. Any laboratories wishing to contribute are encouraged to log on to the OSF website, or to contact Dr. Tremblay by email directly (strem@pennmedicine.upenn.edu). This resource has already generated important information that can be used to inform new optogenetics approaches in NHPs. With more data, more factors influencing the success and failure of optogenetics experiments in NHP can be identified, and interaction effects between multiple factors can be modeled. We encourage investigators around the world to continue expanding and relying on this public resource, including those working with rodents and humans.
In addition to presenting the database, the current manuscript includes a series of comments with advice from contributing laboratories tailored to investigators who wish to undertake optogenetics experiments in their lab. This advice was provided to answer the following question: “If there was one thing you wish you had known before starting your optogenetics experiments, what would that be?” The goal of this exercise was to identify challenges and caveats that investigators experience in practice and that may not always be included in published articles. The detailed answer from each laboratory is included in raw format in the STAR Methods section at the end of the manuscript. We encourage the reader to consult these comments directly in the STAR Methods section below and to communicate with the respective laboratories for more details on how to overcome these practical challenges. We will try to summarize some of the main insights of this section in the following paragraphs.
Optogenetics experiments in NHP share many technical similarities with gene therapy experiments in NHP. Namely, the same viral vectors are used by both fields to deliver transgenes to cells in the primate nervous system (i.e. AAV). To obtain FDA approval for novel treatments, gene therapists had to undertake thorough investigations of the safety of gene transfer using these vectors and their impact on the immune system. Several tests are routinely conducted in gene therapy trials for this purpose, namely neutralizing antibodies assays from cerebrospinal fluid and blood serum, as well as ELISPOT assays to measure T-cell responses to the capsid and the transgene. Pre-existing neutralizing antibodies to the viral capsid could severely impede viral transduction, and many monkeys will naturally present these antibodies before the start of the experiment. The current initiative revealed that fewer than 5% of optogenetic injection experiments included neutralizing antibody testing in their animals, which could partly explain experimental problems with opsin expression encountered by some investigators. There is currently insufficient data to conclude whether or not pre-existing neutralizing antibodies to the viral capsid (detected in the serum) impede viral transduction in the CNS. We encourage investigators to contact their local gene therapy core before initiating their experiment to set up a battery of immunological assays for their animals in order to accumulate more data on this topic.
Many contributing laboratories stressed the importance of histological confirmation of opsin expression in animals before conducting electrophysiological and behavioral testing. Investigators note important variability in the expression of the same viral construct across species (rodents vs primates), animals of the same species, and brain region. Moreover, some investigators report that different opsins (e.g. ChR2 vs C1V1) will localize to different parts of the neuron (Fortuna et al., 2020). This variability begs for thorough histological confirmation that the opsin is expressing correctly in the target neuronal population and that the cellular integrity of the expressing neurons is preserved. The latter can be performed using standard hematoxylin and eosin staining (H&E stain) and interpreted by a professional pathologist from a collaborating veterinary hospital. Once safe and efficient transduction is confirmed post-mortem, in vivo follow-up experiments should also strive to confirm successful expression of the opsin. Several laboratories suggest the use of inexpensive, commercially available fluorescent lights and filter goggles to visualize in vivo epifluorescence through an artificial dura or directly from the cortex in the operating room. Such confirmation is not easily attained from sub-cortical or sulcal injection sites, but new developments in PET-visible opsin variants show promise as a new, non-invasive way to track opsin expression across the entire brain in vivo (Michaelides et al., SFN abstract, 2019).
Investigators also highlighted several surgical challenges inherent to intraparenchymal delivery of viral vectors to the brain. Spatial targeting of injections to specific sites through a recording chamber is non-trivial. Relying only on coordinates established during previous neural recordings can be problematic because of possible movement of the brain tissue over time under the implant, and the buildup of granulation tissue that can deflect the trajectory of cannula and electrodes. Many investigators recommended using an injectrode, a combination of an injection cannula with an electrode, to avert these problems (Chen et al., 2001). Using an injectrode, the injection site is identified first based on its electrophysiological properties, and the attached cannula can deliver the viral solution directly at the site without requiring separate penetrations. Moreover, the exact location of the delivered solution can be imaged in 3D by mixing manganese salt with the viral solution and performing a T1 MRI after the injection (Fredericks et al., 2020). The precise volume of diffusion can also be quantitatively measured with this technique. Finally, when targeting sub-cortical areas that don’t have clear electrophysiological signatures, an MRI-guided neurosurgical system is recommended (e.g. Brainsight Vet). Other groups stressed the importance of closely monitoring the flow of the viral solution through the syringe, tubing and cannula to make sure there are no leakages or clogging along the injection line. The use of inert dyes has been suggested to visually map the flow of liquid inside the injection apparatus, but also to confirm that the solution is not refluxing out of the cannula tract and exiting the brain. Several designs of reflux-resistant cannula have been developed to address this issue in convection-enhanced delivery, but may also be useful for conventional injection methods (Yazdan-Shahmorad et al., 2016). We encourage the reader to consult the STAR Method for more details on the surgical solutions proposed by contributing laboratories.
In closing, we hope this resource will be used not only by basic scientists trying to uncover the workings of the primate brain using optogenetics, but also by translational scientists hoping to bring this powerful technology to the service of patients with neurological or psychiatric conditions. The therapeutic potential of optogenetics for human brain disorders has been recognized for many years (Chow and Boyden, 2013; Deisseroth, 2012), and excitement is growing for applications in Parkinson’s disease, targeted brain reorganization for neurorehabilitation (Yazdan-Shahmorad et al., 2018a) and new-generation sensory implants (El-Shamayleh and Horwitz, 2019). Moreover, recent breakthroughs in gene therapy demonstrate the safety and efficacy of delivering transgenes to the human nervous system using the same viral vector technologies used in optogenetics (Wang et al., 2019). The way is paved for the development of clinical technologies relying on optogenetics to control neural populations and pathways with unprecedented precision. We hope the NHP Optogenetics Open Database will provide a strong basis for those looking to make evidence-based decisions in the development of future pre-clinical and clinical trials.
STAR METHODS
RESOURCE AVAILABILITY
LEAD CONTACT
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Sébastien Tremblay (strem@pennmedicine.upenn.edu).
MATERIALS AVAILABILITY
This study did no generate new unique reagents.
DATA AND CODE AVAILABILITY STATEMENT
Original data for all results in this paper is available at https://osf.io/mknfu/. For a non-anonymized version of the database including names of laboratories who conducted each test, please reach out to the lead contact.
METHOD DETAILS
In this section, we have collected a series of short pieces of advice geared towards investigators wanting to launch new optogenetics experiments in NHP. The advice was authored by individual laboratories wanting to share personal experiences using a specific technique or tool in optogenetics, and its associated outcomes and pitfalls that are rarely shared in published manuscripts. We also include relevant surgical advice by professional surgeons and advice in immunology by professional gene therapists. We hope this series of notes will be useful in identifying unwritten practices that yield the highest chances of success in NHP optogenetics, as well as practices to avoid.
Advice from gene therapy
Although AAV vectors are extensively used in optogenetics, evaluation of host immune responses to AAV capsids and transgenes are not performed routinely. Natural exposure to AAV leads to preexisting humoral and cellular immunity, which can diminish vector efficacy. In addition, invasive intra-parenchymal administration of the vector may trigger an innate immune response that can lead to inflammation which can also have an impact on gene transfer outcome. Pre-existing neutralizing antibodies (NAbs) to AAV are very prevalent in the monkey population. AAV seroprevalence is AAV serotype-, monkey species- and colony-specific. In some colonies, the frequency of NAbs can be as high as 100% (Gao et al., 2003). Although the impact of systemic NAbs on brain gene transfer has not been properly studied, it is known that low levels of NAbs can completely abolish systemic gene transfer (Wang et al., 2011). Thus, NAbs specific to AAV capsids used in brain studies should be evaluated in serum and in CSF before vector administration. In addition, acute innate immune responses after vector administration should be evaluated by analyzing levels of inflammatory cytokines and chemokines also in serum and CSF (Calcedo et al., 2018). Acute innate immune responses are fundamental for the development of efficient cellular immune responses, which can reduce or eliminate transgene expression. T-cells specific to the transgene or to the AAV capsid can recognize transduced neurons and eliminate them. To monitor these responses, blood samples can be collected and peripheral blood mononuclear cells (PBMCs) isolated and tested on an enzyme-linked immunospot (ELISPOT) assay that will quantify the number of T-cells that release IFN-γ after stimulation with AAV or transgene antigens. Additional samples from draining lymph nodes at the time of the necropsy will inform of the presence of local T-cells not detected in blood. If innate and adaptive immune responses present a problem for the long-term expression of the transgene of interest, prophylactic administration of immunosuppressive drugs should be considered. Proper evaluation of different immunosuppression regimens should be carefully evaluated to minimize duration and reduce toxicity associated with a compromised immune system.
- Dr. Roberto Calcedo, Director of Immunology Core at the Gene Therapy Program, University of Pennsylvania
Advice from surgical experts in NHP
The optimal method of surgical access is determined by whether the target brain region is sub-cortical or cortical. When targeting sub-cortical structures, a direct visualized approach is generally not possible. In this case, accuracy in placing the injection may be increased by using MRI-guided stereotactic procedures, or by injecting through a grid in an implanted chamber. The accuracy of the injection can be assessed post-operatively by co-infusing a MR contrast agent (e.g. manganese) and performing an MRI within 24 hours. Injections into dorsal/lateral cortical surface can be made under direct visualization. Targets can be identified using cortical landmarks (e.g. sulci) after a bone flap is turned and a dura flap reflected to provide access. Injections are placed with the aid of an operating microscope or surgical loupes. After completion of the injection series, the dura and bone flap are sewn back into place. For optogenetic applications, a second surgery can be performed to implant a device to deliver light after waiting for the opsin to express. In this case if you place artificial dura (e.g. Tecoflex) between the pial surface and the dura during the first surgery, it will reduce the number of adhesions encountered during the second dural opening. The benefit of the two-step approach is that the region of expression can be visualized, and the light source positioned for maximal effect. Targeting regions on the ventral cortical surface/within a sulcus is more challenging. With a stereotactic approach, the damage caused by multiple needle tracks necessary for complete coverage of the target region is unavoidable and may compromise the experiment. With a visually-guided approach, the brain must be carefully manipulated to provide access to the target region without causing mechanical damage (i.e. brain lifting for ventral access, and sulcal opening for intra-sulcal access). Injections can be performed using a hand-held approach, in which 1 – 2 μL is infused per site, or using devices custom-designed to provide access to the target tissue that can be left in place while larger volumes of virus are infused at a controlled rate (Fredericks et al., 2020). Tissue coverage will be affected by the rate and volume of virus infusion, in concert with local anatomical structure. Infusing at high speeds/’CED’ (> 0.5 μL/min), combined with reflux-resistant needles (Debinski and Tatter, 2009) can produce widespread coverage. However, it can also cause reflux (‘back-flow’) up the needle (see (Mehta et al., 2017) for detailed discussion on the benefits and drawbacks of CED).
- Drs. Richard Saunders & Mark Eldridge, Surgery Instructors, NIMH
Advice from contributing laboratories
When artificial dura is used to replace native dura, we recommend taking photographs of the cortical surface to use for marking the location of viral injections relative to vascular landmarks (Nurminen et al., 2018). This can greatly aid in identifying these locations weeks/months after the injections, particularly for small or deep layer injections, which cannot be visualized using wave-length specific goggles or a fluorescent microscope. Large and superficial injections can be easily identified by the fluorescent protein expression using goggles. Our lab purchased an expensive Zeiss surgical fluorescent stereomicroscope, but found this does not provide significantly better visualization of fluorescent protein expression than the much cheaper goggles.
When using combinations of different viral vectors, e.g. one expressing Cre-recombinase and a Cre-dependent vector, it is of crucial importance to be aware of vector-specific differences in optimal expression times. For example, CAV2 expression peaks at 7 days post-injection, but AAVs require much longer post-injection survival times; therefore multiple surgeries are required for injecting the different vectors at appropriate time intervals.
- Alessandra Angelucci lab
We recommend doing electrophysiology recording before viral vector injection if a standard recording chamber is being used in the experiment. The piloted recordings should identify several sites that give the precise coordinates of the target area where abundant neurons are located. Injection in these sites ensures that the virus is not delivered to tissue with few neurons. Second, we recommend doing multiple injections in nearby sites, which should effectively increase the success rate and the expression extents. In terms of behavioral effect, the property of the neurons being stimulated should be carefully characterized, as we only expect specific and often subtle behavioral effects by stimulating neurons relevant to the current behavioral task. In addition, we think targeting a particular pathway or specific cell type rather than stimulating universally will be more effective in driving behavioral effects. To this end, developing a potent anterograde or retrograde AAV is very useful in targeting a specific pathway. In particular, we recommend using a retrograde AAV, as this approach entails injection at the terminal end and optical stimulation at the soma. Theoretically, it should be easier to activate the soma than the terminal by light stimulation.
- Ji Dai lab & David Sheinberg lab
In initial tests of viral injections through an artificial dura, we made separate injections of the same construct (AAV8.hSyn.Jaws-KGC.GFP.ER2-WPRE.hGH) obtained from two different vector cores. Despite identical injection procedures, expression (as indicated by GFP epifluorescence) was strongly present in the sites injected with the construct from one vector core but not the other. This suggests that the outcome of viral expression may be strongly dependent on either the specific vector core that produced the construct, or the specific batch that the construct was produced in. The use of an artificial dura implant for the above tests was crucial, since it allowed us to confirm viral expression a few weeks after each injection test - by visualization of epifluorescence through the artificial dura - without the need for histological sectioning. This allowed us to sequentially perform several independent tests on the same animal.
- Robert Desimone lab
We recommend injecting virus in the operating room, following a craniotomy and durotomy. At each injection site, we typically inject at multiple depths (approximately 0.5–1 mm apart), starting from the deepest level pulling up gradually. If coverage of large regions is desired, this procedure is time-intensive when injections are done serially. In that case, we recommend using multiple syringes mounted on multiple stereotax manipulators, each targeting one injection site. To cover the target region, we recommend using a thin plastic grid overlay with holes to guide the injection needles through. This grid can be loosely affixed to the bone, to ensure approximately uniform coverage of injections. When possible, we strongly recommend verifying viral expression using fluorescence imaging (e.g. in surgery, using LEDs & filters) prior to beginning any time-intensive behavioral or neurophysiological experiments.
- James DiCarlo lab
Expanding on the advice from the Platt lab for standard chamber recordings, we would recommend using a cell growth inhibitor (like 5-flurouracil) inside the chamber between the time of injection and the start of the recordings. We typically allow 4–6 weeks for opsin expression. If granulation tissue is allowed to proliferate unchecked during this time, the extra tissue is sufficient to slightly alter electrode trajectories, making it more difficult to locate the injection site even when the same grid has been used for injections and recordings (Andrei et al., 2019). Also, since it is less often mentioned in the literature these days, we would also like to emphasize that light artifacts will be present on typical electrodes, and they can often be mistaken for real spikes, though more typically they are confined to local field potentials. Since the artifacts depend on several factors (distance from the light source to the electrode, the electrode material, amplifier cut-off frequencies, etc.), we strongly recommend testing your recording system to identify conditions where artifacts are present.
- Valentin Dragoi lab
To minimize tissue damage, we recently changed the usage of 32G needles of stainless steel (~300 μm diameter) to beveled glass capillaries (~50 μm diameter). This change did not influence the efficiency of viral transduction. A part of our electrophysiological data with photo-stimulations is available at neurotycho.org.
- Naotaka Fujii lab
We have successfully transduced opsins in several regions of the basal ganglia using standard extracellular electrophysiology methods to map the region of interest before selecting the area of injection. To further confirm the injection location, the virus infusions are conducted using a combination of electrode and injection system (Kliem and Wichmann, 2004). During optogenetic stimulation experiments, we have found that tissue damage from the use of optrodes made with a 200-μm core optic fiber is substantial, effectively preventing further penetrations at the same site. Thus, we suggest the use of optrodes built with smaller-diameter optic fibers. Careful histological examination of opsin expression at the injection sites and in brain regions connected to the injection site (to assess potential expression of the opsin after anterograde and/or retrograde expression) is essential to properly interpret the results of these experiments. Ideally, ultrastructural studies to confirm the plasma membrane location of the opsins should be also conducted (Galvan et al., 2012).
- Galvan, Smith and Wichmann labs
The following two methods are useful for successful optogenetic manipulation in NHPs. The first method is to locate viral injection sites using an injectrode, consisting of a microelectrode joined to a silica tube. Injection depth is critical especially for deep brain sites and difficult to determine through other means, as the consistency of brain tissue is nonrigid. By characterizing the physiological properties of neurons at the injection site, the injectrode allows the experimenter to account for movement of brain tissue (for instance caused by insertion of the probe itself). The second method is to confirm neuronal modulation by optical stimulation using an optrode, consisting of a microelectrode joined to a fiber-optic cable. As neuronal activity can be directly or indirectly modulated by photostimulation, measuring physiological responses (inhibitory or excitatory) concomitant with behavioral effects of optical stimulation is useful to constrain mechanistic neural models.
- Okihide Hikosaka lab
To ensure that the virus is physically delivered into the brain as intended, take steps to minimize the probability of equipment failure. First, thoroughly practice the procedure to get an intuitive sense for the components of the injection system (tubing, connectors, syringes, pumps, etc.), which can be fragile. Second, test the equipment beforehand in a configuration as close as possible to that used during the actual injection. We learned the hard way that damage to fittings is not always obvious. Third, check continuously for leaks and blockages during all steps of the procedure. Finally, in case a problem does arise during the injection, have pre-assembled (and tested!) backups of as many of the components as is feasible.
- Mehrdad Jazayeri lab
The importance of negative controls in science cannot be overstated. Monkey electrophysiologists are acutely aware of this; however, optogenetics presents a new set of questions and confounds that must be considered. This is especially true in monkey optogenetics experiments that examine physiological and/or behavioral responses. In behavioral experiments, there are a number of confounds introduced by stimulation equipment. Is your optic fiber entirely shielded, such that no aberrant light escapes the fiber or fiber couplings? Shutters and acousto-optic modulators both often emit faint noises when triggered. These are among the confounds that could easily be noticed by an NHP in a dark, quiet room. Ensuring that these confounds have no effect on behavior must be verified using negative control conditions. In optogenetic tests for physiological (firing rate) responses, one must ensure that increased temperature or altered PH at the optic fiber tip are not modulating neural activity. Thus, it is critical to replicate your recording and stimulation protocols at nearby, non-transfected sites. Furthermore, one should titrate luminance of the photostimulator down at the transfected site to the point where triggering no longer produces behavioral or physiological effect. Lastly, if available, luminance-matched photostimulation with a wavelength that is outside of your opsin’s excitation range can provide valuable insight.
- Julio Martinez-Trujillo lab
Before doing viral injections, I recommend electrophysiological characterization of brain tissue to precisely localize the electrode’s tip in the desired brain area. When you are ready to inject a virus, use the same chamber and approach that you used for the initial functional mapping. Make the viral injection using an injectrode: a combination of an electrode and an injection cannula (Cavanaugh et al., 2012; Monosov and Thompson, 2009). This allows for the characterization of neuronal activity in the brain and therefore a verification of the electrode’s location before the viral injection is made. During viral injection, I recommend a very slow flow rate (for details see (Cavanaugh et al.,2012)), and also very importantly a very slow retraction of the injectrode so that you do not pull up the virus to other nearby brain areas. To manipulate brain activity, I recommend using an optrode: a combination of a fiber and an electrode. This will prevent you from missing the transfected tissue because you can functionally identify the transfected region of interest before performing optogenetic manipulations of neuronal activity.
- Ilya Monosov lab
One of the major challenges to investigate optogenetic in a large brain such as NHP, is to be able to induce a comprehensive opsin expression within a whole brain circuit. Combining optogenetics and tractography may enable anatomo-functional characterization of large brain cortico-subcortical neural pathways. For the proof-of-concept this approach was used in the NHP brain to characterize the motor cortico-subthalamic pathway by utilizing A rabies-G-pseudotyped EIAV lentiviral vector encoding the opsin ChR2 gene stereotaxically injected into a deep brain nucleus (Senova et al., 2018). The opsin gene was retrogradely transported to the layer of the entire cortex projecting to the nucleus. A precise anatomical mapping of this pathway was then performed using histology-guided high angular resolution MRI tractography guiding accurately cortical photostimulation. Thus, optogenetic tractography might help design translational neuromodulation studies in NHP models of neuropsychiatric diseases choosing the most appropriate target for the tested hypothesis.
- Stéphane Palfi lab
Addressing viral vector transport issues: We are based in the UK and in the past have relied on the established US Vector Cores for our constructs. An issue that has affected our approach has been reliable delivery of viable constructs due to dry ice partly or fully melting, high costs of reputable couriers for on time delivery and unreliability of the customs process completing in a timely and consistent manner. We plan to purchase also from EU sites (Leuven Vector Core) but the community might consider establishing additional vector core “hubs”, such that laboratories can have access to more “locally-grown” viral constructs with standardized quality control of purity etc.
Growing the resource with rodent and human data: Since every single animal counts and thus failures can jeopardize projects in NHP work, this resource could grow with data from rodent and human data to allow viral construct efficacy to be compared, for example, to identify species differences or lack-there-of in immune response and viral vector efficacy.
Which is best: anesthetized or awake injections? We are also conducting injections under anesthesia and in awake conditions and are not yet sure which approach is better and for which reasons. This resource as it expands will allow additional comparisons such as these to be made.
- Chris Petkov lab
In one of our first attempts at optogenetics, we injected the virus in the operating room following a large craniotomy and durotomy. The craniotomy was closed with the bone flap after the surgery and reopened a month later to install a recording chamber over the injection site to perform stimulation. Because we could not locate the precise injection sites with reference to the chamber recording grid, we spent months fishing for the expressing neurons with our optrode, not knowing whether daily negative results were explained by choosing wrong recording grid coordinates, or overall failed opsin expression. We recommend that, if stimulation is to be performed using a standard recording chamber, that injection be performed beforehand using the same chamber and grid rather than intraoperatively through a new craniotomy/duratomy.
- Michael Platt lab
We developed methods for conducting optogenetics through cranial windows in non-human primates, with targeted injection of virus to imaged functional columns with fine glass pipettes without damage to cortex, easy periodic observation of expression, and optical stimulation using fiber optics or LEDs (Ruiz et al., 2013b). For large areas of transfection (up to several millimeters), we used Hamilton syringes with fine glass tips. For targeting single columns, we chose lentiviral constructs for small focal injections (<1 mm dia.) and used small injection volumes (1–2 μl) with 20 μm tip glass pipettes. Due to frequent clogging, we resorted to 1:2 dilution of the stock viral titers. Injections were made at 3 different cortical depths, which resulted in transfection primarily in layer IVc, V and VI, with little in the superficial layers; this stratified expression may be due to virus serotype. It is unclear why, but typically only half our injections worked. Even so, this strategy proved successful. We were able to demonstrate intensity and wavelength-dependent activation of single nodes (columns) and selective activation/modulation of their associated (e.g. ocular dominance-specific, orientation-specific) columnar networks (Chernov et al., 2018). This approach is compatible with behavioral work (Nassi et al., 2015).
- Anna Roe lab
We performed immunohistochemistry analysis on all animals (Yazdan-Shahmorad et al., 2016; 2018b). Except for a few failed injections, we observed high levels of reporter expression at the site of injection and at distal locations. NeuN and Nissl staining did not show any neuronal toxicity either in the injection site or distal structures for all animals. We were able to record light-evoked activity in all but one of the animals that has positive histological results. In this animal (Monkey B), we injected a vector with a neuro- and gliotropic affinity (AAV2.9) into thalamus, and saw major histocompatibility complex marker (MHC-II) upregulation at the injection site (Yazdan-Shahmorad et al., 2018b). This indicated an ongoing immune reaction in this animal, with mixed cell transduction. Although the animal didn’t present any clinical signs throughout the study, this inflammation could have been responsible for our inability to obtain optically-stimulated neural activity. In another animal (Monkey H) however, we injected the same virus into a different brain area (primary somatosensory cortex) and observed strong expression through both histology and neurophysiology around the site of infusion. We did not perform MHC-II on this animal. This discrepancy could be an indication that either the location of injection or the animal’s immune response to the virus (as suggested by other labs) play important roles in the success of transduction.
- Philip Sabes lab
Our lab can report very good experiences from combining optogenetics with magnetic resonance imaging (MRI and fMRI) to plan injections and to test the effectiveness of optogenetic stimulation with BOLD-fMRI prior to further electrophysiological assessment. However, we find that careful controls are important. For experiments involving subcortical structures, such as lateral geniculate nucleus (Klein et al., Neuron, 2015), we found electrophysiological assessment more accurate to identify the injection target. For the assessment of optogenetic activation effects at the cortical surface, we found that large volume illumination using LED stimulation through the dura can work very well in combination with fMRI, but that this is much more difficult to achieve with electrophysiology, for which we found stimulation in the cortical tissue, very close to the electrode contact, more effective. Particularly for experiments with opto-fMRI, control experiments with other wavelengths and in non-transfected tissue are important in order to rule out artefacts, such as heating-related modulation of the hemodynamic response. As in-vivo fluorescence was not visible in our case, likely due to dural tissue growth, we believe that dural replacement strategies might be beneficial for effective stimulation and electrophysiological targeting.
- Michael C. Schmid lab
When injecting virus into the cortex where visual access is available, the location of the injection site may be clear, but tracking the spread and backflow of the clear virus solution during injection is difficult, if not impossible. To correct this, we add a blue dye (trypan blue) to the virus solution which allows us to visualize the flow of virus (Seidemann et al., 2016). With the addition of the blue dye, we can both 1) ensure that the injected virus diffuses into the surrounding tissue and 2) identify when the virus has leaked from our intended injection site. Additionally, the dye remains visible for a few weeks after injection, which we can use to assess the spread of virus after injection and visually localize the injection sites more easily. We usually find that if we cannot clearly see the blue dye after injection, we will likely not find expression of the intended protein at that site.
- Eyal Seidemann lab
We find it useful to advance a recording electrode alongside (even glued to) the injection cannula, to improve targeting of injections based on functional properties and to avoid leakage of virus into white matter or extra-pial space. Other suggestions include tracking the progress of an injection using a dye (in the case where tubing connects syringe to cannula), and in doing so be wary of inconsistent flow rates which may indicate possibly tissue-damaging buildup and release of pressure. We typically keep the cannula in place for at least 8–10 minutes after each injection, to limit the degree to which viral solution travels up or down the penetration track. Lastly, the virtues of sharpened/tapered fibers have been extolled elsewhere but are worth repeating (see (Dai et al., 2015; Fetsch et al., 2018) for two different sharpening methods). Indeed, none of these suggestions are new and we are grateful to the researchers who published or mentioned them previously.
- Michael Shadlen lab
It is often helpful to pair optogenetic stimulation with electrophysiology to understand the response of the local circuit to stimulation. Typically, in awake behaving experiments, this requires many repeated penetrations in the transfected region. Also, the brain can translate multiple millimeters with respect to the chamber across days with typical burr hole and craniotomy setups. Consequently, for cortical experiments, we recommend injecting in a grid pattern at the site of interest to ensure that a broad enough patch of cortex will express opsin. This greatly increases the reliability of finding the transfected site and allows for penetrations to be spaced out to minimize damage to the cortical tissue. We have also employed a translucent silicone artificial dura within a chamber optimized for two-photon imaging with virally-delivered GCaMP6 (Trautmann et al., 2019). The additional maintenance and cleaning required to maintain healthy, clear access to brain is considerable. However this approach enable glass pipette injections that avoid vasculature and widefield epifluorescence imaging for localizing transgene expression. Broad illumination or highly-localized light delivery may also be performed in a more precise manner, guided by vascular fiducial markers.
- Krishna Shenoy and Karl Deisseroth labs
To help surmount the problems identified in this report, we have been exploring the usefulness of chimeric capsids created through rational design or directed evolution. Such designer vectors will undoubtedly benefit primate optogenetics, but their impact will likely remain limited if tradition holds and they are developed in rodents, for rodents, and then used in primates with fingers crossed that they will work. Even within rodents, specificity of a capsid can be so dramatic that it may behave as expected only within the strain the capsid was developed in, as was the case with AAV-PHP.B (Hordeaux et al., 2018). We encourage all NHP researchers who want to translate new vectors from rodents to monkeys to invest in histological confirmation of efficacy and functionality, first. In our own lab, we have been surprised at how differently chimeric capsids can behave in their patterns of transduction when injected in rodent vs. primate CNS. Without systematic assessment of how a particular vector functions in the physiological and immunological environment of the primate brain, one risks wasting resources in the training and testing of monkeys, only to find that the vector failed to transduce the target or caused off-target transduction and major confounds for data interpretation.
- Marc Sommer lab
It’s worth noting that, by combining optogenetics with calcium imaging, the implementation of all-optical interrogation (AOI) allows simultaneous optogenetic manipulation and noninvasive two-photon readout (Ju et al., 2018; Packer et al., 2015). To achieve AOI in NHPs at current stage, we suggest to infect cortical neurons with C1V1-ts-EYFP and GCaMP5G/GCamP6s. Though ChR2 is commonly used as the optogenetic actuator, C1V1 is highly recommended due to its high conductance and red-shifted absorption spectrum, which makes it a preferable choice for AOI experiments, since C1V1’s excitation spectrum is well separated from that of GCaMPs. Moreover, in spite of the neighboring spectrum between the fluorescence of GCaMPs and EYFP, they could nevertheless be distinguished by using respective excitation laser and filter (a 920 nm excitation laser with a 500 ± 12.5 nm filter for imaging GCaMPs, and a 1040 nm excitation laser with a 525 ± 35 nm filter for EYFP). On the basis of current AOI achievements in NHPs, it still requires a mass of practice to achieve high efficiency of co-expression of C1V1 and GCaMPs, and to develop soma-targeted C1V1-EYFP for both high efficiency photoactivation and in-vivo quantification of expression.
- Shiming Tang lab
We would like to underscore the benefit of histological verification of the transfection results in rhesus monkeys. We looked at the transduction spread at injection sites, expression specificity, and opsin trafficking into long-range axonal projections for AAV2/5-CaMKIIα-eNpHR3.0-mCherry, -ChR2-eYFP, -C1V1-mCherry. The different constructs displayed different phenotypes in terms of opsin trafficking and cellular expression despite the same promoter and serotype of the virus used. Among the three opsins, only eNpHR3.0-mCherry and ChR2-eYFP were present in axonal projections to downstream target areas. The intracellular distribution of opsin proteins differed: ChR2-eYFP had almost exclusive plasma membrane localization, while eNpHR3.0-mCherry and C1V1-mCherry showed additional intracellular accumulations, which might affect neuronal survival in the long-term. Standard hematoxylin and eosin staining in two out of three tested animals showed an increased number of eosinophilic granulocytes and mononuclear perivascular cuffing, mainly in the vicinity of the injection locations. Since these two animals had an extended history (several years) of extracellular recordings with acute electrodes, we could not conclude whether the signs of pathology were a result of the virus injections and following opsin expression, or of any other previous experimental procedure. The results of our histological analysis have been recently published.
- Stefan Treue and Alexander Gail labs
To substantially increase the success of optogenetic NHP experiments during perceptual and cognitive tasks, we guide trajectories and targets by fMRI and electrophysiology. For example, we found that very small deviations from fMRI-defined task-related activations already resulted in negative optogenetic results, both at behavioral and physiological levels. Currently, we use fMRI in all stages of the experiment: for task optimization, target localization, trajectory calculations, and for measuring functional optogenetic-induced stimulation’s effects (Gerits et al., 2012). In addition, we aim to avoid major blood vessels and ventricles via CT- or MR-based angiography and MRI. In addition, we successfully used MnCl-based visualization of the vector injections using anatomical MRI. We also found that custom-designed optrodes remain a major culprit for NHP optogenetics as they cause too much local tissue damage. We also experienced that identical vectors (even from the same batch) can yield substantially different results in rodents (Scheyltjens et al., 2015) and macaques (Gerits et al., 2015), and even between different cortical regions of macaques (unpublished results).
- Wim Vanduffel lab
To inject viral vectors in deep structures, we found it useful to implant first a recording chamber and, after a post-operative MRI, perform a series of recordings to verify the depth of potential injection sites, measuring known response properties of the area. We employed the same microdrive, grid and guide tube to perform the injections simply by replacing the microelectrode with the injection needle. Also, we found it useful to co-inject manganese(II) chloride tetrahydrate, which allowed visualization of the injection site with MRI after the injection. One major challenge when performing photostimulation of deep brain structures is to ensure that a sufficient amount of light reaches the neurons in order to activate the opsins. In our experiences, we found it easier to produce a photostimulation effect by using a red-shifted opsin, since red wavelengths are propagated better in brain tissue compared with shorter wavelengths.
- Rufin Vogels lab
The goal of our experiments was to verify that ArchT could be used in the superior colliculus and thalamus to inactivate the corollary discharge pathway to cerebral cortex. Optogenetic inactivation permits active and control trials to be interdigitated, whereas the GABA agonist muscimol acts on all trials within the hour or so it is active. Interdigitation greatly reduces the variance because it controls for changes in a monkey’s behavior over time. We made injections in SC because we knew exactly what effects muscimol has on saccades and we could therefore see if ArchT would produce the same deficits. It did: a shift in saccadic end point, increase in latency, and a decrease in velocity. But the problem was that the effects were small compared to previous muscimol injections in SC. The reason might be the relatively small area of inactivation area or the fact that that archT acts on ion pumps rather than ion channels. In any case the effect was too small to use ArchT in an untested region such as MD thalamus.
-Bob Wurtz lab
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| NHP Optogenetics Open Database | This paper | https://osf.io/mknfu/ |
| Other | ||
| Addgene website | Addgene | https://www.addgene.org |
Highlights.
Openly shared, non-human primate optogenetics data resource
Over 500 entries of unpublished data
Efficiency analysis of experimental parameters
Advice from leading NHP optogenetics labs
ACKNOWLEDGEMENTS
This manuscript was inspired by the PRIME-DE NeuroResource authored by Milham et al. (Milham et al., 2018). We would like to thank the Jazayeri lab for the initial impetus for this collaboration, and the MIT group (DiCarlo, Desimone & Jazayeri labs) for providing a first in-house database of optogenetics experiments in NHP that was used as a backbone for the current initiative. We are grateful to the Open Science Framework for hosting the database.
Primary support for the work by M.L.P. and S.T. is provided by the Charles E. Kaufman Foundation and the Canada Banting Fellowship.
Primary support for AA’s lab is provided by the NIH (grants R01 EY026812, R01 EY019743, BRAIN U01 NS099702) and the NSF (IOS 1755431). Support for LN is additionally provided by an NIH K99 grant (EY029374).
The support for the work by A.A (Arash Afraz) (in part) came from the Intramural Research Program of the NIMH (ZIAMH002958).
Support for the work contributed by M.A.B was provided by NIH R01 EY013962 and R21 EY024516
R.D. acknowledges funding from NIH Grant R01EY029666
Primary support for the work by J.D is provided by the National Natural Science Foundation of China (31600870), the Youth Innovation Promotion Association CAS (2017120) and the Chinese Academy of Sciences International Partnership Program (172644KYSB20170004).
Primary support for the work by D.L.S. and J.D. is provided by Defense Advanced Research Projects Agency (DARPA) Biological Technology Office (BTO) “REPAIR” award N66001-10-C-2010 and NIH-EY014681.
Support for the work from the Dragoi/Janz/Spudich labs is provided by the BRAIN Initiative (1U01MH109146) to V.D., R.J., and J.L.S.
Primary support for the work by N.F. and M.K. is provided by the Brain/MINDS (JP19dm0207001) and the KAKENHI (17H06034).
Support for the work by the Treue, Gail, and Scherberger labs was provided by the Leibniz Association (WGL SAW-2014-DPZ-1) and the German Research Foundation (FOR 1847).
Support for the work by D.L.A., M.J.C., X.H., A.G., Y.S. and T.W. at Emory University is provided by NIH/NINDS (P50 NS098685 [Udall Center grant]), and a grant from NIH/ORIP to the Yerkes Center (P51 OD011132).
Primary support for the work by H.A., K.M., and O.H. is provided by National Eye Institute (project number: 1ZIAEY000415-16).
Primary support for the work by E.D.R., S.W.E., E.H., and M.J. is provided by the National Institute of Health (NINDS-NS078127).
Primary support for the work by AA, AK and SSS was provided by the NIH (EY026240), Fight for Sight, and Research to Prevent Blindness.
Primary support for the work by S.G. and J.H.R.M. is provided by NIH R01EY005911.
Primary support for the work by S.P. and S.S. is provided by Fondation de France, Oxford Biomedica (Oxford, UK), ARSC (France).
Primary support for the work by R.S.M. and C.I.P is provided by Wellcome Trust Investigator Award, European Research Council Consolidator Award (MECHIDENT) and NIH NIDCD R01 (with M. Howard at University of Iowa).
Development of this technique was supported by NIH EY022853 to AWR and Gene Stoner.
Primary support for the work by C.K., M.O.-R. and M.C.S. is provided by DFG Emmy Noether grant SCHM 2806/1-1 and European Research Council grant OptoVision 637638
This work was supported by the Wellcome Trust (095495), the European Research Council (293549), and NIH Caltech Conte Center (P50MH094258).
Primary support for the work by G. B., S.C., Y.C., M.P.W and E.S. is provided by NIH-BRAIN U01NS099720, NEI R01EY016454 and DARPA-NESD-N66001-17-C-4012.
Primary support for the work by C.R.F. and M.N.S. was provided by the Howard Hughes Medical Institute and the National Eye Institute (R01 EY11378).
Primary support for the work by D.J.O., K.D., and K.V.S. is provided by an NSF Graduate Research Fellowship, Dr. Regina Casper Stanford Graduate Fellowship, Defense
Advanced Research Projects Agency (DARPA) Biological Technology Office (BTO) “REPAIR” award N66001-10-C-2010 (K.V.S. and K.D.), DARPA BTO “NeuroFAST” award W911NF-14-2-0013 (K.D. and K.V.S.), the Simons Foundation Collaboration on the Global Brain awards 325380 and 543045 (K.V.S.), NIH Director’s Pioneer Award 8DP1HD075623 (K.V.S.), and the Howard Hughes Medical Institute (K.D. and K.V.S.).
Primary support for the work of KI, MM, MT was provided by the Ministry of Education, Science, Sports, Culture, and Technology of Japan, by the Japan Science and Technology Agency and by Japan Agency for Medical Research and Development.
Primary support for the work by N. J., R. J. and S. T. is provided by the National Natural Science Foundation of China and the National Basic Research Program of China.
Primary support for the work by M.A.G. and A.T. is provided by the Medical Research Council
Primary support for the work by W.V. and P.F.B. is provided by the Research Foundation Flanders (FWO-Flanders), Internal KU Leuven funds (C14/17/109), and the European Union’s Horizon 2020 Framework Programme for Research and Innovation under Grant Agreement No 785907
Primary support for the work by R.V and F.F. by Fonds voor Wetenschappelijk Onderzoek Vlaanderen and Internal KU Leuven funds (C14/17/109).
Primary support for the work by RHW and JC was provided by the intramural research program at NIH.
Primary support for the work by A.Y. and P.N.S. was supported by an American Heart Association Western States Affiliate Postdoctoral Fellowship (A.Y.-S.), Defense Advanced Research Projects Agency (DARPA) Reorganization and Plasticity to Accelerate Injury Recovery (REPAIR; N66001-10-C-2010), and by the UCSF Neuroscience Imaging Center
The work by S.W.E, E.A.H., E.D.R. and M.J. was partially supported by NIH R01 R01NS078127.
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes the NHP Optogenetics Open Database and its associated files which can be found on the Open Science Framework at https://osf.io/mknfu/.
DECLARATION OF INTERESTS
S.Pa. received consulting honorarium from Oxfordbiomedica.
K.V.S. is a consultant for Neuralink Corp. and is on the scientific advisory board for CTRL-Labs Inc., MIND-X Inc., Inscopix Inc. and Heal Inc. These entities did not support this work.
P.N.S. has financial interest in Neuralink Corp., a company that is developing clinical therapies using brain stimulation.
REFERENCES
- Acker L, Pino EN, Boyden ES, and Desimone R (2016). FEF inactivation with improved optogenetic methods. Proceedings of the National Academy of Sciences 113, E7297–E7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afraz A, Boyden ES, and DiCarlo JJ (2015). Optogenetic and pharmacological suppression of spatial clusters of face neurons reveal their causal role in face gender discrimination. Proceedings of the National Academy of Sciences 112, 6730–6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amita H, Kim HF, Inoue K-I, Takada M, and Hikosaka O (2020). Optogenetic manipulation of a value-coding pathway from the primate caudate tail facilitates saccadic gaze shift. Nat Commun 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei AR, Pojoga S, Janz R, and Dragoi V (2019). Integration of cortical population signals for visual perception. Nat Commun 10, 3832–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson-Hooper ZB, Fragiadakis GK, Spitzer MH, Madhireddy D, McIlwain D, and Nolan GP (2019). A comprehensive atlas of immunological differences between humans, mice and non-human primates. bioRxiv 173173, 1166–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen MO, El-Nahal HG, and Sommer MA (2019). Transduction of Craniofacial Motoneurons Following Intramuscular Injections of Canine Adenovirus Type-2 (CAV-2) in Rhesus Macaques. Front Neuroanat 13, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, and Deisseroth K (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nature Neuroscience 8, 1263–1268. [DOI] [PubMed] [Google Scholar]
- Calcedo R, Chichester JA, and Wilson JM (2018). Assessment of Humoral, Innate, and T-Cell Immune Responses to Adeno-Associated Virus Vectors. Human Gene Therapy Methods 29, 86–95. [DOI] [PubMed] [Google Scholar]
- Calcedo R, Franco J, Qin Q, Richardson DW, Mason JB, Boyd S, and Wilson JM (2015). Preexisting Neutralizing Antibodies to Adeno-Associated Virus Capsids in Large Animals Other Than Monkeys May Confound In Vivo Gene Therapy Studies. Human Gene Therapy Methods 26, 103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh J, Monosov IE, McAlonan K, Berman R, Smith MK, Cao V, Wang KH, Boyden ES, and Wurtz RH (2012). Optogenetic Inactivation Modifies Monkey Visuomotor Behavior. Neuron 76, 901–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Goffart L, and Sparks DL (2001). A simple method for constructing microinjectrodes for reversible inactivation in behaving monkeys. Journal of Neuroscience Methods 107, 81–85. [DOI] [PubMed] [Google Scholar]
- Chernov MM, Friedman RM, Chen G, Stoner GR, and Roe AW (2018). Functionally specific optogenetic modulation in primate visual cortex. Proceedings of the National Academy of Sciences 115, 10505–10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow BY, and Boyden ES (2013). Optogenetics and translational medicine. Sci Transl Med 5, 177ps5–177ps5. [DOI] [PubMed] [Google Scholar]
- Dai J, Brooks DI, and Sheinberg DL (2014). Optogenetic and electrical microstimulation systematically bias visuospatial choice in primates. Curr Biol 24, 63–69. [DOI] [PubMed] [Google Scholar]
- Dai J, Ozden I, Brooks DI, Wagner F, May T, Agha NS, Brush B, Borton D, Nurmikko AV, and Sheinberg DL (2015). Modified toolbox for optogenetics in the nonhuman primate. Neurophotonics 2, 031202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De A, El-Shamayleh Y, and Horwitz GD (2020). Fast and reversible neural inactivation in macaque cortex by optogenetic stimulation of GABAergic neurons. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debinski W, and Tatter SB (2009). Convection-enhanced delivery for the treatment of brain tumors. Expert Rev Neurother 9, 1519–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K (2010). Optogenetics. Nat Meth 8, 26–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K (2012). Optogenetics and psychiatry: applications, challenges, and opportunities. Biol. Psychiatry 71, 1030–1032. [DOI] [PubMed] [Google Scholar]
- Deyle DR, and Russell DW (2009). Adeno-associated virus vector integration. Curr. Opin. Mol. Ther 11, 442–447. [PMC free article] [PubMed] [Google Scholar]
- Diester I, Kaufman MT, Mogri M, Pashaie R, Goo W, Yizhar O, Ramakrishnan C, Deisseroth K, and Shenoy KV (2011). An optogenetic toolbox designed for primates. Nature Neuroscience 14, 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodiya HB, Bjorklund T, Stansell J, Mandel RJ, Kirik D, and Kordower JH (2010). Differential transduction following basal ganglia administration of distinct pseudotyped AAV capsid serotypes in nonhuman primates. Mol. Ther 18, 579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina T, Obara K, Watakabe A, Masamizu Y, Terada S-I, Matoba R, Takaji M, Hatanaka N, Nambu A, Mizukami H, et al. (2019). Arm movements induced by noninvasive optogenetic stimulation of the motor cortex in the common marmoset. Proc Natl Acad Sci USA 90, 201903445–201903447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Editorial (2019). The importance of no evidence. Nature Human Behaviour 3, 197–197. [DOI] [PubMed] [Google Scholar]
- El-Shamayleh Y, and Horwitz GD (2019). Primate optogenetics: Progress and prognosis. Proceedings of the National Academy of Sciences 116, 26195–26203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shamayleh Y, Kojima Y, Soetedjo R, and Horwitz GD (2017). Selective Optogenetic Control of Purkinje Cells in Monkey Cerebellum. Neuron 95, 51–62.e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbrini F, Van den Haute C, De Vitis M, Baekelandt V, Vanduffel W, and Vogels R (2019). Probing the Mechanisms of Repetition Suppression in Inferior Temporal Cortex with Optogenetics. Current Biology 29, 1988–1998.e4. [DOI] [PubMed] [Google Scholar]
- Fetsch CR, Odean NN, Jeurissen D, El-Shamayleh Y, Horwitz GD, and Shadlen MN (2018). Focal optogenetic suppression in macaque area MT biases direction discrimination and decision confidence, but only transiently. Elife 7, 1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortuna MG, Hüer J, Guo H, Gruber J, Gruber-Dujardin E, Staiger JF, Scherberger H, Treue S, and Gail A (2020). Histological assessment of optogenetic tools to study fronto-visual and fronto-parietal cortical networks in the rhesus macaque. Sci Rep 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericks JM, Dash KE, Jaskot EM, Bennett TW, Lerchner W, Dold G, Ide D, Cummins AC, Minassian, Der VH, Turchi JN, et al. (2020). Methods for Mechanical Delivery of Viral Vectors into Rhesus Monkey Brain. Journal of Neuroscience Methods 108730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A, Homma D, Gibb LG, Amemori K-I, Rubin SJ, Hood AS, Riad MH, and Graybiel AM (2015). A Corticostriatal Path Targeting Striosomes Controls Decision-Making under Conflict. Cell 161, 1320–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan A, Hu X, Smith Y, and Wichmann T (2016). Effects of Optogenetic Activation of Corticothalamic Terminals in the Motor Thalamus of Awake Monkeys. J Neurosci 36, 3519–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan A, Caiola MJ, and Albaugh DL (2017). Advances in optogenetic and chemogenetic methods to study brain circuits in non-human primates. Journal of Neural Transmission 125, 547–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan A, Hu X, Smith Y, and Wichmann T (2012). In vivo optogenetic control of striatal and thalamic neurons in non-human primates. PLoS ONE 7, e50808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Alvira MR, Somanathan S, Lu Y, Vandenberghe LH, Rux JJ, Calcedo R, Sanmiguel J, Abbas Z, and Wilson JM (2003). Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc Natl Acad Sci USA 100, 6081–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerits A, Farivar R, Rosen BR, Wald LL, Boyden ES, and Vanduffel W (2012). Optogenetically Induced Behavioral and Functional Network Changes in Primates. Curr Biol 22, 1722–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerits A, Vancraeyenest P, Vreysen S, Laramée M-E, Michiels A, Gijsbers R, Van den Haute C, Moons L, Debyser Z, Baekelandt V, et al. (2015). Serotype-dependent transduction efficiencies of recombinant adeno-associated viral vectors in monkey neocortex. Neurophotonics 2, 031209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, et al. (2014). Natural neural projection dynamics underlying social behavior. Cell 157, 1535–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurda BL, De Guilhem De Lataillade A, Bell P, Zhu Y, Yu H, Wang P, Bagel J, Vite CH, Sikora T, Hinderer C, et al. (2016). Evaluation of AAV-mediated Gene Therapy for Central Nervous System Disease in Canine Mucopolysaccharidosis VII. Mol. Ther 24, 206–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Chow BY, Zhou H, Klapoetke NC, Chuong A, Rajimehr R, Yang A, Baratta MV, Winkle J, Desimone R, et al. (2011). A High-Light Sensitivity Optical Neural Silencer: Development and Application to Optogenetic Control of Non-Human Primate Cortex. Front. Syst. Neurosci 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Qian X, Bernstein JG, Zhou H-H, Franzesi GT, Stern P, Bronson RT, Graybiel AM, Desimone R, and Boyden ES (2009). Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron 62, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hordeaux J, Wang Q, Katz N, Buza EL, Bell P, and Wilson JM (2018). The Neurotropic Properties of AAV-PHP.B Are Limited to C57BL/6J Mice. Mol. Ther 26, 664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K-I, Takada M, and Matsumoto M (2015). Neuronal and behavioural modulations by pathway-selective optogenetic stimulation of the primate oculomotor system. Nat Commun 6, 8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri M, Lindbloom-Brown Z, and Horwitz GD (2012). Saccadic eye movements evoked by optogenetic activation of primate V1. Nature Neuroscience 15, 1368–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju N, Jiang R, Macknik SL, Martinez-Conde S, and Tang S (2018). Long-term all-optical interrogation of cortical neurons in awake-behaving nonhuman primates. PLoS Biol. 16, e2005839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kells AP, Hadaczek P, Yin D, Bringas J, Varenika V, Forsayeth J, and Bankiewicz KS (2009). Efficient gene therapy-based method for the delivery of therapeutics to primate cortex. Proc Natl Acad Sci USA 106, 2407–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khateeb K, Griggs DJ, Sabes PN, and Yazdan-Shahmorad A (2019). Convection Enhanced Delivery of Optogenetic Adeno-associated Viral Vector to the Cortex of Rhesus Macaque Under Guidance of Online MRI Images. J Vis Exp 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C, Evrard HC, Shapcott KA, Haverkamp S, Logothetis NK, and Schmid MC (2016). Cell-Targeted Optogenetics and Electrical Microstimulation Reveal the Primate Koniocellular Projection to Supra-granular Visual Cortex. Neuron 90, 143–151. [DOI] [PubMed] [Google Scholar]
- Kliem MA, and Wichmann T (2004). A method to record changes in local neuronal discharge in response to infusion of small drug quantities in awake monkeys. Journal of Neuroscience Methods 138, 45–49. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Sugano E, Tomita H, and Fujii N (2017). A Chronically Implantable Bidirectional Neural Interface for Non-human Primates. Front Neurosci 11, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauze MT, Saito R, Noble C, Tamas M, Bringas J, Park JW, Berger MS, and Bankiewicz K (2005). Reflux-free cannula for convection-enhanced high-speed delivery of therapeutic agents: Technical note. J. Neurosurg 103, 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauze MT, Vandenberg SR, Yamashita Y, Saito R, Forsayeth J, Noble C, Park J, and Bankiewicz KS (2008). Safety of real-time convection-enhanced delivery of liposomes to primate brain: a long-term retrospective. Exp. Neurol 210, 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledochowitsch P, Yazdan-Shahmorad A, Bouchard KE, Diaz-Botia C, Hanson TL, He J-W, Seybold BA, Olivero E, Phillips EAK, Blanche TJ, et al. (2015). Strategies for optical control and simultaneous electrical readout of extended cortical circuits. Journal of Neuroscience Methods 256, 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerchner W, Corgiat B, Minassian, Der V, Saunders RC, and Richmond BJ (2014). Injection parameters and virus dependent choice of promoters to improve neuron targeting in the nonhuman primate brain. Gene Ther. 21, 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, and Tonegawa S (2012). Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 484, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Truccolo W, Wagner FB, Vargas-Irwin CE, Ozden I, Zimmermann JB, May T, Agha NS, Wang J, and Nurmikko AV (2015). Optogenetically induced spatiotemporal gamma oscillations and neuronal spiking activity in primate motor cortex. J Neurophysiol 113, 3574–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall M, Nummela SU, Coop S, Disney A, Mitchell JF, and Miller CT (2016). Optogenetic manipulation of neural circuits in awake marmosets. J Neurophysiol 116, 1286–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimon BE, Diaz M, Revol ECM, Schneider AM, Leaker B, Varela CE, Srinivasan S, Weber MB, and Herr HM (2018). Optogenetic Peripheral Nerve Immunogenicity. Sci Rep 8, 14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markakis EA, Vives KP, Bober J, Leichtle S, Leranth C, Beecham J, Elsworth JD, Roth RH, Samulski RJ, and Redmond DE (2010). Comparative transduction efficiency of AAV vector serotypes 1–6 in the substantia nigra and striatum of the primate brain. Mol. Ther 18, 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T, Ozden I, Brush B, Borton D, Wagner F, Agha N, Sheinberg DL, and Nurmikko AV (2014). Detection of optogenetic stimulation in somatosensory cortex by non-human primates--towards artificial tactile sensation. PLoS ONE 9, e114529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor JE, Godat T, Dhakal KR, Parkins K, Strazzeri JM, Bateman BA, Fischer WS, Williams DR, and Merigan WH (2020). Optogenetic restoration of retinal ganglion cell activity in the living primate. Nat Commun 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AM, Sonabend AM, and Bruce JN (2017). Convection-Enhanced Delivery. Neurotherapeutics 14, 358–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza SD, El-Shamayleh Y, and Horwitz GD (2017). AAV-mediated delivery of optogenetic constructs to the macaque brain triggers humoral immune responses. J Neurophysiol 117, 2004–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milham MP, Ai L, Koo B, Xu T, Amiez C, Balezeau F, Baxter MG, Blezer ELA, Brochier T, Chen A, et al. (2018). An Open Resource for Non-human Primate Imaging. Neuron 100, 61–74.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monosov IE, and Thompson KG (2009). Frontal eye field activity enhances object identification during covert visual search. J Neurophysiol 102, 3656–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi Y, Okubo K, Sato T, Hashimoto M, and Tanifuji M (2019). Optical intrinsic signal imaging with optogenetics reveals functional cortico-cortical connectivity at the columnar level in living macaques. Sci Rep 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandy A, Nassi JJ, Jadi MP, and Reynolds J (2019). Optogenetically induced low-frequency correlations impair perception. Elife 8, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassi JJ, Avery MC, Cetin AH, Roe AW, and Reynolds JH (2015). Optogenetic Activation of Normalization in Alert Macaque Visual Cortex. Neuron 86, 1504–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurminen L, Merlin S, Bijanzadeh M, Federer F, and Angelucci A (2018). Top-down feedback controls spatial summation and response amplitude in primate visual cortex. Nat Commun 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguchi M, Okajima M, Tanaka S, Koizumi M, Kikusui T, Ichihara N, Kato S, Kobayashi K, and Sakagami M (2015). Double Virus Vector Infection to the Prefrontal Network of the Macaque Brain. PLoS ONE 10, e0132825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohayon S, Grimaldi P, Schweers N, and Tsao DY (2013). Saccade modulation by optical and electrical stimulation in the macaque frontal eye field. J Neurosci 33, 16684–16697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozden I, Wang J, Lu Y, May T, Lee J, Goo W, O’Shea DJ, Kalanithi P, Diester I, Diagne M, et al. (2013). A coaxial optrode as multifunction write-read probe for optogenetic studies in non-human primates. Journal of Neuroscience Methods 219, 142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Russell LE, Dalgleish HWP, and Häusser M (2015). Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat Meth 12, 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz O, Lustig BR, Nassi JJ, Cetin A, Reynolds JH, Albright TD, Callaway EM, Stoner GR, and Roe AW (2013a). Optogenetics through windows on the brain in the nonhuman primate. J Neurophysiol 110, 1455–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz O, Lustig BR, Nassi JJ, Cetin A, Reynolds JH, Albright TD, Callaway EM, Stoner GR, and Roe AW (2013b). Optogenetics through windows on the brain in the nonhuman primate. J Neurophysiol 110, 1455–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheyltjens I, Laramée M-E, Van den Haute C, Gijsbers R, Debyser Z, Baekelandt V, Vreysen S, and Arckens L (2015). Evaluation of the expression pattern of rAAV2/1, 2/5, 2/7, 2/8, and 2/9 serotypes with different promoters in the mouse visual cortex. J. Comp. Neurol 523, 2019–2042. [DOI] [PubMed] [Google Scholar]
- Seidemann E, Chen Y, Bai Y, Chen SC, Mehta P, Kajs BL, Geisler WS, and Zemelman BV (2016). Calcium imaging with genetically encoded indicators in behaving primates. Elife 5, 3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senova S, Poupon C, Dauguet J, Stewart HJ, x000E9 GPD, Jan C, Hosomi K, Ralph GS, Barnes L, Drouot X, et al. (2018). Optogenetic Tractographyfor anatomo-functional characterization of cortico-subcortical neural circuits in non-human primates. Sci Rep 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea DJOX, Kalanithi P, Ferenczi EA, Hsueh B, Chandrasekaran C, Goo W, Diester I, Ramakrishnan C, Kaufman MT, Ryu SI, et al. (2018). Development of an optogenetictoolkit for neural circuit dissectionin squirrel monkeys. Sci Rep 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer WR, Lak A, Yang A, Borel M, Paulsen O, Boyden ES, and Schultz W (2016). Dopamine Neuron-Specific Optogenetic Stimulation in Rhesus Macaques. Cell 166, 1564–1571.e1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Ohashi Y, Tsubota T, Takeuchi D, Hirabayashi T, Yaguchi M, Matsuyama M, Sekine T, and Miyashita Y (2012). Journal of Neuroscience Methods. Journal of Neuroscience Methods 211, 49–57. [DOI] [PubMed] [Google Scholar]
- Tamura K, Takeda M, Setsuie R, Tsubota T, Hirabayashi T, Miyamoto K, and Miyashita Y (2017). Conversion of object identity to object-general semantic value in the primate temporal cortex. Science 357, 687–692. [DOI] [PubMed] [Google Scholar]
- Trautmann EM, O’Shea DJ, Sun X, Marshel JH, Crow A, Hsueh B, Vesuna S, Cofer L, Bohner G, Allen W, et al. (2019). Dendritic calcium signals in rhesus macaque motor cortex drive an optical brain-computer interface. bioRxiv 6, 780486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunematsu T, Kilduff TS, Boyden ES, Takahashi S, Tominaga M, and Yamanaka A (2011). Acute optogenetic silencing of orexin/hypocretin neurons induces slow-wave sleep in mice. J Neurosci 31, 10529–10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varenika V, Kells AP, Valles F, Hadaczek P, Forsayeth J, and Bankiewicz KS (2009). Controlled dissemination of AAV vectors in the primate brain. Prog. Brain Res 175, 163–172. [DOI] [PubMed] [Google Scholar]
- Wang D, Tai PWL, and Gao G (2019). Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov 18, 358–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Calcedo R, Bell P, Lin J, Grant RL, Siegel DL, and Wilson JM (2011). Impact of pre-existing immunity on gene transfer to nonhuman primate liver with adeno-associated virus 8 vectors. Hum. Gene Ther 22, 1389–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Sano H, Chiken S, Kobayashi K, Fukata Y, Fukata M, Mushiake H, and Nambu A (2020). Forelimb movements evoked by optogenetic stimulation of the macaque motor cortex. Nat Commun 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JJ, Watson AM, Vazquez AL, and Schwartz AB (2019). Viral-Mediated Optogenetic Stimulation of Peripheral Motor Nerves in Non-human Primates. Front Neurosci 13, 1821–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, and Südhof TC (2013). A neural circuit for memory specificity and generalization. Science 339, 1290–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdan-Shahmorad A, Diaz-Botia C, Hanson TL, Kharazia V, Ledochowitsch P, Maharbiz MM, and Sabes PN (2016). A Large-Scale Interface for Optogenetic Stimulation and Recording in Nonhuman Primates. Neuron 89, 927–939. [DOI] [PubMed] [Google Scholar]
- Yazdan-Shahmorad A, Silversmith DB, Kharazia V, and Sabes PN (2018a). Targeted cortical reorganization using optogenetics in non-human primates. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdan-Shahmorad A, Tian N, Kharazia V, Samaranch L, Kells A, Bringas J, He J, Bankiewicz K, and Sabes PN (2018b). Widespread optogenetic expression in macaque cortex obtained with MR-guided, convection enhanced delivery (CED) of AAV vector to the thalamus. Journal of Neuroscience Methods 293, 347–358. [DOI] [PubMed] [Google Scholar]
- Youssef J, Novosad SA, and Winthrop KL (2016). Infection Risk and Safety of Corticosteroid Use. Rheum. Dis. Clin. North Am 42, 157–76–ix–x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Original data for all results in this paper is available at https://osf.io/mknfu/. For a non-anonymized version of the database including names of laboratories who conducted each test, please reach out to the lead contact.


