Abstract
Background: Glaucoma is a leading cause of irreversible blindness. Remodeling of the scleral extracellular matrix (ECM) plays an important role in the development of glaucoma. The aim of this study was to identify the key genes and pathways for the ECM remodeling of sclera in glaucoma by bioinformatics analysis and to explore potential therapeutic agents for glaucoma management.
Methods: Genes associated with glaucoma, sclera and ECM remodeling were detected using the text mining tool pubmed2ensembl, and assigned Gene Ontology (GO) biological process terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using the GeneCodis program. A protein-protein interaction (PPI) network was constructed by STRING and visualized in Cytoscape, module analysis was performed using the Molecular Complex Detection (MCODE) plugin, and GO and KEGG analyses of the gene modules were performed using the Database of Annotation, Visualization and Integrated Discovery (DAVID) platform. The genes that clustered in the significant module were selected as core genes, and functions and pathways of the core genes were visualized using ClueGO and CluePedia. Lastly, the drug-gene interaction database was used to explore drug-gene interactions of the core genes to find drug candidates for glaucoma.
Results: We identified 125 genes common to “Glaucoma”, “Sclera”, and “ECM remodeling” by text mining. Gene functional enrichment analysis yielded 30 enriched GO terms and 20 associated KEGG pathways. A PPI network that included 60 nodes with 249 edges was constructed, and three gene modules were obtained using the MCODE. We selected 13 genes that clustered in module 1 as core candidate genes that were associated mainly with ECM degradation and cell proliferation and division. The HIF-1 signaling pathway, FOXO signaling pathway, PI3K-Akt signaling pathway and TGFB signaling pathway were found to be enriched. We found that 11 of the 13 selected genes could be targeted by 26 existing drugs.
Conclusions: The results showed that VEGFA, TGFB1, TGFB2, TGFB3, IGF2, IGF1, EGF, FN1, KNG1, TIMP1, SERPINE1, THBS1, and VWF were potential key genes involved to scleral ECM remodeling. Furthermore, 26 drugs were identified as potential therapeutic agents for glaucoma treatment and management.
Keywords: glaucoma, sclera, extracellular matrix, text mining, drug discovery
Introduction
Glaucoma is a progressive optic neuropathy characterized by progressive degeneration of retinal ganglion cells and optic nerve head (ONH) axons 1. The global prevalence of glaucoma is 3.54% in the population aged 40-80 years, and glaucoma was estimated to have affected 76 million people worldwide in 2020 2. Glaucoma is a leading cause of irreversible blindness, and it was estimated that 9.4 million people are bilaterally blind as a result of glaucoma worldwide 3. Glaucoma-related visual impairment can significantly reduce the quality of life, causing heavy economic burden to individuals and by extension to the society at large. With an aging global population, the number of patients with glaucoma is projected to grow to 111.8 million by 2040 2, and the overall burden of glaucoma treatment will continue to increase.
Currently, lowering of intraocular pressure (IOP) (by medication, laser/or filtering surgery) is the only available option to delay the development and progression of glaucoma 4. However, treatments that reduce IOP prevent vision loss in only 50%-70% of patients with glaucoma, and thus nearly a half of the patients will experience glaucomatous progression despite the IOP within the normal range 5, 6. Clearly, a more effective therapy for the treatment and prevention of glaucoma is urgently needed.
Retinal ganglion cell axons converge at the ONH, pass through the laminar cribrosa, enter the scleral canal and exit the eye 7. The biomechanical transmission of stress from IOP produces strain in the ONH and leads to retinal ganglion cell axonal injury and vascular perfusion deficiency, eventually leading to glaucomatous optic neuropathy 8, 9. ONH is the principal site of glaucomatous damage 10, and the IOP-related biomechanical response of ONH is determined by the mechanical properties of sclera, especially the adjacent peripapillary sclera 11.
Stiffening of the sclera leading to scleral rigidity is one of the primary structural changes in glaucoma 12, 13. Sclera consists of fibroblasts and extracellular matrix (ECM), which is made up of collagen, elastin, and proteoglycans 8, 14. Scleral fibroblasts are mechanosensitive, and elevated IOP can induce them to differentiate into myofibroblasts, resulting in ECM remodeling, which involves scleral fibroblast proliferation, followed by fibrosis of the sclera with alterations in the biomechanical properties 14, 15. Finite element modeling and ex vivo studies have shown that the biomechanical strain within the laminar cribrosa depends on the stiffness of the sclera, and that increased peripapillary sclera stiffness can reduce the laminar cribrosa strain 16, 17. In an in vivo experiment, mice with microfibril deficiency were shown to have increased susceptibility to glaucomatous damage 18, whereas increased mechanical stiffness of the sclera was found to result in increased glaucomatous damage in a mouse model 19. Therefore, scleral ECM remodeling might be a suitable drug target for preventing the biomechanical damage associated with glaucoma.
The discovery of new drug therapies using conventional approaches can be time-consuming and costly, whereas repurposing an existing drug to treat a condition beyond its original intent may be a more effective and less expensive alternative 20. Text mining of biomedical literature is as an effective method for hypothesis generation, because it can reveal novel associations between genes and pathologies 21. By combining text mining with biological knowledge and other analytical tools, new information about the potential to repurpose existing drugs can be obtained 22. The aim of this study was to investigate therapeutic targets and new drug therapies for glaucoma by mining the available published literature combined with biological databases and other analytical tools.
Methods
Text mining
Text mining was performed using pubmed2ensembl (http://pubmed2ensembl.ls.manchester.ac.uk/), a publicly available resource that links over 2,000,000 articles in PubMed to nearly 150,000 genes from 50 species in Ensembl 23. We used the search terms “Glaucoma”, “Sclera” and “ECM remodeling” from 100,000 relevant document IDs to produce a list of genes. “Homo sapiens (GRCh37)” was selected as the species dataset and “filter on MEDLINE: PubMed ID” was set to constrain the query result. The unduplicated genes were extracted and the intersection of gene hits from the three gene sets was retrieved as the text mining genes (TMGs).
Biological process and pathway enrichment analysis of TMGs
We used GeneCodis (http://genecodis.cnb.csic.es/) to perform an enrichment analysis of the TMGs related to scleral ECM remodeling in glaucoma. GeneCodis is a web‑based server for functional analysis of gene lists that integrates different sources of information 24. The TMGs were used as the input set and analyzed using the gene ontology (GO) biological process categories, and genes with significantly enriched biological processes were selected and used for further analysis of enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations. P =1.00E-07 was set as the cutoff. Genes involved in the significantly enriched KEGG pathways were used for further analysis.
Integration of protein-protein interaction (PPI) network and module analysis
The Search Tool for the Retrieval of Interacting Genes (STRING, https://string-db.org/cgi/input.pl) is an online database resource that covers approximately 24.6 million proteins and more than 3.1 billion interactions originating from 5090 organisms 25. We used STRING (version 11.0) to construct PPI networks of the selected genes. The highest confidence score (0.900) was set as the minimum required interaction score. Molecular interaction networks were visualized using Cytoscape 26, and significant gene modules (clusters) from the PPI network were detected using the Molecular Complex Detection (MCODE) built in Cytoscape, with the following criteria: degree cutoff =2, node score cutoff=0.2, k-core=2, max depth=100 27.
Gene ontology and KEGG pathway enrichment analysis of module genes
The GO functional and KEGG pathway enrichment analyses of significant module genes were performed using Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/), an online tool for gene functional analysis 28. P < 0.05 was set as the cutoff. The GO (http://www.geneontology.org) database contains terms for the functional classification for genomic data under three main categories: biological processes, cellular component, and molecular function 29. KEGG (http://www.genome.ad.jp/kegg/) is a knowledge base for systematic analysis, annotation, and visualization of gene functions 30. The GO functional and KEGG pathway enrichment analyses of the core genes was performed and visualized using the Cytoscape plugins ClueGO (version 2.5.7), and CluePedia (version 1.5.7) 31. P < 0.01 was considered statistically significant.
Drug-gene interactions
The drug-gene interaction database (DGIdb, www. dgidb.org) is a web resource that consolidates disparate data sources that describe drug-gene interactions and gene druggability 32. We used DGIdb (version 3.0) to explore drug-gene interactions in the significant module genes that were identidied as the potential targets for existing drugs or compounds.
Results
Identification of text mining genes (TMGs)
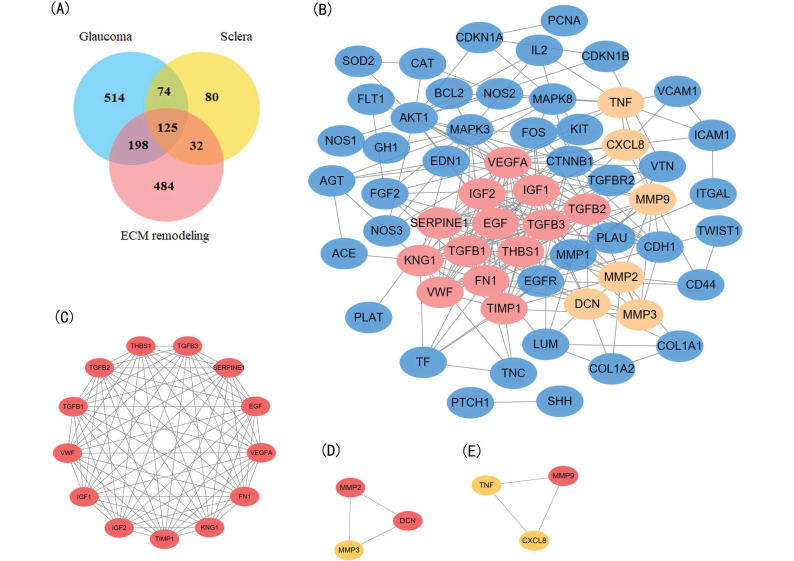
Using the text mining strategy described in the methods section (Figure 1), we obtain 911 unique genes related to glaucoma, 839 related to ECM remodeling, 311 related to sclera; among them, 125 genes were related to all three, and thus we considered them to be involved in scleral ECM remodeling in glaucoma (Figure 2A, Table S1).
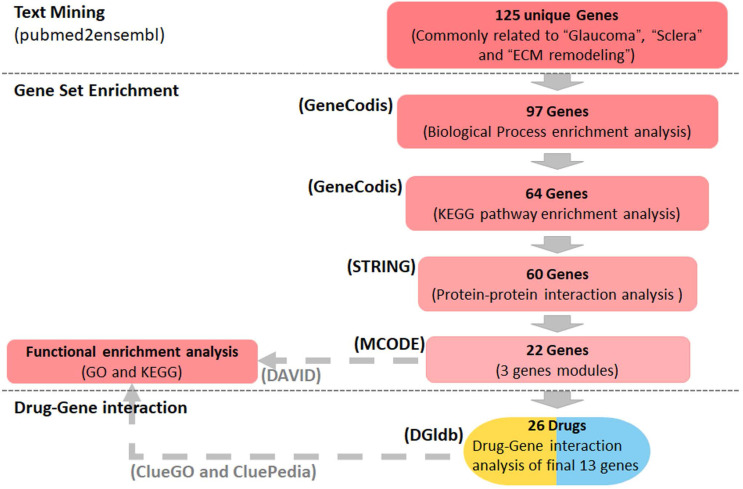
Figure 1.
Summary of the study design. Text mining was conducted using pubmed2ensembl to identify genes associated with glaucoma, sclera, and extracellular matrix (ECM) remodeling. Gene set enrichment was performed using GeneCodis to detect genes enriched in gene ontology (GO) biological process terms and KEGG pathways. STRING and MCODE were used to construct a protein-protein interaction network and identify modules. The GO biological process terms and KEGG pathways were analyzed using DAVID and ClueGO. The drug list was obtained based on the gene-drug interaction analysis conducted using the drug-gene interaction database (DGIdb). KEGG: Kyoto Encyclopedia of Genes and Genomes.
Figure 2.
Identification and enrichment analysis of the text mining genes (TMGs). (A) Venn diagram of the TMGs related to glaucoma, extracellular matrix (EMC) remodeling, and sclera. The 125 genes that were common were considered to be related to scleral EMC remodeling. (B) The protein-protein interaction (PPI) network of the 64 target TMGs was visualized using Cytoscape. (C-E) The three modules were obtained from PPI network using MCODE. (C) Module 1, the most significant module with 13 nodes; (D) Module 2; (E) Module 3.
Enrichment analysis of TMGs
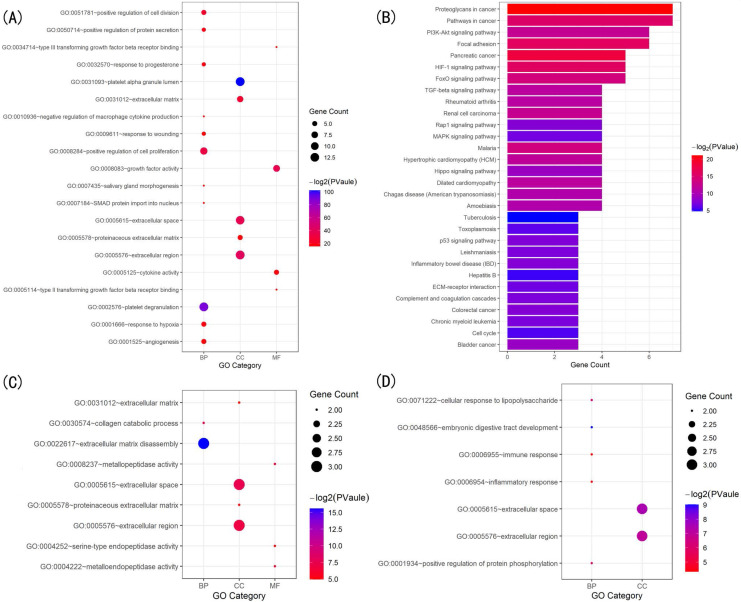
The GO biological process terms and KEGG pathway enrichment analyses using GeneCodis (with P=1.00E-07 as the cutoff) to identified the most enriched terms closely related to the pathology of scleral ECM remodeling in glaucoma. Thirty significantly enriched GO biological process annotations of 97 unique genes were identified. Among them, the five most enriched terms were “extracellular matrix organization” (P=3.47306E-27), “platelet degranulation” (P=2.44362E-19), “cytokine-mediated signaling pathway” (P=3.99419E-18), “response to drug” (P=3.82E-14), and “positive regulation of cell population proliferation” (P=1.45E-13), for 31, 20, 25, 22, and 27 TMGs, respectively (Table 1). Other highly enriched biological process terms included “response to hypoxia”, “wound healing”, “regulation of cell population proliferation”, “response to mechanical stimulus”, “extracellular matrix disassembly”, and “aging” (Table S2).
Table 1.
Top 15 enriched gene ontology (GO) biological process terms assigned to the text mining genes
| Process | Genes in query set |
Total genes in genome | Corrected hypergeometric P value | Genes |
|---|---|---|---|---|
| Extracellular matrix organization | 31 | 257 | 3.47E-27 | CDH1, CD44, VWF, VTN, VCAM1, TNF, THBS1, TGFBI, SPARC, BGN, SERPINE1, NID1, MMP14, MMP9, MMP3, MMP2, MMP1, MFAP2, LUM, ITGAL, ICAM1, TNC, FN1, FGF2, FBN1, ELN, COL18A1, DCN, VCAN, COL1A2, COL1A1. |
| Platelet degranulation | 20 | 125 | 2.44E-19 | CLU, MMRN1, VWF, VEGFA, TIMP1, THBS1, TGFB3, TGFB2, TGFB1, TF, SPARC, SOD1, SERPINE1, KNG1, IGF2, IGF1, APOA1, FN1, ALB, EGF. |
| Cytokine-mediated signaling pathway | 25 | 290 | 3.99E-18 | CDKN1A, CD4, VIM, VEGFA, VCAM1, TWIST1, TNF, TIMP1, TGFB1, BCL2, NOS2, MMP9, MMP3, MMP2, MMP1, KIT, IL8, IL2, ICAM1, FOS, FN1, FGF2, AKT1, F3, COL1A2. |
| Response to drug | 22 | 305 | 3.82E-14 | CDKN1B, CDKN1A, CDH1, CAT, TIMP2, THBS1, TGFBR2, TGFB2, SST, SOD1, BCL2, REN, PTCH1, APOA1, ICAM1, APEX1, FOS, ENG, EDN1, COL18A1, CTNNB1, COL1A1. |
| Positive regulation of cell population Proliferation | 27 | 557 | 1.45E-13 | CDKN1B, CNOT8, VEGFA, TIMP1, THBS1, TGFBR2, TGFB3, TGFB2, TGFB1, SHH, RPS4X, BCL2, KIT, IL2, IGF2, IGF1, TNC, FN1, FLT1, FGF2, AKT1, AGT, EGFR, EGF, EDN1, COL18A1, CTNNB1. |
| Response to hypoxia | 17 | 174 | 7.16E-13 | CDKN1B, CAT, VEGFA, THBS1, TGFBR2, TGFB3, TGFB2, BMP2, PLAU, PLAT, NOS2, NOS1, MMP14, MMP2, ICAM1, ENG, EDN1. |
| Wound healing | 14 | 103 | 1.99E-12 | TIMP1, TGFBR2, TGFB3, TGFB2, SPARC, IGF1, TNC, GSN, FN1, FGF2, ENG, EGFR, DCN, COL1A1. |
| Regulation of cell population Proliferation | 18 | 222 | 2.05E-12 | CLU, CDKN1B, TNF, TGFBR2, TGFB3, TGFB2, TGFB1, SPARC, SHH, PTCH1, PLAU, NOS2, KIT, TNC, ENG, AGT, EGFR, CTNNB1. |
| Negative regulation of gene expression | 20 | 310 | 4.16E-12 | CDKN1A, CD34, VEGFA, TNF, TGFB2, TGFB1, BMP2, SHH, SERPINF1, NOS2, MAX, IL8, IGF1, FGF2, AKT1, ENG, AGT, EDN1, ACE, CTNNB1. |
| Positive regulation of gene expression | 24 | 498 | 4.56E-12 | CLU, CD34, VIM, VEGFA, TWIST1, TNF, TGFB1, BMP2, SHH, MAPK8, MAPK3, NOS3, KIT, IL8, IGF1, TNC, GSN, GJA1, FN1, AKT1, F3, ENG, EGF, CTNNB1. |
| Positive regulation of cell migration | 18 | 255 | 1.66E-11 | VEGFA, THBS1, TGFB1, BMP2, SOD2, PLAU, MMP14, MMP9, KIT, IGF1, HSPA5, FLT1, F3, EGFR, EGF, EDN1, COL18A1, COL1A1. |
| Aging | 15 | 178 | 1.57E-10 | CAT, CALCA, TIMP2, TIMP1, TGFBR2, TGFB3, SOD1, MAPK3, SERPINF1, APEX1, GSN, FOS, AKT1, AGT, DCN. |
| Cellular protein metabolic process | 16 | 226 | 3.55E-10 | CALCA, TIMP1, TGFBI, TF, MMP2, MMP1, KNG1, IGF2, IGF1, APOA1, TNC, GSN, FN1, ALB, FBN1, VCAN. |
| Regulation of blood pressure | 10 | 63 | 1.50E-09 | CD34, CALCA, SOD2, SOD1, REN, NOS3, AGT, EDN1, ACE, COL1A2. |
| Positive regulation of MAP kinase activity | 10 | 63 | 1.50E-09 | VEGFA, TNF, TGFB1, KIT, GH1, FLT1, FGF2, EGFR, EGF, EDN1. |
The KEGG pathway enrichment analysis identified 20 significant pathways that involved 64 TMGs. The five most significantly enriched pathways were “AGE-RAGE signaling pathway in diabetic complications” (P=1.04E-22), “Proteoglycans in cancer” (P=3.04E-19), “Pathways in cancer” (P=1.88327E-16), “HIF-1 signaling pathway” (P=1.17E-12), and “Relaxin signaling pathway” (P= 1.34E-12), involving 23, 26, 34, 16, and 17 TMGs, respectively (Table 2). Other highly enriched pathways included “PI3K-Akt signaling pathway”, “MAPK signaling pathway”, “Rheumatoid arthritis”, “FOXO signaling pathway”, “Focal adhesion” and “Endocrine resistance” (Table S3).
Table 2.
Top 10 enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways assigned to the text mining genes
| Process | Genes in query set | Total genes in genome | Corrected hypergeometric P value | Genes |
|---|---|---|---|---|
| AGE-RAGE signaling pathway in diabetic complications | 23 | 100 | 1.04E-22 | CDKN1B, VEGFA, VCAM1, TNF, TGFBR2, TGFB3, TGFB2, TGFB1, BCL2, MAPK8, MAPK3, SERPINE1, NOS3, MMP2, IL8, ICAM1, FN1, AKT1, F3, AGT, EDN1, COL1A2, COL1A1. |
| Proteoglycans in cancer | 26 | 205 | 3.04E-19 | CDKN1A, CD44, VTN, VEGFA, TWIST1, TNF, THBS1, TGFB2, TGFB1, SHH, PTCH1, MAPK3, PLAU, MMP9, MMP2, LUM, IGF2, IGF1, FN1, FGF2, AKT1, EGFR, DCN, CTNNB1, COL1A2, COL1A1. |
| Pathways in cancer | 34 | 531 | 1.88E-16 | CDKN1B, CDKN1A, CDH1, VEGFA, TGFBR2, TGFB3, TGFB2, TGFB1, BMP2, SHH, BCL2, PTCH1, MAPK8, MAPK3, NOS2, MMP9, MMP2, MMP1, MAX, KNG1, KIT, IL8, IL2, IGF2, IGF1, FOS, FN1, FGF2, AKT1, AGT, EGFR, EGF, EDN1, CTNNB1. |
| HIF-1 signaling pathway | 16 | 109 | 1.17E-12 | CDKN1B, CDKN1A, VEGFA, TIMP1, TF, BCL2, MAPK3, SERPINE1, NOS3, NOS2, IGF1, FLT1, AKT1, EGFR, EGF, EDN1. |
| Relaxin signaling pathway | 17 | 129 | 1.34E-12 | VEGFA, TGFBR2, TGFB1, MAPK8, MAPK3, NOS3, NOS2, NOS1, MMP9, MMP2, MMP1, FOS, AKT1, EGFR, EDN1, COL1A2, COL1A1. |
| Chagas disease (American trypanosomiasis) | 15 | 102 | 6.57E-12 | TNF, TGFBR2, TGFB3, TGFB2, TGFB1, MAPK8, MAPK3, SERPINE1, NOS2, KNG1, IL8, IL2, FOS, AKT1, ACE. |
| Bladder cancer | 11 | 41 | 1.05E-11 | CDKN1A, CDH1, VEGFA, THBS1, MAPK3, MMP9, MMP2, MMP1, IL8, EGFR, EGF. |
| PI3K-Akt signaling pathway | 23 | 354 | 5.42E-11 | CDKN1B, CDKN1A, VWF, VTN, VEGFA, THBS1, BCL2, MAPK3, NOS3, KIT, IL2, IGF2, IGF1, TNC, GH1, FN1, FLT1, FGF2, AKT1, EGFR, EGF, COL1A2, COL1A1. |
| Colorectal cancer | 13 | 86 | 1.41E-10 | CDKN1A, TGFBR2, TGFB3, TGFB2, TGFB1, BCL2, MAPK8, MAPK3, FOS, AKT1, EGFR, EGF, CTNNB1. |
| Prostate cancer | 13 | 97 | 6.18E-10 | CDKN1B, CDKN1A, BCL2, MAPK3, PLAU, PLAT, MMP9, MMP3, IGF1, AKT1, EGFR, EGF, CTNNB1. |
PPI network construction, modular analysis, and identification of core candidate genes
A PPI network was constructed for the 64 target genes using STRING with confidence score >0.900. The network had a total of 60 nodes with 249 edges (Figure 2B). Among the 60 nodes, 22 hub node genes were identified with filtering node degree ≥10 (Table 3). Modular analysis was performed using the MCODE built in Cytoscape, and three modules were obtained (Figure 2C-E).
Table 3.
Hub node genes in the protein-protein interaction network identified with filtering node degree ≥10
| Number | Genes | Degree | MCODE Cluster | MCODE Node Status |
|---|---|---|---|---|
| 1 | VEGFA | 24 | Module 1 | Seed |
| 2 | IGF1 | 21 | Module 1 | Clustered |
| 3 | EGF | 20 | Module 1 | Clustered |
| 4 | FN1 | 20 | Module 1 | Clustered |
| 5 | TGFB1 | 19 | Module 1 | Clustered |
| 6 | KNG1 | 19 | Module 1 | Clustered |
| 7 | TIMP1 | 18 | Module 1 | Clustered |
| 8 | TGFB3 | 17 | Module 1 | Clustered |
| 9 | AKT1 | 16 | -- | Unclustered |
| 10 | TGFB2 | 16 | Module 1 | Clustered |
| 11 | IGF2 | 16 | Module 1 | Clustered |
| 12 | SERPINE1 | 15 | Module 1 | Clustered |
| 13 | THBS1 | 15 | Module 1 | Clustered |
| 14 | MAPK3 | 15 | -- | Unclustered |
| 15 | EGFR | 14 | -- | Unclustered |
| 16 | CTNNB1 | 13 | -- | Unclustered |
| 17 | MMP2 | 13 | Module 2 | Clustered |
| 18 | VWF | 13 | Module 1 | Clustered |
| 19 | MMP9 | 12 | Module 3 | Clustered |
| 20 | DCN | 11 | Module 2 | Seed |
| 21 | CDH1 | 10 | -- | Unclustered |
| 22 | EDN1 | 10 | -- | Unclustered |
The GO enrichment analysis showed that the three modules were related primarily to ECM degrading, cell proliferation and division which play crucial role in scleral ECM remodeling (Figure 3A,C,D). The pathway enrichment analysis showed that the genes in module 1 were associated with the HIF-1 signaling pathway, FOXO signaling pathway, PI3K-Akt signaling pathway and TGFB signaling pathway (Figure 3B). No specific KEGG pathways were associated with the genes in module 2, and the genes in module 3 were significantly associated with metallopeptidase or metalloendopeptidase activity, NOD-like receptor or RIG-I-like receptor signaling pathway, and NF-kappa B signaling pathway (Table 4).
Figure 3.
Gene ontology (GO) and KEGG pathway analysis of the genes in the three modules. (A) Top 20 significantly enriched GO terms in module 1. (B) Significantly enriched KEGG pathways in module 1. (C) Significantly enriched GO terms in module 2. (D) Significantly enriched GO terms in module 3. The functional and pathway enrichment analyses were performed using DAVID. KEGG: Kyoto Encyclopedia of Genes and Genomes.
Table 4.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the genes in module 3
| Term | Description | Count | P value |
|---|---|---|---|
| hsa05161 | Hepatitis B | 3 | 4.41E-04 |
| hsa05219 | Bladder cancer | 2 | 0.011886 |
| hsa05144 | Malaria | 2 | 0.014197 |
| hsa05134 | Legionellosis | 2 | 0.015639 |
| hsa04621 | NOD-like receptor signaling pathway | 2 | 0.016216 |
| hsa04622 | RIG-I-like receptor signaling pathway | 2 | 0.02025 |
| hsa05133 | Pertussis | 2 | 0.021688 |
| hsa04064 | NF-kappa B signaling pathway | 2 | 0.025136 |
| hsa05323 | Rheumatoid arthritis | 2 | 0.025423 |
| hsa05142 | Chagas disease | 2 | 0.030011 |
| hsa04620 | Toll-like receptor signaling pathway | 2 | 0.030583 |
| hsa05146 | Amoebiasis | 2 | 0.030583 |
| hsa04668 | TNF signaling pathway | 2 | 0.030869 |
| hsa05160 | Hepatitis C | 2 | 0.038297 |
| hsa04932 | Non-alcoholic fatty liver disease (NAFLD) | 2 | 0.043423 |
| hsa05202 | Transcriptional misregulation in cancer | 2 | 0.047968 |
| hsa05164 | Influenza A | 2 | 0.049953 |
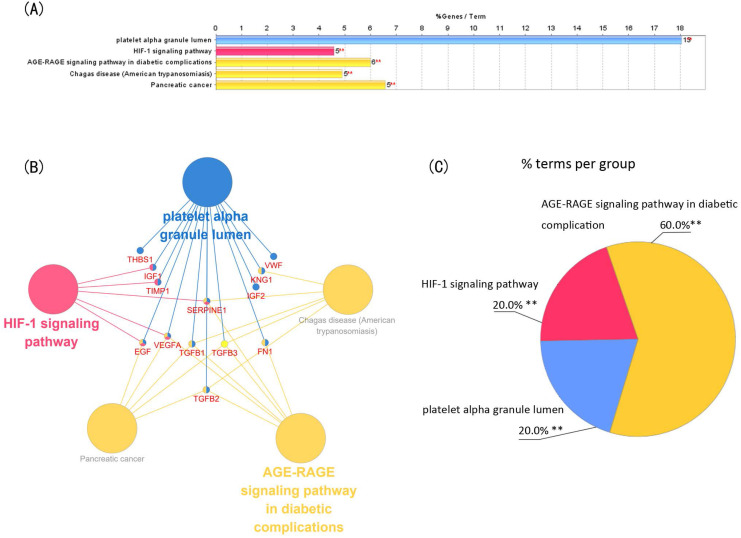
Module 1 contained 13 genes with 88 edges, all of which were hub genes indicating that module 1 has a vital role in the PPI network. We selected the 13 hub genes, TGFB1, TGFB2, TGFB3, VEGFA, IGF2, IGF1, EGF, FN1, KNG1, TIMP1, SERPINE1, THBS1 and VWF, as core candidate genes of the PPI network. The enrichment analysis indicated that these genes were significantly enriched in platelet alpha granule lumen, HIF-1 signaling pathway, and AGE-RAGE signaling pathway in diabetic complication (P <0.01, Figure 4).
Figure 4.
Function analysis of the 13 core genes in module 1. (A) Enriched gene ontology (GO) terms and KEGG pathways. (B) Functions and pathways of the core genes were visualized using ClueGO. (C) Distribution of the functions and pathways among the core genes. Each function or pathway is color coded. Corrected P <0.01 was considered statistically significant. KEGG: Kyoto Encyclopedia of Genes and Genomes.
Drug-gene interaction analysis of core genes
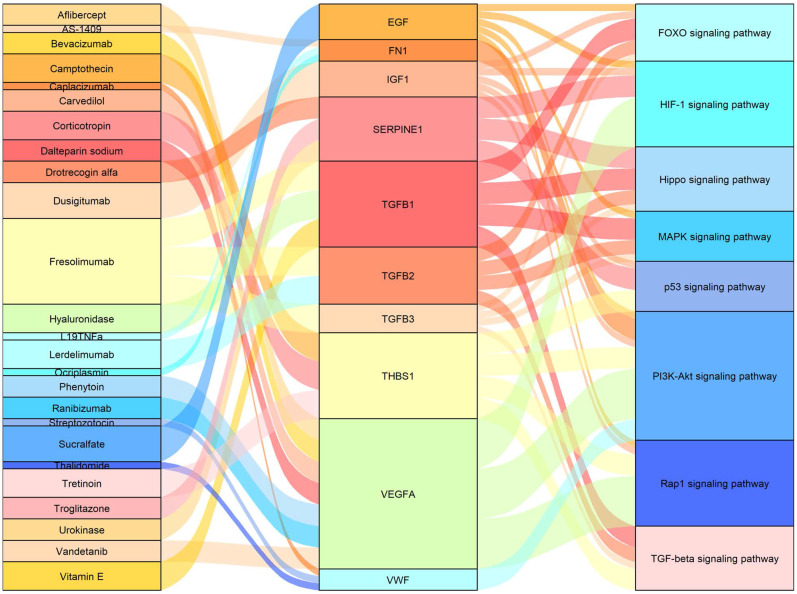
We used the 13 core genes as potential targets in a drug-gene interaction analysis, and obtained an initial list of 26 drugs (Table 5). Eleven of the 13 potential gene targets (the exceptions were IGF2 and TIMP1) were predicted to be involved in drug-gene interactions. The primary connection among drugs, genes and pathways are shown in Figure 5.
Table 5.
Details of the 26 drugs that potentially target 11 of the 13 core genes
| Number | Drug | Genes | Interaction | Score | Drug Class | Approved? | Reference (PubMed ID) |
|---|---|---|---|---|---|---|---|
| 1 | Ranibizumab** | VEGFA | inhibitor | 14 | antineoplastic agents, ocular vascular disorder agents | Yes* | 18046235 |
| 2 | Bevacizumab | VEGFA | inhibitor | 10 | antineoplastic agent, immunomodulating agents | Yes* | 15705858 |
| 3 | Aflibercept** | VEGFA | inhibitor | 7 | antineoplastic agent, ocular vascular disorder agents | Yes* | 22813448 |
| 4 | Carvedilol | VEGFA | other | 5 | vasodilator agents, neurotransmitter agents | Yes* | 15071347 |
| 5 | Dalteparin sodium | VEGFA | inhibitor | 4 | hematologic agents, fibrinolytic agents | Yes | 21091776 |
| 6 | Phenytoin | VEGFA | N/A | 2 | anticonvulsants | Yes* | 15458527 |
| 7 | Vandetanib | VEGFA | inhibitor | 1 | antineoplastic and immunomodulating agent | Yes* | None found |
| 8 | Dusigitumab | IGF1 | inhibitor | 1 | not available | No | None found |
| 9 | Sucralfate | EGF | inducer | 4 | antiulcer agent, antimuscarinics | Yes* | 8578218 |
| 10 | Ocriplasmin** | FN1 | cleavage | 3 | not available | Yes | 23193358 |
| 11 | AS-1409 | FN1 | binder | 2 | antineoplastic agent | No | None found |
| 12 | L19TNFa | FN1 | N/A | 1 | fibronectin binding agent | No | None found |
| 13 | Urokinase | SERPINE1 | N/A | 7 | thrombolytic agents, fibrinolytic agents | Yes* | 12709915 |
| 14 | Troglitazone | SERPINE1 | antagonist | 6 | hypoglycemic agents | No | 10871202 |
| 15 | Drotrecogin alfa | SERPINE1 | N/A | 5 | antithrombotic agents, fibrinolytic agents, | Yes* | 12004248 |
| 16 | Hydrochlorothiazide | KNG1 | inhibitor | 1 | antihypertensive agents, hypotensive agents | Yes* | None found |
| 17 | Tretinoin | THBS1 | N/A | 3 | antineoplastic agents, dermatologic agents, immunosuppressive agents | Yes* | 9447832 |
| 18 | Corticotropin | THBS1 | N/A | 2 | immunosuppressive agents | Yes* | 8698834 |
| 19 | Camptothecin | THBS1 | N/A | 2 | antineoplastic agents | Yes* | 16962673 |
| 20 | Caplacizumab | VWF | inhibitor | 3 | antithrombotic | No | None found |
| 21 | Streptozotocin | VWF | inhibitor | 3 | antibiotics, antineoplastic agents, immunosuppressive agents | Yes* | 16422885 |
| 22 | Thalidomide | VWF | N/A | 2 | antineoplastic agents | Yes* | 12871448 |
| 23 | Hyaluronidase | TGFB1 | inhibitor | 5 | not available | Yes* | 9435505 |
| 24 | Vitamin E | TGFB1 | N/A | 3 | antioxidants | Yes* | 1505665 |
| 25 | Lerdelimumab | TGFB2 | inhibitor | 2 | monoclonal antibody | No | None found |
| 26 | Fresolimumab | TGFB1, TGFB2, TGFB3 | inhibitor | 2 | antineoplastic agents | No | None found |
**Drugs that have been used to treat ophthalmic disease.
*Drugs that have been approved by the US Food and Drug Administration.
Figure 5.
Sankey diagram showing the primary connections among drugs, genes, and pathways.
Discussion
Glaucoma is the leading cause of irreversible blindness globally. Reduction of IOP is the only proven method to treat glaucoma 7. However, many patients who are treated with IOP-lowering therapies have poor prognosis, and nearly a half of patients with glaucoma experience glaucomatous progression despite IOP reduction 33. Therefore, more therapeutic targets and prognostic biomarkers need to be identified. Stiffening of the sclera, particularly in peripapillary sclera, is one of the primary structural changes in glaucoma. It has been shown that elevated IOP can induce scleral fibroblasts to differentiate into myofibroblasts, resulting in ECM remodeling, followed by stiffening of the sclera 14, 15. The ONH is the primary site of damage in glaucoma, and stiffening of the peripapillary sclera significantly impacts the biomechanics of the ONH that eventually influence the susceptibility to glaucomatous damage 16-19. Therefore, ECM remodeling might be an attractive drug target for the prevention and treatment of glaucoma.
In the present study, we identified 125 genes that might be involved in scleral ECM remodeling associated with glaucoma using text mining strategy (Figure 1). The enriched GO biological process terms assigned to these genes were associated mainly with scleral ECM remodeling, including “positive regulation of cell population Proliferation”34, “response to hypoxia” 35, “aging” 36, “wound healing” 37, “response to mechanical stimulus” 38 and “positive regulation of MAP kinase activity” 39 (Table 1). The PPI network and enrichment analysis identified 13 core genes, TGFB1, TGFB2, TGFB3, VEGFA, IGF2, IGF1, EGF, FN1, KNG1, TIMP1, SERPINE1, THBS1 and VWF, that were involved in the HIF-1 signaling pathway, TGFB signaling pathway, FOXO signaling pathway, and PI3K-Akt signaling pathway.
Transforming growth factor β (TGFB) belongs to a family of fibrogenic cytokine that includes TGFB1, TGFB2 and TGFB3 40. Inhibition of TGFB reduces α-SMA expression, myofibroblast differentiation, and proliferation of scleral fibroblasts in experimental glaucoma 41. Vascular endothelial growth factor A (VEGFA) is a central regulator of angiogenesis and vascular permeability, and is involved in angiogenesis, inflammatory cell migration, and fibroblast activity 42.VEGF-A can be induced by TGFB1 through the TGFB1-VEGF-A pathway, resulting in fibrosis in patients who undergo peritoneal dialysis 43. Inhibition of VEGF was shown to improve the surgical outcome of glaucoma surgery by reducing angiogenesis and fibrosis in experimental models 44. Insulin-like growth factor 1 (IGF1) is a pro‐fibrotic growth factor that stimulates fibroblast proliferation and collagen synthesis in idiopathic pulmonary fibrosis 45. IGF2 is involved in inflammation and fibrosis, and the IGF2 plasma levels were closely associated with the stages of liver fibrosis 46. Epidermal growth factor (EGF) is an essential growth factor in stimulating cell proliferation, which plays a role in the progression of fibrosis in the liver 47. Fibronectin 1(FN1) is involved in cell adhesion and migration, which is required for the accumulation of ECM components 48, and the interaction of FN1 with the cell surface is essential for TGFB-mediated activation of fibrogenic cells 49. SERPINE1, also called plasminogen activator inhibitor type 1 (PAI‐1), is a fibrinolysis inhibitor that contributes to fibrogenesis by decreasing the degradation of fibrin and other ECM proteins through the uPA/tPA/plasmin/MMP proteolytic system 50. Kininogen-1 (KNG1) is a multifunctional protein that plays an important role in fibrinolysis and inflammation 51. Tissue inhibitor of metalloproteinase 1 (TIMP1) is a glycoprotein in the TIMP family that plays a role in matrix remodeling by activating macrophages and inhibiting matrix metalloproteinases (MMPs) 52. Thrombospondin-1 (THBS1) belongs to the thrombospondin family of glycoproteins that are important components of the ECM 53. Gao et al. 54 demonstrated that THBS1 expression was positively correlated with scleral ECM remodeling. Von Willebrand factor (VWF), a glycoprotein that functions to bridge platelets with an exposed collagen surface, is associated with the severity of organ fibrosis 55. These 13 core genes are the potential key genes that may be involved in scleral ECM remodeling.
The HIF-1 signaling pathway, one of the enriched KEGG pathways, has been shown to promote scleral myofibroblast transdifferentiation by regulating the actin cytoskeleton pathway, as well as ECM remodeling via the ECM receptor interaction pathway 35. HIF-1α was shown to enhance VEGF-A expression and promote fibrinogenesis via the HIF-1α-VEGF pathway in bronchiolitis obliterans 56. The TGFB pathway is acts as a primary signaling pathway in fibrogenesis by modulating the fibroblast phenotype and function, inducing myofibroblast transdifferentiation and α-SMA expression, and promoting matrix accumulation 57. TGFB-signaling and the abundances of TGFB1, TGFB2 and TGFB-activating protein were found to be elevated in the sclera of glaucomatous eyes versus the control sclera 41. Proteins in the FOXO family are growth factors and stress-regulated transcription factors that are involved in cellular differentiation, apoptosis, and cell proliferation. FOXO proteins can interact with TGFB-activated proteins to prevent fibrosis through β-catenin/FOXO1 signaling 58. The PI3K-Akt signaling pathway is an important regulatory pathway that is involved in the regulation of cell proliferation, migration, differentiation and angiogenesis 59. The PI3K-Akt signaling pathway also is involved in the pathological processes of fibrosis by regulating fibroblast migration and differentiation to myofibroblasts 60. Inhibition of the PI3K/AKT signaling pathway was shown to prevent scarring of the filtering bleb of glaucoma filtration surgery by inhibiting human conjunctival fibroblast migration, proliferation and ECM synthesis 61. These signaling pathways may play essential roles in the scleral stiffening associated with glaucoma.
Potential drugs identified by the drug-gene interaction search were classified as mainly TGFB inhibitors, VEGFA inhibitors, IGF inhibitors, fibrinolytic agents, antineoplastic agents, or immunomodulating agents. The TGFB inhibitors were mainly of hyaluronidase, lerdelimumab and fresolimumab. Hyaluronidases are enzymes that regulate the remodeling of ECM. Hyaluronidases have been shown to inhibit the growth of pulmonary fibrosis and decrease TGFB production and collagen deposition in mice 62. Lerdelimumab is a monoclonal immunoglobulin antibody that specifically and potently neutralizes human TGFB2. Lerdelimumab was found to reduce subconjunctival fibrosis by inhibiting TGFB2-induced Tenon's fibroblast migration and proliferation in in vivo animal experiments 63. Previous phase I and phase II clinical studies demonstrated that lerdelimumab reduced subconjunctival fibrosis in patients with primary glaucoma who had undergone trabeculectomy 64. Fresolimumab is a monoclonal antibody of TGFB that reduced myofibroblast infiltration and inhibited TGFB-regulated gene expression in systemic sclerosis 65.
More than a quarter of identified drugs target VEGFA, including ranibizumab, bevacizumab, aflibercept, carvedilol, dalteparin sodium, phenytoin and vandetanib. Ranibizumab, a monoclonal antibody fragment against VEGF-A, inhibits vascular permeability and has antiproliferative and antimetabolic effects on various cell lines. Several studies have indicated that ranibizumab may prevent scarring after glaucoma filtering surgery through the VEGF pathway 66. Bevacizumab is an IgG1 humanized monoclonal antibody that has been used to negate the effect of VEGF-A. Bevacizumab is approved by the US Food and Drug Administration. In clinical ophthalmology, bevacizumab has been used widely in the management of ophthalmic diseases, including choroidal neovascularization, diabetic macular edema and macular edema 67. Aflibercept is a human fusion protein of the IgG Fc region and VEGF-receptor VEGF-A that also has been used widely in the management of ophthalmic diseases 68. Carvedilol, is a nonselective-blocker that also has α1-adrenergic blocking, antioxidant and antiproliferative effects. Carvedilol was shown to attenuate liver cirrhosis by inhibiting angiogenesis through the VEGF-Src-ERK signaling pathway in human umbilical vascular endothelial cells 69. Dalteparin sodium is a low molecular weight heparin that is used widely in the treatment of thromboembolisms. In rat, dalteparin sodium minimized hepatic fibrogenesis caused by chronic carbon tetrachloride treatment 70. Vandetanib is an oral receptor tyrosine kinase inhibitor that potently inhibits VEGF receptor (VEGFR) tyrosine kinase activity. Vandetanib inhibits not only growth factor-induced phosphorylation of VEGFR and epidermal growth factor receptor (EGFR), but also mitogen-activated protein kinase (MAPK) and protein kinase B (Akt) 71. In mice, vandetanib reduced the degree of fibrosis on cutaneous wound healing 72.
Dusigitumab is a human antibody of IGF1 and IGF2. An in vivo experiment demonstrated that dusigitumab inhibited tumor growth of mouse embryonic fibroblast 73. Sucralfate is a complex salt of sucrose sulfate and aluminum hydroxide that induces significant increases in the expression of EGF and TGFA in gastric mucosal cells 74. Ocriplasmin, a recombinant protein with intrinsic action on collagen and fibronectin, is an effective nonsurgical treatment for vitreomacular traction. In rat, the intravitreal administration of microplasmin degraded fibronectin in the vitreoretinal interface and outer retina 75. Urokinase, also known as urokinase-type plasminogen activator (uPA), is a strong plasminogen activator that is involved in cell migration, wound healing and tissue remodeling. The action of uPA on podocytes was found to be associated with the decreased prevalence of tubulointerstitial fibrosis 76. Hydrochlorothiazide is a thiazide diuretic that is used in the treatment of hypertension and edema. In rat, hydrochlorothiazide was shown to reduce ischemic heart failure-induced fibrosis remodeling by inhibiting the angiotensin II signaling pathway 77. Tretinoin, a retinoid metabolite of naturally occurring vitamin A, stimulated the formation of new collagen, stimulated fibroblasts, prevented collagen loss, and inhibited the induction of metalloproteinases 78. Camptothecin is a DNA topoisomerase I inhibitor that blocks DNA synthesis and down-regulates THBS1 expression in human thyroid carcinoma FTC-133 cells through the JNK/ATF-2 pathway 79. These drugs may have possible uses in the prevention of the scleral ECM remodeling in glaucoma. Further research is needed to confirm their possible new function.
Scleral ECM remodeling also is a key factor in the pathogenesis of myopia. In myopic human eyes, the sclera was found to be thinner, and the scleral ECM was decreased, compared with those in healthy eyes 80. The thinner sclera and the associated decrease in the synthesis of collagen, proteoglycan, and other scleral matrix components also have been observed in animal models of myopia 81-83. During myopia development, scleral ECM remodeling resulted in thinning of the sclera and reduced scleral resistance to IOP-related expansion which eventually contributed to excessive elongation of the ocular globe 84, 85. Scleral stiffening has been proposed as a therapy for myopia, and the 26 drugs identified in this study may also be potential therapeutic agents for the treatment of myopia.
In conclusion, we identified 13 hub genes, VEGFA, TGFB1, TGFB2, TGFB3, IGF2, IGF1, EGF, FN1, KNG1, TIMP1, SERPINE1, THBS1 and VWF, that may be involved in the scleral ECM remodeling associated with glaucoma. These genes were enriched in the HIF-1 signaling pathway, FOXO signaling pathway, PI3K-Akt signaling pathway and TGFB signaling pathway. We also identified 26 potential drugs that may be help to guide future glaucoma therapies. The absence of experimental validation is a limitation of this study, and further experimental studies are required to verify the results.
Supplementary Material
Supplementary tables.
Acknowledgments
We thank Dr. Bin Zhao (Official Wechat Account: SCIPhD) of Sheng Xin Zhu Shou. We thank Margaret Biswas, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding Support
This study was supported by Natural Science Foundation of Shanghai (18ZR1406000), National Science Foundation China (81790641).
Authors' Contributions
D.H., J.H.J and S.H.Q. conceived the study. D.H. and J.H.J collected the data. Z.L. and C.Z. performed the statistical analysis. D.H., J.H.J and Nived Moonasar wrote the paper. All authors read and approved the final manuscript.
Availability of data and materials
The authors declare that the data supporting the findings of this study are available within the article.
References
- 1.Sun X, Dai Y, Chen Y. et al. Primary angle closure glaucoma: What we know and what we don't know. Prog Retin Eye Res. 2017;57:26–45. doi: 10.1016/j.preteyeres.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Tham YC, Li X, Wong TY. et al. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121:2081–90. doi: 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–7. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leske MC, Heijl A, Hussein M. et al. Factors for glaucoma progression and the effect of treatment: the early manifest glaucoma trial. Arch Ophthalmol. 2003;121:48–56. doi: 10.1001/archopht.121.1.48. [DOI] [PubMed] [Google Scholar]
- 5.Drance SM. Some factors in the production of low tension glaucoma. Br J Ophthalmol. 1972;56:229–42. doi: 10.1136/bjo.56.3.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flammer J, Mozaffarieh M. What is the present pathogenetic concept of glaucomatous optic neuropathy? Surv Ophthalmol. 2007;52(Suppl 2):S162–73. doi: 10.1016/j.survophthal.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Weinreb RN, Aung T, Medeiros FA. The pathophysiology and treatment of glaucoma: a review. Jama. 2014;311:1901–11. doi: 10.1001/jama.2014.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernandez MR. The optic nerve head in glaucoma: role of astrocytes in tissue remodeling. Prog Retin Eye Res. 2000;19:297–321. doi: 10.1016/s1350-9462(99)00017-8. [DOI] [PubMed] [Google Scholar]
- 9.Hiraoka M, Inoue K, Ninomiya T, Takada M. Ischaemia in the Zinn-Haller circle and glaucomatous optic neuropathy in macaque monkeys. Br J Ophthalmol. 2012;96:597–603. doi: 10.1136/bjophthalmol-2011-300831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quigley HA, Addicks EM, Green WR, Maumenee AE. Optic nerve damage in human glaucoma. II. The site of injury and susceptibility to damage. Arch Ophthalmol. 1981;99:635–49. doi: 10.1001/archopht.1981.03930010635009. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen C, Midgett D, Kimball EC. et al. Measuring Deformation in the Mouse Optic Nerve Head and Peripapillary Sclera. Invest Ophthalmol Vis Sci. 2017;58:721–33. doi: 10.1167/iovs.16-20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebneter A, Wagels B, Zinkernagel MS. Non-invasive biometric assessment of ocular rigidity in glaucoma patients and controls. Eye (Lond) 2009;23:606–11. doi: 10.1038/eye.2008.47. [DOI] [PubMed] [Google Scholar]
- 13.Coudrillier B, Tian J, Alexander S. et al. Biomechanics of the human posterior sclera: age- and glaucoma-related changes measured using inflation testing. Invest Ophthalmol Vis Sci. 2012;53:1714–28. doi: 10.1167/iovs.11-8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oglesby EN, Tezel G, Cone-Kimball E. et al. Scleral fibroblast response to experimental glaucoma in mice. Mol Vis. 2016;22:82–99. [PMC free article] [PubMed] [Google Scholar]
- 15.Cone-Kimball E, Nguyen C, Oglesby EN. et al. Scleral structural alterations associated with chronic experimental intraocular pressure elevation in mice. Mol Vis. 2013;19:2023–39. [PMC free article] [PubMed] [Google Scholar]
- 16.Sigal IA, Flanagan JG, Tertinegg I, Ethier CR. Finite element modeling of optic nerve head biomechanics. Invest Ophthalmol Vis Sci. 2004;45:4378–87. doi: 10.1167/iovs.04-0133. [DOI] [PubMed] [Google Scholar]
- 17.Coudrillier B, Campbell IC, Read AT. et al. Effects of Peripapillary Scleral Stiffening on the Deformation of the Lamina Cribrosa. Invest Ophthalmol Vis Sci. 2016;57:2666–77. doi: 10.1167/iovs.15-18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu HJ, Kuchtey J, Kuchtey RW. Increased Susceptibility to Glaucomatous Damage in Microfibril Deficient Mice. Invest Ophthalmol Vis Sci. 2020;61:28. doi: 10.1167/iovs.61.10.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimball EC, Nguyen C, Steinhart MR. et al. Experimental scleral cross-linking increases glaucoma damage in a mouse model. Exp Eye Res. 2014;128:129–40. doi: 10.1016/j.exer.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moosavinasab S, Patterson J, Strouse R, 'RE:fine drugs': an interactive dashboard to access drug repurposing opportunities. Database (Oxford). 2016. 2016. [DOI] [PMC free article] [PubMed]
- 21.Mosca E, Bertoli G, Piscitelli E. et al. Identification of functionally related genes using data mining and data integration: a breast cancer case study. BMC Bioinformatics. 2009;10(Suppl 12):S8. doi: 10.1186/1471-2105-10-S12-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berg EL. Systems biology in drug discovery and development. Drug Discov Today. 2014;19:113–25. doi: 10.1016/j.drudis.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Yu S, Tranchevent LC, De Moor B, Moreau Y. Gene prioritization and clustering by multi-view text mining. BMC Bioinformatics. 2010;11:28. doi: 10.1186/1471-2105-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nogales-Cadenas R, Carmona-Saez P, Vazquez M. et al. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317–22. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Mering C, Huynen M, Jaeggi D. et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–61. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smoot ME, Ono K, Ruscheinski J. et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandettini WP, Kellman P, Mancini C. et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson. 2012;14:83. doi: 10.1186/1532-429X-14-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang DW, Sherman BT, Tan Q. et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dennis G Jr, Sherman BT, Hosack DA. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 30.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bindea G, Mlecnik B, Hackl H. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cotto KC, Wagner AH, Feng YY. et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46:D1068–d73. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chauhan BC, Drance SM. The relationship between intraocular pressure and visual field progression in glaucoma. Graefes Arch Clin Exp Ophthalmol. 1992;230:521–6. doi: 10.1007/BF00181772. [DOI] [PubMed] [Google Scholar]
- 34.Christian PG, Harkin DG, Schmid KL. GABAergic agents modify the response of chick scleral fibroblasts to myopic and hyperopic eye cup tissues. Curr Eye Res. 2014;39:172–87. doi: 10.3109/02713683.2013.834941. [DOI] [PubMed] [Google Scholar]
- 35.Wu H, Chen W, Zhao F. et al. Scleral hypoxia is a target for myopia control. Proc Natl Acad Sci U S A. 2018;115:E7091–e100. doi: 10.1073/pnas.1721443115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girard MJ, Suh JK, Bottlang M. et al. Scleral biomechanics in the aging monkey eye. Invest Ophthalmol Vis Sci. 2009;50:5226–37. doi: 10.1167/iovs.08-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grytz R, Yang H, Hua Y. et al. Connective Tissue Remodeling in Myopia and its Potential Role in Increasing Risk of Glaucoma. Curr Opin Biomed Eng. 2020;15:40–50. doi: 10.1016/j.cobme.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui W, Bryant MR, Sweet PM, McDonnell PJ. Changes in gene expression in response to mechanical strain in human scleral fibroblasts. Exp Eye Res. 2004;78:275–84. doi: 10.1016/j.exer.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 39.Huo L, Cui D, Yang X. et al. All-trans retinoic acid modulates mitogen-activated protein kinase pathway activation in human scleral fibroblasts through retinoic acid receptor beta. Mol Vis. 2013;19:1795–803. [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Zhang Y, Xu X. et al. Transforming growth factor-β is involved in maintaining oocyte meiotic arrest by promoting natriuretic peptide type C expression in mouse granulosa cells. Cell Death Dis. 2019;10:558. doi: 10.1038/s41419-019-1797-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pitha I, Oglesby E, Chow A. et al. Rho-Kinase Inhibition Reduces Myofibroblast Differentiation and Proliferation of Scleral Fibroblasts Induced by Transforming Growth Factor β and Experimental Glaucoma. Transl Vis Sci Technol. 2018;7:6. doi: 10.1167/tvst.7.6.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krenn K, Klepetko W, Taghavi S. et al. Vascular endothelial growth factor increases pulmonary vascular permeability in cystic fibrosis patients undergoing lung transplantation. Eur J Cardiothorac Surg. 2007;32:35–41. doi: 10.1016/j.ejcts.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Kariya T, Nishimura H, Mizuno M. et al. TGF-β1-VEGF-A pathway induces neoangiogenesis with peritoneal fibrosis in patients undergoing peritoneal dialysis. Am J Physiol Renal Physiol. 2018;314:F167–f80. doi: 10.1152/ajprenal.00052.2017. [DOI] [PubMed] [Google Scholar]
- 44.Van Bergen T, Vandewalle E, Van de Veire S. et al. The role of different VEGF isoforms in scar formation after glaucoma filtration surgery. Exp Eye Res. 2011;93:689–99. doi: 10.1016/j.exer.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 45.Uh ST, Inoue Y, King TE Jr. et al. Morphometric analysis of insulin-like growth factor-I localization in lung tissues of patients with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 1998;158:1626–35. doi: 10.1164/ajrccm.158.5.9804025. [DOI] [PubMed] [Google Scholar]
- 46.Giraudi PJ, Gambaro SE, Ornelas Arroyo S. et al. A simple in silico strategy identifies candidate biomarkers for the diagnosis of liver fibrosis in morbidly obese subjects. Liver Int. 2018;38:155–63. doi: 10.1111/liv.13505. [DOI] [PubMed] [Google Scholar]
- 47.Xu H, Liu L, Cong M. et al. EGF neutralization antibodies attenuate liver fibrosis by inhibiting myofibroblast proliferation in bile duct ligation mice. Histochemistry and cell biology. 2020;154:107–16. doi: 10.1007/s00418-020-01867-9. [DOI] [PubMed] [Google Scholar]
- 48.Kadler KE, Hill A, Canty-Laird EG. Collagen fibrillogenesis: fibronectin, integrins, and minor collagens as organizers and nucleators. Curr Opin Cell Biol. 2008;20:495–501. doi: 10.1016/j.ceb.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomasek JJ, Gabbiani G, Hinz B. et al. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. 2002;3:349–63. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 50.Tuan TL, Hwu P, Ho W. et al. Adenoviral overexpression and small interfering RNA suppression demonstrate that plasminogen activator inhibitor-1 produces elevated collagen accumulation in normal and keloid fibroblasts. Am J Pathol. 2008;173:1311–25. doi: 10.2353/ajpath.2008.080272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colman RW. Biologic activities of the contact factors in vivo-potentiation of hypotension, inflammation, and fibrinolysis, and inhibition of cell adhesion, angiogenesis and thrombosis. Thromb Haemost. 1999;82:1568–77. [PubMed] [Google Scholar]
- 52.Dong J, Ma Q. TIMP1 promotes multi-walled carbon nanotube-induced lung fibrosis by stimulating fibroblast activation and proliferation. Nanotoxicology. 2017;11:41–51. doi: 10.1080/17435390.2016.1262919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams JC, Lawler J. The thrombospondins. Int J Biochem Cell Biol. 2004;36:961–8. doi: 10.1016/j.biocel.2004.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao H, Frost MR, Siegwart JT Jr, Norton TT. Patterns of mRNA and protein expression during minus-lens compensation and recovery in tree shrew sclera. Mol Vis. 2011;17:903–19. [PMC free article] [PubMed] [Google Scholar]
- 55.Plompen EP, Darwish Murad S, Hansen BE. et al. Prothrombotic genetic risk factors are associated with an increased risk of liver fibrosis in the general population: The Rotterdam Study. J Hepatol. 2015;63:1459–65. doi: 10.1016/j.jhep.2015.07.026. [DOI] [PubMed] [Google Scholar]
- 56.Xu H, Abuduwufuer A, Lv W. et al. The role of HIF-1α-VEGF pathway in bronchiolitis obliterans after lung transplantation. J of Cardiothoracic Surgery. 2019;14:27. doi: 10.1186/s13019-019-0832-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duncan MR, Frazier KS, Abramson S. et al. Connective tissue growth factor mediates transforming growth factor beta-induced collagen synthesis: down-regulation by cAMP. Faseb j. 1999;13:1774–86. [PubMed] [Google Scholar]
- 58.Rao P, Pang M, Qiao X. et al. Promotion of β-catenin/Foxo1 signaling ameliorates renal interstitial fibrosis. Lab Invest. 2019;99:1689–701. doi: 10.1038/s41374-019-0276-z. [DOI] [PubMed] [Google Scholar]
- 59.Zhang H, Bajraszewski N, Wu E. et al. PDGFRs are critical for PI3K/Akt activation and negatively regulated by mTOR. J Clin Invest. 2007;117:730–8. doi: 10.1172/JCI28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li G, Li YY, Sun JE. et al. ILK-PI3K/AKT pathway participates in cutaneous wound contraction by regulating fibroblast migration and differentiation to myofibroblast. Lab Invest. 2016;96:741–51. doi: 10.1038/labinvest.2016.48. [DOI] [PubMed] [Google Scholar]
- 61.Liang L, Wang X, Zheng Y, Liu Y. All-trans-retinoic acid modulates TGF-β-induced apoptosis, proliferation, migration and extracellular matrix synthesis of conjunctival fibroblasts by inhibiting PI3K/AKT signaling. Mol Med Rep. 2019;20:2929–35. doi: 10.3892/mmr.2019.10507. [DOI] [PubMed] [Google Scholar]
- 62.Bitencourt CS, Pereira PA, Ramos SG. et al. Hyaluronidase recruits mesenchymal-like cells to the lung and ameliorates fibrosis. Fibrogenesis Tissue Repair. 2011;4:3. doi: 10.1186/1755-1536-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cordeiro MF, Gay JA, Khaw PT. Human anti-transforming growth factor-beta2 antibody: a new glaucoma anti-scarring agent. Invest Ophthalmol Vis Sci. 1999;40:2225–34. [PubMed] [Google Scholar]
- 64.Siriwardena D, Khaw PT, King AJ. et al. Human antitransforming growth factor beta(2) monoclonal antibody-a new modulator of wound healing in trabeculectomy: a randomized placebo controlled clinical study. Ophthalmology. 2002;109:427–31. doi: 10.1016/s0161-6420(01)00997-6. [DOI] [PubMed] [Google Scholar]
- 65.Rice LM, Padilla CM, McLaughlin SR. et al. Fresolimumab treatment decreases biomarkers and improves clinical symptoms in systemic sclerosis patients. J Clin Invest. 2015;125:2795–807. doi: 10.1172/JCI77958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kahook MY. Bleb morphology and vascularity after trabeculectomy with intravitreal ranibizumab: a pilot study. Am J Ophthalmol. 2010;150:399–403.e1. doi: 10.1016/j.ajo.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 67.Chakravarthy U, Harding SP, Rogers CA. et al. Alternative treatments to inhibit VEGF in age-related choroidal neovascularisation: 2-year findings of the IVAN randomised controlled trial. Lancet. 2013;382:1258–67. doi: 10.1016/S0140-6736(13)61501-9. [DOI] [PubMed] [Google Scholar]
- 68.Sangroongruangsri S, Ratanapakorn T, Wu O. et al. Comparative efficacy of bevacizumab, ranibizumab, and aflibercept for treatment of macular edema secondary to retinal vein occlusion: a systematic review and network meta-analysis. Expert Rev Clin Pharmacol. 2018;11:903–16. doi: 10.1080/17512433.2018.1507735. [DOI] [PubMed] [Google Scholar]
- 69.Ding Q, Tian XG, Li Y. et al. Carvedilol may attenuate liver cirrhosis by inhibiting angiogenesis through the VEGF-Src-ERK signaling pathway. World J Gastroenterol. 2015;21:9566–76. doi: 10.3748/wjg.v21.i32.9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abe W, Ikejima K, Lang T. et al. Low molecular weight heparin prevents hepatic fibrogenesis caused by carbon tetrachloride in the rat. J Hepatol. 2007;46:286–94. doi: 10.1016/j.jhep.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 71.Sarkar S, Mazumdar A, Dash R. et al. ZD6474, a dual tyrosine kinase inhibitor of EGFR and VEGFR-2, inhibits MAPK/ERK and AKT/PI3-K and induces apoptosis in breast cancer cells. Cancer Biol Ther. 2010;9:592–603. doi: 10.4161/cbt.9.8.11103. [DOI] [PubMed] [Google Scholar]
- 72.Ko J, Ross J, Awad H. et al. The effects of ZD6474, an inhibitor of VEGF signaling, on cutaneous wound healing in mice. J Surg Res. 2005;129:251–9. doi: 10.1016/j.jss.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 73.Gao J, Chesebrough JW, Cartlidge SA. et al. Dual IGF-I/II-neutralizing antibody MEDI-573 potently inhibits IGF signaling and tumor growth. Cancer Res. 2011;71:1029–40. doi: 10.1158/0008-5472.CAN-10-2274. [DOI] [PubMed] [Google Scholar]
- 74.Tarnawski A, Tanoue K, Santos AM, Sarfeh IJ. Cellular and molecular mechanisms of gastric ulcer healing. Is the quality of mucosal scar affected by treatment? Scand J Gastroenterol Suppl. 1995;210:9–14. doi: 10.3109/00365529509090261. [DOI] [PubMed] [Google Scholar]
- 75.Chen W, Mo W, Sun K. et al. Microplasmin degrades fibronectin and laminin at vitreoretinal interface and outer retina during enzymatic vitrectomy. Curr Eye Res. 2009;34:1057–64. doi: 10.3109/02713680903308487. [DOI] [PubMed] [Google Scholar]
- 76.Lee JH, Oh MH, Park JS. et al. Urokinase, urokinase receptor, and plasminogen activator inhibitor-1 expression on podocytes in immunoglobulin A glomerulonephritis. Korean J Intern Med. 2014;29:176–82. doi: 10.3904/kjim.2014.29.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo J, Chen X, Luo C, Hydrochlorothiazide modulates ischemic heart failure-induced cardiac remodeling via inhibiting angiotensin II type 1 receptor pathway in rats. Cardiovasc Ther. 2017. 35. [DOI] [PubMed]
- 78.Baldwin HE, Nighland M, Kendall C. et al. 40 years of topical tretinoin use in review. J Drugs Dermatol. 2013;12:638–42. [PubMed] [Google Scholar]
- 79.El Btaouri H, Morjani H, Greffe Y. et al. Role of JNK/ATF-2 pathway in inhibition of thrombospondin-1 (TSP-1) expression and apoptosis mediated by doxorubicin and camptothecin in FTC-133 cells. Biochim Biophys Acta. 2011;1813:695–703. doi: 10.1016/j.bbamcr.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 80.Avetisov ES, Savitskaya NF, Vinetskaya MI, Iomdina EN. A study of biochemical and biomechanical qualities of normal and myopic eye sclera in humans of different age groups. Metab Pediatr Syst Ophthalmol. 1983;7:183–8. [PubMed] [Google Scholar]
- 81.McBrien NA, Gentle A. Role of the sclera in the development and pathological complications of myopia. Prog Retin Eye Res. 2003;22:307–38. doi: 10.1016/s1350-9462(02)00063-0. [DOI] [PubMed] [Google Scholar]
- 82.Rada JA, Nickla DL, Troilo D. Decreased proteoglycan synthesis associated with form deprivation myopia in mature primate eyes. Invest Ophthalmol Vis Sci. 2000;41:2050–8. [PubMed] [Google Scholar]
- 83.McBrien NA, Cornell LM, Gentle A. Structural and ultrastructural changes to the sclera in a mammalian model of high myopia. Invest Ophthalmol Vis Sci. 2001;42:2179–87. [PubMed] [Google Scholar]
- 84.Grytz R, Siegwart JT Jr. Changing material properties of the tree shrew sclera during minus lens compensation and recovery. Invest Ophthalmol Vis Sci. 2015;56:2065–78. doi: 10.1167/iovs.14-15352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sergienko NM, Shargorogska I. The scleral rigidity of eyes with different refractions. Graefes Arch Clin Exp Ophthalmol. 2012;250:1009–12. doi: 10.1007/s00417-012-1973-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the article.