Abstract
The cell membrane possesses an extensive library of proteins, carbohydrates, and lipids that control a significant portion of inter- and intracellular functions, including signaling, proliferation, migration, and adhesion, among others. Augmenting the cell surface composition would open possibilities for advances in therapy, tissue engineering, and probing fundamental cell processes. While genetic engineering has proven effective for many in vitro applications, these techniques result in irreversible changes to cells and are difficult to apply in vivo. Another approach is to instead attach exogenous functional groups to the cell membrane without changing the genetic nature of the cell. This review focuses on more recent approaches of non-genetic methods of cell surface modification through metabolic pathways, anchorage by hydrophobic interactions, and chemical conjugation. Benefits and drawbacks of each approach are considered, followed by a discussion of potential applications for non-genetic cell surface modification and an outlook on the future of the field.
Keywords: Click chemistry, bioconjugation, affinity guided, metabolic labeling, T-cell therapy
Graphical Abstract
Introduction
Decorating cellular surfaces with exogenous functionalities has garnered significant interest in recent years to aid with delivering antibody-drug conjugates,1 mediating extracellular communications,2 and conjugating fluorophores to the cell surface3 for probing and imaging cellular and pathological function.4 Originally, specific receptors on the cell surface were directly targeted by antibodies or other moieties with high binding specificity.5 However, because these interactions are inherently transient,6 focus has shifted to introducing cell surface functionalities through more permanent means. Genetic modification of cellular proteins and receptors is a common tool in vitro, although this approach can lead to unintended changes to the cells.7 Translation of gene delivery in vivo has also been notoriously challenging, as targeted viral vectors often elicit immune responses in patients, even leading to patient death and the halting of clinical trials.8 Less immunogenic, non-viral methods have thus far shown poor transfection.9 Third, the success of any application depending on genetic modification of cellular surfaces relies heavily on the expression levels of the epitopes that are being expressed. These expression levels can be unpredictable and inconsistent across samples, and thus normalizing the expression levels becomes very important in these circumstances to get reliable data.10
As an alternative to genetic engineering, natural components of the cell membrane such as lipids and glycans can be modified through various non-genetic pathways. This approach not only produces more permanent methods of cellular probing than antibody targeting but also provides more reliable control over cellular functions.11 Ideal cell surface engineering methods should introduce functionalities without affecting cell fate and function such as cell survival, proliferation and other cellular activities.12 This review will mainly focus on current non-genetic methods of cell surface modification, including metabolic approaches, lipid-based insertions, and targeted chemical modification.
Conjugation to Functional Groups Introduced Metabolically
Most cells are coated with a dense layer of glycan sugar derivatives that play essential roles in biological processes. The cell’s own machinery involving glycan’s metabolic pathway can be utilized to introduce various biorthogonal functionalities such as azides, alkynes, or ketones on the cell surface.13 Once on the surface, these functionalities can be selectively conjugated to phosphine or hydrazide labeled fluorophores or antibodies through a Staudinger ligation.14 This two-step click chemistry pathway, known as metabolic glycan labeling (MGL), depends on the expression of natural glycans on the cell surface using their sugar analogues. The biochemical mechanism of how unnatural sialic acids become displayed on cell surfaces has been well-reviewed elsewhere.15 Sialic acid, or more commonly N-acetyl-neuraminic acid (Neu5Ac), is a nine carbon sugar that is the most common cellular glycan that is biosynthesized from its precursor N-acetylmannosamine (ManNAc).13 Reutter and coworkers have demonstrated that unnatural mannosamine derivatives, (where the N-acetyl groups have been replaced by N-azido or N-propanoyl groups), can be incorporated on the cell surface through the sialic acid pathway that involves a variety of enzymes including the nuclear cytidine monophosphate-Neu5Ac (CMP-Neu5Ac) synthase, cytosolic epimerase, cytosolic CMP-Neu5Ac hydroxylase, Golgi sialyltransferases, and sialidases. Thus, the azido or the keto functionalities are incorporated as glycoconjugates on the cell surface.16 In a landmark study, Bertozzi et al introduced N-levulinoylsialic acid (SiaLev) in Jurkat cells using N-levulinoylmannosamine (ManLev) containing a ketone functional group substituting for the N-acetyl group in the natural sugar. After expression on the cell surface through glycan metabolism, the ketone was then conjugated to a biotinamidocaproylhydrazide and then tagged with FITC-avidin (Figure 1).17 In a remarkable followup, Bertozzi and coworkers introduced an azido functional group through N-acetylazidosialic acid (SiaNAz) in mice splenocytes in vivo using N-acetylazidomannosamine (ManNAz) and the glycan metabolic pathways. The acetyl groups were cleaved in the cell by naturally-occurring carboxyesterases, and once presented on the cell membrane the azido group was conjugated to a FLAG tag (Phos-FLAG) and labeled with a FITC conjugated anti-FLAG antibody.18 Finally, Bertozzi and Francis also reported ssDNA conjugated to phosphines introduced on the surface of the cells to study cellular adhesion based on DNA hybridization using the Staudinger ligation reactions between phosphines and metabolically driven glycan introduced azides.19
Figure 1.
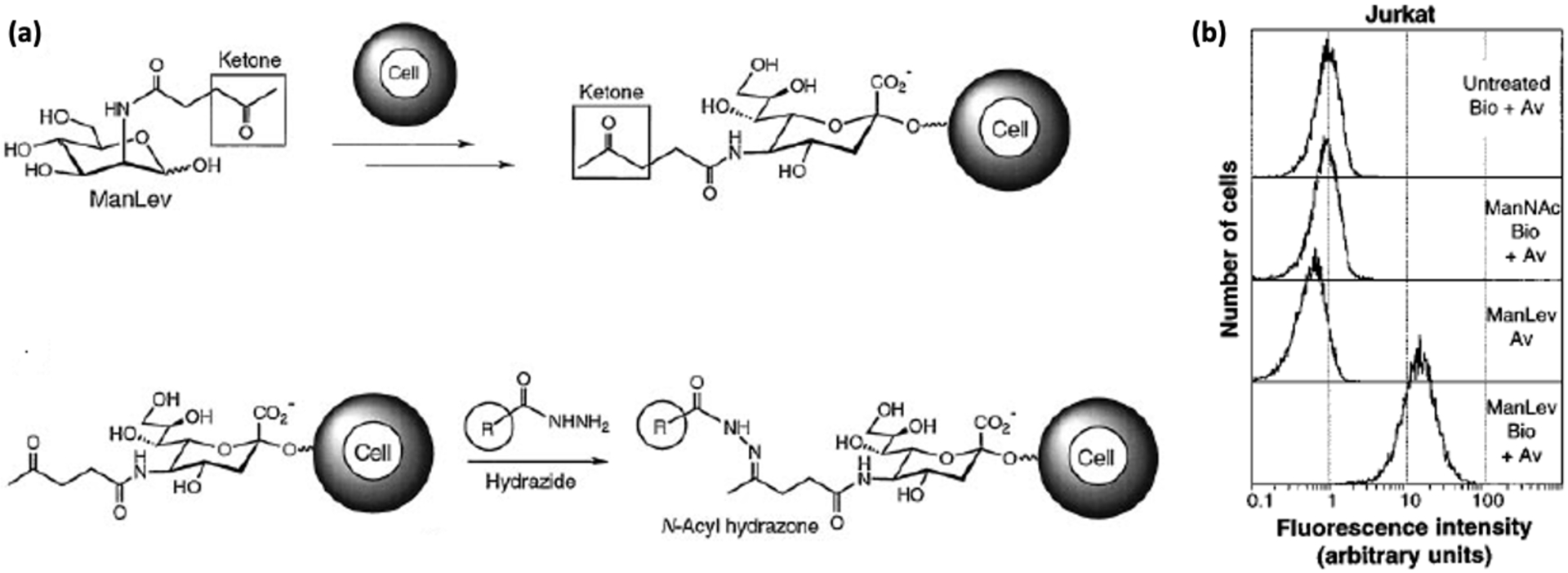
(a) Schematic for incorporating ketone groups on a cell surface using a glycan metabolic pathway for Staudinger ligation with biotinylated hydrazides for detection with FITC-avidin. (b) Flow cytometry of Jurkat cells reveal that the addition of azido sugars (ManLev-Bio+Av) increase the fluorescence 30-fold as compared to the controls [18]. Reproduced with permission from Mahal, L. K.; Yarema, K. J.; Bertozzi, C. R. Engineering Chemical Reactivity on Cell Surfaces through Oligosaccharide Biosynthesis. Science 1997, 276 (5315), 1125–1128.
Because the Staudinger ligation reactions between azides and phosphines can exhibit slow kinetics, they are not suited for detection of rapid biological processes on a minute time scale.20 Hence, Finn et al used ManNAz to incorporate azido groups on CHO and Hela cells in the form of SiaNAz by the same glycan metabolic pathway, then used copper-catalyzed azide-alkyne cycloaddition (CuAAC) for surface labeling of live cells using AF488-alkyne (Figure 2).21 Unfortunately, the Cu/ascorbate catalyst system is toxic to cells due to the generation of reactive oxidative species (ROS); although ROS generation should not be an issue for fixed cells, it poses a problem for live cell imaging.22 In order to address this issue, the authors also added tris(hydroxypropyltriazolyl)methylamine (THPTA) as a scavenger for ROS and promotes Cu-assisted peroxide degradation. The dehydroascorbate that is produced in this process is then captured by aminoguanidine and thus the combined effect of these two additives led to a robust method of labeling live cell surfaces.21
Figure 2.
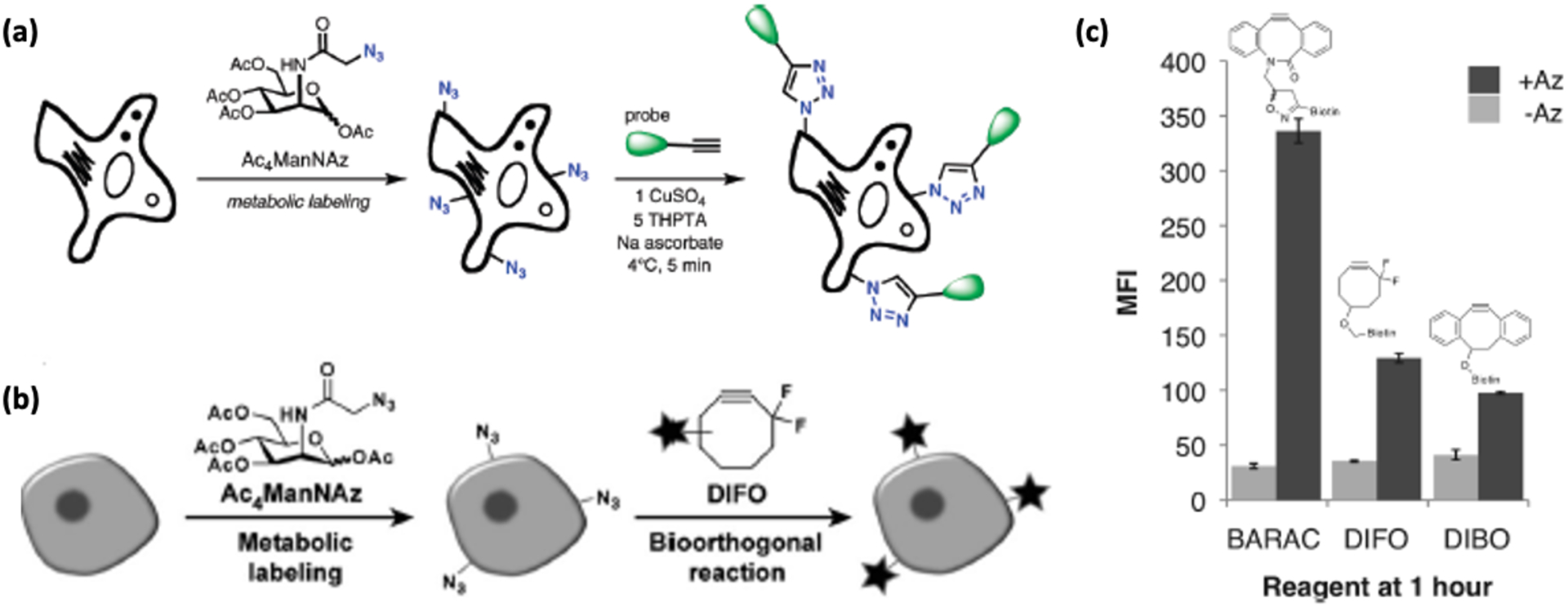
(a) Strategies for cell labeling with azides using metabolic expression of non-natural sugars, followed by Cu-catalyzed click chemistry labeling [21]. (b) Cu-free methods of cell labeling with strained cyclooctynes [23]. Reproduced with permission from: Baskin, J. M.; Prescher, J. A.; Laughlin, S. T.; Agard, N. J.; Chang, P. V.; Miller, I. A.; Lo, A.; Codelli, J. A.; Bertozzi, C. R. Copper-Free Click Chemistry for Dynamic in Vivo Imaging. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (43), 16793–16797. (c) Comparison of Jurkat cell labeling with a biarylazacyclooctyne (BARAC), fluorinated cyclooctyne (DIFO), and non-fluorinated cyclooctyne (DBCO) [26].
In another approach towards copper-free conjugation, Bertozzi and coworkers introduced ring strain and electron withdrawing groups to enhance the reactivity of cycloalkynes to azides.23 These modified cyclooctynes (OCT), including difluorinated cyclooctyne (DIFO), showed >10-fold faster reaction kinetics (k = 0.076 M−1 s−1)24 than their unmodified counterparts (k = 0.0024 M−1 s−1)25 without needing toxic metal catalysts. Adding additional ring strain as a biarylazacyclooctyne (BARAC) improved kinetics by another order of magnitude (k = 0.96 M−1 s−1) (Figure 2).26 In an example of applications of strain promoted azide alkyne cycloaddition (SPAAC), Kang et al incubated lung carcinoma cells A549 with peracetylated ManNAz (Ac4ManNAz) to introduce the azide groups as unnatural sialic acid residues on the cell surface. The azide labeled cells were then implanted in the liver athymic nude mice and dibenzocyclooctyne (DBCO)-Cy5 was intravenously injected into the mice. DBCO conjugation was shown to be successful by near infrared fluorescence intravital imaging.27
In order to further improve the reaction kinetics of CuAAC for cell labeling, Wu and co-workers showed that replacing THPTA with tris(tert-butyl triazolylmethyl) amine based ligands in the copper free reaction increased cycloaddition yields multi-fold in living cells. The tert-buytl and the hydrogen sulphate groups present in bis((tert-butyl triazolylmethyl) amino methyl) ethyl hydrogen sulphate (BTTES) provided a delicate balance of solubility and reactivity to these ligands but did not reduce the required Cu concentration below 50 μM.28 To further confer biocompatibility to these methods, the authors replaced the hydrogen sulphate group in BTTES to an acetic acid group to produce a new additive bis((tert-butyl triazolylmethyl) amino methyl) acetic acid (BTTAA). The acetic acid ionizes at physiological pH to acetate, which acts as a weak donor to the CuI ions and helps the formation of strained copper triazole intermediates by increasing the electron density over the metal ion. This ligand switch not only accelerates the rate of cycloaddition but also allows lower Cu concentrations (30 μM).29
Finally, because cell display of azides is required for both the Staudinger ligation and the strain-promoted click reaction, other click chemistries have been devised to allow dual labeling of different cell receptors.30 For example, recent efforts have employed the use of inverse electron demand Diels-Alder reactions for incorporating cyclooctynes,31 bicyclononynes,32 and norbornenes33 directly on the cell surface through glycan metabolism. However, these structures are too large to be efficiently incorporated into their corresponding sialic acids from their mannose amine counterparts.30 Thus, researchers turned towards ring-strained scaffolds like methyl substituted cyclopropenes (Cp), which are inherently absent in eukaryotes and can be easily ingested by the cells owing to their small size.34 These alkenes are incorporated on the cell surface through the metabolism of N-acetylneuraminic acid (Cp-NeuAc), then ligated to electron deficient tetrazine conjugated probes such as tetrazine-Cy3 (Tz-Cy3)35 or indirectly via tetrazine-biotin (Tz-bio).36 Conjugation was further improved by the group of Xin-Shan Ye, who demonstrated that (N-(cycloprop-2-ene-1-ylcarbonyl)) sugars (NCp) have higher stability, better metabolic efficiency, and faster tagging kinetics to FITC-Tz both in vitro and in vivo.37 Through these processes, cells fed both azide and alkene-derived sugars can impart a membrane with orthogonal chemical reactivity.
Like glycans, phospholipids are integral parts of the cell membrane and the most common phospholipid headgroup in eukaryotic cells is choline (Cho). Cho-containing phospholipids play an important role in both intra- and intercellular signaling and undergo cellular uptake by various membrane transporters, followed by incorporation as phospholipids by the high energy intermediate CDP-Cho.38 Salic et al utilized this metabolic pathway to incorporate propargylcholine (PCho), an alkyne bearing moiety, into phosphatidylcholine (PC). These modified phospholipids were then labled by azide-modified Alexa568 and fluorescein dyes through CuAAC between the terminal alkyne group of PCho and the azide labeling probes, followed by visualization in the kidney, liver, and spleen cells of three week old mice.39 Griese et al demonstrated the transport of phospholipids in ATP binding cassette sub family A member 3 (ABCA3) in lamellar bodies by tracking the choline phospholipids incorporated into the pulmonary cells. The authors first transfected the human carcinoma A549 lung cancer cells with HA-tagged ABCA3, which was detected on cell membranes by a series of antibodies and conjugation of TAMRA-azide.40 Thus, novel assays of cellular imaging and treatment can be developed by tracking the propargyl-Cho labeled lipids inside cells which are introduced using the Cho phospholipid metabolic pathway.
Membrane Insertion with Hydrophobic Groups
Rather than relying on a cell feedstock to supply a clickable functional group, another option is to incorporate the exogenous biomolecule into the membrane with a hydrophobic tag.41, 42 The interactions between the hydrophobic part of the conjugate and the lipid bilayer membrane causes the functional groups to be anchored to the cell surface. These interactions are weaker than covalent conjugation and thus are easily released from the cell surface, but they do not disturb cellular functions and are non-toxic to the cells.43 To demonstrate this approach, Iwata et al designed several lipid-poly(ethylene glycol)-amine (PEG-lipid) conjugates with different alkyl chain lengths to study the interactions between the lipid tails and the membrane surface.44 After the insertion of the PEG-lipid conjugates onto the human T-lymphocyte cells (CCRF-CEM) and subsequent reaction with fluorescein isothiocyanate-N-hydroxyl succinimide (FITC-NHS), the cells were imaged by confocal microscopy. The highest fluorescence and surface density was observed for distearoyl-phosphoethanolamine-PEG (DSPE-PEG), compared to PEG chains with dioleoyl, dipalmitoyl, or dimiristoyl lipids (DOPE, DPPE, and DMPE, respectively); therefore, longer, saturated hydrophobic tails produced stronger retention in the cell membrane (Figure 3). Lim et al built upon this approach by modifying the DSPE-PEG with homing peptides to improve the targeting specificity of the conjugates. The DSPE-PEGs were modified with CRPPR peptide to bind to cysteine-rich protein 2 (CRIP2) as well as a FITC-labeled super paramagnetic iron oxide nanoparticles, which showed attachment to a variety of human cell lines.45
Figure 3.
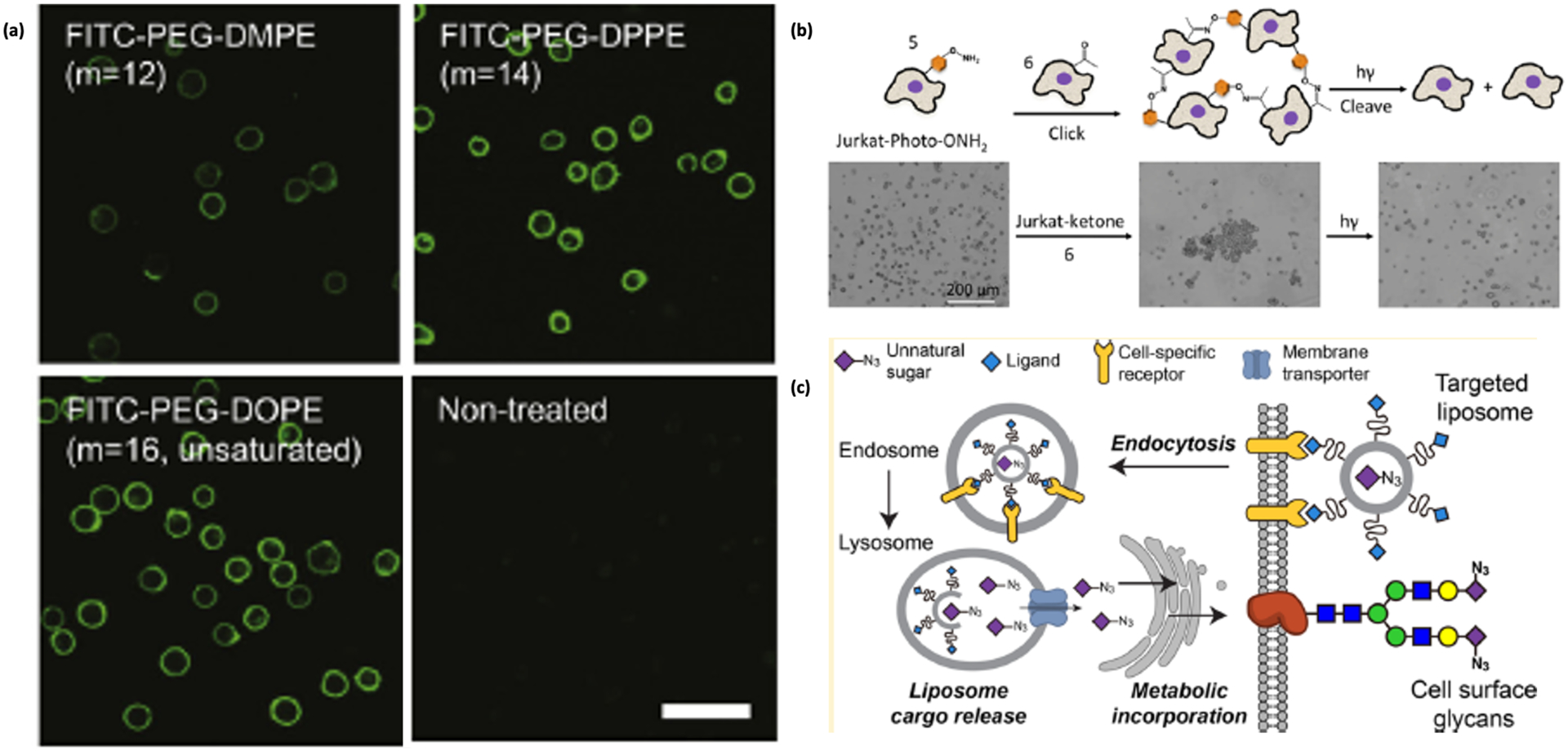
(a) Confocal fluorescence images of CCRF-CEM cells after incubating with lipids of different lengths for 30 min [44]. Reproduced with permission from: Itagaki, T.; Arima, Y.; Kuwabara, R.; Kitamura, N.; Iwata, H. Interaction between Cells and Poly (Ethylene Glycol) -Lipid Conjugates. Colloids Surf. B Biointerfaces 2015, 135, 765–773. (b) Schematic showing labeling of cells with oxyamines and their use in cell assembly, with images showing the sequential formation and disassembly of Jurkat cells by this technique [52]. Reproduced with persmission via Creative Commons from: Luo, W.; Pulsipher, A.; Dutta, D.; Lamb, B. M.; Yousaf, M. N. Remote Control of Tissue Interactions via Engineered Photo-Switchable Cell Surfaces. Sci. Rep. 2014, 4, 1–8. (c) Schematic of azide delivery by liposome assisted bioorthogonal reporter (LABOR) [53].
For cell assembly applications, Gartner and coworkers anchored DNA-conjugated C16–18 dialkylphosphoglycerides directly to cell surfaces. These DNA-decorated non-adherent (Jurkat) and adherent cells (HeLa, MCF-10A) could be assembled into oligomeric structures using DNA hybridization.46 Building upon this work, Gartner et al increased the yield of DNA-lipids on the surface by using a combination of a 100 base anchor and a 20 base complementary co-anchor connected to fatty acid amines. The retention of oligonucleotides in the cell membrane was seven to eight times higher than DNA anchored with dialkylphosphoglycerides or doubly anchored cholesterol. The anchor and the co-anchor strands hybridized in a complementary “lock” region that increased the hydrophobicity of the duplex and increased residence in the membrane.47
While these lipid-based methods are straightforward, inexpensive, and can target a variety of cell lines,48 ultimately these approaches remain nonspecific in terms of labeling. In order to use these lipid conjugates in a more specific manner, Yousaf et al developed liposomes tethered with mutually recognizable molecular pairs like ketones and oxyamines. When added to the cell surface, this pair reacts to form a stable oxime bond, allowing conjugation of fluorophores or other probes. In these studies, two sets of large unilamellar vesicles (LUVs) were synthesized, one containing an oxyamine and a FRET donor dye, and the other possessing a ketone and an acceptor dye. Upon mixing, the liposomes aggregated, adhered, and finally fused due to the formation of stable oxime linkages between them. This same strategy was employed to attach cells to surfaces. To decorate the cell surfaces of fibroblasts with fluorophores, the liposomes were first adhered to the cell surface using a cation liquid. Once the keto or oxy group was anchored on the cell surface, rhodamine-oxyamines and fluorescein-ketones were added to the fibroblast cell cultures that resulted in fluorescent labeling of the cells by chemoselective oxime formation.49
Yousaf and co-workers also extended their work to both attachment of cells to substrates and self-assembly into tissue-like structures. First, cells were attached to gold substrates that were preprinted with oxyamine and aldehyde groups. This approach resulted in the patterning of fibroblasts with the complementary lipid functionalities on their surface. The remainder of the gold substrates were coated with tetraethylene glycol to prevent non-specific cell adhesion through integrin-extracellular matrix (ECM) interactions.50 This idea was further utilized in another work from the same group, in which a UV-labile nitrophenyl oxyamine group51 was introduced on non-adherent Jurkat cells. When conjugated with cells possessing the complementary keto group, the cells formed multi-cellular spheroid assemblies due to the development of oxime linkages (Figure 3). The oxime linkages were cleaved rapidly upon exposure to UV light for 5 min, which resulted in the complete disassembly of tissue-like spheroid structures into individual cells. This photoswitchable tissue assembly technique can be controlled by simple cell irradiation because of the photocleavable group on the lipid molecules anchored on the cell surface, producing both cell-surface adhesion and cell-cell aggregates.52
While useful in vitro, one disadvantage of the methods discussed above is that the conjugates by themselves are not capable of in vivo delivery for cell augmentation in live animals. Liposome-assisted bioorthogonal reporters (LABORs), developed by Chen and coworkers, will help to expand the utility of hydrophobic delivery to various applications.53 Chen and coworkers used LABORs to deliver azido sialic acid on cell membranes using folate targeting to HeLa cells (Figure 3). Azido sialic acid was encapsulated within a liposome layer constructed of folic acid-DSPE-PEG2000. The liposome layer would bind to folate receptors and the liposome encapsulating the azido sugar would be endocytosed. Endocytic degradation would release the azido sugar, which would then be transported to the surface through the sialin biosynthetic pathways. In order to confirm the presence of the azido sugars on the surface, the authors tagged the cells with alkyne-biotin and then labeled them with streptavidin-AF488 (SA-AF488).
Finally, Bertozzi and coworkers designed glycoproteins to mimic cell surface mucins that play a significant role in controlling cell-cell interactions. The mucin mimics were composed of a poly(methyl vinyl ketone) with synthetic glycans appended via oxime linkages and end-functionalized with lipids so that they could be directly anchored onto cell surfaces. These mucin mimics were then anchored onto Chinese Hamster Ovary (CHO) cells deprived of any endogenous mucins. Mucin density on the CHO cells was quantified and their intracellular location and dynamics were studied.54
Affinity-Mediated Covalent Conjugation
Covalent chemical reactions can also be applied to surface-exposed functional groups, usually amino acid side chains, on existing membrane proteins or polysaccharides.55 Initial efforts in membrane protein conjugation utilized a non-specific approach to react with functional groups common to amino acid sidechains. N-hydroxysuccinimidyl (NHS)-ester modified single-stranded DNA (ssDNA) was conjugated to the amine groups of the lysine residues on human T-lymphocytes (Jurkat cells). Because Jurkat cells are non-adherent, the conjugated ssDNA would be responsible for adhering the cells to the glass surfaces treated with the complementary DNA sequence (Figure 4).56 However, the trypsin required to detach cells for further study also tended to cleave lysine (and arginine) residues.57 Hence, Tusnady et al described a method to react the carboxyl acid side chains of aspartic and glutamic acid with ethyl (dimethylaminopropyl) carbodiimide (EDC) and sulfo-NHS, followed by introduction of biotinylated cysteamine. The displayed biotin group was then treated with streptavidin or avidin conjugates for cell surface labeling and elaboration of human leukemia cell line HL60. Similarly, maleimide-conjugated biomolecules can target exposed thiols. For example, Irvine et al designed nanocapsules (NC) loaded with SN-38, the active metabolite of irinotecan, an FDA-approved inhibitor of tropoisomerase I. The NC’s were decorated with maleimide groups to target the thiols on the surface of T-lymphocytes. The polyclonal T-cells were expanded ex vivo under conditions that would retain their adhesion receptors required for homing to lymphoid tissues, followed by delivery of a payload to lymphoma in a mouse model.58
Figure 4.
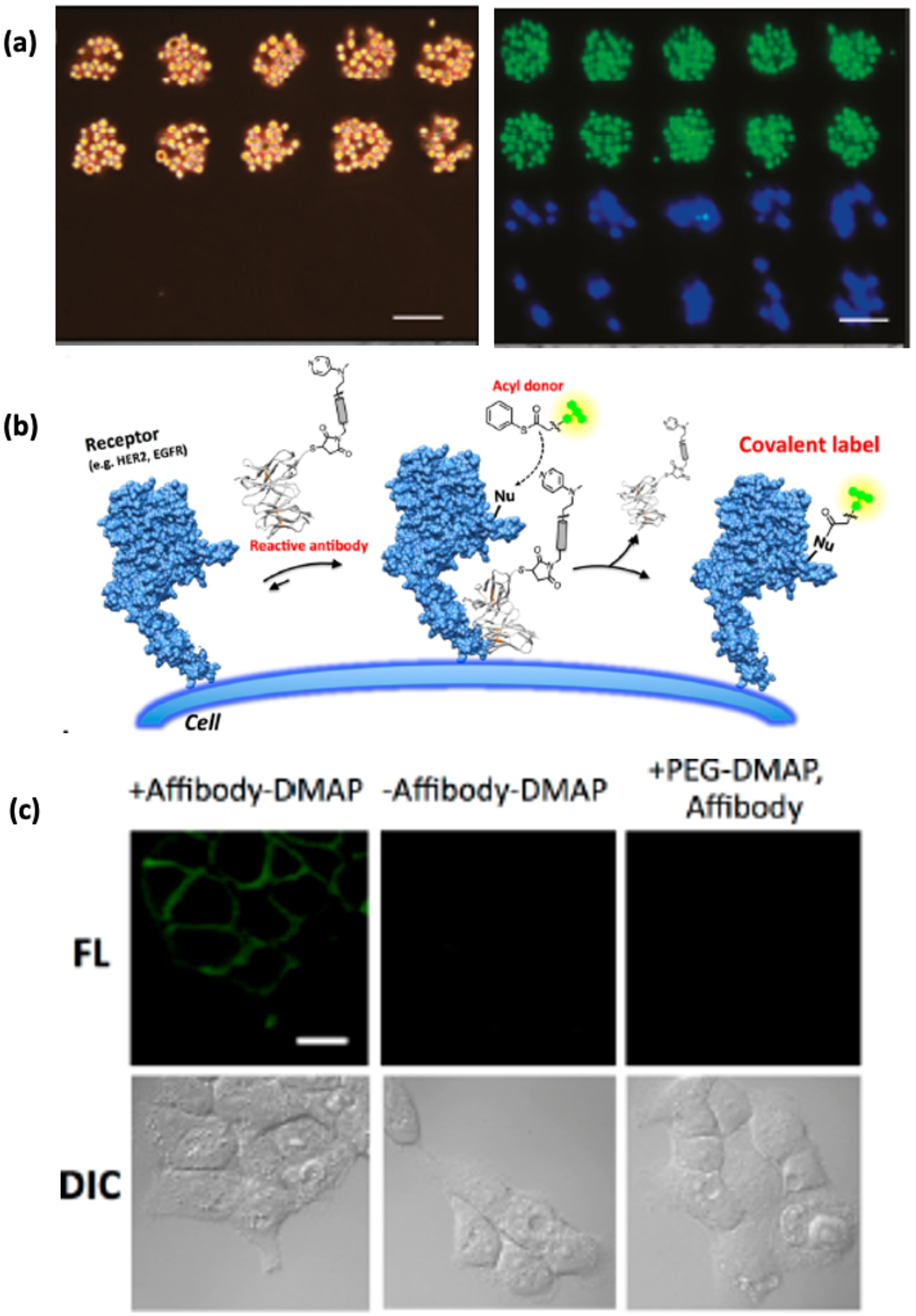
(a) Left: Immobilization of DNA-conjugated MCF-7 cells onto a glass surface with a microarray of a complementary DNA sequence. The neighboring areas on the glass surface having a non-complementary DNA sequence is left untouched. Right: selective immobilization of Jurkat (green) and MDA-MB-231 cells (blue) by DNA hybridization [56]. (b) Schematic of DMAP-tethered antibody labeling of cell surface receptors [60]. (c) Fluorescence images of EGFR expressive human epidermal carcinoma A431 cells labeled by DMAP-tethered anti EGFR affibody conjugated to fluorescein dye, compared to cells (−) affibody-DMAP and cells reacted with non-affibody conjugated PEG-DMAP [60].
Another approach is to add an affinity group to target specific membrane proteins on the cell. Notably, Hamachi and coworkers utilized dimethylaminopyridine (DMAP), a common acyl transfer catalyst, conjugated to a saccharide ligand to ensure efficient and selective acylation of lectins in the presence of thiophenyl ester acyl donors.59 The idea behind this affinity-guided DMAP (AGD) conjugation is that a non-covalently binding affinity ligand binds selectively to the target of interest and the DMAP catalyzes acyl transfer to a nucleophilic group on the target protein (Figure 4). Hamachi et al utilized an anti-epidermal growth factor receptor (EGFR) affibody-DMAP molecule appended with fluorescein or Alexa-Fluor 488 labeled acyl donors to EGFR (Figure 4).60 The AGD approach appears to be quite general and could provide valuable information on protein-protein interactions towards a deeper understanding of cell receptor function.
One of the more recent works of Hamachi and coworkers described the design of ligand-directed N-acyl-N-alkyl sulfonamide (LDNASA) derivatives as an electrophilic reactive group that can be attacked by a nucleophilic amino acid located on the protein surface through a proximity effect.61 In this study, the authors demonstrated that the reaction kinetics of the LDNASA method is faster than other chemistries like ligand-directed tosyl (LDT)62 and alkyloxyacyl imidazole (LDAI).63 The authors also demonstrated successful covalent conjugation of the natural inhibitor PU-H71 causing successful suppression of ATPase activity of Hsp90, which plays a significant role in tumorigenesis. In a similar experiment, Ploegh et al used a LPXTG peptide-tagged probe and transpeptidase enzyme sortase A from Gram-positive bacteria Staphylococcus aureus. The chemistry involves the attack of the enzyme at the amide bond between threonine (T) and glycine (G) to form an acyl-enzyme intermediate, which is then attacked by a nucleophile, preferably exposed N-terminal glycines on the cell surface, to transfer the probe onto the surface for real time imaging.64 In each of these examples, non-covalent ligand-protein binding interactions were converted to permanent covalent conjugations using a chemical reaction. Such reactions can be used to deliver a fluorophore or an antibody depending upon the purpose and application under study.
Finally, more recent developments have utilized photocrosslinking of small molecules like single chain variable fragment (scFV)65 and affibodies66 specifically to cellular receptors as an approach for conjugation.67 As discussed above, NHS-amine and maleimide-thiol chemistries suffer from a lack of control over the location and position of the conjugation. Certain applications like FRET are heavily dependent on a proper control of fluorophore labeling on the cell surface.68 Therefore, Tsourkas et al introduced site-specific mutations on affibodies from Protein Z to incorporate a photocrosslinking agent like the unnatural amino acid (UAA) benzoyl-phenylalanine (BPA) through amber codon suppression. The amber codon was mutated in the Protein Z at different locations using site directed mutagenesis to create ten different mutants and then identified the mutants that only bind to different human and rat immunoglobulins (IgGs) but also show a fast crosslinking reaction. This method comprises of two steps: the non-covalent binding of Protein Z to IgG followed by the covalent UV cross-linking of the affibody to IgG.69
Taking a cue from this work, Goodwin and Cha et al developed anti- EGFR affibodies that can not only bind to these receptors but also can crosslink to them when exposed to UV irradiation. A set of site-directed mutagenesis studies revealed that by mutating the N23 of the native affibody to a cysteine, the photocrosslinker could be incorporated by reacting the mutant with maleimide benzophenone. The modified affibody (N23BP) could then successfully crosslinked to soluble EGFR, as detected by SDS-PAGE. Next, a rhodamine-tagged N23BP affibody mutant was conjugated to EGFR-expressing 4T1 mouse cells, resulting in enhanced retention of affibody expression for up to 24 h in both 2D cell monolayers and 3D tumor spheroid models. In contrast, the wild type affibody (WT) that lacked BP had disappeared after 24 h. The hypothetical mechanism for this behavior was that the permanent conjugation between a site-specific benzophenone on the affibody molecule and the targeted receptor prevented proteolytic degradation when the receptors were endocytosed.70 This hypothesis was tested by the same group when they tracked the local environment of a N23BP affibody-enzyme fusion protein before and after photocrosslinking using a pH-sensitive dye. After photocrosslinking, the affibodies were found to first show exposure to an acidic pH environment (endosome) followed by a neutral environment (endosomal escape) in MDA-MB-468 breast cancer cells (Figure 5). More importantly, the affibody-enzyme was found to retain its enzymatic activity after recycling to the surface, and thus this approach could be used to increase the efficacy of enzyme-prodrug therapy treatments.71
Figure 5.
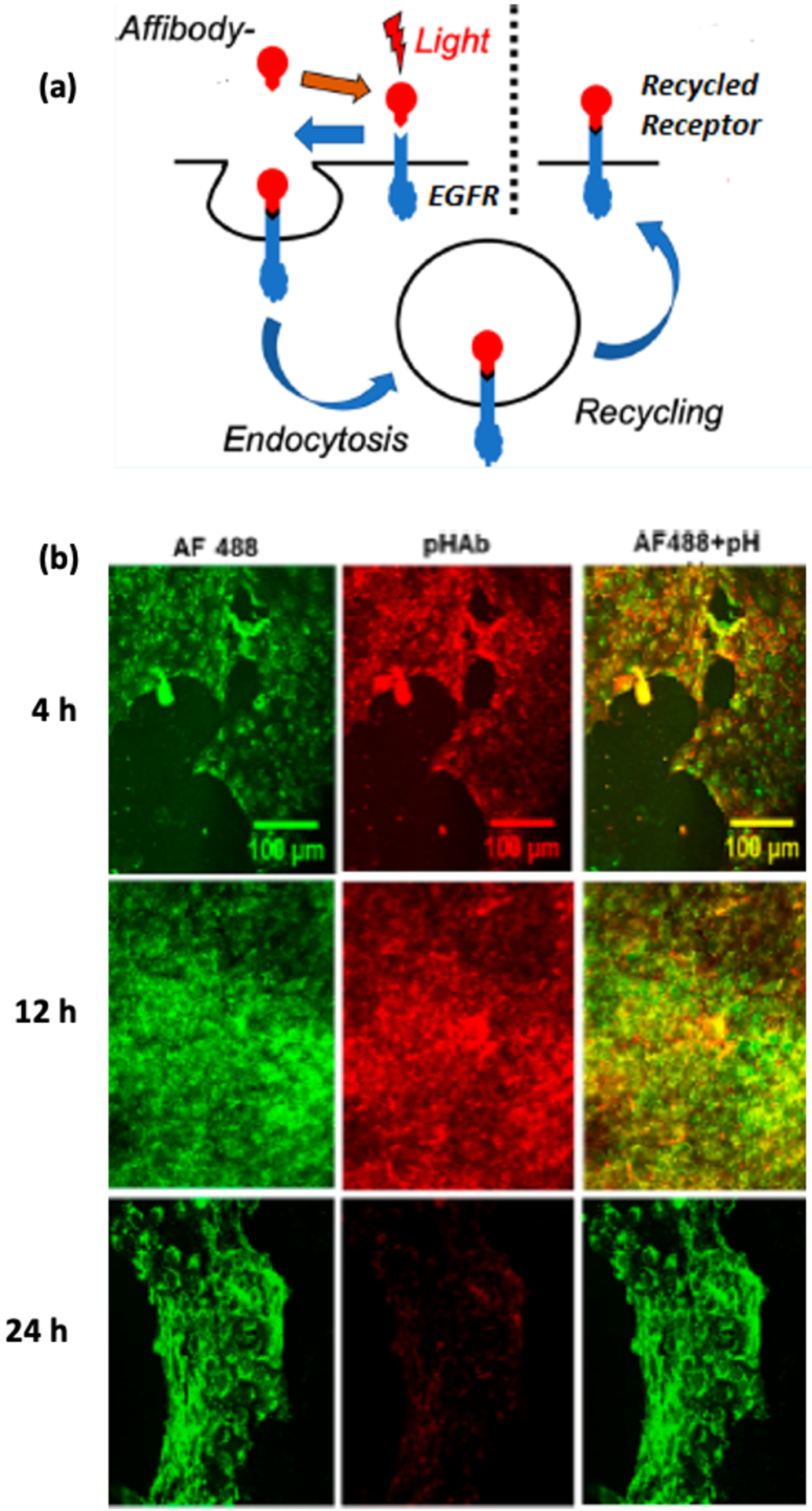
(a) Schematic depicting how affibody photocrosslinked to EGFR causes recycling of the the affibody to the cell surface [63]. (b) Confocal images showing pH-stable Alexafluor 488 and pH-sensitive pHAb. The photocrosslinked affibody-enzyme is internalized in endosomes at 4 h and 12 h but returns to the surface after 24 h [63].
Non-Genetic Cell Modification for Cell and Tissue Based Therapeutics
Cell surface modification is a powerful tool, particularly for applications in which cell contacts are desired. This section will focus on ways in which investigators have leveraged some of the beneficial aspects of non-genetic modification for different applications, primarily in therapy or tissue engineering. For further reading beyond the scope of this review, we recommend a review by Won et al72 concerning the treatment of cardiac diseases and Langer et al73 for a thorough breakdown of approaches by cell type. A review by Wagner et al74 also provides a good perspective on the promise for clinical translation.
1. Increased Cell Adhesion Strength.
Some applications, for example those in flow conditions, might require stronger adhesion between a conjugate and its cell target. As an example, surface engineering of Mesenchymal Stem Cells (MSCs) is gaining increasing importance for ensuring efficient delivery of MSCs to injured sites within the body. Stem cells play a vital role in repairing damaged tissue and suppressing inflammation on endothelial cells (EC) but lack adhesion ligands on their surface that would help them to target the appropriate site of injury. During an inflammatory response,75 ECs express lectins like P-selectin/L-selectin and E-selectin on their surfaces. Modifying the surfaces of the MSCs to attach these epitopes to the surface of ECs would result in efficient homing of MSCs towards the endothelial lining, not only in static conditions but also in dynamic flow environments.76 For example, Karp et al developed a method for guiding MSCs towards damaged tissue by incorporating biotin vesicles on the surface of the MSCs, followed by the addition of streptavidin and then biotinylated sialylated glycocalyx or SialylLewisX (sLeX).77 The MSCs would then exhibit a rolling action toward a P-selectin glycoprotein ligand 1 (PSGL-1)78 coated substrate under a shear stress condition such as that found in normal blood flow. Similar studies by Dennis et al have incorporated antibodies through lipid insertions on MSCs to bind ICAM-1 epitopes on Human Umbilical Vein Endothelial Cells (HUVECs) that are upregulated by the local immune response to cytokines like TNF-α. The adhesion of MSCs to the HUVECs was tested both in a static in vitro study and a dynamic flow chamber by fluorescence microscopy and flow cytometry.79 Based on this work, Dennis and co-workers showed that MSCs modified with homing peptides through lipid interactions exhibited higher MSC localization in mice with myocardial infarction (MI) than untreated MSC or MSC modified by non-specific peptides, with negligible toxicity.80 In another application, Jiang et al utilized cell conjugation to form mimics of blood vessels and other tubular structures. First, smooth muscle cells (SMC) and HUVECs were functionalized with biotin and streptavidin. Using stress induced rolling membranes (SRIM), the resultant tubular structure had a bilayer of cells with the muscle cells on the outside and the HUVECs on the inside.81
These cell surface modifications provide a means to control cell-cell interactions, which in turn can lead to various strategies for cellular therapies and regenerative medicine. For example, Wagner et al developed chemically self assembled nanorings (CSAN), which when inserted onto T-cells target MCF-7 human breast cancer cells through ligand-receptor specific interactions.82 These dihydrofolate reductase (DHFR) nanorings were recombinantly expressed to target epithelial cell adhesion molecules (EpCAM) and placed into cells through interactions with biotin-lipids or Cu-free SPAAC with DBCO-azide groups. When inserted into T-cells, EpCAM (+) MCF-7 cells could be targeted in the presence of EpCAM (−) U87 glioblastoma cells for selective cancer targeting.83 Importantly, these CSANs can be disassembled by treating the cells with an FDA approved antibiotic trimethoprim to reverse detach targeting ligands from cell surfaces.
2. Reversible Cell-Cell and Cell-Substrate Interactions for Tunable Cellular Assembly
Reversing covalent conjugation should allow a cell to its original state after being used for a given application. One method is to destroy the binding groups. For example, Gartner et al formed cellular structures through DNA hybridization by complementary oligonucleotides. These structures could then return to nonbinding cells upon addition of DNAse enzymes.84 Similarly, Tan et al developed DNA aptamers that can anchor themselves onto CCRF-CEM T cells through lipid interactions. These TDO5 aptamers can target the immunoglobulin chains of Ramos B cells and similarly Sgc8 aptamers anchored on Ramos cells can target the tyrosine kinase 7 on CCRF-CEM cells, thereby causing an aggregation of various lymphocytes that can be used in various cell based therapies in treating several cancer cell lines. These lymphocytic aggregates were then disintegrated by a DNAse enzyme that can cleave both single and double stranded DNA.85
Reversibility can also be conferred by temperature changes. Building on their previous work, Tan et al used the same DNA aptamers to capture T cell and B cells but now with more specificity and higher control. The aptamers anchored on CCRF-CEM cells could capture tetravalent SA, which can then capture a biotinylated TDO5 aptmaer targeting the Ramos cells through a SA-biotin bridge at 4°C. The aggregates disassociate when temperature is raised to 37°C due to the melting of the oligonucleotides, thereby disintegrating the aggregates.86 Similarly, Pasparikar et al developed a co-polymer of thermosensitive di(ethylene glycol) methyl ether methacrylate (DEGMA) and amine-reactive NHS-methacrylate (NHS-MA) to covalently bind to the amino acid residues on the membrane proteins. Above the LCST of the co-polymer, intercellular aggregation is promoted by the hydrophobic interactions of the DEGMA residues, driven by the the coil to globule polymer transformataion. Below the LCST, the cells disassemble due to steric repulsion. These aggregates disintegrate when the cells are warmed to 37°C, indicating that reversibility in cellular aggregation can be modulated by temperature changes.87
Finally, reversibility can be built into the functional groups themselves. For example, Yousaf et al synthesized hydroxyquinone (HQ, ‘off state’) functionalized liposomes that can be tethered to cell surface and converted to a quinone (Q, ‘on state’) under slightly oxidative conditions.88 Quinone-modified human MSCs and fibroblasts were then covalently conjugated to amino-oxy (AO) alkanes modified cells.The chemo-selective binding between Q and AO leads to a stable oxime linkage between the cells produced a 3D cell assembly. Because of the reversibility of the quinone redox chemistry, the linkage can be broken down under slightly reductive conditions to restore HQ and release the oxime. Yousaf et al also synthesized a ketone-bearing calcein dye liposome conjugated to a fluorescence quencher dabcyl through a hydrazone linkage.89 Under normal conditions, this conjugate does not fluoresce and is in the ‘off state’ but a ligand exchange reaction replaces the hydrazone with an oxime, releasing the dye and turning on fluorescence. This reversible covalent bond approach was extended for reversible cellular aggregation and can serve as inspiration for several studies involving other biochemical processes that involve cell proliferation and growth and requires dynamic cellular imaging.
3. Increased Duration of Expression on the Cell Membrane Surface.
One question is how long an unnatural conjugation can remain on the cell surface. In one example, Wagner et al showed that the insertion of CSANs through hydrophobic insertions remain stable on the cell surfaces for more than 72 h.90 Goodwin et al also showed that photocrosslinked anti-EGFR targeting N23CD-BP affibodies are maintained on the cell surface for about 72 h. This residence time was measured through the conversion of prodrug 5-fluorocytosine to 5-fluorouracil and killing of cancer cells.71 Boons et al demonstrated that the enzymatic transfer of CMP-Neu5Ac using the sialyltransferase ST6GAL1 to N-acteyllactoseamine (LacNAc) can stably express the modified glycans for ~ 72 h. The CMP-sialic acid derivative has a modified dual functionality with a biotin and heparan sulphate (HS) that binds to a variety of proteins. This enzymatic transfer of CMP-Neu5Ac to cells deficient in HS led to a long lived display of biotin, as observed from the avidin AlexaFluor 488 labeling and phosphorylated ERK activity, that is a consequence of the recruitment and binding of the FGF-2 protein by the cells.91
Conclusions and Outlook
In this review, we focused mainly on various techniques of non-genetic cell surface modification and how these types of conjugations enable applications such as in vivo imaging, controlling cell-cell and cell-substrate interactions, and promoting cellular adhesion. One possible future direction for these approaches is to reduce or control cell signaling on a temporary basis. If a receptor or set of receptors could be blocked, the cell could be made dormant without inducing apoptosis. Reversing or removing the conjugation could then allow the cells to grow again. For example, quiescence is desired in stem cells so that they do not differentiate into a defined type prior to use, but it can be difficult to maintain in culture.92,93 The ability to target specific receptors could provide a more stable quiescent state without changing the cell permanently. Pursuing this goal would require further study on the duration of expression on the cell membrane surface to obtain a more precise rate of receptor regeneration, either by swapping with membrane proteins already present in the endomembrane system, the digestion of tagged receptors and synthesis of new ones, or some combination of these mechanisms. While the rate of receptor degradation and regeneration would certainly depend on the type of targeted receptor, it would be interesting to determine if receptor modification led to changes in lifetime.
Lastly, covalent cell surface modification can potentially allow increased mechanical loading on a cell by stabilizing interactions that are often provided by biotin-streptavidin or DNA hybridization. Retraction experiments on live cells can be used to gain fundamental knowledge of cell-ligand interactions.72 For example, Salaita et al has used DNA for measuring mechanical forces on the cellular surface.94 These nucleic acids were also conjugated with a fluorophore and a natural cell receptor. Upon ligand-receptor recognition through a non-covalent mechanism, the cells transmitted pico-Newton (pN) forces through the receptors. This mechanism can cause DNA melting and other conformational changes that can be tracked using the fluorophore signal and can be correlated with the cellular force. Now these pN forces can be transmitted through the receptors to the ligands and then to DNA molecules.
Of course, the biggest question whether non-genetic cell modification approaches can be translated to the clinic. Although exogenous materials may not be toxic, a question remains as to whether are they necessarily innocuous.95 Glycans, lipids, and proteins are the three most abundant cellular components and hence exogenous materials made from these precursors should not be toxic to the cells by themselves;96 however, care should be taken to not cause steric hindrance or a permanent change in the lipid structure of the cellular membrane, thereby irreversibly affecting biochemical signaling.97 Along with biocompatibility, solubility of lipo-conjugates can be difficult to optimize so that they maintain binding to the membrane but do not adhere non-specifically to other cell membranes,98 which in turn would lead to rapid endocytosis and removal from circulation.99 While non-genetic modification of cell surfaces is still in a nascent stage and has enjoyed some preclinical success,100 its success in human trials will depend on the innocuity and stability of therapeutic conjugates.
ACKNOWLEDGMENT
The authors acknowledge the National Institutes of Health (R21GM135668) for support.
References
- (1).Oakley RH; Laporte SA; Holt JA; Barak LS; Caron MG Molecular Determinants Underlying the Formation of Stable Intracellular G Protein-Coupled Receptor-β-Arrestin Complexes after Receptor Endocytosis. J. Biol. Chem 2001, 276 (22), 19452–19460. 10.1074/jbc.M101450200. [DOI] [PubMed] [Google Scholar]
- (2).Müller P; Schier AF Extracellular Movement of Signaling Molecules. Dev. Cell 2011, 21 (1), 145–158. 10.1016/j.devcel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Sridharan R; Zuber J; Connelly SM; Mathew E; Dumont ME Fluorescent Approaches for Understanding Interactions of Ligands with G Protein Coupled Receptors. Biochim. Biophys. Acta - Biomembr 2014, 1838 (1 PARTA), 15–33. 10.1016/j.bbamem.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Daly CJ; McGrath JC Fluorescent Ligands, Antibodies, and Proteins for the Study of Receptors. Pharmacol. Ther 2003, 100 (2), 101–118. 10.1016/j.pharmthera.2003.08.001. [DOI] [PubMed] [Google Scholar]
- (5).Nath N; Godat B; Zimprich C; Dwight SJ; Corona C; McDougall M; Urh M Homogeneous Plate Based Antibody Internalization Assay Using PH Sensor Fluorescent Dye. J. Immunol. Methods 2016, 431, 11–21. 10.1016/j.jim.2016.02.001. [DOI] [PubMed] [Google Scholar]
- (6).Havel RJ; Hamilton RL Hepatocytic Lipoprotein Receptors and Intracellular Lipoprotein Catabolism. Hepatology 1988, 8 (6), 1689–1704. 10.1002/hep.1840080637. [DOI] [PubMed] [Google Scholar]
- (7).Yano Y; Matsuzaki K Tag-Probe Labeling Methods for Live-Cell Imaging of Membrane Proteins. Biochim. Biophys. Acta - Biomembr 2009, 1788 (10), 2124–2131. 10.1016/j.bbamem.2009.07.017. [DOI] [PubMed] [Google Scholar]
- (8).Rosenbaum L Tragedy, Perseverance, and Chance — The Story of CAR-T Therapy. N. Engl. J. Med 2017, 377 (14), 1313–1315. 10.1056/NEJMp1711886. [DOI] [PubMed] [Google Scholar]
- (9).Douglas KL Toward Development of Artificial Viruses for Gene Therapy: A Comparative Evaluation of Viral and Non-Viral Transfection. Biotechnol. Prog 2008, 24 (4), 871–883. 10.1021/bp.070319o. [DOI] [PubMed] [Google Scholar]
- (10).Pai AA; Pritchard JK; Gilad Y The Genetic and Mechanistic Basis for Variation in Gene Regulation. PLoS Genet. 2015, 11 (1). 10.1371/journal.pgen.1004857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Custódio CA; Mano JF Cell Surface Engineering to Control Cellular Interactions. ChemNanoMat 2019, 2 (5), 376–384. 10.1002/cnma.201600047.Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kellam B; De Bank PA; Shakesheff KM Chemical Modification of Mammalian Cell Surfaces. Chem. Soc. Rev 2003, 32 (6), 327–337. 10.1039/b211643j. [DOI] [PubMed] [Google Scholar]
- (13).Wen L; Liu D; Zheng Y; Huang K; Cao X; Song J; Wang PG A One-Step Chemoenzymatic Labeling Strategy for Probing Sialylated Thomsen-Friedenreich Antigen. ACS Cent. Sci 2018, 4 (4), 451–457. 10.1021/acscentsci.7b00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Saxon E; Bertozzi CR Cell Surface Engineering by a Modified Staudinger Reaction. Science 2000, 287 (5460), 2007–2010. 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- (15).Cheng B; Xie R; Dong L; Chen X Metabolic Remodeling of Cell-Surface Sialic Acids: Principles, Applications, and Recent Advances. ChemBioChem 2016, 17 (1), 11–27. 10.1002/cbic.201500344. [DOI] [PubMed] [Google Scholar]
- (16).Keppler OT; Horstkorte R; Pawlita M; Schmidt C; Reutter W Biochemical Engineering of the N-Acyl Side Chain of Sialic Acid: Biological Implications. Glycobiology 2001, 11 (2), 11–18. 10.1093/glycob/11.2.11R. [DOI] [PubMed] [Google Scholar]
- (17).Prescher JA; Dube DH; Bertozzi CR Chemical Remodelling of Cell Surfaces in Living Animals. Nature 2004, 430 (7002), 873–877. 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- (18).Mahal LK; Yarema KJ; Bertozzi CR Engineering Chemical Reactivity on Cell Surfaces through Oligosaccharide Biosynthesis. Science 1997, 276 (5315), 1125–1128. 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- (19).Chandra RA; Douglas ES; Mathies RA; Bertozzi CR; Francis MB Programmable Cell Adhesion Encoded by DNA Hybridization. Angew. Chem. - Int. Ed 2006, 45 (6), 896–901. 10.1002/anie.200502421. [DOI] [PubMed] [Google Scholar]
- (20).Sletten EM; Bertozzi CR From Mechanism to Mouse: A Tale of Two Bioorthogonal Reactions. Acc. Chem. Res 2011, 44 (9), 666–676. 10.1021/ar200148z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Hong V; Steinmetz NF; Manchester M; Finn MG Labeling Live Cells by Copper-Catalyzed Alkyne-Azide Click Chemistry. Bioconjug. Chem 2010, 21 (10), 1912–1916. 10.1021/bc100272z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Rostovtsev VV; Green LG; Fokin VV; Sharpless KB A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angew. Chem. - Int. Ed 2002, 41 (14), 2596–2599. . [DOI] [PubMed] [Google Scholar]
- (23).Baskin JM; Prescher JA; Laughlin ST; Agard NJ; Chang PV; Miller IA; Lo A; Codelli JA; Bertozzi CR Copper-Free Click Chemistry for Dynamic in Vivo Imaging. Proc. Natl. Acad. Sci. U. S. A 2007, 104 (43), 16793–16797. 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Agard NJ; Prescher JA; Bertozzi CR A Strain-Promoted [3 + 2] Azide-Alkyne Cycloaddition for Covalent Modification of Biomolecules in Living Systems. J. Am. Chem. Soc 2004, 126 (46), 15046–15047. 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- (25).Agard NJ; Baskin JM; Prescher JA; Lo A; Bertozzi CR A Comparative Study of Bioorthogonal Reactions with Azides. ACS Chem. Biol 2006, 1 (10), 644–648. 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- (26).Jewett JC; Sletten EM; Bertozzi CR Rapid Cu-Free Click Chemistry with Readily Synthesized Biarylazacyclooctynones. J. Am. Chem. Soc 2010, 132 (11), 3688–3690. 10.1021/ja100014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kang SW; Lee S; Na JH; Yoon HI; Lee DE; Koo H; Cho YW; Kim SH; Jeong SY; Kwon IC; et al. Cell Labeling and Tracking Method without Distorted Signals by Phagocytosis of Macrophages. Theranostics 2014, 4 (4), 420–431. 10.7150/thno.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Soriano Del Amo D; Wang W; Jiang H; Besanceney C; Yan AC; Levy M; Liu Y; Marlow FL; Wu P Biocompatible Copper(I) Catalysts for in Vivo Imaging of Glycans. J. Am. Chem. Soc 2010, 132 (47), 16893–16899. 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Besanceney-Webler C; Jiang H; Zheng T; Feng L; Soriano Del Amo D; Wang W; Klivansky LM; Marlow FL; Liu Y; Wu P Increasing the Efficacy of Bioorthogonal Click Reactions for Bioconjugation: A Comparative Study. Angew. Chem. - Int. Ed 2011, 50 (35), 8051–8056. 10.1002/anie.201101817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Niederwieser A; Späte AK; Nguyen LD; Jüngst C; Reutter W; Wittmann V Two-Color Glycan Labeling of Live Cells by a Combination of Diels-Alder and Click Chemistry. Angew. Chem. - Int. Ed 2013, 52 (15), 4265–4268. 10.1002/anie.201208991. [DOI] [PubMed] [Google Scholar]
- (31).Lang K; Davis L; Wallace S; Mahesh M; Cox DJ; Blackman ML; Fox JM; Chin JW Genetic Encoding of Bicyclononynes and Trans-Cyclooctenes for Site-Specific Protein Labeling in Vitro and in Live Mammalian Cells via Rapid Fluorogenic Diels-Alder Reactions. J. Am. Chem. Soc 2012, 134 (25), 10317–10320. 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Agarwal P; Beahm BJ; Shieh P; Bertozzi CR Systemic Fluorescence Imaging of Zebrafish Glycans with Bioorthogonal Chemistry. Angew. Chem. - Int. Ed 2015, 54 (39), 11504–11510. 10.1002/anie.201504249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Plass T; Milles S; Koehler C; Szymański J; Mueller R; Wießler M; Schultz C; Lemke EA Amino Acids for Diels-Alder Reactions in Living Cells. Angew. Chem. - Int. Ed 2012, 51 (17), 4166–4170. 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]
- (34).Yang J; Šečkutė J; Cole CM; Devaraj NK Live-Cell Imaging of Cyclopropene Tags with Fluorogenic Tetrazine Cycloadditions. Angew. Chem 2012, 124 (30), 7594–7597. 10.1002/ange.201202122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Patterson DM; Nazarova LA; Xie B; Kamber DN; Prescher JA Functionalized Cyclopropenes as Bioorthogonal Chemical Reporters. J. Am. Chem. Soc 2012, 134 (45), 18638–18643. 10.1021/ja3060436. [DOI] [PubMed] [Google Scholar]
- (36).Späte AK; Bußkamp H; Niederwieser A; Schart VF; Marx A; Wittmann V Rapid Labeling of Metabolically Engineered Cell-Surface Glycoconjugates with a Carbamate-Linked Cyclopropene Reporter. Bioconjug. Chem 2014, 25 (1), 147–154. 10.1021/bc4004487. [DOI] [PubMed] [Google Scholar]
- (37).Xiong DC; Zhu J; Han MJ; Luo HX; Wang C; Yu Y; Ye Y; Tai G; Ye XS Rapid Probing of Sialylated Glycoproteins in Vitro and in Vivo via Metabolic Oligosaccharide Engineering of a Minimal Cyclopropene Reporter. Org. Biomol. Chem 2015, 13 (13), 3911–3917. 10.1039/c5ob00069f. [DOI] [PubMed] [Google Scholar]
- (38).Jao CY; Roth M; Welti R; Salic A Biosynthetic Labeling and Two-Color Imaging of Phospholipids in Cells. ChemBioChem 2015, 16 (3), 472–476. 10.1002/cbic.201402149. [DOI] [PubMed] [Google Scholar]
- (39).Jao CY; Roth M; Welti R; Salic A Metabolic Labeling and Direct Imaging of Choline Phospholipids in Vivo. Proc. Natl. Acad. Sci. U. S. A 2009, 106 (36), 15332–15337. 10.1073/pnas.0907864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Li Y; Kinting S; Höppner S; Forstner ME; Uhl O; Koletzko B; Griese M Metabolic Labelling of Choline Phospholipids Probes ABCA3 Transport in Lamellar Bodies. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 2019, 1864 (12), 158516. 10.1016/j.bbalip.2019.158516. [DOI] [PubMed] [Google Scholar]
- (41).Xie R; Hong S; Feng L; Rong J; Chen X Cell-Selective Metabolic Glycan Labeling Based on Ligand-Targeted Liposomes. J. Am. Chem. Soc 2012, 134 (24), 9914–9917. 10.1021/ja303853y. [DOI] [PubMed] [Google Scholar]
- (42).Du Y; Xie R; Sun Y; Fan X; Chen X Liposome-Assisted Metabolic Glycan Labeling With Cell and Tissue Selectivity, 1st ed.; Elsevier Inc., 2018; Vol. 598. 10.1016/bs.mie.2017.06.037. [DOI] [PubMed] [Google Scholar]
- (43).Kato K; Itoh C; Yasukouchi T; Nagamune T Rapid Protein Anchoring into the Membranes of Mammalian Cells Using Oleyl Chain and Poly(Ethylene Glycol) Derivatives. Biotechnol. Prog 2004, 20 (3), 897–904. 10.1021/bp0342093. [DOI] [PubMed] [Google Scholar]
- (44).Itagaki T; Arima Y; Kuwabara R; Kitamura N; Iwata H Interaction between Cells and Poly (Ethylene Glycol) -Lipid Conjugates. Colloids Surf. B Biointerfaces 2015, 135, 765–773. 10.1016/j.colsurfb.2015.08.014. [DOI] [PubMed] [Google Scholar]
- (45).Lim KS; Lee DY; Valencia GM; Won YW; Bull DA Cell Surface-Engineering to Embed Targeting Ligands or Tracking Agents on the Cell Membrane. Biochem. Biophys. Res. Commun 2017, 482 (4), 1042–1047. 10.1016/j.bbrc.2016.11.155. [DOI] [PubMed] [Google Scholar]
- (46).Selden NS; Todhunter ME; Jee NY; Liu JS; Broaders KE; Gartner ZJ Chemically Programmed Cell Adhesion with Membrane-Anchored Oligonucleotides. J. Am. Chem. Soc 2012, 134 (2), 765–768. 10.1021/ja2080949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Weber RJ; Liang SI; Selden NS; Desai TA; Gartner ZJ Efficient Targeting of Fatty-Acid Modified Oligonucleotides to Live Cell Membranes through Stepwise Assembly. Biomacromolecules 2014, 15 (12), 4621–4626. 10.1021/bm501467h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Contreras JL; Xie D; Mays J; Smyth CA; Eckstein C; Rahemtulla FG; Young CJ; Anthony Thompson J; Bilbao G; Curiel DT; et al. A Novel Approach to Xenotransplantation Combining Surface Engineering and Genetic Modification of Isolated Adult Porcine Islets. Surgery 2004, 136 (3), 537–547. 10.1016/j.surg.2004.05.031. [DOI] [PubMed] [Google Scholar]
- (49).Dutta D; Pulsipher A; Luo W; Mak H; Yousaf MN Engineering Cell Surfaces via Liposome Fusion. Bioconjug. Chem 2011, 22 (12), 2423–2433. 10.1021/bc200236m. [DOI] [PubMed] [Google Scholar]
- (50).Park S; Yousaf MN An Interfacial Oxime Reaction to Immobilize Ligands and Cells in Patterns and Gradients to Photoactive Surfaces. Langmuir 2008, 24 (12), 6201–6207. 10.1021/la8005663. [DOI] [PubMed] [Google Scholar]
- (51).Kloxin AM; Kasko AM; Salinas CN; Anseth KS Photodegradable Hydrogels for Dynamic Tuning of Physical and Chemical Properties. Science 2009, 324 (5923), 59–63. 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Luo W; Pulsipher A; Dutta D; Lamb BM; Yousaf MN Remote Control of Tissue Interactions via Engineered Photo-Switchable Cell Surfaces. Sci. Rep 2014, 4, 1–8. 10.1038/srep06313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Sun Y; Hong S; Xie R; Huang R; Lei R; Cheng B; Sun DE; Du Y; Nycholat CM; Paulson JC; et al. Mechanistic Investigation and Multiplexing of Liposome-Assisted Metabolic Glycan Labeling. J. Am. Chem. Soc 2018, 140 (10), 3592–3602. 10.1021/jacs.7b10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Rabuka D; Forstner MB; Groves JT; Bertozzi CR Noncovalent Cell Surface Engineering: Incorporation of Bioactive Synthetic Glycopolymers into Cellular Membranes. J. Am. Chem. Soc 2008, 130 (18), 5947–5953. 10.1021/ja710644g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Mizusawa K; Takaoka Y; Hamachi I Specific Cell Surface Protein Imaging by Extended Self-Assembling Fluorescent Turn-on Nanoprobes. J. Am. Chem. Soc 2012, 134 (32), 13386–13395. 10.1021/ja304239g. [DOI] [PubMed] [Google Scholar]
- (56).Hsiao SC; Shum BJ; Onoe H; Douglas ES; Gartner ZJ; Mathies RA; Bertozzi CR; Francis MB Direct Cell Surface Modification with DNA for the Capture of Primary Cells and the Investigation of Myotube Formation on Defined Patterns. Langmuir 2009, 25 (12), 6985–6991. 10.1021/la900150n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Müller A; Langó T; Turiák L; Ács A; Várady G; Kucsma N; Drahos L; Tusnády GE Covalently Modified Carboxyl Side Chains on Cell Surface Leads to a Novel Method toward Topology Analysis of Transmembrane Proteins. Sci. Rep 2019, 9 (1), 1–11. 10.1038/s41598-019-52188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Huang B; Abraham WD; Zheng Y; Bustamante López SC; Luo SS; Irvine DJ Active Targeting of Chemotherapy to Disseminated Tumors Using Nanoparticle-Carrying T Cells. Sci. Transl. Med 2015, 7 (291). 10.1126/scitranslmed.aaa5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Rana MS; Wang X; Banerjee A An Improved Strategy for Fluorescent Tagging of Membrane Proteins for Overexpression and Purification in Mammalian Cells. Biochemistry 2018, 57 (49), 6741–6751. 10.1021/acs.biochem.8b01070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Hayashi T; Yasueda Y; Tamura T; Takaoka Y; Hamachi I Analysis of Cell-Surface Receptor Dynamics through Covalent Labeling by Catalyst-Tethered Antibody. J. Am. Chem. Soc 2015, 137 (16), 5372–5380. 10.1021/jacs.5b02867. [DOI] [PubMed] [Google Scholar]
- (61).Tamura T; Ueda T; Goto T; Tsukidate T; Shapira Y; Nishikawa Y; Fujisawa A; Hamachi I Rapid Labelling and Covalent Inhibition of Intracellular Native Proteins Using Ligand-Directed N-Acyl-N-Alkyl Sulfonamide. Nat. Commun 2018, 9 (1), 1–12. 10.1038/s41467-018-04343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Tsukiji S; Miyagawa M; Takaoka Y; Tamura T; Hamachi I Ligand-Directed Tosyl Chemistry for Protein Labeling in Vivo. Nat. Chem. Biol 2009, 5 (5), 341–343. 10.1038/nchembio.157. [DOI] [PubMed] [Google Scholar]
- (63).Fujishima SH; Yasui R; Miki T; Ojida A; Hamachi I Ligand-Directed Acyl Imidazole Chemistry for Labeling of Membrane-Bound Proteins on Live Cells. J. Am. Chem. Soc 2012, 134 (9), 3961–3964. 10.1021/ja2108855. [DOI] [PubMed] [Google Scholar]
- (64).Swee LK; Lourido S; Bell GW; Ingram JR; Ploegh HL One-Step Enzymatic Modification of the Cell Surface Redirects Cellular Cytotoxicity and Parasite Tropism. ACS Chem. Biol 2015, 10 (2), 460–465. 10.1021/cb500462t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Nautiyal K; Kibria MG; Akazawa-Ogawa Y; Hagihara Y; Kuroda Y Design and Assessment of an Active Anti-Epidermal Growth Factor Receptor (EGFR) Single Chain Variable Fragment (ScFv) with Improved Solubility. Biochem. Biophys. Res. Commun 2019, 508 (4), 1043–1049. 10.1016/j.bbrc.2018.11.170. [DOI] [PubMed] [Google Scholar]
- (66).Tolmachev V; Tran TA; Rosik D; Sjoberg A; Abrahmsen L; Orlova A Tumor Targeting Using Affibody Molecules: Interplay of Affinity, Target Expression Level, and Binding Site Composition. J. Nucl. Med 2012, 53 (6), 953–960. 10.2967/jnumed.111.101527. [DOI] [PubMed] [Google Scholar]
- (67).Brasino M; Cha JN Real-Time Femtomolar Detection of Cancer Biomarkers from Photoconjugated Antibody-Phage Constructs. Analyst 2017. 10.1039/c6an01904h. [DOI] [PubMed] [Google Scholar]
- (68).Grunbeck A; Huber T; Sachdev P; Sakmar TP Mapping the Ligand-Binding Site on a G Protein-Coupled Receptor (GPCR) Using Genetically Encoded Photocrosslinkers. Biochemistry 2011. 10.1021/bi200214r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Hui JZ; Tsourkas A Optimization of Photoactive Protein Z for Fast and Efficient Site-Specific Conjugation of Native IgG. Bioconjug. Chem 2014, 25 (9), 1709–1719. 10.1021/bc500305v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Brasino M; Roy S; Erbse AH; He L; Mao C; Park W; Cha JN; Goodwin AP Anti-EGFR Affibodies with Site-Specific Photo-Cross-Linker Incorporation Show Both Directed Target-Specific Photoconjugation and Increased Retention in Tumors. J. Am. Chem. Soc 2018, 140 (37), 11820–11828. 10.1021/jacs.8b07601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Roy S; Brasino M; M. Beirne J; Harguindey A; A. Chapnick D; Liu X; N. Cha J; P. Goodwin A Enzymes Photo-Cross-Linked to Live Cell Receptors Retain Activity and EGFR Inhibition after Both Internalization and Recycling. Bioconjug. Chem 2019, 0 (0). 10.1021/acs.bioconjchem.9b00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Lee DY; Cha BH; Jung M; Kim AS; Bull DA; Won YW Cell Surface Engineering and Application in Cell Delivery to Heart Diseases 09 Engineering 0903 Biomedical Engineering. J. Biol. Eng 2018, 12 (1), 1–11. 10.1186/s13036-018-0123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Wang Q; Cheng H; Peng H; Zhou H; Li PY; Langer R Non-Genetic Engineering of Cells for Drug Delivery and Cell-Based Therapy. Adv. Drug Deliv. Rev 2015, 91, 125–140. 10.1016/j.addr.2014.12.003. [DOI] [PubMed] [Google Scholar]
- (74).Csizmar CM; Petersburg JR; Wagner CR Programming Cell-Cell Interactions through Non-Genetic Membrane Engineering. Cell Chem. Biol 2018, 25 (8), 931–940. 10.1016/j.chembiol.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Zhao W; Loh W; Droujinine IA; Teo W; Kumar N; Schafer S; Cui CH; Zhang L; Sarkar D; Karnik R; et al. Mimicking the Inflammatory Cell Adhesion Cascade by Nucleic Acid Aptamer Programmed Cell‐cell Interactions. FASEB J. 2011, 25 (9), 3045–3056. 10.1096/fj.10-178384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Merzaban JS; Imitola J; Starossom SC; Zhu B; Wang Y; Lee J; Ali AJ; Olah M; Abuelela AF; Khoury SJ; Sackstein R Cell Surface Glycan Engineering of Neural Stem Cells Augments Neurotropism and Improves Recovery in a Murine Model of Multiple Sclerosis. Glycobiology 2015, 25 (12), 1392–1409. 10.1093/glycob/cwv046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Sarkar D; Vemula PK; Zhao W; Gupta A; Karnik R; Karp JM Engineered Mesenchymal Stem Cells with Self-Assembled Vesicles for Systemic Cell Targeting. Biomaterials 2010, 31 (19), 5266–5274. 10.1016/j.biomaterials.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Tivadar ST; McIntosh RS; Chua JX; Moss R; Parsons T; Zaitoun AM; Madhusudan S; Durrant LG; Vankemmelbeke M Monoclonal Antibody Targeting Sialyl-Di-LewiSA–Containing Internalizing and Noninternalizing Glycoproteinswithcancerimmunotherapydevelopment Potential. Mol. Cancer Ther 2020, 19 (3), 790–801. 10.1158/1535-7163.MCT-19-0221. [DOI] [PubMed] [Google Scholar]
- (79).Ko IK; Kean TJ; Dennis JE Targeting Mesenchymal Stem Cells to Activated Endothelial Cells. Biomaterials 2009, 30 (22), 3702–3710. 10.1016/j.biomaterials.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Kean TJ; Duesler L; Young RG; Dadabayev A; Olenyik A; Penn M; Wagner J; Fink DJ; Caplan AI; Dennis JE Development of a Peptide-Targeted, Myocardial Ischemia-Homing, Mesenchymal Stem Cell. J. Drug Target 2012, 20 (1), 23–32. 10.3109/1061186X.2011.622398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Gong P; Zheng W; Huang Z; Zhang W; Xiao D; Jiang X A Strategy for the Construction of Controlled, Three-Dimensional, Multilayered, Tissue-like Structures. Adv. Funct. Mater 2013, 23 (1), 42–46. 10.1002/adfm.201201275. [DOI] [Google Scholar]
- (82).Csizmar CM; Petersburg JR; Perry TJ; Rozumalski L; Hackel BJ; Wagner CR Multivalent Ligand Binding to Cell Membrane Antigens: Defining the Interplay of Affinity, Valency, and Expression Density. J. Am. Chem. Soc 2019, 141 (1), 251–261. 10.1021/jacs.8b09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Gabrielse K; Gangar A; Kumar N; Lee JC; Fegan A; Shen JJ; Li Q; Vallera D; Wagner CR Reversible Re-Programing of Cell-Cell Interactions. Angew. Chem. - Int. Ed 2014, 53 (20), 5112–5116. 10.1002/anie.201310645. [DOI] [PubMed] [Google Scholar]
- (84).Gartner ZJ; Bertozzi CR Programmed Assembly of 3-Dimensional Microtissues with Defined Cellular Connectivity. Proc. Natl. Acad. Sci. U. S. A 2009, 106 (12), 4606–4610. 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Xiong X; Liu H; Zhao Z; Altman MB; Lopez-Colon D; Yang CJ; Chang LJ; Liu C; Tan W DNA Aptamer-Mediated Cell Targeting. Angew. Chem. - Int. Ed 2013, 52 (5), 1472–1476. 10.1002/anie.201207063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Altman MO; Chang YM; Xiong X; Tan W Modifying Cellular Properties Using Artificial Aptamer-Lipid Receptors. Sci. Rep 2013, 3 (November). 10.1038/srep03343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Amaral AJR; Pasparakis G Macromolecular Cell Surface Engineering for Accelerated and Reversible Cellular Aggregation. ChemComm 2015, 6 (35), 1–5. 10.1039/x0xx00000x. [DOI] [PubMed] [Google Scholar]
- (88).Pulsipher A; Dutta D; Luo W; Yousaf MN Cell-Surface Engineering by a Conjugation-and-Release Approach Based on the Formation and Cleavage of Oxime Linkages upon Mild Electrochemical Oxidation and Reduction. Angew. Chem. - Int. Ed 2014, 53 (36), 9487–9492. 10.1002/anie.201404099. [DOI] [PubMed] [Google Scholar]
- (89).Luo W; Westcott N; Dutta D; Pulsipher A; Rogozhnikov D; Chen J; Yousaf MN A Dual Receptor and Reporter for Multi-Modal Cell Surface Engineering. ACS Chem. Biol 2015, 10 (10), 2219–2226. 10.1021/acschembio.5b00137. [DOI] [PubMed] [Google Scholar]
- (90).Csizmar CM; Petersburg JR; Hendricks A; Stern LA; Hackel BJ; and Wagner CR Engineering Reversible Cell-Cell Interactions with Lipid Anchored Prosthetic Receptors. Bioconjug. Chem 2018, 29 (4), 1291–1301. 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Capicciotti CJ; Zong C; Sheikh MO; Sun Tiantian, Wells L; Boons G Cell-Surface Glyco-Engineering by Exogenous Enzymatic Transfer Using a Bifunctional CMP-Neu5Ac Derivative. J Am Chem Soc 2017, 139 (38), 13342–13348. 10.1021/jacs.7b05358.Figures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Cheung TH; Rando TA Molecular Regulation of Stem Cell Quiescence. Nat. Rev. Mol. Cell Biol 2013, 14 (6), 329–340. 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Sousa-Victor P; Gutarra S; García-Prat L; Rodriguez-Ubreva J; Ortet L; Ruiz-Bonilla V; Jardí M; Ballestar E; González S; Serrano AL; et al. Geriatric Muscle Stem Cells Switch Reversible Quiescence into Senescence. Nature 2014, 506 (7488), 316–321. 10.1038/nature13013. [DOI] [PubMed] [Google Scholar]
- (94).Stabley DR; Jurchenko C; Marshall SS; Salaita KS Visualizing Mechanical Tension across Membrane Receptors with a Fluorescent Sensor. Nat. Methods 2012, 9 (1), 64–67. 10.1038/nmeth.1747. [DOI] [PubMed] [Google Scholar]
- (95).Lichtenstein RG; Rabinovich GA Glycobiology of Cell Death: When Glycans and Lectins Govern Cell Fate. Cell Death Differ. 2013, 20 (8), 976–986. 10.1038/cdd.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Ohtsubo K; Marth JD Glycosylation in Cellular Mechanisms of Health and Disease. Cell 2006, 126 (5), 855–867. 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- (97).Paszek MJ; Dufort CC; Rossier O; Bainer R; Mouw JK; Godula K; Hudak JE; Lakins JN; Wijekoon AC; Cassereau L; et al. The Cancer Glycocalyx Mechanically Primes Integrin-Mediated Growth and Survival. Nature 2014, 511 (7509), 319–325. 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Palte MJ; Raines RT Interaction of Nucleic Acids with the Glycocalyx. J. Am. Chem. Soc 2012, 134 (14), 6218–6223. 10.1021/ja2106477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Woods EC; Kai F; Barnes JM; Pedram K; Pickup MW; Hollander MJ; Weaver VM; Bertozzi CR A Bulky Glycocalyx Fosters Metastasis Formation by Promoting G1 Cell Cycle Progression. eLife 2017, 6, 1–15. 10.7554/eLife.25752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Kinnaird T; Stabile E; Burnett MS; Shou M; Lee CW; Barr S; Fuchs S; Epstein SE Local Delivery of Marrow-Derived Stromal Cells Augments Collateral Perfusion Through Paracrine Mechanisms. Circulation 2004, 109 (12), 1543–1549. 10.1161/01.CIR.0000124062.31102.57. [DOI] [PubMed] [Google Scholar]


