Abstract
Methanotrophic bacteria represent a potential route to methane utilization and mitigation of methane emissions. In the first step of their metabolic pathway, aerobic methanotrophs use methane monooxygenases (MMOs) to activate methane, oxidizing it to methanol. There are two types of MMOs: a particulate, membrane-bound enzyme (pMMO) and a soluble, cytoplasmic enzyme (sMMO). The two MMOs are completely unrelated, with different architectures, metal cofactors, and mechanisms. The more prevalent of the two, pMMO, is copper-dependent, but the identity of its copper active site remains unclear. By contrast, sMMO uses a diiron active site, the catalytic cycle of which is well understood. Here we review the current state of knowledge for both MMOs, with an emphasis on recent developments and emerging hypotheses. In addition, we discuss obstacles to developing expression systems, which are needed to address outstanding questions and to facilitate future protein engineering efforts.
Graphical Abstract
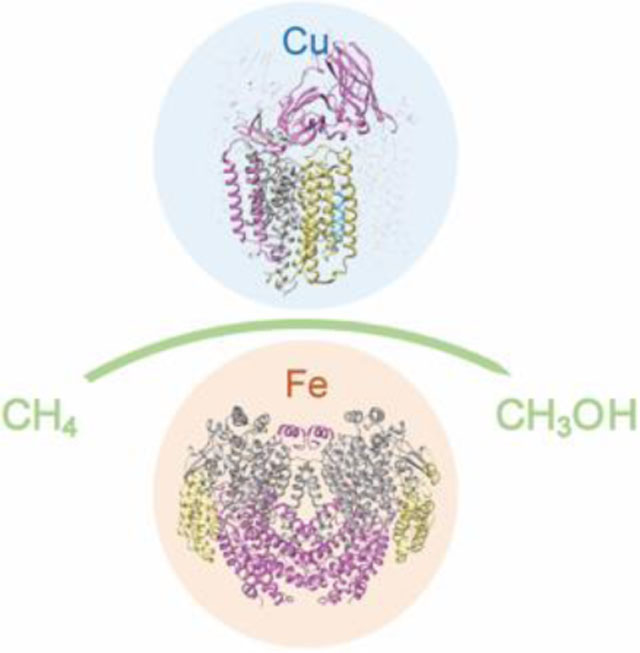
Methane monooxygenase enzymes use metal cofactors to activate methane under ambient, aerobic conditions. This review highlights recent progress in understanding the structure and activity of the copper-containing, membrane-bound and iron-containing, soluble methane monooxygenases.
1. Introduction
During an era of unprecedented climate change, increasing atmospheric methane concentrations are a constant source of concern and debate.1,2 Anthropogenic methane emissions from oil and gas production, livestock, and agriculture threaten to fuel unstoppable global warming if left unchecked. Methane is the second most abundant greenhouse gas after carbon dioxide and accounts for at least 25% of current global warming. Moreover, its superior heat-trapping capacity confers a global warming potential 84 times that of carbon dioxide over a 20-year period. The effectiveness of methane as a greenhouse gas combined with its ~10 year lifetime means that immediate small reductions in atmospheric methane can have a large impact on climate change.3
Methane can be converted to desirable fuels and chemicals, which could simultaneously mitigate global warming and meet increasing energy demands.4 However, activation of the 105 kcal mol−1 methane C-H bond5 presents a significant challenge. Industrial methods rely on the conversion of methane and water to a mixture of carbon monoxide and hydrogen (syngas) followed by Fischer-Tropsch synthesis of longer-chain hydrocarbons.6 This technically complex and expensive process must be carried out in large refineries to capture economies of scale.4 The high capital expenses combined with the abundance of methane in remote locations have renewed interest in biological methane activation, which offers the possibility of deployable smaller-scale methods with higher conversion efficiencies and less environmental impact.7–9 Aerobic biological methane activation occurs in a group of methane-consuming bacteria called methanotrophs.10,11 In contrast to energyintensive industrial processes, methanotrophs activate methane at ambient temperature and pressure. Harnessing this unique ability could dramatically alter industrial methane processing practices.12–14
Aerobic methanotrophs were historically divided into two groups based on metabolic pathways, membrane lipid content, and physical characteristics. Gammaproteobacteria comprise the first group, commonly referred to as type I. These methanotrophs are distinguished by their use of the ribulose monophosphate (RuMP) pathway for carbon assimilation,15,16 membranes comprising mainly 14–16 carbon phospholipids,17 and disc-shaped intracytoplasmic membrane (ICM) structures. The second group, type II, consists of Alphaproteobacteria that use the serine pathway,16 have primarily 18-carbon membrane phospholipids in their membranes,18 and exhibit ICMs along the periphery of the cells. As new species have been characterized, types I and II have become synonymous with Gammaproteobacteria and Alphaproteobacteria.19 A third group of methanotrophs, the Verrucomicrobia, was discovered recently.20 These methanotrophs live at extreme temperatures, use the Calvin-Benson-Bassham cycle, and possess mainly saturated phospholipids.21,22 Methanotrophs of all types are common in soils, rice paddies, swamps, and lakes where they have access to anthropogenic and bacterial sources of methane.10 In addition, methanotrophs inhabit the tundra,23 salt lakes,24,25 and volcanic soil26–29 where they oxidize methane under extreme conditions of temperatures, salinity, and acidity. Understanding the enzymes involved in methane activation under these wide-ranging conditions is critical to realizing the biotechnological potential of methanotrophs.
Methane activation is accomplished by methane monooxygenase enzymes (MMOs), which convert methane to methanol in the first step of methanotroph metabolism (eq. 1).
| (eq. 1) |
Methanol is then converted to formaldehyde by methanol dehydrogenase (MDH), after which the metabolic pathways diverge depending on the type of methanotroph.16 There are two types of MMO: the copper-dependent, membrane-bound or particulate methane monooxygenase (pMMO) and the iron-dependent, soluble methane monooxygenase (sMMO).30–32 pMMO is the primary MMO in nature and is present in all methanotrophs except a few species from the Methylocella33,34 and Methyloferula35 genera, which only possess sMMO.11 pMMO is typically housed within extensive ICM structures,36,37 and is highly abundant, representing ~20% of the total protein in the cell.38,39 Under copper-starved conditions, some methanotrophs utilize sMMO, expression of which is regulated by a poorly-understood mechanism called the copper switch.40,41 Once copper becomes available, sMMO expression is downregulated by 2–3 orders of magnitude, ICMs form, and pMMO is mildly upregulated.42
Although pMMO is more prevalent across methanotroph species, progress toward elucidating its active site and mechanism has been hampered by challenges intrinsic to handling integral membrane proteins. By contrast, sMMO, which is more amenable to study due to its soluble nature, is better understood. One challenge common to the two MMOs is the lack of a recombinant expression system.43–46 The catalytic components of sMMO and pMMO must instead be isolated and purified directly from native methanotrophs, precluding generation of site-directed mutants. Nevertheless, each MMO has been studied extensively by biochemical spectroscopic, and structural techniques. Several key aspects of both MMOs have been revisited in the past five years, altering and enriching our understanding of how these enzymes work. Here we review the current state of knowledge with an emphasis on recent developments and areas of controversy.
2. Particulate methane monooxygenase
2.1. Overall architecture
pMMO consists of three subunits encoded by the pmoCAB operon, which is present in up to three copies in the methanotroph genome, depending on the species.27,47–55 Most methanotrophs also possess up to two extra copies of pmoC that are relatively divergent from those in the pmoCAB operons.56,57 Since methanotrophs are frequently identified by detection of the pmoA gene in soil or marine bacterial populations using mRNA probes, pmoA genes are highly represented in databases.58
Crystal structures of pMMOs from five species have been determined.59–63 The three subunits, PmoA (α), PmoB (β), and PmoC (γ), form an αβγ protomer, and three protomers related by a threefold axis of symmetry form the full (αβγ)3 oligomer (Fig. 1). This trimer, observed in all pMMO structures, adopts a cylindrical shape approximated 90 Å in diameter and 105 Å in length. PmoA (24 kDa) and PmoC (22 kDa) are composed of seven and five transmembrane helices, respectively, and are almost entirely embedded in the lipid bilayer. PmoB (42 kDa) comprises a soluble domain of two cupredoxin-like folds that are connected by two transmembrane helices.59 In crystal structures of pMMO from type II methanotrophs,60–62 an ~20 residue helix cocrystallizes with the full complex, adjacent to each PmoC subunit with a lipid mediating the interaction.62 This helix has yet to be identified, and is reminiscent of supernumerary subunits found in the crystal structures of some respiratory complexes.64 Additional lipids have been observed in the pMMO crystal structures, and may regulate activity and assembly.62
Fig. 1.
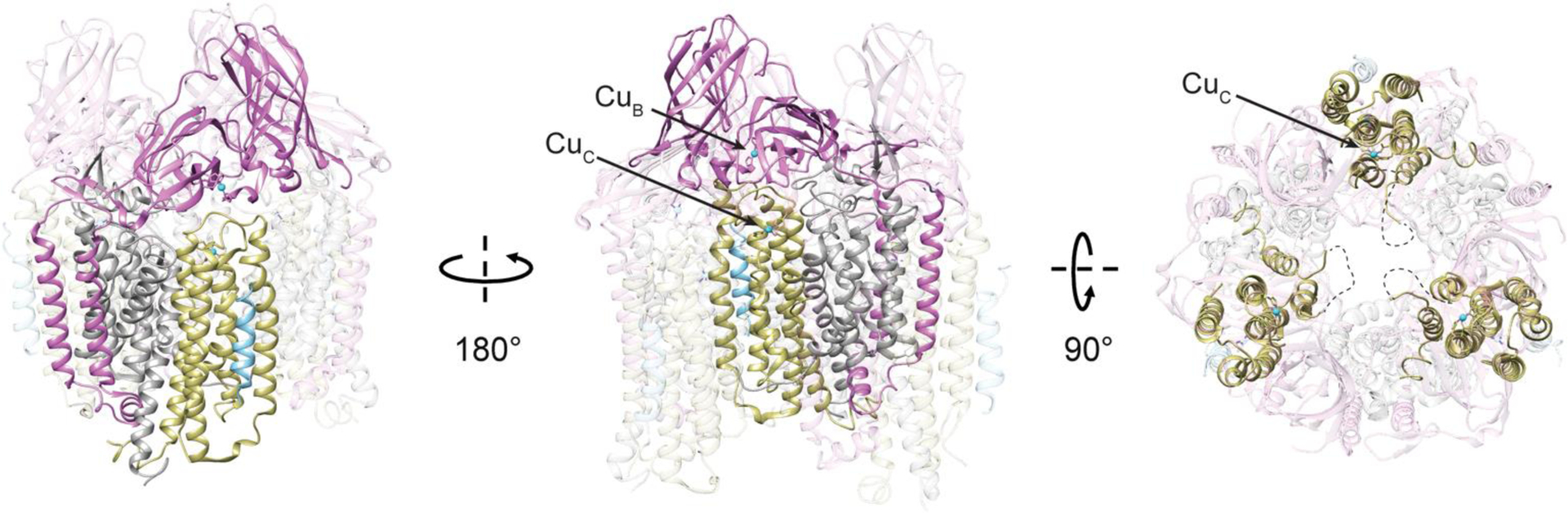
Crystal structure of pMMO from Mc. sp. Rockwell (PDB accession code 4PI0). The PmoB subunits are shown in pink, the PmoA subunits are shown in gray, the PmoC subunits are shown in yellow, and the unidentified helix is shown in blue. Copper ions are shown as cyan spheres. In the two left images, a single αβγ protomer is highlighted. In the right top-down image, the PmoC subunits are highlighted and the location of the unmodeled, highly conserved region is marked with a dashed line.
An ~25 residue region of the PmoC subunit facing the interior of the trimer is missing in every pMMO structure (Fig. 1). This sequence, corresponding to residues 200–223 in Methylocystis species strain (Mc. sp.) Rockwell pMMO,62 is not observed in the electron density maps and therefore cannot be modeled. Importantly, these residues correspond to the most highly conserved part of the PmoC sequence65 and contain multiple potential metal-binding residues (Fig. 2). In the case of Methylomicrobium (Mm.) alcaliphilum 20Z pMMO, 60% of the PmoC subunit could not be modeled.63 The disorder may be related to destabilization and loss of pMMO activity upon detergent solubilization and purification (~100-fold less than in vivo).31,63 Activity of purified pMMO can be restored or improved by reconstitution into bicelles or nanodiscs,63 suggesting that structural studies of pMMO in a native-like lipid environment could resolve the nature of this region.
Fig. 2.
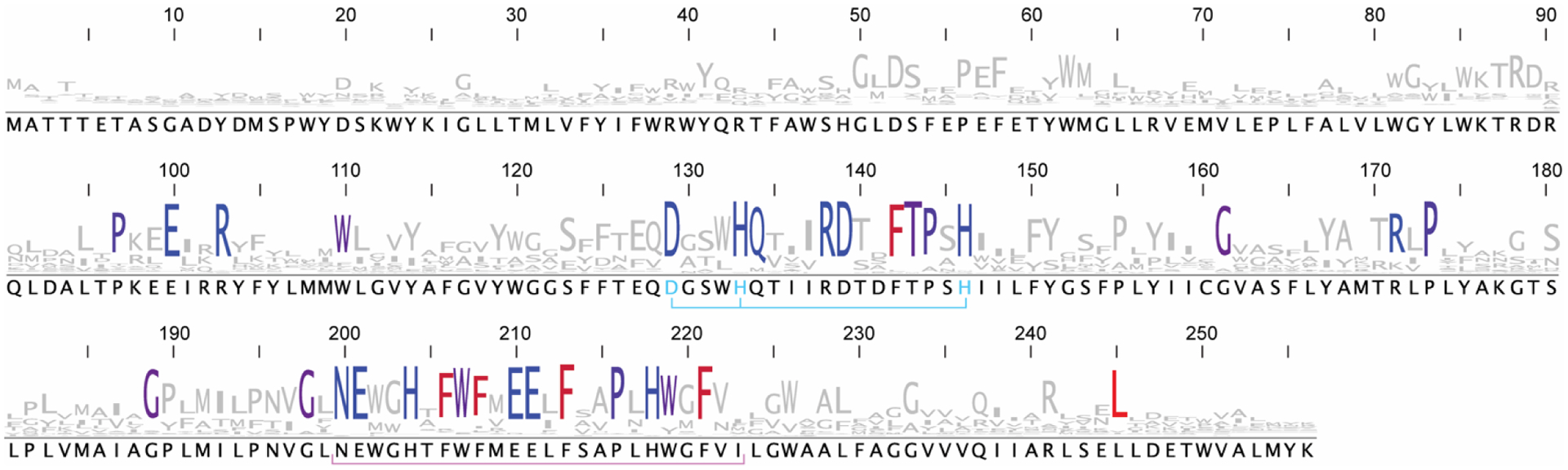
Sequence conservation logo for PmoC. The logo was generated from an alignment of 451 representative sequences >170 amino acids in length from PF04896 and TIGR03078 clustered at 100% identity against a hidden Markov model constructed from 132 representative nodes clustered at 50% identity using the EFI-EST webtool.65 Residues that are conserved in 80% of sequences or more are colored from blue to red in increasing hydrophobicity. The conserved CuC-coordinating residues are in cyan while the flexible loop is underlined in pink. The sequence is numbered using the Mc. sp. Rockwell PmoC numbering (Met49242_1455) and sequences were downloaded from the JGI/IMG database.
2.2. Metal binding sites
The pMMO crystal structures reveal up to three copper binding sites per protomer. These sites are referred to as the bis-His site, the CuB site, and the CuC site. The residues coordinating the CuB and CuC sites are conserved in almost all pMMOs as well as in the homologous enzyme ammonia monooxygenase (AMO) and related hydrocarbon monooxygenases.66–68 X-ray absorption spectroscopy (XAS), advanced paramagnetic resonance spectroscopies, and mass spectrometry have also been used extensively to probe the pMMO copper centers, with a focus on correlating spectroscopic and crystallographic data.
The bis-His site is located in the soluble region of PmoB between the two cupredoxin domains and coordinated by two histidines, His 48 and His 72 in Methylococcus (M.) capsulatus (Bath) pMMO. This site is only occupied in the M. capsulatus (Bath) pMMO structure,59 and His 48 is replaced with asparagine in pMMOs from type II methanotrophs. Notably, the site is also devoid of metal in the Mm. alcaliphilum 20Z pMMO structure despite the presence of both histidine ligands.63 It remains unclear whether the bis-His site is a crystallographic artifact or is biologically relevant, but these observations suggest that it is not essential for methane oxidation. In M. capsulatus (Bath) whole-cell EPR studies, a single Cu(II) signal attributable to the CuB site is observed,69,70 indicating that the bis-His site is either unoccupied by copper in vivo or contains Cu(I).
The CuB site is located in the soluble region of the PmoB subunit, and is occupied by copper in all structurally characterized pMMOs.59–63 Ligands include two histidine side chains derived from an HxH motif and the N-terminal histidine along with its amino group (His29, His 133, and His 135 in Mc. sp. Rockwell pMMO) (Fig. 3A). These residues are conserved in all pMMOs except those from Verrucomicrobia, in which the equivalent residues are methionine, proline, and glycine, which are unlikely to bind metal.20,47,71 The CuB site has been suggested to be analogous to the histidine brace copper active site of lytic polysaccharide monooxygenases (LPMOs) (Fig. 3B). However, there are several important differences. First, CuB has an additional histidine ligand, of which the coordinating nitrogen occupies the axial bonding position that is open in LPMO.72,73 Second, the non-amino-terminal histidine ligand in LPMOs coordinates copper with its ε nitrogen atom whereas all the histidines in CuB use their δ nitrogen atoms. Third, the LPMO site has a tyrosine side chain oxygen within 2.6 Å of the copper ion while the shortest tyrosine-to-CuB distance is 5 Å (Fig. 3). Finally, the coordinating amino group is well ordered in the LPMO structures,72 but may adopt fluctuating orientations in the CuB site as evidenced by poor electron density in every structure.
Fig. 3.
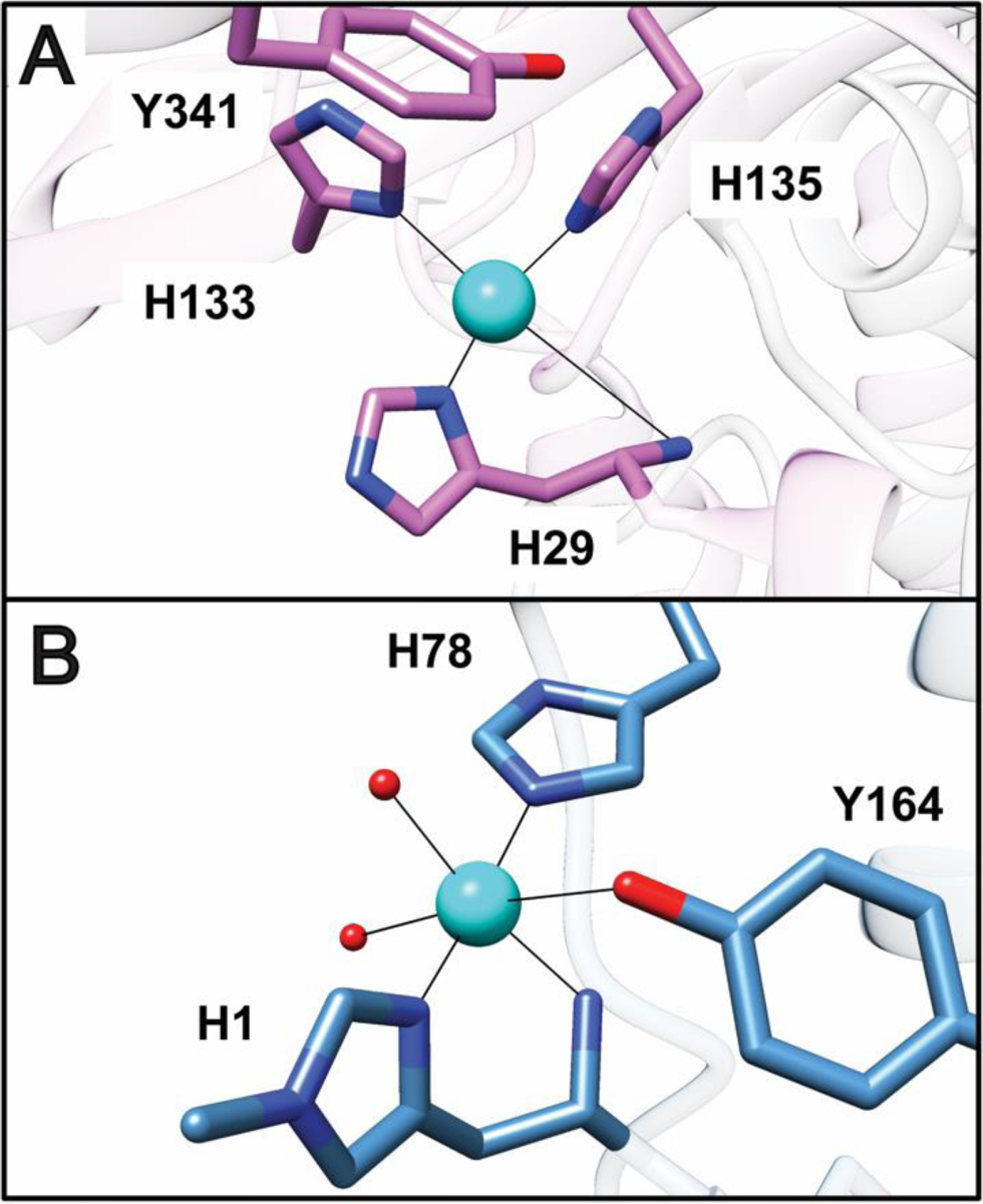
Comparison of the pMMO Cub site to the active site of LPMO. (A) The Mc. sp. Rockwell pMMO Cub site (PDB accession code 4PI0). The shortest distance from Tyr341 to the copper ion is 5 Å. (B) The Lentinus similis LPMO copper active site (PDB accession code 5ACG). Solvent ligands are shown as red spheres. Methylation of the amino-terminal histidine is not a universal feature of LPMO active sites.
The nuclearity of the Cub site has been discussed for more than a decade. In the original crystal structure of pMMO from M. capsulatus (Bath), it could be modeled with either one or two copper ions, sparking the debate over its nuclearity.59 The dinuclear model was inspired by extended X-ray absorption fine structure (EXAFS) data indicating the presence of a 2.5–2.6 Å Cu-Cu scattering interaction.74,75 However, a computational study using quantum methods to further refine the original structure indicated that monocopper is favored at this site,76 consistent with later and higher quality crystal structures also showing a mononuclear CuB site.61–63
Additional strong support for this model comes from a recent electron paramagnetic resonance (EPR) study of M. capsulatus (Bath) whole cells.70 A single signal was observed for 15N/63Cu labeled cells with 15N hyperfine splitting indicative of four equatorial nitrogen ligands. These ligands were assigned by electron nuclear double resonance (ENDOR) measurements to three histidyl imidazole side chains, a ligand assemblage only possible in the CuB site. Thus, not only is CuB mononuclear, but these data confirm that the structures recapitulate pMMO in the cell and are biologically relevant. The EPR data indicate that CuB is unchanged upon purification, and 17O ENDOR of samples incubated with H217O revealed the presence of an axial Hx17O ligand not resolved in the crystal structures, of which the highest resolution is 2.68 Å.62 The CuB site is therefore best described as a mononuclear Cu(II) site with square pyramidal geometry. These results are corroborated by recent native top-down mass spectrometric (nTDMS) analysis of purified pMMO reconstituted into nanodiscs, which clearly showed the presence of a single copper ion in PmoB.77 Nevertheless, the source of the EXAFS Fourier transform feature that was fit as a Cu-Cu interaction remains unclear and must be reinvestigated in order to fully reconcile all the data.
The CuC site, which is conserved in all pMMOs, is located within the transmembrane domain of the PmoC subunit and includes two histidines and an aspartic acid as ligands (Asp 129, His 133, and His 146 in Mc. sp. Rockwell pMMO62) (Fig. 4). If pMMO is crystallized in zinc-containing buffer or if zinc is soaked into the crystals, zinc is present in this site.59,61,62 In the absence of zinc, the site is occupied by copper,60,62 and the occupancy can be increased by soaking in extra copper.62 An EPR signal observed in purified pMMO samples has been assigned to the CuC site on the basis of Cu-Cu distances obtained from double electron-electron resonance (DEER) measurements.70 The signal is not observed in whole cells, presumably because the CuC site is Cu(I) in vivo. Since the EPR signal assigned to the CuC site overlaps with that of the CuB site, its coordination environment is less readily determined. However, 15N ENDOR is consistent with histidine ligation and 1H and 17O ENDOR indicate the presence of a solvent ligand.70 nTDMS analysis of pMMO in nanodiscs also indicates binding of a single copper ion to the PmoC subunit, and copper supplementation during nanodisc formation increases the copper occupancy of PmoC.77 Fragmentation data localizing the bound copper ion to the crystallographically-observed CuC site could not be obtained, however.
Fig. 4.
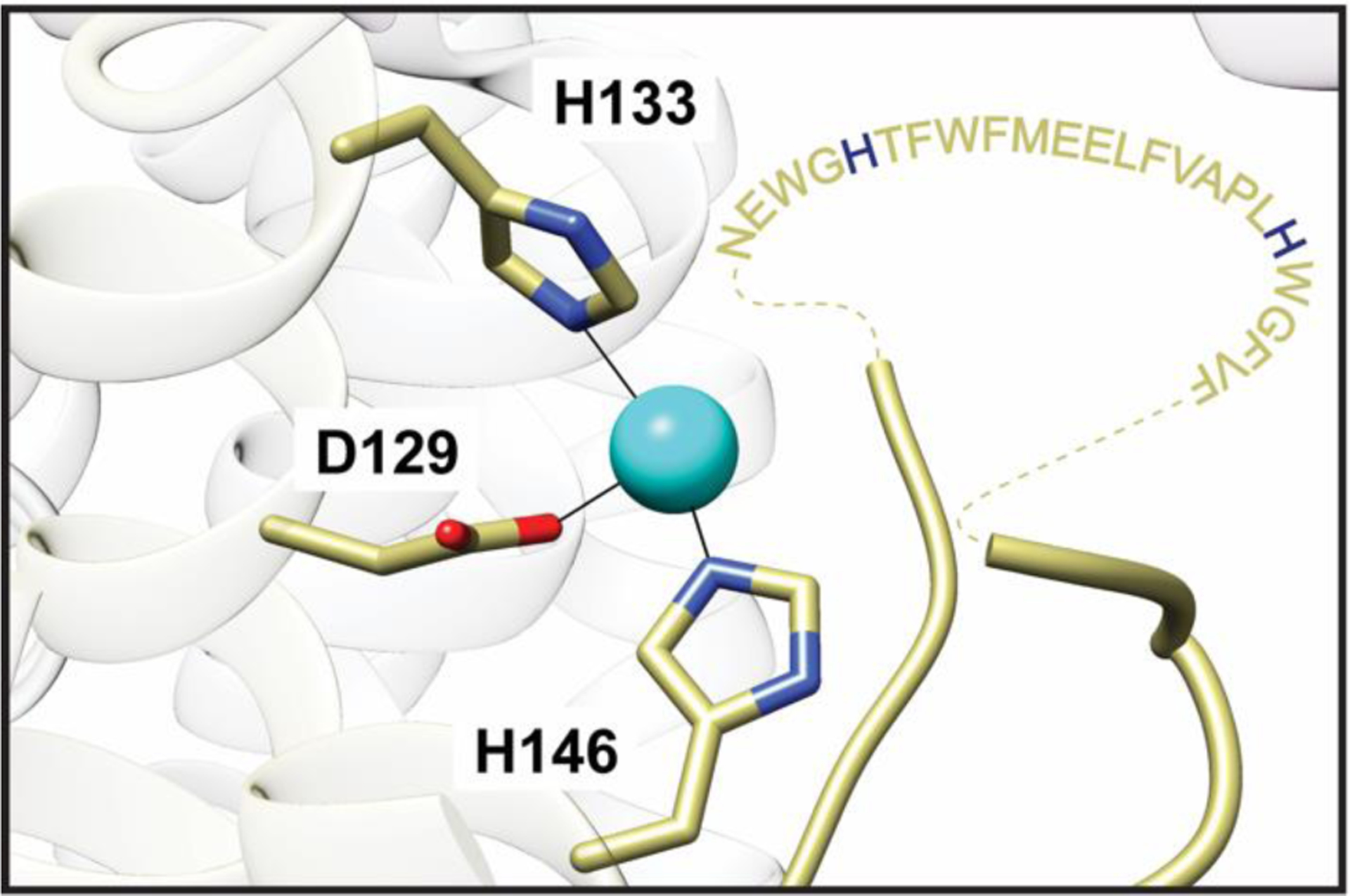
The Mc. sp. Rockwell pMMO CuC site (PDB accession code 4PI0). The unmodeled highly conserved region spanning residues 200–223 is shown as amino acid one-letter codes connected to the model by dashed lines. Potential metal binding residues include two strictly conserved histidines highlighted in blue.
While the advanced paramagnetic resonance analysis is consistent with the crystallographic model for the CuC site, the coordination is not a foregone conclusion. The site directly abuts the unmodeled, highly conserved region of the PmoC subunit (Figs. 1, 4),59–62 and is not even visible in the Mm. alcaliphilum 20Z pMMO structure.63 Weak density suggestive of a fourth ligand has been modeled as a water molecule in the Mc. sp. strain M61 and Mc. sp. Rockwell pMMO structures, while soaking Mc. sp. Rockwell pMMO crystals in zinc led to ordering of 10 additional residues and modeling of a glutamic acid ligand not observed in the structures with copper.62 Given the striking conservation of this region and the presence of potential metal-binding residues, it is imperative to determine its structure. It remains possible that the CuC EPR signal derives from a copper site that has not yet been structurally unveiled.
2.3. Identity of the active site
The location and molecular details of the pMMO active site represent one of the most important outstanding questions in biological methane activation. Methane oxidation assays using samples of pMMO demetallated and inactivated by cyanide treatment indicate that 2–3 copper ions are required for activity. Addition of excess copper inhibits activity, albeit in a reversible fashion.46,62 While a diiron center was once proposed to be the pMMO active site,39 there is no iron in the structures and only copper has been linked to activity. Of the crystallographically-observed copper sites, the bis-His site has been discounted because the coordinating residues are not conserved and the site is only occupied in one crystal structure.59
Instead, the CuB site has been the focus of most discussion. Computational studies have suggested that both a dinuclear78–80 and a mononuclear76 copper site at this location would be capable of methane oxidation. The possibility of a CuB active site was investigated using recombinantly expressed proteins that contain the two PmoB cupredoxin domains, form a CuB site similar to that in native pMMO by EPR,70 and convert methane to methanol.46,70 However, extensive evaluation of these proteins revealed that the methanol is not produced by their CuB-like site, and likely involves adventitious chemistry from the reduction of dioxygen by duroquinol, which is used as a reductant in pMMO activity assays.70 As such, the basis for recently reported enzymatic activity of pMMO mimetics utilizing the PmoB soluble domains tethered to apo ferritin81 is unclear. The argument for a CuB active site is further diminished by the replacement of the three CuB-coordinating histidines in the verrucomicrobial PmoB sequences with methionine, proline, and glycine.29,47,48
In the absence of evidence for CuB being the site of methane activation, attention has turned toward the CuC site. Two recent experimental findings are consistent with a CuC active site. First, nitrite, a known inhibitor of methane oxidation,82,83 perturbs the CuC EPR signal, and 15N ENDOR upon the addition of 15N-nitrite is consistent with NO2− binding to Cu(II) via the oxygen atom(s).70 Second, an increase in the copper occupancy of PmoC detected by nTDMS was correlated with increased pMMO activity.77 These combined data suggest that copper in PmoC, regardless of whether it is located at the crystallographic CuC site, is the site of methane oxidation. In support of this model, mutation of the CuC site in a homologous hydrocarbon monooxygenase completely abrogated activity whereas mutation of the CuB site reduced activity, but maintained affinity for alkane substrate.67,84 Thus, CuB may still play a functional or stabilizing role, accounting for the requirement of more than one copper ion for activity.46,62 Importantly, the CuC ligands along with the neighboring disordered region of PmoC are conserved in all pMMOs, including those from Verrucomicrobia.29,47,48 This observation underscores the potential importance of the CuC site, but characterization of the verrucomicrobial pMMOs, which lack the CuB site ligands, is imperative. Verrucomicrobia grow at high temperatures and low pH, conditions that may affect the structure and metal binding properties of pMMO.
2.4. Protein partners
Another unresolved issue regarding pMMO is the potential existence of larger protein complexes containing pMMO or transient protein-protein interactions between pMMO and partners. Whereas catalysis by sMMO requires interaction of the hydroxylase component (MMOH) with a reductase (MMOR) and a regulatory protein (MMOB) (vide infra),32 pMMO activity can be obtained in vitro by addition of the reductants NADH and duroquinol to membrane-bound and detergent-solubilized samples, respectively.85,86 Duroquinol is believed to mimic a native quinol present in the membranes, presumably ubiquinol, which directly reduces pMMO. NADH is thought to reduce the native quinol via a type 2 NADH:quinone oxidoreductase (NDH-2) that has been observed to copurify with pMMO,38,87,88 although there is no evidence for direct interactions between pMMO and NDH-2. Reduction of pMMO by quinols using this pathway has been referred to as the “redox-arm mode” of electron delivery.89
Electron delivery to pMMO has also been proposed to occur via methanol oxidation to formaldehyde by MDH.90 In this “direct coupling mode,” electrons are funneled to pMMO through the cytochrome c electron acceptor of MDH. Support for both the redox arm and direct coupling models as well as a combination thereof, “uphill electron transfer,” has been obtained from genome-wide metabolic modeling for several methanotrophs.89,91–95 Direct coupling would be facilitated by complexation with MDH, allowing direct transfer of methanol from pMMO to the MDH active site, and a pMMO-MDH complex has been proposed on the basis of low resolution cryoelectron microscopy studies.96 While this complex has not been isolated biochemically, transient interactions between several pMMOs and their cognate MDHs have been detected.97,98 How the MDH dimer (Fig. 5A) interacts with the pMMO trimer is not clear. Resolving the nature of Pmmo’s interaction with electron donor proteins is an important goal for future studies.
Fig. 5.
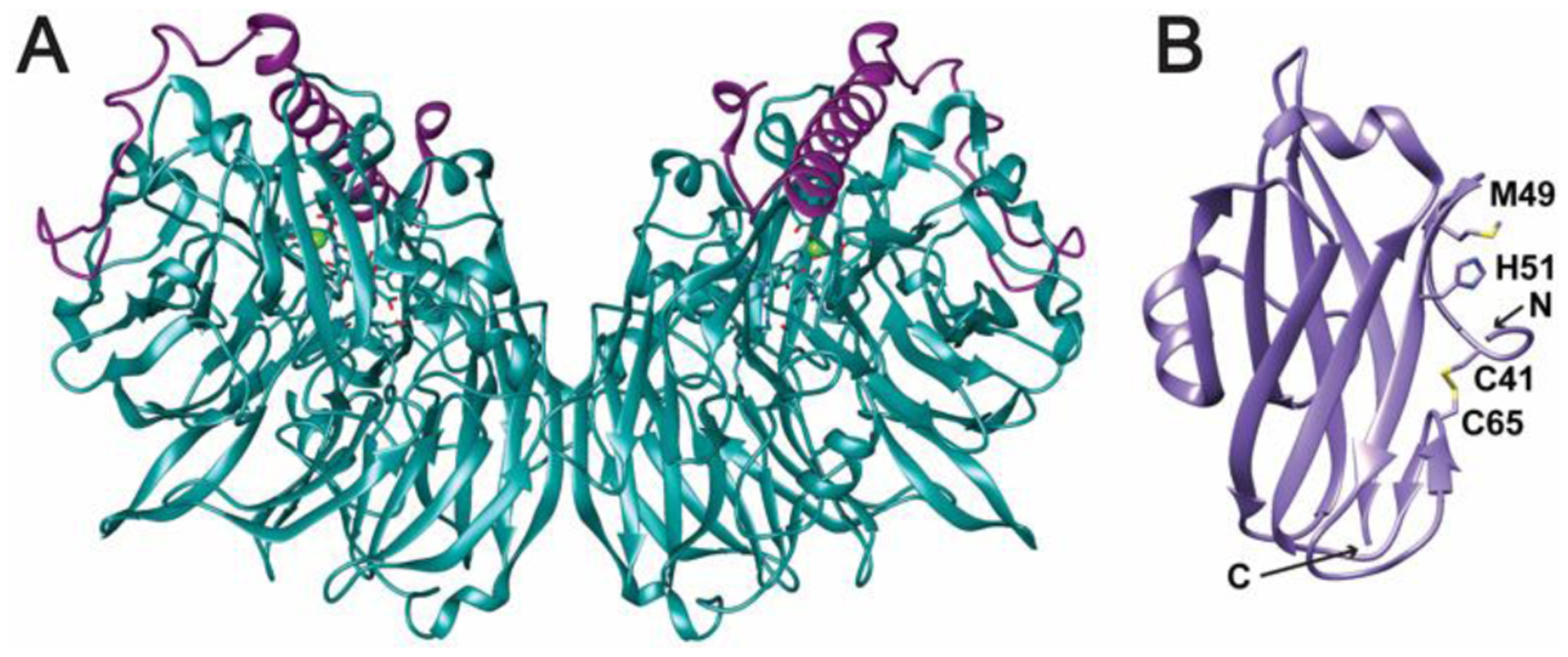
Potential protein interaction partners of pMMO. (A) M. capsulatus (Bath) MDH (PDB accession code 4TQO). The α subunits are shown in teal and the β subunits are shown in violet. The pyrroloquinoline quinone (PQQ)/calcium ion cofactor is shown as sticks and a green sphere. (B) The periplasmic domain of PmoD from Mc. sp. Rockwell (PDB accession code 6CPD). Conserved residues involved in formation of a CuA-like site between two monomers are highlighted. In this structure, the two conserved cysteines form a disulfide bond. A predicted transmembrane helix at the C-terminus is not present in this structure.
Finally, the loss of pMMO activity upon solubilization and purification may be due in part to separation from other protein components necessary for stabilization, copper loading, and delivery of electrons and/or protons. Recent work has suggested that the PmoD protein, found exclusively in methane- and ammonia-oxidizing bacteria and sometimes encoded within the same operon as the pMMO subunits, is one such component.99 Genetic disruption experiments in Methylosinus (Ms.) trichosporium OB3b indicate that PmoD is important for growth under pMMO-utilizing conditions. PmoD consists of a cupredoxin-like periplasmic domain (Fig. 5B) followed by a C-terminal transmembrane helix. In isolation, the periplasmic domain forms an unusual CuA-like site with a symmetric ligand set derived from a PmoD homodimer; this form of the protein has not been crystallographically characterized.100 It remains unclear whether this site forms in vivo in the presence of the transmembrane domain. Direct interactions between PmoD and pMMO have not been established, but the role of this protein in biological methane activation warrants further study.
3. Soluble methane monooxygenase
3.1. Overall architecture
While most methanotrophs rely solely on pMMO for methane oxidation, some have the ability to express sMMO under copper-starvation conditions.11,37,40 sMMO consists of multiple proteins encoded within the mmoXYBZDC operon.101 The mmoX, mmoY, and mmoZ genes encode the α, β, and γ subunits of the hydroxylase component (MMOH), which houses a nonheme diiron active site. The mmoB gene encodes the regulatory protein, MMOB, and mmoC encodes the reductase, MMOR, which reduces the diiron site via NADH and contains FAD and a [2Fe-2S] cluster as cofactors. MMOB increases the reaction rate of MMOH with dioxygen by 1000-fold.102,103 MMOD is not necessary for sMMO function and instead has an inhibitory effect on activity.104,105 Like pMMO, MMOH must be isolated from the native organism for biochemical studies, but MMOB, MMOR, and MMOD can be expressed in E. coli.
sMMO systems from M. capsulatus (Bath) and Ms. trichosporium OB3b have been structurally characterized. MMOH is a dimer comprising the three subunits in an (αβγ)2 arrangement (Fig. 6A).106,107 The diiron active site, housed within a four-helix bundle, is coordinated by two histidine and four carboxylate ligands from the α subunit (Glu 114, His 147 to Fe1, Glu 209, Glu 243, His 246 to Fe2, Glu 144 bridging). In the resting Fe(III)Fe(III) state, the iron ions are bridged by two hydroxo groups (Fig. 7A). Upon reduction to the Fe(II)Fe(II) state, Glu 243 displaces a bridging hydroxo group, bridging the two irons, while coordinating Fe2 in a bidentate fashion (Fig. 7B).108,109 A number of MMOH structures have been determined in the presence of hydrocarbon substrate and product analogs, xenon gas, and different metal ions.31 The MMOB and MMOC structures have been determined by NMR, with the [2Fe-2S] and FAD/NADH domains of MMOC characterized separately.110–113
Fig. 6.
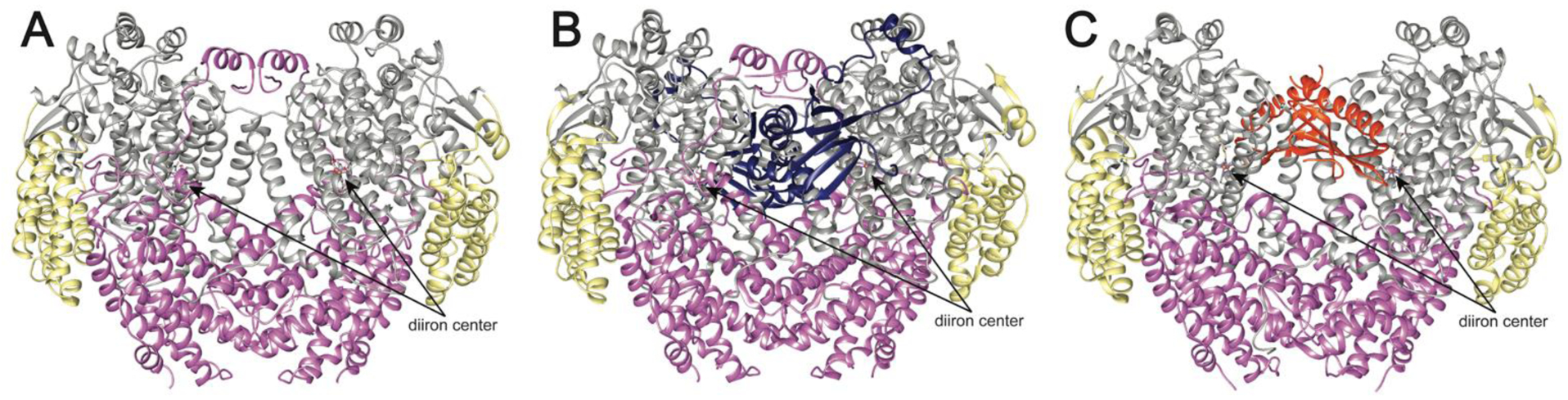
Structures of sMMO. (A) M. capsulatus (Bath) MMOH (PDB accession code 1MTY). The α subunits are shown in gray, the β subunits are shown in pink, and the γ subunits are shown in yellow. The iron ions are shown as orange spheres with the ligands shown as sticks. (B) M. capsulatus (Bath) MMOH-MMOB complex (PDB accession code 4GAM). MMOB is shown in dark blue. (C) M. sporium strain 5 MMOH-MMOD complex (PDB accession code 6D7K). MMOD is shown in orange.
Fig. 7.
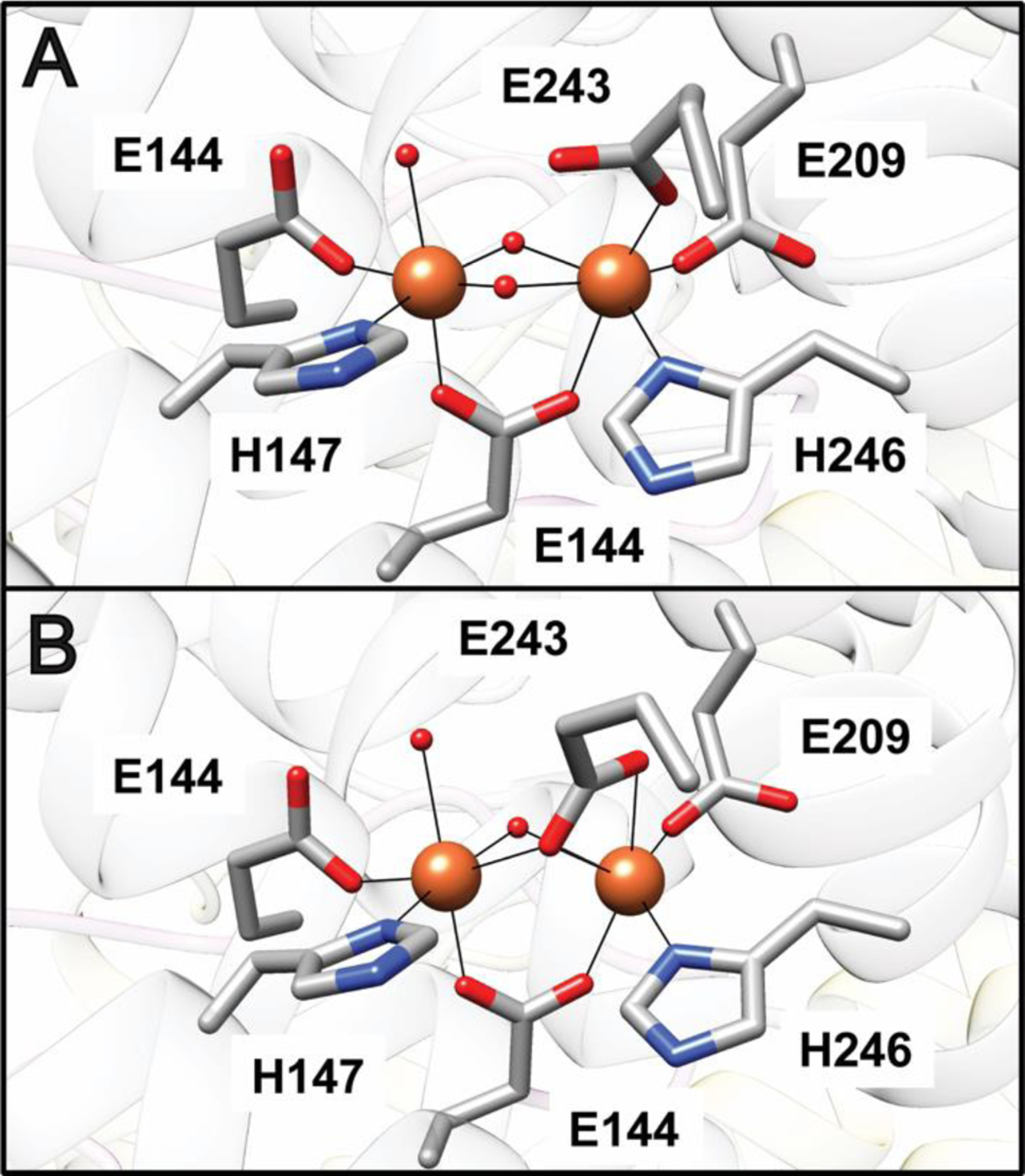
The sMMO active site. (A) The Fe(III)Fe(III) site in M. capsulatus (Bath) MMOHox (PDB accession code 1MTY). (B) The Fe(II)Fe(II) site in M. capsulatus (Bath) MMOHred (PDB accession code 1FYZ). Iron ions are shown as orange spheres and solvent ligands are shown as red spheres.
In addition, crystal structures of MMOH in complex with MMOB109,114 and in complex with MMOD115 have been reported. Both MMOB and MMOD bind symmetrically to MMOH, with one molecule interacting with each αβγ protomer (Figs. 6B,C). Upon binding to MMOH, primarily to the α subunit, MMOB’s flexible N-terminal tail of ~35 residues forms a ring-like structure on the MMOH surface. The C-terminal region of MMOB becomes ordered as well. Several MMOH helices are reorganized, and the resultant side chain rearrangements directly affect the active site.109,114 In the initial M. capsulatus (Bath) MMOH-MMOB structure, Glu 243 shifts to its reduced conformation,114 but this change is not observed in a recent X-ray free electron laser (XFEL) structure of Ms. trichosporium MMOH-MMOB determined at room temperature, suggesting that photoreduction, rather than MMOB binding, caused the shift.109 The binding site for MMOD, as observed in the Ms. sporium strain 5 MMOH-MMOD complex structure, overlaps with that of MMOB. MMOD disrupts the N-terminus of the β subunit, which wraps around the α subunit in each protomer, and affects Fe1, causing dissociation of His 147 and a shift of Glu 114 to bidentate.115
There is no structure of the MMOH-MMOR complex, but the MMOR binding site mapped by hydrogen-deuterium exchange, chemical crosslinking, and computational docking116–119 overlaps with that of MMOB. Overlapping sites are consistent with inhibition of MMOR binding by MMOB,114,120 and suggest a model in which MMOR and MMOB compete for binding to MMOH, although an MMOH-MMOB-MMOR ternary complex has been detected.118 In this model, MMOR binds first and reduces the diiron site, which causes MMOR to lose affinity for MMOH and be replaced by MMOB. Reduced MMOH (MMOHred) has higher affinity for MMOB and binding of MMOB allows formation of reactive intermediates and substrate oxidation. MMOB is then released followed by product egress from the active site. It is possible that MMOB dissociates entirely or remains tethered with its N-terminal ring region to allow the cycle to begin again with MMOR binding.116 However, this model is not compatible with the observation that MMOB decreases the redox potential of MMOH,121,122 necessitating that it bind oxidized MMOH (MMOHox) with a higher affinity than MMOHred. In addition, fluorescence titrations using labeled MMOB indicate a higher affinity for MMOHox than MMOred.123 These discrepancies have been suggested to derive from hysteretic effects on the redox potential.32
3.2. Pores and cavities
While the most intense discussions surrounding pMMO structure have focused on the metal centers and their possible roles in catalysis, the diiron active site of sMMO has been well established for >30 years.124,125 Ongoing debate has instead focused on substrate specificity and access to the active site and the role of MMOB therein. Access to the active site has been considered in the context of three distinct cavities observed in the MMOH crystal structures. The first cavity contains the diiron active site, is ~12 Å from the bulk solvent, and is connected via a pore to the surface of the enzyme. This pore is gated by residues Thr 213, Asn 214, and Glu 240.126 The second cavity is adjacent to the first, but is blocked by residues Phe 188 and Leu 110 in structures of MMOH alone. This cavity is connected to a third large cavity, and all three cavities have been shown to bind halogenated alkanes and xenon gas, suggesting that they form routes for methane, dioxygen, and product access.127,128
Crystal structures of MMOH-MMOB revealed shifts in residues gating the pore and cavities. Upon MMOB binding, new interactions involving MMOH residues Ser 111, Asn 214, Thr 213, and Glu 240 close the pore. In the original M. capsulatus (Bath) MMOH-MMOB structure, shifts in residues Phe 188 and Leu 110 connect the first and second cavities, and were proposed to allow methane and dioxygen to access the reduced diiron site.114 However, kinetic data are not consistent with methane traversing 35–40 Å in cavities prior to reaction at the active site.129,32,130 Moreover, in recent high resolution structures of the Ms. trichosporium OB3b MMOH-MMOB complex, the shift in Phe 188 is only observed upon binding of bulky substrates. The cavities remain separated in MMOHred as well as in the XFEL structures of MMOHox-MMOB and MMOHred-MMOB, suggesting that MMOB may actually serve to close, rather than open, this bottleneck.131 An alternative path, designated the W308-tunnel, has been detected using a probe with solvent radius 1.1 Å instead of the previously used water solvent radius of 1.4 Å. This tunnel, gated by Trp 308 and Pro 215 (Fig. 8), is lined with highly conserved hydrophobic residues, and is open in the MMOHred-MMOB structure. The use of this narrow tunnel is more consistent with the accumulated kinetic data, and variants of MMOB designed to block this tunnel significantly affect the rate of diiron active site oxidation, supporting its role in dioxygen access.131
Fig. 8.
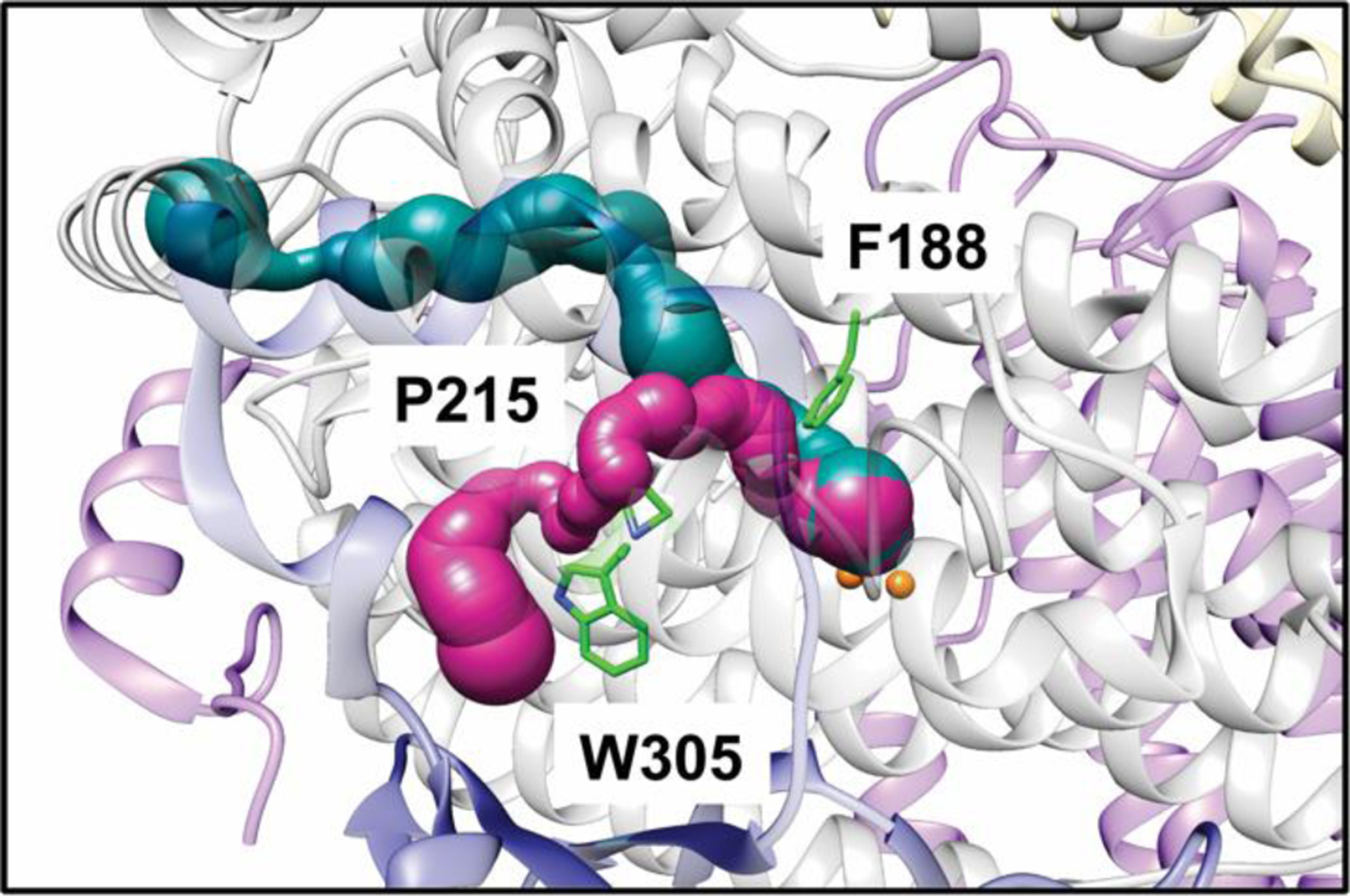
Cavities and tunnels in the Ms. trichosporium OB3b MMOHred-MMOB XFEL structure (PDB accession code 6YDI). The α subunits are shown in gray, the β subunits are shown in pink, the γ subunits are shown in yellow, and MMOB is shown in dark blue. The W308 tunnel is shown in magenta and the previously identified chain of cavities is shown in teal. Key gating residues are highlighted in green sticks.
3.3. Dioxygen and methane activation
In contrast to pMMO, the mechanisms of dioxygen and methane activation by sMMO have been studied in detail. The catalytic cycle of MMOH in the presence of MMOB has been probed extensively by single-turnover kinetics and spectroscopy using chemical reductants, and has been recently reviewed elsewhere.32,132 In brief, reaction of MMOHred with dioxygen yields intermediate O,102,103,133 an Fe(II)Fe(II) species that converts sequentially to intermediates P*, P, and Q.102,134–136 Intermediate P* is also an Fe(II)Fe(II) species,136 while P is an Fe(III)Fe(III) peroxo species,134,135 the exact structure of which remains unclear.32 As the species that reacts with methane, intermediate Q has been the subject of much discussion. Two antiferromagnetically coupled high-spin Fe(IV) ions are present,137–139 but the geometry remains under debate, with evidence supporting both diamond and open core geometries.139–143 Once Q reacts with methane, intermediate T is formed, followed by methanol release and return to MMOHox.139
The mechanism of methane activation by Q has also been investigated extensively, both computationally144,145 and experimentally. Experimental approaches have leveraged sMMO’s wide substrate range (cyclic hydrocarbons and cubanes, linear alkanes C1–C8)146–148 to assess reactions with chiral149,150 and radical clock151–154 substrates, which provide insight into the possible existence of radical intermediates. These studies are consistent with a hydrogen abstraction mechanism,32 but are complicated by the use of nonnatural substrates. Notably, studies using deuterated methane revealed a remarkable kinetic isotope effect (KIE) of 50, which is not observed with other substrates155,156 and is suggestive of hydrogen tunneling. Conformational changes upon MMOB binding have been proposed to selectively guide methane to the active site in such a way as to optimize tunneling.32
4. Protein engineering
Many of the unresolved questions surrounding both MMOs could be addressed by site-directed mutagenesis. For pMMO, mutation of the residues coordinating the crystallographically-observed metal centers would go a long way toward establishing functional relevance as would alteration of the highly conserved part of PmoC that is not observed crystallographically. Substrate specificity and mechanistic studies would also be facilitated. For sMMO, the ability to heterologously generate MMOB variants has helped detect intermediates129 and has provided insight into control of substrate access to intermediate Q,129,130,156 but the roles of the different cavities and channels can only be probed in a systematic fashion by using MMOH variants. Moreover, commercially viable biological methane activation may require engineering of either MMO to have increased methane oxidation rates,8 which would be more feasible in a heterologous expression system.
However, attempts at heterologous expression of MMOH or pMMO have been largely unsuccessful. Recombinant versions of the PmoB subunit recapitulate some of the spectroscopic properties of pMMO, but are unstable in the absence of fusion proteins and are not catalytically active.46,70 The N-terminal cupredoxin domain of the homologous AmoB subunit from Nitrosocaldus yellowstonii, which lacks the C-terminal cupredoxin domain, has been recombinantly expressed and crystallographically characterized, but does not exhibit methane oxidation activity.68 Expression of intact pMMO has been reported in Rhodococcus erythropolis LSSE8–1, albeit with whole cell activity 340 times less than that of Ms. trichosporium OB3b.8,45 Besides likely issues involving insertion of the pMMO subunits into the membrane and proper trimer assembly, an obstacle specific to expression of functional pMMO may be the lack of ICMs in heterologous hosts. In most methanotrophs, pMMO is localized to these intracellular structures,17,37,40,157 which could play a role in structuring the enzyme or concentrating methane.8 The presence of ICMs and the location of pMMO in the verrucomicrobial methanotrophs remain unclear, however.22
While enzymes related to MMOH such as toluene and phenol hydroxylases have been expressed in E. coli,158,159 the only reported heterologous expression of MMOH is in Pseudomonas, although methane oxidation by these strains has not been reported.43 Neither this system nor the pMMO Rhodococcus erythropolis LSSE8–1 expression system45 resulted in the ability to generate variants or isolate recombinant MMOs for further study. As an alternative approach, Ms. trichosporium OB3b MMOH genes with mutations of interest have been reintroduced into a Ms. trichosporium OB3b strain with the smmo operon partially deleted. This strain can be grown using pMMO, with expression of the variant MMOHs occurring as copper levels decline.160,161 Several variants have been studied using this method, primarily focusing on residues in the α subunit surrounding the diiron center.161–163 However, this system requires optimization to be practical for generation of large numbers of variants for protein purification and characterization.
An analogous strategy for pMMO is more complicated because most methanotroph genomes contains multiple copies of the pmo operon. Genetic tools have been developed for the methanotroph Methylotuvimicrobium buryatense 5GB1C (formerly Methylomicrobium buryatense 5GB1C164),165,166 which has been used for metabolic engineering16 and only contains one copy of the pMMO genes. In principle, it should be possible to obtain point mutants of pMMO using these tools and growing under sMMO-producing conditions to mitigate any growth defects from an impaired pMMO. However, such attempts have not been successful, perhaps because for unknown reasons, pMMO remains important for cell growth under sMMO-producing conditions. This notion is consistent with the observation that pMMO is still expressed under conditions of copper starvation and only mildly upregulated when copper is available.42 CRISPR-Cas9, which has been developed for methanotrophs, presents another potential route to variants, but the current method has very low editing efficiency and has not yet been demonstrated to be effective for this purpose.167
Conclusions
The unique lifestyle of methanotrophs presents intriguing possibilities for methane utilization and bioremediation. Their wide variety of habitats highlights the effectiveness of the MMO-based carbon assimilation strategy. For pMMO, progress has been made in understanding the nuclearity of the CuB site and the importance of the CuC site. Further studies of pMMO in a lipid environment and of the verrucomicrobial pMMOs promise to shed light on the active site location and structure, bringing the field closer to determining the reaction mechanism. Beyond the pMMO trimer itself, the possibilities of PmoD and MDH as interaction partners represent an important future direction. For sMMO, new structures of MMOH complexes, some determined using an XFEL, have revealed in detail the effects of MMOB and MMOD on MMOH structure and have delineated new pathways for substrate access to the diiron active site. The geometry of intermediate Q, the relative importance of different cavities in the MMOH structures, and the sequence of events involving MMOB and MMOR binding require additional studies. Studies of both MMOs await development of effective heterologous expression systems, which could be used to generate variants and as a platform for protein engineering. The combination of detailed chemical understanding and protein engineering could finally realize the potential of methanotrophs to add value14 and even help save the planet.
Acknowledgements
The pMMO and PmoD projects in the Rosenzweig laboratory are supported by National Institutes of Health Grant R35 GM118035 and United States Department of Energy Grant DE-SC0016284, respectively.
Biographies

Christopher W. Koo received a BS in biochemistry from the University of California, Los Angeles. He is currently a PhD candidate in the Interdisciplinary Biological Sciences program (IBiS) at Northwestern University. In the Rosenzweig lab, his thesis work is focused on the structure and function of particulate methane monooxygenase in lipid nanodiscs using cryo-electron microscopy and cell-free protein synthesis. He is a former trainee of the NIH-funded Molecular Biosciences Training Program.

Amy C. Rosenzweig received a BA in chemistry from Amherst College and a PhD in inorganic chemistry from Massachusetts Institute of Technology. After postdoctoral research at Harvard Medical School, she joined the faculty of Northwestern University where she is the Weinberg Family Distinguished Professor of Life Sciences. She is a fellow of the American Academy of Arts and Sciences and a member of the National Academy of Sciences. The Rosenzweig laboratory uses structural, biochemical, and biophysical approaches to attack problems at the forefront of bioinorganic chemistry. Their work has been honored recently by the Royal Society of Chemistry Joseph Chatt Award and the American Chemical Society Alfred Bader Award in Bioinorganic or Bioorganic Chemistry.
Footnotes
Conflicts of interest
There are no conflicts to declare.
References
- 1.Fletcher SEM and Schaefer H, Science, 2019, 364, 932–933. [DOI] [PubMed] [Google Scholar]
- 2.Saunois M, Jackson RB, Bosquet P, Poulter B and Canadell JG, Environ. Res. Lett, 2016, 11, 120207. [Google Scholar]
- 3.Myhre G, Shindell D, Bréon F-M, Collins W, Fuglestvedt J, Huang J, Koch D, Lamarque J-F, Lee D, Mendoza B, Nakajima T, Robock A, Stephens G, Takemura T and Zhang H, in Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, ed. Stocker TF, Qin D, Plattner G-K, Tignor M, Allen SK, Boschung J, Nauels A, Xia Y, Bex V and Midgley PM Cambridge University Press, 2013. [Google Scholar]
- 4.Haynes CA and Gonzalez R, Nat. Chem. Biol, 2014, 10, 331–339. [DOI] [PubMed] [Google Scholar]
- 5.Blanksby SJ and Ellison GB, Acc Chem Res, 2003, 36, 255–263. [DOI] [PubMed] [Google Scholar]
- 6.Wood DA, Nwaoha C and Towler BF, J. Nat. Gas Sci. Eng, 2012, 9, 196–208. [Google Scholar]
- 7.Ge XM, Yang LC, Sheets JP, Yu ZT and Li YB, Biotech. Adv, 2014, 32, 1460–1475. [DOI] [PubMed] [Google Scholar]
- 8.Lawton TJ and Rosenzweig AC, Curr. Opin. Chem. Biol, 2016, 35, 142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clomburg JM, Crumbley AM and Gonzalez R, Science, 2017, 355. [DOI] [PubMed] [Google Scholar]
- 10.Hanson RS and Hanson TE, Microbiol. Rev, 1996, 60, 439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Semrau JD, Dispirito AA and Yoon S, FEMS Microbiol. Lett, 2010, 34, 496–531. [DOI] [PubMed] [Google Scholar]
- 12.Kwon M, Ho A and Yoon S, Appl. Microbiol. Biotechnol, 2019, 103, 1–8. [DOI] [PubMed] [Google Scholar]
- 13.Henard CA, Franklin TG, Youhenna B, But S, Alexander D, Kalyuzhnaya MG and Guarnieri MT, Front. Microbiol, 2018, 9, 2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strong PJ, Xie S and Clarke WP, Environ. Sci. Technol, 2015, 49, 4001–4018. [DOI] [PubMed] [Google Scholar]
- 15.Bowman JP, Sly LI and Stackebrandt E, Int. J. Syst. Bacteriol, 1995, 45, 182–185. [DOI] [PubMed] [Google Scholar]
- 16.Kalyuzhnaya MG, Puri AW and Lidstrom ME, Metab. Eng, 2015, 29, 142–152. [DOI] [PubMed] [Google Scholar]
- 17.Davies SL and Whittenbury R, J. Gen. Microbiol, 1970, 61, 227–232. [DOI] [PubMed] [Google Scholar]
- 18.Makula RA, J. Bacteriol, 1978, 134, 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knief C, Front. Microbiol, 2015, 6, 1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Op den Camp HJM, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, Jetten MSM, Birkeland N-K, Pol A and Dunfield PF, Environ. Microbiol. Rep, 2009, 1, 293–306. [DOI] [PubMed] [Google Scholar]
- 21.Khadem AF, Pol A, Wieczorek A, Mohammadi SS, Francoijs KJ, Stunnenberg HG, Jetten MS and Op den Camp HJM, J. Bacteriol, 2011, 193, 4438–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Teeseling MC, Pol A, Harhangi HR, van der Zwart S, Jetten MS, Op den Camp HJM and van Niftrik L, Appl. Environ. Microbiol, 2014, 80, 6782–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowman JP, McCammon SA and Skerratt JH, Microbiology, 1997, 143, 1451–1459. [DOI] [PubMed] [Google Scholar]
- 24.Kalyuzhnaya MG, Kmelenina VN, Starostina NG, Baranova SB, Suzina NE and Trotsenko YA, Mikrobiologiia, 1998, 67, 438–444. [Google Scholar]
- 25.Khmelenina VN, Kalyuzhnaya MG, Starostina NG, Suzina NE and Trotsenko YA, Curr. Microbiol, 1997, 35, 257–261. [Google Scholar]
- 26.Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MSM and Op den Camp HJM, Nature, 2007, 450, 874–878. [DOI] [PubMed] [Google Scholar]
- 27.Hou SB, Makarova KS, Saw JHW, Senin P, Ly BV, Zhou ZM, Ren Y, Wang JM, Galperin MY, Omelchenko MV, Wolf Y, Yutin N, Koonin EV, Stott MB, Mountain BW, Crowe MA, Smirnova AV, Dunfield PF, Feng L, Wang L and Alam M, Biol. Direct, 2008, 3, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, Hou S, Ly B, Saw JH, Zhou Z, Ren Y, Wang J, Mountain BW, Crowe MA, Weatherby TM, Bodelier PLE, Liesack W, Feng L, Wang L and Alam M, Nature, 2007, 450, 879–882. [DOI] [PubMed] [Google Scholar]
- 29.Islam T, Jensen S, Reigstad LJ, Larsen O and Birkeland NK, Proc. Natl. Acad. Sci. USA, 2008, 105, 300–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross MO and Rosenzweig AC, J. Biol. Inorg. Chem, 2017, 22, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sirajuddin S and Rosenzweig AC, Biochemistry, 2015, 54, 2283–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banerjee R, Jones JC and Lipscomb JD, Annu. Rev. Biochem, 2019, 88, 409–431. [DOI] [PubMed] [Google Scholar]
- 33.Dunfield PF, Khmelenina VN, Suzina NE, Trotsenko YA and Dedysh SN, Int. J. Syst. Evol. Microbiol, 2003, 53, 1231–1239. [DOI] [PubMed] [Google Scholar]
- 34.Dedysh SN, Knief C and Dunfield PF, J. Bacteriol, 2005, 187, 4665–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vorobev AV, Baani M, Doronina NV, Brady AL, Liesack W, Dunfield PF and Dedysh SN, Int. J. Syst. Evol. Microbiol, 2011, 61, 2456–2463. [DOI] [PubMed] [Google Scholar]
- 36.Whittenbury R, Phillips KC and Wilkinson JF, J. Gen. Microbiol, 1970, 61, 205–218. [DOI] [PubMed] [Google Scholar]
- 37.Prior SD and Dalton H, J. Gen. Microbiol, 1985, 131, 155–163. [Google Scholar]
- 38.Choi DW, Kunz RC, Boyd ES, Semrau JD, Antholine WE, Han JI, Zahn JA, Boyd JM, de la Mora AM and DiSpirito AA, J. Bacteriol, 2003, 185, 5755–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinho M, Choi DW, DiSpirito AA, Antholine WE, Semrau JD and Munck E, J. Am. Chem. Soc, 2007, 129, 15783–15785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanley SH, Prior SD, Leak DJ and Dalton H, Biotechnol. Lett, 1983, 5, 487–492. [Google Scholar]
- 41.Nielsen AK, Gerdes K and Murrell JC, Mol. Microbiol, 1997, 25, 399–409. [DOI] [PubMed] [Google Scholar]
- 42.Kenney GE, Sadek M and Rosenzweig AC, Metallomics, 2016, 8, 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jahng D and Wood TK, Appl. Environ. Microbiol, 1994, 60, 2473–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murrell JC, Gilbert B and McDonald IR, Arch. Microbiol, 2000, 173, 325–332. [DOI] [PubMed] [Google Scholar]
- 45.Gou Z, Xing X-H, Luo M, Jiang H, Han B, Wu H, Wang L and Zhang MS Microbiol. Lett, 2006, 263, 136–141. [DOI] [PubMed] [Google Scholar]
- 46.Balasubramanian R, Smith SM, Rawat S, Stemmler TL and Rosenzweig C, Nature, 2010, 465, 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kruse T, Ratnadevi CM, Erikstad HA and Birkeland NK, BMC Genomics, 2019, 20, 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anvar SY, Frank J, Pol A, Schmitz A, Kraaijeveld K, den Dunnen JT and Op den Camp HJ, BMC Genomics, 2014, 15, 914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.del Cerro C, Garcia JM, Rojas A, Tortajada M, Ramón D, Galán B, Prieto MA and Garcia JL, J. Bacteriol, 2012, 194, 5709–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vuilleumier S, Khmelenina VN, Bringel F, Reshetnikov AS, Lajus S Mangenot, Rouy Z, Op den Camp HJM, Jetten MSM, Dispirito AA, Dunfield P, Klotz MG, Semrau JD, Stein LY, Barbe V, Medigue C, Trotsenko YA and Kalyuzhnaya MG, J. Bacteriol, 2012, 194, 551–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stein LY, Bringel F, DiSpirito AA, Han S, Jetten MSM, Kalyuzhnaya M, Kits KD, Klotz MG, den Camp H, Semrau JD, Vuilleumier S, Bruce DC, Cheng JF, Davenport KW, Goodwin L, Han SS, Hauser L, Lajus A, Land ML, Lapidus A, Lucas S, Medigue C, Pitluck S and Woyke T, J. Bacteriol, 2011, 193, 2668–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svenning MM, Hestnes AG, Wartiainen I, Stein LY, Klotz MG, Kalyuzhnaya MG, Spang A, Bringel F, Vuilleumier S, Lajus A, Medigue C, Bruce DC, Cheng JF, Goodwin L, Ivanova N, Han J, Han CS, Hauser L, Held B, Land ML, Lapidus A, Lucas S, Nolan M, Pitluck S and Woyke T, J. Bacteriol, 2011, 193, 6418–6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y, Crombie A, Rahman MT, Dedysh SN, Liesack W, Stott M, Alam M, Theisen AR, Murrell JC and Dunfield PF, J. Bacteriol, 2010, 192, 3840–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stein LY, Yoon S, Semrau JD, DiSpirito AA, Crombie A, Murrell JC, Vuilleumier S, Kalyuzhnaya MG, Op den Camp HJ, Bringel D Bruce, Cheng JF, Copeland A, Goodwin L, Han S, Hauser L, Jetten MS, Lajus A, Land ML, Lapidus A, Lucas S, Médigue, Pitluck S, Woyke T, Zeytun A and Klotz MG, J. Bacteriol, 2010, 192, 6497–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward N, Larsen O, Sakwa J, Bruseth L, Khouri H, Durkin AS, Dimitrov L Jiang, Scanlan D, Kang KH, Lewis M, Nelson KE, Metheacute B, Wu M, Heidelberg JF, Paulsen IT, Fouts D, Ravel J, Tettelin H, Ren Q, Read T, DeBoy RT, Seshadri R, Salzberg SL, Jensen HB, Birkeland NK, Nelson WC, Dodson RJ, Grindhaug SH, Holt I, Eidhammer I, Jonason I, Vanaken S, Utterback T, Feldblyum TV, Fraser CM, Lillehaug JR and Eisen JA, PLoS Biol., 2004, 2, e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stolyar S, Costello AM, Peeples TL and Lidstrom ME, Microbiol., 1999, 145, 1235–1244. [DOI] [PubMed] [Google Scholar]
- 57.Semrau JD, Chistoserdov A, Lebron J, Costello A, Davagnino J, Kenna E, Holmes AJ, Finch R, Murrell JC and Lidstrom ME, J. Bacteriol, 1995, 177, 3071–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McDonald IR and Murrell JC, FEMS Microbiol. Lett, 1997, 156, 205–210. [DOI] [PubMed] [Google Scholar]
- 59.Lieberman RL and Rosenzweig AC, Nature, 2005, 434, 177–182. [DOI] [PubMed] [Google Scholar]
- 60.Hakemian AS, Kondapalli KC, Telser J, Hoffman BM, Stemmler TL and Rosenzweig AC, Biochemistry, 2008, 47, 6793–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith SM, Rawat S, Telser J, Hoffman BM, Stemmler TL and Rosenzweig AC, Biochemistry, 2011, 50, 10231–10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sirajuddin S, Barupala D, Helling S, Marcus K, Stemmler TL and Rosenzweig AC, J. Biol. Chem, 2014, 289, 21782–21794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ro SY, Ross MO, Deng YW, Batelu S, Lawton TJ, Hurley JD, Stemmler TL, Hoffman BM and Rosenzweig AC, J. Biol. Chem, 2018, 293, 10457–10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu J, King MS, Yu M, Klipcan L, Leslie AG and Hirst J, Proc. Natl. Acad. Sci. USA, 2015, 112, 12087–12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater R and Whalen KL, Biochim. Biophys. Acta, 2015, 1854, 1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arp DJ, Sayavedra-Soto LA and Hommes NG, Arch. Microbiol, 2002, 178, 250–255. [DOI] [PubMed] [Google Scholar]
- 67.Liew EF, Tong DC, Coleman NV and Holmes AJ, Microbiology, 2014, 160, 1267–1277. [DOI] [PubMed] [Google Scholar]
- 68.Lawton TJ, Ham J, Sun T and Rosenzweig AC, Proteins, 2014, 82, 2263–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lemos SS, Collins MLP, Eaton SS, Eaton GR and Antholine WE, Biophys. J, 2000, 79, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ross MO, MacMillan F, Wang J, Nisthal A, Lawton TJ, Olafson BD, Mayo SL, Rosenzweig AC and Hoffman BM, Science, 2019, 364, 566–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koo CW and Rosenzweig AC, in Encylopedia of Inorganic and Bioinorganic Chemistry, John Wiley & Sons, Ltd., 2020, vol. DOI: 10.1002/9781119951438.eibc2740. [DOI] [Google Scholar]
- 72.Ciano L, Davies GJ, Tolman WB and Walton PH, Nat. Catal, 2018, 1, 571–577. [Google Scholar]
- 73.Walton PH and Davies GJ, Curr. Opin. Chem. Biol, 2016, 31, 195–207. [DOI] [PubMed] [Google Scholar]
- 74.Lieberman RL, Shrestha DB, Doan PE, Hoffman BM, Stemmler TL and Rosenzweig AC, Proc. Natl. Acad. Sci. USA, 2003, 100, 3820–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lieberman RL, Kondapalli KC, Shrestha DB, Hakemian AS, Smith SM, Telser J, Kuzelka J, Gupta R, Borovik AS, Lippard SJ, Hoffman BM, Rosenzweig AC and Stemmler TL, Inorg. Chem, 2006, 45, 8372–8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao LL, Caldararu O, Rosenzweig AC and Ryde U, Angew. Chem. Int. Ed, 2018, 57, 162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ro SY, Schachner LF, Koo CW, Purohit R, Remis JP, Kenney GE, Liauw BW, Thomas PM, Patrie SM, Kelleher NL and Rosenzweig AC, Nat. Commun, 2019, 10, 2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shiota Y, Juhasz G and Yoshizawa K, Inorg. Chem, 2013, 52, 7907–7917. [DOI] [PubMed] [Google Scholar]
- 79.Shiota Y and Yoshizawa K, Inorg. Chem, 2009, 48, 838–845. [DOI] [PubMed] [Google Scholar]
- 80.Yoshizawa K and Shiota Y, J. Am. Chem. Soc, 2006, 128, 9873–9881. [DOI] [PubMed] [Google Scholar]
- 81.Kim HJ, Huh J, Kwon YW, Park D, Yu Y, Jang YE, Lee BR, Jo, Lee EJ, Heo Y, Lee W and Lee J, Nat. Catal, 2019, 2, 342–353. [Google Scholar]
- 82.Nyerges G and Stein LY, FEMS Microbiol. Lett, 2009, 297, 131–136. [DOI] [PubMed] [Google Scholar]
- 83.Stein LY and Arp DJ, Appl. Environ. Microbiol, 1998, 64, 4098–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coleman NV, Le NB, Ly MA, Ogawa HE, McCarl V, Wilson NL and Holmes AJ, Isme J., 2012, 6, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith DD and Dalton H, Eur. J. Biochem, 1989, 182, 667–671. [DOI] [PubMed] [Google Scholar]
- 86.Shiemke AK, Cook SA and Miley T, J. Inorg. Biochem, 1995, 59, 385. [DOI] [PubMed] [Google Scholar]
- 87.Heinrich H and Werner S, Biochemistry, 1992, 31, 11413–11419. [DOI] [PubMed] [Google Scholar]
- 88.Cook SA and Shiemke AK, Arch. Biochem. Biophys, 2002, 398, 32–40. [DOI] [PubMed] [Google Scholar]
- 89.de la Torre A, Metivier A, Chu F, Laurens LML, Beck DAC, Pienkos PT, Lidstrom ME and Kalyuzhnaya MG, Microb. Cell Fact, 2015, 14, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leak DJ and Dalton H, Appl. Microbiol. Biotechnol, 1986, 23, 477–481. [Google Scholar]
- 91.Lieven C, Petersen LAH, Jorgensen SB, Gernaey KV, Herrgard MJ and Sonnenschein N, Front. Microbiol, 2018, 9, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bordel S, Rodriguez Y, Hakobyan A, Rodriguez E, Lebrero R and Munoz R, Metab. Eng, 2019, 54, 191–199. [DOI] [PubMed] [Google Scholar]
- 93.Bordel S, Rojas A and Munoz R, Microb. Cell Fact, 2019, 18, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nguyen AD, Park JY, Hwang IY, Hamilton R, Kalyuzhnaya MG, Kim D and Lee EY, Metab. Eng, 2020, 57, 1–12. [DOI] [PubMed] [Google Scholar]
- 95.Naizabekov S and Lee EY, Microorganisms, 2020, 8, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Myronova N, Kitmitto A, Collins RF, Miyaji A and Dalton H, Biochemistry, 2006, 45, 11905–11914. [DOI] [PubMed] [Google Scholar]
- 97.Culpepper MA and Rosenzweig AC, Biochemistry, 2014, 53, 6211–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Deng YW, Ro SY and Rosenzweig AC, J. Biol. Inorg. Chem, 2018, 23, 1037–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fisher OS, Kenney GE, Ross MO, Ro SY, Lemma BE, Batelu S, Thomas PM, Sosnowski VC, DeHart CJ, Kelleher NL, Stemmler TL, Hoffman BM and Rosenzweig AC, Nat. Commun, 2018, 9, 4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ross MO, Fisher OS, Morgada MN, Krzyaniak MD, Wasielewski MR, Vila AJ, Hoffman BM and Rosenzweig AC, J. Am. Chem. Soc, 2019, 141, 4678–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Murrell JC, Biodeg., 1994, 5, 145–159. [DOI] [PubMed] [Google Scholar]
- 102.Lee S-K, Nesheim JC and Lipscomb JD, J. Biol. Chem, 1993, 268, 21569–21577. [PubMed] [Google Scholar]
- 103.Liu Y, Nesheim JC, Lee SK and Lipscomb JD, J. Biol. Chem, 1995, 270, 24662–24665. [DOI] [PubMed] [Google Scholar]
- 104.Merkx M and Lippard SJ, J. Biol. Chem, 2002, 277, 5858–5865. [DOI] [PubMed] [Google Scholar]
- 105.Sazinsky MH, Merkx M, Cadieux E, Tang SY and Lippard SJ, Biochemistry, 2004, 43, 16263–16276. [DOI] [PubMed] [Google Scholar]
- 106.Rosenzweig AC, Frederick CA, Lippard SJ and Nordlund P, Nature, 1993, 366, 537–543. [DOI] [PubMed] [Google Scholar]
- 107.Elango N, Radhakrishnan R, Froland WA, Wallar BJ, Earhart CA, Lipscomb JD and Ohlendorf DH, Protein Sci., 1997, 6, 556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rosenzweig AC, Nordlund P, Takahara PM, Frederick CA and Lippard SJ, Chem. Biol, 1995, 2, 409–418. [PubMed] [Google Scholar]
- 109.Srinivas V, Banerjee R, Lebrette H, Jones JC, Aurelius O, Kim IS, Pham CC, Gul S, Sutherlin K, Bhowmick A, John J, Bozkurt E, Fransson T, Aller P, Butryn A, Bogacz I, Simon PS, Keable S, Britz A, Tono K, Kim KS, Park SY, Lee SJ, Park J, Alonso-Mori R, Fuller F, Batyuk A, Brewster A, Bergmann U, Sauter N, Orville AM, Yachandra VK, Yano J, Lipscomb JD, Kern JF and Hogbom M, J. Am. Chem. Soc, 2020, 142, 14249–14266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Walters KJ, Gassner GT, Lippard SJ and Wagner G, Proc. Natl. Acad. Sci. USA, 1999, 96, 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chang SL, Wallar BJ, Lipscomb JD and Mayo KH, Biochemistry, 1999, 38, 5799–5812. [DOI] [PubMed] [Google Scholar]
- 112.Muller J, Lugovskoy AA, Wagner G and Lippard SJ, Biochemistry, 2002, 41, 42–51. [DOI] [PubMed] [Google Scholar]
- 113.Chatwood LL, Muller J, Gross JD, Wagner G and Lippard SJ, Biochemistry, 2004, 43, 11983–11991. [DOI] [PubMed] [Google Scholar]
- 114.Lee SJ, McCormick MS, Lippard SJ and Cho U-S, Nature, 494, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim H, An S, Park YR, Jang H, Yoo H, Park SH, Lee SJ and Cho US, Sci Adv, 2019, 5, eaax0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang WX, Iacob RE, Luoh RP, Engen JR and Lippard SJ, J. Am. Chem. Soc, 2014, 136, 9754–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kopp DA, Berg EA, Costello CE and Lippard SJ, J. Biol. Chem, 2003, 278, 20939–20945. [DOI] [PubMed] [Google Scholar]
- 118.Fox BG, Liu Y, Dege JE and Lipscomb JD, J. Biol. Chem, 1991, 266, 540–550. [PubMed] [Google Scholar]
- 119.Liu Y, Nesheim JC, Paulsen KE, Stankovich MT and Lipscomb JD, Biochemistry, 1997, 36, 5223–5233. [DOI] [PubMed] [Google Scholar]
- 120.Acheson JF, Bailey LJ, Elsen NL and Fox BG, Nat Commun, 5, 5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu KE and Lippard SJ, J. Biol. Chem, 1991, 266, 12836–12839, 24859. [PubMed] [Google Scholar]
- 122.Paulsen KE, Liu Y, Fox BG, Lipscomb JD, Munck E and Stankovich MT, Biochemistry, 1994, 33, 713–722. [DOI] [PubMed] [Google Scholar]
- 123.Brazeau BJ, Wallar BJ and Lipscomb JD, Biochem. Biophys. Res. Commun, 2003, 312, 143–148. [DOI] [PubMed] [Google Scholar]
- 124.Ericson A, Hedman B, Hodgson KO, Green J, Dalton H, Bentsen JG, Beer RH and Lippard SJ, J. Am. Chem. Soc, 1988, 110, 2330. [Google Scholar]
- 125.Fox BG, Surerus KK, Munck E and Lipscomb JD, J. Biol. Chem, 1988, 263, 10553–10556. [PubMed] [Google Scholar]
- 126.Wang W, Liang AD and Lippard SJ, Acc. Chem. Res, 2015, 48, 2632–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Whittington DA, Rosenzweig AC, Frederick CA and Lippard SJ, Biochemistry, 2001, 40, 3476–3482. [DOI] [PubMed] [Google Scholar]
- 128.Sazinsky MH and Lippard SJ, J. Am. Chem. Soc, 2005, 127, 5814–5825. [DOI] [PubMed] [Google Scholar]
- 129.Wallar BJ and Lipscomb JD, Biochemistry, 2001, 40, 2220–2233. [DOI] [PubMed] [Google Scholar]
- 130.Brazeau BJ and Lipscomb JD, Biochemistry, 2003, 42, 5618–5631. [DOI] [PubMed] [Google Scholar]
- 131.Jones JC, Banerjee R, Shi K, Aihara H and Lipscomb JD, Biochemistry, 2020, 59, 2946–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jasniewski AJ and Que L Jr., Chem. Rev, 2018, 118, 2554–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu KE, Valentine AM, Qiu D, Edmondson DE, Appelman EH, Spiro TG and Lippard SJ, J. Am. Chem. Soc, 1995, 117, 4987–4990. [Google Scholar]
- 134.Lee S-K and Lipscomb JD, Biochemistry, 1999, 38, 4423–4432. [DOI] [PubMed] [Google Scholar]
- 135.Tinberg CE and Lippard SJ, Biochemistry, 2009, 48, 12145–12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Banerjee R, Meier KK, Münck E and Lipscomb JD, Biochemistry, 2013, 52, 4331–4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee S-K, Fox BG, Froland WA, Lipscomb JD and Münck E, J. Am. Chem. Soc, 1993, 115, 6450–6451. [Google Scholar]
- 138.Liu KE, Valentine AM, Wang D, Huynh BH, Edmondson DE, Salifoglou A and Lippard SJ, J. Am. Chem. Soc, 1995, 117, 10174–10185. [Google Scholar]
- 139.Banerjee R, Proshlyakov Y, Lipscomb JD and Proshlyakov DA, Nature, 2015, 518 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Castillo RG, Banerjee R, Allpress CJ, Rohde GT, Bill E, Que L, Lipscomb JD and DeBeer S, J. Am. Chem. Soc, 2017, 139, 18024–18033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shu L, Nesheim JC, Kauffmann K, Münck E, Lipscomb JD and Que L Jr., Science, 1997, 275, 515–518. [DOI] [PubMed] [Google Scholar]
- 142.Cutsail GE III, Banerjee R, Zhou A, Que L Jr., Lipscomb JD and DeBeer S, J. Am. Chem. Soc, 2018, 140, 16807–16820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xue G, De Hont R, Munck E and Que L Jr., Nature chemistry, 2010, 2, 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gherman BF, Lippard SJ and Friesner RA, J. Am. Chem. Soc, 2005, 127, 1025–1037. [DOI] [PubMed] [Google Scholar]
- 145.Huang SP, Shiota Y and Yoshizawa K, Dalton Trans., 2013, 42, 1011–1023. [DOI] [PubMed] [Google Scholar]
- 146.Burrows KJ, Cornish A, Scott D and Higgins IJ, J. Gen. Microbiol, 1984, 130, 327–3333. [Google Scholar]
- 147.Green J and Dalton H, J. Biol. Chem, 1989, 264, 17698–17703. [PubMed] [Google Scholar]
- 148.Colby J, Stirling DI and Dalton H, Biochem. J, 1977, 165, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Priestley ND, Floss HG, Froland WA, Lipscomb JD, Williams PG and Morimoto H, J. Am. Chem. Soc, 1992, 114, 7561–7562. [Google Scholar]
- 150.Valentine AM, Wilkinson B, Liu KE, Komar-Panicucci S, Priestley ND, Williams PG, Morimoto H, Floss HG and Lippard SJ, J. Am. Chem. Soc, 1997, 119, 1818–1827. [Google Scholar]
- 151.Liu KE, Johnson CC, Newcomb M and Lippard SJ, J. Amer. Chem. Soc, 1993, 115, 939–947. [Google Scholar]
- 152.Jin Y and Lipscomb JD, Biochemistry, 1999, 38, 6178–6186. [DOI] [PubMed] [Google Scholar]
- 153.Valentine AM, LeTadic-Biadatti MH, Toy PH, Newcomb M and Lippard SJ, J. Biol. Chem, 1999, 274, 10771–10776. [DOI] [PubMed] [Google Scholar]
- 154.Brazeau BJ, Austin RN, Tarr C, Groves JT and Lipscomb JD, J. Am. Chem. Soc, 2001, 123, 11831–11837. [DOI] [PubMed] [Google Scholar]
- 155.Nesheim JC and Lipscomb JD, Biochemistry, 1996, 35, 10240–10247. [DOI] [PubMed] [Google Scholar]
- 156.Brazeau BJ, Wallar BJ and Lipscomb JD, J. Am. Chem. Soc, 2001, 123, 10421–10422. [DOI] [PubMed] [Google Scholar]
- 157.Brantner CA, Remsen CC, Owen HA, Buchholz LA and Collins MLP, Arch. Microbiol, 2002, 178, 59–64. [DOI] [PubMed] [Google Scholar]
- 158.Cafaro V, Izzo V, Scognamiglio R, Notomista E, Capasso P, Casbarra A, Pucci P and Di Donato A, Appl. Environ. Microbiol, 2004, 70, 2211–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pikus JD, Studts JM, Achim C, Kauffmann KE, Münck E, Steffan RJ, McClay K and Fox BG, Biochemistry, 1996, 35, 9106–9119. [DOI] [PubMed] [Google Scholar]
- 160.Smith TJ and Murrell JC, Methods Enzymol., 2011, 495, 135–147. [DOI] [PubMed] [Google Scholar]
- 161.Smith TJ and Nichol T, in Methane Biocatalysis: Paving the Way to Sustainability, eds. Kalyuzhnaya MG and Xing X-H, Springer International Publishing, Cham, 2018, DOI: 10.1007/978-3-319-74866-5_10, pp. 153–168. [DOI] [Google Scholar]
- 162.Borodina E, Nichol T, Dumont MG, Smith TJ and Murrell JC, Appl. Environ. Microbiol, 2007, 73, 6460–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lock M, Nichol T, Murrell JC and Smith TJ, FEMS Microbiol. Lett, 2017, 364, fnx137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Orata FD, Meier-Kolthoff JP, Sauvageau D and Stein LY, Front. Microbiol, 2018, 9, 3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Puri AW, Owen S, Chu F, Chavkin T, Beck DAC, Kalyuzhnaya MG and Lidstrom ME, Appl. Environ. Microbiol, 2015, 81, 1766–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yan X, Chu F, Puri AW, Fu YF and Lidstrom ME, Appl. Environ. Microbiol, 2016, 82, 2062–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tapscott T, Guarnieri MT and Henard CA, Appl. Environ. Microbiol, 2019, 85, e00340–00319. [DOI] [PMC free article] [PubMed] [Google Scholar]


