Abstract
Plant dwarfism is a desirable agronomic trait in non-timber trees, but little is known about the physiological and molecular mechanism underlying dwarfism in woody plants. Castor bean (Ricinus communis) is a typical woody oilseed crop. We performed cytological observations within xylem, phloem and cambia tissues, revealing that divergent cell growth in all tissues might play a role in the dwarf phenotype in cultivated castor bean. Based on bulked segregant analyses for a F2 population generated from the crossing of a tall and a dwarf accession, we identified two QTLs associated with plant height, covering 325 candidate genes. One of these, Rc5NG4-1 encoding a putative IAA transport protein localized in the tonoplast was functionally characterized. A non-synonymous SNP (altering the amino acid sequence from Y to C at position 218) differentiated the tall and dwarf plants and we confirmed, through heterologous yeast transformation, that the IAA uptake capacities of Rc5NG4-1Y and Rc5NG4-1C were significantly different. This study provides insights into the physiological and molecular mechanisms of dwarfing in woody non-timber economically important plants, with potential to aid in the genetic breeding of castor bean and other related crops.
Subject terms: Agricultural genetics, Plant breeding
Introduction
Dwarfism of various crops triggered the Green Revolution allowing agricultural modernization and therefore unprecedentedly increased crop production1, 2. For woody non-timber economic plants such as fruit tree and woody oilseed crops, dwarfism is a critical way to breed varieties because dwarfed varieties often not only bring higher yields, but also allow compact planting and convenient management in the field3, 4. The genetic basis of plant height is complex, involved in multiple regulatory mechanisms at the physiological and molecular levels.
Usually, auxin is considered a key regulator to control plant height because of its critical roles in regulating cell division, elongation, and differentiation at the meristem such as apical meristem and vascular cambia5, 6, and orchestrating plant growth7–9. When auxin biosynthesis or its cellular transport is interrupted, apical growth is often repressed, leading to enhanced branch growth and development, resulting in a dwarfed and typically high yielding phenotype10–12. Loss-of-function mutations of genes such as TAA1 (Trytophan aminotransferase of Arabidospsis) and YUCs (encoding YUCCA flavin monooxygenase-like enzymes) involved in regulating IAA biosynthesis7, 13, and TIR1 (encoding transport inhibitor response1) and TMKs (encoding transmembrane kinase auxin) involved in sensing auxin signals, often cause a low content of IAA and a disruption of the IAA signal network, resulting in a dwarf phenotype in Arabidopsis14–16. Intercellular transport of auxin involves various transport proteins. Functional loss of transporters such as Arabidopsis auxin resistant1 (AUX1, a polar imported transporter), PIN-FORMED (PIN) 1–3, 6 and 7 (polar exported transporters), and the ATP-binding cassette subfamily B1 (ABCB1) and 19 (lateral efflux transporters) also lead to a dwarf phenotype or shorter hypocotyl in Arabidopsis and rice17–23. The intracellular transport of auxin often maintains or changes intracellular homoeostasis of free IAA in the cytoplasm and storage conjugated IAA in the vacuole or endoplasmic reticulum (ER)24, 25. When disfunction of cellular IAA transporters located in ER including PIN5, PIN-LIKE2 (PILS2) and PILS5 give rise to a reduced content of free IAA in cytoplasm, it results in a dwarf phenotype24, 26. Functional loss of WAT1 (Walls are thin 1 in Arabidopsis, a member of the Medicago truncatula NODULIN21 (MtN21) gene family), an IAA efflux transporter located in the tonoplast, can cause a dwarf phenotype25, 27, but its potential mechanism appears uncertain. Several studies report that intracellular auxin transporters share conserved transmembrane helices, though their amino acid sequences are quite divergent25, 26, 28.
For woody plants, height largely relies on secondary growth of vascular tissue in the stem. The biosynthesis of the three main components of a secondary cell wall, cellulose, hemicellulose and lignin, is the key factor affecting plant height29, 30. In poplar, disfunctions of the genes CesA7A and CesA3D encoding cellulose synthases, GAUT12 encoding galacturonosyltransferase, and LTF1 encoding a transcription factor involved in regulation of lignin biosynthesis, gives rise to dwarfism because of the disruption of normal cellulose, hemicellulose and lignin formation, respectively31–33. However, the molecular mechanisms behind dwarfism in woody plants remain largely unknown.
Castor bean (Ricinus communis L., 2n = 20) is, a typical woody oilseed crop, and it is economically important due to its seed oils being abundant in ricinoleic acids as well as being broadly used in industry for making aviation oil, lubricants, nylon, inks, adhesives and biodiesel34–36. Owing to the increased demand on castor oils supply, there is an immediate need to create improved varieties that can give higher yields in agriculture. Dwarfism of castor is a crucial direction to enhance yield in breeding4, 37. In particular, most cultivated castor varieties are 1.5–2 m in height, domesticated from the wild perennial woody tree (up to 5–7 m in height). Dwarfism is one of the main drivers of artificial selection during castor cultivation and domestication38. However, the physiological and molecular basis of dwarfism remains unknown in castor bean. Castor could be an ideal system to dissect the physiological and molecular mechanism underlying height and dwarf formation in woody plants.
Based on the construction of recombination inbred lines (RILs) and the segregation of given traits, the identification of key genes responsible for a given traits has been applied in diverse crops using a high throughput sequencing technology39–41. In particular, combining RIL population construction and the high-density SNP association analysis, bulked segregant analysis (BSA) has provided an efficient and accurate method to identify key target genes responsible for a given traits in diverse plants42–45. Generally, F2 progeny is sufficient for segregant bulk construction and analysis, therefore the BSA method is often considered as rapid and efficient compared to quantitative trait locus (QTL) mapping and (GWAS)42, 46. In this study, we compared cell size within xylem and phloem tissues and cell division within cambia tissue, and performed a comprehensive BSA analysis, detecting several genes that may be responsible for controlling plant height and therefore the dwarfing trait in castor bean. In particular, we found that the gene Rc5NG4-1, which we show controls IAA transport, might have been targeted by selection and is responsible in part for plant height variation in castor bean. This study provided novel insights into the physiological and molecular mechanisms of the dwarfing trait for a woody non-timber economic plant, with implications for breeding dwarf varieties in castor bean.
Results
Phenotypic variation of plant height in castor bean
The height of cultivated castor varieties usually varies from 1.5 to 2 m. During a broad field survey and castor bean germplasm collection, we collected a dwarf germplasm (CBd) with the average plant height (PH) of 45.67 ± 23.76 cm at maturity. To dissect the potential physiological and molecular mechanism of dwarfing in castor bean, we first compared CBd and a common castor variety (ZB159), called CBt here, with the average height of 236.5 ± 19.68 cm, at maturity (Fig. 1A). We measured several important traits related to height, including vertical height of the secondary branching (VHSB), height of primary raceme (HPR), number of node on main stem (NN), diameter of main stem (DMS), and average length of internodes (ALI). As shown in Fig. 1B and Table S1, the values of VHSB, HPR, DMS and ALI in CBt were greater than that in CBd, but CBd has more nodes on the main stem than CBt. ALI was substantially and significantly difference (p = 1.02e-6) between CBt and CBd, therefore the length of internodes is one of the main differences underlying height difference between two varieties.
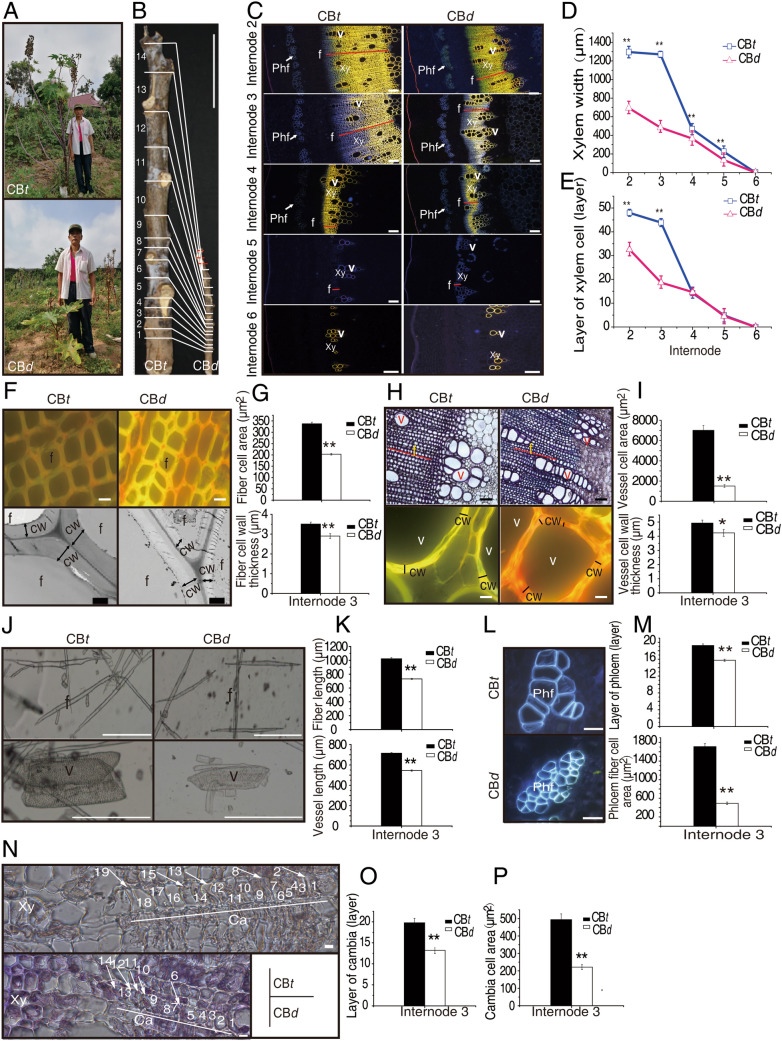
Figure 1.
Phenotypic variation between CBt and CBd. (A) Plant height of CBt and CBd at maturity. (B) Differences in height of the primary raceme and number of nodes on main stem between CBt and CBd. Internodes numbers are marked from root end to shoot end. The red lines in CBd indicate extra internodes absent from CBt. Scale bar = 20 cm. (C) Fluorescence micrographs of cross section of Internodes 2–6 stained with 1% saffron and observed under ultraviolet scanning. Scale bar = 200 μm. (D) and (E) The xylem width and number of xylem layers using (C). (F) Fluorescence micrographs of fiber cell (upper panel) and transmission electron micrographs of fiber cell wall (lower panel) in cross section of Internode 3. The upper panels were obtained as in (C), scale bar = 10 μm. In the lower panels, black lines with double arrows indicate fiber cell wall thickness of certain fiber cells. Scale bar = 2 μm. (G) Fiber cell area and fiber cell wall thickness from (F). (H) Micrographs of vessel cells (upper panel) and fluorescence micrographs of vessel cell wall (lower panel) in cross section of Internode 3. In the upper panel, the graphs were observed as (C) but scanning under bright optic, scale bar = 100 μm. In the lower panel, black lines indicate the cell wall thickness of single vessel cell, scale bar = 10 μm. (I) The vessel cell area and vessel cell wall from (H). (J) Micrographs of fiber and vessel cell separated by 50% acetate acid from Internode 3. Scale Bar = 500 μm. (K) The length of fiber and vessel cell from (J). (L) Fluorescence micrographs of phloem fiber cells of Internode 3. The graphs scanned as (C), scale bar = 50 μm. (M) Measurement of phloem cell area and the cell layers within Internode 3 were depended on (L). (N) Cross section of cambia within Internode 3 scanned by fluorescence microscopy under bright optic. Cell layers are numbered from phloem end to xylem end. Scale bar = 10 μm. (O) and (P) Number and area of cambia cell from (N). In figure C-P, CBt and CBd seedlings with seven internodes were used cytological observation. Xy: xylem, v: vessels, f: fibers, Phf: phloem fibers, cw: cell wall. Results are means ± SD of at least three independent plants from each variety, and significance determined by Student’s t-test, * p ≤ 0.05, ** p ≤ 0.01.
We conducted cytological observations of cell size, cell number and cell layers within xylem, cambia and phloem tissues from developing stems of CBt and CBd within seven internodes from the bottom of the stem (Figure S1). Since it is sometime variable and difficult to delimit Internode 1 and Internode 7, we excluded these and examined Internode 2 to Internode 6. Xylem tissue was not developed at the younger internodes (Internodes 5 and 6). The more visible xylem tissues including the width and cell layers were observed in Internodes 2 to 4, with visible in both CBt and CBd (Fig. 1C).
The width and cell layers of xylem tissues at Internodes 2 and 3 exhibited substantial and significant differences between CBt and CBd (Fig. 1D,E), whereas these figures were very similar at Internodes 4–6, suggesting that developmental difference between CBt and CBd might start in Internode 3. Comparing the cell size and cell wall thickness of fibers and vessels in xylem tissues at Internode 3 revealed that cell size, cell wall thickness and longitudinal cell length of fibers and vessels were significantly different between the two accessions (Fig. 1F–K). The cell size of phloem fibers was greater and the number of cell layers of phloem more numerous in CBt than in CBd at Internode 3 (Fig. 1L,M). Similarly, the cell size and number of cell layers of cambia tissue were significant greater in CBt compared to CBd at Internode 3 (Fig. 1N–P). These observations clearly suggest that the difference in cell size across xylem, cambia and phloem as well as difference of meristematic capacity of cambia tissue might all be important factors that cause the variation in plant height.
BSA-seq identified QTL regions associated with plant height
To identify genes potentially responsible for the variation in height in castor bean, CBd (♀) and CBt (♂) were crossed to generate F1 hybrids. The average height of F1 hybrids was 123.2 cm. After selfing the F1, we obtained an F2 population comprising 330 individuals. Plant height varied from 34 to 286 cm in F2 population and exhibited a normal distribution (Fig. 2A), suggesting polygenic control of plant height in this population. We measured five traits in addition to PH (VHSB, HPR, NN, DMS and ALI) in the F2 population. ALI (r = 0.8443) and DMS (r = 0.7762) were tightly correlated with plant height, whereas NN was not correlated with plant height (r = 0.0186) (Fig. 2B). These observations were consistent with our above findings that greater PH is caused in part by the larger cell size in the xylem and phloem tissues, and the stronger meristematic capacity of cambia tissue within internodes.
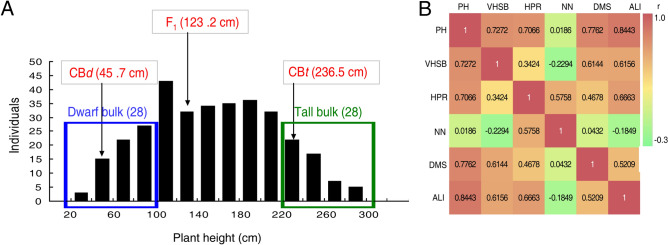
Figure 2.
The segregation of plant height in the F2 population and the correlation between six height-related phenotypes. (A) Frequency distribution of plant height in 330 F2 individuals at maturity. The average values of plant height for CBt, CBd and F1 hybrids are shown in red text. The 28 shortest individuals (< 100 cm) were selected as the dwarf bulk and the 28 tallest individuals (> 220 cm) were selected as the tall bulk. (B) Correlation coefficient (r) among six traits in the F2 population. PH: plant height, VHSB: vertical height of the secondary branching, HPR: height of primary raceme, NN: number of nodes on main stem, DMS: diameter of main stem, ALI: average length of internodes on main stem.
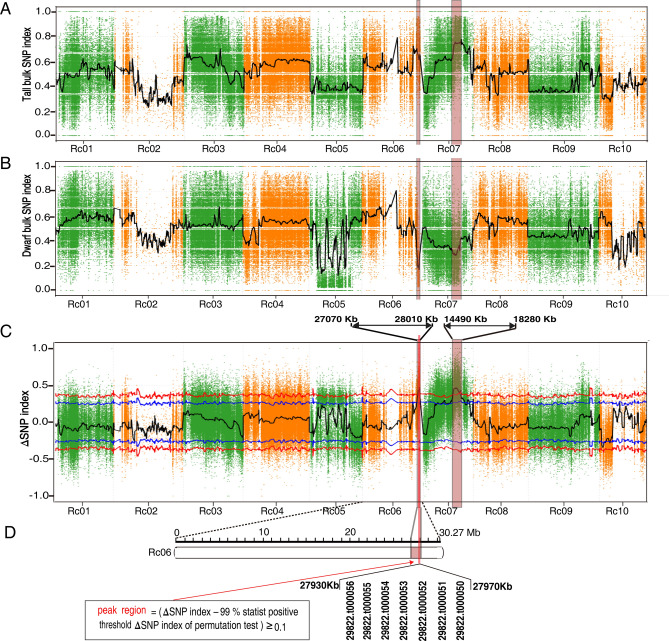
Based on the segregation of phenotype, we selected the 28 shortest individuals (average height 71.32 ± 20.14 cm) as the dwarf bulk and the 28 tallest individuals (average height 254.54 ± 21.44 cm) as the tall bulk (Fig. 2A) for genome sequencing to identify loci potentially associated with plant height. The sequencing generated 83,983,462 and 79,741,716 high quality clean reads for the dwarf bulk and tall bulk, respectively, a depth exceeding 32-fold of the genome (Table S2). After aligning the clean reads to the castor reference genome (http://castorbean.jcvi.org) and variant calling using GATK 3.4, 1,027,623 genomic SNPs were identified from the parents and two bulks. The physical positions of SNP were aligned and annotated by ANNOVAR according to our previously constructed castor chromosomes41. In total, 300,225 SNPs homozygous within but different between the parents were identified, and their ΔSNP index and the average ΔSNP index within each 1 Mb window (with 10 kb step size) were calculated. Based on the 99% statistical confidence intervals (permutation tests under the null hypothesis of no QTLs), two regions, QTL1 (0.94 Mb in length; 27.07–28.01 Mb on chromosome Rc06) and QTL2 (3.79 Mb in length; 14.49–18.28 Mb on chromosome Rc07), were identified as genomic regions associated with height in castor bean (Fig. 3A–C). We chose to focus on the region on Rc06 (27.93–27.97 Mb) where the difference was greatest between the ΔSNP index (0.463) and the 99% statistical threshold (0.350) based on the recommendation of reference42 (Fig. 3D).
Figure 3.
BSA for QTLs contributed to plant height using 1 Mb sliding windows with a step size of 10 Kb. SNP-index of (A) tall bulk and (B) dwarf bulk. The black curve line represents the average SNP index of each window. (C) ΔSNP index. The ΔSNP index of each SNP was computed as the difference between the tall SNP index and the dwarf SNP index. The black curve represents the average of ΔSNP index of each window, blue and red lines represents the 95% and 99% statistical confidence intervals, respectively, based on 1000 permutation tests to each window under the null hypothesis of no QTL. The pink rectangles in (A), (B) and (C) represent the QTLs (p value < 0.01). The red vertical line in (C) is the QTL1 peak region. Rc01-Rc10 in abscissa are the 10 castor bean chromosomes according to41. (D) The peak candidate region located under QTL1 covers seven genes. The pink rectangle and red vertical line are the same as in (C).
Candidate genes associated with plant height were identified
In total, 487 genes were identified in the QTL1 and QTL2 regions (Table 1). After filtering out the 78 genes without ΔSNP index (≥ 0.5) and the 84 genes with SNPs in intergenic regions, we obtained 325 genes with SNPs located within the gene or within 2000 bp of the coding region (Figure S2A–B). The 325 genes were considered candidate genes associated with castor bean height. Gene ontology (GO) analysis showed 146 of 325 genes were functionally involved in cell component (Figure S2C). Based on the p value of GO enrichment analysis, we found that these candidate genes were mainly enriched in lignin metabolism, secondary metabolism and phenylalanine metabolism (Figure S2D). KEGG enrichment analysis found that the majority of candidate genes were enriched in hormone signal transduction and secondary metabolism processes (Figure S2E).
Table 1.
Details of two QTL associated with plant height.
| Position in chromosomes | Interval/bp | Region/Mb | Scaffold/No | SNP/No | Gene/No | |
|---|---|---|---|---|---|---|
| QTL1 | Rc06 | 27,070,001–8,010,001 | 0.94 | 1 | 1501 | 140 |
| QTL2 | Rc07 | 14,490,001–8,280,001 | 3.79 | 29 | 5006 | 347 |
We identified where in the gene the SNPs were located and found non-synonymous polymorphisms in 118 genes that differentiated CBd and CBt (Figure S2B). To reduce the potentially spurious associations between SNPs and height as far as possible we applied a strict criterion with a ΔSNP index ≥ 0.6, as suggested by Takagi et al.42, to sort out target candidate genes. Based on properties of the amino acids polymorphisms, 29 target candidate genes with non-synonymous SNPs (ΔSNP index ≥ 0.6) were selected for further investigation, including 6 and 23 genes located at QTL1 and QTL2, respectively (Table 2).
Table 2.
The candidate genes potentially controlling plant height in castor bean identified in QTL.
| Gene ID | QTL region | ΔSNP index ≥ 0.6 | Functional annotation |
|---|---|---|---|
| 29,822.t000132 | QTL1 | 0.668 | Conserved hypothetical protein |
| 29,822.t000126 | QTL1 | 0.682 | HIPL1 protein precursor, putative |
| 29,822.t000107 | QTL1 | 0.653 | Nop14, putative |
| 29,822.t000095 | QTL1 | 0.62 | Mannosidase alpha class 2a, putative |
| 29,822.t000076 | QTL1 | 0.707 | Peroxidase 31 precursor, putative |
| 29,822.t000050 | QTL1 | 0.628 | Auxin-induced protein 5NG4, putative |
| 29,767.t000004 | QTL2 | 0.707;0.607 | NADPH fad oxidoreductase, putative |
| 29,767.t000005 | QTL2 | 0.616 | Pentatricopeptide repeat-containing protein, putative |
| 29,846.t000009 | QTL2 | 0.617 | Pectin acetylesterase, putative |
| 29,846.t000008 | QTL2 | 0.674 | Protein phosphatase 2c, putative |
| 29,846.t000007 | QTL2 | 0.727 | Conserved hypothetical protein |
| 29,846.t000002 | QTL2 | 0.695 | Ubiquitin ligase E3 alpha, putative |
| 27,798.t000017 | QTL2 | 0.643 | Conserved hypothetical protein |
| 27,798.t000018 | QTL2 | 0.808 | Conserved hypothetical protein |
| 27,798.t000020 | QTL2 | 0.671 | Conserved hypothetical protein |
| 29,973.t000023 | QTL2 | 0.681 | ATP binding protein, putative |
| 29,973.t000005 | QTL2 | 0.629 | Cytoplasmic dynein light chain, putative |
| 29,969.t000005 | QTL2 | 0.654 | Conserved hypothetical protein |
| 28,308.t000003 | QTL2 | 0.658 | Transcription factor, putative |
| 29,568.t000010 | QTL2 | 0.630 | Amino acid transporter, putative |
| 29,568.t000002 | QTL2 | 0.684;0.610 | Cyclin d, putative |
| 29,701.t000006 | QTL2 | 0.700 | Symplekin, putative |
| 29,701.t000011 | QTL2 | 0.611 | Translation initiation factor if-2, putative |
| 29,701.t000013 | QTL2 | 0.637 | Isoamyl acetate-hydrolyzing esterase, putative |
| 29,701.t000019 | QTL2 | 0.667 | Conserved hypothetical protein |
| 28,345.t000007 | QTL2 | 0.740 | Tropinone reductase, putative |
| 28,345.t000005 | QTL2 | 0.631 | Ribulose bisphosphate carboxylase/oxygenase activase 1 |
| 28,345.t000004 | QTL2 | 0.634 | Transferase, transferring glycosyl groups, putative |
| 28,345.t000003 | QTL2 | 0.697 | Hypothetial protein |
Rc5NG4-1 was demonstrated as a candidate gene controlling castor height
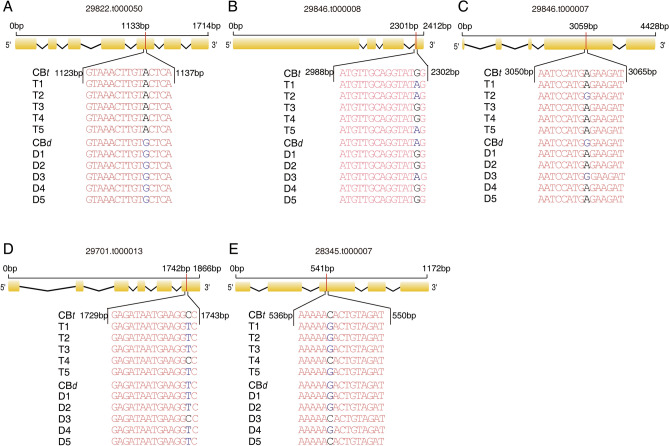
To investigate whether the SNP variants were associated with plant height among different germplasms, we determined the genotypes of five wild castor bean accessions with average height of 539.2 ± 31.76 cm, and five cultivated castor bean accessions with average height of 153.4 ± 13.16 cm (Table S3). Among the 29 candidate genes, based on their putative functional annotation directly or indirectly related to cell wall formation (tightly associated with dwarf phenotype), five (29,822.t000050, 29,846.t000008, 29,846.t000007, 29,701.t000013 and 28,345.t000007) were selected to further investigate the genotypes of non-synonymous SNP with ΔSNP index ≥ 0.6 in this extended germplasm (Table 2). As shown in Fig. 4, we found only the non-synonymous A/G SNP at position 1133 bp in gene 29822.t000050 to be fixed between the tall and dwarf germplasm. These results strongly suggest that gene 29822.t000050 might be an important candidate associated with height. However, we noted that the ΔSNP index of 29,822.t000050 was not the highest among identified candidate genes based on the statistic analysis (see Table 2).
Figure 4.
ΔSNP index ≥ 0.6 non-synonymous SNPs identification for five candidate genes obtained from 10 castor varieties. (A)–(E) The exon (box) and intron (line) structure of 29,822.t000050, 29,846.t000008, 29,846.t000007, 29,701.t000013 and 29,345.t000007 and the genotypes of non-synonymous SNPs with ΔSNP index ≥ 0.6 in tall (T) and dwarf (D) accessions determined using PCR and sequencing. Collection location and plant height of the 10 varieties are shown in Table S4.
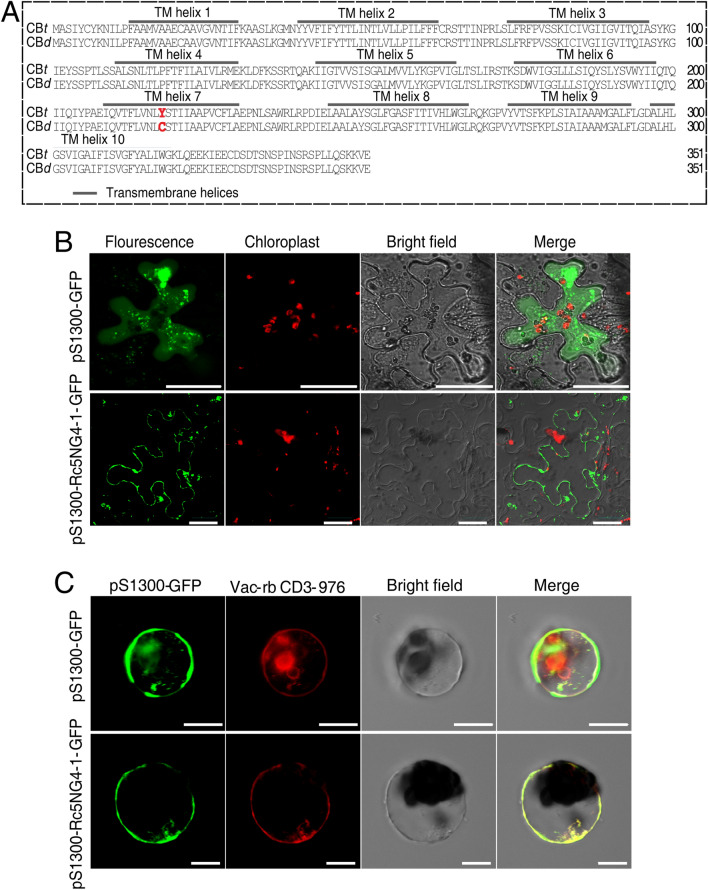
According to the annotation of 29,822.t000050, it is predicted to be an auxin induced protein 5NG4, a member of the MtN21 family47. As the first 5NG4-like gene to be reported in castor bean, we named 29,822.t000050 as Rc5NG4-1. The non-synonymous SNP resulted in the substitution of the amino acid 218th from tyrosine (Y) in CBt to cysteine (C) in CBd (Fig. 5A). Previous studies have found that the protein of 5NG4 members have 10 transmembrane (TM) helices that often form several transmembrane secondary loop structures for facilitating IAA transportation. The Arabidopsis orthologue AtWAT1 (At1g75500) functions as an auxin efflux transporter localized on tonoplast, facilitating IAA transportation from vacuole to cytosol25. We predicted the protein secondary structure of Rc5NG4-1 using TMHMM Server v. 2.0, found it had 10 TM domains and the amino acid substitution described above was located in TM helix 7, though did not result in a change to the TM helix number or number of hydrophilic loops (Fig. 5A). Inspection of the subcellular localization of Rc5NG4-1 in tobacco epidermal cells and Arabidopsis protoplasts, revealed Rc5NG4-1 was not localized in the plasma membrane (Fig. 5B) but was instead co-localized with the tonoplast marker (Fig. 5C). Thus, we reasonably infer that the function of Rc5NG4-1 is involved in IAA transportation from vacuole to cytosol in castor bean. Further, while inspecting the expressional profiles of Rc5NG4-1 among different tissues we found Rc5NG4-1 have relatively higher expressions in root and stem, compared to that in leaf and seed (Figure S3).
Figure 5.
Protein structure and subcellular localization of Rc5NG4-1. (A) Protein sequence structures and transmembrane (TM) domains of Rc5NG4-1 in CBt and CBd predicted using TMHMM. The 218th amino acid change are marked red, which induced by the non-synonymous SNP located in 1133 bp position of Rc5NG4-1. (B) Rc5NG4-1-fused GFP was expressed in hypoepidermal cell of tobacco leaves. Scale bar = 30 μm. (C) Rc5NG4-1-fused GFP was transiently co-expressed with the vacuolar membrane marker in protoplasts. Empty vector pS1300-GFP was expressed in protoplasts as a control (top panels). Vac-rb CD3-976, the vector carries tonoplast marker γ-TIP coding sequence (aquaporin with tonoplast localization). Images in bottom panels show colocalization of Rc5NG4-1 and γ-TIP. The fluorescence graphs were detected by confocal microscope in (B) and (C). Scale bar = 10 μm.
Functional confirmation of Rc5NG4-1 involved in IAA uptake
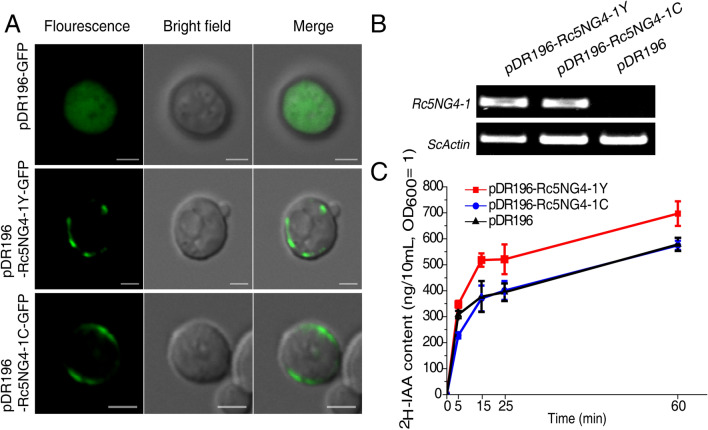
To inspect whether the non-synonymous SNP that resulted in the substitution of amino acid 218 from tyrosine (Y) in CBt to cysteine (C) in CBd gave rise to functional difference in auxin transport, we performed a heterologous expression of Rc5NG4-1 in yeast (Saccharomyces cerevisiae). First, we checked the subcellular localization of Rc5NG4-1Y and Rc5NG4-1C in transformed yeast strains using pDR196-Rc5NG4-1Y-GFP or pDR196-Rc5NG4-1C-GFP vectors. As shown in Fig. 6A, Rc5NG4-1Y and Rc5NG4-1C were localized in the membrane. Second, Rc5NG4-1Y and Rc5NG4-1C were heterologously expressed in yeast cells and expression of Rc5NG4-1Y and Rc5NG4-1C was confirmed using RT-PCR (Fig. 6B). The uptake capacity of free IAA (as 2H-IAA in the growth medium) in transformed yeast strains was assayed. The uptake rates of 2H-IAA were higher in yeast transformed with Rc5NG4-1Y than yeast transformed with Rc5NG4-1C from 5 to 60 min of 2H-IAA supply (Fig. 6C). The uptake rates of 2H-IAA in transformed Rc5NG4-1C yeast strains was almost identical to the control, suggesting Rc5NG4-1C may have lost the capability to uptake free IAA. These results clearly show that Rc5NG4-1Y and Rc5NG4-1C lead to functional protein divergence resulting in a difference in IAA uptake.
Figure 6.
Examination of IAA uptake based on heterologously transformed yeast. (A) Rc5NG4-1Y- or Rc5NG4-1C-fused GFP were expressed in yeast cell and fluorescence observed by confocal microscopy. Scale bar = 2 μm. (B) Rc5NG4-1Y and Rc5NG4-1C expression in Rc5NG4-1Y transformed yeast, Rc5NG4-1C transformed yeast and empty vector pDR196 transformed yeast detected by RT-PCR. The upper group and lower group of blots were cropped from different part of same gels, and full-length blots are presented in Figure S4. (C) 2H-IAA uptake of the different transformed yeast strains. The values are means ± SD of three replications (transformations carried out once for each and then grow in triplicate).
Content of free IAA in CBd and CBt
Content of free IAA in plant tissues is usually determined by intracellular IAA transport between organelle and cytosol, cellular influx and efflux transport, and long distance transport among tissues23, 25, 48. To test whether the differences between the tall and dwarf phenotypes related to free IAA, we measured the content of free IAA in apical buds and Internode 3 tissues in CBt and CBd. As shown in Fig. 7, the content of free IAA did not differ between CBd and CBt in the apical bud, whereas there was significantly lower free IAA in Internode 3 of CBd than in CBt (p value = 0.0013, t-test). This result suggests that the biosynthesis of free IAA in apical bud might be similar between the two accessions, but the transport capability of free IAA from the apical bud to node is weaker in CBd.
Figure 7.
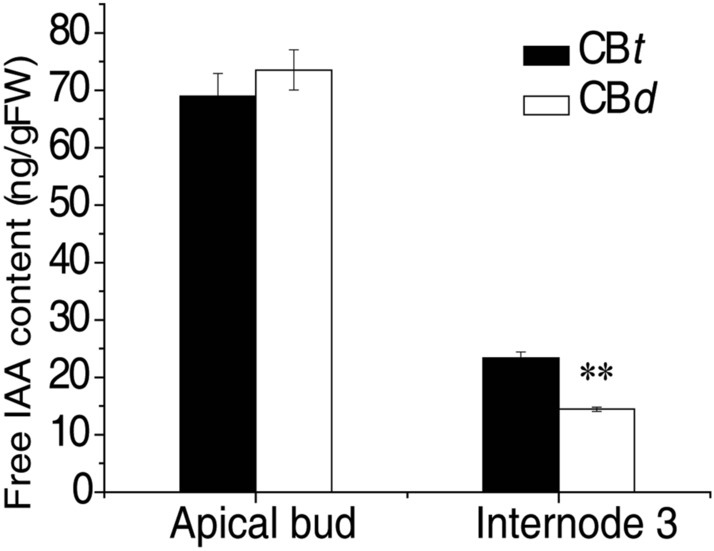
Measurement of free IAA concentration in CBt and CBd. The free IAA concentration within apical bud and Internode 3 were measured using CBt and CBd seedlings with seven internodes. Values are means ± SD of three replicate determinations, **p value ≤ 0.01(Student’s t-test).
Discussion
As a key component of tree architecture, plant height is a critical agronomic trait in non-timber trees such as fruit trees and woody oilseed crops. Dwarfism is beneficial for reducing canopy size, allowing increased planting density and greater yields, lowering costs and increasing the orchard’s lifespan of non-timber trees. Dwarfism in plants is a complex trait and relatively little is known about the physiological and genetic factors that regulate dwarfing in non-timber trees. As mentioned above, castor bean is an ideal system to dissect the potentially physiological and molecular mechanisms underlying dwarfism in woody plants because of its domestication from a tall wild perennial woody tree to the shorter annual oilseed crop38.
Based on our phenotypic observations, dwarfism is clearly caused by shorter internodes and thinner stems, implying that stem growth of CBd might be developmentally repressed. Cytological observations revealed that cell growth in xylem and phloem tissues and the cell division of cambia tissue were repressed in CBd, resulting in smaller cells within the xylem and phloem tissues and fewer cell layer in cambia tissue compared to CBt. Similarly, in poplar, larger phloem and vessel tissues lead to greater plant height33, 49. Since cell division in cambia tissue is an important driver of radial and longitudinal growth in woody plants50, we infer that the capability of cell division within cambia tissue is probably a critical factor affecting the radial and longitudinal growth in castor bean internodes.
BSA is an efficient and rapid method to detect plausible candidate genes responsible for a given trait in plants from the F2 generation. Based on the segregation of plant height in the F2 generation, we detected two QTLs associated with castor bean height. Within the two identified QTLs, 325 candidate genes associated with the plant height were mainly enriched in cell component, functionally involved in hormone signal transduction and secondary metabolism processes, consistent with the requirement of plant growth and development in woody plants. Based on the non-synonymous polymorphisms and a strict criterion (ΔSNP index ≥ 0.6) we sorted out 29 candidate genes that were likely participated in regulating the formation of plant height in castor bean. Within these identified candidate genes, we sorted out a candidate gene Rc5NG4-1 that was functionally involved in the regulation of cellular auxin transport. However, according to current analyses it is difficult to determine or rule out which candidate genes are the causal genes controlling the formation of plant height in castor bean.
Many studies have revealed that the GA signaling network is a critical factor that regulates plant growth, in some cases underlying dwarf phenotypes2, 51, 52. Indeed, the physiological and molecular basis of plant growth regulation and dwarfing are complex and varied across plant species. Our current study demonstrated that Rc5NG4-1 is located in the tonoplast, and is an auxin transporter, likely capable of transporting IAA between vacuole and cytosol. Based on expressing Rc5NG4-1 heterologously in yeast, it is clear that the uptake capacity of free IAA between Rc5NG4-1Y (tall allele) and Rc5NG4-1C (dwarf allele) is significantly different validating the different uptake and transport capacities of Rc5NG4-1Y and Rc5NG4-1C. It is likely that Rc5NG4-1 functions in a similar way to its putative orthologue AtWAT1 encoding an IAA transport protein in Arabidopsis thaliana25 by transferring IAA between the vacuole and cytosol to maintain the free IAA content in cytosol. It appears that the Rc5NG4-1C allele, whilst being expressed, was non-functional. Using Rc5NG4-1 as a query in a BLAST search against the castor bean genome reference we found several putative paralogues (data not shown), suggesting that other copies of Rc5NG4-like genes might compensate for this loss-of-function, thus partially maintaining the free IAA content in cytosol. Due to the free IAA content in the apical bud being similar between CBt and CBd, and the difference in the internode tissues, we infer that long-distance free IAA transport capability from the apical bud to the internode might vary between two varieties. Since auxin affects cell division in cambia tissue and cell growth in xylem and phloem tissues53, 54, we reasonably infer that our observed reduction in IAA in the internode tissues in CBd might be an important physiological difference resulting in the dwarfing of castor bean. Based on the sequence similarity we found the orthologue AtWAT1 of Rc5NG4 in Arabidopsis. The function of AtWAT1 has been verified to control plant height by affecting cellular auxin transport in Arabidopsis25, 27. Our results indicated that Rc5NG4 is likely a causal gene that gives rise in phenotypic variation of plant height in woody castor bean. However, such study is very limited in other plants. Whether or how the mutant Rc5NG4-1C directly influences cellular influx and efflux of IAA and the long distance transportation of free IAA remain unknown.
Conclusions
In this study, we first found divergent cell growth in xylem and phloem tissues and divergent cell division in cambia tissues, suggesting the potential physiological basis of dwarfing in castor bean. Two QTLs associated with plant height which cover 325 candidate genes were identified using bulked segregant analysis sequencing from an F2 population derived from a tall castor variety CBt and a dwarf castor variety CBd. In particular, we detected a gene Rc5NG4-1 (encoding an IAA transporter protein localized in tonoplast) as a responsible candidate for controlling dwarfism formation in woody oilseed crop castor bean. This study provided novel insights into the physiological and molecular mechanisms of dwarfing in woody non-timber economic plants, facilitating the genetic improvement to create dwarf varieties in castor bean.
Materials and methods
Plant materials and phenotype survey
The castor bean varieties CBd (dwarf) and CBt (tall) were developed by self-pollination controlled for at least three generations, provided by Zibo Academy of Agriculture Sciences, Shandong Province. The varieties CBt and CBd were genetically independent, and were applied as parents for creating the F2 population in 2013–2016. The F1 hybrids were produced by crossing between CBt (♂) and CBd (♀) in 2016, and F2 recombinant population that derived from selfing the F1 hybrids. Parents and two populations of offspring were grown in the experiment field managed by Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences in Mengla County, Xishuangbanna, Yunnan province (101° 25′ E, 21 41′ N) in 2017, each planted 1 m apart. Six traits, including plant height (PH), vertical height of the secondary branching (VHSB), height of primary raceme (HPR), number of node on main stem (NN), diameter of main stem (DMS), and average length of internodes on main stem (ALI), were measured after seed ripening for parent lines and each individual of the F2 population. Only PH was measured for F1 hybrids. The DMS was the diameter of the middle internode of main stem. ALI was defined as the ratio of HPR to NN in main stem.
The observations of cytology
The CBt and CBd seedlings were grown in a plastic basin (the ratio of nutrient soil, soil and perlite was 3:1:1, v/v) in the greenhouse (30 °C, light/dark was 12 h/12 h) and individuals at the same maturity stage (seven internodes) were used in the anatomical survey, transmission electron microscopy and maceration of fibers and vessels within stem.
The stem from CBt and CBd seedlings were split into 1 cm sections manually using a razor blade, immersed in 30% glycerin, infiltrated in a vacuum environment (0.08 MPa) for 30 min, and sustained in 30% glycerin. The 1 cm samples were then cut into blocks (2 mm × 2 mm) and fixed into the chuck with refrigerant. Then, sections of 15–25 μm thickness were sliced continuously using freezing microtome (Leica, CM3050S, Germany), transferred onto microslides and rinsed until the refrigerant was removed completely. After this, the sections were stained with 1% saffron (1 g saffron dissolved in 100 ml 50% ethanol, m/v) for one minute, and the residual dye was removed using 50% ethanol. After rehydration with sterile water, the sections were inspected with a fluorescence microscope (Leica Microsystems, DM5508, Germany) under white light or ultraviolet light to scan the xylem tissue and the phloem fibers55. Images were captured using a Leica camera (Leica Microsystems, DFC450C, Germany), and cell area, cell layer and cell wall thickness were measured with Image J software (version 1.46). All cells from each image were measured, and the values from at least five images of at least three plants were measured.
Transmission electron microscopy was used for inspecting cell thickness of xylem in Internode 3. The xylem tissue was shaped to 1 mm × 1 mm blocks, fixed in 2.5% glutaric dialdehyde for at least 24 h and immersed in 1% osmium acid for 12 h under 4 °C. After dehydration in an acetone series (final concentration of 90% acetone) and embedment in resin spur, the 40 nm-thick sections were sliced by microtome (Leica-R, Germany). The sections were dried at room temperature and stained using uranylacetate and lead citrate. The stained sections were inspected under an electron microscope (JEOL Ltd, Tokyo, Japan), and images captured by an Olympus-SIS Megaview camera (Olympus Soft Imaging Solutions GmbH, Münster, Germany). Measurement of the cell wall followed the above method.
The xylem in the Internode 3 of seedlings was macerated to separate fibers and vessels for measuring length. The maceration method was similar to that of Biswal et al.33. The tissue was macerated for 10 h at 90 °C with buffer (50% acetate acid and 3% hydrogen peroxide) in 2 mL polypropylene tubes. After washing with sterile water, the fibers and vessels were observed by microscope (Leica Microsystems, DM5508, Germany). Image capturing and trait measurements were followed above protocols.
DNA extraction and BSA analysis
DNA were isolated from young leaves of single plants in parents, the 28 tallest plants and the 28 most dwarf plants using a plant genomic DNA extraction Kit (TIANGEN, Beijing, China). The tall bulk and dwarf bulk were constructed by mixing equivalent DNA amounts from each of 28 tall plants and each of 28 dwarf plants, respectively. Pair-end sequencing libraries were generated for the two parents, the tall bulk and the dwarf bulk and sequenced using Illumina HiSeq™ 2500 (Illumina Inc, San Diego, CA, United States). High quality clean 150 bp reads (HQ clean reads) were obtained by filtering out reads with adapters, with > 10% unidentified nucleotides (N) and with quality scores (Q) ≤ 20 bases accounting for more than 50%. The depth for two parents and two bulks were above 32-fold (Table S2).
HQ clean reads were aligned against the reference genome (http://castorbean.jcvi.org) using the Burrows–Wheeler Aligner (BWA) software (settings: mem 4 -k 32 -M). After duplicates marking by picard (1.129), variants (SNPs) were identified using UnifiedGenotyper of GATK (3.4–46) and filtered using VariantFiltration (settings: -Window 4, -filter “QD < 4.0 || FS > 60.0 || MQ < 40.0”, -G filter “GQ < 20”) of GATK. The physical positions of variants were aligned and annotated by ANNOVAR according to the 10 chromosomes constructed by Yu et al.41. The SNP index of the tall bulk and dwarf bulk were calculated using homozygous and biallelic SNPs that differentiate the two parental genomes and were found at depth > 2. ΔSNP index were calculated by subtracting SNP index of dwarf bulk from SNP index of tall bulk, after filtering out SNPs with read depth < 6 and SNP index < 3 in the two bulks. The average SNP index and ΔSNP index were computed using 1 Mb sliding window with 10 kb step size, and the 95% and 99% statistical confidence intervals of ΔSNP index were obtained from 1000 permutation tests of each window under the null hypothesis of no QTLs. Windows with < 10 SNPs were ignored. Nr description, GO term and KEGG pathway were performed to candidate genes.
Association analysis of SNP genotype and plant height phenotype
Five tall (wild varieties) and five dwarf (cultivars) castor bean accessions collected from different areas were examined for their SNP genotypes (see Table S3). They were planted in an experiment field at Kunming Institute of Botany, Chinese Academy of Sciences in Kunming, Yunnan Province (102° 10′ E, 26 22′ N). We surveyed the plant height of each variety after maturity and collected young leaves of each and performed genomic DNA extraction as above. To obtained the genotype information of non-synonymous SNP with ΔSNP index ≥ 0.6 located at five target genes in each variety, normal PCR was carried out for each genomic DNA sample using specific primers (listed in Table S4) binding with the flanking sequence of each SNP, with reaction protocol 95 °C for 2 min, 30 cycles of 95 °C for 30 s, 55 °C for 15 s, and 72 °C for 30 s. Amplicons were purified using a gel extraction kit (OMEGA BIO-TEK, Norcross, USA). The genotypes of SNP were captured via Sanger sequencing using the same primers as the PCR amplification.
Construction of subcellular localization and over-expression vectors
The cDNA (1062 bp, sequence see Supplementary file 1) of Rc5NG4-1Y and Rc5NG4-1C were amplified from Internode 3 of CBt or CBd seedlings, respectively, at the seven internode stage, using primers 5NG4-1-cDNA-F (5′-ATGGCTTCAATATACTGTTACAAG-3′) and 5NG4-1-cDNA-R (5′-TTATGCATCTTCAACTTTTTTGCTT-3′). The cDNA sequences from different castor varieties were recovered from 1% (m/v) agarose gels and purified with a Gel Extraction kit (OMEGA BIO-TEK, Norcross, USA). The cDNA sequences of Rc5NG4-1 from CBt and CBd were cloned into pEASY-T1 Cloning vector (Transgen, Beijing, China) to construct pEASY-T1-5NG4-1Y and pEASY-T1-5NG4-1C.
pSuper1300-GFP (pS1300-GFP) used in subcellular localization research in tobacco and protoplast of Arabidopsis thaliana was a kind gift of Professor Pengtao Wang (State Key Laboratory of Cotton Biology, Henan Key Laboratory of Plant Stress Biology, College of Life Sciences, Henan University). When the pS1300-5NG4-1-GFP was constructed, the coding sequence of Rc5NG4-1 in CBt was amplified from pEASY-T1-5NG4-1Y with primers 5′-CTGCAGGGGCCCGGGGTCGACATGGCTTCAATATACTGTTACAAG-3′ and 5′-CATGGTACCGGATCCACTAGTTGCATCTTCAACTTTTTTGCTT-3′ and cloned into pS1300-GFP vector cut with SalΙ and SpeΙ. The cDNA of Rc5NG4-1Y-GFP and Rc5NG4-1C-GFP (GFP fused in carboxy terminal) were amplified from pS1300-5NG4-1-GFP and pS1300-5NG4-1C-GFP (the methods followed pS1300-5NG4-1-GFP construction, but amplified 5NG4-1 from pEASY-T1-5NG4-1C) with primers 5′-AGTGGATCCCCCGGGCTGCAGATGGCTTCAATATACTGTTACAAG-3′ and 5′-GGGCCCCCCCTCGAGGTCGACTTACTTGTACAGCTCGTCCATG-3′, then were cloned into the pDR196 yeast over-expression vector (Transgen, Beijing, China) and then cut with PstΙ and SalΙ, respectively, to construct the vectors of pDR196-5NG4-1Y-GFP and pDR196-5NG4-1C-GFP. When pDR196-5NG4-1Y and pDR196-5NG4-1C were constructed, the coding sequences of Rc5NG4-1Y and Rc5NG4-1C were amplified from pEASY-T1-5NG4-1Y and pEASY-T1-5NG4-1C, respectively, with primers 5′-AGTGGATCCCCCGGGCGCAGATGGCTTCAATATACTGTTACAAG-3′ and 5′-GGGCCCCCCCTCGAGGTCGACTTATGCATCTTCAACTTTTTTGCTT-3′, and were cloned into pDR196 cut with PstΙ and SalΙ.
The transformation of tobacco leaves, Arabidopsis protoplast and yeast
The pS1300-5NG4-1-GFP vector was transformed into Agrobacterium (GV3101). The transient expression in Nicotiana benthamiana by the Agrobacterium-mediation followed Chatre et al.56. pS1300-5NG4-1-GFP vectors (1 μg/μl) were transiently expressed in Arabidopsis protoplast with the Vac-rb CD3-976 vector57, following Yoo et al.’s method of transient gene expression in mesophyll protoplasts58. The preparation of yeast competent cells (INVSc1 strain), and yeast transformation of pDR196 vectors were carried out using Frozen-ER Yeast Transformation ΙΙ kit (ZYMO RESEARCH, Beijing, China) following the manufacturers protocol. The fluorescence of transformed tobacco leaves, protoplasts and yeast were examined using a confocal microscope (Fluoview fv1000, OLYMPUS).
RNA extraction and RT-PCR in yeast cell
RNA extraction from yeast cells followed the manufacturer’s protocol of Yeast RNA kit (OMEGA BIO-TEK, Norcross, USA) using 4 mL yeast cells, OD600 = 1.0. Genomic DNA removal and cDNA synthesis used the One-Step gDNA Removal and cDNA synthesis SuperMix (Transgen, Beijing, China). The semi-quantitative reverse transcription PCR (RT-PCR) was used to detect the expressional level of 5NG4-1Y and 5NG4-1C in yeast cell. The protocol of RT-PCR was 95 °C for 2 min, 30 cycles of 95 °C for 30 s, 60 °C for 15 s, and 72 °C for 10 s. ScACTIN was treated as the reference gene25. The primer sequences of RT-PCR are listed in Table S2.
IAA testing after yeast uptake and IAA concentration determination in castor bean
Before the yeast uptake experiments, the Rc5NG4-1Y over-expression strain, Rc5NG4-1C over-expression strain and pDR196 transformed yeast strain were cultivated in SD/-Ura liquid medium (0.67% nitrogen, 0.077% SD/-Ura, 2% glucose, PH = 5.8) to OD600 = 1. Each sample of the three yeast strains consisted of 10 mL yeast cell (OD600 = 1). The yeast cells in each sample were collected by centrifugation (6,000 g, 5 min), washed with 10 mL sodium citrate buffer (10 mM, pH 4.5) and resuspended in 10 mL transport buffer (10 mM sodium citrate buffer, 2 mM glucose, 20 mM ammonium sulphate, 20 μM 2H-IAA, pH 4.5)25. 2H-IAA (Mr = 177.19) was gifted by Professor Shihong Luo (College of Bioscience and Biotechnology, Shenyang Agricultural University). At four time points (5, 15, 25 and 60 min after sample were resuspended in 10 mL transport buffer), the yeast cells in each sample were collected and washed as above, then were re-collected (6,000 g, 10 min) and the supernatant removed gently. The cell walls of these yeast cells were dissolved in Lysing Enzymes (75 μL yeast lytic enzyme, 15 μL β-mercaptoethanol, 1410 μL sorbitol buffer) (Solarbio, Beijing, China) and placed at 30 °C for 2 h, then stored in -80 °C until determination of 2H-IAA concentration. Three independent replicates were used in the uptake test.
The apical buds and Internode 3 were sliced from seedlings of CBt and CBd (with seven internodes) at the same maturity stage using a razor. Seedlings were grown in greenhouse as above. Three independent samples of each tissue of CBt and CBd were collected and immersed in liquid nitrogen. The sample was ground into a powder using pestle and mortar. 100 mg of powder from each sample were placed in cryogenic vials.
The 2H-IAA uptake by yeast cell and free IAA concentrations in apical buds and Internode 3 were detected and quantified with ultra-high performance liquid chromatography (Shimadzu, LC-20AD, Japan) and QQQ-triple quadrupole mass spectrometer (Shimadzu, LCMS-8040, Japan) (LC–MS) using a positive electrospray ionization (ESI) detection system. 5 ng N5-IAA was used as the internal standard and 1 ml ethyl acetate was added to each sample. After vortex for 10 min, the sample was centrifuged (13,000 g, 10 min) and the supernatants transferred to new 2 mL polypropylene tubes. Residual pellets were extracted again but with 0.5 mL of ethyl acetate and the supernatants were combined. Next, the supernatant was evaporated on a vacuum concentrator (Eppendorf) until dry, 0.5 mL of 70% methanol was added to resuspended the residue, samples were centrifuged and the supernatants transferred to glass vials, for analysis by LC–MS using multiple reaction monitoring (MRM) scan with a windows of 50 s and a target scan time of 3 s. The 2H-IAA and free IAA concentrations were quantified as 5 ng (the weight of internal standards) multiplied the ratio of their peak area to the peak area of N5-IAA.
Informed consent for publication
Informed consent for publication is obtained from the participant for the identifiable image in Fig. 1A. The authors confirm that the Scientific Reports and Springer Nature Press have unrestricted copyright to publish or re-publish the figure (with the photographic portrait) for scientific and educational purposes.
Supplementary Information
Acknowledgements
This work was jointly fund by National Natural Science Foundation of China [31661143002, 31771839, 31701123 and 31501034], Yunnan Applied Basic Research Projects [2016FB060 and 2016FB040] and the Youth Innovation Promotion Association of CAS (2020389 to W.X.). We thank Prof. Pengtao Wang from Henan University for providing the pSuper 1300 and Vac-rb CD3-976 vectors, and Prof. Shihong Luo from Shenyang Agricultural University for providing 2H-IAA. We also thank Dr. Mark Chapman from University of Southampton for language editing.
Author contributions
Z.W. and A.L. conceived and designed the experiments; Z.W., A.Y., F.L. and X.C. constructed the hybrid populations of castor bean; Z.W., A.Y, W.X and B.H. performed the experiments and data analyses; Z.W. and A.L. wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-85644-1.
References
- 1.Khush GS. Green revolution: the way forward. Nat. Rev. Genet. 2001;2:815–822. doi: 10.1038/35093585. [DOI] [PubMed] [Google Scholar]
- 2.Avila LM, Cerrudo D, Swanton C, Lukens L. Brevis plant1, a putative inositol polyphosphate 5-phosphatase, is required for internode elongation in maize. J. Exp. Bot. 2016;67:1577–1588. doi: 10.1093/jxb/erv554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma Y, et al. Involvement of auxin and brassinosteroid in dwarfism of autotetraploid apple (Malus x domestica) Sci. Rep. 2016;6:26719. doi: 10.1038/srep26719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu W, et al. Morphological, physiological and proteomic analyses provide insights into the improvement of castor bean productivity of a dwarf variety in comparing with a high-stalk variety. Front. Plant Sci. 2016;7:1473. doi: 10.3389/fpls.2016.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazur E, Kurczynska EU, Friml J. Cellular events during interfascicular cambium ontogenesis in inflorescence stems of Arabidopsis. Protoplasma. 2014;251:1125–1139. doi: 10.1007/s00709-014-0620-5. [DOI] [PubMed] [Google Scholar]
- 6.Fendrych M, Leung J, Friml J. TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife. 2016 doi: 10.7554/eLife.19048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stepanova AN, et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 8.Zhao B, et al. Identification and characterization of a new dwarf locus DS-4 encoding an Aux/IAA7 protein in Brassica napus. Theor. Appl. Genet. 2019;132:1435–1449. doi: 10.1007/s00122-019-03290-8. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, et al. Enhancing auxin accumulation in maize root tips improves root growth and dwarfs plant height. Plant Biotechnol. J. 2018;16:86–99. doi: 10.1111/pbi.12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kebrom TH. A growing stem inhibits bud outgrowth—the overlooked theory of apical dominance. Front. Plant Sci. 2017;8:1874. doi: 10.3389/fpls.2017.01874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbier FF, Dun EA, Beveridge CA. Apical dominance. Curr. Biol. 2017;27:R864–R865. doi: 10.1016/j.cub.2017.05.024. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Y, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- 13.Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray WM, Muskett PR, Chuang HW, Parker JE. Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell. 2003;15:1310–1319. doi: 10.1105/tpc.010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Yang Z. Novel ABP1-TMK auxin sensing system controls ROP GTPase-mediated interdigitated cell expansion in Arabidopsis. Small GTPases. 2014 doi: 10.4161/sgtp.29711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giri J, et al. Rice auxin influx carrier OsAUX1 facilitates root hair elongation in response to low external phosphate. Nat. Commun. 2018;9:1408. doi: 10.1038/s41467-018-03850-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noh B, Murphy AS, Spalding EP. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell. 2001;13:2441–2454. doi: 10.1105/tpc.010350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones SE, Demeo JS, Davies NW, Noonan SE, Ross JJ. Stems of the Arabidopsis pin1-1 mutant are not deficient in free indole-3-acetic acid. Planta. 2005;222:530–534. doi: 10.1007/s00425-005-1561-1. [DOI] [PubMed] [Google Scholar]
- 20.Inahashi H, et al. OsPIN2, which encodes a member of the auxin efflux carrier proteins, is involved in root elongation growth and lateral root formation patterns via the regulation of auxin distribution in rice. Physiol. Plant. 2018;164:216–225. doi: 10.1111/ppl.12707. [DOI] [PubMed] [Google Scholar]
- 21.Keuskamp DH, Pollmann S, Voesenek LA, Peeters AJ, Pierik R. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc. Natl. Acad. Sci. U.S.A. 2010;107:22740–22744. doi: 10.1073/pnas.1013457108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haga K, Sakai T. Differential roles of auxin efflux carrier PIN proteins in hypocotyl phototropism of etiolated Arabidopsis seedlings depend on the direction of light stimulus. Plant Signal. Behav. 2013;8:e22556. doi: 10.4161/psb.22556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon S, et al. PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol. 2016;211:65–74. doi: 10.1111/nph.14019. [DOI] [PubMed] [Google Scholar]
- 24.Mravec J, et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 2009;459:1136–1140. doi: 10.1038/nature08066. [DOI] [PubMed] [Google Scholar]
- 25.Ranocha P, et al. Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 2013;4:2625. doi: 10.1038/ncomms3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbez E, et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature. 2012;485:119–122. doi: 10.1038/nature11001. [DOI] [PubMed] [Google Scholar]
- 27.Ranocha P, et al. Walls are thin1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 2010;63:469–483. doi: 10.1111/j.1365-313X.2010.04256.x. [DOI] [PubMed] [Google Scholar]
- 28.Feraru E, Vosolsobě S, Feraru MI, Petrášek J, Kleine-Vehn J. Evolution and structural diversification of PILS putative auxin carriers in plants. Front. Plant Sci. 2012;3:227. doi: 10.3389/fpls.2012.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellerowicz EJ, Baucher M, Sundberg B, Boerjan W. Unravelling cell wall formation in the woody dicot stem. Plant Mol. Biol. 2001;47:239–274. doi: 10.1023/A:1010699919325. [DOI] [PubMed] [Google Scholar]
- 30.Shinya T, et al. Transcriptional profiles of hybrid eucalyptus genotypes with contrasting lignin content reveal that monolignol biosynthesis-related genes regulate wood composition. Front. Plant Sci. 2016;7:443. doi: 10.3389/fpls.2016.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xi W, Song D, Sun J, Shen J, Li L. Formation of wood secondary cell wall may involve two type cellulose synthase complexes in Populus. Plant Mol. Biol. 2017;93:419–429. doi: 10.1007/s11103-016-0570-8. [DOI] [PubMed] [Google Scholar]
- 32.Gui J, et al. Phosphorylation of LTF1, an MYB transcription factor in Populus, acts as a sensory switch regulating lignin biosynthesis in wood cells. Mol. Plant. 2019;12:1325–1337. doi: 10.1016/j.molp.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Biswal AK, et al. Downregulation of GAUT12 in Populus deltoides by RNA silencing results in reduced recalcitrance, increased growth and reduced xylan and pectin in a woody biofuel feedstock. Biotechnol. Biofuels. 2015;8:41. doi: 10.1186/s13068-015-0218-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chauke NP, Mukaya HE, Nkazi DB. Chemical modifications of castor oil: a review. Sci. Prog. 2019;102:199–217. doi: 10.1177/0036850419859118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng Q, Dong G, Martin JM. Green superlubricity of Nitinol 60 alloy against steel in presence of castor oil. Sci. Rep. 2016;6:29992. doi: 10.1038/srep29992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogunniyi DS. Castor oil: a vital industrial raw material. Biores. Technol. 2006;97:1086–1091. doi: 10.1016/j.biortech.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 37.Oswalt JS, et al. Plant height and seed yield of castor (Ricinus communis L.) sprayed with growth retardants and harvest aid chemicals. Ind. Crops Prod. 2014;61:272–277. doi: 10.1016/j.indcrop.2014.07.006. [DOI] [Google Scholar]
- 38.Xu W, et al. Genomic analysis reveals rich genetic variation and potential targets of selection during domestication of castor bean from perennial woody tree to annual semi-woody crop. Plant Direct. 2019;3:e00173. doi: 10.1002/pld3.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buckler ES, et al. The genetic architecture of maize flowering time. Science. 2009;325:714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- 40.Hu J, et al. Genetic properties of a nested association mapping population constructed with semi-winter and spring oilseed rapes. Front. Plant Sci. 2018;9:1740. doi: 10.3389/fpls.2018.01740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu A, et al. Application of a high-resolution genetic map for chromosome-scale genome assembly and fine QTLs mapping of seed size and weight traits in castor bean. Sci. Rep. 2019;9:11950. doi: 10.1038/s41598-019-48492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takagi H, et al. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013;74:174–183. doi: 10.1111/tpj.12105. [DOI] [PubMed] [Google Scholar]
- 43.Haase NJ, et al. Shared genomic regions between derivatives of a large segregating population of maize identified using bulked segregant analysis sequencing and traditional linkage analysis. G3. 2015;5:1593–1602. doi: 10.1534/g3.115.017665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue H, et al. Interval mapping for red/green skin color in Asian pears using a modified QTL-seq method. Hortic. Res. 2017;4:17053. doi: 10.1038/hortres.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu X, et al. The major-effect quantitative trait locus CsARN6.1 encodes an AAA ATPase domain-containing protein that is associated with waterlogging stress tolerance by promoting adventitious root formation. Plant J. 2018;93:917–930. doi: 10.1111/tpj.13819. [DOI] [PubMed] [Google Scholar]
- 46.Wang C, et al. Dissecting a heterotic gene through GradedPool-Seq mapping informs a rice-improvement strategy. Nat. Commun. 2019;10:2982. doi: 10.1038/s41467-019-11017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Busov VB, et al. An auxin-inducible gene from loblolly pine (Pinus taeda L.) is differentially expressed in mature and juvenile-phase shoots and encodes a putative transmembrane protein. Planta. 2004;218:916–927. doi: 10.1007/s00425-003-1175-4. [DOI] [PubMed] [Google Scholar]
- 48.Habets ME, Offringa R. PIN-driven polar auxin transport in plant developmental plasticity: a key target for environmental and endogenous signals. New Phytol. 2014;203:362–377. doi: 10.1111/nph.12831. [DOI] [PubMed] [Google Scholar]
- 49.Zhao Y, Song D, Sun J, Li L. Populus endo-beta-mannanase PtrMAN6 plays a role in coordinating cell wall remodeling with suppression of secondary wall thickening through generation of oligosaccharide signals. Plant J. 2013;74:473–485. doi: 10.1111/tpj.12137. [DOI] [PubMed] [Google Scholar]
- 50.Robert EM, et al. Successive cambia: a developmental oddity or an adaptive structure? PLoS ONE. 2011;6:e16558. doi: 10.1371/journal.pone.0016558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Zhao J, Lu W, Deng D. Gibberellin in plant height control: old player, new story. Plant Cell Rep. 2017;36:391–398. doi: 10.1007/s00299-017-2104-5. [DOI] [PubMed] [Google Scholar]
- 52.Liao Z, et al. SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat. Commun. 2019;10:2738. doi: 10.1038/s41467-019-10667-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nilsson J, et al. Dissecting the molecular basis of the regulation of wood formation by auxin in hybrid aspen. Plant Cell. 2008;20:843–855. doi: 10.1105/tpc.107.055798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lucas WJ, et al. The plant vascular system: evolution, development and functions. J. Integr. Plant Biol. 2013;55:294–388. doi: 10.1111/jipb.12041. [DOI] [PubMed] [Google Scholar]
- 55.Liu C, Yu H, Li L. SUMO modification of LBD30 by SIZ1 regulates secondary cell wall formation in Arabidopsis thaliana. PLoS Genet. 2019;15:e1007928. doi: 10.1371/journal.pgen.1007928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chatre L, Brandizzi F, Hocquellet A, Hawes C, Moreau P. Sec22 and Memb11 are v-SNAREs of the anterograde endoplasmic reticulum-Golgi pathway in tobacco leaf epidermal cells. Plant Physiol. 2005;139:1244–1254. doi: 10.1104/pp.105.067447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 58.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.