Abstract
We hypothesized that the correlation of the whole transcriptome with quantifiable phenotypes may unveil genes contributing to the regulation of the corresponding response. We tested this hypothesis in cultured fibroblasts exposed to diverse pharmacological and biological agents, to identify genes influencing chemoattraction of breast cancer cells. Our analyses revealed several genes that correlated, either positively or negatively with cell migration, suggesting that they may operate as activators or inhibitors of this process. Survey of the scientific literature showed that genes exhibiting positive or negative association with cell migration had frequently been linked to cancer and metastasis before, while those with minimal association were not. The current methodology may formulate the basis for the development of novel strategies linking genes to quantifiable phenotypes.
Keywords: Transcriptional reprogramming, expression correlation, cancer, metastasis
Introduction
The association between specific transcripts and quantifiable phenotypes remains a major challenge in biology, especially in today’s era of big data in genomics and transcriptomics. Conventional approaches for target identification are based on expression analyses and usually involve the assessment of differential expression between different experimental groups (1,2). Among the limitations of such strategies is that they usually rely on the magnitude of differential expression, ignoring the fact that the degree of overexpression or underexpression may not necessarily reflect biological impact (3–7). Screening strategies involving genetic manipulation may overcome such limitations, however, by inducing dramatic changes, qualitative or quantitative, in gene expression they may overestimate the role of the corresponding targets that are under interrogation (8,9). Thus, a system that readily allows the association between transcripts and phenotypes in a biologically relevant manner would be highly desirable.
We have hypothesized that in a diverse set of experimental samples, the correlation between levels of expression at the whole transcriptome level, with specific, quantifiable phenotypes of interest, may unveil unrecognized and/or underappreciated associations between transcripts and phenotypes (10,11). Essential for the predictive power of such strategy would be to utilize samples differing drastically in their expression profile in a manner that may not be mechanistically linked to the phenotype evaluated. A strategy of choice, to increase the diversity of the transcriptomic profiles in the samples analyzed, would be to expose them to pharmacological agents that alter drastically gene expression by distinct mechanisms. Such changes may affect highly, or alternatively minimally only, the phenotype of interest facilitating the exclusion of the irrelevant gene targets that would only increase the “noise” of the experimental system.
In the present study, we tested the validity of such approach by testing for paracrine regulators of cell migration. Stromal fibroblasts are known modulators of cell migration by expressing soluble factors that influence cancer cell chemoattraction (12, 13). By exposing fibroblasts to different pharmacological and biological agents we drastically altered their expression profile and then we evaluating if and how such changes in the fibroblasts influenced cancer cell chemoattraction, in a non-cell-autonomous manner. Subsequently, the whole transcriptome was determined, and the abundance of all transcripts were correlated with the migration of the cancer cells. The transcripts associated or not with migration were then surveyed in the scientific literature to confirm that genes identified by this strategy have been linked to cancer and metastasis before.
Results & Discussion
Stromal fibroblasts are well known regulators of chemoattraction operating by paracrine mechanisms and involving the secretion of soluble factors which modulate the motility and migration of adjacently located cancer cells (12, 13). It is plausible that pharmacological or biological manipulation of the expression profile of fibroblasts could change directly or indirectly by their ability to chemoattract cancer cells. We tested this by exposing human HFFF2 fibroblasts cultured in vitro to diverse pharmacological or biological agents. We then measured how the chemoattraction of MDA-MB-231 human breast cancer cells in a transwell system were affected. The agents used were: the inhibitor of protein glycosylation, tunicamycin that induces endoplasmic reticulum stress (14), the cytotoxic anticancer drug doxorubicin, lipopolysacharite (LPS) that induces inflammation (15), and finally the cytokine interleukin 6 (IL-6) and the chemokine CCL8 that act in tumor stroma and modulate cancer cell migration (16–18). In addition to those, MDA-MB-231 conditioned media, serum free media was included and fibroblasts growing under regular media. All these pharmacological or biological agents and cell culture conditions can affect the expression profile of the fibroblasts by various mechanisms and at the same time they may also influence, directly or indirectly, the ability of the fibroblasts to induce the chemoattraction of the cancer cells. The cells that adhered to the lower surface of the transwell chamber (Figure 2a) and those that migrated to the lower chamber (Supplementary figure 1) were evaluated. Both the numbers of the latter and the variation in the measurements were much higher than those of the former. Therefore, in our subsequent analyses we focused on the measurements that reflected the number of cells localized in and on the membrane.
Figure 2.
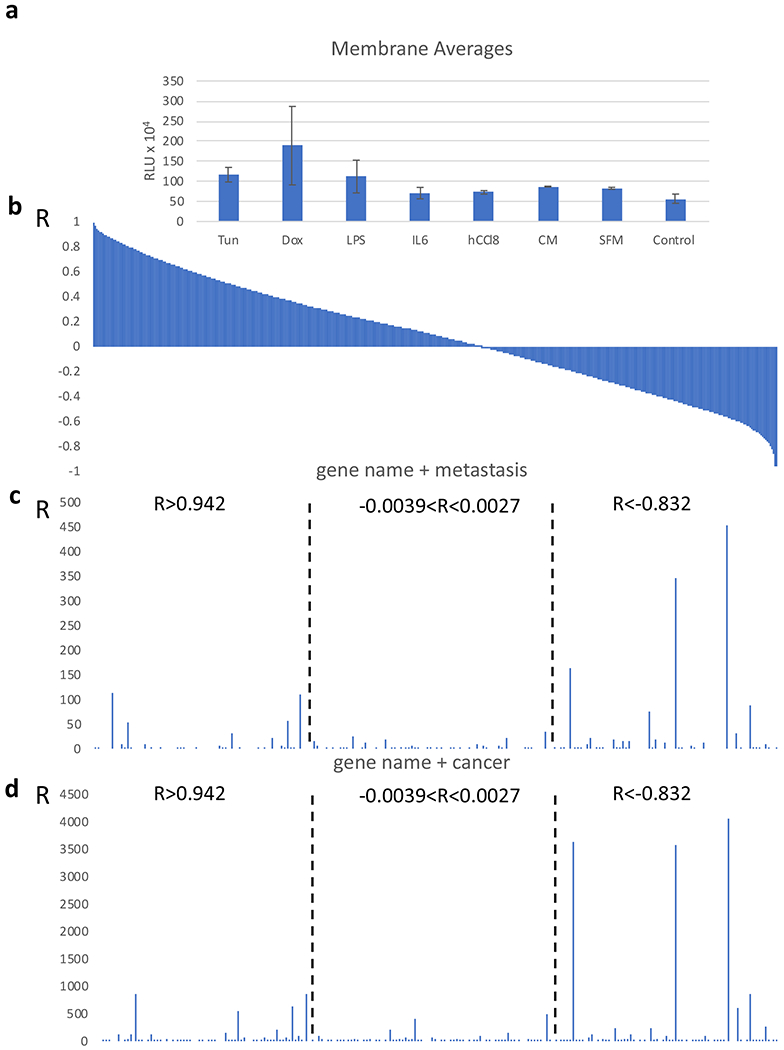
Identification of paracrine regulators of cell migration. a. Migration of MDA-MB-231 cells in a transwell chamber. The conditions at which the fibroblasts in the lower chamber were exposed to are indicated. MDA-MB-231 cell number is expressed as relative luciferase units (RLU). Data are expressed as mean ± SEM. b. Correlation coefficient (R) between expression of each gene in the transcriptome in the fibroblasts and migration of MDMB-231 cells. c and d. Number of hits in PubMed between the gene name and the search term metastasis (c) or cancer (d). Groups of genes (n=70) were selected exhibiting the highest, lowest or close to 0 R, as described in the text.
After evaluating the chemoattraction of MDA-MB-231 cells towards the fibroblasts (Figure 2a) we recorded the transcriptome of the fibroblasts by RNA sequencing. We then calculated the correlation coefficient (R, Pearson’s) between each transcript in the fibroblasts and the migration activity of the cancer cells (Figure 2b). We hypothesized that the R value calculated for each gene would provide a reflection of its ability to modulate the migration of the cancer cells under the given conditions: The transcripts exhibiting positive R values would operate as activators and those showing negative R value would function as inhibitors of chemoattraction. Figure 2b shows the distribution of the R values for the whole transcriptome in relation to chemoattraction. A trend for an increased number of transcripts exhibiting positive R values were revealed, which is likely related to the fact that all experimental conditions induced chemoattraction and thus, the activators of the process were surpassing the inhibitors in this experimental setup.
In order to validate the proposed approach, we performed PubMed database searches, by using the gene name and the terms “cancer” or “metastasis”, to record the hits in genes exhibiting highly positive, highly negative or no correlation with chemoattraction. We postulated that genes with higher number of hits for these terms that are inherently associated with chemoattraction would cluster more tightly in the groups exhibiting either positive or negative correlations, while those not implicated in cell migration would be less abundant and found primarily in the group of genes exhibiting R values around zero. We have applied this step to the top 70 (higher R, range 0.988 to 0.942) and lower 70 (lower R, range −0.966 to −0.832) transcripts. As a control we also included a set of 70 genes exhibiting an R value of around 0 (−0.0039 to 0.0027) corresponding to those genes with presumably no correlation with cancer cell migration. As shown in Figure 1 and Supplementary Table 1, the number of hits in the groups exhibiting strong correlation, either negative or positive, was considerably increased for both the search term “metastasis” and “cancer”. As compared to the group of genes with a correlation coefficient with chemoattraction being around 0, it validates the ability of the proposed strategy to identify regulators of cell migration. Similar results were also obtained when PubMed searches were performed by using the terms “stromal fibroblasts” (Supplementary Figure 2).
Figure 1.
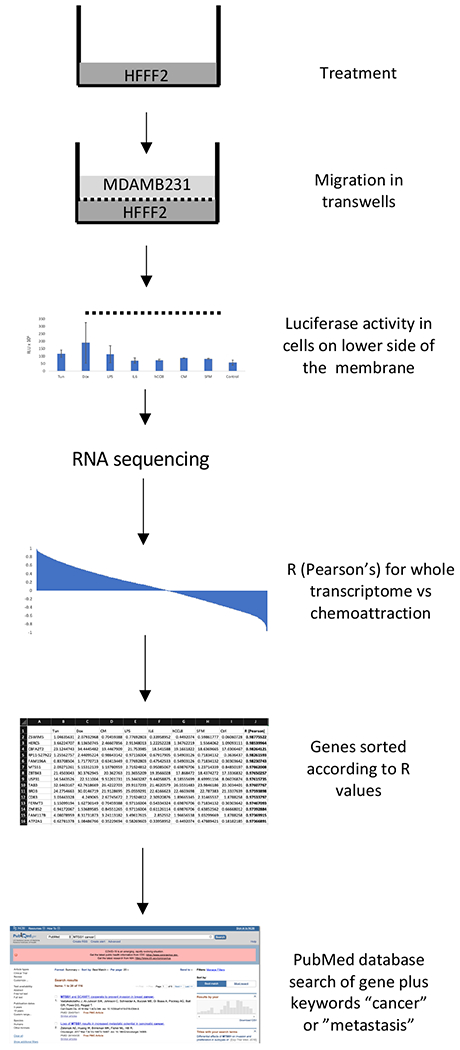
Flowchart of process and analysis followed.
Among the genes correlating strongly with chemoattraction, several have established roles in metastasis; for example, glial cell line-derived neurotrophic factor (GDNF) (R= 0.951) is a soluble ligand that can activate motility and metastasis in cancer cells (19–21) and BMP2 (R=0.942) also encodes for a secreted protein with a role in cancer (22,23). Among the genes with negative correlation WNT10B (R= −0.849) encodes for a ligand with a well-established role in metastasis (24,25) and AES (R=−0.875) has been described as an endogenous metastasis suppressor (26,27). An unexpected target was O6-methylguanine-DNA-methyltransferase (MGMT) MGMT (R= −0.841). It is involved in DNA repair and its expression may reflect the induction of paracrine signaling in fibroblasts that undergo stress due to DNA damage (28,29).
Noteworthy, the number of hits for both the terms “cancer” and “metastasis” was increased for the genes exhibiting negative correlation with chemoattraction as compared to those with positive correlation, suggesting an inhibitory role in this process. A plausible explanation is that in the context of the experimental conditions that stimulated chemoattraction, the activators of cell migration were more abundant than the inhibitors. Therefore, the negative regulators of this process could be identified more stringently.
The present study describes a novel strategy for the identification of regulators of cell migration that operate by paracrine mechanisms. The inclusion of additional experimental conditions at which the expression profile of the fibroblasts could be modulated, it is likely to increase sensitivity and specificity of this approach. It is plausible that this strategy could be modified accordingly to identify regulators of cell migration that operate in a cell-autonomous mechanism as well as other traits that can be quantified and correlation analyses could be performed at a whole transcriptome scale.
Methods
Cell Culture.
MDA-MB-231/Luc cells were obtained from Cell Biolabs. HFFF2 cells were obtained as cryopreserved cells from Sigma-Aldrich (St Louis, MO, USA) MDA-MB-231/Luc and HFFF2 cell lines were cultured in DMEM with 10% Fetal Bovine Serum (Corning). Cell lines were frequently tested for mycoplasma contamination using commercially available Mycoplasma detection kit (Myco Alert kit, Lonza, Walkersville, MD, USA).
Cell treatment.
About 1.25 x 105 HFFF2 cells were seeded on cell culture plate (6-well format, Costar, Waltham, MA, USA) in DMEM with 10% FBS. After 24 h, media was removed, and cells were washed with PBS. Then the cells were treated with 3.0 μg/ml tunicamycin (Sigma-Aldrich), 0.20 μM doxorubicin (Sigma-Aldrich), conditioned media from MDA-MB-231/Luc (cells were plated on a petri dish at a starting density of 8 x 105 cells for 48 h, conditioned media was collected and centrifuged for 5 min at 1200 RPM to remove cell debris), 10 ng/ml IL-6 (Cell Guidance Systems St Louis, MO, USA), 1 ng/ml human CCL8 (Cell Guidance Systems St Louis, MO, USA), and 50 μg/ml LPS (Sigma-Aldrich), for 24 h. Control HFFF2 cells were treated with DMSO. Experiments were performed in duplicates. After treatment, cells were washed with PBS and medium supplemented with 10% FBS was added. Transwell compartment was placed in each well and migration assay was performed immediately after.
Migration Assay.
About 1.4 x 105 MDA-MB-231/Luc cells were seeded on the top chamber of transwells (6-well format, with 8-μm pore size insert, Costar, Waltham, MA, USA) in serum free media and inserted in the plates containing DMEM supplemented with 10% FBS. After 24 h, the transwells were removed and the inside of the chambers was scraped gently with a cotton swab to remove cells that did not migrate. MDA-MB-231/Luc cells on the lower side of the membrane were lysed using cell culture lysis reagent (Promega). HFFF2 cells on bottom of the plate were trypsinised and one part was lysed cell culture lysis reagent (Promega) for luciferase assay and one part for RNA extraction. Luciferase assay was performed using Luciferase Assay System kit (Promega) according to manufacturer’s protocol.
RNA sequencing.
HFFF2 cells were washed with PBS and trypsinised after. Duplicate samples were combined and centrifuged for 5 min at 1200 RPM. Total RNA extraction from cells was performed using RNeasy Micro Kit (Qiagen, Valencia, CA) as per manufacturer’s recommendations. RNA samples were cleaned using RNA clean and Concentrator-5 (Zymo Research, Irvine, CA) as per manufacturer’s recommendations. RNA integrity was assessed using the Agilent Bioanalyzer and samples had a quality score ⩾ 9.6. RNA libraries were prepared using established protocol with NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB, Lynn, MA). Each library was made with one of the TruSeq barcode index sequences and pooled together into one sample to be sequenced on the HiSeq 2x150bp pair-ended sequencing platform (Genewiz). Sequences were aligned to the human reference genome (Home_sapiens.GRCh38, release-98 ensembl.org) using STAR v2.7.2 (30). Reads were counted using the featureCounts function of the Subreads package (31) using Ensembl GTF and summarized at exon, transcript, or gene level. Only reads that were mapped uniquely to the genome were used. Differential expression analysis was performed in R using the edgeR (v3.6) package (32).
Analyses.
The Pearson’s correlation was calculated between the whole transcriptome as obtained by the RNAseq analysis and the average migration data from the transwell membrane in the two duplicate runs, by using Excel. All transcripts were sorted according to their R values and genes which showed an expression value of zero were removed from further analysis. 70 genes from the top, middle and bottom of the transcriptome were compiled into a list to test their association with cancer migration. The gene name and the terms “cancer” or “metastasis” were used as keywords for PubMed database searching, and the number of hits from each search was recorded.
Supplementary Material
Supplementary Table 1. List of genes exhibiting highest, lowest or around 0 R (Pearson’s) values (n=70 per group).
Supplementary Figure 1. Migration of MDA-MB-231 cells in the bottom well of a transwell chamber.
Supplementary Figure 2. Number of hits in PubMed between the gene name and the search term “stromal fibroblasts”.
Acknowledgements
This study was supported by NSF (Award Number: 1736150).
References
- 1.Zhang ZH, Jhaveri DJ, Marshall VM, et al. A comparative study of techniques for differential expression analysis on RNA-Seq data. PLoS One. 2014;9(8):e103207. Published 2014. August 13. doi: 10.1371/journal.pone.0103207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang M, Sun J, Shimizu K et al. Evaluation of methods for differential expression analysis on multi-group RNA-seq count data. BMC Bioinformatics 16, 360 (2015). 10.1186/s12859-015-0794-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moriya H Quantitative nature of overexpression experiments. Mol Biol Cell. 2015;26(22):3932–3939. doi: 10.1091/mbc.E15-07-0512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez-Esteban R, Jiang X Differential gene expression in disease: a comparison between high-throughput studies and the literature. BMC Med Genomics 10, 59 (2017). 10.1186/s12920-017-0293-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Kim Y, Tsang E et al. The impact of rare variation on gene expression across tissues. Nature 550, 239–243 (2017). 10.1038/nature24267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uygun S, Peng C, Lehti-Shiu MD, Last RL, Shiu SH. Utility and Limitations of Using Gene Expression Data to Identify Functional Associations. PLoS Comput Biol. 2016;12(12):e1005244. Published 2016. December 9. doi: 10.1371/journal.pcbi.1005244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crow M, Lim N, Ballouz S, Pavlidis P, Gillis J. Predictability of human differential gene expression. Proc Natl Acad Sci U S A. 2019;116(13):6491–6500. doi: 10.1073/pnas.1802973116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim PM, Tidor B. Limitations of quantitative gene regulation models: a case study. Genome Res. 2003;13(11):2391–2395. doi: 10.1101/gr.1207003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia HG, Phillips R. Quantitative dissection of the simple repression input-output function. Proc Natl Acad Sci U S A. 2011;108(29):12173–12178. doi: 10.1073/pnas.1015616108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Lucius MD, Altomare D, Havighorst A, Farmaki E, Chatzistamou I, Shtutman M, Kiaris H. Coordination Analysis of Gene Expression Points to the Relative Impact of Different Regulators During Endoplasmic Reticulum Stress. DNA Cell Biol. 2019. September;38(9):969–981. doi: 10.1089/dna.2019.4910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Chatzistamou I, Kiaris H. Identification of frailty-associated genes by coordination analysis of gene expression. Aging (Albany NY). 2020. February 29; 12. doi: 10.18632/aging.102875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiaris H, Trimis G, Papavassiliou AG. Regulation of tumor-stromal fibroblast interactions: implications in anticancer therapy. Curr Med Chem. 2008;15(29):3062–7 [DOI] [PubMed] [Google Scholar]
- 13.Kiaris H, Chatzistamou I, Kalofoutis C et al. Tumour–stroma interactions in carcinogenesis: Basic aspects and perspectives. Mol Cell Biochem 261, 117–122 (2004). 10.1023/B:MCBI.0000028746.54447.6c [DOI] [PubMed] [Google Scholar]
- 14.Dorner AJ, Wasley LC, Raney P, Haugejorden S, Green M, Kaufman RJ. The stress response in Chinese hamster ovary cells. Regulation of ERp72 and protein disulfide isomerase expression and secretion. J Biol Chem. 1990. December 15;265(35):22029–34. [PubMed] [Google Scholar]
- 15.Vischer TL, Bretz U, Baggiolini M. In vitro stimulation of lymphocytes by neutral proteinases from human polymorphonuclear leukocyte granules. J Exp Med. 1976;144(4):863–872. doi: 10.1084/jem.144.4.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uciechowski P, Dempke WCM. Interleukin-6: A Masterplayer in the Cytokine Network. Oncology. 2020;98(3):131–137. doi: 10.1159/000505099. Epub 2020 [DOI] [PubMed] [Google Scholar]
- 17.Farmaki E, Chatzistamou I, Kaza V, Kiaris H. A CCL8 gradient drives breast cancer cell dissemination. Oncogene. 2016. December 8;35(49):6309–6318. doi: 10.1038/onc.2016.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farmaki E, Kaza V, Chatzistamou I and Kiaris H. CCL8 promotes postpartum breast cancer by recruiting M2 macrophages. iScience. 2020. In Press. DOI: 10.1016/j.isci.2020.101217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang SM, Chen TS, Chiu CM, Chang LK, Liao KF, Tan HM, Yeh WL, Chang GR, Wang MY, Lu DY. GDNF increases cell motility in human colon cancer through VEGF-VEGFR1 interaction. Endocr Relat Cancer. 2013. December 20;21(1):73–84. doi: 10.1530/ERC-13-0351. [DOI] [PubMed] [Google Scholar]
- 20.Mulligan LM. GDNF and the RET Receptor in Cancer: New Insights and Therapeutic Potential. Front Physiol. 2019. January 7;9:1873. doi: 10.3389/fphys.2018.01873. eCollection 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Funahashi H1, Okada Y, Sawai H, Takahashi H, Matsuo Y, Takeyama H, Manabe T. The role of glial cell line-derived neurotrophic factor (GDNF) and integrins for invasion and metastasis in human pancreatic cancer cells. J Surg Oncol. 2005. July 1;91(1):77–83. [DOI] [PubMed] [Google Scholar]
- 22.Wang MH, Zhou XM, Zhang MY, et al. BMP2 promotes proliferation and invasion of nasopharyngeal carcinoma cells via mTORC1 pathway. Aging (Albany NY). 2017;9(4):1326–1340. doi: 10.18632/aging.101230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bach DH, Park HJ, Lee SK. The Dual Role of Bone Morphogenetic Proteins in Cancer. Mol Ther Oncolytics. 2017;8:1–13. Published 2017. October 24. doi: 10.1016/j.omto.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El Ayachi I, Fatima I, Wend P, et al. The WNT10B Network Is Associated with Survival and Metastases in Chemoresistant Triple-Negative Breast Cancer. Cancer Res. 2019;79(5):982–993. doi: 10.1158/0008-5472.CAN-18-1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wend P1, Runke S, Wend K, Anchondo B, Yesayan M, Jardon M, Hardie N, Loddenkemper C, Ulasov I, Lesniak MS, Wolsky R, Bentolila LA, Grant SG, Elashoff D, Lehr S, Latimer JJ, Bose S, Sattar H, Krum SA, Miranda-Carboni GA. WNTIOB/β-catenin signalling induces HMGA2 and proliferation in metastatic triple-negative breast cancer. EMBO Mol Med. 2013. February;5(2):264–79. doi: 10.1002/emmm.201201320. Epub 2013 Jan 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonoshita M1, Aoki M, Fuwa H, Aoki K, Hosogi H, Sakai Y, Hashida H, Takabayashi A, Sasaki M, Robine S, Itoh K, Yoshioka K, Kakizaki F, Kitamura T, Oshima M, Taketo MM. Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer Cell. 2011. January 18;19(1):125–37. doi: 10.1016/j.ccr.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Okada Y, Sonoshita M, Kakizaki F, et al. Amino-terminal enhancer of split gene AES encodes a tumor and metastasis suppressor of prostate cancer. Cancer Sci. 2017;108(4):744–752. doi: 10.1111/cas.13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cesinaro AM, Sartori G, Migaldi M, Schirosi L, Pellacani G, Collina G, Maiorana A. Prognostic significance of MGMT gene promoter methylation in differently treated metastatic melanomas. Pathology. 2012. June;44(4):313–7. doi: 10.1097/PAT.0b013e328353a0ff. [DOI] [PubMed] [Google Scholar]
- 29.Sinha R, Hussain S, Mehrotra R, et al. Kras gene mutation and RASSF1A, FHIT and MGMT gene promoter hypermethylation: indicators of tumor staging and metastasis in adenocarcinomatous sporadic colorectal cancer in Indian population. PLoS One. 2013;8(4):e60142. doi: 10.1371/journal.pone.0060142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao Y, Smyth GK, Shi W (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. List of genes exhibiting highest, lowest or around 0 R (Pearson’s) values (n=70 per group).
Supplementary Figure 1. Migration of MDA-MB-231 cells in the bottom well of a transwell chamber.
Supplementary Figure 2. Number of hits in PubMed between the gene name and the search term “stromal fibroblasts”.


