Abstract
The crRNA-guided nuclease Cas13 recognizes complementary viral transcripts to trigger the degradation of both host and viral RNA during the type VI CRISPR-Cas antiviral response. How viruses can counteract this immunity is not known. We describe a listeriophage (ϕLS46) encoding an anti-CRISPR protein (AcrVIA1) that inactivated the type VI-A CRISPR system of Listeria seeligeri. Using genetics, biochemistry and structural biology we found that AcrVIA1 interacted with the guide-exposed face of Cas13a, preventing access to the target RNA and the conformational changes required for nuclease activation. Unlike inhibitors of DNA-cleaving Cas nucleases, which cause limited immunosuppression and require multiple infections to bypass CRISPR defenses, a single dose of AcrVIA1 delivered by an individual virion could completely dismantle type VI-A CRISPR-mediated immunity.
CRISPR-Cas systems are prokaryotic adaptive immune systems that protect their hosts from invasion by viruses (1) and plasmids (2). CRISPR loci contain short DNA repeats separated by spacer sequences of foreign origin (3-5). To achieve immunity, the locus is transcribed and processed into small crRNAs, which associate with RNA-guided Cas nucleases (6) to locate and cleave complementary nucleic acid sequences (protospacers) (7). CRISPR systems are categorized into six types (I-VI) which differ in their cas gene content and mechanism of immunity (8). While most types neutralize invaders through destruction of their DNA, Cas13, the RNA-guided nuclease of type VI systems, unleashes non-specific RNA degradation (trans-RNase activity) upon recognition of a phage target transcript (9-11). The cleavage of host transcripts leads to a growth arrest that prevents further propagation of the phage, allowing the uninfected cells in the population to survive and proliferate (11). Because the phage genome is not directly affected by Cas13, it continues to produce target transcripts, leading to a persistent activation of the nuclease and growth arrest (11).
Identification of the Cas13a inhibitor AcrVIA1
Presumably in response to the pressure imposed by CRISPR-Cas immunity, phages evolved anti-CRISPR (Acr) proteins, small proteins (usually <150 aa) that are produced during infection and inactivate Cas nucleases (12). Acrs also exhibit exceptional diversity of sequences and mechanisms and, with few exceptions, specifically inhibit one CRISPR subtype (12-17). Recently, inhibitors of Cas13a were independently reported (18); however, how they allow phages to overcome the type VI-A CRISPR-Cas response is not known. To investigate the molecular mechanisms used by Acr-carrying phages to inhibit Cas13 during infection of a natural host, we first obtained temperate phages from a collection of 62 environmental isolates of Listeria spp., an organism that commonly harbors type VI-A CRISPR-Cas systems. We induced prophages with mitomycin C and isolated phages that infected a mutant of Listeria seeligeri SLCC3954 (19) lacking its two restriction-modification systems and the type VI-A CRISPR array (L. seeligeri ΔRM Δspc), Fig. S1A). We isolated 15 phages (Fig. S1B), which we used to infect wild type L. seeligeri (WT) and obtain 10 lysogens carrying different prophages in their genomes (Fig. S1C). We then tested each lysogen for its ability to disable Cas13a-mediated immunity against plasmid conjugation (Fig. S1D). Only the ϕLS46 lysogen exhibited a high efficiency of plasmid transfer (Fig. 1A and S1D), suggesting the possibility that this prophage harbored a Cas13a inhibitor.
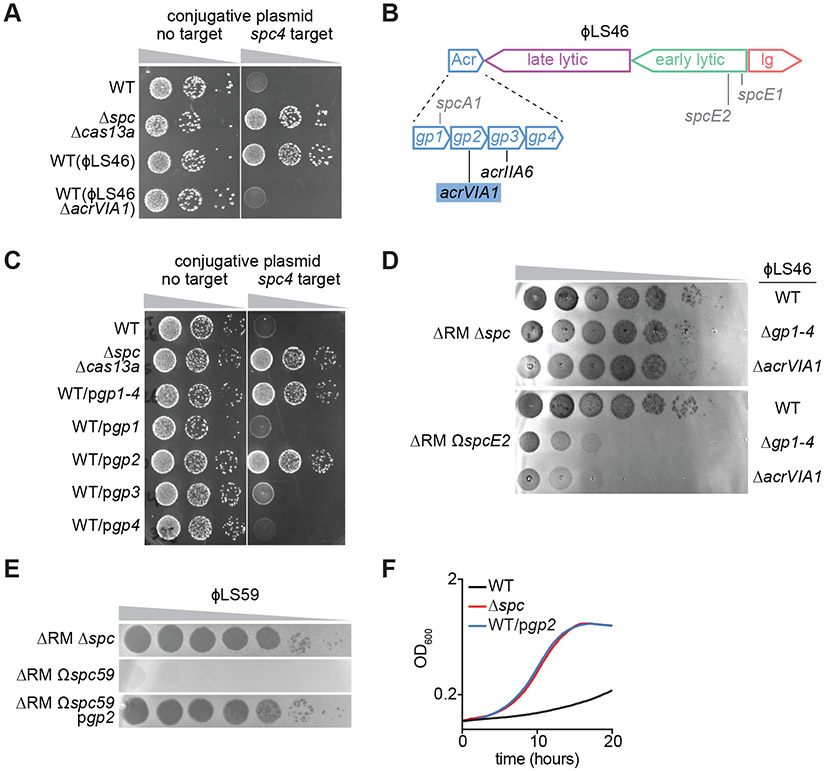
Figure 1. AcrVIA1 inhibits type VI-A CRISPR-Cas immunity against plasmids and phages.
(A) Transfer of a conjugative plasmid with or without the spc4 target of the L. seeligeri type VI-A CRISPR-Cas system into different strains: wild-type (WT), Δspc Δcas13a or WT harboring the ϕLS46 or ϕLS46 ΔacrVIA1 prophages. (B) Schematic of the ϕLS46 genome showing the four main transcription units (acr, lysogeny, early- and late-lytic genes). gp2 was renamed acrVIA1. The locations of the targets of spacers used in this study are shown in grey. (C) Same as (A) but using strains carrying plasmids to express different acr genes from ϕLS46. (D) Detection of phage propagation after spotting ten-fold dilutions of WT, Δgp1-4 or ΔacrVIA1 phage ϕLS46, on lawns of L. seeligeri ΔRM Δspc or ΔRM ΩspcE2. (E) Same as (D) but spotting ϕLS59 into lawns of L. seeligeri ΔRM Δspc, ΔRM Ωspc59 or ΔRM Ωspc59/pgp2. (F) Growth of WT, Δspc and WT/pgp2 L. seeligeri strains expressing a spc4 target RNA under the control of an anhydrotetracycline-inducible promoter, after addition of the inducer.
Sequencing of the ϕLS46 genome revealed a similar organization to a previously characterized temperate phage of L. seeligeri, ϕRR4 (11), which harbors a region containing six genes, two of them with homology to the Cas9 inhibitors AcrIIA1 and AcrIIA2 (Fig. S1E). In ϕLS46, however, this region contains four genes, none of which displayed strong homology to known inhibitors (Figs. 1B and S1E). To investigate if this region contains a type VI Acr, we cloned the operon with its native promoter into a plasmid (pgp1-4), introduced it into wild-type L. seeligeri, and tested for Cas13a-mediated immunity against plasmid conjugation as in Figure 1A. Indeed, the presence of pgp1-4 allowed plasmid conjugation even in the presence of Cas13a targeting, and cloning each individual gene allowed us to identify gp2 as the gene responsible for this anti-CRISPR phenotype (Fig. 1C). Accordingly, we renamed gp2 “type VI-A anti-CRISPR 1”, or AcrVIA1. AcrVIA1 is a protein of 232 amino acids, considerably larger than most previously discovered Acrs. Gp1, AcrVIA1, and Gp4 exhibit no detectable homology to proteins of known function, but we noted that Gp3 contains a helix-turn-helix (HTH) domain with limited similarity to AcrIIA6 from Streptococcus phage DT1, suggesting that it may be an inhibitor of type II-A CRISPR-Cas systems. Many listeriaphages harbor the HTH-containing AcrIIA1, which serves dual roles as an Acr and a transcription autorepressor of the acr cassette during late lytic infection (20). To test whether Gp3 plays a similar role in ϕLS46, we fused the acrVIA1 promoter to a lacZ reporter and measured β-galactosidase activity in the presence and absence of pgp3 (Fig. S2). We observed a ~10-fold reduction in transcription from the acr promoter in the presence of Gp3, a result that confirmed its role as a regulator of AcrVIA1 expression.
Next we tested if AcrVIA1 was necessary for inhibition of Cas13a during ϕLS46 infection. We created a derivative of the ΔRM Δspc strain in which we ectopically integrated different spacer sequences, with their transcription controlled by the native CRISPR promoter (strain ΔRM ΩspcX, Fig. S1A). First, we inserted spacers targeting transcripts of either a conjugative plasmid or phage ϕLS59, whose genome lacks acr genes, and confirmed that they can provide efficient immunity in this experimental system (Figs. S3A-B). We then cloned 10 spacers targeting different transcript regions of ϕLS46 (Fig. S3C), none of which conferred immunity (Fig. S3D). Finally, we isolated phage mutants in the Acr region of ϕLS46 and tested the same spacers for immunity against them. While none of the spacers protected against wild-type ϕLS46, both Δgp1-4 and ΔacrVIA1 mutants exhibited 1-6 orders of magnitude of sensitivity to Cas13a interference (Figs. 1D and S3D). We expressed AcrVIA1 using the pgp2 plasmid and found that it inhibited targeting of the Cas13a-susceptible phage ϕLS59 (Fig. 1E). Finally, the inhibition of anti-plasmid immunity observed in the WT(ϕLS46) lysogen was abolished when we performed the conjugation assay using an WT(ϕLS46 ΔacrVIA1) lysogen (Fig. 1A). To explore the effect of AcrVIA1 on the Cas13a-induced host cell dormancy that is fundamental for type VI-A immunity, we induced the expression of spc4 target RNA, which was previously shown to cause a severe growth defect as a result of nonspecific host transcript degradation (11). Expression of the inhibitor using the pgp2 plasmid, however, reverted this growth defect (Fig. 1F). Thus, acrVIA1 is necessary and sufficient to inhibit Cas13a-induced growth arrest and thereby thwart type VI CRISPR immunity against plasmids and phages.
AcrVIA1 binds Cas13a and inhibits cis- and trans- RNase activities
The inhibition of Cas13a-induced growth arrest suggested that AcrVIA1 could inhibit the trans-RNase activity of Cas13a. To investigate this, we purified both proteins (Fig. S4A, B) and tested their activities using in vitro RNA protospacer cleavage assays. A radiolabeled target RNA was used to investigate inhibition of Cas13a’s cis-RNase activity. Purified L. seeligeri Cas13acrRNA catalyzed rapid RNA cleavage of protospacer RNA, and this activity was gradually decreased in the presence increasing concentrations of AcrVIA1 (Figs. 2A, B). Similarly, AcrVIA1 inhibited Cas13a-mediated trans-cleavage of a labeled non-target RNA upon addition of unlabeled protospacer RNA (Fig. 2C). To investigate the presence of an interaction between the nuclease and its inhibitor, we added C-terminal hexa-histidine and 3xFLAG tags to Cas13a and AcrVIA1, respectively, and confirmed that both are functional in L. seeligeri (Fig. S4C) and in vitro (Figs. 2A, C). We expressed Cas13a-His6 either alone or in the presence of AcrVIA1-3xFLAG, then performed immunoprecipitation with anti-Flag antibody resin. Analysis of the input, unbound, and immunoprecipitated fractions by immunoblot using antibodies against His6, FLAG, and the L. seeligeri housekeeping sigma factor σA as a control showed a specific co-immunoprecipitation of Cas13a-His6 with AcrVIA1-3xFLAG Fig. 2D). Finally, we investigated whether the interaction of AcrVIA1 with Cas13a prevents binding of the Cas13acrRNA complex to its complementary target RNA. To test this, we performed an electrophoretic mobility shift assay (EMSA) to measure the association of labeled protospacer RNA with a nuclease-dead dCas13acrRNA complex (Fig. 2E). In the presence of this complex, the majority of the target RNA was shifted to multiple higher molecular weight species: one corresponding to a crRNA-protospacer RNA duplex, and higher species representing the dCas13acrRNA-protospacer ternary complex. In contrast, in the presence of equimolar AcrVIA1, the target RNA remained mostly unbound and unassociated with dCas13acrRNA. Collectively, these results indicate that AcrVIA1 forms a complex with Cas13acrRNA that prevents binding of the complementary target RNA and therefore inhibits both its cis- and trans-RNase activities.
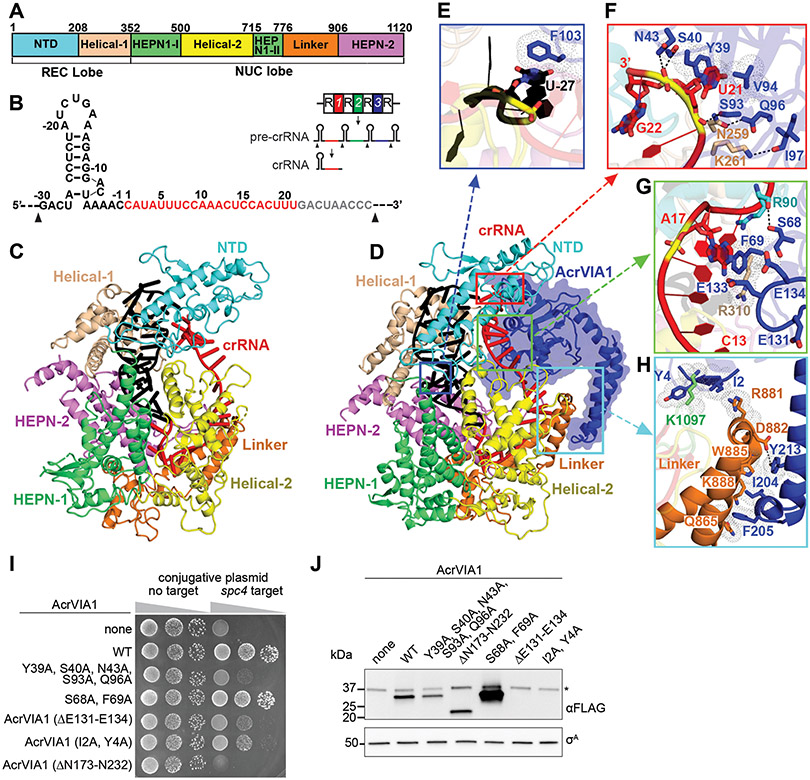
Figure 2. AcrVIA1 interacts with Cas13acRNA to prevent binding of the target RNA and RNase activation.
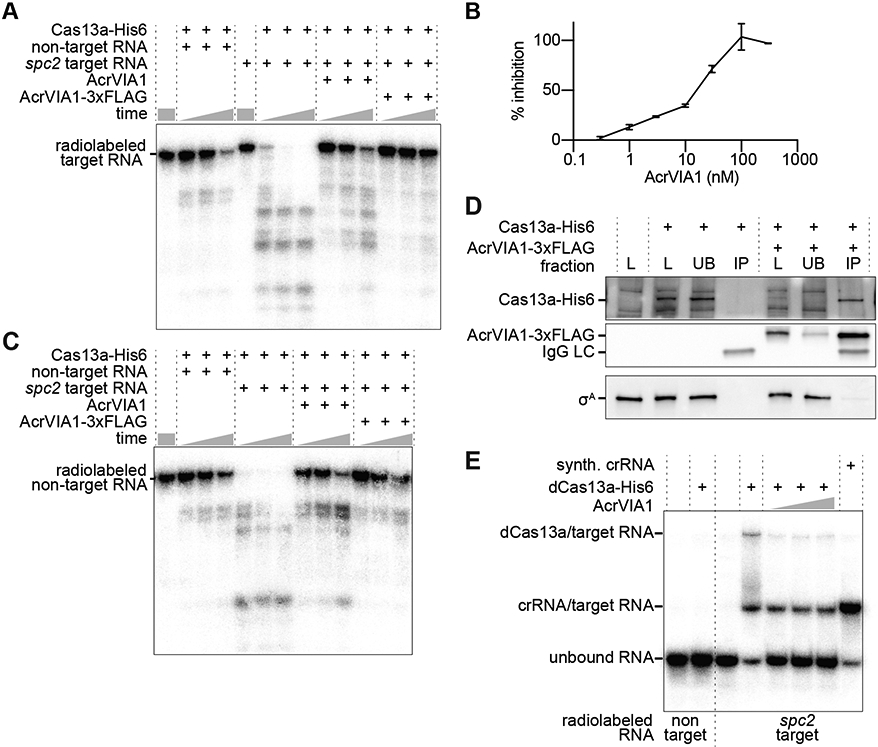
(A) cis-RNA cleavage time course with purified L. seeligeri Cas13a-His6, AcrVIA1 and/or AcrVIA1-3xFLAG using radiolabeled non-target or spc2-target RNA substrates. Reactions were analyzed after 5, 10, or 20 minutes of incubation. (B) trans-RNA cleavage time course as in (A) but using a radiolabeled non-target RNA substrate in the presence of unlabeled non-target or spc2-target RNA. (C) Dose response of Cas13a cis-RNase inhibition by AcrVIA1-3xFLAG (D) Anti-FLAG immunoprecipitation using protein extracts from L. seeligeri cells expressing either Cas13a-His6 alone or co-expressing AcrVIA1-3xFLAG. The His6 and FLAG epitopes, as well as the σA protein were detected via immunoblot. (E) EMSA of radiolabeled non-target or spc2-target RNAs in the presence of dCas13a-His6, with 2:1, 1:1, or 1:2 equivalents of AcrVIA1-3xFLAG.
Structure of the AcrVIA1-Cas13acrRNA complex
To further investigate how AcrVIA1 suppressed Cas13acrRNA activity, we isolated stable homogeneous AcrVIA1-Cas13acrRNA complex (Fig. S5) and determined its cryo-EM structure along with that of Cas13acrRNA alone, at 3.0 Å and 3.2 Å resolution, respectively (Fig. S6 and Table S1). Cas13acrRNA adopts a bilobed architecture, consisting of recognition [REC; N-terminal (NTD) and Helical-1 domains] and nuclease (NUC; Helical-2 and two HEPN domains connected by a Linker element) lobes (Fig. 3A, C), similar to previously reported structures of Cas13a from other species (21, 22). The complex contained a natural 51-nt mature crRNA, which was processed and loaded in the native host (Fig. 3B and S7A). Nucleotides 8-12 and 13-19 in the crRNA spacer region adopt an approximately A-form helical conformation, with their outwards direction positioned to pair with the target RNA (Fig. S7B), a feature different from the crRNA alignment in Leptotrichia buccalis Cas13a (22). The 5’ end of the repeat is located in the cleft between the Helical-1 and HEPN-2 domains (Fig. S7A). Mutations in residues within this region of Cas13a (R1048A, K1049A) abrogate type VI interference against plasmid conjugation (Fig. S7C), revealing their importance for crRNA maturation.
Figure 3. Cryo-EM structures of Cas13acrRNA and AcrVIA1-Cas13acrRNA complexes.
(A) Domain organization of L. seeligeri Cas13a. (B) Schematic representation of the crRNA sequence. The repeat and spacer regions within crRNA are shown in black and red, respectively. The disordered region is shown in gray. The black arrow shows the cleavage site of the pre-crRNA. Inset: crRNA maturation pathway; repeats are represented as “R”, spacers as numbers. (C) Ribbon representation of the structure of Cas13acrRNA. (D) Ribbon and surface (AcrVIA1) representations of AcrVIA1-Cas13acrRNA complex. (E-H) Detailed interactions between AcrVIA1 and Cas13acrRNA in the complex. (I) Transfer of conjugative plasmid with or without spc4 target of the L. seeligeri type VI CRISPR-Cas system into WT L. seeligeri harboring plasmid-borne wild-type or mutant alleles of acrVIA1-3xflag. (J) Anti-Flag immunoblot of AcrVIA1 mutants tested in (I), and anti-σA loading control, *cross-reacting protein.
In the AcrVIA1-Cas13acrRNA complex, the inhibitor is positioned on the crRNA-exposed face of Cas13a and directly interacts with the crRNA and residues in the Helical-1, NTD, HEPN-1 and Linker domains of Cas13a (Figs. 3D-H, S8A-D), with a buried interface area of ~1800 Å2. There is no structural similarity between AcrVIA1 and reported structures upon a DALI search, indicating a novel fold for this inhibitor. Residues in AcrVIA1 form extensive hydrophobic and hydrophilic interactions with the crRNA, which has been shown previously to undergo significant conformational changes, especially at its 3’ end, upon target RNA binding, to activate Cas13a RNase activity (22). Notably, the 3’ end of the crRNA in the AcrVIA1-Cas13acrRNA complex is stabilized by hydrogen bonds formed by N43, S40 and S93, and stacking interactions between U21 in crRNA and Y39 and V94 in AcrVIA1 (Fig. 3F), F69 stacks on A17 in the middle spacer region of crRNA (Fig. 3G), and F103 stacks on U27 in the 5’ repeat region of crRNA (Fig. 3E). In addition, the acidic loop E131-E134 in the inhibitor points towards and blocks access to the central C13 to A17 region of the crRNA (Fig. 3G). This region has been shown previously to be critical for target RNA binding and to turn on Cas13a RNase activity (9, 22).
AcrVIA1 also interacts directly with Cas13a, making several intermolecular contacts in the complex. While residues S93, Q96 and I97 in the inhibitor form hydrogen bonds with N259 and K261 in the Helical-1 domain of the nuclease (Fig. 3F), residue F69 in AcrVIA1 stacks on R310 of the same domain (Fig. 3G). S68 in AcrVIA1 forms hydrogen bonds with R90 in the NTD domain (Fig. 3G), while residues I2 and Y4 in the inhibitor stack on K1097 in the HEPN-1 domain (Fig. 3H). Notably, the two C-terminal helices of AcrVIA1 form extensive interactions with the Linker domain of Cas13a, locking it in place (Fig. 3H) and thereby preventing the significant conformational changes reported to occur upon target RNA binding (22). Finally, we detected only minimal structural changes of Cas13a upon AcrVIA1 binding (Fig. S9A). By contrast, the 3’ end of crRNA undergoes a significant conformational rearrangement (Fig. S9B, C). Thus, AcrVIA1 prevents conformational changes in the crRNA that occur upon target RNA binding, which are required for activation of Cas13a. To investigate the importance of the observed contacts between the nuclease and its inhibitor, we mutated the relevant residues in AcrVIA1-3xFLAG and tested their importance to inhibit Cas13a immunity against plasmid conjugation (Fig. 3I), as well as their impact on protein stability by immunoblot (Fig. 3J). Mutations in I2A, Y4A or deletion of the E131-E134 loop affected both the stability and the function of the inhibitor. In contrast, the quintuple mutant Y39A, S40A, N43A, S93A, Q96A, and the truncation mutant lacking the two AcrVIA1 C-terminal helices (ΔN173-N232) caused nearly complete loss of function with little or no effect on protein expression, corroborating their importance for Cas13a inhibition. When tested individually, none of the five substitutions affected inhibition (Fig. S7D), a result that suggests a very strong association between AcrVIA1 and Cas13a that does not rely on a single interaction. The S68A, F69A double mutant retained full function, suggesting that the interaction with Cas13a is unperturbed in this mutant. However, the mutation also led to an increase in expression levels, which could compensate for a partial loss of function.
AcrVIA1 had no effect on protospacer RNA cleavage by purified L. buccalis Cas13acrRNA (Figure S10A). We performed a structural comparison of L. seeligeri Cas13a with L. buccalis Cas13a (PDB 5XWY). Superposition of L. seeligeri Cas13acrRNA and L. buccalis Cas13acrRNA revealed differences in the NTD domain (Fig. S10B) and the 3’ end of crRNA (Fig. S10C), that generate obvious clashes between L. buccalis Cas13acrRNA and AcrVIA1 (Fig. S10D, E). In addition, there are no identifiable overall structural similarities with the other subtype family members Cas13b (23, 24) or Cas13d (25). Thus, our structural analysis and biochemical tests suggest that AcrVIA1 is limited to neutralizing only the L. seeligeri type VI-A CRISPR-Cas immune response.
AcrVIA1 enables complete evasion of type VI-A CRISPR-Cas immunity
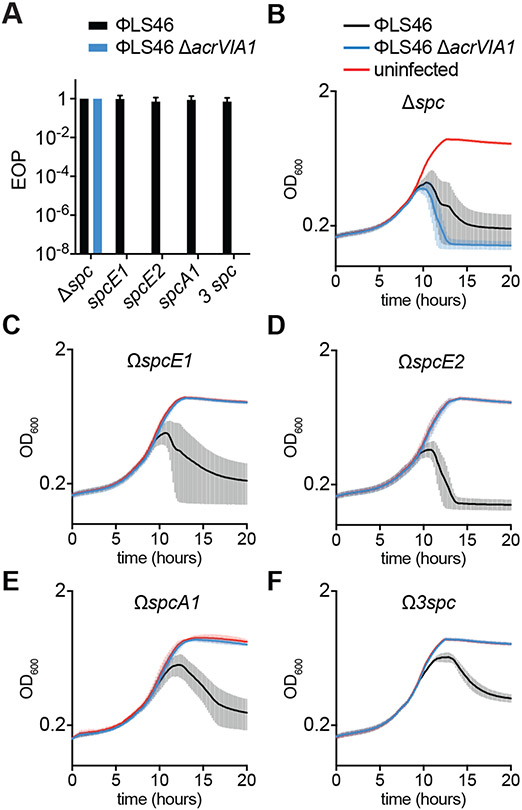
Previously described anti-CRISPRs that inhibit type I and II CRISPR systems require multiple rounds of infection to completely inhibit anti-phage immunity, and fail in conditions of strong CRISPR defense or low viral load (26, 27). To investigate if AcrVIA1 also displayed limited inhibition capabilities, we first tested its efficacy in conditions of weak or strong type VI-A CRISPR-Cas immunity, by infecting cells harboring either one or three targeting spacers, respectively (Fig. 1B). As a control we performed infections with the ϕLS46 ΔacrVIA1 mutant phage and measured the efficiency of plaquing (EOP) in the different host backgrounds. When compared to infection of hosts without targeting spacers, all three individual spacers as well as the triple combination provided efficient immunity against this mutant, reducing the EOP by at least eight orders of magnitude, below our limit of detection. In contrast, immunity was completely abrogated (~100% EOP) during infections with wild-type ϕLS46, when plating on either the single-spacer or triple-spacer strains (Fig. 4A). The 100% EOP value obtained indicated that each viral particle was able to inhibit Cas13a and form a visible plaque. To test this further, we performed infection of liquid cultures of L. seeligeri ΔRM at an extremely low MOI, 0.000001, and followed their growth over time. In the absence of a targeting spacer, both wild-type and ΔacrVIA1 mutant phage led to the lysis of the bacteria in the culture (Fig. 4B). While L. seeligeri strains harboring a single ϕLS46-targeting spacer were immune to the ΔacrVIA1 mutant phage, wild-type ϕLS46 was able to lyse the cultures (Figs. 4C-E, S3C, D, S11), showing that AcrVIA1 efficiently inhibits type VI-A CRISPR immunity even in conditions of low MOI. Infection of the strain containing three targeting spacers resulted in a delay in lysis (Fig. 4F), consistent with the stronger immunity provided by the presence of multiple spacers, yet still led to inhibition of the type VI-A CRISPR immune response. To explore the mechanism of inhibition further, we performed RNA-seq over the course of ϕLS46 infection to determine the timing and expression of each protospacer. We found that many of the spacers used in this study target phage transcripts that were abundantly produced within 10 minutes of infection (those targeted by spcA1, spcE1, and spcE2 for example, Fig. S12) and yet were unable to provide Cas13a-mediated immunity in the presence of AcrVIA1. Thus, AcrVIA1 overcame immunity against protospacers transcribed very early (even earlier than the inhibitor itself as in the case of target A1) during the phage lytic cycle. Altogether, these results demonstrate that AcrVIA1 can enable viral propagation in conditions that are extremely unfavorable for the success of Cas13a inhibition (rapid targeting, low MOI and multiple Cas13a targeting spacers) that normally would lead to the failure of type I and II Acrs (26, 27).
Figure 4. AcrVIA1 enables full phage escape from type VIA CRISPR-Cas immunity.
(A) Efficiency of plaquing (relative to the number of plaques formed in lawns of L. seeligeri ΔRM Δspc) of phages ϕLS46 or ϕLS46 ΔacrVIA1 in lawns of bacteria expressing spcA1, spcE1, spcE2 or all three (3 spc). Error bars represent SEM from 3 biological replicates. (B-F) Growth of L. seeligeri ΔRM Δspc (B), ΔRM ΩspcE1 (C), ΔRM ΩspcE2 (D), ΔRM ΩspcA1 (E) and ΔRM Ω3spc (F), measured as OD600 over time, infected with ϕLS46 or ϕLS46 ΔacrVIA1 phages, or uninfected. The average curves of three different replicates are reported, with +/− SEM values shown.
Discussion
AcrVIA1 inhibits Cas13a by interacting with the crRNA-exposed face of nuclease and making specific contacts with both protein and guide RNA residues that prevent the binding of a complementary target RNA and activation of Cas13a RNase function. In heterologous hosts, AcrVIA1 could be a useful component of the Cas13 toolbox, allowing control of this nuclease during editing, knock-down and/or visualization of RNA molecules (28, 29), as is the case for other recently found type VI-A anti-CRISPRs (18). More importantly, in its natural host, AcrVIA1 can completely neutralize type VI-A CRISPR-Cas immunity against ϕLS46, even in unfavorable conditions for inhibition such as multiple protospacer targeting and low viral load. We believe this to be a consequence of the lack of phage DNA clearance during the type VI response (11). This would lead to a continuous transcription and translation of AcrVIA1 and progressive neutralization of Cas13a. Assuming that the collateral RNA degradation generated by activation of Cas13a in Listeria hosts allows a low level of AcrVIA1 transcription and translation, enough inhibitor will accumulate to inactivate all the Cas13a molecules inside the bacterial cell. This is in contrast to type I and II Acrs, whose initial production inhibits only a fraction of Cascade-Cas3 and Cas9 molecules, respectively, and the Acr-harboring phage is destroyed by the nucleases that remain active (26, 27). Gradual inhibition of Cas13a after phage infection would require AcrVIA1 to constantly capture the Cas13acrRNA molecules that disengage from the target RNA and prevent them from finding their targets again. Alternatively, the inhibitor could displace the target RNA molecules from activated Cas13acrRNA nucleases. Such a mechanism would be especially effective when the target RNA is a transcript that is produced, and therefore activates Cas13a, before AcrVIA1 is generated. Finally, the genetic, biochemical and structural findings of this work highlight the astounding diversity of molecular strategies at play during the host-viral evolutionary arms race.
Supplementary Material
ACKNOWLEDGEMENTS.
We thank all members of the Marraffini lab for helpful discussions, Richard Calendar (UC Berkeley) for A118 and U153 listeriophages, Samuel Kilcher (ETH Zürich) for plasmid pSK1, David Rudner (Harvard Medical School) for anti-σA antibody, and Michael Jason De La Cruz for assistance in data collection on the Sloan Kettering Titan KRIOS cryo-electron microscope and Ed Eng of the New York Structural Biology Center for assistance in cryo-EM data processing.
Funding. Support for this work comes from the National Institute of Health Director’s Pioneer Award 1DP1GM128184-01 (to LAM), funds from the Geoffrey Beene Cancer Research Center and NIH GM129430 to DJP, and by Memorial Sloan-Kettering Cancer Center Core Grant (P30CA008748). LAM is an investigator of the Howard Hughes Medical Institute. AJM is a Helen Hay Whitney postdoctoral fellow.
Footnotes
Competing interests. LAM and AJM are co-inventors on a patent application filed by The Rockefeller University relating to work in this study. LAM is a founder and advisor of Intellia Therapeutics and Eligo Biosciences.
Data and materials availability: The L. seeligeri strain LS46 and LS59 draft genome assemblies have been deposited at GenBank (Accession JAAIYQ000000000). Raw genome sequencing reads and RNA-seq reads have been deposited at the Sequence Read Archive under accession number (PRJNA607241). The atomic coordinates have been deposited in the Protein Data Bank with the codes 6VRC (Cas13acrRNA) and 6VRB (AcrVIA1-Cas13acrRNA). The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with the codes EMD-21367 (Cas13acrRNA) and EMD-21366 (AcrVIA1-Cas13acrRNA).
References and notes
- 1.Barrangou R et al. , CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Marraffini LA, Sontheimer EJ, CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD, Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E, Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol 60, 174–182 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Pourcel C, Salvignol G, Vergnaud G, CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Brouns SJ et al. , Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson RN, van Erp PB, Sternberg SH, Wiedenheft B, Conformational regulation of CRISPR-associated nucleases. Curr. Opin. Microbiol 37, 110–119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makarova KS et al. , Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol 18, 67–83 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abudayyeh OO et al. , C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.East-Seletsky A et al. , Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meeske AJ, Nakandakari-Higa S, Marraffini LA, Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature 570, 241–245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR, Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rauch BJ et al. , Inhibition of CRISPR-Cas9 with Bacteriophage Proteins. Cell 168, 150–158 e110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pawluk A et al. , Naturally Occurring Off-Switches for CRISPR-Cas9. Cell 167, 1829–1838 e1829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhoobalan-Chitty Y, Johansen TB, Di Cianni N, Peng X, Inhibition of Type III CRISPR-Cas Immunity by an Archaeal Virus-Encoded Anti-CRISPR Protein. Cell 179, 448–458 e411 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Marino ND et al. , Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science 362, 240–242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watters KE, Fellmann C, Bai HB, Ren SM, Doudna JA, Systematic discovery of natural CRISPR-Cas12a inhibitors. Science 362, 236–239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin P et al. , CRISPR-Cas13 Inhibitors Block RNA Editing in Bacteria and Mammalian Cells. Mol. Cell, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rocourt J, Schrettenbrunner A, Hof H, Espaze EP, [New species of the genus Listeria: Listeria seeligeri]. Pathol Biol (Paris) 35, 1075–1080 (1987). [PubMed] [Google Scholar]
- 20.Osuna BA et al. , Critical Anti-CRISPR Locus Repression by a Bi-functional Cas9 Inhibitor. Cell Host Microbe, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L et al. , The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Cell 170, 714–726 e710 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Liu L et al. , Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities. Cell 168, 121–134 e112 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Slaymaker IM et al. , High-Resolution Structure of Cas13b and Biochemical Characterization of RNA Targeting and Cleavage. Cell Rep 26, 3741–3751 e3745 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang B et al. , Structural insights into Cas13b-guided CRISPR RNA maturation and recognition. Cell Res. 28, 1198–1201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang C et al. , Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d. Cell 175, 212–223 e217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borges AL et al. , Bacteriophage Cooperation Suppresses CRISPR-Cas3 and Cas9 Immunity. Cell 174, 917–925 e910 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landsberger M et al. , Anti-CRISPR Phages Cooperate to Overcome CRISPR-Cas Immunity. Cell 174, 908–916 e912 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abudayyeh OO et al. , RNA targeting with CRISPR-Cas13. Nature 550, 280–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox DBT et al. , RNA editing with CRISPR-Cas13. Science 358, 1019–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon R, Priefer U, Puhler A, A Broad Host Range Mobilization System for Invivo Genetic-Engineering - Transposon Mutagenesis in Gram-Negative Bacteria. Bio-Technology 1, 784–791 (1983). [Google Scholar]
- 31.Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R, Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol 184, 4177–4186 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meeske AJ, Marraffini LA, RNA Guide Complementarity Prevents Self-Targeting in Type VI CRISPR Systems. Mol. Cell 71, 791–801 e793 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demarre G et al. , A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli xhost strains. Res. Microbiol 156, 245–255 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Fujita M, Temporal and selective association of multiple sigma factors with RNA polymerase during sporulation in Bacillus subtilis. Genes Cells 5, 79–88 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheres SH, RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan YZ et al. , Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersen EF et al. , UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Adams PD et al. , PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemaitre JP, Duroux A, Pimpie R, Duez JM, Milat ML, Listeria phage and phage tail induction triggered by components of bacterial growth media (phosphate, LiCl, nalidixic acid, and acriflavine). Appl. Environ. Microbiol 81, 2117–2124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.