SUMMARY
Objective:
To determine the contribution of the gut microbiota to the development of injury-induced osteoarthritis (OA).
Design:
OA was induced using the destabilized medial meniscus (DMM) model in 20 germ-free (GF) C57BL/6J male mice housed in a gnotobiotic facility and 23 strain-matched specific pathogen free (SPF) mice in 2 age groups −13.5 weeks avg age at DMM (17 SPF and 15 GF) and 43 weeks avg age at DMM (6 SPF and 5 GF). OA severity was measured using scores for articular cartilage structure (ACS), loss of safranin O (SafO) staining, osteophyte size, and synovial hyperplasia. Microbiome analysis by 16S rRNA amplicon sequencing was performed on stool samples and LPS and LPS binding protein (LBP) were measured in plasma.
Results:
Compared to the SPF DMM mice, the maximum (MAX) ACS score per joint was 28% lower (p = 0.036) in GF DMM mice while the SafO sum score of all sections evaluated per joint was decreased by 31% (p = 0.009). The differences between SPF and GF mice in these scores were greater when only the younger mice were included in the analysis. The younger GF DMM mice also had significant reductions in osteophyte size (36%, P = 0.0119) and LBP (27%, P = 0.007) but not synovial scores or LPS. Differences in relative abundance of a number of Operational Taxonomic Units (OTUs) were noted between SPF mice with high vs low maximum ACS scores.
Conclusions:
These results suggest factors related to the gut microbiota promote the development of OA after joint injury.
Keywords: Osteoarthritis, Cartilage, Microbiome, Germ-free, DMM, LBP
Introduction
A number of risk factors play an important role in osteoarthritis (OA), including age, obesity, joint trauma, and genetic predisposition1. There is increasing evidence for an inflammatory component to OA that promotes joint tissue destruction by causing an imbalance in anabolic and catabolic processes within the affected joint2,3. The gut microbiota plays a key role in the development and function of the immune system and allergic and inflammatory responses4–7. Microbiome alterations can activate the innate immune system resulting in increased production of proinflammatory cytokines that could affect multiple organs, including the joint8,9.
A number of inflammatory diseases, including ankylosing spondylitis (AS) and rheumatoid arthritis (RA) have been linked to an altered composition of the microbiota10,11. Recent work also suggests a role for the microbiome in OA. In a rat model of diet-induced obesity, a link was made between the severity of cartilage damage and the amount of fat mass, serum lipopolysaccharide (LPS) levels and presence of a number of bacterial species in fecal samples, including Lactobacillus and Methanobrevibacter12. In a metabolomics study of overweight and obese humans with knee OA, higher levels of urinary hippurate, a mammalian-microbial cometabolite, were noted in the participants who progressed radiographically over an 18-month period13.
The gut microbiota can be a source of factors such as lipopolysaccharide (LPS) that promote inflammation. An association was noted between serum levels of LPS and LPS binding protein (LBP) and the severity of radiographic OA and joint symptoms14. A two-hit model of OA pathogenesis8,14 was proposed in which bacterial products that function as toll-like receptor (TLR) ligands, including LPS, act as one hit by activating the innate immune response. Another hit results from joint damage that releases a number of damage associated molecular patterns (DAMPS), such as hyaluronan and fibronectin fragments that also activate the innate immune system, resulting in a synergistic activation of inflammatory pathways leading to OA.
We hypothesized that in the presence of joint injury as one hit, the gut microbiota could provide a source of microbial products that activate innate immune responses to promote a chronic low-grade pro-inflammatory state, serving as the other hit that contributes to the development of OA. To test this hypothesis, we measured the severity of OA induced after joint injury by destabilized medial meniscus (DMM) surgery in germ-free (GF) mice that lack a gut microbiota compared to specific pathogen-free (SPF) mice.
Materials and methods
Experimental animals and study design
Mice used in this study were purchased from Jackson Labs but had been maintained as separate colonies. Male C57BL/6J GF mice were born, raised and maintained inside sterile isolators at the National Gnotobiotic Rodent Resource Center at the University of North Carolina (UNC), Chapel Hill, NC. Male C57BL/6J SPF mice were born, raised and maintained in a standard SPF facility at the same institution. Animals had access to water and food ad libitum. All animal experiments were approved by the UNC Animal Care and Use Committee and followed the recommendations from the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
The study was designed to determine if male GF mice develop less severe histologic OA 8 weeks after DMM surgery when compared to SPF mice. Based on a previous study from our group using DMM surgery to induce OA and the articular cartilage structure score (ACS) as the outcome measure15, we determined that 12 mice per group would provide greater than 80% power to detect at least a 50% difference in ACS scores between GF and SPF mice. GF mice have been reported to be very sensitive to anesthesia and so we decided to increase the number of mice per group to 25 in case GF mice died during surgery or in the postoperative period and to increase the power of the study. The initial plan was to use mice between 12 and 16 weeks of age at the time of surgery. However, due to competing demands for GF mice by other investigators at our gnotobiotic facility, some of the mice provided to us were older. Therefore, the study was performed with two age groups of mice: a larger, younger group with ages between 12 and 18 weeks at the time of DMM surgery (17 SPF and 19 GF), and a smaller, somewhat older group, containing animals with ages between 37 and 48 weeks (6 SPF and 7 GF) at the time of DMM surgery. Six GF mice died prior to necropsy: four in the young and one in the old GF group did not recover from anesthesia and one was found dead 5 weeks after surgery.
Destabilization of the medial meniscus (DMM) surgery
DMM was induced by transecting the medial meniscotibial ligament in the right (R) knee, as described previously15–18. Details of the surgical procedure are provided in the supplemental methods. Due to the limited availability of GF mice described above and our desire to maximize the number of mice in the DMM groups, we performed the DMM procedure on one knee in all mice and used the unoperated left (L) leg as a control instead of performing sham surgery in a separate group of animals. In our previous studies, we had not seen any differences in OA severity between sham operated and age-matched unoperated control joints15. The animals were sacrificed 8 weeks post-DMM and knee joints, blood and stool samples were collected.
Histology assessment of OA severity
All DMM operated legs (23 SPF, 20 GF) were evaluated histologically, while a subset of the contralateral control legs (17 SPF and 15 GF) were evaluated to confirm the expected lack of OA in unoperated joints. Details of the processing of the joints are provided in the Supplemental methods.
All histological scores were obtained by an evaluator (VU) blinded to the group assignment (all mice were given a numerical code). The ACS score20 was used to grade the cartilage lesions. The ACS score is similar to the Osteoarthritis Research Society International (OARSI) scoring system but has a wider range of scores, from 0 to 12 rather than 0–6. ACS scores were analyzed in all four joint quadrants, on two sections, one located in the mid-coronal region (stained with H&E) and another one taken ~80 μm posteriorly (stained with safranin O/fast green). We reported the ACS scores as either MAX (MAX score for the four joint quadrants for two sections), SUM (SUM score for all four joint quadrants for two sections) or MT-MC (score for the medial tibia (MT) in the mid-coronal (MC) section). In addition to the ACS score, we analyzed the safranin O staining score (SafO score) using a previously published scoring system20 that was modified to follow the same 0–12 scale as the ACS score. The SafO scores were reported as MAX and SUM scores calculated as for the ACS scores. The thickness of AC in the MT region was also measured in 3 different areas (medial, lateral and center of the tibial plateau) using ImageJ and the results were reported as average (AVG).
Osteophyte and synovial evaluation
Osteophyte size was measured in the medial joint compartment using a previously described scale from 0–318,21, and the scores were reported separately for MT and medial femur (MF) and as a MAX score of the two. Osteophyte maturity was also scored on a 0–3 scale as described21.
The synovial lining was evaluated for cellular hyperplasia using a score from 0–319 (0 = 1 cell layer in synovium, 1 = 2–3 cell layers, 2 = 4–5 cell layers, and 3 = 5 or more cell layers) in the region between meniscus and either tibia or femur, without scoring the cells in the area attached to the tibia, femur or meniscus. Each of the four joint quadrants was evaluated separately, and the scores were reported either as a MAX for the entire joint or MAX for the medial side (MF and MT), the side most affected after DMM. Images were taken using an Olympus microscope (BX60), camera (DP73) and software (cellSens). In addition, the synovial thickness was measured between medial meniscus (MM) and MT and between MM and MF in the region adjacent to meniscus and at the maximum thickness point.
Plasma analysis for IL-6, LBP and LPS
Blood samples were collected from 17 SPF and 13 GF younger mice. Plasma was obtained from blood collected in EDTA-treated tubes. Details on the ELISA assays for IL-6 and LPS binding protein and the LPS detection assay are provided in the Supplemental methods.
Microbiome analysis
Stool samples were collected from 12 SPF and 9 GF younger mice at necropsy, 8 weeks after DMM, and then processed and analyzed in the Microbiome Core Facility at UNC-Chapel Hill, as previously described22. Briefly, total DNA was isolated from stool samples using the E.Z.N.A. Stool DNA Kit (Omega Bio-Tek, Norcross, GA). Amplicon libraries were generated using primers specific for the V3–V4 hypervariable region of the bacterial 16S rRNA gene. Amplicon sequencing was done on the Illumina MiSeq platform. Details of the microbiome data analysis are provided in the Supplemental methods.
Statistical analysis
Unless otherwise mentioned, the data were analyzed using GraphPad Prism 7.03 (La Jolla California USA). For the OA severity scores, when comparing the SPF and GF mice, we used one-way ANOVA followed by Tukey’s multiple comparisons post-hoc test (to control the Type I statistical error rate among these pairwise comparisons), and a P-value of ≤0.05 was considered statistically significant. Because in previous studies we had noted a difference in OA severity when comparing young adult to older adult mice15 and because the original study design was to use mice between the ages of 12–16 weeks, we decided a priori to do a stratified analysis of the mice by age group. For the analysis of LBP, LPS and IL6 levels in plasma of SPF and GF mice, we used Student’s t-test. OA severity scores and LBP levels were also analyzed using analysis of covariance (ANCOVA), adjusting for weight at necropsy and % weight change. There were five GF mice with missing values for weight at necropsy; sensitivity analyses indicated that OA severity scores and LBP levels were similar for mice with complete weight at necropsy and mice, who did not. Therefore, analyses were carried out based on a complete-case dataset where all mice had complete information for weight at necropsy. ANCOVA analyses were conducted out using SAS 9.4 (SAS Institute Inc., Cary, NC). For the correlation of LBP levels with OA severity scores, we used Pearson’s correlation coefficient. Results are presented as mean ± S.E.M.
Results
Decreased DMM-induced cartilage damage in GF mice
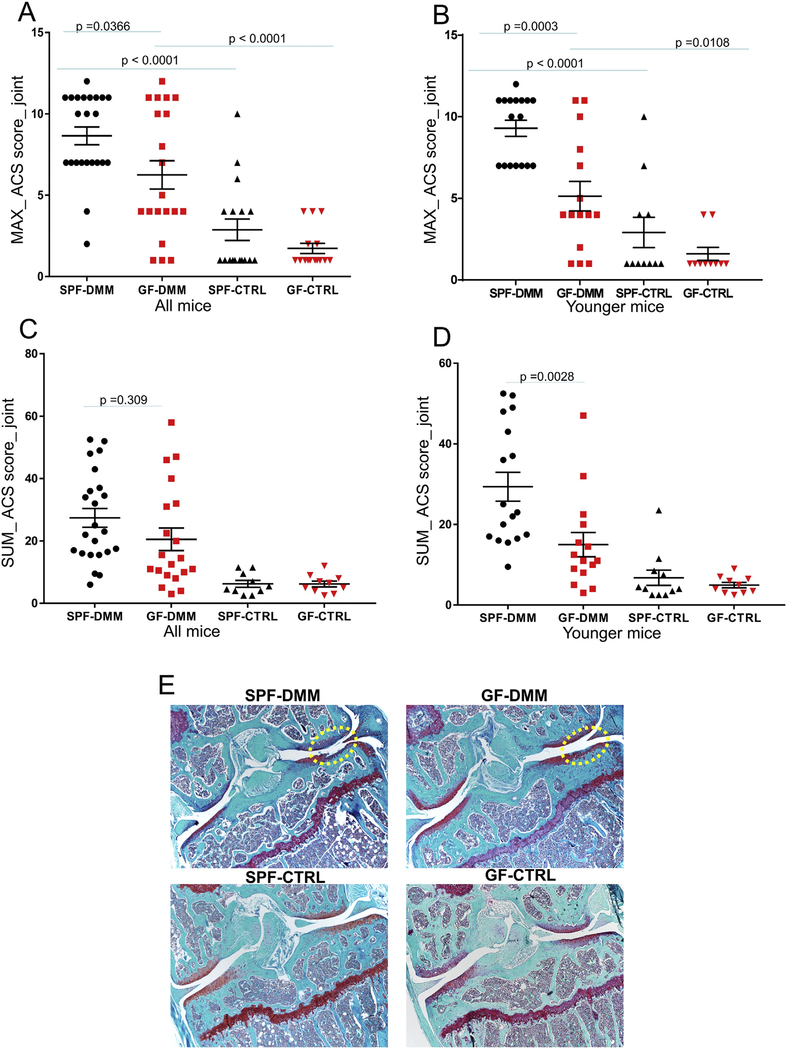
Overall, the ACS scores showed a reduction in cartilage damage after DMM surgery in GF compared to SPF mice, particularly in the group of younger adult mice. As we have seen in other studies using the DMM model, there was some variability in the severity of OA in the DMM joints and no significant cartilage damage was observed in the unoperated control joints [Fig. 1(A), (B)]. The MAX_ACS score in the DMM knees was 28% lower in GF than in SPF mice when all ages were included (P = 0.037) [Fig. 1(A)] and was 45% lower in GF mice when only the younger adult mice were analyzed (P = 0.0003) [Fig. 1(B)]. For the SUM_ACS scores, the results were statistically significant (49% lower in GF, P = 0.003) only for the SPF vs GF comparison in the younger group [Fig. 1(C), (D)]. The only result that was more severe in the GF mice was the ACS score in the older mice when the MT-MC region was analyzed separately (Fig. S1(A)). All scores were lower in the control leg in both GF and SPF mice compared to the DMM operated side [Fig. 1(A)–(D)]. Fig. 1(E) and Figures S2(A) and S2(B) show representative images from SafO-stained joints from DMM and control joints.
Fig. 1.
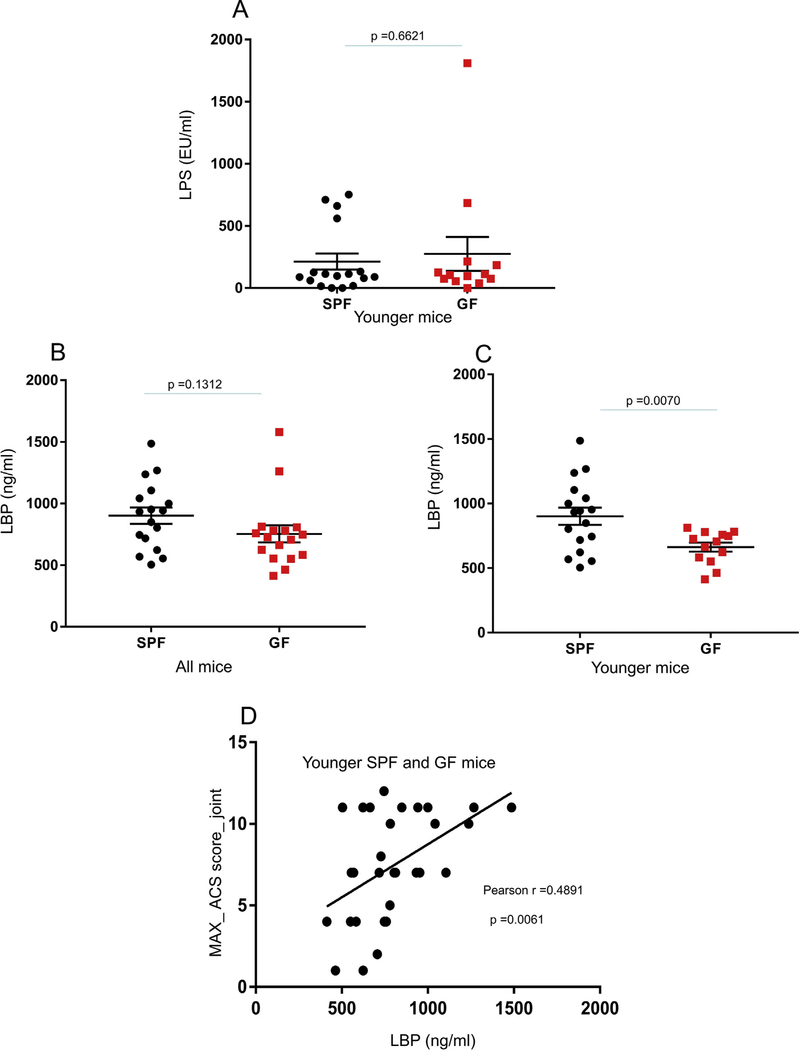
Comparisons of articular cartilage structure scores (ACS) between specific pathogen free (SPF) and germ-free (GF) mice. The ACS_MAX (A,B) and SUM (C,D) scores were derived as detailed in the Methods. Results are shown for the destabilized medial meniscus (DMM) operated and unoperated control (CTRL) joints from mice of all ages (A,B) and when only the larger group of young mice were analyzed (B,D). Representative images from SafO stained joints from DMM and control knees (E). Dot plots represent the mean ± S.E.M.
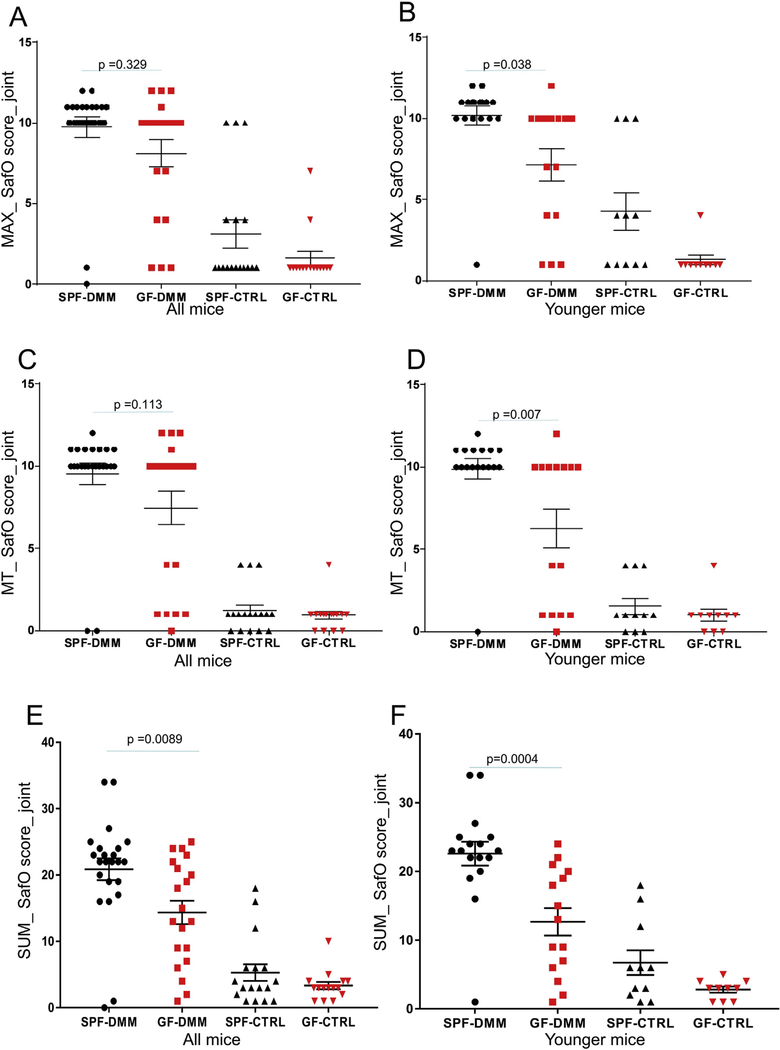
In addition to AC structural changes, we analyzed proteoglycan (PG) loss using the SafO score. Just as with the ACS scores, we noticed less PG loss (lower SafO staining scores) after DMM surgery in the GF mice compared to the SPF mice when reported as either MAX score or SUM score for the joint or when analyzed specifically for MT (Fig. 2). The results were statistically different only when the younger group was analyzed for the SafO_MAX [Fig. 2(B)] and SafO_MT [Fig. 2(D)] scores. The SafO_SUM score was 31% lower in the DMM knees of the GF compared to the SPF mice when all ages were analyzed and 44% lower in young GF-DMM knees compared to the young SPF-DMM [Fig. 2(E), (F)]. In addition, AC-AVG thickness was increased by 30% in the younger GF mice compared to SPF and it was higher in the SPF control compared to the SPF DMM knees (Fig S1(E)).
Fig. 2.
Evaluation of proteoglycan loss by comparing safranin O (SafO) scores in SPF and germ-free (GF) mice. The SafO staining scores are shown for the DMM operated and unoperated control (CTRL) joints and were reported as either MAX scores (A,B), SUM scores (E,F) for the joint or specifically for medial tibia (MT) (C,D). Panels A,C,E are showing results from mice of all ages and B,D,F from the younger mouse group. Dot plots of the mean ± S.E.M.
Differences in osteophyte scores and synovial changes in young GF mice compared to SPF mice after DMM surgery
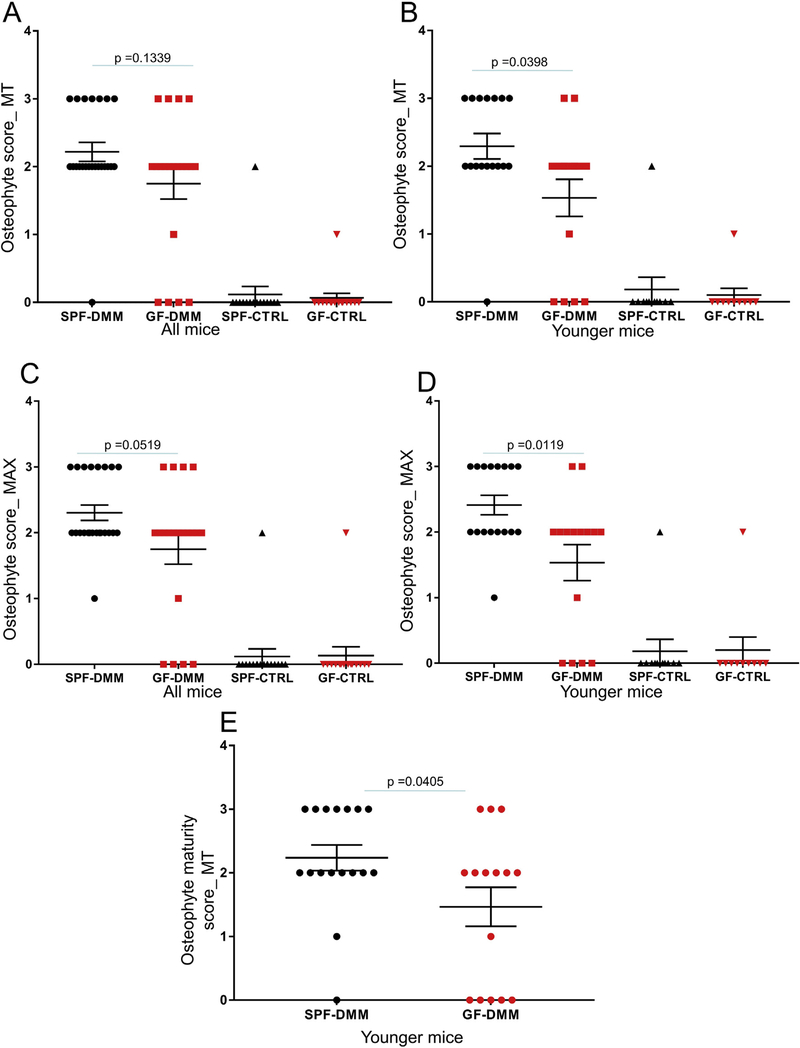
We also observed decreased osteophyte scores in the younger GF mice after DMM surgery compared to SPF mice, at the MT [Fig. 3(A), (B)] or as MAX score for the entire joint [Fig. 3(C), (D)] but not for the MF (Fig.S1(C), (D)). The osteophyte maturity scores were also 34% lower in the younger GF mice compared to SPF mice [Fig. 3(E)].
Fig. 3.
Comparison of osteophyte scores between SPF and germ-free (GF) mice. The osteophyte scores are shown for the DMM operated and unoperated control (CTRL) joints and were reported either separately for MT (A,B) or as a MAX score for the medial side (C,D). Panels A,C are showing results from mice of all ages and B,D, from the younger mouse group. The osteophyte maturity scores are presented in the DMM knees of younger SPF and GF mice (E). Dot plots of the mean ± S.E.M.
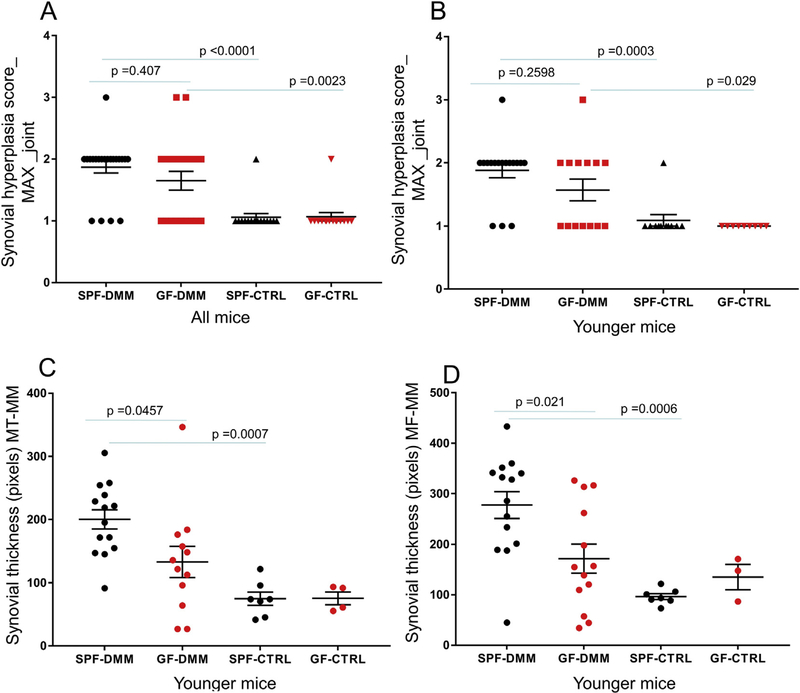
We did not see any differences in synovial hyperplasia scores (MAX score for the entire joint) [Fig. 4(A)] between the GF and SPF mice in the DMM knees, even when specifically analyzed in the younger group [Fig. 4(B)]. As expected, synovial hyperplasia was less in the control unoperated knees compared to DMM knees for both SPF and GF mice [Fig. 4(A), (B)]. We saw similar results when analyzing the scores specifically for the medial side of the joint, which is the most affected side following DMM surgery (MAX_-medial compartment) (Fig. S1(B)). We also measured synovial thickness and found that it was 34% lower in the DMM knee of younger GF vs SPF mice as well as in the unoperated knee of the SPF mice when compared to the DMM knee [Fig. 4(C), (D)].
Fig. 4.
Synovial hyperplasia scores in SPF and germ-free (GF) mice. Synovial hyperplasia was evaluated in DMM operated and unoperated control (CTRL) joints and the scores were reported as MAX score for the entire joint, either for mice of all ages (A) or separately for the younger group of mice (B). Synovial thickness was measured between medial meniscus (MM) and MT and between MT and medial femur (MF) in the DMM and CTRL knees of younger SPF and GF mice (C,D). Dot plots of the mean ± S.E.M.
Plasma LPB, LPS, and IL6 levels in GF and SPF mice
No differences in plasma LPS levels were noted between GF and SPF mice [Fig. 5(A)]. However, measuring LPS in biological samples is known to be challenging due to the presence of inhibitors that interfere with the assay14. We analyzed samples diluted 1000× that exhibited the best LPS spike recovery (lower inhibitors) out of the 4 dilutions tested, however this was still suboptimal (8.4% spike recovery). Most readings were within the range of the lower assay standard, which hindered the utility of further dilutions.
Fig. 5.
Comparisons of LPS and LPS binding protein (LBP) between SPF and germ-free (GF) mice. Concentrations of LPS in plasma samples collected from the younger group of SPF and GF mice (A). Concentrations of LBP in SPF and GF mouse plasma samples were reported for mice of all ages (B) or specifically for the younger mouse group (C). Dot plots of the mean ± S.E.M. (A–C). Scatterplot with overlaid regression line shows the association between ACS_MAX scores and LBP concentration in plasma samples from the SPF and GF younger mouse group (D).
Due to the presence of LPS assay inhibitors in our samples, we analyzed the levels of LBP for which an association with OA severity in humans was also previously reported14. LBP levels were 27% lower in plasma from GF mice compared to SPF controls when analyzed exclusively in the younger animals [Fig. 5(B), (C)]. LBP levels also showed a positive correlation (r = 0.4891, P = 0.0061) with the ACS_MAX score [Fig. 5(D)], in the younger group.
IL6 levels were undetectable in many of the plasma SPF and GF samples. When measureable, we noticed a trend for decreased IL6 levels in the GF mice, but the results were not statistically significant (Fig.S1(F)).
Weight-adjusted results
Previous studies reported that GF mice gain less weight over time when compared to SPF controls23. We also observed this difference in body weight in our current study, in the younger mouse group. The mean (±S.E.M.) body weight at necropsy for the 17 younger SPF mice was 33.3 ± 0.74 g and for the 14 younger GF mice (where we had body weights) was 29 ± 0.64 g (P = 0.0005). The GF mice also gained less weight (8.1%) compared to the SPF mice (11%) over the course of our study. Therefore, to determine whether the reduced OA severity observed in the GF mice might be due to their lower body weight, the previously reported scores were weight-adjusted and re-analyzed. All the scores remained statistically different between GF and SPF mice after adjusting for either body weight at necropsy or for percentage weight change (Table I).
Table I.
Weight-adjusted results in the younger mouse groups
| Scores | Unadjusted | Weight adjusted |
Weight adjusted |
|---|---|---|---|
| Weight at necropsy | % weight change | ||
| ACS_MAX joint score | |||
| Germ-free | 5.00 (0.83) | 4.32 (0.95) | 4.71 (0.80) |
| SPF | 9.29 (0.64) | 9.69 (0.68) | 9.46 (0.61) |
| Difference-mean (SE) | 4.29 (l.04) | 5.37 (1.29) | 4.75 (1.01) |
| P-value* | 0.0002 | 0.0004 | <0.0001 |
| ACS_SUM joint score | |||
| Germ-free | 15.65 (4.40) | 9.58 (4.72) | 13.29 (3.81) |
| SPF | 29.38 (3.37) | 32.95 (3.40) | 30.77 (2.90) |
| Difference-mean (SE) | 13.73 (5.54) | 23.37 (6.43) | 17.49 (4.86) |
| P-value* | 0.0039 | 0.0013 | 0.0014 |
| SafO_SUM joint score | |||
| Germ-free | 13.10 (2.26) | 10.61 (2.52) | 12.18 (2.12) |
| SPF | 22.59 (1.74) | 24.05 (1.82) | 23.13 (1.61) |
| Difference-mean (SE) | 9.49 (2.85) | 13.45 (3.44) | 10.95 (2.70) |
| P-value* | 0.0005 | 0.0007 | 0.0004 |
| MAX-osteophyte size | |||
| Germ-free | 1.40 (0.28) | 1.01 (0.29) | 1.30 (0.26) |
| SPF | 2.41 (0.21) | 2.64 (0.21) | 2.47 (0.20) |
| Difference-mean (SE) | 1.01 (0.35) | 1.63 (0.40) | 1.17 (0.33) |
| P-value* | 0.0054 | 0.0005 | 0.0017 |
| LBP levels | |||
| Germ-free | 621.7 (75.88) | 646.4 (87.94) | 611.4 (75.51) |
| SPF | 901.6 (55.21) | 888.5 (59.88) | 907.0 (54.66) |
| Difference-mean (SE) | 279.9 (93.84) | 242.0 (116.1) | 295.6 (93.91) |
| P-value* | 0.0055 | 0.0484 | 0.0045 |
P-values adjusted for multiple testing using Tukey’s procedure.
Microbiome analysis
As an exploratory analysis, we looked for a relationship between composition of the gut microbiota, identified by fecal microbiome analysis, and OA severity in the DMM knees of the SPF mice (as expected the GF mice had essentially no gut microbiota, data not shown). ACS scores were dichotomized into low and high scores based on scores being above or below the mean, which, for the ACS_MAX scores, gave good separation into two groups (5 SPF with low scores-below the 9.2 avg and 7 SPF with high scores-above the 9.2 avg).
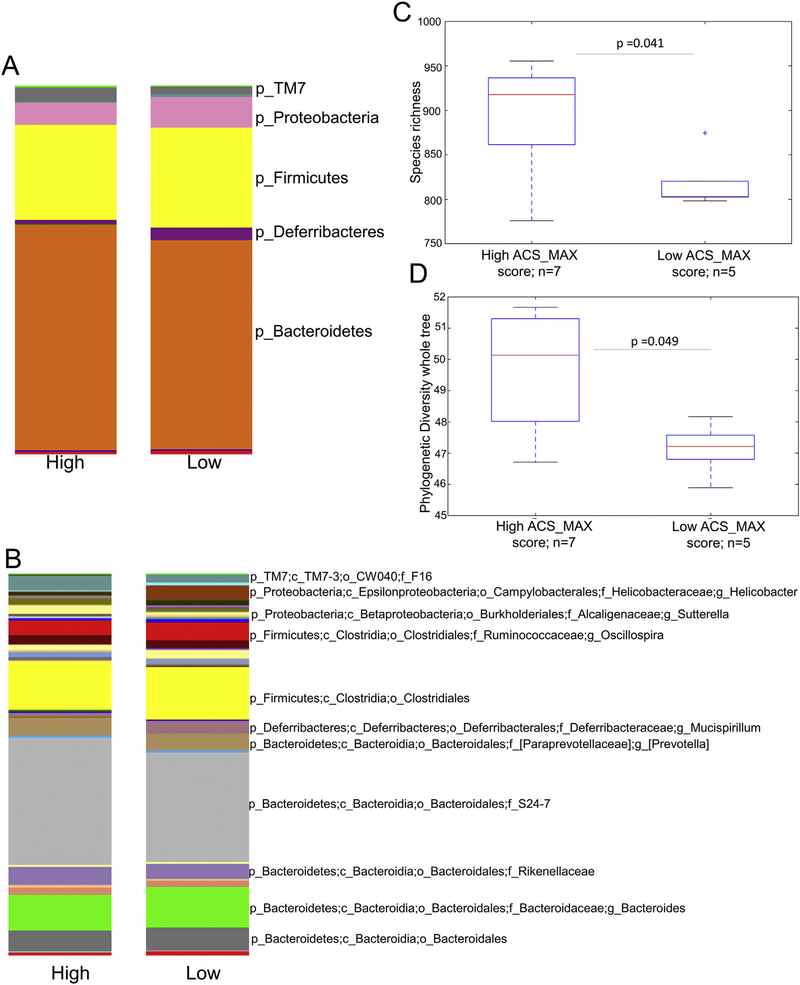
Alpha diversity metrics showed a minor but statistically significant increase in the number of observed species in the SPF mice with higher ACS_MAX scores (P = 0.041) [Fig. 6(C)] and a higher Phylogenetic Diversity whole tree (PD) index (P = 0.049) [Fig. 6(D)]. Principal Coordinate Analysis (PCoA) plots generated by weighted UniFrac analysis showed significant cage and collection date effects (not shown). Overall the differences in the composition of the microbiota of high vs low LBP categories and of high vs low ACS_MAX and SUM categories, as determined by weighted UniFrac analysis were not significant.
Fig. 6.
Taxa summary plots at L2 (Phylum) (A) and L6 (Genus) (B) levels showing Operational Taxonomic Units (OTU) relative abundance in mice with low and high ACS_MAX scores. Box and whisker plots show comparisons of species richness (C) and Phylogenetic Diversity whole tree (D) indexes between samples with high and low ACS-MAX scores.
Taxa summary plots showed minor differences in the abundance of specific taxa between high and low ACS_MAX scores [Fig. 6(A), (B)]. To determine whether the differences were statistically significant, we used the QIIME otu_category_significance.py. script, which compares the relative abundance of each microbial member between the two groups. Twenty-eight Operational Taxonomic Units (OTUs) were significantly over- or- under represented in high ACS_MAX score samples (Mann–Whitney-U P-value < 0.05). When the P-values were adjusted for multiple comparisons via FDR, the statistical significance was lost, possibly due to the limited sample size. The data are presented in the order of the raw P-values for all the taxa showing unadjusted P-values <0.05 (Table II).
Table II.
Comparison of relative OTU (category significance) abundance in mice with high and low ACS_MAX scores (higher mean values are shown in pink, lower mean values, in blue)
| OTU | Taxonomy | P | high_mean | low_mean |
|---|---|---|---|---|
| denovo3997 | k_Bacteria; p_TM7; c_TM7-3; o_CW040; f_F16 | 0.0080 | 1.429 | 0 |
| denovo878 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_Porphyromonadaceae; g_Parabacteroides | 0.0088 | 1.714 | 0 |
| denovo2931 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales | 0.0108 | 0.429 | 4.80 |
| denovo7506 | k_Bacteria; p_TM7; c_TM7-3; o_CW040; f_F16 | 0.0115 | 3.714 | 1.00 |
| denovo720 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae | 0.0181 | 0.143 | 1.80 |
| denovo2798 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_S24-7 | 0.0183 | 0.286 | 2.80 |
| denovo6242 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_S24-7 | 0.0206 | 1.000 | 0 |
| denovo6449 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_S24-7 | 0.0206 | 0.857 | 0 |
| denovo7823 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_S24-7 | 0.0210 | 2.000 | 0.20 |
| denovo5522 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_Rikenellaceae | 0.0217 | 2.143 | 0.20 |
| denovo1978 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_S24-7 | 0.0221 | 1.000 | 0 |
| denovo8512 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae | 0.0221 | 1.000 | 0 |
| denovo8352 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae | 0.0224 | 1.143 | 0 |
| denovo4948 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales | 0.0224 | 0.143 | 2.20 |
| denovo3088 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_S24-7 | 0.0230 | 1.429 | 0 |
| denovo3285 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_S24-7 | 0.0230 | 1.429 | 0 |
| denovo7209 | k_Bacteria; p_Actinobacteria; c_Coriobacteriia; o_Coriobacteriales; f_Coriobacteriaceae | 0.0235 | 0 | 0.60 |
| denovo6944 | k_Bacteria; p_Proteobacteria; c_Betaproteobacteria; o_Burkholderiales; f_Comamonadaceae; g_Hydrogenophaga | 0.0235 | 0 | 0.60 |
| denovo2919 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales | 0.0248 | 0 | 0.80 |
| denovo7361 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_S24-7 | 0.0248 | 0 | 1.00 |
| denovo6837 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Oscillospira | 0.0281 | 85.000 | 7.00 |
| denovo6160 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_S24-7 | 0.0286 | 2.286 | 0.40 |
| denovo1851 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_S24-7 | 0.0326 | 1.429 | 4.20 |
| denovo3650 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_S24-7 | 0.0420 | 88.143 | 24.60 |
| denovo2811 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae | 0.0465 | 2.714 | 14.20 |
| denovo7155 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_Bacteroidaceae; g_Bacteroides; s_acidifaciens | 0.0475 | 0.571 | 0 |
| denovo973 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_S24-7 | 0.0475 | 0.571 | 0 |
| denovo5174 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_Rikenellaceae | 0.0475 | 0.429 | 1.00 |
An example of an OTU that showed both an increase in abundance in mice with high scores at taxonomic level L6 (Genus) and was also found to be statistically significant (before adjusting for multiple comparisons) is represented by: k_Bacteria; p_TM7; c_TM7–3; o_CW040; f_F16 (Fig. 6(B), Table II). In contrast, an OTU higher in abundance in mice with low scores is k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; F_ (Fig. 6(B), Table II).
Discussion
The literature evaluating the role of the microbiota in different chronic conditions has exploded. Many factors affecting the intestinal (gut) microbiota have been reported (e.g., diet, genetics, host immune response, drugs, etc.) and the state of dysbiosis in many chronic intestinal and systemic inflammatory, metabolic and neurodevelopmental disorders has been described7,24. There is evidence that the gut microbiota may contribute to the development of arthritis, including AS, RA, and psoriatic arthritis10,11,25–29. The results shown here, demonstrating decreased severity of injury-induced OA in younger adult mice raised in a GF environment compared to a standard SPF environment, suggest that microbiota also contributes to OA. Since GF mice lack microorganisms at all sites within the body, it cannot be concluded with certainty that only the gut microbiota are responsible. However, the gut represents the site with the largest collection of microbes and the vast majority of studies comparing GF mice to SPF mice have implicated the gut microbiota7.
A number of measures of histological OA severity (ACS, SafO, and osteophyte scores) were decreased in GF mice after DMM surgery but the effects were mostly significant when analyzed in the younger adult mice. The study was not initially designed to compare two age groups of mice but rather two age groups were used as a matter of necessity to obtain the numbers of mice we anticipated were necessary to test our hypothesis (25 mice per housing condition). Mice in our gnotobiotic facility are in high demand and so we used those available at the time of our study. Because age is an important factor in the development of OA, we age-matched the GF and SPF groups and analyzed the results for all mice as well as when the mice were separated by age group.
A possible explanation for age-related differences is that age may have a confounding effect on the development of OA after DMM surgery. In a previous study with male C57BL/6 SPF mice that were either 12 week-old or 12 month-old at the time of DMM surgery, we found striking differences in the patterns of gene expression analyzed using microarrays suggesting age-related differences in mechanisms can be present in this model15. Aging-related changes in bone density are much more severe in GF than SPF mice, which could also influence the development of OA in older GF mice30. Unfortunately, the older adult group was much smaller in number than the younger group making direct statistical comparisons problematic.
To begin to explore mechanisms potentially responsible for the effects of the microbiota on OA severity, we investigated a specific bacterial product, LPS, recently reported to be increased in serum and synovial samples of people with OA14. Studies have shown that gut bacterial products, including LPS, can enter the systemic circulation and affect a number of organs, potentially including the joint, by causing low-grade inflammation14,31. In the circulation, LPS binds to its partner LBP, which is produced mainly in the liver and enhances the binding of LPS with the CD14 receptor and further binding to TLR4, promoting subsequent effects on the innate immune system32. Similar to LPS, LBP was shown to be associated with increased human knee OA severity13. We did not detect a difference in LPS levels between the SPF and GF mice, though the presence of inhibitors appeared to interfere with the LPS assay. However, LBP levels were significantly lower in the younger group of GF mice when compared to SPF controls. Importantly, LBP levels correlated with OA severity scores in these mice.
Since our results suggested a connection between the microbiome and OA severity, we examined differences in OTU abundance between SPF mice with low and high OA scores (e.g., ACS_MAX scores). Despite the small numbers of samples, we feel that dissemination of these results is important to help guide future investigations of microbiome changes in OA. We did not identify any changes in the previously reported12 bacterial families/species associated with OA, such as Lactobacillus and Methanobrevibacter, which could be due to differences in sample size and type of OA models (injury-induced vs obesity-associated OA).
There are differences between mice raised in GF and SPF facilities, other than the lack of a microbiota in GF mice, that could have contributed to the differences noted in OA severity and represent a limitation to our study. This includes use of slightly different cages and cage enrichment that could result in different activity levels that in turn could influence the development of OA after DMM surgery. We do not have activity measures from our facilities but a study from another gnotobiotic facility reported that GF mice have increased locomotor activity rather than decreased activity33. Another difference is that GF mice are known to gain less weight and have a lower body fat composition compared to conventionally raised mice23,34. We did not have body composition data on our mice but did include body weight and % weight change as potential co-founders in the analysis of OA severity measures and this did not change the results. Although the two groups of mice were on the same genetic background and obtained from the same vendor, they had been maintained as separate colonies, which could have resulted in some genetic drift. Though no differences in wound healing were noticed between SPF and GF, we could not exclude the effect of tissue healing as a confounding factor in our data.
In this study, we used the GF mouse model to test the contribution of the gut microbiota to the development of OA after joint injury rather than altering the composition of the microbiota in SPF mice with antibiotics, as an initial step to evaluate the contribution of an intact microbiota to the development of OA. By showing differences in OA severity between mice raised in a conventional facility and mice raised in GF conditions, these findings suggest that targeting the microbiota in OA could be an important therapeutic strategy. Further experiments should be designed to discover the specific microorganisms and factors produced by them that promote OA and those that may be protective. In addition, it would be important to evaluate other time points following DMM surgery to determine if the differences noted between GF and SPF mice are present at earlier and later stages of the OA process.
Supplementary Material
Acknowledgments
We would like to thank Dr. Virginia Kraus for advice using the LPS assay, Zhao Yiwen for help with histology, Dr. Ian Carroll for discussions regarding body fat composition and activity in GF mice and the staff of the Gnotobiotic Core Facility for housing GF mice.
Role of the funding source
This project was founded by a Translational Team Science Award (SOM/CTSA UL1TR001111) from UNC-Chapel Hill, by the Arthritis Foundation and by NIH grant P60 AR064166. The Microbiome and Gnotobiotic Core Facilities are supported in part by the NIH/National Institute of Diabetes and Digestive and Kidney Diseases grant P30 DK34987. Gnotobiotic studies were also supported by NIH grant P40 OD010995.
Footnotes
Conflict of interest
Authors have no conflict of interest to disclose.
Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.joca.2018.05.016.
References
- 1.Johnson VL, Hunter DJ. The epidemiology of osteoarthritis. Best Pract Res Clin Rheumatol 2014;28:5–15. [DOI] [PubMed] [Google Scholar]
- 2.Houard X, Goldring MB, Berenbaum F. Homeostatic mechanisms in articular cartilage and role of inflammation in osteoarthritis. Curr Rheumatol Rep 2013;15:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu-Bryan R, Terkeltaub R. Emerging regulators of the inflammatory process in osteoarthritis. Nat Rev Rheumatol 2015;11: 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science 2012;336:1262–7. [DOI] [PubMed] [Google Scholar]
- 5.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature 2011;474:327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blander JM, Longman RS, Iliev ID, Sonnenberg GF, Artis D. Regulation of inflammation by microbiota interactions with the host. Nat Immunol 2017;18:851–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology 2017;152:327–39. e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Z, Kraus VB. Does lipopolysaccharide-mediated inflammation have a role in OA? Nat Rev Rheumatol 2016;12: 123–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arend WP, Firestein GS. Pre-rheumatoid arthritis: predisposition and transition to clinical synovitis. Nat Rev Rheumatol 2012;8:573–86. [DOI] [PubMed] [Google Scholar]
- 10.Costello ME, Elewaut D, Kenna TJ, Brown MA. Microbes, the gut and ankylosing spondylitis. Arthritis Res Ther 2013;15: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013;2:e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins KH, Paul HA, Reimer RA, Seerattan RA, Hart DA, Herzog W. Relationship between inflammation, the gut microbiota, and metabolic osteoarthritis development: studies in a rat model. Osteoarthr Cartil 2015;23:1989–98. [DOI] [PubMed] [Google Scholar]
- 13.Loeser RF, Pathmasiri W, Sumner SJ, McRitchie S, Beavers D, Saxena P, et al. Association of urinary metabolites with radiographic progression of knee osteoarthritis in overweight and obese adults: an exploratory study. Osteoarthr Cartil 2016;24:1479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang ZY, Stabler T, Pei FX, Kraus VB. Both systemic and local lipopolysaccharide (LPS) burden are associated with knee OA severity and inflammation. Osteoarthr Cartil 2016;24: 1769–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loeser RF, Olex AL, McNulty MA, Carlson CS, Callahan MF, Ferguson CM, et al. Microarray analysis reveals age-related differences in gene expression during the development of osteoarthritis in mice. Arthritis Rheum 2012;64:705–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glasson SS, Askew R, Sheppard B, Carito B, Blanchet T, Ma HL, et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature 2005;434: 644–8. [DOI] [PubMed] [Google Scholar]
- 17.Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthr Cartil 2007;15:1061–9. [DOI] [PubMed] [Google Scholar]
- 18.Longobardi L, Temple JD, Tagliafierro L, Willcockson H, Esposito A, D’Onofrio N, et al. Role of the C-C chemokine receptor-2 in a murine model of injury-induced osteoarthritis. Osteoarthr Cartil 2017;25:914–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowe MA, Harper LR, McNulty MA, Lau AG, Carlson CS, Leng L, et al. Reduced osteoarthritis severity in aged mice with deletion of macrophage migration inhibitory factor. Arthritis Rheumatol 2017;69:352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNulty MA, Loeser RF, Davey C, Callahan MF, Ferguson CM, Carlson CS. A Comprehensive histological assessment of osteoarthritis lesions in mice. Cartilage 2011;2:354–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Little CB, Barai A, Burkhardt D, Smith SM, Fosang AJ, Werb Z, et al. Matrix metalloproteinase 13-deficient mice are resistant to osteoarthritic cartilage erosion but not chondrocyte hypertrophy or osteophyte development. Arthritis Rheum 2009;60:3723–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allali I, Arnold JW, Roach J, Cadenas MB, Butz N, Hassan HM, et al. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol 2017;17:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabot S, Membrez M, Bruneau A, Gerard P, Harach T, Moser M, et al. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEBJ 2010;24:4948–59. [DOI] [PubMed] [Google Scholar]
- 24.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med 2016;375:2369–79. [DOI] [PubMed] [Google Scholar]
- 25.Scher JU, Littman DR, Abramson SB. Microbiome in inflammatory arthritis and human rheumatic diseases. Arthritis Rheumatol 2016;68:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaahtovuo J, Munukka E, Korkeamaki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol 2008;35:1500–5. [PubMed] [Google Scholar]
- 27.Liu X, Zeng B, Zhang J, Li W, Mou F, Wang H, et al. Role of the gut microbiome in modulating arthritis progression in mice. Sci Rep 2016;6:30594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costello ME, Ciccia F, Willner D, Warrington N, Robinson PC, Gardiner B, et al. Brief report: intestinal dysbiosis in ankylosing spondylitis. Arthritis Rheumatol 2015;67:686–91. [DOI] [PubMed] [Google Scholar]
- 29.Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol 2015;67:128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan J, Herzog JW, Tsang K, Brennan CA, Bower MA, Garrett WS, et al. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc Natl Acad Sci U S A 2016;113:E7554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Creely SJ, McTernan PG, Kusminski CM, Fisher f M, Da Silva NF, Khanolkar M, et al. Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am J Physiol Endocrinol Metab 2007;292: E740–7. [DOI] [PubMed] [Google Scholar]
- 32.Iizasa S, Iizasa E, Matsuzaki S, Tanaka H, Kodama Y, Watanabe K, et al. Arabidopsis LBP/BPI related-1 and −2 bind to LPS directly and regulate PR1 expression. Sci Rep 2016;6: 27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A 2007;104:979–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caesar R, Reigstad CS, Backhed HK, Reinhardt C, Ketonen M, Lunden GO, et al. Gut-derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut 2012;61: 1701–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.