Abstract
Transcript labeling in intact tissues using in situ hybridization chain reaction has potential to provide vital spatiotemporal information for molecular characterization of heterogeneous neuronal populations. However, large tissue labeling in non-perfused or fresh-frozen rodent and postmortem human samples, which provide more flexible utilization than perfused tissues, is largely unexplored. In the present study, we optimized the combination of in situ hybridization chain reaction in fresh-frozen rodent brains and then evaluated the uniformity of neuronal labeling between two clearing methods, CLARITY and iDISCO+. We found that CLARITY yielded higher signal-to-noise ratios but more limited imaging depth and required longer clearing times, whereas, iDISCO+ resulted in better tissue clearing, greater imaging depth and a more uniform labeling of larger samples. Based on these results, we used iDISCO+-cleared fresh-frozen rodent brains to further validate this combination and map the expression of a few genes of interest pertaining to mood disorders. We then examined the potential of in situ hybridization chain reaction to label transcripts in cleared postmortem human brain tissues. The combination failed to produce adequate mRNA labeling in postmortem human cortical slices but produced visually adequate labeling in the cerebellum tissues. We next, investigated the multiplexing ability of in situ hybridization chain reaction in cleared tissues which revealed inconsistent fluorescence output depending upon the fluorophore conjugated to the hairpins. Finally, we applied our optimized protocol to assess the effect of glucocorticoid receptor overexpression on basal somatostatin expression in the mouse cortex. The constitutive glucocorticoid receptor overexpression resulted in lower number density of somatostatin-expressing neurons compared to wild type. Overall, the combination of in situ hybridization chain reaction with clearing methods, especially iDISCO+, may find broad application in the transcript analysis in rodent studies, but its limited use in postmortem human tissues can be improved by further optimizations.
Keywords: Fluorescence in situ hybridization, Hybridization chain reaction, Fresh-frozen brain, Postmortem human brain, CLARITY, iDISCO+
Introduction
Ribonucleic acids, especially mRNA and non-coding RNAs, give cells their molecular identity through their unique combination and can serve as important markers of activity-dependent gene expression and disease conditions. Since its first introduction, in situ hybridization has been a valuable tool to study RNA biology (Pardue and Gall 1969). Fluorescence in situ hybridization (FISH), in particular, has potential to provide high resolution spatiotemporal information for the molecular characterization of heterogeneous cell populations in the brain. Notably, recent advances in RNA/DNA hybridization techniques allow mapping of multiple mRNA targets simultaneously and with subcellular-specificity at both organ level (e.g. brain, kidney, liver) as well as organism (e.g., zebrafish) level. One such approach is the ‘in situ hybridization chain reaction’ based FISH (HCR FISH) (Dirks and Pierce 2004; Choi et al. 2010; Choi et al. 2018). In principle, self-assembly of metastable fluorophore-labeled DNA hairpins into tethered fluorescent amplification polymers through a chain reaction, is triggered by the presence of an initiator strand (Dirks and Pierce 2004). Applying several of these custom probes to tissue concurrently allows for multiplexing of up to five targets with high specificity. Moreover, the relatively short lengths of probes and hairpins make this technique particularly favorable for intact tissue transcriptional analysis.
The advent of the HCR FISH method alongside the recent development of intact tissue clearing and high-throughput microscopic imaging methods has broadened the path to obtain unprecedented information about the molecular identity, interactions, and activity-dependent functions in the brain. To date, several tissue clearing approaches have been used to achieve rodent brain transparency but only a few have proved to be versatile and successful across a wide range of applications (Silvestri et al. 2016; Azaripour et al. 2016; Wan et al. 2018). Based on factors like clearing capability, target biomolecule fixation, ability to perform ex-vivo labeling, fluorescence retention and imaging depth, we chose to explore two methods 1) tissue lipid matrix replacement-based CLARITY (Chung et al. 2013) with modifications (Tomer et al. 2014; Yang et al. 2014; Zheng and Rinaman 2016; Krolewski et al. 2018) and 2) non-aqueous solvent-based iDISCO+ (Renier et al. 2014). There has been pioneering work done (Sylwestrak et al. 2016; Kramer et al. 2018; Park et al. 2018) but the combination of HCR FISH with clearing techniques still remains relatively unexplored especially with fresh-frozen tissues. In comparison to perfusion fixation, fresh-frozen brain tissue provides flexibility for studying a wide array of biomolecules (e.g., RNA, protein, metabolite) using multiple methods, including in situ hybridization, immunohistochemistry, PCR, western blotting, and spectroscopy. Moreover, postmortem human brains, an invaluable source of information for studying psychiatric and neurodegenerative disorders, are routinely collected without any perfusion fixation and then stored frozen. Therefore, optimization of large tissue clearing and transcript labeling techniques in unfixed-frozen human and rodent brain tissue is needed. Hence, we decided to investigate the compatibility and efficiency of HCR FISH with two clearing approaches, CLARITY and iDISCO+ in fresh-frozen tissues. Thus, the following experiments were designed to 1) evaluate and optimize the combination of HCR FISH with CLARITY and iDISCO+ in fresh-frozen rodent brains and semi-quantitatively analyze the somatostatin (Sst) FISH signal for the uniformity of cellular labeling between the two clearing methods, 2) use HCR FISH to validate and map the expression of a few genes of our interest in fresh-frozen rodent and postmortem human brain tissues, 3) investigate the efficiency of HCR FISH to detect multiple genes in cleared fresh-frozen samples, and 4) apply an optimized volumetric quantitation pipeline of HCR FISH in cleared tissues to study the expression of Sst mRNA in the cortex of a glucocorticoid receptor overexpressing (GRov) mice, a model of increased emotional lability (Wei et al. 2004; Wei et al. 2007; Hebda-Bauer et al. 2010; Wei et al. 2012).
Experimental Procedures
Animal care
All animal care and experimental procedures followed the guide for the Care and Use of Laboratory Animals: Eighth Edition (revised in 2011, published by the National Academy of Sciences), and approved by the University of Michigan Committee on the Use and Care of Animals. The generation of GRov mice has been described previously (Wei et al. 2004). The GRov mouse line was established in the Akil laboratory by breeding founders and their progeny to C57BL/6J mice, and all transgenic mice are maintained as hemizygotes. Adult male Sprague-Dawley rats (3–4 months old) were obtained from Charles River Laboratories (Wilmington, MA) and acclimated to their cages for one week. Mice and rats were housed on a 14:10 and 12:12 light/dark cycle, respectively, with ad libitum access to food and water.
Rodent brain collection
Fresh-frozen brain
Adult male, GRov and WT littermate mice (2–4.5 months old) and Sprague-Dawley rats (3–4 months old) were sacrificed by rapid decapitation, and their brains removed, snap frozen in 2-methyl butane (Isopentane, cooled with dry ice), and stored at −80°C prior to use.
Perfused brain
Adult male C57BL/6J mice were deeply anesthetized with sodium pentobarbital (150 mg/kg) and perfused transcardially with 1x phosphate buffer-saline (PBS) followed by 4% paraformaldehyde (PFA) dissolved in PBS. The brains were removed and post-fixed in PBS containing 4% PFA for overnight and subsequently used for the supplemental experiments.
Human brain tissue acquisition
Blocks of temporal cortex and cerebellum from postmortem human brains were used, which were collected by the Brain Donor Program at the University of California, Irvine with the consent of the relatives of the deceased. Brains were removed at autopsy, quickly chilled to approximately 4°C and then cut into series of 0.75 cm thick coronal slices that were quickly frozen and stored at −80°C as previously described (Jones et al. 1992). Slabs were then placed on dry ice blocks and dissected using a fine-toothed saw to generate tissue blocks of approximately 4×3 cm in size that were then stored at −80°C until further use. Information regarding physical health, medication use, psychopathology, substance use, and details about the final hours of the deceased was obtained from medical records, the coroner’s investigation, the medical examiner’s conclusions and interviews with relatives. RNA integrity number (RIN) and demographics of the subjects used in the study are listed in the supplemental table ESM_14_1.
Sample preparation
Immersion-fixation in paraformaldehyde (PFA)
Fresh-frozen rodent brains and postmortem human brain blocks were immersion-fixed in 4% PFA at 4°C for 12–36 h.
CLARITY sample preparation
Tissues were prepared as described in (Tomer et al. 2014; Krolewski et al. 2018) with modifications to immersion-fixation and the FISH protocol. After fixation, tissues were rinsed in boric acid buffer (0.2 M borate/0.1% Tween-20, pH 8.5) and then immersed in 2% hydrogel solution (2% acrylamide, 0.25% VA-044 initiator, 1x phosphate buffer-saline (PBS), 4% PFA) for 24 h at 4°C. Tubes containing tissues were vacuum degassed then incubated for hydrogel polymerization at 37°C for 3–5 h. Fixed tissues were washed in boric acid buffer for 3 × 2 h each then sectioned using a vibratome (500–1200 μm thick) or a rodent brain matrix (1–4 mm thick). Cut slices were then passively cleared in 4% sodium dodecyl sulfate (SDS) at 45°C for 1–3 weeks and rinsed in boric acid buffer with 0.1 % tween-20 for 3 × 2 h before processing for FISH.
iDISCO+ sample preparation
Tissues were prepared as described in (Renier et al. 2014) with modifications for immersion-fixation and the FISH protocol. Fixed samples were washed in 0.01M PBS/0.1% tween-20 (PBSTw) for 3 × 1 h, with an additional overnight wash at room temperature. Tissues were then sectioned using a vibratome (500–1200 μm thick) or a brain matrix (1–4 mm). Additionally, intact hippocampi from both hemispheres were dissected from a rat brain after the PFA fixation and rinsing. Tissues were then dehydrated in 20%, 40%, 60%, and 80% methanol (in ddH2O) for 1–2 h each, and 100% methanol for 2 × 1–3 h each. Samples were bleached with 5% H2O2 in ice cold methanol at 4°C overnight, and then rehydrated in 80% methanol/H2O for 1 h, 50% methanol/H2O for 1 h, and finally in PBSTw for 1 h twice. Samples were then used for HCR FISH.
RNA cross-linker based fixation
Fixation of fresh-frozen slices with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) was performed according to the method previously described (Sylwestrak et al. 2016). Briefly, iDISCO+ slices were transferred to EDC solution after immersion-fixation in 4% PFA and CLARITY slices after hydrogel embedding and prior to clearing in SDS. First, samples were incubated for 30–90 minutes in methylimidazole buffer (0.1M, pH 8.5) and then transferred to EDC fixative solution (0.1M EDC, 0.1M 5- Ethylthio-1H-tetrazole, in 0.1M methylimidazole buffer, pH 8.5) and incubated overnight at 37°C in the dark on shaker. Following a quick rinse, samples were processed for clearing or dehydration, depending upon the clearing methods.
Slice volume measurements
The effects of CLARITY and iDISCO+ processing on the slice volume was estimated by measurement of dimensions as length (dorso-ventral) × width (medio-lateral) × thickness using an electronic digital caliper (Kobalt 293883, Lowe’s, Mooresville, NC). Opposing hemisected slices (4 mm) from the fresh-frozen mouse brains (n=3) were divided between CLARITY and iDISCO+ groups. Volume estimates were represented as percentage of volume change and compared between 1) pre-cleared, 2) CLARITY (after incubation in histodenz), and 3) iDISCO+ (after incubation in DBE) samples.
Probe design and synthesis
Split-initiator DNA probes for the following genes were used 1) Rat: somatostatin (Sst, NM_012659.2), parvalbumin (Pvalb, NM_022499.2), dopamine-beta-hydroxylase (Dbh, NM_013158.2), tyrosine hydroxylase (Th, NM_012740.3), solute carrier family 6 member 3 (dopamine transporter, Slc6a3/Dat, NM_012694.2), adrenergic receptor alpha-1A (Adra1a, NM_017191.2), −1B (Adra1b, NM_016991.2), phenylethanolamine-N-methyltransferase (Pnmt, NM_031526.1); 2) Mouse: somatostatin (Sst, NM_009215.1), parvalbumin (Pvalb, NM_001330686.1), solute carrier family 17 member 7 (vesicular glutamate transporter 1, Slc17a7/Vglut1, NM_182993.2), glutamate decarboxylase 1 (Gad 67/Gad 1, NM_008077.5), glutamate decarboxylase 2 (Gad 65/Gad 2, NM_008078.2); 3) Human: somatostatin (SST, NM_001048.4), parvalbumin (PVALB, NM_002854.2) and calbindin-1 (CALB, NM_001366795.1). Probes were designed in our lab based on Choi et al. 2018 (listed as table, supplemental material ESM_14_2) and synthesized by Integrated DNA Technologies (Coralville, Iowa, USA), except for the mouse Pvalb which was purchased from Molecular Instruments, Inc. (Los Angeles, California, USA). Depending upon the gene, we excluded the probe-pairs from the pool which showed homologies to non-target regions for as low as 2–4 bases, which decreased the chances of cross- and weak-hybridizations. In this study, probes for DNA hairpins conjugated with AlexaFluor-488 (AF-488), AF-594 and AF-647 were purchased from Molecular Instruments, Inc. (Los Angeles, California, USA). Additionally, DNA hairpins conjugated with fluorescein isothiocynate (FITC), cyanine-3 (Cy3), and AF-647 were designed based on Choi et al. 2014 and synthesized and conjugated by Integrated DNA Technologies (IDT).
HCR FISH
We modified the HCR FISH method as described by Choi et al. 2014, 2018 for thick fresh-frozen samples. CLARITY- and iDISCO+-processed samples were equilibrated with 5x sodium citrate/0.01% tween-20 (SSCTw) buffer for 1 h, then acetylated with 0.25% v/v acetic anhydride solution for 30–60 min. After rinsing with ddH2O, samples were then equilibrated in hybridization buffer (30% deionized formamide, 5x SSC, 0.5 mg/ml yeast tRNA, 10% dextran sulfate) for 1 −2 h. Tissues were then incubated in the hybridization buffer containing 1–6 nM initiator-labeled probes at 37°C for 12–72 h depending upon the tissue thickness. Following hybridization, samples were washed at 37°C with probe wash buffer (30% formamide, 5x SSC, 9 mM citric acid, 0.1% tween 20) for 3 times and then twice with 5x SSCTw for 1 h each. For samples thicker than 1 mm, an additional rinse of 12 h in 5x SSCTw at room temperature was also performed. Tissues were equilibrated in amplification buffer for 1–3 h (5x SSC, 10% dextran sulfate, and 0.1% Tween 20). Fluorophore- labeled hairpins were diluted separately from 3 μM stock to a 2.25 μM final concentration in 20x SSC, heated at 90°C for 90 seconds, and then cooled to room temperature for 30 min in the dark. Cooled hairpins were diluted to a 20–60 nM final concentration in amplification buffer. Tissues were incubated in amplification buffer with hairpins for 1–3 days at room temperature. Finally, tissues were washed in 5x SSCTw for 3 × 1 h (overnight 1 x wash for sections over 1 mm). For Sst quantitation studies, we used a final concentration of 2 nM probe and 20 nM hairpins based on concentration gradient study which enabled us to acquire signal in the form of unsaturated intensity for majority of the pixels within individual Sst+ neurons. Specificity of the Sst probe set was determined by assessing cell-type and anatomical labeling using 20μm thin fresh-frozen mouse brain sections (supplemental figure ESM_13_1).
Refractive index (RI) matching
After FISH, CLARITY samples were optically matched by incubating in 88% histodenz (Sigma-Aldrich, St. Louis, Missouri, USA) at 37°C overnight (Yang et al. 2014). iDISCO+ samples were dehydrated in 20%, 40%, 60%, 80%, and 100% methanol (in ddH2O) for 1 h each with an additional 1–2 h or overnight dehydration in 100% methanol. Samples were then incubated in 1 volume of methanol and 2 volumes of dichloromethane (DCM, Sigma-Aldrich) until they sank in glass vials. Residual methanol from tissues was then washed by two changes of 100% DCM for 30 min. each. Finally, samples were incubated (without shaking) in dibenzyl ether (DBE, Sigma-Aldrich) for 30–60 min to match the RI.
Confocal Microscopy- Imaging and visualization
Samples were placed in 88% histodenz- or DBE- filled chambers made with 3M™ mounting putty (Scotch brand, St. Paul, Minnesota, USA) on a glass slide and cover-slipped. Image stacks were acquired on an Olympus Fluoview 1000 confocal microscope using a 10x magnification/0.4 numerical aperture (N.A.)/ 2.2 mm working distance (WD) objective lens at 405 nm (DAPI), 488 nm, 594 nm, and 647 nm excitation. Image dimension are 1024 × 1024 (1.24 μm/pixel x 1.24 μm/pixel) in the xy-plane with z-step of 4.26 μm. For comparative quantitation of mouse Sst signal, images were acquired within a region of interest (ROI) consisting of cingulate and primary/secondary motor cortex (rostral) to retrosplenial agranular and retrosplenial granular cortex (caudal) and the ROI was kept consistent for all the samples. The image acquisition settings in the Fluoviewer software and the laser power of the confocal microscope were kept constant for the quantitative comparison study. Three-dimensional volume rendering and ortho-slice/xy-plane mode visualization was performed in Amira (ThermoFisher Scientific, Waltham, Massachusetts, USA) and Imaris (Bitplane Inc., Concord, Massachusetts, USA) software. ImageJ was used to extract histogram data from the images.
Light sheet microscopy- Imaging and visualization
Imaging of large intact samples was performed on the CLARITY-optimized light-sheet microscope (COLM) (Tomer et al. 2014; Krolewski et al. 2018). RI-matched samples were mounted in a quartz cuvette and filled with 88% histodenz or DBE. The entire coronal slice was acquired in a series of tiles with a 10x/0.6-N.A. objective (Olympus XLPLN10XSVMP; WD - 8mm). Voxel dimension of the image stacks are 0.595 μm/pixel x 0.595 μm/pixel in xy-plane and 5 μm/slice in the z-direction and acquired image resolution was 2048 × 2048. The number of tiles was specified by defining the coordinates of the two opposing corners of the entire tissue slice and 15–20% tile overlap. Following the optimization of light-sheet and focal-plane alignment parameters over the entire sample space, final image acquisition was initiated and data collected. Tiles of acquired image stacks were stitched together using an in-house modified version of Terastitcher (Bria and Iannello 2012). Stitched coronal image volumes were down-sampled for 3D rendering and visualization using Amira and Imaris. ImageJ was used to extract histogram data from the COLM-acquired images.
Semi-quantitation and statistical analysis
Semi-quantitative analysis of Sst transcript expression in the mouse cortex was performed on the confocal microscope-acquired image stacks using Amira software. Imaging was systematically randomized among the samples to minimize the variation resulting from longer incubation in the 88% histodenz or DBE. The analysis pipeline involved noise removal, thresholding, and quantitation. Image stacks were first processed to achieve optimal thresholding by applying a median filter to both CLARITY and iDISCO+ images. Individual xy-planes of both CLARITY and iDISCO+ stacks were median filtered to reduce the contrast and soften the edges of objects. Although this process reduces the contrast and tends to defocus the image, it helps to eliminate the background noise which substantially improves the thresholding. Processed images were then locally segmented through each xy-plane to extract the target neurons. The extent of thresholding was proportionally determined based on the mean tissue background intensity and validated manually across z-planes and samples to make sure that the majority of the segmented objects are neuronal cell bodies. The thresholding cut off value ranged between 15–20 % and 20–25 % of the maximum pixel intensity value for the CLARITY and iDISCO+ samples, respectively. In order to exclude ‘poorly’ fragmented objects, intact cells were filtered by a combination of morphometric parameters: area, gray level intensity, perimeter, length, breadth, width, and circularity. These morphometric cutoff values were manually determined and were different for CLARITY and iDISCO+ samples due to tissue expansion and shrinkage, respectively, but kept consistent across all the samples in each clearing group. Quantitative data are compared as mean neuronal number per unit ROI volume (number density) and mean number of pixels in Sst+ neurons, whereas semi-quantitative measurements of RNA level is shown as mean intensity of Sst+ neurons, along with mean tissue intensity (background intensity with or without Sst signal), and intensity count or the number of different gray levels in the intensity image associated to each object. Whole ROI was thresholded as single object to estimate total ROI volume and then used to calculate the number density of Sst+ neurons. Finally, independent sample t-tests (to compare the means between 2 groups) or one-way ANOVAs with Tukey-alpha post hoc tests (to compare the means between multiple groups) were performed. Data are represented as mean ± SD.
Results
Optimization and comparison of HCR FISH signal between CLARITY and iDISCO+
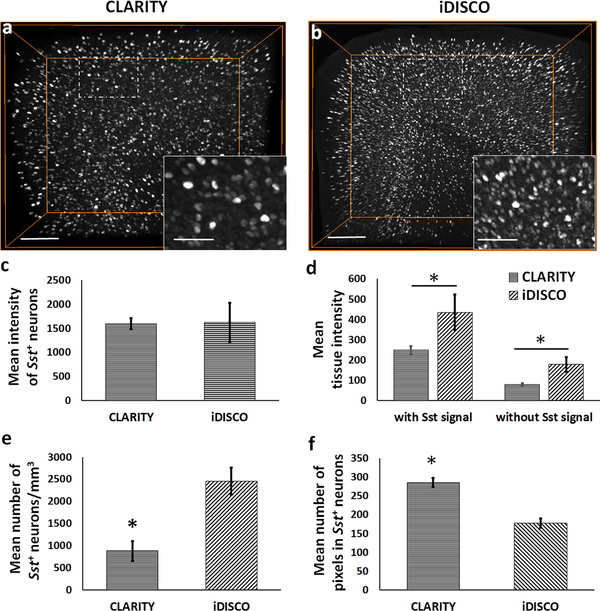
The HCR FISH method as described by Choi et al. 2014, 2018, was modified to include an incubation step of acetylation with acetic anhydride solution which improved the signal to noise (S/N) ratio (data not shown). For iDISCO+ processing, we omitted the tissue permeabilization step with dimethylsulfoxide (DMSO)/glycine/Triton X-100 as mentioned in Renier et al. 2014. This step did not improve the probe permeability, but instead appeared to adversely affect the hybridization step. It is possible that it could be related to the ability of DMSO to form hydrogen bonds with nucleobases and promote destacking, especially in single stranded RNAs (Lee et al. 2013). Following the successful optimization, we compared the efficiency of HCR FISH between CLARITY (n=4) and iDISCO+ (n=5) samples by analyzing the Sst mRNA labeling in the cortex of WT mice (Fig. 1a vs. 1b; supplemental movie ESM_1). We found that the mean intensity of detected Sst+ neurons is similar between the CLARITY and iDISCO+, with relatively higher variation in the iDISCO+ group (Fig. 1c). However, the mean tissue intensity is significantly lower in CLARITY samples vs. iDISCO+ (Fig. 1d) when calculated as background intensity with (two-tailed t(7) = −4.08, p = 0.005) or without (two-tailed t(7) = −5.33, p = 0.001) Sst signal. Lower background intensity in CLARITY samples could help achieve better thresholding outputs than iDISCO+. Within consistent acquired ROI volumes, the CLARITY image stacks comprised a significantly lower number of Sst+ neurons per unit volume (two-tailed t(6) = −8.32, p < 0.001; Fig. 1e) vs. iDISCO+. To control for effect of the acquired ROI volume differences on the number of Sst+ neurons, we evaluated the use of nuclear stain (DAPI), and total RNA staining dyes (acridine orange and SYTO RNASelect) as potential secondary markers. We found that DAPI and acridine orange (data not shown due to negligible signal) are not compatible with iDISCO+ whereas SYTO RNASelect stain, a proprietary dye, has poor compatibility with the CLARITY processed fresh-frozen samples (supplemental figure ESM_13_2). Due to lack of a suitable secondary marker, we normalized the neuronal number data by the total ROI volume and represented it as number of Sst+ neurons per unit volume (number density). In addition, mean number of pixels were significantly higher in the detected Sst+ neurons in CLARITY vs. iDISCO+ (two-tailed t(7) = 13.16, p < 0.001) (Fig. 1f). The above measurements of neuronal number density and pixel coverage, reproduced and validated the previously known expansion and shrinkage caused by CLARITY and iDISCO+ methods, respectively.
Fig. 1. Quantitative differences in the HCR FISH signal (Sst transcript expression) between CLARITY and iDISCO+.
(a-b) 3D volume-rendered images of representative CLARITY and iDISCO+ samples, respectively, showing the observed differences in the detected neuronal density and mean tissue intensity. (c) Bar-diagram shows comparable mean intensity of the detected Sst+ neurons in the acquired cortical ROI between CLARITY and iDISCO+. (d) Mean background tissue intensity, with or without including the Sst signal, and (e) mean number of Sst+ neurons per unit volume were significantly lower in CLARITY samples as compared to the iDISCO+ samples, for consistent cortical ROIs. (f) Bar-diagram shows a significantly higher mean number of pixels in the detected Sst+ neurons in CLARITY images vs. iDISCO+. *p< 0.05. Scale bars- 200 μm (Insets-75 μm).
In a parallel experiment using mouse prefrontal cortical slices, we sought to determine the percent change in tissue volume following CLARITY and iDISCO+ processing. Our measurements showed significant shrinkage in the iDISCO+ samples compared to pre-cleared (two-tailed t(4) = 4.366, p=0.012) and CLARITY processed (two-tailed t(4) = 4.499, p=0.011) samples. However, CLARITY samples did show significant expansion (two-tailed t(4) = −1.771, p=0.177) in comparison to the pre-cleared volume (supplemental figure ESM_13_3).
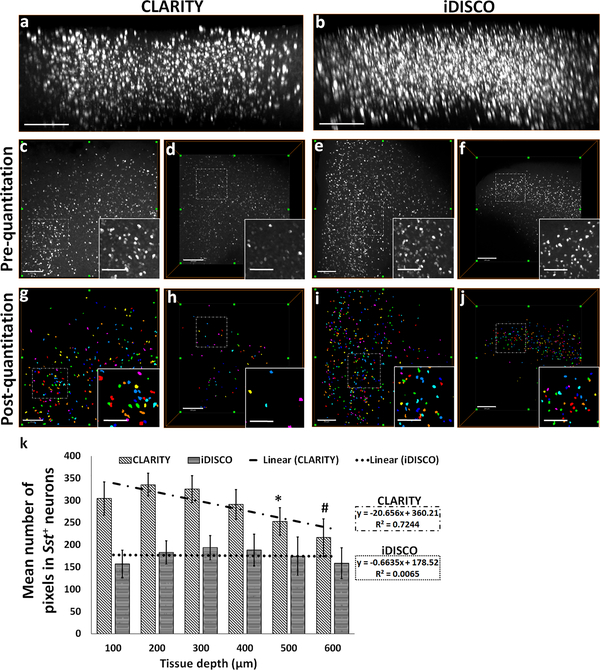
We next compared the fluorescence signal as a function of z-depth to assess any difference which could result from the combination of tissue permeability for DNA probes and optical transparency for imaging between CLARITY and iDISCO+ samples. Confocal microscope-acquired image stacks revealed an attenuation of signal in CLARITY samples with increasing depth (Figs. 2a; 2c vs. 2d; left panel in supplemental movie ESM_1), whereas iDISCO+ samples had a relatively uniform labeling across the z-depth (Figs. 2b; 2e vs. 2f; right panel in supplemental movie ESM_1). In CLARITY samples, thresholded neurons in deeper layers consist of a significantly lower number of pixels in comparison to those in upper layers (F (5, 47) = 17.806; 100 μm vs. 500 μm: p = 0.008, 100 μm vs. 600 μm: p = 0.001; Figs. 2g vs. 2h; 2k). In contrast, the mean number of pixels remained relatively consistent among the thresholded top and bottom layer neurons in the iDISCO+ samples (Figs. 2i vs. 2j; 2k). In summary, the decrease in the optical accessibility and, in turn, the reduced pixel intensity with z-depth potentially result in the detection of fewer numbers of neurons in CLARITY compared to iDISCO+ samples.
Fig. 2. Attenuation of the HCR FISH signal in CLARITY samples measured as a function of z-depth.
xz-plane view of 3D rendered confocal microscope-acquired volumes of (a) CLARITY and (b) iDISCO+ showing greater attenuation of HCR FISH signal with depth. Representative xy-plane images at 100 μm z-depth: CLARITY- (c) pre-quantitation, (g) post-quantitation, iDISCO+- (e) pre-quantitation, (i) post-quantitation, respectively. Representative xy-plane images at 500 μm z-depth: CLARITY- (d) pre-quantitation, (h) post-quantitation, iDISCO+- (f) pre-quantitation, (j) post-quantitation, respectively. (k) Bar-diagram shows mean number of pixels in the detected neurons as a function of increasing z-depth (100 to 600 μm) in CLARITY and iDISCO+ samples. Following the thresholding and quantitation, each neuron is identified and assigned a unique index and displayed using a cyclic colormap so that cells in close proximity are more likely to be shown in a different color. For CLARITY samples, *p<0.05 for comparisons between 100/200/300 vs. 500 μm and #p<0.05 for 100/200/300/400 vs. 600 μm. Scale bar- (a-b) 300 μm; (c-j) 250 μm (Inset bars-100 μm).
HCR FISH in large iDISCO+-cleared fresh-frozen rodent and postmortem human brain
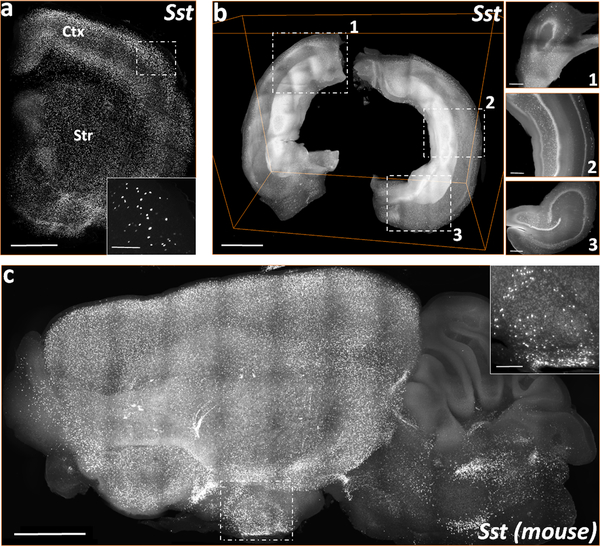
High resolution volumes acquired on COLM allowed us to visualize and reproduce the previously reported anatomical distribution of a few genes of interest to our lab. Layer specific expression of Sst in cortical slices of rat (Fig. 3a, supplemental movie ESM_2) shows a relatively higher density of Sst interneurons (Martinotti cells) in the superficial (L2/3) and deep (L5/6) cortical layers. Mid-layer (L4) contains a sparse number of Sst+ neurons mainly corresponding to small basket cells (Iritani and Satoh 1991; Kawaguchi and Kubota 1997; Markram et al. 2004; Urban-Ciecko and Barth 2016). Fig. 3b and supplemental movie ESM_3 show the Sst mRNA expression in a COLM-acquired volume of an intact rat hippocampus which was dissected from the brain post-PFA fixation and then processed through iDISCO+ and HCR FISH. Fig. 3c and supplemental movie ESM_4 shows the Sst mRNA expression in a representative COLM-acquired volume of a whole mouse hemisphere, with supplemental movie ESM_5 showing a zoomed-in view of the expression in the brainstem region.
Fig. 3. 3D visualizations of COLM acquired stacks of Sst mRNA labeling in the rodent brain cleared using iDISCO+.
(a) A representative stack of rat forebrain hemi-slice corresponding to a 2 mm thick region between bregma = 2.00 mm and 0.00 mm, rostro-caudally. (b) Volume rendering of an intact rat hippocampus. Inset, xy-plane views of 1) dorsal, 2) medio-lateral, and 3) ventral hippocampus. (c) 3D volume rendering of a mouse left hemisphere (~4 mm thick, sagittal view). Scale bars (μm) - (a) 300 (Inset-100); (b-c) 250 (Inset-100); (d) 500 (Inset-200); (e) 1000 (Inset-300); (f) 1000 (Inset-200).
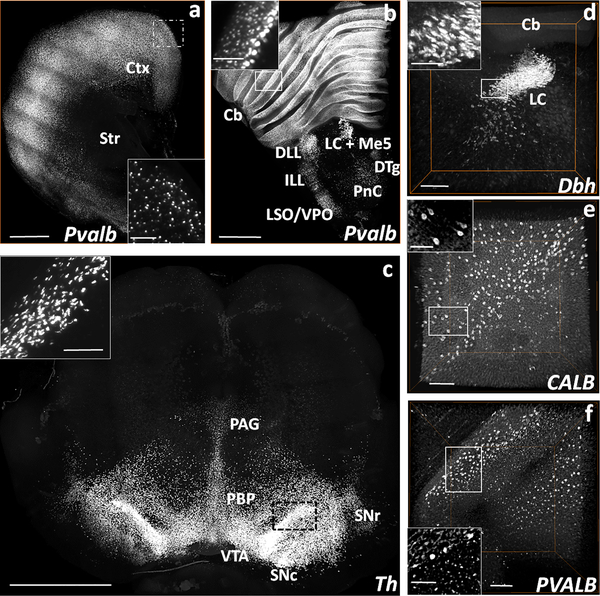
A COLM-acquired volume of a rat forebrain (Figs. 4a, supplemental movie ESM_6) and brainstem hemi-slices (Fig. 4b, supplemental movie ESM_7) show the expression pattern of Pvalb mRNA. The mRNA expression pattern of Th in the ventral tegmental and substantia nigra regions of the rat brain slice can be seen in the COLM-acquired volume (Fig. 4c, supplemental movie ESM_8). The confocal microscope-acquired hemisphere volume in Fig. 4d, supplemental movie ESM_9 shows the Dbh mRNA expression in the caudal half volume of the rat locus coeruleus (LC) consisting of the posterior pole and the middle part spanning the dorso-ventral axis. Following the successful validation in fresh-frozen rodent brain samples, we tested HCR FISH in iDISCO+-processed postmortem human brain tissues (~1mm thick). We observed insufficient labeling for PVALB (supplemental figure ESM_13_4), CALB and SST (data not shown) transcripts in the temporal cortex of a representative control subject (#2292, supplemental material ESM_14_1). However, the 1200 μm deep stacks of cerebellum samples from the same subject showed appreciable signal for transcripts CALB and PVALB. CALB mRNA labeling was observed in most of the Purkinje cells (Fig. 4e, supplemental movie ESM_10) while the PVALB mRNA labeling was seen in the Purkinje cells as well as in the basket and stellate cells of the molecular layer (Fig. 4f, supplemental movie ESM_11).
Fig. 4. Volume rendered visualizations of Pvalb, Dbh, and Th mRNA expressing neurons in the rat brain, and CALB+ and PVALB+ neurons in the postmortem human cerebellum following HCR FISH and iDISCO+.
(a) COLM-acquired volume of Pvalb expressing neurons in a 2 mm thick rat cortex (Ctx) and striatum (Str) hemi-slice between bregma = 2.00 mm and 0.00 mm. (b) COLM-acquired image of Pvalb expression pattern in a 2 mm rat brainstem hemi-slice between bregma = −8.44 mm and −10.44 mm. (c) COLM-acquired image of Th expression in a 2.5 mm thick rat mid-brain volume between bregma = −4.40 mm and −6.90 mm. (d) Confocal microscope-acquired image stack of Dbh expression in the locus coeruleus (LC) of rat brainstem hemi-slice between bregma = −9.72 mm and −10.32 mm. (e) CALB and (f) PVALB expression in the postmortem human cerebellum of a control subject (#2292) acquired on confocal microscope (1200 μm deep stacks). Cb- cerebellum, DTg- dorsal tegmental nucleus, ILL/DLL- intermediate/dorsal lateral lemniscus, LC+Me5- locus coeruleus + mesencephalic trigeminal nucleus, PnC- caudal pontine reticular nucleus, LSO/VPO- lateral/ventral superior olive, PAG- periaqueductal gray, PBP- parabrachial pigmented nucleus of the VTA, SNc/r- substantia nigra compacta/reticulata, VTA-ventral tegmental area. Scale bars (μm) - (a-b) 1000 (Inset-400); (c) 500 (Inset-100); (d) 1000 (Inset-200); (e-f) 500 (Inset-250).
Inadequate FISH signal in postmortem cortical tissues prompted us to explore the potential benefits of RNA cross-linker EDC. Interestingly, our qualitative study using mouse CLARITY and iDISCO+ samples showed weak Sst signal in the PFA+EDC fixed tissues (supplemental figure ESM_13_5) compared to PFA fixed samples. Similarly, we did not observe any improvement in the FISH signal for receptor transcripts like Adra1a/1b in rodent tissue or PVALB, CALB and SST transcripts in postmortem human cortical tissues (data not shown).
Multiplexing ability of HCR FISH in cleared iDISCO+ tissues
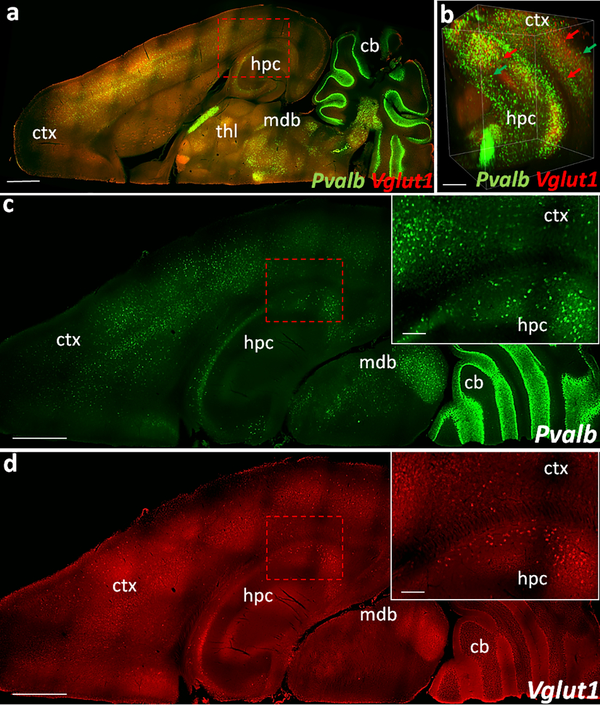
Next, we evaluated the ability of HCR FISH to detect multiple genes in cleared fresh-frozen samples. Our initial experiments using both 10 μm thin sections as well as CLARITY- and iDISCO+-processed thicker samples showed inconsistency in the fluorescence signal with the use of different fluorophore-tagged hairpins. Fig.5 and supplemental movie ESM_12 show dual HCR FISH labeling for Pvalb and Vglut1 in an iDISCO+ processed WT mouse hemisphere. Under basal conditions, Vglut1 expressing glutamatergic neuronal population is known to be significantly greater than the Pvalb expressing interneuron population. However, we detected fewer Vglut1+ neurons with high background signal using the hairpins conjugated with AF-594 in comparison to the Pvalb+ neurons detected with AF-647 dyes. To determine whether these variations were fluorophore-based, we designed a set of probes with the same mRNA detecting sequence but with different initiators, e.g., probe1-B1 and probe1-B2 (Choi et al. 2010). Using multiple initiator-specific DNA hairpins tagged with different fluorophores (e.g., B1H1-AF647:B1H2-AF647; B2H1-AF488:B2H2-AF488), we were able to detect and directly compare the HCR signal of the same gene in different laser channels. Fig. 6 shows confocal microscope-acquired HCR FISH images of Dbh mRNA in the LC region using hairpins conjugated with AF-488, AF-594, and AF-647 in 20 μm thin sections. Hairpins tagged with AF-647 produced a high S/N ratio (Fig. 6d), whereas hairpins tagged with AF-488 or AF-594 showed low S/N ratio (Fig. 6b and 6c). COLM-acquired images of iDISCO+-processed thicker tissues showed similar differences in fluorescence signal (Fig. 7a vs. 7b; 7c vs. 7d). Altogether these indicate a possible difference in the stability of fluorophores AF-488 and AF-594 or their conjugation stability with DNA hairpins, in comparison to the AF-647 which consistently showed higher S/N ratio across the experiments. We also tested the efficiency of DNA hairpins conjugated with alternative fluorophores FITC and Cy3. However, both Cy3 (Fig. 7e vs. 7f) and FITC-conjugated hairpins (data not shown) failed to produce a consistent and equivalent S/N ratio compared to that of AF-647 in the iDISCO+ samples.
Fig. 5. 3D visualization of COLM acquired dual HCR labeling in iDISCO+ tissue.
(a) Dorsal view of a representative mouse hemisphere showing Pvalb (green, AF-647) and Vglut1 (red, AF-594) labeling in down-sampled image volume (b) Volume rendered view showing cellular resolution in ventral hippocampus and cortex (parts of lateral entorhinal cortex) in an extracted sub-volume. (c-d) Dorsal views of a different z-plane than (a) showing Pvalb and Vglut1 FISH signal. Insets, zoomed-in view. Images are pseudocolored for the overlaying purpose. Scale bars (μm) - (a) 1500; (b) 300; (c-d) 1000 (Inset-200).
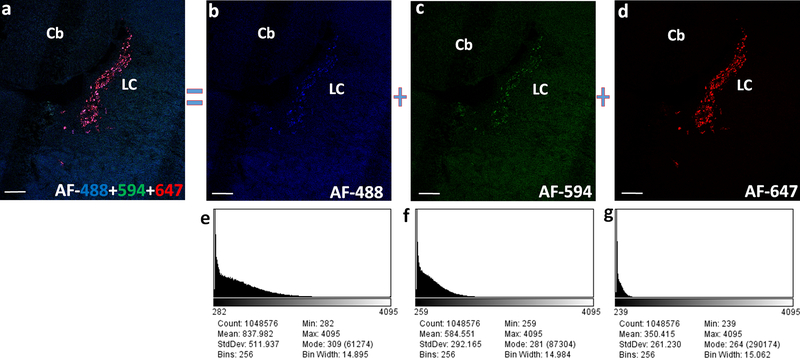
Fig. 6. Fluorophore based differences in the fluorescence output imaged using confocal microscope.
(a) Dbh expression in the rat locus coeruleus (LC) following multiplexed HCR FISH using a set of common mRNA binding probes and 3 different hairpins AF-488, AF-594 and AF-647. Relatively weak fluorescence and high background recorded with the use of AF-488 (b) and AF-594 (c) tagged DNA hairpins in comparison to the strong fluorescence with low background following the use of hairpins conjugated with AF-647 (d). Screenshots of the histogram (e-g) for their respective z-projection images (b-d), generated in the ImageJ software depict the poor S/N ratio resulting from the use of AF-448 and AF-594 compared to a better S/N ratio from the use of AF-647. Images are pseudocolored for the overlaying purpose. Scale bars- 200 μm.
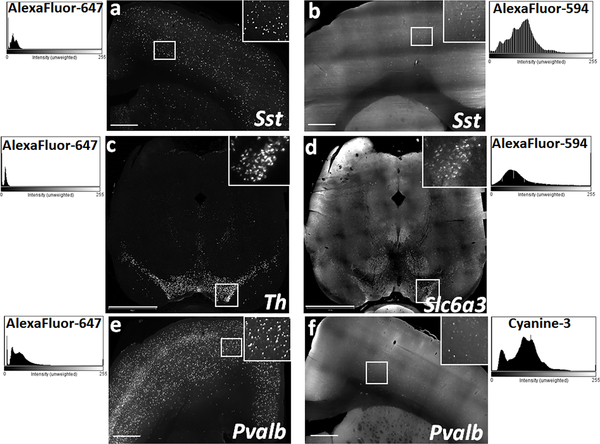
Fig. 7. Fluorophore based differences in the quality of fluorescence output imaged using light sheet microscope.
Representative xy-plane images of Sst labeled neurons in rat cortical slices using DNA hairpins conjugated with (a) AF-647 and (b) AF-594. Representative xy-plane images of rat mid-brain slices following multiplexed HCR FISH for two abundantly expressed genes in the substantia nigra and ventral tegmental area - (c) Th, detected with AF-647 and (d) Dat (Slc6a3) detected with AF-594. Comparison of the Pvalb signal in rat cortical tissue using DNA hairpins conjugated with (e) AF-647 and (f) Cy3. ImageJ generated histograms on the respective sides, show consistently high S/N ratio achieved with the use of AF-647 in comparison to the low S/N ratio with AF-594 or Cy3. Scale bars- (a, b, e, f) 500 μm; (c-d) 1200 μm.
Application of volumetric HCR FISH to semi-quantitatively assess the effect of mouse GR-overexpression on the basal cortical expression of Sst
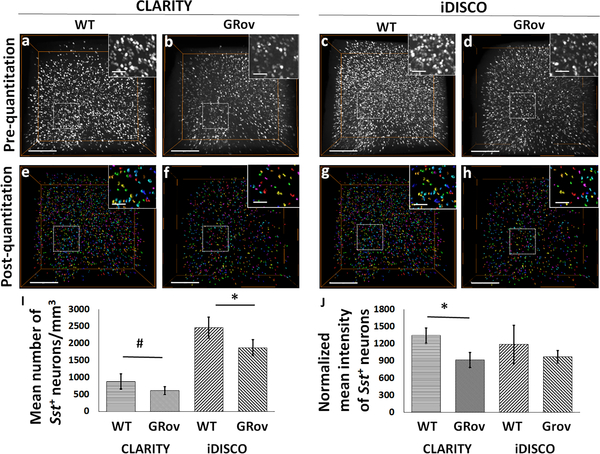
To establish a quantitative application of HCR FISH in combination with clearing methods, we compared the cortical expression of Sst transcripts between WT (n=5) and GRov (n=5) mice. Estimation of Sst+ neurons per mm3 (neuronal volume density) showed a significantly lower number of detected neurons in the cortex of GRov vs. WT mice in both CLARITY (two-tailed t(7) = 2.35, p = 0.051; Fig. 8a vs. 8b; 8e vs. 8f; 8i), and iDISCO+ samples (two-tailed t(10) = 2.84, p = 0.017; Fig. 8c vs. 8d; 8g vs. 8h; 8i). Normalized mean intensity of the detected Sst+ neurons was significantly lower in GRov cortex comparison to WT in CLARITY samples (two-tailed t(7) = 4.82, p = 0.002) but not in iDISCO+-processed samples (two-tailed t(3.5) = 1.67, p = 0.179; Fig. 8j).
Fig. 8. GR overexpression negatively affects the expression of Sst in the analyzed cortical ROI of mice.
3D volume-rendered confocal images show qualitative visualization of Sst+ neuronal density in CLARITY- (a) WT, (b) GRov; and iDISCO+-(c) WT, (d) GRov. Surface-rendered images show post-quantitation volume density of Sst+ neurons in CLARITY- (e) WT, (f) GRov; and iDISCO+-(g) WT, (h) GRov. (i) Bar-diagram shows the significant difference in the number of Sst+ neurons between WT and GRov samples processed through CLARITY (cWT vs. cGRov) or iDISCO+ (iWT vs. iGRov). (j) Bar-diagram shows significant difference in the normalized mean intensity of Sst+ neurons between cWT and cGRov. #p=0.051; *p<0.05. Scale bars-300 μm (Inset bars-100 μm).
Discussion
In the current study, we sought to assess the compatibility and efficiency of HCR FISH in fresh-frozen brain samples cleared using different approaches and select the suitable platform to map and analyze transcripts. Traditionally, fresh-frozen tissues have been preferentially used as starting material for both radioactive- and coloric-based digoxigenin-based in situ hybridization. Non-perfused frozen tissues provide flexible utilization for a wide range of biochemical and molecular studies. It is the most routinely used method for postmortem human brain collection. On the other hand, fixed tissues from live animal perfusions have superior retention of selective biomolecules especially proteins. However, perfusion-induced excellent cross-linking might affect the utilization of these tissue with numerous techniques. Therefore, we explored the differences in fluorescence output between perfused and fresh-frozen iDISCO+-cleared wild-type rodent brain samples by comparing the Pvalb and Dbh labeling (supplemental figure ESM_13_6). Qualitatively, perfused samples had substantially weaker fluorescent intensity for both Pvalb and Dbh transcripts compared to fresh-frozen tissues. Similarly, a weak fluorescence signal for total RNA staining using SYTO RNASelect dye was observed in perfused iDISCO+ slices (supplemental figure ESM_13_2). We believe an extensive cross-linking within the perfused tissues potentially renders mRNAs less accessible to the probes or dyes. However, these observations need further validation using a larger sample size and quantitative measurements. Altogether, based on the unique opportunity fresh-frozen tissues provide, we decided to investigate the compatibility and efficiency of HCR FISH with two clearing approaches, CLARITY and iDISCO+. Subsequently, we explored the optimization of RNA fixation, tissue permeability and clearing, hybridization and semi-quantitative analysis in large tissue samples.
Comparison of HCR FISH in fresh-frozen brain tissues cleared using CLARITY or iDISCO+
The time and efficiency of tissue clearing using CLARITY was dependent upon the origin of tissue and sample thickness, whereas, iDISCO+ processing yielded optimal transparency within a standard time period (Renier et al. 2014) in the fresh-frozen brain samples irrespective of tissue origin or size. Typically, under passive clearing conditions in 4% SDS (Tomer et al. 2014), lipid removal time for a typical 2–4 mm thick slice of 2% acrylamide + 4% PFA perfused rodent brain ranges from 1–2 weeks. In our experience, it could take up to 2–3 weeks to achieve a comparable transparency in fresh-frozen immersion-fixed rodent slices. Additional clearing time is required for postmortem human brain tissues of similar thickness mainly due to the higher tissue density and extensive myelination. In terms of imaging depth capability, we consistently achieved depth of 1500–2500 μm in iDISCO+ samples whereas in most of the CLARITY samples, mRNA labeling could be recorded up to only 700–1000 μm deep.
Semi-quantitatively, both CLARITY- and iDISCO+-processed mouse cortical samples showed a comparable mean signal intensity (Sst mRNA expression), suggesting a similar mRNA preservation and efficacy for the combination of HCR FISH and fresh-frozen immersion-fixed samples. However, the mean tissue background intensity (with or without including the Sst signal) was significantly lower in CLARITY compared to iDISCO+ samples. The final steps of the iDISCO+ method (including dehydration with methanol, de-lipidation with DCM, and RI matching with DBE) shrink and harden the tissue. These modifications in the cleared iDISCO+ samples cause local variations in the RI and contribute to higher tissue background depending upon the regional content and morphology (e.g., ventricles, edges, and fiber dense regions) compared to CLARITY samples. Analysis of the HCR FISH signal as a function of image z-depth revealed a significant attenuation of the signal in CLARITY samples. Mean pixel number quantitation showed that neurons located at deep layers were represented by significantly fewer pixels than those located towards top layers. This attenuation is potentially caused by inadequate sample transparency in the fresh-frozen CLARITY tissues. In contrast, fresh-frozen samples with similar thickness when processed through iDISCO+ showed relatively uniform labeling of Sst+ neurons across the z-depth. The combination of sample volume change and signal attenuation are responsible for the observed difference in the neuronal number density between CLARITY and iDISCO+ methods. Furthermore, CLARITY processing-induced expansion causes the fluorescent signal from Sst+ neurons to be recorded on significantly more pixels. In contrast, iDISCO+ processing-induced tissue shrinkage allowed for Sst+ neurons to be recorded on fewer pixels. Analysis of frequency distribution for the range of different gray-levels associated with individual neurons (also referred to as intensity counts in Amira software) indicated that a higher percent of CLARITY neurons (recorded on more pixels than iDISCO+) have wide range of gray-levels associated with them (supplemental figure ESM_13_7). This could mean that CLARITY may provide more accurate intensity-dependent measurements than iDISCO+ where the majority of neurons have lower range of gray-levels (supplemental figure ESM_13_7). Overall, the shrinkage in iDISCO+ samples does not confound quantitation, especially when comparing samples processed via the same clearing method, either iDISCO+ or CLARITY. Nevertheless, this iDISCO+ limitation is offset by requiring less time and computational resources to acquire larger samples and perform volume analysis. Depending upon the image sensor, acquisition of high-resolution images (for example, using COLM system to acquire images with greater-than or equal to 2048 × 2048 pixels, less-than 0.595 μm/pixel x 0.595 μm/pixel, and less-than 5 μm z-spacing on) could theoretically allow a greater range of pixel numbers associated with each object of interest. However, management, processing, and analysis limitations of high-resolution large data sets present a challenge, often requiring some down-sampling of large acquired volumes, which mitigate any advantage of this alternative. Altogether, considering the tissue shrinkage, better transparency, and resulting ability to image deeper, the iDISCO+ method allowed the acquisition of larger tissue volumes with comparable signal intensity.
One major problem we faced performing quantitative expression analysis is the limitation of 10x magnification and 0.4 (confocal) or 0.6 (COLM) N.A. objectives and relative fluorescence measurements in form of semi-quantitative analysis. Absolute quantitation of mRNA expression in the form of grain counting is the most accurate and sensitive approach, which would have allowed us to do more precise comparison of mRNA visualization and analysis between the two clearing methods. Quantitation at grain-level is only possible through the use of high N.A. (≥1.0), 25–100x magnification and long working distance lenses suitable for CLARITY or iDISCO+ imaging (e.g., multiphoton emission (MPE) objectives) and similar to high resolution imaging, acquisition of thicker samples at such high magnification will generate large amounts of data whose management and processing will presents additional challenges. Exploration of software tools, Matlab blockproc and chunking with natural language toolkit (NLTK) to facilitate challenging jobs of processing and analyzing large data sets in the biological studies are needed. Nonetheless, lenses and system configurations suitable for imaging RNA signal at large scale are currently unavailable or underdeveloped, which limited our fluorescence estimations to the approach we have implemented in the current study. We reduced the uncertainty in total fluorescence measurement by optimizing the hairpin amplification, and imaging parameters to acquire signal in the form of unsaturated intensity for majority of the pixels within an individual cell. Normalization of the intensity variations within a sample (using tissue background signal, as used in our analysis) can provide better control in large tissue analyses in comparison to the intensity normalization across the group (Sylwestrak et al. 2016). Furthermore, we picked a set of highly specific 6 probe-pairs for the mouse Sst studies as compared to our standard 20 probe-pair list. Optimizing the HCR FISH with fewer probes was one of the original goals of our study which was aimed to substantially reduce the cost of probe production without compromising the signal which we have demonstrated in the present study. Ability to work with fewer probes provided us the unique opportunity to select the most specific probes which minimizes the randomness associated with probe binding and manage the signal saturation, both necessary for intensity based semi-quantitative analysis of high copy number genes like Sst. Testing the hybridization efficiency of individual probe-pairs in a large probe-pool required for moderate to low copy numbers genes is practically challenging. In such a case, one can generate an intensity and or grain number curve against increasing number of probe-pairs which can be then used as a standard to validate and estimate the number of bound probe-pairs responsible for acquired signal in the study.
It is also important to highlight the fact that quality of thresholding and quantitative analysis can be affected by the differential gene expression between neurons which is evident from the difference in the number of Sst neurons accounted in analysis. More neurons can be visually perceived in inset of Fig. 2 c/d compared to fewer thresholded neurons in the inset of Fig. 2 g/h. The applied threshold value was chosen after manually exploring various thresholding levels in combination with morphometric filters to ensure that majority of the neurons included in the quantitation are properly segmented. We realized that there will always be a certain level of trade-off between the inclusion of whole cells vs. fragmented cells. However, consistent thresholding values across the samples and groups provide adequate control for the relative comparison in the current study. Such limitations can be minimized by using a combinatorial approach with the use of high N.A. objectives, smaller z-steps, dynamic range imaging, blind deconvolution using practically calculated PSF, multiple iterations of thresholding and implementation of deep learning tools like Deep-Z (Wu et al. 2019). The application of above discussed approaches would minimize the intensity variations and ensure that data from one experiment can be correlated to another and still have the ability to use intensity as a semi-quantitative measure of relative gene expression.
Validation of HCR FISH in iDISCO+-cleared rodent and postmortem human brain
We successfully validated our optimized combination of HCR FISH and iDISCO+ methods by examining the expression pattern of a few genes pertaining to the GABAergic and monoaminergic systems. The selected genes are implicated in the pathophysiology of anxiety, stress and mood disorders which are the primary areas of interest in our laboratory. Sst and Pvalb represent two major populations of interneurons whereas Th and Dbh represent distinct populations in SN/VTA and noradrenergic areas including locus coeruleus, respectively. The combination of HCR FISH and iDISCO+ methods produced excellent signal for the above selected genes (under basal conditions, these genes can be categorized as abundant- to moderately-expressing genes) in intact fresh-frozen rodent brain tissues. However, HCR FISH failed to detect basal level low copy number transcripts (e.g., adrenergic receptor alpha-1A and 1B, Adra1a/1b) or genes expressed in sparse neuronal populations with moderate copy numbers (e.g., phenylethanolamine-N-methyltransferase, Pnmt). This indicated a low sensitivity of HCR FISH towards genes with low expression, especially in the large tissues subjected to a clearing procedure. Sensitivity and signal amplification can be improved by increasing the number of probe-pairs to hybridize a larger portion of the mRNA length and by optimizing the fixation conditions. Our attempts to use an RNA cross-linker like EDC did not show any advantage over the use of PFA for the genes of our interest. Moreover, EDC-fixed CLARITY samples required extended clearing time (e.g., slices of 2–4 mm thickness might require an additional 1 week for rodent tissues and 1–2 weeks for human tissues). We observed relatively poor Sst FISH signal in the EDC fixed CLARITY and iDISCO+ samples compare to non- EDC fixed (supplemental figure ESM_13_5). Together, these finding suggest that the additional cross-linking could negatively impact the tissue clearing, permeability and optical transparency. Such RNA cross-linking, however, might help with the fixation of short RNAs like microRNA as shown by (Sylwestrak et al. 2016).
Next, we investigated the potential of HCR FISH based transcript labeling in the cleared postmortem human brain tissues. Our laboratory primarily studies the mechanisms of psychiatric disorders in postmortem human brain. Previous studies from our lab and others implicate the prefrontal cortex in schizophrenia, bipolar, and major depressive disorders based on microarray and radioactive in-situ hybridization analyses. Since the prefrontal cortex plays a critical role in such disorders, it is a commonly sought-after region studied within the Pritzker Consortium, limiting its availability and use for pilot optimization experiments. Therefore, we decided to evaluate and optimize these methods in temporal cortex tissue which is more readily available and has comparable gross anatomical and histochemical features as the prefrontal cortex. We also used cerebellum as a positive control for the overall methodology, mainly due to its efficient clearing and abundant expression of several genes of interest. We examined the expression pattern of genes belonging to the GABAergic system SST, PVALB, and CALB (which are known to play an important role in the pathophysiology of mood disorders) in cleared temporal cortex and cerebellum slices. We detected sparse signal for PVALB in the iDISCO+ cleared cortical slices. In contrast, adequate labeling for PVALB and CALB transcripts was detected in the cerebellum tissues. We found that significantly greater autofluorescence and tissue density in the cortical regions as compared to cerebellum likely contributes to observed differences in the sensitivity of HCR FISH between these two brain regions. Unfortunately, the presence of lipofuscin-induced autofluorescence across a wide spectral range further limits the multiplexing ability and reduces the HCR FISH sensitivity for less abundant genes. Alternatively, we subtracted the lipofuscin signal (acquired using a different laser (e.g., 488 nm) from the target signal to extract only the transcript specific signal (AF-647). However, this approach did not improve the sparse and weak signal of cortical PVALB labeling in our study. The poor detectability shown in the supplemental image Fig.13_4, highlights the challenge associated with the combination of clearing and FISH methods in the postmortem human cortical tissues. In addressing the challenge, it’s worth noting that lipofuscin excitation profiles can vary across the spectrum. An ideal subtraction experiment would be to include lipofuscin signal acquired before labelling to use on the same target wavelength as the HCR FISH fluorophore (e.g., lipofuscin signal in 647 nm before labeling with hairpins conjugated with AF-647). However, these experiments present practical challenges when scaling to larger samples and accommodating different clearing techniques. Alternatively, the accuracy of spectral subtraction can be improved by combining the lipofuscin signal recorded in more than two channels, especially adjacent to the target fluorophore channel. Further optimization of HCR FISH and clearing methods is still needed to improve the labeling in postmortem human brain tissues. RIN values often only provide a general assessment of RNA quality and may not necessarily indicate transcript-specific information. It also has a limited predictability of postmortem human brain tissue quality and mRNA integrity within the tissue. For example, Sonntag et al. 2016 showed that “the RIN reflects a decrease in full-length mRNA quantity, rather than integrity”. Hence, we believe that our probe design/selection strategy in combination with sample selection based on higher RIN scores, and transcript-specific quality information (Wang et al. 2016), could substantially improve the hybridization efficiency in the postmortem human tissues. Furthermore, utilization of thin slices (less than 500 μm) to optimize the fixation, de-lipidation and clearing, probe and hairpin penetration, could improve the quality of labeling that is needed for the visualization transcripts in the postmortem human tissues. Tissue fixation with low concentration of PFA (e.g., 2%) might provide optimum RNA preservation and yet enable mRNA accessibility to the probes by limiting the cross-linking within the tissue. Extensive myelination in human brain tissue certainly requires longer dehydration and de-lipidation with methanol and dichloromethane to ensure maximum probe permeability and optical transparency. Shorter versions of probes and hairpins, in theory, can maximize the permeability, whereas probe coverage for the full length of mRNA would ensure a greater sensitivity and signal amplification. Lipofuscin-induced autofluorescence can be best circumvented by applying a spectral subtraction method and use of far-red to near-infra red fluorescent tagged hairpins (e.g., AF-750) to avoid the emission range of lipofuscin (Yin 1996; Biesemeier et al. 2011).
Fluorophore based differences in the HCR FISH signal output
Evaluation of multiplexed probes in large fresh-frozen samples was the primary aim of this study. We discovered that effectiveness of the HCR FISH technique was critically dependent on which fluorophores we chose. For example, AF-647 tagged hairpins produced the most consistent fluorescence signal with highest S/N ratio and proved to be the most resistant to photobleaching under extended imaging sessions using both confocal and light sheet microscopes. In contrast, hairpins tagged with AF-488 and AF-594 failed to produce as consistent and comparable S/N ratio across the experiments. One possible reason for the differences in the fluorescent signal output could be the relative instability of AF-488 and AF-594 molecules themselves or their conjugation to the DNA hairpin when compared to that of AF-647. According to the proprietary manufacturer Thermofisher Scientific, the AF-647 fluorophore has superior photostability in buffers and mounting media when compared with both AF-488 and AF-594. This could be potentially explained by the differences in the chemical structure of the fluorophores’ core region (PubChem-AF488; PubChem-AF594; PubChem-AF647). We found that fluorophore-based differences in the output signal were most evident when HCR FISH was performed on low expressing genes, and also when in combination with tissue clearing methods that involve longer incubation in high salt concentraSution buffer (5x SSCTw) and tissue clearing solvents, DCM and DBE (both potent oxidizing chemicals). It is also possible that the higher quantum efficiency of the sCMOS cameras (COLM) or photomultiplier tubes (confocal microscope) in the spectral region of 450 to 600 nm contributes to the observed noise as seen with AF-488 or AF-594, which could substantially confound any weak target signal. Further experiments are required to establish the exact cause of such fluorophore-based differences in signal output as there appear to be several possibilities. In addition to the z-signal attenuation in the CLARITY samples, the inconsistent and weak fluorescence from the use of AF-488/594 conjugated hairpins in both CLARITY and iDISCO+, limits the possible use of genes, vesicular GABA transporter (vGat) and Gad 67/65, that label the GABAergic neuronal population, as secondary markers to control for any volume differences in the quantitative study, as it requires adequate multiplexing of vGat or Gad with Sst.
In order to find an alternative approach, we explored the efficiency of DNA hairpins conjugated with non-AF fluorophores, FITC and Cy3, but these failed to produce a comparable signal to that of AF-647. We compared the HCR FISH signal using AF-647-conjugated hairpins purchased from either IDT or Molecular Instruments, Inc. to test whether vendor differences in conjugation chemistry in generating DNA hairpins makes a difference. We observed less consistency in the fluorescence signal across the FISH experiments using IDT-synthesized AF-647- conjugated hairpins (data not shown), as compared to hairpins from Molecular Instruments, indicating that different conjugation chemistry may be important. Moreover, when immunochemistry was performed using secondary antibodies tagged with the same fluorophores in combination with tissue clearing methods, the above mentioned differences in fluorescence output were not observed (Krolewski et al. 2018). This further indicates that conjugation chemistry likely plays a significant role in the stability of the fluorophores tagged on to the DNA hairpins vs. antibodies.
Forebrain glucocorticoid receptor overexpression reduces the basal level of cortical Sst mRNA expression
Compromised cortical interneurons and a critical role for Sst have been implicated in anxiety, depression and psychiatric illness (Fung et al. 2010; Lin and Sibille 2013; Lin and Sibille 2015; Fee et al. 2017). However, the mechanistic role of Sst, one of most widely distributed GABAergic peptide, in anxiety- and depression-like behaviors remains largely unexplored. We decided to use the optimized volumetric HCR FISH to explore the basal expression of Sst transcripts in the cortex of GRov mice which exhibit higher levels of total GR mRNA and approximately 78% more GR protein in the forebrain leading to a significant increase in anxiety-like and depressant-like behaviors relative to wild type (WT) controls (Wei et al. 2004). Our lab has previously shown that transient early-life, as well as constitutive, GR overexpression in forebrain causes profound long-term alterations in the region-specific transcriptome and emotional reactivity (Wei et al. 2004; Wei et al. 2007; Hebda-Bauer et al. 2010; Wei et al. 2012). The GRov brains used in the present study were from mice that had GR overexpressed in forebrain from early life throughout adulthood (from when the Ca2+/calmodulin-dependent protein kinase promoter becomes active around the 2nd week of life until the animals were sacrificed). Our findings showed that GR overexpression beginning in early life negatively affects the basal expression of Sst in the adult cortex, as shown by a significantly lower Sst+ neuronal number density when compared to the WT samples (processed either through CLARITY or iDISCO+). Based on our semi-quantitative analysis, a similar trend in the normalized mean intensity suggests low Sst transcript copy number in GRov cortices compared to WT. Our observation is consistent with previous studies showing that GR overexpression may amplify the overall glucocorticoid response in the cortex which, in turn, negatively affects the Sst expression in the rodent cortex (Papachristou et al. 1994). The semi-quantitative comparison of Sst expression between WT and GRov groups using both, CLARITY and iDISCO, validated that the differences are indeed biological and not confounded by the clearing method. These findings also suggest that CLARITY would be particularly suited to studies using relatively thin rodent samples (≤1 mm) under circumstances where tissue expansion would provide better resolution and a higher S/N ratio (e.g., densely packed regions of substantia nigra and locus coeruleus).
Conclusion
HCR FISH is compatible with both CLARITY and iDISCO+ clearing techniques for the detection of abundant to moderately expressed mRNAs. The iDISCO+ method provided the more suitable and versatile platform for analyzing transcripts in fresh-frozen and immersion-fixed tissues. iDISCO+ processing is the least time-consuming clearing method and produces excellent tissue clearing and permeability to achieve suitable signal amplification for the imaging of large samples at the cellular level resolution. Tissue shrinkage with iDISCO+ enables imaging of large samples in the form of more manageable image volumes which in turn allow efficient data processing with less down-sampling required. On the other hand, CLARITY images were better resolved with higher S/N ratios than iDISCO+ images likely due to tissue expansion and improved probe and hairpin penetration. The superior resolution of CLARITY images also helps minimize the variations in intensity-based comparisons. However, less efficient clearing and restricted imaging depth in fresh-frozen samples limit the use of CLARITY to relatively small tissues sizes. HCR FISH in cleared samples provides a unique opportunity to study postmortem human brains, which are a valuable resource for understanding the underlying mechanisms of psychiatric disorders. Nonetheless, postmortem human brain presents unique challenges (e.g., tissue and RNA quality as a function of postmortem interval, higher tissue density mainly due to extensive myelination and presence of lipofuscin-induced autofluorescence) which demand further optimization of HCR FISH and clearing methods. Quantitative fluorescence analysis remains limited to semi-quantitation approaches due to the requirement and unavailability of high N.A. (>1.0), 60x/100x magnification and long working distance lenses suitable for CLARITY or iDISCO+. Relative, yet semi-quantitative analysis requires gene- and tissue-specific optimization of signal amplification, image acquisition with nonsaturated-pixels, and normalization of intensity to warrant that data can be correlated between the study groups or different experiments. Overall, we conclude that the benefits of iDISCO+ make it more suitable for HCR FISH based transcript analysis in intact rodent and postmortem human brain samples with further optimizations needed to enhance the sensitivity of HCR FISH.
Supplementary Material
3D visualization of differences in the HCR-FISH labeled Sst signal between CLARITY (left panel) and iDISCO+ (right panel) processed mouse cortical slices. First half of this movie shows a series of xy-plane images moving in z-direction of confocal acquired representative stacks, followed by maximum intensity projection (mip) 3D pre-quantitation volume and post-quantitation surface rendering of Sst neurons. Second half of the movie shows a series of xz-plane images moving in y-direction, followed by mip 3D pre-quantitation volume and post-quantitation surface renderings of Sst neurons. This animation highlights the higher number density of detected Sst neurons and uniform labeling across the z-plane in iDISCO+ compared to CLARITY.
3D visualization of HCR FISH-labeled Sst expressing neurons in a rat forebrain hemi-slice cleared using iDISCO+. This movie first shows a series of xy-plane images acquired on a COLM followed by a mip 3D volume rendering. The characteristic layer specific expression pattern of Sst in rat cortex can be observed in this movie.
Volume rendering of a rat hippocampus showing Sst expression pattern in the iDISCO+ cleared intact tissue and labeled using HCR FISH method. This movie shows a series of xy-plane images acquired on a COLM followed by 3D mip volume rendering.
3D visualization of a mouse left hemisphere showing Sst expressing neurons across the entire rostro-caudal extent of the brain. Post HCR FISH labeling, brain tissue was cleared using the iDISCO+ method and imaging was performed on a COLM.
Sst expression in an iDISCO+ cleared sample of a mouse left hemi-brainstem. COLM-acquired images are first rendered as a series of xy-planes progressing in the medio-lateral direction and subsequently as mip 3D volume rendering.
Volume rendering of HCR FISH-labeled Pvalb expressing neurons in a rat forebrain hemi-slice. This representative COLM-acquired image stack of an iDISCO+-cleared brain shows the predominant cortical expression of Pvalb.
3D visualization of HCR FISH-labeled Pvalb expressing neurons in a rat brainstem hemi-slice showing the expression pattern in cerebellum and several nuclei. iDISCO+-cleared and COLM-acquired image stacks are first projected as a series of xy-planes moving in a rostral-caudal-rostral direction, then as a mip 3D volume rendering.
3D visualization of an iDISCO+ cleared rat mid-brain slice showing the expression of Th mRNA in the SNr/c, VTA, PBP and PAG regions. This movie first shows the rostro-caudal progression of a series of xy-plane images acquired on the COLM followed by a mip 3D volume rendering.
Dbh expressing neurons in the rat locus coeruleus are shown here first as a series of xy-plane images acquired on a confocal microscope followed by a mip 3D volume. This representative brainstem hemi-slice was processed through HCR FISH and cleared using the iDISCO+ method.
3D visualization of HCR FISH-labeled CALB expressing neurons in the postmortem human cerebellum block. Confocal microscope-acquired image stacks are rendered first as a series of xy-plane images followed by a mip 3D volume rendering.
PVALB expressing neurons in the postmortem human cerebellum block are visualized in this movie as a series of xy-plane images then by a mip 3D volume. This representative block was cleared using iDISCO+ and images were acquired on a confocal microscope.
3D visualization of COLM acquired dual HCR FISH labeling in iDISCO+ cleared mouse hemisphere. Movie shows labeling of Pvalb (green, AF-647) and Vglut1 (red, AF-594) mRNAs in a series of dorso-ventral planes from a down-sampled image volume. Left pane shows progression of z-planes from an intact hemisphere whereas right pane shows a zoomed-in view of ventral hippocampus and cortex (regions of lateral entorhinal cortex) whose z-plane progression is synced anatomically with the whole hemisphere in the left pane.
Acknowledgements
This work was supported by the Pritzker Neuropsychiatric Research Consortium, the Hope for Depression Research Foundation, National Institute of Health R01MH104261, Office of Naval Research Grant N00014–12-1–0366 and National Institute on Drug Abuse U01DA043098. The authors have no conflicts of interest to declare. The authors would like to thank Ms. Jennifer Fitzpatrick, Mr. Evan Hughes, Ms. Claire Barcelo, and Mr. Hui Li for their technical assistance.
Conflict of Interest
V.K., D.M.K., E.K.H., B.M., M.F., H.A., and S.J.W are members of the Pritzker Neuropsychiatric Research Consortium, which is supported by the Pritzker Neuropsychiatric Disorders Research Fund, LLC (Fund). There exists a shared intellectual property agreement between the academic and philanthropic entities of the Consortium. The Fund has no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Compliance with Ethical Standards
Ethical approval
“All animal care and procedures performed in studies involving animals were in accordance with the ethical standards of the guide for the Care and Use of Laboratory Animals: Eighth Edition (revised in 2011, published by the National Academy of Sciences), and approved by the University of Michigan Committee on the Use and Care of Animals. “This article does not contain any studies with human participants performed by any of the authors.”
Informed consent
Postmortem human brains were collected by the Brain Donor Program at the University of California, Irvine with the consent of the relatives of the deceased.
References
- Azaripour A, Lagerweij T, Scharfbillig C, Jadczak AE, Willershausen B, Van Noorden CJF (2016) A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Prog Histochem Cytochem 51:9–23. doi: 10.1016/J.PROGHI.2016.04.001 [DOI] [PubMed] [Google Scholar]
- Biesemeier A, Schraermeyer U, Eibl O (2011) Chemical composition of melanosomes, lipofuscin and melanolipofuscin granules of human RPE tissues. Exp Eye Res 93:29–39. doi: 10.1016/j.exer.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Bria A, Iannello G (2012) TeraStitcher - A tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. BMC Bioinformatics 13:316. doi: 10.1186/1471-2105-13-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Beck VA, Pierce NA (2014) Next-Generation in Situ Hybridization Chain Reaction: Higher Gain, Lower Cost, Greater Durability. ACS Nano 8:4284–4294. doi: 10.1021/nn405717p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Chang JY, Trinh LA, Padilla JE, Fraser SE, Pierce NA (2010) Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Biotechnol 28:1208–1212. doi: 10.1038/nbt.1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, Pierce NA (2018) Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145:dev165753. doi: 10.1242/dev.165753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim S-Y, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, Pak S, Bernstein H, Ramakrishnan C, Grosenick L, Gradinaru V, Deisseroth K (2013) Structural and molecular interrogation of intact biological systems. Nature 497:332–337. doi: 10.1038/nature12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks RM, Pierce NA (2004) From The Cover: Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci 101:15275–15278. doi: 10.1073/pnas.0407024101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fee C, Banasr M, Sibille E (2017) Somatostatin-positive GABA Interneuron Deficits in Depression: Cortical Microcircuit and Therapeutic Perspectives. Biol Psychiatry 82:549. doi: 10.1016/J.BIOPSYCH.2017.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung SJ, Webster MJ, Sivagnanasundaram S, Duncan C, Elashoff M, Weickert CS (2010) Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. Am J Psychiatry 167:1479–1488. doi: 10.1176/appi.ajp.2010.09060784 [DOI] [PubMed] [Google Scholar]
- Hebda-Bauer EK, Pletsch A, Darwish H, Fentress H, Simmons TA, Wei Q, Watson SJ, Akil H (2010) Forebrain glucocorticoid receptor overexpression increases environmental reactivity and produces a stress-induced spatial discrimination deficit. Neuroscience 169:645–653. doi: 10.1016/j.neuroscience.2010.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iritani S, Satoh K (1991) Distribution of somatostatin-immunoreactive cell bodies and fibers in the neocortex of Macaca fuscata. Synapse 9:50–59. doi: 10.1002/syn.890090108 [DOI] [PubMed] [Google Scholar]
- Jones EG, Hendry SHC, Liu X-B, Hodgins S, Potkin SG, Tourtellotte WW (1992) A method for fixation of previously fresh-frozen human adult and fetal brains that preserves histological quality and immunoreactivity. J Neurosci Methods 44:133–144. doi: 10.1016/0165-0270(92)90006-Y [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kubota Y (1997) GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex 7:476–86. doi: 10.1093/cercor/7.6.476 [DOI] [PubMed] [Google Scholar]
- Kramer EE, Steadman PE, Epp JR, Frankland PW, Josselyn SA (2018) Assessing Individual Neuronal Activity Across the Intact Brain: Using Hybridization Chain Reaction (HCR) to Detect Arc mRNA Localized to the Nucleus in Volumes of Cleared Brain Tissue. Curr Protoc Neurosci 84:e49. doi: 10.1002/cpns.49 [DOI] [PubMed] [Google Scholar]
- Krolewski DM, Kumar V, Martin B, Tomer R, Deisseroth K, Myers RM, Schatzberg AF, Lee FS, Barchas JD, Bunney WE, Akil H, Watson SJ (2018) Quantitative validation of immunofluorescence and lectin staining using reduced CLARITY acrylamide formulations. Brain Struct Funct 223:987–999. doi: 10.1007/s00429-017-1583-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Vogt CE, McBrairty M, Al-Hashimi HM (2013) Influence of Dimethylsulfoxide on RNA Structure and Ligand Binding. Anal Chem 85:9692–8. doi: 10.1021/AC402038T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LC, Sibille E (2015) Somatostatin, neuronal vulnerability and behavioral emotionality. Mol Psychiatry 20:377–387. doi: 10.1038/mp.2014.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LC, Sibille E (2013) Reduced brain somatostatin in mood disorders: A common pathophysiological substrate and drug target? Front. Pharmacol 2013;4:110. Published 2013 Sep 9. doi: 10.3389/fphar.2013.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C (2004) Interneurons of the neocortical inhibitory system. Nat Rev Neurosci 5:793–807. doi: 10.1038/nrn1519 [DOI] [PubMed] [Google Scholar]
- Papachristou DN, Liu JL, Patel YC (1994) Glucocorticoids regulate somatostatin peptide and steady state messenger ribonucleic acid levels in normal rat tissues and in a somatostatin-producing islet tumor cell line (1027B2). Endocrinology 134:2259–2266. doi: 10.1210/endo.134.5.7908873 [DOI] [PubMed] [Google Scholar]
- Pardue ML, Gall JG (1969) Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc Natl Acad Sci 64:600–604. doi: 10.1073/pnas.64.2.600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y-G, Sohn CH, Chen R, McCue M, Yun DH, Drummond GT, Ku T, Evans NB, Oak HC, Trieu W, Choi H, Jin X, Lilascharoen V, Wang J, Truttmann MC, Qi HW, Ploegh HL, Golub TR, Chen S-C, Frosch MP, Kulik HJ, Lim BK, Chung K (2018) Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat Biotechnol 37:73–83. doi: 10.1038/nbt.4281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- PubChem-AF488 5-Alexa Fluor 488 TFP ester | C27H12F4Li2N2O11S2 - PubChem. https://pubchem.ncbi.nlm.nih.gov/compound/135564337. Accessed 13 Feb 2020
- PubChem-AF594 Alexa Fluor 594 meta-isomer | C39H37N3O13S2 - PubChem. https://pubchem.ncbi.nlm.nih.gov/compound/25137945. Accessed 13 Feb 2020
- PubChem-AF647 Alexa Fluor 647 | C41H59N3O17PS4+ - PubChem. https://pubchem.ncbi.nlm.nih.gov/compound/102227060. Accessed 13 Feb 2020
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M (2014) iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159:896–910. doi: 10.1016/j.cell.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Silvestri L, Costantini I, Sacconi L, Pavone FS (2016) Clearing of fixed tissue: a review from a microscopist’s perspective. J Biomed Opt 21:081205. doi: 10.1117/1.JBO.21.8.081205 [DOI] [PubMed] [Google Scholar]
- Sonntag K-C, Tejada G, Subburaju S, Berretta S, Benes FM, Woo T-UW (2016) Limited predictability of postmortem human brain tissue quality by RNA integrity numbers. J Neurochem 138:53–59. doi: 10.1111/jnc.13637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylwestrak EL, Rajasethupathy P, Wright MA, Jaffe A, Deisseroth K (2016) Multiplexed Intact-Tissue Transcriptional Analysis at Cellular Resolution. Cell 164:792–804. doi: 10.1016/j.cell.2016.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomer R, Ye L, Hsueh B, Deisseroth K (2014) Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc 9:1682–1697. doi: 10.1038/nprot.2014.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban-Ciecko J, Barth AL (2016) Somatostatin-expressing neurons in cortical networks. Nat Rev Neurosci 17:401–409. doi: 10.1038/nrn.2016.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan P, Zhu J, Xu J, Li Y, Yu T, Zhu D (2018) Evaluation of seven optical clearing methods in mouse brain. Neurophotonics 5:1. doi: 10.1117/1.NPh.5.3.035007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Nie J, Sicotte H, Li Y, Eckel-Passow JE, Dasari S, Vedell PT, Barman P, Wang L, Weinshiboum R, Jen J, Huang H, Kohli M, Kocher J-PA (2016) Measure transcript integrity using RNA-seq data. BMC Bioinformatics 17:58. doi: 10.1186/s12859-016-0922-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Fentress HM, Hoversten MT, Zhang L, Hebda-Bauer EK, Watson SJ, Seasholtz AF, Akil H (2012) Early-Life Forebrain Glucocorticoid Receptor Overexpression Increases Anxiety Behavior and Cocaine Sensitization. Biol Psychiatry 71:224–231. doi: 10.1016/j.biopsych.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Hebda-Bauer EK, Pletsch A, Luo J, Hoversten MT, Osetek AJ, Evans SJ, Watson SJ, Seasholtz AF, Akil H (2007) Overexpressing the glucocorticoid receptor in forebrain causes an aging-like neuroendocrine phenotype and mild cognitive dysfunction. J Neurosci 27:8836–44. doi: 10.1523/jneurosci.0910-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Lu X-Y, Liu L, Schafer G, Shieh K-R, Burke S, Robinson TE, Watson SJ, Seasholtz AF, Akil H (2004) Glucocorticoid receptor overexpression in forebrain: a mouse model of increased emotional lability. Proc Natl Acad Sci U S A 101:11851–6. doi: 10.1073/pnas.0402208101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Rivenson Y, Wang H, Luo Y, Ben-David E, Bentolila LA, Pritz C, Ozcan A (2019) Three-dimensional virtual refocusing of fluorescence microscopy images using deep learning. Nat Methods. doi: 10.1038/s41592-019-0622-5 [DOI] [PubMed] [Google Scholar]
- Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen C-K, Lubeck E, Shah S, Cai L, Gradinaru V (2014) Single-Cell Phenotyping within Transparent Intact Tissue through Whole-Body Clearing. Cell 158:945–958. doi: 10.1016/j.cell.2014.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D (1996) Biochemical basis of lipofuscin, ceroid, and age pigment-like fluorophores. Free Radic. Biol. Med 21:871–888 [DOI] [PubMed] [Google Scholar]
- Zheng H, Rinaman L (2016) Simplified CLARITY for visualizing immunofluorescence labeling in the developing rat brain. Brain Struct Funct 221:2375–2383. doi: 10.1007/s00429-015-1020-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3D visualization of differences in the HCR-FISH labeled Sst signal between CLARITY (left panel) and iDISCO+ (right panel) processed mouse cortical slices. First half of this movie shows a series of xy-plane images moving in z-direction of confocal acquired representative stacks, followed by maximum intensity projection (mip) 3D pre-quantitation volume and post-quantitation surface rendering of Sst neurons. Second half of the movie shows a series of xz-plane images moving in y-direction, followed by mip 3D pre-quantitation volume and post-quantitation surface renderings of Sst neurons. This animation highlights the higher number density of detected Sst neurons and uniform labeling across the z-plane in iDISCO+ compared to CLARITY.
3D visualization of HCR FISH-labeled Sst expressing neurons in a rat forebrain hemi-slice cleared using iDISCO+. This movie first shows a series of xy-plane images acquired on a COLM followed by a mip 3D volume rendering. The characteristic layer specific expression pattern of Sst in rat cortex can be observed in this movie.
Volume rendering of a rat hippocampus showing Sst expression pattern in the iDISCO+ cleared intact tissue and labeled using HCR FISH method. This movie shows a series of xy-plane images acquired on a COLM followed by 3D mip volume rendering.
3D visualization of a mouse left hemisphere showing Sst expressing neurons across the entire rostro-caudal extent of the brain. Post HCR FISH labeling, brain tissue was cleared using the iDISCO+ method and imaging was performed on a COLM.
Sst expression in an iDISCO+ cleared sample of a mouse left hemi-brainstem. COLM-acquired images are first rendered as a series of xy-planes progressing in the medio-lateral direction and subsequently as mip 3D volume rendering.
Volume rendering of HCR FISH-labeled Pvalb expressing neurons in a rat forebrain hemi-slice. This representative COLM-acquired image stack of an iDISCO+-cleared brain shows the predominant cortical expression of Pvalb.
3D visualization of HCR FISH-labeled Pvalb expressing neurons in a rat brainstem hemi-slice showing the expression pattern in cerebellum and several nuclei. iDISCO+-cleared and COLM-acquired image stacks are first projected as a series of xy-planes moving in a rostral-caudal-rostral direction, then as a mip 3D volume rendering.
3D visualization of an iDISCO+ cleared rat mid-brain slice showing the expression of Th mRNA in the SNr/c, VTA, PBP and PAG regions. This movie first shows the rostro-caudal progression of a series of xy-plane images acquired on the COLM followed by a mip 3D volume rendering.
Dbh expressing neurons in the rat locus coeruleus are shown here first as a series of xy-plane images acquired on a confocal microscope followed by a mip 3D volume. This representative brainstem hemi-slice was processed through HCR FISH and cleared using the iDISCO+ method.
3D visualization of HCR FISH-labeled CALB expressing neurons in the postmortem human cerebellum block. Confocal microscope-acquired image stacks are rendered first as a series of xy-plane images followed by a mip 3D volume rendering.
PVALB expressing neurons in the postmortem human cerebellum block are visualized in this movie as a series of xy-plane images then by a mip 3D volume. This representative block was cleared using iDISCO+ and images were acquired on a confocal microscope.
3D visualization of COLM acquired dual HCR FISH labeling in iDISCO+ cleared mouse hemisphere. Movie shows labeling of Pvalb (green, AF-647) and Vglut1 (red, AF-594) mRNAs in a series of dorso-ventral planes from a down-sampled image volume. Left pane shows progression of z-planes from an intact hemisphere whereas right pane shows a zoomed-in view of ventral hippocampus and cortex (regions of lateral entorhinal cortex) whose z-plane progression is synced anatomically with the whole hemisphere in the left pane.