SARS-CoV-2 receptor binding domain and S2GΔHR2 spike elicited potent immune responses when displayed on protein nanoparticles.
Abstract
Vaccination against SARS-CoV-2 provides an effective tool to combat the COVID-19 pandemic. Here, we combined antigen optimization and nanoparticle display to develop vaccine candidates for SARS-CoV-2. We first displayed the receptor-binding domain (RBD) on three self-assembling protein nanoparticle (SApNP) platforms using the SpyTag/SpyCatcher system. We then identified heptad repeat 2 (HR2) in S2 as the cause of spike metastability, designed an HR2-deleted glycine-capped spike (S2GΔHR2), and displayed S2GΔHR2 on SApNPs. An antibody column specific for the RBD enabled tag-free vaccine purification. In mice, the 24-meric RBD-ferritin SApNP elicited a more potent neutralizing antibody (NAb) response than the RBD alone and the spike with two stabilizing proline mutations in S2 (S2P). S2GΔHR2 elicited twofold higher NAb titers than S2P, while S2GΔHR2 SApNPs derived from multilayered E2p and I3-01v9 60-mers elicited up to 10-fold higher NAb titers. The S2GΔHR2-presenting I3-01v9 SApNP also induced critically needed T cell immunity, thereby providing a promising vaccine candidate.
INTRODUCTION
Three coronaviruses (CoVs) have caused widespread outbreaks in humans, including severe acute respiratory syndrome CoV-1 (SARS-CoV-1), Middle East respiratory syndrome CoV (MERS-CoV), and SARS-CoV-2, which is the causative agent of CoV disease 2019 (COVID-19) (1–3) and has resulted in more than 2.4 million deaths worldwide (4). Enormous efforts are being undertaken to develop effective therapeutics and prophylactics for SARS-CoV-2. Small molecules that can block the host receptor, angiotensin-converting enzyme 2 (ACE2), and the transmembrane protease serine 2 (TMPRSS2) (5), which is required to process the spike protein, are being considered as treatments in addition to other interventions (6). While the immunology underlying COVID-19 is still being intensively studied (6–8), various vaccine candidates are now in clinical development (9–12). Inactivated virus vaccines have exhibited robust neutralizing antibody (NAb) responses in animals (13, 14), whereas viral vector vaccines based on human adenovirus (Ad5 and Ad26) and chimpanzee ChAdOx1 have been evaluated in nonhuman primates and human trials (15–18). Both DNA (19–21) and mRNA (22, 23) vaccines have been rapidly developed, with moderate NAb titers observed for the mRNA vaccine in medium- and high-dose groups (22). A recombinant spike protein adjuvanted with lipid nanoparticles (NPs), NVX-CoV2373, elicited high NAb titers in a human trial that were, on average, fourfold greater than in convalescent patients (24, 25). Efficacy was recently reported for a vector vaccine (AZD1222: 70.4%) (26) and two mRNA vaccines (mRNA-1273: 94.1%; BNT162b2: 95%) (27, 28). In December 2020, the U.S. Food and Drug Administration (FDA) issued the emergency use authorization (EUA) for the two mRNA vaccines.
The SARS-CoV-2 spike protein is a trimer of S1-S2 heterodimers. The S1 subunit contains a receptor-binding domain (RBD) that binds to ACE2 on host cells to initiate infection. The S2 subunit consists of a fusion peptide (FP) and heptad repeat regions 1 and 2 (HR1 and HR2). Upon endocytosis of the virion, the S1 subunit is cleaved off to facilitate FP insertion into the host cell membrane, while the remaining S2 refolds to bring HR1 and HR2 together to fuse the viral and host cell membranes (29). The spike protein harbors all NAb epitopes and is the main target for vaccine development against SARS-associated CoVs (30). Convalescent plasma has been used to treat COVID-19 patients with severe conditions (31), highlighting the importance of NAbs in protection (32). Because of moderate sequence conservation of the RBDs (~73%), some previously identified NAbs targeting the SARS-CoV-1 RBD have been shown to bind and cross-neutralize SARS-CoV-2 (33, 34). Using single-cell technologies and the SARS-CoV-2 RBD or spike as bait, potent NAbs have now been isolated from COVID-19 patients (35–41). Camelid-derived single-chain NAbs have also been obtained by panning naïve or immune llama single-domain antibody (VHH) libraries (42, 43). Structures of the SARS-CoV-2 spike and RBD in unliganded (44, 45), ACE2-bound (46–48), and antibody-bound (49–51) states determined by x-ray crystallography and cryo–electron microscopy (cryo-EM) have paved the way for rational vaccine design. Cryo-EM and cryo–electron tomography (ET) have revealed the inherent spike metastability and the coexistence of pre-/postfusion spikes on virions (52). A double-proline mutation (52) has been used in most soluble spike (S2P) constructs and all but inactivated vaccines, although a HexaPro version with greater yield and stability is now available (53). Cryo-ET has also uncovered a dynamic, triple-hinged HR2 stalk that facilitates viral entry and immune evasion (54–56).
In this study, we designed and optimized SARS-CoV-2 antigens for multivalent display on self-assembling protein NPs (SApNPs) (57–59), including a ferritin (FR) 24-mer and two 60-mers (E2p and I3-01v9) containing an inner layer of locking domains (LDs) and a cluster of “T cell” epitopes (60). To facilitate tag-free vaccine purification, we developed an immunoaffinity column based on antibody CR3022 that binds to both SARS-CoV-1/2 RBDs (34, 50). We first designed a scaffolded RBD trimer construct to mimic the “RBD-up” spike conformation. The SARS-CoV-1/2 RBDs were attached to SApNPs using the SpyTag/SpyCatcher system (61), providing a robust strategy for developing RBD-based NP vaccines. We then probed the spike metastability by comparing two uncleaved spike antigens, S2P (K986P/V987P) and S2G (K986G/V987G). The SARS-CoV-2 S2G spike exhibited abnormal behavior, suggesting that an unidentified facet of the spike can promote conformational change and block antibody access to the RBD. An HR2-deleted spike, S2GΔHR2, produced high-purity trimers, suggesting that the HR2 stalk may be a trigger of spike metastability, consistent with recent findings (54–56). We next displayed S2GΔHR2 on three SApNPs by gene fusion, resulting in spike SApNPs with high yield, purity, and antigenicity. In mouse immunization, the S2P spike protein elicited the lowest level of NAb response. In contrast, the scaffolded RBD trimer registered two- to threefold higher NAb titers, with another fivefold increase in NAb titer achieved by multivalent display on FR. S2GΔHR2 elicited up to sevenfold higher NAb titers, while the large, multilayered S2GΔHR2 E2p and I3-01 SApNPs induced up to 10-fold higher NAb titers than S2P. Further analysis indicated that the S2GΔHR2-presenting I3-01v9 SApNP can elicit a strong T helper 1 (TH1) response and other types of T cell responses needed for protective cellular immunity. Our study thus identifies the HR2 stalk as a major source of spike metastability, validates an HR2-deleted spike design, and provides a set of RBD- and spike-based virus-like particles (VLPs) as effective protein vaccine candidates against SARS-CoV-2.
RESULTS
Rational design of scaffolded RBD trimers and RBD-presenting SApNPs
RBD binding to the ACE2 receptor initiates the membrane fusion process (5). The crystal structure of SARS-CoV-2 RBD/ACE2 complex revealed the atomic details of receptor recognition (62). The SARS-CoV-2 RBD has been used as bait to isolate monoclonal antibodies (mAbs) from patient samples (35–41). For SARS-CoV-1 and MERS-CoV, RBD-based vaccines have induced potent NAbs that effectively block viral entry (30). Therefore, the RBD represents a major target for humoral responses following infection and can be used to develop epitope-focused vaccines.
We first hypothesized that RBD attached to a trimeric scaffold could mimic the RBD-up spike conformation and elicit NAbs that block ACE2 binding. To test this possibility, we designed a fusion construct containing SARS-CoV-1/2 RBD, a short 5–amino acid G4S linker (with a 2–amino acid restriction site), and a trimeric viral capsid protein, SHP [Protein Data Bank (PDB): 1TD0] (Fig. 1A). Structural modeling showed that the three tethered RBDs form a triangle of 92 Å (measured at L492), which is 14 and 18 Å wider than the SARS-CoV-1 “two-RBD-up” spike (PDB: 6CRX, measured at L478) (63) and the MERS-CoV “all-RBD-up” spike (PDB: 5X59, measured for L506) (64), respectively, allowing NAb access to each RBD. We then developed an immunoaffinity chromatography (IAC) column to facilitate tag-free purification. Previously, NAb-derived IAC columns have been used to purify HIV-1 Env trimers/NPs (58, 59, 65, 66), hepatitis C virus (HCV) E2 cores/NPs (57), and Ebola virus (EBOV) glycoprotein (GP) trimers/NPs (60). Tian et al. (34) reported that a SARS-CoV-1 NAb, CR3022, can bind SARS-CoV-2 RBD. The SARS-CoV-2 RBD/CR3022 structure revealed a conserved cryptic epitope that is shared by the two SARS-CoVs, suggesting that transient breathing motions of the spike protein enabled CR3022 binding to the RBD (50). Here, we examined the utility of CR3022 in IAC columns. The SARS-CoV-1/2 RBD-5GS-1TD0 constructs were transiently expressed in 100-ml ExpiCHO cells and purified on a CR3022 column before size exclusion chromatography (SEC) using a Superdex 200 10/300 GL column. While the SARS-CoV-1 RBD construct showed both aggregate (~8.6 ml) and trimer (~12.7 ml) peaks in the SEC profile, the SARS-CoV-2 RBD construct produced a single, pure trimer peak at ~12.8 ml (Fig. 1B). In SDS–polyacrylamide gel electrophoresis (PAGE), a monomer band of ~37 kDa and a trimer band of ~100 kDa were observed under reducing and nonreducing conditions, respectively (fig. S1A). Antigenicity was assessed for the two scaffolded RBD trimers in the enzyme-linked immunosorbent assay (ELISA) after CR3022/SEC purification (Fig. 1C). RBD-specific NAbs targeting SARS-CoV-1 [CR3022 (67), m396 (68), 80R (69), and S230 (70)] and SARS-CoV-2 [B38 (38), CB6 (37), S309 from a SARS survivor (33), and P2B-2F6 (36)] were tested in the ELISA. Overall, similar half-maximal effective concentration (EC50) values were observed for the two RBD trimers binding to their respective NAbs (Fig. 1C). The SARS-CoV-1 RBD trimer showed greater affinity for CR3022 than its SARS-CoV-2 counterpart, with a 1.3-fold difference in EC50 values, consistent with previous findings (34, 50). Of the SARS-CoV-2 NAbs, B38 yielded a similar EC50 value to CR3022, which can bind but not neutralize SARS-CoV-2. Antibody binding kinetics were measured by biolayer interferometry (BLI) (Fig. 1, B and D). Overall, all tested mAbs exhibited a fast on-rate, but with visible differences in their off-rates. B38 showed a faster off-rate than other SARS-CoV-2 NAbs, while CR3022, the mAb used to purify SARS-CoV-1/2 RBD proteins, exhibited a comparable kinetic profile.
Fig. 1. Rational design of SARS-CoV-1/2 RBD-based vaccines.
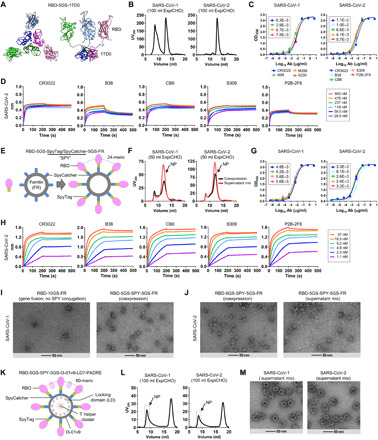
(A) Structural model of RBD-5GS-1TD0 in an extended RBD-up conformation (top view and side view). 1TD0 is a trimerization scaffold of viral origin. (B) SEC profiles of SARS-CoV-1/2 scaffolded RBD trimers following ExpiCHO expression and CR3022 purification. UV280, ultraviolet absorbance at 280 nm. (C) ELISA binding of SARS-CoV-1/2 scaffolded RBD trimers to a panel of mAbs. EC50 (μg/ml) values are labeled. Ab, antibody. (D) Octet binding of the SARS-CoV-2 scaffolded RBD trimer to five mAbs. Sensorgrams were obtained from an Octet RED96 instrument at six antigen concentrations from 950 to 29.5 nM by twofold dilutions. (E) Diagram of conjugating RBD to the 24-meric FR SApNP using the SpyTag/SpyCatcher (SPY) system. (F) SEC profiles of SARS-CoV-1/2 RBD-5GS-SPY-5GS-FR SApNPs produced in ExpiCHO by coexpression (black line) and supernatant mix (red line). (G) ELISA binding of SARS-CoV-1/2 RBD-FR SApNPs to a panel of mAbs. EC50 (μg/ml) values are labeled. (H) Octet binding of the SARS-CoV-2 RBD-FR SApNP to five mAbs. Sensorgrams were obtained from an Octet RED96 instrument at six antigen concentrations from 37 to 1.1 nM by twofold dilutions. (I) EM images of SARS-CoV-1 RBD-10GS-FR (gene fusion) and RBD-5GS-SPY-5GS-FR (coexpression). (J) EM images of SARS-CoV-2 RBD-5GS-SPY-5GS-FR produced by coexpression and supernatant mix. (K) Diagram of conjugating RBD to the 60-meric multilayered I3-01v9 SApNP using the SPY system. Locking domains (LDs) and T helper epitopes within the multilayered SApNP are depicted. (L and M) SEC profiles and EM images of SARS-CoV-1/2 RBD-5GS-SPY-5GS-I3-01v9-LD7-PADRE (or I3-01v9-L7P) SApNPs produced by supernatant mix. SEC profiles of scaffolded RBD trimers in (B) and SPY-linked RBD SApNPs in (F) and (L) were obtained from a Superdex 200 10/300 GL column and a Superose 6 10/300 GL column, respectively.
We then hypothesized that the SpyTag/SpyCatcher (termed SPY) system could be used to conjugate RBD to SApNPs to create multivalent RBD vaccines capable of eliciting a more potent NAb response (Fig. 1E). The 13–amino acid SpyTag spontaneously reacts with the SpyCatcher protein to form an irreversible isopeptide bond (61). The SPY system has been successfully used to attach antigens to VLPs (71). Here, SpyTag was fused to the C terminus of RBD, while SpyCatcher was fused to the N terminus of an SApNP subunit, both with a 5–amino acid G4S linker. This design was first tested for the 24-meric FR. Here, we compared two production strategies, coexpression of RBD-5GS-SpyTag and SpyCatcher-5GS-FR versus supernatant mix after separate expression, both followed by purification on a CR3022 column. Protein obtained from transient transfection in 50-ml ExpiCHO cells was analyzed by SEC on a Superose 6 10/300 GL column (Fig. 1F). Both production strategies produced a peak (12 ml) corresponding to SApNPs. While the SARS-CoV-2 construct outperformed its SARS-CoV-1 counterpart in particle yield (0.6 to 1.0 mg versus 0.3 to 0.5 mg after CR3022/SEC), the supernatant mix appeared to be superior to coexpression for yield in both cases. The results thus suggest that both strategies can be used to produce RBD SApNPs in Chinese hamster ovary (CHO) cells in an industrial setting, as this mammalian expression system has been widely used to manufacture therapeutic glycoproteins under good manufacturing practice (GMP) conditions (72, 73). Antigenicity was assessed for SEC-purified RBD-5GS-SPY-5GS-FR SApNPs. In the ELISA, RBD SApNPs showed slightly improved mAb binding compared to the RBD trimers, as indicated by EC50 values (Fig. 1G). In BLI, a more pronounced effect of multivalent display on antigenicity was observed, showing notably increased binding signals and plateaued dissociation (Fig. 1, C and H). Structural integrity of various RBD SApNPs was analyzed by negative-stain EM (nsEM) (Fig. 1, I and J). For SARS-CoV-1, a genetically fused RBD-10GS-FR construct produced very few, albeit highly pure, SApNPs (Fig. 1I, left). In contrast, the RBD-5GS-SPY-5GS-FR construct produced a high yield of SApNPs with visible surface decorations (Fig. 1I, right). For SARS-CoV-2, the purified RBD-5GS-SPY-5GS-FR SApNPs, regardless of the production strategy, showed morphology of well-formed particles (Fig. 1J). Previously, we genetically fused a small LD protein and a T cell epitope to each subunit of the E2p and I3-01v9 60-mers, resulting in “multilayered” SApNPs (60). PADRE, a 13–amino acid pan-DR epitope that activates CD4+ T cells (74), was used to promote B cell development toward NAbs. Here, two of the best designs, E2p-LD4-PADRE (or E2p-L4P) and I3-01v9-LD7-PADRE (or I3-01v9-L7P), were tested for their ability to display SARS-CoV-1/2 RBDs. Following the strategy established for FR, SARS-CoV-1/2 RBDs were attached to the I3-01v9-L7P SApNP using the SPY system (Fig. 1K). Despite the modest yield in ExpiCHO cells (Fig. 1L), large and pure particles were observed in the EM images (Fig. 1M). However, some impurities were noted for the RBD-presenting E2p-L4P SApNPs (fig. S1D). Nonetheless, we illustrated the utility of the SPY system for rapid development of SARS-CoV-1/2 RBD vaccines based on three different SApNP platforms.
Recently, RBDs were displayed on various SApNPs using the SPY system as MERS-CoV and SARS-CoV-2 vaccine candidates (75, 76). Walls et al. (77) also reported a SARS-CoV-2 RBD vaccine candidate based on the two-component NP platform, which requires a “connector” component to facilitate NP assembly (78), likely resulting in suboptimal stability. In addition, all these vaccine candidates require a complex production process involving two expression systems, separate purification steps, and an in vitro assembly step before final purification. In contrast, our SPY-linked RBD SApNPs are single component by nature and can be produced in CHO cells with a simple purification scheme, offering unique advantages in stability and manufacturability.
Rational design of prefusion spike through minimizing metastability
In addition to the RBD, the SARS-CoV-1/2 spikes contain other NAb epitopes (30), which are all presented in a trimeric context (Fig. 2A). A double proline (2P) mutation between HR1 and the central helix (CH) has been used to stabilize the MERS-CoV (79) and SARS-CoV-1 spikes (63). A similar 2P mutation (K986P/V987P) was introduced into the SARS-CoV-2 spike (termed S2P), which has been used to isolate and characterize NAbs (33, 35, 40, 42–45, 49) and is the antigen in almost all vaccine candidates in clinical development (11, 12). However, a recent cryo-EM study revealed an unexpected packing of S1 in the S2P spike, positioned ~12 Å outward, compared to the full-length native spike, as well as a more ordered FP proximal region (FPPR) in S2 (52). New designs have been generated to control the spike conformation (80) or to further stabilize it with more prolines (HexaPro) (53). Recent cryo-EM and cryo-ET studies revealed that the SARS-CoV-2 spikes could adopt diverse orientations on native virions due to the highly flexible HR2 stalk (54–56). Previously, we identified an HR1 bend as the cause of HIV-1 Env metastability (58, 81) and examined the role of an equivalent HR1 bend and the HR2 stalk in EBOV GP metastability (60). This understanding of metastability proved critical for designing stable trimers and trimer-presenting SApNP vaccines for both viruses (58–60, 81). It is therefore imperative to explore the cause(s) of spike metastability to facilitate rational vaccine design for SARS-CoV-2.
Fig. 2. Rational design of SARS-CoV-2 spike antigens.
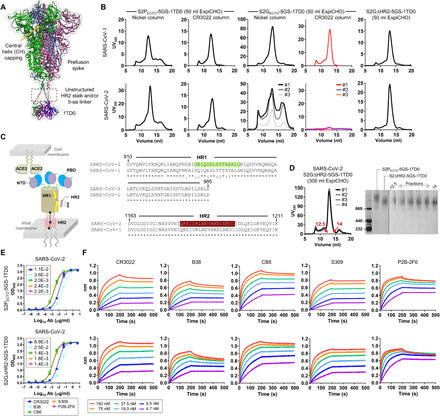
(A) Structural model of prefusion S spike linked to the C-terminal trimerization domain (1TD0) with a 5GS linker in transparent molecular surface. The approximate position for the unstructured HR2 stalk, or, in this case, a 5–amino acid (aa) G4S linker, is highlighted with a dashed line box. (B) SEC profiles of SARS-CoV-1 (top) and SARS-CoV-2 (bottom) spikes. Left to right panels: S2PECTO-5GS-1TD0 purified on nickel and CR3022 columns (1 and 2), S2GECTO-5GS-1TD0 on nickel and CR3022 columns (3 and 4), and S2GΔHR2-5GS-1TD0 on a CR3022 column (5). All spike constructs tested in (B) contain a C-terminal His6 tag to facilitate nickel column purification. Results from three separate production runs are shown for SARS-CoV-2 S2GECTO-5GS-1TD0. (C) Schematic representation of a full-length SARS-CoV-2 spike on the virus membrane surface in the presence of host ACE2 and the HR2 region from a neighboring spike (left), and sequence alignment of SARS-CoV-1/2 HR1 and HR2 regions (right, top, and bottom). The HR1 and HR2 segments that form a six-helix bundle in the postfusion state are highlighted in green and brown shade, respectively. (D) Left: SEC profiles of S2GΔHR2-5GS-1TD0, showing results from four separate production runs. Right: BN-PAGE analysis of S2PECTO-5GS-1TD0 and S2GΔHR2-5GS-1TD0. SEC fractions (12.5 to 14.0) are shown for S2GΔHR2-5GS-1TD0 on the gel. SEC profiles in (B) and (D) were obtained from a Superose 6 10/300 GL column. (E) ELISA binding of two SARS-CoV-2 spikes (S2PECTO-5GS-1TD0 and S2GΔHR2-5GS-1TD0) to five mAbs. EC50 (μg/ml) values are labeled. (F) Octet binding of two SARS-CoV-2 spikes (S2PECTO-5GS-1TD0 and S2GΔHR2-5GS-1TD0) to five mAbs. Sensorgrams were obtained from an Octet RED96 instrument at six antigen concentrations from 150 to 4.7 nM by twofold dilutions. The spike constructs tested in (D) to (F) do not contain a C-terminal His6 tag.
We first created uncleaved spike ectodomain (SECTO) constructs for SARS-CoV-1/2, both containing the 2P mutation (K968P/V969P and K986P/V987P, respectively), a 5–amino acid G4S linker, a trimerization motif (PDB: 1TD0), and a C-terminal His6 tag. The two constructs were transiently expressed in 50-ml ExpiCHO cells followed by purification on either a Nickel column or a CR3022 column. The S2PECTO-5GS-1TD0-His6 protein was characterized by SEC on a Superose 6 10/300 GL column (Fig. 2B, panels 1 and 2). After the nickel column, both S2PECTO constructs showed a trimer peak (~12 ml), with the left and right shoulders indicative of aggregate and dimer/monomer species, respectively. CR3022 purification resulted in a consistent trimer peak and reduction in dimer/monomer species. We then compared a pair of SECTO constructs for SARS-CoV-1/2, both containing a double glycine (2G) mutation, K968G/V969G and K986G/V987G, respectively. The 2G mutation had little effect on the SARS-CoV-1 spike but produced abnormal SEC profiles and showed no yield for the SARS-CoV-2 spike after purification by the nickel and CR3022 columns, respectively (Fig. 2B, panels 3 and 4). Last, we tested a pair of S2G constructs without the HR2 stalk (E1150-Q1208), termed S2GΔHR2. Deletion of the HR2 stalk restored the SARS-CoV-2 trimer peak and reduced aggregates for both SARS-CoV-1/2, as shown by the SEC profiles upon CR3022 purification (Fig. 2B, panel 5). Because the triple-hinged HR2 stalk can generate diverse spike orientations on native virions (54–56), and the fusion core is formed by HR1 and HR2, we hypothesized that HR2 may be a key determinant of SARS-CoV-2 spike metastability (Fig. 2C, left). It is plausible that the interactions between HR1 and HR2 of two neighboring spikes may facilitate the pre- to postfusion transition in addition to ACE2 binding and S1 dissociation. Given the extensive sequence difference in HR1 (9 amino acids in total) compared with SARS-CoV-1 (Fig. 2C, right), we sought to examine the contribution of HR1 to SARS-CoV-2 spike metastability with two HR1-swapped (HR1S) spike constructs. While HR1 swapping proved ineffective, deletion of the HR2 stalk once again restored the trimer peak, suggesting a more important role for HR2 (fig. S2, A to C). Therefore, S2GΔHR2 appeared to be a general spike design for SARS-CoV-1/2 and perhaps other CoVs. Four separate production runs of SARS-CoV-2 S2GΔHR2-5GS-1TD0 in 300-ml ExpiCHO cells resulted in nearly identical SEC profiles with trimer yields of 0.8 to 1.0 mg (Fig. 2D, left). Consistently, blue native PAGE (BN-PAGE) showed high trimer purity across SEC fractions (Fig. 2D, right). Antigenicity was assessed for CR3022/SEC-purified SARS-CoV-2 S2PECTO and S2GΔHR2 spike proteins. In ELISA, the S2GΔHR2 spike showed slightly higher affinity for the five representative mAbs than did the S2PECTO spike (Fig. 2E). When tested against three newly identified human NAbs, C105 (49) and CC12.1/CC12.3 (41), the two spikes yielded similar EC50 values (fig. S2D). In BLI, the S2GΔHR2 spike showed higher binding signals than the S2PECTO spike at the highest concentration while exhibiting similar binding kinetics (Fig. 2F). The use of NAb P2B-2F6 (36) for spike purification resulted in higher trimer yield with similar purity to the CR3022 column across SEC fractions (fig. S2E). Thermostability was assessed by differential scanning calorimetry (DSC) for CR3022/SEC-purified SARS-CoV-2 S2PECTO and S2GΔHR2 spikes (fig. S2F). Upon the 2P-to-2G substitution and HR2 deletion, a small increase in thermal denaturation midpoint (Tm) was observed (46.8°C versus 47.6°C), with notably higher onset temperature (Ton), 35.2°C versus 40.0°C, and narrower half width of the peak (ΔT1/2), 4.7°C versus 3.9°C. Together, we demonstrated that the HR2 stalk is a major source of spike metastability, and S2GΔHR2 presents an alternative spike design to S2P, although more stabilizing mutations may be required to achieve greater thermostability.
Rational design of single-component self-assembling spike NPs
Although it was possible to conjugate trimeric SARS-CoV-2 spikes to an SApNP using the SPY system (82), the random, irreversible linking will likely result in irregular display with unoccupied but spatially occluded anchoring sites on the surface. The SPY system is perhaps more suitable for small individual antigens (e.g., the RBD). Using the gene fusion approach, we previously designed single-component SApNPs displaying stabilized HIV-1 Env trimers (58, 59) and optimized HCV E2 cores (57). Recently, we reengineered E2p and I3-01v9 60-mers to incorporate an inner layer of LDs to stabilize the noncovalently formed NP shell, in addition to a cluster of T cell epitopes (60). This multilayered design may be essential to the stability of resulting SApNP vaccines when they are used to display large, complex viral antigens such as the SARS-CoV-2 spike.
Native SARS-CoV-2 virions present both pre- and postfusion spikes (52, 54, 55) (Fig. 3A, top), while our vaccine strategy aims to develop single-component SApNPs that each present 8 or 20 stable prefusion S2GΔHR2 spikes to the immune system (Fig. 3A, bottom). As demonstrated in our previous studies (58, 59), different linker lengths may be needed to connect a trimer to the SApNP surface, as the spacing between the N termini of NP subunits around each threefold axis varies: FR (20 Å), E2p (9 Å), and I3-01v9 (50.5 Å). On the basis of this consideration, we displayed the S2GΔHR2 spike on FR with a 5–amino acid G4S linker, on E2p with a 5–amino acid G4S linker, and on I3-01v9 with a 10–amino acid (G4S)2 linker, resulting in SApNPs with diameters of 47.9, 55.9, and 59.3 nm, respectively (Fig. 3B). The multilayered E2p-L4P and I3-01v9-L7P (60), which were validated for presenting RBDs (Fig. 1, K to L, and fig. S1D), were used here as NP carriers of the S2GΔHR2 spike. Together, three S2GΔHR2 SApNP constructs were transiently expressed in 400-ml ExpiCHO cells, followed by CR3022 purification and SEC on a Superose 6 10/300 GL column (Fig. 3C). Three production runs for each of the three constructs generated highly consistent SEC profiles, despite the variation of low-molecular-weight impurities observed for FR and E2p-L4P. After CR3022 and SEC purification, we obtained 0.3 to 0.4, 0.5 to 1.0, and 0.8 to 1.2 mg of protein for S2GΔHR2-5GS-FR, S2GΔHR2-5GS-E2p-L4P, and S2GΔHR2-10GS-I3-01v9-L7P, respectively. Overall, the I3-01v9–derived S2GΔHR2 SApNP appeared to perform best in terms of particle yield, purity, and stability. Structural integrity was analyzed by nsEM, which showed well-formed particles of 45 to 65 nm with recognizable protrusions on the surface (Fig. 3D). The varying shape of these profusions may correspond to S2GΔHR2 spikes with “open” RBDs, in contrast to an array of “closed” HIV-1 and EBOV trimers on the SApNP surface (58–60). This open conformation may facilitate induction of a strong RBD-specific NAb response in vivo. Antigenicity of S2GΔHR2 SApNPs was assessed using the same panel of mAbs. In the ELISA, three SApNPs showed slightly improved binding to some, but not all, mAbs compared with the individual spike (Fig. 3E). In BLI, we observed a clear correlation between peak mAb binding signal and antigen valency, with E2p/I3-01v9 > FR > spike (Fig. 3F). Multivalent display on the two 60-mers substantially increased mAb binding compared with the FR 24-mer. In previous studies, we observed a similar correlation for HIV-1 Env trimer and HCV E2 core versus their SApNPs (57, 58). In summary, these VLP-size SApNPs with 8 or 20 spikes on the surface provide promising vaccine candidates for in vivo evaluation.
Fig. 3. Rational design of SARS-CoV-2 spike-presenting NP vaccines.
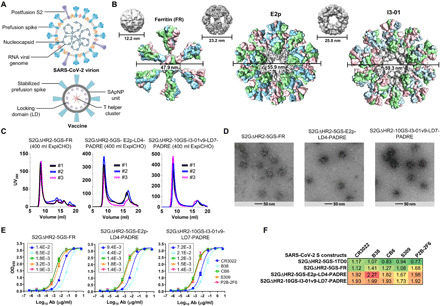
(A) Schematic representation of SARS-CoV-2 virion (top) and spike-presenting SApNP vaccine (bottom). For the SARS-CoV-2 virion, pre-/postfusion S, nucleocapsid, and RNA viral genome are shown, while for the vaccine, stabilized spike and multilayered SApNP composition are depicted. (B) Colored surface models of SApNP carriers (top) and spike-presenting SApNP vaccines (bottom). The three SApNP carriers used here are FR 24-mer and multilayered E2p and I3-01v9 60-mers (the LD layer and PADRE cluster are not shown). Particle size is indicated by diameter (in nanometers). (C) SEC profiles of three SApNPs presenting the SARS-CoV-2 S2GΔHR2 spike, from left to right, S2GΔHR2-5GS-FR, S2GΔHR2-5GS-E2p-LD4-PADRE (or E2p-L4P), and S2GΔHR2-10GS-I3-01v9-LD7-PADRE (or I3-01v9-L7P). SEC profiles were obtained from a Superose 6 10/300 GL column, each showing results from three separate production runs. (D) EM images of three SARS-CoV-2 S2GΔHR2 SApNPs. (E) ELISA binding of three SARS-CoV-2 S2GΔHR2 SApNPs to five mAbs. EC50 (μg/ml) values are labeled. (F) Antigenic profiles of SARS-CoV-2 S2GΔHR2 spike and three SApNPs against five mAbs. Sensorgrams were obtained from an Octet RED96 using six antigen concentrations (150 to 4.6 nM for spike, 9 to 0.27 nM for FR SApNP, and 3.5 to 0.1 nM for E2p and I3-01v9 SApNPs, respectively, all by twofold dilutions) and AHQ biosensors, as shown in fig. S3B. The peak binding signals (in nanometers) at the highest concentration are listed. Color coding indicates the signal strength measured by Octet (green to red: low to high).
SARS-CoV-1/2 vaccine-induced binding antibody response
Selected SARS-CoV-1/2 RBD- and spike-based vaccine constructs were assessed in BALB/c mice (Fig. 4A). We adopted a similar immunization protocol to be consistent with our previous studies on HIV-1, HCV, and EBOV SApNPs (57, 58, 60). Briefly, groups of five mice were immunized four times at 3-week intervals. All antigens (50 μg per dose) were formulated with AddaVax, an oil-in-water emulsion adjuvant, except for I3-01v9, which was mixed with aluminum phosphate (83). We first analyzed the binding antibody (bAb) responses, as measured by EC50 titers, in the two SARS-CoV-2 RBD vaccine groups (Fig. 4B and fig. S4). The SPY-linked RBD SApNP (RBD-5GS-SPY-5GS-FR) elicited significantly higher bAb titers than the scaffolded RBD trimer (RBD-5GS-1TD0) at week 2 (w2) and w5, regardless of the coating antigen, and yielded a significant P value at w8 when the RBD was coated. Compared with the S2PECTO spike (S2PECTO-5GS-1TD0), the RBD SApNP elicited significantly higher bAb titers against the RBD at w2, w5, and w8 (Fig. 4B, right), demonstrating a strong epitope-focusing effect. Mouse sera bound the SARS-CoV-1 spike with lower EC50 titers than the SARS-CoV-2 spike but with similar patterns (fig. S4A). We then analyzed the bAb responses induced by SARS-CoV-2 spikes S2PECTO-5GS-1TD0 and S2GΔHR2-5GS-1TD0, as well as three SApNPs each displaying 8 or 20 S2GΔHR2 spikes (Fig. 4C and fig. S5). The S2GΔHR2 spike showed two- to threefold higher average EC50 titers than the S2PECTO spike, regardless of the coating antigen (of note, to facilitate a fair comparison, mouse sera from these two groups were tested against their respective spike antigens). Three SApNPs exhibited different temporal patterns depending on the coating antigen. In terms of spike-specific response, the I3-01v9 group registered a steady increase in average EC50 titer over time, showing the highest bAb titers at w2 and w8 and significantly outperforming the S2PECTO group at all time points. The I3-01v9 group also yielded higher EC50 titers than the S2GΔHR2 group throughout, although not with significant P values. The FR SApNP group exhibited a similar temporal pattern with lower EC50 titers, which were still significantly higher than the S2PECTO group. Among the three SApNPs, E2p exhibited the lowest average EC50 titer at w2 and reached the highest at w5, which then decreased slightly at w8. In terms of the RBD-specific response, the five groups showed a clear ranking based on their average EC50 titers, which remained consistent across time points. At w2, I3-01v9 showed an average EC50 titer of 175, whereas all other spike-based vaccines induced little RBD-specific bAb response. At w5 and w8, S2GΔHR2 elicited higher bAb titers (on average, by twofold) than S2PECTO, while all three SApNPs outperformed the individual S2GΔHR2 spike with a ranking of average EC50 titers that correlated with their size (FR < E2p < I3-01v9). Sera reacted with the SARS-CoV-1 spike similarly, albeit at a lower level (fig. S5A). Last, we compared the bAb responses induced by three SARS-CoV-1 vaccines, S2PECTO spike (S2PECTO-5GS-1TD0), scaffolded RBD trimer (RBD-5GS-1TD0), and SPY-linked RBD SApNP (RBD-5GS-SPY-5GS-FR) (Fig. 4D and fig. S6). On the basis of average EC50 titers, the SARS-CoV-1 S2PECTO spike appeared to have elicited a more robust bAb response than the SARS-CoV-2 S2GΔHR2 spike, whereas the SARS-CoV-1 RBD SApNP was less advantageous than its SARS-CoV-2 counterpart. Serum reactivity with the SARS-CoV-2 S2PECTO spike was observed for all three SARS-CoV-1 vaccine groups (fig. S6A). In summary, RBD SApNPs can elicit higher titers of RBD-specific bAbs than the scaffolded RBD trimer and S2PECTO spike, albeit at different levels for the two SARS-CoVs. The S2GΔHR2 spike is more immunogenic than the S2PECTO spike, showing, on average, twofold higher bAb titers. The S2GΔHR2-presenting E2p and I3-01v9 SApNPs are the best performers among all the spike-based vaccines, consistent with our previous findings for these two large SApNPs (57, 58, 60).
Fig. 4. SARS-CoV-1/2 vaccine-induced bAb responses in BALB/c mice.
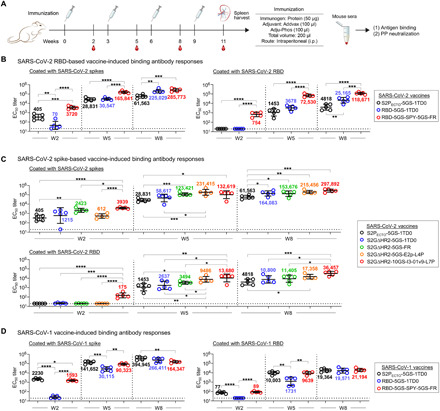
(A) Schematic representation of the mouse immunization protocol. (B) SARS-CoV-2 RBD and RBD-SApNP vaccine-induced bAb responses. Coating antigens: SARS-CoV-2 S2GΔHR2-5GS-foldon (left) and RBD (right). (C) SARS-CoV-2 S2GΔHR2 spike and S2GΔHR2-SApNP vaccine-induced bAb responses. Coating antigens: SARS-CoV-2 S2GΔHR2-5GS-foldon (top) and RBD (bottom). In (B) and (C), the SARS-CoV-2 S2PECTO-5GS-1TD0 group is included for comparison, with S2PECTO-5GS-foldon used as the coating antigen. (D) SARS-CoV-1 RBD and RBD-SApNP vaccine-induced bAb responses. Coating antigens: SARS-CoV-1 S2PECTO-5GS-foldon (left) and RBD (right). In (D), the SARS-CoV-1 S2PECTO-5GS-1TD0 group is included for comparison, with S2PECTO-5GS-foldon used as the coating antigen. In (B) to (D), EC50 titers derived from the ELISA binding of mouse plasma to coating antigens are plotted, with average EC50 values labeled on the plots. The P values were determined by an unpaired t test in GraphPad Prism 8.4.3, with (*) indicating the level of statistical significance. Detailed ELISA data are shown in figs. S4 to S6.
SARS-CoV-1/2 vaccine-induced NAb response
One major goal in COVID-19 vaccine development is to generate a potent NAb response that can protect against SARS-CoV-2 infection. Pseudoparticle (SARS-CoV-1/2-pp) neutralization assays (84) were used to evaluate serum NAb responses elicited by different vaccines. We first analyzed the NAb responses, as measured by 50% inhibitory dilution (ID50) titers, in the two SARS-CoV-2 RBD vaccine groups (Fig. 5A and fig. S7). The SPY-linked RBD SApNP elicited a NAb response against autologous SARS-CoV-2 as early as w2, albeit with low titers, and retained its advantage at w5 and w8, suggesting that these RBD SApNP vaccines can elicit a rapid NAb response. The scaffolded RBD trimer group showed higher average ID50 titers than the S2PECTO spike group at both w5 and w8. A somewhat different pattern was observed in the SARS-CoV-1-pp assay. At w2, no vaccine group showed detectable heterologous NAb response. At w5 and w8, the S2PECTO spike elicited a more potent anti–SARS-CoV-1 NAb response than the scaffolded RBD trimer, suggesting that non-RBD epitopes on the spike may contribute to cross-neutralization. We then analyzed the NAb responses induced by five spike-based vaccines (Fig. 5B and fig. S8). In terms of autologous neutralization, no spike-based vaccine elicited any NAb response that blocks SARS-CoV-2-pps after one dose, but a consistent pattern of serum neutralization was observed at w5 and w8 (Fig. 5B, upper panel). Specifically, the S2PECTO spike showed the lowest average ID50 titers, 879 and 2481 at w5 and w8, respectively, whereas the S2GΔHR2 spike induced a stronger NAb response with 2.8- to 6.7-fold higher average ID50 titers, which did not reach P ≤ 0.05 due to within-group variation. Nonetheless, this result confirmed the beneficial effect of the 2G mutation and the HR2 stalk deletion on NAb elicitation. Among the three SApNPs, E2p was the best performer at w5, showing an average ID50 titer of 8435 that is 9.6-fold higher than S2PECTO and 1.4-fold higher than S2GΔHR2, while I3-01v9 showed the most potent NAb response at w8 with an average ID50 titer of 17,351 that is about 7-fold and 2.5-fold higher than S2PECTO and S2GΔHR2, respectively. A similar temporal pattern was observed in the heterologous SARS-CoV-1-pp assay (Fig. 5B, lower panel). It is worth noting that the multilayered I3-01v9 SApNP elicited a SARS-CoV-1 NAb response with an average ID50 titer of 351 at w2, whereas all other groups showed no detectable serum neutralization. These results suggest that a well-designed SARS-CoV-2 S2GΔHR2 SApNP vaccine may provide protection against both SARS-CoVs. Last, we analyzed the NAb responses induced by three SARS-CoV-1 vaccines (Fig. 5C and fig. S9). In the autologous SARS-CoV-1-pp assay, the S2PECTO spike and the RBD SApNP induced significantly higher NAb titers than the scaffolded RBD trimer at w2 and w5, and all three vaccine groups showed similar ID50 titers at w8. However, heterologous SARS-CoV-2 neutralization was below or at the baseline level for three SARS-CoV-1 vaccines at w2, w5, and w8. In this study, the pseudovirus neutralization assay was validated using a panel of known SARS-CoV-1/2 NAbs (fig. S9C). As a negative control, the w8 mouse sera were tested against pseudoparticles bearing the murine leukemia virus (MLV) Env, MLV-pps, showing no detectable reactivity (fig. S9D). In summary, these results demonstrate an advantage in NAb elicitation by the SARS-CoV-2 S2GΔHR2 spike and its SApNPs compared with the widely used S2PECTO spike. Although SARS-CoV-2 RBD– and S2GΔHR2-presenting SApNPs are both effective at eliciting an autologous NAb response, the latter can induce high NAb titers to SARS-CoV-1 and may provide broader protection against SARS-associated CoVs.
Fig. 5. SARS-CoV-1/2 vaccine-induced NAb responses in BALB/c mice.
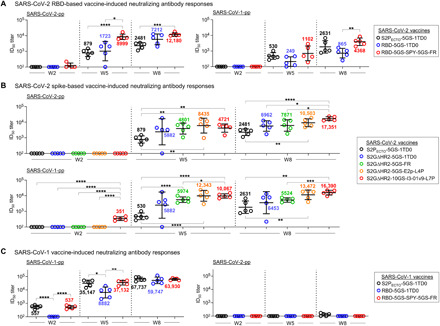
(A) SARS-CoV-2 RBD and RBD-SApNP vaccine-induced NAb responses to autologous SARS-CoV-2 (left) and heterologous SARS-CoV-1 (right). (B) SARS-CoV-2 S2GΔHR2 spike and S2GΔHR2-SApNP vaccine-induced NAb responses to SARS-CoV-2 (top) and SARS-CoV-1 (bottom). In (A) and (B), the SARS-CoV-2 S2PECTO-5GS-1TD0 group is included for comparison. (C) SARS-CoV-1 RBD and RBD-SApNP vaccine-induced NAb responses to autologous SARS-CoV-1 (left) and heterologous SARS-CoV-2 (right). In (C), the SARS-CoV-1 S2PECTO-5GS-1TD0 group is included for comparison. In (A) to (C), ID50 titers derived from the SARS-CoV-1/2-pp neutralization assays are plotted, with average ID50 values labeled on the plots. The P values were determined by an unpaired t test in GraphPad Prism 8.4.3, with (*) indicating the level of statistical significance. Detailed SARS-CoV-1/2-pp neutralization data are shown in figs. S7 to S9.
SARS-CoV-2 vaccine-induced T cell response
While humoral immunity is required to block host-virus interaction and prevent viral infection, cellular immunity is essential for eliminating infected host cells to control viral infection (85–88). Emerging evidence indicates that an early T cell response (89, 90), as well as T cell memory (91), is critical for protection against SARS-CoV-2. However, COVID-19 vaccines must induce a CD4+ TH1, but not TH2-type, T cell response, as the latter has been implicated in vaccine-associated enhancement of respiratory disease (10). Additionally, T follicular helper cells (Tfh) play an important role in the maturation and production of NAbs. Therefore, understanding T cell responses is crucial for development of an effective and safe COVID-19 vaccine.
Interferon-γ (IFN-γ)–producing TH1 cells are important for generating an optimal antibody response and for the induction of cellular immunity (85–87). We first examined various SARS-CoV-2 vaccines on the induction of CD4+ TH1 responses specific to the vaccine antigen at w11, 2 weeks after the fourth immunization, when memory T cells had already developed in spleen (88). Mouse splenocytes from the S2PECTO group and two S2GΔHR2 SApNP groups (E2p and I3-01v9) were analyzed by flow cytometry using naïve samples as a negative control. The I3-01v9 group induced about 1.5- and 2.3-fold higher frequency of IFN-γ–producing CD4+ TH1 cells than the S2PECTO and E2p groups, respectively (Fig. 6A). Notably, after restimulation with the respective antigens for as few as 4 hours, both SApNP groups produced ~2-fold higher frequency of CD107a-producing cytolytic CD4+ T cells than the S2PECTO and naïve groups (Fig. 6B). IFN-γ/interleukin-4 (IL-4) double-positive cells are memory CD4+ T cells that have acquired the ability to produce IL-4 while still retaining the ability to produce IFN-γ under TH1 conditions (92). It appeared that I3-01v9 induced three- and fivefold more IFN-γ/IL-4 double-positive memory CD4+ T cells than S2PECTO and E2p (Fig. 6A). These results suggest that I3-01v9 can induce both CD4+ TH1 cells and IFN-γ/IL-4 double-positive memory CD4+ T cells. In addition, I3-01v9 induced more IFN-γ/granulocyte-macrophage colony-stimulating factor (GM-CSF) double-positive CD8+ effector T cells than S2PECTO and E2p (Fig. 6C), suggesting that protective CD8+ T cells were also generated in mice immunized with I3-01v9. Notably, CD8+ T cells derived from mice immunized with I3-01v9, rather than those with S2PECTO and E2p, acquired the ability to rapidly produce IFN-γ upon antigen restimulation (Fig. 6D), suggesting the generation of I3-01v9–responsive effector/memory T cells. Our results indicate that the S2GΔHR2 I3-01v9 SApNP can induce robust T cell responses consisting of CD4+ TH1 cells, IFN-γ/IL-4 double-positive memory CD4+ T cells, and effector CD8+ T cells, thus providing protective cellular immunity in addition to a potent NAb response. Because T cell immunity against the SApNP backbone cannot be ruled out, a more detailed T cell analysis using soluble spikes, spike peptides, and naked SApNPs for restimulation may be warranted.
Fig. 6. SARS-CoV-2 vaccine-induced T cell responses in BALB/c mice.
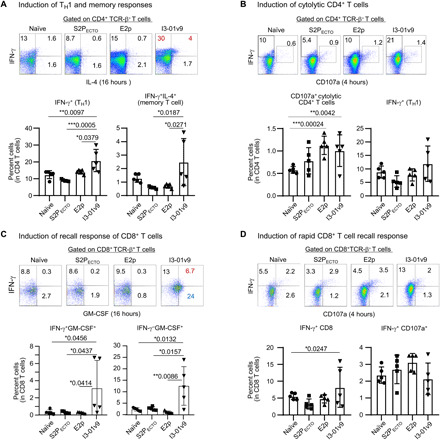
Splenocytes from mice (n = 5 in each group), immunized with S2PECTO spike, S2GΔHR2 E2p SApNP, and S2GΔHR2 I3-01v9 SApNP, were isolated at w11 and cultured in the presence of IL-2 and dendritic cells (DCs) pulsed with S2PECTO (1 × 10−7 μM), S2GΔHR2 E2p (1 × 10−7 μM), and S2GΔHR2 I3-01v9 (1 × 10−7 μM), respectively. Splenocytes from five naïve mice were used as control samples and cultured in the presence of DCs without antigen pulsing. Cells were assessed after 16 hours (A and C) and 4 hours (B and D) of culture. (A and B) Vaccine-induced CD4+ T cell immunity. (C and D) Vaccine-induced CD8+ T cell immunity. Plots show the frequencies of cell fraction. The P values were determined by one-way ANOVA analysis. *P < 0.05, **P < 0.01, and ***P < 0.001.
DISCUSSION
COVID-19 is the first worldwide pandemic of this scale since the infamous Spanish influenza over a century ago (93), which caused ~50 million deaths worldwide and remains a painful reminder of our vulnerability to a new virus without a protective vaccine. The rapid spread of SARS-CoV-2 therefore demanded rapid vaccine development (10). Operation Warp Speed (OWS) was launched in May 2020 with the initial goal of delivering 300 million doses of safe and effective vaccines by January 2021 (94), although that goal has not yet been realized. Nevertheless, in December 2020, while other vaccine candidates were still being evaluated in clinical trials, two OWS-supported mRNA vaccines were approved for EUA, marking an important turning point in the battle against the pandemic. However, vaccine development during a pandemic against a new virus poses unique challenges, one of which is how to balance public health needs versus scientific rigor (95–97). The global vaccine campaign also provides a unique opportunity to compare different vaccine design strategies and platforms—especially new ones—against a common target. While mRNA and viral vector vaccines remain the front-runners, protein-based vaccines are highlighted in a recent review with the prediction that they will eventually reach a larger fraction of the global population (98). As the NAb titers induced by the first-generation nucleic acid vaccines wane over time, effective protein vaccines will be needed to sustain long-term immunity against SARS-CoV-2.
Here, we approached SARS-CoV-2 vaccine with a rational design strategy and set out to develop protein NP vaccines that can be used alone or as a booster vaccine. To this end, a panel of vaccine candidates has been generated based on three SApNP platforms with different in vitro and in vivo attributes (table S1). Our comparative analysis of these vaccine constructs has offered some valuable insights. First, the choice of antigen is critical to the success of SARS-CoV-2 vaccine regardless of the delivery platform. Most vaccine antigens, including OWS’s vaccine candidates, are based on S2P, which produces a spike structure that differs in detail from the full-length wild-type spike, e.g., in FPPR of S2 and in the relative dispositions of the S1 domains (52). These differences may complicate interpretation of vaccine outcome. S2P and other empirical spike designs (53) have attempted to constrain the spike conformation and increase trimer yield. However, as we previously demonstrated for HIV-1 Env and EBOV GP (58, 60, 81), it is important to identify and eliminate (if possible) the cause of spike metastability. During antigen screening, we found that deletion of the HR2 stalk with a 2P-to-2G substitution renders a more stable spike, which is consistent with recent reports on a highly flexible HR2 stalk in the native spikes on SARS-CoV-2 virions (54–56). Thus, S2GΔHR2 would seem to represent a major advance in spike design. Second, single-component SApNPs provide a powerful platform for vaccine development against diverse viral pathogens (57, 58, 60). Here, S2GΔHR2 was genetically fused, rather than chemically linked, to three SApNPs, including two multilayered 60-meric carriers with enhanced stability and an embedded T help signal. These protein vaccines should be more effective in eliciting a potent NAb response and less likely to induce adverse responses (98, 99). An epitope-focused vaccine strategy was also explored by designing scaffolded RBD trimers and SPY-linked RBD SApNPs. Third, to achieve high efficacy and ensure safety, vaccine-induced NAb and T cell responses must be evaluated in animals before clinical trials. In our mouse study, S2GΔHR2 appeared to be more effective than S2P in NAb elicitation, both alone and displayed on SApNPs. Notably, the multilayered S2GΔHR2 I3-01v9 SApNP elicited not only high NAb titers but also desired T cell responses. In addition to CD4+ TH1 cells and memory CD4+ T cells, this SApNP also induced CD107a-producing cytolytic CD4+ T cells, which may directly kill infected host cells, and GM-CSF–producing CD8+ effector T cells, which may promote the generation of macrophages and functional dendritic cells (DCs) to facilitate the clearance of infected cells. It is also worth noting that the NAb response to the large SApNPs plateaued after three doses, suggesting that the number of injections and dosage can be reduced without diminishing vaccine-induced responses. Last, expression of vaccine antigens in CHO cells followed by purification using an antibody column, such as CR3022, would allow rapid and industrial-scale vaccine production. In summary, our study provides promising COVID-19 vaccine candidates for evaluation in clinical trials.
MATERIALS AND METHODS
Design, expression, and purification of SARS-CoV-1/2 RBD and spike antigens
The spike (S) genes of the SARS-CoV-1 isolate Tor2 (GenBank accession number: NC_004718) and the SARS-CoV-2 isolate Wuhan-Hu-1 (GenBank accession number: MN908947) were used to design all the RBD and spike constructs following codon optimization for expression in mammalian cells. The RBD sequence is defined as P317-D518 and P330-N532 for SARS-CoV-1 and SARS-CoV-2, respectively. The SECTO sequence is defined as M1-Q1190 and M1-Q1208 for SARS-CoV-1 and SARS-CoV-2, respectively. To remove the S1/S2 cleavage site, an R667G mutation and a 682GSAGSV687 modification were introduced in the SARS-CoV-1 and SARS-CoV-2 spikes, respectively. The 2P (or 2G) mutation was made to K968/V969 and K986/V987 in the SARS-CoV-1 and SARS-CoV-2 spikes, respectively. The SARS-CoV-2 C-terminal region (E1150-Q1208) containing the HR2 stalk was removed from S2GECTO, resulting in an HR2-deleted spike construct, termed S2GΔHR2. The viral capsid protein SHP (PDB: 1TD0) was used as a trimerization motif in spike constructs for immunization, whereas the foldon domain from the bacteriophage T4 fibritin (PDB: 1RFO) was used in coating spike antigens for the ELISA to mask the 1TD0-derived antibody response. All constructs were transiently expressed in ExpiCHO cells (Thermo Fisher Scientific). Briefly, ExpiCHO cells were thawed and incubated with ExpiCHO Expression Medium (Thermo Fisher Scientific) in a shaker incubator at 37°C, 135 rpm, and 8% CO2. When the cells reached a density of 10 × 106 ml−1, ExpiCHO Expression Medium was added to reduce cell density to 6 × 106 ml−1 for transfection. The ExpiFectamine CHO/plasmid DNA complexes were prepared for 100-ml transfection in ExpiCHO cells following the manufacturer’s instructions. For a given construct, 100 μg of plasmid and 320 μl of ExpiFectamine CHO reagent were mixed in 7.7 ml of cold OptiPRO medium (Thermo Fisher Scientific). After the first feed on day 1, ExpiCHO cells were cultured in a shaker incubator at 33°C, 115 rpm, and 8% CO2 following the Max Titer protocol with an additional feed on day 5 (Thermo Fisher Scientific). Culture supernatants were harvested 13 to 14 days after transfection, clarified by centrifugation at 4000 rpm for 25 min, and filtered using a 0.45-μm filter (Thermo Fisher Scientific). The CR3022 antibody column was used to extract SARS-CoV-1/2 antigens from the supernatants, which was followed by SEC on a Superdex 200 10/300 GL column (for scaffolded RBD trimers) or a Superose 6 10/300 GL column (for SPY-linked RBD SApNPs, spikes, and S2GΔHR2 SApNPs). A human NAb, P2B-2F6 (36), was also used to pack antibody columns for purification of the SARS-CoV-2 S2GΔHR2 spike, which was followed by SEC on a HiLoad 16/600 Superose 6 column. For comparison, His6-tagged SECTO-5GS-1TD0 spike protein was extracted from the supernatants using an immobilized Ni Sepharose Excel column (GE Healthcare) and eluted with 500 mM imidazole before SEC. Protein concentration was determined using UV280 absorbance with theoretical extinction coefficients.
Blue native PAGE
SARS-CoV-2 spikes and spike-presenting SApNPs were analyzed by blue native PAGE and stained with Coomassie blue. The proteins were mixed with sample buffer and G250 loading dye and added to a 4 to 12% Bis-Tris NativePAGE gel (Life Technologies). BN-PAGE gels were run for 2 to 2.5 hours at 150 V using the NativePAGE running buffer (Life Technologies) according to the manufacturer’s instructions.
Enzyme-linked immunosorbent assay
Each well of a Costar 96-well assay plate (Corning) was first coated with 50 μl of phosphate-buffered saline (PBS) containing 0.2 μg of the appropriate antigens. The plates were incubated overnight at 4°C and then washed five times with wash buffer containing PBS and 0.05% (v/v) Tween 20. Each well was then coated with 150 μl of a blocking buffer consisting of PBS and blotting-grade blocker (40 mg ml−1) (Bio-Rad). The plates were incubated with the blocking buffer for 1 hour at room temperature and then washed five times with wash buffer. For antigen binding, antibodies [in the immunoglobulin G (IgG) form] were diluted in the blocking buffer to a maximum concentration of 5 μg ml−1 followed by a 10-fold dilution series. For each antibody dilution, a total of 50-μl volume was added to the appropriate wells. For mouse sample analysis, plasma was diluted by 20-fold in the blocking buffer and subjected to a 10-fold dilution series. For each sample dilution, a total of 50-μl volume was added to the wells. Each plate was incubated for 1 hour at room temperature and then washed five times with PBS containing 0.05% Tween 20. For antibody binding, a 1:5000 dilution of goat anti-human IgG antibody (Jackson ImmunoResearch Laboratories Inc.), or, for mouse sample analysis, a 1:3000 dilution of horseradish peroxidase–labeled goat anti-mouse IgG antibody (Jackson ImmunoResearch Laboratories), was then made in the wash buffer (PBS containing 0.05% Tween 20), with 50 μl of this diluted secondary antibody added to each well. The plates were incubated with the secondary antibody for 1 hour at room temperature and then washed six times with PBS containing 0.05% Tween 20. Lastly, the wells were developed with 50 μl of trimethylboron (Life Sciences) for 3 to 5 min before stopping the reaction with 50 μl of 2 N sulfuric acid. The resulting plate readouts were measured at a wavelength of 450 nm. Notably, the w2 serum binding did not reach the plateau (or saturation) to allow for accurate determination of EC50 titers. Nonetheless, the EC50 values at w2 were derived by setting the lower/upper constraints of OD450 (optical density at 450 nm) at 0.0/3.2 to facilitate the comparison of different vaccine groups at the first time point.
Biolayer interferometry
The kinetics of SARS-CoV-1/2 vaccine antigens, RBD versus RBD-presenting SApNPs as well as spike versus spike-presenting SApNPs, binding to a panel of known antibodies (in the IgG form), was measured using an Octet RED96 instrument (FortéBio, Pall Life Sciences). All assays were performed with agitation set to 1000 rpm in FortéBio 1× kinetic buffer. The final volume for all the solutions was 200 μl per well. Assays were performed at 30°C in solid black 96-well plates (Geiger Bio-One). For all antigens except for S2GΔHR2 SApNPs, antibody (5 μg ml−1) in 1× kinetic buffer was loaded onto the surface of anti-human Fc Capture Biosensors (AHC) for 300 s. For S2GΔHR2 SApNPs, anti-human Fc Quantitation Biosensors (AHQ) were used. A 60-s biosensor baseline step was applied before the analysis of the association of the antibody on the biosensor to the antigen in solution for 200 s. A twofold concentration gradient of antigen, starting at 950 nM for scaffolded RBD trimers, 37 nM for SPY-linked RBD FR SApNP (RBD-5GS-SPY-5GS-FR), 150 nM for soluble spikes, and 9/3.5/3.5 nM for S2GΔHR2 presented on FR/E2p/I3-01v9 SApNPs, was used in a titration series of six. The dissociation of the interaction was followed for 300 s. Correction of baseline drift was performed by subtracting the mean value of shifts recorded for a sensor loaded with antibody, but not incubated with antigen, and for a sensor without antibody but incubated with antigen. Octet data were processed by FortéBio’s data acquisition software v.8.1. Experimental data were fitted with the binding equations describing a 2:1 interaction to achieve optimal fitting. Of note, the S2GΔHR2 spike was also measured using AHQ biosensors to facilitate comparison of mAb binding with the S2GΔHR2-presenting SApNPs.
Differential scanning calorimetry
Thermal melting curves of SARS-CoV-2 S2PECTO-5GS-1TD0 and S2GΔHR2-5GS-1TD0 spike proteins following CR3022 and SEC purification were obtained from a MicroCal PEAQ-DSC Man instrument (Malvern). The purified spike trimer protein produced from ExpiCHO cells were buffer-exchanged into 1× PBS and concentrated to 0.8 μM before analysis by the instrument. Melting was probed at a scan rate of 60°C hour−1 from 20° to 100°C. Data processing, including buffer correction, normalization, and baseline subtraction, was conducted using the MicroCal PEAQ-DSC software. Gaussian fitting was performed using the Origin 9.0 software.
EM assessment of NP constructs
The EM analysis of various RBD and S2GΔHR2-presenting SApNPs was performed by the Core Microscopy Facility at The Scripps Research Institute. All SApNP samples were prepared at the concentration of 0.01 to 0.05 mg/ml. Carbon-coated copper grids (400 mesh) were glow-discharged, and 8 μl of each sample was adsorbed for 2 min. Excess sample was wicked away, and grids were negatively stained with 2% uranyl formate for 2 min. Excess stain was wicked away, and the grids were allowed to dry. Samples were analyzed at 80 kV with a Talos L120C transmission electron microscope (Thermo Fisher Scientific), and images were acquired with a CETA 16M CMOS (complementary metal-oxide semiconductor) camera.
Animal immunization and sample collection
Similar immunization protocols have been reported in our previous vaccine studies (57, 58, 60). Briefly, the Institutional Animal Care and Use Committee (IACUC) guidelines were followed with animal subjects tested in the immunization study. Eight-week-old BALB/c mice were purchased from The Jackson Laboratory and housed in ventilated cages in environmentally controlled rooms at The Scripps Research Institute, in compliance with an approved IACUC protocol and AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care) International guidelines. Mice were immunized at w0, w3, w6, and w9 with 200 μl of antigen/adjuvant mix containing 50 μg of vaccine antigen and 100 μl of adjuvant, AddaVax or Adju-Phos (InvivoGen), via the intraperitoneal route. Blood was collected 2 weeks after each immunization. All bleeds were performed through the retro-orbital sinus using heparinized capillary tubes into EDTA-coated tubes. Samples were spun at 1200 rpm for 10 min to separate plasma (top layer) and the rest of the whole-blood layer. Upon heat inactivation at 56°C for 30 min, the plasma was spun at 2000 rpm for 10 min to remove precipitates. The rest of the whole-blood layer was diluted with an equal volume of PBS and then overlaid on 4.5 ml of Ficoll in a 15-ml SepMate tube (STEMCELL Technologies) and spun at 1200 rpm for 10 min at 20°C to separate peripheral blood mononuclear cells (PBMCs). Cells were washed once in PBS and then resuspended in 1 ml of ACK Red Blood Cell lysis buffer (Lonza). After washing with PBS, PBMCs were resuspended in 2 ml of Bambanker Freezing Media (Lymphotec). Spleens were harvested at w11 and ground against a 70-μm cell strainer (BD Falcon) to release the splenocytes into a cell suspension. Splenocytes were centrifuged, washed in PBS, treated with 5 ml of ammonium-chloride-potassium (ACK) lysing buffer (Lonza), and frozen with 3 ml of Bambanker freezing media. Plasma was used for ELISA and neutralization assays to determine binding and NAb responses in mouse serum.
SARS-CoV-1/2 pseudovirus neutralization assay
Pseudoparticle (SARS-CoV-1/2-pp) neutralization assays were used to assess the neutralizing activity of previously reported antibodies and vaccine-induced murine antibody response. SARS-CoV-1/2-pps were generated by cotransfection of human embryonic kidney (HEK) 293T cells with the HIV-1 pNL4-3.lucR-E- plasmid (obtained from the National Institutes of Health AIDS reagent program: www.aidsreagent.org/) and the expression plasmid encoding the S gene of SARS-CoV-1 isolate Tor2 (GenBank accession number: NC_004718) and the SARS-CoV-2 isolate Wuhan-Hu-1 (GenBank accession number: MN908947) at a 4:1 ratio by Lipofectamine 3000 (Thermo Fisher Scientific). After 48 to 72 hours, SARS-CoV-1/2-pps were collected from the supernatant by centrifugation at 4000 rpm for 10 min, aliquoted, and stored at −80°C before use. The mAbs at a starting concentration of 0.1 to 10 μg/ml, or mouse plasma at a starting dilution of 100-fold, were mixed with the supernatant containing SARS-CoV-1/2-pps and incubated for 1 hour at 37°C in white solid-bottom 96-well plate (Corning). A threefold dilution series was used in the assay. The HEK293T-hACE2 cell line (catalog number: NR-52511) and the vector pcDNA3.1(−) containing the SARS-CoV-2 S gene (catalog number: NR52420) were obtained from BEI RESOURCES (www.beiresources.org/) and used in pseudovirus neutralization assays (84). Briefly, HEK293T-hACE2 cells at 1 × 104 were added to each well, and the plate was incubated at 37°C for 48 hours. After incubation, overlying media were removed, and cells were lysed. The firefly luciferase signal from infected cells was determined using the Bright-Glo Luciferase Assay System (Promega) according to the manufacturer’s instructions. Data were retrieved from a BioTek microplate reader with Gen 5 software, the average background luminescence from a series of uninfected wells was subtracted from each well, and neutralization curves were generated using GraphPad Prism 8.4.3, in which values from wells were compared against a well containing SARS-CoV-1/2-pp only. The same HIV-1 vectors pseudotyped with the MLV Env gene, termed MLV-pps, were produced in HEK293T cells and included in the neutralization assays as a negative control. As the NAb titers plateaued after w8, the w11 results were not shown in Fig. 5 but included in figs. S7 to S9 for comparison.
DC production
Mouse bone marrow was cultured in RPMI 1640 medium containing 10% fetal bovine serum and recombinant mouse Flt3 ligand (Flt3L; 50 ng/ml) and stem cell factor (SCF; 10 ng/ml) for 9 days as described (100). To induce DC activation, immature DCs were incubated with lipopolysaccharide (100 ng/ml) and R848 (resiquimod; 100 ng/ml) overnight, which activated Toll-like receptor 4 (TLR4), TLR7/8, or TLR9 signaling, respectively. Cells were harvested for the experiments. DCs were sorted to isolate CD11c+ cells using magnetic beads (Miltenyi Biotec, CA).
Antibodies and flow cytometry analysis
All antibodies used for immunofluorescence staining were purchased from eBioscience (San Diego, CA), BioLegend (San Diego, CA), or BD Biosciences (San Jose, CA). Magnetic microbead-conjugated antibodies and streptavidin were purchased from Miltenyi Biotec (Auburn, CA). Recombinant human IL-2 protein was purchased from R&D Systems (Minneapolis, MN). Recombinant mouse Flt3L and mouse SCF were purchased from Shenandoah Biotech (Warwick, PA). Cells were stained with appropriate concentrations of mAbs. Dead cells were excluded using Fixable Viability Dye from eBioscience (San Diego, CA). Flow cytometry analyses were performed using LSRII (BD Biosciences, CA) and Canto cytometers (Becton Dickinson, NJ). Cells were sorted on BD FACSAria II (BD Biosciences, CA).
T cell culture and activation
Splenic mononuclear cells from each group of immunized mice were cultured in the presence of DCs pulsed with or without S2PECTO, multilayered S2GΔHR2-presenting E2P, or I3-01v9 SApNP (1 × 10−7 μM) in complete Iscove’s Modified Dulbecco’s Medium (IMDM) containing IL-2 (5.0 ng/ml). Cells were collected 16 and 4 hours later for intracellular cytokine staining and flow cytometric analysis.
Statistics
For the antibody analysis, comparisons of different vaccine groups were performed in GraphPad Prism 8.4.3 using the two-tailed unpaired Student’s t test. For the T cell analysis, comparisons of means were done using two-tailed unpaired Student’s t test, analysis of variance (ANOVA), and then post hoc t test. P values of 0.05 or less were considered significant.
Supplementary Material
Acknowledgments
Funding: This work was funded by NIH grants AI129698 and AI140844 (to J.Z.), Ufovax/SFP-2018-0416, Ufovax/SFP-2018-1013, and Ufovax/SFP-2020-0111 (to J.Z.). Author contributions: Project design by L.H., Y.Z., I.A.W., and J.Z.; construct design of RBD and spike-based immunogens by L.H. and J.Z.; plasmid design and processing by L.H. and C.S.; antigen production, purification, and basic characterization by L.H., X.L., and T.N.; antibody production and column packing by L.H., X.L., and T.N.; antibody-antigen ELISA and BLI by L.H. and C.S.; DSC by L.H. and J.Z.; negative-stain EM by L.H. and J.Z.; plasma-antigen ELISA by L.H., X.L., and C.S.; antibody and mouse plasma neutralization by L.H. and X.L.; vaccine-induced T cell response analysis by Y.W., C.A., and Y.Z.; manuscript written by L.H., Y.Z., I.A.W., and J.Z. All authors were asked to comment on the manuscript. The TSRI manuscript number is 30028. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the corresponding author.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/12/eabf1591/DC1
REFERENCES AND NOTES
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W.; China Novel Coronavirus Investigating and Research Team , A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., Zhang Y.-Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong E. S., Du H. R., Gardner L., An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533–534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tay M. Z., Poh C. M., Rénia L., MacAry P. A., Ng L. F. P., The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 20, 363–374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zohar T., Alter G., Dissecting antibody-mediated protection against SARS-CoV-2. Nat. Rev. Immunol. 20, 392–394 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vabret N., Britton G. J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M. D., Pia L., Risson E., Saffern M., Salomé B., Selvan M. E., Spindler M. P., Tan J., van der Heide V., Gregory J. K., Alexandropoulos K., Bhardwaj N., Brown B. D., Greenbaum B., Gümüş Z. H., Homann D., Horowitz A., Kamphorst A. O., Curotto de Lafaille M. A., Mehandru S., Merad M., Samstein R. M.; Sinai Immunology Review Project , Immunology of COVID-19: Current state of the science. Immunity 52, 910–941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lurie N., Saville M., Hatchett R., Halton J., Developing Covid-19 vaccines at pandemic speed. N. Engl. J. Med. 382, 1969–1973 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Graham B. S., Rapid COVID-19 vaccine development. Science 368, 945–946 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Krammer F., SARS-CoV-2 vaccines in development. Nature 586, 516–527 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Le T. T., Cramer J. P., Chen R., Mayhew S., Evolution of the COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 19, 667–668 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., Gao H., Ge X., Kan B., Hu Y., Liu J., Cai F., Jiang D., Yin Y., Qin C., Li J., Gong X., Lou X., Shi W., Wu D., Zhang H., Zhu L., Deng W., Li Y., Lu J., Li C., Wang X., Yin W., Zhang Y., Qin C., Development of an inactivated vaccine candidate for SARS-CoV-2. Science 369, 77–81 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., Xu W., Zhao Y., Li N., Zhang J., Liang H., Bao L., Xu Y., Ding L., Zhou W., Gao H., Liu J., Niu P., Zhao L., Zhen W., Fu H., Yu S., Zhang Z., Xu G., Li C., Lou Z., Xu M., Qin C., Wu G., Gao G. F., Tan W., Yang X., Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell 182, 713–721.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J. N., Port J. R., Avanzato V. A., Bushmaker T., Flaxman A., Ulaszewska M., Feldmann F., Allen E. R., Sharpe H., Schulz J., Holbrook M., Okumura A., Meade-White K., Pérez-Pérez L., Edwards N. J., Wright D., Bissett C., Gilbride C., Williamson B. N., Rosenke R., Long D., Ishwarbhai A., Kailath R., Rose L., Morris S., Powers C., Lovaglio J., Hanley P. W., Scott D., Saturday G., de Wit E., Gilbert S. C., Munster V. J., ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 586, 578–582 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Folegatti P. M., Ewer K. J., Aley P. K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E. A., Dold C., Faust S. N., Finn A., Flaxman A. L., Hallis B., Heath P., Jenkin D., Lazarus R., Makinson R., Minassian A. M., Pollock K. M., Ramasamy M., Robinson H., Snape M., Tarrant R., Voysey M., Green C., Douglas A. D., Hill A. V. S., Lambe T., Gilbert S. C., Pollard A. J.; Oxford COVID Vaccine Trial Group , Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 396, 467–478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mercado N. B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., Mahan K. M., Tostanoski L. H., He X., Martinez D. R., Rutten L., Bos R., van Manen D., Vellinga J., Custers J., Langedijk J. P., Kwaks T., Bakkers M. J. G., Zuijdgeest D., Rosendahl Huber S. K., Atyeo C., Fischinger S., Burke J. S., Feldman J., Hauser B. M., Caradonna T. M., Bondzie E. A., Dagotto G., Gebre M. S., Hoffman E., Jacob-Dolan C., Kirilova M., Li Z., Lin Z., Mahrokhian S. H., Maxfield L. F., Nampanya F., Nityanandam R., Nkolola J. P., Patel S., Ventura J. D., Verrington K., Wan H., Pessaint L., Van Ry A., Blade K., Strasbaugh A., Cabus M., Brown R., Cook A., Zouantchangadou S., Teow E., Andersen H., Lewis M. G., Cai Y., Chen B., Schmidt A. G., Reeves R. K., Baric R. S., Lauffenburger D. A., Alter G., Stoffels P., Mammen M., Van Hoof J., Schuitemaker H., Barouch D. H., Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 586, 583–588 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu F.-C., Li Y.-H., Guan X.-H., Hou L.-H., Wang W.-J., Li J.-X., Wu S.-P., Wang B.-S., Wang Z., Wang L., Jia S.-Y., Jiang H.-D., Wang L., Jiang T., Hu Y., Gou J.-B., Xu S.-B., Xu J.-J., Wang X.-W., Wang W., Chen W., Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: A dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 395, 1845–1854 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith T. R. F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E. N., Walker S. N., Schultheis K., Purwar M., Xu Z., Walters J., Bhojnagarwala P., Yang M., Chokkalingam N., Pezzoli P., Parzych E., Reuschel E. L., Doan A., Tursi N., Vasquez M., Choi J., Tello-Ruiz E., Maricic I., Bah M. A., Wu Y., Amante D., Park D. H., Dia Y., Ali A. R., Zaidi F. I., Generotti A., Kim K. Y., Herring T. A., Reeder S., Andrade V. M., Buttigieg K., Zhao G., Wu J.-M., Li D., Bao L., Liu J., Deng W., Qin C., Brown A. S., Khoshnejad M., Wang N., Chu J., Wrapp D., McLellan J. S., Muthumani K., Wang B., Carroll M. W., Kim J. J., Boyer J., Kulp D. W., Humeau L. M. P. F., Weiner D. B., Broderick K. E., Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 11, 2601 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J., Tostanoski L. H., Peter L., Mercado N. B., Mahan K. M., Mahrokhian S. H., Nkolola J. P., Liu J., Li Z., Chandrashekar A., Martinez D. R., Loos C., Atyeo C., Fischinger S., Burke J. S., Slein M. D., Chen Y., Zuiani A., Lelis F. J. N., Travers M., Habibi S., Pessaint L., Van Ry A., Blade K., Brown R., Cook A., Finneyfrock B., Dodson A., Teow E., Velasco J., Zahn R., Wegmann F., Bondzie E. A., Dagotto G., Gebre M. S., He X., Jacob-Dolan C., Kirilova M., Kordana N., Lin Z., Maxfield L. F., Nampanya F., Nityanandam R., Ventura J. D., Wan H., Cai Y., Chen B., Schmidt A. G., Wesemann D. R., Baric R. S., Alter G., Andersen H., Lewis M. G., Barouch D. H., DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369, 806–811 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A. Patel, J. Walters, E. L. Reuschel, K. Schultheis, E. Parzych, E. N. Gary, I. Maricic, M. Purwar, Z. Eblimit, S. N. Walker, D. Guimet, P. Bhojnagarwala, A. Doan, Z. Xu, D. Elwood, S. M. Reeder, L. Pessaint, K. Y. Kim, A. Cook, N. Chokkalingam, B. Finneyfrock, E. Tello-Ruiz, A. Dodson, J. Choi, A. Generotti, J. Harrison, N. J. Tursi, V. M. Andrade, Y. Dia, F. I. Zaidi, H. Andersen, M. G. Lewis, K. Muthumani, J. J. Kim, D. W. Kulp, L. M. Humeau, S. Ramos, T. R. F. Smith, D. B. Weiner, K. E. Broderick, Intradermal-delivered DNA vaccine provides anamnestic protection in a rhesus macaque SARS-CoV-2 challenge model. bioRxiv 2020.07.28.225649 [Preprint]. 29 July 2020. 10.1101/2020.07.28.225649. [DOI]
- 22.Jackson L. A., Anderson E. J., Rouphael N. G., Roberts P. C., Makhene M., Coler R. N., McCullough M. P., Chappell J. D., Denison M. R., Stevens L. J., Pruijssers A. J., Dermott A. M., Flach B., Doria-Rose N. A., Corbett K. S., Morabito K. M., O’Dell S., Schmidt S. D., Swanson P. A. II, Padilla M., Mascola J. R., Neuzil K. M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J. E., Graham B. S., Beigel J. H.; mRNA-1273 Study Group , An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 383, 1920–1931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh E. E., Frenck R. W. Jr., Falsey A. R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M. J., Bailey R., Swanson K. A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Tompkins K. R., Lyke K. E., Raabe V., Dormitzer P. R., Jansen K. U., Şahin U., Gruber W. C., Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 383, 2439–2450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian J.-H., Patel N., Haupt R., Zhou H., Weston S., Hammond H., Lague J., Portnoff A. D., Norton J., Guebre-Xabier M., Zhou B., Jacobson K., Maciejewski S., Khatoon R., Wisniewska M., Moffitt W., Kluepfel-Stahl S., Ekechukwu B., Papin J., Boddapati S., Wong C. J., Piedra P. A., Frieman M. B., Massare M. J., Fries L., Bengtsson K. L., Stertman L., Ellingsworth L., Glenn G., Smith G., SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat. Commun. 12, 372 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J. S., Zhu M., Cloney-Clark S., Zhou H., Smith G., Patel N., Frieman M. B., Haupt R. E., Logue J., Grath M. M., Weston S., Piedra P. A., Desai C., Callahan K., Lewis M., Price-Abbott P., Formica N., Shinde V., Fries L., Lickliter J. D., Griffin P., Wilkinson B., Glenn G. M., Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 383, 2320–2332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voysey M., Clemens S. A. C., Madhi S. A., Weckx L. Y., Folegatti P. M., Aley P. K., Angus B., Baillie V. L., Barnabas S. L., Bhorat Q. E., Bibi S., Briner C., Cicconi P., Collins A. M., Colin-Jones R., Cutland C. L., Darton T. C., Dheda K., Duncan C. J. A., Emary K. R. W., Ewer K. J., Fairlie L., Faust S. N., Feng S., Ferreira D. M., Finn A., Goodman A. L., Green C. M., Green C. A., Heath P. T., Hill C., Hill H., Hirsch I., Hodgson S. H. C., Izu A., Jackson S., Jenkin D., Joe C. C. D., Kerridge S., Koen A., Kwatra G., Lazarus R., Lawrie A. M., Lelliott A., Libri V., Lillie P. J., Mallory R., Mendes A. V. A., Milan E. P., Minassian A. M., Gregor A. M., Morrison H., Mujadidi Y. F., Nana A., O’Reilly P. J., Padayachee S. D., Pittella A., Plested E., Pollock K. M., Ramasamy M. N., Rhead S., Schwarzbold A. V., Singh N., Smith A., Song R., Snape M. D., Sprinz E., Sutherland R. K., Tarrant R., Thomson E. C., Török M. E., Toshner M., Turner D. P. J., Vekemans J., Villafana T. L., Watson M. E. E., Williams C. J., Douglas A. D., Hill A. V. S., Lambe T., Gilbert S. C., Pollard A. J.; Oxford COVID Vaccine Trial Group , Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 397, 99–111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polack F. P., Thomas S. J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J. L., Marc G. P., Moreira E. D., Zerbini C., Bailey R., Swanson K. A., Roychoudhury S., Koury K., Li P., Kalina W. V., Cooper D., Frenck R. W. Jr., Hammitt L. L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D. B., Mather S., Dormitzer P. R., Şahin U., Jansen K. U., Gruber W. C.; C4591001 Clinical Trial Group , Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baden L. R., El Sahly H. M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S. A., Rouphael N., Creech C. B., Gettigan J. M., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B. S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T.; COVE Study Group , Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F., Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 117, 11727–11734 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S., The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 7, 226–236 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., Zhang J., Wu X., Li B., Xu Y., Chen W., Peng Y., Hu Y., Lin L., Liu X., Huang S., Zhou Z., Zhang L., Wang Y., Zhang Z., Deng K., Xia Z., Gong Q., Zhang W., Zheng X., Liu Y., Yang H., Zhou D., Yu D., Hou J., Shi Z., Chen S., Chen Z., Zhang X., Yang X., Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U.S.A. 117, 9490–9496 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang S. B., Hillyer C., Du L. Y., Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 41, 355–359 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinto D., Park Y.-J., Beltramello M., Walls A. C., Tortorici M. A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J. B., Chen R. E., Havenar-Daughton C., Snell G., Telenti A., Virgin H. W., Lanzavecchia A., Diamond M. S., Fink K., Veesler D., Corti D., Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T., Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 9, 382–385 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zost S. J., Gilchuk P., Chen R. E., Case J. B., Reidy J. X., Trivette A., Nargi R. S., Sutton R. E., Suryadevara N., Chen E. C., Binshtein E., Shrihari S., Ostrowski M., Chu H. Y., Didier J. E., MacRenaris K. W., Jones T., Day S., Myers L., Lee F. E.-H., Nguyen D. C., Sanz I., Martinez D. R., Rothlauf P. W., Bloyet L.-M., Whelan S. P. J., Baric R. S., Thackray L. B., Diamond M. S., Carnahan R. H., Crowe J. E. Jr., Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 26, 1422–1427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., Tang X., Yu J., Lan J., Yuan J., Wang H., Zhao J., Zhang S., Wang Y., Shi X., Liu L., Zhao J., Wang X., Zhang Z., Zhang L., Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115–119 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W. J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.-S., Wang Q., Gao G. F., Yuan Z., Yan J., A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., Gong Y., Xiao H., Fan Z., Tan S., Wu G., Tan W., Lu X., Fan C., Wang Q., Liu Y., Zhang C., Qi J., Gao G. F., Gao F., Liu L., A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 368, 1274–1278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao Y. L., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X.-D., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X. S., Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 182, 73–84.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robbiani D. F., Gaebler C., Muecksch F., Lorenzi J. C. C., Wang Z., Cho A., Agudelo M., Barnes C. O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T. Y., Viant C., Hurley A., Hoffmann H.-H., Millard K. G., Kost R. G., Cipolla M., Gordon K., Bianchini F., Chen S. T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A. W., Waltari E., Pak J. E., Huey-Tubman K. E., Koranda N., Hoffman P. R., West A. P. Jr., Rice C. M., Hatziioannou T., Bjorkman P. J., Bieniasz P. D., Caskey M., Nussenzweig M. C., Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogers T. F., Zhao F., Huang D., Beutler N., Burns A., He W.-T., Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R. K., Callaghan S., Garcia E., Hurtado J., Parren M., Peng L., Ramirez S., Ricketts J., Ricciardi M. J., Rawlings S. A., Wu N. C., Yuan M., Smith D. M., Nemazee D., Teijaro J. R., Voss J. E., Wilson I. A., Andrabi R., Briney B., Landais E., Sok D., Jardine J. G., Burton D. R., Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huo J. D., Bas A. L., Ruza R. R., Duyvesteyn H. M. E., Mikolajek H., Malinauskas T., Tan T. K., Rijal P., Dumoux M., Ward P. N., Ren J., Zhou D., Harrison P. J., Weckener M., Clare D. K., Vogirala V. K., Radecke J., Moynié L., Zhao Y., Gilbert-Jaramillo J., Knight M. L., Tree J. A., Buttigieg K. R., Coombes N., Elmore M. J., Carroll M. W., Carrique L., Shah P. N. M., James W., Townsend A. R., Stuart D. I., Owens R. J., Naismith J. H., Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 27, 846–854 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Wrapp D., De Vlieger D., Corbett K. S., Torres G. M., Wang N., Van Breedam W., Roose K., van Schie L.; VIB-CMB COVID-19 Response Team, Hoffmann M., Pöhlmann S., Graham B. S., Callewaert N., Schepens B., Saelens X., McLellan J. S., Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell 181, 1004–1015.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., McLellan J. S., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walls A. C., Park Y.-J., Tortorici M. A., Wall A., McGuire A. T., Veesler D., Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q., Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F., Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou T., Tsybovsky Y., Gorman J., Rapp M., Cerutti G., Chuang G.-Y., Katsamba P. S., Sampson J. M., Schön A., Bimela J., Boyington J. C., Nazzari A., Olia A. S., Shi W., Sastry M., Stephens T., Stuckey J., Teng I.-T., Wang P., Wang S., Zhang B., Friesner R. A., Ho D. D., Mascola J. R., Shapiro L., Kwong P. D., Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe 28, 867–879.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barnes C. O., West A. P. Jr., Huey-Tubman K. E., Hoffmann M. A. G., Sharaf N. G., Hoffman P. R., Koranda N., Gristick H. B., Gaebler C., Muecksch F., Lorenzi J. C. C., Finkin S., Hägglöf T., Hurley A., Millard K. G., Weisblum Y., Schmidt F., Hatziioannou T., Bieniasz P. D., Caskey M., Robbiani D. F., Nussenzweig M. C., Bjorkman P. J., Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 182, 828–842.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan M., Wu N. C., Zhu X., Lee C.-C. D., So R. T. Y., Lv H., Mok C. K. P., Wilson I. A., A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan M., Liu H., Wu N. C., Lee C.-C. D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., Rogers T. F., Landais E., Sok D., Jardine J. G., Burton D. R., Wilson I. A., Structural basis of a shared antibody response to SARS-CoV-2. Science 369, 1119–1123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai Y., Zhang J., Xiao T., Peng H., Sterling S. M., Walsh R. M. Jr., Rawson S., Rits-Volloch S., Chen B., Distinct conformational states of SARS-CoV-2 spike protein. Science 369, 1586–1592 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsieh C.-L., Goldsmith J. A., Schaub J. M., Di Venere A. M., Kuo H.-C., Javanmardi K., Le K. C., Wrapp D., Lee A. G., Liu Y., Chou C.-W., Byrne P. O., Hjorth C. K., Johnson N. V., Ludes-Meyers J., Nguyen A. W., Park J., Wang N., Amengor D., Lavinder J. J., Ippolito G. C., Maynard J. A., Finkelstein I. J., McLellan J. S., Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z., Cheng L., Shi D., Lu X., Lei J., Crispin M., Shi Y., Li L., Li S., Molecular architecture of the SARS-CoV-2 virus. Cell 183, 730–738.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ke Z., Oton J., Qu K., Cortese M., Zila V., Keane L. M., Nakane T., Zivanov J., Neufeldt C. J., Cerikan B., Lu J. M., Peukes J., Xiong X., Kräusslich H.-G., Scheres S. H. W., Bartenschlager R., Briggs J. A. G., Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588, 498–502 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turoň ová B., Sikora M., Schürmann C., Hagen W. J. H., Welsch S., Blanc F. E. C., von Bülow S., Gecht M., Bagola K., Hörner C., van Zandbergen G., Landry J., de Azevedo N. T. D., Mosalaganti S., Schwarz A., Covino R., Mühlebach M. D., Hummer G., Locker J. K., Beck M., In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 370, 203–208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He L., Tzarum N., Lin X., Shapero B., Sou C., Mann C. J., Stano A., Zhang L., Nagy K., Giang E., Law M., Wilson I. A., Zhu J., Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines. Sci. Adv. 6, eaaz6225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He L., Kumar S., Allen J. D., Huang D., Lin X., Mann C. J., Saye-Francisco K. L., Copps J., Sarkar A., Blizard G. S., Ozorowski G., Sok D., Crispin M., Ward A. B., Nemazee D., Burton D. R., Wilson I. A., Zhu J., HIV-1 vaccine design through minimizing envelope metastability. Sci. Adv. 4, aau6769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He L., de Val N., Morris C. D., Vora N., Thinnes T. C., Kong L., Azadnia P., Sok D., Zhou B., Burton D. R., Wilson I. A., Nemazee D., Ward A. B., Zhu J., Presenting native-like trimeric HIV-1 antigens with self-assembling nanoparticles. Nat. Commun. 7, 12041 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.L. He, A. Chaudhary, X. Lin, C. Sou, S. Kumar, T. Ngo, E. Kosviner, R. L. Stanfield, I. A. Wilson, J. Zhu, Single-component multilayered self-assembling nanoparticles presenting rationally designed glycoprotein trimers as Ebola virus vaccines. bioRxiv 2020.08.22.262634 [Preprint]. 22 August 2020. 10.1101/2020.08.22.262634. [DOI]
- 61.Zakeri B., Fierer J. O., Celik E., Chittock E. C., Schwarz-Linek U., Moy V. T., Howarth M., Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U.S.A. 109, E690–E697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X., Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Kirchdoerfer R. N., Wang N., Pallesen J., Wrapp D., Turner H. L., Cottrell C. A., Corbett K. S., Graham B. S., McLellan J. S., Ward A. B., Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 8, 15701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., Zhang X., Gao G. F., Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 8, 15092 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanders R. W., Derking R., Cupo A., Julien J.-P., Yasmeen A., de Val N., Kim H. J., Blattner C., de la Peña A. T., Korzun J., Golabek M., de los Reyes K., Ketas T. J., van Gils M. J., King C. R., Wilson I. A., Ward A. B., Klasse P. J., Moore J. P., A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLOS Pathog. 9, e1003618 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pugach P., Ozorowski G., Cupo A., Ringe R., Yasmeen A., de Val N., Derking R., Kim H. J., Korzun J., Golabek M., de los Reyes K., Ketas T. J., Julien J.-P., Burton D. R., Wilson I. A., Sanders R. W., Klasse P. J., Ward A. B., Moore J. P., A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J. Virol. 89, 3380–3395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.ter Meulen J., van den Brink E. N., Poon L. L. M., Marissen W. E., Leung C. S. W., Cox F., Cheung C. Y., Bakker A. Q., Bogaards J. A., van Deventer E., Preiser W., Doerr H. W., Chow V. T., de Kruif J., Peiris J. S. M., Goudsmit J., Human monoclonal antibody combination against SARS coronavirus: Synergy and coverage of escape mutants. PLOS Med. 3, e237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prabakaran P., Gan J., Feng Y., Zhu Z., Choudhry V., Xiao X., Ji X., Dimitrov D. S., Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 281, 15829–15836 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hwang W. C., Lin Y., Santelli E., Sui J., Jaroszewski L., Stec B., Farzan M., Marasco W. A., Liddington R. C., Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 8oR. J. Biol. Chem. 281, 34610–34616 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walls A. C., Xiong X., Park Y.-J., Tortorici M. A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., Zambon M., Rey F. A., Corti D., Veesler D., Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 176, 1026–1039.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bruun T. U. J., Andersson A. M. C., Draper S. J., Howarth M., Engineering a rugged nanoscaffold to enhance plug-and-display vaccination. ACS Nano 12, 8855–8866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chu L., Robinson D. K., Industrial choices for protein production by large-scale cell culture. Curr. Opin. Biotechnol. 12, 180–187 (2001). [DOI] [PubMed] [Google Scholar]
- 73.Durocher Y., Butler M., Expression systems for therapeutic glycoprotein production. Curr. Opin. Biotechnol. 20, 700–707 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Alexander J., Sidney J., Southwood S., Ruppert J., Oseroff C., Maewal A., Snoke K., Serra H. M., Kubo R. T., Sette A., Grey H. M., Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity 1, 751–761 (1994). [DOI] [PubMed] [Google Scholar]
- 75.Okba N. M. A., Widjaja I., van Dieren B., Aebischer A., van Amerongen G., de Waal L., Stittelaar K. J., Schipper D., Martina B., van den Brand J. M. A., Beer M., Bosch B.-J., Haagmans B. L., Particulate multivalent presentation of the receptor binding domain induces protective immune responses against MERS-CoV. Emerg. Microbes Infect. 9, 1080–1091 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma X., Zou F., Yu F., Li R., Yuan Y., Zhang Y., Zhang X., Deng J., Chen T., Song Z., Qiao Y., Zhan Y., Liu J., Zhang J., Zhang X., Peng Z., Li Y., Lin Y., Liang L., Wang G., Chen Y., Chen Q., Pan T., He X., Zhang H., Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity 53, 1315–1330.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walls A. C., Fiala B., Schäfer A., Wrenn S., Pham M. N., Murphy M., Tse L. V., Shehata L., O’Connor M. A., Chen C., Navarro M. J., Miranda M. C., Pettie D., Ravichandran R., Kraft J. C., Ogohara C., Palser A., Chalk S., Lee E.-C., Guerriero K., Kepl E., Chow C. M., Sydeman C., Hodge E. A., Brown B., Fuller J. T., Dinnon K. H. III, Gralinski L. E., Leist S. R., Gully K. L., Lewis T. B., Guttman M., Chu H. Y., Lee K. K., Fuller D. H., Baric R. S., Kellam P., Carter L., Pepper M., Sheahan T. P., Veesler D., King N. P., Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell 183, 1367–1382.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bale J. B., Gonen S., Liu Y., Sheffler W., Ellis D., Thomas C., Cascio D., Yeates T. O., Gonen T., King N. P., Baker D., Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 353, 389–394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pallesen J., Wang N., Corbett K. S., Wrapp D., Kirchdoerfer R. N., Turner H. L., Cottrell C. A., Becker M. M., Wang L., Shi W., Kong W.-P., Andres E. L., Kettenbach A. N., Denison M. R., Chappell J. D., Graham B. S., Ward A. B., McLellan J. S., Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U.S.A. 114, E7348–E7357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Henderson R., Edwards R. J., Mansouri K., Janowska K., Stalls V., Gobeil S. M. C., Kopp M., Li D., Parks R., Hsu A. L., Borgnia M. J., Haynes B. F., Acharya P., Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 27, 925–933 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kong L., He L., de Val N., Vora N., Morris C. D., Azadnia P., Sok D., Zhou B., Burton D. R., Ward A. B., Wilson I. A., Zhu J., Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat. Commun. 7, 12040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang B., Chao C. W., Tsybovsky Y., Abiona O. M., Hutchinson G. B., Moliva J. I., Olia A. S., Pegu A., Phung E., Stewart-Jones G. B. E., Verardi R., Wang L., Wang S., Werner A., Yang E. S., Yap C., Zhou T., Mascola J. R., Sullivan N. J., Graham B. S., Corbett K. S., Kwong P. D., A platform incorporating trimeric antigens into self-assembling nanoparticles reveals SARS-CoV-2-spike nanoparticles to elicit substantially higher neutralizing responses than spike alone. Sci. Rep. 10, 18149 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hotez P. J., Corry D. B., Strych U., Bottazzi M. E., COVID-19 vaccines: Neutralizing antibodies and the alum advantage. Nat. Rev. Immunol. 20, 399–400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crawford K. H. D., Eguia R., Dingens A. S., Loes A. N., Malone K. D., Wolf C. R., Chu H. Y., Tortorici M. A., Veesler D., Murphy M., Pettie D., King N. P., Balazs A. B., Bloom J. D., Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12, 513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rosenberg E. S., Billingsley J. M., Caliendo A. M., Boswell S. L., Sax P. E., Kalams S. A., Walker B. D., Vigorous HIV-1-specific CD4+T cell responses associated with control of viremia. Science 278, 1447–1450 (1997). [DOI] [PubMed] [Google Scholar]
- 86.Snell L. M., Osokine I., Yamada D. H., De la Fuente J. R., Elsaesser H. J., Brooks D. G., Overcoming CD4 Th1 cell fate restrictions to sustain antiviral CD8 T cells and control persistent virus infection. Cell Rep. 16, 3286–3296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu J. F., Yamane H., Paul W. E., Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28, 445–489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Laidlaw B. J., Craft J. E., Kaech S. M., The multifaceted role of CD4+T cells in CD8+T cell memory. Nat. Rev. Immunol. 16, 102–111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grifoni A., Weiskopf D., Ramirez S. I., Mateus J., Dan J. M., Moderbacher C. R., Rawlings S. A., Sutherland A., Premkumar L., Jadi R. S., Marrama D., de Silva A. M., Frazier A., Carlin A. F., Greenbaum J. A., Peters B., Krammer F., Smith D. M., Crotty S., Sette A., Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181, 1489–1501.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jeyanathan M., Afkhami S., Smaill F., Miller M. S., Lichty B. D., Xing Z., Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 20, 615–632 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peng Y., Mentzer A. J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., López-Camacho C., Slon-Campos J., Zhao Y., Stuart D. I., Paesen G. C., Grimes J. M., Antson A. A., Bayfield O. W., Hawkins D. E. D. P., Ker D.-S., Wang B., Turtle L., Subramaniam K., Thomson P., Zhang P., Dold C., Ratcliff J., Simmonds P., de Silva T., Sopp P., Wellington D., Rajapaksa U., Chen Y.-L., Salio M., Napolitani G., Paes W., Borrow P., Kessler B. M., Fry J. W., Schwabe N. F., Semple M. G., Baillie J. K., Moore S. C., Openshaw P. J. M., Ansari M. A., Dunachie S., Barnes E., Frater J., Kerr G., Goulder P., Lockett T., Levin R., Zhang Y., Jing R., Ho L.-P.; Oxford Immunology Network Covid- Response T cell Consortium; ISARICC Investigators, Cornall R. J., Conlon C. P., Klenerman P., Screaton G. R., Mongkolsapaya J., Michael A. M., Knight J. C., Ogg G., Dong T., Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19 patients. Nat. Immunol. 21, 1336–1345 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krawczyk C. M., Shen H., Pearce E. J., Functional plasticity in memory T helper cell responses. J. Immunol. 178, 4080–4088 (2007). [DOI] [PubMed] [Google Scholar]
- 93.Taubenberger J. K., Morens D. M., 1918 influenza: The mother of all pandemics. Emerg. Infect. Dis. 12, 15–22 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O’Callaghan K. P., Blatz A. M., Offit P. A., Developing a SARS-CoV-2 vaccine at warp speed. JAMA 324, 437–438 (2020). [DOI] [PubMed] [Google Scholar]
- 95.Poland G. A., Tortoises, hares, and vaccines: A cautionary note for SARS-CoV-2 vaccine development. Vaccine 38, 4219–4220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Corey L., Mascola J. R., Fauci A. S., Collins F. S., A strategic approach to COVID-19 vaccine R&D. Science 368, 948–950 (2020). [DOI] [PubMed] [Google Scholar]
- 97.Diamond M. S., Pierson T. C., The challenges of vaccine development against a new virus during a pandemic. Cell Host Microbe 27, 699–703 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.DeFrancesco L., Whither COVID-19 vaccines? Nat. Biotechnol. 38, 1132–1145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moore J. P., Klasse P. J., COVID-19 vaccines: “Warp Speed” needs mind melds, not warped minds. J. Virol. 94, e01083-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mochizuki K., Meng L., Mochizuki I., Tong Q., He S., Liu Y., Purushe J., Fung H., Zaidi M. R., Zhang Y., Reshef R., Blazar B. R., Yagita H., Mineishi S., Zhang Y., Programming of donor T cells using allogeneic δ-like ligand 4-positive dendritic cells to reduce GVHD in mice. Blood 127, 3270–3280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/12/eabf1591/DC1


