Abstract
Motivation
Next-generation deep sequencing of viral genomes, particularly on the Illumina platform, is increasingly applied in HIV research. Yet, there is no standard protocol or method used by the research community to account for measurement errors that arise during sample preparation and sequencing. Correctly calling high and low-frequency variants while controlling for erroneous variants is an important precursor to downstream interpretation, such as studying the emergence of HIV drug-resistance mutations, which in turn has clinical applications and can improve patient care.
Results
We developed a new variant-calling pipeline, hivmmer, for Illumina sequences from HIV viral genomes. First, we validated hivmmer by comparing it to other variant-calling pipelines on real HIV plasmid datasets. We found that hivmmer achieves a lower rate of erroneous variants, and that all methods agree on the frequency of correctly called variants. Next, we compared the methods on an HIV plasmid dataset that was sequenced using Primer ID, an amplicon-tagging protocol, which is designed to reduce errors and amplification bias during library preparation. We show that the Primer ID consensus exhibits fewer erroneous variants compared to the variant-calling pipelines, and that hivmmer more closely approaches this low error rate compared to the other pipelines. The frequency estimates from the Primer ID consensus do not differ significantly from those of the variant-calling pipelines.
Availability and implementation
hivmmer is freely available for non-commercial use from https://github.com/kantorlab/hivmmer.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Several next-generation sequencing (NGS) instruments are now used to study pathogens and viruses (Chabria et al., 2014; Quiñones-Mateu et al., 2014). Of the many NGS platforms and approaches that have been developed over the past two decades, Illumina’s sequencing-by-synthesis technology has come to dominate the market, in large part due to increasing yields and decreasing costs (Goodwin et al., 2016). Deep sequencing of HIV samples with Illumina technology is frequently used in studies of viral epidemiology, clinical genotyping and antiretroviral drug resistance. For example, deep sequencing can provide for a more sensitive assay of drug-resistance mutations (Brumme and Poon, 2016); and Sanger sequencing, the current clinical standard, cannot reliably detect mutations at frequencies below 20%, which might be clinically relevant (Ávila Ríos et al., 2016). A common concern in studies using NGS, and also in establishing clinical standards for these new approaches, is the measurement error of their sequencing protocols. Measurement errors can arise in sample preparation (including reverse transcription of RNA genomes to cDNA and amplification of viral genomes), library preparation, sequencing and base calling.
Measurement error creates uncertainty in downstream analyses. For example, errors introduced during genome amplification are difficult to distinguish from real mutations since they are introduced in the early steps of the process, are exponentially amplified and may occur at high frequency in the later steps. Recombination during polymerase chain reaction (PCR) is difficult to distinguish from clinically relevant ‘real’ viral recombination. Mutations at low frequencies can be difficult to distinguish from sequencing and base calling errors, and can confound read alignment, assembly and haplotype reconstruction methods that rely on accurately identifying exact sequence overlaps among sequence reads. Beerenwinkel et al. (2012) speculated that artifacts introduced during the reverse transcription polymerase chain reaction (RT-PCR) step are likely the biggest challenge to accurately estimating viral diversity through reconstructing individual haplotypes for deeply sequenced HIV data.
Many HIV studies in recent years have addressed Illumina sequencing errors by applying a global frequency threshold—typically 1%—below which variants are excluded with the reasoning that they are indistinguishable from amplification or sequencing errors. This approach requires establishing a conservative estimate of the typical error rate for the sequencing protocol, which is then used as a threshold during variant-calling.
The most common approach to estimating sequencing error rates is to analyze reads that come from known sequences, by aligning the reads to the known sequence and counting the frequency of mismatches in the alignment. In the context of HIV, this can be accomplished by sequencing mixtures of HIV plasmids with known sequences. In this study, we use this approach to introduce a new pipeline for analysis of HIV pol sequences from the Illumina MiSeq platform, hivmmer, and compare it to other variant-calling pipelines. While existing pipelines use short-read aligners to align Illumina reads in nucleotide space against an HIV reference (such as HXB2; accession K03455) or a de novo assembly, hivmmer instead uses a probabilistic aligner, HMMER (Eddy, 2011), to achieve a more sensitive alignment in amino acid space.
2 Methods and data
2.1 Pipelines
We created a new pipeline, hivmmer (version 0.1.2), based on the probabilistic aligner HMMER (Eddy, 2011), that consists of the following steps:
Constructs an amino acid profile Hidden Markov Model (pHMM) from a multiple sequence alignment of all HIV-1 Group M amino acid sequences publicly available in the Los Alamos HIV Sequence Database (http://www.hiv.lanl.gov) for the pol gene.
Pre-processes the NGS data using the paired-end read-merging tool PEAR (Zhang et al., 2014) and consolidates duplicate sequences using the FASTQ/A Collapser tool from the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). The number of duplicates is tracked to enable correct inference of frequencies later in the pipeline. Duplicates are consolidated primarily as a performance optimization, as it reduces the computational burden of the later steps.
Translates each de-duplicated sequence into all six possible frames (forward and reverse), retaining only the translated sequences that contain no stop codons (although hivmmer does provide an option to allow stop codons to support analyses of non-coding regions or of degenerate sequences that contain premature stop codons).
Aligns the translated reads to the reference pHMM with hmmsearch from HMMER, producing a multiple sequence alignment of translated reads.
Constructs a sample-specific amino acid pHMM from the multiple sequence alignment of translated reads.
Repeats the hmmsearch alignment against the sample-specific pHMM to yield additional sequences that may have been too divergent from the reference sequences to align in the first hmmsearch alignment.
Maps the translated amino acid coordinates in the alignment to the original frame and coordinates in the nucleotide reads to construct a codon frequency table (adjusting the counts for duplicate reads).
We compared hivmmer to two of the existing pipelines, HyDRA [Ji et al. (2015); version 0.3.1] and shiver [Wymant et al. (2018); version 1.4.1], both of which use the short-read aligner bowtie2 (Langmead and Salzberg, 2012). HyDRA aligns the reads to the HXB2 reference, while shiver uses an iterative alignment to a de novo assembly of the reads. We chose HyDRA and shiver as representatives of a broader group of HIV alignment pipelines such as PASeq (https://paseq.org) and MiCall (http://cfe-lab.github.io/MiCall), which are also based on bowtie2 [for a recent comparison of these methods, see Noguera-Julian et al. (2017)]. As additional robustness checks, we included a naive implementation of a bowtie2 alignment without any additional filtering or quality control (the ‘bowtie2’ method), and the same naive bowtie2 alignment run only on successfully merged reads from PEAR (the ‘bowtie2-pear’ method).
2.2 Data
Our study uses four publicly available HIV plasmid datasets:
5VM (accession SRR961514), a ‘5 virus mix’ of plasmid sequences (89.6, HXB2, JRCSF, NL4.3, YU2) in equal proportions (20%) sequenced by Di Giallonardo et al. (2014);
PL1:1 (accession SRR6725661), a mixture of two plasmid sequences (89.6 and NL4.3) in 1:1 proportion generated in our lab and introduced in this study (described below);
PL1:9 (accession SRR6725662), the same mixture as PL1:1, but in 1:9 proportion;
PID (accessions SRR2097103-8), the same mixture as 5VM, but sequenced using the Primer ID protocol by Seifert et al. (2016).
For PL1:1 and PL1:9, plasmids 89.6 (U39362) and NL4.3 (AF324493.2), obtained from the NIH AIDS Reagent Program (https://www.aidsreagent.org/), were mixed as 1:1 or 1:9 ratios, respectively, followed by amplification of the pol region using primers previously described by Winters et al. (1998), and proof reading polymerase Phusion (Thermofisher). Nextera XT DNA Library Prep chemistry (Illumina) was used to fragment and add adapter sequences onto template DNA to generate multiplexed sequencing libraries that were sequenced on Illumina’s MiSeq platform generating 2 × 250 bp paired-end reads.
5VM contains near-full-length HIV genomes, although for this study we considered their alignment and variants only within the first 1044 nt of the pol region (HXB2 coordinates 2253–3296). This is also the region contained in PL1:1 and PL1:9, and is a genomic region that is clinically relevant for drug-resistance mutations. PID is a restricted fragment within this region, with length 471 nt starting at HXB2 coordinate 2736.
Note that both plasmids present in PL1:1 and PL1:9 (NL4.3 and 89.6) are also present in 5VM and PID. The 5VM dataset included an RT step, as RNA was extracted from viral particles obtained following the transfection of the plasmids into 293 T cells. PL1:1 and PL1:9 were PCR amplified from plasmid DNA and did not include an RT step.
The PID dataset comes from a study of the Primer ID method by Seifert et al. (2016). We compared the variants in the consensus sequences from their Primer ID consensus caller, called pidalyse, to those from running each of the pipelines on the original reads with the Primer ID barcodes removed. That is, we tested the pipelines under the condition where the Primer ID is unknown.
2.3 Reproducibility
All scripts required to reproduce the results presented here are available from https://github.com/kantorlab/hiv-measurement-error and can be executed using the SCons build system (http://scons.org). Compiled versions of all software dependencies for 64-bit Linux and Anaconda Python (https://www.anaconda.com) are available from the kantorlab conda channel (http://anaconda.org/kantorlab). The hivmmer source code is available from (https://github.com/kantorlab/hivmmer) and a pre-compiled Docker (https://docker.com) image is available from DockerHub at https://hub.docker.com/r/kantorlab/hivmmer. A pre-compiled Docker image with all dependencies required to run the analyses described in the paper is also available at https://hub.docker.com/r/kantorlab/hiv-measurement-error.
3 Results
We analyzed the coverage and fragment sizes of the Illumina reads. Figure 1 shows an overview of fragment sizes in 5VM after alignment to the pol region with bowtie2; PL1:1 and PL1:9 have similar fragment size distributions (data not shown). Typically, fragment sizes follow a skewed distribution centered around the read length, 250 nt. Fragments shorter than the read length are fully overlapping, and yield reads that in practice can be treated as technical replicates. Fragments sized between the read length and twice the read length yield partially overlapping reads that can be combined into a single sequence using a read-merging tool like PEAR (Zhang et al., 2014). Finally, fragments larger than twice the read length yield separate read pairs, with a positive insert size between the reads.
Fig. 1.

Histogram of fragment sizes in the 5VM data set showing the proportion of reads that are either technical replicates (fragment is less than the maximum read length; leftmost bars), overlapping (fragment is between the read length and twice the read length; middle bars), or paired-end (fragment is larger than twice the read length; rightmost bars)
To compare methods, we first identified both the correct and erroneous variants in the underlying alignments from each method. We defined erroneous variants as codons with >0 frequency, but which do not exist at that position in any of the known plasmid sequences for the given datasets. Supplementary Figures S1–S4 show a detailed picture of this for each dataset and method. As expected, nearly all of the erroneous variants are at frequencies below 1%, which is a widely accepted global threshold. For the hivmmer method, no erroneous variants occurred at frequencies above 1% for any of the datasets. Similarly, no erroneous variants occurred at frequencies above 1% for the shiver, hydra and bowtie2-pear methods on PL11, and for shiver and hydra on PL19. However, for other methods and datasets, erroneous variants occur above a frequency of 1%, as seen in Table 1. The erroneous variants with the highest frequency for each method and dataset are shown in Supplementary Table S1.
Table 1.
Number of erroneous variants occurring above 1% frequency
| hivmmer | shiver | HyDRA | bowtie2 | bowtie2-pear | pidalyse | |
|---|---|---|---|---|---|---|
| 5VM | 0 | 4 | 9 | 15 | 94 | — |
| PL11 | 0 | 0 | 0 | 1 | 0 | — |
| PL19 | 0 | 1 | 0 | 2 | 0 | — |
| PID | 0 | 53 | — | 69 | — | 2 |
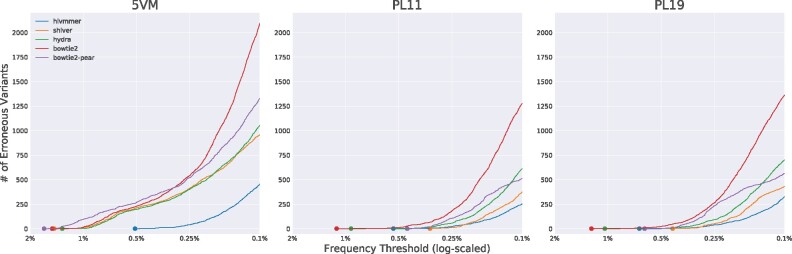
To measure the overall effectiveness of each method, we plotted the cumulative number of erroneous variants as we lowered the global frequency threshold from 2 to 0.1% (Fig. 2). From this plot, we find that hivmmer alignments accumulate fewer errors across all datasets. Both shiver and HyDRA perform better than the naive implementations of bowtie2 alignments, likely due to their additional filtering and quality control strategies. The bowtie2-pear method performs better than bowtie2 in almost all cases, except on 5VM. Supplementary Table S2 reports the number of reads retained in each method’s alignment.
Fig. 2.
The accumulation of erroneous variants at decreasing thresholds, across datasets and alignment methods. For 5VM, hivmmer alignments display the lowest cumulative error rate. For PL1:1 and PL1:9, the cumulative error rates are closer among the methods, but hivmmer alignments display fewer errors at thresholds below 0.25%
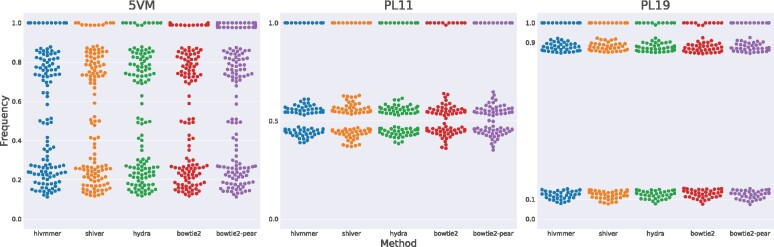
Next, we considered the frequencies of correct variants after thresholding at 1%, and compared their distribution across methods (Fig. 3). While we expect the frequencies to follow the mixture proportions (e.g. multiples of 20% for 5VM, 1:1 for PL1:1, and 1:9 for PL1:9), in reality the frequencies deviate from these expected values. This could be due to sample preparation or preferential primer amplification.
Fig. 3.
The distribution of frequencies for correct variants after thresholding at 1%. Each point represents the frequency of a codon in the plasmid sequences. The overlapping codons with frequency near 1.0 represent identical sites across all plasmid sequences. At sites where the plasmid sequences differ (points not near 1.0), we expect the frequencies to follow the mixture proportions of the plasmid sequences in the dataset. Statistical tests of matched comparisons among the methods show no significant differences, except for bowtie2-pear on the 5VM dataset
Although the true proportions of plasmid sequences in each dataset are unknown, the primer ID data analyzed with the pidalyse method is arguably their best empirical measurement. Under this assumption, we calculate the mean squared error of the variant frequencies from the pidalyse method compared to those from the hivmmer, hydra and bowtie2 methods to assess their accuracy. The mean squared error is lowest for hivmmer (), followed by hydra () and bowtie2 ().
On all datasets, we can test the null hypothesis that at least one of the distributions is significantly different from the others using the Friedman test, a non-parametric analog to the repeated measures ANOVA. This test fails to reject the null for PL1:1 (P = 1.000) and PL1:9 (P = 0.998). The Friedman test is significant for 5VM (P < 0.001), and we performed a post-hoc analysis of pairwise Wilcoxon tests, adjusting the P-values for multiple comparisons using the Holm–Bonferroni method (Supplementary Table S3). The adjusted pairwise tests find that only the bowtie2-pear method differs from the others. Therefore, none of the three pipelines (hivmmer, HyDRA or shiver) differed significantly in their measurement of the variant frequencies for the correctly called variants (all of which are above the range of frequencies for the erroneous variants). No correct variants were missing or occurring at frequency ¡1% for any of the methods or datasets.
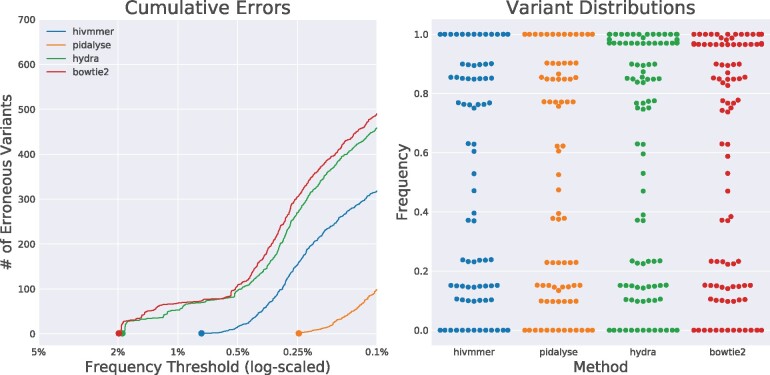
The Primer ID protocol was designed to control for the artifacts during sample preparation that could be potentially skewing our recovered frequencies of correctly called variants. We compared the cumulative error rate and distribution of variant frequencies between three of the pipelines (HyDRA, hivmmer and bowtie2, as the initial de novo assembly of the PID dataset failed for shiver, and the PID dataset has no overlapping reads that would benefit from read-merging in bowtie2-pear) and the pidalyse method for calling the consensus sequence of each Primer ID template (Seifert et al., 2016). Because these consensus sequences should represent individual templates, we expect that the frequency of variants across templates would correspond to the plasmid mixture proportions (e.g. multiples of 20% for 5VM). The Primer ID method does indeed reduce the accumulation of erroneous variants, and hivmmer better approaches this performance than HyDRA (Fig. 4a). The Primer ID consensus sequences, however, also do not recover the correct frequency proportions as one might expect (Fig. 4b). This is consistent with the results reported by Seifert et al. (2016), and they ascribe the discrepancy to ‘noisy RT qPCR quantification.’
Fig. 4.
The cumulative error rates (a) and distribution of correctly-called variants (b) in alignments for the PID data set. The pidalyse method exhibits the fewest alignment errors, since it uses the primer IDs to call consensus sequences for each fragment; hivmmer performs closer to pidalyse than HyDRA or bowtie2
4 Discussion
We have introduced a new variant-calling pipeline, hivmmer, whose alignments exhibit lower error rates than existing pipelines on deep Illumina sequencing of HIV plasmid data.
4.1 Global thresholding
Our results validate that in some cases the 1% global thresholding method will work as expected, as measured on plasmid datasets and assuming the variant-calling pipeline has similar accuracy to the pipelines tested here. Some studies have conducted their own validation of a global threshold. For example, one of the earliest studies to use the global thresholding approach with Illumina MiSeq data was conducted by Dudley et al. (2014), who analyzed HXB2 plasmid sequences to determine a higher threshold of 2%. Fisher et al. (2015) conducted additional validation for the occurrence of non-nucleoside reverse transcriptase inhibitor drug-resistance mutations at frequencies over 1% in their study using 250 clonal sequences. They also presented a method for error correction using a Bayesian Dirichlet mixture of multinomial probabilistic model to distinguish sequencing error from true low-frequency variants at posterior probabilities 99.99%.
Ode et al. (2015) developed an adaptive threshold approach for Illumina MiSeq sequencing based on per-site quality scores and demonstrated that it could reduce mismatches to below a frequency of 1% at most sites on varying mixtures of pNL4-3 and pNL101 plasmid sequences. Their method computes an average quality score across all reads at a reference site and they threshold the variant calls at that site with average score 20 and frequency 1%.
Others, however, have applied thresholding without validation. Studies by Ekici et al. (2014), Pessôa et al. (2014) and Pessôa et al. (2016) applied thresholds of 1% without providing any citation or methodological justification for this approach to error correction. A review of clinical applications of deep HIV sequencing by Casadellà and Paredes (2017) proposed a rule of thumb of a 1% threshold without citation. However, they emphasized that the frequency threshold is likely limited by the number of RNA molecules in the sample, and caution that the frequencies that are retained between 1 and 100% are likely skewed by many sources of bias during sequencing. This is confirmed in our results where we do not recover the expected frequencies given the plasmid mixture proportions.
Our results show that erroneous variants can occur at frequencies above 1 for some methods and datasets. Thus, the rule of thumb of a 1% threshold that is currently explored in the literature can actually identify erroneous variants. The number of erroneous variants varied across methods and datasets, and was higher for the bowtie2 method and for the 5VM dataset (Table 1). Ode et al. (2015) claimed in their analysis that they found mismatches occurring at as high as 6.4% frequency at some sites. Both their results and ours clearly establish the heterogeneity in error profiles, and that a global threshold is overly conservative at most sites.
Going forward, studies using deep Illumina sequencing of HIV to analyze variants at low frequencies should include control datasets and detailed analysis of the error profile, such as the one we have presented in this study. One potential study design is to use a PhiX control library that can be readily incorporated into Illumina sequencing runs (http://www.illumina.com/products/phix_control_v3.html). The control can then be used to establish an error profile for the HIV samples of interest in that lane.
4.2 Overlapping reads as technical replicates
One potential reason why hivmmer outperforms the other methods is that it more closely models the fragment distribution through its use of the PEAR read merger. As shown in Figure 1, the majority of reads are completely overlapping (e.g. technical replicates) or partially overlapping in deep Illumina sequencing of HIV. In particular, the fragment distribution is non-normal, while many short-read aligners, including bowtie2, assume a normal distribution of fragments. Read-merging in the bowtie2-pear method also improves the performance of the naive bowtie2 alignment; however, the performance is still not comparable to hivmmer’s, indicating that read-merging alone is not sufficient and other factors such as the alignment and filtering strategies contribute to the final performance.
PEAR is able to use this replicate information to correct errors at sites where the replicates disagree, by comparing quality scores. Read-merging based on quality scores has been used previously in studies of HIV (Lapointe et al., 2015; Leeet al., 2017), as well as in other non-HIV contexts. For example, it was tested by Chen-Harris et al. (2013) in a study with 1 kb regions of the rabies and BCV viruses. They showed that the PCR error rate exceeds the sequencing error rate at high enough quality scores, and they called variants using a position-dependent model to determine an optimal quality score threshold. Preston et al. (2016) developed a similar protocol called Paired-End Low Error Sequencing that combines barcoding with overlapping read pairs to correct for both PCR and sequencing error and accurately detect rare variants. Although that specific protocol has only been tested with Escherichia coli and nematode DNA samples, the concept is directly relevant to HIV, where barcoding is already in use through the Primer ID protocol.
4.3 Primer ID
Our results confirm that the consensus sequences generated by the Primer ID method do achieve lower error rates than any of the pipelines. Primer ID is an area of active research, and most recently Boltz et al. (2016) extended the existing methods by using shorter PCR primers and more stringent consensus criteria, in a method they call ultrasensitive single-genome sequencing (uSGS). In comparisons with the earlier methods from Jabara et al. (2011), Zhou et al. (2015) and Seifert et al. (2016), they found that the uSGS technique yielded more unique Primer IDs and overall consensus sequences.
However, an important limitation of all of the Primer ID techniques is the difficulty of multiplexing multiple samples in the same lane, which is a common practice to reduce sequencing cost. In fact, because of the short length of the HIV genome, sufficient depth of coverage can be achieved with many fewer reads than a full lane of Illumina sequencing provides. In the extreme case, this was demonstrated with the successful application of ‘wide’ sequencing by Lapointe et al. (2015) to sequence a region of the pol gene from 1143 patient samples in a single Illumina MiSeq run.
In situations where the cost of Primer ID is prohibitive, there are still other avenues for controlling RT-PCR error. Orton et al. (2015) developed a computational model for the accumulation of errors following multiple PCR cycles. They validated this model using Illumina GAIIx sequencing of foot-and-mouth disease virus (FMDV) (not HIV) plasmid sequences with varying rounds of PCR amplification, including a condition with no amplification, and found that RT-PCR errors were concentrated in specific areas related to known variability in the FMDV genome, and not evenly distributed across the genome. They also found that most of the errors came from the PCR amplification rather than the RT step in sample preparation. Overall, their recommendation is to use the highest fidelity enzymes and minimize the number of PCR cycles.
Zanini et al. (2017) presented an Illumina MiSeq protocol with single-round PCR and a new primer design for HIV, and found an error rate of 0.1% that they attribute to PCR error, after removing low quality base calls. They validated the correlation between base calling errors and quality scores with a PhiX spike-in. Furthermore, they tested for in-vitro recombination and found it in nearly 10% of reads generated from nested PCR, but almost none in those from single-round PCR.
Thus, a viable alternative to Primer ID in future experiments may be to combine a sequencing protocol using high fidelity enzymes and single-round PCR with hivmmer.
5 Conclusion
The ideal sequencing technology for genomic studies of HIV would generate full-length reads, without error, of individual virus particles from a patient. Although this is not technically possible with today’s technology, understanding the causes and corrections for measurement errors and optimizing ways to avoid them will get us closer to that goal. Newer, longer-read and single-molecule sequencing technologies such as PacBio and Oxford Nanopore also hold promise in addressing these limitations, although they currently have much higher error rates than Illumina sequencing (Goodwin et al., 2016). Thus, even with newer and improved sequencing technology, understanding measurement error will still be a priority for making robust inferences from HIV sequencing data.
We have introduced a new variant-calling pipeline, hivmmer, whose alignments exhibit lower error rates than existing pipelines on deep Illumina sequencing of HIV plasmid data. One limitation of this study is that plasmid datasets contain limited viral populations that do not represent the much larger diversity of quasispecies in clinical HIV samples. Though plasmid datasets are less ambiguous about what variants are present in the sample population, allowing validation of NGS pipelines, further research is required to demonstrate pipelines’ effectiveness on well-described HIV clinical samples.
A second limitation is that hivmmer, like other methods, cannot differentiate erroneous variants that arise from experimental errors (in amplification or sequencing), analytic errors or natural variation. A remaining challenge is to develop methodologies that better differentiate these processes and prevent discarding potentially clinically significant variants.
A third limitation is that our study focuses on the pol region, which is a relatively conserved region of the HIV genome. Future work on measurement error will also need to consider more variable regions, such as env, which will require a careful inspection of insertions and deletions.
Another area for future research is the use of machine-learning techniques to tune error correction methods to the biases of a specific sequencing run using control data from that run. As noted above, a PhiX control library could provide this source of control data per run. However, a concern with both machine-learning methods, and with the widely-used thresholding approach to variant-calling analyzed here, is the potential of overfitting to the control data. Any robust method will need to be cross-validated on a wide range of HIV data before it can be trusted as a general-purpose tool. It will be important to benchmark error correction methods on datasets from different labs that use varying protocols. This can be facilitated by more public sharing of HIV datasets, several of which have already been deposited with the Sequence Read Archive.
Further refinement of error correction methods for deep Illumina sequencing of HIV that combine protocols to reduce PCR errors with machine-learning classification of sequencing errors will be a valuable and important step toward more robust, and ultimately clinically-trusted, tools for HIV genotyping. Overall, these clear directions for future work will benefit the HIV research community by enabling more robust inference. In the specific context of drug-resistance mutations, more robust error correction will allow for more sensitive detection of emerging resistance at very low variant frequencies and the continued exploration of their significance.
Supplementary Material
Acknowledgements
This research was conducted using computational resources and services at the Center for Computation and Visualization, Brown University.
Funding
This work was facilitated by the National Institutes of Health, including [R01AI108441] and the Providence/Boston Center for AIDS Research [P30AI042853].
Conflict of Interest: none declared.
Contributor Information
Mark Howison, Watson Institute for International and Public Affairs.
Mia Coetzer, Division of Infectious Diseases, The Alpert Medical School, Brown University, Providence, RI, USA.
Rami Kantor, Division of Infectious Diseases, The Alpert Medical School, Brown University, Providence, RI, USA.
References
- Ávila Ríos S. et al. (2016) Pretreatment HIV-drug resistance in Mexico and its impact on the effectiveness of first-line antiretroviral therapy: a nationally representative 2015 WHO survey. Lancet HIV, 3, e579–e591. [DOI] [PubMed] [Google Scholar]
- Beerenwinkel N. et al. (2012) Challenges and opportunities in estimating viral genetic diversity from next-generation sequencing data. Front. Microbiol., 3, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boltz V.F. et al. (2016) Ultrasensitive single-genome sequencing: accurate, targeted, next generation sequencing of HIV-1 RNA. Retrovirology, 13, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumme C.J., Poon A.F.Y. (2016) Promises and pitfalls of Illumina sequencing for HIV resistance genotyping. Virus Res., 239, 97–105. [DOI] [PubMed] [Google Scholar]
- Casadellà M., Paredes R. (2017) Deep sequencing for HIV-1 clinical management. Virus Res., 239, 69–81. [DOI] [PubMed] [Google Scholar]
- Chabria S.B. et al. (2014) Deep sequencing of HIV: clinical and research applications. Annu. Rev. Genomics Hum. Genet., 15, 295–325. [DOI] [PubMed] [Google Scholar]
- Chen-Harris H. et al. (2013) Ultra-deep mutant spectrum profiling: improving sequencing accuracy using overlapping read pairs. BMC Genomics, 14, 96.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giallonardo F.D. et al. (2014) Full-length haplotype reconstruction to infer the structure of heterogeneous virus populations. Nucleic Acids Res., 42, e115–e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley D.M. et al. (2014) Cross-clade simultaneous HIV drug resistance genotyping for reverse transcriptase, protease, and integrase inhibitor mutations by Illumina MiSeq. Retrovirology, 11, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. (2011) Accelerated profile HMM searches. PLoS Comput. Biol., 7, e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekici H. et al. (2014) Cost-efficient HIV-1 drug resistance surveillance using multiplexed high-throughput amplicon sequencing: implications for use in low- and middle-income countries. J. Antimicrob. Chemother., 69, 3349–3355. [DOI] [PubMed] [Google Scholar]
- Fisher R.G. et al. (2015) Next generation sequencing improves detection of drug resistance mutations in infants after PMTCT failure. J. Clin. Virol., 62, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S. et al. (2016) Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet., 17, 333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabara C.B. et al. (2011) Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc. Natl. Acad. Sci. USA, 108, 20166–20171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H. et al. (2015). Establishment of an Illumina MiSeq-based HIV drug resistance testing platform. In: The 8th International AIDS Society Conference on HIV Pathogenesis, Treatment and Prevention. Vancouver, Canada.
- Langmead B., Salzberg S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe H.R. et al. (2015) HIV drug resistance testing by high-multiplex “Wide” sequencing on the MiSeq instrument. Antimicrob. Agents Chemother., 59, 6824–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.Q. et al. (2017) Prevalence and clinical impacts of HIV-1 intersubtype recombinants in Uganda revealed by near-full-genome population and deep sequencing approaches. AIDS, 31, 2345.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguera-Julian M. et al. (2017) Next-generation human immunodeficiency virus sequencing for patient management and drug resistance surveillance. J. Infect. Dis., 216, S829–S833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ode H. et al. (2015) Quasispecies analyses of the HIV-1 near-full-length genome with Illumina MiSeq. Front. Microbiol., 6, 1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orton R.J. et al. (2015) Distinguishing low frequency mutations from RT-PCR and sequence errors in viral deep sequencing data. BMC Genomics, 16, 229.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessôa R. et al. (2014) Deep sequencing of HIV-1 near full-length proviral genomes identifies high rates of BF1 recombinants including two novel circulating recombinant forms (CRF) 70_BF1 and a disseminating 71_BF1 among blood donors in Pernambuco, Brazil. PLoS One, 9, e112674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessôa R. et al. (2016) Ultra-deep sequencing of HIV-1 near full-length and partial proviral genomes reveals high genetic diversity among Brazilian blood donors. PLoS One, 11, e0152499.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J.L. et al. (2016) High-specificity detection of rare alleles with paired-end low error sequencing (PELE-Seq). BMC Genomics, 17, 464.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones-Mateu M.E. et al. (2014) Deep sequencing: becoming a critical tool in clinical virology. J. Clin. Virol., 61, 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert D. et al. (2016) A comprehensive analysis of primer IDs to study heterogeneous HIV-1 populations. J. Mol. Biol., 428, 238–250. [DOI] [PubMed] [Google Scholar]
- Winters M.A. et al. (1998) A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J. Clin. Invest., 102, 1769–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymant C. et al. (2018) Easy and accurate reconstruction of whole HIV genomes from short-read sequence data with shiver. Virus Evol., 4, vey007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanini F. et al. (2017) Error rates, PCR recombination, and sampling depth in HIV-1 whole genome deep sequencing. Virus Res., 239, 106–114. [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. (2014) PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics, 30, 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S. et al. (2015) Primer ID validates template sampling depth and greatly reduces the error rate of next-generation sequencing of HIV-1 genomic RNA populations. J. Virol., 89, 8540–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.