Abstract
Many individuals mount nearly identical antibody responses to SARS-CoV-2. To gain insight into how the viral spike (S) protein receptor-binding domain (RBD) might evolve in response to common antibody responses, we studied mutations occurring during virus evolution in a persistently infected immunocompromised individual. We use antibody Fab/RBD structures to predict, and pseudotypes to confirm, that mutations found in late-stage evolved S variants confer resistance to a common class of SARS-CoV-2 neutralizing antibodies we isolated from a healthy COVID-19 convalescent donor. Resistance extends to the polyclonal serum immunoglobulins of four out of four healthy convalescent donors we tested and to monoclonal antibodies in clinical use. We further show that affinity maturation is unimportant for wild-type virus neutralization but is critical to neutralization breadth. Because the mutations we studied foreshadowed emerging variants that are now circulating across the globe, our results have implications to the long-term efficacy of S-directed countermeasures.
Keywords: SARS-CoV-2, antibody neutralization, neutralization escape, COVID-19, immunocompromised host, variants of concern, affinity maturation
Graphical abstract
Structural and functional analysis of the evolution of SARS-CoV-2 in a persistently infected immunocompromised individual yields insights into mutations that foreshadow emerging viral variants worldwide.
Introduction
SARS-CoV-2 has infected over 110 million individuals worldwide, resulting in over 2.4 million deaths to date. The SARS-CoV-2 spike protein (S) is a central target for vaccine and drug design efforts (Abraham, 2020; Krammer, 2020). S is heavily glycosylated and forms trimers of heterodimers on the virion surface. Each S protomer has two functional subunits; S1, which contains a receptor-binding domain (RBD) that binds the cellular receptor, ACE2 (Hoffmann et al., 2020; Zhou et al., 2020), and S2, which mediates fusion of the viral and host cell membranes during viral entry. Epitopes for neutralizing antibodies include non-overlapping sites on the RBD and the S1 N-terminal domain (NTD) (Chi et al., 2020; Du et al., 2020; Hansen et al., 2020; Liu et al., 2020; Robbiani et al., 2020; Wu et al., 2020). VH3-53 and VH3-66 antibody genes are identical except for a single amino acid change in an antibody framework region (FWR) (Lefranc and Lefranc, 2001), and potent neutralizing antibodies derived from these germline genes have been isolated from multiple COVID-19 convalescent individuals (Du et al., 2020; Robbiani et al., 2020; Rogers et al., 2020; Seydoux et al., 2020; Shi et al., 2020; Wu et al., 2020; Yuan et al., 2020a). All engage the RBD and interfere with viral entry by blocking ACE2 engagement.
Coronaviruses encode a viral exonuclease that increases replication fidelity (Denison et al., 2011), which probably makes antigenic drift in SARS-CoV-2 less significant than in other enveloped RNA viruses. Changes in SARS-CoV-2 S have nonetheless occurred rover time and become fixed among circulating variants; the D614GS mutation is a prime example (Yurkovetskiy et al., 2020). This mutation, however, does not seem to impact the activity of RBD-targeting neutralizing antibodies (Yurkovetskiy et al., 2020). Ultimately, evolution of S antibody escape mutations could impact the long-term effectiveness of vaccines and monoclonal antibody-based therapeutics that target S.
In our efforts to study SARS-CoV-2 antibody neutralization and to predict escape mutations, we examined sequences of S variants that evolved in a persistently infected individual receiving B cell depleting therapy (Choi et al., 2020). We show that mutations acquired in S during persistent infection confer pseudotype resistance to a large panel of clonally related VH3-53-derived neutralizing antibodies we isolated from a healthy COVID-19 convalescent donor. Resistance also extended to B38 (Wu et al., 2020) and CC12.1 (Rogers et al., 2020), two VH3-53-derived antibodies isolated from other COVID-19 convalescent donors, to components of the REGN-COV2 antibody cocktail (Baum et al., 2020; Hansen et al., 2020), and to the polyclonal immunoglobulins (IgG) of four out of four healthy convalescent donors we tested. Antibody affinity enhancements, which we performed based on X-ray crystal structures we determined of VH3-53-derived antibody Fabs bound to the RBD, can, in part, counter neutralization escape caused by S changes that occurred in the immunocompromised host. Notably, the S mutations we studied foreshadowed the appearance of emerging SARS-CoV-2 variants.
Results
Isolated VH3-53-derived neutralizing antibodies bind the RBD with varying affinity
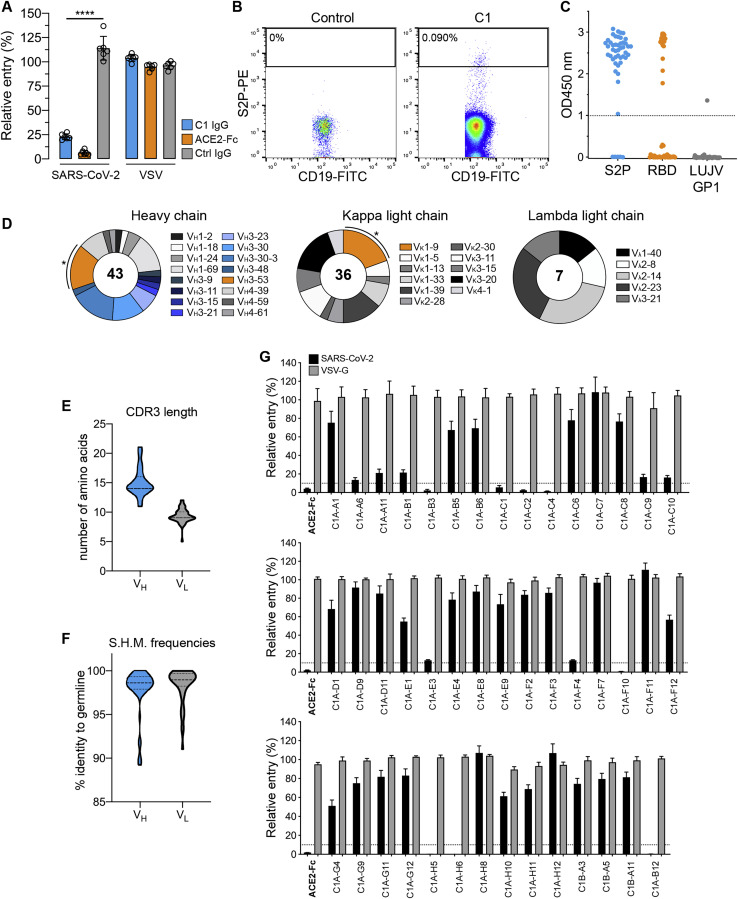
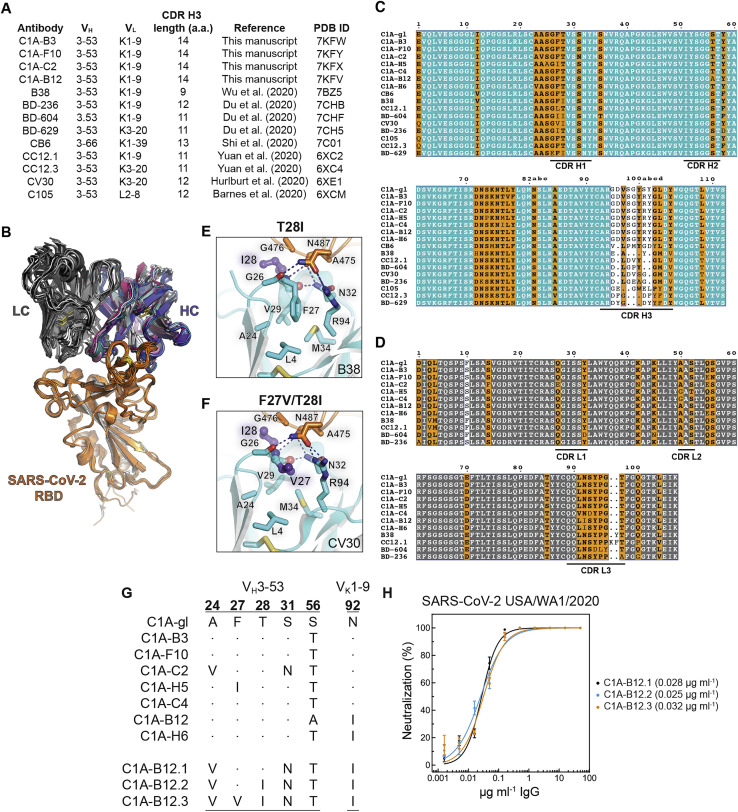
To study neutralizing antibody responses to SARS-CoV-2, we obtained a peripheral blood sample from a healthy adult male individual (“C1”) who had been infected by SARS-CoV-2 5 weeks prior to sampling. Polyclonal IgG purified from the blood of this individual neutralized SARS-CoV-2 lentivirus pseudotype (Figure S1 A). We generated a soluble SARS-CoV-2 S construct that is stabilized through mutations and the addition of a trimerization tag to remain in the S “pre-fusion” conformation (“S2P”) (Wrapp et al., 2020) and used it as an antigen to isolate 116 memory B cells (CD19+, IgG+) by fluorescence-activated cell sorting (FACS) (Figure S1B). We could produce 48 recombinant monoclonal antibodies in sufficient amount for further characterization; forty-three of these bound S2P by ELISA, and 18 also bound the RBD (Figure S1C; Table S1). Most antibodies were derived from the VH3 heavy chain subgroup and had kappa light chains (Figure S1D). Antibody CDR H3 and CDR L3 loops had an average length of 15 and 9 amino acids, respectively, with low frequencies of somatic hypermutation in variable heavy and light chain sequences (Figures S1E and S1F; Table S1).
Figure S1.
Isolation of SARS-CoV-2 S-reactive antibodies from a COVID-19 convalescent individual, related to Table 1
(A) Entry levels of SARS-CoV-2 or vesicular stomatitis virus (VSV) lentivirus pseudotypes after pre-incubation with polyclonal immunoglobulins (IgG) purified from the plasma of a COVID-19 convalescent individual (“C1”), a non-immune control donor (“ctrl”), or with an ACE2-Fc fusion protein all at a concentration of 316 μg ml-1. Data are normalized to a no antibody control. Means ± standard deviation from two experiments performed in triplicate (n = 6) are shown. One-way ANOVA with Tukey’s multiple comparisons test. ∗∗∗∗p < 0.0001.
(B) Density plot from a FACS experiment to isolate memory B cells that bind phycoerythrin (PE)-labeled streptavidin tetramers coupled to a prefusion-stabilized SARS-CoV-2 S construct (S2P-PE). The approximate location of the sorting gate is shown as a box, and the percentage of cells that fall within the gate is indicated. The left panel is for a control donor and the right panel is for COVID-19 convalescent donor C1. CD19 is a B cell marker.
(C) Whisker plot showing ELISA values for IgG binding to S2P, the SARS-CoV-2 RBD, or a control protein Lujo virus (LUJV) GP1. Antibodies were added at a single concentration of 100 μg ml-1. Dashed line represents the cut off for our definition of antibodies that bind the respective protein.
(D) Antibody heavy and light chain gene usage for SARS-CoV-2 S-reactive monoclonal antibodies. Asterisks indicate clonally related VH3-53/VK1-9 antibodies (referred to as “C1A-VH3-53 antibodies” in the text).
(E-F) Violin plots showing CDR3 loop lengths and somatic hypermutation frequencies (S.H.M.) for S-reactive monoclonal antibodies. The median and quartiles are shown as dashed and dotted lines, respectively. For CDR3 loop lengths, the median and first quartile marker overlap.
(G) SARS-CoV-2 or VSV lentivirus pseudotypes were pre-incubated with 100 μg ml-1 of the indicated monoclonal antibody or an ACE2-Fc fusion protein (indicated in bold) and the mixture was used to infect HEK293T-hACE2 cells. Entry levels were quantified 48 h later using FACS. Data are normalized to a no antibody control. Dashed line indicates 10% relative entry. Means ± standard deviation from two experiments performed in triplicate (n = 6) are shown.
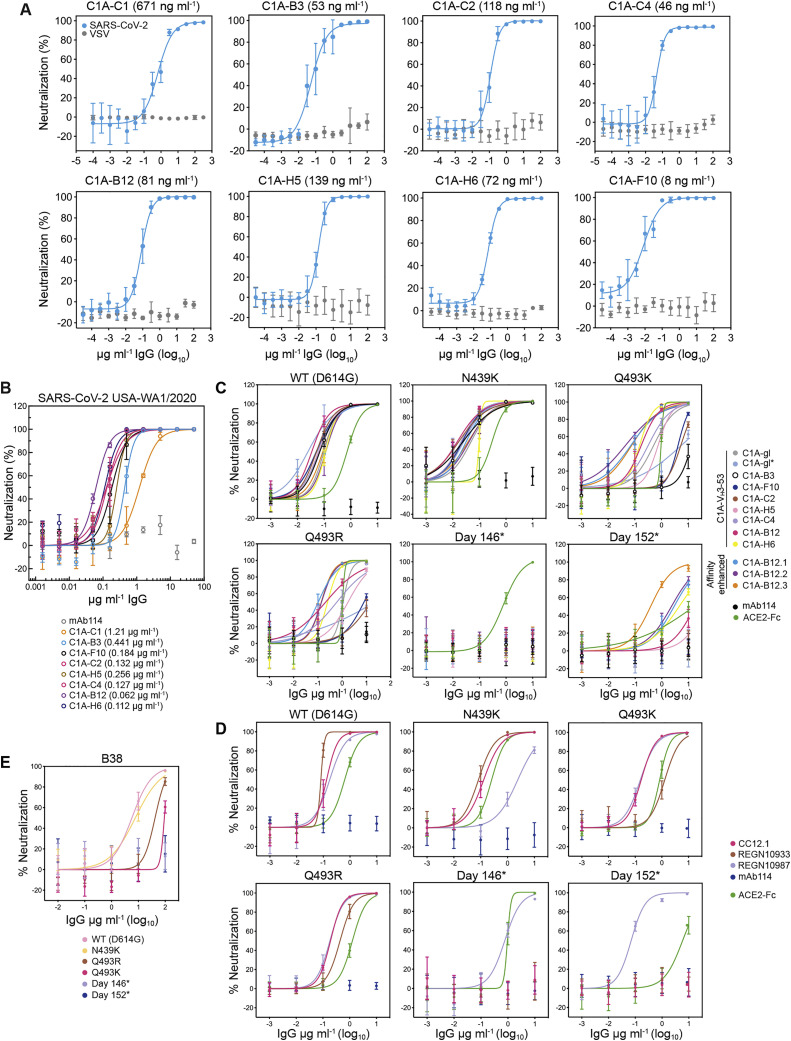
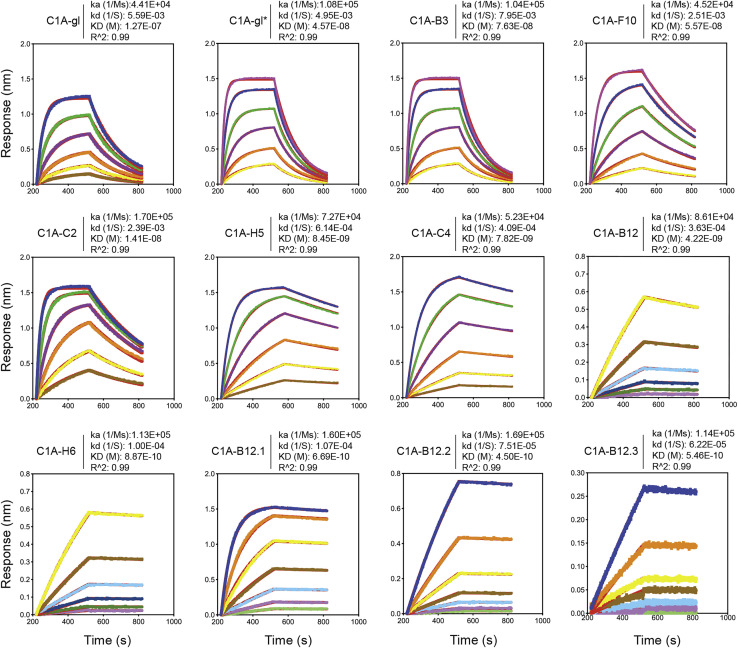
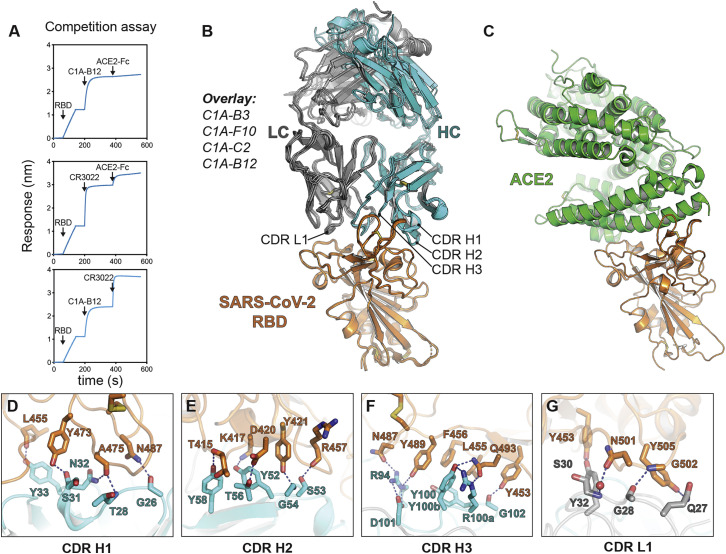
Of the 43 antibodies we tested, only eight neutralized SARS-CoV-2 pseudotype with greater than 90% reduction in entry at a screening concentration of 100 μg mL−1 (Figure S1G). The only potent neutralizing antibodies (defined here as having an IC50 value of less than 0.5 μg mL−1 against infectious SARS-CoV-2) were somatic variants of the same VH3-53/VK1-9-derived antibody (referred to as “C1A-VH3-53 antibodies” throughout the rest of the paper) (Figures 1 A, 1B, S2 A, and S2B; Table 1 ). Monomeric Fabs derived from these antibodies bound tightly to the RBD, with affinities ranging from 76 nM to 0.9 nM (Figure S3 ; Table 1). C1A-B12 prevented an ACE2-Fc fusion protein from binding to the RBD in a biolayer interferometry (BLI)-based competition assay but did not affect the binding of the Fab of a control antibody (CR3022) that does not interfere with ACE2 recognition (Yuan et al., 2020b) (Figure S4 A).
Figure 1.
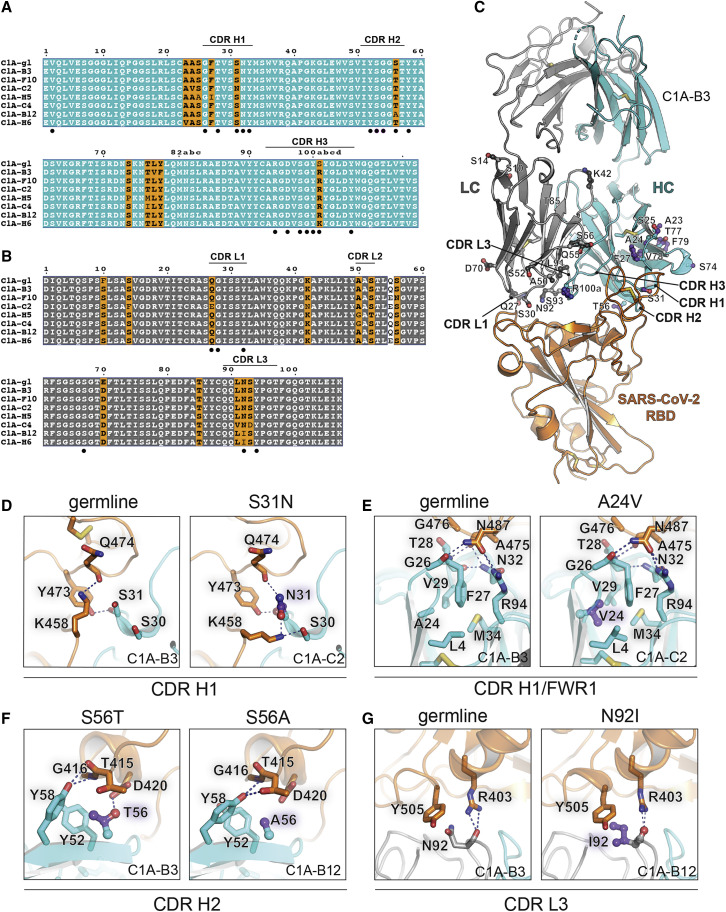
Affinity maturation of C1A-VH3-53 antibodies
(A and B) Alignment of antibody variable heavy (A) or light chain (B) gene sequences. The Kabat numbering scheme is used. The C1A-gl sequences shown are germline revertant sequences designed using IMGT/V-QUEST (Brochet et al., 2008). Note in (A) that CDR H3 germline sequences are impossible to predict but we could identify a possible substitution (see Figure 2A). Panels were generated using ESPrit3 (Robert and Gouet, 2014) and modified. RBD contacting residues are indicated with a filled black circle.
(C) Ribbon diagram of the crystal structure of the C1A-B3 Fab/RBD complex showing the location of somatic mutations. See also Figure S4.
(D) Interactions for CDR H1 residue 31 with the RBD are shown for C1A-B3 (left panel) or C1A-C2 (right panel) showing the effects of the S31NVH substitution.
(E) Interactions occurring at the base of CDR H1 near the framework regions are shown for C1A-B3 (left panel) or C1A-C2 (right panel) showing the effects of the A24VVH mutation.
(F) Interactions of CDR H2 residue 56 with the RBD are shown for C1A-B3 (left panel) or C1A-B12 (right panel) showing the effects of the S56T/AVH mutations. Both sets of interactions shown occur after somatic mutations; we did not visualize germline interactions at this position.
(G) Interactions of CDR L3 residue 92 with the RBD are shown for C1A-B3 (left panel) or C1A-B12 (right panel) showing the effects of the N92IVL substitution.
For (D), (E), and (G), “germline” indicates baseline interactions occurring when a given residue is not somatically mutated.
Figure S2.
SARS-CoV-2 pseudotype and infectious virus neutralization assays, related to Figure 5 and Table 1
(A) SARS-CoV-2 lentivirus pseudotypes were pre-incubated with monoclonal antibodies at the indicated concentrations and the mixture was used to infect HEK293T-hACE2 cells. Entry levels were quantified 48 h later using FACS. VSV pseudotypes are included as a negative control. Data are normalized to a no antibody control. Means ± standard deviation from two experiments performed in triplicate (n = 6) are shown. For some data points, error bars are smaller than symbols. IC50 values are shown in parentheses.
(B) Infectious SARS-CoV-2 (strain USA-WA1/2020) was incubated with monoclonal antibodies at the indicated concentration with infection of Vero E6 cells subsequently measured in a PRNT assay (Zhang et al., 2020). Means ± standard deviation from three experiments performed in triplicate (n = 9) are shown. Data are normalized to a no antibody control. For some data points, error bars are smaller than symbols. IC50 values are shown in parentheses.
(C) Dose response neutralization assays of C1A-VH3-53 and affinity enhanced versions of C1A-B12 with the indicated S pseudotypes. Data are normalized to a no antibody control. Means ± standard deviation from two experiments performed in triplicate (n = 6) are shown. IC50 values are shown in Figures 5A and 5B.
(D) Dose response neutralization assays of REGN10933 and REGN10987 (Baum et al., 2020; Hansen et al., 2020) and CC12.1 (Rogers et al., 2020) with the indicated S pseudotypes. Data are normalized to a no antibody control. Means ± standard deviation from two experiments performed in triplicate (n = 6) are shown. IC50 values are shown in Figure 5C.
(E) Dose response neutralization assays of monoclonal antibody B38 (Wu et al., 2020). Means ± standard deviation from two experiments performed in triplicate (n = 6) are shown. IC50 values are shown in Figure 5C.
Table 1.
Summary of C1A-VH3-53 antibody binding and neutralization data
| Antibody | Number of a.a. changes (VH, VL) | Kinetic analysis |
SARS-CoV-2 pseudotype |
SARS-CoV-2 WA1/2020 |
||
|---|---|---|---|---|---|---|
| ka (1/Ms) | kd (1/S) | KD (nM) | IC50 μg mL−1 | IC50 μg mL−1 | ||
| C1A-gl | – | 4.41E+04 | 5.59E−03 | 127 | 0.098a | 0.126 |
| C1A-gl∗ | – | 1.08E+05 | 4.95E−03 | 45.7 | 0.025a | 0.102 |
| C1A-B3 | 3, 2 | 1.04E+05 | 7.95E−03 | 76.3 | 0.053 | 0.441 |
| C1A-F10 | 1, 4 | 4.52E+04 | 2.51E−03 | 55.7 | 0.008 | 0.184 |
| C1A-C2 | 3, 6 | 1.70E+05 | 2.39E−03 | 14.1 | 0.118 | 0.132 |
| C1A-H5 | 5, 5 | 7.27E+04 | 6.14E−04 | 8.5 | 0.139 | 0.256 |
| C1A-C4 | 2, 5 | 5.23E+04 | 4.09E−04 | 7.8 | 0.046 | 0.127 |
| C1A-B12 | 1, 3 | 8.61E+04 | 3.63E−04 | 4.2 | 0.081 | 0.062 |
| C1A-H6 | 2, 4 | 1.13E+05 | 1.00E−04 | 0.9 | 0.072 | 0.112 |
| C1A-B12.1 | 3, 3 | 1.60E+05 | 1.07E−04 | 0.7 | 0.054a | 0.028 |
| C1A-B12.2 | 4, 3 | 1.69E+05 | 7.51E−05 | 0.5 | 0.115a | 0.025 |
| C1A-B12.3 | 5, 3 | 1.14E+05 | 6.23E−05 | 0.5 | 0.090a | 0.032 |
a.a., amino acids; ND, not determined.
Measured with D614GS lentivirus pseudotypes as reported in Figure 4.
Figure S3.
Fab binding kinetics to the SARS-CoV-2 receptor-binding domain, related to Table 1
Fab affinities for the SARS-CoV-2 RBD were measured using biolayer interferometry. Red lines represent the fit for a 1:1 binding model, and alternate colors represent response curves measured at varying concentrations. Binding kinetics were measured for six concentrations of Fab at twofold dilutions ranging from 500 to 15.6 nM (C1A-B3, C1A-F10, C1A-gl, C1A-gl∗), 250 to 7.8 nM (C1A-C2, C1A-H5, C1A-C4), and from 15.6 to 0.49 nM (C1A-B12 and C1A-H6). For affinity enhanced antibodies, binding kinetics were measured at seven concentrations of Fab at twofold dilution ranging from 100 to 1.56 nM (C1A-B12.1) or from 10 to 0.16 nM (C1A-B12.2 and C1A-B12.3). Each experiment was performed at least twice, and representative data are shown.
Figure S4.
SARS-CoV-2 receptor-binding domain recognition by C1A-VH3-53 antibodies, related to Figure 1
(A) BLI-based competition assay for C1A-B12 Fab, CR3022 Fab, and human ACE2 ectodomain Fc fusion protein (ACE2-Fc) binding to the SARS-CoV-2 RBD. Arrows show the time point at which the indicated protein was added. Representative results of two replicates for each experiment are shown.
(B) Overlay of ribbon diagrams for X-ray crystal structures of Fab/RBD complexes. CDR loops contacting the RBD are indicated.
(C) Ribbon diagram of the X-ray crystal structure of the RBD bound to the ACE2 ectodomain (PDB ID: 6M0J) (Lan et al., 2020) with the RBD in the same orientation as shown in (B) for comparison.
(D-G) Details of the RBD/antibody interface for C1A-B3. The panels show significant contacts made by antibody CDR loops.
Structural basis for affinity maturation of C1A-VH3-53 antibodies
To better understand the basis for affinity maturation of C1A-VH3-53 antibodies, we determined X-ray crystal structures of four Fab/RBD complexes (Figure S4B). As with other VH3-53/3-66-derived antibodies that neutralize SARS-CoV-2 (Du et al., 2020; Hurlburt et al., 2020; Shi et al., 2020; Wu et al., 2020; Yuan et al., 2020a), CDR loops H1, H2, H3, and L1 make the most significant contacts with the RBD on a surface that overlaps with the ACE2 binding site (Figures S4B and S4C). Interactions involve an extensive network of mostly polar contacts mediated by main chain and side chain atoms (Figures S4D–S4G).
High-resolution X-ray crystal structures of different clonotypes allowed us to examine the effects of somatic mutations on the interaction interface (Figure 1). The S31NVH mutation in C1A-C2 provides new contacts with Q474RBD and K458RBD (Figures 1A and 1D). Although the A24VVH mutation is not at the RBD/Fab interface, it is a pocket-filling mutation that, through interactions with the side chain of F27VH, would rigidify CDR H1 in a conformation that is compatible with RBD binding (Figures 1A, 1C, and 1E). The S56TVH mutation, found in most of the antibodies, provides additional hydrophobic contacts with the RBD and with neighboring tyrosines on the antibody, and the S56AVH mutation in C1A-B12 removes a polar contact with D420RBD (Figures 1A and 1F). The N92IVL substitution in the two highest affinity binding antibodies, C1A-H6 and C1A-B12, provides a new hydrophobic contact with Y505RBD (Figures 1B and 1G).
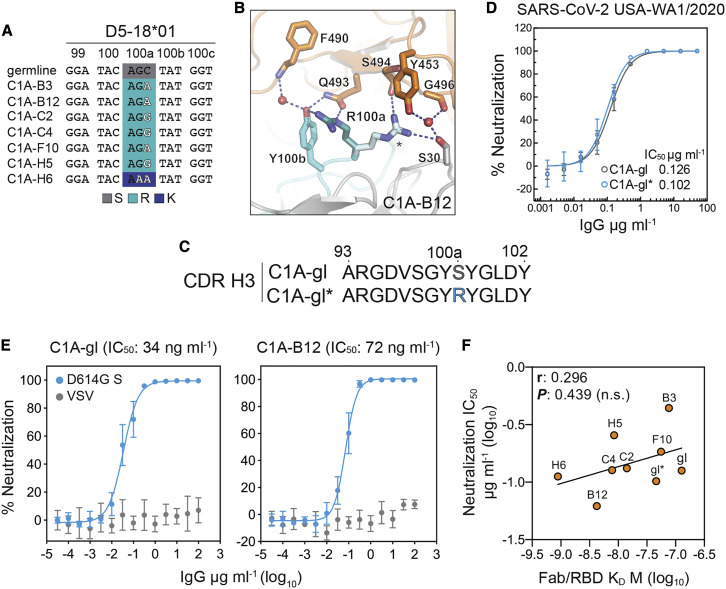
Although it is impossible to predict germline CDR H3 sequences, we identified a potential mutation located centrally in the D5-18∗01 gene segment from which the CDR H3 loop could be derived (Figure 2 A) (Brochet et al., 2008). In six out of seven of our clonally related antibodies, the inferred mutation replaces a germline serine with an arginine, for which two rotamers anchor an extensive network of polar interactions with the RBD (Figure 2B). This network includes Q493RBD, a residue that is relevant to antibody neutralization escape (see below). To determine the potential contribution of the S100RaVH change to affinity maturation, we generated germline revertant antibodies that contain germline VH and VL sequences but vary with either having a serine or an arginine at this CDR H3 position (C1A-gl and C1A-gl∗, respectively) (Figures 1A, 1B, and 2C). C1A-gl and C1A-gl∗ Fabs bound the RBD with affinities of 127 nM and 46 nM, respectively (Figure S3; Table 1). Despite the difference in RBD affinity, C1A-gl and C1A-gl∗ neutralized infectious SARS-CoV-2 with comparable IC50 values (Figure 2D; Table 1). Furthermore, C1A-gl and C1A-B12, the most potent neutralizing antibody against infectious SARS-CoV-2, had similar activity against D614GS pseudotypes (Figure 2E). Although RBD affinities varied over a 100-fold for the antibodies we studied (Table 1), we ultimately observed no statistically significant correlation between RBD binding affinity and infectious SARS-CoV-2 USA-WA1/2020 neutralization (Figure 2F).
Figure 2.
Affinity maturation plays a limited role in potent SARS-CoV-2 neutralization
(A) Nucleotide sequences of the D segment of C1A-VH3-53 antibodies. Changes that likely occurred at CDR H3 position 100a (S100aR or S100aK) during somatic hypermutation are highlighted.
(B) C1A-B12/RBD complex showing interactions occurring with alternate side chain conformers of CDR H3 residue R100a (one conformer is labeled with an asterisk).
(C) Amino acid sequences for CDR H3 loops of C1A-gl and C1A-gl∗.
(D) Results of a PRNT assay with infectious SARS-CoV-2 and the indicated antibodies. Data are normalized to a no antibody control. Means ± standard deviation from three experiments performed in triplicate (n = 9) are shown. Error bars indicate standard deviation. For some data points, error bars are smaller than symbols.
(E) Dose response neutralization assay results with SARS-CoV-2 D614GS pseudotype. Data are normalized to a no antibody control. Means ± standard deviation from two experiments performed in triplicate (n = 6) are shown. For some data points, error bars are smaller than symbols.
(F) Correlation analysis of Fab/RBD antibody affinity measurements for the indicated antibodies and SARS-CoV-2 USA/WA1/2020 neutralization IC50 values. r, Pearson correlation coefficient; n.s., not significant.
Structural predictions of antibody neutralization escape
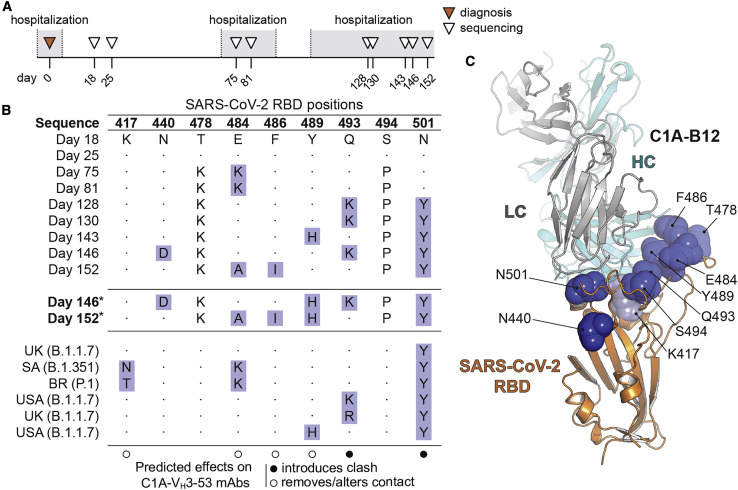
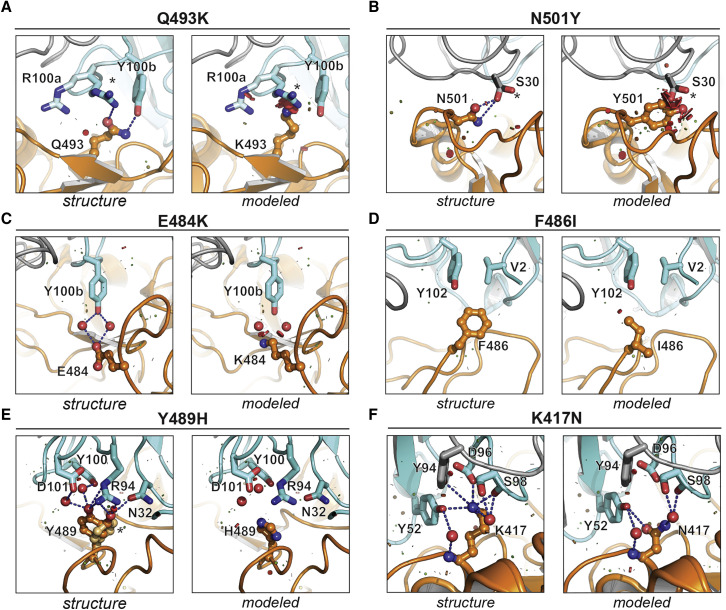
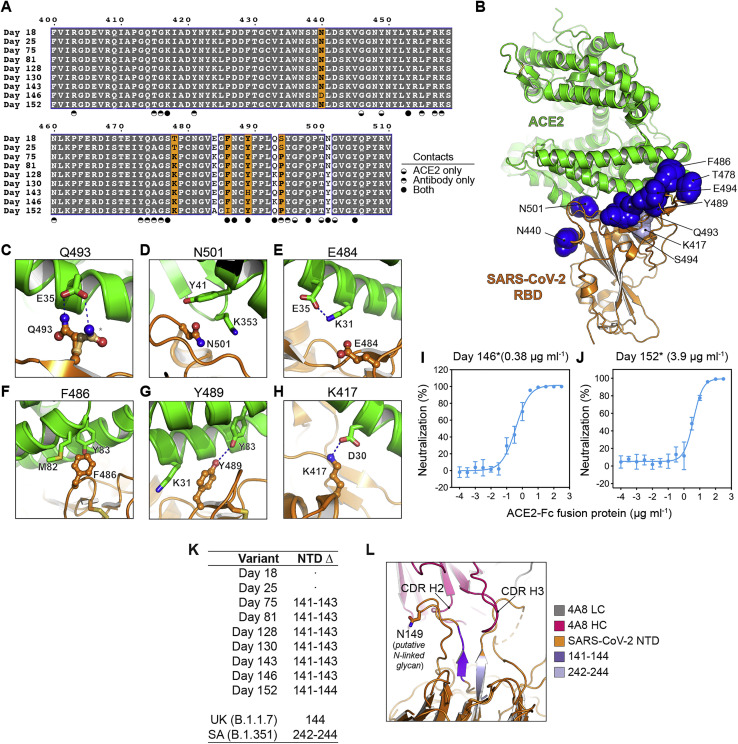
The convergence of nearly identical responses against the RBD in multiple COVID-19 convalescent individuals (Figures S5 A and S5B) led us to hypothesize that SARS-CoV-2 could evolve resistance to VH3-53/3-66 antibodies as the virus continues to circulate in humans. A recent report described significant evolution of SARS-CoV-2 in an individual receiving profound immunosuppression (Choi et al., 2020). The individual had antiphospholipid syndrome complicated by diffuse alveolar hemorrhage and received glucocorticoids, cyclophosphamide, rituximab, and eculizumab as part of their immunosuppression; they ultimately experienced multiple episodes of symptomatic disease (Choi et al., 2020). COVID-19 was diagnosed on day 0 of infection by RT-PCR, and SARS-CoV-2 whole genome viral sequencing was performed from nasopharyngeal specimens at various time points and up to day 152 (Figure 3 A) (Choi et al., 2020). There was evidence of pronounced RBD sequence evolution by the later time points, with a total of eight mutations (Figures 3B and 3C). We predicted that five of these eight mutations would impact C1A-VH3-53 antibody binding (Figures 3B and 3C). The Q493KRBD mutation would introduce a substantial clash with CDR H3 residue R100aVH found in most of the antibody clones (Figures 1A and 4A). C1A-H6 is the only antibody clone that contains a lysine at position 100aVH (Figure 1A); although we did not obtain a crystal structure of the RBD bound to the C1A-H6 Fab to visualize its contacts, K100aVH would probably also clash with K493RBD. The N501YRBD mutation would introduce minor clashes with CDR L1 residue S30V L, a VK1-9 germline residue (Figures 1B and 4B). This germline residue is conserved in the other VH3-53-derived neutralizing antibodies we examined that also contain the VK1-9 light chain and for which structures are available (Figures S5A and S5D). Alternate rotamers we observed in our structures for residues R100aVH and S30VL would partially accommodate the Q493KRBD and N501YRBD mutations (Figures 4A and 4B). The other mutations (E484K/ARBD, F486IRBD, and Y489HRBD) would alter polar or hydrophobic antibody contacts (Figures 4C–4E). In particular, the Y489HRBD change detected on day 128 sequencing would alter an extensive network of polar interactions with antibody residue R94VH, a germline antibody residue that is conserved in all VH3-53/3-66-derived antibodies (Figures 4E and S5C). Mutations at position E484RBD, however, would be better tolerated, because the network of polar interactions with the antibodies, which includes water molecules, would either be lost (E484ARBD) or possibly be reorganized (E484KRBD) (Figure 4C).
Figure S5.
Structure-guided affinity maturation of C1A-B12, related to Figures 1 and 5
(A) Examples of gene usage and CDR H3 lengths for other VH3-53/3-66 antibodies for which structures are available. All antibodies, which were isolated from COVID-19 convalescent donors, engage the RBD with an essentially identical binding mode. CDR H3 length was determined using IMGT/V-QUEST definitions (Brochet et al., 2008). aa: amino acids. PDB ID: protein data bank identification code.
(B) Structural alignment of variable heavy (HC) and light chain (LC) Fab portions of VH3-53/3-66 antibodies bound to the RBD. Antibodies included in the alignment are listed in (A).
(C-D) Alignment of variable heavy chain (C) and VK1-9-derived light chain (D) gene sequences of VH3-53/3-66-derived antibodies reported here and elsewhere. Antibody sequences were obtained from the RCSB record and protein data bank (PDB) IDs listed in (A). Panels were generated using ESPrit3 (Robert and Gouet, 2014) and modified. The Kabat numbering scheme is used.
(E-F) Interactions occurring at the base of CDR H1 with framework regions are shown for the B38 Fab/RBD complex (PDB: 7BZ5) (Wu et al., 2020) (E) or CV30 Fab/RBD complex (PDB: 6XE1) (Hurlburt et al., 2020) (F). The T28IVH mutation adds a hydrophobic contact with G476RBD, and the F27VVH mutation probably makes CDR H1 more flexible, allowing local polar contacts to be optimized.
(G) Partial sequence alignment of C1A-VH3-53 and affinity enhanced antibodies C1A-B12.1, C1A-B12.2, and C1A-B12.3.
(H) Infectious SARS-CoV-2 (strain USA-WA1/2020) was incubated with monoclonal antibodies at the indicated concentrations with infection of Vero E6 cells subsequently measured in a PRNT assay. Means ± standard deviation from three experiments performed in triplicate (n = 9) are shown. Data are normalized to a no antibody control. For some data points, error bars are smaller than symbols. IC50 values are in parentheses.
Figure 3.
RBD sequence evolution during persistent SARS-CoV-2 infection
(A) Timeline and sequencing interval during persistent SARS-CoV-2 infection of an immunocompromised individual as reported by Choi et al. (2020). The individual was admitted three times between days 6 and 68; prolonged hospitalizations are shown in gray. Sequencing on days 18 and 25 was obtained during shorter hospitalizations, which are not shown.
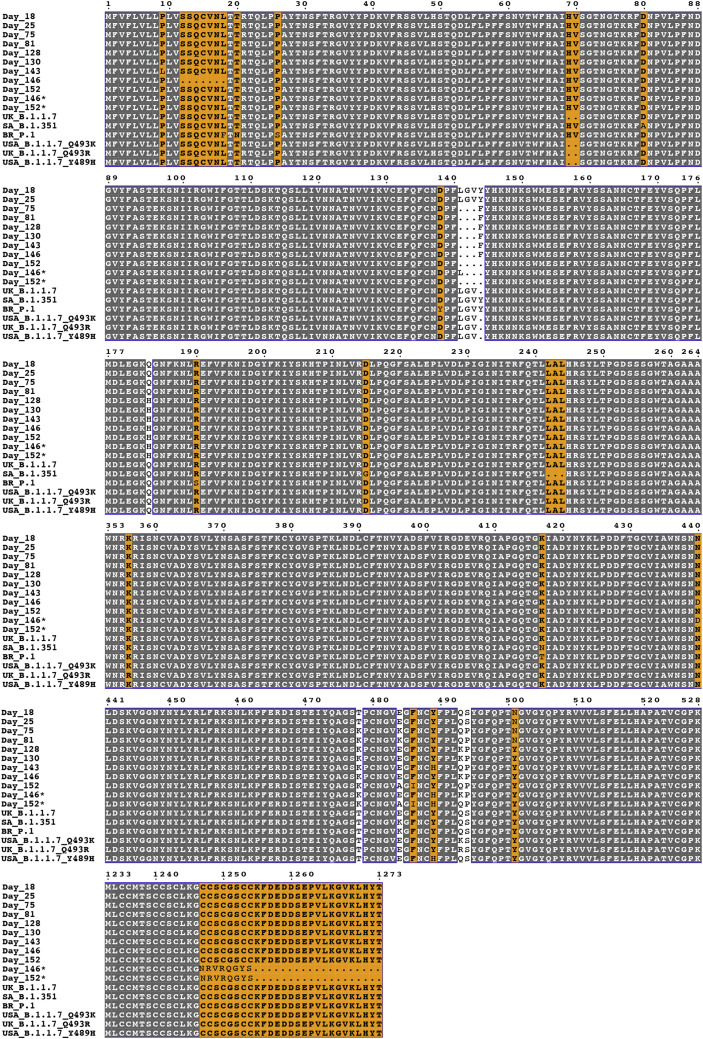
(B) Table showing SARS-CoV-2 S RBD mutations occurring during persistent infection (Choi et al., 2020). Predicted effects of substitutions on binding of the C1A-VH3-53 antibodies are shown in the legend. Mutations that are the focus of our analysis are highlighted. For pseudotyping, we generated S mutants for day 146 and 152 S sequences that retain the Y489HRBD mutation that occurred on day 143 (these are labeled “day 146∗” and “day 152∗”). Sequences from variants that were first detected in the United Kingdom (“UK,” B.1.1.7), South Africa (“SA,” B.1.351), and Brazil (“BR,” P.1), and additional human-derived S sequences containing relevant mutations from samples collected in the United States (USA), are also included for comparison (see Figure S6 and Table S3).
(C) Structure of the C1A-B12 Fab/RBD complex with mutated residues indicated in (B) shown as spheres. Residues mutated during SARS-CoV-2 evolution in the immunocompromised individual are shown as dark blue spheres, and a residue mutated in the B.1.351 and P.1 variants (K417) is shown as a light blue sphere.
See also Figure S7.
Figure 4.
Predicted effects of RBD mutations on C1A-VH3-53 neutralizing antibody binding
(A–F) For each indicated mutation, interactions observed in the C1A-B12/RBD complex structure are shown in the left panels (labeled “structure”) and predicted effects of mutations based on modeling are shown in the right panels (labeled “modeled”). PyMol was used to model mutations and visualize steric clashes; short green lines or small green disks are present when nearby atoms are almost in contact, and large red disks indicate significant van der Waals overlap. For modeling, only residues on the RBD were modified, and all RBD residue rotamers in the rotamer library were checked and the one that caused the least clashes was chosen. Alternate side chain rotamers for R100aVH in (A), S30VL in (B), and for Y489RBD in (E) are indicated with an asterisk.
Evolved spike variants escape VH3-53 antibody neutralization
We next sought to validate our structural predictions for the effects of RBD mutations on VH3-53 antibody neutralization using lentivirus pseudotypes bearing variant S proteins. Because variants detected at later time points contained the most RBD mutations, we generated pseudotypes with S proteins that contain mutations observed on days 143–152 (Figures 3B and S6 ). The original day 146 sequence contains a seven-residue deletion at the N terminus of S1 near the expected signal peptide juncture that is of unclear significance, so we preserved this segment as wild-type for viral pseudotyping (Figure S6). We also retained the Y489HRBD mutation found on day 143 given its predicted impact on the antibody-RBD interface and because it has also been detected in additional human-derived SARS-CoV-2 S sequences (Figures 3B and 4E; Table S3), generating S mutants denoted “day 146∗” and “day 152∗” (Figure S6). We used D614GS pseudotypes as the wild-type control for these experiments because all sequences recovered from the immunocompromised individual included it, suggesting that the initial infecting SARS-CoV-2 virus contained this substitution. Day 146∗ and day 152∗ pseudotypes were neutralized by an ACE2-Fc fusion protein (Figures S7 I and S7J) but were resistant to neutralization by C1A-VH3-53 antibodies (Figures 5 A, 5B, and S2C). C1A-H6, whose Fab binds the tightest to the RBD (0.9 nM; Table 1), had some activity against the day 152∗ S pseudotype, but with a 40-fold decrease in potency (IC50 value of 4.5 μg mL− 1) (Figures 5A and S2C).
Figure S6.
Alignment of SARS-CoV-2 S sequences, related to Figure 3
Alignment of SARS-CoV-2 sequences. The following sequences were used for the alignment: Day 18: hCoV-19/USA/MA-JLL-D18/2020 (EPI_ISL_593478); Day 25: hCoV-19/USA/MA-JLL-D25/2020 (EPI_ISL_593479); Day 75: hCoV-19/USA/MA-JLL-D75/2020 (EPI_ISL_593480); Day 81: hCoV-19/USA/MA-JLL-D81/2020 (EPI_ISL_593553); Day 128: hCoV-19/USA/MA-JLL-D128/2020 (EPI_ISL_593554); Day 130: hCoV-19/USA/MA-JLL-D130/2020 (EPI_ISL_593555); Day 143: hCoV-19/USA/MA-JLL-D143/2020 (EPI_ISL_593556); Day 146: hCoV-19/USA/MA-JLL-D146/2020 (EPI_ISL_593557); Day 152: hCoV-19/USA/MA-JLL-D152/2020 (EPI_ISL_593558). Sequences from United Kingdom (“UK”) B.1.1.7 hCoV-19/England/205261299/2020 (EPI_ISL_754289), South Africa (“SA”) B.1.351 hCoV-19/South Africa/Tygerberg-461/2020 (EPI_ISL_745186), Brazil (“BR”) P.1 hCoV-19/Brazil/AM-20143138FN-R2/2020, United States (USA) B.1.1.7 Q493K hCoV-19/USA/FL-CDC-STM-0000013-F04/2021 (EPI_ISL_884605), UK B.1.1.7 Q493R hCoV-19/England/MILK-11C2FCD/2021 (EPI_ISL_1006449), and USA B1.1.7 Y489H hCoV-19/USA/CA-CDC-STM-A100413/2021 (EPI_ISL_850699) are included for comparison. The “day 146∗” sequence shown is a version of the day 146 sequence that retains wild-type residues at positions 12-18, contains an NTD deletion spanning residues 142-144 (instead of 141-143), and contains the Y489HRBD mutation. The “day 152∗” sequence shown is a version of the day 152 sequence that contains the Y489HRBD mutation. Both day 146∗ and day 152∗ sequences contain mutations in the C-terminal cytoplasmic tail to allow for efficient lentivirus pseudotyping. The figure was generated using ESPrit3 (Robert and Gouet, 2014).
Figure S7.
RBD sequence variation in relation to ACE2 interactions and predicted NTD deletion effects, related to Figures 3 and 4
(A) Sequence alignment for S residues spanning the RBD in an immunocompromised individual (Choi et al., 2020) at the indicated time points. RBD residues that interact with ACE2 only, C1A-VH3-53 antibodies only, or both, are indicated.
(B) Ribbon diagram of the X-ray crystal structure of an ACE2 ectodomain/RBD complex (PDB ID: 6M0J) (Lan et al., 2020). Residues that are mutated during SARS-CoV-2 persistent infection are shown as dark blue spheres. The K417RBD residue, which is mutated in the B.1.351 and P.1 variants (see Figure 3B), is shown as light blue spheres.
(C-H) Views highlighting where select RBD mutations (see Figures 3D–3I) fall with respect to the ACE2 interface.
(I-J) SARS-CoV-2 day 146∗ (I) or day 152∗ (J) S pseudotypes were pre-incubated with an ACE2-Fc fusion protein at the indicated concentrations and the mixture was used to infect HEK293T-hACE2 cells. Entry levels were quantified 48 h later using FACS. Data are normalized to a no antibody control. Means ± standard deviation from two experiments performed in triplicate (n = 6) are shown. IC50 values are shown in parentheses.
(K) Summary of SARS-CoV-2 S N-terminal domain (NTD) deletions occurring during persistent infection of an immunocompromised individual (Choi et al., 2020). Deletions found in United Kingdom (“UK”) B.1.1.7 (hCoV-19/England/205261299/2020, EPI_ISL_754289) and South Africa (“SA”) B.1.351 (hCoV-19/South Africa/Tygerberg-461/2020, EPI_ISL_745186) variants are also included for comparison.
(L) Ribbon diagram of the 4A8 Fab:NTD interface (PDB: 7C2L) (Chi et al., 2020). Residues 141-144, which contain mutations starting on day 75, are shown in dark purple, and residues 242-244, which are mutated in the “SA” B.1.351 SARS-CoV-2 variant (Figure S6), are shown in light purple. The 141-144 deletion would reposition a putative N-linked glycosylation site (N149) and potentially block epitope access.
Figure 5.
Neutralization escape from monoclonal antibodies
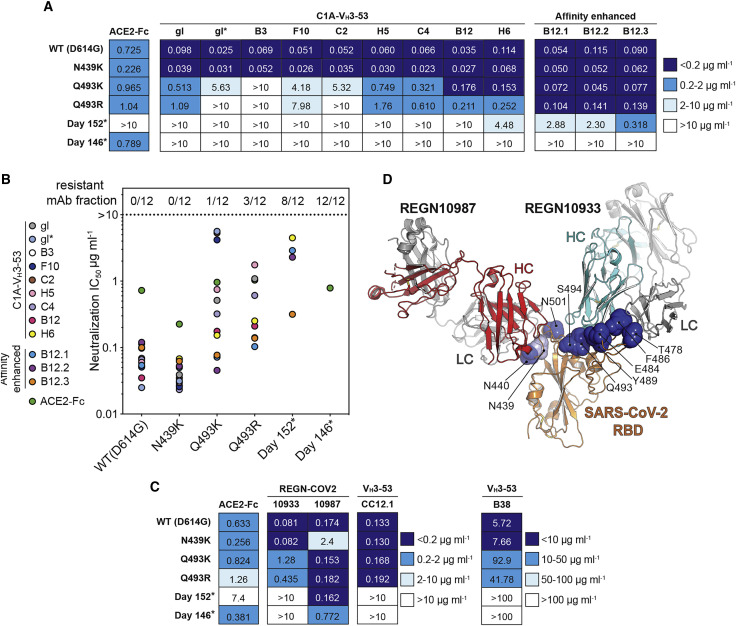
(A) Table showing IC50 values for pseudotype neutralization tests with the indicated SARS-CoV-2 S pseudotypes. Monoclonal antibody names are abbreviated (e.g., C1A-gl is “gl” and C1A-B3 is “B3”). Antibodies are listed, left to right, in order of increasing affinity. IC50 values for an ACE2-Fc neutralization assay done as part of the same experiment are shown. See also Figure S2C.
(B) Summary of results shown in (A) highlighting the fraction of resistant monoclonal antibodies for each S pseudotype.
(C) Table showing IC50 values for SARS-CoV-2 S pseudotype neutralization tests with the indicated monoclonal antibodies. IC50 values for an ACE2-Fc neutralization assay done as part of the same experiment are shown. See also Figures S2D and S2E.
(D) Ribbon diagram of the SARS-CoV-2 RBD bound to Fabs for antibodies REGN10987 and REGN10933 (PDB: 6XDG) (Hansen et al., 2020). Mutated residues of interest are shown as in Figure 3C, with the exception of residue N439RBD (shown as light blue spheres).
The Q493KRBD mutation, which was observed in sequences obtained on days 128, 130, and 146 (Figure 3B), has previously been described through in vitro resistance mapping efforts with recombinant vesicular stomatitis virus expressing SARS-CoV-2 S (rVSV-S) (Weisblum et al., 2020). The Q493KRBD change or a similar mutation at the same position (Q493RRBD) has also been recently described in other human-derived SARS-CoV-2 sequences (Figure 3B; Table S3). To determine the role of the Q493K/RRBD mutations in resistance to C1A-VH3-53 antibodies, we generated pseudotypes containing either mutation in addition to the D614GS change. We also included an N439KRBD variant, a recently described antibody neutralization escape mutant (Thomson et al., 2021). The Q493KRBD mutation caused substantial resistance to the C1A-VH3-53 antibodies that bind the most weakly to the RBD (Figures 5A and 5B). We observed similar findings with the Q493RRBD pseudotypes, although the decrease in neutralization sensitivity was more severe. An exception to the general observation that weaker binding antibodies are less active against mutants was C1A-gl, which neutralized Q493K/RRBD pseudotypes better that C1A-gl∗. This is likely because C1A-gl contains a serine instead of an arginine at CDR H3 position 100a, which would better accommodate these RBD mutations (Figure 4A). The N439KRBD mutation had no effect on pseudotype neutralization by C1A-VH3-53 antibodies (Figure 5A), which was expected, because this mutation falls outside of the VH3-53 antibody epitope on the RBD.
To determine if resistance extends to VH3-53-derived antibodies isolated from different COVID-19 convalescent donors, we also tested antibodies B38 (Wu et al., 2020) and CC12.1 (Rogers et al., 2020; Yuan et al., 2020a). The Q493K/RRBD mutations conferred decreased sensitivity to B38 but had no effect on neutralization by CC12.1 (Figures 5C, S2D, and S2E). Day 146∗ and day 152∗ S pseudotypes, however, were completely resistant to both of these monoclonal antibodies (Figures 5C, S2D, and S2E).
Resistance to therapeutic antibodies
The monoclonal antibody cocktail REGN-COV2 comprises two antibodies that bind non-overlapping sites on the RBD to suppress the emergence of antibody neutralization escape mutations (Baum et al., 2020; Hansen et al., 2020). REGN10933 binds a region of the RBD that overlaps significantly with the ACE2-binding site, while REGN10987 binds a region that has little to no overlap (Figure 5D). Of the S mutations that evolved during persistent SARS-CoV-2 infection in the immunocompromised individual, the Q493KRBD change, found in day 146 sequencing, was previously detected in tissue cell culture passaging experiments using REGN10933 and rVSV-S (Baum et al., 2020). In our experiments, the Q493KRBD mutation decreased REGN10933 pseudotype neutralization potency by 15-fold (Figures 5C and S2D). Day 146∗ and day 152∗ S pseudotypes, however, were completely resistant (Figures 5C and S2D). Notably, the day 152∗ variant lacks the Q493KRBD substitution, but its F486IRBD mutation is similar to a known REGN10933 resistance mutation (F486VRBD) (Figures 3B and 4D) (Baum et al., 2020).
The N440DRBD mutation, which was only detected on day 146 sequencing (Figure 3B), falls on the REGN10987 RBD-binding site. It is adjacent to a N439KRBD mutation that is found in circulating variants with reported REGN10987 resistance (Thomson et al., 2021) (Figure 5D). The day 146∗ variant had a 4-fold decrease in REGN10987 neutralization sensitivity, whereas the N439KRBD mutation caused a 14-fold decrease in sensitivity (Figures 5C and S2D). REGN10987, therefore, is the only antibody we tested that had demonstrable activity against the day 146∗ S pseudotypes, but a single substitution at an adjacent position (N439KRBD) found in circulating variants (Thomson et al., 2021) could result in additional neutralization escape.
Evolved S variants are resistant to convalescent donor polyclonal IgG
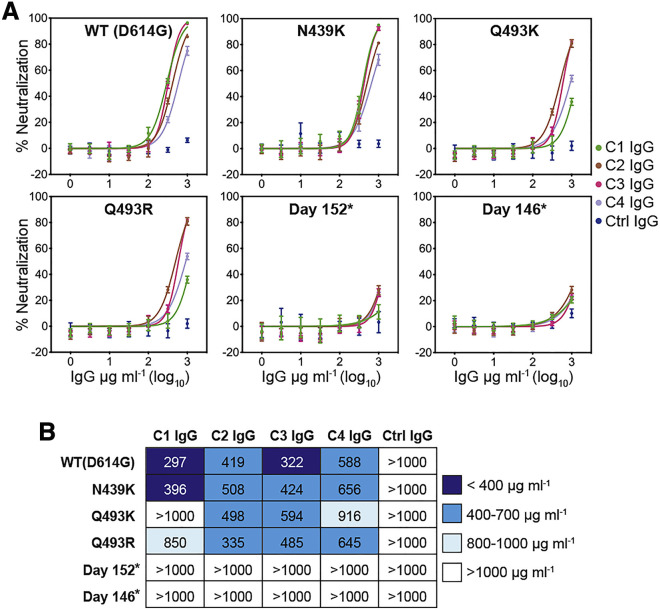
All of the potent SARS-CoV-2 neutralizing antibodies we isolated from the healthy COVID-19 convalescent donor (C1) were clonotypes of a single VH3-53/VK1-9 antibody, suggesting that this individual’s memory B cell response was narrowly focused on this class of neutralizing antibodies (Figure S1; Table S1). Although purified C1 IgG could neutralize WT (D614GS) pseudotypes, days 146∗ and 152∗ S pseudotypes were resistant to C1 serum IgG neutralization (Figures 6A and 6B). The Q493K/RRBD mutations also conferred near complete resistance to C1 IgG (Figures 6A and 6B). The N439KRBD mutation (Thomson et al., 2021), an escape mutation that falls outside of the RBD epitope for VH3-53-derived neutralizing antibodies, had no effect on C1 polyclonal IgG neutralization (Figures 6A and 6B). To determine whether our findings extended to other COVID-19 convalescent donors that may have less epitope biased antibody responses, we performed similar experiments with purified IgG from three additional donors (“C2,” “C3,” and “C4”). The neutralizing activity of purified IgG from these donors was mostly unaffected by the single mutations (Q493K/RRBD or N439KRBD), but day 146∗ and day 152∗ S pseudotypes were resistant to neutralization (Figures 6A and 6B).
Figure 6.
Neutralization escape from convalescent donor polyclonal serum IgG
(A) Dose response neutralization assay with the indicated SARS-CoV-2 S pseudotypes with polyclonal serum IgG of four COVID-19 convalescent donors (C1, C2, C3, and C4) or that of a control, non-immune donor (“ctrl”). Means ± standard deviation from two experiments performed in triplicate (n = 6) are shown.
(B) Table showing IC50 values for pseudotype neutralization tests shown in (A).
See also Table S4.
VH3-53 antibody affinity maturation partially overcomes neutralization escape
Although the benefits of antibody RBD affinity are limited in SARS-CoV-2 USA-WA1/2020 neutralization (Figure 2F), affinity gains, in principle, could compensate for losses of contacts or potential clashes that are caused by escape mutations. Indeed, the highest affinity binding antibodies were seemingly the least impacted by neutralization escape mutations (Figures 5A, 5B, and S2C). We selected C1A-B12, our most potent neutralizing antibody against infectious SARS-CoV-2 (Table 1), to directly test whether additional affinity enhancing mutations could overcome neutralization escape. To generate affinity enhanced versions of C1A-B12, we introduced into its sequence somatic hypermutation changes found in other VH3-53/3-66 antibodies, including antibodies described elsewhere (Hurlburt et al., 2020; Wu et al., 2020) (Figures S5A–S5G). Fabs for the resulting antibodies (C1A-B12.1, C1A-B12.2, and C1A-B12.3) bound to the RBD with a 6- to 10-fold increase in affinity as compared to the parent C1A-B12 Fab (Figure S3; Table 1). Affinity enhanced variants potently neutralized D614GS pseudotypes and infectious SARS-CoV-2 (Figures 5A, 5B, S2C, and S5H; Table 1). Remarkably, although C1A-B12 had no activity against day 152∗ S pseudotype, all three affinity optimized versions were active; the antibody containing the most mutations, C1A-B12.3, was the most potent (IC50 <0.5 μg mL− 1) (Figures 5A, 5B, and S2C). Day 146∗ S pseudotype, however, was still resistant to neutralization by the affinity enhanced antibodies (Figures 5A, 5B, and S2C).
Discussion
Our finding that the day 146∗ and day 152∗ S pseudotypes escape neutralization by all unmodified VH3-53 antibodies we tested and REGN10933, an antibody derived from a different germline gene (VH3-11) (Baum et al., 2020; Hansen et al., 2020), suggest that SARS-CoV-2 can evolve solutions to bind ACE2 while escaping neutralization by major classes of human neutralizing antibodies. Perhaps the most striking finding is that the polyclonal antibody response in a convalescent individual we studied (C1) is so focused on an RBD epitope that single mutations (Q493K/RRBD) can confer substantial resistance to serum IgG neutralization (Figures 6A and 6B). The Q493K/RRBD mutations, however, had less of an effect on the serum IgG of three additional COVID-19 convalescent donors (Figures 6A and 6B), suggesting that the C1 donor may be a rare example of an overly focused antibody response.
The immunocompromised individual we studied received REGN-COV2 (REGN10933 and REGN10987) on day 145 of their illness, so the S mutations detected on days 146 and 152 could have been influenced by selective pressure from this therapeutic antibody cocktail, as described in a recent report (Starr et al., 2021). Nonetheless, several of the RBD mutations we studied were detected before day 146 (Figure 3B), suggesting that they could have arisen through selective pressure from the individual’s weakened neutralizing antibody response (Choi et al., 2020). Furthermore, based on publicly available sequences as of February 19, 2021 in the GISAID database (Elbe and Buckland-Merrett, 2017), seven of the eight RBD mutations we examined in the immunocompromised individual have been detected in additional human-derived SARS-CoV-2 sequences (Figure 3B; Table S3). The only unique mutation is F486IRBD, although a nearly identical mutation, F486LRBD, has been observed in another human-derived SARS-CoV-2 S sequence (Table S3). Importantly, Q493KRBD/N501YRBD, Q493RRBD/N501YRBD, and Y489HRBD/N501YRBD S variants have also recently been reported in the GISAID database, albeit with very low frequency for the time being (Figure 3B; Table S3). Additional mutations that we did not study directly but that could also substantially impact neutralization by VH3-53-derived antibodies are the K417N/TRBD mutations observed in S variants initially detected in South Africa (B.1.351) and Brazil (P.1); like the Y489HRBD mutation, these changes would alter an extensive network of polar contacts with VH3-53/VK1-9 antibodies (Figures 3B, 4E, and 4F).
A detailed understanding of the human antibody response to SARS-CoV-2 and of virus-host co-evolution will be required to design countermeasures that anticipate changes in the virus as it continues to circulate in humans. The portion of the coronavirus S RBDs that interacts with ACE2, called the “receptor-binding motif,” can be thought of as a hypervariable region within an otherwise conserved domain (Li et al., 2005a). The RBD of the closely related virus SARS-CoV, within its receptor-binding motif, contains two “hotspots” for host co-adaptation that are centered on N479RBD and T487RBD (SARS-CoV numbering). Mutations at these positions regulate cross-species transmission and neutralizing antibody escape (Li et al., 2005a; Sui et al., 2008; Wu et al., 2012). Interestingly, two of the residues we pinpointed in our analysis (Q493RBD and N501RBD) are in the equivalent hotspot positions on the SARS-CoV-2 S RBD (Wan et al., 2020).
The N501YRBD mutation, in particular, is involved in SARS-CoV-2 adaptation to murine ACE2 binding (Gu et al., 2020) and has been observed with increasing frequency among circulating variants originally detected in the United Kingdom (B.1.1.7), South Africa (B.1.351), and Brazil (P.1) (Figure 3B). Examination of the structure of an RBD/ACE2 ectodomain complex (Shang et al., 2020) suggests that the N501YRBD change could introduce favorable hydrophobic contacts with Y41ACE2 and K353ACE2 (Figures S7A and S7D). The SARS-CoV-2 Q493KRBD change is also involved in adaptation to murine ACE2 (Leist et al., 2020) and is analogous to the SARS-CoV N479KRBD mutation, which allows preferential engagement of palm civet ACE2 (host reservoir) over human ACE2 (Li et al., 2005b; Wu et al., 2012). The RBD sequence changes we studied, therefore, are likely a combination of neutralizing antibody escape mutations and adaptations to human ACE2 binding.
In certain instances, antibody escape mutations could also negatively impact ACE2 binding. Although D614GS and day 146∗ S pseudotypes had comparable IC50 values in neutralization tests with an ACE2-Fc fusion protein, day 152∗ S pseudotype typically had a more than a 10-fold increase, with some variability in the absolute value depending on the experiment (Figures 5A, 5C, S7I, and S7J). These observations suggest that the affinity of the day 152∗ S for ACE2 may have been compromised. A loss in receptor-binding affinity for the day 152∗ S may be explained by the F486IRBD mutation it contains, which would remove prominent hydrophobic contacts with ACE2 (Figure S7F). Although we used pseudotypes for our studies and focused on the RBD, additional studies with authentic viruses will be required to determine the consequences of the S mutations we studied on viral fitness and potential for transmission. For example, it is unclear how a deletion detected at the S1 N terminus/signal peptide juncture on the original day 146 sequence (Figure S6) would impact S processing, and whether potentially decreased ACE2 binding by a day 152∗-like S variant would affect viral replication and transmission.
Although these were not the focus of our studies, non-RBD binding neutralizing antibodies can target the SARS-CoV-2 S1 NTD. 4A8, an antibody isolated from a COVID-19 convalescent individual, is a representative member of this class (Chi et al., 2020). Examination of late-stage evolved S sequences reveal that they contain internal deletions within the S NTD (spanning residues 141–144) that would disrupt part of 4A8’s epitope (Figures S6, S7K, and S7L). The deletions would also reposition a nearby N-linked glycan and potentially block 4A8 epitope access (Figure S7L). A recent report described persistent SARS-CoV-2 infection in another immunocompromised individual who had acquired hypogammaglobulinemia, with detectable viral RNA more than 100 days after infection (Avanzato et al., 2020). In this individual, S evolution also led to a deletion of a similar segment in the S1 NTD (spanning residues 139–145) (Avanzato et al., 2020). Notably, S1 NTD deletions found in the B.1.1.7 and B.1.351 S variants are in, or near, the NTD deletion found in the S mutants we studied (Figures S6, S7K, and S7L). Although NTD internal deletions could substantially impact 4A8 neutralization, we could not directly test this hypothesis, because 4A8 has very weak neutralizing activity against S lentivirus pseudotypes on HEK293T cells overexpressing human ACE2 (Chi et al., 2020), which we used in our assays.
The value of our study and additional studies examining immune responses in immunocompromised individuals (Kemp et al., 2021; Starr et al., 2021) is obtaining insight from a dynamic immune system over time as opposed to only studying viral escape mutations in vitro. In vitro studies have indeed been highly informative in predicting the range of S mutations that can be acquired for antibody neutralization escape, but studying S sequence evolution during persistent SARS-CoV-2 infection may help highlight the mutations that have the most potential to spread in emerging variants.
Although single mutations are unlikely to confer substantial resistance to polyclonal antibody responses in many individuals, multiple mutations, as are observed in the late stage evolved S variants we studied, are likely to have an impact. To fully understand the consequences of SARS-CoV-2 S genetic drift, including its potential implication to ongoing vaccination campaigns, our study underscores the importance of studying multiple mutations that can concomitantly be found in S, as opposed to single S mutations in isolation.
Limitations of study
The S mutations we studied are only from one immunocompromised individual (n = 1) (Choi et al., 2020). Although some of the S mutations we mention in our work are also now found in other human-derived SARS-CoV-2 sequences available in public databases (e.g., B.1.1.7 variants containing the Q493K/RRBD mutations; see Table S3), these variants for the time being are rare, and the context in which they arose is also not defined based on public information (e.g., whether they occurred in a healthy person or an immunocompromised individual, or whether the individual received treatment with convalescent plasma or therapeutic antibodies prior to sampling, etc.). Although we used infectious SARS-CoV-2 in some assays, we used replication-defective pseudotyped lentiviruses as a surrogate system for studying the effects of S mutations. The replication fitness of infectious SARS-CoV-2 carrying the S mutations we studied remains to be determined. We used multiple VH3-53-derived monoclonal antibodies isolated from a healthy donor (n = 1), and single antibodies from two additional donors (B38 from Wu et al. [2020] and CC12.1 from Rogers et al. [2020]) but did not test all VH3-53-derived monoclonal antibodies identified to date. Other VH3-53-derived antibodies may be differently impacted by specific mutations because of differences in their light chain genes and CDR H3 loops. Last, although we predicted that residue R100aVH was a serine in the germline C1A-VH3-53 antibody based on our analysis using the IMGT/V-QUEST database (Brochet et al., 2008), this database is likely missing alleles. To prove that our D gene assignment was accurate, we would have had to sequence D gene segments in PBMC donor C1, and we did not perform this analysis. There is, therefore, the possibility that an arginine or lysine would be found at position 100aVH in a germline C1A-VH3-53 antibody.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-IgG-APC | BD Biosciences | Cat# 550931; RRID: AB_398478 |
| anti CD19-FITC | BD Biosciences | Cat# 340864; RRID: AB_400152 |
| anti-human Fc-HRP | Sigma-Aldrich | Cat# A0170; RRID: AB_257868 |
| Bacterial and virus strains | ||
| SARS-CoV-2 USA-WA1/2020 | Harcourt et al., 2020 | N/A |
| Biological samples | ||
| PBMC and plasma from SARS-CoV-2 convalescent donors | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant SARS-CoV-2 spike (“S2P”) | This paper | N/A |
| Recombinant SARS-CoV-2 RBD | This paper | N/A |
| TEV protease | This paper | N/A |
| BirA ligase | This paper | N/A |
| Streptavidin-PE | Invitrogen | Cat# S866 |
| Ni Sepharose Excel | GE Healthcare | Cat# 17-3712-02 |
| Human ACE2-Fc fusion protein | This study | N/A |
| MabSelect SuRE Resin | GE Healthcare | Cat# 97067-944 |
| Kifunensine | Sigma-Aldrich | Cat# K1140 |
| Peg/Ion Screen | Hampton | HR2-126 |
| Peg/Ion 2 Screen | Hampton | HR2-098 |
| Pierce Protein G Ultra Link Resin | Thermo Fisher Scientific | Cat# 53126 |
| PEGRx 1 Screen | Hampton | HR2-082 |
| PEGRx 2 Screen | Hampton | HR2-084 |
| Critical commercial assays | ||
| SuperScriptTM III reverse transcriptase | Invitrogen | Cat# 18080044 |
| Expifectamine 293 Transfection Kit | Thermo Fisher Scientific | Cat# A14525 |
| Lipofectamine 3000 | Thermo Fisher Scientific | Cat# L3000150 |
| Deposited data | ||
| Crystal structure of C1A-B3/RBD complex | This paper | PDB: 7KFW |
| Crystal structure of C1A-F10/RBD complex | This paper | PDB: 7KFY |
| Crystal structure of C1A-C2/RBD complex | This paper | PDB: 7KFX |
| Crystal structure of C1A-B12/RBD complex | This paper | PDB: 7KFV |
| Experimental models: Cell lines | ||
| HEK293T cells | ATCC | CRL-11268; RRID:CVCL_1926 |
| HEK293T-hACE2 | Michael Farzan | N/A |
| HEKS293S Gnti−/− cells | ATCC | CRL-3022; RRID:CVCL_A785 |
| Vero E6 C1008 cells | ATCC | ATCC CRL-1586; RRID:CVCL_0574 |
| Expi293F cells | ThermoFisher | Cat#A14527; RRID: CVCL_D615 |
| Expi293F SARS-CoV-2 S2P-His6 | Bing Chen | N/A |
| Oligonucleotides | ||
| Primers for B cell receptor nested PCR and sequencing | Scheid et al., 2011 | N/A |
| Recombinant DNA | ||
| SARS-CoV-2 S2P | This paper | N/A |
| CR3022 Fab | Galit Alter | GenBank: DQ168569.1 and DQ168570.1 |
| 4A8 | Chi et al., 2020 | Sequence from PDB: 7C2L |
| B38 | Wu et al., 2020 | Sequence from PDB: 7BZ5 |
| CC12.1 | (Yuan et al., 2020a) | Sequence from PDB: 6XC2 |
| REGN10933 | Hansen et al., 2020 | Sequence from PDB: 6XDG |
| REGN10987 | Hansen et al., 2020 | Sequence from PDB: 6XDG |
| Human ACE2 | Michael Farzan, Hyeryun Choe | N/A |
| Codon optimized SARS-CoV-2 S pCAGGS | This paper | N/A |
| Codon optimized SARS-CoV-2 S D614G pCAGGS | This paper | N/A |
| Codon optimized SARS-CoV-2 S D614G N439K pCAGGS | This paper | N/A |
| Codon optimized SARS-CoV-2 S D614G Q493K pCAGGS | This paper | N/A |
| Codon optimized SARS-CoV-2 S D614G Q493R pCAGGS | This paper | N/A |
| Codon optimized SARS-CoV-2 S day 146∗ pCAGGS | This paper | N/A |
| Codon optimized SARS-CoV-2 day 152 pCAGGS | This paper | N/A |
| VSV G pCAGGS | Radoshitzky et al., 2007 | N/A |
| psPAX2 | Didier Trono | Addgene Cat#63592 |
| lentiCas9-EGFP | Chen et al., 2015 | Addgene Cat#63592 |
| Software and algorithms | ||
| IMGT/V-QUEST | http://www.imgt.org | N/A |
| ForteBio data analysis software | ForteBio | N/A |
| FlowJo version 10.6.2 | FlowJo | https://www.flowjo.com; RRID:SCR_008520 |
| XDS build 202 00131 | Kabsch, 2010 | https://xds.mr.mpg.de/;RRID:SCR_015652 |
| AIMLESS v0.5.32 | Evans and Murshudov, 2013 |
http://legacy.ccp4.ac.uk/html/aimless.html; RRID:SCR_015747 |
| Phaser v2.8.3 | McCoy et al., 2007 | https://www.phenix-online.org/documentation/reference/phaser.html; RRID:SCR_014219 |
| Phenix v1.18.2-3874 | Adams et al., 2010 | https://www.phenix-online.org; RRID: SCR_014224 |
| Buster v2.10.3 | Bricogne et al., 2017 | https://www.globalphasing.com/buster/; RRID: SCR_015653 |
| Prism v8.4.3 | GraphPad Software | https://www.graphpad.com:443/; RRID: SCR_002798 |
| O v15.0 | Jones et al., 1991 | N/A |
| Other | ||
| anti-CD20 MicroBeads | Miltenyi Biotec | Cat# 130-091-104 |
| MACS LS column | Miltenyi Biotec | Cat# 130-042-401 |
| anti-FLAG M2 Affinity Gel | Sigma-Aldrich | Cat# A2220; RRID: AB_10063035 |
| Protein G UltraLink | Thermo Fisher Scientific | Cat#: 53125 |
| MabSelect SuRE Resin | GE Healthcare | Cat# 17547401 |
| Streptavidin biosensor | ForteBio | Cat# 18-5020 |
| MoFlo Astrios EQ Cell Sorter | Beckman Coulter | N/A |
| iQue Screener PLUS | Intellicyt | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jonathan Abraham (jonathan_abraham@hms.harvard.edu).
Materials availability
Unique reagents generated in this study are available from the Lead Contact upon request with completed material transfer agreements (MTA). GISAID accession numbers for the sequences analyzed in Figure 3 are provided in Figure S6 and Table S3.
Data and code availability
All relevant data are available from the authors upon request. Protein Data Bank (PBD) identification numbers for the C1A-B3/RBD, C1A-F10/RBD, C1A-C2/RBD, and C1A-B12 RBD complexes are PDB: 7KFW, 7KFY, 7KFX, and 7KFV, respectively.
Experimental model and subject details
Donors
This study was approved by the Harvard Medical School Office of Human Research Administration Institutional Review Board (IRB20-0365) as was the use of healthy donor control blood (IRB19-0786). We received informed, written consent from healthy adult males (n = 3) and a female (n = 1) participants who recovered from confirmed SARS2-CoV-2 infection, with mild illness not requiring hospitalization more than five weeks before blood donation. We isolated C1, C2, C3, C4 and control donor plasma and PBMCs by Ficoll-Plaque (GE Healthcare) density centrifugation. C1 is an adult male who was infected with SARS-CoV-2 five weeks prior to blood donation.
Cells and viruses
We maintained HEK293T cells (ATCC CRL-11268; sex/gender: female; RRID: CVCL_1926) in Dulbecco’s Modified Eagle’s Medium (DMEM) (Thermo Fisher Scientific Cat# 10313039) supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% (v/v) penicillin-streptomycin (Thermo Fisher Scientific Cat# 15140163) and Expi293F™ cells (Thermo Fisher Scientific Cat# A14527; sex/gender: female; RRID: CVCL_D615) in Expi293™ expression medium (Thermo Fisher Scientific Cat# A1435102) supplemented with 1% (v/v) penicillin-streptomycin at 37°C. We maintained HEK293T cells grown in suspension in FreeStyle 293 Expression Medium (Thermo Fisher Scientific Cat# 12338026) and HEK293S GnTI−/− cells (ATCC CRL-3022; sex/gender: female; RRID: CVCL_A785) in Freestyle 293 Expression Medium supplemented with 2% (v/v) ultra-low IgG FBS (Thermo Fisher Scientific Cat# 16250078) at 37°C. A HEK293T-hACE2 stable cell line (sex/gender: female) was a gift from Huihui Mou and Michael Farzan and we maintained these cells in DMEM supplemented with 10% (v/v) FBS, 25 mM HEPES, 1% (v/v) penicillin-streptomycin and 1 μg ml-1 puromycin at 37°C. An Expi293F-His6-tagged SARS-CoV-2 S2P stable cell line (sex/gender: female) was a gift from Bing Chen. We maintained these cells as adherent cells in DMEM supplemented with 1% (v/v) GlutaMax (Thermo Fisher Scientific Cat# 35050079), 1% (v/v) penicillin-streptomycin, 10% (v/v) FBS and 1 μg ml-1 puromycin (Thermo Fisher Scientific Cat# A11138-03) at 37°C. The cell line was then adapted to suspension culture and maintained in Expi293™ expression medium supplemented with 1% (v/v) penicillin-streptomycin and 1 μg ml-1 puromycin at 37°C. All cell lines were obtained from commercial vendors with the exception of the Expi293F-His6-tagged SARS-CoV-2 S2P and HEK293T-hACE2 stable cell lines and were not authenticated. We confirmed the absence of mycoplasma in all cell lines through monthly testing using an e-Myco PCR detection kit (Bulldog Bio Cat# 25234).
Passage 4 SARS-CoV-2 USA-WA1/2020 (Harcourt et al., 2020) was received from the University of Texas Medical Branch. A T225 flask of Vero E6 cells was inoculated with 90 μl starting material in 15 mL DMEM containing 2% (v/v) of heat inactivated FBS (HI-FBS) and incubated in a humidified incubator at 37°C with periodic rocking for 1 h. After 1 h, 60 mL of DMEM / 2% (v/v) HI-FBS was added without removing the inoculum and incubated again at 37°C. The flask was observed daily for progression of cytopathic effect and stock was harvested at 66 h post-inoculation. Stock supernatant was harvested and clarified by centrifugation at 5,250 relative centrifugal field (RCF) at 4°C for 10 min and the HI-FBS concentration was increased to a final concentration of 10% (v/v).
Method details
Single B cell sorting and antibody cloning
We stained and sorted single memory B cells as previously described (Scheid et al., 2009) using a MoFlo Astrios EQ Cell Sorter (Beckman Coulter). Briefly, we enriched B cells by incubating PBMCs with anti-CD20 MicroBeads (Miltenyi Biotec Cat# 130-091-104) followed by magnetic separation on a MACS LS column (Miltenyi Biotec Cat# 130-042-401) according to the manufacturer’s instructions. We washed, counted, and resuspended the B cells in phosphate buffered saline (PBS) containing 2% (v/v) FBS. We adjusted the B cells to a density of 1x107 cells and incubated cells with biotinylated SARS CoV-2 spike (S2P) at a concentration of 5 μg ml-1 on ice for 30 min. After washing three times and resuspending the cells, we added anti-IgG-APC antibody (BD Biosciences Cat# 550931; RRID: AB_398478), anti-CD19-FITC antibody (BD Biosciences Cat# 340864; RRID: AB_400152), and streptavidin-PE (Invitrogen Cat# S866). After incubating the cells on ice for 30 min, we washed the cells three times in PBS containing 2% (v/v) FBS and passed the suspension through a cell strainer before sorting.
We performed single cell cDNA synthesis using SuperScript™ III reverse transcriptase (Invitrogen Cat# 18080044) followed by nested PCR amplification to obtain the IgH, Igλ, and IgΚ variable segments from memory B cells as previously described (Scheid et al., 2011). We used IMGT/V-QUEST (Brochet et al., 2008) (http://www.imgt.org) to analyze IgG gene usage and the extent of variable segment somatic hypermutation. The variable segments were cloned into the pVRC8400 vector for expression of the IgG and Fab constructs as previously described (Clark et al., 2018).
Protein production
For single B cell sorting we cloned a construct comprising human codon optimized SARS-CoV-2 S (GenBank ID: QHD43416.1 residues 16-1208) with a “GSAS” substitution at the furin cleavage site (residues 682-685), stabilized in the prefusion conformation through proline substitutions at residues 986 and 987 (Wrapp et al., 2020), and a C-terminal foldon trimerization motif followed by a BirA ligase site, a Tobacco Etch Virus (TEV) protease site, a FLAG tag, and a His6-tag into the pHLsec vector (Aricescu et al., 2006), which contains its own secretion signal sequence. We note that two N-terminal S residues (residues 14 and 15) downstream of the native S signal peptide were inadvertently omitted from the S2P construct during subcloning. We transfected Expi293F™ cells using an ExpiFectamine™ transfection kit (Thermo Fisher Scientific Cat# A14525) according to the manufacturer’s protocol. We purified the protein using anti-FLAG M2 Affinity Gel (Sigma-Aldrich Cat# A2220; RRID: AB_10063035) according to manufacturer’s protocol and removed the FLAG tag and His6-tag with TEV digestion followed by reverse nickel affinity purification and size-exclusion chromatography on a Superose 6 Increase column (GE Healthcare Life Sciences). We biotinylated the protein with BirA ligase as previously described (Clark et al., 2018).
To obtain recombinant S2P for ELISAs, we used Ni Sepharose® Excel (GE Healthcare Life Sciences Cat# 17-3712-02) to purify His6-tagged SARS-CoV-2 S2P from the supernatant of Expi293F cells stably expressing this protein (a gift of Bing Chen). We further purified the protein using size exclusion chromatography on a Superpose 6 Increase column.
We synthesized human codon optimized cDNA for antibodies based on publicly available sequences; 4A8 (Chi et al., 2020) (PDB: 72CL), B38 (PDB:7BZ5) (Wu et al., 2020), CC12.1 (Yuan et al., 2020a) (PDB: 6XC2), and REGN10933 and REGN10987 (Hansen et al., 2020) (PDB 6XDG). Control Fab CR3022 cDNA (GenBank IDs: DQ168569.1 and DQ168570.1) was a gift from Galit Alter. All C1A-VH3-53 and control antibody variable heavy and light chain gene regions were cloned into the pVRC8400 vector. We transfected Expi293F™ cells using an ExpiFectamine™ transfection kit according to the manufacturer’s protocol. We affinity purified the IgG and Fabs using MabSelect SuRE Resin (GE Healthcare Cat# 17547401) using the manufacturer’s protocol. We further purified all Fabs by size exclusion chromatography on a Superdex 200 Increase column (GE Healthcare Life Sciences), which eluted as single peaks at the expected retention volume.
We subcloned constructs for the SARS-CoV-2 S RBD (GenBank ID: QHD43416.1 residues 319-541) into the pHLsec (Aricescu et al., 2006) vector for use in ELISAs, BLI binding studies, and X-ray crystallography. For ELISAs and crystallography the construct includes an N-terminal His6-tag, a TEV protease site and a short linker (amino acids SGSG). For BLI-binding assays, the construct includes an N-terminal His6-tag, followed by a TEV protease site, a BirA ligase site, and a 7-residue linker (amino acids GTGSGTG). We produced proteins for ELISAs and BLI-binding assays by using linear polyethylenimine (PEI) MAX (Polysciences Cat# 24765-1) to transfect HEK293T cells grown in suspension and purified by nickel affinity purification. For BLI-binding assays the protein was digested with TEV protease to remove the His6-tag followed by reverse nickel affinity purification. We biotinylated proteins with BirA ligase as previously described (Mahmutovic et al., 2015), followed by a reverse nickel affinity purification step to remove BirA ligase, which contains a His6-tag and cannot be separated by size exclusion chromatography from the SARS-CoV-2 RBD due to its similar size. For crystallography, we produced RBDs by PEI MAX transfection of GnTI−/− HEK293S cells grown in suspension or HEK293T cells grown in suspension and also in the presence of kifunensine (Sigma-Aldrich Cat# K1140) at 5 μM, purified these by nickel affinity, and removed the His6-tag by TEV digestion followed by reverse nickel affinity purification. As a final step, we used size exclusion on a Superdex 200 Increase column, in which each recombinant RBD protein ran as a single peak at the expected retention volume.
We subcloned the ectodomain of human ACE2 (GenBank ID: BAB40370.1) residues 18-740, with cDNA obtained as a gift from Michael Farzan, with a C-terminal Fc tag into a pVRC8400 vector containing human IgG1 Fc (a gift from Aaron Schmidt). We expressed the protein in Expi293F™ cells using an ExpiFectamine™ transfection kit according to the manufacturer’s protocol and purified the protein with MabSelect SuRE Resin using the manufacturer’s protocol, followed by size exclusion chromatography on a Superose 6 Increase column, with the protein eluting at the expected retention volume.
Protein crystallization
We prepared each Fab:SARS-CoV-2 RBD complex by mixing RBD with 1.5 molar excess of Fab. The mixtures were incubated at 4°C for 1 h prior to purification on a Superdex 200 Increase column in buffer containing 150mM NaCl, 25 mM Tris-HCl, pH 7.5. Each complex co-eluted as a single peak at the expected retention volume. We adjusted the concentration of each complex to 13 mg ml-1 and screened for crystallization conditions in hanging drops containing 0.1 μl of protein and 0.1 μl of mother liquor using a Mosquito protein crystallization robot (SPT Labtech) with commercially available screens (Hampton Research) (see Key resources table). Crystals grew within 24 h for the C1A-B12 Fab:RBD complex in 0.1 M Bicine pH 8.5, 20% (w/v) polyethylene glycol 10,000, for the C1A-B3 Fab:RBD complex in 0.2 M Ammonium phosphate dibasic, 20% (w/v) polyethylene glycol 3,350; for the C1A-C2 Fab:RBD complex in 0.03 M citric acid, 0.07M BIS-TRIS propane pH 7.6, 20% (w/v) polyethylene glycol 3,350, and for C1A-F10 Fab:RBD complex in 0.10% (w/v) n-Octyl-B-glucoside, 0.1 M Sodium citrate tribasic dihydrate pH 4.5, and 22% (w/v) polyethylene glycol 3,350.
Structure determination
All crystals were flash frozen in mother liquor supplemented with 15% (v/v) glycerol as cryoprotectant. We collected single crystal X-ray diffraction data on Eiger X 16M pixel detectors (Dectris) at a wavelength of 0.979180 Å at the Advanced Photon Source (APS, Argonne, IL) NE-CAT beamline 24-ID-E for the C1A-B12 Fab:RBD and C1A-B3 Fab:RBD complexes and NE-CAT beamline 24-ID-C for the C1A-C2 Fab:RBD and C1A-F10 Fab:RBD complexes. Diffraction data were indexed and integrated using XDS (build 202 00131) (Kabsch, 2010) and merged using AIMLESS (v0.5.32) (Evans and Murshudov, 2013). The structure of the C1A-B12 Fab:RBD complex (space group P212121) was determined by molecular replacement using Phaser (v2.8.3) (McCoy et al., 2007), with coordinates for the B38 Fab variable domain, constant domain and RBD (PDB ID: 7BZ5) (Wu et al., 2020) used as search models. Three copies were found in the asymmetric unit (ASU). We performed iterative model using O (Jones et al., 1991) and refinement in Phenix (v1.18.2-3874) (Adams et al., 2010) and Buster (v2.10.3) (Bricogne et al., 2017), during which we also built alternative conformations for residue side chains where density was apparent. During refinements, we updated TLS groups calculated using Phenix (Adams et al., 2010) and a python script, as well as occupancy restraints calculated in Buster. During model building, we also customized geometry restraints to prevent large displacement of unambiguous contacts in poor regions; the restraints were released once refinements became stable. Water molecules were automatically picked and updated in Buster, followed by manual examination and adjustment till late-stage refinement. The structures of the C1A-B3:RBD complex (space group P212121, three copies per ASU), C1A-C2:RBD complex (space group C2221, one copy per ASU) and C1A-F10:RBD complex (space group C2221, one copy per ASU) were determined using RBD and the C1A-B12 Fab variable and constant domains as search ensembles with CDR and flexible loops truncated, with iterative model building and refinement as described above. Data collection, processing and refinement statistics are summarized in Table S2. PDB validation reports are included as Data S1.
Structural Analysis
We analyzed the structures and generated figures using PyMOL (Schrödinger).
Lentivirus pseudotype production
Human codon optimized SARS-CoV-2 S (GenBank ID: QJR84873.1 residues 1-1246) with a modified cytoplasmic sequence that includes HIV gp41 residues (NRVRQGYS) replacing C-terminal residues 1247-1273 of the S protein (a gift from Nir Hacohen) was subcloned into the pCAGGS expression vector. With this human codon optimized modified S construct as a starting point, we used Gibson assembly to generate the D614GS, D614GS/N439KRBD, D614GS/Q493KRBD, D614GS/Q493RRBD, day 146∗, and day 152∗ S variants. Day 146∗ S is derived from hCoV-19/USA/MA-JLL-D146/2020 (EPI_ISL_593557) but contains WT sequences at positions 12-18, a deletion spanning NTD residues 142-144, and the additional Y489HRBD mutation (Figure S6). Day 152∗ S is derived from hCoV-19/USA/MA-JLL-D152/2020 (EPI_ISL_593558) and contains the additional Y489HRBD mutation (Figure S6). A pCAGGS expressor plasmid for VSV G was previously described (Radoshitzky et al., 2007). To package lentivirus, we co-transfected HEK293T cells using lipofectamine™ 3000 (Thermo Fisher Scientific Cat# L3000015) with an envelope gene-encoding pCAGGS vector, a packaging vector containing HIV Gag, Pol, Rev, and Tat (psPAX2, provided by Didier Trono, Addgene #12260), and a transfer vector containing GFP (lentiCas9-EGFP, a gift from Phillip Sharp and Feng Zhang, Addgene #63592) (Chen et al., 2015) in which we deleted Cas9. After 18 h, we changed the supernatant to DMEM containing 2% FBS (v/v). We harvested supernatants after 48 and 72 h, centrifuged at 3000 x g for 5 min, and filtered the supernatants through a 0.45 μm filter. All pseudotypes except those containing day 152∗ S were concentrated. To concentrate the lentivirus pseudotypes, we layered the supernatant on top of a 10% (v/v) sucrose cushion in 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 0.5 mM EDTA and spun samples at 10,000 x g for 4 h at 4°C. We removed supernatants and resuspended virus pellets in Opti-MEM (Thermo Fisher Scientific Cat# 31985070) containing 25 mM HEPES and 5% (v/v) FBS and stored these at −80°C.
Pseudotype neutralization experiments
We purified polyclonal IgG from human plasma samples using Pierce™ Protein G UltraLink™ Resin (Thermo Fisher Scientific Cat# 53126) following the manufacturer’s protocol. We pre-incubated polyclonal serum IgG, monoclonal antibodies, or an ACE2-Fc fusion protein with SARS-CoV-2 S, SARS-CoV-2 S mutants, or VSV G lentivirus pseudotypes in the presence of 0.5 μg ml-1 of polybrene for 1 h at 37°C. Virus antibody mixtures were added to HEK293T-hACE2 with incubation on cells at 37°C for 24 h, and the media replaced with DMEM containing 10% (v/v) FBS, 1% (v/v) penicillin-streptomycin (v/v), and 1 μg ml-1 puromycin. We determined the percent of GFP positive cells by FACS with an iQue Screener PLUS (Intellicyt) 48 h after initial infection. We calculated percent relative entry by using the following equation: Relative entry (%) = (%GFP-positive cells in the presence of antibody/%GFP-positive cells in the absence of antibody) × 100. Antibody neutralization of pseudotypes was calculated as follows: Neutralization (%) = [1−(%GFP-positive cells in the presence of antibody/%GFP-positive cells in the absence of antibody)] × 100.
Live virus PRNT experiments
Monoclonal antibody samples were serially diluted in Dulbecco’s Phosphate-Buffered Saline (DPBS, Thermo Fisher Scientific Cat# 14190144) using half-log dilutions starting at a concentration of 50 μg ml-1. Dilutions were prepared in triplicate for each sample and plated in triplicate. Each dilution was incubated at 37°C for 1 h with 1,000 plaque-forming units ml-1 (PFU ml-1) of SARS-CoV-2 USA-WA1/2020. 200 μl of each dilution was added to the confluent monolayers of NR-596 Vero E6 C1008 cells (ATCC CRL-1586; RRID:CVCL_0574) in triplicate and incubated in a 5% CO2 incubator at 37°C for 1 h. The cells were rocked gently every 15 min to prevent monolayer drying. Cells were then overlaid with a 1:1 solution of 2.5% (v/v) Avicel® RC-591 microcrystalline cellulose and carboxymethylcellulose sodium (DuPont Nutrition & Biosciences) and 2x Modified Eagle Medium (MEM - Temin’s modification, Thermo Fisher Cat# 10370088) supplemented with 100 x antibiotic-antimycotic (Thermo Fisher Scientific Cat# 15240062) and 100 X GlutaMAX both to a final concentration of 2 x, and 10% (v/v) FBS. The plates were then incubated at 37°C for two days. After two days, the monolayers were fixed with 10% (v/v) neutral buffered formalin (NBF) (Fisher Scientific Cat# LC146705) for at least 6 h and stained with 0.2% (v/v) aqueous Gentian Violet (Fisher Scientific Cat# 3233-16) in 10% (v/v) NBF for 30 min, followed by rinsing and plaque counting.
ELISA experiments
We coated NUNC MaxiSorp plates (Thermo Fisher Scientific Cat# 44-2404) with His6-tagged SARS-CoV-2 S2P, SARS-CoV-2 RBD, or LUJV GP1 (produced as previously described (Mahmutovic et al., 2015)) in PBS overnight at 4°C, followed by a blocking step with PBS containing 3% (v/v) and BSA 0.02% (v/v) Tween. We incubated monoclonal antibodies at a concentration of 100 μg ml-1 for one hour. We then washed samples three times with PBS containing 0.02% (v/v) Tween. We detected bound antibody with horseradish peroxidase (HRP)-coupled anti-human (Fc) antibody (Sigma-Aldrich Cat# A0170).
Biolayer interferometry assays
We performed BLI experiments with an Octet RED96e (Sartorius). For affinity measurements, biotinylated SARS-CoV-2 RBD was loaded onto a streptavidin (SA) sensor (ForteBio) at 1.5 μg ml-1 in kinetic buffer (PBS containing 0.02% Tween and 0.1% BSA) for 100 s. After a baseline measurement for 60 s in kinetic buffer, antibody Fabs were associated for 300 s followed by a 300 s dissociation step. We used ForteBio data analysis software to determine kinetics of binding using a 1:1 binding model.
For ACE2-Fc competition experiments, we loaded biotinylated SARS-CoV-2 RBD onto SA sensors (ForteBio) at 1.5 μg ml-1 for 80 s. We associated C1A-B12 Fab or CR3022-Fab at 250 nM or buffer for 180 s followed by an association with ACE2-Fc or CR3022 Fab at a concentration of 250 nM for 180 s.
Quantification and statistical analysis
All statistical analysis was performed using Prism v8.4.3 (GraphPad Software, https://www.graphpad.com; RRID: SCR_002798). Statistical details for experiments are found in figure legends, including the statistical tests used, the exact value of n, and what n represents.
Acknowledgments
We thank Bing Chen for providing a SARS-CoV-2 S2P expressing stable cell line, Michael Farzan and Huihui Mou for providing HEK293T-hACE2 cells, and Hyeryun Choe for providing hACE2 cDNA. We also thank Lindsey Baden, Michael Mina, and Brendan Blumenstiel for help with sample acquisition and SARS-CoV-2 testing. This work is based upon research conducted at the Northeastern Collaborative Access Team (NE-CAT) beamlines, which are funded by the National Institute of General Medical Sciences from the NIH (P30 GM124165). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract DE-AC02-06CH11357. We thank the staff at NE-CAT for assistance with X-ray data collection. Funding was provided by the Massachusetts Consortium on Pathogen Readiness (MassCPR) and the China Evergrande Group (to J.A.). We acknowledge support for COVID-19 related structural biology research at Harvard from the Nancy Lurie Marks Family Foundation and the Massachusetts Consortium on Pathogen Readiness. The project described was also supported by the National Institute of General Medical Sciences (T32GM007753). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Author contributions
Conceptualization, S.A.C., L.E.C., J.P., A.C., S.S., A.G., and J.A.; Investigation, S.A.C., L.E.C., J.P., A.C., L.G.A.M., S.S., R.I.J., V.B., M.C.C., J.R., J.Z.L., A.G., and J.A.; Writing – Original Draft: J.A.; Writing – Review & Editing, S.A.C., L.E.C., J.P., A.C., L.G.A.M., S.S., R.I.J., A.G., and J.A.; Funding acquisition, JA.
Declarations of interest
A provisional application protecting various aspects of inventions described in this publication has been filed by the President and Fellows of Harvard University.
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. We worked to ensure that the study questionnaires were prepared in an inclusive way. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper received support from a program designed to increase minority representation in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: March 16, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.03.027.
Supplemental information
References
- Abraham J. Passive antibody therapy in COVID-19. Nat. Rev. Immunol. 2020;20:401–403. doi: 10.1038/s41577-020-0365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., Barbian K., Judson S.D., Fischer E.R., Martens C., et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell. 2020;183:1901–1912. doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricogne G., Blan E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O.S., Vonrhein C., et al. Global Phasing Ltd.; 2017. BUSTER version 2.10.3. [Google Scholar]
- Brochet X., Lefranc M.P., Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkn316. W503-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Sanjana N.E., Zheng K., Shalem O., Lee K., Shi X., Scott D.A., Song J., Pan J.Q., Weissleder R., et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.H., Boucau J., Bowman K., et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L.E., Mahmutovic S., Raymond D.D., Dilanyan T., Koma T., Manning J.T., Shankar S., Levis S.C., Briggiler A.M., Enria D.A., et al. Vaccine-elicited receptor-binding site antibodies neutralize two New World hemorrhagic fever arenaviruses. Nat. Commun. 2018;9:1884. doi: 10.1038/s41467-018-04271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S., Cao Y., Zhu Q., Yu P., Qi F., Wang G., Du X., Bao L., Deng W., Zhu H., et al. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell. 2020;183:1013–1023.e13. doi: 10.1016/j.cell.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J., et al. Severe Acute Respiratory Syndrome Coronavirus 2 from patient with coronavirus disease, United States. Emerg. Infect. Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlburt N.K., Seydoux E., Wan Y.H., Edara V.V., Stuart A.B., Feng J., Suthar M.S., McGuire A.T., Stamatatos L., Pancera M. Structural basis for potent neutralization of SARS-CoV-2 and role of antibody affinity maturation. Nat. Commun. 2020;11:5413. doi: 10.1038/s41467-020-19231-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A.T.M., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., et al. CITIID-NIHR BioResource COVID-19 Collaboration. COVID-19 Genomics UK (COG-UK) Consortium SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021 doi: 10.1038/s41586-021-03291-y. Published online February 5, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lefranc M.-P., Lefranc G. Elsevier; 2001. The Immunoglobulin FactsBook. [Google Scholar]
- Leist S.R., Dinnon K.H., 3rd, Schäfer A., Tse L.V., Okuda K., Hou Y.J., West A., Edwards C.E., Sanders W., Fritch E.J., et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;183:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N., et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Mahmutovic S., Clark L., Levis S.C., Briggiler A.M., Enria D.A., Harrison S.C., Abraham J. Molecular basis for antibody-mediated neutralization of New World Hemorrhagic fever mammarenaviruses. Cell Host Microbe. 2015;18:705–713. doi: 10.1016/j.chom.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshitzky S.R., Abraham J., Spiropoulou C.F., Kuhn J.H., Nguyen D., Li W., Nagel J., Schmidt P.J., Nunberg J.H., Andrews N.C., et al. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature. 2007;446:92–96. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku316. W320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid J.F., Mouquet H., Feldhahn N., Walker B.D., Pereyra F., Cutrell E., Seaman M.S., Mascola J.R., Wyatt R.T., Wardemann H., Nussenzweig M.C. A method for identification of HIV gp140 binding memory B cells in human blood. J. Immunol. Methods. 2009;343:65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid J.F., Mouquet H., Ueberheide B., Diskin R., Klein F., Oliveira T.Y., Pietzsch J., Fenyo D., Abadir A., Velinzon K., et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.H., Feng J., et al. Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity. 2020;53:98–105.e5. doi: 10.1016/j.immuni.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Aird D.R., Tamin A., Murakami A., Yan M., Yammanuru A., Jing H., Kan B., Liu X., Zhu Q., et al. Broadening of neutralization activity to directly block a dominant antibody-driven SARS-coronavirus evolution pathway. PLoS Pathog. 2008;4:e1000197. doi: 10.1371/journal.ppat.1000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., et al. ISARIC4C Investigators. COVID-19 Genomics UK (COG-UK) Consortium Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. e00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Peng G., Wilken M., Geraghty R.J., Li F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2012;287:8904–8911. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Honko A., Zhou J., Gong H., Downs S.N., Vasquez J.H., Fang R.H., Gao W., Griffiths A., Zhang L. Cellular nanosponges inhibit SARS-CoV-2 infectivity. Nano Lett. 2020;20:5570–5574. doi: 10.1021/acs.nanolett.0c02278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available from the authors upon request. Protein Data Bank (PBD) identification numbers for the C1A-B3/RBD, C1A-F10/RBD, C1A-C2/RBD, and C1A-B12 RBD complexes are PDB: 7KFW, 7KFY, 7KFX, and 7KFV, respectively.