Abstract
Objective
High-throughput electronic phenotyping algorithms can accelerate translational research using data from electronic health record (EHR) systems. The temporal information buried in EHRs is often underutilized in developing computational phenotypic definitions. This study aims to develop a high-throughput phenotyping method, leveraging temporal sequential patterns from EHRs.
Materials and Methods
We develop a representation mining algorithm to extract 5 classes of representations from EHR diagnosis and medication records: the aggregated vector of the records (aggregated vector representation), the standard sequential patterns (sequential pattern mining), the transitive sequential patterns (transitive sequential pattern mining), and 2 hybrid classes. Using EHR data on 10 phenotypes from the Mass General Brigham Biobank, we train and validate phenotyping algorithms.
Results
Phenotyping with temporal sequences resulted in a superior classification performance across all 10 phenotypes compared with the standard representations in electronic phenotyping. The high-throughput algorithm’s classification performance was superior or similar to the performance of previously published electronic phenotyping algorithms. We characterize and evaluate the top transitive sequences of diagnosis records paired with the records of risk factors, symptoms, complications, medications, or vaccinations.
Discussion
The proposed high-throughput phenotyping approach enables seamless discovery of sequential record combinations that may be difficult to assume from raw EHR data. Transitive sequences offer more accurate characterization of the phenotype, compared with its individual components, and reflect the actual lived experiences of the patients with that particular disease.
Conclusion
Sequential data representations provide a precise mechanism for incorporating raw EHR records into downstream machine learning. Our approach starts with user interpretability and works backward to the technology.
Keywords: phenotyping, electronic health records, sequential pattern mining, temporal data representation
INTRODUCTION
Biomedical researchers are progressively applying modern machine learning (ML) algorithms to data from electronic health records (EHRs). Despite the natural excitement about the large amount of information presented by EHRs, daunting challenges remain. As the primary impetus for EHR implementation has been clinical care, EHR observations reflect a complex set of processes that further obscure their utility in research. Dimensionality, sparsity, heterogeneity, and quality issues present significant impediments for secondary use of EHR data.1,2 In particular, the EHR observation records are often not direct indicators of a patient’s true health state, but rather reflect the clinical processes (eg, policies and workflows of the provider and payor organizations), the patient’s interaction with the system, and the recording process.3–5
As a result of the biases inherent in EHR data, identifying cohorts of patients with certain health conditions can become complex. In order to make precise assumptions about the presence of a disease, we would need to perform phenotyping. The key task in phenotyping is to identify patient cohorts with (or without) a certain phenotype or clinical condition of interest.6,7 Developing specialized phenotypic definitions from EHR data can be expensive and often requires involvement by domain experts.8–11 Nevertheless, owing to its critical role in reusing EHR data from research, several healthcare institutions are actively involved in constructing and validating electronic phenotyping algorithms. Efforts to curate computational phenotypes and discover clinical knowledge from EHR observations must account for the potential biases introduced through the recording process.4
Another underutilized aspect of electronic medical records is their temporal dimension. EHRs contain important temporal information about disease progression and treatment outcomes. However, EHR observations are often acquired asynchronously across time (ie, measured at different time instants and sampled irregularly in time) and include sparse and heterogeneous data.12–17 These properties challenge the application of standard temporal analysis methods to clinical data recorded in EHRs.
The record of the EHR diagnosis and its timestamp may not give the true disease state or the actual onset of the disease. In this paper, we utilize a novel sequential pattern mining (SPM) algorithm to construct temporal data representations from EHR data. Using a high-throughput feature selection algorithm, we then utilize the temporal sequential representations to develop computational phenotyping algorithms for 10 phenotypes. We demonstrate that the temporal sequential features significantly outperform raw EHR features that are commonly used in computational phenotyping algorithms. Unlike most deep learning approaches that have been developed to improve prediction, our approach starts with user interpretability and works backward to the technology.
BACKGROUND
Although EHRs often include incomplete, inaccurate, or even biased data, the wealth of information that they provide is sufficient for constructing clinically relevant sets of observable characteristics that define a disease or phenotype.4,18 The task in electronic phenotyping is to determine patients with (or without) certain phenotypic characteristics based on data from electronic medical records,7 which is challenging due to the heterogeneity and complexity of multimodal EHR data.6 As a result, developing specialized phenotypic definitions from EHR data is generally expensive.8,9
Approaches to electronic phenotyping include rule-based methods and computational methods, which can be broadly characterized under text processing and supervised, semi-supervised, and unsupervised statistical learning techniques.6,19 Applying supervised and semi-supervised machine learning to EHR data for identifying cohorts with clinical phenotypes is rapidly prevailing.11,19–37 However, such phenotyping models require a human-annotated gold-standard training set, which remains a bottleneck.20 In addition, defining clinically meaningful EHR features for computational phenotyping relies on a heavy dose of domain expert involvement, using complex ad hoc procedures that are often hard to generalize and scale.8–11 Despite the cost, several academic medical centers are actively involved in constructing and validating EHR phenotyping algorithms. Some of the notable efforts include the i2b2-centered efforts led by Harvard University and Mass General Brigham,21–29 the BioVU led by Vanderbilt University,30,31 and the multicenter eMERGE (Electronic Medical Records and Genomics) Network consortium,32–35 the PheKB (Phenotype Knowledgebase) website,36 and the Observational Medical Outcomes Partnership–centered APHRODITE (Automated PHenotype Routine for Observational Definition, Identification, Training and Evaluation),38 to name a few.
For developing computational phenotyping algorithms with EHR data, features are typically constructed by identifying relevant clinical events (eg, diagnoses or medication records from structured data or certain keywords from the clinical notes). A vector of these records is obtained from patient-level marginal counts and cohort-level aggregation. This approach is rudimentary and misses potentially useful information that is available in the electronic medical records. Of note, EHRs provide a wealth of longitudinal information that can be leveraged to improve computational phenotyping algorithms.18 Temporal representation mining methods offer technical solutions that can account for this aspect of the EHR.
Temporal representation mining involves providing a machine-readable representation that formalizes the concept of time as it is relates to a set of events and temporal relationships.39 In biomedical research, development and evolution of temporal representation mining approaches has been largely focused on the temporal abstraction40 of data from continuous clinical measurements.41 Yet, discrete clinical data, such as diagnoses, medications, and procedures, are poor candidates for the temporal abstraction approach. While numerical observations, such as laboratory test results, have explicit timestamps, precise numeric timestamped information is often unavailable in discrete clinical data.
SPM42 is a viable alternative approach for discrete data. The goal in SPM is to discover “relevant” subsequences from a large set of sequences (of events or items) with time constraints. The relevance is often determined by a user-specified occurrence frequency, known as the minimum support.43 The frequent sequential pattern problem is to find the frequent sequences among all sequences.44 A priori–based SPM methods, such as SPADE (sequential pattern discovery using equivalence classes)45 and SPAM (sequential pattern mining),46 are popular in the healthcare domain. For example, Perer et al (2015) used the SPAM algorithm for mining long sequences of events.25 The a priori property is that if a sequence cannot pass the minimum support test (ie, is not assumed frequent), all of its subsequences will be ignored. However, for various reasons, temporal patterns that are mined based on frequency may not make clinical sense. For example, low blood pressure readings after the administration of a specific, but irrelevant, medication may be frequently observed yet have no real clinical meaning.47
Recently, we introduced the transitive SPM (tSPM) algorithm along with an early implementation of the minimize sparsity and maximize relevance (MSMR) algorithm.48 The tSPM algorithm provides a modified sequencing procedure to address some of the issues caused by the recording processes in EHRs. The MSMR algorithm offers a high-throughput feature engineering technique to improve the frequency-based a priori property in the traditional SPM approach. As a proof of concept, we showed that the sequencing approach improves disease prediction and classification in a single disease. In this study, we apply the tSPM algorithm—with an improved MSMR algorithm—to computational phenotyping in EHR data. We perform a comprehensive comparison of the transitive and traditional sequential representations with the conventional way of using EHR observations as features for computational phenotyping in 10 phenotypes. We also compare the phenotyping performance with the state-of-the-art phenotyping algorithms published in research informatics literature.
MATERIALS AND METHODS
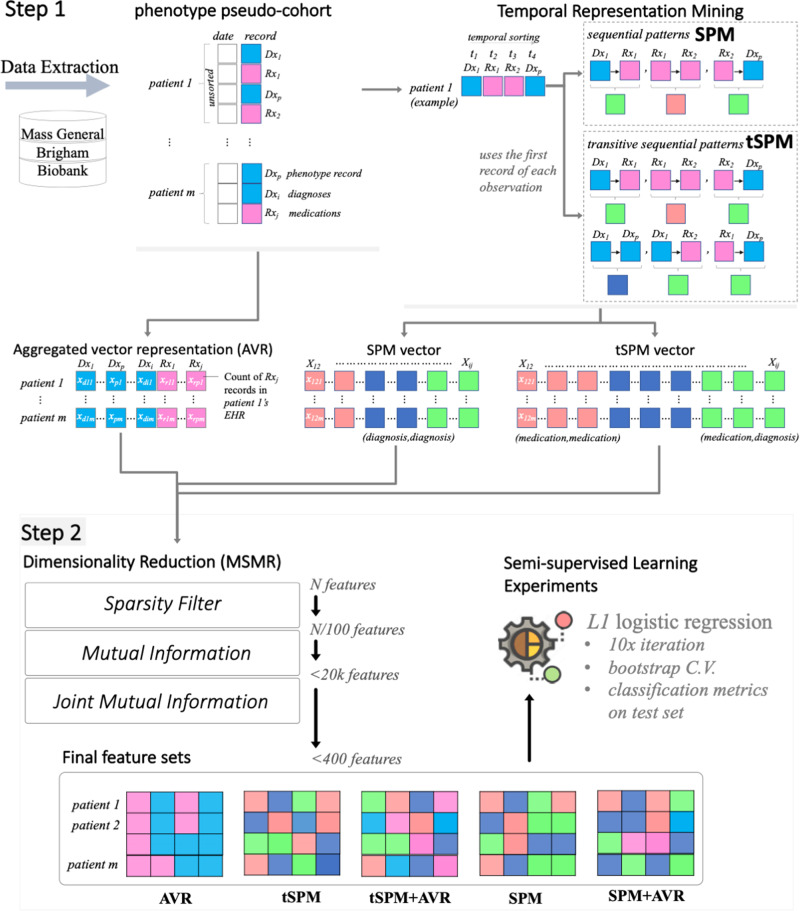
We aimed to answer a principle question: can temporal representations mined through SPM improve computational phenotyping with EHR data? Developing the computational phenotyping algorithms with temporal sequences follows 2 steps: (1) a phenotype-agnostic representation mining and (2) a semi-supervised phenotype-specific dimensionality reduction, the MSMR algorithm (Figure 1). We use terms feature and data representation interchangeably.
Figure 1.
The 2 steps involved in the high-throughput phenotyping with temporal sequences. AVR: aggregated vector representation; C.V.: cross validation; MSMR: minimize sparsity and maximize relevance; SPM: sequential pattern mining; tSPM: transitive sequential pattern mining.
Representation mining
We only used the medication and diagnosis records data. For the diagnosis records, we used the International Classification of Diseases–Ninth and Tenth Revisions–Clinical Modification. For medications, we use RxNorm codes. Given a list of diagnosis or medication records, we mined 3 vectors of data representations:
First, we constructed a baseline representation that applies the conventional approach for using EHRs as features for computational phenotyping. We henceforth call this the aggregated vector representation (AVR) approach.
Second, we mined a set of temporal sequential representations by sequencing the medication and diagnosis records in electronic medical records. For the temporal sequencing, we utilized the traditional SPM schema, in which immediate sequences are mined.
Third, and to account for irregularity of clinical records and the recording processes, we mined the novel tSPM schema,48 in which sequences of unlimited lengths are possible.
AVR representations
In the AVR approach, which is the conventional approach for using EHR data as records for computational phenotyping, the marginal count of a vector of a selected medical record (often diagnosis codes) is calculated for each patient. The patient , is represented by a vector of the length equal to the number of unique events in their medical records. The initial set of AVR representations are all possible records, and for each patient, we record only the numbers of each record. For each and patient , we think of the ’s as samples of a random variable . Our goal is then to predict the class label , given .
For example, when type 2 diabetes mellitus (T2DM) is a feature in the AVR approach, the number of times the diagnosis record for T2DM is recorded in a patient’s electronic record is used as the classifier for training and testing.
SPM representations
In the traditional SPM approach, for each patient , we recorded the times at which the record was logged. The SPM features are all possible pairs of distinct records . To count the frequency of SPM representations, for a given patient and a given time , we let be minimal such that for some and some , . That is, is the first time strictly bigger than at which a record is logged for the patient (it is possible to have multiple records at the same timestamp). For patient, and a given index , let be the set of all pairs , with and , such that the record is logged right after record at time . Formally, if and only if and there exists such that
For each , let . We think of the s as samples of a random variable and the goal is to predict the class label given .
tSPM representations
In the tSPM algorithm, the features are again all possible pairs of distinct medical records . For a fixed patient , and, we set to be 1 if , and 0 otherwise. In words, is 1 if and only if both records and were logged for the patient, and the first record of medical record was before, or at the same time as, the first record of medical record . Here, for each fixed , we think of the s as samples of a random variable . Then our goal is to predict the class label given
The use of the first record (rather than all records) is a specification difference in the way the sequential patterns are organized in tSPM compared with SPM. This difference helps to address the issue of repeated problem list entries.48
Dimensionality reduction
If all pairs of sequences in the transitive sequencing approach exist, there will be exactly pairs with . To extract features for phenotyping, we applied a formal dimensionality reduction procedure that aims to minimize sparsity and maximize relevance (MSMR)48 to all 3 feature vectors. To minimize sparsity, MSMR removes any feature that has a prevalence smaller than 0.5%. For maximizing relevance, MSMR is principally a semi-supervised dimensionality reduction algorithm that takes a silver-standard class label to compute information gain metrics for all features. MSMR is able to effectively scale to large dimensionality spaces, and thus is a high-throughput algorithm.
For the remaining features, MSMR computes the empirical mutual information using an estimation of the entropy of the empirical probability distribution.49,50 Mutual information provides a measurement of the mutual dependence between 2 random variables, which unlike most correlation measures can capture nonlinear relationships.50,51 We ranked the data representations based on the computed mutual information with the silver-standard labeled outcome (in ties, we used prevalence to determine the ranking) and conventionally select the top 20 000 data representations from each approach.
We further dissected the relevance property by applying a filter-type feature extraction method using joint mutual information (JMI).52 The algorithm starts with a set containing the top feature according to mutual information, then iteratively adds to the feature maximizing the JMI score:
Here, denotes the mutual information between random variables and (a measure of the information shared by and —it can be expressed as the entropy of minus the entropy of given ). The random variable is simply the random variable corresponding to the joint distribution of and . In the end, we select the top features that were added to the set . The joint mutual information score also takes into account the redundancy between the features: 2 features not only could each be highly relevant on their own, but also could be strongly correlated. Brown et al53 suggested that the JMI score provides the “best trade-off […] of accuracy and stability.”
The JMI step allows for integration of representations from different types. To obtain combined feature sets (AVR + sequential patterns), we also compute the joint mutual information when the AVR and SPM/tSPM representations are available.
We used high-performance computing resources environment at Mass General Brigham for representation mining (R code available at github.com/hestiri/TSPM) and MSMR (R package available at github.com/hestiri/MSMR) algorithms, both of which are implemented to leverage parallel computing.
Study populations
We used data on 10 phenotypes from the Mass General Brigham Biobank: Alzheimer’s disease, chronic obstructive pulmonary disease (COPD), congestive heart failure, coronary artery disease (CAD), stroke, rheumatoid arthritis, T1DM, T2DM, ulcerative colitis, and atrial fibrillation. For each phenotype, 2 pseudo-cohorts are available. Based on a list of International Classification of Diseases–Ninth and Tenth Revisions codes for the respective phenotype, a given patient in the pseudo-cohort has at least 1 record of the respective diagnosis code(s), as well as an outcome label, which determines the “true” presence of the phenotype. We use the term pseudo-cohort to distinguish these patient cohorts from validated disease cohorts.
For each of the phenotypes, a small dataset included patient pseudo-cohorts with gold-standard outcome labels. To create gold-standard labels, teams of board-certified clinicians or nurses reviewed clinical notes and other required data of samples of patients selected from patients consented into the Mass General Brigham Biobank between April 2012 and April 2017 and with 1 or more diagnosis record of the phenotype. The gold-standard phenotype pseudo-cohort datasets include labels for an average of 351 patients (ranging from 163 to 700 patients). For the 10 phenotypes, larger pseudo-cohorts were also pulled from the Biobank, with an average population of over 6000 patients. For these patients, silver-standard labels are curated using a generative transfer learning algorithm.54 The average number of phenotype records was 28 in the gold-standard datasets and 20 in the silver-standard datasets. Supplementary Table S1 provides descriptive data on each of the phenotype pseudo-cohorts. The use of data for this study was approved by the Mass General Brigham Institutional Review Board (2017P000282).
Model training and evaluation
We applied logistic regression classifiers with L1 regularization to the training sets (with silver-standard labels) for developing the computational phenotyping algorithms, using bootstrap cross-validation. Regularized logistic regression classifiers are the most popular classifiers in EHR phenotyping.8,11,38,55–57 From each data representation class (AVR, SPM, and tSPM), a nested set of the top 50-200 representations are extracted from the MSMR algorithm for phenotyping. In addition, using the MSMR algorithm, we extracted hybrid feature sets that included both the AVR and sequential representations. This resulted in 2 additional feature sets combining the top AVR representations with the SPM and tSPM representations. Overall, we trained computational phenotyping algorithms on 5 classes of feature sets: (1) AVR, (2) SPM, (3) tSPM, (4) AVR+SPM, and (5) AVR+tSPM.
We evaluated the phenotyping algorithms against the gold-standard labels available in the held-out test sets to compute the areas under the receiver-operating characteristic curve. Furthermore, we iterated the training process 10 times with bootstrap sampling and use the median performance metrics for comparing the feature sets. Overall, for each phenotype, we trained 50 classifiers (5 feature sets × 10 bootstrap cross-validation iterations). All features are scaled and centered. Finally, we evaluated the clinical meaning of the top transitive sequences used in the phenotyping algorithms.
RESULTS
As expected, we mined millions of tSPM sequences. Table 1 presents the number of unique representations by type before and after MSMR’s sparsity reduction. On average, we used over 7000 unique medication and diagnosis codes, from which we mined, on average, over 322 000 SPM and about 6 000 000 tSPM sequential representations. Removing sparse representations (prevalence smaller than 0.5%) resulted in on average over 4000, 28 000, and 1 000 000 unique AVR, SPM, and tSPM features, respectively. Using the mutual information and the JMI filters, the MSMR algorithm further shrunk these features to a final vector of between 50 and 200 features for each phenotype.
Table 1.
The number of unique representations by type before and after MSMR’s sparsity reduction
| Phenotype | Mined unique representations |
Representations after sparsity screening |
||||
|---|---|---|---|---|---|---|
| AVR | SPM | tSPM | AVR | SPM | tSPM | |
| AD | 6193 | 209 389 | 3 844 039 | 1661 | 28 672 | 1 211 853 |
| AFIB | 9050 | 172 134 | 3 190 557 | 5460 | 23 957 | 795 316 |
| CAD | 14 406 | 476 617 | 9 142 990 | 5599 | 23 480 | 810 862 |
| CHF | 6857 | 391 679 | 7 419 996 | 6415 | 39 265 | 1 349 704 |
| COPD | 6284 | 369 062 | 6 527 854 | 5412 | 37 721 | 1 556 750 |
| RA | 5203 | 305 787 | 5 782 618 | 4517 | 19 157 | 790 214 |
| Stroke | 5573 | 281 298 | 4 692 549 | 3395 | 24 186 | 839 268 |
| T1DM | 6673 | 385 626 | 7 086 684 | 3334 | 46 260 | 1 538 308 |
| T2DM | 10 439 | 429 115 | 8 376 105 | 5887 | 26 331 | 920 251 |
| UC | 4904 | 207 256 | 3 920 849 | 1893 | 14 651 | 603 902 |
AD: Alzheimer’s disease; AFIB: atrial fibrillation; AVR: aggregated vector representation; CAD: coronary artery disease; CHF: congestive heart failure; COPD: chronic obstructive pulmonary disease; MSMR: minimize sparsity and maximize relevance; RA: rheumatoid arthritis; SPM: sequential pattern mining; T1DM: type 1 diabetes mellitus; T2DM: type 2 diabetes mellitus; tSPM: transitive sequential pattern mining; UC: ulcerative colitis.
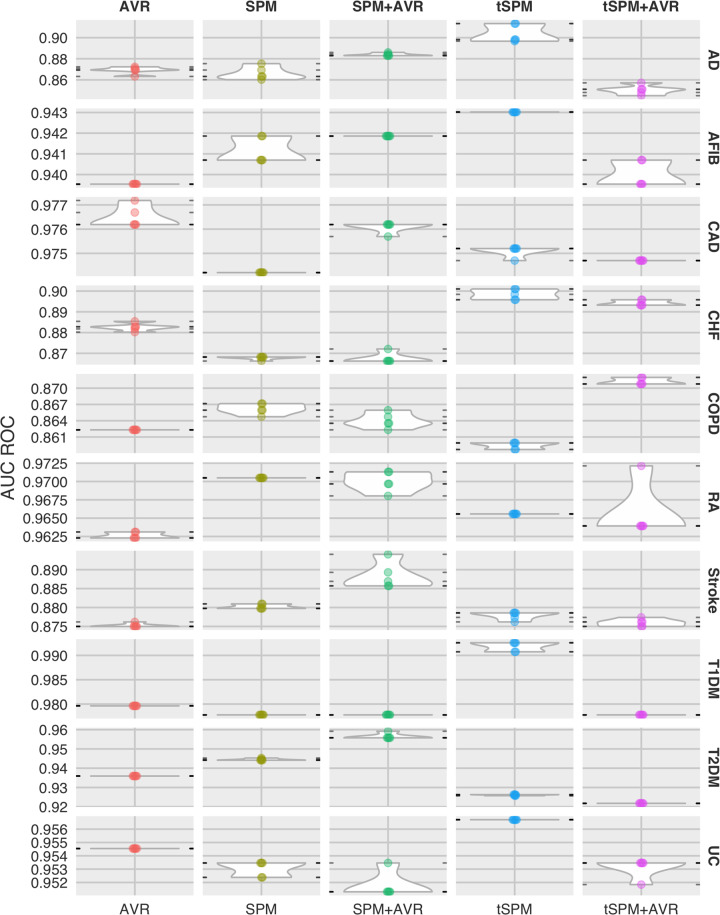
To address the research question, phenotyping results are presented in Table 2 (also illustrated in Figure 2). Overall, we found that temporal sequences provided the best phenotyping performances across all 10 phenotypes (except in CAD, in which we had a tie). Combining sequences with AVR features only resulted in the best overall performance in COPD and rheumatoid arthritis. Among the sequential representations, in an overwhelming majority of the phenotypes, transitive sequences were included in the best results. The 2 exception to this were in CAD (in which the difference was 0.001) and T2DM. For 6 of the 10 phenotypes, we were able to find areas under the receiver-operating characteristic curve reported from other published phenotyping studies. In 5 of the 6 phenotypes, the performances we obtained from temporal sequences was substantially superior.
Table 2.
Area under the receiver-operating characteristic curve
| Data representation | Phenotype | Median AUC | Top AUC | Literature | Phenotype | Median AUC | Top AUC | Literature |
|---|---|---|---|---|---|---|---|---|
| AVR | AD | 0.869 | 0.872 | NA | RA | 0.962 | 0.963 | 0.933-0.96111,58,59 |
| SPM | 0.863 | 0.875 | 0.970a | 0.970 | ||||
| SPM+AVR | 0.883 | 0.886 | 0.970a | 0.971 | ||||
| tSPM | 0.898a | 0.913a | 0.966 | 0.966 | ||||
| tSPM+AVR | 0.851 | 0.857 | 0.964 | 0.972a | ||||
| AVR | AFIB | 0.940 | 0.940 | NA | Stroke | 0.875 | 0.876 | NA |
| SPM | 0.941 | 0.942 | 0.880 | 0.881 | ||||
| SPM+AVR | 0.942 | 0.942 | 0.887 | 0.894a | ||||
| tSPM | 0.943a | 0.943a | 0.879a | 0.879 | ||||
| tSPM+AVR | 0.940 | 0.941 | 0.876 | 0.877 | ||||
| AVR | CAD | 0.976a | 0.976a | 0.896-0.9311,58,59 | T1DM | 0.980 | 0.980 | 0.98159 |
| SPM | 0.974 | 0.974 | 0.978 | 0.978 | ||||
| SPM+AVR | 0.976a | 0.976a | 0.978 | 0.978 | ||||
| tSPM | 0.975 | 0.975 | 0.993a | 0.993a | ||||
| tSPM+AVR | 0.975 | 0.975 | 0.978 | 0.978 | ||||
| AVR | CHF | 0.883 | 0.885 | 0.72-0.879,60,61 | T2DM | 0.936 | 0.936 | 0.99,59 |
| SPM | 0.868 | 0.868 | 0.944 | 0.945 | ||||
| SPM+AVR | 0.866 | 0.872 | 0.956a | 0.959a | ||||
| tSPM | 0.898a | 0.901a | 0.926 | 0.926 | ||||
| tSPM+AVR | 0.893 | 0.896 | 0.922 | 0.922 | ||||
| AVR | COPD | 0.862 | 0.862 | NA | UC | 0.955 | 0.955 | 0.87-0.9759,20,58,59 |
| SPM | 0.866 | 0.867 | 0.953 | 0.953 | ||||
| SPM+AVR | 0.864 | 0.866 | 0.951 | 0.953 | ||||
| tSPM | 0.860 | 0.860 | 0.957a | 0.957a | ||||
| tSPM+AVR | 0.871a | 0.872a | 0.953 | 0.953 |
AUC: area under the curve; AVR: aggregated vector representation; CAD: coronary artery disease; CHF: congestive heart failure; COPD: chronic obstructive pulmonary disease; NA: we were not able to find AUC ROC in the published literature; RA: rheumatoid arthritis; SPM: sequential pattern mining; T1DM: type 1 diabetes mellitus; T2DM: type 2 diabetes mellitus; tSPM: transitive sequential pattern mining; UC: ulcerative colitis.
Top within-phenotype performance.
Figure 2.
Distribution of phenotyping areas under the receiver-operating characteristic curve (AUC ROCs) by phenotype and data representation. AD: Alzheimer’s disease; AFIB: atrial fibrillation; AVR: aggregated vector representation; CAD: coronary artery disease; CHF: congestive heart failure; COPD: chronic obstructive pulmonary disease; MSMR: minimize sparsity and maximize relevance; RA: rheumatoid arthritis; SPM: sequential pattern mining; T1DM: type 1 diabetes mellitus; T2DM: type 2 diabetes mellitus; tSPM: transitive sequential pattern mining; UC: ulcerative colitis.
Clinical evaluation of the top transitive sequences
From a clinical standpoint, the important transitive sequences can be subdivided into specific categories. Many of the most common sequences were the diagnosis code paired with a risk factor, symptom, complication, or treatment for that disease. It was also common to see the disease code paired with the influenza or pneumococcal 23 vaccination (PPSV23). Table 3 shows a sampling of the identified components that were found to be in sequence with the respective disease for the given phenotype. Each of these components, when in sequence with the diagnosis code, has increased accuracy for identifying the phenotype. For example, in the case of CAD, the diagnosis code on its own only identifies true coronary artery disease 34% of the time (ie, based on the chart reviews, only 34% of the patients that had the diagnosis code actually had a confirmed case of CAD). However, when the risk factor, hypertension, precedes the diagnosis code, the accuracy increases to 68%. If the symptom chest pain precedes CAD, the sequence accuracy increases to 70%. If the complication cardiac dysrhythmia precedes the diagnosis, the accuracy of the sequence increases to 71%. And if the treatment clopidogrel precedes the diagnosis, the accuracy of the sequence is 97%.There are also cases in which the diagnosis code is in sequence with a vaccination or a need for a vaccination. This also leads to increased accuracy for the sequence compared with the components. For example, the sequence “PPSV23 Rheumatoid arthritis” is 75% accurate for rheumatoid arthritis. However, “PPSV23” is only 22% accurate and “Rheumatoid Arthritis” is only 68% accurate.
Table 3.
Common components of sequences associated with each phenotype
| Risk factor | Symptoms | Complications | Medications | Vaccination | |
|---|---|---|---|---|---|
| AD |
−Hypertension −High cholesterol |
−Donepezil −Memantine |
−Need for Influenza vaccination | ||
| AF | −Hyperlipidemia | −Palpitations | −Cardiomegaly |
−Warfarin −Digoxin |
|
| CAD |
−Hyperlipidemia −Hypertension |
−Chest pain | −Cardiac dysrhythmia |
−Nitroglycerin −Clopidogrel |
−Need for influenza vaccination |
| CHF | −Hypertension |
−Shortness of Breath −Pulmonary congestion |
−Pulmonary congestion −Chronic kidney disease |
−Furosemide −Spironolactone |
|
| COPD |
−Shortness of breath −Cough |
−Other nonspecific abnormal finding of lung field |
−Tiotropium −Albuterol |
−Need for influenza vaccination | |
| RA | −Pain in limb | −Screening exam for pulmonary tuberculosis |
−Hydrochloro −Etanercept |
PPSV23 | |
| Stroke |
−Cardiac dysrhythmia −Hyperlipidemia |
−Cognitive deficits as late effect of cerebrovascular disease | −Other late effects of cerebrovascular disease |
−Lovenox −Statin |
−Need for influenza vaccination |
| T1DM |
−Obesity −High cholesterol |
−Insulin | −Influenza vaccination | ||
| T2DM |
−Obesity −High cholesterol |
−Other malaise and fatigue |
−Metformin −Atorvastatin |
−Need for PPSV23 | |
| UC |
−Abdominal pain −Diarrhea |
−Anemia |
−Prednisone −Mesalamine |
AD: Alzheimer’s disease; AF: atrial fibrillation; CAD: coronary artery disease; CHF: congestive heart failure; COPD: chronic obstructive pulmonary disease; PPSV23: pneumococcal 23 vaccination; RA: rheumatoid arthritis; T1DM: type 1 diabetes mellitus; T2DM: type 2 diabetes mellitus; UC: ulcerative colitis.
In some cases, the diagnosis code it not even included in the sequence. Instead, the sequence is composed exclusively of risk factors, symptoms, complications, treatments, or vaccines and still offers a high level of accuracy for identifying the specific phenotype. For example, in the case of COPD, “Cough Tiotropium” is 49% accurate for identifying COPD and only includes a symptom and a treatment. Sometimes both components of the sequence come from the same category. For example, in the case of T1DM, “Insulin Glucagon” is a sequence of 2 treatments and accurately identifies the phenotype 63% of the time.
DISCUSSION
We argue that sequences present more precise information by reducing some of the noise in the EHR data. For instance, we might have observations of the diagnosis code in patient ’s medical record. When the diagnosis code is deemed a relevant feature for phenotype (whether through our proposed MSMR or expert ascertion), in the conventional (AVR) approach for computational phenotyping is directly incorporated (as a feature) into the classification algorithm. Sequential data representations, instead, provide a more precise way for incorporating the record into downstream modeling, in that only a proportion of the record may hold useful information for classification that precedes another record (eg, ) or follows another record (). The MSMR algorithm allows for seamless discovery of such precise sequential record combinations. Our approach starts with user interpretability and works backward to the technology.
Furthermore, the sequences not only lead to more precise phenotyping, but also uniquely capture 2 distinct events that reflect the patient’s experiences of those events. For example, “Cardiac dysrhythmia -> Cognitive deficit as a late effect of cerebrovascular disease” is an important sequence for identifying the stroke phenotype. This is likely a common narrative for a stroke patient. The individual develops a cardiac dysrhythmia that leads to the formation of a clot in the heart, that then causes a cerebrovascular accident that leads to cognitive deficits. By sequencing the diagnoses and medications, we developed a rich feature set in which individual labels can accurately tell a patient’s story. Analyzing such sequences could give new insight into disease trajectories. Applying this method to new and emerging diseases (eg, COVID-19 [coronavirus disease 2019]) and then analyzing the sequence features could help us to understand how the disease progresses.
The diagnostic labels and medications listed in Table 3 were all selected by the proposed MSMR algorithm as the significant features for identifying the given phenotype. Each of these labels when in sequence with the disease also has clinical meaning. For example, “Hypertension -> Alzheimer’s Disease” suggests that hypertension precedes Alzheimer’s disease. The scientific literature supports this relationship. Several longitudinal studies have shown that midlife hypertension is consistently associated with the development of Alzheimer’s disease, implying that hypertension is a risk factor.62–64 Another example is the sequence “Congestive heart failure -> Chronic kidney disease.” Again, the literature supports this relationship. In a systematic review of 16 studies with more than 80 000 patients with congestive heart failure, 29% had moderate-to-severe kidney impairment.65 Therefore, congestive heart failure preceding chronic kidney disease makes sense because chronic kidney disease is a known complication. These sequences not only are more accurate, but also tell a clinical narrative that corresponds with the patient’s experience.
While risk factors, symptoms, complications, and medications paired with the disease may make intuitive sense, vaccinations in sequence with the disease are less obvious. Their inclusion in such sequences may be a result of the specific criteria for receiving the vaccination increasing the probability that the disease code matches the phenotype. The Centers for Disease Control and Prevention recommends PPSV23 for patients under 65 years of age with chronic diseases such as heart disease, lung disease, and DM and all adults over 65 years of age.66 And while influenza is recommended yearly to all adults, it is very important for those at high risk for serious complications to influenza.67 The sequence of a vaccination with the disease code may be an accurate label because the vaccination’s presence in the chart further verifies that the patient has a chronic disease.
More complex algorithms such as recurrent neural networks (RNNs)68 and RNN-based models such as long short-term memory69 and gated recurrent unit70 have been used to account for time.71–78 These algorithms often result in highly predictive models, but they are hard to understand, limiting their utility in healthcare settings. The transitive sequences are similar to simple forms of recurrent events in RNN-based models. The difference is that we do not provide any gate or memory constraint and would accept all possible sequences. This resulted in a large dimensional space. The MSMR algorithm is allowed to pick up what is relevant to the outcome of interest. However, in this article, we only studied 2-deep sequences. We envision extracting deeper sequences, which would further increase dimensionality. In that case, future research may need to apply memory constraints on what to remember from the past.
Despite the billions of dollars that have been spent to institute meaningful use of EHR systems over the past several decades, challenges still remain for using EHR data to rapidly address pressing health issues including the COVID-19 pandemic. This machine learning pipeline, which includes both representation mining and the MSMR algorithm, is capable of engineering predictive features without the need for expert involvement to model different phenotypes and outcomes. Without that bottleneck, this method provides a much faster way for extracting meaningful data from the EHR.
CONCLUSION
We presented a high-throughput approach for computational phenotyping using temporal sequential data representations. In contrast to the popular deep learning approaches, our approach started with user interpretability and worked backward to the technology. Feature engineering in this approach is fully automated using silver-standard labels. We also demonstrated that using transitive sequences of EHR diagnosis and medication records as features for computational phenotyping yields improved phenotyping performance compared with the timeless raw EHR records. Sequential data representations provide a precise mechanism for incorporating raw EHR records into downstream machine learning. Together, the temporal sequences and the machine learning pipeline can be rapidly deployed to develop computational models for identifying and validating novel disease markers and advancing medical knowledge discovery.
AUTHOR CONTRIBUTIONS
HE and SNM were involved in conceptualization. HE contributed to the methodology and formal analysis. HE, ZHS, and SNM conducted the investigation. HE and ZHS wrote the original draft and HE, ZHS, and SNM reviewed and edited the manuscript. HE was involved in visualization. SNM was involved in funding acquisition.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr Sebastien Vasey for invaluable contribution to the MSMR algorithm.
CONFLICT OF INTEREST STATEMENT
The authors declare no competing interests.
REFERENCES
- 1. Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 2012; 13 (6): 395–405. [DOI] [PubMed] [Google Scholar]
- 2. Zhao J Papapetrou P Asker L Boström H. Learning from heterogeneous temporal data in electronic health records. J Biomed Inform 2017; 65: 105–19. [DOI] [PubMed] [Google Scholar]
- 3. Hripcsak G Albers DJ Perotte A. Exploiting time in electronic health record correlations. J Am Med Inform Assoc 2011; 18 (Suppl 1): i109–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hripcsak G Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc 2013; 20 (1): 117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Agniel D, Kohane IS, Weber GM. Biases in electronic health record data due to processes within the healthcare system: retrospective observational study. BMJ 2018; 361: k1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Banda JM, Seneviratne M, Hernandez-Boussard T, et al. Advances in electronic phenotyping: from rule-based definitions to machine learning models. Annu Rev Biomed Data Sci 2018; 1 (1): 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ding DY, Simpson C, Pfohl S, et al. The effectiveness of multitask learning for phenotyping with electronic health records data. Pac Symp Biocomput 2019; 24: 18–29. [PMC free article] [PubMed] [Google Scholar]
- 8. Agarwal V, Podchiyska T, Banda JM, et al. Learning statistical models of phenotypes using noisy labeled training data. J Am Med Inform Assoc 2016; 23 (6): 1166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miotto R, Li L, Kidd BA, et al. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep 2016; 6 (1): 26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu S, Chakrabortty A, Liao KP, et al. Surrogate-assisted feature extraction for high-throughput phenotyping. J Am Med Inform Assoc 2017; 24 (e1): e143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu S, Liao KP, Shaw SY, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc 2015; 22 (5): 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moskovitch R, Shahar Y. Classification-driven temporal discretization of multivariate time series. Data Min Knowl Disc 2015; 29 (4): 871–913. [Google Scholar]
- 13. Batal I, Valizadegan H, Cooper GF, et al. A temporal pattern mining approach for classifying electronic health record data. ACM Trans Intell Syst Technol 2013; 4 (4): 10.1145/2508037.2508044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Z, Wu L, Hauskrecht M. Modeling clinical time series using Gaussian process sequences. In: Proceedings of the 2013 SIAM International Conference on Data Mining; 2013. 10.1137/1.9781611972832.69
- 15. Madkour M, Benhaddou D, Tao C. Temporal data representation, normalization, extraction, and reasoning: A review from clinical domain. Comput Methods Programs Biomed 2016; 128: 52–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moskovitch R, Polubriaginof F, Weiss A, et al. Procedure prediction from symbolic Electronic Health Records via time intervals analytics. J Biomed Inform 2017; 75: 70–82. doi : 10.1016/j.jbi.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 17. Albers DJ, Hripcsak G. Estimation of time-delayed mutual information and bias for irregularly and sparsely sampled time-series. Chaos Solitons Fractals 2012; 45 (6): 853–60. doi : 10.1016/j.chaos.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frey LJ, Lenert L, Lopez-Campos G. EHR big data deep phenotyping. Yearb Med Inform 2014; 23 (1): 206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc 2014; 21 (2): 221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu S, Ma Y, Gronsbell J, et al. Enabling phenotypic big data with PheNorm. J Am Med Inform Assoc 2018; 25 (1): 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liao KP, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res 2010; 62 (8): 1120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ananthakrishnan AN Cai T Savova G, et al. Improving Case Definition of Crohn's Disease and Ulcerative Colitis in Electronic Medical Records Using Natural Language Processing. Inflamm Bowel Dis 2013; 19 (7): 1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xia Z, Secor E, Chibnik LB, et al. Modeling disease severity in multiple sclerosis using electronic health records. PLoS One 2013; 8 (11): e78927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kumar V Liao K Cheng S-C, et al. Natural language processing improves phenotypic accuracy in an electronic medical record cohort of type 2 diabetes and cardiovascular disease. J Am Coll Cardiol 2014; 63 (12): A1359. [Google Scholar]
- 25. Murphy SN, Mendis ME, Berkowitz DA, et al. Integration of clinical and genetic data in the i2b2 architecture. AMIA Annu Symp Proc 2006; 2006: 1040. doi : 85881 [pii] [PMC free article] [PubMed]
- 26. Castro VM Minnier J Murphy SN, et al. Validation of Electronic Health Record Phenotyping of Bipolar Disorder Cases and Controls. Am J Psychiatry 2015; 172 (4): 363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu S, Kumamaru KK, George E, et al. Classification of CT pulmonary angiography reports by presence, chronicity, and location of pulmonary embolism with natural language processing. J Biomed Inform 2014; 52: 386–93. doi : 10.1016/j.jbi.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liao KP, Ananthakrishnan AN, Kumar V, et al. Methods to develop an electronic medical record phenotype algorithm to compare the risk of coronary artery disease across 3 chronic disease cohorts. PLoS One 2015; 10 (8): e0136651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liao KP, Cai T, Savova GK, et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ 2015; 350: h1885. doi : 10.1136/bmj.h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008; 84 (3): 362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bowton EA, Collier SP, Wang X, et al. Phenotype-driven plasma biobanking strategies and methods. J Pers Med 2015; 5 (2): 140–52. doi : 10.3390/jpm5020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011; 4 (1): 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) network: past, present, and future. Genet. Med 2013; 15 (10): 761–71. doi : 10.1038/gim.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pathak J, Wang J, Kashyap S, et al. Mapping clinical phenotype data elements to standardized metadata repositories and controlled terminologies: the eMERGE Network experience. J Am Med Inform Assoc 2011; 18 (4): 376–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med 2011; 3 (79): 79re1. doi : 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: A catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 2016; 23 (6): 1046–52. doi : 10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anderson AE, Kerr WT, Thames A, et al. Electronic health record phenotyping improves detection and screening of type 2 diabetes in the general United States population: a cross-sectional, unselected, retrospective study. J Biomed Inform 2016; 60: 162–8. doi : 10.1016/j.jbi.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Banda JM, Halpern Y, Sontag D, et al. Electronic phenotyping with APHRODITE and the Observational Health Sciences and Informatics (OHDSI) data network. AMIA Jt Summits Transl Sci Proc 2017; 2017: 48–57. [PMC free article] [PubMed] [Google Scholar]
- 39. Sun W, Rumshisky A, Uzuner O. Temporal reasoning over clinical text: the state of the art. J Am Med Inform Assoc 2013; 20 (5): 814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shahar Y Musen MA. Knowledge-based temporal abstraction in clinical domains. Artif Intell Med 1996; 8 (3): 267–98. [DOI] [PubMed] [Google Scholar]
- 41. Stacey M, McGregor C. Temporal abstraction in intelligent clinical data analysis: a survey. Artif Intell Med 2007; 39 (1): 1–24. doi : 10.1016/j.artmed.2006.08.002 [DOI] [PubMed] [Google Scholar]
- 42. Agrawal R, Srikant R. Mining sequential patterns. In: ICDE ’95: Proceedings of Eleventh International Conference on Data Engineering; 1995: 3–14. doi : 10.1109/icde.1995.380415
- 43. Mabroukeh NR, Ezeife CI. A taxonomy of sequential pattern mining algorithms. ACM Comput Surv 2010; 43 (1): 1–41. [Google Scholar]
- 44. Berlingerio M, Bonchi F, Giannotti F, et al. Mining clinical data with a temporal dimension: a case study. In: 2007 IEEE International Conference on Bioinformatics and Biomedicine (BIBM 2007); 2007: 429–36. doi : 10.1109/BIBM.2007.42
- 45. Zaki MJ. Parallel sequence mining on shared-memory machines. J Parallel Distrib Comput 2001; 61 (3): 401–26. [Google Scholar]
- 46. Ayres J, Flannick J, Gehrke J, et al. Sequential PAttern mining using a bitmap representation. In: KDD ’02: Proceedings of the Eighth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2002: 429–35. doi : 10.1145/775047.775109
- 47. Shknevsky A, Shahar Y, Moskovitch R. Consistent discovery of frequent interval-based temporal patterns in chronic patients’ data. J Biomed Inform 2017; 75: 83–95. [DOI] [PubMed] [Google Scholar]
- 48. Estiri H, Strasser ZH, Klann JG, et al. Transitive sequencing medical records for mining predictive and interpretable temporal representations. Patterns 2020; 1 (4): 100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meyer PE. Information-Theoretic Variable Selection and Network Inference From Microarray Data [PhD thesis]. Brussels, Belgium, Université Libre Bruxelles; 2008.
- 50. Cover TM, Thomas JA. Elements of Information Theory. 2nd ed. Hoboken, NJ: Wiley; 2012. [Google Scholar]
- 51. Paninski L. Estimation of entropy and mutual information. Neural Comput 2003; 15 (6): 1191–253. [Google Scholar]
- 52. Yang HH, Moody J. Feature selection based on joint mutual information. In: proceedings of International ICSC Symposium on Advances in Intelligent Data Analysis; 1999.
- 53. Brown G, Pocock A, Zhao MJ, et al. Conditional likelihood maximisation: A unifying framework for information theoretic feature selection. J Mach Learn Res 2012; 13: 27–66. [Google Scholar]
- 54. Estiri H, Vasey S, Murphy SN. Generative transfer learning for measuring plausibility of EHR diagnosis records. J Am Med Inform Assoc 2020. Oct 12 [E-pub ahead of print]. doi : 10.1093/jamia/ocaa215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chiu PH, Hripcsak G. EHR-based phenotyping: bulk learning and evaluation. J Biomed Inform 2017; 70: 35–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zheng T, Xie W, Xu L, et al. A machine learning-based framework to identify type 2 diabetes through electronic health records. Int J Med Inform 2017; 97: 120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kartoun U, Aggarwal R, Beam AL, et al. Development of an algorithm to identify patients with physician-documented insomnia. Sci Rep 2018; 8 (1): 7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ning W, Chan S, Beam A, et al. Feature extraction for phenotyping from semantic and knowledge resources. J Biomed Inform 2019; 91: 103122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liao KP, Sun J, Cai TA, et al. High-throughput multimodal automated phenotyping (MAP) with application to PheWAS. J Am Med Inform Assoc 2019; 26 (11): 1255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu J, Roy J, Stewart WF. Prediction modeling using EHR data: challenges, strategies, and a comparison of machine learning approaches. Med Care 2010; 48: S106–13. [DOI] [PubMed] [Google Scholar]
- 61. Liu C, Wang F, Hu J, et al. Temporal phenotyping from longitudinal electronic health records. In: KDD ’15: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2015: 705–14. doi : 10.1145/2783258.2783352
- 62. Gottesman RF, Albert MS, Alonso A, et al. Associations between midlife vascular risk factors and 25-year incident dementia in the Atherosclerosis Risk in Communities (ARIC) cohort. JAMA Neurol 2017; 74 (10): 1246–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Whitmer RA, Sidney S, Selby J, et al. Midlife cardiovascular risk factors and risk of dementia in late life. Neurology 2005; 64 (2): 277–81. [DOI] [PubMed] [Google Scholar]
- 64. Freitag MH, Peila R, Masaki K, et al. Midlife pulse pressure and incidence of dementia: the Honolulu-Asia aging study. Stroke 2006; 37 (1): 33–7. [DOI] [PubMed] [Google Scholar]
- 65. Smith GL, Lichtman JH, Bracken MB, et al. Renal impairment and outcomes in heart failure. systematic review and meta-analysis. J Am Coll Cardiol 2006; 47 (10): 1987–96. [DOI] [PubMed] [Google Scholar]
- 66.Centers for Disease Control and Prevention. Pneumococcal vaccine timing for adults. U.S. Department of Health & Human Services. https://www.cdc.gov/vaccines/vpd/pneumo/downloads/pneumo-vaccine-timing.pdf Accessed August 01, 2020.
- 67.Centers for Disease Control and Prevention. Key facts about seasonal flu vaccine. U.S. Department of Health & Human Services. https://www.cdc.gov/flu/prevent/keyfacts.htm Accessed August 01, 2020.
- 68. Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature 1986; 323 (6088): 533–6. [Google Scholar]
- 69. Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997; 9 (8): 1735–80. doi : 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 70. Cho K, van Merriënboer B, Gulcehre C, et al. Learning phrase representations using RNN encoder–decoder for statistical machine translation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP); 2014: 1724–34. [Google Scholar]
- 71. Choi E, Schuetz A, Stewart WF, et al. Using recurrent neural network models for early detection of heart failure onset. J Am Med Inform Assoc 2017; 24 (2): 361–70. doi : 10.1093/jamia/ocw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Che Z, Purushotham S, Cho K, et al. Recurrent neural networks for multivariate time series with missing values. Sci Rep 2018; 8 (1): 6085. doi : 10.1038/s41598-018-24271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lipton ZC, Kale DC, Elkan C, Wetzel R. Learning to diagnose with LSTM recurrent neural networks. arXiv: 1511.03677; 2015.
- 74. Pham T, Tran T, Phung D, et al. Predicting healthcare trajectories from medical records: a deep learning approach. J Biomed Inform 2017; 69: 218–29. doi : 10.1016/j.jbi.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 75. Jin B, Che C, Liu Z, et al. Predicting the risk of heart failure with EHR sequential data modeling. IEEE Access 2018; 6: 9256–61. [Google Scholar]
- 76. Jagannatha AN, Yu H. Bidirectional RNN for medical event detection in electronic health records. Proc Conf 2016; 2016: 473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Choi E, Bahadori MT, Schuetz A, et al. Doctor AI: predicting clinical events via recurrent neural networks. In: Proceedings of the 1st Machine Learning for Healthcare Conference; 2016: 301–18. http://proceedings.mlr.press/v56/Choi16.pdf. [PMC free article] [PubMed]
- 78. Zhang J, Kowsari K, Harrison JH, et al. Patient2Vec: a personalized interpretable deep representation of the longitudinal electronic health record. IEEE Access. 2018; 6: 65333–46. doi : 10.1109/ACCESS.2018.2875677. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.