Abstract
The fracture resistance of cortical bone and matrix hydration are known to decline with advanced aging. However, the underlying mechanisms remain poorly understood, and so we investigated levels of matrix proteins and post-translational modifications (PTM) of collagen I in extracts from the tibia of 6-mo. and 20-mo. old BALB/c mice (female and male analysis done separately). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis revealed that the levels of collagen I deamidation at specific asparagine (Asn) and glutamine (Gln) residues significantly increased with age. Other non-enzymatic PTMs such as carboxymethylation of lysine (CML) were detected as well, but the relative abundance did not vary with age. No significant age-related differences in the abundance of hydroxylysine glycosylation sites were found, but hydroxylation levels at a few of the numerous lysine and proline hydroxylation sites significantly changed by a small amount with age. We performed molecular modeling and dynamics (MD) simulations for three triple helical fragments representing collagen I regions with prominent age-dependent increases in deamidation as identified by LC-MS/MS of male extracts. These 3 fragments included deamidated Asn and Gln residues as follows: 1) an Asn428 site of the α2(I) chain in which deamidation levels increased from 4.4% at 6-mo. to 8.1% at 20-mo., 2) an Asn983 site of the α2(I) chain with a deamidation increase from 18.3% to 36.8% with age and an Asn1052 site of the α1(I) chain with consistent deamidation levels of ~60% across the age groups, and 3) a Gln410 site of the α1(I) chain that went from no detectable deamidation at 6-mo. to 2.7% at 20-mo. and a neighboring Asn421 site of the same chain with an age-related deamidation increase from 3.6% to 16.3%. The deamidation levels at these sites inversely correlated with an estimate of toughness determined from three-point bending tests of the femur mid-diaphysis. MD revealed that the sidechains become more negatively charged at deamidated sites and that deamidation alters hydrogen bonding with water along the collagen backbone while increasing water interactions with the aspartic and glutamic acid sidechains. Our findings suggest a new mechanism of the age-dependent reduction in the fracture resistance of cortical bone whereby deamidation of Asn and Glu residues redistributes bound water within collagen I triple helix.
Keywords: post-translational modifications, deamidation, aging, molecular dynamics, molecular modeling, mass spectrometry, preclinical model, advanced glycation end-products
1. Introduction:
The ability of bone to resist fracture is dependent on its hierarchical organization that spans multiple length-scales [1]. At the ultrastructural level, the extracellular matrix (ECM) of bone is a composite of collagen type I, mineral, water, and various non-collagenous proteins (NCPs), all arranged to prevent microdamage from culminating into a fracture [2]. In addition to an overall loss in bone mass and a deterioration in the micro-structure of cortical and trabecular bone, disease- and age-related changes to the ECM also occur that contribute to bone fragility [3]. Because of the complexity in the heterogeneous, hierarchical organization of bone, understanding how aging affects the fracture resistance remains a challenge. This limited knowledge is an impediment to the identification of novel targets for treating the age-related decline in bone tissue quality.
Consisting of three polypeptide chains arranged in a triple helical structure, the collagen I protomer undergoes a series of post-translation modifications (PTMs) in order to form fibrils that mineralize. When enzymatic activity causing PTMs is impeded in vivo (e.g., treating rodents with β-aminopropionitrile to inhibit lysyl oxidase or causing a deficiency in vitamin C to impair hydroxylation), a decrease in collagen stability, bone strength, and bone toughness accompanies a reduction in enzymatic collagen crosslinks such as hydroxylysylpyridinoline [4–7]. Perhaps the most striking clinical example pointing to the importance of PTMs to the fracture resistance of bone is the genetic disease osteogenesis imperfecta (OI). Presenting as low-energy, brittle-like fractures early in life, OI can arise from mutations in COL1A1 and COL1A2 that encode the collagen α1(I) and α2(I) polypeptide chains. More recently, mutations causing OI have been identified in genes involved in collagen I processing via PTM formation (e.g., P3H1, CRTAP, LEPRE1) [8], thus suggesting collagen I PTMs play an important role in disease pathogenesis. While enzymatic PTMs are clearly important to the strength and toughness of bone, the specific age-related changes to the ECM that make bone less resistant to fracture are still poorly understood.
The thermal stability, tensile strength, and overall integrity of demineralized human cortical bone (i.e., the organic matrix) decreases with advanced aging [9–11]. Moreover, the temperature at which the organic matrix of bone starts to uncoil (shrinkage begins between 60 °C and 70 °C) and the change in stress per change in temperature (max-slope) during isometric tension of the organic matrix directly correlated with work-to-fracture and fracture toughness of human cortical bone from male donors [11]. In our recent study involving fracture toughness testing of human cortical bone from 50+ heterogeneous donors (females and males including those with disease) and similar hydrothermal isometric tension (HIT) testing of demineralized bone that neighbored the single-edge notched beam specimen, the max-slope was a significant contributor to fracture toughness helping independent measurements of volumetric bone mineral density and matrix-bound water explain the variance in crack initiation toughness [12]. Given this independence of max-slope and bound water, both a loss in network integrity of the organic matrix and a loss in ECM-bound water may contribute to the age-related decline in the fracture resistance of bone.
PTMs affect both the stability of collagen type I and the multiple interactions among collagen I, NCPs, mineral, and water within the ECM of bone [13–15]. Therefore, the age-related decrease in the thermal stability of the organic matrix and in the overall fracture resistance of cortical bone may be due to changes in enzymatic PTMs such as hydroxylation and glycosylation and/or non-enzymatic PTMs such as oxidation, advanced glycation end-products (AGEs), and deamidation of asparagine (Asn) and glutamine (Gln) residues. Of these modifications, AGE accumulation has garnered the most attention in the search for causes of age-related bone fragility with respect to the ECM. This is understandable since fluorescent AGEs are relatively easy to quantify, since the AGE crosslink pentosidine increases with age in bone [16,17], and since pentosidine and fluorescent AGEs in general are inversely associated with the post-yield toughness and fracture toughness of human cortical bone [10,18–20]. Paradoxically though, AGE crosslinks (e.g., pentosidine and glucosepane) increase the thermal stability of collagen I [21], which presumably confers greater resistance to deformation, and the accumulation of AGEs has been suggested to compensate for a loss of stabilizing enzymatic collagen crosslinks in OI bone, specifically oim/oim mouse [22]. Thus, other non-crosslinking modifications such as carboxymethyllysine (CML) may impair ECM integrity of bone. In support of this supposition, CML in human cortical bone has been observed to increase with age and inversely correlate with fracture toughness [23]. Also, incubating rat tail tendon in methylglyoxal to induce AGE accumulation was found to reduce relative sliding between adjacent collagen fibrils and reduce its stress relaxation behavior while stiffening the matrix (i.e., a loss of an energy dissipation mechanism) [24,25]. At present, the extent to which specific PTMs such as hydroxylation, different AGEs, and deamidation change with age in collagen type I of bone is not known. Moreover, the accumulation and structural impact of these PTMs at specific sites within the collagen I triple helix has not been assessed with respect to aging.
In our previous study of the BALB/c mouse model of aging, we observed that ultimate stress, toughness, and fracture toughness of the femur mid-shaft during three-point bending was lower in 20-mo. vs. 6-mo. old female and male mice [26]. Matrix-bound water of the femur also decreased with age. As a possible explanation of the age-related decrease in the toughness of bone at the material level, we hypothesized that age-related changes in PTM levels at specific sites would perturb interactions between water and collagen I triple helical structure. To test this hypothesis, we extracted proteins from the tibia of the same young adult and aged mice for liquid chromatography-tandem mass spectrometry analysis (LC-MS/MS). This allowed us to determine sequence positions and relative abundance (i.e., modified peptide per sum of modified and unmodified peptide) of multiple enzymatic and non-enzymatic PTMs in collagen I as a function of mouse age (i.e., 6-mo. vs. 20-mo.) so that molecular modeling and molecular dynamics (MD) simulations could be developed to illustrate the impact of an observed age-related change in PTMs on hydrogen bonding with water.
2. Materials and Methods
2.1. Animal care and tissue collection
BALB/c female and male mice were acquired from Charles River where the National Institute on Aging (USA) maintains a colony of aging mice and housed until 6-months or 20-months when the mice were deeply anesthetized and then euthanatized by cervical dislocation as previously described [26]. Upon removing the soft tissue from the tibia, the bones were flash-frozen in liquid nitrogen and stored at −80 °C.
2.2. Protein extraction from tibiae
Left tibiae (n=6 per age group per sex) were thawed on ice. After removing the proximal and distal ends, the bone marrow was flushed out in 2 steps: centrifugation at 4000 rpm for 5 min and repeated injections of phosphate buffered saline into the medullary canal until the liquid was clear. The flushed bones were then stored overnight at −80 °C before subsequently being pulverized using a freezer-mill (6770, SPEX SamplePrep, Metuchen, NJ). Powdered samples were placed in 0.3 mL of 10 mM Tris HCl buffer (pH of 7.5) containing 6 M guanidine hydrochloride (GdnCl), 50 mM ethylenediamine-tetraacetic acid (EDTA) and Halt’s protease inhibitor cocktail. Proteins were extracted from the samples for 72 hours at 4 °C under continuous rotation. Each sample was then centrifuged at 13,000 g for 15 minutes at 4 °C. The supernatant was collected and concentrated using a 3 kDa centrifugal filtration unit (Amicon® Ultra-4, Merck Millipore Ltd, Burlington, MA) to remove GdnCl and EDTA. After dilution with 10 mM Tris HCl buffer, the filtered concentration step was repeated three times to achieve complete buffer exchange. The protein concentration in the samples was measured using a Pierce™ BCA protein assay kit after diluting the samples at a 1:5 ratio (Thermo Fisher Scientific, Waltham, MA).
2.3. Analysis of bone matrix protein levels and post-translational modifications
The abundancies of major bone matrix proteins and the site-specific modifications in bone collagen I were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Samples were diluted with 100 mM ammonium bicarbonate, treated with dithiothreitol (DTT) to reduce disulfide bonds, cysteine residues were carbamidomethylated with iodoacetamide, and proteins were digested with sequencing-grade trypsin overnight at 37°C. Following digestion, peptides were diluted with 0.1% formic acid prior to LC-MS/MS. As previously described in [27], samples were spiked with solutions of three synthetic peptides to be used as internal reference standards during data analysis. The three peptides, bradykinin (Sigma B4184), angiotensin I (Sigma A9650), and angiotensin II (Sigma A8846) were reconstituted in 0.1% formic acid, and serial dilutions were made to generate 5 pmol/μL solutions of the individual peptides. Using the individual peptide solutions, a combined peptide stock solution was then generated and spiked into each of the samples to arrive at approximately 50 fmol/μL of each peptide synthetic standard. An analytical column was packed with 20 cm of C18 reverse phase material (Jupiter, 3 μm beads, 300Å, Phenomenox). Peptides were loaded onto this capillary, reverse phase, analytical column (360 μm O.D. × 100 μm I.D.) using a Dionex Ultimate 3000 nanoLC and autosampler. The mobile phase solvents consisted of 0.1% formic acid, 99.9% water (solvent A) and 0.1% formic acid, 99.9% acetonitrile (solvent B). Peptides were gradient-eluted at a flow rate of 350 nL/min using a 95-minute gradient. The gradient consisted of the following steps: 2%−45% B from 1–80 min; 45%−90% B from 80–85 min; constant 90% B from 85–86 min; 90% B back to 2% B from 86–87 min; constant 2% B from 87–95 min (column re-equilibration).
A Q Exactive Plus mass spectrometer (Thermo Scientific), equipped with a nanoelectrospray ionization source, was used to mass analyze the eluting peptides using a data-dependent method. The instrument method consisted of MS1 using an MS AGC target value of 3 × 106, followed by up to 20 MS/MS scans of the most abundant ions detected in the preceding MS scan. A maximum MS/MS ion time of 100 ms was used with a MS2 AGC target of 1 × 105. Higher-energy collisional dissociation was set to 28, and peptide match and isotope exclusion were enabled.
Database searches were performed using SEQUEST as previously described [28], and results were assembled in Scaffold v 4.3.2 (Proteome Software) with minimum filtering criteria of 95% peptide probability and 99% protein probability. Searches were configured to use variable protein residue modifications, as follows: carbamidomethylation of cysteine (ΔM = 57.0215); oxidation of methionine, proline and lysine (ΔM = 15.9949); galactosylation of hydroxylysine (ΔM = 178.0477); glycosylgalactosylation of hydroxylysine (ΔM = 340.1006); glycation of lysine (ΔM = 162.0528); carboxymethylation of lysine (ΔM = 58.0055); 5-hydro-imidazolone (G-H1) on arginine (ΔM = 39.9949); 5-hydro-5-methyl-imidazolone (MG-H1) on arginine (ΔM = 54.0106) and deamidation of asparagine and glutamine (ΔM = 0.9840). Sites of modification were confirmed by manual interpretation of the raw tandem mass spectra using QualBrowser software (Xcalibur 2.2, Thermo Scientific).
Accurate mass measurements of the tryptic and internal standard peptides were used to generate extracted ion chromatograms (XICs). A window of 10 ppm around the theoretical monoisotopic m/z values of the observed precursor ions was utilized for making XICs of the standards and the unmodified and modified peptide forms. Using QualBrowser (Xcalibur2.1.0), the integrated area under the curve (AUC) for each XIC peak was determined. Peptide peak areas were then normalized to the average of the peak areas of the three internal standards. To determine the relative abundance of modified collagen I peptides, XICs were similarly generated for the identified modified and the corresponding unmodified collagen I peptide forms. The relative abundance of each modified peptide was then calculated as a percentage of the summed AUC obtained for the modified and unmodified peptides. Because our LC-MSMS methodology has not been optimized for separation of aspartate from iso-aspartate or α-glutamate from γ-glutamate, when detected as separate peaks, the AUCs for these deamidation products were added together for quantitation of a total deamidation product.
2.4. Statistical analysis
Due the sample size being less than 30 mice, differences between young (n=6) and old mice (n=6) were tested within each sex for statistical significance by individual Mann-Whitney U tests (Prism 8, GraphPad Software, LLC) unless otherwise noted in the supplemental materials. To determine whether the relative abundance of deamidation at selected sites was linearly related to the toughness of cortical bone as determined by three-point bending tests of the left femur mid-diaphysis reported in our previous study [26], ordinary least squares regressions were performed within each sex. In the event that the residuals did not pass the Anderson-Darling normality test or the test of homoscedasticity (Prism 8, GraphPad Software, LLC), the regression was re-analyze using bootstrap with 1000 replicates (STATA v11).
2.5. Molecular dynamics simulations and modeling
MD simulations utilized the AMBER 16 program suite [29] and the ff14SB parameter set [30,31]. Starting coordinates were derived from the X-ray structure of a hydrated collagen peptide (1CGD) [32], and the MD simulations were based on 3 selected 30-amino acid collagen I triple helix regions where age-related differences in the relative abundance of deamidation sites were detected in α1 and/or α2 collagen I chains (Dashed boxes in Fig. 1). Mutations were introduced with PyMOL (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.) to generate a total of 6 individual MD systems: 3 for each MS identified sequence in the native state and 3 for the corresponding deamidated sequence. Following convention, N and Q represent the original asparagine and glutamine residues, respectively, in an unmodified molecule, and n(D) or q(E) represent their respective deamidation products. Charge derivation of the identified hydroxylysine modifications (Fig. 1) was determined by the Restrained Electro Static Potential (RESP) method [33] using Gaussian 03 software (Gaussian, Inc., Wallingford, CT). Each system was charge neutralize with the addition of either sodium or chloride counter ions along a 1.0 Å Coulombic potential grid. Monovalent ion parameter sets were used as described previously [34]. MD systems were solvated in a truncated octahedral box using the TIP3P water model to a distance of 8.0 Å (Fig. S1). Energy minimization and solvent equilibration were achieved under periodic boundary conditions. To avoid dissociation and fraying of the helices, artificial distance restraints were introduced between planer oxygen and nitrogen atoms of terminal residues on different strands between 1.3 to 3.4 Å with a force constant of 32 kcal/mol. Restraint files are provided in supplemental materials. Production calculations were conducted at constant pressure for a time period of 100 ns. Temperature was maintained at 300 K by a Langevin coupling algorithm using a collision frequency of 0.5 ps−1 [35,36]. Electrostatic interactions were treated with the particle mesh Ewald (PME) method [37], and the SHAKE algorithm [38] was used to constrain bond lengths involving hydrogen atoms. A non-bonded cutoff of 9.0 Å was set during all minimization, equilibration, and production stages. Resultant trajectories were analyzed using the CPPTRAJ module of AMBER16. Molecular modeling and trajectory visualization were performed with PyMOL. The Adaptive Poisson-Boltzmann Solver (APBS) was used to generate electrostatic topologies for visualization with PyMOL [39].
Figure 1. Sites of post-translational modifications (PTMs) in mouse type I collagen molecule as determined from extracts of homogenized tibia that were acquired from (A) male and (B) female BALB/c mice.
The α1(I) chain and α2(I) chain amino acid sequences come from UniProtKB - P11087; CO1A1_MOUSE and UniProtKB - Q01149; CO1A2_MOUSE (uniprot.org). Residue numbering for α1(I) is above the sequence, and numbering for α2(I) is below the sequence in grey font. N-terminal and C-terminal telopeptides are surrounded by rectangular boxes. Deamidation sites (N → n(D) or Q → q(E)) are shown in red font and highlighted in yellow; hydroxylation sites are shown in cyan blue with either Pro (P) or Lys (K) underlined; Glycosylated (glucosyl- and glucosylgalactosyl-) hydroxylysine sites (GHL and GGHL) are marked in purple font; Carboxymethylated lysine (CML) sites are marked in green font; 5-hydro-5-methyl-imidazolone-modified arginine sites (MG-H1) are marked in blue font; and Sites with a PTM are bolded if located on the α1(I). If a residue is underlined and marked by a color different than cyan blue, then overlapping peptide sequences were detected in which a residue was either hydroxylated (OH) or had another enzymatic PTM. *p<0.05 for significant age-related differences in the relative abundance. The font color of the star (*) matches the font color of the PTM unless a deamidation site was only detected in the bones of old mice in which case the font is black.
3. Results:
3.1. Overall protein composition in the extracts of mouse bone by LC-MS/MS
Across all the analyzed samples (N=24), ~360 proteins were identified in the bone extracts based on the identification of at least two unique peptides per protein with a >95% probability. Based on total spectrum count, the relative levels of matrix and matrix-bound proteins constituted about 28% of the total amount of all the extracted bone protein. The major non-matrix proteins identified in mouse bone extracts were actin, myosin, troponin, and serum albumin which were not further analyzed.
As expected, the most abundant protein identified in the bone extracts was collagen I (both α1 and α2 chains), which constituted about 65% of total organic matrix proteins based on total spectrum counts. Non-collagenous matrix proteins (NCPs) constituted an additional 10%. Prominent NCPs, osteopontin and osteonectin, constituted 1.0% to 1.5% of the spectrum counts of major ECM proteins (listed in Table S1). Another prominent NCP, osteocalcin, had to be manually identified and was not included in the total spectrum count. The remaining 25% of the matrix protein levels included mostly collagens type II, III, V, VI, XI and XII.
There were some age-related differences in the matrix protein composition as determined by total spectral counts (Table S1). Most notable changes involved collagen type II in males and periostin in females, both showing decreased levels with aging (Table S1). Because these results are not direct measurements of protein levels (e.g., mol of protein per mass of organic matrix), they provide only a general outline of bone matrix composition and require further studies. In this paper, we focused our investigation on collagen I, the major bone matrix constituent.
Based on the abundance of selected tryptic peptides from different sequence regions of collagen I normalized to internal standards, the levels of collagen type I did not vary with the age of the BALB/c mice (Table S2). Since the triple helical region is the basic structural unit of mineralized collagen fibrils, we focused on the quantitation of PTMs within this region of collagen I. Our extraction method and LC-MS/MS analysis resulted in identification of collagen I peptides that covered 91% to 95% of the amino acid sequence of the triple helical region in both male (Fig. 1A) and female (Fig. 1B) bone extracts.
Protein extraction protocols may introduce experimental bias due to preferential extraction of specific matrix components. To address this potential bias, following the initial extraction, the pellets from the female bones were demineralized (0.5 M EDTA for 6 days), heat-denatured (85 °C for 1 h), re-extracted (GdnCl for 72 h at 5 °C), and analyzed by LC-MS/MS. Analysis of these extracts showed essentially the same results in matrix composition and major PTMs as with the initial extracts, thus suggesting adequate representative extraction of matrix components.
3.2. Age-related site-specific increase in deamidation in collagen I.
Compared to sex- and strain-matched young adult mice, deamidation levels at specific Asn (N) and Gln (Q) residues were significantly higher on both α1(I) and α2(I) chains with age (Table 1). For the majority of deamidated sites, the relative abundance was ~0.5% for 6-mo. old mice and ~2.5% for 20-mo. old mice (Table S3). In the extracts from female mouse tibiae, Gln446 of the α1(I) chain [see peptide GEPGATGVq446GPPGPAGEEGKR in Table 1] and four Asn sites of the α2(I) chain [e.g., GEPGAPGEn213GTPGQAGAR] were the most abundantly modified residues exhibiting modification levels >60%. However, the levels of deamidation at these sites did not significantly change between the age groups (Table 1). Other highly abundant deamidated sites of the α1(I) chain in the male extracts [n218GDDGEAGKPGR and n1052GDRGETGPAGPAGPIGPAGAR] and three Asn sites of the α2(I) chain [e.g., GEVGPAGPn729GFAGPAGAAGQPGAK], also did not significantly vary between the two age groups (Table 1). Rather, significant age-related changes in deamidation occurred at a Gln1091 residue of α1(I) [GDKGETGEq1091GDR] and two Asn residues of α2(I) [AGVMGPPGn428R and HGn983RGEPGPAGSVGPVGAVGPR] in the bone extracts of both female and male tibia (Table 1). There were a few sex-related differences in the deamidation sites for which an age-related increase was detected, namely the following α1(I) sites: n218GDDGEAGKPGR and GNSGEPGAPGn430KGDTGAK in females and Gn421SGEPGAPGNK in males (Table 1).
Table 1.
Quantitation of deamidated tryptic peptides of collagen I with a relative abundance (100% × modified peptide / (modified peptide + unmodified peptide)) >5% in bone matrix of either 6-month or 20-month old BALB/c mice (mean ± SD).
| Peptide sequence | Female | Male | ||||
|---|---|---|---|---|---|---|
| 6-mo. | 20-mo. | Fold a | 6-mo. | 20-mo. | Fold | |
| Collagen α1(I) chain b | ||||||
| n218GDDGEAGKPGR | 32.9±3.4 | 40.4±4.5* | 1.23 | 76.2±1.3 | 75.7±2.6 | 0.99 |
| Gn421SGEPGAPGNK c | 5.7±0.8 | 5.3±0.8 | 0.94 | 3.6±1.0 | 16.3±2.8* | 4.56 |
| GNSGEPGAPGn430KGDTGAK | 18.9±6.7 | 40.4±10.1* | 2.14 | Not detected | ||
| GEPGATGVq446GPPGPAGEEGKR | 64.8±3.6 | 64.7±5.8 | 1.00 | 0.7±0.05 | 1.7±0.2* | 2.55 |
| n1052GDRGETGPAGPAGPIGPAGAR d | 42.6±4.3 | 48.2±5.4 | 1.13 | 64.7±3.7 | 62.2±3.3 | 0.96 |
| GDKGETGEq1091GDR | 1.6±0.3 | 6.8±0.9* | 4.19 | 1.9±0.3 | 7.3±1.0* | 3.92 |
| GDKGETGEq1091GDR | ||||||
| Collagen α2(I) chain | ||||||
| GEPGAPGEn213GTPGQAGAR | 90.0±1.1 | 86.8±2.7 | 0.96 | Not detected | ||
| AGVMGPPGn428R e | 2.8±0.4 | 6.3±0.9* | 2.28 | 4.4±0.6 | 8.1±0.8* | 1.86 |
| GPn441GDAGRPGEPGLMGPR | 83.9±2.8 | 86.5±2.5 | 1.03 | 89.7±3.6 | 88.3±2.0 | 0.98 |
| GLPGSPGn464VGPSGK | Not detected | 2.8±0.3 | 6.1±0.4* | 2.22 | ||
| GAPGPDGNn534GAQGPPGPQGVQGGK | 84.3±1.6 | 85.9±1.3 | 1.02 | 86.3±3.0 | 85.2±1.4 | 0.99 |
| GEVGPAGPn729GFAGPAGAAGQPGAK | Not detected | 70.8±7.8 | 68.7±3.6 | 0.97 | ||
| GPPGAVGSPGVn924GAPGEAGR | 61.1±5.3 | 60.3±1.8 | 0.99 | Not detected | ||
| HGn983RGEPGPAGSVGPVGAVGPR | 18.8±7.0 | 35.4±6.5* | 1.89 | 18.3±1.5 | 36.8±5.7* | 2.01 |
| HGn983RGEPGPAGSVGPVGAVGPR d | ||||||
Fold is the relative change as the mean of 20-mo. per mean of 6-mo.
The amino acid positions are given for full α1(I) chain and α2(I) chain sequences (UniProtKB - P11087, CO1A1_MOUSE and UniProtKB - Q01149, CO1A2_MOUSE); The superscript indicates the position number; Hydroxylated Pro (P) and Lys (K) residues are underlined, and deamidated Asn (N) and Gln (Q) residues are indicated by a corresponding lower-case letter in bold font.
p<0.05 from Mann-Whitney test comparing 20 mo. old vs. 6 mo. old (n=6/group).
3.3. A small number of AGE-modified and glycosylated sites were detected in collagen I
We detected two AGEs in helical collagen I from male bone extracts. Methylglyoxal-derived hydroimidazolone-1 (MG-H1) was identified at Arg1025 of α1(I) chain and Arg681 of α2(I) chain (blue font in Fig. 1). Since corresponding unmodified peptides were not detected, the relative abundance of this AGE modification could not be determined. Another AGE, carboxymethyllysine (CML), was detected at Lys731 and Lys740 of the α1(I) chain (green font in Fig. 1). The relative abundance however was less than 4% and did not significantly vary with age (Fig. S2). For both male and female mouse bone extracts, enzymatic PTMs galactosyl-hydroxylysine (GHL) at Lys254 and Lys266 of α1(I) and at Lys270 of α2(I) and glucosyl-galactosyl-hydroxylysine (GGHL) at Lys254 of α1(I) were detected (purple font in Fig. 1). The relative abundance of GHL sites was <50% and did not significantly vary with age (Tables S5-S8).
3.4. Hydroxylation of lysine residues at specific collagen I sites.
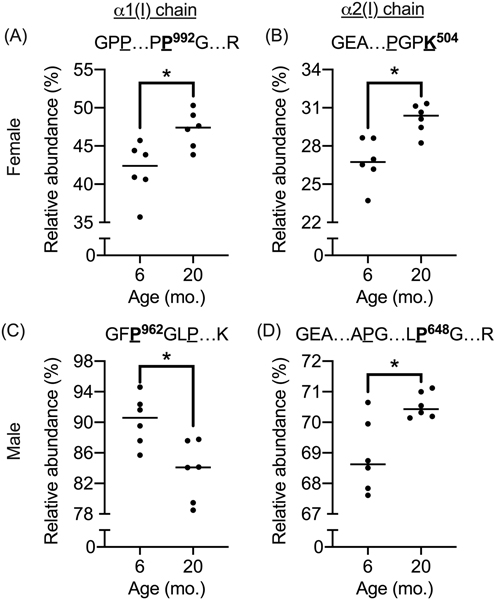
Multiple hydroxylation sites were identified in collagen I extracts from bone (cyan font in Fig. 1). Most of the detected hydroxylation sites had an apparent relative abundance of 100% in both age groups (i.e., no corresponding unmodified peptides were detected). However, at several sites, the relative abundance of hydroxyproline (Hyp) and hydroxylysine (Hyl) was variable primarily ranging from 10.7% to 98.5% (Tables S4-S7). For a small number of these sites (e.g., Pro992 and Pro962 on α1(I) and Lys504 and Pro648 on α2(I)), relative hydroxylation levels were significantly different between the age groups (Fig. 2).
Figure 2. Age-related differences in hydroxylation abundance.
Male: The relative abundance in hydroxylation (OH) of proline at position 962 of the α1(I) chain was lower (A), while the abundance in OH of proline at position 648 of the α2(I) chain was higher (B) with age. Female: The relative abundance in OH of proline at position 992 of the α1(I) chain (C) and in OH of lysine at 504 were both higher with age.
3.5. Toughness inversely correlated with the relative abundance in deamidation.
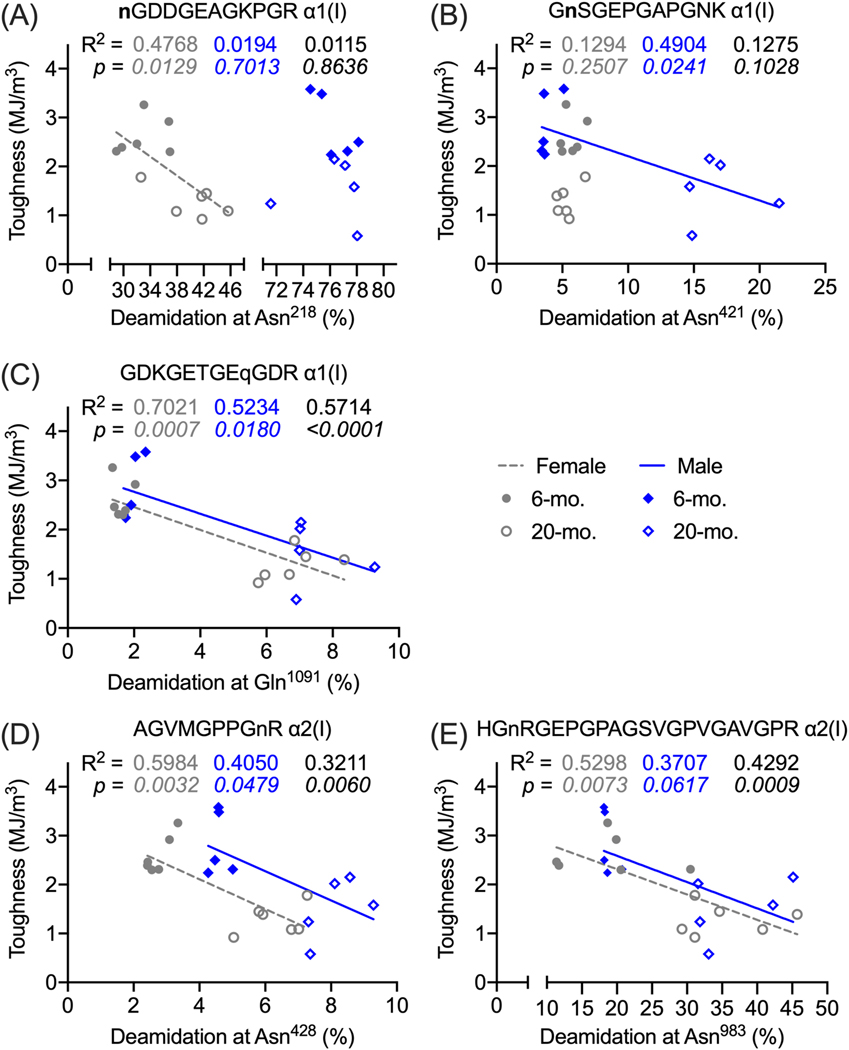
As determined from quasi-static, load-to-failure, three-point bending of the left femur [26], toughness was inversely related to deamidation levels at specific collagen I sites that were quantitated in extracts of the left tibia (Fig. 3A-E and Table S8). The strengths of these linear relationships for Gln1091 of α1(I), Asn428 of α2(I), and Asn983 of α2(I) were moderate, except for the weak association between toughness and relative abundance of Asn983 in male mice (R2 = 0.3707, p=0.0617). When pooling male and female, the slope of this inverse relationship was significantly different than zero (Fig. 3E). The slope of the linear relationship appears to be similar between the sexes, but there is a positive offset in the y-intercept at 0 going from female to male bone (Fig. 3C-E). Whereas the relative abundance of deamidation at Asn421 of α1(I) was linearly related to toughness in the male bone, no such relationship was apparent in the female bone (Fig. 3B). In contrast, for deamidation levels at Asn218 of α1(I), the negative linear relationship was significant in the female but not in male bone (Fig. 3A).
Figure 3. Linear regressions between bone toughness and levels of collagen I deamidation at selected deamidation sites in male and female mice.
Toughness was determined from three-point bending tests of the left femur mid-diaphysis, and the relative abundance [100 × modified peptide \ (unmodified peptide + modified peptide) > 5%] was determined from quantitation of deamidated tryptic peptides of collagen I from the left tibia. Toughness decreased as the relative abundance increased for Asn218 (A), Asn421 (B), and Gln1091 (C) on the α1(I) chain and for Asn428 (D) and Asn983 (E) on the α2(I) chain. In each the graph, the coefficient of determination (R2) above and the p-value of the null hypothesis (slope = 0) below are given for female, male, and combined female and male. The slope of the linear line was not different from zero for Asn421 (female) and Asn218 (male). For other major and minor (<5%) deamidation sites in which relative abundance changed with age, see Table S8.
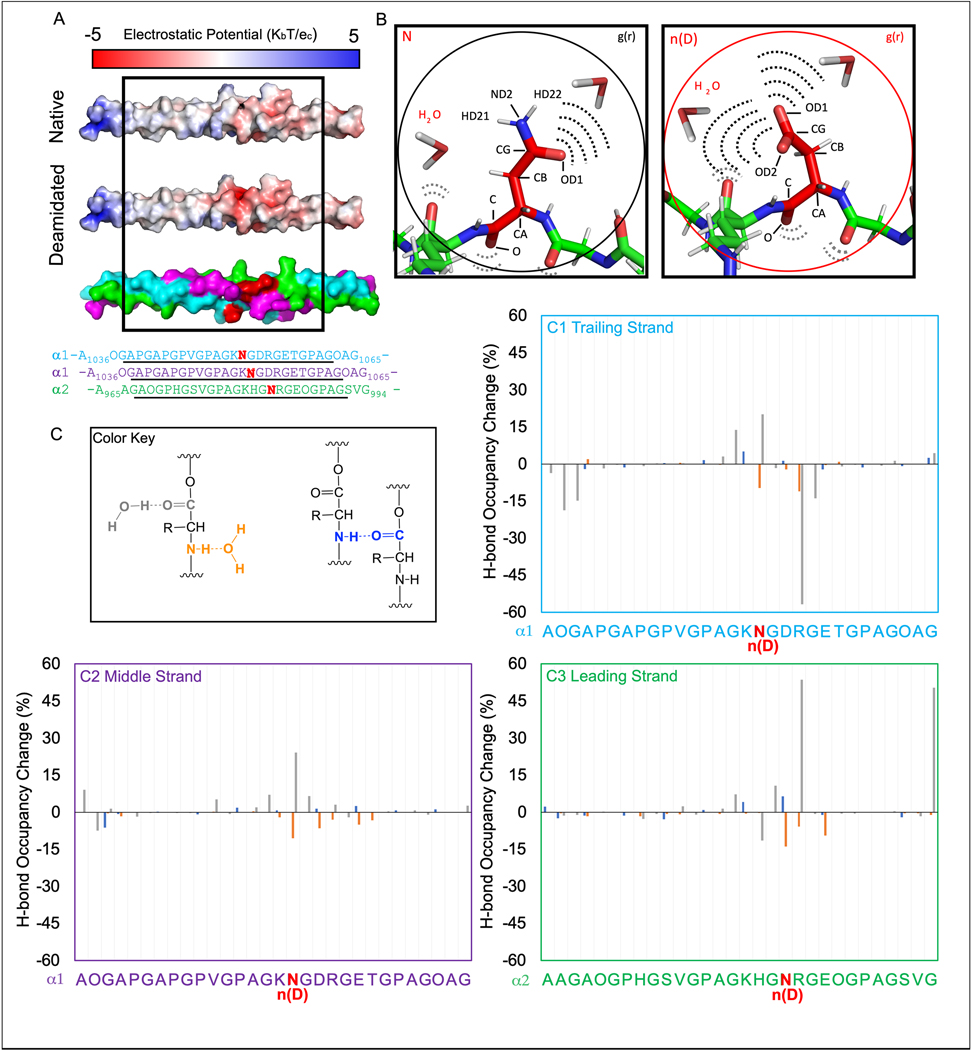
3.6. Molecular modeling and molecular dynamics simulations indicate a rearrangement of hydrogen bonding patterns at deamidation sites
To determine the impact of deamidation on collagen interactions, we employed molecular modeling and dynamics (MD) simulations of three triple helical fragments of collagen I with significant age-dependent changes in deamidation levels as determined by LC-MS/MS analysis in explicit solvent. Molecular modeling revealed that deamidation at Asn1052 of the α1(I) chains and Asn983 of the α2(I) chain produced an estimated 2-fold change in electrostatic potential from a mildly negative to a highly negative surface (Fig. 4A). A complete charge reversal occurred upon deamidation at Asn428 of α2(I) causing a 5-fold change in electrostatic potential from positive to negative (Fig. S3A). A similar charge reversal was observed near the deamidation site at Asn421 of the α1(I) chains, while there was a minimal change from neutral to negative electrostatic surface near deamidated Gln410 of the same α1 chains (Fig. S4A).
Figure 4. Age-dependent deamidation of specific asparagine residues can alter water distribution in bone collagen I.
A comparative modeling analysis demonstrated that deamidation (red bold font in the sequence) significantly impacted the electrostatic topology of the triple helix (A). An illustration of the native (left) and deamidated sidechain (right) shows the labeled atoms (e.g., CG) interacting with water (B). In quantitating hydrogen bonding patterns, a color key was used (C) to indicate amide-to-H2O (orange), carboxyl-to-H2O (grey), and intermolecular amide-carboxyl (blue) motifs in the 24 core residues of the trailing (C1), middle (C2), and leading strands (C3). The backbone hydrogen bonding values are reported as percent bond occupancy of the deamidated MD system minus the corresponding percent occupation of the native MD system. As such, negative values represent a loss of hydrogen bonding upon deamidation while positive values represent a gain of hydrogen bonding.
We then employed MD to predict how deamidation affects collagen structural dynamics and hydrogen bonding patterns. This involved radial distribution analyses centered on the CG atom (labeled in Fig. 3B) of either a native N sidechain or a deamidated n(D) sidechain. We consistently observed altered water distribution within a 2.5 – 8 Å radius from the CG atom in N vs. n(D) sidechain (Fig. S5), while beyond a radial distance of 10 Å from the CG atom, the atomic density of water molecules was similar between N and n(D) sidechains (Fig. S5B-D). In summary, all three MD simulations predicted that deamidation altered the hydrogen bonding patterns between water and the peripheral solute atoms of sidechains. Specifically, there was a loss of solute atomic density in the 2.5 Å range and redistribution of solvent (water) molecules in the 2.5–8 Å range relative to the sidechain.
To gain more specific molecular insights, we quantitated MD predicted bonding occupancies of inter- and intra-backbone carboxyl and amide groups (Fig. 4C, Fig. S3C, and Fig. S4C) as well as bonding occupancies of sidechain functional groups with water (Table 3, Table S9, and Table S10). MD predicted altered backbone hydrogen bonding patterns centered around the deamidated residues and their nearest neighbor residues. There were comparatively small changes (<10%) in hydrogen bonding between amide (NH) and carboxyl group (CO) on an adjacent strand (blue bars in Fig. 4C). The NH group of the backbone amide to water hydrogen bonding patterns (orange bars in Fig. 4C) were similar in magnitude (< 15%) but were all negative indicating that the backbone amide groups lost coordinated water contacts near the deamidation site. Larger perturbations (up to 60%) were observed for carboxyl to water hydrogen bonds (grey bars in Fig. 4C), representing both gains and losses of coordinated water molecules, depending on the collagen chain. Similar patterns were observed in the model of the single deamidated Asn site, designated as n(D), in the α2 chain; however, the magnitudes of the perturbations were much smaller (Fig. S3C). The model of deamidated Asn and Gln sites, n(D) and q(E), in the α1 chains revealed positive and negative changes in hydrogen bonding, which were spread somewhat more broadly along the triple helix (Fig. S4C). In all three samples, the deamidation sites appeared to be the origin of altered binding patterns. (Note: Because artificial restraints were used to constrain the triple helices from fraying, results for the terminal three residues are considered biased and are not reported.)
Table 3.
MDASI-I Scores by TUGT groups at discharge and post-operative follow-up time points.
| Time points (average days from surgery) | MDASI-I | Mean MDASI-I Scores by TUGT grouping |
P-value | |||||
|---|---|---|---|---|---|---|---|---|
| Normal + Frail (<20 s) |
Prolonged (>20 s) |
|||||||
| N | Mean | SD | N | Mean | SD | |||
| Discharge (3.1 ± 1.1 days) | Activity | 99 | 5.14 | 3.24 | 27 | 6.26 | 2.51 | 0.1020 |
| Work | 95 | 5.45 | 3.89 | 27 | 6.59 | 3.89 | 0.1544 | |
| Walking | 99 | 2.79 | 2.46 | 27 | 4.89 | 2.64 | 0.0004 | |
| Did not complete TUGT due to patient refusal | Completed TUGT | |||||||
|---|---|---|---|---|---|---|---|---|
| Discharge (3.1 ± 1.1 days) | Activity | 24 | 7 | 2.84 | 126 | 5.38 | 3.12 | 0.0204 |
| Work | 23 | 6.17 | 3.86 | 122 | 5.7 | 3.91 | 0.6635 | |
| Walking | 25 | 3.72 | 3.01 | 126 | 3.24 | 2.63 | 0.4719 | |
| Time points (Average Days from Surgery) | MDASI-I | Mean MDASI-I Scores by TUGT grouping |
P-value | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Did not complete TGUT | Normal |
Frail |
Prolonged |
|||||||||||
| (≤10 s) |
(11 to 20 s) |
(>20 s) |
||||||||||||
| N | Mean | SD | N | Mean | SD | N | Mean | SD | N | Mean | SD | |||
| Discharge (3.1 ± 1.1 days) | Activity | 40 | 6.08 | 3.28 | 7 | 3.86 | 3.29 | 92 | 5.24 | 3.23 | 27 | 6.26 | 2.51 | 0.1474 |
| Work | 37 | 5.89 | 3.93 | 6 | 2.33 | 1.75 | 89 | 5.66 | 3.91 | 27 | 6.59 | 3.89 | 0.1261 | |
| Walking | 41 | 3.46 | 2.96 | 7 | 0.71 | 1.25 | 92 | 2.95 | 2.46 | 27 | 4.89 | 2.64 | 0.0006 | |
| Post-operative follow-up (29.1 ± 10.8 days) | Activity | 34 | 3.06 | 3.12 | 77 | 2.32 | 2.48 | 56 | 2.73 | 2.59 | 2 | 3.50 | 4.95 | 0.0331 |
| Work | 34 | 3.35 | 2.84 | 76 | 2.36 | 2.62 | 56 | 2.95 | 2.75 | 2 | 4.00 | 5.66 | 0.0823 | |
| Walking | 34 | 2.18 | 2.92 | 77 | 1.18 | 1.9 | 56 | 1.86 | 2.2 | 2 | 3.50 | 4.95 | 0.0544 | |
MDASI-I: interference items of MD Anderson Symptom Inventory module, TUGT: time up & go test.
Next, we quantitated the occupancy of hydrogen bonds between sidechain R group donors/acceptors (OD and HD atoms in Fig. 4B) and water for both N and n(D) residues. When three deamidated residues were analyzed, the native sidechains of Asn1052 in the α1(I) trailing and middle strands and Asn983 in the α2(I) leading strand each increased significantly relative to the deamidated systems (n(D)). A simple interpretation of these data is that N residues typically coordinated 1– 2 water molecules per donor/acceptor (i.e., occupancy ~ 1, for a sum of ~3 waters per N residue), while the more electronegative n(D) sidechains are predicted to coordinate a total of 4–5 water molecules, (occupancy > 1 in Table 2). Similarly, in the single site model the sidechain of Asn429 in the leading strand went from interacting on average with 2 water molecules to interacting with 4–5 water molecules upon deamidation (Table S9). The same effect was observed at Asn421 and Gln411 sidechains in the trailing and middle strands when 4 sites were deamidated in the model (Table S10).
Table 2.
Hydrogen bond occupancies and frame counts for native and deamidated sidechains at Asn1052 of α1(I) and Asn983 of α2(I).
| Acceptor/Donor Atoma | Trailing Strand | Middle Strand | Leading Strand | |||
|---|---|---|---|---|---|---|
| Frames | Occupancy | Frames | Occupancy | Frames | Occupancy | |
| N OD1 | 20781 | 0.83 | 21762 | 0.87 | 31502 | 1.26 |
| N HD21 | 9753 | 0.39 | 9921 | 0.40 | 9129 | 0.37 |
| N HD22 | 11447 | 0.46 | 11256 | 0.45 | 12395 | 0.50 |
| n(D) OD1 | 57158 | 2.29 | 49515 | 1.98 | 61687 | 2.47 |
| n(D) OD2 | 47961 | 1.92 | 54397 | 2.18 | 64314 | 2.57 |
Panel B in Fig. 4 shows the locations of hydrogen bonding
The frames count is the number of instances in which the hydrogen bonding was found during the trajectory. Occupancy is defined as the percentage one hydrogen bond donor is within a distance of 3.0 Å and an angle cutoff of 135° of one hydrogen bond acceptor. Occupancies >1 occur when an atom hydrogen bonds to multiple waters at the same time satisfying the above criteria.
4. Discussion:
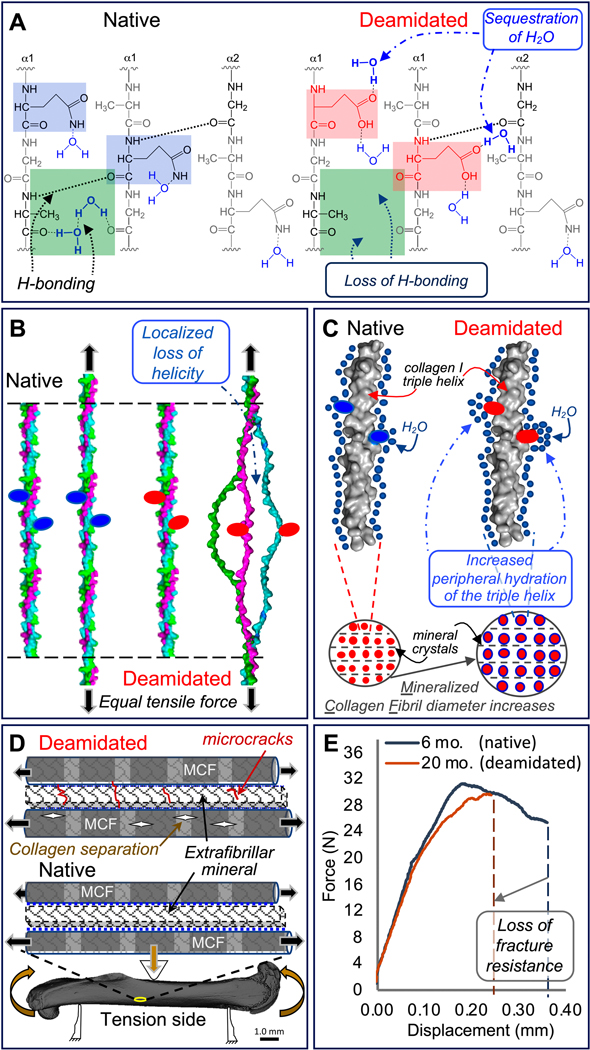
Traditionally, studies of bone matrix PTMs have been focused on glycation-mediated, non-enzymatic collagen crosslinks that are thought to adversely affect bone quality by making the matrix more susceptible to microdamage [40]. Significantly less attention has been paid to non-crosslinking PTMs, and so their role in the mechanical behavior of bone is poorly understood. To this end, we analyzed non-crosslinking modifications to the bone matrix in a mouse model of aging and identified multiple locations of both enzymatic and non-enzymatic PTMs in collagen I of bone. Specifically, we found regions within the collagen triple helix where age-related increases in the levels of deamidation, a non-enzymatic reaction that occurs over time, rearranges the hydrogen bonding patterns with water such that the number of water molecules at the helix periphery increases. Since the toughness of cortical bone decreases as the relative abundance of deamidation at multiple sites increases with advanced aging in BALB/c mice (Fig. 3, Table S3, and Table S8), we propose a new mechanism of bone fragility whereby significant increases in collagen I deamidation may trigger a loss of bone fracture resistance with age due to altered hydrogen bonding between collagen I and water (Fig. 5).
Figure 5. Proposed mechanism for how deamidation affects bone toughness.
Because deamidated sites (pink squares) sequester water molecules altering hydrogen-bonding within the collagen I triple helix (green squares), the stability of the triple helix possibly declines (A). With less stability arising from a loss of optimal hydrogen bonding, the triple helix unravels in the vicinity of deamidated sites (red ovals) at lower tensile forces compared to the native triple helix (B). Deamidation increases local hydration (blue circles) of the triple helix surface and periphery. This results in “swelling” of the mineralized collagen fibrils (MCF) and an increase in MCF diameter (C). Both lower triple helix stability and higher MCF diameter increase the likelihood of collagen ruptures within the MCF and microcrack accumulation in the extrafibrillar mineral when a bone is loaded through the yield point of the tissue (D). As a result, deamidated bone has less capacity than native bone to deform before fracture as depicted by force vs. displacement curves from three-point bending tests of femurs from 6-mo. and 20-mo. old BALB/c mice (E).
The comprehensive mapping of PTM sites within the triple helical region of mouse collagen I revealed numerous enzymatic PTMs (Fig. 1). Glycosylation of lysine consistently occurred at Lys254 and Lys266 of the α1(I) chain and Lys270 of the α2(I) chain. Hydroxylation predominately occurred on proline residues with close to 100% relative abundance (Tables S4-S7), which is expected for this enzymatic PTM that is necessary for collagen stability [41]. The relative abundance of hydroxylation at most sites did not change between 6-mo. and 20-mo. of age. For those few sites where hydroxylation levels significantly differed between the age groups, the differences were relatively small (Fig. 2). Therefore, age-related changes in hydroxylation are unlikely to affect collagen type I in a way that affects the fracture resistance of bone.
The detection of non-enzymatic deamidation in bone is not new. Deamidation occurs when nitrogen (NH) on the collagen backbone attacks the carbon of a carbonyl (C=O) sidechain to form an intermediate, 5-membered ring that then hydrolyzes altering the functional groups of the sidechain. Its quantification in bone by mass spectrometry has been used to assess the degradation of collagen in fossils since deamidation levels increase with postmortem age of bone [42,43]. This non-enzymatic PTM also occurs in vivo; and as such, deamidation levels are in a sense a marker of tissue age [44]. Deamidation can potentially occur during sample processing at basic pH with rates depending on specific protein sites. However, this artifactual deamidation was not detectable in fibrillar proteins upon overnight digestion with trypsin as reported by previous study using a similar extraction protocol [45]. Moreover, in the current study, we detected multiple Asn and Gln sites with minor or no deamidation. Also, significant age-dependent differences were detected in mouse samples which were processed under identical conditions. These results are indicative of minimal, if any, artifactual deamidation occurring during our sample preparation.
While there were a number of similarities in deamidation between the female and male BALB/c mice (Table 1 and Table S3), we observed sex-related differences in the locations of Asn and Gln residues that were deaminated. The present study was not specifically designed to assess whether the age-related increase in deamidation levels depends on the sex of the animal as such a site-specific deamidation pattern was not known a priori. Nonetheless, it is well-known that the likelihood of any given Asn or Gln residue undergoing deamidation depends on microenvironment (i.e., steric hindrance, oxidative stress, and pH). Therefore, sex-related differences in microenvironment may explain why a few sites were heavily deamidated in female bone while a few different sites were heavily deamidated in male bone. This may, for example, be related to differences in how sex steroids affect oxidative stress or extracellular pH. Further studies are necessary to address the causes of the apparent sex-related differences in deamidation pattern. Although the collagen I deamidation profile may vary between the sexes, the ability of deamidation levels to explain the variance in toughness was similar between female and male bone with relative abundance being a somewhat stronger contributor to toughness in females (see standardized β of the slope in Table S8).
To gain insight into how site-specific, localized deamidation affects collagen I, we performed molecular modeling and MD simulations for three different model segments of the triple helix (see dashed boxes in Fig. 1A for location). Specifically, we focused on how deamidation impacts hydrogen bonding patterns because such interactions influence the stability of the collagen molecule and ultimately the ability of collagen I to resist deformation. As illustrated by molecular modeling, the magnitude of the deamidation-induced shift to a more negatively charged sidechain depended on the location of deamidation site(s) along the collagen I sequence. This shift in charge was additive when multiple deamidation sites were clustered together within the sequence.
Given the absence of high-resolution collagen fibrillar structures, our modeling was limited to the context of 30-amino acid triple helices and did not include the mineral component, NCPs or the neighboring collagen molecules. Therefore, we were unable to formulate predictions regarding the impact of deamidation on the stability of higher order structures such as fibrils. Regardless, the significant alterations to the electrostatic topology caused by deamidation suggest that its impact is not limited to the triple helix and would likely affect neighboring collagen molecules. A closer analysis of the affected hydrogen bonds predicted that, in a triple helix, none of the native (N and Q) or respective deamidated n(D) and q(E) residues participated in intramolecular bonding, one of the more stabilizing interactions in a collagen molecule. Rather, the amino acid sidechains exclusively hydrogen bond with water molecules. In fact, the acidic sidechains of Asp and Glu residues are predicted to bind more water molecules, bind them more tightly, and more often compared to native Asn and Gln side chains. In combination with the increased binding of water by the sidechains, MD simulations predicted localized disruption of hydrogen bonding along the backbone of the polypeptide chains. Specifically, there were subtle losses and gains of amide (NHδ+) to carboxyl (COδ-) hydrogen bonding. Such hydrogen bonding between strands are significant factors in helix stability [46] indicating that deamidation redistributes the stability of the triple helix away from the native molecule (Fig. 5A). More striking was the disruption and reshuffling of water hydrogen bonds with either COδ- or NHδ+ of the amide backbone groups localized to the sites of deamidation. Collectively, the MD data suggest that the known age-related decline in collagen integrity, toughness, and matrix-bound water may be due, in part, to deamidated (i.e., more acidic) sidechains attracting water molecules from preferred sites of stabilization such that the stability of the triple helix with deamidated sites is different (Fig. 5B) and no longer in cooperation with neighboring native collagen molecules. The increase in the amount of water molecules interacting with the deamidated sidechains may also cause mineralized collagen fibrils (MCFs) to swell (i.e., increase in diameter) as a compact packing pattern of water molecules within the backbone is perturbed (Fig. 5C). This, in turn, could reduce the interstitial space for the water (blue dashed lines in Fig. 5D) that is thought to reside between mineral crystal surfaces and collagen [47]. The loss of tiple helix stability could favor collagen fibril rupture for a given stress below the normal yield stress, while the increase in MCF diameter pushing into extrafibrillar mineral could disrupt the normal mechanical coupling of water between mineral and MCF such that microcracks more readily form (Fig. 3D). The combined effect of these molecular changes due to deamidation would be lower fracture resistance of bone (Fig. 5E). Additional work using synthetic collagen peptides and larger scale MD simulations are necessary to determine whether deamidation reduces stability of collagen I.
We did not observe age-related changes in the relative abundance of AGEs, namely CML and MG-H1 (Fig. 1). In our previous study of the femurs from the BALB/c mouse model of aging, there was a significant increase in pentosidine, a fluorescent AGE crosslink that can be detected and quantified by high performance liquid chromatography. Thus, the expectation was that non-crosslinking AGEs would increase with the age of the mice. In the study by Thomas et al. quantifying CML in human cortical bone, detection of matrix-bound CML in all donors required an extraction protocol in which homogenized bone was first demineralized and then the pellet was subjected to 2 cycles of denaturation (85 °C) followed by trypsin digestion [23]. Our second extraction – which incorporated demineralization followed by heat denaturation – of the remaining pellet from the first extraction did not reveal any age-related changes in collagen CML levels. Possibly, further optimization of the extraction technique may identify non-crosslinking AGEs that accumulate with age, but the short lifespan of mice compared to humans may simply preclude an appreciable accumulation of CML within the bone as this non-enzymatic modification is a slow forming AGE that requires an oxidative step.
5. Conclusions
We identified sequence position and abundance of multiple post-translational modifications in the collagen type 1 of mouse bone with respect to aging. The primary change between 6-mo. and 20-mo. of age was deamidation levels of specific asparagine and glutamine residues in collagen I. Based on the data about sequence positions in which the abundance of deamidation sites significantly increased with age, MD simulations and models predicted an increase in local negative charge at the surface of the collagen I triple helix around a deamidation site, perturbation of water interactions with collagen I backbone, and attraction of water molecules toward the side chains. Such changes may alter the stability of the triple helix, thereby diminishing its ability to confer fracture resistance to the bone.
Supplementary Material
Highlights.
The degree to which collagen I is deamidated increases with advanced aging in mice.
Deamidation redistributes hydrogen bonding interactions with water.
The level of hydroxylation sites within collagen I is relatively stable with age.
Acknowledgements
This work was funded in part by a training grant from NIDDK (DK101003), by a grant from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (AR067871) and by the VA Office of Research and Development (BX004297). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or other funding agencies. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University, Nashville, TN.
Footnotes
Declaration of competing interests
There authors do not have conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Reznikov N, Shahar R, Weiner S, Bone hierarchical structure in three dimensions, Acta Biomater. 10 (2014) 3815–3826. doi: 10.1016/j.actbio.2014.05.024. [DOI] [PubMed] [Google Scholar]
- [2].Unal M, Creecy A, Nyman JS, The Role of Matrix Composition in the Mechanical Behavior of Bone, Curr Osteoporos Rep. 16 (2018) 205–215. doi: 10.1007/s11914-018-0433-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chavassieux P, Seeman E, Delmas PD, Insights into material and structural basis of bone fragility from diseases associated with fractures: how determinants of the biomechanical properties of bone are compromised by disease, Endocr. Rev 28 (2007) 151–164. doi: 10.1210/er.2006-0029. [DOI] [PubMed] [Google Scholar]
- [4].Paschalis EP, Tatakis DN, Robins S, Fratzl P, Manjubala I, Zoehrer R, et al. , Lathyrism-induced alterations in collagen cross-links influence the mechanical properties of bone material without affecting the mineral, 49 (2011) 1232–1241. doi: 10.1016/j.bone.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yamauchi M, Sricholpech M, Lysine post-translational modifications of collagen, Essays Biochem. 52 (2012) 113–133. doi: 10.1042/bse0520113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Terajima M, Perdivara I, Sricholpech M, Deguchi Y, Pleshko N, Tomer KB, et al. , Glycosylation and cross-linking in bone type I collagen, J Biol Chem. 289 (2014) 22636–22647. doi: 10.1074/jbc.M113.528513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].McNerny EMB, Gong B, Morris MD, Kohn DH, Bone Fracture Toughness and Strength Correlate With Collagen Cross-Link Maturity in a Dose-Controlled Lathyrism Mouse Model, J Bone Miner Res. 30 (2015) 446–455. doi: 10.1002/jbmr.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Forlino A, Marini JC, Osteogenesis imperfecta, Lancet. 387 (2016) 1657–1671. doi: 10.1016/S0140-6736(15)00728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Danielsen CC, Bollerslev J, Mosekilde L, Thermal stability of cortical bone collagen in relation to age in normal individuals and in individuals with osteopetrosis, 15 (1994) 91–96. [DOI] [PubMed] [Google Scholar]
- [10].Wang X, Shen X, Li X, Agrawal CM, Age-related changes in the collagen network and toughness of bone, 31 (2002) 1–7. [DOI] [PubMed] [Google Scholar]
- [11].Zioupos P, Currey JD, Hamer AJ, The role of collagen in the declining mechanical properties of aging human cortical bone, J Biomed Mater Res. 45 (1999) 108–116. [DOI] [PubMed] [Google Scholar]
- [12].Willett TL, Dapaah DY, Uppuganti S, Granke M, Nyman JS, Bone collagen network integrity and transverse fracture toughness of human cortical bone, Bone. 120 (2019) 187–193. doi: 10.1016/j.bone.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Notbohm H, Mosler S, Bodo M, Yang C, Lehmann H, Bätge B, et al. , Comparative study on the thermostability of collagen I of skin and bone: influence of posttranslational hydroxylation of prolyl and lysyl residues, J. Protein Chem. 11 (1992) 635–643. [DOI] [PubMed] [Google Scholar]
- [14].Shoulders MD, Raines RT, Collagen structure and stability, Annu. Rev. Biochem 78 (2009) 929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Panwar P, Lamour G, Mackenzie NCW, Yang H, Ko F, Li H, et al. , Changes in Structural-Mechanical Properties and Degradability of Collagen during Aging-associated Modifications, J Biol Chem. 290 (2015) 23291–23306. doi: 10.1074/jbc.M115.644310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Odetti P, ROSSI S, Monacelli F, POGGI A, CIRNIGLIARO M, FEDERICI M, et al. , Advanced glycation end products and bone loss during aging, Ann. N. Y. Acad. Sci 1043 (2005) 710–717. doi: 10.1196/annals.1333.082. [DOI] [PubMed] [Google Scholar]
- [17].Nyman JS, Roy A, Acuna RL, Gayle HJ, Reyes MJ, Tyler JH, et al. , Age-related effect on the concentration of collagen crosslinks in human osteonal and interstitial bone tissue, Bone. 39 (2006) 1210–1217. doi: 10.1016/j.bone.2006.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nyman JS, Roy A, Tyler JH, Acuna RL, Gayle HJ, Wang X, Age-related factors affecting the postyield energy dissipation of human cortical bone, 25 (2007) 646–655. doi: 10.1002/jor.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zimmermann EA, Schaible E, Bale H, Barth HD, Tang SY, Reichert P, et al. , Age-related changes in the plasticity and toughness of human cortical bone at multiple length scales, Proc Natl Acad Sci U S A. 108 (2011) 14416–14421. doi: 10.1073/pnas.1107966108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Poundarik AA, Wu P-C, Evis Z, Sroga GE, Ural A, Rubin M, et al. , A direct role of collagen glycation in bone fracture, J Mech Behav Biomed Mater. 52 (2015) 120–130. doi: 10.1016/j.jmbbm.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Trębacz H, Wójtowicz K, Wlizło-Dyś E, Dyś W, Effect of “in vitro” induced glycation on thermostability of bone tissue, Int J Biol Macromol. 51 (2012) 561–565. doi: 10.1016/j.ijbiomac.2012.06.003. [DOI] [PubMed] [Google Scholar]
- [22].Carriero A, Zimmermann EA, Paluszny A, Tang SY, Bale H, Busse B, et al. , How tough is brittle bone? Investigating osteogenesis imperfecta in mouse bone, J Bone Miner Res. 29 (2014) 1392–1401. doi: 10.1002/jbmr.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Thomas CJ, Cleland TP, Sroga GE, Vashishth D, Accumulation of carboxymethyl-lysine (CML) in human cortical bone, Bone. 110 (2018) 128–133. doi: 10.1016/j.bone.2018.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li Y, Fessel G, Georgiadis M, Snedeker JG, Advanced glycation end-products diminish tendon collagen fiber sliding, Matrix Biol. 32 (2013) 169–177. doi: 10.1016/j.matbio.2013.01.003. [DOI] [PubMed] [Google Scholar]
- [25].Fessel G, Li Y, Diederich V, Guizar-Sicairos M, Schneider P, Sell DR, et al. , Advanced glycation end-products reduce collagen molecular sliding to affect collagen fibril damage mechanisms but not stiffness, PLoS ONE. 9 (2014) e110948. doi: 10.1371/journal.pone.0110948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Creecy A, Uppuganti S, Girard MR, Schlunk SG, Amah C, Granke M, et al. , The agerelated decrease in material properties of BALB/c mouse long bones involves alterations to the extracellular matrix, Bone. 130 (2020) 115126. doi: 10.1016/j.bone.2019.115126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Madu H, Avance J, Chetyrkin S, Darris C, Rose KL, Sanchez OA, et al. , Pyridoxamine protects proteins from damage by hypohalous acids in vitro and in vivo, Free Radic. Biol. Med 89 (2015) 83–90. doi: 10.1016/j.freeradbiomed.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Brown KL, Darris C, Rose KL, Sanchez OA, Madu H, Avance J, et al. , Hypohalous Acids Contribute to Renal Extracellular Matrix Damage in Experimental Diabetes, Diabetes. 64 (2015) 2242–2253. doi: 10.2337/db14-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, et al. , The Amber biomolecular simulation programs, J. Comput. Chem 26 (2005) 1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, et al. , A second generation force field for the simulation of proteins, nucleic acids, and organic molecules (vol 117, pg 5179, 1995), Journal of the American Chemical Society. 118 (1996) 2309–2309. doi: 10.1021/ja00124a002. [DOI] [Google Scholar]
- [31].Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C, ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB, J Chem Theory Comput. 11 (2015) 3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bella J, Brodsky B, Berman HM, Hydration structure of a collagen peptide, Structure. 3 (1995) 893–906. doi: 10.1016/S0969-2126(01)00224-6. [DOI] [PubMed] [Google Scholar]
- [33].Bayly CI, Cieplak P, Cornell WD, Kollman PA, A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges - the Resp Model, Journal of Physical Chemistry. 97 (1993) 10269–10280. doi: 10.1021/j100142a004. [DOI] [Google Scholar]
- [34].Joung IS, Cheatham TE, Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations, J Phys Chem B. 112 (2008) 9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Larini L, Mannella R, Leporini D, Langevin stabilization of molecular-dynamics simulations of polymers by means of quasisymplectic algorithms, J Chem Phys. 126 (2007) 104101. doi: 10.1063/1.2464095. [DOI] [PubMed] [Google Scholar]
- [36].Wu X, Brooks BR, Vanden-Eijnden E, Self-guided Langevin dynamics via generalized Langevin equation, J. Comput. Chem 37 (2016) 595–601. doi: 10.1002/jcc.24015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Essmann U, PERERA L, BERKOWITZ ML, DARDEN T, Lee H, PEDERSEN LG, A Smooth Particle Mesh Ewald Method, J Chem Phys. 103 (1995) 8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- [38].Ryckaert JP, Ciccotti G, Berendsen HJC, Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes, Journal of Computational Physics. 23 (1977) 327–341. [Google Scholar]
- [39].Jurrus E, Engel D, Star K, Monson K, Brandi J, Felberg LE, et al. , Improvements to the APBS biomolecular solvation software suite, Protein Sci. 27 (2018) 112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Saito M, Marumo K, Collagen cross-links as a determinant of bone quality: a possible explanation for bone fragility in aging, osteoporosis, and diabetes mellitus, Osteoporos Int. 21 (2010) 195–214. doi: 10.1007/s00198-009-1066-z. [DOI] [PubMed] [Google Scholar]
- [41].Rappu P, Salo AM, Myllyharju J, Heino J, Role of prolyl hydroxylation in the molecular interactions of collagens, Essays Biochem. 63 (2019) 325–335. doi: 10.1042/EBC20180053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hurtado PP, O’Connor PB, Deamidation of collagen, Anal. Chem 84 (2012) 3017–3025. doi: 10.1021/ac202980z. [DOI] [PubMed] [Google Scholar]
- [43].van Doorn NL, Wilson J, Hollund H, Soressi M, Collins MJ, Site-specific deamidation of glutamine: a new marker of bone collagen deterioration, Rapid Commun. Mass Spectrom 26 (2012) 2319–2327. doi: 10.1002/rcm.6351. [DOI] [PubMed] [Google Scholar]
- [44].Robinson NE, Robinson AB, Deamidation of human proteins, Proc Natl Acad Sci USA. 98 (2001) 12409–12413. doi: 10.1073/pnas.221463198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Araki N, Moini M, Age estimation of museum wool textiles from Ovis aries using deamidation rates utilizing matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, Rapid Commun. Mass Spectrom 25 (2011) 3396–3400. doi: 10.1002/rcm.5237. [DOI] [PubMed] [Google Scholar]
- [46].Bella J, Collagen structure: new tricks from a very old dog, Biochem. J 473 (2016) 1001–1025. doi: 10.1042/BJ20151169. [DOI] [PubMed] [Google Scholar]
- [47].Wilson EE, Awonusi A, Morris MD, Kohn DH, Tecklenburg MM, Beck LW, Highly ordered interstitial water observed in bone by nuclear magnetic resonance, J Bone Miner Res. 20 (2005) 625–634. doi: 10.1359/JBMR.041217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.