Abstract
The outer membrane (OM) is a defining feature of Gram-negative bacteria that serves as a permeability barrier and provides rigidity to the cell. Critical to OM function is establishing and maintaining an asymmetrical bilayer structure with phospholipids in the inner leaflet and the complex glycolipid lipopolysaccharide (LPS) in the outer leaflet. Cells ensure this asymmetry by regulating the biogenesis of lipid A, the conserved and essential anchor of LPS. Here we review the consequences of disrupting the regulatory components that control lipid A biogenesis, focusing on the rate-limiting step performed by LpxC. Dissection of these processes provides critical insights into bacterial physiology and potential new targets for antibiotics able to overcome rapidly spreading resistance mechanisms.
Functions of Lipopolysaccharide
Gram-negative bacteria are diderms, possessing both an inner membrane (IM) and an outer membrane (OM). These two membranes define a cellular compartment called the periplasm in which the peptidoglycan (PG) cell wall resides. The OM is unlike any other biological membrane. It is a lipid bilayer, but it is asymmetric. Phospholipids (PLs) make up the inner leaflet while the surface-exposed outer leaflet is composed of lipopolysaccharide (LPS) [1],
LPS is a glucosamine (see Glossary) disaccharide with four to seven acyl chains (lipid A), an inner and outer core oligosaccharide, and a long polysaccharide chain known as the O antigen [2]. Certain bacteria attach a low-molecular-weight polysaccharide to lipid A and these molecules are referred to as lipooligosaccharide (LOS) [2]. LPS was first characterized in 1892 by Richard Friedrich Johannes Pfeiffer, a student of Robert Koch, who called the heat-stable poison produced by many different pathogenic Gram-negative bacteria 'endotoxin'. Some 50 years later, Arthur Felix and Edmund Weil showed the relationship between endotoxin and the O antigen (for historical perspective see [3]), and we now know that endotoxin is the lipid A portion of LPS [2].
The acyl chains of lipid A are saturated and the negatively charged phosphates on lipid A and the inner core sugars are bridged by divalent cations like Mg2+. This allows tight packing of the LPS molecules and this contributes substantially to the distinctive properties and essential functions of the OM. The first is a remarkable permeability barrier. The IM and other biological membranes are easily disrupted by detergents and they are impermeable even to protons. By contrast, the OM is highly resistant to detergents because of its tightly packed sugar-coated surface and it is freely permeable to small water-soluble molecules with a molecular mass of less than approximately 700 daltons because a major class of OM proteins function as porins [4]. Because of its selective barrier function, the OM confers a significant level of innate resistance to many antibiotics. This tight LPS packing also contributes substantially to the stiffness and strength of the bacterial cell, a second critical OM function that compensates for the very thin PG cell wall present in Gram-negative bacteria [5,6].
The OM is an essential organelle and, with few known exceptions, lipid A is essential as well [7-9]. In well studied Gram-negative bacteria, such as Escherichia coli, LPS levels are carefully regulated; too much ortoo little leads to cell death. Indeed, E. coli employs three essential IM proteins to maintain proper LPS levels. Here we summarize recent work that sheds light on the novel regulatory mechanisms that monitor both the synthesis of LPS and its transport to the OM.
LPS Synthesis and Transport
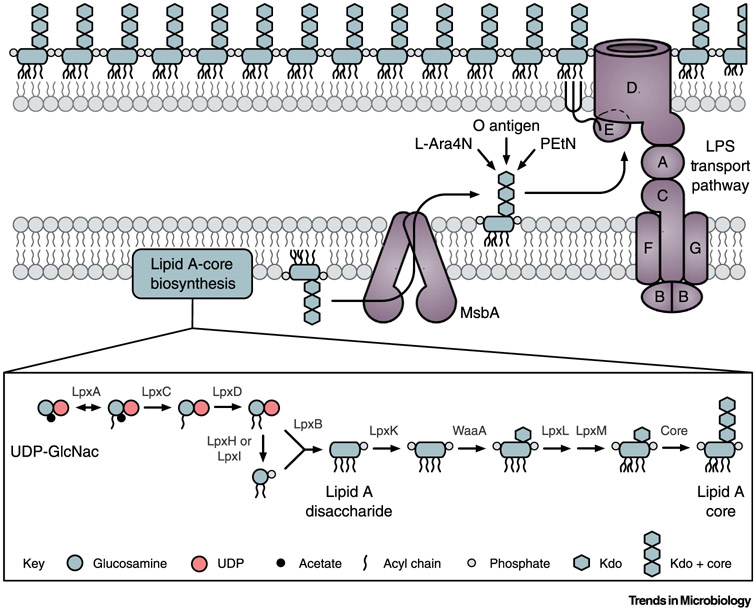
Although LPS is destined for the outer leaflet of the OM, its synthesis is initiated in the cytoplasm. The essential enzymatic steps responsible for assembly of the conserved lipid A anchor, called the Raetz pathway, have all been determined [2,10-12] (Figure 1). The first reaction in this pathway, catalyzed by LpxA, exhibits an unfavorable equilibrium constant which can be overcome by the enzymatic activity of LpxC in the second step of lipid A synthesis [13,14]. Thus, LpxC-catalyzed formation of UDP-monoacyl-glucosamine (UDP-monoacyl-GlcN) is the first committed step in lipid A synthesis and, consequently, LpxC is a regulatory focal point for LPS biogenesis [14,15]. A subsequent critical step and regulatory node in lipid A synthesis is the conversion of lipid A disaccharide to lipid IVA by LpxK [15,16]. After formation of Kdo2-lipid A, the inner and outer core oligosaccharide, consisting of sugars that can vary in number and composition among species, are appended to form lipid A-core. Unlike lipid A, the core oligosaccharide is not essential for E. coli viability but core oligosaccharide synthesis mutants exhibit OM defects [17,18].
Figure 1. Overview of Lipopolysaccharide (LPS) Biosynthesis and Transport.
Lipid A and the core oligosaccharide are synthesized in the cytoplasm and the cytoplasmic leaflet of the inner membrane (IM). Lipid A-core is flipped to the periplasmic leaflet of the IM by MsbA, and O antigen is then attached to the core oligosaccharide. Additional modifications at various positions on lipid A-core can be made on the periplasmic side of the IM, such as the addition of 4-aminoarabinose (L-Ara4N) and phosphoethanolamine (PEtN). Fully assembled LPS is extracted from the IM and transported to the outer leaflet of the outer membrane (OM) by the LPS transport (Lpt) pathway, which is formed by the proteins LptA-G. Abbreviations: GlcNAc, N-acetylglucosamine; Kdo, 2-keto-3-deoxy-octonate; UDP, uridine diphosphate;. Figure inspired by [133].
The final steps of LPS synthesis occur in the periplasmic leaflet of the IM after lipid A-core is transported by the essential ATP-binding cassette transporter MsbA [19-21] (Figure 1). Repeating oligosaccharide units carried by undecaprenyl phosphate are assembled onto the lipid A-core, forming a long, structurally and compositionally diverse, O antigen [22-24]. Additional reactions, including 4-amino-4-deoxy-L-arabinose and phosphoethanolamine additions to lipid A that provide resistance to polymyxins, the antibiotics of last resort, also occur when LPS resides in the periplasmic leaflet of the IM [25,26]. While these final steps in LPS synthesis are not essential for viability, they are critical for virulence [27,28], antibiotic resistance [25,26], and protection from immune molecules such as antibodies and antimicrobial peptides [29-32].
Completed LPS molecules are shuttled to their final destination in the outer leaflet of the OM by the essential LPS transport (Lpt) system [2,33-37] (Figure 1). In addition to extracting LPS from the IM, ATP hydrolysis by LptB in the LptB2FG complex also drives transport of LPS across the periplasm [38-44]. The hydrophobic lipid A acyl chains presumably pass through the structurally conserved jelly roll domains of LptC, LptA, and the N-terminal domain of LptD that form a bridge across the periplasm [45-47]. The OM β-barrel LptD and the lipoprotein LptE unidirectionally insert LPS exclusively into the OM outer leaflet as the final step in establishing this asymmetrical barrier [48-51].
LpxC Is Degraded by the YciM/FtsH Complex
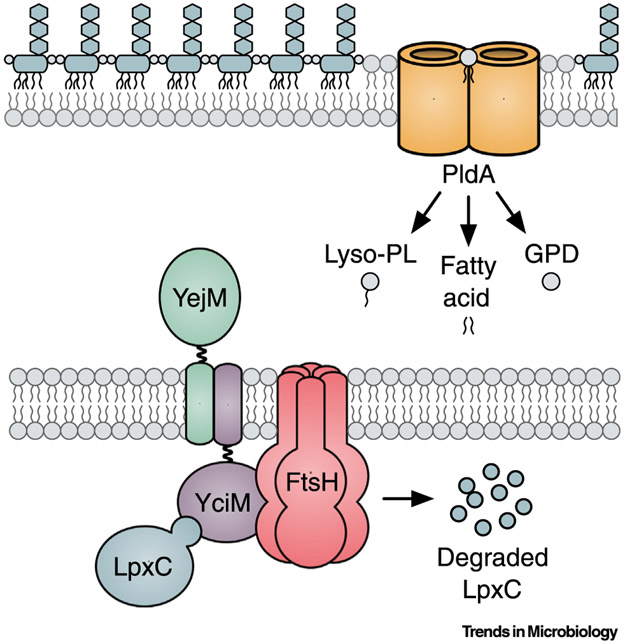
In E. coli and other enteric bacteria, LpxC protein levels are controlled by the YciM/FtsH protease complex (Figure 2). The first hint that LpxC is post-translationally regulated came from a study in the 1990s which showed that inhibiting the early steps of lipid A biosynthesis increased LpxC protein levels without affecting lpxC transcription [52]. In the same decade, Ogura et al. [53] found that LpxC is stabilized in mutants expressing a temperature-sensitive allele of the essential AAA+ protease FtsH (see [54] for a recent review on FtsH). These authors also showed that LpxC is degraded by FtsH in vitro. Nearly 15 years later, several studies showed that the essential IM protein YciM (also known as LapB [55]) is required for FtsH to degrade LpxC in vivo [55,56]. Changes in yciM expression had no effect on FtsH substrates other than LpxC, indicating that YciM does not affect FtsH activity in general. Sequence and structural analysis revealed that the cytoplasmic region of YciM contains several tetratricopeptide repeats [55-59] which are often involved in protein–protein interactions [60]. As such, it was proposed that YciM is an adaptor protein that delivers LpxC to FtsH for proteolysis [56]. In support of this hypothesis, YciM copurifies with both LpxC and FtsH [55,61]. Notably, mutations that decrease lipid A biosynthesis suppress deletion of both ftsH and yciM, indicating that uncontrolled LPS synthesis caused by impaired degradation of LpxC is lethal [53,55,56].
Figure 2. Envelope Proteins That Regulate LpxC.
LpxC is degraded by the inner membrane(IM)-bound protease FtsH. YciM may function as the adaptor that delivers LpxC to FtsH for degradation. YejM directly interacts with YciM and inhibits LpxC degradation. The outer membrane (OM) phospholipase PldA sequentially removes the fatty acids from surface-exposed phospholipids, which produces lysophospholipids (lyso-PL), fatty acids, and glycerophosphodiesters (GPDs). Liberated fatty acids serve as second messengers that inhibit LpxC degradation.
There is evidence to suggest that YciM may alter activity of other LPS biosynthetic enzymes in addition to LpxC [55]. LPS precursors accumulate in the yciM mutant while reduced FtsH activity has no such effect [53,55]. Besides LpxC and FtsH, YciM copurifies with LPS, all proteins of the LPS transport pathway, the glycosyltransferase WaaC, and the chaperones DnaK and DnaJ. YciM is also required for the stability and/or folding of LpxM, WaaC, and WaaO. Accordingly, it was proposed that YciM is an organizing center that plays several roles in LPS biogenesis, including delivering enzymes to the site of lipid A-core biosynthesis, ensuring that these enzymes are properly folded, regulating LpxC protein levels, and coordinating LPS biosynthesis with transport to the OM [55].
The sequence and structure of the C-terminal end of LpxC are both critical for LpxC proteolysis. For LpxC to be degraded by FtsH, the C terminus must be unstructured, consist of at least 20 amino acids, and contain the LpxC degron motif LAXXXXXAVLA [62,63]. These features are thought to help thread LpxC through the FtsH pore. While the C terminus of LpxC is necessary for degradation by FtsH, it is not sufficient [63]. Fusing the C-terminal end of LpxC to glutathione-S-transferase (GST) leads to FtsH-independent degradation. As such, additional features of LpxC are required to direct this protein to FtsH, specifically. The amino acid sequence of the C-terminal end of LpxC varies across Gram-negative bacteria, and LpxC homologs lacking the E. coli degradation tag tend to be regulated in an FtsH-independent manner (Box 1).
Box 1. FtsH-Independent Regulation of LpxC.
LpxC is not always regulated by FtsH. Even in Escherichia coli there is evidence to suggest that another, unidentified protease degrades LpxC [16]. In Agrobacterium tumefaciens and Rhodobacter capsulatus, LpxC turnover is mediated by the AAA+ protease Lon [97]. Intriguingly, LpxC in Pseudomonas aeruginosa is not subject to rapid proteolysis [97]. In support of this finding, FtsH and other ATP-dependent proteases are not essential in this organism [98,99]. Unlike E. coli, overexpression of LpxC in P. aeruginosa is not toxic and does not increase levels of lipid A [97]. As such, it is likely that P. aeruginosa employs a completely novel mechanism to regulate LPS levels. While much of this mechanism remains to be determined, a small RNA that regulates LpxC expression has been identified [100].
Although FtsH is highly conserved, YciM tends to be found in organisms that degrade LpxC in an FtsH-dependent manner [55,56,97]. A notable exception to this is Neisseria meningitidis, which synthesizes LOS rather than LPS and is one of the few Gram-negative organisms that do not require lipid A for growth under standard laboratory conditions [8,101]. N. meningitidis LpxC lacks the C-terminal degradation tag found in E. coli [59], suggesting that regulation is FtsH-independent. However, this organism contains a YciM homolog (named Ght) [59]. While YciM regulates lipid A biosynthesis in both organisms, the phenotypes that arise from loss of YciM are quite different [55,56,59]. In E. coli, reduced YciM activity leads to increased levels of LPS, while LOS is absent in the N. meningitidis yciM mutant. Mutations that decrease LpxC activity suppress deletion of yciM in E. coli, while mutations that increase LpxC expression suppress the defects associated with the N. meningitidis yciM mutant. The reason for these discrepancies is currently unknown but may be related to the different mechanisms used by these organisms to regulate LpxC.
Consequences of Altered LPS Biogenesis
Disruption of lipid A synthesis or LPS transport decreases LPS levels in the outer leaflet of the OM, causing increased permeability [51,64-67]. When there is insufficient LPS, PLs flip into the outer leaflet and disrupt the LPS interaction network that provides rigidity and impermeability to the cell. Consequently, Gram-negative bacteria utilize multiple mechanisms to cope with these mislocalized PLs. PldA, also called OmpLA, is a phospholipase that degrades PLs in the outer leaflet of the OM [68], which ultimately stabilizes LpxC, thus increasing LPS production [69]. Another OM protein, PagP, uses PLs in the OM as substrates for palmitate transfer to LPS [70,71]. PagP-mediated modification of LPS, in addition to eliminating outer-leaflet PLs, also activates the periplasmic σE stress response to mitigate OM disruption [72]. Finally, disrupting lipid A synthesis leads to accumulation of O antigen attached to undecaprenyl phosphate in the periplasm due to depletion of lipid A-core substrate for the O-antigen ligase WaaL and this detrimentally sequesters undecaprenyl phosphate from other essential cell envelope pathways [73,74]. The dire and compounding effects of interfering with LPS synthesis and transport on OM integrity and cell viability have motivated discovery efforts for novel antibacterial molecules targeting this pathway (Box 2).
Box 2. Antibiotic Discovery Targeting LPS Biogenesis.
With the alarming spread of bacterial infections resistant to all clinically available drugs, new antibiotics are desperately needed [102-105]. LPS biogenesis and transport are essential for viability, are conserved, and found only in Gram-negative bacteria. Consequently, multiple steps in this pathway have been explored as possible novel antibacterial targets. Efforts to inhibit lipid A synthesis have focused on targeting LpxC, which performs the first committed step (e.g., CHIR-90, ACHN-975, a pyrrolo-imidazolone scaffold, among others) [2,10,106-111], as well as a dual-targeting of LpxA and LpxD inhibitor (RJPXD33) [112]. In addition to their potential future value to patients, LpxC inhibitors have proved to be useful tools for understanding LPS biogenesis [15,77,111,113,114]. As lipid A and fatty acid biosynthesis are linked [16,61,115] antibacterial FabI inhibitors, which inhibit an essential step in fatty acid biosynthesis [116], could also impact lipid A synthesis. Multiple steps in LPS transport have also been explored as potential antibacterial targets. Inhibitors of MsbA, which transports the lipid A-core to the periplasmic leaflet of the IM, have been described (e.g., G907, G592, and a tetrahydrobenzothiophene scaffold) [21,117]. These inhibitors lead to accumulation of LPS in the IM inner leaflet and have provided mechanistic insight into LPS transport. Later in the pathway, antibacterial compounds that disrupt the periplasmic LPS transport (Lpt) system have been discovered (e.g., IMB-881 and thanatin) [118,119], including the final step of LPS insertion into the OM outer leaflet, performed by LptDE, which has been targeted with molecules derived from the antimicrobial peptide protegrin I (e.g., Murepavadin) [120-122]. Finally, LPS itself is a clinically validated target that is bound by the antibiotics of last resort, the polymyxins. Increased clinical use of polymyxins due to the failure of other classes of antibiotics has led to the spread of lipid A-modifying enzymes that provide resistance to the polymyxins [123,124]. New polymyxin derivatives and LPS-binding scaffolds aim to overcome resistance and mitigate toxicity issues [125-127]. Despite the demonstrated biological importance of targets in the LPS biogenesis and transport pathways, few molecules have been tested in humans, and polymyxins remain the sole clinically relevant antibiotic. However, as a more detailed mechanistic understanding of LPS biogenesis and transport emerges, new strategies to interfere with these critical processes will be revealed.
Overproduction of LPS is also detrimental to OM barrier function and cell viability in Gram-negative bacteria, underscoring the importance of control of LPS biogenesis. Cells appear to counter the consequences of excess LPS by re-establishing the balance with PL biosynthesis. Evidence for this coordination of LPS and PL biosynthesis is a correlation between LpxC and FabZ activity [75], which is required for type II fatty acid biosynthesis [76-78]. This link explains why overproduction of LPS upon deletion of ftsH can be suppressed by a gain-of-function mutation in fabZ that increases fatty acid biosynthesis [53]. However, multiple lines of evidence suggest additional mechanisms are required to manage LPS overproduction. Unlike with FabZ, LpxC levels are negatively correlated with the activities of several critical enzymes in fatty acid biosynthesis, including FabB, FabD, FabF, and FabI, which runs counter to the idea of coordinating synthesis [16,53,61]. Additionally, both an lpp deletion mutant [55,56] and a mutant producing an Lpp variant unable to crosslink to PG suppress the overproduction of LPS in a yciM deletion strain [79]. This suppression is rationalized by the removal of excess LPS by hyper-vesiculation caused by the lpp mutant, thus preventing LPS accumulation in the wrong cellular compartments [79,80]. The explanation for lethality upon LPS overproduction then is not the mismatch in LPS and PL, but likely the aberrant accumulation of LPS in the outer leaflet of the IM. Thus, the complex regulatory pathways used by Gram-negative bacteria are all tuned to ensure that there is a proper balance of LPS–PL not to maintain that balance per se but rather to ensure that LPS remains restricted to the outer leaflet of the OM.
Regulation of LpxC Proteolysis
LpxC proteins levels are affected by the amount of lipid A in the cell [13,52]. To achieve such regulation, the cell must be able to sense the amount of lipid A and adjust activity of the LpxC degradation machinery accordingly. Only recently have the components of this regulatory circuit been identified (Figure 3, Key Figure).
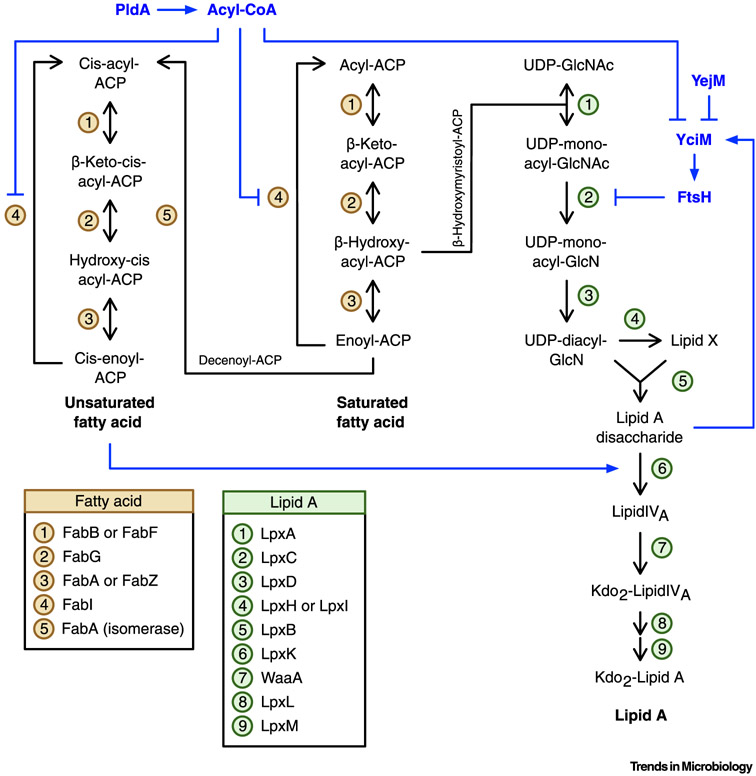
Figure 3. Key Figure. Elongation of Saturated and Unsaturated Fatty Acids, Lipid A Biosynthesis, and Regulation of Lipid Biosynthesis.
Saturated and unsaturated fatty acids are elongated through an iterative cycle of four reactions [134]. Branch points in the elongation of saturated fatty acids lead to the formation of unsaturated fatty acids and lipid A. Unsaturated fatty acids arise from the isomerization of trans-2-decenoyl-ACP to cis-β-decenoyl-ACP by FabA, while β-hydroxymyristoyl-ACP may be attached to UDP-GlcNAc by LpxA to begin synthesis of lipid A. FtsH regulates lipid A biogenesis by degrading LpxC. The adaptor protein YciM enhances LpxC degradation by delivering LpxC to FtsH. Several factors are known to regulate LpxC degradation. First, lipid A disaccharide increases LpxC degradation, possibly by increasing activity of YciM. Unsaturated fatty acids reduce lipid A disaccharide levels by increasing activity of LpxK. Second, PldA stabilizes LpxC by increasing levels of acyl-CoA, which inhibits activity of YciM and FabI. Finally, YejM prevents LpxC degradation by inhibiting YciM and/or FtsH. Blue lines and text indicate pathways that regulate LpxC. Abbreviations: Ac, acetate; ACP, acyl carrier protein; CoA, coenzyme A; GlcN, glucosamine; GlcNAc, N-acetylglucosamine; Kdo, 2-keto-3-deoxy-octonate; UDP, uridine diphosphate.
Lipid A Disaccharide
Early studies on LpxC regulation found that decreased activity of LpxA, LpxC, or LpxD stabilizes LpxC while decreased activity of WaaA does not [52]. These data indicate that the lipid A intermediate responsible for regulating LpxC proteolysis is produced after LpxD but is before lipid IVA (Figure 3). Computational simulations of lipid A biosynthesis found that lipid A disaccharide accumulates in the absence of FtsH, suggesting that lipid A disaccharide may be involved in regulating LpxC degradation [15]. Indeed, reducing lipid A disaccharide levels by overexpressing LpxK stabilizes LpxC [15]. How lipid A disaccharide modulates LpxC degradation is not known. However, LpxC protein levels are not affected by reduced activity of LpxA, LpxC, or LpxD in a yciM mutant [56], suggesting that lipid A disaccharide may regulate LpxC proteolysis by altering activity of YciM (Figure 3).
That LpxC proteolysis is regulated by lipid A disaccharide can explain why fatty acid biosynthetic enzymes have differing effects on LpxC stability [16]. For instance, it is known that reduced FabI activity stabilizes LpxC while reduced FabA activity increases LpxC degradation [16,53]. As FabI catalyzes a committed step in saturated fatty acid biosynthesis, loss of FabI activity increases the abundance of unsaturated fatty acids [16,75]. In consequence, less saturated fatty acids are available for lipid A biosynthesis and the amount of lipid A disaccharide is decreased. Furthermore, unsaturated fatty acids can stimulate activity of LpxK, which would also decrease the amount of lipid A disaccharide [16] (Figure 3). By contrast, loss of FabA activity likely increases the amount of lipid A disaccharide. As FabA processes the lipid A precursor β-hydroxymyristoyl-ACP, reduced FabA activity would increase the amount of precursor available for lipid A biosynthesis [16,75]. This would lead to increased levels of lipid A disaccharide and trigger LpxC degradation.
YejM
YejM is an IM protein with an essential N-terminal transmembrane domain connected to a dispensable C-terminal periplasmic domain by a basic interfacial domain linker [81-84] (Figure 2). The first phenotypes associated with mutations in the periplasmic domain of YejM include increased sensitivity to large or hydrophobic antibiotics, impaired growth at high temperatures, leakage of periplasmic proteins, and a reduced LPS:PL ratio [85,86]. Because many of these phenotypes are shared with lpxA and lpxD mutants, early models proposed that YejM is involved in lipid A biosynthesis.
Several years later, yejM was identified in a screen for mutants that weaken the OM when the PhoPQ two-component system is activated, leading to the alternative name of PbgA for PhoPQ-barrier gene A [82]. Subsequent analysis revealed that the periplasmic domain of YejM is required for the PhoPQ system to increase cardiolipin levels in the OM and binds to cardiolipin in vitro [82,87]. Accordingly, it was proposed that YejM is a transporter that moves cardiolipin from the IM to the OM during stress. However, not all data support this conclusion (Box 3).
Box 3. Transport of Cardiolipin by YejM.
Several studies have shown that YejM influences the cardiolipin content of the OM [82,128], but whether YejM functions as a cardiolipin transporter is debated. Unlike YejM, cardiolipin is not essential under standard laboratory conditions [129]. Mutations within the nonessential periplasmic domain of YejM dramatically weaken the OM, while complete loss of cardiolipin has only a minor effect [84-86,92]. These differences are not due to aberrant accumulation of cardiolipin within the IM of the yejM mutants as the OM defect persists in cells lacking cardiolipin entirely [92]. Moreover, IM cardiolipin levels are not affected by reduced YejM activity [82,128]. In Salmonella, OM cardiolipin levels are altered in yejM mutants only when the PhoPQ system is activated [82]. In the absence of PhoPQ activation, reduced YejM activity does not affect OM cardiolipin levels but still impacts OM integrity. It has also been shown that OM cardiolipin levels are actually increased in the Salmonella yejM mutant during stationary phase [88]. At the very least, these data suggest that YejM is not a cardiolipin transporter exclusively.
Despite its proposed function in cardiolipin transport, YejM shares no sequence or structural similarity to any known transporter. Rather, YejM is homologous to the sulfatases EptA and LtaS, both of which bind and modify lipids [82,84,87,130-132]. Initial studies proposed that a long and deep cleft spanning the length of YejM could serve to transport cardiolipin [83]; however, the negative charge in this cleft would make passage of cardiolipin difficult. The globular region within the periplasmic domain of YejM contains a hydrophobic pocket that binds cardiolipin in vitro, and residues within this pocket are important for YejM function in vivo [87]. How cardiolipin would enter the hydrophobic pocket is not obvious as it is not located near the membrane or within the cleft, indicating a need for substantial structural arrangements to transport cardiolipin [83]. While there was some structural suggestion of a potential YejM interaction with cardiolipin [83,87], a more recent high-resolution structure instead identified LPS bound to the YejM interfacial linker domain [84]. The high-resolution structure combined with molecular dynamic simulations demonstrate that the interfacial domain is not a flexible linker between the transmembrane and periplasmic domains but rather part of a compact YejM structure that does not undergo the significant structural changes that would be required to transport cardiolipin across the periplasm [84].
Recent studies suggest that YejM is a lipid sensor that regulates LPS biosynthesis. lpxC, yciM, and ftsH were identified in screens for mutations that suppress depletion of YejM and/or the OM defect caused by loss of the periplasmic domain, suggesting that YejM may be involved in LpxC proteolysis [79,88-90]. Indeed, partial or complete loss of YejM activity decreases LpxC stability and substantially reduces LPS levels [79,84,89,90]. Together with genetic evidence indicating that YciM is epistatic to YejM [79,89], these findings suggest that YejM regulates LpxC levels by inhibiting activity of the YciM/FtsH protease complex.
Based on the observation that continued LPS synthesis in the absence of transport to the OM is lethal, it was proposed that YejM may sense aberrant accumulation of LPS within the outer leaflet of the IM [79]. While previous structures of YejM suggested the capacity to bind PLs [82,83,87], a high-resolution structure of full-length YejM revealed an LPS molecule bound at the interfacial domain located on the periplasmic surface of the IM [84]. Moreover, in vitro binding assays demonstrated that interfacial domain-derived peptides selectively bind to LPS [84]. In total, a model has emerged in which YejM regulates LpxC levels by inhibiting activity of the YciM/FtsH protease complex in response to the accumulation of LPS in the periplasmic leaflet of the IM.
How YejM communicates the lipid signal to the YciM/FtsH complex is not entirely clear. Given that YejM and YciM directly interact [84,89], it is possible that YejM titrates YciM away from FtsH. Lipid A binding to YejM may relieve the interaction between YciM and YejM, allowing YciM to deliver LpxC to FtsH for degradation. Curiously however, the periplasmic domain of YejM is not required for the protein to interact with YciM yet is required to inhibit LpxC proteolysis [79,84,89,90]. In other words, the failure of YejM lacking the periplasmic domain to properly regulate YciM is not associated with weakened interaction between the two proteins. It has been proposed that the periplasmic domain of YejM may interact with and regulate activity of FtsH [90]. While this hypothesis is not supported by YejM interaction studies [89], it is possible that YejM and FtsH interact under specific, yet-to-be determined conditions.
PldA
Levels of LPS in the OM are indirectly sensed by the phospholipase PldA, which degrades PLs that have been mislocalized to the outer leaflet of the OM [68] (Figure 2). A role for PldA in sensing OM LPS came from studies on the neomorphic mutation mlaA*, which codes for a protein that disrupts OM lipid asymmetry by translocating PLs from the inner leaflet to the outer leaflet [91]. In response to PLs within the outer leaflet, LpxC is stabilized and LPS levels are substantially increased. Such high levels of LPS destabilize the OM and can lead to death in starved cells when the cation concentration is low. As expected, deletion of pldA in the mlaA* background further increases the amount of phospholipids within the outer leaflet. Surprisingly however, deletion of pldA suppresses MlaA*-mediated cell death by reducing LPS levels.
These observations helped to identify PldA as the sentinel in a novel signal transduction pathway that uses outer-leaflet PLs as a proxy for decreased LPS content in the OM [69]. Acyl chains released from PLs that have been degraded by PldA are transported into the cytoplasm and attached to coenzyme A (CoA) by FadD. Acyl-CoA increases LPS levels by inhibiting activity of the YciM/FtsH protease complex (Figure 3). How acyl-CoA inhibits the YciM/FtsH complex is not known. One possibility is that acyl-CoA increases the activity of YejM. However, deletion of pldA exacerbates the growth defect of a yejM mutant [92], suggesting that PldA and YejM may regulate LpxC degradation through independent pathways. Alternatively, PldA-derived acyl-CoA may decrease levels of lipid A disaccharide. Acyl-CoA is known to inhibit FabI [93] (Figure 3), and reduced FabI activity stabilizes LpxC by decreasing levels of lipid A disaccharide [16,53]. Whether lipid A disaccharide is involved in PldA signaling remains to be determined.
Other LpxC Regulators
LpxC protein levels are affected by growth rate. During slow growth, LpxC is rapidly degraded, while during fast growth, LpxC is relatively stable [94]. Growth-dependent proteolysis of LpxC is reversed in mutants unable to synthesize the alarmone (p)ppGpp [94], suggesting that LpxC degradation may be affected by the stringent response.
LpxC is destabilized in mutants overexpressing PyrH [61], which is involved in the synthesis of pyrimidine nucleotides [95]. It was hypothesized that PyrH may regulate LpxC stability by affecting the pool of UDP-GlcNAc available for lipid A biosynthesis [61]. Intriguingly, several studies have shown that PyrH interacts directly with LpxC [61,96]. Whether this interaction influences LpxC stability is unknown.
Concluding Remarks
Maintaining the appropriate level and localization of LPS is critical for establishing a permeability barrier and preserving stiffness of Gram-negative bacterial cells. Because both an excess and a shortage of LPS compromises these critical properties, LpxC protein levels are regulated to ensure the appropriate production of LPS. A variety of signals regulate LPS levels by adjusting activity of the LpxC degradation machinery, including OM LPS via PldA, IM LPS via YejM, and cytoplasmic lipid biosynthesis via lipid A disaccharide. While recent work has deepened our understanding of how LPS levels are monitored and maintained, many questions still remain (see Outstanding Questions). Deciphering the complete regulatory network controlling LPS biogenesis will provide critical insight into Gram-negative bacterial physiology and reveal novel drug targets that can be exploited in the fight against antibiotic resistance.
Outstanding Questions.
How long does LPS reside in the periplasmic leaflet of the IM before it is transported to the OM? Do LPS modifications affect transport?
Why is IM LPS lethal? Does IM LPS form rafts?
How is lipid A disaccharide sensed?
How does YejM regulate the YciM/FtsH complex? Does the periplasmic domain of YejM inhibit FtsH directly?
Does YejM play a direct role in PL sensing, biogenesis, and/or transport?
Does acyl-CoA regulate YciM by inhibiting FabI? Are lipid A disaccharide levels affected in the PldA signaling pathway?
LpxC degradation is regulated by other factors, such as ppGpp and PyrH. How do these factors alter activity of the YciM/FtsH protease complex?
How are LPS levels monitored and maintained in other Gram-negative bacteria, such as Pseudomonas aeruginosa?
Why can some Gram-negative bacteria tolerate loss of lipid A while most cannot?
Highlights.
Lipopolysaccharide (LPS) is found in most Gram-negative bacteria and is critical for outer membrane (OM) barrier function and maintaining cell rigidity.
Gram-negative bacteria precisely regulate LPS levels as imbalances are lethal: too much LPS appears to disrupt inner-membrane integrity while too little weakens the OM.
In Escherichia coli, LPS levels are regulated by the YciM/FtsH protease complex, which degrades the enzyme that catalyzes the first committed step in lipid A biogenesis, LpxC.
Factors in multiple cellular compartments regulate activity of the YciM/FtsH protease complex in response to changes in LPS levels, including the essential inner-membrane protein YejM, the outer-membrane phospholipase PldA, and the lipid A biosynthetic intermediate lipid A disaccharide in the cytoplasm.
Acknowledgments
Work in the Silhavy laboratory is supported by the National Institute of General Medical Sciences of the National Institutes of Health under grant 5R35GM118024. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Glossary
- AAA+ proteases
proteases in the ATPases associated with diverse cellular activities protein superfamily
- Acyl chain
an aliphatic alkyl chain with an acyl group at one end
- Glucosamine
an amino sugar that is a part of lipopolysaccharide, peptidoglycan, and other envelope polysaccharides
- Glutathione-S-transferases
a family of enzymes that attach glutathione to a variety of compounds. Glutathione-S-transferase from Schistosoma japonicum is used as a model substrate to investigate protein degradation by FtsH
- Palmitate
a saturated acyl chain containing 16 carbons
- Saturated fatty acid
a fatty acid in which all of the bonds in the alkyl chain are saturated with hydrogen molecules
- Suppressor mutation
a mutation that masks the phenotype(s) associated with an existing mutation
- Type II fatty acid biosynthesis
a fatty acid biosynthetic pathway that uses multiple proteins as opposed to a single protein
- Undecaprenyl phosphate
an IM lipid carrier involved in the synthesis of peptidoglycan, O antigen, and other envelope polysaccharides
- Unsaturated fatty acid
a fatty acid with an alkyl chain that contains one or more double bonds
- Vesiculation
the act of forming a lipid vesicle derived from the OM
References
- 1.Silhavy TJ et al. , (2010) The bacterial cell envelope. Cold Spring Harb. Perspect. Biol 2, a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitfield C and Trent MS (2014) Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem 83, 99–128 [DOI] [PubMed] [Google Scholar]
- 3.Rietschel ET and Cavaillon J-M (2002) Endotoxin and anti-endotoxin. The contribution of the schools of Koch and Pasteur: life, milestone-experiments and concepts of Richard Pfeiffer (Berlin) and Alexandre Besredka (Paris). J. Endotoxin Res 8, 71–82 [DOI] [PubMed] [Google Scholar]
- 4.Nikaido H (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev 67, 593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rojas ER et al. , (2018) The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559, 617–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jefferies D et al. , (2019) Role of O-antigen in response to mechanical stress of the E. coli outer membrane: insights from coarse-grained MD simulations. J. Phys. Chem. B 123, 3567–3575 [DOI] [PubMed] [Google Scholar]
- 7.Peng D et al. , (2005) Moraxella catarrhalis bacterium without endotoxin, a potential vaccine candidate. Infect. Immun 73, 7569–7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steeghs L et al. (1998) Meningitis bacterium is viable without endotoxin. Nature 392, 449. [DOI] [PubMed] [Google Scholar]
- 9.Moffatt JH et al. , (2010) Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother 54, 4971–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raetz CRH and Whitfield C (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sperandeo P et al. , (2017) Lipopolysaccharide biogenesis and transport at the outer membrane of Gram-negative bacteria. Biochim. Biophys. Acta 1862, 1451–1460 [DOI] [PubMed] [Google Scholar]
- 12.Bertani B and Ruiz N (2018) Function and biogenesis of lipopolysaccharides. EcoSal Plus 8. 10.1128/ecosalplus. ESP-001-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson MS et al. (1993) UDP-N-acetylglucosamine acyl-transferase of Escherichia coli. The first step of endotoxin biosynthesis is thermodynamically unfavorable. J. Biol. Chem 268, 19858–19865 [PubMed] [Google Scholar]
- 14.Emiola A et al. , (2013) A model for the proteolytic regulation of LpxC in the lipopolysaccharide pathway of Escherichia coli. Comput. Biol. Chem 47, 1–7 [DOI] [PubMed] [Google Scholar]
- 15.Emiola A et al. , (2015) A Complete pathway model for lipid A biosynthesis in Escherichia coli. PLoS ONE 10, e0121216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emiola A et al. , (2016) Crosstalk between the lipopolysaccharide and phospholipid pathways during outer membrane biogenesis in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 113, 3108–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z et al. , (2015) Influence of core oligosaccharide of lipopolysaccharide to outer membrane behavior of Escherichia coli. Mar. Drugs 13, 3325–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu A et al. , (2010) Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob. Agents Chemother 54, 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doerrler WT et al. (2004) MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem 279, 45102–45109 [DOI] [PubMed] [Google Scholar]
- 20.Mi W et al. , (2017) Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho H et al. , (2018) Structural basis for dual-mode inhibition of the ABC transporter MsbA. Nature 557,196–201 [DOI] [PubMed] [Google Scholar]
- 22.Bi Y et al. , (2018) Architecture of a channel-forming O-antigen polysaccharide ABC transporter. Nature 553, 361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalynych S et al. (2014) Progress in understanding the assembly process of bacterial O-antigen. FEMS Microbiol. Rev 38, 1048–1065 [DOI] [PubMed] [Google Scholar]
- 24.Han W et al. (2012) Defining function of lipopolysaccharide O-antigen ligase WaaL using chemoenzymatically synthesized substrates. J. Biol. Chem 287, 5357–5365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olaitan AO et al. (2014) Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol 5, 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan A et al. , (2007) An undecaprenyl phosphate-aminoarabinose flippase required for polymyxin resistance in Escherichia coli. J. Biol. Chem 282, 36077–36089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkar S et al. , (2014) Role of capsule and O Antigen in the virulence of uropathogenic Escherichia coli. PLoS ONE 9, e94786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomás JM et al. (1988) Role of lipopolysaccharide and complement in susceptibility of Escherichia coli and Salmonella typhimurium to non-immune serum. Microbiology 134, 1009–1016 [DOI] [PubMed] [Google Scholar]
- 29.Bentley AT and Klebba PE (1988) Effect of lipopolysaccharide structure on reactivity of antiporin monoclonal antibodies with the bacterial cell surface. J. Bacteriol 170, 1063–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Storek KM et al. (2018) Monoclonal antibody targeting the β-barrel assembly machine of Escherichia coli is bactericidal. Proc. Natl. Acad. Sci. U. S. A 115, 3692–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clumeck N et al. , (1982) Serum sensitivity of strains isolated and antibodies against O antigen in gram-negative bacteraemia. Scand. J. Infect. Dis 14, 283–288 [DOI] [PubMed] [Google Scholar]
- 32.Maria-Neto S et al. (2015) Understanding bacterial resistance to antimicrobial peptides: From the surface to deep inside. Biochim. Biophys. Acta 1848, 3078–3088 [DOI] [PubMed] [Google Scholar]
- 33.Okuda S et al. , (2016) Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat. Rev. Microbiol 14, 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.May JM et al. (2015) Lipopolysaccharide transport to the cell surface: periplasmic transport and assembly into the outer membrane. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci 370, 20150027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sperandeo P et al. , (2019) Lipopolysaccharide biosynthesis and transport to the outer membrane of Gram-negative bacteria. In Bacterial Cell Walls and Membranes (Kuhn A, ed.), pp. 9–37, Springer International Publishing; [DOI] [PubMed] [Google Scholar]
- 36.Simpson BW et al. , (2015) Lipopolysaccharide transport to the cell surface: biosynthesis and extraction from the inner membrane. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci 370,20150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman DJ et al. (2018) Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 359, 798–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okuda S et al. (2012) Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338, 1214–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Owens TW et al. , (2019) Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature 567, 550–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang X et al. , (2019) Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism. Nat. Commun 10,4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson BW et al. , (2019) Combining mutations that inhibit two distinct steps of the ATP hydrolysis cycle restores wild-type function in the lipopolysaccharide transporter and shows that ATP binding triggers transport. mBio 10, e01931–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y et al. , (2019) Structural basis of lipopolysaccharide extraction by the LptB 2 FGC complex. Nature 567, 486–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo Q et al. , (2017) Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG. Nat. Struct. Mol. Biol 24,469–474 [DOI] [PubMed] [Google Scholar]
- 44.Sherman DJ et al. , (2014) Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport. Proc. Natl. Acad. Sci. U. S. A 111,4982–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laguri C et al. , (2017) Interaction of lipopolysaccharides at intermolecular sites of the periplasmic Lpt transport assembly. Sci. Rep 7, 9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schultz KM et al. , (2017) Lipopolysaccharide binding to the periplasmic protein LptA. Protein Sci. 26, 1517–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sestito SE et al. , (2014) Functional characterization of E. coli LptC: interaction with LPS and a synthetic ligand. Chembiochem 15, 734–742 [DOI] [PubMed] [Google Scholar]
- 48.Botos I et al. (2016) Structural and functional characterization of the LPS transporter LptDE from Gram-negative pathogens. Structure 24, 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X et al. , (2015) Trapped lipopolysaccharide and LptD intermediates reveal lipopolysaccharide translocation steps across the Escherichia coli outer membrane. Sci. Rep 5, 11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu Y et al. , (2015) Lipopolysaccharide is inserted into the outer membrane through an intramembrane hole, a lumen gate, and the lateral opening of LptD. Structure 23, 496–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malojčić G et al. , (2014) LptE binds to and alters the physical state of LPS to catalyze its assembly at the cell surface. Proc. Natl. Acad. Sci. U. S. A 111, 9467–9472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sorensen PG et al. (1996) Regulation of UDP-3-O-[R-3-hydroxymyristoyl]-N-acetylglucosamine deacetylase in Escherichia coli. The second enzymatic step of lipid a biosynthesis. J. Biol. Chem 271, 25898–25905 [DOI] [PubMed] [Google Scholar]
- 53.Ogura T et al. (1999) Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol. Microbiol 31, 833–844 [DOI] [PubMed] [Google Scholar]
- 54.Bittner L-M et al. (2017) When, how and why? Regulated proteolysis by the essential FtsH protease in Escherichia coli. Biol. Chem 398, 625–635 [DOI] [PubMed] [Google Scholar]
- 55.Klein G et al. (2014) Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J. Biol. Chem 289, 14829–14853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahalakshmi S et al. (2014) yciM is an essential gene required for regulation of lipopolysaccharide synthesis in Escherichia coil. Mol. Microbiol 91, 145–157 [DOI] [PubMed] [Google Scholar]
- 57.Nicolaes V et al. (2014) Insights into the function of YciM, a heat shock membrane protein required to maintain envelope integrity in Escherichia coli. J. Bacteriol 196, 300–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prince C and Jia Z (2015) An unexpected duo: rubredoxin binds nine TPR motifs to form LapB, an essential regulator of lipopolysaccharide synthesis. Structure 23,1500–1506 [DOI] [PubMed] [Google Scholar]
- 59.Putker F et al. (2014) Ght protein of Neisseria meningitidis is involved in the regulation of lipopolysaccharide biosynthesis. J. Bacteriol 196, 780–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.D’Andrea LD and Regan L (2003) TPR proteins: the versatile helix. Trends Biochem. Sci 28, 655–662 [DOI] [PubMed] [Google Scholar]
- 61.Thomanek N et al. (2018) Intricate crosstalk between lipopolysaccharide, phospholipid and fatty acid metabolism in Escherichia coli modulates proteolysis of LpxC. Front. Microbiol 9, 3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Führer F et al. (2006) The C-terminal end of LpxC is required for degradation by the FtsH protease. Mol. Microbiol 59,1025–1036 [DOI] [PubMed] [Google Scholar]
- 63.Führer F et al. (2007) Sequence and length recognition of the C-terminal turnover element of LpxC, a soluble substrate of the membrane-bound FtsH protease. J. Mol. Biol 372, 485–496 [DOI] [PubMed] [Google Scholar]
- 64.Falchi FA et al. (2018) Mutation and suppressor analysis of the essential lipopolysaccharide transport protein LptA reveals strategies to overcome severe outer membrane permeability defects in Escherichia coli. J. Bacteriol 200, e00487–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sampson BA et al. (1989) Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics 122, 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chimalakonda G et al. (2011) Lipoprotein LptE is required for the assembly of LptD by the beta-barrel assembly machine in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 108, 2492–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Young K and Silver LL (1991) Leakage of periplasmic enzymes from envA1 strains of Escherichia coli. J. Bacteriol 173, 3609–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bishop RE (2008) Structural biology of membrane-intrinsic β-barrel enzymes: Sentinels of the bacterial outer membrane. Biochim. Biophys. Acta 1778, 1881–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.May KL and Silhavy TJ (2018) The Escherichia coli phospholipase PldA regulates outer membrane homeostasis via lipid signaling. mBio 9, 00379–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bishop RE et al. (2000) Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 19, 5071–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jia W et al. (2004) Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J. Biol. Chem 279, 44966–44975 [DOI] [PubMed] [Google Scholar]
- 72.Tam C and Missiakas D (2005) Changes in lipopolysaccharide structure induce the sigma(E)-dependent response of Escherichia coli. Mol. Microbiol 55, 1403–1412 [DOI] [PubMed] [Google Scholar]
- 73.Whitfield C et al. (1997) Modulation of the surface architecture of gram-negative bacteria by the action of surface polymer:lipid A-core ligase and by determinants of polymer chain length. Mol. Microbiol 23, 629–638 [DOI] [PubMed] [Google Scholar]
- 74.Jorgenson MA and Young KD (2016) Interrupting biosynthesis of O antigen or the lipopolysaccharide core produces morphological defects in Escherichia coli by sequestering undecaprenyl phosphate. J. Bacteriol 198, 3070–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heath RJ and Rock CO (1996) Roles of the FabA and FabZ β-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J. Biol. Chem 271, 27795–27801 [DOI] [PubMed] [Google Scholar]
- 76.Mohan S et al. (1994) An Escherichia coli gene (FabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase. Relation to fabA and suppression of mutations in lipid A biosynthesis. J. Biol. Chem 269, 32896–32903 [PubMed] [Google Scholar]
- 77.Zeng D et al. (2013) Mutants resistant to LpxC inhibitors by rebalancing cellular homeostasis. J. Biol. Chem 288, 5475–5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mostafavi M et al. (2018) Interplay of Klebsiella pneumoniae fabZ and lpxC Mutations leads to LpxC inhibitor-dependent growth resulting from loss of membrane homeostasis. mSphere 3, e00508–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guest RL et al. (2020) YejM modulates activity of the YciM/FtsH protease complex to prevent lethal accumulation of lipopolysaccharide. mBio 11, e00598–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwechheimer C et al. (2014) Modulation of bacterial outer membrane vesicle production by envelope structure and content. BMC Microbiol. 14, 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De Lay NR and Cronan JE (2008) Genetic interaction between the Escherichia coli AcpT phosphopantetheinyl transferase and the YejM inner membrane protein. Genetics 178,1327–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dalebroux ZD et al. (2015) Delivery of cardiolipins to the Salmonella outer membrane is necessary for survival within host tissues and virulence. Cell Host Microbe 17, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fan J et al. (2020) Structure of an inner membrane protein required for PhoPQ-regulated increases in outer membrane cardiolipin. mBio 11, e03277–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Clairfeuille T et al. Structure of the essential inner membrane lipopolysaccharide-PbgA complex. Nature 584, 479–483 [DOI] [PubMed] [Google Scholar]
- 85.Hirvas L et al. (1997) The lipid a biosynthesis deficiency of the Escherichia coli antibiotic-supersensitive mutant LH530 is suppressed by a novel locus, orf195. Microbiology 143, 73–81 [DOI] [PubMed] [Google Scholar]
- 86.Nurminen M et al. (1997) The outer membrane of lipid A-deficient Escherichia coli mutant LH530 has reduced levels of OmpF and leaks periplasmic enzymes. Microbiology 143, 1533–1537 [DOI] [PubMed] [Google Scholar]
- 87.Dong H et al. (2016) Structural insights into cardiolipin transfer from the Inner membrane to the outer membrane by PbgA in Gram-negative bacteria. Sci. Rep 6, 30815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cian MB et al. (2019) Salmonella enterica serovar Typhimurium uses PbgA/YejM to regulate lipopolysaccharide assembly during bacteremia. Infect. Immun 88, e00758–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fivenson EM and Bernhardt TG (2020) An essential membrane protein modulates the proteolysis of LpxC to control lipopolysaccharide synthesis in Escherichia coli. mBio 11, 00939–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nguyen D et al. (2020) YejM controls LpxC levels by regulatng protease activity of the FtsH/YciM complex of Escherichia coli. J. Bacteriol 202, e00303–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sutterlin HA et al. (2016) Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc. Natl. Acad. Sci. U. S. A 113, E1565–E1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qiu N and Misra R (2019) Overcoming iron deficiency of an Escherichia coli tonB mutant by increasing outer membrane permeability. J. Bacteriol 201, e00340–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bergler H et al. (1996) The enoyl-[acyl-carrier-protein] reductase (FabI) of Escherichia coli, which catalyzes a key regulatory step in fatty acid biosynthesis, accepts NADH and NADPH as cofactors and is inhibited by palmitoyl-CoA. Eur. J. Biochem 242, 689–694 [DOI] [PubMed] [Google Scholar]
- 94.Schäkermann M et al. (2013) FtsH-mediated coordination of lipopolysaccharide biosynthesis in Escherichia coli correlates with the growth rate and the alarmone (p)ppGpp. J. Bacteriol 195, 1912–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Serina L et al. (1995) Escherichia coli UMP-kinase, a member of the aspartokinase family, is a hexamer regulated by guanine nucleotides and UTP. Biochemistry 34, 5066–5074 [DOI] [PubMed] [Google Scholar]
- 96.Butland G et al. (2005) Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433, 531–537 [DOI] [PubMed] [Google Scholar]
- 97.Langklotz S et al. (2011) Control of lipopolysaccharide biosynthesis by FtsH-mediated proteolysis of LpxC is conserved in enterobacteria but not in all gram-negative bacteria. J. Bacteriol 193,1090–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hinz A et al. (2011) Membrane proteases and aminoglycoside antibiotic resistance. J. Bacteriol 193, 4790–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Basta DW et al. (2020) Heat-shock proteases promote survival of Pseudomonas aeruginosa during growth arrest. Proc. Natl. Acad. Sci. U. S. A 117, 4358–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tomaras AP et al. (2014) LpxC inhibitors as new antibacterial agents and tools for studying regulation of lipid A biosynthesis in Gram-negative pathogens. mBio 5, e01551–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kahler CM and Stephens DS (1998) Genetic basis for biosynthesis, structure, and function of meningococcal lipooligosaccharide (endotoxin). Crit. Rev. Microbiol 24, 281–334 [DOI] [PubMed] [Google Scholar]
- 102.Ventola CL (2015) The antibiotic resistance crisis: part 1: causes and threats. P & T 40, 277–283 [PMC free article] [PubMed] [Google Scholar]
- 103.Lewis K (2013) Platforms for antibiotic discovery. Nat. Rev. Drug Discov 12, 371–387 [DOI] [PubMed] [Google Scholar]
- 104.Hofer U (2019) The cost of antimicrobial resistance. Nat. Rev. Microbiol 17, 3. [DOI] [PubMed] [Google Scholar]
- 105.O’Neill J (2014) Antimicrobial resistance: tackling a crisis for the health and wealth of nations, Review on Antimicrobial Resistance, London, UK [Google Scholar]
- 106.Erwin AL (2016) Antibacterial drug discovery targeting the lipopolysaccharide biosynthetic enzyme LpxC. Cold Spring Harb. Perspect. Med 6, a025304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Raetz CRH et al. (2009) Discovery of new biosynthetic pathways: the lipid A story. J. Lipid Res 50, S103–S108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee C-R et al. (2013) Lipid a biosynthesis of multidrug-resistant pathogens – a novel drug target. Curr. Pharm. Des 19, 6534–6550 [DOI] [PubMed] [Google Scholar]
- 109.Surivet J-P et al. (2020) Discovery of novel inhibitors of LpxC displaying potent in vitro activity against Gram-negative bacteria. J. Med. Chem 63, 66–87 [DOI] [PubMed] [Google Scholar]
- 110.Krause KM et al. (2019) Potent LpxC inhibitors with in vitro activity against multidrug-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother 63, e00977–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Barb AW et al. (2007) Inhibition of lipid A biosynthesis as the primary mechanism of CHIR-090 antibiotic activity in Escherichia coli. Biochemistry 46, 3793–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jenkins RJ and Dotson GD (2012) Dual targeting antibacterial peptide inhibitor of early lipid A biosynthesis. ACS Chem. Biol 7, 1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Caughlan RE et al. (2012) Mechanisms decreasing in vitro susceptibility to the LpxC inhibitor CHIR-090 in the Gram-negative pathogen Pseudomonas aeruginosa. Antimicrob. Agents Chemother 56, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wei J-R et al. (2017) LpxK is essential for growth of Acinetobacter baumannii ATCC 19606: relationship to toxic accumulation of lipid A pathway intermediates. mSphere 2, e00199–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Richie DL et al. (2018) A pathway-directed positive growth restoration assay to facilitate the discovery of lipid A and fatty acid biosynthesis inhibitors in Acinetobacter baumannii. PLoS ONE 13, e0193851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yao J and Rock CO (2016) Resistance mechanisms and the future of bacterial enoyl-acyl carrier protein reductase (FabI) antibiotics. Cold Spring Harb. Perspect. Med 6, a027045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang G et al. (2018)Cell-based screenfordiscovering lipopolysaccharide biogenesis inhibitors. Proc. Natl. Acad. Sci. U. S. A 115, 6834–6839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang X et al. (2019) Identification of an anti-Gram-negative bacteria agent disrupting the interaction between lipopolysaccharide transporters LptA and LptC. Int. J. Antimicrob. Agents 53, 442–448 [DOI] [PubMed] [Google Scholar]
- 119.Vetterli SU et al. (2018) Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport in Escherichia coli. Sci. Adv 4, eaau2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Srinivas N et al. (2010) Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327, 1010–1013 [DOI] [PubMed] [Google Scholar]
- 121.Andolina G et al. (2018) A peptidomimetic antibiotic interacts with the periplasmic domain of LptD from Pseudomonas aeruginosa. ACS Chem. Biol 13, 666–675 [DOI] [PubMed] [Google Scholar]
- 122.Werneburg M et al. (2012) Inhibition of lipopolysaccharide transport to the outer membrane in Pseudomonas aeruginosa by peptidomimetic antibiotics. Chembiochem 13, 1767–1775 [DOI] [PubMed] [Google Scholar]
- 123.Trimble MJ et al. (2016) Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb. Perspect. Med 6, a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schaenzer AJ and Wright GD (2020) Antibiotic resistance by enzymatic modification of antibiotic targets. Trends Mol. Med 26, 768–782 [DOI] [PubMed] [Google Scholar]
- 125.Brown P et al. (2019) Design of next generation polymyxins with lower toxicity: The discovery of SPR206. ACS Infect. Dis 5, 1645–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ongwae GM et al. (2020) Broadening activity of polymyxin by quaternary ammonium grafting. ACS Infect. Dis 6, 1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang Y et al. (2020) Evaluation of the in vitro activity of new polymyxin B analogue SPR206 against clinical MDR, colistin-resistant and tigecycline-resistant Gram-negative bacilli. J. Antimicrob. Chemother 75, 2609–2615 [DOI] [PubMed] [Google Scholar]
- 128.Rossi RM et al. (2017) Cardiolipin synthesis and outer membrane localization are required for Shigella flexneri virulence. mBio 8, e01199–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tan BK et al. (2012) Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc. Natl. Acad. Sci. U. S. A 109, 16504–16509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lu D et al. (2009) Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. Proc. Natl. Acad. Sci. U. S. A 106, 1584–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Anandan A et al. (2017) Structure of a lipid A phosphoethanolamine transferase suggests how conformational changes govern substrate binding. Proc. Natl. Acad. Sci. U. S. A 114, 2218–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mackinnon FG et al. (2002) Identification of a gene (lpt-3) required for the addition of phosphoethanolamine to the lipopolysaccharide inner core of Neisseria meningitidis and its role in mediating susceptibility to bactericidal killing and opsonophagocytosis. Mol. Microbiol 43, 931–943 [DOI] [PubMed] [Google Scholar]
- 133.Simpson BW and Trent MS (2019) Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol 17, 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Heath RJ et al. (2002) Chapter 3. Fatty acid and phospholipid metabolism in prokaryotes. In Biochemistry of Lipids, Lipoproteins, and Membranes (4th edn) (Vance JE and Vance D, eds), pp. 55–92, Elsevier Science [Google Scholar]