Abstract
Understanding the mechanism behind probiotic action will enable a rational selection of probiotics, increase the chances of success in clinical studies and make it easy to substantiate health claims. However, most probiotic studies over the years have rather focused on the effects of probiotics in health and disease, whereas little is known about the specific molecules that trigger effects in hosts. This makes it difficult to describe the detailed mechanism by which a given probiotic functions. Probiotics communicate with their hosts through molecular signaling. Meanwhile, since the molecules produced by probiotics under in vitro conditions may differ from those produced in vivo, in vitro mechanistic studies would have to be conducted under conditions that mimic gastrointestinal conditions as much as possible. The ideal situation would, however, be to carry out well-designed clinical trials in humans (or the target animal) using adequate quantities of the suspected probiotic molecule(s) or adequate quantities of isogenic knock-out or knock-in probiotic mutants. In this review, we discuss our current knowledge about probiotic bacteria and yeast molecules that are involved in molecular signaling with the host. We also discuss the challenges and future perspectives in the search for probiotic effector molecules.
Keywords: microbiota, gut barrier functions, immune system, cholesterol reduction, nervous system
Introduction
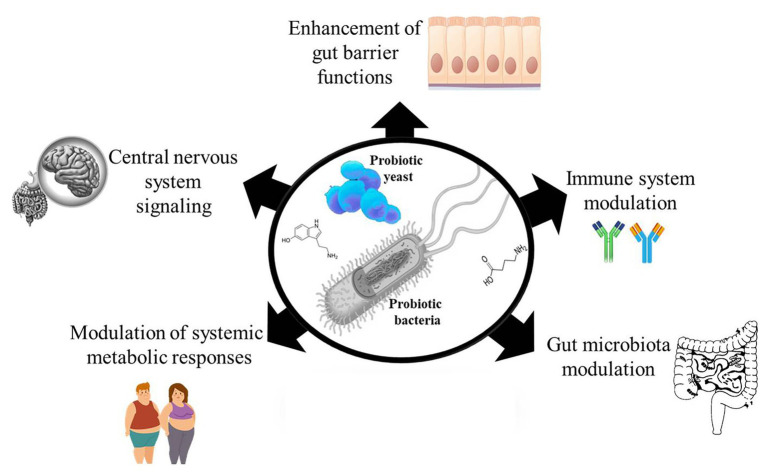
Probiotics are live microorganisms which when administered in adequate quantities provide a beneficial effect to the host (Hill et al., 2014). Over the years, many randomized clinical studies have reported the health benefits of probiotic consumption (Chugh et al., 2020; Tao et al., 2020; Tenorio-Jiménez et al., 2020), yet monitoring targeted health benefits of administered probiotic have rather been difficult to establish. This is because it would require an understanding of the possible metabolic activities of the administered probiotic that distinguishes it from the indigenous microbes in the host that could be eliciting similar effects. The limited knowledge about the mechanisms of probiotic action makes it challenging to rationally select probiotic strains for targeted interventions and also makes reproducibility of results difficult. Meanwhile, a number of mechanisms have been proposed to explain the mode of action behind the health effects of probiotics, such as enhancing gut epithelia barrier functions (Liu et al., 2016), modulating the immune system (Milajerdi et al., 2020; Zhao et al., 2020), modulating the gut microbiota (Botelho et al., 2020; López-Moreno et al., 2020), modulating systemic metabolic responses (van Baarlen et al., 2011; Belguesmia et al., 2016), and signaling host central nervous system (Kong et al., 2020; Figure 1). Since probiotic and host cell interactions are mediated by effector molecules, we discuss our current knowledge on probiotic bioactive molecules that have impact on the host in this review. We also discuss the challenges and future perspectives in the search for probiotic effector molecules.
Figure 1.
Probiotic bacteria and yeast possess and produce many effector molecules that interact with the host to elicit their observed effects. The effector molecules can affect host gut epithelial barrier functions, modulate host immune system, modulate the host’s gut microbiota, modulate the host’s metabolic responses, and affect the host’s central nervous system.
Probiotic Effector Molecules That Affect Gut Barrier Functions
Current scientific evidence show that a disruption of gut epithelial barrier function is important in the pathogenesis of many diseases such as inflammatory bowel disease (IBD; Lee et al., 2018; Soroosh et al., 2019), irritable bowel syndrome (Lee et al., 2020), diabetes (De Kort et al., 2011), and several other diseases. Gut barrier integrity is maintained by tight junction proteins, such as claudins, Zona occludin-1, and occludin, and their levels are significantly reduced during some disease conditions (Lee et al., 2020). This makes the gut epithelium permeable to microbial ligands and harmful metabolites leading to systemic inflammatory responses (Singh et al., 2019). The ability of probiotic effector molecules to protect gut barrier functions has long been reported (Miyauchi et al., 2009; Karczewski et al., 2010; Laval et al., 2015; Martín et al., 2019). Some of the molecules are soluble and therefore are secreted by the bacteria (or fungi) while others are cell wall bound (Delgado et al., 2020). Lactobacillus rhamnosus GG has been shown to secrete protein p40, which stimulates ADAM17 activation and heparin binding-epidermal growth factor (HB-EGF) release. This results in EGF receptor transactivation, apoptosis prevention, and intestinal epithelial function preservation (Yan et al., 2013). Another soluble protein, p75 secreted by both L. rhamnosus and L. casei is known to stimulate EGF receptor activation to prevent apoptosis in intestinal epithelial cells (Bäuerl et al., 2010). More recently, L. rhamnosus GG was shown to secrete protein HM0539, which protects gut barrier functions by promoting the expression of tight junction protein Zona occludin-1 and occluding. The protein also stimulates mucin secretion in intestinal cells (Gao et al., 2019). Meanwhile, the exact molecular mechanism by which HM0539 protects intestinal cells from injury remains to be established. Previous studies have shown that the soluble protein known as TcpC protein produced by Escherichia coli Nissle 1917 can induce protein kinase C-ζ and extracellular-signal-regulated kinase 1/2 phosphorylation to increase the formation of claudin-14, and this could account for the use of the probiotic for gastrointestinal therapy (Hering et al., 2014). Escherichia coli Nissle 1917 also produces 3-hydroxyoctadecanoic acid, which antagonizes peroxisome proliferator activated receptor gamma (PPARγ) to reduce inflammation (Pujo et al., 2020). In addition, probiotics and several other lactic acid bacteria are known to produce conjugated linoleic acid (CLA; Wang et al., 2016), which can upregulate the transcription of E-cadherin 1, claudin-3, ZO-1, and occludin in the gut to protect gut barrier functions (Murphy et al., 2007; Chen et al., 2019). Probiotic CLA can increase the expression of catalase, superoxide dismutase, and glutathione peroxidase, which reduce oxidative stress in colonocytes (Qi et al., 2018; Chen et al., 2019). More so, CLA increases the expression and activity of PPARγ in the gut to inhibit inflammation (Hontecillas et al., 2002). Some structural components, such as pili, of many probiotic bacteria have been shown to play important roles in gut epithelial functions. For instance, the tight adhesion pili of Bifidobacterium breve UCC2003 have been reported to stimulate the proliferation of gut epithelial cells by producing a TadE pseudopilin (O’Connell Motherway et al., 2019). Also, L. plantarum CGMCC 1258 micro integral membrane protein (MIMP) was found to promote the expression of tight junction proteins, such as JAM-1, claudin-1, and occludin, during tight junctional injury (Yin et al., 2018). Meanwhile, the exact mechanism by which MIMPs promotes the upregulation of tight junction proteins remain unestablished.
Probiotic Effector Molecules That Stimulate the Immune System
Many studies have reported the ability of the probiotics to contribute to the maturation of the immune system. It has however been shown that lipopolysaccharide of Gram negative probiotics are responsible for their strong induction of IL-10 in peripheral blood mononuclear cells (Kandasamy et al., 2017). The IL-10 produced contributes to the induction of IgA antibodies at mucosal sites by enhancing isotypic commutation (Cerutti and Rescigno, 2008), and this may account for why Gram negative probiotics induce a stronger antibody response than Gram positive probiotics. This was demonstrated when E. coli Nissle 1917 colonized pigs showed a higher gut IgA response relative to L. rhamnosus GG colonization (Kandasamy et al., 2016). The anti-inflammatory ability of L. plantarum in acute colitis mice (Emília et al., 2018) has at least in part been attributed to the presence of a small domain of the surface layer protein of the bacterium. These surface layer proteins have been shown to bind to a mannose receptor in gut epithelial cells to prevent p38 MAPK phosphorylation by inhibiting the Toll-like receptor (TLR) 5 pathway (Liu et al., 2012) during inflammatory conditions. This suppresses the expression of inflammatory cytokines (IFN-γ, IL-17, and IL-23) and upregulates IL-4 and IL-10 production in the cells (Yin et al., 2018). Another study has shown that sortase-dependent protein on the cell surface of L. plantarum can actively attenuate the NF-kB pathway thereby suppressing inflammation (Marco et al., 2010; Remus, 2012). It is known that different immune-stimulating probiotic strains may however have different cell wall molecules that interact with host immune receptors (Bron et al., 2013; Lee et al., 2013). For instance, the immune stimulating ability of L. plantarum K8 has been partly attributed to the lipotechoic acid (LTA) in its cell wall, which is able to regulate mitogen-activated protein kinase phosphorylation and nuclear factor activation and causes a reduction in IL-8 production in injured intestinal cells (Kim et al., 2017). Meanwhile, LTA from L. rhamnosus GG, L. sakei, and L. delbrueckii did not stimulate IL-8 production under similar conditions (Kim et al., 2017). In probiotic yeast, Saccharomyces cerevisiae cell wall contains β-glucans, which may induce monocyte reprograming via a dectin-1/Raf-1 pathway to enhance cytokine production for protection against Candida albicans infection (Quintin et al., 2012). Similarly, S. cerevisiae chitin has been shown to increase host immune system resistance to C. albicans infection by modulating the production of pro‐ and anti-inflammatory cytokines (Rizzetto et al., 2016). Even the spores of S. cerevisiae contain high amounts of chitin that can trigger inflammatory IL-17 responses in hosts (Rizzetto et al., 2010).
Probiotic Effector Molecules That Modulate Host Central Nervous System
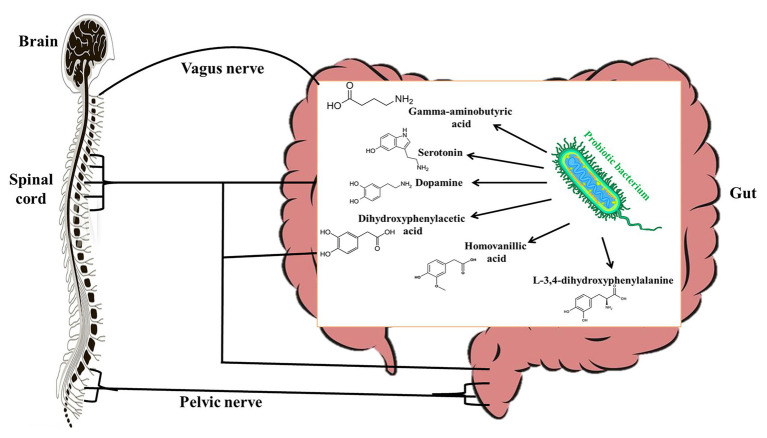
Studies over the years have shown evidence (though mostly indirect) that there is a strong communication between the gut microbiota and the central nervous system and it is mediated by the vagal nerve (Cryan and Dinan, 2012). For this reason, the effects of probiotics on the central nervous system have been studied extensively. However, only several studies have studied the molecular mechanisms by which probiotics affect the central nervous system. Many probiotic bacteria including L. plantarum, L. brevis, L. rhamnosus, and Bifidobacterium bifidum have been shown to produce significant amounts of gamma-aminobutyric acid (GABA) in vitro (Li et al., 2011; Diez-Gutiérrez et al., 2020). It is known that GABA in circulation may act as an autocrine, local paracrine, or gastrointestinal hormone that exerts both stimulatory and inhibitory effects over enteric neuronal activity depending on the type of GABA receptor stimulated (Hardcastle et al., 1991). Interestingly, Koussoulas et al. (2018) have shown that exogenous GABA can bind to GABAA, GABAB, and GABAc receptors to induce calcium [Ca2+]i release by myenteric ganglia. Since glial Ca2+ signaling is a mechanism for integration within glial symplasm and between glial-neuronal circuits (Verkhratsky, 2006), it stands to reason that GABA produced by probiotics could be an effector molecule by which certain probiotics interact with host central nervous system. This could therefore account for the ability of L. rhamnosus JB-1 to alter stress-related disorders via the vagus nerves (Bravo et al., 2011). Other studies have shown that L. helveticus 100ash, L. helveticus NK-1, L. casei K3III24, and L. delbrueckii subsp. bulgaricus produce significant amounts of GABA, L-3,4-dihydroxyphenylalanine (L-DOPA), dopamine, dihydroxyphenylacetic acid (DOPAC), homovanillic acid (HVA), and serotonin (Figure 2) when cultured in milk-containing media (Oleskin et al., 2014). L-DOPA can be transported from the gut through the blood to the brain, where it is converted to dopamine, a neurotransmitter (Disdier and Stonestreet, 2019). Dopamine can be metabolized into DOPAC, which can be degraded to HVA. In the central nervous system, serotonin plays a role in regulating emotions, sleep, and stress (De Deurwaerdère and Di Giovanni, 2020) by influencing the hypothalamic-pituitary-adrenal axis. Though it is likely that probiotics that produce these biogenic amines may have central nervous system modulatory effects, the effector molecules would have to be produced in adequate quantities in the gut of the host to elicit the desired effects. Although some studies have suggested that probiotics may improve diseases associated with the central nervous system by enhancing the production of free tryptophan, which may promote serotonin availability (since serotonin is synthesized from tryptophan; Desbonnet et al., 2008), it is not known whether probiotic tryptophan is used for serotonin synthesis (Daliri et al., 2016).
Figure 2.
Probiotic bacteria produce gamma-aminobutyric acid (GABA), serotonine, dopamine, L-3,4-dihydroxyphenylalanine (L-DOPA), dihydroxyphenylacetic acid (DOPAC), and homovanillic acid (HVA) that modulate the host central nervous system.
Probiotic Effector Molecules That Modulate Host Microbiota
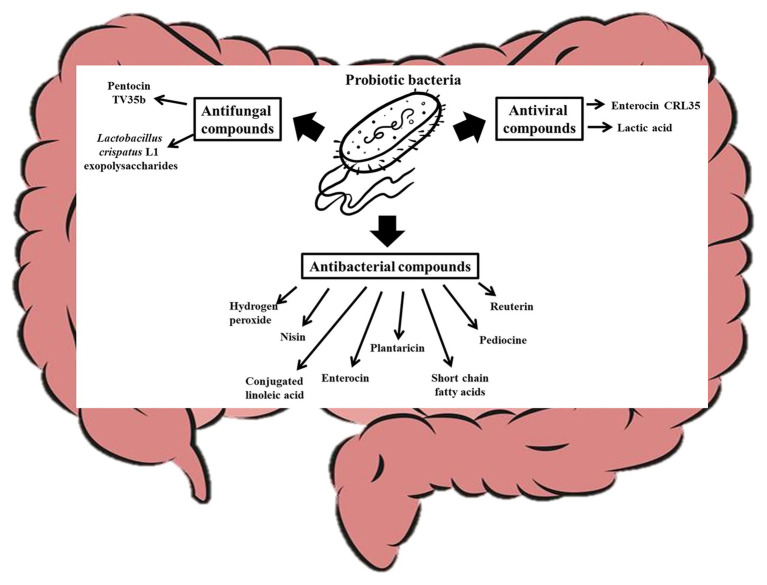
The human body is made of mammalian cells, archaea (Kim et al., 2020), bacteria, viruses, and fungi co-existing in a symbiotic relationship (Daliri et al., 2020). Although host microbes could vary from one person to another (Brooks et al., 2018), recent studies have shown that all these microorganisms in the human body could have impacts on host health and disease (Daliri et al., 2018; Reitmeier et al., 2020) and that probiotic consumption can significantly modulate the host microbiome (Alcon-Giner et al., 2020). In fact, probiotics in food can survive, adapt, and become an established part of the gut microbiome (Pasolli et al., 2020) and have significant influence on the gut microbiome. The mechanisms by which probiotics modulate host microbiome include their effects on the function and composition of the host commensal bacteria and yeast. It is known that probiotics can produce antimicrobial compounds that suppress (O’Shea et al., 2012) or promote (Chen et al., 2019) the growth of certain microorganisms in the gut as shown in Figure 3. Lactococcus lactis for instance produces nisin (a bacteriocin), which can permeate the cell membrane of pathogenic Gram positive-bacteria and bind to Lipid II to inhibit cell wall synthesis (Wiedemann et al., 2001; Breukink and de Kruijff, 2006). Other probiotics such as Pediococcus acidilactici produce pediocine-like bacteriocins, which can bind to mannose phosphotransferase on the cytoplasmic membrane of pathogenic bacteria and penetrate the lipid bilayer resulting in pore formation in the cell (Balandin et al., 2019). This compromise in the cell wall integrity makes the cell extremely vulnerable to the harsh environmental conditions within which the cell is found and can lead to death. Similarly, Plantaricin LPL-1 produced by L. plantarum LPL-1 binds to a receptor on Listeria monocytogenes cell wall, perforate the cell membrane through hydrophobic interactions, and accumulate in the cell membrane by electrostatic interactions. The perforation results in ion leakage and loss of proton motive force, which eventually result in cell death (Wang et al., 2019b). Plantaricins do not only inactivate Gram-positive bacteria, but also Gram-negative bacteria (Zhang et al., 2013; Yang et al., 2019). Wang et al. (2020) showed that Plantaricin BM-1 can inhibit E. coli by acting on the surface of the cell wall to cause cell rapture. The bacteriocin could also permeate and interact with the cell membrane to lead to cell death. A popular probiotic, Lactobacillus reuteri, is known to produce reuterin, which suppresses the expression of Clostridioides difficile exotoxins TcdA and TcdB by inducing reactive oxygen species shifts in carbon metabolism, which may alter gene expression in the pathogen (Engevik et al., 2020). The impaired metabolism of the pathogens decreases their ability to compete for nutrients and eventually result in cell death (Engevik et al., 2020). Certain bacteriocins can inhibit viruses and yeast growth. For instance, it has been shown that Enterocin CRL35 from Enterococcus faecium CRL35 can bind and block the late stages of herpes simplex virus types (HSV) 1 and 2 replication (Wachsman et al., 1999, 2003) while Pentocin TV35b produced by Lactobacillus pentosus TV35b can inhibit the survival of C. albicans (an opportunistic fungus; Okkers et al., 1999). Exopolysaccharides produced by Lactobacillus crispatus L1 also reduces C. albicans adhesion to epithelial cells (Donnarumma et al., 2014) thereby decreasing their tendency to invade host cells. Lactobacillus acidophilus La-5 produces CLA (Macouzet et al., 2009), which can repress virulence genes such as eaeA (an enterohaemorrhagic E. coli invasion lipoprotein gene) and invH (a Staphylococcus typhimirium invasion lipoprotein gene) thereby reducing the ability of these pathogens to attach to the host cells (Peng et al., 2018). Also, probiotic CLA can competitively bind to INT-407 cell surface receptor-like molecules preventing enteric pathogens from binding to the gut (Peng et al., 2018). Consumption of probiotic CLA favored an increase in the levels of Bifidobacterium and Odoribacter while reducing the levels of Bacteroides in the gut (Chen et al., 2019) through an unidentified mechanism. Other probiotics, such as L. brevis CD2, L. salivarius FV2, and L. plantarum FV9, produce lactic acid which impair Herpes simplex virus 2 (HSV-2) protein synthesis and viral replication (Conti et al., 2009), while bacteria including L. paracasei inhibit vesicular stomatitis virus by direct binding with them (Botić et al., 2007). Bacteria with strong mucosal binding abilities, such as L. brevis, CD2 compete with HSV-2 for binding sites thereby preventing the fusion of viral envelope with host cell surface (Conti et al., 2009). Short chain fatty acids (SCFAs), such as butyric acid, produced by probiotics can induce gut epithelia secretion of cathelicidin peptides, which inactivate pathogens such as Shigella (Schauber et al., 2003; Raqib et al., 2006; Termén et al., 2008; Campbell et al., 2012). Some probiotics, including B. bifidum (Kawasaki et al., 2009) and Lactobacillus johnsonii (Pridmore et al., 2008), are known to produce hydrogen peroxide, which can react with O2− and/or iron in pathogenic cells to form toxic hydroxyl radicals which results in cell death (Clifford and Repine, 1982). A probiotic S. cerevisiae isolated from Koumiss has been shown to inhibit E. coli by producing citric acid and propionic acid, which disintegrate the cell membrane and increase cell permeability (Chen et al., 2017). Certain S. cerevisiae strains have been shown to produce killer toxin KHS which inhibits the growth of pathogenic bacteria and yeast (Goto et al., 1991; Younis et al., 2017). These killer toxins inhibit β-glucan synthesis in the cell walls (Muccilli et al., 2013), inhibit DNA synthesis (Klassen and Meinhardt, 2005), cleaves tRNA (Klassen et al., 2008), blocks calcium ion uptake (Brown, 2011), and cause ion leakage from the cytoplasm (Santos et al., 2007) of sensitive cells. Another study showed that chitin from S. cerevisiae can train host immune system to kill S. aureus, E. coli, and C. albicans (Rizzetto et al., 2016), and this shows how yeast molecules can play critical roles in gut microbiota modulation. Many pathogenic bacteria target glycosaminoglycans for attachment and infection of the host (Kawai et al., 2018). Quite recently, it has been shown that probiotics, such as Enterococcus faecium, H57 prevent pathogen adhesion to the gut by degrading GAG (which are major component of extracellular matrix in animals) using KduI and KduD enzymes (Kawai et al., 2018). The probiotic molecules discussed in this section could at least contribute to the gut modulatory ability of probiotic yeast (Adel et al., 2017; Villar-García et al., 2017) and bacteria (Kong et al., 2019; Liu et al., 2020).
Figure 3.
Probiotic bacteria modulate the gut microbiota by producing antifungal, antiviral, and antibacterial compounds that affect the overall gut microbial structure of the host.
Probiotic Effector Molecules That Affect Cholesterol-Lowering
Hypercholesterolemia is a metabolic disorder marked by abnormally high levels of cholesterol in the cells and blood (Wang et al., 2019a). Since high blood cholesterol levels have been strongly associated with an increased risk of coronary heart diseases, several strategies including probiotic consumption have been used to reduce the levels of cholesterol in hypercholesterolemic patients (Jones et al., 2012). However, the mechanism behind the hypocholesterolemic ability of probiotics remains elusive. Yet one of the most reported mechanism has been attributed to the ability of some probiotics to produce bile salt hydrolase (BSH; Jones et al., 2012; Wang et al., 2019a; Huang et al., 2020). Many probiotic bacteria, such as L. plantarum WCFS1, L. plantarum TH1, and Bifidobacteria longum SBT2928, produce BSH (Ishimwe et al., 2015), which can hydrolyze conjugated bile salts, such as taurine-conjugated bile salts and glycine-conjugated bile salts, to release primary bile acids (Li et al., 2020). The resulting deconjugated bile acids are less soluble (Hofmann and Mysels, 1992; Degirolamo et al., 2014), and hence are less reabsorbed into circulation but excreted through feces. Bacteria, such as L. acidophilus, L. bulgaricus, and L. casei ATCC 393, possess intracellular and extracellular cholesterol reductases, which effectively convert cholesterol to coprostanol (Lye et al., 2010). β-glucan from some probiotic lactobacilli (London et al., 2014) and S. cerevisiae (Kusmiati and Dhewantara, 2016) have been reported to reduce serum cholesterol by binding to enteral bile acids to promote their excretion in feces (Sima et al., 2018). Also, probiotic SCFAs, such as butyrate, may inhibit cholesterol biosynthesis by inhibiting DL-3-hydroxy-3-methyl-glutaryl-CoA reductase, which is the enzyme that catalyzes the rate-limiting step of cholesterol biosynthesis (Marcil et al., 2003). Other studies have shown that butyrate significantly induces ATP-binding cassette sub-family A member 1 (ABCA1) via specificity protein 1 (Sp1) in macrophages (Du et al., 2020). ABCA1 is responsible for maintaining lipid homeostasis by regulating cellular cholesterol (He et al., 2020). This may account for the ability of SCFAs to significantly decrease the rate of hepatic and mucosal cholesterol biosynthesis (Hara et al., 1999; Du et al., 2020). Lee et al. (2010) have shown that catabolite control protein A (a membrane associated protein) plays a major role in the ability of L. acidophilus A4 to reduce serum cholesterol. Wild L. acidophilus A4 strains effectively reduced serum cholesterol levels while catabolite control protein A (ccpA) mutants had significantly reduced cholesterol reducing abilities. Although the direct effect of the protein on cholesterol reduction remains unclear, the ccpA gene is known to strongly regulate important cell functions including lipid metabolism, cell envelope biogenesis, carbohydrate transport and metabolism, outer membrane and intracellular cargo trafficking (Zomer et al., 2007), and hence affect cholesterol metabolism. Certain gut bacteria, such as Eubacterium coprostanoligenes, have been identified to possess ismA genes, which express 3β-hydroxysteroid dehydrogenase for converting cholesterol to cholestenone, to coprostanol, and then to coprostanone (Kenny et al., 2020). The bacterium is a good candidate of next generation probiotics as it effectively decreases total cholesterol levels in foods (Madden et al., 1999) and animals (Li et al., 1995, 1996, 1998). However, the ismA gene has not been reported in known probiotics yet.
Challenges and Future Perspectives on Probiotic Effector Molecules
Over the years, server studies have successfully identified as soluble molecules of probiotics in media by using metabolomics approaches (Kahouli et al., 2015; Usta-Gorgun and Yilmaz-Ersan, 2020). After recovery, the molecules are applied in in vitro and in vivo studies to confirm their effects (Wiedemann et al., 2001; Li et al., 2020). For membrane bound molecules, however, the cells are usually broken and the components separated (Urner et al., 2020) and tested for activity. Meanwhile, the metabolic activities of microbes may change when their biological niches are changed (Franzosa et al., 2014), and so the bioactive molecule identified in vitro may not be the cause or the only possible cause of the physiological effect observed in the host after probiotic administration. This has been observed in studies in which knocking out genes suspected to express certain active molecules did not completely attenuate the physiological effects of the bacterium (Lee et al., 2010). Therefore, future studies may have to consider carrying out in vitro tests of the production of probiotic active molecules with gastrointestinal effects under simulated gastrointestinal conditions (mixture of enzymes, acids, salts, mucus, etc.). Such studies may also have to consider the influence of disease conditions on the immune system, host antimicrobial proteins, and gut microbial competition on the probiotic in other to ascertain what genes (and bioactive compounds) are really triggered (and produced) in consumed probiotics. A more plausible way of assessing the probable effects of these stressors on a consumed probiotic would be to collect the microbe from gut samples (or vagina) after the expected physiological effect has occurred and subjected to transcriptomics to ascertain if the mRNA of the suspected bioactive molecule (in case of a protein or peptide) is actively transcribed under those conditions.
For a better understanding of which probiotic molecules trigger a given effect, there is the need to carry out well-designed clinical trials in humans using adequate quantities of the isolated bioactive molecules from probiotics or adequate quantities of isogenic knock-out or knock-in probiotic mutants. This is essential because animals and humans are different and so extrapolating results from animal studies may not always be correct. Meanwhile, such a study would face several ethical and technical hurdles as it involves humans and will have to establish the probiotic strain’s potency, effective dose, targeted host response, targeted host site, and other important parameters.
Author Contributions
ED conceived, designed, and wrote the manuscript. FO, CX, and RC revised and made corrections. D-HO approved the manuscript and provided funding. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Adel M., Lazado C. C., Safari R., Yeganeh S., Zorriehzahra M. J. (2017). Aqualase®, a yeast-based in-feed probiotic, modulates intestinal microbiota, immunity and growth of rainbow trout Oncorhynchus mykiss. Aquac. Res. 48, 1815–1826. 10.1111/are.13019 [DOI] [Google Scholar]
- Alcon-Giner C., Dalby M. J., Caim S., Ketskemety J., Shaw A., Sim K., et al. (2020). Microbiota supplementation with bifidobacterium and lactobacillus modifies the preterm infant gut microbiota and metabolome: an observational study. Cell Rep. Med. 1:100077. 10.1016/j.xcrm.2020.100077, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balandin S. V., Sheremeteva E. V., Ovchinnikova T. V. (2019). Pediocin-like antimicrobial peptides of bacteria. Biochemistry 84, 464–478. 10.1134/S000629791905002X, PMID: [DOI] [PubMed] [Google Scholar]
- Bäuerl C., Pérez-Martínez G., Yan F., Polk D. B., Monedero V. (2010). Functional analysis of the p40 and p75 proteins from Lactobacillus casei BL23. J. Mol. Microbiol. Biotechnol. 19, 231–241. 10.1159/000322233, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belguesmia Y., Domenger D., Caron J., Dhulster P., Ravallec R., Drider D., et al. (2016). Novel probiotic evidence of lactobacilli on immunomodulation and regulation of satiety hormones release in intestinal cells. J. Funct. Foods 24, 276–286. 10.1016/j.jff.2016.04.014 [DOI] [Google Scholar]
- Botelho P. B., Ferreira M. V. R., Araújo A. M., Mendes M. M., Nakano E. Y. (2020). Effect of multi-species probiotic on gut microbiota composition on healthy individuals with intestinal constipation: a double-blind, placebo-controlled randomized trial. Nutrition 78:110890. 10.1016/j.nut.2020.110890, PMID: [DOI] [PubMed] [Google Scholar]
- Botić T., Klingberg T. D., Weingartl H., Cencic A. (2007). A novel eukaryotic cell culture model to study antiviral activity of potential probiotic bacteria. Int. J. Food Microbiol. 115, 227–234. 10.1016/j.ijfoodmicro.2006.10.044, PMID: [DOI] [PubMed] [Google Scholar]
- Bravo J. A., Forsythe P., Chew M. V., Escaravage E., Savignac H. M., Dinan T. G., et al. (2011). Ingestion of lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. PNAS 108, 16050–16055. 10.1073/pnas.1102999108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breukink E., De Kruijff B. (2006). Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 5, 321–323. 10.1038/nrd2004, PMID: [DOI] [PubMed] [Google Scholar]
- Bron P. A., Tomita S., Mercenier A., Kleerebezem M. (2013). Cell surface-associated compounds of probiotic lactobacilli sustain the strain-specificity dogma. Curr. Opin. Microbiol. 16, 262–269. 10.1016/j.mib.2013.06.001, PMID: [DOI] [PubMed] [Google Scholar]
- Brooks A. W., Priya S., Blekhman R., Bordenstein S. R. (2018). Gut microbiota diversity across ethnicities in the United States. PLoS Biol. 16:e2006842. 10.1371/journal.pbio.2006842, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. W. (2011). The KP4 killer protein gene family. Curr. Genet. 57, 51–62. 10.1007/s00294-010-0326-y, PMID: [DOI] [PubMed] [Google Scholar]
- Campbell Y., Fantacone M. L., Gombart A. F. (2012). Regulation of antimicrobial peptide gene expression by nutrients and by-products of microbial metabolism. Eur. J. Nutr. 51, 899–907. 10.1007/s00394-012-0415-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A., Rescigno M. (2008). The biology of intestinal immunoglobulin a responses. Immunity 28, 740–750. 10.1016/j.immuni.2008.05.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. -J., Wang C. -J., Hou W. -Q., Wang X. -S., Gali B. -G., Huasai S. -M. -J. -D., et al. (2017). Effects of antibacterial compounds produced by Saccharomyces cerevisiae in koumiss on pathogenic Escherichia coli Os and its cell surface characteristics. J. Integr. Agric. 16, 742–748. 10.1016/S2095-3119(16)61516-2 [DOI] [Google Scholar]
- Chen Y., Yang B., Ross R. P., Jin Y., Stanton C., Zhao J., et al. (2019). Orally administered CLA ameliorates dss-induced colitis in mice via intestinal barrier improvement, oxidative stress reduction, and inflammatory cytokine and gut microbiota modulation. J. Agric. Food Chem. 67, 13282–13298. 10.1021/acs.jafc.9b05744, PMID: [DOI] [PubMed] [Google Scholar]
- Chugh P., Dutt R., Sharma A., Bhagat N., Dhar M. S. (2020). A critical appraisal of the effects of probiotics on oral health. J. Funct. Foods 70:103985. 10.1016/j.jff.2020.103985 [DOI] [Google Scholar]
- Clifford D. P., Repine J. E. (1982). Hydrogen peroxide mediated killing of bacteria. Mol. Cell. Biochem. 49, 143–149. 10.1007/BF00231175, PMID: [DOI] [PubMed] [Google Scholar]
- Conti C., Malacrino C., Mastromarino P. (2009). Inhibition of herpes simplex virus type 2 by vaginal lactobacilli. J. Physiol. Pharmacol. 60, 19–26. PMID: [PubMed] [Google Scholar]
- Cryan J. F., Dinan T. G. (2012). Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 13, 701–712. 10.1038/nrn3346, PMID: [DOI] [PubMed] [Google Scholar]
- Daliri E. B. -M., Ofosu F. K., Chelliah R., Lee B. H., Oh D. -H. (2020). Health impact and therapeutic manipulation of the gut microbiome. High Throughput 9:17. 10.3390/ht9030017, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daliri E., Oh D., Lee B. (2016). Psychobiotics; a promise for neurodevelopmental therapy. J. Probiotics Health 4:1e4. 10.4172/2329-8901.1000146 [DOI] [Google Scholar]
- Daliri E. B. -M., Tango C. N., Lee B. H., Oh D. -H. (2018). Human microbiome restoration and safety. Int. J. Med. Microbiol. 308, 487–497. 10.1016/j.ijmm.2018.05.002, PMID: [DOI] [PubMed] [Google Scholar]
- De Deurwaerdère P., Di Giovanni G. (2020). Serotonin in health and disease. Int. J. Mol. Sci. 21:3500. 10.3390/ijms21103500, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degirolamo C., Rainaldi S., Bovenga F., Murzilli S., Moschetta A. (2014). Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice. Cell Rep. 7, 12–18. 10.1016/j.celrep.2014.02.032, PMID: [DOI] [PubMed] [Google Scholar]
- De Kort S., Keszthelyi D., Masclee A. (2011). Leaky gut and diabetes mellitus: what is the link? Obes. Rev. 12, 449–458. 10.1111/j.1467-789X.2010.00845.x, PMID: [DOI] [PubMed] [Google Scholar]
- Delgado S., Sánchez B., Margolles A., Ruas-Madiedo P., Ruiz L. (2020). Molecules produced by probiotics and intestinal microorganisms with immunomodulatory activity. Nutrients 12:391. 10.3390/nu12020391, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbonnet L., Garrett L., Clarke G., Bienenstock J., Dinan T. G. (2008). The probiotic Bifidobacteria infantis: an assessment of potential antidepressant properties in the rat. J. Psychiatr. Res. 43, 164–174. 10.1016/j.jpsychires.2008.03.009, PMID: [DOI] [PubMed] [Google Scholar]
- Diez-Gutiérrez L., San Vicente L., Barrón L. J. R., Del Carmen Villarán M., Chávarri M. (2020). Gamma-aminobutyric acid and probiotics: multiple health benefits and their future in the global functional food and nutraceuticals market. J. Funct. Foods 64:103669. 10.1016/j.jff.2019.103669 [DOI] [Google Scholar]
- Disdier C., Stonestreet B. S. (2019). “Chapter 24: Blood-brain barrier: effects of inflammatory stress” in In stress: Physiology, biochemistry, and pathology. ed. Fink G. (Melbourne, Australia: Academic Press; ), 325–336. [Google Scholar]
- Donnarumma G., Molinaro A., Cimini D., De Castro C., Valli V., De Gregorio V., et al. (2014). Lactobacillus crispatus L1: high cell density cultivation and exopolysaccharide structure characterization to highlight potentially beneficial effects against vaginal pathogens. BMC Microbiol. 14:137. 10.1186/1471-2180-14-137, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Li X., Su C., Xi M., Zhang X., Jiang Z., et al. (2020). Butyrate protects against high-fat diet-induced atherosclerosis via up-regulating ABCA1 expression in apolipoprotein E-deficiency mice. Br. J. Pharmacol. 177, 1754–1772. 10.1111/bph.14933, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emília H., Izabela B., Jana Š., Ladislav S., Anna C., Alojz B. (2018). Anti-inflammatory potential of Lactobacillus plantarum LS/07 in acute colitis in rats. Acta Vet. Brno 68, 55–64. 10.2478/acve-2018-0005 [DOI] [Google Scholar]
- Engevik M. A., Danhof H. A., Shrestha R., Chang-Graham A. L., Hyser J. M., Haag A. M., et al. (2020). Reuterin disrupts Clostridioides difficile metabolism and pathogenicity through reactive oxygen species generation. Gut Microbes 12:1795388. 10.1080/19490976.2020.1795388, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa E. A., Morgan X. C., Segata N., Waldron L., Reyes J., Earl A. M., et al. (2014). Relating the metatranscriptome and metagenome of the human gut. PNAS 111, E2329–E2338. 10.1073/pnas.1319284111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Li Y., Wan Y., Hu T., Liu L., Yang S., et al. (2019). A novel postbiotic from Lactobacillus rhamnosus GG with a beneficial effect on intestinal barrier function. Front. Microbiol. 10:477. 10.3389/fmicb.2019.00477, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K., Fukuda H., Kichise K., Kitano K., Hara S. (1991). Cloning and nucleotide sequence of the KHS killer gene of Saccharomyces cerevisiae. Agric. Biol. Chem. 55, 1953–1958. 10.1271/bbb1961.55.1953, PMID: [DOI] [PubMed] [Google Scholar]
- Hara H., Haga S., Aoyama Y., Kiriyama S. (1999). Short-chain fatty acids suppress cholesterol synthesis in rat liver and intestine. J. Nutr. 129, 942–948. 10.1093/jn/129.5.942, PMID: [DOI] [PubMed] [Google Scholar]
- Hardcastle J., Hardcastle P., Mathias W. J. (1991). The influence of the γ-amino butyric acid (GABA) antagonist bicuculline on transport processes in rat small intestine. J. Pharm. Pharmacol. 43, 128–130. 10.1111/j.2042-7158.1991.tb06648.x, PMID: [DOI] [PubMed] [Google Scholar]
- He P., Gelissen I. C., Ammit A. J. (2020). Regulation of ATP binding cassette transporter A1 (ABCA1) expression: cholesterol-dependent and – independent signaling pathways with relevance to inflammatory lung disease. Respir. Res. 21:250. 10.1186/s12931-020-01515-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering N., Richter J., Fromm A., Wieser A., Hartmann S., Günzel D., et al. (2014). TcpC protein from E. coli Nissle improves epithelial barrier function involving PKCζ and ERK1/2 signaling in HT-29/B6 cells. Mucosal Immunol. 7, 369–378. 10.1038/mi.2013.55, PMID: [DOI] [PubMed] [Google Scholar]
- Hill C., Guarner F., Reid G., Gibson G. R., Merenstein D. J., Pot B., et al. (2014). Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. 10.1038/nrgastro.2014.66, PMID: [DOI] [PubMed] [Google Scholar]
- Hofmann A. F., Mysels K. J. (1992). Bile acid solubility and precipitation in vitro and in vivo: the role of conjugation, pH, and Ca2+ ions. J. Lipid Res. 33, 617–626. 10.1016/S0022-2275(20)41426-9, PMID: [DOI] [PubMed] [Google Scholar]
- Hontecillas R., Wannemeulher M. J., Zimmerman D. R., Hutto D. L., Wilson J. H., Ahn D. U., et al. (2002). Nutritional regulation of porcine bacterial-induced colitis by conjugated linoleic acid. J. Nutr. 132, 2019–2027. 10.1093/jn/132.7.2019, PMID: [DOI] [PubMed] [Google Scholar]
- Huang W., Wang G., Xia Y., Xiong Z., Ai L. (2020). Bile salt hydrolase-overexpressing Lactobacillus strains can improve hepatic lipid accumulation in vitro in an NAFLD cell model. Food Nutr. Res. 64:3751. 10.29219/fnr.v64.3751, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimwe N., Daliri E. B., Lee B. H., Fang F., Du G. (2015). The perspective on cholesterol-lowering mechanisms of probiotics. Mol. Nutr. Food Res. 59, 94–105. 10.1002/mnfr.201400548, PMID: [DOI] [PubMed] [Google Scholar]
- Jones M. L., Martoni C. J., Parent M., Prakash S. (2012). Cholesterol-lowering efficacy of a microencapsulated bile salt hydrolase-active Lactobacillus reuteri NCIMB 30242 yoghurt formulation in hypercholesterolaemic adults. Br. J. Nutr. 107, 1505–1513. 10.1017/S0007114511004703, PMID: [DOI] [PubMed] [Google Scholar]
- Kahouli I., Malhotra M., Tomaro-Duchesneau C., Saha S., Marinescu D., Rodes L., et al. (2015). Screening and in-vitro analysis of Lactobacillus reuteri strains for short chain fatty acids production, stability and therapeutic potentials in colorectal cancer. J. Bioequiv. Availab. 7:39. 10.4172/jbb.1000212 [DOI] [Google Scholar]
- Kandasamy S., Vlasova A. N., Fischer D. D., Chattha K. S., Shao L., Kumar A., et al. (2017). Unraveling the differences between gram-positive and gram-negative probiotics in modulating protective immunity to enteric infections. Front. Immunol. 8:334. 10.3389/fimmu.2017.00334, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy S., Vlasova A. N., Fischer D., Kumar A., Chattha K. S., Rauf A., et al. (2016). Differential effects of Escherichia coli Nissle and Lactobacillus rhamnosus strain GG on human rotavirus binding, infection, and B cell immunity. J. Immunol. 196, 1780–1789. 10.4049/jimmunol.1501705, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski J., Troost F. J., Konings I., Dekker J., Kleerebezem M., Brummer R. -J. M., et al. (2010). Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G851–G859. 10.1152/ajpgi.00327.2009, PMID: [DOI] [PubMed] [Google Scholar]
- Kawai K., Kamochi R., Oiki S., Murata K., Hashimoto W. (2018). Probiotics in human gut microbiota can degrade host glycosaminoglycans. Sci. Rep. 8:10674. 10.1038/s41598-018-28886-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S., Satoh T., Todoroki M., Niimura Y. (2009). B-type dihydroorotate dehydrogenase is purified as a H2O2-forming NADH oxidase from Bifidobacterium bifidum. Appl. Environ. Microbiol. 75, 629–636. 10.1128/AEM.02111-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny D. J., Plichta D. R., Shungin D., Koppel N., Hall A. B., Fu B., et al. (2020). Cholesterol metabolism by uncultured human gut bacteria influences host cholesterol level. Cell Host Microbe 28, 245–257.e6. 10.1016/j.chom.2020.05.013, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. W., Kang S. -S., Woo S. -J., Park O. -J., Ahn K. B., Song K. -D., et al. (2017). Lipoteichoic acid of probiotic Lactobacillus plantarum attenuates poly I: C-induced IL-8 production in porcine intestinal epithelial cells. Front. Microbiol. 8:1827. 10.3389/fmicb.2017.01827, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y., Whon T. W., Lim M. Y., Kim Y. B., Kim N., Kwon M. -S., et al. (2020). The human gut archaeome: identification of diverse haloarchaea in Korean subjects. Microbiome 8:114. 10.21203/rs.3.rs-17518/v2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen R., Meinhardt F. (2005). Induction of DNA damage and apoptosis in Saccharomyces cerevisiae by a yeast killer toxin. Cell. Microbiol. 7, 393–401. 10.1111/j.1462-5822.2004.00469.x, PMID: [DOI] [PubMed] [Google Scholar]
- Klassen R., Paluszynski J. P., Wemhoff S., Pfeiffer A., Fricke J., Meinhardt F. (2008). The primary target of the killer toxin from Pichia acaciae is tRNAGln. Mol. Microbiol. 69, 681–697. 10.1111/j.1365-2958.2008.06319.x, PMID: [DOI] [PubMed] [Google Scholar]
- Kong C., Gao R., Yan X., Huang L., Qin H. (2019). Probiotics improve gut microbiota dysbiosis in obese mice fed a high-fat or high-sucrose diet. Nutrition 60, 175–184. 10.1016/j.nut.2018.10.002, PMID: [DOI] [PubMed] [Google Scholar]
- Kong X. -J., Liu J., Li J., Kwong K., Koh M., Sukijthamapan P., et al. (2020). Probiotics and oxytocin nasal spray as neuro-social-behavioral interventions for patients with autism spectrum disorders: a pilot randomized controlled trial protocol. Pilot Feasibility Stud. 6:20. 10.1186/s40814-020-0557-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussoulas K., Swaminathan M., Fung C., Bornstein J. C., Foong J. P. (2018). Neurally released GABA acts via GABAC receptors to modulate Ca2+ transients evoked by trains of synaptic inputs, but not responses evoked by single stimuli, in myenteric neurons of mouse ileum. Front. Physiol. 9:97. 10.3389/fphys.2018.00097, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusmiati, Dhewantara F. X. R. (2016). Cholesterol-lowering effect of beta glucan extracted from Saccharomyces cerevisiae in rats. Sci. Pharm. 84, 153–165. 10.3797/scipharm.ISP.2015.07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval L., Martin R., Natividad J., Chain F., Miquel S., De Maredsous C. D., et al. (2015). Lactobacillus rhamnosus CNCM I-3690 and the commensal bacterium Faecalibacterium prausnitzii A2-165 exhibit similar protective effects to induced barrier hyper-permeability in mice. Gut Microbes 6, 1–9. 10.4161/19490976.2014.990784, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Kim N., Park J. H., Nam R. H., Lee S. M., Song C. -H., et al. (2020). Expression of neurotrophic factors, tight junction proteins, and cytokines according to the irritable bowel syndrome subtype and sex. J. Neurogastroenterol. Motil. 26, 106–116. 10.5056/jnm19099, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kim Y., Yun H. S., Kim J. G., Oh S., Kim S. H. (2010). Genetic and proteomic analysis of factors affecting serum cholesterol reduction by Lactobacillus acidophilus A4. Appl. Environ. Microbiol. 76, 4829–4835. 10.1128/AEM.02892-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I. -C., Tomita S., Kleerebezem M., Bron P. A. (2013). The quest for probiotic effector molecules—unraveling strain specificity at the molecular level. Pharmacol. Res. 69, 61–74. 10.1016/j.phrs.2012.09.010, PMID: [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Wasinger V. C., Yau Y. Y., Chuang E., Yajnik V., Leong R. W. (2018). Molecular pathophysiology of epithelial barrier dysfunction in inflammatory bowel diseases. Proteomes 6:17. 10.3390/proteomes6020017, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Batt S. M., Wannemuehler M., Dispirito A., Beitz D. C. (1998). Effect of feeding of a cholesterol-reducing bacterium, Eubacterium coprostanoligenes, to germ-free mice. Lab. Anim. Sci. 48, 253–255. PMID: [PubMed] [Google Scholar]
- Li L., Baumann C. A., Meling D. D., Sell J. L., Beitz D. C. (1996). Effect of orally administered Eubacterium coprostanoligenes ATCC 51222 on plasma cholesterol concentration in laying hens. Poult. Sci. 75, 743–745. 10.3382/ps.0750743, PMID: [DOI] [PubMed] [Google Scholar]
- Li L., Buhman K. K., Hartman P. A., Beitz D. C. (1995). Hypocholesterolemic effect of Eubacterium coprostanoligenes ATCC 51222 in rabbits. Lett. Appl. Microbiol. 20, 137–140. 10.1111/j.1472-765X.1995.tb00410.x, PMID: [DOI] [PubMed] [Google Scholar]
- Li C., Ji Q., He T., Liu Y., Ma Y. (2020). Characterization of a recombinant bile salt hydrolase (BSH) from Bifidobacterium bifidum for its glycine-conjugated bile salts specificity. Biocatal. Biotransformation. 39, 61–70. 10.1080/10242422.2020.1804881 [DOI] [Google Scholar]
- Li H., Qiu T., Chen Y., Cao Y. (2011). Separation of gamma-aminobutyric acid from fermented broth. J. Ind. Microbiol. Biotechnol. 38, 1955–1959. 10.1007/s10295-011-0984-x, PMID: [DOI] [PubMed] [Google Scholar]
- Liu D., Jiang X. -Y., Zhou L. -S., Song J. -H., Zhang X. (2016). Effects of probiotics on intestinal mucosa barrier in patients with colorectal cancer after operation: meta-analysis of randomized controlled trials. Medicine 95:e3342. 10.1097/MD.0000000000003342, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. F., Kim H. -M., Lim S., Chung M. -J., Lim C. -Y., Koo B. -S., et al. (2020). Effect of probiotic administration on gut microbiota and depressive behaviors in mice. Daru 28, 181–189. 10.1007/s40199-020-00329-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Ma Y., Moyer M. P., Zhang P., Shi C., Qin H. (2012). Involvement of the mannose receptor and p38 mitogen-activated protein kinase signaling pathway of the microdomain of the integral membrane protein after enteropathogenic Escherichia coli infection. Infect. Immun. 80, 1343–1350. 10.1128/IAI.05930-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- London L. E., Kumar A. H., Wall R., Casey P. G., O'sullivan O., Shanahan F., et al. (2014). Exopolysaccharide-producing probiotic Lactobacilli reduce serum cholesterol and modify enteric microbiota in ApoE-deficient mice. J. Nutr. 144, 1956–1962. 10.3945/jn.114.191627, PMID: [DOI] [PubMed] [Google Scholar]
- López-Moreno A., Suárez A., Avanzi C., Monteoliva-Sánchez M., Aguilera M. (2020). Probiotic strains and intervention total doses for modulating obesity-related microbiota dysbiosis: a systematic review and meta-analysis. Nutrients 12:1921. 10.3390/nu12071921, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye H. -S., Rusul G., Liong M. -T. (2010). Removal of cholesterol by lactobacilli via incorporation and conversion to coprostanol. J. Dairy Sci. 93, 1383–1392. 10.3168/jds.2009-2574, PMID: [DOI] [PubMed] [Google Scholar]
- Macouzet M., Lee B., Robert N. (2009). Production of conjugated linoleic acid by probiotic Lactobacillus acidophilus La-5. J. Appl. Microbiol. 106, 1886–1891. 10.1111/j.1365-2672.2009.04164.x, PMID: [DOI] [PubMed] [Google Scholar]
- Madden U. A., Osweiler G. D., Knipe L., Beran G. W., Beitz D. C. (1999). Effects of Eubacterium coprostanoligenes and Lactobacillus on pH, lipid content, and cholesterol of fermented pork and mutton sausage-type mixes. J. Food Sci. 64, 903–908. 10.1111/j.1365-2621.1999.tb15937.x [DOI] [Google Scholar]
- Marcil V. R., Delvin E., Garofalo C., Levy E. (2003). Butyrate impairs lipid transport by inhibiting microsomal triglyceride transfer protein in Caco-2 cells. J. Nutr. 133, 2180–2183. 10.1093/jn/133.7.2180, PMID: [DOI] [PubMed] [Google Scholar]
- Marco M. L., De Vries M. C., Wels M., Molenaar D., Mangell P., Ahrne S., et al. (2010). Convergence in probiotic Lactobacillus gut-adaptive responses in humans and mice. ISME J. 4, 1481–1484. 10.1038/ismej.2010.61, PMID: [DOI] [PubMed] [Google Scholar]
- Martín R., Chamignon C., Mhedbi-Hajri N., Chain F., Derrien M., Escribano-Vázquez U., et al. (2019). The potential probiotic Lactobacillus rhamnosus CNCM I-3690 strain protects the intestinal barrier by stimulating both mucus production and cytoprotective response. Sci. Rep. 9:5398. 10.1038/s41598-019-41738-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milajerdi A., Mousavi S. M., Sadeghi A., Salari-Moghaddam A., Parohan M., Larijani B., et al. (2020). The effect of probiotics on inflammatory biomarkers: a meta-analysis of randomized clinical trials. Eur. J. Nutr. 59, 633–649. 10.1007/s00394-019-01931-8, PMID: [DOI] [PubMed] [Google Scholar]
- Miyauchi E., Morita H., Tanabe S. (2009). Lactobacillus rhamnosus alleviates intestinal barrier dysfunction in part by increasing expression of zonula occludens-1 and myosin light-chain kinase in vivo. J. Dairy Sci. 92, 2400–2408. 10.3168/jds.2008-1698, PMID: [DOI] [PubMed] [Google Scholar]
- Muccilli S., Wemhoff S., Restuccia C., Meinhardt F. (2013). Exoglucanase-encoding genes from three Wickerhamomyces anomalus killer strains isolated from olive brine. Yeast 30, 33–43. 10.1002/yea.2935, PMID: [DOI] [PubMed] [Google Scholar]
- Murphy E. F., Hooiveld G. J., Muller M., Calogero R. A., Cashman K. D. (2007). Conjugated linoleic acid alters global gene expression in human intestinal-like Caco-2 cells in an isomer-specific manner. J. Nutr. 137, 2359–2365. 10.1093/jn/137.11.2359, PMID: [DOI] [PubMed] [Google Scholar]
- O’connell Motherway M., Houston A., O’callaghan G., Reunanen J., O’brien F., O’driscoll T., et al. (2019). A Bifidobacterial pilus-associated protein promotes colonic epithelial proliferation. Mol. Microbiol. 111, 287–301. 10.1111/mmi.14155, PMID: [DOI] [PubMed] [Google Scholar]
- Okkers D. J., Dicks L. M. T., Silvester M., Joubert J. J., Odendaal H. J. (1999). Characterization of pentocin TV35b, a bacteriocin-like peptide isolated from Lactobacillus pentosus with a fungistatic effect on Candida albicans. J. Appl. Microbiol. 87, 726–734. 10.1046/j.1365-2672.1999.00918.x, PMID: [DOI] [PubMed] [Google Scholar]
- Oleskin A. V., Zhilenkova O. G., Shenderov B. A., Amerhanova A. M., Kudrin V. S., Klodt P. M. (2014). Lactic-acid bacteria supplement fermented dairy products with human behavior-modifying neuroactive compounds. J. Pharm. Nutr. Sci. 4, 199–206. 10.6000/1927-5951.2014.04.03.5 [DOI] [Google Scholar]
- O’shea E. F., Cotter P. D., Stanton C., Ross R. P., Hill C. (2012). Production of bioactive substances by intestinal bacteria as a basis for explaining probiotic mechanisms: bacteriocins and conjugated linoleic acid. Int. J. Food Microbiol. 152, 189–205. 10.1016/j.ijfoodmicro.2011.05.025, PMID: [DOI] [PubMed] [Google Scholar]
- Pasolli E., De Filippis F., Mauriello I. E., Cumbo F., Walsh A. M., Leech J., et al. (2020). Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome. Nat. Commun. 11:2610. 10.1038/s41467-020-16438-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M., Tabashsum Z., Patel P., Bernhardt C., Biswas D. (2018). Linoleic acids overproducing Lactobacillus casei limits growth, survival, and virulence of Salmonella Typhimurium and Enterohaemorrhagic Escherichia coli. Front. Microbiol. 9:2663. 10.3389/fmicb.2018.02663, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridmore R. D., Pittet A. -C., Praplan F., Cavadini C. (2008). Hydrogen peroxide production by Lactobacillus johnsonii NCC 533 and its role in anti-Salmonella activity. FEMS Microbiol. Lett. 283, 210–215. 10.1111/j.1574-6968.2008.01176.x, PMID: [DOI] [PubMed] [Google Scholar]
- Pujo J., Petitfils C., Le Faouder P., Eeckhaut V., Payros G., Maurel S., et al. (2020). Bacteria-derived long chain fatty acid exhibits anti-inflammatory properties in colitis. Gut 10.1136/gutjnl-2020-321173 [Epub ahead of print], PMID: [DOI] [PubMed]
- Qi X. -L., Wang J., Yue H. -Y., Wu S. -G., Zhang Y. -N., Ni H. -M., et al. (2018). Trans10, cis12-conjugated linoleic acid exhibits a stronger antioxidant capacity than cis9, trans11-conjugated linoleic acid in primary cultures of laying hen hepatocytes. Poult. Sci. 97, 4415–4424. 10.3382/ps/pey297, PMID: [DOI] [PubMed] [Google Scholar]
- Quintin J., Saeed S., Martens J. H. A., Giamarellos-Bourboulis E. J., Ifrim D. C., Logie C., et al. (2012). Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 12, 223–232. 10.1016/j.chom.2012.06.006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raqib R., Sarker P., Bergman P., Ara G., Lindh M., Sack D. A., et al. (2006). Improved outcome in shigellosis associated with butyrate induction of an endogenous peptide antibiotic. PNAS 103, 9178–9183. 10.1073/pnas.0602888103, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitmeier S., Kiessling S., Clavel T., List M., Almeida E. L., Ghosh T. S., et al. (2020). Arrhythmic gut microbiome signatures predict risk of type 2 diabetes. Cell Host Microbe 28, 258–272.e6. 10.1016/j.chom.2020.06.004, PMID: [DOI] [PubMed] [Google Scholar]
- Remus D. M. (2012). Molecular analysis of candidate probiotic effector molecules of Lactobacillus plantarum. PhD manuscript. Wageningen, The Netherlands: Wageningen University. 187.
- Rizzetto L., Ifrim D. C., Moretti S., Tocci N., Cheng S. -C., Quintin J., et al. (2016). Fungal chitin induces trained immunity in human monocytes during cross-talk of the host with Saccharomyces cerevisiae. J. Biol. Chem. 291, 7961–7972. 10.1074/jbc.M115.699645, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzetto L., Kuka M., De Filippo C., Cambi A., Netea M. G., Beltrame L., et al. (2010). Differential IL-17 production and mannan recognition contribute to fungal pathogenicity and commensalism. J. Immunol. 184, 4258–4268. 10.4049/jimmunol.0902972, PMID: [DOI] [PubMed] [Google Scholar]
- Santos A., San Mauro M., Abrusci C., Marquina D. (2007). Cwp2p, the plasma membrane receptor for Pichia membranifaciens killer toxin. Mol. Microbiol. 64, 831–843. 10.1111/j.1365-2958.2007.05702.x, PMID: [DOI] [PubMed] [Google Scholar]
- Schauber J., Svanholm C., Termén S., Iffland K., Menzel T., Scheppach W., et al. (2003). Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut 52, 735–741. 10.1136/gut.52.5.735, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sima P., Vannucci L., Vetvicka V. (2018). β-Glucans and cholesterol (review). Int. J. Mol. Med. 41, 1799–1808. 10.3892/ijmm.2018.3411, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Chandrashekharappa S., Bodduluri S. R., Baby B. V., Hegde B., Kotla N. G., et al. (2019). Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat. Commun. 10, 1–18. 10.1038/s41467-018-07859-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroosh A., Rankin C. R., Polytarchou C., Lokhandwala Z. A., Patel A., Chang L., et al. (2019). miR-24 is elevated in ulcerative colitis patients and regulates intestinal epithelial barrier function. Am. J. Pathol. 189, 1763–1774. 10.1016/j.ajpath.2019.05.018, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y. -W., Gu Y. -L., Mao X. -Q., Zhang L., Pei Y. -F. (2020). Effects of probiotics on type II diabetes mellitus: a meta-analysis. J. Transl. Med. 18:30. 10.1186/s12967-020-02213-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenorio-Jiménez C., Martínez-Ramírez M. J., Gil Á., Gómez-Llorente C. (2020). Effects of probiotics on metabolic syndrome: a systematic review of randomized clinical trials. Nutrients 12:124. 10.3390/nu12010124, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termén S., Tollin M., Rodriguez E., Sveinsdóttir S. H., Jóhannesson B., Cederlund A., et al. (2008). PU.1 and bacterial metabolites regulate the human gene CAMP encoding antimicrobial peptide LL-37 in colon epithelial cells. Mol. Immunol. 45, 3947–3955. 10.1016/j.molimm.2008.06.020, PMID: [DOI] [PubMed] [Google Scholar]
- Urner L. H., Liko I., Yen H. -Y., Hoi K. -K., Bolla J. R., Gault J., et al. (2020). Modular detergents tailor the purification and structural analysis of membrane proteins including G-protein coupled receptors. Nat. Commun. 11:564. 10.1038/s41467-020-14424-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usta-Gorgun B., Yilmaz-Ersan L. (2020). Short-chain fatty acids production by Bifidobacterium species in the presence of salep. Electron. J. Biotechnol. 47, 29–35. 10.1016/j.ejbt.2020.06.004 [DOI] [Google Scholar]
- Van Baarlen P., Troost F., Van Der Meer C., Hooiveld G., Boekschoten M., Brummer R. J., et al. (2011). Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. PNAS 108, 4562–4569. 10.1073/pnas.1000079107, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhratsky A. (2006). Glial calcium signaling in physiology and pathophysiology. Acta Pharmacol. Sin. 27, 773–780. 10.1111/j.1745-7254.2006.00396.x, PMID: [DOI] [PubMed] [Google Scholar]
- Villar-García J., Güerri-Fernández R., Moya A., González A., Hernández J. J., Lerma E., et al. (2017). Impact of probiotic Saccharomyces boulardii on the gut microbiome composition in HIV-treated patients: a double-blind, randomised, placebo-controlled trial. PLoS One 12:e0173802. 10.1371/journal.pone.0173802, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsman M. B., Castilla V., De Ruiz Holgado A. P., De Torres R. A., Sesma F., Coto C. E. (2003). Enterocin CRL35 inhibits late stages of HSV-1 and HSV-2 replication in vitro. Antivir. Res. 58, 17–24. 10.1016/S0166-3542(02)00099-2, PMID: [DOI] [PubMed] [Google Scholar]
- Wachsman M. B., Farías M. E., Takeda E., Sesma F., De Ruiz Holgado A. P., De Torres R. A., et al. (1999). Antiviral activity of enterocin CRL35 against herpesviruses. Int. J. Antimicrob. Agents 12, 293–299. 10.1016/S0924-8579(99)00078-3, PMID: [DOI] [PubMed] [Google Scholar]
- Wang J., Chen H., Yang B., Gu Z., Zhang H., Chen W., et al. (2016). Lactobacillus plantarum ZS2058 produces CLA to ameliorate DSS-induced acute colitis in mice. RSC Adv. 6, 14457–14464. 10.1039/C5RA24491A [DOI] [Google Scholar]
- Wang G., Huang W., Xia Y., Xiong Z., Ai L. (2019a). Cholesterol-lowering potentials of Lactobacillus strain overexpression of bile salt hydrolase on high cholesterol diet-induced hypercholesterolemic mice. Food Funct. 10, 1684–1695. 10.1039/c8fo02181c, PMID: [DOI] [PubMed] [Google Scholar]
- Wang Y., Qin Y., Zhang Y., Wu R., Li P. (2019b). Antibacterial mechanism of plantaricin LPL-1, a novel class IIa bacteriocin against Listeria monocytogenes. Food Cont. 97, 87–93. 10.1016/j.foodcont.2018.10.025 [DOI] [Google Scholar]
- Wang H., Xie Y., Zhang H., Jin J., Zhang H. (2020). Quantitative proteomic analysis reveals the influence of plantaricin BM-1 on metabolic pathways and peptidoglycan synthesis in Escherichia coli K12. PLoS One 15:e0231975. 10.1371/journal.pone.0231975, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann I., Breukink E., Van Kraaij C., Kuipers O. P., Bierbaum G., De Kruijff B., et al. (2001). Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276, 1772–1779. 10.1074/jbc.M006770200, PMID: [DOI] [PubMed] [Google Scholar]
- Yan F., Liu L., Dempsey P. J., Tsai Y. H., Raines E. W., Wilson C. L., et al. (2013). A Lactobacillus rhamnosus GG-derived soluble protein, p40, stimulates ligand release from intestinal epithelial cells to transactivate epidermal growth factor receptor. J. Biol. Chem. 288, 30742–30751. 10.1074/jbc.M113.492397, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Xie Y., Jin J., Liu H., Zhang H. (2019). Development and application of an active plastic multilayer film by coating a Plantaricin BM-1 for chilled meat preservation. J. Food Sci. 84, 1864–1870. 10.1111/1750-3841.14608, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M., Yan X., Weng W., Yang Y., Gao R., Liu M., et al. (2018). Micro integral membrane protein (MIMP), a newly discovered anti-inflammatory protein of Lactobacillus plantarum, enhances the gut barrier and modulates microbiota and inflammatory cytokines. Cell. Physiol. Biochem. 45, 474–490. 10.1159/000487027, PMID: [DOI] [PubMed] [Google Scholar]
- Younis G., Awad A., Dawod R. E., Yousef N. E. (2017). Antimicrobial activity of yeasts against some pathogenic bacteria. Vet. World 10, 979–983. 10.14202/vetworld.2017.979-983, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Liu L., Hao Y., Zhong S., Liu H., Han T., et al. (2013). Isolation and partial characterization of a bacteriocin produced by Lactobacillus plantarum BM-1 isolated from a traditionally fermented Chinese meat product. Microbiol. Immunol. 57, 746–755. 10.1111/1348-0421.12091, PMID: [DOI] [PubMed] [Google Scholar]
- Zhao W., Liu Y., Kwok L. -Y., Cai T., Zhang W. (2020). The immune regulatory role of Lactobacillus acidophilus: an updated meta-analysis of randomized controlled trials. Food Biosci. 36:100656. 10.1016/j.fbio.2020.100656 [DOI] [Google Scholar]
- Zomer A. L., Buist G., Larsen R., Kok J., Kuipers O. P. (2007). Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189, 1366–1381. 10.1128/JB.01013-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]