Abstract
CRISPR–Cas immune systems process and integrate short fragments of DNA from new invaders as spacers into the host CRISPR locus to establish molecular memory of prior infection, which is also known as adaptation in the field. Some CRISPR–Cas systems rely on Cas1 and Cas2 to complete the adaptation process, which has been characterized in a few systems. In contrast, many other CRISPR–Cas systems require an additional factor of Cas4 for efficient adaptation, the mechanism of which remains less understood. Here we present biochemical reconstitution of the Synechocystis sp. PCC6803 type I-D adaptation system, X-ray crystal structures of Cas1–Cas2–prespacer complexes, and negative stained electron microscopy structure of the Cas4–Cas1 complex. Cas4 and Cas2 compete with each other to interact with Cas1. In the absence of prespacer, Cas4 but not Cas2 assembles with Cas1 into a very stable complex for processing the prespacer. Strikingly, the Cas1-prespacer complex develops a higher binding affinity toward Cas2 to form the Cas1–Cas2–prespacer ternary complex for integration. Together, we show a two-step sequential assembly mechanism for the type I-D adaptation module of Synechocystis, in which Cas4–Cas1 and Cas1–Cas2 function as two exclusive complexes for prespacer processing, capture, and integration.
INTRODUCTION
Bacteria and archaea have evolved an adaptive immune system, known as CRISPR–Cas [Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-CRISPR associated genes (Cas)] system (1). This adaptive immunity is acquired by a three-step mechanism: (i) a short foreign DNA is incorporated into the CRISPR locus as a new spacer, a process known as adaptation; (ii) the CRISPR locus is transcribed and processed into CRISPR RNA (crRNA) to assemble with the Cas proteins, namely expression, maturation and assembly; (iii) guided by the crRNA, the Cas proteins target to and eliminate the invading nucleic acid, termed interference. The expression and maturation stage and the interference stage have been extensively studied over the past decade. However, many of the molecular details for the adaptation process remain elusive (2).
CRISPR–Cas systems can be divided into two classes, six types, and >30 subtypes based on the composition and arrangement of their cas gene loci (3). In spite of the diversity of the CRISPR–Cas systems, they share a universal mechanism for spacer integration, in which the conserved Cas1–Cas2 complex captures and integrates short segments of foreign DNA into the host CRISPR arrays (3). Previous biochemical and structural studies revealed that the Cas1–Cas2 complex carries prespacper with 3′-overhang and integrates the prespacer into the leader-proximal repeat through direct nucleophilic attack (4–7).
Although Cas1 and Cas2 are universally used for catalyzing the integration reaction, many other type-specific factors have been identified to be involved in the adaptation process (8–11).One well-characterized example is the integration host factor (IHF) in the type I-E system. IHF was shown to induce the leader repeat bending and provide additional sequence specificity to Cas1–Cas2 (6,8). Recently, the cas4 genes in some subtypes within type I systems are implicated in adaptation (12–14). In many cases, the cas4 genes are located adjacent to cas1 and cas2, and sometimes even fused to cas1. The biochemical functions of Cas4 in adaptation emerged very recently (13–17). Biochemical and genetic studies revealed that Cas4 recognizes the Protospacer Adjacent Motif (PAM) sequences and processes the prespacer through site-specific endonucleolytic cleavage to ensure the integration of functional spacers into the CRISPR array (13–17). Recent studies on Bacillus halodurans adaptation system uncovered that Cas4 together with Cas1 and Cas2 assembles into a ternary complex and couples the prespacer process and integration, offering the first glimpse into the Cas4–Cas1–Cas2 adaptation systems (13).
Despite the recent inspiring progress in the field, the Cas4–Cas1–Cas2 adaptation systems remain poorly understood at mechanistic level. Here we provide mechanistic insights into the Cas4–Cas1–Cas2 type I-D adaptation system of Synechocystis sp. PCC6803 through biochemical and structural studies. Different from what was discovered in Bacillus halodurans, the adaptation system of Synechocystis acts through a two-step sequential assembly events, uncoupling the prespacer processing and integration processes. The Cas4 and Cas1 assemble into a stable binary complex due to a higher binding affinity, which is responsible for processing the prespacer prior to the integration. The Cas4–Cas1 complex processes the prespacer in a PAM dependent manner. After the prespacer processing, the Cas1–prespacer complex develops a higher affinity toward Cas2 and dissociates from Cas4 to assemble into Cas1–Cas2-prespacer ternary complex for integration.
MATERIALS AND METHODS
Protein preparation
The DNA sequences of cas4, cas1 and cas2 genes of Synechocystis sp. PCC6803 type I-D CRISPR–Cas system were synthesized and cloned into a vector derived from pET-28a (+) (Novagen), which contains an N-terminal His6-MBP (maltose binding protein) tag followed by a tobacco etch virus (TEV) protease recognition site and a linker (ASGSGTGSGS). All the proteins were overexpressed in Escherichia coli strain Rosetta (DE3) (Novagen) at 18°C for 18 h. The cells were harvested by centrifugation and the cell pellets were resuspended in binding buffer (20 mM Tris–HCl pH 8.0, 250 mM NaCl, 10% glycerol). The cells were lysed by an ultrahigh-pressure homogenizer and centrifuged to remove the cell debris. The resultant supernatant was collected and incubated with 1 ml IDA-Nickel magnetic beads (BeaverBeads™, Beaverbio) pre-equilibrated with binding buffer. After washed with binding buffer supplemented with 20 mM imidazole to remove nonspecifically bound proteins, the target protein was eluted using binding buffer supplemented with 250 mM imidazole. The eluted target protein was incubated with TEV protease (100:1) to remove the N-terminal His6-MBP tag. The digested protein passed through a desalting column to remove imidazole and then a Ni-NTA column (GE Healthcare) to remove free His-MBP tag, uncleaved protein and TEV protease. The flow-through was collected and was further purified via a Heparin column (GE Healthcare) and a gel-filtration column (Superdex 75 10/300 GL, GE Healthcare). The purified proteins were concentrated to 10 mg ml−1 and stored at −80°C until use.
DNA substrate preparation
All oligonucleotides were synthesized by Sangon Biotech. The 5′-(Cy3) TGTGCCCCTGGCGGTCGCTTTCTTTTT-3′ alone and annealed with 5′-AAAAAGAAAGCGACCGCCAGGGGCACA-3′ was used as the ssDNA and dsDNA substrates for electrophoretic mobility shift assays, respectively. 5′-(Cy3)TTGTGCCCCTGGCGGTCGCTTTCAATTTTTAACTTTTTTT-3′ or 5′-(Cy3)TTGTGCCCCTGGCGGTCGCTTTCAATTTTTTTTTTTTTTT-3′ was used for Cas4 nuclease activity assays. The 22–7-7 double-forked prespacer (22-bp duplex with 7-nt overhangs) was prepared by annealing 5′-TTTTTTTTGTGCCCCTGGCGGTCGCTTTCTTTTTTT-3′ and 5′-TTTTTTTGAAAGCGACCGCCAGGGGCACATTTTTTT-3′. The 26–5–5 double-forked prespacer (26-bp duplex with 5-nt overhangs) was prepared by annealing 5′-TTTTTTTGTGCCCCTGGCGGTCGCTTTCAAGTTTTT-3′ and 5′-TTTTTCTTGAAAGCGACCGCCAGGGGCACAATTTTT-3′. Annealing was performed by heating DNA oligonucleotides to 95°C for 5min and a gradient cooling (0.1°C/8 s) from 95°C to 25°C in the annealing buffer (20 mM HEPES pH 7.5, 100 mM NaCl and 10% glycerol).
MBP Pull-down assay
MBP or MBP-tagged protein (2mg) was incubated with 100 μl MBP affinity resin (NEB) at 16°C for 30 min in gel-filtration buffer. The proteins were washed with 5 mL buffer followed by incubating with 4 mg untagged Cas proteins at 16°C for 30 min. After washed with 5 ml buffer, the proteins were eluted with elution buffer (20 mM Tris–HCl pH 7.4, 250 mM NaCl, 10% glycerol,1 mM EDTA and 10 mM maltose). Samples were separated by 15% SDS-PAGE. Each experiment was repeated three times and one representative result was shown in the manuscript.
Isothermal titration calorimetry
ITC experiments were performed at 25°C using ITC200 (Microcal). Purified Cas1, Cas2 and Cas4 proteins were desalinated in buffer (20 mM HEPES pH 7.4, 100 mM NaCl, 10% glycerol and 5 mM β-mercaptoethanol). 650 μM Cas2 was titrated with 20 consecutive 4 μl into the cell containing 150 μM Cas1 or Cas1–Cas4. The ITC titration data was analyzed by Origin software (OriginLab).
Electrophoretic mobility shift assays
The ssDNA or dsDNA of 0.2 μM were incubated with different concentrations of Cas proteins (0, 0.2, 0.4, 1 and 4 μM) in annealing buffer (20 mM HEPES pH 7.4, 100 mM NaCl and 10% glycerol) for 30 min at 4°C, respectively. The binding reactions were analyzed on a 6% native polyacrylamide gel with 0.5× TBE buffer (45 mM Tris–HCl, pH 8.3, 45 mM boric acid and 1 mM EDTA). Fluorescent signals of 5′ Cy3 labeled DNA were recorded using the Chemical Imaging System (Bio-Rad).
Surface plasmon resonance (SPR) analysis
The binding of Cas proteins was assessed by SPR on Biacore X100 (GE Healthcare) at room temperature. Purified Cas2 or Cas4 protein was covalently immobilized to the surface of a CM5 sensor chip using the Amine coupling kit (GE Healthcare). Purified Cas1 protein was flowed over the sensor chip in running buffer [10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.05% (v/v) Surfactant P20] at a flow rate of 10 μl min−1. The chip surface was regenerated by 10 mM glycine–HCl, pH 1.7.
Single-particle negative-stain electron microscopy
Cas4–Cas1 complex was diluted to 10 μg ml−1 and 3 μl sample was applied to glow-discharged carbon-coated copper grids. After 30-s adsorption, the grids were stained with 3% (w/v) uranyl acetate solution. The grids were observed with a TF20 electron microscope (FEI Company) operated at 120 keV. Images were acquired at a nominal magnification of 67 000 × (pixel size 1.8 Å). A total of 100 negative staining micrographs were recorded on Gatan CCD camera 895.
A total of 48,355 particles were auto-extracted using a box size of 128 pixels and the stack was binned by a factor of 2 for reference-free 2D alignment with RELION 3.0 (18). After two round iterations of 2D classifications, particles belonging to good classes were pooled for the subsequent 3D classifications.
An ab initio model was created in RELION 3.0 (19) low-pass-filtered to 60 Å and used as the initial model for 3D refinement. A total of 8866 particles were then refined to yield a 3D reconstruction with an estimated resolution of 19.2 Å.
Nuclease activity assays
For nuclease activity assays, 100 pmol of Cas4 protein was incubated with 1 pmol 5′ Cy3 labeled ssDNA at 38°C, in 10 μl reactions containing 20 mM HEPES, pH 7.5, 100 mM KCl buffer. Various divalent ions (Mg2+, Mn2+, Zn2+ or Ca2+) with the final concentration of 10 mM was used in reactions to find the optimal condition for the nuclease activity. The incubation time was 20 min for the endonuclease activity assays. The reaction was quenched with an equal volume of Gel Loading Buffer II (Thermofisher) and 25 mM EDTA. Then the samples were boiled for 10 min and separated by 8 M urea 15% polyacrylamide denaturing gel. DNA was visualized by FluorChem system.
Crystallization and structure determination
The Cas1–Cas2-prespacer complex was prepared by mixing Cas1, Cas2 and prespacer in a 2:1:0.6 molar ratio and further purified by gel filtration. The crystals of Cas1–Cas2-complexed with 22–7–7 or 26–5–5 prespacer were grown using the hanging-drop vapour diffusion method at 20°C, in a buffer consisting of 30% MPD, 0.1 M imidazole pH 6.5, 0.2 M (NH4)2SO4, and 10% (w/v) PEG3350. The Se-Met Cas1 crystals were grown using the hanging-drop vapour diffusion method at 20°C, in a buffer consisting of 25% PEG MME5000, 0.1 M MES pH6.5, 0.2 M (NH4)2SO4 and 20% glycerol.
Crystals were flash frozen in liquid nitrogen directly from the precipitant solutions. Diffraction data were collected on beamline BL19U1 of National Facility for Protein Science Shanghai (NFPS) at Shanghai Synchrotron Radiation Facility. The data collected were processed by the HKL-3000 program suite (20). Details of the data processing and refinement statistics are summarized in Supplementary Table S1. Structures were determined by molecular replacement using the SeMet–Cas1 and Thermus thermophilus Cas2 (PDB ID: 1ZPW) structures as search models. Structure refinement and model building were performed with PHENIX (21) and Coot (22). All models were validated with MolProbity (23). All structure figures were prepared with ChimeraX (24).
RESULTS
Biochemical reconstitution of the adaptation machinery
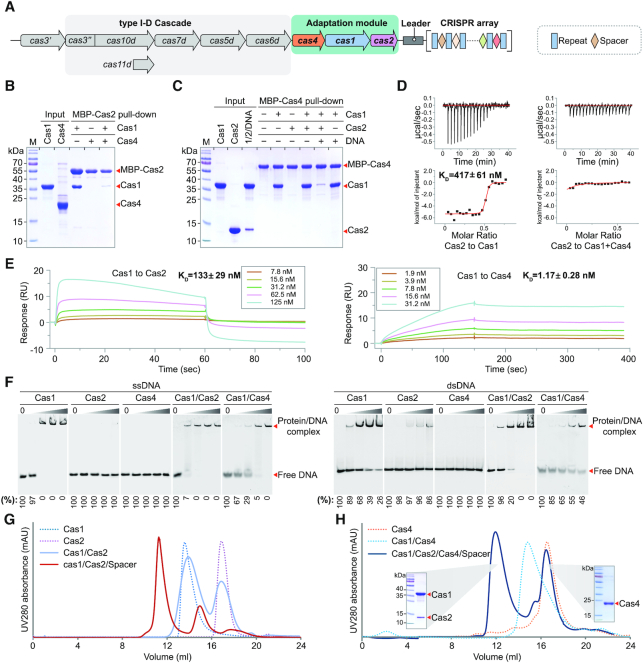
The Synechocystis sp. PCC6803 genome encodes a type I-D CRISPR–Cas system and its adaptation module consists of three genes: cas4, cas1 and cas2 (Figure 1A). To elucidate the molecular mechanism of the type I-D adaptation process in Synechocystis, we biochemically reconstituted the adaptation machinery in vitro. We first expressed and purified Cas1, Cas2 and Cas4 individually. We then identified and characterized the interaction pairs of these three proteins by pull-down assays. The maltose binding protein (MBP) did not bind to tag-free Cas1, Cas2 or Cas4 (Supplementary Figure S1). Using MBP tagged Cas2 as the bait, we clearly showed that Cas2 directly interacted with Cas1 but not Cas4 (Figure 1B). Consistently, MBP tagged Cas4 failed to pull down Cas2 but effectively recruited Cas1 (Figure 1C). More interestingly, our competition assay showed that Cas2 weakly interacted with Cas1 in the presence of Cas4 while Cas4 was able to engage Cas1 in the presence of Cas2 (Figure 1B and C), suggesting that Cas4 and Cas1 assemble into a much more stable complex than that of Cas1 and Cas2. However, in the presence of the prespacer DNA, Cas1 is recruited to Cas2–DNA complex (Figure 1C), indicating that the prespacer DNA enhanced the stability of Cas1 and Cas2 complex (also see below).
Figure 1.
Reconstitution of the type I-D adaptation module in Synechocystis. (A) Architecture of the genomic locus for the type I-D CRISPR–Cas system from Synechocystis. Spacer, repeat and cas genes are shown as rectangle, diamond and arrow, respectively. (B, C) MBP pull-down assays for assessing interactions between purified Cas1, Cas2 and Cas4. The Cas1–Cas2–prespacer complex is labelled as 1/2/DNA for concision. (D) Binding affinity between Cas1 and Cas2, and between Cas2 and Cas4–Cas1 complex measured by ITC. Untagged Cas proteins are used. The baseline-corrected instrumental response is shown in the upper panel, and the integrated data (squares), together with the best fits (solid lines), are shown in the lower panel. Data are representative of three independent experiments. (E) Binding affinity between adaptation Cas proteins measured by SPR. Untagged Cas proteins are used. Cas2 or Cas4 are immobilized on the sensor chip and Cas1 is used as the analyte. The analyte is injected at a series of 2-fold serial dilutions. Data are representative of three independent experiments. (F) DNA binding ability of Ca1, Cas2 and Cas4. EMSA is performed using 5′ Cy3-labeled 36-nt ssDNA or 36-bp dsDNA. Different concentrations of Cas proteins (0, 0.2, 0.4, 1 and 4 μM) were incubated with 0.2 μM ssDNA or dsDNA. Percent of the free DNA is calculated based on the gray scanning analysis. (G, H) Assembly of Cas protein complexes on gel filtration. The two peaks of Cas1/Cas2/Cas4/Spacer are evaluated by SDS-PAGE.
To further validate our conclusion, we performed isothermal titration calorimetry (ITC) and surface plasmon resonance (SPR) experiments to quantify the binding affinity of Cas1 and Cas2, and that of Cas1 and Cas4. While Cas2 binds to Cas1, there is no detectable binding between Cas2 and Cas1–Cas4 complex by ITC (Figure 1D), which is also consistent with our pull-down assay (Figure 1C). In line with our pull-down assay, the measured KD by SPR for Cas1 and Cas4 is about 1 nM in comparison to a KD of ∼100 nM for Cas1 and Cas2 (Figure 1E). Together, these data support that Cas4 binds Cas1 much more tightly than Cas2, suggesting that Cas4 may assemble preferentially with Cas1 in cells.
Since the adaptation machinery functions to process and integrate prespacer into the host CRISPR locus, we next assessed the substrate specificity for individual Cas proteins as well as the assembled Cas complexes using electrophoretic mobility shift assay (EMSA). Cas1 was able to bind both single stranded DNA (ssDNA) and double stranded DNA (dsDNA) with a slight preference on ssDNA (Figure 1F). Cas2 alone failed to bind ssDNA and displayed weak binding affinity to dsDNA (Figure 1F). In contrast, the complex of Cas1 and Cas2 showed much stronger binding affinity towards dsDNA than each of them alone, suggesting that Cas1 and Cas2 cooperatively bind the prespacer dsDNA.
As shown before, Cas4 functions as a nuclease to process the invader DNA for prespacer generation prior to integration (13,15,25). Unexpectedly, Cas4 alone failed to bind to ssDNA and dsDNA (Figure 1F). Since Cas4 was able to form stable complex with Cas1, it is possible that Cas4 may rely on Cas1 for the substrate recognition in cells. The Cas4–Cas1 complex displayed a slightly weaker binding to ssDNA or dsDNA in comparison to Cas1 alone (Figure 1F). Furthermore, we also measured the binding affinity between Cas4–Cas1 complex and the splayed prepsapcer DNA (22-bp duplex with 7-nt overhangs, see below). As expected, the Cas4–Cas1 binding affinity for the splayed DNA is lower than that for ssDNA, but higher than that for dsDNA (Supplementary Figure S2), indicating that the Cas4–Cas1 in complex with mature prespacer are less stable than that with non-processed perspacer.
Together, our binding assays prompted us to hypothesize that Cas4 and Cas1 assembled preferentially for generating the prespacer, the presence of which switched the binding preference of Cas1 from Cas4 to Cas2, leading to the assembly of Cas1–Cas2-prespacer for integration. To further validate this hypothesis, we recapitulated this assembly process using gel filtration. Consistent with our binding assays, Cas1 and Cas4 assembled into a stable complex on gel filtration while Cas1 and Cas2 failed to form a stable complex (Figure 1G and H). In the presence of pre-spacer DNA, Cas1, Cas2 and prespacer assemble into a stable ternary complex (Figure 1G). More strikingly, addition of Cas2 and prespacer to the Cas1–Cas4 complex led to the dissociation of Cas4 and the assembly of the Cas1–Cas2–DNA ternary complex (Figure 1G). These data provided strong evidence to support our hypothesis that the type I-D adaptation in Synechocystis may include two sequential assembling events for the prespacer processing and integration, respectively.
Structure of the Cas1–Cas2–prespacer complex
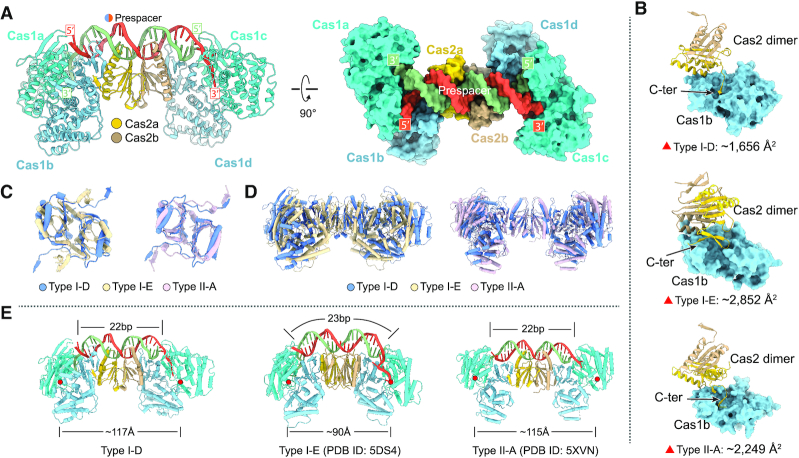
Similar to the type I-E system of E. coli and type II-A system of E. faecalis (5,7,26), Synechocystis CRISPR–Cas system also employs the Cas1–Cas2 complex for carrying and incorporating the prespacer into its CRISPR locus. To elucidate the molecular mechanism of the prespacer capture and integration by the Cas1–Cas2 complex, we assembled the Cas1–Cas2–prespacer ternary complex for structure determination. We obtained the crystals of Cas1–Cas2 complexed with a prespacer containing a 22-bp duplex and 7-nt overhangs for data collection. Initially, we tried to determine the ternary complex by molecular replacement using the published Cas1–Cas2 complex or Cas1–Cas2–DNA ternary complex but failed to get a reasonable phase. We therefore prepared crystals of selenomethionine-containing Cas1 and determined its crystal structure by single-wavelength anomalous diffraction (Supplementary Figure S3). We successfully built the structural model of the Cas1 and utilized this structure and the crystal structure of Thermus thermophilus Cas2 (PDB ID: 1ZPW) as the templates to solve the Cas1–Cas2–prespacer complex structure by molecular replacement (Supplementary Table S1).
The overall architecture of the Cas1–Cas2–prespacer complex of Synechocystis resembles a dumbbell, in which a Cas2 dimer sits in the middle with two Cas1 dimers flanking on each side (Figure 2A). The prespacer DNA duplex runs over the central Cas2 dimer with the 3′ overhangs inserting into the distal Cas1 subunits (Figure 2A). Structure comparison revealed that Cas1 resembles each other, all of which are composed of an N-terminal β-sheet domain and a C-terminal α-helical domain (Supplementary Figure S4a and b). In contrast, the Cas2 of Synechocystis and E. faecalis share a similar fold with one β strand in the very C-termini, which differs from the structure of E. coli Cas2 that is composed of two β strands in the C-terminal domain (Supplementary Figure S4c and d). The differences of monomeric Cas2 across species provide structural basis for their different dimeric assembly, leading to the differences in recruiting Cas1 in the Cas1–Cas2 complex (5,7). Both Synechocystis and E. faecalis utilized only one protomer of Cas2 to engage an adjacent Cas1 subunit to assemble the Cas1–Cas2 complex, while in E. coli Cas1–Cas2 complex the two protomers of Cas2 interact with an adjacent Cas1 subunit (Figure 2B). Despite sharing a similar overall architecture with the Cas1–Cas2-prespacer complexes of E. coli and of E. faecalis, Synechocystis Cas1–Cas2-prespacer display a few unique features. First, the Cas1–Cas2 interface in Synechocystis buries a surface area of ∼1656 Å2, which is much smaller than that of E. coli (∼2852 Å2) and that of E. faecalis (∼2249 Å2). The smaller interaction interface between Synechocystis Cas1 and Cas2 explains why Cas1–Cas2 may have failed to assemble into a stable complex in the absence of prespacer (Figure 1G). Second, Cas1–Cas2 complexes of different species assembled at different angles (Supplementary Figure S5). If one protomer of Synechocystis Cas2 dimer was superimposed with that of E. coli and E. faecalis, the other Cas2 protomer differed by ∼137° with that of E. coli and by ∼23° with that of E. faecalis (Figure 2C). Relative to the Cas2 dimer, more dramatic rotational deviations were observed for Cas1 subunits among the three different species (Figure 2D). These assembling differences contribute to the different distances between the two Cas1 active sites, further defining the preferred repeat lengths in different species (Figure 2E). The repeat length in Synechocystis is 37 bp, longer than that in E. coli (28 bp) and E. faecalis (36 bp). Consistently, the distance between the active sites of the two catalytic Cas1 unites in Synechocystis is ∼117 Å, compared to ∼90 Å in E. coli and ∼115 Å in E. faecalis (Figure 2E). Third, the Synechocystis Cas1–Cas2 complex displayed a few new features in coordinating the prespacer (see next two sections).
Figure 2.
Crystal structure of Synechocystis type I-D Cas1–Cas2–prespacer complex and structural comparison. (A) Ribbon diagram of Synechocystis type I-D Cas1–Cas2–prespacer complex structure. The Cas1a and Cas1c are colored in cyan, Cas1b and Cas1d are colored in light blue, and the two monomers of Cas2 are colored in yellow and brown, respectively. The prespacer DNA is colored in red and blue. (B) The interaction details of Cas1 and Cas2 in various types (type I-D: Synechocystis, type I-E: E. coli, type II-A: E. faecalis). The buried areas of interaction interfaces are indicated. (C) Comparison of Cas2 dimer of various types. One protomer of the Cas2 dimer was aligned, the other displayed obvious rotation as indicated. (D) Comparison of Cas1–Cas2 complex of various types with overlayed Cas2 dimer. (E) Comparison of the DNA binding architecture of Cas1–Cas2 complex from various types. The length of prespacer duplex and the distance between the active sites of the two catalytic Cas1 unites are labeled. The Cas1 active sites are indicated by red circles.
Prespacer coordination by the Cas1–Cas2 complex
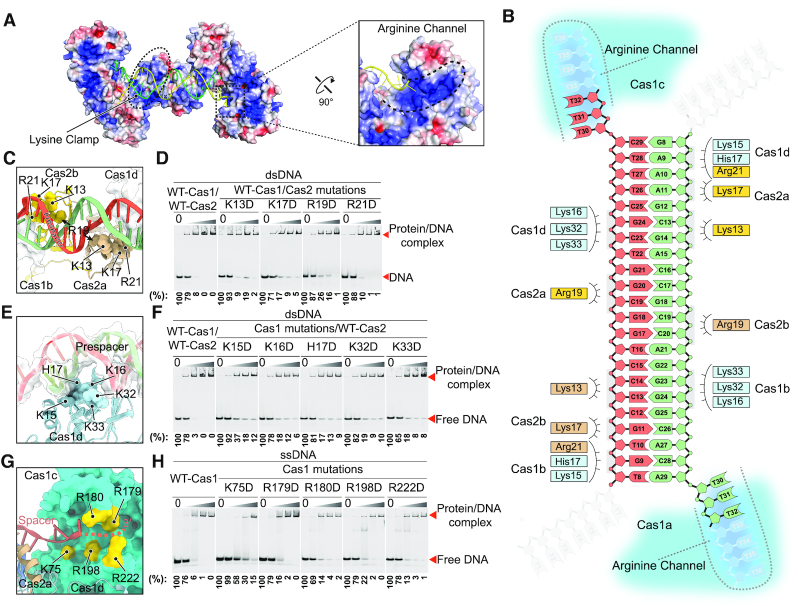
Interactions between Cas1–Cas2 and prespacer DNA come from coordination of the phosphate-backbone rather than base-specific contacts, in line with the non-specific sequence selection of prespacer that is critical for immune resistance toward diverse invader DNA. Earlier studies on E. coli type I-E adaptation system defined two positively charged regions of the Cas1–Cas2 complex for coordinating prespacer, which were coined as the Arginine Clamp and the Arginine Channel (5). Two similar positively charged regions were also observed in Synechocystis Cas1–Cas2 complex. In contrast to the E. coli Cas1–Cas2 that utilized Arginine residues to coordinate back-bone phosphate in both regions, Cas1–Cas2 mainly employed lysine residues in the clamp region while preserved arginine residues in the channel region to interact with the backbone phosphate of the prespacer (Figure 3A). We therefore named these two interaction regions as the lysine clamp and the arginine channel, respectively (Figure 3A and B). The lysine clamp is responsible for coordinating the duplex and is composed of two lysine fingers that are contributed by Cas1 residues K15, K16, H17, K32 and K33, and Cas2 residues K13, K17, R19 and R21, respectively (Figure 3C and E). Charge reversal mutations of these residues significantly reduce the binding affinity between the Cas1–Cas2 complex and dsDNA (Figure 3D and F). Lining in the arginine channel, Cas1 residues K75, R179, R180, R198 and R222 guide the 3′ overhang of the prespacer to enter the active site (Figure 3G). We generated mutations for all these positively charged residues in the arginine channel of Cas1 to assess their binding affinity with ssDNA. Four mutants (K75D, R180D, R198D and R222D) significantly weakened the ssDNA binding ability of Cas1; the mutant R179D moderately decreased Cas1 ssDNA binding ability (Figure 3H). All these mutations have been purified to homogeneity and eluted at the same retention time as the wild type, indicating that these mutations have no effect on protein folding (Supplementary Figure S6).
Figure 3.
Coordination of the prespacer by the Cas1–Cas2 complex. (A) Electrostatic potential surface representation of the Cas1–Cas2 complex with the prespacer colored in green and yellow. Blue and red (±5 kT/e) indicate the positively and negatively charged areas, respectively, of the protein complex. The lysine clamp and arginine channel responsible for coordinating the prespacer are highlighted in dashed line. (B) Diagram of the prespacer and residues coordinating the prespacer. (C) Details of the prespacer coordinated by residues of Cas2 lysine clamp. Coloring is as in (b). (D) EMSA assay for assessing the effect of charge reversal mutation of Cas2 residues involved in prespacer coordination. The protein and DNA concentrations are same as in Figure 1F. Percent of the free DNA is calculated based on the gray scanning analysis. (E) Close up view of the prespacer coordination by Cas1 lysine clamp residues. Coloring is as in (B). (F) Charge reversal mutation of Cas1 lysine clamp residues weakens binding affinity between Cas1–Cas2 complex and the prespacer as shown by EMSA. The protein and DNA concentrations are same as in Figure 1F. (G) Detailed view of the arginine channel that stabilizes the 3′ overhang of the prespacer. (H) Mutation of key arginine channel residues impairs the ssDNA binding ability of Cas1.
Mechanisms of prespacer splay and length determination
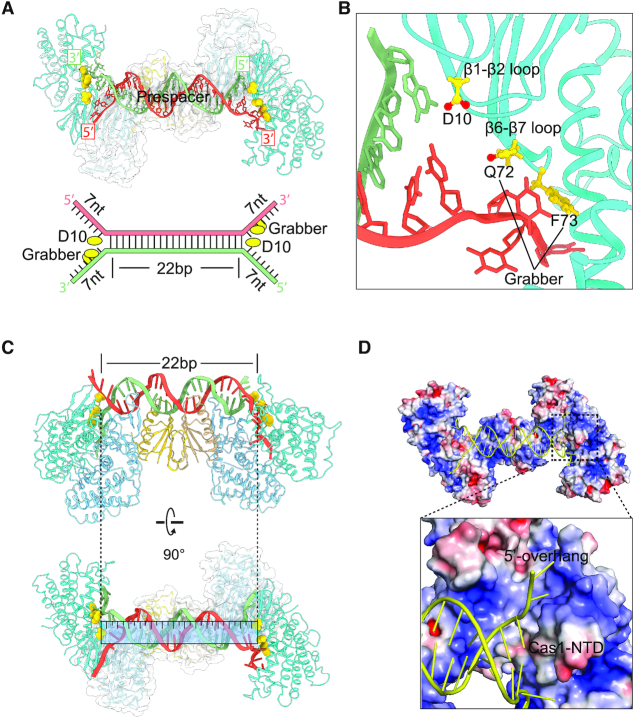
Another intriguing question is how the spacer length was defined by the Cas1–Cas2 complex, which functions as a molecular ruler. Structural analysis showed that residue D10 on the loop connecting β1 and β2 of Cas1 functions as a wedge to terminate the duplex region of the prespacer. Residue D10 on this loop helped to unwind the duplex DNA and directed each single stranded 3′-overhang to dip down into the arginine channel and each single stranded 5′-overhang to tip up towards the non-catalytic Cas1 (Figure 4A). Therefore, the distance between the two symmetric Asp residues on Cas1 loop specified the length of the duplex DNA. Similarly, in E. coli and E. faecalis, two tyrosine residues and two histidine residues from the symmetry-related Cas1 serve as a caliper to measure a 23-bp and a 22-bp duplex segment, respectively (7,26). Moreover, structural comparison allowed us to identify a conserved ‘Grabber’ motif, which gripped and bended the single stranded 3′-overhang sharply into the active channel immediately after the unwinding of the duplex DNA. The Grabber is formed by Q72 and F73 on loop connecting β6 and β7 of Cas1 (Figure 4B), whereas E. coli Grabber and E. faecalis Grabber are composed of Y86 and R84, D69 and R71, respectively (Supplementary Figure S7a and b). Despite the variation of Grabber residues, all of them shared bulk side chains and interacted extensively with the 3′ overhang to bend it ∼90° away from the duplex.
Figure 4.
Mechanism of prespacer splay and length determination. (A) D10 functions as a wedge to terminate the duplex region of the prespacer. The Grabber motif of Q72 and F73 bends and guides the single stranded 3′-overhang to the active site of Cas1. (B) Close up of the splayed prepsapcer showing the wedge residue D10 and the Grabber motif. (C) The 26-bp duplex of 26–5–5 prespacer is unwounded by residue D10 of Cas1, leading to a 22-bp duplex. (D) The displaced 5′ overhang of prepsapcer interacts with the N-terminal domain of Cas1.
To further investigate whether the length of the duplex is restricted to 22-bp by Cas1–Cas2 complex, we further determined the crystal structure of Cas1–Cas2 in complex with a prespacer containing a longer duplex (26-bp) and shorter overhangs (5-nt), which we termed as Cas1–Cas2–26 bp (Supplementary Table S1). As expected, the duplex was unwounded by residue D10 of Cas1, leading to a 22-bp duplex (Figure 4C). After end-splaying, the 5′ overhang tipped up toward an opposite direction from the 3′ overhang. The first three nucleotides of the displaced 5′ overhang established considerable interactions with the N-terminal domain of the catalytic Cas1 subunit (Figure 4D). After the first three nucleotides, we failed to observe clear electron density for the remaining nucleotides of the displaced strand probably due to lack of coordination by the protein subunits.
The assembly of the Cas4–Cas1 complex
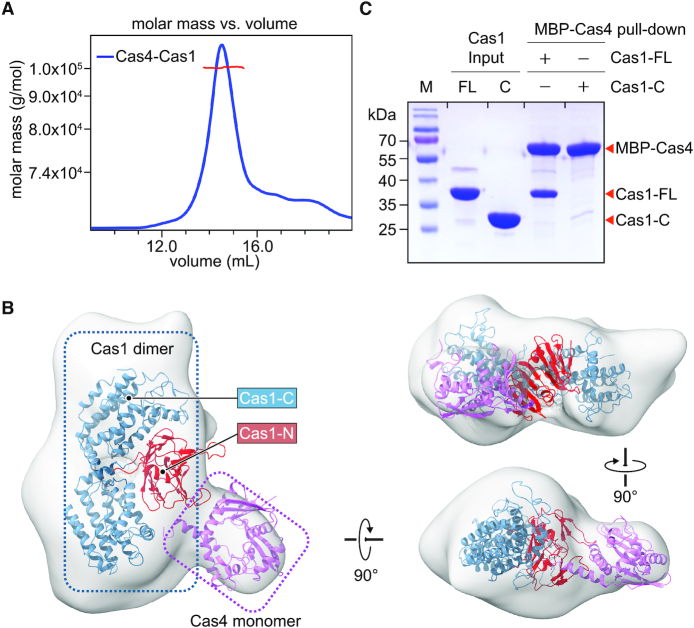
To understand the mechanism of foreign DNA processing by the Cas4–Cas1 complex, we first reconstituted Cas4–Cas1 complex in vitro. We purified Cas1 and Cas4 individually and then incubated them together to assemble the complex. Cas1 (37.5 kDa) assembled with Cas4 (22.7 kDa) into a stable complex with a molecular weight of 98 kDa as estimated by size exclusion chromatography/multi-angle scattering (SEC/MALS) analysis (Figure 5A), which may contain two Cas1 and one Cas4 molecules.
Figure 5.
The assembly of the Cas4–Cas1 complex. (A) SEC/MALS analysis to measure the molecular weight of the Cas4–Cas1 complex. The molecular weight was determined to be 99.8 kDa, with 1% uncertainty. (B) EM envelop of the Cas4–Ca1 complex fitted with crystal structure of Cas1 and structural model of Cas4. Cas1 N-terminal domain, Cas1 C-terminal domain, and Cas4 are colored in red, yellow and blue, respectively. (C) MBP pull-down assays for the interactions between purified full-length Cas1 (Cas1-FL), C-terminal domain of Cas1 (Cas1-C), and MBP-tagged Cas4.
To further probe the assembly of Cas4–Cas1 complex, we decided to determine the structure of Cas4–Cas1 complex. We first tried to crystallize Cas4–Cas1 complex for structural determination. After many trials, we did not get crystals of the complex. We therefore turned to determine the structure of Cas4–Cas1 complex using negative staining electron microscopy (EM). Negative staining EM revealed homogeneous particles with a nonsymmetric shape (Supplementary Figure S8a). A total of 48,355 particles were picked and the 3D reconstruction generated a map of ∼19 Å resolution (Supplementary Figure S8b and c). In order to interpret the map, we first generated a structural model of Cas4 using Saccharolobus solfataricus Cas4 (PDB ID: 4IC1) as a template by SWISS-MODEL (27). We then fitted the Cas4 model and the crystal structure of Cas1 into the EM density. A Cas1 dimer and a Cas4 monomer can be nicely placed into the EM density, consistent with a 2:1 stoichiometry of Cas1 and Cas4 in the complex. The overall architecture of Cas4–Cas1 complex adopted an asymmetric structure and indicated that Cas4 may bind to the N-terminal domain of Cas1 (Figure 5B).
To validate the interaction details between Cas4 and Cas1, we performed pull-down assay. The full-length Cas1 was able to form stable complex with Cas4 while the C-terminal domain of Cas1 failed to interact with Cas4 (Figure 5C), suggesting that the N-terminal domain of Cas1 is responsible for engaging Cas4. As the isolated Cas1 N-terminal domain is not stable, we did not have a chance to further validate our conclusion directly. As both Cas4 and Cas2 interact with the N-terminal domain of Cas1, Cas4 and Cas2 are competing with each other to engage Cas1. Therefore, Cas4–Cas1–Cas2 complex are not compatible due to steric hindrance, which explains our previous biochemical reconstitution results (Figure 1B–D and H).
Cas4 processes prespacer in a PAM-dependent manner
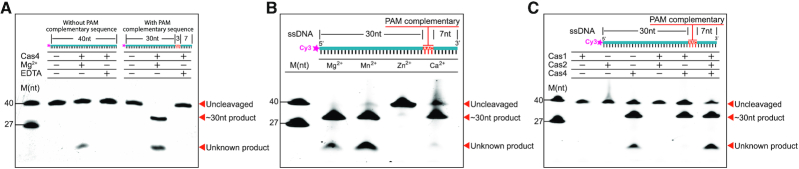
Cas4 was shown to exhibit metal dependent endonuclease and exonuclease activities against ssDNA (11,28,29). Synechocystis Cas4 was able to significantly enrich new spacers with a PAM sequence of ‘GTN’ in cells (14). However, the detailed biochemical characterization of the enzymatic activity of Cas4 is not available. To this end, we assessed the nuclease activity of Synechocystis Cas4 against different 5′-labelled ssDNA substrates under different conditions. We found that Cas4 exhibited PAM-dependent endonuclease activity (Figure 6A). The 5′-labeled PAM-containing ssDNA substrates were processed to ∼30-nt fragments, which is in line with the PAM position. Besides the expected ∼30-nt product, we also observed an unexpected band with shorter length, indicating nonspecific cleavages (Figure 6A). Moreover, this PAM dependent endonuclease activity showed a divalent cation preference of Mg2+ and Mn2+ followed by Ca2+, whereas Zn2+ did not support Cas4 endonuclease activity (Figure 6B).
Figure 6.
The PAM Dependent Endonuclease Activities of Synechocystis Cas4. (A) Cas4 exhibits PAM dependent endonuclease activity. (B) The metal dependent endonuclease activity of Cas4 against 5′ Cy3-labeled ssDNA. (C) Cas1 promotes the PAM dependent endonuclease specificity of Cas4.
To study how Cas1 or Cas2 affects the endonuclease activity of Cas4, we performed the Cas4 cleavage assay in the presence of Cas1 or/and Cas2. Cas1 or Cas2 alone did not display any nuclease activity under our experimental conditions (Figure 6C). Notably, the addition of Cas1 obliterated the nonspecific band observed in Cas4 alone (Figure 6C), indicating that the formation of Cas1–Cas4 complex promote the enzymatic specificity of Cas4. It is likely that the non-specific cleavage sites were protected by Cas1. Unexpectedly, the simultaneous addition of both Cas1 and Cas2 has little effect on the specificity of Cas4, probably due to the prioritized assembly of Cas1–Cas2–DNA complex.
DISCUSSION
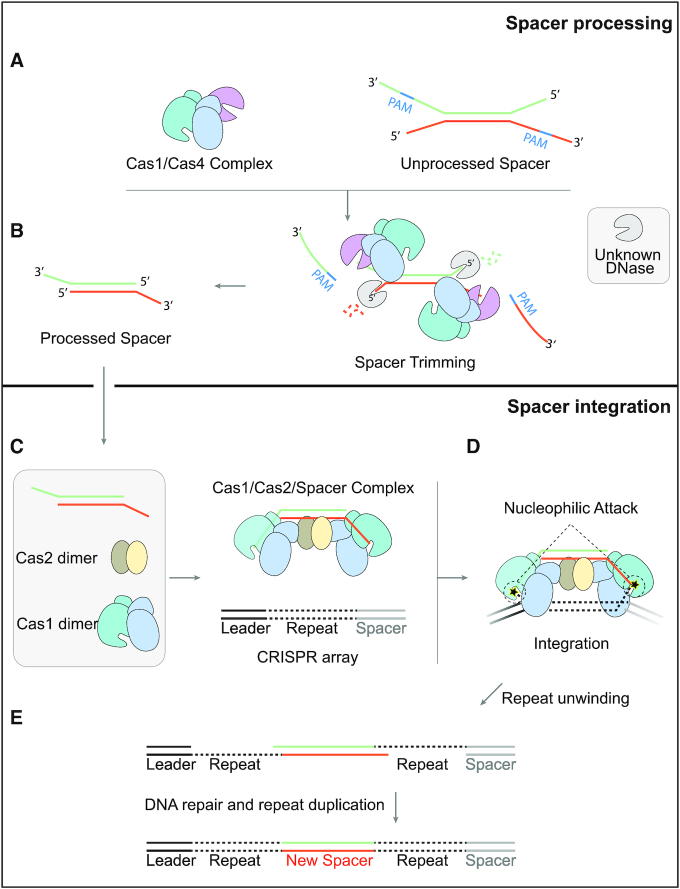
Together, our study enabled us to propose a model for the adaptation process of Synechocystis (Figure 7A–E), which is quite different from what was reported for other species. In this model, two sequential assembly events are included: the assembly of Cas4–Cas1 for prespacer processing and the assembly of Cas1–Cas2–prespacer for prespacer acquisition and integration. As the first step, Cas4 and Cas1 assemble into a stable complex, which contains two copies of Cas1 and one copy of Cas4. With the assistance of Cas1, Cas4 recognizes the PAM sequence of ‘GTN’ and trimmed prespacer precursors into their mature form. The Cas1–Cas4 in complex with mature prespacer becomes less stable than that with unprocessed prespacer, favoring the assembly of Cas1–Cas2–prespacer complex. In the second step, Cas2 comes in and may compete with Cas4 for interacting with the N-terminal domain of Cas1. Although Cas2 and Cas1 proteins displayed relatively weak interactions, they are able to form a much more stable complex in the presence of prespacer DNA. Therefore, in the presence of prespacer generated by the Cas4–Cas1 complex, Cas4 may be kicked away from Cas1 by Cas2, leading to the assembly of the Cas1–Cas2–prespacer ternary complex. Finally, the prespacer carried by the Cas1–Cas2 complex is integrated into the host CRISPR array as a new spacer.
Figure 7.
A two-step sequential assembly model for the type I-D adaptation module of Synechocystis. (A) Synechocystis Cas4 and Cas1 easily assemble into a stable complex with a stoichiometry of 1 Cas4 to 2 Cas1 to recognize the unprocessed prespacer with a PAM sequence. (B) Cas4 trims the PAM-containing 3′ overhang of the prespacer in a PAM dependent manner, while an unknown DNase is thought to process the 5′ overhang. (C) Though Cas1 and Cas2 fail to form stable complex, Cas1, Cas2 and the processed prespacer are able to form a very stable ternary complex for integration. (D) The incoming prespacer is integrated into the CRISPR locus by nucleophilic attack of the two 3′-OH groups. (E) The gap-filling replication are performed by the host DNA replication machinery.
Of note, two key factors dictate this two-step sequential assembly mechanism. One is that the Cas4 and Cas2 proteins compete for a same binding site at Cas1; the other is the binding affinity differences between those Cas proteins. As revealed by our structural study, both Cas4 and Cas2 engage the N-terminal domain of Cas1. Therefore, Cas4 and Cas2 assemble into two mutually exclusive complexes with Cas1, respectively. As uncovered by our binding assays, the complexes of Cas2–Cas1, Cas4–Cas1 and Cas1–Cas2–prespacer display a successively higher affinity, ensuring a hierarchical and stepwise assembly process.
Our structural study also revealed many new features on the assembly of Cas1–Cas2–prespacer complex, highlighting the diversity of the adaptation system. Importantly, structural comparison allowed us to identify a conserved motif of Cas1, termed as Grabber here, for bending and guiding the DNA 3′ overhang (Figure 4A and B), which was not appreciated before.
Furthermore, we found that Cas4 exonuclease activity is ‘PAM’ dependent. The presence of Cas1 helped to minimize the non-specific activity of Cas4 (Figure 6), indicating that Cas1 may bind to the unprocessed prespacer to block the nonspecific cleavage by Cas4.
Despite the progress we made, there are many remaining questions regarding the Cas4–Cas1–Cas2 adaptation system. One important question is what is the substrate of Cas4–Cas1 in cells. Another intriguing question is whether and how Cas4–Cas1 measure the length of prespapcer. After processing, how the Cas4–Cas1 switched to Cas1–Cas2–prepsapcer complex. During this transition, how the prespacer was handed over from Cas4–Cas1 to Cas1–Cas2. Additionally, another important question to address is how the spacer orientation is maintained during the processing and integration steps.
In summary, our findings provide mechanistic understanding on prespacer processing and integration and also open new avenues for future study of the adaptation systems. Our study revealed not only the universal principles that are shared in many adaptation systems but also many new features that are unique to Synechocystis adaptation system. Considering the diversity on the general design of the adaptation systems, we believe that additional variations will be discovered from microbe occupying the extraordinarily diverse ecological niches on earth. The study on these diverse adaptation systems will not only fundamentally deepen our understanding on adaptation systems but also offer new tools for genome editing.
DATA AVAILABILITY
The negative-stain EM volumes for the Cas4–Cas1 complex have been deposited to EMDB under the accession code EMD-30377. The atomic coordinates and structure factors of Cas1–Ca2 complexed with 22-bp-duplex and 26-bp-duplex prespacers have been deposited in the Protein Data Bank under the accession code 7CR6 and 7CR8, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staffs from BL19U1 beamline of National Center for Protein Science Shanghai (NCPSS) at Shanghai Synchrotron Radiation Facility for assistance during data collection.
Author contributions: Q.C., Y.Y. and T.M.F. conceived and designed the experiments. C.W., D.T., J.C. and D.H. performed protein preparation and crystallization. Y.Y., Q.C. and C.W. collected EM and X-ray diffraction data, solved the structures, and conducted the structural analysis. D.T., D.H. and Z.Y. performed MBP pull-down assays. D.H., X.M., H.H. and S.Y. performed DNA binding assays. Q.C. and T.M.F. wrote the manuscript with inputs from all authors.
Contributor Information
Chengyong Wu, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, and Collaborative Innovation Center of Biotherapy, Chengdu 610041, P.R. China.
Dongmei Tang, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, and Collaborative Innovation Center of Biotherapy, Chengdu 610041, P.R. China.
Jie Cheng, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, and Collaborative Innovation Center of Biotherapy, Chengdu 610041, P.R. China.
Daojun Hu, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, and Collaborative Innovation Center of Biotherapy, Chengdu 610041, P.R. China.
Zejing Yang, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, and Collaborative Innovation Center of Biotherapy, Chengdu 610041, P.R. China.
Xue Ma, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, and Collaborative Innovation Center of Biotherapy, Chengdu 610041, P.R. China.
Haihuai He, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, and Collaborative Innovation Center of Biotherapy, Chengdu 610041, P.R. China.
Shaohua Yao, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, and Collaborative Innovation Center of Biotherapy, Chengdu 610041, P.R. China.
Tian-Min Fu, Department of Biological Chemistry and Pharmacology, The Ohio State University, Columbus, OH 43210, USA; The Ohio State University Comprehensive Cancer Center, Columbus, OH 43210, USA.
Yamei Yu, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, and Collaborative Innovation Center of Biotherapy, Chengdu 610041, P.R. China.
Qiang Chen, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, and Collaborative Innovation Center of Biotherapy, Chengdu 610041, P.R. China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31741027, 81672722, 81501368]; Sichuan Science and Technology Program [2019YFH0123, 2019YFH0124]. Funding for open access charge: National Natural Science Foundation of China; Sichuan Science and Technology Program.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wright A.V., Nunez J.K., Doudna J.A.. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell. 2016; 164:29–44. [DOI] [PubMed] [Google Scholar]
- 2. Amitai G., Sorek R.. CRISPR–Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 2016; 14:67–76. [DOI] [PubMed] [Google Scholar]
- 3. Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P.et al.. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020; 18:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nunez J.K., Lee A.S., Engelman A., Doudna J.A.. Integrase-mediated spacer acquisition during CRISPR–Cas adaptive immunity. Nature. 2015; 519:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nunez J.K., Harrington L.B., Kranzusch P.J., Engelman A.N., Doudna J.A.. Foreign DNA capture during CRISPR–Cas adaptive immunity. Nature. 2015; 527:535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wright A.V., Liu J.J., Knott G.J., Doxzen K.W., Nogales E., Doudna J.A.. Structures of the CRISPR genome integration complex. Science. 2017; 357:1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xiao Y., Ng S., Nam K.H., Ke A.. How type II CRISPR–Cas establish immunity through Cas1–Cas2-mediated spacer integration. Nature. 2017; 550:137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nunez J.K., Bai L., Harrington L.B., Hinder T.L., Doudna J.A.. CRISPR immunological memory requires a host factor for specificity. Mol. Cell. 2016; 62:824–833. [DOI] [PubMed] [Google Scholar]
- 9. Heler R., Samai P., Modell J.W., Weiner C., Goldberg G.W., Bikard D., Marraffini L.A.. Cas9 specifies functional viral targets during CRISPR–Cas adaptation. Nature. 2015; 519:199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilkinson M., Drabavicius G., Silanskas A., Gasiunas G., Siksnys V., Wigley D.B.. Structure of the DNA-bound spacer capture complex of a Type II CRISPR–Cas system. Mol. Cell. 2019; 75:90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lemak S., Beloglazova N., Nocek B., Skarina T., Flick R., Brown G., Popovic A., Joachimiak A., Savchenko A., Yakunin A.F.. Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus. J. Am. Chem. Soc. 2013; 135:17476–17487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Z., Pan S., Liu T., Li Y., Peng N.. Cas4 nucleases can effect specific integration of CRISPR spacers. J. Bacteriol. 2019; 201:e00747-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee H., Zhou Y., Taylor D.W., Sashital D.G.. Cas4-Dependent prespacer processing ensures high-fidelity programming of CRISPR arrays. Mol. Cell. 2018; 70:48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kieper S.N., Almendros C., Behler J., McKenzie R.E., Nobrega F.L., Haagsma A.C., Vink J.N.A., Hess W.R., Brouns S.J.J. Cas4 facilitates PAM-Compatible spacer selection during CRISPR adaptation. Cell Rep. 2018; 22:3377–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shiimori M., Garrett S.C., Graveley B.R., Terns M.P.. Cas4 nucleases define the PAM, length, and orientation of DNA fragments integrated at CRISPR Loci. Mol. Cell. 2018; 70:814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Almendros C., Nobrega F.L., McKenzie R.E., Brouns S.J.J.. Cas4–Cas1 fusions drive efficient PAM selection and control CRISPR adaptation. Nucleic Acids Res. 2019; 47:5223–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garrett S., Shiimori M., Watts E.A., Clark L., Graveley B.R., Terns M.P.. Primed CRISPR DNA uptake in Pyrococcus furiosus. Nucleic Acids Res. 2020; 48:6120–6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scheres S.H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012; 415:406–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M.. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D. Biol. Crystallogr. 2006; 62:859–866. [DOI] [PubMed] [Google Scholar]
- 21. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 23. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E.. UCSF chimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018; 27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee H., Dhingra Y., Sashital D.G.. The Cas4–Cas1–Cas2 complex mediates precise prespacer processing during CRISPR adaptation. Elife. 2019; 8:e44248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang J., Li J., Zhao H., Sheng G., Wang M., Yin M., Wang Y.. Structural and mechanistic basis of PAM-Dependent spacer acquisition in CRISPR–Cas systems. Cell. 2015; 163:840–853. [DOI] [PubMed] [Google Scholar]
- 27. Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L.et al.. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018; 46:W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang J., Kasciukovic T., White M.F.. The CRISPR associated protein Cas4 Is a 5′ to 3′ DNA exonuclease with an iron-sulfur cluster. PLoS One. 2012; 7:e47232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lemak S., Nocek B., Beloglazova N., Skarina T., Flick R., Brown G., Joachimiak A., Savchenko A., Yakunin A.F.. The CRISPR-associated Cas4 protein Pcal_0546 from Pyrobaculum calidifontis contains a [2Fe-2S] cluster: crystal structure and nuclease activity. Nucleic Acids Res. 2014; 42:11144–11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The negative-stain EM volumes for the Cas4–Cas1 complex have been deposited to EMDB under the accession code EMD-30377. The atomic coordinates and structure factors of Cas1–Ca2 complexed with 22-bp-duplex and 26-bp-duplex prespacers have been deposited in the Protein Data Bank under the accession code 7CR6 and 7CR8, respectively.