Abstract
Since the emergence of SARS-CoV-2 pandemic, clinical laboratories worldwide are overwhelmed with SARS-CoV-2 testing using the current gold standard: real-time reverse-transcription polymerase chain reaction (RT-PCR) assays. The large numbers of suspected cases led to shortages in numerous reagents such as specimen transport and RNA extraction buffers. We try to provide some answers on how strongly preanalytical issues affect RT-PCR results by reviewing the utility of different transport buffer media and virus inactivation procedures and comparing the literature data with our own recent findings. We show that various viral inactivation procedures and transport buffers are available and are less of a bottleneck for PCR-based methods. However, efficient alternative lysis buffers remain more difficult to find, and several fast RT-PCR assays are not compatible with guanidine-containing media, making this aspect more of a challenge in the current crisis. Furthermore, the availability of different SARS-CoV-2-specific RT-PCR kits with different sensitivities makes the definition of a general cutoff level for the cycle threshold (Ct) value challenging. Only a few studies have considered how Ct values relate to viral infectivity and how preanalytical issues might affect viral infectivity and RNA detection. We review the current data on the correlation between Ct values and viral infectivity. The presence of the SARS-CoV-2 viral genome in its own is not sufficient proof of infectivity and caution is needed in evaluation of the infectivity of samples. The correlation between Ct values and viral infectivity revealed an RT-PCR cutoff value of 34 cycles for SARS-CoV-2 infectivity using a laboratory-developed RT-PCR assay targeting the RdRp gene. While ideally each clinical laboratory should perform its own correlation, we believe this perspective article could be a reference point for others, in particular medical doctors and researchers interested in COVID-19 diagnostics, and a first step toward harmonization.
1. From COVID-19 Symptoms to SARS-CoV-2 RT-PCR
It is now more than a year ago, in December 2019, when health officials in Wuhan, China, reported on a disease outbreak with pneumonia syndromes. Since then, the highly pathogenic 2019 coronavirus disease (COVID-19) linked to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus particles spread worldwide, causing more than 2.19 million fatalities and more than 101 million confirmed cases worldwide (as of January 29, 2021). With a lack of approved antiviral agents, only recent vaccines1,2 and passive immunotherapy approaches using COVID-19 convalescent plasma3,4 at hand, testing has proven to be critical in the management of the pandemic. Target amplification and its subsequent detection is the most widely used approach for viral diagnosis and also employed for the detection of coronaviruses.5 These methods, and, in particular, the real-time reverse-transcription polymerase chain reaction (abbreviated as RT-PCR throughout the article), are based on the use of SARS-CoV-2-specific primers and probes, designed according to the SARS-CoV-2 genome (Figure 1a). SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA virus and possesses a genome approaching 30 kB. It is arranged in the order of a 5′ untranslated region (UTR)–replicase complex [open reading frame(ORF)1ab]–structural proteins [spike (S)–envelope (E)–membrane (M)–nucleocapsid (N)]–3′UTR and nonstructural accessory proteins.6 The RNA-dependent RNA polymerase (RdRp; nsp12) positioned on ORF1ab plays a crucial role in RNA synthesis with features of rapid mutation and recombination.7 Theoretically, the amplification of the target genes using assay-specific primers and probes should have not only high specificity but also high sensitivity. In addition, multiple mutations were detected over the entire genomes of SARS-CoV-2, speculated to influence the rate of disease transmission and mortality but also the false-negative rate, if the mutation occurs at the position of primers or probes of the currently used target genes.8,9
Figure 1.
(a) Genome of SARS-CoV-2 with the most common RT-PCR targets highlighted. (b) Essential steps of the RT-PCR diagnostic workflow including sample collection, storage in a transport medium, lysis and RNA extraction, reverse transcription, amplification, and detection. (c) Example of a real-time RT-PCR amplification curve for SARS-CoV-2 and postrun analysis to interpret results.
Two of the first real-time RT-PCR assays receiving emergency use authorization (EUA) in February 2020 were the COVID-19 RT-PCR panel assay (IDT) as well as the RealStar SARS-CoV-2 RT-PCR (Altona Diagnostics). Since then, a large panel of COVID-19 RT-PCR assays are available (a nonexhaustive selection in Table 1 and reviewed in ref (10)) and allowed not only better management of the crisis but helped in understanding both the disease and its epidemiology.11,12 These assays are all based on some of the essential steps of RT-PCR (Figure 1b), including (i) lysis of SARS-CoV-2 virus particles and purification of nucleic acids from collected samples (oropharyngeal, nasopharyngeal swabs, and/or lower respiratory tract samples, e.g., sputum, tracheal aspirates, or bronchoalveolar lavage specimens) dispersed in a universal or viral transport medium (UTM/VTM), (ii) reverse transcription to complementary DNA (cDNA), (iii) amplification of the specific region of the cDNA, and (iv) optical detection. The assays differ in the primers/probes used to amplify/detect the different target regions of the SARS-CoV-2 genome (Table 1), which consequently affects their sensitivity.13−15 The difference is generally more pronounced for samples with low viral load, with a median cycle threshold (Ct) of >33, as recommended by the “Société Française de Microbiologie”.16
Table 1. Some Selected SARS-CoV-2 RNA Detection Assays.
| developer | kit | extraction | amplification | cycler |
|---|---|---|---|---|
| Thermofisher Scientific | TaqPath COVID-19 CE-IVD RT-PCR Kit | MGI Nucleic Acid Extraction Kit on the MGI SP-960 instrument | ORF1ab, S, N | QuantStudio 5 Real-Time PCR System (Applied Biosystems) |
| Altona Diagnostics | RealStar SARS-CoV-2 RT-PCR Kit 1.0 | AltoStar Purification Kit 1.5 extraction kit on AltoStar Automation System AM16 | E, S | CFX96 Touch Deep Well Real-Time PCR Detection System (Bio-Rad Laboratories) |
| Beijing Genomics Institute | Real-Time Fluorescent RT-PCR Kit for Detecting SARS-CoV-2 | MGI Nucleic Acid Extraction Kit on the MGI SP-960 instrument | ORF1 | QuantStudio 5 Real-Time PCR System (Applied Biosystems) |
| DiaSorin Molecular | Simplexa COVID-19 direct | None | S, ORF1ab | LIAISON MDX |
| BD | BD SARS-CoV-2 reagents for BD MAX system | BD MAX ExK TNA-3 | N1, N2 | BD MAX |
| Seegene | AllPlex 2019-nCoV assay | StarMag 96 Extraction Kit on Microlab Nimbus | E, N, RdRp | CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories) |
| Qiagen | QIAstat-Dx respiratory panel | Included automated RNA extraction | E, ORF1b | QIAstat |
| Cepheid | Xpert Xpress SARS-CoV-2 | Included automated RNA extraction | N2, E | GeneXpert Infinity |
| Institut Pasteur Paris | laboratory developed17 | QIAamp Viral RNA Mini Kit (Qiagen, France) | RdRp (two target regions, named IP2, IP4) | 7500 Real-Time PCR System (Applied Biosystems) |
If SARS-CoV-2 RNA is present, then each round of amplification results in a doubling of the amount of the target. As a result, amplification occurs exponentially, producing an exponential curve of amplification (Figure 1c) visualized by the use of a fluorescent nucleic acid probe. The Ct value is defined as the number of cycles of amplification required for the fluorescence of a PCR product to be detected crossing a threshold, which is above the background signal (a low-level signal in the assay regardless of whether the target is present or not) (Figure 1c).
This rather complex process is based on the use of correct preanalytical procedures, namely, appropriate transport media, storage conditions, and lysis buffers without causing RNA damage as well as on a correct interpretation of the Ct values. The risk of false-positive laboratory tests increases as the probability of COVID-19 infection decreases. One key parameter with infected patients harboring low virus loads (either at the very beginning or at the end of the infection) is high Ct values near the detection limit of the assay (Figure 1c). The risk of false-negative results is correlated with a lack of sensitivity of the analytical method, when the infectious dose is too low but is most strongly affected by the preanalytical steps (storage and transport) and the sampling approach itself (i.e., type and quality of the specimen). Indeed, nasopharyngeal swabs are recommended for screening and need to be correctly performed to contain a sufficient number of infected cells in order to give a right picture of the infection state of the patient. What guidance can be given to enable the discernment of true positives from false positives? How much does sample preparation and the use of lysis buffers and heat inactivation affect the results? Are Ct values correlated to viral infectivity or does a positive result only indicate that the person has come into contact with the genomic sequence at some time in the recent past?
2. Does the Success of RT-PCR Depend on the Sample Quality and Preparation?
Diagnostics of viral infection using PCR or RT-PCR is now well-established for many viruses. Most of the time, these techniques are performed on blood-derived specimens, i.e., whole blood, serum, or plasma. In these cases, there is no need for transport buffers and virus inactivation is not performed. Other preanalytical issues, like storage or lysis, also apply to these specimen types. However, in SARS-CoV-2 diagnostics, two additional challenges apply. First, the specimen type is not blood-derived but, in general, a respiratory swab specimen. Therefore, there is a need to place this specimen in some kind of a transport medium. Second, the pathogen is classified biosafety level 3 (BSL-3) and diagnostic procedures except virus culture can be performed in BSL-2 level facilities with appropriate protective measures.18 Because of high specimen numbers, automatization of diagnostic assays is necessary, and due to the severity of the illness and high infectiousness of the virus, many laboratories, including ours, perform virus inactivation before analysis. The success of the diagnosis of SARS-CoV-2 depends largely on the quality of the collected specimens and the conditions under which they are transported and stored before being processed in the laboratory. The most important point is that the specimen is of good quality, i.e., it contains virus/virus-infected cells. This is especially critical for nasopharyngeal swab specimens. Amplification of a cellular gene, e.g., G6PDH, enables monitoring of the cellular content as a surrogate marker of the specimen quality. However, in routine practice, most commercial assays include instead an internal control, e.g., an artificial RNA that is added to the specimen prior to RNA extraction, which only monitors extraction and amplification steps. In the case of SARS-CoV-2, specimens are mostly dispersed in nutrient solutions such as a viral transport medium (VTM) or a universal transport medium (UTM). The constituents of an appropriate viral transport medium are selected to create an isotonic solution containing proteins to protect the viral structure, antibiotics to inhibit microbial contamination, and one or more buffers to control the pH. Often, VTM/UTM are based on Hanks balanced salt solutions, calcium and magnesium, proteins (fetal bovine serum and albumin), as well as antibiotics such as gentamycin and amphotericin. Viral transport media are prepared with the idea of sustaining the viability of the viruses for detection in cell culture. While these complex solutions present a challenge for molecular SARS-CoV-2 diagnostics such as electrochemical-,19 electrical-20 and/or optical-based biosensors11,21,22 due to eventual surface antifouling,19,20 they are less of a bottleneck for PCR-based methods.
For RT-PCR, effective RNA extraction remains one of the main issues. The key chemical constituent of the lysis buffer is guanidium thiocyanate, a chaotropic agent that denatures proteins, helps the liberation of nuclear RNA and genomic DNA, and protects against RNAse enzyme degradation. In standard formulations, such as those commercialized by, e.g., Qiagen (AVL buffer), up to 6 M guanidium thiocyanate is used.23 With the epidemic doubling in size every 3–7 days and the increased need of RNA extraction reagents such as guanidium thiocyanate, this chemical became scarce and alternatives were sought after. Scallen and her colleagues validated that a lysis buffer containing only 4 M guanidinium thiocyanate and Triton X-100 (3% v/v) was effective for the extraction of SARS-CoV-2 RNA from respiratory specimens and allowed easing the burden on hospital laboratories.23 The inclusion of a detergent such as Triton X-100 enhances the disintegration of the viral envelope during extraction. We have recently demonstrated the utility of a specimen transport buffer containing next to Tris-EDTA 10 × (100 mM Tris/10 mM EDTA), only 2.53 M guanidine thiocyanate (30%), and Triton X-100 (2.5% v/v, 34.4 mM).24 When respiratory specimens were diluted in this buffer, the quantity of viral RNA for two SARS-CoV-2 targets was stable at room temperature until at least day 5. There was a slight increase in Ct values for the SARS-CoV-2 targets after day 5, whereas Ct values for the cellular control gene G6PDH remained stable until day 30. After 30 days of storage at room temperature, SARS-CoV-2 RNA could be detected consistently in the case of high virus loads (i.e., Ct values of up to 30) but not in the case of low virus loads. These results confirm that lower guanidine concentrations had no negative effect on stabilizing viral RNA. This transport buffer could solve two issues: the problems of shortages in a viral transport medium and limited cold room capacities. However, it has to be kept in mind that some rapid RT-PCR assays are not compatible with guanidine-containing media as guanidine thiocyanate is unstable in a sodium hypochlorite solution, commonly known as bleach and a widely used cleaning reagent in molecular testing workflows and is also an inhibitor of the Taq polymerase, which leads to uninterpretable results when using assays without an RNA extraction step.24−26
3. Importance of SARS-CoV-2 Inactivation and Its Consequences for RT-PCR
To allow safe transportation of clinical specimens from collection points to laboratories, as well as for performing experiments involving SARS-CoV-2 in general, inactivation procedures are performed. The efficiency of SARS-CoV-2 inactivation depends on several factors, including next to its concentration, the sample matrix and contact time. Up to date, only limited data on the efficacy of SARS-CoV-2-specific inactivation approaches are reported in the literature (Table 2),27,28 as risk assessments have largely been based upon virus inactivation information from related coronaviruses such as SARS-CoV-1.29,30
Table 2. Inactivation Procedures of SARS-CoV-2.
| method | conditions | inactivation | refs |
|---|---|---|---|
| heat | 100 °C (5 min) | yes | (31) |
| 80 °C (60 min) | yes | (29) | |
| 60 °C (30 min) | yes | (32) | |
| 56 °C (45 min) | yes | (31) | |
| 56 °C (30 min) | no | (31) | |
| UV irradiation | 0.01 J cm–2 | no | (29) |
| 0.04 J cm–2 | yes | (29) | |
| chemical | sodium dodecyl sulfate (1 h, 0.5%) | yes | (28) |
| Triton X-100 (1 h, 0.5%) | yes | (28) | |
| NP-40 (1 h, 0.5%) | yes | (28) | |
| Tween 20 (1 h, 0.5%) | no | (28) | |
| guanidine thiocyanate (1 day, 1.27 M) Triton X-100 (1 day, 2.5% v/v) | no | (24) | |
| TRIzol (10 min,10%) | yes | (31) | |
| formalin (10 min, 0.5–2%) | yes | (31) |
Heat inactivation is the most widely employed method for SARS-CoV-2.28 Patterson et al. heated SARS-CoV-2 at 80 °C for 1 h and demonstrated complete virus inactivation. The complete inactivation of SARS-CoV-2 viral particles upon heating at 60 °C for 30 min was confirmed by us recently.32 These results are in line with the data by Jureka et al.,31 where 5 min at 100 °C was sufficient for virus inactivation, while 45 min at 56 °C was required for complete inactivation. We recently confirmed that virus inactivation can be achieved upon heating at 55 °C for 45 min.32
It is important to note that heat inactivation at 60 °C for 30 min preserved the antigenic spike protein function (Figure 1a), used currently in several point-of-care antigenic tests.19,20 This was confirmed by affinity studies between heated and nonheated SARS-CoV-2 receptor-binding domain (RBD) and SARS-CoV-2 surface receptor using surface plasmon resonance (SPR) binding curves (Figure 2a). Heating the RBD at 100 °C for 60 min showed no binding affinity to SPR chips modified with RBD-specific VHH-72 surface ligands.33 A lower binding efficiency was still observed for RBD heated for 45 min at 60 °C. This contrasts with RBD treatment for 30 or 15 min at 60 °C, where only a slight degradation of the protein seems to have taken place and binding events are comparable to those recorded for non-heat-treated specimens. In addition, heat treatment at 60 °C did not have a negative effect on the RNA stability (Figure 2b–d), making it an ideal and clean inactivation process.
Figure 2.
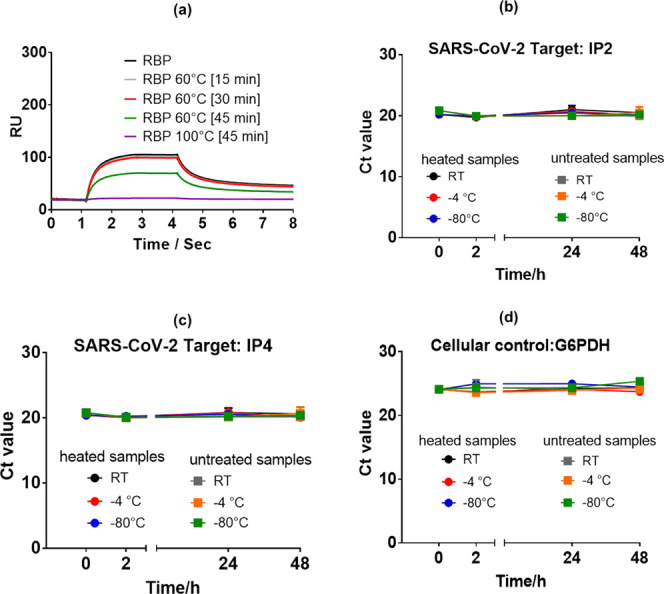
Quantification of SARS-CoV-2 following exposure to different inactivation conditions: (a) surface plasmon resonance (SPR) binding curves recorded on a T200 Biacore for the receptor-binding domain (RBD, 200 nM) and RBD for different time intervals. The interface was modified with SARS-CoV-2-specific VHH-72 nanobodies.33 (b–d) RNA stability after heat inactivation: SARS-CoV-2 RNA-positive nasopharyngeal swab specimens were pooled and divided into equal volumes and heated for 30 min at 60 °C. The aliquots were either kept at room temperature (RT), 4 °C, or −80 °C for the indicated times before RNA extraction and RT-PCR was performed in triplicate.32 As a control, untreated specimens were included. Ct values are indicated for the SARS-CoV-2 targets IP2 (b), IP4 (c), and the cellular control G6PDH (d) (Figure 2−2d report mean values of three samples).
Next to heat, UV irradiation, based on the modification of the nucleic acid structure, proved to be effective in SARS-CoV-2 inactivation.29 The approach is dependent on the exposed light energy with 0.01 J cm–2, leading to partial viral inactivation, while light energy values higher than 0.04 J cm–2 resulted in complete inactivation.
Chemical inactivation using various detergents27,28,31 and chemicals, such as formaldehyde34 and β-propionolactone,35−37 was also evaluated. In the case of sodium dodecyl sulfate (SDS), Triton 126X-100, and NP-40, a commercially available ethoxylated nonylphenol surfactant, complete inactivation of SARS-CoV-2 was achieved.28 However, upon treatment with Tween 20 (0.5%) for 1 h, all specimens remained infectious. This was also the case using our proposed transport buffer based on Tris-EDTA 10 × (100 mM Tris/10 mM EDTA), guanidine thiocyanate (30%, 2.53 M), and Triton X-100 (2.5% v/v, 34.4 mM) at a dilution of 1/2 and 9/10. In contrast, SARS-CoV-2 treatment with TRIzol (10%) and a formalin (0.5–2%) solution for 1 h at room temperature proved to be effective in SARS-CoV-2 inactivation.31 The same was true when SARS-CoV-2 was exposed to β-propiolactone (0.5%) for 16 h at 4 °C, followed by 2 h at 37 °C.31 While heating of specimens is usually performed in the diagnostic laboratory, chemical inactivation using appropriate specimen transport buffers would enable virus inactivation before transport and thus facilitates transport and preanalytical procedures.38
4. Predicting Infectious SARS-CoV-2 from Diagnostic Specimens
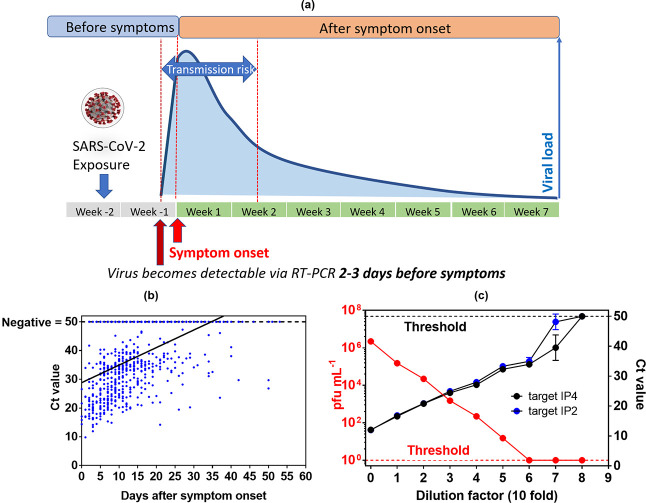
Next to the sampling and specimen preparation, the use of an appropriate cutoff value for the proper interpretation of SARS-CoV-2 PCR results is of paramount importance. However, these values can only be interpreted correctly by having an idea about the health history of the patient, as pointed out lately by communication by Public Health England.39 Byrne et al.40 published one of the first overviews of infectious periods of SARS-CoV-2 up to April 1, 2020. Substantial variations were observed with mean infectious period of asymptomatic cases between 6.5 and 9.5 days, shortening in presymptomatic cases to 1–4 days, with 13.4 days estimated from the onset of the symptoms to a negative RT-PCR.40 This review was complemented more recently by a review and meta-analyses by Weiss and co-workers41 and Cevik et al.42 with data on nasopharyngeal swab samples taken in the first week after the onset of COVID-19 symptoms.41,42 These specimens revealed high viral loads in both studies.41,42Figure 3a schematically illustrates the viral load dynamics from the symptom onset to recovery for symptomatic cases. It is believed that infectiousness begins 2–3 days prior to symptoms’ onset and peaks around the symptom onset.43 The infectious period is thus approximately 9–10 days. In fact, virus load is already high before the symptom onset and almost half of transmissions to secondary cases occur in the presymptomatic period.44−46 The timeline of SARS-CoV-2 RNA was lately confirmed by some of us47 using data from 520 COVID-19 patients. The Ct values of nasopharyngeal swab specimens were plotted against the time after the symptom onset (Figure 3b). The lowest Ct values, corresponding to the highest virus loads, were found early after the symptom onset, followed by a decline in virus load with increasing time after the symptom onset.
Figure 3.
Clinical significance of SARS-CoV-2 RT-PCR results. (a) Timeline of SARS-CoV-2 infectivity taking into account our own findings and those of others.39,40 (b) Ct values (target IP2) as a function of time after the symptom onset in nasopharyngeal swab specimens of COVID-19 patients. Ct of specimens with undetectable SARS-CoV-2 RNA were set to 50. (c) Correlation of Ct values with SARS-CoV-2 infectivity. Vero E6 cells were infected with 10-fold dilutions of a SARS-CoV-2 isolate. The plates were incubated for 6 days in 5% CO2 at 37 °C and examined daily using an inverted microscope (ZEISS Primovert) to evaluate the extent of the virus-induced cytopathic effect in cell culture. The calculation of the estimated virus concentration was carried out by the Spearman and Karber method48,49 and expressed as TCID50/mL (50% tissue culture infectious dose). TCID50/mL values were transformed to PFU mL–1 using the formula PFU mL–1 = TCID50/mL × 0.7.50 RNA extraction and RT-PCR (target IP4 and target IP2) were performed in duplicate for each dilution. Ct of dilutions with undetectable SARS-CoV-2 RNA were set to 50.
There is a general agreement that Ct values are strongly associated with SARS-CoV-2 viral load and infectivity. While a threshold cutoff of Ct 33–35 is associated with low infectivity, a Ct value of 20 is linked to a highly infective person. Ct values are semiquantitative, with a 3-point increase in the Ct value being roughly equivalent to a 10-fold decrease in the quantity of viral genetic material. While clinical laboratories often use Ct cutoff values provided by the RT-PCR kit manufacturers, which are between 37 and 40 cycles, to define a specimen with positive SARS-CoV-2 RNA detection, correlation between viral RNA load with patient specimen infectivity can only be determined in BLS-3 settings.
Some studies have been conducted in an attempt to correlate Ct values with SARS-CoV-2 virus infectivity.46,51−53 While being a laborious process in BLS-3 settings, viability of virus can be determined by inoculating cultured Vero cells and assessing evidence of the cytopathic effect. Data in Figure 3c were obtained by first determining the Ct value of a SARS-CoV-2 clinical isolate, followed by a serial dilution of this specimen in 10-fold steps. As expected, a linear relation between the Ct values and serial dilution was observed for both RT-PCR targets, IP4 and IP2. The same dilutions were further used for infecting Vero cells and determining the infectivity of each dilution. While the initial specimens with a Ct value of 12 correspond to a viral load of 2.2 × 106 pfu mL–1, at a Ct of 33, only 15 pfu mL–1 are detected. Our system correlates with a cutoff value of 34 cycles for SARS-CoV-2 infectivity. The relationship between RT-PCR values and culture positivity was also validated by Singanayagam et al.54 who found that culturing virus declined to 8% in specimens with Ct > 35.
A study by La Scola and colleagues examined 3790 positive specimens with known Ct values and found that 70% of these specimens could be cultured if having a Ct ≤ 25, but less than 3%, if having Ct > 35.52 Bullard tested 90 samples from Covid-19-positive specimens through incubation with Vero cells to assess their infectivity.53 They concluded that specimens with Ct > 24 or taken >8 days after the symptom onset were not able to infect cells. Indeed, positive RT-PCR tests are not correlated to the infection state. Wölfel et al. demonstrated that after 8 days from the symptom onset, virus could not be isolated in cell culture, despite ongoing high viral load in RT-PCR results.55 Indeed, other studies concluded that SARS-CoV-2 RNA can still be detected 6–8 weeks after the symptom onset and after clearance of symptoms, with no correlated cell culture infectivity.45,56 So, could Ct values help doctors to indicate if the patient is highly infectious or not and should be isolated and contacts tracked down? Ct values are believed by many to be an imperfect measure and laboratory-dependent. Furthermore, infectivity, measured by infection of cell culture, can only be a surrogate marker for infectivity. A virus might be more effective in infecting its natural host than cell culture. The real answer to the question of infectivity can only be provided by epidemiological studies, correlating the number of secondary cases infected by a person with a given Ct value at a given time. This kind of study would be extremely valuable but would need exact contact tracing to identify contacts that had only contact on the same day when the Ct value was obtained because nobody can predict what Ct the person’s specimen harbored the day before or after.
5. Conclusions and Future Critical Outlook
With the rapid diagnosis of COVID-19-infected patients using RT-PCR, clinical laboratories played a key role in controlling the spread of SARS-CoV-2 virus. Since the beginning of the pandemic, there are controversial views about the infectiousness of patients as a positive RT-PCR test cannot tell whether the person is infectious or not or when the infection started. In most cases, virus load quickly decreases after the symptom onset, and most patients will no further have SARS-CoV-2 RNA detection in nasopharyngeal swabs in a median of 14–25 days after the symptom onset. However, in elderly, severely ill, or immunosuppressed patients, nasopharyngeal swabs and lower respiratory tract specimens can still be positive for SARS-CoV-2 RNA detection several weeks after the symptom onset.45,57 In general, these specimens harbor high Ct values corresponding to low virus load. The question arose whether this corresponds to infectious virus or only viral genomic material. The detection of the presence of a viral genome on its own is therefore not sufficient. Ct values are thus believed by many to be an imperfect measure, laboratory-dependent, and not very useful. However, several studies that aimed at quantifying the relationship between Ct and the likelihood of culturing live viruses suggest that the probability to recover a live virus from specimens with Ct > 34 is low. Defining a general Ct cutoff is obviously a real challenge. Infectivity and RT-PCR Ct cutoff values also depend on the time span, which evolved between sampling and analysis. Indeed, samples are transported and stored at 4 °C for several hours prior to analysis. The storage at 4 °C for up to 5 days is recommended by WHO and the use of storage buffers is believed to conserve the sample quality.58
However, using the Ct value as a surrogate marker for virus load in a clinical routine will need the harmonization of the numerous SARS-CoV-2 assays. Harmonization of virus load measurements between different laboratories and assays is a general challenge that has not been completely resolved even for virus load assessments that are routinely performed for several years. Development of WHO international standards for quantification is one step to try to harmonize virus load determination.59 Defining RT-PCR cutoff levels is furthermore necessary for evaluating the performance of other SARS–CoV-2 tests compared to RT-PCR.60
The current gold standard for SARS-CoV-2 diagnostics is real-time RT-PCR. Classical assays take 4–6 h while rapid molecular assays have been developed and yield results in less than 2 h.10 However, these need specific instruments and the price per test is high. While clinical laboratories still play the dominant role in patient specimen screening, integrated point-of-care (POC) platforms are believed to hold a central place in the future. While RT-PCR approaches will remain the gold standard for viral testing and thus for SARS-CoV-2, POC devices will allow overcoming the bottleneck of widespread nucleic acid testing due to their intrinsic characteristics being simple, faster, and cheap with the ability of COVID-19 diagnostic in the doctor’s office as well as at home. Such platforms will ensure reduced turnaround time, unlimited retesting, and a potentially lower cost by reducing reagent consumptions. Currently, such platforms mostly detect SARS-CoV-2 antigens and severely lack sensitivity compared to RT-PCR.61−63 Recent developments include loop-mediated isothermal amplification (LAMP) POC tests. These are cost-effective, rapid molecular assays with satisfying specificity and sensitivity. However, they generally require a sample preparation step and a specific instrument.10 Currently available POC tests detect viral components but not the entire virus particle. It would be, however, much more elegant and eventually more efficient to detect the infectivity status of the patient by sensing the presence of the S1 protein on the surface of an intact SARS-CoV-2 particle. We are currently working on the development of such a test. We could indeed discriminate COVID-19-negative from COVID-19-positive patients upon the use of electrodes functionalized with engineered nanobodies in an electrochemical sensing format.19,64 The developed sensor was tested in a clinical trial on 100 nasal swab specimens and showed a 88% positive percentage agreement (PPA) and a 88% negative percentage agreement (NPA), as compared to RT-PCR with an analysis time of 10 min without any pretreatment step.
The use of POC testing also means treating a diagnostic assay as an integrated unit from specimen collection and processing to final analysis. As these tests are designed to be done on site and immediately, no viral transportation media nor viral inactivation protocols are required, making the assays much easier and faster to perform. This implies moving from nasopharyngeal specimens, which have to be taken by professionals, to alternative specimens such as saliva and exhaled breath condensate (EBC) that can easily be collected by the patient at home. The EasyCov saliva-based screening test, developed jointly by CNRS researchers in Montpellier, France, and two private sector companies (Alcediag and SkillCell), is one of the examples where COVID-19 diagnostics will stand in the future. Easycov enables mass testing via loop-mediated isothermal amplification (LAMP), resulting in a colorimetric readout. The test that is completed in 40 min succeeded in detecting 87% of positive cases among 220 people who had been tested at the Montpellier University Hospital since September 6, 2020.
Breath sampling might be another approach in the future as it does not require trained medical personnel and can be done essentially anywhere in any time frame. Exhaled breath contains tiny aerosols and the collection of these aerosol particles, produced by patients during coughing or breathing, potentially provides a noninvasive method for collection of diagnostic specimens of SARS-CoV-2. It has been indeed postulated lately that the exhaled breath is a significant source of SARS-CoV-2 emission.65
In conclusion, the ongoing COVID-19 crisis revealed that complex issues such as viral transportation, viral inactivation, and viral diagnostics require knowledge and expertise in different topics and domains. It is through the tight interplay between biologists, chemists, nanotechnologists, and medical doctors that COVID-19 diagnostics will ultimately allow controlling the worldwide pandemic. The enormous know-how gained with SARS-CoV-2 sensing can often be easily adapted to other viral infection assays and researchers are urged to continue their strong involvement in the actual sanitary crisis. It is timely to put the importance of research and innovation at the forefront to make society understand that money is not wasted while doing research with positive input to society at large.
Acknowledgments
This work was financially supported by the Centre National de la Recherche Scientifique (CNRS), the University of Lille, I-SITE via the COVID task force, and the Hauts-de-France region via ANR Resilience (CorDial-FLU). The project is funded by the Horizon 2020 framework programme of the European Union under grant agreement no 101016038. The authors thank all technicians of the virology laboratory of the Lille University Hospital (CHU Lille) for their excellent technical assistance under extremely challenging conditions. They also acknowledge the valuable help of medical and pharmaceutical students.
The authors declare no competing financial interest.
References
- Voysey M.; Costa Clemens S. A.; Madhi S.; Weckx L. Y.; Folegatti P. M.; Aley P. K.; Angus B.; VBaillie V. L.; Barnabas S. L.; Bhorat Q. E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2020, 397, 99–111. 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L. A.; Anderson E. J.; Rouphael N. G.; Roberts P. C.; Makhene M.; Coler R. N.; McCullough M. P.; Chappell J. D.; Denison M. R.; Stevens L. J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libster R.; Pérez Marc G.; Wappner D.; Coviello S.; Bianchi A.; Bram V.; Estebah I.; Caballero M. T.; Wood C.; Berrueta M.; et al. Early High-Titer Plasma Therapy to Prevent Severe Covid-19 in Older Adults. N. Eng. J. Med. 2021, 384, 610–618. 10.1056/NEJMoa2033700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner M. J.; Carter R. E.; Senefeld J. W.; Klassen S. A.; Mills J. R.; Johnson P. W.; Theel E. S.; Wiggins C. C.; Bruni K. A.; Klompas A. M.; et al. Convalescent Plasma Antibody Levels and the Risk of Death from Covid-19. N. Eng. J. Med. 2021, 10.1056/NEJMoa2031893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M.; Zhou Y.; Ye J.; AL-maskri A. A. A.; Kang Y.; Zeng S.; Cai S. Recent advances and perspectives of nucleic acid detection for coronavirus. J. Pharm. Anal. 2020, 10, 97–101. 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel M.; Ibrahim A.; Fayez M.; Al-Nazawi M. From SARS and MERS CoVs to SARS-CoV-2: moving toward more biased codon usage in viral structural and non-structural genes. J. Med. Virol. 2020, 92, 660–666. 10.1002/jmv.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020, 253, 117592 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect., Genet. Evol. 2020, 81, 104260 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachetti M.; Marini B.; Benedetti F.; Giudici F.; Mauro E.; Storici P.; Masciovecchio C.; Angeletti S.; Ciccozzi M.; Gallo R. C.; et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020, 18, 179. 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C. Y.; Chan K. G.; Yean C. Y.; Ang G. Y. Nucleic Acid-Based Diagnostic Tests for the Detection SARS-CoV-2: An Update. Diagnostics 2021, 11, 53 10.3390/diagnostics11010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama B.; Kadhiresan P.; Kozlowski H. N.; Malekjahani A.; Osborne M.; Li V. Y. C.; Chen H.; Mubareka S.; Gubbay J. B.; Chan W. C. W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Yuan X.; Yang C.; He Q.; Chen J.; Yu D.; Li J.; Zhai S.; Qin Z.; Du K.; Chu Z.; et al. Current and Perspective Diagnostic Techniques for COVId-19. ACS Infect. Dis. 2020, 6, 1998–2016. 10.1021/acsinfecdis.0c00365. [DOI] [PubMed] [Google Scholar]
- Uhteg K.; Jarrett J.; Richards M.; Howard C.; Morehead E.; Geahr M.; Gluck L.; Hanlon A.; Ellis B.; Kaur H.; et al. Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays. J. Clin. Virol. 2020, 127, 104384 10.1016/j.jcv.2020.104384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels C. B. F.; Brito A. F.; Wyllie A. L.; Fauver J. R.; Ott I. M.; Kalinich C. C.; Petrone M. E.; Casanovas-Massana A.; Muenker M. C.; Moore A. J.; et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020, 5, 1299–1305. 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalla A. K.; Casto A. M.; Huang M.-L.; Perchetti G. A.; Sampoleo R.; Shrestha L.; Wei Y.; Zhu H.; Jerome K. R.; Greninger A. L. Comparative Performance of SARS-CoV-2 Detection Assays Using Seven Different Primer-Probe Sets and One Assay Kit. J. Clin. Microbiol. 2022, 58, e00557-20 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisdu 25 septembre 2020 de la Société Française de Microbiologie (SFM) relatifà l’interprétation de la valeur de Ct (estimation de la charge virale)obtenue en cas de RT-PCR SARS-CoV-2 positive sur les prélèvements cliniquesréalisés à des fins diagnostiques ou de dépistage. https://www.sfm-microbiologie.org/covid-19-fiches-et-documents-sfm/ (Version 3 du 7 octobre, 2020).
- Institut Pasteur, Paris. Protocol: Real-time RT-PCR Assays for the Detection of SARS CoV-2. [Internet] [cited 2020 Aug 28]. https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf?sfvrsn=3662fcb6_2.
- Arrêté du 18 décembre 2020 relatif à la classification du coronavirus SARS-CoV-2 dans la liste des agents biologiques pathogènes. https://www.legifrance.gouv.fr/jorf/article_jo/JORFARTI000042840497.
- Szunerits S.; Pagneux Q.; Swaidan A.; Mishyn V.; Roussel A.; Cambillau C.; Devos D.; Engelmann I.; Alidjinou E. K.; Happy H.. et al. The role of the surface ligand on the performance of electrochemical SARS-CoV-2 antigen biosensors. Anal. Bioanal. Chem. 2021. 10.1007/s00216-020-03137-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G.; Lee G.; Kim M. J.; Baek S.-H.; Choi M.; Ku K. B.; Lee C.-S.; Jun S.; Park D.; Kim H. G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Kilic T.; Weissleder R.; Lee H. Molecular and Immunological Diagnostic Tests of COVID-19: Current Status and Challenges. iScience 2020, 23, 101406 10.1016/j.isci.2020.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayamohan H.; Lambert C. J.; Sant H. J.; Jafek A.; Patel D.; Feng H.; Beeman M.; Mahmood T.; Nze U.; Gale B. K. SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal. Bioanal. Chem. 2020, 1–23. 10.1007/s00216-020-02958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan M. F.; Dempsey C.; MacSharry J.; O’Callaghan I.; O’Connor P. M.; Horgan C. P.; Durack E.; Cotter P. D.; Hudson S.; Moynihan H. A.; et al. Validation of a lysis buffer containing 4 M guanidinium thiocyanate (GITC)/Triton X-100 for extraction of SARS-CoV-2 RNA for COVID-19 testing: comparison of formulated lysis buffers containing 4 to 6 M GITC, Roche external lysis buffer and Qiagen RTL lysis buffer. bioRxiv 2020, 026435 10.1101/2020.04.05.026435. [DOI] [Google Scholar]
- Engelmann I.; Benhalima I.; Ouafi M.; Pagneux Q.; Boukherroub R.; Hober D.; Alidjinou E. K.; Szunerits S.. Validation of a specimen transport buffer for stabilization of SARS-CoV-2 RNA at room temperature for molecular diagnostics, 2020, unpublished results.
- Auerswald H.; Yann S.; Dul S.; In S.; Dussart P.; Martin N. J.; Karlsson E. A.; Garcia-Rivera J. A. Assessment of Inactivation Procedures for SARS-CoV-2. BioRxiv 2020, 20, 533 10.1101/2020.05.28.120444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bockel D.; Munier C. M. L.; Turville S.; Badman S. G.; Walker G.; Stella A. O.; Anupriya Aggarwal A.; Yeang M.; Condylios A.; Kelleher A. D.; et al. Evaluation of Commercially Available Viral TransportMedium (VTM) for SARS-CoV-2 Inactivation and Usein Point-of-Care (POC) Testing. Viruses 2020, 12, 1208 10.3390/v12111208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch S. R.; Davies K. A.; Buczkowski H.; Hettiarachchi N.; Green N.; Arnold U.; Jones M.; Hannah M. J.; Evans R.; Burton C.; et al. Analysis of Inactivation of SARS-CoV-2 by Specimen Transport Media, Nucleic Acid Extraction Reagents, Detergents, and Fixatives. J. Clin. Microbiol. 20220, e01713–e01720. 10.1128/JCM.01713-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson E.; Prince T.; Anderson E. R.; Casas-Sanchez A.; Smith S. L.; Cansado-Utrilla C.; Solomon T.; Griffith M.; Acosta-Serrano A.; Turtle L.; et al. Methods of Inactivation of SARS-CoV-2 for Downstream Biological Assays. J. Infect. Dis. 2020, 222, 1462–1467. 10.1093/infdis/jiaa507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell M. E.; Subbarao K.; Feinstone S. M.; Taylor D. R. Inactivation of the coronavirus that induces severe acute respiratory syndrome, SARS-CoV. J. Virol. Methods 2004, 121, 85–91. 10.1016/j.jviromet.2004.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabenau H. F.; Biesert L.; Schmidt T.; Bauer G.; Cinatl J.; Doerr H. W. SARS-coronavirus (SARS-CoV) and the safety of a solvent/detergent (S/D) treated immunoglobulin preparation. Biologicals 2005, 33, 95–99. 10.1016/j.biologicals.2005.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jureka A.; Silvas J. A.; Basler C. F. Propagation, Inactivation, and Safety Testing of SARS-CoV-2. Viruses 2020, 12, 622 10.3390/v12060622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann I.; Benhalima I.; Ouafi M.; Pagneux Q.; Boukherroub R.; Hober D.; Alidjinou E. K.; Szunerits S., Validation of a specimen transport buffer for stabilization of SARS-CoV-2 RNA at room temperature for molecular diagnostics. Diagnostics 2021, submitted. [Google Scholar]
- Wrapp D.; De Vlieger D.; et al. Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell 2020, 181, 1004–1015. 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z.; Oton J.; Qu K.; Cortese M.; Zila V.; McKeane L.; Nakane T.; Zinanov J.; Neufeldt C. J.; Cerikan B.; et al. Structures and distribution of SARS-CoV-2 spike proteins on inatact virions. Nature 2020, 588, 498–502. 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.; Mendonca L.; Yang Y.; Gao Y.; Shen G.; Liu J.; Ni T.; Liu C.; Tang X.; Wei J.; et al. The Architecture of Inactivated SARS-CoV-2 with Postfusion Spikes Revealed by Cryo-EM and Cryo-ET. Structure 2020, 28, 1218.e4–1224.e4. 10.1016/j.str.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q.; Bao L.; Mao H.; Wang L.; Xu K.; Yang M.; Li Y.; Zhu L.; Wang N.; Lv Z.; et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020, 369, 77–81. 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q.; Bao L.; Mao H.; Wang L.; Xu K.; Yang M.; Li Y.; Zhu L.; Wang N.; Lv Z.; et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020, 369, eabc1932 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar R.; Baunoch D.; Wojno K.; Parkash V.; Khosravi-Far R. Viral transportation in covid-19 pandemic: Inactivated virus transportation should be implemented for safe transportation and handling at diagnostics laboratories. Arch. Pathol. Lab. Med. 2020, 16, 32298137 10.5858/arpa.2020-0175-L. [DOI] [PubMed] [Google Scholar]
- Understanding Cycle Threshold Ct in SARS-CoV-2 RT-PCR. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/926410/Understanding_Cycle_Threshold__Ct__in_SARS-CoV-2_RT-PCR_.pdf (last downloaded Jan 4, 2021).
- Byrne A. W.; McEveoy D.; Collins A. B.; Hunt K.; Casey M.; Barber A.; Butler F.; Griggin J.; Lane E. A.; McAloon C.; et al. Inferred duraiton of infectious periode of SARS-CoV-2: rapid scoping review and analysis of avalaible evidence for asymptpomatic and symptomatic CAOVID-19 cases. BMJ Open 2020, 10, e039856 10.1136/bmjopen-2020-039856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A.; Jellingsø M.; Sommer M. O. A. Spatial and temporal dynamics of SARS-CoV-2 in COVID-19 patients: A systematic review and meta-analysis. EBioMedicine 2020, 58, 102916 10.1016/j.ebiom.2020.102916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik M.; Tate M.; Lloyd O.; Maraolo A. E.; Schafers J.; Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2020, 2, e13–e22. 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.; Lau E. H. Y.; Wu P.; Deng X.; Wang J.; Hao X.; Lau Y. C.; Wong J. Y.; Guan Y.; Tan X.; et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020, 26, 672–675. 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Kim S. E.; Jeong H.; Yu Y.; Shin S. U.; Kim S. I.; Oh T. H.; Kim U. J.; Kang S.-J.; Jang H.-C.; Jung S.-I.; et al. Viral kinetics of SARS-CoV-2 in asymptomatic carriers andpresymptomatic patients. Int. J. Infect. Dis. 2020, 95, 441–443. 10.1016/j.ijid.2020.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K. A.; Jordan K.; Clyne B.; Rohde D.; Drummond L.; Bryne P.; Ahern S.; Carty P. G.; O’Brien K. K.; O’Murchu E.; et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J. Infect. 2020, 81, 357–371. 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson T.; Spencer E. A.; Brassey J.; Heneghan C. Viral cultures for COVID-19 infectivity assessment – a systematic review (Update 4. medRxiv 2020, 10.1101/2020.08.04.20167932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann I.; Alidjinou E. K.. Spatial and temporal virus load dynamics of SARS-CoV-2: A single center cohort study, 2020, submitted for publication. [DOI] [PMC free article] [PubMed]
- Spearman C. The Method of “Right and Wrong Cases” (Constant Stimuli) without Gauss’s Formula. Br. J. Psychol. 1908, 2, 227–242. 10.1111/j.2044-8295.1908.tb00176.x. [DOI] [Google Scholar]
- Kärber G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch. Exp. Pathol. Pharmakol. 1931, 162, 480–483. 10.1007/BF01863914. [DOI] [Google Scholar]
- https://www.lgcstandards-atcc.org/support/faqs/48802/Converting%20TCID50%20to%20plaque%20forming%20units%20PFU-124.aspx.
- Singanayagam A.; Patel M.; Charlett A.; Bernal J. L.; Silba V.; Ellis J.; Ladhani S.; Zambon M.; Gopal R. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Eurosurveillance 2020, 25, 2001483 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B.; Le Bideau M.; Andreani J.; Hoang V. T.; Grimaldier C.; Colson P.; Gautret P.; Raolt D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1059–1061. 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard J.; Dust K.; Funk D.; Strong J. E.; Alexander D.; Garnett L.; Boodman C.; Bello A.; Hedley A.; Schiffman Z.; et al. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin. Infect. Dis. 2020, 71, 2663–2666. 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singanayagam A.; Patel M.; Charlett A.; Lopez Bernal J.; Saliba V.; Ellis J.; Ladhani S.; Zambon M.; Gopal R. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Eurosurveillance 2020, 25, 2001483 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R.; Corman V. M.; Guggemos W.; Seilmaier M.; Zange S.; Müller M. A.; Niemeyer D.; Jones T. C.; Vollmar P.; Rothe C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Xiao A. T.; Tong Y. X.; Zhang S. Profile of RT-PCR for SARS-CoV-2: A Preliminary Study From 56 COVID-19 Patients. Clin. Infect. Dis. 2020, 71, 1149–2251. 10.1093/cid/ciaa460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar S.; Bartz C.; Becker S.; Basenach S.; Pfeifer S.; Trapp C.; Hamm H.; Schlichting H. C.; Friederichs M.; Koch U.; et al. Duration of SARS-CoV-2 RNA detection in COVID-19 patients in home isolation, Rhineland-Palatinate, Germany, 2020 – an interval-censored survival analysis. Eurosurveillance 2020, 25, 2001292 10.2807/1560-7917.ES.2020.25.30.2001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Laboratory Testing for 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases. 2020, https://www.who.int/publications/i/item/10665-3315012020.
- Hayden R. T.; Caliendo A. M. Persistent Challenges of Interassay Variability in Transplant Viral Load Testing. J. Clin. Microbiol. 2020, 58, 00782-20 10.1128/JCM.00782-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann I.; Alidjinou E. K.; Lazrek M.; Ogiez J.; Pouillaude J. M.; Chazard E.; Dewilde A.; Hober D. Comparison of two commercial quantitative PCR assays and correlation with the first WHO International Standard for human CMV. Diagn. Microbiol. Infect. Dis. 2018, 91, 27–33. 10.1016/j.diagmicrobio.2017.12.021. [DOI] [PubMed] [Google Scholar]
- Okoye N. C.; Adam P Barker A. P.; Curtis K.; Orlandi R. R.; Snavely E. A.; Wright C.; Hanson K. E.; Pearson L. N. Performance Characteristics of BinaxNOW COVID-19 Antigen Card for Screening an Asymptomatic Individuals in a University Setting. J. Clin. Microbiol. 2021, 10.1128/JCM.03282-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremmels H.; Winkel B. M. F.; Schuurman R.; Rosingh A.; Rigter N. A. M.; Rodriguez O.; Ubijaan J.; Wensing A. M. J.; Bonten M. J. M.; Hofstra L. M. Real-life validation of the Panbio COVID-19 antigen rapid test (Abbott) in community-dwelling subjects with symptoms of potential SARS-CoV-2 infection. EClinicalMedicine 2020, 10.1016/j.eclinm.2020.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohmer N.; Toptan T.; Pallas C.; Karaca O.; Pfeiffer A.; Westhaus S.; Widera M.; Berger A.; Hoehl S.; Kammel M.; et al. The Comparative Clinical Performance of Four SARS-CoV-2 Rapid Antigen Tests and Their Correlation to Infectivity In Vitro. J. Clin. Med. 2021, 10, 328. 10.3390/jcm10020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel A.; Pagneux Q.; Cambillau C.; Engelmann I.; Alidjinou E. K.; Ogiez J.; Boukherroub R.; Devos D.; Szunerits S.. A fast, cheap and reliable SARS-CoV-2 detection method based on a portable voltammetric device functionalized with nanobodies, 2021, submitted for publication.
- Ryan D. J.; Toomey S.; Madden S. F.; Casey M.; Breathnach O. S.; Morris P. G.; Grogan L.; Branagan P.; Costello R. W.; De Barra E.; et al. Use of exhaled breath condensate (EBC) in the diagnosis of SARS-COV-2 (COVID-19). Thorax 2021, 76, 86–88. 10.1136/thoraxjnl-2020-215705. [DOI] [PubMed] [Google Scholar]