Amide proton transfer–weighted imaging (APTWI) is a novel molecular MR imaging technique developed to detect and quantitatively visualize endogenous proteins and peptides. APTWI is usually reported in terms of asymmetry in the magnetization transfer ratio at 3.5 ppm. This study included 42 patients with low-grade (n = 28) or high-grade (n = 14) glioma, all of whom underwent conventional MR imaging, proton MR spectroscopy imaging, and amide proton transfer–weighted imaging on the same 3T scanner within 2 weeks before surgery. The asymmetric magnetization transfer ratio at 3.5 ppm values measured by different readers showed good concordance and were significantly higher in high-grade gliomas than in low-grade gliomas, with sensitivity and specificity values of 92.9% and 71.4%, respectively, at a cutoff value of 2.93%. The asymmetric magnetization transfer ratio at 3.5 ppm may serve as a potential biomarker not only for assessing proliferation, but also for predicting histopathologic grades in gliomas.
Abstract
BACKGROUND AND PURPOSE:
Prognosis in glioma depends strongly on tumor grade and proliferation. In this prospective study of patients with untreated primary cerebral gliomas, we investigated whether amide proton transfer–weighted imaging could reveal tumor proliferation and reliably distinguish low-grade from high-grade gliomas compared with Ki-67 expression and proton MR spectroscopy imaging.
MATERIALS AND METHODS:
This study included 42 patients with low-grade (n = 28) or high-grade (n = 14) glioma, all of whom underwent conventional MR imaging, proton MR spectroscopy imaging, and amide proton transfer–weighted imaging on the same 3T scanner within 2 weeks before surgery. We assessed metabolites of choline and N-acetylaspartate from proton MR spectroscopy imaging and the asymmetric magnetization transfer ratio at 3.5 ppm from amide proton transfer–weighted imaging and compared them with histopathologic grade and immunohistochemical expression of the proliferation marker Ki-67 in the resected specimens.
RESULTS:
The asymmetric magnetization transfer ratio at 3.5 ppm values measured by different readers showed good concordance and were significantly higher in high-grade gliomas than in low-grade gliomas (3.61% ± 0.155 versus 2.64% ± 0.185, P = .0016), with sensitivity and specificity values of 92.9% and 71.4%, respectively, at a cutoff value of 2.93%. The asymmetric magnetization transfer ratio at 3.5 ppm values correlated with tumor grade (r = 0.506, P = .0006) and Ki-67 labeling index (r = 0.502, P = .002). For all patients, the asymmetric magnetization transfer ratio at 3.5 ppm correlated positively with choline (r = 0.43, P = .009) and choline/N-acetylaspartate ratio (r = 0.42, P = .01) and negatively with N-acetylaspartate (r = −0.455, P = .005). These correlations held for patients with low-grade gliomas versus those with high-grade gliomas, but the correlation coefficients were higher in high-grade gliomas (choline: r = 0.547, P = .053; N-acetylaspartate: r = −0.644, P = .017; choline/N-acetylaspartate: r = 0.583, P = .036).
CONCLUSIONS:
The asymmetric magnetization transfer ratio at 3.5 ppm may serve as a potential biomarker not only for assessing proliferation, but also for predicting histopathologic grades in gliomas.
Gliomas are the most common primary cerebral tumors and are classified into 4 grades based on histology and biologic behavior.1 The median survival time depends greatly on tumor grade, being longer for patients with lower-grade tumors.2–4 Another important marker of prognosis in glioma is Ki-67, a protein reflecting cellular proliferation and malignancy; higher Ki-67 values correspond to greater malignancy. Indeed, some investigators have proposed that the Ki-67 labeling index could be superior to histologic grading as an indicator of prognosis.5,6 A noninvasive imaging method that could reveal proliferative features at the molecular level and enable accurate tumor grading before surgery would be of great importance for the clinical management of gliomas.
MR imaging is widely used in the clinical management of gliomas. However, conventional MR imaging is not sufficient for accurate assessments of tumor grade and proliferation. 1H proton MR spectroscopy imaging (MRSI) allows noninvasive measurement of metabolites in tissues, among the most common of which are choline and N-acetylaspartate (NAA). Choline is a metabolic marker of cell membrane density and integrity, reflecting cellular proliferation and tumor progression.7,8 NAA is a marker of neuronal health, viability, and number9; NAA levels are inversely associated with glioma grade and are considered a useful indicator of secondary neuronal damage caused by tumor growth.10 MRSI can detect regions within gliomas that contain aggressive growth or upgrading because of strong correlations between choline and NAA levels and the Ki-67 histopathologic proliferation index.11 Therefore, MRSI can provide important supplementary information for the management of gliomas.
Amide proton transfer–weighted imaging (APTWI) is a novel molecular MR imaging technique developed to detect and quantitatively visualize endogenous proteins and peptides.12 APTWI is usually reported in terms of asymmetry in the magnetization transfer ratio at 3.5 ppm [MTRasym(3.5ppm)] and is measured as a reduction in bulk water intensity caused by chemical exchange with magnetically labeled backbone amide protons (at ∼3.5 ppm downfield of the water resonance) of endogenous mobile proteins and peptides. Previous studies have demonstrated that APTWI can detect response to therapy in glioma13–15 and correlates with grade in adult diffuse gliomas.16 However, a further consideration sustained by multiple methods with bigger samples is required to investigate whether APTWI accurately reflects cellular proliferation in patients with glioma.
We previously observed that choline and NAA levels measured by MRSI correlated with MTRasym(3.5ppm) in a patient with anaplastic astrocytoma. Because APTWI and MRSI can produce similar information regarding tumor proliferation, we hypothesized that MTRasym(3.5ppm) could correlate with the metabolic profiles from MRSI. The purpose of this study was to prospectively explore the utility of the amide proton transfer (APT)–derived measurement MTRasym(3.5ppm) as a noninvasive biomarker of glioma proliferation and histopathologic grade by comparing APTWI with Ki-67 expression and with MRSI by using voxel-to-voxel analysis.
Materials and Methods
Patients
This prospective study was initiated after approval by the institutional ethics committee of Tongji Hospital, and all activities abided by the statement of ethical standards. From June 23, 2015 to January 24, 2016, we identified 66 consecutive patients suspected of having a primary cerebral tumor; none had had chemotherapy or radiation treatment within 6 months before surgery, and all provided informed consent before MR imaging scanning. Inclusion criteria were as follows: 1) undergoing routine MR imaging, APTWI, and MRSI in the same scanner and 2) histopathologic confirmation of glioma. Exclusion criteria were 1) not having surgery or having surgery more than 2 weeks after the MR imaging; 2) having nonglioma tumors; 3) having motion artifacts on imaging; and 4) having missing or unstable baseline MRSI data. Of the 66 patients originally identified, 24 were excluded (6 for not having tumor resection, 14 for having lesions other than glioma, 2 for head motion, 1 for massive intratumoral hemorrhage, and 1 for having a pilocytic astrocytoma with a large cyst and minimal solid tumor). The remaining 42 patients, all of whom underwent tumor resection, were the subjects of this analysis. According to the World Health Organization (WHO) criteria, lesions in 28 patients were low-grade glioma (19 astrocytoma [1 grade I, 18 grade II], 7 oligodendroglioma, and 2 oligoastrocytoma) and lesions in 14 patients were high-grade glioma (1 anaplastic oligodendroglioma, 5 anaplastic astrocytoma [grade III], and 8 glioblastoma [grade IV]). Of these 42 patients, 6 were excluded from the voxel-to-voxel analysis (described under “Data Processing and Analysis”) because of poor-quality MRSI. Patient and treatment characteristics are shown in Table 1.
Table 1:
Patient characteristics in the 2 correlational analyses
| Characteristics | Value or No. of Patients (%) |
|||||
|---|---|---|---|---|---|---|
| MTRasym(3.5ppm) vs Ki-67 |
MTRasym(3.5ppm) vs MR Spectroscopy |
|||||
| Low-Grade Glioma (n = 28) | High-Grade Glioma (n = 14) | P Value | Low-Grade Glioma (n = 23) | High-Grade Glioma (n = 13) | P Value | |
| Age, ya | 44.00 ± 2.81 | 44.64 ± 3.70 | .89 | 42.96 ± 3.04 | 43.31 ± 3.73 | .94 |
| Sex, no. (%) | .82 | .97 | ||||
| Male | 19 (68) | 9 (64) | 14 (61) | 8 (57) | ||
| Female | 9 (32) | 5 (36) | 9 (39) | 5 (43) | ||
| Tumor grade, no. (%)b | ||||||
| II | 28 (100) | 0 | 23 (100) | 0 | ||
| III | 0 | 6 (43) | 0 | 6 (46) | ||
| IV | 0 | 8 (57) | 0 | 7 (54) | ||
Mean ± standard deviation.
Confirmed on histopathologic examination.
Data Acquisition
All patients underwent MR imaging before surgery, with images obtained with a Discovery MR750 3T scanner (GE Healthcare, Milwaukee, Wisconsin). Routine and gadolinium contrast-enhanced MR imaging were included in the basic scanning protocol, which consisted of transverse T1 fluid-attenuated inversion recovery, transverse T2 fast spin-echo, and transverse T2 fluid-attenuated inversion recovery. All routine sequences had the same section thickness, spacing, and FOV (5 mm/1.5 mm, 20 sections, and 240 × 240 mm2, respectively). All sequences had the same scan coverage, and the range covered the entire brain. Routine and contrast-enhanced images were used as anatomic references for functional imaging scan localization.
APT data were collected during the routine MR imaging scanning session before the intravenous bolus injection of chelated gadolinium. Any other unrelated contrast-enhancement scanning was to be done at least 24 hours before or after amide proton scanning to avoid interference with the APT signal.17 Single-section transverse imaging was used at the center of the tumor for the APT data, with TR/TE, 3000/22.6 ms; matrix size, 128 × 128; FOV, 240 × 240 mm2; and section thickness, 5 mm. The power of the radiofrequency saturation pulse for the APT sequence was 2.0 μT and the length was 400 ms. Data were acquired with 2 NEX in a saturation frequency list of 15.6, ±6, ±5, ±4.5, ±4, ±3.75, ±3.5, ±3.25, ±3, ±2.5, ±2, ±1.5, ±1, ±0.75, ± 0.5, ±0.25, and 0 ppm and 1 no-saturation map, resulting in 66 images being acquired in 3 minutes 18 seconds. The MTRasym(3.5ppm) was calculated at 3.5 ppm, and B0 correction was performed by shifting the minimum signal of the z spectrum to 0 Hz.
For MRSI, we used 2D multivoxel 1H imaging based on the point-resolved spectroscopy pulse sequence (PRESS). VOIs were centered on the same section as the position for APTWI based on anatomic reference images, with the following parameters: TR, 1000 ms; TE, 144 ms; FOV, 240 mm2; section thickness, 14 mm; NEX, 0.8; and pixel dimension, 7.5 mm × 7.5 mm × 14 mm with a total scanning time of 4 minutes 20 seconds. The VOIs were chosen carefully to avoid areas of hemorrhage, calcification, cystic components, large vessels, and strong interference from subcutaneous fat and lipid of the skull; saturation slabs were applied in 6 directions to further reduce potential artifacts.
Data Processing and Analysis
All raw data were transferred to an AW4.6 workstation (GE Healthcare). To study the ability of MTRasym(3.5ppm) to reflect tumor proliferation, we compared APT MTRaysm(3.5ppm) with MRSI and Ki-67, and the ROIs were drawn on the APT images in 2 ways. First, for the voxel-to-voxel correlation of MTRasym(3.5ppm) with MRSI, spectral analyses were done. The following steps were included: 1) filtering of the signal to reduce noise, 2) zero-filling of the signal, 3) the suppression of water molecules signal, 4) translating of the signal from the time domain to the frequency domain with fast Fourier transformation, 5) baseline correction and phase correction. The calculation of quantitative parameters was based on curve fitting, and information on the location of each VOI was collected on the anatomic images. Then, a parametric MTRasym(3.5ppm) map was created and automatically matched with the anatomic images. The grids that come with the postprocessing FuncTool software (GE Healthcare) were used to segment the APT maps based on anatomic images, and the size of each grid was adjusted in the same way as for the MR spectroscopy (On-line Fig 1). The grid with the largest mean APT asymmetry value was selected as the target to analyze. In the second approach, to evaluate potential correlations between Ki-67 expression and APTWI, 2 readers who were blinded to the histopathologic diagnosis independently drew 4 circular ROIs on the APT images, taking care to place them so as to include solid tumor in the area with the highest MTRasym(3.5ppm) but to avoid cystic, large necrotic, or hemorrhagic components of the tumor with reference to conventional MR imaging. The measured MTRasym(3.5ppm) in the 4 ROIs were averaged to represent the tumor, as shown in On-line Fig 2.
Surgery and Pathologic Evaluation
Tumor grade was assigned based on the pathologic diagnosis of the surgical specimens according to the 2007 WHO classification; the Ki-67 labeling index was also measured by using standard immunohistochemical staining procedures in samples from 37 patients (2 samples from the other 5 patients were too small). Tumor sections were reviewed and Ki-67 quantified based on the percentage of positive cells in the highest density of the stained areas; all cells with nuclear staining of any intensity were considered positive, and the Ki-67 labeling index was defined as the percentage of positive cells among the total number of counted cells.
Statistical Analysis
All values are expressed as means ± standard deviations. Interobserver agreement on tumor MTRasym(3.5ppm) values made by the 2 readers was analyzed by calculating the intraclass correlation coefficient with MedCalc (MedCalc Software, Ostend, Belgium). Intraclass correlation coefficients >0.74 indicate excellent correlation. Overall correlations between the MTRasym(3.5ppm) values and the MRSI features as well as subgroup correlations were analyzed with the Pearson correlation coefficient. The correlation between Ki-67 expression and MTRasym(3.5ppm) was also assessed with the Pearson correlation analysis. Independent sample t tests were used to compare differences in MTRasym(3.5ppm) and Ki-67 between the high- and low-grade gliomas. Receiver operating characteristic curve analysis was used to evaluate the accuracy of MTRasym(3.5ppm) for determining glioma grade and to establish optimal cutoff values. A default α level of .05 was used for all tests, and all tests were 2-tailed. Statistical analyses were done with SPSS (IBM, Armonk, New York), IBM 19 (IBM), or Prism 5.0 GraphPad software (GraphPad Software, San Diego, California).
Results
MTRasym(3.5ppm) and Glioma Grade
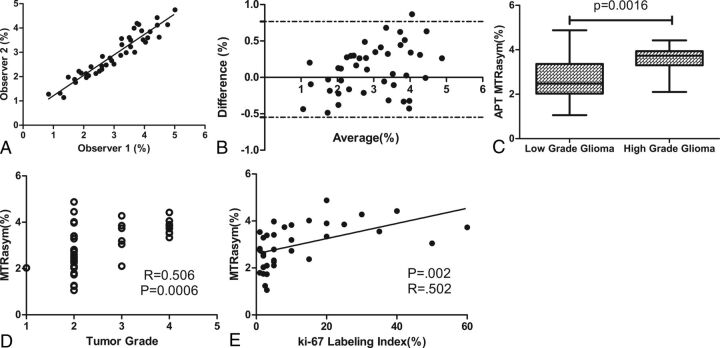
Both readers found that the APT MTRasym(3.5ppm) values were lower in low-grade gliomas than in high-grade gliomas (reader 1, 2.65% ± 0.20 versus 3.74% ± 0.17, P = .001 by Student t test; reader 2, 2.63% ± 0.18 versus 3.47% ± 0.16, P = .004 by Student t test) (Table 2). The sensitivity and specificity values for discriminating low-grade from high-grade gliomas were 92.9% and 71.4%, respectively, when cutoff values for MTRasym(3.5ppm) were 3.11% for reader 1 and 2.85% for reader 2. Areas under the curve of MTRasym(3.5ppm) were 0.813 (95% CI, 0.675–0.95) for reader 1 and 0.770 (95% CI, 0.623–0.918) for reader 2 (Table 2). Interobserver agreement between the 2 readers was excellent, with an intraclass correlation coefficient of 0.94 (95% CI, 0.89–0.96) and a regression equation of y = 0.8928x + 0.3550 (P < .0001) (Fig 1A). The Bland-Altman analysis of the difference in MTRasym(3.5ppm) assessed by the 2 readers also showed excellent concordance, with only 1 value beyond the 95% limits of agreement (Fig 1B). The mean MTRasym(3.5ppm) values of both readers' measurements were significantly lower for low-grade gliomas than for high-grade gliomas (2.64% ± 0.185 versus 3.61% ± 0.155, P = .0016 by Student t test) (Fig 1C, Table 2). Correlation analysis further demonstrated that MTRasym(3.5ppm) values correlated with tumor grade (r = 0.506, P = .0006) (Fig 1D).
Table 2:
Independent t test and receiver operating characteristic curve analysis of low-/high-grade glioma
| MTRasym(3.5ppm) | Low-Grade Gliomas (n = 28) | High-Grade Gliomas (n = 14) | P Value | Area Under the Curvea | Optimal Threshold (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|
| Reader 1 | 2.65% ± 0.20 | 3.74% ± 0.17 | .001 | .813 (.675, .95) | 3.11 | 92.9 | 71.4 |
| Reader 2 | 2.63% ± 0.18 | 3.47% ± 0.16 | .004 | .770 (.623, .918) | 2.85 | 92.9 | 71.4 |
| Mean MTRasym(3.5ppm) | 2.64% ± 0.18 | 3.61% ± 0.155 | .002 | .791 (.650, .931) | 2.93 | 92.9 | 71.4 |
| Mean Ki-67 labeling index | 4.98% ± 1.02 (n = 24) | 24.15% ± 5.04 (n = 13) | .003 | .867 (.702, 1.00) | 6.5 | 84.6 | 83.3 |
Data in parentheses are 95% CIs.
Fig 1.
A, Intraclass agreement for the 2 readers who evaluated the MTRasym(3.5ppm) of the solid parts of tumors in 42 patients was excellent, with an intraclass correlation coefficient of 0.94 (95% CI, 0.89–0.96). B, Bland-Altman analysis of the difference in MTRasym(3.5ppm) by the 2 readers also showed excellent concordance. C, MTRasym(3.5ppm) values were higher for high-grade gliomas than for low-grade gliomas (P = .0016, Student t test). D, The correlation between MTRasym(3.5ppm) and tumor grade was moderate (Pearson r = 0.506; P = .0006). E, The correlation between the Ki-67 staining index and the APT MTRasym(3.5ppm) was also moderate (Pearson r = 0.502; P = .002).
MTRasym(3.5ppm) and Ki-67 Expression
The mean Ki-67 labeling index was significantly lower in low-grade gliomas than in high-grade gliomas (4.98% ± 1.02 versus 24.15% ± 5.04, P = .003) (Table 2). Correlation analysis revealed that the MTRasym(3.5ppm) values were moderately positively correlated with Ki-67 expression levels (r = 0.502, P = .002) (Fig 1E), with a higher MTRasym(3.5ppm) indicating greater proliferative activity and greater malignancy. Findings from APTWI, Ki-67 staining, and MRSI from 2 representative patients, one with low-grade glioma and the other with high-grade glioma, are shown separately in Figs 2 and 3.
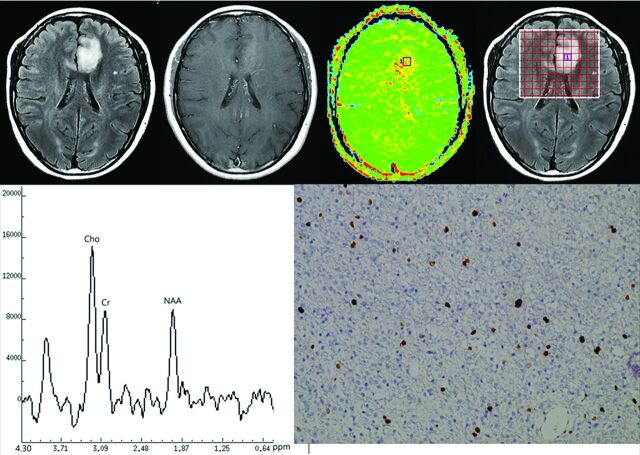
Fig 2.
Top row, left to right: Transverse T2-FLAIR MR image (left), postgadolinium T1-weighted MR image, APT image, and the location of MR spectroscopy voxel of a 45-year-old woman with diffuse astrocytoma (grade II). Bottom row, left to right: MRSI indicating peaks for choline (Cho), creatine (Cr) and N-acetylaspartate (NAA); Ki-67 immunohistochemical stain for the same patient. The MTRasym(3.5ppm) value for this patient was 2.83%; the choline peak was slightly increased and the NAA peak slightly decreased; and the Ki-67 labeling index was 5%.
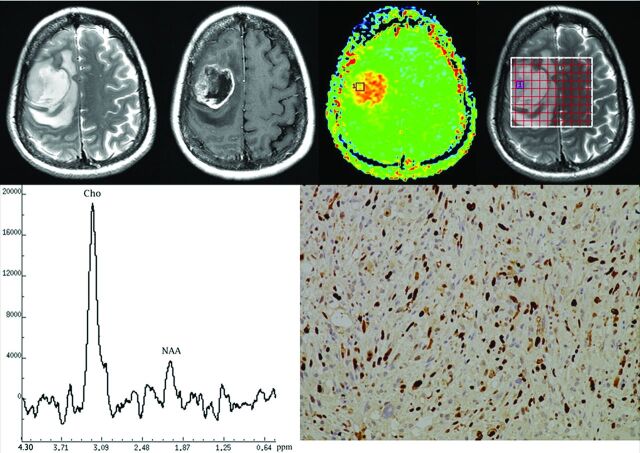
Fig 3.
Top row, left to right: T2 FSE MR image, postgadolinium T1-weighted MR image, APT image, and the location of MR spectroscopy voxel of a 46-year-old woman with anaplastic oligodendroma (grade III). Bottom row, left to right: MRSI indicating peaks for choline (Cho) and N-acetylaspartate (NAA), and Ki-67 immunohistochemical staining for the same patient. The MTRasym(3.5ppm) value for this patient was 4.39%; the choline peak was obviously increased and the NAA peak obviously decreased; and the Ki-67 labeling index was 30%.
Correlation of MTRasym(3.5ppm) with Metabolite Measurements on MRSI
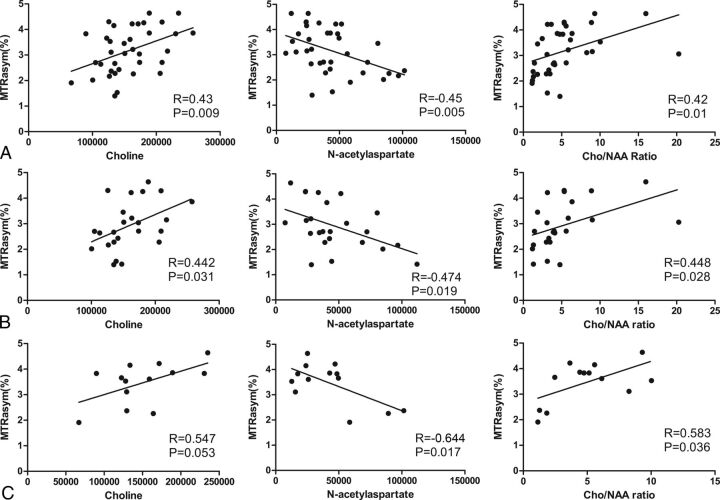
Overall Pearson correlation analysis showed that the MTRasym(3.5ppm) values were moderately positively correlated with choline (r = 0.429, P = .009) and the choline/NAA ratio (r = 0.423, P = .01) and were moderately negatively correlated with NAA (r = −0.455, P = .005) (Fig 4A). This correlation pattern was similar in the subgroup analysis of low- and high-grade gliomas. In low-grade gliomas, MTRasym(3.5ppm) showed moderate correlation with metabolite measurements (choline, r = 0.442, P = .031; NAA, r = –0.474, P = .019; choline/NAA ratio, r = 0.448, P = .028) (Fig 4B). The correlation was similar in high-grade gliomas, but the absolute correlation coefficients were greater (choline, r = 0.547, P = .053; NAA, r = –0.644, P = .017; choline/NAA ratio, r = 0.583, P = .036) (Fig 4C).
Fig 4.
The voxel-to-voxel correlational analyses of MTRasym(3.5ppm) with choline (Cho), N-acetylaspartate (NAA), and Cho/NAA ratio in patients with glioma. A, For all patients (regardless of glioma grade), MTRasym(3.5ppm) showed mildly positive correlations with Cho (r = 0.43, P = .009) and the Cho/NAA ratio (r = 0.42, P = .01) and a slightly stronger negative correlation with NAA (r = −0.45, P = .005). B, Subgroup analyses of low-grade gliomas revealed that MTRasym(3.5ppm) correlated with Cho (r = 0.442, P = .031), NAA (r = −0.474, P = .019), and the Cho/NAA ratio (r = 0.448, P = .028). C, Subgroup analyses of high-grade gliomas revealed a similar pattern of correlation, though the absolute values of the correlation coefficients were higher for the high-grade tumors (Cho: r = 0.547, P = .053; NAA: r = −0.644, P = .017; Cho/NAA: r = 0.583, P = .036).
Discussion
Our key findings were as follows. First, the MTRasym(3.5ppm) values correlated with glioma grades, and MTRasym(3.5ppm) could reliably distinguish low-grade from high-grade gliomas. Moreover, these MTRasym(3.5ppm) values also correlated with choline and NAA levels on MRSI in both low- and high-grade gliomas, with stronger correlation in high-grade gliomas. Finally, the MTRasym(3.5ppm) values correlated moderately with the Ki-67 labeling index. Collectively, these findings suggest that MTRasym(3.5ppm) may be useful as a noninvasive biomarker of glioma proliferation and grade.
We further found that high-grade gliomas showed significantly higher MTRasym(3.5ppm) values than low-grade gliomas and that the increase of MTRasym(3.5ppm) correlated positively with the cellular proliferation marker Ki-67 and glioma grade. Our findings were partly consistent with those results of another group,16 who reported that the amide protons of endogenous mobile proteins and peptides in the cytoplasm are the main source of the APT signal. To further clarify the value of MTRasym(3.5ppm) with regard to tumor proliferation, we compared MTRasym(3.5ppm) with MRSI variables and observed modest correlations between the 2 types of measurements. High-grade gliomas are characterized by particularly rapid proliferation and atypical cytoarchitecture, as well as the presence of necrotic areas, vascular thrombosis, and microvascular proliferation. Because higher-grade gliomas have more actively proliferating tumor cells, and because the concentrations of mobile macromolecules (such as proteins and peptides) increase with the tumor cell density and glioma grades,18 one could expect that the MTRasym(3.5ppm) values would be different in high-grade versus low-grade gliomas. Factors other than intracellular components can also affect APT MTRasym(3.5ppm); for example, tumors with necrosis showed higher MTRasym(3.5ppm) values than tumors without necrosis.16 Highly concentrated mobile proteins and peptides in the extracellular space, such as microscopic necrosis or fluid collection in the microcystic space, may also increase MTRasym(3.5ppm) in tumors. In addition, the amide proton exchange rate is base-catalyzed in the physiologic pH range. Tissue pH in gliomas has been shown to be higher than that in normal tissue (up to 0.2 pH units)19,20; a higher pH increases the proton exchange rate, which in turn increases MTRasym(3.5ppm). These findings, in combination with our own discovery that MTRasym(3.5ppm) correlates positively with the Ki-67 labeling index and glioma grade, provide further support for the concept that APT MTRasym(3.5ppm) may be valuable as a noninvasive biomarker of tumor proliferation.
In our study, MTRasym(3.5ppm) values correlated positively with choline level and choline/NAA ratio and negatively with NAA level, which suggests that the MTRasym(3.5ppm) of the solid parts of gliomas may reflect both the proliferative activity of the tumor and the extent of neuron damage caused by tumor growth. As an important imaging method based on the metabolic characteristics of tissues,21,22 MRSI has been used to map changes in phospholipid metabolism in brain tumors. Increases in total choline have been linked with increases in membrane turnover and cellular density7,8 and with the extent of fiber destruction caused by gliomas.23 Conversely, NAA levels reflect neuronal activity and, thus, decrease in parallel with the growth of brain tumors such as gliomas.21 Our findings on the correlations between MTRasym(3.5ppm) and choline/NAA ratios were consistent in part with those reported by Park et al,15 who used single-voxel MR spectroscopy, and their VOIs were somewhat larger than our multivoxel MRSI findings. Because gliomas are often heterogeneous, using smaller VOIs carries a lower risk of accidentally including cystic and necrotic areas and allows more precise measurement of the tumor areas of interest, which could provide an important part of a comprehensive comparison between MR spectroscopy and APT imaging. Our voxel-to-voxel correlation results, combined with the findings reported by Park et al,15 indicate that MTRasym(3.5ppm) could be a noninvasive means of assessing not only tumor proliferation, but also the extent of secondary neuronal injury in vivo, which would be very important for targeted biopsy and local therapy.
Our findings also demonstrated that APT MTRasym(3.5ppm) provided better sensitivity (93%) and specificity (71.4%) with a cutoff value of 2.93% in distinguishing low- from high-grade gliomas, and the MTRasym(3.5ppm) values correlated with metabolite levels from MRSI. Previous studies10,24 have shown that MRSI is useful for distinguishing among types and grades of brain tumors. However, the reported accuracy in identifying glioma grades by choline levels was not sufficient, having high sensitivity but low specificity. Further, MRSI is limited by heterogeneities in the tumor.11 With the exception of a uniform magnetic field, accurate and reliable measurements also require not only excellent scanning done by a skilled MR imaging technologist, but also careful placement of ROIs to avoid areas of calcification, cysts, hemorrhage, and adjacent bone structures. As for APTWI scanning, these disadvantages are not a major problem. Moreover, in addition to the ability to distinguish malignant brain tumors from peritumoral edema and to distinguish low-grade from high-grade tumors,12,25,26 MTRasym(3.5ppm) can also be used to depict tumor core areas based on its ability to visualize amide proton contrast. Our results also showed that MTRasym(3.5ppm) values correlated positively with glioma grade, with higher MTRasym(3.5ppm) associated with greater malignancy. Sakata et al27 reported that APTWI analysis of a single representative image section could distinguish low-grade from high-grade gliomas as accurately as could APTWI of the entire tumor, and single sections can be acquired within a reasonably short scan time. Collectively, these findings imply that APT can be an efficient tool for assessing glioma malignancy in a clinical setting.
This study had 2 major limitations. First, although the ROIs for the APT measurements were roughly in the same areas as those used to assess Ki-67, a point-to-point match was not done. Resected specimens for Ki-67 examination possess little locational information, and without intraoperative MR imaging, the accuracy of a direct match between MR imaging and samples for immunochemical staining would be a challenge, especially when a single-axis section is used for imaging. Second, relatively few patients in this study had high-grade gliomas, though the correlations were statistically significant.
Conclusions
Comparing APTWI with immunohistochemical staining of Ki-67 and MRSI by using voxel-to-voxel analysis in patients with glioma, we found that MTRasym(3.5ppm) correlated with Ki-67 labeling index, with histopathologic grade, and with key metabolites from MR spectroscopy. We concluded that MTRasym(3.5ppm) may be a useful biomarker not only for assessing tumor proliferation, but also for predicting histopathologic grade, which could have an important impact on the clinical therapy strategy for patients with glioma.
Supplementary Material
ABBREVIATIONS:
- APT
amide proton transfer
- APTWI
amide proton transfer–weighted imaging
- MRSI
1H proton MR spectroscopy imaging
- MTRasym(3.5ppm)
asymmetric magnetization transfer ratio at 3.5ppm
- NAA
N-acetylaspartate
- WHO
World Health Organization
Footnotes
This work was supported by grants from the National Program of the Ministry of Science and Technology of China during the “12th Five-Year Plan” (ID: 2011BAI08B10) and the National Natural Science Foundation of China (No. 81171308 and No. 81570462).
References
- 1. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007;114:97–109 10.1007/s00401-007-0243-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med 2008;359:492–507 10.1056/NEJMra0708126 [DOI] [PubMed] [Google Scholar]
- 3. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987–96 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 4. Cancer Genome Atlas Research Network, Brat DJ, Verhaak RG, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 2015;372:2481–98 10.1056/NEJMoa1402121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hoshino T, Ahn D, Prados MD, et al. Prognostic significance of the proliferative potential of intracranial gliomas measured by bromodeoxyuridine labeling. Int J Cancer 1993;53:550–55 10.1002/ijc.2910530404 [DOI] [PubMed] [Google Scholar]
- 6. Sallinen PK, Haapasalo HK, Visakorpi T, et al. Prognostication of astrocytoma patient survival by Ki-67 (MIB-1), PCNA, and S-phase fraction using archival paraffin-embedded samples. J Pathol 1994;174:275–82 10.1002/path.1711740407 [DOI] [PubMed] [Google Scholar]
- 7. Tamiya T, Kinoshita K, Ono Y, et al. Proton magnetic resonance spectroscopy reflects cellular proliferative activity in astrocytomas. Neuroradiology 2000;42:333–38 10.1007/s002340050894 [DOI] [PubMed] [Google Scholar]
- 8. Tedeschi G, Lundbom N, Raman R, et al. Increased choline signal coinciding with malignant degeneration of cerebral gliomas: a serial proton magnetic resonance spectroscopy imaging study. J Neurosurg 1997;87:516–24 10.3171/jns.1997.87.4.0516 [DOI] [PubMed] [Google Scholar]
- 9. Moffett JR, Ross B, Arun P, et al. N-Acetylaspartate in the CNS: from neurodiagnostics to neurobiology. Prog Neurobiol 2007;81:89–131 10.1016/j.pneurobio.2006.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stadlbauer A, Gruber S, Nimsky C, et al. Preoperative grading of gliomas by using metabolite quantification with high-spatial-resolution proton MR spectroscopic imaging. Radiology 2006;238:958–69 10.1148/radiol.2382041896 [DOI] [PubMed] [Google Scholar]
- 11. Bulik M, Jancalek R, Vanicek J, et al. Potential of MR spectroscopy for assessment of glioma grading. Clin Neurol Neurosurg 2013;115:146–53 10.1016/j.clineuro.2012.11.002 [DOI] [PubMed] [Google Scholar]
- 12. Zhou J, Lal B, Wilson DA, et al. Amide proton transfer (APT) contrast for imaging of brain tumors. Magn Reson Med 2003;50:1120–26 10.1002/mrm.10651 [DOI] [PubMed] [Google Scholar]
- 13. Sagiyama K, Mashimo T, Togao O, et al. In vivo chemical exchange saturation transfer imaging allows early detection of a therapeutic response in glioblastoma. Proc Natl Acad Sci U S A 2014;111:4542–47 10.1073/pnas.1323855111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou J, Tryggestad E, Wen Z, et al. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nat Med 2011;17:130–34 10.1038/nm.2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park JE, Kim HS, Park KJ, et al. Pre- and posttreatment glioma: comparison of amide proton transfer imaging with MR spectroscopy for biomarkers of tumor proliferation. Radiology 2016;278:514–23 10.1148/radiol.2015142979 [DOI] [PubMed] [Google Scholar]
- 16. Togao O, Yoshiura T, Keupp J, et al. Amide proton transfer imaging of adult diffuse gliomas: correlation with histopathological grades. Neuro Oncol 2014;16:441–48 10.1093/neuonc/not158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tee YK, Donahue MJ, Harston GW, et al. Quantification of amide proton transfer effect pre- and post-gadolinium contrast agent administration. J Magn Reson Imaging 2014;40:832–38 10.1002/jmri.24441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Howe FA, Barton SJ, Cudlip SA, et al. Metabolic profiles of human brain tumors using quantitative in vivo 1H magnetic resonance spectroscopy. Magn Reson Med 2003;49:223–32 10.1002/mrm.10367 [DOI] [PubMed] [Google Scholar]
- 19. Maintz D, Heindel W, Kugel H, et al. Phosphorus-31 MR spectroscopy of normal adult human brain and brain tumours. NMR Biomed 2002;15:18–27 10.1002/nbm.735 [DOI] [PubMed] [Google Scholar]
- 20. Ha DH, Choi S, Oh JY, et al. Application of 31P MR spectroscopy to the brain tumors. Korean J Radiol 2013;14:477–86 10.3348/kjr.2013.14.3.477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Howe FA, Opstad KS. 1H MR spectroscopy of brain tumours and masses. NMR Biomed 2003;16:123–31 10.1002/nbm.822 [DOI] [PubMed] [Google Scholar]
- 22. Nelson SJ. Multivoxel magnetic resonance spectroscopy of brain tumors. Mol Cancer Ther 2003;2:497–507 [PubMed] [Google Scholar]
- 23. Stadlbauer A, Hammen T, Buchfelder M, et al. Differences in metabolism of fiber tract alterations in gliomas: a combined fiber density mapping and magnetic resonance spectroscopic imaging study. Neurosurgery 2012;71:454–63 10.1227/NEU.0b013e318258e332 [DOI] [PubMed] [Google Scholar]
- 24. Zeng QS, Li CF, Liu H, et al. Distinction between recurrent glioma and radiation injury using magnetic resonance spectroscopy in combination with diffusion-weighted imaging. Int J Radiat Oncol Biol Phys 2007;68:151–58 10.1016/j.ijrobp.2006.12.001 [DOI] [PubMed] [Google Scholar]
- 25. Zhou J, Blakeley JO, Hua J, et al. Practical data acquisition method for human brain tumor amide proton transfer (APT) imaging. Magn Reson Med 2008;60:842–49 10.1002/mrm.21712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou J, Zhu H, Lim M, et al. Three-dimensional amide proton transfer MR imaging of gliomas: initial experience and comparison with gadolinium enhancement. J Magn Reson Imaging 2013;38:1119–28 10.1002/jmri.24067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sakata A, Okada T, Yamamoto A, et al. Grading glial tumors with amide proton transfer MR imaging: different analytical approaches. J Neurooncol 2015;122:339–48 10.1007/s11060-014-1715-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.