Abstract
A race is currently being launched as a result of the international health situation. This race aims to find, by various means, weapons to counter the Covid-19 pandemic now widespread on all continents. The aquatic world and in particular that of photosynthetic organisms is regularly highlighted but paradoxically little exploited in view of the tremendous possibilities it offers. Computational tools allow not only to clear the existence and activity of many molecules but also to model their relationships with receptors identified in potential hosts. On a routine basis, our laboratory carries out a research activity on functionalities of molecules derived from algae using in silico tools. We have implemented our skills in algae biology and in modeling, as tests in order to identify molecules expressed by the genus Arthrospira showing an antiviral potential and more particularly anti-SARS-CoV-2. Using consensus docking and redocking with Autodock Vina and SwissDock, we were able to identify several promising molecules from Arthrospira: phycocyanobilin, phycoerythrobilin, phycourobilin, and folic acid. These four compounds showed reliable binding energies comprised between − 6.95 and − 7.45 kcal.mol−1 in Autodock Vina and between − 9.285 and − 10.35 kcal.mol−1 with SwissDock. Toxicity prediction as well as current regulations provided promising arguments for the inclusion of these compounds in further studies to assess their ability to compete with the SARS-CoV-2/ACE2 complex both in vitro and in vivo.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10811-021-02372-9.
Keywords: Covid-19, SARS-CoV-2, Arthrospira, Cyanobacteria, Antiviral, Docking
Introduction
Covid-19 is probably the most written, published acronym in this year 2020. This is for objective and obvious reasons. Covid-19 is the pandemic caused by SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) and it has overturned the entire planet as few pandemics have done, and this, in a modern era which has deployed modern and high-level medicine. Research on these pandemics takes multiple paths whether on technical, chemical, or biological levels. We then attack the biology of viruses or the defense mechanisms of their hosts. Different methods can be used for antiviral research to act against SARS-CoV-2. We can first target structural proteins in view to inhibit virus entry in human host cells or looking for functional internal viral protein inhibitors to block its replication for example. A third method would be to target human receptor for virus entry prevention. In view to limit the impact on the human metabolism of such antivirals, we rather focused on the first critical step for the virus, its recognition by the host cell, mediated via specific structural proteins, spike proteins. As a food tech company, Algama produces innovative food ingredients made with microalgae. As part of our daily research, we developed a tool enabling to select the most relevant molecules from microalgae for different food applications according to various criteria (bioavailability, safety, drug interaction…). In the context of the Covid-19 health crisis and to contribute to the collective effort, we tested our methodology toward antiviral compound identification from microalgae. Based on a literature framework about antivirals from Arthrospira spp, the aim of the present study was to identify the most relevant SARS-CoV-2 antiviral molecules thanks to docking, and to conduct an in silico toxicity assessment of the most efficient antiviral compounds. The first part of this work focuses on the description of the virus and presents a state of the art of Arthrospira use as an antiviral agent. The second part of the article deals with the evaluation of the anti-SARS-CoV-2 activity by docking Arthrospira compounds. Finally, an in silico study of the selected molecules toxicity will be proposed.
SARS-CoV-2
SARS-CoV-2 belongs to the coronavirus family, composed of enveloped virus with positive strain of RNA. Its genome is contained in a capsid formed by nucleocapsid proteins, itself included in an envelope. Three structural proteins are characteristic of coronavirus: membrane protein, envelope protein, and spike protein (S), a glycoprotein responsible for virus host cell attachment (Li 2016). Coronavirus took their name from “corona” (crown in Latin), formed by S protein protuberances on their surface.
The spike (S) glycoprotein ectodomain consists in a trimeric structure. Each monomer is composed of two subunits: S1, involved in the host cell receptor recognition; and S2, responsible for the membrane-fusion mechanism. S1 subunit contains two major domains, N-terminal domain (S1-NTD) and C-terminal domain (S1-CTD) (Li 2016) (Fig. 1). The latter constitutes the receptor binding domain (RBD) and is composed of two subdomains: a core structure and a receptor-binding motif (RBM) (Li et al. 2005).
Fig. 1.
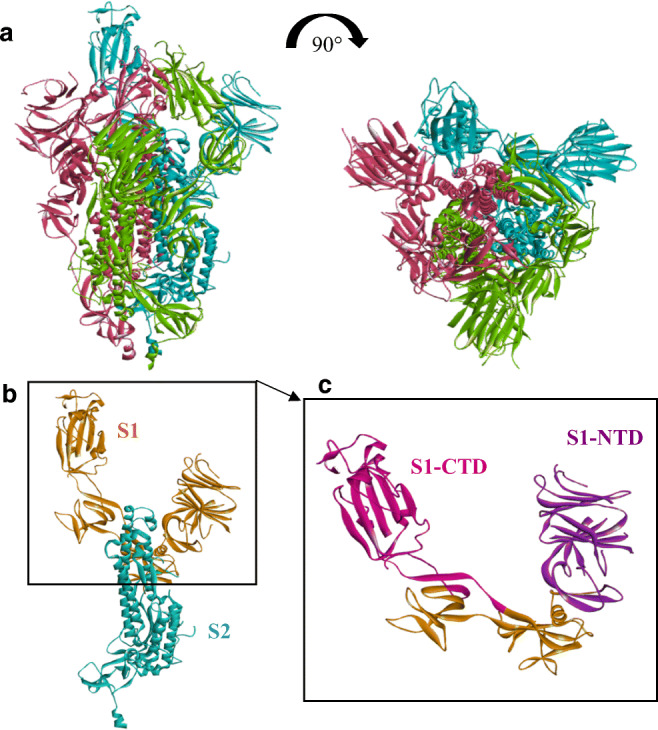
a Cryo-electron microscopy structure of prefusion trimeric SARS-CoV-2 spike protein (from PDB: 6LZG). Three monomers are identified (pink, green, and blue) from two angles of view. b Monomer structure of SARS-CoV-2 spike protein, subunit S1 is represented in orange and S2 in blue. c RBD localization in S1 subunit: S1 C-terminal domain (S1-CTD = RBD) appears in pink and S1 N-terminal domain (S1-NTD) in purple
Zhou et al. (2020) recently determined the ability of SARS-CoV-2 to use human angiotensin-converting enzyme 2 (ACE2) as receptor to engage virus attachment to the host cell via its RBD, as for SARS-CoV. However, they excluded binding with other coronavirus receptors such as APN and DPP4 (Li 2016). ACE2 identification as main receptor of SARS-CoV-2 in human has been confirmed shortly after by two other research teams (Hoffmann et al. 2020; Ou et al. 2020).
ACE2 is a metallocarboxypeptidase involved in arterial pressure system via cleavage of angiotensin peptides (Donoghue et al. 2000). It has been shown to be expressed in lung alveolar epithelial cells, in enterocytes of small intestine and in vascular endothelium (Hamming et al. 2004), explaining the location of Covid-19 pathologic symptoms. ACE2 binding site to SARS-CoV-2 was found to be within its extracellular peptidase domain (Yan et al. 2020).
Once coronavirus RBD is linked to ACE2 peptidase domain, the prefusion S protein becomes unstable and is submitted to a first proteolytic activation via a host cell protease, between S1 and S2 subunits (Walls et al. 2017). In addition to S1/S2 cleavage, a second site, critical for viral fusion, has been determined within the S2 domain, on the S2′ site (Belouzard et al. 2009). This second proteolysis results in the shedding of S1 subunit and exposes an internal fusion peptide. This one is essential for fusion of the virus and host membrane via S2 subunit, leading to the viral genome release in ACE2. This peptide is highly conserved among coronavirus (Madu et al. 2009) and composed of serine residues 798 to phenylalanine 815 in SARS-CoV.
TMPRSS2 was identified in lung cells as the main human protease used by SARS-CoV-2 for S protein modification (Hoffmann et al. 2020). TMPRSS2 belongs to the type II transmembrane serine proteases (TTSPs) which have been involved in the spread of various respiratory viruses (Choi et al. 2009). Expressed in airway epithelial cells (Böttcher et al. 2006), Shulla et al. (2011) further pointed the colocalization of TMPRSS2 and ACE2 during an immunoprecipitation assay. In this context, SARS-CoV-2 using the same receptor (ACE2) and host cell protease (TMPRSS2) as SARS-CoV, this colocalization appears to sorely facilitate its entry and spreading in human host cells.
Gui et al. (2017) determined different conformational prefusion states of SARS-CoV S protein. Each Spike RBD on the S1 subunit of SARS-CoV can be found in two conformational states, one “up” and one “down.” In down position, SARS-CoV RBD was reported as inaccessible for ACE2 recognition due to steric clashes. On the contrary, when a trimer is in “up” position, RBD is then exposed, allowing its binding with host cell ACE2 (Gui et al. 2017; Yuan et al. 2017).
At this day, two conformational states of the S protein have been found in SARS-CoV-2 (Wrapp et al. 2020; Walls et al. 2020). As for SARS-CoV, one presents all RBDs in down state (not completely closed according the Cryo-EM assays) and the second shows one of the three RBDs exposed in an open state. Like for its predecessor, SARS-CoV-2 RBD binding with ACE2 seems to be impossible when the Spike trimer is entirely in down conformation. On the contrary, with one RBD in up position, no steric clash with S protein was detected, suggesting that an open state is required for Spike protein binding with its receptor (Yan et al. 2020).
As crystal structure of RBD has been defined with a better resolution than the open state of the SARS-CoV-2 Spike ectodomain structure, and as no complete complex of Spike protein with ACE2 was available, RBD section of the S protein was chosen for this work. This further ensured to use a valid conformational state of the RBD.
Key binding residues involved in the Spike/ACE2 link were elucidated in several papers. Final consensus sequence for binding site on ACE2 was established from residues highlighted by the several teams (Lan et al. 2020; Shang et al. 2020; Wang et al. 2020; Yan et al. 2020): S19, Q24, F28, D30, K31, H34, E35, D38, Y41, Q42, L79, M82, Y83, K353, D355, and R357. Regarding Spike glycoprotein, consensus for ACE2 binding residues has been established as follows: K417, G446, Y449, Y453, L455, A475, F486,N487, Y489, Q493, G496, Q498, T500, N501, and G502 (Lan et al. 2020; Shang et al. 2020; Walls et al. 2020; Wang et al. 2020; Yan et al. 2020) (Table 3).
Table 3.
Binding residues involved in Spike RBD/ACE2 link; bold residues correspond to consensus sequences
| ACE2 | References |
|---|---|
| S19, Q24, K31, E35, D38, L79, M82, Y83, K353 | Shang et al. 2020 |
| Q24, D30, H34, Y41, Q42, M82, K353, R357 | Yan et al. 2020 |
| S19, Q24, F28, D30, K31, H34, D38, Y41, Q42, L79, M82, Y83, K353, D355 | Wang et al. 2020 |
| Q24, T27, F28, D30, K31, H34, E35, E37, D38, Y41, Q42, L79, M82, Y83, N330, K353, G354, D355, R357, R393 | Lan et al. 2020 |
| Spike RBD | References |
| L455, F486, Q493, S494, N501. | Shang et al. 2020 |
| K417, Y453, Q474, F486, Q498, T500, N501. | Yan et al. 2020 |
| K417, G446, Y449, Y453, A475, E484, F486, N487, Y489, G496, Q498, T500, G502. | Wang et al. 2020 |
|
T402, Y436, N439, Y440, L455, N473, F486, G488, Y491, Q493, Q498, N501. |
Walls et al. 2020 |
| K417, G446, Y449, Y453, L455, F456, A475, F486, N487, Y489, Q493, G496, Q498, T500, N501, G502, Y505. | Lan et al. 2020 |
State of the art on antivirals derived from algae, in particular Arthrospira (Tables 1 and 2)
Table 1.
In vitro evaluation of Arthrospira antiviral activity: state of the art
| Model | Targeted virus | Extract/compound | Antiviral activity | Control | Reference |
|---|---|---|---|---|---|
| Hep-2 cells | VACV | TK V2 (intracellular), exopolysaccharides TK V3 | Decrease of viral hepatic replication. Replication inhibition: EC50(TK V3) = 0.78 μg.mL−1 | Untreated control | (Radonic et al. 2010) |
| MDCK cells | Influenza | Cold water extract of A. platensis | Inhibition of viral plaque formation (EC50 = 0.58 ± 0.02 mg.mL−1) | Untreated control | (Chen et al. 2016) |
| Hep-2 cells | Adenovirus type 40 | Methanolic extract of A. platensis |
50% reduction of viral titer (EC50 = 0.8–3.1 mg.mL−1) |
NA | (Abdo et al. 2012) |
| Hep-2, BGM, and MA104 cell lines | Adenovirus type 7, CVB4, astrovirus type 1, RV Wa strain, and adenovirus type 40 | Ethanolic extract of A. platensis | 53.3%, 66.7%, 76.7%, 56.7%, and 50% reductions of viral titer response; dose: 1.6–1.9 mg.mL−1 | Untreated control | (El-Baz et al. 2013) |
| Vero cells | CVB3 and RV | Methanolic extract of A. platensis | TI(CVB3) = 30, TI(RV) = 45 | Untreated control | (Deyab et al. 2020) |
| MA104, Hep-2, and BGM cell lines | RV Wa strain, adenovirus type 7, adenovirus type 40, CVB4 | 70% methanol and n-hexane extract of A. platensis | Inhibition of 56.7% and 66.7% against RV Wa strain; 60% and 63.3% against adenovirus type 7; 53.3% and 50% against adenovirus type 40, respectively, and 50% for both extracts against CVB4 (0.5 mg.mL−1) | Untreated control | (Hetta et al. 2014) |
| HFFs | HCMV | Polysaccharide fractions isolated from A. platensis (spirulan-like molecules) | HCMV inhibitory effect: EC50 = 1.4 ± 0.3 μg.mL−1 with preincubation, EC50 = 93.3 ± 0.1 μg.mL−1 with post-incubation | Reference drug: ganciclovir (GCV) | (Rechter et al. 2006) |
| Vero cells | HSV 1 and 2 | Partial desulfated and oversulfated sodium spirulan (Na-SP) from A. platensis | HSV-1: EC50 (Na-SP) = 0.63 μg.mL−1, EC50 (Os-SP-2) = 0.46 μg.mL−1, HSV-2: EC50 (Na-SP) = 0.41 μg.mL−1; EC50 (Os-SP-2) = 0.46 μg.mL−1 | Reference drug: acyclovir | (Lee et al. 2007) |
| RC-37 cells | HSV | Cold and hot water (2.5 mg.mL−1), phosphate buffer (10 mg.mL−1) extracts of A. fusiformis | Virus infectivity reduction of 54.9%, 64.6%, and 99.8% | Untreated control and reference drug: acyclovir | (Sharaf et al. 2013) |
| Vero cells | HSV-2 | Hot water extract of A. maxima | Adsorption and penetration inhibition: selectivity index of 128, EC50 = 0.069 mg.mL−1 | Uninfected cells treated with the same extract | (Hernández-Corona et al. 2002) |
| Vero cells | HSV-1 | Sulphoquinovosyl diacylglycerol isolated from A. platensis | HSV-1 inhibition: EC50 = 6.8 μg.mL−1 | Untreated control and reference drug: acyclovir | (Chirasuwan et al. 2009) |
| Vero cell and HepG2 cells | HAV-MBB strain, HSV-1 | Phosphate buffer and water extract of A. platensis | 60% inhibition of hepatitis A virus with 50 μg.mL−1 water extract; 98% inhibition of HSV-1 with 50 μg.mL−1 of water and phosphate buffer extract | 0.02 M NaCl | (Shalaby and Shanab 2010) |
| Burkitt’s lymphoma (BL) cell lines: Akata, B95-8, and P3HR-1 | EBV | Methanolic extract of A. platensis | Effect on virus load: EC50 = 0.021 μg.mL−1, CC50 = 166 μg.mL−1, TI = 7905 in the case of B95-8 cells | Untreated control and reference drugs: acyclovir, foscarnet | (Kok et al. 2011) |
| Human T-cell line MT4 | HIV-1 | Peptide isolated from Spirulina maxima (SM-peptide) | Inhibit induced cell lysis: EC50 = 0.475 mg.mL−1, CC50 = 1.457 mM; inhibition of HIV-1 reverse transcriptase (0.75 mg.mL−1) and p24 antigen production > 95% | Untreated control | (Jang and Park 2016) |
| Human T-cell lines, peripheral blood mononuclear (PBMC) and Langerhans cells (LC) | HIV-1 | Aqueous extract of A. platensis | Reduce viral production in PBMCs: EC50 = 0.3–1.2 μg.mL−1 | Uninfected cells treated with the same extract | (Ayehunie et al. 1998) |
VACV: Vaccinia virus, CV: coxsackievirus, RV: rotavirus, HCMV: human cytomegalovirus, HAV-MBB: hepatitis A virus type MBB, EBV: Epstein–Barr virus, Vero cells: African green monkey kidney, HIV: human immunodeficiency virus, NA: not available
Table 2.
Clinical study evaluation of Arthrospira antiviral activity: state of the art
| Model | Targeted virus | Extract/compound | Antiviral activity | Beneficial effect | Duration | Study parameters | Reference |
|---|---|---|---|---|---|---|---|
| 73 HIV-infected adult females | HIV | 5 g.day−1 of A. platensis | No effect on the viral load and/or the CD4 T cells | Improvement of anemia status, good nutritional rehabilitation effects | 3 months | Placebo, randomized, double-blind | (Winter et al. 2014) |
| 11 antiretroviral-naïve | HIV-1 | 5 g.day−1 of dried A. platensis | No significant effect on CBC, metabolic and lipid panel; stable CD4 and virus load | Clinically significant improvement in CD4 (> 100 cells.mL−1), decreased HIV viral load of 0.5 log10 for 1 subject | 3 months | Placebo, randomized | (Teas and Irhimeh 2012) |
| 320 naïve HIV-1 patients | HIV-1 | 10 g.day−1 of dried A. platensis | Significant increase of CD4 count cells, decrease of viral load level and higher hemoglobin level | Improve immune system and prevent opportunistic diseases | 12 months | Randomized, single-blind, control without spirulina | (Ngo-Matip et al. 2015) |
| 52 HIV-infected | HIV-1 | 0.2 to 0.37 g.kg−1.day−1 of A. platensis | Significantly lower viral load, higher CD4 count, increase in hemoglobin level (1.6 g.dL−1) | Increase quality of weight gain | 3 months | Randomized, single-blind, control group | (Azabji-Kenfack et al. 2011) |
| 25 thalassemic children HCV-infected | HCV | 250 mg.kg−1.day−1 of A. platensis | Significant increase of CD4 (from 19.56 ± 7.8 to 32.2 ± 13.5 × 103 cells.100 mL−1) and CD8 (from 17.68 ± 6.88 to 26.44 ± 9.08 × 103 cells.100 mL−1) after 6 months | Immune stimulation | 6 months | NA | (Gomaa et al. 2017) |
| 66 HCV-infected | HCV | 3 × 500 mg.day−1 of A. platensis | No significant effect on virus load; ALT, CLDG, and ASEX improved by spirulina | Loss or reduction of detectable hepatitis C virus RNA for 6 patients | 6 months | Randomized, double-blind, control group (3 × 140 g of silymarin/day) | (Yakoot and Salem 2012) |
HCV: hepatitis C virus, HIV: human immunodeficiency virus, CBC: complete blood count; ALT: alanine transaminase, CLDG: Chronic Liver Disease Questionnaire, ASEX: Arizona Sexual Experiences Scale, NA: not available
The term “algae” actually includes a wide variety of organisms with a complex evolutionary history. These organisms have features in common such as the ability to do photosynthesis and being predominantly aquatic. We can distinguish two main categories of algae: macroalgae, multicellular eukaryotes; and microalgae, in the largest sense of the word, including unicellular eukaryotes and prokaryotes. Most macroalgae are found in seawater and are called “algae,” but some can also be found in freshwater (e.g., Oedogonium, Cladophora and Spirogyra). Microalgae are found in both environments, but the most consumed species (Arthrospira and Chlorella) generally grow in freshwater.
These aquatic plants are known to be rich in proteins and lipids, but also offer a wide range of immunostimulatory, antioxidant, and antiviral properties. Numerous publications have reviewed the generic properties of algae in these areas (Mimouni et al. 2012). For example, the immune response in the cytokines which is tested by Talukdar et al. (2020a, 2020b) with astaxanthin. Griffithsin, a lectin isolated the from red alga Griffithsia, has shown activity on protein S of MERS-CoV (Middle East Respiratory Syndrome Coronavirus) (Millet et al. 2016). As for microalgae, the very numerous studies have been compiled by De la Jara et al. (2018) and Perumal and Sundararaj (2020). In addition, few studies have been identified in the literature as dealing with the antiviral potential of microalgae compounds against SARS-CoV-2. For instance, astaxanthin as well as linolenic acid (C18:00) have been mentioned as potential candidates for the treatment or the prevention of Covid-19 (Talukdar et al. 2020b). However, these are pre-published studies and the confirmation of the robustness will be given upon the final peer-reviewed publications.
The genus Arthrospira is a genus of prokaryotic filamentous cyanobacteria. The best-known species, Arthrospira platensis and Arthrospira maxima, are mostly used as food supplement due to their nutritional properties: high protein content, vitamins, minerals, etc. Moreover, Arthrospira also contains various metabolites with beneficial health properties such as polyphenols, carotenoids, and sterols (de la Jara et al. 2018). Due to drug-resistant virus strains emergence, and because of adverse effects caused by usual synthetic antiviral treatments, increasingly researchers are looking into more natural approaches. This explains a growing interest in antivirals from natural sources, in algae in general and in Arthrospira for example in particular. Hence, in vitro and in vivo studies have been conducted to evaluate Arthrospira compounds as antiviral against various viruses such as influenza, herpes simplex virus (HSV), and hepatitis C (HCV) as presented in Tables 1 and 2.
Data from scientific literature were critically reviewed in order to highlight results of the most rigorous studies: presence of controls, replicates, measurement of toxicity, and inhibition efficacy. The following information were collected and analyzed: CC50, cytotoxic concentration of the compounds decreasing in vitro cell viability to 50%; EC50, effective concentration needed to inhibit 50% of virus; and therapeutic index (TI or SI), referring to the ratio CC50/EC50. A high therapeutic index (TI), or selectivity index (SI), is preferable for a drug to have a favorable safety and efficacy profile.
Radonic et al. (2010) evaluate the effect of sulfur-containing exopolysaccharides from A. platensis (TKV3). According to their study, anionic polysaccharides from Arthrospira showed antiviral activity against enveloped virus such as VACV (EC50 (TK V3) = 0.78 μg.mL−1) via interactions with viral membrane glycoproteins and thus inhibition of virus binding to host cells. In the work of Chen et al. (2016), Arthrospira extract was capable of inhibiting in vitro influenza viral replication and plaque formation (Table 1), by targeting hemagglutinin, an influenza virus surface glycoprotein.
Abdo et al. (2012), El-Baz et al. (2013), Deyab et al. (2020), and Hetta et al. (2014) focused on A. platensis antiviral effect on adenovirus and Coxsackievirus (CV). Most of them evaluate Arthrospira methanol extract activity on Hep-2 cell in vitro model. In all cases, Arthrospira appears as a good antiviral agent: reducing virus titer from 50% and preventing virus from attaching to host cell receptor when used as pre-treatment with TI(CVB3) = 30 and TI(RV) = 45 (Deyab et al. 2020). Efficiency of methanolic extract is attributed to the presence of polar compounds binding to viral capsid. Moreover, El-Baz et al. (2013) found that Arthrospira ethanolic extract was active against non-enveloped RNA and DNA viruses. However, authors did not compare Arthrospira effect with a control drug since there is no available drug against enteric virus according to them.
Elsewhere, Rechter et al. (2006) compared spirulan-like substance to ganciclovir (GCV), a reference drug used as anti-herpesvirus. According to their in vitro experiments, preincubation of substances enhances antiviral activity via virus entry and replication inhibition: EC50 (TK-V3a) = 1.4 ± 0.3 μg.mL−1, close to drug reference EC50 = 0.7 ± 0.1 μg.mL−1 in post-incubation (Table 1).
Due to the emergence of drug-resistant Herpes simplex strains, several studies have focused on the search for alternatives to synthetical acyclovir in the prevention and treatment of Herpes simplex virus (HSV) 1 and 2. For instance, hot water extract of A. maxima provided EC50 of 0.069 mg.mL−1 against HSV-2. The authors then attributed this antiviral activity to high polar compounds (Hernández-Corona et al. 2002). In a second study, phosphate buffer extract of A. fusiformis showed a viral infection inhibition almost as high as acyclovir (85% against 99%) during the intracellular replication period (Sharaf et al. 2013). In the same way, Chirasuwan et al. (2009) evaluated in vitro A. platensis extract as potential HSV antiviral agent to propose an alternative to the synthetic anti-herpes drug acyclovir. Sulfoquinovosyl diacylglycerol (SQDG) was responsible for antiviral activity of A. platensis lipid fraction (EC50 = 6.8 μg.mL−1) with an effect comparable to the reference drug (EC50 = 1.5 μg.mL−1), without toxicity for cells. This activity is due to DNA polymerase inhibition thanks to SQDG interaction with different regions of the enzyme. Furthermore, Lee et al. (2007) highlighted antiviral potency of spirulan molecules from A. platensis. Best in vitro results were obtained when compounds were added to the medium during HSV-1 and 2 infections. Na-Sp then showed the highest selectivity indexes of 13,000 and 17,000, with very low ECs50 (0.63 and 0.41 μg.mL−1). Otherwise, Shalaby and Shanab (2010) attributed the observed hepatitis A (RNA virus) and HSV-1 antiviral effect of aqueous and phosphate buffer extracts to the sulfated polysaccharides and tannins. According to them, these compounds may interfere at different viral stages such as attachment and penetration of the virus.
In the case of Kok et al.’s (2011) study, different extracts from microalgae were compared to conventional drugs (acyclovir and foscarnet) for their antiviral capacity against Epstein–Barr virus (EBV). Methanol extract from A. platensis reduced the cell-free EBV DNA load in B95-8 cells with an EC50 of 0.021 μg.mL−1 with an CC50 of 166 μg.mL−1 (TI = 7905) (Kok et al. 2011).
Peptides were also evaluated as antiviral agent. Indeed, Jang and Park (2016) found that a peptide isolated from A. maxima displayed in vitro antiviral activity since it inhibited reverse transcriptase activity in HIV-1–infected cell by 90% compared to the no-peptide assay. Moreover, 0.75 mg.mL−1 of this peptide inhibited HIV-1 p24 antibody production by more than 95%, without toxicity for the cells. Similarly, Ayehunie et al. (1998) evaluate HIV antiviral activity of Arthrospira. According to their results, both polysaccharide fraction and the fraction depleted in polysaccharide and tannin of A. platensis inhibited in vitro HIV-1 replication. Preincubation of polysaccharide fraction extract with cells showed better results due to polysaccharides binding to CD4 receptors, thus preventing virus attachment to the host cell through its gp120 envelope glycoprotein.
According to in vitro studies presented above (Table 1), Arthrospira has antiviral properties against different types of viruses: Herpes simplex, hepatitis, vaccinia, coxsakievirus…. It has no, or very little, cytotoxicity to cells, and low doses (EC50) are sufficient to induce an effect (1–10 μg.mL−1) with high therapeutic indices. It seems that Arthrospira compounds act during the early stages of viral infection. According to the main hypotheses evoked, compounds of polar nature and mainly sulfated polysaccharides and peptides are responsible for interactions with viral envelope glycoproteins, thus preventing binding to the host cell. Calcium spirulan, a sulfated polysaccharide from Arthrospira, is the most cited compound with antiviral activity against various enveloped viruses. The sulfate groups as well as the carboxyl groups of polysaccharides, such as Ca-Sp isolated from the extract, have negative charges which can react with the basic amino acids of viral proteins and block the interaction with cellular receptors.
In order to confirm in vitro results, a second state of the art was carried out focusing on human clinical studies involving the use of Arthrospira as an antiviral agent (Table 2).
Most of the in vivo studies focused on Arthrospira supplementation effect on HIV-infected patients. Today, there is no treatment capable of eradicating AIDS. HIV-infected patients still carry the virus and they can spread it to others. So far, there are five major classes of anti-HIV drugs that target distinct steps in the virus life cycle: reverse transcriptase inhibitors, protease inhibitors, fusion inhibitors, integrase inhibitors, and multidrug combinations (Jang and Park 2016). To improve therapeutic potential of existing medicines, research on Arthrospira-based novel treatment have been conducted.
In this context, Winter et al. (2014) conducted an in vivo study on 73 HIV-infected women to evaluate Arthrospira supplementation effect. The study was a 3-month pilot, randomized, double-blind, and placebo-controlled intervention. No significant clinical effect of Arthrospira supplementation was observed regarding viral load and CD4 cells; nevertheless, Arthrospira supplementation showed a positive effect on weight stabilization and protection against opportunistic infections. Similarly, Teas and Irhimeh (2012) found that Arthrospira strengthen the immune system of affected patients. Unfortunately, sample size of selected patients for the present study was too small (11) to conclude that there were significant effects of Arthrospira supplementation. Ngo-Matip et al. (2015) conducted a single-blind, randomized, multicenter study on 320 HIV-1 ARV-naïve participants for 12 months to analyze Arthrospira supplementation effect compared to local diet, both with standard HIV therapy. This work confirmed previous results, showing that Arthrospira supplementation intake positively and significantly stimulated the immune system and inhibited virus replication of HIV-infected subjects. As in the previous cases, Arthrospira contributed to the proper functioning of immune system and thus helped to limit the appearance of opportunistic diseases. Similar results were also obtained by Azabji-Kenfack et al. (2011), indicating that Arthrospira supplementation helps HIV-infected and malnourished adults to improve their immune defenses (Azabji-Kenfack et al. 2011). For this experiment, authors compared effects of Arthrospira and soya bean supplementation. Like other studies, Arthrospira supplement improved general state of health by promoting weight gain of malnourished adults infected with HIV. Regarding the baseline and the results obtained with soya beans, Arthrospira consumption significantly reduced viral load and increased CD4 count (Azabji-Kenfack et al. 2011).
Two in vivo studies listed in Table 2 focused on Arthrospira effect in the case of hepatitis C infection (HCV). In the work of Gomaa et al. (2017), Arthrospira was added to usual thalassemic drug for 6 months to evaluate the effect on thalassemic HCV-infected children. As patients are suffering from two different pathologies, it was difficult to affirm that Arthrospira was responsible for health improvement. However, the authors conclude that Arthrospira stimulated the immune system of thalassemic children infected with HCV since CD4 and CD8 number was increased after 6 months of Arthrospira intake. They also state that phycocyanin from Arthrospira stimulated hematopoiesis, by inducing erythropoietin hormone (EPO) release, thus helping thalassemia children. Moreover, Yakoot and Salem (2012) conducted an in vivo study comparing the effect of Arthrospira and silymarin against hepatitis C. For the present study, 66 patients with chronic hepatitis C virus infection had been randomized, double-blinded, and treated with Arthrospira or silymarin for a period of 6 months of treatment. According to their results, Arthrospira helped to improve the general well-being of patients and has led to a loss or reduction of detectable hepatitis C virus RNA for six patients. Compared to the second group treated with silymarin, these results were no significant (0.12) (Table 2). Nevertheless, the present study did not include a placebo control since silymarin possess itself some benefits in the treatment of viral hepatitis. Thus, it does not allow to conclude on these results.
According to these in vivo studies, it appears that Arthrospira improves the general health of AIDS and HCV-infected patients by contributing to restore their body’s defense mechanism against infectious immune system disorder, thus limiting opportunistic diseases. According to these results, Arthrospira may significantly affect virus progression according to some studies. However, most studies conducted in vivo on human patients have evaluated the effect of daily Arthrospira supplementation in addition to the usual antiviral treatment. These studies were mostly randomized and double-blind, but the study duration was often short with a small number of patients and did not always present a placebo control. It could be interesting to evaluate the effect of Arthrospira alone, without other antiviral treatment, in a rigorous long-term study to confirm its antiviral potential.
According to this literature review, Arthrospira acts as antiviral against various type of viruses. Indeed, significant antiviral effects were observed in vitro, and in vivo studies (Table 1 and 2) confirmed that Arthrospira is well tolerated by patients and even improved their general state of health by increasing immune response. Arthrospira can thus be administered in addition to current gold standard therapy in order to improve its effects, or it can also be considered as an interesting alternative to certain drugs with serious adverse effects. Many compounds such as sulfated polysaccharides (calcium-spirulan), fatty acids (Hetta et al. 2014), and proteins (C-phycocyanin, cyanovirin-N, microvirin) have been identified as responsible for this antiviral activity. Indeed, these compounds can bind to glycoprotein virus envelope, and thus hinder virus attachment to its host cell. Nevertheless, according to Ayehunie et al. (1998), it appears that compounds other than polysaccharides and tannins, present in the aqueous extract of Arthrospira, may also have an antiviral activity. To go further, it might therefore be interesting to evaluate the antiviral potential of other Arthrospira metabolites.
A pre-published work publicly available highlighted Arthrospira potential on two viral proteins of SARS-CoV. However, it was based on SARS-CoV-1 database, and it is pending for publication (DOI: 10.26434/chemrxiv.12051927). Very few authors have taken an interest in the specific case of Arthrospira against SARS-CoV-2, and most of the articles present a literature survey without in silico, in vitro, or in vivo evaluation. The use of docking in order to test Arthrospira as a candidate against SARS-CoV-2 is thus original.
In order to propose new antiviral leads from Arthrospira, the present study evaluates in silico the antiviral potential of Arthrospira metabolites using an updated database specific to SARS-CoV-2 thanks to docking tools. Molecular docking is a computational method which simulates different orientations of a receptor and a ligand to predict the most stable complex, with a minimal free energy. Widely used to new drug discovery, it is composed of two main steps. First, programs perform a conformational sampling of ligands in the receptor active site. Then, an algorithm (scoring function) scores each position. The latter predicts binding energy of compounds and ranks all the docking positions by increasing energy. The lower the binding energy, the greater the affinity between the ligand and its receptor.
Autodock Vina, on one hand, a popular program for molecular docking, involves a hybrid scoring function based on both empirical scoring functions and knowledge-based potentials. Indeed, it is largely inspired of X-score which considers van der Waals (VDW) interactions, hydrogen bonds, hydrophobic effects, and effective rotatable bonds to determine a binding score (Huang et al. 2010; Trott and Olson 2010). On the other hand, SwissDock is based on the EADock DSS (Dihedral Space Sampling), a forcefield scoring function. Indeed, it involves CHARMM22 protein forcefield using VDW and electrostatic grids to determine the interaction energy between the protein and ligands (Grosdidier et al. 2011a).
In order to contribute to the collective effort to find a solution to the current global Covid-19 crisis and to further explore the antiviral potential of Arthrospira, this article investigates antiviral activity of Arthrospira molecules from Algama internal database. This database is composed of 48 high-value molecules from Arthrospira genus. As control molecules, we choose an antiviral (remdesivir) and two molecules with high docking score according to the literature (linoleic acid and pavetannin C1) (Prasanth et al. 2020; Toelzer et al. 2020).
Materials and methods
Protein–ligand docking
Two crystal structures of the receptor-binding domain (RBD) of SARS-CoV-2 Spike protein bound to the cell receptor ACE2 have been determined (Lan et al. 2020; Wang et al. 2020) and deposited on the Protein Data Bank under the respective codes 6M0J and 6LZG. All the docking assays were executed with the firstly deposited crystal structure of the complex, corresponding to the 6LZG identifier (Wang et al. 2020). Complete RBD part (with all residues) was extracted from this crystal complex using Discovery Studio Visualizer, thus expecting having the most relevant conformation of the RBD in the Spike/ACE2 complex (Forli et al. 2016). Receptor structure was then prepared, heteroatoms (water included) were deleted, and hydrogens were added. Protonation states were assigned thanks to H++ 3.2v at physiological pH (7.4) with AMBER ff 14SB parameters (Gordon et al. 2005; Myers et al. 2006; Anandakrishnan et al. 2012). Finally, protein structure was energy minimized with Chiron tool (Ramachandran et al. 2011) and the file was saved as PDB file.
Ligands were divided into three categories: small molecules (mainly secondary metabolites), Arthrospira peptides, and the antiviral drug remdesivir. Small molecule and drug structures were obtained from PubChem and downloaded as SDF format. Peptide sequences were taken from literature and converted in SMILES format via NIH Cactus translator, and finally in SDF format in Open Babel (O’Boyle et al. 2011). Ligands were then prepared for docking with both Autodock Vina and SwissDock programs according to the following protocol. Each molecule was redocked with both software in view to discriminate false-positive results.
Autodock Vina
All the 52 ligands from the Algama database were energy minimized before being finally converted in PDBQT format on PyRx (Dallakyan and Olson 2015). Docking was executed with Autodock Vina (Trott and Olson 2010) implemented in the PyRx platform (Dallakyan and Olson 2015). The grid box was manually adjusted around active residues identified above, with the coordinates X: − 37.348, Y: 29.6843, Z: 3.1984 and dimensions X: 29.0036 Å, Y: 56.1876 Å, Z: 24.9881 Å for the first docking; and coordinates X: − 36.652, Y: 29.8116, Z: 4.1656 and dimensions X: 23.2983 Å, Y: 43.1925 Å, Z: 24.8652 Å for the second docking. Exhaustiveness was set on 8, equivalent to the short mode, since no statistically significant difference in accuracy was detected using the short, medium, or long mode of Autodock Vina (Nguyen et al. 2020).
Binding affinity result files were exported as CSV for analysis and docked structures were visualized in Discovery Studio Visualizer in view to obtain a complete 3D structure of each complex. Ligand interactions were determined with this same tool and analyzed to target better candidates for SARS-CoV-2 RBD binding. In literature, amino acids of Spike RBD are classically numbered from 333 to 527, as mentioned in Table 3. Nevertheless, during docking preparation of the protein, amino acids numbers were modified by the H++ program, thus going from 1 to 195. Interaction prediction files were also generated with these new numbers (Figs. 2, 3, 4, 5, 6, 7, 8, 9, and 10). However, for a better understanding and being aligned with other papers, residue numbers were post-docking reassigned from 333 to 527 in the results and discussion parts, thanks to a translation table (Table 9 Supplemental Data).
Fig. 2.
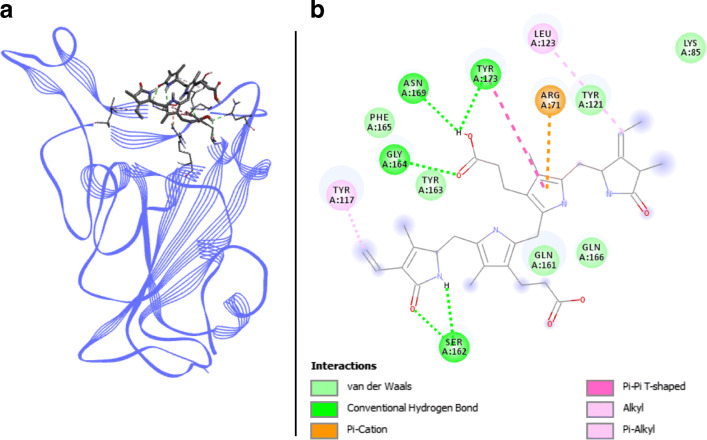
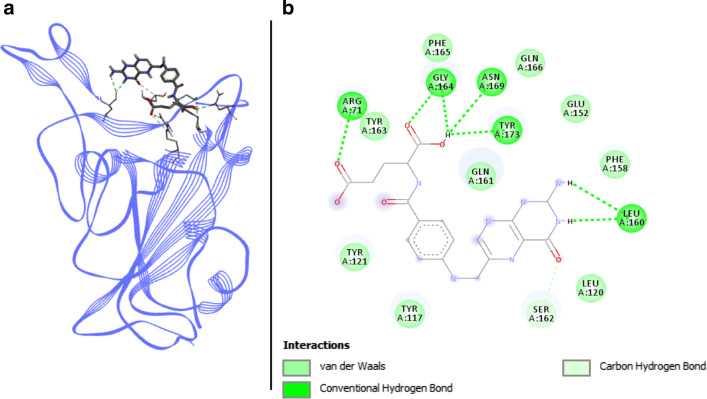
Phycoerythrobilin docked with spike RBD of SARS-CoV-2. a 3D structure of the complex, phycoerythrobilin (ligand) is represented in sticks, Spike RBD (receptor) is in blue. b Interaction map of the ligand/receptor complex
Fig. 3.
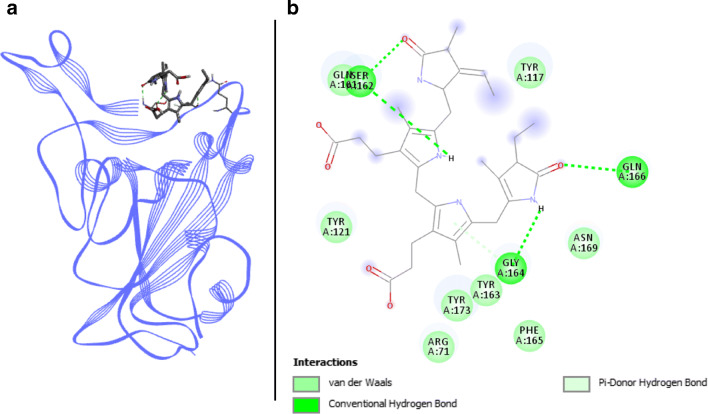
Phycocyanobilin docked with spike RBD of SARS-CoV-2. a 3D structure of the complex, phycocyanobilin (ligand) is represented in sticks, Spike RBD (receptor) is in blue. b Interaction map of the ligand/receptor complex
Fig. 4.
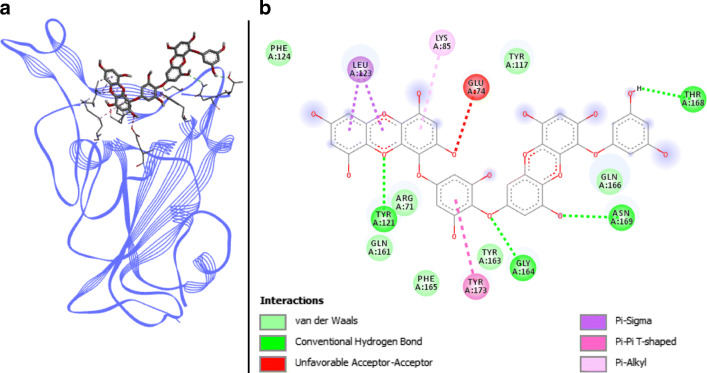
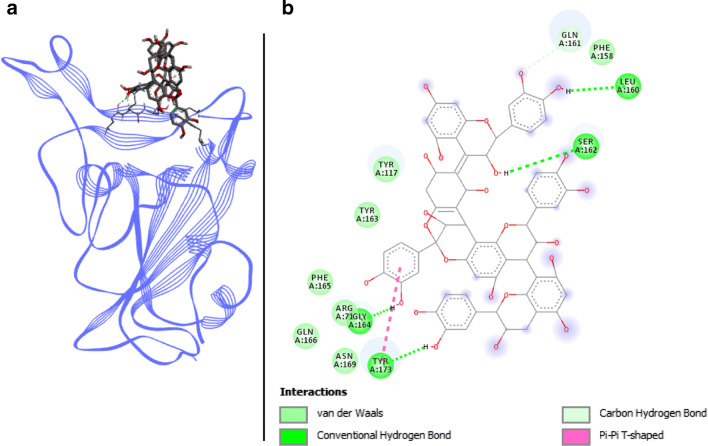
Dieckol docked with spike RBD of SARS-CoV-2. a 3D structure of the complex, dieckol (ligand) is represented in sticks, Spike RBD (receptor) is in blue. b Interaction map of the ligand/receptor complex
Fig. 5.
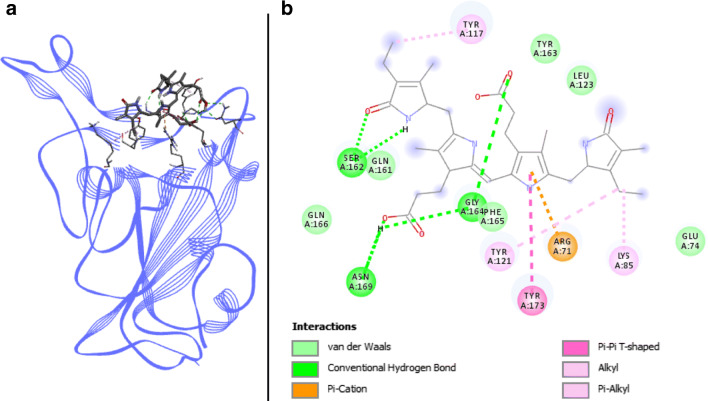
Phycourobilin docked with spike RBD of SARS-CoV-2. a 3D structure of the complex, phycourobilin (ligand) is represented in sticks, Spike RBD (receptor) is in blue. b Interaction map of the ligand/receptor complex
Fig. 6.
Folic acid docked with spike RBD of SARS-CoV-2. a 3D structure of the complex, folic acid (ligand) is represented in sticks, Spike RBD (receptor) is in blue. b Interaction map of the ligand/receptor complex
Fig. 7.
Pavetannin C1 docked with spike RBD of SARS-CoV-2. a 3D structure of the complex, Pavetannin C1 (ligand) is represented in sticks, Spike RBD (receptor) is in blue. b Interaction map of the ligand/receptor complex.
Fig. 8.
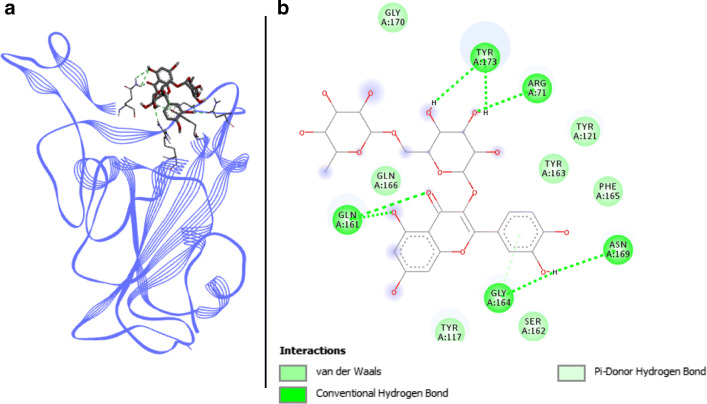
Rutin docked with spike RBD of SARS-CoV-2. a 3D structure of the complex, rutin (ligand) is represented in sticks, Spike RBD (receptor) is in blue. b Interaction map of the ligand/receptor complex
Fig. 9.
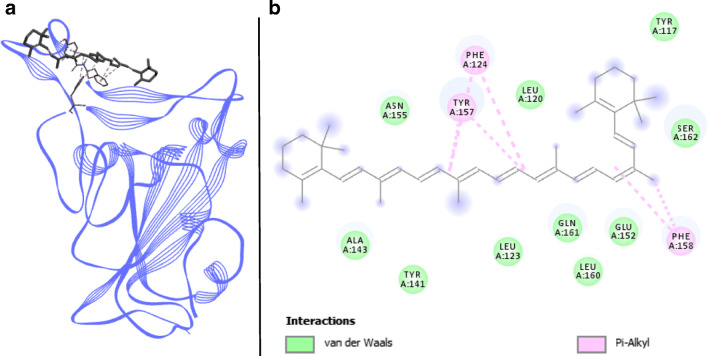
β -carotene docked with spike RBD of SARS-CoV-2. a 3D structure of the complex, β -carotene (ligand) is represented in sticks, Spike RBD (receptor) is in blue. b Interaction map of the ligand/receptor complex
Fig. 10.
Astaxanthin docked with spike RBD of SARS-CoV-2. a 3D structure of the complex, astaxanthin (ligand) is represented in sticks, Spike RBD (receptor) is in blue. b Interaction map of the ligand/receptor complex
SwissDock
SDF files were converted into .mol2 format with 3D coordinates and protonation at physiological pH with Open Babel software (O’Boyle et al. 2011). All ligands were submitted to a control with SwissParam (Zoete et al. 2011) before docking on SwissDock (Grosdidier et al. 2011a, 2011b). Previous PDB file of Spike RBD was used as receptor file again. Docking was set in accurate mode with the same grid box coordinates and dimensions used for the Autodock Vina assays. Pose with the best binding energy was selected for each ligand and respective result file was exported as CSV format.
In silico toxicity assessment
Toxicity of selected molecules was assessed via the AdmetSAR 2 prediction tool (Yang et al. 2019), using canonical smiles from PubChem for compound identification. SwissADME and Osiris platforms were also used to complete toxicity data (Daina et al. 2017).
Acute oral toxicity was determined through toxicity class. Mutagenicity, carcinogenicity, hepatotoxicity, and reprotoxicity were also predicted. In order to avoid interaction with other potential treatment, study of the main cytochrome as well as P-glycoprotein interactions was needed. Finally, druglikeness parameters were assessed such as gastrointestinal absorption, blood–brain barrier permeability, or Lipinski’s rule.
Results
Docking
After docking twice the 52 molecules from Algama’s internal database with Spike RBD in both Autodock Vina and SwissDock, the first mode was selected for each compound, corresponding to the best binding affinity (Table 8 Supplemental data). Both their RMSD/ub and RMSD/lb were lower than 2 Å (Trott and Olson 2010).
In order to identify best compounds and false-positive results, averages, SDs as well as coefficients of variation were calculated for the two assays of each program (Table 8 Supplemental data). Thus, for one molecule, binding energies from Autodock Vina trials were averaged, like those of SwissDock. Two lists of averaged binding energies were then obtained and ranked from the smallest to the largest. In general, redocking confirmed the results since coefficients of variation were almost all behind 5% with both Autodock Vina and SwissDock. Redocking can be therefore considered as a reliable method regarding the two conducted assays for each program.
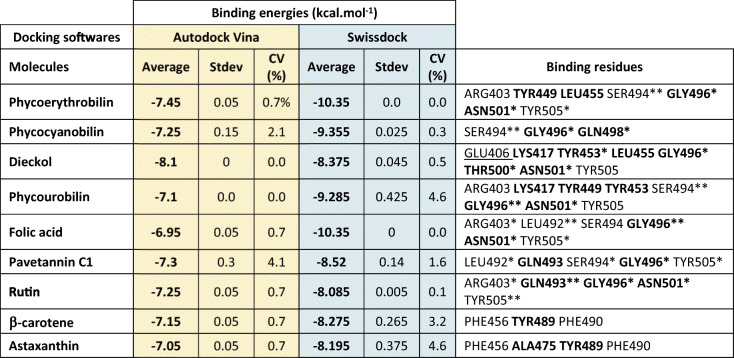
The first 15 compounds were selected on each list and then compared to each other. Autodock Vina provided the following 15 compounds: dieckol, phycoerythrobilin, pavetannin C1, phycocanobilin, rutin, catechin, β-carotene, phycourobilin, rosmarinic acid, epicatechin, astaxanthin, apigenin, folic acid, remdesivir, and poriferasterol (Table 4). Averaged binding energies were comprised between − 6.7 and − 8.1 kcal.mol−1. Regarding substances with lowest binding energies obtained with SwissDock, the first 15-molecule list was slightly different, consisting of peptide GGTCVIRGCVPKKLM, folic acid, phycoerythrobilin, phycocyanobilin, phycourobilin, nervonic acid, pavetannin C1, dieckol, eicosadienoic acid, peptide LDAVNR, β-carotene, astaxanthin, rutin, arachidonic acid, and dihomo-gamma-linolenic acid (Table 5). Binding scores went from − 7.9 to − 11.5 kcal.mol−1. Consensus docking method aimed to identify false-positive results. To do this, common molecules of both tops were selected and grouped in a unique list. Table 6 therefore presents the nine best molecules regarding their binding energy selected for the rest of the study, according to docking results. Among them can be found eight Arthrospira molecules: phycoerythrobilin, phycocyanobilin, dieckol, phycourobilin, folic acid, rutin, β-carotene, and astaxanthin. Pavetannin C1, which was a positive control found to bind with Spike RBD in a previous paper (Prasanth et al. 2020), also showed competitive values with low coefficients of variation (< 5%) in the present study, thus supporting experimental results. The 12 remaining compounds were considered as probably false-positive values. Nine common compounds stand out among the top 15 molecules with the best binding affinity to the RBD domain for Autodock Vina and SwissDock. The remaining 12 compounds from the two software programs are therefore considered false positives. These are catechin, rosmarinic acid, apigenin, remdesivir, and poriferasterol, which are ranked high with Autodock Vina, but have high binding energies in the case of the SwissDock models. Similarly, pGGTCVIRGCVPKKLM, nervonic acid, eicosadienoic acid and pLDAVNR, arachidonic acid, and dihomo-gamma-linolenic acid present in the top 15 molecules with Swissdock are considered poor candidates in the case of Autodock Vina because of their low binding energies (higher than − 5 kcal.mol−1). Analysis of specific interactions between Spike RBD and selected substances was then performed. Binding residues are recorded in Table 6.
Table 4.
15 best docking results for small molecules vs. Spike RBD of SARS-CoV-2 with Autodock Vina
| Binding energies (kcal.mol−1) | |||||
|---|---|---|---|---|---|
| Molecules for docking | Assay 1 | Assay 2 | Average | SD | CV (%) |
| Dieckol | − 8.1 | − 8.1 | − 8.1 | 0 | 0.0 |
| Phycoerythrobilin | − 7.4 | − 7.5 | − 7.45 | 0.05 | 0.7 |
| Pavetannin C1 | − 7.6 | − 7 | − 7.3 | 0.3 | 4.1 |
| Phycocyanobilin | − 7.4 | − 7.1 | − 7.25 | 0.15 | 2.1 |
| Rutin | − 7.2 | − 7.3 | − 7.25 | 0.05 | 0.7 |
| Catechin | − 7.2 | − 7.2 | − 7.2 | 0 | 0.0 |
| β-Carotene | − 7.1 | − 7.2 | − 7.15 | 0.05 | 0.7 |
| Phycourobilin | − 7.1 | − 7.1 | − 7.1 | 0 | 0.0 |
| Rosmarinic acid | − 7.1 | − 7.1 | − 7.1 | 0 | 0.0 |
| Epicatechin | − 7.1 | − 7.1 | − 7.1 | 0 | 0.0 |
| Astaxanthin | − 7 | − 7.1 | − 7.05 | 0.05 | 0.7 |
| Apigenin | − 7 | − 7 | − 7 | 0 | 0.0 |
| Folic acid | − 7 | − 6.9 | − 6.95 | 0.05 | 0.7 |
| Remdesivir | − 7 | − 6.8 | − 6.9 | 0.1 | 1.4 |
| Poriferasterol | − 6.7 | − 6.8 | − 6.75 | 0.05 | 0.7 |
SD: Standard deviation; CV: coefficient of variation
Table 5.
15 best docking results for small molecules vs. Spike RBD of SARS-CoV-2 with Swissdock
| Binding energies (kcal.mol−1) | |||||
|---|---|---|---|---|---|
| Molecules for docking | Assay 1 | Assay 2 | Average | SD | CV (%) |
| pGGTCVIRGCVPKKLM | − 11.57 | − 11.49 | − 11.53 | 0.04 | 0.3 |
| Folic acid | − 10.35 | − 10.35 | − 10.35 | 0 | 0.0 |
| Phycoerythrobilin | − 9.63 | − 9.11 | − 9.37 | 0.26 | 2.8 |
| Phycocyanobilin | − 9.38 | − 9.33 | − 9.355 | 0.025 | 0.3 |
| Phycourobilin | − 9.71 | − 8.86 | − 9.285 | 0.425 | 4.6 |
| Nervonic acid | − 8.48 | − 9.14 | − 8.81 | 0.33 | 3.7 |
| Pavetannin C1 | − 8.66 | − 8.38 | − 8.52 | 0.14 | 1.6 |
| Dieckol | − 8.33 | − 8.42 | − 8.375 | 0.045 | 0.5 |
| Eicosadienoic acid | − 8.71 | − 8.04 | − 8.375 | 0.335 | 4.0 |
| pLDAVNR | − 8.52 | − 8.05 | − 8.285 | 0.235 | 2.8 |
| β-Carotene | − 8.54 | − 8.01 | − 8.275 | 0.265 | 3.2 |
| Astaxanthin | − 7.82 | − 8.57 | − 8.195 | 0.375 | 4.6 |
| Rutin | − 8.08 | − 8.09 | − 8.085 | 0.005 | 0.1 |
| Arachidonic acid | − 7.96 | − 8.08 | − 8.02 | 0.06 | 0.7 |
| Dihomo-gamma-linolenic acid | − 7.96 | − 7.90 | − 7.93 | 0.03 | 0.4 |
SD: Standard deviation; CV: coefficient of variation
Table 6.
Docking results for small molecules vs Spike RBD of SARS-CoV-2
Underlined residue reflects unfavorable interactions between the molecule studied and the spike RBD, residues in bold are directly involved in the link between ACE2 and spike RBD. Stars were added to amino acids involved in one (*) or two (**) H-bond(s)
CV: Coefficient of variation; Stdev: Standard deviation
Phycoerythrobilin
Phycoerythrobilin binds to Spike RBD via six van der Waals interactions, with the following residues: LYS417, TYR453, GLN493, TYR495, PHE497, and GLN498. In addition, 2 alkyl and π-alkyl interactions engage TYR449 and LEU455, and a π-cation interaction binds phycoerythrobilin with ARG403 of Spike RBD. TYR505 is involved in two different kinds of bonds: one π–π T-shaped bond and one H-bond. Two other H-bonds link respectively GLY496 and ASN501 of Spike RBD. The last two H-bonds are located on the SER494 residue (Fig. 2). Thus, with three key binding amino acids involved in different interactions including two H-bonds on them, phycoerythrobilin shows interesting binding affinities from − 7.4 to − 9.63 kcal.mol−1 with coefficients of variation < 1% for AutodockVina and Swissdock, respectively. These results provide us very good arguments for deeper research on a phycoerythrobilin-based antiviral.
Phycocyanobilin
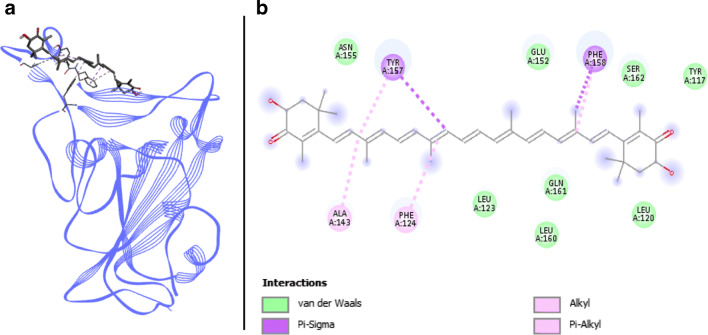
Phycocyanobilin/Spike RBD complex is composed of eight van der Waals interactions, with residues ARG403, TYR449, TYR453, GLN493, TYR495, PHE497, ASN501, and TYR505 (Fig. 3). Four H-bonds are involved between phycocyanobilin and Spike RBD, distributed between SER494 (two H-bonds), GLY496, and GLN498. GLY496 is also linked to phycocyanobilin via a π-donor hydrogen bond. Binding affinity for this compound reaches respectively − 7.4 and − 9.38 kcal.mol−1 with Autodock Vina and SwissDock. This compound does not seem to be impeded by unfavorable strength and its two H-bonds engaged with key binding residues highlight a good potential for antiviral activity.
Dieckol
Dieckol has seven van der Waals interactions spread on residues ARG403, TYR449, PHE456, GLN493, TYR495, PHE497, and GLN498. Two π-sigma, one π-alkyl as well as one π–π T-shaped bonds engage respectively LEU455, LYS417, and TYR505 (Fig. 4). The four present H-bonds involve key binding residues of Spike RBD namely TYR453, GLY496, THR500, and ASN501. Despite an unfavorable acceptor–acceptor interaction on GLU406, dieckol has six different bonds with Spike RBD and possesses the lowest binding energy in Autodock Vina program with 8.1 kcal.mol−1. SwissDock values are also low with −8.375 kcal.mol−1, thus positioning this molecule as one of the best candidates resulting from the present study.
Phycourobilin
Six van der Waals as well as five alkyl, π-alkyl, π-cation, and π–π T-shaped interactions link Spike RBD to phycourobilin, involving residues ARG403, GLU406, LYS417, TYR449, TYR453, LEU455, GLN493, TYR495, PHE497, GLN498, and TYR505. In addition, five H-bonds interact with the compound and SER494, GLY496, and ASN501 amino acids (Fig. 5). Three of them are directly in the binding pocket of Spike RBD, leading to great binding energies with − 7.1 kcal.mol−1 for Autodock Vina and − 9.3 kcal.mol−1 with SwissDock.
Folic acid
Three kinds of interactions were predicted between folic acid and Spike RBD. It accounts the greater number of H-bonds with a total of seven, engaged respectively with ARG403, LEU492, GLY496, ASN501, and TYR505. Among them, three link directly with key binding residues. Moreover, eight VDW and one carbon hydrogen bonds were also detected, in connection with residues TYR449, LEU452, TYR453, GLU484, PHE490, GLN493, SER494, TYR495, PHE497, and GLN498 (Fig. 6). Binding energies went from − 6.9 to − 10.35 kcal.mol−1 with both coefficients of variation lesser than 1%, making folic acid a reliable candidate for Covid-19 solutions.
Pavetannin C1
Pavetannin C1 presents seven VDW bonds as well as one carbon–hydrogen bond and one π–π T-shaped interaction with respectively ARG403, TYR449, PHE490, TYR495, PHE497, GLN498, ASN501, GLN493, and TYR505. The latter residue can also bind with pavetannin C1 thanks to a H-bond, with three others on LEU492, SER494, and GLY496 (Fig. 7). One of them is engaged with key binding residues, namely GLN496, and GLY493 is connected thanks to a carbon hydrogen bond. Binding scores go from − 7.3 to − 8.52 kcal.mol−1 with coefficients of variation larger than for other compounds but still lesser than 5%.
Rutin
Rutin binds to Spike RBD via seven van der Waals interactions, with the following residues: TYR449, TYR453, SER494, TYR495, PHE497, GLN498, and GLY502. In addition, six H-bonds link respectively ARG403, GLN493, GLY496, ASN501, and TYR505 of Spike RBD (Fig. 8). Those located on the GLN493, GLY496, and ASN501 residues belong to the binding pocket of Spike RBD (Table 3). Thus, with three key binding amino acids involved in four different H-bonds on them, rutin shows interesting binding affinities from − 7.25 to − 8.08 kcal.mol−1 with coefficients of variation < 1%. These results provide us good arguments for deeper research on this compound for the elaboration of a new antiviral.
β-Carotene
β-Carotene does not present any H-bond as interacting mode with Spike RBD. Nevertheless, it has many VDW bonds as well as four π-alkyl interactions. The 10 VDW interactions engage the following residues: TYR449, LEU452, LEU455, TYR473, ALA475, GLU484, ASN487, LEU492, GLN493, and SER494, whereas π-alkyl interactions are spread on PHE456, TYR489, and PHE490 residues. Binding energies are close to those obtained for rutin and for astaxanthin, with − 7.15 kcal.mol−1 with Autodock Vina and − 8.275 kcal.mol−1 for SwissDock.
Astaxanthin
In the same way as β-carotene, the algorithm does not predict any H-bond for astaxanthin. Astaxanthin/Spike RBD complex is then composed of eight VDW interactions, with residues TYR449, LEU452, LEU455, GLU484, ASN487, LEU492, GLN493, and SER494. Two π-sigma interactions are also involved between the protein and the ligand, on TYR489 and PHE490 residues. Finally, four alkyl and π-alkyl interactions are located on PHE456, ALA475, TYR489, and PHE490. Only ALA475 and TYR489 share bonds with astaxanthin among key binding residues. This compound therefore appears to have less strong bonds with Spike RBD, but it still has good binding energies. Indeed, as mentioned previously, binding energies calculated for astaxanthin are similar to those for β-carotene and rutin with values included between −7.05 and − 8.19 kcal.mol−1.
Toxicity assessment
After docking assays and binding interaction analysis, an in silico toxicity study was conducted on the top 10 molecules with AdmetSAR 2, SwissADME, and Osiris tools. Substances were analyzed regarding specific toxicity criteria including both general toxicity criteria (mutagenicity, acute oral toxicity, carcinogenicity, hepatotoxicity, and reprotoxicity), druglikeness parameters (GI absorption, BBB permeability, or Lipinski’s rule), and ability of each compound to inhibit human cytochromes, thus identifying possible risk of drug interactions. Each toxicity tool provided different types of results, which are compiled in Table 7. AdmetSAR 2 focused on toxicity risks, cytochrome inhibitions, and blood–brain barrier permeability. A positive or negative result was provided with a p value for each compound. When a negative result was obtained, corresponding to a low-risk probability, “low” was recorded in the table. In the same way, when a criterion responded positively but with a p value < 0.7, it was considered as not enough statistical significance so the risk was determined as “mid,” and finally, in case of positive response combined with a p value > 0.7, a high risk is registered in Table 7. For SwissADME, no p value was provided; results were treated on a binary way. Therefore, a positive result was associated with a “high” risk whereas a negative result gave a “low” risk. Finally, Osiris results consisted in a range of colors: red orange or green. These colors were gradually assigned to risk levels: red color, “high” risk/probability; orange color, “mid” risk/probability; and green color, “low” risk/probability. Some criteria were covered by two toxicity tools. In that case, the higher risk was kept and recorded in Table 7. Nevertheless, detailed data are available in Table 10 (Supplemental data).
Table 7.
Toxicity parameters of selected molecules obtained with AdmetSAR 2, SwissADME, and Osiris tools
| Astaxanthin | β-Carotene | Dieckol | Folic acid | Pavetannin C1 | PC | PE | PCOU | Rutin | |
|---|---|---|---|---|---|---|---|---|---|
| Toxicity risks | |||||||||
| Acute oral toxicity class | III | III | III | III | IV | III | III | III | III |
| Mutagenicity | Low | Low | Mid | Low | Mid | Low | Low | Low | High |
| Carcinogenicity | Low | Low | Low | Low | Low | Low | Low | Low | Low |
| Hepatotoxicity | Low | Mid | Mid | High | Mid | Mid | Mid | Mid | High |
| Reprotoxicity | High | Low | Low | Low | High | Low | Low | Low | Low |
| Absorption | |||||||||
| GI absorption | Low | Low | Low | Low | Low | Low | Low | Low | Low |
| BBB permeability | High | High | Low | High | Low | High | High | High | Low |
| Lipinski's rule | No | No | No | No | No | Yes | Yes | Yes | No |
| Drug likeness | High | Low | Low | Low | High | High | Mid | High | High |
| Inhibition risks | |||||||||
| CYP1A2 | Low | Low | High | Low | Low | Mid | Low | Low | Low |
| CYP2C19 | Low | Low | High | Low | Low | Low | Low | Low | Low |
| CYP2C9 | Low | Low | High | Low | High | Mid | Low | Low | Low |
| CYP2D6 | Low | Low | Low | Low | Low | Low | Low | Low | Low |
| CYP3A4 | Low | Low | Low | Low | Low | Low | Low | Low | Low |
| P-gp | High | High | High | Low | High | High | High | High | Low |
PC: phycocyanobilin, PE: phycoerythrobilin, PCOU: phycourobilin, GI: gastrointestinal, BBB: blood–brain barrier, CYP: cytochrome, P-gp: P-glycoprotein
All of the compounds presented acute oral toxicities included in the third class of toxicity except pavetannin C1 which belongs in the fourth class. The third class corresponds to LD50 that comprised between 500 and 5000 mg.kg−1, whereas the fourth class included LD50 greater than 5000 mg.kg−1. These ones are respectively equivalent to the fourth and fifth classes of toxicity in the Hodge and Sterner scale. Selected molecules can be therefore considered as safe for this criterion. Regarding other toxicity risks, none of the tested compounds presented carcinogenicity, but rutin responds positively to the mutagenicity and hepatotoxicity criteria with high probability levels. Hepatotoxicity risk is also detected for folic acid while astaxanthin and pavetannin C1 were predicted to be potential reprotoxic compounds. It is thus necessary to remain vigilant regarding these parameters.
Except folic acid and rutin, most of the compounds may cause P-glycoprotein inhibition, a major actor in drug metabolism (Rautio et al. 2006). Such inhibition may enhance bioavailability of drugs by accumulation in cells which, if it is not the wanted effect, could lead to toxic effects. Thus, particular attention should be paid to drug interactions with the seven concerned molecules, and more particularly with dieckol and pavetannin C1. Indeed, both substances show significant risk of interaction with human cytochromes CYP1A2, CYP2C19, and CYP2C9 for dieckol and CYP2C9 for pavetannin C1. Also involved in drug metabolism, interactions could modulate activation of other drugs, their absorption, or their elimination. Nevertheless, all the tested compounds showed no relevant probability of inhibition of cytochromes CYP2D6 and CYP3A4. The latter is the most represented of CYP450s in human, distributed in both hepatocytes and enterocytes. Involved in the metabolism of almost half of drugs, it is therefore essential (Martínez-Jiménez et al. 2007). Moreover, none of folic acid and rutin present significant risk of cytochrome inhibition which make these compounds safe for a potential administration in patients already consuming other treatments.
In another way, all the candidates were predicted to be absorbed by the gastrointestinal tract but at a low level. Six molecules showed favorable parameters regarding druglikeness, namely astaxanthin, pavetannin C1, phycocyanobilin, phycoerythrobilin, phycourobilin, and rutin. Among them, three compounds also responded to Lipinski’s rule: phycocyanobilin, phycoerythrobilin, and phycourobilin. Combined with their docking results, these candidates seem to be among the most relevant for the fight against SARS-CoV-2. Nevertheless, in vitro and in vivo studies remain essential to complete these results.
Discussion
In the global context of the Covid-19 pandemic, joint advances in research are essential. Spike protein, a major structural protein of the SARS-CoV-2, constitutes a privileged target for antiviral research, as the first interactions between virus and human host cell take place via the Spike protein RBD, interacting with human ACE2 receptor.
Taking advantage of our skills with in silico studies, we choose to apply this knowledge as a test in the Covid-19 frame. Using consensus docking and redocking with largely widespread software (Autodock Vina and SwissDock), 8 Arthrospira molecules among 48 were identified as potential candidates with antiviral activity against SARS-CoV-2: phycoerythrobilin, phycocyanobilin, dieckol, phycourobilin, folic acid, rutin, β-carotene, and astaxanthin. Thanks to these methods, false-positive results have been discarded to keep the most reliable results.
Phycocyanobilin, phycourobilin, and phycoerythrobilin are phycobilins. The tetrapyrrolic structure of these chromophores is involved in light harvesting of cyanobacteria such as Arthrospira (Akimoto et al. 2012; Rodrigues et al. 2018). Great binding energies were obtained for these compounds from Autodock Vina and SwissDock with respectively − 7.45 and − 10.35 kcal.mol−1, − 7.25 and − 9.355 kcal.mol−1, and − 7.1 and − 9.285 kcal.mol−1 for phycoerythrobilin, phycocyanobilin, and phycourobilin. Low toxicity risks were also predicted with in silico methods for these three compounds. Moreover, phycocyanin and phycoerythrin are authorized by both FDA and European Commission and are usually used as food coloring (Bratinova 2015), thus substantiating previous results. No clinical trial on the website ClinicalTrials.com or docking study directly reported the use of those compounds with the aim to cure or prevent viral infections, thus opening a new way of research. Phycocyanins are mainly known for their great antioxidant properties (Zhou et al. 2005; Wu et al. 2016). An in vitro study showed that phycocyanobilin represents the main Arthrospira compound responsible for its antioxidant effect (Hirata et al. 2000). Phycocyanobilin was shown to have a similar structure to biliverdin, an NADPH inhibitor (Zheng et al. 2013). Phycocyanobilin is converted into phycocyanorubin by biliverdin reductase (Terry et al. 1993) with a structure close to bilirubin (McCarty 2007). In other in vivo studies, both phycocyanin and phycocyanobilin were able to normalize oxidative stress markers and expression of components from NADPH pathway (Zheng et al. 2013). McCarty et al. (2010) suggested NADPH inhibition as a way of treatment in influenza viral infection. Indeed, it has been identified as the main oxidative stress source in lung epithelial cells, participating in viral symptoms. Thus, phycobilins and especially phycocyanobilin present additional arguments to support their antiviral potential.
One clinical study focuses on dieckol. This trial in progress relies on safety issues of several phlorotannins including dieckol (NCT04335045). It should provide further information to confirm or infirm our in silico toxicity assessment. Drug interaction study provided high risk of inhibition of several cytochromes. No scientific paper studied specific inhibition of CYP1A2, 2C9, and 2C19 by dieckol. However, Sadeeshkumar et al. (2017) found that dieckol could reduce activities of liver cytochromes P450 after being previously increased with N-nitrosodiethylamine. Antiviral potential of dieckol has been studied in several papers. Kwon et al. (2013) showed that this phlorotannin was able to block viral entry of porcine epidemic diarrhea virus via hemagglutination and replication inhibitions, with an IC50 of 14.6 μM. Kim (2013) reported an in vitro activity of phlorotannins, especially dieckol, against influenza virus neuraminidase with about 72% inhibition at 30 μg.L−1. Other studies highlighted its in vitro antiviral activity against both human papilloma virus and murine norovirus (Eom et al. 2015; Kim and Kwak 2015). Regarding coronaviruses, dieckol was submitted to an in vitro study wherein it showed an IC50 of 2.7 μM against 3CLPro of SARS-CoV-1 (Park et al. 2013. Moreover, in a recent docking paper using Autodock Vina, dieckol bonds the main protease (Mpro) of SARS-CoV-2 with a binding energy of − 12.0 kcal.mol−1, a greater result than those obtained with Autodock Vina and SwissDock on Spike RBD (− 8.1 and − 8.375 kcal.mol−1) (Gentile et al. 2020). Dieckol therefore arouses the interest of other researchers in the global context of Covid-19 and would be probably a more potent inhibitor against Mpro than Spike RBD but could be limited by its low bioavailability according to absorption prediction data.
Folic acid, also known as vitamin B9, is a largely used molecule, approved and under health claims by both FDA and EFSA (Electronic Code of Federal Regulation 2000; EFSA 2010). Unlike our absorption prediction results, EFSA reported a great bioavailability of different forms of folic acid (EFSA 2014). Indeed, folic acid is usually used during pregnancy to prevent the risk of neural tube defects (Greenberg et al. 2011). It is also widely used as concomitant treatment with methotrexate to offset its side effects including hepatotoxicity. Indeed, methotrexate (an antirheumatic drug) tends to exert a competitive activity against folic acid, necessitating a substantial supplementary administration of this folic acid (Shea et al. 2013). In this context, folic acid has been included in two clinical studies: one in HIV-infected patients taking both methotrexate and folic acid (Clinical trial no. NCT01949116) and a second one on hepatitis C–infected patients (Clinical trial no. NCT02150291). The latter studied how folic acid could act as prophylactic treatment in a viral infection to counteract side effects of the main treatment. It should be noted that predicted hepatotoxicity risk was not verified in literature. Coupled with the fact that folic acid is used to counteract the hepatotoxicity effects of methotrexate, this suggests hepatotoxicity risk is actually not relevant. Besides, folic acid provided strong results during docking assays, with binding energies of − 6.95 and − 10.35 kcal.mol−1 with Autodock Vina and SwissDock. These arguments are in favor of a safety and well-known consumption of folic acid which thus can be seriously considered as a candidate in the fight against Covid-19.
Rutin has already been studied for its potential as antiviral for SARS-CoV-2. Indeed, several docking papers mentioned its promising effect as inhibitor of some virus targets, especially against the main protease (Mpro). Das et al. (2020a) explored the efficiency of Calendula officinalis compounds as ligand for Mpro, using Autodock Vina. With a binding energy of − 8.8 kcal.mol−1, rutin was the best molecule of the assay. Similar results have been obtained in the scientific literature, still with Autodock Vina, with − 9.0 kcal.mol−1 (Huynh et al. 2020). Two other teams confirmed these results, within rutin remained the best tested compound among tens, including antivirals (Das et al. 2020b; Hu et al. 2020). Only one study (pending for publication) focused on the interaction between rutin and ACE2/Spike complex. Among an FDA-approved molecule list, rutin was ranked in the best results. Moreover, molecular dynamic simulation (MD) showed its ability to destabilize the ACE2/Spike complex, thus consolidating our results (binding energies of − 7.2 and − 8.085 kcal.mol−1 with Autodock Vina and SwissDock) (https://europepmc.org/article/ppr/ppr188118). Elsewhere, several studies showed in vitro effects of rutin against other viruses belonging to enterovirus, norovirus, or hepatitis virus families (Lin et al. 2012; Chéron et al. 2015; Parvez et al. 2019. These data highlight the rise of rutin in the field of antivirals. Regarding hepatotoxicity prediction, literature infirmed this result since several studies showed a protective effect of rutin on hepatotoxicity such as paracetamol-induced hepatotoxicity (Janbaz et al. 2002; Reddy et al. 2017). However, controversial studies underlined mutagenic activity of rutin and its main metabolite quercetin, corroborating in silico prediction of our study (Crebelli et al. 1987; Okamoto 2005). If the conclusion of the debate is still unclear, it is yet necessary to use this compound with precautions and further safety assessment would be required to clarify this point.
At this day, no clinical study assesses antiviral abilities of β-carotene and few papers mention it. Nevertheless, this molecule has been included in a placebo-controlled study where it was given in supplementation in HIV patients to analyze enzymatic antioxidant system response. Like selenium, also tested in the experiments, positive conclusions have been made with an improvement of antioxidant enzyme activities (Delmas-Beauvieux et al. 1996). More recently, a docking study has been conducted on interactions between this compound and hepatitis C virus (HCV). However, no interaction was highlighted with the active site of the HCV helicase (Kaneez et al. 2014). In SARS-CoV-2 context, Lianhua Qingwen Capsule (LQC) showed some therapeutic effects on Covid-19 patients (Li et al. 2020). In view to understand the molecular mechanism of such activity, docking analysis has been executed with Autodock Vina (Xia et al. 2020). Among the six active compounds of LQC with great binding energies, a − 6.5 kcal.mol−1 binding score was predicted for β-carotene, showing its ability to bind with the active pocket of Akt1. This human kinase has been suggested as a potential therapeutic target for Covid-19 treatment. Indeed, its overexpression could promote viral protein synthesis and thus help in virus spreading. Regarding Spike RBD interaction, β-carotene presented greater binding energies than with Akt1, with − 7.15 and − 8275 kcal.mol−1, suggesting a complementary activity of this molecule on the two targets. As folic acid previously mentioned, β-carotene is well known as a nutritive molecule and precursor of vitamin A. Used in plenty of clinical trials and widely commercialized as a dietary supplement, β-carotene consumption is generally assumed as safe. Nevertheless, EFSA highlighted some studies with preoccupying side effects on the lung, especially in heavy smokers (EFSA 2006). Moreover, according to our experiments and literature, this compound shows a low bioavailability due to its long carbon chain (EFSA 2006). Thus, an important dose would be required for this compound to act as antiviral compound and inhibit SARS-CoV-2 Spike protein.
Astaxanthin is not a well-documented substance regarding antiviral activities since no clinical study deals with this topic. Indeed, astaxanthin is mainly known for its antioxidant and anti-inflammatory properties. In this way, it has been included in two clinical trials (NCT03945526 and NCT03906825). The first one is related to effects of astaxanthin on antioxidant parameters in stroke patients, whereas the second one relied on effects of a dietary supplement containing astaxanthin on inflammation markers (Birudaraju et al. 2020). Talukdar et al. (2020a) brought arguments to consider this compound as a serious natural molecule to alleviate the risk of cytokine storm in some Covid-19 patients thanks to its anti-inflammatory properties. Elsewhere, in a study conducted in vitro on human papilloma virus (HPV), astaxanthin prevented HPV L1 protein (capsid protein) by binding on human sperm membrane. Such antiviral activity could protect men from idiopathic infertility caused by HPV (Donà et al. 2018). In silico toxicity study highlighted a high risk of reprotoxicity, but this has not been verified in literature. Indeed, acute and subchronic toxicity studies showed no adverse effects of astaxanthin (Stewart et al. 2008; Katsumata et al. 2014) and no reprotoxicity study has been conducted yet to our knowledge. Despite interesting binding energies (− 7.05 and − 8.195 kcal.mol−1), astaxanthin is, such as β-carotene, a low bioavailable molecule (Mercke Odeberg et al. 2003). For the same reasons previously mentioned, these data therefore suggest that astaxanthin would be more effective as an anti-inflammatory agent than a direct antiviral compound in Covid-19 treatment.
Many articles referred to docking techniques in view to assess the potential of a plethora of molecules as inhibitors against different SARS-CoV-2 targets (Elfiky 2020; Quimque et al. 2020; Ton et al. 2020). Nevertheless, except for preprint publications, few papers refer to in silico research of Spike RBD inhibitors. Sinha et al. (2020a) studied interactions of saikosaponins, bioactive molecules from Traditional Chinese Medicine plants, with the entire Spike protein in open conformation. Their results highlighted three saikosaponins (V, U, C) showing binding energies that comprised between − 7.2 and −8.4 kcal.mol−1. Excepting saikosaponin V which interacted with one key binding residue of the Spike protein/ACE2 link (475), none of the two others interacted with key binding residues involved in this critical bind. Another publication presented hesperidin as the only potent inhibitor of Spike/ACE2 interface despite a quite large list of compounds with high binding energies (Wu et al. 2020). This result was consolidated by another article pending publication where hesperidin has been tested on a closed state of Spike protein (https://www.preprints.org/manuscript/202004.0102/v3). The ligand obtained a binding energy of − 10.4 kcal.mol−1 with Autodock Vina software. Nevertheless, this compound, as mentioned by the authors, is characterized by a low oral bioavailability and then does not constitute the ideal candidate for a new antiviral development.
SARS-CoV-2 can recognize several cellular receptors such as ACE2 or HSPA5 (Heat Shock Protein A5) also known as GRP78. Souza et al. (2020) found several compounds among estrogenic compounds capable of competing with SARS-CoV-2 on the substrate binding domain of HSPA5. Autodock Vina provided great binding energies of about − 7.8 kcal.mol−1 for some compounds. Nevertheless, phytoestrogens are controversial molecules, suspected of being involved in some estrogen-dependent cancers (Kwack et al. 2009; Touillaud et al. 2019). Moreover, consequences of human receptor inhibition are unknown, which is why we preferred working on viral protein instead of human targets.
Glycyrrhizic acid has been elsewhere identified after a docking assay with Vina on Spike protein, with a − 9.2 kcal.mol−1 binding energy (Sinha et al. 2020b). Despite this promising result, analysis of binding residues showed that engaged amino acids were not located on key binding residues. Indeed, they were positioned on HIS625, GLU298, ARG319, GLN321, VAL320, TYR343, THR323, ILE624, and ASN317 and thus do not belong to the Spike RBD (i.e., residues 333–527). This suggests that such interactions between glycyrrhizic acid and Spike may not impede the link between Spike and ACE2.
Spike RBD has been isolated from the Spike/ACE2 complex for docking studies in two papers, thus proposing a similar method to this publication. The first one analyzed essential oil components and no compounds showed binding energy greater than − 5.5 kcal.mol−1. Moreover, few H-bonds were predicted and no key binding residues were engaged (Kulkarni et al. 2020). Schrödinger software was used in the second study, focused on Indian ginger compounds (Chikhale et al. 2020). The authors highlighted the potential of QGRG (quercetin-3-o-galactosyl-rhamnosyl-glucoside) for Spike RBD inhibition, with a − 9.25 kcal.mol−1 binding score. This compound presented three H-bonds with Spike RBD, located on GLU340, GLN245, and VAL339 which are not among the key binding residues, suggesting better candidates could be found for Spike RBD inhibition. During cryo-electron microscopy analysis, Toelzer et al. (2020) found that linoleic acid was bound to the Spike RBD in the closed conformation. The authors hypothesized that the fatty acid could stabilize the closed state of RBD and thus limiting ACE2 binding for which open state is required. In the same study, the antiviral remdesivir was able to inhibit RNA polymerase of SARS-CoV-2 in vitro. Because of possible adverse effects, WHO does not recommend conducting clinical trials on hospitalized patients who are weakened. However, clinical trials are still encouraged in populations with non-severe symptoms to identify effective antiviral therapy. As such, remdesivir has been included in several clinical trials (NCT04365725, NCT04582266, NCT04431453). To analyze the potential of linoleic acid as inhibitor of Spike RBD in an open state as well as the impact of an important antiviral molecule, both compounds were added to our docking study. Results revealed that linoleic acid weakly bonded with Spike RBD, with binding energies of − 4.85 and − 7.025 kcal.mol−1 via Autodock Vina and SwissDock (Table 8 Supplemental Data). This suggests the exposition of key binding residues is modified between the two conformational states and is therefore no more favorable for linoleic acid binding. This corroborates observations of Toelzer since no fatty acid has been recovered in the open state during their study. Regarding remdesivir docking, moderate binding affinities were obtained with respectively − 6.9 and − 8.02 kcal.mol−1 with Autodock Vina and SwissDock (Table 8 Supplemental Data). Chikhale et al. (2020) also included remdesivir in their docking study with Schrodinger software. Similar to our results, it presented a low/moderate binding energy of − 4.65 kcal.mol−1, thus suggesting that its antiviral activity is not based on Spike RBD inhibition.
Pavetannin C1 has been confirmed twice by docking assays as potential SARS-CoV-2 antiviral and has been thus chosen as positive control for the present study. This compound has been highlighted with MPro bonding abilities (− 9.5 kcal.mol−1 with Autodock Vina) in a study on phytochemicals from Indian cuisine. Rajendran et al. (2020). Another study presented pavetannin C1 as a great Spike inhibitor (Prasanth et al. 2020). Docking assay with Autodock 4.0 provided binding energy of − 11.1 kcal.mol−1 and different interactions were predicted with Spike RBD region. Indeed, four hydrophobic interactions engaged THR347, TRP349, GLU398, and TYR510, as well as nine H-bonds located on residues SER44, HIS345, ALA348, ASP350, GLU375, GLY395, ASN397, ARG514, and TYR515. However, none of the detected interactions engaged key binding residues. Results obtained with our experiments provided binding energies of − 7.3 and − 8.52 kcal.mol−1 with Autodock Vina and SwissDock and allowed to select this compound among the best candidates for antiviral development, thus corroborating literature results. Moreover, two interactions including one H-bond was predicted on key binding residues (GLN493 and GLY496). The slight variation of binding energies with our study could be explained in part by the use of different docking programs. Moreover, in Prasanth’s work, docking has been executed with the same initial PDB file: 6LZG as in our work, but Spike RBD has not been isolated and thus remained complexed with human ACE2. This could explain the difference between predicted interactions. Pavetannin C1 could not bind directly key binding residues as they were still bonded with human ACE2. However, despite promising binding scores, little is known about this molecule. Indeed, no toxicity data would allow us to infirm or confirm toxicity prediction, especially regarding reprotoxicity and cytochrome interactions. It would be therefore wise to promote molecules on which with have more hindsight.
On the basis of these in silico results, our work provides a list of four reliable compounds with potential antiviral activity via Spike RBD inhibition (phycoerythrobilin, phycocyanobilin, phycourobilin, and folic acid). Interactions such as H-bonds with RBD key binding residues constitute the strength of these results since such bonds could destabilize or impede the link between SARS-CoV-2 and one of its human receptors, ACE2. It will be more interesting to follow the future work that will deal with the interaction of our four molecules in viral episodes and further in silico studies will improve and guide bioactive molecule selection for antiviral compound research.
Conclusion
Arthrospira is one of the most well-known genera in the world of microalgae. In several articles reviewed here, Arthrospira or Arthrospira extracts presented significant antiviral activities against many virus families. In the context of the Covid-19 pandemic, we were interested in the potential of this microalgae to fight against SARS-CoV-2. The present study considered docking techniques on a selection of 48 Arthrospira components which were submitted to two different docking software (Autodock Vina and SwissDock). The main purpose of the present paper was to test their ability to inhibit Spike protein, a structural protein of the virus. In a second time, in silico toxicity analysis has been conducted to assess the safety of selected compounds.
After docking assays, eight Arthrospira molecules have been identified for their great aptitude to link with Spike RBD. Among them, four provided good arguments to be considered for an oral administration according to absorption prediction and existing literature. These compounds were namely phycoerythrobilin, phycocyanobilin, phycourobilin, and folic acid. The other compounds have been discarded due to their low oral bioavailability or potential toxicity risks. Consensus docking and redocking methods allowed us to obtain reliable results by eliminating false-positive results. Our results were therefore consistent with those currently found in the literature.
Our study provides promising arguments for the inclusion of these four compounds in further studies to assess their ability to compete with the SARS-CoV-2/ACE2 complex both in vitro and in vivo.
Supplementary Information
(DOCX 147 kb)
Footnotes
The original online version of this article was revised: Table 6 in the original version of this article contained an error. The corrected table and table legend are presented below.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/10/2021
A Correction to this paper has been published: 10.1007/s10811-021-02476-2
References
- Abdo SM, Hetta MH, El-Senousy WM, El Din RAS, Ali GH. Antiviral activity of freshwater algae. J Appl Pharm Sci. 2012;02:21–25. [Google Scholar]
- Akimoto S, Yokono M, Hamada F, Teshigahara A, Aikawa S, Kondo A. Adaptation of light-harvesting systems of Arthrospira platensis to light conditions, probed by time-resolved fluorescence spectroscopy. Biochim Biophys Acta BBA - Bioenerg. 2012;1817:1483–1489. doi: 10.1016/j.bbabio.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Anandakrishnan R, Aguilar B, Onufriev AV. H++ 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012;40:W537–W541. doi: 10.1093/nar/gks375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayehunie S, Belay A, Baba TW, Ruprecht RM. Inhibition of HIV-1 replication by an aqueous extract of Spirulina platensis (Arthrospira platensis) J Acquir Immune Defic Syndr. 1998;18:7–12. doi: 10.1097/00042560-199805010-00002. [DOI] [PubMed] [Google Scholar]
- Azabji-Kenfack M, Dikosso SE, Loni EG, Onana EA, Sobngwi E, Gbaguidi E, Kana AL, Nguefack-Tsague G, Von der Weid D, Njoya O, Ngogang J, et al. Potential of Spirulina platensis as a nutritional supplement in malnourished HIV-infected adults in Sub-Saharan Africa: a randomised, single-blind study. Nutr Metab Insights. 2011;4:29–37. doi: 10.4137/NMI.S5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birudaraju D, Cherukuri L, Kinninger A, Chaganti BT, Shaikh K, Hamal S, Flores F, Roy SK, Budoff MJ. A combined effect of cavacurcumin, eicosapentaenoic acid (omega-3s), astaxanthin and gamma-linoleic acid (omega-6) (CEAG) in healthy volunteers – a randomized, double-blind, placebo-controlled study. Clin Nutr ESPEN. 2020;35:174–179. doi: 10.1016/j.clnesp.2019.09.011. [DOI] [PubMed] [Google Scholar]
- Böttcher E, Matrosovich T, Beyerle M, Klenk HD, Garten W, Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol. 2006;80:9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratinova S (2015) Provision of scientific and technical support with respect to the classification of extracts/concentrates with colouring properties either as food colours (food additives falling under Regulation (EC) No 1333/2008) or colouring foods. Report EUR 27425 EN, Joint Research Centre – Institute for Reference Material and Measurement, European Comission, Louxembourg. 83 pp
- Chen CC, Hsu LW, Nakano T, Huang KT, Chen KD, Lai CY, Goto S, Chen CL. DHL-HisZn, a novel antioxidant, enhances adipogenic differentiation and antioxidative response in adipose-derived stem cells. Biomed Pharmacother. 2016;84:1601–1609. doi: 10.1016/j.biopha.2016.10.066. [DOI] [PubMed] [Google Scholar]
- Chéron N, Yu C, Kolawole AO, Shakhnovich EI, Wobus CE. Repurposing of rutin for the inhibition of norovirus replication. Arch Virol. 2015;160:2353–2358. doi: 10.1007/s00705-015-2495-y. [DOI] [PubMed] [Google Scholar]
- Chikhale RV, Gurav SS, Patil RB, Sinha SK, Prasad SK, Shakya A, Shrivastava SK, Gurav NS, Prasad R Sal (2020) Sars-cov-2 host entry and replication inhibitors from Indian ginseng: an in-silico approach. J Biomol Struct Dyn. 10.1080/07391102.2020.1778539 [DOI] [PMC free article] [PubMed]
- Chirasuwan N, Chaiklahan R, Kittakoop P, Chanasattru W, Ruengjitchatchawalya M, Tanticharoen M, Bunnag B. Anti HSV-1 activity of sulphoquinovosyl diacylglycerol isolated from Spirulina platensis. ScienceAsia. 2009;35:137–141. doi: 10.2306/scienceasia1513-1874.2009.35.137. [DOI] [Google Scholar]
- Choi S-Y, Bertram S, Glowacka I, Park YW, Pöhlmann S. Type II transmembrane serine proteases in cancer and viral infections. Trends Mol Med. 2009;15:303–312. doi: 10.1016/j.molmed.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Crebelli R, Aquilina G, Falcone E, Carere A. Urinary and faecal mutagenicity in Sprague-Dawley rats dosed with the food mutagens quercetin and rutin. Food Chem Toxicol. 1987;25:9–15. doi: 10.1016/0278-6915(87)90301-2. [DOI] [PubMed] [Google Scholar]
- Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- Das P, Majumder R, Mandal M, Basak P (2020a) In-Silico approach for identification of effective and stable inhibitors for COVID-19 main protease (Mpro) from flavonoid based phytochemical constituents of Calendula officinalis. J Biomol Struct Dyn 0:1–16. [DOI] [PMC free article] [PubMed]
- Das S, Sarmah S, Lyndem S, Roy AS (2020b) An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J Biomol Struct Dyn 0:1–11 [DOI] [PMC free article] [PubMed]
- de la Jara A, Ruano-Rodriguez C, Polifrone M, Assunçao P, Brito-Casillas Y, Wägner AM, Serra-Majem L. Impact of dietary Arthrospira (Spirulina) biomass consumption on human health: main health targets and systematic review. J Appl Phycol. 2018;30:2403–2423. doi: 10.1007/s10811-018-1468-4. [DOI] [Google Scholar]
- Delmas-Beauvieux MC, Peuchant E, Couchouron A, Constans J, Sergeant C, Simonoff M, Pellegrin JL, Leng B, Conri C, Clerc M. The enzymatic antioxidant system in blood and glutathione status in human immunodeficiency virus (HIV)-infected patients: effects of supplementation with selenium or beta-carotene. Am J Clin Nutr. 1996;64:101–107. doi: 10.1093/ajcn/64.1.101. [DOI] [PubMed] [Google Scholar]
- Deyab M, Mofeed J, El-Bilawy E, Ward F. Antiviral activity of five filamentous cyanobacteria against coxsackievirus B3 and rotavirus. Arch Microbiol. 2020;202:213–223. doi: 10.1007/s00203-019-01734-9. [DOI] [PubMed] [Google Scholar]
- Donà G, Andrisani A, Tibaldi E, Brunati AM, Sabbadin C, Armanini D, Ambrosini G, Ragazzi E, Bordin L. al (2018) Astaxanthin prevents human papillomavirus L1 protein binding in human sperm membranes. Mar Drugs 16:427. [DOI] [PMC free article] [PubMed]
- Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R, Breitbart RE, Acton S. A novel angiotensin-converting enzyme–related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res. 2000;87:e1–e9. doi: 10.1161/01.RES.87.5.e1. [DOI] [PubMed] [Google Scholar]
- EFSA (2006) Tolerable upper intake levels for vitamins and minerals. https://www.efsa.europa.eu/sites/default/files/efsa_rep/blobserver_assets/ndatolerableuil.pdf
- EFSA (2010) Scientific Opinion on the substantiation of health claims related to folate and contribution to normal psychological functions (ID 81, 85, 86, 88), maintenance of normal vision (ID 83, 87), reduction of tiredness and fatigue (ID 84), cell division (ID 195. EFSA J 8 (10):1760
- EFSA (2014) Scientific Opinion on Dietary Reference Values for folate. EFSA J 12:3893
- El-Baz FK, El-Senousy WM, El-Sayed AB, Kamel MM (2013) In vitro antiviral and antimicrobial activities of Spirulina platensis extract. J Appl Pharm Sci:052–056
- Electronic Code of Federal Regulations (ECFR) (2000), 65 FR 58918 §101.79
- Elfiky AA. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom S-H, Moon S-Y, Lee D-S, Kim H-J, Park K, Lee E-W, Kim T-H, Chung Y-H, Lee M-S, Kim Y-M. In vitro antiviral activity of dieckol and phlorofucofuroeckol-A isolated from edible brown alga Eisenia bicyclis against murine norovirus. Algae. 2015;30:241–246. doi: 10.4490/algae.2015.30.3.241. [DOI] [Google Scholar]
- Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016;11:905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile D, Patamia V, Scala A, Sciortino MT, Piperno A, Rescifina A. Putative inhibitors of SARS-CoV-2 main protease from a library of marine natural products: a virtual screening and molecular modeling study. Mar Drugs. 2020;18:225. doi: 10.3390/md18040225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomaa M, El-Sheekh MM, El-Shafey AS, Metwally MAA, El-Shanshory MR, Eid MAW. Immunological studies in Egyptian thalassemic children infected with hepatitis C virus. Am J Microbiol Biotechnol. 2017;4:20–26. [Google Scholar]
- Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005;33:W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JA, Bell SJ, Guan Y, Yu Y. Folic acid supplementation and pregnancy: more than just neural tube defect prevention. Rev Obstet Gynecol. 2011;4:52–59. [PMC free article] [PubMed] [Google Scholar]
- Grosdidier A, Zoete V, Michielin O. Fast docking using the CHARMM force field with EADock DSS. J Comput Chem. 2011;32:2149–2159. doi: 10.1002/jcc.21797. [DOI] [PubMed] [Google Scholar]
- Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–W277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M, Song W, Zhou H, Xu J, Chen S, Xiang Y, Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I, Timens W, Bulthuis MLC, Lely AT, Navis GJ, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Corona A, Nieves I, Meckes M, Chamorro G, Barron BI. Antiviral activity of Spirulina maxima against herpes simplex virus type 2. Antiviral Res. 2002;56:279–285. doi: 10.1016/S0166-3542(02)00132-8. [DOI] [PubMed] [Google Scholar]
- Hetta M, Mahmoud R, El-Senousy W, Ibrahim M, El-Taweel G, Ali G. Antiviral and antimicrobial activities of Spirulina platensis. World J Pharm Pharm Sci. 2014;3:31–39. [Google Scholar]
- Hirata T, Tanaka M, Ooike M, Tsunomura T, Sakaguchi M. Antioxidant activities of phycocyanobilin prepared from Spirulina platensis. J Appl Phycol. 2000;12:435–439. doi: 10.1023/A:1008175217194. [DOI] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271-280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Cai X, Song X, Li C, Zhao J, Luo W, Zhang Q, Ekumi IO, He Z. Possible SARS-coronavirus 2 inhibitor revealed by simulated molecular docking to viral main protease and host toll-like receptor. Future Virol. 2020;15:359–368. doi: 10.2217/fvl-2020-0099. [DOI] [Google Scholar]
- Huang S-Y, Grinter SZ, Zou X. Scoring functions and their evaluation methods for protein–ligand docking: recent advances and future directions. Phys Chem Chem Phys. 2010;12:12899–12908. doi: 10.1039/c0cp00151a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T, Wang H, Luan B. Structure-based lead optimization of herbal medicine rutin for inhibiting SARS-CoV-2’s main protease. Phys Chem Chem Phys. 2020;22:25335–25343. doi: 10.1039/D0CP03867A. [DOI] [PubMed] [Google Scholar]
- Janbaz KH, Saeed SA, Gilani AH. Protective effect of rutin on paracetamol- and CCl4-induced hepatotoxicity in rodents. Fitoterapia. 2002;73:557–563. doi: 10.1016/S0367-326X(02)00217-4. [DOI] [PubMed] [Google Scholar]
- Jang I-S, Park SJ. A Spirulina maxima-derived peptide inhibits HIV-1 infection in a human T cell line MT4. Fish Aquat Sci. 2016;19:37. doi: 10.1186/s41240-016-0039-3. [DOI] [Google Scholar]
- Kaneez F, Shilu M, Mohd S, Ashraf A, Ghazi D, Esam A, Ishtiaq Q, Shama A (2014) Docking studies of Pakistani HCV NS3 helicase: a possible antiviral drug target. PLoS One 9(9):e106339 [DOI] [PMC free article] [PubMed]
- Katsumata T, Ishibashi T, Kyle D (2014) A sub-chronic toxicity evaluation of a natural astaxanthin-rich carotenoid extract of Paracoccus carotinifaciens in rats. Toxicol Rep 1:582–588 [DOI] [PMC free article] [PubMed]
- Kim S-K, editor. Marine biomaterials: characterization, isolation and applications. Boca Raton: CRC Press; 2013. [Google Scholar]
- Kim EB, Kwak JH. Antiviral phlorotannin from Eisenia bicyclis against human papilloma virus in vitro. Planta Med. 2015;81:PW_22. doi: 10.1055/s-0035-1565646. [DOI] [Google Scholar]
- Kok YY, Chu WL, Phang SM, Mohamed SM, Naidu R, Lai PJ, Ling SN, Mak JW, Lim PK, Balraj P, Khoo AS, et al. Inhibitory activities of microalgal extracts against Epstein-Barr virus DNA release from lymphoblastoid cells. J Zhejiang Univ Sci B. 2011;12:335–345. doi: 10.1631/jzus.B1000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni SA, Nagarajan SK, Ramesh V, Palaniyandi V, Selvam SP, Madhavan T. Computational evaluation of major components from plant essential oils as potent inhibitors of SARS-CoV-2 spike protein. J Mol Struct. 2020;1221:128823. doi: 10.1016/j.molstruc.2020.128823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwack SJ, Kim KB, Kim HS, Yoon KS, Lee BM. Risk assessment of soybean-based phytoestrogens. J Toxicol Environ Health A. 2009;72:1254–1261. doi: 10.1080/15287390903212212. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Ryu YB, Kim YM, Song N, Kim CY, Rho MC, Jeong JH, Cho KO, Lee WS, Park SJ. In vitro antiviral activity of phlorotannins isolated from Ecklonia cava against porcine epidemic diarrhea coronavirus infection and hemagglutination. Bioorg Med Chem. 2013;21:4706–4713. doi: 10.1016/j.bmc.2013.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lee J-B, Hou X, Hayashi K, Hayashi T. Effect of partial desulfation and oversulfation of sodium spirulan on the potency of anti-herpetic activities. Carbohydr Polym. 2007;69:651–658. doi: 10.1016/j.carbpol.2007.01.024. [DOI] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li LC, Zhang ZH, Zhou WC, Chen J, Jin HQ, Fang HM, Chen Q, Jin YC, Qu J, Kan LD. Lianhua Qingwen prescription for Coronavirus disease 2019 (COVID-19) treatment: advances and prospects. Biomed Pharmacother. 2020;130:110641. doi: 10.1016/j.biopha.2020.110641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YJ, Chang YC, Hsiao NW, Hsieh JL, Wang CY, Kung SH, Tsai FJ, Lan YC, Lin CW. Fisetin and rutin as 3C protease inhibitors of enterovirus A71. J Virol Methods. 2012;182:93–98. doi: 10.1016/j.jviromet.2012.03.020. [DOI] [PubMed] [Google Scholar]
- Madu IG, Roth SL, Belouzard S, Whittaker GR. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Jiménez CP, Jover R, Donato MT, et al. Transcriptional regulation and expression of CYP3A4 in hepatocytes. Curr Drug Metab. 2007;8:185–194. doi: 10.2174/138920007779815986. [DOI] [PubMed] [Google Scholar]
- McCarty MF. Clinical potential of Spirulina as a source of phycocyanobilin. J Med Food. 2007;10:566–570. doi: 10.1089/jmf.2007.621. [DOI] [PubMed] [Google Scholar]
- McCarty MF, Barroso-Aranda J, Contreras F (2010) Practical strategies for targeting NF-kappaB and NADPH oxidase may improve survival during lethal influenza epidemics. Med Hypotheses 74:18–20 [DOI] [PubMed]
- Mercke Odeberg J, Lignell Å, Pettersson A, Höglund P. Oral bioavailability of the antioxidant astaxanthin in humans is enhanced by incorporation of lipid based formulations. Eur J Pharm Sci. 2003;19:299–304. doi: 10.1016/S0928-0987(03)00135-0. [DOI] [PubMed] [Google Scholar]
- Millet JK, Séron K, Labitt RN, Danneels A, Palmer KE, Whittaker GR, Dubuisson J, Belouzard S. Middle East respiratory syndrome coronavirus infection is inhibited by griffithsin. Antiviral Res. 2016;133:1–8. doi: 10.1016/j.antiviral.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimouni V, Ulmann L, Pasquet V, Mathieu M, Picot L, Bougaran G, Cadoret JP, Morant-Manceau A, Schoefs B. The potential of microalgae for the production of bioactive molecules of pharmaceutical interest. Curr Pharm Biotechnol. 2012;13:2733–2750. doi: 10.2174/138920112804724828. [DOI] [PubMed] [Google Scholar]
- Myers J, Grothaus G, Narayanan S, Onufriev A. A simple clustering algorithm can be accurate enough for use in calculations of pKs in macromolecules. Proteins Struct Funct Bioinformat. 2006;63:928–938. doi: 10.1002/prot.20922. [DOI] [PubMed] [Google Scholar]
- Ngo-Matip ME, Pieme CA, Azabji-Kenfack M, Moukette BM, Korosky E, Stefanini P, Ngogang JY, Mbofung CM. Impact of daily supplementation of Spirulina platensis on the immune system of naïve HIV-1 patients in Cameroon: a 12-months single blind, randomized, multicenter trial. Nutr J. 2015;14:70. doi: 10.1186/s12937-015-0058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NT, Nguyen TH, Pham TNH, Huy NT, Bay MV, Pham MQ, Nam PC, Vu VV, Ngo ST. Autodock Vina adopts more accurate binding poses but Autodock4 forms better binding affinity. J Chem Inf Model. 2020;60:204–211. doi: 10.1021/acs.jcim.9b00778. [DOI] [PubMed] [Google Scholar]
- O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchinson GR. Open Babel: an open chemical toolbox. J Cheminformatics. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T. Safety of quercetin for clinical application (review) Int J Mol Med. 2005;16:275–278. [PubMed] [Google Scholar]
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Kim JH, Kwon JM, Kwon HJ, Jeong HJ, Kim YM, Kim D, Lee WS, Ryu YB. Dieckol, a SARS-CoV 3CLpro inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorg Med Chem. 2013;21:3730–3737. doi: 10.1016/j.bmc.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez MK, Tabish Rehman M, Alam P, Al-Dosari MS, Alqasoumi SI, Alajmi M. Plant-derived antiviral drugs as novel hepatitis B virus inhibitors: cell culture and molecular docking study. Saudi Pharm J. 2019;27:389–400. doi: 10.1016/j.jsps.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal E, Sundararaj R. Algae: a potential source to prevent and cure the novel coronavirus – a review. Int J Emerg Technol. 2020;11:479–483. [Google Scholar]
- Prasanth DSNBK, Murahari M, Chandramohan V, Panda SP, Atmakuri LR, Guntupalli C. In silico identification of potential inhibitors from Cinnamon against main protease and spike glycoprotein of SARS CoV-2. J Biomol Struct Dyn. 2020;0:1–15. doi: 10.1080/07391102.2020.1779129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quimque MTJ, Notarte KIR, Fernandez RAT, Mendoza MAO, Liman RAD, Lim JAK, Pilapil LAE, Ong JKH, Pastrana AM, Khan A, Wei DQ, Macabeo APG (2020) Virtual screening-driven drug discovery of SARS-CoV2 enzyme inhibitors targeting viral attachment, replication, post-translational modification and host immunity evasion infection mechanisms. J Biomol Struct Dyn:1–18 [DOI] [PMC free article] [PubMed]
- Radonic A, Thulke S, Achenbach J, Kurth A, Vreemann A, König T, Walter C, Possinger K, Nitsche A. Anionic polysaccharides from phototrophic microorganisms exhibit antiviral activities to Vaccinia virus. J Antivir Antiretrovir. 2010;02:051–055. doi: 10.4172/jaa.1000023. [DOI] [Google Scholar]
- Rajendran M, Roy S, Ravichandran K, Mishra B, Gupta DK, Nagarajan S, Arul Selvaraj RC, Provaznik I (2020) In silico screening and molecular dynamics of phytochemicals from Indian cuisine against SARS-CoV-2 MPro. J Biomol Struct Dyn. 10.1080/07391102.2020.1845980 [DOI] [PubMed]
- Ramachandran S, Kota P, Ding F, Dokholyan NV. Automated minimization of steric clashes in protein structures. Proteins Struct Funct Bioinformat. 2011;79:261–270. doi: 10.1002/prot.22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautio J, Humphreys JE, Webster LO, Balakrishnan A, Keogh JP, Kunta JR, Serabjit-Singh CJ, Polli JW. In vitro P-glycoprotein inhibition assays for assessment of clinical drug interaction potential of new drug candidates: a recommendation for probe substrates. Drug Metab Dispos. 2006;34:786–792. doi: 10.1124/dmd.105.008615. [DOI] [PubMed] [Google Scholar]
- Rechter S, König T, Auerochs S, Thulke S, Walter H, Dörnenburg H, Walter C, Marschall M. Antiviral activity of Arthrospira-derived spirulan-like substances. Antiviral Res. 2006;72:197–206. doi: 10.1016/j.antiviral.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Reddy MK, Reddy AG, Kumar BK, Madhuri D, Boobalan G, Reddy M. Protective effect of rutin in comparison to silymarin against induced hepatotoxicity in rats. Vet World. 2017;10:74–80. doi: 10.14202/vetworld.2017.74-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues RDP, de Castro FC, de Santiago-Aguiar RS, Rocha MVP. Ultrasound-assisted extraction of phycobiliproteins from Spirulina (Arthrospira) platensis using protic ionic liquids as solvent. Algal Res. 2018;31:454–462. doi: 10.1016/j.algal.2018.02.021. [DOI] [Google Scholar]
- Sadeeshkumar V, Duraikannu A, Ravichandran S, Kodisundaram P, Fredrick WS, Gobalakrishnan R. Modulatory efficacy of dieckol on xenobiotic-metabolizing enzymes, cell proliferation, apoptosis, invasion and angiogenesis during NDEA-induced rat hepatocarcinogenesis. Mol Cell Biochem. 2017;433:195–204. doi: 10.1007/s11010-017-3027-8. [DOI] [PubMed] [Google Scholar]
- Shalaby EA, Shanab SMM. Salt stress enhancement of antioxidant and antiviral efficiency of Spirulina platensis. J Med Plants Res. 2010;4:2622–2632. doi: 10.5897/JMPR09.300. [DOI] [Google Scholar]
- Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharaf M, Amara A, Aboul-Enein A, Helmi S, Ballot A, Schnitzler P (2013) Antiherpetic efficacy of aqueous extracts of the cyanobacterium Arthrospira fusiformis from Chad. Pharmazie:376–380 [PubMed]
- Shea B, Swinden MV, Tanjong Ghogomu E, Ortiz Z, Katchamart W, Rader T, Bombardier C, Wells GA, Tugwell P (2013) Folic acid and folinic acid for reducing side effects in patients receiving methotrexate for rheumatoid arthritis. Cochrane Database Syst Rev 2013(5):CD000951 [DOI] [PMC free article] [PubMed]
- Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha SK, Shakya A, Prasad SK, Singh S, Gurav NS, Prasad RS, Gurav SS (2020a) An in-silico evaluation of different saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. J Biomol Struct Dyn. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed]
- Sinha, Saurabh K., Satyendra K. Prasad, Md Ataul Islam, Shailendra S. Gurav, Rajesh B. Patil, Nora Abdullah AlFaris, Tahany Saleh Aldayel, Nora M. AlKehayez, Saikh Mohammad Wabaidur, and Anshul Shakya. (2020b). Identification of bioactive compounds from Glycyrrhiza glabra as possible inhibitor of SARS-CoV-2 spike glycoprotein and non-structural protein-15: a pharmacoinformatics study. J Biomolec Struct Dyn. 10.1080/07391102.2020.1779132 [DOI] [PMC free article] [PubMed]
- Souza PFN, Lopes FES, Amaral JL, Freitas CDT, Oliveira JTA. A molecular docking study revealed that synthetic peptides induced conformational changes in the structure of SARS-CoV-2 spike glycoprotein, disrupting the interaction with human ACE2 receptor. Int J Biol Macromol. 2020;164:66–76. doi: 10.1016/j.ijbiomac.2020.07.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JS, Lignell Å, Pettersson A, Elfving E, Soni MG (2008) Safety assessment of astaxanthin-rich microalgae biomass: Acute and subchronic toxicity studies in rats. Food Chem Toxicol 46(9):3030-3036 [DOI] [PubMed]
- Talukdar J, Bhadra B, Dattaroy T, Nagle V, Dasgupta S. Potential of natural astaxanthin in alleviating the risk of cytokine storm in COVID-19. Biomed Pharmacother. 2020;132:110886. doi: 10.1016/j.biopha.2020.110886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar J, Dasgupta S, Nagle V, Bhadra B (2020b) COVID-19: Potential of microalgae derived natural astaxanthin as adjuntive supplement in alleviating cytokine storm. SSRN. 10.2139/ssrn.3579738
- Teas J, Irhimeh MR. Dietary algae and HIV/AIDS: proof of concept clinical data. J Appl Phycol. 2012;24:575–582. doi: 10.1007/s10811-011-9766-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MJ, Maines MD, Lagarias JC. Inactivation of phytochrome- and phycobiliprotein-chromophore precursors by rat liver biliverdin reductase. J Biol Chem. 1993;268:26099–26106. doi: 10.1016/S0021-9258(19)74286-0. [DOI] [PubMed] [Google Scholar]
- Toelzer C, Gupta K, Yadav SKN, Borucu U, Davidson AD, Kavanagh Williamson M, Shoemark DK, Garzoni F, Staufer O, Milligan R, Capin J, Mulholland AJ, Spatz J, Fitzgerald D, Berger I, Schaffitzel C. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science. 2020;370:725–730. doi: 10.1126/science.abd3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton A-T, Gentile F, Hsing M, Ban F, Cherkasov A (2020) Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol Inform 39:2000028 [DOI] [PMC free article] [PubMed]
- Touillaud M, Gelot A, Mesrine S, Bennetau-Pelissero C, Clavel-Chapelon F, Arveux P, Bonnet F, Gunter M, Boutron-Ruault MC, Fournier A. Use of dietary supplements containing soy isoflavones and breast cancer risk among women aged >50 y: a prospective study. Am J Clin Nutr. 2019;109:597–605. doi: 10.1093/ajcn/nqy313. [DOI] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Tortorici MA, Snijder J, Xiong X, Bosch BJ, Rey FA, Dal V. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181:281–292 [DOI] [PMC free article] [PubMed]
- Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q, Zhou H, Yan J, Qi J (2020) Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181:894–904 [DOI] [PMC free article] [PubMed]
- Winter FS, Emakam F, Kfutwah A, Hermann J, Azabji-Kenfack M, Krawinkel MB. The effect of Arthrospira platensis capsules on CD4 T-cells and antioxidative capacity in a randomized pilot study of adult women infected with human immunodeficiency virus not under HAART in Yaoundé, Cameroon. Nutrients. 2014;6:2973–2986. doi: 10.3390/nu6072973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-L, Wang G-H, Xiang W-Z, Li T, He H. Stability and antioxidant activity of food-grade phycocyanin isolated from Spirulina platensis. Int J Food Prop. 2016;19:2349–2362. doi: 10.1080/10942912.2015.1038564. [DOI] [Google Scholar]
- Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M, Chen L, Li H (2020) Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B 10:766–788 [DOI] [PMC free article] [PubMed]
- Xia QD, Xun Y, Lu JL, Lu YC, Yang YY, Zhou P, Hu J, Li C, Wang SG (2020) Network pharmacology and molecular docking analyses on Lianhua Qingwen capsule indicate Akt1 is a potential target to treat and prevent COVID-19. Cell Prolif 53:e12949 [DOI] [PMC free article] [PubMed]
- Yakoot M, Salem A. Spirulina platensis versus silymarin in the treatment of chronic hepatitis C virus infection. A pilot randomized, comparative clinical trial. BMC Gastroenterol. 2012;12:32. doi: 10.1186/1471-230X-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Lou C, Sun L, Li J, Cai Y, Wang Z, Li W, Liu G, Tang Y. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019;35:1067–1069. doi: 10.1093/bioinformatics/bty707. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Cao D, Zhang Y, Ma J, Qi J, Wang Q, Lu G, Wu Y, Yan J, Shi Y, Zhang X, Gao GF. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Inoguchi T, Sasaki S, Maeda Y, McCarty MF, Fujii M, Ikeda N, Kobayashi K, Sonoda N, Takayanagi R. Phycocyanin and phycocyanobilin from Spirulina platensis protect against diabetic nephropathy by inhibiting oxidative stress. Am J Physiol Regul Integr Comp Physiol. 2013;304:R110–R120. doi: 10.1152/ajpregu.00648.2011. [DOI] [PubMed] [Google Scholar]
- Zhou Z-P, Liu L-N, Chen X-L, Wang J-X, Chen M, Zhang Y-Z, Zhou B-C. factors that affect antioxidant activity of C-phycocyanins from Spirulina platensis. J Food Biochem. 2005;29:313–322. doi: 10.1111/j.1745-4514.2005.00035.x. [DOI] [Google Scholar]
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:(7798)270–273 [DOI] [PMC free article] [PubMed]
- Zoete V, Cuendet MA, Grosdidier A, Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 147 kb)