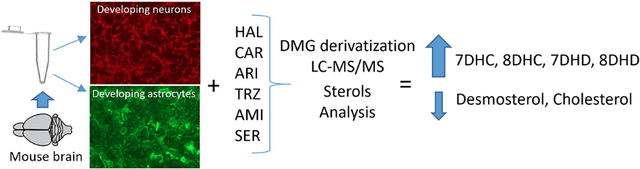
Abstract
Mouse brain contains over 100 million neuronal, glial, and other support cells. Developing neurons and astrocytes synthesize their own cholesterol, and disruption of this process can occur by both genetic and chemical mechanisms. In this study we have exposed cultured murine neurons and astrocytes to six different prescription medications that cross the placenta and blood–brain barriers and analyzed the effects of these drugs on cholesterol biosynthesis by an LC–MS/MS protocol that assays 14 sterols and 7 oxysterols in a single run. Three antipsychotics (haloperidol, cariprazine, aripiprazole), two antidepressants (trazodone and sertraline), and an antiarhythmic (amiodarone) inhibited one or more sterol synthesis enzymes. The result of the exposures was a dose-dependent increase in levels of various sterol intermediates and a decreased level of cholesterol in the cultured cells. Four prescription medications (haloperidol, aripiprazole, cariprazine, and trazodone) acted primarily on the DHCR7 enzyme. The result of this exposure was an increase in 7-dehydrocholesterol in neurons and astrocytes to levels that were comparable to those found in cultured neurons and astrocytes from transgenic mice that carried a Dhcr7 pathogenic mutation modeling the neurodevelopmental disorder Smith–Lemli–Opitz syndrome.
Keywords: DMG method, cholesterol, desmosterol, 7-DHC, pharmaceuticals, DHCR7
Graphical Abstract
INTRODUCTION
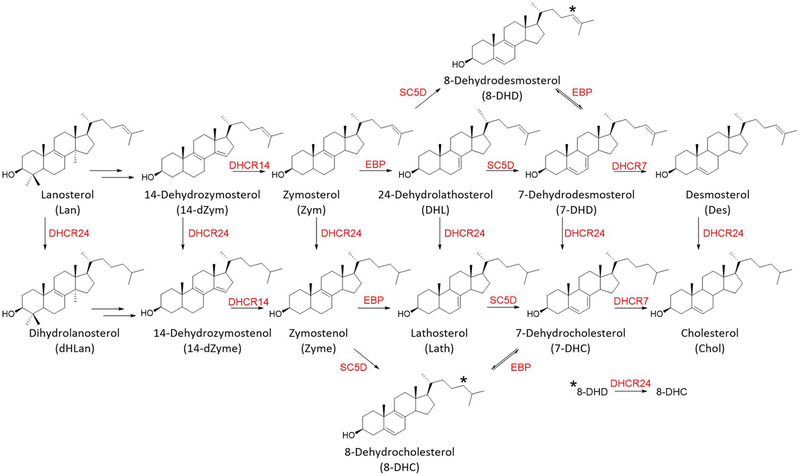
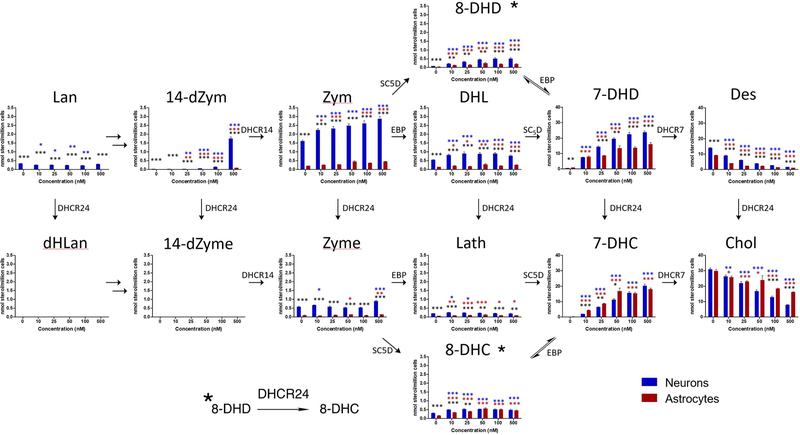
Cholesterol biosynthesis begins in developing neurons during the embryonic period and continues to increase postnatally through the end of myelination. This period of extensive cholesterol synthesis by neurons occurs during a critical period for the proper establishment of neuronal architecture. As a consequence, disturbances in cholesterol biosynthesis, either by genetic mutations in sterol synthesis genes or by pharmacological inhibition of sterol synthesis enzymes (see Figure 1), can have deleterious consequences on neurodevelopment. One example of a genetically induced error in cholesterol biosynthesis is the neurodevelopmental disorder Smith–Lemli–Opitz syndrome (SLOS) with the genetic error leading to mutations in DHCR7, the enzyme that promotes the reduction of 7-dehydrocholesterol (7-DHC) to cholesterol, the last step in the biosynthesis. The result of a pathological mutation in DHCR7 is significantly elevated levels of 7-DHC and a modestly decreased amount of cholesterol in all cells having active cholesterol synthesis.
Figure 1.
Postlanosterol cholesterol biosynthesis pathway. Only chemical structures of sterols analyzed by LC–MS/MS in this study are shown. The enzymes are shown in red. The ∗ identifies conversion of the 8-DHD double bond to form 8-DHC, promoted by the DHCR24 enzyme.
Small molecules, including many highly prescribed medications, can lead to perturbations of sterol homeostasis. The DHCR7 enzyme, in particular, can be inhibited by dozens of medications, including highly prescribed antipsychotics and antidepressants.1–4 Screening studies of libraries of FDA approved drugs in cultures of Neuro2a cells led to the conclusion that over 5% of the compounds tested caused increased levels of 7-DHC in the cells.1,2 Assays of banked human sera from groups prescribed various psychoactive medications connect only those samples from donors taking “DHCR7 active” drugs with elevated levels of 7-DHC found in the samples, providing a link between the in vitro screening studies and the general population.5,6
Studies in rodents exposed to “DHCR7 active” drugs also link the cell culture screening studies with effects on sterol homeostasis. For example, cariprazine (CAR), a recently approved antipsychotic marketed as Vraylar, was found to be one of the most potent inhibitors of DHCR7 tested in Neuro2a cells, and this compound also affects sterol levels in the brain and serum of mice treated with the drug.7 It is notable that the experimental compound AY9944 affects DHCR7 in Neuro2a with about the same potency as cariprazine since AY9944 has been used for nearly 2 decades as a pharmacological model for SLOS.8–12 Rat pups exposed to AY9944 in utero have elevated levels of 7-DHC at birth, and they subsequently display some of the attending deficits associated with SLOS, the genetic disorder.13–20
The fact that drugs having a significant inhibitory effect on DHCR7 account for over 150 million U.S. prescriptions annually along with the potential risk associated with exposure to these drugs5,6 stimulated us to examine the effects of a set of FDA approved drugs on sterol homeostasis in cultured murine cortical neurons and astroglial cells. Thus, cortical neurons and astrocytes were cultured in the presence of three antipsychotic medications (haloperidol (HAL), aripiprazole (ARI), cariprazine (CAR)), two antidepressants (trazodone (TRZ) and sertraline (SERT)), and an antiarrhythmic medication, amiodarone (AMIOD). We report here the results of that effort. As a part of the study, an LC/MS method was developed that includes the assay of 14 sterols in the cholesterol biosynthesis pathway along with another seven cholesterol-derived oxysterols. The method, which involves the derivatization of alcohol functional groups to dimethylglycyl esters, includes assay of the oxysterols and parent sterols in a short LC/MS run.
RESULTS AND DISCUSSION
Prescription Medications Tested in Neuron and Astrocyte Cultures.
All of the compounds tested in this study cross the blood–brain barrier, have the potential to affect brain cholesterol biosynthesis, and therefore present a risk to a developing fetal nervous system. Studies have shown, for example, that serum samples from individuals taking ARI, HAL, and TRZ have increased levels of 7-DHC compared to controls,5,6 as do the brains of fetal mice exposed to these drugs during pregnancy.21,22 CAR was recently approved by the FDA, has a substructure similar to ARI, and is a potent inhibitor of DHCR7 in Neuro2a cells.7 SERT has been found to affect sterol homeostasis in cell culture but only at much higher concentrations than was the case for ARI, HAL, CAR, and TRZ. AMIOD has been shown to affect DHCR24 and EBP enzymes in neuronal cultures, but its effect on the complete sterol set has not been reported.23 The six drugs included in this study were prescribed over 71 million times in the U.S. in 2017. Figure S1 shows the chemical structures of the drugs tested and their prescription information.
Sterol and Oxysterol Analysis as N,N-Dimethylglycyl (DMG) Esters.
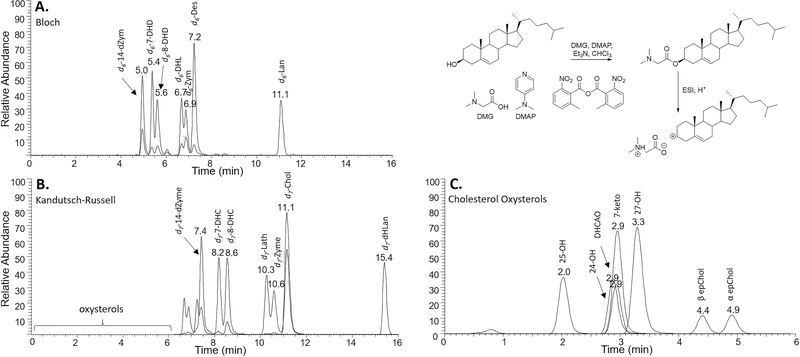
The important post-lanosterol sterols in the biosynthetic sequence along with several cholesterol-derived oxysterols were analyzed in a single LC/MS run after conversion of OH functional groups to the DMG esters. Sterols were derivatized at the C-3 alcohol (Figure 2), while for some oxysterols, two OH groups were converted; see Figure S3. We found that C-3 alcohol derivatizations of sterols bearing 4,4-gemdimethyl substituents (Lan and dHLan) were generally sluggish and required modestly elevated temperatures for complete conversion. Our conditions were chosen to maximize derivatization under the mildest conditions possible in order to minimize decomposition of sensitive oxysterols. The DMG sterol protocol is derived from established approaches24 for chemical derivatization of oxysterol hydroxy groups and extends the protocol to include 14 sterols that are formed in the biosynthetic pathway between lanosterol and cholesterol. DMG derivatizations chemically tag hydroxyls with a group that provides a site for charge introduction in short reaction times at room temperature. Chromatography of the DMG sterol esters is excellent, and MS fragmentation patterns are informative. DMG fragmentation of all sterols that contain a Δ5–6 double bond, for example, occurs with loss of a neutral DMG group, leaving the positive charge on the stable homoallylic sterol carbocation; see Figure 2. We note that this protocol adds to other chemical derivatization approaches reported25 as well as methods utilizing a charge-tagging enzyme-mediated approach26 and a combination ESI/APCI method for analysis of underivatized sterols and oxysterols.27
Figure 2.
LC–MS/MS separation of DMG-derivatized deuterated (d6 and d7) sterols and oxysterols. (A) Bloch and (B) Kandutsch–Russell sterols and (C) cholesterol oxysterols were separated on an Agilent Poroshell EC-C18 (10 cm × 2.1 mm, 1.9 μm) column using a solvent mixture of CH3CN:MeOH:H2O, 70:25:5 (0.01% (v) formic acid, 1 mM NH4OAc) with a column temperature of 40 °C. A TSQ Quantum Ultra tandem mass spectrometer (ThermoFisher) was used for MS detections, and data were acquired with a Finnigan Xcalibur software package. Selected reaction monitoring (SRM) of the DMG derivatives was acquired in the positive ion mode using electrospray ionization (ESI).
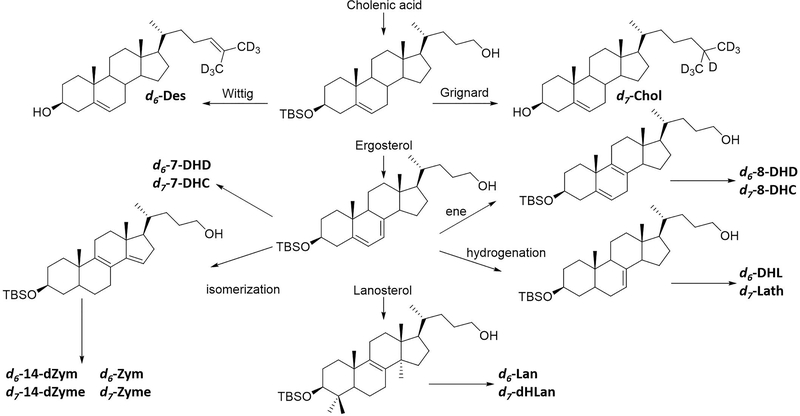
Matched deuterated standards for all of the sterols (d6 for those sterols with a Δ24 double bond in the Bloch pathway and d7 for those in the Kandutsch–Russell sequence) were prepared by independent synthesis to further minimize any error introduced by incomplete reaction of Lan and dHLan. We assumed that the endogenous and matched deuterated standard have the same reactivity, compensating for any reduced reactivity relative to other sterols. We note that 4,4-gemdimethyl steric hindrance may be a problem for other analytical protocols that involve reaction at C-3 if isotopically labeled standards are not used in the protocol. The general synthetic approach to the isotopically labeled standards is shown in Figure 3, and the details are presented in Supporting Information. The syntheses started from cholenic acid, ergosterol, and lanosterol and generally involved manipulation of the ring double bonds and/or side chain to generate the prerequisite aldehyde or alcohol. The resultant aldehyde or alcohol was converted to the d6- and d7-deuterated sterols with Wittig or organometallic coupling reactions.
Figure 3.
General strategy for synthesis of deuterated standards. All standards were synthesized from cholenic acid, ergosterol, or lanosterol. The deuterium was incorporated utilizing a Wittig or Grignard reaction to generate the Bloch (d6) or Kandutsch–Russel (d7) sterols, respectively.
The DMG moiety provides a site of protonation, which allowed the sterols to be readily detected by electrospray ionization (ESI). Furthermore, collision induced dissociation of the DMG-derivatized sterols gave characteristic fragments with SRMs determined for each sterol that are presented in Table 1. Sterols containing a 5,6-double bond fragmented to generate the sterol moiety as the major ion detected, while those sterols lacking the 5,6-double bond gave the N,N-dimethylglycine as the major fragment ion. The DMG-sterols were separated on an Agilent Poroshell EC-C18 column coupled with ESI in the positive mode. All of the sterol intermediates could be assayed by comparison with a known amount of added deuterated derivative and separation by chromatography and/or by characteristic SRMs. In addition, the DMG derivative gave sensitivity such that all of the sterols could be detected at pmol levels in biological samples.
Table 1.
MS Parameters for Sterol Analysis
| metabolite | SRM parent (M + H) (m/z) | SRM fragment (m/z) | CE (eV) | rt (min) |
|---|---|---|---|---|
| 7-DHD | 468.30 | 365.30 | 15 | 5.6 |
| 14d-Zym | 468.31 | 104.10 | 21 | 5.1 |
| 8-DHD | 468.32 | 365.30 | 20 | 5.8 |
| 7-DHC | 470.30 | 367.30 | 13 | 8.4 |
| 8-DHC | 470.31 | 367.31 | 20 | 8.8 |
| Des | 470.31 | 367.31 | 20 | 7.6 |
| DHL | 470.32 | 104.10 | 21 | 7.4 |
| Zym | 470.32 | 104.10 | 21 | 6.9 |
| 14d-Zyme | 470.32 | 104.10 | 21 | 7.1 |
| Chol | 472.30 | 369.30 | 20 | 11.3 |
| Lath | 472.31 | 104.10 | 21 | 10.5 |
| Zyme | 472.31 | 104.10 | 21 | 10.8 |
| Lan | 512.31 | 104.10 | 21 | 11.3 |
| diHLan | 514.31 | 104.10 | 21 | 15.6 |
| d6-7-DHD | 474.30 | 371.30 | 15 | 5.4 |
| d6-14d-Zym | 474.31 | 104.10 | 21 | 4.9 |
| d6-8-DHD | 474.32 | 371.30 | 20 | 5.6 |
| d7-7-DHC | 477.30 | 374.30 | 13 | 8.2 |
| d7-8-DHC | 477.31 | 374.31 | 20 | 8.6 |
| d7-14d-Zyme | 477.32 | 104.10 | 21 | 7.4 |
| d6-Des | 476.30 | 373.30 | 20 | 7.2 |
| d6-DHL | 476.31 | 104.10 | 21 | 6.7 |
| d6-Zym | 476.31 | 104.10 | 21 | 6.9 |
| d7-Chol | 479.30 | 376.30 | 20 | 11.1 |
| d7-Lath | 479.31 | 104.10 | 21 | 10.3 |
| d7-Zyme | 479.31 | 104.10 | 21 | 10.6 |
| d6-Lan | 518.31 | 104.10 | 21 | 11.1 |
| d7-diHLan | 521.31 | 104.10 | 21 | 15.4 |
Cholesterol-derived oxysterols28–32 were included in the DMG derivatization scheme and were chromatographed under the same conditions used for sterol analysis. We included in this assay only those cholesterol-derived oxysterols for which pure standards are commercially available or for oxysterols that can be synthesized by known methods. This list now includes 7-ketocholesterol (7-keto), 24-hydroxycholesterol (24-OH), 25-hydroxycholesterol (25-OH), 27-hydroxycholesterol (27-OH), 3,5-dihydroxycholestanone (DHCAO), 5,6-α epoxycholestanol (α-epChol) and 5,6-β epoxycholestanol (β-epChol). Table 2 presents SRM fragmentation patterns for each oxysterol, including those obtained for DMG derivatization at one or two hydroxyl groups when that occurs under standard derivatization conditions. The structures of parent DMG derivatives monitored by SRM are presented in Figure S2. Response factors for each oxysterol SRM were measured relative to d7-cholesterol, and those factors were used to determine levels of each oxysterol assayed. Efforts are underway to prepare isotopically labeled derivatives of the important set of oxysterols that are not available from commercial sources.
Table 2.
MS Parameters for Cholesterol Oxysterol Analysis
| metabolite | SRM parent (m/z) | SRM center (m/z) | CE (eV) | rt (min) | rf (d7-Chol) |
|---|---|---|---|---|---|
| 7-keto-Chol | 486.31 | 383.31 | 20 | 2.9 | 1.3 |
| 25-OH-Chol | 488.30 | 367.30 | 20 | 2.0 | 6.0 |
| β-epChol | 488.30 | 367.30 | 20 | 4.4 | 4.4 |
| α-epChol | 488.30 | 367.30 | 20 | 4.9 | 14.0 |
| DHCAO | 504.32 | 365.32 | 30 | 2.9 | 45.0 |
| 24-OH-Chol | 573.30 | 470.30 | 20 | 2.9 | 3.1 |
| 27-OH-Chol | 573.30 | 470.30 | 20 | 3.3 | 5.3 |
Figure 2 shows chromatograms for the sterols and oxysterols assayed under standard conditions. Note that the three chromatograms presented in the figure were obtained from the same LC/MS run but they are differentiated according to the particular SRMs monitored in the run. All of the sterols eluted in under 15 min under isocratic conditions, and it was possible to reduce cycle times to 12 min, with appropriate caution, since dHLan is normally not found at significant levels in most cellular lipidomes.33
Comparison of DMG Ester and PTAD Sterol Assays.
4-Phenyl-1,2,4-triazoline-3,5-dione (PTAD) derivatization of sterols followed by LC/MS analysis has been used to develop screens of drugs that affect cellular levels of 7-DHC, desmosterol, lanosterol, and cholesterol.1,2,34–36 7-DHC readily undergoes a Diels–Alder reaction with PTAD, and the adduct is detected at pmol levels by LC/MS. Desmosterol, lanosterol, and other sterols in the Bloch pathway react with PTAD in an “enetype” reaction giving adducts having characteristic fragmentation patterns that are also readily detected by LC/MS.34 The reaction of these sterols with PTAD is rapid, the adducts chromatograph with run times of less than a minute, and the derivatization and workup can be carried out in a 96-well format, making high throughput screening possible. There are limitations to the PTAD method for assay of other sterols however since PTAD reacts with most sterols, giving some adducts that do not readily separate by chromatography. Notably, 8-DHC and 7-DHD react with PTAD giving adducts that have similar chromatographic and MS fragmentation characteristics excluding those sterols from assay by the PTAD method.
We have assayed the sterols in neurons and astrocytes by both the DMG and PTAD methods for several of the drug treatments, and the methods are in good agreement for lanosterol, desmosterol, 7-DHC, and cholesterol. Comparison data for one such sterol mixture analyzed by both methods are provided in Supporting Information; see Figure S4. We conclude that the PTAD method does provide high sensitivity and speed for a limited set of sterols if high throughput is important. Regardless of the method of analysis, the use of isotopically labeled standards provides a clear advantage over other methods of quantification. We have undertaken the synthesis of many of the labeled analytes and have made them generally available for use. Details of the synthetic methods are presented in Supporting Information.
Sterol and Oxysterol Profiles in Neurons and Astrocytes.
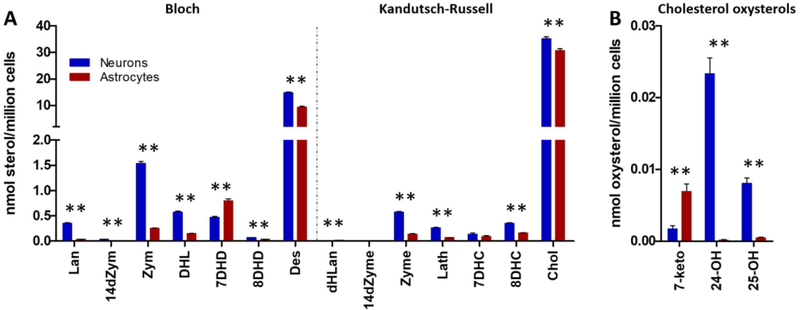
Primary cortical neuronal and astrocytes cultures were prepared from E18 C57BL/6J mice as previously described.22,37 Briefly, both neurons and astrocytes were plated in 96-well plates, grown for 9 days, and analyzed for sterols. Figure 4 compares the levels in neurons and astrocytes in nmol/million cells of the 14 sterols and 7 oxysterols assayed by the DMG method. Note that the Bloch sterols are present in higher concentrations than those along the Kandutsch–Russell pathway, with the exception of desmosterol/cholesterol pair. Sterol levels found in nmol/million cells are generally greater in neurons than in astrocytes with the one exception of 7-DHD, which is present at higher levels in astrocytes. 7-Ketocholesterol (7-keto) was found at significantly higher levels in astrocytes than neurons, while 24-hydroxycholesterol (24-OH) and 25-hydroxycholesterol (25-OH) were the major oxysterols detected in neurons.38 The four other oxysterols included in the DMG assay (see Figure 2) were found at much lower levels than 7-keto, 24-OH and 25-OH in both neurons and astrocytes.
Figure 4.
Bloch and Kandutsch–Russell sterols in neurons and astrocytes. Fourteen sterols (A) and cholesterol oxysterols (B) were analyzed by DMG method: blue, neurons; red, astrocytes. Neurons have higher levels of all sterols except for 7DHD which is present at higher levels in the astrocytes. The most abundant sterols in both cell types are cholesterol and desmosterol. N = 44; **p < 0.001.
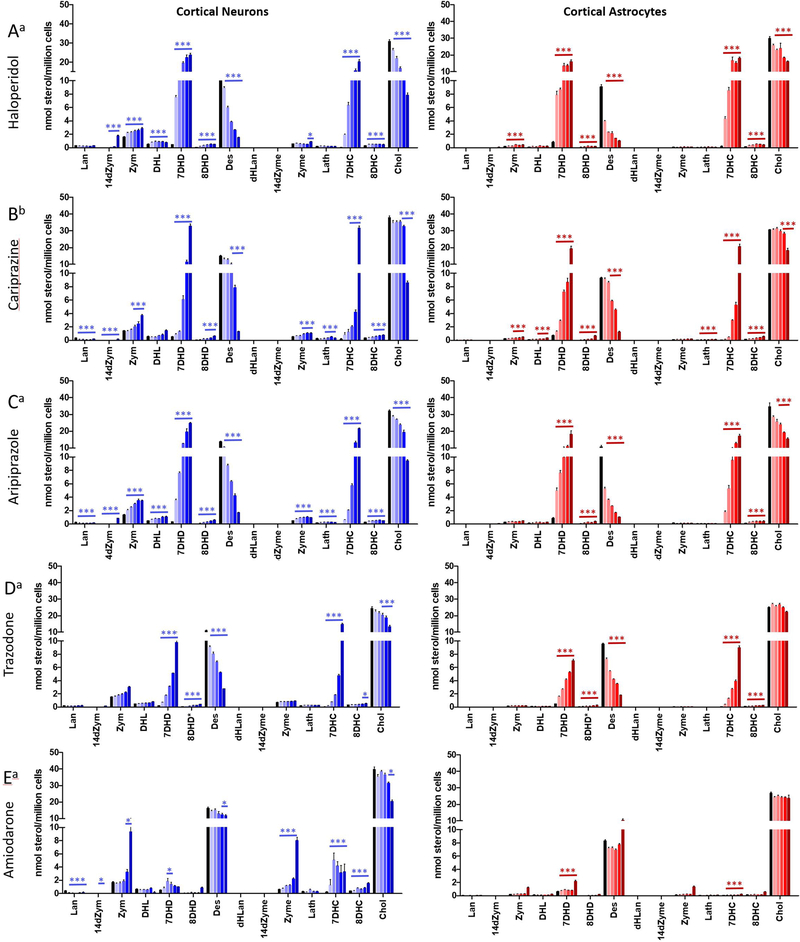
Neurons and astrocytes were treated with different concentrations of pharmaceuticals for 6 days. Haloperidol, aripiprazole, trazodone, sertraline, and amiodarone were used at concentrations ranging from 10 to 500 nM; cariprazine was generally more potent than the other compounds, and concentrations of 1–100 nM were used for this drug. After incubation, deuterated standards were added, and lipids were extracted and converted to the DMGesters at room temperature in 30 min. Both neurons and astrocytes showed altered sterol composition in response to each of the drugs tested with the exception of sertraline, where differences were found only at higher concentrations (Supporting Information).
Figure 5 presents the sterol levels assayed for incubations of neurons and astrocytes with 10–500 nM haloperidol. The figure is oriented to match the sterol cellular concentrations with the biosynthetic scheme, with the Bloch sterols along the top row, those in the Kandutsch–Russell pathway along the bottom, and the overall transformation from lanosterol on the top left to cholesterol on the bottom right as in Figure 1. The scales for all of the sterols in Figure 5 before 7-DHD and 7-DHC in the biosynthetic sequence are set with a maximum of 3.5 nmol/million cells so that the differences in steady state levels are apparent. Desmosterol, 7-DHD, 7-DHC, and cholesterol are also scaled identically at 40 nmol/million cells for comparison purposes.
Figure 5.
Haloperidol treatment inhibits DHCR7 enzyme in neurons and astrocytes. Cortical neuronal and astrocytes cultures were treated with haloperidol (10–500 nM). Fourteen sterols were analyzed by the DMG method. The data are presented to coincide with the sterol biosynthesis scheme presented in Figure 1. The scale for 7-DHD, Des, 7-DHC, and Chol is 0–40 nmol/million cells. The scale of all other sterols shown is 0–3.5 nmol/million cells. Statistical significance between neurons and astrocytes is shown in black, and significance between control and treatment is shown in blue for neurons and in red for astrocytes: *p < 0.05; **p < 0.01; ***p < 0.001.
Haloperidol treatment led to significant concentration dependent increases in 7-DHC in both neurons and astrocytes as expected for a DHCR7 inhibitor. It is notable that increases in levels of 7-DHD were also observed, with the increases found for this sterol being more prominent in neurons than astrocytes. Indeed, the Bloch sterols were generally found to have higher cellular steady state levels than the Kandutsch–Russell counterparts in neurons at all concentrations of haloperidol, the exception being desmosterol/cholesterol, the final pair in the sequence. At 100 nM haloperidol, 7-DHD is the major sterol found in neurons followed in order by 7-DHC, cholesterol, and desmosterol. In addition, Bloch sterols were found to be higher in neurons than astrocytes at every concentration of haloperidol tested. At concentrations of haloperidol of 50 nM and above, desmosterol was found at concentrations lower than 7-DHC and 7-DHD in both neurons and astrocytes. The increase in levels of 14-dZym found at 500 nM haloperidol should also be noted and suggests the possibility of an effect of haloperidol on the 14-reductase enzyme.
We note that sterol levels measured are at a steady-state condition and no conclusions about the relative kinetics of individual steps in the biosynthesis can be drawn from these data. It is notable, however, that Kandutsch–Russell sterols are not found at appreciable levels in the biosynthetic sequence before zymostenol, suggesting that the first significant Bloch/Kandutsch–Russell crossover point in the biosynthesis is at the zymosterol–zymostenol pair for both neurons and astrocytes. This mechanism, the modified Kandutsch–Russell pathway, is in accord with previous detailed studies of sterol biosynthetic flux that identified the zymosterol DHCR24 crossover point as being dominant in the brain.39
Figure 6 presents the sterol analysis data for comparison of five of the six drugs assayed (HAL, CAR, ARI, TRZ, and AMIOD) that had significant activity as inhibitors of late-stage sterol biosynthesis. The sterol profiles of function of inhibitor for HAL, CAR, ARI, and TRZ were remarkably similar, the effect of compound on sterol levels found principally as an increase in 7-DHD and 7-DHC and a decrease in desmosterol. Cholesterol levels were minimally affected except at the highest concentration of these DHCR7 inhibitors. AMIOD had a minimal effect on 7-DHD and 7-DHC, but as expected for an EBP inhibitor, levels of Zym and Zyme increased with increasing AMIOD concentrations. SERT showed no measurable effect on post-lanosterol sterol levels at concentrations of 1 μM or below; see Figure S5.
Figure 6.
Sterol profile in neurons and astrocytes exposed to three antipsychotics, an antidepressant, and an antiarrhythmic: (A) haloperidol; (B) cariprazine; (C) aripiprazole; (D) trazodone; (E) amiodarone; blue = neurons; red = astrocytes. Cells were exposed to increasing concentrations of (a). Concentrations for treatments with haloperidol, aripiprazole, trazodone, and amiodarone were control (DMSO), 10, 25, 50, 100, and 500 nM. (b) Concentrations for treatment with cariprazine were control (DMSO), 5, 10, 25, 50, and 100 nM. Statistical significance is marked by *p < 0.01.
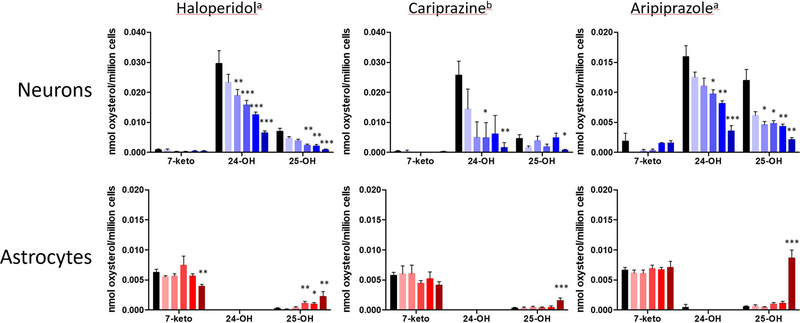
Oxysterol Profiles in Neurons and Astrocytes.
Oxysterol profiles assayed by the DMG method in neurons and astrocytes differed significantly. As shown in Figure 4B, 24-hydroxycholesterol (24-OH) and 25-hydroxycholesterol (25-OH) were the major oxysterols found in neurons; their levels were much higher than those found for 7-ketocholesterol (7-keto) in these cells. Treatment of neurons with the DHCR7 inhibitors haloperidol, cariprazine, and aripiprazole resulted in levels of 24-OH and 25-OH that decreased with increasing drug concentrations, as shown in Figure 7. The decrease of these oxysterols at higher levels of the DHCR7 inhibitors paralleled the decrease in cholesterol found for treatment of neurons at the higher drug levels; see Figure 5. So one explanation for the observed trend is simply the lower substrate availability for the P450 enzymes that promote the conversion of cholesterol to the oxysterols.26,28,29,31,32,40 In contrast to the results in neurons, 25-OH levels increased at the highest doses of haloperidol, cariprazine, and aripiprazole in astrocytes.
Figure 7.
Profile of cholesterol oxysterols in neurons and astrocytes exposed to three antipsychotics haloperidol, cariprazine, and aripiprazole: blue = neurons; red = astrocytes. Neurons and astrocytes were exposed to increasing concentrations of compounds for 6 days, and seven oxysterols were analyzed by the DMG method. n = 11 for control; n = 6 for drug. The major oxysterols found are shown here. Statistical significance is marked by *p < 0.05; **p < 0.01; ***p < 0.001. (a) Concentrations for treatments with haloperidol and aripiprazole were control (DMSO), 10, 25, 50, 100, and 500 nM. (b) Concentrations for treatment with cariprazine were control (DMSO), 5, 10, 25, 50, and 100 nM.
Consequences of Exposures.
All of the drugs in this study that have been classified are listed in the pregnancy category C (see Table S1), indicating that animal reproduction studies have shown an adverse effect on the fetus but that potential benefits may warrant use of the drug in pregnant women. Indeed, a recent review has linked DHCR7 inhibitors to significant adverse fetal outcomes, including unexpected loss, deformaties, and malformations.41 These facts raise the possibility that fetal exposure to DHCR7 inhibitors will ultimately have significant neurodevelopmental consequences. The clinical consequence of mutations in the gene that codes DHCR7 is Smith–Lemli–Opitz syndrome (SLOS), a devastating disorder characterized by elevated serum levels of 7-DHC similar to those found in patients taking cariprazine, aripiprazole, and other drugs that inhibit DHCR7. SLOS is a neurodevelopmental disorder that is linked to intellectual disabilities and behavioral problems, including autistic features42 and hyperactivity, raising the possibility that fetal exposure to DHCR7-active drugs may have similar associated risks.
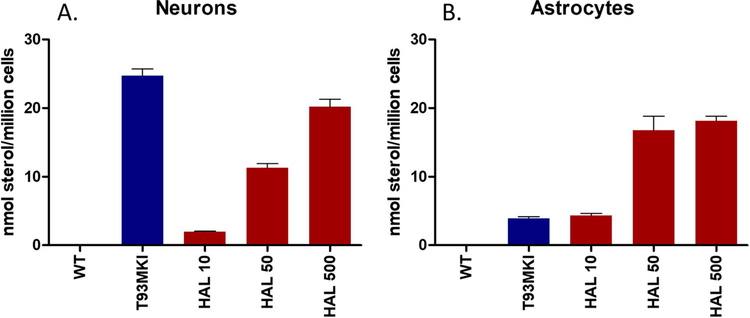
It is of some note then that aripiprazole,21 haloperidol,6 cariprazine,22 and trazodone43 cross the rodent placenta and reach the brain of fetuses during embryonic development. The current work shows that the drugs have a significant and parallel effect on sterol homeostasis in both neurons and astrocytes, leading to increased levels of both 7-DHD and 7-DHC, highly oxidizable lipids44–47 that affect neuronal arborization, cell viability, and gene expression changes.48,49 Indeed, as shown in Figure 8, levels of 7-DHC assayed in WT neurons at 500 nM haloperidol approach those found in neurons from a T93MKI50 SLOS mouse and the treated WT astrocytes have 7-DHC levels that exceed those of the T93MKI astrocytes. The results support the notion that screening for DHCR7 inhibition in the assessment of drug toxicity may help to prevent long-term adverse neurodevelopmental outcomes.
Figure 8.
Comparison of 7-DHC in SLOS T93MKI and WT cells treated with haloperidol. (A) WT neurons and (B) WT astrocytes were treated with haloperidol at 10, 50, and 500 nM and compared with neurons and astrocytes from a SLOS mouse.
METHODS
Materials.
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO). HPLC grade solvents were purchased from Thermo Fisher Scientific Inc. (Waltham, MA). The derivatizing reagents 2-methyl-6-nitrobenzoic anhydride and N,N-dimethylglycine were purchased from Combi-Blocks (San Diego, CA). The synthesis of d7-7-DHC and d7-8-DHC has been reported,36 and the remaining deuterated standards are described in the Supporting Information. The oxysterols 24-OH, 25-OH, and 27-OH cholesterol are available from Sigma-Aldrich. DHCAO,51 7-keto cholesterol,52 and epChol53 were synthesized according to literature procedures. The ratio of α- and β-epChol was determined by 1H NMR. All deuterated sterol standards are available from Kerafast, Inc. (Boston, MA).
Primary Neuronal and Astrocytes Cultures.
Primary cortical neuronal cultures were prepared from E18 C57BL/6J (The Jackson Laboratory stock 000664) mice as previously described.54 All mice were housed under a 12 h light–dark cycle at constant temperature (25 °C) and humidity with ad libitum access to food (Teklad LM-485 mouse/rat irradiated diet 7912) and water in Comparative Medicine at UNMC, Omaha, NE. Embryonic and newborn mice were used for the study. All procedures were performed in accordance with the Guide for the Humane Use and Care of Laboratory Animals. T93M mice were obtained from FD Porter.50 These mice have a mutation in both Dhcr7 alleles at c.278C>T, a mutation equivalent to the human T93M missense mutation. The T93M is the most common missense mutation described in patients with SLOS.50 These mice survive into adulthood and breed. The use of mice in this study was approved by the Institutional Animal Care and Use Committee of UNMC. Briefly, the brain was placed in prechilled HBSS solution (without Ca2+ or Mg2+), and two cortexes were dissected and cut with scissors into small chunks of similar sizes and transferred to trypsin/EDTA (0.25%) for 25 min at 37 °C. Trypsin was inactivated by adding trypsin inhibitor (Sigma T6522) for 5 min. The tissue was rinsed in neurobasal medium with B-27 supplement (Gibco no. 17504–044) and then triturated with a fire-polished Pasteur pipet. The cells were pelleted by centrifugation for 5 min at 80g. The cell pellet was resuspended in neurobasal medium with B-27 supplement, and the cells were counted. The cells were plated on poly-D-lysine coated 96-well plates at 70 000 cells/well. The growth medium was neurobasal medium with B-27 supplement and Glutamax. After the incubation of cells at 37 °C, 5% CO2 cell culture incubator for 3 days, the cells were treated with six different concentration of drugs. For each condition there were 6–12 technical replicates. Each 96-well plate contained 24 control wells without drug. The incubation time with different drugs was 6 days. At the end point of the incubation, Hoechst dye was added to all wells in the 96-well plate and the total number of cells counted using an ImageXpress Pico and cell counting algorithm in CellReporterXpress. After removal of the medium, wells were rinsed twice with 1× PBS, treated with an antioxidant mixture (10 μL per well), and stored at −80 °C for sterol analysis. The antioxidant mixture contained BHT (2.5 mg/mL) and PPh3 (1 mg/mL) in EtOH. All samples were analyzed within 2 weeks of freezing.
After plating of the required number of cells for neuronal cultures, leftover cells were plated in 100 mm dishes at a density of 10 × 106 per tissue culture plate in DMEM with 10% FBS. Under these conditions, astrocytes adhere and divide and completely populate the plate within 10–14 days. Once the plates were full, they were rinsed using the cold jet method.55 The astrocytes were trypsinized and plated in 96-well plates in DMEM plus 10% FBS at 30 000 cells/well. The following day the medium was replaced with neurobasal medium with B-27 supplement (same medium as neuronal cells) ± drugs. Cells were incubated at 37 °C, 5% CO2 for 6 days in the presence of drugs. For each condition there were 6–12 technical replicates. Each 96-well plate contained 24 control wells without drug. At the end point, cultures were processed the same way as neuronal cultures. The preparation of primary cortical neuronal and astrocytes cultures was repeated from five different mouse litters, and each time all drugs were tested and their effects on sterols assayed by LC–MS/MS. The results were concordant across an independent set of experiments.
Extraction of Sterols.
A stock solution of deuterated standards was made containing 30 μM d7-Chol, d6-Lan, d7-dHLan and 3 μM d7-7-DHC, d7-8-DHC, d6-Des, d6-7-DHD, d6-8-DHD, d7-Lath, d6-DHL, d7-Zyme, d6-Zym, d7-14d-Zyme, d6-14d-Zym in MeOH. The standards stock solution contained 1% (v/v) Et3N and antioxidant mixture (BHT and PPh3) described above to prevent isomerization and/or oxidation. To each well of the 96-well plate were added the standards mixture (10 μL) and MeOH (100 μL). The plate was agitated on a shaker for 20 min and then allowed to rest to settle cell debris. The MeOH was transferred to an analysis plate, dried under vacuum, and derivatized as described below.
Derivatization and LC–MS/MS Analysis of Sterols.
Derivatizing reagent was freshly prepared with 2-methyl-6-nitrobenzoic anhydride (20 mg), N,N-dimethylglycine (14 mg), DMAP (6 mg), and Et3N (0.1 mL) in anhydrous CHCl3 (0.9 mL). To each sample was added derivatizing reagent (100 μL) and allowed to react at room temperature for 30 min. The samples were dried under vacuum and subsequently dissolved in MeOH (100 μL) for LC–MS/MS analysis. Samples were analyzed on an Acquity UPLC system equipped with ANSI-compliant well plate holder. The sterols (10 μL injection) were analyzed on an Agilent Poroshell EC-C18 (10 cm × 2.1 mm, 1.9 μm) with CH3CN:MeOH:H2O, 70:25:5 (0.01% (v), formic acid, 1 mM NH4OAc) mobile phase at a column temperature of 40 °C. The flow rate was 400 μL/min for 11.5 min, then ramped to 600 μL/min at 11.6 min with a total run time of 16 min. A TSQ Quantum Ultra tandem mass spectrometer (ThermoFisher) was used for MS detections, and data were acquired with a Finnigan Xcalibur software package. Selected reaction monitoring (SRM) of the DMG derivatives was acquired in the positive ion mode using electrospray ionization (ESI). MS parameters were optimized using DMG-Chol and were as follows: spray voltage at 4500 V, capillary temperature at 300 °C, auxiliary nitrogen gas pressure at 55 psi, and sheath gas pressure at 60 psi. Collision energy (CE) was optimized for each sterol and oxysterol under a collision gas pressure of 1.5 mTorr. The monitored transitions are listed in Tables 1 and 2. Endogenous sterol levels were quantified based on the known matched deuterated standard amount and then normalized to cell count. Oxysterol levels were quantified using d7-Chol as standard and a response factor that was determined for each compound, then also normalized to cell count. Since the DMG derivatization is the same reaction among all of the sterols, it does lack some specificity when compared to the PTAD method. To compensate for this, SRMs were optimized for each sterol so that its signal was enhanced relative to other sterols. In cases where the SRMs were similar, the sterols were separated chromatographically.
Statistical Analysis.
Statistical analyses were performed using GraphPad Prism 8 for Windows. Unpaired two-tailed t tests were performed for individual comparisons between two groups. Discovery was determined using the two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli, with Q = 5%. Each analysis was performed individually, without assuming a consistent SD. Statistical tests showed a normal distribution in all comparison groups. The p values for statistically significant differences are highlighted in the figure legends.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Forbes Porter (NIH) for donating the T93M mice used in this study.
Funding
This work was supported by the National Institutes of Health Grants NICHD HD064727 (N.A.P.), NIMH R01 MH110636 (K.M., N.A.P.), and R01 MH067234 (K.M.).
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschemneuro.0c00765.
Complete contact information is available at: https://pubs.acs.org/10.1021/acschemneuro.0c00765
Additional figures showing prescribed medications used in the study; cholesterol oxysterols included in this LC–MS/MS analysis; comparison of PTAD and DMG methods; neurons and astrocytes exposed to sertraline; profile of cholesterol oxysterols in neurons and astrocytes; ratio of 7-DHD/Des and 7-DHC/Chol for neurons and astrocytes; representative mass spectrum and fragmentations for DMG-derivatized sterols; synthesis of d6-Lan, d7-dHLan, d6-14d-Zym, d7-14d-Zyme, d6-Zym, d7-Zyme, d6-DHL, d7-Lath, d6-7-DHD, d6-8-DHD, d6-Des, d7-Chol; statistics (PDF)
Contributor Information
Keri A. Tallman, Department of Chemistry, Vanderbilt Institute of Chemical Biology and Vanderbilt Kennedy Center for Research on Human Development, Vanderbilt University, Nashville, Tennessee 37235, United States
Luke B. Allen, Munroe-Meyer Institute for Genetics and Rehabilitation and Department of Biochemistry and Molecular Biology, College of Medicine, University of Nebraska Medical Center, Omaha, Nebraska 68105, United States
Korinne B. Klingelsmith, Munroe-Meyer Institute for Genetics and Rehabilitation, University of Nebraska Medical Center, Omaha, Nebraska 68105, United States
Allison Anderson, Munroe-Meyer Institute for Genetics and Rehabilitation, University of Nebraska Medical Center, Omaha, Nebraska 68105, United States.
Thiago C. Genaro-Mattos, Munroe-Meyer Institute for Genetics and Rehabilitation, University of Nebraska Medical Center, Omaha, Nebraska 68105, United States
Károly Mirnics, Munroe-Meyer Institute for Genetics and Rehabilitation and Department of Biochemistry and Molecular Biology, College of Medicine, University of Nebraska Medical Center, Omaha, Nebraska 68105, United States.
Ned A. Porter, Department of Chemistry, Vanderbilt Institute of Chemical Biology and Vanderbilt Kennedy Center for Research on Human Development, Vanderbilt University, Nashville, Tennessee 37235, United States
Zeljka Korade, Department of Pediatrics and Department of Biochemistry and Molecular Biology, College of Medicine, University of Nebraska Medical Center, Omaha, Nebraska 68198, United States.
REFERENCES
- (1).Kim HY, Korade Z, Tallman KA, Liu W, Weaver CD, Mirnics K, and Porter NA (2016) Inhibitors of 7-Dehydrocholesterol Reductase: Screening of a Collection of Pharmacologically Active Compounds in Neuro2a Cells. Chem. Res. Toxicol. 29, 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Korade Z, Kim HY, Tallman KA, Liu W, Koczok K, Balogh I, Xu L, Mirnics K, and Porter NA (2016) The Effect of Small Molecules on Sterol Homeostasis: Measuring 7-Dehydrocholesterol in Dhcr7-Deficient Neuro2a Cells and Human Fibroblasts. J. Med. Chem. 59, 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Lauth M, Rohnalter V, Bergström A, Kooshesh M, Svenningsson P, and Toftgård R (2010) Antipsychotic Drugs Regulate Hedgehog Signaling by Modulation of 7-Dehydrocholesterol Reductase Levels. Mol. Pharmacol. 78, 486–496. [DOI] [PubMed] [Google Scholar]
- (4).Horling A, Müller C, Barthel R, Bracher F, and Imming P (2012) A New Class of Selective and Potent 7-Dehydrocholesterol Reductase Inhibitors. J. Med. Chem. 55, 7614–7622. [DOI] [PubMed] [Google Scholar]
- (5).Hall P, Michels V, Gavrilov D, Matern D, Oglesbee D, Raymond K, Rinaldo P, and Tortorelli S (2013) Aripiprazole and trazodone cause elevations of 7-dehydrocholesterol in the absence of Smith–Lemli–Opitz Syndrome. Mol. Genet. Metab. 110, 176–178. [DOI] [PubMed] [Google Scholar]
- (6).Korade Z, Liu W, Warren EB, Armstrong K, Porter NA, and Konradi C (2017) Effect of psychotropic drug treatment on sterol metabolism. Schizophr Res. 187, 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Genaro-Mattos TC, Tallman KA, Allen LB, Anderson A, Mirnics K, Korade Z, and Porter NA (2018) Dichlorophenyl piperazines, including a recently-approved atypical antipsychotic, are potent inhibitors of DHCR7, the last enzyme in cholesterol biosynthesis. Toxicol. Appl. Pharmacol. 349, 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Fliesler SJ, and Bretillon L (2010) The ins and outs of cholesterol in the vertebrate retina. J. Lipid Res. 51, 3399–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Fliesler SJ, Peachey NS, Herron J, Hines KM, Weinstock NI, Rao SR, and Xu L (2018) Prevention of Retinal Degeneration in a Rat Model of Smith-Lemli-Opitz Syndrome. Sci. Rep. 8, 1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Pfeffer B, Xu L, Porter N, Rao S, and Fliesler S (2016) Differential cytotoxic effects of 7-dehydrocholesterol derived oxysterols on cultured retina derived cells: Dependence on sterol structure, cell type, and density. Exp. Eye Res. 145, 297–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Xu L, Liu W, Sheflin LG, Fliesler SJ, and Porter NA (2011) Novel oxysterols observed in tissues and fluids of AY9944-treated rats - a model for Smith-Lemli-Opitz Syndrome. J. Lipid Res. 52, 1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Xu L, Sheflin LG, Porter NA, and Fliesler SJ (2012) 7-Dehydrocholesterol-derived oxysterols and retinal degeneration in a rat model of Smith-Lemli-Opitz syndrome. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 1821, 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Porter FD (2003) Human malformation syndromes due to inborn errors of cholesterol synthesis. Curr. Opin. Pediatr. 15, 607–613. [DOI] [PubMed] [Google Scholar]
- (14).Porter FD (2006) Cholesterol precursors and facial clefting. J. Clin. Invest. 116, 2322–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Porter FD (2008) Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur. J. Hum. Genet. 16, 535–541. [DOI] [PubMed] [Google Scholar]
- (16).Porter FD, and Herman GE (2011) Malformation syndromes caused by disorders of cholesterol synthesis. J. Lipid Res. 52, 6–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Tint GS, Irons M, Elias ER, Batta AK, Frieden R, Chen TS, and Salen G (1994) Defective cholesterol biosynthesis associated with the Smith-Lemli-Opitz syndrome. N. Engl. J. Med. 330, 107–113. [DOI] [PubMed] [Google Scholar]
- (18).Waterham HR, and Hennekam RCM (2012) Mutational spectrum of Smith-Lemli-Opitz syndrome. Am. J. Med. Genet., Part C 160C, 263–284. [DOI] [PubMed] [Google Scholar]
- (19).Witsch-Baumgartner M, Fitzky BU, Ogorelkova M, Kraft HG, Moebius FF, Glossmann H, Seedorf U, Gillessen-Kaesbach G, Hoffmann GF, Clayton P, Kelley RI, and Utermann G (2000) Mutational spectrum in the Delta7-sterol reductase gene and genotype-phenotype correlation in 84 patients with Smith-Lemli-Opitz syndrome. Am. J. Hum. Genet. 66, 402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Witsch-Baumgartner M, Schwentner I, Gruber M, Benlian P, Bertranpetit J, Bieth E, Chevy F, Clusellas N, Estivill X, Gasparini G, Giros M, Kelley RI, Krajewska-Walasek M, Menzel J, Miettinen T, Ogorelkova M, Rossi M, Scala I, Schinzel A, Schmidt K, Schonitzer D, Seemanova E, Sperling K, Syrrou M, Talmud PJ, Wollnik B, Krawczak M, Labuda D, and Utermann G (2007) Age and origin of major Smith-Lemli-Opitz syndrome (SLOS) mutations in European populations. J. Med. Gen 45, 200–209. [DOI] [PubMed] [Google Scholar]
- (21).Genaro-Mattos TC, Allen LB, Anderson A, Tallman KA, Porter NA, Korade Z, and Mirnics K (2019) Maternal aripiprazole exposure interacts with 7-dehydrocholesterol reductase mutations and alters embryonic neurodevelopment. Mol. Psychiatry 24, 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Genaro-Mattos TC, Anderson A, Allen LB, Tallman KA, Porter NA, Korade Z, and Mirnics K (2020) Maternal cariprazine exposure inhibits embryonic and postnatal brain cholesterol biosynthesis. Mol. Psychiatry 25, 2685–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Allen LB, Genaro-Mattos TC, Anderson A, Porter NA, Mirnics K, and Korade Z (2020) Amiodarone Alters Cholesterol Biosynthesis through Tissue-Dependent Inhibition of Emopamil Binding Protein and Dehydrocholesterol Reductase 24. ACS Chem. Neurosci. 11, 1413–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Pataj Z, Liebisch G, Schmitz G, and Matysik S (2016) Quantification of Oxysterols in Human Plasma and Red Blood Cells by Liquid Chromatography High-Resolution Tandem Mass Spectrometry. J. Chromatogr A 1439, 82–88. [DOI] [PubMed] [Google Scholar]
- (25).Honda A, Yamashita K, Hara T, Ikegami T, Miyazaki T, Shirai M, Xu G, Numazawa M, and Matsuzaki Y (2009) Highly Sensitive Quantification of Key Regulatory Oxysterols in Biological Samples by LC-ESI-MS/MS. J. Lipid Res. 50, 350–357. [DOI] [PubMed] [Google Scholar]
- (26).Griffiths WJ, Abdel-Khalik J, Crick PJ, Ogundare M, Shackleton CH, Tuschl K, Kwok MK, Bigger BW, Morris AA, Honda A, Xu L, Porter NA, Bjorkhem I, Clayton PT, and Wang Y (2017) Sterols and oxysterols in plasma from Smith-Lemli-Opitz syndrome patients. J. Steroid Biochem. Mol. Biol. 169, 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).McDonald JG, Smith DD, Stiles AR, and Russell DW (2012) A comprehensive method for extraction and quantitative analysis of sterols and secosteroids from human plasma. J. Lipid Res. 53, 1399–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lutjohann D, Breuer O, Ahlborg G, Nennesmo I, Siden A, Diczfalusy U, and Bjorkhem I (1996) Cholesterol homeostasis in human brain: evidence for an age-dependent flux of 24S-hydroxycholesterol from the brain into the circulation. Proc. Natl. Acad. Sci. U. S. A. 93, 9799–9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Bjorkhem I, Leoni V, and Svenningsson P (2019) On the fluxes of side-chain oxidized oxysterols across blood-brain and blood-CSF barriers and origin of these steroids in CSF (Review). J. Steroid Biochem. Mol. Biol. 188, 86–89. [DOI] [PubMed] [Google Scholar]
- (30).Brown AJ, and Galea AM (2010) Cholesterol as an Evolutionary Response to Living with Oxygen. Evolution 64, 2179–2183. [DOI] [PubMed] [Google Scholar]
- (31).Brown AJ, and Jessup W (2009) Oxysterols: Sources, Cellular Storage and Metabolism, and New Insights Into Their Roles in Cholesterol Homeostasis. Mol Aspects Med 2009, 30 (3), 111–122. Mol. Aspects Med. 30, 111–122. [DOI] [PubMed] [Google Scholar]
- (32).Testa G, Staurenghi E, Zerbinati C, Gargiulo S, Iuliano L, Giaccone G, Fantò F, Poli G, Leonarduzzi G, and Gamba P (2016) Changes in Brain Oxysterols at Different Stages of Alzheimer’s Disease: Their Involvement in Neuroinflammation. Redox Biol. 10, 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Sharpe LJ, Coates HW, and Brown AJ (2020) Post-Translational Control of the Long and Winding Road to Cholesterol. J. Biol. Chem. 295, 17549–17559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Liu W, Xu L, Lamberson C, Haas D, Korade Z, and Porter NA (2014) A highly sensitive method for analysis of 7-dehydrocholesterol for the study of Smith-Lemli-Opitz syndrome. J. Lipid Res. 55, 329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wages PA, Kim HH, Korade Z, and Porter NA (2018) Identification and characterization of prescription drugs that change levels of 7-dehydrocholesterol and desmosterol. J. Lipid Res. 59, 1916–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Wages PA, Joshi P, Tallman KA, Kim HH, Bowman AB, and Porter NA (2020) Screening ToxCast for Chemicals That Affect Cholesterol Biosynthesis: Studies in Cell Culture and Human Induced Pluripotent Stem Cell-Derived Neuroprogenitors. Environ. Health Perspect. 128, 017014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Xu L, Mirnics K, Bowman AB, Liu W, Da J, Porter NA, and Korade Z (2012) DHCEO accumulation is a critical mediator of pathophysiology in a Smith-Lemli-Opitz syndrome model. Neurobiol. Dis. 45, 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Sun MY, Linsenbardt AJ, Emnett CM, Eisenman LN, Izumi Y, Zorumski CF, and Mennerick S (2016) 24(S)-Hydroxycholesterol as a Modulator of Neuronal Signaling and Survival. Neuroscientist 22, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Mitsche MA, McDonald JG, Hobbs HH, and Cohen JC (2015) Flux Analysis of Cholesterol Biosynthesis in Vivo Reveals Multiple Tissue and Cell-Type Specific Pathways. eLife 4, e07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Crick PJ, Yutuc E, Abdel-Khalik J, Saeed A, Betsholtz C, Genove G, Bjorkhem I, Wang Y, and Griffiths WJ (2019) Formation and metabolism of oxysterols and cholestenoic acids found in the mouse circulation: Lessons learnt from deuterium-enrichment experiments and the CYP46A1 transgenic mouse. J. Steroid Biochem. Mol. Biol. 195, 105475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Boland MR, and Tatonetti NP (2016) Investigation of 7-dehydrocholesterol reductase pathway to elucidate off-target prenatal effects of pharmaceuticals: a systematic review. Pharmacogenomics J. 16, 411–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bukelis I, Porter FD, Zimmerman AW, and Tierney E (2007) Smith-Lemli-Opitz syndrome and autism spectrum disorder. Am. J. Psychiatry 164, 1655–1661. [DOI] [PubMed] [Google Scholar]
- (43).Korade Z, Allen LB, Anderson A, Tallman KA, Genaro-Mattos TC, Porter NA, and Mirnics K (2021) Trazodone effects on developing brain. Transl. Psychiatry 11, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Xu L, and Porter NA (2014) Reactivities and products of free radical oxidation of cholestadienols. J. Am. Chem. Soc. 136, 5443–5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Xu L, and Porter NA (2915) Free radical oxidation of cholesterol and its precursors: Implications in cholesterol biosynthesis disorders. Free Radical Res. 49, 835–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Porter NA, Xu L, and Pratt DA (2020) Reactive Sterol Electrophiles: Mechanisms of Formation and Reactions with Proteins and Amino Acid Nucleophiles. Chemistry 2, 390–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Xu L, Davis TA, and Porter NA (2009) Rate Constants for Peroxidation of Polyunsaturated Fatty Acids and Sterols in Solution and in Liposomes. J. Am. Chem. Soc. 131, 13037–13044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Korade Z, Xu L, Shelton R, and Porter NA (2010) Biological Activities of 7-Dehydrocholesterol-Derived Oxysterols: Implications for Smith-Lemli-Opitz Syndrome. J. Lipid Res. 51, 3259–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Sever N, Mann RK, Xu L, Snell WJ, Hernandez-Lara CI, Porter NA, and Beachy PA (2016) Endogenous B-ring oxysterols inhibit the Hedgehog component Smoothened in a manner distinct from cyclopamine or side-chain oxysterols. Proc. Natl. Acad. Sci. U. S. A. 113, 5904–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Correa-Cerro LS, Wassif CA, Kratz L, Miller GF, Munasinghe JP, Grinberg A, Fliesler SJ, and Porter FD (2006) Development and characterization of a hypomorphic SLOS mouse model and efficacy of simvastatin therapy. Hum. Mol. Genet. 15, 839–851. [DOI] [PubMed] [Google Scholar]
- (51).Rosado-Abon A, Dios-Bravo G. d., Rodriguez-Sotres R, and Iglesias-Arteaga M (2012) A Synthesis and plant growth promoting activity of polyhydroxylated ketones bearing the 5α-hydroxy-6-oxo moiety and cholestane side chain. Steroids 77, 461–466. [DOI] [PubMed] [Google Scholar]
- (52).Cui J-G, Fan L, Huang L-L, Liu H-L, and Zhou A-M (2009) Synthesis and evaluation of some steroidal oximes as cytotoxic agents: Structure/activity studies. Steroids 74, 62–72. [DOI] [PubMed] [Google Scholar]
- (53).Zielinski ZAM, and Pratt DA (2019) H-Atom Abstraction vs Addition: Accounting for the Diverse Product Distribution in the Autoxidation of Cholesterol and Its Esters. J. Am. Chem. Soc. 141, 3037–3051. [DOI] [PubMed] [Google Scholar]
- (54).Allen LB, Genaro-Mattos TC, Porter NA, Mirnics K, and Korade Z (2019) Desmosterolosis and desmosterol homeostasis in the developing mouse brain. J. Inherited Metab. Dis. 42, 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Goudriaan A, Camargo N, Carney KE, Oliet SH, Smith AB, and Verheijen MH (2014) Novel cell separation method for molecular analysis of neuron-astrocyte co-cultures. Front. Cell. Neurosci. 8, 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.