Abstract
Nanopore technology has been employed as a powerful tool for DNA sequencing and analysis. To extend this method to peptide sequencing, a necessary step is to profile individual amino acids (AAs) through their nanopore stochastic signals, which remains a great challenge because of the low signal-to-noise ratio and unpredictable conformational changes of AAs during their translocation through nanopores. We showed that the combination of an N-terminal derivatization strategy of AAs with nanopore technology could lead to effective in situ differentiation of AAs. Four different derivatization reactions have been tested with five selected AAs: Ala, Phe, Tyr, His, and Asp. Using an α-hemolysin nanopore, we demonstrated the feasibility of derivatization-assisted identification of AAs regardless of their charge composition and polarity. The method was further applied to discriminate each individual AA in testing data sets using their established nanopore profiles from training data sets. We envision that this proof-of-concept study will not only pave a way for identification of individual AAs but also lead to future applications in protein/peptide sequencing using the nanopore technology.
Keywords: nanopore, amino acid, identification, derivatization, proteomics
Graphical Abstract
Emerging resistive pulse nanopore sensing technology, ranging from biological protein to artificial solid-state nanometer-scale pores, makes it possible to detect, analyze, manipulate, and characterize a variety of analytes at the single-molecule level.1,2 In general, nanopore sensing operates on a basic structure with a thin membrane containing a single nanopore that separates an ionic solution into two compartments. A transmembrane bias is applied to capture and transport analytes from one side of the membrane to the other through the nanopore.3–6 The entry of a molecule into a nanopore could cause a reduction in the latter’s ionic conductance. The resulting ionic current blockade depth and the residence time have been shown to provide detailed information on the size, adsorbed charge, and other properties of the molecule.7 Consequently, nanopore-based nucleic acid sequencing technology has been successfully commercialized with single base resolution, label-free detection, and long-read capability.8–13 However, to extend this method to peptide sequencing, a large hurdle is to differentiate individual amino acids (AAs) through their nanopore stochastic signals because of the low signal-to-noise ratio and unpredictable conformational changes of AAs during their translocation through nanopores.
To mitigate these challenges, different measurement methods on various nanopores have been developed in an attempt to achieve higher sensitivity. In some pioneer studies, the identification of some specific AAs or short peptides by monitoring the ion translocation in perpendicular nano-channel,14 in recognition tunneling,15 and in sputtered sub-nanometer pores16 made on solid-state materials were first demonstrated. Later on, identification of peptides with different lengths by one AA was achieved with wild-type aerolysin nanopore,17 α-hemolysin (α-HL) nanopore,18 and viral DNA packaging motor,19 whereas recognition of proteins and peptides with minor sequence differences was accomplished using FraC nanopores.20,21
Despite the solid foundation laid by these investigations, identification of AAs by nanopore technology is still limited by the lack of characteristics in the interaction with nanopores because of the much smaller size and the fast translocation rate of the AAs. To utilize the robust structure of biological nanopores, alternative methods such as decreasing the diameter of the pore lumen22 or increasing the volume of AAs23 by efficient and versatile chemical modifications were used to achieve more AA-pore interactions during translocation. A simulation study has shown that nanoporous single-layer MoS2 can detect individual AAs in a polypeptide chain, but the results have not been experimentally proven yet.8 An elegant design by Bayley and others incorporated the usage of metal–organic complexes into the biological nanopore, which could effectively differentiate AA enantiomers.22,24 Recently, aerolysin with a narrow constriction of ~1.0 nm was favored as a biological nanopore for recognizing AAs because of its highly charged sensing interface.25 Lu et al. found that the cyclization of cysteine and homocysteine into thiazolidines could enhance the signal differentiation through an aerolysin nanopore.23 Ying et al. first reported the detection of a single cysteine molecule using the interaction between the aerolysin sensing interface and the analyte.26 An encouraging study by Oukhaled et al. reported identification of all proteinogenic AAs using an aerolysin nanopore with the help of a short peptide carrier. However, translation of this method to practical protein sequencing seems overwhelmingly challenging.27
We envision that an efficient universal conjugation strategy of AAs could augment the interaction of AA derivatives with the pore lumen surface and may be readily applicable toward sequencing. Considering the size mismatch between AAs and the α-HL nanopore, N-terminal conjugation can increase the aspect ratio of AAs, leading to a prolonged interaction with the nanopore. Meanwhile, most positively charged AA or peptides can be neutralized by N-terminal conjugation to form more negatively charged final conjugates. Moreover, for future applications in nanopore protein sequencing, the quantitative nature of the N-terminal conjugation method with similar reactivity toward different AAs and peptides is desired to avoid introducing interfering impurities and any further purification. Among various N-terminal derivatization methods of AAs,28–30 in situ ortho-phthalaldehyde (OPA) and phenyl isothiocyanate (PITC) derivatizations are two of the most widely studied because of their high reaction rate and efficiency.31,32 As shown in Figure 1a,b, four modifiers including OPA, 2,3-naphthalenedicarboxaldehyde (NDA), PITC, and 2-naphthylisothiocyanate (NITC) were employed in the N-terminal derivatization of five representative AAs in this study. The ionic current blockade events of each AA-derivative were characterized using α-HL nanopores, and each derivatization method was evaluated for its ability to distinguish AAs via nanopores. Our results show that NDA and NITC derivatizations were able to facilitate the discrimination of five AAs individually. This proof-of-concept study revealed the possibility of individual AA differentiation using α-HL nanopore technology, which paves the way for future investigations in nanopore-based peptide sequencing and analysis.
Figure 1.
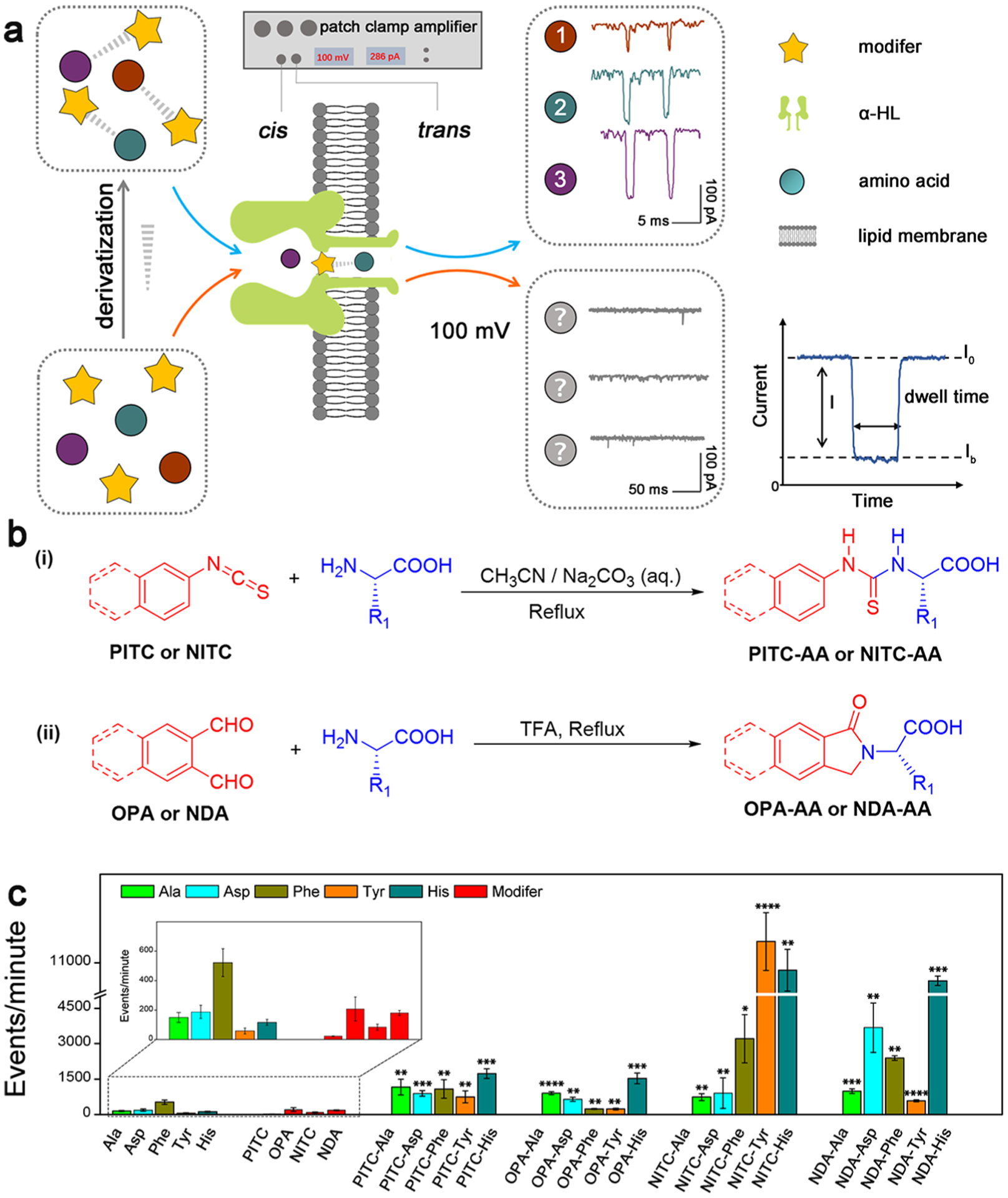
(a) Schematic illustration of the identification strategy of single AA derivatives through α-HL nanopore. The characteristic signals 1, 2, and 3 are from NITC-Ala, NITC-His, and NITC-Tyr, respectively. (b) Synthetic routes of different AA derivatives. Route i is for PITC/NITC derivatives; route ii is for OPA/NDA derivatives. (c) Translocation events frequency of raw materials and various derivatives through α-HL nanopores under the same experimental condition. Statistical significance is shown between derivatives and corresponding single AA (*: 0.01 < p <0.05; **: 0.001 < p < 0.01; ***: 0.0001 < p < 0.001; and ****: p < 0.0001).
MATERIALS AND METHODS
Materials.
Derivative reagents OPA, PITC, NITC, and all AAs were purchased from Sigma-Aldrich and used without further purification. The derivative reagent NDA was synthesized according to the reference method.33 The KCl working solution was prepared using deionized water from a Milli-Q water purification system (resistivity of 18.2 MΩ/cm, 25 °C, Millipore Corporation) and was filtered through 0.02 μm filter before use. α-HL from Staphylococcus aureus (lyophilized powder, Protein ~60% by Lowry, ≥10,000 units/mg protein) was purchased from Sigma-Alrich.
General Procedure for Preparing PITC/NITC Derivatives.
PITC/NITC derivatives were synthesized according to the route i, as shown in Figure 1b. AA (1.2 equiv) was dissolved in the mixture of acetonitrile and 0.1 M Na2CO3 solution, followed by a dropwise addition of PITC/NITC (1.0 equiv). The reaction mixture was stirred and refluxed overnight. The solvent was then removed, and the precipitate was collected as the crude product, which was washed with 1 M HCl and methanol and dried to afford the target product.
General Procedure for Preparing OPA/NDA Derivatives.
OPA/NDA derivatives were synthesized according to the route ii as shown in Figure 1b. AA (1.0 equiv) and OPA/NDA (1.2 equiv) were mixed in acetonitrile together with the catalyst trifluoroacetic acid (1.4 equiv). The reaction mixture was then refluxed for about 3 h before it was cooled down to room temperature. The yellow precipitate was collected and washed with acetonitrile to afford corresponding product.
Characterization of AA Derivatives.
The 1H and 13C nuclear magnetic resonance (NMR) spectra were recorded at 298 K in deuterated solvents using Bruker AVANCE 400 MHz spectrometer. Data is presented as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet), coupling constants in Hertz (Hz), and integration. High-resolution-mass-spectra of AA derivatives were recorded on Thermo Velos Pro Orbitrap liquid chromatography–mass spectrometry (MS). High performance liquid chromatography (HPLC) of AA derivatives were recorded on an Agilent 1100 HPLC equipped with a ZORBAX SB-C18 column (see Supporting Information).
Nanopore Fabrication and Low-Noise Electrical Recording.
All electrophysiology experiments were performed on the Planar Lipid Bilayer Workstation (Warner Instruments) at room temperature (~23 °C). Fabrication of α-HL nanopore devices follows a traditional method previously reported. Briefly, an orifice (200 μm in diameter) punctured on a 25 μm thick Delrin wall that separates the cis (grounded) and the trans chambers of the flow cell was precoated with 1:10 hexadecane/pentane (Sigma-Aldrich). Then, both chambers were filled with 1 mL of 3 M KCl solution buffered in 10 mM Tris-HCl (pH 8). To form a lipid bilayer membrane in the orifice, 20 μL (10 mg/mL) of 1,2 diphytanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids) dissolved in pentane (Sigma-Aldrich) was added to the cis side of chambers to allow self-assembly. Following this, electrical potential was applied to the trans side using Ag/AgCl electrodes and slowly ramped up to examine the stability of the membrane at ±200 mV. The membrane capacitance was maintained between 160 and 170 pF with various voltage bias values throughout each experiment.
To insert a single nanopore channel into the lipid bilayer, trans voltage was changed to 100 mV and a small amount (~0.05 μg) of α-HL protein (Sigma-Aldrich) was added from a monomeric stock solution made in 3 M KCl to the cis compartment. To ensure consistency of testing conditions, the direction of each α-HL nanopore was examined by comparing the value of the channel current under positive and negative voltages after its insertion into the lipid bilayer. A properly inserted α-HL pore exhibits larger ionic current under a positive trans voltage than it is under a negative voltage (Figure S1a).34 After a stable α-HL protein nanopore was inserted and confirmed by an open pore current, an analyte was added to the cis chamber at a bulk concentration of 200 μM from a 10 mM stock solution made in dimethyl sulfoxide.
Data Collection and Analysis.
Ionic current recordings were collected using a patch clamp amplifier (Warner Instruments) with a built-in high-pass Bessel filter (cutoff: 5 kHz) at a holding potential of 100 mV. After sample addition to the cis chamber, magnetic stirring was used to disperse the sample before a characteristic signal was recorded. The magnetic stirring was performed at the bottom of the cis side to avoid any impact on the stability of the membrane and the nanopore (Figure S1b,c). Each sample was measured in three replicates with a 30 min total duration. The ionic current was sampled at 100 kHz using a Digidata 1440A analog-to-digital converter (Molecular Devices) and processed with pClamp10 software (Molecular Devices).
A fresh α-HL protein nanopore was used for each replicate. The raw data was analyzed using an in-house MATLAB based algorithm to find the current blockade and the dwell time of each eligible event, which are two commonly used properties for discriminating different molecules when they translocate nanopores. The current blockade that represents the capture of single molecules and their translocation through the nanopore is defined as I/I0 (I = I0 − Ib, Ib: the average current measured with the analytes inside the pore; I0: the average baseline current in absence of analytes). Dwell time (i.e., duration) represents the effective interaction time between nanopores and single molecule analyst (Figure 1a). Results processed by the MATLAB algorithm were confirmed by manual inspection.
Statistical Analysis.
To profile each analyte, current blockade was plotted against dwell time using Python. The python modules used for scatter plots and contour plots were Matplotlib and Seaborn’s bivariate kernel density estimator. Contours were created according to the density of the data points in the logarithmic duration fractional blockade space, based on a kernel density function, whereby every data point contributes a two-dimensional Gaussian to the cumulative contour, which was then normalized in z such that the entire volume of all of the contributing data integrated to one.16
For discriminant analysis, multiple parallel experiments were performed for each NITC-AA to collect >1000 events as the training data sets, which were used to calculate the similarity relationships with testing data sets of each NITC-AAs. All analyses were performed using Mahalanobis distance matrices and histogram binning methods with an in-house MATLAB-based program. The Mahalanobis distance is a measure of the distance between a point and a distribution. In this case, Mahalanobis distances for points in the testing data sets were calculated against each of the training data sets, and a smaller distance indicates a greater similarity. The same training data sets and testing data sets were used for the histogram binning technique. The training data sets were binned in two dimensions-residual current and dwell time measurements-with 50 bins in each dimension. Afterward, the centroid (i.e., average residual current value and average dwell time of the entire cluster) was binned into the 2D histograms formed from the training data sets. An index was generated from the number of data points within the specific bin in the training data set that would house the centroid of the testing data sets. A larger index represents greater similarity between the clusters. The binning was first conducted with the auto-binning capability in MATLAB and then with 25, 50, 100, and 250 bins. After generating similar data tables for the different binning schemes and manually inspecting the binning schemes of the training set, it was found that the auto-binning capabilities did not generate consistent or sufficient bin numbers. For 25 bins, there was an inflated number of measurements in each bin and reduced the sensitivity of the technique. For 250 bins, there was a deflated number of measurements in each bin and led to a greater number of anomalous measurements. Finally, we chose 50 bins to perform the comparison.
Molecular Modeling.
Geometry optimization of AAs and their derivatives was calculated using Q-chem 4.3.35 Unrestricted B3LYP function was employed to describe our system, making use of the 6–31++G basis sets for C, H, O, N, and S atoms. Solvent effects (KCl aqueous solution with a dielectric constant of 55) were included using the PCM implicit solvation model. The VDW radius was calculated by Multiwfn program,36 in which VDW surface is defined by the lengths of the three sides of the cube.
Data Availability.
The authors declare that the data supporting the findings of this study is available within the article and its Supporting Information files or from the corresponding authors upon reasonable request.
RESULTS
AA Derivatization and Characterization.
Five characteristic AAs with different size, charge, polarity, and hydrophobicity were selected in our initial study: alanine (Ala), phenylalanine (Phe), tyrosine (Tyr), aspartic acid (Asp), and histidine (His). Each unmodified AA was first analyzed using an α-HL nanopore for extended recording time, and no obvious current blockade signal was observed at applied potential bias (Figure S2a). Similarly, no characteristic signal was recorded for the four derivatization reagents selected in this study (i.e., OPA, NDA, PITC, and NITC) through the nanopore (Figure S2b). The low frequency of current blockade events for the selected AAs and modifiers further demonstrates their weak interactions with the lumen of the α-HL nanopore (Figure 1c), which can be attributed to the smaller van der Waals radii of the AAs (~0.3–0.4 nm for all AA based on Spartan calculation, Figure S3) compared to the dimension of the constriction region of the α-HL nanopore (1.4 nm).37
To increase effective interaction between the nanopore and analytes, the N-terminals of the aforementioned AAs were readily modified with PITC, OPA, NITC, and NDA (Figure 1b and Table S1). The structure and the purity (>99%) of all the derivatization products were confirmed by HPLC, MS, and NMR analyses (Figures S7–S81). Purified AA derivatives were employed in our nanopore analysis. As shown in Figure 1c, all four series of AA derivatives, especially the NITC and NDA derivatives, exhibited significant increase in the translocation event frequency through α-HL nanopores under the same conditions in comparison with the unmodified AAs, revealing improved interaction between AAs and the α-HL pore after derivatization.
Translocation Profiles of AA Derivatives.
To assess the ability of PITC derivatization for distinguishing the five different AAs, a contour plot was generated for each of the five PITC-derivatives from its average current blockade and dwell time values to profile its translocation behavior (Figure 2a). In combination with the frequency distribution of current blockade and dwell time (Figure 2c), four out of the five different AAs were distinguishable from each other by their current blockade profiles, including Ala, Asp, Tyr, and His, even though they showed similar dwell time distributions. Unfortunately, the PITC-Phe derivative had significant overlaps with other AA derivatives in both current blockade and dwell time distributions, making it difficult to distinguish Phe from other AAs, especially Tyr and His. Similarly, OPA derivatization was tested. Although the formation of a five-membered ring between the AA and OPA should increase the rigidity of the AA derivatives (Figure S4) and may improve their interactions with the nanopore during translocation, the distinguishability across all AAs was reduced except for the OPA-Phe (Figure 2b,d).
Figure 2.
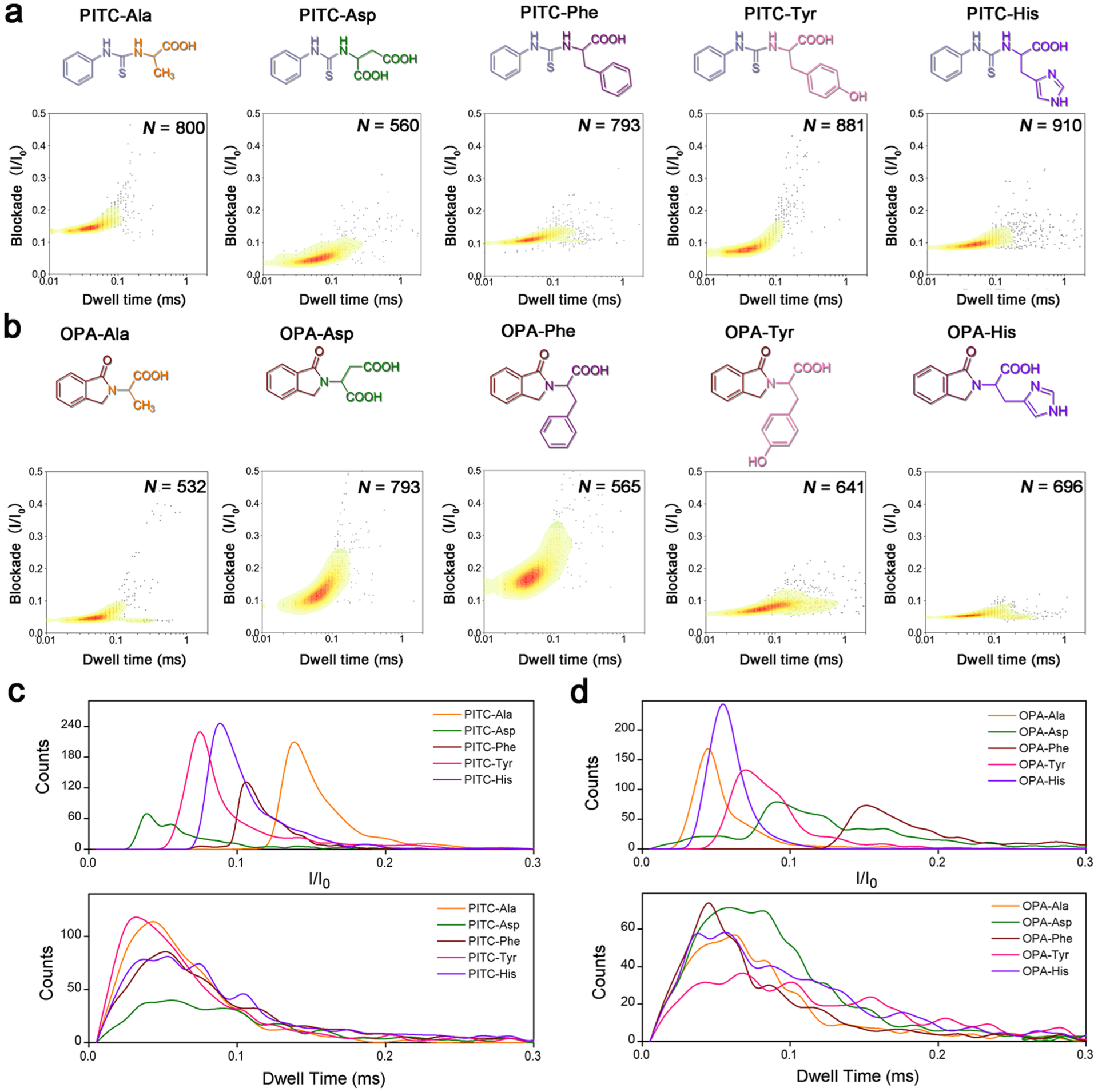
(a,b) Molecular structures and corresponding contour plots depicting the blockade (I/I0) vs dwell time distribution at the same applied potential for different AA derivatives: (a) PITC-derivatives, (b) OPA-derivatives. N value indicates the total number of stochastic signal events included in each analysis. (c,d) Translocation event frequency distribution of blockade (I/I0) and dwell time for (c) PITC and (d) OPA derivatives.
We next tested the NITC derivatization, which adds one more benzene ring to the structures. A clear differentiation among the five AAs could be observed through current blockade and dwell time analysis (Figure 3a). In addition, stochastic event frequency was also increased significantly by NITC modification compared with PITC and OPA modifications (Figure 1c). Despite the similar dwell time centered from 0.05 to 0.1 ms for NITC-Ala, NITC-Asp, and NITC-Phe, their current blockade (I/I0) values concentrated at 0.06, 0.12, and 0.15, respectively, with narrow distribution and minimal overlaps provided excellent distinguishability of these three AAs. NITC-Tyr and NITC-His derivatives exhibited larger current blockade values concentrated around 0.45 and 0.28, respectively, with wider distribution. Together with their longer dwell times (>0.1 ms), these two NITC-derivatives can be effectively distinguished from NITC-Ala, NITC-Asp, and NITC-Phe, as well as from each other (Figure 3c). In addition, the reproducibility of the contour profiles of NITC-derivatives (Figure S5) revealed a much more stable and uniform translocation behaviors, which is essential for future practical applications. With minimal data processing, NITC derivatization of each AAs could be readily distinguished.
Figure 3.
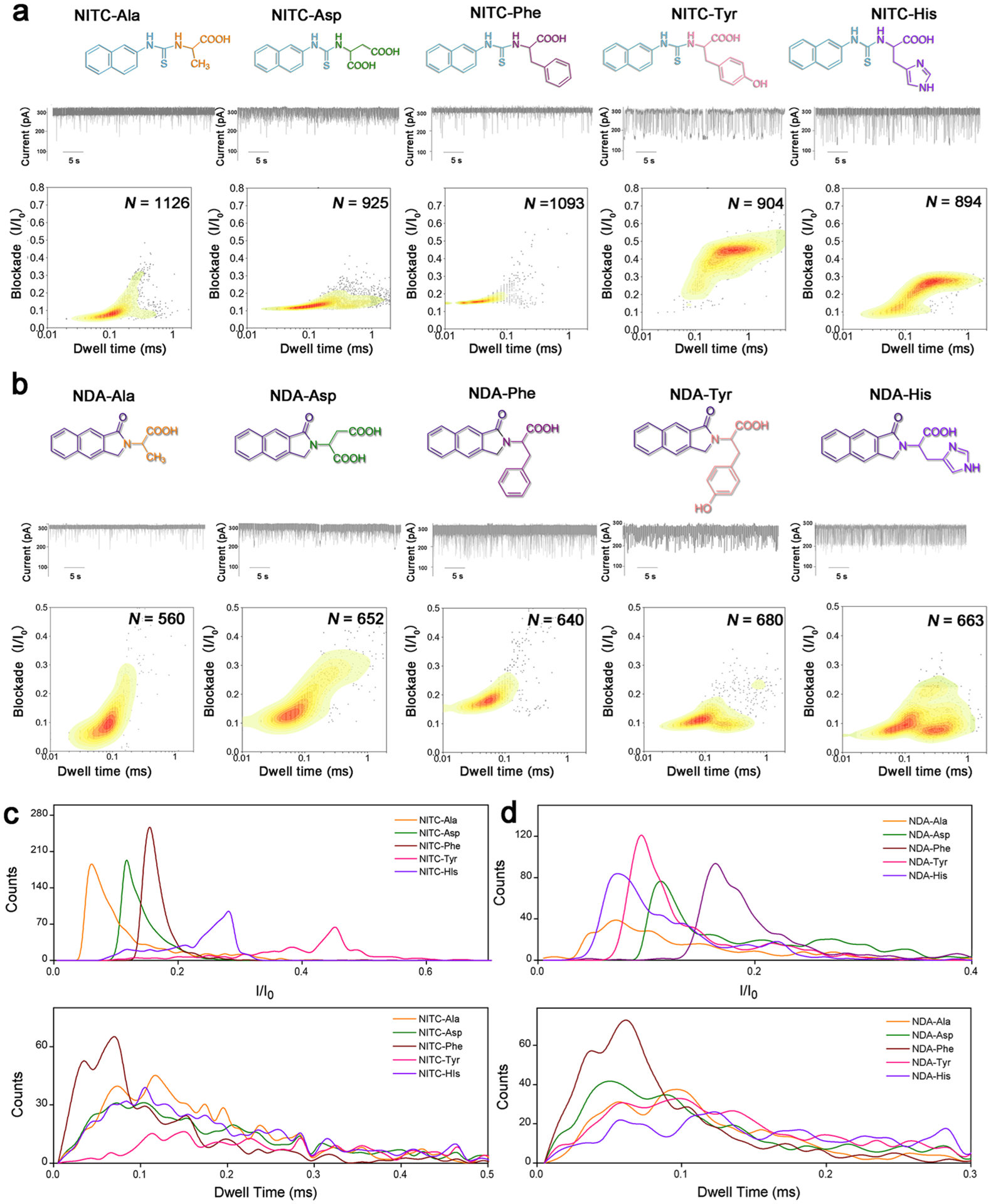
(a,b) From top to bottom: molecular structure, representative current trace of translocations, and corresponding contour plots of different (a) NITC-derivatives and (b) NDA-derivatives. N value indicates the total number of stochastic signal events included in each analysis. (c,d) Translocation event frequency distribution of blockade (I/I0) and dwell time for (c) NITC-derivatives and (d) NDA-derivatives.
The current blockade and dwell time profiles of NDA-derivatives were similarly measured (Figure 3b), which show wider distributions of events in general and significant higher signal overlaps than NITC-derivatives (Figure 3d). This could be attributed to the much more rigid benzoisoindolone structure (Figure S6) resulting in smaller structural differences among different NDA-AAs. Interestingly, multiple peak areas were observed in the contour plots of NDA-Tyr and NDA-His, possibly due to multiple spatial orientations while passing through the nanopore. This phenomenon will be further investigated in our future studies.
Derivatization-Assisted Identification of AAs.
Finally, to assess the discriminatory power of our method for the differentiation of AAs, three different AAs (Ala, His, and Tyr) derivatized with NITC were added to the cis compartment of the nanopore flow chamber sequentially at the same final concentration (200 μM), while translocation signals were recorded simultaneously. As shown in Figure 4, the corresponding fingerprint of each compound can be clearly differentiated in the contour map of the mixtures. Despite the interaction and competitive translocation between different derivatives, each AA derivative exhibited reproducible translocation behavior (current blockade vs dwell time distributions) comparing to its fingerprint obtained separately in our previous experiments (Figure 3a). However, fingerprints of NITC-Ala, NITC-Asp, and NITC-Phe would easily merge into each other on the qualitative contour plot. To efficiently identify AAs, quantitative biostatistics analysis with excellent inter-pore reproducibility is needed to recognize AA derivatives from unknown samples using their established translocation profiles. Discriminant analyses of five NITC-AAs were performed using both Mahalanobis distance matrices and histogram binning method. A combination of multiple replicates of signal event clusters was chosen as the training data sets based on their experimental reliability and reproducibility. The Mahalanobis distances were calculated for the testing data sets against each of the training data sets. A smaller distance indicates a greater similarity (Table 1). The same training data sets and testing data sets were used for the histogram binning technique, and 50 bins were used for the calculation. A larger number indicates a greater similarity (Table 2). For each sample, at least 500 events were analyzed.
Figure 4.
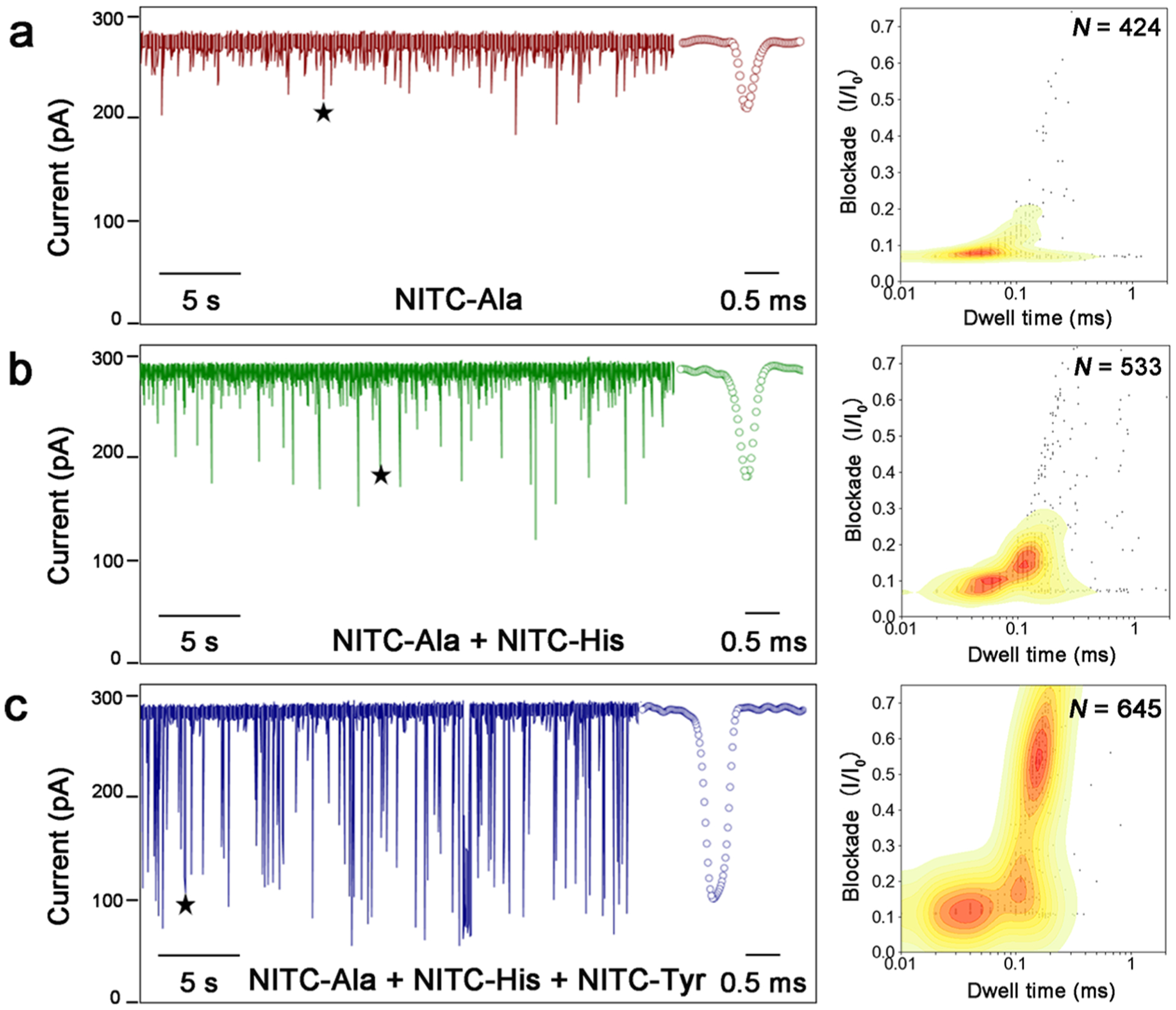
Discrimination of each AA in a mixture sample. After a single α-HL nanopore was obtained in the working buffer (3 M KCl, 10 mM Tris-HCl, pH 8.0), 200 μM NITC-Ala, 200 μM NITC-His, and 200 μM NITC-Tyr were sequentially added to the cis compartment with 5 min intervals. (a–c) Representative current trace of translocations (left) obtained and contour plots depicting current blockade vs dwell time distributions of all translocation events (right) for (a) NITC-Ala, (b) NITC-Ala and NITC-His, and (c) NITC-Ala, NITC-His, NITC-Tyr. The stars denote the typical spike signals shown in the inset.
Table 1.
Discriminant Analysis of Five NITC-AAs Using Mahalanobis Distance Matrices
| training data set | |||||
|---|---|---|---|---|---|
| testing data set | NITC-Ala | NITC-Asp | NITC-Phe | NITC-His | NITC-Tyr |
| NITC-Ala | 0.25 | 1.24 | 2.55 | 1.63 | 2.94 |
| NITC-Asp | 0.92 | 0.17 | 4.41 | 1.17 | 2.50 |
| NITC-Phe | 3.95 | 4.07 | 1.35 | 0.68 | 0.77 |
| NITC-His | 2.34 | 2.05 | 3.97 | 0.37 | 1.58 |
| NITC-Tyr | 5.46 | 5.62 | 11.54 | 1.33 | 0.10 |
Table 2.
Discriminant Analysis of Five NITC-AAs Using the Histogram Binning Method
| training data set | |||||
|---|---|---|---|---|---|
| testing data set | NITC-Ala | NITC-Asp | NITC-Phe | NITC-His | NITC-Tyr |
| NITC-Ala | 2106 | 55 | 56 | 178 | 4 |
| NITC-Asp | 383 | 546 | 3 | 82 | 9 |
| NITC-Phe | 71 | 7 | 122 | 72 | 23 |
| NITC-His | 109 | 65 | 4 | 117 | 16 |
| NITC-Tyr | 0 | 1 | 0 | 27 | 52 |
DISCUSSION
Revealing the primary sequence of a protein or peptide is essential to its identification and function. Traditionally, the most common method for protein sequencing is MS, a technique that involves fractionating the protein into many smaller peptides and then obtaining the mass-to-charge ratio of each new peptide from the mass spectrometer. However, sequencing is sometimes impossible with this technology because of low abundance of precursor peptide and poor fractionation efficiency. The sensitivity of MS also varies among different analytes and between instrument models.38,39 Matrix-assisted laser desorption/ionization-time of flight MS suffers from significant reduced sensitivity on samples with high concentration of salts.40 Although many high-resolution MS have been developed recently and the combination with HPLC may improve the sensitivity, it is still a laborious process to profile the complete sequence of a unknown protein. In addition, MS instruments are too costly and complex to be developed into portable devices.41 A portable, accurate, and easy-to-use protein sequencer will engender future implications in personalized medicine, especially in self-testing, resource-limited settings, disease outbreaks, and novel theranostics concepts. Resistive pulse sensing using biological nanopores shows atomic precision because of extremely small dimension of their sensing regions in the pore lumen (1–4 nm) and has been demonstrated with excellent sensitivity and accuracy in DNA and RNA sequencing.10 Recently, the focus of efforts has been directed toward AA identification and sequencing of proteins and peptides, which holds great promise for the advancement of proteomics.42 Comparing to MS, the nanopore technology has several advantages: (1) long-reads that are not limited by precursor peptide fractionation;10 (2) high tolerance to contaminations such as salts and polymers; and (3) simplicity and cost efficiency.
Pioneering studies have demonstrated that a nanopore is able to detect certain single AA26 and differentiate certain peptides with one AA difference in length.41 However, sequencing of random peptides with single AA resolution is still extremely challenging to realize, likely due to the zwitterionic form, the non-uniform translocation rate, and low signal-to-noise ratio and low distinguishability of most AAs caused by the mismatch between the diameter of AAs (0.6–0.8 nm) and the nanopore (1.4–3 nm).43–45 Inspired by the Edman degradation,28 we developed a new nanopore method to identify a single AA using its N-terminal derivative as a surrogate. Four derivative regents were employed in this study to modify five AAs. Our results indicate that the derivatization afforded a fingerprint on stochastic signals when each AA translocating the α-HL nanopore due to the increased interaction. Importantly, these derivatization methods were efficient and reproducible under simple one-pot mild reaction conditions, affording a reliable strategy for the formation of a structurally diverse array of AA derivatives.
We assessed four series of AA derivatives for their translocation behaviors through the α-HL nanopore, including PITC-derivatives, OPA-derivatives, NITC-derivatives, and NDA-derivatives of five different AAs (Ala, Asp, Phe, Tyr, and His) with various polarity, charge, and size, representing different types of 20 AAs. Significantly increased translocation event frequency of derivatives comparing to unmodified AAs indicates enhanced interaction between the nanopore lumen and analytes. Detailed investigation on the translocation signals of all derivatives by estimating the distribution of current blockade and dwell time revealed best distinguishability among Ala, Asp, Phe, Tyr, and His by the NITC-derivatization. As confirmed by the molecular structure modeling for each AA and AA derivative (Figures S3, S4 and S6), some derivatives with larger cubic volume, such as NITC-His and NITC-Tyr, produced deep current blockades even larger than that of ssDNAs. However, exceptions such as NITC-Phe indicate that other interactions (i.e., charge, polarity, intra-, and intermolecular interactions etc.) also play a role during the translocation. Although it is difficult to quantitatively determine the impact of these interactions on the event frequency and the changing trend of fingerprint of each derivative at the moment, further investigations using suitable mutant hemolysin nanopores and other types of biological nanopores may provide more insights.
While this proof-of-concept study clearly demonstrates that derivatization is a feasible way to identify single AAs with biological nanopores, we do recognize the overall complexity of protein sequencing using nanopores. Although the technology has been proven successful for nucleic acid sequencing, it is considerably more challenging to differentiate 20 AAs than 4 nucleotides. In addition to sensitivity enhancement via different modifications to nanopores and analytes through biochemistry methods, more advanced data analysis technology (e.g., machine learning, pattern recognition, etc.) is in urgent need to improve resolution through novel characteristics other than the traditional blockade and dwell time from the stochastic signals. Similar to the MS technology, the data readout from nanopore also needs sophisticated bioinformatics database and algorithm to be interpreted into sequences and protein identifications. Therefore, the success of nanopore-based protein sequencing no doubt requires multi-disciplinary efforts.
Inspired by the protein ladder sequencing technique46 and the identification of single AA differences in length of peptides,41 we envision a potential “sequencing-by-hydrolysis” method, in which a nanopore will be used to identify the N-terminal AA of each peptide fragment in a peptide ladder generated from a peptide analyte, and then, bioinformatics methods will be applied to reconstitute its full-length sequence.
CONCLUSIONS
We demonstrated that N-terminus derivatization is an effective way to differentiate individual AAs using α-HL nanopore technology. Among four derivatization reagents applied in our work, NITC-derivatization of five typical AAs afforded significantly enhanced distinguishability based on the translocation signals. While we are working on developing more effective N-terminus modification strategies and optimizing the modifier’s structure, more advanced data analysis technology is in urgent need to improve resolution through novel characteristics other than the traditional blockade and dwell time from the stochastic signals. Finally, further simulation work is undergoing to better model the conformational changes of each derivative inside the lumen of the α-HL and to understand the complexity of the interactions between each AA derivative and the lumen of the α-HL protein.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to the invaluable guidance of Dr. Edsel A. Pena in statistical analysis. C.L. acknowledges supports from the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under award number K22AI136686, and the South Carolina IDeA Networks of Biomedical Research Excellence Developmental Research Project funded by the National Institute of General Medical Sciences (NIGMS) of the NIH under award number P20RR016461. Q.W. acknowledges partial support from the Big Data Health Science Center (BDHSC) initiative of the University of South Carolina. D.M. acknowledges the support (201806310084) from the State Scholarship Fund of the China Scholarship Council.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssensors.0c00345.
Change of open pore current value when changing voltage direction after one α-HL nanopore inserted into the lipid membrane and the current trace during the stirring process; representative current trace of the translocation of the raw materials through α-HL nanopores; the space-filling structures of different AAs and AA derivatives calculated using the Q-chem 4.3 software package; reproducibility study of NITC-derivatives; table including structures of different AA derivatives; and general procedure and characterizations of different derivatives (PDF)
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acssensors.0c00345
Contributor Information
Xiaojun Wei, Biomedical Engineering Program and Department of Chemical Engineering, University of South Carolina, Columbia, South Carolina 29208, United States.
Dumei Ma, Department of Chemistry and Biochemistry, University of South Carolina, Columbia, South Carolina 29208, United States;; Department of Chemical and Biochemical Engineering, College of Chemistry and Chemical Engineering, Xiamen University, Xiamen 361005, Fujian, China
Zehui Zhang, Biomedical Engineering Program, University of South Carolina, Columbia, South Carolina 29208, United States.
Leon Y. Wang, Department of Chemical Engineering University of South Carolina, Columbia, South Carolina 29208, United States
Jonathan L. Gray, Biomedical Engineering Program, University of South Carolina, Columbia, South Carolina 29208, United States
Libo Zhang, Department of Chemistry and Biochemistry, University of South Carolina, Columbia, South Carolina 29208, United States.
Tianyu Zhu, Department of Chemistry and Biochemistry, University of South Carolina, Columbia, South Carolina 29208, United States.
Xiaoqin Wang, Department of Chemical Engineering, University of South Carolina, Columbia, South Carolina 29208, United States.
Brian J. Lenhart, Department of Chemical Engineering, University of South Carolina, Columbia, South Carolina 29208, United States
Yingwu Yin, Department of Chemical and Biochemical Engineering, College of Chemistry and Chemical Engineering, Xiamen University, Xiamen 361005, Fujian, China.
Qian Wang, Department of Chemistry and Biochemistry, University of South Carolina, Columbia, South Carolina 29208, United States.
Chang Liu, Biomedical Engineering Program and Department of Chemical Engineering, University of South Carolina, Columbia, South Carolina 29208, United States.
REFERENCES
- (1).Robertson JWF; Reiner JE The Utility of Nanopore Technology for Protein and Peptide Sensing. Proteomics 2018, 18, 1800026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Lee K; Park K-B; Kim H-J; Yu J-S; Chae H; Kim H-M; Kim K-B Recent Progress in Solid-State Nanopores. Adv. Mater 2018, 30, 1704680. [DOI] [PubMed] [Google Scholar]
- (3).Wilson J; Sloman L; He Z; Aksimentiev A Graphene Nanopores for Protein Sequencing. Adv. Funct. Mater 2016, 26, 4830–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kasianowicz JJ; Robertson JWF; Chan ER; Reiner JE; Stanford VM Nanoscopic Porous Sensors. Annu. Rev. Anal. Chem 2008, 1, 737–766. [DOI] [PubMed] [Google Scholar]
- (5).Howorka S; Siwy Z Nanopore analytics: sensing of single molecules. Chem. Soc. Rev 2009, 38, 2360–2384. [DOI] [PubMed] [Google Scholar]
- (6).Wanunu M Nanopores: A journey towards DNA sequencing. Phys. Life Rev 2012, 9, 125–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Asandei A; Chinappi M; Lee J.-k.; Seo CH; Mereuta L; Park Y; Luchian T Placement of oppositely charged aminoacids at a polypeptide termini determines the voltage-controlled braking of polymer transport through nanometer-scale pores. Sci. Rep 2015, 5, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Farimani AB; Heiranian M; Aluru NR Identification of amino acids with sensitive nanoporous MoS2: towards machine learning-based prediction. npj 2D Mater. Appl 2018, 2, 14. [Google Scholar]
- (9).Callaway E Flu virus finally sequenced in its native form. Nature 2018, 556, 420. [DOI] [PubMed] [Google Scholar]
- (10).Howorka S; Cheley S; Bayley H Sequence-specific detection of individual DNA strands using engineered nanopores. Nat. Biotechnol 2001, 19, 636–639. [DOI] [PubMed] [Google Scholar]
- (11).Garalde DR; Snell EA; Jachimowicz D; Sipos B; Lloyd JH; Bruce M; Pantic N; Admassu T; James P; Warland A; Jordan M; Ciccone J; Serra S; Keenan J; Martin S; McNeill L; Wallace EJ; Jayasinghe L; Wright C; Blasco J; Young S; Brocklebank D; Juul S; Clarke J; Heron AJ; Turner DJ Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 2018, 15, 201. [DOI] [PubMed] [Google Scholar]
- (12).Lu H; Giordano F; Ning Z Oxford Nanopore MinION Sequencing and Genome Assembly. Genomics, Proteomics Bioinf. 2016, 14, 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Deamer D; Akeson M; Branton D Three decades of nanopore sequencing. Nat. Biotechnol 2016, 34, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Boynton P; Di Ventra M Sequencing proteins with transverse ionic transport in nanochannels. Sci. Rep 2016, 6. DOI: 10.1038/srep25232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhao Y; Ashcroft B; Zhang P; Liu H; Sen S; Song W; Im J; Gyarfas B; Manna S; Biswas S; Borges C; Lindsay S Single-molecule spectroscopy of amino acids and peptides by recognition tunnelling. Nat. Nanotechnol 2014, 9, 466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Kennedy E; Dong Z; Tennant C; Timp G Reading the primary structure of a protein with 0.07 nm3 resolution using a subnanometre-diameter pore. Nat. Nanotechnol 2016, 11, 968–976. [DOI] [PubMed] [Google Scholar]
- (17).Piguet F; Ouldali H; Pastoriza-Gallego M; Manivet P; Pelta J; Oukhaled A Identification of single amino acid differences in uniformly charged homopolymeric peptides with aerolysin nanopore. Nat. Commun 2018, 9. DOI: 10.1038/s41467-018-03418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Chavis AE; Brady KT; Hatmaker GA; Angevine CE; Kothalawala N; Dass A; Robertson JWF; Reiner JE Single Molecule Nanopore Spectrometry for Peptide Detection. ACS Sens. 2017, 2, 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Ji Z; Kang X; Wang S; Guo P Nano-channel of viral DNA packaging motor as single pore to differentiate peptides with single amino acid difference. Biomaterials 2018, 182, 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Huang G; Willems K; Soskine M; Wloka C; Maglia G Electro-osmotic capture and ionic discrimination of peptide and protein biomarkers with FraC nanopores. Nat. Commun 2017, 8. DOI: 10.1038/s41467-017-01006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Restrepo-Pérez L; Huang G; Bohländer PR; Worp N; Eelkema R; Maglia G; Joo C; Dekker C Resolving Chemical Modifications to a Single Amino Acid within a Peptide Using a Biological Nanopore. ACS Nano 2019, 13, 13668–13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Boersma AJ; Bayley H Continuous Stochastic Detection of Amino Acid Enantiomers with a Protein Nanopore. Angew. Chem., Int. Ed 2012, 51, 9606–9609. [DOI] [PubMed] [Google Scholar]
- (23).Lu Y; Wu X-Y; Ying Y-L; Long Y-T Simultaneous single-molecule discrimination of cysteine and homocysteine with a protein nanopore. Chem. Commun 2019, 55, 9311–9314. [DOI] [PubMed] [Google Scholar]
- (24).Guo Y; Niu A; Jian F; Wang Y; Yao F; Wei Y; Tian L; Kang X Metal-organic complex-functionalized protein nanopore sensor for aromatic amino acids chiral recognition. Analyst 2017, 142, 1048–1053. [DOI] [PubMed] [Google Scholar]
- (25).Ying Y-L; Long Y-T Nanopore-Based Single-Biomolecule Interfaces: From Information to Knowledge. J. Am. Chem. Soc 2019, 141, 15720–15729. [DOI] [PubMed] [Google Scholar]
- (26).Yuan B; Li S; Ying Y-L; Long Y-T The analysis of single cysteine molecules with an aerolysin nanopore. Analyst 2020, 145, 1179–1183. [DOI] [PubMed] [Google Scholar]
- (27).Ouldali H; Sarthak K; Ensslen T; Piguet F; Manivet P; Pelta J; Behrends JC; Aksimentiev A; Oukhaled A Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore. Nat. Biotechnol 2020, 38, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Edman P; Högfeldt E; Sillén LG; Kinell P-O Method for Determination of the Amino Acid Sequence in Peptides. Acta Chem. Scand 1950, 4, 283–293. [Google Scholar]
- (29).Molnar-Perl I In Quantitation of Amino Acids and Amines by Chromatography: Methods and Protocols; Molnar-Perl I, Ed.; Elsevier, 2005; Vol. 70, pp 163–198. [Google Scholar]
- (30).Checa-Moreno R; Manzano E; Mirón G; Capitán-Vallvey LF Revisitation of the phenylisothiocyanate-derivatives procedure for amino acid determination by HPLC-UV. J. Sep. Sci 2008, 31, 3817–3828. [DOI] [PubMed] [Google Scholar]
- (31).Fountoulakis M; Lahm H-W Hydrolysis and amino acid composition analysis of proteins. J. Chromatogr. A 1998, 826, 109–134. [DOI] [PubMed] [Google Scholar]
- (32).Woo K-L; Ahan Y-K Determination of protein amino acids as benzylthiocarbamyl derivatives compared with phenylthiocarbamyl derivatives by reversed-phase high-performance liquid chromatography, ultraviolet detection and precolumn derivatization. J. Chromatogr. A 1996, 740, 41–50. [Google Scholar]
- (33).Mallouli A; Lepage Y Convenient Syntheses of Naphthalene-, Anthracene-, and Naphthacene-2,3-dicarboxaldehydes. Synthesis 1980, 689. [Google Scholar]
- (34).Wei X; Zhang Z; Wang X; Lenhart B; Gambarini R; Gray J; Liu C Insight into the effects of electrochemical factors on host-guest interaction induced signature events in a biological nanopore. Nanotechnol. Precis. Eng 2020, 3, 2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Shao YH; Gan ZT; Epifanovsky E Advances in molecular quantum chemistry contained in the Q-Chem 4 program package. Mol. Phys 2015, 113, 184–215. [Google Scholar]
- (36).Lu T; Chen F Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem 2012, 33, 580–592. [DOI] [PubMed] [Google Scholar]
- (37).Di Muccio G; Rossini AE; Di Marino D; Zollo G; Chinappi M Insights into protein sequencing with an α-Hemolysin nanopore by atomistic simulations. Sci. Rep 2019, 9. DOI: 10.1038/s41598-019-42867-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Świder P; Lewtak JP; Gryko DT; Danikiewicz W Comparison of the sensitivity of mass spectrometry atmospheric pressure ionization techniques in the analysis of porphyrinoids. J. Mass Spectrom 2013, 48, 1116–1124. [DOI] [PubMed] [Google Scholar]
- (39).Murray KK; Boyd RK; Eberlin MN; Langley GJ; Li L; Naito Y Definitions of terms relating to mass spectrometry (IUPAC Recommendations 2013). IUPAC Recommendations 2013, 85, 1515. [Google Scholar]
- (40).Lubec G; Afjehi-Sadat L Limitations and pitfalls in protein identification by mass spectrometry. Chem. Rev 2007, 107, 3568–3584. [DOI] [PubMed] [Google Scholar]
- (41).Piguet F; Ouldali H; Pastoriza-Gallego M; Manivet P; Pelta J; Oukhaled A Identification of single amino acid differences in uniformly charged homopolymeric peptides with aerolysin nanopore. Nat. Commun 2018, 9, 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Timp W; Timp G Beyond mass spectrometry, the next step in proteomics. Sci. Adv 2020, 6, No. eaax8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Kasianowicz JJ; Brandin E; Branton D; Deamer DW Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. U.S.A 1996, 93, 13770–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Wilson J; Sloman L; He Z; Aksimentiev A Graphene Nanopores for Protein Sequencing. Adv. Funct. Mater 2016, 26, 4830–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Kennedy E; Dong Z; Tennant C; Timp G Reading the primary structure of a protein with 0.07 nm3 resolution using a subnanometre-diameter pore. Nat. Nanotechnol 2016, 11, 968. [DOI] [PubMed] [Google Scholar]
- (46).Zhong H; Zhang Y; Wen Z; Li L Protein sequencing by mass analysis of polypeptide ladders after controlled protein hydrolysis. Nat. Biotechnol 2004, 22, 1291–1296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study is available within the article and its Supporting Information files or from the corresponding authors upon reasonable request.


